- 1Department for Innovation Biological, Agro-Food and Forest System, University of Tuscia, Tuscia, Italy
- 2U.S. Department of Energy Joint Genome Institute, Walnut Creek, CA, USA
- 3HudsonAlpha Institute for Biotechnology, Huntsville, AL, USA
- 4School of Environmental and Forest Sciences, University of Washington, Seattle, WA, USA
- 5Adaptive Symbiotic Technologies, Seattle, WA, USA
Background
Here we present the genome sequence of Rhodotorula graminis WP1, a pink-pigmented, encapsulated yeast strain belonging to the Basidiomycota phylum that was isolated from within stems of Populus trichocarpa growing in its native riparian environment alongside the Snoqualmie River in Western Washington state(Xin et al., 2009). Although numerous bacterial endophytes, the microorganisms living fully within plants, have been characterized, relatively few endophytic yeast strains have been studied (Doty, 2011). The genome of strain WP1 was the first endophytic yeast to be sequenced, and has been included in several genomic analyses (Spatafora et al., 2013; Nagy et al., 2014; Pendleton et al., 2014). Although originally isolated from poplar, WP1 has a broad host range, providing growth benefits not only to poplar (Knoth et al., 2014; Khan et al. in review) but also to grasses and agricultural crop species (Khan et al., 2012; Knoth et al., 2013). Strain WP1 improves plant vigor and has useful biochemical properties (Doty, 2014) including the ability to ferment both pentoses and hexoses and to degrade fermentation inhibitors (Xu et al., 2011). Genomic analysis of bacterial endophytes has revealed certain features in common including genes for phytohormone synthesis, adhesion, colonization, stress reduction, and iron and phosphate acquisition (Fouts et al., 2008; Taghavi et al., 2010; Sessitsch et al., 2012; Witzel et al., 2012). We analyzed the genome of WP1 with a focus on genes potentially involved in plant-microbe interactions.
Methods
DNA Purification
Rhodotorula graminis strain WP1 was isolated from surface-sterilized shoot cuttings of poplar (P. trichocarpa) collected at the Three Forks Natural Area in King County, WA in the riparian zone of the Snoqualmie River (+47° 31′ 14.30″, −121° 46′ 28.32″) in August 2002, and glycerol stocks were frozen at −70°C. All subsequent studies were done using samples from these cryo-stocks. For DNA extraction, WP1 was grown in 300 ml YPD (yeast extract, peptone, dextrose) broth, pelleted, washed twice in sterile water, frozen, and freeze-dried. Cells were lysed with glass beads and the genomic DNA purified as previously described (Pitkin et al., 1996).
RNA Extraction
A single WP1 colony from solid NFMS [Nitrogen-Free Murashige and Skoog medium (Murashige and Skoog, 1962); Caisson] was used to inoculate 100 ml of liquid NFMS, and grown with agitation (200–250 rpm) for 24 h at RT. Cells were harvested and resuspended in 100 ml YPD broth and grown with agitation at RT. After 2 days, cells were centrifuged at 1300 × g and washed twice with NFMS medium. Cell density was adjusted to an OD600 0.4 in fresh NFMS and grown with agitation (200–250 rpm) at RT until the cell density reached an OD600 of 1.2. Cells were centrifuged at 1300 × g, washed twice in NFMS media, frozen at −80°C overnight, and lyophilized. Cells were ground in liquid nitrogen until a fine powder was achieved, and RNA extracted using standard protocols as suggested by the manufacturers of the reagent TRlzol (Life Technologies).
Genome Sequencing
The Rhodotorula graminis strain WP1 genome was sequenced by the Joint Genome Institute (JGI) using the Sanger whole genome shotgun approach. Three (3, 6, and 33.8 kb insert size) libraries were sequenced. Sequenced reads were QC filtered for vector sequence, mitochondria, unanchored rDNA, and assembled with the Arachne assembler (Jaffe et al., 2003). The 21,013,998 bp assembly resulted in 26 scaffolds comprising 323 contigs with average read depth coverage of 8.55x (Table 1).
Transcriptome Sequencing
For analysis of the R. graminis transcriptome, polyA mRNA was used to construct cDNA libraries and these were sequenced using the Roche-454 GS-FLX platform. The 1.9 million reads were filtered and screened for quality and contamination and were assembled into contigs using Newbler (v2.3-PreRelease-6/30/2009) with default parameters.
Genome Annotation
The R. graminis genome assembly was annotated using the JGI Annotation Pipeline (Grigoriev et al., 2006), which combines several gene prediction and functional annotation methods, and integrates the annotated genome into JGI web-based resource for fungal comparative genomics, MycoCosm (http://jgi.doe.gov/fungi) (Grigoriev et al., 2014). Before gene prediction, assembly scaffolds were masked using RepeatMasker (Smit et al., 2010), RepBase library (Jurka et al., 2005), and the most frequent (>150 times) repeats recognized by RepeatScout (Price et al., 2005). The following combination of gene predictors was run on the masked assembly: ab initio including Fgenesh (Salamov and Solovyev, 2000) and GeneMark (Ter-Hovhannisyan et al., 2008); homology-based including Fgenesh+ (Salamov and Solovyev, 2000) and Genewise (Birney and Durbin, 2000) seeded by BLASTx alignments against the NCBI NR database; and transcriptome-based using Fgenesh package. In addition to protein-coding genes, tRNAs were predicted using tRNAscan-SE (Lowe and Eddy, 1997). For each genomic locus, the best representative gene model was selected based on a combination of protein homology and transcriptome support. All predicted proteins were functionally annotated using SignalP (Nielsen et al., 1997) for signal sequences, TMHMM (Melén et al., 2003) for transmembrane domains, interProScan (Quevillon et al., 2005) for integrated collection of functional and structural protein domains, and protein alignments to NCBI nr, SwissProt (Boeckmann et al., 2003), KEGG (Kanehisa et al., 2004) for metabolic pathways, and KOG (Koonin et al., 2004) for eukaryotic clusters of orthologs. InterPro and SwissProt hits were used to map Gene Ontology terms (Ashburner et al., 2000).
Results
Genome Characteristics
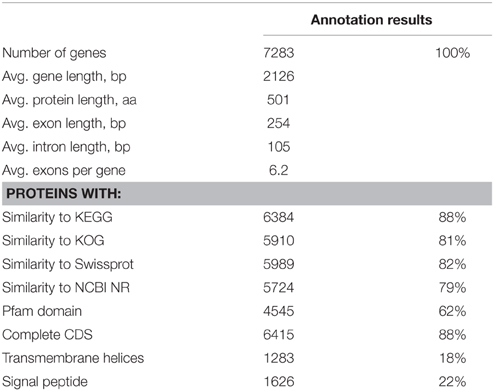
The 21.01 Mbp genome of Rhodotorula graminis strain WP1 was assembled in 28 scaffolds and 323 contigs with 1.13% in scaffold gaps (Table 1). The genome contains 3.63% repetitive DNA and 7283 predicted genes, supported by transcriptomics and homology to proteins from other fungi (Table 2). Three thousand five hundred and fifty-two predicted proteins form 929 multigene families based on MCL clustering, the largest of which include protein kinases, transporters, and transcription factors (http://genome.jgi.doe.gov/Rhoba1_1).
R. graminis has one of the highest GC-rich genomes (67%) among the all publicly available fungal genomes and while expectedly its intron content is also GC-rich, a significant imbalance in the C (42.7%) and G (23%) content of introns is surprising. This 19.7% deviation is the largest asymmetry in C/G content observed among the introns of all fungal genomes in MycoCosm (Grigoriev et al., 2014), where the average asymmetry is 1.8 ± 3.1%, and the second highest asymmetry of 7.3% (29.5% C and 22.2% G) was identified in the related Pucciniomycotina yeast Sporobolomyces roseus. Additionally, an extended donor splice consensus observed in introns of R. graminis is likely due to the high C content of introns. Most eukaryotes have a conserved consensus at positions +1 to +6 of introns (Rogozin and Milanesi, 1997; Bhasi et al., 2007). In R. graminis, positions +7 to +10 have a predominantly C nucleotide, with positions +3 and +4 also having stronger consensus relative to what is observed in other basidiomycetes.
Phytohormones, Volatile Organic Compounds (VOCs), Capsule Production and Small Secreted Proteins (SSPs)
A common feature of endophytic bacteria and fungi is the production of phytohormones (Hardoim et al., 2008; Bulgarelli et al., 2013; Sukumar et al., 2013; Duca et al., 2014). Many of the beneficial plant-associated microorganisms that produce auxin rely on plant-exuded tryptophan as the precursor for biosynthesis of the auxin, indole-3-acetic acid (IAA) (Hardoim et al., 2008). Since auxins stimulate plant growth and there is a higher prevalence of auxin-producing microorganisms within plants than in the rhizosphere, it has been proposed that the plant host environment selects for endophytes with this trait (Patten and Glick, 2002). Genomic analysis of several bacterial endophytes, including Enterobacter from poplar stems (Taghavi et al., 2010), Gluconacetobacter diazotrophicus from sugarcane (Bertalan et al., 2009), and the endophyte community of rice (Sessitsch et al., 2012) provided evidence of multiple microbial pathways for auxin biosynthesis. Although less is known about endophytic yeast than endophytic bacteria, there is evidence of plant growth promotion by auxin-producing yeasts (Doty, 2011). Eight of the Williopsis saturnus endophytic yeast strains of maize roots produced the auxins IAA and IPYA (Nassar et al., 2005). Since WP1 has strong root-promoting activity on recalcitrant poplar clones (Doty, unpublished) and overall plant growth-promoting activity (Khan et al., 2012; Knoth et al., 2014), we analyzed the genome for evidence of auxin and other phytohormone biosynthesis capabilities, putative effectors involved in plant-microbe interaction, and antitoxin systems.
The WP1 genome lacks the standard genes encoding for proteins involved in the biosynthesis of IAA via indole-3-pyruvate (KEGG Entry: R00677; R00684) and indole-3-acetamide (KEGG Entry: R00679). However, three putative proteins, an aromatic-L-amino-acid decarboxylase (Protein ID: 35429), a monoamine oxidase (Protein ID: 54216) and an indol-3-acetaldehyde dehydrogenase (Protein ID: 14581) can be involved in the conversion of L-tryptophan to IAA via tryptamine. In this pathway, trp is first decarboxylated to tryptamine (KEGG Entry: R00685) which is subsequently oxidized into indole-3-acetaldehyde and then to IAA through two consecutive oxidation steps carried out by an amine oxidase (KEGG Entry: R02173) and an aldehyde dehydrogenase, respectively (KEGG Entry: R02678). Furthermore, two putative hydrolases (Protein ID: 34153; 66162) belonging to the nitrilase superfamily, which are important for the microbial colonization of plants due to their role in nitriles detoxification and utilization of plant nitriles as carbon and nitrogen source (Howden and Preston, 2009; Howden et al., 2009), can also convert the indole-3-acetonitrile (IAN) into IAA (KEGG Entry: R03093). However, IAN synthesis occurring in microbes is still unclear (Fu and Wang, 2011).
From research for functional domains, 15 proteins in WP1 were annotated as containing a 2OG-Fe(II) oxygenase domain (IPR005123) which are primarily involved in the biosynthesis of gibberellins and other plant hormones (Prescott and Lloyd, 2000; Zhao et al., 2013; Farrow and Facchini, 2014). WP1 was shown to produce the phytohormones GA3, IAA, JA, ABA, and Br in vitro (Khan et al. in review).
R. graminis WP1 has a set of genes involved in the synthesis of (R)-acetoin and (R,R)-2,3-butandiol, two well-known VOCs that increase resistance to plant pathogens and also act as growth promoting factors (Johnston-Monje and Raizada, 2011; D'Alessandro et al., 2014; Taghavi et al., 2015). In WP1, a putative metabolic pathway which leads to the synthesis of (R)-acetoin and (R,R)-2,3-butandiol starts with the decarboxylation of pyruvate into 2-acetolactate (KEGG Entry: R00006) by a putative acetolactate synthase (Protein ID: 32290; 35922). Under aerobic conditions, the synthesis of (R,R)-2,3-butandiol (KEGG Entry: R02946) from (R)-acetoin by two putative NADH-dependent dehydrogenase (Protein ID: 39181; 46342) occurs through the spontaneous decarboxylation of 2-acetolactate into (R)-acetoin (Atsumi et al., 2009).
In order to offer a more in depth overview about the presence of putative SSPs (aminoacidic seq. < 300) encoding genes, as supposed effectors involved in plant-microbe interactions (Rafiqi et al., 2013), we data mined the annotation file “SignalP” as described in Pendleton et al. (2014) but with slight modification. The following file, accessible from the download section through the sub-directory path: Files > Annotation > Filtered Models > Functional Annotation > Signalp, were analyzed through the TargetP 1.1 Server (Emanuelsson et al., 2000). Among 208 putative SSPs, two coding sequences (Protein ID: 43059 and 46692) for extracellular membrane proteins with a cysteine-rich domain (CFEM domain: IPR008427) were detected. Moreover, through the Search tool, three secreted proteins (Protein ID: 55573, 55528, and 50767) with a Carbohydrate-binding WSC domain (IPR013994) were also detected.
Inoculation of roots with WP1 results in colonization of the plant, including the shoots, possibly through the formation of a yeast-form biofilm (unpublished data). Unlike bacteria that could colonize plants using flagella, most yeasts, just like filamentous fungi, may colonize, and interact with the plant through a filamentous form. The passage from a yeast-form to a filamentous stage can be triggered by a wide range of environmental stimuli or as a result of the interactions with other microorganisms within multispecies biofilms (Lengeler et al., 2000). In a study on the sexuality and life cycle in Rhodotorula glutinis strains, the capability to form filamentous mycelia was reported (Banno, 1967). Although we have not seen a filamentous form of WP1 under culture conditions, we cannot exclude the possibility that WP1 is able to form mycelia under specific environmental conditions.
An interesting feature of WP1 is the presence of a polysaccharidic capsule that surrounds the cellular body (Figure 1). A well-studied encapsulated yeast is represented by Cryptococcus neoformans, an opportunistic pathogen that causes meningoencephalitis in immunocompromised patients (Mitchell and Perfect, 1995). C. neoformans is ubiquitous in nature and the survival under different environmental conditions can be due to his biofilmogenic property. Within biofilms, microbial cells not only have an increased resistance and tolerance against a wide range of biotic and abiotic stress but also, under specific physiological and environmental conditions, can disperse and colonize new ecological niches (Ramage et al., 2009). The capability of C. neoformans to form a biofilm is dependent on the presence of the capsule. Deletion mutants in cap59 and cap10, two genes involved in the capsule synthesis and virulence, are unable to form biofilms, implicating that this structure exerts an important role in the adhesion and subsequent formation of cells aggregates (Garcia-Rivera et al., 2004; Martinez and Casadevall, 2005). Since a putative CAP59 (Protein ID: 4796) and CAP10 (Protein ID: 7100) were detected in WP1, the genome sequence of non-pathogenic encapsulated yeast would be interesting for a comparative analysis between the capsule synthesis in WP1 and C. neoformans.
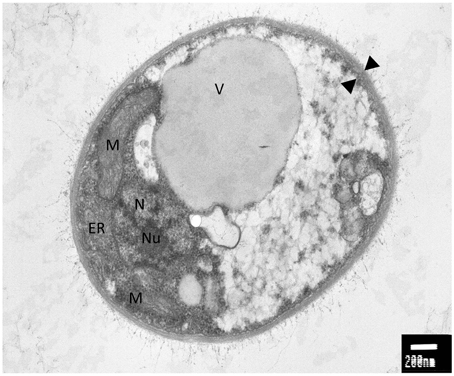
Figure 1. Electron microscopy analysis of Rhodotorula graminis strain WP1; M, mitochondria; ER, endoplasmic reticulum; N, nucleus; Nu, nucleulus; V, vacuole; black triangles, capsule. Photo credit: Prof. Jimmie Lara, Department of Microbiology, University of Washington.
Finally, the WP1 genome was data-mined for functional domains related to notable antitoxin systems. A set of antitoxin systems including multi antimicrobial extrusion protein (MATE), multidrug resistance efflux transporters (EmrE) and putative proteins with a multidrug resistance protein MdtG (IPR001958) domain was observed in the WP1 genome. The WP1 capsule in conjunction with these antitoxin systems represent important features that may be important for competition against other endophytes, plant colonization, and survival under different biotic and abiotic stresses.
Other Symbiotic Traits
Endophytes often share in common sets of genes that are thought to confer symbiotic abilities (Sessitsch et al., 2012; Bulgarelli et al., 2013). These beneficial genes could be acquired by horizontal gene transfer (HGT) (Taghavi et al., 2005; Aminov, 2011). There has been some indication of HGT between bacteria and fungi (Marcet-Houben and Gabaldon, 2010). However, when compared via BLAST against other databases of endophyte-associate genes (Sessitsch et al., 2012), WP1 shares only minimal homology with a few highly conserved bacterial endophyte genes such as catalase (Protein ID: 47984), as well as a gene involved in cofactor-A transport (Protein ID: 66591), and a glutathione S-transferase family protein (Protein ID: 54692). All of these matches, however, are all core metabolism/housekeeping proteins, and had less than 50% identity to the bacterial query proteins. Additionally, the WP1 genome was run through an in-house pipeline designed to identify genes acquired via transdomain HGT (Thomas, unpublished), but the results were negative. This lack of significant evidence of HGT could suggest that this endophytic yeast evolved most of its plant interaction and symbiosis genes independently from its bacterial counterparts. The well-studied symbiotic Basidiomycota, Laccaria bicolor, begins the colonization of poplar roots using an array of effector proteins known as mycorrhiza-induced-cysteine-rich SSPs (MiSSPs) (Martin et al., 2008). A domain of the WP1 mRNA splicing factor Prp8 (ProteinID 49541) is partially homologous to the L. bicolor LbMiSSP17 effector protein (Protein ID: 332226). Otherwise, no homology with the other 22 L. bicolor MiSSPs exists. Despite the fact that both WP1 and L. bicolor are both Basidiomycetes which symbiotically colonize poplar, this lack of homology could suggest WP1 uses an entirely different signaling pathway to communicate with the host.
Conclusions
There is a growing interest in the plant microbiome and its impacts on plant health and growth. With the sequence of the poplar genome (Tuskan et al., 2006) and multiple studies of the poplar microbiome (Hacquard and Schadt, 2015), poplar can become a model system for studying tree-microbiome interactions. The Laccaria-Populus interaction is a well-studied mycorrhizal mutualism at the molecular level (Podila et al., 2009; Aminov, 2011; Larsen et al., 2011; Plett et al., 2015). With comparative genomics studies of the bacteria, mycorrhizae, and yeast associated with Populus, an understanding of common themes in plant-mutualist interactions may emerge.
Data Access
The genome version discussed in this paper is the Rhodotorula graminis strain WP1 v1.1. Assembled scaffolds and all predicted genes and annotations are available at JGI fungal genome portal Mycocosm (http://genome.jgi.doe.gov/Rhoba1_1) and deposited to GenBank under accession JTAO00000000.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The work conducted by the U.S. Department of Energy Joint Genome Institute is supported by the Office of Science of the U.S. DOE under contract DE-AC02-05 CH11231. Partial research support was provided by NSF SGER # 0838091, NIFA grant # 2012-00931, AFRI grant # 2010-05080, and the Byron and Alice Lockwood Professorship. We thank Prof. Jim Lara for the electron microscopy figure, Shaoqun (Simon) Zhou and Dr. James H. Thomas for helpful discussions, and Dr. Stephen Mondo for assistance with the GenBank submission.
References
Aminov, R. I. (2011). Horizontal gene exchange in environmental microbiota. Front. Microbiol. 2:158. doi: 10.3389/fmicb.2011.00158
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat. Genet. 25, 25–29. doi: 10.1038/75556
Atsumi, S., Li, Z., and Liao, J. C. (2009). Acetolactate synthase from Bacillus subtilis serves as a 2-ketoisovalerate decarboxylase for isobutanol biosynthesis in Escherichia coli. Appl. Environ. Microbiol. 75, 6306–6311. doi: 10.1128/AEM.01160-09
Bertalan, M., Albano, R., de, P. V., Rouws, L., Rojas, C., Hemerly, A., et al. (2009). Complete genome sequence of the sugarcane nitrogen-fixing endophyte Gluconacetobacter diazotrophicus Pal5. BMC Genomics 10:450. doi: 10.1186/1471-2164-10-450
Bhasi, A., Pandey, R. V., Utharasamy, S. P., and Senapathy, P. (2007). EuSplice: a unified resource for the analysis of splice signals and alternative splicing in eukaryotic genes. Bioinformatics 23, 1815–1823. doi: 10.1093/bioinformatics/btm084
Birney, E., and Durbin, R. (2000). Using GeneWise in the Drosophila annotation experiment. Genome. Res. 10, 547–548. doi: 10.1101/gr.10.4.547
Boeckmann, B., Bairoch, A., Apweiler, R., Blatter, M. C., Estreicher, A., Gasteiger, E., et al. (2003). The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 31, 365–370. doi: 10.1093/nar/gkg095
Bulgarelli, D., Schlaeppi, K., Spaepen, S., Ver Loren van, T. E., and Schulze-Lefert, P. (2013). Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 64, 807–838. doi: 10.1146/annurev-arplant-050312-120106
D'Alessandro, M., Erb, M., Ton, J., Brandenburg, A., Karlen, D., Zopfi, J., et al. (2014). Volatiles produced by soil-borne endophytic bacteria increase plant pathogen resistance and affect tritrophic interactions. Plant Cell Environ. 37, 813–826. doi: 10.1111/pce.12220
>Doty, S. L. (2011). “Growth-promoting endophytic fungi of forest trees,” in Endophytes of Forest Trees: Biology and Applications, eds A. M. Pirttilä and A. C. Carolin (Dordrecht; Heidelberg; London, UK; New York, NY: Springer), 151–156.
Doty, S. L. (2014). Endophytic Yeast Strains, Methods for Ethanol and Xylitol Production, Methods for Biological Nitrogen Fixation, and a Genetic Source for Improvement of Industrial Strains. Patent No. 8,728,781.
Duca, D., Lorv, J., Patten, C. L., Rose, D., and Glick, B. R. (2014). Indole-3-acetic acid in plant-microbe interactions. Anton. Leeuwen. 106, 85–125. doi: 10.1007/s10482-013-0095-y
Emanuelsson, O., Nielsen, H., Brunak, S., and von, H. G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016. doi: 10.1006/jmbi.2000.3903
Farrow, S. C., and Facchini, P. J. (2014). Functional diversity of 2-oxoglutarate/Fe(II)-dependent dioxygenases in plant metabolism. Front. Plant Sci. 5:524. doi: 10.3389/fpls.2014.00524
Fouts, D. E., Tyler, H. L., DeBoy, R. T., Daugherty, S., Ren, Q., Badger, J., et al. (2008). Complete genome sequence of the N2-fixing broad host range endophyte Klebsiella pneumoniae 342 and virulence predictions verified in mice. PLoS. Genet. 4:e1000141. doi: 10.1371/journal.pgen.1000141
Fu, J., and Wang, S. (2011). Insights into auxin signaling in plant-pathogen interactions. Front. Plant Sci. 2:74. doi: 10.3389/fpls.2011.00074
García-Rivera, J., Chang, Y. C., Kwon-Chung, K. J., and Casadevall, A. (2004). Cryptococcus neoformans CAP59 (or Cap59p) is involved in the extracellular trafficking of capsular glucuronoxylomannan. Eukaryot. Cell 3, 385–392. doi: 10.1128/EC.3.2.385-392.2004
Grigoriev, I. V., Martinez, D. A., and Salamov, A. A. (2006). Fungal genomic annotation. Appl. Mycol. Biotechnol. 6, 123–142. doi: 10.1016/S1874-5334(06)80008-0
Grigoriev, I. V., Nikitin, R., Haridas, S., Kuo, A., Ohm, R., Otillar, R., et al. (2014). MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res. 42, D699–D704. doi: 10.1093/nar/gkt1183
Hacquard, S., and Schadt, C. W. (2015). Towards a holistic understanding of the beneficial interactions across the Populus microbiome. New Phytol. 205, 1424–1430. doi: 10.1111/nph.13133
Hardoim, P. R., van Overbeek, L. S., and Elsas, J. D. (2008). Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol. 16, 463–471. doi: 10.1016/j.tim.2008.07.008
Howden, A. J., Harrison, C. J., and Preston, G. M. (2009). A conserved mechanism for nitrile metabolism in bacteria and plants. Plant J. 57, 243–253. doi: 10.1111/j.1365-313X.2008.03682.x
Howden, A. J., and Preston, G. M. (2009). Nitrilase enzymes and their role in plant-microbe interactions. Microb. Biotechnol. 2, 441–451. doi: 10.1111/j.1751-7915.2009.00111.x
Jaffe, D. B., Butler, J., Gnerre, S., Mauceli, E., Lindblad-Toh, K., Mesirov, J., et al. (2003). Whole-genome sequence assembly for mammalian genomes: arachne 2. Genome. Res. 13, 91–96. doi: 10.1101/gr.828403
Johnston-Monje, D., and Raizada, M. N. (2011). Conservation and diversity of seed associated endophytes in Zea across boundaries of evolution, ethnography and ecology. PLoS ONE 6:e20396. doi: 10.1371/journal.pone.0020396
Jurka, J., Kapitonov, V. V., Pavlicek, A., Klonowski, P., Kohany, O., and Walichiewicz, J. (2005). Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome. Res. 110, 462–467. doi: 10.1159/000084979
Kanehisa, M., Goto, S., Kawashima, S., Okuno, Y., and Hattori, M. (2004). The KEGG resource for deciphering the genome. Nucleic Acids Res. 32, D277–D280. doi: 10.1093/nar/gkh063
Khan, Z., Guelich, G., Phan, H., Redman, R. S., and Doty, S. L. (2012). Bacterial and yeast endophytes from poplar and willow promote growth in crop plants and grasses. ISRN Agron. 2012:890280. doi: 10.5402/2012/890280
Knoth, J., Kim, S.-H., Ettl, G., and Doty, S. L. (2013). Effects of cross host species inoculation of nitrogen-fixing endophytes on growth and leaf physiology of maize. GCB Bioenery 5, 408–418. doi: 10.1111/gcbb.12006
Knoth, J. L., Kim, S. H., Ettl, G. J., and Doty, S. L. (2014). Biological nitrogen fixation and biomass accumulation within poplar clones as a result of inoculations with diazotrophic endophyte consortia. New Phytol. 201, 599–609. doi: 10.1111/nph.12536
Koonin, E. V., Fedorova, N. D., Jackson, J. D., Jacobs, A. R., Krylov, D. M., Makarova, K. S., et al. (2004). A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome. Biol. 5:R7. doi: 10.1186/gb-2004-5-2-r7
Larsen, P. E., Sreedasyam, A., Trivedi, G., Podila, G. K., Cseke, L. J., and Collart, F. R. (2011). Using next generation transcriptome sequencing to predict an ectomycorrhizal metabolome. BMC Syst. Biol. 5:70. doi: 10.1186/1752-0509-5-70
Lengeler, K. B., Davidson, R. C., D'souza, C., Harashima, T., Shen, W. C., Wang, P., et al. (2000). Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64, 746–785. doi: 10.1128/MMBR.64.4.746-785.2000
Lowe, T. M., and Eddy, S. R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964. doi: 10.1093/nar/25.5.0955
Marcet-Houben, M., and Gabaldon, T. (2010). Acquisition of prokaryotic genes by fungal genomes. Trends Genet. 26, 5–8. doi: 10.1016/j.tig.2009.11.007
Martin, F., Aerts, A., Ahren, D., Brun, A., Danchin, E. G., Duchaussoy, F., et al. (2008). The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature 452, 88–92. doi: 10.1038/nature06556
Martinez, L. R., and Casadevall, A. (2005). Specific antibody can prevent fungal biofilm formation and this effect correlates with protective efficacy. Infect. Immun. 73, 6350–6362. doi: 10.1128/IAI.73.10.6350-6362.2005
Melén, K., Krogh, A., and von, H. G. (2003). Reliability measures for membrane protein topology prediction algorithms. J. Mol. Biol. 327, 735–744. doi: 10.1016/S0022-2836(03)00182-7
Mitchell, T. G., and Perfect, J. R. (1995). Cryptococcosis in the era of AIDS–100 years after the discovery of Cryptococcus neoformans. Clin. Microbiol. Rev. 8, 515–548.
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Nagy, L. G., Ohm, R. A., Kovács, G. M., Floudas, D., Riley, R., Gácser, A., et al. (2014). Latent homology and convergent regulatory evolution underlies the repeated emergence of yeasts. Nat. Commun. 5, 4471. doi: 10.1038/ncomms5471
Nassar, A. H., El-Tarabily, K. A., and Sivasithamparam, K. (2005). Promotion of plant growth by an auxin-producing isolate of the yeast Williopsis saturnus endophytic in maize (Zea mays L.) roots. Biol. Fertil. Soils 42, 97–108. doi: 10.1007/s00374-005-0008-y
Nielsen, H., Engelbrecht, J., Brunak, S., and von, H. G. (1997). Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10, 1–6. doi: 10.1093/protein/10.1.1
Patten, C. L., and Glick, B. R. (2002). Role of Pseudomonas putida indoleacetic acid in development of the host plant root system. Appl. Environ. Microbiol. 68, 3795–3801. doi: 10.1128/AEM.68.8.3795-3801.2002
Pendleton, A. L., Smith, K. E., Feau, N., Martin, F. M., Grigoriev, I. V., Hamelin, R., et al. (2014). Duplications and losses in gene families of rust pathogens highlight putative effectors. Front. Plant Sci. 5:299. doi: 10.3389/fpls.2014.00299
Pitkin, J. W., Panaccione, D. G., and Walton, J. D. (1996). A putative cyclic peptide efflux pump encoded by the TOXA gene of the plant-pathogenic fungus Cochliobolus carbonum. Microbiology 142(Pt 6), 1557–1565. doi: 10.1099/13500872-142-6-1557
Plett, J. M., Tisserant, E., Brun, A., Morin, E., Grigoriev, I. V., Kuo, A., et al. (2015). The mutualist laccaria bicolor expresses a core gene regulon during the colonization of diverse host plants and a variable regulon to counteract host-specific defenses. Mol. Plant Microbe Interact. 28, 261–273. doi: 10.1094/MPMI-05-14-0129-FI
Podila, G. K., Sreedasyam, A., and Muratet, M. (2009). Populus rhizosphere and the ectomycorrhizal interactome. Crit. Rev. Plant Sci. 28, 359–367. doi: 10.1080/07352680903241220
Prescott, A. G., and Lloyd, M. D. (2000). The iron(II) and 2-oxoacid-dependent dioxygenases and their role in metabolism. Nat. Prod. Rep. 17, 367–383. doi: 10.1039/a902197c
Price, A. L., Jones, N. C., and Pevzner, P. A. (2005). De novo identification of repeat families in large genomes. Bioinformatics 21(Suppl. 1), i351–i358. doi: 10.1093/bioinformatics/bti1018
Quevillon, E., Silventoinen, V., Pillai, S., Harte, N., Mulder, N., Apweiler, R., et al. (2005). InterProScan: protein domains identifier. Nucleic Acids Res. 33, W116–W120. doi: 10.1093/nar/gki442
Rafiqi, M., Jelonek, L., Akum, N. F., Zhang, F., and Kogel, K. H. (2013). Effector candidates in the secretome of Piriformospora indica, a ubiquitous plant-associated fungus. Front. Plant Sci. 4:228. doi: 10.3389/fpls.2013.00228
Ramage, G., Mowat, E., Jones, B., Williams, C., and Lopez-Ribot, J. (2009). Our current understanding of fungal biofilms. Crit. Rev. Microbiol. 35, 340–355. doi: 10.3109/10408410903241436
Rogozin, I. B., and Milanesi, L. (1997). Analysis of donor splice sites in different eukaryotic organisms. J. Mol. Evol. 45, 50–59. doi: 10.1007/PL00006200
Salamov, A. A., and Solovyev, V. V. (2000). Ab initio gene finding in Drosophila genomic DNA. Genome. Res. 10, 516–522. doi: 10.1101/gr.10.4.516
Sessitsch, A., Hardoim, P., Döring, J., Weilharter, A., Krause, A., Woyke, T., et al. (2012). Functional characteristics of an endophyte community colonizing rice roots as revealed by metagenomic analysis. Mol. Plant Microbe Interact. 25, 28–36. doi: 10.1094/MPMI-08-11-0204
Smit, A. F. A., Hubley, R., and Green, P. (2010). Repeat Masker Open-3.0. Available online at: http://www.repeatmasker.org
Spatafora, J. W., Stajich, J. E., and Grigoriev, I. (2013). 1000 fungal genomes project. Phytopathology 103, 137. Available online at: http://1000.fungalgenomes.org/home/webcite
Sukumar, P., Legué, V., Vayssierès, A., Martin, F., Tuskan, G. A., and Kalluri, U. C. (2013). Involvement of auxin pathways in modulating root architecture during beneficial plant-microorganism interactions. Plant Cell Environ. 36, 909–919. doi: 10.1111/pce.12036
Taghavi, S., Barac, T., Greenberg, B., Borremans, B., Vangronsveld, J., and van der Lelie, D. (2005). Horizontal gene transfer to endogenous endophytic bacteria from poplar improves phytoremediation of toluene. Appl. Environ. Microbiol. 71, 8500–8505. doi: 10.1128/AEM.71.12.8500-8505.2005
Taghavi, S., van der Lelie, D., Hoffman, A., Zhang, Y. B., Walla, M. D., Vangronsveld, J., et al. (2010). Genome sequence of the plant growth promoting endophytic bacterium Enterobacter sp. 638. PLoS. Genet. 6:e1000943. doi: 10.1371/journal.pgen.1000943
Taghavi, S., Wu, X., Ouyang, L., Zhang, Y. B., Stadler, A., McCorkle, S., et al. (2015). Transcriptional responses to sucrose mimic the plant-associated life style of the plant growth promoting endophyte Enterobacter sp. 638. PLoS ONE 10:e0115455. doi: 10.1371/journal.pone.0115455
Ter-Hovhannisyan, V., Lomsadze, A., Chernoff, Y. O., and Borodovsky, M. (2008). Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome. Res. 18, 1979–1990. doi: 10.1101/gr.081612.108
Tuskan, G. A., Difazio, S., Jansson, S., Bohlmann, J., Grigoriev, I., Hellsten, U., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604. doi: 10.1126/science.1128691
Witzel, K., Gwinn-Giglio, M., Nadendla, S., Shefchek, K., and Ruppel, S. (2012). Genome sequence of Enterobacter radicincitans DSM16656(T), a plant growth-promoting endophyte. J. Bacteriol. 194, 5469. doi: 10.1128/JB.01193-12
Xin, G., Glawe, D., and Doty, S. L. (2009). Characterization of three endophytic, indole-3-acetic acid-producing yeasts occurring in Populus trees. Mycol. Res. 113, 973–980. doi: 10.1016/j.mycres.2009.06.001
Xu, P., Bura, R., and Doty, S. L. (2011). Genetic analysis of D-xylose metabolism by endophytic yeast strains of Rhodotorula graminis and Rhodotorula mucilaginosa. Genet. Mol. Biol. 34, 471–478. doi: 10.1590/S1415-47572011000300018
Keywords: symbiosis, endophytes, populus, plant-microbe interactions, endophytic yeast, endophyte genomics, microbiome, phytobiome
Citation: Firrincieli A, Otillar R, Salamov A, Schmutz J, Khan Z, Redman RS, Fleck ND, Lindquist E, Grigoriev IV and Doty SL (2015) Genome sequence of the plant growth promoting endophytic yeast Rhodotorula graminis WP1. Front. Microbiol. 6:978. doi: 10.3389/fmicb.2015.00978
Received: 30 May 2015; Accepted: 03 September 2015;
Published: 17 September 2015.
Edited by:
Carolin Frank, University of California, Merced, USAReviewed by:
Eric Kemen, Max Planck Institute for Plant Breeding Research, GermanyWei Qian, Institute of Microbiology, Chinese Academy of Sciences, China
Copyright © 2015 Firrincieli, Otillar, Salamov, Schmutz, Khan, Redman, Fleck, Lindquist, Grigoriev and Doty. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sharon L. Doty, sldoty@u.washington.edu
 Andrea Firrincieli
Andrea Firrincieli Robert Otillar
Robert Otillar Asaf Salamov
Asaf Salamov Jeremy Schmutz
Jeremy Schmutz Zareen Khan
Zareen Khan Regina S. Redman5
Regina S. Redman5 Neil D. Fleck
Neil D. Fleck Erika Lindquist
Erika Lindquist Igor V. Grigoriev
Igor V. Grigoriev Sharon L. Doty
Sharon L. Doty