Focused Review: Cytotoxic and Antioxidant Potentials of Mangrove-Derived Streptomyces
- 1Biomedical Research Laboratory, Jeffrey Cheah School of Medicine and Health Sciences, Monash University Malaysia, Bandar Sunway, Malaysia
- 2UKM Medical Molecular Biology Institute–UKM Medical Centre, Universiti Kebangsaan Malaysia, Kuala Lumpur, Malaysia
- 3Division of Genetics and Molecular Biology, Institute of Biological Sciences, Faculty of Science, University of Malaya, Kuala Lumpur, Malaysia
Streptomyces pluripotens MUSC 137 was isolated from mangrove soil obtained from Tanjung Lumpur, Pahang, Malaysia. We investigated the phylogenetic, genomic, biochemical, and phenotypic characteristics of this strain. Uniquely adapted microorganisms from mangrove habitats have previously yielded compounds of biopharmaceutical interest. In order to examine the bioactivities possessed by the strain, fermentation extract was prepared through solvent extraction method prior to bioactivities screenings. Antioxidant activity was examined via DPPH assay while the cytotoxic effect was assessed by means of examining the activity of the extract against selected human cancer cell lines, namely colon cancer cells (HCT-116, Caco-2, SW480, and HT-29), breast cancer cell (MCF-7), lung cancer cell (A549), prostate cancer cell (DU145), and cervical cancer cell (Ca Ski). The results revealed MUSC 137 possesses significant antioxidant activity and demonstrates cytotoxic effect against several cancer cell lines tested. The results indicated MCF-7 cells were most susceptible to the extract with the lowest IC50 (61.33 ± 17.10 μg/mL), followed by HCT-116 and A549. Additionally, selective index (SI) showed that MUSC 137 extract was less toxic against normal cell lines when compared to MCF-7 and HCT-116 cells. The extract was further subjected to chemical analysis using GC–MS and revealed the presence of deferoxamine and pyrrolizidines related compounds which may account for the antioxidant and cytotoxic properties observed.
Introduction
Mangrove is a special woody plant area of intertidal coasts in tropical and subtropical coastal regions. Mangrove ecosystems are habitats of various flora and fauna of marine, freshwater, and terrestrial species (Jennerjahn and Ittekkot, 2002). The constant changes in salinity and tidal gradient in the mangrove ecosystem have become driving forces for metabolic pathway adaptations that could direct to the production of useful metabolites (Hong et al., 2009; Lee et al., 2014a). Therefore, the growing interest in the utilization of mangrove microorganism resources have indirectly led to the discovery of Streptomyces from the different mangrove environments globally, such as the isolation of Streptomyces sanyensis (Sui et al., 2011), S. shenzhenensis (Hu et al., 2011), S. qinglanensis (Hu et al., 2012), S. pluripotens (Lee et al., 2014b), S. gilvigriseus (Ser et al., 2015a), and S. mangrovisoli (Ser et al., 2015b).
The genus Streptomyces was proposed by Waksman and Henrici (1943) and at the time of writing (July 2015), the genus Streptomyces is comprised of ca. 600 species with validly published names1 Over the years, through traditional cultivation methods, many of the bioactive compounds have been isolated from Streptomyces species, including antibacterial, antifungal, antioxidant, and antitumor compounds (Hong et al., 2009; Qin et al., 2011; Kawahara et al., 2012; Lee et al., 2014a,b; Kumar et al., 2014; Tan et al., 2015). The importance of these organisms is clearly seen from the fact that approximately 70% of the antibiotics in use were derived from actinobacteria (Subramani and Aalbersberg, 2012), of which 75% were derived from genus Streptomyces (Berdy, 2005; Rehm et al., 2009; Crnovcic et al., 2013).
In our screening program to explore the potential biological activity possessed by the strain MUSC 137, the extract was prepared and the free radical scavenging assay was used to evaluate the antioxidant activity. Different type of human cancer cell lines derived from colon, breast, cervical, prostate, and lung were selected for the screening of cytotoxic activity. These results indicated that MUSC 137 extract exhibited a significant antioxidant and cytotoxic properties. The extract was then subjected to chemical analysis with the use of GC–MS, which eventually led to the identification of chemical constituents present in the extract of MUSC 137. Taken all together, the well characterized strain, S. pluripotens MUSC 137 isolated from mangrove soil in Tanjung Lumpur could be a potential good source of antioxidative and chemopreventive agents.
Materials and Methods
Sample Collection and Isolation of Strain Streptomyces pluripotens MUSC 137
Strain MUSC 137 was isolated from a soil sample collected in December 2012 from site MUSC-TLS4 (3° 48′ 21.3″ N 103° 20′ 3.3″ E) located in the Tanjung Lumpur mangrove forest in the state of Pahang, Peninsular Malaysia. Topsoil samples of the upper 20-cm layer (after removing the top 2–3 cm) were collected and sampled into sterile plastic bags using an aseptic metal trowel, and stored at –20°C. Air-dried soil samples were ground with a mortar and pestle. Selective pretreatment of soil samples was performed using wet heat in sterilized water (15 min at 50°C; Takahashi et al., 1996). Five grams of the pretreated air-dried soil was mixed with 45 ml sterilized water and mill ground, spread onto the isolation medium ISP 2 (Shirling and Gottlieb, 1966) supplemented with cycloheximide (25 μg/ml) and nystatin (10 μg/ml), and incubated at 28°C for 14 days. Pure cultures of strain MUSC 137 were maintained on slants of ISP 2 agar at 28°C and as glycerol suspensions (20%, v/v) at –20°C.
Genomic and Phylogenetic Characterization
The bioactive-producing strain S. pluripotens MUSC 137 was characterized based on phylogenetic, genomic, biochemical, and phenotypic approaches. The extraction of genomic DNA and PCR was performed as described by Hong et al. (2009). The amplification of 16S rRNA gene was performed according to Lee et al. (2014b). The 16S rRNA gene sequence of strain MUSC 137 was aligned with representative sequences of related type strains of the genus Streptomyces retrieved from the GenBank/EMBL/DDBJ databases using CLUSTAL-X software (Thompson et al., 1997). The alignment was verified manually and adjusted prior to the reconstruction of phylogenetic trees. Phylogenetic trees were constructed with the maximum-likelihood algorithm (Felsenstein, 1981), (Figure 1) using MEGA version 6.0 (Tamura et al., 2013). The EzTaxon-e server2 (Kim et al., 2012) was used for calculation of sequence similarity. The stability of the resultant trees topologies were evaluated by using the bootstrap resampling method of Felsenstein (1985). The BOX-PCR fingerprint analysis was used to characterize strain MUSC 137 and the closely related strains using the primer BOX-A1R (5′-CTACGGCAAGGCGACGCTGACG-3′; Lee et al., 2014c). The PCR condition for BOX-PCR was performed as described by Lee et al. (2014d). The PCR products were visualized by 2% agarose gel electrophoresis.

FIGURE 1. Maximum-likelihood phylogenetic tree based on almost complete 16S rRNA sequences (1491 nucleotides) showing the relationship between strain MUSC 137 and representatives of some other related taxa. Numbers at nodes indicate percentages of 1000 bootstrap re-samplings, only values above 50% are shown. Bar, 0.002 substitutions per site.
The genomic DNA extraction for DNA-DNA hybridization of strain MUSC 137 with S. pluripotens MUSC135T was performed according to protocol of Cashion et al. (1977). The DNA–DNA hybridization was carried out by the Identification Service of the DSMZ, Braunschweig, Germany following the protocol of De Ley et al. (1970) under consideration of the modifications described by Huss et al. (1983).
Phenotypic Characterization
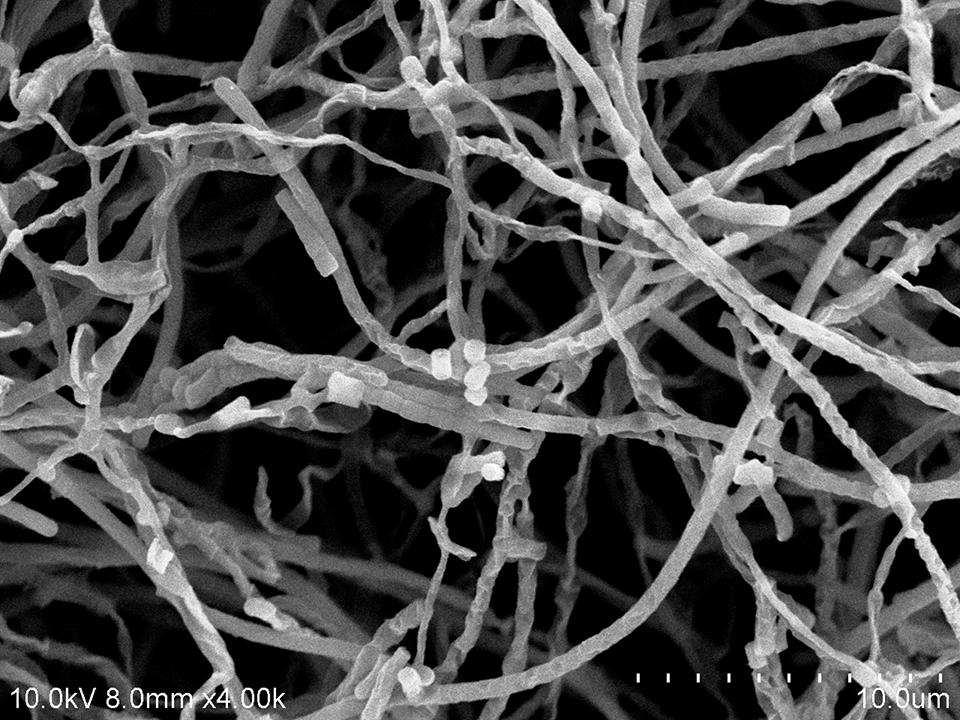
The cultural characteristics of strain MUSC 137 were determined following growth on ISP2 and ISP7 agar (Shirling and Gottlieb, 1966), starch casein agar (SCA; Küster and Williams, 1964), Streptomyces agar (SA; Atlas, 1993), Actinomycetes isolation agar (AIA; Atlas, 1993), and nutrient agar (Macfaddin, 2000) for 7–14 days at 28°C. Light microscopy (80i, Nikon) and scanning electron microscopy (JEOL-JSM 6400) were used to observe the morphology of the strain after incubation on ISP2 agar at 28°C for 7–14 days (Figure 2). The designation of colony color was determined by using the ISCC-NBS color charts (Kelly, 1964). Gram staining was performed by standard Gram reaction and confirmed by using KOH lysis (Cerny, 1978). The NaCl tolerance [0–10% (w/v)] and pH range (pH 4.0–10.0) for growth was tested in TSB. The growth temperature range was tested at 4–40°C at intervals of 4°C on ISP2 agar. The responses to temperature, pH and NaCl were observed for 14 days. Hemolytic activity was assessed on blood agar medium containing 5% (w/v) peptone, 3% (w/v) yeast extract, 5% (w/v) NaCl, and 5% (v/v) horse blood (Carrillo et al., 1996). The plates were examined for hemolysis after incubation at 28°C for 7–14 days. Catalase activity and production of melanoid pigments were determined following protocols described by Lee et al. (2014e). Amylolytic, cellulase, chitinase, lipase, protease, and xylanase activities were determined on ISP2 agar as described by Meena et al. (2013). Antibiotic susceptibility tests were performed by the disk diffusion method as described by Shieh et al. (2003). Carbon-source utilization and chemical sensitivity assays were determined using Biolog GenIII MicroPlates (Biolog, USA) according to the manufacturer’s instructions. Antimicrobials used and their concentrations are as follows: ampicillin (10 μg), ampicillin sulbactam (30 μg), cefotaxime (30 μg), cefuroxime (30 μg), cephalosporin (30 μg), chloramphenicol (30 μg), ciprofloxacin (10 μg), erythromycin (15 μg), gentamicin (20 μg), nalidixic acid (30 μg), Penicillin G (10 μg), streptomycin (10 μg), tetracycline (30 μg), and vancomycin (30 μg).
Extract Preparation of MUSC 137
Strain MUSC 137 was grown in seed medium (TSB) for 14 days prior to fermentation. The fermentation medium, FM3 was sterilized by autoclaving at 121°C for 15 min prior to experiment (Hong et al., 2009; Lee et al., 2014a). Fermentation was performed in shaking 500 mL Erlenmeyer flask containing 200 mL of FM3, for 7–10 days at 28°C. The resulting FM3 medium was recovered by centrifugation at 12000 g for 15 min. The supernatant was filtered and subjected to freeze dry process. Once freeze-dried, the sample was extracted thrice with methanol. The extracting solvent was removed and concentrated by rotary vacuum evaporator at 40°C. The extract of MUSC 137 was collected and suspended in dimethyl sulfoxide (DMSO) as vehicle reagent prior to assay.
Investigation of Antioxidant Activity of MUSC 137 Extract Using 2,2-Diphenyl-1-Picrylhydrazyl (DPPH) Radical Scavenging Method
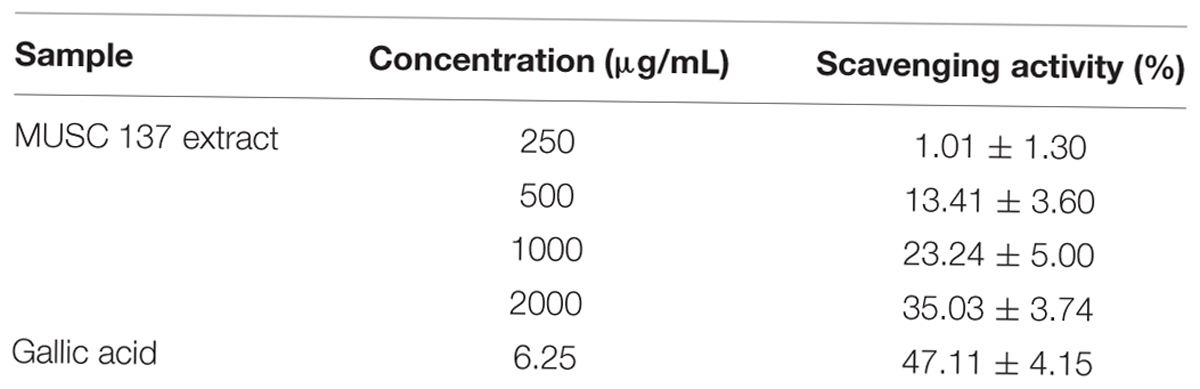
Antioxidant activity of MUSC 137 extract was examined by measuring radical scavenging ability using DPPH (Sigma–Aldrich). Scavenging activity on DPPH free radicals was determined using previous protocol with minor modifications (Ling et al., 2009). Reduction in the amount of free radicals present was measured via a decrease in absorbance of 515 nm. Volume of 195 μL of 0.016% DPPH ethanolic solution was added to 5 μL of extract solution to make up final volume of 200 μL. For control, DMSO was added in place of extract, while gallic acid was included as positive control. Reactions were carried out in the dark at room temperature for 20 min before measurement with spectrophotometer at 515 nm. DPPH scavenging activity was calculated as follows:
Determination of Cytotoxicity Activity of MUSC 137 Using 3-(4,5-Dimethylthazol-2yl)-2,5-Diphenyl Tetrazolium-Bromide (MTT) Assay
For evaluation of cytotoxicity, a variety of human derived cancer cell lines were selected for testing: (a) colon cancer cell, HCT-116, HT-29, Caco-2 and SW480; (b) breast cancer cell, MCF-7; (c) cervical cancer cell, Ca Ski; (d) lung carcinoma cell line, A549; (e) prostate cancer cell, DU 145. Cells were grown in RPMi 1640 supplemented with 10% FBS in humidified incubator (5% CO2 in air at 37°C). Cells were seeded into a sterile flat bottom 96-well plate at a density of 5 × 103 cells/well and allowed to adhere overnight. MUSC 137 extract was added to the cells (0–400 μg/mL) and further incubated for 72 h before performing MTT assay. Curcumin (1.56–25 μg/mL) was included as positive control. Twenty microliter of 5 mg/mL of MTT (Sigma) was then added to each well and the plates were incubated at 37°C in a humid atmosphere with 5% CO2, 95% air for 4 h. The medium was then gently aspirated, and 100 μL of DMSO was added to dissolve the formazan crystals. The amount of formazan product was determined spectrophotometrically at 570 nm (with 650 nm as reference wavelength) using a microplate reader. The percentage of cell viability was calculated with follows:
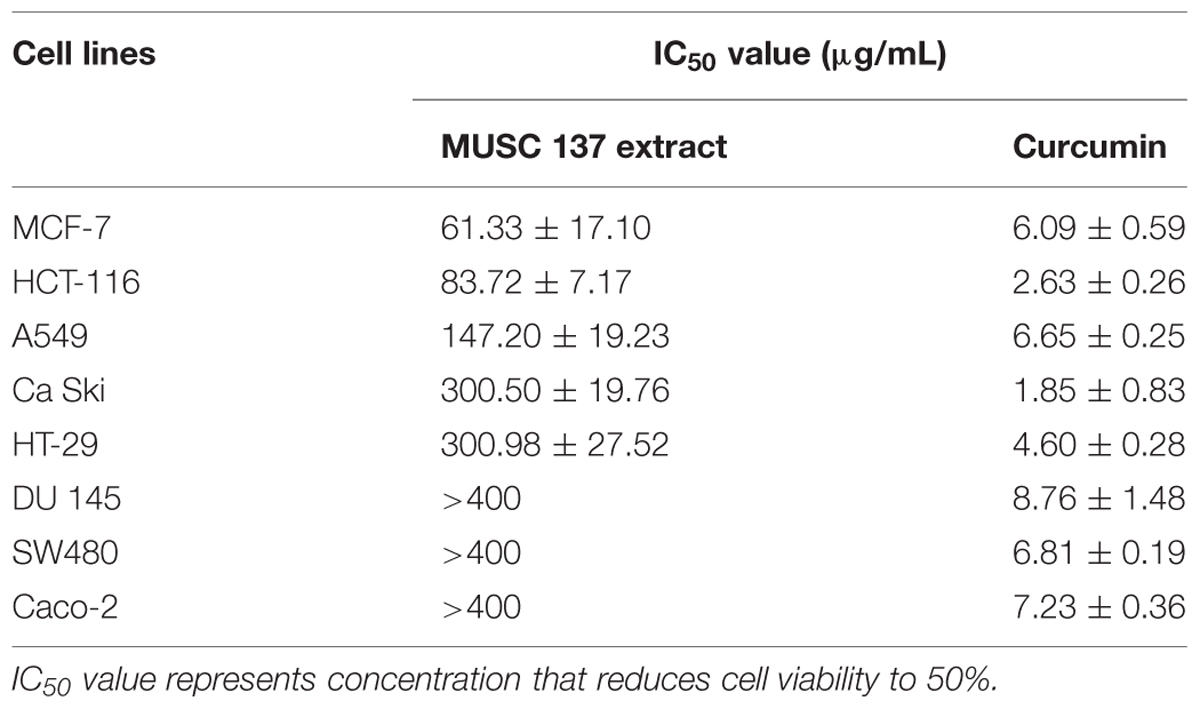
The 50% inhibitory concentration of extract (IC50) was derived from the plotted graphs for each cell line. VERO cells were used as a normal cell model for determination of selective index (SI). SI value was calculated by dividing IC50 of cancer cell line with IC50 value of normal cell line.
Gas Chromatography–Mass Spectrometry (GC–MS) Analysis
Gas chromatography–mass spectrometry analysis was performed as previously described (Ser et al., 2015b). The equipment used was Agilent Technologies 6980N (GC) equipped with 5979 Mass Selective Detector (MS), HP-5MS (5% phenyl methyl siloxane) capillary column of dimensions 30.0 m × 250 μm × 0.25 μm and used helium as carrier gas at 1 mL/min. The column temperature was programmed initially at 40°C for 10 min, followed by an increase of 3°C/min to 250°C and was kept isothermally for 5 min. The MS was operating at 70 eV. The constituents were identified by comparison of their mass spectral data with those from NIST 05 Spectral Library.
Statistical Analysis
Experiments to investigate antioxidant and cytotoxic activities were done in quadruplicate. Data analysis was performed with SPSS statistical analysis software and the results were expressed as mean ± standard deviation (SD). One-way analysis of variance (ANOVA) was used for comparison of more than two means. A difference was considered statistically significant when p ≤ 0.05.
Results
Phenotypic Analyses of Strain S. pluripotens MUSC 137
Strain MUSC 137 was Gram-positive and aerobic. The strain grew well on ISP2 medium, SA and SCA after 1–2 weeks at 28°C, it grew moderately on Luria Bertani agar, ISP7 medium and nutrient agar, whereas it grew poorly on AIA. The morphological observation of the 15-day-old culture grown on ISP2 medium showed a smooth spore surface, with aerial and vegetative hyphae which was well developed and not fragmented. These morphological characteristics were consistent with its assignment to the genus Streptomyces (Williams et al., 1989), (Figure 2). The colors of the aerial and substrate mycelium were yellowish gray and brilliant yellow. Growth occurred at pH 5.0–9.0 (optimum pH 5.0–8.0), with 0–6% NaCl tolerance (optimum 0–4%) and at 24–40°C (optimum 28–32°C). MUSC 137 was positive for catalase but negative for hemolytic activity and melanoid pigment production. Hydrolysis of soluble starch and carboxymethylcellulose were positive; but negative for hydrolysis of tributyrin (lipase), casein, chitin, and xylan. Strain MUSC 137 was found to be able to utilize various compounds as carbon sources (Supplementary Table S1). The strain was sensitive to ampicillin, ampicillin sulbactam, cefotaxime, cefuroxime, cephalosporin, chloramphenicol, ciprofloxacin, erythromycin, gentamicin, nalidixic acid, Penicillin G, streptomycin, tetracycline, and vancomycin.
Phylogenetic and Genomic Analyses
The almost-complete 16S rRNA gene sequences were determined for strain MUSC 137 (1491 bp) and aligned manually with the corresponding partial 16S rRNA gene sequences of the type strains of representative members of the genus Streptomyces retrieved from GenBank/EMBL/DDBJ databases. A phylogenetic tree constructed based on the 16S prRNA gene sequences showed that strain MUSC 137 (Figure 1) formed a distinct clade with type strains S. pluripotens MUSC 135T, S. mexicanus NBRC 100915T, S. cinereospinus NBRC 15397T, S. coeruleofuscus NBRC 12757T, and S. chromofuscus NBRC 12851T. Strain MUSC 137 and S. pluripotens MUSC 135T formed a distinct subclade at 100% bootstrap value, indicating the high confidence level of the association (Figure 1). Strain MUSC 137 exhibited highest 16S rRNA gene sequence similarity to S. pluripotens MUSC 135T (100%), followed by S. cinereospinus NBRC 15397T (99.18%), which corresponds to 12 nucleotide differences at 1462 locations with gaps, and lower similarity values to S. mexicanus NBRC 100915T (99.17%) and S. coeruleofuscus NBRC 12757T (98.97%).
The DNA–DNA relatedness values between strain MUSC 137 and S. pluripotens MUSC 135T were 83 ± 3.2%, which were significantly higher that 70%, the threshold value for the delineation of bacterial species (Wayne et al., 1987). These results supported the notion that strain MUSC 137 belonged to the same species as S. pluripotens MUSC 135T. The BOX-PCR results exhibited that strain MUSC 137 presented a unique BOX-PCR fingerprint as compared to MUSC 135T and other closely related type strains (Supplementary Figure S1).
Antioxidant and Cytotoxic Activity of MUSC 137 Extract
The results obtained from DPPH free radical scavenging assay showed significant antioxidant activity of MUSC 137 extract with 35.03 ± 3.74% scavenging activity at 2 mg/mL (Table 1). In addition, the extract of MUSC 137 displayed variable levels of cytotoxic activity against all the tested human cancer cell lines, with our work recording viability of tested cells ranging from 18.5 to 76.2% (Figure 3). A dose response pattern was evaluated using different doses which indicated that the extract may be effective against some cancer cell lines even at the lowest tested concentration (25 μg/mL). The lowest IC50 value of MUSC 137 extract was recorded at 61.33 ± 17.10 μg/mL for MCF-7 cells, followed by HCT-116 (83.72 ± 7.17 μg/mL), A549 (147.20 ± 19.23 μg/mL) as shown in Table 2. Higher IC50 values of MUSC 137 extract were observed for Ca Ski and HT-29, at 300.50 ± 19.76 μg/mL and 300.98 ± 27.52 μg/mL, respectively.
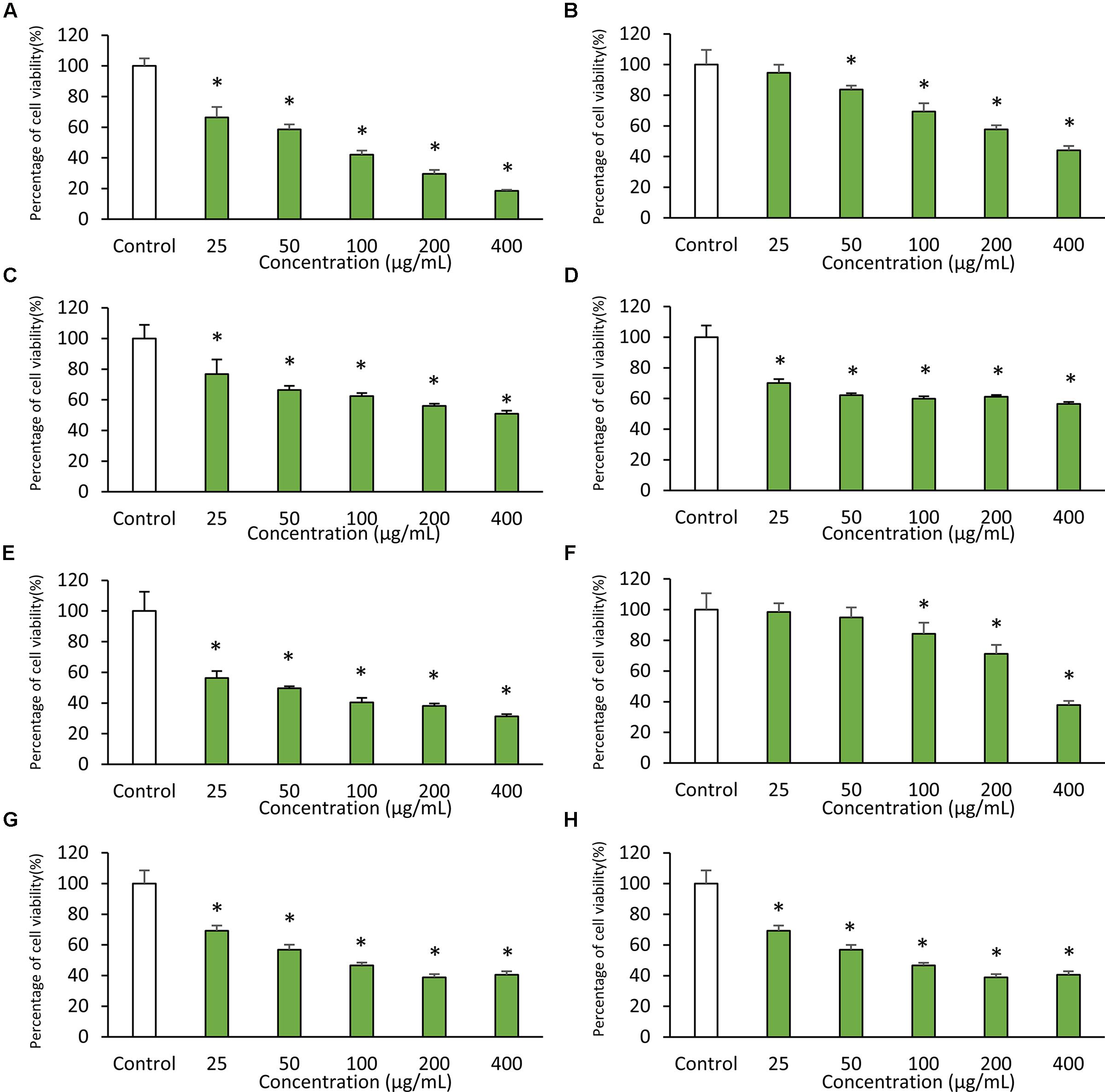
FIGURE 3. Cytotoxic activity of MUSC 137 extract against human cancer cell lines. Cell viability was measured using MTT assay. The graphs show cytotoxicity effects of MUSC 137 extract against (A) HCT-116, (B) HT-29, (C) Caco-2, (D) SW480, (E) MCF-7, (F) Ca Ski, (G) A549, (H) DU 145. * indicates significant difference when compared with control (p value < 0.05).
Of particular note, the extract showed higher level of cytotoxicity against some cancer cell lines with low cytotoxic effects in normal cell lines. The calculation of selectivity index (SI) allows comparison of cytotoxicity by dividing IC50 value of each cancer cell lines and IC50 value of normal cell line. The extract was less cytotoxic to normal cells as indicated by the SI value of 4.22 when compared to MCF-7 cells, followed by HCT-116 with value of 3.09 (Table 3).
GC–MS Analysis of MUSC 137 Extract
Gas chromatography–mass spectrometry analysis identified nine compounds present in the methanolic extract of MUSC 137 (Table 4) and the chemical structures (Figure 4) as 2,2-dimethoxybutane (1), Benzeneacetamide (2), Phenol, 2,5-bis(1,1-dimethylethyl)- (3), (3R,8aS)-3-methyl-1,2,3,4,6,7,8,8a-octahydropyrrolo[1,2-a]pyrazine-1,4-dione (4), 2,5-cyclohexadiene-1,4-dione (5), Pyrrolo[1,2-a]pyrazine-1,4-dione, hexahydro- (6), 1,4-diaza-2,5-dioxo-3-isobutyl bicyclo[4.3.0]nonane (7), Deferoxamine (8), Pyrrolo[1,2-a]pyrazine-1,4-dione, hexahydro-3-(2-phenylmethyl)- (9).
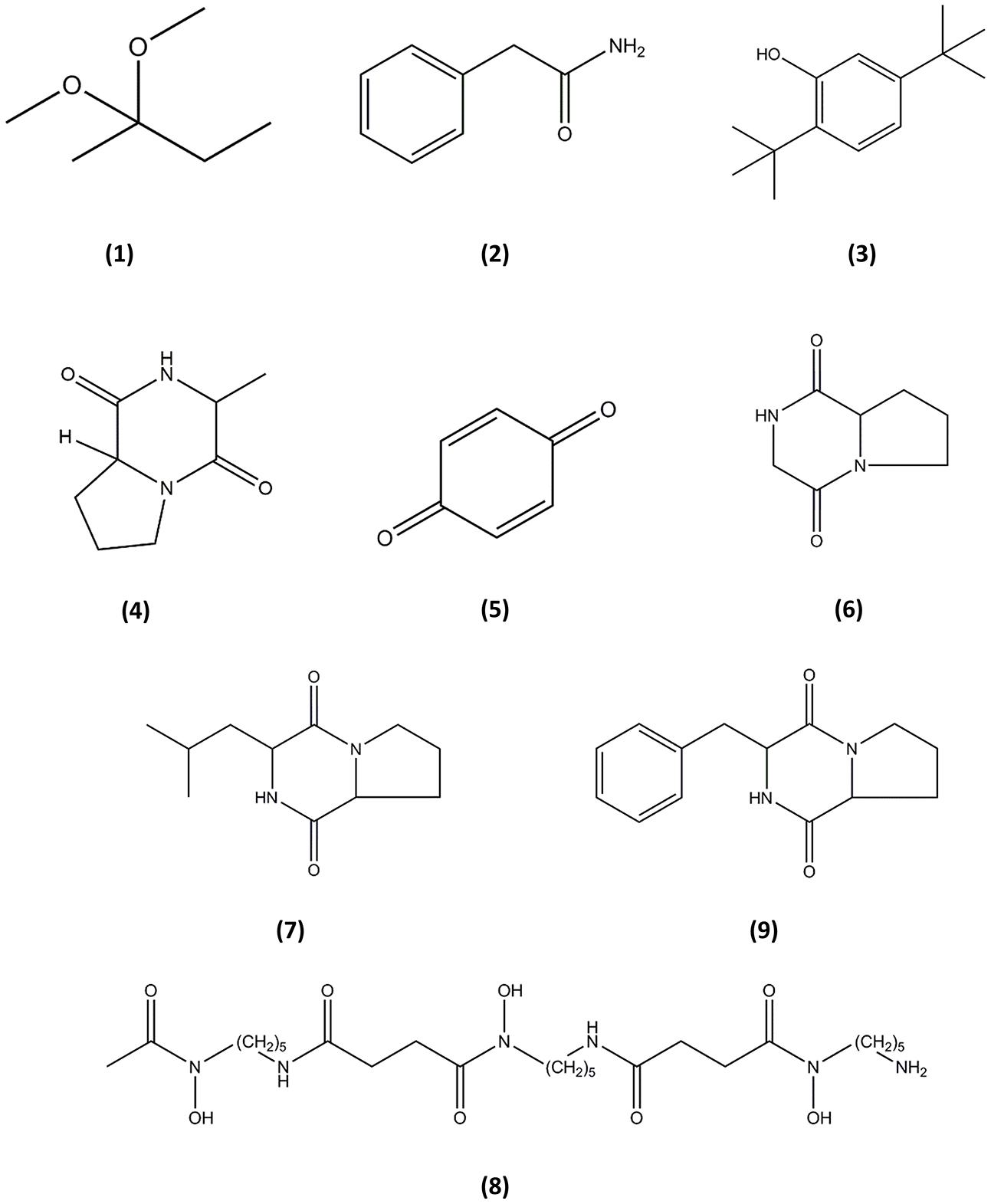
FIGURE 4. Chemical structures of the identified compounds from MUSC 137. (1), 2,2-dimethoxybutane; (2), Benzeneacetamide; (3), Phenol, 2,5-bis(1,1-dimethylethyl)-; (4), (3R,8aS)-3-methyl-1,2,3,4,6,7,8,8a-octahydropyrrolo[1,2-a]pyrazine-1,4-dione; (5), 2,5-cyclohexadiene-1, 4-dione; (6), Pyrrolo[1,2-a]pyrazine-1,4-dione, hexahydro-; (7), 1,4-diaza-2,5-dioxo-3-isobutyl bicyclo[4.3.0]nonane; (8), Deferoxamine; (9), Pyrrolo[1,2-a]pyrazine-1,4-dione, hexahydro-3-(2-phenylmethyl)-.
Discussion
Microorganisms have served as a promising source for the majority of the drugs in use today (Berdy, 2005; Azman et al., 2015). Their ability to produce a diverse array of bioactive small molecule natural products has made them among the key players in the search for novel therapeutic or preventive agents against diseases including cancer, neurodegenerative diseases and diabetes (Berdy, 2005; Chin et al., 2006). The phylum Actinobacteria produces approximately 10,000 clinically important compounds, of which about 75% are isolated from the genus Streptomyces (Watve et al., 2001; Berdy, 2005). These filamentous bacteria under the genus Streptomyces are economically important from a biotechnological and pharmacological point of view as they seem to be a virtually inexhaustible source of bioactive secondary metabolites with potent antibacterial, antifungal, antitumor and antioxidant activity (Kim et al., 2008; Olano et al., 2009a; Saurav and Kannabiran, 2012; Wang et al., 2013). In the current study, S. pluripotens MUSC 137 was isolated from a soil sample from a mangrove forest in Pahang, Peninsular Malaysia. Results from phylogenetic and genomic analyses indicated that strain MUSC 137 is closely related to S. pluripotens MUSC 135T as they formed a distinct subclade at 100% bootstrap value. Furthermore, the DNA–DNA relatedness values between MUSC 137 and MUSC 135T were 83 ± 3.2%, which were significantly higher that 70%, the threshold value for the delineation of bacterial species (Wayne et al., 1987). Also the BOX-PCR results exhibited that strain MUSC 137 presented a unique BOX-PCR fingerprint as compared to MUSC 135T and other closely related type strains, indicating that MUSC 137 and MUSC 135T are not clones. Overall, these results support the notion that strain MUSC 137 belonged to the same species as S. pluripotens MUSC 135T but has sufficient unique characteristics to be designated as a separate strain.
Cancer initiation and progression is associated with accumulation of free radicals or oxidative stress (Reuter et al., 2010). In general, free radicals cause various modifications or cause damage to biological macromolecules such as proteins, lipids and DNA. These deleterious effects then compromise functioning of important pathways including the DNA repair system which eventually results in an increased mutation rate (Pacifici and Davies, 1991; Bartsch and Nair, 2006). As antioxidants are capable of scavenging free radicals, these molecules could potentially affect the activation of signaling pathways which are crucial for survival of cancer cells (Lopaczynski and Zeisel, 2001; Chen et al., 2009). The bioactive potential of S. pluripotens strain MUSC 137 was explored using DPPH free radical scavenging assay to determine antioxidant activity. In the presence of antioxidants, the odd electron of DPPH radical is paired off which causes discoloration of the solution. This important characteristic has enabled researchers to assess the antioxidant strength of the sample(s) of interest. In this current study, DPPH assay revealed significant antioxidant activity of MUSC 137 extract (Table 1), which suggests that the strain may produce valuable compound(s) that could potentially reduce cancer occurrence and be further developed as chemopreventive drugs.
The cytotoxicity of MUSC 137 extract against several human cancer cell lines was examined with MTT assay. This tetrazolium-based colorimetric assay measures only in vitro living cells and is often employed in cytotoxicity studies to evaluate drug efficiency (Rubinstein et al., 1990; Ciapetti et al., 1993; Fotakis and Timbrell, 2006; Chan et al., 2015). The extract of MUSC 137 displayed varying levels of cytotoxicity against the tested cancer cell lines. MCF-7 was found to be the most vulnerable to the treatment extract with the lowest IC50 (61.33 ± 17.10 μg/mL), followed by HCT-116 (83.72 ± 7.17 μg/mL) and A549 (147.20 ± 19.23 μg/mL). Among the tested colon cancer cell lines, the treatment of MUSC 137 extract at the highest tested concentration (400 μg/mL) showed strongest growth inhibition against HCT-116 (18.5 ± 7.8%), while higher percentages of cell viability were still observed in HT-29 (44.1 ± 2.9%), Caco-2 (50.8 ± 2.1%), and SW480 (56.4 ± 1.3%). The tested colon cancer cell lines are known to possess dysfunctional p53 tumor protein, except HCT-116. Therefore, the varied levels of cytotoxic activity among these colon cancer cell lines exhibited by extract of MUSC 137 might be due to the p53 tumor suppressor protein status of these cancer cell lines (Petitjean et al., 2007; Goh et al., 2014). These findings have led to the speculation that MUSC 137 extract might be able to induce cancer cell death via p53 dependent cell death signaling pathway. Elucidation of the underlying mechanism of MUSC 137 extract on cancer cell lines may be a valuable area of future research as these information could potentially assist in its development as chemopreventive drug.
An ideal chemotherapy drug should have high specificity and should be able to discriminate between cancer and normal cells. However, many of the anticancer drugs in use are still lacking in the drug specificity as they kill both cancer cells as well as normal cells (Chabner and Roberts, 2005; Chari, 2007; Fernandez, 2014). Tremendous efforts have been invested to search for novel chemotherapy drugs with high potency and specificity. This current study investigating specificity of MUSC 137 extract revealed that the extract was less toxic against normal cell lines when compared to MCF-7 and HCT-116 cancer cell lines as shown by the SI value of 4.22 and 3.09, respectively. These crucial findings have provided new insight into the cytotoxic potential of MUSC 137 against cancer cells, particularly against MCF-7 and HCT-116 with high specificity. Further analysis investigating the target action of the bioactive compounds could provide helpful information for future development as an anticancer drug. In summary, the results of both the DPPH assay and in vitro anticancer screening suggests the presence of potent biologically active compounds in the extract of MUSC 137, which then prompted subsequent chemical analysis using GC–MS to identify the chemical constituents present in the extract.
A powerful analytical tool, GC–MS combines the separation power of GC and detection power of MS, generating robust and reliable data (Hites, 1997; Stein, 1999). GC–MS plays an important role in natural product discovery, including bioactive compounds derived from Streptomyces species (Pollak and Berger, 1996; Sudha and Masilamani, 2012; Ara et al., 2014; Jog et al., 2014). In the current study, GC–MS detected nine compounds in the extract of MUSC 137, with the majority of the compounds having been detected in marine-derived microorganisms including Streptomyces species (Hong et al., 2008; Gao et al., 2014; Narendhran et al., 2014). The analysis detected several members of pyrrolizidines which are known to exhibit a diverse array of bioactivities including antimicrobial, antitumor, anti-angiogenesis and antioxidant activities (Olano et al., 2009b; Melo et al., 2014; Robertson and Stevens, 2014). The pyrrolizidines in the extract of MUSC 137 include 1,4-diaza-2,5-dioxo-3-isobutyl bicyclo[4.3.0]nonane, Pyrrolo[1,2-a]pyrazine-1,4-dione, hexahydro-3-(2-phenylmethyl)-, Pyrrolo[1,2-a]pyrazine-1,4-dione, hexahydro- and (3R,8aS)-3-methyl-1,2,3,4,6,7,8,8a-octahydropyrrolo[1,2-a]pyrazine-1,4-dione. As a matter of fact, Pyrrolo[1,2-a]pyrazine-1,4-dione, hexahydro-3-(2-phenylmethyl)- has been shown to inhibit expression of serine/threonine kinase Akt which involves in regulation cell proliferation and apoptosis (Hong et al., 2008). Furthermore, the chemical compound 1,4-diaza-2,5-dioxo-3-isobutyl bicyclo[4.3.0]nonane has also been associated with the cytotoxic activity observed on human cervical cancer cell line (Narendhran et al., 2014). Therefore, the presence of these compounds in the extract of MUSC 137 could account for the observed cytotoxicity against the tested cancer cell lines.
In addition, GC–MS analysis also detected compounds that may account for the antioxidant activity observed. In the current study, Pyrrolo[1,2-a]pyrazine-1,4-dione, hexahydro- and (3R,8aS)-3-methyl-1,2,3,4,6,7,8,8a-octahydropyrrolo[1,2-a]pyrazine-1,4-dione were also found in other Streptomyces and Bacillus strains which exhibited antioxidant activity (Gopi et al., 2014; Ser et al., 2015b). As antioxidants are suggested to affect survival of cancer cells (Lopaczynski and Zeisel, 2001; Chen et al., 2009), these pyrrolizidines with antioxidant activity in MUSC 137 could be contributing to the observed cytotoxic effects. Besides pyrrolizidines, GC–MS has also revealed the presence of deferoxamine in MUSC 137. Deferoxamine (with the prescription name of Desferal) is listed on World Health Organization’s List of Essential Medicines due to its medical importance as iron chelator and to protect against iron-induced oxidative stress (World Health Organization [WHO], 2015). This trihydroxamate molecule can reduce accumulation of free radicals and inhibit growth of various cancer cell lines and tumors (Tomoyasu et al., 1992; Shimoni et al., 1994; Melillo et al., 1997; Buss et al., 2004; Yamasaki et al., 2011; Salis et al., 2014). The detection of deferoxamine in MUSC 137 might be associated with the antioxidant activity. Also, it may affect the expression of crucial genes and signaling pathways responsible for the survival of cancer cells, subsequently leading to a reduction in viability of cancer cell as shown in MTT assay.
Conclusion
This study has revealed the biopharmaceutical potential possessed by S. pluripotens strain MUSC 137 as this strain has demonstrated its ability to produce bioactive compounds which may account for its antioxidant and cytotoxic activities. Future studies should focus on the elucidation of mechanism action of cell death to provide more convincing evidence of selective cytotoxic activity observed and work investigating this phenomenon is in fact currently underway.
Author Contributions
H-LS, B-HG, and L-HL contributed to the data analyses and writing of the manuscript. N-SA, W-FY, and K-GC contributed by providing vital technical support for the project. K-GC, B-HG, and L-HL contributed to the inception of the experimental design and funding of the project.
Funding
This work was supported by a University of Malaya for High Impact Research Grant (UM-MOHE HIR Nature Microbiome Grant No. H-50001-A000027 and No. A000001-50001) awarded to K-GC., a MOSTI eScience Fund (06-02-10-SF0300) awarded to L-HL and a MOSTI eScience Fund (02-02-10-SF0215) awarded to B-HG.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
The authors are thankful to Professor Bernhard Schink for the support in the Latin etymology of the new species name.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.01398
Footnotes
References
Ara, I., Bukhari, N. A., Aref, N., Shinwari, M. M., and Bakir, M. (2014). Antiviral activities of streptomycetes against tobacco mosaic virus (TMV) in Datura plant: evaluation of different organic compounds in their metabolites. Afr. J. Biotechnol. 11, 2130–2138.
Azman, A. S., Othman, I., Velu, S. S., Chan, K. G., and Lee, L. H. (2015). Mangrove rare actinobacteria: taxonomy, natural compound, and discovery of bioactivity. Front. Microbiol. 6:856. doi: 10.3389/fmicb.2015.00856
Bartsch, H., and Nair, J. (2006). Chronic inflammation and oxidative stress in the genesis and perpetuation of cancer: role of lipid peroxidation, DNA damage, and repair. Langenbeck’s Arch. Surg. 391, 499–510. doi: 10.1007/s00423-006-0073-1
Buss, J. L., Greene, B. T., Turner, J., Torti, F. M., and Torti, S. V. (2004). Iron chelators in cancer chemotherapy. Curr. Topics Med. Chem. 4, 1623–1635. doi: 10.2174/1568026043387269
Carrillo, P., Mardaraz, C., Pitta-Alvarez, S., and Giulietti, A. (1996). Isolation and selection of biosurfactant-producing bacteria. World J. Microbiol. Biotechnol. 12, 82–84. doi: 10.1007/BF00327807
Cashion, P., Holder-Franklin, M. A., McCully, J., and Frankiln, M. (1977). A rapid method for the base ratio determination of bacterial DNA. Anal. Biochem. 81, 461–466. doi: 10.1016/0003-2697(77)90720-5
Cerny, G. (1978). Studies on the aminopeptidase test for the distinction of gram-negative from gram-positive bacteria. Eur. J. Appl. Microbiol. Biotechnol. 5, 113–122. doi: 10.1007/BF00498805
Chabner, B. A., and Roberts, T. G. (2005). Chemotherapy and the war on cancer. Nat. Rev. Cancer 5, 65–72. doi: 10.1038/nrc1529
Chan, C. K., Supriady, H., Goh, B. H., and Kadir, H. A. (2015). Elephantopus scaber induces apoptosis through ROS-dependent mitochondrial signaling pathway in HCT116 human colorectal carcinoma cells. J. Ethnopharmacol. 168, 291–304. doi: 10.1016/j.jep.2015.03.072
Chari, R. V. (2007). Targeted cancer therapy: conferring specificity to cytotoxic drugs. Acct. Chem. Res. 41, 98–107. doi: 10.1021/ar700108g
Chen, H. M., Wu, Y. C., Chia, Y. C., Chang, F. R., Hsu, H. K., Hsieh, Y. C., et al. (2009). Gallic acid, a major component of Toona sinensis leaf extracts, contains a ROS-mediated anti-cancer activity in human prostate cancer cells. Cancer Lett. 286, 161–171. doi: 10.1016/j.canlet.2009.05.040
Chin, Y.-W., Balunas, M. J., Chai, H. B., and Kinghorn, A. D. (2006). Drug discovery from natural sources. AAPS J. 8, E239–E253. doi: 10.1007/BF02854894
Ciapetti, G., Cenni, E., Pratelli, L., and Pizzoferrato, A. (1993). In vitro evaluation of cell/biomaterial interaction by MTT assay. Biomaterials 14, 359–364. doi: 10.1016/0142-9612(93)90055-7
Crnovcic, I., Vater, J., and Keller, U. (2013). Occurrence and biosynthesis of C-demethyl actinomycins in actinomycin-producing Streptomycesc hrysomallus and Streptomyces parvulus. J. Antibiot. 66, 211–218. doi: 10.1038/ja.2012.120
De Ley, J., Cattoir, H., and Reynaerts, A. (1970). The quantitative measurement of DNA hybridization from renaturation rates. Eur. J. Biochem. 12, 133–142. doi: 10.1111/j.1432-1033.1970.tb00830.x
Felsenstein, J. (1981). Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17, 368–376. doi: 10.1007/BF01734359
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–789. doi: 10.2307/2408678
Fernandez, A. (2014). Synergizing immunotherapy with molecular-targeted anticancer treatment. Drug Dis. Today 19, 1427–1432. doi: 10.1016/j.drudis.2014.03.022
Fotakis, G., and Timbrell, J. A. (2006). In vitro cytotoxicity assays: comparison of LDH, neutral red, MTT and protein assay in hepatoma cell lines following exposure to cadmium chloride. Toxicol. Lett. 160, 171–177. doi: 10.1016/j.toxlet.2005.07.001
Gao, C. H., Chen, Y. N., Pan, L. X., Lei, F., Long, B., Hu, L. Q., et al. (2014). Two new cyclic tetrapeptides from deep-sea bacterium Bacillus amyloliquefaciens GAS 00152. J. Antibiot. 67, 541–543. doi: 10.1038/ja.2014.27
Goh, B. H., Chan, C. K., Kamarudin, M. N. A., and Kadir, H. A. (2014). Swietenia macrophylla King induces mitochondrial-mediated apoptosis through p53 upregulation in HCT116 colorectal carcinoma cells. J. Ethnopharmacol. 153, 375–385. doi: 10.1016/j.jep.2014.02.036
Gopi, M., Dhayanithi, N. B., Devi, K. N., and Kumar, T. T. A. (2014). Marine natural product, Pyrrolo [1,2-a] pyrazine-1,4-dione, hexahydro-(C7H10N2O2) of antioxidant properties from Bacillus species at Lakshadweep archipelago. J. Coast. Life Med. 2, 632–637. doi: 10.12980/JCLM.2.201414J40
Hites, R. A. (1997). “Gas chromatography mass spectrometry,” in Handbook of Instrumental techniques for analytical chemistry, ed. F. Settle (Upper Saddle River, N.J.: Prentice Hall), 609–626.
Hong, K., Gao, A.-H., Xie, Q.-Y., Gao, H. G., Zhuang, L., Lin, H.-P., et al. (2009). Actinomycetes for marine drug discovery isolated from mangrove soils and plants in China. Mar. drugs 7, 24–44. doi: 10.3390/md7010024
Hong, S., Moon, B.-H., Yong, Y., Shin, S. Y., Lee, Y. H., and Lim, Y. (2008). Inhibitory effect against Akt of cyclic dipeptides isolated from Bacillus sp. J. Microbiol. Biotechnol. 18, 682–685.
Hu, H., Lin, H.-P., Xie, Q., Li, L., Xie, X.-Q., and Hong, K. (2012). Streptomyces qinglanensis sp. nov., isolated from mangrove sediment. Int. J. Syst. Evol. Microbiol. 62, 596–600. doi: 10.1099/ijs.0.032201-0
Hu, H., Lin, H.-P., Xie, Q., Li, L., Xie, X.-Q., Sun, M., et al. (2011). Streptomyces shenzhenensis sp. nov., a novel actinomycete isolated from mangrove sediment. Antonie van Leeuwenhoek 100, 631–637. doi: 10.1007/s10482-011-9618-6
Huss, V. A. R., Festl, H., and Schleifer, K. H. (1983). Studies on the spectrophotometric determination of DNA hybridization from renaturation rates. Syst. Appl. Microbiol. 4, 184–192. doi: 10.1016/S0723-2020(83)80048-4
Jennerjahn, T. C., and Ittekkot, V. (2002). Relevance of mangroves for the production and deposition of organic matter along tropical continental margins. Naturwissenschaften 89, 23–30. doi: 10.1007/s00114-001-0283-x
Jog, R., Pandya, M., Nareshkumar, G., and Rajkumar, S. (2014). Mechanism of phosphate solubilization and antifungal activity of Streptomyces spp. isolated from wheat roots and rhizosphere and their application in improving plant growth. Microbiology 160, 778–788. doi: 10.1099/mic.0.074146-0
Kawahara, T., Izumikawa, M., Otoguro, M., Yamamura, H., Hayakawa, M., Takagi, M., et al. (2012). JBIR-94 and JBIR-125, antioxidative phenolic compounds from Streptomyces sp. R56-07. J. Nat. Prod. 75, 107–110. doi: 10.1021/np200734p
Kelly, K. L. (1964). Inter-Society Color Council–National Bureau of Standards Color Name Charts Illustrated with Centroid Colors. Washington, DC: US Government Printing Office.
Kim, K.-J., Kim, M.-A., and Jung, J.-H. (2008). Antitumor and antioxidant activity of protocatechualdehyde produced from Streptomyces lincolnensis M-20. Arch. Pharm. Res. 31, 1572–1577. doi: 10.1007/s12272-001-2153-7
Kim, O. S., Cho, Y. J., Lee, K., Yoon, S. H., Kim, M., Na, H., et al. (2012). Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 62, 716–721. doi: 10.1099/ijs.0.038075-0
Kumar, P. S., Duraipandiyan, V., and Ignacimuthu, S. (2014). Isolation, screening and partial purification of antimicrobial antibiotics from soil Streptomyces sp. SCA 7. Kaohsiung J. Med. Sci. 30, 435–446. doi: 10.1016/j.kjms.2014.05.006
Küster, E., and Williams, S. (1964). Media for the isolation of streptomycetes: starch casein medium. Nature 202, 928–929. doi: 10.1038/202928a0
Lee, L.-H., Zainal, N., Azman, A.-S., Eng, S.-K., Goh, B.-H., Yin, W.-F., et al. (2014a). Diversity and antimicrobial activities of actinobacteria isolated from tropical mangrove sediments in Malaysia. Sci. World J. 2014:14. doi: 10.1155/2014/698178
Lee, L.-H., Zainal, N., Azman, A.-S., Eng, S.-K., Ab Mutalib, N.-S., Yin, W.-F., et al. (2014b). Streptomyces pluripotens sp. nov., a bacteriocin-producing streptomycete that inhibits meticillin-resistant Staphylococcus aureus. Int. J. Syst. Evol. Microbiol. 64, 3297–3306. doi: 10.1099/ijs.0.065045-0
Lee, L.-H., Azman, A.-S., Zainal, N., Eng, S.-K., Fang, C.-M., Hong, K., et al. (2014c). Novosphingobium malaysiense sp. nov. isolated from mangrove sediment. Int. J. Syst. Evol. Microbiol. 64, 1194–1201. doi: 10.1099/ijs.0.059014-0
Lee, L.-H., Azman, A.-S., Zainal, N., Eng, S.-K., Ab Mutalib, N.-S., Yin, W.-F., et al. (2014d). Microbacterium mangrovi sp. nov., an amylotytic actinobacterium isolated from mangrove forest soil. Int. J. Syst. Evol. Microbiol. 64, 3513–3519. doi: 10.1099/ijs.0.062414-0
Lee, L.-H., Zainal, N., Azman, A.-S., Ab Mutalib, N.-S., Hong, K., and Chan, K.-G. (2014e). Mumia flava gen. nov., sp. nov., an actinobacterium of the family Nocardioidaceae. Int. J. Syst. Evol. Microbiol. 64, 1461–1467. doi: 10.1099/ijs.0.058701-0
Ling, L. T., Yap, S.-A., Radhakrishnan, A. K., Subramaniam, T., Cheng, H. M., and Palanisamy, U. D. (2009). Standardised Mangifera indica extract is an ideal antioxidant. Food Chem. 113, 1154–1159. doi: 10.1016/j.foodchem.2008.09.004
Lopaczynski, W., and Zeisel, S. H. (2001). Antioxidants, programmed cell death, and cancer. Nutr. Res. 21, 295–307. doi: 10.1016/S0271-5317(00)00288-8
Macfaddin, J. (2000). Biochemical Tests for Identification of Medical Bacteria. Philadelphia: Lippincott Williams and Wilkins.
Meena, B., Rajan, L. A., Vinithkumar, N. V., and Kirubagaran, R. (2013). Novel marine actinobacteria from emerald Andaman & Nicobar Islands: a prospective source for industrial and pharmaceutical byproducts. BMC Microbiol. 13:145. doi: 10.1186/1471-2180-13-145
Melillo, G., Taylor, L. S., Brooks, A., Musso, T., Cox, G. W., and Varesio, L. (1997). Functional requirement of the hypoxia-responsive element in the activation of the inducible nitric oxide synthase promoter by the iron chelator desferrioxamine. J. Biol. Chem. 272, 12236–12243. doi: 10.1074/jbc.272.18.12236
Melo, I. S., Santos, S. N., Rosa, L. H., Parma, M. M., Silva, L. J., Queiroz, S. C., et al. (2014). Isolation and biological activities of an endophytic Mortierella alpina strain from the Antarctic moss Schistidium antarctici. Extremophiles 18, 15–23. doi: 10.1007/s00792-013-0588-7
Narendhran, S., Rajiv, P., Vanathi, P., and Sivaraj, R. (2014). Spectroscopic analysis of bioactive compounds from Streptomyces cavouresis KU-V39: evaluation of antioxidant and cytotoxicity activity. Int. J. Pharm. Pharmaceut. Sci. 6:322.
Olano, C., Méndez, C., and Salas, J. A. (2009a). Antitumor compounds from actinomycetes: from gene clusters to new derivatives by combinatorial biosynthesis. Nat. Prod. Rep. 26, 628–660. doi: 10.1039/b822528a
Olano, C., Méndez, C., and Salas, J. A. (2009b). Antitumor compounds from marine actinomycetes. Mar. Drugs 7, 210–248. doi: 10.3390/md7020210
Pacifici, R. E., and Davies, K. J. A. (1991). Protein, lipid and DNA repair systems in oxidative stress: the free-radical theory of aging revisited. Gerontology 37, 166–180. doi: 10.1159/000213257
Petitjean, A., Mathe, E., Kato, S., Ishioka, C., Tavtigian, S. V., Hainaut, P., et al. (2007). Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum. Mutat. 28, 622–629. doi: 10.1002/humu.20495
Pollak, F. C., and Berger, R. G. (1996). Geosmin and related volatiles in bioreactor-cultured Streptomyces citreus CBS 109.60. Appl. Environ. Microbiol. 62, 1295–1299.
Qin, S., Xing, K., Jiang, J. H., Xu, L. H., and Li, W. J. (2011). Biodiversity, bioactive natural products and biotechnological potential of plant-associated endophytic actinobacteria. Appl. Microbiol. Biotechnol. 89, 457–473. doi: 10.1007/s00253-010-2923-6
Rehm, S., Han, S., Hassani, I., Sokocevic, A., Jonker, H. R., Engels, J. W., et al. (2009). The high resolution NMR structure of parvulustat (Z-2685) from Streptomyces parvulus FH-1641: comparison with tendamistat from Streptomyces tendae 4158. ChemBioChem 10, 119–127. doi: 10.1002/cbic.200800547
Reuter, S., Gupta, S. C., Chaturvedi, M. M., and Aggarwal, B. B. (2010). Oxidative stress, inflammation, and cancer: how are they linked? Free Radical Biol. Med. 49, 1603–1616. doi: 10.1016/j.freeradbiomed.2010.09.006
Robertson, J., and Stevens, K. (2014). Pyrrolizidine alkaloids. Nat. Prod. Rep. 31, 1721–1788. doi: 10.1039/c4np00055b
Rubinstein, L., Shoemaker, R., Paull, K., Simon, R., Tosini, S., Skehan, P., et al. (1990). Comparison of in vitro anticancer-drug-screening data generated with a tetrazolium assay versus a protein assay against a diverse panel of human tumor cell lines. J. Natl. Cancer Inst. 82, 1113–1117. doi: 10.1093/jnci/82.13.1113
Salis, O., Bedir, A., Kilinc, V., Alacam, H., Gulten, S., and Okuyucu, A. (2014). The anticancer effects of desferrioxamine on human breast adenocarcinoma and hepatocellular carcinoma cells. Cancer Biomark. 14, 419–426. doi: 10.3233/CBM-140422
Saurav, K., and Kannabiran, K. (2012). Cytotoxicity and antioxidant activity of 5-(2,4-dimethylbenzyl)pyrrolidin-2-one extracted from marine Streptomyces VITSVK5 spp. Saudi J. Biol. Sci. 19, 81–86. doi: 10.1016/j.sjbs.2011.07.003
Ser, H.-L., Zainal, N., Palanisamy, U. D., Goh, B.-H., Yin, W.-F., Chan, K.-G., et al. (2015a). Streptomyces gilvigriseus sp. nov., a novel actinobacterium isolated from mangrove forest soil. Antonie van Leeuwenhoek 107, 1369–1378. doi: 10.1007/s10482-015-0431-5
Ser, H.-L., Palanisamy, U. D., Yin, W.-F., Abd Malek, S. N., Chan, K.-G., Goh, B.-H., et al. (2015b). Presence of antioxidative agent, Pyrrolo[1,2-a]pyrazine-1,4-dione,hexahydro- in newly isolated Streptomyces mangrovisoli sp. nov. Front. Microbiol. 6:854. doi: 10.3389/fmicb.2015.00854
Shieh, W. Y., Chen, Y.-W., Chaw, S.-M., and Chiu, H.-H. (2003). Vibrio ruber sp. nov., a red, facultatively anaerobic, marine bacterium isolated from sea water. Int. J. Syst. Evol. Microbiol. 53, 479–484. doi: 10.1099/ijs.0.02307-0
Shimoni, E., Armon, R., and Neeman, I. (1994). Antioxidant properties of deferoxamine. J. Amer. Oil Chemists’ Soc. 71, 641–644. doi: 10.1007/BF02540593
Shirling, E. B., and Gottlieb, D. (1966). Methods for characterization of Streptomyces species. Int. J. Syst. Evol. Microbiol. 16, 313–340. doi: 10.1099/00207713-16-3-313
Stein, S. E. (1999). An integrated method for spectrum extraction and compound identification from gas chromatography/mass spectrometry data. J. Am. Soc. Mass Spectro. 10, 770–781. doi: 10.1016/S1044-0305(99)00047-1
Subramani, R., and Aalbersberg, W. (2012). Marine actinomycetes: an ongoing source of novel bioactive metabolites. Microbiol. Res. 167, 571–580. doi: 10.1016/j.micres.2012.06.005
Sudha, S., and Masilamani, S. M. (2012). Characterization of cytotoxic compound from marine sediment derived actinomycete Streptomyces avidinii strain SU4. Asian Pac. J. Trop. Biomed. 2, 770–773. doi: 10.1016/S2221-1691(12)60227-5
Sui, J.-L., Xu, X.-X., Qu, Z., Wang, H.-L., Lin, H.-P., Xie, Q.-Y., et al. (2011). Streptomyces sanyensis sp. nov., isolated from mangrove sediment. Int. J. Syst. Evol. Microbiol. 61, 1632–1637. doi: 10.1099/ijs.0.023515-0
Takahashi, Y., Matsumoto, A., Seino, A., Iwai, Y., and Omura, S. (1996). Rare actinomycetes isolated from desert soils. Actinomycetologica 10, 91–97. doi: 10.3209/saj.10_91
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tan, L. T.-H., Ser, H.-L., Yin, W.-F., Chan, K.-G., Lee, L.-H., and Goh, B.-H. (2015). Investigation of antioxidative and anticancer potentials of Streptomyces sp. MUM256 isolated from Malaysia mangrove soil. Front. Microbiol. 6:1316. doi: 10.3389/fmicb.2015.01316
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nuc. Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Tomoyasu, S., Fukuchi, K., Yajima, K., Watanabe, K., Suzuki, H., Kawakami, K., et al. (1992). Suppression of HL-60 cell proliferation by deferoxamine: changes in c-myc expression. Anticancer Res. 13, 407–410.
Waksman, S. A., and Henrici, A. T. (1943). The nomenclature and classification of the actinomycetes. J. Bacteriol. 46, 337–341.
Wang, C., Wang, Z., Qiao, X., Li, Z., Li, F., Chen, M., et al. (2013). Antifungal activity of volatile organic compounds from Streptomyces alboflavus TD-1. FEMS Microbiol. Lett. 341, 45–51. doi: 10.1111/1574-6968.12088
Watve, M. G., Tickoo, R., Jog, M. M., and Bhole, B. D. (2001). How many antibiotics are produced by the genus Streptomyces? Arch. Microbiol. 176, 386–390. doi: 10.1007/s002030100345
Wayne, L. G., Brenner, D. J., Colwell, R. R., Grimont, P. A. D., Kandler, O., Krichevsky, M. I., et al. (1987). Report of the Ad Hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37, 463–464. doi: 10.1099/00207713-37-4-463
World Health Organization [WHO] (2015). WHO model list of essential medicines: 19th list (updated) April 2015.
Williams, S. T., Goodfellow, M., and Alderson, G. (1989). “Genus streptomyces waksman and henrici 1943, 339AL,” in Bergey’s Manual of Systematic Bacteriology, eds S. T. Williams, M. E. Sharpe, and J. G. Holt (Baltimore: Williams & Wilkins), 2452–2492.
Keywords: Streptomyces pluripotens, cytotoxic, antioxidative, mangrove, actinobacteria
Citation: Ser H-L, Ab Mutalib N-S, Yin W-F, Chan K-G, Goh B-H and Lee L-H (2015) Evaluation of Antioxidative and Cytotoxic Activities of Streptomyces pluripotens MUSC 137 Isolated from Mangrove Soil in Malaysia. Front. Microbiol. 6:1398. doi: 10.3389/fmicb.2015.01398
Received: 06 October 2015; Accepted: 24 November 2015;
Published: 16 December 2015.
Edited by:
Dongsheng Zhou, Beijing Institute of Microbiology and Epidemiology, ChinaCopyright © 2015 Ser, Ab Mutalib, Yin, Chan, Goh and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Learn-Han Lee, lee.learn.han@monash.edu; leelearnhan@yahoo.com; Bey-Hing Goh, goh.bey.hing@monash.edu
 Hooi-Leng Ser
Hooi-Leng Ser Nurul-Syakima Ab Mutalib
Nurul-Syakima Ab Mutalib Wai-Fong Yin
Wai-Fong Yin Kok-Gan Chan
Kok-Gan Chan Bey-Hing Goh
Bey-Hing Goh Learn-Han Lee
Learn-Han Lee