- 1Division of Genetics and Molecular Biology, Institute of Biological Sciences, Faculty of Science, University of Malaya, Kuala Lumpur, Malaysia
- 2Biomedical Research Laboratory, Jeffrey Cheah School of Medicine and Health Sciences, Monash University Malaysia, Bandar Sunway, Malaysia
High consumer demand for shellfish has led to the need for large-scale, reliable shellfish supply through aquaculture or shellfish farming. However, bacterial infections which can spread rapidly among shellfish poses a major threat to this industry. Shellfish farmers therefore often resort to extensive use of antibiotics, both prophylactically and therapeutically, in order to protect their stocks. The extensive use of antibiotics in aquaculture has been postulated to represent a major contributing factor in the rising incidence of antimicrobial resistant pathogenic bacteria in shellfish. This study aimed to investigate the incidence of pathogenic Vibrio parahaemolyticus and determine the antibiotic resistance profile as well as to perform plasmid curing in order to determine the antibiotic resistance mediation. Based on colony morphology, all 450 samples tested were positive for Vibrio sp; however, tox-R assay showed that only 44.4% (200/450) of these were V. parahaemolyticus. Out of these 200 samples, 6.5% (13/200) were trh-positive while none were tdh-positive. Antibiotic resistance was determined for all V. parahaemolyticus identified against 14 commonly used antibiotics and the multiple antibiotic resistance index (MAR) was calculated. The isolates demonstrated high resistance to several antibiotics tested- including second and third-line antibiotics- with 88% resistant to ampicillin, 81% to amikacin,70.5% to kanamycin, 73% to cefotaxime, and 51.5% to ceftazidime. The MAR index ranged from 0.00 to 0.79 with the majority of samples having an index of 0.36 (resistant to five antibiotics). Among the 13 trh-positive strains, almost 70% (9/13) demonstrated resistance to 4 or more antibiotics. Plasmid profiling for all V. parahaemolyticus isolates revealed that 86.5% (173/200) contained plasmids - ranging from 1 to 7 plasmids with DNA band sizes ranging from 1.2 kb to greater than 10 kb. 6/13 of the pathogenic V. pathogenic strains contained plasmid. After plasmid curing, the plasmid containing pathogenic strains isolated in our study have chromosomally mediated ampicillin resistance while the remaining resistance phenotypes are plasmid mediated. Overall, our results indicate that while the incidence of pathogenic V. parahaemolyticus in shellfish in Selangor still appears to be at relatively reassuring levels, antibiotic resistance is a real concern and warrants ongoing surveillance.
Introduction
Vibrio parahaemolyticus is a Gram-negative bacterium that is widely disseminated in marine and estuarine environments worldwide (Su and Liu, 2007; Ceccarelli et al., 2013; Zhang and Orth, 2013; Letchumanan et al., 2014; Velazquez-Roman et al., 2014; Wu et al., 2014). While the majority of strains isolated from environmental sources are innocuous members of marine microbiota, a small number of V. parahaemolyticus strains are capable of causing human illness and are often associated with food borne gastroenteritis or diarrhea (Nair et al., 2007; Hazen et al., 2015; Raghunath, 2015). The virulent strains are discerned from avirulent strains by the presence of toxigenic genes namely, thermostable direct hemolysin (tdh) and/or tdh-related (trh) hemolysin genes (Letchumanan et al., 2014).
Although V. parahaemolyticus is commonly present in seafood, most of these isolates are regarded as non-pathogenic to human (Nishibuchi and Kaper, 1995; Velazquez-Roman et al., 2012; Raghunath, 2015). The strains isolated from environmental samples usually lack the pathogenic genes thermostable direct hemolysin (tdh) and/or TDH-related hemolysin (trh) which are responsible for causing diseases in human and marine animals (Deepanjali et al., 2005; Canizalez-Roman et al., 2011; Gutierrez West et al., 2013). Previous study reported, around 0–6% of the environmental V. parahaemolyticus strains carry tdh and/or trh genes (DePaola et al., 2000; Vuddhakul et al., 2000; Wong et al., 2000; Alam et al., 2002; Hervio-Heath et al., 2002; Haley et al., 2014). These two genes are considered major virulence factors of V. parahaemolyticus (Kaysner and DePaola, 2001; Zhang and Austin, 2005; Xu et al., 2014).
There are many methods utilized for the detection of V. parahaemolyticus. The standard method for detection and identification of V. parahaemolyticus uses microbiological media enrichments such as Thiosulfate Citrate Bile Salt (TCBS) agar, Alkaline Peptone Water (APW) along with a range of biochemical tests (Vincent et al., 2015). These methods are valuable for estimation of the total load of V. parahaemolyticus in a sample. This then enables estimation of potential risk for presence of pathogenic strains (Malcolm et al., 2015). Routine phenotyping and biochemical identification methods of V. parahaemolyticus are often complicated when the strains are isolated from seafood or marine surroundings (Nishibuchi, 2006). In order to enable rapid and accurate identification of V. parahaemolyticus, a combination of conventional and molecular approaches using PCR assay is often adapted by researchers. At present, pathogenic V. parahaemolyticus isolates in seafood and environmental samples are identified using PCR-based methods that amplify tox-R (the toxin operon gene that is well conserved among V. parahaemolyticus), tdh and trh gene sequences (Panicker et al., 2004; Yamamoto et al., 2008; Paydar et al., 2013; Law et al., 2015; Malcolm et al., 2015). PCR primers can be multiplexed in a single reaction to increase the detection limit or tailored as real-time PCR assay to provide rapid results (Grant et al., 2006; Zhang et al., 2014).
An increasing population with increased purchasing power in globally has increased the demand for and export potential of seafood; resulting in steady expansion of the Asian aquaculture industry (Rico et al., 2012). However, as aquaculture practices have intensified, the sector has been constantly challenged by aquatic animal health problems which are a major constraint to the development and expansion of the aquaculture sector (Bondad-Reantaso et al., 2005). Hence, aquaculture farmers rely on a wide range of antibiotics to prevent (prophylactic use) and treat (therapeutic use) bacterial infections in fish and other invertebrates (Cabello et al., 2013). Oxytetracycline, tetracycline, quinolones, sulphonamides and trimethoprim are among the antimicrobials permitted and utilized in the Asian aquaculture industry (Rico et al., 2012; Yano et al., 2014). The extensive use of antibiotics and other chemotherapeutics in aquaculture fields has caused the emergence of antibiotic resistant strains in the environment. Every year, more and more pathogenic Vibrio species have been reported to develop increasing levels of resistance toward most of the clinically used antibiotics (Letchumanan et al., 2015b).
Generally, Vibrio species are known to be highly susceptible to most clinically used antibiotics (Mala et al., 2014; Shaw et al., 2014; Letchumanan et al., 2015a). It is noted that most of the genetic determinants that confer antibiotic resistance are located in the plasmid. Plasmids are one of the important mediators that facilitate the transfer of antibiotic resistant genes and can be transmitted to the next generation via vertical gene transfer or exchanged with other bacteria via horizontal gene transfer (Okamoto et al., 2009; Manjusha and Sarita, 2011). Plasmid profile determination represents one of the earliest DNA based assay used for epidemiological studies and remains useful today (Meyer, 1988). Previous studies have reported that Vibrio sp. contain plasmids and there is a correlation between possessions of plasmid with antibiotic resistance (Molina-Aja et al., 2002; Zulkifli et al., 2009). Plasmid curing of bacteria is a possible way to eliminate the plasmids which may then allow determination of the mode of antibiotic resistance mediation. Plasmid curing protocols for Vibrio sp. involves chemical agents such as ethidium bromide (EB), acridine orange (AO) and sodium dodecyl sulphate (SDS), and physical agents such as treatment with ultraviolet and growth at elevated temperature (Liu et al., 2012).
Shellfish mainly resides in coastal and estuarine environments. Due to the nature of their habitat, shellfish contains diverse bacterial microbiota including Vibrio sp. (Romalde et al., 2014). Previous studies have found high prevalence of V. parahaemolyticus in shellfish in Malaysia (Zulkifli et al., 2009; Al-Othrubi et al., 2014; Sahilah et al., 2014; Tang et al., 2014; Malcolm et al., 2015). The presence of potentially pathogenic Vibrio species in shellfish in Malaysia highlights the need for continuous monitoring as well as consumer education on food safety. Considering these factors, the present study aimed to investigate the incidence of pathogenic and multidrug resistant strains of V. parahaemolyticus in shellfish by both conventional and molecular methods, as well as investigating the antibiotic resistance profiles of these organisms and to attempt to determine mediations via plasmid curing.
Materials and Methods
Sampling
The study focused mainly on five type of shellfish; mud crab (Scylla serrate), flower crab (Portunus pelagicus), carpet clam (Paphia textile), hard shell clam (Meretrix meretrix), and mud creeper (Cerithidea obtuse). A total of 450 shellfish samples were purchased from three selected local wetmarkets and three local supermarkets in Selangor between January 2014 and June 2014. All the samples were sealed and transported in an ice box to the laboratory for analysis on the day of purchase.
Enumeration and Isolation of Vibrio sp. in Shellfish Samples
Twenty-grams of samples (without shell) were homogenized for 60 s in a stomacher (Bagmixer 400W, Interscience, St Nom, France) with 225 mL of alkaline peptone water with 2% NaCl, pH 8.5, giving a first 10-1 dilution. The homogenates (1:10, 1:100, and 1:1000) were analyzed by spread-plate technique for total Vibrio sp. counts on Thiosulphate Citrate Bile Salt Sucrose (TCBS) agar (HiMedia, India) and incubated at 37°C for 18 h. After incubation, the total colony count and their concentrations in the original shellfish in cfu/mL was calculated.
The isolation of V. parahaemolyticus was carried out by incubating the homogenate at 37°C under aerobic conditions for 18 h. After incubation, a loopful of sample was streaked onto selective media, Thiosulfate Citrate Bile Salts Sucrose (TCBS) agar (HiMedia, India). Plates were then incubated at 37°C for 18 h. Characteristic green colonies (sucrose negative) on TCBS agar were considered presumptive V. parahaemolyticus and picked. The colonies were purified on Tryptic Soy Agar (TSA; HiMedia, India) plates supplemented with 2% w/v sodium chloride (NaCl; Vivantis, USA) and incubated at 37°C under aerobic conditions for 18 h. A loopful of pure colony was inoculated into semi-solid nutrient agar and Tryptic Soy Broth (TSB) with 30% glycerol, incubated at 37°C for 18 h and then stored until further analysis (Zarei et al., 2012; Letchumanan et al., 2015a).
DNA Extraction
Genomic DNA of presumptive V. parahaemolyticus colonies was extracted using direct boiled cell lysate method (Vengadesh et al., 2012; Letchumanan et al., 2015a). In brief, V. parahaemolyticus colonies from semisolid nutrient agar are revived in TSB (HiMedia) with 2% w/v NaCl (Vivantis, USA) and incubated in a shaker incubator at 220 rpm for 37°C for 18 h. 1.5 mL of overnight culture suspension was subjected to centrifugation at 10,000 rpm for 5 min. The supernatant was carefully discarded, leaving the pellet dry as possible. The pellet was re-suspended in 1 mL of sterile ultrapure water, vortexed and boiled at 100°C for 7 min. The boiled cell lysate was immediately cooled on ice for 5 min before being centrifuged at 13,000 rpm for 1 min to separate the debris and DNA containing supernatant. The supernatant was carefully transferred into a new micro-centrifuge tube. The boiled cell lysate was used as the DNA template for PCR assay.
Identification of Vibrio parahaemolyticus using toxR-Based PCR Assay
PCR amplification for detection of V. parahaemolyticus was performed in a final volume of 20 μL, containing 2 μL of DNA template, 10 μL of 2x Taq PLUS PCR Smart mix 1 (SolGentTM, Korea), 6 μL of sterile distilled water and 1 μL of each primer, toxR-F (5′-ATA CGA GTG GTT GCT GTC ATG-3′) and toxR-R (5′-GTC TTC TGA CGC AAT CGT TG-3′) with the expected amplicon size of 368 bp (Kim et al., 1999). The PCR amplification were performed using PCR thermocycler (Kyratec, SuperCycler Thermal Cycler, Australia) with the following cycling conditions: initial denaturation at 95°C for 4 min, 35 cycles of 94°C for 1 min, 68°C for 1 min and 72°C for 30 s, and a final elongation at 72°C for 5 min. PCR products were visualized by using 1.5% agarose gel. V. parahaemolyticus type strain (V. parahaemolyticus NBRC12711) and V. vulnificus type strain (V. vulnificus NBRC15645) was used as a positive control and negative control in every PCR reaction.
Detection of Virulence Gene
PCR amplification for detection of V. parahaemolyticus virulence genes, thermostable direct haemolysin (tdh) and thermostable-related direct haemolysin (trh) was performed in duplex PCR using specific primers adapted from Bej et al. (1999). The PCR was carried out in a final volume of 20 μL, containing 2 μL of DNA template, 10 μL of 2x Taq PLUS PCR Smart mix 1 (SolGentTM, Korea), 4 μL of sterile distilled water, and 1 μL of each primer. The PCR amplifications were performed using a Thermocycler (Kyratec, SuperCycler Thermal Cycler, Australia) with the following cycling conditions: initial denaturation at 94°C for 3 min, 30 cycles of 94°C for 1 min, 58°C for 1 min and 72°C for 1 min, and a final elongation at 72°C for 5 min. All PCR products were visualized by using 1.5% agarose gel. The V. parahaemolyticus type strain (V. parahaemolyticus NBRC12711) was used as a positive control and V. vulnificus type strain (V. vulnificus NBRC15645) was used as a negative control in every PCR reaction.
Antibiotic Susceptibility Test
Antibiotic disks (Oxoid, UK) infused with 14 antibiotics namely ampicillin (10μg), ampicillin/sulbactam (30 μg), amikacin (30 μg), cefotaxime (30 μg), ceftazidime (30 μg), chloramphenicol (30 μg), gentamicin (30 μg), imipenem (10 μg), kanamycin (30 μg), levofloxacin (5 μg), nalidixic acid (30 μg), oxytetracycline (30 μg), Sulfamethoxazole/trimethoprim (25 μg), and tetracycline (30 μg) were used in this study. The antibiotic susceptibility of Vibrio sp. isolates were studied using the disk diffusion method (Yano et al., 2014). The antibiotic disks were dispensed on Mueller Hilton agar (HiMedia, India) supplemented with 2% w/v NaCl (Vivantis, USA) plates with bacterial lawn. After incubation at 37°C for 18 h, the inhibition zone was measured and interpreted based on guidelines of the Clinical and Laboratory Standards Institute (CLSI) M45-A2 (Clinical Laboratory Standard Institute [CLSI], 2010).
Plasmid Profiling
Plasmid profiling was carried out following an adaptation of the method from Devi et al. (2009). Briefly, V. parahaemolyticus cell were grown in TSB containing 2% w/v sodium chloride and incubated at 37°C in a shaker incubator (220 rpm) for 18 h. About 1.5 mL of the culture was transferred into a micro-centrifuge tube followed by centrifugation (10,000 rpm for 2 min at 4°C). The supernatant was removed by aspiration leaving the cell pellet as dry as possible. The pellet was resuspended in ice-cold 100 μl alkaline lysis solution I (Glucose 50 mM; Tris Cl 25 mM; EDTA 10 Mm) by vigorous vortexing followed by addition of freshly prepared 200 μL alkaline lysis solution II (NaOH 2N; SDS 2% w/v). The contents were mixed by vortexing rapidly after which 150 μl ice-cold solution III (Potassium acetate 5 M: 60 ml; Glacial acetic acid 11.5 ml; dissolved in 28.5 m sterile distilled water) was added to it. The tube was closed and gently vortexed for 10 s to disperse solution III through the viscous bacterial lysate.
Then the tubes were stored in ice for 5 min before being centrifuged at 12,000 rpm for 2 min at 4°C. An equal volume of phenol-chloroform (1:1, w/v) was added to the supernatant in a fresh tube, by vortexing. The contents in the micro-centrifuge tube were centrifuged at 8,000 rpm for 3 min at 4°C and the supernatant was transferred into a fresh tube. This was repeated with chloroform: isoamyl-alcohol (24:1, v/v) for removing the phenol. The double stranded DNA was precipitated with two volumes of ethanol at room temperature, followed by vortexing before it was allowed to stand for 5 min at room temperature. The aliquot was centrifuged at 12,000 rpm for 12 min at 4°C and the supernatant was removed by gentle aspiration. The pellet of double stranded DNA was rinsed with ethanol (1 ml, 70% v/v) at 4°C and centrifuged. The supernatant was removed leaving the pellet dry as possible. The pellet was air-dried before it was re-dissolved in 30 μl ultrapure water. Electrophoresis was performed using 1% agarose gel.
Plasmid Curing
All the antibiotic resistant V. parahaemolyticus strains were subjected to a plasmid curing assay using AO. Isolates were grown in TSB supplemented with 0.2 mg/mL AO. The tubes were then incubated at 37°C for 24 h under constant agitation. After treatment with AO, the antibiotic susceptibility profile of the resistant phenotypes was examined. (Reboucas et al., 2011). V. parahaemolyticus VP152 and VP103 strains were used as a positive control in every plasmid curing assay (Letchumanan et al., 2015a).
Statistical Analysis
The experimental data was analyzed by using SPSS software version 20. Statistical analysis was performed in order to determine whether there were any significant differences in the incidence of V. parahaemolyticus in the varying species of crustaceans studied and also to analyze the MAR index of resistant isolates using the Anova. The significance level was set at p-value of <0.05.
Results
Prevalence of Vibrio parahaemolyticus in shellfish
The conventional method based on the colonial appearance (green or bluish green colonies on TCBS agar) detected 450 presumptive V. parahaemolyticus isolates. To further confirm their identity, PCR was performed on all the presumptive V. parahaemolyticus isolates in order to confirm their identity by targeting species level toxin operon gene (toxR), 368bp. Only 44% (200/450) of the isolates were toxR-positive (Table 1). The number of toxR-positive isolates was similar at all sampling sites.
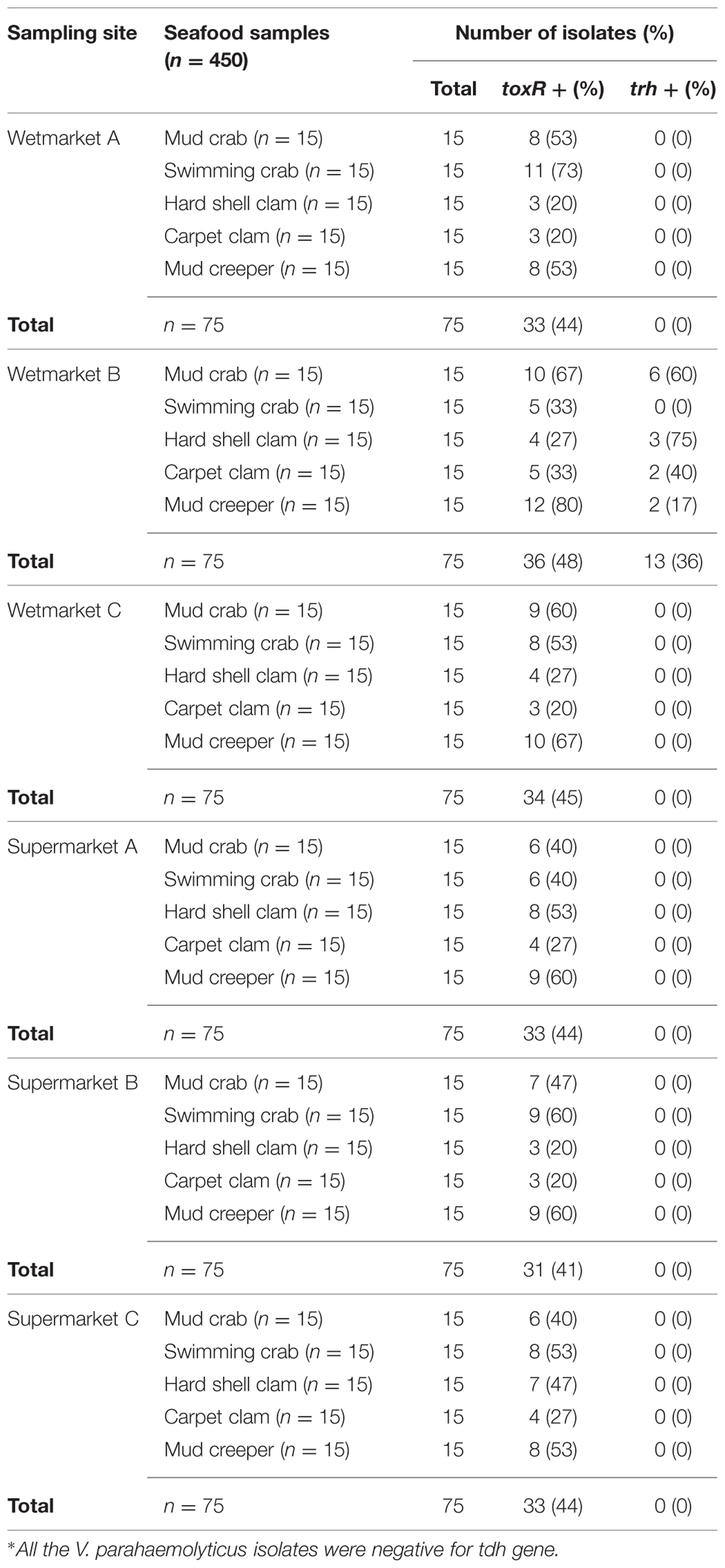
TABLE 1. The identification and detection of toxR+, trh+, and tdh+∗ in Vibrio parahaemolyticus isolates.
The density of total Vibrio count in the shellfish samples are shown in Table 2. The shellfish samples analyzed had a mean total Vibrio count range of 2.45–6.63 log cfu/g. A bacterial load of 5–7 log cfu/g is generally considered to be the level necessary to cause clinically significant infection in humans. Out of the 30 categories, 50% (15/30) were found to be >5 log cfu/g; however, this is with reference to Vibrio sp, not specifically V. parahaemolyticus. Of these 15 samples, eight were from wetmarkets and seven were from supermarkets. Mudcrab from all the locations sampled exceeded 5 log cfu/g. Carpet clams from the three wet markets all exceeded 6 log cfu/g while only one sample from the supermarket exceeded 5 log cfu/g. This pattern was reversed in mud creepers, however, with the supermarket samples showing higher Vibrio density. Overall, swimming crab showed the lowest density of Vibrio sp. density with four of the six sites sampled having mean densities of less than 3 log cfu/g.
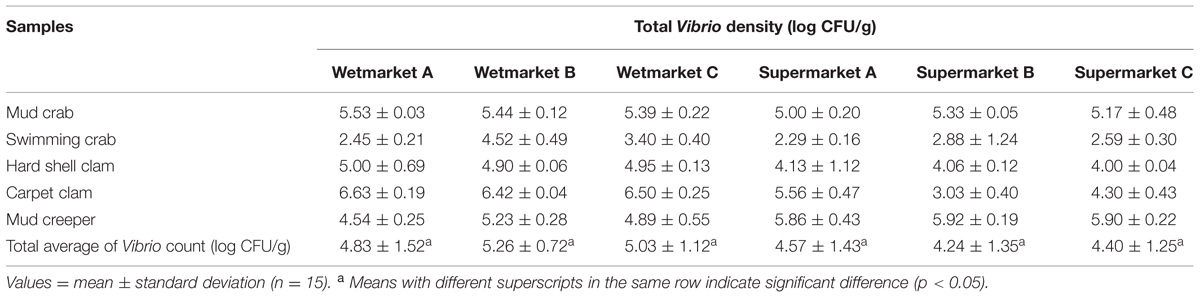
TABLE 2. The mean of total Vibrio counts (log CFU/g) of each shellfish samples from respective sampling site.
Prevalence of Thermostable Direct Hemolysin (tdh) and TDH-Related Hemolysin (trh) Positive Vibrio parahaemolyticus Isolates
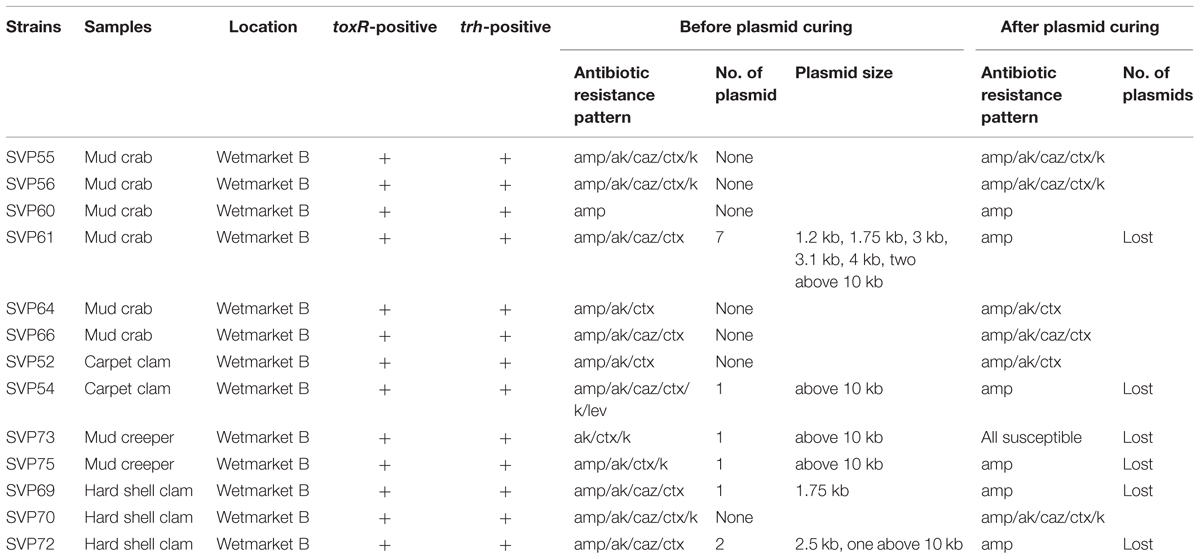
To detect pathogenic isolates, tdh and trh genes were amplified using PCR-based assay. The results of this analysis are summarized in Table 3. 13/200 (6.5%) trh-positive isolates were identified – all 13 were from shellfish samples collected from wetmarket B. Of these, six samples were mud crab, two samples were carpet clam, two samples were mud creeper and three samples were hard shell clam (Table 1). No tdh-positive V. parahaemolyticus isolates were detected.
Antimicrobial Susceptibilities of Vibrio parahaemolyticus Isolates
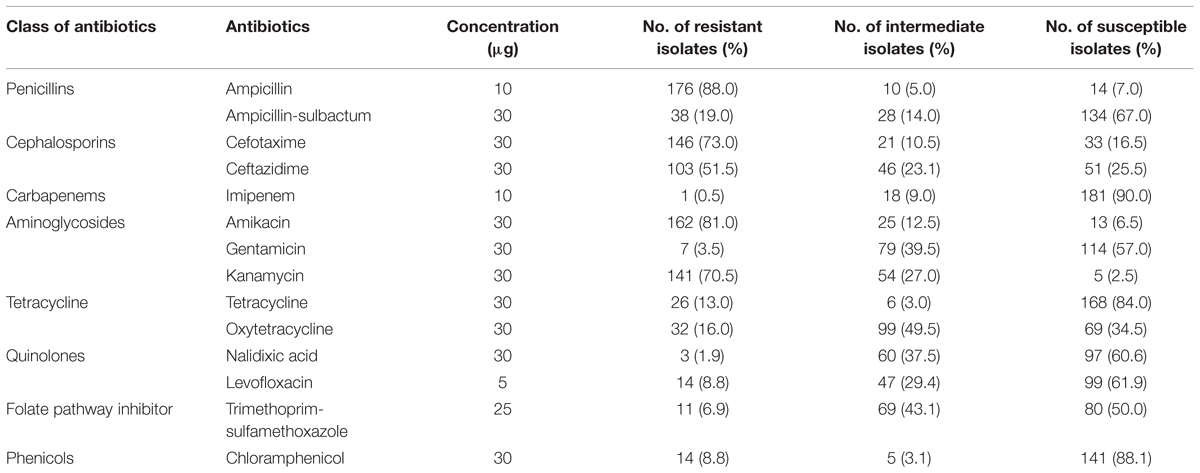
Vibrio parahaemolyticus isolates from shellfish samples were further tested for antimicrobial susceptibilities. All 14 antibiotics used in this study are among the antibiotics recommended by Centre for Disease Control and Prevention (CDC) for the treatment of Vibrio sp. infections that include fluoroquinolones (levofloxacin), cephalosporins (cefotaxime and ceftazidime), aminoglycosides (amikacin and gentamicin) and folate pathway inhibitors (trimethoprim-sulfamethoxazole; Daniels et al., 2000; Shaw et al., 2014). The results of this test are summarized in Table 4. With regard to CDC recommended antimicrobial agents, the isolates exhibited resistance to ampicillin (88.0%), amikacin (81.0%), and kanamycin (70.5%). Within the third generation cephalosporin, isolates exhibited resistance to cefotaxime (73%) and ceftazidime (51.5%). Reassuringly, the isolates tested were still susceptible to several antibiotics tested, imipenem (90.0%), chloramphenicol (88.0%), tetracycline (84.0%), ampicillin/sulbactam (67.0%), levofloxacin (61.5%) and trimethoprim-sulfamethoxazole (50%). With reference to the pathogenic strains, all the thirteen trh-positive isolates were resistant to ampicillin, 12/13 isolates expressed resistance to amikacin and cefotaxime, and 8/13 isolates were resistant to ceftazidime.
Eighty-five percentage of the isolates had multiple antibiotic resistance (MAR) index more than 0.2. Gwendelynne et al. (2005) stated that MAR indices higher that 0.2 could be a marker of contamination from high risk sources, thus indicating a potential human health risk. MAR index in this study ranged from 0.00 to 0.79, with the highest MAR index (0.79) found in a non-pathogenic isolate (SVP129) of carpet clam from supermarket B which expressed resistance to 11/14 antibiotic tested. The majority of V. parahaemolyticus isolates studied showed a MAR index of 0.36 (resistant to five antibiotics tested). 15% of the isolates had MAR index of 0.00–0.07, indicating the isolates were resistant to none or at least one type of the antibiotic tested.
Plasmid Profiling and Plasmid Curing of Vibrio parahaemolyticus Isolates
All the V. parahaemolyticus isolates were tested for the presence of plasmid before curing. Plasmid profiles of the 200 V. parahaemolyticus isolates revealed that 173/200 isolates contained one to seven plasmids ranging from 1.2 kb to above 10 kb in size while the remaining 27 isolates did not have any plasmids. 6/13 trh-positive V. parahaemolyticus isolates (Table 3) contained plasmids, with all six containing at least one plasmid above 10 kb. (SVP61); trh-positive isolate) expressed seven plasmids with sizes of 1.2, 1.75, 3, 3.1, 4, and 2 kb above 10 kb size. SVP61 was resistant to 4/14 antibiotics tested. The study revealed that (SVP129; non-pathogenic isolate) of carpet clam from supermarket B has one plasmid above 10 kb size and is resistant to 11/14 antibiotics tested.
Plasmid curing is a method which potentially enables the elimination of plasmids from the organisms being studied which can then allow the determination of antibiotic resistance mediation by retesting the organisms after plasmid curing. Antibiotic resistance of all V. parahaemolyticus isolates which had underdone plasmid curing was tested by antibiotic disk diffusion method. The resistance exhibited by isolates to ampicillin and tetracycline did not vary even after curing of plasmids. With reference to the 13 trh-positive strains, the antibiotic resistance profile of the six plasmid containing strains altered after curing while the remaining seven were unchanged. All six were ampicillin resistant initially, and after curing, one strain became susceptible to all antibiotics tested while five of the strains (SVP61, SVP54, SVP75, SVP69, VP72) remained resistant to ampicillin and became susceptible to the other antibiotics tested. This suggests that while antibiotic resistance is mediated by both plasmid and chromosomes in pathogenic V. parahaemolyticus isolates, in plasmid containing strains, aside from ampicillin resistance, most of the remaining resistance phenotypes are plasmid mediated.
SVP129 isolate contained one plasmid profile with size more than 10 kn and expressed resistance to 11/14 antibiotics tested. After plasmid curing, SVP129 isolate lost its plasmid and changed its antibiotic resistance phenotype. SVP129 isolate remained resistant to 5/14 antibiotic tested namely ampicillin, oxytetracycline, chloramphenicol, tetracycline, and sulfamethoxazole/trimethoprim. The isolate showed intermediate resistance to amikacin, ceftazidime, cefotaxime, and kanamycin, while it was susceptible to gentamycin and ampicillin/sulbactam after plasmid curing assay.
Discussion
The presence of pathogenic strains of V. parahaemolyticus in the shellfish we studied does raise concern as these organisms are known to be a frequent cause of food-borne gastroenteritis in humans. However, while conventional colony morphology found all samples to be contaminated with Vibrio sp. only 44% (200/450) of these were confirmed to be V. parahaemolyticus based on tox-R assay; and only 6.5% (13/200) of these were pathogenic strains (trh-positive). Conventional bacteriological methods have demonstrated a highly variable occurrence of V. parahaemolyticus in seafood around the world. Our results are in close agreement with those reported by Zhao et al. (2011) in China, who isolated V. parahaemolyticus from 59.7% shellfish samples. Our study samples exhibited a mean total Vibrio count range of 2.45–6.63 log cfu/g. The mean total Vibrio count of the shellfish samples from wetmarkets were higher compared to shellfish samples from supermarkets. However, the results obtained from one-way analysis of variance (Table 2), showed there were no significant difference (p > 0.05) between the total Vibrio counts of the shellfish samples from all the sampling sites. The difference in the incidence of V. parahaemolyticus among samples from both sampling sites could possibly be contributed by the original geographic source from which the shellfish were collected, post-harvest practices and hygiene standards applied during handling, transportation and storage of seafood products. A study in Cochin, India, reported higher Vibrio sp. contamination in shellfish from roadside stalls compared to markets (Sudha et al., 2014). Other studies have reported lack of hygiene, improper handling, cross contamination and difference in storage temperature as the possible cause of variation in V. parahaemolyticus incidences in samples from supermarket (Yang et al., 2008; Tunung et al., 2010; Sudha et al., 2014).
In order to assess the actual risk to human health posed by the presence of V. parahaemolyticus in seafood, incidence of pathogenic strains need to be determined via detection of the toxigenic genes responsible for causing disease in humans. Of the 200 isolates tested in this study, only 13 (6.5%) were trh-positive and there were no tdh-positive strains. This incidence is similar to the results demonstrated in Malaysia by Paydar et al. (2013) who detected 6/50 (12%) trh-positive V. parahaemolyticus among the food samples tested. In another study, a much higher incidence of tdh-positive and trh-positive in shrimp and cockles samples was reported in Malaysia with twenty six isolates were trh-positive and eight tdh-positive (Al-Othrubi et al., 2014). Normally, only 1–2 % of the environmental strains harbor the tdh and trh genes (Wong et al., 2000; Alam et al., 2002; Hervio-Heath et al., 2002; Velazquez-Roman et al., 2012; Haley et al., 2014). Studies in the USA and in Japan reported that only 3% of V. parahaemolyticus strains isolated from seafood and environment samples were pathogenic (DePaola et al., 2003; Mahmoud et al., 2006; Abd-Elghany and Sallam, 2013). It is also reported that environmental factors including interaction with different hosts plays an important impact in the evolution of specific pathogens (Wilson and Salyers, 2003).
The continuous and extensive use of antibiotics in the aquaculture industry favors the development of a variety of resistant isolates and the dissemination of resistance genes within the bacterial population in the environment which reflects the pattern of drug use (Tendencia and Pena, 2002; Reboucas et al., 2011). All the antibiotics tested in this study are recommended antimicrobial agents used in the treatment of Vibrio sp. infections, including the tetracycline, levofloxacin, cefotaxime, ceftazidime, amikacin, gentamicin, and trimethoprim-sulfamethoxazole (Daniels et al., 2000; Shaw et al., 2014). Some of these antibiotics are widely used in aquaculture industry as antimicrobial agents including oxytetracycline and chloramphenicol (Dang et al., 2007). In this study, 88% of the V. parahaemolyticus isolates from shellfish exhibited resistance to ampicillin. Our results are in close agreement with other studies that reported resistance to ampicillin among the V. parahaemolyticus isolates from seafood samples (Okuda et al., 1997; Han et al., 2007; Al-Othrubi et al., 2014; Letchumanan et al., 2015a). First generation antibiotics including ampicillin are extensively used in the aquaculture thus reducing to susceptibility and resulting in low efficacy of ampicillin for Vibrio sp. treatment (Sudha et al., 2014).
Resistance to third generation cephalosporin, was observed in our isolates of V. parahaemolyticus, with 73% resistant to cefotaxime and 51.5% resistant to ceftazidime. This is slightly less compared to 80% in a study by Jun et al. (2012) studying the resistance to third generation cephalosporin among V. parahaemolyticus isolates from Korean seafood. Our study is in agreement with Sahilah et al. (2014) in Terengganu, Malaysia, who reported presence of V. parahaemolyticus isolates from shellfish to be resistant to ceftazidime and cefuroxime. However, a study by Shaw et al. (2014) reported low percentage of V. parahaemolyticus from USA to be resistant to cefotaxime. The discrepancies in the literature regarding the resistance phenotype of V. parahaemolyticus to third generation cephalosporin could possibly due to difference in test methodology or geographical variation. In the present study, high susceptibility to imipenem (90%), chloramphenicol (88.1%) and tetracycline (84%) was observed among the V. parahaemolyticus isolates, much in agreement with other publications (Han et al., 2007; Sahilah et al., 2014; Sudha et al., 2014).
High MAR indices were detected in this study, ranging from 0.00 to 0.79, with 85% of the isolates having a MAR index value more than 0.2. MAR indices higher than 0.2 are markers of high risk sources, which may represent a potential human health risk (Gwendelynne et al., 2005). The most frequent MAR index of the resistant V. parahaemolyticus isolates tested was 0.36, indicating that the strains were resistant toward five different antibiotics. The MAR index of V. parahaemolyticus isolates in present study varied significantly (ANOVA, p < 0.05) between wetmarket and supermarket. Our results are in agreement with a study by Elexson et al. (2014) in Malaysia which reported 97.2% of V. parahaemolyticus isolates had MAR index more than 0.2, however, a lower percentage (14.5%) with MAR index more than 0.2 was reported by Tang et al. (2014). The huge variation in the MAR index of the V. parahaemolyticus isolates observed in present and past studies in Malaysia may be influenced by the variance in the resistance levels depending on the source of sample collection (Khan et al., 2007; Tunung et al., 2012). Lesley et al. (2011) suggested that the difference in geographical locations may have differential selective pressures for the antibiotic resistance level.
Among the 200 V. parahaemolyticus isolates, only 173 isolates (86.5%) harbored between one to seven plasmid DNA bands, which range in size from 1.2 kb to above 10 kb and the rest did not exhibit the presence of plasmid DNA. The study findings correlates with results of previous studies, which have reported that V. parahaemolyticus harbored plasmids (Kagiko et al., 2001; Kaufman et al., 2002; Molina-Aja et al., 2002; Manjusha et al., 2005; Zulkifli et al., 2009). The isolate (SVP129) had only one plasmid above 10 kb band size and showed resistance to 11/14 antibiotics tested. With reference to trh-positive strains, seven of the isolates did not have any plasmid but were resistant to more than one antibiotic tested.
When submitted to plasmid curing, (SVP129) isolate lost its plasmid and altered its resistance phenotype to the antibiotics tested. The isolate remained resistant to sulfamethoxazole/trimethoprim, ampicillin, oxytetracycline, chloramphenicol, and tetracycline. SVP129 isolate showed intermediate resistant to amikacin, ceftazidime, cefotaxime, and kanamycin, while it was susceptible to gentamicin and ampicillin/sulbactam after plasmid curing assay. AO acts as an intercalating agent that inhibits plasmid replications (Letchumanan et al., 2015b). The resistance phenotype exhibited by SVP129 could be chromosomal mediated. Meanwhile, 6/13 trh-positive isolates lost their plasmids after curing assay and altered their resistance phenotype to amikacin, ceftazidime, cefotaxime, kanamycin, and levofloxacin (Table 2).
All the ampicillin resistant isolates (176/200) and tetracycline resistant isolates (26/200) remain resistant to the respective antibiotics after plasmid curing. This suggests that the resistance phenotype to ampicillin and tetracycline expressed by the isolates could be chromosomally mediated. All the ampicillin/sulbactam resistant strains lost their plasmid after the curing assay and subsequently were susceptible to ampicillin/sulbactam suggesting that the resistance was plasmid mediated. Our results are closely in agreement with other studies that reported Vibrio sp., strains lost their plasmids when treated with concentration of 0.2 mg/ml AO and the isolates demonstrated changes in their resistance profile (Molina-Aja et al., 2002; Barman et al., 2010; Reboucas et al., 2011; Carvalho et al., 2013; Costa et al., 2015). A study by Reboucas et al. (2011) in Brazil, reported AO was successfully used to cure multi-resistant Vibrio isolates from marine shrimp and concluded the ampicillin resistance strains in study are plasmid mediated. In contrast, another study reported their isolates resistance was chromosomal mediated after AO curing treatment (Costa et al., 2014). The loss of phenotype in these studies suggest that AO produce an immediate and complete inhibition of plasmid replication, thus able to act as reliable plasmid curing agent.
Plasmid curing assay may be used to eliminate bacterial plasmids and determine antibiotic resistance mediation. The assay provides vital information which would be beneficial in the global surveillance management of environmental multidrug resistance. Reducing and improving the use of antibiotics in the aquatic environment can reduce resistance and allow the antibiotic to resurface eventually as an effective therapy (Barbosa and Levy, 2000). The management of antibiotic resistance may vary depending on the resistance location, plasmidial, or chromosomal mediated resistance. Plasmidial mediated resistance can be controlled by alternating the antibiotics used in the environments. The aquaculture industry could adapt the method of switching antibiotics used in the aquatic field from time to time in order to allow withdrawal of antibiotic resistance profile in strains (Letchumanan et al., 2015b). However, it would be a challenge to control chromosomal mediated resistance because the resistant genes reside in the chromosomes of the bacteria. Hence, non-antibiotic management strategies using bacteriophages may help to control chromosomal mediated resistance. Bacteriophages are found in abundance in the aquatic environment and may play a vital role in controlling microbial populations. Many studies have demonstrated the potential of bacteriophages to control bacterial diseases and have been shown to be effective in reducing pathogen levels in aquaculture species, such as shrimp (Karunasagar et al., 2007) and finfish (Park and Nakai, 2003).
Quorum sensing and quorum quenching may be used as an alternative method to manage antibiotic resistance. This strategy has been widely proposed to control infections in aquaculture environments by detecting bacterial cell-to-cell communication thru small signal molecules of quorum sensing (Defoirdt et al., 2004). Quorum sensing is a mechanism of gene regulation in which bacteria coordinate the expression of certain genes in response to the presence or absence of small signal molecules. The bacterial communication is then stopped through quorum quenching and hence the expression of genes is disrupted (Defoirdt et al., 2008). Hence, this knowledge and information gathered would be useful to manage the emergence of antibiotic resistance.
In summary, this study showed that only 2.8% (13/450) of the seafood samples studied contained pathogenic strains of V. parahaemolyticus. These 13 strains were deemed pathogenic based on trh-positive status. These pathogenic strains all exhibited antibiotic resistance, with almost 70% (9/13) demonstrating resistance to 4 or more antibiotics tested. Out of the 200 V. parahaemolyticus isolates analyzed, there was a high level of resistance to ampicillin and tetracycline as well as to some third generation cephalosporin. While the overall incidence of pathogenic strains of V. parahaemolyticus in Selangor is relatively reassuring, the high MAR of all the V.parahaemolytics tested – including the pathogenic strains- is a definite cause for concern and warrants ongoing surveillance.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by a University of Malaya for High Impact Research Grant (UM-MOHE HIR Nature Microbiome Grant No. H-50001-A000027 and No. A000001-50001) awarded to K-GC and External Industry Grants from Biotek Abadi Sdn Bhd (vote no. GBA-808138 & GBA-808813) awarded to L-HL.
References
Abd-Elghany, S. M., and Sallam, K. I. (2013). Occurrence and molecular identification of Vibrio parahaemolyticus in retail shellfish in Mansoura. Egypt. Food Cont. 33, 399–405. doi: 10.1016/j.foodcont.2013.03.024
Alam, M. J., Tomochika, K. I., Miyoshi, S. I., and Shinoda, S. (2002). Environmental investigation of potentially pathogenic Vibrio parahaemolyticus in the Seto-Inland Sea, Japan. FEMS Microbiol. Lett. 208, 83–87. doi: 10.1111/j.1574-6968.2002.tb11064.x
Al-Othrubi, S. M. Y., Kqueen, C. Y., HMirhosseini, C. Y., Hadi, Y. A., and Radu, S. (2014). Antibiotic Resistance of Vibrio parahaemolyticus isolated from cockles and shrimp sea food marketed in Selangor. Malaysia. Clin. Microbiol. 3, 148–154.
Barbosa, T. M., and Levy, S. B. (2000). The impact of antibiotic use on resistance development and persistence. Drug Resist. Update 3, 303–311. doi: 10.1054/drup.2000.0167
Barman, S., Chatterjee, S., Chowdhury, G., Ramamurthy, T., Niyogi, S. K., Kumar, R., et al. (2010). Plasmid mediated streptomycin and sulfamethoxazole resistance in Shigella flexneri 3a. Int. J. Antimicrob. Agents 36, 348–351. doi: 10.1016/j.ijantimicag.2010.06.037
Bej, A. K., Patterson, D. P., Brasher, C. W., Vickery, M. C., Jones, D. D., and Kaysner, C. A. (1999). Detection of total and hemolysin-producing Vibrio parahaemolyticus in shell fish using multiplex PCR amplification of tl, tdh and trh. J. Microbiol. Methods 36, 215–225. doi: 10.1016/S0167-7012(99)00037-8
Bondad-Reantaso, M. G., Subasinghe, R. P., Arthur, J. R., Ogawa, K., Chinabut, S., Adlard, R., et al. (2005). Disease and health management in Asian aquaculture. Vet. Parasitol. 132, 249–272. doi: 10.1016/j.vetpar.2005.07.005
Cabello, F. C., Godfrey, H. P., Tomova, A., Ivanova, L., Dölz, H., Millanao, A., et al. (2013). Antimicrobial use in aquaculture re-examined: its relevance to antimicrobial resistance and to animal and human health. Appl. Eviron. Microbiol. 15, 1917–1942. doi: 10.1111/1462-2920.12134
Canizalez-Roman, A., Flores-Villasenor, H., Zazueta-Beltran, J., Muro-Amador, S., and Leon-Sicairos, N. (2011). Comparative evaluation of a chromogenic agar medium-PCR protocol with a conventional method for isolation of Vibrio parahaemolyticus strains from environmental and clinical samples. Can. J. Microbiol. 57, 136–142. doi: 10.1139/w10-108
Carvalho, F. C. T., Sousa, O. V., Carvalho, E. M. R., Hofer, E., and Vieira, R. H. S. F. (2013). Antibiotic resistance of Salmonella spp. isolated from shrimp farming fresh water environment in Northeast region of Brazil. J. Pathol. 2013:685193. doi: 10.1155/2013/685193
Ceccarelli, D., Hasan, N. A., Hug, A., and Colwell, R. R. (2013). Distribution and dynamics of epidemic and pandemic Vibrio parahaemolyticus virulence factors. Front. Cell. Infect. Microbiol. 3:97. doi: 10.3389/fcimb.2013.00097
Clinical Laboratory Standard Institute [CLSI] (2010). Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria, Approved Guideline, 2nd Edn. Wayne: Clinical Laboratory Standard Institute.
Costa, R. A., Araujo, R. L., Souza, O. V., and Viera, R. H. S. F. (2015). Antibiotic-resistant Vibrios in farmed shrimp. BioMed Res. Int. 2015:505914. doi: 10.1155/2015/505914
Costa, R. A., Colares, L. P., Lima, R. A., Vieira, R. H. S. D. F., and de Sousa, S. V. (2014). Effect of seawater on the activity of antibiotics against Vibrios isolated from the hemolymph of cultured pacific white shrimp. J. World Aqua. Soc. 43, 727–732.
Dang, H. Y., Zhang, X. X., Song, L. S., Chang, Y. Q., and Yang, G. P. (2007). Molecular determination of oxytetracycline-resistant bacteria and their resistance genes from mariculture environments of China. J. Appl. Microbiol. 103, 2580–2592. doi: 10.1111/j.1365-2672.2007.03494.x
Daniels, N. A., MacKinnon, L., Bishop, R., Altekruse, S., Ray, B., Hammond, R. M., et al. (2000). Vibrio parahaemolyticus infection in the United States, 1973-1998. J. Infect. Dis. 181, 1661–1666. doi: 10.1086/315459
Deepanjali, A., Kumar, H. S., Karunasagar, I., and Karunasagar, I. (2005). Seasonal variation in abundance of total and pathogenic Vibrio parahaemolyticus bacteria in oysters along the southwest coast of India. Appl. Environ. Microbiol. 71, 3575–3580. doi: 10.1128/AEM.71.7.3575-3580.2005
Defoirdt, T., Boon, N., Bossier, P., and Verstraete, W. (2004). Disruption of bacterial quorum sensing: an unexplored strategy to fight infections in aquaculture. Aquaculture 240, 69–88. doi: 10.1016/j.aquaculture.2004.06.031
Defoirdt, T., Boon, N., Sorgeloos, P., Verstraete, W., and Bossier, P. (2008). Quorum sensing and quorum quenching in Vibrio harveyi: lessons learned from in vivo work. Int. Soc. Microb. Eco. 2, 19–26. doi: 10.1038/ismej.2007.92
DePaola, A., Kaysner, C. A., Bowers, J., and Cook, D. W. (2000). Environmental investigations of Vibrio parahaemolyticus in oysters after outbreaks in Washington, Texas, and NewYork (1997 and 1998). Appl. Environ. Microbiol. 66, 4649–4654. doi: 10.1128/AEM.66.11.4649-4654.2000
DePaola, A., Ulaszek, J., Kaysner, C. A., Tenge, B. J., Nordstrom, J. L., Well, J., et al. (2003). Molecular, serological, and virulence characteristics of Vibrio parahaemolyticus isolated from environmental, food, and clinical sources in North America and Asia. Appl. Environ. Microb. 69, 3999–4005. doi: 10.1128/AEM.69.7.3999-4005.2003
Devi, R., Surendran, P. K., and Chakraborty, K. (2009). Antibiotic resistance and plasmid profiling of Vibrio parahaemolyticus isolated from shrimp farms along the coast of India. World J. Microbiol. Biotech. 25, 2005–2012. doi: 10.1007/s11274-009-0101-8
Elexson, N., Afsah-Hejri, L., Rukayadi, Y., Soopna, P., Lee, H. Y., Tuan Zainazor, T. C., et al. (2014). Effect of detergents as antibacterial agents on biofilm of antibiotics-resistant Vibrio parahaemolyticus isolates. Food Cont. 35, 378–385. doi: 10.1016/j.foodcont.2013.07.020
Grant, M. A., Hu, J., and Jinneman, K. C. (2006). Multiplex real-time PCR detection of heatlabile and heat-stable toxin genes in enterotoxigenic Escherichia coli. J. Food Protect. 69, 412–416.
Gutierrez West, C. K., Klein, S. L., and Lovell, C. R. (2013). High frequency of virulence factor genes tdh, trh, and tlh in Vibrio parahaemolyticus strains isolated from a pristine estuary. Appl. Environ. Microbiol. 79, 2247–2252. doi: 10.1128/AEM.03792-12
Gwendelynne, B. T., Son, R., Nishibuchi, M., Raha, A. R., Suhaimi, N., Lesley, M., et al. (2005). Characterization of Vibrio parahaemolyticus isolated from coastal seawater in Peninsular Malaysia. Southeast Asian J. Trop. Med. Pub. Health 36, 940–945.
Haley, B. J., Kokashvili, T., Tskshvediani, A., Janelidze, N., Mitaishvili, N., Grim, C. J., et al. (2014). Molecular diversity and predictability of Vibrio parahaemolyticus along the Georgian coastal zone of the Black Sea. Front. Microbiol. 5:45. doi: 10.3389/fmicb.2014.00045
Han, F., Walker, R. D., Janes, M. E., Prinyawinwatkul, W., and Ge, B. (2007). Antimicrobial susceptibilities of Vibrio parahaemolyticus and Vibrio vulnificus isolates from Louisiana Gulf and retail raw oysters. Appl. Environ. Microbiol. 73, 7096–7098. doi: 10.1128/AEM.01116-07
Hazen, T. H., Lafon, P. C., Garrett, N. M., Lowe, T. M., Silberger, D. J., Rowe, L. A., et al. (2015). Insights into the environmental reservoir of pathogenic Vibrio parahaemolyticus using comparative genomics. Front. Microbiol. 6:204. doi: 10.3389/fmicb.2015.00204
Hervio-Heath, D., Colwell, R. R., Derrien, J. M., Fournier, J. M., and Pommepuy, M. (2002). Occurrence of pathogenic Vibrio in coastal areas of France. J. Appl. Microbiol. 92, 1123–1135. doi: 10.1046/j.1365-2672.2002.01663.x
Jun, J. W., Kim, J. H., Choresca, C. H., Shin, S. P., Han, J. E., Han, S. Y., et al. (2012). Isolation, molecular characterization and antibiotic susceptibility of Vibrio parahaemolyticus in Korean seafood. Foodborne Pathog. Dis. 9, 224–231. doi: 10.1089/fpd.2011.1018
Kagiko, M. M., Damiano, W. A., and Kayihura, M. M. (2001). Characterization of Vibrio parahaemolyticus isolated from fish in Kenya. East African Med. J. 78, 124–127. doi: 10.4314/eamj.v78i3.9076
Karunasagar, I., Shivu, M. M., Girisha, S. K., Krohne, G., and Karunasagar, I. (2007). Biocontrol of pathogens in shrimp hatcheries using bacteriophages. Aquaculture 268, 288–292. doi: 10.1016/j.aquaculture.2007.04.049
Kaufman, G. F., Myers, M. I., Pass, C. L., Bej, A. K., and Kaysner, C. A. (2002). Molecular analysis of Vibrio parahaemolyticus isolated from human patients and shellfish during US Pacific north-west outbreaks. Lett. Appl. Microbiol. 34, 155–161. doi: 10.1046/j.1472-765x.2002.01076.x
Kaysner, C. A., and DePaola, A. J. (2001). “Vibrio,” in Compendium of Methods for the Microbiological Examination of Food, eds F. P. Downes and K. Ito (Washington, DC: American Public Health Association).
Khan, A. W., Hossain, S. J., and Uddin, S. N. (2007). Isolation, identification and determination of antibiotics susceptibility of Vibrio parahaemolyticus from shrimps at Khulna Region of Bangladesh. Res. J. Microbiol. 2, 216–227. doi: 10.3923/jm.2007.216.227
Kim, Y. B., Okuda, J., Matsumoto, C., Takahashi, N., Hashimoto, S., and Nishibuchi, M. (1999). Identification of Vibrio parahaemolyticus strains at the species level by PCR targeted to the toxR gene. J. Clin. Microbiol. 37, 1173–1177.
Law, J. W.-F., Mutalib, N.-S., Chan, K.-G., and Lee, L.-H. (2015). Rapid methods for the detection of foodborne bacterial pathogens: principles, applications, advantages and limitations. Front. Microbiol. 5:770. doi: 10.3389/fmicb.2014.00770
Lesley, M. B., Velnetti, L., Cheah, Y. K., Son, R., Kasing, A., Samuel, L., et al. (2011). Antibiotic resistance and plasmid profiling of Vibrio parahaemolyticus isolated from cockles (Anadara granosa) at Tanjung Karang, Kuala Selangor. Int. Food Res. J. 18, 1183–1188.
Letchumanan, V., Chan, K., and Lee, L. (2014). Vibrio parahaemolyticus: a review on the pathogenesis, prevalence and advance molecular identification techniques. Front. Microbiol. 5:705. doi: 10.3389/fmicb.2014.00705
Letchumanan, V., Yin, W.-F., Lee, L.-H., and Chan, K.-G. (2015a). Prevalence and antimicrobial susceptibility of Vibrio parahaemolyticus isolated from retail shrimps in Malaysia. Front. Microbiol. 6:33. doi: 10.3389/fmicb.2015.00033
Letchumanan, V., Chan, K.-G., and Lee, L.-H. (2015b). An insight of traditional plasmid curing in Vibrio species. Front. Microbiol. 6:735. doi: 10.3389/fmicb.2015.00735
Liu, X., Wang, D., Wang, H., Feng, E., Zhu, L., and Wang, H. (2012). Curing of plasmid pX01 from Bacillus anthracis using plasmid incompatibility. PLoS ONE 7:e29875. doi: 10.1371/journal.pone.0029875
Mahmoud, Z. H., Kassu, A., Mohammad, A., Yamato, M., Bhuiyan, N. A., Nair, B., et al. (2006). Isolation and molecular characterization of toxigenic Vibrio parahaemolyticus from the Kii Channel, Japan. Microbial. Res. 161, 25–37. doi: 10.1016/j.micres.2005.04.005
Mala, E., Oberoi, A., and Alexander, V. S. (2014). Vibrio isolates from cases of acute diarrhea and their antimicrobial susceptibility pattern in a tertiary care hospital. Int. J. Basic Appl. Sci. 3, 35–37.
Malcolm, T. T. H., Cheah, Y. K., Radzi, C. W. J. W. M., Kasim, F. A., Kantilal, H. K., John, T. Y. H., et al. (2015). Detection and quantification of pathogenic Vibrio parahaemolyticus in shellfish by using multiplex PCR and loop-mediated isothermal amplification assay. Food Cont. 47, 664–671. doi: 10.1016/j.foodcont.2014.08.010
Manjusha, S., and Sarita, G. B. (2011). Plasmid associated antibiotic resistance in Vibrio isolated from coastal waters of Kerala. Int. Food Res. J. 18, 1171–1181.
Manjusha, S., Sarita, G. B., Elyas, K. K., and Chandrasekaran, M. (2005). Multiple antibiotic resistances of Vibrio isolates from coastal and brackish water areas. Am. J. Biochem. Biotechnol. 1, 201–206. doi: 10.3844/ajbbsp.2005.201.206
Meyer, L. W. (1988). Use of plasmid profiles in epidemiologic surveillance of disease outbreaks and in tracing the transmission of antibiotic resistance. Clin. Microbiol. Rev. 1, 228–243.
Molina-Aja, A., Garcia-Gasca, A., Abreu-Grobois, A., Bolan-Mejia, C., Roque, A., and Gomez-Gil, B. (2002). Plasmid profiling and antibiotic resistance of Vibrio strains isolated from cultured penaeid shrimp. FEMS Microbiol. Lett. 213, 7–12. doi: 10.1111/j.1574-6968.2002.tb11278.x
Nair, G. B., Ramamurthy, T., Bhattacharya, S. K., Dutta, B., Takeda, Y., and Sack, D. A. (2007). Global dissemination of Vibrio parahaemolyticus serotype O3:K6 and its serovariants. Clin. Microbiol. Rev. 20, 39–48. doi: 10.1128/CMR.00025-06
Nishibuchi, M., and Kaper, J. B. (1995). Thermostable direct hemolysin gene of Vibrio parahaemolyticus: avirulence gene acquired by a marine bacterium. Infect. Immun. 63, 2093–2099.
Okamoto, A. S., Andreatti Filho, R. L., Rocha, T. S., Menconi, A., and Marietto-Goncalves, G. A. (2009). Detection and transfer of antimicrobial resistance gene integron in Salmonella Enteritidis derived from avian material. Rev. Bras. Ciên. Avíc. 11, 195–201.
Okuda, J., Ishibashi, M., Hayakawa, E., Nishino, T., Takeda, Y., Mukhopadhyay, A. K., et al. (1997). Emergence of a unique O3:K6 clone of Vibrio parahaemolyticus in Calcutta, India and isolation of strains from the same clonal group from Southeast Asian travelers arriving in Japan. J. Clin. Microbiol. 35, 3150–3155.
Panicker, G., Myers, M. L., and Bej, A. K. (2004). Rapid detection of Vibrio vulnificus in shellfish and Gulf of Mexico water by real-time PCR. Appl. Environ. Microbiol. 70, 498–507. doi: 10.1128/AEM.70.1.498-507.2004
Park, S. C., and Nakai, T. (2003). Bacteriophage control of Pseudomonas plecoglossicida infection in ayu Plecoglossus altivelis. Dis. Aquat. Organ. 53, 33–39. doi: 10.3354/dao053033
Paydar, M., Teh, C. S. J., and Thong, K. L. (2013). Prevalence and characterisation of potentially virulent Vibrio parahaemolyticus in seafood in Malaysia using conventional methods. PCR and REP-PCR. Food Cont. 32, 13–18. doi: 10.1016/j.foodcont.2012.11.034
Raghunath, P. (2015). Roles of thermostable direct hemolysin (TDH) and TDH-related hemolysin (TRH) in Vibrio parahaemolyticus. Front. Microbiol. 5:805. doi: 10.3389/fmicb.2014.00805
Reboucas, R. H., De Sousa, O. V., Lima, A. S., Vasconcelos, F. R., and de Carvalho, P. B. (2011). Antimicrobial resistance profile of Vibrio species isolated from marine shrimp farming environments (Litopenaeus vannamei) at Ceara, Brazil. Environ. Res. 111, 21–24. doi: 10.1016/j.envres.2010.09.012
Rico, A., Satapornvanit, K., Haque, M. M., Min, J., Nguyen, P. T., Telfer, T., et al. (2012). Use of chemicals and biological products in Asian aquaculture and their potential environmental risks: a critical review. Rev. Aqua. 4, 75–93. doi: 10.1111/j.1753-5131.2012.01062.x
Romalde, J. L., Diéguez, A. L., Lasa, A., and Balboa, S. (2014). New Vibrio species associated to molluscan microbiota: a review. Front. Microbiol. 4:413. doi: 10.3389/fmicb.2013.00413
Sahilah, A. M., Laila, R. A., Sallehuddin, H. M., Osman, H., Aminah, A., and Ahmah Azuhairi, A. (2014). Antibiotic resistance and molecular typing among cockle (Anadara granosa) strains of Vibrio parahaemolyticus by polymerase chain reaction (PCR)-based analysis. World J. Microbiol. Biotechnol. 30, 649–659. doi: 10.1007/s11274-013-1494-y
Shaw, K. S., Rosenberg Goldstein, R. E., He, X., Jacobs, J. M., Crump, B. C., and Sapkota, A. R. (2014). Antimicrobial susceptibility of Vibrio vulnificus and Vibrio parahaemolyticus recovered from recreational and commercial areas of Cheaspeake Bay and Maryland coastal bay. PLoS ONE 9:89616. doi: 10.1371/journal.pone.0089616
Su, C. Y., and Liu, C. (2007). Vibrio parahaemolyticus: a concern of seafood safety. Food Microbiol. 24, 549–558.
Sudha, S., Mridula, C., Silvester, R., and Hatha, A. A. M. (2014). Prevalence and antibiotic resistance of pathogenic Vibrios in shellfishes from Cochin market. Indian J. Mar. Sci. 43, 815–824.
Tang, J. Y. H., Wan-Rosli, W. F., Abdul-Razak, N. H., Yeo, C. C., Abu Bakar, C. A., and Son, R. (2014). Incidence and antibiogram of Vibrio parahaemolyticus in processed and frozen bivalve mollusks in Kuala Terengganu. Malaysia. Int. Food Res. J. 21, 1349–1353.
Tendencia, E. A., and Pena, L. D. (2002). Level and percentage recovery of resistance to oxytetracycline and oxolic acid of bacteria from shrimp ponds. Aquaculture 213, 1–13. doi: 10.1016/S0044-8486(02)00017-0
Tunung, R., Jeyaletchumi, P., Noorlis, A., Tang, Y. H., Sandra, A., Ghazali, F. M., et al. (2012). Biosafety of Vibrio parahaemolyticus from vegetables based on antimicrobial sensitivity and RAPD profiling. Int. Food Res. J. 19, 467–474.
Tunung, R., Margaret, S. P., Jeyaletchumi, P., Chai, L. C., Zainazor, T. C., Ghazali, F. M., et al. (2010). Prevalence and quantification of Vibrio in raw salad vegetables at retail level. J. Microbiol. Biotechnol. 20, 391–396.
Velazquez-Roman, J., León-Sicairos, N., de Jesus Hernández-Díaz, L., and Canizalez-Roman, A. (2014). Pandemic Vibrio parahaemolyticus O3:K6 on the American continent. Front. Cell Infect. Microbiol. 3:110. doi: 10.3389/fcimb.2013.00110
Velazquez-Roman, J., Leon-Sicairos, N., Flores-Villasenor, H., Villafana-Rauda, S., and Canizalez-Roman, A. (2012). Association of pandemic Vibrio parahaemolyticus O3: K6 present in the coastal environment of Northwest Mexico with cases of recurrent diarrhea between 2004 and 2010. Appl. Environ. Microbiol. 78, 1794–1803. doi: 10.1128/AEM.06953-11
Vengadesh, L., Son, R., and Yoke-Kqueen, C. (2012). Molecular quantitation and characterization of Vibrio cholera from different seafood obtained from wetmarket and supermarket. Int. Food Res. J. 19, 45–50.
Vincent, M., Chan, C. S. W., and Apun, K. (2015). Molecular confirmation and characterization of Vibrio parahaemolyticus from retailed fish. Inter. Food Res. J. 22, 1705–1710.
Vuddhakul, V., Chowdhury, A., Laohaprertthisan, V., Pungrasamee, P., Patararungrong, N., and Thianmontri, P. (2000). Isolation of a pandemic O3:K6 clone of a Vibrio parahaemolyticus strain from environmental and clinical sources in Thailand. Appl. Environ. Microbiol. 66, 2685–2689. doi: 10.1128/AEM.66.6.2685-2689.2000
Wilson, B. A., and Salyers, A. A. (2003). Is the evolution of bacterial pathogens an out-of-body experience? Trends Microbiol. 11, 347–350. doi: 10.1016/S0966-842X(03)00179-3
Wong, H. C., Liu, S. H., Ku, L. W., Lee, I. Y., Wang, T. K., Lee, Y. S., et al. (2000). Characterization of Vibrio parahaemolyticus isolates obtained from foodborne illness outbreaks during 1992 through 1995 in Taiwan. J. Food Protect. 63, 900–906.
Wu, Y., Wen, J., Ma, Y., Ma, X., and Chen, Y. (2014). Epidemiology of foodborne disease outbreaks caused by Vibrio parahaemolyticus, China, 2003–2008. Food Cont. 46, 197–202. doi: 10.1016/j.foodcont.2014.05.023
Xu, X., Wu, Q., Zhang, J., Cheng, J., Zhang, S., and Wu, K. (2014). Prevalence, pathogenicity and serotypes of Vibrio parahaemolyticus in shrimp from Chinese retail markets. Food Cont. 46, 81–85. doi: 10.1016/j.foodcont.2014.04.042
Yamamoto, A., Iwahori, J., Vuddhakul, V., Charernjiratrag, W., Vose, D., Osaka, K., et al. (2008). Quantitative modeling for risk assessment of Vibrio parahaemolyticus in bloody clams in southern Thailand. Int. J. Food Microbiol. 124, 70–78. doi: 10.1016/j.ijfoodmicro.2008.02.021
Yang, Z., Jiao, X., Zhou, X., Cao, G., Fang, W., and Gu, R. (2008). Isolation and molecular characterization of Vibrio parahaemolyticus from fresh, low-temperature preserved, dried and salted seafood products in two coastal areas of eastern China. Int. J. Food Microbiol. 125, 279–285. doi: 10.1016/j.ijfoodmicro.2008.04.007
Yano, Y., Hamano, K., Satomi, M., Tsutsui, I., Ban, M., and Aue-umneoy, D. (2014). Prevalence and antimicrobial susceptibility of Vibrio species related to food safety isolated from shrimp cultured at inland ponds in Thailand. Food Cont. 38, 30–45. doi: 10.1016/j.fm.2014.11.003
Zarei, M., Borujeni, M. P., Jamnejad, A., and Khezrzadeh, M. (2012). Seasonal prevalence of Vibrio species in retail shrimps with an emphasis on Vibrio parahaemolyticus. Food Cont. 25, 107–109. doi: 10.1016/j.foodcont.2011.10.024
Zhang, L., and Orth, K. (2013). Virulence determinants for Vibrio parahaemolyticus infection. Curr. Opin. Microbiol. 16, 70–77. doi: 10.1016/j.mib.2013.02.002
Zhang, X. H., and Austin, B. (2005). Haemolysins in Vibrio species. J. App. Microbiol. 98, 1011–1019. doi: 10.1111/j.1365-2672.2005.02583.x
Zhang, Z., Xiao, L., Lou, Y., Jin, M., Liao, C., Malakar, P. K., et al. (2014). Development of a multiplex real-time PCR method for simultaneous detection of Vibrio parahaemolyticus, Listeria monocytogenes and Salmonella spp. in raw shrimp. Food Cont. 51, 31–36. doi: 10.1016/j.foodcont.2014.11.007
Zhao, F., Zhou, D., Cao, H.-H., Ma, L.-P., and Jiang, Y.-H. (2011). Distribution, serological and molecular characterization of Vibrio parahaemolyticus from shellfish in the eastern coast of China. Food Cont. 22, 1095–1100. doi: 10.1016/j.foodcont.2010.12.017
Keywords: Vibrio parahaemolyticus, shellfish, MAR index, antibiotic resistance, plasmid curing, plasmid profile
Citation: Letchumanan V, Pusparajah P, Tan LT-H, Yin W-F, Lee L-H and Chan K-G (2015) Occurrence and Antibiotic Resistance of Vibrio parahaemolyticus from Shellfish in Selangor, Malaysia. Front. Microbiol. 6:1417. doi: 10.3389/fmicb.2015.01417
Received: 20 August 2015; Accepted: 27 November 2015;
Published: 15 December 2015.
Edited by:
Dongsheng Zhou, Beijing Institute of Microbiology and Epidemiology, ChinaReviewed by:
Qiaobin Xiao, Cornell University, USABo Pang, Chinese Center for Disease Control and Prevention, China
Copyright © 2015 Letchumanan, Pusparajah, Tan, Yin, Lee and Chan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Learn-Han Lee, lee.learn.han@monash.edu; leelearnhan@yahoo.com
 Vengadesh Letchumanan
Vengadesh Letchumanan Priyia Pusparajah
Priyia Pusparajah Loh Teng-Hern Tan
Loh Teng-Hern Tan Wai-Fong Yin
Wai-Fong Yin Learn-Han Lee
Learn-Han Lee Kok-Gan Chan
Kok-Gan Chan