- Department of Microbiology, Institute for Water and Wetland Research, Radboud University, Nijmegen, Netherlands
Methane oxidation is an important process to mitigate the emission of the greenhouse gas methane and further exacerbating of climate forcing. Both aerobic and anaerobic microorganisms have been reported to catalyze methane oxidation with only a few possible electron acceptors. Recently, new microorganisms were identified that could couple the oxidation of methane to nitrate or nitrite reduction. Here we investigated such an enrichment culture at the (meta) genomic level to establish a metabolic model of nitrate-driven anaerobic oxidation of methane (nitrate-AOM). Nitrate-AOM is catalyzed by an archaeon closely related to (reverse) methanogens that belongs to the ANME-2d clade, tentatively named Methanoperedens nitroreducens. Methane may be activated by methyl-CoM reductase and subsequently undergo full oxidation to carbon dioxide via reverse methanogenesis. All enzymes of this pathway were present and expressed in the investigated culture. The genome of the archaeal enrichment culture encoded a variety of enzymes involved in an electron transport chain similar to those found in Methanosarcina species with additional features not previously found in methane-converting archaea. Nitrate reduction to nitrite seems to be located in the pseudoperiplasm and may be catalyzed by an unusual Nar-like protein complex. A small part of the resulting nitrite is reduced to ammonium which may be catalyzed by a Nrf-type nitrite reductase. One of the key questions is how electrons from cytoplasmically located reverse methanogenesis reach the nitrate reductase in the pseudoperiplasm. Electron transport in M. nitroreducens probably involves cofactor F420 in the cytoplasm, quinones in the cytoplasmic membrane and cytochrome c in the pseudoperiplasm. The membrane-bound electron transport chain includes F420H2 dehydrogenase and an unusual Rieske/cytochrome b complex. Based on genome and transcriptome studies a tentative model of how central energy metabolism of nitrate-AOM could work is presented and discussed.
Introduction
Methane is an important greenhouse gas that is produced by microbiological processes, mainly methanogenesis in freshwater and marine ecosystems (Thauer et al., 2008) and the demethylation of methylphosphonates in the ocean (Metcalf et al., 2012). Part of the produced methane is oxidized by methanotrophic microorganisms leading to a reduced amount of methane escaping into the atmosphere. Methanotrophic microorganisms can be divided into two classes. Aerobic methanotrophs are bacteria that make use of the enzyme methane monooxygenase to activate the inert methane molecule (Sirajuddin and Rosenzweig, 2015). Anaerobic methanotrophs use an external electron acceptor other than oxygen and can be found in both prokaryotic domains, Bacteria and Archaea (Boetius et al., 2000; Ettwig et al., 2010; Haroon et al., 2013). Nitrite-dependent anaerobic oxidation of methane (AOM) is catalyzed by the anaerobic bacterium Methylomirabilis oxyfera that belongs to the NC10 phylum (Ettwig et al., 2010). After reduction of nitrite to NO it presumably dismutates NO to N2 and O2 to subsequently make use of the produced oxygen for an aerobic-type methane activation reaction via methane monooxygenase (Ettwig et al., 2010, 2012). All other anaerobic methanotrophs have been reported to belong to the domain Archaea and presumably use the reverse reaction of methyl-coenzyme M reductase—the key enzyme in methanogenesis—for the activation of methane (Krüger et al., 2003; Scheller et al., 2010). So far, enrichment cultures were reported to couple the oxidation of methane to the reduction of sulfate or nitrate (Boetius et al., 2000; Raghoebarsing et al., 2006; Haroon et al., 2013). Sulfate-dependent AOM seems to be catalyzed by the symbiotic association of an anaerobic methanotrophic archaeon (ANME) with a bacterial sulfate reducing partner (Knittel and Boetius, 2009; Ruff et al., 2015). Nitrate-dependent AOM, in contrast, seems to be catalyzed by an archaeal methanotroph alone that was named Methanoperedens nitroreducens and is affiliated to the ANME-2d clade (Raghoebarsing et al., 2006; Haroon et al., 2013). In this study, we report on the environmental genome and transcriptome of a Methanoperedens-like archaeon that was found in an enrichment culture performing nitrate-dependent AOM. This draft genome was used to establish a putative model of how nitrate-dependent methanotrophy could work. We discuss how the cytoplasmic process of methane oxidation via reverse methanogenesis may be coupled to the pseudoperiplasmically located reduction of nitrate to nitrite and ammonium by Nar- and Nrf-type nitrogen cycle enzymes. Several cytoplasmic and membrane-bound enzyme complexes homologous to enzymes in methanogens were found and are apparently combined with several metabolic traits not previously found in methanogenic or methanotrophic archaea.
Materials and Methods
Biological Source Material
An initial enrichment culture that contained M. oxyfera and Anaerobic oxidation of methane Associated Archaea (AAA) (Raghoebarsing et al., 2006) was further enriched with the effluent of another reactor that was dominated by M. oxyfera. It contained mineral medium saturated with CH4, low nitrite (50 μM) and high nitrate (2–3 mM). After about 1 year of enrichment, the reactor was uncoupled from the M. oxyfera reactor, and kept flushed with CH4-CO2 (v:v; 95:5). Nitrate was added daily as sole electron acceptor (1–3 mM final concentration) for over 2 years. The reactor was operated in batch mode. Every 2 weeks, approximately 30% of the supernatant was removed and the reactor replenished with fresh anoxic mineral medium (as previously described by Ettwig et al., 2009, omitting nitrate and nitrite). The AAA microbe in the reactor was closely related to M. nitroreducens identified by Haroon et al. (2013) and will subsequently be referred to as M. nitroreducens MPEBLZ whereas the strain identified by Haroon et al. will be referred to as M. nitroreducens ANME2D.
Metagenome and -Transcriptome Sequencing
DNA of the M. nitroreducens MPEBLZ enrichment culture was isolated with a method based on bead beating and SDS lysis as described previously (Ettwig et al., 2009). Total RNA was isolated with the Ambion RiboPureTM Bacteria Kit (MO BIO Laboratories, Uden, The Netherlands) according to the manufacturer's manual. DNA and RNA quality was checked by agarose gel electrophoresis, and concentrations were measured in triplicate with the NanoDrop (ND-1000; Isogen Life Science, The Netherlands). All kits used in library preparation and sequencing were obtained from Life technologies (Life Technologies, Carlsbad, CA, USA). Genomic DNA was sheared for 5 min using the Ion Xpress™ Plus Fragment Library Kit. Further library preparation was performed using the Ion Plus Fragment Library Kit following manufacturer's instructions. Size selection of the library was performed using an E-gel 2% agarose gel. The library was used for two sequencing runs. For both runs, emulsion PCR was performed using the OneTouch 200 bp kit and sequencing was performed on an IonTorrent PGM with the Ion PGM 200 bp sequencing kit and an Ion 318 chip, resulting in a total of 10 million reads with an average length of 170 bp. RNA was sequenced after removal of ribosomal RNA using the MicrobExpress kit (Thermo Scientific, Amsterdam, The Netherlands). The library for RNA-seq was prepared using the RNA-seq kit v2 (Life Technologies, Carlsbad, CA, USA) and two sequencing runs were performed as described above for the metagenomic library.
Assembly, Binning, and Annotation of the Methanoperedens nitroreducens MPEBLZ Draft Genome
For the construction of the environmental genome, reads were trimmed on quality and length (>100 bp). The remaining 8.1 million reads, average length 196 bp, were assembled de novo using the CLC genomics workbench (v6.5.1, CLCbio, Aarhus, Denmark) with word size 31 and bubble size 5000. Contigs were assigned to M. nitroreducens MPEBLZ based on coverage and GC content. The obtained 514 contigs were annotated using Prokka (version 1.10, Seemann, 2014) using an additional custom database containing the genomes of methanogens Methanosarcina barkeri str. Fusaro (NC_007355), Methanosarcina mazei Gö1 (NC_003901) and Methanosarcina acetivorans C2A (NC_003552). After annotation, a round of manual curation was performed to correct detected frameshifts and the contigs were re-annotated. Of the total 4528 ORF's identified 2004 were marked as hypothetical proteins after manual curation. CLC genomics workbench and the sequence visualization and annotation tool Artemis was used to analyse the features of annotated contigs (Rutherford et al., 2000). Initially, reference protein sequences belonging to several methanogenic archaea were retrieved in CLC genomics workbench and homologous protein sequences from M. nitroreducens MPEBLZ were identified through local BLASTp. Next, BLASTp was used to identify homologs of M. nitroreducens target proteins in strain ANME2D. Results were analyzed based on % sequence identity and expectation value (e-value). Homologs to strain ANME2D were defined as exhibiting an e-value < 10−10 and a sequence identity higher than 40%. Signal peptides were predicted with the PRED-SIGNAL tool (Bagos et al., 2009). This Whole Genome Shotgun project has been deposited at DDBJ/EMBL/GenBank under the accession LKCM00000000. The version described in this paper is version LKCM01000000.
Transcriptome Analysis
The draft genome sequence of the M. nitroreducens MPEBLZ was used as the template for the transcriptome analysis. Expression analysis was performed with the RNA-Seq Analysis tool from the CLC Genomic Workbench software (version 8.0, CLC-Bio, Aarhus, Denmark) and values are expressed as RPKM [Reads Per Kilobase of exon model per Million mapped reads (Mortazavi et al., 2008)].
Cofactor Analysis of Methanoperedens nitroreducens MPEBLZ
For the analysis of lipid soluble electron carriers, reactor biomass was first investigated with fluorescence in situ hybridization (FISH) at the time of sampling to quantify the relative amount of M. nitroreducens. FISH was performed as described in Daims et al. (2005) with the probes Arch915 and Eubmix (Stahl and Amann, 1991; Daims et al., 1999). About 50% archaea were found with M. nitroreducens as the only archaeon present as found by metagenome sequencing. Subsequently, 50 mg freeze dried cells were grinded with a 5/32″ steel ball with a Retsch MM 300 mixer mill at 60 Hz for 2 min. To each of the grinded samples 1 mL water and 500 μL pentane (GC grade) was added. The samples were vortexed for 2 min at maximum speed, placed in an ultrasonic bath for 2 min and centrifuged for 5 min at 12,000 × g. The upper pentane phase was transferred to a new tube. Another 500 μL pentane was added to the lower (water) phase and the extraction procedure was repeated as described above. Both pentane phases were combined and evaporated to dryness under a nitrogen flow. The dried extracts were dissolved in 200 μL methanol/ethanol (80:20) and aliquots of 90 μL were injected on an Agilent 1100 HPLC containing a Merck LiChrospher 100 RP-18 (5 μm) column (250 × 4.6 mm; flow 0.750 mL/min, peak detection by diode array detector [DAD], integration wavelength 248 nm). The DAD was also used to obtain UV/Vis spectra from 200 to 600 nm. After 3 min isocratic elution with 20% ethanol in methanol a linear gradient to 100% ethanol in 12 min followed by 10 min isocratic elution with 100% ethanol was used for separation. Methanol and ethanol were of HPLC grade. Ubiquinone 10 (UQ10, Sigma-Aldrich), menaquinone 4 (MK4, Supelco) and a methanophenazine standard provided by Uwe Deppenmeier (University of Bonn, Germany) were used as reference compounds. Phase contrast and fluorescent micrographs were taken with a Leitz Dialux microscope according to the method by Doddema and Vogels (1978).
Results and Discussion
Re-construction of the Methanoperedens nitroreducens MPEBLZ Genome
The here presented genome was reconstructed from a metagenome dataset of a bioreactor enrichment culture that coupled the anaerobic oxidation of methane to nitrate reduction to nitrite and ammonium (Zhu, 2014). Contigs from a de novo metagenome assembly were binned based on coverage and GC-content (Supplementary Figure S1). The community in the reactor was dominated by two organisms, M. nitroreducens MPEBLZ and an organism closely related to M. oxyfera that will not be discussed here (the analysis of all 16S rRNA gene reads of the metagenome is displayed in Supplementary Table 1). The 16S rRNA gene of M. nitroreducens MPEBLZ and M. nitroreducens ANME2D were 95% identical. Manual curation of the binned contigs resulted in a 3.74 Mb draft genome of M. nitroreducens MPEBLZ, on 514 contigs longer than 500 bp. Based on variation present in the reads supporting the draft genome, it is a consensus genome composed of (at least) two very closely related strains. The draft genome contains homologs of all 103 proteins used by Haroon et al. to assess the completeness of the M. nitroreducens ANME2D draft genome (Haroon et al., 2013). Additionally, we have assessed completeness of the draft genome using the lineage specific workflow of checkM, resulting in an estimated completeness higher than 96%, based on 228 markers (Parks et al., 2015). Although the coverage of two contigs that encoded the nitrate reductases is higher than that of the contigs containing the core proteins, the sequence composition and gene content of these contigs support their inclusion in the M. nitroreducens MPEBLZ draft genome sequence.
The Core Pathway of Methanotrophy is Well Conserved and Located in the Cytoplasm
We found a full reverse methanogenesis pathway in the M. nitroreducens MPEBLZ genome which is in accordance with the study of Haroon et al. (2013) and Wang et al. for ANME-2a (Wang et al., 2014). Methane is probably activated by methyl-coenzyme M reductase (Krüger et al., 2003; Shima and Thauer, 2005; Scheller et al., 2010) which was also the most highly transcribed gene cluster detected in the transcriptome. The methyl group is then transferred to the cofactor methanopterin by the action of a Na+ translocationg methyl transferase (Hallam et al., 2004). In methanotrophy, this reaction may dissipate part of the membrane potential (Becher et al., 1992); the sodium/proton gradient to drive this reaction has to be built up in the subsequent steps of methanotrophy. After the transfer to methanopterin, the methyl group is oxidized to CO2 by the reverse reaction of methanogenic enzymes (Hallam et al., 2004; Scheller et al., 2010; Thauer, 2011). The reverse methanogenesis pathway also contained a mer gene which encodes the F420-dependent 5, 10-methenyltetrahydromethanopterin reductase. Within the ANME archaea, this gene seems to be confined to members of the ANME-2 clade (Haroon et al., 2013; Wang et al., 2014) and is missing in ANME-1 clade archaea.
We investigated whether biosynthesis pathways for the crucial C1 carrier molecules were encoded in the genome. For the first acceptor of the methyl group, coenzyme M (2-mercaptoethanesulfonate), we observed that the canonical pathway employing the enzymes ComABCDEF was not encoded but instead the pathway as described for Methanosarcinales and Methanomicrobiales that consists of ComDEF together with cysteate synthase (Graham et al., 2009). Both the biosynthesis pathways for methanofuran and methanopterin are not fully resolved in methanogenic archaea; we could however assign putative biosynthesis proteins according to the report of Kaster et al. (2011a) (Supplementary Table 2).
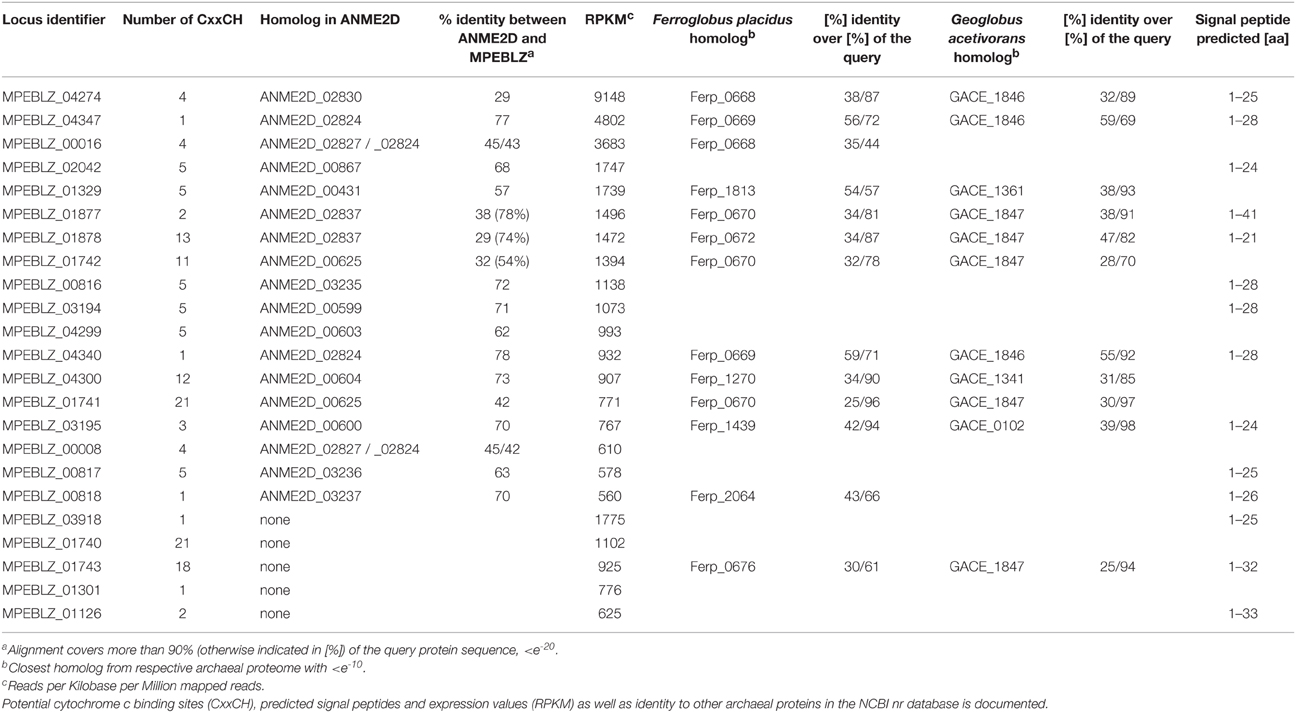
According to our model, electrons from the core methanotrophic pathway are transferred to the cytoplasmic cofactors F420, coenzyme B, and ferredoxin. The biosynthetic pathways for coenzyme B and cofactor F420 were, as far as resolved for methanogens (Kaster et al., 2011a), also encoded in the Methanoperedens genomes (Supplementary Table 2). Cells sampled from the bioreactor showed typical F420 fluorescence as also found in methanogens (Figure 1). The M. nitroreducens MPEBLZ genome harbored nine genes encoding soluble [4Fe4S] ferredoxins whereas Methanosarcina spp. encode up to 20 (Welte and Deppenmeier, 2011a).
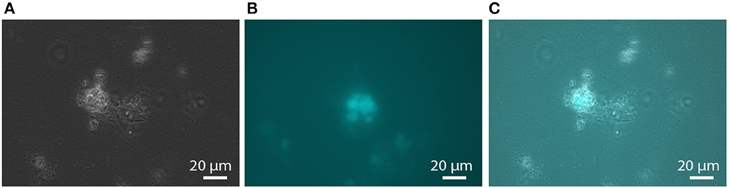
Figure 1. F420 fluorescence of aggregated biomass in the nitrate-AOM enrichment culture. (A) phase contrast micrograph, (B) fluorescence micrograph with an excitation wavelength of 390 nm and an emission wavelength of 420 nm, (C) overlay of the phase contrast and the fluorescence micrograph showing that not all cells in the aggregates exhibit F420 fluorescence.
Nitrate and Nitrite Reducing Enzymes are Predicted to be Located in the Pseudoperiplasm
Nitrate reduction in archaea is a process that is not yet well characterized (Martinez-Espinosa et al., 2007). Bacterial Nar-type nitrate reductase has its active site directed toward the cytoplasm, whereas archaeal nitrate reduction by the Nar enzyme seems to take place at the extracellular side of the cytoplasmic membrane (Yoshimatsu et al., 2000; Martinez-Espinosa et al., 2007; de Vries et al., 2010). If, or how, this process is coupled to the build-up of a proton motive force is not yet known. Different protein interaction partners were suggested to anchor the soluble subunits NarGH to the cytoplasmic membrane (Martinez-Espinosa et al., 2007; Yoshimatsu et al., 2007). de Vries et al. (2010) co-purified a protein designated NarM together with NarGH from the Pyrobaculum aerophilum membrane fraction and consequently suggested that it forms the membrane anchor spanning the cytoplasmic membrane with one transmembrane helix. It is encoded in an operon with narGH and is conserved in most archaeal nar operons (de Vries et al., 2010) but not in M. nitroreducens MPEBLZ and ANME2D. The M. nitroreducens MPEBLZ genome contains two nitrate reductase operons, whereas in the genome of the culture investigated by Haroon et al. (2013) only one was found. One narG copy appeared to be part of a conserved gene cluster between the two methanotrophs (MPEBLZ_02035-02041, Table 1) and the respective protein was 24 % identical on amino acid level to the second copy in the M. nitroreducens MPEBLZ genome (RPKM value 342). The narG operon conserved in the two methanotrophs was further investigated. The gene cluster comprises seven genes with the alpha and beta subunits of the nitrate reductase (NarG and NarH, respectively) encoded in the beginning of the cluster. The NarG protein contains an N-terminal TAT signal peptide for translocation across the cytoplasmic membrane (Table 1); the M. nitroreducens genomes also encoded the biosynthetic proteins needed for production and insertion of the molybdopterin cofactor (Supplementary Table 2, Vergnes et al., 2004). In P. aerophilum and other archaea, narGH are followed by the narM gene encoding the putative membrane anchor (de Vries et al., 2010). In Methanoperedens, we could not find a homolog to narM in the respective nar gene cluster or elsewhere in the genome indicating that this organism contains a nitrate reductase with an unusual subunit composition. Other proteins encoded in the gene cluster comprised the chaperone NarJ and a pseudoperiplasmic b-type cytochrome homologous to the Haloferax mediterranei Orf7 protein which was hypothesized to interact with NarGH in this organism (Martinez-Espinosa et al., 2007). Furthermore, a protein homologous to NapH was encoded in the gene cluster. This protein is a membrane integral subunit with four transmembrane helices of some periplasmic nitrate reductases in bacteria (Brondijk et al., 2002, 2004; Kern and Simon, 2008). It usually co-occurs with NapG that mediates the electron transfer to the catalytic subunits NapAB (Brondijk et al., 2004). In M. nitroreducens MPEBLZ and ANME2D, we could not find homologs to NapG or NapAB. The C-terminus of the NapH-like protein is instead extended and contains five additional transmembrane helices. Two other proteins that were encoded in the same gene cluster were homologous to subunit II of heme copper oxidases (Pereira et al., 2001). As all of the genes encoding these proteins were considerably expressed (Table 1) they may form an unusual nitrate reducing (transient) membrane-bound protein complex (Figure 2), a possibility that has to be addressed by further biochemical studies.
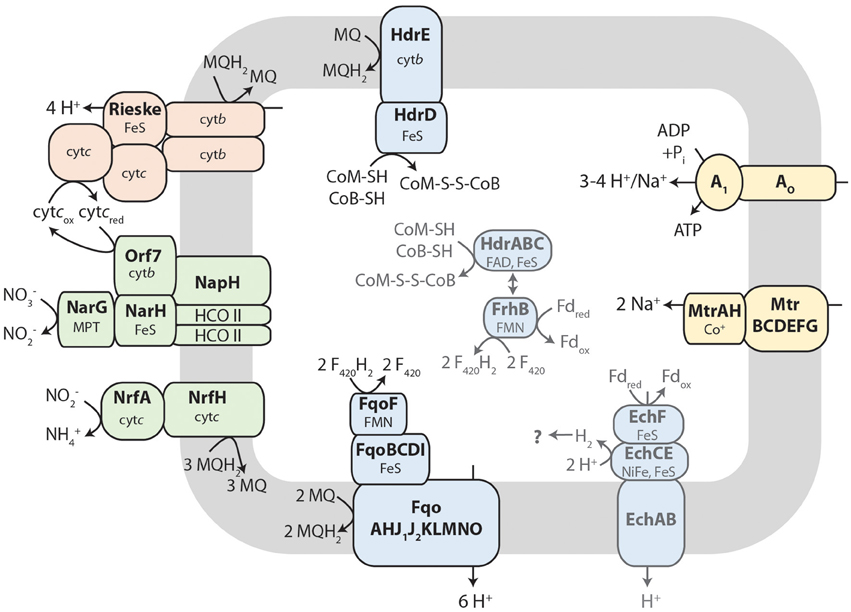
Figure 2. Tentative metabolic pathway model of membrane-bound electron transport in Methanoperedens. Reverse methanogenesis produces F420H2 and the thiol cofactors CoM-SH and CoB-SH as well as reduced ferredoxin (Fdred). F420H2 may be oxidized by the F420H2 dehydrogenase (Fqo) and electrons transferred to menaquinone (MQ, menaquinone; MQH2, menaquinol). The heterodisulfide reductase (Hdr) reaction is reversed resulting in quinone reduction upon CoM-S-S-CoB (heterodisulfide) production. Menaquinol can be oxidized by a Rieske-cytochrome b complex comprising two additional cytochrome c subunits. Electrons are transferred to an unusual nitrate reductase (Nar) complex, presumably via soluble cytochrome c (cytcox/red, oxidized/reduced cytochrome c), to reduce nitrate to nitrite. A small part of the nitrite can further be reduced to ammonium by nitrite reductase (Nrf) with menaquinol as electron donor. The fate of reduced ferredoxin is unclear. It could either be oxidized by Ech hydrogenase, by FrhB or FqoF (homologous to each other) alone or by the hypothesized confurcating HdrABC-FrhB enzyme complex. For more details, see text. Methyltransferase (Mtr) and A1AO ATP synthase make use of the proton motif force built up by the respiratory chain. This metabolic construction is solely based on genome analysis. HCO II, heme copper oxidase subunit II like proteins; cytb, cytochrome b; cytc, cytochrome c; FeS, iron-sulfur cluster; FMN, flavin mononucleotide; FAD, flavin adenine dinucleotide; MPT, molybdopterin; NiFe, nickel-iron center.
In the bioreactor that contained the enriched M. nitroreducens MPEBLZ culture, part of the nitrite was further reduced to ammonium (Zhu, 2014). It is evident that not all nitrite was reduced to ammonium as so far M. nitroreducens was always co-enriched with dedicated nitrite utilizers like M. oxyfera (Haroon et al., 2013; Zhu, 2014) or anaerobic ammonium oxidizers (Haroon et al., 2013) that reduced nitrite to dinitrogen gas. In Escherichia coli, the activities of nitrate and nitrite reductases are concerted by a complicated regulatory network (Rabin and Stewart, 1993; Tyson et al., 1993; Chiang et al., 1997; Wang and Gunsalus, 2000; Noriega et al., 2010). Besides the oxygen availability, one of the key factors in E. coli for the regulation of transcription of nar and nrf genes seems to be the concentration of nitrate and nitrite in the culture medium (Wang et al., 1999; Wang and Gunsalus, 2000). In the same studies it was demonstrated that not all nitrite was converted to ammonium under all experimental conditions but is instead excreted into the medium. As a similar finding was observed in the here presented enrichment culture it is probable that also the archaeon M. nitroreducens contains regulatory mechanisms for gene expression of nitrogen cycle enzymes.
When we searched the Methanoperedens protein complement for enzymes potentially responsible for nitrite-dependent ammonium production, we found proteins that were homologous to the NrfAH type cytochrome c nitrite reductase (Figure 2), an enzyme complex that is well characterized in δ- and ε-proteobacteria (Simon et al., 2000; Simon, 2002; Rodrigues et al., 2008). The catalytic subunit NrfA (MPEBLZ_01114, ANME2D_3312) contains—like in bacteria—a signal peptide to be translocated across the cytoplasmic membrane and therefore resides in the archaeal pseudoperiplasm. The amino acid sequence encodes four canonical CxxCH and one CxxCK heme c binding motifs for the coordination of five heme c. Amino acids required for catalysis and binding of the Ca2+ ion were conserved both in MPEBLZ_01114 and the homologous ANME2D_3312 protein [Lys126, Arg106, Tyr216, Gln263, His264, Glu215, Lys 261; E. coli NrfA numbering (Bamford et al., 2002)]. The nrfA gene is encoded in an operon next to a gene homologous to nrfH (MPEBLZ_1115; ANME2D_3311). The corresponding protein NrfH contains the canonical four CxxCH heme c binding sites as well as the conserved amino acid residues Lys82 and Asp89. It anchors the NrfAH complex in the cytoplasmic membrane and allows the interaction with the quinone pool. As all amino acids known to be involved in catalysis as well as in cofactor coordination are conserved between the bacterial and the here presented archaeal proteins, and furthermore both proteins were expressed (RPKM value of 102 and 113, respectively), this enzyme complex is the best candidate to catalyze the reduction of nitrite to ammonium also in Methanoperedens species.
Membrane-Bound, Quinone-Dependent Electron Transport Proteins May Couple Reverse Methanogenesis to Nitrate Reduction
During reverse methanogenesis, electrons are probably transferred to cytoplasmic electron carriers to yield reduced cofactor F420 (F420H2) and reduced ferredoxin. As nitrate reduction seems to take place in the pseudoperiplasm, one of the key questions in generating a metabolic model for nitrate-dependent AOM is how electrons travel across the cytoplasmic membrane and reach the nitrate reductase in the pseudoperiplasm. In the M. nitroreducens MPEBLZ genome, we identified several membrane-integral electron transport proteins that may be involved in this process (Figure 2, Supplementary Table 2). We found an F420H2 dehydrogenase closely related to the F420H2 dehydrogenase of methanogenic archaea that couples the oxidation of F420H2 to the build-up of a proton gradient (Welte and Deppenmeier, 2014). All subunits of this complex are well conserved and expressed which strongly indicates that F420H2 is oxidized by this complex in Methanoperedens. The genomic arrangement of the corresponding gene cluster in Methanoperedens resembles the one found in M. mazei: the F420H2 interacting subunit FpoF/FqoF is encoded apart from the core F420H2 dehydrogenase gene cluster at a different location on the chromosome. The F420H2 dehydrogenase gene cluster also comprises the gene fpoO which is only found in Methanosarcinales but for which there is no known function. The F420H2 dehydrogenase in Methanosarcinales transfers electrons to the membrane integral electron carrier methanophenazine. The enzyme is also found in the related Euryarchaeal lineage Archaeoglobales where a homologous complex mediates electron transport to menaquinone (Kunow et al., 1994; Brüggemann et al., 2000). As methanophenazine (E −165 mV, Tietze et al., 2003) and menaquinone (E −80 mV, Tran and Unden, 1998) have considerably different redox potentials that have implications for subsequent electron transport pathways in nitrate-dependent AOM, we investigated which of the two lipid-soluble electron acceptors was present in M. nitroreducens MPEBLZ. As the biosynthesis pathway for methanophenazine is not known, we could not mine the genome for presence or absence of these genes. In contrast, there is a complete menaquinone biosynthesis pathway known for Archaeoglobus (Hemmi et al., 2005; Hiratsuka et al., 2008; Nowicka and Kruk, 2010), which is not present in Methanosarcinales. We found that this pathway was present in both M. nitroreducens genomes (Haroon et al., 2013). To obtain further experimental evidence, we extracted quinones and phenazines from the bioreactor biomass that was dominated by cells of strain MPEBLZ and analyzed this fraction with high performance liquid chromatography coupled to UV/Vis spectroscopy (Figure 3). The elution profile of the HPLC chromatogram is shown in Figure 3A. The main peaks in the chromatogram were analyzed with UV/Vis spectroscopy and showed either a ubiquinone-like spectrum (peak 2, comparable to the spectrum in the right panel of Figure 3B) or a menaquinone-like spectrum (peaks 1, 3, 4, 5, 6, 7, comparable to the spectrum in the middle panel of Figure 3B). We could, however, not detect any fraction that showed the characteristic UV/Vis spectrum of methanophenazine (spectrum in the left panel of Figure 3B). As the culture is an enrichment culture it was not possible to assign one of the quinone fractions to M. nitroreducens MPEBLZ. From these experiments we conclude that it is highly likely that M. nitroreducens MPEBLZ uses menaquinone and not methanophenazine in membrane-bound electron transport (Figure 3).
Figure 3. Analysis of the lipid-soluble electron carriers of the nitrate-AOM enrichment culture. (A) HPLC elution profile as visualized by the absorption at 248 nm. Numbers indicate the peaks that were further characterized by UV/Vis spectroscopy. (B) UV-Vis spectra of standard compounds (left, methanophenazine; middle, menaquinone-4; right, ubiquinone-4) for comparison with spectra obtained from fractions separated by HPLC. Based on the comparison of the experimental spectra to the spectra obtained from the HPLC fractions, the different peaks were assigned to contain a representative from the classes of ubiquinones, menaquinones, or methanophenazines. None of the spectra resembled the standard spectrum for methanophenazine (left), seven spectra (obtained from peaks 1, 3, 4, 5, 6, 7,) resembled the standard spectrum of menaquinone-4 (middle) and one spectrum (obtained from peak 2) resembled the standard spectrum of ubiquinone-4. Different retention times within one molecule class indicate a difference in the prenoid chain length. Experimental spectra are displayed in Supplementary Figure S2.
Besides F420H2 dehydrogenase, a membrane-bound heterodisulfide reductase (HdrDE, Figure 2) may contribute to the reduction of the quinone pool. In the M. nitroreducens MPEBLZ genome we were able to locate the gene for the membrane-integral b-type cytochrome subunit HdrE as well as the hydrophilic subunit HdrD. This complex may couple the oxidation of the reduced thiol cofactors CoM-SH and CoB-SH to form the heterodisulfide CoM-S-S-CoB (Figure 2). Electrons may be transferred to the membrane-integral electron carrier menaquinone. As the redox potential E of the CoM-S-S-CoB/CoM-SH+CoB-SH redox couple is −143 mV (Tietze et al., 2003), electron transfer to menaquinone (E80 mV) would be exergonic whereas electron transfer to methanophenazine (E165 mV) would be endergonic.
The reduced quinone pool can subsequently be used by membrane-bound oxidoreductases. We could not find membrane-bound quinol oxidases. Instead, we identified an unusual Rieske/cytochrome b (Rieske/cytb) complex encoded in the M. nitroreducens genomes which is homologous to the cytochrome bc1 and b6f complexes of chemotrophs and phototrophs, respectively. These complexes couple the oxidation of quinones to the reduction of periplasmic cytochrome c and the translocation of protons via the Q-cycle (Berry et al., 2000). The canonical cytochrome bc1/b6f complex contains a membrane-integral cytochrome b, a periplasmic Rieske iron-sulfur protein and another cytochrome (cytochrome c1 or f) at the periplasmic face, all of which are also encoded by the M. nitroreducens genome (Supplementary Table 2, MPEBLZ_00818 and MPEBLZ_00820 to 00822). The Rieske/cytb gene cluster also comprises two pentaheme c-type cytochromes (MPEBLZ_00816 and 00817) and two hypothetical proteins (MPEBLZ_00819 and 00823), all of which are conserved between the strains MPEBLZ and ANME2D. All eight genes are expressed at similar levels in M. nitroreducens MPEBLZ. This transcriptional pattern combined with the gene cluster arrangement suggests that the proteins MPEBLZ_00818 to 00823 may form a non-canonical Rieske/cytb complex which is in line with the finding that those complexes often harbor additional subunits to the canonical ones (Ten Brink et al., 2013). For the related haloarchaea there is strong indication that the Rieske/cytb complex and nitrate reductase are interacting as they are encoded in the same gene cluster (Martinez-Espinosa et al., 2007; Yoshimatsu et al., 2007; Bonete et al., 2008) and the nitrate reductase reaction is inhibited by a the Rieske/cytb complex inhibitor Antimycin A (Martinez-Espinosa et al., 2007). In M. nitroreducens, electrons from the reduced quinone pool may travel via the Rieske/cytb complex and the pentaheme c-type cytochromes to the pseudoperiplasmic space and at that place they may be used by the nitrate reductase to reduce the external electron acceptor nitrate.
Ferredoxin and the Heterodisulfide may be Re-cycled by a Novel Electron-Confurcating Enzyme Complex
The oxidized cofactors ferredoxin and heterodisulfide are needed as electron acceptors during reverse methanogenesis and thus have to be re-oxidized by cytoplasmic or membrane-bound electron transport processes to become available for a new round of methane oxidation. Members of the Methanosarcinales use membrane proteins for heterodisulfide reduction and ferredoxin oxidation (Welte and Deppenmeier, 2014). The genome of Methanoperedens encodes a membrane-bound heterodisulfide reductase (HdrDE, see above) that is highly expressed (RPKM values 946 and 1396, Supplementary Table 2) and therefore presumably the primary CoM-SH/CoB-SH oxidizing enzyme. For ferredoxin oxidation, we did not find an Rnf complex but identified an Ech hydrogenase lacking the subunit EchD (Supplementary Table 2). In the six-subunit Ech hydrogenase, EchD is the only hydrophilic subunit without prosthetic groups and is missing in Ech hydrogenases of some methanogens (Friedrich and Scheide, 2000). This indicates that the Ech hydrogenase of Methanoperedens may be functional. Expression values are, however, low (RPKM 52–80, Supplementary Table 2) so it is unclear whether Ech hydrogenase is used by M. nitroreducens under the investigated growth conditions. In hydrogenotrophic methanogens, the cytoplasmic electron bifurcating enzyme complex heterodisulfide reductase (HdrABC) coupled to a hydrogenase (MvhABG) serves as ferredoxin reducing enzyme (Kaster et al., 2011b): electrons from molecular hydrogen are used to reduce the heterodisulfide in an exergonic reaction which in turn drives the endergonic reduction of ferredoxin. When we analyzed the genome for the presence of this enzyme complex, we found that it encodes three copies of hdrABC that are significantly expressed (RPKM values 137–482, Supplementary Table 2) but we could not detect the mvhABG gene cluster encoding the hydrogenase used in electron bifurcation by methanogenic archaea. In M. nitroreducens, the thiols CoM-SH and CoB-SH (E143 mV) are produced in reverse methanotrophy and would therefore donate electrons to the HdrABC complex; however, the redox potential is too high to allow for a direct reduction of any of the cytoplasmic electron carriers F420, ferredoxin, H2, or NAD(P)+. Instead, the oxidation of the CoM-SH and CoB-SH thiols may be coupled to an electron confurcation reaction in which ferredoxin (E ≈ −500 mV, Thauer et al., 2008) would be oxidized concomitantly and F420 (E = −360 mV, Walsh, 1986) reduced. The overall reaction would turn thermodynamically favorable and allow the backwards electron flow from the CoM-SH and CoB-SH thiols to F420. A possible protein mediating the interaction with F420 and ferredoxin has been described (FpoF, Welte and Deppenmeier, 2011b) and a gene encoding a homologous protein (FrhB) was found in proximity of one of the hdrABC copies, suggesting that an HdrABC-FrhB complex may mediate the above described reaction (Figure 4) in Methanoperedens species. FrhB or FpoF alone might also couple the oxidation of ferredoxin to the reduction of F420 as observed in the cytoplasm of M. mazei (Welte and Deppenmeier, 2011b). In this case, the energy liberated by the redox reaction (ΔE ≈ 140 mV) would not be harnessed but released as heat.
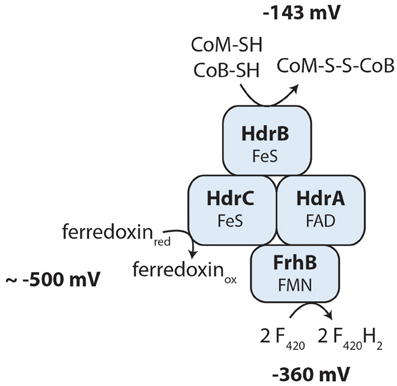
Figure 4. Hypothesis of a novel cytoplasmic electron confurcating heterodisulfide reductase complex for ferredoxin and CoM-S-S-CoB recycling. The exergonic reduction of F420 (E = −360 mV) with reduced ferredoxin (E ≈−500 mV) may be coupled to the endergonic electron transfer from the thiol cofactors CoM-SH and CoB-SH (E = −143 mV) to F420. This hypothesis is based on the metabolic reconstruction using genome and transcriptome sequencing and is not yet supported by biochemical experiments. FAD, flavin adenine dinucleotide; FMN, flavin mononucleotide; FeS, iron-sulfur cluster; FrhB, F420-reducing hydrogenase subunit B; HdrABC, heterodisulfide reductase subunits A, B, C.
Hydrogen Does not Seem to be an Intermediate in the Electron Transport Chain
In many Methanosarcina sp., hydrogen is an intermediate in anaerobic respiration. It is produced either by the action of Ech hydrogenase during ferredoxin oxidation or by the action of the cytoplasmic F420 hydrogenase (FrhABG) during oxidation of F420H2 (Meuer et al., 1999; Kulkarni et al., 2009). In both cases, molecular hydrogen is subsequently oxidized by a membrane-bound hydrogenase (VhoACG/VhtACG/VhxACG) to feed electrons into the methanophenazine pool (Welte and Deppenmeier, 2014). In the M. nitroreducens MPEBLZ and ANME2D genomes, genes encoding the Vho/Vht/Vhx hydrogenase were not found indicating that electrons from molecular hydrogen cannot enter the membrane-bound anaerobic respiratory chain of Methanoperedens species. Furthermore, the M. nitroreducens MPEBLZ genome only contained an frhB gene but not frhABG indicating that the F420 hydrogenase is not functional.
Methanoperedens nitroreducens Lacks Genes Required for Methanogenesis
As methanotrophic archaea are closely related to methanogens we investigated whether all proteins necessary for one of the methanogenesis pathways were encoded in the genome. For methylotrophic methanogenesis, members of the Methanosarcinales contain substrate-specific methyl transferases (Krzycki, 2004). In the genome of M. nitroreducens MPEBLZ we could not find such methyl transferases which makes it unlikely that this species is able to perform methanogenesis from methylated compounds. For hydrogenotrophic methanogenesis, dedicated hydrogenases have to be encoded in the genome (Thauer et al., 2010). For classical hydrogenotrophic methanogenesis, the HdrABC-MvhABG electron bifurcating complex supplies reduced ferredoxin for CO2 reduction (Kaster et al., 2011b). As the MvhABG complex was not encoded in the here investigated genome, classical hydrogenotrophic methanogenesis does not seem to be possible. Methanosarcina sp. make use of membrane-bound hydrogenases to eventually reduce CO2 to CH4. The responsible Vho/Vht/Vhx hydrogenase (subunits) could not be found in the Methanoperedens genomes which strongly indicates that neither form of hydrogenotrophic methanogenesis is possible.
In case of aceticlastic methanogenesis, acetate first has to be activated to acetyl-CoA. In Methanosarcina sp., this happens via acetate kinase and phosphotransacetylase whereas Methanosaeta sp. use AMP-dependent acetyl-CoA synthetases (ACS) (Jetten et al., 1992; Welte and Deppenmeier, 2014). In the genome of M. nitroreducens MPEBLZ we could not detect genes encoding the first acetate activation system. We found one gene encoding an ADP-dependent ACS. Methanosaeta sp. contain several ACS enzymes that are either used in lipid metabolism or aceticlastic methanogenesis; as we only found one ACS enzyme encoded in the M. nitroreducens MPEBLZ genome which was more related to those enzymes involved in fatty acid metabolism it is unlikely that Methanoperedens archaea can make methane from acetate.
It cannot be excluded that hitherto unknown mechanisms lead to an ability for methanogenesis in Methanoperedens species but regarding well investigated metabolic pathways for methane formation this metabolic trait is not likely to be found in these organisms.
Unusual High Number of c-Type Cytochromes May Reflect Adaption to the Metabolic Trait of Nitrate Reduction
The genomes of Methanoperedens species encode an unusually high number of c-type cytochromes (Haroon et al., 2013; Kletzin et al., 2015). The occurrence of c-type cytochromes in archaea was recently reviewed (Kletzin et al., 2015) and it was highlighted that the ANME-2 clade contains apparently the highest number of c-type cytochromes encoded in an archaeal genome. We identified 87 proteins containing at least one CxxCH heme c binding motif. Of these 87 proteins, 68 were homologous to proteins found in M. nitroreducens ANME2D. Kletzin et al. (2015) analyzed this protein subset and came to the conclusion that only 43 of these 68 were likely to be true c-type cytochromes. Nineteen open reading frames in our M. nitroreducens MPEBLZ genome assembly encoded CxxCH motif(s) but did not have a homolog in the strain ANME2D. A total of 23 CxxCH motif containing proteins were abundantly expressed (RPKM value >500) and more closely investigated, 18 with homologs in the ANME2D genome and five without (Table 2). The annotation of these proteins gave no indication on their function as they were all annotated either as hypothetical protein or cytochrome c protein. Most of the proteins (70%, Table 2) contained three or more CxxCH motifs (even up to 21) and can therefore be regarded as multiheme c-type cytochromes. Several of those harbored an additional CxxxCH (MPEBLZ_00816, MPEBLZ_00817, MPEBLZ_03194, MPEBLZ_04300) or CxxxxCH motif (MPEBLZ_01743, MPEBLZ_03195). c-type cytochromes generally reside in the periplasm or archaeal pseudoperiplasm (Thöny-Meyer, 1997) and most of the here investigated c-type cytochromes contained a putative signal peptide (Table 2) or an N-terminal transmembrane helix (MPEBLZ_01329, MPEBLZ_04299, MPEBLZ_04300). In addition, several of the proteins (e.g., MPEBLZ_00008, MPEBLZ_00016, MPEBLZ_04347) were homologous to each other. Recently, McGlynn and co-workers identified several large multiheme c-type cytochromes in ANME archaea that may bridge the S-layer to donate electrons to extracellular partner organisms or metal oxides (Mcglynn et al., 2015). The respective c-type cytochromes are (at least in part) conserved in the MPEBLZ genome (MPEBLZ_02503 and 02608) but were hardly expressed under our experimental conditions (RPKM values of 27 and 37, respectively).
To get insight into the potential function of the 23 abundantly expressed, uncharacterized cytochrome c proteins, we looked for homologs in the nr database using BLASTp (Altschul et al., 1990). For the majority of proteins, no close homologs (besides in strain ANME2D) could be found. An exception to this formed MPEBLZ_02042 which had many homologs in the bacterial and archaeal domain albeit with only moderate sequence identity (= 37%). Homologs that were found for many of the other proteins comprised proteins from the Fe(III) reducing Euryarchaeota Ferroglobus placidus and Geoglobus acetivorans (Mardanov et al., 2015; Smith et al., 2015). The sequence identity was generally low and spanned only part of the protein (Table 2). For both the proteins from F. placidus as well as G. acetivorans, there are no biochemical data available as to their function. However, a recent genomic survey (Mardanov et al., 2015) proposed a potential involvement of several c-type cytochromes in Fe(III) reduction which were encoded by four gene clusters (gace_0099 to 0102 and gace_1341 to _1344, gace_1360 to _1361, gace_1843 to _1847). Many of the c-type cytochromes identified in the Methanoperedens genomes showed homology to proteins encoded in the first two gene clusters. The identity of the amino acid sequences was too weak to allow for a functional comparison between the proteins but it indicates that these proteins (which are also highly expressed) may have a function in pseudoperiplasmic electron transport, possibly to nitrate reductase that resides in the pseudoperiplasm. The protein GACE_1847 is speculated to be the direct electron transfer protein from the cytoplasmic membrane to extracellular hematite crystals in G. acetivorans (Fe2O3, Mardanov et al., 2015). It seems to be anchored in the cytoplasmic membrane, span the pseudoperiplasm with an array of c-type hemes (16 CxxCH binding motifs) and then bridge the S-layer to make contact to hematite crystals (Fe2O3) making use of hematite binding motifs at the C-terminus of the protein ([ST]-[AVILMFYW]-[ST]-P-[ST], Lower et al., 2008; Mardanov et al., 2015). The Methanoperedens homologs, however, lack the C-terminal amino acid sequence, both for crossing of the S-layer as well as the hematite binding motifs. It is thus unlikely that these proteins are involved in binding to Fe(III) minerals.
Three of the c-type cytochrome encoding genes (MPEBLZ_00816 to MPEBLZ_00818) were in the same gene cluster as other subunits for a Rieske/cytb complex indicating their involvement in electron transport from the cytoplasm to the pseudoperiplasm where nitrate and nitrite reductases reside.
The metabolically and phylogenetically closely related methanogens of the order Methanosarcinales only contain few c-type cytochromes whose functions are largely unknown. The surprisingly large number of c-type cytochromes encoded by ANME-2d archaea may thus be connected to electron transfer from reverse methanogenesis to nitrate reductase. This is in accordance with the anticipated redox potentials of part of these cytochromes and the apparent low number of encoded ferredoxins in the genome: whereas bioenergetics in methanogenic archaea spans the redox range from about −500 mV (E for CO2/CO and at the same time midpoint potential of ferredoxins used in this process) to −143 mV (E (CoM-S-S-CoB/CoM-SH+CoB-SH)), nitrate-dependent methanotrophic archaea operate at a wider redox potential range (E (NO/NO) = +433 mV) that requires electron carriers with a redox potential of up to +400 mV. c-type cytochromes are ideal candidates to operate in the redox potential range of −143 to +433 mV and therefore act as redox carriers in nitrate-dependent anaerobic methanotrophy.
Bioenergetics of Nitrate-Dependent AOM
The free energy change associated to nitrate-dependent AOM is high with a Gibbs free energy of ΔG0' = −523 kJ per mol CH4 oxidized [calculated with the standard potentials E (NO/NO) = +433 mV and E (CO2/CH4) = −244 mV (Thauer et al., 1977)]. According to our metabolic reconstruction (Figure 2), the mechanism of energy conservation in Methanoperedens is electron transport phosphorylation and not substrate level phosphorylation. During reverse methanogenesis, 2 mol Na+ per mol of methane are translocated into the cytoplasm and dissipate the proton/sodium motive force. At the same time, electrons are transferred to the cytoplasmic cofactors F420, ferredoxin and CoM-S-S-CoB. F420H2 is recycled via the membrane-bound F420H2 dehydrogenase complex and electrons are transferred to a membrane-bound electron carrier, probably menaquinone. In Ms. mazei, two protons are translocated across the membrane in the course of this reaction (Bäumer et al., 2000); as Methanoperedens probably uses the more electropositive menaquinone instead of methanophenazine this reaction yields a higher ΔE and subsequently may catalyze the translocation of up to 3 H+/2 e−. Quinols may be oxidized by the Rieske/cytb complex that transfers electrons to pseudoperiplasmic cytochrome c; in the course of this reaction, usually 4 H+/2 e− are released at the extracellular side of the membrane via a Q-cycle mechanism (Berry et al., 2000). As for CoM-SH+CoB-SH and ferredoxin also cytoplasmic possibilities to be re-oxidized exist (Figure 4) it is unclear whether their re-oxidation is associated to a build-up of proton motive force. For nitrate reduction in the pseudoperiplasm, it is also not known whether this process is associated to the membrane and thus could contribute to the generation of a proton motive force. Taking together the above mentioned observations, it becomes clear that the oxidation of the four electrons of F420H2 reduced during reverse methanogenesis leads to proton translocation that by far compensates for the 2 Na+ per mol methane that are imported during reverse methanogenesis and allows for the build-up of a proton motive force. This can in turn be used by an A1AO ATP synthase to produce ATP for cellular metabolism and growth. The c-subunit of the M. nitroreducens ATP synthase (MPEBLZ_01699) resembles the M. acetivorans c-subunit; all residues required for Na+ and H+ binding are conserved (except for the replacement of Thr-67 by Ser which is a common feature of Na+ translocating ATP synthases) and may therefore drive ATP synthesis by both a proton and a sodium ion gradient (Schlegel et al., 2012).
Author Contributions
CW, HO, and MJ designed research. AA, DS, and RD collected data and all authors performed parts of the data analysis. CW wrote the paper with contributions from all authors.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the ERC AG 339880 and the SIAM Gravitation Grant 24002002 (Netherlands Organisation for Scientific Research). The authors thank Rolf Thauer for helpful discussions, Uwe Deppenmeier (University of Bonn) for providing a methanophenazine standard and Geert Cremers, Katinka van de Pas-Schoonen and Theo van Alen for their technical assistance. Baoli Zhu and Simon Guerrero are thankfully acknowledged for providing bioreactor samples for sequencing and cofactor analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.01423
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Bagos, P. G., Tsirigos, K. D., Plessas, S. K., Liakopoulos, T. D., and Hamodrakas, S. J. (2009). Prediction of signal peptides in archaea. Protein Eng. Des. Sel. 22, 27–35. doi: 10.1093/protein/gzn064
Bamford, V. A., Angove, H. C., Seward, H. E., Thomson, A. J., Cole, J. A., Butt, J. N., et al. (2002). Structure and spectroscopy of the periplasmic cytochrome c nitrite reductase from Escherichia coli. Biochemistry 41, 2921–2931. doi: 10.1021/bi015765d
Bäumer, S., Ide, T., Jacobi, C., Johann, A., Gottschalk, G., and Deppenmeier, U. (2000). The F420H2 dehydrogenase from Methanosarcina mazei is a redox-driven proton pump closely related to NADH dehydrogenases. J. Biol. Chem. 275, 17968–17973. doi: 10.1074/jbc.M000650200
Becher, B., Müller, V., and Gottschalk, G. (1992). N5-methyl-tetrahydromethanopterin:coenzyme M methyltransferase of Methanosarcina strain Gö1 is an Na+-translocating membrane protein. J. Bacteriol. 174, 7656–7660.
Berry, E. A., Guergova-Kuras, M., Huang, L. S., and Crofts, A. R. (2000). Structure and function of cytochrome bc complexes. Annu. Rev. Biochem. 69, 1005–1075. doi: 10.1146/annurev.biochem.69.1.1005
Boetius, A., Ravenschlag, K., Schubert, C. J., Rickert, D., Widdel, F., Gieseke, A., et al. (2000). A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407, 623–626. doi: 10.1038/35036572
Bonete, M. J., Martinez-Espinosa, R. M., Pire, C., Zafrilla, B., and Richardson, D. J. (2008). Nitrogen metabolism in haloarchaea. Saline Syst. 4:9. doi: 10.1186/1746-1448-4-9
Brondijk, T. H., Fiegen, D., Richardson, D. J., and Cole, J. A. (2002). Roles of NapF, NapG and NapH, subunits of the Escherichia coli periplasmic nitrate reductase, in ubiquinol oxidation. Mol. Microbiol. 44, 245–255. doi: 10.1046/j.1365-2958.2002.02875.x
Brondijk, T. H., Nilavongse, A., Filenko, N., Richardson, D. J., and Cole, J. A. (2004). NapGH components of the periplasmic nitrate reductase of Escherichia coli K-12: location, topology and physiological roles in quinol oxidation and redox balancing. Biochem. J. 379, 47–55. doi: 10.1042/bj20031115
Brüggemann, H., Falinski, F., and Deppenmeier, U. (2000). Structure of the F420H2: quinone oxidoreductase of Archaeoglobus fulgidus - Identification and overproduction of the F420H2-oxidizing subunit. Eur. J. Biochem. 267, 5810–5814. doi: 10.1046/j.1432-1327.2000.01657.x
Chiang, R. C., Cavicchioli, R., and Gunsalus, R. P. (1997). ‘Locked-on’ and ‘locked-off’ signal transduction mutations in the periplasmic domain of the Escherichia coli NarQ and NarX sensors affect nitrate- and nitrite-dependent regulation by NarL and NarP. Mol. Microbiol. 24, 1049–1060. doi: 10.1046/j.1365-2958.1997.4131779.x
Daims, H., Bruhl, A., Amann, R., Schleifer, K. H., and Wagner, M. (1999). The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22, 434–444. doi: 10.1016/S0723-2020(99)80053-8
Daims, H., Stoecker, K., and Wagner, M. (2005). “Fluorescence in situ hybridization for the detection of prokaryotes,” in Molecular Microbial Ecology, eds A. M. Osborn and C. J. Smith (New York, NY: Taylor & Francis Group), 213–239.
de Vries, S., Momcilovic, M., Strampraad, M. J., Whitelegge, J. P., Baghai, A., and Schroder, I. (2010). Adaptation to a high-tungsten environment: Pyrobaculum aerophilum contains an active tungsten nitrate reductase. Biochemistry 49, 9911–9921. doi: 10.1021/bi100974v
Doddema, H. J., and Vogels, G. D. (1978). Improved identification of methanogenic bacteria by fluorescence microscopy. Appl. Environ. Microbiol. 36, 752–754.
Ettwig, K. F., Butler, M. K., Le Paslier, D., Pelletier, E., Mangenot, S., Kuypers, M. M., et al. (2010). Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464, 543–548. doi: 10.1038/nature08883
Ettwig, K. F., Speth, D. R., Reimann, J., Wu, M. L., Jetten, M. S., and Keltjens, J. T. (2012). Bacterial oxygen production in the dark. Front. Microbiol. 3:273. doi: 10.3389/fmicb.2012.00273
Ettwig, K. F., Van Alen, T., Van De Pas-Schoonen, K. T., Jetten, M. S., and Strous, M. (2009). Enrichment and molecular detection of denitrifying methanotrophic bacteria of the NC10 phylum. Appl. Environ. Microbiol. 75, 3656–3662. doi: 10.1128/AEM.00067-09
Friedrich, T., and Scheide, D. (2000). The respiratory complex I of bacteria, archaea and eukarya and its module common with membrane-bound multisubunit hydrogenases. FEBS Lett. 479, 1–5. doi: 10.1016/S0014-5793(00)01867-6
Graham, D. E., Taylor, S. M., Wolf, R. Z., and Namboori, S. C. (2009). Convergent evolution of coenzyme M biosynthesis in the Methanosarcinales: cysteate synthase evolved from an ancestral threonine synthase. Biochem. J. 424, 467–478. doi: 10.1042/BJ20090999
Hallam, S. J., Putnam, N., Preston, C. M., Detter, J. C., Rokhsar, D., Richardson, P. M., et al. (2004). Reverse methanogenesis: testing the hypothesis with environmental genomics. Science 305, 1457–1462. doi: 10.1126/science.1100025
Haroon, M. F., Hu, S., Shi, Y., Imelfort, M., Keller, J., Hugenholtz, P., et al. (2013). Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 500, 567–570. doi: 10.1038/nature12375
Hemmi, H., Takahashi, Y., Shibuya, K., Nakayama, T., and Nishino, T. (2005). Menaquinone-specific prenyl reductase from the hyperthermophilic archaeon Archaeoglobus fulgidus. J. Bacteriol. 187, 1937–1944. doi: 10.1128/JB.187.6.1937-1944.2005
Hiratsuka, T., Furihata, K., Ishikawa, J., Yamashita, H., Itoh, N., Seto, H., et al. (2008). An alternative menaquinone biosynthetic pathway operating in microorganisms. Science 321, 1670–1673. doi: 10.1126/science.1160446
Jetten, M. S. M., Stams, A. J. M., and Zehnder, A. J. B. (1992). Methanogenesis from acetate - a comparison of the acetate metabolism in Methanothrix soehngenii and Methanosarcina spp. FEMS Microbiol. Rev. 88, 181–197. doi: 10.1111/j.1574-6968.1992.tb04987.x
Kaster, A. K., Goenrich, M., Seedorf, H., Liesegang, H., Wollherr, A., Gottschalk, G., et al. (2011a). More than 200 genes required for methane formation from H(2) and CO(2) and energy conservation are present in Methanothermobacter marburgensis and Methanothermobacter thermautotrophicus. Archaea 2011:973848. doi: 10.1155/2011/973848
Kaster, A. K., Moll, J., Parey, K., and Thauer, R. K. (2011b). Coupling of ferredoxin and heterodisulfide reduction via electron bifurcation in hydrogenotrophic methanogenic archaea. Proc. Natl. Acad. Sci. U.S.A. 108, 2981–2986. doi: 10.1073/pnas.1016761108
Kern, M., and Simon, J. (2008). Characterization of the NapGH quinol dehydrogenase complex involved in Wolinella succinogenes nitrate respiration. Mol. Microbiol. 69, 1137–1152. doi: 10.1111/j.1365-2958.2008.06361.x
Kletzin, A., Heimerl, T., Flechsler, J., van Niftrik, L., Rachel, R., and Klingl, A. (2015). Cytochromes c in Archaea: distribution, maturation, cell architecture, and the special case of Ignicoccus hospitalis. Front. Microbiol. 6:439. doi: 10.3389/fmicb.2015.00439
Knittel, K., and Boetius, A. (2009). Anaerobic oxidation of methane: progress with an unknown process. Annu. Rev. Microbiol. 63, 311–334. doi: 10.1146/annurev.micro.61.080706.093130
Krüger, M., Meyerdierks, A., Glöckner, F. O., Amann, R., Widdel, F., Kube, M., et al. (2003). A conspicuous nickel protein in microbial mats that oxidize methane anaerobically. Nature 426, 878–881. doi: 10.1038/nature02207
Krzycki, J. A. (2004). Function of genetically encoded pyrrolysine in corrinoid-dependent methylamine methyltransferases. Curr. Opin. Chem. Biol. 8, 484–491. doi: 10.1016/j.cbpa.2004.08.012
Kulkarni, G., Kridelbaugh, D. M., Guss, A. M., and Metcalf, W. W. (2009). Hydrogen is a preferred intermediate in the energy-conserving electron transport chain of Methanosarcina barkeri. Proc. Natl. Acad. Sci. U.S.A. 106, 15915–15920. doi: 10.1073/pnas.0905914106
Kunow, J., Linder, D., Stetter, K. O., and Thauer, R. K. (1994). F420H2:quinone oxidoreductase from Archaeoglobus fulgidus - characterization of a membrane-bound multisubunit complex containing FAD and iron-sulfur clusters. Eur. J. Biochem. 223, 503–511. doi: 10.1111/j.1432-1033.1994.tb19019.x
Lower, B. H., Lins, R. D., Oestreicher, Z., Straatsma, T. P., Hochella, M. F. Jr., Shi, L., et al. (2008). In vitro evolution of a peptide with a hematite binding motif that may constitute a natural metal-oxide binding archetype. Environ. Sci. Technol. 42, 3821–3827. doi: 10.1021/es702688c
Mardanov, A. V., Slododkina, G. B., Slobodkin, A. I., Beletsky, A. V., Gavrilov, S. N., Kublanov, I. V., et al. (2015). The Geoglobus acetivorans genome: Fe(III) reduction, acetate utilization, autotrophic growth, and degradation of aromatic compounds in a hyperthermophilic archaeon. Appl. Environ. Microbiol. 81, 1003–1012. doi: 10.1128/AEM.02705-14
Martinez-Espinosa, R. M., Dridge, E. J., Bonete, M. J., Butt, J. N., Butler, C. S., Sargent, F., et al. (2007). Look on the positive side! The orientation, identification and bioenergetics of ‘Archaeal’ membrane-bound nitrate reductases. FEMS Microbiol. Lett. 276, 129–139. doi: 10.1111/j.1574-6968.2007.00887.x
Mcglynn, S. E., Chadwick, G. L., Kempes, C. P., and Orphan, V. J. (2015). Single cell activity reveals direct electron transfer in methanotrophic consortia. Nature 526, 531–535. doi: 10.1038/nature15512
Metcalf, W. W., Griffin, B. M., Cicchillo, R. M., Gao, J., Janga, S. C., Cooke, H. A., et al. (2012). Synthesis of methylphosphonic acid by marine microbes: a source for methane in the aerobic ocean. Science 337, 1104–1107. doi: 10.1126/science.1219875
Meuer, J., Bartoschek, S., Koch, J., Künkel, A., and Hedderich, R. (1999). Purification and catalytic properties of Ech hydrogenase from Methanosarcina barkeri. Eur. J. Biochem. 265, 325–335. doi: 10.1046/j.1432-1327.1999.00738.x
Mortazavi, A., Williams, B. A., Mccue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Noriega, C. E., Lin, H. Y., Chen, L. L., Williams, S. B., and Stewart, V. (2010). Asymmetric cross-regulation between the nitrate-responsive NarX-NarL and NarQ-NarP two-component regulatory systems from Escherichia coli K-12. Mol. Microbiol. 75, 394–412. doi: 10.1111/j.1365-2958.2009.06987.x
Nowicka, B., and Kruk, J. (2010). Occurrence, biosynthesis and function of isoprenoid quinones. Biochim. Biophys. Acta 1797, 1587–1605. doi: 10.1016/j.bbabio.2010.06.007
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P., and Tyson, G. W. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055. doi: 10.1101/gr.186072.114
Pereira, M. M., Santana, M., and Teixeira, M. (2001). A novel scenario for the evolution of haem-copper oxygen reductases. Biochim. Biophys. Acta 1505, 185–208. doi: 10.1016/S0005-2728(01)00169-4
Rabin, R. S., and Stewart, V. (1993). Dual response regulators (NarL and NarP) interact with dual sensors (NarX and NarQ) to control nitrate- and nitrite-regulated gene expression in Escherichia coli K-12. J. Bacteriol. 175, 3259–3268.
Raghoebarsing, A. A., Pol, A., van de Pas-Schoonen, K. T., Smolders, A. J., Ettwig, K. F., Rijpstra, W. I., et al. (2006). A microbial consortium couples anaerobic methane oxidation to denitrification. Nature 440, 918–921. doi: 10.1038/nature04617
Rodrigues, M. L., Scott, K. A., Sansom, M. S., Pereira, I. A., and Archer, M. (2008). Quinol oxidation by c-type cytochromes: structural characterization of the menaquinol binding site of NrfHA. J. Mol. Biol. 381, 341–350. doi: 10.1016/j.jmb.2008.05.066
Ruff, S. E., Biddle, J. F., Teske, A. P., Knittel, K., Boetius, A., and Ramette, A. (2015). Global dispersion and local diversification of the methane seep microbiome. Proc. Natl. Acad. Sci. U.S.A. 112, 4015–4020. doi: 10.1073/pnas.1421865112
Rutherford, K., Parkhill, J., Crook, J., Horsnell, T., Rice, P., Rajandream, M. A., et al. (2000). Artemis: sequence visualization and annotation. Bioinformatics 16, 944–945. doi: 10.1093/bioinformatics/16.10.944
Scheller, S., Goenrich, M., Boecher, R., Thauer, R. K., and Jaun, B. (2010). The key nickel enzyme of methanogenesis catalyses the anaerobic oxidation of methane. Nature 465, 606–608. doi: 10.1038/nature09015
Schlegel, K., Leone, V., Faraldo-Gómez, J. D., and Müller, V. (2012). Promiscuous archaeal ATP synthase concurrently coupled to Na+ and H+ translocation. Proc. Natl. Acad. Sci. U.S.A. 109, 947–952. doi: 10.1073/pnas.1115796109
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Shima, S., and Thauer, R. K. (2005). Methyl-coenzyme M reductase and the anaerobic oxidation of methane in methanotrophic Archaea. Curr. Opin. Microbiol. 8, 643–648. doi: 10.1016/j.mib.2005.10.002
Simon, J. (2002). Enzymology and bioenergetics of respiratory nitrite ammonification. FEMS Microbiol. Rev. 26, 285–309. doi: 10.1111/j.1574-6976.2002.tb00616.x
Simon, J., Gross, R., Einsle, O., Kroneck, P. M., Kroger, A., and Klimmek, O. (2000). A NapC/NirT-type cytochrome c (NrfH) is the mediator between the quinone pool and the cytochrome c nitrite reductase of Wolinella succinogenes. Mol. Microbiol. 35, 686–696. doi: 10.1046/j.1365-2958.2000.01742.x
Sirajuddin, S., and Rosenzweig, A. C. (2015). Enzymatic oxidation of methane. Biochemistry 54, 2283–2294. doi: 10.1021/acs.biochem.5b00198
Smith, J. A., Aklujkar, M., Risso, C., Leang, C., Giloteaux, L., and Holmes, D. E. (2015). Mechanisms involved in Fe(III) respiration by the hyperthermophilic archaeon Ferroglobus placidus. Appl. Environ. Microbiol. 81, 2735–2744. doi: 10.1128/AEM.04038-14
Stahl, D. A., and Amann, R. (1991). “Development and application of nucleic acid probes in bacterial systematics,” in Sequencing and Hybridization Techniques in Bacterial Systematics, eds E. Stackebrandt and M. Goodfellow (Chichester: John Wiley & Sons), 205–248.
Ten Brink, F., Schoepp-Cothenet, B., van Lis, R., Nitschke, W., and Baymann, F. (2013). Multiple Rieske/cytb complexes in a single organism. Biochim. Biophys. Acta 1827, 1392–1406. doi: 10.1016/j.bbabio.2013.03.003
Thauer, R. K. (2011). Anaerobic oxidation of methane with sulfate: on the reversibility of the reactions that are catalyzed by enzymes also involved in methanogenesis from CO2. Curr. Opin. Microbiol. 14, 292–299. doi: 10.1016/j.mib.2011.03.003
Thauer, R. K., Jungermann, K., and Decker, K. (1977). Energy conservation in chemotrophic anaerobic bacteria. Bacteriol. Rev. 41, 100–180.
Thauer, R. K., Kaster, A. K., Goenrich, M., Schick, M., Hiromoto, T., and Shima, S. (2010). Hydrogenases from methanogenic archaea, nickel, a novel cofactor, and H2 storage. Annu. Rev. Biochem. 79, 507–536. doi: 10.1146/annurev.biochem.030508.152103
Thauer, R. K., Kaster, A. K., Seedorf, H., Buckel, W., and Hedderich, R. (2008). Methanogenic archaea: ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 6, 579–591. doi: 10.1038/nrmicro1931
Thöny-Meyer, L. (1997). Biogenesis of respiratory cytochromes in bacteria. Microbiol. Mol. Biol. Rev. 61, 337–376.
Tietze, M., Beuchle, A., Lamla, I., Orth, N., Dehler, M., Greiner, G., et al. (2003). Redox potentials of methanophenazine and CoB-S-S-CoM, factors involved in electron transport in methanogenic archaea. Chembiochem 4, 333–335. doi: 10.1002/cbic.200390053
Tran, Q. H., and Unden, G. (1998). Changes in the proton potential and the cellular energetics of Escherichia coli during growth by aerobic and anaerobic respiration or by fermentation. Eur. J. Biochem. 251, 538–543. doi: 10.1046/j.1432-1327.1998.2510538.x
Tyson, K. L., Bell, A. I., Cole, J. A., and Busby, S. J. (1993). Definition of nitrite and nitrate response elements at the anaerobically inducible Escherichia coli nirB promoter: interactions between FNR and NarL. Mol. Microbiol. 7, 151–157. doi: 10.1111/j.1365-2958.1993.tb01106.x
Vergnes, A., Gouffi-Belhabich, K., Blasco, F., Giordano, G., and Magalon, A. (2004). Involvement of the molybdenum cofactor biosynthetic machinery in the maturation of the Escherichia coli nitrate reductase A. J. Biol. Chem. 279, 41398–41403. doi: 10.1074/jbc.M407087200
Walsh, C. (1986). Naturally occurring 5-deazaflavin coenzymes - biological redox roles. Acc. Chem. Res. 19, 216–221. doi: 10.1021/ar00127a004
Wang, F. P., Zhang, Y., Chen, Y., He, Y., Qi, J., Hinrichs, K. U., et al. (2014). Methanotrophic archaea possessing diverging methane-oxidizing and electron-transporting pathways. ISME J. 8, 1069–1078. doi: 10.1038/ismej.2013.212
Wang, H., and Gunsalus, R. P. (2000). The nrfA and nirB nitrite reductase operons in Escherichia coli are expressed differently in response to nitrate than to nitrite. J. Bacteriol. 182, 5813–5822. doi: 10.1128/JB.182.20.5813-5822.2000
Wang, H., Tseng, C. P., and Gunsalus, R. P. (1999). The napF and narG nitrate reductase operons in Escherichia coli are differentially expressed in response to submicromolar concentrations of nitrate but not nitrite. J. Bacteriol. 181, 5303–5308.
Welte, C., and Deppenmeier, U. (2011a). Membrane-bound electron transport in Methanosaeta thermophila. J. Bacteriol. 193, 2868–2870. doi: 10.1128/JB.00162-11
Welte, C., and Deppenmeier, U. (2011b). Re-evaluation of the function of the F420 dehydrogenase in electron transport of Methanosarcina mazei. FEBS J. 278, 1277–1287. doi: 10.1111/j.1742-4658.2011.08048.x
Welte, C., and Deppenmeier, U. (2014). Bioenergetics and anaerobic respiratory chains of aceticlastic methanogens. Biochim. Biophys. Acta 1837, 1130–1147. doi: 10.1016/j.bbabio.2013.12.002
Yoshimatsu, K., Araya, O., and Fujiwara, T. (2007). Haloarcula marismortui cytochrome b-561 is encoded by the narC gene in the dissimilatory nitrate reductase operon. Extremophiles 11, 41–47. doi: 10.1007/s00792-006-0016-3
Keywords: methanotrophy, anaerobic respiration, archaea, methanogenesis, ANME, cytochrome c, heterodisulfide reductase, electron transport
Citation: Arshad A, Speth DR, de Graaf RM, Op den Camp HJM, Jetten MSM and Welte CU (2015) A Metagenomics-Based Metabolic Model of Nitrate-Dependent Anaerobic Oxidation of Methane by Methanoperedens-Like Archaea. Front. Microbiol. 6:1423. doi: 10.3389/fmicb.2015.01423
Received: 13 October 2015; Accepted: 30 November 2015;
Published: 18 December 2015.
Edited by:
Roland Hatzenpichler, California Institute of Technology, USAReviewed by:
Shawn E. McGlynn, Tokyo Metropolitan University, JapanAnne-Kristin Kaster, Stanford University, USA
Grayson L. Chadwick, California Institute of Technology, USA
Copyright © 2015 Arshad, Speth, de Graaf, Op den Camp, Jetten and Welte. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cornelia U. Welte, c.welte@science.ru.nl
 Arslan Arshad
Arslan Arshad Daan R. Speth
Daan R. Speth Rob M. de Graaf
Rob M. de Graaf Huub J. M. Op den Camp
Huub J. M. Op den Camp Mike S. M. Jetten
Mike S. M. Jetten Cornelia U. Welte
Cornelia U. Welte