- 1Department of Microbiology and Immunology, Montana State University, Bozeman, MT, USA
- 2Energy Research Institute, Montana State University, Bozeman, MT, USA
- 3Department of Civil, Environmental and Architectural Engineering, University of Colorado, Boulder, CO, USA
- 4Plant Biology Division, The Samuel Roberts Noble Foundation, Ardmore, OK, USA
- 5Department of Chemistry and Biochemistry, Utah State University, Logan, UT, USA
- 6Center for Biofilm Engineering, Montana State University, Bozeman, MT, USA
- 7Department of Chemical and Biological Engineering, Montana State University, Bozeman, MT, USA
Algal biofuels and valuable co-products are being produced in both open and closed cultivation systems. Growing algae in open pond systems may be a more economical alternative, but this approach allows environmental microorganisms to colonize the pond and potentially infect or outcompete the algal “crop.” In this study, we monitored the microbial community of an outdoor, open raceway pond inoculated with a high lipid-producing alkaliphilic alga, Chlorella vulgaris BA050. The strain C. vulgaris BA050 was previously isolated from Soap Lake, Washington, a system characterized by a high pH (∼9.8). An outdoor raceway pond (200 L) was inoculated with C. vulgaris and monitored for 10 days and then the culture was transferred to a 2,000 L raceway pond and cultivated for an additional 6 days. Community DNA samples were collected over the 16-day period in conjunction with water chemistry analyses and cell counts. Universal primers for the SSU rRNA gene sequences for Eukarya, Bacteria, and Archaea were used for barcoded pyrosequence determination. The environmental parameters that most closely correlated with C. vulgaris abundance were pH and phosphate. Community analyses indicated that the pond system remained dominated by the Chlorella population (93% of eukaryotic sequences), but was also colonized by other microorganisms. Bacterial sequence diversity increased over time while archaeal sequence diversity declined over the same time period. Using SparCC co-occurrence network analysis, a positive correlation was observed between C. vulgaris and Pseudomonas sp. throughout the experiment, which may suggest a symbiotic relationship between the two organisms. The putative relationship coupled with high pH may have contributed to the success of C. vulgaris. The characterization of the microbial community dynamics of an alkaliphilic open pond system provides significant insight into open pond systems that could be used to control photoautotrophic biomass productivity in an open, non-sterile environment.
Introduction
The cultivation of algal biomass has many industrial applications ranging from health-care products to biodiesel (Fields et al., 2014). As the number of applications and subsequent demand for biomass increases, a challenge will be to exponentially increase production cost-effectively. The use of alkaliphilic photoautotrophs may help overcome some of the constraints associated with large-scale biomass production in open systems due to limited niche accessibility caused by higher pH values. Algal production of triacylglycerol (TAG) and other lipids are of substantial interest because of being biodiesel precursors that can be transesterified into fatty acid methyl esters (FAMEs; Sheehan et al., 1998; Dismukes et al., 2008; Scott et al., 2010). Many studies have screened algal species for biodiesel applications based on high lipid content (Sheehan et al., 1998; Dismukes et al., 2008; Griffiths and Harrison, 2009; Scott et al., 2010; Griffiths et al., 2011), and Chlorella vulgaris has been identified as one such species (Li et al., 2008; Griffiths et al., 2011; Duong et al., 2012).
Many studies on C. vulgaris and other species have been conducted in closed photobioreactors (PBRs) whose overall costs are considered a major constraint in the scale-up of algal biodiesel production (Smith et al., 2010; Kazamia et al., 2012a; Chisti, 2013; Shurin et al., 2013). However, in an effort to scale up production at lower capital investments, open pond systems have been shown to be a viable alternative (Smith et al., 2010). These open systems are, however, prone to colonization by environmental microbes spanning all three domains, and may contain hundreds of distinct taxa whose relative abundances vary by orders of magnitude (Fulbright et al., 2014). By employing parameters from “extreme” environments observed in natural systems, such as high pH, unwanted colonization (e.g., invasion) may be limited (Georgianna and Mayfield, 2012; Wang et al., 2013; Selvaratnam et al., 2014). In addition, alkaline systems favor higher dissolved inorganic carbon (DIC) from atmospheric CO2 thereby providing increased carbon delivery for primary producers. Moreover, some of the highest primary production rates have been reported for microbial, alkaline systems (Melack and Kilham, 1974; Hem, 1985).
Bacterial colonization has benefits and drawbacks for biomass production that are determined by the system’s community structure and composition. Numerous studies have documented positive, symbiotic relationships between algal taxa and bacteria (Croft et al., 2005; Watanabe et al., 2005; Sapp et al., 2007; Xie et al., 2013). Specifically, different species of Pseudomonas have been observed living in association with algae, including C. vulgaris (Sapp et al., 2007). The physical association, in which the bacteria live in the exopolymeric substances (EPS) or “phycosphere” of the alga, has been shown to increase C. vulgaris growth (Guo and Tong, 2013). The alga not only benefits from exchange of growth promoting and antibacterial metabolites in the niche space of the phycosphere, but also via the exclusion of potential opportunistic pathogens (Kazamia et al., 2012a,b; Smith and Crews, 2013). Additional work has shown symbiosis to be critical for adaptation to thermal stress resulting in higher algal biomass (Xie et al., 2013). These mutual positive relationships may have benefits for biotechnological applications offering the potential to artificially select microbial consortia that promote the growth of desired species (Kazamia et al., 2012a, 2014; Ortiz-Marquez et al., 2012; Natrah et al., 2013; Santos and Reis, 2014).
In the described study, we utilized pyrosequencing to monitor fluctuations in the community structure of an outdoor raceway pond inoculated with C. vulgaris during scale-up from 200 to 2,000 L. The results indicated that the inoculated algal population could maintain predominance under alkaline conditions, and that bacterial diversity increased while archaeal diversity decreased over time. In addition, particular populations could be correlated with C. vulgaris.
Materials and Methods
Site Description and Raceway Pond Conditions
Outdoor ponds were located in Logan, Utah (July 2011) approximately 40 km west of the northern arm of the Great Salt Lake (GSL). The 200 L oblong pond manufactured by Separations Engineering Inc. was lined with fiberglass and equipped with a paddle wheel promoting gas exchange with ambient air (Separatons Engineering Inc., San Diego, CA, USA). Initially, a 200 L raceway was inoculated with C. vulgaris (10% v/v), maintained at a culture depth of 13 cm, and monitored for 10 days. On day 10, the entire culture was transferred into an adjacent 2,000 L raceway and maintained at a culture depth of 20 cm for the remaining 6 days of the experiment. The 2,000 L raceways were constructed of cinder blocks stacked two high with 46 mil EPDM rubber pond liner creating the pond. Marine board was used to divide the pond into a circulating raceway with a paddlewheel providing circulation. The ponds were inoculated with Chlorella vulgaris BA050 that was previously isolated from Soap Lake, Washington, which is characterized by growth at high pH (∼9.8) (Dimitriu et al., 2008). The isolate was maintained on agar plates and was streak isolated during each subsequent plating every 2 months. The 18S rRNA gene sequences obtained from isolated DNA confirmed the presence of a single eukaryotic microorganism. A single 200 L raceway was inoculated with a 20 L culture (10% volume) that had been previously cultivated in shaker flasks bubbled with 1% CO2. A more saline version of Bold’s Basal Medium, consistent with the salinity of seawater (35 ppt), at pH 8.7 was prepared under non-sterile conditions with the addition of dry salts and concentrated solutions (Nichols and Bold, 1965). Inoculation resulted in a cell density of 3.6E+6 cells/mL with the addition of sufficient medium to bring the total volume to 200 L. The pH was not controlled in the pond. Unfiltered tap water was added each day to replace measured evaporative loss. After 10 days, repeating methods from the initial inoculation, the 2,000 L pond was inoculated by transferring 200 L (10%) of culture from the first pond (200 L).
Sample Collection
Samples were collected twice daily for cell density ascertained by OD750 and direct cell counts via optical microscopy (cells/mL). Additionally, 500 mL samples were collected and frozen at -80°C for DNA extraction and 454 sequencing.
DNA Extraction and Sequencing
DNA Extraction
Samples were slowly thawed at 4°C and microbial biomass was collected via filtration through 0.22 μm polyethersulfone membrane filters. The solids were then suspended in the MO BIO PowerMaxTM Soil DNA Isolation Kit PowerBead Solution, and the cells were lysed via three cycles of liquid nitrogen freeze–thaw and ground with a mortar and pestle aided by sterile sand (Zhou et al., 1996) (MO BIO Laboratories Inc., Carlsbad, CA, USA). The DNA was cleaned and concentrated with the Wizard® SV Gel and PCR Clean-Up System (Promega Corporation, Madison, WI, USA) according to the manufacturer’s protocol.
Bar-Coded Pyrosequencing
Pyrosequencing was utilized to characterize the microbial population of the ponds. PCR was used to increase the DNA concentration needed for pyrosequencing analysis. Each sample was labeled with a unique 10 nucleotide-barcode for multiplexing. The SSU rRNA gene sequences for Eukarya and Bacteria were amplified via 25 cycles of PCR with the following barcoded primers; 7F (5′-ACCTGGTTGATCCTGCCAG-3′) and 591R (5′-GGAGCTGGAATTACCG-3′) for Eukarya and FD1 (5′-AGAGTTTGATCCTGGCTCAG-3′) and 529R (5′-CGCGGCTGCTGGCAC-3′), which targeted the V1–V3 region of Bacteria (Bowen De León et al., 2012). Archaeal sequences were amplified separately from Bacteria using a nested approach with non-barcoded 21F (5′-TTCYGGTTGATCCYGCCRGA-3′) and 1492R (5′-CGGTT ACCTTGTTACGACTT-3′) for 20 cycles followed by an additional 20 cycles with barcoded 751F (5′-CCGACGGTGAGRGRYGAA-3′) and 1204R (5′-TTMGGGGCATRCNKACCT-3′) (Baker et al., 2003; Barnhart et al., 2013). PCR products of the correct size were confirmed using a 1% agarose gel. Products were cut from the gel and pooled using an Ultrafree-DNA gel extraction column (Millipore Corporation, Bedford, MA, USA). The gel extract was cleaned and concentrated using the Wizard® SV Gel and PCR Clean-Up System, and dsDNA was quantified with a Qubit fluorometer (Invitrogen, Carlsbad, CA, USA). Adaptors for 454 sequencing were ligated to the amplicons and were pyrosequenced on a 454 GS-Junior (454 Life Sciences, Branford, CT, USA). Roche’s image analysis separated sequences by barcode. Sequences were trimmed to one standard deviation below the mean length or removed if shorter. Employing the Phred score filter, 15% of the nucleotides were allowed to be below Q27, and removed if primer errors or Ns were observed.
Bioinformatic Sequence and Community Analysis
Data analysis was performed using the Quantitative Insights into Microbial Ecology (QIIME) software package, version 1.4.0 (Caporaso et al., 2010b). Parameter settings for demultiplexing were at a default level of 200 and 1000 bp in length. Metadata files were prepared according to a QIIME compatible template taking into account environmental sampling data on pH, temperature, and ionic concentrations. Libraries were split according to barcode for each of the respective domains (Archaea, Bacteria, and Eukarya). Sequences were then concatenated for data normalization needed in downstream analysis. Operational taxonomic units (OTUs) were assigned using the closed reference OTU picking protocol. Clusters were referenced against the Silva 108 database and pre-clustered at 97% identity using UCLUST (Edgar, 2010).
Sequence reads that matched a Silva reference sequence at 97% identity were clustered within an OTU defined by a reference sequence. OTU assignment (and all subsequent steps) was performed for the combined Archaea, Bacteria and Eukarya reads. The singleton OTUs were discarded. The centroid sequence in every cluster was selected to represent the cluster and aligned with the Silva core set using PyNAST (Caporaso et al., 2010a). Chimeric sequences, identified with Chimera Slayer (Haas et al., 2011) and reads that failed to align with PyNAST were excluded from subsequent analyses. PyNAST (v1.1) was used for sequence alignment and filtering through QIIME using default parameters.
Taxonomic assignments were additionally assigned using the retrained RDP Classifier (Wang et al., 2007) on the Silva 108 database for phylogenetic resolution at the genus level. Taxonomic summary for Archaea, Bacteria, and Eukarya was plotted to the genus level with a given relative abundance based on diversity and distribution pattern per domain. Taxa distribution was also summarized by time (sample day). The biodiversity analysis downstream between samples was derived using UniFrac (Lozupone and Knight, 2005) that took into account the phylogenetic structure of the algal pond microbial communities. Taxonomic richness was calculated by a rarefaction analysis based upon OTU tables that were rarefied at an even sampling threshold value. Richness was measured on the basis of the Chao index (Chao et al., 2010).
The co-occurrence of community members was illustrated in a heat-map using the R vegan package version 2.0-10 (Oksanen, 2011). Due to the fact that some taxa had a 0% relative abundance at certain time points, 0.1 was added to all values in order to be log transformed. In an effort to enhance the visual distribution of taxa, log transformed values were cubed and resulting values plotted (log(relative abundance + 0.1)3). A SparCC analysis was subsequently used to construct community correlation networks by estimating linear correlation values between log transformed abundances based on the absolute number of sequences for an OTU rather than a relative abundance (Friedman and Alm, 2012; Berry and Widder, 2014). The key advantage of this analysis was that, for instance, the ratio of the fractions of two OTUs was independent of the fluctuations in other OTUs included in the analysis (i.e., subcompositional coherence) (Friedman and Alm, 2012). Archaeal taxa were not included in this analysis due to the sharp decline in relative abundance between days 1 and 3 and near absence by the end of the pond experiment. We observed that the drastic decline in Archaeal relative abundance would have produced deceptive relationships in the network model.
Statistical Analysis
A principal coordinate analysis (PCA) was used to reduce dimensionality and give structure to the water chemistry variables obtained from each time point (Legendre and Gallagher, 2001). In order to incorporate taxonomic data, we used the direct-gradient ordination technique, Canonical Correspondence Analysis (CCA), which concurrently showed pond taxa, time points, and water chemistry (Hall and Smol, 1992). This kind of ordination is appropriate when assessing community dynamics because it does not use Euclidean based metrics that assume linear trends in community change. Not only does the ordination show environmental factors influencing community change, but results suggest potential interactions between taxa (Amaral-Zettler et al., 2010). The first two axes, CCA1 and CCA2, typically account for the majority of observed variation. All axes are constrained to present a linear combination of the water chemistry that maximizes the dispersion of taxa (Hall and Smol, 1992).
Results and Discussion
Environmental Variables
Water chemistry was monitored daily over the course of the experiment and fluctuations were used to draw correlations to community structure (Table 1).
The nitrate concentration was lowest on day 8 as C. vulgaris achieved stationary phase in the 200 L raceway. Changes in fluoride and chloride anion concentrations at this time may have been due to the use of tap water to compensate for evaporative loss. Nitrogen concentrations recover on day 10 when the culture is transferred to the 2,000 L raceway and combined with new media. The pH values remained high over time and may have benefited C. vulgaris (R2 = 0.2). The decline in pH from 10.7 to 8.39 on day 11 corresponded with the transfer of the pond from the 200 to 2,000 L raceway.
Community Composition and Interaction
The SSU rRNA for Bacteria, Archaea, and Eukarya were amplified and sequenced for each of the six sample days. After screening sequences for errors (see Materials and Methods), 161,731 quality gene sequences with a valid barcode were retrieved. Sequences with a 97% identity were clustered within an OTU totaling 1,349 observed OTUs composed of 748, 249, and 352 OTUs of Bacteria, Archaea, and Eukarya respectively for all sampled days (Table 2).
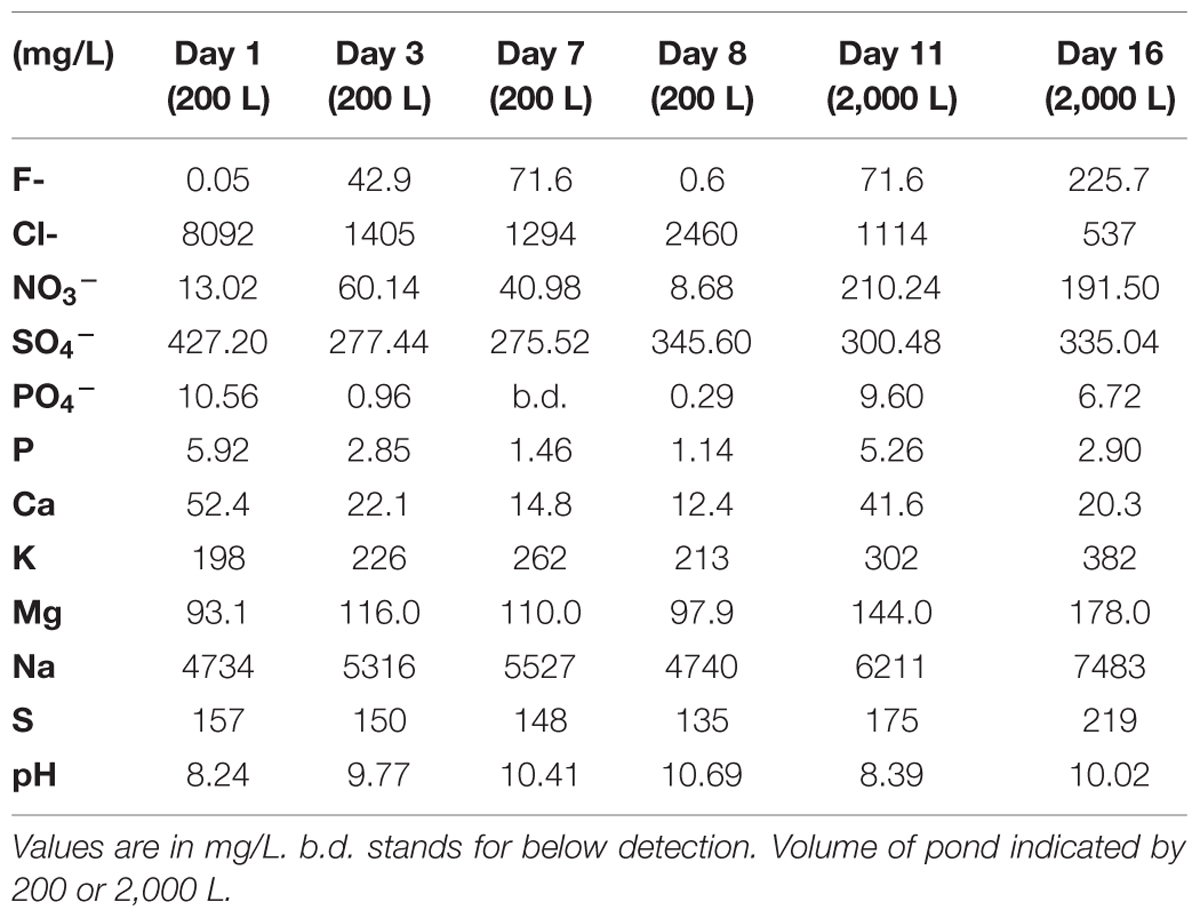
Figure 1 shows the relative abundances for the observed archaeal and bacterial taxa.
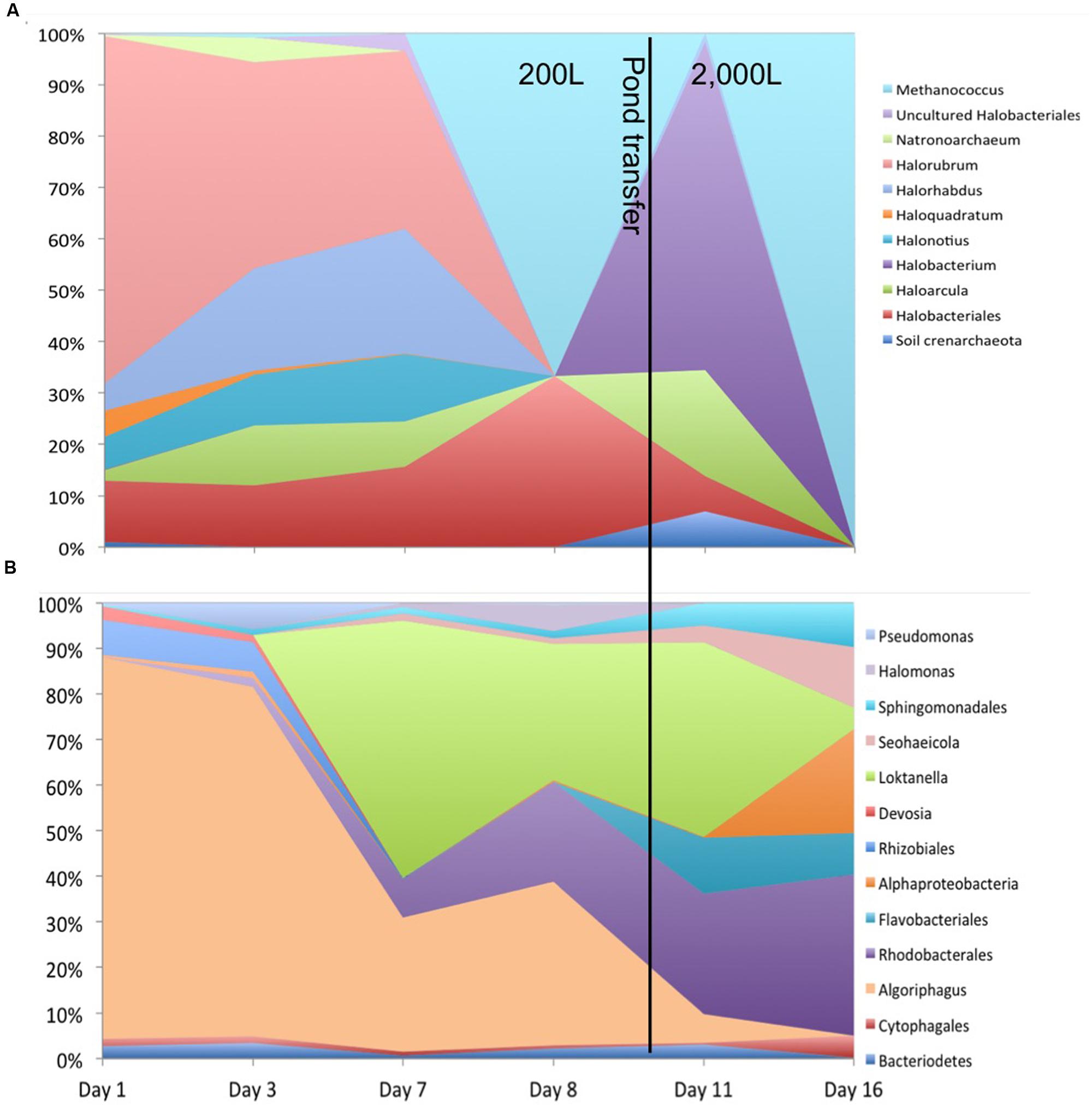
FIGURE 1. Total relative abundances of (A) archaeal and (B) bacterial taxa by order (as order, genus, or unknown) with >2% relative abundance over the 16 day sample period.
Figure 2 shows phosphate limitation in the plateau of C. vulgaris observed from day 3–8, emulating nutrient conditions frequently observed in wastewater and some natural systems. Nitrogen availability has more than doubled due to anthropogenic inputs facilitated by the Haber–Bosch process entering aquatic systems through precipitation, runoff, and dust deposition (Baron et al., 2000; Wolfe et al., 2001; Fenn et al., 2003; Gardner et al., 2008; Miller and McKnight, 2012; Brahney et al., 2015). Thus, phytoplankton is typically phosphorus limited giving a competitive advantage to taxa that can quickly scavenge phosphorus (Baron et al., 2000; Wolfe et al., 2001; Elser et al., 2009). In addition, some C. vulgaris strains have been shown to accumulate polyphosphate (Aitchison and Butt, 1973). Considering that nitrogen inputs are nearly unavoidable in the majority of open systems, most communities will be phosphorus limited, the effects of which are pertinent to endeavors such as commercial algae production in open systems. Moreover, photoautotrophs that have the ability to accumulate and/or scavenge phosphorus will be more competitive in open, mixed systems in which low phosphate levels can be used. Combined, these attributes reduce the need for higher levels of phosphorus and/or the use of low-quality phosphorus.
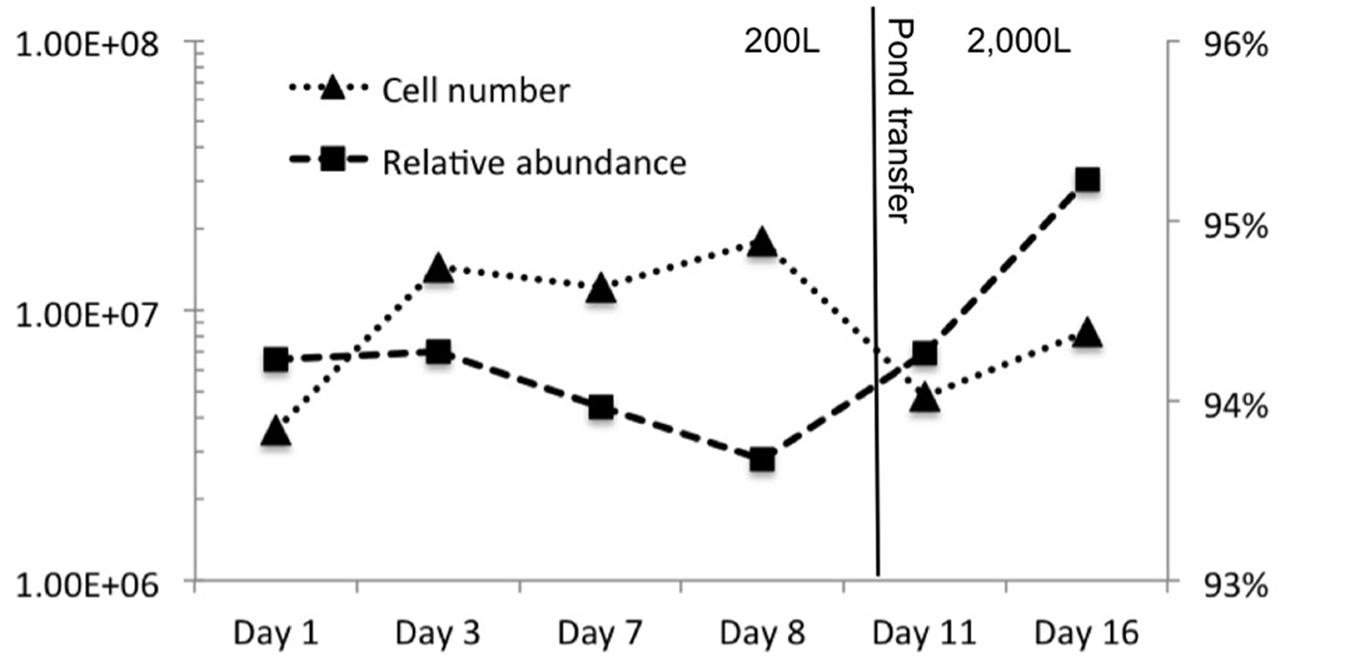
FIGURE 2. Cell concentration (log cells/mL) and the relative abundance of C. vulgaris over 16-day sample period. The transfer from the 200 to 2,000 L raceway is indicated by the vertical line.
Effects of phosphorus limitation on C. vulgaris did not appear to detrimentally affect ability to compete and grow under the tested conditions as it was observed to be the single dominant Eukaryote during the 16-day experiment. Figure 2 shows that C. vulgaris recovered from phosphorus limitation upon transfer to the 2,000 L pond, and throughout the pond experiment it was the dominant eukaryotic taxon and did not decline below 93% of eukaryotic relative abundance. The other 7% was composed of pine pollen, insects, and fungi. While it is difficult to ascertain physiology from phylogeny, sequences indicative of Psuedomonas were inversely correlated to high phosphate levels (Istvánovics, 2008). Organisms from this genus can be phosphate-accumulating and/or grow under low-phosphate conditions (Sidat et al., 1999). It is likely that phosphate-accumulating bacterial populations would be selected as overall P levels are depleted. As expectations for inexpensive biomass and feedstock become greater, we will need improved insight into biological responses to low-level and low-quality phosphorus.
Other organisms propagated in the pond but did not appear to have a detrimental effect on the alga population. Several halophilic archaeal taxa were observed in the first three time points, especially day 1 leading to the greatest observed diversity that declined over time (Figure 3). In addition to the Archaea, several bacteria were present in the first sample. The high archaeal and bacterial diversity did not appear to have a negative effect on C. vulgaris, which was the predominant eukaryote, and minimal fluctuation was observed in the eukaryotic diversity. As the Archaeal taxa diversity declined, the bacterial diversity increased (Figure 3). It is not known if the decline in archaeal populations was related to the increase in bacterial populations or an independent process, such as lower salinity.
FIGURE 3. Chao diversity for each domain plotted over time. Archaea started with high diversity but quickly declined. Eukarya maintained a steady diversity level composed almost entirely of C. vulgaris. Bacterial diversity steadily increased with time.
The transitory presence of halophilic taxa could have been due to suboptimal conditions in the ponds. These halophilic microorganisms have an optimal pH range from 6.8 to 9, NaCl concentration between 3.4 and 5.1 M, and generally require at least 0.85 to 3.4 M in order to maintain osmotic pressure for cell integrity and prevent lysis (DasSarma and DasSarma, 2006; Oren, 2006; Bowers and Wiegel, 2011; Cui et al., 2012). Their presence in the ponds could potentially be due to the proximity of the pond to the GSL that is 40 km to the west and characterized by high salinity and a neutral pH (Post, 1977; Oren, 1994, 2006; Baxter et al., 2005; Tazi et al., 2014). Wind dispersal and precipitation events may have been responsible for the presence of these populations in the pond. We observed numerous haloarchaeal genera including Halorubellus, Haloquadrata, Halalkalicoccus, Candidatus, Halomonas, Halobacterium, and Haloarcula and the halobacterial genera Devosia, Aliihoeflea, Halomonas sp., Seohicola, Eruthromicrobium, Aquiflexum, and Rhodobacterales which have also been found in GSL samples (Post, 1977; Oren, 1994, 2006; Baxter et al., 2005; Tazi et al., 2014). However, another possibility is that these taxa were already present in the salts used to make the medium. The vast majority of these taxa subsisted for only the first two time points (Figure 3). It is unknown if the detected sequences were indicative of populations that survived for a given time in the test pond or simply were static and/or dead cells that were transported to the ponds.
In contrast to the halophilic microorganisms, C. vulgaris has been shown to be inhibited by concentrations greater than 1 M NaCl and showed substantial declines in cell concentration at 0.5 M (Alyabyev et al., 2007). The highest recorded salinity in the ponds was 0.7 M with an average 0.2 M. Thus, the success of C. vulgaris in the ponds further supports that salinity was below the presumptive optima of the halophiles. The changes in archaeal and bacterial community structure did not appear detrimental to C. vulgaris and a high relative abundance (>93%) was observed throughout the course of the pond experiment. A decrease in cell number was observed immediately after pond transfer and was likely due to dilution of cells during the transfer of the 200 L inoculum to the 2,000 L pond; however the relative abundance of C. vulgaris remained consistent (94–95% relative abundance).
Correlation of Environmental Variables and Community Structure
Environmental factors could have also played a role in the success of C. vulgaris. We observed high pH values which ranged between 8.2 and 10.7 (Table 1), which may have prevented other algal taxa from successfully colonizing the pond. This finding demonstrates that the cultivation of a single algal strain in an open alkaline pond without the addition of antibiotics or herbicides can be successful (Lundquist et al., 2010; Smith et al., 2010; Smith and Crews, 2013; McBride et al., 2014).
Using CCA, the most significant water chemistry variables correlated with fluctuations in archaeal taxa were plotted in Figure 4. Taxa and time points were correlated to pH, nitrate, and phosphate (vectors in Figure 4), and the temporal variation in archaeal taxa was observed as the pH increased on day 8 and 16 most likely as a consequence of photosynthesis. Not only does CCA provide insight into the environmental factors influencing the community structure, but it also suggests potential interactions occurring between taxa. The appearance and subsequent decline of plotted taxa was the most influential variable on discrepancies between the time points. For instance, day 8 is more similar to day 16 than other time points due to the loss of three archaeal taxa from day 7 to 8. As discussed, the low levels of salinity were an influential factor associated to the rapid decline of halophilic taxa.
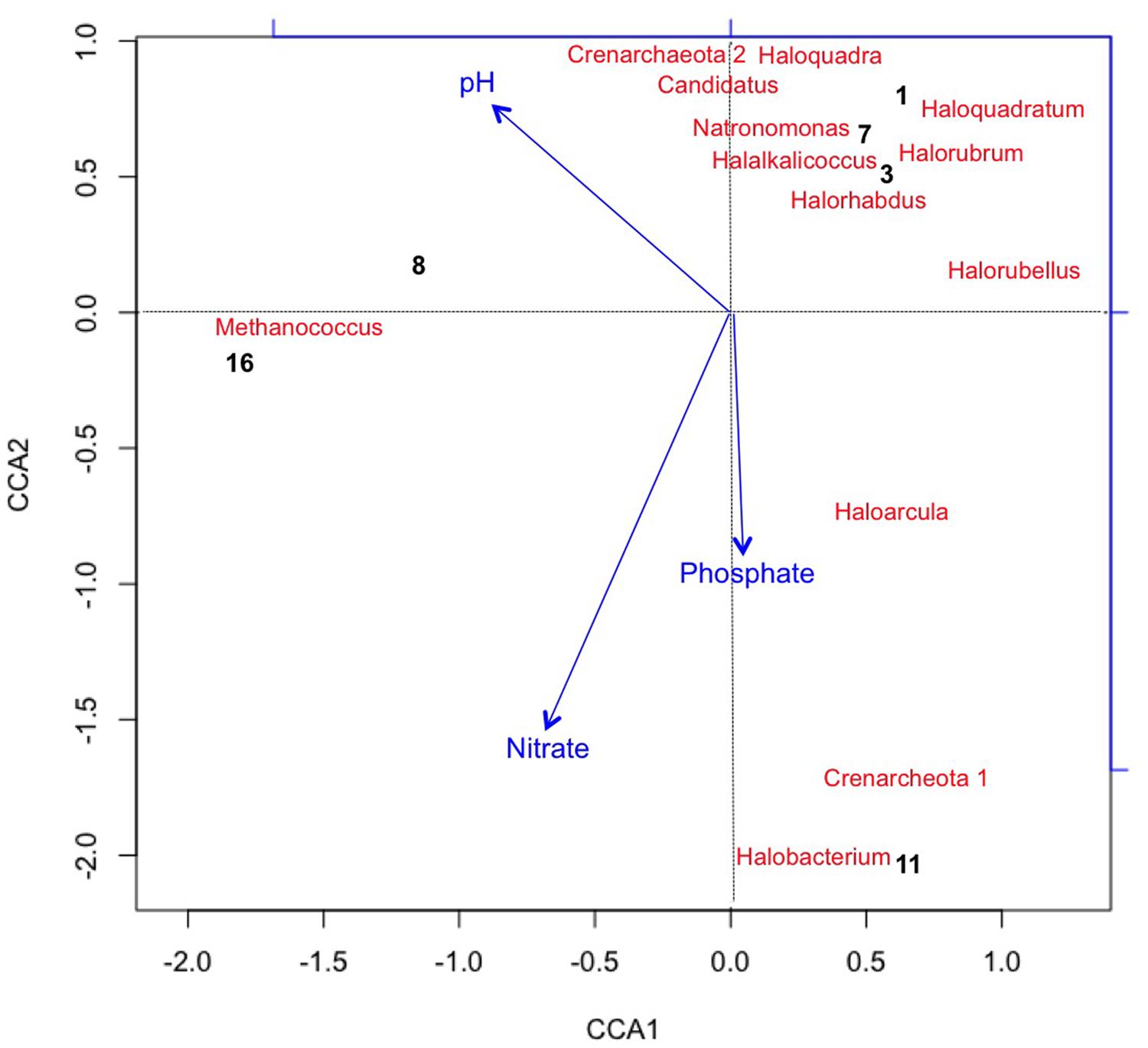
FIGURE 4. Canonical Correspondence Analysis (CCA) plotting chemical variables that correlated with variation in Archaeal taxa (with >2% relative abundance) and sample points. Archaea generally lacked long term survivability in the pond. CCA1 = 81.8% and CCA2 = 57.3%.
Figure 4 also shows that Methanococcus maripaludis was most associated with the variation in CCA1 as it was the only detectable archaeal taxa remaining at the last time point on day 16. M. maripauldis was detected on days 1, 3, 8, and 16, but not detected on days 7 and 11. While known Methanococci are strict anaerobes, there could be micro-anaerobic niches in the raceway pond related to biomass turnover. It is also possible that the sequences are detected at later time points due to PCR biases. Recent research has shown M. maripauldis can survive in anaerobic biofilms (Brileya et al., 2013); and therefore, it is possible that a small population was able to survive within a biofilm matrix on the walls of the raceway or paddle wheel. Its sporadic appearance may also be the result of collection methods that could have disturbed the biofilm or biofilm detachment. CCA2 was most influenced by nitrate and phosphate, which explained 38.4% of the variation in archaeal taxa.
We also applied the same CCA metrics for the bacterial taxa to visualize variance in taxa and time points as correlating with changes in chemical variables (Figure 5). CCA1 was predominately influenced by increased nitrate concentrations accounting for about 57.2% of the variance in bacterial taxa distribution. The first two time points were correlated with the initial halophilic bacterial taxa that were unable to maintain a population due to low salinity, aerobic conditions, and/or increasing pH (Post, 1977; Oren, 1994, 2006; Baxter et al., 2005; Tazi et al., 2014). The decrease in phosphate contributed approximately 30.5% of the observed variation in CCA2 and correlated with the increase in some of the most abundant taxa at day 16 [for example Flavobacteriales (R2 = 0.3) and Proteobacteria (R2 = 0.35)].
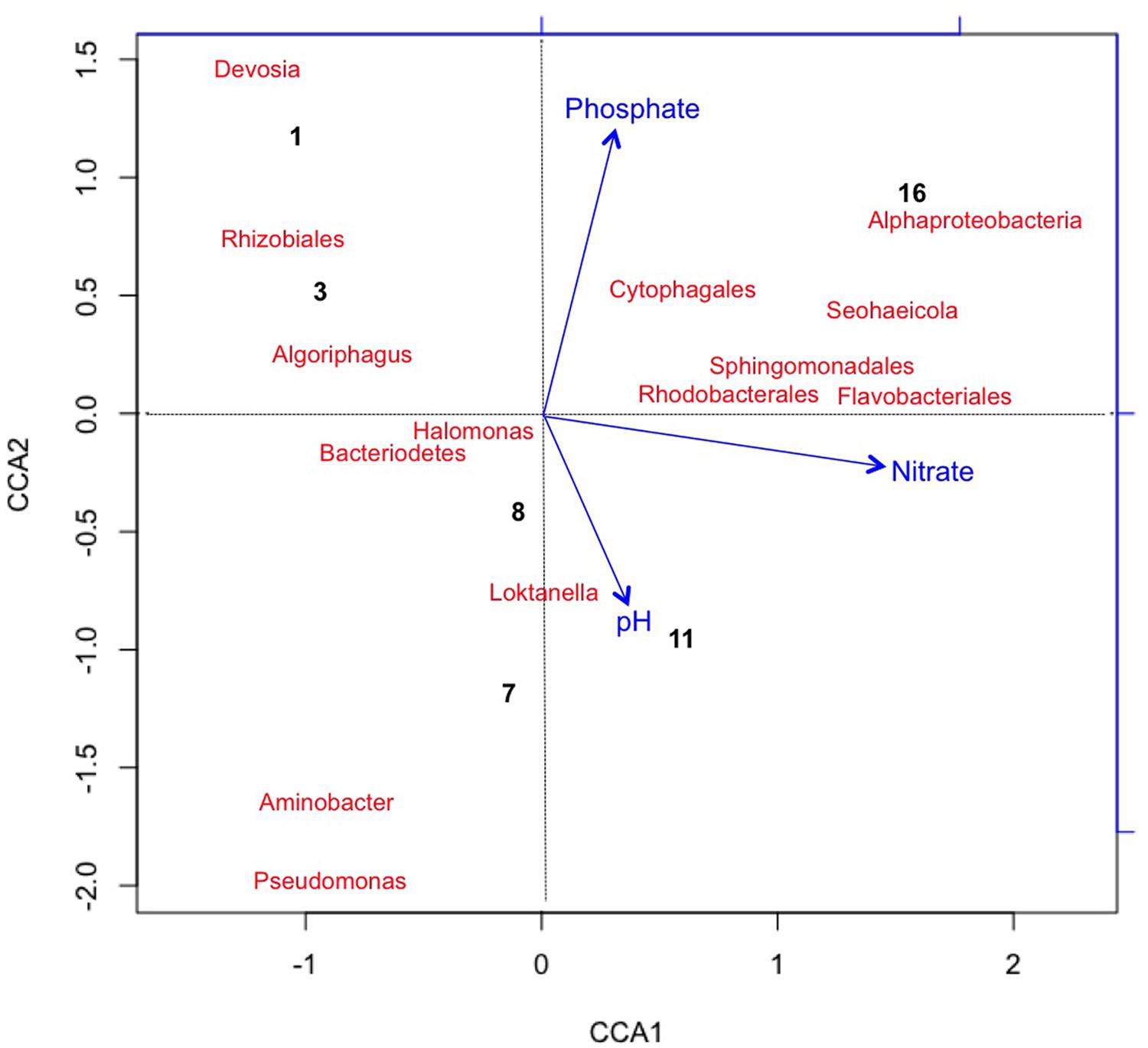
FIGURE 5. Canonical Correspondence Analysis plotting chemical variables that correlated with variation in Bacterial taxa (with >2% relative abundance) and sample points. Bacteria generally trended with an increase nitrate toward end of the experiment, possibly due to an increase in nitrifying taxa. Pseudomonas sp. was associated with the early time points (day 1 and 3). CCA1 = 60% and CCA2 = 8%.
Correlations Between Community Members
The distribution of community member occurrence is shown in Figure 6, illustrating the persistent C. vulgaris population. The upper dendogram clusters taxa by percent relative abundance and frequency of co-occurrence. As observed via CCA, the time points are grouped by declining archaeal sequences with the exception of Methanococcus at day 8 and 16. Post-transfer to 2,000 L, several bacterial OTUs clustered with day 11 and 16. Flavobacterium and Erythromicrobium are common groundwater/tap water organisms that were likely introduced during volume scale-up, but it is not known if the co-occurrence is direct or indirect. In addition, sequences indicative of Loktanella and Roseicyclus correlated with Chlorella on day 8, 11, and 16 during cultivation scale-up. Sequences indicative of Algoriphagus correlated with Chlorella during the 200 L cultivation but declined during the 2,000 L cultivation.
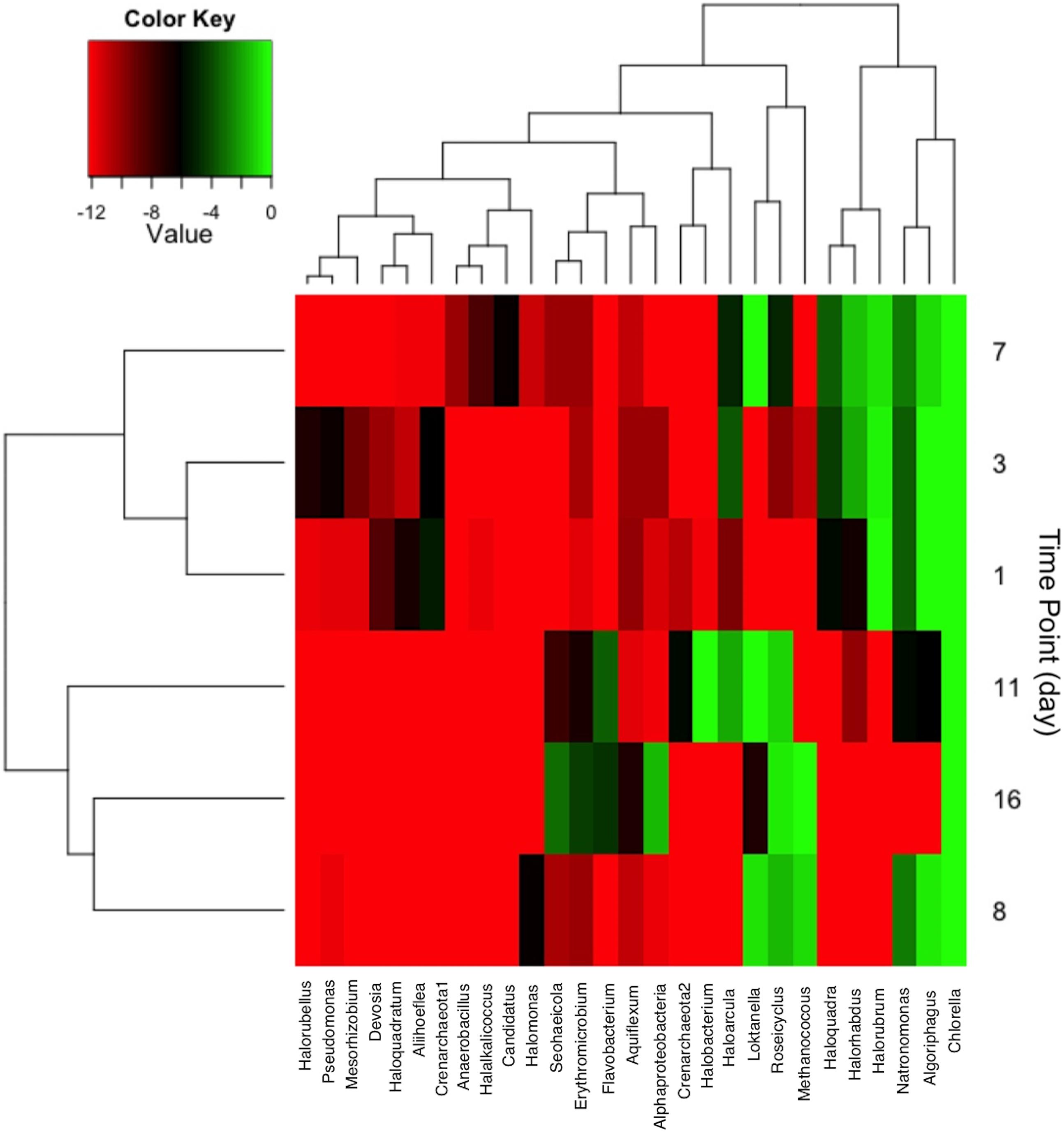
FIGURE 6. Heat map showing taxa with greater than 2% relative abundance at each sampling time point. Taxa co-occurrence and each time points were correlated and clustered according to relatedness as shown in the dendogram.
However, co-occurrence does not infer a statistical correlation between taxa. In order to investigate possible correlations we used SparCC to construct community correlation networks. Figure 7 shows a community network map of correlations between community members (excluding archaea due to their general absence after day 3) (Friedman and Alm, 2012). The most salient of these relationships is the positive 0.85 correlation between C. vulgaris and Pseudomonas sp. (p < 0.05). No other bacteria correlated with C. vulgaris, which was the predominant eukaryote. Previous studies have observed different species of Pseudomonas living in association with algae including C. vulgaris (Sapp et al., 2007). A symbiotic relationship between these organisms was described by Guo and Tong (2013) finding that Pseudomonas sp. fostered the growth of C. vulgaris. When in co-culture with Pseudomonas sp., the cell concentration of C. vulgaris was 1.4 times greater than that of axenic cultures under the same conditions. Scanning electron microscope (SEM) images revealed that the bacteria were living in the EPS or “phycosphere” of C. vulgaris (Guo and Tong, 2013).
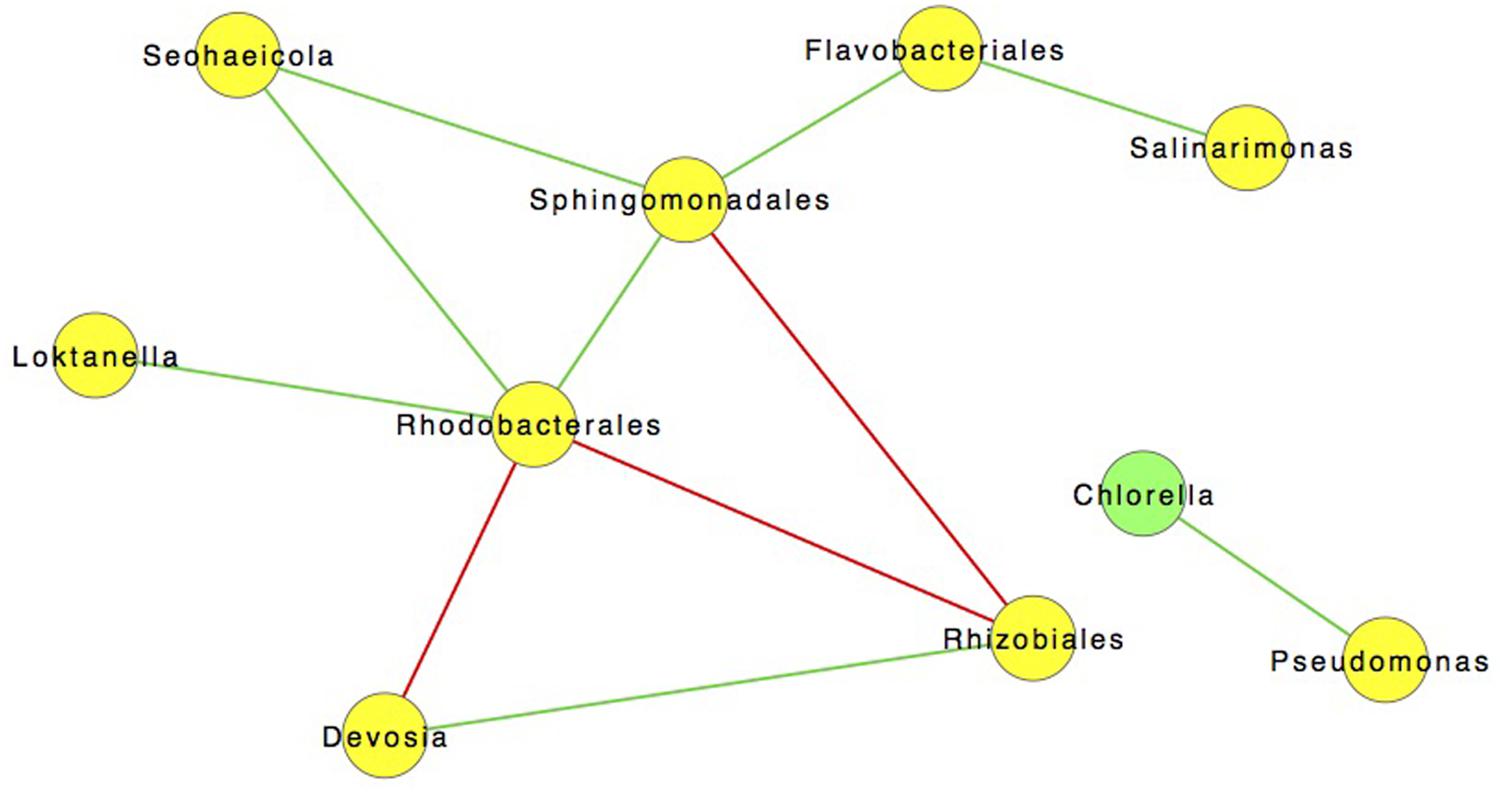
FIGURE 7. SparCC network map showing significant (p < 0.05) of 122 Interactions with a 0.85 correlation between different OTUs incorporating all time points. Green lines are indicative of positive interactions while red lines are negative.
The phycosphere, coined by Bell and Mitchell in 1972, is often colonized by bacteria (Bell and Mitchell, 1972; Sapp et al., 2007; Goecke et al., 2013). This specific niche facilitates a tight exchange of oxygen, carbon, and metabolites minimizing dilution (Sapp et al., 2008; Bruckner et al., 2011; Gärdes et al., 2012; Paul et al., 2012; Martin et al., 2014). Bacteria can provide the alga with sources of growth promoters (e.g., indole-3-acetic acid), and essential vitamins (e.g., cobalamin), while discouraging colonization by other potentially harmful microorganisms with antimicrobial metabolites (Gonzalez and Bashan, 2000; Croft et al., 2005, 2006). In return, bacteria have immediate access to algal exudates that can be a key source of fixed carbon (Bell and Mitchell, 1972; Sapp et al., 2007; Goecke et al., 2013). The correlation of Pseudomonas populations with C. vulgaris throughout the course of the pond experiment, even following the transfer to the larger raceway, may have contributed to the predominance of the algal culture under open conditions (Figure 7). The results suggest that symbiotic-associations could have relevant industrial applications that could result in increased biomass yields (Imase et al., 2008; Natrah et al., 2013). Further work is needed to discern the mechanism(s) of interactions that impact algal biomass and/or lipid accumulation in addition to confirmation of a direct and/or indirect relationship between these two organisms under the tested growth conditions.
Conclusion
Our work demonstrated that the cultivation of a single algal strain in an open pond without the addition of antibiotics or herbicides can be successful. The use of high pH systems and alkaline adapted algal taxa could be a successful strategy for overcoming some of the constraints associated with large-scale biomass production in open systems. Furthermore, certain phycosphere associations could enhance biomass yields and deter colonization by detrimental populations. Further work is needed to determine the longevity and stability of open, outdoor cultivation systems for the production of algal biomass and/or biomolecules.
Funding
This work was financially supported by the NSF IGERT Program in Geobiological Systems (DGE 0654336) in addition to the US Department of Energy-Advancements in Sustainable Algal Production (ASAP) program under contract DE-EE0005993, the National Science Foundation Sustainable Energy Pathways (SEP) Program under NSF CHE-1230632, and the US Department of Energy Bioenergy Technologies Office grant DE-FG36-08GO18161.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank the members of the Center for Biofilm Engineering, especially Ann Willis, for their support. We would also like to acknowledge Dr. Kara Bowen DeLeón for her guidance and all members of the MSU Algal Biofuels Group for their helpful discussions.
References
Aitchison, P. A., and Butt, V. S. (1973). The relation between the synthesis of inorganic polyphosphate and phosphate uptake by Chlorella vulgaris. J. Exp. Bot. 24, 497–510. doi: 10.1093/jxb/24.3.497
Alyabyev, A. J., Loseva, N. L., Gordon, L. K., Andreyeva, I. N., Rachimova, G. G., Tribunskih, V. I., et al. (2007). The effect of changes in salinity on the energy yielding processes of Chlorella vulgaris and Dunaliella maritima cells. Thermochim. Acta 458, 65–70. doi: 10.1016/j.tca.2007.03.003
Amaral-Zettler, L. A., Zettler, E. R., Theroux, S. M., Palacios, C., Aguilera, A., and Amils, R. (2010). Microbial community structure across the tree of life in the extreme Río Tinto. ISME J. 5, 42–50. doi: 10.1038/ismej.2010.101
Baker, G. C., Smith, J. J., and Cowan, D. A. (2003). Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 55, 541–555. doi: 10.1016/j.mimet.2003.08.009
Barnhart, E. P., De León, K. B., Ramsay, B. D., Cunningham, A. B., and Fields, M. W. (2013). Investigation of coal-associated bacterial and archaeal populations from a diffusive microbial sampler (DMS). Int. J. Coal Geol. 115, 64–70. doi: 10.1016/j.coal.2013.03.006
Baron, J. S., Rueth, H. M., Wolfe, A. M., Nydick, K. R., Allstott, E. J., Minear, J. T., et al. (2000). Ecosystem responses to nitrogen deposition in the colorado front range. Ecosystems 3, 352–368. doi: 10.1007/s100210000032
Baxter, B. K., Litchfield, C. D., Sowers, K., Griffith, J. D., DasSarma, P. A., and DasSarma, S. (2005). “Microbial diversity of Great Salt Lake,” in Adaptation to Life at High Salt Concentrations in Archaea, Bacteria, and Eukarya, Vol. 9 eds N. Gunde-Cimerman, A. Oren, A. Plemenitaš, (Dordrecht: Springer Science+Business Media), 9–25.
Bell, W., and Mitchell, R. (1972). Chemotactic and growth responses of marine bacteria to algal extracellular products. Biol. Bull. 143, 265–277. doi: 10.2307/1540052
Berry, D., and Widder, S. (2014). Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Front. Microbiol. 5:219. doi: 10.3389/fmicb.2014.00219
Bowen De León, K., Ramsay, B. D., and Fields, M. W. (2012). Quality-score refinement of SSU rRNA gene pyrosequencing differs across gene region for environmental samples. Microb. Ecol. 64, 499–508. doi: 10.1007/s00248-012-0043-9
Bowers, K. J., and Wiegel, J. (2011). Temperature and pH optima of extremely halophilic archaea: a mini-review. Extremophiles 15, 119–128. doi: 10.1007/s00792-010-0347-y
Brahney, J., Ballantyne, A. P., and Kociolek, P. (2015). Ecological changes in two contrasting lakes associated with human activity and dust transport in western Wyoming. Limnol. Oceanogr. 60, 678–695. doi: 10.1002/lno.10050
Brileya, K. A., Connolly, J. M., Downey, C., Gerlach, R., and Fields, M. W. (2013). Taxis toward hydrogen gas by Methanococcus maripaludis. Sci. Rep. 3, 3140. doi: 10.1038/srep03140
Bruckner, C. G., Rehm, C., Grossart, H. P., and Kroth, P. G. (2011). Growth and release of extracellular organic compounds by benthic diatoms depend on interactions with bacteria. Environ. Microbiol. 13, 1052–1063. doi: 10.1111/j.1462-2920.2010.02411.x
Caporaso, J. G., Bittinger, K., Bushman, F. D., DeSantis, T. Z., Andersen, G. L., and Knight, R. (2010a). PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26, 266–267. doi: 10.1093/bioinformatics/btp636
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010b). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Chao, A., Chiu, C. H., and Jost, L. (2010). Phylogenetic diversity measures based on Hill numbers. Philos. Trans. R. Soc. B Biol. Sci. 365, 3599–3609. doi: 10.1098/rstb.2010.0272
Chisti, Y. (2013). Constraints to commercialization of algal fuels. J. Biotechnol. 167, 201–214. doi: 10.1016/j.jbiotec.2013.07.020
Croft, M. T., Lawrence, A. D., Raux-Deery, E., Warren, M. J., and Smith, A. G. (2005). Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature 438, 90–93. doi: 10.1038/nature04056
Croft, M. T., Warren, M. J., and Smith, A. G. (2006). Algae need their vitamins. Eukaryot. Cell 5, 1175–1183. doi: 10.1128/EC.00097-06
Cui, H. L., Mou, Y. Z., Yang, X., Zhou, Y. G., Liu, H. C., and Zhou, P. J. (2012). Halorubellus salinus gen. nov., sp. nov. and Halorubellus litoreus sp. nov., novel halophilic archaea isolated from a marine solar saltern. Syst. Appl. Microbiol. 35, 30–34. doi: 10.1016/j.syapm.2011.08.001
Dimitriu, P. A., Pinkart, H. C., Peyton, B. M., and Mormile, M. R. (2008). Spatial and temporal patterns in the microbial diversity of a meromictic soda lake in Washington state. Appl. Environ. Microbiol. 74, 4877–4888. doi: 10.1128/AEM.00455-08
Dismukes, G. C., Carrieri, D., Bennette, N., Ananyev, G. M., and Posewitz, M. C. (2008). Aquatic phototrophs: efficient alternatives to land-based crops for biofuels. Curr. Opin. Biotechnol. 19, 235–240. doi: 10.1016/j.copbio.2008.05.007
Duong, V. T., Li, Y., Nowak, E., and Schenk, P. M. (2012). Microalgae isolation and selection for prospective biodiesel production. Energies 5, 1835–1849. doi: 10.3390/en5061835
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Elser, J. J., Andersen, T., Baron, J. S., Bergström, A.-K., Jansson, M., and Kyle, M. (2009). Shifts in lake N: P stoichiometry and nutrient limitation driven by atmospheric nitrogen deposition. Science 326, 835–837. doi: 10.1126/science.1176199
Fenn, M. E., Baron, J. S., Allen, E. B., Rueth, H. M., Nydick, K. R., Geiser, L., et al. (2003). Ecological effects of nitrogen deposition in the western United States. Bioscience 53, 404–420. doi: 10.1016/j.envpol.2009.11.028
Fields, M. W., Hise, A., Lohman, E. J., Bell, T., Gardner, R. D., Corredor, L., et al. (2014). Sources and resources: importance of nutrients, resource allocation, and ecology in microalgal cultivation for lipid accumulation. Appl. Microbiol. Biotechnol. 98, 4805–4816. doi: 10.1007/s00253-014-5694-7
Friedman, J., and Alm, E. J. (2012). Inferring correlation networks from genomic survey data. PLoS Comput. Biol. 8:e1002687. doi: 10.1371/journal.pcbi.1002687
Fulbright, S. P., Dean, M. K., Wardle, G., Lammers, P. J., and Chisholm, S. (2014). Molecular diagnostics for monitoring contaminants in algal cultivation. Algal Res. 4, 41–51. doi: 10.1016/j.algal.2013.11.008
Gärdes, A., Ramaye, Y., Grossart, H. P., Passow, U., and Ullrich, M. S. (2012). Effects of Marinobacter adhaerens HP15 on polymer exudation by Thalassiosira weissflogii at different N:P ratios. Mar. Ecol. Prog. Ser. 461, 1–14. doi: 10.3354/meps09894
Gardner, E. M., McKnight, D. M., Lewis, W.M., and Millon, M. P. (2008). Effects of nutrient enrichment on phytoplankton in an Alpine Lake, Colorado, U.S.A. Arct. Antarct. Alp. Res. 40, 55–64.
Georgianna, D. R., and Mayfield, S. P. (2012). Exploiting diversity and synthetic biology for the production of algal biofuels. Nature 488, 329–335. doi: 10.1038/nature11479
Goecke, F., Thiel, V., Wiese, J., Labes, A., and Imhoff, J. F. (2013). Algae as an important environment for bacteria-phylogenetic relationships among new bacterial species isolated from algae. Phycologia 52, 14–24. doi: 10.2216/12-24.1
Gonzalez, L. E., and Bashan, Y. (2000). Increased growth of the microalga Chlorella vulgaris when coimmobilized and cocultured in alginate beads with the plant-growth-promoting bacterium Azospirillum brasilense. Appl. Environ. Microbiol. 66, 1527–1531. doi: 10.1128/AEM.66.4.1527-1531.2000
Griffiths, M. J., and Harrison, S. T. L. (2009). Lipid productivity as a key characteristic for choosing algal species for biodiesel production. J. Appl. Phycol. 21, 493–507. doi: 10.1007/s10811-008-9392-7
Griffiths, M. J., Hille, R. P., and Harrison, S. T. L. (2011). Lipid productivity, settling potential and fatty acid profile of 11 microalgal species grown under nitrogen replete and limited conditions. J. Appl. Phycol. 24, 989–1001. doi: 10.1007/s10811-011-9723-y
Guo, Z., and Tong, Y. W. (2013). The interactions between Chlorella vulgaris and algal symbiotic bacteria under photoautotrophic and photoheterotrophic conditions. J. Appl. Phycol. 26, 1483–1492. doi: 10.1007/s10811-013-0186-1
Haas, B. J., Gevers, D., Earl, A. M., Feldgarden, M., Ward, D. V., Giannoukos, G., et al. (2011). Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21, 494–504. doi: 10.1101/gr.112730.110
Hall, R. I., and Smol, J. P. (1992). A weighted-averaging regression and calibration model for inferring total phosphorus concentration from diatoms in British Columbia (Canada) lakes. Freshw. Biol. 27, 417–434. doi: 10.1111/j.1365-2427.1992.tb00551.x
Hem, J. D. (1985). Study and Interpretation of the Chemical Characteristics of Natural Water. Department of the Interior, U.S. Geological Survey. Avaialble at: http://pubs.water.usgs.gov/wsp2254
Imase, M., Watanabe, K., Aoyagi, H., and Tanaka, H. (2008). Construction of an artificial symbiotic community using a Chlorella-symbiont association as a model. FEMS Microbiol. Ecol. 63, 273–282. doi: 10.1111/j.1574-6941.2007.00434.x
Istvánovics, V. (2008). The role of biota in shaping the phosphorus cycle in lakes. Freshw. Rev. 1, 143–174. doi: 10.1608/FRJ-1.2.2
Kazamia, E., Aldridge, D. C., and Smith, A. G. (2012a). Synthetic ecology: a way forward for sutainable algal biofuel production? J. Biotechnol. 162, 163–169. doi: 10.1016/j.jbiotec.2012.03.022
Kazamia, E., Czesnick, H., Nguyen, T. T., Croft, M. T., Sherwood, E., Sasso, S., et al. (2012b). Mutualistic interactions between vitamin B12-dependent algae and heterotrophic bacteria exhibit regulation. Environ. Microbiol. 14, 1466–1476. doi: 10.1111/j.1462-2920.2012.02733.x
Kazamia, E., Riseley, A. S., Howe, C. J., and Smith, A. G. (2014). An engineered community approach for industrial cultivation of microalgae. Ind. Biotechnol. 10, 184–190. doi: 10.1089/ind.2013.0041
Legendre, P., and Gallagher, E. (2001). Ecologically meaningful transformations for ordination of species data. Oecologia 129, 271–280. doi: 10.1007/s004420100716
Li, Y., Horsman, M., Wang, B., Wu, N., and Lan, C. Q. (2008). Effects of nitrogen sources on cell growth and lipid accumulation of green alga Neochloris oleoabundans. Appl. Microbiol. Biotechnol. 81, 629–636. doi: 10.1007/s00253-008-1681-1
Lozupone, C., and Knight, R. (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71, 8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005
Lundquist, T. J., Woertz, I., and Benemann, J. R. (2010). A Realistic Technology and Engineering Assessment of Algae Biofuel. Berkeley, CA: Energy Biosciences Institute University of California.
Martin, M., Portetelle, D., Michel, G., and Vandenbol, M. (2014). Microorganisms living on macroalgae: diversity, interactions, and biotechnological applications. Appl. Microbiol. Biotechnol. 98, 2917–2935. doi: 10.1007/s00253-014-5557-2
McBride, R. C., Lopez, S., Meenach, C., Burnett, M., Lee, P. A., Nohilly, F., et al. (2014). Contamination management in low cost open algae ponds for biofuels production. Ind. Biotechnol. 10, 221–227. doi: 10.1089/ind.2013.1614
Melack, J. M., and Kilham, P. (1974). Photosynthetic rates of phytoplankton in East African alkaline, saline lakes. Limnol. Oceanogr. 19, 743–755. doi: 10.4319/lo.1974.19.5.0743
Miller, M. P., and McKnight, D. M. (2012). Limnology of the Green Lakes Valley: phytoplankton ecology and dissolved organic matter biogeochemistry at a long-term ecological research site. Plant Ecol. Divers. 5, 1–14. doi: 10.1080/17550874.2012.738255
Natrah, F. M. I., Bossier, P., Sorgeloos, P., Yusoff, F. M., and Defoirdt, T. (2013). Significance of microalgal-bacterial interactions for aquaculture. Rev. Aquac. 6, 48–61. doi: 10.1111/raq.12024
Nichols, H. W., and Bold, H. C. (1965). Trichosarcina polymorpha gen. et sp. nov. J. Phycol. 1, 34–38. doi: 10.1111/j.1529-8817.1965.tb04552.x
Oksanen, J. (2011). Multivariate Analysis of Ecological Communities in R: Vegan Tutorial. R Package Version 2.0–1. Available at: http://cc.oulu.fi/~jarioksa/opetus/metodi/vegantutor.pdf
Oren, A. (1994). The ecology of the extremely halophilic archaea. FEMS Microbiol. Rev. 13, 415–439. doi: 10.1111/j.1574-6976.1994.tb00060.x
Ortiz-Marquez, J. C., Do Nascimento, M., Dublan, M. L., and Curatti, L. (2012). Association with an ammonium-excreting bacterium allows diazotrophic culture of oil-rich eukaryotic microalgae. Appl. Environ. Microbiol. 78, 2345–2352. doi: 10.1128/AEM.06260-11
Paul, C., Mausz, M. A., and Pohnert, G. (2012). A co-culturing/metabolomics approach to investigate chemically mediated interactions of planktonic organisms reveals influence of bacteria on diatom metabolism. Metabolomics 9, 349–359. doi: 10.1007/s11306-012-0453-1
Post, F. J. (1977). The microbial ecology of the Great Salt Lake. Microb. Ecol. 3, 143–165. doi: 10.1007/BF02010403
Santos, C. A., and Reis, A. (2014). Microalgal symbiosis in biotechnology. Appl. Microbiol. Biotechnol. 98, 5839–5846. doi: 10.1007/s00253-014-5764-x
Sapp, M., Gerdts, G., Wellinger, M., and Wichels, A. (2008). Consuming algal products: trophic interactions of bacteria and a diatom species determined by RNA stable isotope probing. Helgol. Mar. Res. 62, 283–287. doi: 10.1007/s10152-008-0110-2
Sapp, M., Schwaderer, A. S., Wiltshire, K. H., Hoppe, H. G., Gerdts, G., and Wichels, A. (2007). Species-specific bacterial communities in the phycosphere of microalgae? Microb. Ecol. 53, 683–699. doi: 10.1007/s00248-006-9162-5
Scott, S. A., Davey, M. P., Dennis, J. S., Horst, I., Howe, C. J., Lea-Smith, D. J., et al. (2010). Biodiesel from algae: challenges and prospects. Curr. Opin. Biotechnol. 21, 277–286. doi: 10.1016/j.copbio.2010.03.005
Selvaratnam, T., Pegallapati, A. K., Montelya, F., Rodriguez, G., Nirmalakhandan, N., Van, Voorhies W, et al. (2014). Evaluation of a thermo-tolerant acidophilic alga, Galdieria sulphuraria, for nutrient removal from urban wastewaters. Bioresour. Technol. 156, 395–399. doi: 10.1016/j.biortech.2014.01.075
Sheehan, J., Dunahay, T., Benemann, J., and Roessler, P. (1998). A Look Back at the US Department of Energy’s Aquatic Species Program: Biodiesel from Algae. Golden, CO: U.S. Department of Energy, National Renewable Energy Laboratory, 328.
Shurin, J. B., Abbott, R. L., Deal, M. S., Kwan, G. T., Litchman, E., McBride, R. C., et al. (2013). Industrial-strength ecology: trade-offs and opportunities in algal biofuel production. Ecol. Lett. 16, 1393–1404. doi: 10.1111/ele.12176
Sidat, M., Bux, F., and Kasan, H. C. (1999). Polyphosphate accumulation by bacteria isolated from activated sludge. Water SA 25, 175–180.
Smith, V. H., and Crews, T. (2013). Applying ecological principles of crop cultivation in large-scale algal biomass production. Algal Res. 4, 23–34. doi: 10.1016/j.algal.2013.11.005
Smith, V. H., Sturm, B. S., Denoyelles, F. J., and Billings, S. A. (2010). The ecology of algal biodiesel production. Trends Ecol. Evol. 25, 301–309. doi: 10.1016/j.tree.2009.11.007
Tazi, L., Breakwell, D. P., Harker, A. R., and Crandall, K. A. (2014). Life in extreme environments: microbial diversity in Great Salt Lake, Utah. Extremophiles 18, 525–535. doi: 10.1007/s00792-014-0637-x
Wang, H., Zhang, W., Chen, L., Wang, J., and Liu, T. (2013). The contamination and control of biological pollutants in mass cultivation of microalgae. Bioresour. Technol. 128, 745–750. doi: 10.1016/j.biortech.2012.10.158
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Watanabe, K., Takihana, N., Aoyagi, H., Hanada, S., Watanabe, Y., Ohmura, N., et al. (2005). Symbiotic association in Chlorella culture. FEMS Microbiol. Ecol. 51, 187–196. doi: 10.1016/j.femsec.2004.08.004
Wolfe, A. P., Baron, J. S., and Cornett, R. J. (2001). Anthropogenic nitrogen deposition induces rapid ecological changes in alpine lakes of the Colorado Front Range (USA). J. Paleolimnol. 25, 1–7. doi: 10.1023/A:1008129509322
Xie, B., Bishop, S., Stessman, D., Wright, D., Spalding, M. H., and Halverson, L. J. (2013). Chlamydomonas reinhardtii thermal tolerance enhancement mediated by a mutualistic interaction with vitamin B. ISME J. 7, 1544–1555. doi: 10.1038/ismej.2013.43
Keywords: algal biomass, algal biofuel, bio-oil, bio-diesel, phycosphere
Citation: Bell TAS, Prithiviraj B, Wahlen BD, Fields MW and Peyton BM (2016) A Lipid-Accumulating Alga Maintains Growth in Outdoor, Alkaliphilic Raceway Pond with Mixed Microbial Communities. Front. Microbiol. 6:1480. doi: 10.3389/fmicb.2015.01480
Received: 02 August 2015; Accepted: 08 December 2015;
Published: 07 January 2016.
Edited by:
William James Hickey, University of Wisconsin–Madison, USAReviewed by:
François Perreault, Arizona State University, USAJennifer Stewart, University of Delaware, USA
Copyright © 2016 Bell, Prithiviraj, Wahlen, Fields and Peyton. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Brent M. Peyton, bpeyton@coe.montana.edu
 Tisza A. S. Bell
Tisza A. S. Bell Bharath Prithiviraj
Bharath Prithiviraj Brad D. Wahlen5
Brad D. Wahlen5 Matthew W. Fields
Matthew W. Fields Brent M. Peyton
Brent M. Peyton