- 1Shandong Provincial Key Laboratory for Biology of Vegetable Diseases and Insect Pests, College of Plant Protection, Shandong Agricultural University, Tai’an, China
- 2Key Laboratory of Zoological Systematics and Evolution, Institute of Zoology, Chinese Academy of Sciences, Beijing, China
Wolbachia-bacteriophage WO is a good model system for studying interactions between bacteria and viruses. Previous surveys of insect hosts have been conducted via sampling from open or semi-open communities; however, no studies have reported the infection patterns of phage WO of insects living in a closed community. Figs and fig wasps form a peculiar closed community in which the Ficus tree provides a compact syconium habitat for a variety of fig wasp. Therefore, in this study, we performed a thorough survey of Wolbachia and bacteriophage WO infection patterns in a total of 1406 individuals from 23 fig wasps species living on three different fig tree species. The infection rates of Wolbachia and phage WO were 82.6% (19/23) and 39.1% (9/23), respectively. Additionally, phage WO from fig wasps showed strong insect host specificity based on orf7 sequences from fig wasps and 21 other insect species. Probably due to the physical barrier of fig syconium, most phage WO from fig wasps form a specific clade. Phylogenetic analysis showed the absence of congruence between WO and host Wolbachia, WO and insect host, as well as Wolbachia and fig wasps, suggesting that both Wolbachia and phage WO exchanged frequently and independently within the closed syconium. Thus, the infection pattern of bacteriophage WO from fig wasps appeared quite different from that in other insects living outside, although the effect and the transfer routes of phage WO are unclear, which need to be investigated in the future.
Introduction
Wolbachia (Alphaproteobacteria) are maternally inherited obligatory intracellular symbionts that are found in a wide range of arthropods and filarial nematodes (Stouthamer et al., 1999; Taylor et al., 2005), at rates ranging from 20 to 76% (Hilgenboecker et al., 2008; Fenton et al., 2011). The success of Wolbachia in achieving this high prevalence is associated with its ability to induce a variety of phenotypes, from mutualism in nematodes to various reproductive manipulations in arthropods, including cytoplasmic incompatibility (O’Neill and Karr, 1990), parthenogenesis (Stouthamer et al., 1990), male killing (Jiggins et al., 2001), feminization (Bouchon et al., 1998), and even speciation (Rokas, 2000). Bacteriophages are the most abundant organisms in the biosphere (Plett et al., 2011) and play important roles in bacterial genome evolution. The temperate phage WO was first detected in 2000 and is the only one bacteriophage known to infect Wolbachia (Masui et al., 2000). Accompanied by the widespread distribution of Wolbachia, it was reported that about 89% of Wolbachia strains are infected with WO (Bordenstein and Wernegreen, 2004). Polymerase chain reaction (PCR) amplification of the minor capsid gene orf7 has shown that the phage occurs in the majority of the parasitic A and B Wolbachia supergroups (Bordenstein and Wernegreen, 2004; Gavotte et al., 2007). Moreover, most phage-infected Wolbachia strains display low numbers of phage types, with 85% showing only one or two different phage types (Gavotte et al., 2007; Tanaka et al., 2009).
The tripartite insect host–Wolbachia–phage WO is an ideal model system for studying interactions among viruses, bacteria, and eukaryotes (Bordenstein et al., 2006). The phage WO is the only known mobile genetic element that may transform the genome of Wolbachia (Metcalf and Bordenstein, 2012). In Wolbachia, prophage regions can comprise more than 20% of mobile DNA genes and account for the largest fraction of absent/divergent genes between closely related strains (Chafee et al., 2010). Some researchers have investigated the relationship between Wolbachia and WO in such insect species as Drosophila simulans, Ephestia kuehniella, Nasonia vitripennis, Culex pipiens, and Gryllus pennsylvanicus; moreover, no phylogenetic congruence between Wolbachia and WO has been shown, suggesting that the lateral transfer of WO in Wolbachia is not unusual (Masui et al., 2000; Bordenstein and Wernegreen, 2004; Chafee et al., 2010). Thus, unraveling the infection status and evolutionary dynamics of Wolbachia and WO may be the key to understanding the interactions among these organisms and methods for exploiting such interactions.
Previous surveys of insect hosts have been conducted via sampling from open and semi-open communities; however, no studies have reported the infection patterns of bacteriophage WO from insects living in a closed community. Thus, it is particularly interesting to consider the infection patterns of WO in fig wasps and the relationship with Wolbachia as this system occurs within an enclosed syconia. Figs and fig wasps constitute a well-known system of mutualism (Weiblen, 2002): figs (Angiospermae, Dicotyledoneae, Urticales, Moraceae) are pollinated on their inflorescences by their obligate fig wasps (Insecta, Hymenoptera, Chalcidoidea), and the fig wasps lay their eggs in the figs, wherein the eggs develop (Rønsted et al., 2005). The inflorescences, called syconia, provide fig wasps with a compact habitat that is isolated from the outside world (Schiffler, 2002). Besides pollinating wasps, some non-pollinators do not enter the syconia, but inject eggs through the fig wall (Schiffler, 2002). Both the seeds and the offspring of fig wasps develop in the fig syconium until the fig reaches maturity. Previous surveys have shown that the incidence of Wolbachia in fig wasps is up to 59–67%, markedly higher than that in other insects (Shoemaker et al., 2002; Haine and Cook, 2005; Chen et al., 2010).
The syconia provide fig wasps with a compact habitat, in which Wolbachia horizontal transfer has been shown to be more likely to occur in this closed system than in other open and semi-open systems (Yang et al., 2012). Thus, in this study, we sought to determine the bacteriophage WO infection patterns in fig wasps, as well as phage diversity within Wolbachia strains and within different fig wasps. We investigated 23 fig wasp species from three fig species to elucidate the phage infection patterns. Furthermore, we want to find whether the horizontal transfer of Wolbachia has an effect on the diversity and evolutionary dynamics of the temperate bacteriophage WO.
Materials and Methods
Identification of Fig Wasps and DNA Extraction
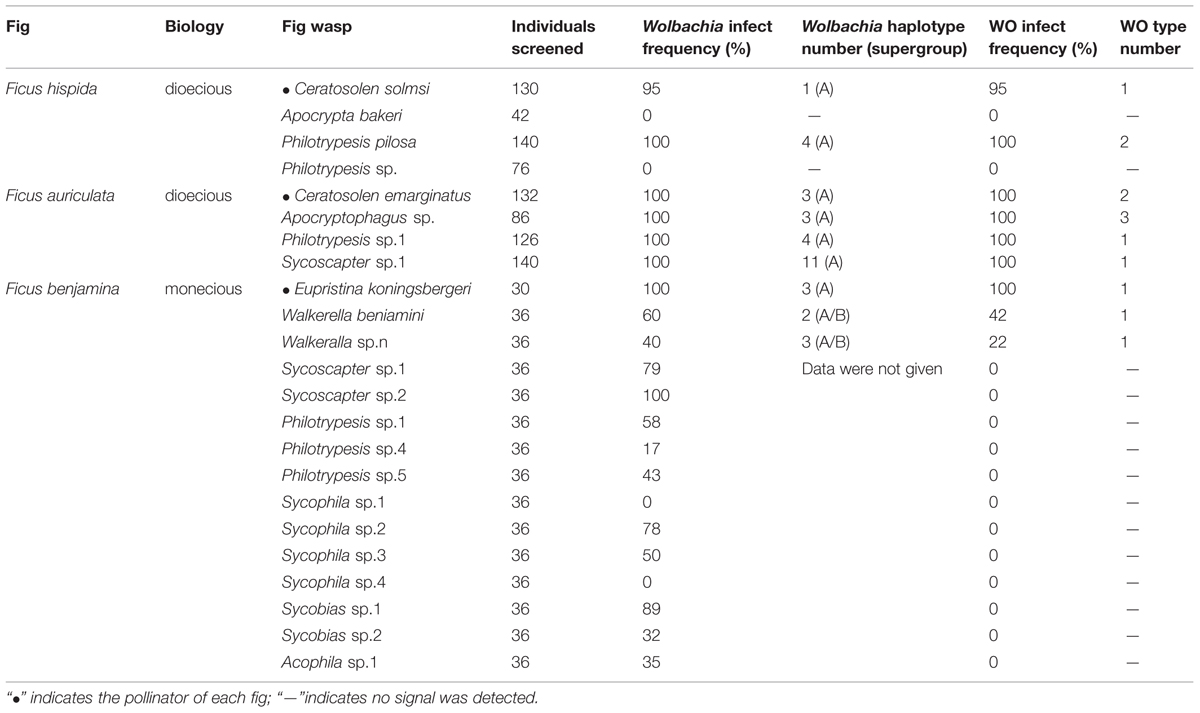
All fig wasps were collected from fig trees in Hainan Province, China, from 2005 to 2013. We collected fig fruits in period D (late in the fruiting cycle but before the period in which fig wasps emerge) and then cultivated the fruits until the fig wasps emerged. The fig wasps were then collected, deposited in 95% alcohol, and stored at –20°C for later use. Different species were morphologically classified under a Nikon SMZ80 microscope. The fig wasp species identified in this study are listed in Table 1.
Total genomic DNA was extracted from each individual sample using EasyPure Genomic DNA Extraction Kits (TransGen, Beijing, China). Initially, genomic DNA was screened for the quality of the template using mitochondrial cytochrome c oxidase 1 (CO1) (Chambers et al., 2011; Dyer et al., 2011) and nuclear ribosomal DNA internal transcribed spacer 2 (ITS2) (Partensky and Garczarek, 2011) via PCR (Kraemer and Velicer, 2011). Poor quality DNA templates were discarded.
PCR and Sequencing
The samples were first screened for Wolbachia infection by PCR amplification with the primers wsp81F (5′-TGG TCC AAT AAG TGA TGA AGA AAC-3′) and wsp691R (5′-AAA AAT TAA ACG CTA CTC CA-3′), which amplified a portion of the Wolbachia surface protein gene (wsp) (Zhou et al., 1998). If the amplification of wsp did not yield a sufficient band on agarose gels, another two pairs of primers were used, ftsZ-F/R for amplification of the Wolbachia cell division gene ftsZ and 16SwolF/R for amplification of the Wolbachia 16sRNA gene (O’Neill et al., 1992; Jeyaprakash and Hoy, 2000). WO was screened by the primers orf7F (5′-GTC TGG AAA GCT TAC AAA AAG-3′) and orf7R (5′-GCT CTA TAA ATT CTC CTA T-3′), and samples that were negative for orf7 were rechecked by another two pairs of gene primers, ORF2F/R and WD0633F/R (Masui et al., 2000). ddH2O was used as a blank control for all amplifications. PCR amplification was performed in a volume of 25 μL, containing 2.5 μL 10× buffer, 0.2 mM dNTPs, 0.5 μM of each primer, and 0.5 U of Trans Taq Enzyme (TransGen Biotech, Beijing, China).
Polymerase chain reaction products were purified using an EasyPure PCR purification Kit (TransGen) and then sent to Beijing Genomic Institute for sequencing. When multiple peaks appeared, the products were cloned in the pEasy-T5 vector (TransGen), and 3–7 positive clones were picked for sequencing.
Sequence Analysis
Raw Sequence Treatments
Sequence homology analysis was performed with the BLAST program in NCBI Web. Haplotypes are defined as having greater than 1.5% nucleotide diversity in the orf7 gene (Chafee et al., 2010). Wolbachia strains that were not infected by phage WO were not included in our study in order to improve readability. If multiple identical sequences were obtained from each species, we chose only one to represent the species. We reserved different sequences and removed identical sequences, yielding 34 wsp sequences and 12 orf7 sequences. The sequences have been deposited in GenBank under the following accession numbers: KT355405–KT355450.
Phylogenetic Analyses
The wsp and orf7 sequences were aligned to relevant sequences previously published on NCBI with Clustal W in BioEdit (Hall, 1999). Maximum likelihood (ML) was carried out to construct the phylogenetic tree using MEGA 6 (Tamura et al., 2013). Model selection for the ML analysis was estimated using the Akaike information criterion in Modeltest v3.7. The DNA substitution model was the general time reversible (GTR) model in which the gamma distribution and invariant sites were estimated from the data (GTR+I+G). ML bootstrap values were generated from 1000 bootstrap replicates.
Results
Wolbachia Infection Patterns in Fig Wasps
We screened a total of 1406 wasps of 23 species for Wolbachia and WO infection. The results are listed in Table 1. Nineteen out of 23 fig wasp species were shown to be infected with Wolbachia. This infection incidence (83%) was higher than in wider screenings of fig wasps in Panama (59%) and Australia (67%) (Shoemaker et al., 2002). The Wolbachia infection incidences in the three syconia in our survey differed, ranging from 100% in Ficus auriculata (4/4) to 86.7% in F. benjamina (13/15) and 50% in F. hispida (2/4). The Wolbachia-infected fig wasp species from F. hispida and F. auriculata showed high infection rates, nearly 100%, which agreed with the reported “most or few” (>90% or<10%) infection pattern within one species (Hilgenboecker et al., 2008). However, except for Eupristina koningsbergeri and Sycoscapter sp.2, which had 100% infection rates, while Sycophila sp.1 and Sycophila sp.4 were completely uninfected, most species in F. benjamina had moderate infection rates, ranging from 17 to 89%, which is similar to our previous survey in F. benjamina (Yang et al., 2012).
After removing repeated identical sequences within each species, 34 wsp haplotypes from 19 wasp species were obtained. Besides five common Wolbachia strains (wHaw, wMel, wUni, wMors, and wCon), three other Wolbachia strains were first detected in fig wasps, named wFig-1, wFig-2 and wFig-3, respectively (Figure 1). wCon belonged to Wolbachia supergroup B, while all the others belonged to Wolbachia supergroup A. wHaw and wMel were widely distributed in F. hispida and F. auriculata, while wCon were only detected in F. benjamina. Among nine Wolbachia-infected species, Ceratosolen solmsi from F. hispida was infected by wHaw only and Walkerella beniamini from F. benjamina were double infected, while all the other seven fig species (P. pilosa from F. hispida, Ceratosolen emarginatus, Apocryptophagus sp., Philotrypesis sp.1, and Sycoscapter sp.1 from F. auriculata, and Euprisina koningsbergeri and Walkerella sp. from F. benjamina) were multiple infected. Noteworthy, Sycoscapter sp.1 from F. auriculata were infected by up to six different bacteria.
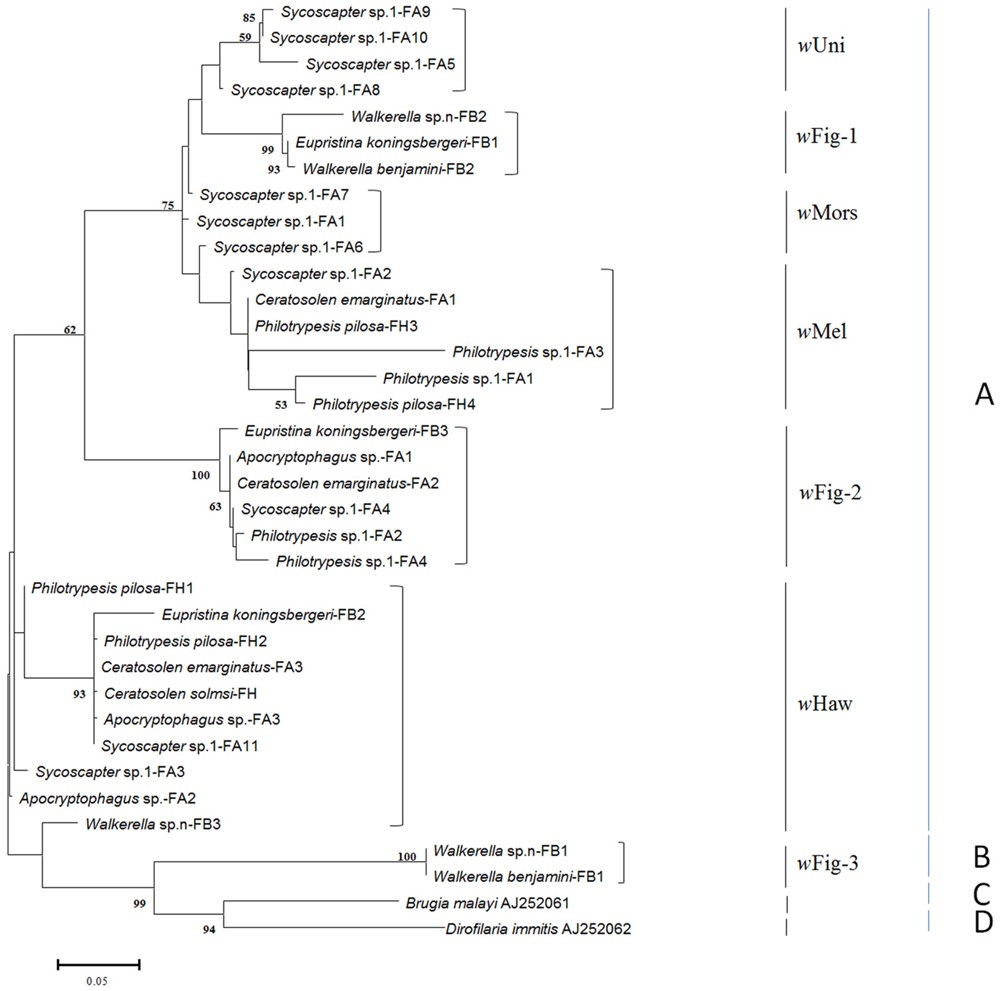
FIGURE 1. ML tree constructed with wsp sequences from fig wasps. Two sequences from supergroup C and D are used as outgroups. The supergroups (A–D) are listed on the right, while the strains previously reported and newly found are also indicated. Each OUT is named as its insect host (fig wasp) name followed by the fig name (FA: Ficus auriculata, FH: Ficus hispida, and FB: Ficus benjamina), and the numbers at last mean different haplotypes of wsp sequences.
WO Infection Patterns Among Wolbachia Strains Associated with Fig Wasps
No bacteriophage WO was detected in fig wasps that were not infected with Wolbachia, as has been reported in previous surveys (Fujii et al., 2004; Braquart-Varnier et al., 2005; Sanogo et al., 2005; Gavotte et al., 2007). Among the 23 fig wasp species tested, only 39% (9/23) were found to harbor phages. Importantly, all Wolbachia-infected fig wasps harbored phage WO. Notably, only three of 15 fig wasp species infected by Wolbachia harbored WO in F. benjamina, which was obviously lower than the infection rate in the other two fig species. Among the WO-infected species, most (7/9) had 100% infection rates, with the exception of Walkerella beniamini (42%) and Walkerella sp.n (22%) from F. benjamina. All the pollinators (3/3) were infected by WO, while only 30% (6/20) nonpollinators harbored phage WO.
Twelve orf7 sequences were gained from the nine phage-harbored fig wasps. Most fig wasps harbored only one bacteriophage WO type while Ceratosolen emarginatus and Apocryptophagus sp., both of which were from Ficus auriculata, harbored two and three WO types, respectively. Eupristina koningsbergeri, Walkerella benjamini, and Walkerella sp.n from F. benjamina, Philotrypesis sp.1, Sycoscapter sp.1 and Apocryptophagus sp. from F. auriculata shared one common WO type, while the other two fig wasps from F. hispida, Ceratosolen solmsi and Philotrypesis pilosa shared another WO type.
Interestingly, the number and type of WO infections was not related to the Wolbachia host. On one hand, the same number of Wolbachia strains that infected different fig wasps harbored different WO types. For example, Ceratosolen emarginatus, which was infected by three different Wolbachia strains, harbored two types of WO phages, while Apocryptophagus sp., which was also infected by three different Wolbachia strains, harbored three different WO. On the other hand, one WO phage type could be found in many different Wolbachia strains. For example, Sycoscapter sp.1, which was infected by up to six different Wolbachia strains, was detected to harbor only one WO type.
Bacteriophage WO Phylogeny
The phylogenetic ML tree of phage WO orf7 sequences of fig wasps and 21 other insect species was constructed (Figure 2). Interestingly, most orf7 fragments from fig wasps tested in our survey bunched together, belonging to one clade (group IV), only three (two from Apocryptophagus sp. and one from Ceratosolen emarginatus) orf7 sequences located in other clades (group I, II, III) constituted by orf7 sequences from the other 21insect species. The profile of the ML tree exhibited strong insect host specificity and suggested that WO in fig wasps had a special origin, which was quite different from previously surveyed insects from open and semi-open environments.
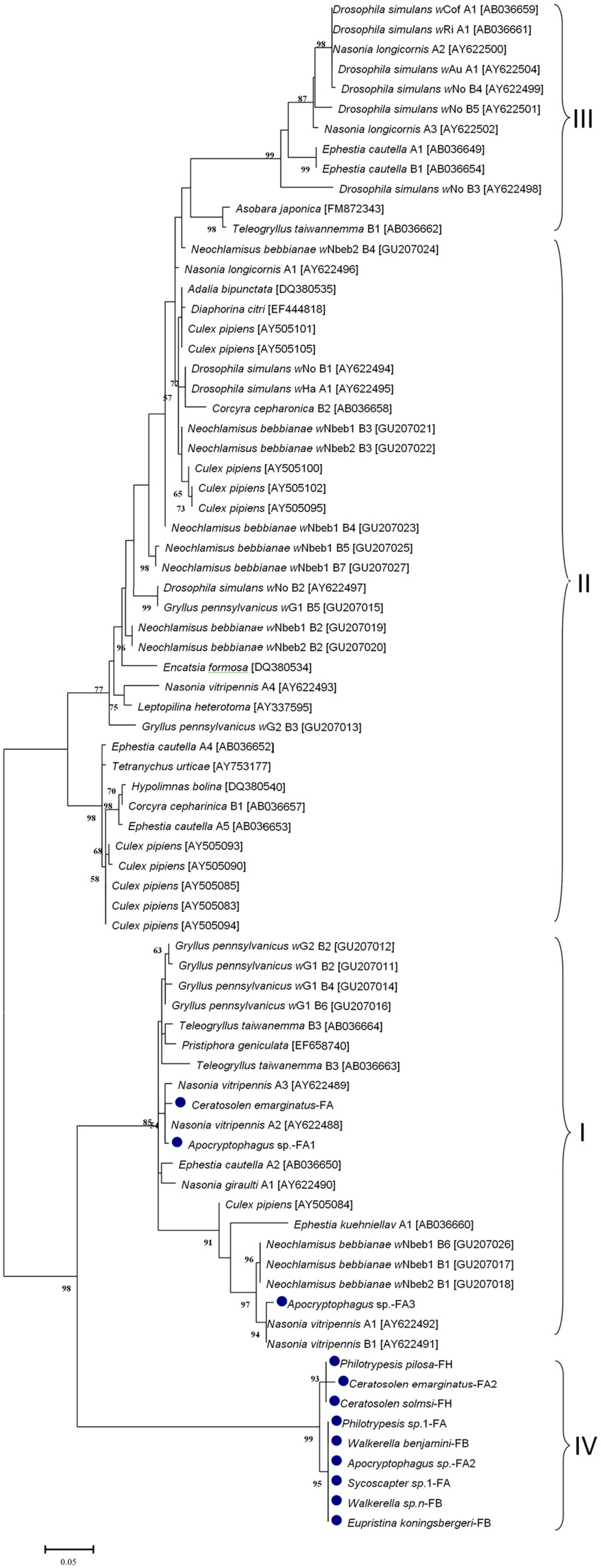
FIGURE 2. Maximum likelihood phylogenetic tree of the bacteriophage orf7 nucleotide sequences. The sequences of fig wasps tested in our survey are marked with •. Some related sequences were downloaded from GenBank, and the accession IDs of the downloaded sequences are annotated with brackets. Different groups are indicated with I, II, III, IV. Samples tested in our survey are named by insect host name followed by the fig name (FA: Ficus auriculata, FH: Ficus hispida, FB: Ficus benjamina) and haplotype numbers.
Phage and Wolbachia Horizontal Transfer
Comparisons of bacteriophage WO phylogeny, based on the orf7 gene, and Wolbachia phylogeny, based on the wsp gene, were performed for 13 species infected by both WO and Wolbachia (Figure 3). Obviously, no congruence was found between phage WO and its host Wolbachia phylogenies, which indicated that phages do not cospeciate with their hosts. Moreover, there was also no congruence between phylogenies between phage WO and fig wasps (Figure 4), as well as between Wolbachia and its fig wasp hosts (Figure 5). No phylogenetic correlation was found between phages and bacteria, bacteria and insect hosts, or phages and insect hosts, as in some reported insects from open and semi-open environments (Masui et al., 2000; Bordenstein and Wernegreen, 2004; Gavotte et al., 2007). The absence of an evolutionary correlation between WO and Wolbachia and between WO and insect host phylogenies indicated that WO can be transferred horizontally by itself between different Wolbachia endosymbionts or even insect hosts.
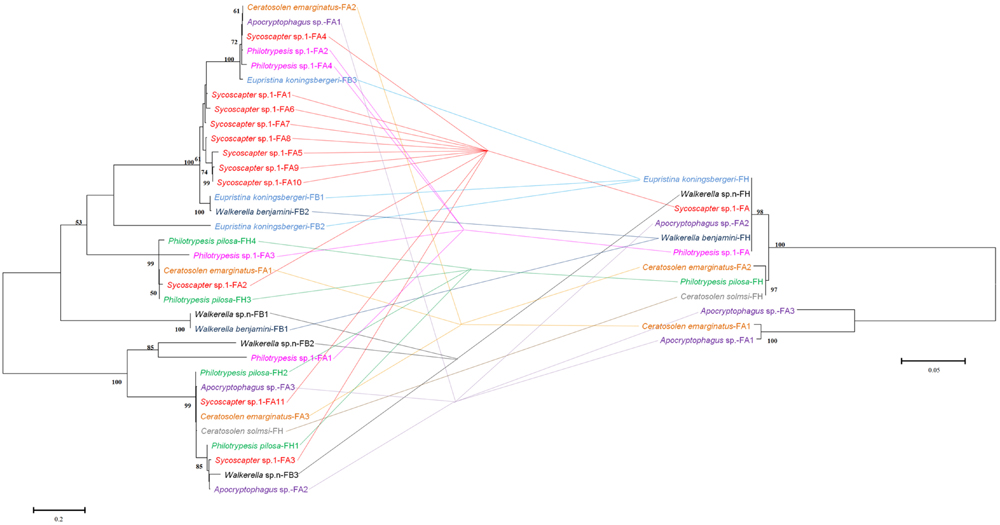
FIGURE 3. Comparison between phylogenies of bacteriophage WO based on the orf7 sequence (right) and Wolbachia based on the wsp sequence (left). Samples tested in our survey are named by insect host name followed by the fig name (FA: Ficus auriculata, FH: Ficus hispida, FB: Ficus benjamina) and haplotype numbers.
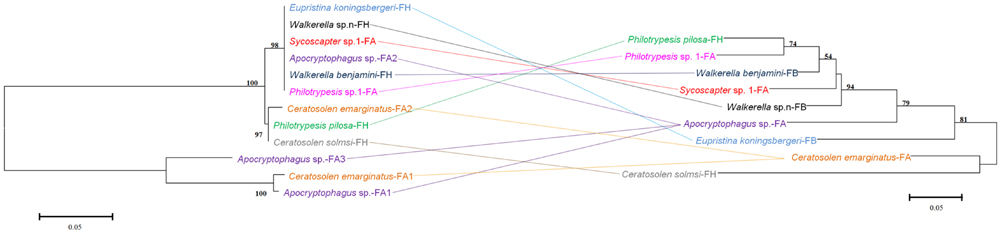
FIGURE 4. Comparison between phylogenies of bacteriophage WO based on the orf7 sequence (left) and fig wasps based on the COI sequence (right). Samples tested in our survey are named by insect host name followed by the fig name (FA: Ficus auriculata, FH: Ficus hispida, FB: Ficus benjamina), and haplotype numbers.
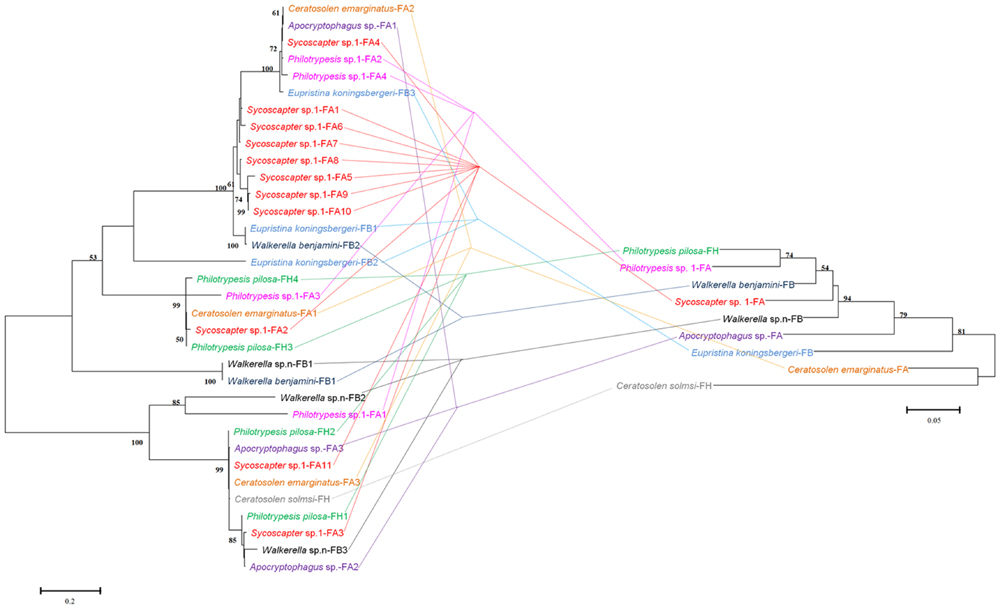
FIGURE 5. Comparison between phylogenies of Wolbachia based on the wsp sequence (left) and fig wasps based on the COI sequence (right). Samples tested in our survey are named by insect host name followed by the fig name (FA: Ficus auriculata, FH: Ficus hispida, FB: Ficus benjamina) and haplotype numbers.
In addition to this lack of congruence between WO and Wolbachia, we found other evidence of phage WO horizontal transfer. Distant phylogenetically related Wolbachia strains (supergroup A and B) shared the same WO type. For example, Walkerella benjamini, which was double infected by Wolbachia strains from different supergroups, harbored only one WO type. Moreover, different insect hosts from different figs (Ficus auriculata and F. benjamina), were found to harbor only one phage type.
Discussion
Host–microbe–phage symbiosis comprises some of the most intimate and long-lasting associations on the planet. In order to determine the relationships between each pair of organisms, it is necessary to first determine the patterns of infection. In this study, we examined the relationships among Wolbachia and phage WO in fig wasp species within a closed system. Our data provided insights into the complex relationships of this symbiosis.
In this study, we report the detailed analysis and detection of Wolbachia and phage WO in 23 fig wasps from three different compact syconia. This system is notable because it is considered a closed community, different from other insects living in open or semi-open environments. In our study, the Wolbachia infection rate (83%) in our tested fig wasps was much higher than fig wasps in Australia (67%) and Panama (59%) (Shoemaker et al., 2002; Haine and Cook, 2005), which showed that fig wasps may have the highest known incidences of Wolbachia amongst all insects (Haine and Cook, 2005). As for bacteriophage WO, the infection rate in our tested fig wasps is 39%, which was unexpectedly lower than that in previous reports 89% (Chauvatcharin et al., 2006; Gavotte et al., 2007). The low WO infection rate (20%, 3/15) of fig wasps from F. benjamina and the high representation of fig wasps from F. benjamina in these samples (15/23) may contribute to the overall infection pattern. Interestingly, despite living in the same syconia, most Wolbachia-infected species did not harbor phage WO in F. benjamina. However, whether these fig wasps were infected previously or have never been infected is unclear. We suggest three possible explanations for this phenomenon. Firstly, the method used to screen for the presence of phage WO may not have detected all phage WO. Although several primer pairs were used to check the infection patterns repeatedly, the orf7F/R primers were not sufficiently degenerate to detect all orf7 variants. Previously, only a single WO haplotype was detected in D. simulans infected with Wolbachia strain wRi (Gavotte et al., 2007). However, the genome sequence confirmed four prophage copies in the genome (Klasson et al., 2009). Moreover, Metcalf proposed that single-gene PCR should not be used to rule out the presence of phage WO in Wolbachia (Metcalf et al., 2014). Second, the particular environment in which fig wasps live in may limit their physical interaction with the outside world. Unlike insects living open environments, chalcidoid wasps live almost all of their lives inside the syconia, which act as a physical barrier to prevent the insects from exchange with other insects. Third, there may be other unknown factors that isolate the insects or bacteria from phage WO infection. Therefore, future studies are needed to examine these possibilities.
Among the infections examined in this study, most bacteriophage WO types were first detected in fig wasps and showed strong insect host specificity based on the phylogeny of orf7, when considering 21 other insect species besides our fig wasps. However, it is not known what determines this specificity; specifically, we suspect that this specificity may be determined by a factor inherent in the chalcidoid and Wolbachia or within the unique living environment. As shown in the phylogenic tree, even species that were closely related with fig wasps exhibited substantial differences in orf7 sequences. For example, Nasonia (Nasonia vitripennis and N. longicornis) and fig wasps are all hymenopterans, but their orf7 genes are located distant from each other in the tree. Moreover, Philotrypesis sp.1, Sycoscapter sp.1, and Nasonia all belong to Pteromalidae, but their orf7 genes are scattered in the tree. Therefore, the most probable explanation of specificity is the enclosed syconium in which the fig wasps live, which may result in a unique lineage for the bacteriophage WO. Importantly, the specificity is not limited to fig wasps living in one fig fruit, but is observed at the level of the fig wasp species.
Multiple infections are common in Wolbachia; however, six out of nine WO-infected species harbored only one phage type, which was confirmed by direct sequencing of the PCR product of orf7 sequences, even when wasps were sampled from different syconia and different fig trees, indicating that they were not occasional infection events. In addition, the complete genome sequence of prophage WO (WOSol) in Wolbachia strain wSol, which infected the fig wasps Ceratosolen solmsi, also showed the presence of only one WO phage in the wSol genome (Wang et al., 2013). Furthermore, a survey of phage WO showed that 85% (28/34) of Wolbachia strains harbor only one or two different WO types (Gavotte et al., 2007).
Wolbachia spreads across hosts through both vertical and horizontal transfer. Vertical transmission is thought to be the predominant mode of Wolbachia transmission within a host (Werren et al., 2008), and horizontal transfer of Wolbachia has also been detected both within and among different host species in many cases (Baldo et al., 2006, 2008; Frost et al., 2010). Considerable horizontal transfer of Wolbachia has been detected within F. benjamina, and researchers have proposed that the syconium may provide a platform for horizontal transfer (Yang et al., 2012). We confirmed this by analyzing the discordance between the phylogenies of Wolbachia and fig wasps from different figs (Figure 5). Similarly, we found abundant horizontal transfer of phage WO in Wolbachia associated with fig wasps, although the exact horizontal transfer routes were uncertain. From our data and previous studies, there may be several possible mechanisms for such transfer. First, we observed multiple horizontal transfer of phage WO of Wolbachia in fig wasps from two pieces of evidence: 1) there was phylogenetic discordance between Wolbachia and phage WO (Figures 2 and 3) based on the orf7 sequences, the same phage type was shared by phylogenetically distant Wolbachia strains of fig wasps from the same and different syconia. Thus, the horizontal transfer of both Wolbachia and WO was common in figs. Second, the horizontal transfer of WO was completely independent of the horizontal transfer of Wolbachia. If the horizontal transfer of WO depends on the transfer of Wolbachia, then the insects infected by the same Wolbachia strains would harbor the same WO types, and congruence between the two phylogenies would be found. However, this was not the case in our data. Instead, we found that WO could be successfully transferred by itself horizontally without its bacterial host between Wolbachia endosymbionts or insects. This has also been confirmed in other studies (Chafee et al., 2010). This independence may be possible because of the WO lysozymes, which could lyse bacterial cell walls (Fischetti, 2010). Third, the horizontal transfer of Wolbachia may facilitate the horizontal transfer of WO. The enclosed syconium provided a platform for horizontal transfer of the endosymbiont Wolbachia; this may also be beneficial for the transfer of the phage to some extent. Each of these potential mechanisms may contribute to the phage transfer.
Recently, genome sequence data shows that some mobile elements are present at sometimes high frequency in obligate intracellular bacteria, including Rickettsia, Phytoplasma, and Wolbachia (Cordaux et al., 2008; Baldridge et al., 2010; Chung et al., 2013). Bacteriophage WO is the only element that has been shown to move horizontally in obligate bacterial endosymbiont Wolbachia (Bordenstein and Wernegreen, 2004). WO is a dynamic element that has a marked effect on the genetic diversity of Wolbachia and could explain some of the interactions with Wolbachia genes and factors, without participating directly in the reproductive manipulations of Wolbachia induced in arthropods (Sanogo et al., 2005; Gavotte et al., 2007).
Conclusion
We provided important insights into the horizontal transfer and interactions of fig wasps, Wolbachia, and phage WO in enclosed syconia. Some evidence of how bacteriophage WO work on Wolbachia or even insect hosts is another focus of our following research. This may facilitate the use of WO DNA-delivery vectors as tools for genetic manipulation of insects in the future.
Author Contributions
NXW and DWH designed the study. NXW, SSJ, and HX performed the analyses. SSJ performed experiments. NXW, SSJ, YL, and DWH wrote the manuscript. All authors revised the manuscript and approved the final version.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Science Foundation of China (NSFC grant nos 31210103912, 31101634), partially by a grant (O529YX5105) from the Key Laboratory of the Zoological Systematics and Evolution of the Chinese Academy of Sciences.
References
Baldo, L., Ayoub, N. A., Hayashi, C. Y., Russell, J. A., Stahlhut, J. K., and Werren, J. H. (2008). Insight into the routes of Wolbachia invasion: high levels of horizontal transfer in the spider genus Agelenopsis revealed by Wolbachia strain and mitochondrial DNA diversity. Mol. Ecol. 17, 557–569. doi: 10.1111/j.1365-294X.2007.03608.x
Baldo, L., Dunning Hotopp, J. C., Jolley, K. A., Bordenstein, S. R., Biber, S. A., Choudhury, R. R., et al. (2006). Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl. Environ. Microbiol. 72, 7098–7110. doi: 10.1128/AEM.00731-06
Baldridge, G. D., Burkhardt, N. Y., Labruna, M. B., Pacheco, R. C., Paddock, C. D., Williamson, P. C., et al. (2010). Wide dispersal and possible multiple origins of low-copy-number plasmids in rickettsia species associated with blood-feeding arthropods. Appl. Environ. Microbiol. 76, 1718–1731. doi: 10.1128/AEM.02988-09
Bordenstein, S. R., Marshall, M. L., Fry, A. J., Kim, U., and Wernegreen, J. J. (2006). The tripartite associations between bacteriophage, Wolbachia, and arthropods. PLoS Pathog. 2:e43. doi: 10.1371/journal.ppat.0020043
Bordenstein, S. R., and Wernegreen, J. J. (2004). Bacteriophage flux in endosymbionts (Wolbachia): infection frequency, lateral transfer, and recombination rates. Mol. Biol. Evol. 21, 1981–1991. doi: 10.1093/molbev/msh211
Bouchon, D., Rigaud, T., and Juchault, P. (1998). Evidence for widespread Wolbachia infection in isopod crustaceans: molecular identification and host feminization. Proc. R. Soc. Lond. B Biol. Sci. 265, 1081–1090. doi: 10.1098/rspb.1998.0402
Braquart-Varnier, C., Greve, P., Felix, C., and Martin, G. (2005). Bacteriophage WO in Wolbachia infecting terrestrial isopods. Biochem. Biophys. Res. Commun. 337, 580–585. doi: 10.1016/j.bbrc.2005.09.091
Chafee, M. E., Funk, D. J., Harrison, R. G., and Bordenstein, S. R. (2010). Lateral phage transfer in obligate intracellular bacteria (Wolbachia): verification from natural populations. Mol. Biol. Evol. 27, 501–505. doi: 10.1093/molbev/msp275
Chambers, E. W., Hapairai, L., Peel, B. A., Bossin, H., and Dobson, S. L. (2011). Male mating competitiveness of a Wolbachia-introgressed Aedes polynesiensis strain under semi-field conditions. PLoS Negl. Trop. Dis. 5:e1271. doi: 10.1371/journal.pntd.0001271
Chauvatcharin, N., Ahantarig, A., Baimai, V., and Kittayapong, P. (2006). Bacteriophage WO-B and Wolbachia in natural mosquito hosts: infection incidence, transmission mode and relative density. Mol. Ecol. 15, 2451–2461. doi: 10.1111/j.1365-294X.2006.02947.x
Chen, L. L., Cook, J. M., Xiao, H., Hu, H. Y., Niu, L. M., and Huang, D. W. (2010). High incidences and similar patterns of Wolbachia infection in fig wasp communities from three different continents. Insect Sci. 17, 101–111. doi: 10.1111/j.1744-7917.2009.01291.x
Chung, W. C., Chen, L. L., Lo, W. S., Lin, C. P., and Kuo, C. H. (2013). Comparative analysis of the peanut witches’-broom phytoplasma genome reveals horizontal transfer of potential mobile units and effectors. PLoS ONE 8:e62770. doi: 10.1371/journal.pone.0062770
Cordaux, R., Pichon, S., Ling, A., Perez, P., Delaunay, C., Vavre, F., et al. (2008). Intense transpositional activity of insertion sequences in an ancient obligate endosymbiont. Mol. Biol. Evol. 25, 1889–1896. doi: 10.1093/molbev/msn134
Dyer, K. A., Burke, C., and Jaenike, J. (2011). Wolbachia-mediated persistence of mtDNA from a potentially extinct species. Mol. Ecol. 20, 2805–2817. doi: 10.1111/j.1365-294X.2011.05128.x
Fenton, A., Johnson, K. N., Brownlie, J. C., and Hurst, G. D. D. (2011). Solving the Wolbachia paradox: modeling the tripartite interaction between host, Wolbachia, and a natural enemy. Am. Nat. 178, 333–342. doi: 10.1086/661247
Fischetti, V. A. (2010). Bacteriophage endolysins: a novel anti-infective to control Gram-positive pathogens. Int. J. Med. Microbiol. 300, 357–362. doi: 10.1016/j.ijmm.2010.04.002
Frost, C. L., Fernández-Marín, H., Smith, J. E., and Hughes, W. O. H. (2010). Multiple gains and losses of Wolbachia symbionts across a tribe of fungus-growing ants. Mol. Ecol. 19, 4077–4085. doi: 10.1111/j.1365-294X.2010.04764.x
Fujii, Y., Kubo, T., Ishikawa, H., and Sasaki, T. (2004). Isolation and characterization of the bacteriophage WO from Wolbachia, an arthropod endosymbiont. Biochem. Biophys. Res. Commun. 317, 1183–1188. doi: 10.1016/j.bbrc.2004.03.164
Gavotte, L., Henri, H., Stouthamer, R., Charif, D., Charlat, S., Bouletreau, M., et al. (2007). A survey of the bacteriophage WO in the endosymbiotic bacteria Wolbachia. Mol. Biol. Evol. 24, 427–435. doi: 10.1093/molbev/msl171
Haine, E. R., and Cook, J. M. (2005). Convergent incidences of Wolbachia infection in fig wasp communities from two continents. Proc. R. Soc. Lond. B Biol. Sci. 272, 421–429. doi: 10.1098/rspb.2004.2956
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Hilgenboecker, K., Hammerstein, P., Schlattmann, P., Telschow, A., and Werren, J. H. (2008). How many species are infected with Wolbachia? – a statistical analysis of current data. FEMS Microbiol. Lett. 281, 215–220. doi: 10.1111/j.1574-6968.2008.01110.x
Jeyaprakash, A., and Hoy, M. A. (2000). Long PCR improves Wolbachia DNA amplification: wsp sequences found in 76% of sixty-three arthropod species. Insect Mol. Biol. 9, 393–405. doi: 10.1046/j.1365-2583.2000.00203.x
Jiggins, F. M., Hurst, G. D., Schulenburg, J. H., and Majerus, M. E. (2001). Two male-killing Wolbachia strains coexist within a population of the butterfly Acraea encedon. Heredity 86, 161–166. doi: 10.1046/j.1365-2540.2001.00804.x
Klasson, L., Westberg, J., Sapountzis, P., Näslund, K., Lutnaes, Y., Darby, A. C., et al. (2009). The mosaic genome structure of the Wolbachia wRi strain infecting Drosophila simulans. Proc. Natl. Acad. Sci. U.S.A. 106, 5725–5730. doi: 10.1073/pnas.0810753106
Kraemer, S. A., and Velicer, G. J. (2011). Endemic social diversity within natural kin groups of a cooperative bacterium. Proc. Natl. Acad. Sci. U.S.A. 108, 10823–10830. doi: 10.1073/pnas.1100307108
Masui, S., Kamoda, S., Sasaki, T., and Ishikawa, H. (2000). Distribution and evolution of bacteriophage WO in Wolbachia, the endosymbiont causing sexual alterations in Arthropods. J. Mol. Evol. 51, 491–497.
Metcalf, J. A., and Bordenstein, S. R. (2012). The complexity of virus systems: the case of endosymbionts. Curr. Opin. Microbiol. 15, 546–552. doi: 10.1016/j.mib.2012.04.010
Metcalf, J. A., Jo, M., Bordenstein, S. R., Jaenike, J., and Bordenstein, S. R. (2014). Recent genome reduction of Wolbachia in Drosophila recens targets phage WO and narrows candidates for reproductive parasitism. PeerJ 2, e529. doi: 10.7717/peerj.529
O’Neill, S. L., Giordano, R., Colbert, A., Karr, T. L., and Robertson, H. M. (1992). 16S rRNA phylogenetic analysis of the bacterial endosymbionts associated with cytoplasmic incompatibility in insects. Proc. Natl. Acad. Sci. U.S.A. 89, 2699–2702. doi: 10.1073/pnas.89.7.2699
O’Neill, S. L., and Karr, T. L. (1990). Bidirectional incompatibility between conspecific populations of Drosophila simulans. Nature 348, 178–180. doi: 10.1038/348178a0
Partensky, F., and Garczarek, L. (2011). Microbiology: arms race in a drop of sea water. Nature 474, 582–583. doi: 10.1038/474582a
Plett, J. M., Kemppainen, M., Kale, S. D., Kohler, A., Legué, V., Brun, A., et al. (2011). A secreted effector protein of Laccaria bicolor is required for symbiosis development. Curr. Biol. 21, 1197–1203. doi: 10.1016/j.cub.2011.05.033
Rokas, I. I. (2000). Wolbachia as a speciation agent. Trends Ecol. Evol. 15, 44–45. doi: 10.1016/S0169-5347(99)01783-8
Rønsted, N., Weiblen, G. D., Cook, J. M., Salamin, N., Machado, C. A., and Savolainen, V. (2005). 60 million years of co-divergence in the fig–wasp symbiosis. Proc. R. Soc. Lond. B Biol. Sci. 272, 2593–2599. doi: 10.1098/rspb.2005.3249
Sanogo, Y. O., Eitam, A., and Dobson, S. L. (2005). No evidence for bacteriophage WO orf7 correlation with Wolbachia-induced cytoplasmic incompatibility in the Culex pipiens complex (Culicidae: Diptera). J. Med. Entomol. 42, 789–794. doi: 10.1603/0022-2585(2005)042[0789:NEFBWO]2.0.CO;2
Schiffler, G. (2002). Fig wasps (Hymenoptera: Agaonidae) associated to Ficus mexiae Standl (Moraceae) in Lavras, Minas Gerais, Brazil. Neotrop. Entomol. 31, 653–655. doi: 10.1590/S1519-566X2002000400022
Shoemaker, D. D., Machado, C. A., Molbo, D., Werren, J. H., Windsor, D. M., and Herre, E. A. (2002). The distribution of Wolbachia in fig wasps: correlations with host phylogeny, ecology and population structure. Proc. R. Soc. Lond. B Biol. Sci. 269, 2257–2267. doi: 10.1098/rspb.2002.2100
Stouthamer, R., Breeuwer, J. A., and Hurst, G. D. (1999). Wolbachia pipientis: microbial manipulator of arthropod reproduction. Annu. Rev. Microbiol. 53, 71–102. doi: 10.1146/annurev.micro.53.1.71
Stouthamer, R., Luck, R. F., and Hamilton, W. D. (1990). Antibiotics cause parthenogenetic Trichogramma (Hymenoptera/Trichogrammatidae) to revert to sex. Proc. Natl. Acad. Sci. U.S.A. 87, 2424–2427. doi: 10.1073/pnas.87.7.2424
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tanaka, K., Furukawa, S., Nikoh, N., Sasaki, T., and Fukatsu, T. (2009). Complete WO phage sequences reveal their dynamic evolutionary trajectories and putative functional elements required for integration into the Wolbachia genome. Appl. Environ. Microbiol. 75, 5676–5686. doi: 10.1128/AEM.01172-09
Taylor, M. J., Bandi, C., and Hoerauf, A. (2005). Wolbachia bacterial endosymbionts of filarial nematodes. Adv. Parasitol. 60, 245–284. doi: 10.1016/S0065-308X(05)60004-8
Wang, G. H., Xiao, J. H., Xiong, T. L., Li, Z., Murphy, R. W., and Huang, D. W. (2013). High-efficiency thermal asymmetric interlaced PCR (hiTAIL-PCR) for determination of a highly degenerated prophage WO genome in a Wolbachia strain infecting a fig wasp species. Appl. Environ. Microbiol. 79, 7476–7481. doi: 10.1128/AEM.02261-13
Weiblen, G. D. (2002). How to be a fig wasp. Annu. Rev. Entomol. 47, 299–330. doi: 10.1146/annurev.ento.47.091201.145213
Werren, J. H., Baldo, L., and Clark, M. E. (2008). Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Microbiol. 6, 741–751. doi: 10.1038/nrmicro1969
Yang, C. Y., Xiao, J. H., Niu, L. M., Ma, G. C., Cook, J. M., Bian, S. N., et al. (2012). Chaos of Wolbachia sequences inside the compact fig syconia of Ficus benjamina (Ficus: moraceae). PLoS ONE 7:e48882. doi: 10.1371/journal.pone.0048882
Keywords: fig wasps, Wolbachia, bacteriophage WO, horizontal transfer, specificity, fig syconia
Citation: Wang NX, Jia SS, Xu H, Liu Y and Huang DW (2016) Multiple Horizontal Transfers of Bacteriophage WO and Host Wolbachia in Fig Wasps in a Closed Community. Front. Microbiol. 7:136. doi: 10.3389/fmicb.2016.00136
Received: 02 November 2015; Accepted: 25 January 2016;
Published: 15 February 2016.
Edited by:
Robert Brucker, Harvard University, USAReviewed by:
Jason Metcalf, Vanderbilt University, USANaruo Nikoh, The Open University of Japan, Japan
Copyright © 2016 Wang, Jia, Xu, Liu and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dawei Huang, huangdw@ioz.ac.cn
†Present address: Sisi Jia, Taian Entry-Exit Inspection and Quarantine Bureau, Tai’an, China
‡These authors have contributed equally to this work.
 Ningxin Wang
Ningxin Wang Sisi Jia1†‡
Sisi Jia1†‡