Corrigendum: Exploring the Secretomes of Microbes and Microbial Communities Using Filamentous Phage Display
- 1Institute of Fundamental Sciences, Massey University, Palmerston North, New Zealand
- 2Animal Science, Grasslands Research Centre, AgResearch Ltd, Palmerston North, New Zealand
Microbial surface and secreted proteins (the secretome) contain a large number of proteins that interact with other microbes, host and/or environment. These proteins are exported by the coordinated activities of the protein secretion machinery present in the cell. A group of bacteriophage, called filamentous phage, have the ability to hijack bacterial protein secretion machinery in order to amplify and assemble via a secretion-like process. This ability has been harnessed in the use of filamentous phage of Escherichia coli in biotechnology applications, including screening large libraries of variants for binding to “bait” of interest, from tissues in vivo to pure proteins or even inorganic substrates. In this review we discuss the roles of secretome proteins in pathogenic and non-pathogenic bacteria and corresponding secretion pathways. We describe the basics of phage display technology and its variants applied to discovery of bacterial proteins that are implicated in colonization of host tissues and pathogenesis, as well as vaccine candidates through filamentous phage display library screening. Secretome selection aided by next-generation sequence analysis was successfully applied for selective display of the secretome at a microbial community scale, the latter revealing the richness of secretome functions of interest and surprising versatility in filamentous phage display of secretome proteins from large number of Gram-negative as well as Gram-positive bacteria and archaea.
Introduction
Microbial secretome is a portion of the proteome comprising proteins that are targeted to the envelope of microbial cells, or are secreted into extracellular milieu. Microbial surface proteins mediate adhesion to other microbes or environmental surfaces, to facilitate colonization of an environment. They include secreted enzymes involved in breaking up various polymeric molecules to produce mono- or oligomeric foodstuffs that microbes can absorb and use as carbon and nitrogen sources for growth. Secretome harbors dominant targets of the host immune responses and are therefore of interest for vaccine development. In commensal and pathogenic bacteria these proteins are also involved in manipulating the innate and adaptive immune system’s signaling pathways. Although many surface proteins that mediate these functions have been identified in individual cultivated bacteria through genetic screens, identities of proteins that bind to specific targets of interest from archaea or from yet uncultivated microbes in complex microbial communities are largely unknown. Partial reason for this is that, with respect to primary sequence conservation, the secretome proteins are most variable group in microbial proteomes; it is rarely possible to predict the binding specificity of annotated secretome proteins using bioinformatics. In the absence of cultivation or genetic manipulation methods for a microbe, affinity screening of recombinant libraries is a suitable approach to identify proteins implicated in interactions of microbes with their hosts or environment. Phage display is a powerful combinatorial technology for affinity-selection of rare variants in vast libraries; due to the physical link between coding sequence inside the virion and protein displayed on the surface of the virion, large number of individual recombinant clones (up to 1012 per mL) can be affinity screened against complex baits of interest, such as tissues or extracellular matrix (ECM). Ff (f1, fd, or M13) phage whose virion proteins belong to the secretome and the virion itself is secreted out of Escherichia coli, is ideally suited for capture, correct folding and display of the secretome proteins. This article reviews the secretome and phage display technology applications in discovery of eubacterial and archaeal secretome proteins and emerging applications in functional metagenomics of the microbial communities’ secretomes.
Eubacterial Secretome and Mechanisms of Secretion
The term ‘secretome’, coined by Tjalsma et al. (2000), was originally proposed to refer to both the secreted proteins and components of the protein secretion machineries in bacteria. Today, the secretome is broadly described as a subset of bacterial proteome, containing the extracellular proteome (exoproteome), released to the extracellular milieu and the surface-associated proteome, either exposed to the bacterial surface or intrinsic to the external side of plasma membrane and the cell wall, but excluding integral membrane proteins and proteins intrinsic to the internal side of the plasma membrane (Desvaux et al., 2009; Zhou et al., 2010).
Secretome proteins (e.g., receptors, transporters, adhesins, complex cell structures, secreted enzymes, toxins, and virulence factors) allow bacteria to interact with, and adapt to their environment. Bacterial secretory proteins are known to be involved in processes such as: provision of nutrients through recognition; binding, degradation, and uptake of complex extracellular molecules; communication between bacterial cells; detoxification of the environment; attachment to host cells and signal transduction; while in pathogenic bacteria they also play critical roles in virulence and immunogenicity (Walsh, 2000; Antelmann et al., 2001; Tjalsma et al., 2004; Wooldridge, 2009; Dalbey and Kuhn, 2012). Secretome proteins have been reported to occupy 10–30% of the total coding capacity of bacterial genomes (Wallin and Heijne, 1998; Kudva et al., 2013).
Secretome includes multi-protein surface appendages such as pili and flagellae, which have pivotal roles in bacterial attachment, horizontal gene transfer, and motility (Van Gerven et al., 2011). The pathways for membrane targeting are also “hijacked” by the filamentous phage or inoviruses, whose virion and assembly proteins belong to the secretome and which are secreted from bacteria without killing the host (Russel and Model, 2006; Rakonjac et al., 2011; Mai-Prochnow et al., 2015).
Secretion Pathways of Bacteria
The cell envelope of Gram-positive bacteria consists of a single, cytoplasmic membrane and a cell wall, comprised of a thick peptidoglycan layer cross-linked with different molecules, such as capsular polysaccharides, cell wall teichoic acids, and proteins (Freudl, 2013). In contrast, Gram-negative bacteria are enveloped by inner (cytoplasmic) and outer membranes. The presence of two membranes defines an additional subcellular compartment (the periplasmic space), containing a thin meshwork of peptidoglycans. Some Gram-positive bacteria also have a distinctive thin granular layer (inner wall zone) between the membrane and the mature cell wall, equivalent to the periplasmic space in Gram-negative bacteria (Zuber et al., 2006).
In order to be anchored to the cell surface or released into the extracellular milieu, secretome proteins must be translocated across one or more biological membranes (Desvaux et al., 2006). Transport of proteins into or across biological membranes (translocation), catalyzed by membrane-bound proteinaceous transport machineries, is a universal event in the protein secretion mechanism, and it can occur several times during the course of secretion (Desvaux et al., 2009). Once a secreted protein is translocated across the outermost membrane, it can remain anchored (covalently or non-covalently associated with cell-wall components in Gram-positive bacteria or outer membrane components in Gram-negative bacteria), assemble into macromolecular structures on the cell surface (flagella, pili), be injected into host cells, or released to the extracellular milieu.
A remarkable array of systems for export of proteins have been described in Gram-positive and Gram-negative bacteria. Descriptive names are used in the nomenclature of systems involved in protein translocation across cytoplasmic membranes of both Gram-negative and Gram-positive bacteria, while an alphanumerical system has been adopted for naming protein secretion systems of Gram-negative bacteria (Desvaux et al., 2009).
Systems that are universally involved in protein translocation across the cytoplasmic membrane, and encoded in both Gram-positive and Gram-negative bacteria are: the conserved general secretion (Sec) system, YidC insertase, the twin-arginine translocation (Tat) system and hole-forming pathway via holins (Desvaux et al., 2009).
The Sec system is a major secretory pathway for protein insertion into the inner (cytoplasmic) membrane, and is conserved in all eubacteria. It is also ubiquitous in archaea, and the membranes of eukaryotic endoplasmic reticulum and chloroplasts (Szabo and Pohlschroder, 2012). This system also plays a key role in further transport of some proteins into the periplasmic space, outer membrane (e.g., lipoproteins and beta barrel proteins), or their assembly into the surface-associated structures (e.g., pili subunits). Furthermore, some of the components of the specialized secretion systems in Gram-negatives and their substrates (proteins transported via these secretion systems) are initially transported across the inner membrane by the SecYEG translocon (Beckwith, 2013; Kudva et al., 2013).
In bacteria, the Sec system is composed of the SecYEG translocon and three major accessory systems that target the secretome proteins to the translocon: SecB/A, SRP/FtsY, and YidC. SecYEG is an evolutionarily conserved heterotrimeric protein complex, and its SecY subunit forms an hourglass-shaped aqueous protein transport channel embedded in the inner membrane (Dalbey and Kuhn, 2012; Kudva et al., 2013). The translocon transiently interacts with different proteins during the transport process (e.g., SecA, FtsY, SecDF). SecA, a post-translational pathway motor protein accepts the substrate protein delivered by the cytosolic targeting factor SecB, and pushes it through the translocon in a stepwise and ATP-dependent manner (Lycklama et al., 2012). FtsY, the SRP-receptor, occupies the ribosome binding site (RBS) of SecY until its displacement by the translating ribosome during co-translational targeting (Kudva et al., 2013). The membrane-integrated SecDF chaperone uses proton-motive force to power ATP-independent protein translocation through the SecYEG channel (Tsukazaki et al., 2011).
In addition to universal secretion systems, Gram-positive bacteria possess Wss (WXG100 secretion systems), accessory Sec systems (SecA2-only and SecA2/SecY2 export pathways), flagella export apparatus (FEA), the fimbrilin-protein exporter (FPE), ABC protein exporter and Sec-dependent sortases. In Gram-positive bacteria, secreted proteins have several different fates. They are transported across the cytoplasmic membrane and then secreted into the extracellular milieu by SecYEG, Tat, holin, or Wss, in addition to being attached (covalently or non-covalently) to the cell wall using the sortase or assembled into the cell surface appendages via Sec pathway (e.g., cellulosomes or pili), via FPE (e.g., competence pseudo-pili), or via FEA (e.g., flagella).
Due to the added complexity of their cell envelope, at least two additional systems for targeting proteins to the outer membrane and eight additional systems for secretion of proteins outside of the cell have been described in Gram-negative bacteria. After Sec- or Tat- dependent translocation across the inner membrane, outer membrane-specific lipoproteins and unfolded β-barrel proteins are targeted to the outer membrane via the Lol pathway and β-barrel assembly machinery (BAM) pathway, respectively (Dalbey and Kuhn, 2012).
Secreted proteins targeted to the extracellular milieu, or to another cell, can be exported out of the cell directly, or by a two-step secretion process via type 1–6 secretion systems (T1SS–T6SS). In addition, the chaperone-usher system (CU or T7SS), the extracellular nucleation-precipitation mechanism (ENP or T8SS) system, as well as type IV pilus biogenesis (T4PBS) and tight-adherence (Tad) piliation systems are dedicated to exporting different types of pili subunits across the outer membrane (Chagnot et al., 2013).
The direct (Sec pathway-independent) secretion in Gram-negatives exports proteins through a contiguous secretion machinery spanning two membranes and the periplasm (T1SS, T3SS, T4SS, and T6SS systems). The two-step secretion process involves protein export to the periplasm by the Sec or, less frequently the Tat pathway, followed by export across outer membrane via T2SS, T5SS, T7SS, or T8SS systems. T1SS and T5SS are relatively simple systems involving few proteins, while T2SS, T3SS, T4SS, and T6SS are complex structures composed of large number of subunits, and spanning the entire bacterial cell envelope (Chagnot et al., 2013).
The filamentous phage assembly system follows a variant of the two-step secretion process, where all virion proteins are first inserted into the inner membrane, followed by the export-coupled assembly of the phage. The assembly is initiated by minor proteins pVII and pIX that interact with a specific phage genome sequence called the packaging signal, followed by rapid elongation by addition of major coat (pVIII) subunits (Rakonjac et al., 2011; Marvin et al., 2014). The virion proteins egress from the inner membrane to form a filament by attaching to double-helical DNA genome via few positively charged C-terminal residues that face the cytoplasm prior to assembly. The assembly process is catalyzed by phage-encoded inner membrane ATPase and requires ATP and proton motive force (Feng et al., 1997); the filament is released from the assembly site by two minor proteins, pIII and pVI (which themselves are integral membrane proteins) when the DNA is completely covered with pVIII (Rakonjac et al., 1999). The resulting filament has no phospholipids and has high temperature (70°C) and detergent (1% Sarkosyl) resistance (Crissman and Smith, 1984; Branston et al., 2013). Whereas over 70 filamentous phage of Gram-negative bacteria have been identified, only two were found in Gram-positives (Day, 2011).
Secretion and Membrane Targeting Signals
The first stage in the secretome protein export is sorting and targeting of proteins to the cytoplasmic membrane, followed by membrane crossing and maturation/release of the translocated protein. The sorting process, through which proteins are directed to their specific subcellular compartments, is based on localization information contained in a short amino acid sequence that acts as a protein sorting signal (‘zip code’) governing protein traffic, transport, and localization in the cell (Blobel and Sabatini, 1971). Discrimination between secreted and cytoplasmic proteins is based on the presence of membrane-targeting sequences, such as signal sequences and transmembrane α-helices that are recognized by distinct secretory pathway-associated molecular chaperones, and are necessary for correctly targeting secreted proteins to the translocation pathway.
Most secreted proteins contain N-terminal signal sequences that are cleavable. The function and overall structure of the signal sequence, transmembrane alpha helix targeted to the SecYEG translocon, are conserved in all domains of life; however, these peptides lack primary sequence homology even within a species (Rusch and Kendall, 2007). Several types of signal sequences have been described: type I (classic) signal sequence, type II (lipoprotein) signal sequence, Tat signal sequence, type IV (pseudopilin-like) signal sequence and bacteriocin/pheromone signal sequence. Based on hydrophobicity and charge, most signal sequences have a conserved overall tripartite organization consisting of an hydrophobic core (h-domain), flanked by hydrophilic positively charged N-terminal region (n-domain) and a polar C-terminal region (c-domain) with cleavage/retention sites (Rusch and Kendall, 2007; Driessen and Nouwen, 2008; Zhou et al., 2008). However, the type IV and bacteriocin/pheromone signal sequences do not precisely follow such a structural layout (Zhou et al., 2008).
Signal sequences are usually removed during or shortly after their translocation across the membrane by several types of membrane-associated signal peptidases (SPases), which also have a role in quality control and regulated turnover of exported proteins (Dalbey et al., 2012). In bacteria, precursor proteins that are translocated through the Sec and Tat-pathways apart from pre-proteins (Lüke et al., 2009) are proteolytically processed by a ‘general’ type I signal peptidase (SPaseI) (Lüke et al., 2009; Auclair et al., 2012). Processing of the lipoprotein signal sequences is performed by type II lipoprotein signal peptidase (SPaseII). The lipoproteins are transported across the inner membrane in a Sec-dependent manner (Okuda and Tokuda, 2011). The Tat-dependent export of lipoproteins has only been demonstrated in streptomycetes (Thompson et al., 2010; Dalbey and Kuhn, 2012). The prepilin signal peptidase (SPaseIV) is responsible for processing proteins containing type IV signal sequence, such as pilins and related pseudopilins, that have mainly Sec-dependent export across the inner membrane (Peabody et al., 2003; Arts et al., 2007; Francetic et al., 2007).
Among the Ff (f1, M13, or fd) filamentous phage virion proteins, major coat protein pVIII that forms body of the filament (Figure 1) has a type I signal sequence and is secreted by SecYEG translocons and, YidC (Samuelson et al., 2000); it is targeted to the inner membrane by a C-terminal transmembrane helix. The length of the mature pVIII protein (after signal sequence cleavage) is 50 amino acids. Once assembled into the viral particle, it is DNA-bound and helically arranged to form the shaft of the filament (Figure 1). The four minor proteins are all integral membrane proteins, however, only the largest, pIII (406 aa) has a type I signal sequence and its membrane targeting is SecYEG/SecAB-dependent (Chang et al., 1978). Minor protein pVI (112aa) has three predicted transmembrane helices, but no signal sequence. The remaining two minor proteins, pVII (33 aa) and pIX (32 aa) are very small and hydrophobic, each containing a transmembrane helix, but no signal sequences.
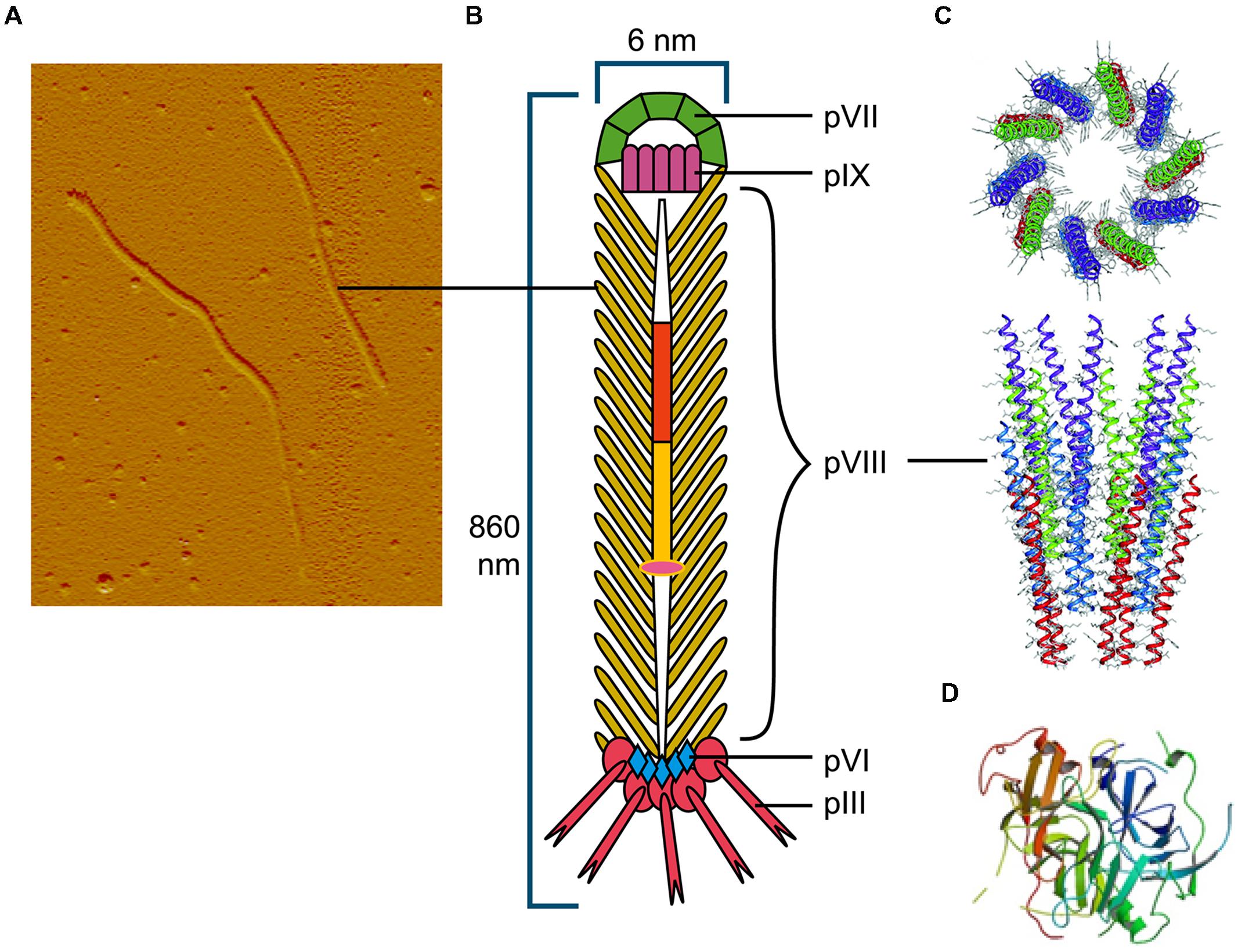
FIGURE 1. The Ff bacteriophage structure and virion proteins used most commonly in phage display. (A) Ff virion visualized by atomic force microscope (M. Russel and P. Model, sample prepared by J. Rakonjac). (B) Schematic diagram of Ff bacteriophage. (C) Ribbon representations (top and side view) of the pVIII coat protein (RCSB PDB database accession number 2cOw; (Marvin et al., 2006) arranged around bacteriophage single stranded DNA (not shown). (D) Ribbon representation of N1 and N2 domains of pIII (RCSB PDB database accession number1g3p; Lubkowski et al., 1998).
Methods for Study of the Secretome
Mining bacterial secretomes is important for a range of applications, including identification of novel enzymes, understanding bacterial adhesion and their interactions with the environment, investigating pathogenic mechanisms, epitope mapping and identification of new vaccine candidates. Secretomes are traditionally studied in vitro, using biochemical and proteomics approaches, and in silico, using bioinformatic tools. Surface display screening methods and reporter fusion systems (Georgiou et al., 1997; Chen and Georgiou, 2002; Åvall-Jääskeläinen et al., 2003; Lee et al., 2003; Wernérus and Ståhl, 2004; Liu et al., 2013), as well as phage-display based systems (Rosander et al., 2002, 2011; Jacobsson et al., 2003; Bjerketorp et al., 2004), described in detail in section 3, have also been used for screening, identifying, and characterizing secretome proteins.
Secretomes are studied in vitro using high-resolution separation (2D gel electrophoresis and/or liquid chroma tography) of secreted or extracted membrane proteins, coupled with mass spectrometric methods for the identification of peptides and proteins in the sample (Yang et al., 2012). Biochemical approaches for elucidating the secretome of a microorganism allow direct functional characterization of identified proteins; however, they are very tedious and limited only to cultivable bacteria. Furthermore, construction of a proteome map of surface-associated and membrane proteins can be hindered by technical limitations of protein extraction from the membranes. In the absence of experimental data, a secretome can be deduced from a completely sequenced genome in silico, using bioinformatic tools for the prediction of secretome proteins based on their specific conserved features.
Computational methods for secretome protein prediction are based on weight matrices, sequence alignment or machine learning algorithms, and can be roughly grouped into global tools for subcellular protein localization prediction, and specialized tools for the prediction of signal sequences (Goudenège et al., 2010; Caccia et al., 2013). More sophisticated machine learning algorithms, based on neural networks and decision trees, support vector machines, Bayesian networks, HMMs, or their combination, are now more prominently used for discriminating secreted and non-secreted proteins (Zhou et al., 2008; Choo et al., 2009). During the training phase, typical signal and non-signal peptides are presented to the algorithm, and a classification model is subsequently built. Tools for signal sequence prediction such as SignalP (Petersen et al., 2011), LipoP (Juncker et al., 2003), TMHMM (Krogh et al., 2001), PRED-LIPO (Bagos et al., 2008), PRED-TAT (Bagos et al., 2010), SecretomeP (Bendtsen et al., 2005), and tools for subcellular protein localization prediction, such as PSORTb (Yu et al., 2010) or TargetP (Emanuelsson et al., 2000) belong to this class.
In bacteria, classically secreted proteins can be predicted based on recognition of the tripartite organization of their N-terminal, cleavable signal sequences, and conserved amino acid residues at the –3 and –1 positions relative to the cleavage sites. In addition to these, the lipobox of type II signal sequences and the Tat motif in Tat signal sequences are highly amenable to identification by bioinformatic tools, while transmembrane α-helices can be identified based on their hydrophobicity (Rahman et al., 2008; Goudenège et al., 2010). Recognition of the SPaseIV cleavage motif is not sufficient for the accurate detection of type IV signal sequences, since these have no tripartite structure like other Sec-dependent substrates. It was demonstrated that the specificity of searches for type IV pilin-like proteins may be enhanced by including additional search requirements, such as the presence of 14 sequential uncharged amino acid residues immediately after the cleavage motif or presence of a single transmembrane helix within 50 amino acid residues of the N-terminus, since true pilins contain only one transmembrane helix, typically close to the cleavage motif (Imam et al., 2011).
Cleavable N-terminal signal peptides of secreted proteins are readily distinguishable from longer hydrophobic N-terminal transmembrane helices of transmembrane proteins. In contrast, their discrimination from uncleaved N-terminal signal anchors, which tether some of these Sec-exported proteins to the membrane, is often problematic (Natale et al., 2008; Zhou et al., 2008; Caccia et al., 2013). However, tools such as SignalP 4.0 are trying to overcome this challenge by combining predictions of transmembrane protein topology with signal sequence identification.
The disadvantages of in silico secretome analysis is that it can be only applied to organisms with sequenced genomes; its accuracy depends on prediction algorithm performance, as well as on genomic annotation accuracy. Therefore, to improve the identification of secretome proteins, genomic predictions need to be integrated with transcriptomics and proteomics data (Caccia et al., 2013). The task of predicting the metasecretomes of complex environmental microbial communities is even more challenging. This is due to current limitations in the identification of complete genes via sequence-based metagenomics approaches from low-coverage metagenomic assemblies derived through next-generation sequencing of complex environmental microbial communities, often containing numerous closely related microbial species (Hess et al., 2011; Luo et al., 2011).
Despite the versatility of bioinformatics approaches in predicting the targeting sequences and cellular location of proteins, these methods are not capable of predicting exact functions that are essential for understanding vital and specific functions such as interacting with partners in the microbes’ surroundings. One experimental method that is well-suited for finding genes that fulfill the functions of interest is expression library screening and display of secretome proteins on the surface of bacteria or filamentous phage as recombinant fusion proteins. This method ensures that protein folding occurs under similar conditions to those where these proteins naturally fold – on cell surfaces. Bacterial cell surface display and yeast surface display are described elsewhere (Georgiou et al., 1997; Chen and Georgiou, 2002; Åvall-Jääskeläinen et al., 2003; Lee et al., 2003; Wernérus and Ståhl, 2004; Liu et al., 2013); this review focuses on filamentous phage display.
Phage Display
The physical link between phenotype and genotype of a (poly) peptide displayed on the surface of the virion, the high replication capacity of bacteriophage and subsequent affinity selection are the elements that underpin phage display technology. In phage display, a very large repertoire of recombinant phage particles displaying (poly) peptides can be generated (1012–1013 different clones) at very low cost using simple methods of microbiology and molecular biology. Nucleotide sequence repertoires such as cDNAs, synthetic oligonucleotides, genomic DNA fragments derived from single organisms or metagenomes, and mRNAs are cloned directly into phage display vectors. Display of peptides encoded by cloned sequences is achieved by translational fusion of a protein or a library of proteins of interest to any of the five structural virion proteins, pVIII, pIII, pVI, pVII, or pIX at N- or C-terminus, although the pIII and pVIII proteins are used most frequently (Russel et al., 2004; Kehoe and Kay, 2005). Peptides fused to the Ff virion proteins fold in the periplasm of E. coli (Figure 2A), therefore display on filamentous phage is suitable for surface and secreted proteins.
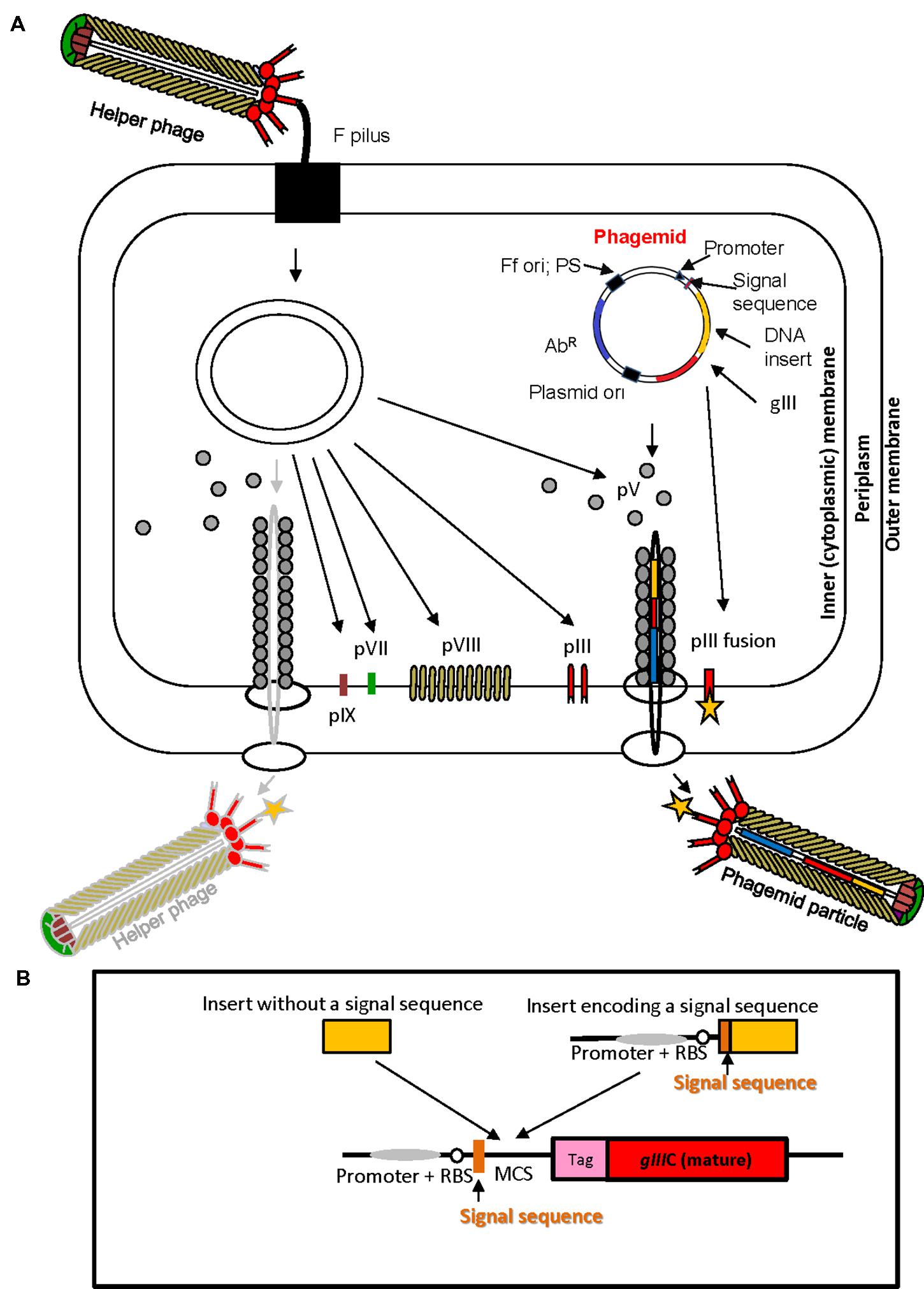
FIGURE 2. Ff virion protein targeting, virion assembly, phagemid display system, and display cassette in phagemid vectors. (A) In Escherichia coli phagemids can be replicated as plasmids or alternatively, in the presence of a helper phage, packaged as “transducing” or “phagemid” particles (PPs). Phagemid encodes phage protein pIII as a fusion partner for display. The resulting phagemid particles may incorporate either pIII derived from the helper phage (red lollipop-like structure) or the polypeptide-pIII fusion protein (red lollipop-like structure decorated with a yellow star), encoded by the phagemid. AbR, antibiotic resistance marker, Ff ori; PS, filamentous phage origin of replication and packaging signal; Plasmid ori, plasmid origin of replication; gIII, gene III; pIII, pV, pVI, pVII, pVIII, and pIX, filamentous phage proteins. Phagemid particles are produced at a 10–100-fold excess over the helper phage (denoted by solid vs. faded lines for phagemid particle vs. helper phage. (B) Typical phage display cloning cassette in a phagemid vector: promoter, a ribosome binding site (RBS), signal sequence (commonly used PelB signal sequence from Erwinia carotovora (Lei et al., 1987); multiple cloning site (MCS), affinity tag (tag), and sequence encoding the mature portion (C-terminal domain) of pIII (gIIIC; required for assembly of the fusion into the virion). Two types of inserts derived from fragmented bacterial or archaeal genomic DNA are shown above the cloning site. Insert without signal sequence, in order to result in displayed peptide, has to correspond to a CDS that is in frame with the upstream signal sequence and downstream gIII (encoding the C-terminal domain). It is typically truncated at the 5′ and 3′ ends to avoid stop codons that terminate translation. A second type of insert, that contains signal sequence, can be displayed if the CDS is truncated at the 3′ end, and is in frame with gIII. The latter type of inserts typically carries its own promoter and RBS. E. coli host strains that contain suppressor mutations (such as supE44) can read through amber stop codons at 50% efficiency and result in display of fusions that include this codon.
In contrast to Ff phage proteins, “tailed” bacteriophages λ, T7, T4, and P4 fold in E. coli cytoplasm and are therefore most commonly used for the display of cytoplasmic proteins. The distinction between cytoplasmic and extracytoplasmic environments mostly refers to oxidative state of Cys residues; which are nearly always reduced in cytoplasmic (or nuclear) proteins, whereas they are oxidized in the periplasm of Gram-negative bacteria or membranous organelles in eukaryotic cells. The readers are referred to several reviews for the latest developments in tailed phage display (Krumpe et al., 2006; Beghetto and Gargano, 2011; Gamkrelidze and Dabrowska, 2014).
Despite the enormous body of reports describing display of a variety of proteins on pIII and pVIII display platforms, there are still many proteins that are recalcitrant to functional display due to host restrictions such as codon usage, protein folding and toxicity to the E. coli host. To some degree folding constraints can be alleviated by introducing changes to the host periplasm environment, for example using ΔdsbA E. coli strains that do not form S–S bridges between Cys residues to allow folding and display of cytoplasmic proteins, or co-expressing with periplasmic chaperones such as Skp and FkpAto facilitate folding of the fusion counterpart (Hayhurst and Harris, 1999; Gunnarsen et al., 2013), or using alternative signal sequences or translocation routes via SRP-dependent or indirect Tat phage display systems (Steiner et al., 2008; Speck et al., 2011).
The non-specific binding to matrices where bait is immobilized can be decreased by “wrapping” the phage in desired charge that is repulsive to the matrix. For example, fusing Lys8 peptide to all major coat protein subunits can be used for positively charged surfaces (Lamboy et al., 2008). To overcome limitations of using only 20 amino acids for the construction of peptide or protein libraries, phage display technology was adapted to incorporate unnatural amino acids by using host strains that express mutant tRNAs and aminoacyl-tRNA synthetases (Tian et al., 2004; Bratkovic, 2010; Bernard and Francis, 2014).
Recently, the scope of phage display extended toward the display of polypeptides containing posttranslational modifi cations, including phosphorylation, phosphopantetheinylation (Yen and Yin, 2007) and glycosylation (Celik et al., 2010; Dürr et al., 2010; Ng S. et al., 2015). As glycosylation is the most frequent posttranslational modification [>50% of total eukaryotic proteins and an increasing number of archaeal and bacterial proteins (Abu-Qarn et al., 2008)], modification of displayed proteins has promising future applications.
Filamentous Phage Display Types
There are two general types of filamentous phage display based on whether the library is constructed in Ff phage vectors or in specially modified plasmids called phagemids in conjunction with helper phage. The advantage of a phage vector system is in its simplicity. The library constructed in a simple Ff phage vector results in fusion gene products being displayed by all copies of a particular virion protein. Given that some protein fusions in a library are likely to interfere with assembly and infectivity, they are reported to be counter-selected during the phage life cycle and are lost from the library (Rodi and Makowski, 1999; Derda et al., 2011). Insertion of a second copy of a particular virion protein-encoding gene is one strategy to overcome the interference (Barbas et al., 2001). For pIII fusions, the inter-domain display is solution to prevent censorship due to proteolytic degradation of peptides (Tjhung et al., 2015).
Another type of phage display system that allows more flexibility and provides independence from phage assembly in the amplification phase is based on phagemid vectors (Figure 2). Phagemids are plasmid vectors that contain the Ff origin of replication, packaging signal, a plasmid origin of replication, and a selective (antibiotic resistance) marker. Phage display phagemid vectors in addition contain a display cassette, allowing construction of fusion to a virion protein. The Ff origin of replication, packaging signal and the display cassette allow packaging of the phagemid genome into phage-like “phagemid particles” (PPs) and display of phagemid-encoded protein fusion on the surface of the particle (Figure 2). A helper phage is the obligatory source of proteins necessary for replication from Ff origin and assembly of a complete virion.
Helper phage typically have an interference-resistant Ff origin of replication and/or truncated packaging signal, or additional (plasmid) origin of replication, allowing preferential replication and packaging of the phagemid single stranded DNA so that the majority (~90%) of secreted virions are PPs rather than the helper phage (Barbas et al., 2001). Helper phage that encode all virion proteins and contribute them to the PPs are herein referred to as a wild-type helpers. The phagemid/wild-type helper system typically results in monovalent display of recombinant protein fusions to minor virion proteins (pIII, pVII, pIX, and pVI); i.e., PPs are mosaic for recombinant and wild-type capsid proteins produced by phagemid encoding the fusion protein and by a helper, respectively (Figure 2A). To increase the copy number of displayed fusion proteins, helper phage containing deletion of the gene encoding the virion protein that is expressed from the phagemid can be used. In the case of most frequently used phagemid vectors that encode virion protein pIII, helper phage that carry deletion of corresponding gene (e.g., ΔgIII) are used. The obtained virions exhibit polyvalent display (Griffiths et al., 1993; Rakonjac et al., 1997; Joo et al., 2008). Additional refinements of some phage display phagemid vectors are peptide tags (e.g., c-myc or E-tag), followed by an amber stop codon between the insert and downstream pIII. These additions allow a soluble version of foreign protein to be produced if phagemids are transformed into an appropriate E. coli strain that does not contain an amber codon suppressor (Hoogenboom et al., 1991).
Affinity Selection of Binding Targets – Bio-Panning
When a library of fusion proteins is constructed and displayed on the phage surface, a recombinant phage clone displaying a certain binding affinity can be selected from the majority of other (non-binding) recombinant phages present in a library, by an affinity selection procedure known as bio-panning (Figure 3) (Parmley and Smith, 1988). Through successive rounds of binding, washing, elution, and amplification, an originally very diverse phage display library, up to 1012 variants (Schier et al., 1996) is increasingly enriched for the phage library clones with a propensity to bind to the target molecule. Ultimately, monoclonal phage populations with desired specificities can be identified using sequencing and analyzed using affinity binding assays.
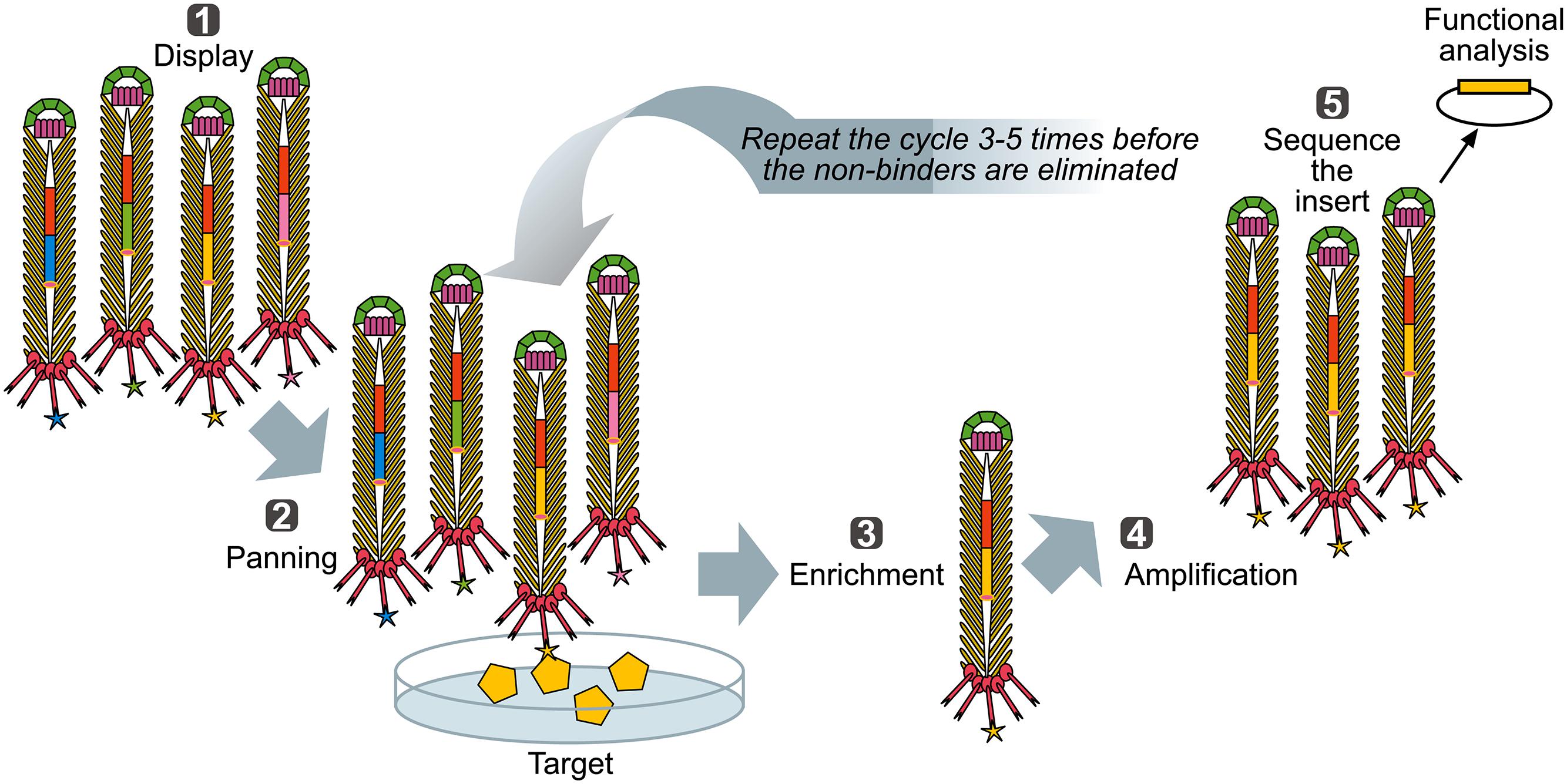
FIGURE 3. Biopanning – a basic selection for binding peptides. (1) Display: Filamentous phage displaying variants of proteins/peptides/antibodies is created, cloned into phage, or phagemid vectors as fusions to a coat protein gene(s) and displayed on the surface of the virions. (2) Panning: The phage library displaying variant peptides or proteins (different-color stars) is exposed to immobilized ligand (yellow pentagon) and phage with appropriate binding specificity is captured (yellow star). (3) Enrichment: Non-binding phages are washed off and bound phage(s) is (are) eluted by conditions that disrupt the peptide-ligand interactions, leading to enrichment for a specific binder. (4) Amplification: Eluted phage is then amplified by infection of a suitable E. coli strain. This amplified phage population is greatly enriched in recombinant phage clones displaying peptides that bind to the target. The biopanning steps (two to four) are repeated for several (three to five) rounds, ultimately resulting in a clonal population of recombinant phages that bind to the target used for affinity panning of the library. Captured putative binder can then be identified by sequencing (5) and functionally analyzed.
Biopanning has potential to identify proteins that bind to enormous diversity of ligands, from extremely complex, including the whole animal (Arap et al., 1998), ex vivo tissues (Antonara et al., 2007), complex mixture of organisms (Ng F. et al., 2015) and whole cells (Fevre et al., 2014) to very simple, such as purified proteins, peptides, nucleic acids, carbohydrates (Zwick et al., 1998; Rodi and Makowski, 1999) or inorganic powders (Mao et al., 2004; Ploss et al., 2014). The outcome of biopanning, however, depends on library complexity (or primary size, equivalent to the number of different variants or recombinant inserts), functionality of displayed fusions and affinity of interaction with the bait used in the biopanning. For isolation of high-affinity binders, monovalent phage display libraries are usually used because they allow selection based on strong affinity. In contrast, multivalent display, in particular on the major coat protein (pVIII), is more suitable for selection of low-affinity binders since the avidity of the virion compensates for low affinity of individual peptides (Shallom and Shoham, 2003).
Phage Display Applications in Functional Identification of Microbial Secretome Proteins
Microbial interactions with the environment have beneficial or detrimental effects on microorganisms involved in interactions or/and the host. In general, these interactions are mediated via microbial secretome proteins that interact with receptor molecules on host cells or other microbes, or with the host cell surface or proteins of the host signaling pathways involved in the immune response. Phage display has been used extensively for identification of bacterial proteins that interact with the host surfaces or are dominant immunogens recognized by host antibodies (Mullen et al., 2006). Bacterial and more recently archaeal phage display libraries are constructed in phagemid vectors, as fusions to pIII or pVIII. Given the lack of introns in bacterial coding sequences, fragmented genomic DNA is used for construction of libraries. These are ligated to a vector that contains a multiple cloning site (MCS) between the signal sequence and the mature portion of gene III or VIII (Figure 2B). If a fragment is ligated such that the coding sequence is in the same direction and frame with the vector sequence encoding both the upstream signal sequence and downstream mature portion of protein, the encoded peptide will be displayed on the surface of phage. This type of fusion can display both the cytosolic and secretome proteins. Alternatively, if a CDS segment encoding a signal sequence is inserted in frame with the mature portion of pIII or pVIII (but not the vector-encoded signal sequence), it is also displayed on Ff. This type of recombinant clone will often contain the native promoter and RBS derived from the genomic DNA. Given the higher probability of a single in-frame joint than that of double, the secretome clones are expected to be reasonably frequent in the shot-gun filamentous phage display libraries. All clones that have inserts whose translated peptide is displayed on the phage surface can be pre-selected by virtue of peptide tags engineered into the mature portion of pIII (Figure 2B). Both types of fusions can be selected in the shot-gun phage display library screenings (Gagic et al., 2013; Ng F. et al., 2015).
Phagemids that rely on pIII for bacterial phage display library construction contain only the coding sequence for the C-terminal domain. The reason for this is that the pIII N-terminal domain expression results in resistance of phagemid-containing cells to helper phage infection, if a promoter sequence is present in the insert (Jankovic et al., 2007).
In the past two decades, particularly in the area of infectious diseases, phage display has proven to be a powerful technology for identification of genes encoding bacterial adhesins and characterization of adhesin domains that mediate interactions with host cells and ECM components such as fibronectin, fibrinogen, collagens, vitronectin, laminin, and heparin sulfate (Mullen et al., 2006). In pathogenic bacteria, microbial adhesion precedes colonization and internalization into the host cell and increases resistance to host defenses. Global spread of antibiotic-resistant pathogenic bacteria commands increased efforts to find new antigenic epitopes for vaccine development and alternative therapies that include interference with microbial adhesion or prevention of immune manipulation by pathogenic bacteria. Shot-gun filamentous phage display (display of random fragments of bacterial genomic DNA) was first exploited by Jacobsson and Frykberg (1996) to identify Staphylococcus aureus proteins that interact with ECM components and host serum or plasma. Following this initial publication many genes encoding proteins involved in host-microbial interactions from a number of bacterial species have been identified using phage display [Table 1; (Ausmees et al., 2001; Lauterbach et al., 2003; Benedek et al., 2005; Mullen et al., 2006; Antonara et al., 2007; Posadas et al., 2012; Evangelista K. V. et al., 2014]. A shot-gun phage display library of Borrelia burgdorferi was used for the in vivo screening for potential adhesins, resulting in discovery of at least five new adhesion proteins (Antonara et al., 2007). Phage display allows not only identification of a gene of interest, but also domains that mediate binding, by identifying a consensus among the library inserts that bind to a bait. For example, this approach was used to identify and map von Willebrand factor binding protein from pathogenic Staphylococcus aureus (Bjerketorp et al., 2002) and novel neutrophil-binding proteins of S. aureus that inhibit neutrophil recruitment to the site of infection (Fevre et al., 2014). Phage display has recently been used for identification of antigenic determinants of Salmonella enterica sv. Typhimurium (Meyer et al., 2012).
Most bacteria in the environment, as well as commensal bacteria that colonize eukaryotic organisms, use adhesins for interactions with inorganic environment, other microbes and multicellular hosts. In comparison to the plethora of adhesins from pathogenic bacteria that we recognize today, adhesins from non-pathogenic bacteria are as numerous; however, their interacting partners are largely unknown. In one example of phage display application in symbiotic bacteria, it was used to identify and map the cell-surface-associated agglutinin (RapA) from legume root nitrogen-fixing bacterium, Rhizobium leguminosarum (Ausmees et al., 2001). Another large bacterial group of interest are probiotics. Although benefits of probiotic bacteria to human health are still debated, adhesion to gut mucosal surface is a mechanism by which they can persist in gut and also may preclude attachment of enteropathogenic bacteria and viruses to the host’s intestinal epithelial cells, possibly leading to beneficial effect. Several proteins from probiotic Lactobacillus casei binding to collagen and fibronectin have been identified (Munoz-Provencio and Monedero, 2011), as well as a Lactobacillus rhamnosus surface protein complex have been identified by affinity-screeing of phage display libraries (Gagic et al., 2013). Surface proteins from probiotic bacteria Lactobacillus reuteri and Lactobacillus rhamnosus at genome level have also been displayed using secretome-specific phage display method to obtain clone banks of displayed surface and secreted proteins suitable for binding and functional assays (Wall et al., 2003; Jankovic et al., 2007; Gagic et al., 2013).
Microbe-microbe interactions in complex environments and secretome proteins involved in these interactions are another research area of interest in recent years. For example in fermentative forestomach (rumen) of ruminants protozoa and specific group of archaea, methanogens, can be found in association. This interaction may facilitate hydrogen transfer from protozoa (hydrogen producers) to archaea, which use hydrogen to produce methane (Ushida et al., 1997; Belanche et al., 2014). Recently Ng F. et al. (2015) used shot-gun phage display in a phagemid/helper phage platform to identify a secretome protein Mru_1499, from rumen methanogenic archaeon Methanobrevibacte rruminantium M1 (Leahy et al., 2010) as a binder to a broad range of rumen protozoa and rumen bacterium Butyrivibrio proteoclasticus, suggesting a broad adhesion spectrum for this protein. Interestingly, the library insert contained an archaeal signal sequence and the 5′ portion of the Mru_1499 gene fused to the mature portion of pIII. Display and selection of this recombinant phagemid from the library therefore showed that archaeal signal sequences are functional in E. coli. Protozoa isolated from the sheep rumen were used as bait for the library panning to identify Mru_1499, and in turn the phagemid particles displaying Mru_1499 were used as bait to identify, among rumen protozoans, those that interact with this protein, by sequencing the 18S rRNAgene from the captured protozoa (Ng F. et al., 2015).
Selective Display of Bacterial Secretome
A standard strategy for identifying targets for vaccine development in the genomics era is bioinformatic identification followed by expression of each individual bacterial secretome protein. This involves cloning and expression of up to several hundred surface proteins and testing their immunogenicity. Selective display of the secretome was proposed as an alternative to this path to allow, in one cloning step, the production of a library and/or clone bank of those recombinant phagemids that display surface-associated and secreted proteins. Secretome-selective phage display methods exploit the requirement of a signal sequence to target a fusion protein that is in frame with the mature portion of pIII to E. coli inner membrane (Figure 4). The phagemids used in these conditional display strategies contain C-terminal domain of pIII without a signal sequence. For assembly of signal sequence-encoding inserts (Rosander et al., 2002) used gIII-positive helper phage and for selection, biopanning based on the presence of a vector-encoded affinity tag incorporated into the fusions that were displayed on the phagemid particles thanks to a functional signal sequence. In the second method, a helper phage with gIII deletion was used for assembly of the particles (Jankovic et al., 2007). Here, the selection was based on removal of pIII-deficient phagemid particles which are structurally unstable and easily disassembled by detergent sarkosyl (Rakonjac and Model, 1998; Rakonjac et al., 1999). This approach was further used to identify six secretome proteins from Mycobacterium tuberculosis by subtractive panning between the sera of M. tuberculosis patients and BCG-vaccinated healthy subjects (Liu et al., 2011), three of which have not been identified prior to this study and which therefore were novel vaccine candidates. Interestingly, the breakdown of targeting sequences that were able to guide the fusion to the inner E. coli membrane and allow assembly of the virion included not only type I signal sequences, but also type II (lipoprotein) and type IV pre-pilin signal sequences as well as transmembrane helices. Moreover, a few “moonlighting” proteins that do not contain SecYEG-dependent signal sequences were identified among the selected fusions (Jankovic et al., 2007; Liu et al., 2011). Together with chance identification of an archaeal-signal-sequence-driven secretome protein fusion reported by Ng F. et al. (2015), the findings that signal sequences and transmembrane helices from Gram-positive bacteria, Mycobacteria and Archaea guide the pIII fusion protein to correctly insert into the inner membrane of E. coli and be processed to be displayed on the surface of the virion, indicate great plasticity in processing of the SecYEG-translocated pIII fusions. With respect to transmembrane helices and signal sequences other than type I, this process most likely involves periplasmic proteases other than signal peptidases, and in the case of moonlighting proteins it could involve SecYEG- and signal-sequence-independent translocons such as ABC transporters. These hypotheses remain to be experimentally verified.
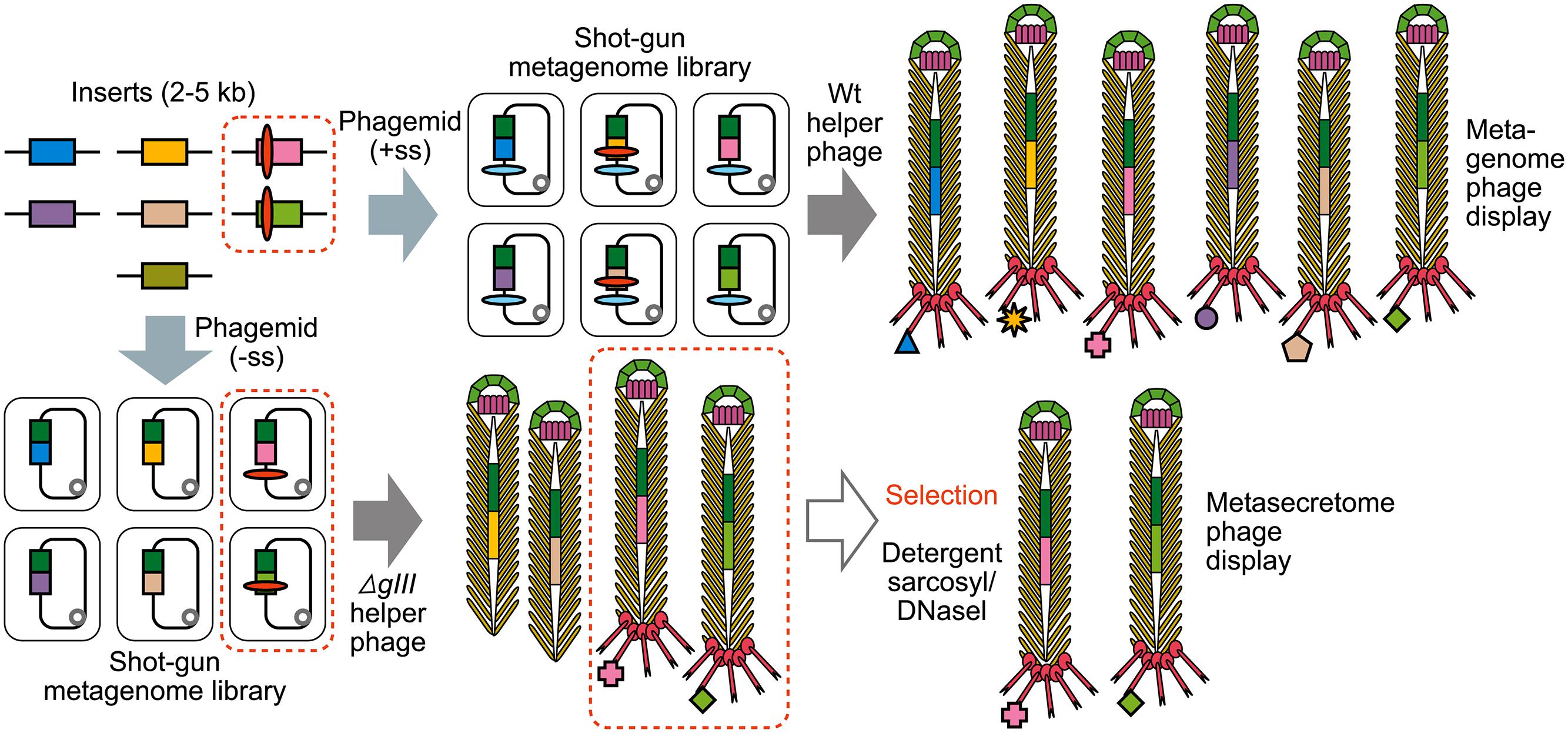
FIGURE 4. The construction of shotgun metagenomic and metasecretome filamentous phage display libraries. Metagenomic DNA is randomly sheared and cloned into phagemid that contains signal sequence (+ss) or as in case for metasecretome into phagemid without signal sequence (-ss). In both cases constructed metagenomic inserts contain endogenous signal sequences, represented by red ovals. Depending on helper phage used [wild-type (wt) or gIII-deleted helper phage] for aid in replication and assembly of recombinant virions, the library will contain virions displaying the whole metaproteome (metagenome phage display) or it will consist of virions capped by insert-pIII fusion proteins (signal sequence-positive clones) that are resistant to sarkosyl (SarkosylR, virions inside the dotted line) and uncapped virions (signal sequence-negative clones) that are sensitive to sarkosyl (SarkosylS). Sarkosyl resistance is used as a basis for selection in metasecretome phage display. Single stranded DNA (ssDNA) purified from SarkosylR virions after the selection can be used to obtain the amplified metasecretome plasmid library for preliminary assessment of metasecretome diversity by next-generation sequencing [Taken from (Ciric et al., 2014) with permission].
Although in some cases secretome display was used for screening of libraries to identify binders to targets of interest (Fevre et al., 2014), this method is very strongly biased toward display of N-terminal portions of the secretome proteins, thereby eliminating binding domains encoded toward the C-terminal end of the large adhesins. Furthermore, with a few exceptions listed above this strategy is not expected to display important SecYEG-independent virulence factors, such as those secreted by the Type 1, 3, 4, and 6 secretion systems, which are predominant in enteric pathogens including Yersinia, E. coli, and S. enterica (Chagnot et al., 2013). Therefore, shot-gun phage display is more suitable than secretome display for screening to identify binding domains for novel ligands.
Filamentous Phage Display in Functional Metagenomics
It is clear from the vast number of research reports that phage display technology has been used successfully to identify and characterize microbial proteins that interact with complex and simple targets as well as to build clone banks of secretome proteins from single organisms. Currently, the large volumes of data generated from fast improving next-generation-sequencing (NGS) technologies and metagenomics research are mainly used for cataloging and comparing the composition of microbial communities. An interesting question is whether the power of phage display and NGS could be combined to identify specific ligand-binding or representative (meta) secretome protein libraries from complex microbial communities.
Metagenomic approaches for analysis of microbial DNA recovered directly from the environment (Handelsman, 2004) have been used over the past decade to uncover extraordinary functional potential of complex microbial communities. Major strategies applied for identification of genes encoding “high-value targets” (enzymes for synthesis of novel bioactive molecules or novel biological activities) from metagenome are sequence-based bioinformatics analyses of the gene content, or function-based, by functional screening of metagenomic expression libraries (Tuffin et al., 2009; Ekkers et al., 2012; Culligan et al., 2013). Sequence-based metagenome mining aims to identify in silico high-value target gene or gene cluster candidates for heterologous expression, while function-based screens aim to phenotypically detect biosynthetically or enzymatically active clones (Charlop-Powers et al., 2014). Use of next generation sequencing (NGS) technologies in metagenomics enables significantly higher resolution and throughput in gene discovery from environmental microbes compared to traditional genomic approaches.
Genetic diversity of most microbial communities is tremendous. This implies an immense DNA sequencing volume is required to find, very sparse high-value target genes and gene clusters. A variety of enrichment strategies ranging from the whole-cell approach based on nutritional, chemical or physical selection, to increasing the frequency and diversity of genes likely to encode novel bioactive molecules, have been applied to increase the screening hit rate and speed up the process of gene discovery using functional metagenomic approaches (Cowan et al., 2005; Banik and Brady, 2010; Xing et al., 2012). DNA stable-isotope probing (Chen and Murrell, 2010; Mazard and Schäfer, 2014), complementation (Charlop-Powers et al., 2012), affinity capture and a number of strategies based on PCR and/or DNA hybridization, such as suppression subtractive hybridization (Galbraith et al., 2004; Meyer et al., 2007; Meiring et al., 2010), differential display, sequence tag interrogation approach (Owen et al., 2013) and metagenome arrays (He et al., 2007; Park et al., 2008; He et al., 2010) have been explored for enriching environmental DNA samples for genes of interest.
Although successful affinity screening of metagenomic Ff phage display libraries is yet to be reported, the precedents for this have been published in the form of selective metasecretome display and NGS-facilitated phage display library screening.
Phage Display Combined with Next-Generation Sequencing
The traditional approach for screening of phage display libraries, consisting of 3–5 rounds of affinity selection (biopanning) followed by sequencing of inserts from limited number of clones (Figure 3), is laborious and unsuitable for screening of metagenomes of diverse microbial community or complex antibody repertoires of higher organisms. In contrast, use of NGS technologies, typically delivering >106 of sequencing reads, is well suited for high-throughput exploration of the diversity of phage binding variants enriched after one or two rounds of panning on ligand of interest. The NGS analysis after limited panning allows high sequence coverage of binding variants, thereby overcoming a problem of competition between high-affinity and low-affinity binders and reducing false positive hits that often arise as a result of binding to non-bait materials present in the selection system (e.g., plastics, BSA) and propagation advantages (Vodnik et al., 2011).
Dias-Neto et al. (2009) proposed the use of NGS in combination with real-time PCR to improve phage quantification and analysis of the library inserts encoding phage-displayed variants. The authors adapted pyrosequencing for deep-sequencing of amplicons derived from phage ssDNA, retrieved directly after two rounds of panning of combinatorial library of random heptapeptides in vivo from four human tissues biopsies, using primers flanking the library insert within the fusion. This approach was applied to obtain sequencing reads directly from tissue-selected and unselected phage display libraries (library before and after panning) in a single run.
Di Niro et al. (2010) screened an ORF-filtered (Zacchi et al., 2003) cDNA phage display library, obtained from mRNA derived from several human cell lines, to identify proteins interacting with tissue transglutaminase 2, an enzyme implicated in different pathological conditions. In this study the authors combined phage display and NGS, enabling at least two orders of magnitude increase in the number of affinity-selected clones compared to traditional affinity screening, with two rounds of panning, to achieve an optimal balance between high numbers of positive clones and broad diversity. This approach led to identification of a “landscape” of binding variants from the phage display library, three of which had been previously characterized as transglutaminase 2-interacting proteins, with the remainder being novel proteins that were subsequently confirmed by functional assays. Based on the ranking of the most frequently selected ORFs within the selected phage population, detected through the high-throughput sequencing, it was estimated that at least 1000 clones would have to be picked and analyzed using traditional screening approach in order to capture the top five most frequent clones after two rounds of selection with 99% probability.
Recently, t Hoen et al. (2012) demonstrated that NGS can improve and accelerate finding of specific binders by screening of the phage display libraries, while reducing the number of false positive hits. Illumina platform was used to compare diversity of Ph.D.-7 M13 peptide phage display library before panning and phage display library retrieved after each of several rounds of panning on osteoblasts, performed to select heptapeptides that mediate binding to and uptake into osteoblasts. It was demonstrated that deep sequencing of the phage pool obtained after the first round of biopanning on osteoblasts was sufficient to identify positive hits. Selection of peptides with high binding to and uptake into osteoblasts was confirmed by confocal microscopy and live cell imaging. In addition, by sequencing the starting phage display library before screening (after one round of amplification in bacterial host), authors identified propagation advantage as an important source of false positive hits (t Hoen et al., 2012).
Due to the high diversity of antibody phage display libraries (typically between 107 and 1011 variants) and observed correlation between the size of the repertoire and the antibody affinities isolated from it (Hust and Dubel, 2004), filamentous phage display coupled with NGS is routinely used for in-depth characterization of immunoglobulin antibody repertoires of different organisms and high-throughput screening of ligand-antibody interactions (Ravn et al., 2010; Reddy et al., 2010; Saggy et al., 2012). This advance in technology can be readily applied to other high complexity samples, such as metagenome shot-gun phage display libraries.
Selective Metasecretome Phage Display
At present, published data on applications of phage display to aid metagenomic gene discovery is limited to “proof-of-principle” studies. T7 phage display and affinity capture was employed for enrichment and identification of genes encoding acyl carrier proteins and peptidyl carrier proteins from soil metagenome (Zhang et al., 2009). These proteins are essential components of biosynthetic enzymes (polyketide synthases and non-ribosomal peptide synthetases) that synthesize natural products with proposed antibiotic, immunosuppressant, and anticancer activities. After six iterative rounds of selection for enrichment of carrier proteins, a limited number (60) of phage clones were sequenced to evaluate usefulness of this approach for identification of carrier protein genes from large metagenomic DNA libraries. Display of the same soil metagenomic DNA library on the surface of filamentous phage and subsequent five rounds of panning did not yield any enrichment. Authors hypothesized that T7 phage display system is more suitable for functional selection of metagenome-encoded cytoplasmic enzymes because the T7 phage capsid protein is displayed on virions that are assembled in the cytoplasm and released by host cell lysis. In contrast, expression and display on filamentous phage depends on host secretion systems for translocation to the periplasm that is an oxidizing environment and therefore not suitable for most cytosolic enzymes which commonly contain an active site Cys residues.
Ciric et al. (2014) combined secretome-selective filamentous phage display (Jankovic et al., 2007) and NGS for the enrichment and sequence-based mining of bovine rumen plant-adherent microbial metagenome for fibrolityc enzymes (Ciric et al., 2014) (Figure 4). This approach enabled enrichment of metagenomic genes encoding surface, transmembrane and secreted proteins (metasecretome). Given that the selection relies on E. coli SecYEG translocons, this publication gave the first assessment of the selection system’s promiscuity, in particular for Gram-positive vs. Gram-negative secretion signals. Whereas secretome proteins from both types of bacteria have been selected for, taxonomic assignment of the sequenced inserts showed an increased frequency of secretome proteins from Gram-negative bacteria in the phylum Bacteroidetes (Figure 5B) relative to those from random shotgun sequence dataset. In terms of the type of secretion signals selected for at the metasecretome scale, inserts containing recognition sites for type I signal sequences were the most prominent, similar to what was reported for the genome-scale secretome selection (Figure 5A).
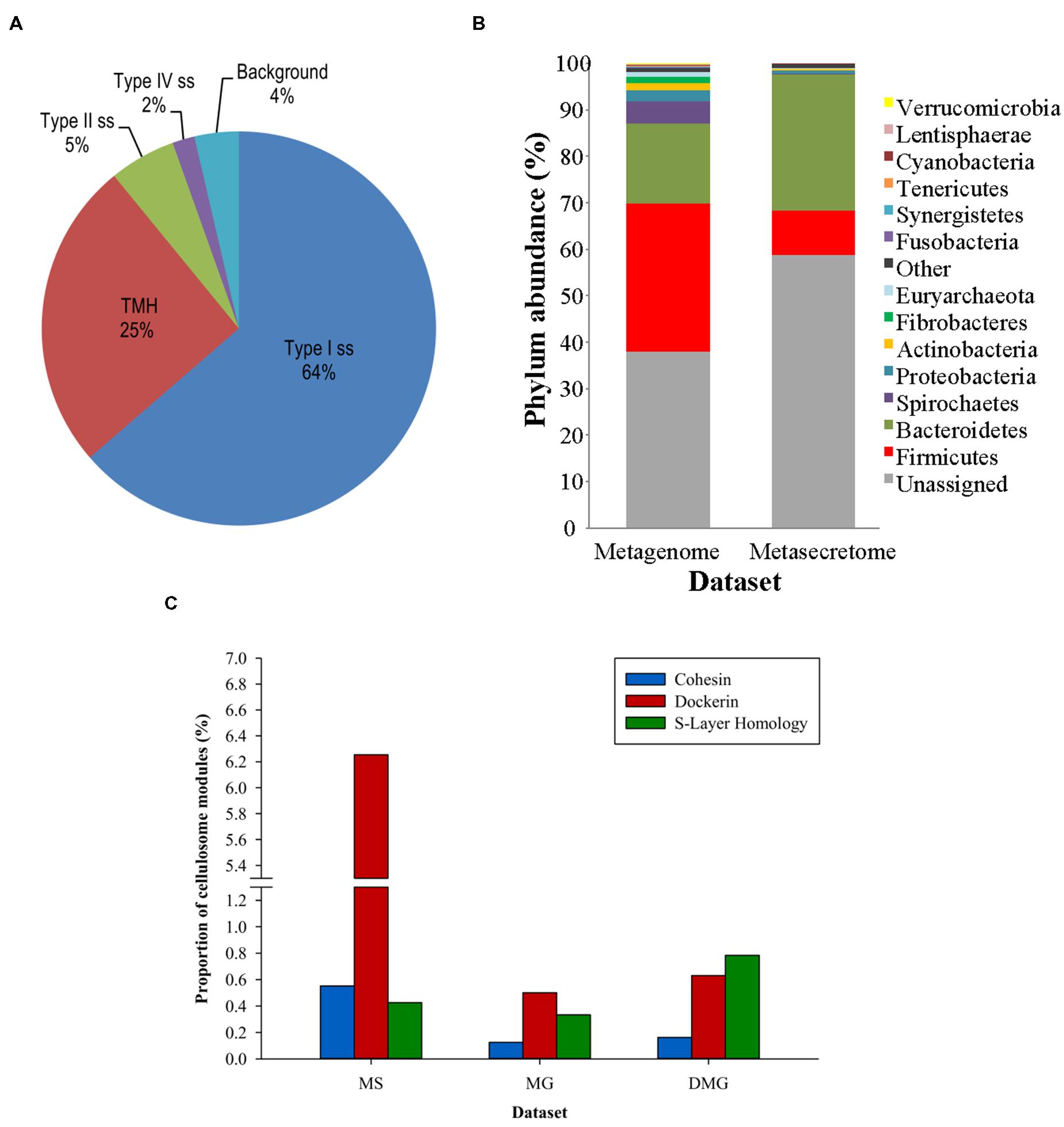
FIGURE 5. Taxonomic distribution, signal sequence types, and cellulosome components enrichment in metasecretome phage display of fiber-adherent rumen microbial community. (A) Distribution of signal sequence types and transmembrane helices in selected metasecretome recombinants; Abbreviations: Type I ss, classical signal sequence; Type II ss, lipoprotein signal sequence; Type IV ss, prepilin-like signal sequence; TMH, N-terminal or internal transmembrane α- helix/helices; background, ORFs encoding putative proteins without a predicted membrane-targeting signal/non-classical secretion, or ORFs encoding putative proteins and peptides ≤24 amino acid residues. (B) Taxonomic distribution of the phage-display-selected metasecretome; The taxonomic assignments, at the phylum level, were based on the distribution of the best BLASTP hits at a 30% amino acid sequence identity threshold for protein-coding genes predicted in metagenome and metasecretome datasets. Each section of the stacked columns represents the percentage of total protein-coding genes assigned to the corresponding phylum. The section labeled ‘Other’ contains putative protein-coding genes assigned to a phylogenetic group with low abundance in the dataset (<0.1%), while the section labeled ‘Unassigned’ corresponds to putative protein-coding genes with best BLASTP hit below 30% identity cut-off. (C) Enrichment for the components of cellulosome; Frequency of three putative distinct ‘signature’ cellulosome modules: cohesins (blue); dockerins (red) and surface S-layer homology (SLH) domains (green) in three datasets: MS, metasecretome dataset; MG, metagenome dataset and published deep-sequenced metagenome (DMG). The latter dataset is from (Hess et al., 2011). (A,C) are taken from (Ciric et al., 2014) and (B) from (Ciric, 2014) with permission.
Next-generation-sequencing analysis of the metasecretome identified an increased frequency of putative ORFs encoding catalytic and binding modules of fibrolytic enzymes, as well as large numbers of putative modules (cohesins and dockerins) that are constructing blocks of cell-surface organelles called cellulosomes (Bayer et al., 2004), specialized for recognition and degradation of plant fiber, were detected (Figure 5C). A high proportion and taxonomic variety of cellulosomal modules, particularly those from cohesins and dockerins has not been reported in previously published metagenomic studies of the rumen microbiome, suggesting that phage display could be a powerful method for enrichment, display and identification of these modules which subsequently can be used in building of microbial “designer” fiber-degrading hyper structures. Genes encoding putative fibrolytic enzyme modules and cellulosome modules are candidates for functional characterization via affinity screening of methagenomic shot-gun phage display libraries.
Despite its power, there are several shortcomings of the metasecretome phage display approach, some of which could potentially be overcome with new and upcoming technologies. As is the case with the genome-scale secretome phage display, the metasecretome library is biased toward the sequences encoding the N-terminal portion of the secretome proteins, hence representation of the C-terminal domains is low. This can in principle be overcome by constructing fosmid- or cosmid-based phage display vectors that would have a capacity for large inserts (up to 40 kb) and would therefore be able to accommodate large portions of secretome proteins fused to pIII.
Next-generation sequencing using 454 or Illumina technologies is powerful in covering the totality of the meta secretome library inserts. However, due to the short lengths of templates required for these sequencing technologies and a PCR amplification step that is necessary in order to produce the sequencing library, sequence information is disconnected from the physical recombinant clones from which it was derived (Ciric et al., 2014), making it impossible to form a clone library without the standard Sanger sequencing. This issue, however, can be overcome by using novel sequencing technologies, like PACBio, which use long templates (Fichot and Norman, 2013; Wang et al., 2015) and could, in principle, be used in an approach that would allow tracking of a sequence to the template. This approach for sequencing could be combined with subtractive or normalization methods to enrich the library for the rare recombinant sequences (Gagic et al., 2015), and to build a balanced metasecretome clone bank that can be used as a resource for expression and purification of large number of secretome proteins from a microbial community, by virtue of their display on PPs.
A shot-gun metagenomic phage display libraries in Ff and T7 have potential to provide the widest possible coverage of SecYEG-dependent and –independent secretome proteins for affinity selection against complex or simple targets of interest to identify adhesins, secreted virulence factors or enzymes, whereas the metasecretome libraries provide a good source of data for large scale identification of immunogenic peptides. Amenability of phage display to large-scale, high-throughput screening using NGS and emerging phage display-based methods for enrichment of target genes in metagenomes, indicate that this is just a beginning of wider use of phage display to accelerate target gene discovery in metagenomics.
Concluding Remarks
Phage display on Ff, combined with recent developments in sequencing technologies, provides a powerful approach for discovery of novel secretome proteins in variety of microorganisms and in microbial communities.
Author Contributions
DG (50%), MC (30%), WW (5%) FN (5%), and JR (10%) have written the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the New Zealand Ministry for Business, Innovation and Employment grant (contract # C10X0803) and The Royal Society of New Zealand through a Marsden Fast Start grant (contract AGR1003), JR laboratory was supported by Anonymous Donor, Massey University Research Fund, Palmerston North Medical Research Foundation and Institute of Fundamental Sciences, Massey University. FN was supported by a Commonwealth Ph.D. Fellowship and WXW by an Institute of Fundamental Sciences (Massey University) Ph.D. scholarship. FN and WXW were also supported by a Completion Bursary Scholarship by Massey University.
References
Abruquah, H. H. (2009). Identification of Host-pathogen Interacting Molecules of Campylobacter Jejuni Using Phage Display Technology and In silico Sequence Analysis. Ph.D. thesis, submitted to the University of Nottingham for the degree of Doctor of Philosophy Microbiology, The University of Nottingham, Nottingham.
Abu-Qarn, M., Eichler, J., and Sharon, N. (2008). Not just for Eukarya anymore: protein glycosylation in Bacteria and Archaea. Curr. Opin. Struct. Biol. 18, 544–550. doi: 10.1016/j.sbi.2008.06.010
Antelmann, H., Tjalsma, H., Voigt, B., Ohlmeier, S., Bron, S., Van Dijl, J. M., et al. (2001). A proteomic view on genome-based signal peptide predictions. Genome Res. 11, 1484–1502. doi: 10.1101/gr.182801
Antonara, S., Chafel, R. M., Lafrance, M., and Coburn, J. (2007). Borrelia burgdorferi adhesins identified using in vivo phage display. Mol. Microbiol. 66, 262–276. doi: 10.1111/j.1365-2958.2007.05924.x
Arap, W., Pasqualini, R., and Ruoslahti, E. (1998). Cancer treatment by targeted drug delivery to tumor vasculature in a mouse model. Science 279, 377–380 doi: 10.1126/science.279.5349.377
Arts, J., Van Boxtel, R., Filloux, A., Tommassen, J., and Koster, M. (2007). Export of the pseudopilin XcpT of the Pseudomonas aeruginosa type II secretion system via the signal recognition particle-Sec pathway. J. Bacteriol. 189, 2069–2076. doi: 10.1128/JB.01236-06
Auclair, S. M., Bhanu, M. K., and Kendall, D. A. (2012). Signal peptidase I: cleaving the way to mature proteins. Protein Sci. 21, 13–25. doi: 10.1002/pro.757
Ausmees, N., Jacobsson, K., and Lindberg, M. (2001). A unipolarly located, cell-surface-associated agglutinin, RapA, belongs to a family of Rhizobium-adhering proteins (Rap) in Rhizobium leguminosarum bv. trifolii. Microbiology 147, 549–559.
Åvall-Jääskeläinen, S., Lindholm, A., and Palva, A. (2003). Surface display of the receptor-binding region of the Lactobacillus brevis S-layer protein in Lactococcus lactis provides nonadhesive lactococci with the ability to adhere to intestinal epithelial cells. Appl. Environ. Microbiol. 69, 2230–2236. doi: 10.1128/AEM.69.4.2230-2236.2003
Bagos, P. G., Nikolaou, E. P., Liakopoulos, T. D., and Tsirigos, K. D. (2010). Combined prediction of Tat and Sec signal peptides with Hidden Markov Models. Bioinformatics 26, 2811–2817. doi: 10.1093/bioinformatics/btq530
Bagos, P. G., Tsirigos, K. D., Liakopoulos, T. D., and Hamodrakas, S. J. (2008). Prediction of lipoprotein signal peptides in Gram-positive bacteria with a Hidden Markov Model. J. Proteome Res. 7, 5082–5093. doi: 10.1021/pr800162c.
Banik, J. J., and Brady, S. F. (2010). Recent application of metagenomic approaches toward the discovery of antimicrobials and other bioactive small molecules. Curr. Opin. Microbiol. 13, 603–609. doi: 10.1016/j.mib.2010.08.012
Barbas, C. F. III, Burton, D. R., Scott, J. K., and Silverman, G. J. (2001). Phage display: a laboratory manual. Cold Spring Harbor. New York, NY: Cold Spring Harbor Laboratory Press.
Bayer, E. A., Belaich, J. -P., Shoham, Y., and Lamed, R. (2004). The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides. Annu. Rev. Microbiol. 58, 521–554. doi: 10.1146/annurev.micro.57.030502.091022
Beckmann, C., Waggoner, J. D., Harris, T. O., Tamura, G. S., and Rubens, C. E. (2002). Identification of novel adhesins from Group B streptococci by use of phage display reveals that C5a peptidase mediates fibronectin binding. Infect. Immun. 70, 2869–2876. doi: 10.1128/IAI.70.6.2869-2876.2002
Beckwith, J. (2013). The Sec-dependent pathway. Res. Microbiol. 164, 497–504. doi: 10.1016/j.resmic.2013.03.007
Beghetto, E., and Gargano, N. (2011). Lambda-display: a powerful tool for antigen discovery. Molecules 16, 3089–3105. doi: 10.3390/molecules16043089
Belanche, A., De La Fuente, G., and Newbold, C. J. (2014). Study of methanogen communities associated with different rumen protozoal populations. FEMS Microbiol. Ecol. 90, 663–677. doi: 10.1111/1574-6941.12423.
Bendtsen, J. D., Kiemer, L., Fausbøll, A., and Brunak, S. (2005). Non-classical protein secretion in bacteria. BMC Microbiol. 5:58. doi: 10.1186/1471-2180-5-58
Benedek, O., Khan, A. S., Schneider, G., Nagy, G., Autar, R., Pieters, R. J., et al. (2005). Identification of laminin-binding motifs of Yersinia pestis plasminogen activator by phage display. Int. J. Med. Microbiol. 295, 87–98. doi: 10.1016/j.ijmm.2005.02.002
Bernard, J. M., and Francis, M. B. (2014). Chemical strategies for the covalent modification of filamentous phage. Front. Microbiol. 5:734. doi: 10.3389/fmicb.2014.00734
Bjerketorp, J., Nilsson, M., Ljungh,Å., Flock, J.-I., Jacobsson, K., and Frykberg, L. (2002). A novel von Willebrand factor binding protein expressed by Staphylococcus aureus. Microbiology 148, 2037–2044.
Bjerketorp, J., Rosander, A., Nilsson, M., Jacobsson, K., and Frykberg, L. (2004). Sorting a Staphylococcus aureus phage display library against ex vivo biomaterial. J. Med. Microbiol. 53, 945–951. doi: 10.1099/jmm.0.45638-0
Blobel, G., and Sabatini, D. D. (1971). “Ribosome-membrane interaction in eukaryotic cells,” in Biomembranes, ed. Hanson (Berlin: Springer), 193–195.
Branston, S. D., Stanley, E. C., Ward, J. M., and Keshavarz-Moore, E. (2013). Determination of the survival of bacteriophage M13 from chemical and physical challenges to assist in its sustainable bioprocessing. Biotechnol. Bioproc. E 18, 560–566. doi: 10.1007/s12257-012-0776–779
Bratkovic, T. (2010). Progress in phage display: evolution of the technique and its application. Cell Mol. Life. Sci. 67, 749–767. doi: 10.1007/s00018-009-0192-2
Caccia, D., Dugo, M., and Callari, M. (2013). Bioinformatics tools for secretome analysis. Biochim. Biophys. Acta 1834, 2442–2453. doi: 10.1016/j.bbapap.2013.01.039
Celik, E., Fisher, A. C., Guarino, C., Mansell, T. J., and Delisa, M. P. (2010). A filamentous phage display system for N-linked glycoproteins. Protein Sci. 19, 2006–2013. doi: 10.1002/pro.472
Chagnot, C., Zorgani, M. A., Astruc, T., and Desvaux, M. (2013). Proteinaceous determinants of surface colonization in bacteria: bacterial adhesion and biofilm formation from a protein secretion perspective. Front. Microbiol. 4:303. doi: 10.3389/fmicb.2013.00303
Chang, C. N., Blobel, G., and Model, P. (1978). Detection of prokaryotic signal peptidase in an Escherichia coli membrane fraction: endoproteolytic cleavage of nascent f1 pre-coat protein. Proc. Natl. Acad. Sci. U.S.A. 75, 361–365. doi: 10.1073/pnas.75.1.361
Charlop-Powers, Z., Banik, J. J., Owen, J. G., Craig, J. W., and Brady, S. F. (2012). Selective enrichment of environmental DNA libraries for genes encoding nonribosomal peptides and polyketides by phosphopantetheine transferase-dependent complementation of siderophore biosynthesis. ACS Chem. Biol. 8, 138–143. doi: 10.1021/cb3004918
Charlop-Powers, Z., Milshteyn, A., and Brady, S. F. (2014). Metagenomic small molecule discovery methods. Curr. Opin. Microbiol. 19, 70–75. doi: 10.1016/j.mib.2014.05.021
Chen, W., and Georgiou, G. (2002). Cell-surface display of heterologous proteins: from high-throughput screening to environmental applications. Biotechnol. Bioeng. 79, 496–503. doi: 10.1002/bit.10407
Chen, Y., and Murrell, J. C. (2010). When metagenomics meets stable-isotope probing: progress and perspectives. Trends Microbiol. 18, 157–163. doi: 10.1016/j.tim.2010.02.002
Ching, A. T., Favaro, R. D., Lima, S. S., Chaves Ade, A., De Lima, M. A., Nader, H. B., et al. (2012). Leptospira interrogans shotgun phage display identified LigB as a heparin-binding protein. Biochem. Biophys. Res. Commun. 427, 774–779. doi: 10.1016/j.bbrc.2012.09.137
Choo, K. H., Tan, T. W., and Ranganathan, S. (2009). A comprehensive assessment of N-terminal signal peptides prediction methods. BMC Bioinform. 10:S2. doi: 10.1186/1471-2105-10-S15-S2
Ciric, M. (2014). Metasecretome Phage Display : a New Approach for Mining Surface and Secreted Proteins from Microbial Communities, Ph.D. thesis, Massey University, Palmerston North.
Ciric, M., Moon, C. D., Leahy, S. C., Creevey, C. J., Altermann, E., Attwood, G. T., et al. (2014). Metasecretome-selective phage display approach for mining the functional potential of a rumen microbial community. BMC Genomics 15:356. doi: 10.1186/1471-2164-15–356.
Coburn, J., Leong, J., and Chaconas, G. (2013). Illuminating the roles of the Borrelia burgdorferi adhesins. Trends Microbiol. 21, 372–379. doi: 10.1016/j.tim.2013.06.005
Cowan, D., Meyer, Q., Stafford, W., Muyanga, S., Cameron, R., and Wittwer, P. (2005). Metagenomic gene discovery: past, present and future. Trends Biotechnol. 23, 321–329. doi: 10.1016/j.tibtech.2005.04.001
Crissman, J. W., and Smith, G. P. (1984). Gene-III protein of filamentous phages: evidence for a carboxyl-terminal domain with a role in morphogenesis. Virology 132, 445–455. doi: 10.1016/0042-6822(84)90049-7
Culligan, E. P., Sleator, R. D., Marchesi, J. R., and Hill, C. (2013). Metagenomics and novel gene discovery: promise and potential for novel therapeutics. Virulence 5, 399–412. doi: 10.4161/viru.27208
Dalbey, R. E., and Kuhn, A. (2012). Protein traffic in Gram-negative bacteria: how exported and secreted proteins find their way. FEMS Microbiol. Rev. 36, 1023–1045. doi: 10.1111/j.1574-6976.2012.00327.x
Dalbey, R. E., Wang, P., and Van Dijl, J. M. (2012). Membrane proteases in the bacterial protein secretion and quality control pathway. Microbiol. Mol. Biol. Rev. 76, 311–330. doi: 10.1128/MMBR.05019-11
Day, L. A. (2011). “Family Inoviridae,” in Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses, eds A. M. Q. King, M. J. Adams, E. B. Carstens and E. J. Lefkowitz (San Diego: Elsevier Academic Press), 375–384.
Derda, R., Tang, S. K., Li, S. C., Ng, S., Matochko, W., and Jafari, M. R. (2011). Diversity of phage-displayed libraries of peptides during panning and amplification. Molecules 16, 1776–1803. doi: 10.3390/molecules16021776
Desvaux, M., Dumas, E., Chafsey, I., and Hebraud, M. (2006). Protein cell surface display in Gram-positive bacteria: from single protein to macromolecular protein structure. FEMS Microbiol. Lett. 256, 1–15. doi: 10.1111/j.1574-6968.2006.00122.x
Desvaux, M., Hébraud, M., Talon, R., and Henderson, I. R. (2009). Secretion and subcellular localizations of bacterial proteins: a semantic awareness issue. Trends Microbiol. 17, 139–145. doi: 10.1016/j.tim.2009.01.004
Di Niro, R., Sulic, A. M., Mignone, F., D’angelo, S., Bordoni, R., Iacono, M., et al. (2010). Rapid interactome profiling by massive sequencing. Nucleic Acids Res. 38:e110. doi: 10.1093/nar/gkq052
Dias-Neto, E., Nunes, D. N., Giordano, R. J., Sun, J., Botz, G. H., Yang, K., et al. (2009). Next-generation phage display: integrating and comparing available molecular tools to enable cost-effective high-throughput analysis. PLoS ONE 4:e8338. doi: 10.1371/journal.pone.0008338
Driessen, A. J., and Nouwen, N. (2008). Protein translocation across the bacterial cytoplasmic membrane. Annu. Rev. Biochem. 77, 643–667. doi: 10.1146/annurev.biochem.77.061606.160747
Dürr, C., Nothaft, H., Lizak, C., Glockshuber, R., and Aebi, M. (2010). The Escherichia coli glycophage display system. Glycobiology 20, 1366–1372. doi: 10.1093/glycob/cwq102
Ekkers, D. M., Cretoiu, M. S., Kielak, A. M., and Van Elsas, J. D. (2012). The great screen anomaly—a new frontier in product discovery through functional metagenomics. Appl. Microbiol. Biotechnol. 93, 1005–1020. doi: 10.1007/s00253-011-3804-3
Emanuelsson, O., Nielsen, H., Brunak, S., and Von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016. doi: 10.1006/jmbi.2000.3903
Evangelista, K., Franco, R., Schwab, A., and Coburn, J. (2014). Leptospira interrogans binds to cadherins. PLoS Negl. Trop Dis. 8:e2672. doi: 10.1371/journal.pntd.0002672
Evangelista, K. V., Hahn, B., Wunder, E. A. Jr., Ko, A. I., Haake, D. A., and Coburn, J. (2014). Identification of cell-binding adhesins of Leptospira interrogans. PLoS Negl. Trop. Dis. 8:e3215. doi: 10.1371/journal.pntd.0003215
Feng, J. N., Russel, M., and Model, P. (1997). A permeabilized cell system that assembles filamentous bacteriophage. Proc. Natl. Acad. Sci. U.S.A. 94, 4068–4073. doi: 10.1073/pnas.94.8.4068
Fevre, C., Bestebroer, J., Mebius, M. M., De Haas, C. J. C., Van Strijp, J. A. G., Fitzgerald, J. R., et al. (2014). Staphylococcus aureus proteins SSL6 and SElX interact with neutrophil receptors as identified using secretome phage display. Cell Microbiol. 16, 1646–1665. doi: 10.1111/cmi.12313
Fichot, E. B., and Norman, R. S. (2013). Microbial phylogenetic profiling with the Pacific Biosciences sequencing platform. Microbiome 1:10. doi: 10.1186/2049-2618-1–10
Francetic, O., Buddelmeijer, N., Lewenza, S., Kumamoto, C. A., and Pugsley, A. P. (2007). Signal recognition particle-dependent inner membrane targeting of the PulG pseudopilin component of a type II secretion system. J. Bacteriol. 189, 1783–1793. doi: 10.1128/JB.01230-06
Freudl, R. (2013). Leaving home ain’t easy: protein export systems in Gram-positive bacteria. Res. Microbiol. 164, 664–674.
Gagic, D., Maclean, P. H., Li, D., Attwood, G. T., and Moon, C. D. (2015). Improving the genetic representation of rare taxa within complex microbial communities using DNA normalization methods. Mol. Ecol. Resour. 15, 464–476. doi: 10.1111/1755-0998.12321
Gagic, D., Wen, W., Collett, M. A., and Rakonjac, J. (2013). Unique secreted-surface protein complex of Lactobacillus rhamnosus, identified by phage display. Microbiologyopen 2, 1–17. doi: 10.1002/mbo3.53
Galbraith, E. A., Antonopoulos, D. A., and White, B. A. (2004). Suppressive subtractive hybridization as a tool for identifying genetic diversity in an environmental metagenome: the rumen as a model. Environ. Microbiol. 6, 928–937. doi: 10.1111/j.1462-2920.2004.00575.x
Gamkrelidze, M., and Dabrowska, K. (2014). T4 bacteriophage as a phage display platform. Arch. Microbiol. 196, 473–479. doi: 10.1007/s00203-014-0989–988
Georgiou, G., Stathopoulos, C., Daugherty, P. S., Nayak, A. R., Iverson, B. L., and Curtiss, R. III (1997). Display of heterologous proteins on the surface of microorganisms: from the screening of combinatorial libraries to live recombinant vaccines. Nat. Biotechnol. 15, 29–34. doi: 10.1038/nbt0197-29
Goudenège, D., Avner, S., Lucchetti-Miganeh, C., and Barloy-Hubler, F. (2010). CoBaltDB: complete bacterial and archaeal orfeomes subcellular localization database and associated resources. BMC Microbiol. 10:88. doi: 10.1186/1471-2180-10-88
Griffiths, A. D., Malmqvist, M., Marks, J. D., Bye, J. M., Embleton, M. J., Mccafferty, J., et al. (1993). Human anti-self antibodies with high specificity from phage display libraries. EMBO J. 12, 725–734.
Gunnarsen, K. S., Kristinsson, S. G., Justesen, S., Frigstad, T., Buus, S., Bogen, B., et al. (2013). Chaperone-assisted thermostability engineering of a soluble T cell receptor using phage display. Sci. Rep. 3:1162. doi: 10.1038/srep01162
Handelsman, J. (2004). Metagenomics: application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 68, 669–685. doi: 10.1128/MMBR.68.4.669-685.2004.
Hayhurst, A., and Harris, W. J. (1999). Escherichia coli skp chaperone coexpression improves solubility and phage display of single-chain antibody fragments. Protein Expr. Purif. 15, 336–343. doi: 10.1006/prep.1999.1035
He, Z., Deng, Y., Van Nostrand, J. D., Tu, Q., Xu, M., Hemme, C. L., et al. (2010). GeoChip 3.0 as a high-throughput tool for analyzing microbial community composition, structure and functional activity. ISME J. 4, 1167–1179. doi: 10.1038/ismej.2010.46
He, Z., Gentry, T. J., Schadt, C. W., Wu, L., Liebich, J., Chong, S. C., et al. (2007). GeoChip: a comprehensive microarray for investigating biogeochemical, ecological and environmental processes. ISME J. 1, 67–77. doi: 10.1038/ismej.2007.2
Heilmann, C., Herrmann, M., Kehrel, B. E., and Peters, G. (2002). Platelet-binding domains in 2 fibrinogen-binding proteins of Staphylococcus aureus identified by phage display. J. Infect. Dis. 186, 32–39. doi: 10.1086/341081
Hess, M., Sczyrba, A., Egan, R., Kim, T. W., Chokhawala, H., Schroth, G., et al. (2011). Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 331, 463–467. doi: 10.1126/science.1200387
Hoogenboom, H. R., Griffiths, A. D., Johnson, K. S., Chiswell, D. J., Hudson, P., and Winter, G. (1991). Multi-subunit proteins on the surface of filamentous phage: methodologies for displaying antibody (Fab) heavy and light chains. Nucleic Acids Res. 19, 4133–4137. doi: 10.1093/nar/19.15.4133
Hust, M., and Dubel, S. (2004). Mating antibody phage display with proteomics. Trends Biotechnol. 22, 8–14. doi: 10.1016/j.tibtech.2003.10.011
Imam, S., Chen, Z., Roos, D. S., and Pohlschröder, M. (2011). Identification of surprisingly diverse type IV pili, across a broad range of Gram-positive bacteria. PLoS ONE 6:e28919. doi: 10.1371/journal.pone.0028919
Ingham, K. C., Brew, S., Vaz, D., Sauder, D. N., and McGavin, M. J. (2004). Interaction of Staphylococcus aureus fibronectin-binding protein with fibronectin: affinity, stoichiometry, and modular requirements. J. Biol. Chem. 279, 42945–42953. doi: 10.1074/jbc.M406984200
Jacobsson, K., and Frykberg, L. (1995). Cloning of ligand-binding domains of bacterial receptors by phage display. Biotechniques 18, 878–885.
Jacobsson, K., and Frykberg, L. (1996). Phage display shot-gun cloning of ligand-binding domains of prokaryotic receptors approaches 100% correct clones. Biotechniques 20, 1080–1071.
Jacobsson, K., Rosander, A., Bjerketorp, J., and Frykberg, L. (2003). Shotgun phage display - selection for bacterial receptins or other exported proteins. Biol. Proced. 5, 123–135. doi: 10.1251/bpo54
Jankovic, D., Collett, M. A., Lubbers, M. W., and Rakonjac, J. (2007). Direct selection and phage display of a Gram-positive secretome. Genome Biol. 8:R266. doi: 10.1186/gb-2007-8-12-r266
Jönsson, K., Guo, B. P., Monstein, H. J., Mekalanos, J. J., and Kronvall, G. (2004). Molecular cloning and characterization of two Helicobacter pylori genes coding for plasminogen-binding proteins. Proc. Natl. Acad. Sci. U.S.A. 101, 1852–1857. doi: 10.1073/pnas.0307329101
Joo, H. Y., Hur, B. U., Lee, K. W., Song, S. Y., and Cha, S. H. (2008). Establishment of a reliable dual-vector system for the phage display of antibody fragments. J. Immunol. Methods 333, 24–37. doi: 10.1016/j.jim.2007.11.015
Juncker, A. S., Willenbrock, H., Von Heijne, G., Brunak, S., Nielsen, H., and Krogh, A. (2003). Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 12, 1652–1662. doi: 10.1110/ps.0303703
Kehoe, J. W., and Kay, B. K. (2005). Filamentous phage display in the new millennium. Chem. Rev. 105, 4056–4072. doi: 10.1021/cr000261r
Krogh, A., Larsson, B., Von Heijne, G., and Sonnhammer, E. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. doi: 10.1006/jmbi.2000.4315
Krumpe, L. R. H., Atkinson, A. J., Smythers, G. W., Kandel, A., Schumacher, K. M., Mcmahon, J. B., et al. (2006). T7 lytic phage-displayed peptide libraries exhibit less sequence bias than M13 filamentous phage-displayed peptide libraries. Proteomics 6, 4210–4222. doi: 10.1002/pmic.200500606
Kudva, R., Denks, K., Kuhn, P., Vogt, A., Müller, M., and Koch, H. -G. (2013). Protein translocation across the inner membrane of Gram-negative bacteria: the Sec and Tat dependent protein transport pathways. Res. Microbiol. 164, 505–534. doi: 10.1016/j.resmic.2013.03.016
Lamboy, J. A., Tam, P. Y., Lee, L. S., Jackson, P. J., Avrantinis, S. K., Lee, H. J., et al. (2008). Chemical and genetic wrappers for improved phage and RNA display. Chembiochem 9, 2846–2852. doi: 10.1002/cbic.200800366
Lauterbach, S. B., Lanzillotti, R., and Coetzer, T. L. (2003). Construction and use of Plasmodium falciparum phage display libraries to identify host parasite interactions. Malar. J. 2:47. doi: 10.1186/1475-2875-2–47.
Leahy, S. C., Kelly, W. J., Altermann, E., Ronimus, R. S., Yeoman, C. J., Pacheco, D. M., et al. (2010). The genome sequence of the rumen methanogen Methanobrevibacter ruminantium reveals new possibilities for controlling ruminant methane emissions. PLoS ONE 5:e8926. doi: 10.1371/journal.pone.0008926
Lee, S. Y., Choi, J. H., and Xu, Z. (2003). Microbial cell-surface display. Trends Biotechnol. 21, 45–52. doi: 10.1016/S0167-7799(02)00006-9
Lei, S. P., Lin, H. C., Wang, S. S., Callaway, J., and Wilcox, G. (1987). Characterization of the Erwinia carotovora pelB gene and its product pectate lyase. J. Bacteriol. 169, 4379–4383.
Lindmark, H., and Guss, B. (1999). SFS, a novel fibronectin-binding protein from Streptococcus equi, inhibits the binding between fibronectin and collagen. Infect. Immun. 67, 2383–2388.
Liu, R., Yang, C., Xu, Y., Xu, P., Jiang, H., and Qiao, C. (2013). Development of a whole-cell biocatalyst/biosensor by display of multiple heterologous proteins on the Escherichia coli cell surface for the detoxification and detection of organophosphates. J. Agric. Food Chem. 61, 7810–7816. doi: 10.1021/jf402999b
Liu, S., Han, W., Sun, C., Lei, L., Feng, X., Yan, S., et al. (2011). Subtractive screening with the Mycobacterium tuberculosis surface protein phage display library. Tuberculosis 91, 579–586. doi: 10.1016/j.tube.2011.07.007
Lubkowski, J., Hennecke, F., Pluckthun, A., and Wlodawer, A. (1998). The structural basis of phage display elucidated by the crystal structure of the N-terminal domains of g3p. Nat. Struct. Biol. 5, 140–147. doi: 10.1038/nsb0298-140
Lüke, I., Handford, J. I., Palmer, T., and Sargent, F. (2009). Proteolytic processing of Escherichia coli twin-arginine signal peptides by LepB. Arch. Microbiol. 191, 919–925. doi: 10.1007/s00203-009-0516-5
Luo, C., Tsementzi, D., Kyrpides, N. C., and Konstantinidis, K. T. (2011). Individual genome assembly from complex community short-read metagenomic datasets. ISME J. 6, 898–901. doi: 10.1038/ismej.2011.147
Lycklama, A., Nijeholt, J. A., and Driessen, A. J. (2012). The bacterial Sec-translocase: structure and mechanism. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 1016–1028. doi: 10.1098/rstb.2011.0201
Mai-Prochnow, A., Hui, J. G., Kjelleberg, S., Rakonjac, J., Mcdougald, D., and Rice, S. A. (2015). ’Big things in small packages: the genetics of filamentous phage and effects on fitness of their host’. FEMS Microbiol. Rev. 39, 465–487. doi: 10.1093/femsre/fuu007
Mao, C., Solis, D. J., Reiss, B. D., Kottmann, S. T., Sweeney, R. Y., Hayhurst, A., et al. (2004). Virus-based toolkit for the directed synthesis of magnetic and semiconducting nanowires. Science 303, 213–217. doi: 10.1126/science.1092740
Marvin, D. A., Symmons, M. F., and Straus, S. K. (2014). Structure and assembly of filamentous bacteriophages. Prog. Biophys. Mol. Biol. 114, 80–122. doi: 10.1016/j.pbiomolbio.2014.02.003
Marvin, D. A., Welsh, L. C., Symmons, M. F., Scott, W. R., and Straus, S. K. (2006). Molecular structure of fd (f1, M13) filamentous bacteriophage refined with respect to X-ray fibre diffraction and solid-state NMR data supports specific models of phage assembly at the bacterial membrane. J. Mol. Biol. 355, 294–309. doi: 10.1016/j.jmb.2005.10.048
Mazard, S., and Schäfer, H. (2014). Stable isotope probing to study functional components of complex microbial ecosystems. Methods Mol. Biol. 668, 169–180.
Meiring, T., Mulako, I., Tuffin, M. I., Meyer, Q., and Cowan, D. A. (2010). Retrieval of full-length functional genes using subtractive hybridization magnetic bead capture. Methods Mol. Biol. 668, 287–297.
Meyer, Q. C., Burton, S. G., and Cowan, D. A. (2007). Subtractive hybridization magnetic bead capture: a new technique for the recovery of full-length ORFs from the metagenome. Biotechnol. J. 2, 36–40. doi: 10.1002/biot.200600156
Meyer, T., Schirrmann, T., Frenzel, A., Miethe, S., Stratmann-Selke, J., Gerlach, G. F., et al. (2012). Identification of immunogenic proteins and generation of antibodies against Salmonella Typhimurium using phage display. BMC Biotechnol. 12:29. doi: 10.1186/1472-6750-12-29
Mullen, L. M., Nair, S. P., Ward, J. M., Rycroft, A. N., and Henderson, B. (2006). Phage display in the study of infectious diseases. Trends Microbiol. 14, 141–147. doi: 10.1016/j.tim.2006.01.006
Mullen, L. M., Nair, S. P., Ward, J. M., Rycroft, A. N., Williams, R. J., and Henderson, B. (2007). Comparative functional genomic analysis of Pasteurellaceae adhesins using phage display. Vet. Microbiol. 122, 123–134. doi: 10.1016/j.vetmic.2006.12.022
Munoz-Provencio, D., and Monedero, V. (2011). Shotgun phage display of Lactobacillus casei BL23 against collagen and fibronectin. J. Microbiol. Biotechnol. 21, 197–203. doi: 10.4014/jmb.1009.09011
Natale, P., Brüser, T., and Driessen, A. J. (2008). Sec- and Tat- mediated protein secretion across the bacterial cytoplasmic membrane-distinct translocases and mechanisms. Biochim. Biophys. Acta 1778, 1735–1756. doi: 10.1016/j.bbamem.2007.07.015
Nilsson, M., Frykberg, L., Flock, J. I., Pei, L., Lindberg, M., and Guss, B. (1998). A fibrinogen-binding protein of Staphylococcus epidermidis. Infect. Immun. 66, 2666–2673.
Nilsson, M., Bjerketorp, J., Guss, B., and Frykberg, L. (2004a). A fibrinogen-binding protein of Staphylococcus lugdunensis. FEMS Microbiol. Lett. 241, 87–93. doi: 10.1016/j.femsle.2004.10.008
Nilsson, M., Bjerketorp, J., Wiebensjö, A., Ljungh, A., Frykberg, L., and Guss, B. (2004b). A von Willebrand factor-binding protein from Staphylococcus lugdunensis. FEMS Microbiol. Lett. 234, 155–161. doi: 10.1111/j.1574-6968.2004.tb09527.x
Ng, F., Kittelmann, S., Patchett, M. L., Attwood, G. T., Janssen, P. H., Rakonjac, J., et al. (2015). An adhesin from hydrogen-utilizing rumen methanogen Methanobrevibacter ruminantium M1 binds a broad range of hydrogen-producing microorganisms. Environ. Microbiol. doi: 10.1111/1462-2920.13155 [Epub ahead of print].
Ng, S., Tjhung, K. F., Paschal, B. M., Noren, C. J., and Derda, R. (2015). Chemical posttranslational modification of phage-displayed peptides. Methods Mol. Biol. 1248, 155–172. doi: 10.1007/978-1-4939-2020-4_11
Okuda, S., and Tokuda, H. (2011). Lipoprotein sorting in bacteria. Annu. Rev. Microbiol. 65, 239–259. doi: 10.1146/annurev-micro-090110-102859
Owen, J. G., Reddy, B. V. B., Ternei, M. A., Charlop-Powers, Z., Calle, P. Y., Kim, J. H., et al. (2013). Mapping gene clusters within arrayed metagenomic libraries to expand the structural diversity of biomedically relevant natural products. Proc. Natl. Acad. Sci. U.S.A. 110, 11797–11802. doi: 10.1073/pnas.1222159110
Park, S. J., Kang, C. H., Chae, J. C., and Rhee, S. K. (2008). Metagenome microarray for screening of fosmid clones containing specific genes. FEMS Microbiol. Lett. 284, 28–34. doi: 10.1111/j.1574-6968.2008.01180.x
Parmley, S. F., and Smith, G. P. (1988). Antibody-selectable filamentous fd phage vectors: affinity purification of target genes. Gene 73, 305–318. doi: 10.1016/0378-1119(88)90495-7
Peabody, C. R., Chung, Y. J., Yen, M.-R., Vidal-Ingigliardi, D., Pugsley, A. P., and Saier, M. H. (2003). Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology 149, 3051–3072. doi: 10.1099/mic.0.26364-0
Petersen, T. N., Brunak, S., Von Heijne, G., and Nielsen, H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786. doi: 10.1038/nmeth.1701
Ploss, M., Facey, S. J., Bruhn, C., Zemel, L., Hofmann, K., Stark, R. W., et al. (2014). Selection of peptides binding to metallic borides by screening M13 phage display libraries. BMC Biotechnol. 14:12. doi: 10.1186/1472-6750-14-12
Posadas, D. M., Ruiz-Ranwez, V., Bonomi, H. R., Martin, F. A., and Zorreguieta, A. (2012). BmaC, a novel autotransporter of Brucella suis, is involved in bacterial adhesion to host cells. Cell Microbiol. 14, 965–982. doi: 10.1111/j.1462-5822.2012.01771.x
Rahman, O., Cummings, S. P., Harrington, D. J., and Sutcliffe, I. C. (2008). Methods for the bioinformatic identification of bacterial lipoproteins encoded in the genomes of Gram-positive bacteria. World J. Microbiol. Biotechnol. 24, 2377–2382. doi: 10.1007/s11274-008-9795-2
Rakonjac, J., Bennett, N. J., Spagnuolo, J., Gagic, D., and Russel, M. (2011). Filamentous bacteriophage: biology, phage display and nanotechnology applications. Curr. Issues Mol. Biol. 13, 51–76.
Rakonjac, J., Feng, J., and Model, P. (1999). Filamentous phage are released from the bacterial membrane by a two-step mechanism involving a short C-terminal fragment of pIII. J. Mol. Biol. 289, 1253–1265. doi: 10.1006/jmbi.1999.2851
Rakonjac, J., Jovanovic, G., and Model, P. (1997). Filamentous phage infection-mediated gene expression: construction and propagation of the gIII deletion mutant helper phage R408d3. Gene 198, 99–103. doi: 10.1016/S0378-1119(97)00298-9
Rakonjac, J., and Model, P. (1998). Roles of pIII in filamentous phage assembly. J. Mol. Biol. 282, 25–41. doi: 10.1006/jmbi.1998.2006
Ravn, U., Gueneau, F., Baerlocher, L., Osteras, M., Desmurs, M., Malinge, P., et al. (2010). By-passing in vitro screening—next generation sequencing technologies applied to antibody display and in silico candidate selection. Nucleic Acids Res. 38:e193. doi: 10.1093/nar/gkq789
Reddy, S. T., Ge, X., Miklos, A. E., Hughes, R. A., Kang, S. H., Hoi, K. H., et al. (2010). Monoclonal antibodies isolated without screening by analyzing the variable-gene repertoire of plasma cells. Nat. Biotechnol. 28, 965–969. doi: 10.1038/nbt.1673
Rodi, D. J., and Makowski, L. (1999). Phage-display technology–finding a needle in a vast molecular haystack. Curr. Opin. Biotechnol. 10, 87–93. doi: 10.1016/S0958-1669(99)80016-0
Rosander, A., Bjerketorp, J., Frykberg, L., and Jacobsson, K. (2002). Phage display as a novel screening method to identify extracellular proteins. J. Microbiol. Methods 51, 43–55. doi: 10.1016/S0167-7012(02)00052-0
Rosander, A., Guss, B., Frykberg, L., Bjorkman, C., Naslund, K., and Pringle, M. (2011). Identification of immunogenic proteins in Treponema phagedenis-like strain V1 from digital dermatitis lesions by phage display. Vet. Microbiol. 153, 315–322. doi: 10.1016/j.vetmic.2011.06.005
Rusch, S. L., and Kendall, D. A. (2007). Interactions that drive Sec-dependent bacterial protein transport. Biochemistry 46, 9665–9673. doi: 10.1021/bi7010064
Russel, M., Lowman, H. B., and Clackson, T. (2004). “Introduction to phage biology and phage display,” in Practical Approach to Phage Display, eds T. Clackson and H. B. Lowman (New York, NY: Oxford University Press Inc), 1–26.
Russel, M., and Model, P. (2006). “Filamentous phage,” in The Bacteriophages, 2nd Edn. ed. R. C. Calendar (New York, NY: Oxford University Press Inc), 146–160.
Saggy, I., Wine, Y., Shefet-Carasso, L., Nahary, L., Georgiou, G., and Benhar, I. (2012). Antibody isolation from immunized animals: comparison of phage display and antibody discovery via V gene repertoire mining. Protein Eng. Des. Sel. 25, 539–549. doi: 10.1093/protein/gzs060
Samuelson, J. C., Chen, M., Jiang, F., Moller, I., Wiedmann, M., Kuhn, A., et al. (2000). YidC mediates membrane protein insertion in bacteria. Nature 406, 637–641. doi: 10.1038/35020586
Schier, R., Bye, J., Apell, G., Mccall, A., Adams, G. P., Malmqvist, M., et al. (1996). Isolation of high-affinity monomeric human anti-c-erbB-2 single chain Fv using affinity-driven selection. J. Mol. Biol. 255, 28–43. doi: 10.1006/jmbi.1996.0004
Shkoporov, A. N., Khokhlova, E. V., Kafarskaia, L. I., Pavlov, K. A., Smeianov, V. V., Steele, J. L., et al. (2008). Search for protein adhesin gene in Bifidobacterium longum genome using surface phage display technology. Bull. Exp. Biol. Med. 146, 782–785. doi: 10.1007/s10517-009-0423-4
Speck, J., Arndt, K. M., and Muller, K. M. (2011). Efficient phage display of intracellularly folded proteins mediated by the TAT pathway. Protein Eng. Des. Sel. 24, 473–484. doi: 10.1093/protein/gzr001
Steiner, D., Forrer, P., and Pluckthun, A. (2008). Efficient selection of DARPins with sub-nanomolar affinities using SRP phage display. J. Mol. Biol. 382, 1211–1227. doi: 10.1016/j.jmb.2008.07.085
Szabo, Z., and Pohlschroder, M. (2012). Diversity and subcellular distribution of archaeal secreted proteins. Front. Microbiol. 3:207. doi: 10.3389/fmicb.2012.00207
t Hoen, P. A., Jirka, S. M., Ten Broeke, B. R., Schultes, E. A., Aguilera, B., Pang, K. H., et al. (2012). Phage display screening without repetitious selection rounds. Analyt. Biochem. 421, 622–631. doi: 10.1016/j.ab.2011.11.005
Thompson, B. J., Widdick, D. A., Hicks, M. G., Chandra, G., Sutcliffe, I. C., Palmer, T., et al. (2010). Investigating lipoprotein biogenesis and function in the model Gram-positive bacterium Streptomyces coelicolor. Mol. Microbiol. 77, 943–957. doi: 10.1111/j.1365-2958.2010.07261.x
Tian, F., Tsao, M. L., and Schultz, P. G. (2004). A phage display system with unnatural amino acids. J. Am. Chem. Soc. 126, 15962–15963. doi: 10.1021/ja045673m
Tjalsma, H., Antelmann, H., Jongbloed, J. D., Braun, P. G., Darmon, E., Dorenbos, R., et al. (2004). Proteomics of protein secretion by Bacillus subtilis: separating the “secrets” of the secretome. Microbiol. Mol. Biol. Rev. 68, 207–233. doi: 10.1128/MMBR.68.2.207-233.2004
Tjalsma, H., Bolhuis, A., Jongbloed, J. D., Bron, S., and Van Dijl, J. M. (2000). Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey of the secretome. Microbiol. Mol. Biol. Rev. 64, 515–547. doi: 10.1128/MMBR.64.3.515-547.2000
Tjhung, K. F., Deiss, F., Tran, J., Chou, Y., and Derda, R. (2015). Intra-domain phage display (ID-PhD) of peptides and protein mini-domains censored from canonical pIII phage display. Front. Microbiol. 6:340. doi: 10.3389/fmicb.2015.00340
Tsukazaki, T., Mori, H., Echizen, Y., Ishitani, R., Fukai, S., Tanaka, T., et al. (2011). Structure and function of a membrane component SecDF that enhances protein export. Nature 474, 235–238. doi: 10.1038/nature09980
Tuffin, M., Anderson, D., Heath, C., and Cowan, D. A. (2009). Metagenomic gene discovery: how far have we moved into novel sequence space? Biotechnol. J. 4, 1671–1683. doi: 10.1002/biot.200900235
Ushida, K., Newbold, C. J., and Jouany, J.-P. (1997). Interspecies hydrogen transfer between the rumen ciliate Polyplastron multivesiculatum and Methanosarcina barkeri. J. Gen. Appl. Microbiol. 43, 129–131. doi: 10.2323/jgam.43.129
Van Gerven, N., Waksman, G., and Remaut, H. (2011). Pili and flagella biology, structure, and biotechnological applications. Prog Mol. Biol. Transl. Sci. 103, 21–72. doi: 10.1016/B978-0-12-415906-8.00005-4
Vasi, J., Frykberg, L., Carlsson, L. E., Lindberg, M., and Guss, B. (2000). M-like proteins of Streptococcus dysgalactiae. Infect. Immun. 68, 294–302. doi: 10.1128/IAI.68.1.294-302.2000
Vodnik, M., Zager, U., Strukelj, B., and Lunder, M. (2011). Phage display: selecting straws instead of a needle from a haystack. Molecules 16, 790–817. doi: 10.3390/molecules16010790
Wall, T., Roos, S., Jacobsson, K., Rosander, A., and Jonsson, H. (2003). Phage display reveals 52 novel extracellular and transmembrane proteins from Lactobacillus reuteri DSM 20016(T). Microbiology 149, 3493–3505. doi: 10.1099/mic.0.26530-0
Wallin, E., and Heijne, G. V. (1998). Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 7, 1029–1038. doi: 10.1002/pro.5560070420
Walsh, C. (2000). Molecular mechanisms that confer antibacterial drug resistance. Nature 406, 775–781. doi: 10.1038/35021219
Wang, M., Beck, C. R., English, A. C., Meng, Q., Buhay, C., Han, Y., et al. (2015). PacBio-LITS: a large-insert targeted sequencing method for characterization of human disease-associated chromosomal structural variations. BMC Genomics 16:214. doi: 10.1186/s12864-015-1370-2
Wernérus, H., and Ståhl, S. (2004). Biotechnological applications for surface-engineered bacteria. Biotechnol. Appl. Biochem. 40, 209–228. doi: 10.1042/BA20040014
Williams, R. J., Henderson, B., and Nair, S. P. (2002a). Staphylococcus aureus fibronectin binding proteins A and B possess a second fibronectin binding region that may have biological relevance to bone tissues. Calcif. Tissue Int. 70, 416–421. doi: 10.1007/s00223-001-2073-z
Williams, R. J., Henderson, B., Sharp, L. J., and Nair, S. P. (2002b). Identification of a fibronectin-binding protein from Staphylococcus epidermidis. Infect. Immun. 70, 6805–6810. doi: 10.1128/IAI.70.12.6805-6810.2002
Wooldridge, K. (ed.). (2009). Bacterial Secreted Proteins: Secretory Mechanisms and Role in Pathogenesis. Norfolk: Caister Academic Press.
Xing, M. N., Zhang, X. Z., and Huang, H. (2012). Application of metagenomic techniques in mining enzymes from microbial communities for biofuel synthesis. Biotechnol. Adv. 30, 920–929. doi: 10.1016/j.biotechadv.2012.01.021
Yang, X.-Y., Lu, J., Sun, X., and He, Q.-Y. (2012). Application of subproteomics in the characterization of Gram-positive bacteria. J. Proteomics 75, ch2803–ch2810. doi: 10.1016/j.jprot.2011.12.027
Yen, M., and Yin, J. (2007). High-throughput profiling of posttranslational modification enzymes by phage display. Biotechniques 43, 31–33. doi: 10.2144/000112502
Yu, N. Y., Wagner, J. R., Laird, M. R., Melli, G., Rey, S., Lo, R., et al. (2010). PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26, 1608–1615. doi: 10.1093/bioinformatics/btq249
Zacchi, P., Sblattero, D., Florian, F., Marzari, R., and Bradbury, A. R. (2003). Selecting open reading frames from DNA. Genome Res. 13, 980–990. doi: 10.1101/gr.861503
Zhang, K., He, J., Yang, M., Yen, M., and Yin, J. (2009). Identifying natural product biosynthetic genes from a soil metagenome by using T7 phage selection. Chembiochem 10, 2599–2606. doi: 10.1002/cbic.200900297
Zhou, M., Boekhorst, J., Francke, C., and Siezen, R. J. (2008). LocateP: genome-scale subcellular-location predictor for bacterial proteins. BMC Bioinform. 9:173. doi: 10.1186/1471-2105-9-173
Zhou, M., Theunissen, D., Wels, M., and Siezen, R. (2010). LAB-Secretome: a genome-scale comparative analysis of the predicted extracellular and surface-associated proteins of Lactic Acid Bacteria. BMC Genomics 11:651. doi: 10.1186/1471-2164-11-651
Zuber, B., Haenni, M., Ribeiro, T., Minnig, K., Lopes, F., Moreillon, P., et al. (2006). Granular layer in the periplasmic space of gram-positive bacteria and fine structures of Enterococcus gallinarum and Streptococcus gordonii septa revealed by cryo-electron microscopy of vitreous sections. J. Bacteriol. 188, 6652–6660. doi: 10.1128/JB.00391-06
Keywords: phage display, secretome, adhesins, metagenomics, next generation sequencing, bacteriophage
Citation: Gagic D, Ciric M, Wen WX, Ng F and Rakonjac J (2016) Exploring the Secretomes of Microbes and Microbial Communities Using Filamentous Phage Display. Front. Microbiol. 7:429. doi: 10.3389/fmicb.2016.00429
Received: 22 September 2015; Accepted: 17 March 2016;
Published: 07 April 2016.
Edited by:
Gilbert Greub, University of Lausanne, SwitzerlandReviewed by:
Bradley D. Jones, The University of Iowa, USANicole Jane Moreland, University of Auckland, New Zealand
Copyright © 2016 Gagic, Ciric, Wen, Ng and Rakonjac. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dragana Gagic, d.gagic@massey.ac.nz
 Dragana Gagic
Dragana Gagic Milica Ciric
Milica Ciric Wesley X. Wen
Wesley X. Wen Filomena Ng
Filomena Ng Jasna Rakonjac
Jasna Rakonjac