- 1Laboratorio de Parasitología Molecular, IIB-INTECH, CONICET-UNSAM, Chascomús, Argentina
- 2Cell Cycle Genomic Instability Laboratory, Fundación Instituto Leloir, IIBBA-CONICET, Chascomús, Argentina
Toxoplasma gondii is an apicomplexan parasite of medical and veterinary importance which causes toxoplasmosis in humans. Great effort is currently being devoted toward the identification of novel drugs capable of targeting such illness. In this context, we believe that the thorough understanding of the life cycle of this model parasite will facilitate the identification of new druggable targets in T. gondii. It is important to exploit the available knowledge of pathways which could modulate the sensitivity of the parasite to DNA damaging agents. The homologous recombination repair (HRR) pathway may be of particular interest in this regard as its inactivation sensitizes other cellular models such as human cancer to targeted therapy. Herein we discuss the information available on T. gondii's HRR pathway from the perspective of its conservation with respect to yeast and humans. Special attention was devoted to BRCT domain-containing and end-resection associated proteins in T. gondii as in other experimental models such proteins have crucial roles in early/late steps or HRR and in the pathway choice for double strand break resolution. We conclude that T. gondii HRR pathway is a source of several lines of investigation that allow to to comprehend the extent of diversification of HRR in T. gondii. Such an effort will serve to determine if HRR could represent a potential targer for the treatment of toxoplasmosis.
Introduction
The protozoan parasite Toxoplasma gondii is a medical and veterinary relevant pathogen (Tenter et al., 2000; Pfaff et al., 2014). Toxoplasma belongs to phylum Apicomplexa among other important human and veterinary parasites such as Plasmodium spp., Cryptosporidium spp., Eimeria spp. Albeit the toxoplasmic infection is usually asymptomatic, severe complications, and even death might occur as a result of a congenital infection or in immunocompromised individuals (e.g., AIDS, transplantation). Congenital toxoplasmosis causes several types of neurological defects, chorioretinitis and in some cases even abortion (Cortés et al., 2012; Moncada and Montoya, 2012; Torgerson and Mastroiacovo, 2013). In immunocompromised patients, the reactivation of the infection may trigger further complications including neurological defects, and encephalitis (Yan et al., 2013).
T. gondii is an intracellular obligated protozoan parasite with a life cycle that includes sexual and asexual stages. Asexual replication occurs in a wide variety of intermediate host species and tissues and is characterized by two stages: the rapidly growing “tachyzoites” which is sensitive to the immune system of the host and several drugs, and the slowly dividing encysted “bradyzoites” which evades both the host immune response and currently available anti-Toxoplasma drugs (Dubey, 1998; Weiss and Kim, 2000). Besides, anti-folate treatment is only effective against the tachyzoite stage, but is toxic in that it causes bone marrow depression; moreover, many patients are allergic to the sulfa drug component (Baatz et al., 2006; Cortés et al., 2012). The pathogenicity of toxoplasmosis has been associated to multiple cycles of host cell invasion, intracellular division of the parasite and release from host cells. T. gondii amplification takes place in any nucleated cell within a parasitophorous vacuole generated by an internal budding process known as endodiogeny (Gubbels et al., 2008; Francia and Striepen, 2014).
While many molecular pathways including cell cycle and cell duplication were thoroughly characterized in the parasite, the molecular signals ruling DNA replication in T. gondii are yet poorly characterized. Notably, after host cell invasion, the tachyzoite replicates with a doubling time of 5–9 h (Radke et al., 2001). We have recently proposed that such fast and uninterrupted rounds of DNA replication during the tachyzoite stage might trigger replication stress. In fact, we have evidenced a striking increase in the levels of a bona-fide replication-stress marker, the phosphorylation at Ser132 of γH2A.X, in T. gondii tachzyoite (Dalmasso et al., 2009). Albeit other replication-associated defects may also trigger γH2A.X activation, the classical interpretation of γH2A.X accumulation is the generation of double strand break (DSB) (Redon et al., 2002; Tu et al., 2013; Turinetto and Giachino, 2015). DSBs are extremely genotoxic DNA lesions capable of impairing central DNA process such as DNA transcription, replication, and segregation. Given that DSBs can be repaired by more than one mechanism, DSBs accumulated during the DNA replication of tachyzoite most likely require a precise choice of DNA repair pathway. A failure or a delay in the repair of DSBs may trigger cell death due to the accumulation of genomic and chromosomic rearrangements as has been showed in cancer cells (Prakash et al., 2015).
If DSBs accumulate during the DNA replication of tachyzoite, it is important to discuss the DNA repair pathways available for the repair of DSBs in T. gondii. In Eukaryotes, two well-characterized pathways are in charge of DSB Repair: Homologous Recombination repair (HRR) and Non-Homologous End Joining (NHEJ). While it is broadly accepted that HRR is error-free and NHEJ is error-prone, new evidence suggests that, at least, under certain cirscutances, HRR can also represent an error-prone mechanism and NHEJ can be very precise depending on the structure of the DNA ends (Betermier et al., 2014; Guirouilh-Barbat et al., 2014).
Intriguingly, while most DNA repairs pathways are conserved in T. gondii, recently reviewed inSmolarz et al. (2014), differences in the HRR cascade have been reported in different organisms (Smith, 2012; Blackwood et al., 2013; Daley et al., 2013; Yoshiyama et al., 2013). Suchdiversification indicates the existence of a window of opportunity for the identification of specific HRR components in T.gondii. If available, such factors could represent attractive candidates for the development of drug against toxoplasmosis. Hence, herein we analyze the extent of conservation between the HRR components of T. gondii and their yeast and human counterparts.
The Homologous Recombination in T. gondii
HRR is preferentially an error-free mechanism which represents the preferred pathway chosen in eukaryotes for the repair of DSBs during the late S/G2-phases of cell cycle. This mechanism has been extensively studied in both yeast and higher eukaryotes (Daley et al., 2013; Jasin and Rothstein, 2013). The restriction of HRR to S/G2 phases is linked to the requirement of homologous sequences as a template for DNA repair (Sancar et al., 2004). Typical substrates for HRR include: (a) direct double-ended DSBs generated by genotoxic agents such as γ-irradiation and X-rays, (b) inter-strand crosslinks generated after exposure to genotoxins such as mitomycin C (MMC), and (c) one-ended DSBs generated after fork collapse resulting from persistent stalling at bulky adduct or at naturally-occurring replication barriers. The resolution of direct DSBs by an HRR subpathway may or may not involve crossing over. One-ended DSBs are expected to be resolved by another HRR sub-pathway involving long range D-loop migration (break-induced repair) (Carr and Lambert, 2013; Malkova and Ira, 2013). If homologous sequences are not available, for example during G1, DSBs are repaired by NHEJ, a pathway that prompts rapid fusion between the ends of double-ended DSBs. In contrast, NHEJ is disfavored during S phase since its activation at one-ended DSBs can jeopardize genomic instability by fusing non-homologous chromosomes. Hence, while NHEJ can function along the cell cycle (Shibata and Jeggo, 2014), NHEJ is the pathway chosen for the repair of DSBs in G1 and HRR is preferentially activated at collapsed replication forks during S and G2 phases (Johnson and Jasin, 2000; Sancar et al., 2004; Blackwood et al., 2013).
Effectors of the HRR and NHEJ pathways were identified in T. gondii (Smolarz et al., 2014). When attempting to establish the hierarchy between both pathways in the parasite, surprising results were obtained. The inoculation of linear plasmid in tachyzoites robustly activates the NHEJ pathway, while gene replacement by HRR was rarely detected (Fox et al., 2009). Notably, these results suggested that, in contrast to yeast and humans, the NHEJ pathway is the pathway preferentially used by T. gondii. It should however be mentioned that HRR can efficiently be activated in T. gondii when NHEJ factors Ku70/Ku80 are eliminated by means of deletion of the Ku80. In such scenario efficient HRR-dependent integration rate at correct locus of different plasmid constructions were observed (Fox et al., 2009; Huynh and Carruthers, 2009). Moreover, when focusing on events such as crossing over, a high efficiency of activation was observed, hence indicating active HRR during sporozoite development (Khan et al., 2014). Together, these evidences demonstrate that T. gondii has an intact and functional HRR molecular pathway.
The Homologous Recombination Basic Machinery is Conserved in T. gondii
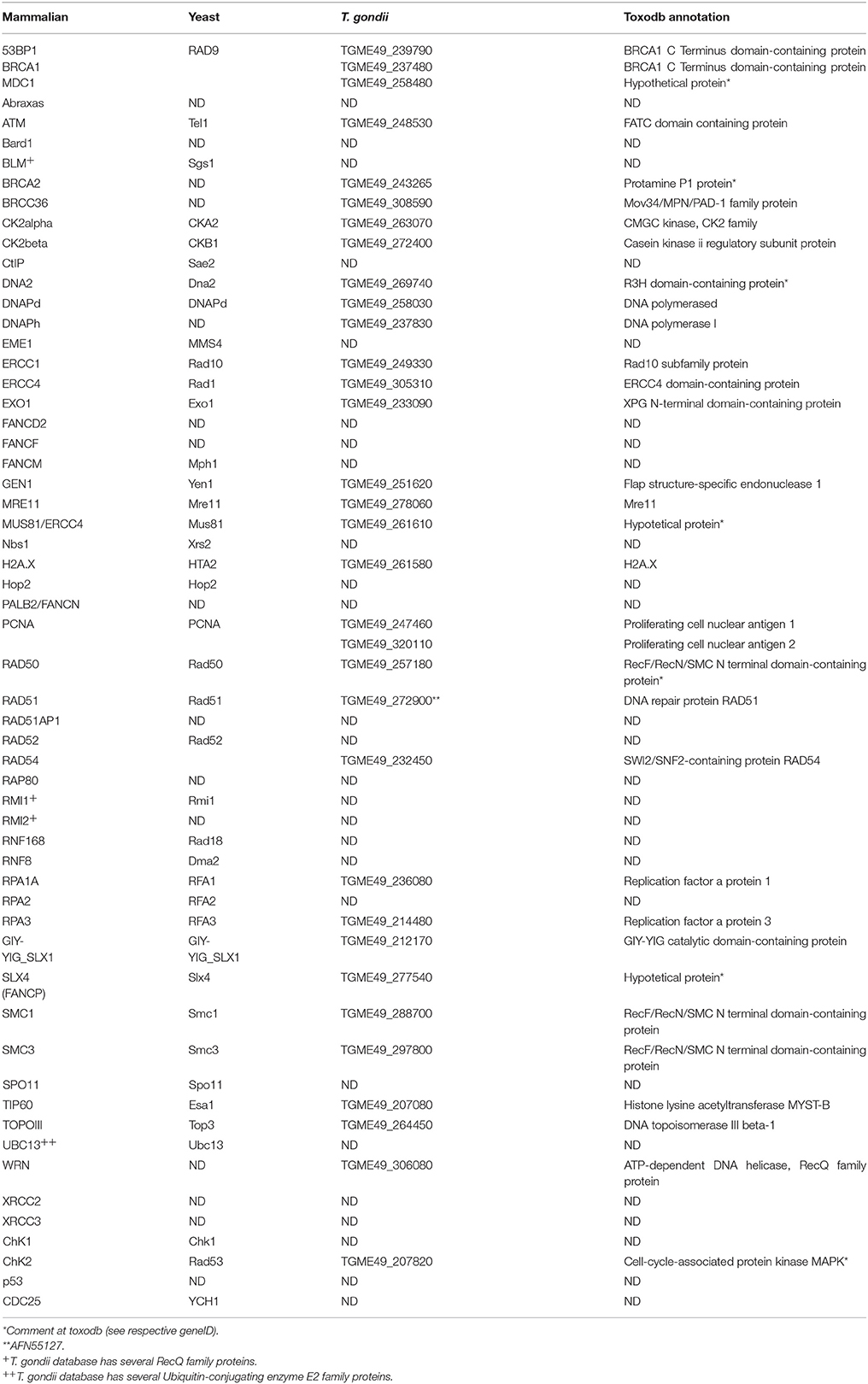
The HRR pathway is activated after DSB recognition by DNA damage sensors (e.g., γH2A.X), and signal transducers (e.g., ATM/Tel1 PIKK4 kinase). The commitment of DSBs to HRR resolution is achieved by mediators/adaptors (e.g., BRCA1 in mammals) and effectors (e.g., Mre11, RAD50, Nbs1/Xrs2 complex; Prakash et al., 2015). HRR core components include many DNA damage repair (DDR) protein (e.g., RAD51, BRCA2, RAD52) that regulate homology search and other downstream events(Jasin and Rothstein, 2013). Herein we evaluate whether mammalian and yeast factors are present in T. gondii by Gene Text Search at Toxodb database (Table S1, Figures S1, S2, and Figure 3). Table S1 also contains putative HRR counterparts from Plasmodium falciparum, another apicomplexan parasite. We found 39 putative HRR components in T. gondii (Table S1). In addition, we have attempted to infer whether the conserved HRR factors retrieved in T. gondii are sufficient to support full HRR activation when establishing a direct comparison with the essential components of the HRR cascade in yeast and humans (Figure S2). As a result we have generated a putative basic model of T. gondii HRR (Figure 1). The more relevant HRR proteins found in T. gondii are listed in Table 1.
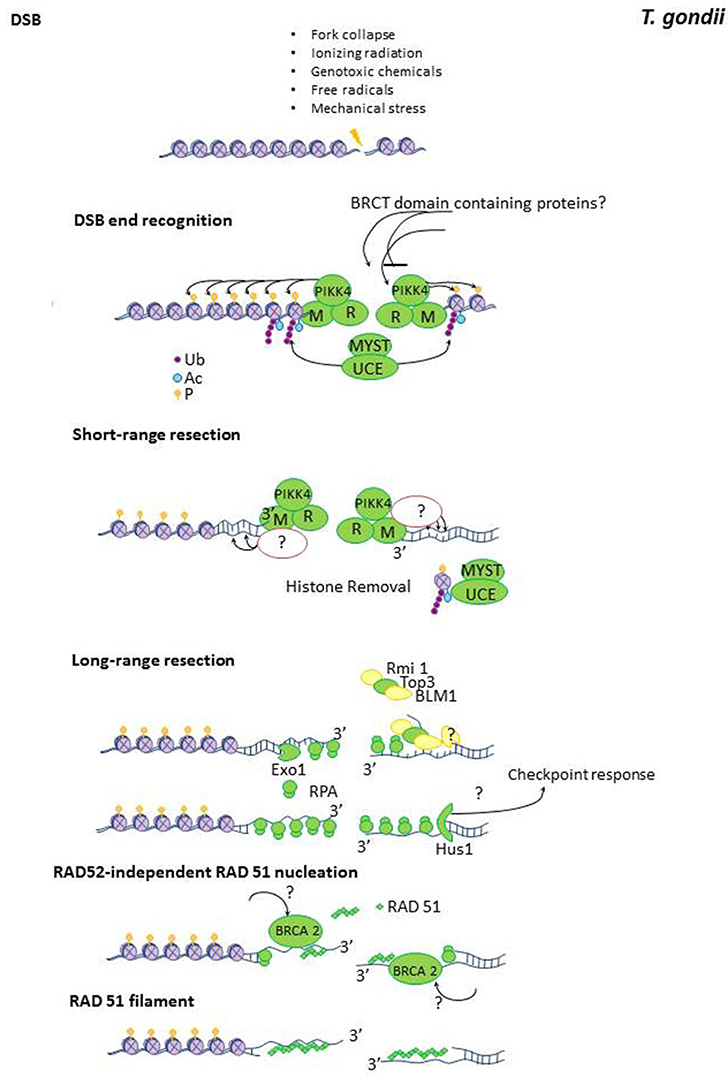
Figure 1. Homologous recombination repair in T. gondii. Potential components of the HRR pathway in T. gondii. The pathway was build by taking the HRR components described in mammals, in yeast as references and using the information regarding the putative HRR protein retrieved from www.toxodb.org. TGME49_258480, TGME49_239790, and TGME49_237480 are putative BRCT domain containing proteins. PIKK4 is the putative ATM/Tel1 kinase TGME49_248530. UCE is a ubiquitin conjugate enzyme. High levels of conservation between T. gondii and humans/yeast protein domains are indicated in green. Proteins which are not detected by annotation but have compatible features with the respective protein are shown as yellow shapes.
Toxoplasma
HRR will be discussed below and will be organized accordingly to the following HRR stages: (A) DSB recognition, (B) end-resection and generation of protruding ends for homologous search, (C) strand invasion, (D) homologous DNA synthesis, and (E) resolution of DNA- repair intermediates.
DSB Recognition
The proteins in charge of DSB recognition are well-conserved in all three kingdoms. In bacteria, DSBs are recognized by SbcD and SbcC while in Archaea and Eukaryota these components are known as Mre11 and RAD50, respectively (Blackwood et al., 2013). Yeast and vertebrates have an additional highly divergent protein, Xrs2 (yeast) and Nbs1 (higher eukaryotes), which along with Mre11 and RAD50 form the MRX/N complex. From T. gondii database analysis it could be inferred that the Mre11 and RAD50 proteins are present, while Nbs1 was not detected in the database (Table 1 and Table S1). Recently, a functional plasmodial Mre11(PF3D7_0107800, Tables S1), similar to putative T. gondii Mre11, was identified (Badugu et al., 2015). The lack of Nbs1/Xrs2 is unexpected since Nbs1/Xsr2 is required for optimal activation of the checkpoint kinase ATM which is required for the arrest of the cell cycle and to trigger DNA damage-induced apoptosis (Difilippantonio and Nussenzweig, 2007). Nbs1 senses the conformation of Mre11 dimer, which is in turn influenced by RAD50-ATP state, promoting the activation of Mre11 (Lafrance-Vanasse et al., 2015). Nbs1 possesses a forkhead associated (FHA) domain and two breast cancer-associated 1C terminus (BRCT) domains known to bind phosphoproteins such as CtIP facilitating its recruitment at DSB (Williams et al., 2009). Moreover, Nbs1 also interacts with ATM through its C-terminal FXF/Y motif promoting its activation (You et al., 2005). We speculate that Nbs1 is not annotated in T. gondii genome database possibly due to its tendency to diverge. However, based on the above-mentioned data we believe that the MR complex, in charge of DSB recognition is mainly conserved in T. gondii (Figure 1).
End-Resection
In order to generate protruding ssDNA ends with invasion capacity, DSBs need to be extensively processed after MRN loading. Central enzymes capable of achieving such processing are the single strand 3′-5′ exonuclease and endonuclease Mre11 and the endonuclease CtIP [CtBP (C-terminal-binding protein)-interacting protein] (Sae2 in yeast). After an initial cleavage by Mre11, a second end-resection in eukaryotes depends mainly upon the Exo1 5′-3′ exonuclease which exerts long end resection forming the protruding DNA ends required for invasion and homologous search. An alternative pathway to end resection involves the Dna2 exonuclease and the BLM helicase (Figures S1, S2). Although CtIP (Sae2) is not identified in T. gondii database (there is a Sae2/CtIP annotated protein [TGVEG_252280] in toxodb but to our knowlege with no BLASTP evidence that support it.), a conserved Mre11 (see above) and a putative Exo1 exonuclease (Table 1) are present. Therefore, the end-resection stage of HRR is potentially conserved in this organism (Figure 1).
Strand-Invasion
In this phase, the protruding ssDNA is coated with a factor known as RecA in bacteria, RAD51 in eukaryotes or RadA in Archae. RAD51 facilitates strand invasion and homology search (Jasin and Rothstein, 2013). To promote RAD51 loading, factors known as mediators facilitate the displacement of the ssDNA coating factor, RPA. In eukaryotes, RAD51 is recruited by RAD52 or BRCA2 (Liu and Heyer, 2011). RAD51-coated ssDNA actively searches for homologus DNA, an event which is facilitated by increased chromosome moving (ICM) promoted by protein such as Rad9, RAD51, RAD54, Mec1/ATR, among others (Mine-Hattab and Rothstein, 2013) The analysis of T. gondii database revealed a putative sequence for RAD51 and BRCA2 but not RAD52 (Table 1). In fact, TgRad51 has been characterized by Achanta et al. (2012). When the authors compared it to a yeast cell model, they concluded that TgRad51 is less efficient in gene targeting and gene conversion than yeast Rad51. We speculate that a slight defect in this particular event may support the puzzling preponderance of NHEJ in T.gondii which has been discussed in previous sections. In fact, in the next section we will present the multiple levels of cross-regulation between HRR and NHEJ mediators and their major influence in the DSBs repair pathway choice.
Homologous DNA Synthesis and Resolution of DNA- Repair Intermediates
From mammals to yeast, once RAD51 bounds to ssDNA it generates a contiguous helical nucleoprotein filament, which searches for an intact homologous dsDNA template (Figure 2 and Figure S1). When the homologous region is found, RAD51 promotes the exchange of DNA strands leading to the formation of joint molecules and D-loops (Mehta and Haber, 2014). RAD54, a member of the Snf2-family of SF2 helicases also binds to RAD51 (Figure 1). Instead of taking part in the separation of the DNA duplex, RAD54 acts as a motor protein that translocates on duplex DNA and remodels specific protein–duplex DNA complexes (Pazin and Kadonaga, 1997; Ceballos and Heyer, 2011). The homology between RAD54 and Snf2/Swi2 further supports a role of RAD54 in chromatin relaxation during HRR, which could facilitate many HRR events such as Rad51 filament assembly, homology search, DNA strand invasion, or even later HRR stages (Ceballos and Heyer, 2011). In fact, RAD54 is crucial to promote branch migration (Mazin et al., 2010) when the DNA polymerase polη extends DNA from D loop recombination intermediates, using an invading strand as a primer, (McIlwraith et al., 2005), that generate a Holliday junction (HJ).
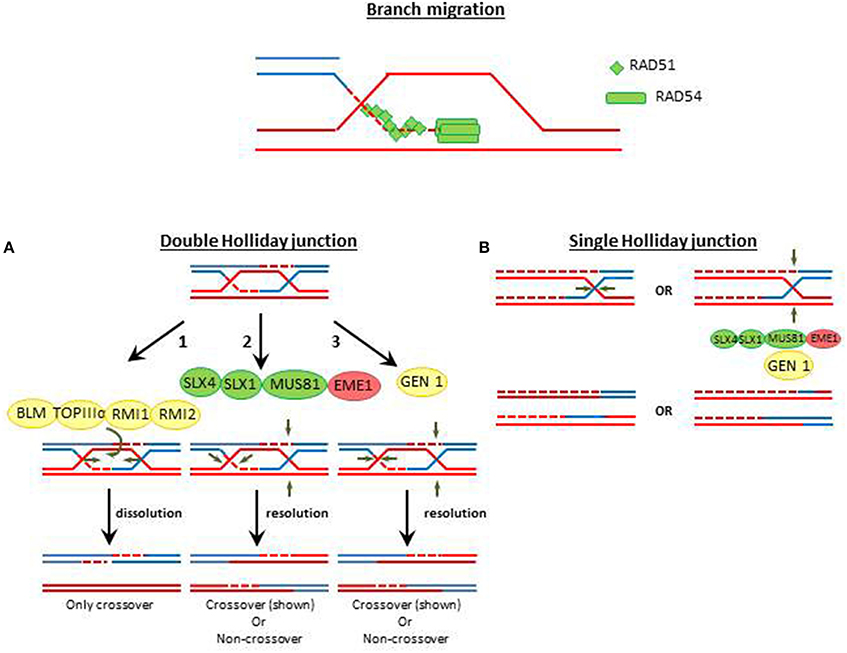
Figure 2. Holliday junction resolution pathways. Double Holliday junction dissolution is a conserved mechanism by which crossover is prevented and noncrossover HRR events are facilitated (Sarbajna and West, 2014). The Double Holliday junction (DHJ) dissolution is promoted by the BLM helicase, the TopoIIIα topoisomerase and the RMI1, which influencing the dynamics of TopIIIα (Bocquet et al., 2014). Double Holliday junctions are resolved by two mechanisms. (A) The nuclease complex MUS81, EME1, SLX1, and SLX4 generate asymmetric cleavage at two positions in the DHJ. (B) The GEN1 resolvase introduces two symmetrical nicks at equivalent positions of the DHJ. In both scenarios non-crossover o crossover resolutions are possible. Single Holliday junctions are intermediates of meiotic recombination. Proteins with high level of conservation are colored in green. Proteins which are not detected by annotation but have compatible features with the respective protein are shown as yellow shapes and factors wich have not been yet identified in in T. gondii are colored in red.
Nucleases in charge of HJ resolution are the ERCC1-XPF/SLX1/SLX4 and the Mus81-EME1/Mms4 complexes (Cejka, 2015). A third complex which may also resolve HJ when SLX4 is absent is the BLM/GEN1 nuclease (Garner et al., 2013). MUS81-EME1/Mms4 are essential components of HJ resolvase (Boddy et al., 2001) and seems to be the preferred nuclease in charge of the processing of crossover events while GEN1 seems to work as a backup pathway (Garner et al., 2013). Such hierarchy is also influenced by the cell cyle. During unperturbed duplication, different kinases (e.g., Mitosis phase CDKs) and phosphatases restrict the activity of MUS81-EME1 and GEN1 to different cell cycle phases. As a consequence of such regulation MUS82-EME/Mms4 are active during pro-metaphase and metaphase whereas GEN1/Yen1 are active during metaphase and anaphase (Matos and West, 2014).
T. gondii possess a conserved machinery responsible of HJ resolution (Table 1 and Figure 2). Still, to this date none of their components were experimentally characterized. Based on T. gondii annotation, RAD54, and TOPOIIIα are potentially expressed in the parasite (Table 1). MUS81 and SLX1 nucleases and the SLX4 scafolding factor are present in T. gondii. The same analysis provided modest evidence supporting the presence of the BLM helicase, the GEN1 nuclease and the RecQ-mediated genome instability protein 1 (RMI1). EME1/Mms4 was not found in T. gondii database (Table S1). Moreover, T. gondii database only retrieved two putative ERCC4 domain containing proteins, one resembling MUS81 and another displaying similatities with the RAD1/ERCC4-XPF endonuclease. The absence of EME1/Mms4 may indicate the existence of divergent proteins which were not yet identified. Alternatively, it is also possible that most crossover events in T. gondii relay exclusively on the GEN1 pathway. Further studies should reveal the mechanism supporting crossover and HJ resolution in the parasite.
The sexual cycle of T. gondii occurs in felines which serve as the definitive host and shed infectious oocysts in their feces. Meiosis events take place after oocyst sheed to generate haploid sporozoites. In a recent study, the mixture of Me49 and VAND strains in cats revealed both conventional and double-crossover HRR events (Khan et al., 2014). Moreover,Khan et al. (2014) has reported elevated frequency of small double-crossover events (less 1000 bp). Interestingly, double crossover events within the 1000 bp are classified as gene conversion, a mechanism associated with HRR-dependent resolution of DSB in other systems (Haber et al., 2004; Chen et al., 2007).
Collectively, the examination of the different HRR steps in T. gondii indicates that the basic HRR machinery is conserved, with the unanticipated exception of few but very important players including Nbs1/Xrs2, CtIP, RAD52 and EME1/Mms4. It is however important to mention that the evidences of a functional HRR pathway in T. gondii is solid (as it will be discussed in the next section). Thereafter we propose that the “missing” HRR components may have diverged to the point of not being recognized by data mining. Alternatively they may have been replaced by functional paralogs. In both scenarios, the identification of those central HRR components may serve as a tool to boost the rational design of drugs that may specifically impair HRR in the parasite.
The DSB Repair Pathway Choice in T. gondii
The current understanding of the DSB repair pathway choice in mammals is summarized in Figure 3 (Ceccaldi et al., 2016). In the case of mammals, the cell cycle majorly influences the DSB pathway choice. While HRR is the preferent choice during S/G2, NHEJ is the best option during the G1 phase(Sancar et al., 2004; Kass and Jasin, 2010). Such a strong influence of the cell cycle is accepted to depend on the availabity of intact sister chromatid during late S and G2 phases of the cell cycle (Kass and Jasin, 2010). Therefore, It's crucial to understand why NHEJ is dominant over HRR in T. gondii. On one hand, T. gondii tachyzoite has a cell cycle with a long G1 and no G2 phase (Radke et al., 2001). On the other hand, as we discuss bellow, the diversification in the molecules in charge of the commitment of a DSB to a given resolution pathway may, at least partially, explain why different pathway choice strategies may have evolved in T. gondii in comparison with mammals.
Figure 3. The DSB pathway choice. DSB could be repaired either by homologous recombination repair (HRR) during S/G2/M phases or by non-homologous end-joining (NHEJ) during G1 phase. The endonuclease CtIP (Sae3 in yeast) is a target of cyclin dependent kinases (CDKs) during G2/S-phase and is recruited to the DSB by the MRN (MRX in yeast) complex. In mammals, CtIP recruits BRCA1 to DSBs, a BRCT-containing factor that releases the NHEJ factor 53BP1 from the DBS site. 53BP1, in opposition to BRCA1, inhibits end resection, and promotes NHEJ activation. In yeast, the BRCT domain containing protein Rad9/Crb2 has shown similar role than 53BP1 in mammals. Phosphorylated CtIP also promotes end resection which evicts Ku70/Ku80 and commits the DSB to HRR. Proteins with high level of domain conservation are represented as green shapes, Proteins that are not present in T. gondii are represented as red shapes.
The generation of a 5′ long end resected DNA, which prevents NHEJ process, is the key event that commits DSBs to HRR (Daley et al., 2013; Daley and Sung, 2014). The end resection requires different exonucleases (Exo1 and DNA2), the exo- and endonuclease Mre11, endonucleases (CtIP/Sae2), and helicases (BLM/Sgs1) (Figures S1, S2). Almost all the above-mentioned nucleases are positively regulated by cyclin dependent kinases (CDKs) mediated phosphorylations during G2/S-phase (Huertas et al., 2008; Huertas and Jackson, 2009; Ferretti et al., 2013). These data consolidate CtIP as a key player which redirects DSBs into HRR (Kakarougkas and Jeggo, 2014). In fact, phosphorylated CtIP cooperate with MRN (MRX in yeast) to facilitate end resection (Lafrance-Vanasse et al., 2015), reducing the chances for NHEJ activation. In mammals, phosphorylated CtIP also favors HRR activation by recruiting the mediator tumor suppressor protein breast cancer 1 (BRCA1) which is a BRCA1 C-terminal (BRCT) domain containing protein. BRCA1 evicts another BRCT-containing mediator, the NHEJ factor 53BP1 from the DSB (Daley and Sung, 2014). Intriguingly, 53BP1 also has a BRCT domain but, in opposition to BRCA1, 53BP1 function is that of inhibiting end resection (Chapman et al., 2012). In fact, 53BP1 binds canonical double-ended DSBs upstream the Ku heterodimer loading during G1, facilitating NHEJ (Bothmer et al., 2010). Interestingly, at one-ended DSBs the depletion of BRCA1 suffices to promote NHEJ while the simultaneous depletion of BRCA1 and 53BP1 restores HRR, therefore demonstrating an exquisite cross-regulation of HRR or NHEJ at DSBs (Bunting et al., 2010).
The lack of conservation in the above mentioned pathway choice step, that we postulate may happen in T. gondii, is intriguing. In fact, in yeast, only one BRCT-containing protein domain associated to HRR was found: Rad9 in Saccharomyces cerevisiae or Crb2 in Schizosaccharomyces pombe. The conservation between Rad9/Crb2 and mammalian BRCT containing proteins such as 53BP1 or BRCA1 is very low. Nevertheless, Rad9/Crb2 and 53BP1 functionally overlap. Similarly to 53BP1, Rad9 blocks end resection, and inhibits Exo1- and RAD50-dependent nucleases therefore inhibiting the formation of HRR-proficient substrates (Lazzaro et al., 2008). In concordance, NHEJ is facilitated when Rad9 is recruited to DSBs by the 9-1-1 checkpoint clamp loader (Ngo and Lydall, 2015). In addition, Rad9 acts as an adaptor that favors the activation of checkpoint kinases Mec1 (ATR) or Tel1 (ATM) and RAD53 and Chk1, which in turn facilitates successful finalization of S phase and promotes cell cycle arrest in G2 and G1, creating a time window for replication-dissociated DNA repair (Gilbert et al., 2001; Blankley and Lydall, 2004; Sweeney et al., 2005). Hence, the pathway choice in yeast may be tilted toward the choice of NHEJ, as we predict it may happen also in T. gondii.
Remarkably, according to the T. gondii database, homologs of BRCA1, 53BP1, or Rad9/Crb2 were so far not reported. We speculate that it is unlikely that such regulatory factors are completely missing in T. gondii. As the degree of conservation is low between yeast and mammals, it is possible that similar diversification may have taken place in T. gondii. In fact, similar functions were showed to be accomplished by different BRCT domains-containing proteins that share only few residues including hydrophobic amino acids which may facilitate the generation of appropriate secondary structure (Bork et al., 1997; Gabrielse et al., 2006). Interestingly, in Toxoplasma database are at least three putatives BRCT domain-containing-protein (Table 1). More work is required to establish whether such proteins are functional during the DSB pathway choice. Remarkably as well, homologous of CtIP and Nbs1 (Sae2 and Xrs2 in yeast, respectively) were also not present in T. gondii database (Table 1). While, Mre11 and RAD50 are conserved in all three domains of life and therefore also in T. gondii (Blackwood et al., 2013), the existence of a functional complex lacking Nsb1 is unconvincing to us. As it was already mentioned in Section DSB Recognition, the absence of Nbs1/Xrs2 annotation in Toxoplasma database, could be explained by the fact that this protein represents a highly divergent component of the MRN complex. Hence, in order to solve the molecular bases for the apparent defect in HRR activation in T. gondii, missing components need to be identified or their absence needs to be actively proved. In any case, the strong diversification of the HRR pathway that may have taken place in T. gondii may provide initial mechanistic bases for the predominant role of NHEJ in the repair of DSBs in the tachyzoite. We still believe that such conclusion may be precipitous since it is still unclear if different sets of proteins associated with the DSBs repair pathway choice has diversified in T. gondii. This is why in our opinion, the identification of mediators that rule the DSB pathway choice in T. gondii is a field that deserves much attention. Future work may ultimately address the role in DDR of TGME49_258480, TGME49_239790, and TGME49_237480, the three putative BRCT domain containing proteins. The search and identification of other putative BRCT containing proteins may also serve to comprehend HRR activation in T. gondii and to identify species-specific druggable targets for the treatment of toxoplamosis.
γH2A.X Spreading and Foci Formation
While H2A.X may be incorporated randomly in the genome of resting cells, its phosphorylated form γH2A.X, which is modified at its C-terminal motif SQEF/Y, can accumulate in discrete subnuclear foci at replication factories. γH2A.X is directly recruited to the site of DSBs or collapsed replication forks, a complex signaling network promotes the spreading of the γH2A.X signal along the chromosome from the damaged site up to 2-Mb (Redon et al., 2002). Such an increase in γH2A.X has also been reported when replication forks collapse in cells undergoing fast replication, such as precancerous and cancerous cells, showing an 8-fold increase both in levels of H2A.X and in γH2A.X when compared to resting cells (Bartkova et al., 2005). T. gondii tachyzoites also undergo fast DNA duplication and, similarly to cancer cells, increase γH2A.X as revealed by Western blot and mass spectrometry analysis (Dalmasso et al., 2009; Nardelli et al., 2013). The phosphorylation of the SQE motif of H2A.X in response to DSB relies on PIKK4 kinases ATM, ATR, or DNA-PK (van Attikum and Gasser, 2009), all of them apparently present in T. gondii (ATM and ATR are shown in Table S1). Intriguingly, while H2A.X is conserved in T. gondii, it is not present in all apicomplexas as well as other protozoan organisms (Dalmasso et al., 2009). This may suggest that the spreading of γH2A.X and foci formation in response of DSBs, while conserved in T. gondii, is not essential for HRR activation in all species.
The function of γH2A.X at DSB site and its spreading to both side of the DSB has been associated with the facilitation of homology search (Renkawitz et al., 2013) and with the recruitment of different components of DDR at the foci (Figures S1, S2). In mammals, one of the proteins that is recruited by γH2A.X is the BRCT-containing sensor MDC1 (mediator of DNA damage checkpoint protein 1), which was initially identified as a positive regulator of cell-cycle checkpoints effectors SMC1 and Chk1 during the S-phase and G2/M phases of the cell cycle (Stewart et al., 2003; Scully and Xie, 2013). MDC1 phosphorylation at its N-terminal region by casein kinase 2 (CK2) increases its interaction with Nbs1, enhancing the recruitment of the MRN complex at DSB site (Stewart et al., 2003; Melander et al., 2008; Spycher et al., 2008; Wu et al., 2008). Similarly, in yeast the checkpoint mediator Rad9/Crb2 relies on its C-terminal BRCT domain to interact with γH2A.X and to be recruited to the DSB (van Attikum and Gasser, 2009). In opposition to mammals, yeast has only one BRCT containing protein and therefore it is likely that a lower eukaryote as T. gondii may also have few BRCT-containing protein with HRR-regulating abilities. As mentioned above, T. gondii has three putative BRCT domain containing protein (TGME49_258480TGME49_239790 and TGME49_237480). In the future it will be important to determine whether these proteins have a function during DNA damage response and if that is the case, whether they act as mediator and/or has the ability to interact with γH2A.X.
RAD52- Independent HR Pathway
Once the DSB is commited to HRR, the ssDNA generated by nucleases is immediately protected by RPA/RFA proteins. An important step is then the removal of RPA/RFA to allow the binding of the homology searching RAD51 recombinase to ssDNA. In Figures S1, S2 we summarize the current understanding of the mechanisms that regulate RAD51 loading to DNA (Zhang et al., 2009; Buisson et al., 2010, 2014; Dray et al., 2010; Ramadan, 2012; Mermershtain and Glover, 2013; Park et al., 2014). RAD52 is crucial both for displacement of RPA by RAD51 and for the stimulation of RAD51-mediated homologous DNA pairing (Baumann and West, 1999; Jackson et al., 2002). In S. cerevisiae, RAD52 plays a key role in HRR, but in vertebrates, RAD52 knockouts only have reduced HRR but do not have hypersensitivity to agents that induce DSBs (Rijkers et al., 1998; Paques and Haber, 1999). However, in vertebrates, the absence of both BRCA2 and RAD52 is synthetic lethal and is associated with severe chromosomal fragility (Feng et al., 2011). Hence, RAD52 and BRCA2 represent alternative pathways that converge to support RAD51-mediated HRR (Liu and Heyer, 2011; Lok and Powell, 2012). Moreover, BRCA2 displaces RPA from ssDNA and promotes RAD51 filament formation and strand exchange more efficiently than yeast and human RAD52 (Jensen et al., 2010). To this date it is unclear if Caenorhabditis elegans and Drosophila melanogaster have a RAD52 homolog but they do have a BRCA2 protein (Liu and Heyer, 2011). In C. elegans, BRCA2 (CeBRC-2) stimulates both RAD51-mediated D-loop formation and single strand annealing of RPA-oligonucleotide complexes (Petalcorin et al., 2006). This suggests that CeBRC-2 may have taken over the role of vertebrate RAD52 in DNA single-strand annealing.
As for other mediators, we and others have found no evidence of RAD52 expression in T. gondii, Plasmodium spp., and trypanosomatids genome (Passos-Silva et al., 2010; Lee et al., 2014; Smolarz et al., 2014). In contrast, RAD52 has been identified in Entamoeba hystolytica and Giardia spp. (Lopez-Camarillo et al., 2009). Hence, it is possible that RAD52 may indeed not be part of the DNA damage response pathway in T. gondii and protozoan parasites. In such scenario, the recruitment of T. gondii RAD51 to the DSB might be controlled by a RAD52-independent mechanism as proposedSmolarz et al. (2014). Interestingly, it is also possible that the putative BRCA2 may represent the sole protein in charge of displacing RPA and recruiting RAD51 to ssDNA in this organisms, a scenario which is not exceptional (Petalcorin et al., 2006; Liu and Heyer, 2011).
BRCA2 is a protein with multiple domains, including oligonucleotide/oligosaccharide-binding (OB) domains, BRC tandem repeats, and TR2 C-terminal domain. In human BRCA2, the three OB repeats are implicated on ssDNA binding, whereas the BRC repeats promote the protein-protein interactions that facilitate the DNA binding and the focal organization of RAD51(Flynn and Zou, 2010). Moreover, the TR2 domain in the BRCA2 C-terminus stabilizes RAD51 nucleoprotein filament (Lee, 2014). Depending on the organisms, the interaction of BRC domains with RAD51 can be weak or strong therefore positively or negatively impacting on the control over RAD51's activities (Davies et al., 2001). For the putative BRCA2 protein, TGME49_243265, that we have found in T. gondii database (Table 1), it could be identified two BRCA2 domains. One at position 2505-2619 (pfam09103) and other at position 760 to 1838. Hence, near 16 repeat sequences in TGME49_243265 presents striking similarities to the BRC repeats present in humans or Trypanosoma brucei (Trenaman et al., 2013; Lee, 2014). Interestingly, in T. brucei, RAD51 encodes a high number of BRC repeats which facilitate RAD51 foci formation. As the number of cells with detectable RAD51 foci is proportional to the number of BRC-repeats (Trenaman et al., 2013), the increased BRC repeats in RAD51, might be relevant when attempting RAD51 loading in the absence of multiple mediators.
In mammals, ubiquitylated H2A.X recruits a complex of proteins which promote BRCA2 loading to DNA (Scully and Xie, 2013). Hence, the analysis of post-translational modifications in histones of T. gondii may be informative. Recent reports indicates that this organism has four detectable ubiquitylation on H2A.X (Silmon de Monerri et al., 2015). It will be of interest to determine the role of these PTMs on T. gondii H2A.X in HRR.
The Role of Chromatin in HRR
In addition to H2A.X, several PTMs of histones such as acetylation, phosphorylation, methylation, and ubiquitination, occur at regions of damaged DNA(Gospodinov and Herceg, 2013). Moreover, chromatin-remodeling complexes INO80, SWR1, SWI/SNF, RSC, and NuRD are all important initiators of the DSB repair pathway in both low and high eukaryotes (van Attikum and Gasser, 2009). Likewise, H2A.Z may have a crucial role in defining the extent of the nucleosome-free DNA regions, restricting DNA resection by CtIP and favoring the recruitment of NHEJ initiators such as the Ku70/Ku80 complex (Xu et al., 2012). The remodeling of chromatin not only facilitates DNA repair but also prevents stalled forks from collapsing and promotes their subsequent restart (Vassileva et al., 2014).
Histone acetylations are generated by histone acetyl transferases (HAT) as the members of the MYST (e.g., Tip60, Esa1) or GCN5 family (Gardner et al., 2011). NuA4-Tip60 complex participates in two major DDR steps which are the remodeling of chromatin at DSBs and the acetylation and activation of the ATM kinase (Sun et al., 2010). The Tip60 chromodomain interacts with H3 trimethylated on lysine 9 (H3K9me3) at DSB and then the NuA4-Tip60 complex acetylates H4 to generate H4K16Ac (Kusch et al., 2004; Daley and Sung, 2014). Once recruited to chromatin, Tip60 also acetylates ATM in the proximity of DSB, facilitating the phosphorylation of several HRR proteins required to achieve efficient HRR (Sun et al., 2007) (Figure S1 and Figure 2). At the same time, the acetylation of lysine 16 on H4 (H4K16Ac) disfavors the recruitment of 53BP1 to H4K20me and prompts HRR by enhancing the loading of BRCA1 to DSB (Tang et al., 2013). A recent report showed that H3K56Ac is important for the activation of a HRR-dependent events known as sister chromatid exchange (Munoz-Galvan et al., 2013).
As mentioned above, H3K9me is crucial to recruit a NuA4-Tip60 complex to DSBs. Defective H3K9 methylation negatively regulates HRR favoring NHEJ (Ayrapetov et al., 2014). Other methylations of histones such as H3K79me and H4K20me2 are also relevant for NHEJ-directed DSB repair, potentially acting as docking sites for the recruitment of DNA repair factors including 53BP1 (Hsiao and Mizzen, 2013). In yeast, Set2-dependent H3K36 methylation (H3K36me) reduces the chromatin accessibility of HRR factors and the resection of DNA-ends promoting NHEJ. In contrast, GCN5-dependent H3K36 acetylation promotes HRR by increasing the chromatin accessibility to HRR factors and the resection of DNA-ends (Pai et al., 2014). As mentioned in previous sections, histone ubiquitination, in particular histone H2A, is another PTM relevant for the recruitment of different HRR associated proteins to DSBs (see above and Figure S1).
In T. gondii, MYST-B were proposed to function as putative TIP60 HATs (Vonlaufen et al., 2010). Furthermore, two GCN5 (isoforms A and B) were also reported in the parasite (Sullivan and Hakimi, 2006). Histones H2A.Z, H2A.X and a novel histone variant H2B.Z, which forms a novel nucleosome integrating a double variant of H2A.Z/H2B.Z were also described in T. gondii (Dalmasso et al., 2009). These observations suggest the conservation in T. gondii of the chromatin remodeling factors required during the onset of DSB repair. As mentioned above, overexpression of tagged TgMYST-B reduces growth rate in vitro and confers protection from the methyl methanesulfonate DNA-alkylating agent (Vonlaufen et al., 2010). These results suggest a role of this HAT in the activation of DNA repair and/or in the prevention of fork collapse. Despite the high homology between the HAT domains, the two TgGCN5s exhibit differential substrate specificities. While TgGCN5-A exclusively targets lysine 18 of H3 (H3K18), TgGCN5-B acetylates multiple lysines in the H3 tail (Bhatti et al., 2006). TgGCN5-A is dispensable for the proliferation of the parasite in vitro, but it is required for the parasite recovery when challenged with alkaline stress (Naguleswaran et al., 2010). In contrast, the expression of a catalytically inactive TgGCN5-B arrests the cell outside S-phase (Wang et al., 2014). Despite the initial evidences discussed above, further investigation is required to determine whether one or both TgGCN5 participate in HRR.
Interestingly, some of the DSB-triggered histone's PTMs are conserved in T. gondii. The phosphorylation of the SQE motif in TgH2A.X was observed in RH strain treated with oxidative stress agents such as H2O2 (Dalmasso et al., 2009). Other conserved PTMs in T. gondii include HRR- (H4K16Ac and H3K9me) and NHEJ-associated marks (H3K36me, H3K79me, and H4K20me2) (Nardelli et al., 2013) T. gondii H4K16me3 was also detected, suggesting a putative regulation of this mark by signals arising from the accumulation of damaged DNA (Nardelli et al., 2013). To note, the PTM map of T. gondii histones was generated from tachyzoites samples grown in unperturbed conditions. In light of these facts, it can be proposed that in T.gondii the remodeling of chromatin and the changes in histones PTMs may also participate in the choice between NHEJ- and HRR-directed repair of DSBs. However, clear differences with other species such as the undetectable acetylation of histone H3 at K3 and 36 were also revealed. Moreover, the identification of a novel H2B.Z isoform specific for T. gondii, confirms a certain level of diversification with other species. We believe that the biological relevance of such differential regulation should be promptly explored as it can provide tools for the design of specific treatments that impair DSBs repair in T. gondii but not in its host.
Conclusions and Future Perspectives for Novel Drug Targets
This review has discussed the multiple evidences that support the conservation of pathways in charge of DSB repair such as HRR in T. gondii and its parent Plasmodium spp. Since HRR requires sister chromatids as a template for DNA repair we reasoned that the highly proliferative stages of T. gondii would highly depend on HRR after DSB accumulation. The tachyzoite stage is characterized by the highest replication rate in T. gondii, while the bradyzoite replicates within the cyst in vivo (Watts et al., 2015). It is therefore expected that at least in the tachyzoite, HRR would be the preferred pathway choice. However, the insertion of DSB-like plasmid suggests that NHEJ is the preferential mechanism of DSBs in tachyzoites while HRR events are evidenced only if NHEJ is blocked by elimination of Ku80 (Fox et al., 2009). Whether NHEJ is always the preferred pathway chosen under all conditions of DSB generation remains to be tested. We anticipate that this is highly unlikely, at least for single-ended DSBs at collapsed forks, as the repair of such lesions by the NHEJ pathway should cause lethal chromosomal fusions.
To improve the understanding of the DSB repair pathway choice in T.gondii, the identification of all parasite factors regulating such decision is required. Our analysis suggests that the basic components of the HRR machinery are conserved in T. gondii. However, the picture is incomplete particularly when focusing on the factors in charge of the choice between HRR/NHEJ (mediators, CtIP, Nbs1, EME1, etc). Similar limitations were reported in Plasmodium spp. (Table S1), an organism that clearly chooses HRR in many instances such as the generation of var gene antigen in the subtelomeric gene family and the sequence diversification and var gene family composition during mitosis (Lee et al., 2014). Hence, it is possible that, by identifying key mediators in T.gondii and Plasmodium spp., specific druggable factors useful for the treatment of toxoplasmosis may be revealed.
Another important subject that requires further investigation is the evaluation of the extent of DSB accumulation and HRR activation during the tachyzoite stage. High rates of DNA replication may increase the rates of replication forks collapse generating one-ended DSB which require the HRR for fork restart and cell survival (Lee et al., 2014). In that context, HRR might be essential for the repair of collapsed forks, in particularly after treatment with anti-tumoral compounds such as topoisomerase I inhibitors including Camptothecin (CPT), irinotecan, topotecan (Tomicic and Kaina, 2013). Interestingly, the combination of treatments that increase one-ended DSB with an HRR defect emerged as a novel, potent and synthetic lethal alternative for cancer treatment (Batey et al., 2013). Moreover, it has been recently suggested that HRR regulators can represent suitable candidates for anti-cancer therapy (Batey et al., 2013; Krajewska et al., 2015). In fact, there are some drugs that target factors such as the MRN/X complex or the ATM kinase that show synergism when used with DNA damage drugs such as cisplatin and PARP inhibitors including Olaparib. Moreover, a repertoire of small and microRNAs and peptides that target different HRR proteins such as BRCA1, BRCA2, RAD51, BLM among others, also cause synthetic lethal effects when combined with PARP inhibitors (Farmer et al., 2005). We postulate that the knowledge of the structural basis of protein-protein interactions required for HRR activation (Mermershtain and Glover, 2013) may serve to design small molecule inhibitors specific for the DSB repair pathway in T.gondii. We therefore consider that it is crucial to promptly identify the missing components of the HRR pathway in T. gondii. If factors that have diversified from humans are validated they may represent a unique source of druggable targets, which could be used along with clasical DNA damaging agents to improve current anti-toxoplasmic therapies.
Last but not least, HRR independent functions of BRCA2, RAD51 and the exonucleases Mre11, DNA2 were recently reported. These factors can initiate and/or regulate the extent of exonucleolytic cleavage of nascent DNA in conditions of persistent stalling of replication forks (Schlacher et al., 2011; Thangavel et al., 2015). Such events are independent from DSBs formation and other HRR factors such as RAD54. The HRR-indpendent function of the above-mentioned factors is required to protect the genomic stability of human cells. Therefore, it will be important to evaluate if there is a HRR-independent contribution of the putative BRCT-containing proteins and the RAD51 of T. gondii in the protection of stalled forks. The evaluation of the level of conservation of such cascade and the evaluation of its contribution to the genomic stability of the parasite will be very important when attempting to design specific targeted theapies for the treatment of toxoplasmosis.
Author Contributions
All of the authors reviewed the data from the literature and organized and wrote the manuscript. IF, SC, and SA were involved in design the figures. IF, VG, and SA were involved in editing the final version of the manuscript. All of the authors read and approved the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Alejandra Goldman for critically reading the manuscript. SA (Researcher), VG (Researcher), IF (Post-doctorla Fellow), SB (Fellow), and SC (Fellow) are members of National Research Council of Argentina (CONICET). This work was supported by ANPCyT PICT 2011 0623 (SA), ANPCyT PICT 2013 1049 (VG), and by UNSAM PUENTE 2014 (IF).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00627
References
Achanta, S. S., Varunan, S. M., Bhattacharyya, S., and Bhattacharyya, M. K. (2012). Characterization of Rad51 from apicomplexan parasite Toxoplasma gondii: an implication for inefficient gene targeting. PLoS ONE 7:e41925. doi: 10.1371/journal.pone.0041925
Ayrapetov, M. K., Gursoy-Yuzugullu, O., Xu, C., Xu, Y., and Price, B. D. (2014). DNA double-strand breaks promote methylation of histone H3 on lysine 9 and transient formation of repressive chromatin. Proc. Natl. Acad. Sci. U.S.A. 111, 9169–9174. doi: 10.1073/pnas.1403565111
Baatz, H., Mirshahi, A., Puchta, J., Gümbel, H., and Hattenbach, L. O. (2006). Reactivation of toxoplasma retinochoroiditis under atovaquone therapy in an immunocompetent patient. Ocul. Immunol. Inflamm. 14, 185–187. doi: 10.1080/09273940600659740
Badugu, S. B., Nabi, S. A., Vaidyam, P., Laskar, S., Bhattacharyya, S., and Bhattacharyya, M. K. (2015). Identification of Plasmodium falciparum DNA repair protein Mre11 with an evolutionarily conserved nuclease function. PLoS ONE 10:e0125358. doi: 10.1371/journal.pone.0125358
Bartkova, J., Horejsí, Z., Koed, K., Krämer, A., Tort, F., Zieger, K., et al. (2005). DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434, 864–870. doi: 10.1038/nature03482
Batey, M. A., Zhao, Y., Kyle, S., Richardson, C., Slade, A., Martin, N. M., et al. (2013). Preclinical evaluation of a novel ATM inhibitor, KU59403, in vitro and in vivo in p53 functional and dysfunctional models of human cancer. Mol. Cancer Ther. 12, 959–967. doi: 10.1158/1535-7163.MCT-12-0707
Baumann, P., and West, S. C. (1999). Heteroduplex formation by human Rad51 protein: effects of DNA end-structure, hRP-A and hRad52. J. Mol. Biol. 291, 363–374. doi: 10.1006/jmbi.1999.2954
Betermier, M., Bertrand, P., and Lopez, B. S. (2014). Is non-homologous end-joining really an inherently error-prone process? PLoS Genet. 10:e1004086. doi: 10.1371/journal.pgen.1004086
Bhatti, M. M., Livingston, M., Mullapudi, N., and Sullivan, W. J. Jr. (2006). Pair of unusual GCN5 histone acetyltransferases and ADA2 homologues in the protozoan parasite Toxoplasma gondii. Eukaryotic Cell 5, 62–76. doi: 10.1128/EC.5.1.62-76.2006
Blackwood, J. K., Rzechorzek, N. J., Bray, S. M., Maman, J. D., Pellegrini, L., and Robinson, N. P. (2013). End-resection at DNA double-strand breaks in the three domains of life. Biochem. Soc. Trans. 41, 314–320. doi: 10.1042/BST20120307
Blankley, R. T., and Lydall, D. (2004). A domain of Rad9 specifically required for activation of Chk1 in budding yeast. J. Cell Sci. 117, 601–608. doi: 10.1242/jcs.00907
Bocquet, N., Bizard, A. H., Abdulrahman, W., Larsen, N. B., Faty, M., Cavadini, S., et al. (2014). Structural and mechanistic insight into Holliday-junction dissolution by topoisomerase IIIalpha and RMI1. Nat. Struct. Mol. Biol. 21, 261–268. doi: 10.1038/nsmb.2775
Boddy, M. N., Gaillard, P. H., McDonald, W. H., Shanahan, P., Yates, J. R. III, and Russell, P. (2001). Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 107, 537–548. doi: 10.1016/S0092-8674(01)00536-0
Bork, P., Hofmann, K., Bucher, P., Neuwald, A. F., Altschul, S. F., and Koonin, E. V. (1997). A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J. 11, 68–76.
Bothmer, A., Robbiani, D. F., Feldhahn, N., Gazumyan, A., Nussenzweig, A., and Nussenzweig, M. C. (2010). 53BP1 regulates DNA resection and the choice between classical and alternative end joining during class switch recombination. J. Exp. Med. 207, 855–865. doi: 10.1084/jem.20100244
Buisson, R., Dion-Cote, A. M., Coulombe, Y., Launay, H., Cai, H., Stasiak, A. Z., et al. (2010). Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination. Nat. Struct. Mol. Biol. 17, 1247–1254. doi: 10.1038/nsmb.1915
Buisson, R., Niraj, J., Pauty, J., Maity, R., Zhao, W., Coulombe, Y., et al. (2014). Breast cancer proteins PALB2 and BRCA2 stimulate polymerase eta in recombination-associated DNA synthesis at blocked replication forks. Cell Rep. 6, 553–564. doi: 10.1016/j.celrep.2014.01.009
Bunting, S. F., Callen, E., Wong, N., Chen, H. T., Polato, F., Gunn, A., et al. (2010). 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141, 243–254. doi: 10.1016/j.cell.2010.03.012
Carr, A. M., and Lambert, S. (2013). Replication stress-induced genome instability: the dark side of replication maintenance by homologous recombination. J. Mol. Biol. 425, 4733–4744. doi: 10.1016/j.jmb.2013.04.023
Ceballos, S. J., and Heyer, W. D. (2011). Functions of the Snf2/Swi2 family Rad54 motor protein in homologous recombination. Biochim. Biophys. Acta 1809, 509–523. doi: 10.1016/j.bbagrm.2011.06.006
Ceccaldi, R., Rondinelli, B., and D'Andrea, A. D. (2016). Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 26, 52–64. doi: 10.1016/j.tcb.2015.07.009
Cejka, P. (2015). DNA End Resection: nucleases team up with the right partners to initiate homologous recombination. J. Biol. Chem. 290, 22931–22938. doi: 10.1074/jbc.R115.675942
Chapman, J. R., Taylor, M. R., and Boulton, S. J. (2012). Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell 47, 497–510. doi: 10.1016/j.molcel.2012.07.029
Chen, J. M., Cooper, D. N., Chuzhanova, N., Ferec, C., and Patrinos, G. P. (2007). Gene conversion: mechanisms, evolution and human disease. Nat. Rev. Genet. 8, 762–775. doi: 10.1038/nrg2193
Cortés, J. A., Gómez Marin, J., Silva, P., Arévalo, L., Arevalo, I., Alvarez, M. I., et al. (2012). Guía de atención integral para la prevención, detección temprana y tratamiento de las complicaciones del embarazo, parto y puerperio: sección toxoplasmosis en el embarazo. Infectio 16, 230–246. doi: 10.1016/S0123-9392(12)70018-8
Daley, J. M., Kwon, Y., Niu, H., and Sung, P. (2013). Investigations of homologous recombination pathways and their regulation. Yale J. Biol. Med. 86, 453–461.
Daley, J. M., and Sung, P. (2014). 53BP1, BRCA1, and the choice between recombination and end joining at DNA double-strand breaks. Mol. Cell. Biol. 34, 1380–1388. doi: 10.1128/MCB.01639-13
Dalmasso, M. C., Onyango, D. O., Naguleswaran, A., Sullivan, W. J. Jr., and Angel, S. O. (2009). Toxoplasma H2A variants reveal novel insights into nucleosome composition and functions for this histone family. J. Mol. Biol. 392, 33–47. doi: 10.1016/j.jmb.2009.07.017
Davies, A. A., Masson, J. Y., McIlwraith, M. J., Stasiak, A. Z., Stasiak, A., Venkitaraman, A. R., et al. (2001). Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol. Cell 7, 273–282. doi: 10.1016/S1097-2765(01)00175-7
Difilippantonio, S., and Nussenzweig, A. (2007). The NBS1-ATM connection revisited. Cell Cycle 6, 2366–2370. doi: 10.4161/cc.6.19.4758
Dray, E., Etchin, J., Wiese, C., Saro, D., Williams, G. J., Hammel, M., et al. (2010). Enhancement of RAD51 recombinase activity by the tumor suppressor PALB2. Nat. Struct. Mol. Biol. 17, 1255–1259. doi: 10.1038/nsmb.1916
Dubey, J. (1998). Advances in the life cycle of Toxoplasma gondii. Int. J. Parasitol. 28, 1019–1024. doi: 10.1016/S0020-7519(98)00023-X
Farmer, H., McCabe, N., Lord, C. J., Tutt, A. N. J., Johnson, D. A., Richardson, T. B., et al. (2005). Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921. doi: 10.1038/nature03445
Feng, Z., Scott, S. P., Bussen, W., Sharma, G. G., Guo, G., Pandita, T. K., et al. (2011). Rad52 inactivation is synthetically lethal with BRCA2 deficiency. Proc. Natl. Acad. Sci. U.S.A. 108, 686–691. doi: 10.1073/pnas.1010959107
Ferretti, L. P., Lafranchi, L., and Sartori, A. A. (2013). Controlling DNA-end resection: a new task for CDKs. Front. Genet. 4:99. doi: 10.3389/fgene.2013.00099
Flynn, R. L., and Zou, L. (2010). Oligonucleotide/Oligosaccharide-Binding (OB) fold proteins: a growing family of genome guardians. Crit. Rev. Biochem. Mol. Biol. 45, 266–275. doi: 10.3109/10409238.2010.488216
Fox, B. A., Ristuccia, J. G., Gigley, J. P., and Bzik, D. J. (2009). Efficient gene replacements in Toxoplasma gondii strains deficient for nonhomologous end joining. Eukaryotic Cell 8, 520–529. doi: 10.1128/EC.00357-08
Francia, M. E., and Striepen, B. (2014). Cell division in apicomplexan parasites. Nat. Rev. Microbiol. 12, 125–136. doi: 10.1038/nrmicro3184
Gabrielse, C., Miller, C. T., McConnell, K. H., DeWard, A., Fox, C. A., and Weinreich, M. (2006). A Dbf4p BRCA1 C-terminal-like domain required for the response to replication fork arrest in budding yeast. Genetics 173, 541–555. doi: 10.1534/genetics.106.057521
Gardner, K. E., Allis, C. D., and Strahl, B. D. (2011). Operating on chromatin, a colorful language where context matters. J. Mol. Biol. 409, 36–46. doi: 10.1016/j.jmb.2011.01.040
Garner, E., Kim, Y., Lach, F. P., Kottemann, M. C., and Smogorzewska, A. (2013). Human GEN1 and the SLX4-associated nucleases MUS81 and SLX1 are essential for the resolution of replication-induced Holliday junctions. Cell Rep. 5, 207–215. doi: 10.1016/j.celrep.2013.08.041
Gilbert, C. S., Green, C. M., and Lowndes, N. F. (2001). Budding yeast Rad9 is an ATP-dependent Rad53 activating machine. Mol. Cell 8, 129–136. doi: 10.1016/S1097-2765(01)00267-2
Gospodinov, A., and Herceg, Z. (2013). Chromatin structure in double strand break repair. DNA Repair (Amst). 12, 800–810. doi: 10.1016/j.dnarep.2013.07.006
Gubbels, M.-J., White, M., and Szatanek, T. (2008). The cell cycle and Toxoplasma gondii cell division: tightly knit or loosely stitched? Int. J. Parasitol. 38, 1343–1358. doi: 10.1016/j.ijpara.2008.06.004
Guirouilh-Barbat, J., Lambert, S., Bertrand, P., and Lopez, B. S. (2014). Is homologous recombination really an error-free process? Front. Genet. 5:175. doi: 10.3389/fgene.2014.00175
Haber, J. E., Ira, G., Malkova, A., and Sugawara, N. (2004). Repairing a double-strand chromosome break by homologous recombination: revisiting Robin Holliday's model. Philos. Trans. R. Soc. Lond. B Biol. Sci. 359, 79–86. doi: 10.1098/rstb.2003.1367
Hsiao, K. Y., and Mizzen, C. A. (2013). Histone H4 deacetylation facilitates 53BP1 DNA damage signaling and double-strand break repair. J. Mol. Cell Biol. 5, 157–165. doi: 10.1093/jmcb/mjs066
Huertas, P., Cortes-Ledesma, F., Sartori, A. A., Aguilera, A., and Jackson, S. P. (2008). CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature 455, 689–692. doi: 10.1038/nature07215
Huertas, P., and Jackson, S. P. (2009). Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J. Biol. Chem. 284, 9558–9565. doi: 10.1074/jbc.M808906200
Huynh, M. H., and Carruthers, V. B. (2009). Tagging of endogenous genes in a Toxoplasma gondii strain lacking Ku80. Eukaryotic Cell 8, 530–539. doi: 10.1128/EC.00358-08
Jackson, D., Dhar, K., Wahl, J. K., Wold, M. S., and Borgstahl, G. E. (2002). Analysis of the human replication protein A:Rad52 complex: evidence for crosstalk between RPA32, RPA70, Rad52 and DNA. J. Mol. Biol. 321, 133–148. doi: 10.1016/S0022-2836(02)00541-7
Jasin, M., and Rothstein, R. (2013). Repair of strand breaks by homologous recombination. Cold Spring Harb. Perspect. Biol. 5:a012740. doi: 10.1101/cshperspect.a012740
Jensen, R. B., Carreira, A., and Kowalczykowski, S. C. (2010). Purified human BRCA2 stimulates RAD51-mediated recombination. Nature 467, 678–683. doi: 10.1038/nature09399
Johnson, R. D., and Jasin, M. (2000). Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 19, 3398–3407. doi: 10.1093/emboj/19.13.3398
Kakarougkas, A., and Jeggo, P. A. (2014). DNA DSB repair pathway choice: an orchestrated handover mechanism. Br. J. Radiol. 87:20130685. doi: 10.1259/bjr.20130685
Kass, E. M., and Jasin, M. (2010). Collaboration and competition between DNA double-strand break repair pathways. FEBS Lett. 584, 3703–3708. doi: 10.1016/j.febslet.2010.07.057
Khan, A., Shaik, J. S., Behnke, M., Wang, Q., Dubey, J. P., Lorenzi, H. A., et al. (2014). NextGen sequencing reveals short double crossovers contribute disproportionately to genetic diversity in Toxoplasma gondii. BMC Genomics 15:1168. doi: 10.1186/1471-2164-15-1168
Krajewska, M., Fehrmann, R. S., de Vries, E. G., and van Vugt, M. A. (2015). Regulators of homologous recombination repair as novel targets for cancer treatment. Front. Genet. 6:96. doi: 10.3389/fgene.2015.00096
Kusch, T., Florens, L., Macdonald, W. H., Swanson, S. K., Glaser, R. L., Yates, J. R. III, et al. (2004). Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science 306, 2084–2087. doi: 10.1126/science.1103455
Lafrance-Vanasse, J., Williams, G. J., and Tainer, J. A. (2015). Envisioning the dynamics and flexibility of Mre11-Rad50-Nbs1 complex to decipher its roles in DNA replication and repair. Prog. Biophys. Mol. Biol. 117, 182–193. doi: 10.1016/j.pbiomolbio.2014.12.004
Lazzaro, F., Sapountzi, V., Granata, M., Pellicioli, A., Vaze, M., Haber, J. E., et al. (2008). Histone methyltransferase Dot1 and Rad9 inhibit single-stranded DNA accumulation at DSBs and uncapped telomeres. EMBO J. 27, 1502–1512. doi: 10.1038/emboj.2008.81
Lee, A. H., Symington, L. S., and Fidock, D. A. (2014). DNA repair mechanisms and their biological roles in the malaria parasite Plasmodium falciparum. Microbiol. Mol. Biol. Rev. 78, 469–486. doi: 10.1128/MMBR.00059-13
Lee, H. (2014). Cycling with BRCA2 from DNA repair to mitosis. Exp. Cell Res. 329, 78–84. doi: 10.1016/j.yexcr.2014.10.008
Liu, J., and Heyer, W. D. (2011). Who's who in human recombination: BRCA2 and RAD52. Proc. Natl. Acad. Sci. U.S.A. 108, 441–442. doi: 10.1073/pnas.1016614108
Lok, B. H., and Powell, S. N. (2012). Molecular pathways: understanding the role of Rad52 in homologous recombination for therapeutic advancement. Clin. Cancer Res. 18, 6400–6406. doi: 10.1158/1078-0432.CCR-11-3150
Lopez-Camarillo, C., Lopez-Casamichana, M., Weber, C., Guillen, N., Orozco, E., and Marchat, L. A. (2009). DNA repair mechanisms in eukaryotes: special focus in Entamoeba histolytica and related protozoan parasites. Infect. Genet. Evol. 9, 1051–1056. doi: 10.1016/j.meegid.2009.06.024
Malkova, A., and Ira, G. (2013). Break-induced replication: functions and molecular mechanism. Curr. Opin. Genet. Dev. 23, 271–279. doi: 10.1016/j.gde.2013.05.007
Matos, J., and West, S. C. (2014). Holliday junction resolution: regulation in space and time. DNA Repair (Amst). 19, 176–181. doi: 10.1016/j.dnarep.2014.03.013
Mazin, A. V., Mazina, O. M., Bugreev, D. V., and Rossi, M. J. (2010). Rad54, the motor of homologous recombination. DNA Repair (Amst). 9, 286–302. doi: 10.1016/j.dnarep.2009.12.006
McIlwraith, M. J., Vaisman, A., Liu, Y., Fanning, E., Woodgate, R., and West, S. C. (2005). Human DNA polymerase eta promotes DNA synthesis from strand invasion intermediates of homologous recombination. Mol. Cell 20, 783–792. doi: 10.1016/j.molcel.2005.10.001
Mehta, A., and Haber, J. E. (2014). Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 6:a016428. doi: 10.1101/cshperspect.a016428
Melander, F., Bekker-Jensen, S., Falck, J., Bartek, J., Mailand, N., and Lukas, J. (2008). Phosphorylation of SDT repeats in the MDC1 N terminus triggers retention of NBS1 at the DNA damage-modified chromatin. J. Cell Biol. 181, 213–226. doi: 10.1083/jcb.200708210
Mermershtain, I., and Glover, J. N. (2013). Structural mechanisms underlying signaling in the cellular response to DNA double strand breaks. Mutat. Res. 750, 15–22. doi: 10.1016/j.mrfmmm.2013.07.004
Mine-Hattab, J., and Rothstein, R. (2013). DNA in motion during double-strand break repair. Trends Cell Biol. 23, 529–536. doi: 10.1016/j.tcb.2013.05.006
Moncada, P. A., and Montoya, J. G. (2012). Toxoplasmosis in the fetus and newborn: an update on prevalence, diagnosis and treatment. Expert Rev. Anti Infect. Ther. 10, 815–828. doi: 10.1586/eri.12.58
Munoz-Galvan, S., Jimeno, S., Rothstein, R., and Aguilera, A. (2013). Histone H3K56 acetylation, Rad52, and non-DNA repair factors control double-strand break repair choice with the sister chromatid. PLoS Genet. 9:e1003237. doi: 10.1371/journal.pgen.1003237
Naguleswaran, A., Elias, E. V., McClintick, J., Edenberg, H. J., and Sullivan, W. J. Jr. (2010). Toxoplasma gondii lysine acetyltransferase GCN5-A functions in the cellular response to alkaline stress and expression of cyst genes. PLoS Pathog. 6:e1001232. doi: 10.1371/journal.ppat.1001232
Nardelli, S. C., Che, F.-Y., de Monerri, N. C. S., Xiao, H., Nieves, E., Madrid-Aliste, C., et al. (2013). The histone code of Toxoplasma gondii comprises conserved and unique posttranslational modifications. MBio 4, e00922–e00913. doi: 10.1128/mBio.00922-13
Ngo, G. H., and Lydall, D. (2015). The 9-1-1 checkpoint clamp coordinates resection at DNA double strand breaks. Nucleic Acids Res. 43, 5017–5032. doi: 10.1093/nar/gkv409
Pai, C. C., Deegan, R. S., Subramanian, L., Gal, C., Sarkar, S., Blaikley, E. J., et al. (2014). A histone H3K36 chromatin switch coordinates DNA double-strand break repair pathway choice. Nat. Commun. 5, 4091. doi: 10.1038/ncomms5091
Paques, F., and Haber, J. E. (1999). Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63, 349–404.
Park, J. Y., Zhang, F., and Andreassen, P. R. (2014). PALB2: the hub of a network of tumor suppressors involved in DNA damage responses. Biochim. Biophys. Acta 1846, 263–275. doi: 10.1016/j.bbcan.2014.06.003
Passos-Silva, D. G., Rajao, M. A., Nascimento de Aguiar, P. H., Vieira-da-Rocha, J. P., Machado, C. R., and Furtado, C. (2010). Overview of DNA repair in Trypanosoma cruzi, Trypanosoma brucei, and Leishmania major. J. Nucleic Acids 2010:840768. doi: 10.4061/2010/840768
Pazin, M. J., and Kadonaga, J. T. (1997). What's up and down with histone deacetylation and transcription? Cell 89, 325–328.
Petalcorin, M. I., Sandall, J., Wigley, D. B., and Boulton, S. J. (2006). CeBRC-2 stimulates D-loop formation by RAD-51 and promotes DNA single-strand annealing. J. Mol. Biol. 361, 231–242. doi: 10.1016/j.jmb.2006.06.020
Pfaff, A. W., de-la-Torre, A., Rochet, E., Brunet, J., Sabou, M., Sauer, A., et al. (2014). New clinical and experimental insights into Old World and neotropical ocular toxoplasmosis. Int. J. Parasitol. 44, 99–107. doi: 10.1016/j.ijpara.2013.09.007
Prakash, R., Zhang, Y., Feng, W., and Jasin, M. (2015). Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb. Perspect. Biol. 7:a016600. doi: 10.1101/cshperspect.a016600
Radke, J. R., Striepen, B., Guerini, M. N., Jerome, M. E., Roos, D. S., and White, M. W. (2001). Defining the cell cycle for the tachyzoite stage of Toxoplasma gondii. Mol. Biochem. Parasitol. 115, 165–175. doi: 10.1016/S0166-6851(01)00284-5
Ramadan, K. (2012). p97/VCP- and Lys48-linked polyubiquitination form a new signaling pathway in DNA damage response. Cell Cycle 11, 1062–1069. doi: 10.4161/cc.11.6.19446
Redon, C., Pilch, D., Rogakou, E., Sedelnikova, O., Newrock, K., and Bonner, W. (2002). Histone H2A variants H2AX and H2AZ. Curr. Opin. Genet. Dev. 12, 162–169. doi: 10.1016/S0959-437X(02)00282-4
Renkawitz, J., Lademann, C. A., and Jentsch, S. (2013). gammaH2AX spreading linked to homology search. Cell Cycle 12, 2526–2527. doi: 10.4161/cc.25836
Rijkers, T., Van Den Ouweland, J., Morolli, B., Rolink, A. G., Baarends, W. M., Van Sloun, P. P., et al. (1998). Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol. Cell. Biol. 18, 6423–6429. doi: 10.1128/MCB.18.11.6423
Sancar, A., Lindsey-Boltz, L. A., Unsal-Kacmaz, K., and Linn, S. (2004). Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73, 39–85. doi: 10.1146/annurev.biochem.73.011303.073723
Sarbajna, S., and West, S. C. (2014). Holliday junction processing enzymes as guardians of genome stability. Trends Biochem. Sci. 39, 409–419. doi: 10.1016/j.tibs.2014.07.003
Schlacher, K., Christ, N., Siaud, N., Egashira, A., Wu, H., and Jasin, M. (2011). Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 145, 529–542. doi: 10.1016/j.cell.2011.03.041
Scully, R., and Xie, A. (2013). Double strand break repair functions of histone H2AX. Mutat. Res. 750, 5–14. doi: 10.1016/j.mrfmmm.2013.07.007
Shibata, A., and Jeggo, P. A. (2014). DNA double-strand break repair in a cellular context. Clin. Oncol. (R. Coll. Radiol). 26, 243–249. doi: 10.1016/j.clon.2014.02.004
Silmon de Monerri, N. C., Yakubu, R. R., Chen, A. L., Bradley, P. J., Nieves, E., Weiss, L. M., et al. (2015). The ubiquitin proteome of Toxoplasma gondii reveals roles for protein ubiquitination in cell-cycle transitions. Cell Host Microbe 18, 621–633. doi: 10.1016/j.chom.2015.10.014
Smith, G. R. (2012). How RecBCD enzyme and Chi promote DNA break repair and recombination: a molecular biologist's view. Microbiol. Mol. Biol. Rev. 76, 217–228. doi: 10.1128/MMBR.05026-11
Smolarz, B., Wilczynski, J., and Nowakowska, D. (2014). DNA repair mechanisms and Toxoplasma gondii infection. Arch. Microbiol. 196, 1–8. doi: 10.1007/s00203-013-0944-0
Spycher, C., Miller, E. S., Townsend, K., Pavic, L., Morrice, N. A., Janscak, P., et al. (2008). Constitutive phosphorylation of MDC1 physically links the MRE11-RAD50-NBS1 complex to damaged chromatin. J. Cell Biol. 181, 227–240. doi: 10.1083/jcb.200709008
Stewart, G. S., Wang, B., Bignell, C. R., Taylor, A. M., and Elledge, S. J. (2003). MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 421, 961–966. doi: 10.1038/nature01446
Sullivan, W. J. Jr., and Hakimi, M. A. (2006). Histone mediated gene activation in Toxoplasma gondii. Mol. Biochem. Parasitol. 148, 109–116. doi: 10.1016/j.molbiopara.2006.03.010
Sun, Y., Jiang, X., and Price, B. D. (2010). Tip60: connecting chromatin to DNA damage signaling. Cell Cycle 9, 930–936. doi: 10.4161/cc.9.5.10931
Sun, Y., Xu, Y., Roy, K., and Price, B. D. (2007). DNA damage-induced acetylation of lysine 3016 of ATM activates ATM kinase activity. Mol. Cell. Biol. 27, 8502–8509. doi: 10.1128/MCB.01382-07
Sweeney, F. D., Yang, F., Chi, A., Shabanowitz, J., Hunt, D. F., and Durocher, D. (2005). Saccharomyces cerevisiae Rad9 acts as a Mec1 adaptor to allow Rad53 activation. Curr. Biol. 15, 1364–1375. doi: 10.1016/j.cub.2005.06.063
Tang, J., Cho, N. W., Cui, G., Manion, E. M., Shanbhag, N. M., Botuyan, M. V., et al. (2013). Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat. Struct. Mol. Biol. 20, 317–325. doi: 10.1038/nsmb.2499
Tenter, A. M., Heckeroth, A. R., and Weiss, L. M. (2000). Toxoplasma gondii: from animals to humans. Int. J. Parasitol. 30, 1217–1258. doi: 10.1016/S0020-7519(00)00124-7
Thangavel, S., Berti, M., Levikova, M., Pinto, C., Gomathinayagam, S., Vujanovic, M., et al. (2015). DNA2 drives processing and restart of reversed replication forks in human cells. J. Cell Biol. 208, 545–562. doi: 10.1083/jcb.201406100
Tomicic, M. T., and Kaina, B. (2013). Topoisomerase degradation, DSB repair, p53 and IAPs in cancer cell resistance to camptothecin-like topoisomerase I inhibitors. Biochim. Biophys. Acta 1835, 11–27. doi: 10.1016/j.bbcan.2012.09.002
Torgerson, P. R., and Mastroiacovo, P. (2013). The global burden of congenital toxoplasmosis: a systematic review. Bull. World Health Organ. 91, 501–508. doi: 10.2471/BLT.12.111732
Trenaman, A., Hartley, C., Prorocic, M., Passos-Silva, D. G., van den Hoek, M., Nechyporuk-Zloy, V., et al. (2013). Trypanosoma brucei BRCA2 acts in a life cycle-specific genome stability process and dictates BRC repeat number-dependent RAD51 subnuclear dynamics. Nucleic Acids Res. 41, 943–960. doi: 10.1093/nar/gks1192
Tu, W.-Z., Li, B., Huang, B., Wang, Y., Liu, X.-D., Guan, H., et al. (2013). γH2AX foci formation in the absence of DNA damage: Mitotic H2AX phosphorylation is mediated by the DNA-PKcs/CHK2 pathway. FEBS Lett. 587, 3437–3443. doi: 10.1016/j.febslet.2013.08.028
Turinetto, V., and Giachino, C. (2015). Multiple facets of histone variant H2AX: a DNA double-strand-break marker with several biological functions. Nucleic Acids Res. 43, 2489–2498. doi: 10.1093/nar/gkv061
van Attikum, H., and Gasser, S. M. (2009). Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol. 19, 207–217. doi: 10.1016/j.tcb.2009.03.001
Vassileva, I., Yanakieva, I., Peycheva, M., Gospodinov, A., and Anachkova, B. (2014). The mammalian INO80 chromatin remodeling complex is required for replication stress recovery. Nucleic Acids Res. 42, 9074–9086. doi: 10.1093/nar/gku605
Vonlaufen, N., Naguleswaran, A., Coppens, I., and Sullivan, W. J. Jr. (2010). MYST family lysine acetyltransferase facilitates ataxia telangiectasia mutated (ATM) kinase-mediated DNA damage response in Toxoplasma gondii. J. Biol. Chem. 285, 11154–11161. doi: 10.1074/jbc.M109.066134
Wang, J., Dixon, S. E., Ting, L. M., Liu, T. K., Jeffers, V., Croken, M. M., et al. (2014). Lysine acetyltransferase GCN5b interacts with AP2 factors and is required for Toxoplasma gondii proliferation. PLoS Pathog. 10:e1003830. doi: 10.1371/journal.ppat.1003830
Watts, E., Zhao, Y., Dhara, A., Eller, B., Patwardhan, A., and Sinai, A. P. (2015). Novel approaches reveal that Toxoplasma gondii bradyzoites within tissue cysts are dynamic and replicating entities in vivo. MBio 6, e01155–e01215. doi: 10.1128/mBio.01155-15
Weiss, L. M., and Kim, K. (2000). The development and biology of bradyzoites of Toxoplasma gondii. Front. Biosci. 5, D391–D405. doi: 10.2741/Weiss
Williams, R. S., Dodson, G. E., Limbo, O., Yamada, Y., Williams, J. S., Guenther, G., et al. (2009). Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell 139, 87–99. doi: 10.1016/j.cell.2009.07.033
Wu, L., Luo, K., Lou, Z., and Chen, J. (2008). MDC1 regulates intra-S-phase checkpoint by targeting NBS1 to DNA double-strand breaks. Proc. Natl. Acad. Sci. U.S.A. 105, 11200–11205. doi: 10.1073/pnas.0802885105
Xu, Y., Ayrapetov, M. K., Xu, C., Gursoy-Yuzugullu, O., Hu, Y., and Price, B. D. (2012). Histone H2A. Z controls a critical chromatin remodeling step required for DNA double-strand break repair. Mol. Cell 48, 723–733. doi: 10.1016/j.molcel.2012.09.026
Yan, J., Huang, B., Liu, G., Wu, B., Huang, S., Zheng, H., et al. (2013). Meta-analysis of prevention and treatment of toxoplasmic encephalitis in HIV-infected patients. Acta Trop. 127, 236–244. doi: 10.1016/j.actatropica.2013.05.006
Yoshiyama, K. O., Sakaguchi, K., and Kimura, S. (2013). DNA damage response in plants: conserved and variable response compared to animals. Biology (Basel) 2, 1338–1356. doi: 10.3390/biology2041338
You, Z., Chahwan, C., Bailis, J., Hunter, T., and Russell, P. (2005). ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol. Cell Biol. 25, 5363–5379. doi: 10.1128/MCB.25.13.5363-5379.2005
Keywords: Toxoplasma, DNA damage, homologous recombination repair, chromatin, fork collapse, double strand break
Citation: Fenoy IM, Bogado SS, Contreras SM, Gottifredi V and Angel SO (2016) The Knowns Unknowns: Exploring the Homologous Recombination Repair Pathway in Toxoplasma gondii. Front. Microbiol. 7:627. doi: 10.3389/fmicb.2016.00627
Received: 16 November 2015; Accepted: 18 April 2016;
Published: 03 May 2016.
Edited by:
Yuji Morita, Aichi Gakuin University, JapanReviewed by:
Jorge Enrique Gomez-Marin, Universidad del Quindio, ColombiaSirinart Ananvoranich, University of Windsor, Canada
Renata Maria Augusto Da Costa, Universidade Federal do ABC, Brazil
Copyright © 2016 Fenoy, Bogado, Contreras, Gottifredi and Angel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vanesa Gottifredi, vgottifredi@leloir.org.ar;
Sergio O. Angel, sangel@intech.gov.ar
 Ignacio M. Fenoy
Ignacio M. Fenoy Silvina S. Bogado1
Silvina S. Bogado1 Susana M. Contreras
Susana M. Contreras Sergio O. Angel
Sergio O. Angel