- Department of Plant Pathology, College of Plant Protection, Anhui Agricultural University, Hefei, China
Protein O-mannosylation is a type of O-glycosylation that is characterized by the addition of mannose residues to target proteins, and is initially catalyzed by evolutionarily conserved protein O-mannosyltransferases (PMTs). In this study, three members of PMT were identified in Magnaporthe oryzae, and the pathogenic roles of MoPmt2, a member of PMT2 subfamily, were analyzed. We found that MoPmt2 is a homolog of Saccharomyces cerevisiae Pmt2 and could complement yeast Pmt2 function in resistance to CFW. Quantitative RT–PCR revealed that MoPmt2 is highly expressed during conidiation, and targeted disruption of MoPmt2 resulted in defects in conidiation and conidia morphology. The MoPmt2 mutants also showed a distinct reduction in fungal growth, which was associated with severe alterations in hyphal polarity. In addition, we found that the MoPmt2 mutants severely reduced virulence on both rice plants and barley leaves. The subsequent examination revealed that the fungal adhesion, conidial germination, CWI and invasive hyphae growth in host cells are responsible for defects on appressorium mediated penetration, and thus attenuated the pathogenicity of MoPmt2 mutants. Taken together, our results suggest that protein O-mannosyltransferase MoPmt2 plays essential roles in fungal growth and development, and is required for the full pathogenicity of M. oryzae.
Introduction
The filamentous fungus Magnaporthe oryzae, which causes rice blast disease, is the most destructive pathogen of cultivated rice plants worldwide (Howard and Valent, 1996). It has been developed as a model system to study fungus–plant interactions due to its economic and scientific importance (Talbot, 2003; Xu et al., 2007; Dean et al., 2012). During growing seasons, like many other phytopathogens, asexual spores play an important role in the disease cycle of M. oryzae (Lee et al., 2006). When landed on the plant surface, asexual spores immediately secrets conidial tip mucilage to adhere themselves on rice leaves. Under suitable condition, conidia begin to germinate, and four to 6 h later, a dome-shaped infection structure known as appressorium differentiates at the tip of the germ tube. Rice blast fungus generates enormous amount of turgor pressure (up to 8 MPa) within appressorium to penetrate the plant cuticle layer (Howard et al., 1991; Howard and Valent, 1996; Talbot, 2003), and after penetration, the fungus develops bulbous biotrophic infectious hyphae in the rice leaf cells and typical necrotic lesions on the leaf surface (Kankanala et al., 2007). After 5–7 days, newly formed pyriform conidia differentiate from the hyphae on the lesion, and serve as inocula for secondary infection cycles (Talbot, 2003). These findings suggest that the sporulation and appressorium formation are essential for successful disease development. Thus, an understanding of the molecular mechanisms involved in these processes could provide insights into the nature of the plant–fungi interaction and is of great interest in the development of antifungal strategies.
Protein glycosylation is a post-translational modification conserved in organisms from yeasts to humans, and plays a critical role in determining the structure and function of numerous secreted and membrane-bound proteins (Lehle et al., 2006). In eukaryotic cells, there are two types of protein glycosylation (N- and O-glycosylation) that are highly regulated by the activity of protein- and site-specific enzymes (Fernández-Álvarez et al., 2009). O-mannosylation is a type of O-glycosylation that is characterized by the addition of mannose residues to target proteins, and is initially catalyzed by protein O-mannosyltransferases (PMTs), a family of proteins that are evolutionarily conserved from yeast to humans (Strahl-Bolsinger et al., 1999; Willer et al., 2003). In fungi, the PMTs are grouped into Pmt1, Pmt2, and Pmt4 subfamilies based on phylogeny, with each subfamily having three to seven members (Girrbach et al., 2000). In Saccharomyces cerevisiae, seven homologous PMT proteins have been identified and divided into the Pmt1, Pmt2, and Pmt4 subfamilies, which differ in their number of genes and the protein substrate specificity (Girrbach and Strahl, 2003). However, in many other filamentous fungi, including M. oryzae, only a single member in each Pmt subfamily was identified (Dean et al., 2005; Fernández-Álvarez et al., 2009; Goto et al., 2009; Mouyna et al., 2010; Gonzalez et al., 2013). In the past two decades, studies of the functions of fungal PMTs has been increasing, and now it is clear that protein glycosylation contributes to the modification of proteins involved in important processes, such as cell wall integrity (CWI), sensing of environmental signals, morphogenesis and the virulence of fungal pathogens (Gentzsch and Tanner, 1996; Prill et al., 2005; Olson et al., 2007; Zhou et al., 2007; Fernández-Álvarez et al., 2009, 2012). Simultaneous disruptions of three different types of PMT genes in Schizosaccharomyces pombe were lethal (Willer et al., 2005), suggesting that each class provided a unique function for O-mannosylation. In S. cerevisiae, the PMT genes are not individually essential for viability, probably as a result of gene redundancy (Gentzsch et al., 1995). Deletion of PMT1 does not affect viability but leads to cells that tend to aggregate. Inactivation of both PMT1 and PMT2 causes defects in growth and resistance to antifungal drug (Lussier et al., 1995), whereas triple mutants are not viable, indicating that PMT protein activity is essential in S. cerevisiae, although individual genes are dispensable (Tanner et al., 1996; Loibl and Strahl, 2013). In Candida albicans and Cryptococcus neoformans, Pmt disruption affects morphogenesis and virulence (Prill et al., 2005; Olson et al., 2007). In filamentous fungus Aspergillus fumigatus, deletion of Pmt4 results in abnormal growth, defective conidiation and associated proteomic changes, while disruption of Pmt2 results in lethal growth (Mouyna et al., 2010). In Aspergillus nidulans, all the single Pmt disruption mutants were viable, but defective in cell wall integrity, hyphal growth and asexual development (Kriangkripipat and Momany, 2009). In Ustilago maydis, the three Pmt orthologs play diverse roles in fungal development. The deletion of Pmt1 doesn't affect the fungal growth and plant infection, while the mutation in Pmt2 is not viable, indicating an essential role in fungal development. By contrast, the disruption of Pmt4 specifically affected appressorium formation, penetration and tumor formation in maize (Fernández-Álvarez et al., 2009). In Botrytis cinerea, PMTs are individually required for morphogenesis, fungal adherence, cell wall integrity and virulence on plants, and deletion of PMT2 results in defects on the stability of the cell wall, poor sporulation and attenuated virulence on plants (Gonzalez et al., 2013). In Beauveria bassiana, PMTs play crucial roles on fungal development, and individual Pmt gene deletion results in defects on growth, conidiation, stress tolerance and virulence (Wang et al., 2014). In Penicillium digitatum, the disruption of Pmt2 causes pleiotropic effects, including defects on cell wall integrity, conidiogenesis, virulence and resistance to the antifungal peptide PAF26 (Harries et al., 2015). Based on the above facts, it is therefore evident that the O-mannosyltransferases Pmts play important roles in fungal development and pathogenesis in pathogenic fungi.
Recently, evidence that the α-1, 3-mannosyltransferase ALG3 from M. oryzae play a critical role in mediating the glycosylation of secreted effectors, and thus required for fungal pathogenicity on host (Chen et al., 2014), suggest that protein glycosylation may be important for the pathogenic development of M. oryzae. However, till now, enzymes involving O-mannosylation pathway has not been characterized in the rice blast fungus. Here, we describe a detailed characterization of the role of MoPmt2 in M. oryzae, and our results showed that MoPmt2 contribute to fungal morphology, growth, CWI and virulence on host plants.
Materials and Methods
Fungal Strains and Culture Conditions
The M. oryzae Guy11 was used as wild-type strains throughout this work. Fungal mycelia grown in liquid complete media at 28°C for 2 days were harvested and used for genomic DNA and RNA extractions. For observing the mycelial growth, strains were inoculated in liquid CM as described in the reference (Guo et al., 2015). For conidiation, mycelial plugs were inoculated on RDC agar plates (Guo et al., 2011) and maintained at 28°C for 7 days in the dark followed 3–5 days constant fluorescent light condition to promote conidiation. For medium containing cell wall-perturbing agents, the final concentrations were 50, 100, 200 μg/mL for Congo red (CR, 860956, Sigma, China), and/or for Calcofluor white (CFW, F3543, Sigma, China), respectively. The inhibition rate was calculated by the method described in the reference (Guo et al., 2015).
Yeast Pmt2 Mutant Complementation
S. cerevisiae BY4741ΔYAL023c (ΔPmt2) and the strain from which it was derived, BY4741 (MATa; ura3Δ0; leu2Δ0; his3Δ1; met15Δ0) were purchased from Euroscarf (http://www.uni-frankfurt.de/fb15/mikro/euroscarf/). The full-length of MoPmt2 cDNA (XM_003715348.1) from M. oryzae was amplified using primer pairs Pmt2-YC1/ Pmt2-YC2. The PCR products, digested with HindIII and XhoI, were cloned into pYES2 (Invitrogen) and transformed into Δpmt2 mutant (BY4741, ΔYAL023c::kanMX4). Positive transformants were selected on SD medium lacking uracil. For complementation assays, all tested strains were grown and treated as described byHarries et al. (2015). The cells of the tested strains were diluted to an OD600 of 0.1 and 10 μL of ten-fold serial dilutions were spotted onto SC-Ura plates containing 2% glucose with and without 12.5 μg/mL CFW or 2% galactose with 12.5 μg/mL CFW, respectively, and grown for 3 days at 30°C. The wild type strain BY4741 as well as the Pmt2 deletion mutant BY4741ΔYAL023c expressed empty pYES2 vector were used as a control.
Multiple Sequence Alignment and Phylogenetic Analysis
The Pmt proteins from different organisms were obtained from NCBI database (http://www.ncbi.nlm.nih.gov/) using the BLAST algorithm (McGinnis and Madden, 2004). Sequence alignments were performed using the Clustal_W program (Thompson et al., 1994) and a phylogenetic tree was created with the Mega 4.0 Beta program (Tamura et al., 2007). Domain architecture was automatically provided by the SMART online software program (Letunic et al., 2012) or TMHMM Server v. 2.0 (Krogh et al., 2001).
Gene Disruption and Complementation
The gene deletion mutants were generated using the standard one step gene replacement strategy. For gene replacement construct, DNA fragments of the 1.0 kb flanking regions of the target gene were amplified with the primers Pmt2-1F/Pmt2-1R and Pmt2-2F/ Pmt2-2R (Table S1), then a ~2-kb fragment containing the two flanking sequences was amplified by overlap PCR with primers Pmt2-1F / Pmt2-2R (Table S1), and then was inserted into the pMD19-T vector (Takara Co., Dalian, China) to generate plasmid pMDT–MoPmt2. The hygromycin B-resistance cassette, which was amplified with primers HPH-F/HPH-R by KOD-Plus-Neo (KOD-401, TOYOBO, Shanghai, China), was purified to insert into the PmeI site in plasmid pMDT–MoPmt2 to generate deletion construct pMDT-MoPmt2-hph. A ~3.4-kb fragments, which was amplified from the deletion construct with primers Pmt2-1F/Pmt2-2R, was transformed into protoplasts of wild type Guy11. To obtain MoPmt2 null mutants, all hygromycin B-resistant transformants were screened by PCR using the primer pairs P1/P2 and P3/P4 (Figure S1), respectively, and were further validated by Southern blot hybridization and RT-PCR (Figures S2D–F). For complementation of MoPmt2, the fragments containing the 1.5-kb native promoter region of the gene, MoPmt2 full-length coding region and a 0.5-kb terminator sequence were amplified using primers Pmt2-C1/Pmt2-C2 and then inserted into the pMo1102 vector (unpublished plasmid) using the yeast gap repair approach (Park et al., 2004) to generate pMo1102::MoPmt2. The complemental fragment was then reintroduced into the MoPmt2-6 mutant by the Agrobacterium mediated transformation of M. oryzae, and the complemented strains were validated by semiquantitative RT-PCR.
Phenotypic Characterization of Mutants
To assess the growth rate, mycelia plugs of Guy11 and its derivative mutants were obtained from the edge of 5-day-old cultures and placed onto fresh media (CM, MM, OM, and RDC), followed by incubation in the dark condition at 28°C for 5 days (Guo et al., 2015). The conidia production of the tested strains was carried out by counting the number of conidia harvested with 5 ml of sterilized distilled water from 10-day-old RDC agar plates using a haemocytometer (Zhang et al., 2009). Conidium size was measured under a microscope as previously described (Guo et al., 2011). For conidium germination and appressorium formation, drops (20 μl) of conidial suspension (1.0 × 105 spores/ml) were inoculated onto a hydrophobic coverslip and placed under humid conditions at 28°C. Conidia germination and appressoria formation were observed by microscopic examination of at least 99 conidia per replicate at each time point. Appressorium turgor was measured by incipient cytorrhysis assay using a 1-4 molar concentration of glycerol solution (Zhang et al., 2009). For conidium adhesion assay, drops (30 μl) of conidial suspension (1.0 × 105 spores/ml) were placed on the hydrophobic surface of coverslips for 2 h in a humid box, then the coverslips were washed by pipetting 1 ml distilled water three times (each washing takes 3 s)., and the percentage of conidium adhesion was assessed as described previously (Han et al., 2015). All above experiments were independently repeated three times with three replicate each, and representative results from one experiment are shown in this study.
Pathogenicity Assay and Infectious Growth Observation
For plant infection assays, conidial suspension (4 ml, 1.0 × 105 spores/ml) containing gelatin (2%, wt/vol) were sprayed on 14-day-old rice seedlings (Oryza sativa cv CO-39) or drops of conidial suspension (10 μL, 1.0 × 105 spores/ml) were placed on 7-day-old barley leaves (Hordeum vulgare cv Golden Promise). Inoculated plants were kept in a moist chamber in dark at 28°C for first 2 h, and then were moved to a moist chamber with a photoperiod of 12 h under fluorescent lights (Guo et al., 2011). The disease development on plants was assessed at 5 days after inoculation. For pathogenicity assay on abraded rice leaves, drops (10 μl) of conidia suspension (1.0 × 105 ml−1) of the tested strains were placed on wounded rice leaves and kept in a moist chamber as described above, and their virulence was evaluated at 5 days after incubation. Conidiation on the lesion was treated as described above and observed 15 days after incubation. Plant penetration assays were carried out using 7-day-old barley leaves (Chen et al., 2014). Conidial suspension (1 × 104 spores/ml) was dropped onto barley leaves and incubated at 28°C in a moistened chamber. Invasive growth inside plant cells was examined after at 48 and 72 h using a light microscopy. These experiments were repeated three times, and representative results from one experiment are shown.
FITC-ConA and CFW Staining
FITC staining was performed with conidia collected from RDC agar plates (Zhang et al., 2009). Conidia suspension (1 × 105 conidia ml−1) was inoculated on the hydrophobic surface of coverslips and then treated as described in reference (Hamer et al., 1988) at 0.5 and 24 h, respectively. Microscopic examination of stained conidia and mature appressorium (>100 conidia or appressorium) was carried out using a Nikon inverted Ti-S epifluorescence microscope (Nikon Co., Tokyo, Japan). For CFW staining, conidia suspension (1 × 104 conidia ml−1) was inoculated on glassslides covered with a thin layer of water agar and kept in a moist plate for 48 h or on hydrophobic coverslips and kept in a moist plate for 24 h, then both the vegetative hyphae and appressorium stained by CFW was observed using the Nikon inverted Ti-S epifluorescence microscope (Nikon Co., Tokyo, Japan), respectively.
Nucleic Acid Manipulation and Southern Blotting
The standard method was used to prepare genomic DNA for both diagnostic PCR and Southern blot hybridization (Sambrook et al., 1989). A DNA hybridization probe, which is amplified by primer set Pmt2-P1/Pmt2-P2, were labeled with digoxigenin-11-dUTP using DIG-High prime according to the manufacturer's instructions (11745832910, Roche, China). Southern hybridization experiments, including restriction enzyme digestion, DNA gel blotting and hybridization were performed as described byGuo et al. (2015). Total RNA samples from mycelia, conidia and plants infectious stages (8, 24, 48, and 72 h) were extracted using the methods described in E.Z.N.A. Total RNA Kit I (R6834-01, Omega Bio-Tek, Norcross, USA).
qRT–PCR, RT–PCR, and Gene Expression Analysis
For RT-PCR and qRT-PCR, 5 μg of total RNA were reversely transcribed into first-strand cDNA using the ReverTra Ace® qPCR-RT Master Mix (FSQ-301, TOYOBO, Shanghai, China). Confirmation of deletions and reintroduction of MoPmt2 gene was made with primer pairs Pmt2-S1/Pmt2-S2 (Table S2). The stable expression actin gene (MGG_03982.5) and β-tubulin gene (MGG_00604.5) amplified by primer pairs Actin-F/Actin-R and β-tubF/β-tubR, respectively, (Table S2) was used as internal control (Guo et al., 2015).
qRT-PCR were performed using a BIO-RAD CFX96 touch q-PCR system (BIO-RAD, Hercules, California, USA), following previously established procedures: 3 min at 95°C (one cycle) followed by 15 s at 95°C, 30 s at 60°C, and 30 s at 72°C (40 cycles). To measure the relative abundance of MoPmt2 transcripts, the average threshold cycle (Ct) was normalized to that of actin gene (MGG_03982.5) and expressed as 2−ΔCt, where −ΔCt = (Ct, target gene − Ct, actin gene). Fold changes in expression during fungal development and infectious growth were calculated as 2−ΔΔCt, where −ΔΔCt = (Ct, target gene − Ct, actin gene)testcondition − (Ct, target gene − Ct, actin gene)CM (Livak and Schmittgen, 2001). Three independent pools of tissues in three sets of experimental replicates were performed and primer pairs used in this section were listed in Table S2.
Measurement of Enzyme Activity in Extracellular Culture Filtrate
Enzyme activity was assayed using culture filtrate from 2-day old CM liquid culture. For measurement of peroxidase and laccase activity, a reaction mixture (1 ml) containing 50 mM acetate buffer (pH 5.0) and 10 mM 2, 2′- azino-di-3-ethylbenzthiazoline-6-sulphonate (ABTS, Sigma, A1888) was mixed with the culture filtrate (200 μl) and incubated at 25°C for 5 min with or without 3 mM of H2O2. Absorbance was evaluated at 420 nm.
Transmission Electron Microscopy
Transmission electron microscopy was carried out using hyphae grown in liquid CM for 48 h. The hyphae were fixed in 2.5% (v/v) glutaraldehyde and 1% (v/v) osmium tetroxide. Sections were prepared and visualized using a H-7650 transmission electron microscope (Hitachi, Tokyo, Japan) as described byXu et al. (2010).
Results
Identification of Three Pmt Genes in M. oryzae
In our previous study, the Moap1 mutants were identified as non-pathogenic to rice plants (Guo et al., 2011). To investigate the possible reason for this, a serial analysis of gene expression (SAGE) libraries for the wild type Guy11 and Moap1 mutant were generated. In the SAGE libraries, three O-Mannosyltransferases encoding genes were identified with different expression patterns, with MGG_02954 and MGG_04427 showing 0.35- and 1.0-fold increased expression, respectively, while MGG_07190 showing 1.35-fold reduced expression in the Moap1 mutants. Sequence alignment using amino acid sequences of the three O-Mannosyltransferases revealed the MGG_02954, a protein of 998 amino acids, showed an amino acid identity of 39% to S. cerevisiae Pmt1p; MGG_07190, of 737 amino acids, showed 47% identity to S. cerevisiae Pmt2p; and MGG_04427, of 775 amino acids, showed 45% identity to S. cerevisiae Pmt4p (Table S1; Gentzsch and Tanner, 1996). The hydropathy analysis of M. oryzae Pmt sequences revealed the presence of two large transmembrane domains, located in their N- and C-terminal regions and separated by a central hydrophilic region (Strahl-Bolsinger and Scheinost, 1999; Figure 1A). In addition, analysis of the candidate proteins showed the presence of two characteristic domains of the PMT family: the PMT domain, a region implicated in PMT complex formation and O-mannosyltransferase activity, found within the N-terminal transmembrane region, and the mannosyltransferase IP3 ryanodine receptor domain (MIR) composed of three submotifs, located in the hydrophilic central region (Girrbach et al., 2000; Figure 1B; Table S1). The assignment of these identified genes as protein O-mannosyltransferases was further confirmed by phylogenetic reconstruction with homologous fungal genes, and they were grouped into each PMT subfamily (Lehle et al., 2006; Figure S1). Thus, consistent with their hydropathy profiles, conserved sequence motifs, and our phylogenetic analysis, we can provisionally named MGG_02954, MGG_07190 and MGG_04427 as MoPmt1, MoPmt2, and MoPmt4 respectively.
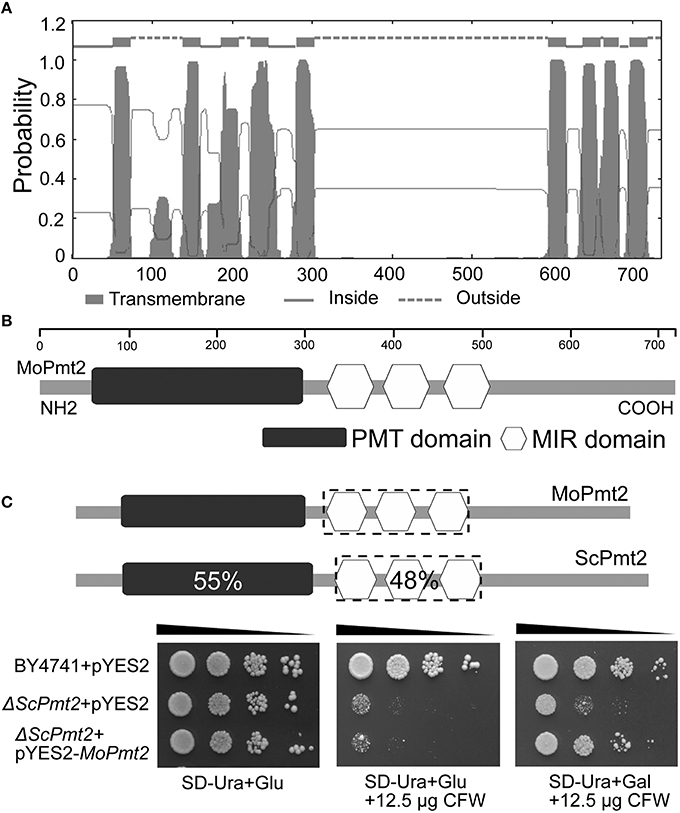
Figure 1. M. oryzae MoPmt2 encodes a functional homolog of S. cerevisiae Pmt2. (A) Prediction of transmembrane structure in MoPmt2. The position of each transmembrane domain was generated by the TMHMM Server v. 2.0 online program, and the MoPmt2 possess the typical hydropathy profiles of O-mannosyltransferases. (B) Conservation of the Pmt sequence motifs in MoPmt2. Physical map of MoPmt2 revealed two conserved motifs (Pmt and MIR) of the PMT family at the N-terminal transmembrane region and central hydrophilic region. (C) Functional complementation of S. cerevisiae Pmt2 mutant. Serial 10-fold dilutions of cells from wild-type strain BY4741 transformed with pYES2 and the Pmt2 strain transformed with either pYES2 or pYES2-MoPmt2 are dotted onto SC-Ura plates with Glu or Gal supplemented with 12.5 μg/mL CFW as indicated, and incubated at 30°C for 72 h.
MoPmt2 Encodes Protein O-mannosyltransferases
The SAGE libraries revealed that MoPmt2 showed transcriptional reduction in the Moap1 mutant, indicating its potential roles in regulating fungal development and pathogenicity in M. oryzae. Therefore, the functional roles of MoPmt2 were characterized in this study. In S. cerevisiae, the pmt2 gene is responsible for cell wall integrity, and Δpmt2 mutant has defects in cell wall and exhibit increased sensitivity to the chitin-binding fluorophore CFW (Harries et al., 2013). To provide experimental evidence that MoPmt2 encodes a functional homolog of S. cerevisiae Pmt2 with O-mannosyltransferase activity, we tested the ability of MoPmt2 to complement a yeast strain lacking pmt2. Our results showed that the S. cerevisiae Δpmt2 mutants transformed with the expression vector pYES2-MoPmt2 restored the resistance to CFW under induction conditions (Figure 1C), indicating that M. oryzae Pmt2 is functional homolog of the S. cerevisiae Pmt2. Moreover, it also confirms our phylogenetic-based assignment of PMT family members.
The Expression and Disruption of MoPmt2 Gene in M. oryzae
To gain insight into the possible function of MoPmt2 in M. oryzae, the changes in MoPmt2 gene expression were analyzed by quantitative RT-PCR (qRT-PCR), and found that MoPmt2 gene is expressed during fungal vegetative growth and plant infection, with the highest induction at conidia stage (7.4-fold; Figure S2A), compared with the stable expression of actin gene.
We generated MoPmt2 mutants using the MoPmt2 deletion vector which contains the hygromycin resistance gene (Figure S2B). The transformants that confers resistance to the antibiotic hygromycin B were verified by diagnostic PCR (Figure S2C). The putative mutants were further confirmed by Southern blot and semiquantitative RT-PCR (RT-PCR) analysis (Figures S2D–F), and two deletion mutants, MoPmt2-6 and MoPmt2-8, were obtained for further analysis. To ascertain that the observed phenotypes of the MoPmt2 mutants are caused by the deletion of MoPmt2, a complemented strain MoPmt2c was generated and verified by RT-PCR (Figure S2F). As expected, the complemented strain recovered all the defects described in this study (Figures 2–8).
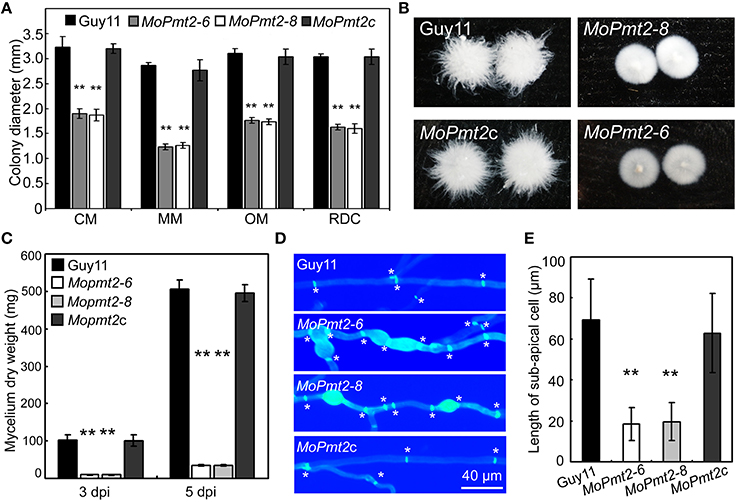
Figure 2. The effect of MoPmt2 on mycelia growth of M. oryzae. (A) Mycelial growth is altered in the MoPmt2 mutant. Colony diameters of the tested strains on different media were measured and statistically analyzed by Duncan analysis. (B) Phenotype of mycelia grown in liquid CM. All tested strains were inoculated in liquid CM for 48 h at 28°C in darkness and then photographed. (C) Measurement of fungal biomass. The fungal mycelia of the tested strains, grown in liquid CM for 3 and 5 days respectively, were collected and freezing dried, and were statistically analyzed. (D) Mycelia growth on water agar media. CFW staining of mycelia is used to show the distance of septa and swollen of mycelia. Asterisks in white color indicate septa. (E) The average length of the sub-apical cell of vegetative hyphae in Guy11, MoPmt2-6, MoPmt2-8, and MoPmt2c. The mean and standard deviations were calculated based on three independent experiments by measuring at least 50 sub-apical cells in each replicate. Error bar represents standard deviation, and asterisks in this figure indicate significant differences from the control (P < 0.01).
MoPmt2 Deletion Leads to Retarded Vegetative Growth
To investigate the role of MoPmt2 on mycelium growth in M. oryzae, mycelial growth of wild type strain Guy11, the MoPmt2 mutants and complemented strain MoPmt2c was compared on complete media (CM), minimal media (MM), oatmeal medium (OM), and RDC medium (Guo et al., 2015). Mycelial growth of the MoPmt2 mutants differed from that of Guy11, with a significant reduction in radial growth on the four media, compared to Guy11 and MoPmt2c (Figure 2A; Figure S3). In liquid CM media, we found that mycelial growth of the MoPmt2 mutants was more compact than that of Guy11 and MoPmt2c (Figure 2B), and a subsequent assay indicated significantly less fungal biomass for the MoPmt2 mutants than for Guy11 and MoPmt2c after incubating in liquid CM for 72 and 120 h, respectively (Figure 2C). In view of these phenotypes, we examined the morphology of MoPmt2 mutants by microscopy after CFW staining. Our results showed that MoPmt2 mutants presented more septa, which were intensively stained and had shorter interseptal distances than those of Guy11 and complemented strain MoPmt2c (Figures 2D,E). In addition, The MoPmt2 mutants also showed defects in polarity, with more branching hyphae and globular balloon-like structures (Figure 2D; Figure S4A).
MoPmt2 Plays Critical Role in Asexual Spore Development
It is clear that asexual spores play an important role in the disease cycle of M. oryzae (Lee et al., 2006), thus, sporulation of the Guy11, MoPmt2 mutants (MoPmt2-6 and MoPmt2-8), and the complemented strain MoPmt2c was compared on 14 day old RDC cultures. Our findings revealed that conidiation was dramatically reduced by approximately 12 to 14-fold in MoPmt2 deletion mutants (MoPmt2-6, 12-fold; MoPmt2-8, 14-fold), compared with Guy11 and complemented strains MoPmt2c (Figures 3A,B). In addition, of the spores that formed in MoPmt2-6, most exhibited abnormal, with 49.9% in large size and 43.7% in small size, compared with Guy11 and MoPmt2c (Figures 3C–E; Figure S5). Combined with gene expression profiles that MoPmt2 showed a much higher level of expression in conidia (Figure S2A), we concluded that MoPmt2 plays an important role in conidial development.
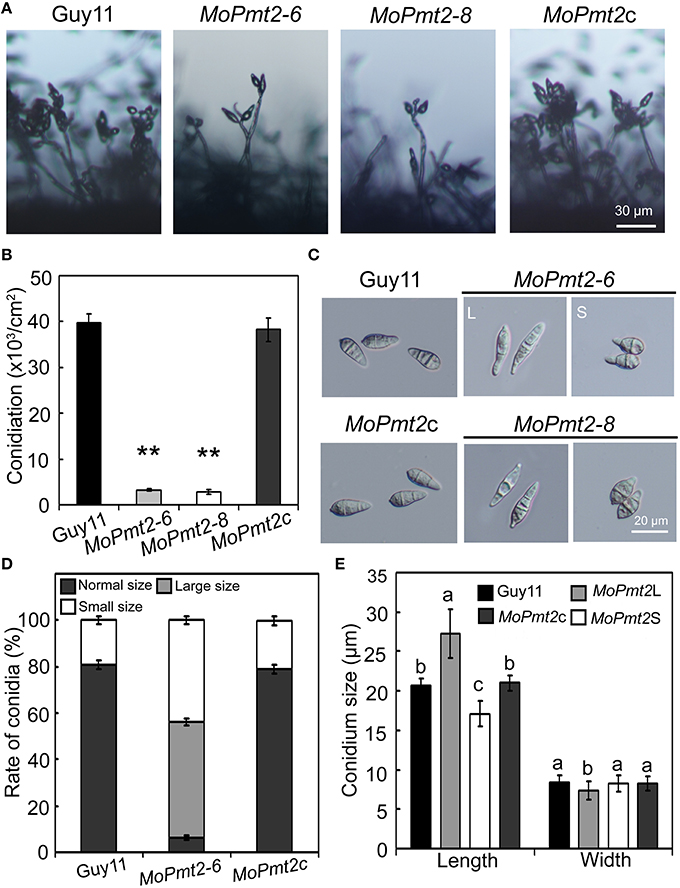
Figure 3. The effect of MoPmt2 on conidiation and conidial morphology of M. oryzae. (A) MoPmt2 deletion results in defects on conidiation. The development of conidia on conidiophores was examined by light microscope using strains grown on RDC medium for 7 days. Scale bar = 30 μm. (B) Statistical analysis of conidiation. The conidia produced by the tested strains grown on RDC medium for 14 days were measured and analyzed by Duncan analysis (p < 0.01). Asterisks indicate significant differences of conidiation among tested strains. Error bar represents standard deviation. (C) Conidial morphology comparison. Conidia of tested strains were collected from 14-day-old cultures, and then photographed under light microscope. Bars = 20 μm. (D) The percentage of each type of abnormal conidia. Conidia of tested strains were harvested from 14-day-old cultures, and the rate of large and small conidia was calculated and statistically analyzed, respectively. (E) Conidia sizes comparison. The conidia sizes were determined as width by length from 297 conidia of each strain.
MoPmt2 is Responsible for Conidia Germination and Appressorium Formation
Conidia germination is the vital step during M. oryzae infection (Howard et al., 1991). Therefore, we measured the germination ability of conidia on a hydrophobic surface of coverslips. Our results showed that conidial germination was significantly delayed in the MoPmt2 mutant when compared with that of Guy11 and MoPmt2c. By 2 h, only 4.7% of MoPmt2 conidia germinated compared with 85.9% of the Guy11 and 85% of the MoPmt2c. When prolonged the inoculation time to 8 h, even though the conidial germination of the mutant gradually increased to 92.3%, it was still significantly less germination by the mutant than that of by Guy11 (Figures 4A,B).
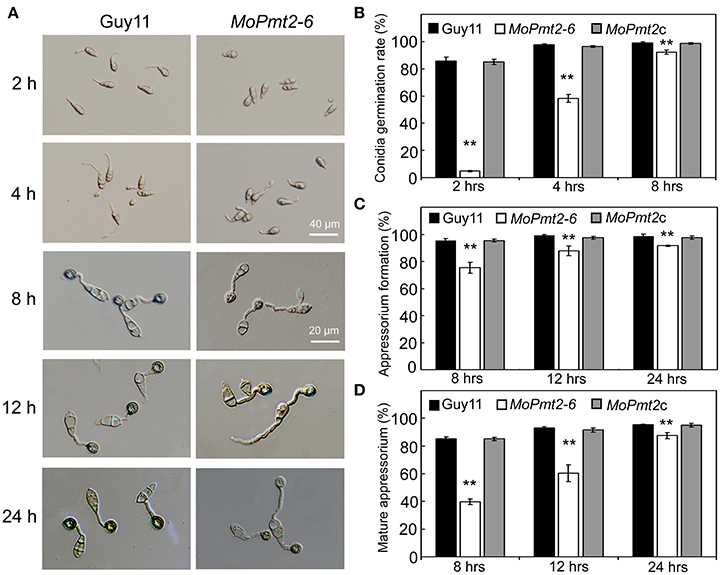
Figure 4. Conidial germination and appressorium formation. (A) Delayed conidial germination and appressorium formation of the MoPmt2 mutant. Conidia of indicated strains were harvested from 14-day-old cultures. Droplets of conidial suspension (1 × 105 ml−1) were inoculated on the hydrophobic surface of the coverslips for indicated time, and then photographed. Scale bars are indicated in the figure. (B) Statistical analysis of conidial germination. Germinated conidia were counted at each indicated time under a light microscope, and statistically analyzed by Duncan analysis. (C) Statistical analysis of appressorium formation. Appressorium formed at the germ tube were counted and statistically analyzed. (D) Measurement of mature appressorium. Appressorium formed at the germ tube with melanin were counted, and the ratio of mature appressorium was statistically analyzed. Asterisks in this figure indicate significant differences from the control (p < 0.01). Error bar represents standard deviation.
The appressorium is a typical penetration structure that enables the M. oryzae to invade into host cells (Howard and Valent, 1996). Thus, we also evaluated the ability of appressorium formation by the mutants, and found that appressoria developed by the MoPmt2 mutants were 75.4% by 8 h, 87.9% by 12 h, and 91.6% by 24 h, with a dramatic decrease compared with that of by Guy11 and MoPmt2c (Figures 4A,C). Meanwhile, of those appressoria formed by the mutant at each time point, only 39.7, 60.3, and 87.4% were melanic and non-reduced in size, compared with 85.2, 92.9, and 95.3% of that by Guy11 (Figure 4D). In addition, we also find that over 53% of the conidia were bipolar germination, compared with less than 8% of that by Guy11 and MoPmt2c strain at 24 hpi (Figure 4A; Figures S4B,C). Based on the above, we conclude that the MoPmt2 is associated with both conidia germination and appressorium development in M. oryzae.
MoPmt2 is Essential for Pathogenic Development
To investigate the role of MoPmt2 in pathogenic development, conidial suspension (1 × 105 conidia ml−1) of the tested strains were inoculated on 14-day-old susceptible rice seedlings (Oryza sativa cv CO-39) and/or 7-day-old barley leaves by spraying. The wild type strain Guy11 caused numerous typical necrotic lesions, whereas the MoPmt2 mutants developed small and restricted lesions on host plants (Figures 5A,B). Meanwhile, most of those lesions caused by the MoPmt2 mutants seldom produced conidia, compared to wild type Guy11 and MoPmt2c. When the incubation time was extending to 15 days after inoculation, only few conidia were observed on the larger lesions (Figure 5C). To further determine the role of MoPmt2 in pathogenicity, conidial suspension of the tested strains was inoculated on wounded rice leaves. The results revealed that the wild type Guy11 were fully pathogenic, and developed extendible and necrotic lesions, in contrast, the MoPmt2 mutant only causes small lesions on the inoculation sites (Figure 5D). When the MoPmt2 gene was reintroduced in the mutant, it recovered the pathogenicity on both rice and barley plants (Figures 5A–D), suggesting a potential role for the MoPmt2 in plant invasion and colonization.
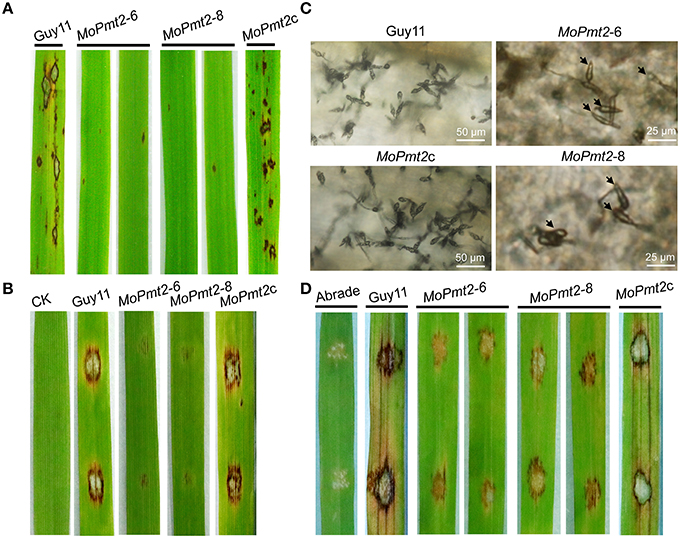
Figure 5. Pathogenicity assays. (A) The pathogenic development of the mutants on rice leaves. The MoPmt2 deletion attenuated pathogenicity on rice leaves. Diseased leaves were harvested 5 days after inoculation. (B) Pathogenicity assays on barley leaves. The MoPmt2 deletion attenuated virulence on barley leaves. Diseased leaves were harvested 5 days after inoculation. (C) Microscopic examination of conidia produced on the lesions. Diseased leaves were harvested 15 days after inoculation and observed under a light microscope. (D) Pathogenicity assay on abraded rice leaves. Drops (10 μL) of conidia suspension from tested strains were inoculated on abraded rice leaves and a virulence defect was indicated in the mutants, compared with the control strains at 5 dpi.
MoPmt2 Deletion Results in Defects on Fungal Adhesion
In M. oryzae, the persistent adhesion of the three-celled conidia to the rice leaf by means of the spore tip mucilage is a prerequisite for pathogenic development (Hamer et al., 1988). Thus, to identify the reason for attenuated virulence of MoPmt2 mutants, we firstly tested conidium adhesion to a hydrophobic surface due to the fact that the spore tip mucilage could stick to hydrophobic coverslips (Hamer et al., 1988; Han et al., 2015). Our results showed that more than 63% of conidia of the MoPmt2 mutant were washed away from the hydrophobic surface of coverslips, whereas only 15% of those by the wild type and MoPmt2c (Figures 6A,B), indicating that MoPmt2 plays a critical role in fungal adhesion in M. oryzae. As the lectin concanavalin A (ConA) conjugated to fluorescein isothiocyanate (FITC) could be used for detecting STM (Hamer et al., 1988), thus, STM secreted by Guy11, MoPmt2 mutant and complemental strain MoPmt2c were compared on hydrophobic glass cover slips. At 0.5 hpi, conidia of Guy11 showed a very strong signal for STM compared with those of MoPmt2 mutant, with 93.9 and 91.8% conidia from Guy11 and MoPmt2c strain, respectively, showing fluorescence, in contrast to 10.9% conidia from MoPmt2 mutant (Figures 6C,D). When the incubation time was prolonged to 24 h, however, no obvious differences were observed by comparison of fluorescence signal from mature appressoria of Guy11 and MoPmt2 mutant (Figure S6A).
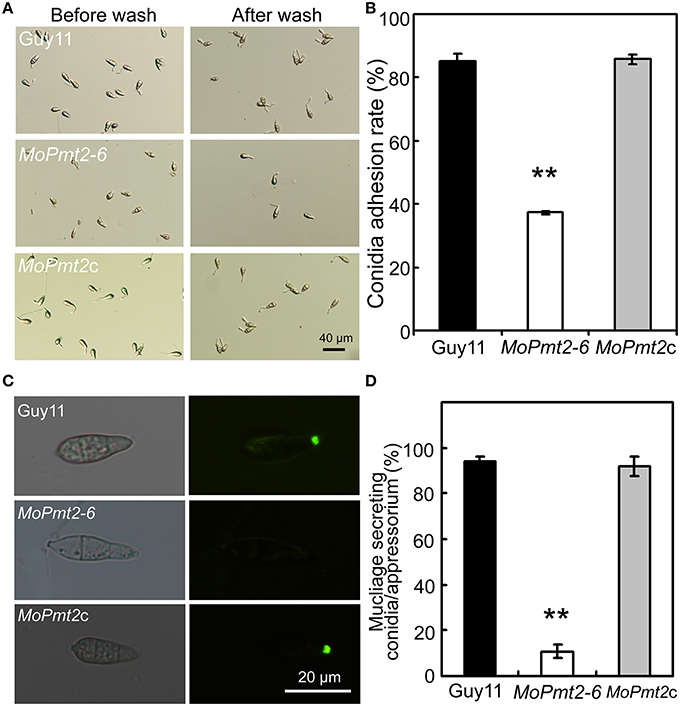
Figure 6. The effect of MoPmt2 on conidia adhesion. (A) Inability of MoPmt2 conidia to adhere to a hydrophobic surface. Conidia suspension of tested strains grew on a hydrophobic coverslips for 2 h was washed with sterilized water by pipetting and then visualized with brightfield optics of Nikon inverted Ti-S microscope. Scale bar = 40 μm. (B) Statistical analysis of conidia adhesion to hydrophobic coverslips. The numbers of conidia adhesive to the coverslips after wash were counted and statistically analyzed by Duncan's analysis (p < 0.01). Asterisks indicate significant differences of conidia adhesion among strains. Error bar represents standard deviation. (C) STM secretion test. STM from the conidia of Guy11, MoPmt2 mutant and MoPmt2c were stained with FITC-ConA, and the fluorescence signal from the germinated conidia were visualized by a Nikon inverted Ti-S epifluorescence microscope. (D) Statistical analysis of conidia stained by FITC-ConA. The numbers of conidia stained by FITC-ConA were counted and statistically analyzed by Duncan's analysis (p < 0.01). Asterisks indicate significant differences among strains. Error bar represents standard deviation.
MoPmt2 is Required for Penetration and Invasive Hyphae Growth
To further understand the role of MoPmt2 in disease development, we carried out penetration assays on the barley leaves (Figure 7A). The appressoria formed by MoPmt2 mutants could less effectively penetrate into the epidermal cells of barley leaves at 48 hpi when compared to Guy11 and MoPmt2c. In addition, most of the invasive hyphae developed by the penetrated appressorium of the mutants displayed extremely retarded growth in the host cells (Figure 7A). When infected leaves were examined after 72 hpi, most invasive hyphae of MoPmt2 mutant had still failed to colonize the leaf epidermis beyond the first cell (Figure 7B). In contrast, the wild type and MoPmt2c could freely expand and successfully colonized new cells across the cell walls of the initially invaded cell (Figures 7A,C). These results indicated that MoPmt2 is required for appressorium-mediated penetration as well as invasive hyphae growth in host cells, and the defect on these aspects might be responsible, at least in part, for the reduction of pathogenicity.

Figure 7. The deletion of MoPmt2 results in impaired appressorium penetration. (A) The MoPmt2 mutants are defective on appressorium-mediated penetration. The MoPmt2 mutants exhibited penetration defects on barley leaves after 48 and 72 h, respectively, in contrast with control strains. Scale bar = 50 μm. (B) Statistical analysis of appressorium penetrated in host cells. The appressoria that penetrated into barley epidermal cells were counted at 48 hpi, and the data were statistically analyzed. Asterisks indicate significant differences among strains. Error bar represents standard deviation. (C) Quantification of hyphae infection on barley epidermal cells. The percentage of infected cells occupied by infectious hyphae of tested strains at 72 hpi were measured and statistically analyzed.
MoPmt2 is Required for Turgor Generation and Cell Wall Integrity
In M. oryzae, the defects in cell wall composition can affect the accumulation of turgor pressure and impair the successful infection of rice plants (Howard et al., 1991; Howard and Valent, 1996; Thines et al., 2000). Based on penetration-defective phenotypes of the MoPmt2 mutants, appressorium turgor pressure of the MoPmt2 mutant was compared to wild-type Guy11, and the results showed that appressoria of the MoPmt2 mutant were in a tendency to collapse at lower glycerol concentrations, compared to those of by the Guy11 and MoPmt2c (Figure 8A), suggesting a defect in maintaining appressorium cell wall integrity in MoPmt2 mutant. In M. oryzae, an intact cell wall is the guarantee of full virulence on rice plants (Jeon et al., 2008), we thus examined the sensitivity of the transformants to different cell wall inhibitor, and the results showed that MoPmt2 mutants showed significantly increased sensitivity to cell wall damaging agents CFW and CR, respectively (Figures 8B,C). Meanwhile, CFW staining of appressorium also revealed that much stronger fluorescence signal was observed around the appressorium of Guy11 and MoPmt2c, compared to MoPmt2 mutant (Figure S6B), indicating less chitin accumulated on the cell wall of MoPmt2 mutant. Moreover, protoplast release by the MoPmt2 mutant was much faster than that of Guy11 and MoPmt2c after enzyme treatment for 30, 60, 90, and 120 min (Figure 8D), suggesting that cell wall integrity was impaired in the MoPmt2 mutants. TEM assay further confirmed this phenotype and the cell wall ultrastructures of MoPmt2 mutants were much thinner than wild type Guy11 (Figure 8E), indicating CWI defect may be results in failure of accumulation of appressorium turgor pressure, and thus attenuation of pathogenicity on rice plants.
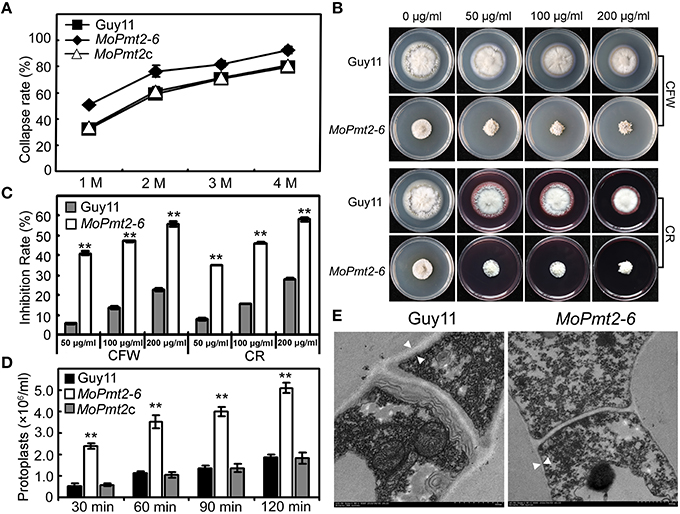
Figure 8. The deletion of MoPmt2 impaired cell wall integrity of M. oryzae. (A) Measurement of collapsed appressoria. The collapsed appressoria (>100) were observed at each glycerol concentration and statistically analyzed. (B) Sensitivity of MoPmt2 mutants to cell wall damaging agents. All tested strains were inoculated on CM containing cell wall-perturbing agents (CR and CFW) with final concentrations of 50, 100, 200 mg/mL, respectively. (C) Statistical analysis of mycelial growth of the tested strains under CR and CFW. (D) Protoplast release assay. Protoplasts released under the treatment of lysing enzymes were counted and statistically analyzed at each indicated time. Asterisks in this figure indicate significant differences among the strains (p < 0.01). Error bar represents standard deviation. (E) Ultrastructure of the cell wall in the MoPmt2 mutant. Cell wall ultrastructures of mycelia of wild type strain Guy11 and the MoPmt2 mutant were observed by sectioning and TEM. Distances between the white triangles indicated the width of cell wall. Three independent experiments were carried out and representative images from one experiment were shown.
MoPmt2 Deletion Attenuates the Activity of Extracellular Peroxidases and Laccases
In M. oryzae, the secreted extracellular peroxidases were presumed to be responsible for CR degradation, which, as a result, could generate a bright halo around the colony (Guo et al., 2011). In the examination of cell wall integrity of the MoPmt2 mutants, we found that the degradation halo of CR by the MoPmt2 mutants was not as apparent as the wild type (Figure 8B), indicating a deficiency of the CR-degrading activity in the MoPmt2 mutants. An enzyme activity assay using ABTS as substrate revealed that the MoPmt2 mutant severely reduced its peroxidase activity in the extracellular culture filtrate (Figure S7A). In addition, the activity of additional extracellular enzyme laccases was determined and found that the decreased laccase activity was also observed in the MoPmt2 mutant, with lower levels of laccase activity in the culture filtrate, compared with the wild-type strain (Figure S7B).
Discussion
In this study, three putative O-mannosyltransferases, with one member in each Pmt subfamily, were identified in rice blast fungus M. oryzae, which is consistent with findings that showed the existence of only one ortholog for each Pmt subfamily in other filamentous fungi and in the fission yeast (Willer et al., 2005; Fernández-Álvarez et al., 2009; Goto et al., 2009; Gonzalez et al., 2013; Harries et al., 2015). Amino acid sequence alignment showed that, similar to other members of the Pmt2 family in fungi, MoPmt2 contains a conserved the PMT domain in the N-terminal transmembrane region, and a MIR domain in the hydrophilic central region, which share 55 and 48% amino acid identity to the respective domains of Pmt2p from S. cerevisiae (Gentzsch and Tanner, 1996). Compared to MoPmt1 and MoPmt4, the MoPmt2 showed reduced transcriptional expression in the Moap1 mutant, indicating distinctive roles during fungal development and pathogenicity. The functional analysis by complementation of yeast Pmt2 deletion mutant demonstrated that MoPmt2 is homologous to Pmt2 from S. cerevisiae. These findings are also consistent with previous findings in P. digitatum, and suggest a conserved role of MoPmt2 in the regulation of growth and development in M. oryzae (Harries et al., 2015).
Previous studies revealed that the O-mannosyltransferase encoding gene Pmt2 had diverse roles in fungal development (Fernández-Álvarez et al., 2009; Goto et al., 2009; Fang et al., 2010; Shimizu et al., 2014; Wang et al., 2014). In M. oryzae, our findings reveal that the MoPmt2 deletion mutants are viable, but show severe defects in polarity, with more branching hyphae and septa, and globular balloon-like structures as compared to Guy11, which is similar to previous findings in P. digitatum and B. cinerea (Gonzalez et al., 2013; Harries et al., 2015). Previous studies in null chs mutants of P. digitatum revealed that the globular structures are resulted from abnormal fungal cell wall synthesis (Gandia et al., 2014). We therefore stained the cell wall of mycelia with CFW and found that these morphological defects are ascribed to production of cell enlargements with altered chitin content (Figure 2D), similar to results identified in A. nidulans (Shaw and Momany, 2002) and P. digitatum (Harries et al., 2015), suggesting an important role of MoPmt2 in regulating of the distribution of chitin content in M. oryzae. In addition, pathogenic test also revealed that MoPmt2 mutants showed restricted invasive growth in host cells. Most invasive hyphae of MoPmt2 mutant were confined to the host cells at early infection stage and the disease lesions (< 10 days) of the MoPmt2 mutant seldom produce conidia, indicating a possible necrotic reaction and low level invasive hyphae growth of MoPmt2 mutants in plant cells. These above results are similar to the previous findings that deletion of a α-1, 3-Mannosyltransferase encoding gene ALG3, which is essential for N-glycosylation of secreted effector like Slp1, resulted in the arrest of secondary infection hyphae and a significant reduction in virulence, and thus made us presume that the constrained growth of MoPmt2 mutants might be ascribed to the failure of evading host innate immunity and thus attenuated virulence on host. However, in view of the facts that the MoPmt2 mutant is defective in polarity, and some large lesions of MoPmt2 mutant can produce conidia, we couldn't exclude that the hindered hyphae growth might also partly affect the proliferation of invasive hyphae and lesion development by the MoPmt2 mutants.
In M. oryzae, conidiogenesis is a complex process that involves a series of morphological events (Liu et al., 2010). In field condition, the severity of the disease epidemic lies on the quantity of conidia produced in the rice blast lesion, suggesting an important role of conidiation in the disease cycle. Previous studies in filamentous fungus showed that disruption of Pmt2 genes in A. nidulans, A. fumigatus and P. digitatum equally led to a significant reduction of conidiation (Goto et al., 2009; Fang et al., 2010; Harries et al., 2015), whereas in the B. cinerea, deletion of Pmt2 gene resulted in a complete loss of sporulation (Gonzalez et al., 2013), indicating a potential role of Pmt2 gene in conidial development. In this study, we observed an increased expression level of MoPmt2 in conidia, which is similar to the transcriptional pattern of Moap1 in M. oryzae, demonstrating a potential role in asexual spore development (Guo et al., 2011). This hypothesis was confirmed by the analysis of MoPmt2 mutants, with phenotypes of significant reduction of conidiation, abnormal conidia size and delayed conidial germination, which is consistent with the identification in other fungi (Gonzalez et al., 2013; Wang et al., 2014; Harries et al., 2015), suggesting a conserved role of MoPmt2 protein in asexual spore development in M. oryzae. In particular, it is pointed out that conidial morphology of large conidia from the MoPmt2 mutants, was similar to those observed in Moap1 mutants, demonstrating that the MoPmt2 protein might function downstream of the MoAp1 and is responsible for conidial development (Guo et al., 2011).
The initial stages of infection by M. oryzae usually require the immediate and persistent adhesion of a conidium to the rice leaf by means of the spore tip mucilage released from the spore apex (Jelitto et al., 1994). Cell wall glycoproteins have been identified as fungal adhesives and have been implicated in host cell adhesion in many organisms (Gaur and Klotz, 1997; Frieman et al., 2002; Harries et al., 2015). O-mannosylation, a type of protein O-glycosylation with the capacity of addition of mannose residues to target proteins, have been described for fungal development, including in cell wall integrity, cell morphology and cell adhesion (Fernández-Álvarez et al., 2009; Kriangkripipat and Momany, 2009; Lommel and Strahl, 2009; Harries et al., 2015). In this study, we identified MoPmt2 as a homolog of the yeast Pmt2 protein, an O-mannosyltransferase from PMT2 subfamily, in M. oryzae. Deletion of MoPmt2 resulted in reduced ability of conidial adhesion to hydrophobic surfaces, and thus attenuated virulence on rice plants, which agreed with the findings in B. cinerea, A. fumigatus (Kriangkripipat and Momany, 2009; Gonzalez et al., 2013), indicating a conserved role of MoPmt2 in fungal adhesion during plant infection. Our subsequent FITC-ConA staining revealed that STM secretion was not detected in the MoPmt2 mutants, compared to wild-type Guy11, making us further confirm that MoPmt2 might be involved in synthesis and secretion of STM in M. oryzae. However, this seems paradoxical to the result that secretion of STM from mature appressoria of MoPmt2 mutant was comparable to that of Guy11. In M. oryzae, previous studies showed that STM is stored at the conidial apex of dormant conidia, and its release is an early event before germ tube emergence (Hamer et al., 1988). Therefore, we speculated that MoPmt2 deletion might be lead to defects on storage and secretion of STM in M. oryzae, and our subsequent staining of mature appressoria with FITC-ConA might be not reflected the true STM, but newly synthesized glycosylated-proteins that were secreted outside of the appressorium cell wall of MoPmt2 mutants and wild-type Guy11.
In M. oryzae, the fungal cell wall provides mechanical protection against attack from the host during host–pathogen interactions, and an intact cell wall is the guarantee for full virulence on rice plants (Jeon et al., 2008; Guo et al., 2015). In this study, we identified that defects on appressorium-mediated penetration was one of the main reasons for attenuated virulence of MoPmt2 mutants (Figure 7B). Our subsequent assay revealed that the MoPmt2 mutants showed increased sensitivity to CFW and CR (Figure 8), and more protoplast was released in the mutants as compared to Guy11, suggesting a defective CWI in the MoPmt2 mutants (Figure 8). This deduction was supported by the analysis of turgor pressure, with more collapsed appressorium being observed in the MoPmt2 mutants at different glycerol solutions. Together with the altered distribution of chitin content in the cell wall of both mycelia and appressorium, we concluded that the defective CWI in the mutants might be responsible for their inability to accumulate sufficient turgor pressure in appressorium, thus leading to failed penetration of host cells and attenuated pathogenicity on rice plants. Our results is consistent with the biological roles of Pmt2 in phytopathogen like B. cinerea, B. bassiana and P. digitatum, which is critical for the stability of cell wall integrity (Gonzalez et al., 2013; Wang et al., 2014; Harries et al., 2015).
In phytopathogens, secreted peroxidases are regarded to help pathogens to detoxify host-derived ROS during plant-microbe interactions (Chi et al., 2009; Guo et al., 2010, 2011). We identified an attenuation of secreted peroxidase activity in the MoPmt2 mutant by comparing CR discoloration and assaying the peroxidase activity in culture filtrates. Meanwhile, the secreted laccases were also reduced in enzyme activity in the culture filtrates of MoPmt2 mutant. In M. oryzae, a previous study revealed that hundreds of putatively secretory proteins possessed Ser/Thr-rich regions and could be potentially O-glycosylated in protein posttranslational modification (Gonzalez et al., 2012), thus, combined with the observed phenotypes, we presumed that MoPmt2 may be required for the O-glycosylation of those secreted peroxidases and laccases, and its deletion may result in proteins modification defects, which thus reduced the enzymatic activity of the MoPmt2 mutants.
In summary, our result reveals that the MoPmt2 play a critical role during the development of M. oryzae. The deletion of MoPmt2 could result in defects on conidiation, fungal adhesion, conidia germination, CWI and invasive hyphae growth, and thus attenuated the pathogenicity of M. oryzae on rice plants.
Author Contributions
Conceived and designed the experiments: MG, ZG. Performed the experiments: MG, LT, XN. Analyzed the data: MG, XZ, XN, YP. Wrote the paper: MG.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Natural Science Foundations of China (Grant No: 31101401), the Foundation for the Author of National Excellent Doctoral Dissertation of PR China (Grant No: 201470), the Foundation for the Excellent Talents of Anhui Agricultural University (Grant No: RC2015002), and the Key Grant for Excellent Young Talents of Anhui Higher Education Institutions (gxyqZD2016037). We gratefully acknowledge Professor Zhengguang Zhang for providing the plasmids pYES2.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00630
Figure S1. The dendrogram of PMT proteins from different organisms. A phylogenetic tree of MoPmts homologs was created by the distance based minimum evolution method, based on 1000 bootstraps. M. oryzae sequences characterized in this study are highlighted in dotted box. Pmt proteins from different fungal organisms were obtained from NCBI database (http://www.ncbi.nlm.nih.gov/) and the accession numbers of all the sequences used in this analysis are as followings: M. oryzae (XP_003720753, ELQ38305, XP_003713520), S. cerevisiae (NP_010188, NP_009379, NP_014966, NP_012677, NP_010190, NP_011715), C. albicans (XP_716993, XP_719907, XP_714280, XP_719311, XP_717283), S. pombe (NP_593237, NP_594135, NP_596807), U. maydis (XP_762320, XP_761621, XP_761580), Drosophila melanogaster (NP_524025, NP_569858), Homo sapiens (NP_009102.3, NP_037514.2), C. neoformans (XP_570521, XP_567365, XP_570292), B. cinerea (XP_001548518, XP_001558317, XP_001558914), A. nidulans (XP_662365, XP_662709, XP_659063), B. bassiana (EJP63368, EJP63582, EJP70423), Metarhizium anisopliae (EFY94173, EFZ02257, EFZ00337), Aspergillus niger (XP_001394947, XP_001392110, XP_001398147), A. fumigates (EAL92923, XP_754961, XP_747257), Penicillium mameffei (EEA19578, EEA22196), Neurospora crassa (XM_960450, XM_951177, XM_958833), P. digitatum (KC757712, KC757713, KC757714), and Penicillium crysogenum (AM920435, AM920427, AM920436).
Figure S2. The relative expression, targeted gene replacement and complementation of MoPmt2. (A) MoPmt2 expression at different developmental stage. Significant differences are presented in the figure (P < 0.01), and the error bar represents the standard deviation. (B) Targeted gene replacement strategies. The DNA fragment deleted from the MoPmt2 region was used as the probe to validate the deletion of MoPmt2 by Southern blot (Scale bar = 1 kb). (C) Validation of the transformants by PCR amplification. Primer pairs (p1 to p4) showed in this figure were used to validate the transformants by PCR amplification. (D,E). Southern blot analysis. To confirm the copy number of MoPmt2 gene in Guy11 and the deletion of MoPmt2 gene in the mutants, both the genomic DNA of Guy11 and the mutants, which were digested with SalI, respectively, were hybridized with probe of MoPmt2 left flank. To validate the integration of a single copy of HPH gene in the mutants, genomic DNA of Guy11 and MoPmt2 mutants were digested with SalI, and hybridized with HPH probe. (F) Semiquantitative RT-PCR. RNA samples from Guy11, MoPmt2 mutants and MoPmt2c were reversely transcripted and used to confirm the deletion and reintroduction of the MoPmt2 gene by PCR amplification.
Figure S3. Mycelial growth of the MoPmt2 mutants on different media. The wild-type strain Guy11, MoPmt2 mutants and complemented strain MoPmt2c was inoculated on CM, MM, OM, and RDC, and cultured at 28°C for 5 days.
Figure S4. Conidia bipolar germination in MoPmt2 mutants. (A) Polarity growth defects of MoPmt2 mutants. CFW staining of mycelia show polarity growth defects and swollen of mycelia of MoPmt2 mutants. (B) Conidial suspension (1 × 105 ml−1) of Guy11, MoPmt2-6 mutant and MoPmt2c, harvested from 14-day-old cultures, were inoculated on the hydrophobic surface of the coverslips for 24 h, and then observed under light microscope. (C) Statistical analysis of conidia with bipolar germination. The percentage of conidia with unipolar and bipolar germination was calculated and statistically analyzed, respectively.
Figure S5. Conidial morphology of MoPmt2 mutants on conidiophores. The development of conidia on conidiophores was examined by light microscope using strains grown on RDC medium for 7 days. Scale bar = 30 μm.
Figure S6. The staining of appressorium with FITC-ConA or CFW. (A) Mucilage secreted from the appressorium of Guy11 and MoPmt2 mutants were stained with FITC-ConA. The fluorescence signal from mature appressoria were visualized by a Nikon inverted Ti-S epifluorescence microscope. (B) The CFW staining of appressorium of Guy11, MoPmt2 mutant and MoPmt2c. The fluorescence signal from mature appressoria were captured by a Nikon inverted Ti-S epifluorescence microscope.
Figure S7. Decreased extracellular laccase and peroxidase activities in the MoPmt2 mutants. Guy11, MoPmt2 mutants and MoPmt2c were inoculated in CM liquid medium and the peroxidase activity (A) and laccase activity (B) were measured in the filtrate cultures through ABTS oxidization test with or without H2O2. Error bars represent the standard deviations and asterisks represent significant differences among the strains tested (p < 0.01).
Table S1. Bioinformatics of putative M. oryzae PMT proteins.
Table S2. Primers used in this paper.
References
Chi, M. H., Park, S. Y., Kim, S., and Lee, Y. H. (2009). A novel pathogenicity gene is required in the rice blast fungus to suppress the basal defenses of the host. PLoS Pathog. 5:e1000401. doi: 10.1371/journal.ppat.1000401
Chen, X. L., Shi, T., Yang, J., Shi, W., Gao, X., Chen, D., et al. (2014). N-glycosylation of effector proteins by an alpha-1,3-mannosyltransferase is required for the rice blast fungus to evade host innate immunity. Plant Cell 26, 1360–1376. doi: 10.1105/tpc.114.123588
Dean, R., Van Kan, J. A., Pretorius, Z. A., Hammond-Kosack, K. E., Di Pietro, A., Spanu, P. D., et al. (2012). The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. doi: 10.1111/j.1364-3703.2011.00783.x
Dean, R. A., Talbot, N. J., Ebbole, D. J., Farman, M. L., Mitchell, T. K., Orbach, M. J., et al. (2005). The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434, 980–986. doi: 10.1038/nature03449
Fang, W., Ding, W., Wang, B., Zhou, H., Ouyang, H., Ming, J., et al. (2010). Reduced expression of the O-mannosyltransferase 2 (AfPmt2) leads to deficient cell wall and abnormal polarity in Aspergillus fumigatus. Glycobiology 20, 542–552. doi: 10.1093/glycob/cwp206
Fernández-Álvarez, A., Elias-Villalobos, A., and Ibeas, J. I. (2009). The O-mannosyltransferase PMT4 is essential for normal appressorium formation and penetration in Ustilago maydis. Plant Cell 21, 3397–3412. doi: 10.1105/tpc.109.065839
Fernández-Álvarez, A., Marin-Menguiano, M., Lanver, D., Jimenez-Martin, A., Elias-Villalobos, A., Perez-Pulido, A. J., et al. (2012). Identification of O-mannosylated virulence factors in Ustilago maydis. PLoS Pathog. 8:e1002563. doi: 10.1371/journal.ppat.1002563
Frieman, M. B., McCaffery, J. M., and Cormack, B. P. (2002). Modular domain structure in the Candida glabrata adhesin Epa1p, a beta1,6 glucan-cross-linked cell wall protein. Mol. Microbiol. 46, 479–492. doi: 10.1046/j.1365-2958.2002.03166.x
Gandia, M., Harries, E., and Marcos, J. F. (2014). Identification and characterization of chitin synthase genes in the postharvest citrus fruit pathogen Penicillium digitatum. Fungal Biol. 116, 654–664. doi: 10.1016/j.funbio.2012.03.005
Gaur, N. K., and Klotz, S. A. (1997). Expression, cloning, and characterization of a Candida albicans gene, ALA1, that confers adherence properties upon Saccharomyces cerevisiae for extracellular matrix proteins. Infect. Immun. 65, 5289–5294.
Gentzsch, M., Immervoll, T., and Tanner, W. (1995). Protein O-glycosylation in Saccharomyces cerevisiae: the protein O-mannosyltransferases Pmt1p and Pmt2p function as heterodimer. FEBS Lett. 377, 128–130. doi: 10.1016/0014-5793(95)01324-5
Gentzsch, M., and Tanner, W. (1996). The PMT gene family: protein O-glycosylation in Saccharomyces cerevisiae is vital. EMBO J. 15, 5752–5759.
Girrbach, V., and Strahl, S. (2003). Members of the evolutionarily conserved PMT family of protein O-mannosyltransferases form distinct protein complexes among themselves. J. Biol. Chem. 278, 12554–12562. doi: 10.1074/jbc.M212582200
Girrbach, V., Zeller, T., Priesmeier, M., and Strahl-Bolsinger, S. (2000). Structure-function analysis of the dolichyl phosphate-mannose: protein O-mannosyltransferase ScPmt1p. J. Biol. Chem. 275, 19288–19296. doi: 10.1074/jbc.M001771200
Gonzalez, M., Brito, N., and Gonzalez, C. (2012). High abundance of Serine/Threonine-rich regions predicted to be hyper-O-glycosylated in the secretory proteins coded by eight fungal genomes. BMC Microbiol. 12:213. doi: 10.1186/1471-2180-12-213
Gonzalez, M., Brito, N., Frias, M., and Gonzalez, C. (2013). Botrytis cinerea protein O-mannosyltransferases play critical roles in morphogenesis, growth, and virulence. PLoS ONE 8, e65924. doi: 10.1371/journal.pone.0065924
Goto, M., Harada, Y., Oka, T., Matsumoto, S., Takegawa, K., and Furukawa, K. (2009). Protein O-mannosyltransferases B and C support hyphal development and differentiation in Aspergillus nidulans. Eukaryotic Cell 8, 1465–1474. doi: 10.1128/EC.00371-08
Guo, M., Chen, Y., Du, Y., Dong, Y., Guo, W., Zhai, S., et al. (2011). The bZIP transcription factor MoAP1 mediates the oxidative stress response and is critical for pathogenicity of the rice blast fungus Magnaporthe oryzae. PLoS Pathog. 7:e1001302. doi: 10.1371/journal.ppat.1001302
Guo, M., Gao, F., Zhu, X., Nie, X., Pan, Y., and Gao, Z. (2015). MoGrr1, a novel F-box protein, is involved in conidiogenesis and cell wall integrity and is critical for the full virulence of Magnaporthe oryzae. Appl. Microbiol. Biotechnol. 99, 8075–8088. doi: 10.1007/s00253-015-6820-x
Guo, M., Guo, W., Chen, Y., Dong, S., Zhang, X., Zhang, H., et al. (2010). The basic leucine zipper transcription factor Moatf1 mediates oxidative stress responses and is necessary for full virulence of the rice blast fungus Magnaporthe oryzae. Mol. Plant Microbe Interact. 23, 1053–1068. doi: 10.1094/MPMI-23-8-1053
Hamer, J. E., Howard, R. J., Chumley, F. G., and Valent, B. (1988). A mechanism for surface attachment in spores of a plant pathogenic fungus. Science 239, 288–290. doi: 10.1126/science.239.4837.288
Han, J. H., Lee, H. M., Shin, J. H., Lee, Y. H., and Kim, K. S. (2015). Role of the MoYAK1 protein kinase gene in Magnaporthe oryzae development and pathogenicity. Environ Microbiol. 17, 4672–4689. doi: 10.1111/1462-2920.13010
Harries, E., Carmona, L., Munoz, A., Ibeas, J. I., Read, N. D., Gandia, M., et al. (2013). Genes involved in protein glycosylation determine the activity and cell internalization of the antifungal peptide PAF26 in Saccharomyces cerevisiae. Fungal Genet. Biol. 58–59, 105–115. doi: 10.1016/j.fgb.2013.08.004
Harries, E., Gandia, M., Carmona, L., and Marcos, J. F. (2015). The Penicillium digitatum protein O-mannosyltransferase Pmt2 is required for cell wall integrity, conidiogenesis, virulence and sensitivity to the antifungal peptide PAF26. Mol. Plant Pathol. 16, 748–761. doi: 10.1111/mpp.12232
Howard, R. J., Ferrari, M. A., Roach, D. H., and Money, N. P. (1991). Penetration of hard substrates by a fungus employing enormous turgor pressures. Proc. Natl. Acad. Sci. U.S.A. 88, 11281–11284. doi: 10.1073/pnas.88.24.11281
Howard, R. J., and Valent, B. (1996). Breaking and entering: host penetration by the fungal rice blast pathogen Magnaporthe grisea. Annu. Rev. Microbiol. 50, 491–512. doi: 10.1146/annurev.micro.50.1.491
Jelitto, T., Page, H., and Read, N. (1994). Role of external signals in regulating the pre-penetration phase of infection by the rice blast fungus, Magnaporthe grisea. Planta 194, 471–477. doi: 10.1007/BF00714458
Jeon, J., Goh, J., Yoo, S., Chi, M. H., Choi, J., Rho, H. S., et al. (2008). A putative MAP kinase kinase kinase, MCK1, is required for cell wall integrity and pathogenicity of the rice blast fungus, Magnaporthe oryzae. Mol. Plant Microbe Interact. 21, 525–534. doi: 10.1094/MPMI-21-5-0525
Kankanala, P., Czymmek, K., and Valent, B. (2007). Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell 19, 706–724. doi: 10.1105/tpc.106.046300
Kriangkripipat, T., and Momany, M. (2009). Aspergillus nidulans protein O-mannosyltransferases play roles in cell wall integrity and developmental patterning. Eukaryotic Cell 8, 1475–1485. doi: 10.1128/EC.00040-09
Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. doi: 10.1006/jmbi.2000.4315
Lee, K., Singh, P., Chung, W. C., Ash, J., Kim, T. S., Hang, L., et al. (2006). Light regulation of asexual development in the rice blast fungus, Magnaporthe oryzae. Fungal Genet. Biol. 43, 694–706. doi: 10.1016/j.fgb.2006.04.005
Lehle, L., Strahl, S., and Tanner, W. (2006). Protein glycosylation, conserved from yeast to man: a model organism helps elucidate congenital human diseases. Angew. Chem. Int. Ed Engl. 45, 6802–6818. doi: 10.1002/anie.200601645
Letunic, I., Doerks, T., and Bork, P. (2012). SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 40, D302–D305. doi: 10.1093/nar/gkr931
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Liu, W., Xie, S., Zhao, X., Chen, X., Zheng, W., Lu, G., et al. (2010). A homeobox gene is essential for conidiogenesis of the rice blast fungus Magnaporthe oryzae. Mol. Plant Microbe Interact. 23, 366–375. doi: 10.1094/MPMI-23-4-0366
Loibl, M., and Strahl, S. (2013). Protein O-mannosylation: what we have learned from baker's yeast. Biochim. Biophys. Acta 1833, 2438–2446. doi: 10.1016/j.bbamcr.2013.02.008
Lommel, M., and Strahl, S. (2009). Protein O-mannosylation: conserved from bacteria to humans. Glycobiology 19, 816–828. doi: 10.1093/glycob/cwp066
Lussier, M., Gentzsch, M., Sdicu, A. M., Bussey, H., and Tanner, W. (1995). Protein O-glycosylation in yeast. The PMT2 gene specifies a second protein O-mannosyltransferase that functions in addition to the PMT1-encoded activity. J. Biol. Chem. 270, 2770–2775. doi: 10.1074/jbc.270.6.2770
McGinnis, S., and Madden, T. L. (2004). BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 32, W20–W25. doi: 10.1093/nar/gkh435
Mouyna, I., Kniemeyer, O., Jank, T., Loussert, C., Mellado, E., Aimanianda, V., et al. (2010). Members of protein O-mannosyltransferase family in Aspergillus fumigatus differentially affect growth, morphogenesis and viability. Mol. Microbiol. 76, 1205–1221. doi: 10.1111/j.1365-2958.2010.07164.x
Olson, G. M., Fox, D. S., Wang, P., Alspaugh, J. A., and Buchanan, K. L. (2007). Role of protein O-mannosyltransferase Pmt4 in the morphogenesis and virulence of Cryptococcus neoformans. Eukaryotic Cell 6, 222–234. doi: 10.1128/EC.00182-06
Park, G., Bruno, K. S., Staiger, C. J., Talbot, N. J., and Xu, J. R. (2004). Independent genetic mechanisms mediate turgor generation and penetration peg formation during plant infection in the rice blast fungus. Mol. Microbiol. 53, 1695–1707. doi: 10.1111/j.1365-2958.2004.04220.x
Prill, S. K., Klinkert, B., Timpel, C., Gale, C. A., Schroppel, K., and Ernst, J. F. (2005). PMT family of Candida albicans: five protein mannosyltransferase isoforms affect growth, morphogenesis and antifungal resistance. Mol. Microbiol. 55, 546–560. doi: 10.1111/j.1365-2958.2004.04401.x
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Shaw, B. D., and Momany, M. (2002). Aspergillus nidulans polarity mutant swoA is complemented by protein O-mannosyltransferase pmtA. Fungal Genet. Biol. 37, 263–270. doi: 10.1016/S1087-1845(02)00531-5
Shimizu, K., Imanishi, Y., Toh-e, A., Uno, J., Chibana, H., Hull, C. M., et al. (2014). Functional characterization of PMT2, encoding a protein-O-mannosyltransferase, in the human pathogen Cryptococcus neoformans. Fungal Genet. Biol. 69, 13–22. doi: 10.1016/j.fgb.2014.05.007
Strahl-Bolsinger, S., Gentzsch, M., and Tanner, W. (1999). Protein O-mannosylation. Biochim. Biophys. Acta 1426, 297–307. doi: 10.1016/S0304-4165(98)00131-7
Strahl-Bolsinger, S., and Scheinost, A. (1999). Transmembrane topology of pmt1p, a member of an evolutionarily conserved family of protein O-mannosyltransferases. J. Biol. Chem. 274, 9068–9075. doi: 10.1074/jbc.274.13.9068
Talbot, N. J. (2003). On the trail of a cereal killer: Exploring the biology of Magnaporthe grisea. Annu. Rev. Microbiol. 57, 177–202. doi: 10.1146/annurev.micro.57.030502.090957
Tamura, K., Dudley, J., Nei, M., and Kumar, S. (2007). MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599. doi: 10.1093/molbev/msm092
Tanner, G. A., Gretz, N., Connors, B. A., Evan, A. P., and Steinhausen, M. (1996). Role of obstruction in autosomal dominant polycystic kidney disease in rats. Kidney Int. 50, 873–886. doi: 10.1038/ki.1996.387
Thines, E., Weber, R. W., and Talbot, N. J. (2000). MAP kinase and protein kinase A-dependent mobilization of triacylglycerol and glycogen during appressorium turgor generation by Magnaporthe grisea. Plant Cell 12, 1703–1718. doi: 10.1105/tpc.12.9.1703
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Wang, J. J., Qiu, L., Chu, Z. J., Ying, S. H., and Feng, M. G. (2014). The connection of protein O-mannosyltransferase family to the biocontrol potential of Beauveria bassiana, a fungal entomopathogen. Glycobiology 24, 638–648. doi: 10.1093/glycob/cwu028
Willer, T., Brandl, M., Sipiczki, M., and Strahl, S. (2005). Protein O-mannosylation is crucial for cell wall integrity, septation and viability in fission yeast. Mol. Microbiol. 57, 156–170. doi: 10.1111/j.1365-2958.2005.04692.x
Willer, T., Valero, M. C., Tanner, W., Cruces, J., and Strahl, S. (2003). O-mannosyl glycans: from yeast to novel associations with human disease. Curr. Opin. Struct. Biol. 13, 621–630. doi: 10.1016/j.sbi.2003.09.003
Xu, J. R., Zhao, X., and Dean, R. A. (2007). From genes to genomes: a new paradigm for studying fungal pathogenesis in Magnaporthe oryzae. Adv. Genet. 57, 175–218. doi: 10.1016/S0065-2660(06)57005-1
Xu, Y., Li, H., Zhang, J., Song, B., Chen, F., Duan, X., et al. (2010). Disruption of the chitin synthase gene CHS1 from Fusarium asiaticum results in an altered structure of cell walls and reduced virulence. Fungal Genet. Biol. 47, 205–215. doi: 10.1016/j.fgb.2009.11.003
Zhou, H., Hu, H., Zhang, L., Li, R., Ouyang, H., Ming, J., et al. (2007). O-Mannosyltransferase 1 in Aspergillus fumigatus (AfPmt1p) is crucial for cell wall integrity and conidium morphology, especially at an elevated temperature. Eukaryot. Cell 6, 2260–2268. doi: 10.1128/EC.00261-07
Keywords: Magnaporthe oryzae, O-mannosylation, conidia germination, appressoria formation, cell wall integrity, pathogenicity
Citation: Guo M, Tan L, Nie X, Zhu X, Pan Y and Gao Z (2016) The Pmt2p-Mediated Protein O-Mannosylation Is Required for Morphogenesis, Adhesive Properties, Cell Wall Integrity and Full Virulence of Magnaporthe oryzae. Front. Microbiol. 7:630. doi: 10.3389/fmicb.2016.00630
Received: 22 February 2016; Accepted: 18 April 2016;
Published: 02 May 2016.
Edited by:
Pietro Daniele Spanu, Imperial College London, UKReviewed by:
Yasin Fatih Dagdas, The Sainsbury Laboratory, UKMichael John Kershaw, University of Exeter, UK
Copyright © 2016 Guo, Tan, Nie, Zhu, Pan and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Min Guo, kandylemon@163.com
†These authors have contributed equally to this work.
 Min Guo
Min Guo Leyong Tan†
Leyong Tan†