- 1Laboratory of Microbiology, Department of Biochemistry and Microbiology, Ghent University, Ghent, Belgium
- 2Leibniz Institute DSMZ–German Collection of Microorganisms and Cell Cultures GmbH, Braunschweig, Germany
- 3Department of Microbiology and Molecular Genetics, University of Pittsburgh School of Medicine, Pittsburgh, PA, USA
- 4BCCM/LMG Bacteria Collection, Department of Biochemistry and Microbiology, Ghent University, Ghent, Belgium
Partial gyrB gene sequence analysis of 17 isolates from human and environmental sources revealed 13 clusters of strains and identified them as Burkholderia glathei clade (BGC) bacteria. The taxonomic status of these clusters was examined by whole-genome sequence analysis, determination of the G+C content, whole-cell fatty acid analysis and biochemical characterization. The whole-genome sequence-based phylogeny was assessed using the Genome Blast Distance Phylogeny (GBDP) method and an extended multilocus sequence analysis (MLSA) approach. The results demonstrated that these 17 BGC isolates represented 13 novel Burkholderia species that could be distinguished by both genotypic and phenotypic characteristics. BGC strains exhibited a broad metabolic versatility and developed beneficial, symbiotic, and pathogenic interactions with different hosts. Our data also confirmed that there is no phylogenetic subdivision in the genus Burkholderia that distinguishes beneficial from pathogenic strains. We therefore propose to formally classify the 13 novel BGC Burkholderia species as Burkholderia arvi sp. nov. (type strain LMG 29317T = CCUG 68412T), Burkholderia hypogeia sp. nov. (type strain LMG 29322T = CCUG 68407T), Burkholderia ptereochthonis sp. nov. (type strain LMG 29326T = CCUG 68403T), Burkholderia glebae sp. nov. (type strain LMG 29325T = CCUG 68404T), Burkholderia pedi sp. nov. (type strain LMG 29323T = CCUG 68406T), Burkholderia arationis sp. nov. (type strain LMG 29324T = CCUG 68405T), Burkholderia fortuita sp. nov. (type strain LMG 29320T = CCUG 68409T), Burkholderia temeraria sp. nov. (type strain LMG 29319T = CCUG 68410T), Burkholderia calidae sp. nov. (type strain LMG 29321T = CCUG 68408T), Burkholderia concitans sp. nov. (type strain LMG 29315T = CCUG 68414T), Burkholderia turbans sp. nov. (type strain LMG 29316T = CCUG 68413T), Burkholderia catudaia sp. nov. (type strain LMG 29318T = CCUG 68411T) and Burkholderia peredens sp. nov. (type strain LMG 29314T = CCUG 68415T). Furthermore, we present emended descriptions of the species Burkholderia sordidicola, Burkholderia zhejiangensis and Burkholderia grimmiae. The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA and gyrB gene sequences determined in this study are LT158612-LT158624 and LT158625-LT158641, respectively.
Introduction
The genus Burkholderia currently comprises 90 validly named species (Euzeby, 1997) and several uncultured Candidatus species (Van Oevelen et al., 2004; Verstraete et al., 2011; Lemaire et al., 2012) which occupy very diverse niches (Coenye and Vandamme, 2003). Many Burkholderia species have thus far only been isolated as free-living organisms but a growing body of literature reveals that they live in close interaction with numerous plant, animal, fungal or even amoebozoan hosts (Marolda et al., 1999; Van Borm et al., 2002; Kikuchi et al., 2011; Verstraete et al., 2013; Stopnisek et al., 2016; Xu et al., 2016). Burkholderia species may be beneficial to their hosts because some strains can fix nitrogen, produce plant hormones or siderophores, or lower pathogen-related ethylene levels; hence they have been exploited for plant growth promotion and biocontrol of plant diseases (Compant et al., 2008; Vial et al., 2011). Yet, other Burkholderia species are notorious pathogens in plants, animals and humans (Mahenthiralingam et al., 2008). This ecological diversity is likely attributed to their large, multireplicon genomes (typically between 6 and 9 Mb) which also confer a metabolic versatility allowing them to degrade a wide range of recalcitrant xenobiotics (Parke and Gurian-Sherman, 2001; Coenye and Vandamme, 2003).
Phylogenetic analyses based on the 16S rRNA and protein-coding genes showed that Burkholderia glathei clade (BGC) species are phylogenetically divergent from other Burkholderia species and form a separate clade (Sawana et al., 2014; Vandamme et al., 2014). Although this clade thus far includes only 12 formally named species, its functional diversity is impressive. In this clade too, most species have been isolated from bulk and rhizosphere soil (Zolg and Ottow, 1975; Viallard et al., 1998; Vandamme et al., 2013; Draghi et al., 2014; Baek et al., 2015), but also from contaminated soil and sludge from a wastewater treatment system (Lu et al., 2012; Vandamme et al., 2013; Liu et al., 2014). Two BGC species were associated with less studied hosts like fungi (Burkholderia sordidicola) and mosses (Burkholderia grimmiae) (Lim et al., 2003; Tian et al., 2013) but numerous, mostly uncultivated BGC species adopted endosymbiotic lifestyles in insect guts (Kikuchi et al., 2011; Tago et al., 2015; Xu et al., 2016) or plant leaf tissue (Verstraete et al., 2013; Carlier et al., 2015) and many additional unclassified B. glathei-like bacteria have been reported (Nogales et al., 2001; Salles et al., 2006; Pumphrey and Madsen, 2008; Draghi et al., 2014; Verstraete et al., 2014; Peeters et al., 2016).
The present study aimed to perform a phylogenomic study of established and novel species in the B. glathei clade, to formally name the latter and to make reference cultures and whole-genome sequences of each of these versatile bacteria publicly available. The genome sequence-based phylogeny was assessed using the Genome Blast Distance Phylogeny (GBDP) method (Meier-Kolthoff et al., 2013) and an extended multilocus sequence analysis (MLSA) approach. For phenotypic characterization, whole-cell fatty acid profiling and biochemical analyses were performed.
Materials and Methods
Bacterial Strains and Growth Conditions
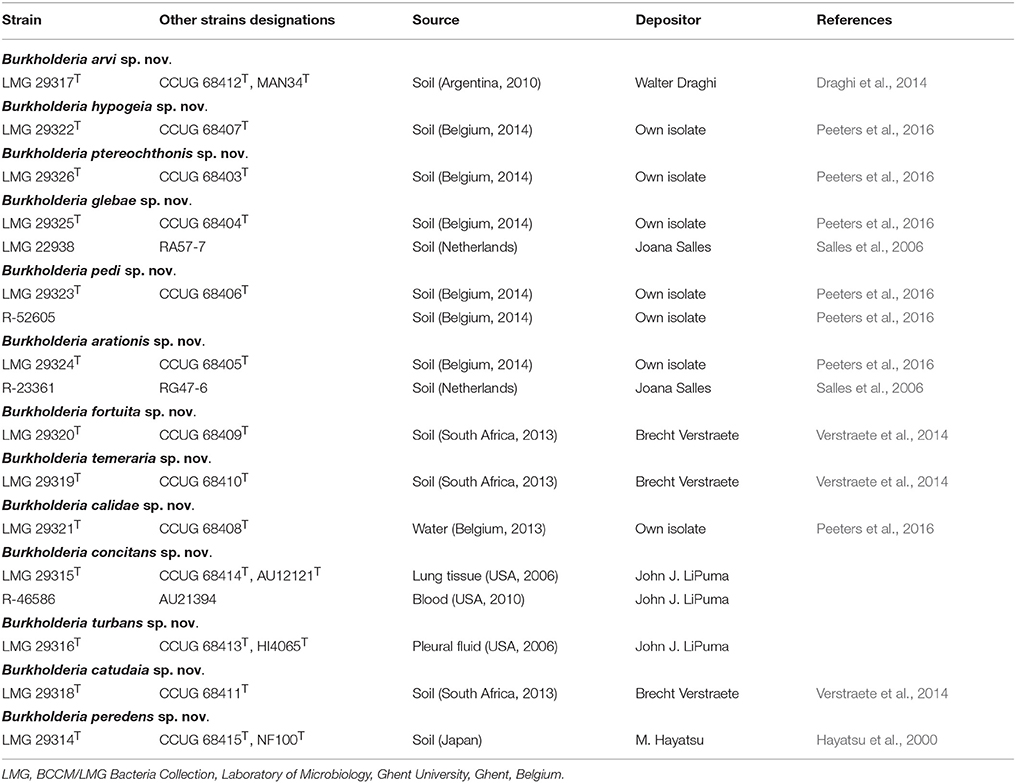
Table 1 lists the sources of the 17 studied isolates. Details of type strains of each of the present BGC species were described previously (Zolg and Ottow, 1975; Lim et al., 2003; Lu et al., 2012; Tian et al., 2013; Vandamme et al., 2013; Draghi et al., 2014; Liu et al., 2014; Baek et al., 2015). Strains were grown aerobically on buffered nutrient agar (Oxoid, pH 6.8) and incubated at 28°C. Cultures were preserved in MicroBankTM vials at −80°C.
16S rRNA Gene Sequence Analysis
Nearly complete sequences were obtained as described previously (Peeters et al., 2013).
gyrB Gene Sequence Analysis
Partial gyrB gene sequences were obtained as described previously (Spilker et al., 2009; Peeters et al., 2013). Sequence assembly was performed using BioNumerics v7.5 (Applied Maths). Sequences (589–1182 bp) were aligned based on amino acid sequences using Muscle (Edgar, 2004) in MEGA6 (Tamura et al., 2013). All positions with less than 95% site coverage were eliminated, resulting in a total of 570 positions in the final dataset. Phylogenetic analysis was conducted in MEGA6 (Tamura et al., 2013).
Whole-Genome Sequencing
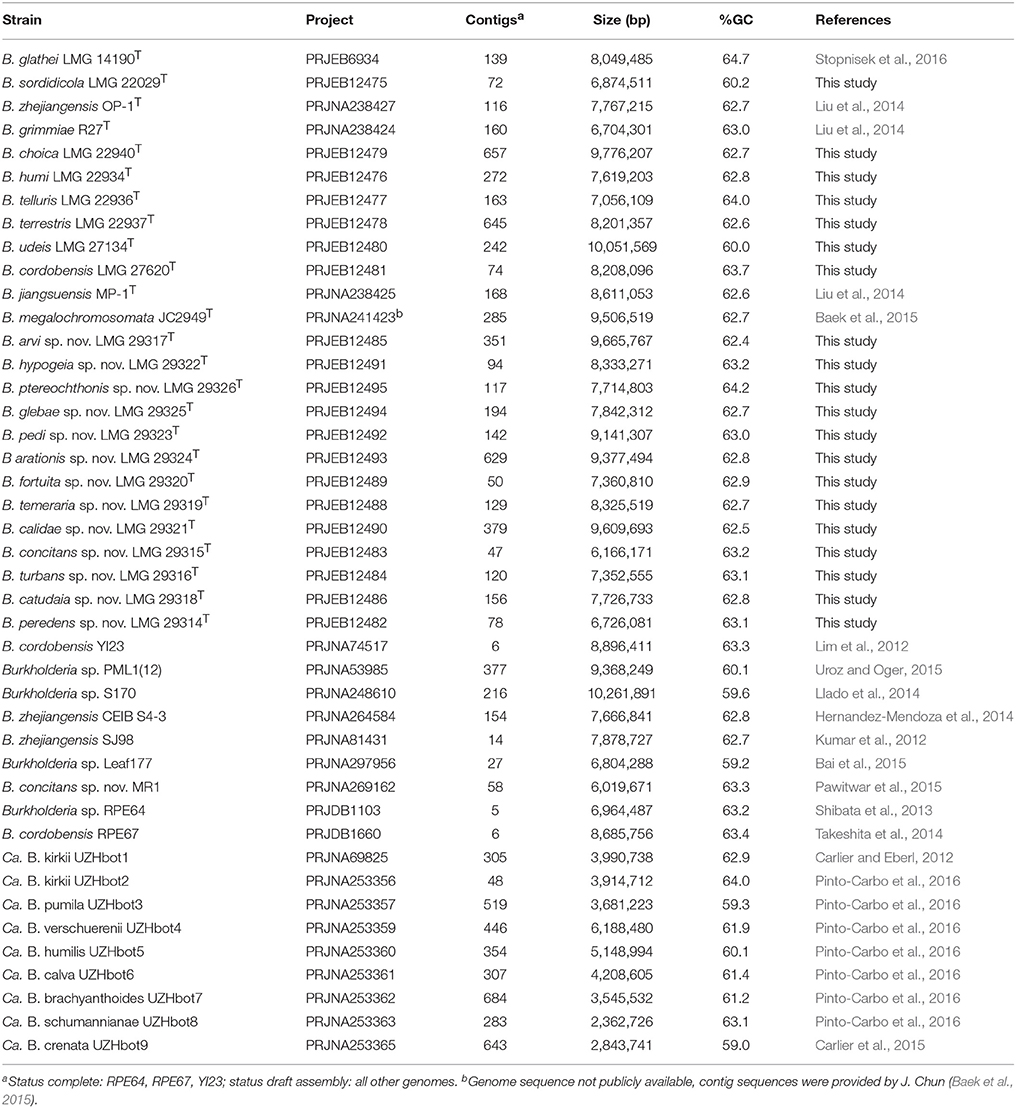
Genomic DNA of 20 strains (Table 2) was prepared as described by Pitcher et al. (1989). Genomic libraries were prepared using the Nextera kit following the methods introduced by Baym et al. (2015) and the 151 bp paired-end libraries were sequenced on the Illumina HiSeq platform of the University of New Hampshire Hubbard Center for Genomics Studies with an average insert size of 386 bp. Quality reports were created by FastQC. Adaptors and low-quality reads were trimmed using Trimmomatic (Bolger et al., 2014) with the following options: ILLUMINACLIP:NexteraPE-PE.fa:2:30:10 MAXINFO:60:0.4 MINLEN:60. Assembly was performed using SPAdes (Bankevich et al., 2012) with default k-mer sizes (21, 33, 55, 77) and mismatch correction (option—careful). Contigs with length <500 bp and coverage <2 were discarded from the resulting assemblies. Raw reads were mapped against the assemblies using bwa-mem (Li, 2013) and contigs were polished using Pilon (Walker et al., 2014) with default parameters. Quast (Gurevich et al., 2013) was used to create quality reports of the resulting assemblies. Annotation was performed using Prokka 1.11 (Seemann, 2014) with a genus-specific database based on reference genomes from the Burkholderia Genome Database (Winsor et al., 2008).
Publicly Available Genomes
Twenty three publicly available whole-genome sequences of BGC bacteria were downloaded from the NCBI database (Table 2). B. gladioli BSR3 (Seo et al., 2011) was used as an outgroup in all phylogenomic analyses. For B. megalochromosomata JC2949T the whole-genome sequence was not publicly available (February 1st, 2016) and the contig sequences were provided by J. Chun (Baek et al., 2015). For B. sordidicola S170, B. zhejiangensis CEIB S4-3 and B. megalochromosomata JC2949T no annotation was available and annotation was performed using Prokka as described above.
Phylogenomic Analysis
The latest version of the Genome Blast Distance Phylogeny (GBDP) approach was applied (Meier-Kolthoff et al., 2013) to calculate the intergenomic distance between each pair of genomes (based on the nucleotide data) and included the calculation of 100 replicate distances to assess pseudo-bootstrap support (Meier-Kolthoff et al., 2014a). Distance calculations were conducted under the recommended settings of the Genome-to-Genome Distance Calculator (GGDC 2.1; http://ggdc.dsmz.de), as described earlier (Meier-Kolthoff et al., 2013). The GBDP trimming algorithm and formula d5 were chosen because of their advantages for phylogenetic inference (Meier-Kolthoff et al., 2014a) and according distance matrices were prepared (a single matrix for the original distances plus 100 matrices containing the replicates). A phylogenomic tree with branch support (Meier-Kolthoff et al., 2014a) was inferred using FastME v2.07 with tree bisection and reconnection post-processing (Lefort et al., 2015). Moreover, pairwise digital DNA-DNA hybridization (dDDH) values and their confidence intervals were also determined using GGDC 2.1 under recommended settings (Meier-Kolthoff et al., 2013). The potential affiliation of the novel strains to existing species was determined by clustering using a 70% dDDH radius around each of the 12 BGC type strains as previously applied (Liu et al., 2015). Visualization and annotation of the phylogenetic tree was performed using iTOL (Letunic and Bork, 2011).
As an alternative for the GBDP method, an extended MLSA analysis was performed in which a whole-genome phylogeny was calculated based on single-copy orthologous genes as described previously (Pinto-Carbo et al., 2016). In short, single-copy orthologs were identified using blastp and OrthoMCL v2.0.9 (with e-value cutoff 1e10−6 and 50% match cutoff; Fischer et al., 2011) and aligned based on their amino acid sequences using MUSCLE. The alignments were trimmed using TrimAl (removing positions with gaps in more than 50% of the sequences) and concatenated to construct a Maximum Likelihood tree using RaXML v7.4.2 (Stamatakis, 2014) with the WAG amino acid substitution model and 100 rapid bootstrap analyses.
Phenotypic Characterization
Phenotypic and cellular fatty acid analyses were performed as described previously (Draghi et al., 2014).
Results
16S rRNA Gene Sequence Analysis
The 16S rRNA gene sequences determined in the present study are publicly available through the GenBank/EMBL/DDBJ accession numbers LT158612-LT158624.
gyrB Gene Sequence Analysis
Partial gyrB gene sequences were compared to those of the type strains of the 12 validly named BGC species (Figure 1). The 17 unclassified isolates represented 13 taxa which showed 83.4–96.2% pairwise identity with the gyrB sequences of the type strains of other BGC species. The gyrB gene sequences determined in the present study are publicly available through the GenBank/EMBL/DDBJ accession numbers LT158625-LT158641.
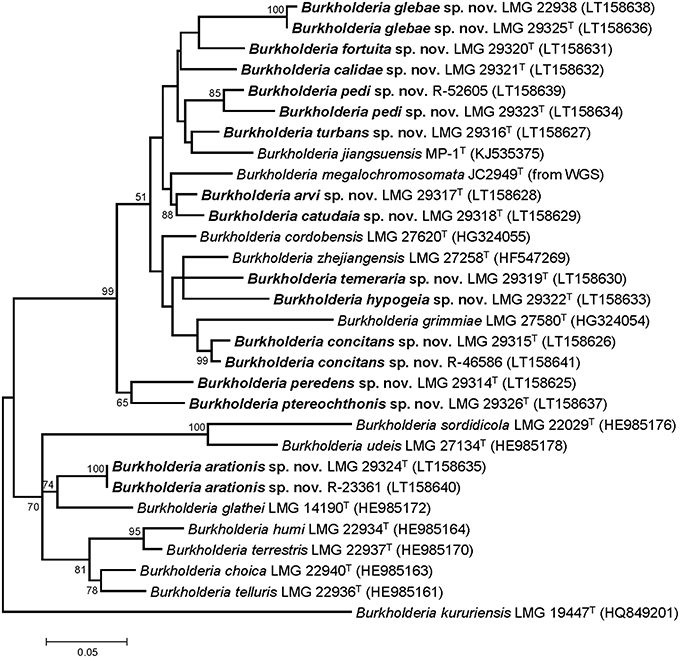
Figure 1. Phylogenetic tree based on partial gyrB sequences of the 17 isolates in this study and type strains of phylogenetically related Burkholderia species. The optimal tree (highest log likelihood) was constructed using the Maximum Likelihood method and General Time Reversible model in MEGA6 (Tamura et al., 2013). A discrete Gamma distribution was used to model evolutionary rate differences among sites [5 categories (+G, parameter = 0.5462)] and allowed for some sites to be evolutionarily invariable ([+I], 37.9331% sites). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches if greater than 50%. For B. megalochromosomata JC2949T the gyrB gene sequence was extracted from the genome sequence. The gyrB sequence of B. kururiensis LMG 19447T was used as outgroup. The scale bar indicates the number of substitutions per site.
Whole-Genome Sequencing
To further characterize the taxonomic status of these 13 taxa, we determined the whole-genome sequence of one strain per gyrB cluster and of B. sordidicola LMG 22029T, B. choica LMG 22940T, B. humi LMG 22934T, B. telluris LMG 22936T, B. terrestris LMG 22937T, B. udeis LMG 27134T, and B. cordobensis LMG 27620T. The assembly of the Illumina HiSeq 150 bp paired end reads resulted in assemblies with 47–657 contigs and a total of 6,166,171–10,051,569 bp (Table 2). The annotated assemblies of these 20 genomes were submitted to the European Nucleotide Archive and are publicly available through the GenBank/EMBL/DDBJ accession numbers listed in Table 2 and the species descriptions. The genome sequences of the remaining five BGC type strains and of 18 additional strains were publicly available (Table 2).
DNA Base Composition
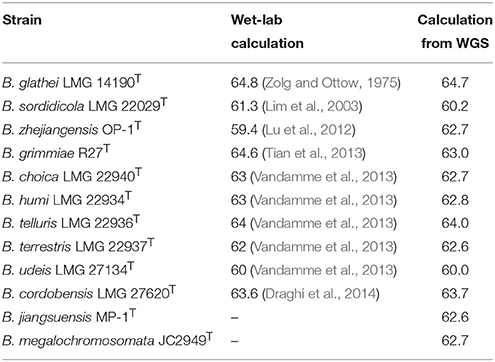
The G+C content of all type strains was calculated from their genome sequences and ranged from 62.4 to 64.2 mol% (Table 2).
Phylogenomic Analysis
The pairwise intergenomic distances and dDDH estimates of the 44 genome sequences are listed in Supplementary Table 1. The phylogenetic tree inferred from the intergenomic distances (Figure 2) was well resolved and most branches showed a very high bootstrap support (average support: 94.8%). Species delineation based on the pairwise dDDH values and a 70% dDDH radius around each type strain yielded 39 species which included the present 12 validly named species as well as the 13 novel species delineated by means of partial gyrB gene sequences (Figure 1).
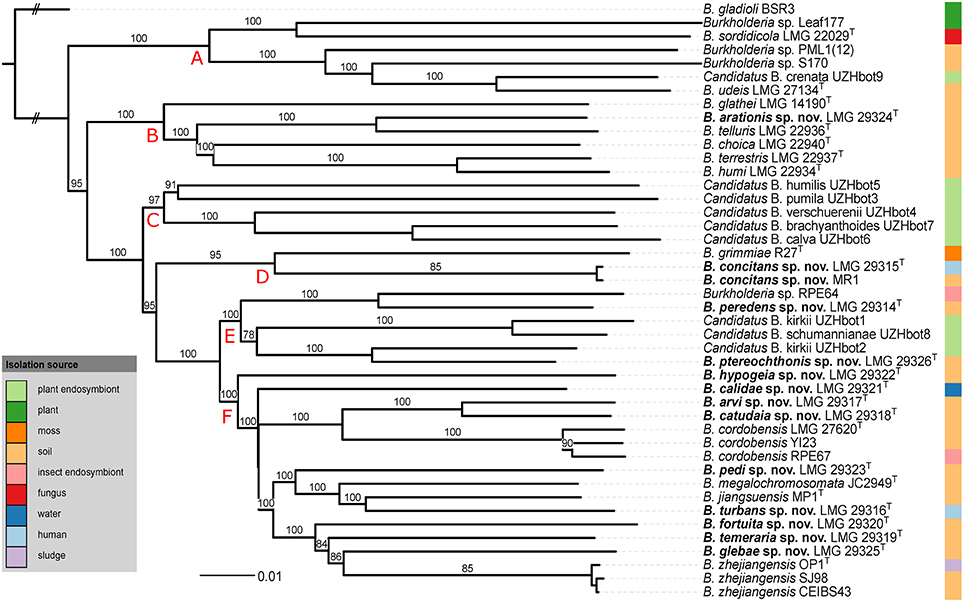
Figure 2. Whole-genome sequence based phylogenomic tree of all BGC genomes inferred by GBDP. The outer column shows the isolation source of the strains. Pseudo-bootstrap support values above 60% are shown. The tree reveals a high average support of 94.8%. Long terminal branches are due to the distinct scaling used by GBDP's formula d5. B. gladioli BSR3 was used as outgroup. Red capital letters define subtrees that also occur in the tree depicted in Figure 3.
For the extended MLSA approach, we identified 332 single-copy orthologs that were present in all 44 genomes. The Maximum-Likelihood phylogenetic tree based on the concatenated amino acid alignment (Figure 3) was well resolved and showed a high bootstrap support on almost all branches.
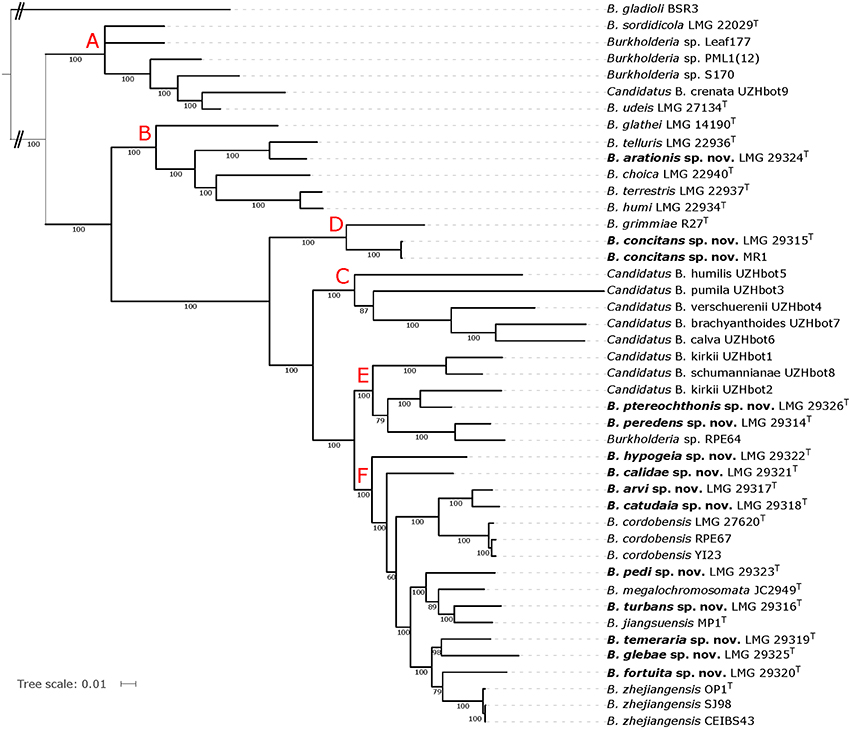
Figure 3. Whole-genome phylogeny based on single-copy orthologs of all BGC genomes. The phylogenetic tree was constructed using the WAG protein substitution model and RAxML and is based on an amino acid alignment with 105,141 positions from 332 single-copy orthologous genes. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (100 replicates) are shown next to the branches if greater than 60%. B. gladioli BSR3 was used as outgroup. Red capital letters define subtrees that also occur in the tree depicted in Figure 2.
The topologies of the two phylogenomic trees (Figures 2, 3) were very similar and both revealed six clusters of species (A-F). The main difference in tree topology related to the phylogenetic position of the Candidatus species in cluster C. This cluster was supported by a 100% bootstrap value in both analyses but its relative position to cluster D species differed in the two trees (Figures 2, 3). Additionally, the internal branching order of cluster C, E and F species differed minimally between both analyses. Both phylogenomic analyses showed that strain MR1 clustered with B. concitans sp. nov. and that strain RPE67 clustered with B. cordobensis. Finally, the large distances between strains PML1(12) and S170, and the type strains of B. glathei and B. sordidicola, respectively, indicated that both strains were misidentified and wrongly annotated in the NCBI database as B. glathei and B. sordidicola, respectively (Figures 2, 3). Both strains occupy unique positions in the phylogenomic trees and represent additional novel BGC species.
Cellular Fatty Acid Analysis
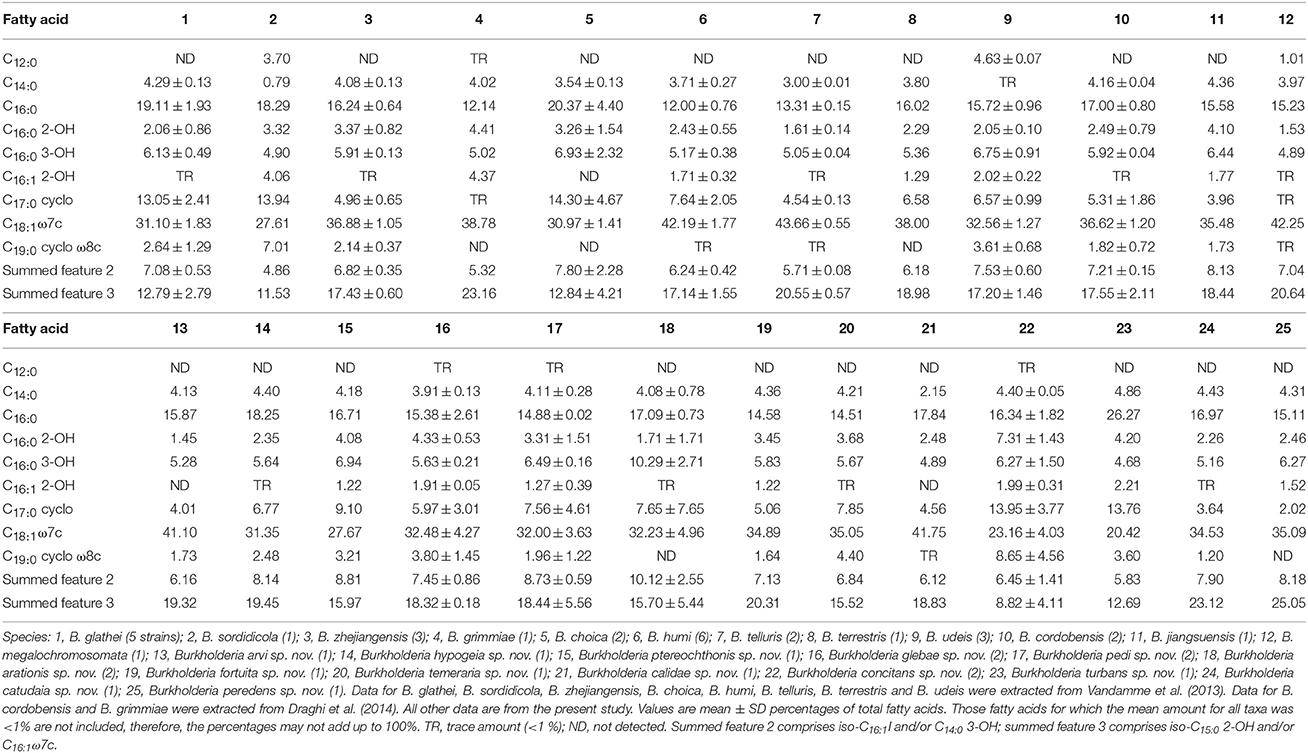
The fatty acid profiles of all strains are shown in Table 3. Branched chain fatty acids have not been reported in members of the genus Burkholderia and therefore summed features 2 and 3 very likely represent C14:0 3-OH and C16:1 ω7c, respectively (Yabuuchi et al., 1992). The main fatty acid components are C16:0, C18:1 ω7c and summed feature 3 (most probably representing C16:1 ω7c).
Biochemical Characterization
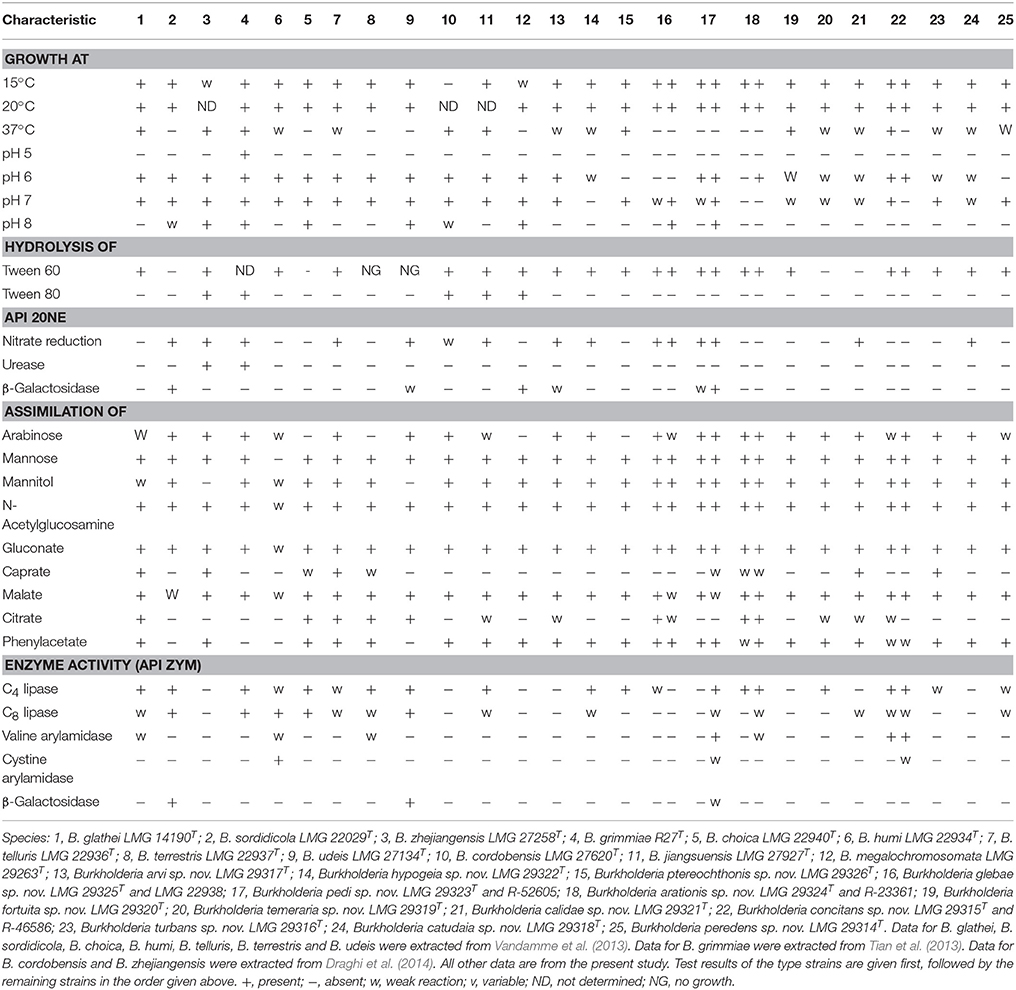
An overview of biochemical characteristics useful for distinguishing the BGC species is shown in Table 4.
Discussion
While soil is a well-known source of free-living Burkholderia species, these organisms often live in close interaction with plants, animals, fungi, or amoebae (Marolda et al., 1999; Van Borm et al., 2002; Kikuchi et al., 2011; Verstraete et al., 2013; Stopnisek et al., 2016; Xu et al., 2016). The BGC represents a poorly known line of descent within the genus Burkholderia and most of the 12 validly named BGC species have been isolated from soil. Yet, publicly available sequence data indicate that the taxonomic diversity in this clade is severely underestimated (Nogales et al., 2001; Salles et al., 2006; Pumphrey and Madsen, 2008; Draghi et al., 2014; Verstraete et al., 2014; Peeters et al., 2016; Xu et al., 2016). In the present study, gyrB gene sequence analysis was used to screen our strain collection and 17 isolates from human and environmental samples were identified as B. glathei-like bacteria. The gyrB sequence similarity levels toward other BGC species suggested that the 17 isolates in this study represented 13 novel species (Figure 1). To further characterize the taxonomic status of these isolates, we analyzed the genome sequence of 13 isolates representative for the 13 gyrB sequence clusters and of 7 BGC type strains and compared those to 23 whole-genome sequences of BGC strains that were publicly available. Additionally, we also studied their chemotaxonomic and biochemical properties to comply with the polyphasic taxonomic consensus approach to bacterial systematics (Vandamme et al., 1996).
In this genomics era, state-of-the-art sequencing technologies enable direct access to the information contained in whole-genome sequences and it is no longer adequate to deduce genome relatedness through traditional DNA-DNA hybridization experiments (Vandamme and Peeters, 2014; Whitman, 2015). Genomic taxonomy can be studied through various parameters including average nucleotide identity (ANI), GBDP, Maximal Unique Matches index (MUMi), and core gene identity (CGI) (Konstantinidis and Tiedje, 2005; Goris et al., 2007; Deloger et al., 2009; Vanlaere et al., 2009; Meier-Kolthoff et al., 2013). Although, there is a general consensus that genome sequencing could revolutionize prokaryotic systematics (Sutcliffe et al., 2013; Meier-Kolthoff et al., 2014b; Rossello-Mora and Amann, 2015; Thompson et al., 2015), traditional DDH experiments are still being performed and new genome-based methods are evaluated in terms of their correspondence to the existing classifications which are based on DDH data (Wayne et al., 1987; Stackebrandt et al., 2002). The GGDC implementation of the GBDP method provides a quick and reliable alternative to the wet-lab DDH technique and its dDDH prediction capability (including confidence intervals) produces classifications which correlate better with the traditional DDH values than do any of the ANI implementations (Meier-Kolthoff et al., 2013). Among several advantages, GBDP is independent from genome annotation, is applicable to both nucleotide and amino acid data and is immune against problems caused by incompletely sequenced or low-quality draft genomes. Finally, GBDP provides branch support values for the resulting phylogenetic trees (Meier-Kolthoff et al., 2013, 2014a).
We complemented the results of the GBDP analysis with a whole-genome-based phylogeny based on the sequence analysis of 332 single-copy orthologous genes in all BGC genomes. This extended MLSA approach takes only the coding part of the genomes into account and is therefore not influenced by non-coding sequences or pseudogenes that might have a different evolutionary history than the rest of the genome. It depends however on genome annotation, is unable to cope with problems caused by incompletely sequenced or low-quality draft genomes, and its calculations are more compute-intensive and cannot be carried out incrementally. Although, the GBDP and extended MLSA methods used different algorithms, the conclusions drawn from their phylogenies were consistent thus illustrating the robustness of whole-genome based taxonomic methods (Colston et al., 2014).
The GGDC dDDH values and the application of the 70% dDDH cut-off for species delineation (Supplementary Table 1) demonstrated that the 13 clusters delineated through gyrB sequence analysis (Figure 1) represented 13 novel BGC species and thus confirmed that gyrB gene sequence analysis is a reliable tool for the identification of Burkholderia species (Tayeb et al., 2008; Vandamme et al., 2013). Both phylogenomic analyses identified strain MR1, which was isolated from Florida golf course soil and which was shown to reduce the herbicide methylarsenate, as B. concitans sp. nov. Next to strain YI23, which was previously identified as B. cordobensis by Draghi et al. (2014), the present study also identified strain RPE67, which was isolated from the gut of a stink bug, as B. cordobensis. Finally, both phylogenomic analyses also showed that strain PML1(12), an ectomycorrhizosphere-inhabiting bacterium with mineral-weathering ability (Uroz and Oger, 2015), strain S170, a potential plant growth promoter isolated from coniferous forest soil (Llado et al., 2014), strain RPE64, a bacterial symbiont of the bean bug Riptortus pedestris (Shibata et al., 2013) and strain Leaf177, an Arabidopsis leaf isolate (Bai et al., 2015) all represent novel BGC species.
Burkholderia genomes vary in size from 3.75 Mb (B. rhizoxinica HKI 454) to 11.3 Mb (B. terrae BS001), are characterized by a high G+C content (60–68%) and consist of multiple replicons (Winsor et al., 2008; Ussery et al., 2009). The DNA G+C content of the 13 novel species was calculated from their genome sequences and was in the range of that reported for other BGC species (60–65 mol%). For 10 of the 12 established BGC species, the G+C content was previously calculated by traditional wet-lab methods and the reported values differed by 0.1–3.3 mol% from the values calculated from their genome sequences (Table 5). As reported by Meier-Kolthoff et al., the G+C content calculations based on genome sequences show a higher precision than calculations based on traditional wet-lab methods because the latter methods do not count nucleotides but estimate the genomic G+C content based on the physical properties of the extracted and/or digested genomic DNA (Meier-Kolthoff et al., 2014b). The difference between literature data (Lim et al., 2003; Lu et al., 2012; Tian et al., 2013) and the genome sequence-based G+C content values of B. sordidicola LMG 22029T, B. zhejiangensis OP-1T and B. grimmiae R27T is larger than 1% and we therefore present emended descriptions of these species. The genome sizes of the type strains of the 13 novel species ranged from 6.2 Mb (B. concitans sp. nov. LMG 29315T) to 9.7 Mb (B. arvi sp. nov. LMG 29317T) and corresponded with the genome sizes of other free-living BGC species (Table 2). Consistent with reductive genome evolution in obligatory symbionts, the smallest BGC genomes belong to the obligatory leaf endosymbionts (2.4–6.2 Mb; Carlier and Eberl, 2012; Carlier et al., 2015; Pinto-Carbo et al., 2016).
Biochemically, these novel species are similar to their nearest neighbors. However, tests particularly useful for distinguishing BGC species are growth at 37°C and at pH 8, hydrolysis of tween 60 and 80, nitrate reduction, assimilation of arabinose, caprate and citrate, beta-galactosidase activity and C4 lipase (Table 4). The most discriminating fatty acids are C16:0 3-OH, C17:0 cyclo, C19:0 cyclo ω8c and summed features 2 and 3 (Table 3). The overall fatty acid profiles of the novel taxa are similar to those of their nearest neighbors and support their placement in the genus Burkholderia (Yabuuchi et al., 1992).
The present study again underscores the multifaceted nature of Burkholderia bacteria (Coenye and Vandamme, 2003; Mahenthiralingam et al., 2005) and highlights that also BGC species have evolved a broad range of interactions with different hosts. B. cordobensis is a striking example of phenotypic and geographic breadth: it was recovered from agricultural soil in Argentina (strain LMG 27620T) (Draghi et al., 2014), from golf course soil in South Korea (strain YI23) (Lim et al., 2012) and from the gut of the bean bug Riptortus pedestris in Japan (strain RPE67) (Takeshita et al., 2014). The two latter strains (YI23 and RPE67) have fenitrothion degrading properties. The former two strains (LMG 27620T and YI23) were free-living but the latter (RPE67) is an endosymbiont of stink bugs that is not vertically transmitted but acquired from soil by the nymphal insect (Kikuchi et al., 2007). The insecticide resistance to fenitrothion in the pest insects was shown to be established by the endosymbiotic Burkholderia strain in the insect gut (Kikuchi et al., 2012) and was shown to emerge as a consequence of repeated insecticide use (Tago et al., 2015). The Riptortus pedestris-B. cordobensis association thus appears to be a rather young endosymbiosis and contrasts with the symbiosis observed between plant species of the Rubiaceae and Primulaceae families and several Candidatus Burkholderia species. The Candidatus designation is a provisional taxonomic status for organisms that have been characterized but that cannot be cultivated at present (Schleifer, 2009). These obligate leaf endosymbionts are vertically transmitted and represent an obligatory symbiosis which was estimated to originate millions of years ago (Lemaire et al., 2011).
BGC species harbor both beneficial and pathogenic strains. Strains PML1(12) and S170 show biotechnological potential for mineral-weathering and plant growth promotion, respectively, and are exemplary for the metabolic versatility of Burkholderia organisms (Llado et al., 2014; Uroz and Oger, 2015). Mineral-weathering bacteria dissolute key nutrients from minerals and thereby increase the bioavailability of chemical nutrients in the environment (Uroz et al., 2009). On the other hand, three strains analyzed in the present study were isolated from human clinical samples, i.e., blood, pleural fluid and lung tissue (Table 1) and were classified as two novel species (Burkholderia concitans sp. nov. and Burkholderia turbans sp. nov.). They represent, to our knowledge, the first examples of human clinical isolates in the B. glathei clade. Strikingly, strain MR1, which was isolated from Florida golf course soil and shown to reduce the herbicide methylarsenate, was also identified as Burkholderia concitans sp. nov., and this species thus represents yet another human clinical Burkholderia species with interesting biotechnological properties (Coenye et al., 2001; Coenye and Vandamme, 2003; Goris et al., 2004; Mahenthiralingam et al., 2005). This study therefore further underscores that there is no phylogenetic subdivision in the genus Burkholderia that distinguishes beneficial from pathogenic strains (Angus et al., 2014; Sawana et al., 2014; Estrada-de los Santos et al., 2016; Dobritsa and Samadpour, 2016).
In summary, the present study provides genotypic, chemotaxonomic and phenotypic data which enable the differentiation of 13 novel species in the genus Burkholderia and we propose the names Burkholderia arvi sp. nov., Burkholderia hypogeia sp. nov., Burkholderia ptereochthonis sp. nov., Burkholderia glebae sp. nov., Burkholderia pedi sp. nov., Burkholderia arationis sp. nov., Burkholderia fortuita sp. nov., Burkholderia temeraria sp. nov., Burkholderia calidae sp. nov., Burkholderia concitans sp. nov., Burkholderia turbans sp. nov., Burkholderia catudaia sp. nov. and Burkholderia peredens sp. nov., with strains LMG 29317T, LMG 29322T, LMG 29326T, LMG 29325T, LMG 29323T, LMG 29324T, LMG 29320T, LMG 29319T, LMG 29321T, LMG 29315T, LMG 29316T, LMG 29318T, and LMG 29314T as type strains, respectively. By making reference cultures and whole-genome sequences of each of these versatile bacteria publicly available, we aim to contribute to future knowledge about the metabolic versatility and pathogenicity of Burkholderia organisms.
Description of Burkholderia arvi sp. nov.
Burkholderia arvi (ar'vi. L. gen. n. arvi of a field).
Cells are Gram-negative, non-motile rods (less than 1 μm wide and about 1 μm long) with rounded ends that occur as single units or in pairs. After 48 h of incubation on trypticase soy agar at 28°C, colonies are round (typically less than 1 mm in diameter), smooth, shiny, non-translucent, with entire margins and a white-creamy color. Grows on MacConkey agar. Growth occurs at 15–37°C and at pH 6–7 in NB at 28°C. Catalase and oxidase activities are present. Hydrolyses tween 60, but not tween 80, starch and casein. When tested using API 20NE strips, positive for nitrate reduction, beta-galactosidase (PNPG) (weak) and assimilation of glucose, arabinose, mannose, mannitol, N-acetyl-glucosamine, gluconate, malate, citrate (weak), and phenylacetate; negative for production of indol, fermentation of glucose, arginine dihydrolase, urease, esculin hydrolysis, gelatin liquefaction and assimilation of maltose, caprate, and adipate. When tested using API ZYM strips, positive for alkaline phosphatase, leucyl arylamidase, acid phosphatase, and phosphoamidase (weak); negative for C4 lipase, C8 lipase, C14 lipase, valine arylamidase, cystine arylamidase, trypsin, chymotrypsin, alpha-galactosidase, beta-galactosidase, beta-glucuronidase, alpha-glucosidase, beta-glucosidase, N-acetyl-beta-glucosaminidase, alpha-mannosidase, and alpha-fucosidase. The following fatty acids are present: C16:0, C16:0 3-OH, C18:1 ω7c, summed feature 2 (most likely C14:0 3-OH), and summed feature 3 (most likely C16:1 ω7c) in moderate amounts (>5%), and C14:0, C16:0 2-OH, C17:0 cyclo, and C19:0 cyclo ω8c in minor amounts (1–5%).
The type strain is LMG 29317T (=CCUG 68412T) and was isolated from agricultural soil in Argentina in 2010 (Draghi et al., 2014). Its G+C content is 62.4 mol% (calculated based on its genome sequence). The 16S rRNA, gyrB and whole-genome sequence of LMG 29317T are publicly available through the accession numbers LT158615, LT158628, and FCOM02000000, respectively.
Description of Burkholderia hypogeia sp. nov.
Burkholderia hypogeia (hy.po.ge'ia. Gr. adj. hypogeios subterraneous; N. L. fem. adj. hypogeia, subterraneous, earth-born).
Cells are Gram-negative, non-motile rods (about 1 μm wide and 1–2 μm long) with rounded ends that occur as single units or in pairs. After 48 h of incubation on trypticase soy agar at 28°C, colonies are round (typically less than 1 mm in diameter), smooth, shiny, non-translucent, with entire margins and a white-creamy color. Grows on MacConkey agar. Growth occurs at 15–37°C and at pH 6 in NB at 28°C. Catalase and oxidase activities are present. Hydrolyses tween 60, but not tween 80, starch and casein. When tested using API 20NE strips, positive for nitrate reduction and assimilation of glucose, arabinose, mannose, mannitol, N-acetyl-glucosamine, gluconate, malate, and phenylacetate; negative for production of indol, fermentation of glucose, arginine dihydrolase, urease, esculin hydrolysis, gelatin liquefaction, beta-galactosidase (PNPG) and assimilation of maltose, caprate, adipate and citrate. When tested using API ZYM strips, positive for alkaline phosphatase (weak), C4 lipase, C8 lipase (weak), leucyl arylamidase, acid phosphatase and phosphoamidase (weak); negative for C14 lipase, valine arylamidase, cystine arylamidase, trypsin, chymotrypsin, alpha-galactosidase, beta-galactosidase, beta-glucuronidase, alpha-glucosidase, beta-glucosidase, N-acetyl-beta-glucosaminidase, alpha-mannosidase, and alpha-fucosidase. The following fatty acids are present: C16:0, C16:0 3-OH, C17:0 cyclo, C18:1ω7c, summed feature 2 (most likely C14:0 3-OH) and summed feature 3 (most likely C16:1 ω7c) in moderate amounts (>5%), and C14:0, C16:0 2-OH and C19:0 cyclo ω8c in minor amounts (1–5%).
The type strain is LMG 29322T (=CCUG 68407T) and was isolated from greenhouse soil in Belgium in 2014 (Peeters et al., 2016). Its G+C content is 63.2 mol% (calculated based on its genome sequence). The 16S rRNA, gyrB and whole-genome sequence of LMG 29322T are publicly available through the accession numbers LT158620, LT158633, and FCOA02000000, respectively.
Description of Burkholderia ptereochthonis sp. nov.
Burkholderia ptereochthonis (pte.re.o.chtho'nis Gr. n. pteris fern; Gr. n. chthon soil; N. L. gen. n. ptereochthonis, from fern soil).
Cells are Gram-negative, non-motile rods (less than 1 μm wide and about 1 μm long) with rounded ends that occur as single units or in pairs. After 48 h of incubation on trypticase soy agar at 28°C, colonies are round (typically less than 1 mm in diameter), smooth, shiny, non-translucent, with entire margins and a white-creamy color. Grows on MacConkey agar. Growth occurs at 15–37°C and at pH 7 in NB at 28°C. Catalase and oxidase activities are present. Hydrolyses tween 60, but not tween 80, starch and casein. When tested using API 20NE strips, positive for the assimilation of glucose, mannose, mannitol, N-acetyl-glucosamine, gluconate, malate, and phenylacetate; negative for nitrate reduction, production of indol, fermentation of glucose, arginine dihydrolase, urease, esculin hydrolysis, gelatin liquefaction, beta-galactosidase (PNPG) and assimilation of arabinose, maltose, caprate, adipate and citrate. When tested using API ZYM strips, positive for alkaline phosphatase, C4 lipase, leucyl arylamidase, acid phosphatase and phosphoamidase (weak); negative for C8 lipase, C14 lipase, valine arylamidase, cystine arylamidase, trypsin, chymotrypsin, alpha-galactosidase, beta-galactosidase, beta-glucuronidase, alpha-glucosidase, beta-glucosidase, N-acetyl-beta-glucosaminidase, alpha-mannosidase, and alpha-fucosidase. The following fatty acids are present: C16:0, C16:0 3-OH, C17:0 cyclo, C18:1ω7c, summed feature 2 (most likely C14:0 3-OH) and summed feature 3 (most likely C16:1 ω7c) in moderate amounts (>5%), and C14:0, C16:0 2-OH, C16:1 2-OH, and C19:0 cyclo ω8c in minor amounts (1–5%).
The type strain is LMG 29326T (=CCUG 68403T) and was isolated from botanical garden soil in Belgium in 2014 (Peeters et al., 2016). Its G+C content is 64.2 mol% (calculated based on its genome sequence). The 16S rRNA, gyrB and whole-genome sequence of LMG 29326T are publicly available through the accession numbers LT158624, LT158637, and FCOB02000000, respectively.
Description of Burkholderia glebae sp. nov.
Burkholderia glebae (gle'bae. L. gen. n. glebae from a lump or clod of earth, soil).
Cells are Gram-negative, non-motile rods (less than 1 μm wide and about 1 μm long) with rounded ends that occur as single units or in pairs. After 48 h of incubation on trypticase soy agar at 28°C, colonies are round, tiny (typically less than 0.5 mm in diameter), non-translucent, with a white-creamy color. Grows on MacConkey agar. Growth occurs at 15–28°C and at pH 7–8 in NB at 28°C (for the type strain only at pH 7). Catalase and oxidase activities are present. Hydrolyses tween 60, but not tween 80, starch and casein. When tested using API 20NE strips, positive for nitrate reduction and assimilation of glucose, arabinose, mannose, mannitol, N-acetyl-glucosamine, gluconate, malate, citrate, and phenylacetate; negative for production of indol, fermentation of glucose, arginine dihydrolase, urease, esculin hydrolysis, gelatin liquefaction, beta-galactosidase (PNPG) and assimilation of maltose, caprate, and adipate. When tested using API ZYM strips, positive for leucyl arylamidase, acid phosphatase and phosphoamidase; negative for C8 lipase, C14 lipase, valine arylamidase, cystine arylamidase, trypsin, chymotrypsin, alpha-galactosidase, beta-galactosidase, beta-glucuronidase, alpha-glucosidase, beta-glucosidase, N-acetyl-beta-glucosaminidase, alpha-mannosidase, and alpha-fucosidase; strain-dependent reactions for alkaline phosphatase (type strain negative) and C4 lipase (type strain weak). The following fatty acids are present in all isolates: C16:0, C16:0 3-OH, C17:0 cyclo, C18:1ω7c, summed feature 2 (most likely C14:0 3-OH), and summed feature 3 (most likely C16:1ω7c) in moderate amounts (>5%), and C14:0, C16:0 2-OH, C16:1 2-OH, and C19:0 cyclo ω8c in minor amounts (1–5%) (mean value of all isolates).
The type strain is LMG 29325T (=CCUG 68404T) and was isolated from botanical garden soil in Belgium in 2014 (Peeters et al., 2016). Its G+C content is 62.7 mol% (calculated based on its genome sequence). The 16S rRNA, gyrB and whole-genome sequence of LMG 29325T are publicly available through the accession numbers LT158623, LT158636, and FCOJ02000000, respectively. An additional strain has been isolated from soil in the Netherlands (Table 1).
Description of Burkholderia pedi sp. nov.
Burkholderia pedi (pe'di. Gr. n. pedon soil, earth; N. L. gen. n. pedi, from soil).
Cells are Gram-negative, non-motile rods (less than 1 μm wide and 1–2 μm long) with rounded ends that occur as single units or in pairs. After 48 h of incubation on trypticase soy agar at 28°C, colonies are round (typically less than 1 mm in diameter), smooth, shiny, non-translucent, with entire margins and a beige color. Grows on MacConkey agar. Growth occurs at 15–28°C and at pH 6–8 in NB at 28°C (type strain only in pH 6–7). Catalase and oxidase activities are present. Hydrolyses tween 60, but not tween 80, starch and casein. When tested using API 20NE strips, positive for nitrate reduction, beta-galactosidase (PNPG) and assimilation of glucose, arabinose, mannose, mannitol, N-acetyl-glucosamine, gluconate, adipate, malate, and phenylacetate; negative for production of indol, fermentation of glucose, urease, esculin hydrolysis, gelatin liquefaction and assimilation of maltose and citrate; strain-dependent reactions for arginine dihydrolase (type strain negative) and the assimilation of caprate (type strain negative). When tested using API ZYM strips, positive for alkaline phosphatase, leucyl arylamidase, acid phosphatase, and phosphoamidase; negative for C14 lipase, trypsin, chymotrypsin, alpha-galactosidase, beta-glucuronidase, alpha-glucosidase, beta-glucosidase, alpha-mannosidase, and alpha-fucosidase; strain-dependent reactions for C4 lipase (type strain negative), C8 lipase (type strain negative), valine arylamidase (type strain negative), cystine arylamidase (type strain negative), beta-galactosidase (type strain negative), and N-acetyl-beta-glucosaminidase (type strain negative). The following fatty acids are present in all isolates: C16:0, C16:0 3-OH, C17:0 cyclo, C18:1ω7c, summed feature 2 (most likely C14:0 3-OH), and summed feature 3 (most likely C16:1ω7c) in moderate amounts (>5%), and C14:0, C16:0 2-OH, C16:1 2-OH, and C19:0 cyclo ω8c in minor amounts (1–5%) (mean value of all isolates).
The type strain is LMG 29323T (=CCUG 68406T) and was isolated from greenhouse soil in Belgium in 2014 (Peeters et al., 2016). Its G+C content is 63.0 mol% (calculated based on its genome sequence). The 16S rRNA, gyrB, and whole-genome sequence of LMG 29323T are publicly available through the accession numbers LT158621, LT158634, and FCOE02000000, respectively. An additional strain has been isolated from the same sample (Table 1).
Description of Burkholderia arationis sp. nov.
Burkholderia arationis (a.ra.ti.o'nis. L. gen. n. arationis from a field).
Cells are Gram-negative, non-motile rods (less than 1 μm wide and about 1 μm long) with rounded ends that occur as single units or in pairs. After 48 h of incubation on trypticase soy agar at 28°C, colonies are round (typically less than 1 mm in diameter), smooth, shiny, translucent, with entire margins and a white-creamy color. Grows on MacConkey agar. Growth occurs at 15–28°C and at pH 6 in NB at 28°C (the type strain did not grow in liquid NB medium). Catalase and oxidase activities are present. Hydrolyses tween 60, but not tween 80, starch and casein. When tested using API 20NE strips, positive for assimilation of glucose, arabinose, mannose, mannitol, N-acetyl-glucosamine, gluconate, caprate (weak), adipate (weak), malate, citrate, and phenylacetate; negative for nitrate reduction, production of indol, fermentation of glucose, arginine dihydrolase, urease, esculin hydrolysis, gelatin liquefaction, beta-galactosidase (PNPG), and assimilation of maltose. When tested using API ZYM strips, positive for alkaline phosphatase, C4 lipase, leucyl arylamidase, acid phosphatase and phosphoamidase; negative for C14 lipase, cystine arylamidase, trypsin, alpha-galactosidase, beta-galactosidase, beta-glucuronidase, alpha-glucosidase, beta-glucosidase, N-acetyl-beta-glucosaminidase, alpha-mannosidase, and alpha-fucosidase; strain-dependent reactions for C8 lipase (type strain negative), valine arylamidase (type strain negative), and chymotrypsin (type strain negative). The following fatty acids are present in all isolates: C16:0, C16:0 3-OH, C18:1ω7c, summed feature 2 (most likely C14:0 3-OH), and summed feature 3 (most likely C16:1 ω7c) in moderate amounts (>5%), and C14:0 in minor amounts (1-5%) (mean value of all isolates).
The type strain is LMG 29324T (=CCUG 68405T) and was isolated from botanical garden soil in Belgium in 2014 (Peeters et al., 2016). Its G+C content is 62.8 mol% (calculated based on its genome sequence). The 16S rRNA, gyrB, and whole-genome sequence of LMG 29324T are publicly available through the accession numbers LT158622, LT158635, and FCOG02000000, respectively. An additional strain has been isolated from soil in the Netherlands (Table 1).
Description of Burkholderia fortuita sp. nov.
Burkholderia fortuita (for.tu.i'ta. L. fem. adj. fortuita accidental, unpremeditated; referring to its fortuitous isolation when searching for Burkholderia caledonica endophytes).
Cells are Gram-negative, non-motile rods (less than 1 μm wide and about 1 μm long) with rounded ends that occur as single units or in pairs. After 48 h of incubation on trypticase soy agar at 28°C, colonies are round (typically less than 1 mm in diameter), smooth, shiny, non-translucent, with entire margins and a beige color. Grows on MacConkey agar. Growth occurs at 15–37°C and at pH 6–7 in NB at 28°C. Catalase and oxidase activities are present. Hydrolyses tween 60, but not tween 80, starch and casein. When tested using API 20NE strips, positive for the assimilation of glucose, arabinose, mannose, mannitol, N-acetyl-glucosamine, gluconate, malate, and phenylacetate; negative for nitrate reduction, production of indol, fermentation of glucose, arginine dihydrolase, urease, esculin hydrolysis, gelatin liquefaction, beta-galactosidase (PNPG) and assimilation of maltose, caprate, adipate, and citrate. When tested using API ZYM strips, positive for alkaline phosphatase (weak), leucyl arylamidase, acid phosphatase, and phosphoamidase (weak); negative for C4 lipase, C8 lipase, C14 lipase, valine arylamidase, cystine arylamidase, trypsin, chymotrypsin, alpha-galactosidase, beta-galactosidase, beta-glucuronidase, alpha-glucosidase, beta-glucosidase, N-acetyl-beta-glucosaminidase, alpha-mannosidase, and alpha-fucosidase. The following fatty acids are present: C16:0, C16:0 3-OH, C17:0 cyclo, C18:1ω7c, summed feature 2 (most likely C14:0 3-OH), and summed feature 3 (most likely C16:1 ω7c) in moderate amounts (>5%), and C14:0, C16:0 2-OH, C16:1 2-OH, and C19:0 cyclo ω8c in minor amounts (1–5%).
The type strain is LMG 29320T (=CCUG 68409T) and was isolated from Fadogia homblei rhizosphere soil in South Africa in 2013 (Verstraete et al., 2014). Its G+C content is 62.9 mol% (calculated based on its genome sequence). The 16S rRNA, gyrB and whole-genome sequence of LMG 29320T are publicly available through the accession numbers LT158618, LT158631, and FCNX02000000, respectively.
Description of Burkholderia temeraria sp. nov.
Burkholderia temeraria (te.me.ra'ri.a. L. fem. adj. temeraria accidental, inconsiderate; referring to its accidental isolation when searching for Burkholderia caledonica endophytes).
Cells are Gram-negative, non-motile rods (less than 1 μm wide and about 1 μm long) with rounded ends that occur as single units or in pairs. After 48 h of incubation on trypticase soy agar at 28°C, colonies are round (typically less than 1 mm in diameter), smooth, shiny, non-translucent, with entire margins and a white-creamy color. Grows on MacConkey agar. Growth occurs at 15–37°C and at pH 6–7 in NB at 28°C. Catalase and oxidase activities are present. Does not hydrolyze tween 60, tween 80, starch and casein. When tested using API 20NE strips, positive for the assimilation of glucose, arabinose, mannose, mannitol, N-acetyl-glucosamine, gluconate, malate, citrate (weak), and phenylacetate; negative for nitrate reduction, production of indol, fermentation of glucose, arginine dihydrolase, urease, esculin hydrolysis, gelatin liquefaction, beta-galactosidase (PNPG) and assimilation of maltose, caprate, and adipate. When tested using API ZYM strips, positive for alkaline phosphatase, C4 lipase, leucyl arylamidase, acid phosphatase, and phosphoamidase (weak); negative for C8 lipase, C14 lipase, valine arylamidase, cystine arylamidase, trypsin, chymotrypsin, alpha-galactosidase, beta-galactosidase, beta-glucuronidase, alpha-glucosidase, beta-glucosidase, N-acetyl-beta-glucosaminidase, alpha-mannosidase, and alpha-fucosidase. The following fatty acids are present: C16:0, C16:0 3-OH, C17:0 cyclo, C18:1ω7c, summed feature 2 (most likely C14:0 3-OH) and summed feature 3 (most likely C16:1 ω7c) in moderate amounts (>5%), and C14:0, C16:0 2-OH, and C19:0 cyclo ω8c in minor amounts (1–5%).
The type strain is LMG 29319T (=CCUG 68410T) and was isolated from Fadogia homblei rhizosphere soil in South Africa in 2013 (Verstraete et al., 2014). Its G+C content is 62.7 mol% (calculated based on its genome sequence). The 16S rRNA, gyrB and whole-genome sequence of LMG 29319T are publicly available through the accession numbers LT158617, LT158630, and FCOI02000000, respectively.
Description of Burkholderia calidae sp. nov.
Burkholderia calidae (ca'li.dae. L. gen. n. calidae from warm water, because this strain was isolated from pond water in a tropical garden).
Cells are Gram-negative, non-motile rods (about 1 μm wide and 1 μm long) with rounded ends that occur as single units or in pairs. After 48 h of incubation on trypticase soy agar at 28°C, colonies are round (typically about 1 mm in diameter), smooth, shiny, non-translucent, with entire margins and a white-creamy color. Grows on MacConkey agar. Growth occurs at 15–37°C and at pH 6–7 in NB at 28°C. Catalase and oxidase activities are present. Does not hydrolyze tween 60, tween 80, starch and casein. When tested using API 20NE strips, positive for nitrate reduction and assimilation of glucose, arabinose, mannose, mannitol, N-acetyl-glucosamine, gluconate, caprate, malate, citrate (weak), and phenylacetate; negative for production of indol, fermentation of glucose, arginine dihydrolase, urease, esculin hydrolysis, gelatin liquefaction, beta-galactosidase (PNPG) and assimilation of maltose and adipate. When tested using API ZYM strips, positive for alkaline phosphatase (weak), C8 lipase (weak), leucyl arylamidase (weak), acid phosphatase and phosphoamidase (weak); negative for C4 lipase, C14 lipase, valine arylamidase, cystine arylamidase, trypsin, chymotrypsin, alpha-galactosidase, beta-galactosidase, beta-glucuronidase, alpha-glucosidase, beta-glucosidase, N-acetyl-beta-glucosaminidase, alpha-mannosidase, and alpha-fucosidase. The following fatty acids are present: C16:0, C18:1ω7c, summed feature 2 (most likely C14:0 3-OH) and summed feature 3 (most likely C16:1 ω7c) in moderate amounts (>5%), and C14:0, C16:0 2-OH, C16:0 3-OH, and C17:0 cyclo in minor amounts (1–5%).
The type strain is LMG 29321T (=CCUG 68408T) and was isolated from greenhouse pond water in Belgium in 2013 (Peeters et al., 2016). Its G+C content is 62.5 mol% (calculated based on its genome sequence). The 16S rRNA, gyrB and whole-genome sequence of LMG 29321T are publicly available through the accession numbers LT158619, LT158632, and FCOX02000000, respectively.
Description of Burkholderia concitans sp. nov.
Burkholderia concitans (con.ci'tans. L. fem. part. pres. concitans disturbing, upsetting; because the isolation of this bacterium from human sources, including blood, further disturbs the image of this lineage of Burkholderia species as benign bacteria).
Cells are Gram-negative, non-motile rods (less than 1 μm wide and about 1 μm long) with rounded ends that occur as single units or in pairs. After 48 h of incubation on trypticase soy agar at 28°C, colonies are round (typically less than 1 mm in diameter), smooth, shiny, non-translucent, with entire margins and a white-creamy color. Grows on MacConkey agar. Growth occurs at 15–28°C (additionally, the type strains grows at 37°C) and at pH 6–7 in NB at 28°C. Catalase and oxidase activities are present. Hydrolyses tween 60, but not tween 80, starch and casein. When tested using API 20NE strips, positive for the assimilation of glucose, arabinose, mannose, mannitol, N-acetyl-glucosamine, gluconate, malate, and phenylacetate; negative for nitrate reduction, production of indol, fermentation of glucose, arginine dihydrolase, urease, esculin hydrolysis, gelatin liquefaction, beta-galactosidase (PNPG) and assimilation of maltose, caprate, and adipate; strain-dependent reactions for the assimilation of citrate (type strain weak). When tested using API ZYM strips, positive for alkaline phosphatase, C4 lipase, C8 lipase (weak), leucyl arylamidase, valine arylamidase, acid phosphatase, and phosphoamidase; negative for C14 lipase, trypsin, chymotrypsin, alpha-galactosidase, beta-galactosidase, beta-glucuronidase, alpha-glucosidase, beta-glucosidase, N-acetyl-beta-glucosaminidase, alpha-mannosidase, and alpha-fucosidase; strain-dependent reactions for cystine arylamidase (type strain negative). The following fatty acids are present in all isolates: C16:0, C16:0 3-OH, C17:0 cyclo, C18:1ω7c, C19:0 cyclo ω8c, summed feature 2 (most likely C14:0 3-OH) and summed feature 3 (most likely C16:1 ω7c) in moderate amounts (>5%), and C14:0, C16:0 2-OH, and C16:1 2-OH in minor amounts (1–5%) (mean value of all isolates).
The type strain is LMG 29315T (=CCUG 68414T) and was isolated from human lung tissue in the USA in 2006. Its G+C content is 63.2 mol%. The 16S rRNA, gyrB, and whole-genome sequence of LMG 29315T are publicly available through the accession numbers LT158613, LT158626 and FCNV02000000, respectively. An additional strain has been isolated from human blood in the USA in 2010 (Table 1).
Description of Burkholderia turbans sp. nov.
Burkholderia turbans (tur'bans. L. fem. part. pres. turbans disturbing, agitating, because the isolation of this bacterium from human pleural fluid further disturbs the image of this lineage of Burkholderia species as benign bacteria).
Cells are Gram-negative, non-motile rods (about 1 μm wide and 1–1.5 μm long) with rounded ends that occur as single units or in pairs. After 48 h of incubation on trypticase soy agar at 28°C, colonies are round (typically less than 1 mm in diameter), smooth, shiny, non-translucent, with entire margins and a white-creamy color. Grows on MacConkey agar. Growth occurs at 15–37°C and at pH 6–7 in NB at 28°C. Catalase and oxidase activities are present. Hydrolyses tween 60, but not tween 80, starch and casein. When tested using API 20NE strips, positive for the assimilation of glucose, arabinose, mannose, mannitol, N-acetyl-glucosamine, gluconate, caprate, malate, and phenylacetate; negative for nitrate reduction, production of indol, fermentation of glucose, arginine dihydrolase, urease, esculin hydrolysis, gelatin liquefaction, beta-galactosidase (PNPG) and assimilation of maltose, adipate and citrate. When tested using API ZYM strips, positive for alkaline phosphatase, C4 lipase (weak), leucyl arylamidase, acid phosphatase, and phosphoamidase (weak); negative for C8 lipase, C14 lipase, valine arylamidase, cystine arylamidase, trypsin, chymotrypsin, alpha-galactosidase, beta-galactosidase, beta-glucuronidase, alpha-glucosidase, beta-glucosidase, N-acetyl-beta-glucosaminidase, alpha-mannosidase, and alpha-fucosidase. The following fatty acids are present: C16:0, C17:0 cyclo, C18:1ω7c, summed feature 2 (most likely C14:0 3-OH) and summed feature 3 (most likely C16:1 ω7c) in moderate amounts (>5%), and C14:0, C16:0 2-OH, C16:0 3-OH, C16:1 2-OH, and C19:0 cyclo ω8c in minor amounts (1–5%).
The type strain is LMG 29316T (=CCUG 68413T) and was isolated from human pleural fluid in the USA in 2006. Its G+C content is 63.1 mol% (calculated based on its genome sequence). The 16S rRNA, gyrB and whole-genome sequence of LMG 29316T are publicly available through the accession numbers LT158614, LT158627, and FCOD02000000, respectively.
Description of Burkholderia catudaia sp. nov.
Burkholderia catudaia (ca.tu.da'ia. Gr. adj. catudaios subterraneous; N. L. fem. adj. catudaia, earth-born).
Cells are Gram-negative, non-motile rods (about 1 μm wide and 1–2 μm long) with rounded ends that occur as single units or in pairs. After 48 h of incubation on trypticase soy agar at 28°C, colonies are round (typically less than 1 mm in diameter), smooth, shiny, non-translucent, with entire margins and a white-creamy color. Grows on MacConkey agar. Growth occurs at 15–37°C and at pH 6–7 in NB at 28°C. Catalase and oxidase activities are present. Hydrolyses tween 60, but not tween 80, starch and casein. When tested using API 20NE strips, positive for nitrate reduction and assimilation of glucose, arabinose, mannose, mannitol, N-acetyl-glucosamine, gluconate, malate, and phenylacetate; negative for production of indol, fermentation of glucose, arginine dihydrolase, urease, esculin hydrolysis, gelatin liquefaction, beta-galactosidase (PNPG) and assimilation of maltose, caprate, adipate, and citrate. When tested using API ZYM strips, positive for alkaline phosphatase (weak), leucyl arylamidase, acid phosphatase, and phosphoamidase (weak); negative for C4 lipase, C8 lipase, C14 lipase, valine arylamidase, cystine arylamidase, trypsin, chymotrypsin, alpha-galactosidase, beta-galactosidase, beta-glucuronidase, alpha-glucosidase, beta-glucosidase, N-acetyl-beta-glucosaminidase, alpha-mannosidase, and alpha-fucosidase. The following fatty acids are present: C16:0, C16:0 3-OH, C18:1ω7c, summed feature 2 (most likely C14:0 3-OH) and summed feature 3 (most likely C16:1 ω7c) in moderate amounts (>5%), and C14:0, C16:0 2-OH, C17:0 cyclo, and C19:0 cyclo ω8c in minor amounts (1–5%).
The type strain is LMG 29318T (=CCUG 68411T) and was isolated from Fadogia homblei rhizosphere soil in South Africa in 2013 (Verstraete et al., 2014). Its G+C content is 62.8 mol% (calculated based on its genome sequence). The 16S rRNA, gyrB and whole-genome sequence of LMG 29318T are publicly available through the accession numbers LT158616, LT158629, and FCOF02000000, respectively.
Description of Burkholderia peredens sp. nov.
Burkholderia peredens (per.e'dens. L. fem. part. pres. peredens consuming, devouring; referring to the capacity of this bacterium to degrade fenitrothion).
Cells are Gram-negative, non-motile rods (about 1 μm wide and 1–2 μm long) with rounded ends that occur as single units or in pairs. After 48 h of incubation on trypticase soy agar at 28°C, colonies are round (typically less than 1 mm in diameter), smooth, shiny, non-translucent, with entire margins and a white-creamy color. Grows on MacConkey agar. Growth occurs at 15–37°C and at pH 7 in NB at 28°C. Catalase and oxidase activities are present. Hydrolyses tween 60, but not tween 80, starch and casein. When tested using API 20NE strips, positive for the assimilation of glucose, arabinose (weak), mannose, mannitol, N-acetyl-glucosamine, gluconate, malate, and phenylacetate; negative for nitrate reduction, production of indol, fermentation of glucose, arginine dihydrolase, urease, esculin hydrolysis, gelatin liquefaction, beta-galactosidase (PNPG) and assimilation of maltose, caprate, adipate, and citrate. When tested using API ZYM strips, positive for alkaline phosphatase, C4 lipase (weak), C8 lipase (weak), leucyl arylamidase, acid phosphatase, and phosphoamidase (weak); negative for C14 lipase, valine arylamidase, cystine arylamidase, trypsin, chymotrypsin, alpha-galactosidase, beta-galactosidase, beta-glucuronidase, alpha-glucosidase, beta-glucosidase, N-acetyl-beta-glucosaminidase, alpha-mannosidase, and alpha-fucosidase. The following fatty acids are present: C16:0, C16:0 3-OH, C18:1ω7c, summed feature 2 (most likely C14:0 3-OH) and summed feature 3 (most likely C16:1 ω7c) in moderate amounts (>5%), and C14:0, C16:0 2-OH, C16:1 2-OH and C17:0 cyclo in minor amounts (1–5%).
The type strain is LMG 29314T (=CCUG 68415T) and was isolated from soil in Japan (Hayatsu et al., 2000). Its G+C content is 63.1 mol% (calculated based on its genome sequence). The 16S rRNA, gyrB and whole-genome sequence of LMG 29314T are publicly available through the accession numbers LT158612, LT158625, and FCOH02000000, respectively.
Emended Description of the Species Burkholderia sordidicola (Lim et al., 2003)
The description of the species Burkholderia sordidicola is the one given by Lim et al. (2003) with the following modification. The G+C content of the type strain is 60.2%.
Emended Description of the Species Burkholderia zhejiangensis (Lu et al., 2012)
The description of the species Burkholderia zhejiangensis is the one given by Lu et al. (2012) with the following modification. The G+C content of the type strain is 62.7%.
Emended Description of the Species Burkholderia grimmiae (Tian et al., 2013)
The description of the species Burkholderia grimmiae is the one given by Tian et al. (2013) with the following modification. The G+C content of the type strain is 63.0%.
Author Contributions
CP carried out the genomic data analysis and drafted the manuscript. JM performed all GBDP-related analyses. BV participated in the ortholog analysis and whole-genome based phylogeny. ED performed the DNA extractions, fatty acid analysis, and biochemical characterization. VC directed the genomic sequencing methods and initial analysis. PV conceived of the study, participated in the design and coordination and helped writing the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
CP is indebted to the Special Research Council of Ghent University. We thank Marcus Dillon for constructing sequencing libraries, Aurélien Carlier for his advice on genome analysis and all strain depositors listed in Table 1 to make this study possible.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00877
Supplementary Table1. Pairwaise GGD and dDDH values. For each pair of genomes the intergenomic distance (GGD), dDDH and 95% confidence interval (CI) is given, sorted from high to low dDDH. dDDH values above the 70% cut-off for species delineation are shown in bold type.
Abbreviations
BGC, Burkholderia glathei clade; GGDC, Genome-to-Genome Distance Calculator; GBDP, Genome Blast Distance Phylogeny; dDDH, digital DNA-DNA hybridization; MLSA, multilocus sequence analysis.
References
Angus, A. A., Agapakis, C. M., Fong, S., Yerrapragada, S., Estrada-de los Santos, P., Yang, P., et al. (2014). Plant-associated symbiotic Burkholderia species lack hallmark strategies required in mammalian pathogenesis. PLoS ONE 9:e83779. doi: 10.1371/journal.pone.0083779
Baek, I., Seo, B., Lee, I., Lee, K., Park, S.-C., Yi, H., et al. (2015). Burkholderia megalochromosomata sp. nov., isolated from grassland soil. Int. J. Syst. Evol. Microbiol. 65, 959–964. doi: 10.1099/ijs.0.000046
Bai, Y., Muller, D. B., Srinivas, G., Garrido-Oter, R., Potthoff, E., Rott, M., et al. (2015). Functional overlap of the Arabidopsis leaf and root microbiota. Nature 528, 364–369. doi: 10.1038/nature16192
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Baym, M., Kryazhimskiy, S., Lieberman, T. D., Chung, H., Desai, M. M., and Kishony, R. (2015). Inexpensive multiplexed library preparation for megabase-sized genomes. PLoS ONE 10:e128036. doi: 10.1371/journal.pone.0128036
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Carlier, A., Fehr, L., Pinto, M., Schaberle, T., Reher, R., Dessein, S., et al. (2015). The genome analysis of Candidatus Burkholderia crenata reveals that secondary metabolism may be a key function of the Ardisia crenata leaf nodule symbiosis. Environ. Microbiol. doi: 10.1111/1462-2920.13184. [Epub ahead of print].
Carlier, A. L., and Eberl, L. (2012). The eroded genome of a Psychotria leaf symbiont: hypotheses about lifestyle and interactions with its plant host. Environ. Microbiol. 14, 2757–2769. doi: 10.1111/j.1462-2920.2012.02763.x
Coenye, T., Laevens, S., Willems, A., Ohlen, M., Hannant, W., Govan, J. R. W., et al. (2001). Burkholderia fungorum sp. nov. and Burkholderia caledonica sp. nov., two new species isolated from the environment, animals and human clinical samples. Int. J. Syst. Evol. Microbiol. 51, 1099–1107. doi: 10.1099/00207713-51-3-1099
Coenye, T., and Vandamme, P. (2003). Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ. Microbiol. 5, 719–729. doi: 10.1046/j.1462-2920.2003.00471.x
Colston, S. M., Fullmer, M. S., Beka, L., Lamy, B., Gogarten, J. P., and Graf, J. (2014). Bioinformatic genome comparisons for taxonomic and phylogenetic assignments using Aeromonas as a test case. MBio 5, e02136–e02114. doi: 10.1128/mBio.02136-14
Compant, S., Nowak, J., Coenye, T., Clement, C., and Ait Barka, E. (2008). Diversity and occurrence of Burkholderia spp. in the natural environment. FEMS Microbiol. Rev. 32, 607–626. doi: 10.1111/j.1574-6976.2008.00113.x
Deloger, M., El Karoui, M., and Petit, M.-A. (2009). A genomic distance based on MUM indicates discontinuity between most bacterial species and genera. J. Bacteriol. 191, 91–99. doi: 10.1128/JB.01202-08
Dobritsa, A. P., and Samadpour, M. (2016). Transfer of eleven Burkholderia species to the genus Paraburkholderia and proposal of Caballeronia gen. nov., a new genus to accommodate twelve species of Burkholderia and Paraburkholderia. Int. J. Syst. Evol. Microbiol. doi: 10.1099/ijsem.0.001065. [Epub ahead of print].
Draghi, W. O., Peeters, C., Cnockaert, M., Snauwaert, C., Wall, L. G., Zorreguieta, A., et al. (2014). Burkholderia cordobensis sp. nov., from agricultural soils. Int. J. Syst. Evol. Microbiol. 64, 2003–2008. doi: 10.1099/ijs.0.059667-0
Edgar, R. C. (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113
Estrada-de los Santos, P., Rojas-Rojas, F. U., Tapia-Garcia, E. Y., Vasquez-Murrieta, M. S., and Hirsch, A. M. (2016). To split or not to split: an opinion on dividing the genus Burkholderia. Ann. Microbiol. doi: 10.1007/s13213-015-1183-1. [Epub ahead of print].
Euzeby, J. P. (1997). List of bacterial names with standing in nomenclature: a folder available on the internet. Int. J. Syst. Evol. Microbiol. 47, 590–592. doi: 10.1099/00207713-47-2-590
Fischer, S., Brunk, B. P., Chen, F., Gao, X., Harb, O. S., Iodice, J. B., et al. (2011). “Using OrthoMCL to assign proteins to OrthoMCL-DB groups or to cluster proteomes into new ortholog groups,” in Current Protocols in Bioinformatics (Hoboken, NJ: John Wiley & Sons, Inc.). 6.12.1–6.12.19.
Goris, J., De Vos, P., Caballero-Mellado, J., Park, J., Falsen, E., Quensen, J. F., et al. (2004). Classification of the biphenyl- and polychlorinated biphenyl-degrading strain LB400T and relatives as Burkholderia xenovorans sp. nov. Int. J. Syst. Evol. Microbiol. 54, 1677–1681. doi: 10.1099/ijs.0.63101-0
Goris, J., Konstantinidis, K. T., Klappenbach, J. A., Coenye, T., Vandamme, P., and Tiedje, J. M. (2007). DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91. doi: 10.1099/ijs.0.64483-0
Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Hayatsu, M., Hirano, M., and Tokuda, S. (2000). Involvement of two plasmids in fenitrothion degradation by Burkholderia sp. strain NF100. Appl. Environ. Microbiol. 66, 1737–1740. doi: 10.1128/AEM.66.4.1737-1740.2000
Hernandez-Mendoza, A., Martinez-Ocampo, F., Lozano-Aguirre Beltran, L. F., Popoca-Ursino, E. C., Ortiz-Hernandez, L., Sanchez-Salinas, E., et al. (2014). Draft genome qequence of the organophosphorus compound-degrading Burkholderia zhejiangensis strain CEIB S4-3. Genome Announc. 2:e01323–14. doi: 10.1128/genomeA.01323-14
Kikuchi, Y., Hayatsu, M., Hosokawa, T., Nagayama, A., Tago, K., and Fukatsu, T. (2012). Symbiont-mediated insecticide resistance. Proc. Natl. Acad. Sci. U.S.A. 109, 8618–8622. doi: 10.1073/pnas.1200231109
Kikuchi, Y., Hosokawa, T., and Fukatsu, T. (2007). Insect-microbe mutualism without vertical transmission: a stinkbug acquires a beneficial gut symbiont from the environment every generation. Appl. Environ. Microbiol. 73, 4308–4316. doi: 10.1128/AEM.00067-07
Kikuchi, Y., Hosokawa, T., and Fukatsu, T. (2011). An ancient but promiscuous host–symbiont association between Burkholderia gut symbionts and their heteropteran hosts. ISME J. 5, 446–460. doi: 10.1038/ismej.2010.150
Konstantinidis, K. T., and Tiedje, J. M. (2005). Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. U.S.A. 102, 2567–2572. doi: 10.1073/pnas.0409727102
Kumar, S., Vikram, S., and Raghava, G. P. S. (2012). Genome sequence of the nitroaromatic compound-degrading bacterium Burkholderia sp. strain SJ98. J. Bacteriol. 194, 3286–3286. doi: 10.1128/JB.00497-12
Lefort, V., Desper, R., and Gascuel, O. (2015). FastME 2.0: a comprehensive, accurate, and fast distance-based phylogeny inference program. Mol. Biol. Evol. 32, 2798–2800. doi: 10.1093/molbev/msv150
Lemaire, B., Vandamme, P., Merckx, V., Smets, E., and Dessein, S. (2011). Bacterial leaf symbiosis in angiosperms: host specificity without co-speciation. PLoS ONE 6:e24430. doi: 10.1371/journal.pone.0024430
Lemaire, B., Van Oevelen, S., De Block, P., Verstraete, B., Smets, E., Prinsen, E., et al. (2012). Identification of the bacterial endosymbionts in leaf nodules of Pavetta (Rubiaceae). Int. J. Syst. Evol. Microbiol. 62, 202–209. doi: 10.1099/ijs.0.028019-0
Letunic, I., and Bork, P. (2011). Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 39, W475–W478. doi: 10.1093/nar/gkr201
Li, H. (2013). Aligning Sequence Reads, Clone Sequences and Assembly Contigs with BWA-MEM. arXiv. Available online at: http://arxiv.org/abs/1303.3997
Lim, J. S., Choi, B. S., Choi, A. Y., Kim, K. D., Kim, D. I., Choi, I. Y., et al. (2012). Complete genome sequence of the fenitrothion-degrading Burkholderia sp. strain YI23. J. Bacteriol. 194, 896–896. doi: 10.1128/JB.06479-11
Lim, Y. W., Baik, K. S., Han, S. K., Kim, S. B., and Bae, K. S. (2003). Burkholderia sordidicola sp. nov., isolated from the white-rot fungus Phanerochaete sordida. Int. J. Syst. Evol. Microbiol. 53, 1631–1636. doi: 10.1099/ijs.0.02456-0
Liu, X.-Y., Li, C. L., Luo, X.-J., Lai, Q.-L., and Xu, J.-H. (2014). Burkholderia jiangsuensis sp. nov., a methyl parathion degrading bacterium, isolated from methyl parathion contaminated soil. Int. J. Syst. Evol. Microbiol. 64, 3247–3253. doi: 10.1099/ijs.0.064444-0
Liu, Y., Lai, Q., Goker, M., Meier-Kolthoff, J. P., Wang, M., Sun, Y., et al. (2015). Genomic insights into the taxonomic status of the Bacillus cereus group. Sci. Rep. 5, 14082. doi: 10.1038/srep14082
Llado, S., Xu, Z., Sorensen, S. J., and Baldrian, P. (2014). Draft genome sequence of Burkholderia sordidicola S170, a potential plant growth promoter isolated from coniferous forest soil in the Czech Republic. Genome Announc. 2:e00810–14. doi: 10.1128/genomeA.00810-14
Lu, P., Zheng, L.-Q., Sun, J.-J., Liu, H.-M., Li, S.-P., Hong, Q., et al. (2012). Burkholderia zhejiangensis sp. nov., a methyl-parathion-degrading bacterium isolated from a wastewater-treatment system. Int. J. Syst. Evol. Microbiol. 62, 1337–1341. doi: 10.1099/ijs.0.035428-0
Mahenthiralingam, E., Baldwin, A., and Dowson, C. G. (2008). Burkholderia cepacia complex bacteria: opportunistic pathogens with important natural biology. J. Appl. Microbiol. 104, 1539–1551. doi: 10.1111/j.1365-2672.2007.03706.x
Mahenthiralingam, E., Urban, T. A., and Goldberg, J. B. (2005). The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev. Microbiol. 3, 144–156. doi: 10.1038/nrmicro1085
Marolda, C. L., Hauroder, B., John, M. A., Michel, R., and Valvano, M. A. (1999). Intracellular survival and saprophytic growth of isolates from the Burkholderia cepacia complex in free-living amoebae. Microbiology 145, 1509–1517.
Meier-Kolthoff, J. P., Auch, A. F., Klenk, H.-P., and Goker, M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. doi: 10.1186/1471-2105-14-60
Meier-Kolthoff, J. P., Auch, A. F., Klenk, H.-P., and Goker, M. (2014a). Highly parallelized inference of large genome-based phylogenies. Concurr. Comput. Pract. Exp. 26, 1715–1729. doi: 10.1002/cpe.3112
Meier-Kolthoff, J. P., Klenk, H.-P., and Goker, M. (2014b). Taxonomic use of DNA G+C content and DNA–DNA hybridization in the genomic age. Int. J. Syst. Evol. Microbiol. 64, 352–356. doi: 10.1099/ijs.0.056994-0
Nogales, B., Moore, E. R. B., Llobet-Brossa, E., Rossello-Mora, R., Amann, R., and Timmis, K. N. (2001). Combined use of 16S ribosomal DNA and 16S rRNA to study the bacterial community of polychlorinated biphenyl-polluted soil. Appl. Environ. Microbiol. 67, 1874–1884. doi: 10.1128/AEM.67.4.1874-1884.2001
Parke, J. L., and Gurian-Sherman, D. (2001). Diversity of the Burkholderia cepacia complex and implications for risk assessment of biological control strains. Annu. Rev. Phytopathol. 39, 225–258. doi: 10.1146/annurev.phyto.39.1.225
Pawitwar, S. S., Utturkar, S. M., Brown, S. D., Yoshinaga, M., and Rosen, B. P. (2015). Draft genome sequence of Burkholderia sp. MR1, a methylarsenate-reducing bacterial isolate from Florida golf course soil. Genome Announc. 3:e00608–15. doi: 10.1128/genomeA.00608-15
Peeters, C., Depoorter, E., Praet, J., and Vandamme, P. (2016). Extensive cultivation of soil and water samples yields various pathogens in patients with cystic fibrosis but not Burkholderia multivorans. J. Cyst. Fibros. doi: 10.1016/j.jcf.2016.02.014. [Epub ahead of print].
Peeters, C., Zlosnik, J. E. A., Spilker, T., Hird, T. J., LiPuma, J. J., and Vandamme, P. (2013). Burkholderia pseudomultivorans sp. nov., a novel Burkholderia cepacia complex species from human respiratory samples and the rhizosphere. Syst. Appl. Microbiol. 36, 483–489. doi: 10.1016/j.syapm.2013.06.003
Pinto-Carbo, M., Sieber, S., Dessein, S., Wicker, T., Verstraete, B., Gademann, K., et al. (2016). Evidence of horizontal gene transfer between obligate leaf nodule symbionts. ISME J. doi: 10.1038/ismej.2016.27. [Epub ahead of print].
Pitcher, D. G., Saunders, N. A., and Owen, R. J. (1989). Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 8, 151–156.
Pumphrey, G. M., and Madsen, E. L. (2008). Field-Based stable isotope probing reveals the identities of benzoic acid-metabolizing microorganisms and their in situ growth in agricultural soil. Appl. Environ. Microbiol. 74, 4111–4118. doi: 10.1128/AEM.00464-08
Rossello-Mora, R., and Amann, R. (2015). Past and future species definitions for Bacteria and Archaea. Syst. Appl. Microbiol. 38, 209–216. doi: 10.1016/j.syapm.2015.02.001
Salles, J. F., Samyn, E., Vandamme, P., van Veen, J. A., and van Elsas, J. D. (2006). Changes in agricultural management drive the diversity of Burkholderia species isolated from soil on PCAT medium. Soil Biol. Biochem. 38, 661–673. doi: 10.1016/j.soilbio.2005.06.018
Sawana, A., Adeolu, M., and Gupta, R. S. (2014). Molecular signatures and phylogenomic analysis of the genus Burkholderia: proposal for division of this genus into the emended genus Burkholderia containing pathogenic organisms and a new genus Paraburkholderia gen. nov. harboring environmental species. Front. Genet. 5:e429. doi: 10.3389/fgene.2014.00429
Schleifer, K. H. (2009). Classification of Bacteria and Archaea: past, present and future. Syst. Appl. Microbiol. 32, 533–542. doi: 10.1016/j.syapm.2009.09.002
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Seo, Y.-S., Lim, J., Choi, B.-S., Kim, H., Goo, E., Lee, B., et al. (2011). Complete genome sequence of Burkholderia gladioli BSR3. J. Bacteriol. 193, 3149–3149. doi: 10.1128/JB.00420-11
Shibata, T. F., Maeda, T., Nikoh, N., Yamaguchi, K., Oshima, K., Hattori, M., et al. (2013). Complete GENOME SEQUENCE of Burkholderia sp. STRAIN RPE64, bacterial symbiont of the bean bug Riptortus pedestris. Genome Announc. 1:e00441–13. doi: 10.1128/genomeA.00441-13
Spilker, T., Baldwin, A., Bumford, A., Dowson, C. G., Mahenthiralingam, E., and LiPuma, J. J. (2009). Expanded multilocus sequence typing for Burkholderia species. J. Clin. Microbiol. 47, 2607–2610. doi: 10.1128/JCM.00770-09
Stackebrandt, E., Frederiksen, W., Garrity, G. M., Grimont, P. A. D., Kampfer, P., Maiden, M. C. J., et al. (2002). Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 52, 1043–1047. doi: 10.1099/ijs.0.02360-0
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stopnisek, N., Zuhlke, D., Carlier, A., Barberan, A., Fierer, N., Becher, D., et al. (2016). Molecular mechanisms underlying the close association between soil Burkholderia and fungi. ISME J. 10, 253–264. doi: 10.1038/ismej.2015.73
Sutcliffe, I. C., Trujillo, M. E., Whitman, W. B., and Goodfellow, M. (2013). A call to action for the international committee on systematics of prokaryotes. Trends Microbiol. 21, 51–52. doi: 10.1016/j.tim.2012.11.004
Tago, K., Kikuchi, Y., Nakaoka, S., Katsuyama, C., and Hayatsu, M. (2015). Insecticide applications to soil contribute to the development of Burkholderia mediating insecticide resistance in stinkbugs. Mol. Ecol. 24, 3766–3778. doi: 10.1111/mec.13265
Takeshita, K., Shibata, T. F., Nikoh, N., Nishiyama, T., Hasebe, M., Fukatsu, T., et al. (2014). Whole-genome sequence of Burkholderia sp. strain RPE67, a bacterial gut symbiont of the bean bug Riptortus pedestris. Genome Announc. 2:e00556–14. doi: 10.1128/genomeA.00556-14
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tayeb, L. A., Lefevre, M., Passet, V., Diancourt, L., Brisse, S., and Grimont, P. A. D. (2008). Comparative phylogenies of Burkholderia, Ralstonia, Comamonas, Brevundimonas and related organisms derived from rpoB, gyrB and rrs gene sequences. Res. Microbiol. 159, 169–177. doi: 10.1016/j.resmic.2007.12.005
Thompson, C. C., Amaral, G. R., Campeao, M., Edwards, R. A., Polz, M. F., Dutilh, B. E., et al. (2015). Microbial taxonomy in the post-genomic era: rebuilding from scratch? Arch. Microbiol. 197, 359–370. doi: 10.1007/s00203-014-1071-2
Tian, Y., Kong, B. H., Liu, S. L., Li, C. L., Yu, R., Liu, L., et al. (2013). Burkholderia grimmiae sp. nov., isolated from a xerophilous moss (Grimmia montana). Int. J. Syst. Evol. Microbiol. 63, 2108–2113. doi: 10.1099/ijs.0.045492-0
Uroz, S., Calvaruso, C., Turpault, M.-P., and Frey-Klett, P. (2009). Mineral weathering by bacteria: ecology, actors and mechanisms. Trends Microbiol. 17, 378–387. doi: 10.1016/j.tim.2009.05.004
Uroz, S., and Oger, P. (2015). Draft genome sequence of Burkholderia sp. strain PML1(12), an ectomycorrhizosphere-inhabiting bacterium with effective mineral-weathering ability. Genome Announc. 3:e00798–15. doi: 10.1128/genomeA.00798-15
Ussery, D. W., Kiil, K., Lagesen, K., Sicheritz-Ponten, T., Bohlin, J., and Wassenaar, T. M. (2009). The genus Burkholderia: analysis of 56 genomic sequences. Microb. Pathog. 6, 140–157. doi: 10.1159/000235768
Van Borm, S., Buschinger, A., Boomsma, J. J., and Billen, J. (2002). Tetraponera ants have gut symbionts related to nitrogen-fixing root-nodule bacteria. Proc. R. Soc. B 269, 2023–2027. doi: 10.1098/rspb.2002.2101
Vandamme, P., De Brandt, E., Houf, K., Salles, J. F., van Elsas, J. D., Spilker, T., et al. (2013). Burkholderia humi sp. nov., Burkholderia choica sp. nov., Burkholderia telluris sp. nov., Burkholderia terrestris sp. nov. and Burkholderia udeis sp. nov.: Burkholderia glathei-like bacteria from soil and rhizosphere soil. Int. J. Syst. Evol. Microbiol. 63, 4707–4718. doi: 10.1099/ijs.0.048900-0
Vandamme, P., and Peeters, C. (2014). Time to revisit polyphasic taxonomy. Antonie Van Leeuwenhoek 106, 57–65. doi: 10.1007/s10482-014-0148-x
Vandamme, P., Pot, B., Gillis, M., De Vos, P., Kersters, K., and Swings, J. (1996). Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol. Rev. 60, 407–438.
Vandamme, P., Verheyde, B., Peeters, C., and Dawyndt, P. (2014). “Genomic taxonomy and biodiversity of the Burkholderia cepacia complex,” in Burkholderia: From Genomes to Function, eds. T. Coenye and E. Mahenthiralingam (Norfolk, UK: Caister Academic Press), 15–29.
Vanlaere, E., Baldwin, A., Gevers, D., Henry, D., De Brandt, E., LiPuma, J. J., et al. (2009). Taxon K, a complex within the Burkholderia cepacia complex, comprises at least two novel species, Burkholderia contaminans sp. nov. and Burkholderia lata sp. nov. Int. J. Syst. Evol. Microbiol. 59, 102–111. doi: 10.1099/ijs.0.001123-0
Van Oevelen, S., De Wachter, R., Vandamme, P., Robbrecht, E., and Prinsen, E. (2004). “Candidatus Burkholderia calva” and “Candidatus Burkholderia nigropunctata” as leaf gall endosymbionts of African Psychotria. Int. J. Syst. Evol. Microbiol. 54, 2237–2239. doi: 10.1099/ijs.0.63188-0
Verstraete, B., Janssens, S., Smets, E., and Dessein, S. (2013). Symbiotic ß-Proteobacteria beyond Legumes: Burkholderia in Rubiaceae. PLoS ONE 8:e55260. doi: 10.1371/journal.pone.0055260
Verstraete, B., Peeters, C., van Wyk, B., Smets, E., Dessein, S., and Vandamme, P. (2014). Intraspecific variation in Burkholderia caledonica: Europe vs. Africa and soil vs. endophytic isolates. Syst. Appl. Microbiol. 37, 194–199. doi: 10.1016/j.syapm.2013.12.001
Verstraete, B., Van Elst, D., Steyn, H., Van Wyk, B., Lemaire, B., Smets, E., et al. (2011). Endophytic bacteria in toxic South African plants: identification, phylogeny and possible involvement in Gousiekte. PLoS ONE 6:e19265. doi: 10.1371/journal.pone.0019265.g001
Vial, L., Chapalain, A., Groleau, M.-C., and Deziel, E. (2011). The various lifestyles of the Burkholderia cepacia complex species: a tribute to adaptation. Environ. Microbiol. 13, 1–12. doi: 10.1111/j.1462-2920.2010.02343.x
Viallard, V., Poirier, I., Cournoyer, B., Haurat, J., Wiebkin, S., Ophel-Keller, K., et al. (1998). Burkholderia graminis sp. nov., a rhizospheric Burkholderia species, and reassessment of [Pseudomonas] phenazinium, [Pseudomonas] pyrrocinia and [Pseudomonas] glathei as Burkholderia. Int. J. Syst. Bacteriol. 48, 549–563.
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 9:e112963. doi: 10.1371/journal.pone.0112963
Wayne, L. G., Brenner, D. J., Colwell, R. R., Grimont, P. A. D., Kandler, O., Krichevsky, M. I., et al. (1987). Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37, 463–464.
Whitman, W. B. (2015). Genome sequences as the type material for taxonomic descriptions of prokaryotes. Syst. Appl. Microbiol. 38, 217–222. doi: 10.1016/j.syapm.2015.02.003
Winsor, G. L., Khaira, B., Van Rossum, T., Lo, R., Whiteside, M. D., and Brinkman, F. S. L. (2008). The Burkholderia Genome Database: facilitating flexible queries and comparative analyses. Bioinformatics 24, 2803–2804. doi: 10.1093/bioinformatics/btn524
Xu, Y., Buss, E. A., and Boucias, D. G. (2016). Culturing and characterization of the gut symbiont Burkholderia from the Southern chinch bug, Blissus insularis (Hemiptera: Blissidae). Appl. Environ. Microbiol. 82, 3319–3330. doi: 10.1128/AEM.00367-16
Yabuuchi, E., Kosako, Y., Oyaizu, H., Yano, I., Hotta, H., Hashimoto, Y., et al. (1992). Proposal of Burkholderia gen. nov. and transfer of seven species of the genus Pseudomonas homology group II to the new genus, with the type species Burkholderia cepacia (Palleroni and Holmes 1981) comb. nov. Microbiol. Immunol. 36, 1251–1275.
Keywords: Burkholderia, genomic taxonomy, GBDP, GGDC, MLSA, phylogenomics
Citation: Peeters C, Meier-Kolthoff JP, Verheyde B, De Brandt E, Cooper VS and Vandamme P (2016) Phylogenomic Study of Burkholderia glathei-like Organisms, Proposal of 13 Novel Burkholderia Species and Emended Descriptions of Burkholderia sordidicola, Burkholderia zhejiangensis, and Burkholderia grimmiae. Front. Microbiol. 7:877. doi: 10.3389/fmicb.2016.00877
Received: 01 April 2016; Accepted: 24 May 2016;
Published: 08 June 2016.
Edited by:
Martha E. Trujillo, Universidad de Salamanca, SpainReviewed by:
Baojun Wu, Wayne State University, USAPaulina Estrada De Los Santos, Instituto Politecnico Nacional, Mexico
Copyright © 2016 Peeters, Meier-Kolthoff, Verheyde, De Brandt, Cooper and Vandamme. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peter Vandamme, peter.vandamme@ugent.be
 Charlotte Peeters
Charlotte Peeters Jan P. Meier-Kolthoff
Jan P. Meier-Kolthoff Bart Verheyde1
Bart Verheyde1 Vaughn S. Cooper
Vaughn S. Cooper Peter Vandamme
Peter Vandamme