- 1Soil and Water Science Department, University of Florida–Institute of Food and Agricultural Sciences, Genetics Institute, Gainesville, FL, USA
- 2Smithsonian Marine Station, Fort Pierce, FL, USA
Dark Spot Syndrome (DSS) is one of the most common diseases of boulder corals in the Caribbean. It presents as sunken brown lesions in coral tissue, which can spread quickly over coral colonies. With this study, we tested the hypothesis that similar to other coral diseases, DSS is a dysbiosis characterized by global shifts in the coral microbiome. Because Black Band Disease (BBD) was sometimes found following DSS lesions, we also tested the hypothesis that DSS is a precursor of BBD. To track disease initiation and progression 24 coral colonies were tagged. Of them five Orbicella annularis corals and three O. faveolata corals exhibited DSS lesions at tagging. Microbiota of lesions and apparently healthy tissues from DSS-affected corals over the course of 18 months were collected. Final visual assessment showed that five of eight corals incurred substantial tissue loss while two corals remained stable and one appeared to recover from DSS lesions. Illumina sequencing of the V6 region of bacterial 16S rRNA genes demonstrated no significant differences in bacterial community composition associated with healthy tissue or DSS lesions. The epimicrobiomes of both healthy tissue and DSS lesions contained high relative abundances of Operational Taxonomic Units assigned to Halomonas, an unclassified gammaproteobacterial genus, Moritella, an unclassified Rhodobacteraceae genus, Renibacterium, Pseudomonas, and Acinetobacter. The relative abundance of bacterial taxa was not significantly different between samples when grouped by tissue type (healthy tissue vs. DSS lesion), coral species, collection month, or the overall outcome of DSS-affected corals (substantial tissue loss vs. stable/recovered). Two of the tagged corals with substantial tissue loss also developed BBD during the 18-month sampling period. The bacterial community of the BBD layer was distinct from both healthy tissue and DSS lesions, with high relative abundances of the presumed BBD pathogen Roseofilum reptotaenium and an unclassified Bacteroidales genus, similar to previous results. Roseofilum was detected in all samples from this study, with the highest relative abundance in healthy tissue from DSS-affected corals sampled in August, suggesting that while DSS is not a precursor to BBD, DSS-affected corals are in a weakened state and therefore more susceptible to additional infections.
Introduction
It is now clear that many, if not all, animals and plants depend on their microbiota for nutrient acquisition and/or assimilation, responses to environmental stressors, resistance to pathogens, and overall health (Bosch and McFall-Ngai, 2011). Destabilization of the coral epimicrobiota or its invasion by opportunistic pathogens has been linked to a number of coral diseases (Bourne et al., 2009). Despite several decades of coral disease research and the description of over a dozen coral diseases and/or syndromes, pathogens associated with these anomalies have been assigned to less than half a dozen coral diseases (Rosenberg and Ben-Haim, 2002; Lesser et al., 2007; Bourne et al., 2009). In the absence of the causative agent, some coral diseases can be classified as dysbioses, characterized by global changes in the composition, structure, and function of the microbiome. The loss of Oceanospirillales and other abundant members of the commensal microbiota has been linked to the appearance of coral disease symptoms (Cardenas et al., 2012; Vezzulli et al., 2013; Meyer et al., 2014), however, it is unclear whether the dysbiosis is the cause or consequence of the host’s degraded health state.
Dark Spot Syndrome (DSS), also known as “Dark Spot Disease”, is the most prevalent coral pathology in the Caribbean, especially on Siderastrea siderea corals. Field studies report that up to 3% of S. siderea colonies in relatively pristine reefs in the Bahamas and up to 80% of corals in more impacted reefs have been affected by this disease (Cervino et al., 2001; Borger, 2003; Gochfeld et al., 2006; Voss and Richardson, 2006; Porter et al., 2011). In some locations, DSS also affects other boulder corals, such as Orbicella (formerly Montastraea) annularis and Stephanocoenia michelenii, impacting between 3 and 46% of coral colonies (Cervino et al., 2001; Porter et al., 2011). DSS is characterized by a brown, sunken appearance of the tissue, which has been observed spreading over coral colonies at a rate of 3–4 cm/month (Cervino et al., 2001). A significant (13–56%) decrease in the number of intracellular algal symbionts (Symbiodinium spp.) and their cell division rates have been reported in DSS-affected Siderastrea and Stephanocoenia coral tissue (Cervino et al., 2001). Previous work has also shown that the diversity of Symbiodinium spp. is lower in DSS-affected polyps than in asymptomatic corals and that certain Symbiodiunium genotypes are less abundant in the disease samples (Correa et al., 2009). Despite the high prevalence of DSS in Caribbean corals, total colony mortality due to DSS was generally low; however, impacted colonies experienced up to 50% net tissue loss (Porter et al., 2011).
Multiple studies demonstrate the impact of DSS on coral reef ecosystems throughout the Caribbean, yet the causes, transmission potential, and the role of environmental factors on the appearance of lesions have been the subject of much debate. DSS is typically not observed at great depths; however, corals of the most susceptible species are also more rare at these depths (Gil-Agudelo and Garzon-Ferreira, 2001). Previous work has shown that the prevalence and cover of DSS in Siderastrea corals is correlated with summer months when surface sea temperatures are highest (Gil-Agudelo and Garzon-Ferreira, 2001; Borger, 2005; Gochfeld et al., 2006), but similar patterns were not detected in DSS-affected Montastraea corals (Gil-Agudelo and Garzon-Ferreira, 2001). While lower DSS prevalence was reported in more pristine ecosystems (Voss and Richardson, 2006) compared to sites more heavily impacted by human activities (Cervino et al., 2001; Borger, 2003; Voss and Richardson, 2006; Porter et al., 2011), lesioned corals were not distributed along a gradient of human impact (Gil-Agudelo and Garzon-Ferreira, 2001). Nutrient enrichment also did not increase prevalence of DSS (Gochfeld et al., 2006). Corals exhibiting DSS lesions have been observed both in a clumped fashion (potentially indicative of a colony-to-colony transmission) and in a randomly distributed fashion (Gil-Agudelo and Garzon-Ferreira, 2001; Voss and Richardson, 2006). In a common garden experiment, three out of five colonies displayed signs of the syndrome after 5 months when placed on a reef where ∼50% of the colonies were diseased (Gochfeld et al., 2006). It is not clear, however, whether these colonies developed DSS due to transmission from nearby colonies or because they experienced the same environmental stress, which led to the observed disease symptoms. In controlled aquarium studies no transmission of the disease was observed over 11 days when water-borne or direct contact transmission modes were tested (Randall et al., 2016).
Characterization of DSS lesions using clone libraries revealed differences in the microbial community composition compared to healthy corals and identified Photobacterium, two Vibrio species (including V. campbellii), Corynebacterium, Acinetobacter, Parvularculaceae, and Oscillatoria as well as a terrestrial phytopathogenic fungus Rhytisma acernum as potential culprits of the disease (Sweet et al., 2013). However, Illumina sequencing and PhyloChip microrray analyses did not reveal significant differences in the microbial community composition of DSS lesions and healthy coral epimicrobiomes (Kellogg et al., 2014; Randall et al., 2016). Nevertheless, PhyloChip microarrays detected higher abundance of the cyanobacterium Pseudoscillatoria corallii (now classified as Roseofilum reptotaenium (Casamatta et al., 2012)) in dark spot lesions than in healthy tissues sampled in multiple locations. However, these differences were not statistically significant (Kellogg et al., 2014).
Because R. reptotaenium has been linked to Black Band Disease (BBD) (Rasoulouniriana et al., 2009; Casamatta et al., 2012; Meyer et al., 2016), this observation suggests that the same pathogen may be responsible for both types of the disease signs. In fact, BBD lesions were sometimes observed surrounded by DSS, and approximately 10% of colonies with DSS later developed BBD (Borger, 2005). Therefore, with this study we aimed to test the overall hypothesis that the DSS is a precursor of BBD and that the same pathogen is responsible for the appearance of both lesion types.
Materials and Methods
Sample Collection
Orbicella annularis and O. faveolata corals with signs of DSS were located and tagged at Carrie Bow Cay, Belize while scuba diving at depths up to 20 m in February 2013. This study was conducted in parallel with the characterization of coral microbiome transitions leading to BBD in the same ecosystem over the same period (Meyer et al., 2016). Twenty-four coral colonies were tagged and followed for ∼31 months, approximately 1/3 of the colonies were asymptomatic, 1/3 had DSS and 1/3 had BBD. Of them, eight colonies had dark spot lesions during the 31-mo observation period. Samples of DSS lesions and surface mucus from healthy tissue on lesioned corals were collected by aspiration with needle-less sterile syringes that were uncapped immediately prior to the sample collection as in (Ritchie, 2006). Tagged corals were re-sampled in July 2013 and August 2014, when possible (Table 1). Most tagged corals were photographed at the time of sample collections as well as in September 2015. In the shore-side laboratory, samples were spun in a microfuge at 4,000 rpm for 5 min, seawater was decanted, and the pelleted mucus was preserved with approximately 5–10 volumes of RNAlater (Qiagen, Germantown, MD, USA) and stored at -20°C until extraction of nucleic acids. Genomic DNA was extracted with a PowerSoil DNA isolation kit according to the manufacturer’s instructions (MoBio, Carlsbad, CA, USA).
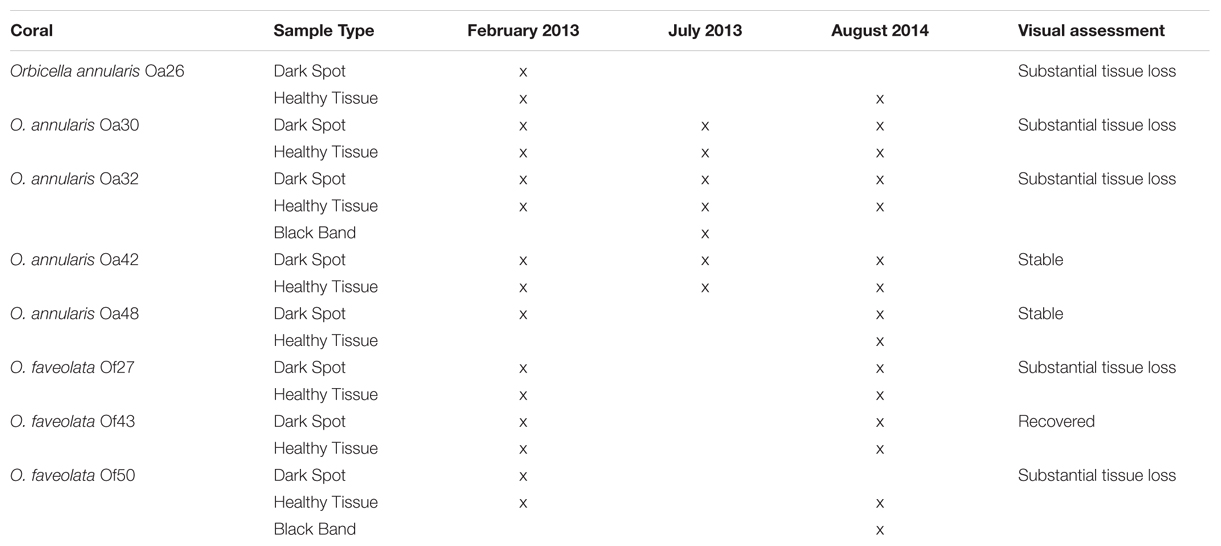
TABLE 1. Summary of coral surface microbiota sampled and the overall health status of the coral colony in August 2014.
PCR Detection of the Rhytisma Fungus
We developed a PCR assay to detect the presence of the fungus Rhytisma that was previously identified in DSS lesions from Caribbean corals (Sweet et al., 2013). The newly developed primer, Rhy_ITS_1F (5′-CCGATTCCACCCTTGATG-3′) was used in conjunction with the previously published primer, ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) (White et al., 1990) to specifically amplify partial ribosomal RNA genes and internal transcribed regions of the coral-associated Rhytisma (GenBank Accession KC521543). M. Sweet provided samples of DNA extracted from DSS lesions from the published study (Sweet et al., 2013) to be used as positive controls for the assay. Samples were amplified in 30 μl reactions containing 1.7U Taq DNA Polymerase with 1X Standard Taq Buffer (New England Biolabs, Ipswich, MA, USA), 0.75 μl DMSO, 0.2 mM each dNTP, 0.5 μM primers, and approximately 10 ng of DNA template. The reaction conditions were an initial denaturation at 94°C for 5 min, followed by 35 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 45 s, and a final elongation at 72°C for 5 min.
16S Illumina Tag Sequencing
The V6 region of bacterial 16S rRNA genes was amplified from whole community DNA in triplicate for each sample with previously published primers (Eren et al., 2013). Samples were amplified in 25 μl reactions containing 0.5 Units Phusion High-Fidelty Polymerase (New England Biolabs, Ipswich, MA, USA), 1X Phusion HF Reaction Buffer, 0.75 μl DMSO, and 0.2 mM each dNTP. Triplicate PCR amplifications were pooled for each sample, cleaned with a MinElute kit (Qiagen, Germantown, MD, USA), visualized on an ethidium bromide stained 1% agarose gel, and quantified by NanoDrop (ThermoScientific, NanoDrop Products, Wilmington, DE, USA). Two hundred nanograms of each cleaned amplicon library was submitted to the Genomics Core Facility at Pennsylvania State University where the pool libraries were size selected with a 2% agarose PippinPrep cassette to produce a narrow range of fragment sizes from 200 to 240 bp for sequencing (confirmed by bioanalyzer) and cleaned again to remove agarose. Sequencing was performed on an Illumina MiSeq with a 150-bp paired-end protocol, using single indexing.
Sequencing reads were parsed by Illumina index at the sequencing center. Reads were then further parsed by the inline barcode, paired reads merged, and primers and adaptors removed using a combination of tools in cutadapt (Martin, 2011), Galaxy (Giardine et al., 2005; Blankenberg et al., 2010; Goecks et al., 2010) and eautils (Aronesty, 2011). Parsed, quality-filtered sequencing reads are publicly available through NCBI’s Sequence Read Archive (SRA) under the BioProject ID PRJNA308473. Sample names were added to the definition lines of sequencing reads using sed and concatenated into one fasta file, to make them compatible for analysis in QIIME v1.8 (Caporaso et al., 2010). Two BBD samples included here were part of a previously published study (Meyer et al., 2016), but were re-analyzed in this study, beginning with OTU clustering, as the samples were from coral colonies also displaying DSS. Clustering of Operational Taxonomic Units (OTUs) at 97% similarity was performed with the subsampled open-reference OTU picking method (Rideout et al., 2014), with no removal of singletons. The Greengenes reference dataset version 13.8 (DeSantis et al., 2006) was used as the reference for OTU picking and for taxonomy assignment with uclust (Edgar, 2010). OTUs identified as mitochondrial DNA or as chloroplasts were removed from further analyses. Community structure was analyzed in R with phyloseq (McMurdie and Holmes, 2013) and plotted with ggplot2 (Wickham, 2009). Analysis of similarities (ANOSIM) was performed in R using VEGAN v2.0–8 (Dixon, 2003). Differences in taxonomic profiles were analyzed by Welch’s t-test (for two groups) or by ANOVA (for multiple groups) with Tukey–Kramer post hoc tests and Benjamini-Hochberg False Discovery Rate, q-value filter > 0.05 with STAMP (Parks et al., 2014).
Results
Overview of Bacterial Community Structure in Healthy and Diseased Corals
Seventeen samples of Orbicella DSS lesions and eighteen samples of healthy tissue on corals with DSS were collected. Between February 2013 and September 2015, substantial tissue loss was observed in five of the eight corals, two corals remained relatively unchanged, and one coral appeared to have recovered from Orbicella DSS lesions (Table 1; Figure 1). Two of the corals that displayed substantial tissue loss also developed BBD during the sampling period. Rhytisma ITS sequences were amplifiable in the positive controls, but were not amplifiable from any of the samples collected for the current study.
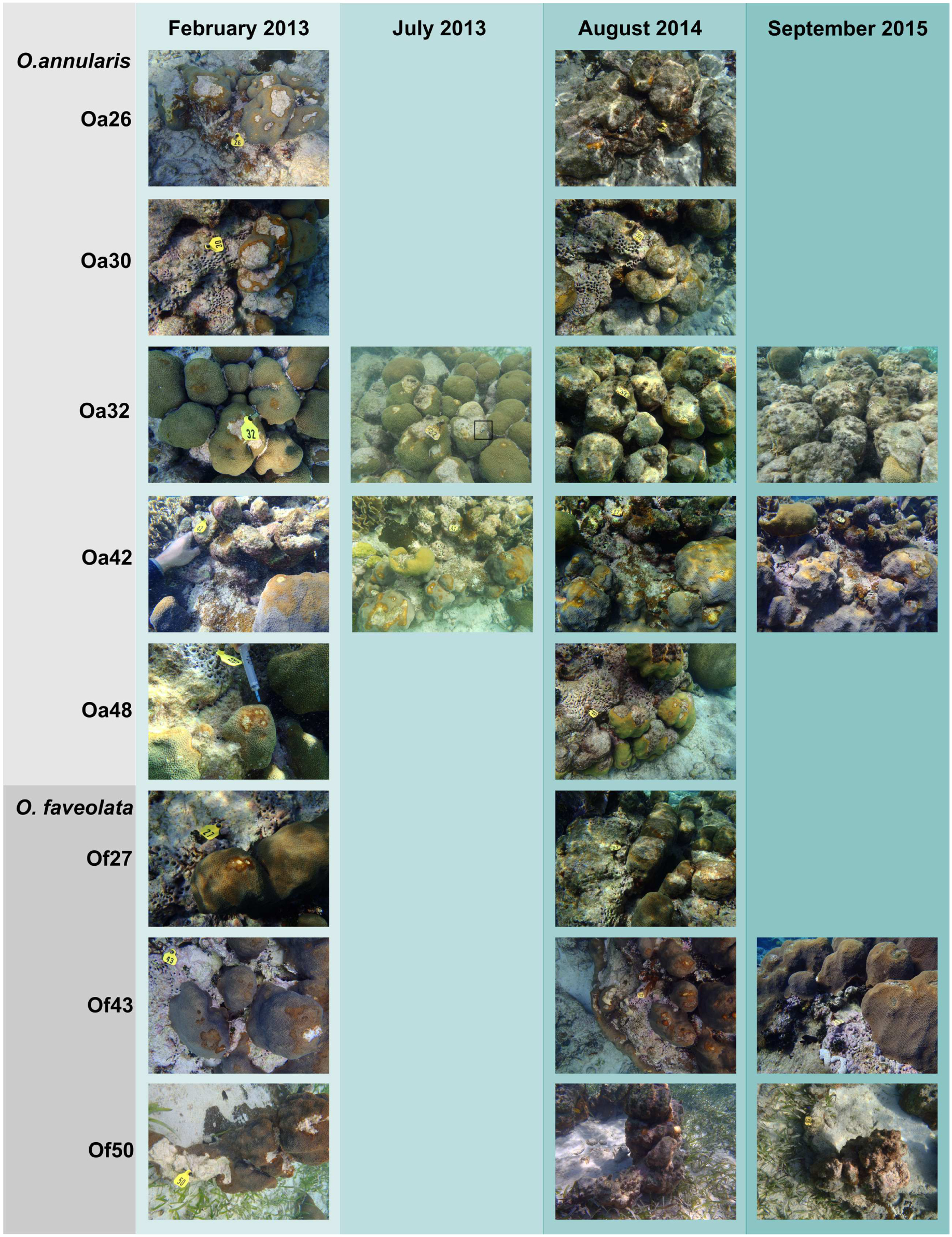
FIGURE 1. Photographic time series of Orbicella corals affected by Dark Spot Syndrome (DSS). Photographs from February 2013 to September 2015 of five Orbicella annularis and three O. faveolata corals at Carrie Bow Cay, Belize. Coral names beginning with Oa are from O. annularis and those beginning with Of are from O. faveolata. Corals Oa26, Oa30, Oa32, Of27, and Of50 exhibited substantial tissue loss, corals Oa42 and Oa48 remained stable, and coral Of43 appeared to recover from DSS over the observational period. Coral Oa32 had Black Band Disease (BBD) in July 2013, indicated by a black box. Coral Of50 had BBD in August 2014, on the lower right side of the coral colony (not visible in the photograph).
To characterize bacterial communities, 10,852,271 quality-filtered sequencing reads were analyzed, with 6,576 to 1,029,276 sequencing reads per sample (SRA BioProject ID PRJNA308473). A total of 36,878 OTUs were detected in the 37 surface microbiota samples. The bacterial communities associated with the two BBD samples were similar to each other and distinct from all other samples (Figure 2A). In contrast, the bacterial community composition was not significantly different between the healthy tissues of DSS-affected corals and DSS lesions (ANOSIM R = 0.016, p = 0.217), and no clustering of bacterial communities was observed for either the health condition or the date of sample collection (Figure 2A). Excluding the two BBD samples, no significant differences were detected in the abundance of bacterial orders, families, or genera when samples were grouped by health condition (Orbicella DSS lesion or healthy tissue), sample date, or the outcome of the infections (substantial tissue loss vs. stable/recovered) (ANOVA with Benjamini–Hochberg False Discovery Rate, q-value filter >0.05). No clustering of bacterial communities was observed based on coral species (Figure 2B). However, some clustering was observed for a few individual coral colonies such as O. faveolata corals Of27 and Of43 (Figure 2B), which reflects the stability of the microbiota associated with these two coral colonies. In addition, four bacterial orders were significantly more abundant in some coral colonies (ANOVA with Benjamini-Hochberg False Discovery Rate, q-value filter >0.05, effect size >0.6), including the betaproteobacterial order Procabacteriales and three gammaproteobacterial orders: Aeromonadales, Pseudomonadales, and an unclassified gammaproteobacterial order discussed in further detail below. O. annularis coral Oa26 and O. faveolata coral Of43 clustered away from the other six coral colonies, due to their higher relative abundance of these four orders (Figure 3).
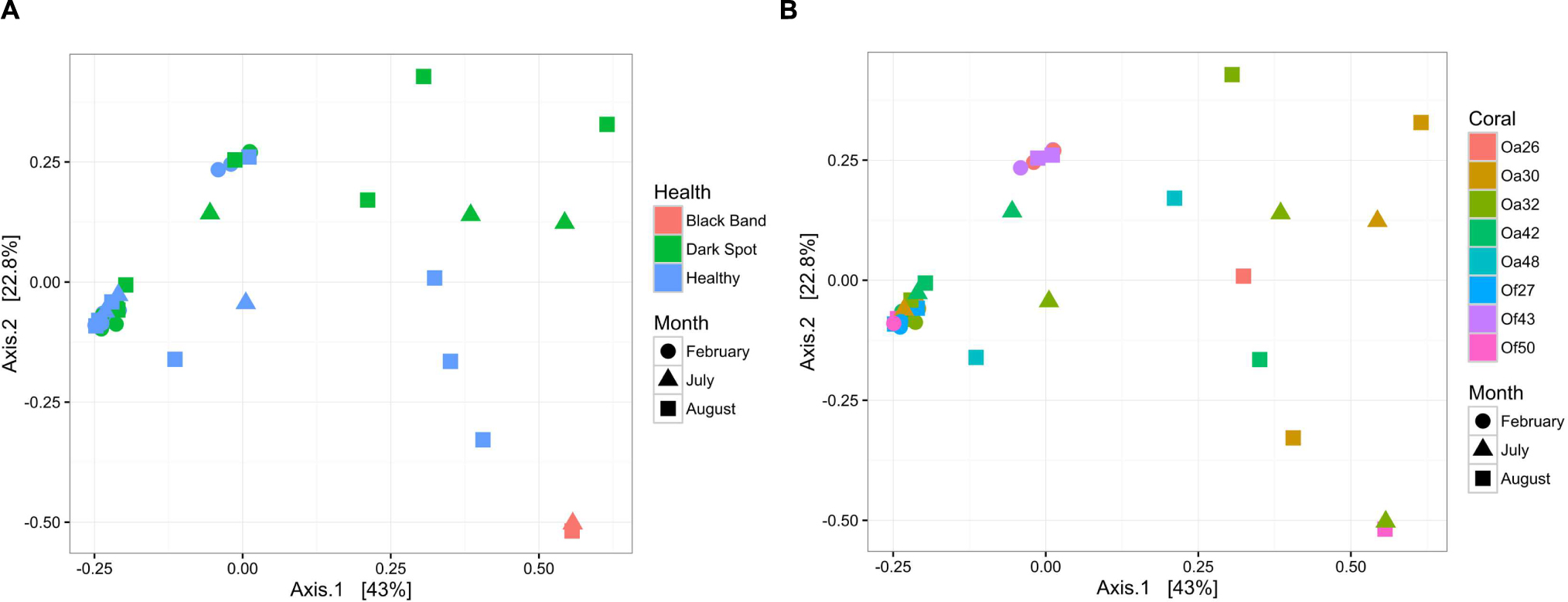
FIGURE 2. Bacterial community structure in healthy and diseased coral surface microbiota. Non-metric multidimensional scaling plot of bacterial community similarity based on Morisita–Horn beta diversity of Illumina MiSeq 16S rRNA gene libraries in the surface mucus layer of corals with DSS. Both healthy tissue and Dark Spot lesions were sampled for each affected coral, with up to three time points sampled (February 2013, July 2013, August 2014). Two of the tagged corals also had BBD at one sampling time. (A) The health state of each sample is indicated by color and the collection date is indicated by shape. (B) The coral colony (Oa for Orbicella annularis and Of for O. faveolata) is indicated by color and the collection date is indicated by shape.
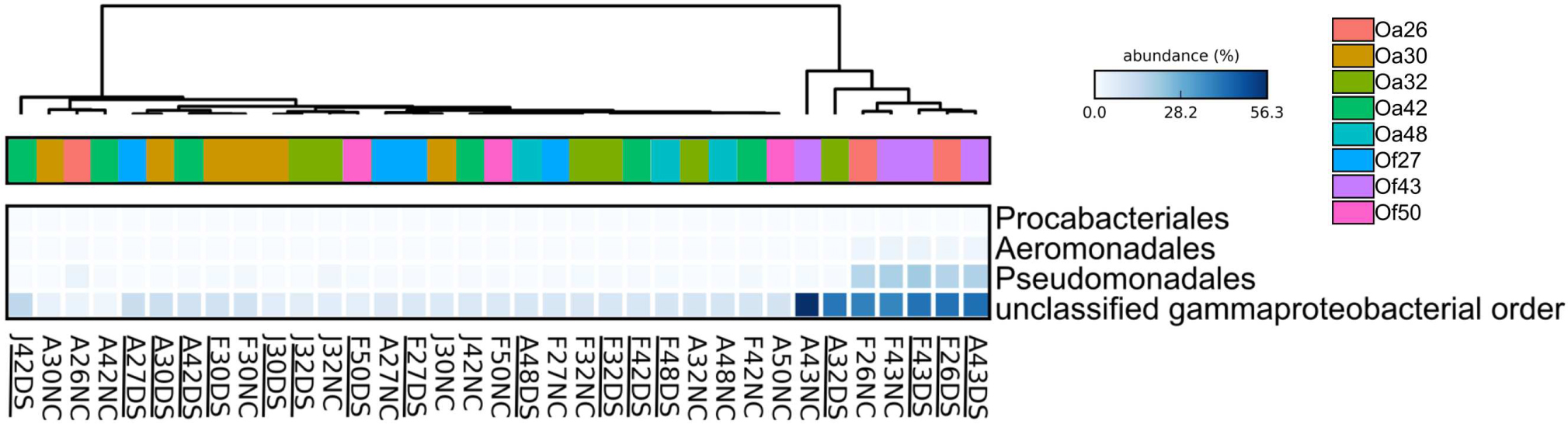
FIGURE 3. Heatmap showing the relative abundance of four bacterial orders that varied across Orbicella coral colonies. Coral names beginning with Oa are from O. annularis and those beginning with Of are from O. faveolata. O. annularis coral Oa26 and O. faveolata coral Of43 clustered away from the other six coral colonies, due to their higher relative abundance of these four orders. Sample names begin with the first letter of the collection month (F, February 2013, J, July 2013, A, August 2014), followed by the tag number, and end with health status (DS, Dark Spot, NC, healthy tissue). Underlined sample names are the samples of Dark Spot lesions. The intensity of the blue color represents abundance of the indicated bacterial orders.
Bacteria Associated with Healthy Tissue, Dark Spot, and Black Band Lesions
The most abundant bacterial genera detected in the healthy and DSS samples were Halomonas, an unclassified gammaproteobacterial genus, Moritella, an unclassified Rhodobacteraceae genus, Renibacterium, Pseudomonas, and Acinetobacter (Figure 4). The ten most abundant OTUs were assigned to these seven genera, as well as to Roseofilum and an unclassified Cytophagales genus (Figure 4). The unclassified gammaproteobacterial genus was represented primarily by a single OTU that was detected in every sample, and the relative abundance of this OTU was not significantly different between healthy tissue and Orbicella DSS lesions or between different sample collection dates. A blastn search of this OTU sequence resulted in equally good hits (100% query coverage with 100% sequence identity) to Vibrio, Serratia, Photobacterium, and Pantoea sequences, indicating that the 60-bp V6 region was not able to resolve the taxonomy of this particular OTU. A second OTU assigned to the unclassified gammaproteobacterial genus was also among the ten most abundant OTUs and had a similarly unresolvable taxonomy with perfect matches to Serratia, Shewanella, and Photobacterium. Rhodobacteraceae appeared to be more abundant in healthy tissues sampled in August 2014, but a two-way ANOVA detected no significant difference in the relative abundance of Rhodobacteraceae related to health and/or sample month. The two BBD consortium samples on DSS-affected corals had high proportions of Roseofilum, unclassified genera of Rhodobacteraceae and Bacteroidales, Fusibacter, and Desulfovibrio. In agreement with the BBD study (Meyer et al., 2016) monitored concurrently with this project, each of the BBD consortium members was detected at lower levels in healthy tissues, as well as in DSS lesions. The relative abundance of Roseofilum was not significantly different between healthy tissues and DSS lesions (Welch’s t-test, p > 0.05). While the two BBD samples were dominated by R. reptotaenium (26 and 36% relative abundance), the highest relative abundance of Roseofilum in non-BBD samples was detected in two healthy tissue samples, both collected in August 2014 from O. annularis corals Oa30 (16%) and Oa48 (9%) (Figure 4). Coral Oa30 experienced substantial tissue loss over the study period, but the lesions on Coral Oa48 appeared to remain stable over the study period (Figure 1).
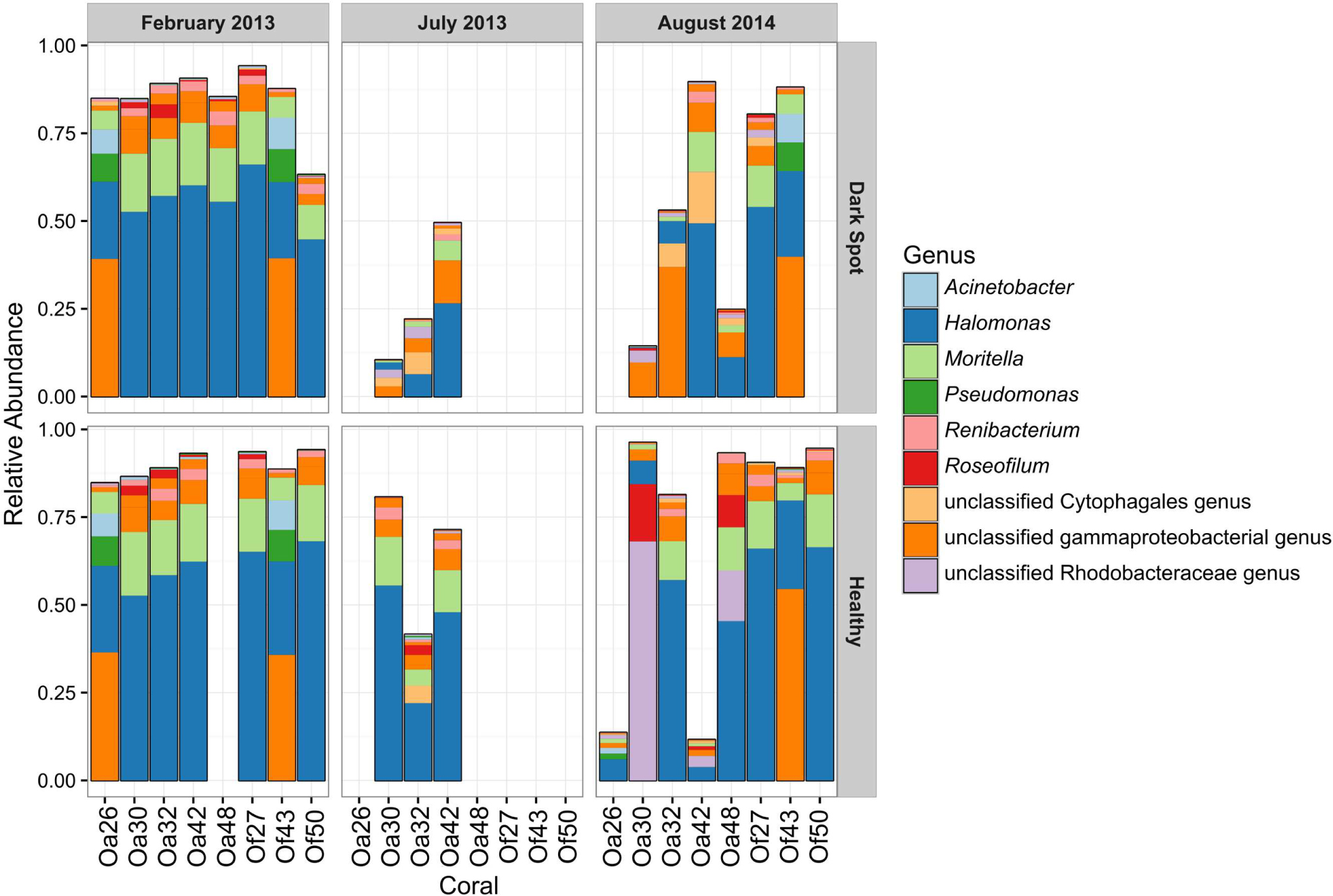
FIGURE 4. Relative abundance of the top ten OTUs in healthy tissue and Dark Spot lesions in Orbicella corals. Relative abundance of sequencing reads for the ten most abundant operational taxonomic units (OTUs) in eight tagged corals, across three sampling dates, in healthy tissue and Dark Spot lesions. Coral names beginning with Oa are from O. annularis and those beginning with Of are from O. faveolata.
Comparison of the Microbiota of Recovered and Declined Corals
Five of the eight corals in this study showed substantial tissue loss over the sampling period (Oa26, Of27, Oa30, Oa32, and Of50) (Figure 1). One coral (Oa32) was well documented with photographs and samples throughout the study period and is representative of the progression of DSS leading to the nearly complete decay of the coral colony. DSS lesions in O. annularis coral Oa32 exhibited a loss of the most abundant genus, Halomonas, and an increase in the unclassified gammaproteobacterial genus over time (Figure 4). The healthy tissue samples in Oa32 retained a high relative abundance of Halomonas throughout the study period.
In contrast, only one coral (O. faveolata coral Of43) appeared to recover from DSS by September 2015 (Figure 1). Coral Of43 sustained high levels of the unclassified gammaproteobacterium throughout the course of the 18-month sampling period, and the overall bacterial community structure did not change over time, as indicated by the clustering of Of43 samples in Figure 1. The dominance of this unclassified gammaproteobacterium in all samples from this coral sets it apart from other coral colonies (Figure 3). Coral Oa26 clustered with Of43 initially, but over time exhibited a loss of the unclassified gammaproteobacterium and the concurrent decay of the coral colony.
Discussion
While DSS has been previously reported in boulder corals (Gil-Agudelo and Garzon-Ferreira, 2001), this is the first study to characterize the bacterial community associated with DSS-affected Orbicella corals. Based on the sequencing of the V6 hypervariable region of 16S rRNA genes, we detected no shift in the bacterial communities between healthy tissue and Orbicella DSS lesions, with all samples containing a high relative abundance of Gammaproteobacteria, especially of the genera Halomonas, Moritella, and Pseudomonas, as well as an unclassifiable gammaproteobacterial genus. These results are consistent with earlier reports (Kellogg et al., 2014; Randall et al., 2016) that detected no significant shift in the microbiome composition of healthy Siderastrea siderea corals compared to those with DSS lesions. Sequencing of the V1–V3 hypervariable region of 16S rRNA genes showed no differences in the overall bacterial community composition of healthy or DSS-affected Siderastrea siderea corals, no significant differences in bacterial species richness or species diversity, and no significant differences in the relative abundance of putative pathogen taxa (Vibrio, Corynebacterium, Acinetobacter, Photobacterium, Parvularculacea, and Oscillatoria) (Randall et al., 2016). However, taxon-specific Kruskal–Wallis rank-sum tests identified 9 taxa that were significantly more abundant in diseased corals than in healthy corals and corals experimentally exposed to DSS, including Alteromonas, Aquabacterium, Arthrobacter, Bermanella, Haliscomenobacter, Litoreibacter, Oscillatoriales, Pseudomonas, and Sorangiineae (Randall et al., 2016). In contrast, an earlier study using PhyloChip G3 microarrays detected no significant differences in the relative abundance of specific bacterial taxa between healthy S. siderea corals and those with dark spot lesions (Kellogg et al., 2014).
In Stephanocoenia intersepta corals, the sequencing of clone libraries of bacterial 16S rRNA genes and fungal ITS region revealed differences in the bacterial community composition of healthy corals versus corals with varying sizes of DSS lesions (Sweet et al., 2013). Burkholderia, Pseudomonas, Parvibaculum, and Ochrobactrum were detected in all healthy samples, but not in DSS lesions. Photobacterium and two Vibrio species (including V. campbellii) were abundant in lesions and absent from healthy samples. Corynebacterium, Acinetobacter, Parvularculaceae, and Oscillatoria were also present in lesions and absent from healthy corals and increased in abundance in apparently healthy tissue adjacent to lesions (Sweet et al., 2013). A fungal ribotype associated with a terrestrial phytopathogenic fungus, Rhytisma acernum, was found in the lesions, but not in healthy samples (Sweet et al., 2013). Our PCR assay did not detect Rhytisma in any of the Orbicella coral samples in this study, but it did positively amplify Rhytisma in the samples of DSS lesions from the Stephanocoenia intersepta corals, suggesting that this fungal pathogen is not associated with DSS on different coral types or that Stephanocoenia DSS is a different disease than Orbicella DSS.
The absence of differences in the microbiomes of apparently healthy tissue or asymptomatic corals and those with DSS in both Orbicella (this study) and Siderastrea (Kellogg et al., 2014) corals suggests that it is not a dysbiosis syndrome or that DSS may not be the same disease in Stephanocoenia corals. The apparent lack of transmissibility of this syndrome supports the hypothesis that it is not infectious disease. In fact, some researchers suggested that DSS is a general stress response, exacerbated by high water temperature or other stressors (Borger, 2005). However, data collected to date does not rule out the possibility that the disease may be caused by a virus that is a normal member of the coral’s virome but under some stress conditions impacts the host’s health. Atypical herpes virus and a megavirus affecting Symbiodinium spp. and leading to the loss of algal symbionts have been reported (Correa et al., 2009).
Recovery of corals from DSS has been previously reported, although DSS tended to re-occur on the same corals in 30% of the cases in some studies (Voss and Richardson, 2006). Recovery rates have been reported from 4 to 9% or as high as 30 to 60%, with the differences in the recovery rates likely due to the definition of “recovery” used by the researchers (Borger, 2005; Porter et al., 2011). In some studies, up to 7.1% of colonies regained tissue following disappearance of the lesions, mostly by re-growth over the former lesion (Porter et al., 2011). In this study, three out of eight corals either completely recovered or the spread of dark sport lesions seized. However, the majority of affected corals reported here suffered substantial tissue loss and looked similar to the progression of DSS in O. annularis previously documented (Porter et al., 2011).
While we found no differences in the microbiota associated with Orbicella DSS lesions and healthy tissue on DSS-affected corals, the BBD layer on two DSS-affected corals hosted a distinct BBD consortium. BBD is caused by R. reptotaenium, which is a normal (albeit, minor) member of the microbiota of Caribbean Orbicella, Montastraea, and Pseudodiploria corals (Meyer et al., 2016) and was detected in every sample in this study. Under some conditions, likely associated with an environmental stress, R. reptotaenium escapes controls imposed on it by the host and/or other members of the microbiome and overtakes the community, leading to the appearance of BBD. Despite the fact that BBD appeared on 2 out of 8 corals with DSS, there was no evidence that DSS was a precursor of BBD. However, the two corals that developed BBD were among those that experienced the most substantial tissue loss over the observational period. Therefore, we hypothesize that either DSS itself or the environmental conditions leading to it were triggers resulting in the take-over of the host microbiota by R. reptotaenium.
Author Contributions
VP and MT conceived and designed the project. MT collected samples. JM, JR, and BD performed molecular work and data analysis. JM and MT wrote the manuscript and all authors contributed to critical revisions.
Funding
Smithsonian George E. Burch Fellowship (MT), L’Oreal Fellowship for Women in Science (JM), Smithsonian Caribbean Coral Reef Ecosystem Program. NIFA REEport project # FLA-SWS-005474.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Belize Fisheries Department for providing research permits for this work. We are grateful to M. Sweet for providing samples, and C. Thacker for field assistance. This is contribution # 989 of the Smithsonian Caribbean Coral Reef Ecosystem and contribution # 1032 of the Smithsonian Marine Station.
References
Aronesty, E. (2011). ea-utils: Command-Line Tools for Processing Biological Sequencing Data. Available at: http://code.google.com/p/ea-utils
Blankenberg, D., Von Kuster, G., Coraor, N., Ananda, G., Lazarus, R., Mangan, M., et al. (2010). Galaxy: a web-based genome analysis tool for experimentalists. Curr. Protoc. Mol. Biol. Chap. 19, Unit 19.10.1–19.10.21. doi: 10.1002/0471142727.mb1910s89
Borger, J. L. (2003). Three scleractinian coral diseases in Dominica, West Indies: distribution, infection patterns and contribution to coral tissue mortality. Rev. Biol. Trop. 51, 25–38.
Borger, J. L. (2005). Dark spot syndrome: a scleractinian coral disease or a general stress response? Coral Reefs 24, 139–144. doi: 10.1007/s00338-004-0434-6
Bosch, T. C., and McFall-Ngai, M. J. (2011). Metaorganisms as the new frontier. Zoology. (Jena). 114, 185–190. doi: 10.1016/j.zool.2011.04.001
Bourne, D. G., Garren, M., Work, T. M., Rosenberg, E., Smith, G. W., and Harvell, C. D. (2009). Microbial disease and the coral holobiont. Trends. Microbiol. 17, 554–562. doi: 10.1016/j.tim.2009.09.004
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Cardenas, A., Rodriguez, R. L., Pizarro, V., Cadavid, L. F., and Arevalo-Ferro, C. (2012). Shifts in bacterial communities of two Caribbean reef-building coral species affected by white plague disease. ISME J. 6, 502–512. doi: 10.1038/ismej.2011.123
Casamatta, D., Stanic, D., Gantar, M., and Richardson, L. L. (2012). Characterization of Roseofilum reptotaenium (Oscillatoriales, Cyanobacteria) gen. et sp nov isolated from Caribbean black band disease. Phycologia 51, 489–499. doi: 10.2216/11-10.1
Cervino, J., Goreau, T. J., Nagelkerken, I., Smith, G. W., and Hayes, R. (2001). Yellow band and dark spot syndromes in Caribbean corals: distribution, rate of spread, cytology, and effects on abundance and division rate of zooxanthellae. Hydrobiologia 460, 53–63. doi: 10.1023/A:1013166617140
Correa, A. M. S., Brandt, M. E., Smith, T. B., Thornhill, D. J., and Baker, A. C. (2009). Symbiodinium associations with diseased and healthy scleractinian corals. Coral Reefs 28, 437–448. doi: 10.1007/s00338-008-0464-6
DeSantis, T. Z., Hugenholtz, P., Larsen, N., Rojas, M., Brodie, E. L., Keller, K., et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072. doi: 10.1128/AEM.03006-05
Dixon, P. (2003). VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14, 927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Eren, A. M., Vineis, J. H., Morrison, H. G., and Sogin, M. L. (2013). A filtering method to generate high quality short reads using Illumina paired-end technology. PLoS ONE 8:e66643. doi: 10.1371/journal.pone.0066643
Giardine, B., Riemer, C., Hardison, R., Burhans, R., Elnitski, L., Shah, P., et al. (2005). Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 15, 1451–1455. doi: 10.1101/gr.4086505
Gil-Agudelo, D. L., and Garzon-Ferreira, J. (2001). Spatial and seasonal variation of dark spots disease in coral communities of the Santa Marta area (Colombian Caribbean). Bull. Mar. Sci. 69, 619–629.
Gochfeld, D. J., Olson, J. B., and Slattery, M. (2006). Colony versus population variation in susceptibility and resistance to dark spot syndrome in the Caribbean coral Siderastrea siderea. Dis. Aquat. Organ. 69, 53–65. doi: 10.3354/dao069053
Goecks, J., Nekrutenko, A., Taylor, J., and Team, T. G. (2010). Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 11:R86. doi: 10.1186/gb-2010-11-8-r86
Kellogg, C. A., Piceno, Y. M., Tom, L. M., DeSantis, T. Z., Gray, M. A., and Andersen, G. L. (2014). Comparing bacterial community composition of healthy and dark spot-affected Siderastrea siderea in Florida and the Caribbean. PLoS ONE 9:e108767. doi: 10.1371/journal.pone.0108767
Lesser, M. P., Bythell, J. C., Gates, R. D., Johnstone, R. W., and Hoegh-Guldberg, O. (2007). Are infectious diseases really killing corals? Alternative interpretations of the experimental and ecological data. J. Exp. Mar. Biol. Ecol. 346, 36–44. doi: 10.1016/j.jembe.2007.02.015
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12. doi: 10.14806/ej.17.1.200
McMurdie, P. J., and Holmes, S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8:e61217. doi: 10.1371/journal.pone.0061217
Meyer, J. L., Gunasekera, S. P., Scott, R. M., Paul, V. J., and Teplitski, M. (2016). Microbiome shifts and the inhibition of quorum sensing by black band disease cyanobacteria. ISME J. 10, 1204–1216. doi: 10.1038/ismej.2015.184
Meyer, J. L., Paul, V. J., and Teplitski, M. (2014). Community shifts in the surface microbiomes of the coral Porites astreoides with unusual lesions. PLoS ONE 9:e100316. doi: 10.1371/journal.pone.0100316
Parks, D. H., Tyson, G. W., Hugenholtz, P., and Beiko, R. G. (2014). STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30, 3123–3124. doi: 10.1093/bioinformatics/btu494
Porter, J. W., Torres, C., Sutherland, K. P., Meyers, M. K., Callahan, M. K., Ruzicka, R., et al. (2011). Prevalence, severity, lethality, and recovery of dark spots syndrome among three Floridian reef-building corals. J. Exp. Mar. Biol. Ecol. 408, 79–87. doi: 10.1016/j.jembe.2011.07.027
Randall, C. J., Jordan-Garza, A. G., Muller, E. M., and van Woesik, R. (2016). Does dark-spot syndrome experimentally transmit among Caribbean corals? PLoS ONE 11:e0147493. doi: 10.1371/journal.pone.0147493
Rasoulouniriana, D., Siboni, N., Ben-Dov, E., Kramarsky-Winter, E., Loya, Y., and Kushmaro, A. (2009). Pseudoscillatoria coralii gen. nov., sp. nov., a cyanobacterium associated with coral black band disease (BBD). Dis. Aquat. Organ. 87, 91–96. doi: 10.3354/dao02089
Rideout, J. R., He, Y., Navas-Molina, J. A., Walters, W. A., Ursell, L. K., Gibbons, S. M., et al. (2014). Subsampled open-reference clustering creates consistent, comprehensive OTU definitions and scales to billions of sequences. PeerJ. 2:e545. doi: 10.7717/peerj.545
Ritchie, K. B. (2006). Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar. Ecol. Prog. Ser. 322, 1–14. doi: 10.3354/meps322001
Rosenberg, E., and Ben-Haim, Y. (2002). Microbial diseases of corals and global warming. Environ. Microbiol. 4, 318–326. doi: 10.1046/j.1462-2920.2002.00302.x
Sweet, M., Burn, D., Croquer, A., and Leary, P. (2013). Characterisation of the bacterial and fungal communities associated with different lesion sizes of dark spot syndrome occurring in the coral Stephanocoenia intersepta. PLoS ONE 8:e62580. doi: 10.1371/journal.pone.0062580
Vezzulli, L., Pezzati, E., Huete-Stauffer, C., Pruzzo, C., and Cerrano, C. (2013). 16SrDNA Pyrosequencing of the mediterranean gorgonian paramuricea clavata reveals a link among alterations in bacterial holobiont members, anthropogenic influence and disease outbreaks. PLoS ONE 8:e67745. doi: 10.1371/journal.pone.0067745
Voss, J. D., and Richardson, L. L. (2006). Coral diseases near lee stocking Island, Bahamas: patterns and potential drivers. Dis. Aquat. Organ. 69, 33–40. doi: 10.3354/dao069033
White, T. J., Bruns, T. D., Lee, S. B., and Taylor, J. W. (1990). “Analysis of phylogenetic relationships by amplification and direct sequencing of ribosomal RNA genes,” in PCR Protocols: A Guide to Methods and Applications, eds M. A. Innis, D. H. Gelfand, J. J. Sninsky and T. J. White (New York, NY: Academic Press), 315–322.
Keywords: coral disease, coral microbiome, Dark Spot Disease, Orbicella annularis, Orbicella faveolata, surface mucus layer, Black Band Disease
Citation: Meyer JL, Rodgers JM, Dillard BA, Paul VJ and Teplitski M (2016) Epimicrobiota Associated with the Decay and Recovery of Orbicella Corals Exhibiting Dark Spot Syndrome. Front. Microbiol. 7:893. doi: 10.3389/fmicb.2016.00893
Received: 15 April 2016; Accepted: 26 May 2016;
Published: 07 June 2016.
Edited by:
Virginia Weis, Oregon State University, USAReviewed by:
Kimberly B. Ritchie, Mote Marine Laboratory, USAFrank O’Neill Aylward, University of Hawaii at Manoa, USA
Michael Sweet, University of Derby, UK
Copyright © 2016 Meyer, Rodgers, Dillard, Paul and Teplitski. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Max Teplitski, maxtep@ufl.edu
 Julie L. Meyer
Julie L. Meyer John M. Rodgers
John M. Rodgers Brian A. Dillard
Brian A. Dillard Valerie J. Paul
Valerie J. Paul Max Teplitski
Max Teplitski