- 1Faculty of Medicine, Institute of Biochemistry, University of Ljubljana, Ljubljana, Slovenia
- 2Department of Biotechnology and Biophysics, Biocenter, Julius Maximilian University of Würzburg, Würzburg, Germany
The basidiomycetous fungus Wallemia ichthyophaga grows between 1.7 and 5.1 M NaCl and is the most halophilic eukaryote described to date. Like other fungi, W. ichthyophaga detects changes in environmental salinity mainly by the evolutionarily conserved high-osmolarity glycerol (HOG) signaling pathway. In Saccharomyces cerevisiae, the HOG pathway has been extensively studied in connection to osmotic regulation, with a valuable knock-out strain collection established. In the present study, we reconstructed the architecture of the HOG pathway of W. ichthyophaga in suitable S. cerevisiae knock-out strains, through heterologous expression of the W. ichthyophaga HOG pathway proteins. Compared to S. cerevisiae, where the Pbs2 (ScPbs2) kinase of the HOG pathway is activated via the SHO1 and SLN1 branches, the interactions between the W. ichthyophaga Pbs2 (WiPbs2) kinase and the W. ichthyophaga SHO1 branch orthologs are not conserved: as well as evidence of poor interactions between the WiSho1 Src-homology 3 (SH3) domain and the WiPbs2 proline-rich motif, the absence of a considerable part of the osmosensing apparatus in the genome of W. ichthyophaga suggests that the SHO1 branch components are not involved in HOG signaling in this halophilic fungus. In contrast, the conserved activation of WiPbs2 by the S. cerevisiae ScSsk2/ScSsk22 kinase and the sensitivity of W. ichthyophaga cells to fludioxonil, emphasize the significance of two-component (SLN1-like) signaling via Group III histidine kinase. Combined with protein modeling data, our study reveals conserved and non-conserved protein interactions in the HOG signaling pathway of W. ichthyophaga and therefore significantly improves the knowledge of hyperosmotic signal processing in this halophilic fungus.
Introduction
The hypersaline environments of salterns are inhabited by fungal species that are well adapted to these extreme environmental conditions (Gostinčar et al., 2011). Studies on these halophilic and halotolerant fungi aim to better understand the molecular mechanisms behind their extreme osmotolerance (Plemenitaš et al., 2014), which might then be applicable to the production of salt and drought resistant crops (Cominelli and Tonelli, 2010; Gašparič et al., 2013). The dominant fungal group in salterns is the polymorphic ascomycetous black yeasts (Zajc et al., 2012). However, the basidiomycete Wallemia ichthyophaga, which was originally isolated from the solar salterns of Sečovlje, Slovenia, is the most halophilic eukaryote (Zalar et al., 2005). This obligate halophilic fungus forms multicellular clumps in liquid media, has a coccoid, sarcina-like, morphology (Kralj Kunčič et al., 2010), and grows in media enriched with NaCl concentrations from 1.7 to 5.1 M. This phenotype is unusual in the fungal kingdom, and makes W. ichthyophaga an interesting model organism.
Recently, the osmoadaptation strategy of W. ichthyophaga has been investigated in detail. The NaCl range from 2.5 to 3.4 M was shown to be optimal for W. ichthyophaga, and associated with its highest dry biomass production (5–6 mg per ml of growth medium, respectively; Zajc et al., 2014). Despite these high external NaCl concentrations, W. ichthyophaga maintains low intracellular sodium concentrations, which only transiently increase upon hypersaline shock (Zajc et al., 2014). In W. ichthyophaga, glycerol is synthesized as a compatible solute for turgor regulation, and its concentration rises with increasing external salinity, and lowers under hypo-osmotic shock (Zajc et al., 2014). Also, at 4.3 M external NaCl concentrations, thickening of the cell wall, increased clump size, and higher abundance of extracellular polysaccharides appear to be important adaptations to the extreme external salinity for this fungus (Kralj Kunčič et al., 2010, 2013). The genome of W. ichthyophaga is at 9.6 Mb highly compact and contains only 4884 predicted protein-coding genes. Of these genes, 425 were differentially expressed when W. ichthyophaga was grown at 1.7 M, and 214 when it was grown at 5.1 M NaCl concentrations. The analysis of the transcriptomic data also revealed enrichment and salt-responsiveness of the hydrophobins, which are small cell-wall proteins usually involved in rigidity, strength and permeability, and have been hypothesized to be important in highly saline environments (Zajc et al., 2013).
For successful osmoadaptation in highly saline environments, it is essential that organisms can sense and respond to increase or decrease in salt concentrations (Plemenitaš et al., 2014). In yeast and fungi, increases in environmental osmolarity are sensed by the evolutionarily conserved high-osmolarity glycerol (HOG) signaling pathway (Krantz et al., 2006b; Bahn, 2008), which regulates the response of cells to new osmotic conditions (Hohmann et al., 2007). The HOG pathway has been extensively studied in Saccharomyces cerevisiae, where the final MAPK ScHog1 is phosphorylated upon hyperosmotic shock, and regulates several intracellular systems (Westfall et al., 2008; Mas et al., 2009; Sole et al., 2011), including, most importantly, glycerol production (Petelenz-Kurdziel et al., 2011). Once the cellular water potential is restored, the phosphatases ScPtc1, ScPtp2, and ScPtp3 can deactivate ScHog1 by its dephosphorylation (Hohmann et al., 2007). In S. cerevisiae, this pathway consists of two branches that are known as the SHO1 branch and the SLN1 branch. For the SHO1 branch, the signal is perceived by the combined actions of the transmembrane co-osmosensor ScSho1, the membrane anchor ScOpy2, and the mucines ScMsb2 and ScHkr1. Moreover, the guanine exchange factor ScCdc24 and the GTPase ScCdc42 are also involved in the SHO1 sensory apparatus. Signal perception leads to the activation of ScSte20 and ScCla4, which consequently activate the MAPKKK ScSte11. Redundantly, the SLN1 branch transduces the signal via the evolutionarily conserved two-component phosphorelay system, which consists of the ScSln1 sensor histidine kinase, the ScYpd1 intermediate protein, and the ScSsk1 response regulator that activates the MAPKKKs ScSsk2/ScSsk22 (Saito and Posas, 2012). The MAPKKKs ScSte11 and ScSsk2/ScSsk22 all activate the scaffold MAPKK ScPbs2. As ScPbs2 is the connecting hub of the two upstream pathway branches (i.e., SHO1, SLN1) and is an activator of ScHog1 MAPK, various interaction motifs, binding regions, localization signals, and the catalytic function are encoded in its sequence (Krantz et al., 2006a).
In filamentous fungi like Aspergillus fumigatus and Aspergillus nidulans, the HOG pathway is also involved in osmotic stress responses (Ma and Li, 2013; de Oliveira Bruder Nascimento et al., 2016). However, in A. nidulans, the ShoA (AnSho1) protein is not involved in osmosensing (Furukawa et al., 2005). In a similar manner, the HOG pathway in the basidiomycetous fungus Cryptococcus neoformans is activated exclusively via the two-component phosphorelay system (Bahn et al., 2006). In addition to osmotic regulation, the two-component branches of the HOG pathway in pathogenic C. neoformans (Kozubowski et al., 2009), Magnaporthe oryzae (Jacob et al., 2015), and Candida albicans (Prieto et al., 2014) are associated with virulence regulation, plant infection, and colonization, respectively. In these pathogens, the pathway is regulated mainly by histidine kinases, which also confer fungicide sensitivity (Kozubowski et al., 2009; El-Mowafy et al., 2013).
The HOG pathway was recently described in detail in halotolerant Hortaea werneckii, where survival of the cells depends on the HOG pathway only at salinities ≥3.0 M NaCl (Kejžar et al., 2015). Initial studies of the HOG pathway in W. ichthyophaga showed that its regulation is controlled by rapid dephosphorylation under both hyperosmolar and hypo-osmolar stress (Konte and Plemenitaš, 2013). Interestingly, the W. ichthyophaga genome contains two HOG1-like genes: WiHOG1A and WiHOG1B, which are up-regulated three- and six-fold, respectively, under both hyper- and hypo-osmolar conditions. Results from our previous complementation studies with the WiHOG1A and WiHOG1B genes in S. cerevisiae showed that while the WiHog1B protein was able to fully rescue the hog1Δ cells, WiHog1A showed a partial rescue of the osmosensitive phenotype (Konte and Plemenitaš, 2013).
In the current study, we further probed the HOG pathway of W. ichthyophaga using S. cerevisiae HOG pathway mutants, similar to our previous investigations (Konte and Plemenitaš, 2013). The S. cerevisiae mutants are easily grown, genetically tractable, and a large library of knock-out mutant strains is available. In particular, the high conservation of the MAPK signaling pathway components makes S. cerevisiae an ideal system for functional investigations of orthologous signaling proteins from fungi (Bansal et al., 2001; El-Mowafy et al., 2013), and even from evolutionarily distant organisms such as plants (Reiser et al., 2003; Jain et al., 2009) and mammals (Levin-Salomon et al., 2009). Here, we have employed the above system to further elucidate the HOG pathway components in halophilic W. ichthyophaga.
Materials and Methods
Strains, Growth Conditions, DNA and RNA Isolation, and cDNA Synthesis
W. ichthyophaga (EXF 994) was provided by the culture collection of the Department of Biology, Biotechnical Faculty, University of Ljubljana (EXF), and was grown at 28°C and 180 rpm in YNB medium supplemented with 3.4 M NaCl, as described previously (Konte and Plemenitaš, 2013). The S. cerevisiae parental strains (here referred to as the wild-types; WT) and mutant strains used in this study are listed in Supplemental Table S1. For the detailed growth conditions of these S. cerevisiae strains, please see the section on “Expression of Genes in the S. cerevisiae Mutants and Halotolerance Assays” below. A method described previously (Rozman and Komel, 1994) was used for isolation of genomic DNA from W. ichthyophaga cells in mid-exponential-phase grown in YNB medium with 3.4 M NaCl, and from the Euroscarf S. cerevisiae Y00000 (S288c) WT strain grown in YNB medium. The construction of pools of uncloned, adaptor-ligated, genomic DNA fragments of W. ichthyophaga was carried out with Genome Walker Universal kits (Clontech). The mid-exponential-phase cells of W. ichthyophaga grown in YNB medium containing 3.4 M NaCl were also used for the RNA extraction, with the TRI Reagent (Sigma Aldrich). The DNA was removed using DNAse I (Fermentas), and the RNA was evaluated spectrophotometrically (A260/A280, BioTek Synergy 2 Take3 plates) and by capillary electrophoresis (Agilent 2100 bioanalyser). SuperScript III First-Strand cDNA Synthesis kits (Invitrogen) and random hexamer primers (Promega) or SMARTer Race cDNA Amplification kits (Clontech) were used for the synthesis of cDNA.
Identification, Amplification, Cloning, and Sequencing of the Genes under Study
A partial sequence of the PBS2-like gene from W. ichthyophaga was cloned in our previous study (Lenassi et al., 2007) using degenerate primers. In the present study, the partial sequence of the WiSTE11 gene was obtained using the same method. The degenerate primers degWiSTE11.f and degWiSTE11.r were constructed based on the highly conserved parts of the WiSte11 kinase domain (GELMAVKQV, NIFLEYVPG) using the iCODEHOP software, and then manually refined. Gene-specific primers were designed (see Supplemental Table S2: the pWiPBS2 and npWiPBS2, and pWiSTE11 and npWiSTE11 primers) based on the partial sequences of the WiSTE11 and WiPBS2 genes. These primers were used in the primary touch-down PCR and the following secondary nested PCR (annealing for 5 cycles at 70°C, 5 cycles at 68°C, and 25 cycles at 68°C), which was performed with High Fidelity Polymerase Mix (Fermentas) and 50 ng cDNA or gDNA template, in 20-μL reactions. The amplicons were purified with EZNA gel extraction kits (Omega Bio-Tek), cloned into the pJET cloning vector (Fermentas), transformed into E. coli XL-1 Blue cells, isolated with GeneJET MiniPrep kits (Fermentas), and sequenced (Macrogen). The sequences were later confirmed in the genome of W. ichthyophaga that became available during the present study (Zajc et al., 2013). The ScPBS2 and ScSTE11 gene sequences were retrieved from the Saccharomyces Genome Database (SGD Project; http://www.yeastgenome.org/; March 2015) and were cloned using the same method. The WiSHO1 gene sequence was obtained using local BLAST searches of the W. ichthyophaga genome (Zajc et al., 2013). The pAZ301 vector (pR316 with SHO1 promoter and GFP tag) containing the ScSHO1 gene was kindly provided by W. Lim (UCSF, San Francisco). The WiHOG1A (GenBank accession number AGG39582), WiHOG1B (GenBank accession number AGG39583) and ScHOG1 (GenBank accession number DAA09427) genes were amplified and cloned in our previous study (Konte and Plemenitaš, 2013). Based on the sequences obtained, new forward and reverse primers were designed for PCR amplification of the whole gene-coding sequences, again with 50 ng template cDNA, High Fidelity Polymerase Mix, and annealing for 30 cycles at 60°C. Again, the amplicons were purified, cloned, and sequenced.
Sequences, Domains, and Phylogenetic Analysis
For protein sequence alignments, the M-Coffee software with default settings was used (http://tcoffee.crg.cat/apps/tcoffee/do:mcoffee; July 2015) and visualization was performed with the Jalview 2.7 software (Waterhouse et al., 2009). Annotation of domains in alignment was based on the literature and data from the UniProt database (Uniprot Consortium, 2012; http://www.uniprot.org/; July 2015) and the SMART software (http://smart.embl-heidelberg.de/; June 2015). The proline-rich motifs of WiPbs2 and ScPbs2 were annotated according to the Eukaryotic Linear Motif Resource (http://elm.eu.org/; July 2015) and the literature. The physicochemical parameters of the protein sequences were obtained with the ProtParam ExPasy program (http://web.expasy.org/protparam/; June 2015; Wilkins et al., 1999). Sequences of the orthologous Pbs2 kinases were retrieved from the National Center for Biotechnology Information (NCBI) database; (http://www.ncbi.nlm.nih.gov/; April 2015). For identification of the W. ichthyophaga HOG pathway components, the W. ichthyophaga genome was searched for orthologous pathway proteins with the local BLAST software (version 2.2.25; http://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=Web&PAGE_TYPE=BlastDocs&DOC_TYPE=Download; November 2010) using as probes the S. cerevisiae HOG pathway proteins, and the C. neoformans histidine kinase Nik1 (see Supplemental Table S3 for GenBank accession numbers). The NCBI Conserved Domain Database (CDD; Marchler-Bauer and Bryant, 2004) was also the source of domain annotations of the identified W. ichthyophaga HOG pathway. The maximum likelihood phylogenetic tree of orthologous proteins was calculated as described before (Konte and Plemenitaš, 2013). The interactions of the Src homology 3 (SH3) domain from WiSho1 and the WiPbs2 proline-rich motif were modeled with the SWISS-MODEL software (Biasini et al., 2014), based on the crystal structure of the ScSho1 SH3 domain complexed with the ScPbs2 proline-rich motif (PDB: 2VKN). In the model, the ScPbs2 proline-rich motif was replaced with the WiPbs2 APLPPNQLR peptide using the WhatIf program (Vriend, 1990). The COCOMAPS software (Vangone et al., 2011) was used to analyze the residue interactions in the models and the PyMOL software was used for visualization of the three-dimensional structures (The PyMOL Molecular Graphics System, Version 1.1, LLC).
Expression of Genes in the S. cerevisiae Mutants, and Halotolerance Assays
The expression of the W. ichthyophaga WiPBS2 and WiSTE11 genes in S. cerevisiae cells was carried out with the corresponding open reading frame amplification from the pJET cloning vectors (see Section “Identification, Amplification, Cloning, and Sequencing of the Genes under Study,” above) with primers containing restriction sites (Supplemental Table S2). The resulting products were cloned into the corresponding restriction sites of the low-copy-number plasmids pYX142 (CEN, LEU, TPI promoter, Ampr), YCplac22 (CEN, TRP, TPI promoter, Ampr), and YCplac 33 (CEN, URA, TPI promoter, Ampr), and transformed into the S. cerevisiae strains using the Quick and Easy LiAc/SS carrier DNA/ polyethylene glycol method (Gietz and Schiestl, 2007). The open reading frame of the WiSHO1 gene was amplified from the pJET cloning vector, with the primers WiSHO1xSHO.r and GFPxWiSHO1.r, which have sequence overhangs that are homologous to the 3′ end of the SHO1 promoter and the 5′ end of the GFP tag. The SHO1 promoter of S. cerevisiae and the GFP tag were amplified separately from the pAZ301 plasmid using the primers (KpnI)SHO.f and WiSHO1xSHO.r, and WiSHO1xGFP.f and (SacI)GFP.r, respectively. The amplified SHO1 promoter, the WiSHO1 gene, and the GFP tag were then combined in the PCR reaction with High Fidelity Polymerase Mix and the primers (KpnI)SHO.f and (SacI)GFP.r, and annealed for 30 cycles at 60°C. The product was confirmed by sequencing, and cloned into pAZ301 (CEN, URA, Ampr), which was previously cut with KpnI and SacI to remove the ScSHO1. In addition to WiSHO1, we also constructed and cloned the hybrid gene between ScSHO1 and WiSHO1 into pAZ301. This hybrid gene was made from ScSHO1, where the part of the sequence that encodes the ScSHO1 SH3 domain was replaced with the sequence of the WiSHO1 SH3 domain [amplified with primers (BamHI)WiSH3.f and (EcoRI)WiSH3.r; Supplemental Table S2], using the BamHI and EcoRI restriction sites already present in ScSHO1. The constructed hybrid gene is named as ScSHO1WiSH3. The resulting plasmid constructs and transformants are given in Supplemental Table S4. For controls, both the mutant strains expressing S. cerevisiae genes (ScPBS2, ScSHO1, ScSTE11, ScHOG1) and WT strains or mutant strains transformed with empty vectors (EVs) were used. The cells were grown on YNB plates with appropriate drop-out selection (Supplemental Table S4), with the presence of the plasmids confirmed by colony PCR.
For the construction of the WiPbs2 protein with the inserted proline-rich motif VNKPLPPLPVAG from ScPbs2, we used the oligonucleotide-directed PCR mutagenesis method. Using the primers WiPbs2(ScPRO).f and r (Supplemental Table S2) and the plasmid YCplac22+WiPBS2, the ScPbs2 proline-rich motif was inserted using a PCR reaction with annealing for 30 cycles at 60°C. The resulting plasmid YCplac22+WiPBS2ScPRO was transformed simultaneously with the plasmid pAZ301+ScSHO1WiSH3 into the sho1Δssk2/22Δpbs2Δ cells. Furthermore, the simultaneous transformation of YCplac22+WiPBS2 and pAZ301+ScSHO1WiSH3, YCplac22+ScPBS2, and pAZ301+ScSHO1, or YCplac22+ScPBS2 and EV pAZ301 into the sho1Δssk2/22Δpbs2Δ cells was also performed. Positive transformants were confirmed by PCR and then used for the halotolerance assays. They were grown to mid-exponential phase in YNB medium containing: 0.08% (w/v) Complete Supplement Mixture with appropriate drop-out (Formedium), 0.16% (w/v) YNB (Formedium), 2.0% (w/v) glucose (Mallinckrodt Baker), 0.5% (w/v) ammonium sulfate (Carlo Erba Reagents), and NaCl (Carlo Erba Reagents) added to different concentrations, at pH 7.0 and 30°C in a rotary shaker (180 rpm). The OD600 was then adjusted to 0.5, and 3.5 μL of 10-fold serial dilutions (1–103 dilutions) were spotted onto the selective plates (YNB with the appropriate drop-out, and 2% (w/v) agar) that contained the indicated NaCl concentrations. We incubated the plates at 30°C for 3–6 days, and then scanned them. All of the experiments were repeated 2–3 times.
The Cross-Talk β-Galactosidase Assay
The activation of Fus3 kinase was followed under salt stress with the induction of the cross-talk mating response of the FUS1-lacZ reporter expression (O'Rourke and Herskowitz, 1998). The FUS1-lacZ reporter system was integrated into the genome of S. cerevisiae pbs2Δhog1Δ cells using yeast integration plasmids pRS306N (URA3 integration) and into the ssk2/22Δpbs2Δ cells using pRS303H (HIS3 integration). The FUS1-lacZ reporter system integration with pRS306N was also performed for the wild-type S1278b cells, which were used as the negative control for pbs2Δhog1Δ FUS1-lacZ. For the negative control of the ssk2/22Δpbs2Δ FUS1-lacZ cells, the W303 WT strain was transformed with the YCplac22 vector containing the FUS1-lacZ reporter. Next, in pbs2Δhog1Δ FUS1-lacZ cells, WiPbs2 or ScPbs2 and WiHog1A, WiHog1B, or ScHog1 were expressed, and in ssk2/22Δpbs2Δ FUS1-lacZ cells WiPbs2 or ScPbs2 were expressed. For both strains, the EV (positive control) was also transformed (see Supplemental Table S4 for detailed description of these transformants). The positive transformants were subjected to 1 M NaCl for 4 h, and the activity of the β-galactosidase enzyme before and after the application of NaCl was measured, as described previously (Konte and Plemenitaš, 2013). We calculated the β-galactosidase values according to the formula (A420× 1000 − 1.75 × OD550)/(time(min)× V(mL)× OD600) (Miller, 1972; Murakami et al., 2008), in triplicate, and the means ± SD were calculated. The graphs were designed using the GraphPad Prism 5 software.
Western Blotting
For the Western blotting, mid-exponential-phase pbs2Δhog1Δ cells expressing WiPbs2 or ScPbs2 and one of the kinases WiHog1A, WiHog1B, or ScHog1 were adjusted to an OD600 of 0.5. The transformants were harvested by centrifugation (6000 × g, 1 min, 25°C) before and 5 min and 10 min after application of 0.7 M NaCl, and frozen in liquid nitrogen. Protein extracts were prepared with the boiling of cells in protein extraction buffer (100 mM Tris-HCl, pH 6.8, 20% glycerol, 2% 2-mercaptoethanol, 4% SDS) containing a cocktail of fungal protease inhibitors (Sigma Aldrich) and phosphatase inhibitors (Thermo Scientific), for 10 min. The BCA protein assay (Pierce) was used to measure protein concentrations, and equal amounts of proteins were boiled for 10 min in 5 × protein-loading buffer before loading (0.313 M Tris-HCl, pH 6.8 at 25°C, 10% SDS, 0.05% bromophenol blue, 50% glycerol; Fermentas). After separation by SDS–PAGE in 12% polyacrylamide gels, the protein extracts were blotted onto Immobilon PVDF membranes (Millipore) and incubated with: anti-phospho-p38 antibodies (Rabbit, Cell Signaling, LOT21, #9211BC) for phosphorylated Hog1; anti-MYC-tag antibodies (Rabbit, Abcam, ab9106) for tagged Pbs2; and anti-rabbit secondary antibodies (Cell Signaling Technology) conjugated with horseradish peroxidase. The Rapid Step ECL Reagent (Calbiochem) and a LAS-1000 camera (Fuji) were used for visualization of the signals on the membranes.
Fluorescence Microscopy
The EVs and plasmids pAZ301+ScSHO1, pAZ301+WiSHO1, and pAZ301+ScSHO1WiSH3 where the proteins were C-terminally tagged with GFP, were transformed into S. cerevisiae sho1Δssk2/22Δ cells. Positive transformants were grown in YNB-URA medium to mid-exponential growth phase in a rotary shaker (180 rpm, 30°C), and the OD600 was adjusted to 0.5. Before the preparation of microscope slides, the cells were exposed to 0.5 M NaCl for 5 min, washed three times in PBS, fixed in 3% paraformaldehyde, and resuspended in distilled water. Then the cells were dried onto poly-D-lysine–coated slides, mounted with Vectashield (Vector Laboratories), and imaged at room temperature with a camera (AxioCam Mrm) on an Axio Imager M2 (Carl Zeiss), equipped with a Plan-Apochromat objective (63 × /1.4), and an illuminator (HXP 120 C). For each field, GFP images (excitation, 488 nm; detection, 505–530 nm) and differential interference contrast images were captured using the ZEN 2012 software (Carl Zeiss).
Fludioxonil Sensitivity Assays
The cells of W. ichthyophaga were grown to mid-exponential phase at 28°C and 180 rpm in YNB medium supplemented with 3.4 M NaCl (Carlo Erba Reagents). Then 30 mL cell culture was harvested by centrifugation (4600 × g, 10 min, 25°C), and the pellet was washed twice with 30 mL modified OS buffer (50 mM succinic acid, 1 M NaCl, 2.75 M sorbitol, pH 7.5) using the same centrifugation parameters. The pellet was then resuspended in 10 mL modified OS buffer that contained 32 mg cell-wall-degrading enzyme cocktail Caylase (Cayla), and incubated at 25°C and 50 rpm for 1.5 h to disintegrate cellular clumps. Next, the cells were harvested and resuspended in 5 mL YNB containing 2.75 M sorbitol, and then they were 10-fold diluted in YNB containing 2.75 M sorbitol, 100 μg/mL fludioxonil (Sigma Aldrich), and 0.75% low-melting point agarose, and poured onto YNB agar plates that contained different NaCl concentrations (i.e., 2.5, 3.4, 4.3 M) and 100 μg/mL fludioxonil. The plates were incubated at 28°C and their growth was scanned after 7–10 days.
Results
Identification of Orthologous Hog-Pathway Proteins in the Genome of W. ichthyophaga
We searched for homologs of the S. cerevisiae HOG pathway in W. ichthyophaga (Figure 1). Additionally we also searched for a homolog of the histidine kinase Tco1 (CnNik1) as it has been shown to be involved in the HOG pathway of C. neoformans (Bahn et al., 2006). We were unable to identify homologs of the sensory mucins ScMsb2 and ScHkr1, and the anchor ScOpy2. However, we did identify homologs of the WiSho1, WiCdc24, WiCdc42, WiSte20, WiCla4, and WiSte11 proteins, which all show similar domain structures to the homologs from S. cerevisiae. Additionally, we determined that WiSte50 contains two SH3 domains, which are absent in ScSte50, in addition to the characteristic Ras-association domain (CDD accession number pfam00788) and Sterile alpha motif (CDD accession number cl15755; Figure 1; for GenBank accession numbers of W. ichthyophaga HOG-pathway proteins please see Supplemental Table S5).
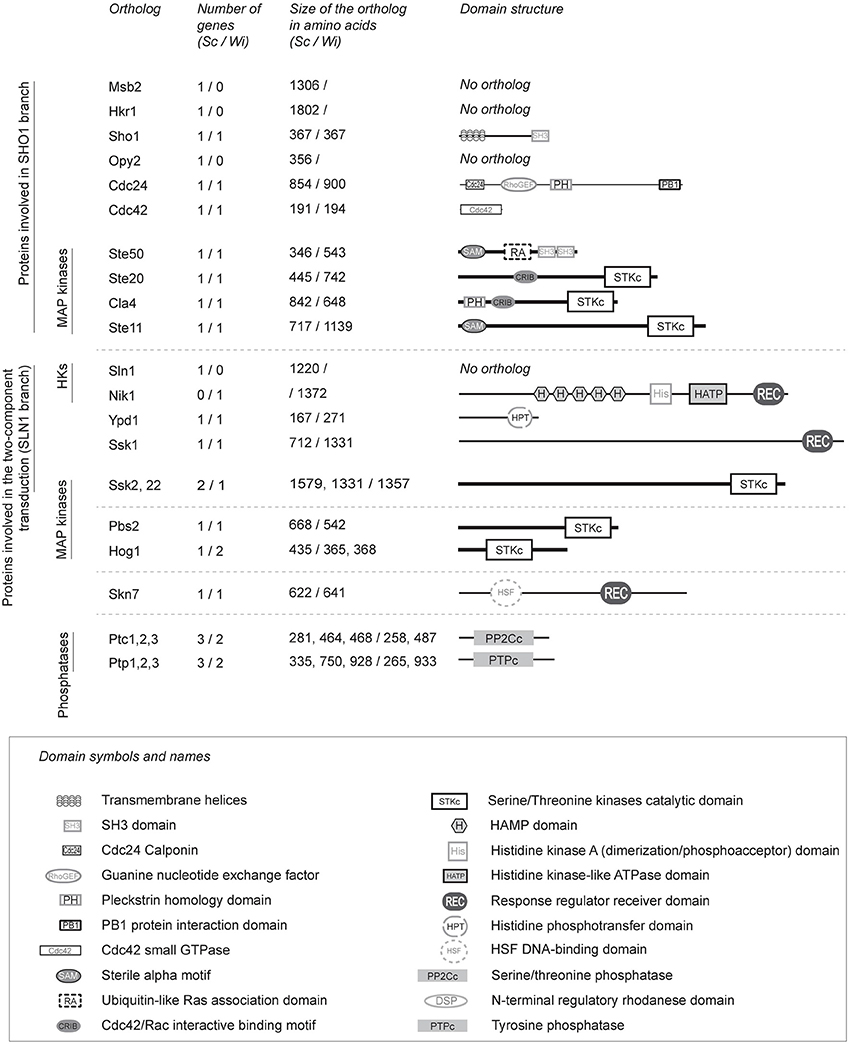
Figure 1. Details of the orthologous proteins of the S. cerevisiae HOG signaling pathway in W. ichthyophaga. The HOG signaling proteins were identified in the genome of W. ichthyophaga with the local BLAST algorithm using known HOG pathway proteins as probes (Supplemental Table S3). Sc (S. cerevisiae) and Wi (W. ichthyophaga) in column headers indicate the source organism of the data. The lengths of the illustrated domain structures correspond to the actual sizes of the orthologs. HKs, histidine kinases. For the GenBank accession numbers of W. ichthyophaga proteins, please see Supplemental Table S5. Domain annotations were retrieved from NCBI CDD (Marchler-Bauer and Bryant, 2004), according to: SH3 domain, pfam00018; Cdc24 calponin, pfam06395; guanine nucleotide exchange factor, cd00160; pleckstrin homology domain, cd13246; PB1 protein interaction domain, cd05992; Cdc42 small GTPase, cd01874; Sterile alpha motif, cd09533; Ubiquitin-like Ras-association domain, cd01786; Cdc42/Rac-interactive binding motif, cd01093; serine/ threonine kinases catalytic domain, smart00220; HAMP domain, cd06225; histidine kinase A dimerization/phosphoacceptor domain, cd00082; histidine kinase-like ATPase domain, cd00075; response regulator receiver domain, pfam00072; histidine phosphotransfer domain, cd00088; HSF DNA-binding domain, pfam00447; serine/ threonine phosphatase, cd00143, tyrosine phosphatase, cd00047.
The results from our BLAST searches showed that the core phosphorelay system of the SLN1-like branch is well conserved in W. ichthyophaga. This system contains WiYpd1 and WiSsk1, the kinase WiSsk2, and the response regulator WiSkn7 (its ortholog, ScSkn7, interacts with ScYpd1, and in S. cerevisiae it is involved in the hypo-osmotic response; Saito and Posas, 2012). However, we were unable to detect an ortholog of ScSln1 in W. ichthyophaga. Instead, we did find the Group III cytosolic histidine kinase, WiNik1, which contains the Histidine kinase, Adenylyl cyclase, Methyl-accepting protein, and Phosphatase (HAMP; CDD accession number cl01054) domain repeats. The genome of S. cerevisiae does not contain Nik1, but its orthologs are involved in osmosensing in other fungi (Bahn, 2008; El-Mowafy et al., 2013).
The HOG pathway in S. cerevisiae is regulated by negative-feedback through the action of phosphothreonine and phosphotyrosine phosphatases: ScPtc1, ScPtp2, and ScPtp3 (Hohmann et al., 2007). In the genome of W. ichthyophaga, we identified two phosphothreonine phosphatases—WiPtc1 (homolog of ScPtc1), and WiPtc3 (homolog of ScPtc3 and ScPtc2), as well as two phosphotyrosine phosphatases—WiPtp1 (homolog of ScPtp1), and WiPtp3 (homolog of ScPtp3, which is a broad-range phosphatase).
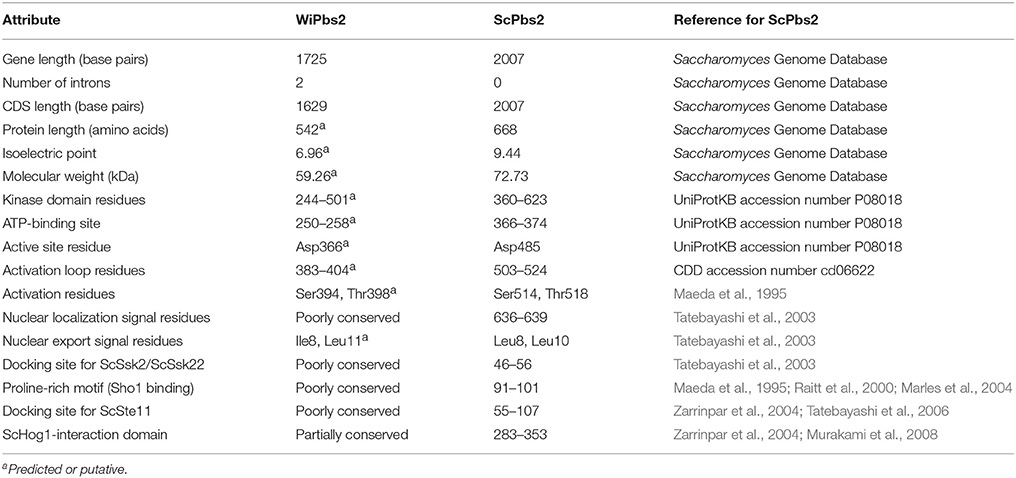
Comparison of WiPbs2 with ScPbs2, and with Other Fungal Pbs2-Like Kinases
As Pbs2-like kinases are the central element of fungal HOG pathways (Krantz et al., 2006b), we further analyzed and compared WiPbs2 with ScPbs2 (the comparison is summarized in Table 1), to identify any conserved elements of this protein. The WiPBS2 gene (GenBank accession number EOR04233) has 1725 nucleotides and contains two introns, one 47-bp-long (from nucleotide 1096–1142) and the other 49-bp-long (from nucleotide 1269–1317). We predict the remaining 1629 nucleotides to encode a 542 amino acid protein with a 542 amino acid protein with a theoretical isoelectric point (pI) of 6.96 and a predicted molecular weight of 59.26 kDa. Amino-acid alignment of the orthologous Pbs2-like kinases reveals poor conservation of the N-terminal region, but higher conservation of the C-terminal region. The C-terminus contains the kinase domain (Supplemental Figure S1, a region located within the two asterisks in the alignment), which starts six amino acids before the well-conserved ATP-binding site. High conservation was observed in the core of the kinase domain, especially in the active site and the activation loop that contains the SLAKT motif. This motif is important for the activation of the kinase, with its Ser and Thr residues phosphorylated by MAPKKK (Maeda et al., 1995; Supplemental Figure S1). Moreover, just after the end of the kinase domain in ScPbs2, there is a nuclear localization signal (Tatebayashi et al., 2003). This YITE sequence is poorly conserved in WiPbs2, as in other selected Pbs2 kinases (Supplemental Figure S1).
The N-terminal region of ScPbs2 contains important motifs for interactions with its protein partners (Figure 2A). ScPbs2 residues 4–18 contain a nuclear export signal, with indicative Leu residues at positions 8 and 10 (Tatebayashi et al., 2003). In WiPbs2, Ile at position 8 and Leu at position 11 might serve the same function. ScPbs2 residues 46–56 are important for SLN1 signaling and contain the ScSsk2/ScSsk22 docking site (Tatebayashi et al., 2003; Figure 2A). In agreement with earlier investigations, we observed that the Ala52, Arg53, and Ala56 motif is conserved in yeast; however, in many filamentous and basidiomycetous fungi it is not (Krantz et al., 2006b; Figure 2A). Moreover, the proline-rich motif of S. cerevisiae represented by residues 91–101, which is crucial for ScSho1 binding and signaling through the SHO1 branch (Maeda et al., 1995; Raitt et al., 2000; Marles et al., 2004; Figure 2A) is also well conserved in the majority of yeast (Krantz et al., 2006a). In contrast, we found that this motif is not conserved in either W. ichthyophaga or in the selected filamentous and basidiomycetous fungi shown in Figure 2A. Other important features in ScPbs2 are the docking site for ScSte11, which has been mapped to residues 55–107 (Zarrinpar et al., 2004; Tatebayashi et al., 2006) and overlaps with the ScSsk2/ScSsk22 docking site, and the ScHog1-interaction domain (Figure 2A). Our alignment revealed that the Ste11 docking site in Pbs2-like kinases from W. ichthyophaga and selected fungi is only poorly conserved (Figure 2A). Similarly, the region proximal to the kinase domain (residues 283–353), which in S. cerevisiae is required for signal transmission from ScPbs2 to ScHog1 (Zarrinpar et al., 2004; Murakami et al., 2008; Figure 2A, ScHog1-interaction domain), is only partially conserved in the aligned Pbs2 kinases. These sequences show notable conservation only downstream of the ScPbs2 Thr297 (Figure 2A).
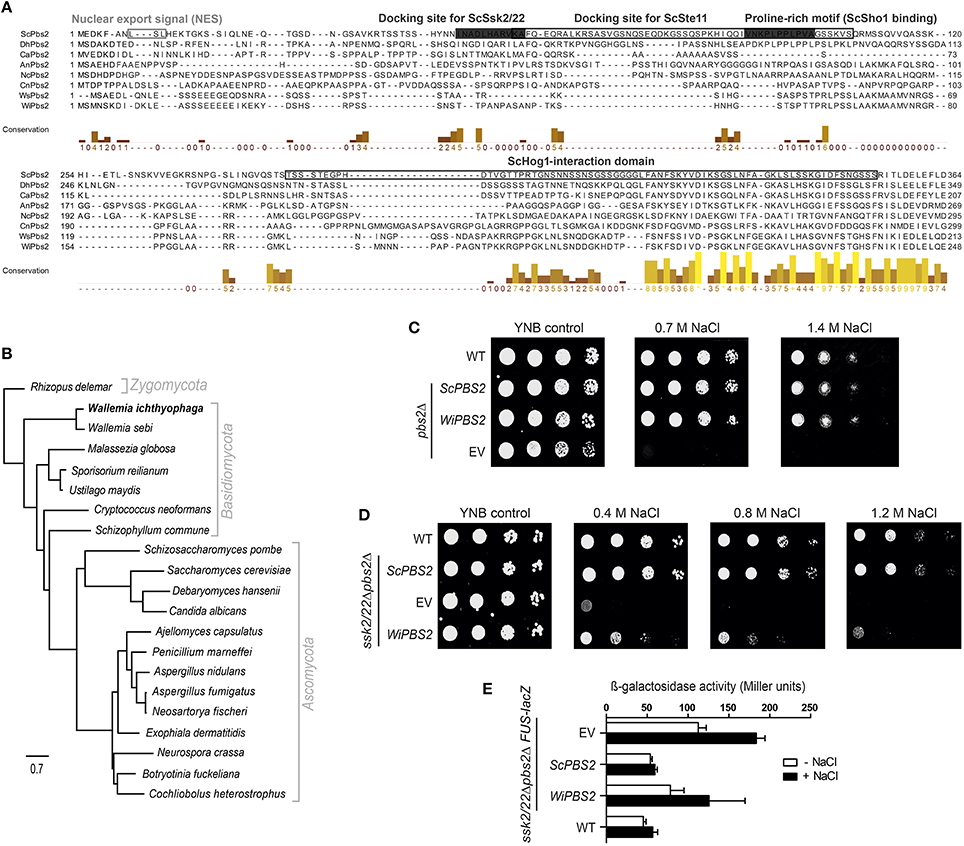
Figure 2. Characterization of WiPbs2. (A) Partial protein alignment of selected Pbs2 kinases. Prefixes indicate the source organism of Pbs2: Sc, S. cerevisiae; Dh, D. hansenii; Ca, C. albicans; An, A. nidulans; Nc, N. crassa; Cn, C. neoformans; Ws, W. sebi; Wi, W. ichthyophaga. Important domains, motifs and sites are highlighted. Framed boxes, ScPbs2 nuclear export signal, docking site for ScSte11, and ScHog1 interaction domain; gray boxes, ScPbs2 docking site for ScSsk2/ScSsk22, and proline-rich motifs for ScSho1 binding. The columns demonstrate conservation of amino acids (higher and lighter, greater conservation). See Supplemental Table S3 for the GenBank accession numbers. (B) Maximum likelihood phylogenetic tree of orthologous Pbs2 kinases arbitrarily rooted to Rhizopus delemar. (C) Representative halotolerance rescue assay. The S. cerevisiae Y00000 WT strain and pbs2Δ strain were transformed as the EV positive control (WT) and pbs2Δ negative control (EV), respectively, and pbs2Δ with ScPBS2 (positive control) or WiPBS2. (D) Representative halotolerance rescue assay with the W303 wild-type strain (WT) and the ssk2/22Δpbs2Δ strain transformed as in (C). (E) Cross-talk activation of the mating pathway, as β-galactosidase activity, in ssk2/22Δpbs2ΔFUS-lacZ cells transformed as in (C). Activities were measured before and after 1 M NaCl for 4 h. Data are means ± SD (n = 3). Box indicating docking site is to dark.
These findings are supported by the maximum likelihood phylogenetic tree in which we compared the identified WiPbs2 sequence to orthologs of other fungi (Figure 2B). This phylogenetic tree was arbitrarily rooted to Rhizopus delemar. The yeasts grouped in a clade that is clearly separated from filamentous and other fungi. On the other hand, the MAPKK Pbs2 from W. ichthyophaga is the most similar to the MAPKK from Wallemia sebi, and as expected, these are both positioned at the base of Basidiomycota, near Zygomycota.
Although the ScPbs2-interaction motifs are not highly conserved in WiPbs2 kinase (Figure 2A) we wanted to examine if WiPbs2 can complement the function of the missing ScPbs2, when expressed in S. cerevisiae pbs2Δ cells (Figure 2C). The growth of the WiPbs2-expressing cells was similar to the cells expressing ScPbs2 and the WT cells, regardless of the salt concentration used (Figure 2C; 0.7 M NaCl, 1.4 M NaCl). The cells transformed with EV were used as the negative control, and as expected, their growth was only evident on the YNB-LEU plates (Figure 2C, YNB control). This showed that in the pbs2Δ background, WiPbs2 kinase can completely replace the function of the missing ScPbs2 when cells are grown in salt.
WiPbs2 Is Not Efficiently Activated by the SHO1 Branch in S. cerevisiae
In S. cerevisiae, the native ScPbs2 kinase can be activated by either the SHO1 or the SLN1 branch of the HOG pathway (Hohmann et al., 2007). Although the results from Figure 2C show that WiPbs2 fits into the HOG pathway of S. cerevisiae with no observed differences compared to ScPbs2, in these pbs2Δ cells it is not possible to discriminate through which branch the WiPbs2 kinase was activated (i.e., SHO1 or SLN1). Therefore, we next expressed the WiPbs2 kinase in S. cerevisiae ssk2/22Δpbs2Δ cells. Due to the Ssk2/Ssk22 deletion in this strain, the SLN1 branch was inactive and the signal was transferred to the WiPbs2 kinase only via the SHO1 branch. We transformed the ssk2/22Δpbs2Δ cells with WiPbs2 or ScPbs2, and plated them on YNB-TRP medium. The strain ssk2/22Δpbs2Δ with EV was used as the negative control, and the WT strain as the positive control. The data presented in Figure 2D show that the growth of these cells expressing ScPbs2 was similar to the WT cells (Figure 2D, ScPBS2, WT) under all of the tested salinities. Compared to the ScPbs2-expressing and WT cells, the growth of the WiPbs2-expressing cells was reduced, and was almost completely inhibited at 1.2 M NaCl (Figure 2D, WiPBS2).
These data were confirmed with the β-galactosidase assay (Figure 2E). In these assays, we monitored the activation of Fus3 kinase in S. cerevisiae under salt stress, because the signal leaks from the HOG pathway to the mating pathway when some of the components are missing or do not fit optimally into the HOG pathway (O'Rourke and Herskowitz, 1998). The induction of the FUS-lacZ reporter upon osmotic shock doubled in the WiPbs2-expressing cells as compared to the cells with ScPbs2 and the WT cells (Figure 2E, WiPBS2, ScPBS2, WT). These data suggested that in this S. cerevisiae system, WiPbs2 was not optimally activated by the SHO1 branch of the pathway. This is most likely due to inefficient tethering or activation by the ScSho1 osmosensor or the ScSte11 MAPKKK, respectively.
Characterization of Putative Osmosensor WiSho1 from W. ichthyophaga
To investigate the differences in the architectures of the SHO1-branch components in W. ichthyophaga, the WiSho1 protein was analyzed. WiSho1 (GenBank accession number KUR62632) is a 367-residue-long protein with a theoretical pI of 6.86 and a molecular weight of 39.22 kDa. This was deduced from the 1104-bp CDS, which is the product of the 1148-bp gene that includes one intron (44 bp long, between nucleotides 406 and 449). The N-terminal part of this WiSho1 protein contains four transmembrane helices and a characteristic SH3 domain (CDD accession number cd11855) at the C-terminus (Figure 3A). In ScSho1, the residues Pro63, Phe65, Tyr106, Thr108, and Pro120 are important for interactions with the putative osmosensing mucines ScHkr1 and ScMsb2 (Tatebayashi et al., 2007). The alterations of these residues resulted in either constitutive signaling activation (for Pro120 variants) or defective signaling (for Pro63, Phe65, Tyr106, and Thr108 variants). However, these residues are not conserved in WiSho1 (Figure 3A, black arrows). In S. cerevisiae, Hog1 phosphorylation of ScSho1 at Ser166 (Figure 3A, black arrow) causes its monomerization and regulates the pathway in a negative-feedback manner (Hao et al., 2007). Although this position is not conserved in WiSho1, there are other Ser residues in this region (i.e., original WiSho1 positions 164, 166, 167, 174, 175), which might serve as phosphorylation sites (Figure 3A). Also, the region that is important for interactions with ScSte11 (residues 172–211; Zarrinpar et al., 2004), is not conserved in the Sho1 orthologs used in our alignment (Figure 3A, Ste11 interaction domain). In contrast, the SH3 domain located at the C-terminus is highly conserved in WiSho1 and in the aligned Sho1-like proteins. Interestingly, there is an insertion of Gly that is only seen for the Sho1 SH3 domains of W. ichthyophaga and W. sebi (Figure 3A, SH3 domains).
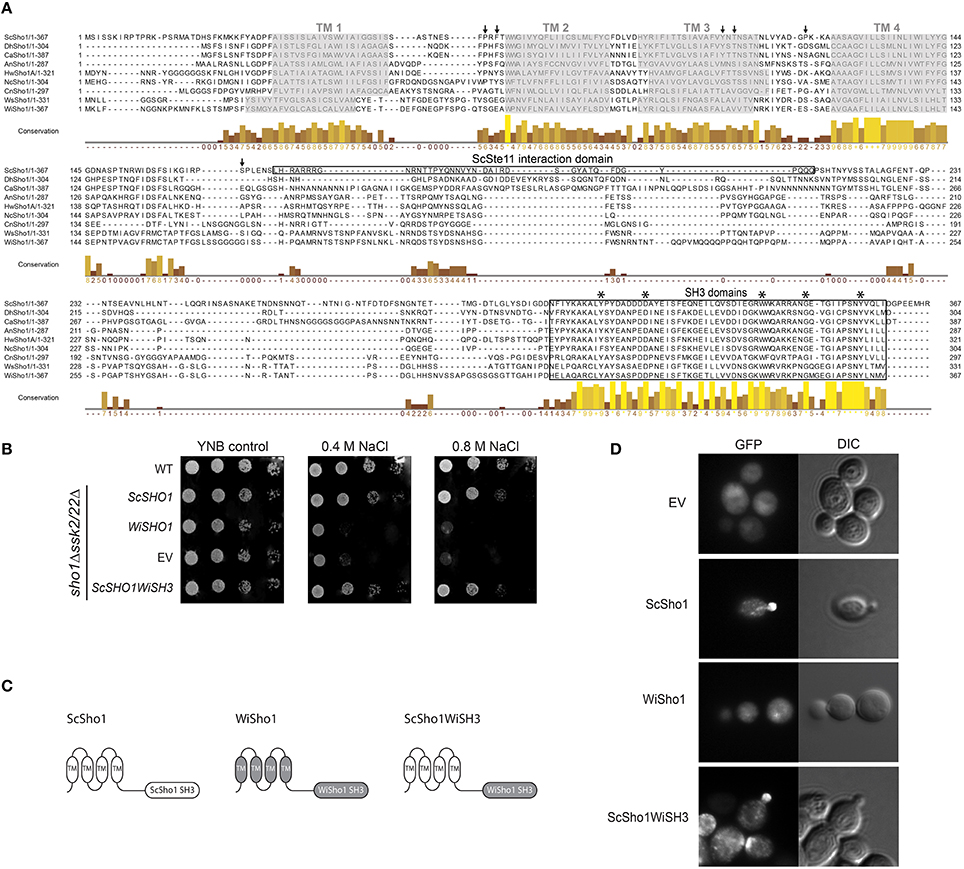
Figure 3. Bioinformatics and functional analysis of WiSho1. (A) Protein alignment of selected orthologous Sho1 proteins. Prefixes indicate the source organism of Sho1, as in Figure 2A. Gray boxes, putative transmembrane helices (TM1-4); black arrows, ScSho1 residues important for interactions with mucines Hkr1 and Msb2 (Pro63, Phe65, Tyr106, Thr108, Pro120) or for monomerisation (Ser166); framed boxes, ScSte11 interaction domain and SH3 domains; asterisks, important residues for ScSho1 SH3 domain function. The columns demonstrate conservation of amino acids (higher and lighter, greater conservation). See Supplemental Table S3 for the GenBank accession numbers. (B) Representative halotolerance rescue assay. The W303 wild-type strain and sho1Δssk2/22Δ strain were transformed as the EV positive control (WT) and sho1Δssk2/22Δ negative control (EV), respectively, and sho1Δssk2/22Δ with ScSHO1 (positive control), WiSHO1, or the ScSHO1WiSH3 hybrid. (C) Cartoons of ScSho1, WiSho1, and the ScSho1WiSH3 hybrid. (D) Representative fluorescence microscopy images (GFP) and differential interference contrast (DIC) of sho1Δssk2/22Δ cells transformed as vector alone (EV, negative control) or for GFP-tagged ScSho1, WiSho1, and ScSho1WiSH3.
To determine whether WiSho1 can complement the function of ScSho1, we expressed the WiSho1 protein in sho1Δssk2/22Δ cells. The data presented in Figure 3B show that the cells that expressed WiSho1 did not grow in the presence of NaCl (Figure 3B, WiSHO1, 0.4 M, 0.8 M NaCl). The growth of these cells was reduced to the same level as the EV cells (Figure 3B, EV).
We hypothesized that the SH3 domain of WiSho1 is functional. Therefore we constructed a chimera of the ScSho1 and WiSho1 proteins. We constructed this ScSho1WiSH3 hybrid protein (Figure 3C, ScSho1WiSH3, right) by replacing the SH3 domain of ScSho1 (Figure 3C, ScSho1 SH3, left) with the WiSho1 SH3 domain (Figure 3C, WiSho1 SH3, middle). Indeed, expression of the hybrid ScSho1WiSH3 protein (Figure 3B, ScSHO1WiSH3) recovered the osmosensitivity of these sho1Δssk2/22Δ cells. This result confirmed that the WiSho1 SH3 domain is functional when it is integrated into ScSho1.
The potential reason for the osmosensitivity of the WiSho1-expressing S. cerevisiae cells is that WiSho1 does not undergo trafficking to the yeast plasma membrane. To investigate this, we tagged the ScSho1, WiSho1, and ScSho1WiSh3 proteins with GFP and analyzed their localizations under fluorescent microscopy. The fluorescence of the WiSho1-GFP was distributed equally within the cell, and resembled that of the EV cells expressing only the GFP tag (Figure 3D, WiSho1, EV). This indicated either false folding of WiSho1-GFP or cleavage of the GFP from the membrane protein. However, the ScSho1WiSH3-GFP hybrid protein was localized predominantly in the plasma membrane at places of polarized growth (Figure 3D, ScSho1WiSH3), which was similar to ScSho1-GFP (used as the positive control; Figure 3D, ScSho1). In conclusion, this functional complementation and appropriate localization of the ScSho1WiSH3 hybrid protein indicates that although full-length WiSho1 did not complement the function of ScSho1 due to its incorrect localization, when the conserved SH3 domain from WiSho1 was inserted into the ScSho1WiSH3 hybrid protein, the HOG activation signal was transmitted to ScPbs2.
Lower Affinity between WiPbs2 and ScSho1WiSH3 Results in Lower Osmotolerance
As the SH3 domain from WiSho1 can interact with ScPbs2, we investigated whether the WiSho1 SH3 domain can also bind to WiPbs2 kinase. We first performed in-silico analysis of the interactions between the WiPbs2 proline-rich motif and the WiSho1 SH3 domain. WiPbs2 was submitted to examination with the Eukaryotic Linear Motif Resource (Dinkel et al., 2012) for identification of proline-rich motifs. Multiple PXXP motifs were identified in the N-terminal part of WiPbs2; however, only the APLPPNQLR peptide (i.e., residues 117–125; E-value, 1.237e−03) met the criteria for being recognized by class I SH3 domains (CDD accession number pfam00018; Saksela and Permi, 2012). Figure 4A shows the alignment of the ScSho1 SH3 domain, the WiSho1 SH3 domain, and the Fyn kinase SH3 domain (Figure 4A, ScSho1 SH3, WiSho1 SH3, Fyn SH3), which highlights the residues that are important for interactions with the proline-rich motif. Alterations of the Tyr309 and Asp317 residues in the RT loop, and the Tyr355 residue, have been reported to lower the binding affinity between the ScSho1 SH3 domain and the ScPbs2 proline-rich motif (Marles et al., 2004; Figure 4A, black framed boxes). Moreover, Trp338 is essential for proline-rich motif binding, while Arg342 and Gly346 replacement causes constitutive activation of the HOG pathway with these altered ScSho1 proteins (Tatebayashi et al., 2006; Figure 4A, gray boxes). All of these residues are well conserved in the WiSho1 SH3 domain. However, in the alignment of the WiPbs2 and ScPbs2 proline-rich motifs shown in Figure 4B (ScPbs2 PRO, WiPbs2 PRO), although the Leu95 and Pro96 of ScPbs2 that have been indicated as essential for signal transduction (Figure 4B, gray-boxed; Maeda et al., 1995; Tatebayashi et al., 2003) are conserved in WiPbs2, the Lys93 of ScPbs2 that has been indicated as important for mediating interactions with acidic residues from SH3 domain (Wu et al., 1995; Marles et al., 2004) is replaced by alanine in WiPbs2.
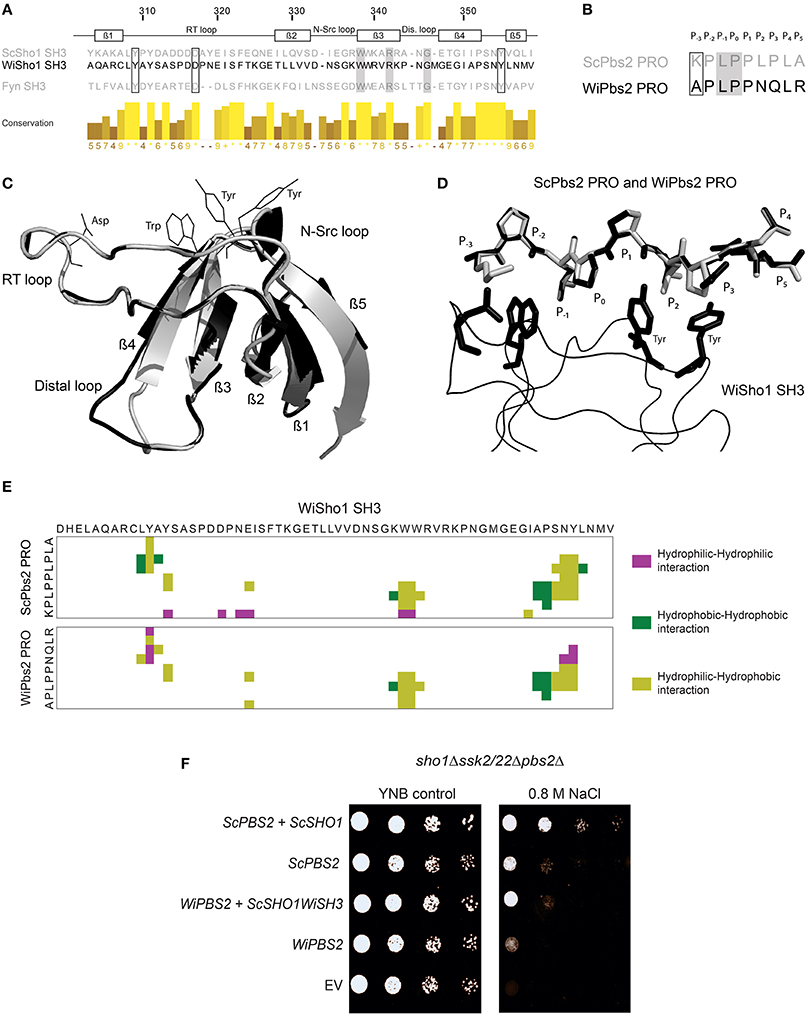
Figure 4. Interaction between WiSho1 and WiPbs2. (A) Alignment of SH3 domains of ScSho1, WiSho1, and the human tyrosine kinase Fyn. β-Sheet and loop regions are denoted above. Framed and gray boxes, important residues for function of the Sho1 SH3 domains (Marles et al., 2004; Tatebayashi et al., 2006). The columns demonstrate conservation of amino acids (higher and lighter, greater conservation). (B) Alignment of proline-rich motifs (PRO) of ScPbs2 and WiPbs2, indicating sites of binding into the SH3 domain pocket (P−3–P5). Framed and gray boxes, important sites for interactions and signal transduction, respectively. (C) Model of superposition of ScSho1 SH3 structure (gray) and WiSho1 SH3 (black). Positions of conserved Asp, Trp, and two Tyr residues of the WiSho1 SH3 domain that are important in the ScSho1 SH3 domain for proline-rich motif binding are shown with black lines. (D) Model of WiSho1 SH3 (black ribbon) interaction with ScPbs2 PRO (gray sticks) and the superimposed WiPbs2 PRO (black sticks). Sites of binding are indicated as P−3–P5 and the important Asp, Trp, and two Tyr residues in WiSho1 SH3 are highlighted (black sticks). (E) COCOMAPS property maps, which show the hydrophilic-hydrophilic (purple), hydrophobic-hydrophobic (dark green), and hydrophilic-hydrophobic (olive) residue interactions in the 7 Å atom-to-atom distance from the model of WiSho1 SH3 interaction with ScPbs2 PRO (upper panel) and WiPbs2 PRO (lower panel). (F) Representative halotolerance rescue assay. The sho1Δssk2/22Δpbs2Δ strain was transformed as the negative control (EV) and with WiPBS2 or ScPBS2, and ScSHO1, WiSHO1, or ScSHO1WiSH3.
To further investigate the similarities between these proteins, we modeled the interactions of the WiSho1 SH3 domain and the WiPbs2 proline-rich motif with the SWISS-MODEL software, based on the crystal structure of the ScSho1 SH3 domain complexed with the ScPbs2 proline-rich motif (PDB: 2VKN). The best model that showed reliable GMQE and QMEAN4 values (Biasini et al., 2014) of 0.78 and −1.85, respectively, is illustrated in Figures 4C,D. Superposition of the WiSho1 SH3 domain model on the ScSho1 SH3 domain from the 2VKN structure in Figure 4C shows good alignment. The only important differences detected for this WiSho1 SH3 domain were shorter β4 and β5 sheets, a longer β2 sheet, and a larger distal loop. However, these differences did not influence the positions of the side chains of the conserved residues in the SH3 domains (Figure 4A) that are important for interactions with the proline-rich motif (Figure 4C, Asp, Trp, two Tyr residues, indicated with black lines).
Figure 4D shows the model of the interaction of the proline-rich motifs from ScPbs2 (Figure 4D, gray sticks) and WiPbs2 (Figure 4D, black sticks) with the WiSho1 SH3 domain (Figure 4D, black ribbon). The conserved homologous residues in the SH3 domain that are important for the interactions with the proline-rich motif are also indicated in Figure 4D by black sticks (i.e., WiSho1 SH3, Asp, Trp, two Tyr residues). The WiPbs2 APLPPNQLR proline-rich motif fits well to the major part of the SH3 hydrophobic surface (Figure 4D, P−2–P2). However, in the WiPbs2 motif, there are the Asn and Gln residues at positions P2 and P3 respectively, whereas in the ScPbs2 motif these are Lys and Pro (Figures 4B,D). As these Lys and Pro residues from ScPbs2 form hydrophobic interactions with the rings of two Tyr residues at the end of the SH3 hydrophobic surface (Wu et al., 1995; Figure 4D), the Asn and Gln might result in a lower affinity for the binding of the WiPbs2 proline-rich motif to the WiSho1 SH3 domain. Moreover, there is a missing interaction with the acidic Asp residue of the WiSho1 SH3 domain due to the Ala residue at position P−3 in the WiPbs2 proline-rich motif (Figure 4D) that might further lower this affinity. However, our estimation of lower affinity only considers the literature-reported residues that are important for interactions between the of SH3 domain and the ScPbs2 proline-rich motif (Wu et al., 1995). We therefore used the COCOMAPS software (Vangone et al., 2011) to cover all possible residue interactions at a 7 Å atom-to-atom distance. In the property maps shown in Figure 4E, the software reports 377 Å2 of interaction area, which is formed by nine residues in the ScPbs2 proline-rich motif that are involved in 44 interactions with 18 residues of the WiSho1 SH3 domain (Figure 4E, upper panel). On the other hand, for WiPbs2 PRO-WiSho1 SH3 model, the interaction area of 337 Å2 was calculated, as formed by nine residues of the WiPbs2 proline-rich motif that make 38 interactions with 14 residues of WiSho1 SH3 domain (Figure 4E, lower panel). Taking these in-silico data together, they predict that the interaction of the proline-rich motif from WiPbs2 with the WiSho1 SH3 domain will be weaker than that between the ScPbs2 proline-rich motif and the WiSho1 SH3 domain.
To determine whether this predicted weaker interaction of the WiPbs2 and the WiSho1 SH3 domain is also reflected in lower osmotolerance of S. cerevisiae, in the sho1Δssk2/22Δpbs2Δ cells we simultaneously expressed the kinase WiPbs2 and ScSho1WiSH3 (Figure 4F). Indeed, the growth of these WiPbs2-expressing and ScSho1WiSH3-expressing cells was reduced in comparison to the positive-control cells expressing ScPbs2 and ScSho1 (Figure 4F, WiPBS2 + ScSHO1WiSH3, ScPBS2 + ScSHO1), and similar to the cells expressing only the ScPbs2 kinase (Figure 4F, ScPBS2). The growth of the cells expressing only WiPbs2 (Figure 4F, WiPBS2) was even more reduced, and in the case of the EV (Figure 4F, EV; negative control), the growth was completely inhibited. This supports the results of the in-silico prediction that the WiSho1 SH3 domain binds WiPbs2 with lower affinity.
Insertion of the ScPbs2 Proline-Rich Motif into WiPbs2 Results in Stronger Interactions with ScSho1WiSH3
Upon SHO1 branch activation in S. cerevisiae, ScSho1 recruits ScPbs2 to the membrane, where it is activated via phosphorylation by the MAPKKK ScSte11. We additionally investigated whether inefficient activation by ScSte11 kinase, rather than weaker binding with the WiSho1 SH3 domain, was the reason for the lower osmotolerance of these cells expressing WiPbs2 and ScSho1WiSH3 (Figure 4F, WiPBS2 + ScSHO1WiSH3). Using oligonucleotide-directed mutagenesis, we constructed an altered WiPbs2 protein (Figure 5A, WiPbs2ScPRO), which contained an artificial insertion of the proline-rich motif VNKPLPPLPVAG from ScPbs2 (Figure 5A, ScPbs2) in the N-terminal portion of WiPbs2 (Figure 5A, WiPbs2). When this WiPbs2ScPRO mutant protein was expressed simultaneously with the ScSho1WiSH3 protein in the sho1Δssk2/22Δpbs2Δ cells, the osmotolerance of these cells was enhanced on 1 M NaCl in comparison to the cells expressing WiPbs2 and ScSho1WiSH3 (Figure 5B, WiPBS2ScPRO + ScSHO1WiSH3, WiPBS2 + ScSHO1WiSH3). This indicates that the insertion of the ScPbs2 proline-rich motif into WiPbs2 leads to stronger interactions with ScSho1WiSH3, and consequently to more efficient pathway activation and greater osmotolerance. Moreover, this also suggests that the interaction between WiPbs2 and the WiSho1 SH3 domain, rather than WiPbs2 and ScSte11, is the limiting step in WiPbs2 activation via the SHO1 branch of the HOG pathway.
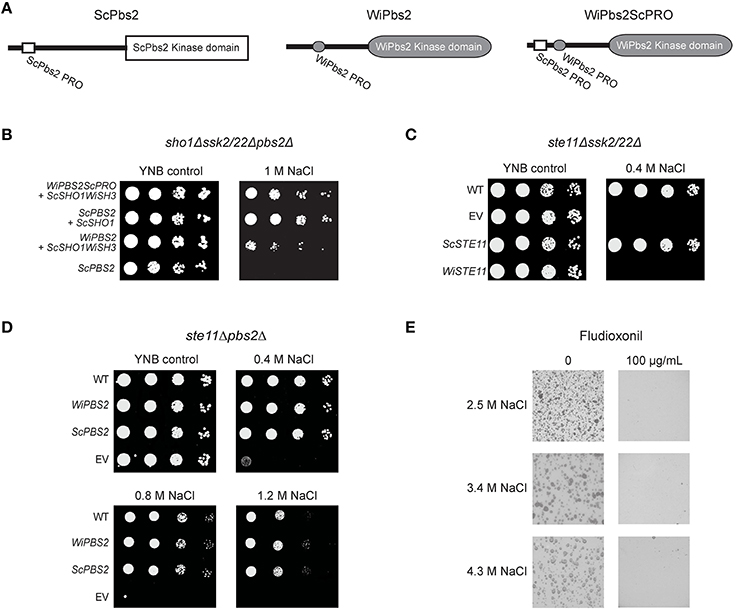
Figure 5. Expression of WiPbs2 with inserted ScPbs2 proline-rich motif. (A) Schematic representation of ScPbs2, WiPbs2, and WiPbs2ScPRO. The proline-rich motif VNKPLPPLPVAG from ScPbs2 was inserted into the N-terminus of WiPbs2 to obtain the WiPbs2ScPRO altered protein. (B–D) Representative halotolerance rescue assays. (B) The sho1Δssk2/22Δpbs2Δ strain was transformed with the negative control (ScPBS2 + EV) and the positive control (ScPBS2 + ScSHO1), and with ScSHO1WiSH3 and WiPBS2ScPRO or WiPBS2. (C) The Y00000 wild-type strain and ste11Δssk2/22Δ strain were transformed as the EV positive control (WT) and ste11Δssk2/22Δ negative control (EV), respectively, and ste11Δssk2/22Δ with ScSTE11 (positive control) or WiSTE11. (D) The W303 wild-type strain and ste11Δpbs2Δ strain were transformed as the EV positive control (WT) and ste11Δpbs2Δ negative control (EV), respectively, and ste11Δpbs2Δ with ScPBS2 (positive control) or WiPBS2. (E) Representative plates of growth of W. ichthyophaga in the absence (left) and presence (right) of 100 μg/mL fludioxonil, on plates containing 1.7, 3.4, and 5.1 M NaCl.
However, despite this indication of efficient WiPbs2 activation by ScSte11 in S. cerevisiae, the expression of the W. ichthyophaga MAPKKK WiSte11 in ste11Δssk2/22Δ cells did not abolish the osmosensitivity of these cells (Figure 5C, WiSTE11). Instead, their growth was completely inhibited already at the low 0.4 M NaCl concentration (Figure 5C, WiSTE11). In contrast, the ScSte11-expressing cells (Figure 5C, ScSTE1; the positive control) were as osmoresistant as the WT cells (Figure 5C, ScSTE1, WT). Although the WiSte11 kinase contains the conserved N-terminal Sterile alpha motif and C-terminal Kinase domain (Supplemental Figure S2), the highly conserved phosphorylation sites for Ste20 (van Drogen et al., 2000; Supplemental Figure S2, Ser281, Ser285, Thr286), and the moderately conserved Sho1 binding region (Tatebayashi et al., 2006; Supplemental Figure S2), it is 422 amino acids larger than ScSte11 (Figure 1). This might be the reason for this inefficient functional complementation that results in high osmosensitivity of the WiSte11-expressing ste11Δssk2/22Δ cells.
Conserved ScSsk2/22-WiPbs2 Interaction and Fludioxonil Sensitivity Indicate the Importance of the Phosphorelay System in W. ichthyophaga
As the growth of the pbs2Δ cells expressing either WiPbs2 or ScPbs2 was identical (Figure 2C), these two kinases likely have similar roles in the SLN1 branch, which masks the less efficient activation of WiPbs2 by the SHO1 branch (Figures 2D, 4F). To confirm this, we expressed WiPbs2 in the ste11Δpbs2Δ cells, where only the two-component SLN1 branch was functional. Indeed, these data show that the growth of these WiPbs2-expressing cells is identical to the ScPbs2-expressing cells at all NaCl concentrations used (Figure 5D). This indiscriminate activation of WiPbs2 and the ScPbs2 kinases by the ScSsk2/ScSsk22 MAPKKK in S. cerevisiae indicates that interactions in this part of the pathway are evolutionarily highly conserved.
In contrast to S. cerevisiae, which has the two paralogs ScSsk2 and ScSsk22, the genome of W. ichthyophaga contains only one ortholog, WiSsk2. This is a 1357-amino-acid protein with a conserved kinase domain at its C terminus (Supplemental Figure S3). The S. cerevisiae ScSsk2 N-terminus contains the ScSsk1 binding domain (Posas et al., 1998; Supplemental Figure S3) and the activation domain (Horie et al., 2008; Supplemental Figure S3), which is also known as the autoinhibitory domain (Tatebayashi et al., 2003). Although the N-terminal sequence of WiSsk2 is not highly conserved (Supplemental Figure S3), the data in Figure 5D show that in S. cerevisiae the WiPbs2 kinase is preferentially activated via ScSsk2/ScSsk22 of the SLN1 branch.
The SLN1 branch in S. cerevisiae is controlled by the transmembrane histidine kinase ScSln1. W. ichthyophaga does not have the ScSln1 ortholog. Possibly, instead, the cytosolic Group III histidine kinase WiNik1 is involved in osmosensing and fludioxonil sensitivity, as has been shown for C. neoformans strains with constitutive CnHog1 phosphorylation (Bahn et al., 2007). W. ichthyophaga also shows constitutive WiHog1 phosphorylation under noninducing conditions (Konte and Plemenitaš, 2013), and this provides the prerequisite to test the sensitivity of cells to fludioxonil. In the presence of fludioxonil, the growth of W. ichthyophaga cells was completely inhibited at all NaCl concentrations used (Figure 5E).
Cells Expressing WiHog1A Have Greater Osmosensitivity and Show Greater Cross-Talk
When expressed in S. cerevisiae, WiHog1A only partially rescued the osmosensitive phenotype of hog1Δ cells (Konte and Plemenitaš, 2013). One of the possible reasons for this poor performance of WiHog1A in S. cerevisiae hog1Δ cells might be weaker activation by ScPbs2, which differs from WiPbs2 in its sequence, and is not the native activating kinase of WiHog1A. To address this, in S. cerevisiae pbs2Δhog1Δ cells, we expressed the MAPKKs WiPbs2 or ScPbs2 together with the MAPKs WiHog1A, WiHog1B, or ScHog1. In this way, we tested whether the salt-tolerance of the pbs2Δhog1Δ cells was improved after activation of the WiHog1 MAPKs by their native activating MAPKK, WiPbs2. The growth of the cells expressing both WiPbs2 and WiHog1A was reduced to the same level as the growth of the cells expressing ScPbs2 and WiHog1A (i.e., used as the control), when compared to the growth of the cells expressing either of the two Pbs2 MAPKKs (i.e., WiPbs2 or ScPbs2) together with WiHog1B or ScHog1 (Figure 6A, 0.5 M NaCl, 1 M NaCl). This is in agreement with our previous data in the S. cerevisiae hog1Δ cells, where only WiHog1B, and not WiHog1A, fully complemented the function of ScHog1 (Konte and Plemenitaš, 2013).
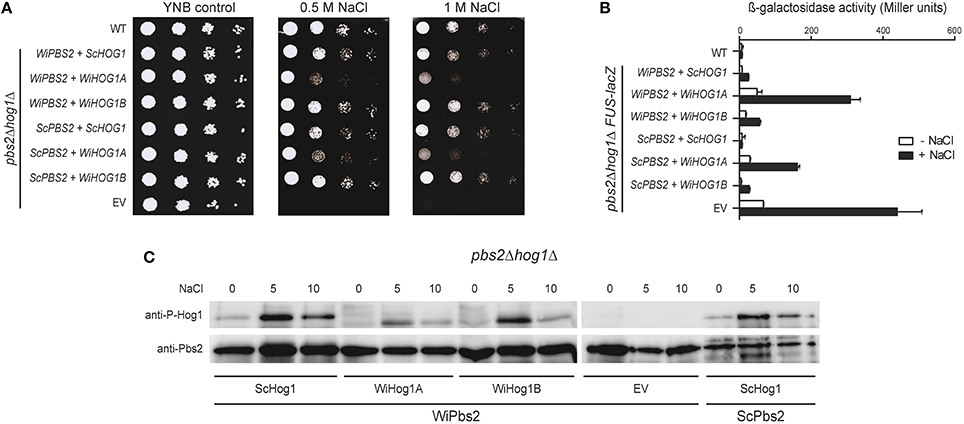
Figure 6. Simultaneous expression of WiPbs2, and WiHog1A or WiHog1B in S. cerevisiae pbs2Δhog1Δ cells. (A) Halotolerance rescue assay. The S1278b WT strain and pbs2Δhog1Δ strain were transformed as the EV positive control (WT) and pbs2Δhog1Δ negative control (EV), respectively, and pbs2Δhog1Δ with the indicated combinations of WiPBS2 or ScPBS2, and ScHOG1, WiHOG1A, or WiHOG1B. (B) Cross-talk activation of the mating pathway, as β-galactosidase activity, in S1278bFUS1lacZ and pbs2Δhog1ΔFUS-lacZ cells transformed as in (A). Activities were measured before and after 1 M NaCl for 4 h. Data are means ± SD (n = 3). (C) Western blotting of pbs2Δhog1Δ cells expressing WiPbs2 and ScHog1, WiHog1A, or WiHog1B. Expression of ScPbs2 and ScHog1 was the positive control, and of WiPbs2 alone (EV) was the negative control. Expressed Pbs2 and phosphorylated Hog1 kinases were immunodetected in lysates before and after (5, 10 min) application of 0.8 M NaCl. The proteins were immunodetected on two separate membranes as indicated by the spliced image.
To further confirm these data, we performed β-galactosidase assays (Miller, 1972) with the FUS-lacZ reporter. We integrated the FUS-lacZ reporter system into the genome of the S. cerevisiae pbs2Δhog1Δ and S1278b WT cells prior to transformation with WiPbs2 or ScPbs2, and WiHog1A, WiHog1B, or ScHog1. Figure 6B shows the results for the activation of this mating pathway upon 1 M NaCl shock, for 4 h. As expected, the cells with EV (Figure 6B, EV), which were the positive control, showed the greatest β-galactosidase activity (i.e., the highest cross-talk), while the WT cells (Figure 6B, WT) and the cells expressing ScPbs2 and ScHog1 (Figure 6B, ScPBS2+ScHOG1), both as negative controls, showed almost no activation when stressed with NaCl. In the cells expressing ScHog1 or WiHog1B, we detected low β-galactosidase activity regardless of the co-expressed MAPKKs ScPbs2 or WiPbs2 (Figure 6B, ScPBS2+WiHOG1B, WiPBS2+ScHOG1, WiPBS2+WiHOG1B). On the other hand, when WiHog1A was expressed simultaneously with ScPbs2, the induction of β-galactosidase reached more than 150 Miller units, and even doubled to 300 units when it was co-expressed with WiPbs2 (Figure 6B; ScPBS2+WiHOG1A, WiPBS2+WiHOG1A).
Finally, we directly determined the levels of activation of the Hog1 MAPKs by WiPbs2, to determine whether the reason for the poor performance of the cells expressing WiHog1A and WiPbs2 was a result of differential phosphorylation by WiPbs2. The cells expressing WiPbs2 and one of WiHog1A, WiHog1B, or ScHog1 were shocked with 1 M NaCl. The phosphorylation patterns were similar for the WiHog1A, WiHog1B, and ScHog1 MAPKs. The greatest intensity of phosphorylation was detected after 5 min of the 1 M NaCl shock (Figure 6C, NaCl 5), and decreased after 10 min (Figure 6C, NaCl 10) for all of the MAPKs, regardless of the MAPKK expressed (Figure 6C; WiPbs2, ScPbs2). This high phosphorylation after 5 min of the 1 M NaCl shock suggests that WiPbs2 efficiently activates WiHog1A, WiHog1B, and ScHog1, with slightly lower intensity in the case of WiHog1A.
Discussion
The obligate halophilic fungus W. ichthyophaga grows in media enriched with NaCl concentrations from 1.7 to 5.1 M (Zajc et al., 2014). This is what makes W. ichthyophaga an interesting model organism for investigating the mechanisms of osmotolerance and osmosensing. Herein, we identified components of the HOG pathway, which is the central osmosensing pathway of W. ichthyophaga (Konte and Plemenitaš, 2013). We analyzed the genome of W. ichthyophaga, heterologously expressed the selected W. ichthyophaga HOG signaling proteins, and studied their functions and interactions to further illucidate the architecture of the HOG pathway in this halophilic basidiomycetous fungus.
S. cerevisiae has previously been used as a system for expression of mammalian MAP kinases, which in yeast lose their ability to function normally and become spontaneously phospohorylated (Levin-Salomon et al., 2009). The turgor sensor function was identified in Arabidopsis thaliana as the plant cytokinin receptor 1 (Cre1) when it replaced the function of ScSln1 histidine kinase at high osmolarity (Reiser et al., 2003), and in a similar manner, A. thaliana ATMEKK1 kinase replaced the function of ScSte11 (Covic et al., 1999). The Cicer arietinum zinc finger protein CaZF was even shown to enhance the osmotolerance of yeast, with effective accumulation of glycerol and induction of stress genes in a HOG-dependant and calcineurin-dependant manner (Jain et al., 2009). S. cerevisiae has also been used for the characterization of many fungal kinases, including the Hog1-like kinases from Zygosaccharomyces rouxii (Iwaki et al., 1999) and the DhPbs2 kinase from Debaryomyces hansenii (Bansal et al., 2001), and recently also for the molecular dissections of the D. hansenii DhNik1 (Furukawa et al., 2012) and C. albicans CaNik1 (El-Mowafy et al., 2013) histidine kinases. As a step further, in our investigation, we comprehensively reconstructed the pathway with the heterologous expression of the core module of the pathway, which consists of the MAPKK WiPbs2 and its interacting proteins, WiSho1, WiSte11, and WiHog1A and B. Moreover, we confirmed the significance of the two-component HOG-pathway system in W. ichthyophaga cells, using the fludioxonil sensitivity assay.
It has been reported recently that within the S. cerevisiae SHO1 branch there are two signaling sub-branches that can be distinguished, known as HKR1 and MSB2 (Tanaka et al., 2014). ScSho1 acts as a true osmosensor only in the HKR1 sub-branch, where upon hyperosmolar stimulus, the ScOpy2-ScSte50 complex recruits ScSte11, the ScHkr1, and ScCdc42 proteins recruit ScSte20/ScCla4, and ScSho1 recruits ScPbs2 to the membrane. ScSho1 also forms a multi-component signaling complex with ScOpy2 and ScHkr1, and the high-osmolarity-dependent structural changes in the ScSho1 transmembrane domains induce its binding with ScSte50 (Tatebayashi et al., 2015). In the genome of W. ichthyophaga, we were not able to identify a considerable number of the orthologs that are involved in the SHO1 branch of the osmosensing apparatus in S. cerevisiae; namely, the mucins Msb2 and Hkr1, and the membrane anchor Opy2. However, other orthologs of this branch are present, with significant size expansion in the case of the kinases WiSte50, WiSte20, and WiSte11. The important interactions for the activation of ScPbs2 by the SHO1 branch also include ScSho1 binding with the ScPbs2 proline-rich motif (Marles et al., 2004) and docking of ScSte11 (Tatebayashi et al., 2006). Although these interaction motifs are poorly conserved in the sequence of WiPbs2, there is an APLPPNQLR motif in the N-terminal part of WiPbs2 (residues 117–125) that represents the only putative proline-rich motif that can be recognized by the class I SH3 domains.
We compared the binding of both ScPbs2 and the WiPbs2 proline-rich motif with the WiSho1 SH3 domain from W. ichthyophaga. The analysis of the model with COCOMAPS predicted weaker interactions for the binding of the WiPbs2 proline-rich motif and the WiSho1 SH3 domain. This prediction of weaker interactions was shown by the lower osmotolerance and the greater cross-talk activation in WiPbs2-expressing ssk2/22Δpbs2Δ cells, and this was additionally confirmed by the lower osmotolerance of the sho1ssk2/22Δpbs2Δ cells that expressed WiPbs2 and the ScSho1WiSH3 hybrid protein. As the insertion of the ScPbs2 proline-rich motif into the N-terminus of WiPbs2 increased the osmotolerance of S. cerevisiae cells, it is possible that the weaker binding of WiPbs2 and ScSho1WiSH3 was the cause of the weak activation of WiPbs2 via the SHO1 branch. This assumption was further supported when the insertion of the ScPbs2 proline-rich motif into the N-terminal part of WiPbs2 increased the osmotolerance of S. cerevisiae cells, most probably by strengthening the binding of the hybrid protein WiPbs2ScPRO to the ScSho1WiSH3 hybrid osmosensor. Although these data also showed that WiPbs2 can be efficiently activated by ScSte11, the WiSte11-expressing S. cerevisiae ste11Δssk2/22Δ cells were osmosensitive, which indicated that WiSte11 cannot complement the function of ScSte11.
This, and the poor interactions between the WiSho1 SH3 domain and the WiPbs2 proline-rich motif that we have shown here, combined with the demonstration that the glycosylated transmembrane orthologs Hkr1, Msb2, and Opy2 (the essential components of osmosensing in S. cerevisiae; Tatebayashi et al., 2007) were not found in W. ichthyophaga, suggest that the SHO1-branch is not involved in HOG signaling in W. ichthyophaga (Figure 7). This is also in line with the results reported for A. nidulans PbsA (AnPbs2; Furukawa et al., 2005). When expressed in S. cerevisiae, AnPbs2 is not activated by ScSho1 in ssk2/22Δpbs2Δ cells, whereas ShoA (AnSho1) is functional in the sho1Δssk2/22Δ background (Furukawa et al., 2005). Similarly, the HwSho1 proteins from the extremely halotolerant H. werneckii can bind ScPbs2 (Fettich et al., 2011), but an interaction of ScSho1 with HwPbs2 kinases in S. cerevisiae was not confirmed (Plemenitaš et al., 2014). Also in Candida sp., which is closely related to S. cerevisiae, Sho1 only has an indirect role in the osmotic stress response, and it is not involved in osmosensing (Jandric et al., 2013; Herrero-de-Dios et al., 2014). On the other hand, the SHO1 and SLN1 branches are functional in Kluyveromyces lactis (Velazquez-Zavala et al., 2015), which suggests that the existence of the sophisticated SHO1-branch osmotic signal perception in not limited to S. cerevisiae. Even though the W. ichthyophaga genome does not contain the orthologs of typical osmosensors, it cannot be excluded that some other unconventional osmosensors and membrane anchors are present that can recruit WiSte50 (e.g., by interacting with its Ras-association domain). Hypothetically, WiSte50 would than bind WiSte11 through Sterile alpha motif interactions, and WiPbs2 through the SH3 domains (for details, see the domain structure of WiSte50 in Figure 1), providing the scaffold for the activation of WiPbs2 by WiSte11, without the need for participation of WiSho1.
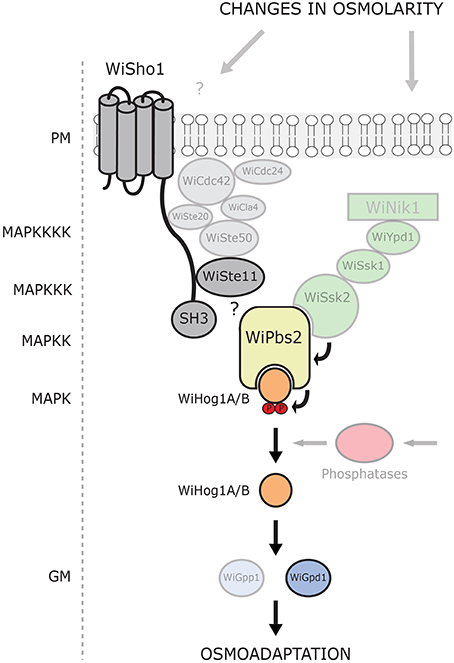
Figure 7. Model of current understanding of the HOG pathway architecture in W. ichthyophaga. Putative HOG-pathway components identified in the genome of W. ichthyophaga are shown. Proteins experimentally characterized to date are in bold. The components of the SHO1-like branch are shown in gray, and the two-component (SLN1-like) branch in green. The MAPK hierarchy is indicated (left). PM, plasma membrane; SH3, Src homology 3 domain; P, phosphate groups; GM, glycerol metabolism.
In contrast to the SHO1 branch, the two-component SLN1 branch orthologs appear to have pivotal roles. Although the Ssk2/Ssk22 binding site of WiPbs2 is not highly conserved, the growth in NaCl of S. cerevisiae pbs2Δ and ste11Δpbs2Δ cells that express WiPbs2 is similar to those expressing ScPbs2. This suggests that WiPbs2 is efficiently activated by ScSsk2/ScSsk22 kinase in S. cerevisiae and has a comparable role to ScPbs2 when activated through the SLN1 branch. All of the orthologs of the S. cerevisiae SLN1 branch, except the Sln1 transmembrane histidine kinase, are present in the genome of W. ichthyophaga (Figures 1, 7). In S. cerevisiae, ScSln1 is the only histidine kinase, but in other fungi there are several groups (Catlett et al., 2003; Lenassi and Plemenitaš, 2007; Ma and Li, 2013) The genome of W. ichthyophaga contains four hybrid histidine kinases that belong to groups I or II, III and VIII. It has been shown for several fungi that the Nik1 orthologs from group III are involved in the two-component HOG-pathway signaling (Bahn, 2008; El-Mowafy et al., 2013). We therefore speculate that the HAMP domain repeats in WiNik1 (Figure 1) are involved in osmosensing in W. ichthyophaga, as has been demonstrated for D. hansenii DhNik1 (Kaur et al., 2014), C. albicans CaNik1 (El-Mowafy et al., 2013), and Candida lusitaniae ClNik1 (Randhawa et al., 2016). Furthermore, it has been shown that the group III histidine kinases are also involved in fungal sensitivity to fludioxonil, which makes this phenylpyrrole fungicide a potent tool for investigations into group III histidine kinase signaling. Interestingly, in the fludioxonil-sensitive C. neoformans serotype A strain H99, CnHog1 is constitutively phosphorylated under normal conditions and dephosphorylated under osmotic shock (Bahn et al., 2007). In C. neoformans, the group III histidine kinase Tco1 (CnNik1) is involved in osmosensing, as well as in fludioxonil sensitivity (Bahn et al., 2006). As W. ichthyophaga also contains the group III histidine kinase WiNik1 (Figures 1, 7) and shows similar constitutive phosphorylation patterns under optimal (noninducing) conditions (Konte and Plemenitaš, 2013), we investigated the sensitivity of W. ichthyophaga cells to fludioxonil. Indeed, the presence of fludioxonil completely inhibited the growth of W. ichthyophaga cells at all NaCl concentrations used. This is also in line with the recently published results of Nik1 kinase involvement in the fungicide sensitivity and osmotic signaling in M. oryzae (Jacob et al., 2015) and C. lusitaniae (Randhawa et al., 2016). As we were not able to produce any HOG pathway knock-out mutants of the halophilic W. ichthyophaga even after multiple attempts, this result of fludioxonil sensitivity indicates the importance of the group III histidine kinase signaling and the two-component signal transduction in W. ichthyophaga, and suggests the involvement of the HOG pathway in survival of W. ichthyophaga, even at its optimal NaCl concentration of 3.4 M.
Unconventional constitutive phosphorylation of Hog1 kinases under nonstress conditions has only been observed in the halophilic W. ichthyophaga (Konte and Plemenitaš, 2013) and the pathogenic C. neoformans serotype A strain H99 (Bahn et al., 2005) to date. Such constitutive phosphorylation patterns of HOG pathway regulation are the reverse to those of yeast and the majority of other fungi, where Hog1 is dephosphorylated under nonstress conditions and phosphorylated upon osmotic shock (Krantz et al., 2006b; Saito and Posas, 2012; Schmidt-Heydt et al., 2013). Interestingly, when the cells of halotolerant H. werneckii are grown in up to 3 M NaCl, the phosphorylation patterns are similar to that of S. cerevisiae, but when they are exposed to higher salinities, HwHog1 is phosphorylated constitutively (Kejžar et al., 2015). These hybrid phosphorylation patterns might represent the foundations of H. werneckii extreme halotolerance.
However, analysis of the HOG pathway components in the closely related C. neoformans strains H99 and JEC21, which show different phosphorylation patterns (constitutive vs. transient), has demonstrated that the change of Phe240 to Leu in the MAPKKK CnSsk2 is the sole reason for constitutive phosphorylation of CnHog1 in the serotype A strain H99 (Bahn et al., 2007). This residue is located in the putative CnSsk1 binding domain on CnSsk2, which most probably changes the mechanism of CnSsk2 regulation by CnSsk1. Interestingly, when compared to other C. neoformans strains, serotype A strain H99 is more resistant to various environmental stresses, including osmotic stress, although it is sensitive to fludioxonil (Kojima et al., 2006; Bahn et al., 2007). As the alignment of CnSsk2 and WiSsk2 revealed the presence of a Leu residue at the homologous site in WiSsk2 (Supplemental Figure S3), we propose that this might be the reason for constitutive phosphorylation of WiHog1 also in W. ichthyophaga. Furthermore, our data suggest that this constitutive phosphorylation originates from a component upstream of WiHog1 and WiPbs2, and not from their autophosphorylation. This assumption is supported by our finding that when expressed in S. cerevisiae pbs2Δhog1Δ cells, WiHog1 and WiPbs2 show normal phosphorylation patterns that are similar to those of ScHog1 and ScPbs2, and can even replace their function.
In a previous study, we showed that the two MAPK paralogs WiHog1A and WiHog1B from W. ichthyophaga differ, as WiHog1A did not complement the function of ScHog1 as efficiently as WiHog1B in S. cerevisiae hog1Δ cells (Konte and Plemenitaš, 2013). This HOG1-like gene duplication is an unusual trait, which has only been identified in W. sebi (Padamsee et al., 2012), H. werneckii (Kejžar et al., 2015), Z. rouxii (Iwaki et al., 1999), A. nidulans (Furukawa et al., 2005), and A. fumigatus (de Oliveira Bruder Nascimento et al., 2016). While the paralogous Hog1-like kinases from H. werneckii and Z. rouxii appear to be redundant, in A. nidulans and A. fumigatus their roles are divergent. The A. fumigatus HogA (AfSakA) is essential for osmosensing, while the A. nidulans AnSakA is not (Ma and Li, 2013; de Oliveira Bruder Nascimento et al., 2016). On the other hand, in A. fumigatus, the paralogous AfMpkC kinase has been linked to the use of polyalcohol sugars (Reyes et al., 2006) and virulence (de Oliveira Bruder Nascimento et al., 2016), while the exact role of A. nidulans AnMpkC has not been defined (Furukawa et al., 2005). In the present study we expressed WiHog1A or WiHog1B together with WiPbs2 in pbs2Δhog1Δ cells. When challenged with NaCl, the cells expressing WiPbs2 and WiHog1A showed greater cross-talk activation, greater osmosensitivity, and slightly lower WiHog1A phosphorylation, as compared to the cells expressing WiPbs2 and WiHog1B. However, the dephosphorylation of WiHog1A and WiHog1B that occurred 10 min after exposure to NaCl show that both of these MAPKs can efficiently activate S. cerevisiae phosphatases for their negative-feedback regulation. While WiHog1A cannot inhibit the cross-talk but efficiently binds to the GPD1 promoter in S. cerevisiae, WiHog1B can inhibit the cross-talk and can also efficiently induce GPD1-lacZ (Konte and Plemenitaš, 2013). This, and that the induction of the WiHOG1B mRNA transcripts doubled in comparison to WiHOG1A transcripts in W. ichthyophaga upon hyperosmotic shock (Lenassi et al., 2011; Konte and Plemenitaš, 2013), indicate that WiHog1B is also the primary HOG pathway kinase in W. ichthyophaga.
One of the major and first Hog1 targets in S. cerevisiae is glycerol transport and metabolism. Upon hyperosmotic shock, the aquaglyceroporin Fps1 is rapidly closed to prevent the efflux of glycerol, and there is strong induction of the expression of glycerol proton symporter Stl, as well as of genes for glycerol synthesis (GPD1, GPP1, and GPP2; Hohmann et al., 2007). The genome of W. ichthyophaga contains three aquaglyceroporins and four putative glycerol Stl1-like transporters (Zajc et al., 2013). Key enzymes involved in glycerol metabolism are also present and conserved in W. ichthyophaga. The expression of the gene encoding WiGpd1 was previously shown to be salt-induced (Lenassi et al., 2011), however, the genome of W. ichthyophaga also contains another paralogous Gpd enzyme. On the other hand, only the ortholog of Gpp1, WiGpp1, is present, without Gpp2 (Zajc et al., 2013). This salt-responsive WiGPD1 expression was much lower compared to HwGPD1 from H. werneckii (Lenassi et al., 2011), which might reflect the halophilic vs. halotolerant natures of W. ichthyophaga and H. werneckii, respectively. Interestingly, in C. neoformans, the genes that encode Stl1-like transporter, aquaglyceroporin, Gpd1, and Gpp1 are all salt responsive and induced in a HOG-dependant manner (Ko et al., 2009). Glycerol accumulation in C. neoformans is regulated through a Hog1-regulated kinase 1 (Hkr1), which is induced via the two-component system (Kim et al., 2011). It has also been shown that fludioxonil promotes glycerol overaccumulation, which causes cell swelling and death (Kojima et al., 2006). Similarly, glycerol overaccumulation might also be the reason for fludioxonil sensitivity of W. ichthyophaga cells.
Taken together, our investigation of the architecture of the W. ichthyophaga HOG pathway shows that the interactions of WiPbs2 kinase and the SHO1-branch orthologs are not conserved (Figure 7), which in turn suggests that these orthologs are not involved in the activation of WiPbs2. On the other hand, the ScSsk2/ScSsk22 activation of WiPbs2 is fully conserved, and W. ichthyophaga cells are sensitive to fludioxonil, which highlights the importance of the two-component HOG pathway signaling in W. ichthyophaga (Figure 7). As demonstrated to be necessary for higher stress-resistance of C. neoformans, the constitutive two-component HOG-pathway phosphorylation could also help W. ichthyophaga to survive in its naturally hypersaline environments. However, the exact mechanism of the hyperosmotic signal processing and the role of phosphatases in pathway activation by dephosphorylation remains the subject of further studies.
Author Contributions
AP provided funding. AP and TK conceived and designed the study and the experiments. TK performed the experiments. TK, AP, and UT analyzed and interpreted the data. TK wrote the manuscript. AP and UT provided critical revision. All of the authors reviewed the results and approved the final version of the manuscript.
Funding
This study was funded in part by Research Grant P1-0170, and in part by a Young Researcher Fellowship, from the Slovenian Research Agency.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Sincere thanks go to the laboratories of Prof. Haruo Saito (IMSUT, Tokyo), Prof. Stefan Hohmann (Chalmers University of Technology, Göteborg), Prof. Francesc Posas (UPF, Barcelona), and Prof. Wendell Lim (UCSF, San Francisco), for sharing of yeast strains and plasmids. The authors also thank Dr. Metka Lenassi for her useful suggestions, Dr. Martin Fettich for his help with illustrations, Prof. Jurij Stojan for help with the protein modeling, and Jernej Praprotnik for technical support.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00901
References
Bahn, Y. S. (2008). Master and commander in fungal pathogens: the two-component system and the HOG signaling pathway. Eukaryot. Cell 7, 2017–2036. doi: 10.1128/EC.00323-08
Bahn, Y. S., Geunes-Boyer, S., and Heitman, J. (2007). Ssk2 mitogen-activated protein kinase kinase kinase governs divergent patterns of the stress-activated Hog1 signaling pathway in Cryptococcus neoformans. Eukaryot. Cell 6, 2278–2289. doi: 10.1128/EC.00349-07
Bahn, Y. S., Kojima, K., Cox, G. M., and Heitman, J. (2005). Specialization of the HOG pathway and its impact on differentiation and virulence of Cryptococcus neoformans. Mol. Biol. Cell 16, 2285–2300. doi: 10.1091/mbc.E04-11-0987
Bahn, Y. S., Kojima, K., Cox, G. M., and Heitman, J. (2006). A unique fungal two-component system regulates stress responses, drug sensitivity, sexual development, and virulence of Cryptococcus neoformans. Mol. Biol. Cell 17, 3122–3135. doi: 10.1091/mbc.E06-02-0113
Bansal, P. K., Sharma, P., and Mondal, A. K. (2001). A PBS2 homologue from Debaryomyces hansenii shows a differential effect on calcofluor and polymyxin B sensitivity in Saccharomyces cerevisiae. Yeast 18, 1207–1216. doi: 10.1002/yea.769
Biasini, M., Bienert, S., Waterhouse, A., Arnold, K., Studer, G., Schmidt, T., et al. (2014). SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258. doi: 10.1093/nar/gku340
Catlett, N. L., Yoder, O. C., and Turgeon, B. G. (2003). Whole-genome analysis of two-component signal transduction genes in fungal pathogens. Eukaryot. Cell 2, 1151–1161. doi: 10.1128/EC.2.6.1151-1161.2003
Cominelli, E., and Tonelli, C. (2010). Transgenic crops coping with water scarcity. N. Biotechnol. 27, 473–477. doi: 10.1016/j.nbt.2010.08.005
Covic, L., Silva, N. F., and Lew, R. R. (1999). Functional characterization of ARAKIN (ATMEKK1): a possible mediator in an osmotic stress response pathway in higher plants. BBA Mol. Cell Res. 1451, 242–254. doi: 10.1016/s0167-4889(99)00096-8
de Oliveira Bruder Nascimento, A. C. M., dos Reis, T. F., de Castro, P. A., Hori, J. I., Bom, V. L. P., de Assis, L. J., et al. (2016). Mitogen activated protein kinases SakAHOG1 and MpkC collaborate for Aspergillus fumigatus virulence. Mol. Microbiol. 100, 841–859. doi: 10.1111/mmi.13354
Dinkel, H., Michael, S., Weatheritt, R. J., Davey, N. E., Van Roey, K., Altenberg, B., et al. (2012). ELM - the database of eukaryotic linear motifs. Nucleic Acids Res. 40, D242–D251. doi: 10.1093/nar/gkr1064
El-Mowafy, M., Bahgat, M. M., and Bilitewski, U. (2013). Deletion of the HAMP domains from the histidine kinase CaNik1p of Candida albicans or treatment with fungicides activates the MAP kinase Hog1p in S. cerevisiae transformants. BMC Microbiol. 13:209. doi: 10.1186/1471-2180-13-209
Fettich, M., Lenassi, M., Veraniè, P., Gunde-Cimerman, N., and Plemenitaš, A. (2011). Identification and characterization of putative osmosensors, HwSho1A and HwSho1B, from the extremely halotolerant black yeast Hortaea werneckii. Fungal. Genet. Biol. 48, 475–484. doi: 10.1016/j.fgb.2011.01.011
Furukawa, K., Hoshi, Y., Maeda, T., Nakajima, T., and Abe, K. (2005). Aspergillus nidulans HOG pathway is activated only by two-component signalling pathway in response to osmotic stress. Mol. Microbiol. 56, 1246–1261. doi: 10.1111/j.1365-2958.2005.04605.x
Furukawa, K., Randhawa, A., Kaur, H., Mondal, A. K., and Hohmann, S. (2012). Fungal fludioxonil sensitivity is diminished by a constitutively active form of the group III histidine kinase. FEBS Lett. 586, 2417–2422. doi: 10.1016/j.febslet.2012.05.057
Gašparič, M. B., Lenassi, M., Gostinčar, C., Rotter, A., Plemenitaš, A., Gunde-Cimerman, N., et al. (2013). Insertion of a specific fungal 3′-phosphoadenosine-5′-phosphatase motif into a plant homologue improves halotolerance and drought tolerance of plants. PLoS ONE 8:e81872. doi: 10.1371/journal.pone.0081872
Gietz, R. D., and Schiestl, R. H. (2007). Quick and easy yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 35–37. doi: 10.1038/nprot.2007.14
Gostinčar, C., Lenassi, M., Gunde-Cimerman, N., and Plemenitaš, A. (2011). Fungal adaptation to extremely high salt concentrations. Adv. Appl. Microbiol. 77, 71–96. doi: 10.1016/B978-0-12-387044-5.00003-0
Hao, N., Behar, M., Parnell, S. C., Torres, M. P., Borchers, C. H., Elston, T. C., et al. (2007). A systems-biology analysis of feedback inhibition in the Sho1 osmotic-stress-response pathway. Curr. Biol. 17, 659–667. doi: 10.1016/j.cub.2007.02.044
Herrero-de-Dios, C., Alonso-Monge, R., and Pla, J. (2014). The lack of upstream elements of the Cek1 and Hog1 mediated pathways leads to a synthetic lethal phenotype upon osmotic stress in Candida albicans. Fungal Genet. Biol. 69, 31–42. doi: 10.1016/j.fgb.2014.05.010
Hohmann, S., Krantz, M., and Nordlander, B. (2007). Yeast osmoregulation. Method Enzymol. 428, 29–45. doi: 10.1016/S0076-6879(07)28002-4
Horie, T., Tatebayashi, K., Yamada, R., and Saito, H. (2008). Phosphorylated Ssk1 prevents unphosphorylated Ssk1 from activating the Ssk2 mitogen-activated protein kinase kinase kinase in the yeast high-osmolarity glycerol osmoregulatory pathway. Mol. Cell Biol. 28, 5172–5183. doi: 10.1128/MCB.00589-08
Iwaki, T., Tamai, Y., and Watanabe, Y. (1999). Two putative MAP kinase genes, ZrHOG1 and ZrHOG2, cloned from the salt-tolerant yeast Zygosaccharomyces rouxii are functionally homologous to the Saccharomyces cerevisiae HOG1 gene. Microbiology 145, 241–248. doi: 10.1099/13500872-145-1-241
Jacob, S., Foster, A. J., Yemelin, A., and Thines, E. (2015). High osmolarity glycerol (HOG) signalling in Magnaporthe oryzae: Identification of MoYPD1 and its role in osmoregulation, fungicide action, and pathogenicity. Fungal Biol. 119, 580–594. doi: 10.1016/j.funbio.2015.03.003
Jain, D., Roy, N., and Chattopadhyay, D. (2009). CaZF, a plant transcription factor functions through and parallel to HOG and calcineurin pathways in Saccharomyces cerevisiae to provide osmotolerance. PLoS ONE 4:e5154. doi: 10.1371/journal.pone.0005154
Jandric, Z., Gregori, C., Klopf, E., Radolf, M., and Schuller, C. (2013). Sorbic acid stress activates the Candida glabrata high osmolarity glycerol MAP kinase pathway. Front. Microbiol. 4:350. doi: 10.3389/fmicb.2013.00350
Kaur, H., Singh, S., Rathore, Y. S., Sharma, A., Furukawa, K., Hohmann, S., et al. (2014). Differential role of HAMP-like linkers in regulating the functionality of the group III histidine kinase DhNik1p. J. Biol. Chem. 289, 20245–20258. doi: 10.1074/jbc.M114.554303
Kejžar, A., Grotli, M., Tamas, M. J., Plemenitaš, A., and Lenassi, M. (2015). HwHog1 kinase activity is crucial for survival of Hortaea werneckii in extremely hyperosmolar environments. Fungal Genet. Biol. 74, 45–58. doi: 10.1016/j.fgb.2014.11.004
Kim, S. Y., Ko, Y. J., Jung, K. W., Strain, A., Nielsen, K., and Bahn, Y. S. (2011). Hrk1 plays both Hog1-dependent and -independent roles in controlling stress response and antifungal drug resistance in Cryptococcus neoformans. PLoS ONE 6:e18769. doi: 10.1371/journal.pone.0018769
Ko, Y. J., Yu, Y. M., Kim, G. B., Lee, G. W., Maeng, P. J., Kim, S., et al. (2009). Remodeling of global transcription patterns of Cryptococcus neoformans genes mediated by the stress-activated HOG signaling pathways. Eukaryot. Cell 8, 1197–1217. doi: 10.1128/EC.00120-09
Kojima, K., Bahn, Y. S., and Heitman, J. (2006). Calcineurin, Mpk1 and Hog1 MAPK pathways independently control fludioxonil antifungal sensitivity in Cryptococcus neoformans. Microbiology 152, 591–604. doi: 10.1099/mic.0.28571-0
Konte, T., and Plemenitaš, A. (2013). The HOG signal transduction pathway in the halophilic fungus Wallemia ichthyophaga: identification and characterisation of MAP kinases WiHog1A and WiHog1B. Extremophiles 17, 623–636. doi: 10.1007/s00792-013-0546-4
Kozubowski, L., Lee, S. C., and Heitman, J. (2009). Signalling pathways in the pathogenesis of Cryptococcus. Cell. Microbiol. 11, 370–380. doi: 10.1111/j.1462-5822.2008.01273.x
Kralj Kunčič, M., Kogej, T., Drobne, D., and Gunde-Cimerman, N. (2010). Morphological response of the halophilic fungal genus Wallemia to high salinity. Appl. Environ. Microbiol. 76, 329–337. doi: 10.1128/AEM.02318-09
Kralj Kunčič, M., Zajc, J., Drobne, D., Pipan Tkalec, Z., and Gunde-Cimerman, N. (2013). Morphological responses to high sugar concentrations differ from adaptation to high salt concentrations in the xerophilic fungi Wallemia spp. Fungal Biol. 117, 466–478. doi: 10.1016/j.funbio.2013.04.003
Krantz, M., Becit, E., and Hohmann, S. (2006a). Comparative analysis of HOG pathway proteins to generate hypotheses for functional analysis. Curr. Genet. 49, 152–165. doi: 10.1007/s00294-005-0039-9
Krantz, M., Becit, E., and Hohmann, S. (2006b). Comparative genomics of the HOG-signalling system in fungi. Curr. Genet. 49, 137–151. doi: 10.1007/s00294-005-0038-x
Lenassi, M., and Plemenitaš, A. (2007). Novel group VII histidine kinase HwHhk7B from the halophilic fungi Hortaea werneckii has a putative role in osmosensing. Curr. Genet. 51, 393–405. doi: 10.1007/s00294-007-0131-4
Lenassi, M., Vaupotiè, T., Gunde-Cimerman, N., and Plemenitaš, A. (2007). The MAP kinase HwHog1 from the halophilic black yeast Hortaea werneckii: coping with stresses in solar salterns. Saline Syst. 3:3. doi: 10.1186/1746-1448-3-3
Lenassi, M., Zajc, J., Gostinčar, C., Gorjan, A., Gunde-Cimerman, N., and Plemenitaš, A. (2011). Adaptation of the glycerol-3-phosphate dehydrogenase Gpd1 to high salinities in the extremely halotolerant Hortaea werneckii and halophilic Wallemia ichthyophaga. Fungal Biol. 115, 959–970. doi: 10.1016/j.funbio.2011.04.001
Levin-Salomon, V., Maayan, I., Avrahami-Moyal, L., Marbach, I., Livnah, O., and Engelberg, D. (2009). When expressed in yeast, mammalian mitogen-activated protein kinases lose proper regulation and become spontaneously phosphorylated. Biochem. J. 417, 331–340. doi: 10.1042/BJ20081335
Ma, D., and Li, R. (2013). Current understanding of HOG-MAPK pathway in Aspergillus fumigatus. Mycopathologia 175, 13–23. doi: 10.1007/s11046-012-9600-5
Maeda, T., Takekawa, M., and Saito, H. (1995). Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science 269, 554–558. doi: 10.1126/science.7624781
Marchler-Bauer, A., and Bryant, S. H. (2004). CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32, W327–W331. doi: 10.1093/nar/gkh454
Marles, J. A., Dahesh, S., Haynes, J., Andrews, B. J., and Davidson, A. R. (2004). Protein-protein interaction affinity plays a crucial role in controlling the Sho1p-mediated signal transduction pathway in yeast. Mol. Cell 14, 813–823. doi: 10.1016/j.molcel.2004.05.024
Mas, G., de Nadal, E., Dechant, R., Rodriguez de la Concepcion, M. L., Logie, C., Jimeno-Gonzalez, S., et al. (2009). Recruitment of a chromatin remodelling complex by the Hog1 MAP kinase to stress genes. EMBO J. 28, 326–336. doi: 10.1038/emboj.2008.299
Miller, J. H. (1972). Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Murakami, Y., Tatebayashi, K., and Saito, H. (2008). Two adjacent docking sites in the yeast Hog1 mitogen-activated protein (MAP) kinase differentially interact with the Pbs2 MAP kinase kinase and the Ptp2 protein tyrosine phosphatase. Mol. Cell. Biol. 28, 2481–2494. doi: 10.1128/MCB.01817-07
O'Rourke, S. M., and Herskowitz, I. (1998). The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev. 12, 2874–2886. doi: 10.1101/gad.12.18.2874
Padamsee, M., Kumar, T. K., Riley, R., Binder, M., Boyd, A., Calvo, A. M., et al. (2012). The genome of the xerotolerant mold Wallemia sebi reveals adaptations to osmotic stress and suggests cryptic sexual reproduction. Fungal Genet. Biol. 49, 217–226. doi: 10.1016/j.fgb.2012.01.007
Petelenz-Kurdziel, E., Eriksson, E., Smedh, M., Beck, C., Hohmann, S., and Goksor, M. (2011). Quantification of cell volume changes upon hyperosmotic stress in Saccharomyces cerevisiae. Integr. Biol. (Camb.) 3, 1120–1126. doi: 10.1039/c1ib00027f
Plemenitaš, A., Lenassi, M., Konte, T., Kejžar, A., Zajc, J., Gostinčar, C., et al. (2014). Adaptation to high salt concentrations in halotolerant/halophilic fungi: a molecular perspective. Front. Microbiol. 5:199. doi: 10.3389/fmicb.2014.00199
Posas, F., Takekawa, M., and Saito, H. (1998). Signal transduction by MAP kinase cascades in budding yeast. Curr. Opin. Microbiol. 1, 175–182. doi: 10.1016/S1369-5274(98)80008-8
Prieto, D., Roman, E., Correia, I., and Pla, J. (2014). The HOG pathway is critical for the colonization of the mouse gastrointestinal tract by Candida albicans. PLoS ONE 9:e87128. doi: 10.1371/journal.pone.0087128
Raitt, D. C., Posas, F., and Saito, H. (2000). Yeast Cdc42 GTPase and Ste20 PAK-like kinase regulate Sho1-dependent activation of the Hog1 MAPK pathway. EMBO J. 19, 4623–4631. doi: 10.1093/emboj/19.17.4623
Randhawa, A., Chawla, S., and Mondal, A. K. (2016). Functional dissection of HAMP domains in NIK1 ortholog from pathogenic yeast Candida lusitaniae. Gene 577, 251–257. doi: 10.1016/j.gene.2015.12.002
Reiser, V., Raitt, D. C., and Saito, H. (2003). Yeast osmosensor Sln1 and plant cytokinin receptor Cre1 respond to changes in turgor pressure. J. Cell Biol. 161, 1035–1040. doi: 10.1083/jcb.200301099
Reyes, G., Romans, A., Nguyen, C. K., and May, G. S. (2006). Novel mitogen-activated protein kinase MpkC of Aspergillus fumigatus is required for utilization of polyalcohol sugars. Eukaryot. Cell 5, 1934–1940. doi: 10.1128/EC.00178-06
Rozman, D., and Komel, R. (1994). Isolation of genomic DNA from filamentous fungi with high glucan level. Biotechniques 16, 382–384.
Saito, H., and Posas, F. (2012). Response to hyperosmotic stress. Genetics 192, 289–318. doi: 10.1534/genetics.112.140863
Saksela, K., and Permi, P. (2012). SH3 domain ligand binding: what's the consensus and where's the specificity? FEBS Lett. 586, 2609–2614. doi: 10.1016/j.febslet.2012.04.042
Schmidt-Heydt, M., Stoll, D. A., Mrohs, J., and Geisen, R. (2013). Intraspecific variability of HOG1 phosphorylation in Penicillium verrucosum reflects different adaptation levels to salt rich habitats. Int. J. Food Microbiol. 165, 246–250. doi: 10.1016/j.ijfoodmicro.2013.05.011
Sole, C., Nadal-Ribelles, M., Kraft, C., Peter, M., Posas, F., and de Nadal, E. (2011). Control of Ubp3 ubiquitin protease activity by the Hog1 SAPK modulates transcription upon osmostress. EMBO J. 30, 3274–3284. doi: 10.1038/emboj.2011.227
Tanaka, K., Tatebayashi, K., Nishimura, A., Yamamoto, K., Yang, H. Y., and Saito, H. (2014). Yeast osmosensors Hkr1 and Msb2 activate the Hog1 MAPK cascade by different mechanisms. Sci. Signal. 7:ra21. doi: 10.1126/scisignal.2004780
Tatebayashi, K., Takekawa, M., and Saito, H. (2003). A docking site determining specificity of Pbs2 MAPKK for Ssk2/Ssk22 MAPKKKs in the yeast HOG pathway. EMBO J. 22, 3624–3634. doi: 10.1093/emboj/cdg353
Tatebayashi, K., Tanaka, K., Yang, H. Y., Yamamoto, K., Matsushita, Y., Tomida, T., et al. (2007). Transmembrane mucins Hkr1 and Msb2 are putative osmosensors in the SHO1 branch of yeast HOG pathway. EMBO J. 26, 3521–3533. doi: 10.1038/sj.emboj.7601796
Tatebayashi, K., Yamamoto, K., Nagoya, M., Takayama, T., Nishimura, A., Sakurai, M., et al. (2015). Osmosensing and scaffolding functions of the oligomeric four-transmembrane domain osmosensor Sho1. Nat. Commun. 6:6975. doi: 10.1038/ncomms7975
Tatebayashi, K., Yamamoto, K., Tanaka, K., Tomida, T., Maruoka, T., Kasukawa, E., et al. (2006). Adaptor functions of Cdc42, Ste50, and Sho1 in the yeast osmoregulatory HOG MAPK pathway. EMBO J. 25, 3033–3044. doi: 10.1038/sj.emboj.7601192
van Drogen, F., O'Rourke, S. M., Stucke, V. M., Jaquenoud, M., Neiman, A. M., and Peter, M. (2000). Phosphorylation of the MEKK Ste11p by the PAK-like kinase Ste20p is required for MAP kinase signaling in vivo. Curr. Biol. 10, 630–639. doi: 10.1016/S0960-9822(00)00511-X
Vangone, A., Spinelli, R., Scarano, V., Cavallo, L., and Oliva, R. (2011). COCOMAPS: a web application to analyze and visualize contacts at the interface of biomolecular complexes. Bioinformatics 27, 2915–2916. doi: 10.1093/bioinformatics/btr484
Velazquez-Zavala, N., Rodriguez-Gonzalez, M., Navarro-Olmos, R., Ongay-Larios, L., Kawasaki, L., Torres-Quiroz, F., et al. (2015). Ineffective phosphorylation of mitogen-activated protein kinase Hog1p in response to high osmotic stress in the yeast Kluyveromyces lactis. Eukaryot. Cell 14, 922–930. doi: 10.1128/EC.00048-15
Vriend, G. (1990). WHAT IF: a molecular modeling and drug design program. J. Mol. Graph. 8, 52–56. doi: 10.1016/0263-7855(90)80070-V
Waterhouse, A. M., Procter, J. B., Martin, D. M., Clamp, M., and Barton, G. J. (2009). Jalview Version 2 - a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. doi: 10.1093/bioinformatics/btp033
Westfall, P. J., Patterson, J. C., Chen, R. E., and Thorner, J. (2008). Stress resistance and signal fidelity independent of nuclear MAPK function. Proc. Natl. Acad. Sci. U.S.A. 105, 12212–12217. doi: 10.1073/pnas.0805797105
Wilkins, M. R., Gasteiger, E., Bairoch, A., Sanchez, J. C., Williams, K. L., Appel, R. D., et al. (1999). Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 112, 531–552.
Wu, X., Knudsen, B., Feller, S. M., Zheng, J., Sali, A., Cowburn, D., et al. (1995). Structural basis for the specific interaction of lysine-containing proline-rich peptides with the N-terminal SH3 domain of c-Crk. Structure 3, 215–226.
Zajc, J., Kogej, T., Galinski, E. A., Ramos, J., and Gunde-Cimerman, N. (2014). Osmoadaptation strategy of the most halophilic fungus, Wallemia ichthyophaga, growing optimally at salinities above 15% NaCl. Appl. Environ. Microbiol. 80, 247–256. doi: 10.1128/AEM.02702-13
Zajc, J., Liu, Y., Dai, W., Yang, Z., Hu, J., Gostinčar, C., et al. (2013). Genome and transcriptome sequencing of the halophilic fungus Wallemia ichthyophaga: haloadaptations present and absent. BMC Genomics 14:617. doi: 10.1186/1471-2164-14-617
Zajc, J., Zalar, P., Plemenitaš, A., and Gunde-Cimerman, N. (2012). The mycobiota of the salterns. Prog. Mol. Subcell. Biol. 53, 133–158. doi: 10.1007/978-3-642-23342-5_7
Zalar, P., Sybren de Hoog, G., Schroers, H. J., Frank, J. M., and Gunde-Cimerman, N. (2005). Taxonomy and phylogeny of the xerophilic genus Wallemia (Wallemiomycetes and Wallemiales, cl. et ord. nov.). Antonie Van Leeuwenhoek 87, 311–328. doi: 10.1007/s10482-004-6783-x
Keywords: signaling, protein-protein interaction, protein phosphorylation, mitogen activated protein kinase (MAPK), high-osmolarity glycerol (HOG) signaling pathway, Saccharomyces cerevisiae, halophilic fungus, Wallemia ichthyophaga
Citation: Konte T, Terpitz U and Plemenitaš A (2016) Reconstruction of the High-Osmolarity Glycerol (HOG) Signaling Pathway from the Halophilic Fungus Wallemia ichthyophaga in Saccharomyces cerevisiae. Front. Microbiol. 7:901. doi: 10.3389/fmicb.2016.00901
Received: 11 March 2016; Accepted: 26 May 2016;
Published: 13 June 2016.
Edited by:
Baolei Jia, Chung-Ang University, South KoreaReviewed by:
James A. Coker, University of Maryland University College, USAOzlem Ates, Nişantaşi University, Turkey
Ana Lúcia Leitão, Universidade Nova de Lisboa, Portugal
Copyright © 2016 Konte, Terpitz and Plemenitaš. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ana Plemenitaš, ana.plemenitas@mf.uni-lj.si
 Tilen Konte
Tilen Konte Ulrich Terpitz
Ulrich Terpitz Ana Plemenitaš
Ana Plemenitaš