Introduction
The threat of climate change has intensified efforts toward the development of safer alternatives to depleting fossil fuels (Cox et al., 2000). Lignocellulosic bioethanol is considered to be a viable and environmentally friendly alternative to fossil fuels. Though lignocellulosic biomass is available in massive quantities and is renewable (Dillon and Dillon, 2003; Lynd et al., 2008; Pauly and Keegstra, 2008; Kricka et al., 2015), the presence of certain barriers makes lignocellulosic bioethanol expensive. Discovery of proteins with novel specificities is necessary to break these barriers and make lignocellulosic bioethanol economically viable (Horn et al., 2012; Ulaganathan et al., 2015). Cellulolytic bacteria isolated from various environments have been explored for proteins of potential use in lignocellulosic bioethanol production (Badger, 2002; Wang et al., 2012; Pinheiro et al., 2015). Bacteria belonging to the genera Bacillus, Bacteroides, Butyrivibrio, Cellulosimicrobium, Citrobacter, Clostridium, Devosia, Dyadobacter, Ensifer, Kaistia, Labrys, Methanobrevibacter, Microbacterium, Ochrobactrum, Paracoccus, Pseudomonas, Rhizobium, Ruminococcus, Shinella, Siphonobacter, Stenotrophomonas, Trichonympha, and Variovorax, were found to be cellulolytic (Saxena et al., 1993; Schwarz, 2001; Gupta et al., 2012; Huang et al., 2012; Yanga et al., 2014). Bacillus pumilus strains are known to produce cellulase enzyme up to a maximum of 11.4 mg/g of cell dry mass (Suzuki and Kaneko, 1976; Kotchoni and Shonukan, 2002; Ariffin et al., 2006). The cellulase enzyme produced by B. pumilus strain EB3 has been found to be superior to fungal cellulases due to its higher optimum pH and temperature (Ariffin et al., 2006). Further it has been shown that the B. pumilus cellulase enzyme could be mutated to remove the catabolite repression (Kotchoni et al., 2003). We have recently isolated bacterial strains from the gut contents of the wood boring Mesomorphus sp. These isolates were screened for cellulolytic and xylose isomerase activities and the isolate ku-bf1 which exhibited maximum cellulolytic and xylose isomerase activities was identified as B. pumilus by 16S rRNA sequencing. The whole genome of this strain has been sequenced. The dataset has been submitted to NCBI and is reported here.
Materials and Methods
Isolation of the Bacterial Strain
Bacterial isolates were made by plating the gut contents of wood boring Mesomorphus sp. on YEP-Agar medium (Yeast extract, peptone and agar). After incubation for 24 h at 25°C, the growing bacterial colonies were sub-cultured. These colonies were tested for cellulolytic and xylose isomerase activities on CMC-Agar medium (NH4H2PO4—1 g/L; KCl—0.2 g/L; MgSO4.7H2 O—1 g/L; Yeast Extract—1 g/L; Carboxymethyl Cellulose—26 g/L; Agar—3 g/L) and YEP-Xylose-Agar medium, respectively (Sapunova et al., 2004; Ponnambalam et al., 2011). The bacterial isolate (ku-bf1) which produced maximum clearance zone in both plate assays was selected for this work.
Genomic DNA Isolation, Library Preparation and Sequencing
Genomic DNA was isolated using a modified Cetyltrimethyl ammonium bromide (CTAB) method (Murray and Thompson, 1980; Zhou et al., 1996). The quality of isolated DNA was checked using a Qubit fluorimeter (Thermo Fisher) and 50 ng of pure genomic DNA was used for library preparation. Genomic DNA was fragmented and adapter-tagged using a Sure Select QXTKit (Agilent Technologies). Fragmented DNA was cleaned using HighPrepBeads (MagBio Genomics). Cleaned and adapter tagged fragments were amplified and indexed. The prepared library was quantified using a Qubit Fluorimeter. The quality of the library was checked by running an aliquot (1 ul) on a High Sensitivity Bioanalyzer DNA Chip (Agilent Technologies). The library showed a size range of ~300–1000 bp in the Bioanalyzer profile. The effective insert size of the library was in the range of ~180–880 bp, Whole genome sequencing was carried out with an IluminaMiseq system (Illumina, San Diego, CA) at Genotypic Technology (P) Ltd., Bangalore
Preprocessing and Genome Assembly
The quality of sequence reads was analyzed using the FastQC tool (Andrews, 2010). Reads were trimmed off adapters using the Fastx-toolkit (Gordon and Hannon, 2010). Reference genome assembly was carried out using the Bowtie2 tool (ver. 2.2.4) (Langmead and Salzberg, 2012). The genome of B. pumilus W3, downloaded from Genbank, was used as the reference genome. Reference based assembly involved indexing of the reference genome and alignment of reads to the reference and creation of a SAM file using SAMtools (ver 0.1.18) (Li et al., 2009). The SAM file was converted to a binary BAM file, sorted and indexed by using the “view,” “sort” and “index” functions of SAMtools, respectively. The BAM file was checked using the BamView tool and used for variation report generation (Carver et al., 2010). The consensus sequence was generated using SAMtools. The variation report in “bcf” format was converted into a “vcf” file using BCFTools.
Results
Whole Genome Sequencing of B. Pumilus ku-bf1
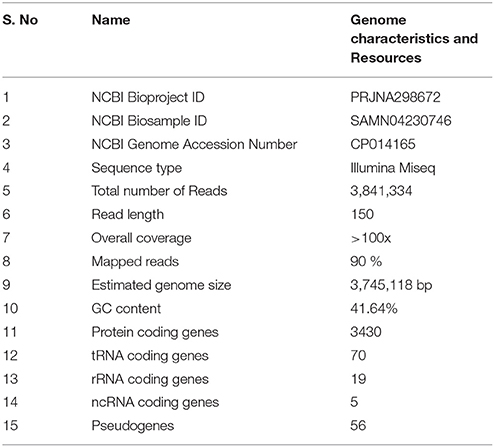
Sequencing the genome of B. pumilus ku-bf1 produced a total of 3,841,334 paired-end reads (150 bp). After removing adapters and low quality reads, the reads were used for reference based genome assembly. These reads were assembled on to the reference genome (B. pumilus W3) using Bowtie-2 (Langmead and Salzberg, 2012). Over 90% of the reads were aligned to the reference genome and the coverage was estimated to be >100x. The BAM file was used for generating the variation report using SAMtools with a mapping quality of >30 and read depth of >20 as cutoffs. The consensus sequence generated was 37,45,118 bp long. NCBI Prokaryotic genome annotation pipeline predicted a total of 3430 protein coding genes, 94 RNA coding genes and 56 pseudogenes. The RNA coding genes predicted include seventy tRNA genes, six 5S rRNA genes, seven 16S rRNA genes, six 23S rRNA genes and five non-coding RNA genes (Table 1).
Direct Link to Deposited Data and Information to Users
The dataset submitted to NCBI include the assembled consensus sequence of B. pumilus ku-bf1 in Fasta format and the Bam file generated by reference based assembly. The genome sequence can be accessed at NCBI using the accession number CP014165. Users can download and use the data freely for research purpose only with acknowledgment to us and quoting this paper as reference to the data.
Author Contributions
Work was planned by KU and executed jointly by KU and JB. SR was associated with isolation of the bacterial strain.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was carried out with financial Assistance from University Grants Commission, Government of India through the University of Potential Excellence (UPE) programm to Osmania University. JB is supported by Junior Research Fellowship from University Grants Commission, Government of India. SR is supported by UGC-UPE Junior Research Fellowship.
References
Andrews, S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc
Ariffin, H., Abdullah, N., Umi Kalsom, M. S., Shirai, Y., and Hassan, M. A. (2006). Production and characterization of cellulase by Bacillus pumilus EB3. Int. J. Eng. Technol. 3, 47–53.
Badger, P. C. (2002). Ethanol from Cellulose: A General Review. Trends in New Crops and New Uses. Alexandria, VA: ASHS Press.
Carver, T., Böhme, U., Otto, T. D., Parkhill, J., and Berriman, M. (2010). BamView: viewing mapped read alignment data in the context of the reference sequence. Bioinformtics 26, 676–677. doi: 10.1093/bioinformatics/btq010
Cox, P. M., Betts, R. A., Jones, C. D., Spall, S. A., and Totterdell, I. J. (2000). Acceleration of global warming due to carbon-cycle feedbacks in a coupled climate model. Nature 408, 184–187. doi: 10.1038/35041539
Dillon, R. J., and Dillon, V. M. (2003). The gut bacteria of insects: nonpathogenic Interactions. Annu. Rev. Entomol. 49, 71–92. doi: 10.1146/annurev.ento.49.061802.123416
Gordon, A., and Hannon, G. (2010). Fastx-Toolkit. FASTQ/A Short-Reads Pre-Processing Tools. Available online at: http://www.hannonlab.cshl.edu.com/fastx_toolkit
Gupta, P., Samant, K., and Sahu, A. (2012). Isolation of cellulose-cegrading cacteria and determination of their cellulolytic potential. Int. J. Microbiol. 2012: 578925. doi: 10.1155/2012/578925
Horn, S. J., Vaaje-Kolstad, G., Westereng, B., and Eijsink, V. G. H. (2012). Novel enzymes for the degradation of cellulose. Biotechnol. Biofuels. 5:45. doi: 10.1186/1754-6834-5-45
Huang, S., Sheng, P., and Zhang, H. (2012). Isolation and identification of cellulolytic bacteria from the Gut of Holotrichia parallela Larvae (Coleoptera: Scarabaeidae). Int. J. Mol. Sci. 13, 2563–2577. doi: 10.3390/ijms13032563
Kotchoni, O. S., and Shonukan, O. O. (2002). Regulatory mutations affecting the synthesis of cellulase in Bacillus pumilus. World J. Biotechnol. 18, 487–491. doi: 10.1023/A:1015571022652
Kotchoni, O. S., Shonukan, O. O., and Gachomo, W. E. (2003). Bacillus pumillus, BpCRI 6, a promising candidate for cellulase production under conditions of catabolite repression. African J. Biotechnol. 2, 140–146. doi: 10.5897/AJB2003.000-1028
Kricka, W., Fitzpatrick, J., and Bond, U. (2015). Challenges for the production of bioethanol from biomass using recombinant yeasts. Adv. Appl. Microbiol. 92, 89–125. doi: 10.1016/bs.aambs.2015.02.003
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). 1000 Genome project data processing subgroup. the sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Lynd, L. R., Laser, M. S., Brandsby, D., Dale, B. E., Davison, B., et al. (2008). How biotech can transform biofuels. Nat. Biotechnol. 2, 169–172. doi: 10.1038/nbt0208-169
Murray, M. G., and Thompson, W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4325. doi: 10.1093/nar/8.19.4321
Pauly, M., and Keegstra, K. (2008). Cell-wall carbohydrates and their modification as a resource for biofuels. Plant. J. 54, 559–568. doi: 10.1111/j.1365-313X.2008.03463.x
Pinheiro, G. L., Correa, R. F., Cunha, R. S., Cardoso, A. M., Chaia, C., Clementino, M. M., et al. (2015). Isolation of aerobic cultivable cellulolytic bacteria from different regions of the gastrointestinal tract of giant land snail Achatina fulica. Front. Microbiol. 20:860. doi: 10.3389/fmicb.2015.00860
Ponnambalam, A. S., Deepthi, R. S., and Ghosh, A. R. (2011). Qualitative display and measurement of enzyme activity of isolated cellulolytic bacteria. Biotechnol. Bioinf. Bioeng. 1, 33–37.
Sapunova, L. I., lobanok, A. G., Kazakevich, I. O., and Evtushenkov, A. N. (2004). A plate method to screen for microorganisms producing xylose isomerase. Microbiology 173, 107. doi: 10.1023/B:MICI.0000016378.43654.0a
Saxena, S., Bahadur, J., and Varma, A. (1993). Cellulose and hemicellulose degrading bacteria from termite gut and mould soils of India. Indian J. Microbiol. 33, 55–60.
Schwarz, W. H. (2001). The cellulosome and cellulose degradation by anaerobic bacteria. Appl. Microbiol. Biotechnol. 56, 634–649. doi: 10.1007/s002530100710
Suzuki, H., and Kaneko, T. (1976). Degradation of barley glucan and lichenan by a Bacillus pumilus enzyme. Agric. Biol. Chem. 40, 577–586. doi: 10.1080/00021369.1976.10862095
Ulaganathan, K., Goud, B. S., Reddy, M. M., Kumar, V. P., Balsingh, J., and Radhakrishna, S. (2015). Proteins for breaking barriers in lignocellulosic bioethanol production. Curr. Protein Peptide Sci. 16, 100–134. doi: 10.2174/138920371602150215165718
Wang, M., Li, Z., Fang, X., Wang, L., and Qu, Y. (2012). Cellulolytic enzyme production and enzymatic hydrolysis for second-generation bioethanol production. Adv. Biochem. Eng. Biotechnol. 128, 1–24. doi: 10.1007/10_2011_131
Yanga, W., Menga, F., Penga, J., Hana, P., Fanga, F., Maa, L., et al. (2014). Isolation and identification of a cellulolytic bacterium from the Tibetan pig's intestine and investigation of its cellulase production. Electron. J. Biotechnol. 17, 262–267. doi: 10.1016/j.ejbt.2014.08.002
Keywords: Bacillus pumilus, genome sequencing, cellulolytic bacteria, xylose isomerase, bioethanol, Mesomorphus
Citation: Balsingh J, Radhakrishna S and Ulaganathan K (2016) Draft Genome Sequence of Bacillus pumilus ku-bf1 Isolated from the Gut Contents of Wood Boring Mesomorphus sp. Front. Microbiol. 7:1037. doi: 10.3389/fmicb.2016.01037
Received: 05 February 2016; Accepted: 20 June 2016;
Published: 30 June 2016.
Edited by:
Vijai Kumar Gupta, National University of Ireland, Galway, IrelandReviewed by:
Sean Cameron Booth, University of Calgary, CanadaJoseph Selvin, Pondicherry University, India
Copyright © 2016 Balsingh, Radhakrishna and Ulaganathan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kandasamy Ulaganathan, kulaganathan123@gmail.com
 Jatoth Balsingh
Jatoth Balsingh Surabhi Radhakrishna
Surabhi Radhakrishna Kandasamy Ulaganathan
Kandasamy Ulaganathan