- 1Department of Food Science and Technology, Yeungnam University, Gyeongsan, South Korea
- 2Food Standard Research Center, Korea Food Research Institute, Seongnam, South Korea
Cronobacter species (Cronobacter spp.) are hazardous foodborne pathogens associated with baby food, powdered infant formula (PIF). To develop a rapid and sensitive method for simultaneous detection of seven Cronobacter spp. in PIF, an indirect non-competitive enzyme-linked immunosorbent assay (INC-ELISA) was developed based on a novel immunoglobulin G (IgG), anti-Cronobacter IgG. The developed INC-ELISA was able to detect seven Cronobacter spp. at concentrations ranging from (5.6 ± 0.30) × 103 to (2.1 ± 0.01) × 105 colony forming unit (CFU)/mL in pure culture. Further, INC-ELISA employing anti-Cronobacter IgG was applicable for analysis of PIF samples contaminated with less than <10 cells of Cronobacter spp. per 25 g of PIF in 36 h. The developed antibody showed slight cross-reactivity with Franconibacter pulveris (LMG 24057) at high concentration (108 CFU/mL). The INC-ELISA method displayed excellent specificity without compromising cross-reactivity with other foodborne pathogens. The INC-ELISA assay method developed in this study using a novel anti-Cronobacter IgG facilitated highly sensitive, efficient, and rapid detection of Cronobacter spp. in baby food.
Introduction
Bacteria of the genus Enterobacter are increasingly important human pathogens that cause neonatal meningitis and sepsis (Hunter and Bean, 2013). Enterobacterial infections have become a worldwide health problem partially due to the lack of development of preventive treatments and the emergence of new multi-drug resistant strains. Enterobacter sakazakii has recently been reclassified as a Cronobacter (Strydom et al., 2012). Cronobacter species (Cronobacter spp.), previously known as E. sakazakii, are foodborne pathogens that pose a high risk of infection to neonates as well as immuno-compromised individuals (World Health Organization [WHO], 2007). Cronobacter spp. affect the central nervous system of infants, and survivors often suffer from severe neurological impairments such as hydrocephalus, quadriplegia, and developmental delays (Lai, 2001; Strydom et al., 2012; Jaradat et al., 2014). Species level classification of the Cronobacter genus, which includes C. sakazakii, C. muytjensii, C. malonaticus, C. turicensis, C. dublinensis, C. universalis, and C. condimenti, was recently defined by Iversen et al. (2008) and Joseph et al. (2012b). Cronobacter spp. have been isolated from plant-based food products, including cereal, fruits, vegetables, legumes, herbs, and spices, as well as from animal-based food materials such as milk, meat, and fish (Friedemann, 2007; Lee et al., 2012).
All Cronobacter spp., except C. condimenti, have been associated with human infections (Cruz-Córdova et al., 2012). Historically, Cronobacter spp. have been implicated in newborn and infant infections, causing meningitis, necrotizing enterocolitis, and bacteremia (Healy et al., 2010). However, not all Cronobacter spp. are linked to infantile infections, and it is thought that virulence among strains may vary. C. sakazakii, C. malonaticus, and C. turicensis are most often isolated from infantile cases (Joseph and Forsythe, 2011). Recent research based on international microbiological standards suggested that all species of Cronobacter must be absent in 10 grams of powdered infant formula (PIF) (Odeyemi and Sani, 2016). For an understanding of recent unresolved issues persisting with respect to taxonomy, sources and clinical relevance, and for suggestions on how to safely feed premature neonates (Holy and Forsythe, 2014), it is plausible that virulence determinants have evolved in certain lineages (Joseph et al., 2012a).
Although reservoirs of Cronobacter spp. and their modes of transmission are still unknown, Cronobacter spp. have been suggested as a source of food contamination, with rodents and flies serving as a secondary route of contamination (Jung and Park, 2006; World Health Organization [WHO], 2007). According to a report by Jung and Park (2006), 20% of PIF samples were found contaminated with Cronobacter in the Republic of Korea. Lee et al. (2012) also reported similar results in which Cronobacter spp. were isolated from 18.6% of detected food samples. Although a number of reported cases of Cronobacter infection are quite low, sequelae can occur with high mortality rates (Lai, 2001; United States Food and Drug Administration [USFDA], 2002; Friedemann, 2007). World Health Organization [WHO] (2007), classified Cronobacter together with Salmonella as group A pathogens associated with PIF with clear evidence of illness in infants. These categories of organisms were based on their risk of illness to infants.
Culture-dependent isolation and assay methods for complete analysis of Cronobacter spp. from PIF usually require 5 to 7 days (United States Food and Drug Administration [USFDA], 2002). A further method was recommended by the International Organization for Standardization and the International Dairy Federation as ISO 22964 (Anonymous, 2006). This method includes a pre-enrichment in buffered peptone water (BPW), a selective enrichment in modified lauryl sulfate tryptose broth containing vancomycin and isolation of presumptive C. sakazakii colonies colored in blue-green on E. sakazakii isolation agar. These colonies should be streaked on tryptic soy agar and resulting yellow colonies are indicatory for C. sakazakii. However, this method was also time- and labor-consuming, as it usually requires 5 to 6 days to obtain a positive result (Norberg et al., 2012).
Lampel and Chen (2009) previously described a new method for the isolation and detection of Cronobacter spp. from PIF using a real-time PCR-based assay and chromogenic agar. In their study, suspended cells were isolated from enrichment culture, streaked onto chromogenic agar, and confirmed by real-time PCR assay. Mullane et al. (2006) also developed a method using cationic-magnetic beads to capture Cronobacter spp., and subsequent identification was performed after washing off bound cells from the capture phase and plating them onto Druggan–Forsythe–Iversen formulation agar to detect 1 to 5 colony forming unit (CFU)/500 g of PIF within 24 h. However, these PCR-based methods have significant technical requirements of ultra-pure reagents and chemicals along with high equipment costs. Therefore, a rapid, sensitive, and inexpensive method is needed for the detection of Cronobacter spp.
Polyclonal antibody can be obtained within a short time (4 to 8 weeks) with minimal financial investment, whereas it takes about 3 to 6 months to produce monoclonal antibodies (Leenaars and Hendriksen, 2005). Polyclonal antibody is commonly used in immunological methods, including enzyme-linked immunosorbent assay (ELISA), for the detection of foodborne pathogens (Brigmon et al., 1992; Kumar et al., 2008; Velusamy et al., 2010). It is also critical for developing a rapid and genus-specific method for identification of Cronobacter spp. in the context of food safety issues. Previously, a sandwich ELISA method was developed for the detection of C. muytjensii with a detection limit of 6.3 × 104 CFU/mL using a species-specific anti-C. muytjensii immunoglobulin G (IgG) (Park et al., 2012). In addition, Xu et al. (2014) also developed a polyclonal and monoclonal antibodies-based indirect ELISA and a sandwich ELISA for the detection of Cronobacter spp. The indirect ELISA detected all species of Cronobacter assayed, and the limit of detection (LOD) was established as 105 CFU/mL. In contrast, sandwich ELISA was specific for C. sakazakii and exhibited greater sensitivity than indirect ELISA (LOD of 2 × 104 CFU/mL). Following 10 h of enrichment, Cronobacter spp. were detected using either of the two analytical methods in samples inoculated with 1 CFU/100 g PIF. The results from this study demonstrated that both of these novel ELISAs were specific, sensitive, and rapid assays for the screening of pathogenic Cronobacter spp. in PIF.
A widely available polyclonal antibody capable of detecting Cronobacter spp. could be used to avoid expensive and time-consuming methods. Thus, we decided to produce and characterize a polyclonal antibody for seven Cronobacter spp. To our knowledge, there has been no report on the ELISA-based detection of multiple Cronobacter spp. In the current work, an indirect non-competitive ELISA (INC-ELISA) method based on anti-Cronobacter IgG was developed in pure culture and applied for the detection of seven Cronobacter spp. in PIF.
Materials and Methods
Strains and Reagents
In the present study, seven Cronobacter strains were used to produce antibody and to assess the cross-reactivity of developed method, while eight other bacterial strains belonging to different genera were selected to check the cross-reactivity of developed assay due to their partial gene sequence similarity and to confirm the accurate detection of pathogen contaminants such as Salmonella spp., Citrobacter spp., and Bacillus cereus (Iversen et al., 2004; World Health Organization [WHO], 2007; Pinto et al., 2013). In the present study, B. cereus and S. Enteritidis were selected since it was found that the chances of contamination in PIF were highly concerned due to the presence of B. cereus and S. Enteritidis (Giammanco et al., 2011; Pinto et al., 2013). Giammanco et al. (2011) also reported that Salmonella spp. and Cronobacter spp. (formerly E. sakazakii) are the microorganisms of greatest concern in PIF. In addition, regarding the use of Citrobacter freundii, Giammanco et al. (2011) observed that molecular epidemiological survey of C. freundii was misidentified as Cronobacter spp. isolated from PIF. The phylogenetic tree also showed that C. freundii and Cronobacter spp. are very closely related species based on their smaller amounts of mahalanobis distances than other species. Hence, it was suggested that C. freundii may also be an under-reported cause of bacterial infection, especially in high risk neonates, due to misidentification. B. cereus (KCCM 40935), Buttiauxella noackiae (ATCC 51713), C. condimenti (LMG 26250), C. dublinensis (LMG 23823), C. malonaticus (LMG 23826), C. muytjensii (CDC 3523-75), C. turicensis (LMG 23827), C. muytjensii (ATCC 51329), C. sakazakii (ATCC 29544), C. sakazakii (ATCC 29004), C. universalis (LMG 26249), C. freundi (ATCC 8090), Escherichia coli (ATCC 39418), Franconibacter helveticus (LMG 23732), Franconibacter pulveris (LMG 24057), and Salmonella Typhimurium (ATCC 13311) were used in this study. C. muytjensii (CDC 3523–75) was donated by Dr. Carol Iversen from University College Dublin, Ireland. Other strains used in this study were purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA) and Korean Culture Center of Microorganisms (KCCM; Seoul, Korea). Strains indicated with LMG were purchased from the Belgian Coordinated Collections of Microorganisms (BCCM; Gent, Belgium). All strains were cultured in nutrient broth (NB) for 18 h at 37°C in a shaking incubator (150 rpm).
NB, nutrient agar (NA), peptone, and skim milk were purchased from Difco (Franklin Lakes, NJ, USA). Enterobacteriaceae enrichment (EE) broth and violet red bile glucose (VRBG) agar were purchased from MB cell (Seoul, Korea). Sodium carbonate, sodium bicarbonate, sodium azide, potassium phosphate monobasic, potassium phosphate dibasic, sodium chloride, caprylic acid, ammonium sulfate, and alkaline phosphatase yellow liquid substrate p-nitrophenyl phosphate (pNPP) were purchased from Sigma (St. Louis, MO, USA). Phosphatase-labeled goat anti-rabbit IgG was purchased from Kierkegaard & Perry Laboratories, Inc. (Gaithersburg, MD, USA). Lipopolysaccharide (LPS) extraction kit was purchased from Intron Biotechnology (Seongnam, Korea).
Animal Care Ethics
Animal use protocol was reviewed by the committee members of Yeungnam University and approved by Korea Food and Drug Administration, Republic of Korea (Animal Ethics License No. 2013-012 and 2012-010).
Preparation of Immunogen and Immunization
Three types of immunogens were prepared for the development of a genus-specific antibody against Cronobacter spp. A sonicated cell protein (SCP) mixture of seven Cronobacter spp. was used in this study. The seven Cronobacter spp. were separately cultured in NB at 37°C for 18 h to a concentration of 1 × 109 CFU/mL. Each culture (30 mL) was centrifuged at 3,000 × g at 4°C for 30 min. The pellets were washed three times with 30 mL of 0.01 M phosphate-buffered saline (PBS) and then suspended in 10 mL of 0.01 M PBS for sonication under 20 KHz power for 5 min on ice. Supernatant was collected after centrifugation at 12,000 × g, 4°C for 20 min. Protein concentrations of supernatants were checked with a Bradford kit, after which SCP solutions were adjusted to a concentration of 0.5 mg/L with 0.01 M PBS. SCP solutions were stored at -20°C before use. For immunization, equal volumes of the seven SCP solutions were mixed for use as an immunogen in New Zealand white rabbits.
LPS mixture of the seven Cronobacter spp. was prepared using an LPS extraction kit following the instructions of the manufacturer. Each Cronobacter spp. culture was centrifuged at 10,000 × g at room temperature for 20 min to harvest bacterial cells. Lysis buffer was then added and vortexed vigorously, followed by addition of chloroform, vortexing for 20 s, and incubated at room temperature for 5 min. The mixture was then centrifuged at 10,000 × g for 10 min at 4°C. An aliquot of supernatant was transferred to a new tube, mixed well with purification buffer, and kept at -20°C for 10 min. The solution was then centrifuged at 10,000 × g for 15 min at 4°C, after which the upper layer was removed to obtain an LPS pellet. The pellet was washed three times with 1 mL of ethanol, centrifuged for 3 min at 10,000 × g at 4°C, and then collected and dried at room temperature. For the LPS solution, 50 μL of Tris-HCl buffer (10 mM, pH 8.0) was used to dissolve the LPS pellet. LPS solution of the seven Cronobacter spp. was prepared by mixing an equal volume of each LPS solution for use as an immunogen in New Zealand white rabbits.
Formalin-killed cell (FKC) mixture was also prepared as an immunogen. Cultures of the seven Cronobacter spp. were applied as treatments with 0.5% formalin for 24 h, followed by centrifugation (3,000 × g, 4°C, for 30 min) according to the method of Song and Kim (2013). First injection was administered with a mixture of immunogen and Freund’s complete adjuvant (1:1, v/v). Second and third injections were administered using the same mixture of immunogen and Freund’s incomplete adjuvant (1:1, v/v) at 4 weeks after the first injection. Immunogen was injected into the back of rabbits at four sites at a concentration of 0.25 mL/site. Blood samples were taken every week until 18 weeks after the first injection.
Preparation and Purification of IgG
Blood samples were centrifuged (10,000 × g) for 30 min at 4°C to separate the anti-sera. Anti-Cronobacter IgG was purified from the anti-sera by caprylic acid and ammonium sulfate precipitation as described by McKinney and Parkinson (1987) and according to further modified methods of Shukla et al. (2012) and Song and Kim (2013).
Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE) for Purified Anti-Cronobacter IgG
The purity of various anti-Cronobacter IgGs preparations was checked using SDS-PAGE (Atto, Tokyo, Japan) under reduced conditions as described by Park et al. (2012) with some modifications.
Development of INC-ELISA for Detection of Cronobacter spp.
Rabbit anti-Cronobacter IgG was used to develop an INC-ELISA method for detection of Cronobacter spp. Standard response curve of the seven Cronobacter spp. was constructed using the following procedure. First, fresh cultures of all tested Cronobacter spp. were diluted decimally with 0.05 M carbonate buffer (pH 9.5) in order to coat 96-well plates (SPL Life Sciences, Gyeonggi-do, Korea), whereas only carbonate buffer was used as a negative control. Simultaneously, coated cultures of Cronobacter spp. were counted on NA and VRBG agar plates for determination of concentration. After coating at 4°C overnight, 96-well plates were washed three times with 0.01 M PBS (pH 7.0) and blocked with 200 μL of 5% skim milk at 37°C for 2 h. Ninety six-well plates were then washed with 0.01 mol/L PBS-0.05% Tween 20 (PBST), added with rabbit anti-Cronobacter IgG, and incubated at 37°C for 1 h. Then, plates were washed again with 0.01 M PBST, after which 100 μL of phosphatase-labeled goat anti-rabbit IgG was added and the plates incubated at 37°C for 1 h. Plates were washed again with 0.01 M PBST, after which 50 μL of pNPP liquid substrate was added to each well for a 30 min enzyme-substrate reaction. Finally, 50 μL of 0.01 M NaOH solution was added to stop the reaction, after which the final reaction mixture was analyzed using an Infinite M200 (Tecan; Seestrasse, Switzerland). A detection limit of the developed INC-ELISA was determined by using a data linearity function and standard deviation of responses for the calibration standard and samples, respectively (Hubaux and Vos, 1970).
Specificity and Sensitivity of INC-ELISA Assay
Seven common Cronobacter foodborne pathogens (Table 1) were used for the specificity test of the developed INC-ELISA. Cultures of tested foodborne pathogens were serially diluted to final concentrations of 100 to 108 CFU/mL and then used to check sensitivity of the developed INC-ELISA. All experiments were repeated three times.
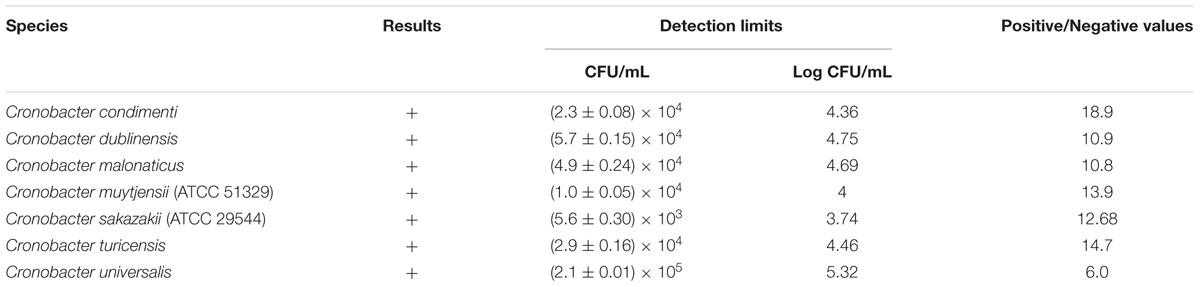
TABLE 1. Detection of Cronobacter species in pure culture using the developed indirect non-competitive enzyme-linked immunosorbent assay.
Food Trial Using Cronobacter-Spiked PIF Sample (Artificial Inoculation of Cronobacter into PIF Sample)
Firstly, the seven Cronobacter strains were separately cultured in NB at 37°C for 20 h, after which the cultures were serially diluted with BPW to obtain the lowest final concentration (<10 CFU/mL) of Cronobacter live cells for use in further experiments. To examine the applicability of the developed anti-Cronobacter IgG against each Cronobacter strain, 25 g of PIF sample was aseptically placed into a flask, mixed with 225 mL of BPW as a pre-enriched broth, and separately spiked with 1 mL (less than 10 cells/25 g of PIF) of each Cronobacter species, followed by pre-enrichment incubation at 37°C for 8 h. Following this, 1 mL of the pre-enrichment culture was inoculated into EE broth and incubated for 8 h of enrichment (Korea Food and Drug Administration [KFDA], 2014). Further, enriched aliquots of pre-treated food samples were tested using the developed INC-ELISA and standard USFDA methods. For control, non-spiked PIF samples were analyzed using the developed INC-ELISA and standard microbiological USFDA methods (United States Food and Drug Administration [USFDA], 2002). All experiments were performed in three replicates in order to maintain reliability of the work.
Statistical Analysis
The detection limit was calculated as the average value of absorbance at zero concentration with three standard deviations (Park et al., 2012). All experiments and results were analyzed in at least three trials for proving reliable and reproducible data.
Results
Characteristics of Developed Antibodies
As shown in Figure 1, anti-serum containing anti-Cronobacter IgG developed by the LPS mixture showed no titer for any Cronobacter species. Figure 1 provides the fact that LPS antigen of Cronobacter is not appropriate for the production of anti-Cronobacter antibody. For antibodies developed using FKC and SCP mixtures of Cronobacter spp., titers of the developed rabbit anti-Cronobacter serum containing IgG were determined by INC-ELISA after 18 weeks of immunization, as shown in Figures 2 and 3. Titers of the developed anti-Cronobacter seru containing IgG increased after the first injection as well as gradually increased after the second and third injections. Titers of anti-Cronobacter serum containing IgG developed from the FKC mixture of Cronobacter spp. (Figure 2) were in the following order: C. muytjensii, C. turicensis, C. malonaticus, C. condimenti, C. sakazakii, C. dublinensis, and C. universalis. Comparatively, anti-Cronobacter serum containing IgG developed from an SCP mixture of the seven Cronobacter spp. showed higher titers against the seven Cronobacter spp. (Figure 3), which were in the following order: C. muytjensii, C. condimenti, C. turicensis, C. sakazakii, C. malonaticus, C. dublinensis, and C. universalis. It is well known that an increased titer of developed antibody results in effective performance during immunoassay. The developed and purified anti-Cronobacter IgG showed very high purity as compared to the commercial rabbit IgG (Figure 4). Developed antibody as well as commercial rabbit IgG (lanes 2, 3, 4, and 5) showed a strong band around 51 kDa and a light band around 25 kDa, respectively. These results confirm that our developed rabbit anti-Cronobacter IgG with high purity is comparable to commercial rabbit IgG.
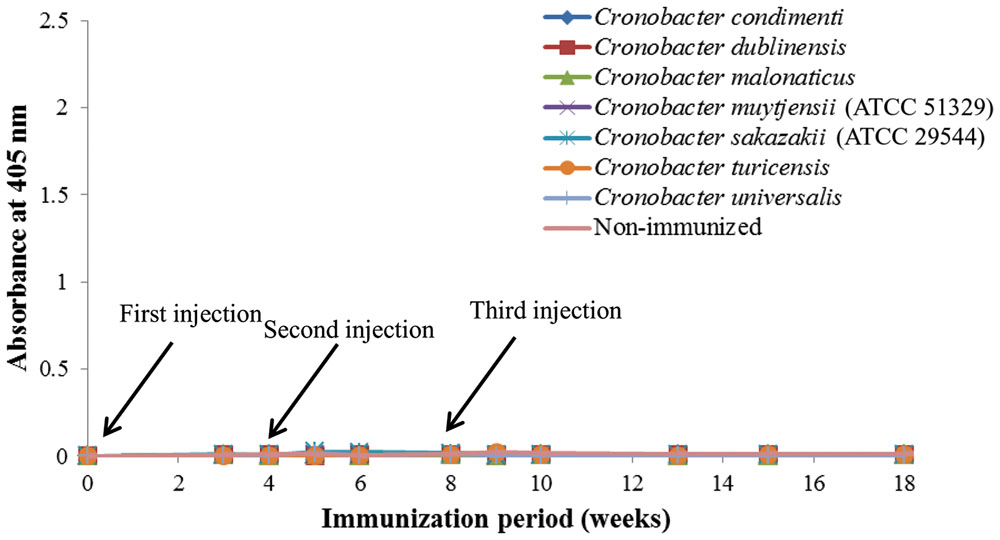
FIGURE 1. Titers of rabbit anti-serum developed by lipopolysaccharide against seven Cronobacter species. All experiments were conducted three times, and data represent mean ± standard deviation.
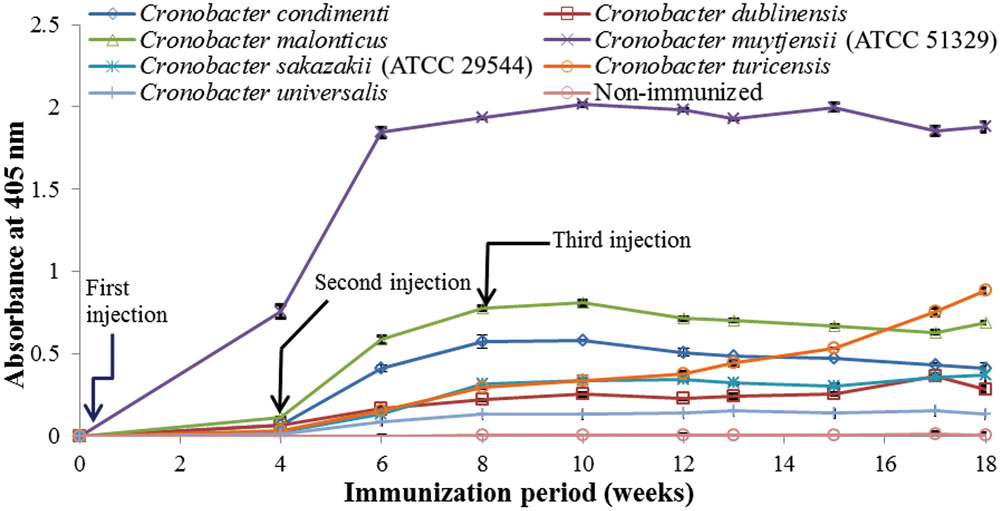
FIGURE 2. Titers of rabbit anti-serum developed from formalin killed cells mixture against seven Cronobacter species. All experiments were conducted three times, and data represent mean ± standard deviation.
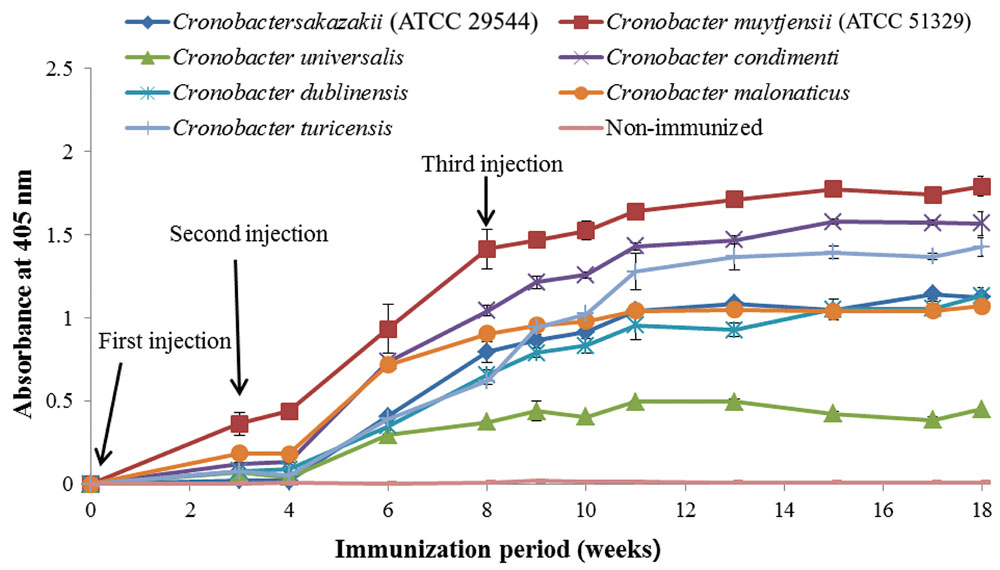
FIGURE 3. Titers of rabbit anti-serum developed from sonicated cell protein mixture against seven Cronobacter species. All experiments were conducted three times, and data represent mean ± standard deviation.
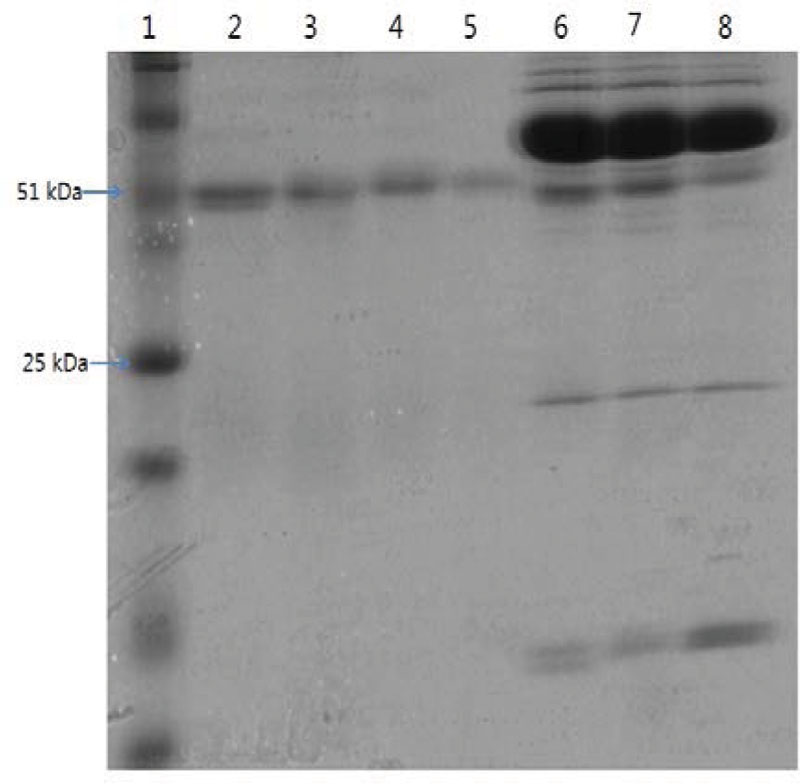
FIGURE 4. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of developed rabbit anti-Cronobacter IgG and anti-serum. Lane 1: standard protein marker, lane 2: commercial rabbit IgG, lane 3: anti-Cronobacter IgG developed from sonicated cell protein mixture of Cronobacter spp., lane 4: anti-Cronobacter IgG developed from formalin killed cells mixture of Cronobacter spp., lane 5: anti-Cronobacter IgG developed from lipopolysaccharide mixture of Cronobacter spp., lane 6: anti-serum developed from sonicated cell protein mixture of Cronobacter spp., lane 7: anti-serum developed from formalin killed cells mixture of Cronobacter spp., lane 8: anti-serum developed from lipopolysaccharide mixture of Cronobacter spp.
INC-ELISA for Detection of Cronobacter spp. Using Developed Anti-Cronobacter IgG
An INC-ELISA method was developed to detect the seven Cronobacter spp. using rabbit anti-Cronobacter IgG developed from the aforementioned SCP mixture. The developed INC-ELISA method showed excellent sensitivity toward Cronobacter spp., including C. condimenti, C. dublinensis, C. malonaticus, C. muytjensii, C. sakazakii, C. turicensis, and C. universalis, confirming its ability to detect all seven Cronobacter spp. in a single test (Table 1). Furthermore, the developed INC-ELISA was highly sensitive to all seven different Cronobacter spp. with a detection limit ranging from (5.6 ± 0.30) × 103 to (2.1 ± 0.01) × 105 CFU/mL (Table 1).
Specificity of Developed INC-ELISA
According to Table 2, the developed INC-ELISA showed no cross-reactivity with any of the other tested bacterial genera. The recognition difference between targeted bacteria (positive value; P value) and non-targeted bacteria (negative value; N value) is an important parameter affecting the results of the immunoassay. In the present study, non-targeted bacteria (non-Cronobacter) showed a P/N value of <2 (Table 2), which is considered as a negative result, whereas targeted strains showed a P/N value of >2, which is considered as a positive result (Table 1). In support of this, the P/N value for F. pulveris was around 1.942 (Table 2) where a P/N value >2 is considered as a positive result. Compared to several other methods, our newly developed INC-ELISA based on rabbit anti-Cronobacter IgG is less expensive and easier to perform since it does not require expensive reagents or probes and has an assay time of only 36 h comprising 8 h of pre-enrichment, 8 h of enrichment, and 20 h of INC-ELISA. During assay development, accuracy of the rapid detection method was confirmed by standard plate colony counting techniques. Moreover, the developed rapid assay is accurate enough to avoid repeated colony counting steps to save detection time.

TABLE 2. Cross-reactivity of the developed indirect non-competitive enzyme-linked immunosorbent assay with non-Cronobacter species in pure culture.
Detection of Cronobacter spp. in Spiked PIF Samples
To detect small amounts of Cronobacter spp. cells using the developed method, PIF samples were separately spiked with each of the seven Cronobacter spp. at low concentrations (<10 cells/25 g). All PIF samples spiked separately with C. condimenti, C. dublinesis, C. malonaticus, C. muytjensii, C. sakazakii, C. turicensis, and C. universalis showed the presence of Cronobacter spp. in both the INC-ELISA and USFDA/KFDA methods, whereas unspiked samples showed the absence of Cronobacter spp. Specific enrichment was carried out in EE broth as per KFDA standard methods (Korea Food and Drug Administration [KFDA], 2014). In the present study, the detection limit of the developed INC-ELISA against Cronobacter spp. was found to be <10 cells/25 g of PIF after 16 h of enrichment (Table 3).
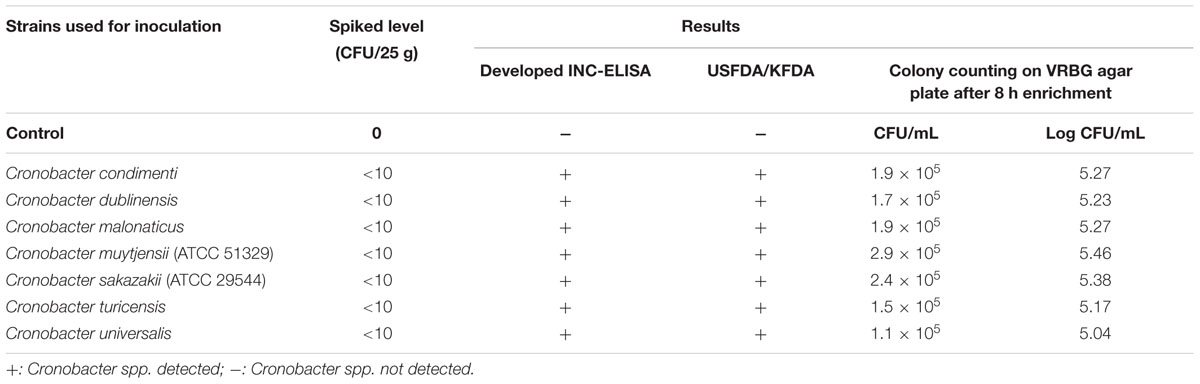
TABLE 3. Food trial for detection of Cronobacter and indirect non-Cronobacter species in artificially contaminated PIF using the developed non-competitive enzyme-linked immunosorbent assay.
Further, to confirm the results obtained using the developed antibody and INC-ELISA for Cronobacter detection, conventional standard plate count technique using the spreading method on NA and VRBG agar plates was carried out. The PIF samples artificially contaminated with each Cronobacter spp. separately (<10 cells/25 g) after 8 h of pre-enrichment and 8 h of enrichment in EE broth showed similar bacterial counts for each Cronobacter spp. in the range of 1.1 × 105 – 2.9 × 105 CFU/mL on specific VRBG agar plates (Table 3). These results can give an idea to reconfirm the detection limit obtained in pure culture of Cronobacter spp. using the developed antibody and INC-ELISA assay (Table 1).
Discussion
In this study, an INC-ELISA method based on our laboratory-produced anti-Cronobacter IgG was developed and applied for the detection of seven Cronobacter spp. in PIF. This study employed three types of immunogens (LPS, FKC, and SCP) to produce three types of antibodies in rabbit for the detection of seven Cronobacter spp. As recommended by the Canadian Council on Animal Care [CCAC] (2002), a second injection of immunogen should be performed 3 to 6 weeks after the first immunization. Therefore, in the present work, two immunogen injections were made after the first injection at an interval of 4 weeks during the immunization period. As a result, IgG developed by LPS immunogen showed no titer for any Cronobacter spp. These results agree with previous findings of Luk and Lindberg (1991) and Jaradat et al. (2011), who were unable to obtain stable antibodies against LPS from Cronobacter and Salmonella due to the structure and composition of LPS obtained from bacterial cells of Cronobacter and Salmonella.
Previously, Hochel and Skvor (2009) developed a serotype-specific polyclonal antibody against E. sakazakii (Cronobacter spp.) capable of detecting 13 E. sakazakii spp. from different isolations. Compared to the results of Hochel and Skvor (2009), the developed rabbit anti-Cronobacter IgG in this study showed a higher titer, purity, and specificity, which makes it suitable for developing a fast and simple immunoassay method for the detection of Cronobacter spp. Therefore, anti-Cronobacter IgG developed from an SCP mixture of the seven Cronobacter spp. was chosen to develop an INC-ELISA method for genus-specific detection of Cronobacter spp. Similarly, Liang et al. (2001) developed a monoclonal antibody for the detection of Bartonella spp. capable of specifically detecting all tested species of Bartonella.
In addition, Huang et al. (2013) designed two pairs of primers based on gyrB sequences for specific identification of C. sakazakii and C. dublinensis. Although these PCR-based methods can be used to rapidly analyze Cronobacter spp., their strict requirements in terms of technique, equipment, and probes limit their application (Ruan et al., 2013). Further, Zhang et al. (2010) reported a cross-priming amplification method combined with immunoblotting analysis for genus-specific detection of Cronobacter spp. under isothermal conditions.
Similarly, we developed a polyclonal antibody for C. muytjensii as well as fluorescence-based liposome immunoassay for the detection of C. muytjensii in our previous study (Song et al., 2015). In this study, the developed antibody was also able to detect other strains of C. muytjensii such as C. muytjensii ATCC 51329 and C. muytjensii CDC3523–75 (Song et al., 2015). Furthermore, as shown in Table 2, F. pulveris (LMG 24057) showed slight cross-reactivity with INC-ELISA at a high bacterial concentration (108 CFU/mL), which might be due to close resemblance between multi-locus sequence typing loci for Cronobacter genus members and Franconibacter genera (previously recognized in Enterobacter genus) (Forsythe et al., 2014). Recently, Xu et al. (2014) developed a polyclonal antibody for Cronobacter spp. by using heat-killed antigen preparation and evaluated its cross-reactivity against various Cronobacter strains of the same species. Their results confirmed cross-reactivity for all Cronobacter strains of the same species based on positive signals. The detection limit was observed in the range of 104–105 CFU/mL (Xu et al., 2014). Therefore, it can be hypothesized that the developed antibody and assay in the present study could be applicable for the detection of several Cronobacter strains of similar species (Xu et al., 2014).
The above findings confirm that the developed assay method in this study can detect Cronobacter spp. in PIF samples with high sensitivity within 36 h. Similarly, Blazkova et al. (2011) developed a method using an immunochromatographic strip for the detection of Cronobacter spp. with a detection limit less than 10 cells/10 g of PIF. In the present study, all PIF samples artificially contaminated with Cronobacter spp. were positively detected using the developed genus-specific anti-Cronobacter IgG antibody in 25 g of PIF after 16 h of sample pre-treatment (8 h of pre-enrichment and 8 h of enrichment) and 20 h of assay time. In contrast, detection and confirmation of this pathogen by the USFDA culture-based method takes up to 3–5 days.
However, the developed method has limited differentiation ability for detection of each Cronobacter spp., which must be overcome to validate its practical and industrial usefulness. Hence, development of additional immunoassays based on the produced Cronobacter genus-specific antibody multiplexing format such as ELISA are in progress in our laboratory for the purpose of developing a novel assay method with reduced detection time, high sensitivity, and multiplexing detection ability.
In summary, this study focused on the development of a simple, quick, and sensitive genus-specific detection assay for Cronobacter spp. using rabbit anti-Cronobacter IgG. For this purpose, three immunogens, LPS mixture of Cronobacter spp., FKC mixture of Cronobacter spp., and SCP mixture of Cronobacter spp., were prepared and used to develop antibodies against Cronobacter spp. The newly developed rabbit anti-Cronobacter IgG was purified using caprylic acid and ammonium sulfate precipitation from anti-sera and showed high purity similar to the commercial rabbit IgG. The rabbit anti-Cronobacter IgG developed from the SCP mixture was used to develop a new INC-ELISA method for the detection of Cronobacter spp. The developed INC-ELISA showed excellent reactivity with all seven Cronobacter spp., excluding other non-Cronobacter foodborne pathogens. These results confirm that the developed INC-ELISA is highly sensitive, efficient, and rapid for the detection of Cronobacter spp. with a markedly reduced total detection time (from 5 days to 36 h), costs, and handling procedure. This is the first report of a genus-specific antibody and immunoassay method for the detection of Cronobacter spp. with the goal of reducing risk of Cronobacter spp. contamination in food.
Author Contributions
XS, GL, and SP performed experiments and drafted manuscript; SS contributed interpretation, analyzed data, and wrote paper, MK contributed for conception, designed experiment, analyzed data, and provided technical support.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by the Basic Science Research Program through the National Research Foundation, Republic of Korea (NRF), funded by the Ministry of Education (NRF- 2014R1A2A1A11053211).
References
Anonymous (2006). Milk and Milk Products – Detection of Enterobacter Sakazakii. Technical Specification ISO/TS 22964. ISO/TS 22964:2006(E) and IDF/RM 210:2006(E), 1st Edn. (Geneva: International Organization for Standardization).
Blazkova, M., Javurkova, B., Fukal, L., and Rauch, P. (2011). Immunochromatographic strip test for detection of genus Cronobacter. Biosens. Bioelectron. 26, 2828–2834. doi: 10.1016/j.bios.2010.10.001
Brigmon, R. L., Zam, S. G., Bitton, G., and Farrah, S. R. (1992). Detection of Salmonella enteritidis in environmental samples by monoclonal antibody-based ELISA. J. Immunol. Method 152, 135–142. doi: 10.1016/0022-1759(92)90096-C
Canadian Council on Animal Care [CCAC] (2002). CCAC guidelines on: antibody production. Ottawa, Canada: The Canadian Council on Animal Care.
Cruz-Córdova, A., Rocha-Ramrez, L. M., Ochoa, S. A., González-Pedrajo, B., Espinosa, N., Eslava, C., et al. (2012). Flagella from five Cronobacter species induce pro-inflammatory cytokines in macrophage derivatives from human monocytes. PLoS ONE 7:e52091. doi: 10.1371/journal.pone.0052091
Forsythe, S. J., Dickens, B., and Jolley, K. A. (2014). Cronobacter, the emergent bacterial pathogen Enterobacter sakazakii comes of age, MLST and whole genome sequence analysis. BMC Genome 15:1115–1121. doi: 10.1186/1471-2164-15-1121
Friedemann, M. (2007). Enterobacter sakazakii in food and beverages (other than infant formula and milk powder). Int. J. Food Microbiol. 116, 1–10. doi: 10.1016/j.ijfoodmicro.2006.12.018
Giammanco, G. M., Aleo, A., Guida, I., and Mamminia, C. (2011). Molecular epidemiological survey of Citrobacter freundii misidentified as Cronobacter spp. (Enterobacter sakazakii) and Enterobacter hormaechei isolated from powdered infant milk formula. Foodborne Pathog. Dis. 8, 517–525. doi: 10.1089/fpd.2010.0719
Healy, B., Cooney, S., O’Brien, S., Iversen, C., Whyte, P., Nally, J., et al. (2010). Cronobacter (Enterobacter sakazakii): an opportunistic foodborne pathogen. Foodborne Pathog. Dis. 7, 339–350. doi: 10.1089/fpd.2009.0379
Hochel, I., and Skvor, J. (2009). Characterisation of antibodies for the immunochemical detection of Enterobacter sakazakii. Czech J. Food Sci. 27, 66–74.
Holy, O., and Forsythe, S. (2014). Cronobacter spp. as emerging causes of healthcare-associated infection. J. Hosp. Infect. 86, 169–177. doi: 10.1016/j.jhin.2013.09.011
Huang, C. H., Chang, M. T., and Huang, L. (2013). Use of the novel species-specific PCR primers targeted to DNA gyrase subunit B (gyrB) gene for species identification of the Cronobacter sakazakii and Cronobacter dublinensis. Mol. Cell Probes 27, 15–18. doi: 10.1016/j.mcp.2012.08.004
Hubaux, A., and Vos, G. (1970). Decision and detection limits for linear calibration curves. Anal. Chem. 42, 849–855. doi: 10.1021/ac60290a013
Hunter, C. J., and Bean, J. F. (2013). Cronobacter: an emerging opportunistic pathogen associated with neonatal meningitis, sepsis and necrotizing enterocolitis. J. Perinatol. 33, 581–585. doi: 10.1038/jp.2013.26
Iversen, C., Lane, M., and Forsythe, S. J. (2004). The growth profile, thermotolerance and biofilm formation of Enterobacter sakazakii grown in infant formula milk. Lett. Appl. Microbiol. 38, 378–382. doi: 10.1111/j.1472-765X.2004.01507.x
Iversen, C., Mullane, N., McCardell, B., and Tall, B. D. (2008). Cronobacter gen. nov., a new genus to accommodate the biogroups of Enterobacter sakazakii, and proposal of Cronobacter sakazakii gen. nov. comb. nov., C. malonaticus sp. nov., C. turicensis sp. nov., C. muytjensii sp. nov., C. dublinensis sp. nov., Cronobacter genomospecies 1, and of three subspecies, C. dublinensis sp. nov. subsp. dublinensis subsp. nov., C. dublinensis sp. nov. subsp. lausannensis subsp. nov., and C. dublinensis sp. nov. subsp. lactaridi subsp. nov. Int. J. Syst. Evol. Microbiol. 58, 1442–1447. doi: 10.1099/ijs.0.65577-0
Jaradat, Z. W., Al Nabulsi, A., and Tall, B. D. (2014). Cronobacter spp. – opportunistic food-borne pathogens. A review of their virulence and environmental-adaptive traits. J. Med. Microbiol. 63, 1023–1037. doi: 10.1099/jmm.0.073742-0
Jaradat, Z. W., Rashdan, A. M., Ababneh, Q. O., and Jaradat, S. A. (2011). Characterization of surface proteins of Cronobacter muytjensii using monoclonal antibodies and MALDI-TOF mass spectrometry. BMC Microbiol. 11:148. doi: 10.1186/1471-2180-11-148
Joseph, S., Cetinkaya, E., Drahovska, H., and Levican, A. (2012a). Cronobacter condimenti sp. nov., isolated from spiced meat and Cronobacter universalis sp. nov., a novel species designation for Cronobacter sp. genomospecies 1, recovered from a leg infection, water, and food ingredients. Int. J. Syst. Evol. Microbiol. 62, 1277–1283. doi: 10.1099/ijs.0.032292-0
Joseph, S., Desai, P., Ji, Y., Cummings, C. A., and Shih, R. (2012b). Comparative analysis of genome sequences covering the seven Cronobacter species. PLoS ONE 7:e49455. doi: 10.1371/journal.pone.0049455
Joseph, S., and Forsythe, S. J. (2011). Predominance of Cronobacter sakazakii sequence type 4 in neonatal infections. Emerg. Infect. Dis. 17, 1713–1715. doi: 10.3201/eid1709.110260
Jung, M. K., and Park, J. H. (2006). Prevalence and thermal stability of Enterobacter sakazakii from unprocessed ready-to-eat agricultural products and powdered infant formulas. Food Sci. Biotechnol. 15, 152–157.
Korea Food and Drug Administration [KFDA] (2014). Korea Food and Drug Administration [KFDA]. Available at: http://www.foodsafetykorea.go.kr/foodcode/menu_01_03.jsp?idx=394
Kumar, S., Balakrishna, K., and Batra, H. V. (2008). Enrichment-ELISA for detection of Salmonella typhi from food and water samples. Biomed. Environ. Sci. 21, 137–143. doi: 10.1016/S0895-3988(08)60019-7
Lai, K. K. (2001). Enterobacter sakazakii infections among neonates, infants, children, and adults. Case reports and a review of the literature. Medicine 80, 113–122. doi: 10.1097/00005792-200103000-00004
Lampel, K. A., and Chen, Y. (2009). Method for the isolation and detection of Enterobacter sakazakii (Cronobacter) from powdered infant formula. Int. J. Food Microbiol. 136, 179–184. doi: 10.1016/j.ijfoodmicro.2009.08.016
Lee, Y. D., Park, J. H., and Chang, H. (2012). Detection, antibiotic susceptibility and biofilm formation of Cronobacter spp. from various foods in Korea. Food Control 24, 225–230. doi: 10.1016/j.foodcont.2011.09.023
Leenaars, M., and Hendriksen, C. F. M. (2005). Critical steps in the production of polyclonal and monoclonal antibodies: evaluation and recommendations. ILAR J. 46, 269–279. doi: 10.1093/ilar.46.3.269
Liang, Z., Scola, B. L., Lepidi, H., and Raoult, D. (2001). Production of Bartonella genus-specific monoclonal antibodies. Clin. Diag. Lab. Immunol. 4, 847–849. doi: 10.1128/CDLI.8.4.847-849.2001
Luk, J. M., and Lindberg, A. A. (1991). Rapid and sensitive detection of Salmonella by immunomagnetic monoclonal antibody-based assays. J. Immunol. Method 137, 1–8. doi: 10.1016/0022-1759(91)90387-U
McKinney, M. M., and Parkinson, A. (1987). A simple, non-chromatographic procedure to purify immunoglobulins from serum and ascetic fluid. J. Immunol. Method 96, 271–278. doi: 10.1016/0022-1759(87)90324-3
Mullane, N. R., Murry, J., Drudy, D., and Prentice, N. (2006). Detection of Enterobacter sakazakii in dried infant milk formula by cationic-magnetic-bead capture. Appl. Environ. Microbiol. 72, 6325–6330. doi: 10.1128/AEM.03056-05
Norberg, S., Stanton, C., Ross, R. P., Hill, C., Fitzgerald, G. F., and Cotter, P. D. (2012). Cronobacter spp. in powdered infant formula. J. Food Prot. 75, 607–620. doi: 10.4315/0362-028X.JFP-11-285
Odeyemi, O. A., and Sani, N. A. (2016). The prevention of Cronobacter infections in hospital neonatal intensive care units. J. Infect. Public Health 9, 110–112. doi: 10.1016/j.jiph.2015.06.012
Park, S., Shukla, S., Kim, Y., Oh, S., Kim, S. H., and Kim, M. (2012). Development of sandwich enzyme-linked immunosorbent assay for the detection of Cronobacter muytjensii (formerly called Enterobacter sakazakii). Microbiol. Immunol. 56, 472–479. doi: 10.1111/j.1348-0421.2012.00466.x
Pinto, A., Bonerba, E., and Bozzo, G. (2013). Occurrence of potentially enterotoxigenic Bacillus cereus in infant milk powder. Euro. Food Res. Technol. 237, 275–279. doi: 10.1007/s00217-013-1988-8
Ruan, J., Li, M., Liu, Y. P., and Li, Y. Q. (2013). Rapid and sensitive detection of Cronobacter spp. (previously Enterobacter sakazakii) in food by duplex PCR combined with capillary electrophoresis-laser-induced fluorescence detector. J Chromatogr. B Analyt. Technol. Biomed. Life Sci. 922, 15–20. doi: 10.1016/j.jchromb.2013.01.008
Shukla, S., Bang, J., Heu, S., and Kim, M. (2012). Development of immunoliposome-based assay for the detection of Salmonella typhimurium. Eur. Food Res. Technol. 234, 53–59. doi: 10.1007/s00217-011-1606-6
Song, X., and Kim, M. (2013). Development of an indirect non-competitive enzyme-linked immunosorbent assay for the detection of Cronobacter muytjensii in infant formula powder. Korean J. Food Nutri. 26, 936–944. doi: 10.9799/ksfan.2013.26.4.936
Song, X., Shukla, S., Oh, S., Kim, Y., and Kim, M. (2015). Development of fluorescence-based liposome immunoassay for detection of Cronobacter muytjensii in pure culture. Curr. Microbiol. 70, 246–252. doi: 10.1007/s00284-014-0708-3
Strydom, A., Cawthorn, D. M., Cameron, M., and Witthuhn, R. C. (2012). Species of Cronobacter-A review of recent advances in the genus and their significance in infant formula milk. Int. Dairy J. 27, 3–12. doi: 10.1016/j.idairyj.2012.06.005
United States Food and Drug Administration [USFDA] (2002). Isolation and Enumeration of Enterobacter sakazakii from Dehydrated Infant Formula. Available at: http://www.fda.gov/Food/ScienceResearch/LaboratoryMethods/ucm114665.htm
Velusamy, V., Arshak, K., Korostynska, O., and Oliwa, K. (2010). An overview of foodborne pathogen detection: in the perspective of biosensors. Biotechnol. Adv. 28, 232–254. doi: 10.1016/j.biotechadv.2009.12.004
World Health Organization [WHO] (2007). Enterobacter sakazakii and Salmonella in Powdered Infant Formula. Geneva: WHO.
Xu, X., Zhang, Y., Shi, M., and Sheng, W. (2014). Two novel analytical methods based on polyclonal and monoclonal antibodies for the rapid detection of Cronobacter spp.: development and application in powdered infant formula. LWT Food Sci. Technol. 56, 335–340. doi: 10.1016/j.lwt.2013.11.028
Keywords: Cronobacter species, immunoglobulin G, indirect non-competitive enzyme-linked immunosorbent assay, genus-specificity, powdered infant formula
Citation: Song X, Shukla S, Lee G, Park S and Kim M (2016) Detection of Cronobacter Genus in Powdered Infant Formula by Enzyme-linked Immunosorbent Assay Using Anti-Cronobacter Antibody. Front. Microbiol. 7:1124. doi: 10.3389/fmicb.2016.01124
Received: 26 April 2016; Accepted: 06 July 2016;
Published: 21 July 2016.
Edited by:
Maria Schirone, University of Teramo, ItalyReviewed by:
Zhao Chen, Clemson University, USAGonçalo Nieto Almeida, Catholic University of Portugal, Portugal
Copyright © 2016 Song, Shukla, Lee, Park and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Myunghee Kim, foodtech@ynu.ac.kr
†These authors have contributed equally to this work.
 Xinjie Song1†
Xinjie Song1† Shruti Shukla1†
Shruti Shukla1† Gibaek Lee
Gibaek Lee Sunhyun Park2
Sunhyun Park2 Myunghee Kim
Myunghee Kim