- 1Belozersky Institute for Physico-Chemical Biology, Lomonosov Moscow State University, Moscow, Russia
- 2Kharkevich Institute for Information Transmission Problems, Russian Academy of Sciences, Moscow, Russia
- 3Institute of Animal Physiology, Biochemistry and Nutrition, Kaluga, Russia
- 4Institute for the Biology of Inland Waters, Russian Academy of Sciences, Borok, Russia
- 5Institute of Gene Pool of Plants and Animals, Uzbek Academy of Sciences, Tashkent, Uzbekistan
- 6National University of Uzbekistan, Tashkent, Uzbekistan
- 7Zoological Institute, Russian Academy of Sciences, St. Petersburg, Russia
- 8St. Petersburg State University, St. Petersburg, Russia
Heterotrophic lineages of Heterokonta (or stramenopiles), in contrast to a single monophyletic group of autotrophs, Ochrophyta, form several clades that independently branch off the heterokont stem lineage. The nearest neighbors of Ochrophyta in the phylogenetic tree appear to be almost exclusively bacterivorous, whereas the hypothesis of plastid acquisition by the ancestors of the ochrophyte lineage suggests an ability to engulf eukaryotic alga. In line with this hypothesis, the heterotrophic predator at the base of the ochrophyte lineage may be regarded as a model for the ochrophyte ancestor. Here, we present a new genus and species of marine free-living heterotrophic heterokont Develorapax marinus, which falls into an isolated heterokont cluster, along with the marine flagellate Developayella elegans, and is able to engulf eukaryotic cells. Together with environmental sequences D. marinus and D. elegans form a class-level clade Developea nom. nov. represented by species adapted to different environmental conditions and with a wide geographical distribution. The position of Developea among Heterokonta in large-scale phylogenetic tree is discussed. We propose that members of the Developea clade represent a model for transition from bacterivory to a predatory feeding mode by selection for larger prey. Presumably, such transition in the grazing strategy is possible in the presence of bacterial biofilms or aggregates expected in eutrophic environment, and has likely occurred in the ochrophyte ancestor.
Introduction
Heterokonta is one of the major groups of eukaryotes and encompasses organisms with a wide variety of life styles – from autotrophic diatom algae and kelps to heterotrophic bicosoecid flagellates and parasitic oomycetes. Autotrophic heterokonts form a bulk of the group’s described species, and are presumed to comprise a single monophyletic cluster Ochrophyta. By contrast, heterotrophic lineages of heterokonts do not form a single group, and are instead found as several clades that branch off the heterokont stem lineage preceding the divergence of ochrophytes (Leipe et al., 1994, 1996; Riisberg et al., 2009; Yubuki et al., 2010, 2015). The evolution of heterokonts, where autotrophs occupy the tree “crown” and heterotrophs form a grade of early-diverging lineages, implies either multiple independent transitions to heterotrophy from the ancestral autotrophic lifestyle assumed by the “Chromalveolata” hypothesis (Cavalier-Smith, 1999) or an event of plastid acquisition in the heterotrophic ancestors of the Ochrophyta. The question of autotrophic versus heterotrophic ancestry of heterokont lineages remains open (Mukhina, 2014; Petersen et al., 2014; Stiller et al., 2014; Archibald, 2015, for recent review). One of the difficulties with the latter evolutionary scenario is the apparent prevalence of bacterivory in close relatives of ochrophytes, which precludes the prerequisite for plastid acquisition – the ability to engulf eukaryotic prey. It was shown experimentally that there are optimum prey size spectra for various species of predatory protists (Hansen, 1992; Jakobsen and Hansen, 1997; Montagnes et al., 2008; Nielsen and Kiørboe, 2015). Among recent heterotrophic and mixotrophic species there are those that are able to consume both bacterial and eukaryotic prey (Strom, 1991; Burkholder et al., 2008; Piwosz and Pernthaler, 2010; Zhang et al., 2013; Lee et al., 2014; Mitra et al., 2016) while others are limited by the size of prey cells, with some eukaryotes lacking the ability to graze on bacteria (Jonsson, 1986; Hansen and Calado, 1999; Montagnes et al., 2008). Bacterivorous flagellates have two alternative feeding strategies: whereas some species are grazing on single bacterial cells only, other species are capable of ingesting aggregates of bacterial cells (Sibbald and Albright, 1988). Flagellates, adhering to the second strategy, are able to consume bacterial biofilms, including those that grow on solid substrates (Böhme et al., 2009; Erken et al., 2012).
Here, we present a new genus and species of marine free-living heterotrophic heterokont Develorapax marinus – an obligatory predator feeding on kinetoplastids and to lesser extent by ingesting bacterial cells. Its 18S and 28S rRNA genes are clustered in the phylogenetic analyses with those of Developayella elegans (Tong, 1995). The bacterivorous marine flagellate Developayella together with few environmental sequences forms a clade with unstable position in the phylogenetic trees (Beakes et al., 2012): it is either grouped with the oomycete/hyphochytrid clade (Leipe et al., 1996; Cavalier-Smith and Chao, 2006; Riisberg et al., 2009; Yubuki et al., 2010), or is nested between the ochrophytes and the oomycete/hyphochytrid clade (Yubuki et al., 2015). Thus, on the one hand, free-living Developayella-like organisms might contain ancestral characters common with parasitic oomycetes (Beakes et al., 2012) or with a larger part of heterokonts (Moriya et al., 2000). On the other hand, Developayella-like organisms might possess features shared by the ancestors of the ochrophyte lineage. We suggest that the transition from the bacterivorous to predatory mode of feeding behavior in the Developayella/Develorapax clade might have paved the way for the development of endosymbiosis with a rhodophyte cell (Bhattacharya and Medlin, 1995; Delwiche et al., 1995; Daugbjerg and Andersen, 1997; Keeling, 2004) in the initially bacterivorous ancestors of ochrophytes.
Results
General morphology of Develorapax marinus (see description in taxonomy section; Figure 1) is similar to that of Developayella elegans, the only other described and studied member of this clade (Tong, 1995). The result of molecular phylogenetic analysis with small and large subunit rRNA gene sequences confirms that Develorapax marinus and Developayella elegans are sisters. Therefore, we describe the light and electron microscopic observations of D. marinus by comparing it with D. elegans. D. marinus differs from D. elegans in the details of morphology and biological peculiarities, which justifies its description as a new genus and species.
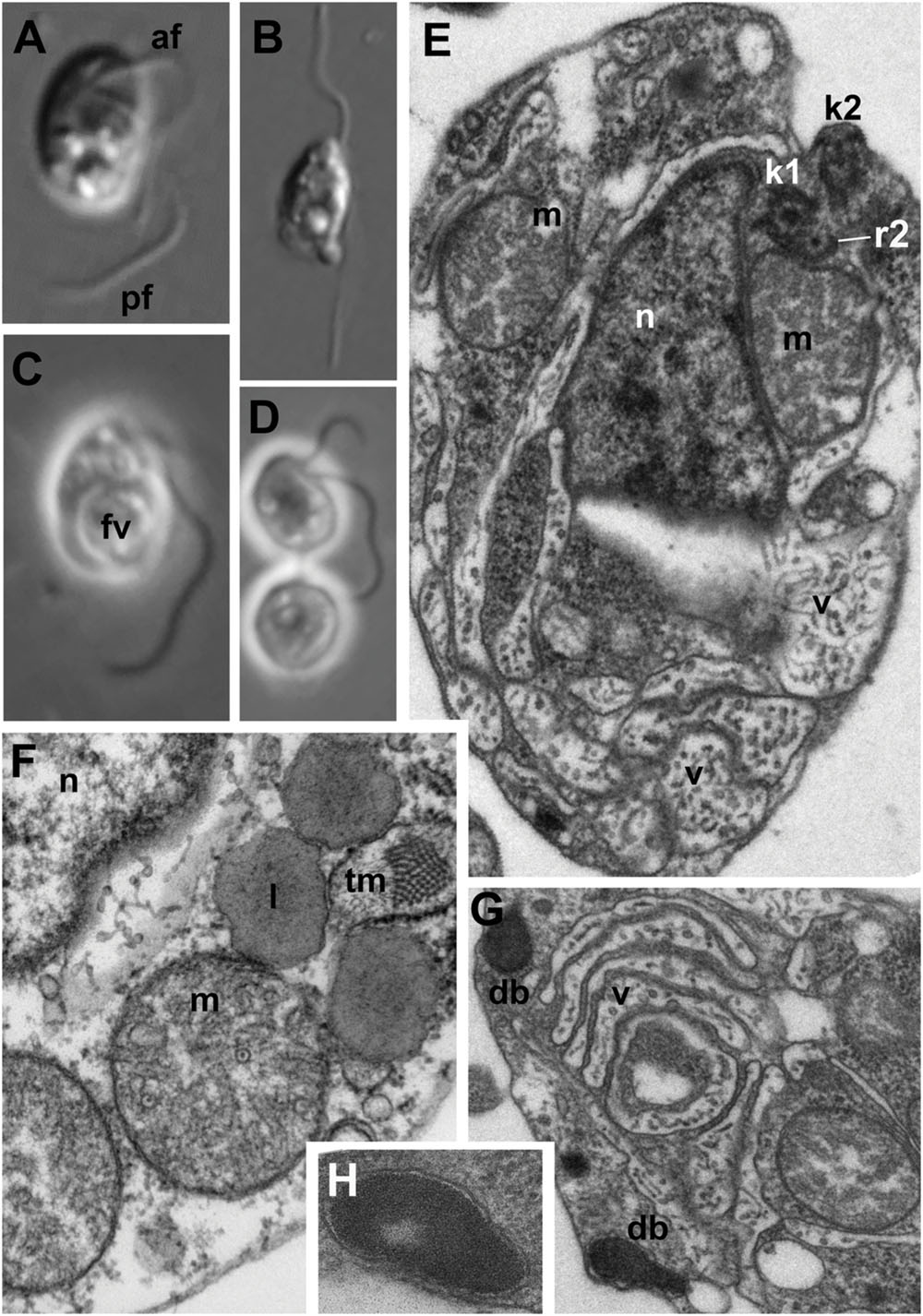
FIGURE 1. Light (A–D) and electron (E–H) microscopic images of Develorapax marinus. (A,C) Trophont, (B) swimming cell, (D) two daughter cells still connected by thin cytoplasmic bridge (arrowhead). (A,B) DIC, (C,D) phase contrast. (E) General view at longitudinal section, (F) some organelles and portion of nucleus at higher magnification (arrow shows an axial filament in the crista), (G) structure of vacuoles and dense bodies, (H) dense body at higher magnification. Scale bar: (A–D) 5 μm; (E,G) 500 nm; (F) 300 nm; (H) 125 nm. Abbreviations: af, anterior flagellum; b, bacteria; db, dense bodies; fv, food vacuole; ga, Golgi apparatus; kp, kinetoplast of the prey; k1, kinetosome of posterior flagellum; k2, kinetosome of anterior flagellum; l, lipid globule; m, mitochondrion; mp, mitochondrion of the prey; n, nucleus; nu, nucleolus; pf, posterior flagellum; pr, prey; ps, pseudopodium; r1–r4, microtubular flagellar roots; smt, secondary microtubules of r3; tm, tubular mastigonemes; tp, transversal plate; v, vacuole.
Light Microscopy
Cells of D. marinus are oval in shape, and measure 7–10 μm in length (vs. 3.5–8.5 μm for D. elegans) and 4–6 μm in width (vs. 2–6 μm for D. elegans). Two flagella are inserted at the deepest part of a pronounced depression on the right-hand side of the anterior half of the cell (Figure 1), similar to Developayella. The anterior flagellum is 1.5 of the cell length, and the posterior flagellum is about twice the cell length. Similar to D. elegans, the posterior flagellum is fastened along the bottom of the depression, whilst the anterior flagellum emerges freely, and nucleus is visible close to the point of flagella insertion.
Unlike the bacterivorous Developayella, D. marinus is able to engulf free-living bodonids Procryptobia sorokini, which were used as a prey for the culture, with the posterior end of the cell. The cells of D. marinus often contain a huge food vacuole with remnants of the bodonid prey (Figures 1C and 2C,D), and the cultures of D. marinus died off when the bodonid prey was depleted. The remnants of the bodonid mitochondrion with flat cristae and the kinetoplast were found in the food vacuole (Figure 2D). Most of the time the predator swims in a zig-zag motion, but sometimes makes broad circles hunting for prey. Sometimes the cells of D. marinus settle down to the bottom of the Petri dish staying immobile, but unlike D. elegans never attach to the substrate with a thread.
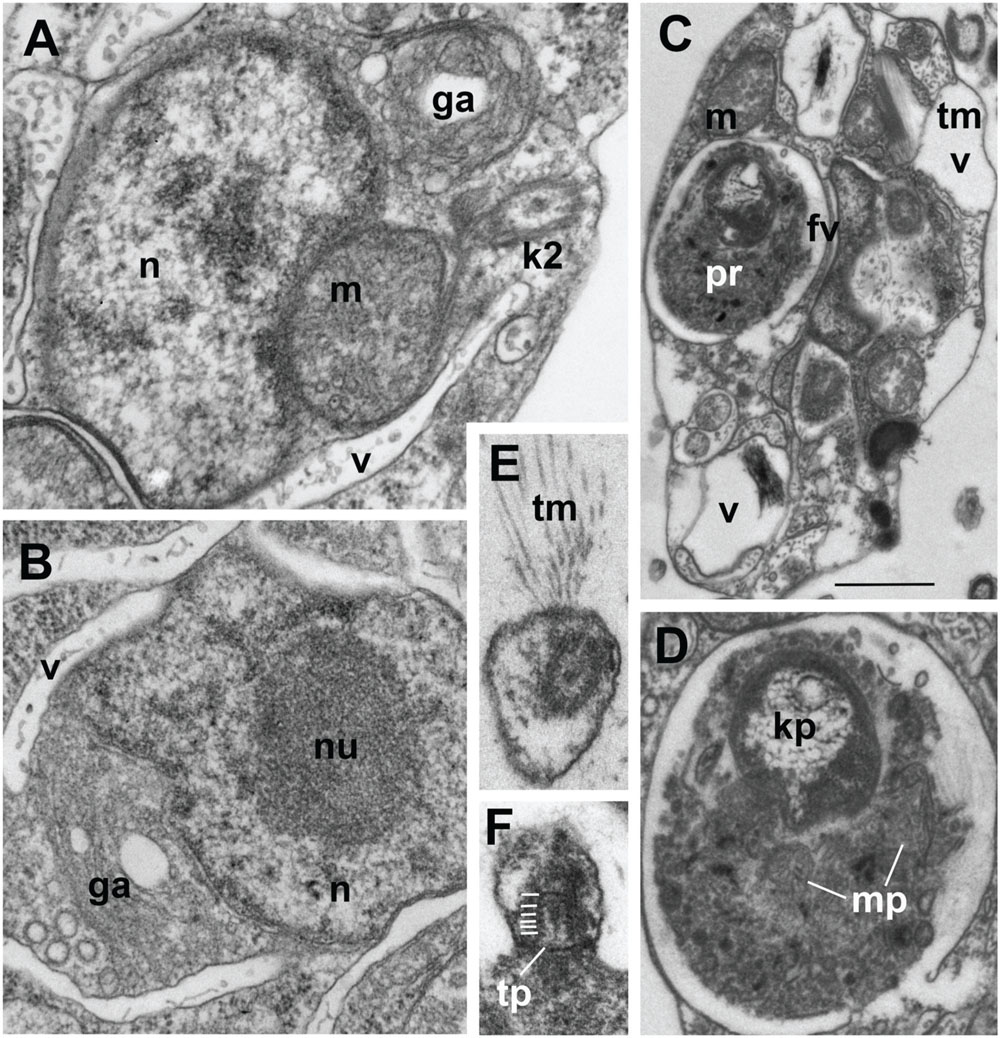
FIGURE 2. Ultrastructure of Develorapax marinus. (A,B) Nucleus and complex of associated organelles. (C) Food vacuole with partly digested kinetoplastid, (D) food vacuole at higher magnification with remained kinetoplast and mitochondrion, (E) unilateral tubular mastigonemes (tm), (F) tangential section of flagellar transition zone with six gyres of transitional helix. Scale bars: (A,B) 500 nm; (C) 1.5 μm; (D–F) 200 nm.
Electron Microscopy
General View
Similar to the cells of D. elegans, the cells of D. marinus were not well preserved in whole mount preparations, e.g., the flagellar mastigonemes were not described by this method. But in the sections the typical heterokont tubular tripartite mastigonemes are present at one side of the anterior flagellum, i.e., anterior flagellum has unilateral mastigonemes (Figures 2E and 3I,J). Vacuoles and dilations of endoplasmic reticulum with tubular mastigonemes are also commonly visible around the nucleus (Figure 1F).
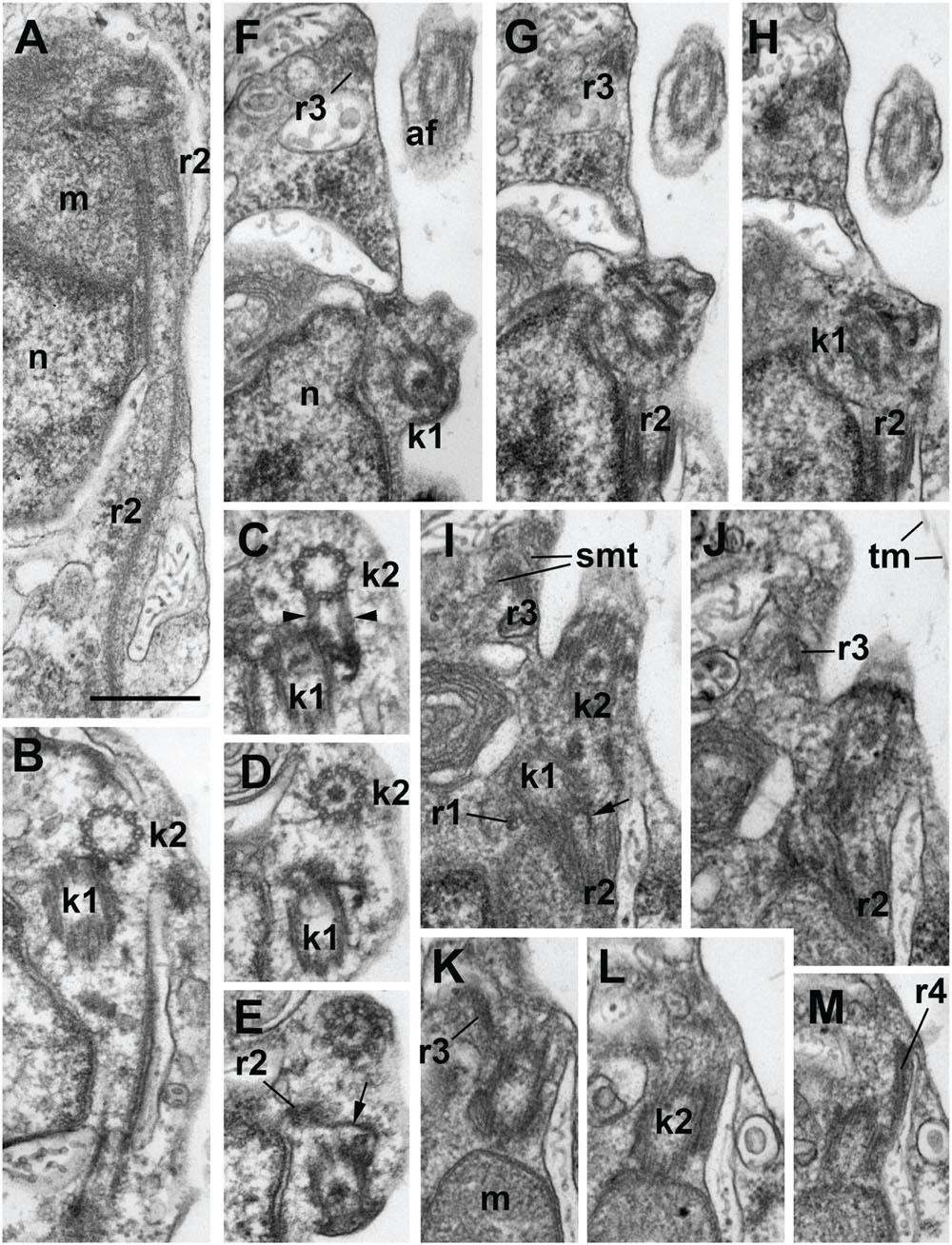
FIGURE 3. Structure of kinetid in Develorapax marinus. (A) Root 2 at longitudinal section. (B–E) Series of consecutive transverse sections of kinetosome 2. Arrowheads on (C) show two connectives between kinetosomes. (F–M) Series of consecutive longitudinal sections of kinetid. Arrows on (E,F) show the fibrillar strand of k1 becoming the core of r2. Scale bar: 400 nm.
The cell body of D. marinus is covered by the plasma membrane that is irregularly underlined by flattened broad vesicles similar to those observed in D. elegans. But unlike D. elegans these alveolar-like vesicles are not restricted to the anterior part of the cell and are also present in its posterior half (Figures 1E and 2C). They are absent in some cytoplasmic regions, but there seems to be no rule for their position. Densely staining bodies are also present under the plasma membrane, but they are bigger, rarer, and have more irregular shape than those in D. elegans, and also occur toward the cell interior. These bodies are certainly membrane bounded, and are reminiscent of extrusomes when located near the plasma membrane (Figures 1G,H), however, we found no extruded bodies. We also did not find microtubules underlying the alveoli, which are observed in D. elegans, other than a few groups that seem to belong to the flagellar roots or their secondary microtubules.
Nucleus and Organelles
Structure and location of the nucleus, mitochondria and the Golgi apparatus are similar to those in D. elegans. Nucleus with prominent central nucleolus and developed heterochromatin is located slightly anterior to the body center (Figures 1E and 2A,B). It is slightly elongated, but often has irregular shape on the sections. Mitochondria profiles with tubular cristae (some cristae contain axial fiber – Figure 1F) can be seen throughout the cell, but two of them are certainly associated with an anterior part of the nucleus: one is dorsal, another – more ventral, always associated with the kinetid (Figures 1E,F and 2A). A Golgi dictyosome lies anterior to the nucleus, and close to the kinetid.
Numerous elongated and rounded vacuoles, the so called multivesicular bodies are located around the nucleus and in the posterior part of the cell, similar to D. elegans (Tong, 1995). Most of them do not contain any food objects, but many small vesicles with cytoplasm, or membranes can be present (Figures 1E,G and 2A–C). Some vacuoles contain one or several bacteria (Figure 2C). Big vacuoles with prey at different stages of digestion also occur in the cell posterior where phagocytosis takes place (Figures 1C and 2C,D). Large food vacuoles with eukaryotic prey occur much more often in D. marinus than small food vacuoles with bacteria. The efficiency of digestion of the ingested bacteria Pseudomonas fluorescens, which are not common to marine communities, remains unknown.
Kinetid Structure
The flagella and their kinetosomes are inserted at an angle of about 60° to each other in D. elegans and up to an angle of 120° in D. marinus (Figures 3F–J). In the latter the kinetosomes lay in different planes (Figures 3B–D) and are slightly apart from each other, while in D. elegans they lay in the same plane, and the proximal end of the posterior kinetosome is attached to the side of the anterior kinetosome. The kinetosomes in both species are connected to each other by several fibers (Figures 3C,D,F–H). They have the usual 9 × 3 structure with a cartwheel pattern at the base. The organelle position around the kinetosomes is characteristic and similar in both species: the kinetosome of the anterior flagellum (K2) terminates close to the edge of a Golgi body, attaching to the mitochondrion (Figures 3K–M), while the kinetosome of the posterior flagellum (K1) passes parallel to the mitochondrion, which lies against the ventral side of the anterior part of the nucleus (Figure 3A) (Tong, 1995).
Develorapax marinus has a flagellum with short transition zone containing a transitional plate at the cell surface level, and six gyres of a double transitional helix above the plate (Figure 2F). D. elegans has the same structure of flagellar transition zone, and the number of gyres is also 6 (Figure 9 in Tong, 1995).
A conspicuous microtubular root arranged in a semicircle around an electron-dense core was found in both species (Figure 1E). This root contains 10 microtubules and passes from kinetosome 1 posterior along the right margin of the ventral depression for about 1.8 μm in D. elegans (Tong, 1995). In D. marinus this root (R2) has eight microtubules. Other elements of the root system were not illustrated for D. elegans.
Kinetid structure was investigated for D. marinus on serial sections (Figure 3), showing organization of the roots. Kinetosome 1 has two microtubular roots: short inconspicuous r1 comprising 1–2 microtubules and passing almost perpendicular from K1 to the plasma membrane, and long and prominent R2 comprising eight microtubules and passing backward nearly parallel to the ventral cell surface. Root 2 forms a semicircle at cross sections and is associated with a fibrillar core, which lies nearly at the center of the semicircle. This core is, in fact, a distal end of the fiber originating from K1 (Figures 3E,I).
Kinetosome 2 also has two microtubular roots: r3, which comprises two microtubules, produces many secondary microtubules supporting the shape of the anterior end of the cell, and a rather short r4 also comprising 1–2 microtubules and passing to the cell anterior (Figures 3J–M).
Several fibrillar interkinetosomal connectives can be noticed (Figures 3A,C–E,G,H). Some of them have rather peculiar shape, other are long and underline the anterior/right depression of the cell, but they were not studied in details.
Kinetid structure of Developayella is similar to that of Develorapax. Having more information on Develorapax we can add that R2 has a reduced number of microtubules (8 vs. 10) and an angle between the kinetosomes is not stable: in trophic cells it is about 60°, but becomes wider during the transformation into the swimming cells where it can reach 120° (Figures 3F–J).
Molecular Phylogeny
The 18S and 28S rRNA gene sequences of Develorapax marinus and Developayella elegans have 94 and 91% identity within 1795 and 3280 overlapping sites, respectively. In Bayesian inference and ML trees the sequences are grouped with a Developayella-like nanoflagellate (del Campo et al., 2013) and a small cluster of environmental sequences (Scheckenbach et al., 2010; Takishita et al., 2010) with 1.0 posterior probability or 100% bootstrap support, respectively (Figure 4 and Supplementary Figure S1). This group occupies an isolated position in the phylogenetic tree: in our analyses its closest relatives are a clade of derived environmental sample sequences from the abyssal plains of Atlantic and Pacific (Scheckenbach et al., 2010; Takishita et al., 2010; Jensen et al., 2012). The rDNA sequences of the latter clade are distinctly divergent and form a tight cluster with the exception of three environmental sequences (GU218726, GU218730, and GU2187370), which group with the “abyssal” clade in 90% of bootstrap replicates. These three almost identical sequences were generated in a single study and they passed a test for chimeric origin (Scheckenbach et al., 2010), but remain problematic in our analyses, as the inclusion of any of these three sequences in the dataset lowers the support for sister group relationship between the Developayella/Develorapax and the “abyssal” clades (Supplementary Figure S2). Ribosomal DNA sequences of the “abyssal” clade representatives are very divergent and can be a source of long branch attraction artifacts. Nevertheless, the removal of these sequences from the dataset has little effect in the overall topology of the tree (Supplementary Figure S3): the only noticeable difference in the tree topology is a switch in the relative positions of the Developayella/Develorapax and the Pirsonia-related branches.
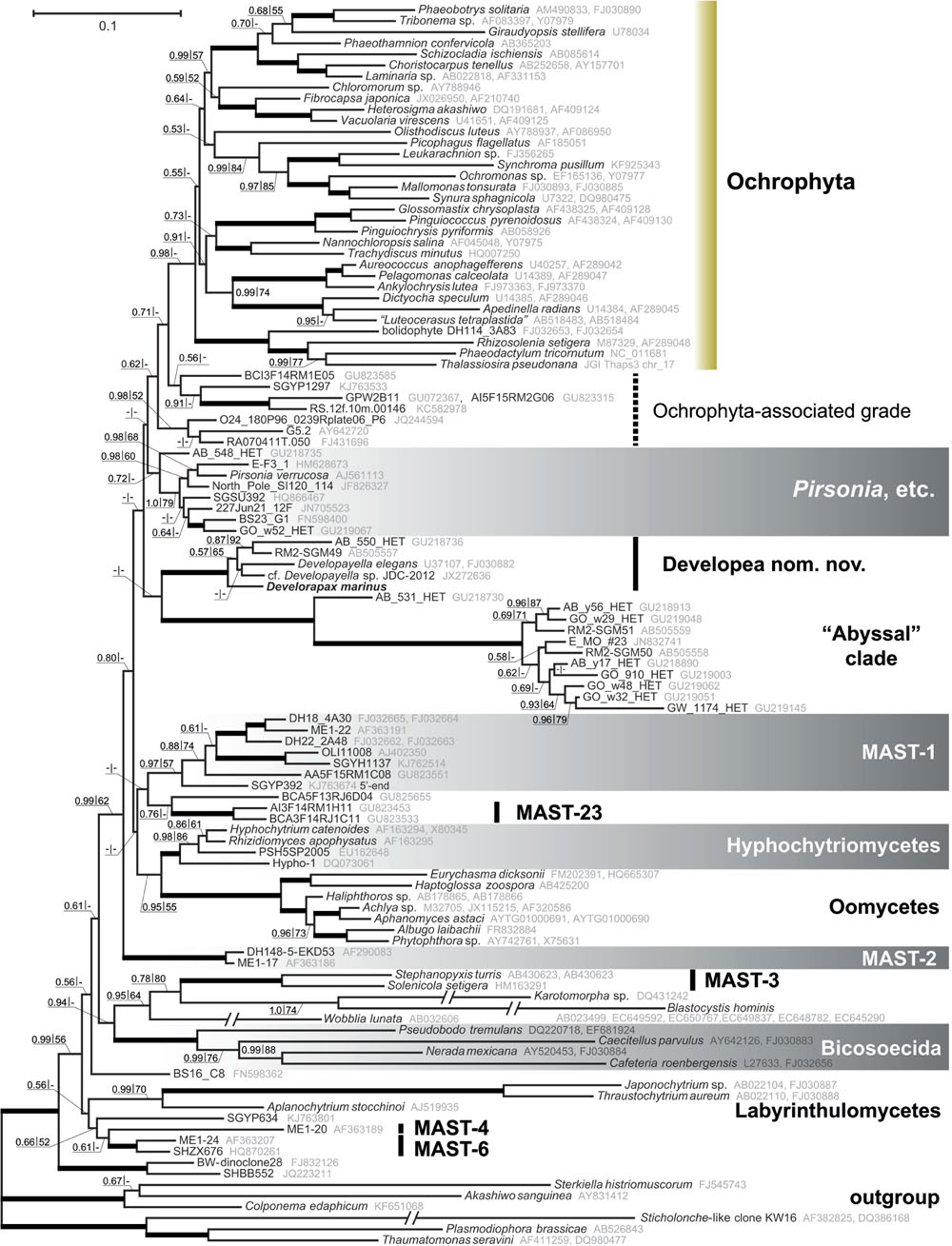
FIGURE 4. Heterokont ribosomal RNA gene phylogeny with focus on the Gyrista clade. Concatenated alignment of 18S and 28S rRNA genes with cumulative length of 4,606 positions was used for the analysis. The tree was reconstructed using the Bayesian Inference approach implemented by PhyloBayes under the CAT+GTR model with eight discrete gamma distributed rate categories. Nodes with Bayesian posterior probability of more than 0.9 and bootstrap support above 90% (RAxML) are marked with thicker lines. Resections mark branches shortened to half of their actual length.
More generally, the Developayella/Develorapax is a subclade within a much larger group of heterokonts – Gyrista sensu Cavalier-Smith and Scoble (2013). Omitting the unnamed clades of environmental sequences, the closest branch to ochrophytes in our analyses is Pirsonia, followed by the Developayella/Develorapax clade. Pseudofungi (oomycetes plus hyphochytriomycetes), MAST-1, and MAST-23 occupy an intermediate position between MAST-2 and the “crown” groups. The MAST-2 group is the first branch within the Gyrista in all analyses, although the support for the partition is only moderate. The support values for any of these partitions are low and insufficient for reaching confident conclusions about the exact branching order.
Another branch of colorless heterokonts, represented by environmental sample sequences and a well known parasitoid of diatom algae – Pirsonia (Schnepf and Schweikert, 1996; Schweikert and Schnepf, 1997; Kühn et al., 2004), diverges in between the Developayella/Develorapax cluster and the Ochrophyta (Figure 4). However, Pirsonia-related and Developayella/Develorapax branches are exchanged in the Bayesian tree when the taxon sample is changed (Supplementary Figure S3). Furthermore, these branches are sometimes grouped into one cluster with low support (not shown).
Discussion
Morphological Traits
Develorapax marinus differs from Developayella elegans by having larger cells, unilateral mastigonemes at the anterior flagellum, different kinetosome disposition: they lie in different planes, and have more varied angles; fewer microtubules (8 vs. 10) in R2; presence of dense bodies of irregular shape, and less ordered vacuoles. Cytoskeletal structures were not studied in detail in D. elegans therefore it is not possible to compare the R1, R3, and R4 roots.
The flagellar roots of Develorapax and obviously of Developayella are more similar to the ochrophyte flagellar root system, where each kinetosome has two microtubular roots. The most prominent long ventral root comprising 8–10 microtubules is, probably, homologous to the ventral root of Wobblia (or Pendulomonas – see Karpov et al., 2001) and Placidia, which are also arc-shaped in cross sections but composed of 10 microtubules with a core structure (Moriya et al., 2002).
One of the most conservative morphological characters for large scale phylogeny is the structure of the flagellar transition zone (Karpov and Fokin, 1995; Karpov, 2000). In this respect, both Developayella and Develorapax having the double helix are more similar to oomycetes than to the Ochrophyta and Pirsonia with an ordinary helix in the transition zone of flagella. A double helix was observed in Placidia flagellar transition zone (Moriya et al., 2002) therefore the state of this character in oomycetes, Developayella, and Develorapax is plesiomorphic relative to Ochrophyta and Pirsonia, and it is consistent with the position of these taxa in the phylogenetic tree (Figure 4).
The nearest described Ochrophyta neighbor is the Pirsonia – an amoeboid parasitoid of diatoms, which has biflagellate zoospores of heterokont type (Schnepf and Schweikert, 1996; Schweikert and Schnepf, 1997; Kühn et al., 2004). Zoospores of Pirsonia demonstrate features common to heterokonts: anterior flagellum with tripartite mastigonemes, tubular mitochondrial cristae, presence of an ordinary transitional helix in both flagella. At the same time their root system is unique: both microtubular roots of the anterior kinetosome (K2) are associated with fibrillar roots, and the posterior kinetosome (K1) produces only two fibrillar roots having no microtubular ones (Schnepf and Schweikert, 1996; Schweikert and Schnepf, 1997). Thus, the cytoskeleton of Pirsonia’s flagellates is rather unusual for heterokonts, and one can suggest, that this organism has reduced roots in specialized cells such as zoospores.
Occurrence and Distribution
Develorapax marinus falls into a somewhat isolated heterokont cluster, along with the marine flagellate Developayella elegans. The flagellate D. elegans is a rare species: Among nearly 766 million raw rDNA sequence reads from 334 plankton samples collected during the circumglobal Tara Oceans expedition only 32 sequences from five samples have been found to belong to D. elegans or its close relatives (de Vargas et al., 2015). Nonetheless, Developayella elegans was registered after the first description (Tong, 1995) from marine sites worldwide (subtropical Australia, England and Korea [Lee, 2007]) at a wide range of depths (from surface water samples [Tong, 1997] to the depth of 467 m [Lee, 2007]) and salinity (from 28‰ [Tong, 1997] or freshwater [Lee and Patterson, 1998] to 67‰ [Patterson and Simpson, 1996; Al-Qassab et al., 2002]). D. elegans has been observed in planktonic and diverse benthic ecosystems (Lee and Patterson, 1998), in association with shallow microbial mats as well as with adjacent sediments (Al-Qassab et al., 2002). It can be hypothesized that a complex of cryptic highly specialized species is concealed under the name of “D. elegans.” Related to D. elegans sequences were found in plankton (de Vargas et al., 2015), microbial mats in deep-sea sulfide rich sediment associated with seeping fluids, Japan (Takishita et al., 2010), abyssal plains of the southeastern Atlantic (Scheckenbach et al., 2010), and from the Mediterranean plankton (del Campo et al., 2013). In the latter case, a stable culture of the bacteriotrophic Developayella-like nanoflagellates was isolated, and differences from the D. elegans in size and shape of the cells were detected, as well as differences in the 18S rRNA gene sequences (del Campo et al., 2013). D. elegans is the only described member of a whole group of morphologically diverged and ecologically specialized heterokonts.
Endosymbiont Acquisition in Ochrophyta-Associated Heterotrophs
Rhodophyte-type plastids are found in many classes of Ochrophyta (Yoon et al., 2002), and the emergence of these plastids can be viewed as a relatively late event in heterokont evolution (Leipe et al., 1994, 1996; Stiller et al., 2014), as the majority of its early diverging lineages are heterotrophic, while autotrophs are located only within its diverse crown (Cavalier-Smith et al., 1994; Leipe et al., 1994, 1996; Guillou et al., 1999; Moriya et al., 2000, 2002; Karpov et al., 2001; Ben Ali et al., 2002; Marande et al., 2009; Riisberg et al., 2009; Yubuki et al., 2010, 2015; Harder et al., 2014). Eukaryotrophy has been documented for heterokonts outside Ochrophyta, the bicosoecids (Cafeteria), which can engulf a tiny Ostreococcus, which has the size of a bacterium (0.8 μm; Christaki et al., 2005). Unidentified thraustochytrids MAST-6 are likely able to ingest both algae (∼70%) and bacteria (up to 20%; Piwosz and Pernthaler, 2010). Other thraustochytrids Amphitremida seem to get the autotrophy independently the ochrophytes by the acquisition of green algal symbionts (Gomaa et al., 2013). All these groups are not close relatives to ochrophytes (Figure 4), and also can be considered as facultative predators, taking into account that the protists normally select the food particles by size and can eat both bacteria and picoeukaryotes. Develorapax is definitely an obligate predator engulfing large kinetoplastid cells by pseudopodia and its gene sequences branch off at the base of the Ochrophyta lineage. Thus, it can be suggested that it is a good candidate for a model of heterotrophic ancestor of the Ochrophyta.
The secondary loss of plastid hypothesis (Cavalier-Smith, 2002) is less parsimonious under the constraints of the topology obtained in our Bayesian tree, which also agrees with the trees obtained in previous studies. The list of heterotrophic lineages, in addition to Pirsonia (Kühn et al., 2004), MAST-1, MAST-2 (Massana et al., 2004, 2006, 2009), Pseudofungi, and Developayella, might also include a number of unnamed clades from the aphotic zones. The likely candidate is the “abyssal” clade, which is closely related to the Developayella/Develorapax group. Members of this clade were found in samples from white-colored microbial mats in deep-sea sulfide rich sediment associated with seeping fluids (Sagami Bay, Japan; Takishita et al., 2010), abyssal plains of the southeastern Atlantic (Scheckenbach et al., 2010), and in plankton from deep-water coral reef, Norwegian Sea (Jensen et al., 2012). Another candidate is MAST-23 (Massana et al., 2014), which are likely related to MAST-1 or Pseudofungi. Members of MAST-23 were found in a micro-oxic water column of the Caribbean Sea at depth of 200–900 m (Edgcomb et al., 2011). Micro-oxic habitats are not common for phototrophic eukaryotes, nor are deep water habitats, which hint at their likely heterotrophic lifestyle. The environmental sequences BCI3F14RM1E05, A95F15RM4A12, and their neighbors, which are grouped next to the ochrophytes (Figure 4), were also recovered from the same site in the Caribbean Sea. Another sequence related to the ochrophytes, GPW2B11, was found in an oxygen-depleted shallow sample from the Arabian Sea (Jebaraj et al., 2010). Two sequences with 100% identity (RA070411T.050 and RA070411T.002) were found in the total surface seawater sample, but were not detected in the sample after enrichment for photosynthetic organisms using flow cytometric cell sorting (Marie et al., 2010), therefore they are likely derived from non-photosynthetic organisms. The sequences of Ochrophyta-associated grade (Figure 4) are few in the environmental samples, and the conditions for the acquisition of some of them do not preclude their autotrophic lifestyle (Lefranc et al., 2005; Acosta et al., 2013; Lie et al., 2014) whereas for the other members of the grads are unambiguous indication of their heterotrophy. We suggest that bacterivory is the likely ancestral mode of phagocytosis as it is very common for the lineages of heterotrophic heterokonts. In this regard, Develorapax might be representative of the possible mechanism for endosymbiont acquisition in the ochrophyte lineage. We observed that the D. marinus tries to engulf the aggregates of bacterial cells in conditions of excessive bacterial mass in the culture medium, but it does not help to survive without eukaryotic prey. The formation of microcolonies or cell aggregates in plankton or on substrates is a common bacterial adaptive trait against protozoan grazing (Hahn et al., 2000; Jürgens and Matz, 2002; Matz et al., 2004; Matz and Kjelleberg, 2005; Jousset, 2012). However bodonids, which are common in eutrophic environments, and some other protozoans overcome this mechanism of bacterial protection by ingesting cell aggregates (Sibbald and Albright, 1988; Böhme et al., 2009; Erken et al., 2012). Members of the Developayella/Develorapax clade were found in the communities associated with bacterial mats (Al-Qassab et al., 2002; Takishita et al., 2010) or coral reefs (Jensen et al., 2012). It can be hypothesized that some bacterivorous members of eutrophic communities have acquired the ability to phagocytize not only individual bacterial cells but also their aggregates and larger eukaryotic prey. Ingestion of detritus is an option for food switching in predatory species (Pratt and Cairns, 1985), therefore bacterivory might reflect their ancestral trophic strategy. For example, MAST-6 strains are mixotrophic (algivorous and bacterivorous) in eutrophic water of the Southern Baltic Sea (Piwosz and Pernthaler, 2010) in contrast to the other known MAST organisms from oligotrophic waters that were shown to be bacterivorous (Massana et al., 2009). According to the rhodoplex (Petersen et al., 2014) or serial endosymbioses (Stiller et al., 2014) hypotheses, the Ochrophyta ancestor is suggested to be able to ingest an eukaryotic-sized cell of the cryptophyte-related endobiont precursor. Other scenarios, which suggest the origin of Ochrophyta by direct symbiosis with red algae, do not require a special adaptation to engulf eukaryotic cells because the cells of red algae can be very small. However, there are additional ways for prey to avoid the predators (Matz and Kjelleberg, 2005; Montagnes et al., 2008; Jousset, 2012). For example, the cells of the red microalgae are encapsulated within a cell wall composed of polysaccharides and glycoproteins, which protect the prey from recognition and digestion. This makes the cells of red algae unsuitable for a broad range of potential predators and may result in predator-prey specificity (Ucko et al., 1997, 1999; Martel, 2009). The scenario involving multiple symbiosis with rhodophytes requires independent symbiosis of one species of red algae with various hosts to explain the monophyletic plastid lineage (Yoon et al., 2002; Stiller et al., 2014) in the phyla Ochrophyta, Haptophyta, and Cryptophyta. Although the mechanisms for endosymbiont acquisition are far from being clear, the aforementioned adaptations of red algae against predation undermine the likelihood of direct symbioses with multiple distinct hosts. Unfortunately, the ability to engulf eukaryotic prey has not been tested experimentally for other members of the Developayella/Develorapax clade or any representatives of the Ochrophyta-associated grade. The Developayella/Develorapax clade is not sister to the ochrophytes in our trees and is unlikely to be representative of their immediate ancestor, but it may indicate the conditions under which an ancestor of ochrophytes acquired the ability to phagocytize eukaryotic prey and subsequently develop symbiotic relationship with a photosynthetic alga.
Taxonomy
Develorapax Karpov et Aleoshin
Free swimming heterotrophic heterokont flagellate, predator. Anterior flagellum with unilateral mastigonemes, kinetosomes lie in different planes, with angles between kinetosomes from 60 to 120°. Root R2 comprises eight microtubules and forms an arc with a core at the proximal end at cross sections. Prominent flat or elongated vesicles with vermiform contents are abundant in cytoplasm; they do not form a layer of subplasmalemmal vesicles. The 18S and 28S sequences of Develorapax marinus have 94 and 91% identity, respectively, to those of Developayella elegans.
D. marinus Karpov et Aleoshin (Figures 1–3)
Cells are oval in shape, measuring 7–10 μm in length and 4–6 μm in width. Two flagella are inserted at the deepest part of a conspicuous depression at the right anterior half of the cell; anterior flagellum is 1.5 of the cell length, posterior flagellum is about twice the cell length; posterior flagellum is fastened along the bottom of the depression, while the anterior flagellum emerges freely. Flagellates are able to engulf free-living bodonids and do not produce a thread for attachment to the substrate.
Clonal culture Colp-4a was produced as a generation of a single cell isolated by a micropipette from a sample taken from the littoral of the Red Sea, 51°08′ northern latitude, 54°59′ eastern longitude, salinity 42‰ on the 4th of November, 2010. The culture is now extinct.
The blocks of epoxy resin with cells of Develorapax marinus are deposited at the culture collection of the Institute for the Biology of Inland Waters of the Russian Academy of Sciences (Borok, Yaroslavl district, Russia).
Earlier, Cavalier-Smith (1997) established class Bigyromonadea in phylum Bigyra to accommodate genus Developayella. The results of our analysis agree with a class level rank for the Developayella/Develorapax clade. However, the later division of the polyphyletic “Bigyra” assemblage sensu Cavalier-Smith (1997) lead to another, still polyphyletic, phylum “Bigyra” sensu Cavalier-Smith and Chao (2006), but now without the Bigyromonadea class. The International Code of Zoological Nomenclature does not enforce priority for higher level taxa, and to avoid possible confusion, caused by the association of the class name with the paraphyletic “Bigyra” nom. ambig., we propose a new class name for the Developayella/Develorapax clade – Developea nom. nov., including the order Developayellales and the family Developayellaceae (Cavalier-Smith, 1997).
The results of our analysis do not support the Developea+Oomycete (Leipe et al., 1996) or Developea+Pseudofungi group hypothesis (Cavalier-Smith and Chao, 2006). Furthermore, the extended sample of heterokont sequences studied here highlights the problems of reconstruction an accurate rDNA-based tree, and as yet insufficient sampling of the heterokont diversity, especially in close relatives of ochrophytes. At the same time, our results support the monophyly of a large group of heterokonts discovered earlier (Cavalier-Smith and Scoble, 2013), which includes Ochrophyta, Pirsonia and related environmental clones, Developea, Pseudofungi (oomycetes plus hyphochytriomycetes), MAST-1, MAST-23, another 3-4 isolated clades of environmental sample sequences, and MAST-2 as the first branch of the assemblage. Cavalier-Smith and Scoble (2013) referred to this large group as Gyrista, introduced earlier as a superphylum to accommodate Ochrophyta and “Bigyra,” and as a likely sister group to Sagenista, which includes Labyrinthulea and its closest relatives (Cavalier-Smith, 1997). The Sagenista were later included in the paraphyletic “Bigyra” (Cavalier-Smith and Chao, 2006; Cavalier-Smith and Scoble, 2013). In the ML tree from an earlier work (Cavalier-Smith and Scoble, 2013), Opalozoa, a part of the paraphyletic “Bigyra,” is sister to Gyrista. The same relationship can be observed in our Bayesian tree, albeit with poor support (Figure 4). Therefore, we consider it justified to keep the name Gyrista for the clade that is sister to Opalozoa, or alternatively recommend a new name, which would not be burdened by a history of previous revisions.
Materials and Methods
Clonal culture of Colp-4a was produced as a generation of a single cell isolated by micropipette from the sample which has been taken from the littoral of the Red Sea, 51°08′ northern latitude, 54°59′ eastern longitude on 04th November 2010. The culture was maintained in Petri dishes with artificial marine medium Schmalz-Pratt (35‰; 28.15 g/l NaCl, 0.67 g/l KCl, 5.51 g/l MgCl2 × 6 H2O, 6.92 g/l MgSO4 × 7 H2O, 1.45 g/l CaCl2 × H2O, 0.1 g/l KNO3, and 0.01 g/l K2HPO4 × 3H2O in deionized water) without vitamins or microelements. The suspension of bacterivorous flagellates Procryptobia sorokini Frolov, Karpov et Mylnikov 2001 (Kinetoplastea) has been used as a food for Colp-4a. P. sorokini was fed by bacteria Pseudomonas fluorescens Migula.
Light microscopic (LM) observations were made with an AxioScope A1 (Carl Zeiss, Germany) using phase (PH) and differential interference contrast (DIC). The objectives of water immersion (40x and 70x) have been used. The microscope had analogous video camera AVT HORN MC-1009/S. The video frames have been directly digitized with tuner Behold TV 409 FM.
For transmission electron microscopy (TEM), cells were centrifuged and fixed in a cocktail of 0.6% glutaraldehyde and 2% OsO4 on Schmaltz-Pratt medium for 15–30 min at 1°C then dehydrated in alcohol and acetone series (30, 50, 70, 96, and 100%) for 10–30 min. Afterward, the cells were embedded in a mixture of Araldite and Epon (Spurr, 1969). Ultrathin sections were obtained with a LKB ultramicrotome and observed at JEM-1011 (Japan).
DNA Extraction and Sequencing
The DNA was extracted with a Diatom DNA Prep kit (IsoGen Lab, Moscow). The rRNA gene fragments were amplified using Encyclo PCR kit (Evrogen, Moscow) and a set of primers (Medlin et al., 1988; Van der Auwera et al., 1994) using the following protocol: initial denaturation at 95°C for 3 min, 35 cycles of 92°C for 30 s, 55°C for 30 s, 72°C for 1.5 min, followed by a final extension period at 72°C for 10 min. PCR products were gel isolated and purified using the Cytokine DNA isolation kit (Cytokine, Russia) and cloned using the InsTAclone PCR Cloning Kit (Thermo Fisher Scientific). Clones containing rRNA gene fragments of Develorapax marinus and a prey Procryptobia sorokini were discriminated by sequencing. The Develorapax marinus rDNA sequences are deposited in GenBank under Accession Numbers KX500025–KX500026.
Data Set Construction
Ribosomal DNA sequences of Develorapax marinus were aligned with 105 other members of Heterokonta found in the GenBank database, including uncultured clone sequences. Sequences were selected using the following scheme. First, we collected all heterokont large subunit rRNA genes outside of the Ochrophyta and included a few representatives from each class of the Ochrophyta, then we collected small subunit rRNA genes for all listed species if they were available. If the small subunit rRNA gene was missing for the selected species, a closely related species was used instead. Second, all fragments of heterokont small subunit rRNA genes outside of Labyrinthulomycetes that occupied isolated positions on the distance tree were selected from cultured strains and environmental samples. Third, two partially overlapping environmental sequences GPW2B11 (Accession Number GU072367) and A95F15RM4A12 (Accession Number GU823490) were concatenated into a single OTU because they both were found to be closely related to environmental sequence RS.12f.10m.00146 (Accession Number KC582978). Fourth, environmental sequence SGYP392 (Accession Number KJ763674), which is derived from a basal member of MAST-1 clade, was trimmed by 488 nucleotides from the 3′-end, which were derived from an ochrophyte species, a member of Dictyochophyceae: Florenciellales. Fifth, we removed the following sets of sequences: redundant sequences in the large clades, highly divergent sequences (Rictus lutensis, Diplophrys spp. etc.) branching far from the Developea, and environmental sequences, which were a suspect of chimeric origin (AY381178, GU219138, HQ867445, JN692703, JQ222877, KJ757848, KJ757946, KJ758005, KJ758134, KJ760760, KJ762852, KJ762875, KJ763015, KJ763027, KJ763520, and KJ763707). Three alveolate and three rhizarian species were chosen as an outgroup. Alignments were generated with MUSCLE (Edgar, 2004) and refined manually using BioEdit (Hall, 1999). After discarding ambiguously aligned nucleotide positions and concatenating the alignments of small and large subunit rRNA genes the alignment consisted of 4606 positions. GenBank Accession Numbers of the aligned sequences are given next to the sequence names in the tree (Figure 4).
Phylogenetic Analysis
Tree search for the concatenated alignment was performed using the Bayesian method implemented by PhyloBayes version 3.2 (Lartillot et al., 2009) under the GTR+CAT+Γ8 model (Lartillot and Philippe, 2004) which was shown to be the least sensitive to systematic errors (Lartillot and Philippe, 2008). Four independent chains were run for 30,000 cycles sampling trees every 10 cycles and the first 2,000 points were discarded as burn-in. Bootstrap support values for the consensus tree reconstructed by PhyloBayes were generated using RAxML v. 7.2.6 (Stamatakis, 2006) on the basis of 1,000 replicates under the GTR+Γ+I model.
Author Contributions
Conceived and designed the experiments: SK, AM, and VA. Performed the experiments: SK, AM, GM, KM, and VA. Analyzed the data: SK, AM, KM, VA. Writing of the manuscript: SK, AM, KM, and VA.
Funding
Cultivation of the flagellates and deposition of the epoxy resin blocks of the hapantotype was supported by the Russian Foundation for Basic Research, grant No 15-29-02734. The phylogenetic analysis in this study was supported by the Russian Scientific Foundation grant No. 14-50-00029. We thank the French Agence Nationale de la Recherche (project ANR-15-CE32-0003 “ANCESSTRAM”) supported morphological study, the Research Resource Center for Molecular and Cell Technologies (RRC MCT) at St. Petersburg State University (SPbSU) for access to the EM facilities and the Supercomputing Center of Lomonosov Moscow State University (http://parallel.ru/cluster) for access to the computation resources.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank T. G. Simdyanov for providing the samples of marine sediment, a source of the Develorapax marinus culture, and the reviewers for critical comments to early version of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01194
References
Acosta, F., Ngugi, D. K., and Stingl, U. (2013). Diversity of picoeukaryotes at an oligotrophic site off the Northeastern Red Sea Coast. Aquat. Biosyst. 9:16. doi: 10.1186/2046-9063-9-16
Al-Qassab, S., Lee, W. J., Murray, S., Simpson, A. G. B., and Patterson, D. J. (2002). Flagellates from stromatolites and surrounding sediments in Shark Bay, Western Australia. Acta Protozool. 41, 91–144.
Archibald, J. M. (2015). Genomic perspectives on the birth and spread of plastids. Proc. Natl. Acad. Sci. U.S.A. 112, 10147–10153. doi: 10.1073/pnas.1421374112
Beakes, G. W., Glockling, S. L., and Sekimoto, S. (2012). The evolutionary phylogeny of the oomycete “fungi.” Protoplasma 249, 3–19. doi: 10.1007/s00709-011-0269-2
Ben Ali, A., De Baere, R., De Wachter, R., and Van de Peer, Y. (2002). Evolutionary relationships among heterokont algae (the autotrophic stramenopiles) based on combined analyses of small and large subunit ribosomal RNA. Protist 153, 123–132. doi: 10.1078/1434-4610-00091
Bhattacharya, D., and Medlin, L. (1995). The phylogeny of plastids: a review based on comparisons of small-subunit ribosomal RNA coding regions. J. Phycol. 31, 489–498. doi: 10.1111/j.1529-8817.1995.tb02542.x
Böhme, A., Risse-Buhl, U., and Küsel, K. (2009). Protists with different feeding modes change biofilm morphology. FEMS Microbiol. Ecol. 69, 158–169. doi: 10.1111/j.1574-6941.2009.00710.x
Burkholder, J. A. M., Glibert, P. M., and Skelton, H. M. (2008). Mixotrophy, a major mode of nutrition for harmful algal species in eutrophic waters. Harmful Algae 8, 77–93. doi: 10.1016/j.hal.2008.08.010
Cavalier-Smith, T. (1997). Sagenista and Bigyra, two phyla of heterotrophic heterokont chromists. Arch. Protistenkunde 148, 253–267. doi: 10.1016/S0003-9365(97)80006-1
Cavalier-Smith, T. (1999). Principles of protein and lipid targeting in secondary symbiogenesis: euglenoid, dinoflagellate, and sporozoan plastid origins and the eukaryote family tree. J. Eukaryot. Microbiol. 46, 347–366. doi: 10.1111/j.1550-7408.1999.tb04614.x
Cavalier-Smith, T. (2002). Chloroplast evolution: secondary symbiogenesis and multiple losses. Curr. Biol. 22, R62–R64. doi: 10.1016/S0960-9822(01)00675-3
Cavalier-Smith, T., Allsopp, M. T. E. P., and Chao, E. E. (1994). Thraustochytrids are chromists, not Fungi: 18S rRNA signatures of Heterokonta. Philos. Trans. R. Soc. Lond. B 346, 387–397. doi: 10.1098/rstb.1994.0156
Cavalier-Smith, T., and Chao, E. E. (2006). Phylogeny and megasystematics of phagotrophic heterokonts (kingdom Chromista). J. Mol. Evol. 62, 388–420. doi: 10.1007/s00239-004-0353-8
Cavalier-Smith, T., and Scoble, J. M. (2013). Phylogeny of Heterokonta: incisomonas marina, a uniciliate gliding opalozoan related to Solenicola (Nanomonadea), and evidence that Actinophryida evolved from raphidophytes. Eur. J. Protistol. 49, 328–353. doi: 10.1016/j.ejop.2012.09.002
Christaki, U., Vazquez-Domınguez, E., Courties, E. C., and Lebaron, P. (2005). Grazing impact of different heterotrophic nanoflagellates on eukaryotic (Ostreococcus tauri) and prokaryotic picoautotrophs (Prochlorococcus and Synechococcus). Environ. Microbiol. 7, 1200–1210. doi: 10.1111/j.1462-2920.2005.00800.x
Daugbjerg, N., and Andersen, R. A. (1997). A molecular phylogeny of the heterokont algae based on analyses of chloroplast-encoded rbcL sequence data. J. Phycol. 33, 1031–1041. doi: 10.1111/j.0022-3646.1997.01031.x
de Vargas, C., Audic, S., Henry, N., Decelle, J., Mahé, F., Logares, R., et al. (2015). Eukaryotic plankton diversity in the sunlit ocean. Science 348, 6237. doi: 10.1126/science.1261605
del Campo, J., Not, F., Forn, I., Sieracki, M. E., and Massana, R. (2013). Taming the smallest predators of the oceans. ISME J. 7, 351–358. doi: 10.1038/ismej.2012.85
Delwiche, C. F., Kuhsel, M., and Palmer, J. D. (1995). Phylogenetic analysis of tufA sequences indicates a cyanobacterial origin of all plastids. Mol. Phylogenet. Evol. 4, 110–128. doi: 10.1006/mpev.1995.1012
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Edgcomb, V., Orsi, W., Bunge, J., Jeon, S., Christen, R., Leslin, C., et al. (2011). Protistan microbial observatory in the Cariaco Basin, Caribbean. I. Pyrosequencing vs Sanger insights into species richness. ISME J. 5, 1344–1356. doi: 10.1038/ismej.2011.6
Erken, M., Farrenschon, N., Speckmann, S., Arndt, H., and Weitere, M. (2012). Quantification of individual flagellate – bacteria interactions within semi-natural biofilms. Protist 163, 632–642. doi: 10.1016/j.protis.2011.10.008
Gomaa, F., Mitchell, E. A. D., and Lara, E. (2013). Amphitremida (Poche, 1913) is a new major, ubiquitous Labyrinthulomycete clade. PLoS ONE 8:e53046. doi: 10.1371/journal.pone.0053046
Guillou, L., Chrétiennot-Dinet, M. J., Boulben, S., Moon-van der Staay, S. Y., and Vaulot, D. (1999). Symbiomonas scintillans gen. et sp. nov. and Picophagus flagellatus gen. et sp. nov. (Heterokonta): two new heterotrophic flagellates of picoplanktonic size. Protist 150, 383–398. doi: 10.1016/S1434-4610(99)70040-4
Hahn, M. W., Moore, E. R., and Höfle, M. G. (2000). Role of microcolony formation in the protistan grazing defense of the aquatic bacterium Pseudomonas sp. MWH1. Microb. Ecol. 39, 175–185. doi: 10.1007/s002480000026
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Hansen, P. J. (1992). Prey size selection, feeding rates and growth dynamics of heterotrophic dinoflagellates with special emphasis on Gyrodinium spirale. Mar. Biol. 114, 327–334. doi: 10.1007/BF00349535
Hansen, P. J., and Calado, A. J. (1999). Phagotrophic mechanisms and prey selection in free-living dinoflagellates. J. Eukaryot. Microbiol. 46, 382–389. doi: 10.1111/j.1550-7408.1999.tb04617.x
Harder, C. B., Ekelund, F., and Karpov, S. A. (2014). Ultrastructure and phylogenetic position of Regin rotiferus and Otto terricolus genera et species novae (Bicosoecida, Heterokonta/Stramenopiles). Protist 165, 144–160. doi: 10.1016/j.protis.2014.01.004
Jakobsen, H. H., and Hansen, P. J. (1997). Prey size selection, grazing and growth response of the small heterotrophic dinoflagellate Gymnodinium sp. and the ciliate Balanion comatum – A comparative study. Mar. Ecol. Prog. Ser. 158, 75–86. doi: 10.3354/meps158075
Jebaraj, C. S., Raghukumar, C., Behnke, A., and Stoeck, T. (2010). Fungal diversity in oxygen-depleted regions of the Arabian Sea revealed by targeted environmental sequencing combined with cultivation. FEMS Microbiol. Ecol. 71, 399–412. doi: 10.1111/j.1574-6941.2009.00804.x
Jensen, S., Bourne, D. G., Hovland, M., and Murrell, J. C. (2012). High diversity of microplankton surrounds deep-water coral reef in the Norwegian Sea. FEMS Microbiol. Ecol. 82, 75–89. doi: 10.1111/j.1574-6941.2012.01408.x
Jonsson, P. R. (1986). Particle size selection, feeding rates and growth dynamics of marine planktonic oligotrichous ciliates (Ciliophora: Oligotrichina). Mar. Ecol. Prog. Ser. 33, 265–277. doi: 10.3354/meps033265
Jousset, A. (2012). Ecological and evolutive implications of bacterial defences against predators. Environ. Microbiol. 14, 1830–1843. doi: 10.1111/j.1462-2920.2011.02627.x
Jürgens, K., and Matz, C. (2002). Predation as a shaping force for the phenotypic and genotypic composition of planktonic bacteria. Antonie Van Leeuwenhoek 81, 413–434. doi: 10.1023/A:1020505204959
Karpov, S. A. (2000). “Flagellate phylogeny: ultrastructural approach,” in The Flagellates. Systematics Association Special Publications, eds B. S. C. Leadbeater and J. C. Green (London: Taylor & Francis), 336–360.
Karpov, S. A., and Fokin, S. I. (1995). The structural diversity of flagellar transitional zone in heterotrophic flagellates and other protists. Tsitologia 37, 1038–1052.
Karpov, S. A., Sogin, M. L., and Silberman, J. D. (2001). Rootlet homology, taxonomy, and phylogeny of bicosoecids based on 18S rRNA gene sequences. Protistology 2, 34–47.
Keeling, P. J. (2004). Diversity and evolutionary history of plastids and their hosts. Am. J. Bot. 91, 1481–1493. doi: 10.3732/ajb.91.10.1481
Kühn, S., Medlin, L., and Eller, G. (2004). Phylogenetic position of the parasitoid nanoflagellate Pirsonia inferred from nuclear-encoded small subunit ribosomal DNA and a description of Pseudopirsonia n. gen. and Pseudopirsonia mucosa (Drebes) comb. nov. Protist 155, 143–156. doi: 10.1078/143446104774199556
Lartillot, N., Lepage, T., and Blanquart, S. (2009). PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25, 2286–2288. doi: 10.1093/bioinformatics/btp368
Lartillot, N., and Philippe, H. (2008). Improvement of molecular phylogenetic inference and the phylogeny of Bilateria. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363, 1463–1472. doi: 10.1098/rstb.2007.2236
Lartillot, N., and Philippe, H. A. (2004). Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol. Biol. Evol. 21, 1095–1109. doi: 10.1093/molbev/msh112
Lee, S. K., Jeong, H. J., Jang, S. H., Lee, K. H., Kang, N. S., Lee, M. J., et al. (2014). Mixotrophy in the newly described dinoflagellate Ansanella granifera: feeding mechanism, prey species, and effect of prey concentration. Algae 29, 137–152. doi: 10.4490/algae.2014.29.2.137
Lee, W. J. (2007). Free-living heterotrophic stramenopile flagellates from Gippsland Basin, South-eastern Australia. Algae 22, 23–29.
Lee, W. J., and Patterson, D. J. (1998). Diversity and geographic distribution of free-living heterotrophic flagellates – analysis by PRIMER. Protist 149, 229–244. doi: 10.1016/S1434-4610(98)70031-8
Lefranc, M., Thénot, A., Lepère, C., and Debroas, D. (2005). Genetic diversity of small eukaryotes in lakes differing by their trophic status. Appl. Environ. Microbiol. 71, 5935–5942. doi: 10.1128/AEM.71.10.5935-5942.2005
Leipe, D. D., Tong, S. M., Goggin, C. L., Slemenda, S. B., Pieniazek, N. J., and Sogin, M. L. (1996). 16S-like rDNA sequences from Developayella elegans, Labyrinthuloides haliotidis, and Proteromonas lacertae confirm that the stramenopiles are a primarily heterotrophic group. Eur. J. Protistol. 32, 449–458. doi: 10.1016/S0932-4739(96)80004-6
Leipe, D. D., Wainright, P. O., Gunderson, J. H., Porter, D., Patterson, D. J., Valois, F., et al. (1994). The stramenopiles from a molecular perspective: 16S-like rRNA sequences from Labyrinthuloides minuta and Cafeteria roenbergensis. Phycologia 33, 369–377. doi: 10.2216/i0031-8884-33-5-369.1
Lie, A. A., Liu, Z., Hu, S. K., Jones, A. C., Kim, D. Y., Countway, P. D., et al. (2014). Investigating microbial eukaryotic diversity from a global census: insights from a comparison of pyrotag and full-length sequences of 18S rRNA genes. Appl. Environ. Microbiol. 80, 4363–4373. doi: 10.1128/AEM.00057-14
Marande, W., López-García, P., and Moreira, D. (2009). Eukaryotic diversity and phylogeny using small- and large-subunit ribosomal RNA genes from environmental samples. Environ. Microbiol. 11, 3179–3188. doi: 10.1111/j.1462-2920.2009.02023.x
Marie, D., Shi, X. L., Rigaut-Jalabert, F., and Vaulot, D. (2010). Use of flow cytometric sorting to better assess the diversity of small photosynthetic eukaryotes in the English Channel. FEMS Microbiol. Ecol. 72, 165–178. doi: 10.1111/j.1574-6941.2010.00842.x
Martel, C. M. (2009). Nitrogen-deficient microalgae are rich in cell-surface mannose: potential implications for prey biorecognition by phagotrophic protozoa. Braz. J. Microbiol. 40, 86–89. doi: 10.1590/S1517-83822009000100014
Massana, R., Castresana, J., Balagué, V., Guillou, L., Romari, K., Groisillier, A., et al. (2004). Phylogenetic and ecological analysis of novel marine stramenopiles. Appl. Environ. Microbiol. 70, 3528–3534. doi: 10.1128/AEM.70.6.3528-3534.2004
Massana, R., del Campo, J., Sieracki, M. E., Audic, S., and Logares, R. (2014). Exploring the uncultured microeukaryote majority in the oceans: reevaluation of ribogroups within stramenopiles. ISME J. 8, 854–866. doi: 10.1038/ismej.2013.204
Massana, R., Terrado, R., Forn, I., Lovejoy, C., and Pedró-Alió, C. (2006). Distribution and abundance of uncultured heterotrophic flagellates in the world oceans. Environ. Microbiol. 8, 1515–1522. doi: 10.1111/j.1462-2920.2006.01042.x
Massana, R., Unrein, F., Rodríguez-Martínez, R., Forn, I., Lefort, T., Pinhassi, J., et al. (2009). Grazing rates and functional diversity of uncultured heterotrophic flagellates. ISME J. 3, 588–596. doi: 10.1038/ismej.2008.130
Matz, C., Bergfeld, T., Rice, S. A., and Kjelleberg, S. (2004). Microcolonies, quorum sensing and cytotoxicity determine the survival of Pseudomonas aeruginosa biofilms exposed to protozoan grazing. Environ. Microbiol. 6, 218–226. doi: 10.1111/j.1462-2920.2004.00556.x
Matz, C., and Kjelleberg, S. (2005). Off the hook–how bacteria survive protozoan grazing. Trends Microbiol. 13, 302–307. doi: 10.1016/j.tim.2005.05.009
Medlin, L., Elwood, H. J., Stickel, S., and Sogin, M. L. (1988). The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 71, 491–499. doi: 10.1016/0378-1119(88)90066-2
Mitra, A., Flynn, K. J., Tillmann, U., Raven, J. A., Caron, D., Stoecker, D. K., et al. (2016). Defining planktonic protist functional groups on mechanisms for energy and nutrient acquisition: incorporation of diverse mixotrophic strategies. Protist 167, 106–120. doi: 10.1016/j.protis.2016.01.003
Montagnes, D. J. S., Barbosa, A. B., Boenigk, J., Davidson, K., Jürgens, K., Macek, M., et al. (2008). Selective feeding behaviour of key free-living protists: avenues for continued study. Aquat. Microb. Ecol. 53, 83–98. doi: 10.3354/ame01229
Moriya, M., Nakayama, T., and Inouye, I. (2000). Ultrastructure and 18S rDNA sequence analysis of Wobblia lunata gen. et sp. nov., a new heterotrophic flagellate (Stramenopiles, incertae sedis). Protist 151, 41–55. doi: 10.1078/1434-4610-00006
Moriya, M., Nakayama, T., and Inouye, I. (2002). A new class of the stramenopiles, Placididea classis nova: description of Placidia cafeteriopsis gen. et sp. nov. Protist 153, 143–156. doi: 10.1078/1434-4610-00093
Nielsen, L. T., and Kiørboe, T. (2015). Feeding currents facilitate a mixotrophic way of life. ISME J. 9, 2117–2127. doi: 10.1038/ismej.2015.27
Patterson, D. J., and Simpson, A. G. B. (1996). Heterotrophic flagellates from coastal marine and hypersaline sediments in Western Australia. Eur. J. Protistol. 32, 423–448. doi: 10.1016/S0932-4739(96)80003-4
Petersen, J., Ludewig, A. K., Michael, V., Bunk, B., Jarek, M., Baurain, D., et al. (2014). Chromera velia, endosymbioses and the rhodoplex hypothesis – plastid evolution in cryptophytes, alveolates, stramenopiles, and haptophytes (CASH lineages). Genome Biol. Evol. 6, 666–684. doi: 10.1093/gbe/evu043
Piwosz, K., and Pernthaler, J. (2010). Seasonal population dynamics and trophic role of planktonic nanoflagellates in coastal surface waters of the Southern Baltic Sea. Environ. Microbiol. 12, 364–377. doi: 10.1111/j.1462-2920.2009.02074.x
Pratt, J. R., and Cairns, J. (1985). Functional groups in the protozoa: roles in differing ecosystems. J. Protozool. 32, 415–423. doi: 10.1111/j.1550-7408.1985.tb04037.x
Riisberg, I., Orr, R. J., Kluge, R., Shalchian-Tabrizi, K., Bowers, H. A., Patil, V., et al. (2009). Seven gene phylogeny of heterokonts. Protist 160, 191–204. doi: 10.1016/j.protis.2008.11.004
Scheckenbach, F., Hausmann, K., Wylezich, C., Weitere, M., and Arndt, H. (2010). Large-scale patterns in biodiversity of microbial eukaryotes from the abyssal sea floor. Proc. Natl. Acad. Sci. U.S.A. 107, 115–120. doi: 10.1073/pnas.0908816106
Schnepf, E., and Schweikert, M. (1996). Pirsonia, phagotrophic nanoflagellates incertae sedis, feeding on marine diatoms: attachment, fine structure and taxonomy. Arch. Protistenkunde 147, 361–371. doi: 10.1016/S0003-9365(97)80060-7
Schweikert, M., and Schnepf, E. (1997). Light and electron microscopical observations on Pirsonia punctigerae spec. nov., a nanoflagellate feeding on the marine centric diatom Thalassiosira punctigera. Eur. J. Protistol. 33, 168–177. doi: 10.1016/S0932-4739(97)80033-8
Sibbald, M. J., and Albright, L. J. (1988). Aggregated and free bacteria as food sources for heterotrophic microflagellates. Appl. Environ. Microbiol. 54, 613–616.
Spurr, A. R. (1969). A low-viscosity epoxy resin embedding medium for electron microscopy. J. Ultrastruc. Res. 26, 31–43. doi: 10.1016/S0022-5320(69)90033-1
Stamatakis, A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. doi: 10.1093/bioinformatics/btl446
Stiller, J. W., Schreiber, J., Yue, J., Guo, H., Ding, Q., and Huang, J. (2014). The evolution of photosynthesis in chromist algae through serial endosymbioses. Nat. Commun. 5, 5764. doi: 10.1038/ncomms6764
Strom, S. L. (1991). Grazing and growth rates of the herbivorous dinoflagellate Gymnodinium sp. from the subarctic Pacific Ocean. Mar. Ecol. Prog. Ser. 78, 103–113. doi: 10.3354/meps078103
Takishita, K., Kakizoe, N., Yoshida, T., and Maruyama, T. (2010). Molecular evidence that phylogenetically diverged ciliates are active in microbial mats of deep-sea cold-seep sediment. J. Eukaryot. Microbiol. 57, 76–86. doi: 10.1111/j.1550-7408.2009.00457.x
Tong, S. M. (1995). Developayella elegans nov. gen., nov. spec., a new type of heterotrophic flagellate from marine plankton. Eur. J. Protistol. 31, 24–31. doi: 10.1016/S0932-4739(11)80352-4
Tong, S. M. (1997). Heterotrophic flagellates and other protists from Southampton Water, U.K. Ophelia 47, 71–131. doi: 10.1080/00785236.1997.10427291
Ucko, M., Elbrachter, M., and Schnepf, E. (1997). A Crypthecodinium cohnii-like dinoflagellate feeding myzocytotically on the uni-cellular red alga Porphyridium sp. Eur. J. Phycol. 32, 133–140. doi: 10.1017/S0967026297001121
Ucko, M., Shrestha, R. P., Mesika, P., Bar-Zvi, D., and Arad, S. (1999). Glycoprotein moiety in the cell wall of the red microalga Porphyridium sp. (Rhodophyta) as the biorecognition site for the Crypthecodinium cohnii-like dinoflagellate. J. Phycol. 35, 1276–1281. doi: 10.1046/j.1529-8817.1999.3561276.x
Van der Auwera, G., Chapelle, S., and De Wachter, R. (1994). Structure of the large ribosomal subunit RNA of Phytophthora megasperma, and phylogeny of the oomycetes. FEBS Lett. 338, 133–136. doi: 10.1016/0014-5793(94)80350-1
Yoon, H. S., Hackett, J. D., Pinto, G., and Bhattacharya, D. (2002). The single, ancient origin of chromist plastids. Proc. Natl. Acad. Sci. U.S.A. 99, 15507–15512. doi: 10.1073/pnas.242379899
Yubuki, N., Leander, B. S., and Silberman, J. D. (2010). Ultrastructure and molecular phylogenetic position of a novel phagotrophic stramenopile from low oxygen environments: Rictus lutensis gen. et sp. nov. (Bicosoecida, incertae sedis). Protist 161, 264–278. doi: 10.1016/j.protis.2009.10.004
Yubuki, N., Pánek, T., Yabuki, A., Čepička, I., Takishita, K., Inagaki, Y., et al. (2015). Morphological identities of two different marine stramenopile environmental sequence clades: Bicosoeca kenaiensis (Hilliard 1971) and Cantina marsupialis (Larsen and Patterson, 1990) gen. nov., comb. nov. J. Eukaryot. Microbiol. 62, 532–542. doi: 10.1111/jeu.12207
Keywords: Develorapax marinus, ochrophyte ancestor, molecular phylogeny, ultrastructure, prey size
Citation: Aleoshin V V, Mylnikov AP, Mirzaeva GS, Mikhailov KV and Karpov SA (2016) Heterokont Predator Develorapax marinus gen. et sp. nov. – A Model of the Ochrophyte Ancestor. Front. Microbiol. 7:1194. doi: 10.3389/fmicb.2016.01194
Received: 05 February 2016; Accepted: 19 July 2016;
Published: 03 August 2016.
Edited by:
Svetlana N. Dedysh, Winogradsky Institute of Microbiology, RussiaReviewed by:
Steven Graham Ball, Lille University of Science and Technology, FranceBaojun Wu, Wayne State University, USA
Shuhei Ota, University of Tokyo, Japan
Copyright © 2016 Aleoshin, Mylnikov, Mirzaeva, Mikhailov and Karpov. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vladimir V. Aleoshin, aleshin@genebee.msu.su Sergey A. Karpov, sakarpov4@gmail.com
 Vladimir V. Aleoshin
Vladimir V. Aleoshin Alexander P. Mylnikov4
Alexander P. Mylnikov4 Sergey A. Karpov
Sergey A. Karpov