- 1Key Laboratory of Agricultural Environmental Microbiology, Ministry of Agriculture, College of Life Sciences, Nanjing Agricultural University, Nanjing, China
- 2Department of Microbiology, College of Life Sciences, Zhejiang University, Hangzhou, China
- 3Department of Environmental Science, College of Environment, Zhejiang University of Technology, Hangzhou, China
Background
Nicotine is a natural alkaloid that is very toxic to humans. To eliminate the harmful effects of nicotine in the environment, biological methods employing microbes to degrade nicotine are required (Brandsch, 2006; Liu et al., 2015). Shinella sp. HZN7 can degrade nicotine efficiently via the variant of a pyridine and pyrrolidine pathways (VPP; Ma et al., 2013; Qiu et al., 2014, 2015). The main intermediates in this pathway include 6-hydroxy-nicotine, 6-hydroxy-N-methylmyosmine, 6-hydroxypseudooxynicotine, 6-hydroxy-3-succinoyl-pyridine, and 2,5-dihydroxypyridine. This strain is the first nicotine-degrading bacterium to be isolated from the genus Shinella.
The genus Shinella was established in 2006 within the “Rhizobiaceae group” of the Alphaproteobacteria (An et al., 2006). Six species were assigned to this genus namely S. daejeonensis, S. fusca, S. granuli, S. kummerowiae, S. yambaruensis, S. zoogloeoides. However, most strains in this genus have not been identified at the species level. Shinella spp. have been isolated from various environmental samples, such as active sludge, zooplankton gut, soils, and water. They also exhibit a range of functional diversity, such as nitrogen fixation (Lin et al., 2008), assimilation of phosphite (Poehlein et al., 2016), and degradation of the toxic pollutants, 4-aminobenzenesulfonate (Biala et al., 2014), chlorothalonil (Liang et al., 2011), and pyridine (Bai et al., 2009). Until now, only three Shinella draft genomes have been deposited in GenBank (http://www.ncbi.nlm.nih.gov/genome/genomes/32494). To further understand the molecular mechanism of nicotine degradation and advance the potential biotechnological applications of Shinella strains, we present the first complete genome sequence of Shinella sp. HZN7 and its features.
Materials and Methods
Bacterial Strain and DNA Purification
Shinella sp. HZN7 was isolated from the active sludge of a wastewater-treatment system of a pesticide manufacturer in Hangzhou City, China. This bacterium was cultured aerobically in LB medium at 30°C with 100 μg/mL ampicillin. Genomic DNA from Shinella sp. HZN7 was extracted and purified using a QIAamp DNA Mini Kit (Qiagen, Germany). The concentration of genomic Genomic DNA was measured using a Qubit 2.0 Fluorometer (Thermo Scientific, USA). Purity of DNAs samples (UV A260/A280) was assessed using a NanoDrop 2000 Spectrophotometer (Thermo Scientific, USA).
Genome Sequencing and Assembly
The genome of strain HZN7 was sequenced using the PacBio RSII platform. A 20-kb DNA library was constructed according to the manufacturer's instructions and sequenced using single-molecule realtime (SMRT) sequencing technology with the P6 DNA polymerase and C4 chemistry. The sequences from two SMRT cells were assembled with SMRT Pipe version 2.1.1 using the hierarchical genome-assembly process (HGAP). The reads were de novo assembled and polished using the PacBio software HGAP3/Quiver (Chin et al., 2013).
Genome Annotation
The coding sequences (CDSs) were predicted using the Prokaryotic Genome Annotation Pipeline (PGAP) version 3.2 software on NCBI (https://www.ncbi.nlm.nih.gov/genome/annotation_prok/). The locus tag prefix was set as “shn.” Additional gene prediction and annotation was performed using the Rapid Annotation Subsystems Technology (RAST) server (Aziz et al., 2008). CRISPR finder (http://crispr.u-psud.fr/Server/) was used for identifying CRISPR/Cas systems (Grissa et al., 2007).
Results
Genome Features
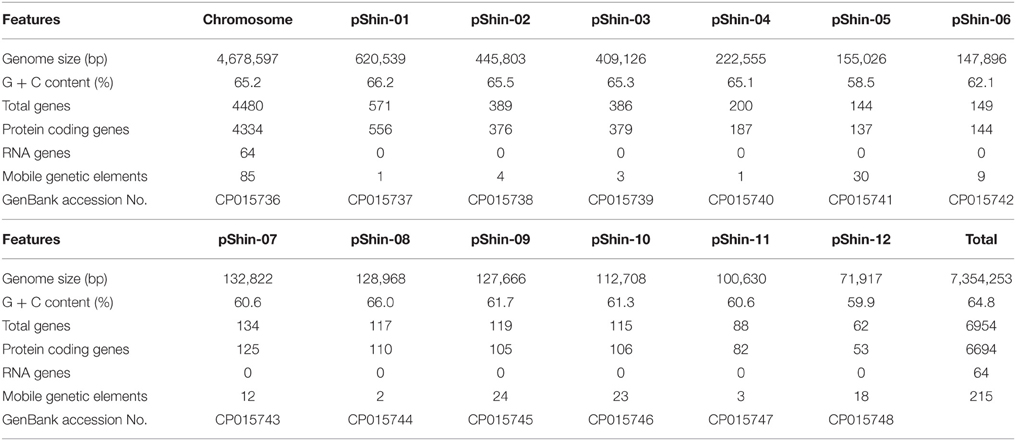
After quality control, ~4 Gb of data was obtained with a 550-fold average coverage. A total of 7,354,253 bp genome sequence was assembled. The features for the complete genome sequence of Shinella sp. HZN7 are summarized in Table 1. The complete genome is composed of one circular chromosome and 12 circular plasmids (designated as plasmid pShin-01 to pShin-12) with an average GC content of 64.8%. The whole genome contains 6954 genes, including 6694 coding sequences, 3 5S rRNAs, 3 16S rRNAs, 3 23S rRNAs, 51 tRNAs, 4 ncRNA, and 196 pseudo genes. Interestingly, 215 mobile genetic elements were predicted, including 96 transposases, 33 integrases, and 86 conjugative transfer proteins. Moreover, one CRISPR gene cluster was identified by the CRISPR finder tool. To the best of our knowledge, there are no bacterial strains containing up to 12 circular plasmids. Previous reports have shown that Shinella zoogloeoides strain BC026 contained 3 or more 200 kb megaplasmids (Bai et al., 2010) and Shinella sp. DD12 contained at least seven plasmids (Poehlein et al., 2016). These results indicate that the possession of multiple plasmids is a common feature in the genus Shinella.
Nicotine-Degrading Gene Cluster
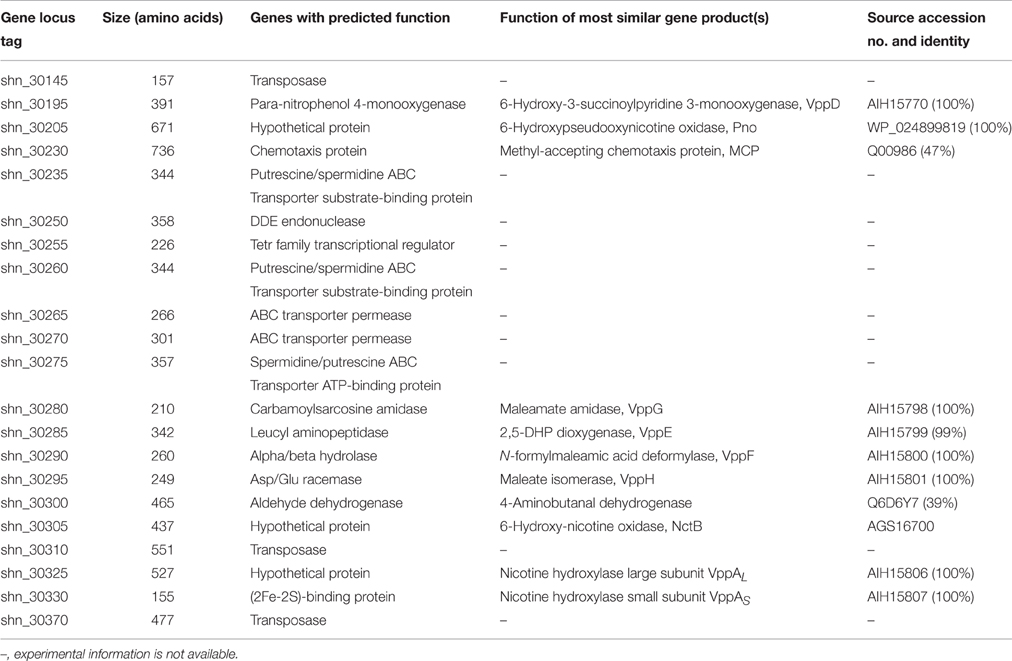
Our previous study showed that the novel 6-hydroxy-nicotine oxidase, NctB, was responsible for the degradation of 6-hydroxy-nicotine to 6-hydroxypseudooxynicotine (Qiu et al., 2014). The nctB gene (locus tag shn_30305) was found on the plasmid pShin-05. The nctB gene, as well as genes homologous to vppA (nicotine hydroxylase gene), vppE (2,5-dihydroxypyridine dioxygenase gene) from Ochrobactrum sp. strain SJY1 (Yu et al., 2015) and pno (6-hydroxypseudooxynicotine oxidase gene) from Agrobacterium tumefaciens S33 (Li et al., 2016), appeared in an 50 kb region of DNA with a GC content of 56.6%. The predicted genes (shn_30145 to shn_30370) in this cluster and their characterized homolog are summarized in Table 2. This cluster was not found in three other Shinella draft genomes (http://www.ncbi.nlm.nih.gov/genome/genomes/32494). In addition, two transposase genes flanked this cluster of DNA, indicating that it may have been acquired by horizontal gene transfer.
In conclusion, we present the first complete genome of Shinella sp. HZN7. We hope this will facilitate a deeper the understanding of the molecular mechanism of nicotine degradation via the VPP pathway, and provide a reference genome for genus Shinella.
Ethics Statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Data Access
The complete genome sequence of Shinella sp. HZN7 has been deposited at the GenBank/EMBL/DDBJ under the accession numbers CP015736-CP015748. The strain is available from the China Center for Type Culture Collection under the accession no CCTCC M 2013060 or from Dr. JQ at Nanjing Agricultural University.
Author Contributions
JQ and ZL conceived and designed the research; YY, JZ, and HW performed experiments and analyzed data; JQ, YM, JH, and ZL analyzed data; JQ and ZL wrote the manuscript; all authors commented on the manuscript and approved the contents.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 31422003 and 31500082) and the Program for New Century Excellent Talents in University (NCET-13-0861). Also thanks Dr. Weijie Song for sequencing at Biology Technology Company of Guhe.
References
An, D.-S., Im, W.-T., Yang, H.-C., and Lee, S.-T. (2006). Shinella granuli gen. nov., sp. nov., and proposal of the reclassification of Zoogloea ramigera ATCC 19623 as Shinella zoogloeoides sp. nov. Int. J. Syst. Evol. Microbiol. 56, 443–448. doi: 10.1099/ijs.0.63942-0
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Bai, Y., Sun, Q., Zhao, C., Wen, D., and Tang, X. (2009). Aerobic degradation of pyridine by a new bacterial strain, Shinella zoogloeoides BC026. J. Ind. Microbiol. Biotechnol. 36, 1391–1400. doi: 10.1007/s10295-009-0625-9
Bai, Y., Sun, Q., Zhao, C., Wen, D., and Tang, X. (2010). Study on the function of plasmid in pyridine-degrading bacteria. Environ. Sci. 31, 1679–1683. doi: 10.13227/j.hjkx.2010.07.039
Biala, S., Chadha, P., and Saini, H. S. (2014). Biodegradation of 4-aminobenzenesulfonate by indigenous isolate Shinella yambaruensis SA1 and its validation by genotoxic analysis. Biotechnol. Bioprocess. Eng. 19, 1034–1041. doi: 10.1007/s12257-013-0801-7
Brandsch, R. (2006). Microbiology and biochemistry of nicotine degradation. Appl. Microbiol. Biotechnol. 69, 493–498. doi: 10.1007/s00253-005-0226-0
Chin, C.-S., Alexander, D. H., Marks, P., Klammer, A. A., Drake, J., Heiner, C., et al. (2013). Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569. doi: 10.1038/nmeth.2474
Grissa, I., Vergnaud, G., and Pourcel, C. (2007). CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35, W52–W57. doi: 10.1093/nar/gkm360
Li, H., Xie, K., Yu, W., Hu, L., Huang, H., Xie, H., et al. (2016). Nicotine dehydrogenase complexed with 6-hydroxypseudooxynicotine oxidase involved in the hybrid nicotine-degrading pathway in Agrobacterium tumefaciens S33. Appl. Environ. Microbiol. 82, 1745–1755. doi: 10.1128/AEM.03909-15
Liang, B., Wang, G., Zhao, Y., Chen, K., Li, S., and Jiang, J. (2011). Facilitation of bacterial adaptation to chlorothalonil-contaminated sites by horizontal transfer of the chlorothalonil hydrolytic dehalogenase gene. Appl. Environ. Microbiol. 77, 4268–4272. doi: 10.1128/AEM.02457-10
Lin, D. X., Wang, E. T., Tang, H., Han, T. X., He, Y. R., Guan, S. H., et al. (2008). Shinella kummerowiae sp. nov., a symbiotic bacterium isolated from root nodules of the herbal legume Kummerowia stipulacea. Int. J. Syst. Evol. Microbiol. 58, 1409–1413. doi: 10.1099/ijs.0.65723-0
Liu, J., Ma, G., Chen, T., Hou, Y., Yang, S., Zhang, K.-Q., et al. (2015). Nicotine-degrading microorganisms and their potential applications. Appl. Microbiol. Biotechnol. 99, 3775–3785. doi: 10.1007/s00253-015-6525-1
Ma, Y., Wei, Y., Qiu, J., Wen, R., Hong, J., and Liu, W. (2013). Isolation, transposon mutagenesis, and characterization of the novel nicotine-degrading strain Shinella sp. HZN7. Appl. Microbiol. Biotechnol. 98, 2625–2636. doi: 10.1007/s00253-013-5207-0
Poehlein, A., Freese, H., Daniel, R., and Simeonova, D. D. (2016). Genome sequence of Shinella sp. strain DD12, isolated from homogenized guts of starved Daphnia magna. Stand. Genomic Sci. 11, 1–8. doi: 10.1186/s40793-015-0129-3
Qiu, J., Liu, M., Wen, R., Zhang, D., and Hong, J. (2015). Regulators essential for nicotine degradation in Shinella sp. HZN7. Process Biochem. 50, 1947–1950. doi: 10.1016/j.procbio.2015.07.013
Qiu, J., Wei, Y., Ma, Y., Wen, R., Wen, Y., and Liu, W. (2014). A novel (S)-6-hydroxynicotine oxidase gene from Shinella sp. strain HZN7. Appl. Environ. Microbiol. 80, 5552–5560. doi: 10.1128/AEM.01312-14
Keywords: Shinella sp. HZN7, PacBio, genome sequence, plasmids, nicotine, biodegradation, reference genome
Citation: Qiu J, Yang Y, Zhang J, Wang H, Ma Y, He J and Lu Z (2016) The Complete Genome Sequence of the Nicotine-Degrading Bacterium Shinella sp. HZN7. Front. Microbiol. 7:1348. doi: 10.3389/fmicb.2016.01348
Received: 15 June 2016; Accepted: 16 August 2016;
Published: 30 August 2016.
Edited by:
Feng Gao, Tianjin University, ChinaReviewed by:
Seong Woon Roh, Korea Basic Science Institute, South KoreaJonathan Badger, National Cancer Institute, USA
Copyright © 2016 Qiu, Yang, Zhang, Wang, Ma, He and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiguo Qiu, qiujiguo@njau.edu.cn
Zhenmei Lu, lzhenmei@zju.edu.cn
 Jiguo Qiu
Jiguo Qiu Youjian Yang1
Youjian Yang1 Zhenmei Lu
Zhenmei Lu