- 1Key Laboratory of Pathogenic Fungi and Mycotoxins of Fujian Province, Key Laboratory of Biopesticide and Chemical Biology of Education Ministry, School of Life Sciences, Fujian Agriculture and Forestry University, Fuzhou, China
- 2State Key Laboratory of Mycology, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China
In Aspergillus nidulans, the nitrogen metabolite repression (NMR) regulator NmrA plays a major role in regulating the activity of the GATA transcription factor AreA during nitrogen metabolism. However, the function of nmrA in A. flavus has not been previously studied. Here, we report the identification and functional analysis of nmrA in A. flavus. Our work showed that the amino acid sequences of NmrA are highly conserved among Aspergillus species and that A. flavus NmrA protein contains a canonical Rossmann fold motif. Deletion of nmrA slowed the growth of A. flavus but significantly increased conidiation and sclerotia production. Moreover, seed infection experiments indicated that nmrA is required for the invasive virulence of A. flavus. In addition, the ΔnmrA mutant showed increased sensitivity to rapamycin and methyl methanesulfonate, suggesting that nmrA could be responsive to target of rapamycin signaling and DNA damage. Furthermore, quantitative real-time reverse transcription polymerase chain reaction analysis suggested that nmrA might interact with other nitrogen regulatory and catabolic genes. Our study provides a better understanding of NMR and the nitrogen metabolism network in fungi.
Introduction
Aspergillus flavus is ubiquitous in soil and can infect or contaminate a wide range of organic nutrient sources, such as economically important commodities, insects, animal carcasses, and even immunocompromised humans and animals (Amaike and Keller, 2011; Yu, 2012; Zhang et al., 2014; Bai et al., 2015a,b; Zhang et al., 2015). Aflatoxins (AFs), mainly produced by A. flavus and A. parasiticus, have been identified as a class of the most toxic and carcinogenic secondary metabolites of fungi. AF contamination of agricultural grains is not only a significant food safety concern but also an economic concern for the agricultural industry worldwide (Yu, 2012; Amaike et al., 2013; Yang et al., 2015). To develop effective and novel strategies against AF contamination, it is a necessity to investigate the molecular mechanisms by which AF biosynthesis is regulated in A. flavus.
It has been proposed that nitrogen limitation induces the expression of infection-related genes in plant pathogenic fungi (Snoeijers et al., 2000; López-Berges et al., 2010). Microorganisms can use a great many nitrogen sources, and it has become widely accepted that glutamine is a key effector of nitrogen metabolite repression (NMR), a regulatory circuit in filamentous fungi that ensures the preferential use of superior nitrogen sources such as ammonium and glutamine over a broad array of non-preferred compounds (Caddick et al., 1994; Magasanik and Kaiser, 2002; Wong et al., 2008; Wiemann and Tudzynski, 2013; Tudzynski, 2014). The signaling pathway that orchestrates NMR is complex and refined. The positive-acting GATA family transcription factor AreA functions in NMR to allow utilization of preferred nitrogen sources ammonium and glutamine (Wong et al., 2008; Fernandez et al., 2012; Tudzynski, 2014). However, AreA loss-of-function mutants are unable to use non-preferential nitrogen sources other than ammonium and glutamine (Arst and Cove, 1973; Marzluf, 1997). In the presence of ammonium and glutamine, the nitrogen metabolite repressor NmrA interacts with AreA to prevent nitrogen catabolic gene expression; however, in the presence of less preferred nitrogen sources such as nitrate, NmrA dissociates from AreA, allowing AreA to activate the expression of genes involved in alternative nitrogen source usage (Caddick et al., 1994; Andrianopoulos et al., 1998; Wilson and Arst, 1998; Fernandez et al., 2012; Tudzynski, 2014).
The function of NmrA in nitrogen source metabolism in A. flavus has not been well characterized (Andrianopoulos et al., 1998; Schönig et al., 2008; Wagner et al., 2010; Zhao et al., 2010). In this study, we identified and cloned the nmrA gene from A. flavus and characterized nmrA deletion and complementation mutants. We demonstrate that nmrA plays a negative role in NMR and nitrogen metabolism. Furthermore, nmrA appears to be involved in AF biosynthesis, conidiation, sclerotia formation, invasive virulence, and stress responses.
Materials and Methods
Fungal Strains and Growth Conditions
Fungal strains and plasmids used in this study are listed in Supplementary Table S1. The A. flavus strain PTSΔku70ΔpyrG (Chang et al., 2010), a uracil auxotroph, was purchased from the Fungal Genetics Stock Center (School of Biological Sciences, University of Missouri, Kansas City, MO, USA) and used for gene deletion (Yang et al., 2016). Wild-type (WT) A. flavus and the transformants generated in this study were grown on yeast extract–sucrose (YES) agar, yeast extract–glucose agar, or yeast extract–glucose agar with trace elements, uracil, and uridine (Yang et al., 2016). YES medium, potato dextrose agar (PDA, Bai and Shaner, 1996), Czapek agar (CA, Becton Dickinson), and glucose minimal medium (GMM, Shimizu and Keller, 2001) were used for mycelial growth assays, sporulation analysis, and AF analysis. All experiments included four replicate plates and were performed at least three times with similar results, and the error was expressed as the standard deviation.
Sequence Analysis of NmrA
The NmrA sequence (EED47920) from A. flavus was originally identified in the NCBI Protein database using BLAST. To verify the existence and the size of the exon in nmrA, we cloned the coding sequence into the pET-28a(+) expression vector and sequenced it. The obtained sequence was compared with that reported in the NCBI database. The protein domains were analyzed using InterPro1. A phylogenetic tree based on NmrA sequences from A. clavatus (UniProt: A1C544), A. flavus (UniProt: B8NR87), A. fumigatus (UniProt: Q4WEG4), A. nidulans (UniProt: Q5AU62), A. niger (G3XN01), A. oryzae (GenBank: XP_001822655.2), and A. terreus (UniProt: Q0CAL7) was constructed with DNAMAN 6.0 software using the neighbor-joining method.
Targeted Deletion and Complementation of the nmrA Gene
To create an nmrA deletion mutant, we constructed an AP-pyrG-BP vector by inserting upstream and downstream flanking sequences of the nmrA gene on either side of the pyrG gene. The upstream and downstream flanking sequences of nmrA were amplified from genomic DNA of A. flavus WT with primer pairs nmrA-P1/nmrA-P2 and nmrA-P3/nmrA-P4, respectively (Supplementary Table S2). The nmrA deletion mutant (ΔnmrA) was complemented with a full-length nmrA gene, a 2869 bp fragment, spanning from 1069 bp upstream of the WT A. flavus nmrA translation initiation codon to 681 bp downstream of the translation termination codon, to confirm that the phenotype of the ΔnmrA mutant was due to deletion of the nmrA gene. The full-length nmrA gene was amplified from genomic DNA of A. flavus WT using the primer pair nmrA-CF/nmrA-CR (Supplementary Table S2), and it contained a constitutive promoter, which controlled nmrA gene expression. The complementation plasmid (pPTR I-nmrA) was constructed using the backbone of the chromosomal integrating shuttle vector pPTR I DNA (Takara, Japan, Kubodera et al., 2000), which contained the same restriction enzyme recognition sites as the full-length nmrA gene and could integrate into A. flavus genome randomly. Six out of 15 pyrithiamine resistance transformants were selected for their wild-type growth phenotype in the presence of GMM supplementing ammonium as the sole nitrogen source. We concluded that these transformants had integrated an intact copy of the A. flavus nmrA gene into the genome after the A. flavus nmrA gene in this plasmid was sequenced to ensure flawlessness of the sequence.
Mycelial Growth and Stress Assays
Mycelial growth assays were performed on YES medium, PDA, and GMM supplemented with 50 mM glutamine, ammonium, proline, alanine, or sodium nitrite (NaNO2). For the stress assays, rapamycin, methyl methanesulfonate (MMS), NaCl, and sorbitol were added to YES medium at the concentrations indicated in the figure legends (Yang et al., 2016). Each plate was inoculated with 1 μL of conidial suspension (4 × 104 conidia/mL) and incubated at 28°C for 4–7 days in the dark. The assays were carried out in triplicate and were repeated three times.
Observation and Counting of Conidia
Glucose minimal medium agar and PDA were point-inoculated with 1 μL of conidial suspension (4 × 104 conidia/mL) and incubated at 28°C for 5 days in the dark. Three plugs were removed from each plate and the conidia produced by A. flavus were suspended in a solution of 0.05% Tween 20 and 7% dimethylsulfoxide (DMSO), and counted in a hemocytometer. Conidial suspensions were viewed with an inverted microscope (Leica Microsystems, Germany).
Analysis of AF Production
For analysis of AF production, 1 mL of spore suspension (1 × 106 spores/mL) was inoculated in YES, PDA or GMM supplemented with 50 mM glutamine, ammonium, proline, alanine, or sodium nitrite and incubated in the dark at 28°C for 7 days. AFs were extracted from 500 μL of culture filtrate with an equal volume of chloroform. The chloroform layer was transferred to a new 1.5 mL tube and evaporated to dryness at 70°C. Thin-layer chromatography was used to identify AFs. A solvent system consisting of acetone and chloroform (1:9, v/v) was used, and the plates were observed under UV light at 365 nm. For quantitative analysis of AF production, GeneTools image analysis software was used.
Sclerotia Assays
Sclerotia formation was measured as previously described (Amaike and Keller, 2011). Briefly, GMM supplemented with the indicated nitrogen sources, 2% agar, and 2% sorbitol was overlaid with 1 × 104 spores/plate. Cultures were grown at 37°C in complete darkness for 7–10 days. Plates were sprayed with 70% ethanol to kill and wash away conidia and exposed sclerotia. The sclerotia were collected from 10 mm cores and counted in triplicate. Experiments were repeated three times.
Seed Infection
Mature live peanut seeds were used to measure the pathogenicity of the WT, ΔnmrA, and complementation (ΔnmrA::nmrA) strains (Duran et al., 2007; Amaike and Keller, 2011). Plates were cultured in the dark at 28°C for 5 days, and the filter paper was moistened daily. Inoculated peanut cotyledons were processed to count conidia and extract AFs, and the methodology of counting conidia and extracting AFs was the same as above described.
Real-Time Quantitative Reverse Transcription Polymerase Chain Reaction (RT-qPCR)
Aspergillus flavus mycelia from the WT, ΔnmrA, and ΔnmrA::nmrA strains were harvested after 48 h of growth. RNA was extracted with TRIzol reagent (Biomarker Technologies, Beijing, China), and then a Nano Drop 2000 and Agilent 2100 were used to evaluate the quality of RNA after total RNA extraction and DNase I treatment. TransScript® All-in-One First-Strand cDNA Synthesis SuperMix was used to synthesize cDNA, and RT-qPCR was performed on a PikoRealTM Real-Time PCR System (Thermo Scientific Inc.) with PikoRealTM 2.2 software using TransStart Top Green qPCR SuperMix (TransGen Biotech, Beijing, China). The RT-qPCR conditions were as follows: 95°C for 7 min and 40 cycles of 95°C for 5 s and 60°C for 30 s. The A. flavus actin gene was used as an reference gene to normalize the expression data. The relative expression of genes was calculated using the 2-ΔΔCt method, and standard deviation was calculated from three biological replicates (Livak and Schmittgen, 2001). The gene-specific primers are shown in Supplementarey Table S3.
Results
Sequence Analysis of NmrA in Aspergillus
In A. nidulans, NmrA can bind nicotinamide dinucleotides and may have a redox-sensing function, since NmrA is bound with NAD+ and NADP+ preferentially to NADH and NADPH. In addition, NmrA interacts directly with AreA zinc fingers (Stammers et al., 2001; Kotaka et al., 2008; Zhao et al., 2010). The A. flavus nmrA ORF consists of 1,119 bp with one introns, and encodes a putative NMR regulator NmrA with 351 amino acids. A. flavus NmrA might function similarly to its ortholog in A. nidulans, since the two protein sequences have a similarity of 88.07%. We downloaded the NmrA protein sequences of seven representative Aspergillus species (A. clavatus, A. flavus, A. fumigatus, A. nidulans, A. niger, A. oryzae, and A. terreus) from FungiDB2 and aligned using DNAMAN software version 6.0.3.99 (Figure 1). The sequence similarity was as high as 94.81%, demonstrating a highly conserved nature of NmrA in Aspergillus. For instance, the NmrA protein sequences in A. flavus and A. oryzae are only differed at one amino acid between the proteins of, reflecting the evolutionary similarity between them. We also identified a canonical Rossmann fold motif in A. flavus NmrA.
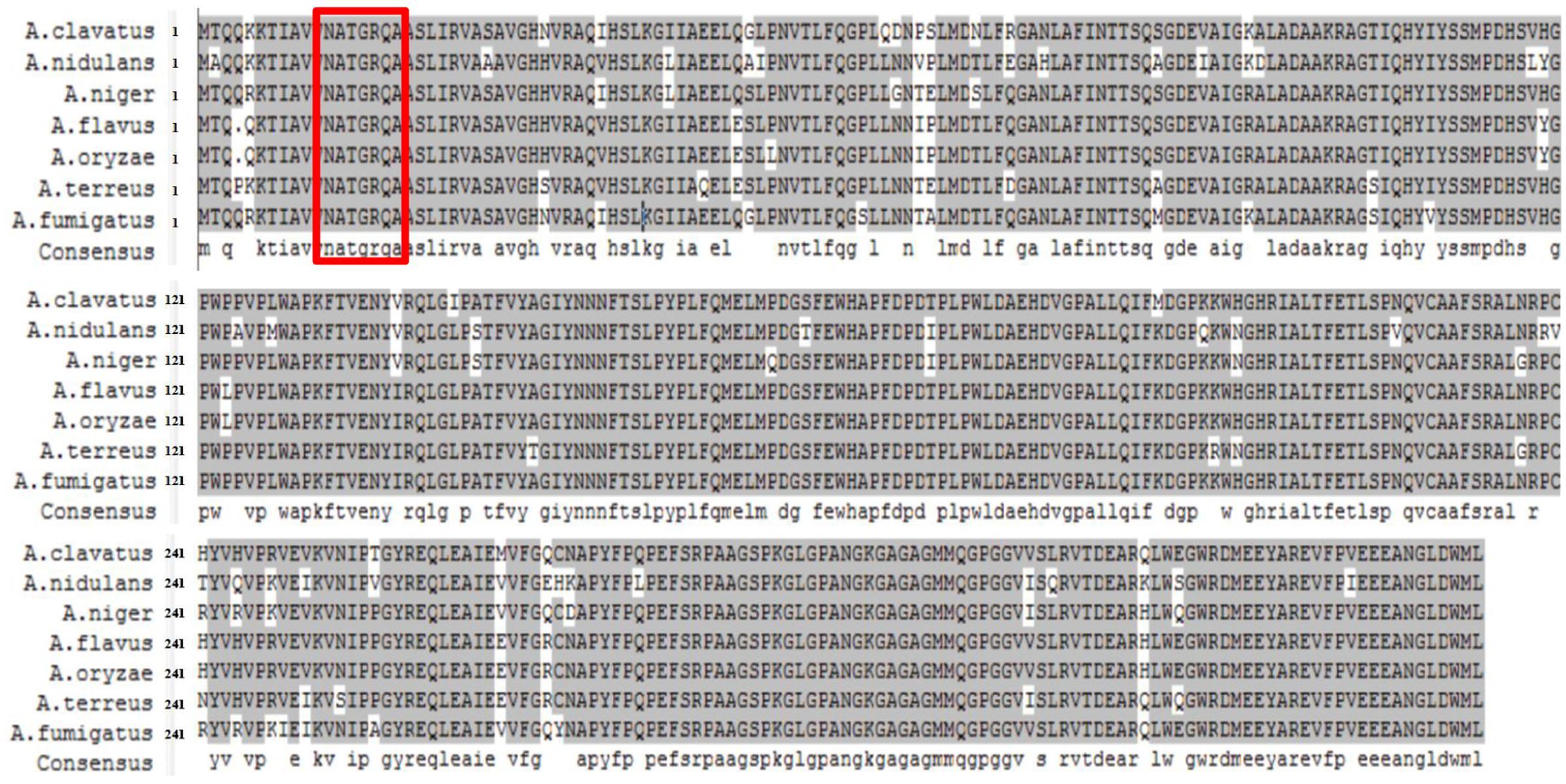
FIGURE 1. Sequence alignment of nitrogen metabolite repressor NmrA proteins in Aspergillus clavatus, A. flavus, A. fumigatus, A. nidulans, A. niger, A. oryzae, and A. terreus. DNAMAN 6.0.3.99 software was used for the alignment and presentation. Residues boxed in red mark the Rossmann fold motif.
Radial Growth on Various Nitrogen Sources
A targeted gene deletion strategy was employed to determine the roles of nmrA in A. flavus (Supplementary Figure S1A). The nmrA gene was successfully replaced with the pyrG cassette (Supplementary Figure S1B), indicating that the nmrA deletion mutant (ΔnmrA) was successfully constructed. The ΔnmrA strain was complemented by introducing a construct consisting of the nmrA open reading frame in the pPTR I vector, which generated the complementary strain ΔnmrA::nmrA. The ΔnmrA and ΔnmrA::nmrA strains were verified by RT-PCR and RT-qPCR (Supplementary Figures S1B,C). The growth rate of the ΔnmrA strain consistently matched that of the WT and ΔnmrA::nmrA strains on YES medium and PDA, but the growth rate was slower than that of the WT and ΔnmrA::nmrA strains on GMM supplemented with glutamine, ammonium, proline, alanine, or sodium nitrite as the sole nitrogen source (the usual nitrogen source in GMM is sodium nitrate, NaNO3) (Figures 2A,B). The edges of the mycelial colonies of the ΔnmrA strain were more irregular on glutamine, ammonium, and proline than on the other nitrogen sources (Figure 2A). It should be mentioned that A. flavus strains could not grow normally on sodium nitrate because of a deficiency of the nitrate reductase gene niaD. Therefore, sodium nitrite was used in place of sodium nitrate; however, the deletion strain showed retarded growth on this nitrogen source as well. These results suggested that nmrA could regulate nitrogen utilization and metabolism.
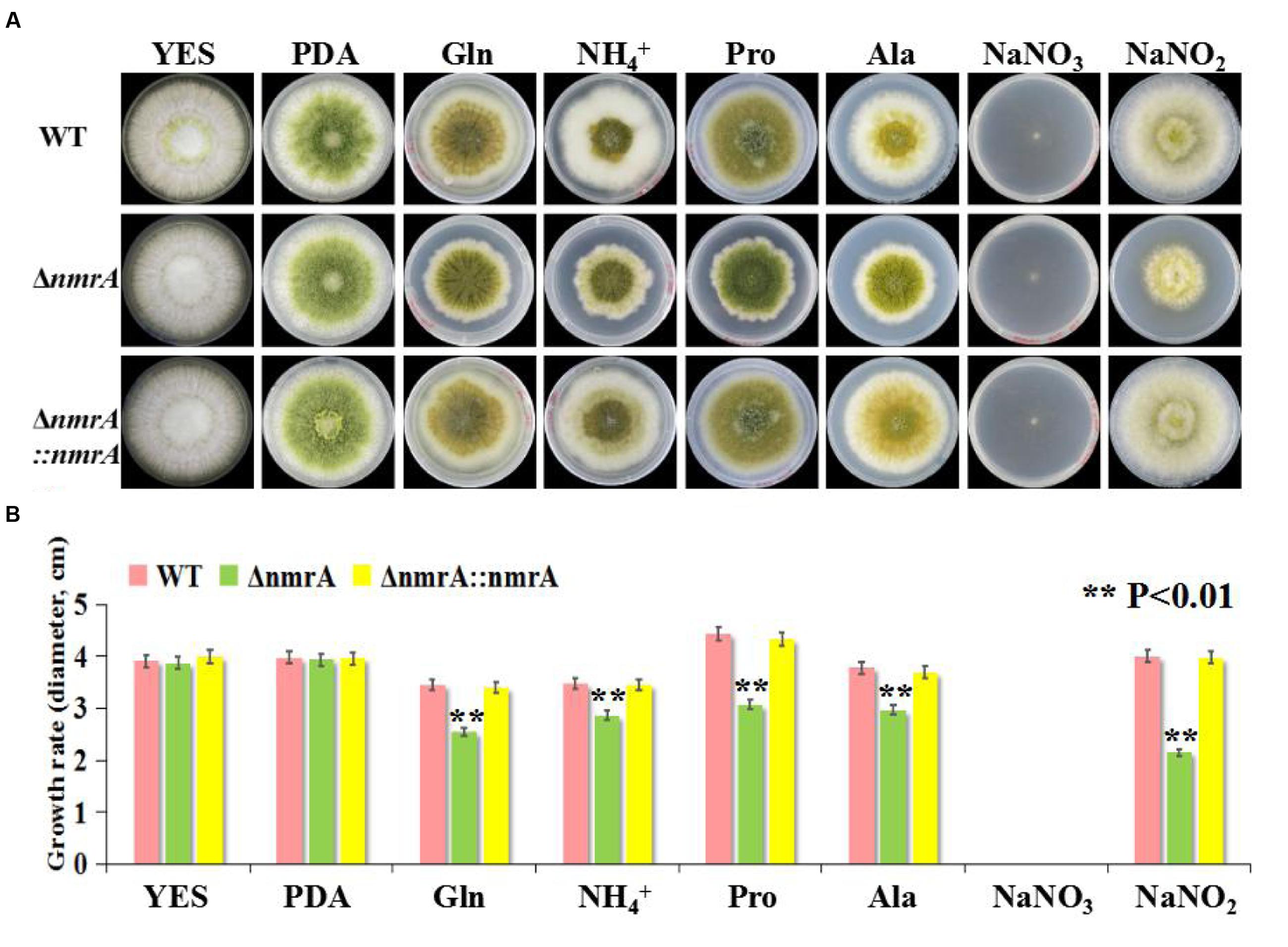
FIGURE 2. Utilization of A. flavus ΔnmrA mutant on different nitrogen sources. (A) Radial growth of mycelia on YES, PDA, and GMM supplemented with 50 mM glutamine, ammonium, proline, alanine, sodium nitrate, and sodium nitrite, respectively, as a sole nitrogen source for 7 days at 28°C in the darkness. The photographs were taken on the 5th day. (B) Growth rate of WT, ΔnmrA and ΔnmrA::nmrA strains vegetated on YES, PDA, and GMM supplemented with different nitrogen sources on the 5th day at 28°C in the dark. Asterisk indicated statistical significance at P < 0.01.
Colony Morphology and Conidiation of the ΔnmrA Mutant
There was little difference in conidia production among strains when they were germinated on PDA, but the ΔnmrA strain produced more spores than the WT and ΔnmrA::nmrA strains when germination occurred on GMM with ammonium (Figures 2A and 3A). The ΔnmrA mutant also produced more conidiophores than the WT and ΔnmrA::nmrA strains on GMM with ammonium (Figure 3B). To gain further insight into the role of NmrA in conidiation, we performed RT-qPCR analysis. The results showed that transcript levels of the conidiation-related genes abaA (AFLA_029620) and brlA (AFLA_082850) were higher in ΔnmrA than in WT A. flavus and ΔnmrA::nmrA when the strains were germinated on GMM with ammonium (Kato et al., 2003; Son et al., 2013), while there was little difference among strains when germination occurred on PDA (Figure 3C). Collectively, these results indicated that nmrA was likely involved in the regulation of conidiation in A. flavus.
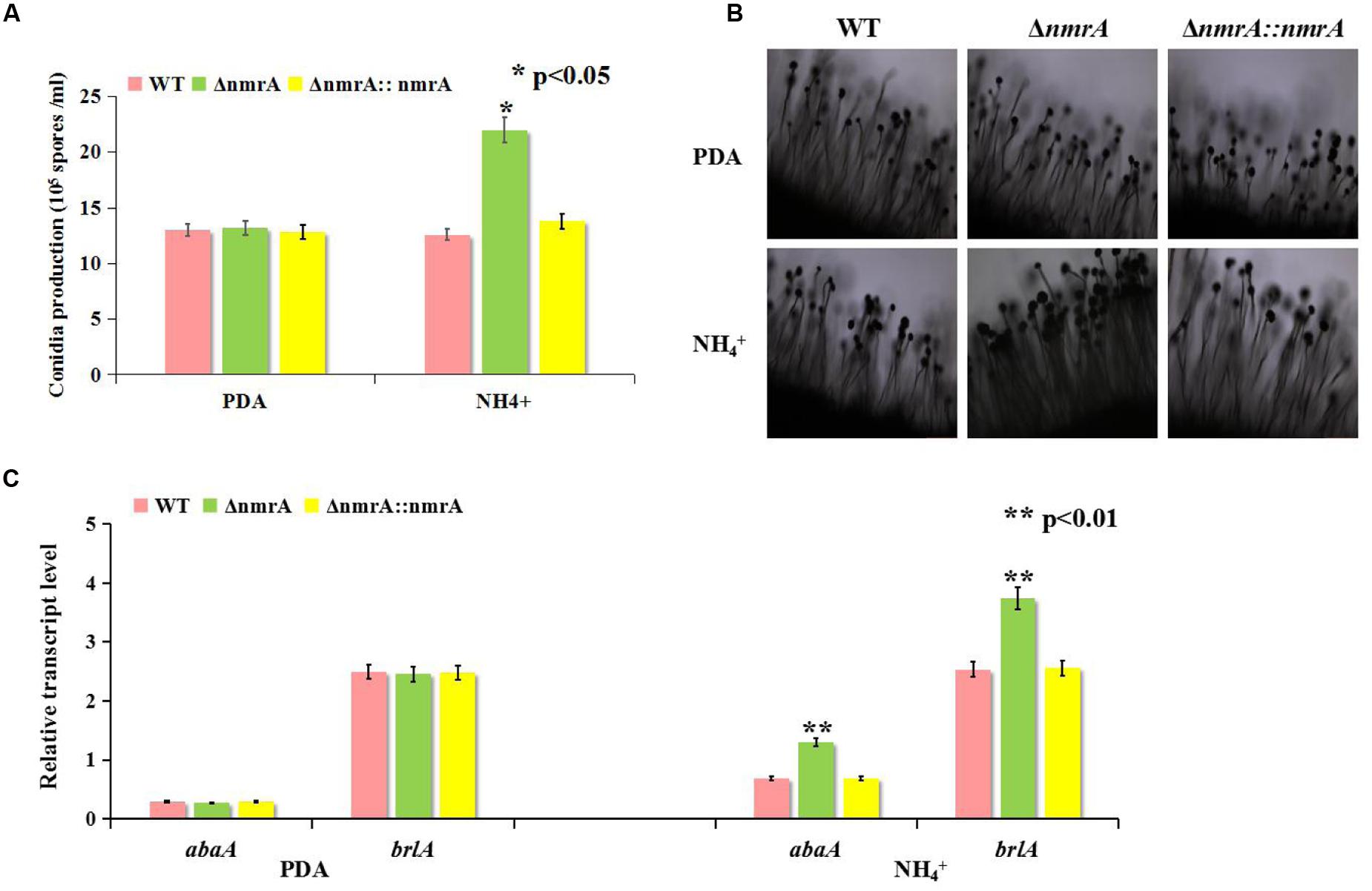
FIGURE 3. Deletion of nmrA affected conidia production. Bars represent SE from three independent experiments with three replicates. (A) Deletion of nmrA resulted in conidiation augment on GMM supplemented with ammonium. Asterisk indicated statistical significance at P < 0.05. (B) Conidiophores were observed under a light microscope at 12 h after induction with illumination. Scale bar: 200 μm. (C) RT-qPCR analysis was performed in the indicated strains germinated on media as described in (A). The asterisks represented a significant difference level of P < 0.01.
AF Production by the ΔnmrA Mutant
Next, we assessed the production of AFs by the WT, ΔnmrA, and ΔnmrA::nmrA strains on the media described in Figure 2. Like the growth rate, AF production was similar among the WT, ΔnmrA, and ΔnmrA::nmrA strains when they were grown on YES or PDA plates (Figure 4). However, there were significant decrease in AF production among the strains when they were germinated on GMM supplemented with glutamine and alanine (Figures 4A,B). We concluded that nmrA might participate in the regulation of AF biosynthesis in A. flavus.
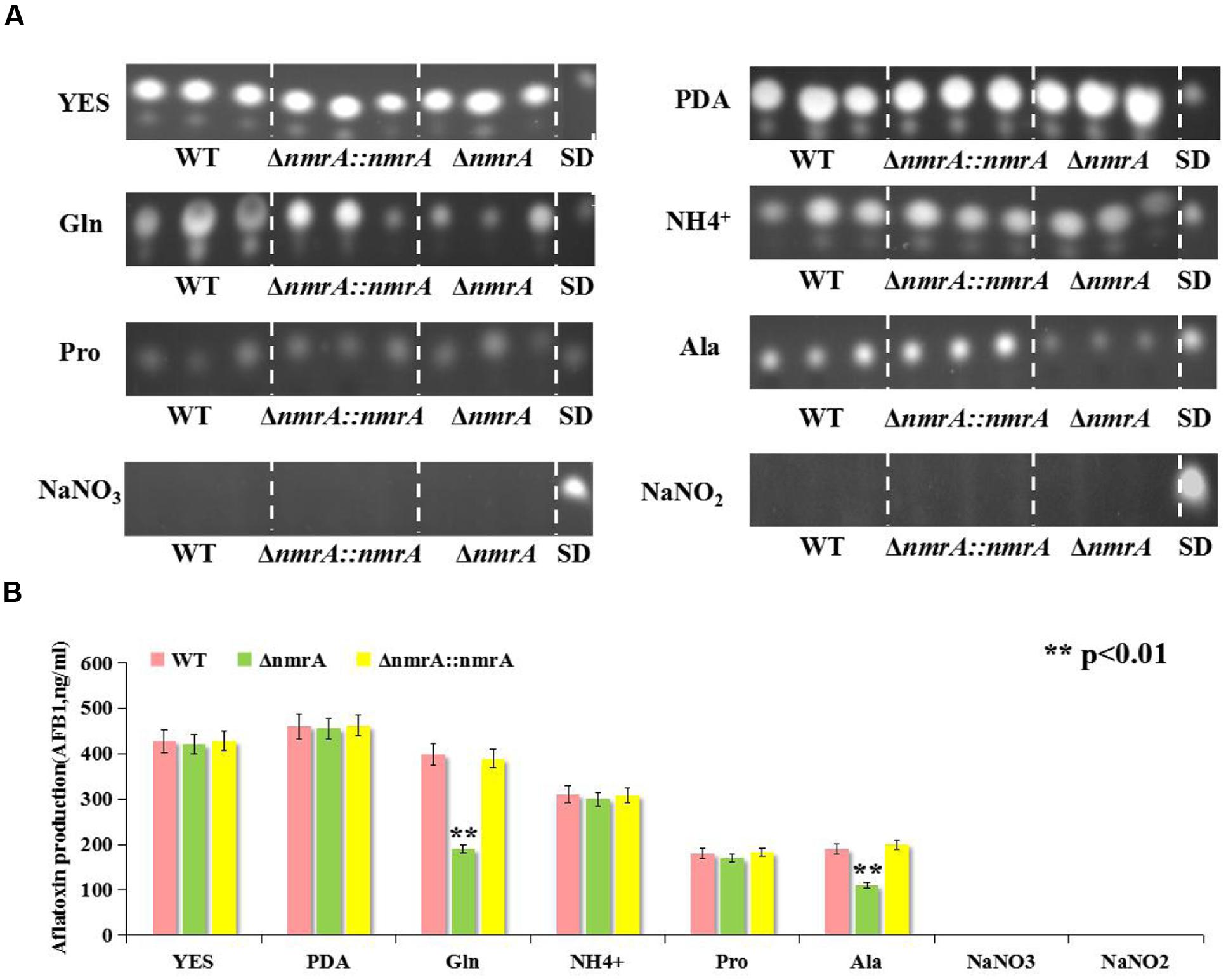
FIGURE 4. Aflatoxin (AFB1) assessment of WT, ΔnmrA and ΔnmrA::nmrA strains grown on different culture media. (A) Aflatoxin production of WT, ΔnmrA and ΔnmrA::nmrA strains grown on media as described in Figure 2. (B) Quantification results of AFB1 of WT, ΔnmrA and ΔnmrA::nmrA strains grown on as (A) by Gene Tools analysis system software. SD means standard AFB1. The asterisks represented a significant difference level of P < 0.01.
Sclerotia Production by the nmrA Deletion Strain
We found that in addition to suppressing AF biosynthesis, nmrA deletion resulted in a concomitant increase in sclerotia production. The ΔnmrA strain displayed a more compact and intensive distribution of sclerotia on the plate than the WT and ΔnmrA::nmrA strains (Figure 5A). To some extent, sclerotia production by the ΔnmrA mutant was greater on glutamine than on ammonium (Figure 5B), indicating that it could be affected by the quality of the nitrogen source. In summary, the effect of the nmrA gene on sclerotia production by A. flavus was modulated by the nitrogen source.
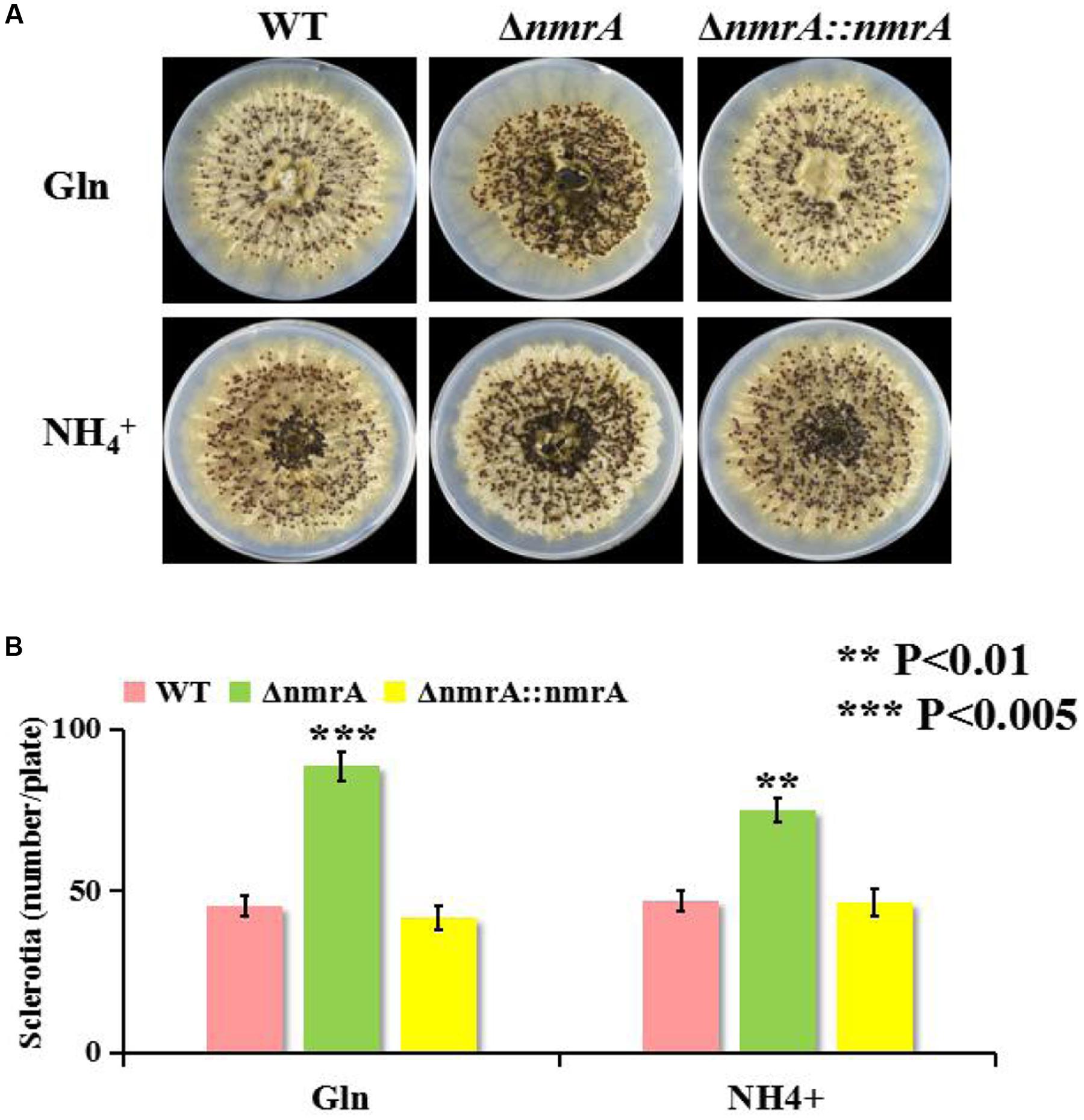
FIGURE 5. Sclerotia production of ΔnmrA mutant. (A) One representative plate for the WT, ΔnmrA and ΔnmrA::nmrA strains grown on GMM supplemented with 50 mM glutamine and ammonium plus 2% sorbitol to induce sclerotia production at 37°C for 7 days in dark condition. (B) Quantity analysis of sclerotia among the strains listed. Asterisks indicated significant differences between each depletion strain relative to the WT and ΔnmrA::nmrA strains as determined by a Student T-test, with ∗∗P < 0.01, ∗∗∗P < 0.005.
Requirement of nmrA for Virulence of A. flavus
Although Wilson examined the role of three NmrA orthologs in Magnaporthe oryzae during infection of rice (Wilson et al., 2010), there has been no report on the role of NmrA in infection by A. flavus. To dissect the role of NmrA in invasive growth, we assessed the growth of A. flavus on peanut seeds. Figure 6A shows that the ΔnmrA strain was crippled in its ability to colonize and sporulate on host seeds compared with WT A. flavus and ΔnmrA::nmrA. Deletion of nmrA also led to statistically significant reductions in conidia production and AF biosynthesis (Figures 6B,C), which resulted in the decrease in virulence. These data suggested that nmrA deletion reduced the virulence of A. flavus.
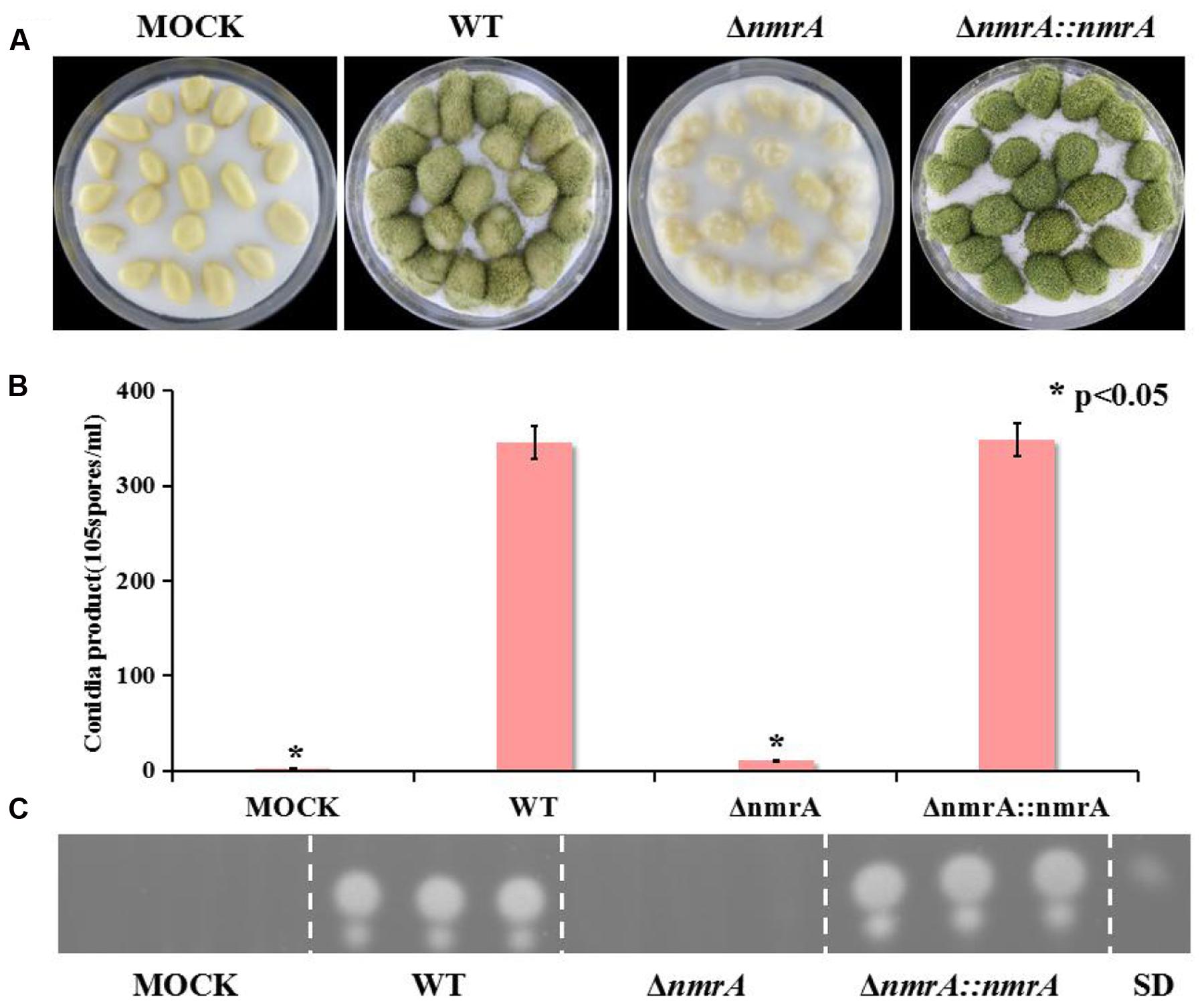
FIGURE 6. Pathogenicity assay of ΔnmrA mutant. (A) Growth of A. flavus WT, ΔnmrA and ΔnmrA::nmrA strains on living peanut cotyledons after 5 days of inoculation. (B) Conidia production on peanut cotyledons after inoculation of 5 days. Asterisk indicates statistical significance at P < 0.05. (C) TLC measurements of aflatoxin (AFB1) extracted from seed in (A). SD means standard AFB1.
Responses of the ΔnmrA Mutant to Multiple Stressors
To explore the potential roles of NmrA in responses to environmental stressors, we tested fungal sensitivity to the target of rapamycin (TOR) inhibitor rapamycin, the alkylating agent MMS, and the osmotic stressors NaCl and sorbitol. Previous research has shown that TOR-mediated repression of nitrogen catabolic genes and virulence occurs through MeaB-dependent and MeaB-independent mechanisms in Fusarium oxysporum (López-Berges et al., 2010), so it seemed possible that an interaction between NmrA and TOR might be involved in nitrogen metabolism regulation in A. flavus. The assay results showed that the ΔnmrA mutant was more sensitive to rapamycin and MMS than WT A. flavus or ΔnmrA::nmrA. However, the ΔnmrA mutant was not more sensitive to the osmotic stressors (Figures 7A,B). Overall, these results indicated that NmrA is associated with the TOR pathway.
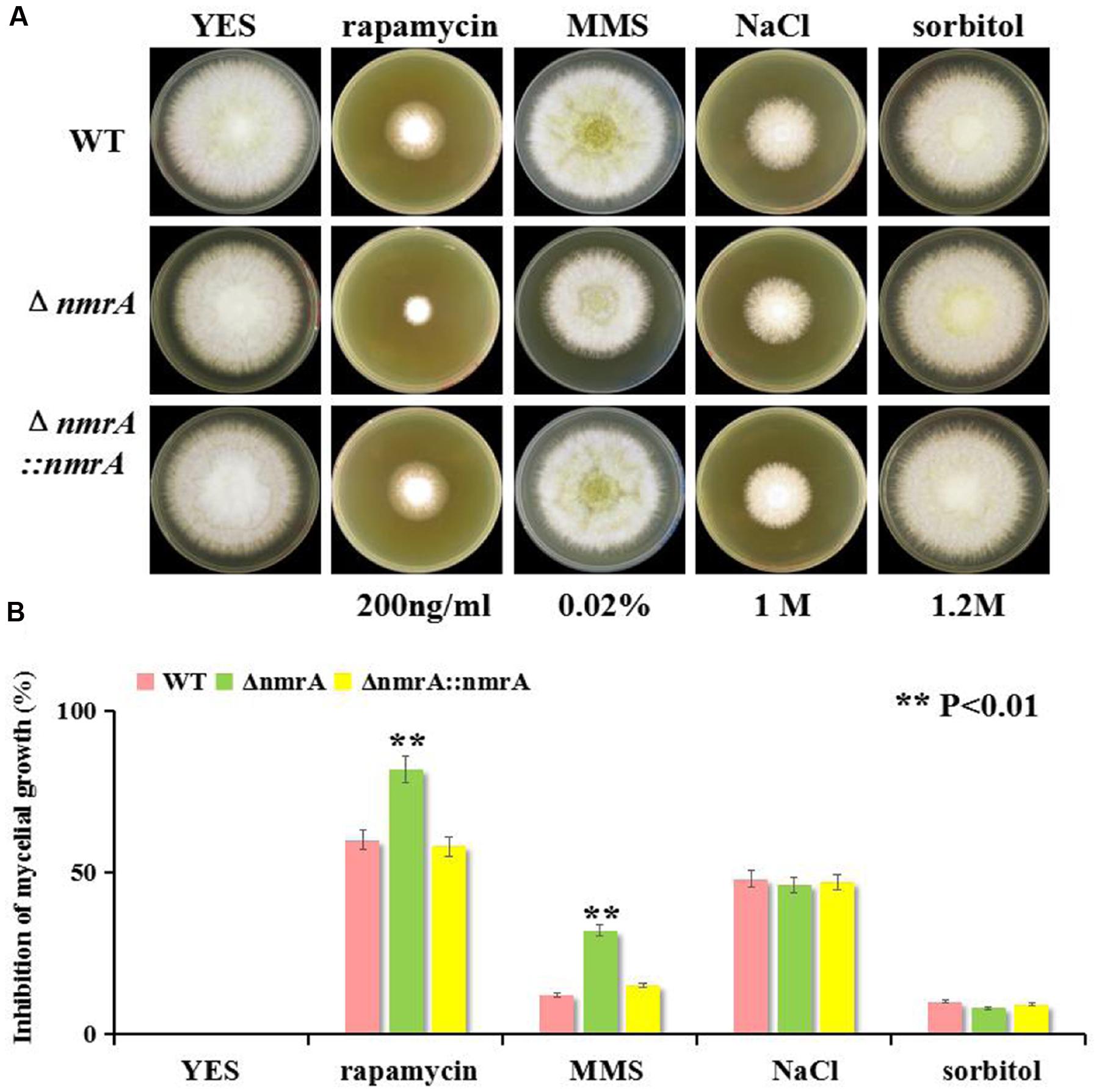
FIGURE 7. Sensitivity of the ΔnmrA strain to multiple stress reagents. (A) Sensitivity of WT, ΔnmrA and ΔnmrA::nmrA strains to 200 ng/mL rapamycin (specific inhibitor of TOR, Target of Rapamycin), 0.02% MMS (methyl methanesulfonate), 1 M NaCl and 1.2 M sorbitol added in YES agar. (B) Inhibition of mycelial growth in the ΔnmrA strain to multiple stress reagents. Asterisk indicates statistical significance at P < 0.01.
Possible Crosstalk between NmrA and Other Nitrogen Regulatory Genes
The potential interactive partners of NmrA were identified using the SMART analysis service3. The identified partners included the GATA transcriptional activator AreA (AFLA_049870), the bZIP transcription factor MeaB (AFLA_031790), the GATA transcription factor AreB (AFLA_136100), the siderophore transcription factor SreA (AFLA_132440), the nitrate reductase NiaD (AFLA_018810), and the C6 transcription factor NirA (AFLA_093040) (Figure 8A). Nitrite reductase NiiA (AFLA_018800) was also analyzed. The possible interactions were analyzed by real-time qRT-PCR. The transcript levels of areA, areB, nmrA, meaB, sreA, and nirA on ammonium were higher than those on nitrite, suggesting that these genes are likely involved in NMR. The finding that the areA expression level on ammonium exceeded that on nitrite was unexpected and was probably correlated with nitrate reductase deficiency and the relative richness of the nitrogen sources. However, transcript levels of niaD and niiA on ammonium were lower than those on nitrite (Figure 8B). Furthermore, on ammonium, all transcript levels except that of sreA were higher in the ΔnmrA strain than in the WT and ΔnmrA::nmrA strains. In contrast, on nitrite, areA, meaB, sreA, and nirA transcript levels were downregulated in the ΔnmrA mutant, while areB, niaD, and niiA transcript levels were upregulated, which was consistent with the postulated reverse roles of areA and areB. Importantly, the finding that the areA, areB, and meaB transcript levels on ammonium were higher in the ΔnmrA mutant than in the WT and ΔnmrA::nmrA strains indicates that the roles of these genes in nitrogen repression might be related to nmrA. Moreover, the observation that niaD and niiA transcript levels on nitrite were significantly higher in the ΔnmrA mutant than in WT A. flavus and ΔnmrA::nmrA demonstrates that NmrA strictly repressed transcriptional activation of genes encoding enzymes required for utilization of less favored nitrogen sources. In conclusion, these findings shed light on the complexity and sophistication of the regulatory mechanisms of NMR.
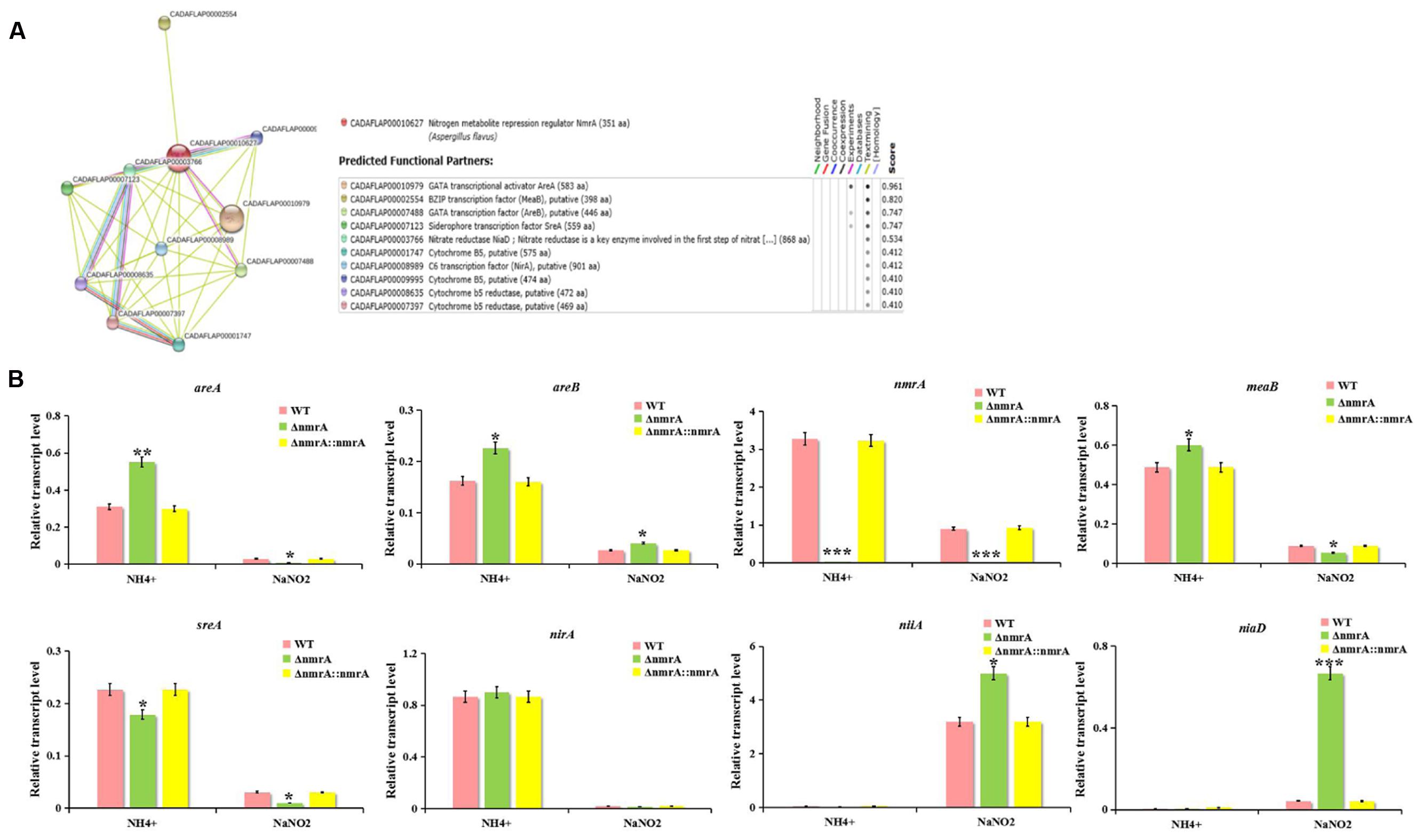
FIGURE 8. Effect of nitrogen source and ΔnmrA on transcript levels of nitrogen regulatory and catabolic genes. (A) Interaction network analyzed by SMART website. (B) RT-qPCR analysis was performed in the indicated strains germinated 30 h on GMM supplemented with 50 mM ammonium or NaNO2 as the sole nitrogen source. Transcript levels of nitrogen regulatory genes areA, areB, nmrA, meaB, sreA, and nirA as well as nitrogen catabolic genes niaD and niiA in ΔnmrA strain (light-green columns) were expressed compared to transcript levels of the WT (pink columns), and ΔnmrA::nmrA strains (yellow columns). The A. flavus actin gene was used as an internal control to normalize the expression data. Bars represent SE from three independent experiments with four replicates each. Asterisks indicated significant differences between each depletion strain relative to the WT and ΔnmrA::nmrA strains as determined by a Student T-test, with ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.005.
Discussion
The protein NmrA was defined as a repressor of the GATA transcription factor AreA, which regulates several genes required for utilization of less preferred nitrogen sources (Caddick et al., 1986; Andrianopoulos et al., 1998; Tudzynski, 2014), and the bZIP protein MeaB was proposed to activate NmrA in A. nidulans (Wong et al., 2007; López-Berges et al., 2010; Amaike et al., 2013). AreA, MeaB, and NmrA and are conserved in filamentous fungi (Wagner et al., 2010). In this study, we established that NmrA is highly conserved among Aspergillus species at the amino acid sequence level, and NmrA of A. flavus has a canonical Rossmann fold motif. We then investigated the roles of NmrA in A. flavus.
Our work showed that nmrA deletion reduced the growth of A. flavus, suggesting that nmrA participated in nitrogen source metabolism and utilization. Moreover, we also found that there was no difference in mycelial growth among the WT, ΔnmrA and ΔnmrA::nmrA strains grown on Czapek Dox media (carbon source is sucrose), regardless of the nitrogen source (Supplementary Figure S2), which was different from the results of GMM (carbon source is glucose), we speculated that NmrA might also participate in carbon catabolite repression (CCR). It has been shown that deletion of nmr (an nmrA homolog) in F. graminearum had little effect on growth or toxin production in the presence of sucrose (Giese et al., 2013). Furthermore, NmrA was suggested to play a role in carbon metabolism (Macios et al., 2012). Therefore, further investigation of the connections of NmrA with nitrogen and carbon metabolism should be carried out. Interestingly, A. flavus WT (functional mutations of nitrate reductase) was incapable of growth on culture medium with nitrate instead of nitrite, but A. flavus NRRL3357 was incapable of growth on culture medium with nitrite (data not shown). Therefore, we doubted that there was preferential utilization of nitrate over nitrite.
Aspergillus flavus differentiates to produce asexual dispersing spores (conidia) or overwintering survival structures called sclerotia (Diener et al., 1987; Horowitz Brown et al., 2008). However, sclerotia are also hypothesized to be degenerate sexual structures and may represent a vestige of cleistothecium production (Geiser et al., 1996; Chang et al., 2001). In this study, nmrA deletion resulted in significant increases in conidiation and sclerotia production compared with the WT and ΔnmrA::nmrA strains. Asexual and sexual processes in filamentous fungi are likely to be oppositely regulated by coordinate mechanisms (Horowitz Brown et al., 2008; Chang et al., 2013; Sarikaya-Bayram et al., 2014), so it was surprising that nmrA deletion stimulated both conidiation and sclerotia production. These results indicate that NmrA is likely involved in asexual and sexual development.
Aflatoxins, the most deleterious of natural products, are biosynthesized through an extremely refined and sophisticated pathway. This pathway could be affected by many biotic and abiotic factors, including nutritional factors such as carbon and nitrogen sources and environmental factors such as water activity and temperature (Yu, 2012; Amare and Keller, 2014; Zhang et al., 2014, 2015; Bai et al., 2015a,b). In this study, deletion of nmrA resulted in reduced AF production only in the presence of glutamine or alanine, suggesting that the effect of nmrA on AF biosynthesis is mediated by nitrogen sources and that additional factors must be involved in nitrogen regulation, particularly in the regulation of AF biosynthesis. Although the influence of nmrA deletion on secondary metabolism has not been studied in filamentous fungi, the ΔnmrA strain of F. fujikuroi did not display differential expression of the gibberellic acid biosynthetic gene (Schönig et al., 2008; Tudzynski, 2014). This was not the case in our study and likely reflected the different expression patterns of Nmr1/NmrA in the regulation of secondary metabolism in Fusarium and Aspergillus.
In some plant pathogenic fungi, AreA is required for full virulence and contributes to pathogenicity, probably because of the failure of mutants to fully adapt to poor nitrogen conditions during infection (Min et al., 2012; Tudzynski, 2014). Deletion of nmrA in this study decreased the virulence of A. flavus on peanut seeds, resulting in decreased colonization as reflected by lower conidia production and AF production. This indicates that NmrA could also contribute to virulence and pathogenicity. As far as we know, this is the first report of the role of NmrA in the virulence and pathogenicity of A. flavus.
We examined the sensitivity of the ΔnmrA strain to several stressors to dissect the role of NmrA in stress responses. The ΔnmrA mutant showed increased sensitivity to rapamycin and MMS, indicating that nmrA might be responsive to TOR signaling and DNA damage. Besides the TOR cascade, other signaling components seem to be involved in nitrogen sensing and subsequent regulation of secondary metabolism (Tudzynski, 2014), so it was not surprising that NmrA might be responsive to TOR. However, deletion of the nmr1 gene in F. fujikuroi resulted in increased rapamycin resistance (Teichert et al., 2006), which was the opposite of what we observed. Therefore, we suspect that the roles of Nmr1/NmrA in nitrogen regulation are regulated differently by TOR signaling in Fusarium and Aspergillus. Zhao et al. (2010) reported that NmrA could discriminate between oxidized and reduced dinucleotides and was positioned close to the GATA motif in DNA, so NmrA might play a role in DNA protection.
In A. nidulans, nmrA transcription is partially regulated by the bZIP transcription factor MeaB (Wong et al., 2007). However, recent studies showed that nmrA expression in A. nidulans and F. fujikuroi is not MeaB-dependent (Wagner et al., 2010; Amaike et al., 2013), and MeaB and AreA mediate nitrogen repression coordinately while also functioning independently (Wong et al., 2007; Wagner et al., 2010). Moreover, the ability of AreA and AreB to respond to carbon status probably depends on NmrA rather than the transcription factor CreA, which mediates CCR in A. nidulans (Macios et al., 2012). Similarly, the sugar sensor Tps1 and the inhibitor proteins Nmr1–3 (orthologs of NmrA) are all regulators of CCR in Magnaporthe oryzae (Fernandez et al., 2012), again illustrating how carbon and nitrogen metabolism are intimately linked. Furthermore, AreA, AreB, MeaB, and Nmr (a homolog of NmrA) have been implicated in the regulation of secondary metabolite production in F. fujikuroi (Andrianopoulos et al., 1998; Mihlan et al., 2003; Schönig et al., 2008; Wagner et al., 2010). Nevertheless, for most nitrogen-regulated secondary metabolites in fungi, the molecular mechanism of the nitrogen dependency is not well understood (Tudzynski, 2014). So we attempted to find links between NmrA and other interactive partners in the study. We searched for possible interactions between nmrA and other nitrogen regulatory and catabolic genes using the SMART website. RT-qPCR showed that the expression of these interactive partners was up- or down-regulated in the ΔnmrA mutant compared with the WT and ΔnmrA::nmrA strains in the presence of ammonium and nitrite. However, the detailed interaction networks of NmrA and its interactive partners are still unknown. Therefore, further investigation of the interaction networks of nitrogen regulatory and catabolic genes should be undertaken.
In this study, the nmrA gene was identified in A. flavus, and characterization of nmrA mutants revealed that NmrA plays crucial roles in radial growth, conidia and sclerotia formation, AF production, virulence, and stress responses. nmrA may also interact with other nitrogen regulatory and catabolic genes. To our knowledge, this is the first report of the function of NmrA in A. flavus. Our study provides new insights into the role of NmrA in regulating nitrogen source metabolism and AF biosynthesis in A. flavus.
Author Contributions
XH, BW, and SW conceived and designed the experiments. XH and W-BY wrote the manuscript. XH and MQ performed the experiments. XH and BW analyzed the data. XN, QQ, SR, KY, FZ, and ZZ contributed reagents/materials/analysis tools. SW supported financially and administratively, final approval of manuscript.
Funding
This research was supported by the National 973 Program (No. 2013CB127802) from the Ministry of Science, Technology of China and by grants (No. 31172297, 31000961 and 31400100) from the National Natural Science Foundation of China (NSFC), and Foundation of The Education Department of Fujian Province for Young and Middle-aged Researchers (JA14115).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer FS and handling Editor declared their shared affiliation, and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
Acknowledgment
We are grateful to Prof. Zhumei He (Sun Yat-sen University, Guangzhou, China) provide the strain A. flavus NRRL3357.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01794/full#supplementary-material
Footnotes
References
Amaike, S., Affeldt, K. J., Yin, W. B., Franke, S., Choithani, A., and Keller, N. P. (2013). The bZIP protein MeaB mediates virulence attributes in Aspergillus flavus. PLoS ONE 8:e74030. doi: 10.1371/journal.pone.0074030
Amaike, S., and Keller, N. P. (2011). Aspergillus flavus. Annu. Rev. Phytopathol. 49, 107–133. doi: 10.1146/annurev-phyto-072910-095221
Amare, M. G., and Keller, N. P. (2014). Molecular mechanisms of Aspergillus flavus secondary metabolism and development. Fungal Genet. Biol. 66, 11–18. doi: 10.1016/j.fgb.2014.02.008
Andrianopoulos, A., Kourambas, S., Sharp, J. A., Davis, M. A., and Hynes, M. J. (1998). Characterization of the Aspergillus nidulans nmrA gene involved in nitrogen metabolite repression. J. Bacteriol. 180, 1973–1977.
Arst, H. N. Jr., and Cove, D. J. (1973). Nitrogen metabolite repression in Aspergillus nidulans. Mol. Gen. Genet. 126, 111–141. doi: 10.1007/BF00330988
Bai, G. H., and Shaner, G. (1996). Variation in Fusarium graminearum and cultivar resistance to wheat scab. Plant Dis. 80, 975–979. doi: 10.1094/PD-80-0975
Bai, Y., Lan, F., Yang, W., Zhang, F., Yang, K., Li, Z., et al. (2015a). sRNA profiling in Aspergillus flavus reveals differentially expressed miRNA-like RNAs response to water activity and temperature. Fungal Genet. Biol. 81, 113–119. doi: 10.1016/j.fgb.2015.03.004
Bai, Y., Wang, S., Zhong, H., Yang, Q., Zhang, F., Zhuang, Z., et al. (2015b). Integrative analyses reveal transcriptome-proteome correlation in biological pathways and secondary metabolism clusters in A. flavus in response to temperature. Sci. Rep. 5:14582. doi: 10.1038/srep14582
Caddick, M. X., Arst, H. N. Jr., Taylor, L. H., Johnson, R. I., and Brownlee, A. G. (1986). Cloning of the regulatory gene areA mediating nitrogen metabolite repression in Aspergillus nidulans. EMBO J. 5, 1087–1090.
Caddick, M. X., Peters, D., and Platt, A. (1994). Nitrogen regulation in fungi. Antonie Van Leeuwenhoek 65, 169–177. doi: 10.1007/BF00871943
Chang, P. K., Bennett, J. W., and Cotty, P. J. (2001). Association of aflatoxin biosynthesis and sclerotial development in Aspergillus parasiticus. Mycopathologia 153, 41–48. doi: 10.1023/A:1015211915310
Chang, P. K., Scharfenstein, L. L., Li, P., and Ehrlichet, K. C. (2013). Aspergillus flavus VelB acts distinctly from VeA in conidiation and may coordinate with FluG to modulate sclerotial production. Fungal Genet. Biol. 58–59, 71–79. doi: 10.1016/j.fgb.2013.08.009
Chang, P. K., Scharfenstein, L. L., Wei, Q., and Bhatnagar, D. (2010). Development and refinement of a high-efficiency gene-targeting system for Aspergillus flavus. J. Microbiol. Methods 81, 240–246. doi: 10.1016/j.mimet.2010.03.010
Diener, U. L., Cole, R. J., Sanders, T. H., Payne, G. A., Lee, L. S., and Klich, M. A. (1987). Epidemiology of aflatoxin formation by Aspergillus flavus. Annu. Rev. Phytopathol. 25, 249–270. doi: 10.1146/annurev.py.25.090187.001341
Duran, R. M., Cary, J. W., and Calvo, A. M. (2007). Production of cyclopiazonic acid, aflatrem, and aflatoxin by Aspergillus flavus is regulated by veA, a gene necessary for sclerotia formation. Appl. Microbiol. Biotechnol. 73, 1158–1168. doi: 10.1007/s00253-006-0581-5
Fernandez, J., Wright, J. D., Hartline, D., Quispe, C. F., Madayiputhiya, N., and Wilson, R. A. (2012). Principles of carbon catabolite repression in the rice blast fungus: Tps1, Nmr1-3, and a MATE–family pump regulate glucose metabolism during infection. PLoS Genet. 8:e1002673. doi: 10.1371/journal.pgen.1002673
Geiser, D. M., Timberlake, W. E., and Arnold, M. L. (1996). Loss of meiosis in Aspergillus. Mol. Biol. Evol. 13, 809–817. doi: 10.1093/oxfordjournals.molbev.a025641
Giese, H., Sondergaarda, T. E., and Sørensenaet, J. L. (2013). The AreA transcription factor in Fusarium graminearum regulates the use of some nonpreferred nitrogen sources and secondary metabolite production. Fungal Biol. 117, 814–821. doi: 10.1016/j.funbio.2013.10.006
Horowitz Brown, S., Zarnowski, R., Sharpee, W. C., and Keller, N. P. (2008). Morphological transitions governed by density dependence and lipoxygenase activity in Aspergillus flavus. Appl. Environ. Microbiol. 74, 5674–5685. doi: 10.1128/AEM.00565-08
Kato, N., Brooks, W., and Calvo, A. M. (2003). The expression of sterigmatocystin and penicillin genes in Aspergillus nidulans is controlled by veA, a gene required for sexual development. Eukaryot. Cell 2, 1178–1186. doi: 10.1128/EC.2.6.1178-1186.2003
Kotaka, M., Johnson, C., Lamb, H. K., Hawkins, A. R., Ren, J., and Stammers, D. K. (2008). Structural analysis of the recognition of the negative regulator NmrA and DNA by the zinc finger from the GATA-type transcription factor AreA. J. Mol. Biol. 381, 373–382. doi: 10.1016/j.jmb.2008.05.077
Kubodera, T., Yamashita, N., and Nishimura, A. (2000). Pyrithiamine resistance gene (ptrA) of Aspergillus oryzae: cloning, characterization and application as a dominant selectable marker for transformation. Biosci. Biotechnol. Biochem. 64, 1416–1426. doi: 10.1271/bbb.64.1416
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real time quantitative PCR and the 2-ΔΔCt Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
López-Berges, M. S., Rispail, N., Prados-Rosales, R. C., and Di Pietro, A. (2010). A nitrogen response pathway regulates virulence functions in Fusarium oxysporum via the protein kinase TOR and the bZIP protein MeaB. Plant Cell 22, 2459–2475. doi: 10.1105/tpc.110.075937
Macios, M., Caddick, M. X., Weglenski, P., Scazzocchio, C., and Dzikowska, A. (2012). The GATA factors AREA and AREB together with the co-repressor NMRA, negatively regulate arginine catabolism in Aspergillus nidulans in response to nitrogen and carbon source. Fungal Genet. Biol. 49, 189–198. doi: 10.1016/j.fgb.2012.01.004
Magasanik, B., and Kaiser, C. A. (2002). Nitrogen regulation in Saccharomyces cerevisiae. Gene 290, 1–18. doi: 10.1016/S0378-1119(02)00558-9
Marzluf, G. A. (1997). Genetic regulation of nitrogen metabolism in the fungi. Microbiol Mol Biol Rev 61, 17–32.
Mihlan, M. V., Homann, V., Liu, T. W., and Tudzynski, B. (2003). AREA directly mediates nitrogen regulation of gibberellin biosynthesis in Gibberella fujikuroi, but its activity is not affected by Nmr. Mol. Microbiol. 47, 975–991. doi: 10.1046/j.1365-2958.2003.03326.x
Min, K., Min, K., Shin, Y., Son, H., Lee, J., Kim, J. C., et al. (2012). Functional analyses of the nitrogen regulatory gene areA in Gibberella zeae. FEMS Microbiol. Lett. 334, 66–73. doi: 10.1111/j.1574-6968.2012.02620.x
Sarikaya-Bayram, O., Bayram, O., Feussner, K., Kim, J. H., Kim, H. S., Kaever, A., et al. (2014). Membrane-bound methyltransferase complex VapA-VipC-VapB guides epigenetic control of fungal development. Dev. Cell 29, 406–420. doi: 10.1016/j.devcel.2014.03.020
Schönig, B., Brown, D. W., Oeser, B., and Tudzynski, B. (2008). Cross-species hybridization with Fusarium verticillioides microarrays reveals new insights into Fusarium fujikuroi nitrogen regulation and the role of AreA and Nmr. Eukaryot Cell 7, 1831–1846. doi: 10.1128/EC.00130-08
Shimizu, K., and Keller, N. P. (2001). Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating molphological and chemical transitions in Aspergillus nidulans. Genetics 157, 591–600.
Snoeijers, S. S., Perez-Garcia, A., Joosten, M. H. A. J., and de Wit, P. J. G. M. (2000). The effect of nitrogen on disease development and gene expression in bacterial and fungal plant pathogens. Eur. J. Plant Pathol. 106, 493–506. doi: 10.1023/A:1008720704105
Son, H., Kim, M. G., Min, K., Seo, Y. S., Lim, J. Y., Choi, G. J., et al. (2013). AbaA regulates conidiogenesis in the Ascomycete Fungus Fusarium graminearum. PLoS ONE 8:e72915. doi: 10.1371/journal.pone.0072915
Stammers, D. K., Ren, J., Leslie, K., Nichols, C. E., Lamb, H. K., Cocklin, S., et al. (2001). The structure of the negative transcriptional regulator NmrA reveals a structural superfamily which includes the short-chain dehydrogenase/reductases. EMBO J. 20, 6619–6626. doi: 10.1093/emboj/20.23.6619
Teichert, S., Wottawa, M., Schönig, B., and Tudzynski, B. (2006). Role of the Fusarium fujikuroi TOR kinase in nitrogen regulation and secondary metabolism. Eukaryot. Cell 5, 1807–1819. doi: 10.1128/EC.00039-06
Tudzynski, B. (2014). Nitrogen regulation of fungal secondary metabolism in Fungi. Front. Microbiol. 5:656. doi: 10.3389/fmicb.2014.00656
Wagner, D., Schmeinck, A., Mos, M., Morozov, I. Y., Caddick, M. X., and Tudzynski, B. (2010). The bZIP transcription factor MeaB mediates nitrogen metabolite repression at specfic loci. Eukaryot. Cell 9, 1588–1601. doi: 10.1128/EC.00146-10
Wiemann, P., and Tudzynski, B. (2013). “The nitrogen regulation network and its impact on secondary metabolism and pathogenicity,” in Fusarium: Genomics, Molecular and Cellular Biology, eds D. W. Brown and R. H. Proctor (Norwich: Caister Academic Press), 111–142.
Wilson, R. A., and Arst, H. N. Jr. (1998). Mutational analysis of AREA, a transcriptional activator mediating nitrogen metabolite repression in Aspergillus nidulans and a member of the “streetwise” GATA family of transcription factors. Microbiol. Mol. Biol. Rev. 62, 586–596.
Wilson, R. A., Gibson, R. P., Quispe, C. F., Littlechild, J. A., and Talbot, N. J. (2010). An NADPH-dependent genetic switch regulates plant infection by the rice blast fungus. Proc. Natl. Acad. Sci. U.S.A. 107, 21902–21907. doi: 10.1073/pnas.1006839107
Wong, K. H., Hynes, M. J., Todd, R. B., and Davis, M. A. (2007). Transcriptional control of nmrA by the bZIP transcriptional factor MeaB reveals a new level of nitrogen regulation in Aspergillus nidulans. Mol. Microbiol. 66, 534–551. doi: 10.1111/j.1365-2958.2007.05940.x
Wong, K. H., Hynes, M. J., and Davis, M. A. (2008). Recent advances in nitrogen regulation: a comparison between Saccharomyces cerevisiae and filamentous fungi. Eukaryot. Cell 7, 917–925. doi: 10.1128/EC.00076-08
Yang, K., Liang, L., Ran, F., Liu, Y., Li, Z., Lan, H., et al. (2016). The DmtA methyltransferase contributes to Aspergillus flavus conidiation, sclerotial production, aflatoxin biosynthesis and virulence. Sci. Rep. 6:23259. doi: 10.1038/srep23259
Yang, K., Zhuang, Z., Zhang, F., Song, F., Zhong, H., Ran, F., et al. (2015). Inhibition of aflatoxin metabolism and growth of Aspergillus flavus in liquid culture by a DNA methylation inhibitor. Food Addit. Contam. A 32(4), 554–563. doi: 10.1080/19440049.2014.972992
Yu, J. (2012). Current understanding on aflatoxin biosynthesis and future perspective in reducing aflatoxin contamination. Toxins (Basel) 4(11), 1024–1057. doi: 10.3390/toxins4111024
Zhang, F., Guo, Z., Zhong, H., Wang, S., Yang, W., Liu, Y., et al. (2014). RNA-Seq-based transcriptome analysis of aflatoxigenic Aspergillus flavus in response to water activity. Toxins (Basel) 6, 3187–3207. doi: 10.3390/toxins6113187
Zhang, F., Zhong, H., Han, X., Guo, Z., Yang, W., Liu, Y., et al. (2015). Proteomic profile of Aspergillus flavus in response to water activity. Fungal Biol. 119, 114–124. doi: 10.1016/j.funbio.2014.11.005
Keywords: Aspergillus flavus, nitrogen metabolism, nmrA, aflatoxins, AreA
Citation: Han X, Qiu M, Wang B, Yin W-B, Nie X, Qin Q, Ren S, Yang K, Zhang F, Zhuang Z and Wang S (2016) Functional Analysis of the Nitrogen Metabolite Repression Regulator Gene nmrA in Aspergillus flavus. Front. Microbiol. 7:1794. doi: 10.3389/fmicb.2016.01794
Received: 31 July 2016; Accepted: 25 October 2016;
Published: 25 November 2016.
Edited by:
Maria De Lourdes Teixeira De Moraes Polizeli, University of São Paulo, BrazilReviewed by:
Fernando Segato, University of São Paulo, BrazilAndré Ricardo Lima Damásio, University of Campinas, Brazil
Maria Celia Bertolini, Instituto de Química, Brazil
Copyright © 2016 Han, Qiu, Wang, Yin, Nie, Qin, Ren, Yang, Zhang, Zhuang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shihua Wang, wshyyl@sina.com
†These authors have contributed equally to this work.
 Xiaoyun Han
Xiaoyun Han Mengguang Qiu
Mengguang Qiu Bin Wang1†
Bin Wang1† Wen-Bing Yin
Wen-Bing Yin Qiuping Qin
Qiuping Qin Kunlong Yang
Kunlong Yang Shihua Wang
Shihua Wang