- 1Key Laboratory of Zoological Systematics and Evolution, Institute of Zoology, Chinese Academy of Sciences, Beijing, China
- 2University of Chinese Academy of Sciences, Beijing, China
- 3Disease Genomics and Individualized Medicine Laboratory, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing, China
- 4College of Life Sciences, Hebei University, Baoding, China
- 5Section of Infectious Diseases, Yale University School of Medicine, New Haven, CT, USA
Phage-mediated horizontal gene transfer (HGT) is common in free-living bacteria, and many transferred genes can play a significant role in their new bacterial hosts. However, there are few reports concerning phage-mediated HGT in endosymbionts (obligate intracellular bacteria within animal or plant hosts), such as Wolbachia. The Wolbachia-infecting temperate phage WO can actively shift among Wolbachia genomes and has the potential to mediate HGT between Wolbachia strains. In the present study, we extend previous findings by validating that the phage WO can mediate transfer of non-phage genes. To do so, we utilized bioinformatic, phylogenetic, and molecular analyses based on all sequenced Wolbachia and phage WO genomes. Our results show that the phage WO can mediate HGT between Wolbachia strains, regardless of whether the transferred genes originate from Wolbachia or other unrelated bacteria.
Introduction
Horizontal gene transfer (HGT), or lateral gene transfer, is the exchange of genetic elements across species. Abundant evidence of HGT has been detected over the last few decades, particularly in prokaryotic organisms (Ortiz et al., 2015). The genes acquired by HGT can provide new activities to a bacterial host (Waldor and Mekalanos, 1996; Brüssow et al., 2004; Rodriguez-Valera et al., 2009; Modi et al., 2013). Additionally, these genes can play a significant role in the ecological and evolutionary adaptation to a new host (Ochman et al., 2000). Bacteriophages, plasmids, and transposons are the typical genetic vehicles that mediate HGT (Brüssow et al., 2004). The global rate of phage-mediated HGT events is estimated to be as much as 2 × 1016 per second (Bushman, 2002). Recently, molecular evidence for HGT in the genomes of several obligate intracellular bacteria has been reported (Gavotte et al., 2004; Ishmael et al., 2009; Chafee et al., 2010). However, the role of phage in such transfers has not been thoroughly investigated and is considered likely to be rare due to the constraints of an intracellular lifestyle (Fineran et al., 2009).
The obligate intracellular bacterium Wolbachia, a cytoplasmically inherited Rickettsiales, has recently attracted increasing attention. As one of the most widespread endosymbionts in nature (Hilgenboecker et al., 2008; Zug and Hammerstein, 2012), Wolbachia can manipulate arthropod hosts' reproductive systems to facilitate their own spread (Werren et al., 2008). Accordingly, there is worldwide interest in using Wolbachia-infected mosquitoes to reduce mosquito populations for the elimination of mosquito-borne pathogens, such as dengue virus (Zabalou et al., 2004; Turley et al., 2009; Walker et al., 2011). Wolbachia also has a mutualistic relationship with filarial nematodes and is a potential drug target for filarial diseases (Nutman, 2001; Taylor et al., 2001). However, studies on the applications of Wolbachia have been seriously hampered due to the lack of in vitro culture methods and genetic transformation tools for testing Wolbachia gene function (Fujii et al., 2004). The phage WO, which can infect Wolbachia, has the potential to mediate gene transfer and thus offers hope for Wolbachia transformation and genetic engineering (Fujii et al., 2004; Metcalf and Bordenstein, 2012).
In this study, we investigated the hypothesis that phage WO might mediate HGT in Wolbachia. Several considerations support this hypothesis. First, as a temperate phage that can shift between the lysogenic and lytic forms, phage WO is a dynamic element in the Wolbachia genome (Masui et al., 2001). Second, phage WO is widespread among Wolbachia genomes (present in about 89%; Bordenstein and Wernegreen, 2004). Nearly all sequenced Wolbachia genomes, if infected with phage WO, have at least one intact WO prophage (Kent et al., 2011a), which has the potential to produce phage particles. Third, the transfer of the phage minor capsid gene (Masui et al., 2001; Bordenstein and Wernegreen, 2004; Gavotte et al., 2007; Chafee et al., 2010) and the complete bacteriophage (Kent et al., 2011b) has been observed between different Wolbachia strains. All of the above indicate that other genetic material associated with phage WO may also be transferred when phage WO transfers between hosts.
We used bioinformatic, molecular, and phylogenetic analyses of all the published Wolbachia and phage WO genomes to investigate the occurrence of phage WO mediated HGT in Wolbachia. We first detected the “alien” genes associated with phage WO through blastp and blastn searches, phylogenetic approaches (genes with restricted distributions) or parametric approaches (genes showing distinct nucleotide composition bias or molecular evolution pattern compared to bacterial host genes). These “alien” genes are shown to be packaged in phage WO by a combination of experimental evidence, molecular experiments of reverse PCR or real-time qPCR, and from literature searching. However, these phylogenetic and parametric approaches do not suggest that these “alien” genes are of virus origin (Azad and Lawrence, 2012). In addition, thorough comparable genomic analyses are used to investigate phage WO horizontal transfer vestiges and their association with mediating transfer of “alien” genes.
Materials and Methods
Data Mining
The complete prophage WOcauB3 (B3gp1–B3gp46), prophage WOcauB2 (B2gp1–B2gp47), the flanking-region genes from the prophage WOVitA1 (VA1gp52–VA1gp63), and two flanking-region genes from the prophage WORiB1 (WRi_005400–WRi_005900) were used as queries in a blastp search of the NCBI non-redundant protein database and a blastn search of the NCBI nucleotide collection (nr/nt) and whole-genome shotgun contigs (wgs) databases. The output E-value (<10−5) of the searches were used as criteria for data parsing. Sequences were aligned with ClustalW in BioEdit (Hall, 1999), and the Gblocks program (ver. 0.91b) (Castresana, 2000) was used to remove poorly aligned positions.
Phylogenetic Analysis
For phylogenetic analyses, ProtTest 3 (for amino acid sequences; Darriba et al., 2011) and jModelTest 2 (for nucleotide sequences; Darriba et al., 2012) were used to determine the best evolution model based on the corrected Akaike information criterion (AICc). PhyML 3.0 (Guindon et al., 2010) and Mrbayes 3.2 (Ronquist et al., 2012) were used to build phylogenetic trees with ML and BI methods respectively. The best models chosen by ProtTest 3, LG + I + G was used to generate the ML and BI tree for B3gp45. The best model chosen by jModelTest 2, GTR+G, was used to generate the ML tree for the Wolbachia MLST phylogeny.
Sequence Analysis
To visualize the general compositional features of the putative horizontally transferred genes using GC-content, a cumulative GC profile was assembled (Gao and Zhang, 2006). The cumulative GC profile can identify genomic islands or HGTs through comparison of nucleotide compositional features (Gao and Zhang, 2006). The halting parameter was set to 7, and the minimum length to segment was set to 100.
Selection Analysis
MEGA6 was used to estimate the mean synonymous divergence for each group of sequences representing potential recent horizontal transfer of WO phages, other WO phages that seem not to results from recent horizontal transfer, and their corresponding Wolbachia hosts (Tamura et al., 2013). For each group of sequences, the Nei-Gojobori method was used to calculate the synonymous rate, and variance was computed using 1000 bootstrap replicates (Nei and Gojobori, 1986).
Sample Collection
Musca domestica and Nasonia vitripennis were used in these experiments. The N. vitripennis populations were the Hangzhou strain (from the Gongyin Ye lab, ZheJiang University) (Zhang et al., 2005) infected with Wolbachia supergroup A (Liu et al., 2014). The housefly larvae were fed bran for 5–6 days until pupation. All wasps were reared on fresh house fly pupae at 25 ± 2°C under a 14 h light cycle in an atmosphere of 50–60% relative humidity, supplemented with a piece of cotton in a soft capsule shell of 10% honey water. The adult houseflies were kept at 25 ± 2°C, but supplied with a sugar/milk powder mixture (25/75%) and water instead. Adults of N. vitripennis were initially immersed in 95% ethanol at −20°C prior to DNA extraction.
DNA Extraction, PCR Amplification and Cloning
Total N. vitripennis genomic DNA was extracted from a single wasp using the DNeasy Tissue Kit (Qiagen, Hilden, Germany) following the manufacturer's recommendations and resuspended in 20 μl double-distilled sterile water. DNA purity and concentration were determined with a NanoDrop 2000 Spectrophotometer (Thermo, Madison, WI, USA), and samples of poor quality were discarded. The identity of the DNA templates was confirmed by wsp 81f and 691r primers to amplify the Wolbachia surface protein gene (Zhou et al., 1998). The PCR reactions were performed using TransTaq DNA Polymerase HiFi Fidelity (TransGen Biotech, Beijing, China) with the recommended conditions and reagents. The resulting amplicons were electrophoresed on a 1% TBE agarose gel and photographed under UV illumination. The amplified PCR products were sequenced directly with an ABI3730 capillary autosequencer (Biosune, Beijing, China) after purification with the EasyPure PCR Purification Kit (TransGen Biotech, Beijing, China). If the products could not be sequenced directly, we cloned them into the pEASY-T5 vector (TransGen Biotech, Beijing, China), and a minimum of three positive clones were sequenced due to transformation-induced mutation. Sequence editing was performed with BioEdit (Hall, 1999).
Real-Time qPCR
Real-time qPCR was performed with a Stratagene Mx3000p qPCR System (Stratagene, La Jolla, CA, USA) (the primers are listed in Table S3). We used real-time qPCR to quantify the DNA copies of a putative transcriptional regulator gene (VA1gp53) and an Hsp20-family heat shock protein gene (VA1gp62) from the flanking region of WOVitA1; an ank gene (VA1gp3) from phage WOVitA1; and a heat-shock protein 60 gene (groEL) (Bordenstein et al., 2006) and cell division gene (ftsZ) from wVitA vs. prepared standard solutions. The amplified PCR products were sequenced directly to confirm the gene identity. A standard 10-fold dilution series from 107 to 103 copies were prepared and used to calculate the copy numbers of the genes. The genes's amplification efficiency in our experiments are 96.6–104.4%. Also, each melting curve showed that the primers amplify a single product.
Statistical Analysis
The average copy number of the integrated phage was compared with the expected number and the difference was analyzed statistically with a two-tailed t-test (SAS Institute, Cary, NC, USA). With a single lysogenic copy of WOVitA1, the expected WOVitA1 number should always equal (no lytic activity) or exceed (with lytic activity producing multiple phage virions) the wVitA copy number. We normalized the small plate effects in real-time qPCR experiments as described previously (Wang et al., 2014). The compared percent nucleotide identity was analyzed by an Mann–Whitney U two-tailed test using Origin8.0.
Nucleotide Sequence Accession Number
De novo nucleotide sequences were deposited in GenBank under accession numbers KP966832–KP966840.
Results
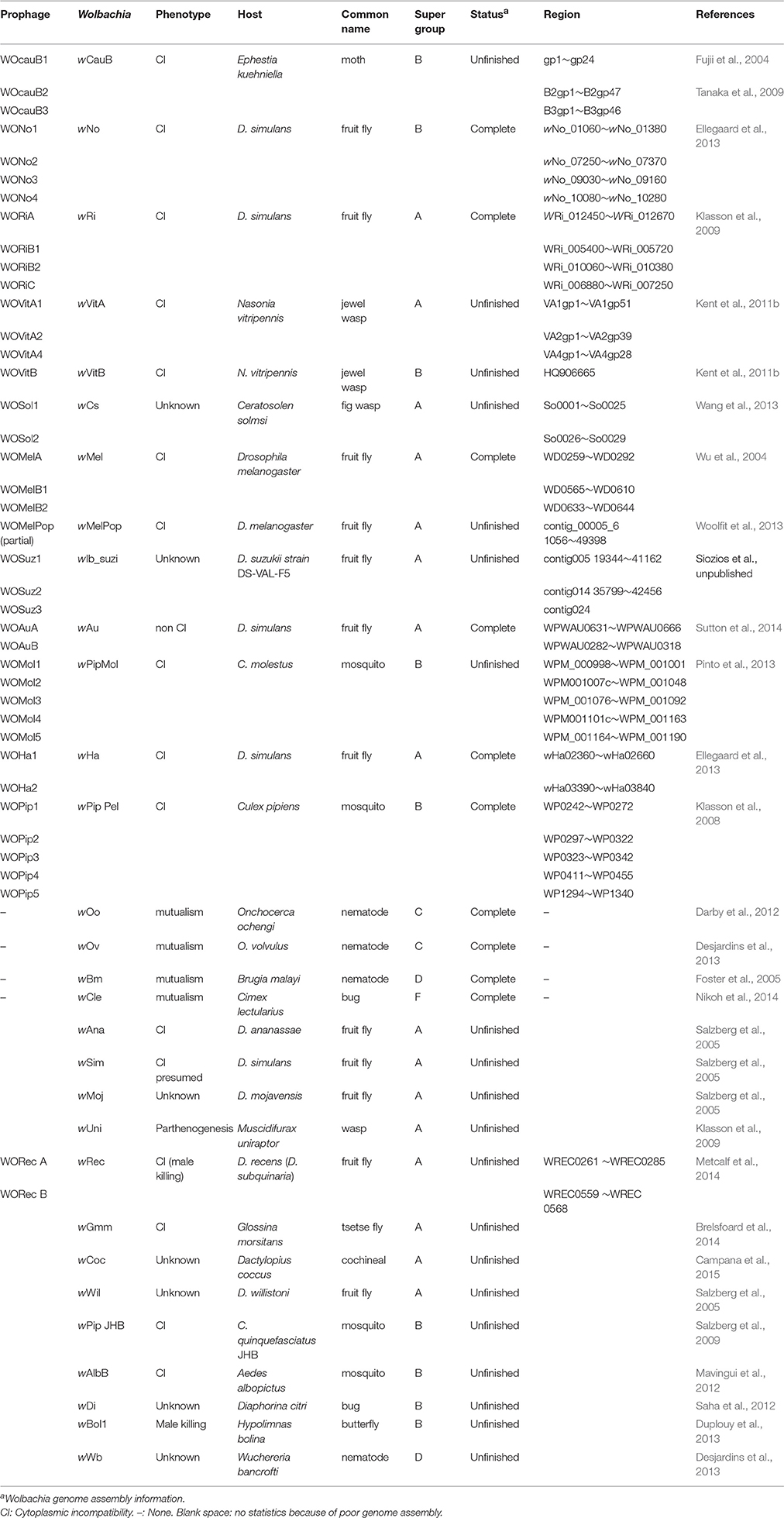
Several previous studies have shown that the phage WO might mediate HGT. The genome of the Wolbachia endosymbiont (wCauB) of the flour moth, Ephestia kuehniella, contains two related prophages, WOcauB2 and WOcauB3 (Table 1), which share high nucleotide sequence identity and conserved gene arrangements (Tanaka et al., 2009). However, there are differences in the 3′ ends of both phages: two ankyrin-domain-containing (ank) genes (B2gp46 and B2gp47) are present in WOcauB2 but absent in WOcauB3. Additionally, WOcauB3 possesses a Salmonella virulence plasmid protein B gene (B3gp45, spvB gene) and a hypothetical protein-encoding gene (B3gp46) that WOcauB2 lacks. These differences indicate that though quite similar, WOcauB2 and WOcauB3 are mobile elements that have experienced dynamic evolutionary trajectories. The genes only present in WOcauB2 (e.g., ank) or WOcauB3 (e.g., spvB) are suggested to have been transduced by phage WO (Tanaka et al., 2009). Occasionally, the transfer of a complete phage can occur between different Wolbachia strains. For example, the WO phage WOVitA1 can transfer between Wolbachia wVitA and wVitB strains hosted in N. vitripennis, and interestingly, the transfer seems to involve not only the phage region (including genes of VA1gp1–VA1gp51) but also the flanking bacterial region (Kent et al., 2011b). In this work, we used a series of stringent filters to identify phage WO mediating HGT events (Figure 1).
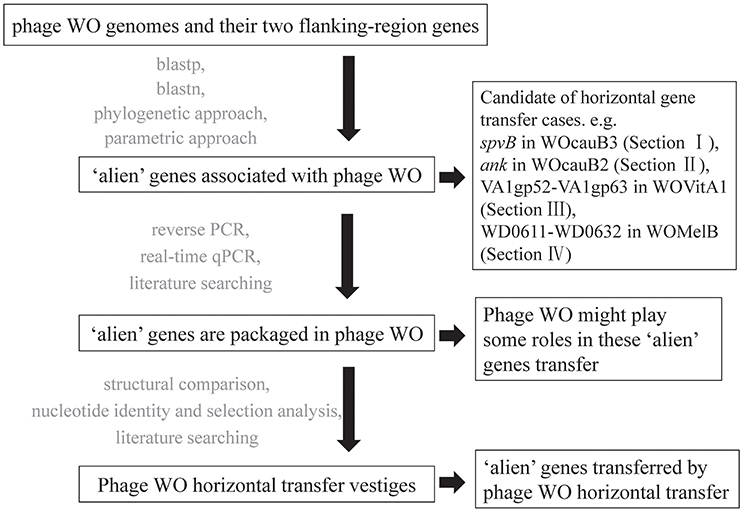
Figure 1. Flow chart of the screening methods and the results of each step used to detect phage WO mediating horizontally transferred genes.
Phage WOcauB3 Has Transferred between WcauB and wNo (A Wolbachia Strain from Supergroup B Infecting D. Simulans) and Mediated Gene Transfer
Previous searches of public databases suggested the transfer of bacterial spvB gene (B3gp45) between an unrelated bacterial genetic lineage and Wolbachia wCauB by WO (Tanaka et al., 2009). In the present study, we expand this finding by conducting a homology search for the complete phage WOcauB3 genes in all of the 32 reported Wolbachia genomes (information on all Wolbachia genomes in this study is listed in Table 1). Of the 32 genomes tested, we detected that only two, wCauB and wNo, have uniquely encoded spvB and nearby gene, B3gp46, which encodes a hypothetical protein that is packaged in phage WOcauB3 particles (Tanaka et al., 2009; Figure 2A). The Wolbachia wNo is a strain from supergroup B infecting D. simulans (Ellegaard et al., 2013).
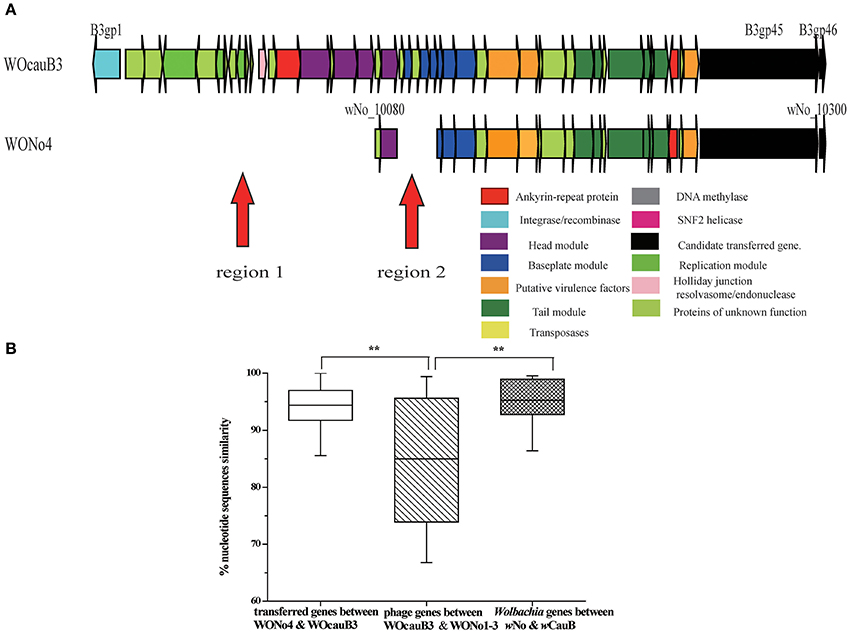
Figure 2. Phage WO transferred between wCauB and wNo. (A) Structural comparison between the WOcauB3 and WONo4 prophage sequences. Red arrows indicate discrepant regions between the prophage genomes. Genes are presented by arrows while psudogenes and non-coding regions are boxes. Genes are colored based on functional type and homology. (B) Percent nucleotide identity between prophage genes encoded on WOcauB3, WONo4, and other phages. Percent nucleotide identity is compared between phage genes transferred from WOcauB3 (between WOcauB3 and WONo4), other phage genes (between WOcauB3 and WONo1, WONo2, WONo3 in wNo that are likely not transferred from WOcauB3), Wolbachia genes (wNo and wCauB previously sequenced protein-coding genes). Error bars represent one standard deviation. The double asterisk indicates a significant difference (P <0.01; Mann–Whitney U, two-tailed test).
To further trace the transfer trajectory of spvB and B3gp46, we compared the divergence between prophages WOcauB3 and WONo1–4 and between their Wolbachia hosts, wCauB and wNo. The Wolbachia strain wNo harbors four WO phages, WONo1–4 (Table 1).
Several lines of evidence support the possibility that WOcauB3 was transferred between wCauB and wNo and mediated the transfer of both genes.
Structural Comparisons of WOcauB3 with WONo4 or with WONo1,3
Structurally, the genes in prophages WOcauB3 and WONo4 are syntenic, except in two regions: region 1 (including B3gp1–B3gp18) and region 2 (B3gp21–B3gp25). These regions are present in WOcauB3 and absent in WONo4 (Figure 2A). However, when WOcauB3 is compared with prophages WONo1 or WONo3 (WONo2 is not included in the analysis due to its short length), the gene orders are only partially conserved, indicating frequent inversion/translocation/recombination events (Figure S1A). This structural pattern indicates a recent transfer between WOcauB3 and WONo4 with the erosion of recombination, replication, head, and baseplate module as few genes exist in these modules in WONo4 while are present in WOcauB3.
Nucleotide Identity and Selection Analyses between WOcauB3 and WONo4, WOcauB3, and WONo1–3, and Their Wolbachia Hosts wCauB and wNo
Overall, prophage WOcauB3 genes are 94.37% identity to those of WONo4 at the nucleotide level (range 83.74–100.00%), which is significantly higher than the average 84.97% nucleotide identity between WOcauB3 and the other phages (WONo1–3) in the wNo genome [range 66.78–99.39%; Mann–Whitney U (MWU), two-tailed, P < 0.01; Figure 2B]. In addition, the synonymous mutation rate between prophage WOcauB3 and WONo4 is 0.07 (range 0.00–0.48), significantly lower than the average 0.27 between WOcauB3 and phages WONo1–3 (range 0.00–0.61; MWU, two-tailed, P < 0.01, data not shown). This also demonstrates a smaller divergence between prophage WOcauB3 and WONo4 than between WOcauB3 and WONo1–3. Additionally, the sequenced Wolbachia protein-coding genes (Table S1) from wCauB and wNo have a significantly higher nucleotide identity (95.74%, range 83.18–99.53%) than the phages WOcauB3 and WONo1–3 (MWU, two-tailed, P < 0.01; Figure 2B). It is noteworthy that the average nucleotide identity of WOcauB3 and WONo4 is not significantly different than the average nucleotide identity of Wolbachia genes from wCauB and wNo (MWU, two-tailed, P >0.05; Figure 2B). Given the 3.5-fold higher sequence diversity between WOcauB3 and WONo1–3 when compared to wCauB and wNo, it conservatively indicates that phage WOcauB3 may transfer directly from wCauB to wNo, or indirectly through other unsequenced Wolbachia hosts to wNo. Also, the synonymous mutation rate between WOcauB3 and WONo4 (0.07) is not significantly different from that between Wolbachia protein-coding genes of wCauB and wNo (0.10, range 0.01–0.29; MWU, two-tailed, P > 0.05, data not shown). However, there is approximately a 3.0-fold higher synonymous mutation rate between WOcauB3 and WONo1–3, when compared with wCauB and wNo protein-coding genes.
Furthermore, a Wolbachia phylogenetic tree constructed using the Multi-Locus Sequence Typing (MLST) method indicates that wNo and wCauB are not closely related Wolbachia strains (Figure S2). If phages WOcauB3 and WONo4 were assumed vertically descended from a recent common ancestor, it would require at least three independent losses (based on Figure S2 phylogenetic tree) of this phage in Wolbachia strains of wVitB, wPipPel (infecting Culex pipiens; Klasson et al., 2008), and wPipMol (infecting Culex molestus; Pinto et al., 2013), which is less parsimonious than a single phage horizontal transfer event. All of the above analyses suggest that WOcauB3 was horizontally transferred. Previous reports have demonstrated that wNo infects D. simulans (Ellegaard et al., 2013) and wCauB infects E. kuehniella (Tanaka et al., 2009). Therefore, it seems likely that these Wolbachia strains infect an intermediate host concurrently to facilitate exchange of the phage WOcauB3.
SpvB and Nearby B3gp46 Gene are Transferred via the Transmission of WOcauB3
Homologs of spvB (B3gp45) and nearby B3gp46 gene from WOcauB3 are present in only two of the 32 sequenced Wolbachia strains, wCauB and wNo (Figure 2A). spvB phylogenetic tree inferred from Maximum Likelihood (ML) and Bayesian Inference (BI) methods (Figure 3) is shown. In public databases, there are no closely related orthologs of B3gp46 except in the wCauB and wNo. These scattered distribution patterns indicate recent transmission of the spvB and B3gp46 genes. Furthermore, both genes are located at the 3′ ends of phages WOcauB3, WONo4 (Figure 2A) and are packaged into WOcauB3 (Tanaka et al., 2009), indicating that phage WO is the vehicle of their transmission.
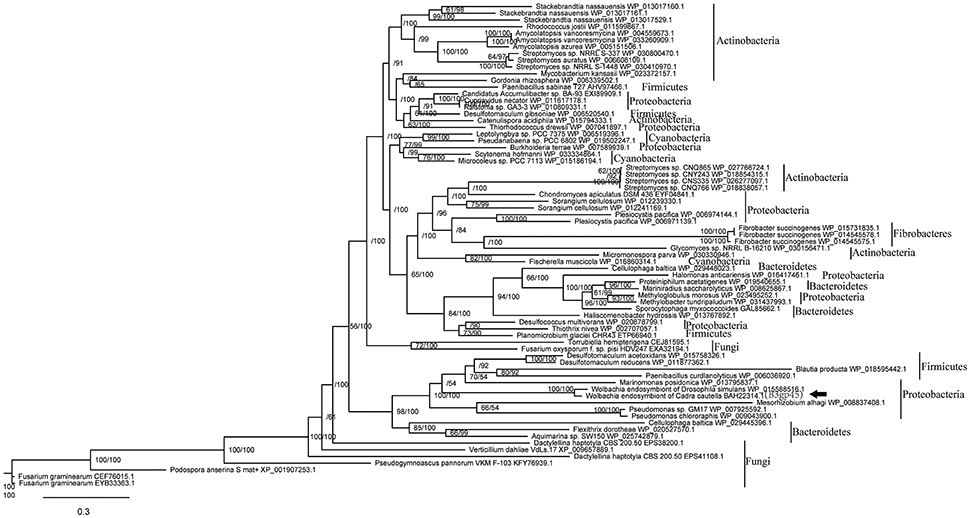
Figure 3. Phylogeny based on the amino acid sequences of phage WOcauB3 spvB gene B3gp45 and its homologs. Phylogeny based on alignment of 766 aa consisting of top E-value (<10−5) hits to blastp using WOcauB3 spvB gene as a query. There is an apparent transfer from unrelated genetic lineages of bacteria to Wolbachia, marked with arrowhead. Each tip is labeled with the species name and the sequence's Genbank accession number. Bootstrap values (maximum likelihood phylogeny) and posterior probability (bayesian phylogeny) higher than 50% are shown. The black box on the right represents the phyla.
Phage WOcauB2 Has Transferred between wCauB and wRi and Mediated the Horizontal Transfer of Two ank Genes
In addition to WOcauB3, there is also evidence to support that WOcauB2 (from the same Wolbachia strain, wCauB) has experienced a transmission event. The transmission likely mediated the horizontal transfer of two associated ank genes.
Structural Comparison of WOcauB2 with WORiC or with WORiA and WORiB1
The prophages WOcauB2, from Wolbachia wCauB, and WORiC, from Wolbachia wRi, are syntenically conserved with the exception of four heterogeneous regions, including a deletion of the B2gp2–B2gp12 region in WORiC (region 1), two insertions of transposase genes (WRi_007230 and WRi_007040; regions 2 and 3), and a deletion of a transposase gene (B2gp35) in WORiC (region 4; Figure 4A). However, gene order between WOcauB2 and WORiA or WORiB1, also from Wolbachia wRi (Klasson et al., 2009), are only partially conserved (Figure S1B; WORiB1 and WORiB2 are identical, so only WORiB1 is used for analysis in this study; Ishmael et al., 2009; Tanaka et al., 2009; Wang et al., 2013). Taken together, these similarities indicate a phage transfer (WOcauB2 and WORiC) between wCauB and wRi.
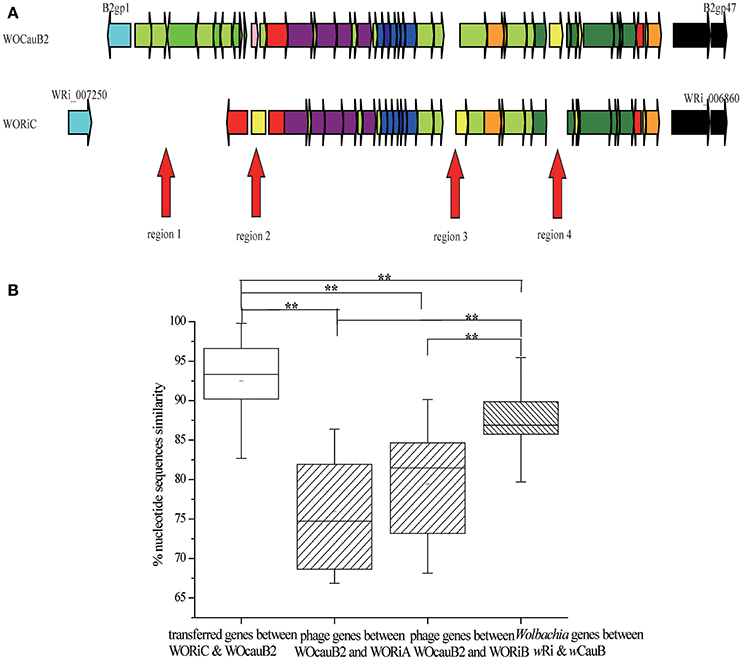
Figure 4. Phage WO transferred between wCauB and wRi. (A) Structural comparison between the WOcauB2 and WORiC prophage sequences. Red arrows indicate discrepant regions between the prophage genomes. Genes are presented by arrows while psudogenes and non-coding regions are boxes. Colors of ORFs are as described in the legend of Figure 2. (B) Percent nucleotide identity between prophage genes encoded on WOcauB2, WORiC, and other phages. Percent nucleotide identity is compared between phage genes transferred from WOcauB2 (between WOcauB2 and WORiC), other phage genes (between WOcauB2 and WORiA and WORiB in wRi that are unlikely transferred from WOcauB2), and Wolbachia genes (previously sequenced protein-coding genes in Wolbachia wCauB and wRi). Error bars represent one standard deviation. The double asterisk indicates a significant difference (P <0.01; Mann–Whitney U, two-tailed test).
Nucleotide Identity and Selection Analysis between WOcaub2 and WORiC, WOcauB2 and WORiA or WORiB1, and Their wolbachia Hosts wCauB and wRi
Comparison of nucleotide identity revealed that, on average, WORiC genes are 92.52% nucleotide identity to genes of the phage WOcauB2 (range 77.98–99.79%), which is significantly higher than the average nucleotide identity between WOcauB2 and the other phages in the wRi genome: WORiA shares 75.94% nucleotide identity with WOcauB2 (range 66.87–86.39%), and WORiB1 shares 79.43% nucleotide identity with WOcauB2 (range 68.15–90.15%; MWU, two-tailed, P < 0.01; Figure 4B). The nucleotide identity between WORiC and WOcauB2 genes is also significantly higher than the average nucleotide identity between the sequenced Wolbachia protein-coding genes (Table S2) from wCauB and wRi (86.78%; range 68.26–98.71%; MWU, two-tailed, P < 0.01; Figure 4B). Additionally, the nucleotide identity between Wolbachia strains (86.78%) is significantly higher than identity between WOcauB2 and WORiA (75.94%) or phage WORiB1 (79.43%; MWU, two-tailed, P < 0.01; Figure 4B). These results suggest that phage WOcauB2 may transfer directly from wCauB to wRi, or indirectly through other unsequenced Wolbachia hosts to wRi. Furthermore, the synonymous mutation rate between prophages WOcauB2 and WORiC is 0.13 (range 0.00–0.53), significantly lower than that between WOcauB2 and the phages WORiA (0.34; range 0.10–0.63) and WORiB1 (0.22; range 0.04–0.58) in the wRi genome (MWU, two-tailed, P < 0.01; data not shown). The synonymous mutation rate between prophages WOcauB2 and WORiC is also lower than that between wCauB and wRi (0.32; range 0.04–0.56; MWU, two-tailed, P < 0.01; data not shown).
Considering that wRi belongs to the Wolbachia supergroup A and wCauB belongs to supergroup B (Figure S2), this distant phylogenetic relationship excludes the possibility that WOcauB2 and WORiC are descended from a recent common ancestor. This further supports the transfer of WOcauB2 between wCauB and wRi. As previously reported, wRi infects D. simulans (Klasson et al., 2009) and wCauB infects E. kuehniella (Tanaka et al., 2009), which suggests that the exchange of phage WOcauB2 may have been facilitated by the coninfection of an intermediate host.
Two ank Genes Are Transferred via the Transmission of WOcauB2
Homologs of two ank genes from WOcauB2, B2gp46, and B2gp47, are present in six of the 32 sequenced Wolbachia strains (Table 2). Homologs of B2gp46 are present in Wolbachia strains wRi, wVitA, and wVitB from N. vitripennis (Kent et al., 2011b), wNo from D. simulans (Ellegaard et al., 2013), wAlbB from Aedes albopictus (Mavingui et al., 2012), and wAna from Drosophila ananassae (Salzberg et al., 2005). B2gp47 has homologs in Wolbachia strains wRi, wVitA, and wVitB. These scattered distribution patterns indicate recent transmission of the two ank genes. The segmentation point by cumulative GC profile of phage WOcauB2 supports that the phage WOcauB2 has acquired the two ank genes, B2gp46 and B2gp47, from a foreign DNA source (Figure S3). Furthermore, both genes are located at the 3′ ends of phages WOcauB2, WORiC, WOVitA1, and WOVitB (Figures 4A, 5) and are packaged into WOcauB2 (Tanaka et al., 2009) and WOVitA1 particles (see following part a), indicating that phage WO is the vehicle of their transmission.
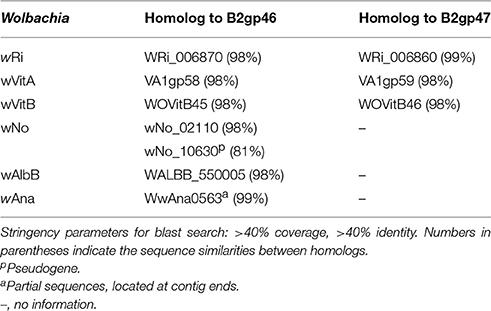
Table 2. Distribution of B2gp46 and B2gp47 genes in WOcauB2 with highly similar positional homologs in other sequenced Wolbachia genomes.
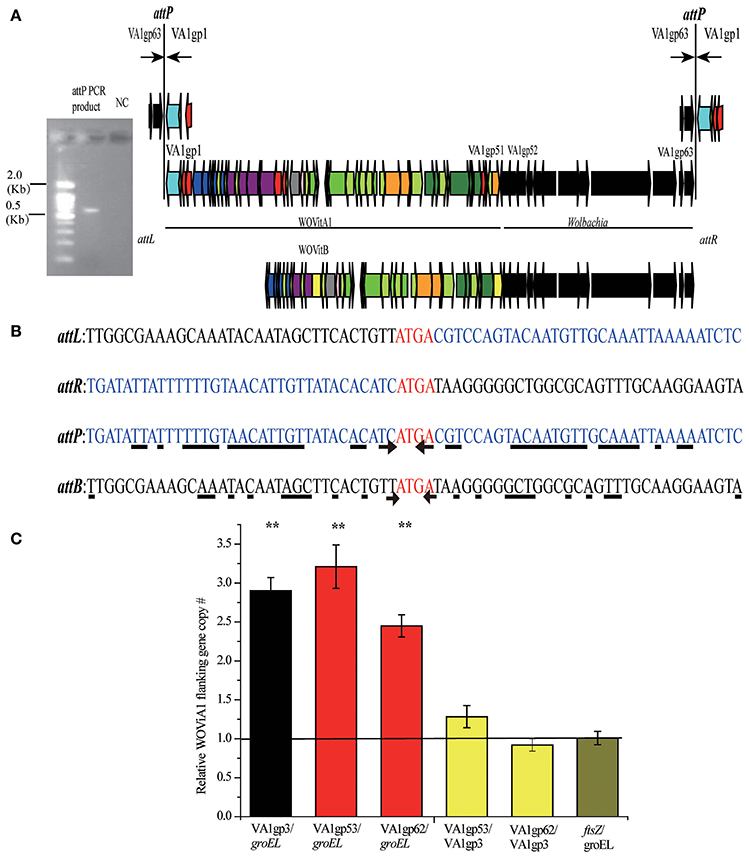
Figure 5. Determination of WOVitA1 prophage region. (A) attP PCR product, a 0.6 kb PCR product containing the WOVitA1 attP region. NC, negative control with distilled water as template. Black arrows show the locations of outward primers. Genes are presented by arrows while psudogenes and non-coding regions are boxes. Colors of ORFs are as described in the legend of Figure 2. (B) Alignment of the attP PCR product sequence and the wVitA sequence. The arrow indicates the beginning of the inverted repeat sequences, the underline shows nucleotide which we can find the corresponding inverted repeat sequences beside the core sequence. (C) Relative copy number of defined prophage WOVitA1 region, prophage WOVitA1 flanking region, and wVitA of N. vitripennis. Relative copy number of ORFs encoding genes groEL and ftsz represented wVitA, VA1gp3 represented prophage WOVitA1, and VA1gp53 and VA1gp62 represented WOVitA1 flanking region were measured by real-time qPCR. The black line (number one) depicts the expected copy number. Error bars represent one standard deviation. The double asterisk indicates a significant difference (P < 0.01; two-tailed t-test).
Phage WOVitA1 Mediates the Transfer of the Flanking Region
In addition to WOcauB2 and WOcauB3, Wolbachia wVitA possesses another active phage, WOVitA1, which has been transferred to Wolbachia wVitB in N. vitripennis (the phage WOVitA1 in wVitB is WOVitB1; Kent et al., 2011b). It is interesting to note that both WOVitA1 and WOVitB not only have nearly identical phage regions (the WOVitA1 region is VA1gp1–VA1gp51) but also highly similar flanking regions with bacterial genes (VA1gp52–VA1gp63 in WOVitA1; Kent et al., 2011b). Here, by analyzing the attachment site (att) of WO phages (attP) and the density correlation of WO with its Wolbachia host, we demonstrate that the active phage WOVitA1 can mediate the transfer of its flanking bacterial region.
The VA1gp52–VA1gp63 Flanking Region Is Packaged into Active WOVitA1 Particles
The core sequence of the attachment site is the region where the phage undergoes site-specific recombination, which occurs when the phage integrates into and excises out of the bacterial host genome (Smith and Thorpe, 2002). Phage WO has a self-ligated circular genome (Tanaka et al., 2009). If the flanking Wolbachia genes, VA1gp52–VA1gp63, are included in phage WOVitA1 particles, PCR with outward primers at the end of the prophage WOVitA1 and the flanking region would be expected to yield an attP site product. Indeed, we obtain the attP site product (Figure 5A) by using these primers (Table S3). By comparing the attP, attB (bacterial att site), attL (left prophage att site), and attR (right prophage att site) sequences, we discovered that the tetranucleotides ATGA are identical among the att sites (Figure 5B). Thus, these sequences are inferred to be the candidate core sequence for WOVitA1. However, for phages WOcauB2 and WOcauB3, the core sequences are only a single nucleotide T and trinucleotides TTG, respectively (Tanaka et al., 2009). Further, the core sequence of WOVitA1 is flanked by a pair of inverted repeat sequences (Figure 5B), where the core sequences of WOcauB2 and WOcauB3 are not (Tanaka et al., 2009).
To assess relative copy number of phage and Wolbachia, we measured the copy number of the wVitA genome (represented by both the single-copy heat-shock protein 60 gene groEL and the cell division gene ftsZ), the phage WOVitA1 genome (represented by the single-copy gene ank, VA1gp3), and the phage WOVitA1 flanking region (represented by both the single-copy transcriptional regulator gene VA1gp53 and the Hsp20-family heat shock protein gene VA1gp62; the primers are listed in Table S3). With a single lysogenic copy of WOVitA1, the WOVitA1 density should always equal (no lytic activity) or exceed (with lytic activity producing multiple phage virions) the wVitA copy number. Additionally, if the region flanking WOVitA1 is packaged into the virion, it should also exceed the genome copy number during lytic replication. The phage to Wolbachia ratio was determined to be 2.90 ± 0.17 for VA1gp3: groEL (p < 0.01; two-tailed t-test), while the ratios of the phage flanking region to Wolbachia were measured as 3.21 ± 0.28 for VA1gp53: groEL and 2.45 ± 0.14 for VA1gp62: groEL (all p < 0.01; two-tailed t-test; Figure 5C). For comparison, the phage flanking region to phage ratios were 1.28 ± 0.14 for VA1gp53: VA1gp3 and 0.92 ± 0.08 for VA1gp62: VA1gp3 and the Wolbachia to Wolbachia ratio is 1.01 ± 0.09 for ftsZ: groEL. These results indicate that the flanking region is part of WOVitA1 and is being replicated extrachromosomally. In addition, in the cumulative GC profile of phage WOVitA1, the segmentation point includes VA1gp52–VA1gp63, further indicating that the phage WO acquired the region from foreign DNA sources (Figure S4). Thus, we propose that Wolbachia genes VA1gp52–VA1gp63 were transmitted along with the transmission of the phage WOVitA1 (VA1gp1–VA1gp51) to wVitB (Kent et al., 2011b).
The Potential Roles of Packaged Genes in Phage WOVitA1 Particles
Because most of the packaged Wolbachia genes (VA1gp52–VA1gp63) are conserved among many bacteria (Kent et al., 2011b), we can predict their functions using blastp search and further trace their origins. The three genes VA1gp52, VA1gp53, and VA1gp56 are transcriptional regulators homologous to wtrM in wPipMol, which is implicated in cytoplasmic incompatibility (CI) in Culex mosquitoes via regulating mosquito gene expression (Pinto et al., 2013). The packaged genes may also encode DNA repair protein RadC (VA1gp55), adaptor protein MutL (VA1gp57), heat shock protein (VA1gp62), and ANK proteins (VA1gp58, VA1gp59, VA1gp60, and VA1gp61). All of these genes function in DNA binding or protein-protein interactions and could be involved in CI (Penz et al., 2012).
Inactive Phages WO Are Transferred and Mediate Gene Transfer
Inactive WO phages may also have been involved in gene transfer events. In eight Wolbachia genomes, we detected a conserved bacterial region extending over 20 kb that is highly homologous (>70% nt identity) to regions in the bacterial plasmids of Rickettsia buchneri sp. nov. and Rickettsia helvetica (Ishmael et al., 2009). These bacteria infect Ixodes scapularis (Kurtti et al., 2015) and Ixodes ricinus (Dong et al., 2012) ticks respectively. Interestingly, except in wNo and wAlb, this region in each of the other Wolbachia genomes is inserted in or near the phage WO, and some of the associated WO phages are degenerate (Figure 6A). We also detect homologs of some of the genes from this region in wBol1 (from Hypolimnas bolina; Duplouy et al., 2013), wVitB, wWil (from Drosophila willistoni) (Craig Venter Institute), wCoc (from Dactylopius coccus; Campana et al., 2015), and wRec (from Drosophila recens; Metcalf et al., 2014). However, all of the homologs are in scaffolds with small sizes, which prevents us from obtaining their flanking regions; therefore, we did not further analyze them. In public databases, there are no closely related orthologs of this bacterial region except in the Wolbachia strains and the two Rickettsia strains as mentioned above. There are at least three possible explanations for the distribution pattern of this region. First, this bacterial region is one of the modules of phage WO. Second, this conserved bacterial region has been frequently and independently inserted into Wolbachia at the same phage WO location, where there exists an active cloning location. Third, phage WO or plasmid mediates the transfer of this bacterial region among different Wolbachia strains, between Rickettsia from I. scapularis and I. ricinus, or between the two bacterial genera. However, the average nucleotide identity of the genes in this region (97.00%) is significantly higher than that of the other Wolbachia genes (93.28%; MWU, two-tailed, p < 0.01; Figure 6B and Table S4) and is also significantly higher than that of WORiB1, WORiB2, WOSol, WOMelB, WOSuz1, and WOAuB (88.67%; MWU, two-tailed, p < 0.01; Figure 6B). Furthermore, the synonymous mutation rate of the genes in the region from wRi, wCs, wMel, wSuz, and wAu (0.02) is significantly lower than the average synonymous mutation rate of the associated phage WO genes from WORiB1, WORiB2, WOSol, WOMelB, WOSuz1, and WOAuB (0.08; MWU, two-tailed, p < 0.01; Figure 6C). The average nucleotide identity and synonymous mutation rate analyses indicate this bacterial region is not phage WO module. Additionally, in the cumulative GC profile of phage WOMelB (Figure S5), the segmentation point, including the putative HGTs, also demonstrates phage WO and this bacterial region are from different DNA sources. Based on these analyses, we exclude the first explanation. However, we cannot exclude the second possibility that this bacterial region has been frequently and independently inserted into Wolbachia at the same phage WO location. Considering that phage and plasmid are two of the most common genetic vectors in nature (Syvanen, 1994; Canchaya et al., 2003), it is thus a more parsimonious explanation that phage WO or Rickettsia plasmid may have recently mediated the transfer of these genes.
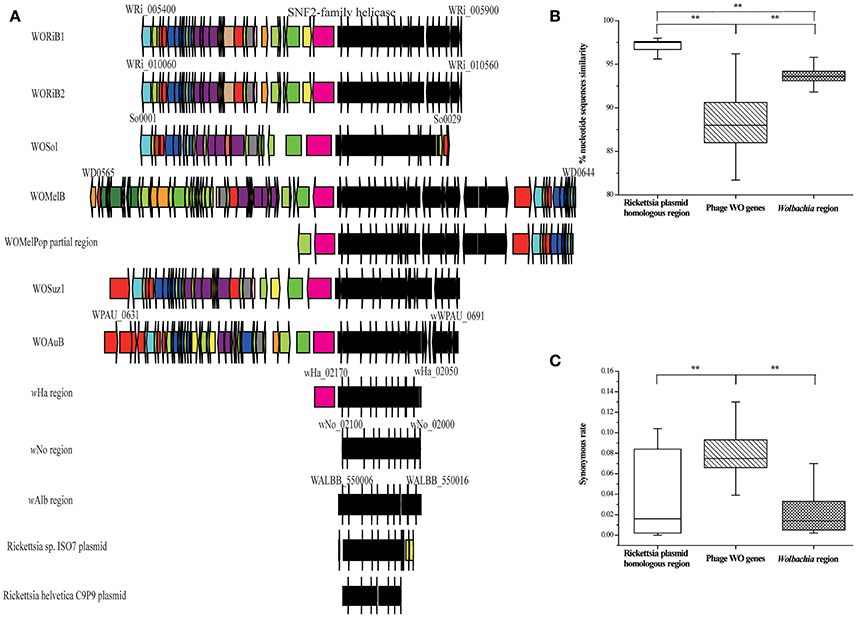
Figure 6. A conserved bacterial region associated with phage WO, Wolbachia strains, and Rickettsia plasmid. (A) Structural comparisons of the gene compositions among prophage WO, Wolbachia, and plasmid of Rickettsia endosymbiont in Ixodes scapularis and I. ricinus, all of which have the same conserved bacterial region genes (WRi_005730–WRi_005830) with WORiB1 flanking region. Genes are presented by arrows while psudogenes and non-coding regions are boxes. Colors of ORFs are as described in the legend of Figure 2. (B) Percent nucleotide identity between genes from the conserved bacterial region, phage WO genes (WORiB1, WORiB2, WOSol, WOMelB, WOSuz1, and WOAuB), and Wolbachia genes. (C) Synonymous rate between genes from the conserved bacterial region, phage WO genes (WORiB1, WORiB2, WOSol, WOMelB, WOSuz1, and WOAuB), and Wolbachia genes. Error bars represent one standard deviation. The double asterisk indicates a significant difference (P < 0.01; Mann–Whitney U, two-tailed test).
Within the transfered region, there are 11 conserved genes (WRi_005730–WRi_005830; Figure 6A). These genes encode an NAD-dependent epimerase/dehydratase family protein, a glycosyltransferase, two putative L-allo-threonine aldolases, an ABC transporter permease, a GlpT/PgpT/UhpT transporter family protein, a UDP-glucose 6-dehydrogenase, and three conserved hypothetical proteins. Many of these proteins play a role in the synthesis and degradation of surface polysaccharides, which could alter the ability of different Wolbachia strains to interact with eukaryotic hosts (Ishmael et al., 2009).
Moreover, this transfered region is located adjacent to a conserved gene encoding a SNF2-family helicase, which was detected in a phage region via genome analysis (Ishmael et al., 2009). In eukaryotes, this gene may function in processes including transcriptional regulation, the maintenance of chromosome stability during mitosis and the processing of DNA damage (Eisen et al., 1995).
Discussion
Effect of Transferred Genes on the Host
Transfer events mediated by phage WO can shape the genome composition of Wolbachia. For example, ANKs are rare in bacteria but common in eukaryotes and viruses (Bork, 1993; Li et al., 2006), while these genes are overrepresented in Wolbachia bacteria. For example, there are typically only 1–3 ank genes in the α-Proteobacteria (Andersson et al., 1998; Caturegli et al., 2000), but there are 60 ank genes in Wolbachia wPip from C. pipiens (the largest number of ank genes in any sequenced bacterial genome; Klasson et al., 2008), 35 in wRi (Klasson et al., 2009), and 23 in wMel (Wu et al., 2004). In this study, we demonstrate that WO phage particles can package and mediate the transfer of “extra” ank genes into Wolbachia genomes, e.g., B2gp46 and B2gp47 in WOcauB2, both of which are of non-Wolbachia origin. Moreover, WO can also mediate the transfer of “extra” ank genes between different Wolbachia strains. These results indicate that the WO-mediated transfer of “extra” ank genes may be a partial explanation for the abundance of ank genes in Wolbachia compared to other closely related bacteria.
The discovery of these horizontal transfer events raises the question of whether transferred genes play a role in Wolbachia or their eukaryotic hosts. Prophage-encoded virulence factors are important for a number of bacterial species, and these genes can increase pathogenicity or result in the emergence of new pathogens (Canchaya et al., 2003; Brüssow et al., 2004). This phenomenon has been recognized for the toxins of Vibrio cholerae (Waldor and Mekalanos, 1996), Streptococcus pyogenes (Broudy et al., 2002), and Hamiltonella defensa (Moran et al., 2005; Oliver et al., 2009), all of which are phage-encoded. Here, the presence of the transferred spvB motif gene in phage WO particles, and the role of this gene in WO infection of Wolbachia and the corresponding eukaryotic hosts needs further study.
Wolbachia mediated mosquito-borne disease control is a hot topic (Dobson et al., 2016; Loreto and Wallau, 2016; O'Neill, 2016; Waltz, 2016). Caged and open-field experiments showed that the wMel Wolbachia strain is able to block dengue transmission (Walker et al., 2011). However, there is also a potential risk that the Wolbachia strains, along with phage WO and other genes, may be transferred to other insects (Loreto and Wallau, 2016). Here we demonstate that phage WO can mediate HGT among different Wolbachia strains. Thus, future studies should also evaluate the biosafety of this phage vector when utilizing Wolbachia-infected mosquitos.
Phage WO Has the Potential to be Reengineered As a Transformation Tool for Wolbachia
The phenomenon of eukaryotic host reproductive manipulation by Wolbachia is compelling, but the underlying mechanism still remains poorly characterized due to the lack of robust tools for transforming Wolbachia (Werren, 1997; LePage and Bordenstein, 2013). The phage WO has been proposed as the only potential transformation tool for Wolbachia (Fujii et al., 2004; Metcalf and Bordenstein, 2012). However, it remains unclear whether phage WO can be successfully used as such a tool, and there is little research concerning this issue (LePage and Bordenstein, 2013). Here, we show that phage WO can mediate gene transfer; the active phage WOVitA1 has typical characteristics of the core sequences. Additionally, the 3′ end of WO prophages sites might be used as multiple-cloning sites. All of these results further support that phage WO has the potential to be utilized as a genetic vector for the study of Wolbachia.
Phage WO Let Us Rethink Endosymbiont Genome Evolution Theory
In the evolution of intracellular endosymbionts, genome reduction is the predominant trend differentiating endosymbionts from free-living bacteria. Additionally, intracellular endosymbionts are strictly constrained to living inside host-derived cells: their effective population size is reduced, which renders selection less efficient; they have limited opportunities to come into contact with other unrelated bacteria and have little chance to exchange genetic material; the stable and rich nutrients of the intracellular environment remove selection constraints on genes (like mobile DNA) that are no longer strictly required (Bordenstein and Reznikoff, 2005; Moya et al., 2008; Moran and Bennett, 2014). However, phage WO is widespread among Wolbachia genomes (present in about 89%; Bordenstein and Wernegreen, 2004) and even can comprise more than 20% of mobile DNA genes in Wolbachia (Chafee et al., 2010). What's more, the active mobile elements located within the genomes of endosymbionts can still mediate the deletion and insertion of genetic components at different locations in the genome. Based on several lines of evidence, the present study shows that the phage WO could mediate HGT between different Wolbachia strains of genes from Wolbachia and unrelated bacterial lineages, which showes Wolbachia genomes are not stable and might gain new genes by phage WO.
Author Contributions
GW, conception and design, acquisition of data, analysis, and interpretation of data, drafting and revising the article; DH, conception and design; JX, analysis and interpretation of data, drafting the article; BS and TX, analysis and interpretation of data; KM, analysis and drafting the article; YW, acquisition of data.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Science Foundation of China (NSFC grant nos 31210103912, 31422050), partially by a grant (O529YX5105) from the Key Laboratory of the Zoological Systematics and Evolution of the Chinese Academy of Sciences, and the National Science Fund for Fostering Talents in Basic Research (Special Subjects in Animal Taxonomy, NSFC-J0930004). We thank Prof. Heidi Goodrich-Blair at University of Wisconsin-Madison, Madison, USA, for critical discussion of revising the article. We thank Dr. Wen Xin and TransGen Biotech for providing most of the reagents used in this study. We thank Prof. Wei-Feng Shi at Tai shan Medical College, Shan Dong, China, for the discussion of data analysis. We also thank the reviewers for their valuable comments and suggestions.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01867/full#supplementary-material
Table S1. Nucleotide identity of Wolbachia protein-coding genes between wCauB and wNo.
Table S2. Nucleotide identity of Wolbachia protein-coding genes between wCauB and wRi.
Table S3. Description of primers used in study.
Table S4. Selected Wolbachia genes in the wRi genome and their homologs in other Wolbachia genomes.
Figure S1. Structural comparison between prophage WO. (A) Gene order comparisons among phage WOcauB3, WONo1, and WONo3 (WONo2 is too short and we didn't take it for further analysis), (B) Gene order comparisons among phage WOcauB2, WORiA, and WORiB1 (WORiB1 and WORiB2 are two identical copies and we just took WORiB1 for analysis). Gray lines connect matched ORFs with E < 1e−15. Colors of ORFs are as described in the legend of Figure 2.
Figure S2. Phylogenetic analysis of Wolbachia MLST genes. Maximum likelihood phylogenetic analyse demonstrates wNo and wCauB, wRi, and wCauB are divergent Wolbachia strains. The name of each sequence is the abbreviation of the Wolbachia strain (Table 1). Capital letters indicate Wolbachia strain supergroup affiliation from the literature. MLST: multi-locus sequence typing (with genes of coxA, fbpA, ftsZ, gatB, and hcpA).
Figure S3. The cumulative GC profile for prophage WOcauB2. (A) Gene presence in prophage WOcauB2. Colors of ORFs are as described in the legend of Figure 2. (B1) z′ curve for prophage WOcauB2. Segmentation points are marked with green squares. Segmentation point coincides with the HGT (B2gp46–B2gp47). (B2) The GC content distribution of prophage WOcauB2, using a 100 bp sliding window.
Figure S4. The cumulative GC profile for prophage WOVitA1. (A) Gene presence in prophage WOVitA1. Colors of ORFs are as described in the legend of Figure 2. (B1) z′ curve for prophage WOVitA1. Segmentation points are marked with green squares. Segmentation point includes the HGT (VA1gp52–VA1gp63). (B2) The GC content distribution of prophage WOVitA1, using a 100 bp sliding window.
Figure S5. The cumulative GC profile for prophage WOMelB. (A) Gene presence in prophage WOMelB. Colors of ORFs are as described in the legend of Figure 2. (B1) z′ curve for prophage WOMelB. Segmentation points are marked with green squares. Segmentation point includes the HGT (WD0611–WD0632). (B2) The GC content distribution prophage WOMelB, using a 100 bp sliding window.
References
Andersson, S. G. E., Zomorodipour, A., Andersson, J. O., Sicheritz-Pontén, T., Alsmark, U. C. M., Podowski, R. M., et al. (1998). The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396, 133–140. doi: 10.1038/24094
Azad, R. K., and Lawrence, J. G. (2012). Detecting laterally transferred genes. Methods Mol. Biol. 855, 281–308. doi: 10.1007/978-1-61779-582-4_10
Bordenstein, S. R., Marshall, M. L., Fry, A. J., Kim, U., and Wernegreen, J. J. (2006). The tripartite associations between bacteriophage, Wolbachia, and arthropods. PLoS Pathog. 2:e43. doi: 10.1371/journal.ppat.0020043
Bordenstein, S. R., and Reznikoff, W. S. (2005). Mobile DNA in obligate intracellular bacteria. Nat. Rev. Microbiol. 3, 688–699. doi: 10.1038/nrmicro1233
Bordenstein, S. R., and Wernegreen, J. J. (2004). Bacteriophage flux in endosymbionts (Wolbachia): infection frequency, lateral transfer, and recombination rates. Mol. Biol. Evol. 21, 1981–1991. doi: 10.1093/molbev/msh211
Bork, P. (1993). Hundreds of ankyrin-like repeats in functionally diverse proteins: mobile modules that cross phyla horizontally? Proteins 17, 363–374. doi: 10.1002/prot.340170405
Brelsfoard, C., Tsiamis, G., Falchetto, M., Gomulski, L. M., Telleria, E., Alam, U., et al. (2014). Presence of extensive Wolbachia symbiont insertions discovered in the genome of its host Glossina morsitans morsitans. PLoS Negl. Trop. Dis. 8:e2728. doi: 10.1371/journal.pntd.0002728
Broudy, T. B., Pancholi, V., and Fischetti, V. A. (2002). The in vitro interaction of Streptococcus pyogenes with human pharyngeal cells induces a phage-encoded extracellular DNase. Infect. Immun. 70, 2805–2811. doi: 10.1128/IAI.70.6.2805-2811.2002
Brüssow, H., Canchaya, C., and Hardt, W. D. (2004). Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68, 560–602. doi: 10.1128/MMBR.68.3.560-602.2004
Bushman, F. (2002). Lateral DNA Transfer: Mechanisms and Consequences. New York, NY: Cold Spring Harbor Laboratory Press.
Campana, M. G., García, N. M. R., and Tuross, N. (2015). America's red gold: multiple lineages of cultivated cochineal in Mexico. Ecol. Evol. 5, 607–617. doi: 10.1002/ece3.1398
Canchaya, C., Fournous, G., Chibani-Chennoufi, S., Dillmann, M. L., and Brüssow, H. (2003). Phage as agents of lateral gene transfer. Curr. Opin. Microbiol. 6, 417–424. doi: 10.1016/S1369-5274(03)00086-9
Castresana, J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552. doi: 10.1093/oxfordjournals.molbev.a026334
Caturegli, P., Asanovich, K. M., Walls, J. J., Bakken, J. S., Madigan, J. E., Popov, V. L., et al. (2000). ankA: an Ehrlichia phagocytophila group gene encoding a cytoplasmic protein antigen with ankyrin repeats. Infect. Immun. 68, 5277–5283. doi: 10.1128/IAI.68.9.5277-5283.2000
Chafee, M. E., Funk, D. J., Harrison, R. G., and Bordenstein, S. R. (2010). Lateral phage transfer in obligate intracellular bacteria (Wolbachia): verification from natural populations. Mol. Biol. Evol. 27, 501–505. doi: 10.1093/molbev/msp275
Darby, A. C., Armstrong, S. D., Bah, G. S., Kaur, G., Hughes, M. A., Kay, S. M., et al. (2012). Analysis of gene expression from the Wolbachia genome of a filarial nematode supports both metabolic and defensive roles within the symbiosis. Genome Res. 22, 2467–2477. doi: 10.1101/gr.138420.112
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2011). ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27, 1164–1165. doi: 10.1093/bioinformatics/btr088
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9, 772–772. doi: 10.1038/nmeth.2109
Desjardins, C. A., Cerqueira, G. C., Goldberg, J. M., Hotopp, J. C. D., Haas, B. J., Zucker, J., et al. (2013). Genomics of Loa loa, a Wolbachia-free filarial parasite of humans. Nat. Genet. 45, 495–502. doi: 10.1038/ng.2585
Dobson, S. L., Bordenstein, S. R., and Rose, R. I. (2016). Wolbachia mosquito control: regulated. Science 352, 526–527. doi: 10.1126/science.352.6285.526-b
Dong, X., El Karkouri, K., Robert, C., Gavory, F., Raoult, D., and Fournier, P. E. (2012). Genomic comparison of Rickettsia helvetica and other Rickettsia species. J. Bacteriol. 194, 2751–2751. doi: 10.1128/JB.00299-12
Duplouy, A., Iturbe-Ormaetxe, I., Beatson, S. A., Szubert, J. M., Brownlie, J. C., McMeniman, C. J., et al. (2013). Draft genome sequence of the male-killing Wolbachia strain wBol1 reveals recent horizontal gene transfers from diverse sources. BMC Genomics 14:20. doi: 10.1186/1471-2164-14-20
Eisen, J. A., Sweder, K. S., and Hanawalt, P. C. (1995). Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 23, 2715–2723. doi: 10.1093/nar/23.14.2715
Ellegaard, K. M., Klasson, L., Näslund, K., Bourtzis, K., and Andersson, S. G. (2013). Comparative genomics of Wolbachia and the bacterial species concept. PLoS Genet. 9:e1003381. doi: 10.1371/journal.pgen.1003381
Fineran, P. C., Petty, N. K., and Salmond, G. P. C. (2009). “Transduction: host DNA transfer by bacteriophages,” in Encyclopedia of Microbiology, 3rd Edn. ed M. Schaechter (Oxford: Academic Press), 666–679.
Foster, J., Ganatra, M., Kamal, I., Ware, J., Makarova, K., Ivanova, N., et al. (2005). The Wolbachia genome of Brugia malayi: endosymbiont evolution within a human pathogenic nematode. PLoS Biol. 3:e121. doi: 10.1371/journal.pbio.0030121
Fujii, Y., Kubo, T., Ishikawa, H., and Sasaki, T. (2004). Isolation and characterization of the bacteriophage WO from Wolbachia, an arthropod endosymbiont. Biochem. Biophys. Res. Commun. 317, 1183–1188. doi: 10.1016/j.bbrc.2004.03.164
Gao, F., and Zhang, C. T. (2006). GC-Profile: a web-based tool for visualizing and analyzing the variation of GC content in genomic sequences. Nucleic Acids Res. 34, W686–W691. doi: 10.1093/nar/gkl040
Gavotte, L., Henri, H., Stouthamer, R., Charif, D., Charlat, S., Boulétreau, M., et al. (2007). A survey of the bacteriophage WO in the endosymbiotic bacteria Wolbachia. Mol. Biol. Evol. 24, 427–435. doi: 10.1093/molbev/msl171
Gavotte, L., Vavre, F., Henri, H., Ravallec, M., Stouthamer, R., and Boulétreau, M. (2004). Diversity, distribution and specificity of WO phage infection in Wolbachia of four insect species. Insect Mol. Biol. 13, 147–153. doi: 10.1111/j.0962-1075.2004.00471.x
Guindon, S., Dufayard, J.-F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Hilgenboecker, K., Hammerstein, P., Schlattmann, P., Telschow, A., and Werren, J. H. (2008). How many species are infected with Wolbachia? – a statistical analysis of current data. FEMS Microbiol. Lett. 281, 215–220. doi: 10.1111/j.1574-6968.2008.01110.x
Ishmael, N., Dunning Hotopp, J. C., Ioannidis, P., Biber, S., Sakamoto, J., Siozios, S., et al. (2009). Extensive genomic diversity of closely related Wolbachia strains. Microbiology 155, 2211–2222. doi: 10.1099/mic.0.027581-0
Kent, B. N., Funkhouser, L. J., Setia, S., and Bordenstein, S. R. (2011a). Evolutionary genomics of a temperate bacteriophage in an obligate intracellular bacteria (Wolbachia). PLoS ONE 6:e24984. doi: 10.1371/journal.pone.0024984
Kent, B. N., Salichos, L., Gibbons, J. G., Rokas, A., Newton, I. L. G., Clark, M. E., et al. (2011b). Complete bacteriophage transfer in a bacterial endosymbiont (Wolbachia) determined by targeted genome capture. Genome Biol. Evol. 3, 209–218. doi: 10.1093/gbe/evr007
Klasson, L., Walker, T., Sebaihia, M., Sanders, M. J., Quail, M. A., Lord, A., et al. (2008). Genome evolution of Wolbachia strain wPip from the Culex pipiens group. Mol. Biol. Evol. 25, 1877–1887. doi: 10.1093/molbev/msn133
Klasson, L., Westberg, J., Sapountzis, P., Näslund, K., Lutnaes, Y., Darby, A. C., et al. (2009). The mosaic genome structure of the Wolbachia wRi strain infecting Drosophila simulans. Proc. Natl. Acad. Sci. U.S.A. 106, 5725–5730. doi: 10.1073/pnas.0810753106
Kurtti, T. J., Felsheim, R. F., Burkhardt, N. Y., Oliver, J. D., Heu, C. C., and Munderloh, U. G. (2015). Rickettsia buchneri sp. nov.: a rickettsial endosymbiont of the blacklegged tick ixodes scapularis. Int. J. Syst. Evol. Microbiol. 65, 965–970. doi: 10.1099/ijs.0.000047
LePage, D., and Bordenstein, S. (2013). Wolbachia: can we save lives with a great pandemic? Trends Parasitol. 29, 385–393. doi: 10.1016/j.pt.2013.06.003
Li, J., Mahajan, A., and Tsai, M.-D. (2006). Ankyrin repeat: a unique motif mediating protein-protein interactions. Biochemistry 45, 15168–15178. doi: 10.1021/bi062188q
Liu, H.-Y., Wang, Y.-K., Zhi, C.-C., Xiao, J.-H., and Huang, D.-W. (2014). A novel approach to eliminate Wolbachia infections in Nasonia vitripennis revealed different antibiotic resistance between two bacterial strains. FEMS Microbiol. Lett. 355, 163–169. doi: 10.1111/1574-6968.12471
Loreto, E. L., and Wallau, G. L. (2016). Risks of Wolbachia mosquito control. Science 351, 1273. doi: 10.1126/science.351.6279.1273-b
Masui, S., Kuroiwa, H., Sasaki, T., Inui, M., Kuroiwa, T., and Ishikawa, H. (2001). Bacteriophage, WO and virus-like particles in Wolbachia, an endosymbiont of arthropods. Biochem. Biophys. Res. Commun. 283, 1099–1104. doi: 10.1006/bbrc.2001.4906
Mavingui, P., Valiente Moro, C., Tran-Van, V., Wisniewski-Dyé, F., Raquin, V., Minard, G., et al. (2012). Whole-genome sequence of Wolbachia strain wAlbB, an endosymbiont of tiger mosquito vector Aedes albopictus. J. Bacteriol. 194, 1840. doi: 10.1128/JB.00036-12
Metcalf, J. A., and Bordenstein, S. R. (2012). The complexity of virus systems: the case of endosymbionts. Curr. Opin. Microbiol. 15, 1–7. doi: 10.1016/j.mib.2012.04.010
Metcalf, J. A., Jo, M., Bordenstein, S. R., Jaenike, J., and Bordenstein, S. R. (2014). Recent genome reduction of Wolbachia in Drosophila recens targets phage WO and narrows candidates for reproductive parasitism. PeerJ. 2:e529. doi: 10.7717/peerj.529
Modi, S. R., Lee, H. H., Spina, C. S., and Collins, J. J. (2013). Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature 499, 219–223. doi: 10.1038/nature12212
Moran, N. A., and Bennett, G. M. (2014). The tiniest tiny genomes. Annu. Rev. Microbiol. 68, 195–215. doi: 10.1146/annurev-micro-091213-112901
Moran, N. A., Degnan, P. H., Santos, S. R., Dunbar, H. E., and Ochman, H. (2005). The players in a mutualistic symbiosis: insects, bacteria, viruses, and virulence genes. Proc Natl. Acad. Sci. U.S.A. 102, 16919–16926. doi: 10.1073/pnas.0507029102
Moya, A., Peretó, J., Gil, R., and Latorre, A. (2008). Learning how to live together: genomic insights into prokaryote-animal symbioses. Nat. Rev. Genet. 9, 218–229. doi: 10.1038/nrg2319
Nei, M., and Gojobori, T. (1986). Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3, 418–426.
Nikoh, N., Hosokawa, T., Moriyama, M., Oshima, K., Hattori, M., and Fukatsu, T. (2014). Evolutionary origin of insect-Wolbachia nutritional mutualism. Proc. Natl. Acad. Sci. U.S.A. 111, 10257–10262. doi: 10.1073/pnas.1409284111
Nutman, T. B. (2001). Lymphatic filariasis: new insights and prospects for control. Curr. Opin. Infect. Dis. 14, 539–546. doi: 10.1097/00001432-200110000-00006
Ochman, H., Lawrence, J. G., and Groisman, E. A. (2000). Lateral gene transfer and the nature of bacterial innovation. Nature 405, 299–304. doi: 10.1038/35012500
Oliver, K. M., Degnan, P. H., Hunter, M. S., and Moran, N. A. (2009). Bacteriophages encode factors required for protection in a symbiotic mutualism. Science 325, 992–994. doi: 10.1126/science.1174463
O'Neill, S. L. (2016). Wolbachia mosquito control: tested. Science 352, 526. doi: 10.1126/science.352.6285.526-a
Ortiz, M. F., Wallau, G. L., Graichen, D. Â., and Loreto, E. L. (2015). An evaluation of the ecological relationship between Drosophila species and their parasitoid wasps as an opportunity for horizontal transposon transfer. Mol. Genet. Genomics 290, 67–78. doi: 10.1007/s00438-014-0900-y
Penz, T., Schmitz-Esser, S., Kelly, S. E., Cass, B. N., Müller, A., Woyke, T., et al. (2012). Comparative genomics suggests an independent origin of cytoplasmic incompatibility in Cardinium hertigii. PLoS Genet. 8:e1003012. doi: 10.1371/journal.pgen.1003012
Pinto, S. B., Stainton, K., Harris, S., Kambris, Z., Sutton, E. R., Bonsall, M. B., et al. (2013). Transcriptional regulation of Culex pipiens mosquitoes by Wolbachia influences cytoplasmic incompatibility. PLoS Pathog. 9:e1003647. doi: 10.1371/journal.ppat.1003647
Rodriguez-Valera, F., Martin-Cuadrado, A. B., Rodriguez-Brito, B., Pasic, L., Thingstad, T. F., Rohwer, F., et al. (2009). Opinion explaining microbial population genomics through phage predation. Nat. Rev. Microbiol. 7, 828–836. doi: 10.1038/nrmicro2235
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Saha, S., Hunter, W. B., Reese, J., Morgan, J. K., Marutani-Hert, M., Huang, H., et al. (2012). Survey of endosymbionts in the Diaphorina Citri metagenome and assembly of a Wolbachia wDi draft genome. PLoS ONE 7:e50067. doi: 10.1371/journal.pone.0050067
Salzberg, S. L., Dunning Hotopp, J. C., Delcher, A. L., Pop, M., Smith, D. R., Eisen, M. B., et al. (2005). Serendipitous discovery of Wolbachia genomes in multiple Drosophila species. Genome Biol. 6:R23. doi: 10.1186/gb-2005-6-3-r23
Salzberg, S. L., Puiu, D., Sommer, D. D., Nene, V., and Lee, N. H. (2009). Genome sequence of the Wolbachia endosymbiont of Culex quinquefasciatus JHB. J. Bacteriol. 191, 1725–1725. doi: 10.1128/JB.01731-08
Smith, M. C., and Thorpe, H. M. (2002). Diversity in the serine recombinases. Mol. Microbiol. 44, 299–307. doi: 10.1046/j.1365-2958.2002.02891.x
Sutton, E. R., Harris, S. R., Parkhill, J., and Sinkins, S. P. (2014). Comparative genome analysis of Wolbachia strain wAu. BMC Genomics 15:928. doi: 10.1186/1471-2164-15-928
Syvanen, M. (1994). Horizontal gene transfer: evidence and possible consequences. Annu. Rev. Genet. 28, 237–261. doi: 10.1146/annurev.ge.28.120194.001321
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tanaka, K., Furukawa, S., Nikoh, N., Sasaki, T., and Fukatsu, T. (2009). Complete WO phage sequences reveal their dynamic evolutionary trajectories and putative functional elements required for integration into the Wolbachia genome. Appl. Environ. Microbiol. 75, 5676–5686. doi: 10.1128/AEM.01172-09
Taylor, M. J., Cross, H. F., Ford, L., Makunde, W. H., Prasad, G. B., and Bilo, K. (2001). Wolbachia bacteria in filarial immunity and disease. Parasite Immunol. 23, 401–409. doi: 10.1046/j.1365-3024.2001.00400.x
Turley, A. P., Moreira, L. A., O'Neill, S. L., and McGraw, E. A. (2009). Wolbachia infection reduces blood-feeding success in the dengue fever mosquito, Aedes aegypti. PLoS Negl. Trop. Dis. 3:e516. doi: 10.1371/journal.pntd.0000516
Waldor, M. K., and Mekalanos, J. J. (1996). Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272, 1910–1914. doi: 10.1126/science.272.5270.1910
Walker, T., Johnson, P. H., Moreira, L. A., Iturbe-Ormaetxe, I., Frentiu, F. D., McMeniman, C. J., et al. (2011). The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature 476, 450–453. doi: 10.1038/nature10355
Waltz, E. (2016). US reviews plan to infect mosquitoes with bacteria to stop disease. Nature 533, 450–451. doi: 10.1038/533450a
Wang, G. H., Niu, L. M., Ma, G. C., Xiao, J. H., and Huang, D. W. (2014). Large proportion of genes in one cryptic WO prophage genome are actively and sex-specifically transcribed in a fig wasp species. BMC Genomics 15:893. doi: 10.1186/1471-2164-15-893
Wang, G. H., Xiao, J. H., Xiong, T. L., Li, Z., Murphy, R. W., and Huang, D. W. (2013). High-efficiency thermal asymmetric interlaced pcr (hitail-pcr) for determination of a highly degenerated prophage wo genome in a wolbachia strain infecting a fig wasp species. Appl. Environ. Microbiol. 79, 7476–7481. doi: 10.1128/AEM.02261-13
Werren, J. H. (1997). Biology of Wolbachia. Annu. Rev. Entomol. 42, 587–609. doi: 10.1146/annurev.ento.42.1.587
Werren, J. H., Baldo, L., and Clark, M. E. (2008). Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Mocrobiol. 6, 741–751. doi: 10.1038/nrmicro1969
Woolfit, M., Iturbe-Ormaetxe, I., Brownlie, J. C., Walker, T., Riegler, M., and Seleznev, A. (2013) Genomic evolution of the pathogenic Wolbachia strain, wMelPop. Genome Biol. Evol. 5, 2189–2204. doi: 10.1093/gbe/evt169
Wu, M., Sun, L. V., Vamathevan, J., Riegler, M., Deboy, R., Brownlie, J. C., et al. (2004). Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2:E69. doi: 10.1371/journal.pbio.0020069
Zabalou, S., Riegler, M., Theodorakopoulou, M., Stauffer, C., Savakis, C., and Bourtzis, K. (2004). Wolbachia-induced cytoplasmic incompatibility as a means for insect pest population control. Proc. Natl. Acad. Sci. U.S.A. 101, 15042–15045. doi: 10.1073/pnas.0403853101
Zhang, Z., Ye, G.-Y., Cai, J., and Hu, C. (2005). Comparative venom toxicity between Pteromalus puparum and Nasonia vitripennis (Hymenoptera: Pteromalidae) toward the hemocytes of their natural hosts, non-target insects and cultured insect cells. Toxicon 46, 337–349. doi: 10.1016/j.toxicon.2005.05.005
Zhou, W., Rousset, F., and O'Neil, S. (1998). Phylogeny and PCR-based classification of Wolbachia strains using wsp gene sequences. Proc. Biol. Sci. 265, 509–515. doi: 10.1098/rspb.1998.0324
Keywords: horizontal gene transfer, bacteriophage WO, Wolbachia, obligate intracellular bacteria, transduction
Citation: Wang GH, Sun BF, Xiong TL, Wang YK, Murfin KE, Xiao JH and Huang DW (2016) Bacteriophage WO Can Mediate Horizontal Gene Transfer in Endosymbiotic Wolbachia Genomes. Front. Microbiol. 7:1867. doi: 10.3389/fmicb.2016.01867
Received: 27 July 2016; Accepted: 07 November 2016;
Published: 29 November 2016.
Edited by:
Thomas Carl Bosch, University of Kiel, GermanyReviewed by:
Colleen A. Burge, University of Maryland, USAYuval Gottlieb, Hebrew University of Jerusalem, Israel
Copyright © 2016 Wang, Sun, Xiong, Wang, Murfin, Xiao and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guan H. Wang, wangguanhong88@163.com
Da W. Huang, huangdw@ioz.ac.cn
Jin H. Xiao, xiaojinhua2015@163.com
†Present Address: Guan H. Wang, Rowland Institute at Harvard University, Cambridge, MA, USA
 Guan H. Wang
Guan H. Wang Bao F. Sun3
Bao F. Sun3 Yan K. Wang
Yan K. Wang Da W. Huang
Da W. Huang