- 1Department of Pharmaceutical Sciences, School of Pharmaceutical Sciences of Ribeirao Preto, University of São Paulo, Ribeirão Preto, Brazil
- 2Department of Biophysics, Paulista Medical School, Federal University of São Paulo, São Paulo, Brazil
- 3Department of Physics and Chemistry, School of Pharmaceutical Sciences of Ribeirao Preto, University of São Paulo, Ribeirão Preto, Brazil
Enzymes have important features that may facilitate their application in industrial processes and have been used as alternatives to chemical catalysts. In particular, proteases can be isolated from microorganisms, which provide important sources of advantageous enzymes for industrial processes. For example, Eupenicillium javanicum is a filamentous fungus that has been shown to express industrially applicable enzymes and chemical components, such as antifungal compounds. The biotechnological potential of E. javanicum and proteases made us search a novel protease from this microorganism. The macromolecule was isolated, the main biochemical properties was evaluated, and the specificity of the protease subsites was determined. The protease was produced under solid-state bioprocess with wheat bran and isolated by two chromatography steps with yield of 27.5% and 12.4-fold purification. The molecular mass was estimated at 30 kDa. The N-terminal sequence of the first 20 amino acid residues was AVGAGYNASVALALEKALNN. The enzyme presented higher proteolytic activity at pH 6.0 and 60°C. The protease is stable at wide range of pH values and temperatures and in the presence of surfactants. The “primed” side of the catalytic site showed the highest catalytic efficiency of the enzyme isolated from E. javanicum. The S′1 subsite is responsible for catalyzing the protease reaction with substrates with tyrosine in P′1. These findings provide important insights into the biochemical characterization of a highly active protease from E. javanicum and may facilitate the development of industrial processes involving this protease.
Introduction
Enzymes present some advantages when compared with chemical catalysts. These macromolecules are able to catalyze a variety of chemical reactions, industrial processes have been shifting to the application of enzymes rather than chemicals as catalysts (Sarrouh et al., 2012). In addition, enzymes have several advantages over chemical catalysts, such as mild reaction conditions, the lack of requirement for protection of substrate functional groups, long half-lives, high stereo selectivity, and the ability to be genetically modified to improve stability, substrate specificity, and specific activity (Adrio and Demain, 2014). The key properties of enzymes, including stable activity over a range of temperatures and pH values and broad catalytic specificity, can be exploited in various industries and processes, such as the food, laundry, detergent, leather, pharmaceutical, and silk industries; for recovery of silver from x-ray film; and for waste management (Nigam, 2013; Hmidet et al., 2015).
Proteases are hydrolytic enzymes that have been used in industrial processes for decades. Microorganisms under bioprocesses can act as sources of various proteases to improve the protease production and activity owing to their rapid growth and potential for genetic modification, thereby allowing scientists to enhance the features of proteases for applications in industrial processes (Souza et al., 2015).
The solid-state bioprocess present potential applications in the production of enzymes and other industrial products. It has been defined as bioprocess in the absence or near-absence of free water, with enough moisture to support the growth and metabolic activity of the microorganism. Other characteristic is the use of low-cost agro-industrial residues, it makes the solid-state fermentation attractive financially and sustainably (Thomas et al., 2013).
Among these microbial sources, the filamentous fungus Eupenicillium javanicum has been shown to produce different products with industrial interests, such as antifungal compounds (Nakadate et al., 2007, 2008), endoglucanase, β-glucosidase, pectinase, xylanase (Tao et al., 2011), and a protease stable to spray dryer process (Hamin Neto et al., 2014).
Therefore, in this study, we aimed to isolate a protease produced in E. javanicum during solid-state fermentation, evaluate the main biochemical properties of this protease, and determine the specificity of its subsites.
Materials and Methods
Isolation, Identification, and Maintenance of E. javanicum
The fungus E. javanicum was isolated from silage and belongs to a collection of microorganisms at the Enzyme Technology Laboratory under the supervision of Dr. Hamilton Cabral (Faculdade de Ciências Farmacêuticas de Ribeirão Preto, Universidade de São Paulo). The fungus could be maintained in Sabouraud medium at 4°C for up to 1 month.
Inoculum Preparation
Eupenicillium javanicum was grown in 250-mL Erlenmeyer flasks with 30 mL Sabouraud culture medium. The inoculum was maintained for 7 days at 30°C, and the mycelial surface was then scraped in presence of 20 mL of saline solution composed by 0.1%[w/v] (NH4)2SO4 + 0.1% [w/v] NH4NO3 + 0.1% [w/v]MgSO4⋅7H2O.
Solid-State Bioprocess (SSB)
The protease were produced by E. javanicum under SSB in 250-mL erlenmeyer flasks containing 5 g wheat bran and 9.0 mL saline solution. The medium was sterilized by autoclaving at 121°C for 40 min. One milliliter of the inoculum was added before incubation at 30°C. After 140 h, the bioprocess was stopped, and 40 mL distilled water (4°C) was added to each flask for extracellular enzyme solubilization. This process was aided by maceration with a plastic rod, and the flasks were then agitated in a shaker at 200 rpm for 30 min at 4°C. The material was filtered and centrifuged at 5,000 × g for 20 min at 4°C. The supernatant was collected as the enzymatic extract (Hamin Neto et al., 2013).
Evaluation of Proteolytic Activity with Casein as Substrate
Proteolytic activity was evaluated using casein substrate according to the protocol described by Sarath et al. (1989), with some modifications. One milliliter of 1% (w/v) casein in 50 mM monobasic sodium phosphate buffer (pH 6.5) was combined with100 μL of 50 mM monobasic sodium phosphate buffer (pH 6.5) and 100 μL enzymatic extract. The reaction mixture was incubated for 60 min at 40°C, and 600 μL of 10% (w/v) trichloroacetic acid (TCA) was then added to stop the reaction. The reaction tubes were centrifuged at 10000 × g for 10 min at 30°C. The absorbance of the supernatants was then measured relative to the blank controls in cuvettes at 280 nm in a spectrophotometer (GENESYS 10S UV Vis; Thermo Fischer Scientific Inc.). One unit of activity was defined as the amount of the enzyme required to cause an increase of 0.001 of absorbance at 280 nm (Gupta et al., 2002).
Enzyme Purification by Chromatography
The enzymatic extract was subjected to gel filtration with a column (100 cm × 4 cm) using Sephadex G-50 resin. The equilibration buffer was 50 mM acetate (pH 5.0) with 50 mM NaCl, and the elution flow rate was 0.62 mL/min, regulated by a peristaltic pump (GE-Healthcare). The resin was equilibrated with five column volumes (CV), and 5 mL of sample was then applied. The gradient was isocratic, and 5-mL fractions were collected.
Enzyme fractions were subjected to dialysis with a 14-kDa membrane and 50 mM Tris-HCl buffer (pH 8.0) for 24 h at 4°C. The dialyzed samples (15 mL) were applied to Tricorn columns with 6 mL Resource-Q resin (anionic properties), pre-equilibrated with five CV of 50 mM Tris-HCl buffer (pH 8.0). After application, the resin was washed with the same buffer (two CV), and a linear salt gradient was then started from 0 to 500 mM NaCl using 20 CV of buffer at an elution flow rate of 2 mL/min. One-milliliter fractions were collected. The process was carried out using an ÄKTA Purifier chromatograph (GE-Healthcare) at 25°C.
Proteolytic activity assays and analysis by determination of absorbance at 280 nm were carried out to determine the enzymatic and protein profiles, respectively, and a NanoVue UV/Visible Spectrophotometer (GE-Healthcare) was used to quantify total protein at 280 nm, after each chromatographic process.
Evaluation of the Purity and Molecular Mass of the Protease
The enzyme fraction purity was evaluated by denaturing polyacrylamide gel electrophoresis (SDS-PAGE) using 12% gels (Laemmli, 1970) stained with silver nitrate (See and Jackowski, 1989). The size of the protease was estimated using Image Lab software version 3.0.
Determination of the N-Terminal Sequence of the Protease
The N-terminal sequence was determined by cleaving the N-terminal amino acids of proteins and peptides using Edman degradation on a Protein Sequencer PPSQ-33A instrument (Shimadzu Corporation). The PTH-amino acids were separated by high-performance liquid chromatography (HPLC), identified, and analyzed by comparing retention times and UV absorption with a previously quantified standard (Graminho et al., 2013).
Peptide Substrate Synthesis and Cleavage Site Determination
Fluorescence resonance energy transfer (FRET) peptides were synthesized in an automatic solid-phase peptide synthesizer (Model PSSM-8; Shimadzu Corporation; Hirata et al., 1995) and purified using semi preparative HPLC. Molecular mass determination was carried out using matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry with a Microflex LT mass spectrometer (Bruker-Daltonics). Peptide solutions were prepared by resuspending the substrate in dimethyl sulfoxide (DMSO), and their concentrations were determined with a molar extinction coefficient at 365 nm of 17,300 M-1 cm-1 in a spectrophotometer (Hirata et al., 1995). The scissile bonds of hydrolyzed peptides were identified by the isolation of fragments using analytical HPLC followed by determination of their molecular mass with an LCMS-2020 equipped with an electrospray ionization (ESI) probe (Shimadzu Corporation; Oliveira et al., 2015).
Biochemical Characterization
Functional biochemical characterization of the purified protease was conducted using FRET peptides. In these peptides, an ortho-aminobenzoic acid (Abz), responsible for the molecular fluorescence emission, was conjugated to the N-terminal amino group, and (2,4-dinitrophenyl)ethylenediamine (EDDnp), a quencher of fluorescence, was conjugated to the C-terminal carboxyl group (Chagas et al., 1991). A fluorescence signal was observed upon cleavage of any peptide bond within the amino acid sequence.
Test Conditions for the Enzymatic Reaction during Functional Biochemical Characterization Using FRET Substrates
The enzymatic reaction was carried out using a Lumina fluorescence spectrometer (Thermo Fischer Scientific Inc.) coupled with a Peltier system 4-Position Cell Holder Fluorescence device to control the agitation speed and assay temperature. Reactions with Abz-KLRSSKQ-EDDnp substrate were carried out in a quartz cuvette with an optical path length of 10 mm. The wavelengths were set to λex: 320 nm and λem: 420 nm. Data were collected and analyzed using Luminous software version 3.0.
Effects of pH and Temperature
The optimum pH was determined by evaluating different pH values from 4.0 to 10.5 with intervals of 0.5 pH units at 40°C. The buffers used in this analysis were acetate (pH 4.0–5.0), MES (pH 5.5–6.0), HEPES (pH 7.0–8.0), BICINE (pH 8.5–9.0), and CAPS (pH 9.5–10.5). After determining the optimum pH, the optimum temperature was determined from 30 to 75°C, with intervals of 5°C, at pH 6.0. The statistical analysis was performed with one-way ANOVA (analysis of variance) and post hoc Tukey, results with p < 0.05 were considered significant.
The stability of the enzyme at different pH values was evaluated by incubating the pure enzyme at 25°C for 60 min at various pH values (4.0–10.5), with intervals of 0.5 pH units, followed by reaction at pH 6.0 and 45°C. The buffers used in this analysis were acetate (pH 4.5–5.0), MES (pH 5.5–6.0), HEPES (pH 7.0–8.0), BICINE (pH 8.5–9.0), and CAPS (pH 9.5–10.0).
Thermal stability at different temperatures (30–60°C) and for different incubation times (5, 15, 30, and 60 min) was evaluated by proteases pre-incubation, followed by reaction at pH 6.0 and 45°C.
Effects of Ions and Inhibitors
The effects of ions on proteolytic activity were analyzed using NaCl, CoCl2, CuCl2, CaCl2, MgCl2, BaCl2, and AlCl3. The inhibitors tested in this analysis were phenylmethylsulfonyl fluoride (PMSF), ethylenediaminetetraacetic acid (EDTA), iodoacetic acid (IAA), and pepstatin (100 mM stock solutions) according to the protocol described by Dunn (1989). Pure enzyme solution and inhibitors or ions were added to each reaction tube at a final concentration of 10 mM for the inhibitors and ions. The effects of these inhibitors and ions were then analyzed by reaction for 5 min at 45°C, followed by further incubation at pH 6.0 at 45°C. The statistical analysis was performed with one-way ANOVA (analysis of variance) and post hoc Tukey, results with p < 0.05 were considered significant.
Effects of Surfactants
The effects of surfactants on proteolytic activity were analyzed using sodium dodecyl sulfate (SDS), cetyltrimethylammonium bromide (CTAB), TritonX-100, and Tween 20 solutions. Pure enzyme solution and surfactants were added to each reaction at final concentrations of 0.1, 0.2, 0.5, or 1.0% for all surfactants except SDS, which was analyzed at 0.02, 0.04, 0.06, 0.08, and 0.1%. The effects of these surfactants were then analyzed by reaction for 5 min at 45°C, followed by further incubation at pH 6.0 at 45°C.
Effects of Urea, Guanidine, and Dithiothreitol (DTT)
Pure enzyme solution and test agents were added to reaction tubes at final concentrations of 10, 25, 50, 100, or 150 mM for urea, guanidine, or DTT. The effects of these three components were then analyzed by reaction for 5 min at 45°C, followed by further incubation at pH 6.0 at 45°C.
Determination of the Molar Concentration of the Purified Enzyme
The molar concentration of the enzyme was determined by active site titration with the inhibitor phosphoramidon, as described by Klemencic et al. (2000) with modifications. Samples were analyzed in a Lumina fluorescence spectrometer (Thermo Fischer Scientific Inc.) using the substrate Abz-KLRSSKQ-EDDnp at pH 6.0 and 45°C. The wavelengths of fluorescence were adjusted to λex: 320 nm and λem: 420 nm.
Kinetic Experiments with Synthetic Substrate
Kinetic assays were performed to study the effects of changes in the positions of the amino acids in the substrate and to determine the substrate preference of the anchoring enzyme according to the distribution of amino acids in the peptide sequence. These variations were defined as P3, P2, P1, P′1, P′2, and P′3, represented as follows in enzymes: S3, S2, S1, S′1, S′2, and S′3 (Berger and Schechter, 1970). The kinetic parameters of substrate (Abz-KLXSSKQ-EDDnp) hydrolysis were evaluated with X variations in the indicated positions (P3, P2, P1, P′1, P′2, and P′3).
Enzymatic kinetic data were obtained by addition the substrate to the reaction cuvette with increasing concentrations. The experiments were performed at pH 6.0 and 45°C, and absorbance was measured using a Lumina fluorescence spectrometer (Thermo Scientific), with control of agitation and reaction temperature using a Fluorescence Peltier 4-Position Cell Holder (Thermo Fischer Scientific Inc.). The wavelengths were set to λex: 320 nm and λem: 420 nm.
The kinetic parameters were obtained from the Michaelis-Menten equation calculated by non-linear regression of data from hydrolysis of the substrate using Grafit version 5.0. We analyzed the KM, kcat, and kcat/KM to determine the preference of the enzyme for different substrates.
Results
Enzyme Purification by Chromatography
The fungus E. javanicum produced a protease that could be reproducibly purified using the steps described in Table 1. A final yield of 27.5% was obtained, with 12.4-fold purification. The elution profile from Sephadex G-50 resin chromatography showed two protein peaks, and peak II (fractions 70–90) exhibited proteolytic activity (Figure 1). These fractions were then subjected to anion exchange chromatography (Resource-Q resin), and the elution profile showed four protein peaks. Peaks II (fractions 61–63) and III (fractions 65–67) showed enzymatic activity (Figure 2). Peak III was used as the purified enzyme in subsequent steps as this peak showed higher specific activity, fold purification, and yield than peak II. Figure 3 shows the purity and estimated molecular weight of the enzyme (30 kDa).

TABLE 1. Summary of the purification steps for the Eupenicillium javanicum protease, produced by a solid-state bioprocess.
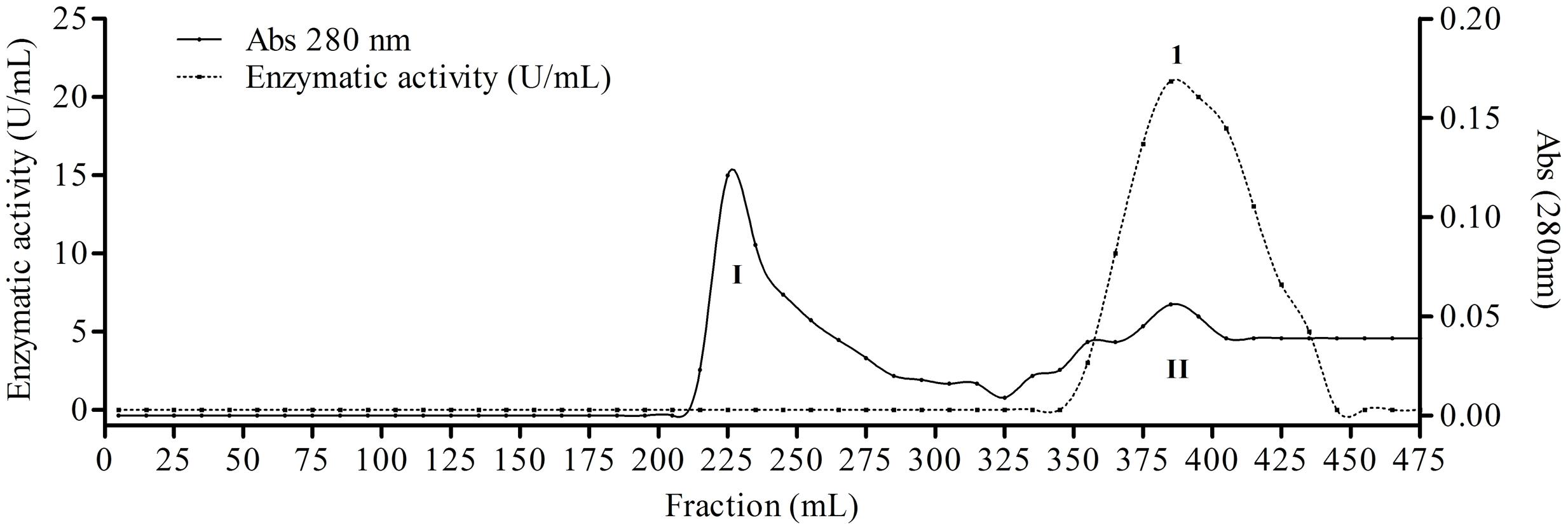
FIGURE 1. Elution profile of proteins and protease from Eupenicillium javanicum solid-state bioprocess in chromatography of mass exclusion with Sephadex g-50 resin. The elution buffer was acetate 50 mM, pH 5.0 and 50 mM NaCl. The flow was maintained in 0.62 mL/min and fractions of 5 mL.
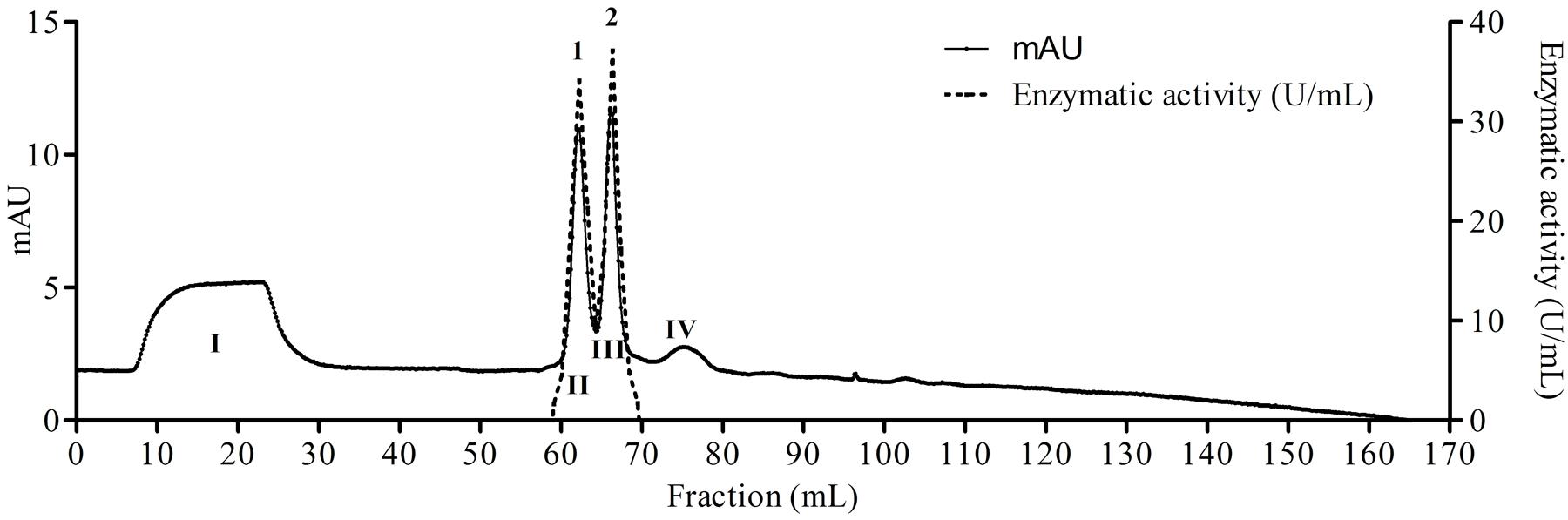
FIGURE 2. Elution profile of proteins and protease from E. javanicum solid-state bioprocess in chromatography of anion exchange with Resource-Q resin. The elution buffer was Tris-HCl 50 mM, pH 8.0 and gradient from 0 to 500 mM of NaCl. The flow was maintained in 2 mL/min and fractions of 1 mL.
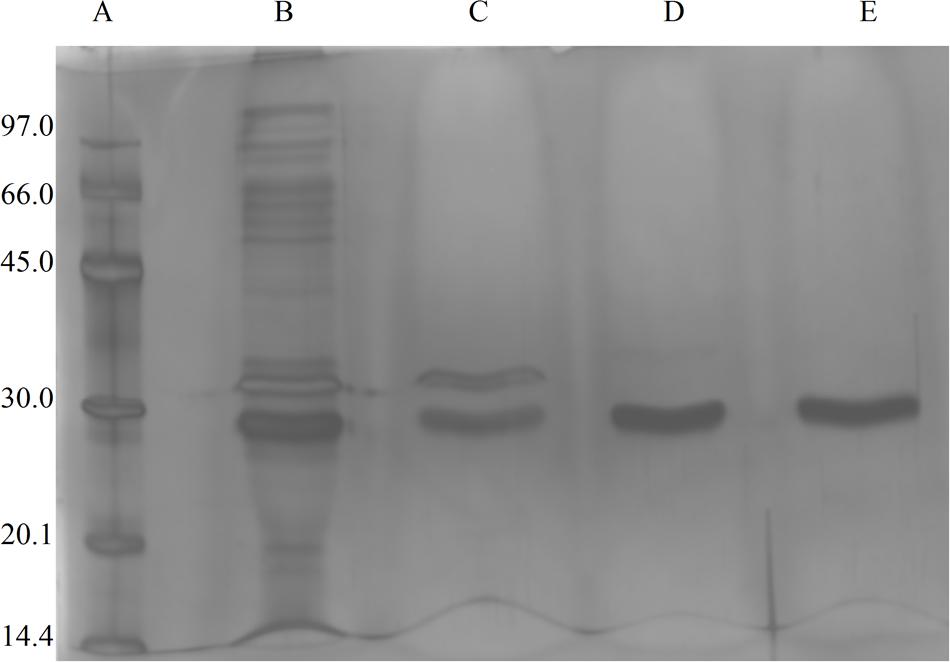
FIGURE 3. Image of SDS-PAGE gels (12%) stained with silver nitrate, the sample were from Eupenicillium javanicum solid-state bioprocess. (A) Molecular weight markers, (B) crude extract, (C) g-50 fraction, (D) Resource-Q peak I, and (E) Resource-Q peak II.
Determination of the N-Terminal Sequence
The N-terminal sequence of the first 20 amino acid residues of the protease from E. javanicum was AVGAGYNASVALALEKALNN. The N-terminal sequence was similar (query cover: 75% and identity: 73%) with other proteases secreted from the filamentous fungi Colletotrichum salicis (KXH28323.1), C. fioriniae PJ7 (XP 007596472.1), C. gloeosporioides Nara gc5 (XP 007281997.1), and C. Simmondsii (KXH42882.1), as shown in Figure 4.

FIGURE 4. N-terminal amino acid sequence of the protease isolated from E. javanicum solid-state bioprocess and comparisons with other proteases from Colletotrichum salicis, C. fioriniae PJ7, C. gloeosporioides Nara gc5, and C. simmondsii.
Biochemical Characterization
Effects of pH and Temperature on Enzyme Activity
For biochemical characterization of the enzyme, we first analyzed the optimum pH. Interestingly, the protease showed about 75% or more residual activity from pH 5.5–8.0, with 97% or more between pH 5.5 and 6.5, the optima pH was at 6.0, with The optimum temperature was 60°C, with statistical difference compared with all pH values except at 6.5, p < 0.05 (Figure 5). Proteolytic activity was lower at more extreme pH values. Enzymatic assays with incubation at different temperatures showed an increase from 30 to 60°C, followed by a drop in activity at temperatures over 60°C. The enzyme maintained and 80% or more activity from 50 to 70°C. The optimum temperature was 60°C, with statistical difference compared with all temperatures tested, p < 0.05 (Figure 6).
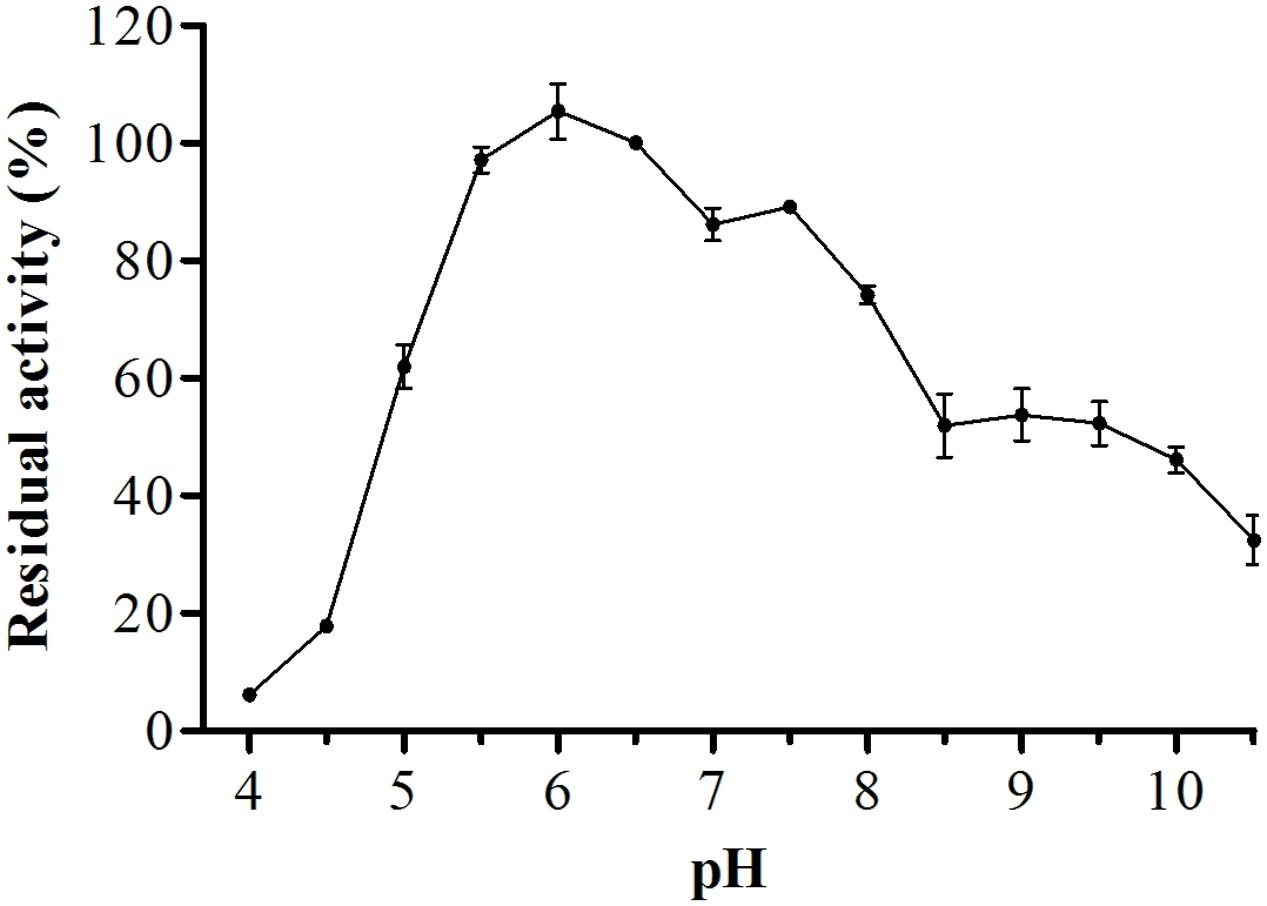
FIGURE 5. Optimum proteolytic activity at different pH values (4.0–10.5), the reaction was conducted at 40°C, using Abz-KLRSSKQ-EDDnp as substrate in fluorescence spectrometer.
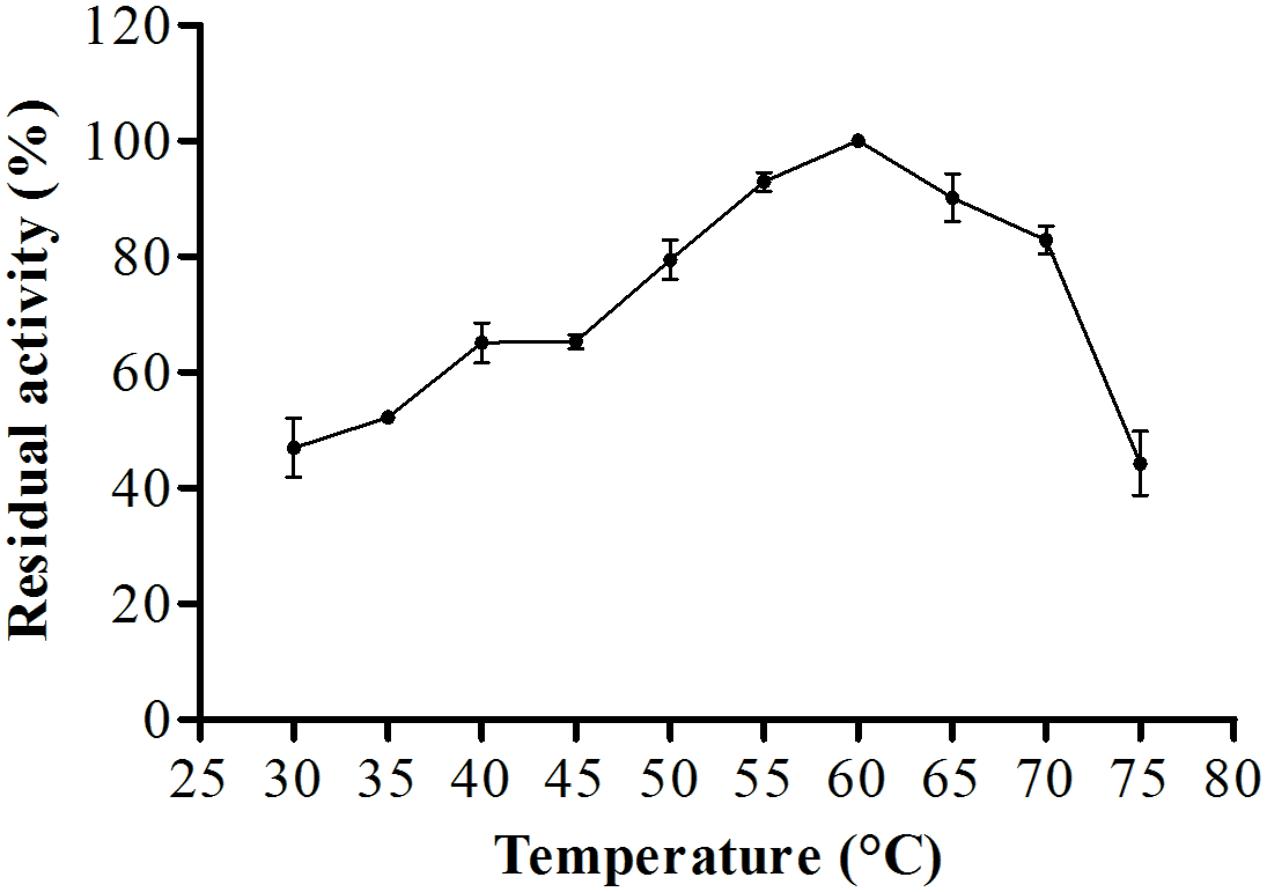
FIGURE 6. Optimum proteolytic activity at different temperatures (30–75°C) the reaction was conducted at pH 6.0, using Abz-KLRSSKQ-EDDnp as substrate in fluorescence spectrometer.
Analysis of protease stability at different pH values showed that the enzyme had greater stability from pH 4.5–10.0 with maintenance of about 67% or more of it stability after 60 min of incubation at 25°C (Figure 7). The protease showed great thermal stability during a 60-min incubation; from 30 to 55°C, the residual activity was about 80% or more, and at 60°C, the residual activity of the enzyme was 50% (Figure 8). Thus, these results suggested that the enzyme was thermostable despite being produced by a mesophilic fungus.
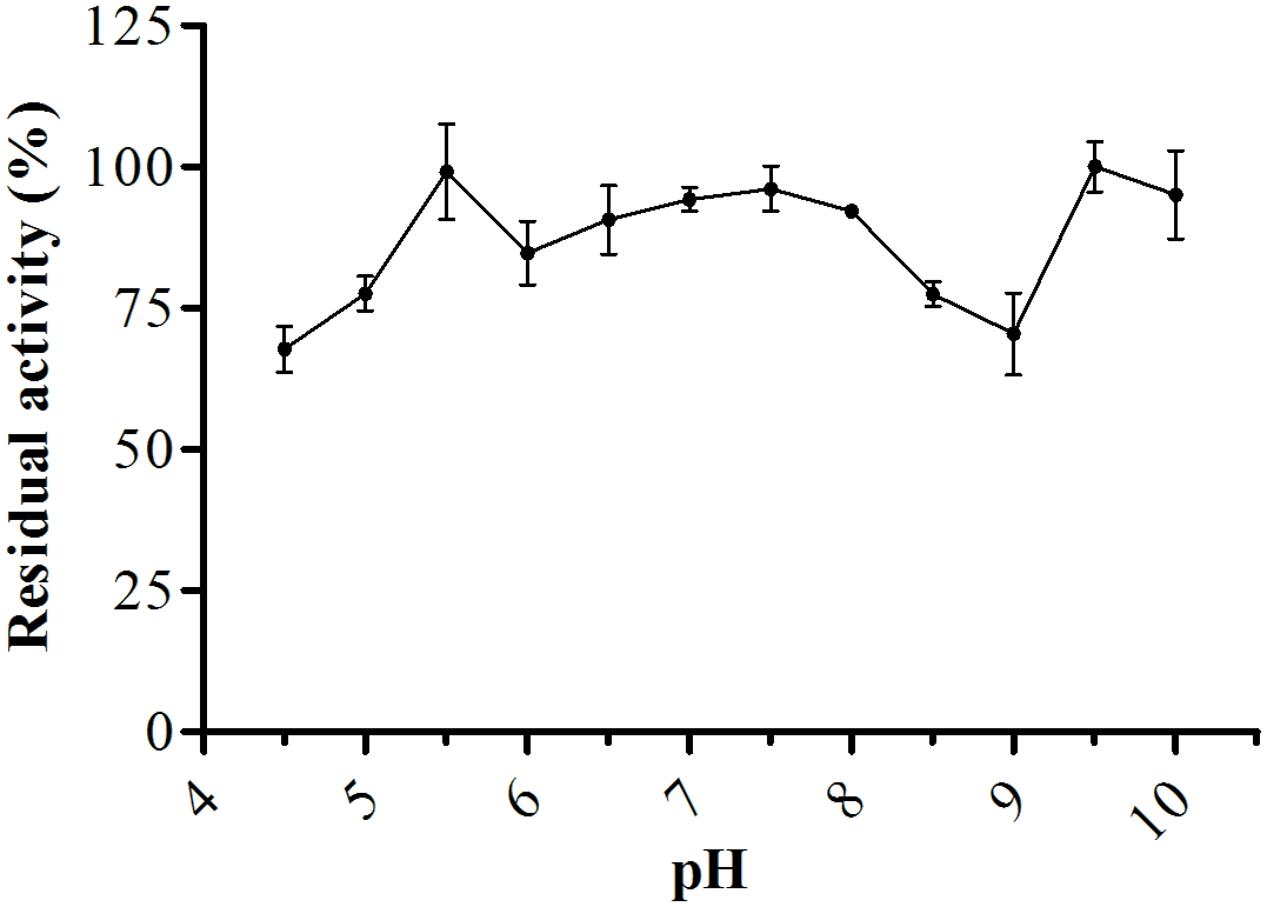
FIGURE 7. Protease stability after pre-incubation at different pH values (4.5–10.0) after 60 min at 25°C, the reaction was conducted at pH 6.0 and 45°C, using Abz-KLRSSKQ-EDDnp as substrate in fluorescence spectrometer.
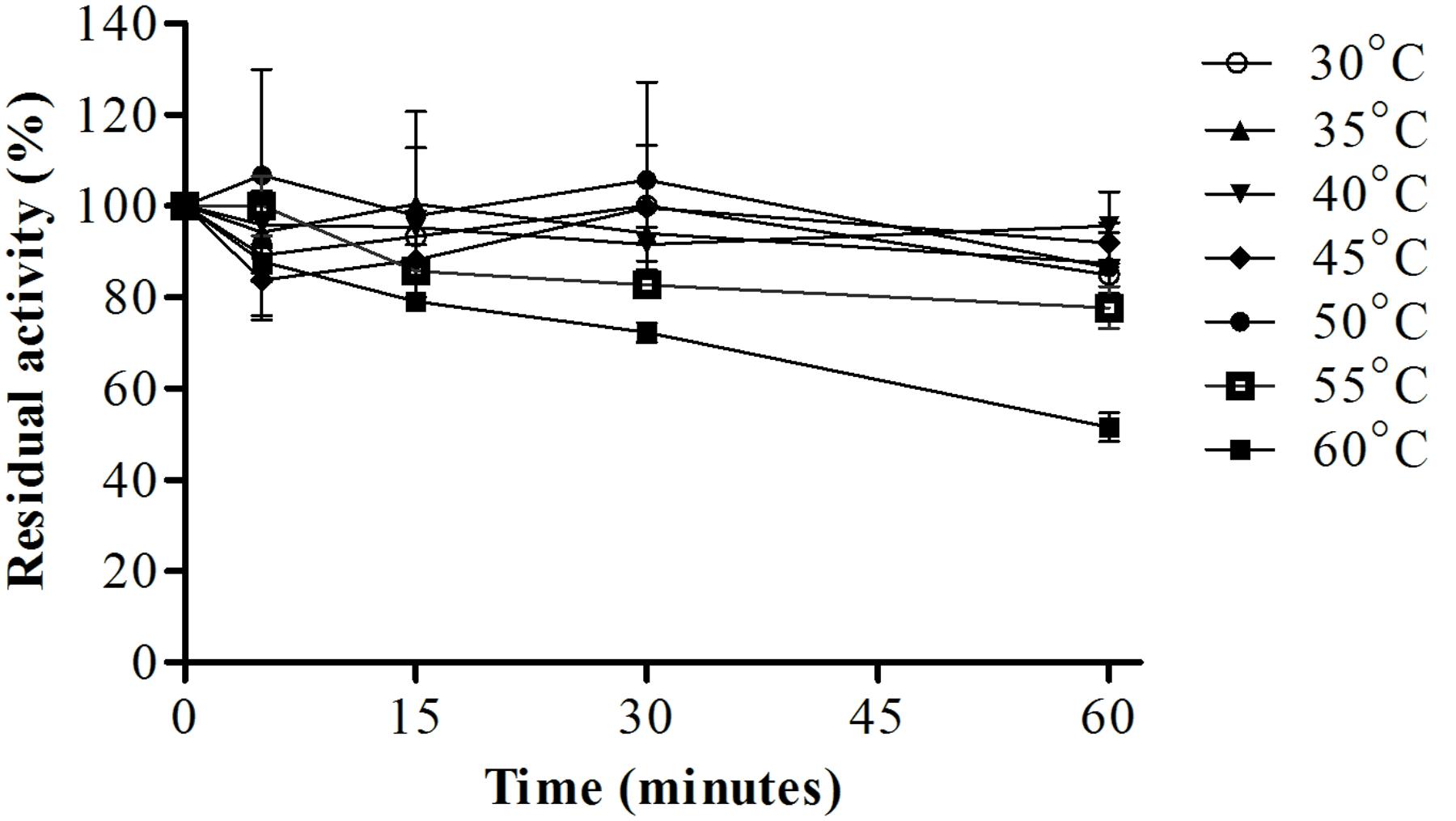
FIGURE 8. Protease stability after pre-incubation at different temperatures (30–60°C) for different times (5, 15, 30, and 60 min), the reaction was conducted at pH 6.0 and 45°C, using Abz-KLRSSKQ-EDDnp as substrate in fluorescence spectrometer.
Effects of Ions and Inhibitors
Metal ions can either positively and negatively affect protease activity. In this study, we found that 10 mM sodium, barium, or cobalt increased proteolytic activity by 52, 24, and 24%, only sodium showed statistical difference compared with control, p < 0.0001 (Table 2), respectively, while aluminum decreased the original proteolytic activity by 16%. Additionally, EDTA inhibited the proteolytic activity by about 69% with statistical difference, p < 0.0001, whereas PMSF, IAA, and pepstatin inhibited proteolytic activity by 30, 27, and 20%, respectively, (Table 3). These results suggested that the enzyme maybe a metalloprotease.
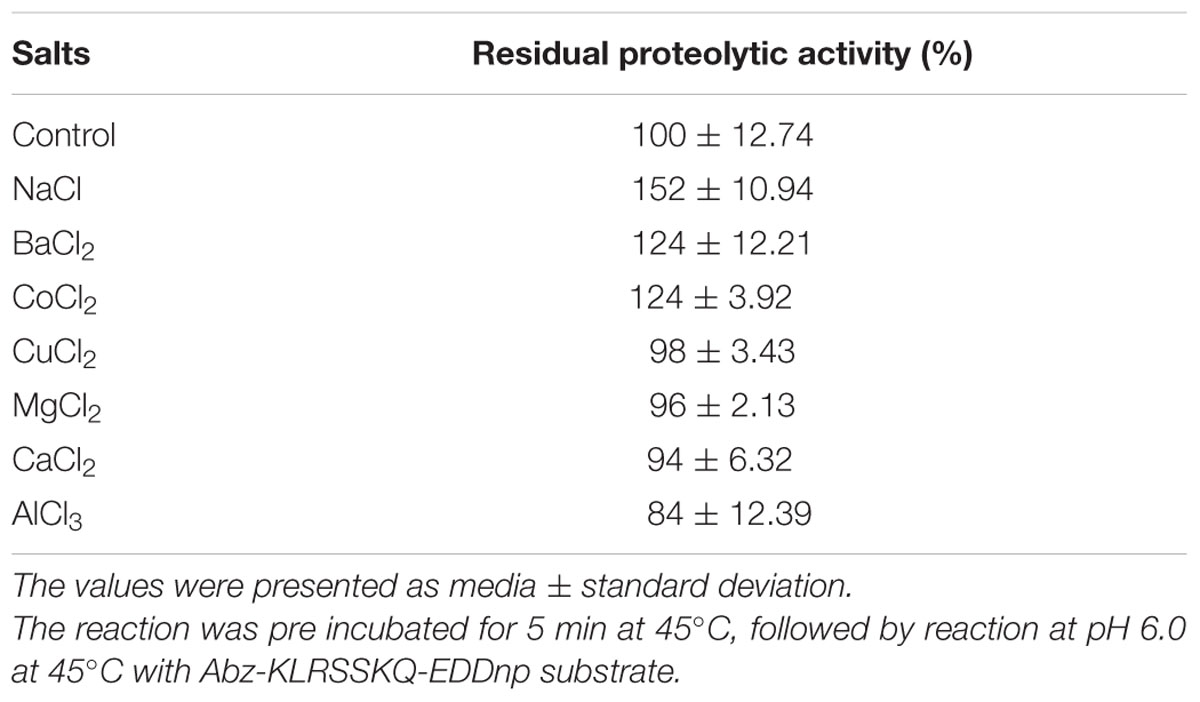
TABLE 2. Effects of 10 mM of different ions on the hydrolysis of Fluorescence resonance energy transfer (FRET) substrate by the purified E. javanicum protease.
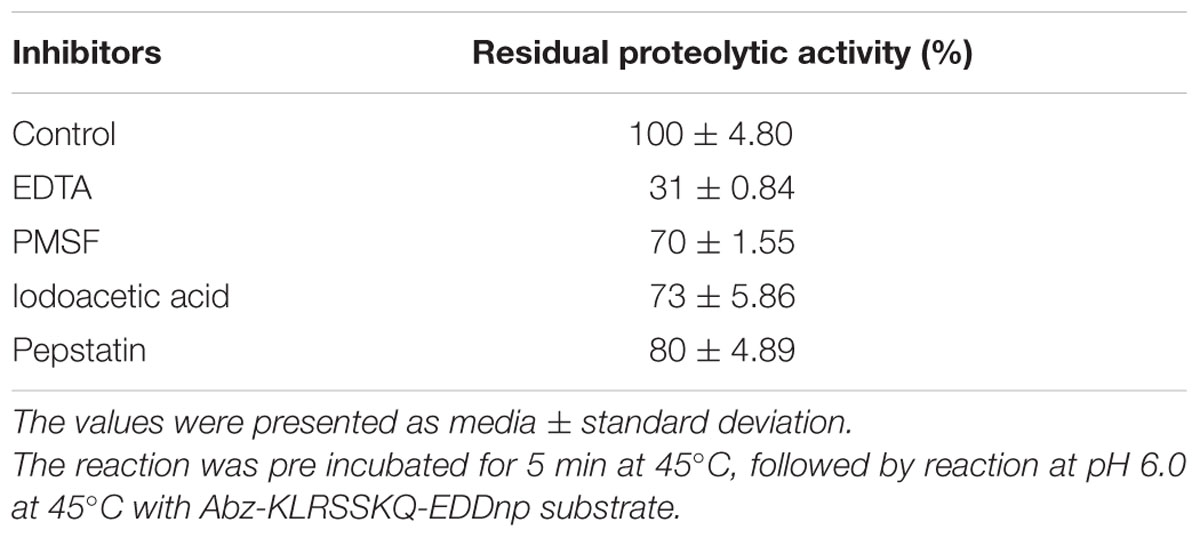
TABLE 3. Effects of 10 mM different inhibitors on the hydrolysis of FRET substrate by the purified E. javanicum protease.
Effects of Surfactants
Figure 9 shows the residual activity of the protease isolated from E. javanicum in the presence of surfactants. The enzyme was stable at different concentrations of Triton X-100 and Tween 20; at the maximum concentration tested (1%), the protease maintained about 68 and 58% of its activity, respectively. Increases in CTAB and SDS concentrations decreased protease activity; specifically, the protease retained about 28% activity in the presence of 0.1% CTAB and about 30% activity in the presence of 0.02% SDS.
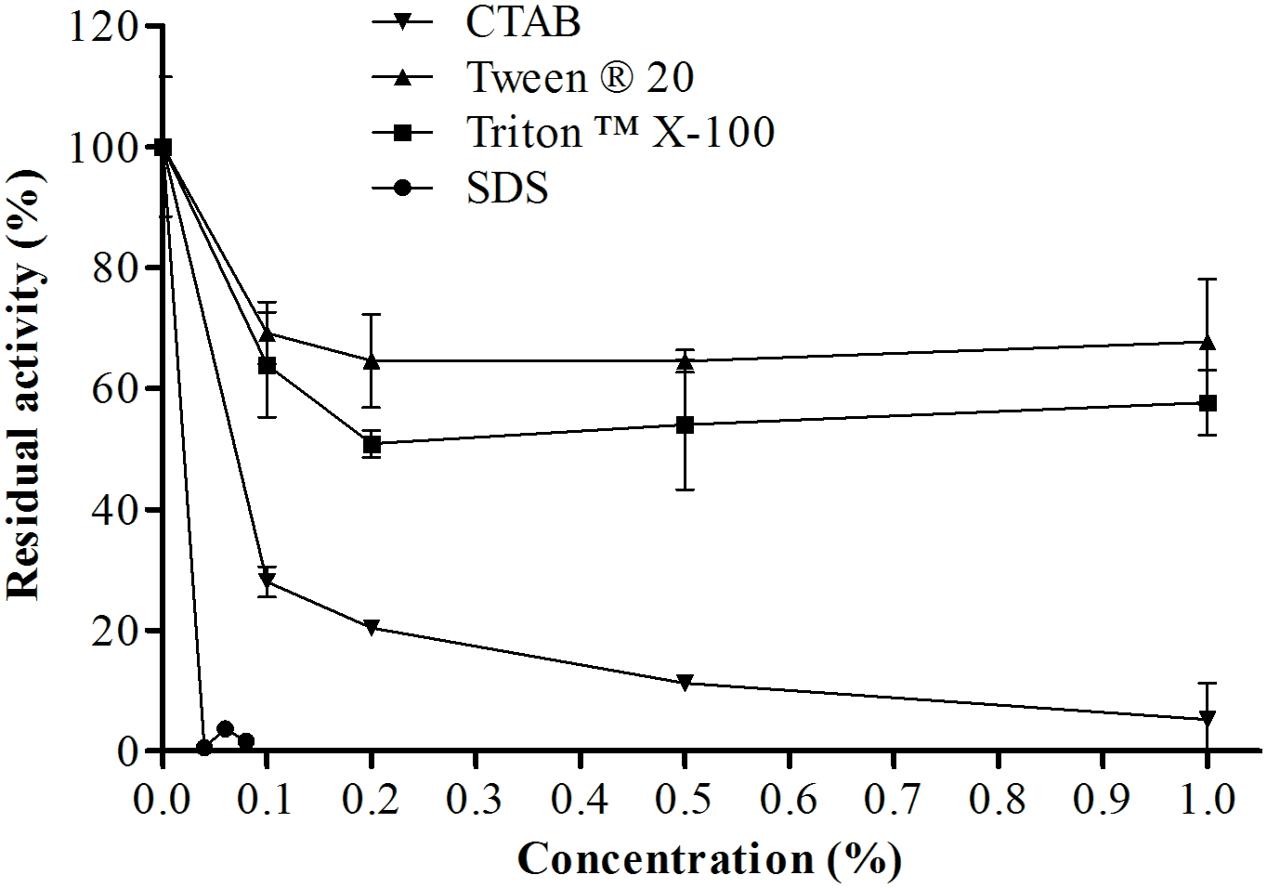
FIGURE 9. Effects of different concentrations of a variety of surfactants (CTAB, SDS, Triton X-100, and Tween 20) on the proteolytic activity of E. javanicum protease, the reaction was conducted at pH 6.0 and 45°C, using Abz-KLRSSKQ-EDDnp as substrate in fluorescence spectrometer.
Effects of Urea, Guanidine, and DTT
As shown in Figure 10, urea, guanidine, and DTT also affected proteolytic activity. Increased concentrations of DTT resulted in decreased enzymatic activity, maintaining about 40% of enzyme activity when the concentration of DTT was 100 mM. In the presence of 150 mM urea, the enzyme maintained about 54% of its original activity. Interestingly, guanidine had a greater effect, reducing the activity of the enzyme by only 10% when used at a concentration of 150 mM.
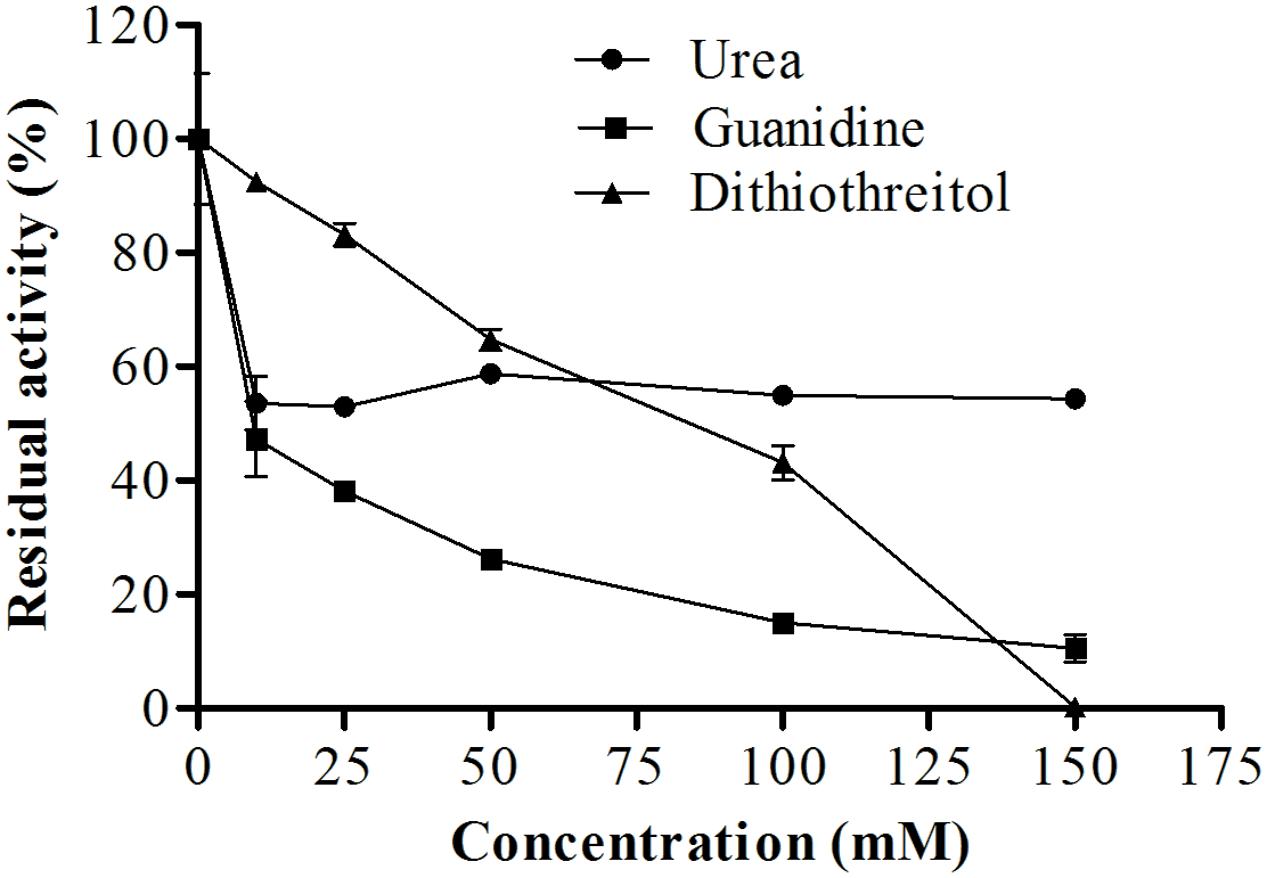
FIGURE 10. Effects of different concentrations of dithiothreitol, guanidine, and urea on the proteolytic activity of E. javanicum protease, the reaction was conducted at pH 6.0 and 45°C, using Abz-KLRSSKQ-EDDnp as substrate in fluorescence spectrometer.
Kinetic Experiments with a Synthetic Substrate
Table 4 shows the catalytic specificity of the protease for S1, S2, and S3 subsites in relation to replacement of amino acids at P1, P2, and P3 positions using a FRET peptide series based on the sequence Abz-KLRSSKQ-EDDnp (considering the cleavage between Arg and Ser). The position P1 provided higher catalytic efficiency with arginine, phenylalanine, and methionine (2,146, 2,043, and 1,843 mM-1⋅seg-1, respectively). Moreover, the protease exhibited higher affinity when glutamine was in this amino acid position, with a KM of 0.004 mM. Most of the substrates were cleaved between P′1 and P′2; in the presence of arginine and glutamine, the substrate was cleaved as follows: P1↓P′1↓P′2. The best values for the S2 subsite were with substrates containing valine and arginine at the P2 substrate position; the catalytic efficiencies were 2,141 and 1,850 mM-1⋅seg-1. The enzyme showed high affinity for phenylalanine, with a KM of 0.012 mM. The substrates were cleaved between P′1 and P′2, except for phenylalanine and tyrosine, which were cleaved as follows: P1↓P′1↓P′2. The kinetic parameters of the protease at S3 showed that the presence of hydrophobic amino acids, such as valine, isoleucine, and alanine, led to the highest catalytic efficiencies (4,539, 3,886, and 2,836 mM-1⋅seg-1, respectively). The enzyme showed high affinity for valine, with a KM of 0.010 mM. All substrates were cleaved at two positions, P1↓P′1↓P′2, except in the presence of serine, where the cleavage pattern was P′1↓P′2; that of arginine was not determined.
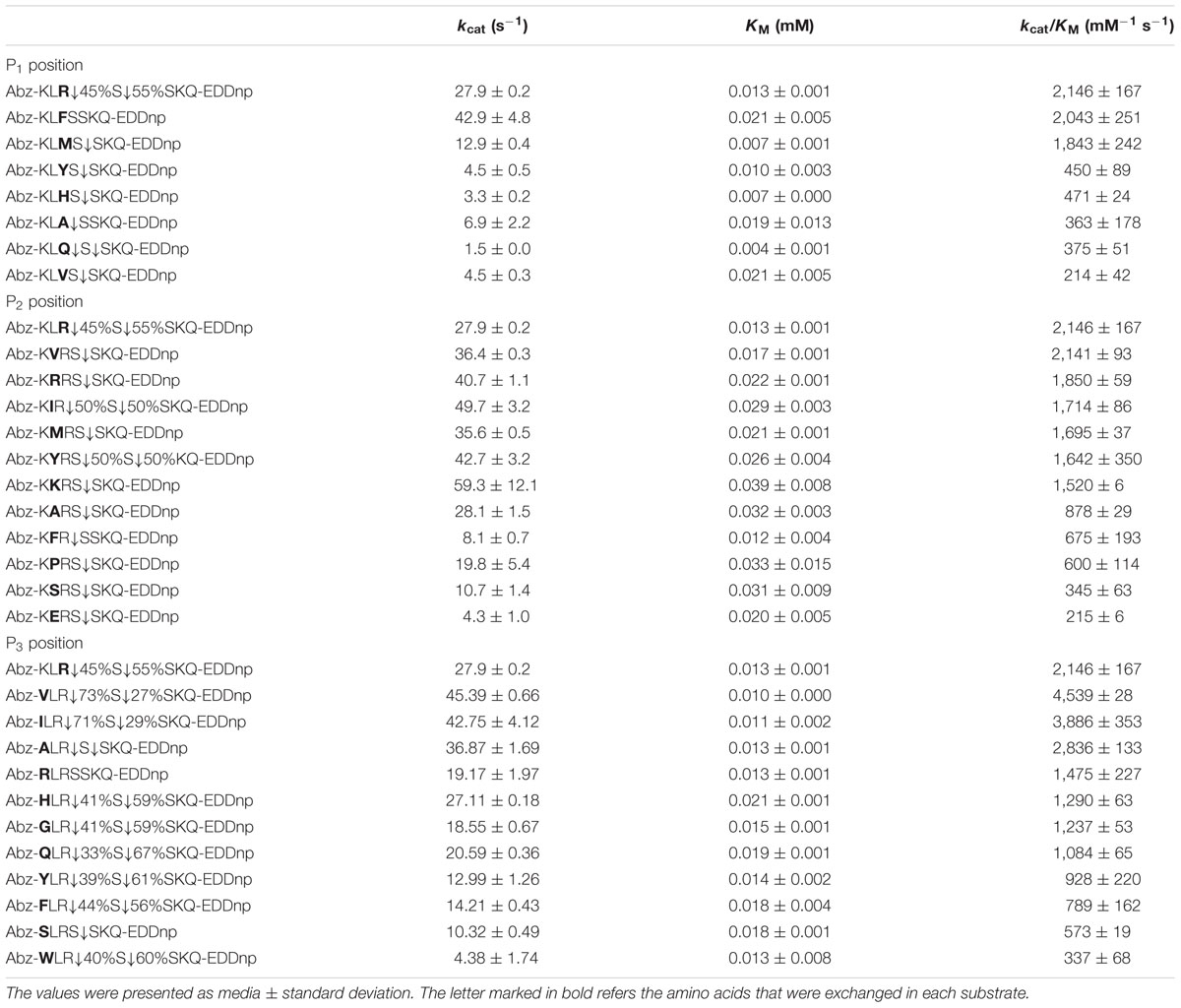
TABLE 4. Kinetic parameters for the hydrolysis by the E. javanicum protease of the peptide series derived from reference peptide Abz-KLRSSKQ-EDDnp modified at P1, P2, and P3 positions.
Table 5 shows values of the protease kinetic parameters at S′1, S′2, and S′3 subsites related to substrate substitution at P′1, P′2, and P′3 positions. Analysis of the S′1 data showed that the substrate containing tyrosine was the most hydrolyzed, with a catalytic efficiency of 87,849 mM-1⋅seg-1, followed by arginine (catalytic efficiency: 18,914 mM-1⋅seg-1) and lysine (14,876 mM-1⋅seg-1). All substrates evaluated could be hydrolyzed by protease, particularly tyrosine, which resulted in the highest enzyme affinity (KM: 0.007 mM). The most frequently cleaved site was P′1↓P′2, except in the presence of valine (P2↓P1), lysine (P′1↓P′2↓P′3), and Arg or Met (P2↓P1P′1↓P′2). The protease kinetic parameters in isoleucine presence at S′2 subsite provided high catalytic efficiency with 13,700 mM-1⋅seg-1. The affinity of the enzyme was higher in the presence of isoleucine and tyrosine, KM 0.003. Additionally, the most frequently cleaved sites were P2↓P1↓P′1 for proline and tyrosine; P1↓P′1↓P′2 for alanine, arginine, glutamine, glutamic acid, and histidine; P2↓P1↓P′1↓P′2 for glycine; and P2↓P1↓P′1↓P′2↓P′3 for histidine. The S′3 subsite had higher catalytic efficiency in the presence of tyrosine and phenylalanine (3,833 and 2,085 mM-1⋅seg-1, respectively). The cleavage was different for all substrates.
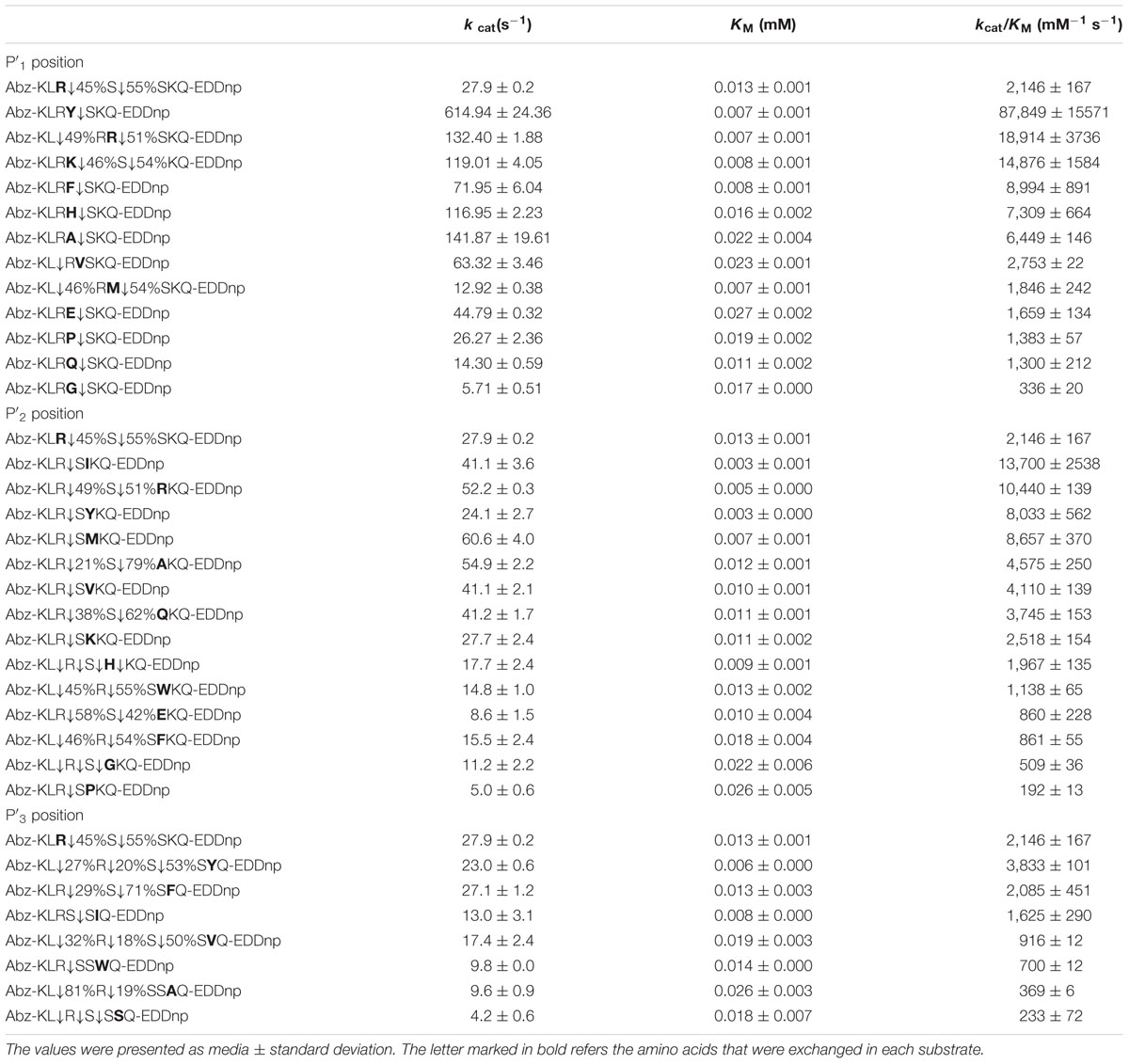
TABLE 5. Kinetic parameters for the hydrolysis by the E. javanicum protease of the peptide series derived from reference peptide Abz-KLRSSKQ-EDDnp modified at P′1, P′2, and P′3 positions.
Discussion
In this study, we evaluated the optimum pH and temperature, stability, and kinetic parameters of a protease isolated from solid-state fermentation of E. javanicum. According to the literature, the Penicillium genus has the potential to produce different proteases; for example, Penicillium waksmanii (Graminho et al., 2013), P. digitatum (Aissaoui et al., 2014), and P. italicum (Abidi et al., 2014) produce serine proteases under submerged bioprocesses, whereas P. waksmanii and P. corylophilum (Da Silva et al., 2013) also produce serine proteases under solid-state fermentation. Moreover, similar to E. javanicum, Penicillium ssp. produces metalloproteases during solid-state fermentation (El-Gendy, 2010).
Enzyme Purification by Chromatography
Purification processes can vary depending on the specific enzyme being isolated. For example, the different chromatography steps can be modified according to the stability of the enzyme in response to process conditions, influencing the recovery and purification fold, that influence directly in cost of industrial use. Previous studies have described appropriate purification parameters for enzymes from P. italicum (Abidi et al., 2014), Beauveria sp. (Shankar et al., 2011), and Trichoderma harzianum (Savitha et al., 2011). Additionally, the protease purified from submerged medium of Aspergillus oryzae KSK-3 had 182.7-fold purification, but a recovery of only 0.005% (Shirasaka et al., 2012). A metalloprotease isolated from Penicillium spp. showed lower recovery and purification than that from E. javanicum (4.93 and 3.56%, respectively; El-Gendy, 2010). Therefore the purification of the metalloprotease from E. javanicum presented results better than some protease purifications.
Previous studies have shown that other metalloproteases exhibit variable molecular weights. For example, Bersanetti et al. (2005) isolated a 51.5-kDa metalloprotease from Serratia proteamaculans culture medium, and a 19-kDa metalloprotease was produced by Penicillium spp. under solid-state fermentation (El-Gendy, 2010). Additionally, Termitomyces clypeatus (Majumder et al., 2015) and Bacillus sp TKU004 (Wang et al., 2006) produce metalloproteases with molecular masses similar to those of the enzyme secreted by E. javanicum in our study (30 kDa).
Biochemical Characterization
Effects of pH and Temperature on Enzyme Activity
Proteases have several properties that may facilitate their use in industrial applications, including optimal temperature, optimal pH, specificity, activity, and stability (Mahajan and Badgujar, 2010). Similar to E. javanicum, many microorganisms are able to produce neutral proteases. For example, P. digitatum (Aissaoui et al., 2014), P. italicum (Abidi et al., 2014), Graphium putredinis, and Trichoderma harzianum (Savitha et al., 2011) produce serine proteases with an optimum pH at 7.0. Additionally, the metalloproteases from Microbacterium sp. (Thys and Brandelli, 2006), Penicillium spp. (El-Gendy, 2010), Thermoascus aurantiacus (Merheb-Dini et al., 2009), and Streptomyces septatus TH-2 (Hatanaka et al., 2005) produce enzymes with a neutral pH optimum.
Additionally, microorganisms are able to secrete enzymes that can function in wide temperature range. As previously reported, P. italicum (Abidi et al., 2014), P. chrysogenum (Chen et al., 2013), Botrytis cinerea (Abidi et al., 2011), Graphium putredinis, and Trichoderma harzianum (Savitha et al., 2011) produce proteases with an optimum temperature of 50°C, whereas the serine protease from P. waksmanii (Graminho et al., 2013) exhibits maximum activity at 35°C. A metalloprotease isolated from Thermoascus aurantiacus was found to have high proteolytic activity at 75°C (Merheb-Dini et al., 2009), and that from Termitomyces clypeatus was found to have high proteolytic activity at 45°C (Majumder et al., 2015).
The stability of enzymes at different pH values and temperatures also affects the applicability of the enzyme in industrial processes. Enzymes that are stable at a wide range of temperatures and pH values may have broad industrial applications. For example, proteases produced by P. italicum (Abidi et al., 2014) and Beauveria sp. (Shankar et al., 2011) exhibit high activity from pH 4.0–11.0 and 3.0–11.0, respectively. In contrast, the protease produced by Botrytis cinerea is stable from pH 6.0–9.0 (Abidi et al., 2011). The metalloproteases from Penicillium spp. show stability in a lower pH range (6.0–8.0 and 6.0–11.0; El-Gendy, 2010). In our study, we found that the enzyme from E. javanicum was stable at a wide pH range. Moreover, the thermal stability of our enzyme was similar to that of proteases from P. italicum, which maintained about 80% activity at 40 and 50°C and 60% activity at 60°C after 60 min (Abidi et al., 2014). In contrast, the proteases from Boritrys cinerea maintained only 10% activity at 60°C after 60 min (Abidi et al., 2011), whereas the proteases from Graphium putredinis and Trichoderma harzianum were stable for only 15 min at 60°C (Savitha et al., 2011). A metalloprotease produced by Microbacterium sp showed similar behavior to that isolated from E. javanicum, maintaining more than 80% activity at 55°C and about 70% activity at 60°C after 60 min (Thys and Brandelli, 2006). Thus, the enzyme produced by E. javanicum was more stable than some proteases from thermophilic fungi. This resistance to high temperatures is interesting for industrial applications of the enzyme.
Some food processes use high temperatures to avoid the contamination with microorganisms. Proteases that act in high temperatures can be applied in meat tenderization, clean up of DNA before PCR (Bruins et al., 2001) and textile industry to remove gum (Srilakshmi et al., 2014).
Effects of Ions and Inhibitors
Some proteases, particularly metalloproteases, may be influenced by the presence of metal ions. Therefore, researchers have evaluated the effects of ions on proteolytic activity. For example, the metalloprotease produced under solid-state fermentation by Bacillus sp. exhibits improved activity in the presence of sodium (Saxena and Singh, 2011). Additionally, proteases from P. waksmanii (Graminho et al., 2013) and Botrytis cinerea (Abidi et al., 2011) show increased activity in the presence of barium. We observed the same tendencies in the metalloprotease isolated in this study. Similarly, a protease from Beauveria sp. exhibits increased activity after pre-incubation with cobalt (Shankar et al., 2011). In contrast, the presence of aluminum decreases the activity of proteases from P. waksmanii (Graminho et al., 2013) and Aspergillus oryzae KSK-3 (Shirasaka et al., 2012). Finally, inhibitors, such as EDTA, can be used to classify the protease according to it catalytic site. EDTA is a metal chelating agent, and proteolytic activity of a metalloprotease is linked to presence of divalent metal ions, which can be directly related to the catalytic site or structure. In our study, EDTA inhibited the activity of our enzyme, whereas barium enhanced the activity of our enzyme, suggesting that the protease isolated from E. javanicum in this study was a metalloprotease.
Effect of Surfactants
In order to utilize proteases in the detergent industry, the enzymes must be highly active and stable at a high pH and temperature and must show compatibility with other chelating and oxidant agents added to the detergent (Rani et al., 2013). Detergents and surfactants may disrupt hydrophobic interactions within the protein structure and negatively or positively affect proteolytic activity. Enzymes produced by P. digitatum maintain about 64% activity in the presence of Tween (Aissaoui et al., 2014). In contrast, enzymes produced by Myceliophthora sp. show increased activity in the presence of Tween (117% of residual activity; Zanphorlin et al., 2011). In addition, the protease produced by P. waksmanii exhibits only 43% activity in the presence of this detergent (Graminho et al., 2013). The protease from P. digitatum exhibits about 70% residual activity in the presence of Triton (Aissaoui et al., 2014), whereas that from Thermoascus aurantiacus has no stability in the presence of the same surfactant (Merheb-Dini et al., 2009). Proteases produced by P. waksmanii have been shown to retain about 43% activity in the presence of 0.8% Triton (Graminho et al., 2013). Some proteases produced by P. waksmanii (Graminho et al., 2013) and Myceliophthora sp (Zanphorlin et al., 2011) have no activity when SDS is added to the solution, whereas the protease produced by Botrytis cinerea (Abidi et al., 2011) shows increased stability in the presence of 1% SDS.
Effects of Urea, Guanidine, and DTT
Reducing agents such as DTT may break disulfide bonds from sulfhydryl groups. The tertiary protein structure depends on these connections and hydrophobic interactions to stabilize the protein (Vieille and Zeikus, 2001). Indeed, disulfide bonds are important for the structure and proteolytic activity of the metalloprotease produced by E. javanicum. Additionally, the enzyme from Thermoascus aurantiacus also showed a decrease in activity as the concentration of DTT increased (Merheb-Dini et al., 2009). Additionally, the enzyme produced by P. waksmanii maintained only 15% activity in the presence of 20 mM DTT (Graminho et al., 2013); thus, this enzyme was less stable than that produced by E. javanicum. Hydrogen bonds also help to maintain the high order structure of proteins. Chaotropic agents, such as urea and guanidine, can disrupt these connections and induce changes in protein conformation (Adrio and Demain, 2014), thereby affecting proteolytic activity, as was observed for the metalloprotease secreted by E. javanicum.
Kinetic Experiments with a Synthetic Substrate
In this study, we performed kinetic analysis to determine the specificity of the protease to certain substrates, which can be targets in industrial applications. Replacement of some amino acids at the P1 and P2 positions of the Abz-KLRSSKQ-EDDnp substrate blocked substrate cleavage, suggesting that the enzyme required specific amino acids at this position. Analysis of the amino acid residues at P3 position and the catalytic efficiency of the enzyme showed that there was a preference for non-polar amino acids. In contrast, all the amino acid residues allowed substrate cleavage at the P3 position; thus, there was lower enzymatic specificity at P3 compared with those at P1 and P2.
Replacement of amino acids at P′1 showed that polar amino acids positively influenced catalytic efficiency, with tyrosine having the greatest effects. At P′2, the subsite alternated between polar and non-polar substrates; therefore, this position was non-specific for the enzyme. Additionally, analysis of the kinetics at the P′3 position showed that there was a preference for amino acids with an aromatic ring. Some amino acids also blocked cleavage completely, demonstrating that this position required specific amino acids. For positions at the “primed” side, the enzyme showed greater catalytic efficiency than when there were changes at the “unprimed” side positions. There were no similarities among the P1, P2, and P3 positions. Additionally, analysis of the specificity of proteases using FRET substrates showed that enzymes secreted by P. waksmanii (Graminho et al., 2013) and Myceliophthora sp. (Zanphorlin et al., 2011) showed the highest catalytic efficiency at the “unprimed” side, with Ile at P1 and Ile at P2, respectively. In contrast, proteases from Myceliophthora thermophila (Hamin Neto et al., 2015) and Aspergillus fumigatus (Da Silva et al., 2014) showed the highest catalytic efficiency at the “primed” side, with Ala at P′2 and Leu at P′3, respectively. Furthermore, different enzymes show specificity to the various types of substrates, highlighting the importance of kinetic assays.
In summary, the metalloprotease isolated from E. javanicum during solid-state fermentation exhibited characteristics that were important and desirable for a variety of industrial applications. These findings may provide important insights into the availability of novel enzyme resources for industrial processes; it is necessary studies in each industrial area to suggest an application.
Author Contributions
Bioprocess, purification, biochemical characterization and kinetic experiment (YH and HC). Peptide substrate synthesis and cleavage site determination (LdO, JdO, MJ, and LJ). Determination the N-terminal sequence (EA).
Funding
The authors would like to acknowledge financial support provided by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP; 2012/24703-8 and 2011/06986-0).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abidi, F., Aissaoui, N., Chobert, J. M., Haertle, T., and Marzouki, M. N. (2014). Neutral serine protease from Penicillium italicum. Purification, biochemical characterization, and use for antioxidative Peptide preparation from Scorpaena notatamuscle. Appl. Biochem. Biotechnol. 174, 186–205. doi: 10.1007/s12010-014-1052-6
Abidi, F., Chobert, J. M., Haertle, T., and Marzouki, M. N. (2011). Purification and biochemical characterization of stable alkaline protease Prot-2 from Botrytis cinerea. Process. Biochem. 46, 2301–2310. doi: 10.1016/j.procbio.2011.09.010
Adrio, J. L., and Demain, A. L. (2014). Microbial enzymes: tools for biotechnological processes. Biomolecules 4, 117–139. doi: 10.3390/biom4010117
Aissaoui, N., Abidi, F., Mahat, S., and Marzouki, M. N. (2014). Purification and biochemical characterization of a novel protease from Penicillium digitatum – Use in bioactive peptides production. J. Basic Microbiol. 54, S178–S189. doi: 10.1002/jobm.201400179
Berger, A., and Schechter, I. (1970). Mapping the active site of papain with the aid of peptide substrates and inhibitors. Philos. Trans. R. Soc. Lond. B Biol. Sci. 257, 249–264. doi: 10.1098/rstb.1970.0024
Bersanetti, P. A., Park, H. Y., Bae, K. S., Son, K. H., Shin, D. H., Hirata, I. Y., et al. (2005). Characterization of arazyme, an exocellular metalloprotease isolated from Serratia proteamaculans culture medium. Enzyme Microb. Technol. 37, 574–581. doi: 10.1016/j.enzmictec.2005.01.041
Bruins, M. E., Janssen, A. E. M., and Boom, R. M. (2001). Thermozymes and their applications a review of recent literature and patents. Appl. Biochem. Biotechnol. 90, 155–186. doi: 10.1385/ABAB:90:2:155
Chagas, J. R., Juliano, L., and Prado, E. S. (1991). Intramolecularly quenched fluorogenic tetrapeptide substrates for tissue and plasma kallikreins. Anal. Biochem. 192, 419–425. doi: 10.1016/0003-2697(91)90558-B
Chen, Z., Ao, J., Yang, W., Jiao, L., Zheng, T., and Chen, X. (2013). Purification and characterization of a novel antifungal protein secreted by Penicillium chrysogenum from an Arctic sediment. Appl. Microbiol. Biotechnol. 97, 10381–10390. doi: 10.1007/s00253-013-4800-6
Da Silva, R. R., Ângelo, T., and Cabral, H. (2013). Comparative evaluation of peptidases produced by Penicillium corylophilum and Penicillium waksmanii in solid-state fermentation using agro-industrial residues. J. Agric. Sci. Technol. B 3, 230–237.
Da Silva, R. R., Caetano, R. C., Okamotoc, D. N., De-Oliveira, L. C. G., Rosa, J. C., and Cabral, H. (2014). The identification and biochemical properties of the catalytic specificity of a serine peptidase secreted by Aspergillus fumigatus Fresenius. Protein Pept. Lett. 21, 663–671. doi: 10.2174/0929866521666140408114646
Dunn, B. M. (1989). “Determination of protease mechanism,” in Proteolytic Enzymes: A Practical Approach, eds R. J. Beynon and J. S. Bond (Oxford: IRL Press), 57–81.
El-Gendy, M. M. A. (2010). Keratinase production by endophytic Penicillium spp. Morsy1 under solid-state fermentation using rice straw. Appl. Biochem. Biotechnol. 162, 780–794. doi: 10.1007/s12010-009-8802-x
Graminho, E. R., da Silva, R. R., de Freitas Cabral, T. P., Arantes, E. C., da Rosa, N. G., Juliano, L., et al. (2013). Purification, characterization, and specificity determination of a new serine protease secreted by Penicillium waksmanii. Appl. Biochem. Biotechnol. 169, 201–214. doi: 10.1007/s12010-012-9974-3
Gupta, R., Beg, Q. K., Khan, S., and Chauhan, B. (2002). An overview on fermentation, downstream processing and properties of microbial alkaline proteases. Appl. Microbiol. Biotechnol. 60, 381–395. doi: 10.1007/s00253-002-1142-1
Hamin Neto, Y. A. A., De Freitas, L. A. P., and Cabral, H. (2014). Multivariate analysis of the stability of spray-dried Eupenicillium javanicum peptidases. Dry. Technol. 32, 614–621. doi: 10.1080/07373937.2013.853079
Hamin Neto, Y. A. A., de Souza Motta, C. M., and Cabral, H. (2013). Optimization of metalloprotease production by Eupenicillium javanicum in both solid-state and submerged bioprocesses. Afr. J. Biochem. Res. 7, 146–157.
Hamin Neto, Y. A. A., De-Oliveira, L. C. G., De-Oliveira, A. H. C., Rosa, J. C., Juliano, M. A., Juliano, L., et al. (2015). Determination of specificity and biochemical characteristics of neutral protease isolated from Myceliophthora thermophila. Protein Pept. Lett. 22, 972–982. doi: 10.2174/0929866522666150817093719
Hatanaka, T., Arima, J., Uesugi, Y., and Iwabuchi, M. (2005). Purification, characterization cloning, and sequencing of metalloendopeptidase from Streptomyces septatus TH-2. Arch. Bioquem. Biophys. 434, 289–298. doi: 10.1016/j.abb.2004.11.018
Hirata, I., Sedenho Cezari, M., Nakaie, C., Boschcov, P., Ito, A., Juliano, M., et al. (1995). Internally quenched fluorogenic protease substrates: solid-phase synthesis and fluorescence spectroscopy of peptides containing ortho-aminobenzoyl/dinitrophenyl groups as donor-acceptor pairs. Lett. Pept. Sci. 1, 299–308. doi: 10.1007/BF00119771
Hmidet, N., Nawani, N., and Ghorbel, S. (2015). Recent development in production and biotechnological application of microbial enzymes. Biol. Med. Res. Int. 2015:280518.
Klemencic, I., Carmona, A. K., Cezari, M. H. S., Juliano, M. A., Juliano, L., Guncar, G., et al. (2000). Biochemical characterization of human cathepsin X revealed that the enzyme is an exopeptidase, acting as carboxymonopeptidase or carboxydipeptidase. Eur. J. Biochem. 267, 5404–5412. doi: 10.1046/j.1432-1327.2000.01592.x
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. doi: 10.1038/227680a0
Mahajan, R. T., and Badgujar, S. B. (2010). Biological aspects of proteolytic enzymes: a review. J. Pharm. Res. 3, 2048–2068.
Majumder, R., Banik, S. P., and Khowala, S. (2015). Purification and characterisation of κ-casein specific milk-clotting metalloprotease from Termitomyces clypeatus MTCC 5091. Food Chem. 173, 441–448. doi: 10.1016/j.foodchem.2014.10.027
Merheb-Dini, C., Cabral, H., Leite, R. S. R., Zanphorlin, L. M., Okamoto, D. N., Bonilla Rodriguez, G. O., et al. (2009). Biochemical and functional characterization of ametalloprotease from the thermophilic fungus Thermoascus aurantiacus. J. Agric. Food Chem. 57, 9210–9217. doi: 10.1021/jf9017977
Nakadate, S., Nozawa, K., Horie, H., Fujii, Y., Nagai, M., Hosoe, T., et al. (2007). Eujavanicols A–C, decalin derivatives from Eupenicillium javanicum. J. Nat. Prod. 70, 1510–1512. doi: 10.1021/np078008a
Nakadate, S., Nozawa, K., Sato, H., Horie, H., Fujii, Y., Nagai, M., et al. (2008). Antifungal cyclic depsipeptide, eujavanicin a, isolated from Eupenicillium javanicum. J. Nat. Prod. 71, 1640–1642. doi: 10.1021/np8002904
Nigam, P. S. (2013). Microbial enzymes with special characteristics for biotechnological applications. Biomolecules 3, 597–611. doi: 10.3390/biom3030597
Oliveira, J. R., Bertolin, T. C., Andrade, D., Oliveira, L. C., Kondo, M. Y., Santos, J. Á, et al. (2015). Specificity studies on kallikrein related peptidase 7 (KLK7) and effects of osmolytes and glycosaminoglycans on its peptidase activity. Biochim. Biophys. Acta 1854, 73–83. doi: 10.1016/j.bbapap.2014.10.018
Rani, K., Rana, R., and Datt, S. (2013). Review on latest overview of proteases. Int. J. Sci. Res. 3, 1–9.
Sarath, G., de la Motte, R. S., and Wagner, F. W. (1989). “Protease assay methods,” in Proteolytic Enzymes: A Practical Approach, eds R. J. Beynon and J. S. Bond (Oxford: IRL Press), 25–56.
Sarrouh, B., Santos, T. M., Miyoshi, A., Dias, R., and Azevedo, V. (2012). Up-to-date insight on industrial enzymes applications and global market. J. Bioprocess Biotechnol. S4:002.
Savitha, S., Sadhasivam, S., Swaminathan, K., and Lin, F. H. (2011). Fungal protease: production, purification and compatibility with laundry detergents and their wash performance. J. Taiwan Inst. Chem. Eng. 42, 298–304. doi: 10.1007/s00253-009-2145-y
Saxena, R., and Singh, R. (2011). Characterization of a metallo-protease produced in solid-state fermentation by a newly isolated Bacillus strain. Acta Biol. Szeged. 55, 13–18.
See, Y. S., and Jackowski, G. (1989). “Estimating molecular weights of polypeptides by SDS gel electrophoresis,” in Protein Structure a Pratical Aprroach, ed. T. E. Creigton (New York: Oxford University Press), 1–19.
Shankar, S., Rao, M., and Laxman, R. S. (2011). Purification and characterization of an alkaline protease by a new strain of Beauveria sp. Process Biochem. 46, 579–585. doi: 10.1016/j.procbio.2010.10.013
Shirasaka, N., Naitou, M., Okamura, K., Kusuda, M., Fukuta, Y., and Terashita, T. (2012). Purification and characterization of a fibrinolytic protease from Aspergillus oryzae KSK-3. Mycoscience 53, 354–364. doi: 10.1007/S10267-011-0179-3
Souza, P. M. D., Bittencourt, M. L. D. A., Caprara, C. C., Freitas, M. D., Almeida, R. P. C. D., Silveira, D., et al. (2015). A biotechnology perspective of fungal proteases. Braz. J. Microbiol. 46, 337–346. doi: 10.1590/S1517-838246220140359
Srilakshmi, J., Madhavi, J., and Ammani, K. (2014). Commercial potential of fungal protease: past, present and future prospects. Int. J. Pharm. Chem. Biol. Sci. 2, 218–234.
Tao, N. G., Shi, W. Q., Liu, Y. J., and Huang, S. R. (2011). Production of feed enzymes from citrus processing waste by solid-state fermentation with Eupenicillium javanicum. Int. J. Food Sci. Technol. 46, 1073–1079. doi: 10.1111/j.1365-2621.2011.02587.x
Thomas, L., Larroche, C., and Pandey, A. (2013). Current developments in solid-state fermentation. Biochem. Eng. J. 81, 146–161. doi: 10.1016/j.bej.2013.10.013
Thys, R. C. S., and Brandelli, A. (2006). Purification and properties of a keratinolytic metalloprotease from Microbacterium sp. J. Appl. Microbiol. 101, 1259–1268. doi: 10.1111/j.1365-2672.2006.03050.x
Vieille, C., and Zeikus, G. J. (2001). Hyperthermophilic enzymes: sources, uses, and molecular mechanisms for thermostability. Microbiol. Mol. Biol. Rev. 65, 1–43. doi: 10.1128/MMBR.65.1.1-43.2001
Wang, S.-L., Kao, T.-Y., Wang, C.-L., Yen, Y.-H., Chern, M.-K., and Chen, Y.-H. (2006). A solvent sTable metalloprotease produced by Bacillus sp TKU004 and its application in the deproteinization of squid pen for beta-chitin preparation. Enzyme Microb. Technol. 39, 724–731. doi: 10.1016/j.enzmictec.2005.12.007
Keywords: biochemical characterization, fluorescence resonance energy transfer peptides, microbial enzyme, protease, solid-state fermentation
Citation: Hamin Neto YAA, de Oliveira LCG, de Oliveira JR, Juliano MA, Juliano L, Arantes EC and Cabral H (2017) Analysis of the Specificity and Biochemical Characterization of Metalloproteases Isolated from Eupenicillium javanicum Using Fluorescence Resonance Energy Transfer Peptides. Front. Microbiol. 7:2141. doi: 10.3389/fmicb.2016.02141
Received: 29 August 2016; Accepted: 19 December 2016;
Published: 09 January 2017.
Edited by:
Martin G. Klotz, Queens College, City University of New York, USAReviewed by:
Susana Rodriguez-Couto, Ikerbasque, SpainGerardo Díaz-Godínez, Autonomous University of Tlaxcala, Mexico
Copyright © 2017 Hamin Neto, de Oliveira, de Oliveira, Juliano, Juliano, Arantes and Cabral. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hamilton Cabral, hamilton@fcfrp.usp.br
 Youssef A. A. Hamin Neto
Youssef A. A. Hamin Neto Lilian C. G. de Oliveira
Lilian C. G. de Oliveira Juliana R. de Oliveira2
Juliana R. de Oliveira2 Luiz Juliano
Luiz Juliano Hamilton Cabral
Hamilton Cabral