- 1Hadal Science and Technology Research Center, Shanghai Ocean University, Shanghai, China
- 2Department of Natural Sciences, Hawaii Pacific University, Honolulu, HI, USA
- 3Department of Marine Biodiversity Research, Japan Agency for Marine-Earth Science and Technology, Yokosuka, Japan
- 4Environmental Management Research Institute, National Institute of Advanced Industrial Science and Technology, Ibaraki, Japan
- 5State Key Laboratory of Marine Geology, Tongji University, Shanghai, China
- 6Kochi Institute for Core Sample Research, Japan Agency for Marine-Earth Science and Technology, Kochi, Japan
- 7Research and Development Center for Ocean Drilling Science, Japan Agency for Marine-Earth Science and Technology, Yokohama, Japan
- 8Research and Development Center for Submarine Resources, Japan Agency for Marine-Earth Science and Technology, Yokosuka, Japan
Phylogenetically diverse microorganisms have been observed in marine subsurface sediments down to ~2.5 km below the seafloor (kmbsf). However, very little is known about the pressure-adapted and/or pressure-loving microorganisms, the so called piezophiles, in the deep subseafloor biosphere, despite that pressure directly affects microbial physiology, metabolism, and biogeochemical processes of carbon and other elements in situ. In this study, we studied taxonomic compositions of microbial communities in high-pressure incubated sediment, obtained during the Integrated Ocean Drilling Program (IODP) Expedition 337 off the Shimokita Peninsula, Japan. Analysis of 16S rRNA gene-tagged sequences showed that members of spore-forming bacteria within Firmicutes and Actinobacteria were predominantly detected in all enrichment cultures from ~1.5 to 2.4 km-deep sediment samples, followed by members of Proteobacteria, Acidobacteria, and Bacteroidetes according to the sequence frequency. To further study the physiology of the deep subseafloor sedimentary piezophilic bacteria, we isolated and characterized two bacterial strains, 19R1-5 and 29R7-12, from 1.9 and 2.4 km-deep sediment samples, respectively. The isolates were both low G+C content, gram-positive, endospore-forming and facultative anaerobic piezophilic bacteria, closely related to Virgibacillus pantothenticus and Bacillus subtilis within the phylum Firmicutes, respectively. The optimal pressure and temperature conditions for growth were 20 MPa and 42°C for strain 19R1-5, and 10 MPa and 43°C for strain 29R7-12. Bacterial (endo)spores were observed in both the enrichment and pure cultures examined, suggesting that these piezophilic members were derived from microbial communities buried in the ~20 million-year-old coal-bearing sediments after the long-term survival as spores and that the deep biosphere may host more abundant gram-positive spore-forming bacteria and their spores than hitherto recognized.
Introduction
The deep subseafloor biosphere is unique in that microorganisms long persist in the high-pressure environment over geologic time. Pressure is one of the important physical properties of subseafloor sedimentary habitats, which most likely affects microbial cell viability, growth, and physiology (Allen et al., 1999; Fang et al., 2004; Fichtel et al., 2015). Pressure ranges over four-orders of magnitude in the surface biosphere (Bartlett et al., 1995) whereas probably more in the deep subseafloor biosphere. In such habitats under the deep-sea bed, there may be microorganisms that require high-pressure conditions for optimal growth, so called “piezophiles” (Yayanos, 1995), whose activity possibly plays important roles in global biogeochemical cycles (e.g., Fichtel et al., 2015).
A better understanding of microbial metabolism of subsurface microorganisms is essential to help delineate their impact on biogeochemical cycling in the deep biosphere. However, we still do not know the metabolic activities of pressure-adapted subsurface microorganisms and the pressure (depth) and habitability limit of the deep biosphere, as the deep subsurface prokaryotic cells are mostly resistant to cultivation and only <0.1% of all microscopically and/or molecular genetically detected cells have been isolated and identified (Batzke et al., 2007; Sass and Parkes, 2011; Ciobanu et al., 2014; Russell et al., 2016). Thus far, studies of the deep subsurface biosphere have yielded relatively few pressure-adapted bacterial isolates for laboratory studies of their physiology, ecology and biogeochemical functions (Bale et al., 1997; Russell et al., 2016). The pressure (depth) and habitability limits of the deep subseafloor life remain poorly constrained.
In 2012, the Integrated Ocean Drilling Program (IODP) Expedition 337 provided an unprecedented opportunity to study piezophilic microbial communities, which may include some spore-formers buried in the deep and old coal-bearing sediment. Using the riser-drilling research vessel Chikyu, we drilled at Site C0020 in a former forearc basin in the north western Pacific off the Shimokita Peninsula, Japan (Inagaki et al., 2012, 2015; Gross et al., 2015; Glombitza et al., 2016; Tanikawa et al., 2016). The primary goal of this study was to investigate microbial communities of three sediment core samples from the depths of ~1.5 to 2.4 km below the ocean floor and, by isolating piezophilic bacterial strains, to study metabolism of the piezophilic isolates in the deep subseafloor biosphere. Our results show that enrichment cultures from the very deep sedimentary microbial communities were dominated by gram-positive endospore-forming piezophilic bacteria. The successful isolation and cultivation of these gram-positive piezophilic bacteria underline that microorganisms were revivable after the long-term burial as spores.
Materials and Methods
Sampling Site, Sedimentological and Geochemical Characteristics of the Sediment Samples
Deep subseafloor sediment samples used in this study were obtained by the riser-drilling technology of the deep-sea drilling research vessel Chikyu during the Integrated Ocean Drilling Program (IODP) Expedition 337 at Site C0020 (41°10.60′ N, 142°12.03′ E; water depth 1,180 m) off the Shimokita Peninsula, Japan (Supplementary Figure 1; Inagaki et al., 2012, 2015). Three sediment samples, 6R-3, 19R-1~25R-3, and 29R-7 were obtained from 1,498 mbsf, 1,951~1,999 mbsf, and 2,406 mbsf immediately after core recovery and the X-ray computed tomography (CT) scan. All sediment samples were blackish and associated with Miocene lignite coals that contain possibly biogenic pyrite veins (Gross et al., 2015; Glombitza et al., 2016).
Lithological and geochemical characteristics of the sediment samples were determined on the Chikyu by combined analysis of cuttings and cores (Inagaki et al., 2012, 2015; Gross et al., 2015; Glombitza et al., 2016; Tanikawa et al., 2016). Generally, two samples at greater depths, 19R-1~25R-3 and 29R-7, had rather different lithological and geochemical characteristics to the shallow sample 6R-3. Sample 19R-1~25R-3 represents deposition in a back-barrier shallow marine environment with wetlands in early middle Miocene (Gross et al., 2015). The sedimentary unit was characterized by sandstones, siltstones and coaly shale with excellent pollen and spore assemblages. Total organic carbon (TOC) content of the sediment was up to 30% (Inagaki et al., 2012). Sample 29R-7 was deposited in late Oligocene in an environment similar to that for 19R-1~25R-3. The lithology includes sandstone intercalated with silt and a thin coal layer. In contrast, the sediment unit represented by 6R-3, with massive sandstone and siltstone and marine fossiliferous material was deposited in an offshore marine environment. Thus, sediments of 19R-1~25R-3 and 29R-7 represent deposition in a transitional depositional environment with obvious terrestrial influence, whereas sample 6R-3 represents a cool-water continental shelf environment (Inagaki et al., 2012; Gross et al., 2015).
High Pressure Enrichment and Isolation of Deep Subseafloor Piezophilic Bacteria
The three sediment samples (about 10 g each), 6R-3, 19R-1~25R-3, and 29R-7, were set up as three high-pressure (HP) enrichment cultures to be used for the downstream isolation of pure cultures, designated as 6R-3HP, 19R-1~25R-3HP, and 29R-7HP, according to the previously published procedure (see Kato, 2011). During Expedition 337 and thereafter, the three HP enrichment cultures were kept under the pressure in situ (i.e., 35 MPa), anoxic, and the respective environmental temperatures at 35, 45, and 55°C, respectively. The HP cultivation was conducted using Marine Broth 2216 (MB 2216, Difco) liquid medium for 3 months. The HP enrichment cultures were then transferred to new MB 2216 (1% seed, v/v) and the procedure was repeated under the same temperature and pressure condition for three times. Finally, the cultures were plated on MB 2216 agar medium (1.2%, w/v) for single colony isolation at atmospheric condition. Further growth experiments of bacterial isolates were performed under anoxic conditions using the AnaeroPack chamber (Mitsubishi Gas Chemical, Tokyo) at the same conditions described above.
Taxonomic Analysis of HP Enrichment Cultures Based on 16S rRNA Gene Sequences
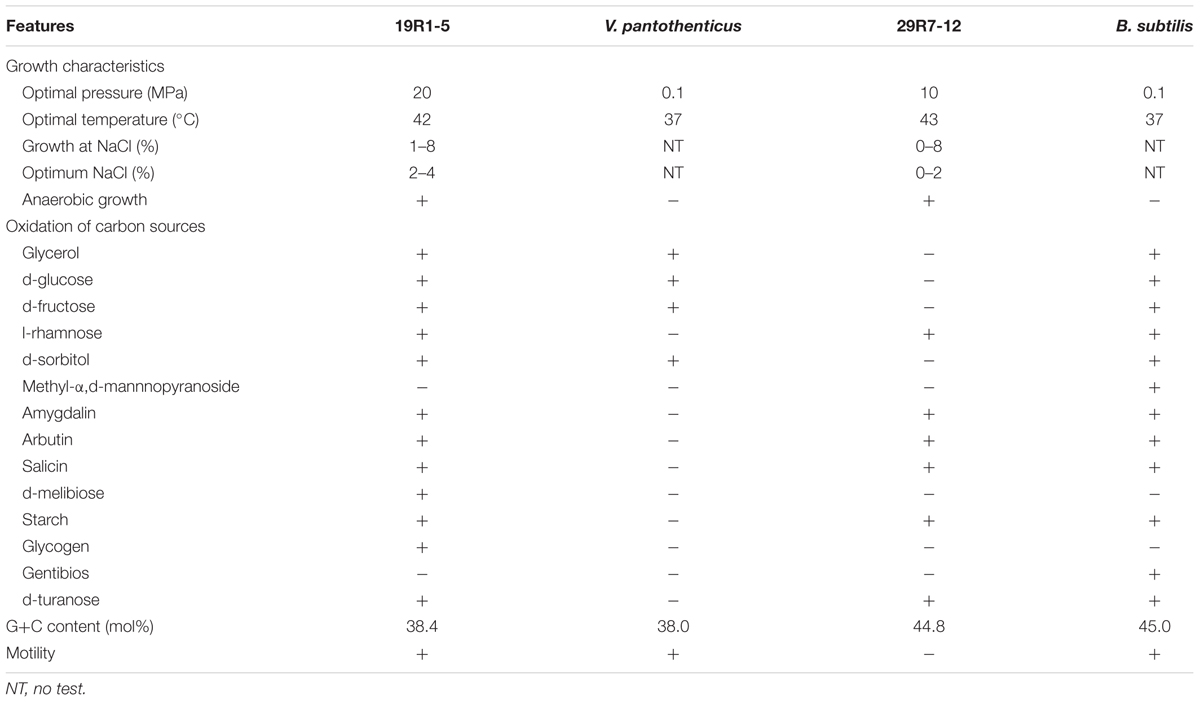
To study taxonomic composition and community structure of HP enrichment culture samples, we extracted DNA from each sediment and aqueous phase of the first enrichment slurry: for the sediment, we designated the sample name as 6R-1 for Core 6R-3, 19R-1 for Core 19R-1~25R-3, and 29R-1 for Core 29R-7; for the medium aqueous phase, 6R-2 for Core 6R-3, 19R-2 for Core 19R-1~25R-3, and 29R-2 for Core 29R-7 (Figure 1).
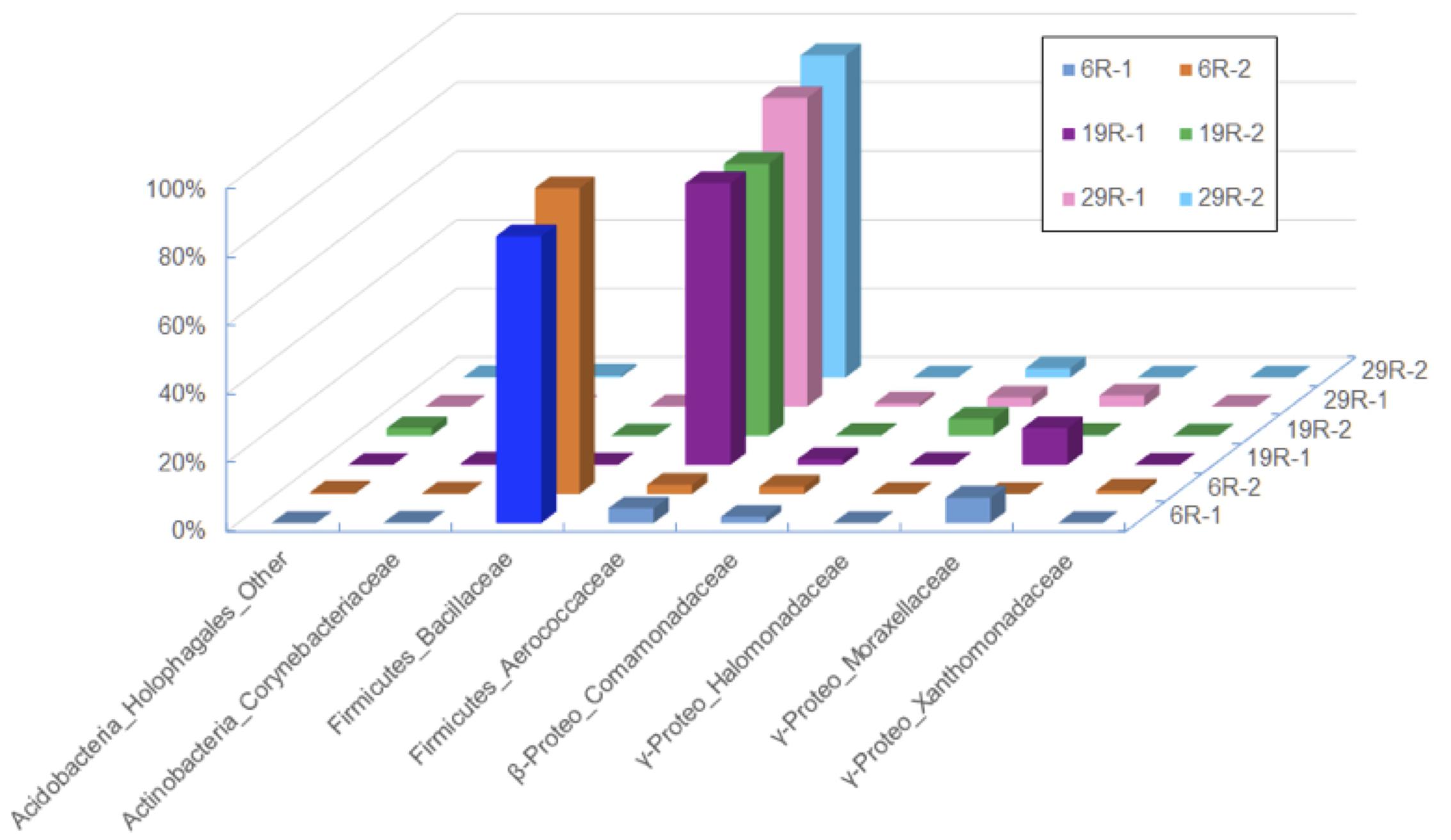
FIGURE 1. Bacterial compositions showing the relative abundance of family level classification among different sediments and aqueous phases after the first high-pressure enrichment cultivation. Only bacterial lineages whose abundances are >1% of reads are shown.
Total DNA was extracted from sediment and aqueous phases in the first HP enrichment cultures using a Power soil DNA extraction kit (MoBio Laboratories, Carlsbad, CA, USA), according to the manufacturer’s protocol. The 16S rRNA gene amplicon sequencing was performed to analyze taxonomic composition of three HP enrichment cultures using MiSeq system (Illumina, USA) as described previously (Aoyagi et al., 2015). Briefly, the primer set used for the amplicon sequencing was 515f/806r, which primer sequences were both modified to include an Illumina adapter region, and the reverse primer was encoded with 12-bp barcodes (Caporaso et al., 2012). PCR condition was as described previously (Navarro et al., 2015). The PCR amplicons were purified, first with an AMPure XP Kit (Beckman Coulter, USA), and the second with a QIAquick gel extraction kit (QIAGEN, USA). The concentration of the purified amplicon DNA was determined with a Quant-iT PicoGreen dsDNA reagent and kit (Life Technologies, USA). An appropriate amount of the purified amplicon and an internal control (PhiX Control V3; Illumina) were subjected to sequencing with a 300-cycles MiSeq Reagent kit (Illumina) and a MiSeq sequencer. The removal of the PhiX, low-quality (Q < 30) and chimeric sequences, and the assembly of the paired-end sequences were discarded as described previously (Itoh et al., 2014). The sequences in the sequence libraries were characterized phylogenetically by using the QIIME software (Caporaso et al., 2010). Alpha-diversity indices (e.g., Chao1, Shannon, and Simpson reciprocal) and the weighted UniFrac distances for principal coordinate analysis (PCoA) were calculated with the QIIME software (Caporaso et al., 2010) on the base of sequences obtained as above.
Characterization of Piezophilic Bacterial Isolates
DNA of piezophilic bacterial isolates was purified using the method of Saito and Miura (1963), and 16S rRNA genes were amplified by PCR using the primers Eubac27F and Eubac1492R (DeLong, 1992). Amplified sequences (~1.5 kb) were determined using a Perkin-Elmer 3730xl DNA sequencer (Perkin-Elmer/Applied Biosystems, USA). PCR amplification was performed with a DNA Thermal Cycler model 9600 (Perkin-Elmer/Cetus, Co.) by using 50 ul of the PCR reaction mixture under the conditions recommended by the manufacturers (Takara, Co.) (Kato et al., 1995). A total of 30 cycles of amplification were performed with temperate DNA denaturation at 95°C for 1.5 min, primer annealing at 55°C for 1.5 min, and primer extension at 72°C for 1.5 min (DeLong, 1992). A phylogenetic tree was constructed based on 16S rRNA gene sequences. Nucleotide substitution rates (Knuc: Kimura, 1980) were determined, and the distance matrix tree was constructed by the neighbor-joining method (Saitou and Nei, 1987) using the Clustal X 2.0 program (Larkin et al., 2007). Cell morphology was observed under scanning electron microscope (SEM; model JSM-6700F, JEOL, Co., Japan) and transmission electron microscope (TEM; model JEM-1210, JEOL, Co., Japan) of the negative stained cells (Nogi et al., 1998) (Supplementary Figure 2). Phenotypic and biochemical characterizations were completed using the API50CHB strips (BioMerieux, Lyon), according to the manufacturer’s instructions. The G+C content (mol%) was measured by reversed-phase high-performance liquid chromatography (Tamaoka and Komagata, 1984). For analysis of relatedness between the isolates and reference species, DND-DNA hybridization was performed at 40°C for 3 h and measured fluorometrically according to the method of Ezaki et al. (1989). The isolated strains were deposited to the Japan Collection of Microorganisms (JCM, Tsukuba-City, Japan), and the strain numbers were JCM31625 (6R3-1), JCM31626 (6R3-15), JCM31627 (19R1-5), and JCM31625 (29R7-12).
Fatty Acids Analyses
Total lipids were extracted from microbial cells using a one-phase solvent system (Fang and Findlay, 1996). The total lipids obtained were separated into neutral lipids, glycolipids, and phospholipids using miniature champagne columns (Supelco, Inc., Bellefonte, PA, USA) (Fang et al., 2007). The phospholipid fraction was subjected to a mild alkaline trans-methylation procedure to produce fatty acid methyl esters (FAMEs). FAMEs were analyzed on an Agilent 6890 GC interfaced with an Agilent 5973N Mass Selective Detector. Individual fatty acids were identified according to their mass spectra.
Growth Experiments
High-pressure cultivation tests of the isolates were performed at various pressures of 0.1–70 MPa with an increment of 10 MPa, according to the procedure described by Kato et al. (1995). Optimal temperature conditions were determined at the range of 10–60°C, and optimal salt conditions were determined at the concentration range of 0–11 ‰ NaCl, using a bio-photo recorder system (model TVS126MA, Advantec, Japan) on the basis of optical density at 600 nm (OD600).
Nucleotide Sequence Accession Numbers
The 16S rRNA gene sequences of isolates reported in this paper were deposited in the DDBJ (DNA Data Bank of Japan) under accession numbers LC155963 – LC155966. The nucleotide sequences obtained from the Illumina sequencing of 16S rRNA genes were deposited in the MG-RAST database as “Isolation and characterization of piezophilic microorganisms from 1.5 and 2.4 km-deep subseafloor sediment core samples” project under accession ID 4695872.3-4695877.3.
Results and Discussion
Taxonomic Composition of High-Pressure Enrichment Communities
Taxonomic classification analysis of HP enrichment cultures clearly indicated that cultivated piezophilic communities were dominated by members of the phylum Firmicutes with >80% of the reads, followed by members of Gamma-proteobacteria, which sequence frequency ranged between 1.2 and 11.6% (Figure 1). No obvious differences in bacterial community compositions between the sediment and aqueous phases were observed after the first HP cultivation (Figure 1), suggesting that no significant microbial fractionation occurred between the sediment (sedimentary microbes) and aqueous phases (free-living microbes) during HP cultivation. Thereafter, we decided to use whole (i.e., mixed) microbial communities for the DNA analysis of HP enrichment cultures.
There were 39,020, 9,899, and 44,365 qualified 16S rRNA gene sequences recovered from HP enrichment samples 6R-1, 19R-1, and 29R-1, respectively. The sequences represented 12 phyla, 39 classes, 47 orders, 91 families, and 129 genera. At the phylum level, the vast majority of the recovered sequences was found to be the phylum Firmicutes, which was 88.8, 83.8, and 91.2% of the total sequence read number for 6R-1, 19R-1, and 29R-1, respectively (Supplementary Figure 3). The next most abundant phyla were Proteobacteria and Actinobacteria. Based on relative abundance, Bacilli (88.7, 83.6, and 91.6%), Gamma-proteobacteria (7.7, 11.6, and 6.8%) and Beta-proteobacteria (2.2, 2.6, and 1.2%) were the most abundant classes in those enrichment samples (data not shown). However, there are significant differences in HP-cultivated bacterial communities between 6R-3 and two deeper samples (19R-1~25R-3 and 29R-7), possibly corresponding to the lithological characteristics (6R-3 was deposited in an offshore marine environment whereas 19R-1~25R-3 and 29R-7 were deposited in a transitional to shallow marine environment) (Figure 1). At the order level, members of the Bacillales dominated enriched bacterial communities in 6R-1 (84.0%), whereas members of the Lactobacillales were predominant in 19R-1 (90.3%) and 27R-1 (82.5%) (Supplementary Figure 3). The difference in cultured bacterial communities between shallow and deep sediment samples could be further illustrated at the genus level. The most abundant genus in samples 19R-1 and 29R-1 was Alkalibacterium (81.0 and 88.8% of the total sequence reads, respectively). In contrast, the most abundant genus in sample 6R-1 was Marinilactibacillus (83.4%), which is a fermentative marine lactic acid bacterium that has been isolated from shallow subsurface sediments in the northwestern Pacific, being either psychrophilic (Ishikawa et al., 2003) or piezophilic (Toffin et al., 2005) (Supplementary Figure 3). However, given the geochemical conditions at Site CC0020, it is unlikely that the Shimokita coalbed sediments host psychro-piezophilic bacteria, but rather, meso- and thermo-piezophilic bacteria (see below). Based on the cut-off value of 97% similarity, a total of 170 OTUs were identified from three HP enrichment culture samples. The relatively low number of OTUs suggests generally low species richness in the cultivated microbial community. The Venn diagram showed that three sediment samples 6R-1, 19R-1 and 29R-1 shared 50 OTUs (29.4%), with each possessing 37 (21.8%), 9 (5.3%), and 28 (16.5%) unique OTUs, respectively (Figure 2). PCoA clearly separated HP-cultured communities in sediment 6R-1 from 19R-1 and 29R-1, with the first coordinate accounting for 70.2% and the second for 17.0% of the samples’ variance (Supplementary Figure 4).
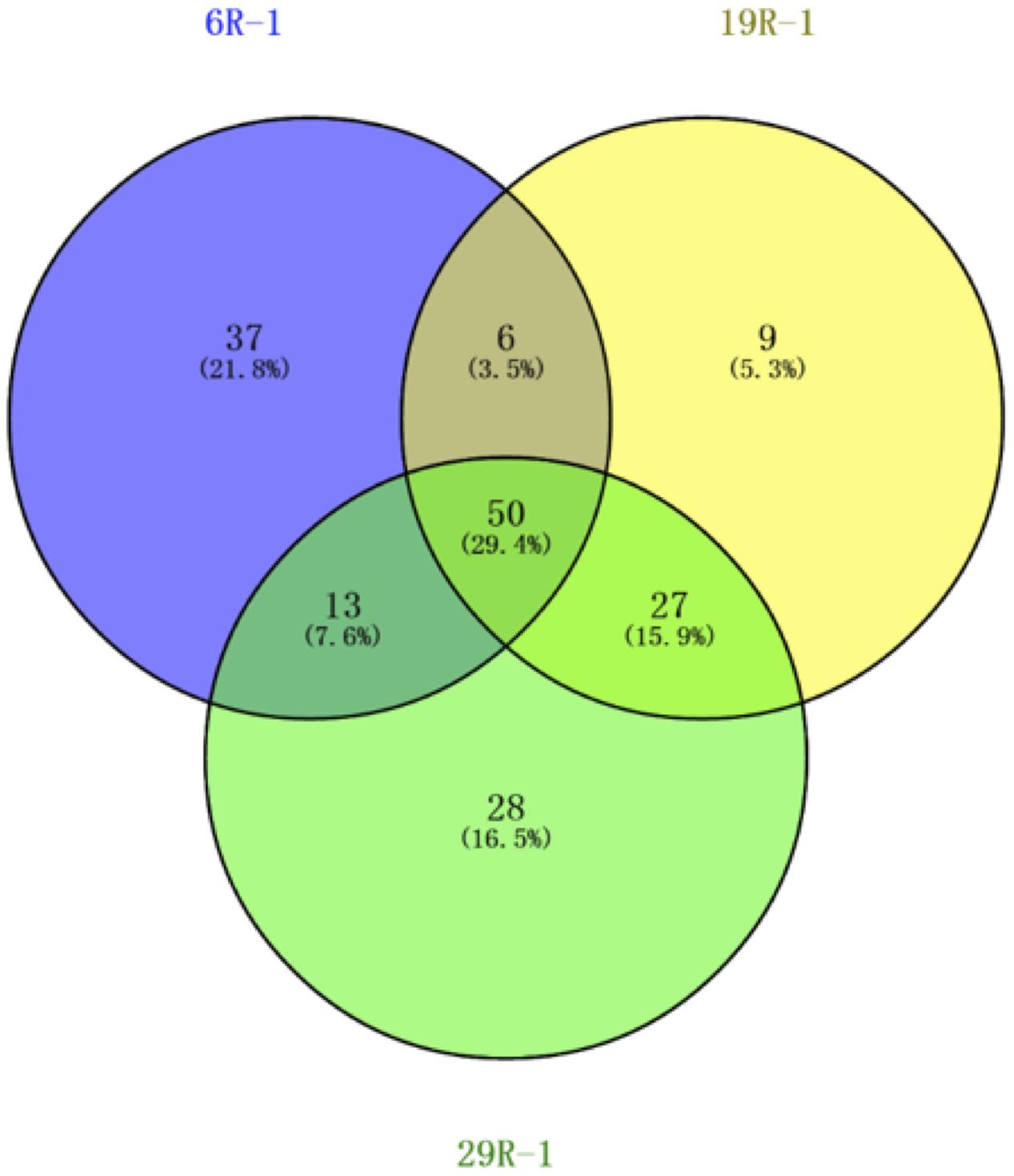
FIGURE 2. Venn diagram of OTUs for bacterial diversity of the first high-pressure enrichment cultures. Unique and shared OTUs between the sample pairs are based on 97% similarity.
It is instructive to note that microbial communities cultivated under the HP condition mainly consisted of gram-positive endospore-forming bacteria. Within the phylum Firmicutes, members of the families Bacillaceae and Aerococcaceae are typical low G+C content gram-positive lineages commonly present in terrestrial soil and sediment, whereas members of the phylum Actinobacteria, detected in these sediments with a range of 0.2~2.8%, are well-known high G+C content gram-positive bacteria in various marine and freshwater environments (e.g., Ventura et al., 2007). This result corroborates that of Inagaki et al. (2015), which showed that Firmicutes and Actinobacteria were detected as indigenous bacterial communities in deep coal-bearing sediments at Site C0020. In addition, we employed the same cultivation method that has been previously used for deep-sea piezophilic bacteria and resulted in successful isolation of both gram-negative and gram-positive piezophilic bacteria (e.g., Kato and Nogi, 2001). These results suggest that gram-positive spore-forming piezophilic bacteria probably constitute a large part of cultivable deep subseafloor bacterial communities at Site C0020.
Isolation and Characterization of Piezophilic Bacteria
To study more about physiology of piezophilic bacteria, we isolated four bacterial strains, named 6R3-1, 6R3-15, 19R1-5, and 29R7-12, from each HP enrichment culture of 6R-3 HP, 19R-1~25R-3 HP, and 29R-7 HP, respectively. Phylogenetic analysis of each16S rRNA gene sequence revealed that bacterial isolates 6R3-1 and 19R1-5 are closely related to Virgibacillus pantothenticus whereas isolates 6R3-15, 29R7-12 are to Bacillus subtilis (Figure 3). DNA–DNA hybridization comparing the isolated strains and reference species V. pantothenticus and B. subtilis suggests that strains 6R3-1 and 19R1-15 are the same species as V. pantothenticus (91.7–100% of homology), and strains 6R3-15 and 29R7-12 are the same as B. subtilis (70.4–90.7% of homology) (Supplementary Table 1). The physiological data further confirmed that isolate 6R3-15 is identical to 19R1-5, and 6R3-15 to 29R7-12 (Supplementary Table 1). Therefore, we selected the bacterial isolates 19R1-5 and 29R7-12 as representative piezophilic isolates for further characterizations.
FIGURE 3. Phylogenetic tree based on 16S rRNA gene sequences showing the relationship among the isolated subsurface bacteria within the phylum Firmicutes. The scale represents the average numbers of nucleotide substitutions per site. Bootstrap values are shown for frequencies per 1,000 times.
Isolates 19R1-5 and 29R7-12 were characterized as gram-positive facultative anaerobic bacteria, able to grow under both anoxic and oxic conditions. The strains were also able to grow not only at high pressures but also at relatively high temperature conditions (Table 1). Strains 19R1-5 and 29R7-12 exhibited optimal growth at 42 and 43°C, respectively, lower than in situ habitat temperatures of 46°C for 19R1-5 and 58°C for 29R7-12 (Figures 4A,B). The isolates also showed piezophilic growth with their optimum growth pressure of 20 and 10 MPa for 19R1-5 and 29R7-12, respectively (Figures 4C,D). It is apparent that the optimal growth pressures for the bacterial isolates are considerably lower than habitat pressures, which are 53 and 65 MPa (as hydrostatic pressure) for 19R1-5 and 29R7-12, respectively. The reason for this large pressure difference is possibly because those piezophilic isolates are derived from spores in coaly shale sediments, which have originally been buried from warm and shallow coastal environments (Gross et al., 2015; Inagaki et al., 2015). Alternatively, it has been observed that the upper cardinal temperature for piezophilic bacterial growth can be extended by high pressure, but the inverse tests have not been done. In our study, the high temperature and high pressure growth experiments were conducted independently; i.e., in determining optimal growth pressure of the isolates, pressure was varied while maintaining the culture at in situ temperature, whereas the growth temperature experiments were carried out at the atmospheric pressure condition, rather than at the in situ pressure. This experimental setup may explain why the optimal growth temperatures for two isolates were lower than the in situ habitat temperatures, but is in contrast to the observation of deep-sea piezophilic bacteria that showed more stenothermal growth for those from deeper depths of the original habitat (e.g., Yayanos, 1986).
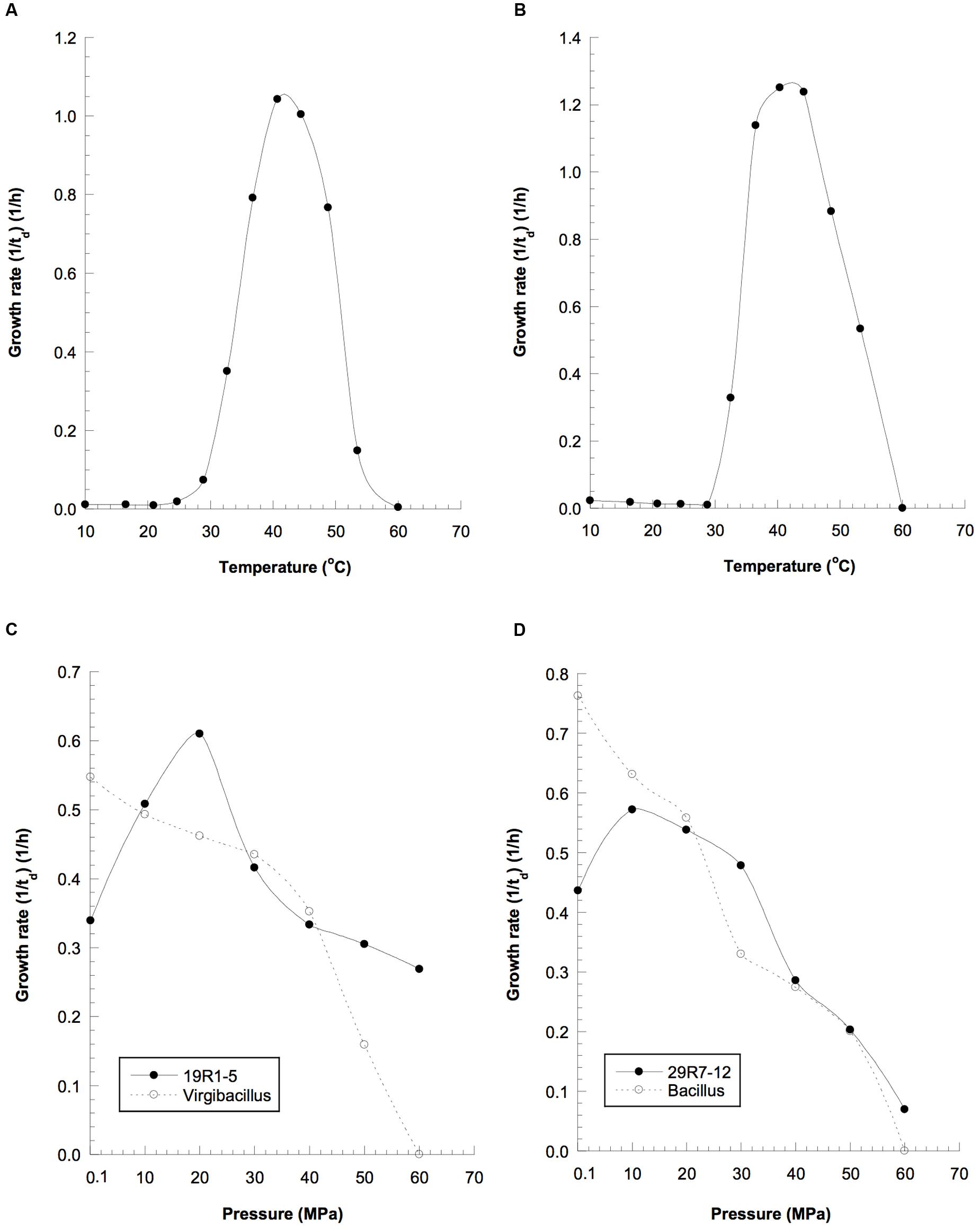
FIGURE 4. Growth properties of the spore-forming piezophilic bacterial isolates and the closely related reference strains at elevated temperatures: (A) strain 19R-1, (B) strain 29R-1; and hydrostatic pressures: (C) solid line, strain 19R1-5; dotted line, Vergibacillus pantothenticus; (D) solid line, strain 29R7-12; dotted line, Bacillus subtilis, td, doubling time (in hours).
The two bacterial isolates 19R1-5 and 29R7-12 exhibited many phenotypic characteristics common to the reference species V. pantothenticus and B. subtilis, such as the G+C content, composition of cellular fatty acids, and utilization of different carbon substrates (Tables 1 and 2). The two strains are facultative anaerobic chemoorganotrophs, displaying both respiratory and fermentative types of metabolism. Iso- and anteiso-branched fatty acids dominate the cellular lipids. The preponderance of methyl-branched fatty acids in both strains is in concordance with gram-positive Firmicutes. Taken together, these results indicate that bacterial isolates 19R1-5 and 29R7-12 are gram-positive spore-forming piezophilic bacteria (Fang et al., 2010), of the same species as V. pantothenticus and B. subtilis, respectively.
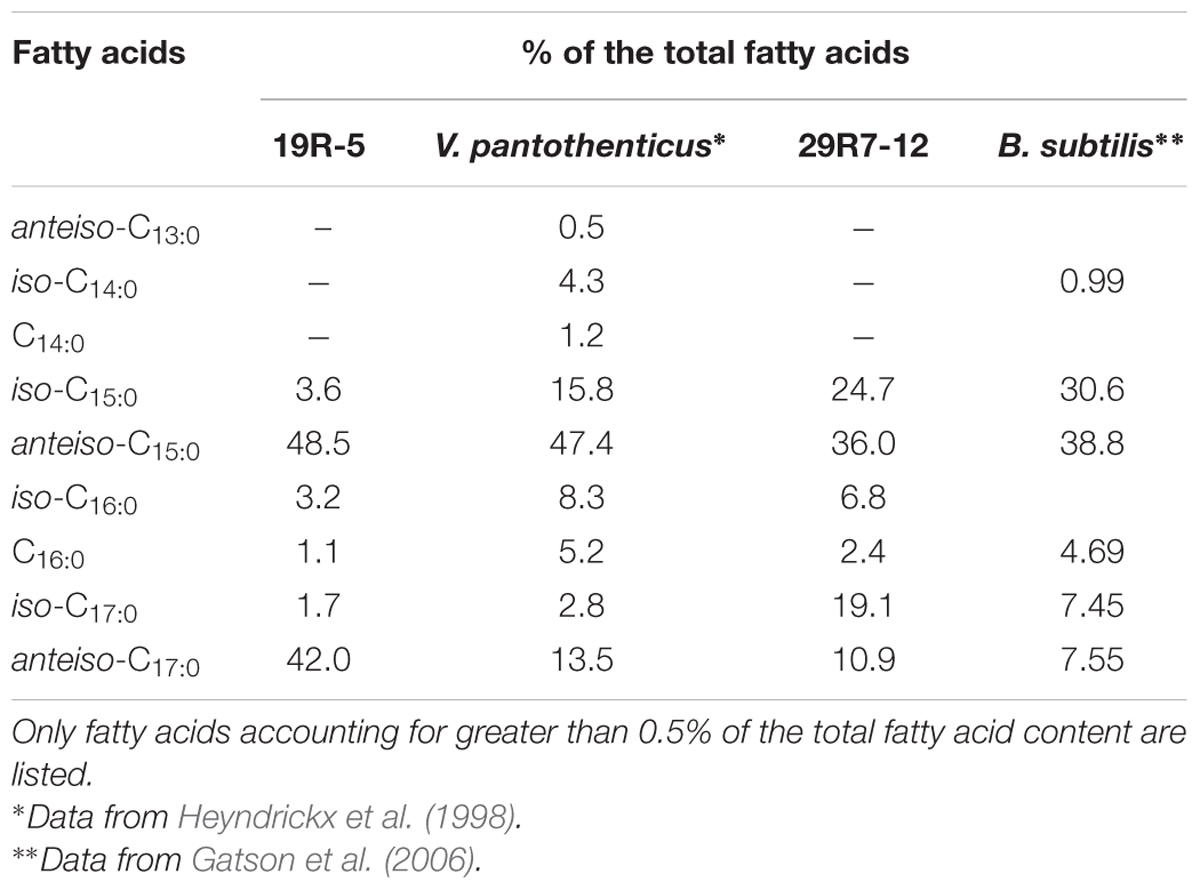
TABLE 2. Comparison of the fatty acid profile of the isolates and the related reference species as measured by GC analysis of their methyl esters.
Implications for Cultivable Piezophilic Bacteria in the Deep Subseafloor Biosphere
Interpreting the origin of cultivable piezophilic bacteria in deep and old subseafloor sediments is a particular challenge for the scientific ocean drilling community. Inagaki et al. (2015) and others suggested that the depositional environment and source of the stratified sediment exert a dominant control on indigenous microbial communities in the deep sedimentary biosphere. Site C0020 is in a former forearc basin where organic-rich terrestrial deposits are expected to be dominant and pervasive in the initial development stage of the basin (Gross et al., 2015). Subsequent subsidence of the basin allowed the deposition of marine sediments overlying the deep terrigenous coal-sand formation. Our results also showed that microbial communities in deep coalbed layers (samples 19R-1 and 29R-1) resemble those commonly found in terrestrial surface soil environment where phyla Firmicutes, Actinobacteria, Proteobacteria, Acidobacteria and Bacteroidetes (designated “FAPAB”) dominate. The Venn diagram for the assemblage of first HP-enrichment cultures clearly shows that sediment samples 19R-1 and 29R-1 from deeper depths shared more OTUs (16%) than each shared with 6R-1, 3.5 and 7.6%, from the shallower depth. We hypothesize that the deposition of terrestrial sediment and the FAPAB buried therein help the organisms persisting in the subsurface for prolonged periods, and the persisted FAPAB species provides a seed bank for populating the deep-biosphere community under the increasing selection pressure with the burial depth and time. As sediment deposition and progressive burial of sediments continues in the basin, temperature and pressure of the buried sediment also increase, which may selectively increase the relative population of cultivable meso- and thermo-pizophilic bacteria in the deep sedimentary biosphere. We further deduce that microbial communities in deeper sedimentary layers may be more or less inherited from the initially buried sediment microbial communities and that the current rare microbial taxa in the rare biosphere may be originated from the former common biosphere and may have served as a seed bank to deeper microbial ecosystems that adapt to geophysical and geochemical changes.
Bacterial sporulation in the natural environment is one of the self-preservation processes that allow bacteria, under unfavorable conditions, to preserve and propagate the genetic information and functional survivability. Endospores are the most resistant cellular structure (Abecasis et al., 2013; Filippidou et al., 2016) and can remain viable for 1000s (Gest and Mandelstam, 1987) or even millions of years (Vreeland et al., 2000). In a previous study, spores of V. pantothentics and B. marismortui were isolated from a 250-million-year-old salt crystal (Vreeland et al., 2000). Recent studies showed that bacterial endospores are globally distributed in shallow to deep subseafloor sediments (Müller et al., 2014), as abundant as vegetative cells (Langerhuus et al., 2012; Lomstein et al., 2012), and generally increase with depth (Batzke et al., 2007; Fichtel et al., 2008). It is likely that the formation of spores and other quiescent cell forms allows bacteria to persist over geologic time, and perhaps more importantly, spores may contribute as a seed bank to the ecosystem development in the deep biosphere, since they are highly resistant and less prone to viral attack or predation (Sogin et al., 2006; Pedrós-Alió, 2007; Hubert et al., 2010). Consequently, they can be expected to accumulate in marine sediments and may contribute greatly to the total biomass of deep subseafloor microbial communities (Lomstein et al., 2012). Our bacterial isolates are gram-positive endospore-forming piezophilic bacteria. Many spores have been observed in the HP enrichment cultures and also in the pure cultures of the bacterial isolates (Figure 5), which is in good agreement with a bioreactor cultivation experiment for ~2 km-deep methanogenic communities associated with lignite coals (Inagaki et al., 2015). The results in this study augment a ‘paleome’ concept, a pool of ancient DNA and/or descendants preserved in sediments buried millions of years, which can provide insights into ancient genetic records and/or microbial ecosystems (Inagaki et al., 2005, 2015; Glombitza et al., 2016). To verify these hypotheses, more intensive cultivation-dependent studies of spores in deep and hot sedimentary systems will be required because it was found to be difficult to obtain metagenomic information from the deeply buried spore pool (Kawai et al., 2015). This is one of our on-going efforts in the exploration of the deep subseafloor biosphere.
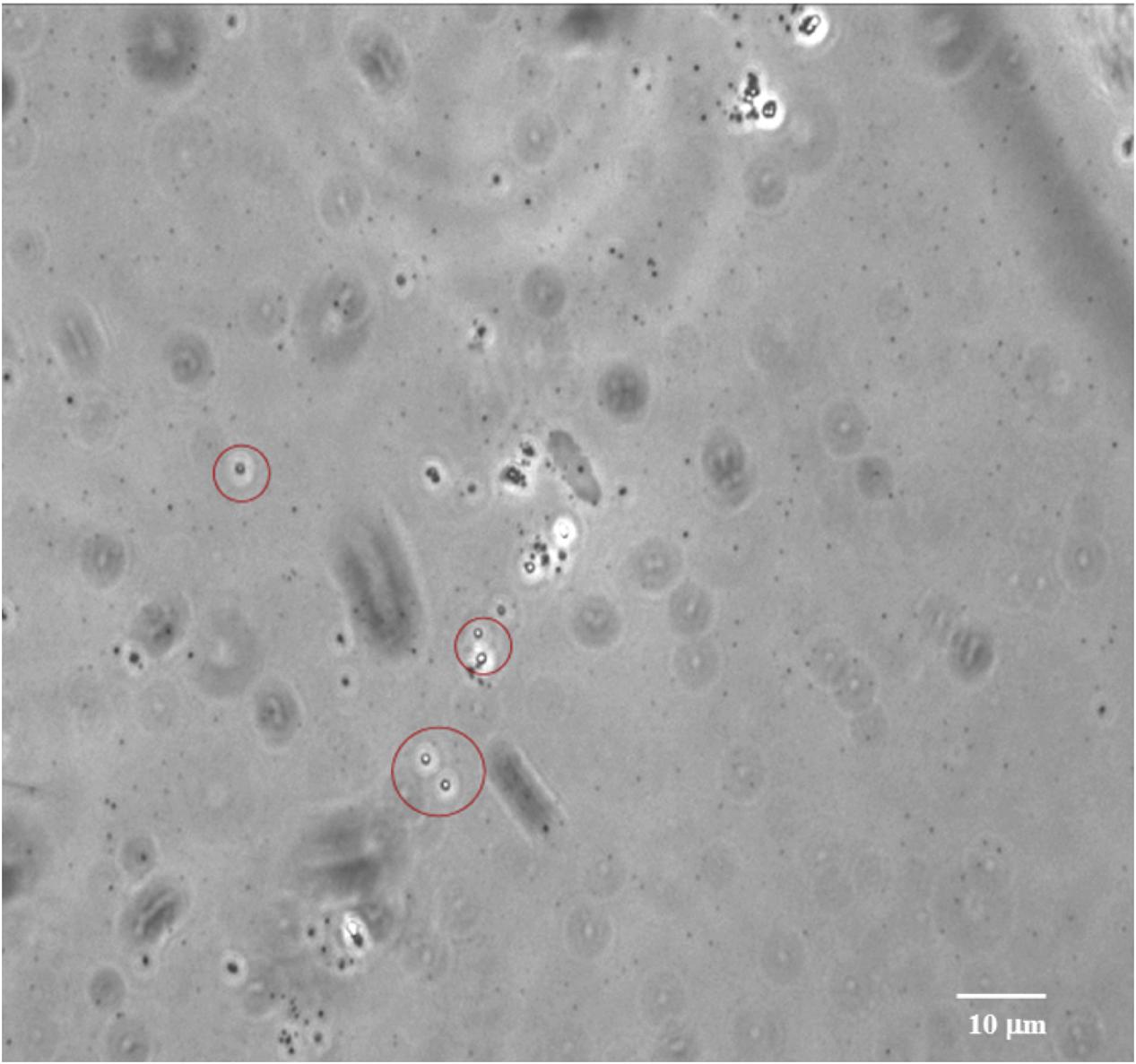
FIGURE 5. Bacterial endospores observed in the subsurface sediment at 2406 mbsf at Site C0020. Image was taken using an optical microscope (Olympus Co., Tokyo, Japan).
Conclusion
Unlike the surface biosphere where gram-negative bacteria are predominant (e.g., Gontang et al., 2007), the deep subseafloor biosphere may host more abundant spore-forming piezophilic bacteria, as demonstrated in this study. Even as spores, the deeply buried quiescent community may have the capacity to impact their surrounding geochemical environment (Hubert et al., 2010), and play an integral role in microbial ecosystem-development as a seed pool in the deeper sedimentary biosphere. It is also clear from this study that bacteria can be readily isolated from the deep subseafloor when a special physiological group (e.g., piezophiles) is targeted and appropriate cultivation methods are applied. Our results underline that the deeply buried microorganisms are still alive and revivable. The continued use of cultivation-dependent approaches will lead to the discovery of additional pressure-adapted gram-positive bacteria and their spores, and provide a direct means to learn more about their adaptation and survival strategies as well as the evolution in the deep subseafloor biosphere.
Author Contributions
JF and CK designed the research. TH, YM, and FI collected the core samples. CK, GR, and YN conducted cultivation experiments. TH and JL performed molecular and electron microscopic analyses. JF and CK wrote the manuscript with significant input from FI. All authors contributed to interpretation of data.
Funding
JF gratefully acknowledges the support by the Trustees’ Scholarly Endeavors Program of Hawaii Pacific University and by the National Natural Science Foundation of China (91328208 and 41373071). This work was supported in part by the Japan Society for the Promotion of Sciences (JSPS) Strategic Fund for Strengthening Leading-Edge Research and Development (to JAMSTEC and FI), the JSPS Funding for Next Generation World-Leading Researchers (no. GR102 to FI), and the JSPS Grants-in-Aid for Science Research (26251041 to FI).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to the Integrated Ocean Drilling Program (IODP) and the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT) for providing an opportunity to explore the deep coal-bed biosphere off Shimokita during Expedition 337. We thank all crews, drilling team members, lab technicians and scientists including the co-chief scientist Kai-Uwe Hinrichs, University of Bremen, Germany, on the drilling vessel Chikyu for supporting core sampling and measurements during Expedition 337. The authors thank Yusuke Kubo, the Expedition Project Manager for Expedition 337, for his coordination of this drilling project.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00137/full#supplementary-material
References
Abecasis, A. B., Serrano, M., Alves, R., Quintais, L., Pereira-Leal, J. B., and Henriques, A. O. (2013). A genomic signature and the identification of new sporulation genes. J. Bacteriol. 195, 2101–2115. doi: 10.1128/JB.02110-12
Allen, E. E., Facciotti, D., and Bartlett, D. H. (1999). Monounsaturated but not polyunsaturated fatty acids are required for growth of the deep-sea bacterium Photobacterium profundum SS9 at high pressure and low temperature. Appl. Environ. Microbiol. 65, 1710–1720.
Aoyagi, T., Kimura, M., Yamada, N., Navarro, R. R., Itoh, H., Ogata, A., et al. (2015). Dynamic transition of chemolithotrophic sulfur-oxidizing bacteria in response to amendment with nitrate in deposited marine sediments. Front. Microbiol. 6:426. doi: 10.3389/fmicb.2015.00426
Bale, S. J., Goodman, K., Rochelle, P. A., Marchesi, J. R., Fry, J. C., Weightman, A. J., et al. (1997). Desulfovibrio profundus sp. nov., a novel barophilic sulfate-reducing bacterium from deep sediment layers in the Japan Sea. Int. J. Syst. Bacteriol. 47, 515–521. doi: 10.1099/00207713-47-2-515
Bartlett, D. H., Kato, C., and Horikoshi, K. (1995). High pressure influences on gene and protein expression. Res. Microbiol. 146, 697–706. doi: 10.1016/0923-2508(96)81066-7
Batzke, A., Engelen, B., Sass, H., and Cypionka, H. (2007). Phylogenetic and physiological diversity of cultured deep-biosphere bacteria from equatorial Pacific Ocean and Peru margin sediments. Geomicrobiol. J. 24, 261–273. doi: 10.1080/01490450701456453
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Huntley, J., Fierer, N., et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624. doi: 10.1038/ismej.2012.8
Ciobanu, M.-C., Burgaud, G., Dufresne, A., Breuker, A., Rédou, V., Maamar, S. B., et al. (2014). Microorganisms persist at record depths in the subseafloor of the Canterbury Basin. ISME J. 8, 1370–1380. doi: 10.1038/ismej.2013.250
DeLong, E. F. (1992). Archaea in coastal marine environments. Proc. Natl. Acad. Sci. U.S.A. 89, 5685–5689. doi: 10.1073/pnas.89.12.5685
Ezaki, T., Hashimoto, Y., and Yabuuchi, E. (1989). Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int. J. Syst. Bacteriol. 39, 224–229. doi: 10.1099/00207713-39-3-224
Fang, J., and Findlay, R. H. (1996). The use of a classic lipid extraction method for simultaneous recovery of organic pollutants and phospholipids. J. Microbiol. Methods 27, 63–71. doi: 10.1016/0167-7012(96)00929-3
Fang, J., Gupta, S. D., Hasiotis, S. T., Brake, S. S., and Bazylinski, D. A. (2007). Microbial community structure of a stromatolite from an acid mine drainage system, implications for the role of microeukaryotes in the formation of ancient Fe-rich stromatolites. Chem. Geol. 243, 191–204. doi: 10.1016/j.chemgeo.2007.06.001
Fang, J., Kato, C., Sato, T., Chan, O., Agarkar, N., and McKay, D. S. (2004). Polyunsaturated fatty acids in piezophilic bacteria: biosynthesis or dietary uptake? Comp. Biochem. Physiol. B 137, 455–461. doi: 10.1016/j.cbpc.2004.01.009
Fang, J., Zhang, L., and Bazylinski, D. A. (2010). The deep-sea piezosphere and piezophiles: geomicrobiology and biogeochemistry. Trends Microbiol. 18, 413–422. doi: 10.1016/j.tim.2010.06.006
Fichtel, J., Koster, J., Rullkotter, J., and Sass, H. (2008). High variations in endospore numbers within tidal flat sediments revealed by quantification of dipicolinic acid. Geomicrobiol. J. 25, 371–380. doi: 10.1080/01490450802402877
Fichtel, J., Logemann, J., Fichtel, J., Rullkötter, J., Cypionka, H., and Engelen, B. (2015). Temperature and pressure adaptation of a sulfate reducer from the deep subsurface. Front. Microbiol. 6:1078. doi: 10.3389/fmicb.2015.01078
Filippidou, S., Wunderlin, T., Junier, T., Jeanneret, N., Dorador, C., Molina, V., et al. (2016). A combination of extreme environmental conditions favor the prevalence of endospore-forming firmicutes. Front. Microbiol. 7:1707. doi: 10.3389/fmicb.2016.01707
Gatson, J. W., Benz, B. F., Chandrasekaran, C., Satomi, M., Venkateswaran, K., and Hart, M. E. (2006). Bacillus tequilensis sp. nov., isolated from a 2000-year-old Mexican shaft-tomb, is closely related to Bacillus subtilis. Int. J. Syst. Evol. Microbiol. 56, 1475–1484. doi: 10.1099/ijs.0.63946-0
Gest, H., and Mandelstam, J. (1987). Longevity of microorganisms in natural environments. Microbiol. Sci. 4, 69–71.
Glombitza, C., Adhikari, R. R., Riedinger, N., Gilhooly, W. P. III, Hinrichs, K.-U., and Inagaki, F. (2016). Microbial sulfate reduction potential in coal-bearing sediments down to ~2.5 km below the seafloor off Shimokita Peninsula, Japan. Front. Microbiol. 7:1576. doi: 10.3389/fmicb.2016.05176
Gontang, E. A., Fenical, W., and Jensen, P. R. (2007). Phylogenetic diversity of gram-positive bacteria cultured from marine sediments. Appl. Environ. Microbiol. 73, 3272–3282. doi: 10.1128/AEM.02811-06
Gross, D., Bachtel, A., and Harrington, G. J. (2015). Variability in coal facies as reflected by organic petrological and geochemical data in Cenozoic coal beds offshore Shimokita (Japan) –IODP Exp. 337. Int. J. Coal Geol. 152, 63–79. doi: 10.1016/j.coal.2015.10.007
Heyndrickx, M., Lebbe, L., Kersters, K., De Vos, P., Forsyth, G., and Logan, N. A. (1998). Virgibacillus: a new genus to accommodate Bacillus pantothenticus (Proom and Knight 1950). Emended description of Virgibacillus pantothenticus. Int. J. Syst. Bacteriol. 48, 99–106. doi: 10.1099/00207713-48-1-99
Hubert, C., Arnosti, C., Brüchert, V., Loy, A., Vandieken, V., and Jørgensen, B. B. (2010). Thermophilic anaerobes in Arctic marine sediments induced to mineralize complex organic matter at high temperature. Environ. Microbiol. 12, 1089–1104. doi: 10.1111/j.1462-2920.2010.02161.x
Inagaki, F., Hinrichs, K.-U., Kubo, Y., Bowles, M. W., Heuer, V. B., Hong, W.-L., et al. (2015). Exploring deep microbial life in coal-bearing sediment down to ~2.5 km below the ocean floor. Science 349, 420–424. doi: 10.1126/science.aaa6882
Inagaki, F., Hinrichs, K.-U., Kubo, Y., and the Expedition 337 Scientists. (2012). Deep coalbed biosphere off Shimokita: microbial processes and hydrocarbon system associated with deeply buried coalbed in the ocean. IODP Prel. Rept. 337. doi: 10.2204/iodp.pr.337.2012
Inagaki, F., Okada, H., Tsapin, A. I., and Nealson, K. H. (2005). Microbial survival: the Paleome: a sedimentary genetic record of past microbial communities. Astrobiology 5, 141–153. doi: 10.1089/ast.2005.5.141
Ishikawa, M., Nakajima, K., Yanagi, M., Yamamoto, Y., and Yamasato, K. (2003). Marinilactibacillus psychrotolerans gen. nov., sp. nov., a halophilic and alkaliphilic marine lactic acid bacterium isolated from marine organisms in temperate and subtropical areas of Japan. Int. J. Syst. Evol. Microbiol. 53, 711–720. doi: 10.1099/ijs.0.02446-0
Itoh, H., Aita, M., Nagayama, A., Meng, X. Y., Kamagata, Y., Navarro, R., et al. (2014). Evidence of environmental and vertical transmission of Burkholderia symbionts in the oriental chinch bug Cavelerius saccharivorus (Heteroptera: Blissidae). Appl. Environ. Microbiol. 80, 5974–5983. doi: 10.1128/AEM.01087-14
Kato, C. (2011). “Cultivation methods for piezophiles,” in Extremophiles Handbook, eds K. Horikoshi, G. Antranikian, A. Bull, F. Robb, and K. Stetter (Tokyo: Springer-Verlag), 719–726.
Kato, C., and Nogi, Y. (2001). Correlation between phylogenetic structure and function: examples from deep-sea Shewanella. FEMS Microbiol. Ecol. 35, 223–230. doi: 10.1111/j.1574-6941.2001.tb00807.x
Kato, C., Sato, T., and Horikoshi, K. (1995). Isolation and properties of barophilic and barotolerant bacteria from deep-sea mud samples. Biodivers. Conserv. 4, 1–9. doi: 10.1007/BF00115311
Kawai, M., Uchiyama, I., Takami, H., and Inagaki1, F. (2015). Low frequency of endospore-specific genes in subseafloor sedimentary metagenomes. Environ. Microbiol. Rep. 7, 1–10. doi: 10.1111/1758-2229.12254
Kimura, M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/BF01731581
Langerhuus, A. T., Røy, H., Lever, M. A., Morono, Y., Inagaki, F., Jørgensen, B. B., et al. (2012). Endospore abundance and D: L-amino acid modeling of bacterial turnover in holocene marine sediment (Aarhus Bay). Geochim. Cosmochim. Acta 99, 87–99. doi: 10.1016/j.gca.2012.09.023
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, M., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Lomstein, B. A., Langerhuus, A. T., D’Hondt, S., Jørgensen, B. B., and Spivack, A. J. (2012). Endospore abundance, microbial growth and necromass turnover in deep sub-seafloor sediment. Nature 484, 101–104. doi: 10.1038/nature10905
Müller, A. L., de Rezende, J. R., Hubert, C. R. J., Kjeldsen, K. U., Lagkouvardos, I., Berry, D., et al. (2014). Endospores of thermophilic bacteria as tracers of microbial dispersal by ocean currents. ISME J. 8, 1153–1165. doi: 10.1038/ismej.2013.225
Navarro, R. R., Aoyagi, T., Kimura, M., Itoh, H., Sato, Y., Kikuchi, Y., et al. (2015). High-resolution dynamics of microbial communities during dissimilatory selenate reduction in Anoxic Soil. Environ. Sci. Technol. 49, 7684–7691. doi: 10.1021/es505210p
Nogi, Y., Kato, C., and Horikoshi, K. (1998). Taxonomic studies of deep-sea barophilic Shewanella strains and description of Shewanella violacea sp. nov. Arch. Microbiol. 170, 331–338. doi: 10.1007/s002030050650
Pedrós-Alió, C. (2007). Ecology. Dipping into the rare biosphere. Science 315, 192–193. doi: 10.1126/science.1135933
Russell, J. A., León-Zayas, R., Wrighton, K., and Biddle, J. (2016). Deep subsurface life from North Pond: enrichment, isolation, characterization and genomes of heterotrophic bacteria. Front. Microbiol. 7:678. doi: 10.3389/fmicb.2016.00678
Saito, H., and Miura, K. (1963). Preparation of transforming deoxyribonucleic acid by phenol treatment. Biochim. Biophys. Acta 72, 612–629. doi: 10.1016/0926-6550(63)90386-4
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Sass, H., and Parkes, R. J. (2011). “Sub-seafloor sediments: an extreme but globally significant prokaryotic habitat (taxonomy, diversity, ecology),” in Extremophiles Handbook, eds K. Horikoshi, G. Antranikian, A. T. Bull, F. T. Robb, and K. O. Stetter (Tokyo: Springer), 1016–1036.
Sogin, M. L., Morrison, H. G., Huber, J. A., Welch, D. M., Huse, S. M., Neal, P. R., et al. (2006). Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc. Natl. Acad. Sci. U.S.A. 103, 12115–12120. doi: 10.1002/9781118010549.ch24
Tamaoka, J., and Komagata, K. (1984). Determination of DNA base composition by reverse-phase high-performance liquid chromatography. FEMS Microbiol. Lett. 25, 125–128. doi: 10.1111/j.1574-6968.1984.tb01388.x
Tanikawa, W., Tadai, O., Morita, S., Lin, W., Yamada, Y., Sanada, Y., et al. (2016). Thermal properties and thermal structure in the deep-water coalbed basin off the Shimokita Peninsula, Japan. Mar. Petroleum Geol. 73, 445–461. doi: 10.1016/j.marpetgeo.2016.03.006
Toffin, T., Zink, K., Kato, C., Pignet, P., Bidault, A., Nadège, B., et al. (2005). Marinilactibacillus piezotolerans sp. nov., a novel marine lactic acid bacterium isolated from deep sub-seafloor sediment of the Nankai Trough. Int. J. Syst. Evol. Microbiol. 55, 345–351. doi: 10.1099/ijs.0.63236-0
Ventura, M., Canchaya, C., Tauch, A., Chandra, G., Fitzgerald, G. F., Chater, K. F., et al. (2007). Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. 71, 495–548. doi: 10.1128/MMBR.00005-07
Vreeland, R. H., Resenzweig, W. D., and Powers, D. W. (2000). Isolation of a 250 million year old halotolerant bacterium from a primary salt crystal. Nature 407, 897–900. doi: 10.1038/35038060
Yayanos, A. A. (1986). Evolutional and ecological implications of the properties of deep-sea barophilic bacteria. Proc. Natl. Acad. Sci. U.S.A. 83, 9542–9546. doi: 10.1073/pnas.83.24.9542
Keywords: deep biosphere, endospore, piezophiles, high pressure, coal beds, marine sediment
Citation: Fang J, Kato C, Runko GM, Nogi Y, Hori T, Li J, Morono Y and Inagaki F (2017) Predominance of Viable Spore-Forming Piezophilic Bacteria in High-Pressure Enrichment Cultures from ~1.5 to 2.4 km-Deep Coal-Bearing Sediments below the Ocean Floor. Front. Microbiol. 8:137. doi: 10.3389/fmicb.2017.00137
Received: 04 November 2016; Accepted: 19 January 2017;
Published: 06 February 2017.
Edited by:
Axel Schippers, Federal Institute for Geosciences and Natural Resources, GermanyReviewed by:
Aude Picard, Harvard University, USACristiana Cravo-Laureau, University of Pau and Pays de l’Adour, France
Copyright © 2017 Fang, Kato, Runko, Nogi, Hori, Li, Morono and Inagaki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chiaki Kato, kato_chi@jamstec.go.jp Jiasong Fang, jfang@hpu.edu
 Jiasong Fang
Jiasong Fang Chiaki Kato
Chiaki Kato Gabriella M. Runko
Gabriella M. Runko Yuichi Nogi
Yuichi Nogi Tomoyuki Hori
Tomoyuki Hori Jiangtao Li
Jiangtao Li Yuki Morono
Yuki Morono Fumio Inagaki
Fumio Inagaki