- 1Department of Genetics, University of Delhi South Campus, New Delhi, India
- 2TERI Deakin Nanobiotechnology Centre, The Energy and Resources Institute, Gurgaon, India
Fluorescent Pseudomonas strain Psd is a soil isolate, possessing multiple plant growth promoting (PGP) properties and biocontrol potential. In addition, the strain also possesses high Zn2+ biosorption capability. In this study, we have investigated the role exopolysaccharides (EPS) play in Zn2+ biosorption. We have identified that alginates are the prime components contributing to Zn2+ biosorption. Deletion of the alg8 gene, which codes for a sub-unit of alginate polymerase, led to a significant reduction in EPS production by the organism. We have also demonstrated that the increased alginate production in response to Zn2+ exposure leads to improved biofilm formation by the strain. In the alg8 deletion mutant, however, biofilm formation was severely compromised. Further, we have studied the functional implications of Zn2+ biosorption by Pseudomonas strain Psd by demonstrating the effect on the PGP and biocontrol potential of the strain.
Introduction
Metals have become integral part of cellular functions, both structural and functional; and have been broadly classified as essential or non-essential metals. While the latter are extremely toxic, the former, including transition elements like Co2+, Mn2+, Ni2+, Fe2+, Cd2+, and Zn2+, can also exert the harmful effects beyond a certain concentration. To tackle this, bacterial systems have evolved a number of mechanisms to maintain a fine-tuned balance between metal deficiency and excess.
Zinc plays an important role as a trace element in organisms belonging to all domains of life, ranging from bacteria to humans, where it is present as the divalent cation (Zn2+; Blindauer, 2015). Zn2+ is unique among transition metals since it is redox inactive under physiological conditions (Mangold et al., 2013). It serves as the cofactor of a large number of the known enzymes (Choudhury and Srivastava, 2001; Blencowe and Morby, 2003; Blindauer, 2015) and is involved in DNA-protein interactions in the form of Zn2+-finger motifs (Matthews and Sunde, 2002). In the context of pathogens, Zn2+ is required for exhibiting full virulence by many pathogenic organisms (Shafeeq et al., 2013). This is the reason why Zn2+ availability is reduced during acute-phase response to a bacterial infection (Corbin et al., 2008; LeGrand and Alcock, 2012). Since Zn2+ is associated with a number of important cellular processes essential for growth and metabolism, its cellular levels need to be maintained in order to ensure proper availability of the metal for various processes, and at the same time preventing the cellular components from the deleterious effects of Zn2+ toxicity. Microbial communities, like all other organisms, have adapted themselves to the metal concentrations encountered by two major strategies, namely avoidance and sequestration. Based on these strategies, the major mechanisms operating in the prokaryotes that maintain cellular Zn2+ concentration can be classified into: (i) physico-chemical interactions (adsorption or biosorption to cell wall and other constituents); (ii) regulated import; (iii) efflux; and (iv) sequestration (Ledin, 2000; Choudhury and Srivastava, 2001; Blencowe and Morby, 2003; Upadhyay and Srivastava, 2014).
Biosorption of metals onto a microbial surface is a function of negatively charged cell surface and is dependent on the surface properties of the cell, such as charge and orientation of metal-binding functional groups, metal speciation and chemistry in aqueous phase (Ledin, 2000). Another important determinant of the biosorption process are the extracellular polymeric substances produced by bacteria, which confer an overall negative charge to the bacterial surface under circumneutral pH conditions, due to the presence of carboxylic and phosphoryl groups (Beveridge, 1988). The main constituents of extracellular polymeric substances include extracellular polysaccharides or exopolysaccharides (EPS), proteins, lipids, and DNA (Flemming and Wingender, 2001; Allesen-Holm et al., 2006). Bacterial EPS are implicated in a number of functions, such as adhesion to substratum, protection against anti-bacterial compounds and binding to organic molecules and inorganic ions (Ma et al., 2009). Additionally, bacterial EPS are also involved in metal adsorption due to the interaction between metal cations and negative functional groups of EPS (Ledin, 2000; Vijayaraghavan and Yun, 2008).
Pseudomonas sp. are known to secrete three major types of EPS, namely alginate, Psl and Pel Conti (Conti et al., 1994; Ma et al., 2009; Franklin et al., 2011; Ghafoor et al., 2011; Yang et al., 2011). Psl is a galactose and mannose-rich polysaccharide, aiding mainly in the initial attachment and mature biofilm formation (Ma et al., 2009). Generally produced during planktonic growth, this EPS mediates attachment to surfaces and formation of micro-colonies. The other polysaccharide, Pel, is a glucose-rich cellulose-like polymer required for pellicle formation at air-liquid interface (Friedman and Kolter, 2004). Alginates are linear EPS consisting of β-1,4-linked β-D-mannuronic acid and its C5 epimer α-L-guluronic acid (Remminghorst and Rehm, 2006). Produced only by two bacterial genera of Pseudomonas and Azotobacter, these EPS are responsible for a mucoid colony phenotype and are also the premier components of bacterial biofilms (Sutherland, 2001; Ghafoor et al., 2011; Whitfield et al., 2015). The alginate biosynthesis operon consists of 12 genes (algD, alg8, alg44, algK, algE, algG, algX, algL, algI, algJ, algF, and algA; Figure S1) under tight control of the promoter upstream of algD (Remminghorst and Rehm, 2006).
A number of pseudomonads, including Pseudomonas aeruginosa, P. syringae, Pseudomonas putida, and Pseudomonas fluorescens, serve as dominant members of Plant Growth Promoting (PGP) bacteria (Vessey, 2003; Couillerot et al., 2009; Bhattacharyya and Jha, 2012). In this context, EPS production and the biofilm formation is expected to play an important role in root colonization and also contribute to the rhizosphere competence of these bacteria. Fluorescent Pseudomonas strain Psd is a rhizosphere isolate, possessing multiple PGP properties and biocontrol potential (Upadhyay and Srivastava, 2008, 2010; Kochar et al., 2011). Besides, we have earlier demonstrated that this strain possesses high resistance toward Zn2+, which emanates from extracellular biosorption (Upadhyay and Srivastava, 2014). We have also established that Zn2+ biosorption is coupled with an increase in the total EPS production. In this communication, we dissect this aspect further and identify the key players that mediate the biosorption process. Since the strain in hand is a PGP bacterium, effect of Zn2+ biosorption and alginate production on the PGP potential of the strain is also demonstrated. These observations may provide important leads in ascertaining if the strain could continue to provide its beneficial effects even in soils contaminated with high levels of Zn2+ and be applied as a bioinoculant.
Materials and Methods
Organism, Culture Conditions, and Chemicals
Fluorescent Pseudomonas strain Psd, isolated from the roots of Vigna mungo, has been characterized as mentioned before (Upadhyay and Srivastava, 2008). The strain was maintained on Gluconate Minimal medium (GMM; Gilotra and Srivastava, 1997) with and without ZnSO4.7H2O. As per the experimental requirement, the media was supplemented with the appropriate concentration of autoclaved metal salt solution (ZnSO4.7H2O). The shake cultures were raised on Controlled Environment Shaker Incubator (Kühner, Switzerland) at 200 rpm at 30°C for the required period of time. Growth was determined turbidometrically at 600 nm, as per the protocol described earlier (Upadhyay and Srivastava, 2014). All chemicals used in the study were of analytical grade and purchased from Sigma Aldrich (St. Louis, MO, USA). All the other strains and plasmids used in this study along with their relevant characteristics are listed in Table S1.
Isolation and Purification of Extracellular Polysaccharides
EPS extraction was carried from 5 day-spent culture filtrate of strain Psd grown at different Zn2+ concentrations in GMM by the method described earlier (Bitton and Freihofer, 1978). Briefly, 2 volumes of 95% ethyl alcohol was added to the culture supernatant and was kept at 4°C overnight. The resulting precipitate was recovered by centrifugation at 8,000 × g for 10 min. The precipitate was dissolved in sterile distilled H2O and dialyzed at 4°C against distilled H2O for desalting. The purified EPS was concentrated under vacuum and stored at 4°C until use.
Transmission Electron Microscopy
Cells grown at different Zn2+ concentrations were analyzed for the ultra-structural changes by Transmission Electron Microscopy (TEM). Pellets of sedimented cells, washed in saline, were fixed with 2.5% gluteraldehyde in 0.1 M phosphate buffer for 6 h at 4°C, and washed thrice with the same buffer. Dehydration was carried out in a graded ascending series of acetone followed by toluene. Specimens were infiltrated with 3:1 (v/v) mixture of toluene and Araldite [50% Epoxyresin + 50% Dodecenyl succinic anhydride (DDSA)] for 2 h, followed by pure Araldite, and were finally embedded in beam capsule with Araldite + Accelerator [(Trimethyl aminomethyl phenol (DMP-30)] and polymerized at 50°C for 24 h and 60°C for 48 h. Thin sections were cut with an Ultracut Microtome E (Ultracut E, Riechert Jung, Germany) and stained with uranyl acetate and lead citrate. The sections were analyzed in Transmission electron Microscope (FEI Electron Optics, USA).
Fourier-Transformed Infrared Spectroscopy
Fourier-Transformed Infrared (FT-IR) spectrum of purified EPS was recorded to elucidate the chemical binding environment of Zn2+. Analyses were performed on an FT-IR spectrophotometer (Tensor 37, Bruker Optics, USA), equipped with total attenuated reflectance (ATR) objective. The purified EPS from strain Psd grown at varying Zn2+ concentrations was scanned over a wave number range of 4,000–1,000 cm−1 with a resolution of 4 cm−1. Samples were placed over a Zn-Se crystal and the peak emanating from the crystal was obtained between wave number 2400–2200 cm−1. Background spectrum of water was collected and normalized prior to measurement of samples. For each sample, 16 scans were collected in order to evaluate the heterogeneity in the sample. All the spectra obtained were smoothened and their baseline was corrected.
Expression Analysis
Total RNA Extraction and cDNA Preparation
Total RNA was isolated from bacterial cultures with the RNeasy Protect Bacteria Mini Kit for total RNA purification (Qiagen, Netherlands), as per manufacturer's instructions. Traces of DNA were removed from RNA preparations using DNase I, (Amplification Grade, Sigma Aldrich). DNase-free RNA (1 μg) was used in a one-step RT-PCR reaction (RevertAid First Strand cDNA Synthesis Kit, Thermo Scientific, USA) performed with the universal hexamer primers provided with the kit. The cDNA was used as a template for semi-quantitative RT-PCR. The primers used are listed in Table 1.
Quantitative PCR
The cDNA prepared as mentioned above was subjected to quantitative PCR (qPCR) using SYBR GREEN reaction mix in a 7900HT Fast Real-Time PCR System (Applied Biosystems, USA). Thermal cycling conditions were as follows: 10 min at 95°C followed by 40 repeats of 30 s at 95°C, 30 s at 58°C. Following PCR amplification, the reactions were subjected to temperature ramping to create the dissociation curve, measured in terms of changes in fluorescence intensity as a function of temperature, by which non-specific products can be detected. The dissociation program was 95°C for 1 min, 60°C for 10 s, and 95°C for 30 s. The experiment included three biological replicates and each biological replicate was evaluated by three technical replicates. The constitutively expressed gene of Pseudomonas sp., for 16s rDNA, was taken as the calibrator in order to normalize gene expression levels. The MIQE guidelines (Bustin et al., 2009) for qPCR recommend the more generic quantification cycle (Cq). However, as Ct and Cq are used interchangeably and refer to the same value, Ct was used in this study. The Ct-values obtained in the experiment were used for quantification of relative change in gene expression, by the 2−ΔΔCt method (Livak and Schmittgen, 2001).
Alginate Quantification
Alginate content in EPS was estimated as per the protocol described by Richardson et al. (2004). Briefly, to 1 mL of purified EPS, 1 mL of 0.8 M NaOH was added and neutralized with 120 μL 2.25 M citric acid after 5 min of incubation. To this, 40 μL of DMMB (1,9-dimethyl methylene blue) was added. The solution was vortexed vigorously and incubated at room temperature for 45 min. Thereafter, UV-visible spectrum of the samples was recorded between 500 and 700 nm using microtiter plate reader (Ultramark, Bioplate Imaging system, Biorad, USA). The absorbance intensities at 520 and 650 nm, signifying alginate-bound and unbound DMMB were used to estimate the respective alginate concentrations.
Generation of alg8 Knockout Mutant
The alg8 gene knockout strategy involved gene replacement by homologous recombination, wherein the native alg8 gene was replaced with an antibiotic resistance (kanr) cassette. For homologous recombination, a construct consisting of kanr gene flanked by the regions upstream and downstream to alg8 gene was generated, providing homology, as shown in Figure S2A. To assemble this construct, primers were designed to amplify ~500 bp upstream and downstream regions of alg8 from strain Psd, as XhoI/HindIII and PstI/BamHI fragments, respectively. The primer sequences have been mentioned in Table 1. These fragments were sequentially cloned in pBlueScript (pBKS+) vector, using the aforementioned restriction sites to generate a construct termed as pBKS_alg8_up/down. Subsequently, kanr, was cloned between the upstream and downstream regions. The recombinant plasmid, pBKSKmΔAp (~3.3 kb) carrying kanamycin resistance gene, kanr (~800 bp) cloned in SacI restriction site, served as the source of antibiotic resistance gene. The vector construct, pBKS_alg8_up/down was digested with EcoRV to generate a blunt-ended linear DNA fragment. Both of these fragments were then ligated and transformed into E. coli XL1-Blue to generate a construct carrying kanr gene flanked by regions upstream and downstream to alg8 (pBKS_alg8_up/kan/down). Screening of transformants was done on LB Agar supplemented with 50 μg mL−1 of kanamycin. This was followed by plasmid isolation and PCR amplification of three regions, viz. upstream, downstream and kanr using their respective primers. Amplification profile confirmed successful cloning of these regions in pBKS+ (Figure S2C). One of the positive constructs was transferred to strain Psd by electroporation and putative transformants were selected on kanamycin-containing agar medium. Due to the narrow host-range of pBKS+, the construct pBKS_alg8_up/kan/down was less likely to be maintained as a plasmid in strain Psd. Hence, growth of clones on kanamycin-supplemented medium is explained by the integration of kanr cassette in place of alg8 gene through homologous recombination (Figure S2B). In order to confirm stable insertion of kanr cassette, positive transformants were transferred to non-selective conditions (without kanamycin) and grown for a few generations. Transformants with a stable integration were able to resist kanamycin when transferred back to the selection medium containing kanamycin.
Assessment of Plant-Growth Promoting and Biocontrol Parameters
Assessment of phosphate solubilization was carried out as per the protocol of Pikovskaya (1948). Extraction and detection of IAA was carried out using Ultra Performance Liquid Chromatography (UPLC), according to the modified protocol of Malhotra and Srivastava (2008). The secretion of siderophores was observed by spectral analysis at 400 nm (Schwyn and Neilands, 1987). Phenazine was extracted and quantified by the method described by Whistler and Pierson (2003). The antifungal activity of the strain was tested against two known plant pathogenic fungi, namely, Fusarium oxysporum and Fusarium graminearum by the following assays:
(a) Dual culture assay
For dual culture assay, the Potato Dextrose Agar (PDA) plate was divided in two-halves. Inhibition assays were carried out by placing an agar-block from fully grown fungal plate on one half of the plate and streaking the biocontrol bacterium on the other half. The plates were incubated at 28°C for 5-days. Fungal inhibition was obtained as a zone of clearance on the plate in the area where bacteria were growing.
The plates with bacterial growth and fungal block alone were taken as reference.
(b) Biomass inhibition
For quantitative evaluation of antifungal activity, ~106 spores of the fungi, F. oxysporum and F. graminearum were inoculated in PD medium diluted with bacterial culture extract in 1:1 ratio. The flasks were incubated at 28°C, 120 rpm for 4-days. Antifungal nature was demonstrated by comparing the percent dry weight of the treated fungal biomass in comparison to the untreated control. All untreated controls were grown in 50% diluted PD medium with standard succinate medium (SSM).
Biofilm Formation
Biofilm formation was measured using microtiter plate biofilm assay (Merritt et al., 2005). Briefly, 1% of the overnight grown cultures were subcultured in 10 mL GMM supplemented with increasing Zn2+ concentrations (0, 1, 2, 5 mM). From this, 100 μL culture was pipetted in a fresh 96-well microtiter plate. The plate was covered and incubated at 30°C for 48 h under static conditions. After incubation, the wells were washed thoroughly with distilled H2O to remove planktonic bacteria. Following this, 125 μL of crystal violet stain (0.1%) was added to each well. The stain was removed after 10 min of incubation at room temperature and plates were allowed to air-dry. Thereafter, 200 μL of 95% ethyl alcohol was added to each stained well in order to solubilize the dye. The contents of each well were mixed well and 125 μL of the crystal violet/ethyl alcohol solution was transferred to a fresh microtiter plate. The absorbance at 560 nm was measured using a microtiter plate reader (Ultramark, Bioplate Imaging system, Biorad, USA). All values were normalized with cellular OD600.
Bacterial Inoculation of Seeds
Wheat (Triticum aestivum var. HD2851, IARI, New Delhi) seeds were surface sterilized with 0.1% (w/v) HgCl2 for 5 min and washed thoroughly with sterile distilled water. For bacterial inoculation, stationary phase bacterial cells, raised in GMM supplemented with different Zn2+ concentrations, were washed twice with saline to remove the residual medium and re-suspended in saline at a cell density of ~108 cfu mL−1. To this suspension, 10 surface-sterilized wheat seeds were added and flasks were incubated at 30°C, 70 rpm for 2 h. After this treatment, seeds were thoroughly rinsed with sterile water. Seeds were sown in pots and growth of seedlings was monitored for 6-days after treatment.
Confocal Laser Scanning Microscopy
Confocal laser scanning microscopy of root samples from 6-day-old plantlets was performed according to the modified protocol by Bianciotto et al. (2001). Briefly, the root samples from 6-day-old plantlets were excised and washed with 100 mM cacodylate buffer (pH 7.40). Thereafter, the samples were stained with LIVE/DEAD® BacLight™ Bacterial Viability Kit (ThermoFisher Scientific, USA) for 25 min at room temperature. The kit contains a mixture of SYTO 9 and propidium iodide stains, with excitation/emission maxima of 480/500 and 490/635 nm, respectively. After incubation at room temperature for 15 min, samples were mounted on clean glass slides using the mounting oil provided in the kit and observed under Leica TCS SP5 confocal microscope (Leica Microsystems, Germany).
Scanning Electron Microscopy
Scanning electron microscopy (SEM) of root samples from 6-day-old plantlets was performed as described by Koul et al. (2015). Briefly, root samples (1–1.5 cm) were fixed with 2.5% gluteraldehyde in 0.1 M phosphate buffer for 6 h at 4°C. Following this, post-fixation was done with osmium tetraoxide (1% v/v in 0.1 M phosphate buffer) for 30 min. Thereafter, the specimens were washed thrice with milliQ water and treated with 2% (w/v) uranyl acetate for 40 min. The samples were dehydrated through a graded ethanol series (30–100%) followed by acetone (100%). The treated specimens were mounted on aluminum stubs, coated with gold-palladium and examined under a scanning electron microscope (EVO MA10, Carl Zeiss).
Zn2+ Estimation
Zn2+ content was estimated using atomic absorption spectrophotometer (Perkin Elemer model AAnalyst400) at 219.86 nm, as described earlier (Upadhyay and Srivastava, 2014). Flow diagram of the detailed protocol is shown in Figure S3.
Statistical Analysis
All bacterial culture experiments were carried out in three independent sets, each consisting of three replicates. Values shown here represent mean ± standard deviation (SD). All bacterial inoculation experiments were carried out in three independent sets containing 10 seeds each and values shown are mean ± SD. Data were tested at a significance level of P < 0.05 using one-way ANOVA followed by Dunnett's t-test and expressed as mean ± SD.
Results
Zn2+ Accumulation Induces Ultrastructural Changes
Fluorescent Pseudomonas strain Psd can sustain an external Zn2+ concentration of 5 mM (Upadhyay and Srivastava, 2014). We have used transmission electron microscopic (TEM) analysis to monitor the changes induced by Zn2+ accumulation by strain Psd. In comparison to cells growing without extra Zn2+ supplementation in the medium (Figure 1A), cells exposed to increasing Zn2+ concentration displayed thickening of the outer membrane, which pointed toward extracellular biosorption of Zn2+ on to the cell surface (Figures 1B,C). In addition to the extracellular accumulation, some electron dense aggregates (EDA) were also found in the cytoplasm of the cells. Although most of the Zn2+ is located on the outer surface, internally cytoplasmic granules may provide additional sites for deposition of the heavy metal.
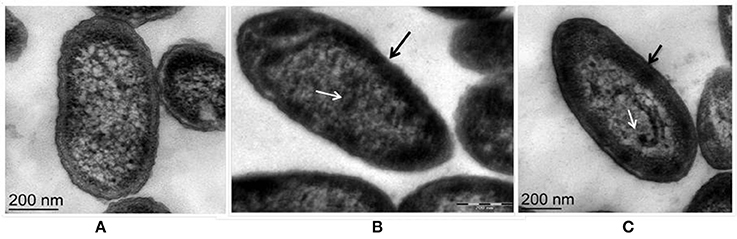
Figure 1. Transmission Electron Microscopy images of Pseudomonas strain Psd exposed to varying Zn2+ concentrations in GMM for 24 h. (A) Untreated control; Cells exposed to (B) 2 mM Zn2+ and (C) 5 mM Zn2+, respectively. Black arrows indicate thickening of cell wall due to metal biosorption. White arrows indicate intracellular accumulation by cytoplasmic granules (scale bar = 200 nm).
FT-IR Spectrum of the EPS Revealed the Presence of Characteristic Functional Groups
For the qualitative analysis of functional groups facilitating Zn2+ biosorption, EPS from the cells of strain Psd grown at increasing Zn2+ concentrations was subjected to FT-IR spectroscopy. The presence of functional groups such as C-O and C-O-C, signified by stretching between 1,200 and 1,000 cm−1 and O-H, signified by elongation between 3,700 and 3,200 cm−1, was indicative of the presence of carbohydrates, which is the major component of the EPS biopolymer. Sharp peaks in the range of 1,125 to 1,000 cm−1 indicated toward the presence of uronic acids in the EPS. The spectrum also revealed the presence of mannose in the EPS, signified by a peak at ~2,900 cm−1. The mannose peak, however, was weaker in comparison to the uronic acids. Besides, the peaks in the range of 1,210 to 1,140 cm−1 revealed the presence of phosphoryl groups in the EPS. The presence of protein in the EPS was also implicated. Generally, proteins are detected by C = O (Amide I) stretching between 1,680 and 1,630 cm−1, N-H bending vibration (Amide II) between 1,650 and 1,550 cm−1 and N–H stretching (Amide A) vibration between 3,290 and 3,300. Peaks corresponding to Amide I and Amide II bonds were observed in untreated control and cells exposed to 2 mM Zn2+ (Figures 2A,B). In the cells exposed to 5 mM Zn2+, however, only peak corresponding to Amide I bond was observed (Figure 2C).
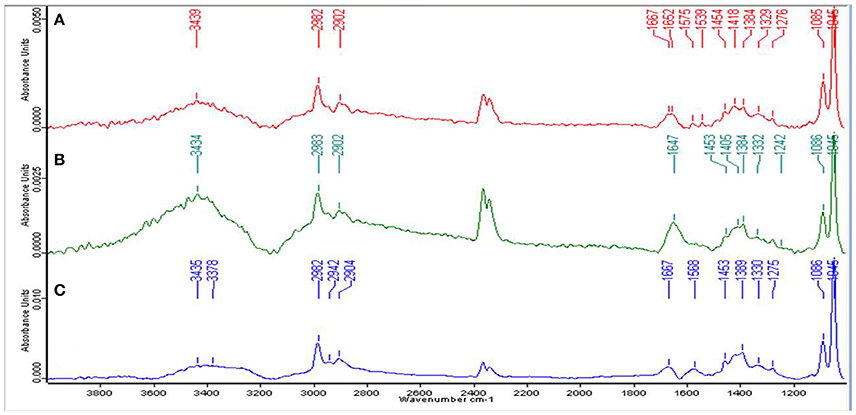
Figure 2. Fourier-transformed infrared (FTIR) spectrum of the exopolysaccharide produced by Psd grown in the presence of varying Zn2+ concentrations in Gluconate Minimal Medium. (A) Untreated control; Cells grown in the presence of (B) 2 mM Zn2+, and (C) 5 mM Zn2+, respectively.
Zn2+ Replete Conditions Affect alg8 Expression
Based on the identification of uronic acids and mannose by FT-IR spectroscopy, the relative expression of two genes, alg8 and pslA, involved in biosynthesis of two principle exopolysaccharides, alginate and Psl, were studied in strain Psd exposed to varying Zn2+ concentrations, by real-time PCR using the gene encoding 16s rRNA as the reference gene or calibrator. The alg8 gene codes for alginate polymerase and pslA, encoding a sugar transferase, is responsible for the production of the mannose and galactose-rich Psl polysaccharide. When compared to the untreated control, a significant up-regulation was obtained in alg8 expression in the presence of Zn2+ (Figures 3A,B). Cells exposed to 5 mM Zn2+ displayed 8-fold increase in alg8 expression when compared to control or untreated cells (Figure 3B). On the other hand, no significant changes in the expression of pslA were obtained between control and Zn2+—grown cells (Figures 3A,C).
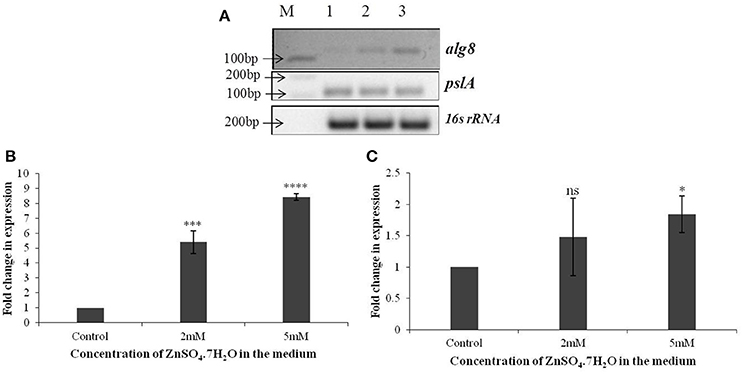
Figure 3. (A) Semi-quantitative-RT PCR of alg8 and pslA involved in exopolysaccharide biosynthesis in Pseudomonas strain Psd exposed to varying Zn2+ concentrations (M-1 kb DNA ladder, Lane 1 represents strain Psd grown in the absence of added Zn2+; Lanes 2 and 3 represent strain Psd grown at 2 and 5 mM Zn2+, respectively); (B) Fold-change in the expression of alg8 in strain Psd in response to exposure to increasing Zn2+ concentrations; (C) Fold-change in the expression of pslA in strain Psd in response to exposure to increasing Zn2+ concentrations. *P < 0.05; ***P < 0.001; ****P < 0.0001; ns, not significant.
Zn2+ Accumulation Is Coupled with Higher Alginate Production
Based on the above results, it was concluded that alginate is the principle component of EPS produced by strain Psd as a direct correlation with its exposure to varying concentrations of Zn2+, and, thus may be involved in Zn2+ biosorption. To confirm this, spectrophotometric quantification of alginates, based on 1,9-dimethyl methylene blue (DMMB) complexation was used to estimate the amount of alginates in the EPS produced by the bacterial cells growing in different Zn2+ concentrations. As shown in Figure S4, strain Psd was able to produce 0.6 mg/mL of alginates after 5-days of incubation. In the presence of Zn2+, up to 3-fold increase was recorded in the amount of alginate produced. This result complied with the earlier hypothesis of the involvement of alginate in Zn2+ biosorption by strain Psd. Further evidence for the same was provided by generation of an alg8 knockout of strain Psd.
Deletion of alg8 Reduces Zn2+ Biosorption Potential
In order to ascertain the involvement of alg8 during Zn2+ biosorption by strain Psd, it was important to generate a mutant strain devoid of this function. The alg8 gene knockout strategy involved gene replacement by homologous recombination, wherein the native alg8 gene was replaced with an antibiotic resistance (kanr) cassette (Figure S2).
The mutant strain Psd Δalg8::kan had a similar growth profile as the wild-type strain Psd in the absence of Zn2+. This showed that growth of the mutant strain was not affected by insertion of the kanamycin cassette. Supplementation of medium with 2 mM Zn2+, however, led to up to 20% drop in cell viability of the mutant. The survival further declined with the increase in Zn2+ concentration to 5 mM, wherein, 44% decrease in cell survival was obtained (Figure 4A). Besides, alg8 deletion resulted in 80% decrease in the levels of EPS synthesized at external Zn2+ concentrations of 2 mM, which increased to 97% at 5 mM Zn2+ (Figure 4B). Further, since the EPS production in the alg8-negative mutant was compromised, Zn2+ accumulation by the mutant was compared with that of the wild-type strain. For this purpose, the mutant strain (Psd Δalg8::kan) was grown in GMM supplemented with different Zn2+ concentrations (1, 2, 5 mM) for 24 h, along with a set containing no Zn2+. Wild-type strain Psd was used as control. It was observed that besides a reduced EPS secretion, the deletion of alg8 also led to a drastic decrease in Zn2+ accumulation potential of the strain (Figure 4C).
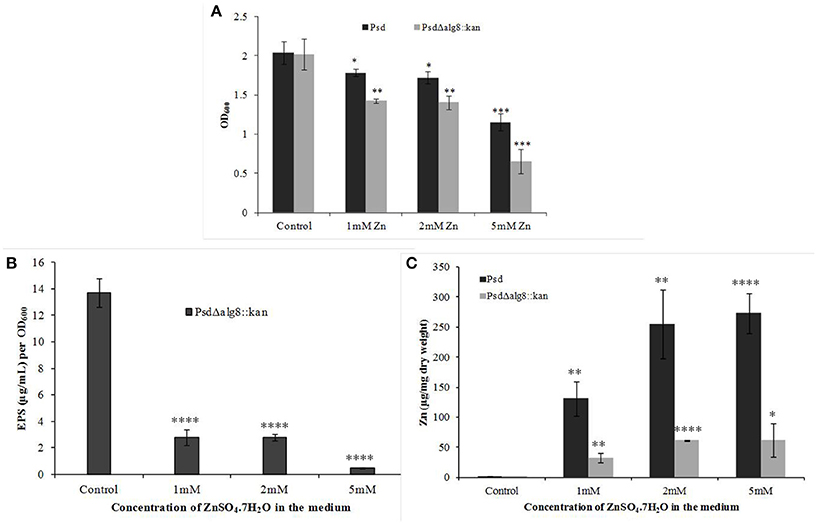
Figure 4. Effect of deletion of alg8 on (A) Net growth (B) EPS production and (C) Zn2+ accumulation by strain Psd (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
Morphological examination of the mutant cells through TEM revealed a comparatively thin outer membrane along with alterations in membrane architecture, supporting the above observations. There was an evident reduction in the cell wall thickening in the untreated control cells as compared to the wild-type strain (Figure 5A, also refer Figure 1). Further, exposure of cells to higher Zn2+ concentrations did not show traces of extracellular accumulation of Zn2+ (Figure 5B). However, cytoplasmic granules were observed in the presence of extracellular Zn2+, indicating that intracellular Zn2+ entry was not affected (Figure 5B). This was suggestive of a regulated intracellular entry of Zn2+ in the strain. When EPS is present, the cell is able to keep the high concentration of Zn2+ outside. Taken together, all these observations supported the fact that alg8 plays an important role in EPS production by strain Psd and is, also, the primary component involved in Zn2+ biosorption by the strain.
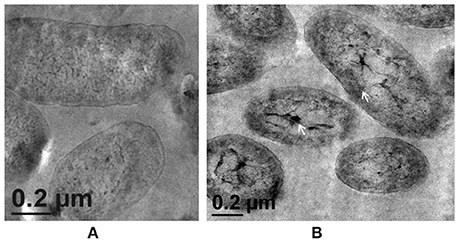
Figure 5. Transmission Electron Microscopy images of strain PsdΔalg8::kan exposed to varying Zn2+ concentrations in Gluconate Minimal Medium. (A) Untreated control; (B) Cells exposed 5 mM Zn2+.The samples were taken after 24 h of growth in Gluconate Minimal Medium. White arrows indicate the intracellular Zn2+ accumulation in the form of cytoplasmic granules (scale bar = 200 nm).
Biofilm Formation and In vitro Root Colonization
Formation of static biofilms was analyzed by crystal violet binding assay. As shown in Figure S5, increased Zn2+ concentration aided in biofilm formation by the strain. This was in accordance with the earlier observation that Zn2+ biosorption led to increased EPS biosynthesis by the strain. On the other hand, PsdΔalg8::kan produced significantly low amount of biofilm as compared to wild-type. This could be attributed to compromised EPS production by the mutant.
In the context of PGP bacteria, biofilm formation aids in effective root colonization, thus playing a crucial role in plant-microbe interactions. The association of strain Psd and its alg8 deletion variant with 6-day-old plantlets of T. aestivum was visualized by means of confocal microscopy, followed by SEM. As shown in Figures 6A, 7A, no bacteria were observed on the control roots. On the other hand, the wild-type strain Psd was able to associate with the roots (Figure 6B), and form thick biofilms associated with EPS matrix (Figure 7B). Additionally, Zn2+ accumulation also aided formation of bacterial aggregates and biofilm (Figures 6C,D, 7C). In case of the mutant strain, however, biofilm formation was drastically reduced, and mostly planktonic cells were observed on the roots (Figures 6E,F, 7D,E). In certain cases, however, presence of bacterial clusters was detected (Figure 7E).
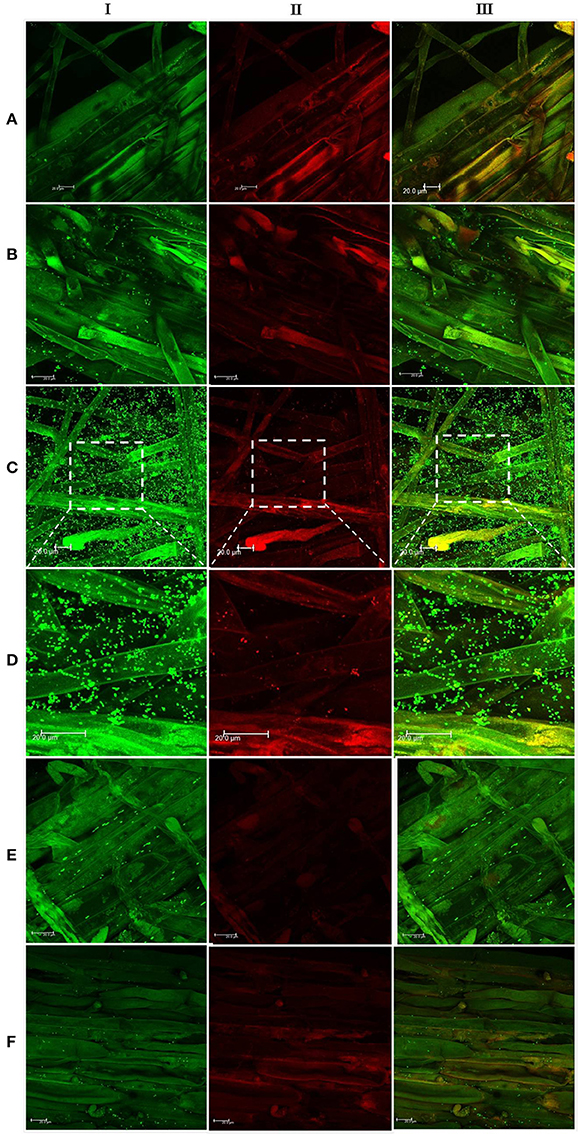
Figure 6. Confocal laser scanning microscopy images of roots from 6-day-old plantlets of Triticum aestivum in the presence of strain Psd and its alg8 deletion mutant. Untreated control (A); roots treated with wild type strain Psd grown in the absence of Zn2+ (B); roots treated with wild type strain Psd grown in the presence of 2 mM Zn2+ (C); enlarged section of panel C (D); roots treated with PsdΔalg8::kan grown in the absence of Zn2+; (E) roots treated with PsdΔalg8::kan grown in the presence of 2 mM Zn2+ (F). The columns I, II, and III represent Syto 9, propidium iodide stained and merged versions of samples. Bacteria are visible as small green dots on the root surface. Scale bar = 20 μm.
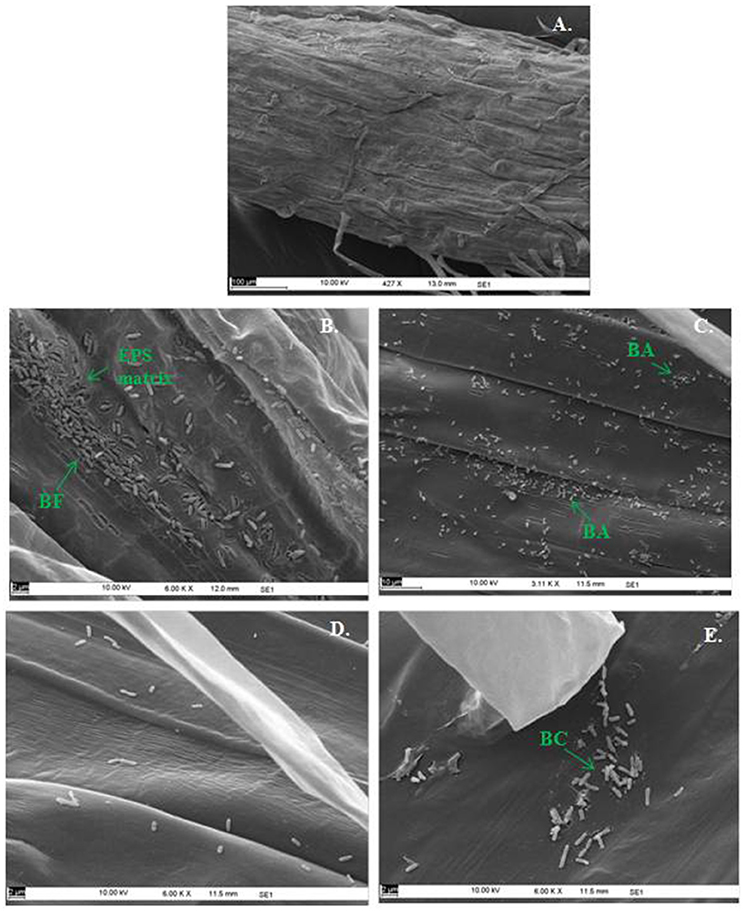
Figure 7. Scanning Electron Microscopy of Triticum aestivum roots inoculated with strain Psd and its alg8 deletion mutant to show the effect on roots: Untreated control (A), roots treated with wild type strain Psd grown in the absence of Zn2+; (B) roots treated with wild type strain Psd grown in the presence of 2 mM Zn2+ (C), roots treated with PsdΔalg8::kan grown in the absence of Zn2+; (D) roots treated with PsdΔalg8::kan grown in the presence of 2 mM Zn2+ (E). BF, Biofilm; BC, Bacterial Clusters; BA, Bacterial aggregates. Scale bars vary from 2 to 100 μm and are shown in each image.
Effect of alg8 Deletion on PGP Potential, Biocontrol Activity, and Antifungal Potential
In continuation with the observation that alg8 deletion severely affects the biofilm formation by the strain Psd, we were interested in observing its subsequent effect on the PGP potential. Increased accumulation of Zn2+ by strain Psd interestingly led to an increase in the phosphate-solubilization and siderophore production by the strain (Figures 8A,B). On the other hand, in strain Psd Δalg8::kan, both these activities were severely compromised. A different response was observed during the assessment of IAA production. In the absence of added Zn2+ in the medium, a 24 h-grown culture of strain Psd produced 164 μg IAA/OD600. With increase in medium Zn2+ concentration, however, there was a significant drop in the amount of IAA produced. Strain Psd grown in the presence of 2 and 5 mM Zn2+ concentrations produced ~70% less IAA than the control. The mutant PsdΔalg8::kan produced ~67% less IAA than the wild-type strain Psd in the absence of Zn2+ (Figure 8C). Zn2+ supplementation, however, did not have a significant effect on IAA produced by the mutant. Increased Zn2+ accumulation also had a stimulatory effect on phenazine production by strain Psd (Figure 8D). This indicated that Zn2+ accumulation by strain Psd will enhance its biocontrol activity. On the other hand, in PsdΔalg8::kan, no phenazine production was detected.
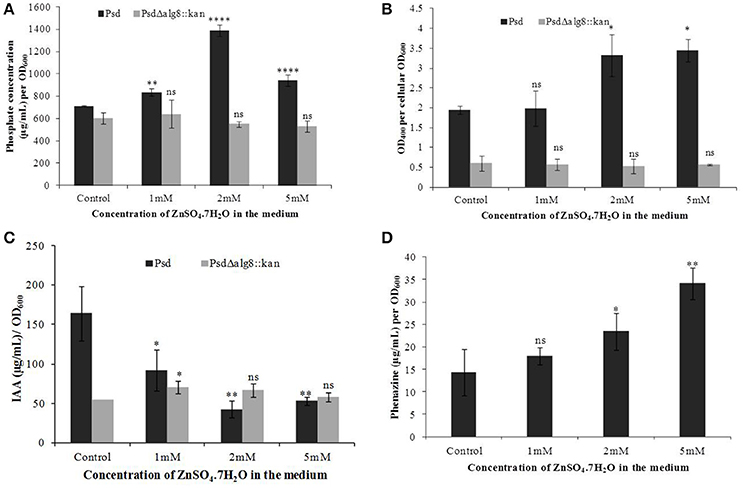
Figure 8. Quantification of (A) Phosphate solubilization (B) Siderophore production, (C) IAA production, and (D) Phenazine biosynthesis by Psd and PsdΔalg8::kan in GMM supplemented with increasing Zn2+ concentrations. *P < 0.05, ***P < 0.001, ****P < 0.0001, ns, not significant.
The secondary metabolites produced by Pseudomonas sp. contribute to their biocontrol properties by inhibition of phytopathogens. Strain Psd showed an increase in the production of siderophores and phenazine in the presence of added Zn2+ in the medium, suggesting that Zn2+ modulates the production of these crucial secondary metabolites. On the contrary, deletion of alg8 resulted in decreased production of Fe3+-chelating siderophores by the strain. Additionally, phenazine production was also hampered in the alg8 deletion mutant. The major implication of these differences was reflected in the antifungal properties of strain Psd and its alg8-negative variant. Antifungal assays to test the ability of the strains to inhibit two phytopathogenic fungi, F. oxysporum, F. graminearum, comprised dual-culture tests followed by biomass inhibition experiments. Strain Psd was able to inhibit the growth of both the fungal strains growing on PDA (Figure 9A). In contrast, PsdΔalg8::kan showed inhibition of only F. oxyporum to some extent. No inhibition, however, was obtained in the case of F. graminearum. This clearly indicated that both strains differed in their antifungal spectrum, and can be explained by the fact that these Pseudomonads are known to produce a variety of antifungal metabolites. To corroborate the above observation, inhibition of fungal biomass was studied. The culture filtrate from wild-type strain Psd led to 70 and 90% reduction in biomass of F. oxysporum and F. graminearum, respectively. The potential of strain Psd as an antifungal agent was supported by the observation that the culture filtrate was able to cause 70 and 90% reduction in biomass of F. oxysporum and F. graminearum, respectively (Figures 9B,C). Inhibition by strain Psd grown in the presence of added Zn2+ in the medium increased up to 80% in case of F. oxysporum (Figure 9B). No significant difference, however, was obtained in case of F. graminearum (Figure 9C). In contrast, inhibition by PsdΔalg8::kan reduced to 60% in F. oxysporum (Figure 9B). The inhibition efficiency declined further with increase in Zn2+ concentration of the medium, with 40% inhibition at 5 mM Zn2+. The mutant strain was not able to inhibit F. graminearum at all (Figure 9C).
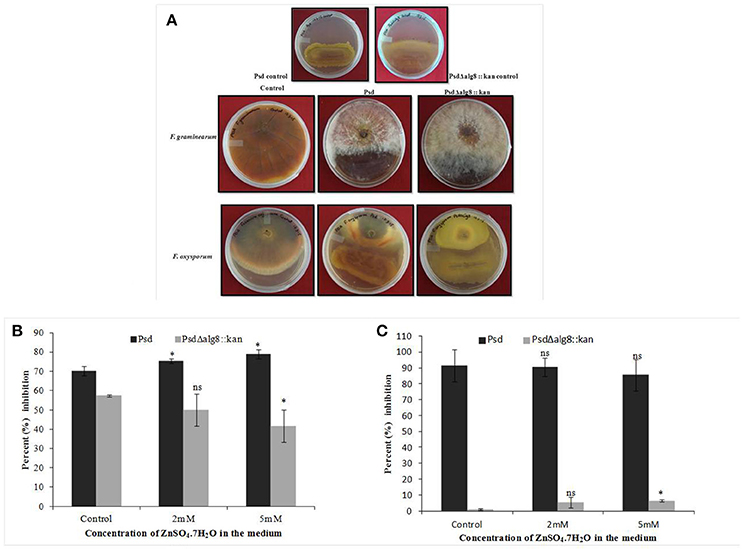
Figure 9. (A) Dual culture assay to show inhibition of Fusarium graminearum and F. oxysporum by strains Psd and PsdΔalg8::kan. Effect of culture filtrate of strains Psd and PsdΔalg8::kan on the biomass growth of (B) F. oxysporum, and (C) F. graminearum (*P < 0.05, ns-not significant).
Effect of alg8 Deletion on Seedlings Growth
The above results highlighted the Zn2+ accumulation potential of strain Psd and, subsequently, its beneficial effect on PGP and biocontrol potential of the strain. Furthermore, it was observed that this effect translated into improved plant growth. The treatment of wheat seeds with strain Psd improved root growth and proliferation, as is evident from Figure S6A. Zn2+-laden biomass of strain Psd led to improved seedlings growth as higher number of root hairs was observed in these cases (Figure S7). On the other hand, PsdΔalg8::kan-treated wheat seeds did not show any differences with respect to the untreated control (Figure S6B), indicating that lack of alginate has affected the PGP potential of the strain.
Discussion
Many members of fluorescent Pseudomonas sp., including P. fluorescens are inhabitants of the rhizosphere and possess the ability to enhance plant growth, via mechanisms such as phytohormone production (Upadhyay and Srivastava, 2010; Kochar et al., 2011; Ahemad and Kibret, 2014), biocontrol of phytopathogens by synthesizing allelopathic factors such as toxins, antibiotics, and siderophores (Duffy and Défago, 1999; Upadhyay and Srivastava, 2008, 2010; Couillerot et al., 2009), and improvement of nutrient acquisition by the plants (Upadhyay and Srivastava, 2008; Sirohi et al., 2015; Wang et al., 2015). Besides, these bacteria have also emerged as promising candidates for in situ bioremediation of various organic and inorganic pollutants, including heavy metals (Zhuang et al., 2007; Upadhyay and Srivastava, 2014). Fluorescent Pseudomonas sp. strain Psd, isolated from the rhizosphere of V. mungo, has been extensively studied and has been demonstrated to possess multiple plant-growth promoting and biocontrol potential (Upadhyay and Srivastava, 2008, 2010; Kochar et al., 2011). Besides, we have reported earlier that the Zn2+ accumulation by strain Psd is accompanied by increased EPS production (Upadhyay and Srivastava, 2014). We decipher here the role EPS plays in the process of biosorption of Zn2+ and also their implications on the PGP and biocontrol potential of the strain.
Exposure to metals induces morphological and ultrastructural changes in the cell. Membrane thickening and protuberances were observed in the cells of strain Psd exposed to high Zn2+ concentrations. These changes are likely to provide increased number of binding sites to accommodate high levels of Zn2+. In such cells, EDA which mediate intracellular accumulation of the metal were also observed. Similar results were obtained in Pseudomonas stutzeri RS34 exposed to Zn2+ (Bhagat and Srivastava, 1994). Changes in cell morphology in response to exposure to heavy metals has been reported in case of extremophiles Acidocella, Acidiphillium, and cyanobacterium, Microcoleus chthonoplastes that show greater excretion of EPS in response to exposure to heavy metals like Cd2+, Cu2+, Ni2+, Pb2+, and Zn2+ (Chakravarty et al., 2007; Diestra et al., 2007; Chakravarty and Banerjee, 2008). Further, profiling of functional groups present in the EPS secreted in response to exposure to Zn2+ revealed the presence of carbohydrates, uronic acids, mannose, and phosphoryl groups. Out of these components, uronic acids have been established as the main constituents of bacterial biofilms (Sutherland, 2001). Additionally, uronic acids were also found to form the major components of the EPS involved in the sequestration of cations such as Pb2+, Cd2+, Co2+, Ni2+, and Zn2+ and Cu2+ in Paenibacillus jamilae (Pérez et al., 2008). Several functional groups present on bacterial cell wall, including carboxyl, phosphonate, amine, and hydroxyl groups, which assist in biosorption have been identified (Ledin, 2000; Vijayaraghavan and Yun, 2008). Ueshima et al. (2008) have demonstrated the presence of carboxyl and phosphoryl groups in the EPS produced by P. putida, which leads to the biosorption of Cd2+. Glucose and mannose pre-dominated the EPS involved in Zn2+ and Cd2+ biosorption by Anoxybacillus sp. and Pb2+ and Hg2+ biosorption in Azotobacter chroococcum (Rasulov et al., 2013; Zhao et al., 2014).
Alginate forms the major component of the biofilms secreted by bacteria (Ghafoor et al., 2011). We found an increase in the total alginate content of the EPS isolated from cells growing at different Zn2+ concentrations, indicative of the role this polysaccharide may play in the process. Several multivalent cations, like Ca2+, Cd2+, Pb2+, and Zn2+, bind to EPS by virtue of electrostatic interactions, thus triggering extensive production of these polysaccharides (Sutherland, 2001). In order to further characterize the role of alginates, we studied a gene from the alginate biosynthesis complex, alg8, which codes for β-glycosyl transferases of the GT-1 family and is a key protein involved in alginate polymerization (Rehm, 2010; Ghafoor et al., 2011). Increased Zn2+ accumulation, accompanied by upregulation of alg8 expression, confirmed that alginate, besides being responsible for biofilm formation, also has a role in Zn2+ biosorption by Psd. Although many recent studies have demonstrated the role of alginates in metal biosorption in algal species (Plazinski, 2013; Bertagnolli et al., 2014), only a few reports are available in the bacterial system for the same (Zhang et al., 2010; François et al., 2012). Further insight into the role of alg8 in Zn2+ biosorption was obtained with the help of a mutant devoid of a functional alg8 gene. Gene replacement via homologous recombination has been employed in the present study to obtain alg8 knockout mutant in strain Psd. Similar methods have been used in other studies to generate alg8 negative mutant (Remminghorst and Rehm, 2006; Ghafoor et al., 2011). These studies have shown that deletion of alg8 results in non-mucoid phenotype, which is deficient in alginate production. None of the studies so far, however, have related deletion of alg8 with metal tolerance. We have shown that the alg8-negative mutant had a compromised EPS production in comparison to the wild-type, ultimately affecting the Zn2+ accumulation potential of the strain. The mutant strain accumulated 80% less Zn2+ than the wild-type. This observation was further substantiated with the ultra-structural analysis, wherein the Zn2+- exposed cells of the mutant exhibited a clear reduction in the extracellular biosorption when compared to wild-type. Cytoplasmic granules, however, were observed in the mutant, indicating that the intracellular accumulation, if any, was not affected.
Biofilm formation by PGP bacteria is an important determinant in plant-microbe association. Formation of an extracellular matrix composed of EPS is the hallmark for biofilm formation (Wozniak et al., 2003). In the present study, we found a positive correlation between Zn2+ biosorption by strain Psd and alginate production. Hence, it was likely that inability of the strain to produce alginates would affect the strain's biofilm formation potential. It was, indeed, found that Zn2+ triggers the formation of biofilms by the strain. Microscopic and ultra-structural analysis revealed effective colonization of wheat roots by strain Psd, where the cells were found embedded in an extracellular matrix. Inability of the mutant PsdΔalg8::kan to synthesize EPS affected its biofilm formation and as a consequence, cells were mostly found to exist as planktonic cells. However, we did find few aggregates on the mutant treated roots. This may be attributed to the fact that mutants unable to synthesize EPS may still attach to the surface and form micro-colonies to a limited extent (Sutherland, 2001).
Since strain Psd is a PGPB, it was essential to ascertain the effect of Zn2+ biosorption on its PGP potential. The present study has shown that the cells of strain Psd growing at high concentration of Zn2+ led to enhanced phosphate solubilization, which could be attributed to either enhanced EPS production at higher Zn2+ concentration which holds free P from insoluble phosphate in the medium (Ahemad and Kibret, 2014) or due to involvement of Zn2+ in conversion of glucose to gluconic acid (Ramachandran et al., 2006). The former is the most plausible explanation for decreased P solubilization activity of the alg8 deletion mutant. Further, analysis of siderophores, which provide a competitive advantage to the biocontrol agents over harmful phytopathogens by limiting the supply of essential trace elements, revealed ~1.8-fold increase in siderophore production in Zn2+-grown cells of strain Psd. Exogenous environmental signals, like carbon sources and minerals, play an important role in modulating secondary metabolite production by microbes. It has been proposed that Zn2+ may hinder cellular iron uptake, leading to higher siderophore production. In P. aeruginosa, Zn2+ supplementation resulted in an overall iron deficiency (Rossbach et al., 2000). Alternatively, Zn2+ may bind to the siderophores, necessitating their enhanced production to chelate the available iron (Höfte et al., 1993). In agreement, significantly low levels of siderophores were detected in the mutant PsdΔalg8::kan, due its compromised Zn2+ accumulation ability.
On the contrary to the above observations, in the present study, a decrease in IAA production was obtained with increased Zn2+ accumulation by strain Psd. This may be the consequence of lower IAA biosynthesis induced by Zn2+ or to auxin degradation by IAA peroxidases, which are up-regulated by metal-catalyzed free radical formation (Dimkpa et al., 2008). Besides, we found an increase in phenazine production with increased Zn2+ accumulation by strain Psd. This was in agreement with the observation of Duffy and Défago (1999), who reported the stimulatory effect of zinc sulfate on PHL and PLT production by P. protegens CHA0. Supplementation with Zn2+ also led to stimulation of phenazine-1-carboxylic acid production in P. fluorescens 2–79, which further improved the biocontrol potential of the strain (Slininger and Jackson, 1992). The exact mechanism for such a response is uncertain. However, it has been proposed that Zn2+ and other mineral nutrients stabilize the regulatory genes critical for antibiotic production in Pseudomonads (Duffy and Défago, 1997). Interestingly the alginate knock-out mutant, PsdΔalg8::kan, which had a compromised Zn2+ accumulation, was not able to produce phenazines, even in the medium devoid of Zn2+. This pointed toward the importance of mineral nutrients in the antibiotic biosynthesis. Alternatively, this observation can be a result of a cross-talk between alginate that helps in better association with plant roots and antibiotic biosynthesis pathways.
A variety of PGP bacterial strains like Azospirillum, Azotobacter, Bacillus, Pseudomonas, and Streptomyces have earlier been implicated in biocontrol of plant pathogens like tomato mottle virus, tobacco necrosis virus, Rhizoctonia bataticola, and Fusarium avenaceum (Bhattacharyya and Jha, 2012 and references therein). In the present study, competence of Zn2+-laden biomass of strain Psd in biocontrol of plant pathogens, F. oxysporum and F. graminearum had a stimulatory effect on biocontrol potential, with up to 80% inhibition obtained in case of F. oxysporum. This finding was in line with the results obtained in tomato, wherein the disease suppression by P. fluorescens CHA0 was augmented upon addition of Zn2+ (Duffy and Défago, 1997). It was proposed that Zn2+ amendment abolished fusaric acid production by F. oxyposrum, reducing the pathogenicity of the fungus (Duffy and Défago, 1997). Application of Zn2+ alone or in combination with the biocontrol agent P. aeruginosa significantly decreased the penetration of the root knot nematode Meloidogyne javanica in tomato (Siddiqui et al., 2002). The reduced pathogenecity can also be attributed to increased production of antifungal metabolites. On the other hand, PsdΔalg8::kan exhibited significantly low biocontrol activity. The strain displayed reduced biomass inhibition of F. oxysporum and was not able to inhibit F. graminearum at all. This observation corroborated inability of the mutant to produce/release phenazines.
The stimulatory effect of Fluorescent Pseudomonas strain Psd on root growth has been studied earlier (Kochar et al., 2011; Sirohi et al., 2015). It has been established that biofilm formation by PGP bacteria leads to improved plant growth by facilitating dense bacterial population to produce various phytohormones, antibiotics, beneficial secondary metabolites and exoenzymes (Danhorn and Fuqua, 2007; Koul et al., 2015). The fact that Zn2+ accumulation triggers an improved PGP response in strain Psd was also observed in terms of improved root growth and proliferation. Bacterial attachment led to increase in formation of root hairs, indicative of better root growth. The functional implication, on the other hand, of decreased EPS production and subsequently, inability to form biofilms by strain PsdΔalg8::kan was reflected in the mutant's inability to lead to a significant improvement in plant growth.
Overall, as sets out in the present study, alginates not only mediate the Zn2+ biosorption by strain Psd, but may also serve as important determinants in plant-growth-promotion. Interestingly, a possible dependence of phenazine biosynthesis with that of alginate production is also indicated in the study. However, this part warrants further investigation. The study also ascertains the rhizosphere competence of the strain in heavy-metal contaminated soils, without jeopardizing their PGP potential.
Author Contributions
AU, MK, SS, and MR conceived and designed the experiments. AU and MK performed the experiments and analyzed the data. SS and MR contributed financial assistance. AU, MK, and MR wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The financial assistance provided to Department of Genetics under UGC-SAP and DST-FIST programs of Govt. of India is gratefully acknowledged. The work of AU has been supported by UGC, Govt. of India and SRF from ICMR, Govt. of India. AU and MK thank Mr. C. K. Tripathi (TERI) for critical suggestions in SEM analysis. Authors also acknowledge Central Instrumentation Facility (CIF), University of Delhi South Campus (UDSC), New Delhi and SAIF, AIIMS, New Delhi for assistance with various analytical techniques.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00284/full#supplementary-material
Figure S1. Schematic representation of the alginate biosynthesis pathway operating in Pseudomonas sp.
Figure S2. Schematic representation of (A) upstream and downstream region of alg8 gene; (B) homologous recombination-based mechanism to replace alg8 gene with kanr cassette; (C) PCR amplification of five putative constructs (pBKS_alg8_up/kan/down) with alg8 upstream, downstream and kanamycin resistance gene specific primers. M-1 kb DNA ladder (NEB, USA); Lanes 1–5 represent amplification of ~500 bp region upstream to alg8; Lanes 6–10 represent amplification of ~500 bp region downstream to alg8 and; Lanes 11–15 represent amplification of the kanr gene.
Figure S3. Flow diagram of the protocol used for Zn2+ estimation in bacterial cells.
Figure S4. Quantitative determination of alginates in EPS secreted from strain Psd grown in different Zn2+ concentrations by DMMB-binding assay (****p < 0.0001).
Figure S5. Biofilm production by Psd and PsdΔalg8::kan in GMM supplemented with increasing Zn2+ concentrations (*P < 0.05, ***P < 0.001, ****P < 0.0001, ns, not significant).
Figure S6. Bioassay showing the effect of treatment of wheat seeds with (A) Pseudomonas strain Psd, and (B) strain PsdΔalg8::kan, on seedling growth after 6-days of inoculation.
Figure S7. Scanning electron microscopy of wheat roots inoculated with Pseudomonas strain Psd grown at increasing Zn2+ concentrations to show the effect on roots proliferation: Untreated control (A), roots treated with wild type strain Psd grown in absence (B), and presence of 2 mM Zn2+ (C). RH, root hair. Scale bars vary from 20 to 100 μm and are shown in each image.
Table S1. List of strains and plasmids used in the study.
References
Ahemad, M., and Kibret, M. (2014). Mechanisms and applications of plant growth promoting rhizobacteria: current perspective. J. King Saud Univ. Sci. 26, 1–20. doi: 10.1016/j.jksus.2013.05.001
Allesen-Holm, M., Barken, K. B., Yang, L., Klausen, M., Webb, J. S., Kjelleberg, S., et al. (2006). A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 59, 1114–1128. doi: 10.1111/j.1365-2958.2005.05008.x
Bertagnolli, C., Uhart, A., Dupin, J. C., da Silva, M. G., Guibal, E., and Desbrieres, J. (2014). Biosorption of chromium by alginate extraction products from Sargassum filipendula: investigation of adsorption mechanisms using X-ray photoelectron spectroscopy analysis. Bioresour. Technol. 164, 264–269. doi: 10.1016/j.biortech.2014.04.103
Beveridge, T. J. (1988). “Wall ultrastructure: how little we know,” in Antibiotic Inhibition of the Bacterial Cell: Surface Assembly and Function, eds P. Actor, L. Daneo-Moore, M. L. Higgins, M. R. J. Salton, and G. D. Shockman (Washington, DC: American Society of Microbiology), 3–20.
Bhagat, R., and Srivastava, S. (1994). Effect of zinc on morphology and ultrastructure of Pseudomonas stutzeri RS34. J. Gen. Appl. Microbiol. 40, 265–270. doi: 10.2323/jgam.40.265
Bhattacharyya, P. N., and Jha, D. K. (2012). Plant growth-promoting rhizobacteria (PGPR): emergence in agriculture. World J. Microbiol. Biotechnol. 28, 1327–1350. doi: 10.1007/s11274-011-0979-9
Bianciotto, V., Andreotti, S., Balestrini, R., Bonfante, P., and Perotto, S. (2001). Mucoid mutants of the biocontrol strain Pseudomonas fluorescens CHA0 show increased ability in biofilm formation on mycorrhizal and nonmycorrhizal carrot roots. Mol. Plant Microbe Interact. 14, 255–260. doi: 10.1094/MPMI.2001.14.2.255
Bitton, G., and Freihofer, V. (1978). Influence of extracellular polysaccharides on the toxicity of copper and cadmium toward Klebsiella aerogenes. Microbial. Ecol. 4, 119–125. doi: 10.1007/BF02014282
Blencowe, D. K., and Morby, A. P. (2003). Zn(II) metabolism in prokaryotes. FEMS Microbiol. Rev. 27, 291–311. doi: 10.1016/S0168-6445(03)00041-X
Blindauer, C. A. (2015). Advances in the molecular understanding of biological zinc transport. Chem. Commun. 51, 4544–4563. doi: 10.1039/C4CC10174J
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE Guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797
Chakravarty, R., and Banerjee, P. C. (2008). Morphological changes in an acidophilic bacterium induced by heavy metals. Extremophiles 12, 279–284. doi: 10.1007/s00792-007-0128-4
Chakravarty, R., Manna, S., Ghosh, A. K., and Banerjee, P. C. (2007). Morphological changes in an Acidocella strain in response to heavy metal stress. Res. J. Microbiol. 2, 742–748. doi: 10.3923/jm.2007.742.748
Choudhury, R., and Srivastava, S. (2001). Zinc resistance mechanisms in bacteria. Curr. Sci. 81, 768–775.
Conti, E., Flaibani, A., O'Regan, M., and Sutherland, I. W. (1994). Alginate from Pseudomonas fluorescens and P. putida: production and properties. Microbiology 140, 1125–1132.
Corbin, B. D., Seeley, E. H., Raab, A., Feldmann, J., Miller, M. R., Torres, V. J., et al. (2008). Metal chelation and inhibition of bacterial growth in tissue abscesses. Science 319, 962–965. doi: 10.1126/science.1152449
Couillerot, O., Prigent-Combaret, C., Caballero-Mellado, J., and Moënne-Loccoz, Y. (2009). Pseudomonas fluorescens and closely-related fluorescent Pseudomonads as biocontrol agents of soil-borne phytopathogens. Lett. Appl. Microbiol. 48, 505–512. doi: 10.1111/j.1472-765X.2009.02566.x
Danhorn, T., and Fuqua, C. (2007). Biofilm formation by plant associated bacteria. Ann. Rev. Microbiol. 61, 401–422. doi: 10.1146/annurev.micro.61.080706.093316
Diestra, E., Esteve, I., Burnat, M., Maldonado, J., and Solé, A. (2007). “Isolation and characterization of a heterotrophic bacterium able to grow in different environmental stress conditions, including crude oil and heavy metals,” in Communicating Current Research and Educational Topics and Trends in Applied Microbiology, Vol. I, ed A. M. Vilas (Extremadura: Formatex Publications), 90–99.
Dimkpa, C. O., Svatoš, A., Dabrowska, P., Schmidt, A., Boland, W., and Kothe, E. (2008). Involvement of siderophores in the reduction of metal-induced inhibition of auxin synthesis in Streptomyces spp. Chemosphere 74, 19–25. doi: 10.1016/j.chemosphere.2008.09.079
Duffy, B. K., and Défago, G. (1997). Zinc improves biocontrol of Fusarium crown and root rot of tomato by Pseudomonas fluorescens and represses the production of pathogen metabolites inhibitory to bacterial antibiotic biosynthesis. Phytopathology 87, 1250–1257. doi: 10.1094/PHYTO.1997.87.12.1250
Duffy, B. K., and Défago, G. (1999). Environmental factors modulating antibiotic and siderophore biosynthesis by Pseudomonas fluorescens biocontrol strains. Appl. Environ. Microbiol. 65, 2429–2438.
Flemming, H. C., and Wingender, J. (2001). Relevance of microbial extracellular polymeric substances (EPSs)-Part I: structural and ecological aspects. Water Sci. Technol. 43, 1–8.
François, F., Lombard, C., Guigner, J. M., Soreau, P., Brian-Jaisson, F., Martino, G., et al. (2012). Isolation and characterization of environmental bacteria capable of extracellular biosorption of mercury. Appl. Environ. Microbiol. 78, 1097–1106. doi: 10.1128/AEM.06522-11
Franklin, M. J., Nivens, D. E., Weadge, J. T., and Howell, P. L. (2011). Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides, alginate, Pel, and Psl. Front. Microbiol. 2:167. doi: 10.3389/fmicb.2011.00167
Friedman, L., and Kolter, R. (2004). Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51, 675–690. doi: 10.1046/j.1365-2958.2003.03877.x
Ghafoor, A., Hay, I. D., and Rehm, B. H. A. (2011). Role of exopolysaccharides in Pseudomonas aeruginosa biofilm formation and architecture. Appl. Environ. Microbiol. 77, 5238–5246. doi: 10.1128/AEM.00637-11
Gilotra, U., and Srivastava, S. (1997). Plasmid-bound copper sequestration in Pseudomonas pickettii strain US321. Curr. Microbiol. 34, 378–381. doi: 10.1007/s002849900199
Höfte, M., Buysens, S., Koedam, N., and Cornelis, P. (1993). Zinc affects siderophore-mediated high affinity iron uptake systems in the rhizosphere Pseudomonas aeruginosa 7NSK2. Biometals 6, 85–91.
Kochar, M., Upadhyay, A., and Srivastava, S. (2011). Indole-3-acetic acid biosynthesis in the biocontrol strain Pseudomonas fluorescens Psd and plant growth regulation by hormone overexpression. Res. Microbiol. 162, 426–435. doi: 10.1016/j.resmic.2011.03.006
Koul, V., Tripathi, C., Adholeya, A., and Kochar, M. (2015). Nitric oxide metabolism and indole acetic acid biosynthesis cross-talk in Azospirillum brasilense SM. Res. Microbiol. 166, 174–185. doi: 10.1016/j.resmic.2015.02.003
Ledin, M. (2000). Accumulation of metals by microorganisms - processes and importance for soil systems. Earth Sci. Rev. 51, 1–31. doi: 10.1016/S0012-8252(00)00008-8
LeGrand, E. K., and Alcock, J. (2012). Turning up the heat: immune brinksmanship in the acute-phase response. Q. Rev. Biol. 87, 3–18. doi: 10.1086/663946
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, L., Conover, M., Lu, H., Parsek, M. R., Bayles, K., and Wozniak, D. J. (2009). Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog. 5:e1000354. doi: 10.1371/journal.ppat.1000354
Malhotra, M., and Srivastava, S. (2008). An ipdC gene knock-out of Azospirillum brasilense strain SM and its implications on indole-3-acetic acid biosynthesis and plant growth promotion. Anton. van Leeuwenhoek 93, 425–433. doi: 10.1007/s10482-007-9207-x
Mangold, S., Potrykus, J., Björn, E., Lövgren, L., and Dopson, M. (2013). Extreme zinc tolerance in acidophilic microorganisms from the bacterial and archaeal domains. Extremophiles 17, 75–85. doi: 10.1007/s00792-012-0495-3
Matthews, J. M., and Sunde, M. (2002). Zinc fingers–folds for many occasions. IUBMB Life 54, 351–355. doi: 10.1080/15216540216035
Merritt, J. H., Kadouri, D. E., and O'Toole, G. A. (2005). Growing and analyzing static biofilms. Curr. Protoc. Microbiol. 1B, 1.1–1.18. doi: 10.1002/9780471729259.mc01b01s00
Pérez, J. A. M., Ribera, R. G., Quesada, T., Aguilera, M., Cormenzana, A. R., and Sánchez, M. M. (2008). Biosorption of heavy metals by the exopolysaccharide produced by Paenibacillus jamilae. World J. Microbiol. Biotechnol. 24, 2699–2704. doi: 10.1007/s11274-008-9800-9
Pikovskaya, R. I. (1948). Mobilization of phosphorus in soil in connection with the vital activity of some microbial species. Microbiology 17, 362–370.
Plazinski, W. (2013). Binding of heavy metals by algal biosorbents. Theoretical models of kinetics, equilibria and thermodynamics. Adv. Colloid Interface Sci. 197–198, 58–67. doi: 10.1016/j.cis.2013.04.002
Ramachandran, S., Fontanille, P., Pandey, A., and Larroche, C. (2006). Gluconic acid: properties, applications and microbial production. Food Technol. Biotechnol. 44, 185–195.
Rasulov, B. A., Yili, A., and Aisa, H. A. (2013). Biosorption of Metal ions by exopolysaccharide produced by Azotobacter chroococcum XU1. J. Environ. Protoc. 4, 989–993. doi: 10.4236/jep.2013.49114
Rehm, B. H. A. (2010). Bacterial polymers: biosynthesis, modifications and applications. Nat. Rev. Microbiol. 8, 578–592. doi: 10.1038/nrmicro2354
Remminghorst, U., and Rehm, B. H. A. (2006). In vitro alginate polymerization and the functional role of alg8 in alginate production by Pseudomonas aeruginosa. Appl. Environ. Microbiol. 72, 298–305. doi: 10.1128/AEM.72.1.298-305.2006
Richardson, J. C., Dettmar, P. W., Hampson, F. C., and Melia, C. D. (2004). A simple, high throughput method for the quantification of sodium alginates on oesophageal mucosa. Eur. J. Pharm. Biopharm. 57, 299–305. doi: 10.1016/S0939-6411(03)00150-4
Rossbach, S., Wilson, T. L., Kukuk, M. L., and Carty, H. A. (2000). Elevated zinc induces siderophore biosynthesis genes and a zntA-like gene in Pseudomonas fluorescens. FEMS Microbiol. Lett. 191, 61–70. doi: 10.1111/j.1574-6968.2000.tb09320.x
Schwyn, B., and Neilands, J. B. (1987). Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160, 47–56. doi: 10.1016/0003-2697(87)90612-9
Shafeeq, S., Kuipers, O. P., and Kloosterman, T. G. (2013). The role of zinc in the interplay between pathogenic streptococci and their hosts. Mol. Microbiol. 88, 1047–1057. doi: 10.1111/mmi.12256
Siddiqui, I. A., Shaukat, S. S., and Hamid, M. (2002). Role of zinc in rhizobacteria-mediated suppression of root-infecting fungi and root-knot nematode. J. Phytopathol. 150, 569–575. doi: 10.1046/j.1439-0434.2002.00805.x
Sirohi, G., Upadhyay, A., Srivastava, P. S., and Srivastava, S. (2015). PGPR mediated zinc biofertilization of soil and its impact on growth and productivity of wheat. J. Soil Sci. Plant Nutr. 15, 115–132. doi: 10.4067/s0718-95162015005000017
Slininger, P. J., and Jackson, M. A. (1992). Nutritional factors regulating growth and accumulation of phenazine 1-carboxylic acid by Pseudomonas fluorescens 2-79. Appl. Microbiol. Biotechnol. 37, 388–392. doi: 10.1007/BF00210998
Sutherland, R. (2001). Biofilm exopolysaccharides: a strong and sticky framework. Microbiology 147, 3–9. doi: 10.1099/00221287-147-1-3
Ueshima, M., Ginn, B. R., Haack, E. A., Szymanowski, J. E. S., and Fein, J. B. (2008). Cd adsorption onto Pseudomonas putida in the presence and absence of extracellular polymeric substances. Geochim. Cosmochim. Acta 72, 5885–5895. doi: 10.1016/j.gca.2008.09.014
Upadhyay, A., and Srivastava, S. (2008). Characterization of a new isolate of a Pseudomonas fluorescens strain Psd as a potential biocontrol agent. Lett. Appl. Microbiol. 47, 98–105. doi: 10.1111/j.1472-765X.2008.02390.x
Upadhyay, A., and Srivastava, S. (2010). Evaluation of multiple plant growth promoting traits of an isolate of Pseudomonas fluorescens strain Psd. Indian J. Exp. Biol. 48, 601–609.
Upadhyay, A., and Srivastava, S. (2014). Mechanism of Zinc resistance in a plant growth promoting Pseudomonas fluorescens strain. World J. Microbiol. Biotechnol. 30, 2273–2282. doi: 10.1007/s11274-014-1648-6
Vessey, J. K. (2003). Plant growth promoting rhizobacteria as biofertilizers. Plant Soil 255, 571–586. doi: 10.1023/A:1026037216893
Vijayaraghavan, K., and Yun, Y. S. (2008). Bacterial biosorbents and biosorption. Biotechnol. Adv. 26, 266–291. doi: 10.1016/j.biotechadv.2008.02.002
Wang, X., Mavrodi, D. V., Ke, L., Mavrodi, O. V., Yang, M., Thomashow, L. S., et al. (2015). Biocontrol and plant growth-promoting activity of rhizobacteria from Chinese fields with contaminated soils. Microb. Biotechnol. 8, 404–418. doi: 10.1111/1751-7915.12158
Whistler, C. A., and Pierson, L. S. III. (2003). Repression of phenazine antibiotic production in Pseudomonas aureofaciens strain 30-84 by RpeA. J. Bacteriol. 85, 3718–3725. doi: 10.1128/JB.185.13.3718-3725.2003
Whitfield, G. B., Marmont, L. S., and Howell, P. L. (2015). Enzymatic modifications of exopolysaccharides enhance bacterial persistence. Front. Microbiol. 6:471. doi: 10.3389/fmicb.2015.00471
Wozniak, D. J., Wyckoff, T. J. O., Starkey, M., Keyser, R., Azadi, P., O'Toole, G. A., et al. (2003). Alginate is not a significant component of the extracellular polysaccharide matrix of PA14 and PAO1 Pseudomonas aeruginosa biofilms. Proc. Natl. Acad. Sci. U.S.A. 100, 7907–7912. doi: 10.1073/pnas.1231792100
Yang, L., Hu, Y., Liu, Y., Zhang, J., Ulstrup, J., and Molin, S. (2011). Distinct roles of extracellular polymeric substances in Pseudomonas aeruginosa biofilm development. Environ. Microbiol. 13, 1705–1717. doi: 10.1111/j.1462-2920.2011.02503.x
Zhang, H. L., Lin, Y. M., and Wang, L. (2010). Cu2+ biosorption by bacterial alginate extracted from aerobic granules and its mechanism investigation. [Article in Chinese] Huan Jing Ke Xue 31, 731–737.
Zhao, S., Cao, F., Zhang, H., Zhang, L., Zhang, F., and Liang, X. (2014). Structural characterization and biosorption of exopolysaccharides from Anoxybacillus sp. R4-33 isolated from radioactive radon hot spring. Appl. Biochem. Biotechnol. 172, 2732–2746. doi: 10.1007/s12010-013-0680-6
Keywords: biosorption, exopolysaccharides, alginates, biofilms, biocontrol
Citation: Upadhyay A, Kochar M, Rajam MV and Srivastava S (2017) Players over the Surface: Unraveling the Role of Exopolysaccharides in Zinc Biosorption by Fluorescent Pseudomonas Strain Psd. Front. Microbiol. 8:284. doi: 10.3389/fmicb.2017.00284
Received: 11 July 2016; Accepted: 10 February 2017;
Published: 24 February 2017.
Edited by:
Abdul Latif Khan, University of Nizwa, OmanReviewed by:
Melanie J. Filiatrault, United States Department of Agriculture-Agricultural Research Service, USAAnushree Malik, Indian Institute of Technology Delhi, India
Copyright © 2017 Upadhyay, Kochar, Rajam and Srivastava. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sheela Srivastava, sslab222012@gmail.com
 Anamika Upadhyay
Anamika Upadhyay Mandira Kochar
Mandira Kochar Manchikatla V. Rajam
Manchikatla V. Rajam Sheela Srivastava
Sheela Srivastava