Erratum: Distribution of cfr in Staphylococcus spp. and Escherichia coli Strains from Pig Farms in China and Characterization of a Novel cfr-Carrying F43:A-:B- Plasmid
- National Reference Laboratory of Veterinary Drug Residues, College of Veterinary Medicine, South China Agricultural University, Guangzhou, China
The multi-resistance gene cfr is widely distributed among various gram-positive and gram-negative species in livestock in China. To better understand the epidemiology of cfr among Staphylococcus spp. and E. coli isolates, 254 Staphylococcus spp. and 398 E. coli strains collected from six swine farms in China were subjected to prevalence and genetic analysis. Forty (15.7%) Staphylococcus spp. isolates, including 38 Staphylococcus sciuri strains, one Staphylococcus chromogenes strain, and one Staphylococcus lentus strain, and two (0.5%) E. coli isolates were found to contain the cfr gene. Most of the 38 S. sciuri strains were clonally unrelated; however, clonal dissemination of cfr-positive S. sciuri was detected at the same farm. In eight randomly selected cfr-positive staphylococci, a cfr-harboring module (IS21-558-cfr-ΔtnpB) was detected in six S. sciuri isolates; cfr was bracketed by two copies of ISEnfa4 or IS256 in the remaining two S. sciuri isolates. In the two E. coli isolates, EP25 and EP28, cfr was flanked by two IS26 elements in the same or opposite orientation, respectively. Complete sequence analysis of the novel F43:A-:B- plasmid pHNEP28 revealed that it contains two multi-resistance regions: cfr together with floR, qnrS1 interspersed with IS26, ΔISCR2 and ISKpn19, and blaTEM-1 together with tet(M) interspersed with IS26, ISApl1, ΔTn2, and ΔIS1B. The coexistence of cfr with other resistance genes on a conjugative plasmid may contribute to the dissemination of these genes by co-selection. Thus, rational drug use and continued surveillance of cfr in swine farms are warranted.
Introduction
The multi-resistance gene cfr encodes a methyltransferase that modifies position A2503 at 23S rRNA and confers resistance to six different classes of antibiotics that bind to the peptidyl transferase center (Kehrenberg et al., 2005; Giessing et al., 2009). Cfr confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics (the PhLOPSA phenotype) and reduces susceptibility to the 16-membered macrolides josamysin and spiramycin (Long et al., 2006), which are used extensively in the treatment of infections in both humans and animals. Oxazolidinones, in particular, are regarded as the last defense against methicillin-resistant Staphylococcus aureus (MRSA) or vancomycin-resistant enterococci in humans (Toh et al., 2007). The cfr gene was first identified in a bovine Staphylococcus sciuri isolate (Stefan et al., 2000) and subsequently in staphylococcal isolates of human and animal origin from around the world (Shen et al., 2013). In addition, various Enterococcus, Bacillus, Macrococcus, Jeotgalicoccus, Streptococcus suis, Proteus vulgaris, and Escherichia coli strains have also been reported to be cfr-positive (Shen et al., 2013; Wang et al., 2013b). The cfr gene is not only widely disseminated in staphylococci from pigs in China (Shen et al., 2013), but also increasingly reported in porcine E. coli isolates in China. To date, a total of 16 cfr-positive E. coli isolates have been identified in the Shandong, Sichuan, Guangxi, and Guangdong provinces, China (Deng et al., 2014; Wang et al., 2012a; Zhang et al., 2014; Zhang R. et al., 2015; Zhang W.J. et al., 2015), suggesting the possibility of the transfer of cfr from Staphylococcus to E. coli. In this study, we identified cfr-positive Staphylococcus spp. and E. coli isolates from the same farm and determined the complete nucleotide sequence of a novel conjugative F43:A-:B- plasmid bearing cfr in an E. coli isolate.
Materials and Methods
Sampling and Bacterial Isolates
A total of 323 pig nasal swab samples and 442 fecal samples were collected in 2013 from six swine farms located in four geographically distinct provinces in China: one farm in Jilin (DBNA), two farms in Guangdong (GDEP and GDYD), one farm in Jiangxi (JXTC), and two farms in Henan (HNYM and HNYC). The usage of antibiotic agents at these farms over the past year (except farm DBNA) was supplied by their veterinarians. All animal studies were performed with the approval of the Animal Care and Use Committee of South China Agriculture University. Nasal swab samples were incubated in 7.5% Sodium Chloride Broth for 16–18 h at 37°C and then streaked onto Mannitol Salt agar plates and grown for 16–18 h at 37°C. Next, one presumptive staphylococcal isolate per sample was randomly selected for further analysis. Fecal samples were used to isolate E. coli as previously described (Chen et al., 2007). Species identification of all cfr-positive strains was determined by the ID32 STAPH system (bioMérieux, Craponne, France) and was further confirmed by sequencing the 16S rRNA using universal prokaryotic primers (Weisburg et al., 1991).
Identification of Resistance Genes and the Genetic Environment of cfr
All Staphylococcus spp. and E. coli isolates were screened for the presence of cfr by polymerase chain reaction (PCR) as previously described (Kehrenberg and Schwarz, 2006). The cfr-positive staphylococci were also PCR-screened for the presence of the fexA, fexB, mecA, and mecC genes (Murakami et al., 1991; Kehrenberg and Schwarz, 2006; Liu et al., 2012; Paterson et al., 2012). Additionally, the cfr-positive E. coli strains were screened for floR (Chen et al., 2004).
The genetic context surrounding cfr was examined by PCR mapping and sequencing in eight randomly selected cfr-positive Staphylococcus spp. and two cfr-positive E. coli isolates. The primers used to determine the regions upstream and downstream of cfr are listed in Supplementary Table S1.
Antimicrobial Susceptibility Testing
All cfr-positive Staphylococcus spp. and E. coli isolates were tested for their antimicrobial susceptibility using the agar dilution method. Antimicrobial susceptibility tests were conducted and evaluated according to the recommendations specified in CLSI documents VET01-S2 (CLSI, 2013) and M100-S25 (CLSI, 2015). Minimal inhibitory concentrations (MICs) of ≥16 mg/L were tentatively considered as criteria of florfenicol resistance (Kehrenberg and Schwarz, 2006). S. aureus ATCC®29213 and E. coli ATCC®25922 served as quality control strains.
Multilocus Sequence Typing (MLST)
Seven housekeeping genes, including adk, fumC, gyrB, icd, mdh, purA, and recA, were used to determine the sequence types (STs) of all the cfr-positive E. coli (Lau et al., 2008). The MLST databases were used for BLAST analysis1.
Pulsed-Field Gel Electrophoresis (PFGE)
Pulsed-field gel electrophoresis (PFGE) was used to determine the clonality of the cfr-positive S. sciuri isolates. Whole-cells were packed with an equal volume of 2% (wt/vol) low melting point agarose (Bio-Rad, Laboratories, Hercules, CA, USA) dissolved in EC buffer (Liu et al., 2014) and poured into plug molds. The frozen plugs were addressed according to the previously described with minor modification. The digested fragments were separated using a CHEF-DRIII system (Bio-Rad) with a clamped homogeneous electric field of 6 V/cm, using a 120° switch angle for 24 h at 14°C, with the pulse time linearly ramped from 3 to 40 s. PFGE was performed for all cfr-positive E. coli isolates using the CHEF Mapper System (Bio-Rad), according to a previously described protocol (Chen et al., 2007). Comparison of PFGE patterns was performed using the BioNumerics software (Applied Maths, Sint-Martens-Latem, Belgium) using the Dice coefficient (1.5% optimization and 1.0% tolerance); a similarity cutoff of 80% was used to identify a PFGE cluster.
S1-Pfge, Conjugation/Transformation, and Replicon Type of cfr-Carrying Plasmids from E. coli
S1 nuclease pulsed-field gel electrophoresis (S1-PFGE) combined with Southern blotting was performed to determine the location of cfr in E. coli and the size of the plasmid according to previous protocol with minor modifications (Barton et al., 1995). Conjugation experiments were conducted using E. coli C600 (streptomycin resistant) as the recipient. Transconjugants were selected on MacConkey agar containing 3000 mg/L streptomycin and 10 mg/L florfenicol. Plasmid DNA was extracted from cfr-carrying E. coli strains and transformed into E. coli recipient strain DH5α (Takara) using the calcium chloride method when the cfr-carrying plasmid could not be transferred to E. coli C600 via conjugation. Transformants were selected on Luria-Bertani agar plates containing 10 mg/L florfenicol. The replicon types of cfr-carrying plasmids were determined by PCR-based replicon typing (PBRT), as previously described (Carattoli et al., 2005). Replicon sequence typing (RST) was performed to further characterize the IncFII plasmid (Villa et al., 2010).
Sequencing of Plasmid pHNEP28
Plasmid pHNEP28 from porcine E. coli isolate EP28 was purified from a transconjugant using a Qiagen plasmid midi kit (Qiagen, Hilden, Germany) and 3 μg DNA was quantified by NanoDropTM 2000/2000c Spectrophotometers from Thermo Scientific. Sequence was performed by the Roche 454 GS-FLX system. Contigs were assembled with the 454 GS de novo assembler (Newbler) v2.8. Gaps between contigs were closed by PCR and sequencing. Annotation of pHNEP28 was performed using the RAST server (Aziz et al., 2008), ISfinder2, and BLAST3.
Statistical Analysis
Statistical significance for the comparison of prevalence data was determined by the x2-test. p-values less than 0.05 were regarded as statistically significant.
Nucleotide Sequence Accession Number
The nucleotide sequence of the plasmid pHNEP28 has been deposited with the GenBank nucleotide sequence database under accession number KT845955.
Results
Detection of cfr and Antimicrobial Susceptibility of cfr-Positive Isolates
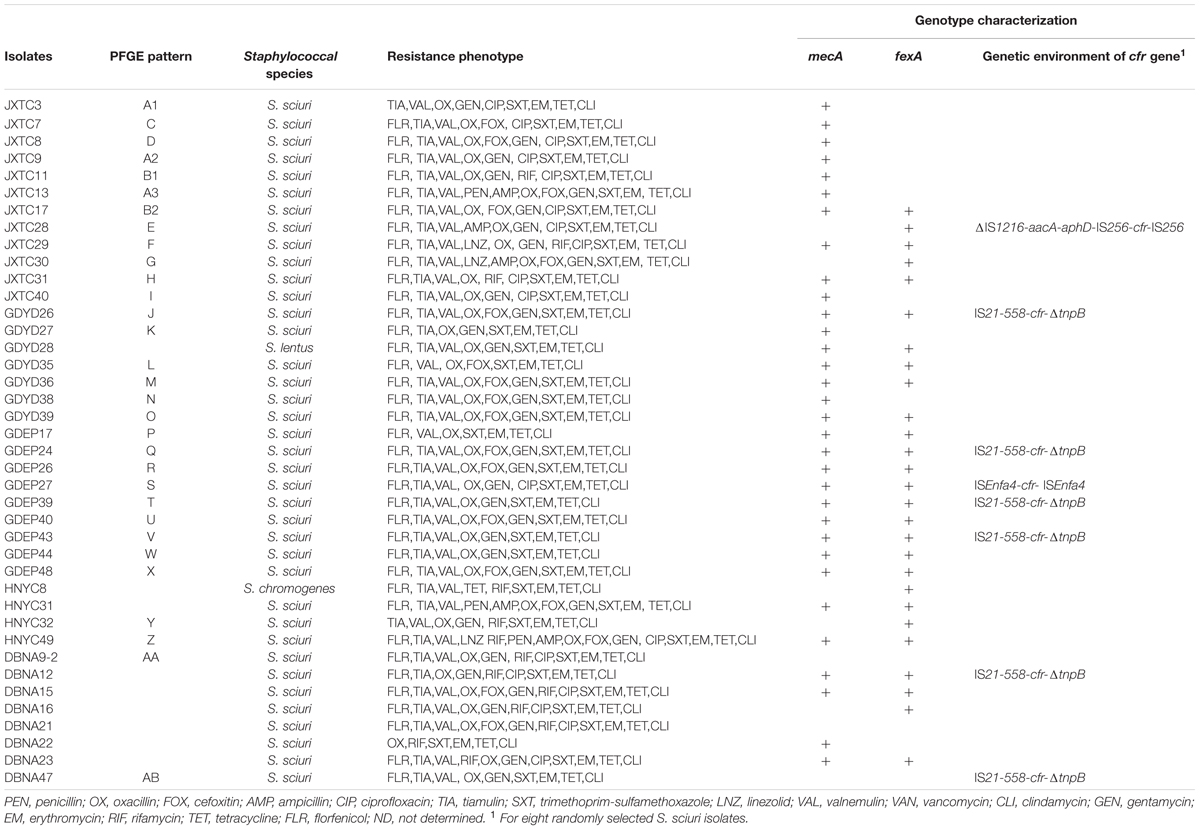
Two-hundred and fifty four Staphylococcus spp. strains and 398 E. coli strains were recovered from the 323 nasal swab and 442 fecal samples, respectively (Table 1). Among them, cfr was detected in 40 (15.7%) Staphylococcus spp. and two (0.5%) E. coli isolates. Farm GDEP in the Guangdong province showed the highest prevalence of cfr-positive (37.5%) Staphylococcus spp. strains and it was also the only farm with cfr-positive E. coli strains (Table 1). The predominant cfr-positive Staphylococcus spp. species was S. sciuri (n = 38); additionally, one S. chromogenes isolate was obtained from farm HNYC and one S. lentus isolate was found at farm GDYD.
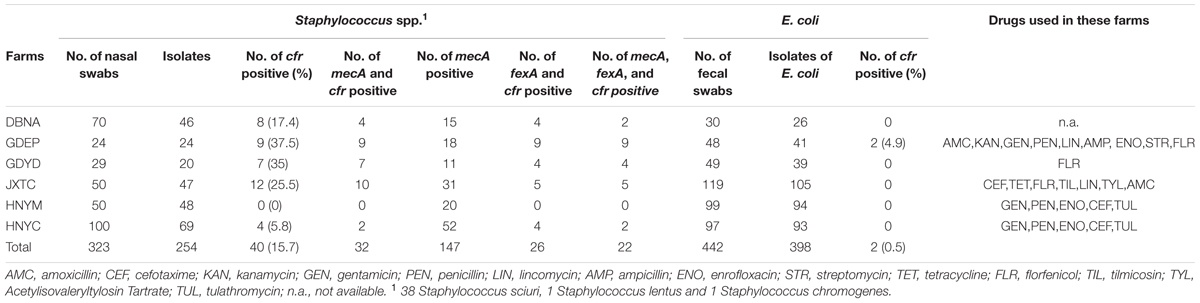
TABLE 1. Detection rates of cfr and fexA among 40 Staphylococcus spp. and 2 Escherichia coli strains from six pig farm.
Antimicrobial Resistance and Resistance Determinants
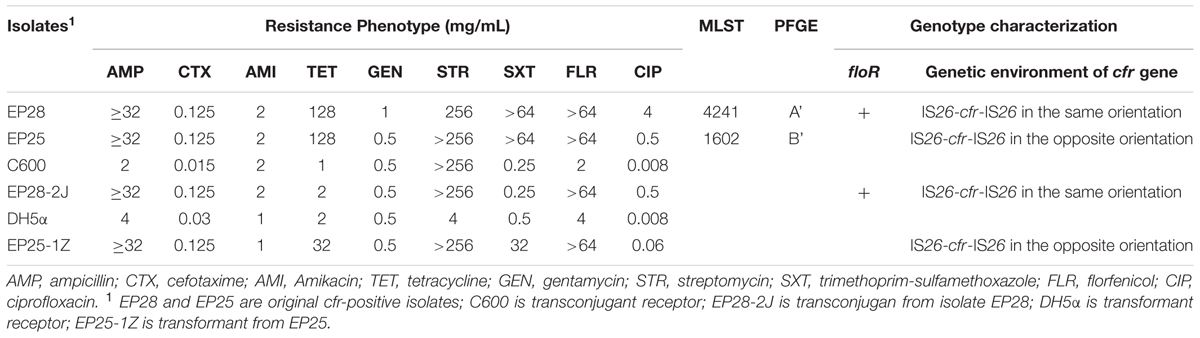
Antimicrobial susceptibility assays showed that all 40 cfr-positive Staphylococcus spp. isolates exhibited resistance to erythromycin, tetracycline, clindamycin, and trimethoprim-sulfamethoxazole. In addition, 39 (97.5%), 37 (92.5%), 36 (90%), 36 (90%), and 34 (85%) cfr-positive Staphylococcus spp. isolates demonstrated resistance to oxacillin, tiamulin, valnemulin, florfenicol, and gentamicin, respectively. Of the 40 cfr-positive staphylococci, 19 (47.5%) and 11 (27.5%) were resistant to ciprofloxacin and rifamycin, while only three (7.5%) isolates were linezolid-resistant (Table 2 and Supplementary Table S2). The two cfr-positive E. coli strains also exhibited a multiresistance phenotype, namely, ampicillin, tetracycline, streptomycin, florfenicol, and trimethoprim-sulfamethoxazole resistance. Strain EP28 was also ciprofloxacin-resistant (Table 3).
Of the 40 cfr-positive staphylococcal isolates, 25 strains carried fexA, while fexB was not detected. Interestingly, 32 of the 39 oxacillin-resistant staphylococci harbored mecA; mecC was not detected in the remaining seven mecA-negative strains. In addition, mecA, fexA, and cfr coexisted in 22 staphylococci (Table 2). The cfr-positive E. coli strain EP28 also carried floR (Table 3).
Molecular Typing
The 38 cfr-positive S. sciuri strains were analyzed by PFGE and 28 major SmaI patterns were observed, whereas seven strains were nontypeable by PFGE using SmaI (Table 2 and Supplementary Figure S1). Although obtained from the same farm, the two cfr-carrying E. coli isolates were not related (Table 3). The cfr-positive E. coli strain EP25 was shown to belong to ST1602, whereas E. coli EP28 represented a novel ST, ST4241 (Table 3). Interestingly, five cfr-positive S. sciuri strains isolated from the same farm located in the Jiangxi province were grouped in two clusters (Dice similarity index was ≥80%): strains with similar PFGE patterns A1, A2, and A3, and two strains showing related PFGE patterns B1 and B2.
Genetic Environment of cfr
The regions surrounding cfr were determined by PCR mapping and sequencing in eight randomly selected Staphylococcus strains. In six isolates, IS21-558 was identified 99% identity with upstream of cfr and truncated transposase gene tnpB was found downstream of cfr (Table 2). This arrangement (IS21-558-cfr-ΔtnpB) is identical 99% identity to the corresponding region of plasmids pSS-02 (accession no. JF834910, S. saprophyticus, swine, China), pHNTLD18 (accession no. KF751702, S. equorum, pork, China), pSA737 (accession no. KC206006, S. aureus, patient, USA), pHK01 (accession no. KC820816, S. cohnii, patient, China), and pHNCR35 (accession no. KF861983, S. simulans, hog market worker, China) (Wang et al., 2015).
In S. sciuri strain GDEP27, cfr was bracketed by two copies of ISEnfa4 in the same orientation, which was identical 98% identity to the corresponding region of E. faecalis pW9-2 (accession no. JQ911741, sewage at pig farm, China) and pHOU-cfr (accession no. JQ660368, patient, Thailand), E. thailandicus pW3 (accession no. JQ911739, sewage at pig farm, China), and p3-38 (accession no. JQ911740, pig, China).
In S. sciuri strain JXTC28, cfr is bracketed by two copies of IS256 in the same orientation, a partial fragment of IS1216 and the aminoglycoside resistance gene aacA-aphD were located downstream of IS256. This ΔIS1216-aacA-aphD-IS256-cfr-IS256 module was 98% identity to that found on plasmid pSS-04 (accession no. KF129410) carried by porcine S. sciuri GN5-1 in Sichuan province, China.
In both cfr-positive E. coli strains, the cfr gene was flanked by two copies of IS26 (Supplementary Figure S2). Strain EP25 had a similar genetic environment to plasmids pEC-01 (accession no. JN982327) identified in a porcine E. coli strain from the Shandong province, China, and pSD7 (accession no. KJ453116) found in a porcine E. coli isolate from the Guangdong province, China; 2 copies of IS26 located in the same orientation, but opposite to cfr gene (Supplementary Figure S2B). In EP28, the 2 copies of IS26 are in opposite orientation relative to one another, with cfr in the same orientation than IS26 right-copy (Supplementary Figure S2A).
Transformation of Plasmids Carrying cfr into E. coli
S1-PFGE and Southern blotting showed that the cfr gene in isolates EP25 and EP28 were located on a plasmid of approximately 95 and 105 kb, respectively. The cfr-carrying plasmids from isolate EP28 and EP25, designated as pHNEP28 and pHNEP25, were successfully transferred to E. coli C600 and DH5α, respectively. Plasmid pHNEP28 was identified as F43:A-:B-, while the replicon type of pHNEP25 could not be determined using the PBRT method. Both the transconjugant EP28-2J and the transformant EP25-1Z exhibited resistance against ampicillin, florfenicol, and elevated MICs of cefotaxime and ciprofloxacin, compared with the recipient E. coli strain (C600 or DH5α; Table 3). In addition, EP25-1Z also showed resistance to tetracycline, streptomycin, and sulfamethoxazole-trimethoprim, whereas the acquisition of pHNEP28 increased tetracycline MIC by only onefold.
Characterization of pHNEP28 from E. coli Strain EP28
Sequence analysis revealed that pHNEP28 is 108,837 bp in size with an average GC content of 49.85% (contigN50: 108837 bp). Plasmid pHNEP28 consisted a typical IncFII-type backbone (83,398 bp) encoding plasmid replication, horizontal transfer, maintenance and stability functions, and two multi-resistance regions (MRR) (Figure 1).
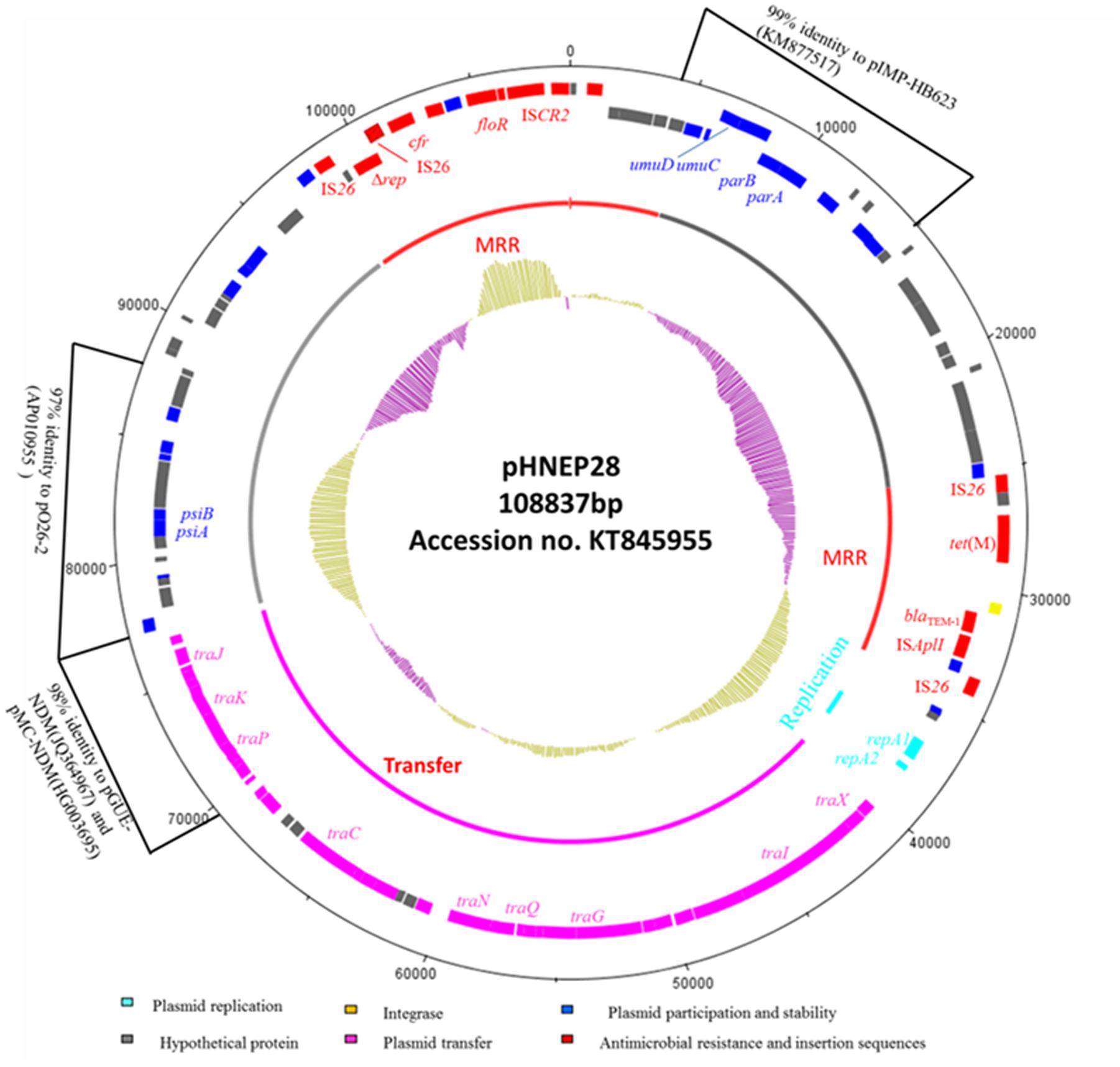
FIGURE 1. Circular map of the IncFII:43 plasmid pHNEP28. The most external circle represents the size of complete plasmid circle in bp. The clockwise orientation open reading frames (ORFs) are presented first and then in the counterclockwise orientation. The inner circle indicates the functional regions. The G+C plot is displayed in the inner circle, with yellow indicating >50% and purple indicating <50%.
The replication region (position 36,852–38,269 bp), consisting of repA2 and repA1, was classified as F43:A-:B:- by RST. This region shared 97% identity to two F2:A-:B- NDM-producing plasmids from E. coli isolates: pMC-NDM (accession no. HG003695) and pGUE-NDM (accession no. JQ364967). The transfer-leading region comprised genes related to plasmid maintenance and stability, including parA, parB, and ssb, as well as many putative genes. Additionally, genes involved in induced mutagenesis and DNA damage tolerance (umuC and umuD) and in the inhibition of the bacterial SOS response (psiA and psiB) were also identified. A section of the plasmid transfer-leading region (position 4,906–15,340 bp) shared 99% nucleotide identity with Enterobacter cloacae plasmid pIMP-HB623 (accession no. KM877517). Another section (position 77,101–88,852 bp) showed 97% identity to enteropathogenic E. coli plasmid p026_2 (accession no. AP010955).
The transfer region of pHNEP28 included 24 tra genes and finO and is less highly related to other IncFII plasmids, with ∼94%-97% identity to this region in E. coli plasmid pEB3 (accession no. CP006001) from Vietnam, which was designed to F30:A-:B:- by RST. Furthermore, the partial tra region of pHNEP28 (position 69475-77025 bp) showed high homology (98% nucleotide identity) with plasmids pMC-NDM and pGUE-NDM.
The pHNEP28 contained two mosaic MRR regions (Figure 2). The first 9,677 bp MRR, located downstream of the replication region, contains two resistance genes (tet(M) and blaTEM-1) and complete or truncated insertion sequences and transposons (IS1B, ΔintI4, ΔTn2, ISAplI, and IS26). The tet(M) gene found in pHNEP28 shared 96–98% nucleotide identity with sequences in S. pneumoniae strain 11930 (accession no. FR671416), Tn916-like transposon Tn6085 from E. faecium (accession no. HM243623), Tn916/Tn1545-like transposon from E. faecalis (accession no. DQ223241), and S. aureus strain TW20 (accession no. NG_048252); whereas it was identical to the tet(M) gene from porcine E. coli isolate CICYT-332 (accession no. KJ755873) found in Spain, although only 1802/1935 bps of CICYT-332 tet(M) were sequenced. While tet(M) demonstrated only 96% identity to that in plasmid pTCY4 (KJ772289, duck E. coli, China), the 1,204-bp module (IS26-orf12-orf13) upstream of tet(M) was identical to the corresponding region of pTCY4. Transposon Tn2 carrying resistance gene blaTEM-1 was interrupted by ISApl1, which generated 2-bp direct repeats (DR) (5′-CT-3′); tnpR of Tn2 was truncated by IS26, which interrupted another Tn2 with an opposite orientation (Figure 2A).
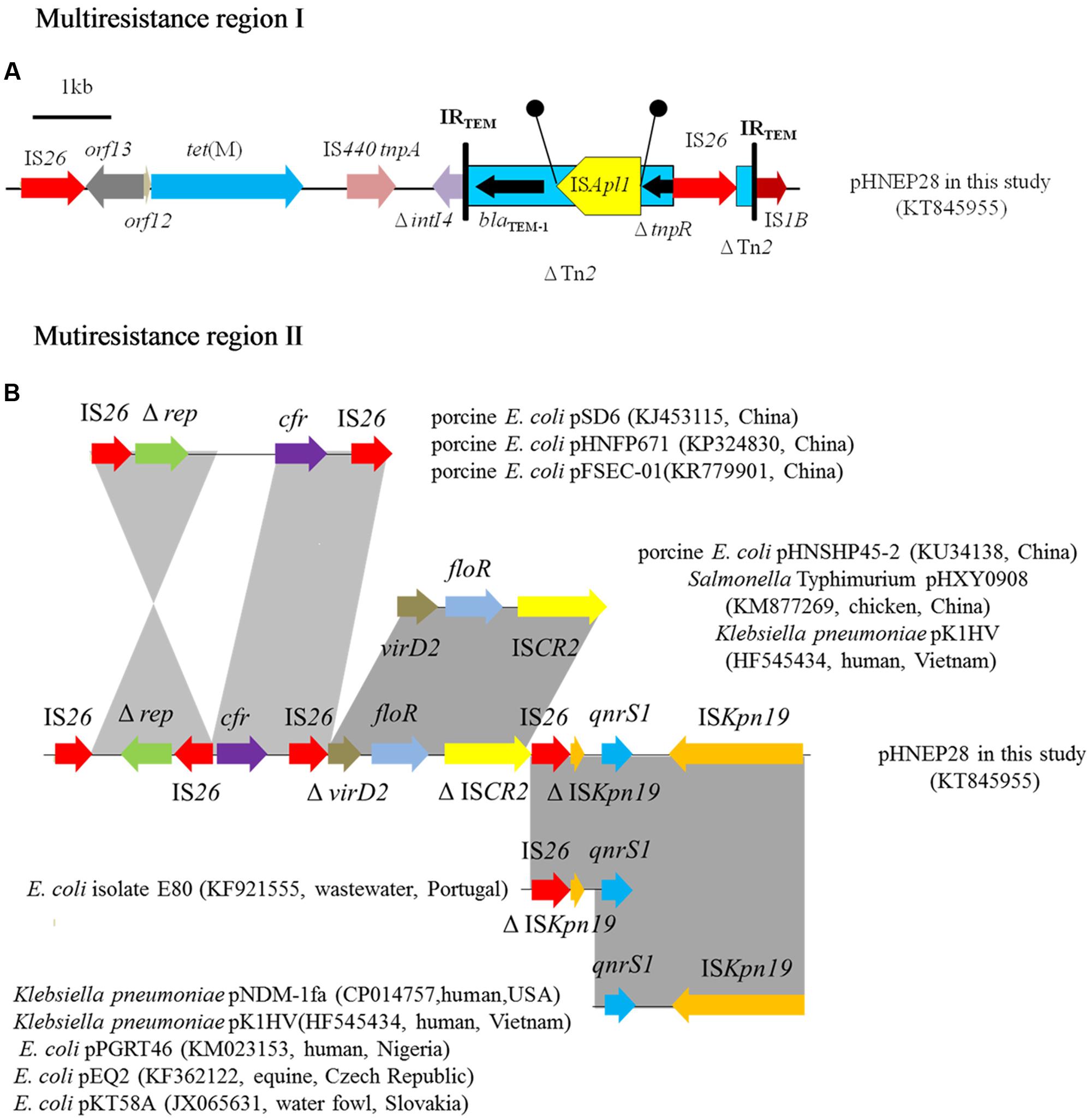
FIGURE 2. Multiresistance regions in plasmid pHNEP28. The arrows indicate the positions and transcription directions of the genes. Regions with >99% homology are shaded in gray. Δ indicates a truncated gene. Black circles mean direct repeats generated by the insertion of ISApl1. (A) Multiresistance region I in plasmid pHNEP28 (B) Mutiresistance region II in plasmid pHNEP28 and the structural comparison with other plasmids.
Another MRR module (14,370 bp), including the multiresistance gene cfr, florfenicol efflux pump gene floR, and quinolone resistance gene qnrS1, was inserted into the plasmid stability region. As shown in Figure 2B, the cfr gene was flanked by two IS26 located in the opposite orientation, in agreement with the result obtained by PCR mapping. In addition, the 2,513-bp module (Δrep-IS26) was identical to the corresponding region in plasmids pHNFP671 (accession no. KP324830), pSD6 (accession no. KJ453115), and pFSEC-01 (accession no. KR779901) from porcine E. coli in China. In plasmid pHNEP28, the Δrep-IS26 region was found in the opposite orientation with a second copy of IS26 present in the downstream region. Furthermore, another IS26 element is present downstream of cfr on plasmids pHNFP671 and pSD6, though the 2,414-bp segment containing cfr and IS26 was identical to plasmid pHNEP28 in our study (Figure 2B).
Moreover, plasmid-mediated quinolone resistance gene qnrS1 was flanked by two copies of ISKpn19 located in the opposite orientation, although the first one was interrupted by IS26 (Figure 2B). The 4615-bp module (qnrS1-ISKpn19) was present in multiple plasmids, such as Klebsiella pneumoniae plasmids pNDM-1fa (CP014757, human, USA) and pK1HV (HF545434, human, Vietnam), and E. coli plasmids pPGRT46 (KM023153, human, Nigeria), pEQ2 (KF362122, equine, Czech Republic), and pKT58A (JX065631, water fowl, Slovakia) (Dolejska et al., 2013). The IS26-ΔISKpn19-ΔqnrS1 module was identical 99% to that in E. coli isolate E80 (KF921555, raw wastewater, Portugal), containing a disrupted qnrS1 gene (Figure 2B). The 4,292-bp segment (ΔvirD2-floR-ΔISCR2) was identical 99% identity to the corresponding region of plasmids pHNSHP45-2 (KU34138, porcine E. coli, China), pHXY0908 (KM877269, chicken Salmonella Typhimurium, China), and pK1HV (HF545434, human K. pneumoniae, Vietnam); however, both virD2 and ISCR2 were truncated by IS26 in plasmid pHNEP28 (Figure 2B).
Discussion
In this study, cfr was identified in 15.7 and 0.5% of Staphylococcus spp. and E. coli isolates from pigs, respectively. The prevalence of cfr among E. coli strains was still low, which is in agreement with a previous report (Deng et al., 2014). However, to the best of our knowledge, this is the first report of cfr being found in Staphylococcus spp. and E. coli strains at the same farm. The presence of cfr-positive E. coli at farm GDEP may not be surprising given the high prevalence of cfr-positive staphylococci at that farm. The prevalence of cfr-positive staphylococci from farms GDEP (37.5%) and GDYD (35.5%) in the Guangdong province and from farm JXTC (25.5%) in the Jiangxi province was significantly higher than that in strains from farms HNYM (0%) and HNYC (5.8%) located in the Henan province (p < 0.5). According to drug usage records, florfenicol is used prophylactically or for treatment of bacterial diseases at farms GDEP, GDYD, and JXTC, but not at farms HNYM and HNYC (Table 1), which may account for the different detection rates. The direct selective pressure imposed by florfenicol appears to be the main driving force promoting the dissemination of cfr among same or different species at the same farm. This hypothesis is further supported by the presence of the florfenicol efflux gene floR in one cfr-positive E. coli isolate and the florfenicol efflux gene fexA in 25 cfr-carrying staphylococcal strains, which is higher than the prevalence of fexA among cfr-positive staphylococci (17/33, 51.5%) obtained from pigs in the Shandong province, China in 2011 (Wang et al., 2012b). In addition, the cfr-carrying isolates may also persist and be co-selected under the pressure of other agents; the results of the antimicrobial susceptibility assay reveal that the cfr-positive staphylococci exhibit a multiresistance phenotype, which is not limited to PhLOPSA.
In the present study, S. sciuri (n = 38) was the predominant species among the 40 cfr-positive staphylococcal strains. Previous report indicates that S. sciuri might be a reservoir for resistance and virulence genes both in veterinary and human medicine (Nemeghaire et al., 2014). cfr was first identified on plasmid pSCFS1 from a bovine S. sciuri in Germany (Stefan et al., 2000) and has since been detected on plasmids or the chromosomal DNA in S. sciuri strains from pigs and cattle in Germany (Kehrenberg and Schwarz, 2006) and from pigs, duck, chickens, and chicken meat in China (Wang et al., 2012a, 2013a; Zeng et al., 2014), indicating that S. sciuri is an important reservoir for the cfr gene, which contributes to the spread of cfr among animals and animal food. SmaI PFGE performed to further elucidate the dissemination of cfr-positive S. sciuri, revealed a variety of patterns. Most of the cfr-carrying S. sciuri isolates were clonally unrelated, indicating that the spread of cfr in pig farms was not due to clonal dissemination. The related PFGE patterns exhibited by a number of strains from farm JXTC, suggest that clonal dissemination of cfr-positive S. sciuri had occurred at this farm. However, the main mechanism for the dissemination of cfr among S. sciuri was probably horizontal transmission mediated by mobile genetic elements. The presence of a similar cfr-harboring module (IS21-558-cfr-ΔtnpB) in six randomly selected S. sciuri, which was identical to corresponding regions of plasmids from animals, humans, and food, may further confirm this hypothesis. In addition, cfr was bracketed by two copies of ISEnfa4 or IS256 in S. sciuri strain GDEP27 and JXTC28, respectively, which was highly similar to that in Enterococcus or Staphylococcus isolates, further suggesting these insertion sequences are associated with the transmission of cfr among same or different species from various sources.
Similarly, the two cfr-carrying E. coli showed different XbaI PFGE patterns and ST types. Additionally, cfr was flanked by two copies of IS26 in both strains, as previously reported in other E. coli plasmids, whereas IS26 in strain EP28 was in the opposite orientation. IS26 is widespread among gram-negative bacteria and can mediate the mobility of cfr in gram-negative isolates (Shen et al., 2013). The cfr-harboring module (IS26-cfr-IS26) was the most prevalent genetic structure among E. coli strains, though cfr was flanked by two copies of IS256 on plasmid pSCEC2, identified in an E. coli isolate (Zhang et al., 2014). IS26 may play an important role in the dissemination of cfr, in accordance with previous reports (Deng et al., 2014; Zhang R. et al., 2015).
Plasmid pHNEP28 obtained in the present study from E. coli isolate EP28 was classified as F43:A-:B- by RST. Because cfr has not been detected previously in an IncFII plasmid, plasmid pHNEP28 was fully sequenced. In addition to the multiresistance gene cfr, pHNEP28 contains four other resistance genes including tet(M), blaTEM-1, floR, and qnrS1. Similar to cfr, tet(M) is widely disseminated among various gram-positive organisms and is commonly related to transposons Tn916 and Tn1545 (Shen et al., 2008; Ye et al., 2008; Sadowy et al., 2013). Although tet(M) found in pHNEP28 showed only 96–98% identity to that from gram-positive isolates and only a slightly increased (onefold) tetracycline MIC, it was identical to the tet(M) identified in porcine E. coli isolate CICYT-332 from Spain, which was classified as a new E. coli tet(M) allele distantly related to enterococcal tet(M) sequences based on phylogenetic analysis; thus, the possibility of gram-positive to gram-negative strain transfer of tet(M) cannot be ruled out. Additionally, the coexistence of cfr and other resistance genes (tet(M), blaTEM-1, floR, qnrS1) on the same plasmid not only confers resistance to multiple agents, but also may allow for co-selection of cfr under selective pressure imposed by other agents, thus facilitating the dissemination of cfr. To date, the complete DNA sequences of five cfr-carrying plasmids from E. coli, including IncA/C plasmid pSCEC2 (Zhang et al., 2014), conjugative plasmid pFSEC-01 (Zhang R. et al., 2015), IncX4 plasmid pSD11 (Sun et al., 2015), and the two similar plasmids pGXEC6 and pGXEC3, have been reported (Zhang W.J. et al., 2015). Our findings not only expand the range of plasmids that capture and spread cfr, but also emphasize the importance of IncFII plasmids, which have been reported to be involved in importance in the spread of many resistance genes, such as floR, blaCTX-M, rmtB, oqxAB (Yang et al., 2015), blaCMY -2, and blaKPC (Del Franco et al., 2015), and blaNDM (Fiett et al., 2014).
Conclusion
This is the first study to demonstrate the presence of cfr among Staphylococcus spp. and E. coli isolates on the same farm and the first report of the complete sequence of a novel F43:A-:B- plasmid carrying cfr. The extensive use of florfenicol as a preventative and treatment agent in swine farms in China most likely promotes and facilitates the spread of cfr across species and genus boundaries, mainly via horizontal transfer mediated by mobile elements including plasmids and insertion sequences such as IS26 and IS256. Thus, rational use of florfenicol and alternating antibiotic therapy are required in order to reduce cfr dissemination, which constitutes a potential public health risk.
Ethics Statement
For the human cells experiments, this study was carried out in accordance with the recommendations of ethical guidelines of Chinese Academy of Sciences with written informed consent from all subjects. All subjects gave written informed consent in accordance with the Declaration of Helsinki. The protocol was approved by the Chinese Academy of Sciences.
For the zebrafish experiments, this study was carried out in accordance with the recommendations of ethical guidelines of Chinese Academy of Sciences. The protocol was approved by the ethical guidelines of Chinese Academy of Sciences.
Author Contributions
Conceived and designed the experiments: J-HL, Z-LZ, and X-QL. Performed the experiments: X-QL, WL, L-QZ, and YL. Analyzed the data: X-QL and JW. Contributed reagents/materials/analysis tools: WL, L-QZ, and YL. Wrote the manuscript: X-QL and JW.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00329/full#supplementary-material
Acknowledgments
This work was supported in part by the National Key Basic Research Program of China (No. 2013CB127200), the Program for Changjiang Scholars and Innovative Research Team in University (No. IRT13063), and the Guangdong Natural Science Foundation (No. S2012030006590).
Footnotes
- ^ http://mlst.warwick.ac.uk/mlst/mlst/dbs/Ecoli
- ^ https://www-is.biotoul.fr//
- ^ http://blast.ncbi.nlm.nih.gov/Blast.cgi
References
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Barton, B. M., Harding, G. P., and Zuccarelli, A. J. (1995). A general method for detecting and sizing large plasmids. Anal. Biochem. 226, 235–240. doi: 10.1006/abio.1995.1220
Carattoli, A., Bertini, A., Villa, L., Falbo, V., Hopkins, K. L., and Threlfall, E. J. (2005). Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63, 219–228. doi: 10.1016/j.mimet.2005.03.018
Chen, L., Chen, Z. L., Liu, J. H., Zeng, Z. L., Ma, J. Y., and Jiang, H. X. (2007). Emergence of RmtB methylase-producing Escherichia coli and Enterobacter cloacae isolates from pigs in China. J. Antimicrob. Chemother. 59, 880–885. doi: 10.1093/jac/dkm065
Chen, S., Zhao, S., White, D. G., Schroeder, C. M., Lu, R., Yang, H., et al. (2004). Characterization of multipe-antimicrobial-resistant salmonella serovars isolated from retail meats. Appl. Environ. Microbiol. 70, 1–7. doi: 10.1128/AEM.70.1.1-7.2004
CLSI (2013). Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals; Approved Standard-fourth Edition and Supplement. Documents VET01-A4E and VET01-S2E. Wayne, PA: Clinical and Laboratory Standards Institute.
CLSI (2015). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fifth Informational Supplement, M100-S25. Wayne, PA: Clinical and Laboratory Standards Institute.
Del Franco, M., Paone, L., Novati, R., Giacomazzi, C. G., Bagattini, M., Galotto, C., et al. (2015). Molecular epidemiology of carbapenem resistant Enterobacteriaceae in Valle d’Aosta region, Italy, shows the emergence of KPC-2 producing Klebsiella pneumoniae clonal complex 101 (ST101 and ST1789). BMC Microbiol. 15:260. doi: 10.1186/s12866-015-0597-z
Deng, H., Sun, J., Ma, J., Li, L., Fang, L. X., Zhang, Q., et al. (2014). Identification of the multi-resistance gene cfr in Escherichia coli isolates of animal origin.PLoS ONE 9:e102378. doi: 10.1371/journal.pone.0102378
Dolejska, M., Villa, L., Hasman, H., Hansen, L. H., and Carattoli, A. (2013). Characterization of IncN plasmids carrying blaCTX-M-1 and qnr genes in Escherichia coli and Salmonella from animals, the environment and humans. J. Antimicrob. Chemother. 68, 333–339. doi: 10.1093/jac/dks387
Fiett, J., Baraniak, A., Izdebski, R., Sitkiewicz, I., Zabicka, D., Meler, A., et al. (2014). The first NDM metallo-beta-lactamase-producing Enterobacteriaceae isolate in Poland: evolution of IncFII-type plasmids carrying the blaNDM-1 gene. Antimicrob. Agents. Chemother. 58, 1203–1207. doi: 10.1128/AAC.01197-13
Giessing, A. M., Jensen, S. S., Rasmussen, A., Hansen, L. H., Gondela, A., Long, K., et al. (2009). Identification of 8-methyladenosine as the modification catalyzed by the radical SAM methyltransferase Cfr that confers antibiotic resistance in bacteria. RNA 15, 327–336. doi: 10.1261/rna.1371409
Kehrenberg, C., and Schwarz, S. (2006). Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob. Agents Chemother. 50, 1156–1163. doi: 10.1128/AAC.50.4.1156-1163
Kehrenberg, C., Schwarz, S., Jacobsen, L., Hansen, L. H., and Vester, B. (2005). A new mechanism for chloramphenicol, florfenicol and clindamycin resistance: methylation of 23S ribosomal RNA at A2503. Mol. Microbiol. 57, 1064–1073. doi: 10.1111/j.1365-2958.2005.04754.x
Lau, S. H., Reddy, S., Cheesbrough, J., Bolton, F. J., Willshaw, G., Cheasty, T., et al. (2008). Major uropathogenic Escherichia coli strain isolated in the northwest of England identified by multilocus sequence typing. J. Clin. Microbiol. 46, 1076–1080. doi: 10.1128/JCM.02065-07
Liu, H., Wang, Y., Wu, C., Schwarz, S., Shen, Z., Jeon, B., et al. (2012). A novel phenicol exporter gene, fexB, found in enterococci of animal origin. J. Antimicrob. Chemother. 67, 322–325. doi: 10.1093/jac/dkr481
Liu, Y., Wang, Y., Dai, L., Wu, C., and Shen, J. (2014). First report of multiresistance gene cfr in Enterococcus species casseliflavus and gallinarum of swine origin. Vet. Microbiol. 170, 352–357. doi: 10.1016/j.vetmic.2014.02.037
Long, K. S., Poehlsgaard, J., Kehrenberg, C., Schwarz, S., and Vester, B. (2006). The Cfr rRNA methyltransferase confers resistance to Phenicols, Lincosamides, Oxazolidinones, Pleuromutilins, and Streptogramin A antibiotics. Antimicrob. Agents Chemother. 50, 2500–2505. doi: 10.1128/AAC.00131-06
Murakami, K., Minamide, W., Wada, K., Nakamura, E., Teraoka, H., and Watanabe, S. (1991). Identification of methicillin-resistant strains of Staphylococci by polymerase chain reaction. J. Clin. Microbiol. 29,2240–2244.
Nemeghaire, S., Argudin, M. A., Fessler, A. T., Hauschild, T., Schwarz, S., and Butaye, P. (2014). The ecological importance of the Staphylococcus sciuri species group as a reservoir for resistance and virulence genes. Vet. Microbiol. 171, 342–356. doi: 10.1016/j.vetmic.2014.02.005
Paterson, G. K., Larsen, A. R., Robb, A., Edwards, G. E., Pennycott, T. W., Foster, G., et al. (2012). The newly described mecA homologue, mecALGA251, is present in methicillin-resistant Staphylococcus aureus isolates from a diverse range of host species. J. Antimicrob. Chemother. 67, 2809–2813. doi: 10.1093/jac/dks329
Sadowy, E., Sieriko, A., Gawryszewska, I., Bojarska, A., Malinowska, K., and Hryniewicz, W. (2013). High abundance and diversity of antimicrobial resistance determinants among early vancomycin-resistant Enterococcus faecium in Poland. Eur. J. Clin. Microbiol. Infect. Dis. 32, 1193–1203. doi: 10.1007/s10096-013-1868-y
Shen, J., Wang, Y., and Schwarz, S. (2013). Presence and dissemination of the multiresistance gene cfr in Gram-positive and Gram-negative bacteria. J. Antimicrob. Chemother. 68, 1697–1706. doi: 10.1093/jac/dkt092
Shen, X., Yang, H., Yu, S., Yao, K., Wang, Y., Yuan, L., et al. (2008). Macrolide-resistance mechanisms in Streptococcus pneumoniae isolates from Chinese children in association with genes of tetM and integrase of conjugative transposons 1545. Microb. Drug. Resist. 14, 155–161. doi: 10.1089/mdr.2008.0773
Stefan, S., Christiane, W., and Corinna, K. (2000). Identification of a plasmid-borne chloramphenicol-florfenicol resistance gene in Staphylococcus sciuri. Antimicrob. Agents Chemother. 44, 2530–2533.
Sun, J., Deng, H., Li, L., Chen, M. Y., Fang, L. X., Yang, Q. E., et al. (2015). Complete nucleotide sequence of cfr-carrying IncX4 plasmid pSD11 from Escherichia coli. Antimicrob. Agents Chemother. 59, 738–741. doi: 10.1128/AAC.04388-14
Toh, S. M., Xiong, L., Arias, C. A., Villegas, M. V., Lolans, K., Quinn, J., et al. (2007). Acquisition of a natural resistance gene renders a clinical strain of methicillin-resistant Staphylococcus aureus resistant to the synthetic antibiotic linezolid. Mol. Microbiol. 64, 1506–1514. doi: 10.1111/j.1365-2958.2007.05744.x
Villa, L., Garcia-Fernandez, A., Fortini, D., and Carattoli, A. (2010). Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J. Antimicrob. Chemother. 65, 2518–2529. doi: 10.1093/jac/dkq347
Wang, J., Lin, D. C., Guo, X. M., Wei, H. K., Liu, X. Q., Chen, X. J., et al. (2015). Distribution of the multidrug resistance gene cfr in Staphylococcus species isolates from swine, farm environment and close contactors in China. Foodborne Pathog. Dis. 12, 598–605. doi: 10.1089/fpd.2014.1891
Wang, Y., He, T., Schwarz, S., Zhao, Q., Shen, Z., Wu, C., et al. (2013a). Multidrug resistance gene cfr in methicillin-resistant coagulase-negative staphylococci from chickens, ducks, and pigs in China. Int. J. Med. Microbiol. 303, 84–87. doi: 10.1016/j.ijmm.2012.12.004
Wang, Y., He, T., Schwarz, S., Zhou, D., Shen, Z., Wu, C., et al. (2012a). Detection of the staphylococcal multiresistance gene cfr in Escherichia coli of domestic-animal origin. J. Antimicrob. Chemother. 67, 1094–1098. doi: 10.1093/jac/dks020
Wang, Y., Li, D., Song, L., Liu, Y., He, T., Liu, H., et al. (2013b). First report of the multiresistance gene cfr in Streptococcus suis. Antimicrob. Agents Chemother. 57, 4061–4063. doi: 10.1128/AAC.00713-13
Wang, Y., Zhang, W., Wang, J., Wu, C., Shen, Z., Fu, X., et al. (2012b). Distribution of the multidrug resistance gene cfr in Staphylococcus species isolates from swine farms in China. Antimicrob. Agents Chemother. 56, 1485–1490. doi: 10.1128/AAC.05827-11
Weisburg, W. G., Barns, S. M., Pelletier, D. A., and Lane, D. J. (1991). 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703. doi: 10.1128/jb.173.2.697-703.1991
Yang, Q. E., Sun, J., Li, L., Deng, H., Liu, B. T., Fang, L. X., et al. (2015). IncF plasmid diversity in multi-drug resistant Escherichia coli strains from animals in China. Front. Microbiol. 6:964. doi: 10.3389/fmicb.2015.00964
Ye, C., Bai, X., Zhang, J., Jing, H., Zheng, H., Du, H., et al. (2008). Spread of Streptococcus suis sequence type 7, China. Emerg. Infect. Dis. 14, 787–791. doi: 10.3201/eid1405.070437
Zeng, Z. L., Wei, H. K., Wang, J., Lin, D. C., Liu, X. Q., and Liu, J. H. (2014). High prevalence of Cfr-producing Staphylococcus species in retail meat in Guangzhou, China. BMC Microbiol. 14:151. doi: 10.1186/1471-2180-14-151
Zhang, R., Sun, B., Wang, Y., Lei, L., Schwarz, S., and Wu, C. (2015). Characterization of a cfr-carrying plasmid from porcine Escherichia coli that closely resembles plasmid pEA3 from the plant pathogen Erwinia amylovora. Antimicrob. Agents Chemother. 60, 658–661. doi: 10.1128/AAC.02114-15
Zhang, W. J., Wang, X. M., Dai, L., Hua, X., Dong, Z., Schwarz, S., et al. (2015). Novel conjugative plasmid from Escherichia coli of swine origin that co-harbors the multi-resistance gene cfr and the extended-spectrum beta-lactamase gene blaCTX-M-14b. Antimicrob. Agents Chemother. 59, 1337–1340. doi: 10.1128/AAC.04631-14
Keywords: cfr, Escherichia coli, F43:A-:B- plasmid, Staphylococcus spp., swine
Citation: Liu X-Q, Wang J, Li W, Zhao L-Q, Lu Y, Liu J-H and Zeng Z-L (2017) Distribution of cfr in Staphylococcus spp. and Escherichia coli Strains from Pig Farms in China and Characterization of a Novel cfr-Carrying F43:A-:B- Plasmid. Front. Microbiol. 8:329. doi: 10.3389/fmicb.2017.00329
Received: 07 September 2016; Accepted: 17 February 2017;
Published: 28 February 2017.
Edited by:
Manuela Caniça, National Institute of Health Dr. Ricardo Jorge, PortugalReviewed by:
Qijing Zhang, Iowa State University, USAJuan Wang, University College Dublin, Ireland
Vera Manageiro, Institute of Agrarian and Agri-Food Sciences and Technologies (CECA), Portugal
Copyright © 2017 Liu, Wang, Li, Zhao, Lu, Liu and Zeng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhen-Ling Zeng, zlzeng@scau.edu.cn
†These authors have contributed equally to this study.
 Xiao-Qin Liu†
Xiao-Qin Liu† Li-Qing Zhao
Li-Qing Zhao Zhen-Ling Zeng
Zhen-Ling Zeng