- 1Department of Food, Nutrition, and Health, University of British Columbia, Vancouver, BC, Canada
- 2Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, BC, Canada
- 3Laboratory for Foodborne Zoonoses, Public Health Agency of Canada, Lethbridge, AB, Canada
- 4Institute for Food Safety and Hygiene, University of Zurich, Zurich, Switzerland
- 5Division for Microbiology and Production, National Food Institute, Technical University of Denmark, Kongens Lyngby, Denmark
The human pathogen Listeria monocytogenes is a large concern in the food industry where its continuous detection in food products has caused a string of recalls in North America and Europe. Most recognized for its ability to grow in foods during refrigerated storage, L. monocytogenes can also tolerate several other food-related stresses with some strains possessing higher levels of tolerances than others. The objective of this study was to use a combination of phenotypic analyses and whole genome sequencing to elucidate potential relationships between L. monocytogenes genotypes and food-related stress tolerance phenotypes. To accomplish this, 166 L. monocytogenes isolates were sequenced and evaluated for their ability to grow in cold (4°C), salt (6% NaCl, 25°C), and acid (pH 5, 25°C) stress conditions as well as survive desiccation (33% RH, 20°C). The results revealed that the stress tolerance of L. monocytogenes is associated with serotype, clonal complex (CC), full length inlA profiles, and the presence of a plasmid which was identified in 55% of isolates. Isolates with full length inlA exhibited significantly (p < 0.001) enhanced cold tolerance relative to those harboring a premature stop codon (PMSC) in this gene. Similarly, isolates possessing a plasmid demonstrated significantly (p = 0.013) enhanced acid tolerance. We also identified nine new L. monocytogenes sequence types, a new inlA PMSC, and several connections between CCs and the presence/absence or variations of specific genetic elements. A whole genome single-nucleotide-variants phylogeny revealed sporadic distribution of tolerant isolates and closely related sensitive and tolerant isolates, highlighting that minor genetic differences can influence the stress tolerance of L. monocytogenes. Specifically, a number of cold and desiccation sensitive isolates contained PMSCs in σB regulator genes (rsbS, rsbU, rsbV). Collectively, the results suggest that knowing the sequence type of an isolate in addition to screening for the presence of full-length inlA and a plasmid, could help food processors and food agency investigators determine why certain isolates might be persisting in a food processing environment. Additionally, increased sequencing of L. monocytogenes isolates in combination with stress tolerance profiling, will enhance the ability to identify genetic elements associated with higher risk strains.
Introduction
Listeria monocytogenes is a ubiquitous bacterial foodborne pathogen that is most recognized for its ability to grow at temperatures as low as −0.4°C (Walker et al., 1990) and cause listeriosis, a serious disease with an average mortality rate of 30% among at-risk people (Yildiz et al., 2007). In addition to possessing cold tolerance, L. monocytogenes is also capable of surviving many other food-related stresses including high osmolarity (Shabala et al., 2008) and low pH (Sorrells et al., 1989), further adding to its hardiness. Additionally, cross contamination of foods is facilitated by biofilm formation (Chavant et al., 2002; Møretrø and Langsrud, 2004; Moltz, 2005; Di Bonaventura et al., 2008; Hingston et al., 2013), and the ability of the organism to survive desiccation for extended periods of time on food contact surfaces (Vogel et al., 2010). Post-processing levels of L. monocytogenes contamination in foods are usually low (Fenlon et al., 1996; Kozak et al., 1996; Cabedo et al., 2008) and unlikely to cause disease (Buchanan et al., 1997; Chen et al., 2003). It is therefore, refrigerated, ready-to-eat (RTE) foods with extended shelf lives and the potential for regrowth that present the largest risk to consumers.
Both Canada and the EU have adopted regulations for the control of L. monocytogenes in RTE foods (Health Canada, 2011; Luber, 2011), allowing up to 100 CFU/g in foods that do not permit growth beyond this level within the shelf-life of the product, and a zero tolerance policy for foods identified as supporting growth. When validating growth inhibition of L. monocytogenes in stabilized RTE foods, it is important that the strains used represent the extremes of L. monocytogenes' stress response behavior. In the US, the zero tolerance policy is applicable for all food products (US FDA, 2016). However, nationwide outbreaks continue to occur in the US. To date, there have been three multistate listeriosis outbreaks in 2016 that were associated with frozen vegetables, packaged salads, and raw milk and resulted in 29 illnesses and 4 deaths (CDC, 2016a). These numbers are yet to exceed that of 2015 where two multistate outbreaks involving soft cheese and ice cream resulted in 40 illnesses and 6 deaths (CDC, 2016a).
In 2013, the US established the Listeria Whole Genome Sequencing (WGS) Project to assist in detecting, investigating, and mitigating foodborne outbreaks (CDC, 2016b). Though valuable for tracing outbreaks, WGS is not routinely used to determine the stress tolerance of outbreak strains. However, WGS provides the information that could potentially lead to identification of molecular biomarkers related to the stress tolerance of L. monocytogenes isolates. Such biomarkers could greatly aid in monitoring the risks of L. monocytogenes contamination and regrowth in food products and processing environments (Jacquet et al., 2004).
Currently, one common molecular biomarker used for L. monocytogenes' virulence is the internalin A encoding gene (inlA) which can contain one of several different premature stop codons producing truncated and secreted proteins associated with attenuated virulence (Jonquieres et al., 1998; Jacquet et al., 2004; Rousseaux et al., 2004; Nightingale et al., 2005; Felicio et al., 2007; Handa-Miya et al., 2007; Roldgaard et al., 2009; Van Stelten et al., 2011). Recently, Kovacevic et al. (2013) discovered that full-length variants of inlA were more prevalent among fast cold-adapting L. monocytogenes strains than intermediate and slow cold-adapting strains, suggesting that inlA profiling may also be suitable for predicting the cold tolerance of strains. Another potential biomarker is the L. monocytogenes stress survival islet 1 (SSI-1). Included in this five gene cluster (lmo0444–lmo0448) are two genes (gadT1 and gadD1) from the glutamate decarboxylase acid resistance system which has been shown to significantly improve the growth of L. monocytogenes in mildly acidic environments (Cotter et al., 2005). Additionally, an L. monocytogenes SSI-1 deletion mutant exhibited impaired growth at low pH (pH 4.8), high salt (7.5% NaCl), and on frankfurters stored at 4°C (Ryan et al., 2010). Further, research with naturally occurring SSI-1 positive and negative strains is needed to determine if this island would be a suitable biomarker for predicting stress tolerance phenotypes.
To date, studies which evaluated the stress tolerances of L. monocytogenes isolates have focused on associating phenotypes with genetic lineages (Bergholz et al., 2010), serotypes (Junttila et al., 1988; Barbosa et al., 1994; Ribeiro et al., 2014), and isolation sources (Begot et al., 1997; Durack et al., 2013). However, few significant differences between these groups were observed, suggesting that the diversity among isolates within these means of classification is not definitive for predicting phenotypic behavior. Instead, stronger phenotype associations might be observed among more closely related isolates, e.g., those sharing the same sequence type (ST) or clonal complex (CC). Additionally, the presence of specific genetic elements (e.g., inlA and SSI-1) may also influence the stress tolerance phenotypes of isolates as well as more minor genetic differences such as single nucleotide variants (SNVs).
The objective of this study was to use a combination of phenotypic analyses and WGS to elucidate novel associations between L. monocytogenes genotypes and food-related stress tolerance phenotypes with the goal of identifying biomarkers that can be used to predict the stress tolerances of food-chain isolates. To accomplish this, 166 L. monocytogenes isolates were evaluated on their ability to grow in cold (4°C), salt (6% NaCl), and acid (pH 5) stress conditions as well as survive desiccation stress (33% RH). Factors investigated for potential associations with the observed phenotypes were: genetic lineage, serotype, CC, inlA profiles, and the presence of a plasmid, SSI-1, unique SNVs, and Listeria genomic island 1 (LGI1).
Materials and Methods
Isolates and Culture Conditions
A collection of 166 Listeria monocytogenes isolates from Canada and Switzerland were used in this collaborative study. This included: (i) 159 food and food processing environment isolates from Canada (n = 139) and Switzerland (n = 20), (ii) six isolates from sporadic human listeriosis cases in Switzerland, and (iii) one isolate from an asymptomatic human (Table S1). All human isolates were anonymized and no ethical approval was required as per the institutional and national guidelines. Isolates were stored at −70°C in brain heart infusion broth (BHIB, Difco, Fisher Scientific, Canada) +20% glycerol and routinely cultured at 30°C on BHI agar (BHIA, Difco, Fisher Scientific) plates.
Whole Genome Sequencing
Genomic DNA was isolated using the PureLink Mini Kit from Life Technologies, Canada. PicoGreen quantification was performed (Invitrogen, Canada) and DNA was assessed using the NanoDrop 2000 (Fisher Scientific). Genomic DNA samples of sufficient quality and quantity were sequenced by Genome Quebec (Montréal, QC, Canada) using TruSeq automated library preparation (Illumina) and paired-end, 100 bp sequencing on the Illumina Hi-Seq. Between 4.9 and 16.5 million high quality reads remained after quality control for each genomic library. Raw FASTQ files were trimmed using Cutadapt in Trim Galore! version 0.4.1 and de novo genome assembly was performed using SPAdes version 3.1.0 (careful option used; Bankevich et al., 2012). Low coverage (<10) and small contigs (<200 bp) were removed from assemblies using a custom perl script. Assemblies were subsequently annotated using Prokka version 1.5.2 (genus Listeria, species monocytogenes; Seemann, 2014). Assembled sequences were deposited into the NCBI Whole Genome Shotgun (WGS) database under Bioproject PRJNA329415.
Lineage Determination
To classify isolates into genetic lineages, a reference free, k-mer based single nucleotide variants (SNV) phylogeny was generated using the kSNP 3.0 program (Gardner et al., 2015) and reference isolates for the major lineages of L. monocytogenes (LI—F2365; LII—EGD-e; LIII—HCC23; LIV—J1-208). The resulting maximum parsimony tree (based on the consensus of 100 trees) clearly segregated the four lineages.
Multi Locus Sequence Typing
To group isolates based on their epidemiological context, in silico MLST was performed using the Center for Genomic Epidemiology's MLST typing tool (https://cge.cbs.dtu.dk/services/MLST/). Clonal Complexes (CCs) were assigned based on the Pasteur Institute schema (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Lmono.html). Novel sequence types (STs) were confirmed using Sanger sequencing and submitted to the Pasteur Institute Database for new assignments (http://bigsdb.pasteur.fr/listeria/listeria.html).
In silico Serogroup/Serotype Assignment
Antibody-based serotyping was conducted on a subset of isolates (n = 91) within both the current study and previous studies (Arguedas-Villa et al., 2010; Kovacevic et al., 2013). Remaining isolates were assigned one or more possible serotypes by performing a MegaBLAST search (>95% nt identity) ncbi-blast+ v. 2.3.0 available at: ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/) for four genes used in a multiplex PCR developed by Doumith et al. (2004). Additionally, predictive serotypes were assigned to isolates with STs that are known to be associated with a specific serotype.
Targeted Genomic Element Screenings
The genes and genomic regions evaluated in this study were (1) the plasmid replicon gene repA, used to indicate the presence of a plasmid (Kuenne et al., 2010), (2) emrE, representing Listeria genomic island 1 (LGI1), a 50 kb island with putative roles in stress tolerance and persistence (Gilmour et al., 2010), and (3) stress survival islet 1 (SSI-1), a five gene cluster previously identified as having a role in L. monocytogenes' response to cold, osmotic, and acid stress conditions (Ryan et al., 2010). Additionally, the coding sequence of inlA was investigated to determine if isolates possessed a full length sequence or a premature stop codon (PMSC) mutation. emrE, SSI-1, and inlA were screened for among isolate sequence assemblies using MegaBLAST (>95% nt identity) and repA was screened for using BLASTP (>30% aa identity over >80% coverage). inlA and repA sequences were then extracted from the isolate assemblies for further analysis.
Identification of Putative Plasmid Contigs
Detection of repA sequences meant that at least one contig belonged to a putative plasmid. To identify additional plasmid associated contigs, isolate assemblies were aligned to the closed genome of L. monocytogenes EDG-e (Accession: NC_003210.1) using Contig Mover in Mauve version 2.3.1 (Rissman et al., 2009). Contigs not aligning to the EDG-e chromosome were compared to published L. monocytogenes plasmids (Kuenne et al., 2010) by BLAST. Contigs were excluded if they displayed open readings frames associated with chromosomal DNA (e.g., rRNA, tRNA) or did not align to any of the Listeria associated plasmids annotated by Kuenne et al. (2010): pLM33, pLM1-2bUG1, pLM5578, pLM80, and pLI100. A summary of the putative plasmid contigs found within each isolate can be found in Table S1.
Cold Tolerance Assay
Overnight cultures grown in BHIB at 37°C were standardized to 109 CFU/ml using spectrophotometric methods, and diluted in pre-chilled BHIB to yield a final density of 103 CFU/ml and stored at 4°C (previously described in Arguedas-Villa et al., 2010). The bacterial density was enumerated daily for the first 4 days and then bi-weekly for up to 5 weeks by plating on tryptic soy agar (BD, Fisher Scientific) +6% yeast extract (BD, Fisher Scientific). The resulting growth curves were fitted using a four parameter logistic model described by Dalgaard and Koutsoumanis (2001).
Salt and Acid Tolerance Assay
Isolates were assessed for salt and acid tolerance using modified versions of published protocols (Cotter et al., 2005; Van Der Veen et al., 2008; Bergholz et al., 2010). In short, overnight cultures grown in BHIB at 30°C were diluted in either BHIB+6% (w/w) NaCl or BHIB adjusted to pH 5 (with 1 M HCl) to achieve a final concentration of 107 CFU/ml. From these cultures, 200 μl was added in duplicate (technical replicates) to 96-well-plates (Costar™ clear polystyrene, Fisher Scientific) that were incubated at 25°C in a microplate reader (Spectramax, V6.3; Molecular Devices, Sunnyvale, CA). A temperature of 25°C was used to assess isolate salt and acid tolerance under non-intracellular or cold stress conditions. The absorbance (A600nm) of each well was recorded every 30 min until all isolates reached stationary phase (~26 h) and the resulting growth curves were fitted to the Baranyi and Roberts model (Baranyi and Roberts, 1994) using DMfit (v3.5) available on the ComBase browser (http://browser.combase.cc/DMFit.aspx).
Desiccation Tolerance Assay
Cultures grown for 24 h in BHIB at 20°C were diluted to 107 CFU/ml in buffered peptone water (BD, Fisher Scientific) and 10 μl (105 CFU) was spotted in duplicate (technical replicates) on the bottom of wells in lid-less 96-well-plates. The plates were then stored for 3 days at 20°C in desiccators (SICCO, Bohlender, Germany) pre-conditioned to 33% relative humidity (RH) using a saturated solution of MgCl2 (protocol adapted from Hingston et al., 2015). The RH of the chambers was monitored throughout the desiccation periods using data loggers (included with desiccators). A temperature of 20°C was used to simulate desiccation conditions that might occur in a food plant. Following desiccation, the plates were rehydrated with 200 μl of BHIB, and incubated at 25°C in a plate reader where the A600nm of each well was recorded every 30 min until all isolates reached stationary phase (~24 h). The resulting growth curves were then fitted to the Baranyi and Roberts model and the model parameters recorded.
Phenotype Designations and Statistical Analyses
For all four stress exposure experiments, a minimum of two biological replicates with two technical replicates each, were conducted for all isolates. Based on the findings of Aryani et al. (2015), the data was standardized for biological variability between replicates by dividing isolate growth parameters by the median value for each experimental run, thereby making the median equal to 1. The median was selected for standardization rather than the mean to avoid the influence of very stress sensitive isolates. Model parameters (LPD, lag phase duration; μmax, maximum growth rate; Nmax, maximum cell density) were averaged across biological replicates and presented as standardized (std) values. For isolates where the average std values had a standard deviation (SD) >0.05, additional replicates were completed to obtain more representative means. Isolates were considered tolerant or sensitive to cold, salt, or acid stress if they had an average std-μmax > or < than 1 SD from the median, respectively. All remaining isolates were considered to have intermediate stress tolerance. For desiccation stress survival the model parameter of most interest was the LPD, indicating the time to (detectable) regrowth (TRG) that is negatively correlated with the number of cells, which survived the desiccation treatment. Isolates were classified as desiccation tolerant or sensitive if they had an average std-TRG < or > than 1 SD from the median, respectively. A standard curve generated using five cell levels (101–105 CFU) produced a correlation of y = −0.25x + 2.07 (R2 = 0.97) where y is the TDR and x is the log10 number of viable cells in each well following desiccation.
To elucidate potential associations between the factors we investigated, statistical tests were performed using IBM SPSS Statistics version 23. Specifically, individual two-tailed T-tests and one-way ANOVAs with Tukey post-hoc tests were used to compare the average standardized stress tolerance model parameters of two (±plasmid, ±SSI-1, ±LGI1, lineage I and II, repA group 1 vs. group 2 isolates) or more groups (serotypes, CCs, inlA profiles, and sensitive, intermediate and stress tolerant groups), respectively. G*Power 3.1.9.2 (Faul et al., 2007, 2009) was used to determine the minimum sample sizes required to ensure a power of 0.80 for all statistical tests. All data sets were accessed for outliers, homogeneity of variances (Levene's test), and normality (Shapiro-Wilk's test). Where homogeneity of variances could not be achieved, Welch T-tests and Welch ANOVAs in combination with Games-Howell post-hoc tests were used. P-values below 0.05 were considered significant for all comparisons.
Phylogenetic Reconstruction Based on Core Genome Single Nucleotide Variants
Parsnp, a tool within Harvest suite of tools (Treangen et al., 2014), was used to perform core genome alignment of all 166 de novo assembled genomes and the reference L. monocytogenes EGD-e strain in order to identify single nucleotide variants (SNVs) within the core genome. SNVs clustered within 20 base pairs were removed as these may indicate repetitive regions containing more erroneous SNV calls. The remaining high quality SNVs were used to generate maximum likelihood trees using the RaxML version 8 (Stamatakis, 2014) on the CIPRES science gateway (Miller et al., 2010) using default parameters (including the GTRCAT nucleotide model and 100 bootstrap replicates). Corresponding heatmaps containing additional genotype and phenotype information were generated in R version 2.15.1 (Team, 2016) using the heatmap.2 function from the gplots library.
SNV Detection
SNVs were also detected against the Listeria monocytogenes EGD-e (NC_003210.1) reference genome. SMALT version 0.7.6 (http://www.sanger.ac.uk/science/tools/smalt-0) with default parameters except “–i 330” was used to first align raw reads against the reference. Samtools version 1.2 (Li, 2011) was used on these assemblies to sort the aligned reads (“samtools sort”), remove potential PCR duplicates (“samtools rmdup”) and call the SNVs (“samtools mpileup”). Additional filtering of SNV calls included removing those with a read depth <50 and heterozygous genotypes (since our genomes are haploid) using the “bcftools filter” command. SNVs found in repetitive regions of the genome as assessed by the index of repetitiveness (Schwender et al., 2004) were also removed manually. The remaining high confidence SNVs were annotated using SNPEff version 4.1 (Cingolani et al., 2012) with the Listeria_monocytogenes_EGD_e_uid61583 annotation. Synonymous SNVs were also removed in the end for identification of non-synonymous or potential regulatory SNVs that may be contributing to phenotypic differences in cold growth.
Statistical Methods for Elucidating SNVs Associated with Stress Tolerance Phenotypes
SNPSift version 4.1 “CaseControl” (Cingolani et al., 2012) was used to run a Fisher Exact test to identify SNVs that were significantly associated with case vs. control groups. To identify SNVs only found in tolerant isolates, these were used as the case group, while all others were used as the control group. Since this did not yield any results, subsequently, the sensitive isolates alone were used as the control group so as to allow SNVs to be seen in intermediate growers. This method has certain limitations in that certain associations may require very large sample sizes to become statistically significant, especially considering genetic heterogeneity leads to the same phenotype or the potential for multiple SNVs to interact. An alternative approach, Random Forests™ (Breiman, 2001), was also used to discover important SNVs in distinguishing stress tolerant and sensitive groups since previous genome wide association studies have shown random forests outperform the Fisher Exact test in these special cases (Lunetta et al., 2004; Schwender et al., 2004; Bureau et al., 2005; De Lobel et al., 2010; Bulinski et al., 2011). The RandomForest™ version 4.6–10 library was used in R with default parameters except “importance = TRUE, proximity = TRUE, ntree = 5000.” This allows the method to run as a classifier that then ranks SNVs on their ability to classify isolates based on their phenotypic designation.
Genomic Islands Analysis
Annotated draft genomes were submitted to IslandViewer 3 (Dhillon et al., 2015) using L. monocytogenes EGD-e (NC_003210.1) as a reference for contig reordering. Genomic islands were predicted using IslandPath-DIMOB (Hsiao et al., 2005) and SIGI-HMM (Waack et al., 2006). Predicted genomic islands positioned on the genome within <10 kb of each other were merged into one single region.
To form groups of similar genomic islands, the genetic distance between genomic island sequences was computing using Mash (parameter –s 2000; Ondov et al., 2016) and groups of similar sequences were identified using hclust and cutree in R.
Results
Genetic Characteristics of L. monocytogenes Isolates Based on WGS Data
The complete sequenced genome assembly sizes of the isolates ranged from 2.56 to 3.13 Mbp with a mean size of 2.97 Mbp (Table S1). Isolates belonged to one of three different lineages: LI (n = 44, serotypes 4b, 1/2b, 3b, and 3c), LII (n = 121, serotypes 1/2a, 1/2c, and 3a), and LIII (n = 1, serotype 4c). The majority of isolates were serotype 1/2a (n = 92), followed by 1/2c and 4b (n = 25 each), 1/2b (n = 18), 3a (n = 2), and 3b and 4c (n = 1 each; Table 1). The exact serotype was not determined for two remaining isolates. Beyond serotypes, our isolates belonged to 36 different known STs and a further nine were assigned novel STs (ST1017-1025). Isolates also belonged to one of 29 different CCs with a further seven isolates being unique non-clonal singletons. The most prevalent CCs in the collection were CCs 9, 8, and 7 (Table 2). Other less common CCs in decreasing prevalence included CCs 11, 155, 1, 3, and 321 (Table 2). Interestingly, only one CC121 isolate existed in our collection. This is surprising given that CC121 is often highly prevalent among L. monocytogenes food-associated isolates (Parisi et al., 2010; Chenal-Francisque et al., 2011; Martín et al., 2014; Ebner et al., 2015; Maury et al., 2016).
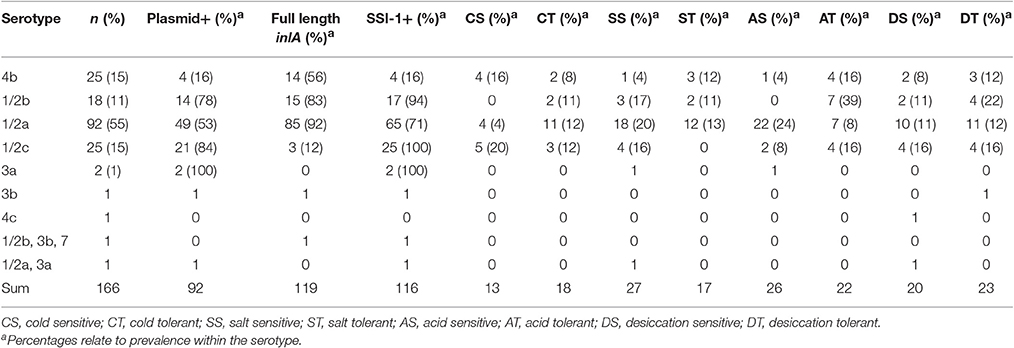
Table 1. Genetic characteristics and prevalence of sensitive and tolerant phenotypes among L. monocytogenes belonging to different serotypes.
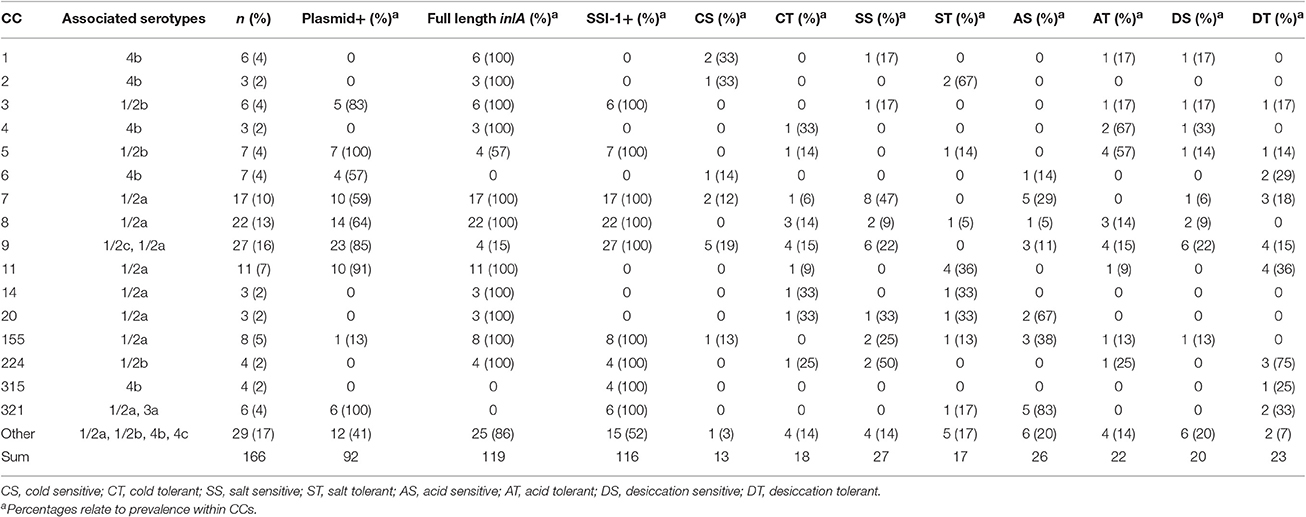
Table 2. Genetic characteristics and prevalence of sensitive and tolerant phenotypes among L. monocytogenes belonging to different clonal complexes.
The plasmid replication gene, repA, was observed in 55% (n = 92) of our isolates, with a prevalence of 41 and 61% among LI and LII isolates, respectively. Notably, one isolate was observed to contain two putative plasmids as indicated by the presence of two different repA containing contigs of 61 and 69 kb. Among serotypes, repA was present in 100% of 3a isolates, 84% of 1/2c isolates, 78% of 1/2b isolates, 53% of 1/2a isolates, and 16% of 4b isolates (Table 1). Among CCs, plasmids were observed in >80% of CC 3, 5, 9, 11, and 321 isolates (Table 2).
A phylogeny, constructed on repA sequences as described in Kuenne et al. (2010), divided the sequences into two groups. Group 1 included isolates from serotypes 1/2a, 1/2b, 1/2c, and 4b with estimated plasmid sizes ranging from 26 to 88 kb. Group 2 included 1/2a, 1/2b, 1/2c, and 3a serotype isolates, harboring significantly larger plasmids (p < 0.0005, 55–100 kb) than those from group 1. These sizes are in line with those observed in Kuenne et al. (2010), supporting the assertion that these contigs belong to plasmids. The most prevalent plasmid size (56553–56554 bp) was observed for 26 isolates from seven different CCs and from both lineages I and II. Also noteworthy is that isolates from Switzerland and Canada contained plasmids with 100% nucleotide identity.
Premature stop codons (PMSCs) in inlA were observed in 20% of our isolates encompassing seven (Table S1) of 19 published PMSCs (Jonquieres et al., 1998; Olier et al., 2002, 2003; Rousseaux et al., 2004; Nightingale et al., 2005, 2008; Orsi et al., 2007; Van Stelten and Nightingale, 2008; Van Stelten et al., 2010; Wu et al., 2016) and one novel PMSC at 760aa, which was identified in two serotype 1/2c isolates. The most common PMSC occurred at 9aa (n = 13) and was associated with CC9, serotype 1/2c isolates. Ten of the 14 remaining CC9 isolates also had one of four inlA PMSCs (326, 576, 685, 760aa) and all CC321 isolates contained inlA PMSC's at 700aa (Table S1). An additional 13 isolates, all from the serotype 4b CCs 6 and 315, contained a three codon deletion mutation previously reported in Kovacevic et al. (2013). With the exception of CCs 5 and 9, all isolates from the same CC either contained full length inlA or a truncated version.
During the screening of the whole genome sequences, the absence of lmo1078 was noted among serotype 4b isolates. This gene, which encodes a UDP-glucose pyrophosphorylase, has been previously demonstrated to have a role in L. monocytogenes cold growth (Chassaing and Auvray, 2007). It was also observed that 70% (n = 116) of strains possessed SSI-1 with this island being most prevalent among serotype 1/2c isolates (100%) followed by 1/2b (94%), 1/2a (71%), and 4b (16%; Table 2). Furthermore, all isolates from CCs 3, 5, 7, 8, 9, 155, 224, 315, and 321 contained SSI-1 (Table 2). All remaining isolates possessed a homolog to F2365_0481 in place of SSI-1 (Ryan et al., 2010), with the exception of the CC121 isolate which possessed lin0464 and lin0465 homologs as reported in Hein et al. (2011).
The LGI1 indicator gene, emrE, was found in 16 of our isolates and as previously reported (Gilmour et al., 2010; Althaus et al., 2014; Kovacevic et al., 2015), all originated from Canada and 14 were serotype 1/2a ST120-CC8. The remaining two isolates represented novel STs (ST1022 and 1025) that also belonged to CC8. All emrE containing isolates also harbored SSI-1 and full length inlA.
Stress Tolerance Distributions among L. monocytogenes Isolates
All L. monocytogenes isolates were evaluated on their ability to grow in cold (4°C), salt (6% NaCl), and acid (pH 5) stress conditions as well as survive desiccation stress (33% RH). The cold growth plate count data was modeled using the Dalgaard and Koutsoumanis (2001) logistic model because it was more accommodating of fewer sampling points [average R2 = 0.998, mean standard error (MSE) = 0.129]. From the std-μmax values, 13 isolates were classified as cold sensitive and 18 were classified as cold tolerant with average std-μmax values of 0.85 ± 0.08 and 1.09 ± 0.02, respectively (Figure 1A). For the salt, acid, and desiccation tolerance assays, the Baranyi and Roberts (1994) was suitable for modeling the spectrophotometrically obtained data with average R2 and MSE-values ranging from 0.997 to 0.998 and 0.003 to 0.017, respectively. Overall, 27 and 17 isolates were classified as salt sensitive and tolerant with average std-μmax values of 0.83 ± 0.05 and 1.16 ± 0.05 (Figure 1B); 26 and 22 isolates were classified as acid sensitive and tolerant with average std-μmax values of 0.64 ± 0.14 and 1.34 ± 0.12 (Figure 1C); and 20 and 23 isolates were identified as desiccation sensitive and tolerant isolates with average std-TRGs of 0.81 ± 0.06 and 1.22 ± 0.11 (Figure 1D), respectively.
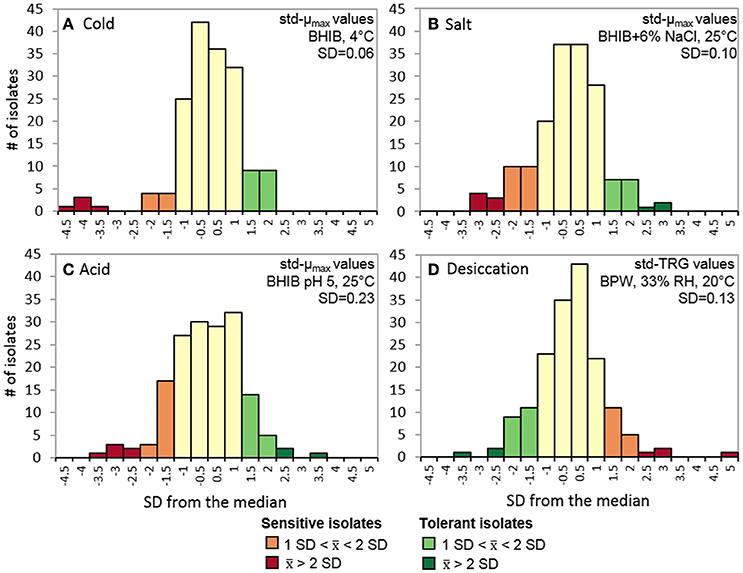
Figure 1. Stress tolerance distributions of 166 L. monocytogenes isolates. Std-μmax of isolates grown in (A) BHIB at 4°C, (B) BHIB+6% NaCl at 25°C, and (C) BHIB pH 5 at 25°C. (D) std-TRG of isolates after being desiccated at 33% RH for 3 days in BPW at 20°C and then rehydrated with BHIB and grown at 30°C. Isolates were classified as sensitive or tolerant if they displayed an average std-μmaxorstd-TRG >1 SD from the median (= 1). std-μmax, standardized maximum growth rate; std-TRG, standardized time to detectable regrowth; BHIB, brain heart infusion broth; BPW, buffered peptone water.
Overlapping Stress-Tolerance Phenotypes
Five isolates were classified as sensitive to three out of four stresses and 10 isolates were sensitive to two stresses, seven of which were salt and acid sensitive (Figure 2A). Only two isolates were classified as tolerant to three out of the four stresses and another 14 isolates were tolerant to two of the four stresses (Figure 2B). Twenty additional isolates displayed a total of 16 combinations of overlapping sensitive and tolerant phenotypes (Table S1). The most common overlapping phenotypes were salt and acid sensitive (n = 10), salt sensitive and desiccation tolerant (n = 6), cold and acid tolerant (n = 5), and cold tolerant and salt sensitive (n = 5).
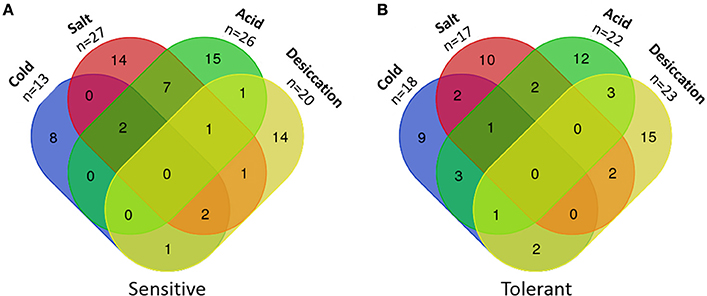
Figure 2. Numbers of L. monocytogenes isolates with multiple sensitivities or tolerances to food-related stresses. (A) Sensitive isolates. (B) Tolerant isolates.
Isolates designated sensitive, intermediate or tolerant to one stress were analyzed to determine if they significantly differed in their tolerances to other stresses. When grown in 6% NaCl, acid tolerant isolates had larger std-μmax values compared to acid sensitive isolates (p = 0.03, x 1.02 vs. 0.95). Similarly, salt sensitive isolates had smaller std-μmax values (x = 0.80) in BHIB pH 5 compared to intermediate (p < 0.0005, x = 1.03) and salt tolerant isolates (p = 0.006, x = 1.00).
Stress Tolerances of L. monocytogenes Lineages, Serotypes, and Clonal Complexes
Between lineages, the only significant difference observed was that LI isolates had significantly larger std-μmax values in BHIB pH 5 than LII isolates (p < 0.0005, x = 1.13 vs. x = 0.94). Additional significant differences were observed between serotypes. At 4°C, serotype 1/2a isolates had significantly larger (p = 0.017) std-μmax values compared to serotype 1/2c isolates (Figure 3). In support of this, serotype 1/2a isolates accounted for 61% of the cold tolerant isolates and only 31% of cold sensitive isolates compared to a 55% prevalence of this serotype in the collection (Table 1). Similarly, serotype 1/2c isolates accounted for 38% of cold sensitive isolates despite a 15% overall prevalence in the collection (Table 1). When isolates were grown in 6% NaCl, no significant differences were observed between serotypes (Figure 3), however, 71% of salt tolerant and 67% of salt sensitive isolates were serotype 1/2a isolates, relative again to a prevalence of 55% in the collection (Table 1).
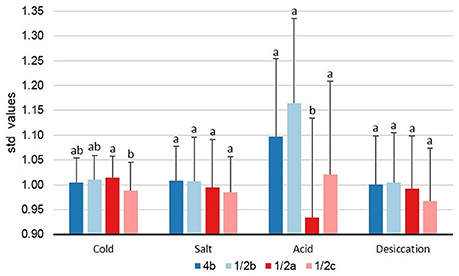
Figure 3. Levels of tolerance to food-related stresses among L. monocytogenes serotypes. Isolates were evaluated on their ability to survive cold (BHIB at 4°C), salt (BHIB+6% NaCl, 25°C), acid (BHIB pH 5, 25°C), and desiccation stress (33% RH for 3 days at 20°C followed by rehydration with BHIB at 30°C). Error bars represent standard deviations (n − 1). Serotypes with different letters within the same stress are significantly different (p < 0.05). std-μmax, standardized maximum growth rate; std-TRG, standardized time to detectable regrowth; BHIB, brain heart infusion broth.
In BHIB pH 5, serotype 1/2a isolates had significantly smaller (p = 0.027) std-μmax values than serotypes 1/2b, 1/2c, and 4b (Figure 3). In agreement with these findings, 85% of acid sensitive isolates were serotype 1/2a whereas only 32% of acid tolerant isolates were serotype 1/2a (Table 1). No significant differences (p > 0.05) were observed between serotypes with respect to desiccation stress std-TRGs (Figure 3).
Beyond the stress tolerances of lineages and serotypes, some significant differences were also detected between CCs. As a minimum of six isolates per CC were needed to ensure a power >0.80 for ANOVA results, statistical analyses were only performed using CCs 1, 3, 5, 6, 7, 8, 9, 11, 155, and 321. Figure 4 shows the average levels of cold (std-μmax), salt (std-μmax), acid (std-μmax), and desiccation (std-TRG) tolerance among CCs with three or more isolates. At At 4°C, no significant differences were found between the growth rates of different CCs, however, it was interestingly to see that CCs associated with 4b isolates had both the lowest and highest average average std-μmax values at 4°C, demonstrating why stress tolerance differences were not observed between this serotype and others at 4°C (Figure 4A).
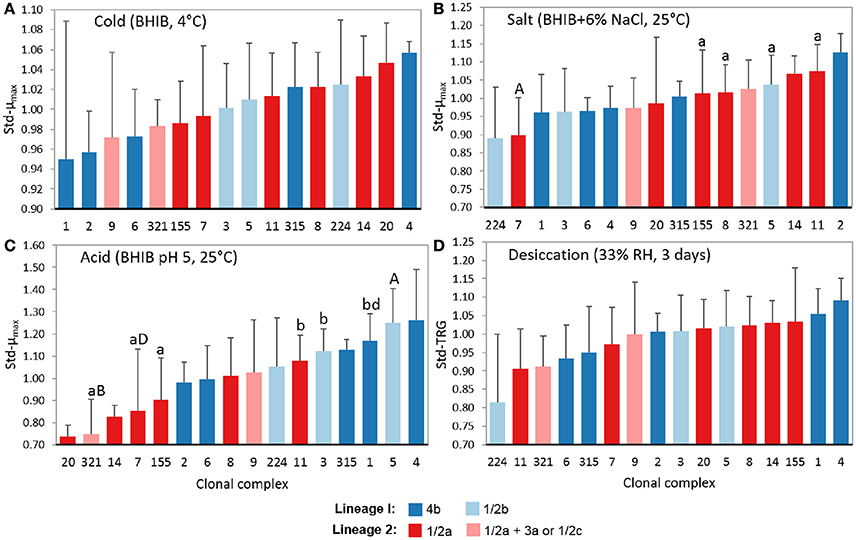
Figure 4. Levels of tolerance to food-related stresses of different L. monocytogenes clonal complexes. Isolates were evaluated on their ability to survive (A) cold (BHIB at 4°C), (B) salt (BHIB+6% NaCl, 25°C), (C) acid (BHIB pH 5, 25°C), and (D) desiccation stress (33% RH for 3 days at 20°C followed by rehydration with BHIB at 30°C). Error bars represent standard deviations (n − 1) of standardized model values. CCs with different cases of the same letter are significantly different (p < 0.05). std-μmax, standardized maximum growth rate; std-TRG, standardized time to detectable regrowth; BHIB, brain heart infusion broth.
In 6% NaCl, CC7 (1/2a) isolates had significantly (p < 0.05) smaller std-μmax values compared to CCs 5 (1/2b), 8 (1/2a), 11 (1/2a), and 155 (1/2a; Figure 4B). This highlights the range of salt tolerances between CCs within the same serotype and again explains why no significant differences were observed at the serotype level for salt tolerance. In support of the results shown in Figure 4B, 67% of CC2 isolates were salt tolerant while 50% of CC224, 47% of CC7, and 22% of CC9 isolates were salt sensitive (Table 2).
In BHIB pH 5, CC5 (1/2b) isolates exhibited significantly (p < 0.05) larger std-μmax values than CCs 7, 155, and 321, and CC321 isolates additionally had smaller (p < 0.05) std-μmax values compared to CCs 1, 3, and 11 (Figure 4C). CC1 (4b) isolates also had significantly (p = 0.02) smaller std-μmax values compared to CC7 (1/2a) isolates. From Figure 4C it can be seen that lineage I isolates were more acid tolerant than LII isolates, as the five CCs with the highest average std-μmax values in BHIB pH 5 were all from LI while the five CCs with the lowest average std-μmax values belonged to LII (predominantly 1/2a isolates). Notably, 57% of CC5 and 67% of CC4 isolates were acid tolerant while 83% of CC321, 67% of CC20, and 29% of CC7 isolates were acid sensitive (Table 2).
No significant differences were found between the desiccation stress std-TRGs of different CCs (Figure 4D). Nevertheless, CC224 (1/2b) had the smallest average std-TRGs and correspondingly, 75% of these isolates were classified as desiccation tolerant. CC11 (1/2a) had the next smallest std-TRGs while CCs 1 and 4 (both 4b) had the two largest average std-TRGs.
Associations between Plasmid Harborage and Stress Tolerances
Although plasmids were identified in 55% of all isolates, a higher percentage of plasmid carriers were observed among acid tolerant (73%), desiccation sensitive (75%), and desiccation tolerant (60%) isolates as compared to cold tolerant (33%) and acid sensitive (46%) isolates (Figure 5). Within LII, plasmid-positive isolates had smaller std-μmax values at 4°C (p = 0.024, x = 1.00 vs. 1.02) and larger std-μmax (p < 0.0005, x = 1.01 vs. 0.86) values when grown in BHIB pH 5 compared to their plasmid-free counterparts. No significant differences were found between the stress tolerance levels of LI plasmid-harboring and plasmid-free isolates. It was, however, observed that isolates containing repA group 1 plasmids; which were significantly (p < 0.0005) smaller than group 2 plasmids, had smaller std-μmax (p = 0.002, x = 0.98 vs. 1.04) values in 6% NaCl.
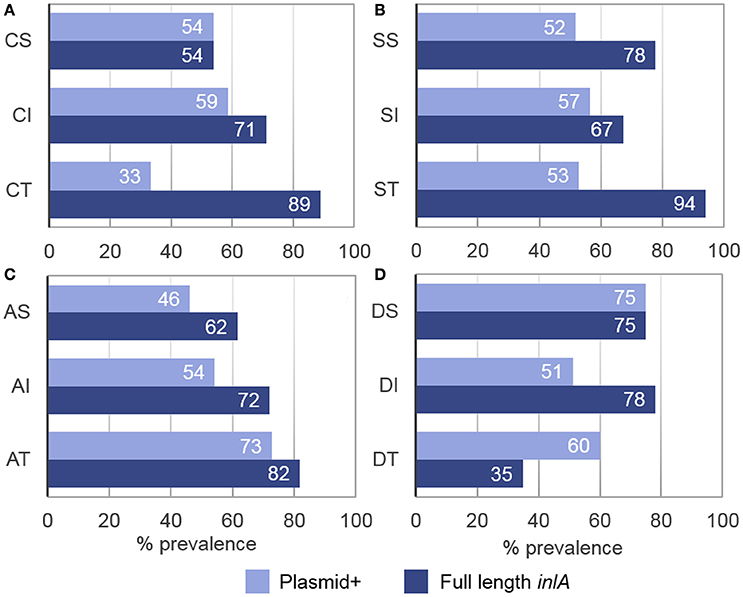
Figure 5. Prevalence of full length inlA and plasmid harborage among L. monocytogenes stress tolerance phenotypes. (A) Cold sensitive (CS), intermediate (CI), and tolerant (CT) isolates. (B) Salt sensitive (SS), intermediate (SI), and tolerant (ST) isolates. (C) Acid sensitive (AS), intermediate (AI), and tolerant isolates (AT). (D) Desiccation sensitive (DS), intermediate (DI), and tolerant (DT) isolates. Full length inlA and the presence of a plasmid were observed in 72 and 55% of all isolates, respectively.
Associations between inlA Profiles and Stress Tolerances
Full length inlA was observed in 72% of isolates, where a higher percentage of the intact gene prevailed among cold (89%), salt (94%), and acid (82%) tolerant isolates while a lower prevalence was detected among desiccation tolerant isolates (35%; Figure 5). Statistically, isolates with full length inlA had significantly larger std-μmax values at 4°C than isolates with an inlA PMSC (p = 0.001, x = 1.01 vs. 0.97). Additionally, serotype 4b isolates possessing a three-codon deletion in inlA, had significantly shorter desiccation stress std-TRGs (p = 0.002 x = 0.94 vs. 1.05) compared to serotype 4b isolates with full length inlA. No significant associations were found between inlA profiles and salt or acid stress tolerance.
Associations between Stress Tolerances and the Presence of SSI1 or LGI1
In 6% NaCl, isolates containing SSI-1 had significantly smaller std-μmax values (p = 0.004, x = 0.98 vs. 1.03) than isolates without SSI-1 though this difference was not large. To determine potential associations between LGI1 harborage and stress tolerance, serotype 1/2a isolates containing the island were compared to other 1/2a LGI1-negative isolates but similar to SSI-1, no significant differences in stress tolerances were detected.
SNV Analyses of Stress-Sensitive and Tolerant Isolates
Figure 6 shows a whole genome SNV phylogeny of all 166 L. monocytogenes isolates with their corresponding genetic and phenotypic properties. In this figure, groups of closely related isolates that share the same phenotypes can be seen. However, also shown are several cases where neighboring isolates have opposing stress tolerances. Of particular interest was whether specific SNVs could be related to isolates possessing the same stress tolerance phenotypes, however, none were detected to be uniquely shared among stress tolerant isolates that weren't also seen in intermediate or sensitive isolates. Among stress sensitive isolates, unique SNVs shared by subsets of isolates were identified, but no single SNV was prevalent among >4 isolates from the same stress sensitive phenotype group. In contrast, a large number of SNVs were uniquely observed for one or two isolates from the same stress sensitive group, causing frameshifts, premature stop codons, loss of start codons, or missense variants. Information regarding the SNVs identified among all sensitive and tolerant isolates are presented in Tables S2–S9. Notably, a number of stress sensitive isolates contained different PMSCs in several σB regulator genes. A cold and desiccation sensitive isolate contained a PMSC in rsbS as did two other desiccation sensitive isolates. Furthermore, an additional cold sensitive isolate contained a PMSC in rsbV, and two desiccation sensitive isolates contained PMSCs in rsbU.
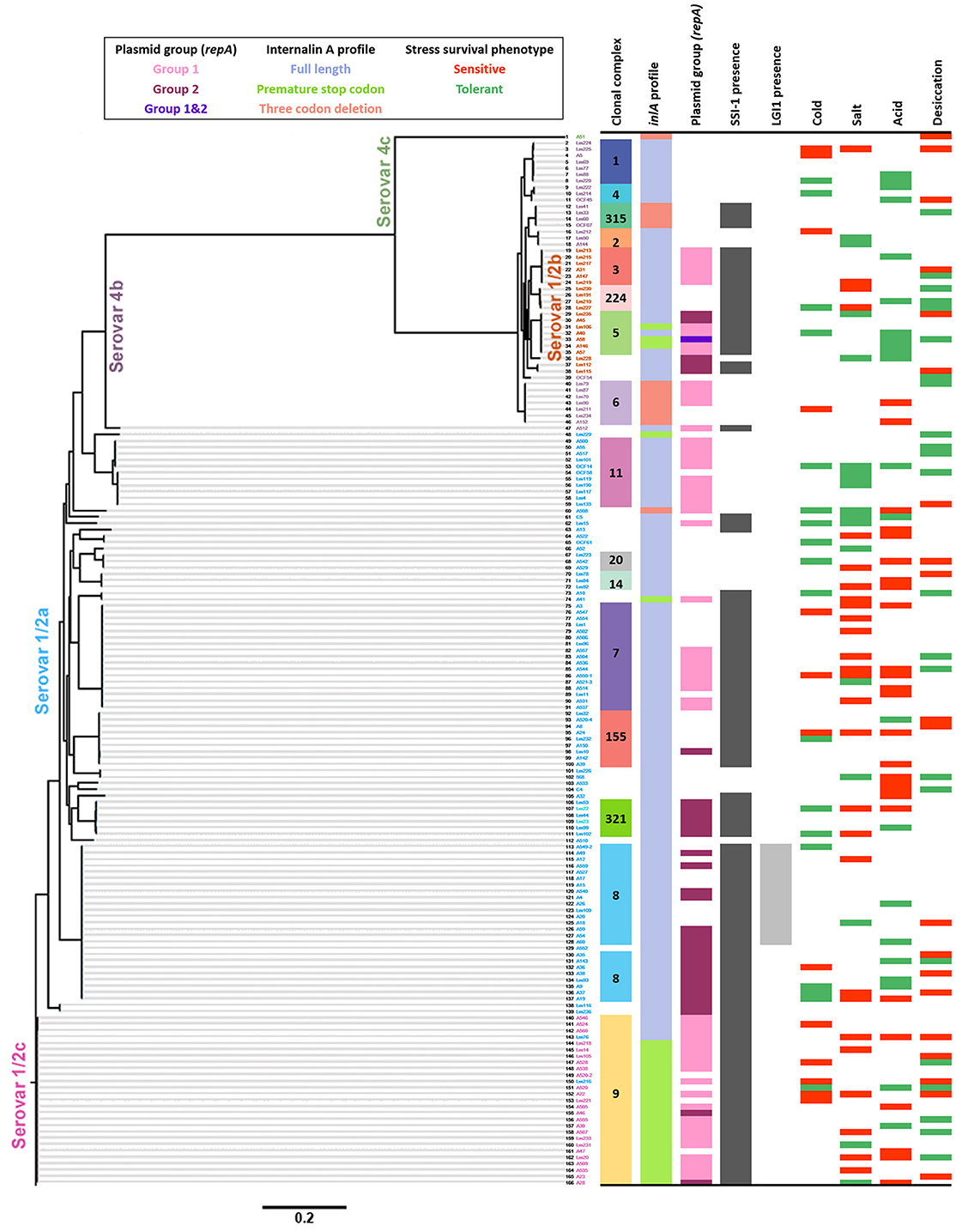
Figure 6. Whole genome single nucleotide variant (SNV) phylogeny of 166 L. monocytogenes isolates and their associated genetic characteristics and stress tolerance phenotypes. The scale at the bottom indicates the substitutions per SNV.
Genomic Islands of Stress-Sensitive and Tolerant Isolates
All L. monocytogenes isolates were predicted to harbor 1,318 genomic islands in total, resulting in an average of eight genomic islands per genome. These islands were clustered into 200 groups of similar sequences. The conservation of genomic island groups across L. monocytogenes lineages ranged from unique to a single isolate to conserved in 97 isolates (58%). Most frequently, intermediate groups conserved in subsets of monophyletic isolates were observed, as can be expected from a combination of vertical inheritance of the genomic islands with further modifications by mutation, insertions, and deletions. The clustering of L. monocytogenes isolates based on the presence or absence of groups of genomic islands reflected their phylogenetic proximity and did not relate particular genomic island content in isolates with the exhibition of similar phenotypes. Indeed, no single genomic island was found to occur in a large proportion of strains with a given phenotype (Figure S1).
Discussion
L. monocytogenes' Tolerance to Food-Related Stresses Differs between and within Lineages, Serotypes, and Clonal Complexes
Cold Stress
L. monocytogenes' ability to grow at refrigeration temperatures highlights this pathogen as a concern for the food industry and consumers alike. However, it is known that L. monocytogenes strains can largely differ in their ability to adapt to cold stress. In the present study, it was found that serotypes 1/2a and 1/2b were on average more cold-tolerant than serotypes 4b and 1/2c. Other cold growth studies have also reported serotype 1/2a strains to be more cold tolerant than serotype 4b strains (Junttila et al., 1988; Buncic et al., 2001; Lianou et al., 2006) though similar to the current findings, many of these differences were not statistically significant due to strain to strain variations. Barbosa et al. (1994) reported that out of 39 L. monocytogenes strains, Scott A, a 4b clinical isolate, grew the slowest at 4°C and that 1/2a strains grew the fastest followed by 1/2b, and 4b. Similarly, in De Jesús and Whiting (2003), LII isolates (all serotype 1/2a) exhibited the shortest LPDs at 5°C followed by LI and then LIII isolates. Researchers have suggested that LII strains may be able to survive better under food-related stresses due to an enhanced ability to acquire advantageous mutations and extrachromosomal DNA compared to LI strains which typically have more conserved genomes (Orsi et al., 2007, 2008, 2011; Ragon et al., 2008; Dunn et al., 2009). Certain stress response genes, predominantly involved in membrane transport and cell wall structure (Doumith et al., 2004), have also been reported to be present in LII isolates but absent among LI isolates (Borucki and Call, 2003; Call et al., 2003; Zhang et al., 2003; Doumith et al., 2004; Chan and Wiedmann, 2008). Given the critical roles of these structures in allowing bacteria to adapt and tolerate numerous stresses (Annous et al., 1997; Verheul et al., 1997; Klein et al., 1999; Weber et al., 2001; Álvarez-Ordóñez et al., 2008), it is not surprising that L. monocytogenes lineages and serotypes can behave differently under certain stresses. The alternative sigma factor, σC, and lmo1078, encoding a UDP-glucose pyrophosphorylase, are examples of genes with reported roles in L. monocytogenes cold tolerance, that are present in LII strains but absent in LI and serotype 4b strains, respectively (Chassaing and Auvray, 2007; Chan and Wiedmann, 2008). These absences may partly explain the overall reduced cold tolerance of serotype 4b isolates respective to 1/2a isolates. However, they do not appear to be necessary for adequate cold growth as in the present study some 4b isolates were also classified as cold tolerant.
Salt Stress
LI strains have been shown to be more salt tolerant than LII strains (Bergholz et al., 2010) and serotype 4b strains to be more salt tolerant than serotype 1/2a and 1/2b strains (Van Der Veen et al., 2008; Bergholz et al., 2010; Ribeiro et al., 2014). In the present study, no significant differences were found between the growth rates of different serotypes in 6% NaCl, however, five times as many 4b isolates were classified as salt tolerant as were classified as salt sensitive. Additionally, despite a 55% prevalence of serotype 1/2a isolates in our collection, isolates of this serotype accounted for 71% of salt tolerant isolates but also for 67% of salt sensitive isolates, indicating a broad range of salt tolerance among isolates from this serotype. These differences were found to be associated with specific 1/2a CCs, notably, CC7 isolates were on average the most salt sensitive CC and significantly differed from the 1/2a CCs 8 and 11 with most 1/2a salt tolerant isolates belonging to CC11.
Acid Stress
LI isolates tolerated acid stress conditions significantly better than LII isolates. Specifically, serotype 1/2a showed higher sensitivity to acidity. This trend was also clear when the acid tolerance of individual CCs was investigated. Van Der Veen et al. (2008), also reported that 4b (LI) isolates had enhanced acid tolerance relative to 1/2a isolates. The authors hypothesized that the increased survival of 4b strains may in part be due to the presence of ORF2110 which encodes a putative serine protease similar to HtrA. This protein has been shown to be important for growth at low pH, high osmolarity, and high temperatures (Wonderling et al., 2004; Stack et al., 2005; Wilson et al., 2006). Though this gene may contribute to the overall acid tolerance of 4b isolates, some acid sensitive 4b isolates were also identified. Similarly, despite serotype 1/2a isolates having relatively low acid tolerance overall, some 1/2a isolates were also acid tolerant, highlighting the importance of not overgeneralizing isolate phenotypes based on the trends seen for their sero- or sequence type.
Desiccation Stress
Desiccation tolerance reflects a bacteria's ability to survive on a surface for extended periods of time with little access to nutrients and water. As so, desiccation tolerance is believed to be associated with L. monocytogenes' ability to persist in food production plants (Vogel et al., 2010) and subsequently cross-contaminate food products. To date, surprisingly little research has been conducted regarding L. monocytogenes' desiccation survival and that which does exist has focused primarily on factors influencing the survival of a small number of isolates (Vogel et al., 2010; Hansen and Vogel, 2011; Takahashi et al., 2011; Hingston et al., 2013; Overney et al., 2016; Zoz et al., 2016). The current study is the first to our knowledge, to compare the desiccation tolerance of L. monocytogenes isolates from multiple serotypes. No significant differences were found between serotypes or CCs however, some prominent trends were observed. Serotypes 1/2c and 1/2b were on average the most desiccation tolerant, followed by 4b and 1/2a. More specifically, CC224 (1/2b) isolates had the highest levels of desiccation survival. Interestingly, a large listeriosis outbreak in Denmark, which resulted in 41 illnesses and 17 deaths, was linked to the consumption of deli meat contaminated with a CC224 strain (Kvistholm Jensen et al., 2016). Though there is not enough evidence to suggest that all CC224 strains are desiccation tolerant, it is possible that long-term desiccation survival may have contributed to the occurrence of this outbreak. Another interesting finding from the present study was that the most desiccation sensitive isolate, deviating more than 4.5 SD from the median, was a CC193 (serotype 1/2a) isolate. Since this was the only CC193 isolate in our collection, it would be interesting to analyze additional isolates from this CC to determine if this sequence type is associated with a high degree of desiccation sensitivity.
Certain Genetic Elements Are Associated with the Stress Tolerance of L. monocytogenes
Plasmids
The presumptive presence of a plasmid(s) was detected in 55% of our isolates which is comparable to other studies where rates of plasmid isolation have ranged from 0 to 79% with an overall average of around 30% (Perez-Diaz et al., 1982; Kolstad et al., 1992; Lebrun et al., 1992; Peterkin et al., 1992; McLauchlin et al., 1997). In agreement with earlier work, we also observed that plasmid DNA was more prevalent among LII isolates than LI isolates (Kolstad et al., 1992; Lebrun et al., 1992; McLauchlin et al., 1997; Margolles and de los Reyes-Gavilán, 1998; Orsi et al., 2011).
Kuenne et al. (2010) discovered that L. monocytogenes plasmids could be divided into two phylogenetic groups based on their repA sequences and that group 2 plasmids (77–83 kb) were larger than those belonging to group 1 (32–57 kb). Here plasmid repA sequences also formed two distinct phylogenetic groups and in agreement with Kuenne et al. (2010), group 2 plasmids were significantly larger (55–100 kb) than group 1 plasmids (26–88 kb). Notably, one serotype 1/2b isolate contained two plasmids of similar sizes (62 and 69 kb) but belonging to different repA groups. Though rare, the presence of two plasmids has been reported in other Listeria spp. isolates (Earnshaw and Lawrence, 1998; Margolles and de los Reyes-Gavilán, 1998).
Our results showed that among LII isolates, which exhibited higher rates of plasmid harborage than LI isolates, plasmid harborage was associated with significantly enhanced acid tolerance but also cold sensitivity. Studies have shown that plasmid harborage and subsequent replication increases the metabolic demands of cells, leading to decreased growth rate relative to plasmid-free strains (reviewed in Diaz Ricci and Hernández, 2000). However, depending on the genes contained on a plasmid, plasmid-harborage can also provide cells with a growth advantage when exposed to certain conditions. In this study, isolates with the larger repA group 2 plasmids were significantly more salt-tolerant than isolates that harbored the smaller repA group 1 plasmids, collectively suggesting that plasmid harborage may be a hindrance to L. monocytogenes during replication at low temperatures but provide an advantage when exposed to acid and salt stress conditions. Furthermore, the observation that isolates containing larger plasmids had higher levels of salt tolerance suggests that these plasmids may contain additional genes that are beneficial for adaptation to high osmolarity environments.
To date, plasmids acquired by L. monocytogenes have been shown to contain genes that confer resistance to benzalkonium chloride (bcrABC; Elhanafi et al., 2010; Rakic-Martinez et al., 2011; Katharios-Lanwermeyer et al., 2012), cadmium (cadA2, cadAC; Lebrun et al., 1992; Rakic-Martinez et al., 2011; Katharios-Lanwermeyer et al., 2012) and antibiotics including chloramphenicol, clindamycin, erythromycin, streptomycin, and tetracycline (Poyart-Salmeron et al., 1990; Hadorn et al., 1993). Listeria spp. plasmids also commonly contain several other uncharacterized efflux pumps (MDR, SMR, MATE; Gibson and Parales, 2000; Masaoka et al., 2000; Boylan et al., 2006; Kuroda and Tsuchiya, 2009), as well as oxidative stress response genes (peroxidases, reductases; Kuenne et al., 2010) but their exact roles in stress tolerance have yet to be investigated. In other bacterial species, multidrug efflux pumps have been linked to stress response, virulence, and quorum sensing (reviewed in Li and Nikaido, 2009). Ma et al. (1995) reported that transcription of a MDR pump (acrAB) in E. coli increased in response to fatty acids, ethanol, high salt, and cellular transitioning into stationary phase. Among the putative plasmid associated contigs a wide variety of different genes were identified including those encoding cell surface proteins, lipoproteins, secretion pathways, heavy metal transporters, transcription regulators, general stress proteins (CplB, ClpL), NADH oxidoreductases, a glycine betaine transport permease (ProW), and the multidrug resistance proteins EbrA and EbrB among others. All group 2 plasmids shared a general secretion pathway protein and a cell surface protein. Other genes identified among many but not all group 2 plasmids included those which encoded DNA topoisomerase III, a membrane-bound protease, a NLP/P60 family lipoprotein, an NADH peroxidase, and a type IV secretory pathway. Further investigations are currently focusing on whether these genes or others with no known function contribute to L. monocytogenes' acid and salt tolerance.
Lastly, it should be highlighted that plasmids with 99–100% nucleotide identity were found in isolates from different serotypes, provinces (Alberta and British Columbia), and countries (Canada and Switzerland). One plasmid was observed in 26 isolates, which strongly suggests that L. monocytogenes' benefits from its presence. The occurrence of the same plasmid in multiple food-related isolates from different regions also suggests that bacteria are frequently transported between places of food production, possibly alongside imported raw materials. On the contrary, it is particularly interesting that other plasmids were conserved among specific CCs, serotypes, provinces, and countries.
Full Length inlA
Full length inlA profiles were observed among 92% of serotype 1/2a isolates, 83% of serotype 1/2b isolates, and 12% of serotype 1/2c isolates, reflecting what has been previously observed (Jonquieres et al., 1998; Jacquet et al., 2004; Rousseaux et al., 2004; Nightingale et al., 2005; Felicio et al., 2007; Orsi et al., 2007; Ragon et al., 2008). Additionally, 44% of 4b isolates contained a 3-codon deletion that unlike inlA PMSCs, is not associated with attenuated virulence (Kovacevic et al., 2013; Kanki et al., 2015).
The present study results showed that full length inlA profiles were more prevalent among cold, salt, and acid tolerant isolates compared to their sensitive counterparts. Also, isolates with full length inlA profiles were significantly more cold tolerant than isolates containing inlA PMSCs. Kovacevic et al. (2013) were the first to report that cold tolerant isolates more likely possess full length inlA than intermediate and cold sensitive isolates. This increased stress tolerance has now been shown to extend to salt and acid tolerance, making it reasonable to hypothesize that full length inlA may participate in L. monocytogenes' stress response. When bacteria are exposed to unfavorable conditions, their cell envelope is the first line of defense. It is possible that the absence of cell wall anchored InlA proteins may alter cell-surface characteristics, leaving cells more susceptible to certain environmental stresses. Interestingly, only a small percentage of desiccation tolerant isolates contained full length inlA while serotype 4b isolates with full length inlA profiles had significantly impaired desiccation survival relative to those with a 3-codon deletion. Again, it is suspected that the structure of inlA may influence L. monocytogenes' desiccation tolerance, this time with the full length form possibly imparting a disadvantage. Other researchers have also detected associations between internalin mutations and certain phenotypes. In Hingston et al. (2015), inlC was identified as the interrupted gene in a desiccation tolerant transposon mutant and Franciosa et al. (2009) found that strains possessing a truncated inlA protein formed increased levels of biofilm. Similarly, transposon mutants containing an interrupted internalin A, B, or H gene, formed thicker biofilms relative to the wildtype (Piercey et al., 2016). Together, these findings along with those presented in this study, emphasize a need for more research regarding the potential roles of internalins in other processes other than virulence.
SSI-1
Stress survival islet 1 (SSI-1) is a five-gene cluster which has previously been shown via mutagenesis studies to enhance L. monocytogenes tolerance to acid, salt, and low temperature conditions (Cotter et al., 2005; Ryan et al., 2010). On the other hand, Arguedas-Villa et al. (2014) found no significant differences in cold tolerance between naturally occurring L. monocytogenes isolates with and without SSI-1. In the present study, it was found that SSI-1-positive isolates showed no enhanced cold, acid, salt, and desiccation stress tolerances relative to SSI-1 negative isolates. It is possible that any positive influence of SSI-1 on the stress tolerance of L. monocytogenes' may be masked by the presence of other genetic elements when comparing large collections of isolates as opposed to a mutant and its wildtype strain.
LGI1
LGI1 is a Listeria 50 kb genomic island that was first identified in Canadian CC8 isolates associated with a large 2008 listeriosis outbreak involving contaminated deli meats and resulting in 22 fatalities (Gilmour et al., 2010). Since then, LGI1 has been identified in other CC8 L. monocytogenes isolates from Canada (Kovacevic et al., 2013) but not from other countries (Althaus et al., 2014). In agreement with these studies, the presence of LGI1 was only detected in Canadian isolates from CC8. However, instead of all LGI1+ isolates being ST120 as previously reported, two novel CC8 STs (ST1022 and 1025) were also associated with LGI1 harborage. The conservation of LGI1 among Canadian isolates and its association with a fatal outbreak has led to heightened interest in the putative functions of the genes located on this island including those encoding putative type II and type IV secretion systems, pilus-like surface structures, a multidrug efflux pump homolog (EmrE), and an alternative sigma factor (Gilmour et al., 2010). Recently, Kovacevic et al. (2015) reported that deletion of LGI1 genes with putative efflux (emrE), regulatory (lmo1852), and adhesion (sel1) functions, had no impact on the tolerance of L. monocytogenes' to acid, cold, or salt, but that deletion of emrE increased susceptibility to quaternary ammonium-based sanitizers. Based on these findings, it was investigated whether the presence or absence of the whole LGI1 island could be associated with stress tolerance differences between CC8 isolates however, no significant differences were identified. Consequently, LGI1 had no major influence on L. monocytogenes' ability to adapt to the food-related stresses evaluated in the present study. However, it is possible that the island contributes in other ways to the persistence of CC8 Canadian isolates in food processing environments in addition to the role of emrE in sanitizer resistance.
SNVs Associated with Stress Tolerance Phenotypes
An important finding from this study was that closely related isolates from within the same clonal complexes exhibited opposing stress tolerances, suggesting that minor genetic differences can also exert great impact on stress tolerance phenotypes. This was observed in a study by Hoffmann et al. (2013), where a single thymine deletion in the σA-like promoter region of betL, encoding an osmolyte transporter specific for betaine uptake, dramatically increased betL transcription, and hence the osmo- and chill-tolerance. Karatzas et al. (2003) reported that a spontaneous high hydrostatic pressure tolerant L. monocytogenes mutant of Scott A, contained a single codon deletion in ctsR, a negative regulator of several heat shock and general stress proteins, that also conferred increased thermo-tolerance and resistance to H2O2. Additionally, in Metselaar et al. (2015) a number of spontaneous acid tolerant mutants were found to contain SNVs in the ribosomal protein gene rpsU. None of these aforementioned mutations were detected in the present study.
Analyses to determine if unique SNVs could be detected among isolates from individual stress tolerance phenotype groups were also performed. Studies identifying the genetic basis of phenotypic traits using the variation within natural populations are known as a genome-wide association studies (GWAS). While GWAS have been effective for identifying mutations responsible for phenotypic traits in humans, the clonal nature of bacterial replication where mutations can reach a high frequency on a single genetic background, makes it difficult to distinguish mutations responsible for an observed phenotype (Read and Massey, 2014; Falush, 2016). As a result, bacterial molecular epidemiology has focused on identifying clonal lineages with particular phenotypic properties rather than identifying the specific genetic variants responsible. Recently, Earle et al. (2016) used a GWAS approach to successfully identify genes and genetic variants underlying resistance to 17 antimicrobials in over 3000 isolates of taxonomically diverse clonal and recombining bacteria. While these results show the potential of bacterial GWAS, antimicrobial resistance is usually gained during antimicrobial exposure and thus it is more likely that the traits evolve on multiple independent backgrounds making them easier to detect (Falush, 2016). To date, the identification of mutations responsible for more complex phenotypes such as those evaluated in the present study, remain challenging.
Our analyses did not detect any unique SNVs among more than one isolate from the same stress tolerant group, suggesting homoplasy among stress tolerant phenotypes where mutations evolve independently to confer tolerance. On the contrary, a few different SNVs were identified among four or fewer isolates from the same stress sensitive groups. Notably, the cold sensitive phenotypes of two isolates may be associated with PMSCs detected in the σB regulator genes rsbS and rsbV (Voelker et al., 1995). In support of this hypothesis, deletion of rsbV in a L. monocytogenes mutagenesis study resulted in impaired cold stress tolerance (Chan et al., 2008). Three desiccation sensitive isolates also contained different PMSCs in rsbS, including one isolate which was also cold sensitive. An additional two desiccation sensitive isolates contained different PMSCs in another σB posttranscriptional regulator, rsbU. Given the importance of σB in L. monocytogenes' adaptation to several environmental stresses (Wiedmann et al., 1998; Kazmierczak et al., 2003), it is possible that these mutations contributed to the reduced desiccation tolerance of these isolates. In fact, in Huang et al. (2015) an L. monocytogenes sigB mutant demonstrated reduced desiccation survival relative to the wildtype strain.
Genomic Islands
In this study, specific genomic islands could not be exclusively associated with a particular stress tolerance phenotype. This is to be expected as genomic islands often contain virulence factors, and in general these are overrepresented in genomic islands as compared to the chromosome (Sui et al., 2009). In L. monocytogenes in particular, genomic islands have been associated with virulence, heavy metal resistance, and benzalkonium chloride efflux (Gilmour et al., 2010; Kuenne et al., 2010; Kovacevic et al., 2015). SSI-1 as described above, has been previously associated with L. monocytogenes' stress response, but was not significantly correlated with the stress tolerance phenotypes examined in the present study.
Conclusions
In summary, L. monocytogenes' tolerances to certain food-related stresses differs between serotypes as well as CCs with the latter being a better predictor of isolate salt and acid tolerance but not of cold and desiccation tolerance. To the best of our knowledge, this is the first study to evaluate the stress tolerance of different L. monocytogenes CCs. Other noteworthy findings include potential relationships between the presence of full length inlA and enhanced cold tolerance and the presence of a plasmid and enhanced acid tolerance. On the contrary, the presence of genomic islands including SSI-1 and LGI1, provided isolates with no noticeable advantages under the stresses evaluated in this study. Additional research is needed to confirm the potential roles of full length inlA and plasmid associated genes in L. monocytogenes' response to various stresses.
A whole genome SNV phylogeny of isolate assemblies identified a number of unique SNVs shared by up to four stress sensitive isolates while no common SNVs were observed among stress tolerant isolates. More specifically, six isolates with sensitivity to cold and/or desiccation stress contained PMSCs in σB regulator genes (rsbS, rsbU, rsbV) that may be contributing to these phenotypes.
A number of novel genetic elements were also elucidated in this study including nine new L. monocytogenes STs, a new inlA PMSC, the absence of a cold stress associated gene (lmo1078) in 4b isolates, and several connections between L. monocytogenes CCs and the presence/absence or variations of specific genetic elements. For example, SSI-1 was detected in 100% of isolates from specific CCs, certain plasmid groups and sizes were conserved among isolates from the same CCs, and plasmids with 100% identity were found in isolates belonging to the same CCs but from very different geological areas. While our isolate collection represented of a number of L. monocytogenes CCs, some of which have been previously identified as common among food-related isolates, other CCs were less prevalent or absent from our study of Canadian and Swiss isolates. This highlights the regional prevalence of certain L. monocytogenes genotypes and emphasizes the need for more international collaborative studies.
Collectively, the results suggest that using whole genome sequencing to (1) determine the STs of L. monocytogenes food-related isolates and to (2) screen for the presence of genetic elements such as full length inlA and a plasmid(s), could help food processors and food agency investigators to quickly identify if isolates are likely to possess enhanced tolerances to certain stresses that may be facilitating their long-term survival/persistence in a food processing environment. The US FDA and CDC are rapidly making whole genome sequencing of foodborne bacterial pathogens a routine part of screening to help link illnesses to contaminated foods and to identify outbreaks earlier. While no one SNV was identified among isolates with the same stress tolerant phenotype, increased sequencing of L. monocytogenes isolates in combination with stress tolerance profiling, will enhance the ability to identify genetic elements associated with more stress tolerant strains.
Author Contributions
PH composed the paper and conducted all experiments and statistics. CL assembled and annotated the whole genome sequences. PH, JC, BD, and CB conducted the bioinformatics analyses. JC, VG, FB, TT, KA, LT, and SW provided guidance on study design, statistics, and assisted in writing the paper.
Funding
This work was funded by an investment agreement between Alberta Innovates—Bio Solutions and the University of British Columbia (FSC-12-030). PH was funded by an NSERC CGS D Scholarship. CB is supported by a fellowship from the Swiss National Science Foundation (P300-PA_164673) and a grant from the Société Académique Vaudoise.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Franco Pagotto and his team at the Listeriosis Reference Service at the Bureau of Microbial Hazards in Ottawa, Ontario for serotyping our isolates.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00369/full#supplementary-material
References
Althaus, D., Lehner, A., Brisse, S., Maury, M., Tasara, T., and Stephan, R. (2014). Characterization of Listeria monocytogenes strains isolated during 2011–2013 from human infections in Switzerland. Foodborne Pathog. Dis. 11, 753–758. doi: 10.1089/fpd.2014.1747
Álvarez-Ordóñez, A., Fernández, A., López, M., Arenas, R., and Bernardo, A. (2008). Modifications in membrane fatty acid composition of Salmonella Typhimurium in response to growth conditions and their effect on heat resistance. Int. J. Food Microbiol. 123, 212–219. doi: 10.1016/j.ijfoodmicro.2008.01.015
Annous, B. A., Becker, L. A., Bayles, D. O., Labeda, D. P., and Wilkinson, B. J. (1997). Critical role of anteiso-C15:0 fatty acid in the growth of Listeria monocytogenes at low temperatures. Appl. Environ. Microbiol. 63, 3887–3894.
Arguedas-Villa, C., Kovacevic, J., Allen, K. J., Stephan, R., and Tasara, T. (2014). Cold growth behaviour and genetic comparison of Canadian and Swiss Listeria monocytogenes strains associated with the food supply chain and human listeriosis cases. Food Microbiol. 40, 81–87. doi: 10.1016/j.fm.2014.01.001
Arguedas-Villa, C., Stephan, R., and Tasara, T. (2010). Evaluation of cold growth and related gene transcription responses associated with Listeria monocytogenes strains of different origins. Food Microbiol. 27, 653–660. doi: 10.1016/j.fm.2010.02.009
Aryani, D. C., Den Besten, H. M. W., Hazeleger, W. C., and Zwietering, M. H. (2015). Quantifying strain variability in modeling growth of Listeria monocytogenes. Int. J. Food Microbiol. 208, 19–29. doi: 10.1016/j.ijfoodmicro.2015.05.006
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comp. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Baranyi, J., and Roberts, T. A. (1994). A dynamic approach to predicting bacterial growth in food. Int. J. Food Microbiol. 23, 277–294. doi: 10.1016/0168-1605(94)90157-0
Barbosa, W. B., Cabedo, L., Wederquist, H. J., Sofos, J. N., and Schmidt, G. R. (1994). Growth variation among species and strains of Listeria in culture broth. J. Food Prot. 57, 765–769. doi: 10.4315/0362-028X-57.9.765
Begot, C., Lebert, I., and Lebert, A. (1997). Variability of the response of 66 Listeria monocytogenes and Listeria innocua strains to different growth conditions. Food Microbiol. 14, 403–412. doi: 10.1006/fmic.1997.0097
Bergholz, T. M., den Bakker, H. C., Fortes, E. D., Boor, K. J., and Wiedmann, M. (2010). Salt stress phenotypes in Listeria monocytogenes vary by genetic lineage and temperature. Foodborne Pathog. Dis. 7, 1537–1549. doi: 10.1089/fpd.2010.0624
Borucki, M. K., and Call, D. R. (2003). Listeria monocytogenes serotype identification by PCR. J. Clin. Microbiol. 41, 5537–5540. doi: 10.1128/JCM.41.12.5537-5540.2003
Boylan, J. A., Hummel, C. S., Benoit, S., Garcia-Lara, J., Treglown-Downey, J., Crane, E. J., et al. (2006). Borrelia burgdorferi bb0728 encodes a coenzyme A disulphide reductase whose function suggests a role in intracellular redox and the oxidative stress response. Mol. Microbiol. 59, 475–486. doi: 10.1111/j.1365-2958.2005.04963.x
Buchanan, R. L., Damert, W. G., Whiting, R. C., and van Schothorst, M. (1997). Use of epidemiologic and food survey data to estimate a purposefully conservative dose-response relationship for Listeria monocytogenes levels and incidence of listeriosis. J. Food Prot. 60, 918–922. doi: 10.4315/0362-028X-60.8.918
Bulinski, A., Butkovsky, O., Shashkin, A., and Yaskov, P. (2011). Statistical methods of SNP data analysis with applications. arXiv:1106.4989
Buncic, S., Avery, S. M., Rocourt, J., and Dimitrijevic, M. (2001). Can food-related environmental factors induce different behaviour in two key serovars, 4b and 1/2a, of Listeria monocytogenes? Int. J. Food Microbiol. 65, 201–212. doi: 10.1016/S0168-1605(00)00524-9
Bureau, A., Dupuis, J., Falls, K., Lunetta, K. L., Hayward, B., Keith, T. P., et al. (2005). Identifying SNPs predictive of phenotype using random forests. Genet. Epidemiol. 28, 171–182. doi: 10.1002/gepi.20041
Cabedo, L., Picart i Barrot, L., and Teixidó i Canelles, A. (2008). Prevalence of Listeria monocytogenes and Salmonella in ready-to-eat food in Catalonia, Spain. J. Food Prot. 71, 855–859. doi: 10.4315/0362-028X-71.4.855
Call, D. R., Borucki, M. K., and Besser, T. E. (2003). Mixed-genome microarrays reveal multiple serotype and lineage-specific differences among strains of Listeria monocytogenes. J. Clin. Microbiol. 41, 632–639. doi: 10.1128/JCM.41.2.632-639.2003
CDC (2016a). Listeria Outbreaks. Available online at: https://www.cdc.gov/listeria/outbreaks/
CDC (2016b). The Listeria Whole Genome Sequencing Project. Available online at: https://www.cdc.gov/listeria/surveillance/whole-genome-sequencing.html
Chan, Y. C., and Wiedmann, M. (2008). Physiology and genetics of Listeria monocytogenes survival and growth at cold temperatures. Crit. Rev. Food Sci. Nutr. 49, 237–253. doi: 10.1080/10408390701856272
Chan, Y. C., Hu, Y., Chaturongakul, S., Files, K. D., Bowen, B. M., Boor, K. J., et al. (2008). Contributions of two-component regulatory systems, alternative sigma factors, and negative regulators to Listeria monocytogenes cold adaptation and cold growth. J. Food Prot. 71, 420–425. doi: 10.4315/0362-028X-71.2.420
Chassaing, D., and Auvray, F. (2007). The lmo1078 gene encoding a putative UDP-glucose pyrophosphorylase is involved in growth of Listeria monocytogenes at low temperature. FEMS Microbiol. Lett. 275, 31–37. doi: 10.1111/j.1574-6968.2007.00840.x
Chavant, P., Martinie, B., Meylheuc, T., Bellon-Fontaine, M. N., and Hebraud, M. (2002). Listeria monocytogenes LO28: surface physicochemical properties and ability to form biofilms at different temperatures and growth phases. Appl. Environ. Microbiol. 68, 728–737. doi: 10.1128/AEM.68.2.728-737.2002
Chen, Y., Ross, W. H., Scott, V. N., and Gombas, D. E. (2003). Listeria monocytogenes: low levels equal low risk. J. Food Prot. 66, 570–577. doi: 10.4315/0362-028X-66.4.570
Chenal-Francisque, V., Lopez, J., Cantinelli, T., Caro, V., Tran, C., Leclercq, A., et al. (2011). Worldwide distribution of major clones of Listeria monocytogenes. Emerg. Infect. Dis. 17, 1110–1112. doi: 10.3201/eid1706.101778
Cingolani, P., Platts, A., Wang, L. L., Coon, M., Nguyen, T., Wang, L., et al. (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6, 80–92. doi: 10.4161/fly.19695
Cotter, P. D., Ryan, S., Gahan, C. G., and Hill, C. (2005). Presence of GadD1 glutamate decarboxylase in selected Listeria monocytogenes strains is associated with an ability to grow at low pH. Appl. Environ. Microbiol. 71, 2832–2839. doi: 10.1128/AEM.71.6.2832-2839.2005
Dalgaard, P., and Koutsoumanis, K. (2001). Comparison of maximum specific growth rates and lag times estimated from absorbance and viable count data by different mathematical models. J. Microbiol. Methods 43, 183–196. doi: 10.1016/S0167-7012(00)00219-0
De Jesús, A. J., and Whiting, R. C. (2003). Thermal inactivation, growth, and survival studies of Listeria monocytogenes strains belonging to three distinct genotypic lineages. J. Food Prot. 66, 1611–1617. doi: 10.4315/0362-028X-66.9.1611
De Lobel, L., Geurts, P., Baele, G., Castro-Giner, F., Kogevinas, M., and Van Steen, K. (2010). A screening methodology based on Random Forests to improve the detection of gene–gene interactions. Eur. J. Hum. Genet. 18, 1127–1132. doi: 10.1038/ejhg.2010.48
Dhillon, B. K., Laird, M. R., Shay, J. A., Winsor, G. L., Lo, R., Nizam, F., et al. (2015). IslandViewer 3: more flexible, interactive genomic island discovery, visualization and analysis. Nucleic Acids Res. 43, W104–W108. doi: 10.1093/nar/gkv401
Di Bonaventura, G., Piccolomini, R., Paludi, D., D'orio, V., Vergara, A., Conter, M., et al. (2008). Influence of temperature on biofilm formation by Listeria monocytogenes on various food-contact surfaces: relationship with motility and cell surface hydrophobicity. J. Appl. Microbiol. 104, 1552–1561. doi: 10.1111/j.1365-2672.2007.03688.x
Diaz Ricci, J. C., and Hernández, M. E. (2000). Plasmid effects on Escherichia coli metabolism. Crit. Rev. Biotechnol. 20, 79–108. doi: 10.1080/07388550008984167
Doumith, M., Buchrieser, C., Glaser, P., Jacquet, C., and Martin, P. (2004). Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 42, 3819–3822. doi: 10.1128/JCM.42.8.3819-3822.2004
Dunn, K. A., Bielawski, J. P., Ward, T. J., Urquhart, C., and Gu, H. (2009). Reconciling ecological and genomic divergence among lineages of Listeria under an “extended mosaic genome concept”. Mol. Biol. Evol. 26, 2605–2615. doi: 10.1093/molbev/msp176
Durack, J., Ross, T., and Bowman, J. P. (2013). Characterisation of the transcriptomes of genetically diverse Listeria monocytogenes exposed to hyperosmotic and low temperature conditions reveal global stress-adaptation mechanisms. PLoS ONE 8:e73603. doi: 10.1371/journal.pone.0073603
Earle, S. G., Wu, C. H., Charlesworth, J., Stoesser, N., Gordon, N. C., Walker, T. M., et al. (2016). Identifying lineage effects when controlling for population structure improves power in bacterial association studies. Nat. Microbiol. 1:16041. doi: 10.1038/nmicrobiol.2016.41
Earnshaw, A., and Lawrence, L. (1998). Sensitivity to commercial disinfectants, and the occurrence of plasmids within various Listeria monocytogenes genotypes isolated from poultry products and the poultry processing environment. J. Appl. Microbiol. 84, 642–648. doi: 10.1046/j.1365-2672.1998.00395.x
Ebner, R., Stephan, R., Althaus, D., Brisse, S., Maury, M., and Tasara, T. (2015). Phenotypic and genotypic characteristics of Listeria monocytogenes strains isolated during 2011–2014 from different food matrices in Switzerland. Food Control 57, 321–326. doi: 10.1016/j.foodcont.2015.04.030
Elhanafi, D., Dutta, V., and Kathariou, S. (2010). Genetic characterization of plasmid-associated benzalkonium chloride resistance determinants in a Listeria monocytogenes strain from the 1998-1999 outbreak. Appl. Environ. Microbiol. 76, 8231–8238. doi: 10.1128/AEM.02056-10
Falush, D. (2016). Bacterial genomics: microbial GWAS coming of age. Nat. Microbiol. 1:16059. doi: 10.1038/nmicrobiol.2016.59
Faul, F., Erdfelder, E., Buchner, A., and Lang, A. (2009). Statistical power analyses using G* Power 3.1: Tests for correlation and regression analyses. Behav. Res. Methods 41, 1149–1160. doi: 10.3758/BRM.41.4.1149
Faul, F., Erdfelder, E., Lang, A., and Buchner, A. (2007). G* Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav. Res. Methods 39, 175–191. doi: 10.3758/BF03193146
Felicio, M. T., Hogg, T., Gibbs, P., Teixeira, P., and Wiedmann, M. (2007). Recurrent and sporadic Listeria monocytogenes contamination in alheiras represents considerable diversity, including virulence-attenuated isolates. Appl. Environ. Microbiol. 73, 3887–3895. doi: 10.1128/AEM.02912-06
Fenlon, D., Wilson, J., and Donachie, W. (1996). The incidence and level of Listeria monocytogenes contamination of food sources at primary production and initial processing. J. Appl. Bacteriol. 81, 641–650. doi: 10.1111/j.1365-2672.1996.tb03559.x
Franciosa, G., Maugliani, A., Scalfaro, C., Floridi, F., and Aureli, P. (2009). Expression of internalin A and biofilm formation among Listeria monocytogenes clinical isolates. Int. J. Immunopathol. Pharmacol. 22, 183–193. doi: 10.1177/039463200902200121
Gardner, S. N., Slezak, T., and Hall, B. G. (2015). kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 31, 2877–2878. doi: 10.1093/bioinformatics/btv271
Gibson, D. T., and Parales, R. E. (2000). Aromatic hydrocarbon dioxygenases in environmental biotechnology. Curr. Opin. Biotechnol. 11, 236–243. doi: 10.1016/S0958-1669(00)00090-2
Gilmour, M. W., Graham, M., Van Domselaar, G., Tyler, S., Kent, H., Trout-Yakel, K. M., et al. (2010). High-throughput genome sequencing of two Listeria monocytogenes clinical isolates during a large foodborne outbreak. BMC Genomics 11:120. doi: 10.1186/1471-2164-11-120
Hadorn, K., Hächler, H., Schaffner, A., and Kayser, F. (1993). Genetic characterization of plasmid-encoded multiple antibiotic resistance in a strain of Listeria monocytogenes causing endocarditis. Eur. J. Hum. Genet. 12, 928–937.
Handa-Miya, S., Kimura, B., Takahashi, H., Sato, M., Ishikawa, T., Igarashi, K., et al. (2007). Nonsense-mutated inlA and prfA not widely distributed in Listeria monocytogenes isolates from ready-to-eat seafood products in Japan. Int. J. Food Microbiol. 117, 312–318. doi: 10.1016/j.ijfoodmicro.2007.05.003
Hansen, L. T., and Vogel, B. F. (2011). Desiccation of adhering and biofilm Listeria monocytogenes on stainless steel: survival and transfer to salmon products. Int. J. Food Microbiol. 146, 88–93. doi: 10.1016/j.ijfoodmicro.2011.01.032
Health Canada (2011). Policy on Listeria monocytogenes in Ready-to-Eat Foods. Available online at: http://www.hc-sc.gc.ca/fn-an/legislation/pol/policy_listeria_monocytogenes_2011-eng.php
Hein, I., Klinger, S., Dooms, M., Flekna, G., Stessl, B., Leclercq, A., et al. (2011). Stress survival islet 1 (SSI-1) survey in Listeria monocytogenes reveals an insert common to Listeria innocua in sequence type 121 L. monocytogenes strains. Appl. Environ. Microbiol. 77, 2169–2173. doi: 10.1128/AEM.02159-10
Hingston, P. A., Piercey, M. J., and Truelstrup Hansen, L. (2015). Genes associated with desiccation and osmotic stress in Listeria monocytogenes as revealed by insertional mutagenesis. Appl. Environ. Microbiol. 81, 5350–5362. doi: 10.1128/AEM.01134-15
Hingston, P. A., Stea, E. C., Knøchel, S., and Hansen, T. (2013). Role of initial contamination levels, biofilm maturity and presence of salt and fat on desiccation survival of Listeria monocytogenes on stainless steel surfaces. Food Microbiol. 36, 46–56. doi: 10.1016/j.fm.2013.04.011
Hoffmann, R. F., McLernon, S., Feeney, A., Hill, C., and Sleator, R. D. (2013). A single point mutation in the Listerial betL σA-dependent promoter leads to improved osmo-and chill-tolerance and a morphological shift at elevated osmolarity. Bioengineered 4, 401–407. doi: 10.4161/bioe.24094
Hsiao, W. W., Ung, K., Aeschliman, D., Bryan, J., Finlay, B. B., and Brinkman, F. S. (2005). Evidence of a large novel gene pool associated with prokaryotic genomic islands. PLoS Genet. 1:e62. doi: 10.1371/journal.pgen.0010062
Huang, Y., Ells, T. C., and Hansen, L. T. (2015). Role of sigB and osmolytes in desiccation survival of Listeria monocytogenes in simulated food soils on the surface of food grade stainless steel. Food Microbiol. 46, 443–451. doi: 10.1016/j.fm.2014.09.007
Jacquet, C., Doumith, M., Gordon, J. I., Martin, P. M., Cossart, P., and Lecuit, M. (2004). A molecular marker for evaluating the pathogenic potential of foodborne Listeria monocytogenes. J. Infect. Dis. 189, 2094–2100. doi: 10.1086/420853
Jonquieres, R., Bierne, H., Mengaud, J., and Cossart, P. (1998). The inlA gene of Listeria monocytogenes LO28 harbors a nonsense mutation resulting in release of internalin. Infect. Immun. 66, 3420–3422.
Junttila, J. R., Niemelä, S., and Hirn, J. (1988). Minimum growth temperatures of Listeria monocytogenes and non−haemolytic Listeria. J. Appl. Bacteriol. 65, 321–327. doi: 10.1111/j.1365-2672.1988.tb01898.x
Kanki, M., Naruse, H., Taguchi, M., and Kumeda, Y. (2015). Characterization of specific alleles in InlA and PrfA of Listeria monocytogenes isolated from foods in Osaka, Japan and their ability to invade Caco-2 cells. Int. J. Food Microbiol. 211, 18–22. doi: 10.1016/j.ijfoodmicro.2015.06.023
Karatzas, K. A., Wouters, J. A., Gahan, C. G., Hill, C., Abee, T., and Bennik, M. H. (2003). The CtsR regulator of Listeria monocytogenes contains a variant glycine repeat region that affects piezotolerance, stress resistance, motility and virulence. Mol. Microbiol. 49, 1227–1238. doi: 10.1046/j.1365-2958.2003.03636.x
Katharios-Lanwermeyer, S., Rakic-Martinez, M., Elhanafi, D., Ratani, S., Tiedje, J. M., and Kathariou, S. (2012). Coselection of cadmium and benzalkonium chloride resistance in conjugative transfers from nonpathogenic Listeria spp. to other Listeriae. Appl. Environ. Microbiol. 78, 7549–7556. doi: 10.1128/AEM.02245-12
Kazmierczak, M. J., Mithoe, S. C., Boor, K. J., and Wiedmann, M. (2003). Listeria monocytogenes sigma B regulates stress response and virulence functions. J. Bacteriol. 185, 5722–5734. doi: 10.1128/JB.185.19.5722-5734.2003
Klein, W., Weber, M. H., and Marahiel, M. A. (1999). Cold shock response of Bacillus subtilis: isoleucine-dependent switch in the fatty acid branching pattern for membrane adaptation to low temperatures. J. Bacteriol. 181, 5341–5349.
Kolstad, J., Caugant, D. A., and Rørvik, L. M. (1992). Differentiation of Listeria monocytogenes isolates by using plasmid profiling and multilocus enzyme electrophoresis. Int. J. Food Microbiol. 16, 247–260. doi: 10.1016/0168-1605(92)90085-H
Kovacevic, J., Arguedas-Villa, C., Wozniak, A., Tasara, T., and Allen, K. J. (2013). Examination of food chain-derived Listeria monocytogenes strains of different serotypes reveals considerable diversity in inlA genotypes, mutability, and adaptation to cold temperatures. Appl. Environ. Microbiol. 79, 1915–1922. doi: 10.1128/AEM.03341-12
Kovacevic, J., Ziegler, J., Walecka-Zacharska, E., Reimer, A., Kitts, D. D., and Gilmour, M. W. (2015). Tolerance of Listeria monocytogenes to quaternary ammonium sanitizers is mediated by a novel efflux pump encoded by emrE. Appl. Environ. Microbiol. 82, 939–953. doi: 10.1128/AEM.03741-15
Kozak, J., Balmer, T., Byrne, R., and Fisher, K. (1996). Prevalence of Listeria monocytogenes in foods: incidence in dairy products. Food Control 7, 215–221. doi: 10.1016/S0956-7135(96)00042-4
Kuenne, C., Voget, S., Pischimarov, J., Oehm, S., Goesmann, A., Daniel, R., et al. (2010). Comparative analysis of plasmids in the genus Listeria. PLoS ONE 5:e12511. doi: 10.1371/journal.pone.0012511
Kuroda, T., and Tsuchiya, T. (2009). Multidrug efflux transporters in the MATE family. Biochim. Biophys. Acta 1794, 763–768. doi: 10.1016/j.bbapap.2008.11.012
Kvistholm Jensen, A., Nielsen, E. M., Bjorkman, J. T., Jensen, T., Muller, L., Persson, S., et al. (2016). Whole-genome sequencing used to investigate a nationwide outbreak of listeriosis caused by ready-to-eat delicatessen meat, Denmark, 2014. Clin. Infect. Dis. 63, 64–70. doi: 10.1093/cid/ciw192
Lebrun, M., Loulergue, J., Chaslus-Dancla, E., and Audurier, A. (1992). Plasmids in Listeria monocytogenes in relation to cadmium resistance. Appl. Environ. Microbiol. 58, 3183–3186.
Li, H. (2011). Improving SNP discovery by base alignment quality. Bioinformatics 27, 1157–1158. doi: 10.1093/bioinformatics/btr076
Li, X., and Nikaido, H. (2009). Efflux-mediated drug resistance in bacteria. Drugs 69, 1555–1623. doi: 10.2165/11317030-000000000-00000
Lianou, A., Stopforth, J. D., Yoon, Y., Wiedmann, M., and Sofos, J. N. (2006). Growth and stress resistance variation in culture broth among Listeria monocytogenes strains of various serotypes and origins. J. Food Prot. 69, 2640–2647. doi: 10.4315/0362-028X-69.11.2640
Luber, P. (2011). The Codex Alimentarius guidelines on the application of general principles of food hygiene to the control of Listeria monocytogenes in ready-to-eat foods. Food Control 22, 1482–1483. doi: 10.1016/j.foodcont.2010.07.013
Lunetta, K. L., Hayward, L., Segal, J., and Van Eerdewegh, P. (2004). Screening large-scale association study data: exploiting interactions using random forests. BMC Genet. 5:32. doi: 10.1186/1471-2156-5-32
Ma, D., Cook, D. N., Alberti, M., Pon, N. G., Nikaido, H., and Hearst, J. E. (1995). Genes acrA and acrB encode a stress−induced efflux system of Escherichia coli. Mol. Microbiol. 16, 45–55. doi: 10.1111/j.1365-2958.1995.tb02390.x
Margolles, A., and de los Reyes-Gavilán, C. G. (1998). Characterization of plasmids from Listeria monocytogenes and Listeria innocua strains isolated from short-ripened cheeses. Int. J. Food Microbiol. 39, 231–236. doi: 10.1016/S0168-1605(97)00132-3
Martín, B., Perich, A., Gómez, D., Yangüela, J., Rodríguez, A., Garriga, M., et al. (2014). Diversity and distribution of Listeria monocytogenes in meat processing plants. Food Microbiol. 44, 119–127. doi: 10.1016/j.fm.2014.05.014
Masaoka, Y., Ueno, Y., Morita, Y., Kuroda, T., Mizushima, T., and Tsuchiya, T. (2000). A two-component multidrug efflux pump, EbrAB, in Bacillus subtilis. J. Bacteriol. 182, 2307–2310. doi: 10.1128/JB.182.8.2307-2310.2000
Maury, M. M., Tsai, Y. H., Charlier, C., Touchon, M., Chenal-Francisque, V., Leclercq, A., et al. (2016). Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat. Genet. 48, 308–313. doi: 10.1038/ng.3501
McLauchlin, J., Hampton, M., Shah, S., Threlfall, E., Wieneke, A., and Curtis, G. (1997). Subtyping of Listeria monocytogenes on the basis of plasmid profiles and arsenic and cadmium susceptibility. J. Appl. Microbiol. 83, 381–388. doi: 10.1046/j.1365-2672.1997.00238.x
Metselaar, K. I., den Besten, H. M., Boekhorst, J., van Hijum, S. A., Zwietering, M. H., and Abee, T. (2015). Diversity of acid stress resistant variants of Listeria monocytogenes and the potential role of ribosomal protein S21 encoded by rpsU. Front. Microbiol. 6:422. doi: 10.3389/fmicb.2015.00422
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2010). Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proc GCE, 1–8. doi: 10.1109/gce.2010.5676129
Moltz, A. G. (2005). Formation of biofilms by Listeria monocytogenes under various growth conditions. J. Food Prot. 68, 92–97. doi: 10.4315/0362-028X-68.1.92
Møretrø, T., and Langsrud, S. (2004). Listeria monocytogenes: biofilm formation and persistence in food-processing environments. Biofilms 1, 107–121. doi: 10.1017/S1479050504001322
Nightingale, K. K., Ivy, R. A., Ho, A. J., Fortes, E. D., Njaa, B. L., Peters, R. M., et al. (2008). inlA premature stop codons are common among Listeria monocytogenes isolates from foods and yield virulence-attenuated strains that confer protection against fully virulent strains. Appl. Environ. Microbiol. 74, 6570–6583. doi: 10.1128/AEM.00997-08
Nightingale, K. K., Windham, K., Martin, K. E., Yeung, M., and Wiedmann, M. (2005). Select Listeria monocytogenes subtypes commonly found in foods carry distinct nonsense mutations in inlA, leading to expression of truncated and secreted internalin A, and are associated with a reduced invasion phenotype for human intestinal epithelial cells. Appl. Environ. Microbiol. 71, 8764–8772. doi: 10.1128/AEM.71.12.8764-8772.2005
Olier, M., Pierre, F., Lemaître, J., Divies, C., Rousset, A., and Guzzo, J. (2002). Assessment of the pathogenic potential of two Listeria monocytogenes human faecal carriage isolates. Microbiology 148, 1855–1862. doi: 10.1099/00221287-148-6-1855
Olier, M., Pierre, F., Rousseaux, S., Lemaitre, J. P., Rousset, A., Piveteau, P., et al. (2003). Expression of truncated Internalin A is involved in impaired internalization of some Listeria monocytogenes isolates carried asymptomatically by humans. Infect. Immun. 71, 1217–1224. doi: 10.1128/IAI.71.3.1217-1224.2003
Ondov, B. D., Treangen, T. J., Melsted, P., Mallonee, A. B., Bergman, N. H., Koren, S., et al. (2016). Mash: fast genome and metagenome distance estimation using MinHash. bioRxiv 029827. doi: 10.1186/s13059-016-0997-x
Orsi, R. H., Borowsky, M. L., Lauer, P., Young, S. K., Nusbaum, C., Galagan, J. E., et al. (2008). Short-term genome evolution of Listeria monocytogenes in a non-controlled environment. BMC Genomics 9:539. doi: 10.1186/1471-2164-9-539
Orsi, R. H., den Bakker, H. C., and Wiedmann, M. (2011). Listeria monocytogenes lineages: genomics, evolution, ecology, and phenotypic characteristics. Int. J. Med. Microbiol. 301, 79–96. doi: 10.1016/j.ijmm.2010.05.002
Orsi, R., Ripoll, D., Yeung, M., Nightingale, K., and Wiedmann, M. (2007). Recombination and positive selection contribute to evolution of Listeria monocytogenes inlA. Microbiology 153, 2666–2678. doi: 10.1099/mic.0.2007/007310-0
Overney, A., Chassaing, D., Carpentier, B., Guillier, L., and Firmesse, O. (2016). Development of synthetic media mimicking food soils to study the behaviour of Listeria monocytogenes on stainless steel surfaces. Int. J. Food Microbiol. 238, 7–14. doi: 10.1016/j.ijfoodmicro.2016.08.034
Parisi, A., Latorre, L., Normanno, G., Miccolupo, A., Fraccalvieri, R., Lorusso, V., et al. (2010). Amplified fragment length polymorphism and multi-locus sequence typing for high-resolution genotyping of Listeria monocytogenes from foods and the environment. Food Microbiol. 27, 101–108. doi: 10.1016/j.fm.2009.09.001
Perez-Diaz, J., Vicente, M., and Baquero, F. (1982). Plasmids in Listeria. Plasmid 8, 112–118. doi: 10.1016/0147-619X(82)90049-X
Peterkin, P. I., Gardiner, M., Malik, N., and Idziak, E. S. (1992). Plasmids in Listeria monocytogenes and other Listeria species. Can. J. Microbiol. 38, 161–164. doi: 10.1139/m92-027
Piercey, M. J., Hingston, P. A., and Hansen, L. T. (2016). Genes involved in Listeria monocytogenes biofilm formation at a simulated food processing plant temperature of 15°C. Int. J. Food Microbiol. 223, 63–74. doi: 10.1016/j.ijfoodmicro.2016.02.009
Poyart-Salmeron, C., Carlier, C., Trieu-Cuot, P., Courvalin, P., and Courtieu, A. (1990). Transferable plasmid-mediated antibiotic resistance in Listeria monocytogenes. Lancet 335, 1422–1426. doi: 10.1016/0140-6736(90)91447-I
Ragon, M., Wirth, T., Hollandt, F., Lavenir, R., Lecuit, M., Le Monnier, A., et al. (2008). A new perspective on Listeria monocytogenes evolution. PLoS Pathog. 4:e1000146. doi: 10.1371/journal.ppat.1000146
Rakic-Martinez, M., Drevets, D. A., Dutta, V., Katic, V., and Kathariou, S. (2011). Listeria monocytogenes strains selected on ciprofloxacin or the disinfectant benzalkonium chloride exhibit reduced susceptibility to ciprofloxacin, gentamicin, benzalkonium chloride, and other toxic compounds. Appl. Environ. Microbiol. 77, 8714–8721. doi: 10.1128/AEM.05941-11
Read, T. D., and Massey, R. C. (2014). Characterizing the genetic basis of bacterial phenotypes using genome-wide association studies: a new direction for bacteriology. Genome Med. 6:109. doi: 10.1186/s13073-014-0109-z
Ribeiro, V., Mujahid, S., Orsi, R., Bergholz, T., Wiedmann, M., Boor, K., et al. (2014). Contributions of σ B and PrfA to Listeria monocytogenes salt stress under food relevant conditions. Int. J. Food Microbiol. 177, 98–108. doi: 10.1016/j.ijfoodmicro.2014.02.018
Rissman, A. I., Mau, B., Biehl, B. S., Darling, A. E., Glasner, J. D., and Perna, N. T. (2009). Reordering contigs of draft genomes using the Mauve aligner. Bioinformatics 25, 2071–2073. doi: 10.1093/bioinformatics/btp356
Roldgaard, B. B., Andersen, J. B., Hansen, T. B., Christensen, B. B., and Licht, T. R. (2009). Comparison of three Listeria monocytogenes strains in a guinea-pig model simulating food-borne exposure. FEMS Microbiol. Lett. 291, 88–94. doi: 10.1111/j.1574-6968.2008.01439.x
Rousseaux, S., Olier, M., Lemaitre, J. P., Piveteau, P., and Guzzo, J. (2004). Use of PCR-restriction fragment length polymorphism of inlA for rapid screening of Listeria monocytogenes strains deficient in the ability to invade Caco-2 cells. Appl. Environ. Microbiol. 70, 2180–2185. doi: 10.1128/AEM.70.4.2180-2185.2004
Ryan, S., Begley, M., Hill, C., and Gahan, C. (2010). A five-gene stress survival islet (SSI-1) that contributes to the growth of Listeria monocytogenes in suboptimal conditions. J. Appl. Microbiol. 109, 984–995. doi: 10.1111/j.1365-2672.2010.04726.x
Schwender, H., Zucknick, M., Ickstadt, K., and Bolt, H. M., and GENICA network. (2004). A pilot study on the application of statistical classification procedures to molecular epidemiological data. Toxicol. Lett. 151, 291–299. doi: 10.1016/j.toxlet.2004.02.021
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Shabala, L., Lee, S. H., Cannesson, P., and Ross, T. (2008). Acid and NaCl limits to growth of Listeria monocytogenes and influence of sequence of inimical acid and NaCl levels on inactivation kinetics. J. Food Prot. 71, 1169–1177. doi: 10.4315/0362-028X-71.6.1169
Sorrells, K. M., Enigl, D. C., and Hatfield, J. R. (1989). Effect of pH, acidulant, time, and temperature on the growth and survival of Listeria monocytogenes. J. Food Prot. 52, 571–573. doi: 10.4315/0362-028X-52.8.571
Stack, H. M., Sleator, R. D., Bowers, M., Hill, C., and Gahan, C. G. (2005). Role for HtrA in stress induction and virulence potential in Listeria monocytogenes. Appl. Environ. Microbiol. 71, 4241–4247. doi: 10.1128/AEM.71.8.4241-4247.2005
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Sui, S. J. H., Fedynak, A., Hsiao, W. W., Langille, M. G., and Brinkman, F. S. (2009). The association of virulence factors with genomic islands. PLoS ONE 4:e8094. doi: 10.1371/journal.pone.0008094
Takahashi, H., Kuramoto, S., Miya, S., and Kimura, B. (2011). Desiccation survival of Listeria monocytogenes and other potential foodborne pathogens on stainless steel surfaces is affected by different food soils. Food Control 22, 633–637. doi: 10.1016/j.foodcont.2010.09.003
Team, R. C. (2016). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. Available online at: http://www.R-project.org
Treangen, T. J., Ondov, B. D., Koren, S., and Phillippy, A. M. (2014). The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 15, 1–15. doi: 10.1186/s13059-014-0524-x
US Food Drug Administration (US FDA) (2016). CPG Sec. 555.320 Listeria monocytogenes. Available online at: http://www.fda.gov/ICECI/ComplianceManuals/CompliancePolicyGuidanceManual/ucm136694.htm
Van Der Veen, S., Moezelaar, R., Abee, T., and Wells-Bennik, M. H. (2008). The growth limits of a large number of Listeria monocytogenes strains at combinations of stresses show serotype-and niche-specific traits. J. Appl. Microbiol. 105, 1246–1258. doi: 10.1111/j.1365-2672.2008.03873.x
Van Stelten, A., and Nightingale, K. K. (2008). Development and implementation of a multiplex single-nucleotide polymorphism genotyping assay for detection of virulence-attenuating mutations in the Listeria monocytogenes virulence-associated gene inlA. Appl. Environ. Microbiol. 74, 7365–7375. doi: 10.1128/AEM.01138-08
Van Stelten, A., Simpson, J. M., Chen, Y., Scott, V. N., Whiting, R. C., Ross, W. H., et al. (2011). Significant shift in median guinea pig infectious dose shown by an outbreak-associated Listeria monocytogenes epidemic clone strain and a strain carrying a premature stop codon mutation in inlA. Appl. Environ. Microbiol. 77, 2479–2487. doi: 10.1128/AEM.02626-10
Van Stelten, A., Simpson, J. M., Ward, T. J., and Nightingale, K. K. (2010). Revelation by single-nucleotide polymorphism genotyping that mutations leading to a premature stop codon in inlA are common among Listeria monocytogenes isolates from ready-to-eat foods but not human listeriosis cases. Appl. Environ. Microbiol. 76, 2783–2790. doi: 10.1128/AEM.02651-09
Verheul, A., Russell, N. J., Van'T Hof, R., Rombouts, F. M., and Abee, T. (1997). Modifications of membrane phospholipid composition in nisin-resistant Listeria monocytogenes Scott A. Appl. Environ. Microbiol. 63, 3451–3457.
Voelker, U., Voelker, A., Maul, B., Hecker, M., Dufour, A., and Haldenwang, W. G. (1995). Separate mechanisms activate sigma B of Bacillus subtilis in response to environmental and metabolic stresses. J. Bacteriol. 177, 3771–3780. doi: 10.1128/jb.177.13.3771-3780.1995
Vogel, B. F., Hansen, L. T., Mordhorst, H., and Gram, L. (2010). The survival of Listeria monocytogenes during long term desiccation is facilitated by sodium chloride and organic material. Int. J. Food Microbiol. 140, 192–200. doi: 10.1016/j.ijfoodmicro.2010.03.035
Waack, S., Keller, O., Asper, R., Brodag, T., Damm, C., Fricke, W. F., et al. (2006). Score-based prediction of genomic islands in prokaryotic genomes using hidden Markov models. BMC Bioinform. 7:142. doi: 10.1186/1471-2105-7-142
Walker, S., Archer, P., and Banks, J. G. (1990). Growth of Listeria monocytogenes at refrigeration temperatures. J. Appl. Bacteriol. 68, 157–162. doi: 10.1111/j.1365-2672.1990.tb02561.x
Weber, M. H., Klein, W., Müller, L., Niess, U. M., and Marahiel, M. A. (2001). Role of the Bacillus subtilis fatty acid desaturase in membrane adaptation during cold shock. Mol. Microbiol. 39, 1321–1329. doi: 10.1111/j.1365-2958.2001.02322.x
Wiedmann, M., Arvik, T. J., Hurley, R. J., and Boor, K. J. (1998). General stress transcription factor σB and its role in acid tolerance and virulence of Listeria monocytogenes. J. Bacteriol. 180, 3650–3656.
Wilson, R. L., Brown, L. L., Kirkwood-Watts, D., Warren, T. K., Lund, S. A., King, D. S., et al. (2006). Listeria monocytogenes 10403S HtrA is necessary for resistance to cellular stress and virulence. Infect. Immun. 74, 765–768. doi: 10.1128/IAI.74.1.765-768.2006
Wonderling, L. D., Wilkinson, B. J., and Bayles, D. O. (2004). The htrA (degP) gene of Listeria monocytogenes 10403S is essential for optimal growth under stress conditions. Appl. Environ. Microbiol. 70, 1935–1943. doi: 10.1128/AEM.70.4.1935-1943.2004
Wu, S., Wu, Q., Zhang, J., Chen, M., and Guo, W. (2016). Analysis of multilocus sequence typing and virulence characterization of Listeria monocytogenes isolates from Chinese retail ready-to-eat food. Front. Microbiol. 7:168. doi: 10.3389/fmicb.2016.00168
Yildiz, O., Aygen, B., Esel, D., Kayabas, U., Alp, E., Sumerkan, B., et al. (2007). Sepsis and meningitis due to Listeria monocytogenes. Yonsei Med. J. 48, 433–439. doi: 10.3349/ymj.2007.48.3.433
Zhang, C., Zhang, M., Ju, J., Nietfeldt, J., Wise, J., Terry, P. M., et al. (2003). Genome diversification in phylogenetic lineages I and II of Listeria monocytogenes: identification of segments unique to lineage II populations. J. Bacteriol. 185, 5573–5584. doi: 10.1128/JB.185.18.5573-5584.2003
Keywords: Listeria monocytogenes, stress tolerance, whole genome sequencing, sequence typing, food safety, plasmids, internalin A
Citation: Hingston P, Chen J, Dhillon BK, Laing C, Bertelli C, Gannon V, Tasara T, Allen K, Brinkman FSL, Truelstrup Hansen L and Wang S (2017) Genotypes Associated with Listeria monocytogenes Isolates Displaying Impaired or Enhanced Tolerances to Cold, Salt, Acid, or Desiccation Stress. Front. Microbiol. 8:369. doi: 10.3389/fmicb.2017.00369
Received: 16 December 2016; Accepted: 22 February 2017;
Published: 08 March 2017.
Edited by:
Jennifer Ronholm, McGill University, CanadaCopyright © 2017 Hingston, Chen, Dhillon, Laing, Bertelli, Gannon, Tasara, Allen, Brinkman, Truelstrup Hansen and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Siyun Wang, siyun.wang@ubc.ca
†Present Address: Jessica Chen, IHRC Inc., Atlanta, GA, USA
 Patricia Hingston
Patricia Hingston Jessica Chen
Jessica Chen Bhavjinder K. Dhillon
Bhavjinder K. Dhillon Chad Laing
Chad Laing Claire Bertelli
Claire Bertelli Victor Gannon
Victor Gannon Taurai Tasara
Taurai Tasara Kevin Allen
Kevin Allen Fiona S. L. Brinkman
Fiona S. L. Brinkman Lisbeth Truelstrup Hansen
Lisbeth Truelstrup Hansen Siyun Wang
Siyun Wang