- 1State Key Laboratory of Respiratory Disease, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou, China
- 2University of Chinese Academy of Sciences, Beijing, China
- 3Key Laboratory of Biotechnology of Antibiotics, Ministry of Health, Institute of Medicinal Biotechnology, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
- 4State Key Laboratory of Respiratory Disease, Department of Clinical Laboratory, Guangzhou Chest Hospital, Guangzhou, China
- 5State Key Laboratory of Virology, Wuhan Institute of Virology, Chinese Academy of Sciences, Wuhan, China
The genetic manipulation of Mycobacterium tuberculosis genome is limited by the availability of selection markers. Spontaneous resistance mutation rate of M. tuberculosis to the widely used kanamycin is relatively high which often leads to some false positive transformants. Due to the few available markers, we have created a cassette containing thiostrepton resistance gene (tsr) for selection in M. tuberculosis and M. bovis BCG, and gentamicin resistance gene (aacC1) for Escherichia coli and M. smegmatis mc2155, flanked with dif sequences recognized by the Xer system of mycobacteria. This cassette adds to the limited available selection markers for mycobacteria.
Introduction
Gene manipulation in mycobacteria is performed using a limited number of selection markers. Mycobacteria are naturally resistant to many antibiotics and requires use of stable drugs with low frequency of spontaneous resistance for selection, hence limiting the alternative choices (Parish and Brown, 2008). The combined use of multiple markers enables more versatile genetic modifications, including the stable maintenance of multiple plasmids and inactivation of multiple genes (Wada et al., 2016).
Aminoglycoside phosphotransferase (aph) genes, conferring resistance to kanamycin (KAN), were the first to be used as selection markers in mycobacteria (Snapper et al., 1988) owing to their stability over the extended periods of incubation for slow-growing mycobacteria. However, their utility is limited by emergence of spontaneous resistance, albeit at low frequencies (Hatfull, 1996). Unlike fast-growing bacteria, slow-growing mycobacteria have a single rRNA operon (Suzuki et al., 1987) which is more prone to mutations conferring resistance to agents such as KAN (Bottger, 1994). Besides, selection using KAN in Mycobacterium w and Mycobacterium vaccae has not been achieved. Radford and Hodgson (1991) first reported the use of hygromycin (HYG) resistance gene (hyg) as a selection marker in M. smegmatis and M. bovis BCG in 1991. Since then, it has been used in other mycobacteria. The use of hyg provides a marker gene which does not provide cross resistance to clinically useful drugs (Garbe et al., 1994). It offers an improved transformation frequency over KAN and is probably more efficiently expressed in mycobacteria than the Escherichia coli-derived aminoglycoside phosphotransferase genes conferring KAN resistance (Garbe et al., 1994). The use of apramycin as a selection marker in both slow- and fast-growing mycobacteria was first reported by Paget and Davies in 1996, following its disapproval for clinical use in humans. However, its utility is limited by the acetylation of other closely related aminoglycoside such as KAN (Davies and O‘Connor, 1978; Consaul and Pavelka, 2004), and low transformation efficiencies. β-lactam-based selection markers such as the ampicillin resistance gene ampr are not useful in mycobacteria since they contain endogenous β-lactamases that confers natural resistance to penicillins (Hatfull, 1996).
Other past explorations have included resistance to chloramphenicol (Das Gupta et al., 1993), but have been limited due to poor stability and high rates of spontaneous mutations, hence unsuitable for slow-growing mycobacteria. Streptomycin, sulfonamide (Gormley and Davies, 1991) and mercury salts (Baulard et al., 1995) have also been explored as possible selectable markers, but to date, KAN, HYG, and GEN resistance genes remain the often widely exploited selectable markers in mycobacterial genetics.
Owing to the limited number of markers and their disadvantages, we hence sought to explore use of methyl-accepting chemotaxis protein I, a serine sensor receptor tsr gene conferring resistance to thiostrepton (TSR), as a selectable marker in M. tuberculosis and M. bovis BCG. TSR, a thiazole antibiotic, was first isolated and characterized from Streptomyces azureus (Cundliffe, 1971) in 1954 at the Squibb Institute (Pagano et al., 1956) and is used in veterinary medicine to treat mastitis, and as a topical agent for dogs. However, it has only found limited applications due to its poor solubility and toxicity (Kuiper and Conn, 2014).
Thiostrepton inhibits protein translation by firmly binding to the complex formed by 23S rRNA and ribosomal protein L11 in bacterial ribosomes (Cannon and Burns, 1971; Cundliffe, 1971). The tsr gene encodes an RNA methyltransferase that prevents TSR from binding to ribosomes by 23S rRNA methylation (Thompson et al., 1982). The tsr confer total resistance to TSR and thus has been the selection marker of choice in many of the Streptomyces spp. cloning vectors (Thompson et al., 1980).
In a recent study on ovarian cancer cell lines, Westhoff et al. (2014) demonstrated that when TSR is used in combination with the standard paclitaxel/cisplatin chemotherapy, it decreases Forkhead box M1 (FOXM1) gene expression besides showing an enhanced synergistic cytotoxicity in ascites cells from platinum-resistant patients. In addition, Wada et al. (2016) also demonstrated tsr as a viable selection marker for the thermophilic Geobacillus kaustophilus besides demonstrating accurate selection as a single copy in Streptomyces strains.
However, only scanty data showed that TSR is active against M. tuberculosis (Vermeulen and Wu, 2004; Lougheed et al., 2009) in drug testing.
Materials and Methods
Strains, Media, and Culture Conditions
Escherichia coli DH5α was grown at 37°C in Luria-Bertani (LB) broth and agar. M. tuberculosis H37Rv, autoluminescent M. tuberculosis H37Ra (Yang et al., 2015), M. bovis BCG Tice and M. smegmatis mc2155 and their recombinants were grown in Middle Brook 7H9 broth (Becton Dickinson, USA) supplemented with 10% oleic acid albumin dextrose catalase (OADC, Becton Dickinson) and 0.05% tween80, or on solid Middle Brook 7H11 medium (Difco) supplemented with 10% OADC. On agar plates, M. tuberculosis H37Rv, M. bovis BCG Tice and M. avium were incubated for 4–5 weeks, and M. abscessus GZ002 and M. smegmatis mc2155 were incubated for 3–4 days in 37°C.
Thiostrepton and GEN were purchased from Sigma–Aldrich (China) and dissolved in dimethyl sulfoxide (DMSO) and double distilled water, respectively. GEN 20 and 5 μg/mL was used for selection of E. coli and M. smegmatis mc2155, respectively, and TSR 5 and 10 μg/mL of both M. tuberculosis H37Rv and M. bovis BCG Tice. LB broth was augmented with 170 μg/mL chloramphenicol Sigma–Aldrich (China).
Drug Susceptibility Testing
We first tested the potential of TSR as a selection antibiotic for M. tuberculosis up to a final concentration of 10 μg/mL in liquid culture of autoluminescent M. tuberculosis H37Ra (AUlRa) (Table 1) as previously described (Zhang et al., 2012). Briefly, 2 mL of AUlRa was inoculated in 50 mL 7H9 plus OADC and tween80 with shaking at 37°C to mid log phase (OD600 = 0.6–0.8) in a flask and then diluted to appropriate concentrations. Drugs (5 μL/drug) were added into the 1.5 mL vial, mixed with 195 μL AUlRa and incubated at 37°C. Controls using 195 μL AUlRa and DMSO (5 μL) or 195 μL AUlRa and water (5 μL) tubes were included. Relative light measurements (RLUs) were monitored starting day 0, day 1, day 3, and day 5 using GloMax 20/20 Luminometer (Promega).
Susceptibilities of M. tuberculosis H37Rv, M. bovis BCG Tice, M. avium and M. abscessus GZ002 to TSR were performed using mid log phase high titer (>107 CFU/plate) cultures on 0, 2, 20, and 100 μg/mL Middle Brook 7H11 TSR agar plates.
Minimum inhibition concentration (MIC) was defined as the lowest concentration of a drug inhibiting 99% of bacterial growth (Zhang et al., 2010). The MIC values for wild-type and recombinant mycobacteria were detected on Middle Brook 7H11 agar plates containing different concentrations of TSR (0–160 μg/mL) and GEN (0–100 μg/mL).
General DNA Techniques
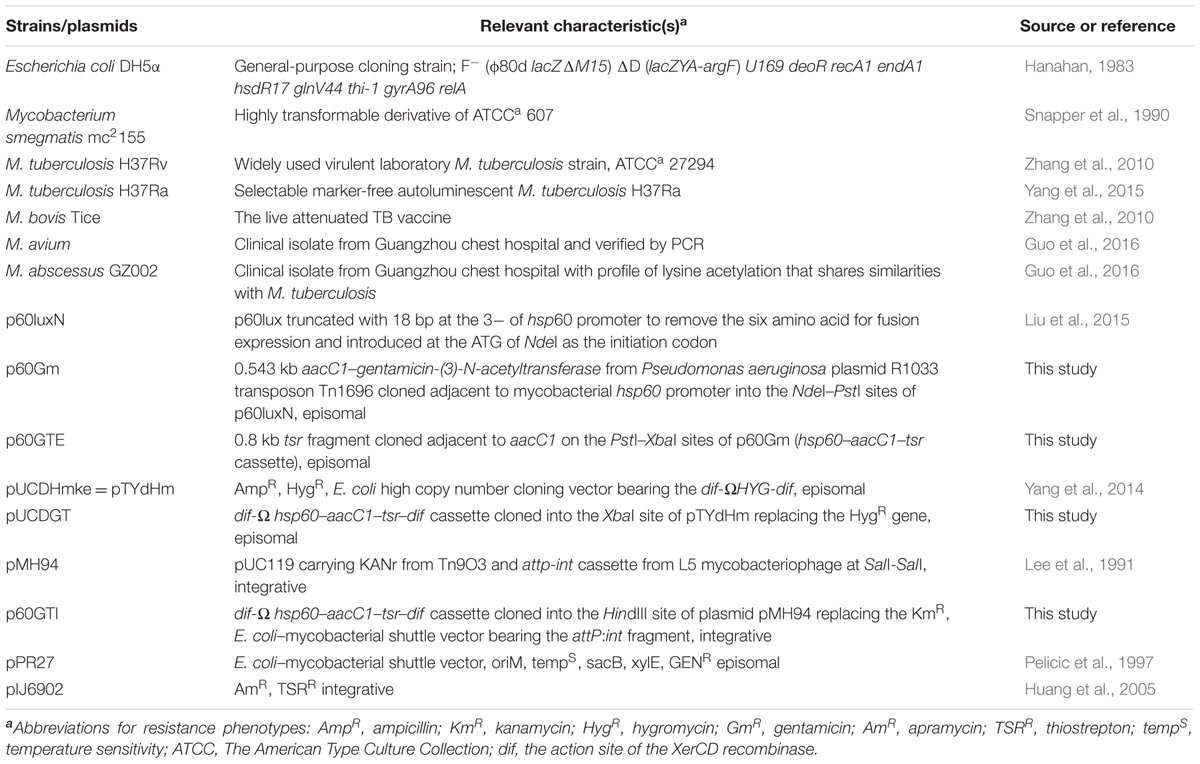
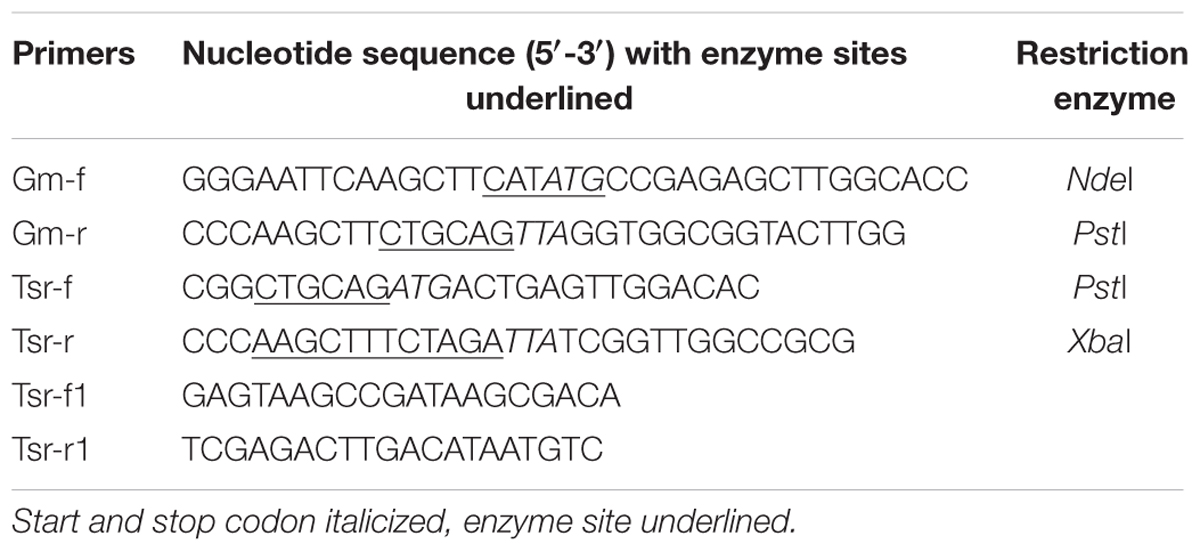
Polymerase chain reaction (PCR) amplification reactions were performed with pfu DNA polymerase (Takara). The PCR products and plasmids were analyzed by electrophoresis in agarose gels and purified using a DNA gel extraction kit (Magen, China). Plasmids were also extracted and purified using kits from the same company. Purified PCR products and plasmids were sequenced (BGI, Shenzhen, China). The aacC1 gene (0.543 kb) was amplified from plasmid pPR27 (Table 1) using primers Gm-f and Gm-r (Table 2) while the 0.8 kb tsr gene was amplified from plasmid pIJ6902 (Table 1) using primers Tsr-f and Tsr-r (Table 2).
Construction of Shuttle Vector Containing tsr + aacC1 Resistance Genes
To construct a vector bearing tsr+aacC1, we arranged the genes into a cassette under the control of the M. tuberculosis hsp60 promoter (Figure 1) in plasmid p60LuxN (Liu et al., 2015) intending the aacC1 gene to be used for selection in E. coli and M. smegmatis mc2155 and the tsr gene to be used in M. tuberculosis and M. bovis BCG. The aacC1 was cloned adjacent to the hsp60 promoter into the NdeI-PstI sites of p60LuxN resulting in plasmid p60Gm. The tsr gene was cloned into the PstI-HindIII sites of plasmid p60Gm to get E. coli-mycobacteria shuttle plasmid p60GTE bearing hsp60-aacC1-tsr cassette.
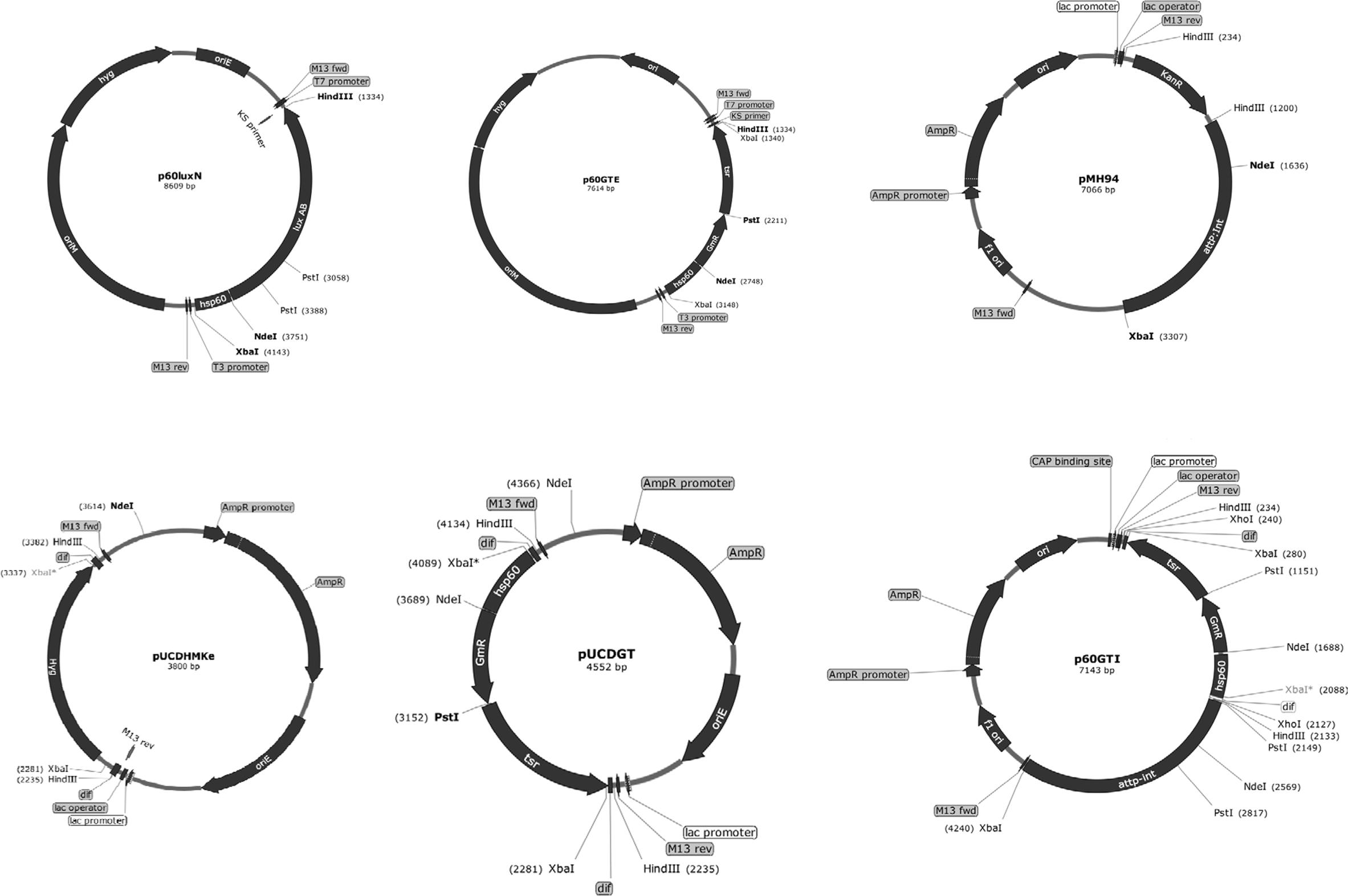
FIGURE 1. Scheme of vector construction. The Escherichia coli-mycobacterial plasmid p60GTE was derived by inserting aacC1 and tsr fragments next to the mycobacterial hsp60 promoter. The hsp60-aacC1-tsr cassette was excised and inserted on to the XbaI sites of pUCDHmke bearing the dif sequences. The dif-hsp60-aacC1-tsr-dif cassette was excised and inserted on the Hind III sites of plasmid pMH94. attP, mycobacteriophage L5 attachment site; int, integrase gene; oriE, origin region of E. coli; oriM, thermosensitive origin region of mycobacteria; KanR, KAN resistance gene, dif: the putative MTB dif sequence. Useful enzyme sites: NdeI; HindIII; PstI and XbaI.
Construction of dif-hsp60-aacC1-tsr-dif Cassette
The hsp60-aacC1-tsr cassette was excised with XbaI from plasmid p60GTE and cloned into the XbaI sites of E. coli pUCDHmke derived from pTYdHm (Yang et al., 2014) plasmid (Table 1) bearing a dif-ΩHYG-dif cassette replacing hyg gene and creating plasmid pUCDGT. The dif-hsp60-aacC1-tsr-dif cassette (Figure 1) was excised by HindIII from pUCDGT and cloned into the integrative plasmid pMH94 (Table 1) replacing the KAN resistance gene and creating plasmid p60GTI.
Transformation
Plasmids p60GTE and p60GTI were used. M. smegmatis was transformed as previously described (Snapper et al., 1990), while M. tuberculosis and M. bovis BCG were transformed as previously described (Wards and Collins, 1996; Yang et al., 2015) with some modifications. The competent M. tuberculosis and M. bovis BCG cells were first incubated at 37°C for 10 min before electroporation and transformation was performed at room temperature. Transformants were selected on plates containing TSR (5 and 10 μg/mL) for both M. bovis BCG and M. tuberculosis while containing GEN (5 μg/mL) for M. smegmatis. Individual transformant colonies of three independent transformations were counted to determine the transformation frequencies per microgram of DNA and tested by PCR with primers Tsr-f and Tsr-r.
Analysis of Unmarked Recombinant M. tuberculosis and M. bovis BCG Transformants
Unmarked recombinant transformants were analyzed according to Yang et al. (2014). Briefly, PCR verified TSR-resistant single p60GTI colonies were individually cultured in 7H9 media to late log phase (OD600 = 0.8–1.0) without selection to allow excision of the dif-hsp60-aacC1-tsr-dif cassette by the endogenous mycobacteria XerC and XerD. Ten-fold serial dilutions of bacterial culture were spread on plain agar plates. The colonies were picked and replica streaked on both plain and 10 μg/mL TSR-containing 7H11 plates. The TSR-sensitive colonies were verified further by PCR amplification of the 1.9 kb cassette using primers Tsr-f1 and Tsr-r1 (Table 2) and the shorter PCR products (∼0.5 kb) bearing one single dif sequence were confirmed by sequencing.
Results and Discussion
TSR as a Potential Selection Antibiotic against Mycobacteria
We first tested the potential use of TSR as a selective antibiotic against mycobacteria. Using liquid culture autoluminescent M. tuberculosis H37Ra, we tested different TSR concentrations up to 10 μg/mL and the relative light units (RLUs) declined sharply within 2 days and continuously till the end of the assay, while those of blank control rose steadily (MIClux = 0.05 μM, ∼ = 0.08 μg/mL). Additional susceptibility testing on 2–50 μg/mL 7H11 TSR plates of M. tuberculosis and M. bovis BCG Tice yielded complete growth inhibition while we observed complete insensitivity even on 7H11 plates containing 100 μg/mL TSR for M. avium, M. abscessus GZ002 and M. smegmatis mc2155 illustrating the unsuitability of TSR as their selection antibiotic. We detected the TSR MICs of M. tuberculosis H37Rv strain and M. bovis BCG Tice as 0.125 and 0.25 μg/mL (Table 3) similar to the 0.08 μM (∼0.133 μg/mL to M. tuberculosis H37Rv) reported by Lougheed et al. (2009) and no mutant resistant colonies were observed.

TABLE 3. Minimum inhibition concentrations (MICs) of TSR for wild-type and recombinant mycobacteria.
Construction of Plasmids p60GTE and p60GTI, Their Transformation Frequencies and MICs in Respective Recombinant Strains
We set out to construct two plasmids expressing tsr and aacC1 genes in both E. coli and mycobacteria. We constructed episomal and integrative E. coli-mycobacterial shuttle plasmids bearing the mycobacterial hsp60 promoter, aacC1 and the tsr gene flanked by dif sequences (Figure 1). Both antibiotic resistance markers, the streptomyces TSR resistance gene, tsr, and the Pseudomonas aeruginosa GEN resistance gene, aacC1, worked in mycobacterial transformants. TSR resistance is not a selectable marker in E. coli due to outer membrane exclusion of TSR by gram-negative bacteria (Gale et al., 1981). To circumvent this, we used GEN for selection in E. coli and supplemented the media with chloramphenicol 170 μg/mL to increase the plasmid copy number. The transformation frequency for H37Rv and M. bovis BCG overexpressed with the episomal plasmid p60GTE were 1.26 × 104 and 4.3 × 103 CFUs and 3.5 × 103 and 2 × 102 CFUs, respectively, with the integrative plasmid p60GTI on TSR 5 μg/mL (Table 4). Both H37Rv and M. bovis BCG recombinant strains increased the MICs by >300-fold (Table 3) while M. smegmatis mc2155 strains increased the MICs by 40-fold (Table 5).
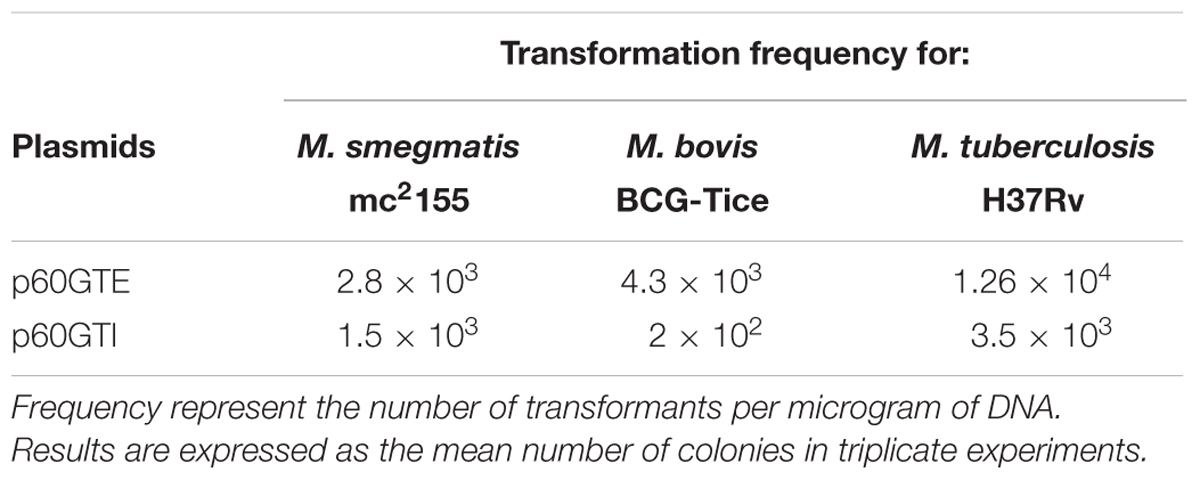
TABLE 4. Transformation frequency for M. bovis BCG Tice and M. tuberculosis H37Rv using TSR and M. smegmatis mc2155 using GEN as a selection marker.

TABLE 5. Minimum inhibition concentrations of GEN for wild-type and recombinant M. smegmatis mc2155.
The loss of the tsr marker gene verified by PCR, yielded ∼0.5 kb products confirmed by sequencing to bear one dif sequence as expected, from 12 and 20 randomly selected recombinant p60GTI containing M. tuberculosis H37Rv and M. bovis BCG colonies. We found that five of each recombinant strain had lost the tsr gene which should be excised by the endogenous mycobacterial recombinase XerCD system expressed by XerC and XerD genes recognizing the Ωdif cassette (Cascioferro et al., 2010; Yang et al., 2014), resulting in selectable marker-free colonies.
Our TSR MICs results concurs with the antimicrobial bactericidal activity reported by others (Vermeulen and Wu, 2004; Lougheed et al., 2009), and to the best of our knowledge this is the first report showing the use of TSR resistance as putative selective marker for gene transfer in mycobacteria.
Conclusion
We have successfully constructed a cassette containing tsr and aacC1 genes flanked by dif sequences for selection in mycobacteria and demonstrated the potential of this cassette for use as a mycobacteria selection marker in M. tuberculosis and M. bovis BCG. The novelty of this work is the introduction and expression of genes in a new cassette and verified by raising of resistance in the corresponding host cells. The new reliable selection marker comes in handy for M. tuberculosis genetic manipulation studies and is a new tool for efficient construction of selection-marker free recombinant strains.
Author Contributions
Conceived and designed research: JM, BW, YC, YZ, and TZ. Performed research: JM, GM, YC, SH, and CC. Co-wrote the manuscript: JM, GM, MN, TZ, YZ, and ST. Contributed reagents/materials and laboratory space for conducting mycobacterial experiments: TZ, YT, XL, JL, YZ, JD, and ST.
Conflict of Interest Statement
The dif-hsp60-aacC1-tsr-dif cassette was filed as a patent for TZ, JM, BW, YC, SH, GM, YZ, and CC.
The other authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81572037), by the Chinese Academy of Sciences Grants (154144KYSB20150045, KFZD-SW-207), by the University of Chinese Academy of Sciences Scholarships (to JM and MN), by the Guangzhou Municipal Industry and Research Collaborative Innovation Program (201508020248, 201604020019), by the Guangzhou Municipal Clinical Medical Center Program (155700012), by the CASTWAS Scholarships (to GM and CC) and by the Key Project Grant (SKLRD2016ZJ003) and the Open Project Grant (2014SKLRD-O06) from the State Key Lab of Respiratory Disease, Guangzhou Institute of Respiratory Diseases, First Affiliated Hospital of Guangzhou Medical University.
References
Baulard, A., Scuyer, V. E., Haddad, N., Kremer, L., Locht, C., and Berche, P. (1995). Mercury resistance as a selective marker for recombinant mycobacteria. Microbiology 141, 1045–1050. doi: 10.1099/13500872-141-4-1045
Bottger, E. C. (1994). Resistance to drugs targeting protein synthesis in mycobacteria. Trends Microbiol. 2, 416–421. doi: 10.1016/0966-842X(94)90622-X
Cannon, M., and Burns, K. (1971). Modes of action of erythromycin and thiostrepton as inhibitors of protein synthesis. FEBS Lett. 18, 1–5. doi: 10.1016/0014-5793(71)80392-7
Cascioferro, A., Boldrin, F., Serafini, A., Provvedi, R., Palù, G., and Manganelli, R. (2010). Xer site-specific recombination, an efficient tool to introduce unmarked deletions into mycobacteria. Appl. Environ. Microbiol. 76, 5312–5316. doi: 10.1128/AEM.00382-10
Consaul, S. A., and Pavelka, M. S. Jr. (2004). Use of a novel allele of the Escherichia coli aacC4 aminoglycoside resistance gene as a genetic marker in mycobacteria. FEMS Microbiol. Lett. 234, 297–301. doi: 10.1111/j.1574-6968.2004.tb09547.x
Cundliffe, E. (1971). The mode of action of thiostrepton in vivo. Biochem. Biophys. Res. Commun. 44, 912–917. doi: 10.1016/0006-291X(71)90798-4
Das Gupta, S. K., Bashyam, M. D., and Tyagi, A. K. (1993). Cloning and assessment of mycobacterial promoters by using a plasmid shuttle vector. J. Bacteriol. 175, 5186–5192. doi: 10.1128/jb.175.16.5186-5192.1993
Davies, J., and O‘Connor, S. (1978). Enzymatic modification of the aminoglycoside antibiotics: 3-N-acetyltransferase with broad specificity that determines resistance to the novel antibiotic apramycin. Antimicrob. Agents Chemother. 14, 69–72. doi: 10.1128/AAC.14.1.69
Gale, E. F., Cundliffe, E., Reynolds, P. E., Richmond, M. H., and Waring, M. J. (1981). The Molecular Basis of Antibiotic Action. Hoboken, NJ: John Wiley & Sons, Inc, 492–500.
Garbe, T. R., Barathi, J., Barnini, S., Zhang, Y., Abou-Zeid, C., Tang, D., et al. (1994). Transformation of mycobacterial species using hygromycin resistance as selectable marker. Microbiology 140, 133–138. doi: 10.1099/13500872-140-1-133
Gormley, E. P., and Davies, J. E. (1991). Transfer of plasmid RSF1010 by conjugation from Escherichia coli to Streptomyces lividans and Mycobacterium smegmatis. J. Bacteriol. 173, 6705–6708. doi: 10.1128/jb.173.21.6705-6708.1991
Guo, J., Changwei, W., Yi, H., Zhiyong, L., Tian, W., Yan, L., et al. (2016). Identification of lysine acetylation in Mycobacterium abscessus using LC-MS/MS after immunoprecipitation. J. Proteome Res. 15, 2567–2578. doi: 10.1021/acs.jproteome.6b00116
Hanahan, D. (1983). Studies on transformation of Escheriehia coli with plasmids. J. Mol. Biol. 166, 557–580. doi: 10.1016/S0022-2836(83)80284-8
Hatfull, G. F. (1996). “The molecular genetics of Mycobacterium tuberculosis,” in Tuberculosis, ed. T. M. Shinnick (Berlin: Springer-Verlag), 29–47.
Huang, J., Shi, J., Molle, V., Sohlberg, B., Weaver, D., Bibb, M. J., et al. (2005). Cross-regulation among disparate antibiotic biosynthetic pathways of Streptomyces coelicolor. Mol. Microbiol. 58, 1276–1287. doi: 10.1111/j.1365-2958.2005.04879.x
Kuiper, E. G., and Conn, G. L. (2014). Binding induced RNA conformational changes control substrate recognition and catalysis by the thiostrepton resistance methyltransferase (Tsr). J. Biol. Chem. 289, 26189–26200. doi: 10.1074/jbc.M114.574780
Lee, M. H., Pascopella, L., Jacobs, W. R. Jr., and Hatfull, G. F. (1991). Site-specific integration of mycobacteriophage Ll5: integration-proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis and Bacille Calmette-Guerin. Proc. Natl. Acad. Sci. U.S.A. 88, 3111–3115. doi: 10.1073/pnas.88.8.3111
Liu, T., Wang, B., Guo, J., Zhou, Y., Julius, M., Njire, M., et al. (2015). Role of folP1 and folP2 genes in the action of sulfamethoxazole and trimethoprim against mycobacteria. J. Microbiol. Biotechnol. 25, 1559–1567. doi: 10.4014/jmb.1503.03053
Lougheed, K. E. A., Debra, L. T., Simon, A. O., Justin, S. B., and Roger, S. B. (2009). New anti-tuberculosis agents amongst known drugs. Tuberculosis 89, 364–370. doi: 10.1016/j.tube.2009.07.002
Pagano, J. F., Weinstein, M. J., Stout, M. A., and Donovick, R. (1956). Thiostrepton, a new antibiotic. I. In vitro studies. Antibiot. Ann. 3, 554–559.
Pelicic, V., Jackson, M., Reyrat, J. M., Jacobs, W. R. Jr., Gicquel, B., and Guilhot, C. (1997). Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 94, 10955–10960. doi: 10.1073/pnas.94.20.10955
Radford, A., and Hodgson, A. L. M. (1991). Construction and characterization of a Mycobacterium-Escherichia coli shuttle vector. Plasmid 25, 149–153. doi: 10.1016/0147-619X(91)90029-V
Snapper, S. B., Lugosi, L., Jekkel, A., Melton, R. E., Kieser, T., Bloom, B. R., et al. (1988). Lysogeny and transformation in mycobacteria: stable expression of foreign genes. Proc. Natl. Acad. Sci. U.S.A. 85, 6987–6991. doi: 10.1073/pnas.85.18.6987
Snapper, S. B., Melton, R. E., Mustafa, S., Kieser, T., and Jacobs, W. R. Jr. (1990). Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4, 1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x
Suzuki, Y., Yoshinaga, K., Ono, Y., Nagata, A., and Yamada, T. (1987). Organization of rRNA genes in Mycobacterium bovis BCG. J. Bacteriol. 169, 839–843. doi: 10.1128/jb.169.2.839-843.1987
Thompson, J., Cundliffe, E., and Stark, M. (1980). Binding of thiostrepton to a complex of 23-S rRNA with ribosomal protein L11. Eur. J. Biochem. 98, 261–265. doi: 10.1111/j.1432-1033.1979.tb13184.x
Thompson, J., Schmidt, F., and Cundliffe, E. (1982). Site of action of a ribosomal RNA methylase conferring resistance to thiostrepton. J. Biol. Chem. 257, 7915–7917.
Vermeulen, M. W., and Wu, J. (2004). Use of Thiostrepton as An Anti-Mycobacterial Agent. US 20040254100A1.
Wada, K., Kobayashi, J., Furukawa, M., Doi, K., Ohshiro, T., and Suzuki, H. (2016). A thiostrepton resistance gene and its mutants serve as selectable markers in Geobacillus kaustophilus HTA426. Biosci. Biotechnol. Biochem. 80, 368–375. doi: 10.1080/09168451.2015.1079478
Wards, B. J., and Collins, D. M. (1996). Electroporation at elevated temperatures substantially improves transformation efficiency of slow-growing mycobacteria. FEMS Microbiol. Lett. 145, 101–105. doi: 10.1111/j.1574-6968.1996.tb08563.x
Westhoff, G. L., Chen, Y., Bieber, M., and Teng, N. N. H. (2014). Forkhead box M1 (FOXM1) gene expression inversely correlates with survival and targeting FOXM1 improves cytotoxicity of paclitaxel and cisplatinum in platinum-resistant ovarian cancer ascites cells ex vivo. Gynecol. Oncol. 133, 2–207. doi: 10.1016/j.ygyno.2014.03.240
Yang, F., Njire, M. M., Liu, J., Wu, T., Wang, B., Liu, T., et al. (2015). Engineering more stable, selectable marker-free autoluminescent mycobacteria by one step. PLoS ONE 10:e0119341. doi: 10.1371/journal.pone.0119341
Yang, F., Yaoju, T., Jia, L., Tianzhou, L., Bangxing, W., Yuanyuan, C., et al. (2014). Efficient construction of unmarked recombinant mycobacteria using an improved system. J. Microbiol. Meth. 103, 29–36. doi: 10.1016/j.mimet.2014.05.007
Zhang, T., Bishai, W. R., Grosset, J. H., and Nuermberger, E. L. (2010). Rapid assessment of antibacterial activity against Mycobacterium ulcerans by using recombinant luminescent strains. Antimicrob. Agents Chemother. 54, 2806–2813. doi: 10.1128/AAC.00400-10
Keywords: mycobacteria, selection marker, thiostrepton, gentamicin
Citation: Mugweru J, Makafe G, Cao Y, Zhang Y, Wang B, Huang S, Njire M, Chhotaray C, Tan Y, Li X, Liu J, Tan S, Deng J and Zhang T (2017) A Cassette Containing Thiostrepton, Gentamicin Resistance Genes, and dif sequences Is Effective in Construction of Recombinant Mycobacteria. Front. Microbiol. 8:468. doi: 10.3389/fmicb.2017.00468
Received: 06 October 2016; Accepted: 07 March 2017;
Published: 24 March 2017.
Edited by:
Awdhesh Kalia, University of Texas MD Anderson Cancer Center, USAReviewed by:
Martin Föge, Leibniz Institute for Natural Product Research and Infection Biology, GermanySubhalaxmi Nambi, University of Massachusetts Medical School, USA
Copyright © 2017 Mugweru, Makafe, Cao, Zhang, Wang, Huang, Njire, Chhotaray, Tan, Li, Liu, Tan, Deng and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tianyu Zhang, zhang_tianyu@gibh.ac.cn
†Present address: Moses Njire Department of Botany, Jomo Kenyatta University of Agriculture and Technology, Juja, Kenya
 Julius Mugweru
Julius Mugweru Gaelle Makafe
Gaelle Makafe Yuanyuan Cao1
Yuanyuan Cao1 Shaobo Huang
Shaobo Huang Chiranjibi Chhotaray
Chiranjibi Chhotaray Jiaoyu Deng
Jiaoyu Deng Tianyu Zhang
Tianyu Zhang