- 1Department of Environment and Energy Systems, Graduate School of Science and Technology, Shizuoka University, Shizuoka, Japan
- 2Department of Biological Science, Faculty of Science, Shizuoka University, Shizuoka, Japan
- 3Department of Geosciences, Faculty of Science, Shizuoka University, Shizuoka, Japan
- 4Research Institute of Green Science and Technology, Shizuoka University, Shizuoka, Japan
The halophilic archaeon Haloarcula hispanica harbors three ribosomal RNA (rRNA) operons (rrnA, rrnB, and rrnC) that contain the 16S rRNA genes rrsA, rrsB, and rrsC, respectively. Although rrsB and rrsC (rrsBC) have almost identical sequences, the rrsA and rrsBC sequences differ by 5.4%, and they differ by 2.5% with respect to guanine-plus-cytosine content (PGC). The strong correlation between the typical growth temperatures of archaea and PGC of their 16S rRNA genes suggests that H. hispanica may harbor different 16S rRNA genes having different PGC to maintain rapid growth in a wide range of temperatures. We therefore performed reverse transcription-coupled quantitative PCR to assess expression levels of rrsA (PGC, 58.9%) and rrsBC (PGC, 56.4–56.5%) at various temperatures. The expression ratio of rrsA to rrsBC increased with culture temperature. Mutants with complete deletions of one or two of the three rRNA operons were constructed and their growth rates at different temperatures compared to that of the wild-type. The growth characteristics of the rRNA operon single-mutant strains were indistinguishable from the wild-type. The rRNA operon double-mutant strains maintained the same temperature range as wild-type but displayed reduced growth rates. In particular, the double-mutant strains grew much slower than wild-type at low temperature related to minimum growth temperature of the wild-type. On the other hand, at physiologically high temperatures the wild-type and the double-mutant strain which harbors only rrnA with high-PGC rrsA grew significantly faster than the double-mutant strain which harbors only rrnC with low-PGC rrsC. These findings suggest the importance of 16S rRNAs transcribed from rrsA with high-PGC in maintaining rapid growth of this halophilic archaeon at raised growth temperatures.
Introduction
Ribosomal RNA (rRNA) genes, especially 16S rRNA genes, are a particularly good marker for phylogenetic analysis of prokaryotes because they are highly conserved in all prokaryotes (Staley, 2002). Many studies have examined the phylogenetic positions of prokaryotic species and microbial diversities in natural environments using 16S rRNA gene sequences (e.g., Hiraoka et al., 2016; Kuechler et al., 2016). On the other hand, recent studies based on genome sequences have demonstrated that the genomes of certain prokaryotes harbor several divergent 16S rRNA genes. Sun et al. (2013) reported that almost half of the complete prokaryotic genomes examined contain a base or more dissimilarity in 16S rRNA genes. Furthermore, >3% differences in 16S rRNA gene sequences have been detected in 14 of the 1690 complete genomes in domain Bacteria (Větrovský and Baldrian, 2013). Within the domain Archaea, some methanogens, e.g., Methanocaldococcus jannaschii and Methanothermobacter thermoautotrophicus, exhibit 16S rRNA gene polymorphisms displaying as much as 0.1% divergence, whereas some halophilic archaea harbor more divergent 16S rRNA gene copies (4.9–9.8%) in their genomes (Acinas et al., 2004; Boucher et al., 2004; Pei et al., 2010; Sun et al., 2013).
Haloarcula strains, halophilic archaea belonging to class Halobacteria, have mainly been isolated from solar salterns and salt lakes (Torreblanca et al., 1986), where large day-night and seasonal temperature variations are observed (Post, 1977; Wieland et al., 2005; Sima et al., 2013; Andrade et al., 2015). So far, 15 Haloarcula strains, including 10 strains named validly, have been reported (Javor et al., 1982; Mizuki et al., 2004; Namwong et al., 2011; Ding et al., 2014; Yun et al., 2015). Complete genome sequences of four strains, i.e., H. hispanica, two strains closely related to H. hispanica, and H. marismortui, have been determined and show that all strains carry three rRNA operons (rrnA, rrnB, and rrnC) (Baliga et al., 2004; Liu et al., 2011; Ding et al., 2014; Yun et al., 2015). Each rRNA operon includes the 5S rRNA gene, the 16S rRNA gene (rrsA, rrsB, or rrsC), and the 23S rRNA gene. The 5S rRNA genes have identical sequences. Although the 23S rRNA gene sequences of rrnA and rrnB are almost identical, the 23S rRNA gene sequence of rrnC is slightly different (∼2%) from those of rrnA and rrnB. Among the 16S rRNA genes, the rrsB and rrsC (rrsBC) sequences are almost identical, whereas the rrsA and rrsBC sequences differ by ∼6% (Dennis et al., 1998; Dennis, 1999).
Previous studies of the expression of 16S rRNA genes in Haloarcula strains under different salinity conditions suggest that variation in salinity does not affect the expression level of each 16S rRNA gene (López-López et al., 2007; Cui et al., 2009). On the other hand, López-López et al. (2007) demonstrated that H. marismortui displays different expression patterns of each 16S rRNA gene under a wide range of temperatures. They also performed cultivation experiments with wild-type H. marismortui and the rRNA operon single-mutant strain, which lacks rrnB containing low guanine-plus-cytosine content (PGC) of 16S rRNA gene, under various temperature conditions (Tu et al., 2005; López-López et al., 2007). They found that growth of the rRNA operon single-mutant strain was slower than that of wild-type at all tested temperatures. López-López et al. (2007) could not determine whether rrnB inactivation or a lower copy number of rRNA operons would affect growth of the mutant strain, because the rRNA operon double-mutant strains that harbor only one rRNA operon containing low- or high-PGC 16S rRNA gene were not constructed and examined. Therefore, the functional importance of rRNA transcribed from each rRNA operon including rrsA, rrsB, or rrsC on growth under different temperature conditions has not been well understood yet.
Previous studies have reported that 16S rRNA gene sequences are naturally inscribed with the thermal features of their prokaryotic hosts (Galtier and Lobry, 1997; Khachane et al., 2005; Kimura et al., 2007, 2010, 2013). The observation was based on a high correlation between the growth temperatures of the prokaryotes and the PGC of their 16S rRNA sequences: 16S rRNA genes of hyperthermophiles and thermophiles tend to have high PGC, whereas 16S rRNA genes of mesophiles and psychrophiles have relatively low PGC. On the basis of the relationship between the growth temperatures and PGC of 16S rRNA gene sequences, we propose that Haloarcula strains express and utilize high PGC of 16S rRNAs at high temperature and low PGC of 16S rRNAs at low temperature, respectively.
In the present study, Haloarcula hispanica, for which the complete genome sequence has been determined, was used. The 16S rRNA genes of H. hispanica were sequenced, and the minimum (Tmin), optimum (Topt), and maximum (Tmax) growth temperatures were estimated based on PGC of the 16S rRNA genes using the microbial molecular thermometer proposed by Kimura et al. (2013). Additionally, expression levels of rrsA and rrsBC between 25°C (actual Tmin of the strain) and 50°C (actual Tmax of the strain) were determined by reverse transcription-coupled quantitative PCR (qPCR) using specific primer sets. Moreover, we constructed rRNA operon single-mutant strains that lack rrnA, rrnB, or rrnC by using wild-type H. hispanica. We further constructed rRNA operon double-mutant strains that harbor only rrnA or rrnC by using the single-mutant strains and assessed their growth in a wide temperature range. The combined results from both gene expression and mutation experiments provide insight into the physiological advantage of harboring 16S rRNA genes of different sequence with respect to the growth of H. hispanica.
Materials and Methods
Strain and Cultivation for DNA Extraction
Haloarcula hispanica JCM8911 was obtained from the Japan Collection of Microorganisms (JCM, Tsukuba, Ibaraki, Japan). The strain was grown in Medium 307, which contained 2 g casamino acid (BD, Franklin Lakes, NJ, USA), 2 g Bacto yeast extract (BD), 1 g sodium glutamate, 3 g trisodium citrate, 10 g MgSO4⋅7H2O, 1 g CaCl2⋅2H2O, 1 g KCl, 200 g NaCl, 0.36 mg FeCl2⋅4H2O, and 0.36 mg MnCl2⋅4H2O per liter of distilled water. After the pH of the medium was adjusted to 7.0, the medium was sterilized by filtration with a polyethersulfone membrane filter (pore size, 0.22 μm; Thermo Fisher Scientific, Waltham, MA, USA) and autoclaving at 121°C for 20 min. Exactly 20 ml of the medium was injected into autoclaved 60-ml screw-cap test tubes and inoculated with cells of H. hispanica in exponential growth phase in pre-culture, and the cultures were incubated in the darkness with shaking at 180 rpm at 37°C. The cultures were centrifuged at 6230 × g for 3 min. The pelleted cells were stored at -25°C until DNA extraction.
Cloning and Sequencing of 16S rRNA Genes
In order to make standards for qPCR described below, we performed cloning and sequencing of 16S rRNA genes of H. hispanica. Bulk DNA was extracted from H. hispanica cells grown in Medium 307 with modifications (Tchinda et al., 2016). Briefly, the pelleted cells were lysed with lysozyme and proteinase K solution. Then the genomic DNA was extracted with successive phenol:chloroform:isoamyl alcohol and chloroform:isoamyl alcohol steps and precipitated with ethanol. Next, 16S rRNA genes were amplified from the bulk DNA using the archaea-specific primer set 8aF/1512uR (Table 1). PCR products were purified with a MicroSpin S-400 HR column (GE Healthcare, Little Chalfont, UK) and cloned using the Zero Blunt TOPO PCR Cloning kit (Life Technologies, Carlsbad, CA, USA). The PCR products were ligated into vector pCR4Blunt-TOPO (Life Technologies). Escherichia coli TOP10 cells (Life Technologies) were transformed with the ligated plasmid to construct a clone library. Insert DNA from selected recombinant colonies was sequenced by the dideoxy cycle-sequencing method using a Model 3730xl DNA analyzer (Applied Biosystems, Foster City, CA, USA). The most similar 16S rRNA gene sequence was determined by the BLAST program (Altschul et al., 1990). The 16S rRNA gene sequences obtained in this study were deposited in the DDBJ/EMBL/GenBank database under accession numbers LC085245, LC085246, and LC085247.
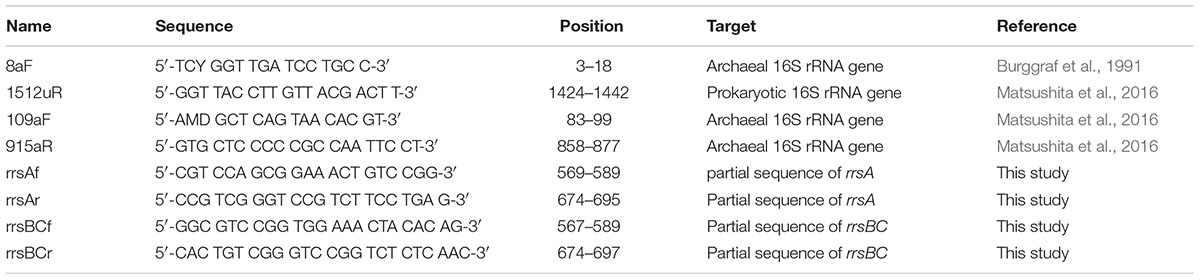
TABLE 1. Primers targeting 16S rRNA genes for PCR, reverse transcription-coupled qPCR, and sequencing in this study.
Estimating Tmin, Topt, and Tmax Based on PGC of 16S rRNA Genes
Because thermophilic and hyperthermophilic archaea have greater PGC values for 16S rRNA genes compared with psychrophilic and mesophilic archaea (Galtier and Lobry, 1997; Khachane et al., 2005; Kimura et al., 2006, 2007, 2010), Kimura et al. (2013) proposed linear regression equations to infer Tmin, Topt, and Tmax of cultured and not-yet cultured archaea based on PGC of 16S rRNA genes. We therefore used these equations to estimate growth temperatures of H. hispanica based on PGC value of each 16S rRNA gene.
Kimura et al. (2013) used partial 16S rRNA gene sequences (ca. 800 bp) between the archaea-specific primers 109aF and 915aR in order to estimate their growth temperatures (Table 1). Thus, we manually selected the internal sequences from the 16S rRNA gene sequences determined in this study. PGC values for the internal sequences were calculated using Genetyx-Mac ver. 17.0.6 (Genetyx, Tokyo, Japan). Tmin, Topt, and Tmax were calculated based on PGC of the respective sequence using Kimura’s equations.
Culture Experiment to Assess the Expression of 16S rRNA Genes
Haloarcula hispanica cells in exponential growth phase in pre-culture were inoculated into 60-ml screw-capped tubes containing 20 ml of Medium 307. The cultures were incubated in the dark with shaking at 180 rpm at 25, 30, 35, 40, 45, and 50°C. The optical density at 660 nm (OD660) of the culture was monitored using a Spectronic 200 spectrophotometer (Thermo Fisher Scientific) with sterilized medium as the negative control. When the cultures reached the early exponential growth phase (OD660 = 0.25–0.50), the cultures were centrifuged at 6230 × g for 3 min. The pelleted cells were mixed with 100 μl RNAlater (Life Technologies) and stored at –85°C until RNA extraction.
RNA Extraction and Complementary DNA (cDNA) Synthesis
Cells were thawed on ice, and the RNAlater was removed and discarded. Total RNA was extracted from the cells using the mirVana miRNA Isolation kit (Ambion, Austin, TX, USA). Contaminating genomic DNA in the extracted RNA samples was removed using the TURBO DNA-Free kit (Life Technologies). Total RNA was purified with the RNeasy MinElute Cleanup kit (Qiagen, Hilden, Germany). The quality and concentration of RNA were verified using a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) and a NanoVue Plus spectrophotometer (GE Healthcare). Single-strand complementary DNA (cDNA) was synthesized from the purified total RNA using the SuperScript III first strand synthesis system (Life Technologies) as following manufacturer’s protocol. The cDNA was purified using QIAquick PCR Purification kit (Qiagen). The purified cDNA was stored at -25°C until qPCR analysis.
qPCR
The specific primer sets rrsAf/rrsAr for rrsA and rrsBCf/rrsBCr for rrsBC were designed using Primer Express 2.0 software ver. 2.0 (Applied Biosystems) (Table 1). To test the specificity of these primer sets, qPCR was performed with PCR products of rrsA, rrsB, and rrsC that were amplified from the clones in the 16S rRNA gene-clone library described above. The PCR products were purified with QIAquick PCR purification kit (Qiagen). The qPCR was performed on an ABI Prism 7300 Real Time PCR System (Applied Biosystems) with PowerUP SYBR Green master mix (Life Technologies). Since the sequences of rrsB and rrsC are nearly identical, it was not possible to design primer sets for specifically amplifying rrsB and rrsC, independently.
Next, rrsA and rrsBC in the cDNA were quantified by qPCR using the ABI Prism 7300 Real Time PCR System. Each PCR mixture contained 2 μl of diluted cDNA template, 2 μl of each designed primer set (each 300 nM), 10 μl of PowerUP SYBR Green PCR master mix (Life Technologies), and 4 μl of nuclease-free water (Ambion). The PCR conditions included an initial step of 50°C for 2 min and 95°C for 2 min followed by 40 cycles of 95°C for 15 s, 58°C for 15 s, and 72°C for 1 min. The standard curves were prepared from diluted PCR products (1/10, 1/100, 1/1000, 1/10000) of rrsA, rrsB, and rrsC that were amplified from the clones in the 16S rRNA gene-clone library described above. The PCR reactions were performed in triplicate for technical repeats and four individuals for biological repeats.
Construction and Cultivation of rRNA Operon Double-Mutant Strains
To assess any advantage to H. hispanica of having multiple distinct 16S rRNA genes, we disrupted the three rRNA operons by using a mutation method (See Supplementary Materials and Methods for details). Briefly, an rRNA operon in wild-type H. hispanica was replaced with novobiocin resistance gene to construct rRNA operon single-mutant strains (HA2, HB2, or HC2) that lack rrnA, rrnB, or rrnC, respectively (Supplementary Table 1 and Figure 1). Furthermore, rrnB in HC2, rrnA in HC2 (or rrnC in HA2), and rrnB in HA2 were replaced with mevinolin resistance genes to construct the rRNA operon double-mutant strains HCB2, HCA2 (or HAC2), or HAB2 that contain only rrnA, rrnB, or rrnC, respectively (Supplementary Table 1 and Figure 2). Disruption of operons rrnA, rrnB, and rrnC in these mutant strains was confirmed by PCR amplification using primer sets, AVF/ACR, BVF/BCR, or CVF/CCR (Supplementary Table 2). The rRNA operon double-mutant strains harboring only rrnB (HCA2 and HAC2) could not be constructed despite repeated attempts (Supplementary Table 1).
The rRNA operon single- and double-mutant strains and wild-type strain were inoculated into Medium 307 and incubated in the dark with shaking at 180 rpm at 25, 30, 35, 40, 45, and 50°C. OD660 of the cultures was monitored using a Spectronic 200 spectrophotometer (Thermo Fisher Scientific), and growth curves were drawn based on the values. The culture experiments were performed in quadruplicate or quintuplicate. The culture experiment at 50°C (actual Tmax) was carried out twice to confirm the growth of the double-mutant and wild-type strains.
To calculate the maximum growth rates at each temperature, we determined a cell number factor to convert from OD660 value to cell density. Briefly, wild-type H. hispanica was grown in Medium 307, and the cultures were diluted with sterilized medium. After OD660 of the cultures was measured using Spectronic 200 spectrophotometer (Thermo Fisher Scientific), the cells in the cultures were fixed in formaldehyde (final concentration 7%) for 16 h at 4°C as described previously (Antón et al., 1999). The cultures were filtered using pre-blackened polycarbonate filters (pore size, 0.2 μm; diameter, 25 mm) (Millipore, Billerica, MA, USA). The cells collected on the filters were stained with SYBR Green I (1:100 dilution) (Life Technologies). The cells were observed under a model BX51 epifluorescence microscope equipped with a U-MNIB3 fluorescence filter (Olympus, Tokyo, Japan), and 50 microscopic fields were counted for each sample. A cell number factor of 2.1 × 109 cells ml-1 per OD660 determined in this study was used to determine cell density in the cultures (Supplementary Figure 3). Growth rate (μ) was calculated between the individual incubation periods (t1 and t2) with an assumption of exponential growth; i.e., μ (h-1) = (ln Nt2 – ln Nt1)/(t2 -t1), where Nt1 and Nt2 are the cell densities. On the basis of the growth rate, Tmin, Topt, and Tmax of the strains were determined.
Results and Discussion
16S rRNA Gene Sequences and Growth Temperature Estimation
A total of 16 clones were randomly selected from a clone library of H. hispanica strain JCM8911, and the sequences of 16S rRNA genes were determined (1440 bp). Three types of 16S rRNA genes were identified and matched rrsA, rrsB, and rrsC in the genome sequence as determined by Liu et al. (2011). The sequences of rrsB and rrsC were 99.6% identical, whereas the sequences of rrsA and rrsBC were 94.6–94.9% identical. These results confirm previous reports of intragenomic polymorphism of 16S rRNA genes in Haloarcula (e.g., Cui et al., 2009).
The sequence regions between the archaea-specific primers 109aF and 915aR (795 bp) were selected from the 16S rRNA gene sequences. The PGC of the internal sequences of rrsA, rrsB, and rrsC were 58.9, 56.5, and 56.4%, respectively (Table 2). The offset between the PGC of rrsA and rrsBC was ∼2.5%. Table 2 summarizes the estimated growth temperatures based on these PGC values. The estimated Tmin, Topt, and Tmax based on the PGC of rrsA were 32.6 ± 16.7, 51.6 ± 11.8, and 59.7 ± 13.1°C, which are much higher than those calculated from the PGC of rrsB and rrsC. The offsets between the estimated growth temperatures based on PGC of rrsA and rrsBC was >10°C. These findings may indicate that harboring 16S rRNA genes with relatively high- and low-PGC values allows H. hispanica to maintain rapid growth over a wide temperature range.
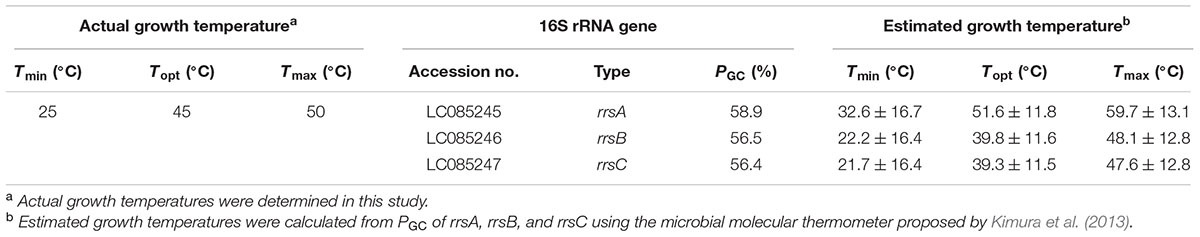
TABLE 2. Actual growth temperatures, 16S rRNA genes, and estimated growth temperatures of Haloarcula hispanica JCM8911.
Survey of Expression of 16S rRNA Genes of H. hispanica
To check the selectivity of the specific primers designed in this study, qPCR was performed with PCR products of rrsA, rrsB, and rrsC that were amplified from the clones in the 16S rRNA gene-clone library. The rrsA-specific primer set, rrsAf/rrsAr, provided the proper products from only diluted PCR products of rrsA as templates at annealing temperature of 58°C, whereas the rrsBC-specific primer set, rrsBCf/rrsBCr, provided the proper products from only diluted PCR products of rrsB and rrsC at annealing temperature of 58°C. These results indicated that these primer sets were sufficiently selective to detect and quantify rrsA and rrsBC, respectively.
Our survey of 16S RNA gene expression demonstrated that rrsA and rrsBC expression varied with temperatures. In particular, the expression ratio of rrsA to rrsBC (rrsA:rrsBC) increased with culture temperature (Figure 1). The ratios at 45 and 50°C exceeded 1.0, which were significantly greater than those in the 25–35°C range (P < 0.05 by Student’s t-test). On the other hand, the ratios at 25, 30, and 35°C were below 1.0. Especially, the ratios at 25 and 30°C were 0.56, which means that total expression of rrsB and rrsC was almost as twice as that of rrsA at the low temperatures. Our results suggest that transcription of high-PGC 16S rRNA gene rrsA and low-PGC 16S rRNA genes rrsBC may be regulated in response to culture temperature.
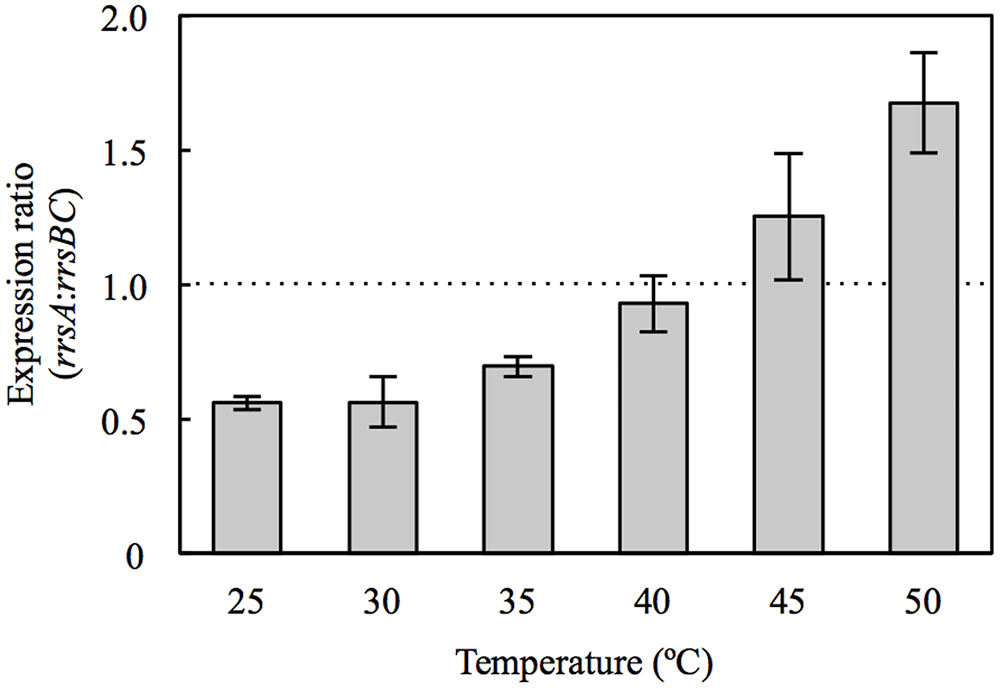
FIGURE 1. Expression ratio of rrsA to rrsBC in Haloarcula hispanica cultured at 25, 30, 35, 40, 45, and 50°C. Error bars denote standard deviation of mean values for triplicate measurements. The dashed line indicates rrsA:rrsBC ratio of 1.0.
Construction and Culture of rRNA Operon Double-Mutants
We constructed rRNA operon single-mutant strains (HA2, HB2, and HC2) that lack rrnA, rrnB, or rrnC, respectively (Supplementary Table 1). We further constructed rRNA operon double-mutant strains, namely HCB2 harboring only rrnA and HAB2 harboring only rrnC (Supplementary Table 1). Operon deletion was confirmed by PCR with specific primer sets and by electrophoresis of the PCR products (Supplementary Figure 4).
The wild-type H. hispanica was able to grow at temperature ranging from 25 to 50°C, with optimum growth at 45°C (Table 2). The fastest growth was 0.16 h-1 at the Topt of 45°C (Supplementary Figure 5). The growth characteristics of the wild-type strain at various temperatures were almost the same as those of the rRNA operon single-mutant strains (data not shown). In the culture experiments using the double-mutant strains, the wild-type grew faster than HCB2 and HAB2 at all tested temperatures (Figure 2). HCB2 and HAB2 were able to grow within the same temperature range (i.e., 25–50°C) as the wild-type strain (Supplementary Figure 5). The HCB2 that harbors only rrnA containing high-PGC rrsA grew optimally at 45°C. On the other hand, optimum growth of HAB2, which harbors only rrnC containing low-PGC rrsC, was slightly shifted to low temperature of 40°C.
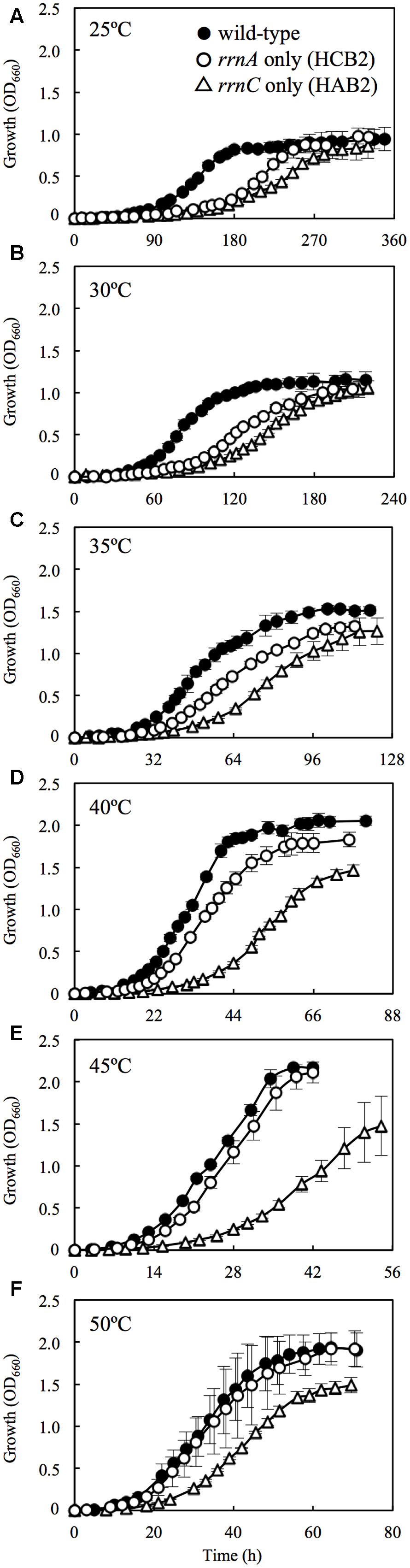
FIGURE 2. Growth curves for wild-type H. hispanica and the rRNA operon double-mutant strains at 25 (A), 30 (B), 35 (C), 40 (D), 45 (E), and 50°C (F). Error bars denote standard deviation of mean values for quadruplicate or quintuplicate measurements.
At 25°C, the wild-type H. hispanica grew significantly faster than HAB2 and HCB2 (Figure 2A). Condon et al. (1995) used Escherichia coli with multiple rRNA operons inactivated by antibiotic cassettes to demonstrate that the number of rRNA operons in the genome affects cell proliferation rate. Yano et al. (2013) found that the existence of multiple rRNA operons underlies the high growth rate of Bacillus subtilis. Another study reported that the copy number of rRNA operons on the genomes is correlated with growth rate of the prokaryotes under optimal culture condition (Vieira-Silva and Rocha, 2010; Roller et al., 2016). Our results support these studies and suggest that the number of rRNA operons influences cell proliferation rate at low temperatures close to actual Tmin (25°C).
At 30°C, HCB2 had slightly higher growth rate than HAB2 (Figure 2B), and this difference increased in cultures at 35 and 40°C (Figures 2C,D). At 45 and 50°C, the growth curves for HCB2 were almost identical with those of the wild-type strain (Figures 2E,F). These findings suggest that the rRNAs transcribed from rrnA, including the high-PGC 16S rRNA gene rrsA, result in more rapid growth of H. hispanica at high temperatures of 45 and 50°C. The expression survey in this study also showed that expression of rrsA was higher than those of rrsBC at 45 and 50°C (Figure 1), which also supports that rRNAs transcribed from rrnA including high-PGC 16S rRNA gene rrsA may be important for growth under the high temperature conditions.
Survival Strategy of Haloarcula Strains
Haloarcula hispanica has been identified in a number of solar salterns and desert salt lakes (e.g., Arahal et al., 1996; Pašić et al., 2005; Tapilatu et al., 2010; Mani et al., 2012). These hyper-saline habitats generally have large daily temperature fluctuations, i.e., temperature can vary by >10°C (Post, 1977; Wieland et al., 2005; Sima et al., 2013; Andrade et al., 2015). Because of this habitat feature, it is predicted that H. hispanica would express the rRNA operon with high-PGC 16S rRNA gene to grow faster in the daytime, when environmental temperatures rise to around Tmax (50°C). On the other hand, H. hispanica would express all of the three rRNA operons to grow in the nighttime and/or early morning, when environmental temperatures drop to around Tmin (25°C).
Fifteen Haloarcula strains have been isolated from hyper-saline environments worldwide (e.g., Juez et al., 1986; Ihara et al., 1997; Oren et al., 1999; Yang et al., 2007). Except for H. aidinensis, 14 Haloarcula strains so far examined harbor several different 16S rRNA gene sequences in the genome (Supplementary Table 3). Thus, intragenomic 16S rRNA gene heterogeneity seems to be common feature in the genus Haloarcula. We further confirmed that 9 of the 14 Haloarcula strains for which sequence was available show a >2.0% difference in PGC among the 16S rRNA genes. The estimated growth temperatures based on PGC values of the respective 16S rRNA genes suggested >10°C differences as well as H. hispanica (Supplementary Table 3). Additionally, previous study using a Haloarcula strain suggested that the sequences of putative promoter regions were obviously different among upstream regions of rRNA operons (Dennis et al., 1998; Dennis, 1999; López-López et al., 2007). These findings suggest that Haloarcula strains may regulate the expression of these 16S rRNA genes in response to culture temperature conditions, and this can be tested in future studies.
Conclusion
In this study, we determined the sequences and PGC values of 16S rRNA genes in the genome of the halophilic archaeon H. hispanica, and growth temperatures of H. hispanica were estimated based on the PGC values. The estimated growth temperatures of cells carrying the high-PGC 16S rRNA gene (rrsA) were approximately 10°C higher than those carrying the low-PGC 16S rRNA genes (rrsB and rrsC), suggesting that H. hispanica harbors different 16S rRNA genes of different PGC to maintain rapid growth in a wide range of temperatures.
We characterized the expression of rrsA and rrsBC of H. hispanica at different growth temperatures. We found that rrsA was expressed at significantly higher levels than rrsBC at higher temperatures such as 45 and 50°C. Our results indicate the importance of a high-PGC 16S rRNA gene at raised growth temperatures in the Haloarcula species. We further constructed rRNA operon double-mutant strains of H. hispanica. Culture experiments showed that the wild-type strain grew faster than the mutant strains at temperatures between 25 and 40°C. At 45 and 50°C, the double-mutant strain harboring only rrnA (including rrsA) grew much faster than the double-mutant strain harboring only rrnC (including rrsC), and the growth rate was similar to that of the wild-type strain. These findings suggest that the copy number of rRNA operons affects the growth rate of H. hispanica under low temperature conditions and that rRNAs transcribed from rrnA, which contains the high-PGC 16S RNA gene rrsA, function to promote rapid growth under high temperature conditions.
Author Contributions
YS and HK conceived this study. YS performed all the experiments and drafted the manuscript. TF helped YS to construct the mutant strains. All authors confirmed and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
This work was supported by Japan Society for the Promotion of Science KAKENHI grant (No. 23657016 and No. 25870295).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00482/full#supplementary-material
References
Acinas, S. G., Marcelino, L. A., Klepac-Ceraj, V., and Polz, M. F. (2004). Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J. Bacteriol. 186, 2629–2635. doi: 10.1128/JB.186.9.2629-2635.2004
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Andrade, K., Logemann, J., Heidelberg, K. B., Emerson, J. B., Comolli, L. R., Hug, L. A., et al. (2015). Metagenomic and lipid analyses reveal a diel cycle in a hypersaline microbial ecosystem. ISME J. 9, 2697–2711. doi: 10.1038/ismej.2015.66
Antón, J., Llobet-Brossa, E., Rodríguez-Valera, F., and Amann, R. (1999). Fluorescence in situ hybridization analysis of the prokaryotic community inhabiting crystallizer ponds. Environ. Microbiol. 1, 517–523. doi: 10.1046/j.1462-2920.1999.00065.x
Arahal, D. R., Dewhirst, F. E., Paster, B. J., Volcani, B. E., and Ventosa, A. (1996). Phylogenetic analyses of some extremely halophilic archaea isolated from Dead Sea water, determined on the basis of their 16S rRNA sequences. Appl. Environ. Microbiol. 62, 3779–3786.
Baliga, N. S., Bonneau, R., Facciotti, M. T., Pan, M., Glusman, G., Deutsch, E. W., et al. (2004). Genome sequence of Haloarcula marismortui: a halophilic archaeon from the Dead Sea. Genome Res. 14, 2221–2234. doi: 10.1101/gr.2700304
Boucher, Y., Douady, C. J., Sharma, A. K., Kamekura, M., and Doolittle, W. F. (2004). Intragenomic heterogeneity and intergenomic recombination among haloarchaeal rRNA genes. J. Bacteriol. 186, 3980–3990. doi: 10.1128/JB.186.12.3980-3990.2004
Burggraf, S., Stetter, K. O., Rouviere, P., and Woese, C. R. (1991). Methanopyrus kandleri: an archaeal methanogen unrelated to all other known methanogens. Syst. Appl. Microbiol. 14, 346–351. doi: 10.1016/S0723-2020(11)80308-5
Condon, C., Liveris, D., Squires, C., Schwartz, I., and Squires, C. L. (1995). rRNA operon multiplicity in Escherichia coli and the physiological implications of rrn inactivation. J. Bacteriol. 177, 4152–4156. doi: 10.1128/jb.177.14.4152-4156.1995
Cui, H.-L., Zhou, P.-J., Oren, A., and Liu, S.-J. (2009). Intraspecific polymorphism of 16S rRNA genes in two halophilic archaeal genera, Haloarcula and Halomicrobium. Extremophiles 13, 31–37. doi: 10.1007/s00792-008-0194-2
Dennis, P. P. (1999). “Expression of ribosomal RNA operons in halophilic archaea,” in Microbiology and Biogeochemistry of Hypersaline Environments, ed. A. Oren (Boca Raton, FL: CRC Press), 319–329.
Dennis, P. P., Ziesche, S., and Mylvaganam, S. (1998). Transcription analysis of two disparate rRNA operons in the halophilic archaeon Haloarcula marismortui. J. Bacteriol. 180, 4804–4813.
Ding, J.-Y., Chiang, P.-W., Hong, M.-J., Dyall-Smith, M., and Tang, S.-L. (2014). Complete genome sequence of the extremely halophilic archaeon Haloarcula hispanica strain N601. Genome Announc. 2:e00178-14. doi: 10.1128/genomeA.00178-14
Galtier, N., and Lobry, J. R. (1997). Relationships between genomic G+C content, RNA secondary structures, and optimal growth temperature in prokaryotes. J. Mol. Evol. 44, 632–636. doi: 10.1007/PL00006186
Hiraoka, S., Yang, C. C., and Iwasaki, W. (2016). Metagenomics and bioinformatics in microbial ecology: current status and beyond. Microbes Environ. 31, 204–212. doi: 10.1264/jsme2.ME16024
Ihara, K., Watanabe, S., and Tamura, T. (1997). Haloarcula argentinensis sp. nov. and Haloarcula mukohataei sp. nov., two new extremely halophilic archaea collected in Argentina. Int. J. Syst. Bacteriol. 47, 73–77. doi: 10.1099/00207713-47-1-73
Javor, B., Requadt, C., and Stoeckenius, W. (1982). Box-shaped halophilic bacteria. J. Bacteriol. 151, 1532–1542.
Juez, G., Rodríguez-Valera, F., Ventosa, A., and Kushner, D. J. (1986). Haloarcula hispanica spec. nov. and Haloferax gibbonsii spec, nov., two new species of extremely halophilic archaebacteria. Syst. Appl. Microbiol. 8, 75–79. doi: 10.1016/S0723-2020(86)80152-7
Khachane, A. N., Timmis, K. N., and Dos Santos, V. A. P. M. (2005). Uracil content of 16S rRNA of thermophilic and psychrophilic prokaryotes correlates inversely with their optimal growth temperatures. Nucleic Acids Res. 33, 4016–4022. doi: 10.1093/nar/gki714
Kimura, H., Ishibashi, J., Masuda, H., Kato, K., and Hanada, S. (2007). Selective phylogenetic analysis targeting 16S rRNA genes of hyperthermophilic archaea in the deep-subsurface hot biosphere. Appl. Environ. Microbiol. 73, 2110–2117. doi: 10.1128/AEM.02800-06
Kimura, H., Mori, K., Tashiro, T., Kato, K., Yamanaka, T., Ishibashi, J., et al. (2010). Culture-independent estimation of optimal and maximum growth temperatures of archaea in subsurface habitats based on the G+C content in 16S rRNA gene sequences. Geomicrobiol. J. 27, 114–122. doi: 10.1080/01490450903456699
Kimura, H., Mori, K., Yamanaka, T., and Ishibashi, J. (2013). Growth temperatures of archaeal communities can be estimated from the guanine-plus-cytosine contents of 16S rRNA gene fragments. Environ. Microbiol. Rep. 5, 468–474. doi: 10.1111/1758-2229.12035
Kimura, H., Sugihara, M., Kato, K., and Hanada, S. (2006). Selective phylogenetic analysis targeted at 16S rRNA genes of thermophiles and hyperthermophiles in deep-subsurface geothermal environments. Appl. Environ. Microbiol. 72, 21–27. doi: 10.1128/AEM.72.1.21-27.2006
Kuechler, S. M., Matsuura, Y., Dettner, K., and Kikuchi, Y. (2016). Phylogenetically diverse Burkholderia associated with midgut crypts of spurge bugs, Dicranocephalus spp. (Heteroptera: Stenocephalidae). Microbes Environ. 31, 145–153. doi: 10.1264/jsme2.ME16042
Liu, H., Wu, Z., Li, M., Zhang, F., Zheng, H., Han, J., et al. (2011). Complete genome sequence of Haloarcula hispanica, a model haloarchaeon for studying genetics, metabolism, and virus-host interaction. J. Bacteriol. 193, 6086–6087. doi: 10.1128/JB.05953-11
López-López, A., Benlloch, S., Bonfá, M., Rodríguez-Valera, F., and Mira, A. (2007). Intragenomic 16S rDNA divergence in Haloarcula marismortui is an adaptation to different temperatures. J. Mol. Evol. 65, 687–696. doi: 10.1007/s00239-007-9047-3
Mani, K., Salgaonkar, B. B., and Braganca, J. M. (2012). Culturable halophilic archaea at the initial and crystallization stages of salt production in a natural solar saltern of Goa, India. Aquat. Biosyst. 8, 15. doi: 10.1186/2046-9063-8-15
Matsushita, M., Ishikawa, S., Nagai, K., Hirata, Y., Ozawa, K., Mitsunobu, S., et al. (2016). Regional variation of CH4 and N2 production processes in the deep aquifers of an accretionary prism. Microbes Environ. 31, 329–338. doi: 10.1264/jsme2.ME16091
Mizuki, T., Kamekura, M., DasSarma, S., Fukushima, T., Usami, R., Yoshida, Y., et al. (2004). Ureases of extreme halophiles of the genus Haloarcula with a unique structure of gene cluster. Biosci. Biotechnol. Biochem. 68, 397–406. doi: 10.1271/bbb.68.397
Namwong, S., Tanasupawat, S., Kudo, T., and Itoh, T. (2011). Haloarcula salaria sp. nov. and Haloarcula tradensis sp. nov., isolated from salt in Thai fish sauce. Int. J. Syst. Evol. Microbiol. 61, 231–236. doi: 10.1099/ijs.0.021790-0
Oren, A., Ventosa, A., Gutiérrez, M. C., and Kamekura, M. (1999). Haloarcula quadrata sp. nov., a square, motile archaeon isolated from a brine pool in Sinai (Egypt). Int. J. Syst. Bacteriol. 49, 1149–1155. doi: 10.1099/00207713-49-3-1149
Pašić, L., Bartual, S. G., Ulrih, N. P., Grabnar, M., and Velikonja, B. H. (2005). Diversity of halophilic archaea in the crystallizers of an Adriatic solar saltern. FEMS Microbiol. Ecol. 54, 491–498. doi: 10.1016/j.femsec.2005.06.004
Pei, A. Y., Oberdorf, W. E., Nossa, C. W., Agarwal, A., Chokshi, P., Gerz, E. A., et al. (2010). Diversity of 16S rRNA genes within individual prokaryotic genomes. Appl. Environ. Microbiol. 76, 3886–3897. doi: 10.1128/AEM.02953-09
Post, F. J. (1977). The microbial ecology of the Great Salt Lake. Microb. Ecol. 3, 143–165. doi: 10.1007/BF02010403
Roller, B. R. K., Stoddard, S. F., and Schmidt, T. M. (2016). Exploiting rRNA operon copy number to investigate bacterial reproductive strategies. Nat. Microbiol. 1:16160. doi: 10.1038/nmicrobiol.2016.160
Sima, S., Ahmadalipour, A., and Tajrishy, M. (2013). Mapping surface temperature in a hyper-saline lake and investigating the effect of temperature distribution on the lake evaporation. Remote Sens. Environ. 136, 374–385. doi: 10.1016/j.rse.2013.05.014
Staley, J. T. (2002). “A microbiological perspective of biodiversity,” in Biodiversity of Microbial Life: Foundation of Earth’s Biosphere, eds J. T. Staley and A.-L. Reysenbach (New York, NY: Wiley-Liss), 3–23.
Sun, D.-L., Jiang, X., Wu, Q. L., and Zhou, N.-Y. (2013). Intragenomic heterogeneity of 16S rRNA genes causes overestimation of prokaryotic diversity. Appl. Environ. Microbiol. 79, 5962–5969. doi: 10.1128/AEM.01282-13
Tapilatu, Y. H., Grossi, V., Acquaviva, M., Militon, C., Bertrand, J. C., and Cuny, P. (2010). Isolation of hydrocarbon-degrading extremely halophilic archaea from an uncontaminated hypersaline pond (Camargue, France). Extremophiles 14, 225–231. doi: 10.1007/s00792-010-0301-z
Tchinda, R. A. M., Boudjeko, T., Simao-Beaunoir, A.-M., Lerat, S., Tsala,É, Monga, E., et al. (2016). Morphological, physiological, and taxonomic characterization of actinobacterial isolates living as endophytes of cacao pods and cacao seeds. Microbes Environ. 31, 56–62. doi: 10.1264/jsme2.ME15146
Torreblanca, M., Rodríguez-Valera, F., Juez, G., Ventosa, A., Kamekura, M., and Kates, M. (1986). Classification of non-alkaliphilic halobacteria based on numerical taxonomy and polar lipid composition, and description of Haloarcula gen. nov. and Haloferax gen. nov. Syst. Appl. Microbiol. 8, 89–99. doi: 10.1016/S0723-2020(86)80155-2
Tu, D., Blaha, G., Moore, P. B., and Steitz, T. A. (2005). Gene replacement in Haloarcula marismortui: construction of a strain with two of its three chromosomal rRNA operons deleted. Extremophiles 9, 427–435. doi: 10.1007/s00792-005-0459-y
Větrovský, T., and Baldrian, P. (2013). The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PLoS ONE 8:e57923. doi: 10.1371/journal.pone.0057923
Vieira-Silva, S., and Rocha, E. P. C. (2010). The systemic imprint of growth and its uses in ecological (meta)genomics. PLoS Genet. 6:e1000808. doi: 10.1371/journal.pgen.1000808
Wieland, A., Zopfi, J., Benthien, M., and Kühl, M. (2005). Biogeochemistry of an iron-rich hypersaline microbial mat (Camargue, France). Microb. Ecol. 49, 34–49. doi: 10.1007/s00248-003-2033-4
Yang, Y., Cui, H.-L., Zhou, P.-J., and Liu, S.-J. (2007). Haloarcula amylolytica sp. nov., an extremely halophilic archaeon isolated from Aibi salt lake in Xin-Jiang, China. Int. J. Syst. Evol. Microbiol. 57, 103–106. doi: 10.1099/ijs.0.64647-0
Yano, K., Wada, T., Suzuki, S., Tagami, K., Matsumoto, T., Shiwa, Y., et al. (2013). Multiple rRNA operons are essential for efficient cell growth and sporulation as well as outgrowth in Bacillus subtilis. Microbiology 159, 2225–2236. doi: 10.1099/mic.0.067025-0
Keywords: 16S rRNA genes, guanine-plus-cytosine content, Haloarcula, temperature, intragenomic heterogeneity
Citation: Sato Y, Fujiwara T and Kimura H (2017) Expression and Function of Different Guanine-Plus-Cytosine Content 16S rRNA Genes in Haloarcula hispanica at Different Temperatures. Front. Microbiol. 8:482. doi: 10.3389/fmicb.2017.00482
Received: 12 December 2016; Accepted: 08 March 2017;
Published: 28 March 2017.
Edited by:
Peter Dunfield, University of Calgary, CanadaReviewed by:
Camilla Lothe Nesbø, University of Alberta, CanadaMike L. Dyall-Smith, University of Melbourne, Australia
Copyright © 2017 Sato, Fujiwara and Kimura. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hiroyuki Kimura, kimura.hiroyuki@shizuoka.ac.jp
 Yu Sato
Yu Sato Taketomo Fujiwara
Taketomo Fujiwara Hiroyuki Kimura1
Hiroyuki Kimura1