- Department of Geological Sciences, University of Texas at Austin, Austin, TX, USA
This study tests the hypothesis that surface composition influences microbial community structure and growth of biofilms. We used laboratory biofilm reactors (inoculated with a diverse subsurface community) to explore the phylogenetic and taxonomic variability in microbial communities as a function of surface type (carbonate, silicate, aluminosilicate), media pH, and carbon and phosphate availability. Using high-throughput pyrosequencing, we found that surface type significantly controlled ~70–90% of the variance in phylogenetic diversity regardless of environmental pressures. Consistent patterns also emerged in the taxonomy of specific guilds (sulfur-oxidizers/reducers, Gram-positives, acidophiles) due to variations in media chemistry. Media phosphate availability was a key property associated with variation in phylogeny and taxonomy of whole reactors and was negatively correlated with biofilm accumulation and α-diversity (species richness and evenness). However, mineral-bound phosphate limitations were correlated with less biofilm. Carbon added to the media was correlated with a significant increase in biofilm accumulation and overall α-diversity. Additionally, planktonic communities were phylogenetically distant from those in biofilms. All treatments harbored structurally (taxonomically and phylogenetically) distinct microbial communities. Selective advantages within each treatment encouraged growth and revealed the presence of hundreds of additional operational taxonomix units (OTU), representing distinct consortiums of microorganisms. Ultimately, these results provide evidence that mineral/rock composition significantly influences microbial community structure, diversity, membership, phylogenetic variability, and biofilm growth in subsurface communities.
Introduction
It is estimated that up to 99.9% of microbial biomass in a subsurface environment is attached to surfaces as biofilms (Madigan et al., 2009). Microbial attachment to natural surfaces is a complex and dynamic process involving interaction between the organism, the surface, and the aqueous phase. The subsurface is a complex and heterogeneous environment where varying mineralogy results in different surface chemistries and microscale spatial heterogeneity that contribute to growth, present challenges, or influence community membership. The purpose of this study is to characterize the mineralogical contribution to microbial diversity, investigating the contribution of different natural surface types under a range of environmental conditions.
Recent studies suggest a link between mineral composition and colonization by specific microbial communities. Factors controlling dynamic diversity, growth, and specific survival strategies include and have greater implications for adjacent studies of physical properties (hydrodynamics) of bulk fluid (Kugaprasatham et al., 1992), physicochemical nature of surfaces (Dalton et al., 1994; Rogers et al., 1998; Rogers and Bennett, 2004; Carson et al., 2009; Sylvan et al., 2012), microbial community composition (Lawrence et al., 1991), and nutrient cycling and availability (primarily carbon, phosphorous, and nitrogen; Ohashi et al., 1995; Huang et al., 1998; Rogers et al., 2001). pH is often identified as a key factor controlling microbial community composition (Fierer and Jackson, 2006; Lauber et al., 2009; Chu et al., 2010; Siciliano et al., 2014; Winsley et al., 2014). Biofilm and extracellular polysaccharides (EPS) production are both influenced by the nutrient content of the growth medium with respect to available carbon, or phosphate limitations (Ellwood et al., 1982; Matin et al., 1989; Wrangstadh et al., 1990). However, few studies examine complex natural communities on natural surfaces. In oligotrophic environments, such as those in the subsurface, microorganisms are likely highly reliant on minerals (“mineraltrophic”) to support various biogeochemical processes (Stevens, 1997; Anderson, 2001; Chapelle et al., 2002; Edwards et al., 2005, 2012).
Previously, we found that, in oligotrophic conditions, specific guilds showed an affinity for specific mineral types according to their metabolic requirements and environmental tolerances (Jones and Bennett, 2014). Sequences from similar surface types (e.g., carbonates, silicates, aluminosilicates) were more taxonomically similar. Specifically, given a choice of surface types, neutrophilic, but acid-producing sulfur-oxidizers (SOB) were dominant on highly-buffering carbonates, acidophiles on non-buffering silicates, Gram-positives on silicates, and aluminum tolerant bacteria on aluminosilicates (Jones and Bennett, 2014). Additionally, mineral-phosphate availability was correlated with biofilm accumulation (Jones and Bennett, 2014). Those experiments were designed to favor growth of autotrophic SOB and to mimic the nutrient-limited environment found within Lower Kane Cave (WY, USA) where sulfidic water serves as the metabolic backbone for a diverse microbial community (Egemeier, 1981; Engel et al., 2004). However, natural environments are subjected to variable geochemical conditions that may influence microbial surface colonization. We hypothesize that surface type is an important variable influencing biofilm community structure (growth, taxonomic, and phylogenetic variability) even under distinct geochemical conditions.
For this study we utilize high-throughput 454-pyrosequencing (Margulies et al., 2005) of bacterial 16S rRNA sequences to examine the response of community structure and diversity to environmental stimuli (pH variability, carbon and phosphate limitations/amendments) as biofilms develop on different mineral surfaces within flow-through biofilm reactors. This allowed us to assess the major biogeochemical reactions in a controlled setting and further constrain the key parameters affecting microbial diversity, not by replicating the complexity of the natural system, but by fine tuning parameters in order to evaluate their influence on microbial communities. We use phylogenetic distance measures (UniFrac), to account for the relationships among populations attached to the various surfaces (Lozupone and Knight, 2005). We use permutational multivariate analysis of variance (PERMANOVA) to evaluate mineralogical and environmental influence on microbial community structure (McArdle and Anderson, 2001). Additionally, we test our hypothesis on the 16S rRNA sequences and growth data from Jones and Bennett (2014) using a more robust methodology. Taxonomic variations among specific guilds (SOB, SRB, Gram-positives, Acidophiles), are identified to interpret the ecological role of the detected taxa.
Materials and Methods
Flow Through Biofilm Reactor
We used a modified CDC biofilm reactor (Biosurface Technologies, Bozeman, MT, USA; see http://biofilms.biz/products/biofilm-reactors): a 1-liter glass vessel with a ported polyethylene top that supports 8 polypropylene rods, each holding up to three coupons (12.7 mm OD disks ~3 mm thick). The reactor was operated as a continuous-flow stirred reactor at 1.5 ml/min liquid medium flow. Consistent shear and mixing at all positions within the reactors was maintained using a stir vane rotated by a magnetic stir plate.
The mixed environmental inoculum was collected in the field at Lower Kane Cave (LKC) (WY, USA) in sterile falcon tubes and was identical to that used in Jones and Bennett (2014). The inoculum is a consortium composed primarily of autotrophic sulfur-oxidizing members of lineages Gammaproteobacteria (34.7%) of the genus Thiothrix, and Epsilonproteobacteria (62.4%) of the genus Sulfurovum (Engel et al., 2003, 2004; Jones and Bennett, 2014), but with many other Bacterial lineages at lower abundance. Approximately 15 ml of the raw mat (inoculum) was added to the sterilized CDC biofilm reactor for each experiment.
Synthetic cave water was prepared by equilibrating DI-H2O water with finely powdered Iceland spar calcite to equilibrium. The solution was filtered to 0.2 μm and 0.1 g MgSO4 and 0.25 g NH4Cl were added per liter and autoclaved at 121°C for 45 min before adding 2 and 5 ml/L of filter-sterilized trace metal solution and Wolfe's Vitamin solution, respectively (Burlage, 1998). The reduced sulfur electron donor was S2 prepared from a stock filter-sterilized 1 M solution of Na2S2O3 mixed in-line via a syringe pump to a final concentration of 0.83 mM.
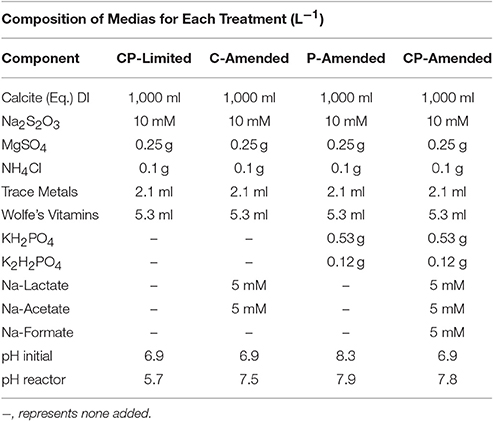
Amendments (P, C, or both) were then added to this basic liquid media or pH adjusted to examine the influence of environmental conditions (Table 1). Specifically, the carbon/phosphorus-limited (CP-Limited) media used in Jones and Bennett (2014) was this basic liquid media with sterile 0.1 N H2SO4 added to achieve a final pH of 6.9 (Jones and Bennett, 2014). The C-Amended medium was prepared by amending the basic medium with 5 mM Na-Acetate, 5 mM Na-Lactate and filter sterilized 0.1 N H2SO4 added to a final pH of 6.9. The P-Amended media was prepared with 0.53 g/L KH2PO4 and 0.12 g/L K2H2PO4 and filter sterilized NaOH added to a final pH of 8.3. The no limitation medium (CP-Amended) was amended with 5 mM Na-Acetate, 5 mM Na-Lactate, 5 mM Na-Formate, 0.53 g/L KH2PO4, 0.12 g/L K2H2PO4, and 0.1 N H2SO4 added to achieve a final pH of 6.9 (Table 1).
Surface Substrata Preparation
Mineral/rock substrata were selected to represent the lithology of a variety of geologic environments [carbonates, silicates, aluminosilicates, planktonic (not attached)] (Table 2). Additionally, some advantages and disadvantages to microorganisms equipped to exploit (or defend against) them are described in Table 2. Specimens of calcite, microcline, albite, basalt, and quartz were obtained from Ward's Natural Science Establishment Incorporated. These materials have been previously characterized (Bennett et al., 2001; Jones and Bennett, 2014). Unaltered Mississippian-age Upper Madison Limestone, Upper Madison Dolostone and the contained chert, were collected from an outcrop near Lower Kane Cave. The Madison Limestone is nearly pure calcite (microsparite) with a minor quartz component, and the Madison Dolostone is nearly pure dolomite also with a minor quartz component (Plummer et al., 1990). Mineral/rock coupons were prepared using previously published methods (Jones and Bennett, 2014).
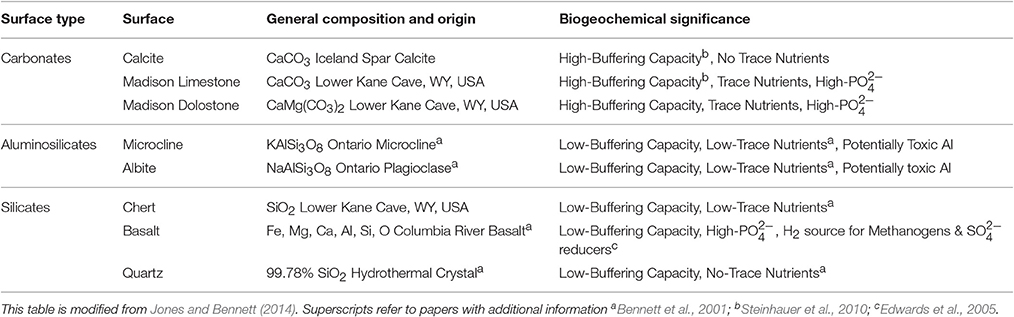
Table 2. Surface types, general compositions, and biogeochemical significance of the rocks/minerals used in these biofilm reactor experimental treatments.
Biomass Measurement and Extraction
To measure biomass accumulation after 3 weeks, triplicate mineral coupons were weighed (wet, biofilm attached), dried overnight at 104°C, weighed again (dry, biofilm attached), processed by 3 × 5-min cycles of alternating sonication and vortexing in a calcite equilibrated (to prevent dissolution) 2% tween 20 solution to remove biomass, dried overnight again, and weighed again (dry, biofilm removed). The dry weight of biomass accumulated is the difference between the final dry weight with biomass and the dry weight after processing.
Using these methods, biofilm growth curves were constructed in order to determine the standard duration (3-weeks) of each experiment. Curves were constructed for both pure and mixed culture treatments under CP-Limited, C-Amended, and P-Amended conditions. For these experiments, limestone was the sole surface type occupying all 24 coupon spaces. Two limestone coupons (chosen randomly) were sacrificed at 48-h intervals and biomass was measured according to the method described above. The resulting curves are shown in Supplementary Figure 1.
For DNA extraction, biomass was aseptically isolated from mineral coupons in 1 mM EDTA and 0.9X phosphate-buffered saline (PBS with physical disruption by freeze-thaw (3 times, −80°–65°C) cycles followed by alternating sonication and vortexing (3 × 5-min) (Jones and Bennett, 2014). The biomass was isolated from solution by centrifugation at 5000 rpm for 10 min, and the supernatant decanted. DNA extraction from biomass was conducted using an Ultraclean Microbial DNA Isolation Kit (MoBio Laboratories, Inc.; Catalog # 12224-50). DNA samples were quantified and qualified using a Nanodrop spectrophotometer (Nyxor Biotech, Paris, France). Bacterial tag-encoded FLX-titanium amplicon pyrosequencing (bTEFAP) was used to evaluate the bacterial populations removed from the mineral surfaces at MR DNA Lab (http://www.mrdnalab.com, Shallowater, TX, USA). The bTEFAP procedures are based on Research and Testing Laboratory protocols http://www.researchandtesting.com and are previously described (Dowd et al., 2008). Briefly, the 16S universal Eubacterial primers 27F (5′-AGRGTTTGATCMTGGCTCAG-3′) and 519R (5′-GTNTTACNGCGGCKGCTG-3′) were used to amplify the v1-v3 region of 16S rRNA genes using 30 cycles of PCR. HotStarTaq Plus Master Mix Kit (Qiagen) was used for PCR under the following conditions: 94°C for 3 min, followed by 28 cycles of 94°C for 30 s; 53°C for 40 s and 72°C for 1 min after which a final elongation step at 72°C for 5 min was performed. After PCR, all amplicon products from the different samples were mixed in equal volumes and purified using Agencourt Ampure Beads (Agencourt Bioscience Corporation, Beverly, Ma). Adaptors and barcodes for 454 pyrosequencing were ligated, and sequencing on a Roche 454 GS-FLX Titanium™ (454 Life Sciences, Branford, CT, USA).
Sequences were quality screened prior to clustering into operational taxonomic units (OTUs) using the open source software package QIIME version 1.9 (http://qiime.sourceforge.net; Caporaso et al., 2010). We removed from further analysis: sequences < 200 or >550 bp, sequences with ambiguous base calls (>6 bp), sequences with homopolymer runs (>6 bp), low quality scores (<25), sequences with primer mismatches, and barcode errors (>1 bp). Additionally, noisy sequences were discarded using the “denoise_wrapper” script (Reeder and Knight, 2010). Chimeric sequences were removed using ChimeraSlayer with the QIIME default settings after OTU-picking and taxonomic assignment. The uclust method was used to pick de novo OTUs at 3% (genus level) divergences. Representative sequences for each OTU were then aligned with PyNAST and taxonomy was assigned with the uclust consensus taxonomy assigner using the greengenes_13_8 reference database. Potential contaminants and OTUs with < 4 members were then filtered from the resulting OTU table. Assigned taxa were compared with extant taxa described in the literature, at the genus level, to infer the putative ecological role of these microorganisms.
All amplicons obtained from this study were submitted to the NCBI Sequence Read Archive (SRA) under the Bioproject PRJNA323607, with the BioSample accession numbers SAMN05181927- SAMN05181927 (http://www.ncbi.nlm.nih.gov). Amplicon data and SRA accesion numbers for each sample are included in Supplemental Table 2.
Biodiversity Metrics and Statistical Analysis
Community diversity was evaluated using both standard statistical analysis on OTUs (e.g., Species Richness, Shannon-Wiener, Simpson's) and phylogenetic analysis (Bohannan and Hughes, 2003). α and β-diversity values were calculated on the resulting datasets using QIIME with selective manual calculations for validation (Hill et al., 2003; Caporaso et al., 2010). Due to the different number of sequences among samples, the data was normalized for diversity analysis using rarefaction curves. Rarefaction curves of the observed richness were calculated in QIIME using 100,000-fold resampling without replacement (Supplementary Figure 2). Rarefactions were performed for each treatment independently at the maximum subsampling depth to include all samples within a treatment. Estimates of α-diversity were based on evenly rarefied OTU abundance matrices and included observed species richness (S), the reciprocal of the Simpson dominance index (D), Shannon-Weiner index (H'), and species evenness (E) (Hill et al., 2003). Sampling effort was estimated using Good's coverage (Good, 1953). The reciprocal Simpson dominance index (D) gives more weight to dominant OTUs by expressing the likelihood that two individuals, chosen at random, will belong to different OTUs. The effect of geochemical (pHin, pHout, C-P Availability) and mineralogical (mineral geochemistry, P-Availability, Buffering Capacity) variations on microbial community diversity was assessed at rarefication depths according to the smallest sample within each subset.
Beta diversity (global diversity) both between communities on multiple surfaces and between treatments was assessed using weighted UniFrac (phylogenetic) distance matrices at sequence divergences of 3% (Lozupone et al., 2011) between samples were calculated after rarefying all samples. The effect of geochemistry on microbial/mineral associations was depicted using UniFrac distance based 3D Principal Coordinate Analysis (PCoA) plots (Borg and Groenen, 2005), and Unweighted Pair Group Method with Arithmetic Mean (UPGMA) trees (Felsenstein, 2004).
To assess the statistical significances in microbial community dissimilarity between particular groups, the UniFrac distance matrix generated in QIIME was analyzed with R statistical software (R foundation for Statistical Computing, Vienna, Austria). Statistical significance was considered at P < 0.05. Alpha diversity metrics were compared within and between treatment types using Wilcoxon signed rank test. Testing for the presence of a significant effect of sample type on beta diversity metrics was done using permutational multivariate analysis of variance (PERMANOVA) and the “Adonis” function from the R package “vegan,” which partitions the distance matrix among sources of variation, fits linear models to distance matrices and uses a permutation test (105) with pseudo-F ratios to obtain P-values (McArdle and Anderson, 2001). Data sets for the effect of phosphate and carbon on microbial biomass were analyzed by analysis of variance (ANOVA) with significance at P < 0.05.
Results
Biomass Abundance
Biomass accumulation on each surfaces was measured (in triplicate) after 3-weeks for each treatment. Total dry mass of biofilm on all minerals for the CP-Limited from Jones and Bennett (2014), C-Amended, P-Amended, and CP-Amended treatments was 50.5, 110.1, 12.8, and 25.4 mg·cm−2, respectively (Figure 1). ANOVA analysis revealed that the addition of phosphate to the media decreased biofilm biomass (P < 0.002), while the addition of available carbon significantly increased biofilm biomass (P < 0.04). In all treatments, P-bearing surfaces (limestone, dolostone, and basalt) had significantly higher biomass (Figure 1). In the CP-Limited treatment, high-P surfaces accumulated up to 40X that of low-P surfaces. In the C-Amended treatment was up to 10X higher (Supplementary Table 1). The P- and CP-Amended treatments had the lowest biofilm accumulations on every surface, but even here the high-P minerals accumulated ~3–60X higher biomass than the low-P surfaces. Although this is the largest relative difference, the actual variation in total biomass (SD ~2.3 mg·cm−2) was lowest in the P-Amended treatment as most of the biomass was planktonic (Supplementary Table 1).
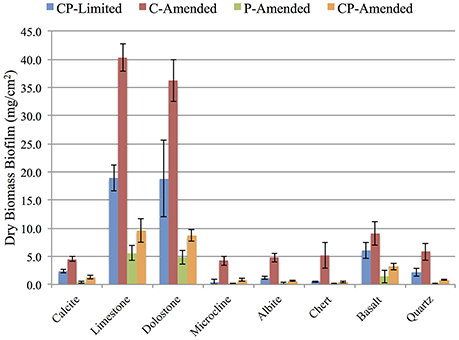
Figure 1. Dry weight (mg/cm2) of biomass accumulation on surfaces for each reactor treatment. Error bars denote standard deviation, n = 3. See Supplementary Table 1 for values.
Effect of Treatment Conditions and Surface Type on Bacterial Diversity
There were 578,969 total raw sequences obtained from all samples in the four treatments. Of those, a total of 209,973 (CP-Limited—27,173 total with an average of 3397 ± 3133 per sample, C-Amended—66,383 total with an average of 7,376 ± 5,662 per sample, P-Amended—34,847 total with an average of 4205 ± 934 per sample, CP-Amended—78,570 total with and average of 8,730 ± 2,900 per sample) bacterial 16S high-quality sequences with an average read length of 440 bp were obtained for the 35 samples. Rarefaction curves for richness of 33 (except CP-Limited microcline and albite) of the 35 samples approach a plateau at their respective maximum sampling depth, indicating an adequate sampling procedure (Supplementary Figure 2). Additionally, the overall average Good's coverage is 98.4 ± 0.9% including CP-Limited microcline and albite with Good's coverage values of 95.1 and 95.7%, respectively, indicating an adequate sampling procedure (Supplementary Table 2). Changes in diversity due to treatment conditions and surface type were evaluated.
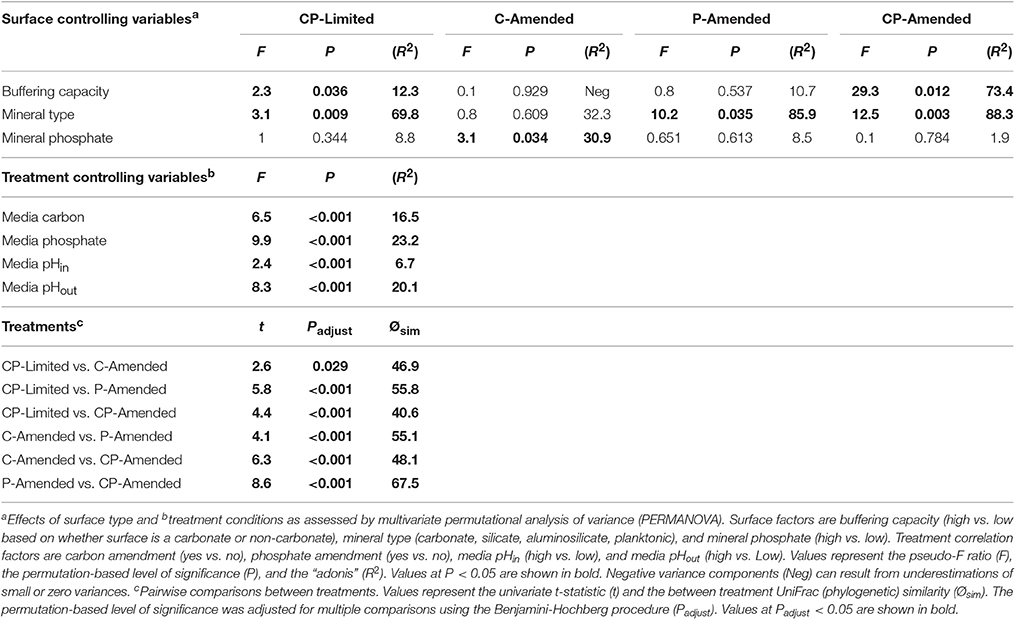
The bacterial communities between treatments formed phylogenetically distinct clusters in ordination space (Figure 2). UniFrac (phylogenetic) differences between treatment communities were significantly distinct from each other (PERMANOVA P < 0.001, R2 = 45.6%,) with overall similarities between 40.6 and 67.5% (Figure 2, Table 3). Each of the treatment variables (carbon, phosphate, pH) contributed significantly (P < 0.001) to phylogenetic differences between reactors (Table 3). Phosphate was the most important treatment controlling variable (R2 = 23.2%), but carbon (R2 = 16.5%), and media pH buffering (R2 = 20.1%) were also significant (P < 0.001; Table 3).
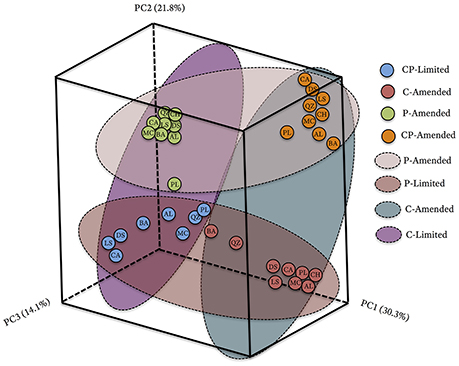
Figure 2. Principal coordinate analysis (PCoA) plot based on the relative abundances and phylogenetic diversity of 16S rRNA gene sequences using a UniFrac weighted distance matrix, colored according to reactor treatment conditions and labeled according to solid substrate type; blue, CP-Limited; green, P-Amended; orange, CP-Amended; red, C-Amended. Percentage of the diversity distribution explained by each axis is indicated in the figure. The colored ellipses encircle variations in reactor treatment conditions.
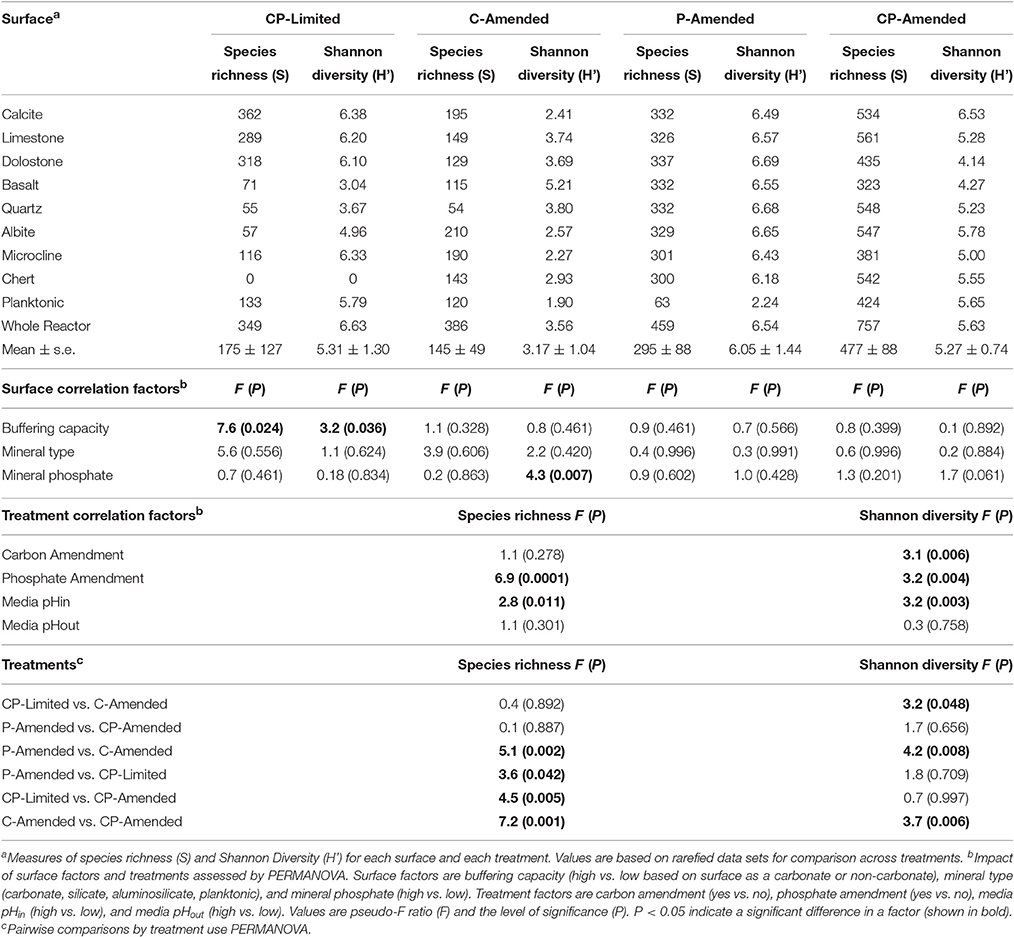
Despite the effects of treatment, PERMANOVA revealed significant effects (P < 0.05) of surface type (carbonate, silicate, aluminosilicate, planktonic) on diversity within each reactor (Table 3). Trees constructed from UPGMA clustering of the UniFrac distance matrices serve to visualize differences in communities between mineral types (Figure 3). We applied these phylogenetic and statistical analysis to the oligotrophic treatment (CP-Limited) data from Jones and Bennett (2014). Here we confirmed that surface type (R2 = 69.8%, P = 0.009) and (R2 = 12.3%, P = 0.036) accounted for a majority of the variability in bacterial communities, while mineral phosphate (significant for growth) had no significant effect on community variability (Table 3). In both P-Amended reactors, surface type controlled >85% (P < 0.05) of the variance in phylogenetic diversity (P < 0.05) despite an overall decrease in phylogenetic variability between surface communities (Figure 3, Table 3). For the C-Amended treatment, mineral phosphate is the only statistically significant controlling variable (R2 = 30.9%, P = 0.034), but some clustering by surface type is apparent (Figure 3B). The C-Amended treatment had a planktonic community that was closely related to that of the surface communities (Figure 3). It should be noted that removal of the planktonic sample from PERMANOVA analysis makes surface type statistically significant (Table 3, Figure 3). Simply put, bacterial communities on similar surface types were statistically more phylogenetically similar for a given treatment (Figure 3).
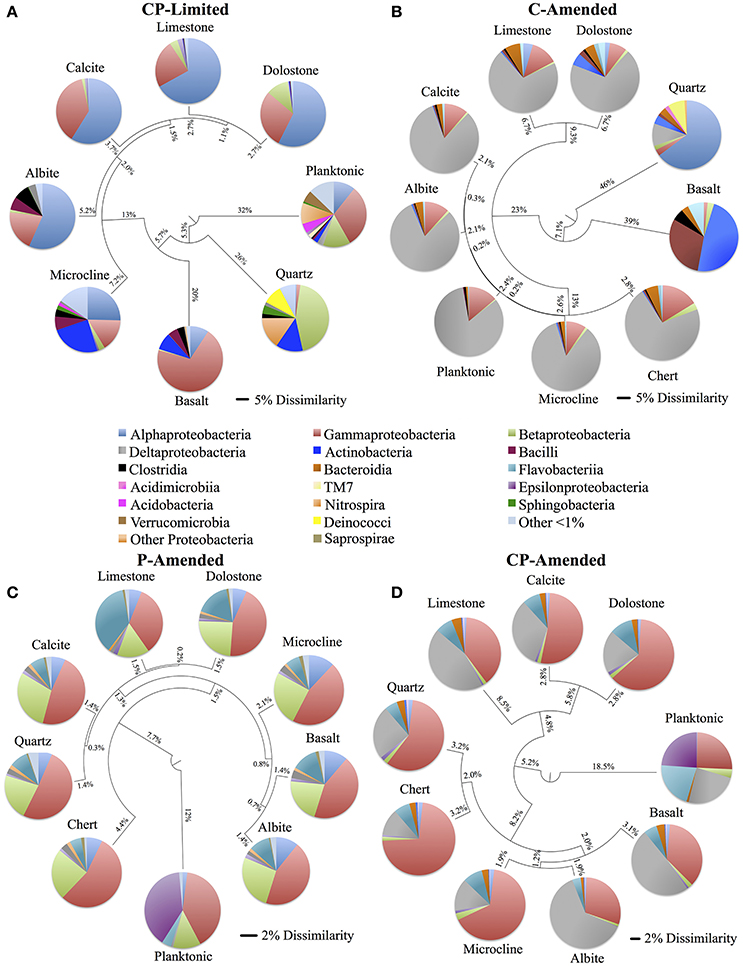
Figure 3. Unweighted pair group method with arithmetic mean (UPGMA) trees constructed using weighted UniFrac (phylogenetic) distance matrix constructed from 16S rRNA sequences clustered at 97% similarity for each of the four reactor treatments (CP-Limited, P-Amended, C-Amended, CP-Amended). The trees display the phylogenetic overlap in bacterial communities colonizing various solid surfaces within each reactor treatment. The key (center) contains the class level taxonomy associated with each mineral surface (see Supplementary Tables 3–6 for proportional abundances). The scale bars for the CP-Limited and C-Amended treatments represents 0.05 (5%) dissimilarity in 16S rRNA sequences isolated from each surface and the scale bars for the P-Amended and CP-Amended treatments 0.02 (2%) dissimilarity. Note that although the taxonomy of the CP-Limited treatment is the same as Jones and Bennett (2014), the UPGMA tree here is based on UniFrac distances, while the tree in Jones and Bennett (2014; Figure 4) used the OTU based Sorenson similarity index.
Additionally, all of the treatment variables contributed significantly (P < 0.01, for all treatments) to the overall Shannon diversity of each treatment (Table 4). The Shannon diversity was significantly higher in C-limited and P-amended treatments, but species richness and number of unique OTUs were both significantly lower in both P-limited treatments (P = 0.0001; Table 4). Within separate treatments, mineral-buffering capacity was correlated with higher species richness (P = 0.024) and Shannon diversity (P = 0.036) in the CP-Limited treatment. In the C-Amended treatment, Shannon diversity was significantly higher on high-P surfaces (P = 0.007, Table 4). Within both the P-Amended and CP-Amended treatments, differences in alpha-diversity values were statistically insignificant (Table 4).
Taxonomic Composition and Condition Sensitive Taxa
Upon taxonomy assignment, OTUs were affiliated with 280 classified bacterial genera. Of those, 73 genera had relative abundances of more than 1%. These microbial communities include 20 bacterial classes: Alphaproteobacteria, Betaproteobacteria, Deltaproteobacteria, Gammaproteobacteria, Epsilonproteobacteria, Acidobacteria, Acidomicrobiia, Actinobacteria, Bacilli, Bacteroidia, Clostridia, Coriobacteriia, Cytophagia, Deinococci, Flavobacteria, Nitrospira, Opitutae, Saprospirae, Sphingobacteria, and TM7 (Supplementary Tables 3–6). Proteobacteria was the most abundant Phylum across treatments. At the genus level there is very little overlap between overall experiment communities and no taxa common to all 4 experimental treatments (Figure 4).
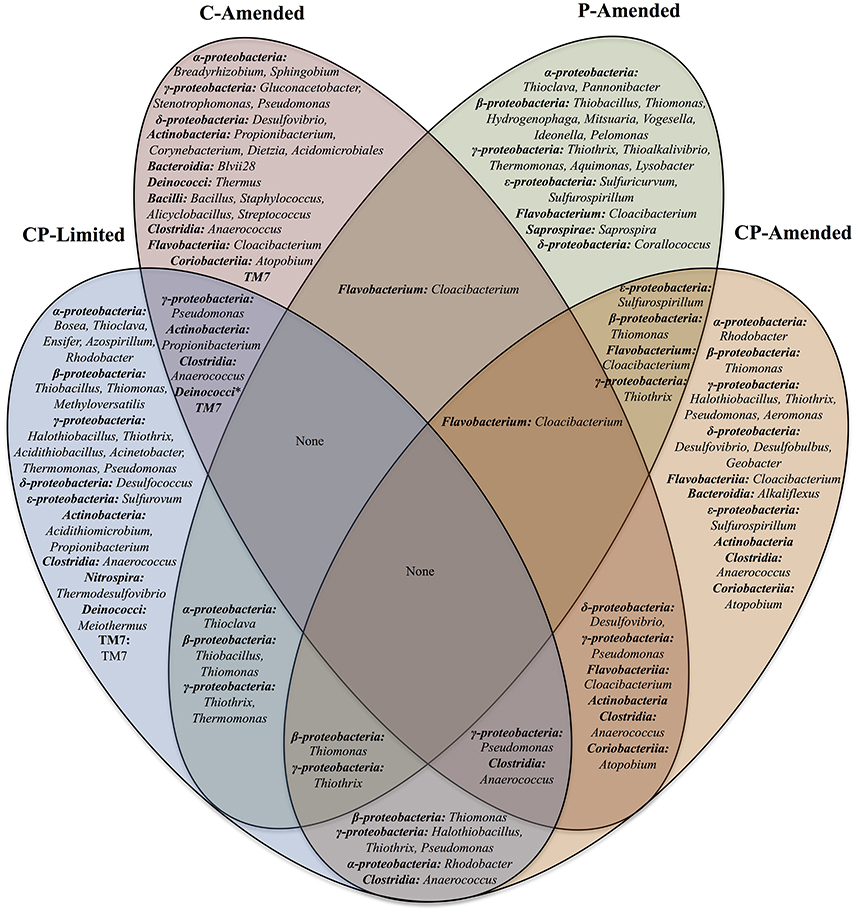
Figure 4. Venn diagram of the shared bacterial genera found in the four reactor treatments (CP-Limited, P-Amended, C-Amended, CP-Amended). Community membership overlaps between the reactors are indicated by overlaps in the diagram.
In the CP-Limited treatment distinct lineages were constrained by mineralogy and mineral buffering capacity (Jones and Bennett, 2014). Briefly, neutrophilic autotrophic sulfur-oxidizing members of the lineages Gammaproteobacteria, Betaproteobacteria, and Alphaproteobacteria, were detected on highly buffering carbonates, Gram-positive Actinobacter, Bacilli, and Clostridia were detected almost exclusively on silicates, and representatives of acidophilic sulfur-oxidizing genera Acidithiomicrobium, Thermodesulfovibrio, and Acidithiobacillus detected exclusively on non-buffering quartz (Supplementary Table 3; Jones and Bennett, 2014). Similarly, samples from the P-Amended and CP-Amended treatments were dominated by autotrophic sulfur-oxidizing members of these lineages. However, these representatives were distributed on all surfaces within these treatments (Supplementary Table 3). P-Amended treatment samples hosted Gammaproteobacteria (34.2–54.9%), Betaproteobacteria (16.0–32.1%), and Alphaproteobacteria (6.3–12.6%); as representatives of the genera Thiothrix, Thiomonas, Thiobacillus, and Thioclava (Supplementary Table 3). In the CP-Amended treatment samples, sulfur-oxidizing Gammaproteobacteria (Halothiobacillus (9.6–49.5%) were most prominent, but lesser quantities of Thiothrix (<3.0%), and Thiomonas (<1.0%) were detected (Supplementary Table 4). Gram-positive bacteria were nearly absent (<1%) on all surfaces in both P-amended treatments, and not detectable in either planktonic sample (Supplementary Tables 4, 6). However, Gram-positives were detected on all surfaces in the C-Amended reactor as members of the lineages Actinobacteria, Bacilli, Clostridia, and TM7 (Supplementary Table 5). Actinobacteria and Bacilli were particularly abundant in the Basalt sample. Additionally, representatives of the only genus detected in 3 treatments (Cloacibacterium) were abundant on all surfaces within the P-Amended (7.1–38.6%) and CP-Amended treatments (3.7–21.4%), but less abundant in the C-Amended samples (Supplementary Tables 4–6; Figure 4).
Although absent from both C-limited treatments, representatives of the heterotrophic sulfur-reducing lineage Deltaproteobacteria (Desulfovibrio) were present in both the C-Amended and CP-Amended treatments (Figure 4). Abundant Desulfovibrio was distributed amongst all samples within the CP-Amended treatment (11.6–61.4%), but not detected in the C-Amended basalt sample although it is found in higher overall abundance (69.1–84.5%) in the remaining C-Amended samples (Supplementary Tables 4, 5). The CP-Amended treatment was the only treatment to host significant quantities of both sulfur-oxiders (i.e., Halothiobacillus) and sulfur-reducers (i.e., Desulfovibrio; Supplementary Table 4).
A majority of the microbes in the inoculant came from two dominant classes (Epsilonproteobacteria and Gammaproteobacteria) representing ~97.1% of the total sequences (Supplementary Table 3; Jones and Bennett, 2014). Despite the dominance of Epsilonproteobacteria in the inoculant, Epsilonproteobacteria were all but absent in surface samples from every treatment, but present in appreciable amounts in the planktonic samples obtained from the P-Amended (39.5% as Sulfuricurvum) and CP-Amended (23.9% as Sulfurospirillum) treatments (Figure 3, Supplementary Tables 3–6). The CP-Limited planktonic sample is both phylogenetically and taxonomically distinct from any of the attached communities (Figure 3A). The primary differences taxonomically are the significant abundance of Acidithiobacillus, Chloroacidobacterium, Acidisphaera, Thiobacillus, and Thermodesulfovibrio).
Discussion
The subsurface environment is a complex blend of rocks and minerals that are generally not considered to play an active role in microbial colonization. Using laboratory flow-through biofilm reactors, our study revealed that the media chemistry and the minerals present in the media significantly affect growth, diversity, and composition of a subsurface microbial community. Remarkably, 16S rRNA pyrosequencing revealed that microbial communities were mainly controlled by the media chemistry at the taxonomic level and by mineral type at the phylogenetic level. This study reveals significant evidence that mineral colonization is a non-random process controlled by both environmental and mineral conditions.
Mineral selectivity and local environmental geochemistry have been shown to impact the structure of attached microbial communities in soils, surface outcrops, and aquatic environments (Bennett et al., 2001; Gleeson et al., 2006; Carson et al., 2009; Hartmann et al., 2015; Uroz et al., 2015). However, it is currently unclear what motivates specific microbial communities to colonize specific surfaces. Here, we show that media pH, phosphate amendments, and carbon amendments significantly impacted the overall community taxonomy, but the phylogenetic distribution of that community among surfaces is ultimately correlated to surface type. Additionally, phosphate amendments, carbon amendments, and surface phosphate availability all significantly impacted biofilm accumulation. In fact, mineral phosphate concentration exerted significant control on biomass accumulation under all treatment conditions (Figure 1). Overall taxonomy and proportional abundance were significantly sensitive to variations in media and surface chemistry with consistent patterns emerging among specific guilds (SOB, SRB, Gram-positives, acidophiles).
Keystone Microorganisms
Looking at the total communities within each treatment, the main factor affecting taxonomy is the presence/absence of carbon amendments. C-Limited treatments were dominated by autotrophic SOB and C-Amended by heterotrophic SRB (Supplementary Tables 3–6). These taxonomic shifts in keystone microorganisms have diametrically opposing geochemical consequences due to differences in metabolic processes. Mineral selection is then a consequence of the reactivity of the surface to the metabolic byproducts of these keystone organisms and their environmental tolerances.
Locally, SOB benefit from attachment to carbonates; buffering the acidity generated by sulfur-oxidation to sulfate:
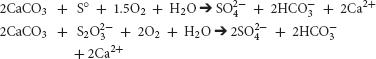
Previously, we demonstrated a preference for carbonates by the genera Thiothrix, Thioclava, Halothiobacillus, Thiobacillus, Bosea, Thiomonas, and Sulfurovum (Jones and Bennett, 2014). Aggressive dissolution of carbonates (Calcite, Madison Limestone, and Madison Dolostone) was confirmed by SEM analysis (Jones and Bennett, 2014).
If SOB-carbonate surface selection was linked to a need for pH buffering, then this preference would be less pronounced in a media buffered environment. Indeed, within the P-Amended treatment, media buffering reduced the dependence of neutrophilic SOB on mineral buffering of metabolically generated acidity (Supplementary Table 6). In the P-Amended treatment, potential SOB were identified ubiquitously on every surface, represented by the genera Thiothrix, Thioalkalivibrio, Thiomonas, and Thiobacillus (Supplementary Table 6). Obligately alkaliphilic Thioalkalivibrio are found exclusively within this treatment (Supplementary Tables 3–6; Sorokin et al., 2006). UniFrac analysis showed that all communities were very similar (>91% similar; Figure 3C). Despite this high degree of similarity, 85.9% (P = 0.035) of the phylogenetic variability in the P-Amended treatment was controlled by overall variations in mineral chemistry. Regardless, both of these treatment communities were composed of chemoautotrophic microorganisms, as the treatment media was carbon limited.
The addition of a carbon source (in the form of acetate, lactate, and formate) to the media promotes chemoheterotrophic growth. Both of the C-Amended treatments have significant quantities of Deltaproteobacteria represented by members of the heterotrophic sulfur-reducing genus Desulfovibrio (Supplementary Tables 4, 5). Desulfovibrio is a motile, vibrio-shaped, heterotrophic, sulfur-reducing bacteria capable of growth on a variety of sulfur substrates (as the terminal electron acceptor) as well as lactate, pyruvate, acetate, propionate, and butyrate (as electron donor and carbon source; Liu and Peck, 1981; Cypionka, 2000).
Metabolic sulfate reduction by SRB generally causes an increase in pH (Lyons et al., 1984; Walter et al., 1993; Van Lith et al., 2003; Dupraz et al., 2009). The geochemical consequences of sulfur-reduction by Desulfovibrio are localized consumption of acidity by paired acetate (shown below), lactate, or formate oxidation with reduction of inorganic sulfur compounds.
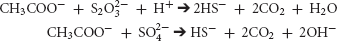
Also, in aerobic environments, several species of Desulfovibrio have been shown to pair sulfur-reduction with O2 reduction to H2O (Cypionka, 2000).
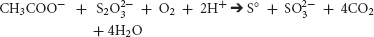
Within the C-Amended treatments, the metabolism of SRB allows more favorable conditions to be established on feldspars (albite & microcline) where consumption of local acidity decreases the mobility of potentially toxic mineral-bound aluminum (Rogers and Bennett, 2004). The relatively high pHs of both P-Amended medias reduces the dependence of neutrophilic, but acid-producing SOB for highly-buffering carbonates as media buffering facilitated acid consumption. Consequently, neutrophilic SOB colonized all surfaces at relatively high proportional abundances (Supplementary Table 6). Additionally, this decreases the overall species richness (alpha-diversity) as well as increases the amount of shared species (beta-diversity) between the surfaces (Figure 3B, Table 4). Overall, the dominance of these organisms was statistically correlated with lower Shannon diversity and β-diversity with less phylogenetic diversity between surfaces within C-Amended treatments (Table 4, Figures 3B,D). Decreased species richness has been documented to be a function of increasing carbon concentration (Larson and Passy, 2013). However, to our knowledge, a decrease in species diversity in attached communities as a function of carbon limitation across multiple surfaces is previously undocumented.
Mineral Specific Accessory Microorganisms
For the oligotrophic (CP-Limited) treatment taxonomy is clearly distinct for every mineral type (Figure 3A). Generally, for the other treatments, taxonomy is nearly identical on every surface. We found that surface type controlled a significant proportion of the variance in phylogenetic β-diversity for each treatment (Table 3). Geochemical conditioning of the near surface habitat by the keystone microorganisms is likely best suited for specific suites of accessory microorganisms.
Gram-positive bacteria are more attracted to silicate surfaces in low pH environments (Gordienko and Kurdish, 2007; Winsley et al., 2014). In both of the low-pH (and low-P) treatments (CP-Limited and C-Amended) there is a clear bias for Gram-positive bacteria on silicate surfaces (Supplementary Tables 3, 5). Furthermore, Gram-positives are negligible on all surfaces in the high-pH/C-limited treatment (P-amended). This suggests that pH and C may be the controlling factors on Gram-positive membership in microbial biofilm communities and additionally explains the lack of Gram-positive organisms within either of the treatments (P-Amended and CP-Amended) where a high pH was sustained (Supplementary Tables 4, 6). In the C-Amended treatment, Gram-positive Actinobacteria, Bacilli, and Clostridia composed a large proportion (86.8%) of the microbial community associated with basalt (Supplementary Table 5). Recent studies demonstrated that the ability of Actinobacteria and other Gram-positive microorganisms to weather basaltic materials in order to access mineral bound nutrients is increased significantly when provided a carbon source (Cockell et al., 2013). In the C-Amended treatment, Alphaproteobacteria with glycosphingolipids (GSLs) (Sphingobium, Blastomonas, and Novosphingobium) show an affinity for the quartz surface (Supplementary Table 5). Members of this lineage are known for strong silicate surface adhesion in oligotrophic and extreme environments and well-modulated cellular pH (Eguchi et al., 1996; Laskin and White, 1999; Yamaguchi and Kasamo, 2002; Sun et al., 2013; Varela et al., 2014). The Quartz surface was the ideal habitat for these and acidophilic microorganisms within the C-Amended treatment due to their unique ability to tolerate low-pH environments without having to compete with the dominant keystone heterotrophic SRB within the reactor.
Additionally, the relatively basic pH of the P-Amended treatment provided an environmental advantage for potentially alkaliphilic microorganisms, constituting a proportional abundance of 13.3–28.1% on all surfaces (Supplementary Table 6). The most abundant of these organisms were members of the class β-proteobacteria. Of this lineage, Hydrogenophaga was the most abundant on all surfaces. Hydrogenophaga is a well-known aerobic, hydrogen-oxidizing microorganism commonly associated with subsurface serpentinization processes. Although many members of this genus were thought to be obligately neutrophilic, recently many members of this lineage have been isolated from and shown to thrive in high-alkalinity environments (Willems et al., 1989; Roadcap et al., 2005; Suzuki et al., 2013).
Biomass Accumulation
Nutrient limitations can stimulate biomass growth and production of EPS (Ellwood et al., 1982; Matin et al., 1989; Wrangstadh et al., 1990; Zisu and Shah, 2003; Eboigbodin et al., 2007). Here, significantly higher biofilm was positively correlated with carbon amendments (P < 0.04) independent of media phosphate, mineral phosphate, and media pH (Figure 1, Supplementary Table 1). Both of the C-Amended treatments accumulated ~2X the total biomass of their respective P-Amended counterparts (CP-Limited 50.5 mg·cm−2 vs. C-Amended 110.1 mg·cm−2, and P-Amended 12.8 mg·cm−2 vs. CP-Amended 25.4 mg·cm−2; Supplementary Table 1). Carbon limitations in the C-Limited treatments decreased the metabolic efficacy of heterotrophic populations (Matin et al., 1989). Previous studies have also demonstrated an increase in biofilm formation by heterotrophic bacteria in response to nutrient limitations (Matin et al., 1989; Wrangstadh et al., 1990). It should be noted that this dramatic increase in biofilm concentration is complex as it is likely tied to dynamic biochemical and biophysical interactions of specific heterotrophic taxa with specific surfaces and treatments (Supplementary Tables 4, 6).
P-Amended treatments had significantly lower biofilm biomass (P < 0.002), but high-phosphate surfaces (limestone, dolostone, basalt) had significantly higher biomass (2–60X; Figure 1). Previously, we found that the primary control on total biomass accumulation was the concentration of mineral bound nutrients, particularly in the form of phosphate (Figure 1; Jones and Bennett, 2014). Several studies have reported that microorganisms exhibit active adhesion/detachment processes that may be a response to local nutrient availability (Dawson et al., 1981; Kjelleberg and Hermansson, 1984; Van Loosdrecht et al., 1990; Wrangstadh et al., 1990; Marshall, 1996; Araújo et al., 2010). In particular, these investigators have noted that starvation or nutrient availability can stimulate a change in the partitioning of a microbial community between the solid and aqueous phases (Ginn et al., 2002). During starvation bacteria show increased levels of adhesion by increasing production of EPS, allowing them to take advantage of organic and inorganic compounds that accumulate at solid-liquid interfaces (Dawson et al., 1981).
These previous investigations speculated that such tactics might be particularly important in oligotrophic waters where bacteria are exposed to conditions of extreme nutrient limitation. We observed that bacteria exploit these tactics in a multitude of nutrient and environmental conditions. In P-Amended treatments the magnitude of actual variation (standard deviation) in total biomass between high-P and low-P surfaces was much lower (Figure 1, Supplementary Table 1). It is easy to intuit that the availability of media phosphate reduces the reliance on surface-bound phosphate for survival, but these surfaces are still favored by non-motile bacteria.
Planktonic vs. Attached Communities
In natural aquifers, microbial populations differentiate between planktonic and surface attached communities (Lehman et al., 2001). Here, for every treatment, the planktonic communities and inoculants were taxonomically and phylogenetically distinct from attached communities (Figure 3, Supplementary Tables 3–6). However, in the P-Amended media these distinctions were less apparent. Unlike oligotrophic media (P-Amended) there is a less significant advantage to attachment. Previous investigations have used this explanation for distinctions between planktonic and attached communities (Hazen et al., 1991; Zhou et al., 2012).
Additionally, this might explain the high degree of phylogenetic similarity (67.5%) between the two P-Amended treatments. Both P-Amended treatments had high proportions of Epsilonproteobacteria found exclusively in these samples (Figure 3, Supplementary Tables 3–6). These lineages were composed entirely of members of the genera Sulfuricurvum and Sulfurospirillum, within the P-Amended and CP-Amended treatments, respectively. Sulfuricurvum is a motile, chemolithoautotrophic SOB which grows best at near neutral pHs (Kodama and Watanabe, 2004). Sulfurospirillum is a motile, chemoheterotrophic, SRB which also grows best at near neutral pHs and is capable of using acetate as a carbon source and acetate and formate as electron donors (Kodama et al., 2007). These common treatment media conditions of phosphate availability and neutral to high pH, combined with the motility of these organisms, favor planktonic existence.
Despite phylogenetic similarities between treatments, there are no taxa common to all 4 experimental treatments (Table 3, Figure 4). This result was unexpected, but serves to underscore the fundamental role that environment plays in community membership. In truly oligotrophic and unfavorable conditions (CP-Limited), specific microorganisms are highly dependent on specific minerals for survival. This leads to taxonomically and phylogenetically distinct communities segregated by mineral type (Figure 3A). Variability in environmental conditions (i.e., pH, C, P) influences viability of specific guilds, which facilitates specific mineral selective strategies. Each experimental treatment and surface type represented a unique habitat that encouraged growth of both major (keystone) and minor (accessory) populations and revealed the presence of hundreds of additional OTUs that were below detection in the original inoculant.
In conclusion, we reported a statistically significant link between phylogenetic diversity of microbial communities and specific natural surface types under a variety of geochemical conditions. This suggests that phylogenetically similar microorganisms are significantly more likely to have similar surface habitat requirements. In resource stressed and harsh environments, minerals act as environmental filters providing specific microhabitats for metabolically similar microorganisms. Successful growth and succession is a function of the capacity of a microorganism, or community, to facilitate, tolerate, or adapt to microenvironmental cultivation and modification of the geochemical conditions at the microbe-mineral-aqueous interface. Environmental pressures such as pH, carbon, and/or phosphate variability will determine the extent of taxonomic disparity between mineral microniches, but surface type ultimately controls the phylogenetic diversity between these microenvironments. Due to their unique and variable chemical compositions, rocks and minerals should be considered as ecosystems that are primarily colonized by uniquely adapted microbes. Further investigation is required to determine if these phylogenetic similarities are the result of ecological shifts caused by environmental stresses imposed by localized surface geochemistry or latent adaptations by specific keystone and accessory organisms.
Author Contributions
AJ conceived and designed the experiments. AJ acquired and processed the data. AJ and PB interpreted the data. AJ drafted the original work as part of his dissertation at The University of Texas at Austin entitled: Mineralogical Controls on Microbial Community Structure and Biogeochemical Processes in Subsurface Environments. AJ and PB revised this work for important intellectual contend and agreed on a final version to be submitted for publication. Therefore, AJ and PB both agree to be accountable for all aspects of this work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Funding for this research was provided in part by The National Science Foundation (grant EAR-0617160) and the Geology Foundation of the University of Texas at Austin. We thank Christopher R. Omelon for valuable critique and discussion.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00491/full#supplementary-material
References
Anderson, M. J. (2001). A new method for non-parametric multivariate analysis of variance. Austra. Ecol. 26, 32–46. doi: 10.1111/j.1442-9993.2001.01070.pp.x
Araújo, E. A., de Andrade, N. J., da Silva, L. H. M., de Carvalho, A. F., de Sá Silva, C. A., and Ramos, A. M. (2010). Control of microbial adhesion as a strategy for food and bioprocess technology. Food Bioprocess Technol. 3, 321–332. doi: 10.1007/s11947-009-0290-z
Bennett, P. C., Rogers, J. R., and Choi, W. J. (2001). Silicates, silicate weathering, and microbial ecology. Geomicrobiol. J. 18, 3–19. doi: 10.1080/01490450151079734
Bohannan, B. J. M., and Hughes, J. (2003). New approaches to analyzing microbial biodiversity data. Curr. Opin. Microbiol. 6, 282–287. doi: 10.1016/S1369-5274(03)00055-9
Borg, I., and Groenen, P. J. F. (2005). Modern Multidimensional Scaling: Theory and Applications. New York, NY: Springer Science & Business Media.
Caporaso, G. J., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Meth. 7, 335–336. doi: 10.1038/nmeth.f.303
Carson, J. K., Campbell, L., Rooney, D., Clipson, N., and Gleeson, D. B. (2009). Minerals in soil select distinct bacterial communities in their microhabitats. FEMS Microbiol. Ecol. 67, 381–388. doi: 10.1111/j.1574-6941.2008.00645.x
Chapelle, F. H., O'Neil, K. O., Bradley, P. M., Methe, B. A., Ciufo, S. A., Knobel, L. L., et al. (2002). A hydrogen-based subsurface microbial community dominated by methanogens. Nature 415, 312–315. doi: 10.1038/415312a
Chu, H., Fierer, N., Lauber, C. L., Caporaso, J. G., Knight, R., and Grogan, P. (2010). Soil bacterial diversity in the Arctic is not fundamentally different from that found in other biomes. Environ. Microbiol. 12, 2998–3006. doi: 10.1111/j.1462-2920.2010.02277.x
Cockell, C. S., Kelly, L. C., and Marteinsson, V. (2013). Actinobacteria—An ancient phylum active in volcanic rock weathering. Geomicrobiol. J. 30, 706–720. doi: 10.1080/01490451.2012.758196
Cypionka, H. (2000). Oxygen respiration by Desulfovibrio species. Annu. Rev. Microbiol. 54, 827–848. doi: 10.1146/annurev.micro.54.1.827
Dalton, H. M., Poulsen, L. K., Halasz, P., Angles, M. L., Goodman, A. E., and Marshall, K. C. (1994). Substratum-induced morphological changes in a marine bacterium and their relevance to biofilm structure. J. Bacteriol. 176, 6900–6906. doi: 10.1128/jb.176.22.6900-6906.1994
Dawson, P. M., Humphrey, B. A., and Marshall, K. C. (1981). Adhesion: a tactic in the survival strategy of a marine vibrio during starvation. Curr. Microbiol. 6, 195–199. doi: 10.1007/BF01566971
Dowd, S. E., Wolcott, R. D., Sun, Y., McKeehan, T., Smith, E., and Rhoads, D. (2008). Polymicrobial nature of chronic diabetic foot ulcer biofilm infections determined using bacterial tag encoded FLX amplicon pyrosequencing (bTEFAP). PLoS ONE 3:e3326. doi: 10.1371/journal.pone.0003326
Dupraz, C., Reid, R. P., Braissant, O., Decho, A. W., Norman, R. S., and Visscher, P. T. (2009). Processes of carbonate precipitation in modern microbial mats. Earth Sci. Rev. 96, 141–162. doi: 10.1016/j.earscirev.2008.10.005
Eboigbodin, K. E., Ojeda, J. J., and Biggs, C. A. (2007). Investigating the surface properties of Escherichia coli under glucose controlled conditions and its effect on aggregation. Langmuir 23, 6691–6697. doi: 10.1021/la063404z
Edwards, K. J., Bach, W., and McCollom, T. M. (2005). Geomicrobiology in oceanography: microbe–mineral interactions at and below the seafloor. Trends Microbiol. 13, 449–456. doi: 10.1016/j.tim.2005.07.005
Edwards, K. J., Becker, K., and Colwell, F. (2012). The deep, dark energy biosphere: intraterrestrial life on earth. Annu. Rev. Earth Planet. Sci. 40, 551–568. doi: 10.1146/annurev-earth-042711-105500
Eguchi, M., Nishikawa, T., MacDonald, K., Cavicchioli, R., Gottschal, J. C., and Kjelleberg, S. (1996). Responses to stress and nutrient availability by the marine ultramicrobacterium Sphingomonas sp. strain RB2256. Appl. Environ. Microbiol. 62, 1287–1294.
Ellwood, D. C., Keevil, C. W., Marsh, P. D., Brown, C. M., Wardell, J. N., and Roux, N. L. (1982). Surface-associated growth [and Discussion]. Philos. Trans. R. Soc. Lond. B Biol. Sci. 297, 517–532. doi: 10.1098/rstb.1982.0058
Engel, A. S., Lee, N., Porter, M. L., Stern, L. A., Bennett, P. C., and Wagner, M. (2003). Filamentous “Epsilonproteobacteria” dominate microbial mats from sulfidic cave springs. Appl. Environ. Microbiol. 69, 5503–5511. doi: 10.1128/AEM.69.9.5503-5511.2003
Engel, A. S., Porter, M. L., Stern, L. A., Quinlan, S., and Bennett, P. C. (2004). Bacterial diversity and ecosystem function of filamentous microbial mats from aphotic (cave) sulfidic springs dominated by chemolithoautotrophic “Epsilonproteobacteria”. FEMS Microbiol. Ecol. 51, 31–53. doi: 10.1016/j.femsec.2004.07.004
Felsenstein, J. (2004). Inferring Phylogenies. Massachusetts, MA: Sinauer Associates Incorporated Sunderland.
Fierer, N., and Jackson, R. B. (2006). The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. U.S.A. 103, 626–631. doi: 10.1073/pnas.0507535103
Ginn, T. R., Wood, B. D., Nelson, K. E., Scheibe, T. D., Murphy, E. M., and Clement, T. P. (2002). Processes in microbial transport in the natural subsurface. Adv. Water Resour. 25, 1017–1042. doi: 10.1016/S0309-1708(02)00046-5
Gleeson, D. B., Kennedy, N. M., Clipson, N., Melville, K., Gadd, G. M., and McDermott, F. P. (2006). Characterization of bacterial community structure on a weathered pegmatitic granite. Microb. Ecol. 51, 526–534. doi: 10.1007/s00248-006-9052-x
Good, I. J. (1953). The population frequencies of species and the estimation of population parameters. Biometrika 40, 237–264. doi: 10.1093/biomet/40.3-4.237
Gordienko, A. S., and Kurdish, I. K. (2007). Electrical properties and interaction with silicon dioxide particles of Bacillus subtilis cells. Biofizika 52, 314–317. doi: 10.1134/s0006350907020121
Hartmann, M., Frey, B., Mayer, J., Mader, P., and Widmer, F. (2015). Distinct soil microbial diversity under long-term organic and conventional farming. ISME J. 9, 1177–1194. doi: 10.1038/ismej.2014.210
Hazen, T. C., Jimenez, L., and de Victoria, G. L. (1991). Comparison of bacteria from deep subsurface sediment and adjacent ground water. Microb. Ecol. 22, 293–304. doi: 10.1007/BF02540231
Hill, T. C. J., Walsh, K. A., Harris, J. A., and Moffett, B. F. (2003). Using ecological diversity measures with bacterial communities. FEMS Microbiol. Ecol. 43, 1–11. doi: 10.1111/j.1574-6941.2003.tb01040.x
Huang, C.-T., Xu, K. D., McFeters, G. A., and Stewart, P. S. (1998). Spatial patterns of alkaline phosphatase expression within bacterial colonies and biofilms in response to phosphate starvation. Appl. Environ. Microbiol. 64, 1526–1531.
Jones, A. A., and Bennett, P. C. (2014). Mineral microniches control the diversity of subsurface microbial populations. Geomicrobiol. J. 31, 246–261. doi: 10.1080/01490451.2013.809174
Kjelleberg, S., and Hermansson, M. (1984). Starvation-induced effects on bacterial surface characteristics. Appl. Environ. Microbiol. 48, 497–503.
Kodama, Y., Ha, L. T., and Watanabe, K. (2007). Sulfurospirillum cavolei sp. nov., a facultatively anaerobic sulfur-reducing bacterium isolated from an underground crude oil storage cavity. Int. J. Syst. Evol. Microbiol. 57, 827–831. doi: 10.1099/ijs.0.64823-0
Kodama, Y., and Watanabe, K. (2004). Sulfuricurvum kujiense gen. nov., sp. nov., a facultatively anaerobic, chemolithoautotrophic, sulfur-oxidizing bacterium isolated from an underground crude-oil storage cavity. Int. J. Syst. Evol. Microbiol. 54, 2297–2300. doi: 10.1099/ijs.0.63243-0
Kugaprasatham, S., Nagaoka, H., and Ohgaki, S. (1992). Effect of turbulence on nitrifying biofilms at non-limiting substrate conditions. Water Res. 26, 1629–1638. doi: 10.1016/0043-1354(92)90162-W
Larson, C. A., and Passy, S. I. (2013). Rates of species accumulation and taxonomic diversification during phototrophic biofilm development are controlled by both nutrient supply and current velocity. Appl. Environ. Microbiol. 79, 2054–2060. doi: 10.1128/AEM.03788-12
Laskin, A. I., and White, D. C. (1999). Preface to special issue on Sphingomonas. J. Ind. Microbiol. Biotechnol. 23, 231–231. doi: 10.1038/sj.jim.2900748
Lauber, C. L., Hamady, M., Knight, R., and Fierer, N. (2009). Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 75, 5111–5120. doi: 10.1128/AEM.00335-09
Lawrence, J. R., Korber, D. F., Hoyle, B. D., Costerton, J. W., and Caldwell, D. E. (1991). Optical sectioning of microbial biofilms. J. Bacteriol. 173, 6558–6567. doi: 10.1128/jb.173.20.6558-6567.1991
Lehman, R. M., Colwell, F. S., and Bala, G. A. (2001). Attached and unattached microbial communities in a simulated basalt aquifer under fracture- and porous-flow conditions. Appl. Environ. Microbiol. 67, 2799–2809. doi: 10.1128/AEM.67.6.2799-2809.2001
Liu, C. L., and Peck, H. D. (1981). Comparative bioenergetics of sulfate reduction in Desulfovibrio and Desulfotomaculum spp. J. Bacteriol. 145:966.
Lozupone, C., and Knight, R. (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71, 8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005
Lozupone, C., Lladser, M. E., Knights, D., Stombaugh, J., and Knight, R. (2011). UniFrac: an effective distance metric for microbial community comparison. ISME J. 5, 169–172. doi: 10.1038/ismej.2010.133
Lyons, W. B., Long, D. T., Hines, M. E., Gaudette, H. E., and Armstrong, P. B. (1984). Calcification of cyanobacterial mats in solar lake, sinai. Geology 12, 623–626. doi: 10.1130/0091-7613(1984)12<623:COCMIS>2.0.CO;2
Madigan, M. T., Martinko, J. M., Dunlap, P. V., and Clark, D. P. (2009). Brock Biology of Microorganisms. San Francisco, CA: Pearson Benjamin Cummings.
Margulies, M., Egholm, M., Altman, W. E., Attiya, S., Bader, J. S., Bemben, L. A., et al. (2005). Genome sequencing in microfabricated high-density picolitre reactors. Nature 437, 376–380. doi: 10.1038/nature03959
Matin, A., Auger, E. A., Blum, P. H., and Schultz, J. E. (1989). Genetic basis of starvation survival in nondifferentiating bacteria. Annu. Rev. Microbiol. 43, 293–314. doi: 10.1146/annurev.mi.43.100189.001453
McArdle, B. H., and Anderson, M. J. (2001). Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology 82, 290–297. doi: 10.1890/0012-9658(2001)082[0290:FMMTCD]2.0.CO;2
Ohashi, A., Viraj de Silva, D. G., Mobarry, B., Manem, J. A., Stahl, D. A., and Rittmann, B. E. (1995). Influence of substrate C/N ratio on the structure of multi-species biofilms consisting of nitrifiers and heterotrophs. Water Sci. Technol. 32, 75–84. doi: 10.1016/0273-1223(96)00010-8
Plummer, L. N., Busby, F., Lee, R. W., and Hanshaw, B. B. (1990). Geochemical modeling of the Madison Aquifer in parts of Montana, Wyoming, and South Dakota. Water Res. Res. 26, 1981–2014. doi: 10.1029/WR026i009p01981
Reeder, J., and Knight, R. (2010). Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nat. Methods 7, 668–669. doi: 10.1038/nmeth0910-668b
Roadcap, G. S., Kelly, W. R., and Bethke, C. M. (2005). Geochemistry of extremely alkaline (pH> 12) ground water in slag-fill aquifers. Groundwater 43, 806–816. doi: 10.1111/j.1745-6584.2005.00060.x
Rogers, J. R., and Bennett, P. C. (2004). Mineral stimulation of subsurface microorganisms: release of limiting nutrients from silicates. Chem. Geol. 203, 91–108. doi: 10.1016/j.chemgeo.2003.09.001
Rogers, J. R., Bennett, P. C., and Choi, W. J. (1998). Feldspars as a source of nutrients for microorganisms Am. Mineral. 83, 1532–1540. doi: 10.2138/am-1998-11-1241
Rogers, J. R., Bennett, P. C., and Choi, W. J. (2001). “Enhanced weathering of silicates by subsurface microorganisms: a strategy to release liminity inorganic nutrients?,” in 10th International Symposium on Water-Rock Interaction. Cagliari.
Siciliano, S. D., Palmer, A. S., Winsley, T., Lamb, E., Bissett, A., Brown, M. V., et al. (2014). Soil fertility is associated with fungal and bacterial richness, whereas pH is associated with community composition in polar soil microbial communities. Soil Biol. Biochem. 78, 10–20. doi: 10.1016/j.soilbio.2014.07.005
Sorokin, D. Y., Banciu, H., Robertson, L. A., and Kuenen, J. G. (2006). “Haloalkaliphilic sulfur-oxidizing bacteria,” in The prokaryotes, eds M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackerbrandt (New York, NY: Springer), 969–984.
Steinhauer, E. S., Omelon, C. R., and Bennett, P. C. (2010). Limestone corrosion by neutrophilic sulfur-oxidizing bacteria: a coupled microbe-mineral system. Geomicrobiol. J. 27, 723–738. doi: 10.1080/01490451003614963
Stevens, T. (1997). Lithoautotrophy in the subsurface. FEMS Microbiol. Rev. 20, 327–337. doi: 10.1111/j.1574-6976.1997.tb00318.x
Sun, W., Liu, W., Cui, L., Zhang, M., and Wang, B. (2013). Characterization and identification of a chlorine-resistant bacterium, Sphingomonas TS001, from a model drinking water distribution system. Sci. Total Environ. 458–460, 169–175. doi: 10.1016/j.scitotenv.2013.04.030
Suzuki, S., Ishii, S. I., Wu, A., Cheung, A., Tenney, A., Wanger, G., et al. (2013). Microbial diversity in the cedars, an ultrabasic, ultrareducing, and low salinity serpentinizing ecosystem. Proc. Natl. Acad. Sci. U.S.A. 110, 15336–15341. doi: 10.1073/pnas.1302426110
Sylvan, J. B., Toner, B. M., and Edwards, K. J. (2012). Life and death of deep-sea vents: bacterial diversity and ecosystem succession on inactive hydrothermal sulfides. MBio 3, e00279–e00211. doi: 10.1128/mBio.00279-11
Uroz, S., Kelly, L. C., Turpault, M.-P., Lepleux, C., and Frey-Klett, P. (2015). The mineralosphere concept: mineralogical control of the distribution and function of mineral-associated bacterial communities. Trends Microbiol. 23, 751–762. doi: 10.1016/j.tim.2015.10.004
Van Lith, Y., Warthmann, R., Vasconcelos, C., and McKenzie, J. A. (2003). Sulphate-reducing bacteria induce low-temperature Ca-dolomite and high Mg-calcite formation. Geobiology 1, 71–79. doi: 10.1046/j.1472-4669.2003.00003.x
Van Loosdrecht, M. C., Lyklema, J., Norde, W., and Zehnder, A. J. (1990). Influence of interfaces on microbial activity. Microbiol. Rev. 54, 75–87.
Varela, A. R. P., Gonçalves da Silva, A. M. P. S., Fedorov, A., Futerman, A. H., Prieto, M., and Silva, L. C. (2014). Influence of intracellular membrane ph on sphingolipid organization and membrane biophysical properties. Langmuir 30, 4094–4104. doi: 10.1021/la5003397
Walter, L. M., Bischof, S. A., Patterson, W. P., Lyons, T. W., O'Nions, R. K., Gruszczynski, M., et al. (1993). Dissolution and recrystallization in modern shelf carbonates: evidence from pore water and solid phase chemistry. Philos. Trans. Phys. Sci. Eng. 344, 27–36. doi: 10.1098/rsta.1993.0072
Willems, A., Busse, J., Goor, M., Pot, B., Falsen, E., Jantzen, E., et al. (1989). Hydrogenophaga, a new genus of hydrogen-oxidizing bacteria that includes Hydrogenophaga flava comb. nov. (formerly Pseudomonas flava), Hydrogenophaga palleronii (formerly Pseudomonas palleronii), Hydrogenophaga pseudoflava (formerly Pseudomonas pseudoflava and Pseudomonas carboxydoflava), and Hydrogenophaga taeniospiralis (formerly Pseudomonas taeniospiralis). Int. J. Syst. Bacteriol. 39, 319–333. doi: 10.1099/00207713-39-3-319
Winsley, T. J., Snape, I., McKinlay, J., Stark, J., van Dorst, J. M., Ji, M., et al. (2014). The ecological controls on the prevalence of candidate division TM7 in polar regions. Front. Microbiol. 5:345. doi: 10.3389/fmicb.2014.00345
Wrangstadh, M., Szewzyk, U., Ostling, J., and Kjelleberg, S. (1990). Starvation-specific formation of a peripheral exopolysaccharide by a marine Pseudomonas sp., strain S9. Appl. Environ. Microbiol. 56, 2065–2072.
Yamaguchi, M., and Kasamo, K. (2002). Modulation of proton pumping across proteoliposome membranes reconstituted with tonoplast H+-ATPase from cultured rice (Oryza sativa L. var. Boro) cells by acyl steryl glucoside and steryl glucoside. Plant Cell Physiol. 43, 816–822. doi: 10.1093/pcp/pcf096
Zhou, Y., Kellermann, C., and Griebler, C. (2012). Spatio-temporal patterns of microbial communities in a hydrologically dynamic pristine aquifer. FEMS Microbiol. Ecol. 81, 230–242. doi: 10.1111/j.1574-6941.2012.01371.x
Keywords: biofilms, microbial communities, cave microbiology, subsurface, bioreactors, microbe/mineral interactions
Citation: Jones AA and Bennett PC (2017) Mineral Ecology: Surface Specific Colonization and Geochemical Drivers of Biofilm Accumulation, Composition, and Phylogeny. Front. Microbiol. 8:491. doi: 10.3389/fmicb.2017.00491
Received: 23 January 2016; Accepted: 09 March 2017;
Published: 28 March 2017.
Edited by:
Kurt O. Konhauser, University of Alberta, CanadaReviewed by:
Alexis Templeton, University of Colorado Boulder, USAJuliane Hopf, University of Notre Dame, USA
Copyright © 2017 Jones and Bennett. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aaron A. Jones, aaajones@utexas.edu
 Aaron A. Jones
Aaron A. Jones Philip C. Bennett
Philip C. Bennett