- 1Laboratorio de Genética y Biología Molecular, IRNASUS–Consejo Nacional de Investigaciones Científicas y Técnicas, Facultad de Ciencias Químicas, Universidad Católica de Córdoba, Córdoba, Argentina
- 2Bodega La Caroyense, Colonia Caroya, Argentina
Grape must harbors a complex community of yeast species responsible for spontaneous alcoholic fermentation. Although there are detailed studies on the microbiota of Vitis vinifera L. grapes, less is known about the diversity and behavior of yeast communities present on fermenting grape must from other species of Vitis. In this work, we used a culture-dependent method to study the identity and dynamics of the indigenous yeast population present during the spontaneous fermentation of Isabella (Vitis labrusca L.) grape must. Alcoholic fermentation was conducted using standard enological practices, and the associated non-Saccharomyces and S. cerevisiae yeast community was analyzed using selective growth media and 5.8-ITS DNA sequencing. Candida californica, Candida hellenica, Starmerella bacillaris (synonym Candida zemplinina), Hanseniaspora uvarum, and Hanseniaspora vineae were the main non-Saccharomyces species identified on Isabella fermenting must. Issatchenkia hanoiensis, a yeast species rarely found on Vitis vinifera L. grapes, was also recognized on Isabella grape must. Candida azymoides, Candida californica and Pichia cecembensis, identified in this work on Isabella fermenting must, have not previously been found on Vitis vinifera L. grape must. Interestingly, C. azymoides, I. hanoiensis and P. cecembensis have recently been isolated from the surface of Vitis labrusca L. grapes from vineyards in the Azores archipelago, suggesting that specific Vitis-yeast species associations are formed independently of geographic origin. We suggest that C. azymoides, C. californica, and P. cecembensis are yeast species preferentially associated with Vitis labrusca L. grapes. Specific biological interactions between grapevines and yeast species may underlie the assembly of differential Vitis-microbial communities.
Introduction
The surface of grapes lodges the microbiota responsible for spontaneous fermentation of grape must into wine (Mortimer and Polsinelli, 1999; Barata et al., 2012). The complexity of indigenous non-Saccharomyces and Saccharomyces yeast microbiota present during spontaneous grape must fermentation has a major impact on the organoleptic and sensory properties of the final wines (Jolly et al., 2013; Padilla et al., 2016b; Varela and Borneman, 2016).
The identity and relative abundance of indigenous yeast species on grapes is considered to be dependent on the terroir (i.e., soil type, annual mean temperature, and rainfall, etc.), the ripeness and health of the grapes, as well as the production procedures in the vineyards (Mortimer and Polsinelli, 1999; Bokulich et al., 2014; Drumonde-Neves et al., 2016; Grangeteau et al., 2016). The resulting grapevine microbiota could specifically identify a vineyard and, accordingly, the identity of wines from that particular terroir (Bokulich et al., 2014; Knight et al., 2015; Capece et al., 2016).
Detailed studies on the microbiota of grapes and grape musts may enable valuable indigenous yeast strains that contribute to the regional character of wines to be identified (Knight et al., 2015; Capece et al., 2016; Padilla et al., 2016b; Varela and Borneman, 2016). Molecular methods permit rapid and precise identification of yeasts at the species or strain level (Barata et al., 2012). Among these, the sequencing of rDNA regions D1–D2 (James et al., 1997; Kurtzman and Robnett, 1998), RFLP analyses (Guillamón et al., 1998) and/or the sequencing of ribosomal 5.8S-ITS are frequently used for rapid identification of indigenous isolated yeasts (Barata et al., 2012).
Non-Saccharomyces are the predominant yeast species isolated at early stages of the spontaneous fermentation of Vitis vinifera L. grape musts (Combina et al., 2005; Jolly et al., 2013; Padilla et al., 2016b). Among these, Hanseniaspora, Candida, Pichia, and Metschnikowia are the most important genera (Jolly et al., 2013; Varela and Borneman, 2016). As fermentation progresses, the population of non-Saccharomyces species decreases and the wine yeast Saccharomyces cerevisiae completes the fermentation process (Albergaria and Arneborg, 2016). The ability of S. cerevisiae to outcompete non-Saccharomyces species is associated with its higher fermentative power as well as its additional advantageous phenotypes that include alcohol tolerance and the secretion of killer-like compounds (Albergaria and Arneborg, 2016). Although there are detailed studies on yeast microbiota from Vitis vinifera L. grape musts (Combina et al., 2005; Zott et al., 2008; Albertin et al., 2014; Padilla et al., 2016a), less is known about the communities of yeast present on grapes from other species of Vitis. Moreover, the potential existence of selective interactions between various grapevine and microbial species, which may underlie the assembly of specific microbial communities (Wolfe and Dutton, 2015), is unknown.
Isabella grapes are one of the main varieties of Vitis labrusca L., a plant originally from the south of the United States, with worldwide distribution (Hernandez et al., 2011; Hurrel et al., 2014; Almanza-Merchán et al., 2015). We hypothesized that the microbial community of Isabella grape must, as well as other poorly explored ecological niches, constitutes a potential source of relevant yeast strains for the winemaking industry. We report here a study of the yeast microbiota present during spontaneous fermentation of Isabella grape must from Colonia Caroya, Argentina.
The Isabella vineyards of Colonia Caroya have remarkable historical value because they are located in a traditional winemaking region in Córdoba, Argentina, initiated four centuries ago by the Jesuit missions. Cultivated grapes in Colonia Caroya are mostly of Vitis vinifera L. (i.e., ∼75% of the vineyards) and Isabella grapes are mainly used for making grape juice. Some local producers and wineries, however, still culture and harvest these grapes to prepare either pure Isabella or Isabella blended wines. Our study is the first analysis of indigenous yeasts on grapes from a Vitis species different from Vitis vinifera L. in Argentina. It is also the first characterization of the indigenous yeasts in fermenting grape must from Córdoba, an emerging winemaking region in Argentina.
Materials and Methods
Spontaneous Fermentation of Isabella Must
Isabella grapes were harvested in Colonia Caroya vineyards (vintage of March 2015), located at 31°02′00″S/64°05′36″O and 491 m above sea level, in the province of Córdoba, Argentina. The region has an annual rainfall of 765 mm and a mean temperature of 15.8°C. Spontaneous fermentation of destemmed and partially crushed Isabella grapes (supplemented with 85 mg/l sodium metabisulfite) was performed in a reference local winery. A must sample of ∼60 lt (an aliquot taken from a total of 15,000 kg of Isabella grapes subjected to spontaneous fermentation) was fermented at 25–28°C in a stainless steel tank located in a room of the winery not previously used for winemaking. Isabella must in the stainless steel tank was punched down twice a day. Aliquots from the must were taken daily for 5 days (i.e., 0, 24, 48, 72, and 96 h) and stored in 30% (v/v) glycerol at -70°C.
Yeast Isolation, Culturing, and Characterization
Appropriate dilutions of fermenting Isabella must samples (i.e., t0, t24, t48, t72, and t96) were plated in duplicate on both YPD-Cm agar [yeast extract 1% (w/v), peptone 2% (w/v), glucose 2% (w/v), agar 2% (w/v), chloramphenicol 10 μg/ml] and YPD-Cm-Cx agar [yeast extract 1% (w/v), peptone 2% (w/v), glucose 2% (w/v), agar 2% (w/v), chloramphenicol 10 μg/ml, cycloheximide 0.5 μg/ml]. Plates were incubated for 5 days at 25°C. Colony counts on YPD-Cm and YPD-Cm-Cx plates were used to determine the relative contribution of total yeasts versus non-Saccharomyces (i.e., cycloheximide resistant) yeast species, respectively (Zott et al., 2008).
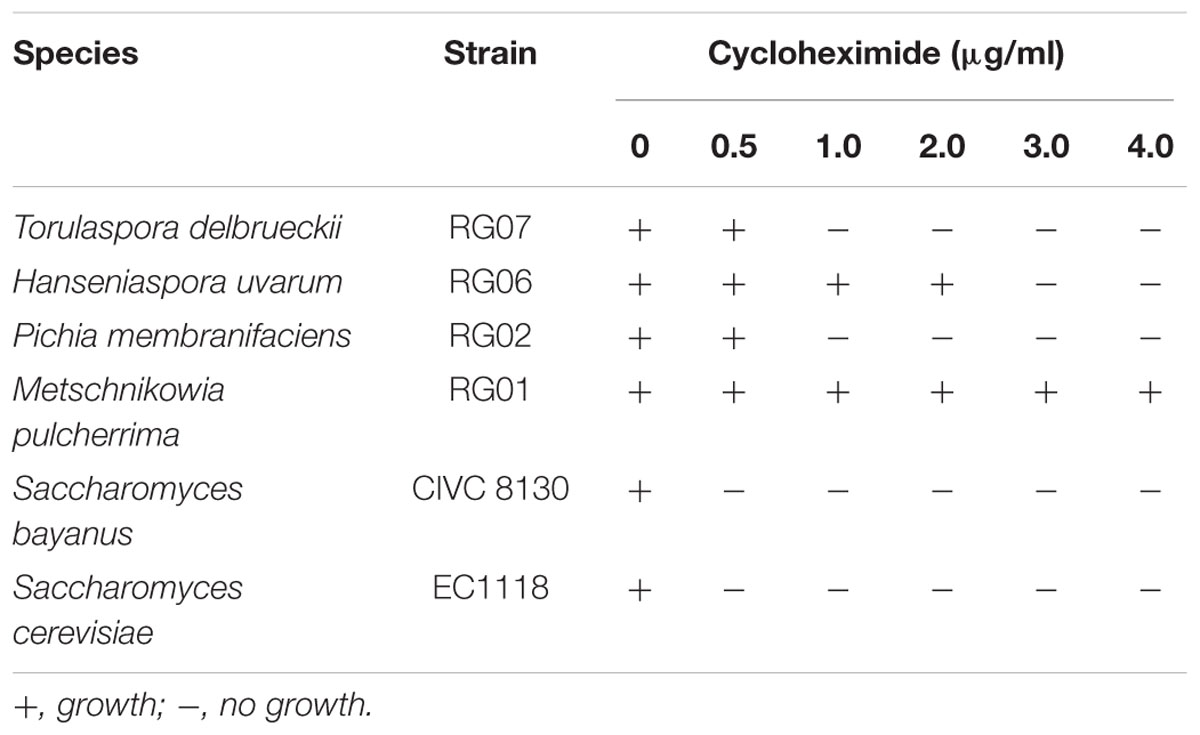
For these studies, we first determined the concentration of cycloheximide able to inhibit the growth of reference Saccharomyces spp. strains (i.e., Saccharomyces bayanus – strain CIVC 8130 – and S. cerevisiae –strain EC1118), while still allowing the growth of reference non-Saccharomyces strains (i.e., Torulaspora delbrueckii –strain RG07–, Metschnikowia pulcherrima –strain RG01–, Pichia membranifaciens –strain RG02– and Hanseniaspora uvarum –strain RG06–). Non-Saccharomyces strains, RG07, RG01, RG02 and RG06, were kindly provided by R. Gonzalez (ICVV; Logroño, Spain). Strains were plated (i.e., ∼50 viable cells) on YPD-Cm agar supplemented with various concentrations of cycloheximide (i.e., between 0 and 4.0 ug/ml). Plates were incubated during 5 days at 25°C and growth was considered positive when colonies larger than 0.25 ± 0.05 mm were observed. Based on the sensitivity of the reference strains to cycloheximide, YPD-Cm agar supplemented with cycloheximide 0.5 μg/ml was used in subsequent studies. Isabella must samples t0, t24, t48, t72, and t96 were also plated in duplicate on WL-Cm Nutrient agar [Oxoid WL Nutrient agar medium 7.5% (w/v), chloramphenicol 10 μg/ml] and incubated for 5 days at 25°C. WL-Cm Nutrient agar was used to recognize alternative colony phenotypes potentially corresponding to different yeast species.
Forty randomly selected yeast colonies were isolated from YPD-Cm agar plates from each sampling time. These colonies enabled strains to be identified representing ≥2.5% of the yeast species present at each sampling time. To assure random isolation, plates (i.e., YPD-Cm agar) with 30–50 independent yeast colonies were superimposed on a design with 20-evenly separated dots. The 20 yeast colonies closest to each dot were selected from two independent YPD-Cm agar plates. In addition, 10 yeast colonies showing different phenotypes (i.e., morphology and/or color) were isolated from WL-Cm Nutrient agar plates. These colonies could correspond to rare yeast species present at each sampling. All isolated yeast strains were streaked on YPD agar, grown for 48 h at 25°C, and stored at -70°C in YPD broth with 30% (v/v) glycerol added. A total of 250 yeast strains were isolated from the five samplings of spontaneously fermenting Isabella grape must.
Molecular Identification of Yeast Species
All isolated strains (i.e., 250) were identified by PCR amplification and sequencing of their 5.8-ITS (Internal Transcribed Spacer) rDNA regions (Granchi et al., 1999), using ITS1 and ITS4 primers (White et al., 1990). Total genomic DNA was extracted from 4.5 ml yeast cultures grown in YPD broth with orbital shaking (200 rpm) during 48 h at 25°C. Cells were collected by centrifugation at 3,000 rpm for 30 s in a bench top centrifuge and washed with sterile distilled water. Yeasts were disrupted in 400 μl of buffer TENT [HCl-Tris 10 mM (pH 8.0), Na-EDTA 1 mM, NaCl 100 mM, Triton X-100 2% (v/v) and SDS 1% (w/v)] using 0.1 ml of acid-washed glass beads (Scientific Industries, Inc.) and a Disruptor Genie (Scientific Industries, Inc.) apparatus. Broken cells had 50 μl of cold sodium acetate 5 M (pH 4.7) added and were centrifuged for 10 min at 10,000 rpm. Supernatants (i.e., 300–350 μl) were carefully transferred into clean tubes and supplemented with 300 μl of cold isopropanol. DNA was recovered by 10 min centrifugation at 10,000 rpm, washed with 1.0 ml of 70% (v/v) ice-cold ethanol and re-centrifuged at 10,000 rpm for 5 min. Pelleted DNA was dried at room temperature and suspended in 50 μl of buffer TE. PCR mixtures contained 100 ng DNA, 1.5 mM MgCl2, Taq polymerase buffer 1X (Invitrogen, USA), 200 μM dNTPs, 10 pmol of each ITS1 and ITS4 and 1.25 units of Taq polymerase (Invitrogen, USA). The amplification reaction was performed in a MJ Mini Bio-Rad thermocycler (Bio-Rad, USA) using an initial denaturation step at 93°C for 3 min, followed by 35 cycles of: 93°C for 30 s, annealing at 52°C for 30 s, extension at 72°C for 1 min followed by a final extension at 72°C for 10 min. PCR products were subjected to DNA sequencing (Macrogen, Korea) and the resulting sequences were analyzed using BLASTN software and GenBank NCBI reference sequences. Species identification was considered valid when the identity of a 5.8-ITS sequence and a reference sequence was ≥98%. Sequences from representative strains were deposited in the GenBank NCBI database with the accession numbers KY693700 (Candida azymoides: IT0-016), KY693709 (Candida californica: IT2-010), KY693702 (H. uvarum: IT0-031), KY693711 (Hanseniaspora vineae: IT2-021), KY693701 (Issatchenkia hanoiensis: IT0-025), KY693704 (Pichia cecembensis: IT0-042), KY693703 (Starmerella bacillaris: IT0-033), KY693705 (S. bacillaris: IT1-027), KY693706 (S. bacillaris: IT1-033), KY693708 (S. cerevisiae: IT2-031), KY693710 (S. cerevisiae: IT4-007), and KY693707 (T. delbrueckii: IT1-039).
Killer Phenotype and Ethanol Tolerance
The killer phenotype was analyzed using the diffusion agar technique (Lopes and Sangorrín, 2010) on YPD-methylene blue (MB) medium [i.e., yeast extract 1% (w/v), peptone 2% (w/v), glucose 2% (w/v), agar 2% (w/v), 0.003% (w/v) methylene blue] buffered at pH 4.6 with 0.1 M citrate-phosphate buffer (Lopes and Sangorrín, 2010). Potential killer yeast strains were grown on YPD broth and spotted (i.e., 3 μl 106 cells/ml) onto a lawn of each tested killer-sensitive strain. Lawns were prepared using molten YPD-MB agar inoculated with 105 cells/ml. Plates were incubated for 72 h at 24°C. S. cerevisiae CLIB154 (K2 killer) and S. cerevisiae BY4742 (K2 sensitive) were used as reference strains. Strains CLIB154 and BY4742 were kindly provided by M. Bely (ISVV; Bordeaux, France).
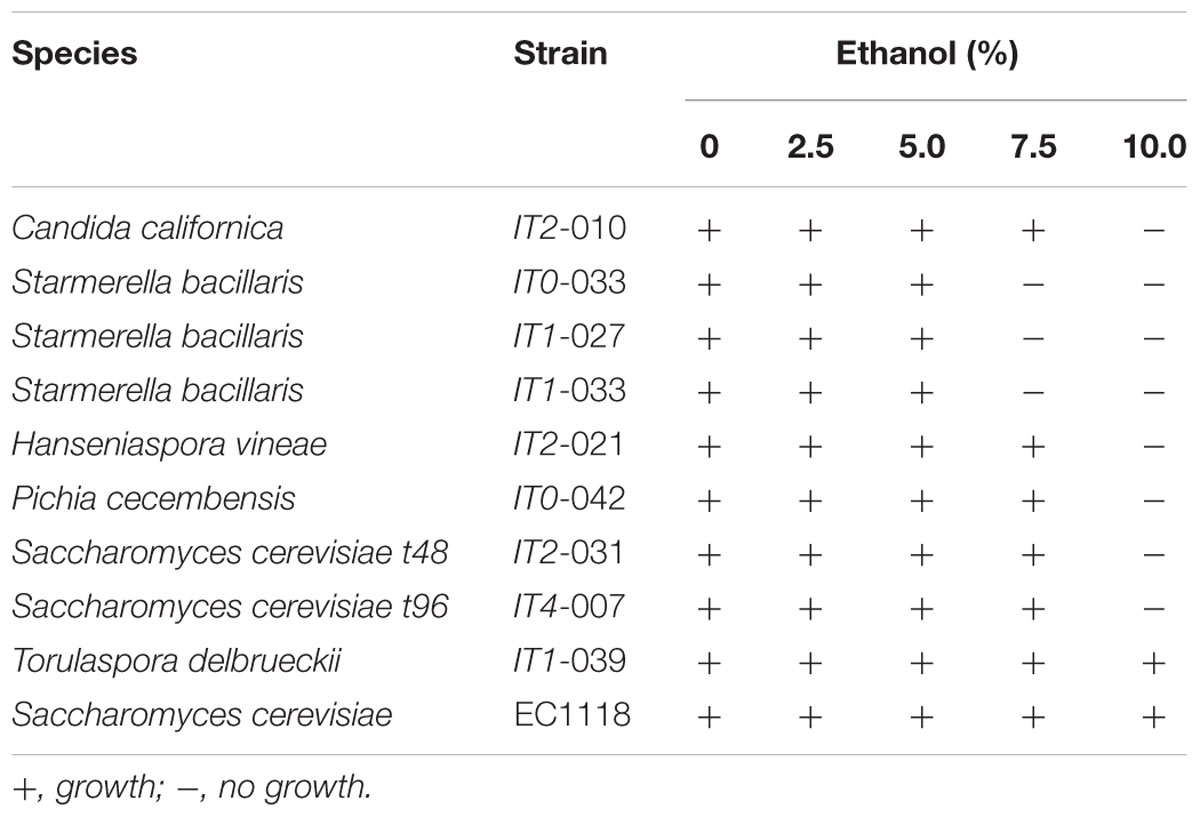
To study ethanol tolerance, we developed a simple spot assay on low dextrose [i.e., glucose 0.5% (w/v)] YPD agar supplemented with either 0, 2.5, 5.0, 7.5, or 10.0% (v/v) ethanol. The test was validated using S. cerevisiae strain EC1118. In these studies, strains were grown overnight at 25°C on YPD broth and cells (i.e., 3 μl of a suspension of 106 cells/ml) were spotted onto the agar. Growth was observed after 48 h at 22°C and considered positive when colonies larger than 0.25 ± 0.05 mm were visible. The following Isabella strains isolated in this work were analyzed: C. californica (strain IT2-010), H. vineae (strain IT2-021), P. cecembensis (strain IT0-042), S. cerevisiae (strains IT2-031 and IT4-007), S. bacillaris (strains IT0-033, IT1-027, and IT1-033), and T. delbrueckii (strain IT1-039).
Results
Dynamics of Non-Saccharomyces and S. cerevisiae Yeast Populations
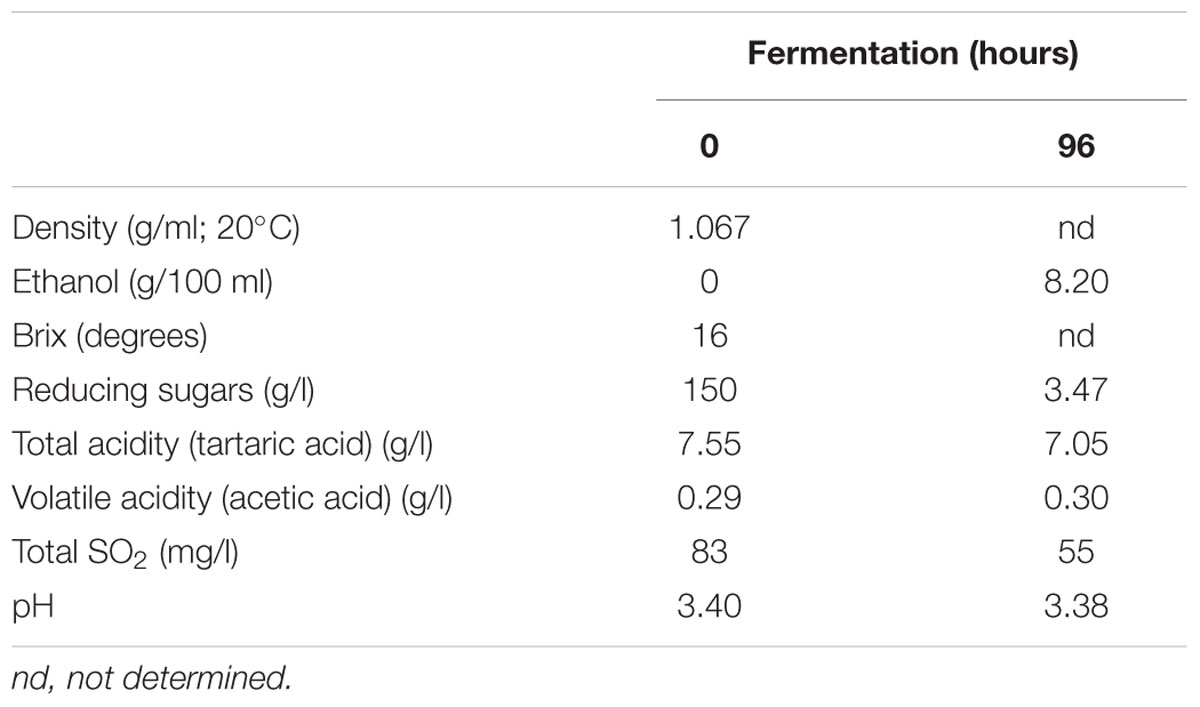
The spontaneous fermentation of Isabella grape must was almost complete in 5 days (i.e., t96). Results from physicochemical analyses of the must are shown in Table 1. After 5 days of fermentation, the concentration of the remaining reducing sugars in the Isabella must was 3.47 g/l. A final concentration of 8.2% ethanol was achieved (Table 1). The final volatile acidity and pH were 0.30 g/l and 3.38, respectively. For yeast population analyses, samples from the Isabella fermenting must were taken every 24 h, from t0 to t96 (see Materials and Methods). The relative contribution of non-Saccharomyces and S. cerevisiae yeasts, among the total yeast population, was determined on YPD-Cm agar media supplemented or not with cycloheximide. Table 2 shows that YPD-agar containing 0.5 ug/ml cycloheximide was able to inhibit the growth of reference Saccharomyces spp. strains, still allowing the growth of reference non-Saccharomyces strains. Figure 1 shows that non-Saccharomyces were the predominant yeast species at early stages of Isabella grape must fermentation (i.e., t0 and t24). In the mid-stages of fermentation (t48 and t72), the total yeast count increased, with S. cerevisiae being the prevalent species (Figure 1). The complete dominance of S. cerevisiae was evident after 5 days of fermentation (t96) when only ≤104 colonies of non-Saccharomyces species developed on YPD-Cm-Cx media (Figure 1). The observed dynamics of the non-Saccharomyces and S. cerevisiae yeast populations are similar to those observed on spontaneously fermenting must of Vitis vinifera L. grapes (Combina et al., 2005; Zott et al., 2008; Jolly et al., 2013).
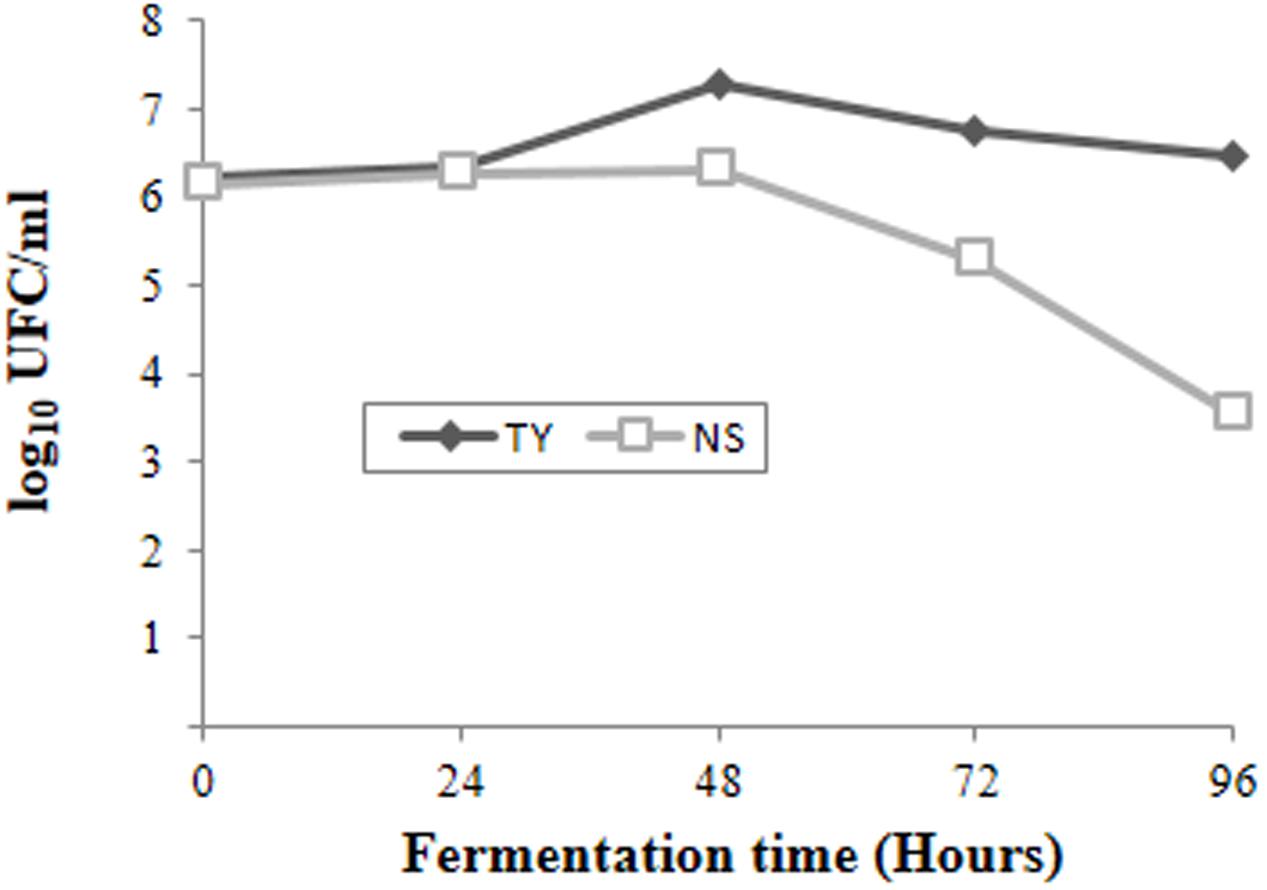
FIGURE 1. Population dynamics of non-Saccharomyces (NS) and total (TY) yeasts at different times during spontaneous fermentation of Isabella grape must.
Identification of Yeast Species of Isabella Fermenting Grape Must
A total of 250 yeast strains were isolated and identified from the fermenting Isabella grape must (see Materials and Methods). These various yeast species are shown in Figures 2A,B. A wide diversity of non-Saccharomyces yeasts was observed at early stages of fermentation (Figures 2A,B; t0 and t24), with S. bacillaris, H. uvarum, and Candida hellenica being the major species at t0 (62.5, 17.5, and 7.5% of the total yeast population, respectively) (Figure 2A). C. californica was observed among colonies isolated from both YPD-Cm agar and WL-Cm Nutrient agar plates (Figures 2A,B). C. azymoides, C. bentonensis, I. hanoiensis, and P. cecembensis, rarely recognized in winemaking ecosystems, were isolated early in fermentation. S. bacillaris was the most abundant species (i.e., 75%) at t24 (Figure 2A). S. cerevisiae, although present at very low percentages during early stages of fermentation, was able to dominate the yeast population as from t48 (Figures 1, 2A). This result was expected, based on the well-known predominance of S. cerevisiae at intermediate and advanced stages of the spontaneous fermentation of grape musts from various origins (Fleet, 2003). H. vineae was the main non-Saccharomyces species coexisting with S. cerevisiae at advanced times of Isabella grape must fermentation (i.e., t48 and t72). No non-Saccharomyces yeast species were observed among the 40 random isolated colonies at t96 (Figure 2A). Moreover, all the 10 yeast colonies selected at t96 on WL Nutrient medium, based on their rare morphological and/or color phenotypes, corresponded to S. cerevisiae strains (Figure 2B).
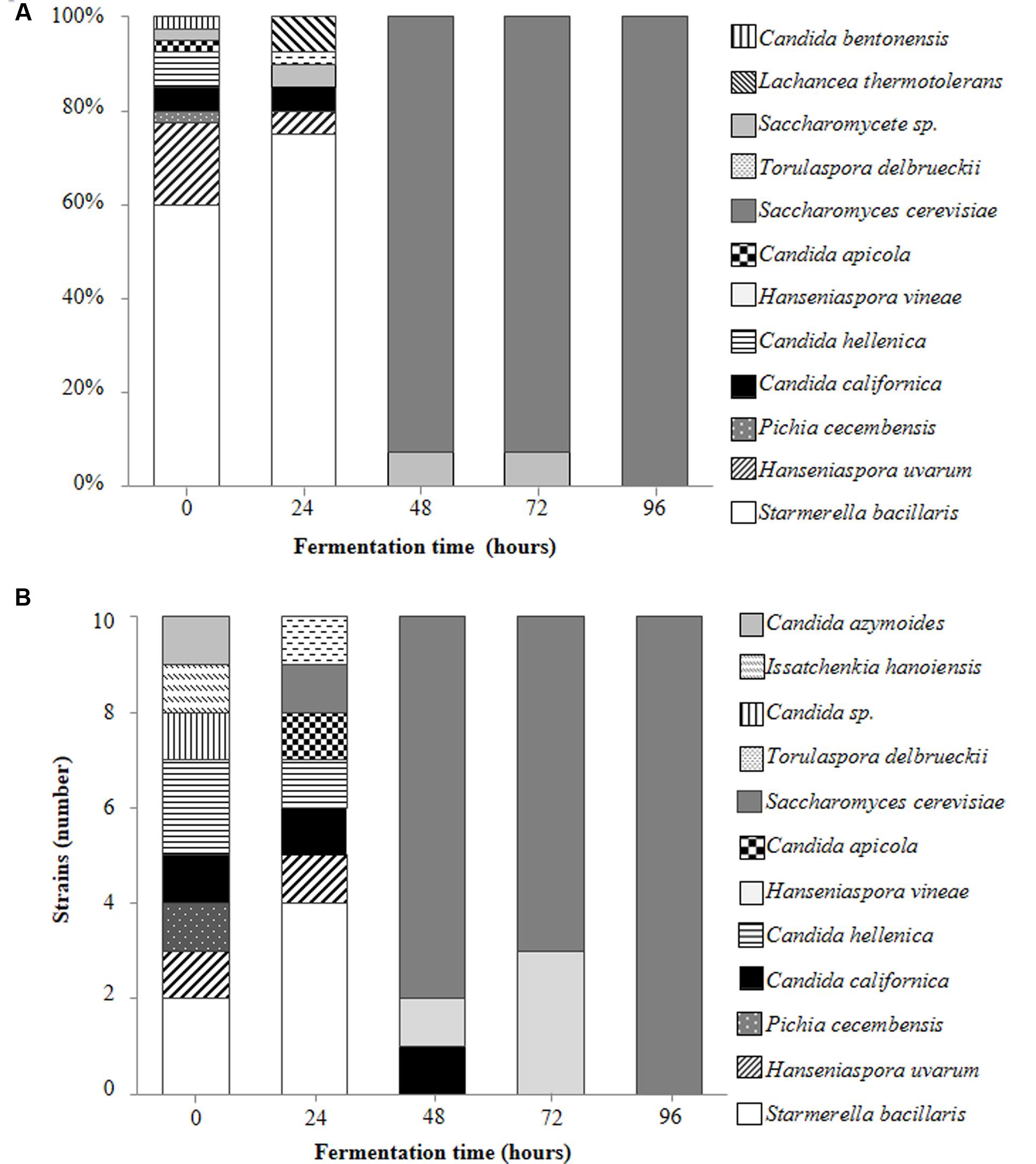
FIGURE 2. Main contributing yeast species during spontaneous fermentation of Isabella grape must. Percentages represent the relative contribution of the indicated yeast species among 40 randomly selected colonies obtained at the indicated times of fermentation (A). Yeast species identified among 10 rare colonies isolated from WL-Cm Nutrient agar plates at the indicated times of fermentation (B).
Killer Sensitivity and Ethanol Tolerance of S. bacillaris Isolates
Based on the relative contribution of S. bacillaris at t0 and t24, we expected to find at least ∼10% of S. bacillaris colonies at t48. This species, however, was not represented among the 50 yeast strains isolated at t48 (Figures 2A,B). We hypothesized that this may result from the sensitivity of this species to unknown indigenous killer strains developing in the interval t24–t48. A similar phenomenon has been described for other non-Saccharomyces species present in mixed fermentations (Pérez-Nevado et al., 2006; de Ullivarri et al., 2014). To study this possibility, isolated S. bacillaris strains were used as lawn in a standard killer-assay (see Materials and Methods). No particular sensitivity of the S. bacillaris isolates was observed when challenged to different non-Saccharomyces or S. cerevisiae isolates as from t48 (not illustrated). We also considered the possibility that the decrease in the population of S. bacillaris at t48 was based on a low tolerance of these strains to ethanol. S. bacillaris strains have been described as tolerant to high levels of ethanol (i.e., 8–12%) (Combina et al., 2005; Tofalo et al., 2011). Table 3 shows that S. bacillaris strains isolated from Isabella fermenting must developed only up to 5% alcohol while other isolated non-Saccharomyces strains tolerated higher levels of ethanol (i.e., 7.5%). This low tolerance to ethanol could explain, at least in part, why S. bacillaris was not isolated at t48, when the ethanol concentration was 4–5% (not shown). These results indicate that ethanol tolerance may dramatically differ between S. bacillaris strains isolated from various geographic origins and/or Vitis species. It was also remarkable from these studies that indigenous Isabella S. cerevisiae strains (Table 3) had relatively low tolerance to ethanol (i.e., <10%) compared with indigenous S. cerevisiae isolates from Vitis vinifera L. (Baǧder Elmac et al., 2014; Capece et al., 2016; Šuranská et al., 2016).
Specific Yeast Microbial Communities of Vitis labrusca L. Grapes
Non-Saccharomyces species recognized on fermenting Isabella must include a large diversity of yeasts (Figures 2A,B). Among these, we isolated I. hanoiensis, which is rarely found in winemaking environments (Hierro et al., 2006). I. hanoiensis has also been recognized in a recent elegant and comprehensive study of the indigenous yeast community present at the surface of Vitis labrusca (L. cultivars and its hybrids) grapes from the Azores archipelago (Drumonde-Neves et al., 2016). We also found C. azymoides, C. californica and P. cecembensis on Isabella grape must, which have not been recognized on Vitis vinifera L. grapes. Interestingly, C. azymoides and P. cecembensis have also been found on Vitis labrusca (L. cultivars and its hybrids) grapes from the Azores archipelago (Drumonde-Neves et al., 2016). It is remarkable that these two yeast species are present on Vitis labrusca L. grapes from two such distant regions in the world (i.e., Azores archipelago and Argentina), corresponding to different geographic regions and production conditions in their respective vineyards. Moreover, C. azymoides and P. cecembensis were not found in grape musts from Vitis vinifera L. vineyards in the Azores archipelago (Drumonde-Neves et al., 2017). In our study, we did not find Barnettozyma californica, an additional potential specific yeast from Vitis labrusca L. grapes (Drumonde-Neves et al., 2016). Taken together, the study of Drumonde-Neves et al. (2016) and the results reported here strongly suggest that at least C. azymoides and P. cecembensis are preferentially associated with the microbiota of Vitis labrusca L. grapes. To our knowledge, these observations constitute the first data about specific associations between Vitis and yeast species. This finding may have significant ecological and/or evolutionary interest as well as for the understanding of the spontaneous assembly of the Vitis microbiota.
Discussion
Many yeast species have been recognized in spontaneously fermenting must from Vitis vinifera L. grapes (Combina et al., 2005; Zott et al., 2008; Aponte and Bialotta, 2016; Padilla et al., 2016a). Interestingly, the yeast microbiota of Isabella grape must reported here, as well as recently elsewhere (Drumonde-Neves et al., 2016), show a remarkable diversity of non-Saccharomyces yeast species. In Vitis vinifera L. grapes, the more relevant non-Saccharomyces species include the apiculate yeasts Hanseniaspora and a large proportion of yeasts from the Metschnikowia and Candida genera (Garofalo et al., 2015; Albergaria and Arneborg, 2016). H. uvarum, present at later stages during fermentation of Vitis vinifera L. grape must (Zott et al., 2008; Padilla et al., 2016a), was also found in spontaneously fermenting Isabella (Baffi et al., 2010; Bezerra-Bussoli et al., 2013; Hong and Park, 2013). Isabella H. uvarum, however, markedly decreased at the mid-stages of fermentation. In fact, H. vineae was the only non-Saccharomyces species able to coexist with S. cerevisiae until relatively advanced stages of fermentation of Isabella grape must.
The main non-Saccharomyces species contributing to the initial stages of Isabella must fermentation was S. bacillaris. This represented 62.5% of the strains isolated at t0, increasing to 75% of the total yeast population at t24, and it was not isolated at t48. This behavior was unexpected, as S. bacillaris appears to develop similarly to other non-Saccharomyces yeasts in mixed fermentations of Vitis vinifera L. musts (Wang et al., 2016). Moreover, similar studies identified S. bacillaris at advanced stages of fermentation coexisting with S. cerevisiae (Zott et al., 2008; Tofalo et al., 2011; Padilla et al., 2016a). Indigenous S. bacillaris and S. cerevisiae strains isolated from Isabella must showed a relatively low tolerance to ethanol. Considering the relatively low average levels of total reducing sugars on Isabella grapes from Colonia Caroya, these findings could highlight a low selective pressure for ethanol tolerance on yeast communities in these vineyards. In the case of S. bacillaris, however, it has been suggested that this species is not under selective pressure in winemaking environments (Masneuf-Pomarede et al., 2015).
Fourteen different non-Saccharomyces yeast species were identified at initial stages of spontaneous fermentation of Isabella must. Although some of these yeast species have been described in winemaking ecosystems (i.e., H. uvarum, S. bacillaris, T. delbrueckii, H. vineae, I. hanoiensis) (Zott et al., 2008; Albertin et al., 2014; Padilla et al., 2016a; Drumonde-Neves et al., 2017), other species such as C. azymoides, C. californica and P. cecembensis, have not previously been isolated from Vitis vinifera L. grapes. This observation reinforces the interest in searching for wine yeast diversity in ecological niches alternative to the traditional Vitis vinifera L. environments (Combina et al., 2005; Knight et al., 2015; Capece et al., 2016; Vigentini et al., 2016). For example, valuable indigenous H. uvarum strains, isolated from Vitis Labrusca (cultivar Campbell Early) grape must, have proved to be useful starters for winemaking (Hong and Park, 2013).
Interestingly, C. azymoides and P. cecembensis have recently been recognized on the surface of Vitis labrusca L. grapes from vineyards in the Azores archipelago (Drumonde-Neves et al., 2016). This could be explained by the fortuitous presence of rare yeast species on Vitis labrusca L. grapes from the Azores Archipelago, appearing because of particular environmental influences and/or vineyard and/or winery production conditions. C. azymoides and P. cecembensis, however, were not present on Vitis vinifera L. grape musts from the same geographic location (Azores Archipelago) (Drumonde-Neves et al., 2017). Moreover, the finding of C. azymoides and P. cecembensis during spontaneous fermentation of Isabella grapes harvested in Argentina strongly suggests that at least these two yeast species are associated with Vitis labrusca L. grapes, independently of their geographic origin and/or the associated human interventions. We propose that C. azymoides, C. californica and P. cecembensis are yeast species preferentially associated with Vitis labrusca L. grapes. Specific Vitis-microbial interactions may underlie the assembly of specific grapevine yeast communities. Modern metagenomic approaches (Wolfe and Dutton, 2015) may permit the composition and dynamics of the microbiota of grapes from various Vitis species to be explored, offering more detailed information on microbial diversity than is retrieved from culture-based studies (Bokulich et al., 2014; Pinto et al., 2014; Setati et al., 2015).
This work reports a detailed analysis on the identity and dynamics of the yeast community present during spontaneous fermentation of grape must from a Vitis cultivar different from Vitis vinifera L. The collection of strains isolated in this work constitutes a potential source of valuable non-Saccharomyces and S. cerevisiae strains for enology research as well as for biological, ecological and evolutionary studies on wine yeasts. The study represents the first characterization of indigenous grapevine yeasts from the province of Córdoba, Argentina.
Author Contributions
Fundamental contributions to the conception and design of the work (MR, CR, SL, AR); acquisition, analysis and interpretation of data (MR, CR, SL, AR); drafting of the work and revising it critically for intellectual content (MR, AR). All authors approved the final version of the manuscript to be submitted for publication and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy and integrity of any part of the work are appropriately investigated and resolved.
Funding
This work was supported by PICT2014-313 from the FONCYT (Argentina) and a grant from the Catholic University of Córdoba. MR is supported by a fellowship from the Argentina National Research Council (CONICET) and AR is a Principal Investigator of CONICET (Argentina).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
The authors wish to thank H. Paci Horno for initial help with yeast strain isolation and C. Gaggero for critical reading and suggestions on the manuscript.
References
Albergaria, H., and Arneborg, N. (2016). Dominance of Saccharomyces cerevisiae in alcoholic fermentation processes; role of physiological fitness and microbial interactions. Appl. Microbiol. Biotechnol. 100, 2035–2046. doi: 10.1007/s00253-015-7255-0
Albertin, W., Miot-Sertier, C., Bely, M., Marullo, P., Coulton, J., Moine, V., et al. (2014). Oenological prefermentation practices strongly impact yeast population dynamics and alcoholic fermentation kinetics in Chardonnay grape must. Int. J. Food Microbiol. 178, 87–97. doi: 10.1016/j.ijfoodmicro.2014.03.009
Almanza-Merchán, P. J., Reyes-M, A. J., Ayala, M. L., Balaguera-L, W., and Serrano-Cely, P. A. (2015). Sensory evaluation of the wine grape Isabella (Vitis labrusca L.) hand crafted. Rev. Cien. Agric. 12, 71–81. doi: 10.19053/01228420.4393
Aponte, M., and Bialotta, G. (2016). Potential role of yeast strains isolated from grapes in the production of taurasi DOCG. Front. Microbiol. 7:809. doi: 10.3389/fmicb.2016.00809
Baffi, M. A., dos Santos, C., Arévalo-Villena, M., Briones-Pérez, A. I., Gomes, E., and Da Silva, R. (2010). Isolation and molecular identification of wine yeasts from a Brazilian vineyard. Ann. Microbiol. 61, 75–78. doi: 10.1007/s13213-010-0099-z
Baǧder Elmac, S., Özçelik, F., Tokatl, M., and Çakr, Ý. (2014). Technological properties of indigenous wine yeast strains isolated from wine production regions of Turkey. Antonie Van Leeuwenhoek 105, 835–847. doi: 10.1007/s10482-014-0138-z
Barata, A., Malfeito-Ferreira, M., and Loureiro, V. (2012). The microbial ecology of wine grape berries. Int. J. Food. Microbiol. 153, 243–259. doi: 10.1016/j.ijfoodmicro.2011.11.025
Bezerra-Bussoli, C., Baffi, M. A., Gomes, E., and Da-Silva, R. (2013). Yeast diversity isolated from grape musts during spontaneous fermentation from a Brazilian winery. Curr. Microbiol. 67, 356–361. doi: 10.1007/s00284-013-0375-9
Bokulich, N. A., Thorngate, J. H., Richardson, P. M., and Mills, D. A. (2014). Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc. Natl. Acad. Sci. U.S.A. 111, 139–148. doi: 10.1073/pnas.1317377110
Capece, A., Granchi, L., Guerrini, S., Mangani, S., Romaniello, R., Vicenzini, M., et al. (2016). Diversity of Saccharomyces cerevisiae strains isolated from two italian wine-producing regions. Front. Microbiol. 7:1018. doi: 10.3389/fmicb.2016.01018
Combina, M., Elía, A., Mercado, L., Catania, C., Ganga, A., and Martinez, C. (2005). Dynamics of indigenous yeast populations during spontaneous fermentation of wines from Mendoza, Argentina. Int. J. Food Microbiol. 99, 237–243. doi: 10.1016/j.ijfoodmicro.2004.08.017
de Ullivarri, M. F., Mendoza, L. M., and Raya, R. R. (2014). Killer activity of Saccharomyces cerevisiae strains: partial characterization and strategies to improve the biocontrol efficacy in winemaking. Antonie Van Leeuwenhoek 106, 865–878. doi: 10.1007/s10482-014-0256-7
Drumonde-Neves, J., Frando-Duarte, R., Lima, T., Schuller, D., and Pais, C. (2016). Yeast biodiversity in vineyard environment is increased by human intervention. PLoS ONE 11:e0160579. doi: 10.1371/journal.pone.0160579
Drumonde-Neves, J., Frando-Duarte, R., Lima, T., Schuller, D., and Pais, C. (2017). Association between grape yeast communities and the vineyard ecosystems. PLoS ONE 12:e0169883. doi: 10.1371/journal.pone.0169883
Fleet, G. H. (2003). Yeast interactions and wine flavour. Int. J. Food Microbiol. 86, 11–22. doi: 10.1016/S0168-1605(03)00245-9
Garofalo, C., Tristezza, M., Griecco, F., Spano, G., and Capozzi, V. (2015). From grape to wine: population dynamics of cultivable yeasts associated to “Nero di Troia” autochtonous grape cultivar. World J. Microbiol. Biotechnol. 32, 59. doi: 10.1007/s11274-016-2017-4
Granchi, L., Bosco, M., Messini, A., and Vicenzini, M. (1999). Rapid detection and quantification of yeast species during spontaneous wine fermentation by PCR-RFLP analysis of the rDNA ITS región. J. Appl. Microbiol. 87, 949–956. doi: 10.1046/j.1365-2672.1999.00600.x
Grangeteau, C., Roullier-Gall, C., Rousseaux, S., Gougeon, R. D., Schmitt-Kopplin, P., Alexandre, H., et al. (2016). Wine microbiology is driven by vineyard and winery anthropogenic factors. Microb. Biotechnol. 10, 354–370. doi: 10.1111/1751-7915.12428
Guillamón, J. M., Sabaté, J., Barrio, E., Cano, J., and Querol, A. (1998). Rapid identification of wine yeast species based on RFLP analysis of the ribosomal internal transcribed spacer (ITS) region. Arch. Microbiol. 169, 387–392. doi: 10.1007/s002030050587
Hernandez, C. J. D., Trujillo, N. Y. Y., and Durán, O. D. S. (2011). Phenolic potential determination and yeasts identification with significant leavens in Isabella grape (Vitis labrusca) from Villa del Rosario (Norte de Santander) for winemaking. Vitae 18, 17–25.
Hierro, N., González, A., Mas, A., and Guillamón, J. M. (2006). Diversity and evolution of non-Saccharomyces yeast populations during wine fermentation: effect of grape ripeness and cold maceration. FEMS Yeast Res. 6, 102–111. doi: 10.1111/j.1567-1364.2005.00014.x
Hong, Y. A., and Park, H. D. (2013). Role of non-Saccharomyces yeasts in Korean wines produced from Campbell Early grapes: potential use of Hanseniaspora uvarum as a starter culture. Food Microbiol. 34, 207–214. doi: 10.1016/j.fm.2012.12.011
Hurrel, J. A., Cabanillas, P., Guerrero, E. L., and Delucchi, G. (2014). Naturalization and ethnobotany of Vitis labrusca L. (Vitaceae) in the Rio de la Plata region, Argentina. Rev. Mus. Argentino cienc. Nat. 16, 13–18.
James, S. A., Cai, J., Roberts, I. N., and Collins, M. D. (1997). A phylogenetic analysis of the genus Saccharomyces based on 18S rRNA gene sequences: description of Saccharomyces kunashirensis sp. nov. and Saccharomyces martiniae sp. nov. Int. J. Syst. Bacteriol. 47, 453–460. doi: 10.1099/00207713-47-2-453
Jolly, N. P., Varela, C., and Pretorius, I. S. (2013). Not your ordinary yeast: non-Saccharomyces yeasts in wine production uncovered. FEMS Yeast Res. 14, 215–237. doi: 10.1111/1567-1364.12111
Knight, S., Klaere, S., Fedrizzi, B., and Goddard, M. R. (2015). Regional microbial signatures positively correlate with differential wine phenotypes: evidence for a microbial aspect to terroir. Sci. Rep. 5:14233. doi: 10.1038/srep14233
Kurtzman, C. P., and Robnett, C. J. (1998). Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek 73, 331–371. doi: 10.1023/A:1001761008817
Lopes, C. A., and Sangorrín, M. P. (2010). Optimization of killer assays for yeast selection protocols. Rev. Argent. Microbiol. 42, 298–306. doi: 10.1590/S0325-75412010000400011
Masneuf-Pomarede, I., Juquin, E., Miot-Sertier, C., Renault, P., Laizet, Y., Salin, F., et al. (2015). The yeast Starmerella bacillaris (synonym Candida zemplinina) shows high genetic diversity in winemaking environments. FEMS Yeast Res. 15:fov045. doi: 10.1093/femsyr/fov045
Mortimer, R., and Polsinelli, M. (1999). On the origins of wine yeast. Res. Microbiol. 150, 199–204. doi: 10.1016/S0923-2508(99)80036-9
Padilla, B., García-Fernández, D., González, B., Izidoro, I., Esteve-Zarozo, B., Beltrán, G., et al. (2016a). Yeast biodiversity from DOQ priorat uninoculated fermentations. Front. Microbiol. 7:930. doi: 10.3389/fmicb.2016.00930
Padilla, B., Gil, J. V., and Manzanares, P. (2016b). Past and future of non-Saccharomyces yeasts: from spoilage microorganisms to biotechnological tools for improving wine aroma complexity. Front. Microbiol. 7:411. doi: 10.3389/fmicb.2016.00411
Pérez-Nevado, F., Albergaria, H., Hogg, T., and Girio, F. (2006). Cellular death of two non-Saccharomyces wine-related yeasts during mixed fermentations with Saccharomyces cerevisiae. Int. J. Food. Microbiol. 108, 336–345.
Pinto, C., Pinho, D., Sousa, S., Pinheiro, M., Egas, C., and Gomes, A. C. (2014). Unravelling the diversity of grapevine microbiome. PLoS ONE 16:e85622. doi: 10.1371/journal.pone.0085622
Setati, M. E., Jacobson, D., and Bauer, F. F. (2015). Sequence-based analysis of the Vitis vinifera L. cv cabernet sauvignon grape must mycobiome in three South African vineyards employing distinct agronomic systems. Front. Microbiol. 6:1358. doi: 10.3389/fmicb.2015.01358
Šuranská, H., Vránová, D., and Omelková, J. (2016). Isolation, identification and characterization of regional indigenous Saccharomyces cerevisiae strains. Braz. J. Microbiol. 47, 181–190. doi: 10.1016/j.bjm.2015.11.010
Tofalo, R., Schirone, M., Torriani, S., Rantsiou, K., Cocolin, L., Perpetuini, G., et al. (2011). Diversity of Candida zemplinina stains from grapes and Italian wines. Food Microbiol. 29, 18–26. doi: 10.1016/j.fm.2011.08.014
Varela, C., and Borneman, A. R. (2016). Yeast found in vineyards and wineries. Yeast 34, 111–128. doi: 10.1002/yea.3219
Vigentini, I., Maghradze, D., Petrozziello, M., Bonello, F., Mezzapelle, V., Valdetara, F., et al. (2016). Indigenous gregorian wine-associated yeasts and grape cultivars to edit the wine quality in a precision oenology perspective. Front. Microbiol. 7:352. doi: 10.3389/fmicb.2016.00352
Wang, C., Mas, A., and Esteve-Zarzoso, B. (2016). The Interaction between Saccharomyces cerevisiae and non-Saccharomyces yeast during alcoholic fermentation is species and strain specific. Front. Microbiol. 7:502. doi: 10.3389/fmicb.2016.00502
White, T., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungi ribosomal RNA genes for phylogenetics,” in PCR Protocols: A Guide to Methods and Applications, eds M. Innis, D. Gelfand, J. Sninsky, and T. White (San Diego, CA: Academic Press), 315–322.
Wolfe, B. E., and Dutton, R. J. (2015). Fermented foods as experimentally tractable microbial ecosystems. Cell 161, 49–55. doi: 10.1016/j.cell.2015.02.034
Keywords: Vitis labrusca L., Isabella, grapes, spontaneous fermentation, wine, microbiota, yeast
Citation: Raymond Eder ML, Reynoso C, Lauret SC and Rosa AL (2017) Isolation and Identification of the Indigenous Yeast Population during Spontaneous Fermentation of Isabella (Vitis labrusca L.) Grape Must. Front. Microbiol. 8:532. doi: 10.3389/fmicb.2017.00532
Received: 23 January 2017; Accepted: 14 March 2017;
Published: 29 March 2017.
Edited by:
Kate Howell, University of Melbourne, AustraliaReviewed by:
Rossana Sidari, Mediterranea University of Reggio Calabria, ItalyBraulio Esteve-Zarzoso, Universitat Rovira i Virgili, Spain
Copyright © 2017 Raymond Eder, Reynoso, Lauret and Rosa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alberto L. Rosa, alberto_l_rosa@yahoo.com.ar
 María L. Raymond Eder
María L. Raymond Eder Cristina Reynoso2
Cristina Reynoso2 Alberto L. Rosa
Alberto L. Rosa