- 1Applied Chemistry and Biochemical Engineering Course, Department of Engineering, Graduate School of Integrated Science and Technology, Shizuoka University, Hamamatsu, Japan
- 2Graduate School of Science and Technology, Shizuoka University, Hamamatsu, Japan
- 3Japan Collection of Microorganisms, RIKEN BioResource Center, Tsukuba, Japan
- 4Department of Applied Chemistry and Biochemical Engineering, Faculty of Engineering, Shizuoka University, Hamamatsu, Japan
- 5Research Institute of Green Science and Technology, Shizuoka University, Shizuoka, Japan
Membrane vesicles (MVs) are secreted from a wide range of microbial species and transfer their content to other cells. Although MVs play critical roles in bacterial communication, whether MVs selectively interact with bacterial cells in microbial communities is unclear. In this study, we investigated the specificity of the MV-cell interactions and evaluated the potential of MVs to target bacterial cells for delivery. MV association with bacterial cells was examined using a fluorescent membrane dye to label MVs. MVs derived from the enterobacterium Buttiauxella agrestis specifically interacted with cells of the parent strain but interacted less specifically with those of other genera tested in this study. Electron microscopic analyses showed that MVs were not only attached on B. agrestis cells but also fused to them. The interaction energy, which was characterized by hydrodynamic diameter and zeta potential based on the Derjaguin–Landau–Verwey–Overbeek (DLVO) theory, was significant low between MVs and cells in B. agrestis, compared to those between B. agrestis MVs and cells of other genera. Similar specific interaction was also occurred between B. agrestis MVs and cells of six other species belonging to Buttiauxella spp. B. agrestis harboring plasmid pBBR1MCS-1 secreted plasmid-containing MVs (p-MVs), and plasmid DNA in p-MVs was transferred to the same species. Moreover, antibiotic-associated MVs enabled effective killing of target species; the survival rate of B. agrestis was lower than those of Escherichia coli and Pseudomonas aeruginosa in the presence of gentamicin-associated MVs derived from B. agrestis. Altogether, we provide the evidence that MVs selectively interact with target bacterial cells and offer a new avenue for controlling specific bacterial species using bacterial MVs in microbial communities.
Introduction
Many bacteria and archaea produce small bilayered particles (20–200 nm) from their surfaces (Mashburn-Warren and Whiteley, 2006; Deatherage and Cookson, 2012). These microbial liposomes are called membrane vesicles (MVs) or outer membrane vesicles (OMVs), particularly in gram-negative bacteria. MVs are composed of proteins, phospholipids, and polysaccharides and contain proteins, DNA, RNA and, in some cases, quorum sensing signals (Mashburn and Whiteley, 2005; Kulp and Kuehn, 2010; Toyofuku et al., 2014; Schwechheimer and Kuehn, 2015; Turnbull et al., 2016). MVs have multifunctional roles, such as transferring nucleic acids or toxic compounds to other cells, promoting the formation and maintenance of biofilm, releasing unnecessary compounds from cells, providing resistance to antibiotics and phages and extending the membrane for extracellular electron transport (Tashiro et al., 2012; Biller et al., 2014; Pirbadian et al., 2014; Schwechheimer and Kuehn, 2015; Toyofuku et al., 2015; Roier et al., 2016). Importantly, substances such as virulence factors and genetic materials are highly concentrated in MVs and remain stable against environmental stresses.
Microbes interact with each other in multicellular communities, and this interaction includes both cooperative and offensive functions for bacterial survival (Shank and Kolter, 2009; Okabe et al., 2010; Tashiro et al., 2013). MVs play important roles in microbial interactions, as they transfer their contents to other microbial cells, even over long distances. Several previous studies have shown that MVs are involved in DNA transfer among bacteria (Dorward et al., 1989; Kolling and Matthews, 1999; Yaron et al., 2000; Klieve et al., 2005; Chiura et al., 2011; Fulsundar et al., 2014; Ho et al., 2015). Gene transfer via MVs has attracted particular interest because DNA encapsulated by MVs is stable against DNase (Renelli et al., 2004), enabling the long-distance transport of cytoplasmic contents and resulting in horizontal gene transfer. Additionally, MVs package QS signals, such as Pseudomonas quinolone signals and hydrophobic homoserine lactones (Mashburn and Whiteley, 2005; Li et al., 2016). QS signals associated with MVs are transferred to bacterial cells and control QS-regulated gene expression (Tashiro et al., 2010). MVs also have roles in cell-cell inhibition and killing among competing species (Berleman and Auer, 2013). Lytic activities have been reported in MVs derived from Pseudomonas aeruginosa, Shigella flexneri, and Myxococcus xanthus (Kadurugamuwa and Beveridge, 1996, 1999; Kadurugamuwa et al., 1998; Evans et al., 2012). Thus, MVs play crucial roles in intra- and interspecies communication; however, in bacterial cell–cell interactions via MVs, the recipients of MVs are not determined or not fully understood (Hasegawa et al., 2015). Elucidating the selectivity in MV interactions with bacterial cells is critical for an improved understanding of bacterial interactions in communities.
Despite the fact that the targeted delivery of signaling and lytic molecules via MVs in microbial communities has not been fully characterized, several studies have indicated that MVs derived from pathogens enable the target delivery of the interior toxins to host cells (Kesty et al., 2004; Bauman and Kuehn, 2009; Parker et al., 2010). Transmembrane or other surface-exposed proteins encode specific peptide sequences and recognize target cells or tissues. The presence of specific peptides on the surface of MVs also enables cell targeting (Alves et al., 2015). For example, addition of the Ail peptide from Yersinia pestis to MVs derived from Escherichia coli promoted delivery of the MV contents to eukaryotic cells in vitro (Kesty and Kuehn, 2004), and E. coli MVs with an anti-HER2 affibody on their surface could selectively deliver their contents to tumor cells. Thus, engineering MVs involves investigating both outer membrane protein adhesion for the host cell interaction with MVs and developing strategies for utilizing MVs as nanomedicines to achieve a cell-specific drug delivery system.
In the current study, we investigated whether bacterial MVs deliver their content to specific bacterial cells. Understanding the selective delivery of MVs to target bacterial cells offers a new avenue for controlling bacterial cells in heterogeneous samples. As a result of this screening, we found that MVs derived from the enterobacterium Buttiauxella agrestis CUETM77-167 specifically interacted with the same species. We used this bacterium as a model organism and characterized the specific interaction of its MVs. First, we examined the electrostatic energy between MVs and cells and showed that there was a low electrostatic energy between them in Buttiauxella spp. Next, we found that MVs attached to bacterial cells and transferred their contents, including a plasmid, to bacterial cells. Finally, we investigated whether the specific interaction of MVs enabled them to deliver an antibiotic to control target cells.
Materials and Methods
Microbial Strains, Plasmids, and Primers
The microbial strains, plasmids, and primers used in this study are listed in Table 1. Microbial cells were grown in tryptic soy broth (TSB: Becton, Dickinson and Company, Franklin Lakes, NJ, USA) medium shaken at 200 rpm. Buttiauxella spp., Corynebacterium glutamicum, Micrococcus luteus, Flavobacterium johnsoniae, Rhizobium spp., and Pseudomonas alcaligenes were grown at 30°C. Bacillus subtilis, Hydrogenophaga pseudoflava, E. coli, Erwinia persicina, and P. aeruginosa were grown at 37°C. For genetic manipulations, LB (Luria-Bertani Lennox: 1% w/v tryptone, 0.5% yeast extract and 0.5% NaCl) was used. When necessary, chloramphenicol was used at a concentration of 20 μg/mL for E. coli and B. agrestis. E. coli β2163 was used for conjugation to transfer the plasmid pBBR1MCS-1 to B. agrestis. DAPA (2,6-diaminopimelic acid) was used at a concentration of 60 μg/mL in LB medium for the growth of E. coli β2163.
Vesicle Extraction and Purification
Membrane vesicle extraction and purification were carried out as previously described (Tashiro et al., 2009). To obtain a sample of MVs without bacterial cells, 100 mL of overnight batch culture was centrifuged for 15 min at 6,000 × g, 4°C. The supernatant was filtered through 0.45 and 0.20 μm membrane filters and ultracentrifuged for 2 h at 100,000 × g and 4°C using an angle rotor (P45AT, Hitachi, Tokyo, Japan). The pellets were resuspended in 200 μL of 50 mM HEPES-0.85% NaCl (HEPES-NaCl buffer). For MV purification, MVs were labeled with 100 μg/mL FM4-64 in HEPES-NaCl buffer and washed using ultracentrifugation. MV samples were adjusted to 1 mL of 45% (w/v) iodixanol (OptiPrep; Axis-Shield Diagnostics Ltd., Dundee, UK) in HEPES-NaCl, transferred to the bottom of ultracentrifuge tubes, and layered with iodixanol-HEPES-NaCl (2 mL of 40, 35, 30, 25, and 20%). The samples were ultracentrifuged for 3 h at 100,000 × g and 4°C using a swing rotor (P40ST, Hitachi). Then, 500 μL fractions were collected from each gradient. To confirm the fraction containing MVs, the phospholipid and protein concentrations in each fraction were measured using the Stewart assay and the bicinchoninic acid (BCA) protein assay. The fraction containing MVs was ultracentrifuged and resuspended in HEPES-NaCl.
Quantification of Vesicle Production
Membrane vesicle production was quantified by determining the phospholipid concentration in the bacterial culture supernatant. The phospholipid concentration was measured using a previously reported method with some modifications (Stewart, 1980). Briefly, 1 mL of the supernatant filtered through a 0.20-μm membrane, 200 μL of concentrated ammonium ferrothiocyanate solution (135 g/L iron (III) chloride hexahydrate and 152 g/L ammonium thiocyanate), and 200 μL of chloroform were vortexed. The absorption of the chloroform layer (lower layer) was measured at 488 nm. To construct a calibration curve, L-α-phosphatidylethanolamine was used as a reference standard. MV production was normalized to the quantity of cell protein. Bacterial cells were lysed by 5% SDS, and the protein concentration was determined by a BCA protein assay (Pierce BCA Protein Assay Kit; Thermo, Rockford, IL, USA).
Vesicle Association Assay
The MV association degree was determined by the relative fluorescence units (RFUs) of fluorescently labeled MVs per concentration of cell protein. The extracted MVs (200 μg phospholipid) were incubated with 5 μg/mL FM4-64 in 1 mL of phosphate-buffered saline (PBS) for 30 min at 30°C and washed three times in PBS. In some cases, 100 μg/mL fluorescein isothiocyanate (FITC) or 5 μM DiO was used instead of FM4-64. Bacterial cells grown to the late exponential phase were washed in PBS and diluted to an OD600 of 1.0. FM4-64-labeled MVs (20 μg of phospholipid) were incubated with 1 mL of bacterial cells (OD600 of 1.0) for 30 min at 30°C. Bacterial cells with FM4-64-labeled MVs were washed in PBS. The fluorescence of FM4-64 was measured using a microplate reader (Infinite M200; TECAN, Männedorf, Switzerland), and the cell protein concentration was measured by a BCA protein assay.
Flow Cytometry Analysis
Flow cytometry analysis using an EPICS Altra AJ49030 system (Beckman Coulter, Brea, CA, USA) was performed to detect cells associated with fluorescence-labeled MVs. MVs (200 μg of phospholipid) were labeled with FM4-64 and washed in PBS by ultracentrifugation. Bacterial cells were adjusted to OD600 = 1.0 in PBS, labeled with 100 μg/mL FITC for 30 min at 30°C and washed three times in PBS. FM4-64-labeled MVs and FITC-labeled bacterial cells were incubated for 30 min at 30°C and washed with PBS three times. The particle number and fluorescence intensity of bacterial cells associated with MVs were analyzed by flow cytometry.
Microscopy Observation
For fluorescence microscopy analysis, the cells were examined using the fluorescence microscope OLYMPUS BX53 (OLYMPUS, Tokyo, Japan), and images were taken with the CCD camera DP72 (OLYMPUS) and the imaging software CellSens (OLYMPUS). Bacterial cells were adjusted to OD600 = 1.0 in PBS, labeled with 100 μg/mL FITC for 30 min at 30°C and washed three times in PBS. FM4-64-labeled MVs and FITC-labeled bacterial cells were incubated for 30 min at 30°C and washed with PBS three times.
For transmission electron microscopy observations of purified MVs, MVs were placed on Cu400 mesh grids, stained with 2% uranyl acetate and visualized by JEM100EX (JEOL, Tokyo, Japan) by the Hanaichi Ultrastructure Research Institute (Okazaki, Aichi, Japan). For other TEM analyses, the samples were placed on Cu200 mesh grids that had been treated with 0.01% poly-L-lysine in PBS. The grids were rinsed with distilled water and stained with 2% phosphotungstic acid neutralized with KOH. After rinsing with distilled water and dried, the samples were visualized by JEM 2000FX-II (JEOL).
Immunogold Labeling
The extracted MVs were labeled in 100 μg/mL FITC and washed with PBS by ultracentrifugation. Bacterial cells were adjusted to OD600 = 1.0 in PBS, incubated with FITC-labeled MVs for 30 min at 30°C and washed with PBS three times. The samples were placed on Cu200 mesh grids that had been treated with 0.01% poly-L-lysine in PBS. The bacteria were fixed with 4% paraformaldehyde in PBS for 30 min and washed with PBS. The specimens were incubated with 1% bovine serum albumin (BSA) in PBS for 5 min and washed again with PBS. The specimens were then incubated with anti-FITC antibody (Sigma–Aldrich) at a dilution of 1:300 in PBS containing 1% BSA for 60 min. The specimens were washed with PBS, incubated with colloidal gold-labeled anti-rabbit immunoglobulin (diameter of colloidal gold particles, 15 nm) at a dilution of 1:1000 in PBS containing 1% BSA for 60 min, and then washed. The grids were fixed with 2% glutaraldehyde in PBS for 15 min and washed with PBS. After being stained with 2% phosphotungstic acid and washed, the samples were observed by JEM 2000FX-II.
Physicochemical Analysis of Particles
The particle size distribution and zeta potential were analyzed with a Zetasizer Nano ZS particle analyzer (Malvern Instruments, Malvern, UK) at 25°C. Purified MVs were adjusted to a concentration of 20 μg of phospholipid/mL in PBS. Cells grown to the stationary phase in TSB were washed with PBS and adjusted to OD600 = 0.1. The hydrodynamic zeta average diameter was calculated by the light scattering method, and the zeta potential was determined by applying the Smoluchowski approximation. The electrostatic charge on the surfaces of MVs and cells was characterized as the zeta potential.
Calculating the Interaction Energy between Cells and Vesicles
The potential interaction energy between cells and vesicles was calculated based on the Derjaguin–Landau–Verwey–Overbeek (DLVO) theory. The interaction energy Vtotal was defined as:
where VA is an attractive London-van der Waals interaction, and VR is an electric repulsive interaction energy. Based on previous studies (Ohki and Ohshima, 1999), VA was defined as follows:
where
where A is the Hamaker constant; a and a′ are the radii of the cells and vesicles, respectively; R is the separation distance between the center of the particles of cells and vesicles; and r is their separation distance. Hamaker constant A is described as follows:
where q is the volume density, and aaa is London-van der Waals constant. A of the phospholipid bilayer was expressed as 4 × 10-19 N (Ohki et al., 1982).
According to a previous report (Ohki and Ohshima, 1999), VR was defined as follows:
where φ and φ′ are the Stern potentials of cells and vesicles, respectively, and κ is the Debye constant. The values of the Stern potentials were used as zeta potentials (ζ) obtained by the experiments. When the absolute values of φ and φ′ were less than 60 mV (κr > 1), VR was approximated as follows:
where 𝜀r is the relative permittivity for the medium, and 𝜀0 is the permittivity of a vacuum. The Debye constant κ was expressed by the following:
where e is the elementary charge, NA is Avogadro’s number, c is the ion concentration, z is the charge of ions and k is the Boltzmann constant.
Vesicle-mediated Gene Transfer
Experiments for vesicle-mediated gene transfer were based on a previously published method (Yaron et al., 2000). The donor strain, B. agrestis CUETM77-167/pBBR1MCS-1, was grown in LB medium containing chloramphenicol at 200 rpm for 16 h at 30°C. MVs were extracted using sterile ultracentrifuge tubes after filtering the supernatant. Extracted MVs were reacted with DNase I in reaction buffer [40 mM Tris-HCl (pH 7.9), 10 mM NaCl, 6 mM MgCl2, and 1 mM CaCl2], and the extracellular DNA around the MVs was degraded. Then, the MVs were washed with PBS by ultracentrifugation. MVs (20 μg of phospholipids) and approximately 1,000 recipient bacterial cells of B. agrestis were mixed in 1 mL of PBS, and the cells were washed with PBS at each time point. The transfer of plasmid DNA was examined by counting the colony forming units (CFUs) on agar medium containing chloramphenicol. As a control experiment, naked plasmid DNA, which was extracted from B. agrestis CUETM77-167/pBBR1MCS-1, was added to the cell suspension (final DNA concentration was 10 ng/mL), and the possibility of natural transformation was examined by CFU counting. PCR analysis with a primer pair (CamR-f and CamR-r, listed in Table 1), and plasmid extraction were conducted to confirm whether the obtained colonies harbored the plasmid pBBR1MCS-1.
Real-time PCR Analysis
The copy number of the plasmid was determined using real-time PCR analysis. The chloramphenicol acyl transferase gene in pBBR1MCS-1 was amplified and quantified using LightCycler FastStart DNA Master SYBR Green I (Roche, Basel, Switzerland) with a primer pair (CamR-f and CamR-r) on a LightCycler 2.0 (Roche) according to the instruction manual.
Nanoparticle Tracking Analysis
The concentration of MVs was measured by nanoparticle tracking analysis using a NanoSight LM10 instrument (Malvern) equipped with sCMOS camera (Andor, Belfast, UK). The samples were diluted to an average concentration of 106–109 particles per milliliter in 50 mM HEPES buffer. Five replicates videos were collected from each sample, and particle movement was analyzed by NTA software (Version 3.1, Malvern). The velocity of particle movement was used to calculate the particle size using the two-dimensional Stokes-Einstein equation.
Vesicle-Mediated Antibiotic Transfer
Gentamicin-containing MVs (g-MVs) were prepared as previously described (Kadurugamuwa and Beveridge, 1995). Briefly, B. agrestis CUETM77-167 was grown in TSB medium until the late stationary phase, and gentamicin was added to a final concentration of 32 μg/mL [four times the minimal inhibitory concentration (MIC) of gentamicin for this strain]. After a further 30 min of cultivation, MVs were extracted from the bacterial culture. The amount of gentamicin in the MV suspension was determined by examining the bacterial killing effect of homogenized MVs. The concentration of gentamicin in MVs was calculated using the number of MVs in the MV suspension. To analyze the killing effect on a single bacterial species, MVs (20 μg of phospholipids) and approximately 1,000 bacterial cells were mixed in 1 mL of PBS, and the cells were washed with PBS at each time point. The surviving cells were counted by CFU. To analyze the killing effect in mixed bacterial culture, MVs (20 μg of phospholipids) and approximately 1,000 cells of each bacterial species (B. agrestis CUETM77-167 and E. coli DH5α) were mixed in 1 mL of PBS, and the cells were washed with PBS at each time point. The surviving cells were counted by CFU on an agar plate containing 200 μg/mL 5-bromo-4-chloro-indolyl β-D-galactopyranoside (X-gal) and 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). B. agrestis and E. coli DH5α showed blue and white colonies, respectively.
Determination of the Antibiotic Concentration in Vesicles
Vesicles associated with gentamicin were homogenized by an ultrasonic disintegrator. The suspended lysed vesicles containing gentamicin were diluted and added to LB agar on a 24-well plate before the agar had gelatinized. A cell suspension of E. coli MG1655 was grown on the plate, and the result was compared to that on an agar plate containing gentamicin. Using the MIC, the gentamicin concentration in the sample was determined. The vesicle particle number and average particle size were measured by nanoparticle tracking analysis, and the gentamicin concentration in the vesicle particle was calculated.
Phylogenetic Analysis
Sequence data were accessed from NCBI (GenBank accession numbers are AJ233400 to AJ233406 and NC_000913). 16S rRNA gene sequences were aligned using the ClustalW program, and distance matrix trees were constructed by the neighbor-joining method. The topology of the trees was evaluated by bootstrapping with 1,000 replicates, and the percentages of values are shown on branching points.
Statistical Analysis
Error bars throughout the figures refer to the standard error for all of the experiments. The statistical significance of the differences between groups was examined using unpaired and one-way Student’s t-tests. A P-value of <0.005 was considered to indicate statistical significance.
Results
Specific Interaction of Vesicles with Bacterial Cells in B. agrestis
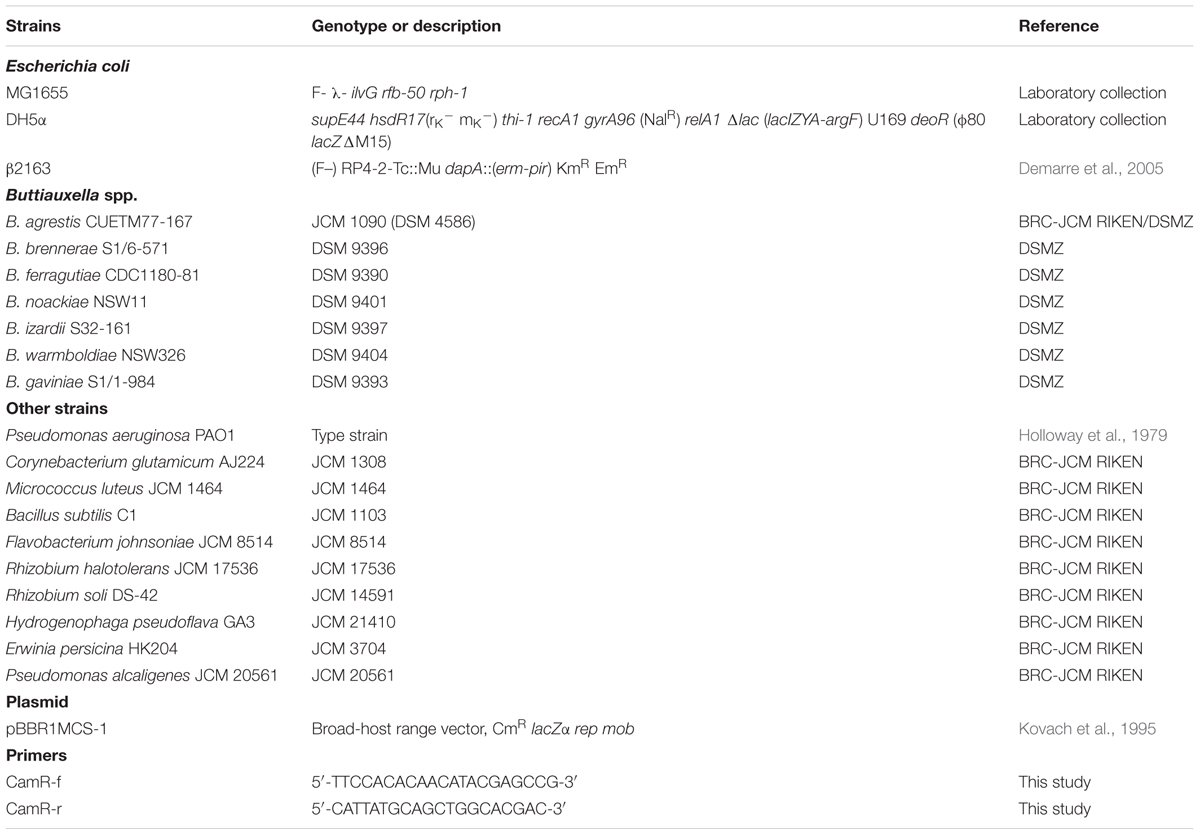
To identify putative high MV-producing bacteria, we examined the phospholipid concentrations in the supernatant of 76 stationary cultures of microbial strains in laboratory stocks. We arbitrarily selected eleven bacterial strains, including C. glutamicum AJ2247, M. luteus JCM 1464, B. subtilis C1, F. johnsoniae JCM 8514, R. halotolerans JCM 17536, R. soli DS-42, H. pseudoflava GA3, B. agrestis CUETM77-167, E. persicina HK204, P. aeruginosa PAO1, and P. alcaligenes JCM 20561, which are supposed to release MVs in the culture at high levels compared to E. coli MG1655 (Supplementary Figure S1), for use in further analyses. Each supernatant from the bacterial culture was ultracentrifuged, and the pellet was labeled with the styryl dye FM4-64, a lipophilic styryl compound that stains the vacuolar membrane. To determine whether each putative MV selectively interacted with bacterial cells, MVs associated with bacterial cells were quantified by fluorescence detection (Supplementary Table S1). The interaction of MVs derived from P. aeruginosa with bacterial cells has been studied well (Kadurugamuwa and Beveridge, 1996, 1997; Li et al., 1998; Tashiro et al., 2010) and showed different levels of association based on bacterial species in this study (Figure 1A). Notably, MVs derived from B. agrestis CUETM77-167 interacted with the parent strain at a significant level but not with other cells (Figure 1B). This specific interaction was not due to the labeling of FM4-64 because similar results were obtained when MVs from B. agrestis were labeled with amino group-reactive FITC or carbocyanine membrane dye DiO (Supplementary Figure S2).
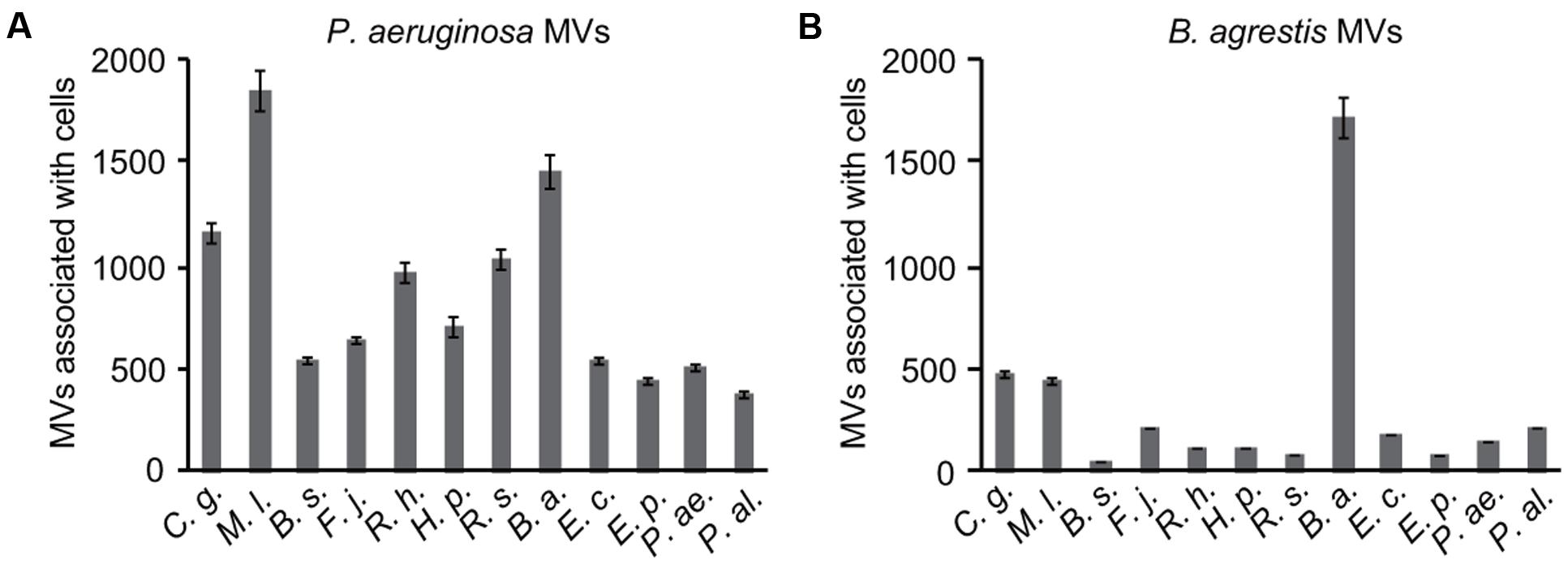
FIGURE 1. Association of membrane vesicles (MVs) with various bacterial species. MVs (20 μg/mL of phospholipids) derived from P. aeruginosa PAO1 (A) and B. agrestis CUETM77-167 (B) were labeled with FM4-64 and incubated with bacterial cells for 30 min at 30°C. Corynebacterium glutamicum AJ2247 (C. g.), Micrococcus luteus JCM 1464 (M. l.), Bacillus subtilis C1 (B. s.), Flavobacterium johnsoniae JCM 8514 (F. j.), Rhizobium halotolerans JCM 17536 (R. h.), R. soli DS-42 (R. s.), Hydrogenophaga pseudoflava GA3 (H. p.), Buttiauxella agrestis CUETM77-167 (B. a.), Escherichia coli MG1655 (E. c.), Erwinia persicina HK204 (E. p.), Pseudomonas aeruginosa PAO1 (P. ae.), and P. alcaligenes JCM 20561 (P. al.) were used as the recipient strains. The MVs associated with the cells were identified by RFUs normalized to the cellular protein concentration (mg/mL). The data are shown as the mean ± standard deviations from three replicates.
To further corroborate the specific interaction of MVs derived from B. agrestis with the same species, the association between MVs and bacterial cells was analyzed at the single-cell level. B. agrestis cells labeled with FITC were incubated with FM4-64-labeled MVs for 30 min, and washed cells were analyzed by flow cytometry. As a control, FITC-labeled cells, which were not treated with MVs, were also assessed by flow cytometry. The fluorescence intensities of both FM4-64 and FITC were detected in MV-reacted cells (approximately 1 × 102 RFU), while only the intensity of FITC was detected in the control sample (Figure 2A). Then, the same experiment was conducted using E. coli and MVs derived from B. agrestis. The fluorescence of FM4-64 was not detected in E. coli either with or without B. agrestis MVs (Figure 2B). Similar results were also obtained by fluorescence microscopy analysis; the fluorescence of both FM4-64 and FITC was observed in MV-treated B. agrestis (Figure 2C), but the fluorescence of FM4-64 was not observed in E. coli, which was treated identically (Figure 2D). These data confirm the hypothesis that MVs derived from B. agrestis specifically interact with bacterial cells of the same species.
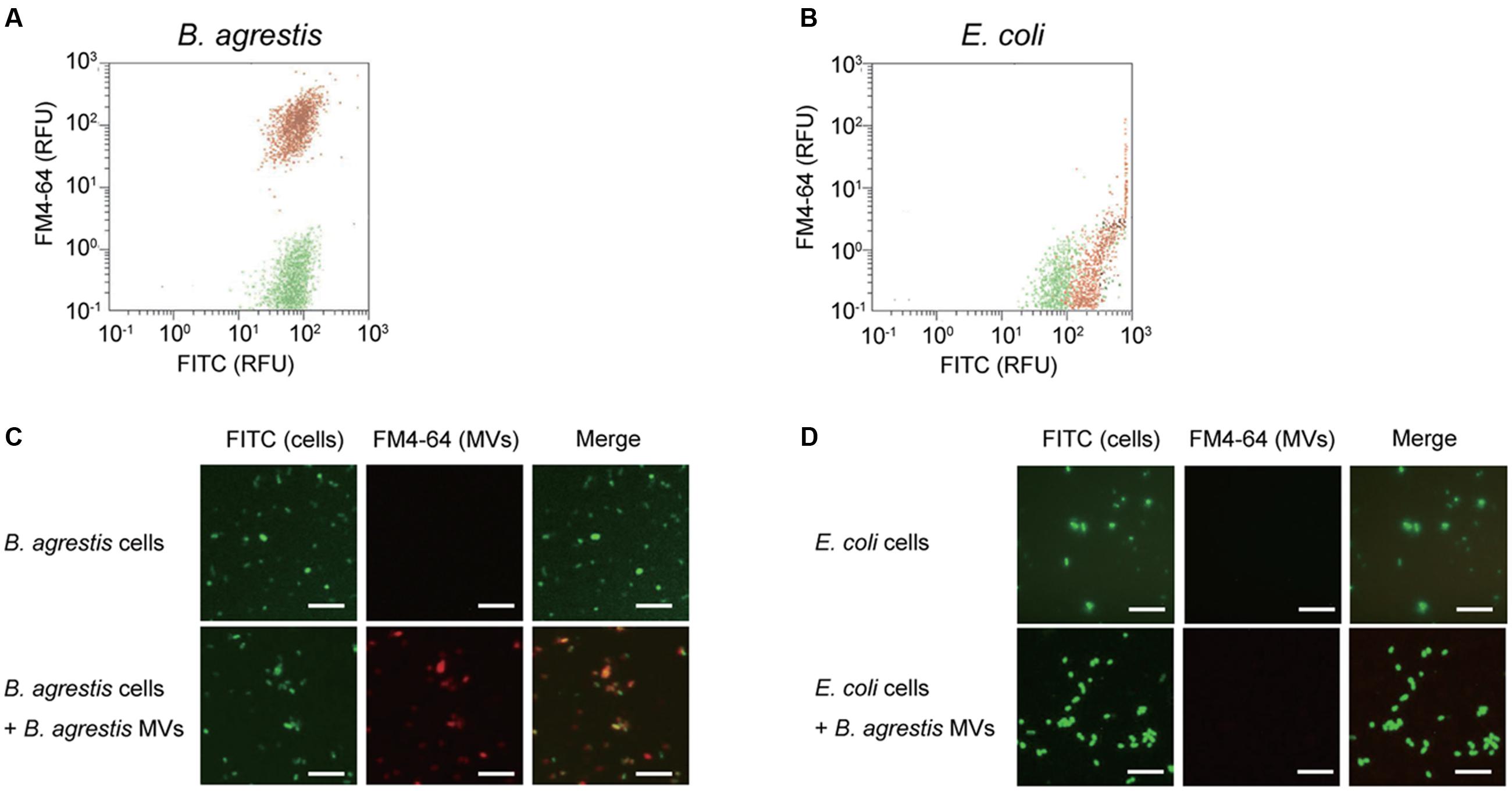
FIGURE 2. Detection and observation of the association of MVs derived from B. agrestis CUETM77-167 with CUETM77-167 and E. coli MG1655 cells. FM4-64-labeled MVs (20 μg/mL of phospholipids) were incubated with FITC-labeled bacterial cells for 30 min at 30°C. (A,B) Flow cytometry analysis of CUETM77-167 (A) and MG1655 cells (B) associated with MVs. FITC-labeled cells (green plots) and FITC-labeled cells incubated with FM4-64-labeled MVs (red plots) underwent flow cytometry independently, and the plots are shown in the same chart. (C,D) Fluorescence microscopy observation of FITC-labeled CUETM77-167 (C) and MG1655 (D) cells incubated with FM4-64-labeled MVs derived from CUETM77-167. The bars indicate 5 μm.
To determine if bacterial viability is essential for MV-cell interactions, the associations of MVs with viable and dead cells were examined. Ultraviolet (UV)-treated cells showed a high association with B. agrestis MVs as well as non-UV treated cells (Supplementary Figure S3), suggesting that the bacterial viability of recipients is not required for the specific MV-cell interaction in B. agrestis.
We confirmed that the extract from the supernatant of B. agrestis culture from ultracentrifugation contained spherical MVs by TEM observation (Figure 3A). Next, we investigated if supplemented MVs interacted with bacterial cells using TEM observation. FM dyes have been used to determine the localization of supplemented vesicles because their photoactivation creates reactive oxygen species, which form an electron-dense precipitate in the vacuoles that is readily visualized under electron microscope (Harata et al., 2001). Then, FM4-64-labeled MVs were incubated with non-labeled cells of B. agrestis, and TEM analysis was conducted to observe the interaction of added MVs with B. agrestis cells. The result showed that MVs attached to the cellular surface were darkly stained (Figure 3B), suggesting that additional MVs were definitely associated with cells of B. agrestis. To further analyze MV-cell interaction, FITC-labeled MVs were incubated with non-labeled cells, and FITC was labeled by gold particles using a gold-conjugate antibody against FITC. The fusion of MVs onto bacterial cells was visualized, and MVs were labeled with gold particles (Figure 3C), suggesting that the MVs in the image were not those being released from cells but rather those being incorporated into cells. When MVs were not added to bacterial suspension before immunolabeling as the control, dense gold particles were not observed around cells (Figure 3D). Thus, the interaction of MVs with bacterial cells is not only the surface attachment but also the membranous fusion.
FIGURE 3. TEM imaging. (A) Image of purified MVs derived from B. agrestis CUETM77-167. (B) Association of FM4-64-labeled MVs with cells. The white arrows indicate MVs, which have a high density due to FM4-64 labeling. (C) Association of FITC-labeled MVs with cells. Cell-associated MVs were detected by small gold particles (black arrows) through the FITC antibody. (D) Cells with no addition of MVs were reacted with FITC antibody. All bars indicate 100 nm.
Interaction Energy between Vesicles and Cells
Because the specific interaction between MVs and cells occurred regardless of the viability of recipient cells, we hypothesized that the physicochemical properties of surfaces affect this association. When MVs and bacterial cells are assumed to be colloid particles, the interaction between two particles is explained by the DLVO theory (Hermansson, 1999; Ohki and Ohshima, 1999; Strevett and Chen, 2003). When the primary maximum energy is positive, there is an energy barrier between MVs and cells (Figure 4A). The interaction energy is the total of the van der Waals’ force and electric repulsion energy, and these depend on the particle size and zeta potential of the particles, respectively. To evaluate the interaction energy between MVs and bacterial cells, their surface thermodynamic properties were measured using a Zeta Sizer (Figure 4B). The interaction energy between B. agrestis MVs and each bacterial cell was calculated based on the DLVO theory (Supplementary Figure S4). Interestingly, the primary energy in the interaction of B. agrestis MVs with the same species was significantly lower than that of other bacterial cells (Figure 4C). These results suggest that one factor in the specific interaction of MVs with the same species in B. agrestis is the low interaction energy based on the DLVO theory.
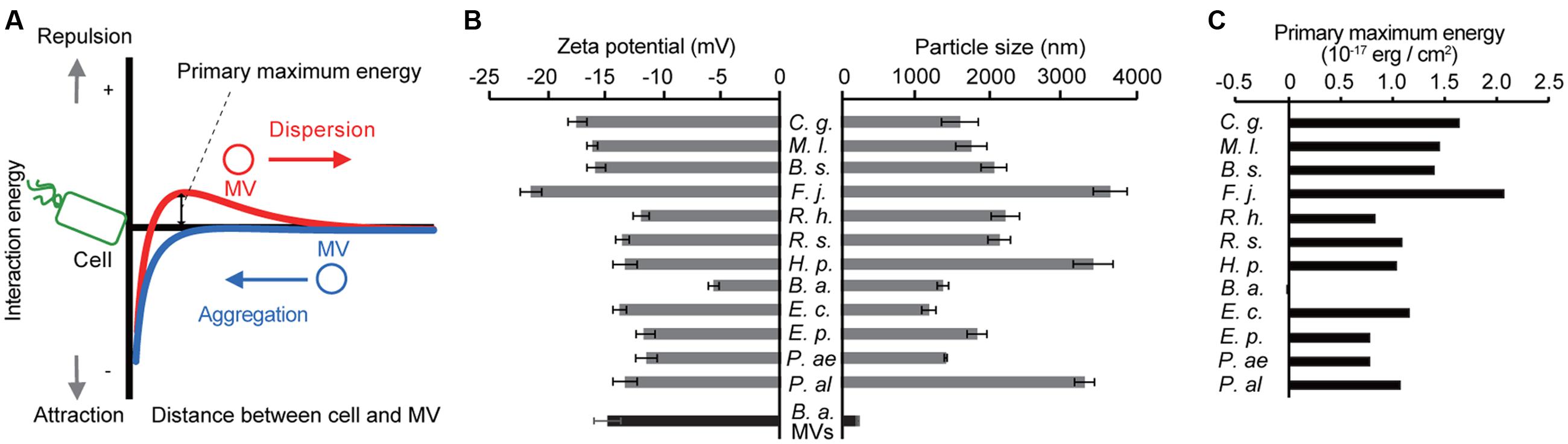
FIGURE 4. Interaction energy between cells and MVs derived from B. agrestis. (A) Model for the free energy profile of the interaction between cells and MVs according to a generalized Derjaguin–Landau–Verwey–Overbeek (DLVO) theory. When the energy barrier is large enough (red line), MVs are prevented from approaching cells by electrostatic repulsion. However, a small energy barrier causes MVs to aggregate with cells (blue line) due to van der Waals attractions. (B) Zeta potential and particle size of each bacterial cell (gray) and B. agrestis MVs (black). C. g., Corynebacterium glutamicum AJ224; M. l., Micrococcus luteus JCM 1464; B. s., Bacillus subtilis C1; F. j., Flavobacterium johnsoniae JCM 8514; R. h., Rhizobium halotolerans JCM 17536; R. s., R. soli DS-42; H. p., Hydrogenophaga pseudoflava GA3; B. a., Buttiauxella agrestis CUETM77-167; E. p., Erwinia persicina HK204; P. ae., Pseudomonas aeruginosa PAO1; P. al., P. alcaligenes JCM 20561. The data are shown as the mean ± standard deviation from three replicates. (C) Primary maximum energy between each cell and B. agrestis MVs based on the DLVO theory.
Specific Vesicle-cell Interaction Is Conserved in Buttiauxella spp.
Although a specific interaction of MVs with the same species was observed in B. agrestis, it was unknown whether this characteristic was confined to this bacterium. We prepared 6 other Buttiauxella strains (B. brennerae S1/6-571, B. ferragutiae CDC1180-81, B. noackiae NSW11, B. izardii S3/2-161, B. warmboldiae NSW326, and B. gaviniae S1/1-984), and the phylogenetic relationship among them and E. coli is shown in Figure 5A. Interestingly, the MVs derived from B. agrestis CUETM77-167 showed a much higher association with cells of Buttiauxella spp. than with E. coli (Figure 5B), suggesting that the specific interaction of MVs with cells is conserved in Buttiauxella spp. To determine whether the high interaction between MVs derived from CUETM77-167 and Buttiauxella strains is due to the physicochemical interaction energy, the zeta potentials and hydrodynamic diameters of each cell were examined. The calculated interaction energy based on the DLVO theory showed that the primary maximum energies of several Buttiauxella cells with MVs derived from CUETM77-167 were significantly lower than that for E. coli (Figure 5C and Supplementary Figure S5). We next evaluated the relationship between the primary maximum energies calculated by DLVO theory and the association values of MVs with bacterial cells. Figure 5D shows that plots of Buttiauxella strains are localized at different positions from those of other strains. Statistical analyses indicated significant differences between Buttiauxella strains and other strains in both association with B. agrestis MVs and primary maximum energies (Supplementary Figure S6), suggesting that interaction energy based on the DLVO theory is one factor explaining why MVs specifically interact with bacterial cells of Buttiauxella spp. However, the correlation between the two parameters is not linear in Figure 5D (the coefficient of determination R2 is 0.59), and other factors besides low interaction energy based on the DLVO theory may affect the specific interaction of MVs in Buttiauxella strains. When MVs and/or cells were treated with proteinase K, the association between MVs and cells in B. agrestis was decreased more than 50% (Supplementary Figure S7), suggesting that proteins localized on the surface of cells and MVs also affect the MV-cell interaction in B. agrestis.
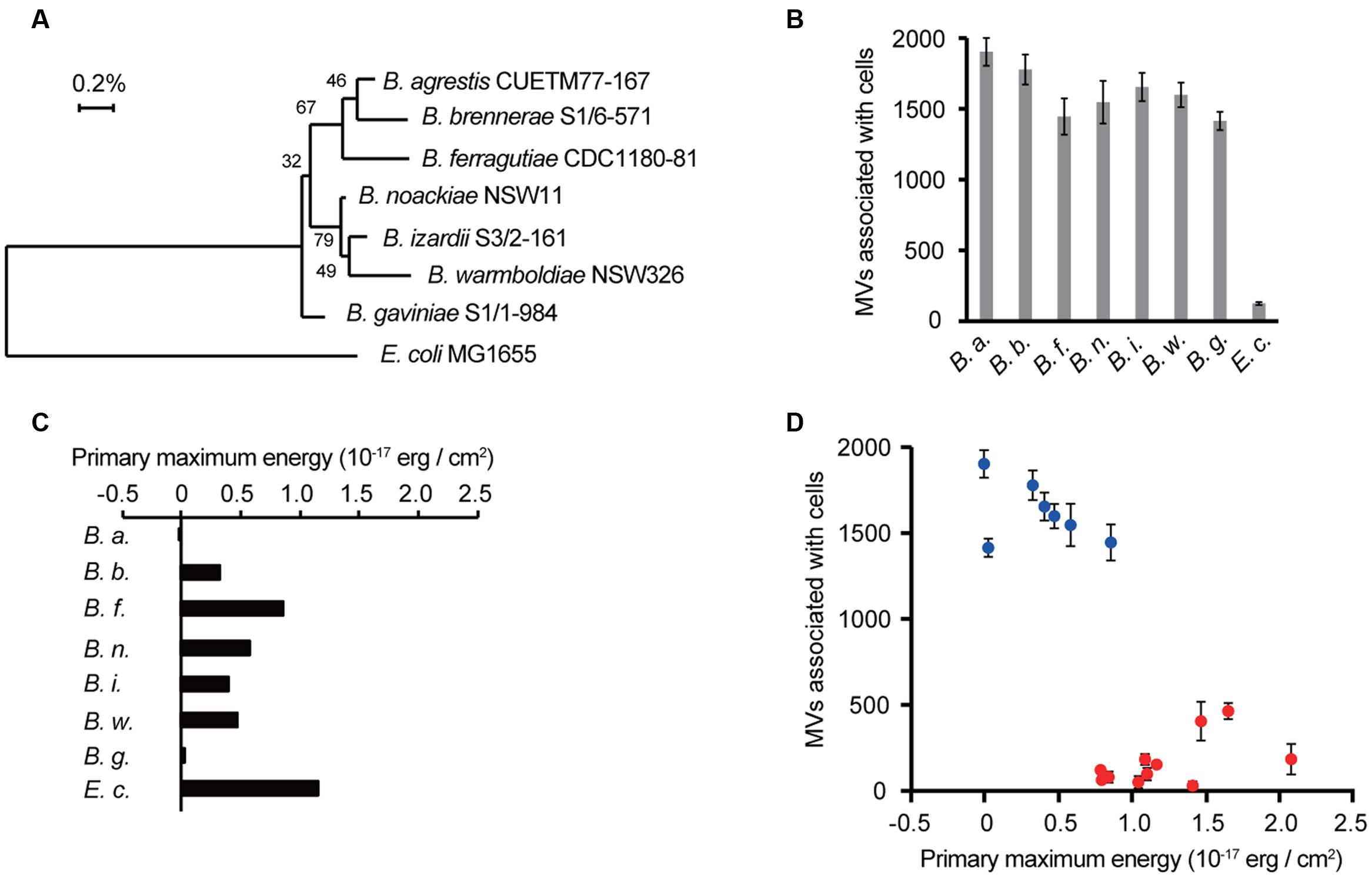
FIGURE 5. Selective interactions between cells and MVs are conserved in Buttiauxella spp. (A) The phylogenetic relationship between Buttiauxella spp. and E. coli. Numbers on the branching points are the percentages of bootstrap values with 1000 replicates. (B) Association of MVs derived from B. agrestis CUETM77-167 with bacterial cells. FM4-64-labeled MVs (20 μg/mL of phospholipids) were incubated with bacterial cells for 30 min at 30°C. MVs associated with cells were identified by RFUs of FM4-64 normalized to the cellular protein concentration (mg/mL). The data are shown as the mean ± standard deviation from three replicates. (C) Primary maximum energy between each cell and B. agrestis MVs. The values were calculated from the zeta potential and particle size based on the DLVO theory. (D) Relationship between the MV association with cells and primary maximum energy. Blue plots show Buttiauxella strains (from B,C), and red plots show other bacterial strains (from Figures 1B, 4C). The coefficient of determination R2 is 0.59.
Vesicle-mediated DNA Transfer
To determine whether the interaction of MVs with bacterial cells contributes to the delivery of the MV contents to bacterial cells, we evaluated vesicle-mediated plasmid DNA transfer. B. agrestis harboring pBBR1MCS-1 was grown in TSB medium to the stationary phase, and plasmid-containing MVs (p-MVs) were extracted from the supernatant. An examination of the pBBR1MCS-1 concentration by quantitative PCR showed 3.11 × 106 and 2.27 × 106 copies/mL in the supernatant before and after the removal of MVs through ultracentrifugation, respectively, suggesting that at least approximately one-third of the plasmid localized in the extracellular milieu was associated with p-MVs in the B. agrestis supernatant. The external DNA surrounding p-MVs was degraded by DNase I treatment, and the plasmid concentration in the p-MVs was calculated by counting the number of MVs using nano tracking analysis. The results showed that p-MVs contain 1.03 × 109 copies/mL of pBBR1MCS-1, indicating that p-MVs maintain a high concentration of plasmid and that the DNA in p-MVs was stable against DNase I treatment. When B. agrestis cells (approximately 1.0 × 103 cells/mL) were treated with an excessive amount of p-MVs, more than 30% B. agrestis transformants were obtained after 3 h of incubation with DNase I-treated p-MVs and non-treated p-MVs (Figure 6), suggesting that the plasmid contained in MVs was transferred to bacterial cells. When naked plasmid DNA instead of p-MVs was added to directly bacterial cell suspension in this experiment (data not shown), indicating that natural transformation was not occurred in the condition. Notably, other methods for DNA transformation into this strain have not yet been established in our experiments, suggesting that DNA transfer via p-MVs is a useful tool to obtain transformants in this strain.
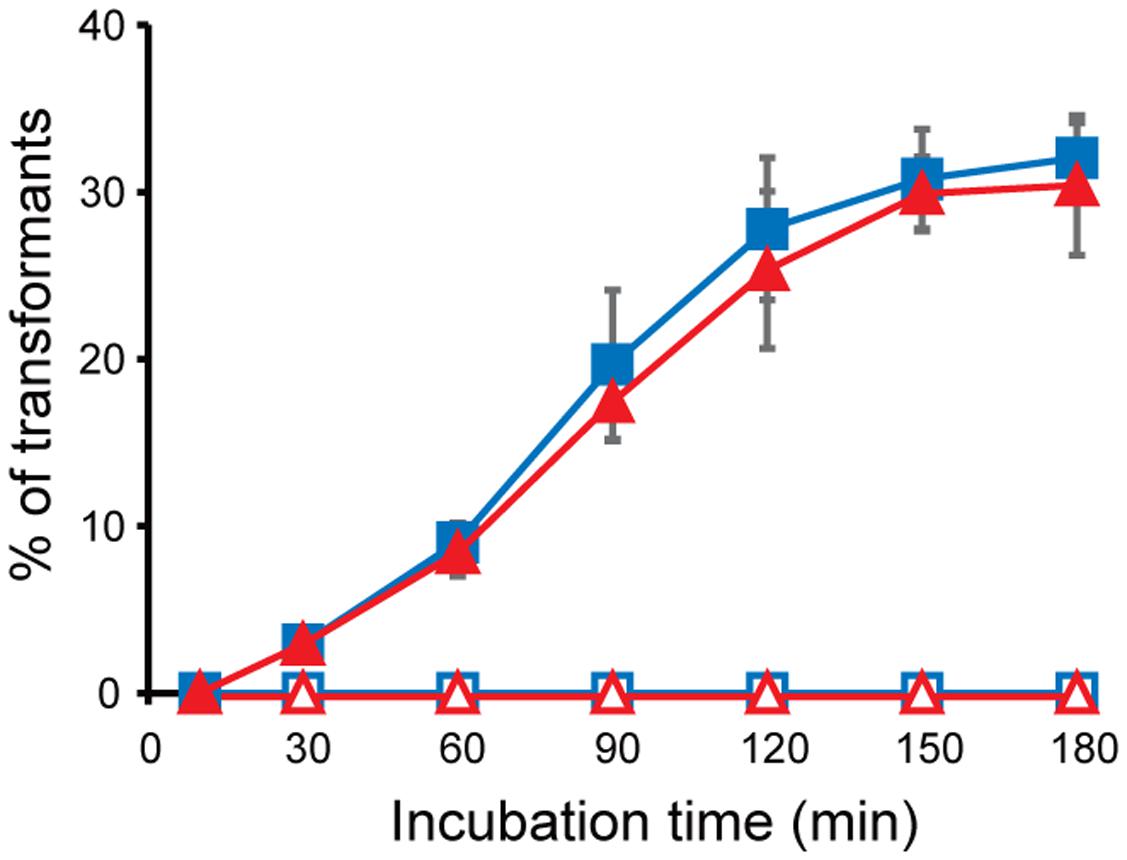
FIGURE 6. Plasmid transfer through MVs derived from B. agrestis. MVs were extracted from the culture of CUETM77-167 (open) or CUETM77-167/pBBR1-MCS (closed). DNase-treated (red triangles) or non-treated (blue squares) MVs (20 μg/mL of phospholipids) were incubated with CUETM77-167 at 30°C. Transformants were calculated by counting the CFUs on agar medium containing chloramphenicol. The data are shown as the mean ± standard deviation from three replicates.
Vesicles Enable the Selective Delivery of an Antibiotic
We expected that the specific interaction of MVs with bacterial cells could be used for the selective control of bacterial cells. Exposure to the antibiotic gentamicin has been shown to increase MV formation, and gentamicin was retained within the MVs (Kadurugamuwa and Beveridge, 1995; Fulsundar et al., 2014). We evaluated the potential of MVs as tools for the delivery of gentamicin to specific bacterial cells. B. agrestis was grown to the stationary phase, and gentamicin was added to the culture at a final concentration of 32 μg/mL (four times the MIC). MVs were extracted, and gentamicin was concentrated in MVs (146 μg/mL) based on the calculated gentamicin concentration and the number of MVs. We prepared gentamicin-associated MVs (g-MVs) and homogenized g-MVs, in which the gentamicin concentration was likely the same as that in the sample of g-MVs. As a negative control, MVs derived from B. agrestis not associated with gentamicin (n-MVs) were also prepared. These MVs were incubated with cells of B. agrestis, E. coli, and P. aeruginosa, and the survival rates were examined. The killing effect of g-MVs on B. agrestis was much greater than that on E. coli and P. aeruginosa, while homogenized g-MVs had a high killing effect on all of the bacteria that were tested in this experiment (Figures 7A–C). Under these experimental conditions, the gentamicin concentration of each sample ranged from 1.3 to 2.3 μg/mL, and this concentration was lower than the MIC of each strain. Subsequently, to determine whether selective antibiotic delivery via MVs occurred in the microbial complexes, the effect of g-MVs in the mixed samples containing B. agrestis and E. coli cells was examined. The results showed that the killing effect of g-MVs on B. agrestis was higher than that on E. coli, even in mixed samples (Figure 7D). These data indicate that the specific interaction of B. agrestis MVs is useful for killing target species in heterogeneous samples.
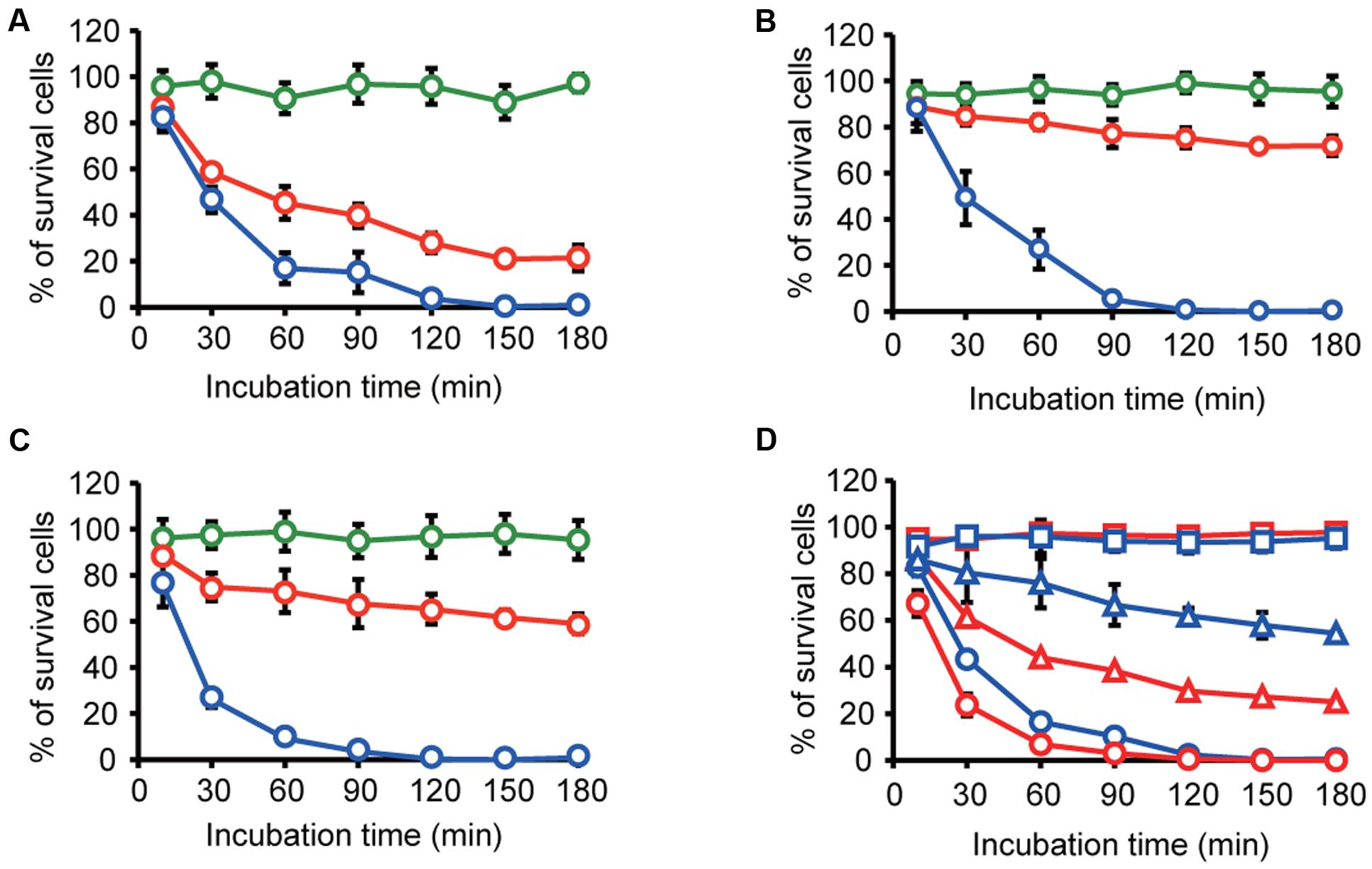
FIGURE 7. Transfer of gentamicin through MVs derived from B. agrestis. (A–C) Survival rate of B. agrestis CUETM77-167 (A), E. coli MG1655 (B) or P. aeruginosa PAO1 (C). Each bacterial cell was incubated with normal MVs (n-MVs: green), gentamicin-harboring MVs (g-MVs: red) and homogenized g-MVs (blue). The data are shown as the mean ± standard deviation from three replicates. (D) Survival rate of bacterial cells in the mixture of CUETM77-167 (red) and E. coli DH5α (blue). The samples were incubated with n-MVs (squares), g-MVs (triangles), and homogenized g-MVs (circles). The data are shown as the mean ± standard deviation from three replicates.
Discussion
Although bacterial cell-cell interaction via MVs has attracted particular attention in the past decade, whether bacterial cells are recipients for secreted MVs is unclear. To our knowledge, this study is the first report that MVs secreted from a certain bacterial species selectively interact with specific cells. The main finding of this study is that MVs have a function of specific delivery of MV contents to intraspecies bacterial cells in Buttiauxella spp. Buttiauxella strains have been isolated from unpolluted soil and drinking water, surface water, sewage, soil, and fecal samples (Müller et al., 1996), but little is understood about their ecological characteristics in the environment. Although Buttiauxella spp. can be a host of plasmid pBP136 in soil bacteria (Shintani et al., 2014), MV production has not been studied in this bacterium.
One important factor in the association between MVs and bacterial cells is electric charges on the cellular surface. The surfaces of bacterial cells are generally negatively charged, and there are many cations, such as Mg2+ and Ca2+, to stabilize the negative charge on the cellular surfaces. These cations play crucial roles in the interaction of MVs with bacterial cells because cations can form a bridge between two negatively charged interfaces (Kadurugamuwa and Beveridge, 1996; Tashiro et al., 2010). The strong MV-cell association in B. agrestis was different from this case; the negative charge of the cellular surfaces in B. agrestis was lower than that of other bacteria, and the interaction energy between MVs and cells in this species was much lower than that in other bacteria based on the DLVO theory (Figures 4C). Because bacterial surface thermodynamic theory is fundamental in interpreting bacterial adhesion, several studies have applied the classic or modified DLVO theory to predict bacterial aggregation and attachment on the surface of substances (Hermansson, 1999; Strevett and Chen, 2003; Wang et al., 2013; Yoshihara et al., 2014), while it has never been used to investigate the MV-bacterial cell interaction. To our knowledge, this is the first report to apply the DLVO theory to explain the interaction between MVs and bacterial cells. It might be that interaction energy, which is characterized by hydrodynamic diameter and zeta potential, based on DLVO theory provides good indication to understand the association between MVs and cells. However, we cannot deny the possibility that factors other than interaction energy based on the DLVO theory are also involved in the specific association between MVs and bacterial cells in Buttiauxella spp. because the association degree of MVs from B. agrestis with Buttiauxella spp. cells was significantly higher than that with other species even when the primary maximum energies were unchanged (Figure 5D and Supplementary Figure S6). Bacterial cells were coated with extracellular polymeric substances (EPS) and lipopolysaccharides (LPS), and these polysaccharides play an important role in bacterial aggregation (Flemming and Wingender, 2010). MV-cell recognition was suggested to occur via LPS in Myxococcus xanthus because the distance between the MVs and the cellular surface was 5–10 nm, corresponding to that of the LPS (Remis et al., 2014). The factor connecting MVs and cells in Buttiauxella spp. remains unknown, but polysaccharides or outer membrane proteins, which are conserved in Buttiauxella spp., may play a role in the specific association between MVs and bacterial cells. Indeed, the proteinase K treatment decreased the MV-cell interaction in B. agrestis (Supplementary Figure S7), suggesting that, at least, proteins localized on the cellular surface is related to the MV-cell association.
As previously reported (Dorward et al., 1989; Kolling and Matthews, 1999; Yaron et al., 2000; Klieve et al., 2005; Chiura et al., 2011; Gaudin et al., 2013; Fulsundar et al., 2014; Ho et al., 2015), MV-mediated gene transfer also occurred in B. agrestis in our study (Figure 6). However, little is understood regarding the detailed mechanism of MV-mediated DNA transfer in any microbe. According to a previous study, DNA transfer did not occur when the recipient cells lacked genes related to natural transformation, including comA and comB-F in A. baylyi, indicating that the competence proteins play a role in the uptake of DNA via MVs (Fulsundar et al., 2014). Indeed, B. agrestis CUETM77-167 has competence-related genes, such as comEA and comEC, as determined by searching the Integrated Microbial Genomes (IMG) data warehouse (Markowitz et al., 2014), but whether these genes are related to the uptake of DNA via MVs in this strain is unknown. Notably, the plasmid contained in MVs was transferred to E. coli in previous reports (Kolling and Matthews, 1999; Fulsundar et al., 2014), while natural transformation was not observed in E. coli, suggesting that factors other than natural competence play a role in MV-mediated DNA uptake. Furthermore, we have never obtained transformants of CUETM77-167 using several transformation methods (the calcium chloride method and electroporation, as well as natural transformation), but approximately 30% of the cells received the plasmid contained in the MVs when the p-MV concentration was high. These results suggest that MV-mediated DNA transfer can be a useful method to obtain transformants in this strain. A comprehensive mechanism for DNA uptake via MVs will be a major breakthrough for developing a method of DNA incorporation through MVs.
Several previous studies have shown that MV production is increased by exposure to antibiotics, including gentamicin, polymyxin B, colistin, ceftazidime, and imipenem (Kadurugamuwa and Beveridge, 1995; Manning and Kuehn, 2011; Koning et al., 2013; Devos et al., 2015). In particular, gentamicin and antimicrobial peptides (polymyxin B and colistin) bound to LPS on bacterial cell surfaces resulted in increased MV formation (Kadurugamuwa and Beveridge, 1995; Manning and Kuehn, 2011). However, several studies have indicated that MV secretion has a protective effect against antibiotics, e.g., MVs can carry active β-lactamase (Ciofu et al., 2000; Schaar et al., 2011) or bind antibiotics in the extracellular milieu (Grenier et al., 1995; Manning and Kuehn, 2011). Thus, previous reports have focused on antibiotic resistance and the association of MVs with antibiotics; however, little is understood regarding the therapeutic potential of MVs containing antibiotics, except for several reports by the Beveridge group indicating that gentamicin-induced MVs derived from P. aeruginosa had a killing effect against both Gram-negative and Gram-positive bacteria (Kadurugamuwa and Beveridge, 1996; MacDonald and Beveridge, 2002). In this study, we investigated whether gentamicin-induced MVs derived from B. agrestis had a selective killing effect on target cells and examined the potential for the biotechnical use of MVs as antibiotic-delivery vehicles to control specific microbial species in heterogeneous microbial communities. We showed that g-MVs derived from B. agrestis had a higher killing effect on B. agrestis than on E. coli and P. aeruginosa (Figures 7A–C); in particular, the number of surviving B. agrestis cells was much lower than that of surviving E. coli cells in the sample including both species (Figure 7D). The results obtained in this study only demonstrate the effective killing of B. agrestis, but further comprehensive analysis of the mechanism underlying the specific association of MVs with bacterial cells, other than interaction energy based on surface charges, will create a new path for developing MVs as a tool to eliminate pathogenic or other bacterial species in heterogeneous microbial communities.
Conclusion
We presented the first example of the delivery of cargo in MVs to specific cells. MVs derived from B. agrestis CUETM77-167 specifically interacted with bacterial cells of Buttiauxella spp. The specific interaction between MVs and Buttiauxella spp. cells was explained in terms of interaction energy based on the DLVO theory, but other as-yet-unknown factors may be involved in this specific interaction. Interaction of MVs with bacterial cells enabled MV-mediated gene transfer. Moreover, the specific interaction of MVs enables their use for the delivery of gentamicin to target cells. These results indicate that MVs selectively associate with bacterial cells and provide a novel model in which MVs effectively deliver their cargo to target microbial cells in microbial communities. We anticipate that studies of MVs will be a valuable contribution to the development of a novel biotechnological tool for controlling target bacterial cells.
Author Contributions
YT and YH planned and designed the experiments. YT, YH, MS, and KT performed the experiments. YT, YH, MS, KT, MO, KK, and HF analyzed the data. YT and YH wrote the manuscript. All of the authors contributed to the discussion and provided comments on the manuscript.
Funding
This research was supported by JSPS KAKENHI Grant Numbers JP15K21043 and JP15H01315 and the Kurita Water and Environmental Foundation to YT.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to George Salmond of the University of Cambridge for providing E. coli β2163. We thank Nobuyuki Mase and Kohei Sato of Shizuoka University for assistance with the nanoparticle tracking analysis. We also thank the Instrumental Research Support Office in the Research Institute of Green Science and Technology for the use of the zetasizer.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00571/full#supplementary-material
References
Alves, N. J., Turner, K. B., Medintz, I. L., and Walper, S. A. (2015). Emerging therapeutic delivery capabilities and challenges utilizing enzyme/protein packaged bacterial vesicles. Ther. Deliv. 6, 873–887. doi: 10.4155/tde.15.40
Bauman, S. J., and Kuehn, M. J. (2009). Pseudomonas aeruginosa vesicles associate with and are internalized by human lung epithelial cells. BMC Microbiol. 9:26. doi: 10.1186/1471-2180-9-26
Berleman, J., and Auer, M. (2013). The role of bacterial outer membrane vesicles for intra- and interspecies delivery. Environ. Microbiol. 15, 347–354. doi: 10.1111/1462-2920.12048
Biller, S. J., Schubotz, F., Roggensack, S. E., Thompson, A. W., Summons, R. E., and Chisholm, S. W. (2014). Bacterial vesicles in marine ecosystems. Science 343, 183–186. doi: 10.1126/science.1243457
Chiura, H. X., Kogure, K., Hagemann, S., Ellinger, A., and Velimirov, B. (2011). Evidence for particle-induced horizontal gene transfer and serial transduction between bacteria. FEMS Microbiol. Ecol. 76, 576–591. doi: 10.1111/j.1574-6941.2011.01077.x
Ciofu, O., Beveridge, T., Kadurugamuwa, J., Walther-Rasmussen, J., and Høiby, N. (2000). Chromosomal beta-lactamase is packaged into membrane vesicles and secreted from Pseudomonas aeruginosa. J. Antimicrob. Chemother. 45, 9–13. doi: 10.1093/jac/45.1.9
Deatherage, B. L., and Cookson, B. T. (2012). Membrane vesicle release in bacteria, eukaryotes, and archaea: a conserved yet underappreciated aspect of microbial life. Infect. Immun. 80, 1948–1957. doi: 10.1128/IAI.06014-11
Demarre, G. L., GueìRout, A.-M., Matsumoto-Mashimo, C., Rowe-Magnus, D. A., MarlieÌRe, P., and Mazel, D. (2005). A new family of mobilizable suicide plasmids based on broad host range R388 plasmid (IncW) and RP4 plasmid (IncPα) conjugative machineries and their cognate Escherichia coli host strains. Res. Microbiol. 156, 245–255. doi: 10.1016/j.resmic.2004.09.007
Devos, S., Van Oudenhove, L., Stremersch, S., Van Putte, W., Derycke, R., Van Driessche, G., et al. (2015). The effect of imipenem and diffusible signaling factors on the secretion of outer membrane vesicles and associated Ax21 proteins in Stenotrophomonas maltophilia. Front. Microbiol. 6:298. doi: 10.3389/fmicb.2015.00298
Dorward, D. W., Garon, C. F., and Judd, R. C. (1989). Export and intercellular transfer of DNA via membrane blebs of Neisseria gonorrhoeae. J. Bacteriol. 171, 2499–2505. doi: 10.1128/jb.171.5.2499-2505.1989
Evans, A. G. L., Davey, H. M., Cookson, A., Currinn, H., Cooke-Fox, G., Stanczyk, P. J., et al. (2012). Predatory activity of Myxococcus xanthus outer-membrane vesicles and properties of their hydrolase cargo. Microbiology 158, 2742–2752. doi: 10.1099/mic.0.060343-0
Flemming, H.-C., and Wingender, J. (2010). The biofilm matrix. Nat. Rev. Microbiol 8, 623–633. doi: 10.1038/nrmicro2415
Fulsundar, S., Harms, K., Flaten, G. E., Johnsen, P. J., Chopade, B. A., and Nielsen, K. M. (2014). Gene transfer potential of outer membrane vesicles of Acinetobacter baylyi and effects of stress on vesiculation. Appl. Environ. Microbiol. 80, 3469–3483. doi: 10.1128/AEM.04248-13
Gaudin, M., Gauliard, E., Schouten, S., Houel-Renault, L., Lenormand, P., Marguet, E., et al. (2013). Hyperthermophilic archaea produce membrane vesicles that can transfer DNA. Environ. Microbiol. Rep. 5, 109–116. doi: 10.1111/j.1758-2229.2012.00348.x
Grenier, D., Bertrand, J., and Mayrand, D. (1995). Porphyromonas gingivalis outer membrane vesicles promote bacterial resistance to chlorhexidine. Oral Microbiol. Immunol. 10, 319–320. doi: 10.1111/j.1399-302X.1995.tb00161.x
Harata, N., Pyle, J. L., Aravanis, A. M., Mozhayeva, M., Kavalali, E. T., and Tsien, R. W. (2001). Limited numbers of recycling vesicles in small CNS nerve terminals: implications for neural signaling and vesicular cycling. Trends Neurosci. 24, 637–643. doi: 10.1016/S0166-2236(00)02030-0
Hasegawa, Y., Futamata, H., and Tashiro, Y. (2015). Complexities of cell-to-cell communication through membrane vesicles: implications for selective interaction of membrane vesicles with microbial cells. Front. Microbiol. 6:633. doi: 10.3389/fmicb.2015.00633
Hermansson, M. (1999). The DLVO theory in microbial adhesion. Colloids Surf. B Biointerfaces 14, 105–119. doi: 10.1016/S0927-7765(99)00029-6
Ho, M.-H., Chen, C.-H., Goodwin, J. S., Wang, B.-Y., and Xie, H. (2015). Functional advantages of Porphyromonas gingivalis vesicles. PLoS ONE 10:e0123448. doi: 10.1371/journal.pone.0123448
Holloway, B. W., Krishnapillai, V., and Morgan, A. F. (1979). Chromosomal genetics of Pseudomonas. Microbiol. Rev. 43, 73–102.
Kadurugamuwa, J., Mayer, A., Messner, P., Sára, M., Sleytr, U., and Beveridge, T. (1998). S-layered Aneurinibacillus and Bacillus spp. are susceptible to the lytic action of Pseudomonas aeruginosa membrane vesicles. J. Bacteriol. 180, 2306–2311.
Kadurugamuwa, J. L., and Beveridge, T. J. (1995). Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: a novel mechanism of enzyme secretion. J. Bacteriol. 177, 3998–4008. doi: 10.1128/jb.177.14.3998-4008.1995
Kadurugamuwa, J. L., and Beveridge, T. J. (1996). Bacteriolytic effect of membrane vesicles from Pseudomonas aeruginosa on other bacteria including pathogens: conceptually new antibiotics. J. Bacteriol. 178, 2767–2774. doi: 10.1128/jb.178.10.2767-2774.1996
Kadurugamuwa, J. L., and Beveridge, T. J. (1997). Natural release of virulence factors in membrane vesicles by Pseudomonas aeruginosa and the effect of aminoglycoside antibiotics on their release. J. Antimicrob. Chemother. 40, 615–621. doi: 10.1093/jac/40.5.615
Kadurugamuwa, J. L., and Beveridge, T. J. (1999). Membrane vesicles derived from Pseudomonas aeruginosa and Shigella flexneri can be integrated into the surfaces of other gram-negative bacteria. Microbiology 145, 2051–2060. doi: 10.1099/13500872-145-8-2051
Kesty, N., and Kuehn, M. (2004). Incorporation of heterologous outer membrane and periplasmic proteins into Escherichia coli outer membrane vesicles. J. Biol. Chem. 279, 2069–2076. doi: 10.1074/jbc.M307628200
Kesty, N., Mason, K., Reedy, M., Miller, S., and Kuehn, M. (2004). Enterotoxigenic Escherichia coli vesicles target toxin delivery into mammalian cells. EMBO J. 23, 4538–4549. doi: 10.1038/sj.emboj.7600471
Klieve, A. V., Yokoyama, M. T., Forster, R. J., Ouwerkerk, D., Bain, P. A., and Mawhinney, E. L. (2005). Naturally occurring DNA transfer system associated with membrane vesicles in cellulolytic Ruminococcus spp. of ruminal origin. Appl. Environ. Microbiol. 71, 4248–4253. doi: 10.1128/AEM.71.8.4248-4253.2005
Kolling, G. L., and Matthews, K. R. (1999). Export of virulence genes and Shiga toxin by membrane vesicles of Escherichia coli O157:H7. Appl. Environ. Microbiol. 65, 1843–1848.
Koning, R. I., De Breij, A., Oostergetel, G. T., Nibbering, P. H., Koster, A. J., and Dijkshoorn, L. (2013). Cryo-electron tomography analysis of membrane vesicles from Acinetobacter baumannii ATCC19606T. Res. Microbiol. 164, 397–405. doi: 10.1016/j.resmic.2013.02.007
Kovach, M. E., Elzer, P. H., Steven Hill, D., Robertson, G. T., Farris, M. A., Roop, R. M., et al. (1995). Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166, 175–176. doi: 10.1016/0378-1119(95)00584-1
Kulp, A., and Kuehn, M. J. (2010). Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu. Rev. Microbiol. 64, 163–184. doi: 10.1146/annurev.micro.091208.073413
Li, J., Azam, F., and Zhang, S. (2016). Outer membrane vesicles containing signalling molecules and active hydrolytic enzymes released by a coral pathogen Vibrio shilonii AK1. Environ. Microbiol. 18, 3850–3866. doi: 10.1111/1462-2920.13344
Li, Z., Clarke, A. J., and Beveridge, T. J. (1998). Gram-negative bacteria produce membrane vesicles which are capable of killing other bacteria. J. Bacteriol. 180, 5478–5483.
MacDonald, K. L., and Beveridge, T. J. (2002). Bactericidal effect of gentamicin-induced membrane vesicles derived from Pseudomonas aeruginosa PAO1 on gram-positive bacteria. Can. J. Microbiol. 48, 810–820. doi: 10.1139/w02-077
Manning, A., and Kuehn, M. (2011). Contribution of bacterial outer membrane vesicles to innate bacterial defense. BMC Microbiol. 11:258. doi: 10.1186/1471-2180-11-258
Markowitz, V. M., Chen, I.-M. A., Palaniappan, K., Chu, K., Szeto, E., Pillay, M., et al. (2014). IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res. 42, D560–D567. doi: 10.1093/nar/gkt963
Mashburn, L. M., and Whiteley, M. (2005). Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature 437, 422–425. doi: 10.1038/nature03925
Mashburn-Warren, L. M., and Whiteley, M. (2006). Special delivery: vesicle trafficking in prokaryotes. Mol. Microbiol. 61, 839–846. doi: 10.1111/j.1365-2958.2006.05272.x
Müller, H. E., Brenner, D. J., Fanning, G. R., Grimont, P. A. D., and Kämpfer, P. (1996). Emended description of Buttiauxella agrestis with recognition of six new species of Buttiauxella and two new species of Kluyvera: Buttiauxella ferragutiae sp. nov., Buttiauxella gaviniae sp. nov., Buttiauxella brennerae sp. nov., Buttiauxella izardii sp. nov., Buttiauxella noackiae sp. nov., Buttiauxella warmboldiae sp. nov., Kluyvera cochleae sp. nov., and Kluyvera georgiana sp. nov. Int. J. Syst. Evol. Microbiol. 46, 50–63. doi: 10.1099/00207713-46-1-50
Ohki, S., Duzgunes, N., and Leonards, K. (1982). Phospholipid vesicle aggregation: effect of monovalent and divalent ions. Biochemistry 21, 2127–2133. doi: 10.1021/bi00538a022
Ohki, S., and Ohshima, H. (1999). Interaction and aggregation of lipid vesicles (DLVO theory versus modified DLVO theory). Colloids Surf. B Biointerfaces 14, 27–45. doi: 10.1016/S0927-7765(99)00022-3
Okabe, S., Oshiki, M., Kamagata, Y., Yamaguchi, N., Toyofuku, M., Yawata, Y., et al. (2010). A great leap forward in microbial ecology. Microbes Environ. 25, 230–240. doi: 10.1264/jsme2.ME10178
Parker, H., Chitcholtan, K., Hampton, M. B., and Keenan, J. I. (2010). Uptake of Helicobacter pylori outer membrane vesicles by gastric epithelial cells. Infect. Immun. 78, 5054–5061. doi: 10.1128/IAI.00299-10
Pirbadian, S., Barchinger, S. E., Leung, K. M., Byun, H. S., Jangir, Y., Bouhenni, R. A., et al. (2014). Shewanella oneidensis MR-1 nanowires are outer membrane and periplasmic extensions of the extracellular electron transport components. Proc. Natl. Acad. Sci. U.S.A. 111, 12883–12888. doi: 10.1073/pnas.1410551111
Remis, J. P., Wei, D., Gorur, A., Zemla, M., Haraga, J., Allen, S., et al. (2014). Bacterial social networks: structure and composition of Myxococcus xanthus outer membrane vesicle chains. Environ. Microbiol. 16, 598–610. doi: 10.1111/1462-2920.12187
Renelli, M., Matias, V., Lo, R. Y., and Beveridge, T. J. (2004). DNA-containing membrane vesicles of Pseudomonas aeruginosa PAO1 and their genetic transformation potential. Microbiology 150, 2161–2169. doi: 10.1099/mic.0.26841-0
Roier, S., Zingl, F. G., Cakar, F., Durakovic, S., Kohl, P., Eichmann, T. O., et al. (2016). A novel mechanism for the biogenesis of outer membrane vesicles in Gram-negative bacteria. Nat. Commun. 7:10515. doi: 10.1038/ncomms10515
Schaar, V., Nordstrom, T., Morgelin, M., and Riesbeck, K. (2011). Moraxella catarrhalis outer membrane vesicles carry β-lactamase and promote survival of Streptococcus pneumoniae and Haemophilus influenzae by inactivating amoxicillin. Antimicrob. Agents Chemother. 55, 3845–3853. doi: 10.1128/AAC.01772-10
Schwechheimer, C., and Kuehn, M. J. (2015). Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat. Rev. Microbiol. 13, 605–619. doi: 10.1038/nrmicro3525
Shank, E. A., and Kolter, R. (2009). New developments in microbial interspecies signaling. Curr. Opin. Microbiol. 12, 205–214. doi: 10.1016/j.mib.2009.01.003
Shintani, M., Matsui, K., Inoue, J.-I., Hosoyama, A., Ohji, S., Yamazoe, A., et al. (2014). Single-cell analyses revealed transfer ranges of IncP-1, IncP-7, and IncP-9 plasmids in a soil bacterial community. Appl. Environ. Microbiol. 80, 138–145. doi: 10.1128/AEM.02571-13
Stewart, J. C. (1980). Colorimetric determination of phospholipids with ammonium ferrothiocyanate. Anal. Biochem. 104, 10–14. doi: 10.1016/0003-2697(80)90269-9
Strevett, K. A., and Chen, G. (2003). Microbial surface thermodynamics and applications. Res. Microbiol. 154, 329–335. doi: 10.1016/S0923-2508(03)00038-X
Tashiro, Y., Ichikawa, S., Shimizu, M., Toyofuku, M., Takaya, N., Nakajima-Kambe, T., et al. (2010). Variation of physiochemical properties and cell association activity of membrane vesicles with growth phase in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 76, 3732–3739. doi: 10.1128/AEM.02794-09
Tashiro, Y., Sakai, R., Toyofuku, M., Sawada, I., Nakajima-Kambe, T., Uchiyama, H., et al. (2009). Outer membrane machinery and alginate synthesis regulators control membrane vesicle production in Pseudomonas aeruginosa. J. Bacteriol. 191, 7509–7519. doi: 10.1128/JB.00722-09
Tashiro, Y., Uchiyama, H., and Nomura, N. (2012). Multifunctional membrane vesicles in Pseudomonas aeruginosa. Environ. Microbiol. 14, 1349–1362. doi: 10.1111/j.1462-2920.2011.02632.x
Tashiro, Y., Yawata, Y., Toyofuku, M., Uchiyama, H., and Nomura, N. (2013). Interspecies interaction between Pseudomonas aeruginosa and other microorganisms. Microbes Environ. 28, 13–24. doi: 10.1264/jsme2.ME12167
Toyofuku, M., Tashiro, Y., Hasegawa, Y., Kurosawa, M., and Nomura, N. (2015). Bacterial membrane vesicles, an overlooked environmental colloid: biology, environmental perspectives and applications. Adv. Colloid Interface Sci. 226, 65–77. doi: 10.1016/j.cis.2015.08.013
Toyofuku, M., Zhou, S., Sawada, I., Takaya, N., Uchiyama, H., and Nomura, N. (2014). Membrane vesicle formation is associated with pyocin production under denitrifying conditions in Pseudomonas aeruginosa PAO1. Environ. Microbiol. 16, 2927–2938. doi: 10.1111/1462-2920.12260
Turnbull, L., Toyofuku, M., Hynen, A. L., Kurosawa, M., Pessi, G., Petty, N. K., et al. (2016). Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nat. Commun. 7:11220. doi: 10.1038/ncomms11220
Wang, H., Sodagari, M., Ju, L.-K., and Zhang Newby, B.-M. (2013). Effects of shear on initial bacterial attachment in slow flowing systems. Colloids Surf. B Biointerfaces 109, 32–39. doi: 10.1016/j.colsurfb.2013.03.016
Yaron, S., Kolling, G., Simon, L., and Matthews, K. (2000). Vesicle-mediated transfer of virulence genes from Escherichia coli O157:H7 to other enteric bacteria. Appl. Environ. Microbiol. 66, 4414–4420. doi: 10.1128/AEM.66.10.4414-4420.2000
Keywords: membrane vesicles, DLVO theory, zeta potential, horizontal gene transfer, Buttiauxella agrestis
Citation: Tashiro Y, Hasegawa Y, Shintani M, Takaki K, Ohkuma M, Kimbara K and Futamata H (2017) Interaction of Bacterial Membrane Vesicles with Specific Species and Their Potential for Delivery to Target Cells. Front. Microbiol. 8:571. doi: 10.3389/fmicb.2017.00571
Received: 04 January 2017; Accepted: 20 March 2017;
Published: 07 April 2017.
Edited by:
Marc Bramkamp, Ludwig-Maximilians-Universität München, GermanyReviewed by:
Simon Ringgaard, Max Planck Society (MPG), GermanyArmen Trchounian, Yerevan State University, Armenia
Copyright © 2017 Tashiro, Hasegawa, Shintani, Takaki, Ohkuma, Kimbara and Futamata. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yosuke Tashiro, tashiro.yosuke@shizuoka.ac.jp
 Yosuke Tashiro
Yosuke Tashiro Yusuke Hasegawa
Yusuke Hasegawa Masaki Shintani
Masaki Shintani Kotaro Takaki4
Kotaro Takaki4 Moriya Ohkuma
Moriya Ohkuma Hiroyuki Futamata
Hiroyuki Futamata