- 1State Key Laboratory of Medicinal Chemical Biology, Key Laboratory of Molecular Microbiology and Technology of the Ministry of Education, Department of Microbiology, College of Life Sciences, Nankai University, Tianjin, China
- 2State Key Laboratory of Medicinal Chemical Biology, College of Life Sciences, Nankai University, Tianjin, China
- 3Department of Molecular Genetics and Microbiology, College of Medicine, University of Florida, Gainesville, FL, USA
Fis is a versatile DNA binding protein in bacteria. It has been demonstrated in multiple bacteria that Fis plays crucial roles in regulating bacterial virulence factors and optimizing bacterial adaptation to various environments. However, the role of Fis in Pseudomonas aeruginosa virulence as well as gene regulation remains largely unknown. Here, we found that Fis was required for the virulence of P. aeruginosa in a murine acute pneumonia model. Transcriptome analysis revealed that expression of T3SS genes, including master regulator ExsA, was defective in a fis::Tn mutant. We further demonstrate that the continuous transcription of exsC, exsE, exsB, and exsA driven by the exsC promoter was required for the activation of T3SS. Fis was found to specifically bind to the exsB-exsA intergenic region and plays an essential role in the transcription elongation from exsB to exsA. Therefore, we found a novel role of Fis in the regulation of exsA expression.
Introduction
Pseudomonas aeruginosa is a wide-spread Gram-negative opportunistic human pathogen that causes hospital-acquired infections especially in patients with burns, surgical wounds, cancer or cystic fibrosis (Williams et al., 2010; Gellatly and Hancock, 2013). P. aeruginosa causes acute and chronic infections by orchestrating the expression of a variety of virulence factors (Turner et al., 2014; Huber et al., 2016), such as type III secretion system (T3SS) (Anantharajah et al., 2016), iron acquisition (Reinhart and Oglesby-Sherrouse, 2016), biofilm formation (Rybtke et al., 2015) etc., and quorum sensing system (QS) dependent virulence factors such as pyocyanin, rhamnolipids, etc., (Goo et al., 2015; Moradali et al., 2017).
The T3SS injectisome of P. aeruginosa plays an important role in acute infections (Hauser, 2009). The T3SS is a syringe-like export machine that injects effectors from the bacteria directly into host cell cytosol, influencing cell signaling or leading to cell death (Cornelis, 2006; Sato and Frank, 2011). Expression of the T3SS genes is directly regulated by ExsA, the activity of which is regulated by a partner-switching mechanism (Diaz et al., 2011). Under non-inducing conditions (high Ca2+ or the absence of eukaryotic host cells), ExsA is bound by ExsD and remains inactive (Rietsch and Mekalanos, 2006). Under inducing conditions (Ca2+ depletion or contact with host cells), ExsE is secreted through the T3SS machinery, releasing its binding partner ExsC. ExsC then binds to ExsD, disrupting the association between ExsD and ExsA. The free ExsA then binds to and activates the promoters of T3SS genes (Hauser, 2009; Anantharajah et al., 2016).
ExsA was identified as the last gene in the operon composed of exsC, exsE, exsB, and exsA (Diaz et al., 2011). The promoter of exsC (PexsC) is regulated by ExsA (Diaz et al., 2011). A RNA-seq analysis under a T3SS non-inducing condition revealed a gap between the transcripts of exsB and exsA, indicating an independent promoter of exsA (Wurtzel et al., 2012). Recently, Marsden et al identified a Vfr dependent promoter of exsA (PexsA) located at the exsB-exsA intergenic region, which regulates exsA expression independent of ExsA (Marsden et al., 2016). However, it remains elusive how the exsC and exsA promoters coordinately regulate the transcription of exsA under T3SS inducing conditions.
Fis is an abundant bacterial nucleoid-associated protein, which influences DNA topology by directly binding and bending DNA (Dillon and Dorman, 2010). It also indirectly affects DNA topological state and supercoiling by regulating DNA gyrase and topoisomerase I genes expression (Travers and Muskhelishvili, 2005). Besides, binding of Fis to DNA can introduce an interwound and branched structure in the DNA which may influence the local transcriptional activity (Schneider et al., 2001; Kahramanoglou et al., 2011). It has been demonstrated that Fis coordinates expression of multiple virulence factors in various bacterial pathogens, such as pathogenic Escherichia coli (Falconi et al., 2001; Goldberg et al., 2001), Salmonella enterica serovar Typhimurium (Schechter et al., 2003; Kelly et al., 2004), Shigella flexneri (Falconi et al., 2001), Dickeya dadantii (Lautier and Nasser, 2007), and Yersinia pseudotuberculosis (Green et al., 2016). S. enterica harbors two distinct T3SSs, namely SPI-1 and SPI-2 T3SS, which play important roles in bacterial invasion of nonphagocytic cells and replication inside host cells, respectively (Ellermeier and Slauch, 2007; Figueira and Holden, 2012). It has been found that Fis regulates the SPI-1 and SPI-2 T3SSs through regulatory genes hilA and ssrB, respectively (Kelly et al., 2004; Wang et al., 2013). In D. dadantii, Fis regulates the T3SS through activation of hrpN (effector of T3SS) (Lautier and Nasser, 2007). In E. coli, the Fis binding sequence has been characterized as GNNBNwwwwwNVNNC (B = not A, V = not T, w = high proportion A or T, and N = any nucleotide) (Cho et al., 2008; Shao et al., 2008; Kahramanoglou et al., 2011; Hancock et al., 2016). In a previous study in P. aeruginosa, a fis::Tn mutant was found to be defective in swarming motility (Liberati et al., 2006; Yeung et al., 2009). Meanwhile, the swimming and twitching motility as well as biofilm formation are normal in the fis::Tn mutant (Yeung et al., 2009). However, the role of Fis in P. aeruginosa virulence as well as its DNA binding sequence has not been well defined.
Here we report that Fis is essential for P. aeruginosa virulence in a murine acute pneumonia model and the expression of T3SS genes is defective in a fis::Tn mutant. We demonstrate that under T3SS inducing condition, transcription of exsA is driven by the PexsC and binding of Fis to the intergenic region between exsB and exsA is essential for the continuation of transcription from exsB to exsA, which is required for the expression of exsA. Therefore, our results reveal a novel role of Fis in the regulation of exsA expression.
Materials and Methods
Bacterial Strains and Plasmids
The bacterial strains, plasmids and primers used in this study are listed in Table S1. E. coli DH5α (TransGen, Beijing, China), S17-1 (Simon et al., 1983) strains used for general cloning conjugal transferring (Chen et al., 2016) were cultured in Luria–Bertani (LB) broth [10 g/l tryptone (Oxoid Ltd., Basingstoke, UK), 5 g/l NaCl (BBI life sciences, Shanghai, China), 5 g/l yeast extract (Oxoid Ltd.), pH 7.0–7.5] or LB agar (LB broth containing 15 g/l agar) under aerobic conditions at 37°C. When needed, the medium was supplemented with tetracycline (50 μg/ml) (BBI life sciences), gentamicin (100 μg/ml) (BBI life sciences), or carbenicillin (150 μg/ml) (BBI life sciences) for P. aeruginosa PA14 (Liberati et al., 2006), and ampicillin (100 μg/ml) (BBI life sciences) or tetracycline (10 μg/ml) for E. coli. When needed, Isopropyl β-D-1-thiogalactopyranoside (IPTG) at indicated concentrations was added to culture mediums.
For DNA manipulation, standard protocols or manufacture instructions of commercial products were followed. For the complementation of the fis gene, the open reading frame of fis and its upstream 486 bp region was amplified by PCR with PA14 chromosomal DNA as the template. The PCR product was cloned into the BamHI-EcoRI sites of pUC18T-mini-Tn7T-Tc (Choi and Schweizer, 2006), resulting in pUC18T-mini-Tn7T-Tc-fis. The pEX18Tc-T0T1 insertion was constructed by cloning the 934 bp upstream and 987 bp downstream fragments of the exsB-exsA intergenic region into the EcoRI-HindIII sites of plasmid pEX18Tc (Hoang et al., 1998), and a 289 bp DNA fragment containing terminators T0T1 was amplified by PCR from pUC18T-mini-Tn7T (Choi and Schweizer, 2006) and inserted in between the two fragments. Chromosomal gene mutations were generated by homologous recombination as described previously (Hoang et al., 1998). To construct the C-terminus His-tagged ExsA driven by its native promoter, a DNA fragment containing the 300 bp fragment upstream of exsA and the exsA coding region was amplified by PCR, the His-tag coding sequence was included in one of the PCR primers. The PCR product was cloned into the XbaI-HindIII sites of a promoterless pUCP20 (Li et al., 2016). To construct the exsA promoter lacZ transcriptional fusion (PexsA-lacZ), the 500 bp fragment upstream of the exsA coding region was amplified by PCR and cloned into the BamHI-EcoRI sites of pDN19lacZΩ. Sequences of the PCR primers were listed in Table S1.
Real-Time PCR
Bacteria were grown at 37°C under indicated conditions to indicated optical density of 600 nm (OD600). Total RNA was isolated with a RNAprep Pure Bacteria Kit (Tiangen Biotech, Beijing, China). The cDNA was synthesized from total RNA using random primers and PrimeScript Reverse Transcriptase (TaKaRa, Dalian, China). Specific Primers (Table S1) were used for reverse transcription (RT) and quantitative PCR. For quantitative PCR, cDNA was mixed with 4 pmol of forward and reverse primers and SYBR Premix Ex Taq™ II (TaKaRa) in a total reaction volume of 20 μl. The 30S ribosomal protein coding gene rpsL was used as an internal control (Li et al., 2013). The results were determined using a CFX Connect Real-Time system (Bio-Rad, USA).
Transcriptome Sequencing and Analysis
The transcriptome sequencing analysis was performed by Beijing Genomics Institute. Total RNA was isolated from bacteria at OD600 of 1.0 with an RNAprep Pure Bacteria Kit (Tiangen). After DNase I (NEB) digestion, rRNA was removed from the total RNA by using Ribo-Zero Magnetic Kit (Bacteria, EPICENTRE). The mRNA was fragmented into short fragments by using fragmentation buffer (Ambion). Then cDNA was synthesized using the mRNA fragments as templates. The purified fragmented cDNA was combined with End Repair Mix and A-Tailing Mix for end reparation and single nucleotide A (adenine) addition. Then, the short fragments were connected with adapters. After agarose gel electrophoresis, the suitable fragments were selected for the PCR amplification as templates. During the quality control steps, Agilent 2100 Bioanaylzer and ABI StepOnePlus Real-Time PCR System were used in quantification and qualification of the sample library. At last, the library was sequenced using Illumina HiSeq™ 2000 or other sequencer when necessary. The RNA expression analysis was based on the predicted genes of strain PA14 (http://www.pseudomonas.com). P-values were calculated referring to “the significance of digital gene expression profiles” (Audic and Claverie, 1997). A strict algorithm to identify differentially expressed genes between two samples is described as follow:
Denote the number of unambiguous clean tags (which means reads in RNA-Seq) from gene A as x, given every gene's expression occupies only a small part of the library, x yields to the Poisson distribution:
The total clean tag number of the sample 1 is N1, and total clean tag number of sample 2 is N2; gene A holds x tags in sample 1 and y tags in sample 2. The probability of gene A expressed equally between two samples can be calculated with:
The full transcriptome sequencing data has been deposited in the NCBI SRA, with accession number SRP099178.
Cell Culture and Cytotoxicity Assay
The bacterial cytotoxicity was determined by the lactate dehydrogenase (LDH) release assay as previously described (Anderson et al., 2010; Li et al., 2016). HeLa cells (Li et al., 2016) were cultured in DMEM medium (Corning, USA) with 10% fetal bovine serum (FBS) supplemented with 1% penicillin/streptomycin (Gibco, USA) at 37°C in 5% CO2. 1.5 × 104 HeLa cells were seeded into each well of a 96-well plate and cultured for 24 h. The medium was replaced with antibiotic and FBS free DMEM before P. aeruginosa infection. Overnight bacterial cultures were subcultured in fresh LB and grown to an OD600 of 1.0. Bacteria were washed once and resuspended in 1 × PBS. HeLa cells were infected with bacteria at a multiplicity of infection (MOI) of 50. The plate was centrifuged at 2,000 g for 10 min to synchronize the bacterial infection. After 3 h, LDH present in the supernatant was measured using the LDH cytotoxicity assay kit (Beyotime, China). Cells treated with 1% Triton X-100 were used as positive control of maximum release (100% percentage of cytotoxicity). The percentage of cytotoxicity was calculated following the manufacturer's instruction.
Murine Acute Pneumonia Model
All animal experiments complied with Nankai University and Chinese National Guidelines regarding the use of animals in research. The protocol was approved by the institutional animal care and use committee of the college of life sciences of Nankai University with permit number: NK-04-2012. Overnight bacterial cultures were subcultured in fresh LB at 37°C to an OD600 of 1.0. The bacterial cultures were centrifuged at 12 000 rpm for 1 min, and the pellets were resuspended in PBS. Each 6-week old female BALB/c mouse (Academy of Military Medical Sciences, Beijing, China) was anesthetized with 100 μL of 7.5% chloral hydrate by intraperitoneal injection. Then each mouse was intranasally inoculated with 1 × 107 CFU bacteria. Bacterial colonization in the lung was determined as described previously (Sun et al., 2014). Briefly, 14 h post infection (hpi), mice were sacrificed by inhalation of CO2. Lungs were isolated and homogenized in 1% proteose peptone (Solarbio, Beijing, China). Bacterial loads were determined by plating serial dilutions and counting colonies. For the survival assay, the mice were intranasally inoculated with 2 × 107 CFU bacteria and monitored for 5 days.
RNA Extraction from In vivo Samples
Six week old female BALB/c mice were intranasally inoculated with 2 × 107 CFU bacteria as described above. Mice were sacrificed by inhalation of CO2 at 6 hpi. Bronchoalveolar lavage fluid (BALF) was obtained by annulation of the trachea followed by twice instillations of 1 ml sterile PBS with 0.5 mM EDTA. 200 μl of the BALF was used for bacterial counting. The remaining BALF was centrifuged at 12,000 rpm for 2 min, and the pellets were resuspended in 200 μl TRIzol reagent (Thermo Fisher Scientific, USA). Total RNA was isolated using a Direct-zol RNA Miniprep kit (ZYMO research, USA).
Electrophoretic Mobility Shift Assay (EMSA)
EMSA was performed as previously described with minor modification (Sun et al., 2014). Briefly, DNA fragments corresponding to sequence upstream of exsA and exsC were synthesized. DNA fragments (200 ng) were incubated with 0 to 6 mM purified recombinant Fis protein at 25°C for 30 min in a 20-μl reaction [10 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 5 mM KCl, 0.1% (v/v) NP-40 (Solarbio), and 1 mM dithiothreitol]. Samples were loaded onto an 8% native polyacrylamide gel in 0.5 × Tris-borate-EDTA (TBE) buffer (0.044 M Tris base, 0.044 M boric acid, 0.001 M EDTA, pH 8.0) that had been prerun for 1 h, electrophoresed on ice at 100 V for 1.5 h, followed by DNA staining in 0.5 × TBE containing 0.5 μg/ml ethidium bromide. Bands were visualized with a molecular imager ChemiDoc™ XRS+ (Bio-Rad).
Western Blotting
Overnight bacterial cultures were subcultured in fresh LB with or without 5 mM EGTA at 37°C for 3 h. The pellets from 1 ml cultures were then resuspended in 100 μl loading buffer (50 mM Tris-HCl, pH 6.8, 2% (w/v) SDS, 0.1% (w/v) BPB, 10% (v/v) Glycerol, 1% (v/v) 2-ME). Protein samples from equivalent amounts of protein were loaded onto a 15% SDS-PAGE gel. Proteins were separated by electrophoresis followed by transferring to a polyvinylidene difluoride (PVDF) membrane (Millipore, USA). The target proteins were hybridized with a rabbit monoclonal anti-His antibody (CST, USA) or a mouse monoclonal anti-Flag antibody (Sigma, USA). Signals were detected with an Immobilon™ Western kit (Millipore).
Transcriptional Reporter Assay
Overnight bacterial cultures were subcultured in fresh LB with or without 5 mM EGTA at 37°C with shaking, and samples were harvested when OD600 of the cultures reached 1.0. The β-galactosidase activity was measured with substrate ortho-nitrophenyl-galactopyranoside (ONPG) (BBI life sciences) as previously described (Ha et al., 2004).
Statistical Analysis
The statistical analyses were performed with the Prism software (Graphpad Software). The real time PCR and β-galactosidase assay results were analyzed by the Student's t-test (two-tailed). Bacterial colonization results were analyzed with the Mann-Whitney test. Survival data were analyzed with the Log-rank (Mantel-Cox) test.
Results
Fis Is Essential for the Virulence of P. aeruginosa in a Mouse Acute Pneumonia Model
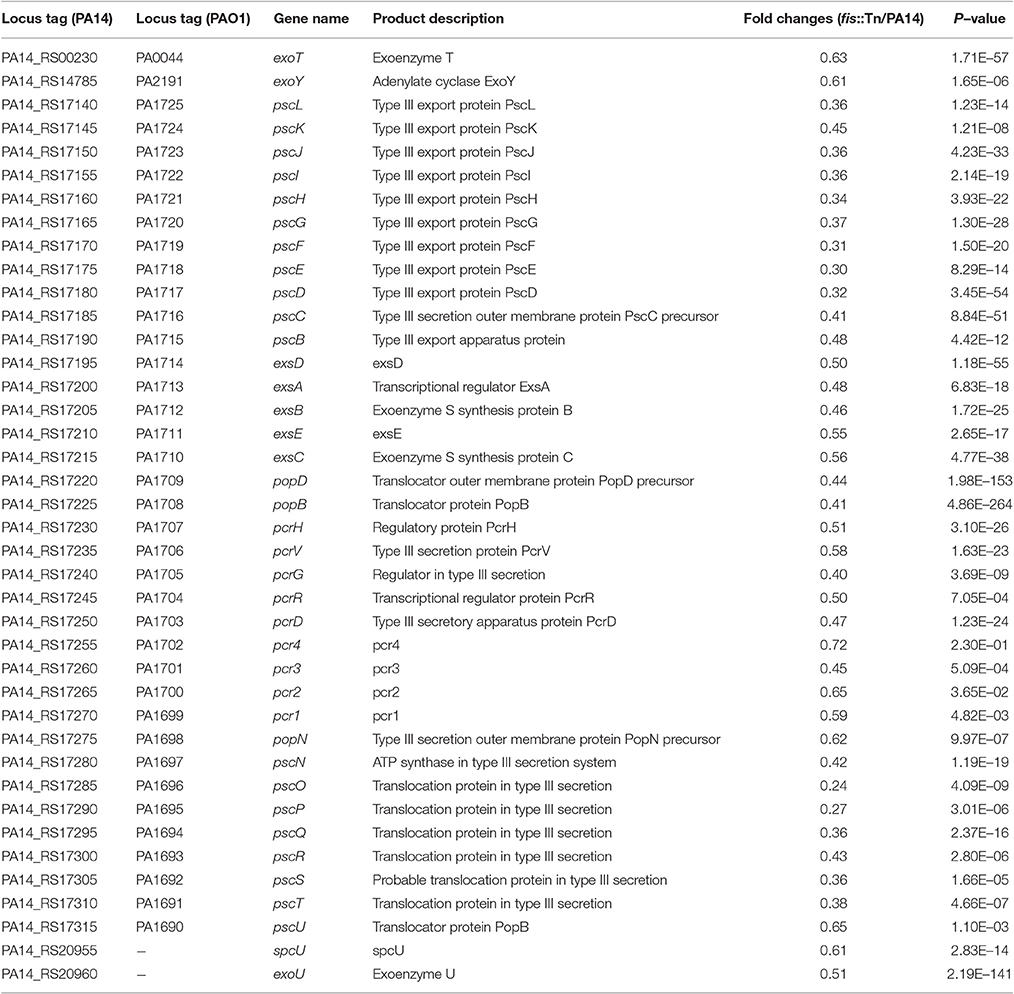
To examine the role of Fis in the virulence of P. aeruginosa, we infected mice with wild type PA14 or a fis::Tn mutant from the nonredundant library of PA14 transposon mutants (Liberati et al., 2006) in an acute pneumonia model. 14 h post infection, lungs were isolated and the bacterial numbers were determined by serial dilution and plating. Compared to the wild type strain, the number of the fis::Tn mutant was significantly lower (Figure 1A). Complementation with a fis gene driven by its native promoter partially restored the bacterial load (Figure 1A). To confirm the role of Fis in virulence, we monitored the survival rate in the acute pneumonia model. Infection with wild type PA14 or the complemented strain caused death in all the infected mice, whereas infection with the fis::Tn resulted in 40% survival rate (Figure 1B).
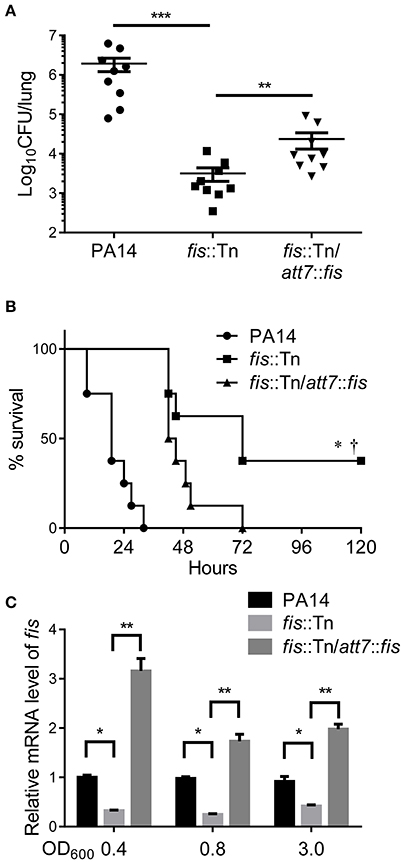
Figure 1. Fis is essential for the bacterial virulence in vivo. (A) Mice were inoculated intranasally with 1 × 107 CFU bacteria of indicated strains. 14 hpi, mice were sacrificed and lungs were isolated and homogenized. Bacterial loads were determined by serial dilution and plating. The central bar indicates the mean, and error bars indicate standard error of the mean. ***p < 0.001; **p < 0.01 by the Mann-Whitney test. (B) Mice were inoculated intranasally with 2 × 107 CFU bacteria of indicated strains. The mice were monitored for 5 days after the infection. *p < 0.001, compared to wild type PA14 by log-rank (Mantel-Cox) test; †p < 0.05, compared to the complemented strain (fis::Tn/att7::fis) by log-rank (Mantel-Cox) test. (C) Total RNA was isolated from bacterial culture at indicated optical densities (OD600). cDNA was synthesized with fis and rpsL specific primers. Relative mRNA levels of fis were determined by quantitative real-time PCR. The 30S ribosomal protein coding gene rpsL was used as an internal control. Data represents the mean ± standard deviation from three samples. *p < 0.05; **p < 0.01 by Student's t-test.
In the PA14 fis::Tn mutant, the Tn was inserted right before the stop codon of the fis gene (Liberati et al., 2006). To examine the effect of Tn insertion on the expression of Fis, we determined the mRNA level of Fis by real time PCR. Compared to wild type PA14, the mRNA level of Fis was lower in the fis::Tn mutant, but higher in the complemented strain in bacteria at early-, mid-log and stationary growth phases (Figure 1C). Thus, the excessive expression of Fis in the complemented strain might affect bacterial virulence, resulting in partial restoration of bacterial number in lung and slower killing of infected mice (Figures 1A,B).
The fis gene is in the same operon with PA4852. Thus, the expression of PA4852 might be affected by the Tn insertion. A real time PCR assay revealed similar mRNA levels of PA4852 in wild type PA14 and the fis::Tn mutant (Figure S1), suggesting that the Tn insertion did not affect the expression of PA4852. It has been demonstrated that the translation of Fis is repressed by the small RNA RgsA (Lu et al., 2016). The RgsA level in the fis::Tn mutant was similar as that in the wild type PA14 (Figure S2), suggesting that the RgsA mediated regulation on Fis might be normal in the fis::Tn mutant. In combination, these results suggest that the lower expression of Fis might result in attenuation of the virulence of PA14. Since fis has been indicated as an essential gene in P. aeruginosa, we used the fis::Tn mutant in our further studies (Liberati et al., 2006; Jakovleva et al., 2012).
Fis Is Required for Expression of the T3SS and for Cytotoxicity
To understand how Fis affects bacterial virulence, we compared transcriptome profiles between the fis::Tn mutant and wild type PA14. Interestingly, the T3SS genes were down regulated in the fis::Tn mutant (Table 1). To confirm the expression levels of T3SS genes, bacteria were cultured under T3SS non-inducing and inducing conditions. Real time PCR assay revealed lower mRNA levels of exoU, pcrV, exsC, and exsA in the fis::Tn mutant, which were partially restored in the complemented strain (Figure 2A). By utilizing a transcriptional fusion of the exsC promoter and a lacZ gene (PexsC-lacZ), we found the exsC promoter activity was lower in the fis::Tn mutant (Figure S3). To confirm the expression level of T3SS genes, a His-tagged exoU (ExoU-His) driven by its native promoter was transferred into the bacteria. As shown in Figure 2B, the ExoU-His level was reduced in the fis::Tn mutant under T3SS inducing condition. Since T3SS plays a major role in bacterial cytotoxicity (Yahr and Wolfgang, 2006; Tan et al., 2016), we infected HeLa cells with PA14, the fis::Tn mutant and the complemented strain. Consistent with the mRNA levels of T3SS genes, the fis::Tn mutant displayed reduced cytotoxicity to HeLa cells (Figure 2C). We then examined the expression levels of T3SS genes in the murine acute pneumonia model. Bacteria were isolated from BALF at 6 hpi. mRNA levels of exoU, pcrV, exsC and exsA were lower in the fis::Tn mutant, which was partly restored in the complemented strain (Figure 2D).
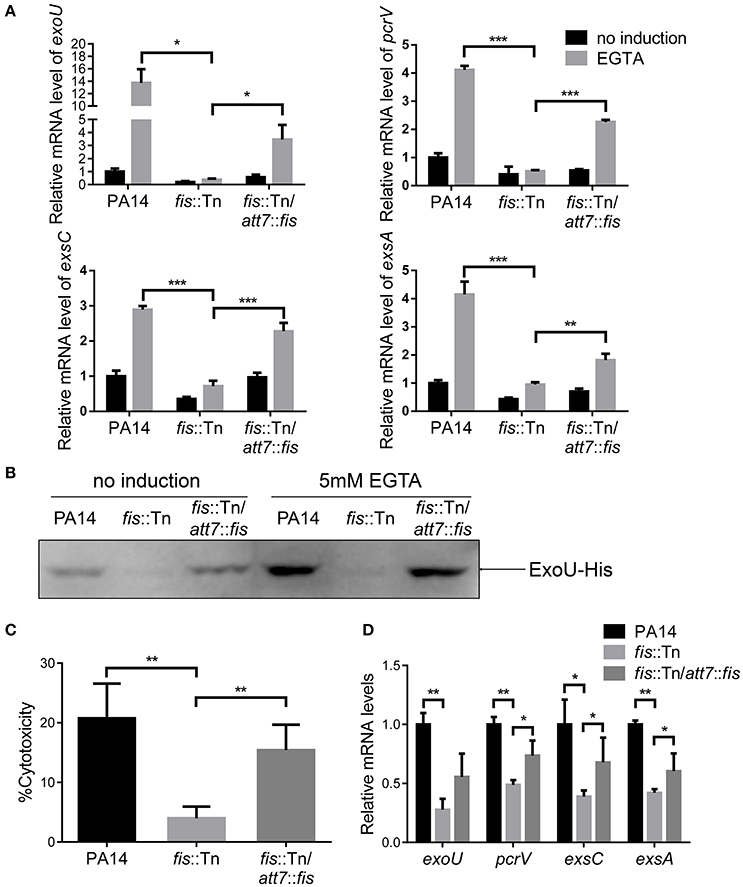
Figure 2. Fis is required for T3SS gene expression and bacterial cytotoxicity. (A) Relative mRNA levels of T3SS genes exoU, pcrV, exsC, exsA. Total RNA was isolated from bacteria grown with or without 5 mM EGTA and relative mRNA levels of these genes were determined by quantitative real-time PCR. Data represents the mean ± standard deviation from three samples. *p < 0.05; **p < 0.01; ***p < 0.001 by Student's t-test. (B) PA14, fis::Tn mutant and fis::Tn/att7::fis carrying an exoU-His driven by its native promoter (PexoU-exoU-His) were grown at 37°C with or without 5 mM EGTA for 3 h. Proteins samples from equal amounts of protein were separated by SDS-PAGE and the ExoU-His levels were determined by western blotting analysis using an anti-His antibody. (C) Bacterial cytotoxicity on HeLa cells. HeLa cells were infected with indicated strains at a MOI of 50 for 3 h. The bacterial cytotoxicity was determined by the LDH release assay. Error bars indicate standard deviations of triplicate assays. **p < 0.01 by Student's t-test. (D) Relative mRNA levels of T3SS genes during lung infection. Mice were infected intranasally with indicated strains. 6 hpi, bacteria from BALF were collected, followed by RNA isolation. Relative mRNA levels of exoU, pcrV, exsC, and exsA were determined by quantitative real-time PCR. Data represents the mean ± standard deviation from three independent experiments. *p < 0.05; **p < 0.01 by Student's t-test.
To further confirm the role of Fis in the regulation of T3SS, we cloned a C-terminus His-tagged fis gene (fis-His) into pMMB67EH (Fürste et al., 1986), where the expression of fis-His is driven by an inducible tac promoter. In the presence of this plasmid, we deleted the chromosomal fis gene, resulting in the strain Δfis/pMMB67EH-fis-His. In the absence of IPTG, the strain could not grow up in LB, confirming the essential role of Fis in P. aeruginosa. For an unknown reason, EGTA significantly repressed the growth of the strain Δfis/pMMB67EH-fis-His in the presence of various concentrations of IPTG. Thus, we only examined the expression of T3SS genes in LB medium. In the presence of 0.02 mM IPTG, the RNA levels of T3SS genes were lower in the Δfis/pMMB67EH-fis-His strain. Increasing amount of IPTG resulted in higher amount of Fis-His protein and up regulation of the T3SS genes (Figures 3A,B). Next, we examined the bacterial cytotoxicity with IPTG in the tissue culture medium. At low concentration of IPTG, the bacteria displayed minimal cytotoxicity, whereas the presence of IPTG increased the bacterial cytotoxicity (Figure 3C). In combination, these results suggest that Fis is required for the activation of the T3SS.
Figure 3. Fis is required for the activation of the T3SS. (A) The strain Δfis/pMMB67EH-fis-His was grown at 37°C with indicated concentrations of IPTG to an OD600 of 1.0. Proteins samples from equal amounts of protein were separated by SDS-PAGE and the Fis-His levels were determined by western blotting analysis using an anti-His antibody. (B) Relative mRNA levels of T3SS genes. Total RNA of indicated strains was isolated from bacteria grown with indicated concentrations of IPTG and mRNA levels of T3SS genes were determined by quantitative real time PCR. Data represents the mean ± standard deviation from three samples. *p < 0.05; **p < 0.01; ***p < 0.001 by Student's t-test. (C) Bacterial cytotoxicity on HeLa cells. HeLa cells were infected with indicated strains with indicated concentration of IPTG at a MOI of 50 for 3 h. The bacterial cytotoxicity was determined by the LDH release assay. Error bars indicate standard deviations of triplicate assays. **p < 0.01, by Student's t-test. (D) HeLa cells were infected with indicated strains at a MOI of 50 for 3 h. The bacterial cytotoxicity was determined by the LDH release assay. Error bars indicate standard deviations of triplicate assays. **p < 0.01; ***p < 0.001 by Student's t-test. (E) Mice were inoculated intranasally with 1 × 107 CFU bacteria of indicated strains. 14 hpi, mice were sacrificed and lungs were isolated and homogenized. Bacterial loads were determined by serial dilution and plating. The central bar indicates the mean, and error bars indicate standard error of the mean. ***p < 0.001 by the Mann-Whitney test.
In LB broth, the growth rate of the fis::Tn mutant was similar as that of the wild type PA14 (Figure S4). However, it has been reported that the fis::Tn mutant grows more slowly in BM2 swarming medium (Yeung et al., 2009), indicating a role of Fis in bacterial growth under certain conditions. Thus, the lower bacterial loads in the lungs from the fis::Tn mutant infected mice might be due to slower bacteria growth rate. To verify the role of T3SS in the reduced colonization of the fis::Tn mutant, we overexpressed exsA in the fis::Tn mutant and examined bacterial cytotoxicity and colonization of lungs. As shown in Figure S5, overexpression of exsA did not affect the growth of the fis::Tn mutant. Overexpression of exsA in the fis::Tn mutant restored bacterial cytotoxicity (Figure 3D) and colonization in vivo (Figure 3E). Therefore, the defective T3SS indeed attributed to the attenuated virulence of the fis::Tn mutant.
Fis Directly Interacts with exsA Promoter Region
T3SS genes are directly regulated by ExsA (Hauser, 2009; Diaz et al., 2011) and we have found that the mRNA level of exsA was lower in the fis::Tn mutant (Figures 2A,D). Since Fis functions as a transcriptional factor (TF) (Muskhelishvili et al., 1995; Schneider et al., 2001; Aiyar et al., 2002; Cho et al., 2008; Kahramanoglou et al., 2011; Prigent-Combaret et al., 2012), we suspected Fis might directly affect the transcription of exsA. By carefully searching for the consensus Fis binding motif (Cho et al., 2008; Shao et al., 2008; Kahramanoglou et al., 2011; Hancock et al., 2016), we identified a possible Fis binding site at the -10 box of the exsA promoter (Figure 4A). An EMSA assay demonstrated an interaction between Fis and this fragment (Figure 4B). As a control, the fragments up stream of the binding site could not bind to Fis at the concentration of 2 mM. Of note, at higher concentrations of Fis, DNA retardation was observed with all the tested probes (Figure 4B), which might be attributed to the nonspecific coating of DNA by Fis, resulting in the formation of Fis-DNA filament referred to as a “low mobility complex (LMC)” (Skoko et al., 2006). To further verify the specific binding between Fis and the -10 box of the PexsA, we mutated the conserved nucleotides of the Fis binding site based on the study in E. coli. The replaced nucleotides were highlighted in Figure 4C. Indeed, alternation of the highly conserved nucleotides abolished the interaction between Fis and the fragment (Figure 4C). Since ExsC positively regulates T3SS (Hauser, 2009; Diaz et al., 2011), we also examined whether Fis specifically binds to the exsC promoter region. However, no specific binding between Fis and the test fragments were observed (Figure S6). These results indicated that Fis directly binds to the -10 box of PexsA.
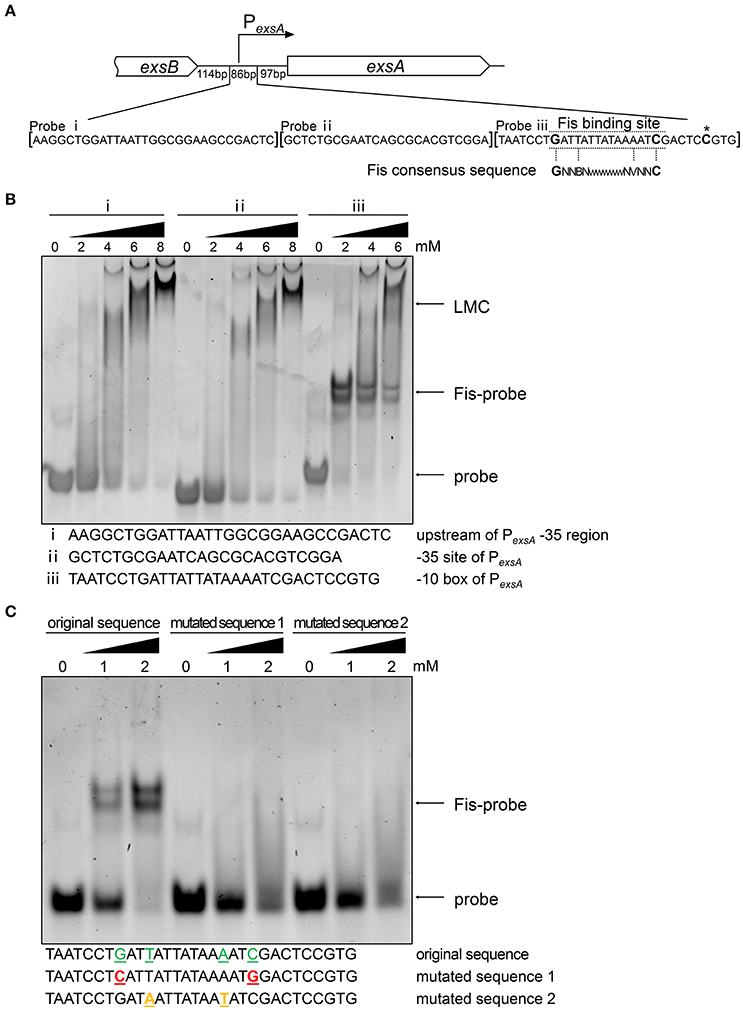
Figure 4. Fis directly interacts with exsA promoter region. (A) Diagram of the exsA promoter region. PexsA is indicated by an arrow. Sequences of the probes used in EMSA and the consensus Fis binding sequence are shown. The transcription start site of exsA is indicated by an asterisk. Fragment i, ii, and iii represent DNA probes used in the EMSA. (B) Fis was incubated with probe i, ii, or iii for 30 min at 25°C. Arrows indicate the positions of unbound probes or the Fis-probe complex and LMC. (C) Binding of the Fis to the DNA fragment containing PexsA -10 box original and mutated sequences. Colored underlined letters represent mutated nucleotides. Arrows indicate positions of the Fis-probe complex and unbound probes, respectively.
Role of Fis in the PexsA Activity
The specific binding between Fis and the PexsA region raised a possibility that Fis affects the activity of PexsA. To test this possibility, we constructed a PexsA-lacZ transcriptional fusion (Figure 5A). The β-galactosidase levels were similar between wild type PA14 and the fis::Tn mutant under either T3SS non-inducing or inducing condition (Figure 5B). To confirm this observation, we constructed a C-terminus His-tagged ExsA driven by the PexsA (Figure 5C). Consistent with the PexsA-lacZ reporter result, similar ExsA-His protein levels were observed in wild type PA14 and the fis::Tn mutant (Figure 5D). These results suggest that Fis might not directly affect the PexsA activity.
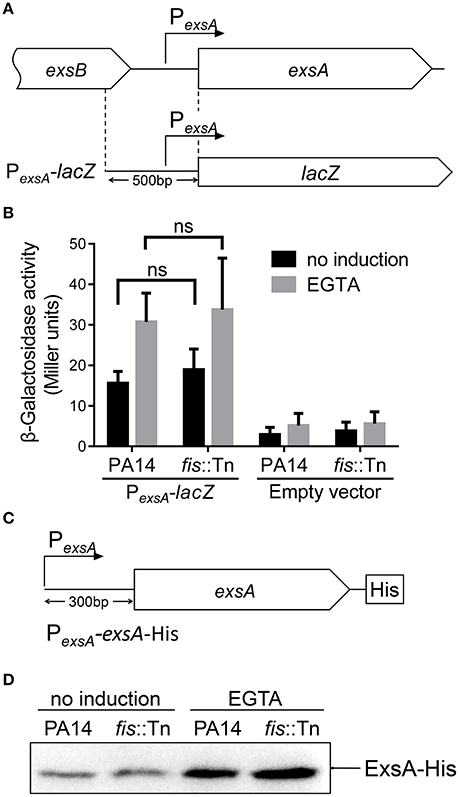
Figure 5. Role of Fis in the regulation of PexsA. (A) Diagram of the PexsA-lacZ transcriptional fusion. (B) PA14 and the fis::Tn mutant carrying the PexsA-lacZ fusion reporter or empty vector (promoterless lacZ) were grown at 37°C with or without 5 mM EGTA for 3 h. The values (Miller units) are the means of three experiments. ns, not significant by Student's t-test. (C) Diagram of the PexsA-exsA-His construct. The exsA open reading frame with its upstream 300 bp region was fused with a 6 × His tag at the C-terminus. (D) PA14 and the fis::Tn mutant carrying an exsA-His driven by its native promoter (PexsA-exsA-His) were grown at 37°C with or without 5 mM EGTA for 3 h. Proteins samples from equal amounts of protein were separated by SDS-PAGE. The ExsA-His levels were determined by western blotting analysis with an anti-His antibody.
exsA Transcription Relies Mainly on the Transcription Initiated from PexsC
Previously studies demonstrated that the PexsA transcriptional activity is much weaker than PexsC (Yahr and Frank, 1994; Marsden et al., 2016). We observed the same results under both T3SS non-inducing and inducing conditions (Figure 6B). It has been speculated that transcription of exsA might be driven by the exsC promoter under T3SS inducing condition (Hauser, 2009; Diaz et al., 2011). If this is the case, Fis might be involved in the regulation of exsA transcription initiated from the exsC promoter. To test this hypothesis, we firstly examined whether exsCEBA are in one transcript. We constructed a PexsC-A-lacZ transcriptional fusion, where the lacZ gene was cloned downstream of a fragment ranging from PexsC to exsA coding region (Figure 6A). In wild type PA14, the β-galactosidase levels were much higher than that driven by PexsA (PexsA-lacZ), indicating a continuous transcription from PexsC to exsA coding region (Figure 6B). We then designed a pair of primers annealing to the coding regions of exsB and exsA, thus the PCR product spans the intergenic region between exsB and exsA (Figure 7A). Total RNA was isolated from PA14 grown under T3SS non-inducing and inducing conditions, followed by RT-PCR. PCR products were observed at both conditions, with higher amount under T3SS inducing condition (Figure 7B), indicating a transcript from exsB to exsA. To further confirm the continuous transcription from exsB to exsA, we performed real time PCR with primers annealing to the coding regions of exsC, exsB, exsA as well as the exsB-exsA intergenic region (Figure 7A). The mRNA levels of all the tested genes and the RNA level of the exsB-exsA intergenic region were induced by EGTA in wild type PA14 (Figure 7C). These results suggest that exsA and exsB are in one transcript and the transcription level responses to the T3SS inducing signal.
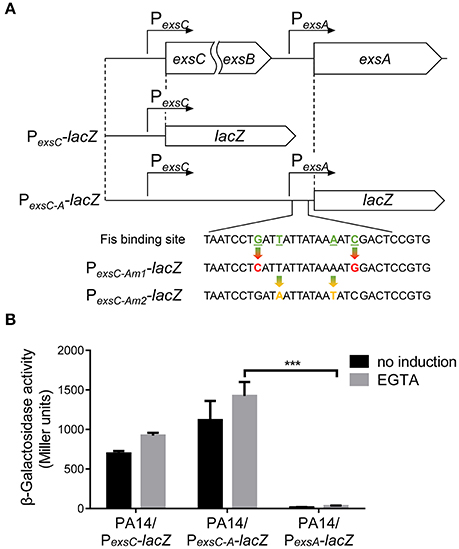
Figure 6. Transcription driven by the exsC or exsA promoter. (A) Diagram of the PexsC-lacZ and PexsC-A-lacZ transcriptional fusions. Point mutations of Fis binding site in PexsC-Am1-lacZ and PexsC-Am2-lacZ transcriptional fusions are indicated by arrows. (B) PA14 carrying PexsC-lacZ, PexsC-A-lacZ or PexsA-lacZ were grown at 37°C with or without 5 mM EGTA and assayed for β-galactosidase activities. The reported values (Miller units) are the means of results of at least three independent experiments. ***p < 0.001 by Student's t-test.
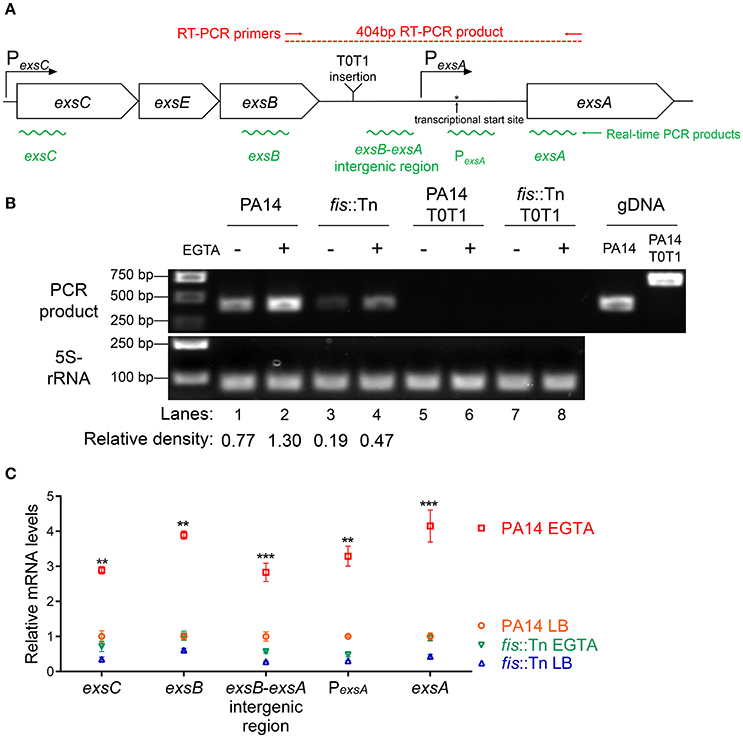
Figure 7. Transcription of exsC, exsE, exsB and exsA. (A) Diagram of the exsCEBA operons. PexsC and PexsA are indicated by arrows. Red arrows indicate the directions and locations of the primers for RT-PCR. Insertion site of the transcriptional terminator T0T1 is indicated. Green wavy lines indicate positions of real time PCR products. (B) Total RNA was isolated from indicated strains grown with or without 5 mM EGTA for 3 h. cDNA was synthesized and used as templates in PCR. The 5S rRNA was used as an internal control. The density of each band in lanes 1–4 was determined with ImageJ (ImageJ software k1.45). The relative density was calculated by dividing the density of each exsB-exsA RT-PCR product by that of the corresponding 5S rRNA RT-PCR product (C) Relative mRNA levels of five regions within the exsCEBA operon. PA14 and fis::Tn mutant were grown in the presence or absence of 5 mM EGTA. The value of each tested fragment represents the RNA level relative to that in wild type PA14 grown in LB medium. Data represents the mean ± standard deviation from three independent experiments. **p < 0.01, ***p < 0.001, compared to wild type PA14 grown in LB medium by Student's t-test.
If the PexsC driven transcription of exsA is required for the activation of the T3SS, interception of the transcription between exsB and exsA coding regions should diminish the expression of exsA and consequently other T3SS genes. To test this hypothesis, we inserted two tandem transcription terminators (T0T1) up stream of the exsA promoter (Figure 7A). RT-PCR confirmed the break between exsB and exsA transcript (Figure 7B, lanes 5–8). Indeed, the exsA mRNA levels were significantly reduced by the insertion of T0T1 under both T3SS inducing and non-inducing conditions (Figure 8A). Consequently, the levels of exsC mRNA and the ExoU protein were reduced by the T0T1 insertion (Figures 8B,C). Therefore, the PexsC driven transcription of exsA plays an essential role in the expression of T3SS genes.
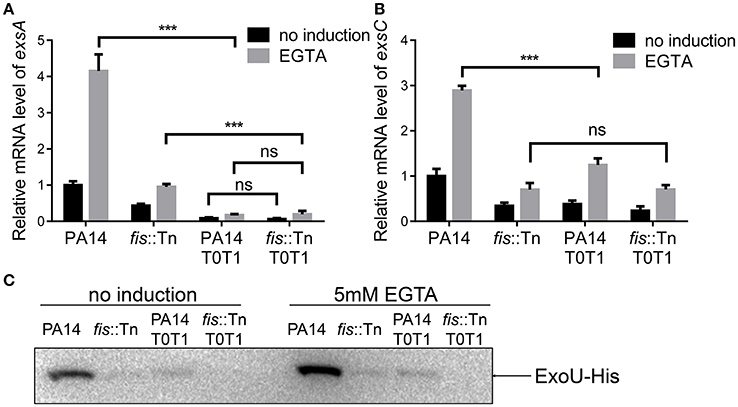
Figure 8. exsA transcription relies mainly on PexsC. (A) Relative mRNA levels of exsA. Total RNA of indicated strains was isolated from bacteria grown with or without 5 mM EGTA and mRNA levels of exsA were determined by quantitative real time PCR. Data represents the mean ± standard deviation from three samples. ***p < 0.001 by Student's t-test. (B) Relative mRNA levels of exsC. Total RNA of indicated strains was isolated from bacteria grown with or without 5 mM EGTA and mRNA levels of exsC were determined by quantitative real time PCR. Data represents the mean ± standard deviation from three samples. ***p < 0.001 by Student's t-test. (C) PA14, fis::Tn mutant, PA14 T0T1 or fis::Tn T0T1 carrying an exoU-His driven by its native promoter (PexoU-exoU-His) were grown at 37°C with or without 5 mM EGTA for 3 h. Proteins samples from equal amounts of protein were separated by SDS-PAGE. ExoU-His levels were determined by western blotting analysis using an anti-His antibody.
Fis Is Required for the Transcription of exsA Driven by PexsC
We have demonstrated that the PexsA activity in the fis::Tn mutant was similar to that in the wild type PA14 (Figure 5B). Consistently, insertion of T0T1 up stream of PexsA in the fis::Tn mutant resulted in similar mRNA level of exsA as that in PA14 with the T0T1 insertion (Figure 8A), confirming that Fis is not involved in the regulation of PexsA activity. Thus, we suspected that Fis might affect the transcription of exsA driven by PexsC and subsequent expression of T3SS genes. To test this possibility, we transferred the PexsC-A-lacZ transcriptional fusion into the fis::Tn mutant. The β-galactosidase level was significantly lower in the fis::Tn mutant than that in the wild type PA14 (Figure 9A). To further verify that Fis affects the transcription of exsA driven by PexsC by directly binding to the exsB-exsA intergenic region, the conserved nucleotides inside the Fis binding sequence were mutated (Figure 6A), which had been shown to significantly reduced the binding between Fis and the fragment (Figure 4C). The mutated PexsC-A-lacZ fusions were designated as PexsC-Am1-lacZ and PexsC-Am2-lacZ, respectively (Figure 6A). Compared to the original PexsC-A-lacZ, the PexsC-Am1-lacZ, and PexsC-Am2-lacZ yielded lower levels of β-galactosidase in wild type PA14 (Figure 9A). In combination, these results suggest that Fis affects the PexsC driven exsA transcription through direct binding to the exsB-exsA intergenic region.
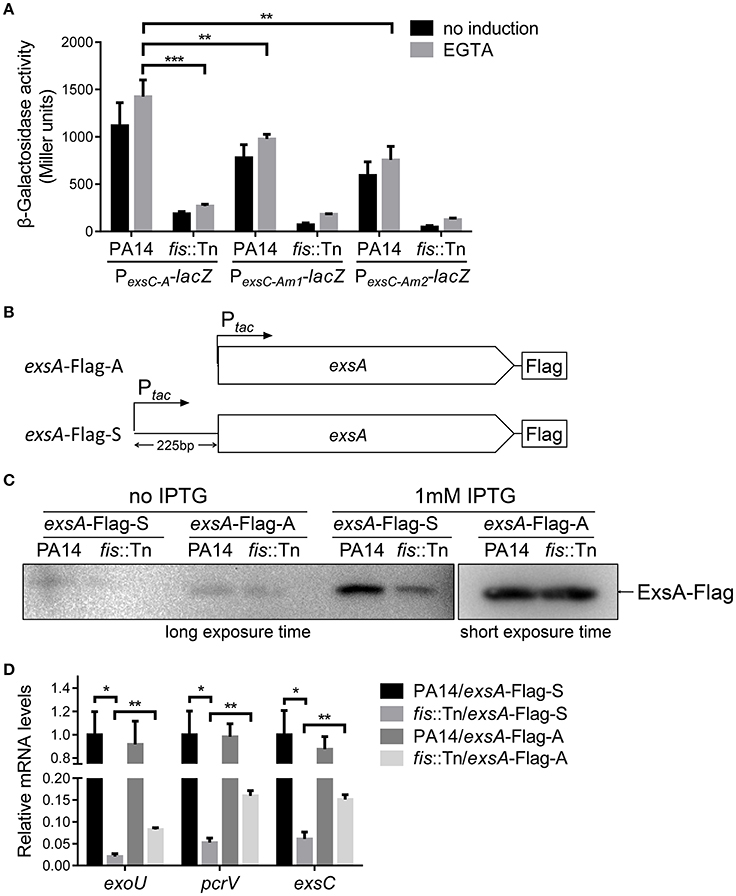
Figure 9. Role of Fis in the transcription elongation from exsB to exsA. (A) PA14 and fis::Tn mutant carrying PexsC-A-lacZ, PexsC-Am1-lacZ, or PexsC-Am2-lacZ transcriptional reporters were grown at 37°C with or without 5 mM EGTA. The values are the means of at least three independent experiments. ***p < 0.001; **p < 0.01 by Student's t-test. (B) Constructs of exsA-Flag-S and exsA-Flag-A. exsA-Flag-S contains exsA ORF and 225 bp upstream fragment fused with tac promoter and exsA-Flag-A contains exsA ORF only fused with tac promoter. (C) PA14 and fis::Tn mutant carrying plasmids exsA-Flag-S or exsA-Flag-A were grown with or without 1 mM IPTG for 3 h. ExsA-Flag levels were determined by western blot using an anti-Flag antibody. The amounts of protein in different samples were equal. (D) Relative mRNA levels of T3SS genes exoU, pcrV, exsC, exsA. Total RNA was isolated from bacteria grown with 1 mM IPTG and relative mRNA levels of these genes were determined by quantitative real-time PCR. Data represents the mean ± standard deviation from three samples. *p < 0.05, **p < 0.01 by Student's t-test.
We then isolated RNA from the fis::Tn mutant under T3SS inducing or non-inducing conditions and performed RT-PCR with the primers annealing to the coding regions of exsB and exsA. The PCR product level was much lower than that in the wild type PA14 (Figure 7B). Consistently, real time PCR results revealed that the RNA spanning the exsB-exsA intergenic region was less in the fis::Tn mutant under T3SS inducing condition (Figure 7C). These results suggest a role of Fis in the continuous transcription from exsB to exsA. Since ExsA activates PexsC (Diaz et al., 2011), our observation might be due to defective translation of the ExsA indirectly affected by Fis. To test this possibility, we utilized two previously constructed C-terminus FLAG-tagged exsA (exsA-Flag) driven by an exogenous tac promoter (Li et al., 2013). In one of the construct, namely exsA-Flag-A, the exsA coding region was directly fused with the tac promoter, whereas in the other construct (exsA-Flag-S), the exsB-exsA intergenic region was included (Figure 9B). Without the exsB-exsA intergenic region, similar levels of ExsA-FLAG were observed in wild type PA14 and the fis::Tn mutant. However, presence of the exsB-exsA intergenic region resulted in less ExsA-FLAG in the fis::Tn mutant (Figure 9C), thus confirming the role of Fis in the continuous transcription from exsB to exsA. Consistently, in the fis::Tn mutant the exsA-Flag-A resulted in higher mRNA levels of exoU, exsC, and pcrV than the exsA-Flag-S (Figure 9D). In wild type PA14, the exsA-Flag-A resulted in higher mRNA levels of exoU, exsC, and pcrV than those in the fis::Tn mutant, which might be due to higher expression of the chromosomal exsA. In combination, these results suggest that Fis is required for the PexsC–dependent transcription of exsA, which is required for the full activation of the T3SS.
Discussion
In this study, we demonstrate that Fis is involved in the regulation of T3SS in P. aeruginosa. Since negative supercoiling of DNA can influence transcription elongation (Baaklini et al., 2004; Travers and Muskhelishvili, 2005) and Fis is considered to be a local topological homeostat (Travers and Muskhelishvili, 2005), Fis might be involved in the regulation of transcription elongation. DNA regulated by Fis might require Fis binding and bending in multiple tandem sites in a non-random fashion (Schneider et al., 2001; Kahramanoglou et al., 2011). In addition, Fis displays a preference for binding particularly at regions upstream of open reading frames (Kahramanoglou et al., 2011). For example, the tyrT promoter is regulated by three Fis dimers binding to the core and upstream regions of the tyrT promoter (Pemberton et al., 2002). Here, we found that Fis specifically interacts with the -10 box of PexsA promoter and directly controls mRNA transcription elongation initiated from PexsC promoter. However, we cannot eliminate the possibility of additional Fis binding sites upstream or inside the exsCEBA operon, i.e., Fis may interact with multiple sites to control the transcription of exsCEBA. It has been found that the expression of exsA is regulated at the post-transcriptional level (Intile et al., 2015; Tan et al., 2016). Besides, multiple genes have been found to affect the expression of exsA (Dong et al., 2013; Marsden et al., 2016; Yu et al., 2016; Zhu et al., 2016a,b). Since Fis functions as a global regulator, it is possible that Fis indirectly regulates ExsA expression at both transcriptional and post-transcriptional level.
ExsA is the master transcriptional activator for the T3SS genes in P. aeruginosa, including exsC (Diaz et al., 2011). Recently, it has been reported that the transcription of exsA is controlled by a Vfr-dependent PexsA promoter located in the exsB-exsA intergenic region (Marsden et al., 2016). Together with our findings in this study, we propose the following regulatory mechanism for the expression of exsA. T3SS inducing signals stimulate the increase of intracellular cAMP level (Fuchs et al., 2010). Subsequently, Vfr binds to cAMP and activates PexsA (Marsden et al., 2016), resulting in higher amount of ExsA, which activates the PexsC. With the help of Fis, the transcription from PexsC extends into exsA, which generates a positive feedback loop, resulting in a quick up regulation of the exsA. In this way, bacteria can quickly express T3SS genes in response to environmental stimulations.
Fis is a growth phase dependent global regulator in Enterobacteriaceae (Mallik et al., 2006; Bradley et al., 2007; Cho et al., 2008). In E. coli, it has been demonstrated that the level of Fis peaks in the early exponential phase, and drops upon entering the stationary phase (Mallik et al., 2006; Bradley et al., 2007). Meanwhile, Fis also responds to the nutritional changes in Enterobacteriaceae (Mallik et al., 2004). However, the Fis expression pattern is not well known in P. aeruginosa. Recently, Lu et al identified a small regulatory RNA (sRNA) named RgsA which regulates Fis expression at the post-transcriptional level in P. aeruginosa (Lu et al., 2016). RgsA is directly controlled by sigma factor σS (RpoS) (Lu et al., 2016). It is well known that RpoS is up regulated when bacteria enter into stationary phase, and plays crucial role in bacterial survival in stationary phase or under various stress conditions (Hengge-Aronis, 2002). So we suspect that when bacteria enter the stationary phase, the increased level of RpoS leads to up regulation of RgsA. Subsequently, RgsA represses Fis expression at the post-transcriptional level. The expression pattern of the Fis in P. aeruginosa warrants further studies. While growing the bacteria, we found that the fis::Tn mutant produced higher amount of pyocyanin at the stationary growth phase (data not shown). These results imply a role of Fis in the quorum sensing system. In addition, RgsA is indirectly regulated by the GacS/GacA two-component system (González et al., 2008), which has been demonstrated to regulate T3SS through small RNAs RsmY and RsmZ (Vakulskas et al., 2009; Chen et al., 2016). Therefore, RgsA and Fis may also participate in the T3SS regulation via GacS/GacA.
Previous studies in various bacteria demonstrate that Fis plays pleiotropic roles in bacterial virulence and response to environmental stresses (Duprey et al., 2014), however, the role of Fis in P. aeruginosa remains largely unknown. Previous studies demonstrated that Fis is required for swarming motility in P. aeruginosa (Yeung et al., 2009; Lu et al., 2016). Other than T3SS, our transcriptome analysis revealed that pyocin biosynthesis genes were up regulated in the fis::Tn mutant (Table S2). Biosynthesis of pyocins has been demonstrated to correlate to bacterial susceptibility to DNA damaging agents (Chen et al., 2017). Indeed, we found that the fis::Tn mutant was more susceptible to quinolone antibiotics such as ciprofloxacin and ofloxacin (data not shown). Further studies are needed to examine the role of Fis in the regulation of pyocin biosynthesis and whether abnormal expression of pyocin genes contributes to the increased susceptibility in the fis::Tn mutant (Agnello et al., 2016). In addition, although overexpression of ExsA in the fis::Tn mutant fully restored the bacterial cytotoxicity, the bacterial virulence was partially restored, indicating additional virulence factors might be regulated by Fis. Therefore, genes directly regulated by Fis as well as the global binding site of Fis in P. aeruginosa warrants further studies.
Author Contributions
Conceived and designed the experiments and wrote the paper: WW, XD, ZC, and SJ. Performed the experiments: XD, ML, XP, RZ, CL, FC, and XL. Analyzed the data: XD, ML, WW, ZC, and SJ.
Funding
This work was supported by National Science Foundation of China (31670130, 31370168, 31370167, and 31600110); Program of international S&T cooperation (2015DFG32500) and Science and Technology Committee of Tianjin (15JCYBJC53900 and 15JCZDJC33000). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00669/full#supplementary-material
Figure S1. Relative mRNA level of PA4852. Total RNA was isolated from bacteria at indicated optical densities (OD600). cDNA was synthesized with PA4852 and rpsL specific primer. Relative mRNA levels of PA4852 were determined by quantitative real-time PCR. rpsL was used as an internal control. Data represents the mean ± standard deviation from three samples.
Figure S2. Relative mRNA level of RgsA. Total RNA was isolated from bacteria at indicated optical densities (OD600). Relative mRNA levels of RgsA were determined by quantitative real-time PCR. rpsL was used as an internal control. Data represents the mean ± standard deviation from three samples.
Figure S3. exsC promoter activity. PA14 and the fis::Tn mutant carrying PexsC-lacZ were grown at 37°C with or without 5 mM EGTA and assayed for β-galactosidase activities. The reported values (Miller units) are the means of results of at least three independent experiments. ***p < 0.001 by Student's t-test.
Figure S4. Growth curves of the fis::Tn mutant and the wild type PA14. Indicated strains were grown at 37°C in LB medium. Bacterial numbers were determined at indicated time by serial dilution and plating. The error bars indicate standard error of the mean.
Figure S5. Growth curves of the wild type PA14, fis::Tn mutant and the exsA overexpression strain. Indicated strains were grown at 37°C in LB medium. Bacterial numbers were determined at indicated time by serial dilution and plating. The error bars indicate standard error of the mean.
Figure S6. EMSA for Fis binding to the PexsC promoter. Fis was incubated with PexsC promoter probes i, ii, iii for 30 min at 25°C. Probes i, ii, iii represent the upstream region of PexsC -35 box, PexsC promoter -35 box, and PexsC promoter -10 box, respectively. Arrows indicate the positions of unbound probes and LMC.
Table S1. Bacterial strains, plasmids, and primers used in this study.
Table S2. PA14 Transcriptome analysis: differentially expressed genes.
References
Agnello, M., Finkel, S. E., and Wong-Beringer, A. (2016). Fitness cost of fluoroquinolone resistance in clinical isolates of Pseudomonas aeruginosa differs by Type III secretion genotype. Front. Microbiol. 7:1591. doi: 10.3389/fmicb.2016.01591
Aiyar, S. E., McLeod, S. M., Ross, W., Hirvonen, C. A., Thomas, M. S., Johnson, R. C., et al. (2002). Architecture of Fis-activated transcription complexes at the Escherichia coli rrnB P1 and rrnE P1 promoters. J. Mol. Biol. 316, 501–516. doi: 10.1006/jmbi.2001.5390
Anantharajah, A., Mingeot-Leclercq, M. P., and Van Bambeke, F. (2016). Targeting the type three secretion system in Pseudomonas aeruginosa. Trends Pharmacol. Sci. 37, 734–749. doi: 10.1016/j.tips.2016.05.011
Anderson, G. G., Yahr, T. L., Lovewell, R. R., and O'Toole, G. A. (2010). The Pseudomonas aeruginosa magnesium transporter MgtE inhibits transcription of the type III secretion system. Infect. Immun. 78, 1239–1249. doi: 10.1128/IAI.00865-09
Audic, S., and Claverie, J. M. (1997). The significance of digital gene expression profiles. Genome Res. 7, 986–995.
Baaklini, I., Hraiky, C., Rallu, F., Tse-Dinh, Y. C., and Drolet, M. (2004). RNase HI overproduction is required for efficient full-length RNA synthesis in the absence of topoisomerase I in Escherichia coli. Mol. Microbiol. 54, 198–211. doi: 10.1111/j.1365-2958.2004.04258.x
Bradley, M. D., Beach, M. B., de Koning, A. P., Pratt, T. S., and Osuna, R. (2007). Effects of Fis on Escherichia coli gene expression during different growth stages. Microbiology 153(Pt 9), 2922–2940. doi: 10.1099/mic.0.2007/008565-0
Chen, F., Chen, G., Liu, Y., Jin, Y., Cheng, Z., Liu, Y., et al. (2017). Pseudomonas aeruginosa Oligoribonuclease contributes to tolerance to Ciprofloxacin by regulating pyocin biosynthesis. Antimicrob. Agents Chemother. 61:e02256–16. doi: 10.1128/AAC.02256-16
Chen, R., Weng, Y., Zhu, F., Jin, Y., Liu, C., Pan, X., et al. (2016). Polynucleotide phosphorylase regulates multiple virulence factors and the stabilities of small RNAs RsmY/Z in Pseudomonas aeruginosa. Front. Microbiol. 7:247. doi: 10.3389/fmicb.2016.00247
Cho, B. K., Knight, E. M., Barrett, C. L., and Palsson, B. Ø. (2008). Genome-wide analysis of Fis binding in Escherichia coli indicates a causative role for A-/AT-tracts. Genome Res. 18, 900–910. doi: 10.1101/gr.070276.107
Choi, K. H., and Schweizer, H. P. (2006). mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat. Protoc. 1, 153–161. doi: 10.1038/nprot.2006.24
Cornelis, G. R. (2006). The type III secretion injectisome. Nat. Rev. Microbiol. 4, 811–825. doi: 10.1038/nrmicro1526
Diaz, M. R., King, J. M., and Yahr, T. L. (2011). Intrinsic and extrinsic regulation of Type III secretion gene expression in Pseudomonas aeruginosa. Front. Microbiol. 2:89. doi: 10.3389/fmicb.2011.00089
Dillon, S. C., and Dorman, C. J. (2010). Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 8, 185–195. doi: 10.1038/nrmicro2261
Dong, Y. H., Zhang, X. F., and Zhang, L. H. (2013). The global regulator Crc plays a multifaceted role in modulation of type III secretion system in Pseudomonas aeruginosa. Microbiologyopen 2, 161–172. doi: 10.1002/mbo3.54
Duprey, A., Reverchon, S., and Nasser, W. (2014). Bacterial virulence and Fis: adapting regulatory networks to the host environment. Trends Microbiol. 22, 92–99. doi: 10.1016/j.tim.2013.11.008
Ellermeier, J. R., and Slauch, J. M. (2007). Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium. Curr. Opin. Microbiol. 10, 24–29. doi: 10.1016/j.mib.2006.12.002
Falconi, M., Prosseda, G., Giangrossi, M., Beghetto, E., and Colonna, B. (2001). Involvement of FIS in the H-NS-mediated regulation of virF gene of Shigella and enteroinvasive Escherichia coli. Mol. Microbiol. 42, 439–452. doi: 10.1046/j.1365-2958.2001.02646.x
Figueira, R., and Holden, D. W. (2012). Functions of the Salmonella pathogenicity island 2 (SPI-2) type III secretion system effectors. Microbiology 158(Pt 5), 1147–1161. doi: 10.1099/mic.0.058115-0
Fuchs, E. L., Brutinel, E. D., Jones, A. K., Fulcher, N. B., Urbanowski, M. L., Yahr, T. L., et al. (2010). The Pseudomonas aeruginosa Vfr regulator controls global virulence factor expression through cyclic AMP-dependent and -independent mechanisms. J. Bacteriol. 192, 3553–3564. doi: 10.1128/JB.00363-10
Fürste, J. P., Pansegrau, W., Frank, R., Blöcker, H., Scholz, P., Bagdasarian, M., et al. (1986). Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene 48, 119–131. doi: 10.1016/0378-1119(86)90358-6
Gellatly, S. L., and Hancock, R. E. (2013). Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog. Dis. 67, 159–173. doi: 10.1111/2049-632X.12033
Goldberg, M. D., Johnson, M., Hinton, J. C., and Williams, P. H. (2001). Role of the nucleoid-associated protein Fis in the regulation of virulence properties of enteropathogenic Escherichia coli. Mol. Microbiol. 41, 549–559. doi: 10.1046/j.1365-2958.2001.02526.x
González, N., Heeb, S., Valverde, C., Kay, E., Reimmann, C., Junier, T., et al. (2008). Genome-wide search reveals a novel GacA-regulated small RNA in Pseudomonas species. BMC Genomics 9:167. doi: 10.1186/1471-2164-9-167
Goo, E., An, J. H., Kang, Y., and Hwang, I. (2015). Control of bacterial metabolism by quorum sensing. Trends Microbiol. 23, 567–576. doi: 10.1016/j.tim.2015.05.007
Green, E. R., Clark, S., Crimmins, G. T., Mack, M., Kumamoto, C. A., and Mecsas, J. (2016). Fis Is essential for Yersinia pseudotuberculosis virulence and protects against reactive oxygen species produced by phagocytic cells during infection. PLoS Pathog. 12:e1005898. doi: 10.1371/journal.ppat.1005898
Ha, U. H., Kim, J., Badrane, H., Jia, J., Baker, H. V., Wu, D., et al. (2004). An in vivo inducible gene of Pseudomonas aeruginosa encodes an anti-ExsA to suppress the type III secretion system. Mol. Microbiol. 54, 307–320. doi: 10.1111/j.1365-2958.2004.04282.x
Hancock, S. P., Stella, S., Cascio, D., and Johnson, R. C. (2016). DNA Sequence determinants controlling affinity, stability and shape of DNA complexes bound by the nucleoid protein Fis. PLoS ONE 11:e0150189. doi: 10.1371/journal.pone.0150189
Hauser, A. R. (2009). The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nat. Rev. Microbiol. 7, 654–665. doi: 10.1038/nrmicro2199
Hengge-Aronis, R. (2002). Signal transduction and regulatory mechanisms involved in control of the sigma(S) (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66, 373–395. doi: 10.1128/MMBR.66.3.373-395.2002
Hoang, T. T., Karkhoff-Schweizer, R. R., Kutchma, A. J., and Schweizer, H. P. (1998). A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212, 77–86. doi: 10.1016/S0378-1119(98)00130-9
Huber, P., Basso, P., Reboud, E., and Attrée, I. (2016). Pseudomonas aeruginosa renews its virulence factors. Environ. Microbiol. Rep. 8, 564–571. doi: 10.1111/1758-2229.12443
Intile, P. J., Balzer, G. J., Wolfgang, M. C., and Yahr, T. L. (2015). The RNA helicase DeaD stimulates ExsA translation to promote expression of the Pseudomonas aeruginosa Type III Secretion System. J. Bacteriol. 197, 2664–2674. doi: 10.1128/JB.00231-15
Jakovleva, J., Teppo, A., Velts, A., Saumaa, S., Moor, H., Kivisaar, M., et al. (2012). Fis regulates the competitiveness of Pseudomonas putida on barley roots by inducing biofilm formation. Microbiology 158(Pt 3), 708–720. doi: 10.1099/mic.0.053355-0
Kahramanoglou, C., Seshasayee, A. S., Prieto, A. I., Ibberson, D., Schmidt, S., Zimmermann, J., et al. (2011). Direct and indirect effects of H-NS and Fis on global gene expression control in Escherichia coli. Nucleic Acids Res. 39, 2073–2091. doi: 10.1093/nar/gkq934
Kelly, A., Goldberg, M. D., Carroll, R. K., Danino, V., Hinton, J. C., and Dorman, C. J. (2004). A global role for Fis in the transcriptional control of metabolism and type III secretion in Salmonella enterica serovar Typhimurium. Microbiology 150(Pt 7), 2037–2053. doi: 10.1099/mic.0.27209-0
Lautier, T., and Nasser, W. (2007). The DNA nucleoid-associated protein Fis co-ordinates the expression of the main virulence genes in the phytopathogenic bacterium Erwinia chrysanthemi. Mol. Microbiol. 66, 1474–1490. doi: 10.1111/j.1365-2958.2007.06012.x
Li, K., Xu, C., Jin, Y., Sun, Z., Liu, C., Shi, J., et al. (2013). SuhB is a regulator of multiple virulence genes and essential for pathogenesis of Pseudomonas aeruginosa. mBio 4, e00419–e00413. doi: 10.1128/mBio.00419-13
Li, M., Long, Y., Liu, Y., Liu, Y., Chen, R., Shi, J., et al. (2016). HigB of Pseudomonas aeruginosa enhances killing of phagocytes by up-regulating the Type III secretion system in ciprofloxacin induced persister cells. Front. Cell. Infect. Microbiol. 6:125. doi: 10.3389/fcimb.2016.00125
Liberati, N. T., Urbach, J. M., Miyata, S., Lee, D. G., Drenkard, E., Wu, G., et al. (2006). An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. U.S.A. 103, 2833–2838. doi: 10.1073/pnas.0511100103
Lu, P., Wang, Y., Zhang, Y., Hu, Y., Thompson, K. M., and Chen, S. (2016). RpoS-dependent sRNA RgsA regulates Fis and AcpP in Pseudomonas aeruginosa. Mol. Microbiol. 102, 244–259. doi: 10.1111/mmi.13458
Mallik, P., Paul, B. J., Rutherford, S. T., Gourse, R. L., and Osuna, R. (2006). DksA is required for growth phase-dependent regulation, growth rate-dependent control, and stringent control of fis expression in Escherichia coli. J. Bacteriol. 188, 5775–5782. doi: 10.1128/JB.00276-06
Mallik, P., Pratt, T. S., Beach, M. B., Bradley, M. D., Undamatla, J., and Osuna, R. (2004). Growth phase-dependent regulation and stringent control of fis are conserved processes in enteric bacteria and involve a single promoter (fis P) in Escherichia coli. J. Bacteriol. 186, 122–135. doi: 10.1128/JB.186.1.122-135.2004
Marsden, A. E., Intile, P. J., Schulmeyer, K. H., Simmons-Patterson, E. R., Urbanowski, M. L., Wolfgang, M. C., et al. (2016). Vfr directly activates exsA transcription to regulate expression of the Pseudomonas aeruginosa Type III secretion system. J. Bacteriol. 198, 1442–1450. doi: 10.1128/JB.00049-16
Moradali, M. F., Ghods, S., and Rehm, B. H. (2017). Pseudomonas aeruginosa Lifestyle: a paradigm for adaptation, survival, and persistence. Front. Cell. Infect. Microbiol. 7:39. doi: 10.3389/fcimb.2017.00039
Muskhelishvili, G., Travers, A. A., Heumann, H., and Kahmann, R. (1995). FIS and RNA polymerase holoenzyme form a specific nucleoprotein complex at a stable RNA promoter. EMBO J. 14, 1446–1452.
Pemberton, I. K., Muskhelishvili, G., Travers, A. A., and Buckle, M. (2002). FIS modulates the kinetics of successive interactions of RNA polymerase with the core and upstream regions of the tyrT promoter. J. Mol. Biol. 318, 651–663. doi: 10.1016/S0022-2836(02)00142-0
Prigent-Combaret, C., Zghidi-Abouzid, O., Effantin, G., Lejeune, P., Reverchon, S., and Nasser, W. (2012). The nucleoid-associated protein Fis directly modulates the synthesis of cellulose, an essential component of pellicle-biofilms in the phytopathogenic bacterium Dickeya dadantii. Mol. Microbiol. 86, 172–186. doi: 10.1111/j.1365-2958.2012.08182.x
Reinhart, A. A., and Oglesby-Sherrouse, A. G. (2016). Regulation of Pseudomonas aeruginosa virulence by distinct iron sources. Genes (Basel). 7:126. doi: 10.3390/genes7120126
Rietsch, A., and Mekalanos, J. J. (2006). Metabolic regulation of type III secretion gene expression in Pseudomonas aeruginosa. Mol. Microbiol. 59, 807–820. doi: 10.1111/j.1365-2958.2005.04990.x
Rybtke, M., Hultqvist, L. D., Givskov, M., and Tolker-Nielsen, T. (2015). Pseudomonas aeruginosa biofilm infections: community structure, antimicrobial tolerance and immune response. J. Mol. Biol. 427, 3628–3645. doi: 10.1016/j.jmb.2015.08.016
Sato, H., and Frank, D. W. (2011). Multi-functional characteristics of the Pseudomonas aeruginosa Type III Needle-Tip protein, PcrV; comparison to orthologs in other gram-negative bacteria. Front. Microbiol. 2:142. doi: 10.3389/fmicb.2011.00142
Schechter, L. M., Jain, S., Akbar, S., and Lee, C. A. (2003). The small nucleoid-binding proteins H-NS, HU, and Fis affect hilA expression in Salmonella enterica serovar Typhimurium. Infect. Immun. 71, 5432–5435. doi: 10.1128/IAI.71.9.5432-5435.2003
Schneider, R., Lurz, R., Lüder, G., Tolksdorf, C., Travers, A., and Muskhelishvili, G. (2001). An architectural role of the Escherichia coli chromatin protein FIS in organising DNA. Nucleic Acids Res. 29, 5107–5114. doi: 10.1093/nar/29.24.5107
Shao, Y., Feldman-Cohen, L. S., and Osuna, R. (2008). Functional characterization of the Escherichia coli Fis-DNA binding sequence. J. Mol. Biol. 376, 771–785. doi: 10.1016/j.jmb.2007.11.101
Simon, R., Priefer, U., and Pühler, A. (1983). A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat. Biotechnol. 1, 784–791. doi: 10.1038/nbt1183-784
Skoko, D., Yoo, D., Bai, H., Schnurr, B., Yan, J., McLeod, S. M., et al. (2006). Mechanism of chromosome compaction and looping by the Escherichia coli nucleoid protein Fis. J. Mol. Biol. 364, 777–798. doi: 10.1016/j.jmb.2006.09.043
Sun, Z., Shi, J., Liu, C., Jin, Y., Li, K., Chen, R., et al. (2014). PrtR homeostasis contributes to Pseudomonas aeruginosa pathogenesis and resistance against ciprofloxacin. Infect. Immun. 82, 1638–1647. doi: 10.1128/IAI.01388-13
Tan, H., Zhang, L., Zhao, Q., Chen, R., Liu, C., Weng, Y., et al. (2016). DeaD contributes to Pseudomonas aeruginosa virulence in a mouse acute pneumonia model. FEMS Microbiol. Lett. 363:fnw227. doi: 10.1093/femsle/fnw227
Travers, A., and Muskhelishvili, G. (2005). DNA supercoiling–a global transcriptional regulator for enterobacterial growth? Nat. Rev. Microbiol. 3, 157–169. doi: 10.1038/nrmicro1088
Turner, K. H., Everett, J., Trivedi, U., Rumbaugh, K. P., and Whiteley, M. (2014). Requirements for Pseudomonas aeruginosa acute burn and chronic surgical wound infection. PLoS Genet. 10:e1004518. doi: 10.1371/journal.pgen.1004518
Vakulskas, C. A., Brady, K. M., and Yahr, T. L. (2009). Mechanism of transcriptional activation by Pseudomonas aeruginosa ExsA. J. Bacteriol. 191, 6654–6664. doi: 10.1371/journal.pone.0064688
Wang, H., Liu, B., Wang, Q., and Wang, L. (2013). Genome-wide analysis of the salmonella Fis regulon and its regulatory mechanism on pathogenicity islands. PLoS ONE 8:e64688. doi: 10.1371/journal.pone.0064688
Williams, B. J., Dehnbostel, J., and Blackwell, T. S. (2010). Pseudomonas aeruginosa: host defence in lung diseases. Respirology 15, 1037–1056. doi: 10.1111/j.1440-1843.2010.01819.x
Wurtzel, O., Yoder-Himes, D. R., Han, K., Dandekar, A. A., Edelheit, S., Greenberg, E. P., et al. (2012). The single-nucleotide resolution transcriptome of Pseudomonas aeruginosa grown in body temperature. PLoS Pathog. 8:e1002945. doi: 10.1371/journal.ppat.1002945
Yahr, T. L., and Frank, D. W. (1994). Transcriptional organization of the trans-regulatory locus which controls exoenzyme S synthesis in Pseudomonas aeruginosa. J. Bacteriol. 176, 3832–3838. doi: 10.1128/jb.176.13.3832-3838.1994
Yahr, T. L., and Wolfgang, M. C. (2006). Transcriptional regulation of the Pseudomonas aeruginosa type III secretion system. Mol. Microbiol. 62, 631–640. doi: 10.1111/j.1365-2958.2006.05412.x
Yeung, A. T., Torfs, E. C., Jamshidi, F., Bains, M., Wiegand, I., Hancock, R. E., et al. (2009). Swarming of Pseudomonas aeruginosa is controlled by a broad spectrum of transcriptional regulators, including MetR. J. Bacteriol. 191, 5592–5602. doi: 10.1128/JB.00157-09
Yu, H., Xiong, J., Zhang, R., Hu, X., Qiu, J., Zhang, D., et al. (2016). Ndk, a novel host-responsive regulator, negatively regulates bacterial virulence through quorum sensing in Pseudomonas aeruginosa. Sci. Rep. 6:28684. doi: 10.1038/srep28684
Zhu, M., Zhao, J., Kang, H., Kong, W., and Liang, H. (2016a). Modulation of type III secretion system in Pseudomonas aeruginosa: Involvement of the PA4857 gene product. Front. Microbiol. 7:7. doi: 10.3389/fmicb.2016.00007
Keywords: Pseudomonas aeruginosa, Fis, type III secretion system, bacterial virulence, exsA transcription
Citation: Deng X, Li M, Pan X, Zheng R, Liu C, Chen F, Liu X, Cheng Z, Jin S and Wu W (2017) Fis Regulates Type III Secretion System by Influencing the Transcription of exsA in Pseudomonas aeruginosa Strain PA14. Front. Microbiol. 8:669. doi: 10.3389/fmicb.2017.00669
Received: 09 January 2017; Accepted: 31 March 2017;
Published: 19 April 2017.
Edited by:
Yuji Morita, Aichi Gakuin University, JapanReviewed by:
Shiyun Chen, Wuhan Institute of Virology (CAS), ChinaThibault Géry Sana, Stanford University, USA
Xiaogang Wu, Guangxi University, China
Neta Sal-Man, Ben-Gurion University of the Negev, Israel
Copyright © 2017 Deng, Li, Pan, Zheng, Liu, Chen, Liu, Cheng, Jin and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhihui Cheng, zhihuicheng@nankai.edu.cn
Shouguang Jin, sjin@ufl.edu
Weihui Wu, wuweihui@nankai.edu.cn
 Xuan Deng
Xuan Deng Mei Li
Mei Li Xiaolei Pan
Xiaolei Pan Ruiping Zheng
Ruiping Zheng Chang Liu
Chang Liu Fei Chen
Fei Chen Xue Liu
Xue Liu Zhihui Cheng
Zhihui Cheng Shouguang Jin
Shouguang Jin Weihui Wu
Weihui Wu