- 1Department of Microbiology, School of Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 2Department of Biology, Faculty of Natural Sciences, University of Tabriz, Tabriz, Iran
- 3Applied Microbiology Research Center, Baqiyatallah University of Medical Sciences, Tehran, Iran
- 4Department of Microbiology, School of Medicine, Tehran University of Medical Sciences, Tehran, Iran
- 5Thoracic Research Center, Imam Khomeini Hospital, Tehran University of Medical Sciences, Tehran, Iran
Infectious diseases caused by clinically important Mycobacteria continue to be an important public health problem worldwide primarily due to emergence of drug resistance crisis. In recent years, the control of tuberculosis (TB), the disease caused by Mycobacterium tuberculosis (MTB), is hampered by the emergence of multidrug resistance (MDR), defined as resistance to at least isoniazid (INH) and rifampicin (RIF), two key drugs in the treatment of the disease. Despite the availability of curative anti-TB therapy, inappropriate and inadequate treatment has allowed MTB to acquire resistance to the most important anti-TB drugs. Likewise, for most mycobacteria other than MTB, the outcome of drug treatment is poor and is likely related to the high levels of antibiotic resistance. Thus, a better knowledge of the underlying mechanisms of drug resistance in mycobacteria could aid not only to select the best therapeutic options but also to develop novel drugs that can overwhelm the existing resistance mechanisms. In this article, we review the distinctive mechanisms of antibiotic resistance in mycobacteria.
Introduction
Infections due to Mycobacterium species are an increasing problem in many countries in the world. This genus contains several important bacteria, which can cause life-threatening infections in human beings. Tuberculosis (TB) is a severe infectious disease caused by Mycobacterium tuberculosis (MTB). Non-tuberculosis Mycobacteria (NTM) are also capable of causing serious illnesses in both immunocompetent and immunocompromised individuals. According to the latest report released by the World Health Organization (WHO), there were an estimated 10.4 million new TB cases and 1.4 million deaths from the disease (World Health Organization, 2016). Till now, TB cases caused by drug susceptible strains of MTB, in most instances, can be cured in 6 months using combinations of first-line anti-TB drugs (Horsburgh et al., 2015). According to WHO guidelines, patients with drug susceptible TB should be treated with standard regimen consisting of an initial intensive phase of 2-months of isoniazid (INH), rifampicin (RIF), ethambutol (EMB), and pyrazinamide (PZA) followed by a continuation phase of 4 months of INH and RIF (World Health Organization, 2010).
However, treatment of drug resistant strains, including those of multidrug-resistant (MDR) (defined as resistance to INH and RIF, the two most potent first-line drugs for TB treatment)—and extensively drug resistant-TB (XDR-TB) (defined as in vitro drug resistance to INH and RIF plus any fluoroquinolone and at least one of the injectable aminoglycosides), is more challenging, requiring prolonged and expensive chemotherapy (World Health Organization, 2014; Millard et al., 2015). Current WHO guidelines for treatment of MDR-TB recommend that at least four second-line drugs that are likely to be effective, as well as PZA, be administered during the intensive phase of the regimen (World Health Organization, 2011). Therapeutic options for XDR-TB are extremely limited because of resistance to the more potent second line drugs (World Health Organization, 2011).
In the other side, based on reports from many countries in the world, the number of diseases caused by NTMs is also rising (Glassroth, 2008; Nasiri et al., 2015; Wu and Holland, 2015). Yet, all mycobacteria are acid-fast; many species cause lung disease that is often indistinguishable from TB and diagnosis of TB in many parts of the world is still only by sputum smear (Shahraki et al., 2015). Unfortunately, due to the lack of standardized or accepted criteria to define diseases caused by NTM, many NTM cases are frequently misdiagnosed as TB and put on anti-TB medications while treatment of NTM disease is not similar to that of MTB (Brown-Elliott et al., 2012). The newer and more expensive macrolides (i.e., azithromycin and clarithromycin) and quinolones (i.e., ofloxacin and ciprofloxacin) have become the cornerstones of therapy for the most clinically important NTM (Brown-Elliott et al., 2012). Recent studies have also shown the high prevalence of drug resistance in NTM species that threatens adequate control of the disease (Brown-Elliott et al., 2012, 2014; Cândido et al., 2014; Cowman et al., 2015).
With recognizing the scale of the problem, a better understanding of the mechanisms of drug resistance in mycobacterial species will have an important impact on the better application of currently available drugs and will also stimulate the exploration of new targets for development of novel classes of anti-mycobacterial compounds.
Drug Resistance Mechanisms in Mycobacteria
Distinctive mechanisms of antibiotic resistance have been described in mycobacteria. Examples of each of these mechanisms are provided in the following paragraphs (Figure 1).
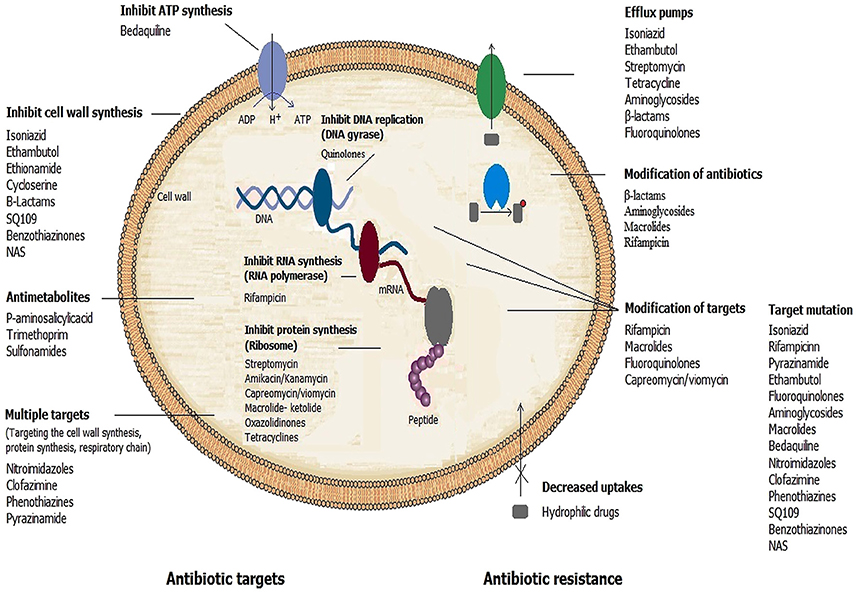
Figure 1. Mechanisms of action of antimicrobial drugs and ways by which mycobacteria become resistant to them.
Intrinsic Resistance
Prevention of Access to Target
Cell wall impermeability
The mycobacterial cell envelope consists of three main structural components: (a) a network of peptidoglycan (PG), (b) the arabinogalactan (AG) polysaccharide, and (c) the long-chain mycolic acids (MA) (Alderwick et al., 2015; Daffé, 2015; Jankute et al., 2015). This cell wall-based permeability barrier shields the organism from environmental stress and contributes to resistance of mycobacteria to many antibiotics. Thus, defects in these lipids would damage the function of the cell wall as a barrier and increase the sensitivity to various anti-mycobacterial drugs (Jackson et al., 2013). The enzymes involved in cell wall integrity play an important role in the development of drug resistance in Mycobacterium (Jackson et al., 2013; Xu et al., 2014). Table 1 shows some important enzymes require for integrity of mycobacterial cell wall.
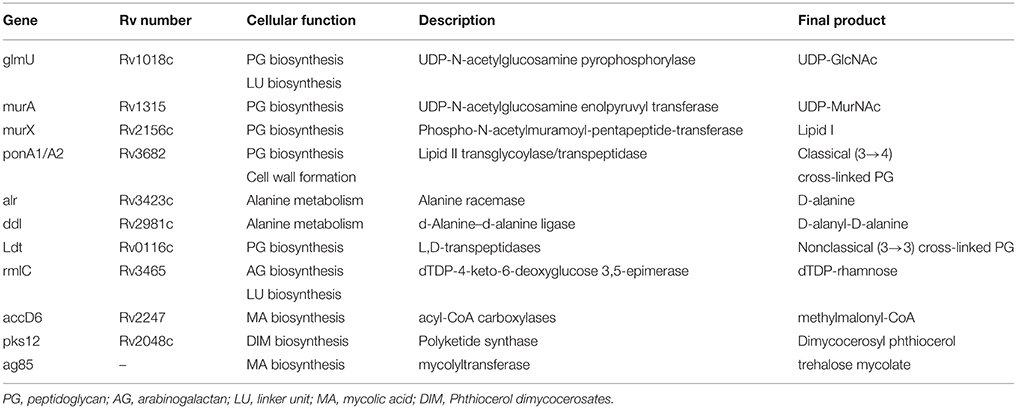
Table 1. Important genes involved in the cell wall integrity of mycobacteria (Philalay et al., 2004; Jackson et al., 2013; Alderwick et al., 2015; Jankute et al., 2015).
MurA and MurB are the key biosynthetic enzymes involved in the formation of UDP-MurNAc, an important step in biosynthesis of PG (Alderwick et al., 2015). The naturally occurring broad-spectrum antibiotic fosfomycin is a well-known inhibitor of MurA (Alderwick et al., 2015; Moraes et al., 2015). It specifically inhibits MurA by forming a covalent adduct with a cysteine residue in the active site (Moraes et al., 2015). However, MTB are intrinsically resistant to fosfomycin, as their corresponding cysteine residue is changed into aspartic acid (Alderwick et al., 2015). Conversely, mutation of the wild-type aspartate residue in the MurA active site to a cysteine renders an enzyme sensitive to fosfomycin in MTB (Castañeda-García et al., 2013).
In most bacteria, β-lactam antibiotics inactivate the essential D,D-transpeptidase activity of classical penicillin-binding proteins (PBPs). These enzymes cross-link glycan chains by forming 4 → 3 peptide bonds connecting residues at the fourth and third positions of stem peptides (Gupta R. et al., 2010). However, MTB has been recently found to have a second class of transpeptidases, the L,D-transpeptidases (Ldt) (Gupta R. et al., 2010; Schoonmaker et al., 2014; Basta et al., 2015; Kieser et al., 2015), which are responsible for resistance to β-lactam antibiotics such as amoxicillin and carbapenems (Gupta R. et al., 2010; Alderwick et al., 2015). Till date, five Ldts (LdtMt1 to LdtMt5) enzymes which form the non-classical 3 → 3 linkages between opposing stem peptides in PG have been identified in MTB (Schoonmaker et al., 2014). MTB strain that lacks both ldtMt1 and ldtMt2 displays enhanced susceptibility to amoxicillin and a glycopeptide drug, vancomycin (Schoonmaker et al., 2014).
Moreover, two proteins, PonA1 and PonA2, are bifunctional penicillin-binding proteins that contribute to the biosynthesis and homeostasis of the cell wall components (Jankute et al., 2015). Recent studies indicated that ponA1 mutant MTB had the same minimum inhibitory concentration (MIC) for b-lactams as wild-type MTB; however, ponA2 mutant cells were four- to eightfold more susceptible to b-lactams (Shi et al., 2011, 2014; Farhat et al., 2013).
Likewise, any defects in enzymes and proteins that are involved in the cell wall integrity can result in increased susceptibility to multiple drugs. Among the various enzymes in the cell envelope biogenesis, one can find the well-known proteins of the antigen 85 (Ag85) complex (Jankute et al., 2015). These proteins, which show mycolyltransferase activity, are involved in the biogenesis of trehalose dimycolates (TDM), a dominant structure necessary for maintaining cell wall integrity (Jankute et al., 2015). It was shown that inactivation of the Ag85 gene profoundly affects the mycolate content and alters the permeability of the MTB cell envelope (Jackson et al., 1999; Ronning et al., 2000). This change in the permeability of the cell envelope can also result in changes in drug resistance phenotype (Jackson et al., 1999; Nguyen et al., 2005). For instance, individual knockout studies on the Ag85 genes (fbpA, fbpB, and fbpC) showed that the mutant strains had 40% less mycolate moieties on the cell wall (Torres et al., 2015; Zhang et al., 2017). Nguyen et al. also indicated that the Ag85 mutants display increased sensitivity to both first-line TB drugs as well as other broad-spectrum antibiotics widely used for antibacterial chemotherapy. In their study, they showed that mutant strains of Mycobacterium smegmatis were 24-, 85-, 8-, and 12-fold more susceptible to erythromycin, imipenem, RIF, and vancomycin, respectively (Lingaraju et al., 2016). Furthermore, recent studies have shown that the production of TDM by Ag85 is essential for the intrinsic antibiotic resistance of some mycobacteria and support the concept that Ag85-specific inhibitors, alone or in combination with other antibiotics, could provide an effective treatment for TB and other mycobacterial diseases (Nguyen et al., 2005).
Examples of other important proteins include: GlmU, MurX, Alr, Ddl, RmlC, accD6, and Pks12 that were reported to be involved in cell wall integrity of mycobacteria (Table 1; Mills et al., 2004; Philalay et al., 2004; Farhat et al., 2013; Calvanese et al., 2014; Schoonmaker et al., 2014; Alderwick et al., 2015; Jankute et al., 2015). These enzymes were shown to be essential and were attractive targets for the development of new antibacterial agents (Alderwick et al., 2015).
Dormancy and latency
Three terms, latency, dormancy and persistence, will be defied here as follow:
Latency refers to a state of asymptomatic infection without apparent disease. Dormancy refers to a non-replicating state with low-to-absent metabolic activity. A subpopulation of dormant cells may be responsible for latency and persistence despite host defense and drug treatment. Persisters, are bacteria that survive antibiotic treatment by remaining dormant even though they are genetically susceptible to these antibiotics (Gomez and McKinney, 2004; Chao and Rubin, 2010).
MTB enters a dormant state during latent infection which is characterized by a shutdown of most of its metabolism causing increased tolerance to antimicrobial agents that are lethal to replicating bacilli a phenomenon referred to as phenotypic drug resistance (Gomez and McKinney, 2004; Gengenbacher and Kaufmann, 2012). In contrast to genotypic drug resistance, this form of drug tolerance is due to reduced metabolic activities or cell division rather than chromosomal resistance mutations (Garton et al., 2008). This is reasoned by the fact that the low metabolic activity in slowly growing or non-replicating dormant MTB is associated with reduced production of antibiotic target proteins or machineries by the bacteria. In fact these bacteria were found to be resensitized upon reentering active growth phase due to resumed production of antibiotic cellular targets (Smith et al., 2012). It's been demonstrated that, in vitro induction of dormant state in MTB by oxygen deprivation is associated with shutdown of DNA, RNA and protein synthesis and subsequently increased resistance to anti-TB drugs such as INH and RIF but susceptibility to the anaerobic bactericidal action of metronidazole, which has no effect on aerobic cultures of MTB (Wayne and Hayes, 1996; Hu et al., 1998).
Another system has used nutrient starvation for induced transition of MTB from active growth into a dormant state and revealed decreased respiration and transcription rate, energy metabolism, lipid biosynthesis, cell division as well as concomitant increased drug tolerance to antibiotics targeting functions required for active growth (Betts et al., 2002; Xie et al., 2005). Therefore, the ability of MTB to enter a metabolically inactive dormant phase can be classified as an intrinsic drug tolerance mechanism. Identifying the factors inducing the dormant state and promoting long term survival or persistence of latent MTB might provide novel targets for development of compounds that can treat TB infections by inhibition of these persistence factors. The Ldts enzymes are among the most important factors that plays a vital role in MTB adaptation to stationary phase (Lavollay et al., 2008). As mentioned above, PG of MTB contains a complex network of classical (4 → 3) and non-classical (3 → 3) transpeptide bonds. However, recent studies have shown that upon MTB entry in to the dormancy phase the number of 3 → 3 cross-linkages significantly increases through PG rearrangement by Ldts (Lavollay et al., 2008; Jankute et al., 2015). The rearrangement of PG may have at least two selective advantages. First, Ldts are the only enzymes able to catalyze the formation of new cross-links in the absence of de novo synthesis of precursors (Lavollay et al., 2008). Second, modification of the cross-links may render the peptidoglycan resistant to the hydrolytic activity of endopeptidases (Lavollay et al., 2008). Thus, the development of Ltd inhibitors is a novel and promising approach to obtain drugs for treatment of TB.
Several MTB protein chaperones may also possibly function as a stress responder and consequently contribute to persistence of MTB (Vaubourgeix et al., 2015; Lupoli et al., 2016; Botella et al., 2017). Many of the stresses that MTB encounters, such as reactive oxygen species, changes in pH and mistranslation caused by antibiotics, can cause reversible or irreversible damage to bacterial proteins (Lupoli et al., 2016). In stationary phase, exposure to an aminoglycoside antibiotic like streptomycin, corrupts ribosomal fidelity and induces mistranslation (Dukan et al., 2000; Ballesteros et al., 2001; Vaubourgeix et al., 2015). Recent works suggest that ClpB/DnaK chaperones play unique roles in mycobacteria (Vaubourgeix et al., 2015; Lupoli et al., 2016). These ATP-powered chaperones have been shown to unfold and refold aggregated proteins to their native structures to restore the function of protein (Lupoli et al., 2016). Recent study indicated that ClpB-deficient MTB had a marked recovery defect from stationary phase or antibiotic exposure and survived poorly in mice (Vaubourgeix et al., 2015). In this regard, ClpB and DnaK chaperones in mycobacteria may serve as a novel class of targets for the development of drugs that can sensitize MTB to other drugs (Vaubourgeix et al., 2015).
Another important mechanism which allows MTB to temporarily survive lethal concentration of antibiotics is stochastic variation in the expression of resistance genes. During this phenomenon, MTB persister cells stochastically enter a dormant state, allowing the population to hedge against the appearance of an antibiotic. It is important to know that transient resistance is not caused by genetic changes, rather MTB induces gene expression to generate a resistant phenotype (Wakamoto et al., 2013; El Meouche et al., 2016). For example, stochastic expression of genes affecting antimicrobial action such as catalase-peroxidase katG, which activates the prodrug INH accounts for differential antibiotic susceptibility of MTB to INH in a subset of cells (El Meouche et al., 2016; Wakamoto et al., 2013).
Porin channels
The permeability of lipid membranes of mycobacteria for hydrophilic solutes is extremely low compared to Gram-negative bacteria. Therefore, hydrophilic agents often utilize channel proteins, such as porins, to cross the outer membrane (Faller et al., 2004; Mailaender et al., 2004; Danilchanka et al., 2008). Porin-like proteins can be detected by channel-forming activities in detergent extracts of MTB and M. smegmatis (Heinz and Niederweis, 2000; Raynaud et al., 2002; Niederweis, 2003; Faller et al., 2004; Nguyen and Thompson, 2006; Danilchanka et al., 2008; Song et al., 2008; Niederweis et al., 2010). One of these proteins, MspA has been well characterized and constitutes the major porin of M. smegmatis (Niederweis et al., 1999). Deletion of the MspA, drastically increased the resistance of M. smegmatis to several agents, indicating that MspA plays an important role in the uptake of hydrophilic antibiotics (Stahl et al., 2001; Stephan et al., 2004). Heterologous expression of the M. smegmatis porin gene mspA in MTB increasesits susceptibility to hydrophilic antibiotics such as β-lactams as well as anti-TB drugs INH, EMB and streptomycin (Stephan et al., 2004). An understanding of the pathways that enable passage of these drugs through the envelope of MTB is essential to the successful design of new therapeutic agents.
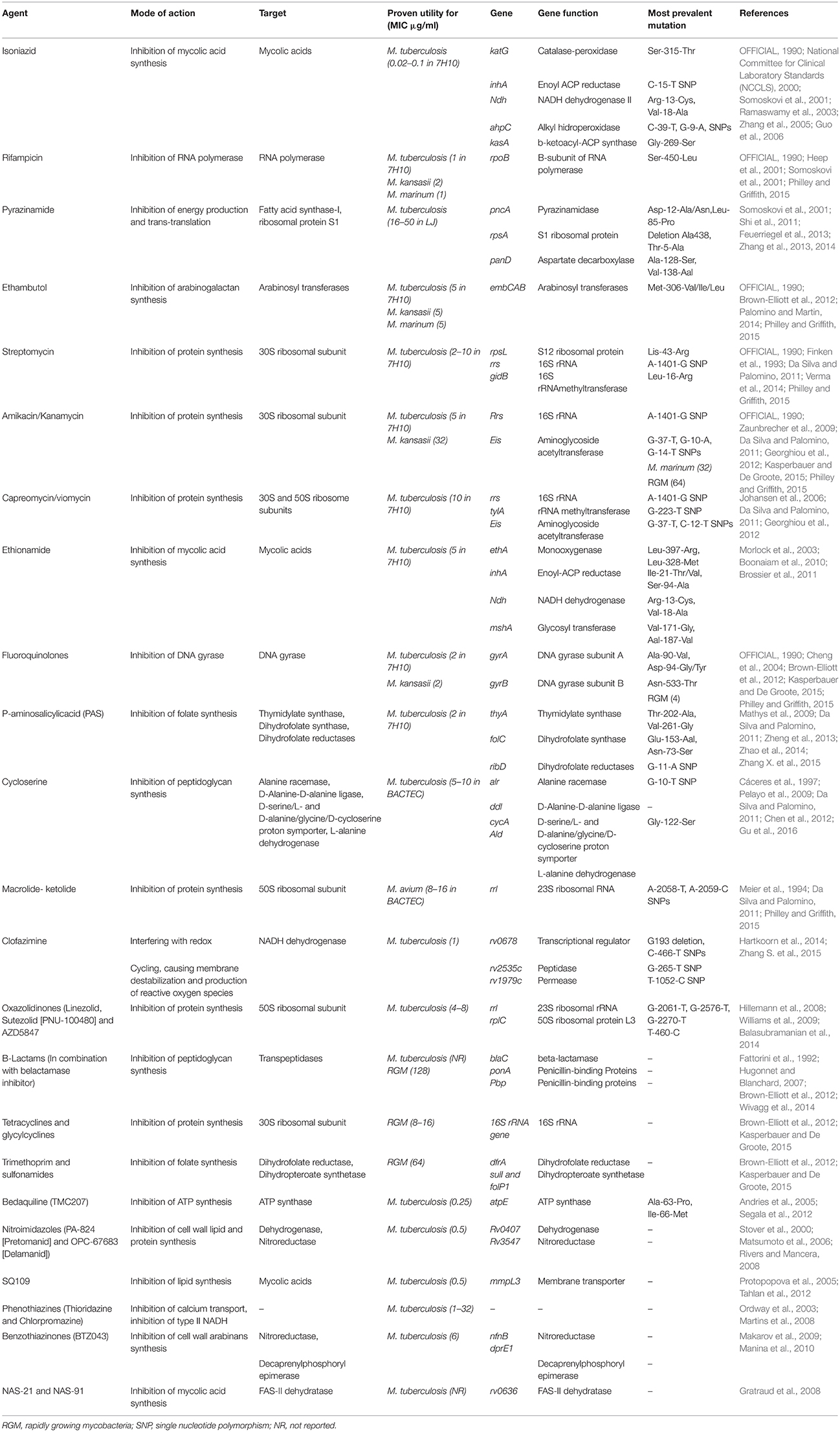
Efflux pumps
The intrinsic resistance of mycobacterial species to most antibiotics is generally attributed to the low permeability of the mycobacterial cell wall (Brennan, 2003). Along with cell wall permeability, active efflux systems also provide resistance by expelling the drug molecules that enter the cell. Several mycobacterial drug efflux pumps have been identified and can be grouped into five different structural families: the major facilitator superfamily (MFS) (Siddiqi et al., 2004; Balganesh et al., 2012; Xu et al., 2015), the small multidrug resistance (SMR) family (De Rossi et al., 2002; Garima et al., 2015), the resistance-nodulation-cell division (RND) superfamily (De Rossi et al., 2006; Murakami et al., 2006), the adenosine triphosphate (ATP)-binding cassette (ABC) superfamily (Bhatt et al., 2000; Choudhuri et al., 2002; Pasca et al., 2004; Lubelski et al., 2007; Caleffi-Ferracioli et al., 2016) and the multidrug and toxic compound extrusion (MATE) family (Table 2) (Mishra and Daniels, 2013).
These efflux pumps actively transport many antibiotics out of the cell and are major contributors to the intrinsic resistance of mycobacteria to many drugs. Some efflux pumps have narrow substrate specificity (such as TetV pumps), but many transport a wide range of structurally dissimilar substrates and are known as MDR efflux pumps. The MFS family of MDR efflux pumps is found in many species of mycobacteria and includes several members that are relevant to antibiotic resistance (Yamchi et al., 2015). When overexpressed, MFS pumps confer clinically relevant levels of MDR and export an extremely wide range of substrates.
For example, the increased transcription of jefA, a well-studied example of MFS pumps in MTB, leads to increased resistance to INH, EMB and streptomycin (Gupta et al., 2010b). Likewise, the best-characterized efflux pumps in NTM include the tap in M. fortuitum conferring resistance to tetracycline and aminoglycoside or lfrA efflux pumps in M. smegmatis that mediate resistance to fluoroquinolones, RIF and INH (Liu et al., 1996; Takiff et al., 1996; Aínsa et al., 1998; Li et al., 2004; Ramón-García et al., 2006; Bowman and Ghosh, 2014). In clinical strains of MTB, increased expression of efflux pumps occurs mainly as a result of induction in response to antibiotic stress (Jiang et al., 2008; Gupta et al., 2010a; Li et al., 2015). For instance, studies that investigated the relevance of active efflux in the drug resistance of clinical strains of MTB indicated that jefA, drrA, drrB, efpA, mmr, and RV1217-Rv1218 efflux pumps were overexpressed under INH and RIF stress (Wang et al., 2013; Blair et al., 2015; Li et al., 2015). Furthermore, several studies indicated that some RIF or INH-resistant strains of MTB did not have sequence alterations in the core region of the drug target encoding genes (e.g., rpoB, katG, inhA; Yamchi et al., 2015; Manson et al., 2017). Consequently, efflux pumps might play an important role in RIF- and INH- resistance in MTB, especially in those strains having no mutation in genes associated with INH and RIF resistance. Similarly, elevated level of expression in efflux pump genes are also demonstrated to be involved in drug resistance development in NTM species (Rodrigues et al., 2009; van Ingen et al., 2012). Understanding the mechanisms controlling the overexpression is important as it is a common mechanism of resistance in clinically important mycobacteria, and also provides promising target for designing novel class of anti-mycobacterial therapeutics that can treat mycobacterial infections by efflux-pump prevention.
Since efflux pumps play a major role in the development of drug resistance in MTB, many studies have focused on identifying potential efflux pump inhibitors. For example, efflux pump inhibitors reserpine, verapamil, 2,4-dinitrophenol (DNP) and pyrrole derivatives have been shown to increase the susceptibility to norfloxacin, RIF and INH by inhibiting efflux pumps (i.e., Rv1258c and MmpL3), in drug-susceptible mycobacteria (Sharma et al., 2010; World Health Organization, 2011; La Rosa et al., 2012; Machado et al., 2012; Willers et al., 2017). Another efflux pump inhibitor is a mammalian efflux pump inhibitor, timcodar (Nasiri et al., 2016). An adjuvant effect was demonstrated in combination with RIF, bedaquiline and clofazimine when MTB was cultured in host macrophages (Nasiri et al., 2016). Recent studies also demonstrate the potential role of efflux pump inhibitors in treatment regimens to improve and shorten anti-TB treatment (World Health Organization, 2010, 2011).
Modification of Targets
Intrinsic resistance to several important antibiotics in mycobacteria can also be achieved by modification of target sites. For example, erythromycin resistance methylase (erm) gene in MTB encodes an enzyme that methylates a specific site in the 23S rRNA and alters the drug-binding site, thus preventing the binding of macrolides (Buriánková et al., 2004). Subinhibitory levels of clarithromycin have been shown to cause induction of erm expression and a 4- to 8-fold increase in MIC values (Andini and Nash, 2006).
These methylases have also been characterized in the clinically important RGM Mycobacterium abscessus, as well as in several clinically less relevant NTM species (Nash et al., 2005, 2009; van Ingen et al., 2012; Maurer et al., 2015). Expression of erm in M. abscessus conferred resistance to clarithromycin, ethromycin and ketolides (Nash et al., 2009; Choi et al., 2012; Stout and Floto, 2012). Thus, macrolide resistance induced by the erm gene expression may explain the inefficiency of macrolide based therapy against M. abscessus (Nash et al., 2009; Choi et al., 2012; Stout and Floto, 2012).
Another identified example is mfpA gene, which confers resistance to quinolones in MTB (Hegde et al., 2005; Tao et al., 2013; Mayer and Takiff, 2014). The mfpA gene encodes pentapeptide repeat proteins, which bind to and protect DNA gyrase from the lethal action of quinolones (Hegde et al., 2005). Three-dimensional structure analysis of MfpA showed that its structure displays size, shape, and electrostatic similarity to B-form DNA, and MfpA has been suggested to interact with DNA gyrase via DNA mimicry (Hegde et al., 2005). By binding to gyrase in DNA's place, MfpA apparently divests fluoroquinolones of their target (Ferber, 2005).
Loss of methylation can also confer resistance to certain antibiotics. A well-known example is the resistance to capreomycin and viomycin that occurs when the methyltransferase tlyA is deactivated (Maus et al., 2005b). This gene codes an rRNA methyltransferase and that loss of methyltransferase activity yields an unmethylated ribosome that is resistant to these drugs (Maus et al., 2005b). Furthermore, inactivation of GidB, which methylates 16S rRNA, confers low-level streptomycin resistance (Verma et al., 2014).
An intrinsic mechanism by which mycobacteria can increase their tolerance to RIF is the RNA polymerase binding protein A (RbpA), which has been characterized in MTB and M. smegmatis. This protein binds to the RNA polymerase, where it hampers binding of RIF (Dey et al., 2010).
Modification of Antibiotics
As well as preventing antibiotics from entering the cell or altering their targets, mycobacteria can also degrade or modify antibiotics.
Enzymatic degradation of antibiotics
Mycobacteria can encode several specific enzymes that have evolved to target and cleave antibiotics of different classes, including β-lactams, aminoglycosides and macrolides (Wang et al., 2006; Da Silva and Palomino, 2011; Wivagg et al., 2014). The β-lactam antibiotics are among the most important classes of antibacterial compounds and act by inhibiting the bacterial transpeptidases that are responsible for the final step of peptidoglycan cross-linking (Wright, 2005). In mycobacteria, β-lactam resistance is primarily due to the production of an Ambler class-A β-lactamase encoded by the blaC gene (Hugonnet and Blanchard, 2007). However, other factors such as low cell envelope permeability and presence of low penicillin-binding protein binding affinity for beta-lactams were also believed to contribute to the ineffectiveness of beta-lactams in mycobacteria (Wang et al., 2006; Jankute et al., 2015). Class A enzymes in MTB include the beta-lactamase encoded by the chromosomal blaC gene, which shares extensive similarity with many eubacterial b-lactamases (Wang et al., 2006; Nampoothiri et al., 2008). These enzymes are large proteins with much greater penicillinase activity than cephalosporinase activity (Wang et al., 2006). Like other Class A β-lactamases, BlaC hydrolysis the β- lactam ring via nucleophilic attack by an active site serine residue to inactive the drug (Tremblay et al., 2010). In addition, MTB BlaC displays relatively broad hydrolysis spectrum to all β-lactam classes, including the members of the carbapenem antibiotics, which are generally resistant to β-lactamases of other pathogenic bacteria (Hugonnet and Blanchard, 2007; Tremblay et al., 2010). Moreover, β-lactamase inhibitors such as clavulanic acid are less effective against BlaC compared to other class A enzymes (Hugonnet and Blanchard, 2007). Although mycobacterial beta-lactamases can hydrolysis carbapenems, reactions proceed slowly, and one member of the carbapenem family, meropenem, has been reported to be active in vitro in association with clavulanic acid against drug resistant strains (Hugonnet et al., 2009; Mainardi et al., 2011). Meropenem may also have potential applications for the treatment of susceptible TB, because meropenem–clavulanic acid is active against nonreplicating forms of bacilli, which are difficult to eradicate even with INH and RIF (Mainardi et al., 2011).
Ambler class A β-lactamases have also been described in M. fortuitum and M. smegmatis (Nguyen and Thompson, 2006; Soroka et al., 2014).
Enzymatic modification of antibiotics
The addition of chemical groups to specific sites on the antibiotic by mycobacterial modifying enzymes causes antibiotic resistance by preventing the antibiotic from binding to its target protein.
Among mycobacterial species, aminoglycoside resistance is sometimes caused by modifying enzymes that are coded by genes on the chromosome (Zaunbrecher et al., 2009; Maurer et al., 2015). Resistance to aminoglycoside antibiotics is also conferred by target mutation, reduced uptake and/or increased efflux (Aínsa et al., 1998; Da Silva and Palomino, 2011). Till now, two main classes of aminoglycoside-modifying enzymes namely, acetyltransferase and phosphotransferase, have been demonstrated in several mycobacterial species (Zaunbrecher et al., 2009; Kim et al., 2011; Ahn, 2013). The best biochemically characterized aminoglycoside-modifying enzyme in mycobacteria is aminoglycoside N-acetyltransferase (AAC 2′) which is capable of acetylating all know aminoglycosides bearing 2′ amino group including neomycin, kanamycin, gentamycin, tobramycin and ribostamycin (Vetting et al., 2002). Distinct N-acetyltransferases have been identified in the genomes of the MTB, M. kansasii, the rapid growers M. fortuitum, M. smegmatis, and M. abscessus (Ho et al., 2000; Ripoll et al., 2009; Ramirez and Tolmasky, 2010; van Ingen et al., 2012).
Another discovery is the presence of a RIF-resistance gene in some mycobacterial species (Baysarowich et al., 2008). In the opportunistic pathogen M. smegmatis, RIF is an ineffective drug because of the presence of a chromosomally encoded RIF ADP-ribosyltransferase (Baysarowich et al., 2008). This specific enzyme, transfers the ADP-ribose unit to a hydroxyl residue at position 23 of RIF, rendering the bacterium resistant to RIFs (Baysarowich et al., 2008).
Activation of a Transcriptional Regulator
Intrinsic resistance to any antimicrobial drugs may also be determined by an interactive network, including effector proteins, regulatory proteins, and inducers (Morris et al., 2005; Burian et al., 2012). WhiB7, is a transcriptional regulator that contributes to intrinsic antibiotic resistance in mycobacteria by activating its own expression and many drug resistance genes (Burian et al., 2012). whiB7 transcription is auto-regulated and its expression can be induced by exposure to sub-inhibitory concentrations of antibiotics as well as a variety of stress conditions such as heat shock, iron starvation, and entry in to stationary phase (Geiman et al., 2006; Burian et al., 2012). Importantly, WhiB7 controls expression of eis gene that plays an important role in mycobacterial survival within macrophages (Wei et al., 2000).
A recent condition-specific model analysis also suggests that knocking out and overexpressing of several transcription factors would cause different phenotypes in MTB. For example, this model predicted overexpression of the transcription factor whiB4 in the presence of ethionamide and INH (Ma et al., 2015).
In addition to WhiB7, MTB also encodes several sigma factors, including SigF that is antibiotic-inducible and plays a part in intrinsic MDR phenotypes (Sharma et al., 2010; La Rosa et al., 2012; Machado et al., 2012).
Other control genes such as dosR, mbtB and hspX in MTB have also been reported to be implicated in various processes ranging from dormancy to drug tolerance/persistence (Timm et al., 2003; Voskuil et al., 2003; Boshoff et al., 2004; Nandakumar et al., 2014; Sharma and Tyagi, 2016). DosR (dormancy transcriptional regulator) is a well-characterized two component system in MTB which is induced in response to hypoxia and multiple stresses (Kendall et al., 2004; Gautam et al., 2014; Mehra et al., 2015; Sharma and Tyagi, 2016). DosR is believed to be one of the key regulators that mediate MTB survival within granulomatous lesions found in TB (Converse et al., 2009). Recent study indicated that treatment of wild-type MTB with INH resulted in increased induction of dosR gene (Nandakumar et al., 2014). These antibiotic-induced responses then may contribute functionally to endogenous antibiotic tolerance in MTB (Nandakumar et al., 2014).
Acquired Resistance
The anti-mycobacterial agents specifically bind to their targets with high affinity, thus preventing the normal activity of the targets. Changes to the target structure that prevent efficient antibiotic binding can confer resistance. Unlike the situation in other bacteria where acquired drug resistance is generally mediated through horizontal transfer by mobile genetic elements, in mycobacterial species, it is caused mainly by spontaneous mutations in chromosomal genes encoding targets. Table 3 provides a summary of drug targets and known or possible mechanisms of resistance in mycobacteria.
In MTB, resistance-associated point mutations, have been described for all first-line drugs (INH, RIF, PZA and EMB), and for several second-line and newer drugs (fluoroquinolones, macrolides and Bedaquiline) (Somoskovi et al., 2001; Da Silva and Palomino, 2011; Segala et al., 2012).
Acquired Resistance to First-Line TB Drugs due to Mutations
This is considered as the principal mechanism of resistance to INH and RIF, the most powerful anti-TB agents (see Table 3). Resistance to INH is a complex process. Mutations in several genes, including katG, inhA, ahpC, kasA, and ndh have all been associated with INH resistance (Da Silva and Palomino, 2011). INH is a pro-drug requiring activation by the catalase/peroxidase enzyme encoded by katG. A reduction in catalase/peroxidase activity as a result of katG mutations is the most common mechanism associated with INH resistance. Other such mechanisms involve mutations in the inhA promoter that result in overexpression of inhA, which confers low-level resistance to INH (Cohen et al., 2014). More than 80% of INH resistance cases could be explained by the two mutations of katG S315T and inhA promoter -15C-T (Torres et al., 2015). Recently, Torres et al, discovered novel mutations that are able to explain 98% of INH resistant phenotypes by a katG, inhA promoter, or a fabG1 mutation (Torres et al., 2015). Another important finding about the mutations associated with INH resistance is that the identification of harbinger mutations, such as katG S315T, may serve as an early warning signal for MDR emergence (Manson et al., 2017). The finding has a major public health impact as it can enable targeted treatment of patients with pre-MDR-TB.
For RIF, more than 95% of resistant strains have a mutation within the 81-bp hotspot region (codons 507–533) of the RNA polymerase beta-subunit gene (rpoB) (Nasiri et al., 2016).
PZA is the backbone of short-course chemotherapy for TB, which is an important drug to shorten treatment regimens. Mutations in the pncA gene result in the loss of pyrazinamidase activity and have strong correlation with PZA resistance (Gu et al., 2016). Studies also indicated that mutations in the rpsA (encode the ribosomal protein S1) and panD (encodes aspartate decarboxylase) genes are responsible for PZA resistance (Shi et al., 2011, 2014). In addition to above mentioned genes, Zhang et al. showed that mutations in a newly identified gene clpC1, which encodes an ATP-dependent ATPase involved in protein degradation is associated with PZA resistance in MTB (Zhang et al., 2017).
EMB, another first-line anti-TB drug, together with INH, RIF, and PZA, currently is used for treatment of TB and prevents the emergence of drug resistance. Several studies have shown that mutations in the embCAB operon (encoding arabinosyltransferases in MTB) particularly the embB gene, are a major cause of EMB resistance in MTB (Lingaraju et al., 2016). Mutations in the ubiA gene also appear to be responsible for high-level EMB resistance in MTB (Safi et al., 2013; He et al., 2015; Lingaraju et al., 2016).
Acquired Resistance to Second-Line TB Drugs due to Mutations
Kanamycin and amikacin are the major aminoglycosides used for treatment of MDR-TB and several other mycobacterial species. These antibiotics inhibit protein synthesis by binding to the 30S subunit of the mycobacterial ribosome. The most common mechanism of resistance to aminoglycosides has been associated with an A1401G mutation in the rrs gene coding for 16S rRNA (Da Silva and Palomino, 2011). Unlike most other bacteria, which have multiple copies of the rrs gene, mycobacteria have a single copy of the gene (Cohen et al., 2014). Consequently, mutations in this gene are usually associated with high-level aminoglycoside resistance. Capreomycin and viomycin are polypeptide antimicrobial agents that are also used in combination therapy for treatment of drug-resistant MTB strains (Maus et al., 2005a). Like the structurally unrelated aminoglycosides, capreomycin and viomycin are bactericidal drugs that inhibit protein synthesis. MTB strains that acquire resistance to kanamycin, usually become resistant to capreomycin or viomycin (Maus et al., 2005a). However, cross-resistance between kanamycin and capreomycin and between kanamycin and viomycin is variable (Musser, 1995). Mutations in the rrs gene have also been implicated in resistance to capreomycin and viomycin (Jnawali et al., 2013).
Resistances to quinolones are also associated mainly with mutations in drug target genes. Fluoroquinolones are second-line anti-TB agents, and they currently form the backbone of MDR-TB therapy. They are also used for treatment of mycobacterial infections caused by M. kansasii, M. simiae, and M. fortuitum (OFFICIAL, 1990). Quinolones target two essential bacterial type II topoisomerases, DNA gyrase (also known as topoisomerase II) and DNA topoisomerase IV, enzymes that regulate the supercoiling of DNA and are thus essential for bacterial DNA replication and cell division (Drlica, 1999). While many bacterial species contain both DNA gyrase and topoisomerase IV, mycobacteria lack topoisomerase IV and contain only DNA gyrase, a tetramer consisting of two A and two B subunits, encoded by the genes gyrA and gyrB, respectively (Cole et al., 1998). Different bactericidal activity of various fluoroquinolones may be explained by their specificity for different enzymes; ciprofloxacin, which is less effective against MTB, preferentially targets topoisomerase IV, which is lacking in MTB, whereas newer-generation fluoroquinolones including moxifloxacin and levofloxacin, has a predilection for DNA gyrase (Ginsburg et al., 2003). Fluoroquinolone resistance in mycobacteria is associated with mutations within a highly conserved region, the quinolone resistance-determining region (QRDR), of the gyrA and gyrB genes (Takiff et al., 1994; Ginsburg et al., 2003). The most common mutation in fluoroquinolone-resistant MTB isolates involves a substitution at codon 90 and 94 of the gyrA gene (Von Groll et al., 2009; Sirgel et al., 2012). Specific amino acid changes in the QRDR can cause distinct levels of quinolone resistance. A high-level resistance is often associated with at least two mutations in gyrA or mutations in gyrA plus gyrB, with double gyrA mutants expressing the highest level of resistance (MIC 20 mg/mL) to sparfloxacin (Ginsburg et al., 2003). As indicated by previous studies, fluoroquinolone resistance is also mediated by efflux mechanism (Pasca et al., 2004; da Silva et al., 2011; Li et al., 2014).
Macrolides are bacteriostatic antibiotics that inhibit protein synthesis in a wide range of bacteria by binding to the 50S ribosomal subunit. Species of mycobacteria belonging to MTB complex (MTC) are intrinsically resistant to macrolides and different members of this class of antibiotics have little or no effect on MTC.
Linezolid, the main oxazolidinone currently in clinical use, is most commonly used to treat drug resistant TB, but its use has been limited by toxicity concerns. Mutations in rplC gene encoding for 50S ribosomal protein L3 and rrl gene encoding for 23S rRNA have been detected in linezolid-resistant clinical isolates (Hillemann et al., 2008; Beckert et al., 2012).
Resistance to new anti-TB drugs has also been documented. Bedaquiline (Sirturo, TMC207), is a diarylquinoline drug that was recently approved for use against MDR-TB. It binds to and inhibits the mycobacterial ATP synthase encoded by the essential gene atpE (Andries et al., 2005). Resistance to Bedaquiline is mediated by mutations in the atpE gene, typically at positions 63 or 66 (Petrella et al., 2006). However, resistant mutants have been identified that lack any mutation in the atpE or in the other genes encoding components of ATP synthase, indicating alternative mechanisms of drug resistance (Huitric et al., 2010). Recently, mutations in transcriptional repressor (Rv0678) in Bedaquiline resistant MTB isolates without atpE mutations have been recognized (Andries et al., 2014; Hartkoorn et al., 2014; Zhang S. et al., 2015). These mutations resulted in the upregulation of mmpL5-mmpS5 expression, thereby leading to increased efflux and cross-resistance to Clofazimine (Andries et al., 2014; Hartkoorn et al., 2014; Zhang S. et al., 2015).
Mutations Conferring Resistance to Drugs Used to Treat NTM
Unlike TB, disease caused by NTM, is rarely treated by first-line anti-TB drugs. For the treatment of NTM infections, the macrolide antibiotics play an important role in the therapeutic regimens (van Ingen et al., 2012). Since the strains from the MTC are intrinsically resistant to macrolides, the problem of emergence of macrolide resistance will be discussed only for NTM (Doucet-Populaire et al., 2002; Buriánková et al., 2004). Single point mutation in position 2058 or 2059 of the 23S rRNA gene (rrl) has been associated with high level macrolide resistance in several clinical isolates of NTM, including M. abscessus, M. avium complex (MAC), M. chelonae, M. fortuitum, and M. kansasii (Meier et al., 1996; Wallace et al., 1996; Burman et al., 1998; Bastian et al., 2011; Brown-Elliott et al., 2012).
Resistance to macrolides in MAC poses a significant challenge to effective prophylaxis and treatment outcome in HIV infected patients (Griffith et al., 2007). In these patients, macrolides are used for MAC infection prophylaxis. Macrolide-resistant isolates of MAC have been found in most patients with unsuccessful macrolide prophylaxis. Emergence of resistance to macrolides during treatment has been also described for other NTM species (Doucet-Populaire et al., 2002). According to the current American Thoracic Society guidelines, macrolide antibiotics (clarithromycin and azithromycin) must be administrated in combination with other drugs to prevent the emergence of macrolide resistance in NTM species (Griffith et al., 2007).
The primary mechanism of acquired resistance to aminoglycosides in NTM is based on mutations in the 16S rRNA gene. A mutation in position 1408 of the 16S ribosomal RNA (rrs) gene is responsible for high-level resistance in both M. abscessus and M. chelonae after therapy as well as in vitro selection (Wallace et al., 1985; Prammananan et al., 1998). Mycobacterium abscessus represents one of the most common antibiotic-resistant RGM species, which is usually resistant to major anti-TB drugs, as well as most antimycobacterial drugs, including tetracycline, fluoroquinolones and sulphonamides (Nessar et al., 2011). However, this species is naturally susceptible to amikacin and clarithromycin/azithromycin, which are used in combination for treatment of infections caused by this bacterium (Petrini, 2006). Thus, emergence of amikacin-resistant isolates of M. abscessus could complicate the management of these infections.
Fluoroquinolones appear to be drugs with therapeutic possibilities against clinical isolates of M. fortuitum, M. chelonae, and M. kansasii (Dıaz et al., 2003). In NTM species, the most common acquired resistance mechanism to fluoroquinolones involves a stepwise accumulation of mutations in the QRDR of gyrA gene (Brown-Elliott et al., 2012; Monego et al., 2012).
RIF is the key component of treatment regimens for diseases causes by M. kansasii and MAC (Klein et al., 2001; Obata et al., 2006; Brown-Elliott et al., 2012). Acquired RIF resistance in M. kansasii and MAC has been documented and is conferred primarily by mutations in the rpoB gene (Klein et al., 2001; Obata et al., 2006). These mutations are identical to those observed in RIF-resistant MTB isolates (Brown-Elliott et al., 2012).
Conclusions
The infections due to Mycobacterium species, particularly strains that are clinically important and resistant to clinical agents, poses significant public health problems. Reports about drug resistant mycobacteria from different countries, suggest that common mycobacterial species may become refractory to any chemotherapeutic agent in the future (Velayati et al., 2009; Migliori et al., 2012; Udwadia et al., 2012; Klopper et al., 2013).
The limited number of new anti- mycobacterial agents coming to market and new threats arising from drug resistant isolates (including MDR and XDR isolates) brings us to the end of the “antibiotics era.” Mycobacterium species including MTB are armed with a wide variety of intrinsic and acquired drug resistance mechanisms: Modification of antibiotic targets mediated by bacterial specific enzymes or mutations and degradation/modification of antibiotics by production of antibiotic inactivating enzymes renders the Mycobacterium species resistant to most classes of antimicrobials. Several mycobacterial drug efflux pumps have been identified in INH and RIF resistant isolates providing resistance by expelling the drug molecules that enter the cell. Besides, unique structural and physiological properties of MTB including low cell envelope permeability, slow growth rate, and the ability to enter a metabolically inactive dormant phase have even exacerbated the drug resistance problem leaving us perilously close to none or very limited number of therapeutic options. This unavoidable circumstance emphasizes the urgent need for the development of multidisciplinary approaches to combat the resistance crisis. Modification of existing antibiotics and screening for new antibiotics from unexplored ecological niches that can act on novel targets and inhibit existing resistance mechanisms such as inhibitors of efflux pumps, factors involved in dormancy induction and bacterial enzymes involved in cell envelope synthesis, target modification or antibiotic inactivation/modification provide promising strategies to disarm the resistant Mycobacterium species. To this end, understanding the mechanisms involved in mycobacterial tolerance to antibiotics could aid not only for planning an appropriate therapeutic regimen but also for development of novel therapeutic agents that can overwhelm existing resistance mechanisms. In recent years, due to the advances in genomics and biology, our knowledge of the remarkable diversity of mechanisms of antimicrobial resistance in Mycobacteria has greatly increased. However, despite substantial progress, there is clearly much work to be done to fully elucidate the molecular basis of antimicrobial resistance in this group of bacteria. The best hope for the future is the development of a greater understanding of exact mechanisms of antimicrobial resistance in mycobacteria to further improve the therapeutic outcomes in Mycobacterium infected patients.
Author Contributions
Conceived and designed the study and wrote the paper: MJN. Participated in manuscript revising and editing: MJN, MH, MG, HG, AP, AAIF, and MMF.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Ahn, J. W. (2013). Rv3168 phosphotransferase activity mediates kanamycin resistance in Mycobacterium tuberculosis. J. Microbiol. Biotechnol. 23, 1529–1535. doi: 10.4014/jmb.1306.06048
Aínsa, J. A., Blokpoel, M. C., Otal, I., Young, D. B., De Smet, K. A., and Martín, C. (1998). Molecular cloning and characterization of Tap, a putative multidrug efflux pump present in Mycobacterium fortuitum and Mycobacterium tuberculosis. J. Bacteriol. 180, 5836–5843.
Alderwick, L. J., Harrison, J., Lloyd, G. S., and Birch, H. L. (2015). The Mycobacterial cell wall—peptidoglycan and Arabinogalactan. Cold Spring Harb. Perspect. Med. 5:a021113. doi: 10.1101/cshperspect.a021113
Andini, N., and Nash, K. A. (2006). Intrinsic macrolide resistance of the Mycobacterium tuberculosis complex is inducible. Antimicrob. Agents Chemother. 50, 2560–2562. doi: 10.1128/AAC.00264-06
Andries, K., Verhasselt, P., Guillemont, J., Göhlmann, H. W., Neefs, J.-M., and Lee, E. (2005). A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 307, 223–227. doi: 10.1126/science.1106753
Andries, K., Villellas, C., Coeck, N., Thys, K., Gevers, T., Vranckx, L., et al. (2014). Acquired resistance of Mycobacterium tuberculosis to bedaquiline. PLoS ONE 9:e102135. doi: 10.1371/journal.pone.0102135
Balasubramanian, V., Solapure, S., Iyer, H., Ghosh, A., Sharma, S., Kaur, P., et al. (2014). Bactericidal activity and mechanism of action of AZD5847, a novel oxazolidinone for treatment of tuberculosis. Antimicrob. Agents Chemother. 58, 495–502. doi: 10.1128/AAC.01903-13
Balganesh, M., Dinesh, N., Sharma, S., Kuruppath, S., Nair, A. V., and Sharma, U. (2012). Efflux pumps of Mycobacterium tuberculosis play a significant role in antituberculosis activity of potential drug candidates. Antimicrob. Agents Chemother. 56, 2643–2651. doi: 10.1128/AAC.06003-11
Balganesh, M., Kuruppath, S., Marcel, N., Sharma, S., Nair, A., and Sharma, U. (2010). Rv1218c, an ABC transporter of Mycobacterium tuberculosis with implications in drug discovery. Antimicrob. Agents Chemother. 54, 5167–5172. doi: 10.1128/AAC.00610-10
Ballesteros, M., Fredriksson, Å., Henriksson, J., and Nyström, T. (2001). Bacterial senescence: protein oxidation in non-proliferating cells is dictated by the accuracy of the ribosomes. EMBO J. 20, 5280–5289. doi: 10.1093/emboj/20.18.5280
Basta, L. A. B., Ghosh, A., Pan, Y., Jakoncic, J., Lloyd, E. P., Townsend, C. A., et al. (2015). Loss of a functionally and structurally distinct ld-Transpeptidase, LdtMt5, compromises cell wall integrity in Mycobacterium tuberculosis. J. Biol. Chem. 290, 25670–25685. doi: 10.1074/jbc.M115.660753
Bastian, S., Veziris, N., Roux, A.-L., Brossier, F., Gaillard, J.-L., Jarlier, V., et al. (2011). Assessment of clarithromycin susceptibility in strains belonging to the Mycobacterium abscessus group by erm (41) and rrl sequencing. Antimicrob. Agents Chemother. 55, 775–781. doi: 10.1128/AAC.00861-10
Baysarowich, J., Koteva, K., Hughes, D. W., Ejim, L., Griffiths, E., Zhang, K., et al. (2008). Rifamycin antibiotic resistance by ADP-ribosylation: structure and diversity of Arr. Proc. Natl. Acad. Sci. U.S.A. 105, 4886–4891. doi: 10.1073/pnas.0711939105
Beckert, P., Hillemann, D., Kohl, T. A., Kalinowski, J., Richter, E., Niemann, S., et al. (2012). rplC T460C identified as a dominant mutation in linezolid-resistant Mycobacterium tuberculosis strains. Antimicrob. Agents Chemother. 56, 2743–2745. doi: 10.1128/AAC.06227-11
Betts, J. C., Lukey, P. T., Robb, L. C., McAdam, R. A., and Duncan, K. (2002). Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43, 717–731. doi: 10.1046/j.1365-2958.2002.02779.x
Bhatt, K., Banerjee, S. K., and Chakraborti, P. K. (2000). Evidence that phosphate specific transporter is amplified in a fluoroquinolone resistant Mycobacterium smegmatis. Eur. J. Biochem. 267, 4028–4032. doi: 10.1046/j.1432-1327.2000.01437.x
Blair, J. M., Webber, M. A., Baylay, A. J., Ogbolu, D. O., and Piddock, L. J. (2015). Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 13, 42–51. doi: 10.1038/nrmicro3380
Boonaiam, S., Chaiprasert, A., Prammananan, T., and Leechawengwongs, M. (2010). Genotypic analysis of genes associated with isoniazid and ethionamide resistance in MDR-TB isolates from Thailand. Clin. Microbiol. Infect. 16, 396–399. doi: 10.1111/j.1469-0691.2009.02838.x
Boshoff, H. I., Myers, T. G., Copp, B. R., McNeil, M. R., Wilson, M. A., and Barry, C. E. (2004). The transcriptional responses of Mycobacterium tuberculosis to inhibitors of metabolism novel insights into drug mechanisms of action. J. Biol. Chem. 279, 40174–40184. doi: 10.1074/jbc.M406796200
Botella, H., Vaubourgeix, J., Lee, M. H., Song, N., Xu, W., Makinoshima, H., et al. (2017). Mycobacterium tuberculosis protease MarP activates a peptidoglycan hydrolase during acid stress. EMBO J. 36, 536–548. doi: 10.15252/embj.201695028
Bowman, J., and Ghosh, P. (2014). A complex regulatory network controlling intrinsic multidrug resistance in Mycobacterium smegmatis. Mol. Microbiol. 91, 121–134. doi: 10.1111/mmi.12448
Brennan, P. J. (2003). Structure, function, and biogenesis of the cell wall of Mycobacterium tuberculosis. Tuberculosis 83, 91–97. doi: 10.1016/S1472-9792(02)00089-6
Brossier, F., Veziris, N., Truffot-Pernot, C., Jarlier, V., and Sougakoff, W. (2011). Molecular investigation of resistance to the antituberculous drug ethionamide in multidrug-resistant clinical isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 55, 355–360. doi: 10.1128/AAC.01030-10
Brown-Elliott, B. A., Nash, K. A., and Wallace, R. J. (2012). Antimicrobial susceptibility testing, drug resistance mechanisms, and therapy of infections with nontuberculous mycobacteria. Clin. Microbiol. Rev. 25, 545–582. doi: 10.1128/CMR.05030-11
Brown-Elliott, B. A., Philley, J. V., Benwill, J. L., and Wallace, R. J. Jr. (2014). Current Opinions in the Treatment of Pulmonary Nontuberculous Mycobacteria in Non-Cystic Fibrosis Patients: Mycobacterium abscessus Group, Mycobacterium avium Complex, and Mycobacterium kansasii. Curr. Treat. Opt. Infect. Dis. 6, 392–408. doi: 10.1007/s40506-014-0032-2
Burian, J., Ramón-García, S., Sweet, G., Gómez-Velasco, A., Av-Gay, Y., and Thompson, C. J. (2012). The mycobacterial transcriptional regulator whiB7 gene links redox homeostasis and intrinsic antibiotic resistance. J. Biol. Chem. 287, 299–310. doi: 10.1074/jbc.M111.302588
Buriánková, K., Doucet-Populaire, F., Dorson, O., Gondran, A., Ghnassia, J.-C., Wieser, J., et al. (2004). Molecular basis of intrinsic macrolide resistance in the Mycobacterium tuberculosis complex. Antimicrob. Agents Chemother. 48, 143–150. doi: 10.1128/AAC.48.1.143-150.2004
Burman, W. J., Stone, B. L., Brown, B. A., Wallace, R. J., and Böttger, E. C. (1998). AIDS-related Mycobacterium kansasii infection with initial resistance to clarithromycin. Diagn. Microbiol. Infect. Dis. 31, 369–371. doi: 10.1016/s0732-8893(98)00013-3
Cáceres, N. E., Harris, N. B., Wellehan, J. F., Feng, Z., Kapur, V., and Barletta, R. G. (1997). Overexpression of the D-alanine racemase gene confers resistance to D-cycloserine in Mycobacterium smegmatis. J. Bacteriol. 179, 5046–5055. doi: 10.1128/jb.179.16.5046-5055.1997
Caleffi-Ferracioli, K. R., Amaral, R. C., Demitto, F. O., Maltempe, F. G., Canezin, P. H., Scodro, R. B., et al. (2016). Morphological changes and differentially expressed efflux pump genes in Mycobacterium tuberculosis exposed to a rifampicin and verapamil combination. Tuberculosis 97, 65–72. doi: 10.1016/j.tube.2015.12.010
Calvanese, L., Falcigno, L., Maglione, C., Marasco, D., Ruggiero, A., Squeglia, F., et al. (2014). Structural and binding properties of the PASTA domain of PonA2, a key penicillin binding protein from Mycobacterium tuberculosis. Biopolymers 101, 712–719. doi: 10.1002/bip.22447
Cândido, P. H. C., Nunes Lde, S., Marques, E. A., Folescu, T. W., Coelho, F. S., de Moura, V. C., et al. (2014). Multidrug-resistant nontuberculous mycobacteria isolated from cystic fibrosis patients. J. Clin. Microbiol. 52, 2990–2997. doi: 10.1128/JCM.00549-14
Castañeda-García, A. Blázquez, J., and Rodríguez-Rojas, A. (2013). Molecular mechanisms and clinical impact of acquired and intrinsic fosfomycin resistance. Antibiotics 2, 217–236. doi: 10.3390/antibiotics2020217
Chao, M. C., and Rubin, E. J. (2010). Letting sleeping dos lie: does dormancy play a role in tuberculosis? Annu. Rev. Microbiol. 64, 293–311. doi: 10.1146/annurev.micro.112408.134043
Chen, J. M., Uplekar, S., Gordon, S. V., and Cole, S. T. (2012). A point mutation in cycA partially contributes to the D-cycloserine resistance trait of Mycobacterium bovis BCG vaccine strains. PLoS ONE 7:e43467. doi: 10.1371/journal.pone.0043467
Cheng, A. F., Yew, W. W., Chan, E. W., Chin, M. L., Hui, M. M., and Chan, R. C. (2004). Multiplex PCR amplimer conformation analysis for rapid detection of gyrA mutations in fluoroquinolone-resistant Mycobacterium tuberculosis clinical isolates. Antimicrob. Agents Chemother. 48, 596–601. doi: 10.1128/AAC.48.2.596-601.2004
Choi, G.-E., Shin, S. J., Won, C.-J., Min, K.-N., Oh, T., Hahn, M. Y., et al. (2012). Macrolide treatment for Mycobacterium abscessus and Mycobacterium massiliense infection and inducible resistance. Am. J. Respir. Crit. Care Med. 186, 917–925. doi: 10.1164/rccm.201111-2005OC
Choudhuri, B., Bhakta, S., Barik, R., Basu, J., Kundu, M., and Chakrabarti, P. (2002). Overexpression and functional characterization of an ABC (ATP-binding cassette) transporter encoded by the genes drrA and drrB of Mycobacterium tuberculosis. Biochem. J 367, 279–285. doi: 10.1042/bj20020615
Cohen, K., Bishai, W., and Pym, A. (2014). Molecular basis of drug resistance in Mycobacterium tuberculosis. Microbiol. Spectr. 2:MGM2–0036–2013. doi: 10.1128/microbiolspec.MGM2-0036-2013
Cole, S., Brosch, R., Parkhill, J., Garnier, T., Churcher, C., Harris, D., Gordon, S., et al. (1998). Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544. doi: 10.1038/31159
Converse, P. J., Karakousis, P. C., Klinkenberg, L. G., Kesavan, A. K., Ly, L. H., Allen, S. S., et al. (2009). Role of the dosR-dosS two-component regulatory system in Mycobacterium tuberculosis virulence in three animal models. Infect. Immun. 77, 1230–1237. doi: 10.1128/IAI.01117-08
Cowman, S., Burns, K., Benson, S., Wilson, R., and Loebinger, M. (2015). The antimicrobial susceptibility of non-tuberculous mycobacteria. J. Infect. 72, 324–331. doi: 10.1016/j.jinf.2015.12.007
Daffé, M. (2015). The cell envelope of tubercle bacilli. Tuberculosis 95, S155–S158. doi: 10.1016/j.tube.2015.02.024
Danilchanka, O., Pavlenok, M., and Niederweis, M. (2008). Role of porins for uptake of antibiotics by Mycobacterium smegmatis. Antimicrob. Agents Chemother. 52, 3127–3134. doi: 10.1128/AAC.00239-08
Da Silva, P. E. A., and Palomino, J. C. (2011). Molecular basis and mechanisms of drug resistance in Mycobacterium tuberculosis: classical and new drugs. J. Antimicrob. Chemother. 66, 1417–1430. doi: 10.1093/jac/dkr173
da Silva, P. E. A., Von Groll, A., Martin, A., and Palomino, J. C. (2011). Efflux as a mechanism for drug resistance in Mycobacterium tuberculosis. FEMS Immunol. Med. Microbiol. 63, 1–9. doi: 10.1111/j.1574-695X.2011.00831.x
De Rossi, E., Aínsa, J. A., and Riccardi, G. (2006). Role of mycobacterial efflux transporters in drug resistance: an unresolved question. FEMS Microbiol. Rev. 30, 36–52. doi: 10.1111/j.1574-6976.2005.00002.x
De Rossi, E., Arrigo, P., Bellinzoni, M., Silva, P. A., Martín, C., Aínsa, J. A., et al. (2002). The multidrug transporters belonging to major facilitator superfamily in Mycobacterium tuberculosis. Mol. Med. 8:714.
De Rossi, E., Branzoni, M., Cantoni, R., Milano, A., Riccardi, G., and Ciferri, O. (1998). mmr, a Mycobacterium tuberculosis gene conferring resistance to small cationic dyes and inhibitors. J. Bacteriol. 180, 6068–6071.
Dey, A., Verma, A. K., and Chatterji, D. (2010). Role of an RNA polymerase interacting protein, MsRbpA, from Mycobacterium smegmatis in phenotypic tolerance to rifampicin. Microbiology 156, 873–883. doi: 10.1099/mic.0.033670-0
Dıaz, López, J. R. M., Ruiz, M., and Royo, G. (2003). In vitro activity of new fluoroquinolones and linezolid against non-tuberculous mycobacteria. Int. J. Antimicrob. Agents 21, 585–588. doi: 10.1016/S0924-8579(03)00048-7
Doran, J. L., Pang, Y., Mdluli, K. E., Moran, A. J., Victor, T. C., Stokes, R. W., et al. (1997). Mycobacterium tuberculosis efpA encodes an efflux protein of the QacA transporter family. Clin. Diagn. Lab. Immunol. 4, 23–32.
Doucet-Populaire, F., Buriankova, K., Weiser, J., and Pernodet, J. (2002). Natural and acquired macrolide resistance in mycobacteria. Curr. Drug Targets Infect. Disord. 2, 355–370. doi: 10.2174/1568005023342263
Drlica, K. (1999). Mechanism of fluoroquinolone action. Curr. Opin. Microbiol. 2, 504–508. doi: 10.1016/S1369-5274(99)00008-9
Dukan, S., Farewell, A., Ballesteros, M., Taddei, F., Radman, M., and Nyström, T. (2000). Protein oxidation in response to increased transcriptional or translational errors. Proc. Natl. Acad. Sci. U.S.A. 97, 5746–5749. doi: 10.1073/pnas.100422497
Dutta, N. K., Mehra, S., and Kaushal, D. (2010). A Mycobacterium tuberculosis sigma factor network responds to cell-envelope damage by the promising anti-mycobacterial thioridazine. PLoS ONE 5:e10069. doi: 10.1371/journal.pone.0010069
El Meouche, I., Siu, Y., and Dunlop, M. J. (2016). Stochastic expression of a multiple antibiotic resistance activator confers transient resistance in single cells. Sci. Rep. 6:19538. doi: 10.1038/srep19538
Faller, M., Niederweis, M., and Schulz, G. E. (2004). The structure of a mycobacterial outer-membrane channel. Science 303, 1189–1192. doi: 10.1126/science.1094114
Farhat, M. R., Shapiro, B. J., Kieser, K. J., Sultana, R., Jacobson, K. R., Victor, T. C., et al. (2013). Genomic analysis identifies targets of convergent positive selection in drug-resistant Mycobacterium tuberculosis. Nat. Genet. 45, 1183–1189. doi: 10.1038/ng.2747
Fattorini, L., Orefici, G., Jin, S. H., Scardaci, G., Amicosante, G., Franceschini, N., et al. (1992). Resistance to beta-lactams in Mycobacterium fortuitum. Antimicrob. Agents Chemother. 36, 1068–1072. doi: 10.1128/AAC.36.5.1068
Ferber, D. (2005). Protein that mimics DNA helps tuberculosis bacteria resist antibiotics. Science 308, 1393–1393. doi: 10.1126/science.308.5727.1393a
Feuerriegel, S., Köser, C. U., Richter, E., and Niemann, S. (2013). Mycobacterium canettii is intrinsically resistant to both pyrazinamide and pyrazinoic acid. J. Antimicrob. Chemother. 68, 1439–1440. doi: 10.1093/jac/dkt042
Finken, M., Kirschner, P., Meier, A., Wrede, A., and Böttger, E. C. (1993). Molecular basis of streptomycin resistance in Mycobacterium tuberculosis: alterations of the ribosomal protein S12 gene and point mutations within a functional 16S ribosomal RNA pseudoknot. Mol. Microbiol. 9, 1239–1246. doi: 10.1111/j.1365-2958.1993.tb01253.x
Garima, K., Pathak, R., Tandon, R., Rathor, N., Sinha, R., Bose, M., et al. (2015). Differential expression of efflux pump genes of Mycobacterium tuberculosis in response to varied subinhibitory concentrations of antituberculosis agents. Tuberculosis 95, 155–161. doi: 10.1016/j.tube.2015.01.005
Garton, N. J., Waddell, S. J., Sherratt, A. L., Lee, S.-M., Smith, R. J., Senner, C., et al. (2008). Cytological and transcript analyses reveal fat and lazy persister-like bacilli in tuberculous sputum. PLoS Med. 5:e75. doi: 10.1371/journal.pmed.0050075
Gautam, U. S., Sikri, K., Vashist, A., Singh, V., and Tyagi, J. S. (2014). Essentiality of DevR/DosR interaction with SigA for the dormancy survival program in Mycobacterium tuberculosis. J. Bacteriol. 196, 790–799. doi: 10.1128/JB.01270-13
Geiman, D. E., Raghunand, T. R., Agarwal, N., and Bishai, W. R. (2006). Differential gene expression in response to exposure to antimycobacterial agents and other stress conditions among seven Mycobacterium tuberculosis whiB-like genes. Antimicrob. Agents Chemother. 50, 2836–2841. doi: 10.1128/AAC.00295-06
Gengenbacher, M., and Kaufmann, S. H. (2012). Mycobacterium tuberculosis: success through dormancy. FEMS Microbiol. Rev. 36, 514–532. doi: 10.1111/j.1574-6976.2012.00331.x
Georghiou, S. B., Magana, M., Garfein, R. S., Catanzaro, D. G., Catanzaro, A., and Rodwell, T. C. (2012). Evaluation of genetic mutations associated with Mycobacterium tuberculosis resistance to amikacin, kanamycin and capreomycin: a systematic review. PLoS ONE 7:e33275. doi: 10.1371/journal.pone.0033275
Ginsburg, A. S., Grosset, J. H., and Bishai, W. R. (2003). Fluoroquinolones, tuberculosis, and resistance. Lancet Infect. Dis. 3, 432–442. doi: 10.1016/S1473-3099(03)00671-6
Glassroth, J. (2008). Pulmonary disease due to nontuberculous mycobacteria. CHEST J. 133, 243–251. doi: 10.1378/chest.07-0358
Gomez, J. E., and McKinney, J. D. (2004). M. tuberculosis persistence, latency, and drug tolerance. Tuberculosis 84, 29–44. doi: 10.1016/j.tube.2003.08.003
Gratraud, P., Surolia, N., Besra, G. S., Surolia, A., Kremer, L., et al. (2008). Antimycobacterial activity and mechanism of action of NAS-91. Antimicrobial Agents Chemother. 52, 1162–1166.
Griffith, D. E., Aksamit, T., Brown-Elliott, B. A., Catanzaro, A., Daley, C., Gordin, F., et al. (2007). An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am. J. Respir. Crit. Care Med. 175, 367–416. doi: 10.1164/rccm.200604-571ST
Gu, Y., Yu, X., Jiang, G., Wang, X., Ma, Y., Li, Y., et al. (2016). Pyrazinamide resistance among multidrug-resistant tuberculosis clinical isolates in a national referral center of China and its correlations with pncA, rpsA, and panD gene mutations. Diagn. Microbiol. Infect. Dis. 84, 207–211. doi: 10.1016/j.diagmicrobio.2015.10.017
Guo, H., Seet, Q., Denkin, S., Parsons, L., and Zhang, Y. (2006). Molecular characterization of isoniazid-resistant clinical isolates of Mycobacterium tuberculosis from the USA. J. Med. Microbiol. 55, 1527–1531. doi: 10.1099/jmm.0.46718-0
Gupta, A. K., Katoch, V. M., Chauhan, D. S., Sharma, R., Singh, M., Venkatesan, K., et al. (2010a). Microarray analysis of efflux pump genes in multidrug-resistant Mycobacterium tuberculosis during stress induced by common anti-tuberculous drugs. Microbial. Drug Resist. 16, 21–28. doi: 10.1089/mdr.2009.0054
Gupta, A. K., Reddy, V. P., Lavania, M., Chauhan, D., Venkatesan, K., Sharma, V., et al. (2010b). jefA (Rv2459), a drug efflux gene in Mycobacterium tuberculosis confers resistance to isoniazid & ethambutol. Indian J. Med. Res. 132, 176–188.
Gupta, R., Lavollay, M., Mainardi, J.-L., Arthur, M., Bishai, W. R., and Lamichhane, G. (2010). The Mycobacterium tuberculosis protein LdtMt2 is a nonclassical transpeptidase required for virulence and resistance to amoxicillin. Nat. Med. 16, 466–469. doi: 10.1038/nm.2120
Hao, P., Shi-Liang, Z., Ju, L., Ya-Xin, D., Biao, H., Xu, W., et al. (2011). The role of ABC efflux pump, Rv1456c-Rv1457c-Rv1458c, from Mycobacterium tuberculosis clinical isolates in China. Folia Microbiol. (Praha) 56, 549–553. doi: 10.1007/s12223-011-0080-7
Hartkoorn, R. C., Uplekar, S., and Cole, S. T. (2014). Cross-resistance between clofazimine and bedaquiline through upregulation of MmpL5 in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 58, 2979–2981. doi: 10.1128/AAC.00037-14
He, L., Wang, X., Cui, P., Jin, J., Chen, J., Zhang, W., et al. (2015). UbiA (Rv3806c) encoding DPPR synthase involved in cell wall synthesis is associated with ethambutol resistance in Mycobacterium tuberculosis. Tuberculosis 95, 149–154. doi: 10.1016/j.tube.2014.12.002
Heep, M., Brandstätter, B., Rieger, U., Lehn, N., Richter, E., Rüsch-Gerdes, S., et al. (2001). Frequency of rpoB mutations inside and outside the cluster I region in rifampin-resistant clinical Mycobacterium tuberculosis isolates. J. Clin. Microbiol. 39, 107–110. doi: 10.1128/JCM.39.1.107-110.2001
Hegde, S. S., Vetting, M. W., Roderick, S. L., Mitchenall, L. A., Maxwell, A., Takiff, H. E., et al. (2005). A fluoroquinolone resistance protein from Mycobacterium tuberculosis that mimics DNA. Science 308, 1480–1483. doi: 10.1126/science.1110699
Heinz, C., and Niederweis, M. (2000). Selective extraction and purification of a mycobacterial outer membrane protein. Anal. Biochem. 285, 113–120. doi: 10.1006/abio.2000.4728
Hillemann, D., Rüsch-Gerdes, S., and Richter, E. (2008). In vitro-selected linezolid-resistant Mycobacterium tuberculosis mutants. Antimicrob. Agents Chemother. 52, 800–801. doi: 10.1128/AAC.01189-07
Ho, I., Chan, C., and Cheng, A. (2000). Aminoglycoside Resistance in Mycobacterium kansasii, Mycobacterium avium-M. intracellulare, and Mycobacterium fortuitum: are aminoglycoside-modifying enzymes responsible? Antimicrob. Agents Chemother. 44, 39–42. doi: 10.1128/AAC.44.1.39-42.2000
Horsburgh, C. R. Jr., Barry, C. III, and Lange, C. (2015). Treatment of tuberculosis. New Engl. J. Med. 373, 2149–2160. doi: 10.1056/NEJMra1413919
Hu, Y. M., Butcher, P. D., Sole, K., Mitchison, D., and Coates, A. (1998). Protein synthesis is shutdown in dormant Mycobacterium tuberculosis and is reversed by oxygen or heat shock. FEMS Microbiol. Lett. 158, 139–145. doi: 10.1111/j.1574-6968.1998.tb12813.x
Hugonnet, J.-E., and Blanchard, J. S. (2007). Irreversible inhibition of the Mycobacterium tuberculosis β-lactamase by clavulanate. Biochemistry 46, 11998–12004. doi: 10.1021/bi701506h
Hugonnet, J.-E., Tremblay, L. W., Boshoff, H. I., Barry, C. E., and Blanchard, J. S. (2009). Meropenem-clavulanate is effective against extensively drug-resistant Mycobacterium tuberculosis. Science 323, 1215–1218. doi: 10.1126/science.1167498
Huitric, E., Verhasselt, P., Koul, A., Andries, K., Hoffner, S., and Andersson, D. I. (2010). Rates and mechanisms of resistance development in Mycobacterium tuberculosis to a novel diarylquinoline ATP synthase inhibitor. Antimicrob. Agents Chemother. 54, 1022–1028. doi: 10.1128/AAC.01611-09
Jackson, M., McNeil, M. R., and Brennan, P. J. (2013). Progress in targeting cell envelope biogenesis in Mycobacterium tuberculosis. Future Microbiol. 8, 855–875. doi: 10.2217/fmb.13.52
Jackson, M., Raynaud, C., Lanéelle, M. A., Guilhot, C., Laurent-Winter, C., Ensergueix, D., et al. (1999). Inactivation of the antigen 85C gene profoundly affects the mycolate content and alters the permeability of the Mycobacterium tuberculosis cell envelope. Mol. Microbiol. 31, 1573–1587. doi: 10.1046/j.1365-2958.1999.01310.x
Jankute, M., Cox, J. A., Harrison, J., and Besra, G. S. (2015). Assembly of the Mycobacterial Cell Wall. Annu. Rev. Microbiol. 69, 405–423. doi: 10.1146/annurev-micro-091014-104121
Jiang, X., Zhang, W., Zhang, Y., Gao, F., Lu, C., Zhang, X., et al. (2008). Assessment of efflux pump gene expression in a clinical isolate Mycobacterium tuberculosis by real-time reverse transcription PCR. Microbial. Drug Resist. 14, 7–11. doi: 10.1089/mdr.2008.0772
Jnawali, H. N., Yoo, H., Ryoo, S., Lee, K.-J., Kim, B.-J., Koh, W.-J., et al. (2013). Molecular genetics of Mycobacterium tuberculosis resistant to aminoglycosides and cyclic peptide capreomycin antibiotics in Korea. World J. Microbiol. Biotechnol. 29, 975–982. doi: 10.1007/s11274-013-1256-x
Johansen, S. K., Maus, C. E., Plikaytis, B. B., and Douthwaite, S. (2006). Capreomycin binds across the ribosomal subunit interface using tlyA-encoded 2′-O-methylations in 16S and 23S rRNAs. Mol. Cell 23, 173–182. doi: 10.1016/j.molcel.2006.05.044
Kasperbauer, S. H., and De Groote, M. A. (2015). The Treatment of Rapidly Growing Mycobacterial Infections. Clin. Chest Med. 36, 67–78. doi: 10.1016/j.ccm.2014.10.004
Kendall, S., Movahedzadeh, F., Rison, S., Wernisch, L., Parish, T., Duncan, K., et al. (2004). The Mycobacterium tuberculosis dosRS two-component system is induced by multiple stresses. Tuberculosis 84, 247–255. doi: 10.1016/j.tube.2003.12.007
Kieser, K. J., Baranowski, C., Chao, M. C., Long, J. E., Sassetti, C. M., Waldor, M. K., et al. (2015). Peptidoglycan synthesis in Mycobacterium tuberculosis is organized into networks with varying drug susceptibility. Proc. Natl. Acad. Sci. U.S.A. 112, 13087–13092. doi: 10.1073/pnas.1514135112
Kim, S., Nguyen, C. M. T., Kim, E. J., and Kim, K. J. (2011). Crystal structure of Mycobacterium tuberculosis Rv3168: a putative aminoglycoside antibiotics resistance enzyme. Proteins 79, 2983–2987. doi: 10.1002/prot.23119
Klein, J. L., Brown, T. J., and French, G. L. (2001). Rifampin resistance in Mycobacterium kansasii is associated with rpoB mutations. Antimicrob. Agents Chemother. 45, 3056–3058. doi: 10.1128/AAC.45.11.3056-3058.2001
Klopper, M., Warren, R. M., Hayes, C., Gey van Pittius, N. C., Streicher, E. M., Müller, B., et al. (2013). Emergence and spread of extensively and totally drug-resistant tuberculosis, South Africa. Emerg. Infect. Dis. 19:449. doi: 10.3201/eid1903.120246
La Rosa, V., Poce, G., Canseco, J. O., Buroni, S., Pasca, M. R., Biava, M., et al. (2012). MmpL3 is the cellular target of the antitubercular pyrrole derivative BM212. Antimicrob. Agents Chemother. 56, 324–331. doi: 10.1128/AAC.05270-11
Lavollay, M., Arthur, M., Fourgeaud, M., Dubost, L., Marie, A., Veziris, N., et al. (2008). The peptidoglycan of stationary-phase Mycobacterium tuberculosis predominantly contains cross-links generated by L, D-transpeptidation. J. Bacteriol. 190, 4360–4366. doi: 10.1128/JB.00239-08
Li, G., Zhang, J., Guo, Q., Jiang, Y., Wei, J., Zhao, L. L., et al. (2015). Efflux pump gene expression in multidrug-resistant Mycobacterium tuberculosis clinical isolates. PLoS ONE 10:e0119013. doi: 10.1371/journal.pone.0119013
Li, J., Gao, X., Luo, T., Wu, J., Sun, G., Liu, Q., et al. (2014). Association of gyrA/B mutations and resistance levels to fluoroquinolones in clinical isolates of Mycobacterium tuberculosis. Emer. Microb. Infect. 3:e19. doi: 10.1038/emi.2014.21
Li, X.-Z., Zhang, L., and Nikaido, H. (2004). Efflux pump-mediated intrinsic drug resistance in Mycobacterium smegmatis. Antimicrob. Agents Chemother. 48, 2415–2423. doi: 10.1128/AAC.48.7.2415-2423.2004
Lingaraju, S., Rigouts, L., Gupta, A., Lee, J., Umubyeyi, A. N., Davidow, A. L., et al. (2016). Geographic differences in the contribution of ubiA mutations to high-level ethambutol resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 60, 4101–4105. doi: 10.1128/AAC.03002-15
Liu, J., Takiff, H. E., and Nikaido, H. (1996). Active efflux of fluoroquinolones in Mycobacterium smegmatis mediated by LfrA, a multidrug efflux pump. J. Bacteriol. 178, 3791–3795. doi: 10.1128/jb.178.13.3791-3795.1996
Lubelski, J., Konings, W. N., and Driessen, A. J. (2007). Distribution and physiology of ABC-type transporters contributing to multidrug resistance in bacteria. Microbiol. Mol. Biol. Rev. 71, 463–476. doi: 10.1128/MMBR.00001-07
Lupoli, T. J., Fay, A., Adura, C., Glickman, M. S., and Nathan, C. F. (2016). Reconstitution of a Mycobacterium tuberculosis proteostasis network highlights essential cofactor interactions with chaperone DnaK. Proc. Natl. Acad. Sci. U.S.A. 113, E7947–E7956. doi: 10.1073/pnas.1617644113
Ma, S., Minch, K. J., Rustad, T. R., Hobbs, S., Zhou, S.-L., Sherman, D. R., et al. (2015). Integrated modeling of gene regulatory and metabolic networks in Mycobacterium tuberculosis. PLoS Comput. Biol. 11:e1004543. doi: 10.1371/journal.pcbi.1004543
Machado, D., Couto, I., Perdigão, J., Rodrigues, L., Portugal, I., Baptista, P., et al. (2012). Contribution of efflux to the emergence of isoniazid and multidrug resistance in Mycobacterium tuberculosis. PLoS ONE 7:e34538. doi: 10.1371/journal.pone.0034538
Mailaender, C., Reiling, N., Engelhardt, H., Bossmann, S., Ehlers, S., and Niederweis, M. (2004). The MspA porin promotes growth and increases antibiotic susceptibility of both Mycobacterium bovis BCG and Mycobacterium tuberculosis. Microbiology 150, 853–864. doi: 10.1099/mic.0.26902-0
Mainardi, J., Hugonnet, J., Gutmann, L., and Arthur, M. (2011). Fighting resistant tuberculosis with old compounds: the carbapenem paradigm. Clin. Microbiol. Infect. 17, 1755–1756. doi: 10.1111/j.1469-0691.2011.03699.x
Makarov, V., Manina, G., Mikusova, K., Möllmann, U., Ryabova, O., Saint-Joanis, B., et al. (2009). Benzothiazinones kill Mycobacterium tuberculosis by blocking arabinan synthesis. Science 324, 801–804. doi: 10.1126/science.1171583
Manina, G., Bellinzoni, M., Pasca, M. R., Neres, J., Milano, A., Ribeiro, A. L., et al. (2010). Biological and structural characterization of the Mycobacterium smegmatis nitroreductase NfnB, and its role in benzothiazinone resistance. Mol. Microbiol. 77, 1172–1185. doi: 10.1111/j.1365-2958.2010.07277.x
Manson, A. L., Cohen, K. A., Abeel, T., Desjardins, C. A., Armstrong, D. T., Barry, C. E. III, et al. (2017). Genomic analysis of globally diverse Mycobacterium tuberculosis strains provides insights into the emergence and spread of multidrug resistance. Nat. Genet. 49, 395–402. doi: 10.1038/ng.3767
Martins, M., Dastidar, S. G., Fanning, S., Kristiansen, J. E., Molnar, J., Pagès, J. M., et al. (2008). Potential role of non-antibiotics (helper compounds) in the treatment of multidrug-resistant Gram-negative infections: mechanisms for their direct and indirect activities. Int. J. Antimicrob. Agents 31, 198–208. doi: 10.1016/j.ijantimicag.2007.10.025
Mathys, V., Wintjens, R., Lefevre, P., Bertout, J., Singhal, A., Kiass, M., et al. (2009). Molecular genetics of para-aminosalicylic acid resistance in clinical isolates and spontaneous mutants of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 53, 2100–2109. doi: 10.1128/AAC.01197-08
Matsumoto, M., Hashizume, H., Tomishige, T., Kawasaki, M., Tsubouchi, H., Sasaki, H., et al. (2006). OPC-67683, a nitro-dihydro-imidazooxazole derivative with promising action against tuberculosis in vitro and in mice. PLoS Med. 3:e466. doi: 10.1371/journal.pmed.0030466
Maurer, F. P., Bruderer, V. L., Castelberg, C., Ritter, C., Scherbakov, D., Bloemberg, G. V., et al. (2015). Aminoglycoside-modifying enzymes determine the innate susceptibility to aminoglycoside antibiotics in rapidly growing mycobacteria. J. Antimicrob. Chemother. 70, 1412–1419. doi: 10.1093/jac/dku550
Maus, C. E., Plikaytis, B. B., and Shinnick, T. M. (2005a). Molecular analysis of cross-resistance to capreomycin, kanamycin, amikacin, and viomycin in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 49, 3192–3197. doi: 10.1128/AAC.49.8.3192-3197.2005
Maus, C. E., Plikaytis, B. B., and Shinnick, T. M. (2005b). Mutation of tlyA confers capreomycin resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 49, 571–577. doi: 10.1128/AAC.49.2.571-577.2005
Mayer, C., and Takiff, H. (2014). The molecular genetics of fluoroquinolone resistance in Mycobacterium tuberculosis. Microbiol. Spectr. 2:MGM2-0009-2013. doi: 10.1128/microbiolspec.MGM2-0009-2013
Mehra, S., Foreman, T. W., Didier, P. J., Ahsan, M. H., Hudock, T. A., Kissee, R., et al. (2015). The DosR regulon modulates adaptive immunity and is essential for Mycobacterium tuberculosis persistence. Am. J. Respir. Crit. Care Med. 191, 1185–1196. doi: 10.1164/rccm.201408-1502OC
Meier, A., Heifets, L., Wallace, R. J., Zhang, Y., Brown, B. A., Sander, P., et al. (1996). Molecular Mechanisms of Clarithromycin Resistance in Mycobacterium avium: Observation of Multiple 238 rDNA Mutations in a clonal population. J. Infect. Dis. 174, 354–360. doi: 10.1093/infdis/174.2.354
Meier, A., Kirschner, P., Springer, B., Steingrube, V. A., Brown, B. A., Wallace, R., et al. (1994). Identification of mutations in 23S rRNA gene of clarithromycin-resistant Mycobacterium intracellulare. Antimicrob. Agents Chemother. 38, 381–384. doi: 10.1128/AAC.38.2.381
Migliori, G. B., Centis, R., D'Ambrosio, L., Spanevello, A., Borroni, E., Cirillo, D. M., et al. (2012). Totally drug-resistant and extremely drug-resistant tuberculosis: the same disease? Clin. Infect. Dis. 54, 1379–1380. doi: 10.1093/cid/cis128
Milano, A., Pasca, M. R., Provvedi, R., Lucarelli, A. P., Manina, G., Ribeiro, A. L., et al. (2009). Azole resistance in Mycobacterium tuberculosis is mediated by the MmpS5–MmpL5 efflux system. Tuberculosis (Edinb). 89, 84–90. doi: 10.1016/j.tube.2008.08.003
Millard, J., Ugarte-Gil, C., and Moore, D. A. (2015). Multidrug resistant tuberculosis. BMJ 350:h882. doi: 10.1136/bmj.h882
Mills, J. A., Motichka, K., Jucker, M., Wu, H. P., Uhlik, B. C., Stern, R. J., et al. (2004). Inactivation of the mycobacterial rhamnosyltransferase, which is needed for the formation of the arabinogalactan-peptidoglycan linker, leads to irreversible loss of viability. J. Biol. Chem. 279, 43540–43546. doi: 10.1074/jbc.M407782200
Mishra, M. N., and Daniels, L. (2013). Characterization of the MSMEG_2631 gene (mmp) encoding a multidrug and toxic compound extrusion (MATE) family protein in Mycobacterium smegmatis and exploration of its polyspecific nature using biolog phenotype microarray. J. Bacteriol. 195, 1610–1621. doi: 10.1128/JB.01724-12
Monego, F., Duarte, R. S., and Biondo, A. W. (2012). gyrA and gyrB gene mutation in ciprofloxacin-resistant Mycobacterium massiliense clinical isolates from Southern Brazil. Microbial. Drug Resist. 18, 1–6. doi: 10.1089/mdr.2011.0047
Moraes, G. L., Gomes, G. C., Monteiro de Sousa, P. R., Alves, C. N., Govender, T., Kruger, H. G., et al. (2015). Structural and functional features of enzymes of Mycobacterium tuberculosis peptidoglycan biosynthesis as targets for drug development. Tuberculosis 95, 95–111. doi: 10.1016/j.tube.2015.01.006
Morlock, G. P., Metchock, B., Sikes, D., Crawford, J. T., and Cooksey, R. C. (2003). ethA, inhA, and katG loci of ethionamide-resistant clinical Mycobacterium tuberculosis isolates. Antimicrob. Agents Chemother. 47, 3799–3805. doi: 10.1128/AAC.47.12.3799-3805.2003
Morris, R. P., Nguyen, L., Gatfield, J., Visconti, K., Nguyen, K., Schnappinger, D., et al. (2005). Ancestral antibiotic resistance in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 102, 12200–12205. doi: 10.1073/pnas.0505446102
Murakami, S., Nakashima, R., Yamashita, E., Matsumoto, T., and Yamaguchi, A. (2006). Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature 443, 173–179. doi: 10.1038/nature05076
Musser, J. M. (1995). Antimicrobial agent resistance in mycobacteria: molecular genetic insights. Clin. Microbiol. Rev. 8, 496–514.
Nampoothiri, K., Rubex, R., Patel, A., Narayanan, S., Krishna, S., Das, S., et al. (2008). Molecular cloning, overexpression and biochemical characterization of hypothetical β-lactamases of Mycobacterium tuberculosis H37Rv. J. Appl. Microbiol. 105, 59–67. doi: 10.1111/j.1365-2672.2007.03721.x
Nandakumar, M., Nathan, C., and Rhee, K. Y. (2014). Isocitrate lyase mediates broad antibiotic tolerance in Mycobacterium tuberculosis. Nat. Commun. 5:4306. doi: 10.1038/ncomms5306
Nash, K. A., Brown-Elliott, B. A., and Wallace, R. J. (2009). A novel gene, erm (41), confers inducible macrolide resistance to clinical isolates of Mycobacterium abscessus but is absent from Mycobacterium chelonae. Antimicrob. Agents Chemother. 53, 1367–1376. doi: 10.1128/AAC.01275-08
Nash, K. A., Zhang, Y., Brown-Elliott, B. A., and Wallace, R. J. (2005). Molecular basis of intrinsic macrolide resistance in clinical isolates of Mycobacterium fortuitum. J. Antimicrob. Chemother. 55, 170–177. doi: 10.1093/jac/dkh523
Nasiri, M. J., Dabiri, H., Darban-Sarokhalil, D., and Shahraki, A. H. (2015). Prevalence of Non-Tuberculosis Mycobacterial Infections among Tuberculosis Suspects in Iran: Systematic Review and Meta-Analysis. PLoS ONE 10:e0129073. doi: 10.1371/journal.pone.0129073
Nasiri, M. J., Darban-Sarokhalil, D., Fooladi, A. A., and Feizabadi, M. M. (2016). katG Ser315 and rpoB 81-bp hotspot region substitutions: reliability for detection of drug-resistant strains of Mycobacterium tuberculosis. J. Glob. Antimicrob. Resist 5, 92–93. doi: 10.1016/j.jgar.2015.12.001
National Committee for Clinical Laboratory Standards (NCCLS) (2000). Susceptibility Testing Mycobacteria, Nocardia, and Other Aerobic Actin omycetes: Tentative Standard. Wayne, PA.
Nessar, R., Reyrat, J. M., Murray, A., and Gicquel, B. (2011). Genetic analysis of new 16S rRNA mutations conferring aminoglycoside resistance in Mycobacterium abscessus. J. Antimicrob. Chemother. 66, 1719–1724. doi: 10.1093/jac/dkr209
Nguyen, L., Chinnapapagari, S., and Thompson, C. J. (2005). FbpA-dependent biosynthesis of trehalose dimycolate is required for the intrinsic multidrug resistance, cell wall structure, and colonial morphology of Mycobacterium smegmatis. J. Bacteriol. 187, 6603–6611. doi: 10.1128/JB.187.19.6603-6611.2005
Nguyen, L., and Thompson, C. J. (2006). Foundations of antibiotic resistance in bacterial physiology: the mycobacterial paradigm. Trends Microbiol. 14, 304–312. doi: 10.1016/j.tim.2006.05.005
Niederweis, M. (2003). Mycobacterial porins–new channel proteins in unique outer membranes. Mol. Microbiol. 49, 1167–1177. doi: 10.1046/j.1365-2958.2003.03662.x
Niederweis, M., Danilchanka, O., Huff, J., Hoffmann, C., and Engelhardt, H. (2010). Mycobacterial outer membranes: in search of proteins. Trends Microbiol. 18, 109–116. doi: 10.1016/j.tim.2009.12.005
Niederweis, M., Ehrt, S., Heinz, C., KloÈcker, U., Karosi, S., Swiderek, K. M., et al. (1999). Cloning of the mspA gene encoding a porin from Mycobacterium smegmatis. Mol. Microbiol. 33, 933–945. doi: 10.1046/j.1365-2958.1999.01472.x
Obata, S., Zwolska, Z., Toyota, E., Kudo, K., Nakamura, A., Sawai, T., et al. (2006). Association of rpoB mutations with rifampicin resistance in Mycobacterium avium. Int. J. Antimicrob. Agents 27, 32–39. doi: 10.1016/j.ijantimicag.2005.09.015
OFFICIAL, T. (1990). Diagnosis and treatment of disease caused by nontuberculous mycobacteria. Am. Rev. Respir. Dis. 142, 940–953. doi: 10.1164/ajrccm/142.4.940
Ordway, D., Viveiros, M., Leandro, C., Bettencourt, R., Almeida, J., Martins, M., et al. (2003). Clinical concentrations of thioridazine kill intracellular multidrug-resistant Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 47, 917–922. doi: 10.1128/AAC.47.3.917-922.2003
Palomino, J. C., and Martin, A. (2014). Drug Resistance Mechanisms in Mycobacterium tuberculosis. Antibiotics 3, 317–340. doi: 10.3390/antibiotics3030317
Pang, Y., Lu, J., Wang, Y., Song, Y., Wang, S., and Zhao, Y. (2013). Study of the rifampin monoresistance mechanism in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 57, 893–900. doi: 10.1128/AAC.01024-12
Pasca, M. R., Guglierame, P., Arcesi, F., Bellinzoni, M., De Rossi, E., and Riccardi, G. (2004). Rv2686c-Rv2687c-Rv2688c, an ABC fluoroquinolone efflux pump in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 48, 3175–3178. doi: 10.1128/AAC.48.8.3175-3178.2004
Pasca, M. R., Guglierame, P., De Rossi, E., Zara, F., and Riccardi, G. (2005). mmpL7 gene of Mycobacterium tuberculosis is responsible for isoniazid efflux in Mycobacterium smegmatis. Antimicrob. Agents Chemother. 49, 4775–4777. doi: 10.1128/AAC.49.11.4775-4777.2005
Pelayo, M. C. G., Uplekar, S., Keniry, A., Lopez, P. M., Garnier, T., Garcia, J. N., et al. (2009). A comprehensive survey of single nucleotide polymorphisms (SNPs) across Mycobacterium bovis strains and M. bovis BCG vaccine strains refines the genealogy and defines a minimal set of SNPs that separate virulent M. bovis strains and M. bovis BCG strains. Infect. Immun. 77, 2230–2238. doi: 10.1128/IAI.01099-08
Petrella, S., Cambau, E., Chauffour, A., Andries, K., Jarlier, V., and Sougakoff, W. (2006). Genetic basis for natural and acquired resistance to the diarylquinoline R207910 in mycobacteria. Antimicrob. Agents Chemother. 50, 2853–2856. doi: 10.1128/AAC.00244-06
Petrini, B. (2006). Mycobacterium abscessus: an emerging rapid-growing potential pathogen. APMIS 114, 319–328. doi: 10.1111/j.1600-0463.2006.apm_390.x
Philalay, J. S., Palermo, C. O., Hauge, K. A., Rustad, T. R., and Cangelosi, G. A. (2004). Genes required for intrinsic multidrug resistance in Mycobacterium avium. Antimicrob. Agents Chemother. 48, 3412–3418. doi: 10.1128/AAC.48.9.3412-3418.2004
Philley, J. V., and Griffith, D. E. (2015). Treatment of slowly growing Mycobacteria. Clin. Chest Med. 36, 79–90. doi: 10.1016/j.ccm.2014.10.005
Prammananan, T., Sander, P., Brown, B. A., Frischkorn, K., Onyi, G. O., Zhang, Y., et al. (1998). A single 16S ribosomal RNA substitution is responsible for resistance to amikacin and other 2-deoxystreptamine aminoglycosides in Mycobacterium abscessus and Mycobacterium chelonae. J. Infect. Dis. 177, 1573–1581. doi: 10.1086/515328
Protopopova, M., Hanrahan, C., Nikonenko, B., Samala, R., Chen, P., Gearhart, J., et al. (2005). Identification of a new antitubercular drug candidate, SQ109, from a combinatorial library of 1, 2-ethylenediamines. J. Antimicrob. Chemother. 56, 968–974. doi: 10.1093/jac/dki319
Ramaswamy, S. V., Reich, R., Dou, S.-J., Jasperse, L., Pan, X., Wanger, A., et al. (2003). Single nucleotide polymorphisms in genes associated with isoniazid resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 47, 1241–1250. doi: 10.1128/AAC.47.4.1241-1250.2003
Ramirez, M. S., and Tolmasky, M. E. (2010). Aminoglycoside modifying enzymes. Drug Resist. Updat. 13, 151–171. doi: 10.1016/j.drup.2010.08.003
Ramón-García, S., Martín, C., Aínsa, J. A., and De Rossi, E. (2006). Characterization of tetracycline resistance mediated by the efflux pump Tap from Mycobacterium fortuitum. J. Antimicrob. Chemother. 57, 252–259. doi: 10.1093/jac/dki436
Raynaud, C., Papavinasasundaram, K., Speight, R. A., Springer, B., Sander, P., Böttger, E. C., et al. (2002). The functions of OmpATb, a pore-forming protein of Mycobacterium tuberculosis. Mol. Microbiol. 46, 191–201. doi: 10.1046/j.1365-2958.2002.03152.x
Ripoll, F., Pasek, S., Schenowitz, C., Dossat, C., Barbe, V., Rottman, M., et al. (2009). Non mycobacterial virulence genes in the genome of the emerging pathogen Mycobacterium abscessus. PLoS ONE 4:e5660. doi: 10.1371/journal.pone.0005660
Rivers, E. C., and Mancera, R. L. (2008). New anti-tuberculosis drugs in clinical trials with novel mechanisms of action. Drug Disc. Tod. 13, 1090–1098. doi: 10.1016/j.drudis.2008.09.004
Rodrigues, L., Machado, D., Couto, I., Amaral, L., and Viveiros, M. (2012). Contribution of efflux activity to isoniazid resistance in the Mycobacterium tuberculosis complex. Infect. Genet. Evol. 12, 695–700. doi: 10.1016/j.meegid.2011.08.009
Rodrigues, L., Sampaio, D., Couto, I., Machado, D., Kern, W. V., Amaral, L., et al. (2009). The role of efflux pumps in macrolide resistance in Mycobacterium avium complex. Int. J. Antimicrob. Agents 34, 529–533. doi: 10.1016/j.ijantimicag.2009.07.010
Rodrigues, L., Villellas, C., Bailo, R., Viveiros, M., and Aínsa, J. A. (2013). Role of the Mmr efflux pump in drug resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 57, 751–757. doi: 10.1128/AAC.01482-12
Ronning, D. R., Klabunde, T., Besra, G. S., Vissa, V. D., Belisle, J. T., and Sacchettini, J. C. (2000). Crystal structure of the secreted form of antigen 85C reveals potential targets for mycobacterial drugs and vaccines. Nat. Struct. Mol. Biol. 7, 141–146. doi: 10.1038/72413
Safi, H., Lingaraju, S., Amin, A., Kim, S., Jones, M., Holmes, M., et al. (2013). Evolution of high-level ethambutol-resistant tuberculosis through interacting mutations in decaprenylphosphoryl-β-D-arabinose biosynthetic and utilization pathway genes. Nat. Genet. 45, 1190–1197. doi: 10.1038/ng.2743
Schoonmaker, M. K., Bishai, W. R., and Lamichhane, G. (2014). Nonclassical transpeptidases of Mycobacterium tuberculosis alter cell size, morphology, the cytosolic matrix, protein localization, virulence, and resistance to β-lactams. J. Bacteriol. 196, 1394–1402. doi: 10.1128/JB.01396-13
Segala, E., Sougakoff, W., Nevejans-Chauffour, A., Jarlier, V., and Petrella, S. (2012). New mutations in the mycobacterial ATP synthase: new insights into the binding of the diarylquinoline TMC207 to the ATP synthase C-ring structure. Antimicrob. Agents Chemother. 56, 2326–2334. doi: 10.1128/AAC.06154-11
Shahraki, A. H., Heidarieh, P., Bostanabad, S. Z., Khosravi, A. D., Hashemzadeh, M., Khandan, S., et al. (2015). “Multidrug-resistant tuberculosis” may be nontuberculous mycobacteria. Eur. J. Intern. Med. 26, 279–284. doi: 10.1016/j.ejim.2015.03.001
Sharma, S., Kumar, M., Sharma, S., Nargotra, A., Koul, S., and Khan, I. A. (2010). Piperine as an inhibitor of Rv1258c, a putative multidrug efflux pump of Mycobacterium tuberculosis. J. Antimicrob. Chemother. 65, 1694–1701. doi: 10.1093/jac/dkq186
Sharma, S., and Tyagi, J. S. (2016). Mycobacterium tuberculosis DevR/DosR dormancy regulator activation mechanism: dispensability of phosphorylation, cooperativity and essentiality of α10 helix. PLoS One 11:e0160723. doi: 10.1371/journal.pone.0160723
Shi, W., Chen, J., Feng, J., Cui, P., Zhang, S., Weng, X., et al. (2014). Aspartate decarboxylase (PanD) as a new target of pyrazinamide in Mycobacterium tuberculosis. Emer. Microb. Infect. 3:e58. doi: 10.1038/emi.2014.61
Shi, W., Zhang, X., Jiang, X., Yuan, H., Lee, J. S., Barry, C. E., et al. (2011). Pyrazinamide inhibits trans-translation in Mycobacterium tuberculosis. Science 333, 1630–1632. doi: 10.1126/science.1208813
Siddiqi, N., Das, R., Pathak, N., Banerjee, S., Ahmed, N., Katoch, V., et al. (2004). Mycobacterium tuberculosis isolate with a distinct genomic identity overexpresses a tap-like efflux pump. Infection 32, 109–111. doi: 10.1007/s15010-004-3097-x
Silva, P. E., Bigi, F., Santangelo, M. P., Romano, M. I., Martın, C., Cataldi, A., et al. (2001). Characterization of P55, a Multidrug Efflux Pump in Mycobacterium bovis and Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 45, 800–804. doi: 10.1128/AAC.45.3.800-804.2001
Sirgel, F. A., Warren, R. M., Streicher, E. M., Victor, T. C., van Helden, P. D., and Böttger, E. C. (2012). gyrA mutations and phenotypic susceptibility levels to ofloxacin and moxifloxacin in clinical isolates of Mycobacterium tuberculosis. J. Antimicrob. Chemother. 67, 1088–1093. doi: 10.1093/jac/dks033
Smith, T., Wolff, K. A., and Nguyen, L. (2012). “Molecular biology of drug resistance in Mycobacterium tuberculosis,” in Pathogenesis of Mycobacterium tuberculosis and Its Interaction with the Host Organism, eds J. Pieters and J. D. McKinney (Berlin; Heidelberg: Springer), 53–80.
Somoskovi, A., Parsons, L. M., and Salfinger, M. (2001). The molecular basis of resistance to isoniazid, rifampin, and pyrazinamide in Mycobacterium tuberculosis. Respir. Res. 2:164. doi: 10.1186/rr54
Song, H., Sandie, R., Wang, Y., Andrade-Navarro, M. A., and Niederweis, M. (2008). Identification of outer membrane proteins of Mycobacterium tuberculosis. Tuberculosis 88, 526–544. doi: 10.1016/j.tube.2008.02.004
Soroka, D., Dubée, V., Soulier-Escrihuela, O., Cuinet, G., Hugonnet, J.-E., Gutmann, L., et al. (2014). Characterization of broad-spectrum Mycobacterium abscessus class A β-lactamase. J. Antimicrob. Chemother. 69, 691–696. doi: 10.1093/jac/dkt410
Stahl, C., Kubetzko, S., Kaps, I., Seeber, S., Engelhardt, H., and Niederweis, M. (2001). MspA provides the main hydrophilic pathway through the cell wall of Mycobacterium smegmatis. Mol. Microbiol. 40, 451–464. doi: 10.1046/j.1365-2958.2001.02394.x
Stephan, J., Mailaender, C., Etienne, G., Daffé, M., and Niederweis, M. (2004). Multidrug resistance of a porin deletion mutant of Mycobacterium smegmatis. Antimicrob. Agents Chemother. 48, 4163–4170. doi: 10.1128/AAC.48.11.4163-4170.2004
Stout, J. E., and Floto, R. A. (2012). Treatment of Mycobacterium abscessus: all macrolides are equal, but perhaps some are more equal than others. Am. J. Respir. Crit. Care Med. 186, 822–823. doi: 10.1164/rccm0.201208-1500ED
Stover, C. K., Warrener, P., VanDevanter, D. R., Sherman, D. R., Arain, T. M., Langhorne, M. H., et al. (2000). A small-molecule nitroimidazopyran drug candidate for the treatment of tuberculosis. Nature 405, 962–966. doi: 10.1038/35016103
Tahlan, K., Wilson, R., Kastrinsky, D. B., Arora, K., Nair, V., Fischer, E., et al. (2012). SQ109 targets MmpL3, a membrane transporter of trehalose monomycolate involved in mycolic acid donation to the cell wall core of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 56, 1797–1809. doi: 10.1128/AAC.05708-11
Takiff, H., Cimino, M., Musso, M., Weisbrod, T., Martinez, R., Delgado, M., et al. (1996). Efflux pump of the proton antiporter family confers low-level fluoroquinolone resistance in Mycobacterium smegmatis. Proc. Natl. Acad. Sci. U.S.A. 93, 362–366. doi: 10.1073/pnas.93.1.362
Takiff, H. E., Salazar, L., Guerrero, C., Philipp, W., Huang, W. M., Kreiswirth, B., et al. (1994). Cloning and nucleotide sequence of Mycobacterium tuberculosis gyrA and gyrB genes and detection of quinolone resistance mutations. Antimicrob. Agents Chemother. 38, 773–780. doi: 10.1128/AAC.38.4.773
Tao, J., Han, J., Wu, H., Hu, X., Deng, J., Fleming, J., et al. (2013). Mycobacterium fluoroquinolone resistance protein B, a novel small GTPase, is involved in the regulation of DNA gyrase and drug resistance. Nucleic Acids Res. 41, 2370–2381. doi: 10.1093/nar/gks1351
Timm, J., Post, F. A., Bekker, L.-G., Walther, G. B., Wainwright, H. C., Manganelli, R., et al. (2003). Differential expression of iron-, carbon-, and oxygen-responsive mycobacterial genes in the lungs of chronically infected mice and tuberculosis patients. Proc. Natl. Acad. Sci. U.S.A. 100, 14321–14326. doi: 10.1073/pnas.2436197100
Torres, J. N., Paul, L. V., Rodwell, T. C., Victor, T. C., Amallraja, A. M., Elghraoui, A., et al. (2015). Novel katG mutations causing isoniazid resistance in clinical M. tuberculosis isolates. Emer. Microb. Infect. 4:e42. doi: 10.1038/emi.2015.42
Tremblay, L. W., Fan, F., and Blanchard, J. S. (2010). Biochemical and structural characterization of Mycobacterium tuberculosis β-lactamase with the carbapenems ertapenem and doripenem. Biochemistry 49, 3766–3773. doi: 10.1021/bi100232q
Udwadia, Z. F., Amale, R. A., Ajbani, K. K., and Rodrigues, C. (2012). Totally drug-resistant tuberculosis in India. Clin. Infect. Dis. 54, 579–581. doi: 10.1093/cid/cir889
van Ingen, J., Boeree, M. J., van Soolingen, D., and Mouton, J. W. (2012). Resistance mechanisms and drug susceptibility testing of nontuberculous mycobacteria. Drug Resist. Updat. 15, 149–161. doi: 10.1016/j.drup.2012.04.001
Vaubourgeix, J., Lin, G., Dhar, N., Chenouard, N., Jiang, X., Botella, H., et al. (2015). Stressed mycobacteria use the chaperone ClpB to sequester irreversibly oxidized proteins asymmetrically within and between cells. Cell Host Microb. 17, 178–190. doi: 10.1016/j.chom.2014.12.008
Velayati, A. A., Masjedi, M. R., Farnia, P., Tabarsi, P., Ghanavi, J., ZiaZarifi, A. H., et al. (2009). Emergence of new forms of totally drug-resistant tuberculosis bacilli: super extensively drug-resistant tuberculosis or totally drug-resistant strains in Iran. Chest J. 136, 420–425. doi: 10.1378/chest.08-2427
Verma, J. S., Gupta, Y., Nair, D., Manzoor, N., Rautela, R. S., Rai, A., et al. (2014). Evaluation of gidB alterations responsible for streptomycin resistance in Mycobacterium tuberculosis. J. Antimicrob. Chemother. 69, 2935–2941. doi: 10.1093/jac/dku273
Vetting, M. W., Hegde, S. S., Javid-Majd, F., Blanchard, J. S., and Roderick, S. L. (2002). Aminoglycoside 2′-N-acetyltransferase from Mycobacterium tuberculosis in complex with coenzyme A and aminoglycoside substrates. Nat. Struct. Mol. Biol. 9, 653–658. doi: 10.1038/nsb830
Von Groll, A., Martin, A., Jureen, P., Hoffner, S., Vandamme, P., Portaels, F., et al. (2009). Fluoroquinolone resistance in Mycobacterium tuberculosis and mutations in gyrA and gyrB. Antimicrob. Agents Chemother. 53, 4498–4500. doi: 10.1128/AAC.00287-09
Voskuil, M. I., Schnappinger, D., Visconti, K. C., Harrell, M. I., Dolganov, G. M., Sherman, D. R., et al. (2003). Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 198, 705–713. doi: 10.1084/jem.20030205
Wakamoto, Y., Dhar, N., Chait, R., Schneider, K., Signorino-Gelo, F., Leibler, S., et al. (2013). Dynamic persistence of antibiotic-stressed mycobacteria. Science 339, 91–95. doi: 10.1126/science.1229858
Wallace, R., Meier, A., Brown, B. A., Zhang, Y., Sander, P., Onyi, G. O., et al. (1996). Genetic basis for clarithromycin resistance among isolates of Mycobacterium chelonae and Mycobacterium abscessus. Antimicrob. Agents Chemother. 40, 1676–1681.
Wallace, R. J. Jr., Hull, S. I., Bobey, D. G., Price, K. E., Swenson, J. M., Steele, L. C., et al. (1985). Mutational resistance as the mechanism of acquired drug resistance to aminoglycosides and antibacterial agents in mycobacterium fortuitum and mycobacterium chelonei: evidence is based on plasmid analysis, mutational frequencies, and aminoglycoside-modifying enzyme assays 1–3. Am. Rev. Respir. Dis. 132, 409–416.
Wang, F., Cassidy, C., and Sacchettini, J. C. (2006). Crystal structure and activity studies of the Mycobacterium tuberculosis β-lactamase reveal its critical role in resistance to β-lactam antibiotics. Antimicrob. Agents Chemother. 50, 2762–2771. doi: 10.1128/AAC.00320-06
Wang, K., Pei, H., Huang, B., Zhu, X., Zhang, J., Zhou, B., et al. (2013). The expression of ABC efflux pump, Rv1217c–Rv1218c, and its association with multidrug resistance of Mycobacterium tuberculosis in China. Curr. Microbiol. 66, 222–226. doi: 10.1007/s00284-012-0215-3
Wayne, L. G., and Hayes, L. G. (1996). An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect. Immun. 64, 2062–2069.
Wei, J., Dahl, J. L., Moulder, J. W., Roberts, E. A., O'Gaora, P., Young, D. B., et al. (2000). Identification of a Mycobacterium tuberculosis gene that enhances mycobacterial survival in macrophages. J. Bacteriol. 182, 377–384. doi: 10.1128/JB.182.2.377-384.2000
Willers, C., Wentzel, J. F., du Plessis, L. H., Gouws, C., and Hamman, J. H. (2017). Efflux as a mechanism of antimicrobial drug resistance in clinical relevant microorganisms: the role of efflux inhibitors. Expert Opin. Ther. Targets 21, 23–36. doi: 10.1080/14728222.2017.1265105
Williams, K. N., Brickner, S. J., Stover, C. K., Zhu, T., Ogden, A., Tasneen, R., et al. (2009). Addition of PNU-100480 to first-line drugs shortens the time needed to cure murine tuberculosis. Am. J. Respir. Crit. Care Med. 180, 371–376. doi: 10.1164/rccm.200904-0611OC
Wivagg, C. N., Bhattacharyya, R. P., and Hung, D. T. (2014). Mechanisms of β-lactam killing and resistance in the context of Mycobacterium tuberculosis. J. Antibiot. 67, 645–654. doi: 10.1038/ja.2014.94
World Health Organization (2011). Guidelines for the Programmatic Management of Drug-Resistant Tuberculosis-2011 Update. Geneva.
World Health Organization (2014). Companion Handbook to the WHO Guidelines for the Programmatic Management of Drug-Resistant Tuberculosis. Geneva.
Wright, G. D. (2005). Bacterial resistance to antibiotics: enzymatic degradation and modification. Adv. Drug Deliv. Rev. 57, 1451–1470. doi: 10.1016/j.addr.2005.04.002
Wu, U.-I., and Holland, S. M. (2015). Host susceptibility to non-tuberculous mycobacterial infections. Lancet Infect. Dis. 15, 968–980. doi: 10.1016/S1473-3099(15)00089-4
Xie, Z., Siddiqi, N., and Rubin, E. J. (2005). Differential antibiotic susceptibilities of starved Mycobacterium tuberculosis isolates. Antimicrob. Agents Chemother. 49, 4778–4780. doi: 10.1128/AAC.49.11.4778-4780.2005
Xu, W.-X., Zhang, L., Mai, J.-T., Peng, R.-C., Yang, E.-Z., Peng, C., et al. (2014). The Wag31 protein interacts with AccA3 and coordinates cell wall lipid permeability and lipophilic drug resistance in Mycobacterium smegmatis. Biochem. Biophys. Res. Commun. 448, 255–260. doi: 10.1016/j.bbrc.2014.04.116
Xu, Y., Zhang, Z., and Sun, Z. (2015). Drug resistance to Mycobacterium tuberculosis: from the traditional Chinese view to modern systems biology. Crit. Rev. Microbiol. 41, 399–410. doi: 10.3109/1040841X.2013.860948
Yamchi, J. K., Haeili, M., Feyisa, S. G., Kazemian, H., Shahraki, A. H., Zahednamazi, F., et al. (2015). Evaluation of efflux pump gene expression among drug susceptible and drug resistant strains of Mycobacterium tuberculosis from Iran. Infect. Genet. Evol. 36, 23–26. doi: 10.1016/j.meegid.2015.08.036
Zaunbrecher, M. A., Sikes, R. D., Metchock, B., Shinnick, T. M., and Posey, J. E. (2009). Overexpression of the chromosomally encoded aminoglycoside acetyltransferase eis confers kanamycin resistance in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 106, 20004–20009. doi: 10.1073/pnas.0907925106
Zhang, M., Yue, J., Yang, Y. P., Zhang, H. M., Lei, J. Q., Jin, R. L., et al. (2005). Detection of mutations associated with isoniazid resistance in Mycobacterium tuberculosis isolates from China. J. Clin. Microbiol. 43, 5477–5482. doi: 10.1128/JCM.43.11.5477-5482.2005
Zhang, S., Chen, J., Cui, P., Shi, W., Zhang, W., and Zhang, Y. (2015). Identification of novel mutations associated with clofazimine resistance in Mycobacterium tuberculosis. J. Antimicrob. Chemother. 70, 2507–2510. doi: 10.1093/jac/dkv150
Zhang, S., Chen, J., Shi, W., Cui, P., Zhang, J., Cho, S., et al. (2017). Mutation in clpC1 encoding an ATP-dependent ATPase involved in protein degradation is associated with pyrazinamide resistance in Mycobacterium tuberculosis. Emer. Microb. Infect. 6:e8. doi: 10.1038/emi.2017.1
Zhang, S., Chen, J., Shi, W., Liu, W., Zhang, W., and Zhang, Y. (2013). Mutations in panD encoding aspartate decarboxylase are associated with pyrazinamide resistance in Mycobacterium tuberculosis. Emer. Microb. Infect. 2:e34. doi: 10.1038/emi.2013.38
Zhang, X., Liu, L., Zhang, Y., Dai, G., Huang, H., and Jin, Q. (2015). Genetic Determinants Involved in p-Aminosalicylic Acid Resistance in Clinical Isolates from Tuberculosis Patients in Northern China from 2006 to 2012. Antimicrob. Agents Chemother. 59, 1320–1324. doi: 10.1128/AAC.03695-14
Zhang, Y., Shi, W., Zhang, W., and Mitchison, D. (2014). Mechanisms of pyrazinamide action and resistance. Microbiol. Spectr. 2:MGM2–0023–2013. doi: 10.1128/microbiolspec.MGM2-0023-2013
Zhao, F., Wang, X.-D., Erber, L. N., Luo, M., Guo, A.-Z., Yang, S.-S., et al. (2014). Binding pocket alterations in dihydrofolate synthase confer resistance to para-aminosalicylic acid in clinical isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 58, 1479–1487. doi: 10.1128/AAC.01775-13
Keywords: Mycobacterium, drug resistance, resistance, tuberculosis, Nontuberculous mycobacteria
Citation: Nasiri MJ, Haeili M, Ghazi M, Goudarzi H, Pormohammad A, Imani Fooladi AA and Feizabadi MM (2017) New Insights in to the Intrinsic and Acquired Drug Resistance Mechanisms in Mycobacteria. Front. Microbiol. 8:681. doi: 10.3389/fmicb.2017.00681
Received: 27 December 2016; Accepted: 04 April 2017;
Published: 25 April 2017.
Edited by:
Miguel Cacho Teixeira, Universidade de Lisboa, PortugalReviewed by:
Shashank Gupta, Brown University, USAParas Jain, Albert Einstein College of Medicine, USA
Copyright © 2017 Nasiri, Haeili, Ghazi, Goudarzi, Pormohammad, Imani Fooladi and Feizabadi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohammad J. Nasiri, mj.nasiri@hotmail.com
Hossein Goudarzi, hgoudarzi@sbmu.ac.ir
 Mohammad J. Nasiri
Mohammad J. Nasiri Mehri Haeili2
Mehri Haeili2