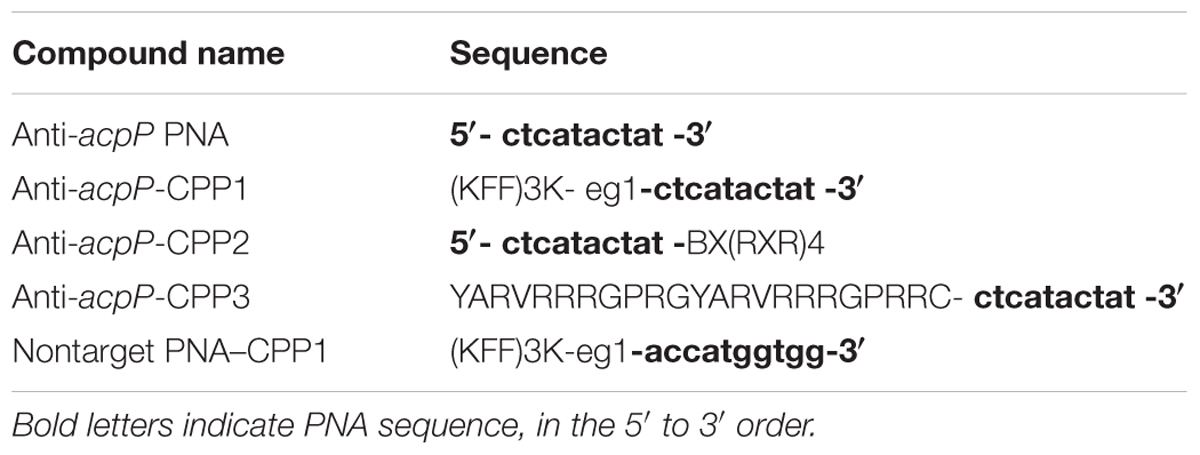
- 1Department of Plant Pathology and Ecology, The Connecticut Agricultural Experiment Station, New Haven, CT, USA
- 2Department of Plant, Soil, and Microbial Sciences, Michigan State University, East Lansing, MI, USA
- 3Department of Biological Sciences, University of Wisconsin–Milwaukee, Milwaukee, WI, USA
- 4Department of Plant Biology, Michigan State University, East Lansing, MI, USA
Erwinia amylovora is a Gram-negative bacterial plant pathogen in the family Enterobacteriaceae and is the causal agent of fire blight, a devastating disease of apple and pear. Fire blight is traditionally managed by the application of the antibiotic streptomycin during bloom, but this strategy has been challenged by the development and spread of streptomycin resistance. Thus, there is an urgent need for effective, specific, and sustainable control alternatives for fire blight. Antisense antimicrobials are oligomers of nucleic acid homologs with antisense sequence of essential genes in bacteria. The binding of these molecules to the mRNA of essential genes can result in translational repression and antimicrobial effect. Here, we explored the possibility of developing antisense antimicrobials against E. amylovora and using these compounds in fire blight control. We determined that a 10-nucleotide oligomer of peptide nucleic acid (PNA) targeting the start codon region of an essential gene acpP is able to cause complete growth inhibition of E. amylovora. We found that conjugation of cell penetrating peptide (CPP) to PNA is essential for the antimicrobial effect, with CPP1 [(KFF)3K] being the most effective against E. amylovora. The minimal inhibitory concentration (MIC) of anti-acpP-CPP1 (2.5 μM) is comparable to the MIC of streptomycin (2 μM). Examination of the antimicrobial mechanisms demonstrated that anti-acpP-CPP1 caused dose-dependent reduction of acpP mRNA in E. amylovora upon treatment and resulted in cell death (bactericidal effect). Anti-acpP-CPP1 (100 μM) is able to effectively limit the pathogen growth on stigmas of apple flowers, although less effective than streptomycin. Finally, unlike streptomycin that does not display any specificity in inhibiting pathogen growth, anti-acpP-CPP1 has more specific antimicrobial effect against E. amylovora. In summary, we demonstrated that PNA–CPP can cause an effective, specific antimicrobial effect against E. amylovora and may provide the basis for a novel approach for fire blight control.
Introduction
Fire blight, caused by the bacterial pathogen Erwinia amylovora, is one of the most serious diseases of apple and pear in the United States and worldwide. Fire blight infection can occur in flowers, leaves, shoots, and fruits, resulting in yield reduction; the pathogen also can spread systemically through trees to the rootstock, ultimately resulting in tree death (Norelli et al., 2003; van der Zwet et al., 2012). Annual losses to fire blight and costs of control in the US are estimated at over $100 million (Norelli et al., 2003).
The management of fire blight is challenged due to the availability of limited control options. As the pathogens enter plants through the natural opening of the flowers, antibiotic spray applications during bloom are the most effective and widely used control method for fire blight in the US (Norelli et al., 2003; Sundin et al., 2016). Streptomycin is the most effective antibiotic targeting E. amylovora, and has been used for fire blight management in the US since the 1950s (Goodman, 1959). The intensive, long-term use of streptomycin, however, has resulted in the development of streptomycin resistance in the E. amylovora population. Since its original report in California in 1972 (Miller and Schroth, 1972), streptomycin resistance has been observed in most major apple-producing regions in the United States (Chiou and Jones, 1995; Evans, 2007; Russo et al., 2008; McGhee et al., 2011). In addition, agricultural application of streptomycin also raises significant concerns for the potential selection of antibiotic resistant bacteria in the environment, and the potential impact to human health (Sundin and Bender, 1996). Besides antibiotics, copper bactericides and other biological control products are also used for fire blight management, however the use of these materials is limited by their inconsistent control efficacy and copper use can also result in phytotoxicity (Tsiantos et al., 2003; Sundin et al., 2009; Johnson and Temple, 2013). Because of these reasons, developing effective control alternatives for fire blight has become an urgent need for sustainable apple and pear production in the US (Khokhani et al., 2013; Yang et al., 2014; Sundin et al., 2016).
RNA silencing is the translational repression of a mRNA caused by the binding of an antisense RNA (Nakashima et al., 2012). In principle, the translation of any mRNA could be silenced by an antisense RNA with sequences complementary to the translational initiation sequences of the target mRNA (Good and Nielsen, 1998; Bennett and Swayze, 2010). The discovery of RNA silencing provides a powerful tool to artificially modulate gene expression. One application of RNA silencing is the synthesis of artificial RNA homologs to silence the expression of essential genes in microbes, and use of these compounds as antimicrobials (Rasmussen et al., 2007).
Antisense antimicrobials are short oligomers of nucleic acid homologs with antisense sequences to the translational initiation sites of essential genes of bacteria (Rasmussen et al., 2007; Bai and Luo, 2012). The binding of the antisense compounds to the translational initiation sites can lead to the silencing of these essential genes and subsequent growth inhibition (Bai and Luo, 2012). Compared to traditional antibiotics, antisense antimicrobials have many unique advantages including: (1) Unlike antibiotics that usually target a universal cellular process and kill bacteria with little selection, antisense antimicrobials can target a specific DNA sequence of the pathogen without affecting the survival of other potentially beneficial, environmental bacteria; (2) Unlike antibiotics that are typically limited to targeting a single cellular process, antisense antimicrobials can target any essential genes through sequence complementation, thus significantly enlarging the target selection; (3) In the case of a pathogen developing resistance to antisense antimicrobials through mutations, the resistance could be overcome by designing new antisense sequences against the mutated sequence.
The fact that DNA and RNA are unstable under UV and can be easily degraded by enzymes makes them undesirable materials for antisense antimicrobials. Improvements designed to modify or replace the sugar-phosphate backbone of DNA/RNA has resulted in nucleic acid homologs with significantly enhanced stability (Bai and Luo, 2012). These nucleic acid homologs include peptide nucleic acids (PNAs), phosphorodiamidate morpholino oligomers (PMOs), and phosphorothioate oligonucleotides (PS-ODNs) (Bai and Luo, 2012). Among them, PNAs have shown promising antimicrobial effects against some animal pathogenic bacterial species (Good and Nielsen, 1998; Nekhotiaeva et al., 2004; Kulyte et al., 2005; Kurupati et al., 2007; Hatamoto et al., 2010). In PNA, the sugar–phosphate backbone of DNA/RNA was replaced with a pseudopeptide backbone, while nearly identical geometry and spacing of the bases was retained (Bai and Luo, 2012). This modification significantly enhances the stability, as the PNA molecules are stable when exposed to bacterial cytoplasmic extracts, human blood serum, and enzymes that degrade DNA and peptide (DNase, protease, et al.) (Demidov et al., 1994; Bai and Luo, 2012). In addition, it also increases the PNA–RNA binding affinity, as RNA is negatively charged but PNA is electrically neutral (Bai and Luo, 2012). However, since PNA oligomers are large molecules, they may not enter the cellular membrane as readily. Thus, the delivery of PNA oligomers into bacterial cells often requires external assistance. Cell penetrating peptides (CPPs) are short peptides of less than 30 amino acids that can penetrate cell membranes and deliver covalently conjugated cargoes into cells (Abes et al., 2007; Bai and Luo, 2012). Conjugation of CPPs with PNAs has been demonstrated to significantly increase PNA entry into bacterial cells (Zatsepin et al., 2005; Rasmussen et al., 2007). Two features of CPPs that are required for cell penetrating ability include amphipathicity and positive charge (Wu et al., 2007; Bai and Luo, 2012). These characteristics are acquired by synthesizing CPPs with an alternation of cationic amino acid residues and nonpolar residues. No obvious toxicity of CPP to animal cells has been observed and CPP is considered safe to use as drug delivery in human medicine (Dinca et al., 2016).
PNA–CPPs have been successfully used as antisense antimicrobials in both in vitro and in vivo trials against animal pathogenic bacteria. For example, CPP-conjugated PNAs targeting essential genes such as acpP, inhA, gyrA, ompA, 16 s rRNA, and adk have shown significant growth inhibition effect against a number of bacteria including Escherichia coli (Good and Nielsen, 1998; Tan et al., 2005), Pseudomonas aeruginosa (Ghosal and Nielsen, 2012), Staphylococcus aureus (Hatamoto et al., 2010), Mycobacterium smegmatis (Kulyte et al., 2005), and Klebsiella pneumoniae (Kurupati et al., 2007). Previous research suggests that the start codon and Shine-Dalgarno (SD) region of the mRNA are the most sensitive sequences for inhibition caused by antisense antimicrobials (Dryselius et al., 2003).
Although PNA–CPPs have shown some promising applications in controlling bacterial infections in animal models, to our knowledge, no research has explored the use of PNA–CPP in controlling plant diseases. Regulatory small RNAs (sRNAs) play important roles in modulating gene expressions in bacteria (Gottesman and Storz, 2010). We previously have identified regulatory small RNAs in the fire blight pathogen E. amylovora and have described the roles of these sRNAs in regulating various virulence and cellular functions (Zeng et al., 2013; Zeng and Sundin, 2014). These findings suggest that RNA silencing is a naturally occurring process in E. amylovora and that it is possible that the expression of a given gene could also be modulated by artificially synthesized RNA homologs. In this research, we explored the proof-of-concept of using PNA–CPP that targets an essential gene acpP in E. amylovora as an antimicrobial, and using this compound to control fire blight. We determined the application conditions (selection of essential genes and CPP), mechanisms of antimicrobial activity, and documented effectiveness of PNA–CPP in limiting pathogen growth on detached apple flowers.
Materials and Methods
Bacterial Strains, Culture Conditions, and PNA–CPP Synthesis
The highly virulent strain E. amylovora Ea110, which was isolated from an apple orchard in Michigan (Zhao et al., 2005), was used in this study. Bacterial strains were stored at -80°C in 15% glycerol and cultured in Luria–Bertani (LB) medium at 28°C. PNA–CPP was synthesized using Bts oligomerization method by Panagene Inc (Daejeon, Korea). Streptomycin and PNA–CPP were implemented at rates indicated in each assay.
Measurement of Bacterial Growth Inhibition
Erwinia amylovora Ea110 was cultured in LB broth overnight and then cell concentrations were adjusted to 5 × 105 CFU/ml in LB broth. A total of 80 μl of the bacterial suspension in LB was added into each well of a 96-well plate. Lyophilized PNA–CPP was resuspended in water to a stock concentration of 100 μM and was serial diluted. Twenty microliter of the diluted PNA–CPP solution was added into each well that contained 80 μl of the bacterial suspension in LB to make the final concentrations indicated in each assay. Water and streptomycin were added to the wells as negative and positive controls. The plate was incubated at 28°C with orbital shaking in a BioTek Synergy H1 microplate reader (BioTek, Winooski, VT, USA) for 20 h. During the incubation, the OD 600 of each well was measured every 10 min. The lowest concentration of a PNA–CPP that prevented growth after 20 h represented the MIC. Three replicates were included in each testing, and the experiment was repeated twice.
Viability Test
Overnight cultures of E. amylovora Ea110 were adjusted to 5 × 105 CFU / ml in LB broth. Forty microliters of anti-acpP-CPP1 (100 μM), streptomycin (100 μM), or H2O were added into 460 μl of the LB broth containing E. amylovora cells to reach a final concentration of 8 μM of the compounds mentioned above. The cultures were incubated at 28°C with 200 rpm of continuous shaking. Samples were taken at 0, 1, 2, 3, 4, 5, and 6 hours post inoculation. Bacterial cells from each sample were collected by centrifugation (6500 rpm for 8 min), washed with sterile water to remove the residual compounds, diluted 102 to 105-fold, and plated on LB agar plates. Colonies formed after 48 hour-incubation at 28°C are counted and original cell concentration was calculated. Five replicates were included in each assay and the experiment was repeated twice with similar results observed.
Fluorescence Microscopy
Bacterial cells were washed with sterile water to remove residual medium from the culture. LIVE/DEAD BacLight bacterial viability kit (Molecular Probes, Eugene, OR, USA) components A and B were added to the cells following the manufacturer’s instructions. Green fluorescence and red fluorescence of cells stained were observed using a Zeiss Scope A1 fluorescence microscope equipped with a FITC filter and a DAPI filter (Oberkochen, Germany). Images were acquired with a Spot RT3 camera using the Spot Advanced software (Sterling Heights, MI, USA). Statistical analysis was performed using ANOVA with α = 0.05. The level of significance labeled in figure (denoted by different letters) was calculated using LSD (α = 0.05). ANOVA and LSD analyses were conducted in R ver. 3.3.1 (R Core Team, 2015).
RNA Isolation
Erwinia amylovora Ea110 was inoculated into LB broth at the final concentration of 5 × 105 CFU/ml, and was treated with various concentrations of PNA–CPP, water, or streptomycin. The inoculated cells were incubated at 28°C with orbital shaking, and were collected for RNA isolation at 15.5 h post inoculation. Total bacterial RNA was isolated using the RNeasy Protect Bacteria Mini Kit (Qiagen, Valencia, CA, USA) following the manufacturer’s instructions. The quality and quantity of RNA was measured with a Nanodrop 2000c (Thermo Scientific, Wilmington, DE, USA).
Northern Blot
The probe for acpP detection in Northern blots was synthesized from polymerase chain reaction (PCR) using acpP primers (forward primer 5′-TGG GCG TTA AGC AGG AAG AAG-3′ and reverse primer 5′-TAC GCC TGG TGA CCA TTG AT-3′). The PCR product was purified using a QIAquick PCR Purification kit (Qiagen) and labeled using a Biotin DecaLabel DNA Labeling Kit (Thermo Fisher Scientific, Grand Island, NY, USA) as per instruction of kit manual. Thirty micrograms of total RNA from different treatments were loaded to a formaldehyde denaturing gel in MOPS buffer. 16s rRNA was quantified using a transluminator as internal control of RNA quantity. BrightStar®-Plus positively charged nylon membranes (Thermo Fisher Scientific) were used for transfer of RNA from gel to membrane. Transfer, pre-hybridization, and hybridization were performed following the manufacturer’s instructions of the Northern Max kit (Thermo Fisher Scientific). The biotin signal was detected using a Biotin Chromogenic Detection Kit (Thermo Fisher Scientific). The band intensity was quantified by ImageJ (Schneider et al., 2012).
Detached Apple Flower Assay
Freshly opened flowers were collected from apple trees Malus × domestica ‘McIntosh’ from the Connecticut Agricultural Experiment Station Lockwood farm in Hamden, CT, USA) in May 2016. The pedicel of the flowers was detached from the flower cluster, and each flower was placed in a 7 ml plastic tube (containing 10% sucrose solution) through a hole created in the plastic cap. One microliter of E. amylovora (Ea110) at the concentration of 107 CFU/ml was inoculated onto the five stigmas of each flower (approximately 0.2 μl per stigma). Twenty microliters of the PNA–CPP at different concentrations were evenly applied onto the stigmas of each flower 2 h before and 19 h after the inoculation. Water and streptomycin (100 μM) were used as negative and positive controls. In the PNA–bacteria mix treatment, PNA at the concentration 20 μM was mixed with an equal volume of E. amylovora cells at the concentration of 107 CFU/ml in a microcentrifuge tube and incubated for 30 min at 22–25°C. Two microliters of the mixture were applied evenly onto the five stigmas of the flowers. The pathogen population on the apple stigmas was quantified at 42 h post inoculation, by dissecting the stigmas from the flowers, and suspending them in 1 ml of 0.5x PBS. A Taqman probe realtime PCR assay was used to quantify the pathogen amount. Primers used in this assay are: amsk120F: 5′-CAT GCA ATT TCC AGT TTC CT-3′; amsk120R: 5′-GCA TGA CGG TTA ACC AAA TC-3′, amsk120Probe: 5′-TGC GTG ACC TGA TTC AGC ACA A-3′. The reaction was performed on a Biorad CFX96 Realtime PCR machine. The cell concentrations were calculated by f (Cq) = –3.47 Cq + 45.513. A standard curve was generated using five predetermined concentrations of Ea110 (107, 106, 105, 104, 103 CFU/ml) cells. Six flowers were used in each treatment. The experiment was repeated twice with similar results observed, and results from one representative experiment are displayed. The box plot was generated by R ver. 3.3.1 (R Core Team, 2015). Statistical analysis was performed using ANOVA with α = 0.05. The level of significance labeled in figure (denoted by different letters) was calculated using LSD (α = 0.05). ANOVA and LSD analyses were conducted in R ver. 3.3.1 (R Core Team, 2015).
Collection of Apple Flower-associated Bacteria
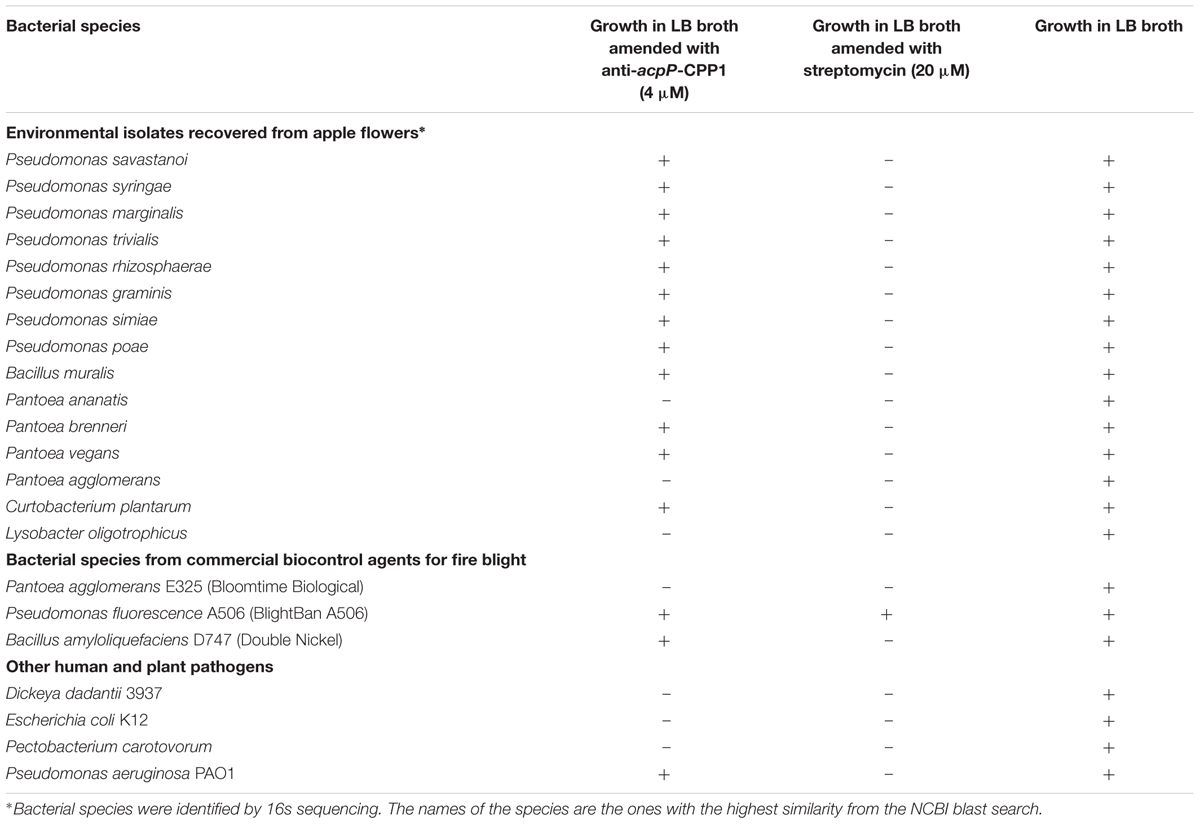
Fifty flowers were collected from apple trees Malus × domestica ‘McIntosh’ from the Hamden, CT Lockwood farm in May 2016. The stigmas from all 50 flowers were removed, placed into 3 ml of 0.5x PBS, vortexed for 20 s, and sonicated for 5 min in a water bath sonicator to release the stigma associated bacteria. PBS buffer containing bacteria cells was serially diluted (10x, 100x, and 1000x) and plated on LB agar medium and incubated at 28°C for 48 h. On the basis of morphological colony characteristics, 200 bacterial colonies were shortlisted, sub-cultured, and identified using16s rRNA sequencing using the forward primer 63f (5′-CAG GCC TAA CAC ATG CAA GTC-3′) and reverse primer 1387r (5′-GGG CGG WGT GTA CAA GGC-3′). Fifteen different species were selected from the 200 bacterial cultures for testing against anti-acpP-CPP1 and streptomycin. Biological control agents Pseudomonas fluorescence A506, Bacillus amyloliquefaciens D747 and Pantoea agglomerans E325 were isolated from the original products on LB agar plate.
Results
PNAs with Antisense Sequence to the Start Codon Region of acpP Caused Growth Inhibition of E. amylovora When Conjugated with CPP
To explore the possibility of using PNAs as antimicrobials against the fire blight pathogen E. amylovora, we first synthesized a 10-nucleotide PNA oligomer with antisense sequence to the start codon region (–5 to +5) of a previously discovered essential gene acpP (encoding an acyl carrier protein), anti-acpP PNA (Table 1) (Zhang and Cronan, 1998). The effect of the anti-acpP PNA on the growth of E. amylovora was evaluated under an in vitro condition. Compared to the water-treated cells, cells treated with anti-acpP PNA did not display any growth inhibition at either concentration tested (Figure 1A).
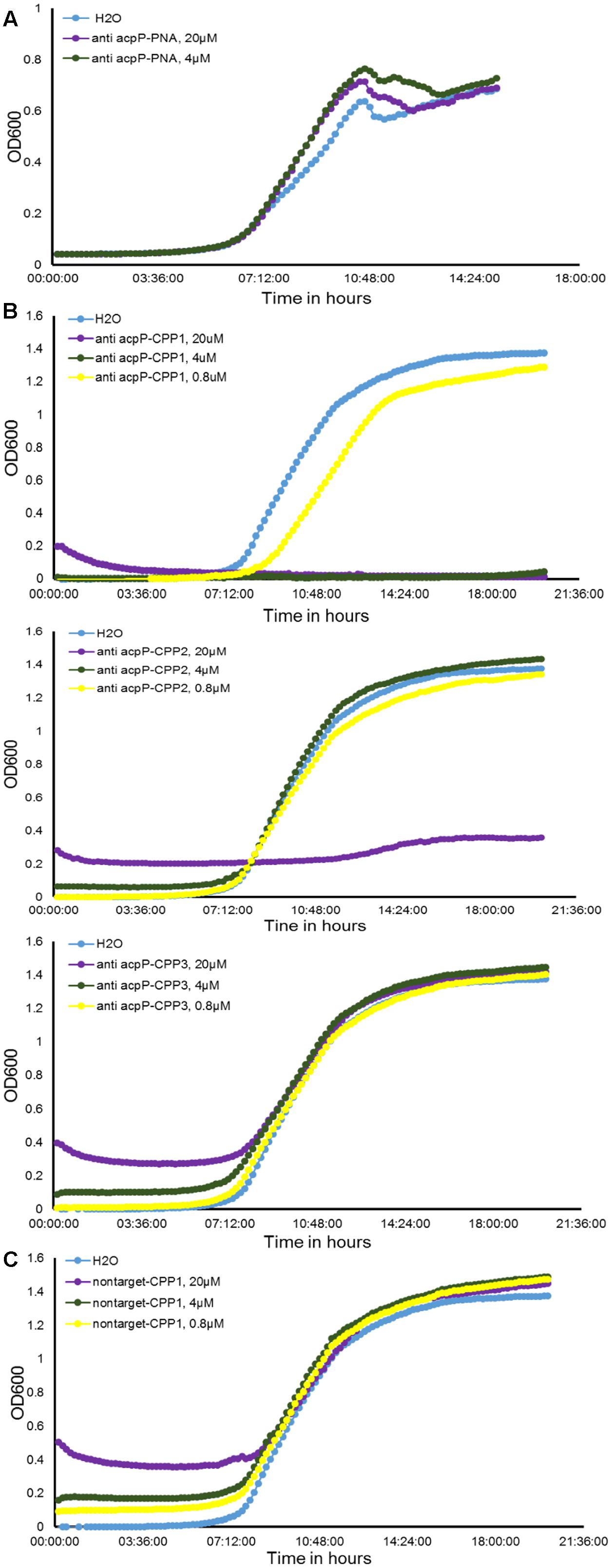
FIGURE 1. Effect of peptide nucleic acid–cell penetrating peptide (PNA–CPP) on the growth of E. amylovora Ea110 in LB broth. (A) Erwinia amylovora treated with anti-acpP PNA. (B) E. amylovora treated with CPP conjugated PNA: anti-acpP-CPP1, anti-acpP-CPP2, and anti-acpP-CPP3. (C) E. amylovora treated with CPP1 conjugated PNA containing random sequence (non-target-CPP1). E. amylovora Ea110 cells were maintained in 100 μl of LB broth supplemented with PNA–CPPs at different concentrations in a 96-well plate. The 96-well plate was maintained at 28°C with orbital shaking in a plate reader. Turbidity (OD600) was measured every 10 min for a period of 20 h. Three replicates were included in the assay and the experiment was repeated twice. Results from one experiment are displayed in this figure.
As the ineffectiveness of the anti-acpP PNA on E. amylovora growth could be due to the inefficient entry of PNA into the cytoplasm, we determined whether conjugation of the anti-acpP PNA with CPP would result in enhanced antimicrobial effect. Three formulations of CPP (CPP1, CPP2, and CPP3) that were demonstrated to have promising delivery efficiency of PNAs in animal pathogenic bacteria were then individually conjugated to the anti-acpP PNA, resulting in anti-acpP-CPP1, anti-acpP-CPP2, and anti-acpP-CPP3 (Table 1). The effect of the CPP-conjugated PNAs on E. amylovora growth was tested under the same conditions. Compared to the water control, addition of anti-acpP-CPP1 or anti-acpP-CPP2 both resulted in complete growth inhibition of E. amylovora at the 20 μM concentration, whereas addition of anti-acpP-CPP3 did not cause any growth inhibition at any concentrations (Figure 1B). The growth inhibition caused by anti-acpP-CPP1 was more potent than the inhibition caused by anti-acpP-CPP2, suggesting that CPP1 [(KFF)3K] was the most efficient CPP in delivering PNA into E. amylovora. To determine if the growth inhibition caused by PNA–CPP was through specific targeting of acpP, a PNA with random nucleotide sequence in conjugation with CPP1 (nontarget PNA–CPP1) was also tested (Table 1). The nontarget-CPP1 did not cause any inhibition of E. amylovora growth (Figure 1C).
We further determined that the minimal inhibitory concentration (MIC) of anti-acpP-CPP1 and streptomycin was 2.5 and 2 μM, respectively (Figure 2A). In addition, we showed that the growth inhibition caused by anti-acpP-CPP1 is positively correlated with the concentrations of anti-acpP-CPP1, in a linear manner (Figure 2B). Taken together, these results suggest that CPP-conjugated PNA with antisense sequence targeting the start codon of acpP is able to cause potent growth inhibition of E. amylovora under in vitro conditions. This inhibition was caused by the antisense sequence of acpP on PNA. CPP was essential in delivering PNA into E. amylovora and causing the antimicrobial effect. Among the three CPPs previously used in animal pathogens, CPP1 was the most efficient for PNA delivery into E. amylovora.
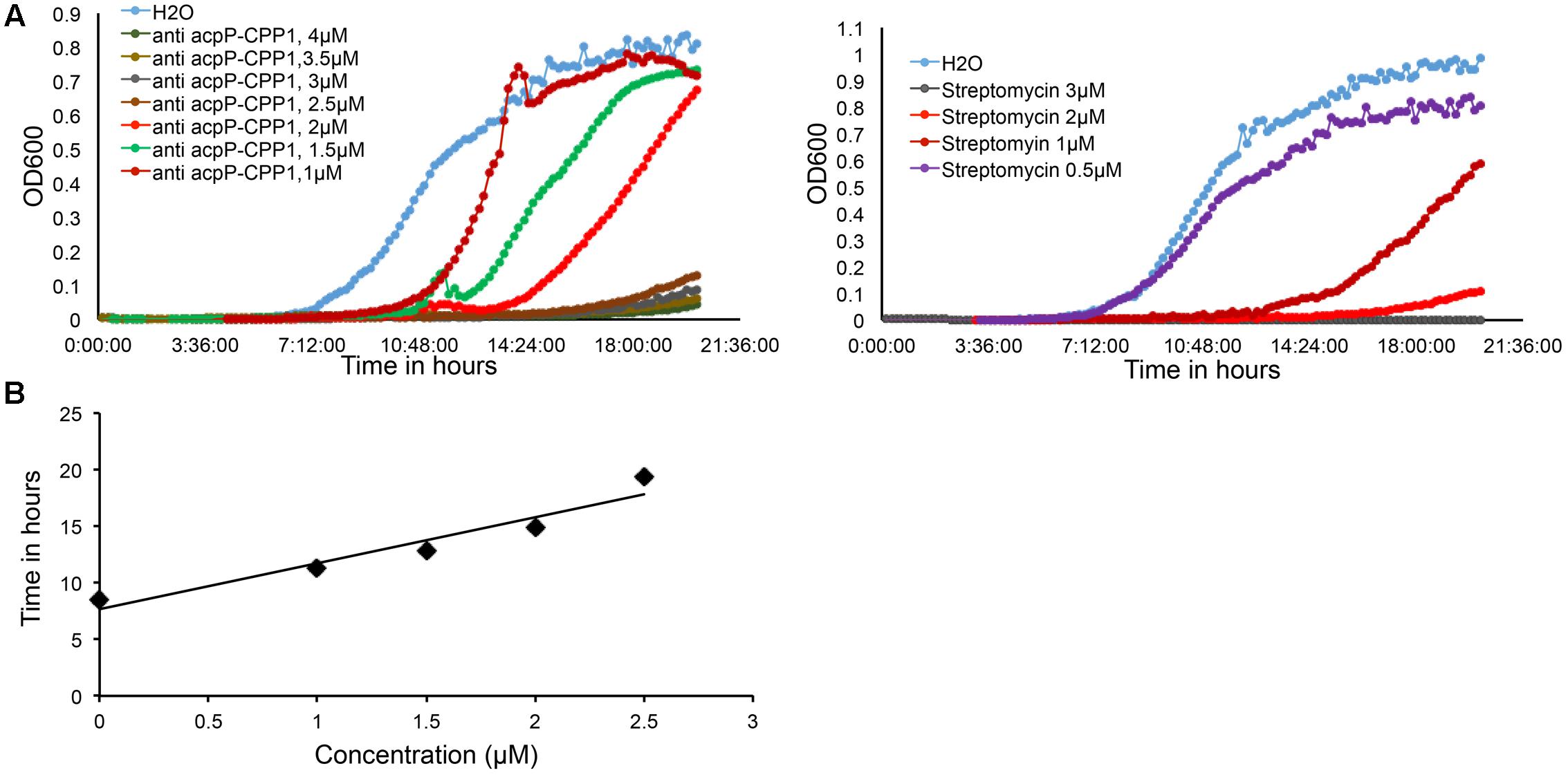
FIGURE 2. Dose dependent growth inhibition of E. amylovora Ea110 caused by anti-acpP-CPP1. (A) E. amylovora Ea110 growth in LB broth in the presence of different concentrations of anti-acpP-CPP1 and streptomycin. (B) Time to reach OD600 = 0.1 of Ea110 growth at different concentrations of anti-acpP-CPP1.
Erwinia amylovora Cells Treated With Anti-acpP-CPP1 Showed Reduced acpP mRNA Levels
After entering the bacterial cytoplasm, PNA–CPP is believed to bind to the start codon region of the target mRNA and block its translation (Good and Nielsen, 1998; Wang and Xu, 2004). However, whether the PNA–mRNA binding would also cause the degradation of the target mRNA is not clear. We compared the levels of acpP mRNA in E. amylovora cells treated with water, anti-acpP-CPP1 at two sub-lethal concentrations (1 and 2 μM), and streptomycin at sub-lethal concentrations (1 and 3 μM) using Northern blot. Compared to the water-treated cells, cells treated with anti-acpP-CPP1 showed a visual reduction in the level of acpP mRNA. This reduction is positively correlated with anti-acpP-CPP1 concentrations (Figure 3). In contrast, streptomycin treatment did not affect the acpP mRNA abundance. These observations suggest that anti-acpP-CPP1 binding to acpP mRNA caused the degradation of acpP mRNA. It also indicates that the growth inhibition caused by anti-acpP-CPP1 is through a gene specific interaction with acpP mRNA.
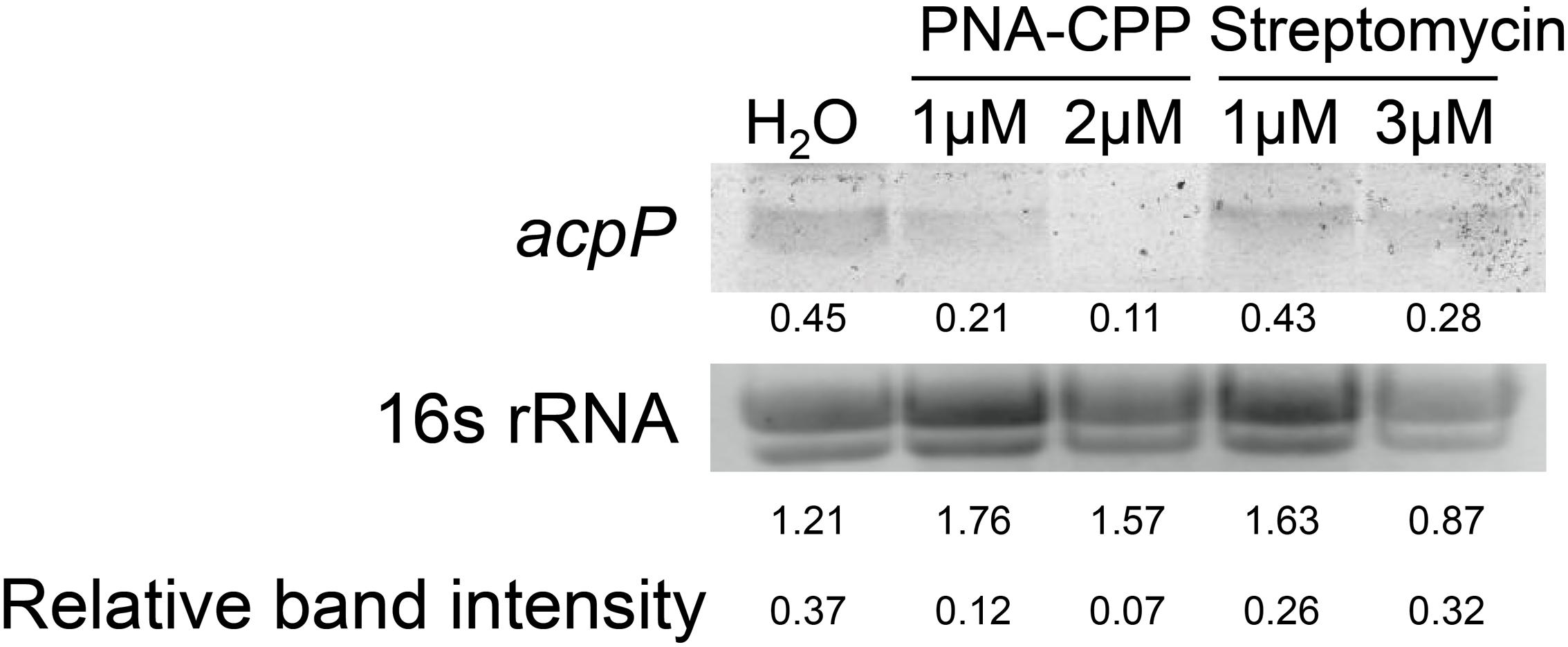
FIGURE 3. Northern blot detection of the expression levels of acpP in E. amylovora Ea110 treated with anti-acpP-CPP1 and streptomycin. E. amylovora was cultured in LB broth supplemented with water, anti-acpP-CPP1 (at final concentrations 1 and 2 μM), and streptomycin (at final concentrations 1 and 3 μM) for 18 h. Total RNA was quantified by Nanodrop. Thirty micrograms of RNA were loaded to a denaturing gel and analyzed for the acpP mRNA amount using a Northern blot assay. The image of 16s rRNA was taken using a transluminator as internal control of RNA quantity between samples. The band intensity was quantified by ImageJ. Relative band intensity was calculated by deviding the intensity of the acpP band with the intensity of the 16S band.
The Growth Inhibition of E. amylovora by Anti-acpP-CPP1 Is Bactericidal
To determine whether the growth inhibition of E. amylovora by anti-acpP-CPP1 was bactericidal or bacteriostatic, we tested the viability of E. amylovora cells in LB broth at different time points after treatment of anti-acpP-CPP1 (8 μM), streptomycin (8 μM), or water, using a serial dilution plating method. In the water-treated sample, we observed an exponential increase in the amount of viable cells after inoculation (Figure 4A). Following treatment with anti-acpP-CPP1, the viable cell count remained the same during the first 3 h, displaying a bacteriostatic effect (Figure 4A). After 3 h, the viability of cells treated with anti-acpP-CPP1 drastically declined, which suggested that anti-acpP-CPP1 caused a bactericidal effect during this period. After 6 h of treatment with anti-acpP-CPP1, the number of viable cells was reduced by more than 1000-fold (log10 CFU/ ml reduced from 5.3 at 0 h to 1.8 at 6 h, Figure 4A), at a slightly faster rate than streptomycin. These observations suggest that the growth inhibition of E. amylovora caused by anti-acpP-CPP1 is bactericidal, similar to the traditional antibiotic streptomycin.
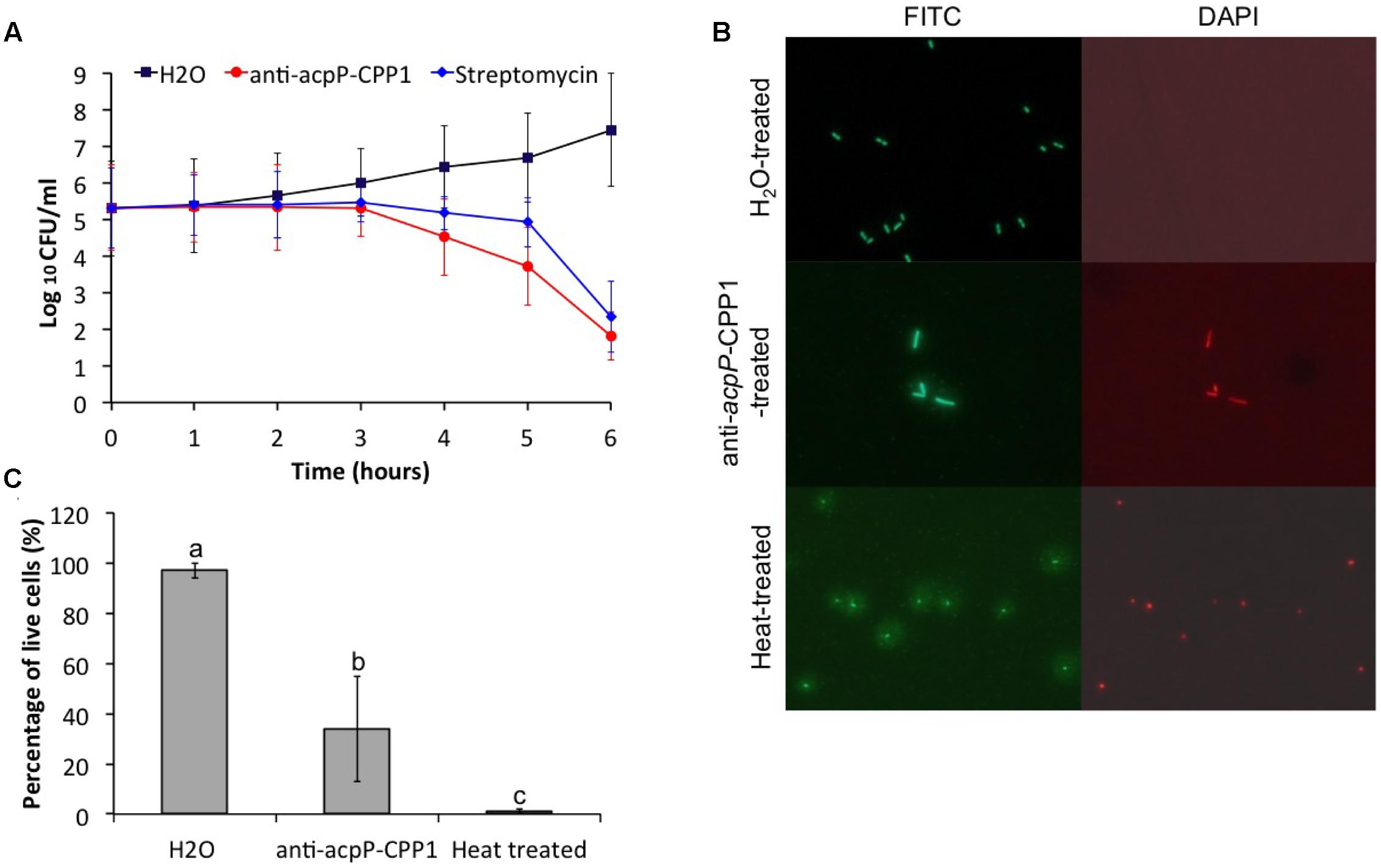
FIGURE 4. Effect of anti-acpP-CPP1 on the viability of E. amylovora Ea110. (A) Number of viable E. amylovora cells recovered on LB plates from the anti-acpP-CPP1 and streptomycin treatment. (B) E. amylovora stained with fluorescent dye SYTO 9 and propidium after 6 hrs of treatment with anti-acpP-CPP1 and water. Heat-treated cells (80°C for 10 mins) were included as a control. (C) Percentage of live cells of the total E. amylovora cells after 6 hrs of treatment with water, anti-acpP-CPP1, and treated with heat (80°C for 10 mins). E. amylvoora Ea110 (5 × 105) were inoculated into LB broth supplemented with anti-acpP-CPP1 or water, and incubated at 28°C with shaking. The viability of E. amylovora cells was tested at different time points by plating on LB agar plates and by fluorescence staining and microscopy. Statistical analysis was performed using ANOVA with α = 0.05. The level of significance was calculated using LSD (α = 0.05).
The mechanism of the growth inhibition caused by anti-acpP-CPP1 was also studied using a bacterial viability kit and fluorescence microscopy. At 6 h after inoculation, cells treated with water were green fluorescent but not red fluorescent when stained with a mixture of fluorescent dye SYTO 9 and propidium iodide, suggesting that these cells were viable (Figure 4B). However, a large number of cells treated with anti-acpP-CPP1 at a concentration above the MIC (8 μM) were both green fluorescent and red fluorescent, suggesting that many of these cells lost viability (Figures 4B,C). Interestingly, cells treated with anti-acpP-CPP1 showed an elongated cell shape compared to the water-treated cells (Figure 4B). No cell lysis (as shown in the heat-treated cells in Figure 4B) was observed following treatment with anti-acpP-CPP1, which suggests that the bactericidal effect may not be caused by physical disintegration of the cell structure (Figure 4B). The fluorescence microscopy observation is consistent with the plating method, together suggesting that the growth inhibition of E. amylovora caused by anti-acpP-CPP1 is bactericidal.
Anti-acpP-CPP1 was Able to Inhibit Pathogen Growth on Apple Stigmas
Following demonstration that anti-acpP-CPP1 has potent growth inhibition of E. amylovora under in vitro conditions, we further evaluated the effectiveness of anti-acpP-CPP1 in inhibiting E. amylovora growth on apple stigmas using a detached flower assay. Compared to the water-treated control, stigmas treated with 100 μM anti-acpP-CPP1 showed a significant reduction of E. amylovora population (>100-fold reduction, Figure 5). However, this reduction was less potent in comparison to the reduction by streptomycin (Figure 5). At lower concentrations, anti-acpP-CPP1 treatment did not result in significant reduction of the pathogen population when directly applied to the apple stigmas (Figure 5). However, premixing anti-acpP-CPP1 (20 μM) with E. amylovora cells for 30 min before applying the cell-compound mixture onto the apple stigmas resulted in excellent reductions of pathogen populations on apple stigmas (Figure 5). The results above suggest that anti-acpP-CPP1 is able to inhibit pathogen growth on apple stigmas when applied at 100 μM. It also suggests that the efficacy of anti-acpP-CPP1 may be improved if optimal cell-compound contact can be established.
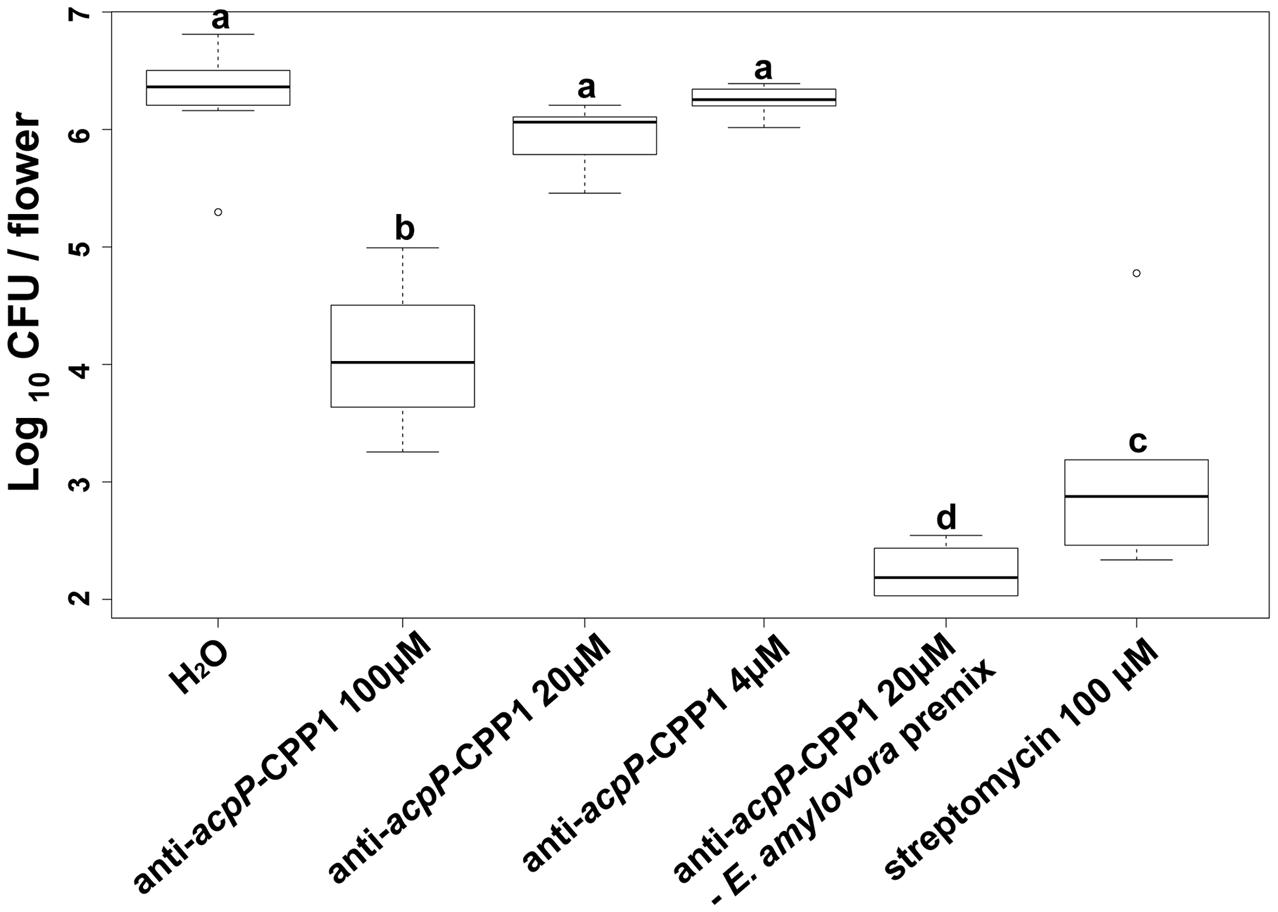
FIGURE 5. Amount of E. amylovora Ea110 cells on detached apple stigmas after treatment with anti-acpP-CPP1 and streptomycin. E. amylovora Ea110 cells (107 CFU/ml) were inoculated on the stigma of freshly opened ‘McIntosh’ flowers (1 μl per flower by a pipette). Twenty microliters of anti-acpP-CPP1 (100, 20, and 4 μM) were evenly applied onto the stigmas of each flower 2 h before and 19 h after the inoculation. Water and streptomycin (100 μM) were used as negative and positive controls. In the PNA–bacteria mix treatment, anti-acpP-CPP1 (20 μM) was mixed with an equal volume of E. amylovora cells (107 CFU/ml) in a microcentrifuge tube and incubated for 30 min at room temperature (22–25°C). Two microliters of the mixture were applied evenly onto the five stigmas of the flowers. All flowers were incubated at room temperature and the pathogen population on the apple stigmas was quantified at 42 h post inoculation, using a Taqman probe realtime PCR assay. Statistical analysis was performed using ANOVA with α = 0.05. The level of significance (denoted by different letters) was calculated using LSD (α = 0.05).
Effect of Anti-acpP-CPP1 on Microbiome Associated with Apple Flowers
Compared to the traditional antibiotics that kill microbes with little selection, antisense antimicrobials inhibit bacterial growth through a sequence specific manner. To determine whether anti-acpP-CPP1 affects the growth of environmental microbes associated with apple flowers, bacteria were first cultured from freshly collected ‘McIntosh’ apple flowers. Two hundred bacterial colonies were subcultured, and the identities of these colonies were determined by 16s sequencing. The sequencing and Blast analyses suggest that these bacteria belong to 15 different species (Table 2). Representative strains from each species were tested for their susceptibility to anti-acpP-CPP1 and streptomycin at concentrations above the MICs for E. amylovora (4 and 20 μM, respectively). Our results suggest that anti-acpP-CPP1 did not affect the growth of most species (12/15), such as Pseudomonas ssp., Bacillus muralis, Curtobacterium plantarum, and multiple species in the Pantoea genus. However, three species, P. ananatis, P. agglomerans, and Lysobacter oligotrophicus displayed susceptibility to anti-acpP-CPP1 similar to E. amylovora. Comparison of the start codon sequences of acpP in species with genomes available in NCBI revealed that many species contain sequence discrepancies with E. amylovora in this region, although some other species contain similar sequences to E. amylovora (Figure 6). P. agglomerans that has an identical sequence as E. amylovora was also susceptible to anti-acpP-CPP1. In addition to the bacterial species isolated from the apple flower, we also tested the susceptibility of three microorganisms from fire blight biological control products to anti-acpP-CPP1. Similar to the environmental isolates, Pseudomonas fluorescens A506 and Bacillus amyloliquefaciens D747 (active ingredients of BlightBan A506 and Double Nickel) were not affected by anti-acpP-CPP1 whereas P. agglomerans E325 (active ingredient of Bloomtime Biological) was susceptible (Table 2). All strains tested are susceptible to streptomycin with the exception of P. fluorescens A506 (Table 2). Finally, some other plant and animal pathogens, such as Dickeya dadantii and Pectobacterium carotovorum are susceptible to anti-acpP-CPP1 (Table 2) and further examination of their sequences suggests they have similar sequences targeted by anti-acpP-CPP1 as E. amylovora.
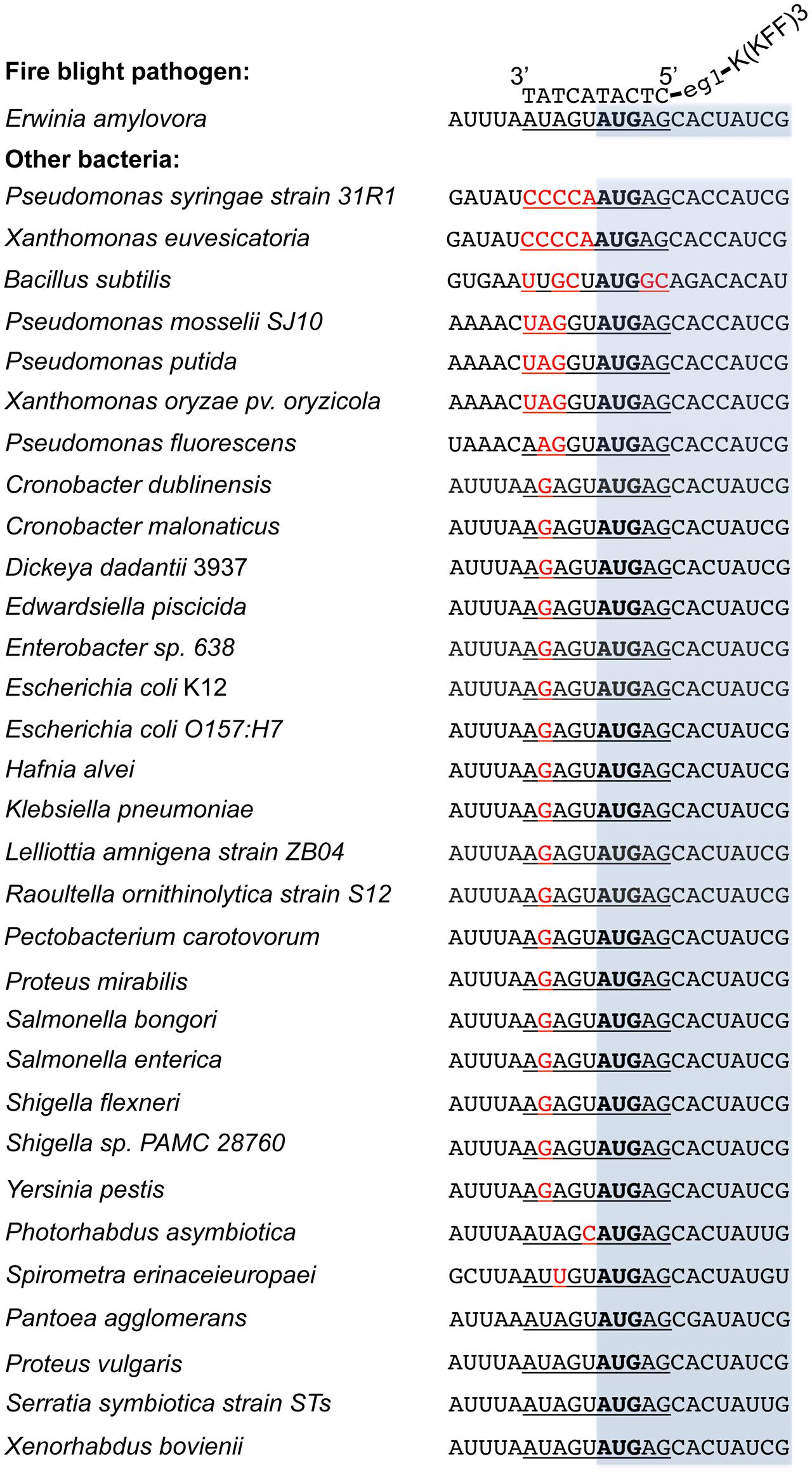
FIGURE 6. Alignment of the acpP start codon sequences of E. amylovora with other bacterial species. Sequences were obtained from NCBI database based on the availability of the bacterial genomes. The –5 to +5 sequences where the PNA–CPP targets are underlined, with mismatches heighted in red and start codon in bold. The open reading frame of acpP was shaded in light blue. The binding of PNA–CPP to acpP in E. amylovora was also illustrated. All sequences were written in a 5′ to 3′ order otherwise labeled.
Discussion
In this study, we demonstrated the proof-of-concept of using PNA–CPPs as antimicrobials against the fire blight pathogen E. amylovora. We showed that a 10-bp oligomer of PNA containing antisense sequence of the essential gene acpP is able to cause complete growth inhibition of E. amylovora at a similar efficacy of streptomycin under in vivo conditions. Although there are two additional sites on the chromosome (in hrcU and EAM_0144) that could be potentially bound by the 10-bp oligomer PNA, hrcU, and EAM_0144 (encoding a T3SS protein and a fatty acid synthesis regulator) are not known as essential genes in bacteria and reduction of acpP mRNA was observed upon treatment of anti-acpP-CPP1 (Figure 3), which suggests that the growth inhibition of anti-acpP-CPP1 should be through the interaction with acpP mRNA. We also showed that conjugation of the PNA with a CPP is essential for the PNA to exert antimicrobial effect in E. amylovora and identified the most effective CPP sequence for E. amylovora. We provided evidence that the antimicrobial effect observed is bactericidal. And finally, no phytotoxicity (e.g., scorching, browning, or any other types of obvious damage) was observed in flowers treated with PNA–CPP molecules only during the flower assay. These qualities suggest that PNA–CPPs are good alternatives for streptomycin in future fire blight management. Conditions generated from this research will add to our knowledge base for the practical development of formulations, rates, and timing for future applications, although many other factors, such as the toxicity and the stability of PNA–CPP in the natural environment, need to be determined before application.
Considerations that are often taken into account when developing antimicrobials include effectiveness, specificity, sustainability, and cost. We demonstrated that anti-acpP-CPP1 has a potent antimicrobial effect against E. amylovora with higher strain specificity than traditional antibiotics. We hypothesize that the antisense antimicrobials may also have good sustainability: if resistance is developed through mutations, the mutated nucleotide(s) could be identified by sequencing, and new antisense molecules could be synthesized to target the mutated sequence. Some bacteria may be able to generate resistance by acquiring multi-drug transporters. However, it is not very likely in this case as previous work demonstrated that PNA–CPPs cannot be readily transported out of the cells due to the large molecular weight (Nikravesh et al., 2007). Furthermore, as PNA–CPP molecules target individual essential genes rather than a conserved biological process like antibiotics, it widens our selection of targets during the development of antimicrobials. Combining PNA–CPP targeting multiple essential genes in E. amylovora may provide more potent and robust growth inhibition, although further actions need to be taken to enhance the entry of PNA–CPP molecules into bacterial cells. Currently, the biggest limitation of agricultural application of PNA–CPP is the cost. With the current synthesis method, the cost of PNA–CPP is about $1000USD/μMol. Thus, research in developing new synthesis technologies and improving the synthesis efficacy is essential for future large-scale application of PNA–CPP.
Prior to this research, the use of PNA–CPPs in controlling bacterial infections has been explored in human medicine. Compared to human medicine application, there are essential differences in agricultural use. First, the delivery method and locations where the antimicrobial activity occurs are different in the two systems. Antimicrobials for human medical purposes are usually delivered through the circulation system and the antimicrobial activity occurs mostly internally in the human body. For agricultural use, the delivery is mostly achieved through aerial spray to the plant surface, and the antimicrobial activity occurs mostly externally on the plant surface. Second, non-pathogenic, beneficial bacteria are not readily used as a disease control strategy in human medicine, but the beneficial, pathogen-antagonistic microbes are of great use in plant disease management as biological controls. This emphasizes a need of selectively targeting the pathogen without affecting the non-pathogenic, beneficial bacteria in agricultural settings.
With the differences between the two systems, our study demonstrated that the application of a PNA–CPP can reduce the bacterial pathogen population on plant surfaces. Our results also emphasized the importance of ensuring the compound-bacteria contact for the optimization of antimicrobial results, as we showed that premixing bacteria with PNA–CPP ensured the full bacteria-compound contact and had the most potent antimicrobial effect. It is possible that the use of a nonionic surfactant may be able to improve the antimicrobial effect of PNA–CPP on plant surfaces. In addition, whether the reduction in pathogen population on stigmas would also result in the reduction in blossom blight infection incidence needs to be further evaluated in future orchard trials. Finally, the stability of PNA–CPP in natural environment, when exposed to UV radiation, rain, and temperature changes, is not known. Understanding the stability will help us formulate the product rates, and timing of sprays to achieve the optimal control efficacy in the field.
Our study also showed that the antimicrobial effect of PNA–CPP molecules is more specific to E. amylovora and less toxic to other environmental bacteria than traditional antibiotics. Not affecting the environmental microbiome has many benefits. First, healthy microbiota serves as competitors of the pathogens for nutrients, space and can have a positive effect in preventing and limiting disease occurrence. Second, the selective pressure on the total microbiome of antibiotics often facilitated the development and spread of antibiotic resistance into pathogen population (Sundin and Bender, 1996). For example, the streptomycin resistance genes strA–strB that confer streptomycin resistance in the fire blight pathogen E. amylovora are believed to be acquired from saprophytic bacteria isolates such as P. agglomerans through horizontal gene transfer (Chiou and Jones, 1991). Thus, compared to streptomycin and other antibiotics, PNA–CPP may be a more sustainable management approach for fire blight.
One strategy of reducing the usage of antibiotics in tree fruit disease management is to combine biocontrol agents with antibiotics. However, as antibiotics and biocontrol agents are often non-compatible, the antibiotics and biocontrol agents often have to be applied separately, which not only adds labor cost and time constraints, but also may reduce the efficiency. Here we showed that anti-acpP-CPP1 is compatible with multiple biocontrol agents such as P. fluorescens and B. subtilis. The combined application of biological control agents with PNA–CPP may further enhance the control efficacy.
We observed that PNA–CPP antisense antimicrobials caused degradation of the target mRNA of an essential gene with a subsequent bactericidal effect against E. amylovora. This suggests that the antimicrobial effect of PNA–CPP on E. amylovora is potent and permanent. It also suggests that besides the potential application in disease management, PNA–CPPs may also be used as an effective approach to modulate gene expression in E. amylovora and potentially other bacteria. As the mRNA inhibition and dose of PNA–CPP is positively correlated, gene expression can be easily repressed at different levels for molecular research purposes. Thus, the gene expression repression by PNA–CPPs may have potential advantages over the gene knockout approach. This is specifically true in situations such as studying the functions of essential genes, when the gene knockout approach is not an option.
In summary, we performed the first exploration of using PNA–CPPs in controlling a bacterial plant disease. Plant diseases caused by bacteria have a long history of control difficulties, and the lack of effective management options results in significant economical losses worldwide (Sundin et al., 2016). The results produced from this work suggest that antisense antimicrobials may be a valuable future choice for bacterial plant disease management.
Author Contributions
RP performed most experiments and analyses. GS and C-HY helped with the experiment design and edited the manuscript. JW, RH, and XY helped with some experiment procedures the analyses. QZ designed this study, performed some experiments, and wrote the manuscript. All authors read and approved the manuscript.
Funding
This study was supported by the USDA-NIFA-AFRI-Exploratory Research program (2016-67030-24856), Northeastern IPM center partnership grant, and USDA-Hatch grant (CONH00650) to QZ, and by Michigan State University AgBioResearch to GS.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank undergraduate assistants M. Hoang (Quinnipiac University), and P. Sim (Southern CT State University) for their excellent technical support. We thank A. L. Arango Velez, T. G. Andreadis, and P. M. Armstrong (Connecticut Agricultural Experiment Station) for sharing fluorescence microscope and other equipment. We also thank W. H. Elmer (Connecticut Agricultural Experiment Station) for reviewing this manuscript.
References
Abes, R., Arzumanov, A. A., Moulton, H. M., Abes, S., Ivanova, G. D., Iversen, P. L., et al. (2007). Cell-penetrating-peptide-based delivery of oligonucleotides: an overview. Biochem. Soc. Trans. 35, 775–779. doi: 10.1042/BST0350775
Bai, H., and Luo, X. (2012). “Antisense antibacterials: from proof-of-concept to therapeutic perspectives,” in A Search for Antibacterial Agents, ed. V. Bobbarala (Rijeka: INTECH Open Access Publisher), 319–344.
Bennett, C. F., and Swayze, E. E. (2010). RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 50, 259–293. doi: 10.1146/annurev.pharmtox.010909.105654
Chiou, C. S., and Jones, A. L. (1991). The analysis of plasmid-mediated streptomycin resistance in Erwinia amylovora. Phytopathology 81, 710–714. doi: 10.1094/PHYTO-03-15-0078-R
Chiou, C. S., and Jones, A. L. (1995). Expression and identification of the strA-strB gene pair from streptomycin-resistant Erwinia amylovora. Gene 152, 47–51. doi: 10.1016/0378-1119(94)00721-4
Demidov, V. V., Potaman, V. N., Frank-Kamenetskil, M. D., Egholm, M., Buchard, O., Sönnichsen, S. H., et al. (1994). Stability of peptide nucleic acids in human serum and cellular extracts. Biochem. Pharmacol. 48, 1310–1313. doi: 10.1016/0006-2952(94)90171-6
Dinca, A., Chien, W.-M., and Chin, M. T. (2016). Intracellular delivery of proteins with cell-penetrating peptides for therapeutic uses in human disease. Int. J. Mol. Sci. 17:263. doi: 10.3390/ijms17020263
Dryselius, R., Aswasti, S. K., Rajarao, G. K., Nielsen, P. E., and Good, L. (2003). The translation start codon region is sensitive to antisense PNA inhibition in Escherichia coli. Oligonucleotides 13, 427–433. doi: 10.1089/154545703322860753
Evans, C. K. (2007). Survey results of Erwinia amylovora in Utah for resistance to streptomycin and investigations comparing kasugamycin (kasumin) to streptomycin and oxytetracycline for control of fire blight. Acta Hortic. 793, 433–437.
Ghosal, A., and Nielsen, P. E. (2012). Potent antibacterial antisense peptide-peptide nucleic acid conjugates against Pseudomonas aeruginosa. Nucleic Acid Ther. 22, 323–334. doi: 10.1089/nat.2012.0370
Good, L., and Nielsen, P. E. (1998). Inhibition of translation and bacterial growth by peptide nucleic acid targeted to ribosomal RNA. Proc. Natl. Acad. Sci. U.S.A. 95, 2073–2076. doi: 10.1073/pnas.95.5.2073
Goodman, R. N. (1959). “The influence of antibiotics on plants and plant disease control,” in Antibiotics: Their Chemistry and Non-Medical Uses, ed. H. S. Goldberg (Princeton, NJ: D. van Nostrand Company, Inc.), 322–448.
Gottesman, S., and Storz, G. (2010). Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb. Perspect. Biol. 3:a003798. doi: 10.1101/cshperspect.a003798
Hatamoto, M., Ohashi, A., and Imachi, H. (2010). Peptide nucleic acids (PNAs) antisense effect to bacterial growth and their application potentiality in biotechnology. Appl. Microbiol. Biotechnol. 86, 397–402. doi: 10.1007/s00253-009-2387-8
Johnson, K. B., and Temple, T. N. (2013). Evaluation of strategies for fire blight control in organic pome fruit without antibiotics. Plant Dis. 97, 402–409. doi: 10.1094/PDIS-07-12-0638-RE
Khokhani, D., Zhang, C., Li, Y., Wang, Q., Zeng, Q., Yamazaki, A., et al. (2013). Discovery of plant phenolic compounds that act as type three secretion system inhibitors or inducers of fire blight pathogen Erwinia amylovora. Appl. Environ. Microbiol. 79, 5424–5436. doi: 10.1128/AEM.00845-13
Kulyte, A., Nekhotiaeva, N., Awasthi, S. K., and Good, L. (2005). Inhibition of Mycobacterium smegmatis gene expression and growth using antisense peptide nucleic acids. J. Mol. Microbiol. Biotechnol. 9, 101–109. doi: 10.1159/000088840
Kurupati, P., Tan, K. S., Kumarasinghe, G., and Poh, C. L. (2007). Inhibition of gene expression and growth by antisense peptide nucleic acids in a multiresistant beta-lactamase-producing Klebsiella pneumoniae strain. Antimicrob. Agents Chemother. 51, 805–811. doi: 10.1128/AAC.00709-06
McGhee, G. C., Guasco, J., Bellomo, L. M., Blumer-Schuette, S. E., Shane, W. W., Irish-Brown, A., et al. (2011). Genetic analysis of streptomycin-resistant (Sm(R)) strains of Erwinia amylovora suggests that dissemination of two genotypes is responsible for the current distribution of Sm(R) E. amylovora in Michigan. Phytopathology 101, 182–191. doi: 10.1094/PHYTO-04-10-0127
Miller, T. D., and Schroth, M. N. (1972). Monitoring the epiphytic populations of Erwinia amylovora on pear with a selective medium. Phytopathology 62, 1175–1182. doi: 10.1094/Phyto-62-1175
Nakashima, N., Goh, S., Good, L., and Tamura, T. (2012). Multiple-gene silencing using antisense RNAs in Escherichia coli. Methods Mol. Biol. 815, 307–319. doi: 10.1007/978-1-61779-424-7_23
Nekhotiaeva, N., Awasthi, S. K., Nielsen, P. E., and Good, L. (2004). Inhibition of Staphylococcus aureus gene expression and growth using antisense peptide nucleic acids. Mol. Ther. 10, 652–659. doi: 10.1016/j.ymthe.2004.07.006
Nikravesh, A., Dryselius, R., Faridani, O. R., Goh, S., Sadeghizadeh, M., Behmanesh, M., et al. (2007). Antisense PNA accumulates in Escherichia coli and mediates a long post-antibiotic effect. Mol. Ther. 15, 1537–1542. doi: 10.1038/sj.mt.6300209
Norelli, J. L., Jones, A. L., and Aldwinckle, H. S. (2003). Fire blight management in the twenty-first century: using new technologies that enhance host resistance in apple. Plant Dis. 87, 756–765. doi: 10.1094/PDIS.2003.87.7.756
R Core Team (2015). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. Available at: https://www.R-project.org/
Rasmussen, L. C., Sperling-Petersen, H. U., and Mortensen, K. K. (2007). Hitting bacteria at the heart of the central dogma: sequence-specific inhibition. Microb. Cell Fact. 6:24. doi: 10.1186/1475-2859-6-24
Russo, N. L., Burr, T. J., Breth, D. I., and Aldwinckle, H. S. (2008). Isolation of streptomycin-resistant isolates of Erwinia amylovora in New York. Plant Dis. 92, 714–718. doi: 10.1094/PDIS-92-5-0714
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. doi: 10.1038/nmeth.2089
Sundin, G. W., and Bender, C. L. (1996). Dissemination of the strA-strB streptomycin-resistance genes among commensal and pathogenic bacteria from humans, animals, and plants. Mol. Ecol. 5, 133–143. doi: 10.1111/j.1365-294X.1996.tb00299.x
Sundin, G. W., Castiblanco, L. F., Yuan, X., Zeng, Q., and Yang, C. H. (2016). Bacterial disease management: challenges, experience, innovation, and future prospects. Mol. Plant Pathol. 17, 1506–1518. doi: 10.1111/mpp.12436
Sundin, G. W., Werner, N. A., Yoder, K. S., and Aldwinckle, H. S. (2009). Field evaluation of biological control of fire blight in the eastern United States. Plant Dis. 93, 386–394. doi: 10.1094/PDIS-93-4-0386
Tan, X. X., Actor, J. K., and Chen, Y. (2005). Peptide nucleic acid antisense oligomer as a therapeutic strategy against bacterial infection: proof of principle using mouse intraperitoneal infection. Antimicrob. Agents Chemother. 49, 3203–3207. doi: 10.1128/AAC.49.8.3203-3207.2005
Tsiantos, J., Psallidas, P., and Chatzaki, A. (2003). Efficacy of alternatives to antibiotic chemicals for the control of fire blight of pears. Annu. Appl. Biol. 143, 319–323. doi: 10.1111/j.1744-7348.2003.tb00300.x
van der Zwet, T., Orolaza-Halbrendt, N., and Zeller, W. (2012). Fire Blight History, Biology, and Management. St. Paul, MN: American Phytopathological Society Press.
Wang, G., and Xu, X. S. (2004). Peptide nucleic acid (PNA) binding-mediated gene regulation. Cell Res. 14, 111–116. doi: 10.1038/sj.cr.7290209
Wu, R. P., Youngblood, D. S., Hassinger, J. N., Lovejoy, C. E., Nelson, M. H., Iversen, P. L., et al. (2007). Cell-penetrating peptides as transporters for morpholino oligomers: effects of amino acid composition on intracellular delivery and cytotoxicity. Nucleic Acids Res. 35, 5182–5191. doi: 10.1093/nar/gkm478
Yang, F., Korban, S. S., Pusey, P. L., Elofsson, M., Sundin, G. W., and Zhao, Y. (2014). Small-molecule inhibitors suppress the expression of both type III secretion and amylovoran biosynthesis genes in Erwinia amylovora. Mol. Plant Pathol. 15, 44–57. doi: 10.1111/mpp.12064
Zatsepin, T. S., Turner, J. J., Oretskaya, T. S., and Gait, M. J. (2005). Conjugates of oligonucleotides and analogues with cell penetrating peptides as gene silencing agents. Curr. Pharm. Des. 11, 3639–3654. doi: 10.2174/138161205774580769
Zeng, Q., McNally, R. R., and Sundin, G. W. (2013). Global small RNA chaperone Hfq and regulatory small RNAs are important virulence regulators in Erwinia amylovora. J. Bacteriol. 195, 1706–1717. doi: 10.1128/JB.02056-12
Zeng, Q., and Sundin, G. W. (2014). Genome-wide identification of Hfq-regulated small RNAs in the fire blight pathogen Erwinia amylovora discovered small RNAs with virulence regulatory function. BMC Genomics 15:414. doi: 10.1186/1471-2164-15-414
Zhang, Y., and Cronan, J. E. Jr. (1998). Transcriptional analysis of essential genes of the Escherichia coli fatty acid biosynthesis gene cluster by functional replacement with the analogous Salmonella typhimurium gene cluster. J. Bacteriol. 180, 3295–3303.
Keywords: antisense antimicrobials, fire blight, plant disease management, peptide nucleic acid, cell penetrating peptide, Erwinia amylovora
Citation: Patel RR, Sundin GW, Yang C-H, Wang J, Huntley RB, Yuan X and Zeng Q (2017) Exploration of Using Antisense Peptide Nucleic Acid (PNA)-cell Penetrating Peptide (CPP) as a Novel Bactericide against Fire Blight Pathogen Erwinia amylovora. Front. Microbiol. 8:687. doi: 10.3389/fmicb.2017.00687
Received: 05 October 2016; Accepted: 04 April 2017;
Published: 19 April 2017.
Edited by:
Benjamin Gourion, CNRS & INRA, FranceCopyright © 2017 Patel, Sundin, Yang, Wang, Huntley, Yuan and Zeng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Quan Zeng, quan.zeng@ct.gov
 Ravi R. Patel
Ravi R. Patel George W. Sundin
George W. Sundin Ching-Hong Yang
Ching-Hong Yang Jie Wang4
Jie Wang4 Regan B. Huntley
Regan B. Huntley Xiaochen Yuan
Xiaochen Yuan Quan Zeng
Quan Zeng