- 1Department of Chemistry, Biotechnology and Food Science, Norwegian University of Life Sciences, Ås, Norway
- 2Department of Biosciences, University of Oslo, Oslo, Norway
- 3Institute of Biochemistry and Biophysics, Polish Academy of Sciences, Warsaw, Poland
Enterocin K1 (EntK1), enterocin EJ97 (EntEJ97), and LsbB are three sequence related leaderless bacteriocins. Yet LsbB kills only lactococci while EntK1 and EntEJ97 target wider spectra with EntK1 being particularly active against Enterococcus faecium, including nosocomial multidrug resistant isolates. NMR study of EntK1 showed that it had a structure very similar to LsbB – both having an amphiphilic N-terminal α-helix and an unstructured C-terminus. The α-helix in EntK1 is, however, about 3–4 residues longer than that of LsbB. Enterococcal mutants highly resistant to EntEJ97 and EntK1 were found to have mutations within rseP, a gene encoding a stress response membrane-bound Zn-dependent protease. Heterologous expression of the enterococcal rseP rendered resistant cells of Streptococcus pneumoniae sensitive to EntK1 and EntEJ97, suggesting that RseP likely serves as the receptor for EntK1 and EntEJ97. It was also shown that the conserved proteolytic active site in E. faecalis RseP is partly required for EntK1 and EntEJ97 activity, since alanine substitutions of its conserved residues (HExxH) reduced the sensitivity of the clones to the bacteriocins. RseP is known to be involved in bacterial stress response. As expected, the growth of resistant mutants with mutations within rseP was severely affected when they were exposed to higher (stressing) growth temperatures, e.g., at 45∘C, at which wild type cells still grew well. These findings allow us to design a hurdle strategy with a combination of the bacteriocin(s) and higher temperature that effectively kills bacteriocin sensitive bacteria and prevents the development of resistant cells.
Introduction
The spread of antibiotic-resistant bacteria poses a great threat to public health and is growing worse since the current progress in developing of new antibiotics is limited (Brown and Wright, 2016). Aside from the introduction of carbapenems in 1985, all new antibiotics between the early 1960s and 2000 were synthetic derivatives of existing scaffolds, which often allow resistant strains to arise rapidly (Fischbach and Walsh, 2009). Only in the USA the economic loss provoked by antibiotic resistance is estimated to be up to 55 billion USD per year with some data suggesting that the total cost can be even higher (Smith and Coast, 2013). Consequently, there is a need for new antimicrobials that can be used as alternatives to conventional antibiotics. Among such alternatives the antimicrobial peptides, the bacteriocins, are potential antibacterial agents to fight infections (Cotter et al., 2013).
Bacteriocins are ribosomally synthesized peptides produced by bacteria to inhibit or kill other bacteria in competition for nutrients or habitats. They are almost with no exception cationic, amphiphilic, and heat-stable. The antimicrobial inhibition spectra and potency (strength) of bacteriocins differ greatly, with the majority being active only against closely related bacteria while some being much broader, targeting cells from different genera (Nes et al., 2014).
Bacteriocins have a number of advantages over conventional antibiotics and chemical food preservatives. Bacteriocins show antimicrobial activity at pico- to nanomolar concentrations and are commonly produced by bacteria found in many fermented foods and in the human gut microflora – properties making them very attractive as natural food preservatives (Nes et al., 1996). Most bacteriocins from gram positive bacteria are known as membrane-active peptides, i.e., they disrupt membrane integrity, leading to leakage of intracellular solutes and cell death (Kjos et al., 2011). This killing mechanism is different from that of most antibiotics, which often act as enzyme inhibitors of key metabolic pathways. Due to their different modes of action, bacteriocins do not discriminate between antibiotic resistant and sensitive bacteria (Cotter et al., 2013).
Most bacteriocins are synthesized as a propeptide with an N-terminal leader which is removed by a dedicated ABC transporter or the Sec system during exporting from cells (Havarstein et al., 1995; Cintas et al., 1997). However, there is a group of so-called leaderless bacteriocins, which are synthesized without an N-terminal leader and the mechanism behind their export is unknown. Leaderless bacteriocins are often relatively small (30–50 amino acids) and contain no modifications. These features make them relatively easy to obtain by synthetic means, thereby opening new opportunities for detailed bacteriocin study and design of new peptides with improved properties (Ovchinnikov et al., 2014).
Enterocin K1 (EntK1) is a leaderless bacteriocin, which belongs to LsbB bacteriocin family. The family presently contains four members: LsbB, EntK1, EntQ, and EntEJ97 (Ovchinnikov et al., 2014), all being leaderless and non-modified peptides. LsbB is a 30 amino acid (aa) residues peptide produced by Lactococcus lactis. It has a very narrow inhibition spectrum which contains only lactococcal strains (Gajic et al., 2003). The remaining three bacteriocins are produced by different enterococcal strains. EntQ (34 aa residues) and especially EntEJ97 (44 aa residues) have broader antimicrobial spectra than LsbB (Galvez et al., 1998; Cintas et al., 2000; Basanta et al., 2008). Less is known about the activity of the newly discovered EntK1 (37 aa residues), although it has been shown to inhibit L. lactis IL1403 (Ovchinnikov et al., 2014).
It is has been shown for an increasing number of bacteriocins that they kill target cells in a receptor-mediated manner, i.e., bacteriocins recognize specific receptors on target cells where they bind and cause membrane-disruption that eventually leads to cell death (Diep et al., 2007; Cotter, 2014). Recently, it has been shown that the lactococcal Zn-dependent metallopeptidase YvjB (also known as RseP) belonging to site-2 protease (S2P) protein family, serves as the receptor for LsbB (Uzelac et al., 2013; Miljkovic et al., 2016). Our previous structure-function study of LsbB provided strong evidence that the peptide binds to its receptor using its unstructured C-terminal part (Ovchinnikov et al., 2014).
In this work we determined the structure of EntK1, its activity spectrum and unravel the details behind target specificity of EntK1 and its sequence related bacteriocin EntEJ97. Based on the fact that RseP is involved in bacterial stress response (Varahan et al., 2013) we developed an efficient strategy to improve the activity of EntK1 and its sequence related bacteriocins. We also showed that at least in Enterococcus faecalis, another membrane protein is responsible for bacterial sensitivity to EntEJ97, besides EntEJ97 receptor RseP.
Materials and Methods
Bacterial Strains, Growth Conditions, Bacteriocins and Antimicrobial Assays
All the strains (see below) used in minimal inhibitory concentration (MIC) assays were grown in BHI medium (Oxoid) at 30°C without shaking. LsbB, EntK1, EntEJ97, and BHT-B bacteriocins were synthesized by Pepmic Co., Ltd, China with 98–99% purity. Synthesized peptides were solubilized to the concentrations of 10.0–0.1 mg/ml in 0.1% (vol/vol) trifluoroacetic acid and stored at -20°C until use. Bacteriocin garvicin ML was purified to 95% as described by (Borrero et al., 2011). Bacteriocin activity was determined by microtiter plate assay as previously described (Holo et al., 1991). The MIC was defined as the minimal bacteriocin concentration that inhibited the growth of the indicator strain by at least 50% (50% of the turbidity of the control culture without bacteriocin) in 200 μl culture.
CD Spectroscopy
Circular dichroism (CD) spectra were recorded using a Jasco J-810 spectropolarimeter (Jasco International Co.) calibrated with D-camphor-10-sulfonate (Icatayama Chemical). All measurements were done using a quartz cuvette (Starna) with 0.1 cm path length at 25°C. Samples were scanned five times with a scanning rate of 50 nm/min with a bandwidth of 0.5 nm and response time of 1 s over the wavelength range 190–250 nm. Spectra were recorded at different (30 and 50%) trifluoroethanol (TFE) (Aldridge) concentrations at 25°C. The approximate α-helical content of the protein was estimated from its molar ellipticity at 222 nm (Scholtz et al., 1991).
NMR Spectroscopy
The experiments were run on a sample containing 1.0 mM EntK1, 50% D3-TFE (99.5% D) (Aldrich), Milli-Q water and 0.2 mM of 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS) (Larodan).
2D NOESY (Jeener et al., 1979), 2D TOCSY (Braunschweiler and Ernst, 1983), 2D DQCOSY, 15N-HSQC (Davis et al., 1992), and13C-HSQC (Hurd, 1991) were recorded. The data was acquired on a 600 MHz Bruker Avance II spectrometer with four channels and a 5 mm TCI cryoprobe (Bruker Biospin). NOESY spectra with mixing times between 120 and 300 ms were obtained for both samples. TOCSY mixing times of 15–80 ms was used. The experiments were run at 298.15 K. Spectra were processed using the Topspin program (Bruker Biospin). DSS was used as a chemical shift standard, and 15N and 13C data were referenced using frequency ratios as described in (Wishart et al., 1995).
For visualization, assignment and integration of the spectra the computer program CARA was used (Keller, 2004). The spectra were assigned using standard methods (Wüthrich, 1986).
Dihedral angle restraints were obtained from the chemical shift values using the program TALOS-N (Shen and Bax, 2013). Nuclear Overhauser effect (NOE) distance restraints were calculated from the peak volumes in the NOESY spectra with NOESY mixing time 200 ms.
All structure calculations were made using the structure calculation program CYANA 2.1 (Guntert et al., 1997; Herrmann et al., 2002). The applied restraints are presented in Table 1. The annealing macro in CYANA calculated 100 structures. The 20 structures with the lowest energy were kept and analyzed further. The root-mean-square deviation (RMSD) was calculated and the structures were visualized using MolMol (Koradi et al., 1996).
Generation of Bacteriocin Resistant Mutants of E. faecalis and E. faecium
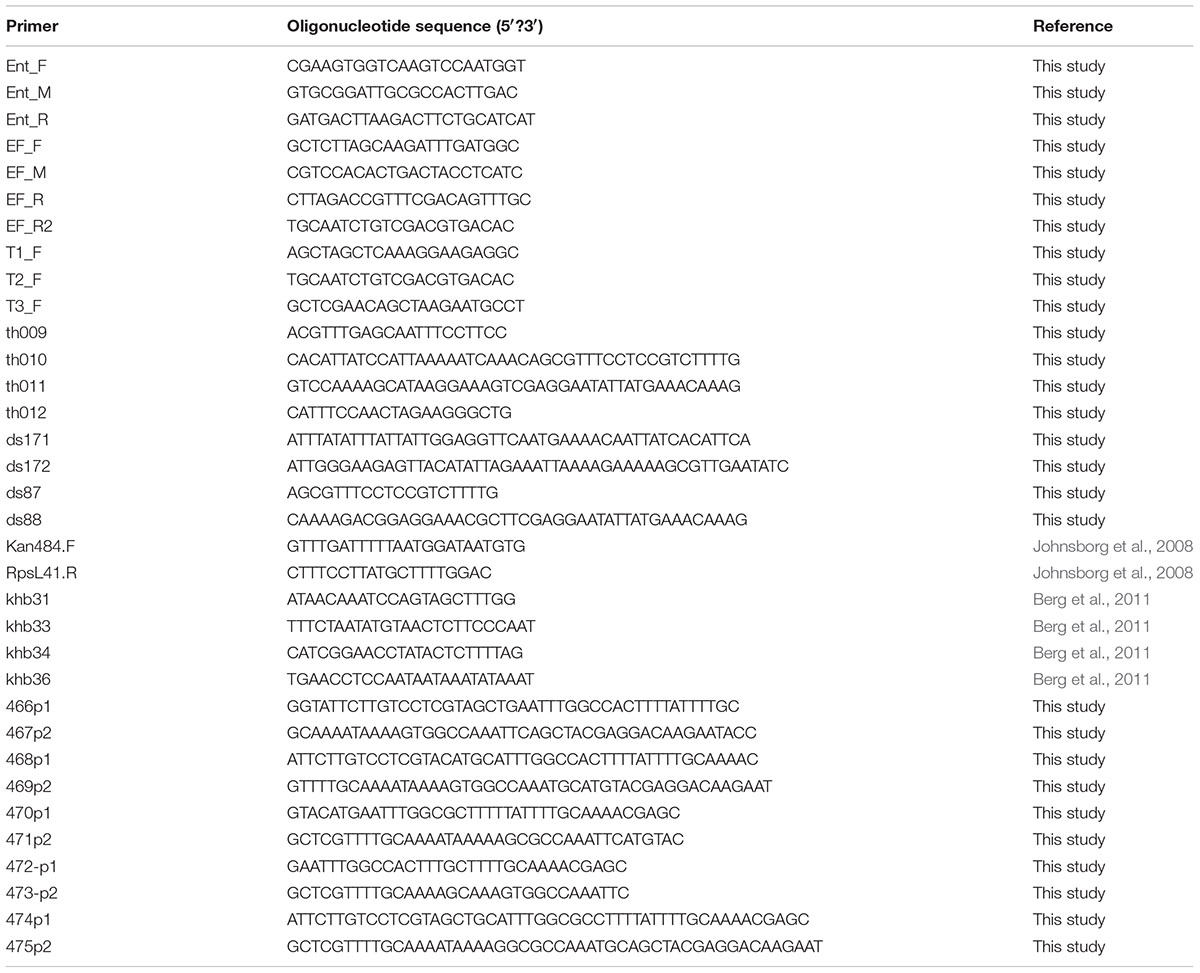
EntEJ97 and EntK1-resistant mutants were obtained by spot-on-lawn assay using 20 μl of bacteriocin with concentration 1.0–0.1 mg/ml. After overnight incubation at 30°C, resistant colonies of E. faecalis LMG3358 (resistant to EntEJ97) and E. faecium LMG2787 (resistant to EntEJ97 and EntK1) appeared within the inhibition zones. Resistant isolates were picked and re-streaked on plates for pure cultures before frozen cultures containing 15% glycerol were made and stored at -80°C. The level of resistance against EntEJ97 and EntK1 was determined by a microtitre plate assay (Holo et al., 1991). DNA from the bacteriocin resistant mutants was extracted from overnight cultures with a GenEluteTM Bacterial Genomic DNA Kit (Sigma–Aldrich) and PCR of the rseP gene was performed using primers Ent, EF (F, M, R) (Table 2). For sequencing of E. faecium LMG2787 mutants with transposons inside rseP, additional primers (EF_R2, T1_F, T2_F, and T3_F) were created (Table 2). PCR products were purified with NucleoSpin Extract II (Macherey-Nagel, Düren, Germany) and sent to GATC Biotech, Germany, for sequencing.
Construction of S. pneumoniae Transformants and Mutants of rseP
To express the E. faecalis rseP gene heterologously in S. pneumoniae, the gene was placed by homologous recombination in the genome of strain SPH131, behind the ComS-inducible PcomX promoter (ComRS system) (Berg et al., 2011). The PcomX-rseP construct was created by overlap extension PCR (Higuchi et al., 1988). First, the E. faecalis LMG3358 rseP was amplified using the primer pair ds171/ds172 with genomic E. faecalis DNA as template. The PcomX promoter and its ∼1000 bp upstream and downstream regions were amplified using the primer pairs khb31/khb36 and khb33/khb34, respectively. Genomic DNA derived from strain SPH131 served as template. The PcomX with its upstream region was fused to the 5′ end of the E. faecalis rseP gene using the primers khb31 and ds172. The PcomX downstream fragment was fused to the 3′ end of the rseP gene using primer pair ds171 and khb34. Finally, these two fragments were fused using primer khb31 and khb34 giving rise to PcomX-rseP. The Janus cassette (Sung et al., 2001) in strain SPH131 was replaced with the PcomX-rseP fragment by natural transformation, giving rise to strain ds218 (Table 3 and Supplementary Figure 1).
Streptococcus pneumoniae has a gene homologous to the lactococcal rseP. To avoid the potential background noise of the S. pneumoniae rseP, this gene was removed from the genome in strain ds218 using the Janus cassette (Sung et al., 2001). An rseP deletion cassette was constructed by amplifying Janus with the primers kan484F and RpsL41.R (Johnsborg et al., 2008) where genomic DNA from strain RH426 (Johnsborg et al., 2008) served as template. The Janus cassette was then fused to the ∼1000 bp upstream [primers th009 and th010 with genomic RH1 (Johnsborg et al., 2008) DNA as template] and downstream region (primers th011 and th012 with genomic RH1 DNA as template) of rseP using primers th009 and th012. The resulting fragment was used to transform strain ds218 resulting in the replacement of S. pneumoniae rseP with Janus generating strain ds219. Janus was then removed from strain ds219 by transforming with a fragment consisting of the rseP flanking regions only. This fragment was constructed by amplifying the rseP ∼1000 bp upstream region using primers th009 and ds87, while the ∼1000 bp downstream region was amplified with the primers ds88 and th012. The upstream and downstream fragments where then fused using the primers pair th009/th012. The resulting fragment was used to replace the Janus in strain ds219 resulting in strain ds220 (Table 3 and Supplementary Figure 1).
To create a strain expressing point mutated rseP genes we replaced the E. faecalis 3358 rseP in strain ds220 with Janus cassette giving rise to strain ds221. This Janus cassette was amplified with the primer pair khb31/khb34 and genomic DNA from strain SPH131 served as template. Selected residues in RseP were substituted with alanines by a PCR approach, using primer pairs listed in Table 2. The resulting DNA pair fragments were subsequently fused using the primers khb31 and khb34. The resulting fragment was used to replace Janus cassette in strain ds221 giving rise to strains ms1-5 (Table 3). Ectopic expression of the rseP gene in strain ds220 and ms1-5 was induced with the synthetic ComS peptide (NH2-LPYFAGCL-COOH) (Genosphere Biotechnologies) as described by Berg et al. (2011).
Results
Structural Analysis of EntK1 by CD and NMR-Spectroscopy
Circular dichroism spectra of EntK1 showed that it was unstructured in water but became structured in TFE solution (Supplementary Figure 2). Maximum structuring was obtained in 50% TFE (55% α-helical content). This concentration was consequently used in the NMR experiment.
The NMR spectra were assigned using standard methods as described in “Materials and Methods.” Supplementary Figure 3 shows the assigned 15N HSQC spectrum of EntK1 in 50% TFE. Figure 1 shows connected strips from NOESY, the torsion angle restraints used in structure calculations and the Hα chemical shift indexes (CSIs). CSI (low Hα ppm values) (Matthews et al., 1992) indicates that there is an α-helix in EntK1 from residue 8 to 27 and TALOS-N torsion angle predictions (Shen and Bax, 2013) indicated backbone torsion angles (ϕ- and ψ-angle) consistent with α-helical regions from residue 8 to 25 (Figure 1).
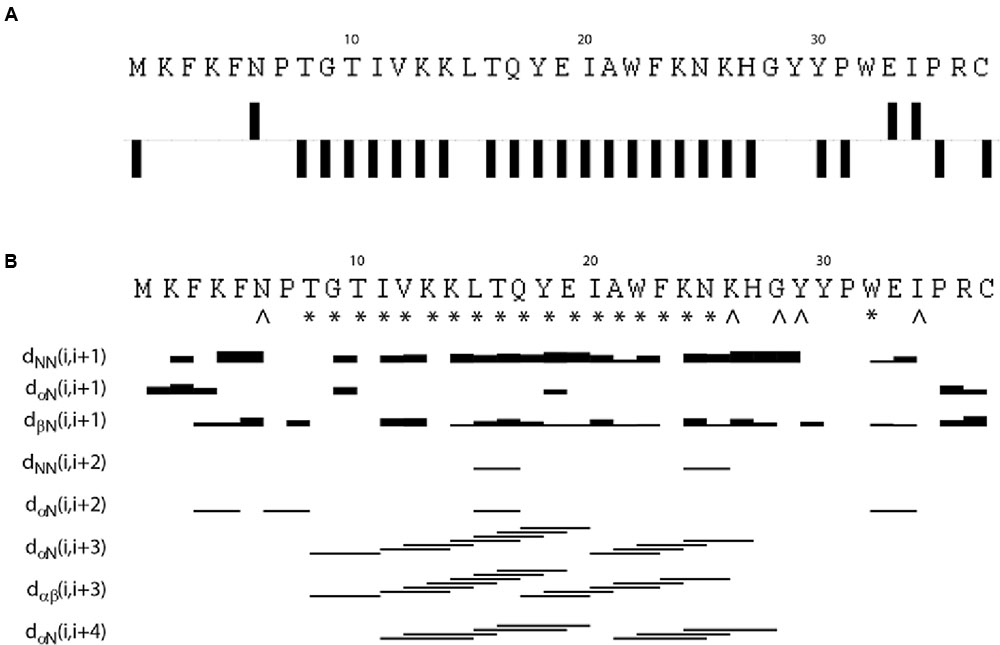
FIGURE 1. Results from analysis of NMR chemical shifts and nuclear Overhauser effects (NOE’s) of EntK1 in TFE. Chemical shift index (CSI) of Hα carbons is shown in (A). Intermediate NOE restraints and residues where determined torsion angles from the chemical shifts are shown in (B). The signs ∗ and ˆ indicate that TALOS determined phi and psi torsion angle restraints, the ∗ are used to symbolize torsion angle restraints in the α-helical region of the Ramachandran plot while ˆ is used for all other torsion angles. The lines indicate typical α-helical restraints obtained from analysis of the NOE spectra. Thicknesses of the lines shown are related to NOE cross peak intensities.
A total of 561 (15.1 per residue) unique NOE connectivities were assigned. The total number of distance restrictions and their classes are shown in Table 1. The mid-range NOE connectivities that are especially important for α-helices, NN(i, i+2), αN(i, i+2), αN(i, i+3), αβ(i, i+3), and αN(i, i+4), are shown in Figure 1. In the presence of TFE, the observed connectivities indicated an α-helical region stretching from residue 8 to 26.
Structures of EntK1 based on the experimentally obtained constrains were calculated using CYANA. A superimposition of the structure ensemble of the 20 lowest energy structures of EntK1 and the cartoon depiction of the lowest energy structure of EntK1 are shown in Figures 2A,B, respectively. The NMR structure of EntK1 contains an α-helix from residue 8 to 24. The structure ensemble has been deposited to the Protein Data Bank with access code: 5L82 and the NMR data have been deposited to the Biomagnetic Resonance Data Bank with the access code 34006.
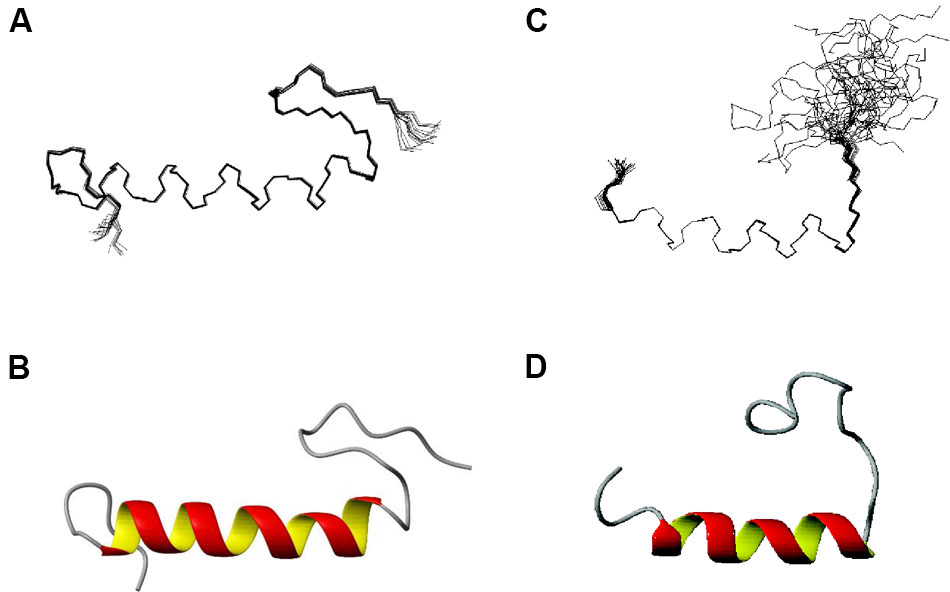
FIGURE 2. NMR structure of EntK1 (A,B) compared with LsbB (C,D) (Ovchinnikov et al., 2014) in 50% TFE. The structures ensembles of the 20 lowest energy structures superimposed are shown in (A,C) and cartoon representations of the lowest energy structures are shown in (B,D).
The Inhibitory Spectra of the EntK1 and Two Sequence Related Bacteriocins
A sizable collection of Gram-positive bacteria from different species and genera was used as indicators to assess the inhibition spectra of the bacteriocins EntK1, EntEJ97, and LsbB (Table 4). EntQ, which is a member of the LsbB family bacteriocins, was not tested because synthetic EntQ peptides had poor activity, likely due to formation of disulphide bridges between and inside the molecules, since adding reducing agents (e.g., DTT) restored some of the activity of EntQ (data not shown). As seen in Table 4, the antimicrobial spectra of EntK1, EntEJ97, and LsbB differ greatly. LsbB was active only against L. lactis IL1403, one of the four strains of L. lactis tested and it had no activity against other genera and species. EntK1 was active mostly against E. faecium and E. hirae and some lactococcal strains. EntEJ97 showed the broadest activity spectrum, inhibiting E. faecium, E. faecalis, L. lactis, Staphylococcus aureus, Lactococcus garvieae, and Listeria monocytogenes. It is noteworthy that EntK1 appeared to have significantly lower MIC values (i.e., being more potent) than EntEJ97 against E. faecium, but was inactive against E. faecalis (Table 4). Similarly, all three bacteriocins were active against the L. lactis IL1403, but LsbB was about 340 and 70 times more potent than EntK1 and EntEJ97, respectively (Table 4).
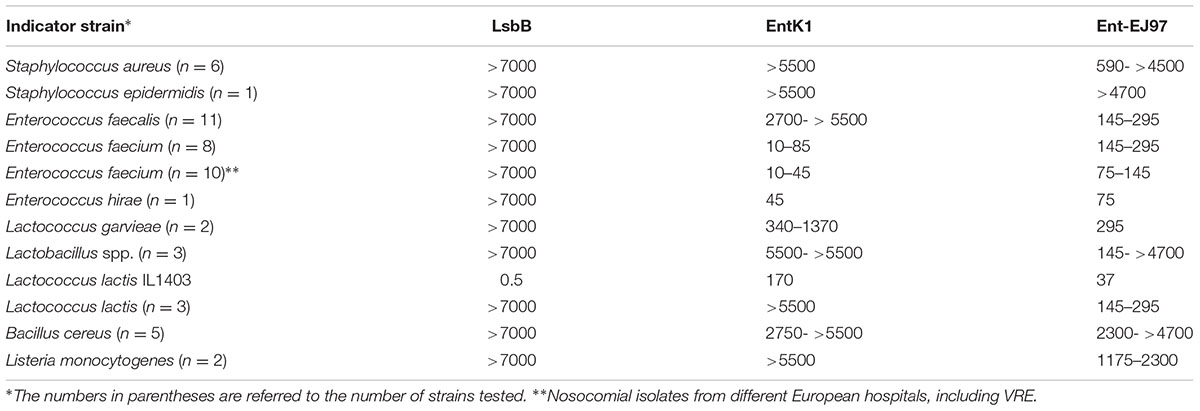
TABLE 4. Minimal inhibitory concentration (MIC) values (nM) of EntK1 and its related bacteriocins against different bacterial species.
Different Levels of Resistance against EntK1 and EntEJ97 in E. faecalis and E. faecium
The strain E. faecium LMG2787 was used to generate mutants resistant to EntK1 and EntEJ97. Since E. faecalis is resistant to EntK1, E. faecalis LMG3358 was used to generate mutants resistant only to EntEJ97. The two enterococcal strains were exposed to various concentrations of EntEJ97 and EntK1 at 30°C and relatively many resistant colonies appeared within the inhibition zones on agar plates (see below). For E. faecalis, 12 EntEJ97 resistant colonies were randomly selected and examined for resistance to EntEJ97 by microtiter assays. Similarly, six EntEJ97 and six EntK1mutants of E. faecium were selected for further analysis.
Two levels of E. faecalis resistance to EntEJ97 were found: seven colonies were highly resistant (HR) – at least 500 times more compared to the wild type (WT) strain. The second set of five colonies showed a lower resistance (LR) level, being about 16–32 times more resistant than the WT E. faecalis. For E. faecium only HR (at least 500 times against EntK1 and EntEJ97, compared to WT) type mutants were found. When all these mutants were challenged with the non-related bacteriocins BHT-B (Hyink et al., 2005) and garvicin ML (Borrero et al., 2011), they were found as sensitive as the WT strains (data not shown). These results clearly imply the involvement of specific resistance mechanism(s) toward EntEJ97/EntK1 amongst these mutants.
Highly Resistant Mutants of E. faecalis and E. faecium Have Mutations in the rseP Gene
As the lactococcal RseP has previously been found to serve as receptor for LsbB, we suspected that the RseP homologous in Enterococcus might serve the same function for the LsbB sequence related EntEJ97/EntK1 bacteriocins. The DNA regions containing rseP in all the EntEJ97/EntK1-resistant mutants (high and low) were therefore obtained by PCR and sequenced. Different types of mutations were found. For E. faecalis, all seven HR mutants contained either one or three consecutive 8-bp repeats of the sequence CAAAAAAT within the rseP gene, while the WT rseP has two such repeats. On the other hand, all E. faecium HR mutants carried a transposon within rseP (Figure 3). In all cases, these mutations caused frameshift in rseP and premature termination, indicating that a functional ResP is necessary for the sensitivity toward EntEJ97/EntK1. Surprisingly, no mutations were found in rseP from all five EntEJ97 LR mutants of E. faecalis, indicating that additional genes can affect the sensitivity of the bacterium to these bacteriocins. This latter aspect will be further mentioned in the “Discussion” section.
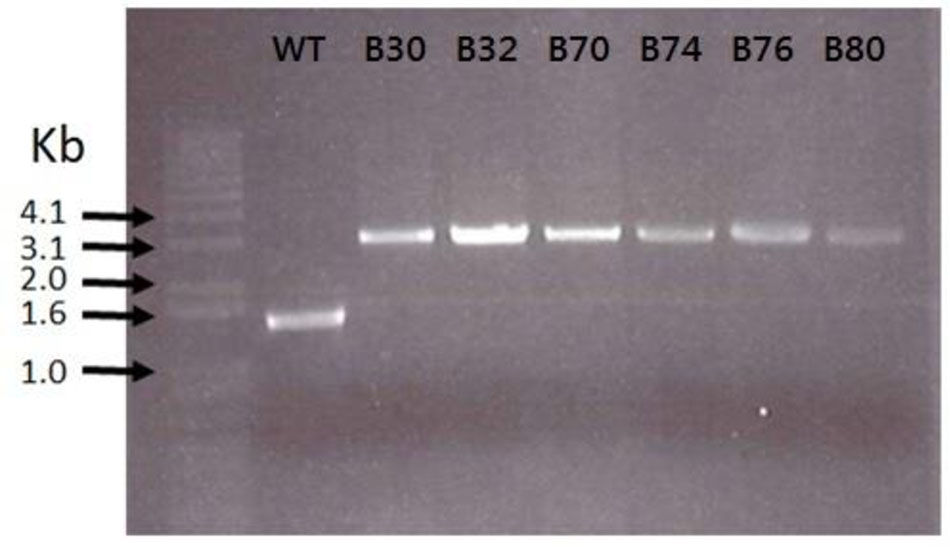
FIGURE 3. Transposon insertions in E. faecium LMG2787 rseP mutants (B30–B80) compared to the wild type (WT).
rseP Expression in S. pneumoniae Confers Sensitivity to EntEJ97/EntK1
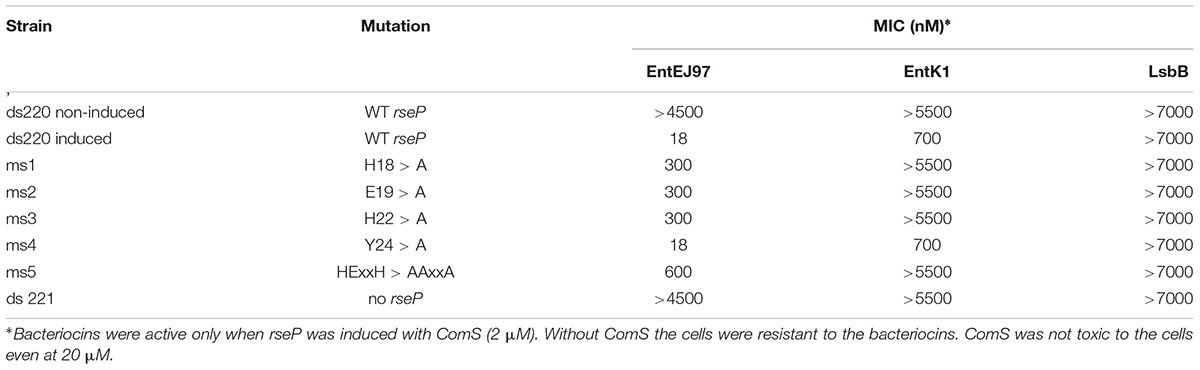
In order to confirm that the expression of rseP is sufficient to confer sensitivity to EntEJ97 and EntK1, the rseP gene from E. faecalis LMG3358 was heterologously expressed in the distantly related and non-sensitive host S. pneumoniae strain SPH131 as described in (Berg et al., 2011). To avoid potential background noise the endogenous rseP of S. pneumoniae SPH131 was removed by homologous recombination, giving rise to the clone S. pneumoniae ds220 with the endogenous rseP deleted but with the enterococcal rseP expressed under the control of the inducible promoter PcomX (Berg et al., 2011). While non-induced cells were not sensitive to EntEJ97 and EntK1, they became sensitive to the bacteriocins upon induction of rseP (Table 5). The results indicate that the enterococcal rseP is indeed directly involved in the sensitivity to EntEJ97 and EntK1. Further, it should be noted that the induced clone (expressing the E. faecalis rseP) was most sensitive to EntEJ97 (MIC of 18 nM), intermediate-sensitive to EntK1 (MIC of 700 nM), and poorly sensitive to LsbB (MIC > 7000 nM). This observation is line with the results presented in Table 4, where the E. faecalis strains showed a corresponding differentiation in sensitivity to the three bacteriocins.
Mutations within the Active Site of RseP Reduce Sensitivity to EntEJ97/EntK1
Members of the RseP protein family have a conserved proteolytic motif (HExxH, where x is any amino acid) located within the first transmembrane helix (Koide et al., 2007). To analyze whether this active site is important for bacteriocin sensitivity, we changed the invariant residues in this motif (i.e., H18, E19, and H22), one by one, and three altogether in the enterococcal RseP to alanine and then assessed for bacteriocin sensitivity. In addition, the non-conserved residue Y24 located nearby the active site was replaced with alanine to serve as a control. The result showed that all clones expressing the RseP protein in which conserved residues were changed to alanine became less sensitive to EntEJ97 and EntK1, especially when all three conserved residues were replaced with alanine (30 times less sensitive to EntEJ97 than the WT) (Table 5). In the control clone (Y24 > A) no changes in the bacteriocin sensitivity were observed. This result demonstrates that the active site of RseP is essential for bacteriocin activity.
The Growth of EntK1/EntEJ97 Resistant Mutants Is Prevented at Elevated Growth Temperature
RseP protein is known to play a key role in bacterial stress response in Escherichia coli, Bacillus subtilis, and other bacteria (Alba et al., 2002; Varahan et al., 2013; Kim, 2015). We therefore examined how elevated growth temperature as a stress factor, influences the development of EntEJ97 and EntK1 resistance in E. faecium and E. faecalis. As expected resistant mutants only appeared at 30°C but not at 45°C (Figure 4). Cell counts of WT cultures grown for 8 h at 30 and 45°C in BHI broth showed that the cell number at 30°C was only about 1.2 times higher than that at 45°C (data not shown), implying that the lack of resistant mutants at the elevated temperature was not due to poor growth of WT cells but rather due to lack of development of resistant mutants. This is further supported by the fact that EntEJ97/EntK1-resistant mutants with a non-functional rseP gene (described above) obtained at 30°C could not grow at 45°C, while WT cells grew well (data not shown).
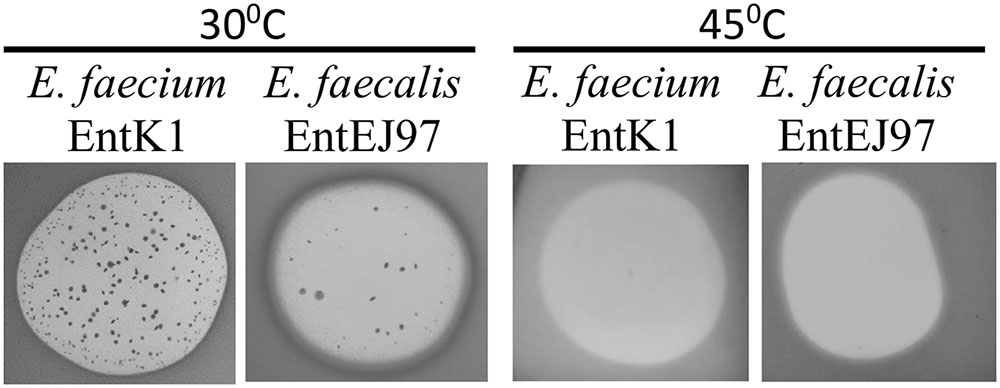
FIGURE 4. EntK1 and EntEJ97 resistant mutants appear on at 30°C but not at the elevated temperature 45°C. Bacteriocin (20 μl of 1 mg/ml) was applied to soft agar containing indicator cells on plates, which was then incubated overnight at indicated temperatures for growth and developing the inhibition zones and resistant colonies.
Discussion
Structural analyses by CD spectroscopy showed that EntK1 was unstructured in water but became structured when exposed to membrane-mimicking environments (DPC-micelles or TFE). This is a common property for the majority of bacteriocins (Montalban-Lopez et al., 2008; Martin-Visscher et al., 2009).
The structure of EntK1 in 50% TFE is very similar to that of LsbB (Figure 2). Both bacteriocins have an N-terminal part mostly composed of an amphiphilic α-helical motif, and an unstructured C-terminal half. EntK1 α-helix is longer than the one of LsbB, being 16–19 residues, versus 13–15 residues in LsbB. A closer look at the α-helices of EntK1 and LsbB showed that both are amphiphilic with the basic amino acids along one side of the helix and non-polar residues along the other side (data not shown). Such amphiphilic distribution is common for many bacteriocins as well as other antimicrobial peptides (Kristiansen et al., 2005; Rogne et al., 2008; Last and Miranker, 2013). The α-helical part of the bacteriocins is probably involved in pore-formation, where the hydrophobic part is facing the hydrophobic membrane or a membrane-located protein (receptor) while the hydrophilic part is facing the inner part of the pore to cause cellular leakage (Kjos et al., 2011).
The C-terminal unstructured parts of EntK1 and LsbB are about the same lengths, 10–13 residues and more similar in sequence. Structure prediction servers (PONDR®VL-XT, JPred 4) indicated that EntEJ97 also contains an α-helix from Lys10 to Gly 35 with an unstructured C-terminal tail of 10 residues, which corresponds with our data on LsbB and EntK1 structures.
Besides EntK1 and LsbB, structures of three other leaderless bacteriocins have been obtained so far (enterocin 7, lacticin Q, and aureocin A53). Enterocin 7 is a two-peptide bacteriocin, where the two peptides constituting the bacteriocin are related in sequence and both have a similar fold, consisting of N-terminal, middle and C-terminal helices (Lohans et al., 2013). Lacticin Q and aureocin A53 both consist of four helices surrounding a hydrophobic core (Acedo et al., 2016). The structures of enterocin 7, lacticin Q, and aureocin A53 resemble the structure of circular bacteriocins, and all of them are highly structured in aqueous solutions. As such, EntK1 and LsbB are very different from these bacteriocins by being smaller (30 and 37 residues for LsbB and K1 versus 43–53 residues for the others), unstructured in water (partly due to the absence of any modifications such as disulphide bridges or circularization in these leaderless bacteriocins) and containing only one helix (Figure 2).
The lactococcal Zn-dependent metallopeptidase RseP has been shown to be the receptor for LsbB in L. lactis IL1403 (Uzelac et al., 2013; Miljkovic et al., 2016). Our present study provides strong evidence that EntEJ97 and EntK1 employ the homologous RseP in enterococci as receptor. This is based on the fact that mutants of E. faecalis LMG3358 and E. faecium LMG2787 that are HR to EntEJ97/EntK1, contain frameshift mutations in rseP and, more conclusively, heterologous expression of the enterococcal rseP rendered resistant pneumococcal cells sensitive to these bacteriocins (Table 5). However, at least in the case of E. faecalis LMG3358, we also found some EntEJ97-resistant mutants, which were less resistant, compared to those with mutated rseP and had intact rseP, suggesting that these probably had mutations in other gene(s), see discussion below. Nevertheless, both types of resistance were specific to EntEJ97 and EntK1 because all resistant cells were sensitive to the non-related bacteriocins BHT-B (leaderless type) and garvicin ML (circular type). Furthermore, both (high and low) types of resistance share one common feature, that they emerged at the standard growth temperature 30°C but not at the elevated 45°C, indicating that both resistance types are linked to stress response (Figure 4). Indeed, RseP has been shown to be involved in stress response in some bacteria. In Escherichia coli, cell envelope stress response leads to proteolytic cleavage of the membrane-bound anti-sigma protein RseA by the RseP protease that eventually leads to σE release in the cytoplasm to transcribe stress response genes (Kroos and Akiyama, 2013; Hizukuri et al., 2014; Akiyama et al., 2015). Similarly, in E. faecalis, RseP (also known as Eep) is involved in the stress response via degradation of the anti-sigma protein RsiV. This degradation leads to activation of SigV – enterococcal extracytoplasmic sigma protein that plays a key role in stress response (Varahan et al., 2013). Furthermore, RseP deletion mutants have shown increased susceptibility toward heat, ethanol, lysozyme, and acid compared to the susceptibility of the WT strain (Varahan et al., 2013).
Interestingly, RseP of E. faecalis is also involved in a proteolysis of the sex pheromone precursor peptide to form an active pheromone (cCF10) which is released by a cognate ABC transporter, called PptAB. It belongs to the EcsAB family and is composed of two proteins – PptA and PptB (Varahan et al., 2014). It has also been shown in Bacillus subtilis that mutations within EcsAB blocked the activity of RasP (Bacillus homolog of enterococcal RseP) and made the cells sensitive to stress (Heinrich et al., 2008). Remarkably, when we did whole genome sequencing of the five E. faecalis LR mutants with intact rseP gene, we discovered frequent mutations within genes pptA and pptB (preliminary results). Moreover, in EntEJ97 LR mutants of Pediococcus acidilactici and Lactococcus garvieae, all with an intact rseP gene also had mutations in ecsA and ecsB (data not shown).
Thus, our results also revealed a functional connection between these two systems: RseP and EcsAB, that both are involved in the sensitivity to our bacteriocins. As mentioned elsewhere, YvjB (a homolog of RseP) serves as receptor for LsbB which binds directly to the receptor upon killing target cells (Miljkovic et al., 2016). At present it is not clear the exact role of EcsAB in the sensitivity to bacteriocins. Whether it is directly involved as a receptor as described for RseP or in regulation of RseP function or gene expression awaits further investigation.
Members of the RseP protein family are membrane-bound proteins, most if not all, containing multiple transmembrane helices. The putative active site of RseP proteases is normally located within the hydrophobic plane of the membrane and contains a consensus sequence motif HExxH, in which the two histidine residues are thought to coordinate a zinc atom together with a conserved glutamate residue (Feng et al., 2007; Koide et al., 2007; Rawson, 2013). Our results indicate that the active site of RseP is somehow involved in the sensitivity to the bacteriocins. This is based on the observation that replacing of the conserved residues with alanine residues in E. faecalis RseP made the cloned cells less sensitive to EntEJ97 and EntK1 especially when all three conserved residues were replaced. This finding is different from other known bacteriocin receptor systems. Substitution mutations of conserved active site residues in LsrS – Zn-dependent membrane protein, serving as receptor for the two-peptide lantibiotic bacteriocin Smb, had no effect on cells sensitivity to the bacteriocin (Biswas and Biswas, 2014). Similarly, the man-PTS membrane-located components IIC and IID can serve as a receptor for pediocin-like bacteriocins even without its cytosolic component (IIAB) which is needed for functional sugar transport (Diep et al., 2007). How the active site of RseP involved in the sensitivity to EntK1/EntEJ95 is still elusive. The structure of RseP homologous protein from archaebacterial species Methanocaldococcus jannaschii showed that it is quite dynamic during the proteolytic reaction. The protein displays structural changes including the movement of the transmembrane helices to bring the substrate (a peptide or part of a protein) into the active site and to release the digested product after proteolysis (Feng et al., 2007). It is tempting to speculate that site-directed mutations in the active site of RseP might have altered its 3D-structure, making it less accessible for the bacteriocins. Other possibilities might also exist. For example, these mutations could somehow create a less stable or incorrectly folded RseP protein that was removed from cells by chaperone proteases.
The study of EntK1 and EntEJ97 inhibition spectrum showed an interesting detail – EntK1 was less active and had generally narrower antibacterial spectrum than EntEJ97. However, against E. faecium strains EntK1 was much more potent (i.e., lower MIC values) than EntEJ97. The latter was equally active against E. faecalis and E. faecium (Table 4). Since EntK1 is produced by E. faecium and EntEJ97 by E. faecalis, this difference is in line with the general characteristic of bacteriocins, namely that they are most active against species closely related to the producers. This narrow-spectrum activity is not well understood in terms of mechanisms, but likely has an ecological meaning, e.g, giving the bacteriocin producer an advantage in competition for nutrients or habitats over its closest competitors. However, multiple sequence alignment analysis (using Clustal Omega) of many RseP (30 from E. faecalis and 30 from E. faecium) did not show any pronounced differences between the two RseP groups (data not shown). Thus, what properties of RseP that define the different levels of sensitivity toward EntEJ97 and EntK1 awaits further investigation.
During the past few decades, enterococci have emerged as important healthcare-associated pathogens due to their resistance to antibiotics and environmental stress factors (Arias and Murray, 2012). The rapid spread of E. faecium with resistance to vancomycin (VRE), ampicillin and high-levels of aminoglycosides is of particular concern (Agudelo Higuita and Huycke, 2014). E. faecium is now a nosocomial pathogen as common as E. faecalis (Arias and Murray, 2012). Recently RseP was shown to be a major virulence determinant in enterococcal rabbit endocarditis model (Frank et al., 2012). The importance of RseP in virulence is most likely due to its role in stress response (Varahan et al., 2013). Therefore, EntK1 and EntEJ97, both targeting enterococcal RseP, can be used in combination with stress factors to effectively kill antibiotic-resistant enterococci including VRE.
Author Contributions
KO: NMR of EntK1, heterologous expression of rseP, MIC assays, and paper writing. PK: NMR of EntK1. DS: heterologous expression of rseP. MJ: rseP mutant characterization, MIC assays. TA-P: paper writing. IN: paper writing. DD: project leadership and paper writing.
Funding
KO and DD were supported by the grant (#254784/O70) from The Research Council of Norway. This work was partially supported by the grant no. 2013/08/M/NZ9/01025 from Polish National Science Centre.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Professor Rob J. L. Willems (University Medical Center Utrecht, Utrecht, the Netherlands) for providing us with E. faecium isolates from human infections.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00774/full#supplementary-material
References
Acedo, J. Z., van Belkum, M. J., Lohans, C. T., Towle, K. M., Miskolzie, M., and Vederas, J. C. (2016). Nuclear magnetic resonance solution structures of lacticin Q and aureocin A53 reveal a structural motif conserved among leaderless bacteriocins with broad-spectrum activity. Biochemistry 55, 733–742. doi: 10.1021/acs.biochem.5b01306
Agudelo Higuita, N. I., and Huycke, M. M. (2014). “Enterococcal disease, epidemiology, and implications for treatment,” in Enterococci: From Commensals to Leading Causes of Drug Resistant Infection, eds M. S. Gilmore, D. B. Clewell, Y. Ike, and N. Shankar (Boston, MA: Massachusetts Eye and Ear Infirmary).
Akiyama, K., Mizuno, S., Hizukuri, Y., Mori, H., Nogi, T., and Akiyama, Y. (2015). Roles of the membrane-reentrant beta-hairpin-like loop of RseP protease in selective substrate cleavage. Elife 4:e08928. doi: 10.7554/eLife.08928
Alba, B. M., Leeds, J. A., Onufryk, C., Lu, C. Z., and Gross, C. A. (2002). DegS and YaeL participate sequentially in the cleavage of RseA to activate the sigma(E)-dependent extracytoplasmic stress response. Genes Dev. 16, 2156–2168. doi: 10.1101/gad.1008902
Arias, C. A., and Murray, B. E. (2012). The rise of the Enterococcus: beyond vancomycin resistance. Nat. Rev. Microbiol. 10, 266–278. doi: 10.1038/nrmicro2761
Basanta, A., Sanchez, J., Gomez-Sala, B., Herranz, C., Hernandez, P. E., and Cintas, L. M. (2008). Antimicrobial activity of Enterococcus faecium L50, a strain producing enterocins L50 (L50A and L50B), P and Q, against beer-spoilage lactic acid bacteria in broth, wort (hopped and unhopped), and alcoholic and non-alcoholic lager beers. Int. J. Food Microbiol. 125, 293–307. doi: 10.1016/j.ijfoodmicro.2008.04.011
Berg, K. H., Biornstad, T. J., Straume, D., and Havarstein, L. S. (2011). Peptide-regulated gene depletion system developed for use in Streptococcus pneumoniae. J. Bacteriol. 193, 5207–5215. doi: 10.1128/jb.05170-11
Biswas, S., and Biswas, I. (2014). A conserved streptococcal membrane protein, LsrS, exhibits a receptor-like function for lantibiotics. J. Bacteriol. 196, 1578–1587. doi: 10.1128/jb.00028-14
Borrero, J., Brede, D. A., Skaugen, M., Diep, D. B., Herranz, C., Nes, I. F., et al. (2011). Characterization of garvicin ML, a novel circular bacteriocin produced by Lactococcus garvieae DCC43, isolated from mallard ducks (Anas platyrhynchos). Appl. Environ. Microbiol. 77, 369–373. doi: 10.1128/aem.01173-10
Braunschweiler, L., and Ernst, R. R. (1983). Coherence transfer by isotropic mixing: application to proton correlation spectroscopy. J. Magn. Reson. 53, 521–528. doi: 10.1016/0022-2364(83)90226-3
Brown, E. D., and Wright, G. D. (2016). Antibacterial drug discovery in the resistance era. Nature 529, 336–343. doi: 10.1038/nature17042
Cintas, L. M., Casaus, P., Havarstein, L. S., Hernandez, P. E., and Nes, I. F. (1997). Biochemical and genetic characterization of enterocin P, a novel sec-dependent bacteriocin from Enterococcus faecium P13 with a broad antimicrobial spectrum. Appl. Environ. Microbiol. 63, 4321–4330.
Cintas, L. M., Casaus, P., Herranz, C., Havarstein, L. S., Holo, H., Hernandez, P. E., et al. (2000). Biochemical and genetic evidence that Enterococcus faecium L50 produces enterocins L50A and L50B, the sec-dependent enterocin P, and a novel bacteriocin secreted without an N-terminal extension termed enterocin Q. J. Bacteriol. 182, 6806–6814. doi: 10.1128/JB.182.23.6806-6814.2000
Cotter, P. D. (2014). An ‘Upp’-turn in bacteriocin receptor identification. Mol. Microbiol. 92, 1159–1163. doi: 10.1111/mmi.12645
Cotter, P. D., Ross, R. P., and Hill, C. (2013). Bacteriocins - a viable alternative to antibiotics? Nat. Rev. Microbiol. 11, 95–105. doi: 10.1038/nrmicro2937
Davis, A. L., Keeler, J., Laue, E. D., and Moskau, D. (1992). Experiments for recording pure absorption heteronuclear correlation spectra using pulsed field gradients. J. Magn. Reson. 98, 207–216. doi: 10.1016/0022-2364(92)90126-r
Diep, D. B., Skaugen, M., Salehian, Z., Holo, H., and Nes, I. F. (2007). Common mechanisms of target cell recognition and immunity for class II bacteriocins. Proc. Natl. Acad. Sci. U.S.A. 104, 2384–2389. doi: 10.1073/pnas.0608775104
Feng, L., Yan, H., Wu, Z., Yan, N., Wang, Z., Jeffrey, P. D., et al. (2007). Structure of a site-2 protease family intramembrane metalloprotease. Science 318, 1608–1612. doi: 10.1126/science.1150755
Fischbach, M. A., and Walsh, C. T. (2009). Antibiotics for emerging pathogens. Science 325, 1089–1093. doi: 10.1126/science.1176667
Frank, K. L., Barnes, A. M., Grindle, S. M., Manias, D. A., Schlievert, P. M., and Dunny, G. M. (2012). Use of recombinase-based in vivo expression technology to characterize Enterococcus faecalis gene expression during infection identifies in vivo-expressed antisense RNAs and implicates the protease Eep in pathogenesis. Infect. Immun. 80, 539–549. doi: 10.1128/iai.05964-11
Gajic, O., Buist, G., Kojic, M., Topisirovic, L., Kuipers, O. P., and Kok, J. (2003). Novel mechanism of bacteriocin secretion and immunity carried out by lactococcal multidrug resistance proteins. J. Biol. Chem. 278, 34291–34298. doi: 10.1074/jbc.M211100200
Galvez, A., Valdivia, E., Abriouel, H., Camafeita, E., Mendez, E., Martinez-Bueno, M., et al. (1998). Isolation and characterization of enterocin EJ97, a bacteriocin produced by Enterococcus faecalis EJ97. Arch. Microbiol. 171, 59–65. doi: 10.1007/s002030050678
Guntert, P., Mumenthaler, C., and Wuthrich, K. (1997). Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273, 283–298. doi: 10.1006/jmbi.1997.1284
Havarstein, L. S., Diep, D. B., and Nes, I. F. (1995). A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol. Microbiol. 16, 229–240. doi: 10.1111/j.1365-2958.1995.tb02295.x
Heinrich, J., Lunden, T., Kontinen, V. P., and Wiegert, T. (2008). The Bacillus subtilis ABC transporter EcsAB influences intramembrane proteolysis through RasP. Microbiology 154(Pt 7), 1989–1997. doi: 10.1099/mic.0.2008/018648-0
Herrmann, T., Guntert, P., and Wuthrich, K. (2002). Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 319, 209–227. doi: 10.1016/S0022-2836(02)00241-3
Higuchi, R., von Beroldingen, C. H., Sensabaugh, G. F., and Erlich, H. A. (1988). DNA typing from single hairs. Nature 332, 543–546. doi: 10.1038/332543a0
Hizukuri, Y., Oda, T., Tabata, S., Tamura-Kawakami, K., Oi, R., Sato, M., et al. (2014). A structure-based model of substrate discrimination by a noncanonical PDZ tandem in the intramembrane-cleaving protease RseP. Structure 22, 326–336. doi: 10.1016/j.str.2013.12.003
Holo, H., Nilssen, O., and Nes, I. F. (1991). Lactococcin A, a new bacteriocin from Lactococcus lactis subsp. cremoris: isolation and characterization of the protein and its gene. J. Bacteriol. 173, 3879–3887. doi: 10.1128/jb.173.12.3879-3887.1991
Hurd, R. E. (1991). Gradient enhanced proton-detected heteronuclear multiple quantum coherence: GE HMQC. J. Magn. Reson. 91, 648–653.
Hyink, O., Balakrishnan, M., and Tagg, J. R. (2005). Streptococcus rattus strain BHT produces both a class I two-component lantibiotic and a class II bacteriocin. FEMS Microbiol. Lett. 252, 235–241. doi: 10.1016/j.femsle.2005.09.003
Jeener, J., Meier, B. H., Bachmann, P., and Ernst, R. R. (1979). Investigation of exchange processes by 2-dimensional NMR-spectroscopy. J. Magn. Reson. 71, 4546–4553. doi: 10.1063/1.438208
Johnsborg, O., and Håvarstein, L. S. (2009). Pneumococcal LytR, a protein from the LytR-CpsA-Psr family, is essential for normal septum formation in Streptococcus pneumoniae. J. Bacteriol. 191, 5859–5864. doi: 10.1128/JB.00724-09
Johnsborg, O., Eldholm, V., Bjornstad, M. L., and Havarstein, L. S. (2008). A predatory mechanism dramatically increases the efficiency of lateral gene transfer in Streptococcus pneumoniae and related commensal species. Mol. Microbiol. 69, 245–253. doi: 10.1111/j.1365-2958.2008.06288.x
Kim, D. Y. (2015). Two stress sensor proteins for the expression of sigmaE regulon: DegS and RseB. J. Microbiol. 53, 306–310. doi: 10.1007/s12275-015-5112-6
Kjos, M., Borrero, J., Opsata, M., Birri, D. J., Holo, H., Cintas, L. M., et al. (2011). Target recognition, resistance, immunity and genome mining of class II bacteriocins from Gram-positive bacteria. Microbiology 157(Pt 12), 3256–3267. doi: 10.1099/mic.0.052571-0
Koide, K., Maegawa, S., Ito, K., and Akiyama, Y. (2007). Environment of the active site region of RseP, an Escherichia coli regulated intramembrane proteolysis protease, assessed by site-directed cysteine alkylation. J. Biol. Chem. 282, 4553–4560. doi: 10.1074/jbc.M607339200
Koradi, R., Billeter, M., and Wuthrich, K. (1996). MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graphics 14, 51–55. doi: 10.1016/0263-7855(96)00009-4
Kristiansen, P. E., Fimland, G., Mantzilas, D., and Nissen-Meyer, J. (2005). Structure and mode of action of the membrane-permeabilizing antimicrobial peptide pheromone plantaricin A. J. Biol. Chem. 280, 22945–22950. doi: 10.1074/jbc.M501620200
Kroos, L., and Akiyama, Y. (2013). Biochemical and structural insights into intramembrane metalloprotease mechanisms. Biochim. Biophys. Acta 1828, 2873–2885. doi: 10.1016/j.bbamem.2013.03.032
Last, N. B., and Miranker, A. D. (2013). Common mechanism unites membrane poration by amyloid and antimicrobial peptides. Proc. Natl. Acad. Sci. U.S.A. 110, 6382–6387. doi: 10.1073/pnas.1219059110
Lohans, C. T., Towle, K. M., Miskolzie, M., McKay, R. T., van Belkum, M. J., McMullen, L. M., et al. (2013). Solution structures of the linear leaderless bacteriocins enterocin 7A and 7B resemble carnocyclin A, a circular antimicrobial peptide. Biochemistry 52, 3987–3994. doi: 10.1021/bi400359z
Martin-Visscher, L. A., Gong, X., Duszyk, M., and Vederas, J. C. (2009). The three-dimensional structure of carnocyclin A reveals that many circular bacteriocins share a common structural motif. J. Biol. Chem. 284, 28674–28681. doi: 10.1074/jbc.M109.036459
Matthews, B., Nicholson, H., and Becktel, W. (1992). The chemical shift index: a fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry 31, 1647–1651. doi: 10.1021/bi00121a010
Miljkovic, M., Uzelac, G., Mirkovic, N., Devescovi, G., Diep, D. B., Venturi, V., et al. (2016). LsbB bacteriocin interacts with the third transmembrane domain of the YvjB receptor. Appl. Environ. Microbiol. 82, 5364–5374. doi: 10.1128/aem.01293-16
Montalban-Lopez, M., Spolaore, B., Pinato, O., Martinez-Bueno, M., Valdivia, E., Maqueda, M., et al. (2008). Characterization of linear forms of the circular enterocin AS-48 obtained by limited proteolysis. FEBS Lett. 582, 3237–3242. doi: 10.1016/j.febslet.2008.08.018
Nes, I. F., Diep, D. B., Havarstein, L. S., Brurberg, M. B., Eijsink, V., and Holo, H. (1996). Biosynthesis of bacteriocins in lactic acid bacteria. Antonie Van Leeuwenhoek 70, 113–128. doi: 10.1007/BF00395929
Nes, I. F., Diep, D. B., and Ike, Y. (2014). “Enterococcal bacteriocins and antimicrobial proteins that contribute to niche control,” in Enterococci: From Commensals to Leading Causes of Drug Resistant Infection, eds M. S. Gilmore, D. B. Clewell, Y. Ike, and N. Shankar (Boston, MA: Massachusetts Eye and Ear Infirmary).
Ovchinnikov, K. V., Kristiansen, P. E., Uzelac, G., Topisirovic, L., Kojic, M., Nissen-Meyer, J., et al. (2014). Defining the structure and receptor binding domain of the leaderless bacteriocin LsbB. J. Biol. Chem. 289, 23838–23845. doi: 10.1074/jbc.M114.579698
Rawson, R. B. (2013). The site-2 protease. Biochim. Biophys. Acta 1828, 2801–2807. doi: 10.1016/j.bbamem.2013.03.031
Rogne, P., Fimland, G., Nissen-Meyer, J., and Kristiansen, P. E. (2008). Three-dimensional structure of the two peptides that constitute the two-peptide bacteriocin lactococcin G. Biochim. Biophys. Acta 1784, 543–554. doi: 10.1016/j.bbapap.2007.12.002
Scholtz, J. M., Qian, H., York, E. J., Stewart, J. M., and Baldwin, R. L. (1991). Parameters of helix-coil transition theory for alanine-based peptides of varying chain lengths in water. Biopolymers 31, 1463–1470. doi: 10.1002/bip.360311304
Shen, Y., and Bax, A. (2013). Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J. Biomol. NMR 56, 227–241. doi: 10.1007/s10858-013-9741-y
Smith, R., and Coast, J. (2013). The true cost of antimicrobial resistance. BMJ 346:f1493. doi: 10.1136/bmj.f1493
Sung, C. K., Li, H., Claverys, J. P., and Morrison, D. A. (2001). An rpsL cassette, janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl. Environ. Microbiol. 67, 5190–5196. doi: 10.1128/aem.67.11.5190-5196.2001
Uzelac, G., Kojic, M., Lozo, J., Aleksandrzak-Piekarczyk, T., Gabrielsen, C., Kristensen, T., et al. (2013). A Zn-dependent metallopeptidase is responsible for sensitivity to LsbB, a class II leaderless bacteriocin of Lactococcus lactis subsp. lactis BGMN1-5. J. Bacteriol. 195, 5614–5621. doi: 10.1128/JB.00859-13
Varahan, S., Harms, N., Gilmore, M. S., Tomich, J. M., and Hancock, L. E. (2014). An ABC transporter is required for secretion of peptide sex pheromones in Enterococcus faecalis. mBio 5:e1726-14. doi: 10.1128/mBio.01726-14
Varahan, S., Iyer, V. S., Moore, W. T., and Hancock, L. E. (2013). Eep confers lysozyme resistance to Enterococcus faecalis via the activation of the extracytoplasmic function sigma factor SigV. J. Bacteriol. 195, 3125–3134. doi: 10.1128/jb.00291-13
Wishart, D. S., Bigam, C. G., Yao, J., Abildgaard, F., Dyson, H. J., Oldfield, E., et al. (1995). 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J. Biomol. NMR 6, 135–140. doi: 10.1007/BF00211777
Keywords: leaderless bacteriocins, NMR, receptors, RseP, enterococci
Citation: Ovchinnikov KV, Kristiansen PE, Straume D, Jensen MS, Aleksandrzak-Piekarczyk T, Nes IF and Diep DB (2017) The Leaderless Bacteriocin Enterocin K1 Is Highly Potent against Enterococcus faecium: A Study on Structure, Target Spectrum and Receptor. Front. Microbiol. 8:774. doi: 10.3389/fmicb.2017.00774
Received: 01 March 2017; Accepted: 18 April 2017;
Published: 03 May 2017.
Edited by:
Bart Devreese, Ghent University, BelgiumReviewed by:
John Christopher Vederas, University of Alberta, CanadaTaoufik Ghrairi, Tunis El Manar University, Tunisia
Copyright © 2017 Ovchinnikov, Kristiansen, Straume, Jensen, Aleksandrzak-Piekarczyk, Nes and Diep. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dzung B. Diep, dzung.diep@nmbu.no
 Kirill V. Ovchinnikov
Kirill V. Ovchinnikov Per Eugen Kristiansen2
Per Eugen Kristiansen2 Tamara Aleksandrzak-Piekarczyk
Tamara Aleksandrzak-Piekarczyk Dzung B. Diep
Dzung B. Diep