- 1Major in Food Biotechnology, Division of Bioindustry, Silla University, Busan, South Korea
- 2Department of Korean Cusine, Jeonju University, Jeonju, South Korea
- 3School of Applied Biosciences, Kyungpook National University, Daegu, South Korea
- 4Research Center for Extremophiles and Marine Microbiology, Silla University, Busan, South Korea
- 5Microbiology and Functionality Research Group, World Institute of Kimchi, Gwangju, South Korea
Salting is one of the oldest food preservation techniques. However, salt is also the source of living halophilic microorganisms that may affect human health. In order to determine the microbial communities of commercial salts, an investigation were done using amplicon sequencing approach in four commercial salts: Ethiopian Afdera salt (EAS), Ethiopian rock salt (ERS), Korean Jangpan salt (KJS), and Korean Topan salt (KTS). Using domain-specific primers, a region of the 16S rRNA gene was amplified and sequenced using a Roche 454 instrument. The results indicated that these microbial communities contained 48.22–61.4% Bacteria, 37.72–51.26% Archaea, 0.51–0.86% Eukarya, and 0.005–0.009% unclassified reads. Among bacteria, the communities in these salts were dominated by the phyla Proteobacteria, Bacteroidetes, Actinobacteria, and Firmicutes. Of the archaea, 91.58% belonged to the class Halobacteria, whereas the remaining 7.58, 0.83, and 0.01% were Nanoarchaea, Methanobacteria, and Thermococci, respectively. This comparison of microbial diversity in salts from two countries showed the presence of many archaeal and bacterial genera that occurred in salt samples from one country but not the other. The bacterial genera Enterobacter and Halovibrio were found only in Korean and Ethiopian salts, respectively. This study indicated the occurrence and diversity of halophilic bacteria and archaea in commercial salts that could be important in the gastrointestinal tract after ingestion.
Introduction
Salting is one of the oldest food preservation methods, which is usually regarded as a technique to control microbial activity. However, some studies revealed that salt itself contains a wide range of halophilic organisms, including bacteria and archaea that are unique to these environments (Henriet et al., 2014) as well as salted and fermented foods (Park et al., 2009; Roh et al., 2010). In recent years, novel halophilic archaea namely Halopiger thermotololerans (Minegishi et al., 2016), Haloparvum alkalitolerans (Kondo et al., 2016), and Halarchaeum grantii (Shimane et al., 2015) were isolated from commercial salts using culture-based methods. Indeed, a variety of halophilic archaea were found in high salt-fermented foods and human intestines, suggesting that halophiles play an important role in food processing and preparation (Lee, 2013). Analogously, high-throughput microbial community studies demonstrated that diverse bacteria and archaea were also found in hypersaline or saline environments such as ancient halite (Schubert et al., 2010), estuarine sediments (Webster et al., 2015), lacustrine sediments (Gugliandolo et al., 2015), saline lakes (Li et al., 2015), saline soil (Walsh et al., 2005), seawater (Bougouffa et al., 2013), and solar salterns (Mutlu and Guven, 2015). In addition, there were isolation of archaea from primary salt crystal (Vreeland et al., 2007), middle late Eocene rock salt (Jaakkola et al., 2014), ancient halite (Schubert et al., 2010), and bacteria from primary salt crystal (Vreeland et al., 2000; Satterfield et al., 2005). In light of this, ecological and microbiological studies of commercial salts are of special interest because these can provide insights into the correlative microbial dynamics between hypersaline environments and salted and fermented foods. However, the whole microbiota of commercial salts still remains unclear.
Korean traditional foods like kimchi, seasoning pastes, soy sauce, and salted seafoods are seasoned exclusively with Shinan solar salt to maintain their taste and flavor. Some physicochemical and sensory characteristics of Korean fermented foods salted with domestic solar salts are distinct from those with imported salts. This might be a result of the specific microbiota in various salts as well as mineral concentration, although this speculation requires further investigation. The Danakil depression is one of the most ancient salt-producing areas in the world, and it is still the source of salt for more than 100 million people in Africa, especially in Ethiopia, Eritrea, and Djibouti. In addition, the area represents an unexplored hypersaline ecosystem that is unique in its geology, climate, and landscape features.
To our knowledge, there is no study on the occurrence and diversity of microbial communities in commercial salts from these two countries. These reasons tempted us to investigate and compare the composition of the microbial community and diversity in salts. To study the composition of microbial community and its diversity, a culture-independent amplicon sequencing approach seems to be more sensitive and effective than a conventional culture-dependent method (Streit and Schmitz, 2004; Bik, 2014). Several metagenomic studies revealed a fair number of microbial communities that were not found in a culture-based approach by the next-generation sequencing with the Roche/454 GSFLX Titanium platform (Streit and Schmitz, 2004; Jung and Kang, 2014; Suh et al., 2015). Such metagenomic studies can provide a comprehensive understanding of microbial survival and adaptation strategies in extreme environments and special ecosystems (Fierer et al., 2012; Neelakanta and Sultana, 2013). Here, we chose four kinds of commercial salts from two different areas in Korea (Topan and Jangpan) and Ethiopia (Afdera Lake and rock salt) to compare their microbial communities using a culture-independent approach and discuss the microbial diversity and community structure in salts from these regions.
Materials and Methods
Site Description and Sample Collection
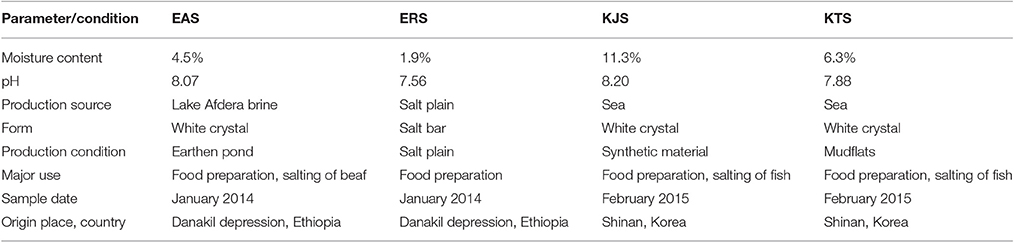
Four commercially available salts, namely, Afdera, rock, Topan, and Jangpan samples, were collected directly from the companies that produce and distribute the salts in Ethiopia (Afar Salt and Bashenfer Salt) and Korea (Sumdleche Co. Korea). They were stored at room temperature in a dark and dry place until they were used in experiments. The Afdera and rock salts were from Ethiopia, and Topan and Jangpan were from Korea. The salts were designated based on the production process, source, and country of origin: Ethiopia Afdera Salt (EAS), Ethiopian Rock Salt (ERS), Korean Topan Salt (KTS), and Korean Jangpan Salt (KJS). The Ethiopian salts were produced in the Danakil depression, and the Korean salts were produced in Shinan, Korea. Shinan is located in the southeast of Korea at latitude 35° 12″ 30′ north and longitude 126° 23″ 00′ east, and has a length from east to west of 79.3 km and south to north of 65.6 km. The average temperature in January is 2–4°C and in August is 24°C. The average annual temperature is 14°C, and average annual rainfall is 800–1,000 mm. Two types of sun-dried salts, KTS and KJS are being produced in Shinan area. KTS is dried on the hardened mud-flat and KJS on the plastic material like polyvinyl chloride (PVC). The Danakil depression is located in Afar, Ethiopia, Africa with coordinates of 14° 14′ 30.1″ (14.2417°) north (latitude) and 40° 18′ (40.3°) east (longitude). It is within the Great Rift Valley in northeastern Ethiopian, to the southeast of Eritrea and of west Djibouti. It covers about 5,000 km2, the center of which is known as the Dallol depression and is 120 m below sea level. It is one of the hottest and most inhospitable places on earth (Morel, 2005). Danakil has the highest average temperatures on Earth, exceeding 34°C every day of the year and reaching 55°C in the summer.
Moisture and pH Determination
The salt samples were ground using a mortar and pestle, and 5 g of the salt powder was placed in an oven at 105°C until it reached a constant weight, and then, it was allowed to cool. The moisture content was calculated based on the weight difference before and after baking. Ten grams of salt was mixed with 50 ml of distilled water (DW) in a ratio of 1:5, and the pH was measured after complete dissolution.
DNA Extraction
Microbial DNA extraction from salt samples was carried out using a slightly modified method as described in Henriet et al. (2014). Afdera salt was used as a model. The three basic extraction steps tested were (i) dissolution of stored salt sample with DW, (ii) cell concentration by centrifugation, and (iii) bead beating, chemical lysis, and DNA recovery. A 10 g sample of salt was dissolved to 50 ml of DW until complete dissolution. As the microbial content in commercial salt is very low, the cells with soils were concentrated by centrifugation (5 min, 10,000 × g and 4°C). Cell bead beating, chemical lysis, and DNA recovery were achieved with PowerSoil DNA isolation kit (MO-Bio Laboratories Inc., Carlsbad, CA, USA) following the manufacturer's instructions. A Quawell Q3000 UV spectrophotometer (San Jose, USA) was used to quantify the DNA.
Polymerase Chain Reaction (PCR) and Pyrosequencing
Standard methods were used to enumerate the approximate bacterial and archaeal loads in the four metagenomic samples. Purified DNA was amplified using primers targeting the V1–V3 regions of the prokaryotic 16S rRNA gene. The primers used for bacteria were V1-9F (5′-CCTATCCCCTGTGTGCCTTGGCAGTCTCAGACGAGTTTGATCMTGGCTCAG-3′); and V3-541R (5′-CCATCTCATCCCTGCGTGTCTCCGACTCAGXACWTTACCGCGGCTGCT-GG-3′) (Chun et al., 2010). The primers used for archaeal organisms were AV1-21F (5′-CCTATCCCCTGTGTGCCTTGGCAGTCTCAGAGTCCGGTTGATCCYGCCGG-3′) and AV3-519R (5′-CCATCTCATCCCTGCGTGTCTCCGACTCAGXGAGGTDTTACCGCGGCK-GCTG-3′) (Hur et al., 2011). The X denotes a 7–11 nucleotide barcode that was unique for each subject, followed by a common linker for bacteria (AC) and archaea (AG), and underlining indicates the gene specific section. PCR amplification was performed in a 50 μl volume containing 0.5 μl of 5 U/μl Taq DNA polymerase, 5 μl of 10 × PCR reaction buffer, 4 μl of 0.2 mM dNTP mix, 2 μl of each primer, and 1 μl of template DNA with the following thermal cycler (PTC-200 DNA Engine; MJ Research, CA, USA) conditions: initial denaturation at 95°C for 5 min; 30 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 60 s; with a final extension at 72°C for 7 min. The size and the homogeneity of PCR products were confirmed by gel electrophoresis. PCR products were purified using a QIAquick PCR Purification Kit (Qiagen, Hilden, Germany). Bands of less than 300 base pairs were removed using a QIAquick Gel Extraction Kit (Qiagen) in a subsequent gel electrophoresis. The DNA sequencing was performed by Chunlab, Inc. (Seoul, Korea) with a Roche/454 GS FLX Titanium platform (454 Life Sciences, Roche Branford, USA) according to the manufacturer's instructions.
Sequence Data Processing and Statistical Analysis
The sequencing reads from the different samples were separated by unique barcodes. The sequences of the barcode, linker, and PCR primers were then removed from each side of the original sequencing reads. The resulting sequences were subjected to a filtering process where only reads containing 0 to 1 ambiguous base calls (Ns) and 300 or more base pairs were selected for the final bioinformatics analyses. Nonspecific PCR amplicons that showed no match with sequences in the 16S rRNA gene database by BLASTN search (expectation value of > e−5) were also removed from later analyses. Chimeric sequences were removed by analyzing the differences in BLASTN-based sequence similarity patterns between the first half and second half of a sequence. When the first and second halves were identified as sequences of different bacterial orders, the sequence was regarded as a chimera and eliminated (Edgar et al., 2011). The filtered reads of the 16S rRNA gene sequences were identified via BLAST search against the EzTaxon-e database (http://www.ezbiocloud.net/) (Chun et al., 2007), which contains 16S rRNA gene sequences of type strains with valid published names and representative species-level phylotypes for either cultured or uncultured entries in the GenBank public database, with complete hierarchical taxonomic classification from the phylum to the species level.
The pyrosequences from the samples were analyzed using CLcommunity 3.0 (Chunlab Inc., Seoul, Korea). Operational taxonomic units (OTUs) were defined with a 3% divergence threshold using the average neighbor-clustering algorithm. The cutoff value for assigning the sequence to a species-level phylotype was ≥97% similarity. The cutoff value for a genus-level cluster was ≥94% similarity. CD-HIT was used for massive clustering of amplified sequences (Fu et al., 2012), and corresponding graphical representations were generated using this program. Alpha diversity indices such as the Abundance-based Coverage Estimator (ACE), Chao1 estimator, Shannon diversity index, and Good's coverage estimator were calculated using Mothur software version 1.28.0 (Schloss et al., 2009) to estimate species richness. The accession numbers of the GenBank sequences with the highest similarity to the sequence cluster were used to assign a tentative genus, if the genus level sequence cluster was not identified as a valid bacterial or archaea genus. The overall phylogenetic distance between each pair of communities was estimated using the Fast UniFrac web interface (Hamady et al., 2010) and visualized by a principal coordinate analysis (PCoA). In addition, similarities with various metagenomic datasets (Asia and Europe seawater) were used to evaluate community composition. The seawater datasets are open resources provided by Chunlab Inc. and show microbial diversity of Asia and European seawater. Metagenomic sequences were deposited in MG-RAST (http://metagenomics.anl.gov/) under deposition numbers EAS (mgm4717086.3 & 4717097.3), ERS (mgm4717082.3 & 4717088), KJS (mgm4717099.3 & 4717080.3), and KTS (mgm4717084.3 & 4717095.3).
Results
Moisture, pH, and Production Process
The Ethiopian Afdera Lake salt (EAS) and rock salt (ERS) were sampled from Ethiopia in January 2014, and Korean Jangpan salt (KJS) and Topan salt (KTS) were sampled from Korea in February 2015. Earthen ponds, salt plains, mudflats, and synthetic materials were used for the production of EAS, ERS, KTS, and KJS, respectively. According to Table 1, KJS had the highest moisture content among the sampled salts, whereas the lowest was recorded for ERS. The pH range of salts was 7.56–8.20, which indicated that they were produced in alkaline environments.
Quantitative Characterization of Microbial Diversity
A total of 65,256 valid reads with an average read length of 414 base pairs were obtained (Table 2). The amplicons comprised 48.22–61.40% bacteria, 37.72–51.26% archaea, and 0.507–0.86% eukarya. A total of 0.005–0.009% of reads were considered unclassified. In prokaryotes, taxonomic classification and quantification of the amplicon sequencing data showed that the bacteria belonged to 24 described phyla and 7 unaffiliated phyla. The bacterial phyla Proteobacteria, Bacteroidetes, Actinobacteria, and Firmicutes were dominant. However, the proportion of each phylum varied greatly among the salts. For example, Proteobacteria were the most abundant, comprising 83.89, 46.55, and 46.54% in KJS, KTS, and EAS, respectively. The second most abundant phylum was Bacteriodetes, which represented a relatively high proportion of bacteria in EAS (39.86%), and smaller proportions in KTS (26.55%) and KJS (8.81%). The third most abundant phylum was Actinobacteria, which ranged in abundance from 27.07, 13.32, 2.06, and 1.79% in ERS, KTS, KJS, and EAS, respectively. In contrast, the Firmicutes represented a large percentage (27.22%) of the bacteria, which, in ERS, was slightly more abundant than the Proteobacteria (26.59%) and Actinobacteriodes (22.07%). Interestingly, ERS and EAS had unique phyla: Streptophyta, Chloropflexi, and Acidobacteria.
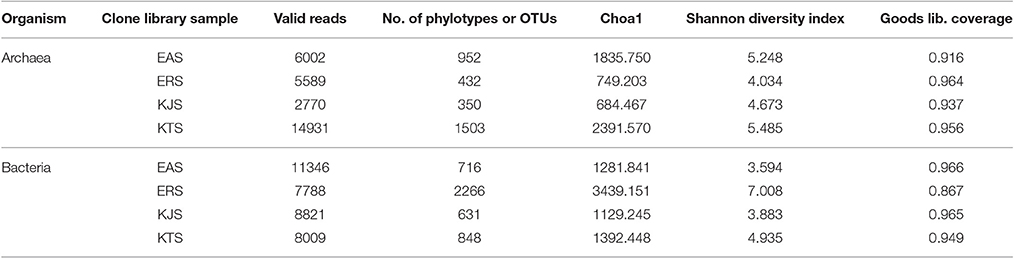
Table 2. Number of sequences and OTUs, Shannon diversity and Choa indices, coverage and valid number of reads of archaeal and bacterial metagenomics from samples.
Further classification at the class level indicated that the bacterial communities varied markedly among the salts. For instance, KTS and EAS were dominated by three classes: Gammaproteobacteria, Deltaproteobacteria, and the unaffiliated Rhodothermus-c. The majority of the reads belonged to the phyla Proteobacteria and Bacteroidetes, although Gammaproteobacteria were heavily represented in KJS (73.22%). In addition, unaffiliated Actinobacteria_uc (10.84%, phylum Actinobacteria) and Sphingobacteria (20.9%, phylum Bacteriodetes) were better represented in KTS and EAS, respectively. Intriguingly, the ERS bacterial community was dominated by unaffiliated Actinobacteria_c (25.23%), Bacilli (16.18%), and Clostridia (10.79%) in the phyla Actinobacteria and Firmicutes. Alphaproteobacteria in the phylum Proteobacteria were heavily represented in ERS (12.46%), KTS (10.21%), and KJS (8.03%). Unaffiliated Bacteria_uc_c (6.6%), Betaproteobacteria (4.55%), and unaffiliated Rhodothermus_c (4.02%) were most common in EAS, ERS, and KJS, respectively. The dominant genus in each sample is presented in Figure 1. Further comparison of the two salt groups (from the Danakil depression in Ethiopia and Shinan in Korea) showed the presence of many bacterial groups unique to each area (Table 3). For instance, Planococcus, Arthrobacter, Brachybacterium, Micrococcus, Thermoleophilia, and Thermomicrobia were occurred only to EAS or ERS, or were present in both salts. Similarly, Salicola, Aeromonas, Spiribacter, Leclercia, Coscinodiscophyceae, and Chlorophyceae were only present in KTS and KJS.
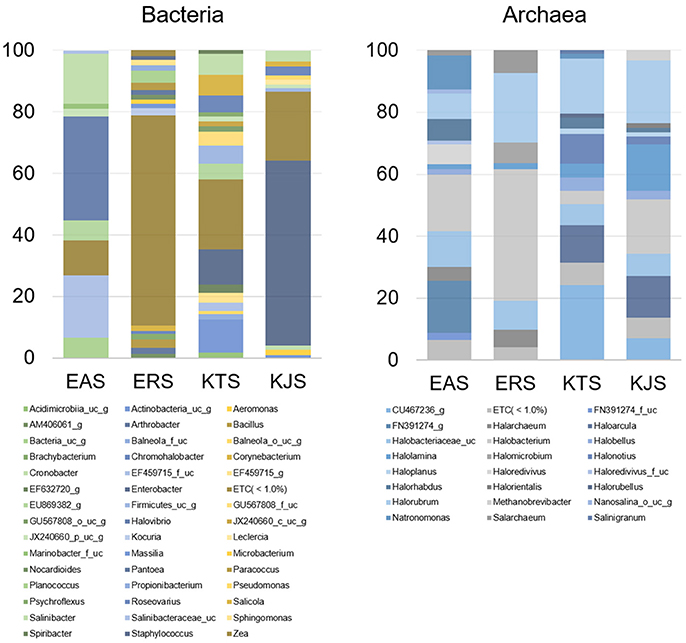
Figure 1. Average composition of selected communities of metagenomically identified bacterial and archaeal genera of salt samples.
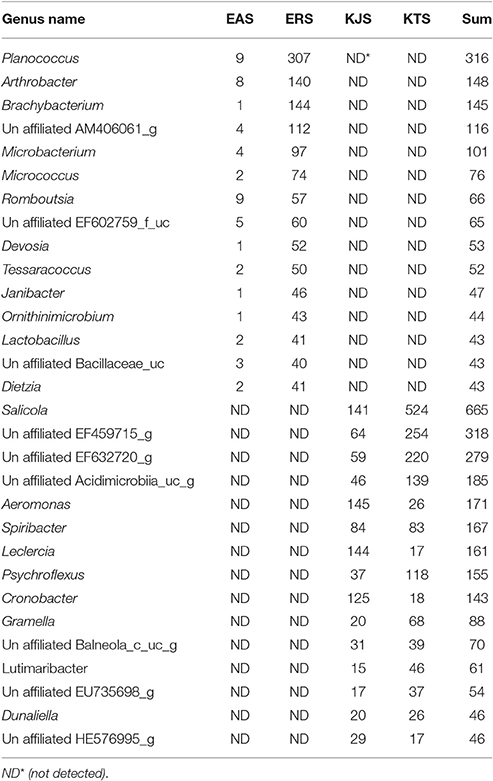
Table 3. Some exclusively recorded bacteria genera identified in the Danakil depression (EAS and ERS) and Shinan (KJS and KTS) salts from the metagenomic dataset.
Within the domain Archaea, all salts were represented by only one archaeal phylum, Euryarchaeota. Of the overall archaeal community, 91.58% belonged to the class Halobacteria, while the remaining 7.58, 0.83, and 0.01% were members of the Nanoarchaea, Methanobacteria, and Thermococci, respectively. Further examination of the salt metagenomic data indicated that the family Halobacteriaceae dominated the archaeal communities, representing 100, 99.18, and 96.68% of archaea in KTS, ERS, and KJS, respectively, and a relatively low percentage (69.88%) of archaea in EAS. Different unaffiliated groups in class Nanohaloarchaea made up more than 30% of the archaea in EAS. In addition, Methanobacteriaceae (3.32%) was only present in KJS. At the genus level, the top five dominant archaeal genera were Halorubrum (20.25%), Halobacterium (17.47%), Halolamina (14.98%), Haloarcula (13.5%), and unaffiliated Halobacteriaceae_uc (7.22%) in KJS; unaffiliated CU467236-g (24.14%), Halorubrum (17.83%), Haloarcula (12.0%), Halonotlus (9.54%), and unaffiliated Halobacteriaceae_uc (6.91%) in KTS; Halobacterium (18.18%), unaffiliated FN391274_g (16.84%), unaffiliated Halobacteriaceae_uc (11.68%), Natronomonas (10.96%), and Halorubrum (8.3%) in EAS; and Halobacterium (42.39%), Halorubrum (22.49%), unaffiliated Halobacteriaceae_uc (9.43%), Salachaeum (7.32%), and Halomicrobium (6.51%) in ERS (Figure 1). In addition, Salinigranum, Halorientalis, unaffiliated EF533953_g, Halomicroarcula, and Halosimplex were some of the genera that were found only in the Shinan salts, whereas unaffiliated FN391274_g, FN391274_f_uc, EU869371_f_uc, and Halorussus were some of the archaeal genera that were only recorded in the Danakil depression salts (Table 4).
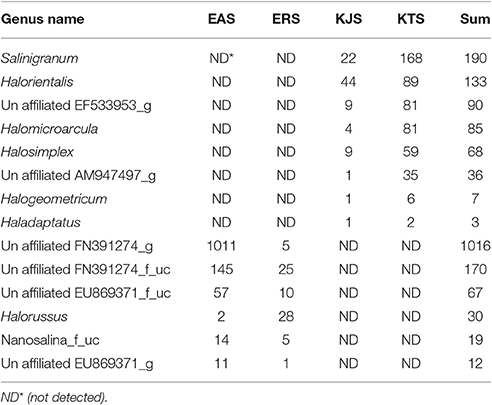
Table 4. Some exclusively identified archaea genera from the Danakil depression (EAS and ERS) and Shinan (KTS and KJS) salts from the metagenomic data set.
A heat map analysis of the bacterial and archaeal communities at the genus level revealed distinctions in diversity in the salts. For example, Enterobacter, the most abundant genus, was detected only in KJS and KTS, whereas Salinibacter, observed in KTS, KJS, and EAS, was more dominant in EAS than in the other two salts (Figure 2). In addition, more sequences associated with Roseovarius, Salicola, and unaffiliated EU869382_g were detected in KTS and EAS than in the other salts. Unaffiliated Actinobacteria_uc_g was mainly present in KTS, and unaffiliated Bacteria_uc_ and Halovibro were mainly present in EAS. Among the archaea, Halorubrum, Halobacterium, Haloarcula, Halonotius, Natromonas, Halarchaeum, Halomicrobium, and Salarchaeum were detected in each salt. Halolamina, Halonotius, Halarchaeum, and Natronomonas were among the genera that were well represented in KJS, KTS, ERS, and EAS respectively. Haloredvivus was well represented only in EAS.
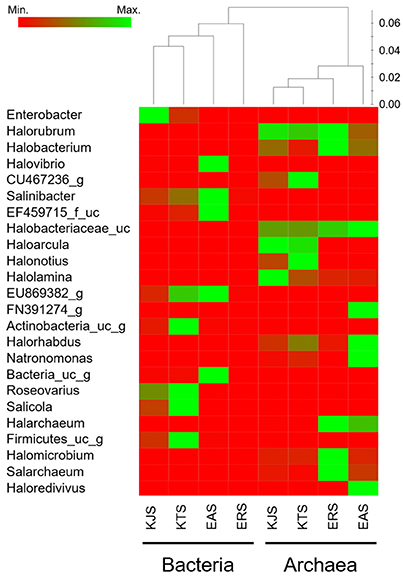
Figure 2. Heat map showing the relative abundances and distribution of representative 16S rRNA gene tag sequences classified at the genus level. The color code indicates the differences in the relative abundance from the mean, ranging from red (negative) through black (the mean) to the green (positive).
Diversity Estimation and Microbial Community Structure
Bacterial and archaeal diversity were estimated using the Shannon index, as well as non-parametric richness (Chao1) and Good's Coverage estimations (Table 2). The alpha diversity (species richness) varied across samples. These results indicated that among the four salts, ERS had the highest bacterial diversity, followed by KTS and EAS (Shannon indices of 7.008, 4.935, and 3.883, respectively). KJS showed the lowest value for bacteria (Shannon index of 3.883). In terms of bacterial richness, ERS (3439.15) had the highest chao1 value, and KJS (1129.245) had the lowest. Similarly, archaeal richness was highest in KTS (2391.57) and lowest in KJS (684.467). Furthermore, rarefaction curves were computed for the four datasets based on the number of reads and species to evaluate the extent of diversity captured (Figure 3).
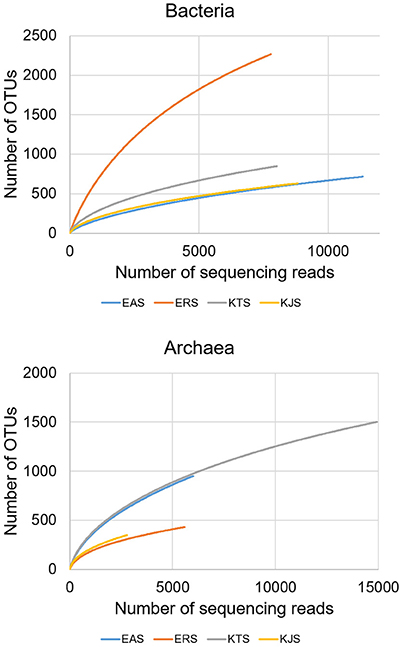
Figure 3. The rarefaction curves of pyrosequenced bacteria (upper panel) and archaea (bottom panel) communities of the four salts.
Comparison Within and Other Biomes
The constitutions of the 16S rRNA gene sequences in salt-based and in other similar environments, were assessed using the UniFrac clustering method. This statistical analysis showed that all of the archaeal communities from the four salt samples clustered together. Interestingly, the bacterial communities in the salts were significantly different from each other and even more different from other communities that were used for comparison; they were placed in different clusters as indicated by the PCoA based on the unweighted UniFrac distances (Figures 4, 5). The plot showed that the bacteria of KTS and KJS clustered into one group. This is because many of the gene categories showed similar abundance levels. However, EAS and ERS did not cluster with KTS and KJS. Furthermore, clustering with similar environmental gene sequences (Asian and European sea bacteria, data taken from the Chunlab database) showed separate clustering of the bacterial communities in the salts. Salinity and other environmental factors may be an important factor in determining the level of similarity between microbial communities. Finally, a taxon exclusive-OR (XOR) analysis showed the presence of many unique genera of archaea and bacteria in each sample. ERS had many unique genera, including Pantoea, Erminia, Weissella, Microvirga, and Caryophanon. In addition, Oceanicella and Oceaniovalibus in KTS, Salmonella and Roseibacterium in KJS, and Dactococcospsis and Halorhodospira in EAS were the genera that were only detected in one salt.
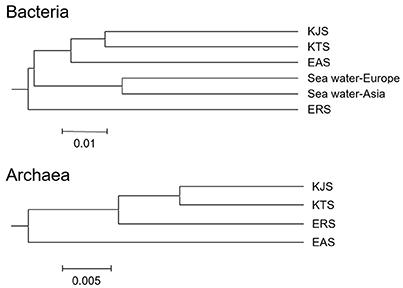
Figure 4. Clustering analysis for the samples classified as archaea and bacteria using the UniFrac service after genome sequencing. The scale bar indicates the distance between clusters in UniFrac units (data generated with CL community, Chun Lab Inc.).
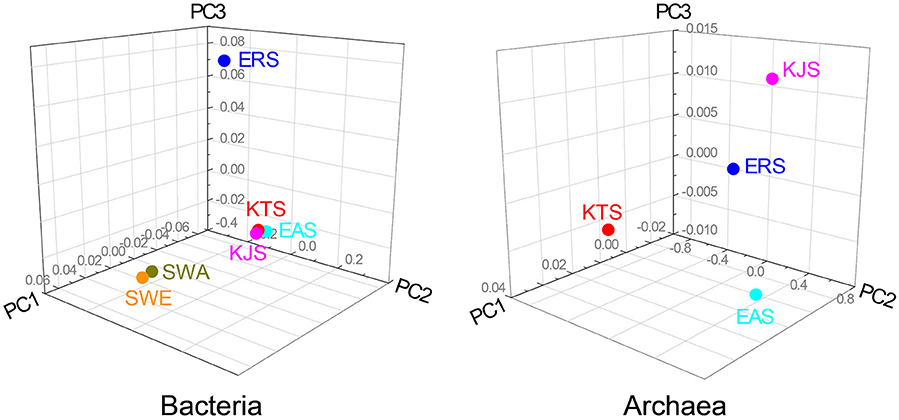
Figure 5. UniFrac distance-based Jackknife clustering of bacteria and archaea communities associated with difference salts. Asian seawater (SWA) and Europian seawater (SWE) samples as the control were also included in this analysis. Unifrac PCoA image were captured from 3D UniFrac PCoA to illustrate differences in the microbiota among the different salts. The following UniFrac analysis were based on the OTU data, with the first three principal coordinates (PCs) shown: unweighted UNiFrac with PC1 (66.71%, 35.71%), PC2 (0.00, 0.00), and PC3 (9.23, 9.23).
Discussion
In this study, we performed amplicon sequencing analyses to survey the microbial content of four commercial salts that were produced by solar evaporation of hypersaline waters and halite mining (Table 1). It is a well-known property of many halophiles to be polyploid and also to harbor multiple unrelated 16S rRNA gene copies in one cell (Oren, 2012). Our previous studies on haloarchaeal taxonomy also revealed that the haloarchaeal taxa such as Halostella salina (Song et al., 2016), Halapricum salinum (Song et al., 2014), and Halorubrum halophilum (Yim et al., 2014), have at least two different copies of the 16S rRNA gene. Because it is accurate to consider all of the reported heterogeneous 16S rRNA gene sequences for the phylogenetic study of the haloarchaea, we used the EzTaxon-e database containing the unrelated 16S rRNA gene copies of the haloarchaea to analyze haloarchaeal community profiles in the commercial salt samples.
Salted, fermented foods and extreme environments have been reported to maintain a high density of halophilic bacteria and archaea dominated primarily by Proteobacteria, Bacteroidetes, Actinobacteria, Firmicutes, and Euryacheota (Keshri et al., 2013; Fernandez et al., 2014). In this study, the microbial diversity of the salts revealed that the bacteria phyla Proteobacteria, Bacteroidetes, Actinobacteria, and Firmicutes were dominant in at least one of the salts, whereas the archaeal phylum Euryacheota was dominant in all salts. Most of extreme halophilic archaea are classified in class Halobacteria under one phylum Euryarchaeota while halophilic bacteria are distributed across several phyla.
In the amplicon sequencing data, we detected 16S rRNA gene sequences related to members of the genera Enterobacter, Halovibrio, Roseovarius, Salicola, and Salinibacter in one or more than one of the salts (Figure 1). Most of these genera are common in salted foods and hypersaline environments (Kim et al., 2012; Fernandez et al., 2014; Pandit et al., 2015). Remarkably, we observed the genus Enterobacter only in Shinan (KTS and KJS); this genus is not common in salted and fermented foods. In some salt samples, the study recorded genus like Salmonella and Entrobacter which are common in domestic wastes (Vrints et al., 2007) needs further study whether they are contaminated from human wastes. Interestingly Enterobacter is much more abundant in KJS than other salts. In order to determine whether the KJS production method is related to Enterobacter flourishing, further study is also needed. On the other hand this genus was almost absent from Danakil depression salts (EAS and ERS), perhaps because the Danakil depression is rather hot for Enterobacter survival, as their optimal growth temperature is 30°C (Grimont and Grimont, 2006). This idea is supported by the finding of Arthrobacter, Choracidobacterium, Thermoleophilia, Thermomicrobia, and Planococcus bacteria in EAS and ERS, which were recorded in hot environments (Pearson and Noller, 2011; Ghati et al., 2013; Stan-Lotter and Fendrihan, 2015). In addition, Brachybacterium, Romboutsia, and many other bacterial genera were only detected in the Danakil depression salts. Similarly, Salicola, Aeomonas, Spiribacter, Leclercia, and Psychroflexus were detected only in Shinan salts (Table 3). It is important to note that the presence of many unique bacterial genera in the salts of the Danakil Depression and Shinan is a result of local and regional environmental factors that influence the assembly of bacterial communities (Lindstrom and Langenheder, 2012).
Within the domain Archaea, the genera Halorubrum, Halobacterium, Haloarcula, Halonotius, Natromonas, Halarchaeum, Halomicrobium, and Salarchaeum were recorded in all salts. Halorubrum, Halobacterium, and Haloarcula also dominated the archaeal communities in different salted seafoods and kimchi (Chang et al., 2008; Roh et al., 2010). In addition, at least one of the archaeal genera has been found to dominate a hypersaline environment (Ochsenreiter et al., 2002; Youssef et al., 2014; Oren, 2015). In contrast, Haloquadratum, which is one of the dominant archaeal genera in salterns (Oh et al., 2010; Dillon et al., 2013), appeared to be a minor member in commercial salts. Despite the geographical separation, EAS from the Danakil depression and KTS from the Shinan share a very similar community structure, as most of the dominant bacterial and archaeal groups were the same. This might be attributed to the production of salts in earthen ponds and mudflats, where salt may be contaminated with soil during the production processes, because most of these bacterial groups have also been detected in saline sediments (Lopez-Lopez et al., 2010; Kim et al., 2012). Comparison of the rarefaction curves with the number of OTUs determined by the Chao1 richness estimator and other diversity indices revealed that the two geographical areas had high microbial diversity, likely because of the intrinsic features of these ecosystems that provide specialized niches for the evolution of unique microbial communities (Smith et al., 2012). A comparison of the salt datasets with those from Asian and European seawater resulted in different clusters in the PCoA plot (Figures 4, 5). The overall microbial community structure of all salts in the current study was similar to that of the Asian and European seawater which were used to evaluate community comparison. This similarity in salt metagenomics is based on similar proportions of Gammaproteobacteria, Deltaproteobacteria, Actinobacteria_uc, and Betaproteobacteria in the bacteria communities. Archaeal communities were also similar, as Halorubrum, Halobacterium, Haloarcula, Halonotius, Natromonas, Halarchaeum, Halomicrobium, and Salarchaeum were detected in each salt.
In conclusion, the exploration of microbial diversity in commercial salts revealed that they contained various bacteria and archaeal taxa. The salts contained surprisingly diverse microbial communities. The bacteria were dominated by the phyla Proteobacteria, Bacteroidetes, Actinobacteria, and Firmicutes. While a substantial amount of the bacteria are predicted to be halophilic, some halotolerant strains producing exopolysaccharides may be able to survive on the crystal salts. The archaea, however, were represented only by Euryacheota. Unique genera were recorded in every salt sample; these merit further study. Recent analyses of the microbiota of patients who were the sources of nosocomial infections such as urinary tract infections, bacteremia, bowel diseases, and human colon diseases showed highly diverse bacteria and archaea from the Enterobacteriaceae, Halobacteriaceae, and Methanobacteriaceae, including close relatives of Enterobacter, Halobacterium, Halorubrum, and Methanobrevibacter (Grimont and Grimont, 2006; Samuel et al., 2007; Oxley et al., 2010). In addition, the genus Romboutsia, a spore-forming obligatory anaerobic bacterium, was isolated from the gastro-intestinal tract of a rat (Gerritsen et al., 2014). These disease causing genera were detected in this study. The first halophile which had been suspected of being pathogenic for man was Pseudomonas enteritis TAKIKAWA. It was isolated from a case of food-poisoning in 1955 (Okudaira et al., 1962). Recently, Bang and Schmitz (2015) reviewed the current knowledge of human mucosa-associated archaeal species, their interaction with the human immune system and their potential contribution to humans' health and disease. However, no concerted efforts have been undertaken to implicate archaea in human diseases. The full spectrum of outcomes from these archaea-human interactions, whether it includes tissue damage, or clinical disease, altered host physiology, remains a mystery. Effects on the health and immune response of haloarchaea are being investigated. Therefore, halophilic bacteria and haloarchaea ingested with salty foods and their roles in human health should be further investigated.
Author Contributions
AG, KP, and MW prepared metagenomic DNA from commertial salts and carried out other experiments. AG and JS(Shin) collected the commercial salts and characterized the physicochemical properties of the salts. AG, DL, JS(Sohn), MS, and SR analyzed the sequencing data and phylogenetical analysis. DL, SR, SL, and HL conceived the study, participated in its design and interpreted the data. HL coordinated the authors and wrote the manuscript with approvals of all authors. All authors are participated in editing of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by the Human Resource Training Program for Regional Innovation & Creativity through the Ministry of Education (MOE) & National Research Foundation (NRF) of Korea (NRF-2014H1C1A1066945), the Basic Science Research Program through the NRF of Korea funded by the Ministry of Education (NRF-2016R1D1A1B04934489), and NRF grant funded by the Ministry of Science, ICT and Future Planning (MSIP), Korea (NRF-2014R1A2A2A01006765).
Abbreviations
EAS, Ethiopian Afdera salt; ERS, Ethiopian rock salt; KJS, Korean Jangpan salt; KTS, Korean Topan salt; PCoA, principal coordinate analysis; OUT, operational taxonomic unit; XOR, exclusive-OR; SWA, seawater in Asia; SWE, seawater in Europe.
References
Bang, C., and Schmitz, R. (2015). Archaea associated with human surfaces: not to be underestimated. FEMS Microbiol. Rev. 39, 631–648. doi: 10.1093/femsre/fuv010
Bik, H. M. (2014). Deciphering diversity and ecological function from marine metagenomes. Biol. Bull. 227, 107–116. doi: 10.1086/BBLv227n2p107
Bougouffa, S., Yang, J. K., Lee, O. O., Wang, Y., Batang, Z., Al-Suwailem, A., et al. (2013). Distinctive microbial community structure in highly stratified deep-sea brine water columns. Appl. Environ. Microbiol. 79, 3425–3437. doi: 10.1128/AEM.00254-13
Chang, H. W., Kim, K. H., Nam, Y. D., Roh, S. W., Kim, M. S., Jeon, C. O., et al. (2008). Analysis of yeast and archaeal population dynamics in kimchi using denaturing gradient gel electrophoresis. Int. J. Food Microbiol. 126, 159–166. doi: 10.1016/j.ijfoodmicro.2008.05.013
Chun, H., Lin, X., Fujii, T., Morimoto, S., Yagi, K., Hu, J., et al. (2007). Soil microbial biomass, dehydrogenase activity, and bacterial community structure in response to long-term fertilizer management. Soil Biol. Biochem. 39, 2971–2976. doi: 10.1016/j.soilbio.2007.05.031
Chun, J., Kim, K. Y., Lee, J. H., and Choi, Y. (2010). The analysis of oral microbial communities of wild-type and toll-like receptor 2-deficient mice using a 454 GS FLX Titanium pyrosequencer. BMC Microbiol. 10:101. doi: 10.1186/1471-2180-10-101
Dillon, J. G., Carlin, M., Gutierrez, A., Nguyen, V., and McLain, N. (2013). Patterns of microbial diversity along a salinity gradient in the Guerrero Negro solar saltern, Baja CA. Sur, Mexico. Front. Microbiol. 4:399. doi: 10.3389/fmicb.2013.00399
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Fernandez, A. B., Ghai, R., Martin-Cuadrado, A. B., Sanchez-Porro, C., Rodriguez-Valera, F., and Ventosa, A. (2014). Prokaryotic taxonomic and metabolic diversity of an intermediate salinity hypersaline habitat assessed by metagenomics. FEMS Microbiol. Ecol. 88, 623–635. doi: 10.1111/1574-6941.12329
Fierer, N., Leff, J. W., Adams, B. J., Nielsen, U. N., Bates, S. T., and Lauber, C. L. (2012). Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc. Natl. Acad. Sci. U.S.A. 109, 21390–21395. doi: 10.1073/pnas.1215210110
Fu, L., Niu, B., Zhu, Z., Wu, S., and Li, W. (2012). CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152. doi: 10.1093/bioinformatics/bts565
Gerritsen, J., Fuentes, S., Grievink, W., van Niftrik, L., Tindall, B. J., Timmerman, H. M., et al. (2014). Characterization of Romboutsia ilealis gen. nov., sp. nov., isolated from the gastro-intestinal tract of a rat, and proposal for the reclassification of five closely related members of the genus Clostridium into the genera Romboutsia gen. nov., Intestinibacter gen. nov., Terrisporobacter gen. nov. and Asaccharospora gen. nov. Int. J. Syst. Evol. Microbiol. 64, 1600–1616. doi: 10.1099/ijs.0.059543-0
Ghati, A., Sarkar, K., and Paul, G. (2013). Isolation, characterization and molecular identification of esterolytic thermophilic bacteria from an indian hot spring. Curr. Res. Microbiol. Biotechnol. 1, 196–202.
Grimont, F., and Grimont, T. A. D. (2006). The genus enterobacter. Prokaryotes 6, 197–214. doi: 10.1007/0-387-30746-x_9
Gugliandolo, C., Michaud, L., Lo Giudice, A., Lentini, V., Rochera, C., Camacho, A., et al. (2015). Prokaryotic community in lacustrine sediments of byers peninsula (Livingston Island, Maritime Antarctica). Microb. Ecol. 71, 387–400. doi: 10.1007/s00248-015-0666-8
Hamady, M., Lozupone, C., and Knight, R. (2010). Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 4, 17–27. doi: 10.1038/ismej.2009.97
Henriet, O., Fourmentin, J., Delince, B., and Mahillon, J. (2014). Exploring the diversity of extremely halophilic archaea in food-grade salts. Int. J. Food Microbiol. 191, 36–44. doi: 10.1016/j.ijfoodmicro.2014.08.019
Hur, M., Kim, Y., Song, H. R., Kim, J. M., Choi, Y. I., and Yi, H. (2011). Effect of genetically modified poplars on soil microbial communities during the phytoremediation of waste mine tailings. Appl. Environ. Microbiol. 77, 7611–7619. doi: 10.1128/AEM.06102-11
Jaakkola, S. T., Zerulla, K., Guo, Q., Liu, Y., Ma, H., Yang, C., et al. (2014). Halophilic archaea cultivated from surface sterilized middle-late Eocene rock salt are polyploid. PLoS ONE 9:e110533. doi: 10.1371/journal.pone.0110533
Jung, S. P., and Kang, H. (2014). Assessment of microbial diversity bias associated with soil heterogeneity and sequencing resolution in pyrosequencing analyses. J. Microbiol. 52, 574–580. doi: 10.1007/s12275-014-3636-9
Keshri, J., Mishra, A., and Jha, B. (2013). Microbial population index and community structure in saline-alkaline soil using gene targeted metagenomics. Microbiol. Res. 168, 165–173. doi: 10.1016/j.micres.2012.09.005
Kim, J. S., Makama, M., Petito, J., Park, N.-H., Cohan, F. M., and Dungan, R. S. (2012). Diversity of Bacteria and Archaea in hypersaline sediment from Death Valley National Park, California. Microbiol. Open 1, 135–148. doi: 10.1002/mbo3.20
Kondo, Y., Minegishi, H., Echigo, A., Shimane, Y., Kamekura, M., Itoh, T., et al. (2016). Haloparvum alkalitolerans sp. nov., alkali-tolerant haloarchaeon isolated from commercial salt. Int. J. Syst. Evol. Microbiol. 66, 5314–5319. doi: 10.1002/mbo3.20
Lee, H.-S. (2013). Diversity of halophilic archaea in fermented foods and human intestines and their application. J. Microbiol. Biotechnol. 23, 1645–1653. doi: 10.4014/jmb.1308.08015
Li, L., Hao, C., Wang, L., and Pei, L. (2015). Microbial diversity of salt lakes in Badain Jaran desert. Wei Sheng Wu Xue Bao 55, 412–424.
Lindstrom, E. S., and Langenheder, S. (2012). Local and regional factors influencing bacterial community assembly. Environ. Microbiol. Rep. 4, 1–9. doi: 10.1111/j.1758-2229.2011.00257.x
Lopez-Lopez, A., Yarza, P., Richter, M., Suarez-Suarez, A., Anton, J., Niemann, H., et al. (2010). Extremely halophilic microbial communities in anaerobic sediments from a solar saltern. Environ. Microbiol. Rep. 2, 258–271. doi: 10.1111/j.1758-2229.2009.00108.x
Minegishi, H., Shimogaki, R., Enomoto, S., Echigo, A., Kondo, Y., Nagaoka, S., et al. (2016). Halopiger thermotolerans sp. nov., a thermo-tolerant haloarchaeon isolated from commercial salt. Int. J. Syst. Evol. Microbiol. 66, 4975–4980. doi: 10.1099/ijsem.0.001455
Morel, V. (2005). Cruelest Place on Earth, The Africa's Danakil Desert. National Geographic, October, 32–53.
Mutlu, M. B., and Guven, K. (2015). Bacterial diversity in camalti saltern, Turkey. Pol. J. Microbiol. 64, 37–45.
Neelakanta, G., and Sultana, H. (2013). The use of metagenomic approaches to analyze changes in microbial communities. Microbiol. Insights 6, 37–48. doi: 10.4137/MBI.S10819
Ochsenreiter, T., Pfeifer, F., and Schleper, C. (2002). Diversity of Archaea in hypersaline environments characterized by molecular-phylogenetic and cultivation studies. Extremophiles 6, 267–274. doi: 10.1007/s00792-001-0253-4
Oh, D., Porter, K., Russ, B., Burns, D., and Dyall-Smith, M. (2010). Diversity of Haloquadratum and other haloarchaea in three, geographically distant, Australian saltern crystallizer ponds. Extremophiles 14, 161–169. doi: 10.1007/s00792-009-0295-6
Okudaira, M., Kawamura, H., Ueno, M., Nakahara, Y., Zen-Yoji, H., Sakai, S., et al. (1962). Food poisoning caused by pathogenic caused by pathogenic halophilic bacteria, (Pseudomonas enteritis TAKIKAWA). Pathol. Int. 12, 299–304. doi: 10.1111/j.1440-1827.1962.tb02727.x
Oren, A. (2012). Taxonomy of the family Halobacteriaceae: a paradigm for changing concepts in prokaryote systematics. Int. J. Syst. Evol. Microbiol. 62, 263–271 doi: 10.1099/ijs.0.038653-0
Oren, A. (2015). Halophilic microbial communities and their environments. Curr. Opin. Biotechnol. 33, 119–124. doi: 10.1016/j.copbio.2015.02.005
Oxley, A. P., Lanfranconi, M. P., Wurdemann, D., Ott, S., Schreiber, S., McGenity, T. J., et al. (2010). Halophilic archaea in the human intestinal mucosa. Environ. Microbiol. 12, 2398–2410. doi: 10.1111/j.1462-2920.2010.02212.x
Pandit, A. S., Joshi, M. N., Bhargava, P., Shaikh, I., Ayachit, G. N., Raj, S. R., et al. (2015). A snapshot of microbial communities from the Kutch: one of the largest salt deserts in the World. Extremophiles 19, 973–987. doi: 10.1007/s00792-015-0772-z
Park, E. J., Chang, H. W., Kim, K. H., Nam, Y. D., Roh, S. W., and Bae, J. W. (2009). Application of quantitative real-time PCR for enumeration of total bacterial, archaeal, and yeast populations in kimchi. J. Microbiol. 47, 682–685. doi: 10.1007/s12275-009-0297-1
Pearson, M. D., and Noller, H. F. (2011). The draft genome of Planococcus donghaensis MPA1U2 reveals nonsporulation pathways controlled by a conserved Spo0A regulon. J. Bacteriol. 193, 6106–6111. doi: 10.1128/JB.05983-11
Roh, S. W., Kim, K. H., Nam, Y. D., Chang, H. W., Park, E. J., and Bae, J. W. (2010). Investigation of archaeal and bacterial diversity in fermented seafood using barcoded pyrosequencing. ISME J. 4, 1–16. doi: 10.1038/ismej.2009.83
Samuel, B. S., Hansen, E. E., Manchester, J. K., Coutinho, P. M., Henrissat, B., and Fulton, R. (2007). Genomic and metabolic adaptations of Methanobrevibacter smithii to the human gut. Proc. Natl. Acad. Sci. U.S.A. 104, 10643–10648. doi: 10.1073/pnas.0704189104
Satterfield, C. L., Lowenstein, T. K., Vreeland, R. H., Rosenzweig, W. D., and Powers, D. W. (2005). New evidence for 250 Ma age of halotolerant bacterium from a Permian salt crystal. Geology 33, 265–268. doi: 10.1130/G21106.1
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., and Hollister, E. B. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Schubert, B. A., Lowenstein, T. K., Timofeeff, M. N., and Parker, M. A. (2010). Halophilic Archaea cultured from ancient halite, Death Valley, California. Environ. Microbiol. 12, 440–454. doi: 10.1111/j.1462-2920.2009.02086.x
Shimane, Y., Minegishi, H., Echigo, A., Kamekura, M., Itoh, T., Ohkuma, M., et al. (2015). Halarchaeum grantii sp. nov., a moderately acidophilic haloarchaeon isolated from a commercial salt sample. Int. J. Syst. Evol. Microbiol. 65, 3830–3835. doi: 10.1099/ijsem.0.000501
Smith, R. J., Jeffries, T. C., Roudnew, B., Fitch, A. J., Seymour, J. R., Delpin, M. W., et al. (2012). Metagenomic comparison of microbial communities inhabiting confined and unconfined aquifer ecosystems. Environ. Microbiol. 14, 240–253. doi: 10.1111/j.1462-2920.2011.02614.x
Song, H. S., Cha, I. T., Rhee, J. K., Yim, K. J., Kim, A. Y., Choi, J. S., et al. (2016). Halostella salina gen. nov., sp. nov., an extremely halophilic archaeon isolated from solar salt. Int. J. Syst. Evol. Microbiol. 66, 2740–2746. doi: 10.1099/ijsem.0.001118
Song, H. S., Cha, I. T., Yim, K. J., Lee, H. W., Hyun, D. W., Lee, S. J., et al. (2014). Halapricum salinum gen. nov., sp. nov., an extremely halophilic archaeon isolated from non-purified solar salt. Antonie Van Leeuwenhoek 105, 979–986. doi: 10.1007/s10482-014-0156-x
Stan-Lotter, H., and Fendrihan, S. (2015). Halophilic archaea: life with desiccation, radiation and oligotrophy over geological times. Life (Basel). 5, 1487–1496. doi: 10.3390/life5031487
Streit, W. R., and Schmitz, R. A. (2004). Metagenomics–the key to the uncultured microbes. Curr. Opin. Microbiol. 7, 492–498. doi: 10.1016/j.mib.2004.08.002
Suh, S. S., Park, M., Hwang, J., Kil, E. J., Jung, S. W., Lee, S., et al. (2015). Seasonal dynamics of marine microbial community in the South Sea of Korea. PLoS ONE 10:e0131633. doi: 10.1371/journal.pone.0131633
Vreeland, R. H., Jones, J., Monson, A., Rosenzweig, W. D., Lowenstein, T. K., Timofeeff, M., et al. (2007). Isolation of live Cretaceous (121-112 million years old) halophilic archaea from primary salt crystals. Geomicrobiol. J. 24, 275–282. doi: 10.1080/01490450701456917
Vreeland, R. H., Rosenzweig, W. D., and Powers, D. W. (2000). Isolation of a 250 million-year-old halotolerant bacterium from a primary salt crystal. Nature 407, 897–900. doi: 10.1038/35038060
Vrints, M., Bertrand, S., and Collard, J.-M. (2007). A bacterial population study of commercialized wastewater inoculants. J. Appl. Microbiol. 103, 2006–2015. doi: 10.1111/j.1365-2672.2007.03444.x
Walsh, D. A., Papke, R. T., and Doolittle, W. F. (2005). Archaeal diversity along a soil salinity gradient prone to disturbance. Environ. Microbiol. 7, 1655–1666.
Webster, G., O'Sullivan, L. A., Meng, Y., Williams, A. S., Sass, A. M., Watkins, A. J., et al. (2015). Archaeal community diversity and abundance changes along a natural salinity gradient in estuarine sediments. FEMS Microbiol. Ecol. 91, 1–18. doi: 10.1093/femsec/fiu025
Yim, K. J., Cha, I. T., Lee, H. W., Song, H. S., Kim, K. N., Lee, S. J., et al. (2014). Halorubrum halophilum sp. nov., an extremely halophilic archaeon isolated from a salt-fermented seafood. Antonie Van Leeuwenhoek 105, 603–612. doi: 10.1007/s10482-014-0115-6
Keywords: commercial salts, halophilic archaea, halophilic bacteria, Ethiopia, Korea, diversity
Citation: Gibtan A, Park K, Woo M, Shin J-K, Lee D-W, Sohn JH, Song M, Roh SW, Lee S-J and Lee H-S (2017) Diversity of Extremely Halophilic Archaeal and Bacterial Communities from Commercial Salts. Front. Microbiol. 8:799. doi: 10.3389/fmicb.2017.00799
Received: 21 September 2016; Accepted: 19 April 2017;
Published: 10 May 2017.
Edited by:
Martin G. Klotz, Queens College (CUNY), USAReviewed by:
Jerry Eichler, Ben-Gurion University of the Negev, IsraelFrank T. Robb, University of Maryland, Baltimore, USA
Copyright © 2017 Gibtan, Park, Woo, Shin, Lee, Sohn, Song, Roh, Lee and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Han-Seung Lee, hanslee@silla.ac.kr
 Ashagrie Gibtan1
Ashagrie Gibtan1 Dong-Woo Lee
Dong-Woo Lee Han-Seung Lee
Han-Seung Lee