- 1National Agricultural Research Laboratories, Kampala, Uganda
- 2Department of Agricultural Production, College of Agricultural and Environmental Sciences, Makerere University, Kampala, Uganda
- 3International Institute of Tropical Agriculture, Nairobi, Kenya
- 4Department of Plant Pathology, University of California, Davis, Davis, CA, USA
Black Sigatoka disease, caused by Pseudocercospora fijiensis is a serious constraint to banana production worldwide. The disease continues to spread in new ecological niches and there is an urgent need to develop strategies for its control. The high osmolarity glycerol (HOG) pathway in Saccharomyces cerevisiae is well known to respond to changes in external osmolarity. HOG pathway activation leads to phosphorylation, activation and nuclear transduction of the HOG1 mitogen-activated protein kinases (MAPKs). The activated HOG1 triggers several responses to osmotic stress, including up or down regulation of different genes, regulation of protein translation, adjustments to cell cycle progression and synthesis of osmolyte glycerol. This study investigated the role of the MAPK-encoding PfHog1 gene on osmotic stress adaptation and virulence of P. fijiensis. RNA interference-mediated gene silencing of PfHog1 significantly suppressed growth of P. fijiensis on potato dextrose agar media supplemented with 1 M NaCl, indicating that PfHog1 regulates osmotic stress. In addition, virulence of the PfHog1-silenced mutants of P. fijiensis on banana was significantly reduced, as observed from the low rates of necrosis and disease development on the infected leaves. Staining with lacto phenol cotton blue further confirmed the impaired mycelial growth of the PfHog1 in the infected leaf tissues, which was further confirmed with quantification of the fungal biomass using absolute- quantitative PCR. Collectively, these findings demonstrate that PfHog1 plays a critical role in osmotic stress regulation and virulence of P. fijiensis on its host banana. Thus, PfHog1 could be an interesting target for the control of black Sigatoka disease in banana.
Introduction
Banana and plantain (Musa sp.) is the eighth most economically staple food crop (Tripathi et al., 2014). Worldwide it is cultivated across tropical and subtropical countries on more than 10 million hectares and total production estimated at 144 million tons (FAOSTAT, 2013). It is one of the fundamental energy sources for millions of people in developing countries and the most significant of all fruits, with world trade total of $2.5 billion annually. It is mainly grown by small holder farmers and only 10% of the annual output reaches the international market (Ploetz, 2001).
In Africa, banana and plantain are mainly used as a staple food, providing more than 25% of the carbohydrate requirements for over 70 million people (IITA, 1998). It is also a critical and often the only source of income for both small and large-scale farmers, thus being an important contributor to local economies. The Great Lakes region including Burundi, Rwanda, Democratic Republic of Congo, Uganda, Kenya and Tanzania is the largest producer and consumer of banana in Africa (Smale et al., 2006), with Uganda ranked the third largest producer of the crop worldwide and largest producer in Africa. Although banana is the most important staple food crop in Uganda, its production is declining since 2010 (FAOSTAT, 2013), probably due to pests and diseases.
The ascomycete fungus Pseudocercospora fijiensis (synonym Mycosphaerella fijiensis) is the causal agent of black Sigatoka disease, one of the important diseases of banana worldwide (Arango Isaza et al., 2016). Black Sigatoka disease can reduce yields by 33 to 76% if left uncontrolled (Gauhl et al., 2000; Marin et al., 2003; Agrios, 2005). The fungus induces necrotic leaf streak and lesion symptoms, which later coalesce into blotches that can cover over 70% of the banana leaf lamina. This significantly reduces the photosynthetic capacity of plants, resulting into poor quality fruits and often premature ripening (Marin et al., 2003). P. fijiensis produces conidia and ascospores, and both can cause disease. They are disseminated by wind, and in the case of conidia, spread can also be by water. Ascospores are more important than conidia in spreading the disease within banana plants and plantations. In developing countries, infected planting material and leaves, which are often used as packing materials, are also responsible for the long-distance spread of the disease (Ploetz, 2001).
Mitogen-activated protein kinases (MAPKs) are conserved eukaryotic serine/threonine protein kinases with vital roles in numerous cellular processes, including gene expression, cell differentiation, mitosis, cell survival and apoptosis. Additionally, MAPKs partake in signal transduction pathways that are activated by osmotic stress and in regulation of growth and development (Alonso-Monge et al., 2001). The high osmolarity glycerol (HOG) pathway in Saccharomyces cerevisiae is well known to respond to changes in external osmolarity. HOG pathway activation leads to phosphorylation, activation and nuclear transduction of the HOG1 MAPK. The activation of HOG1 triggers several responses to osmotic stress, including up or down regulation of different genes, regulation of protein translation, adjustments to cell cycle progression and synthesis of osmolyte glycerol (Hohmann, 2002).
The HOG MAPKs pathway signaling cascade in fungi regulates responses to stress and adaptation to hyperosmotic conditions (Alonso-Monge et al., 2001). For example, HOG1 pathway was found critical in regulating colonization of mouse gastrointestinal tract in Candida albicans (Prieto et al., 2014). In the entomopathogenic fungus Metarhizium acridum, MaHog1, a member of the Hog1/Sty1/p38 MAP-kinase gene family, has been shown to be critical for adaptation to hyper osmolarity, high temperature and oxidative stress (Jin et al., 2012). It is also an important virulence factor as disruption of MaHog1 results in reduced infectivity and growth of the fungus on its insect hosts (Jin et al., 2012). In a similar way, Hog1 is also activated in response to high osmolarity, oxidative stress, and other stress stimuli in S. cerevisiae and the human pathogenic yeast C. albicans (Alonso-Monge et al., 2001; Winkler et al., 2002), whereas in the wheat pathogen Zymoseptoria tritici (synonym M. graminicola), Hog1 regulates dimorphism and pathogenicity (Mehrabi et al., 2006). Given the importance of Hog1 for virulence in fungi and the relatively close phylogenetic relatedness between Z. tritici and P. fijiensis, it is possible that PfHog1 is also an important virulence factor for causing black Sigatoka disease.
Gene specific silencing by RNA interference (RNAi) has been widely used to understand functional genomics of fungi over the last couple of decades (Agrawal et al., 2003). In addition, RNAi can be used in control of fungal diseases in plants through host induced gene silencing (HIGs) (Nowara et al., 2010). Proof of concept for HIGS in plant pathogenic fungi was obtained for barley powdery mildew caused by Blumeria graminis and Fusarium verticillioides (Nowara et al., 2010). Transgenic wheat expressing an RNAi hairpin construct targeting β-1, 3-glucan synthase gene FcGls1 of F. culmorum, showed enhanced resistance to Fusarium head blight disease (Chen et al., 2016). Similarly, host induced post-transcriptional mediated gene silencing of the fungal genes, velvet and F. transcriptional factor 1, showed resistance against F. oxysporum f sp. cubense (Foc) in transgenic banana (Ghag et al., 2014).
The aim of this study was to investigate the role of PfHog1 in adaptation to osmotic stress and virulence of P. fijiensis on its host banana. Silencing of PfHog1 through Agrobacterium tumefaciens-mediated transformation (ATMT) was tested for osmotic stress and for virulence on non-transgenic tissue culture banana plantlets. Virulence was confirmed by visual necrosis, staining with lacto phenol cotton blue (LPCB) and absolute quantitation by qPCR. This study confirmed that silencing of PfHog1 limits P. fijiensis adaptation to osmotic stress and virulence.
Materials and Methods
Isolation of P. fijiensis
Pseudocercospora fijiensis was isolated from infected leaves of banana cultivar ‘Nakitembe’. The infected leaves were collected from the field, cut into pieces of 5 cm and then placed in a moist cotton wool inside a clear polythene bag. The leaf culture was incubated at 25°C overnight, in order to allow maturation of spores. The leaf pieces were recovered, cleaned with 70% ethanol and then sterile water. The mature conidia were collected onto 1% agar (w/v in water) by stapling necrotic leaves under moist blot paper in a petri- dish and incubated for 24 h at 25°C. The conidia discharged on agar were picked under a light microscope using a sterile needle and transferred to V8 juice agar medium [30% (v/v) V8 juice, 3 g CaCo3, 20 g bacterial agar, pH 7.2]. Cultures were incubated at 25°C for 14 days; the resulting pure isolates of P. fijiensis were then stored at room temperature.
Isolation of Genomic DNA of P. fijiensis and PCR Amplification of PfHOG1 Gene
Genomic DNA of P. fijiensis was extracted from the pure culture mycelia as described (Mahuku, 2004) with minor modification, which involved incubation of the sample at room temperature for a period of 12–13 h and then overnight at -20°C.
PfHog1 gene of P. fijiensis was amplified from the genomic DNA extract using PfHog1 specific primers [HOG1F: ACGGAGCTGCGTAACGAATTAG and HOG1R: CTGCGTGTGATCGACTAG]. The primers were designed based on P. fijiensis CIRAD86 MAP kinase sequences (accession number XM_007924474.1). The PCR reaction mixture contained 10 μM each of forward and reverse primers (0.5 μl), AmpliTaq® DNA polymerase (0.25 μl; Applied Biosystems, USA), 10x Buffer with 15 mM MgCl2 (2.5 μl), (Applied Biosystems, USA), 10 μM deoxyribonucleotides (dNTP; 0.5 μl), 1 μl of genomic DNA of P. fijiensis and then adjusted with water to 25 μl final volume. The cycle conditions used were the following: initial denaturation at 95°C for 5 min and then 34 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s and extension at 72°C for 1 min, followed by final extension at 72°C for 5 min and storage at 12°C. The amplified PCR product was separated by electrophoresis on agarose gel.
Preparation of Plasmid Construct
The amplified PfHog1 product was resolved by electrophoresis on 1% agarose gel and then purified by ZymocleanTM Gel DNA recovery kit following the manufacturer’s protocol. The purified DNA was ligated into pGEM®-T easy vector according to the procedure in the manual of Promega, Madison, W1, USA, and incubated at 16°C overnight. The ligated vector was later digested with EcoRI. The product PfHog1 was ligated into RNAi silencing pKOIISD1 fungal transformation vector at EcoRI site (Figure 1). The pKOIISD1 was designed based on pSilent-Dual1 (pSD1) vector with dual promoters (i.e., sense promoter PtrpC and antisense promoter Pgpd), provided by Stergiopoulos laboratory at University of California Davis, USA. Basically, pKOIISD1 is the combination of pSD1 and pBHt2 vectors. Three restriction enzyme sites (ApaI, BglII, and AvrII) were inserted into XmnI site of pBHt2. The pSD1 was digested with ApaI and SacI to obtain DNA fragment containing geneticin-resistant marker, promoter PtrpC and promoter Pgpd. Subsequently, this DNA fragment was ligated into ApaI/SacI digested pBHt2 to generate pKOIISD1.
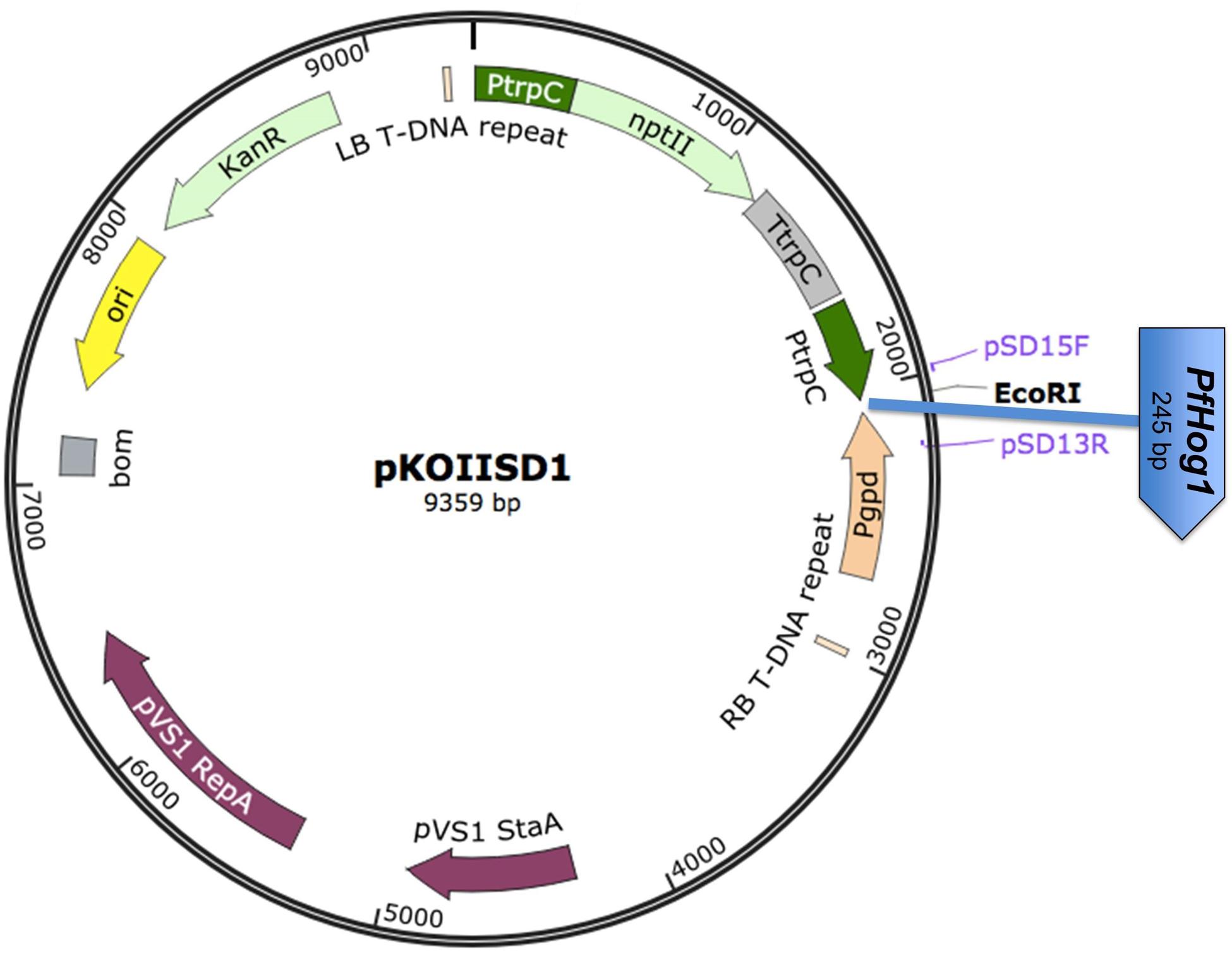
FIGURE 1. Schematic diagram of the RNAi plasmid construct pKOIISD1-PfHog1. PtrpC, promoter trpC of Aspergillus nidulans; Pgpd, promoter gpd of A. nidulans; nptII, neomycin phosphotransferase II gene; PfHog1, Hog1 gene from P. fijiensis; LB- Left Border of T-DNA; RB- Right Border of T- DNA fragment.
The pKOIISD1-PfHog1 plasmid was validated by PCR and sequencing for the presence and orientation of insert. After validation the plasmid was transformed into Escherichia coli strain DHα5 according to high efficiency transformation protocol of the New England Biolabs.
Transformation of P. fijiensis
Culturing of P. fijiensis
A mycelial plug of P. fijiensis was grinded and the fungal suspension (10 ml) added to 200 ml of sterile rich medium (2% yeast and 3% glucose in water) supplemented with 100 μg/ml ampicillin and 100 μg/ml cefotaxim in a flask. The culture was then incubated at 25°C with gentle shaking at 200 rpm for 5 to 7 days at room temperature.
Transformation of Agrobacterium tumefaciens
The electro-competent A. tumefaciens strain AGL1 (40 μl) was dispensed in eppendorf tube and 5 μl pKOIISD1-PfHog1 (500 ng/μl) plasmid DNA was added. The mixture was then electroporated by applying an electric pulse (2.5 kV), capacitance (25 μF) and resistance (400 Ω). Immediately LB medium (300 μl) was added to the transformed cells followed by incubation at 28°C with shaking at 230 rpm for 4 h. Then 60 and 100 μl of the culture were plated on LB agar supplemented with 50 μg/ml kanamycin and incubated at 28°C for 2 days. Single colonies were picked and inoculated into LB broth (10 ml) supplemented with rifampicin and kanamycin at 10 and 50 μg/ml, respectively. The culture was incubated at 25°C with shaking at 230 rpm for 48 h. Thereafter, cells were harvested by centrifugation at 6000 rpm for 10 min. The pellet obtained was re-suspended in 20 ml Agrobacterium induction medium (AIM) and acetosyringone (Sigma Aldrich) was added to 200 μM final concentration. The culture was incubated at 25°C with shaking at 230 rpm for 3 h. The Agrobacterium culture with an O.D600 nm of 0.2–0.5 were used to transform P. fijiensis.
Co-cultivation and Selection of Transformed P. fijiensis
For infection, the liquid culture of P. fijiensis mycelium (200 μl) was mixed with Agrobacterium culture (300 μl) and incubated at room temperature for 10min. Thereafter, an aliquot of the mixture (200 μl) was spread onto Agrobacterium induction agar media plates covered with cellulose membranes and co-cultivated at 25°C for 3 days. The co-cultivated cultures were transferred to standard potato dextrose agar (PDA) medium supplemented with 150 μg/ml geneticin (G148) and 100 μg/ml cefotaxime. After 5 days, single colonies of Agro-infected P. fijiensis were transferred to PDA medium containing geneticin (150 μg/ml).
All the transformants of P. fijiensis were generated using one plasmid construct pKOIISD1-PfHog1, in which the sense strand of PfHog1 gene was driven by PtrpC promoter and anti-sense strand by Pgpd promoter (Figure 1).
Molecular Characterisation of Transformed P. fijiensis
Extraction of Genomic DNA and PCR Analysis of Transformed P. fijiensis
A plug of transformed mycelia was freeze-dried and then lyophilised. The genomic DNA was extracted as previously described (Thon et al., 2000). The primers, pSD15F (CTTTAAGTTCGCCCTTCCTC), pSD13R (GTTGACAAGGTCGTTGCGT), designed from pKOIISD1 vector were used to amplify PfHog1 gene from genomic DNA of transformed P. fijiensis. PCR reaction mixture contained 10 μM primers pSD15F and pSD13R (0.5 μl each). The PCR reaction component and PCR conditions are same as described above.
After validation by PCR, three mutants of P. fijiensis (ΔPfHog1-1, ΔPfHog1-4, and ΔPfHog1-5) were selected randomly from independent transformation experiments for further analysis. All the mutants were generated using same plasmid construct.
Assessment of Gene Expression in Transformed P. fijiensis
RNA Extraction and Complementary DNA (cDNA) Synthesis
The wild type (WT) and transformed P. fijiensis were grown on V8 juice agar medium at 25°C and total RNA was isolated using Trizol® reagent according to the protocol provided by Ambion RNA life technologies. The RNA extract was further purified by cleaning using RNA clean and concentrator-TM kit following the protocol from Zymo research Corp. Then cDNA was synthesized using Maxima first strand cDNA kit for RT-qPCR, Thermo Fishers Scientific, Inc.
Quantitative Real Time-Polymerase Chain Reaction (qRT-PCR) Assay
The qRT-PCR assay was performed using Applied Biosystem 7500 Life Technology machine. Specific primers (TUBF1-ATACACACCGCATCAACGAC and TUBR1-ATGAACGATCTCGCATTC) with amplification product of 114 bp were designed based on β-tubulin gene sequence from P. fijiensis genome. To detect the level of transcript for silencing in P. fijiensis, PfHog1 gene specific primers (HOGF- TGAAAACGGAGCTGCGTAAC and HOGR-TTCTCACGGTTCCGTAATGC) were designed based on P. fijiensis sequence.
A standard curve was made for determining primer specificity, efficiency and for calculation of P. fijiensis DNA and Biomass. A known serial dilution of DNA/cDNA was generated with dilution factor of 1/10. The standard curve was then generated using GraphPad Prism software version 5 and Microsoft Excel 2007.
Quantitation comparative Ct was used in PfHog1 expression or detection level of transcript for silencing in P. fijiensis. The qRT-PCR reaction mixture contained Maxima SYBR Green/ ROX qPCR Master mix (2x; Thermo Scientific), 300 nM primer (i.e., β- tubulin and Hog1), 100 ng/μl cDNA in a total reaction volume of 12 μl. RT- PCR program used was as follows; 40 cycles at 50°C for 2 min, 95°C for 10 min, 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s, followed by melting curve stages.
Three different mutants (ΔPfHog1-1, ΔPfHog1-4, and ΔPfHog1-5) and WT control were used with three technical replicates in each experiment. The experiment was repeated three times.
Osmotic Stress Assay
Freshly grown transformed mycelia were ground and filtered using double layer cheese-cloth under sterile conditions. The mycelia were counted using a haemocytometer and suspension was adjusted to a density of 104–105/ml. About 104–105 mycelia were then used for culturing on each plate. Mycelium mixture was prepared in deionised sterile water in a total volume of 20 ml and 300 μl of the mixture was spread onto PDA medium supplemented with 1 M NaCl. This culture was incubated at 25°C for approximately 10 days and germination of colonies was assessed visually and photographs taken with an ordinary digital camera.
Virulence Assay
Preparation of Inoculums
Freshly ground transformed and non-transformed mycelium fragments was resuspended in 200 ml of rich medium supplemented with 100 μg/ml ampicillin and culture was incubated at room temperature with shaking at 150 rpm for 5–10 days. The fungal culture was filtered using double layer cheese-cloth and mycelium was counted. Then 104–105 mycelia were used for inoculation of each sample. Inoculums were prepared in 10% rich medium containing 1% Tween 20.
Inoculation of Banana Plants
Three months old potted tissue culture banana plants were used for virulence assay. Plants were placed in a locally made inoculation chamber in a contained glasshouse. Three leaves per plant of three replicated plants were inoculated with transformed fungal culture using an art brush size 9. A WT inoculum was included as a positive control. The inoculation chamber was then covered with clear polythene sheet to create a humid condition. Plants were sprayed with water three times a day to maintain humidity levels of 80–90% for 72 h. Polythene sheets were removed from the inoculation chamber after 72 h. Disease severity index was scored according to Alvarado-Capo et al. (2003) with modification, a scale of 1–6, ∗6, and ∗∗6. (i.e., Score 1: No symptom, Score 2: Brown streak visible on underside of leaf but later visible on leaf upper surface as yellow streak; color changes progressively to brown black on upper leaf surface, Score 3: Enlarged stage 2 becomes longer as disease progresses, Score 4: Streak appears on underside as brown spot and black spot on upper leaf surface, Score 5: Elliptical spot totally black on the underside of the leaf surrounded by yellow halo, Score 6: Center of spot dries out turns gray, surrounded by a well defined margin and a bright yellow halo, Score ∗6: leaf dried/dead due to severe disease infection, Score ∗∗6: Leaf died due to normal aging processes, i.e., senescence).
Determining Fungal Mycelium Growth and Symptom Development in Infected Plant Tissues
Leaf disk of approximately 2–3 cm of both inoculated and non-inoculated banana leaves were detached and soaked in 10 ml of absolute HCl for 20 min. The samples were then washed in sterile distilled water for 5 min and soaked twice in 100% ethanol for 1 h to remove chlorophyll. Finally, the leaf samples were rinsed in sterile distilled water for 5 min, dehydrated with 95% ethanol and stored at 4°C until ready for staining.
To observe fungal mycelium growth and development within banana leaf cells, the clear leaves were stained with LPCB for 30 min. Excess stains on leaf disk were blotted and fixed on slides. The slides were observed under COSLAB light microscope and picture was taken using digital camera MDCE- 5C (ISO 9001 Co) and analyzed using Optika Vision Lite 2.1 software.
Detection of P. fijiensis in Plant Tissue
DNA Extraction
DNA was extracted from 1 g infected and non-infected banana leaf samples and pure culture of P. fijiensis as previously described (Mahuku, 2004). In order to remove the RNA contaminants, each of the DNA samples (50 μl) were treated with 1 μl of 10 mg/μl RNase A.
Real Time PCR Assay
Quantitation standard curve was used for P. fijiensis DNA detection and biomass estimation in infected, non-infected and pure fungal culture samples.
The amount of fungal genomic DNA detected in each of the sample analyzed was calculated using sample DNA Cycle threshold (Ct) mean values, correlating to Y concentration values from the regression curve. The RT- PCR reaction mixture contained Maxima SYBR Green/ ROX qPCR Master mix (2x; Thermo Scientific), 300 nM primer (i.e., β- tubulin), 100 ng DNA in a total reaction volume of 12 μl. The program cycle was same as described in Expression assay above.
Each experiment has three technical replicates of three mutants and WT. The experiment was replicated twice.
Statistical Analysis
The data obtained were analyzed using GenStat 7th edition statistical software package employing ANOVA to test significance difference and comparison of means.
Results
Preparation of RNAi Silencing Plasmid Construct
RNAi plasmid construct pKOIISD1 was developed based on plasmid pSD1 with dual convergent opposing RNA polymerase II promoters, PtrpC and Pgpd for filamentous fungi. A PCR amplified fragment of PfHog1 was inserted at the EcoRI cloning site of pKOIISD1 (Figure 1). The RNAi construct was validated by confirming the presence of PfHog1 gene through end point PCR using the primers (pSD15F and pSD13R) designed from the promoters region of pKOIISD1 plasmid construct. The insertion and orientation of insert PfHog1 in pKOIISD1 was further checked by sequencing. The sense strand of PfHog1 gene was driven by PtrpC promoter and its anti-sense strand was regulated by Pgpd promoter (Figure 1). The pSD1 vector system carrying opposing PtrpC and Pgpd promoters have got sense and anti-sense RNA of the target gene PfHog1. This system form dsRNA in the cell and are transcribed independently with control of two opposing RNA polymerase II promoters.
Agrobacterium tumefaciens-Mediated Transformation (ATMT) of P. fijiensis and Molecular Analysis of Transformants
ΔPfHog1 were generated by transformation of mycelial fragments using A. tumefaciens (Figure 2a). Neomycin phosphotransferase II (nptII) gene or (NeoR/KanR) was used as a selectable marker for the selection of transformed P. fijiensis. Agro-infected P. fijiensis were grown and selected on medium containing geneticin. Transformants P. fijiensis were validated by PCR analysis using PfHog1 specific primers. The non-transformed P. fijiensis and pKOIISD1 plasmid were used as negative and positive controls for PCRs, respectively. The expected size (based on plasmid construct map) of amplified fragment of 0.5 kb was obtained from 6 of the 11 putative transformants of P. fijiensis, whereas two bands of 0.5 and 0.3 kb, respectively, were amplified from one of the putative transformants (Figure 2b). No amplification was observed in the non-transformed P. fijiensis that was used as negative control lacking the insert PfHog1.
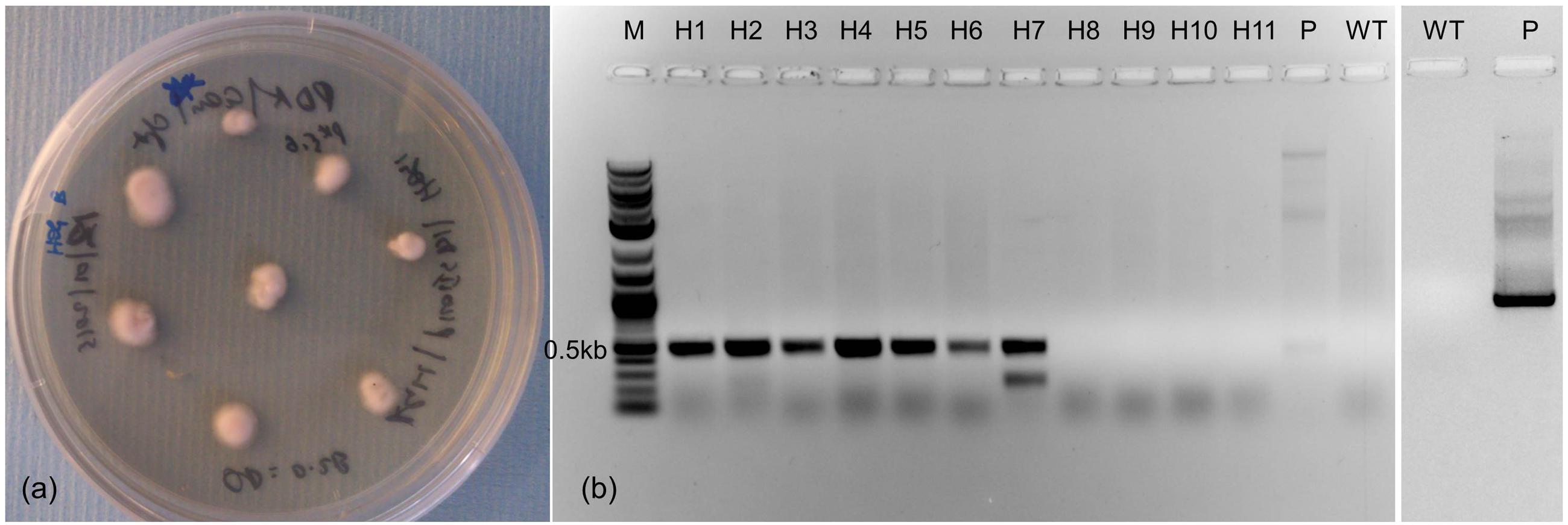
FIGURE 2. Generation of P. fijiensis transformants through Agrobacterium-mediated transformation. (a) Transformed mycelium on selective potato dextrose agar (PDA) medium containing geneticin. (b) Gel showing PCR amplification of PfHog1 gene from genomic DNA of transformants of P. fijiensis. M, molecular marker; P, plasmid pKOIISD1-PfHog1; WT, control non-transformed P. fijiensis.
Expression Analysis of PfHog1 Gene in Transformants of P. fijiensis
The expression of PfHog1 gene was analyzed in the transformants of P. fijiensis by qRT-PCR using primers specific to WT P. fijiensis as a positive control. As low primer specificity can affect the accuracy of qRT-PCR assays, the specificity of the primers used in the qRT-PCR assays was verified by generating a linear regression curve using absolute quantitation. The R square (R2) and efficiency of the primer pair used for the amplification of β-tubulin from P. fijiensis was 0.99981 and 104.8%, respectively, whereas the R2 and efficiency of primer pair used for amplification of the PfHog1 gene was 0.98 and 97.161%, respectively. A high linear correlation (R2> 0.91) suggest that similar experiments can be performed under similar conditions since R2 determines the precision and reproducibility of an experiment. A good qRT-PCR reaction should have an efficiency of 90–110%, which correspond to a slope between -3.58 and -3.10. While the β- tubulin primer had the lowest detection at Ct value of 20.257 and highest detection at Ct value of 33.363, and the PfHog1 primer showed lowest detection at Ct value of 21.943 and highest detection at Ct value of 35.236. The samples with Ct values greater than 35 were considered as negative as this is in line with several q-PCR studies that used Ct value of 35 as the maximum limit for detection (Zhang et al., 2009).
Expression level of PfHog1 gene in the P. fijiensis transformants was performed using Relative (Ct) Quantitation. Only three P. fijiensis mutants (i.e., ΔPfHog1-1, ΔPfHog1-4, and ΔPfHog1-5) were selected for expression in this study. The β-tubulin gene employed as a control of an endogenous housekeeping gene and PfHog1 expression in wild-type non-transformed P. fijiensis as a reference control. Three biological replicate samples of transformed P. fijiensis were used. All the P. fijiensis mutants (i.e., ΔPfHog1-1, ΔPfHog1-4, and ΔPfHog1-5) used in the expression study had silenced PfHog1 gene as shown in Figure 3. Level of ΔPfHog1 gene expression (RQ value) was 0.00131 (0.131%) in ΔPfHog1-1, 0.08891 (8.89%) in ΔPfHog1-4 and 0.00746 (0.746%) in ΔPfHog1-5 as compared to RQ value of 1 (100%) in WT non-transformed P. fijiensis. This result confirmed that mycelium of P. fijiensis can be transformed through A. tumefaciens. About 63% of ΔPfHog1 colonies randomly picked from the generated transformants where found positive.
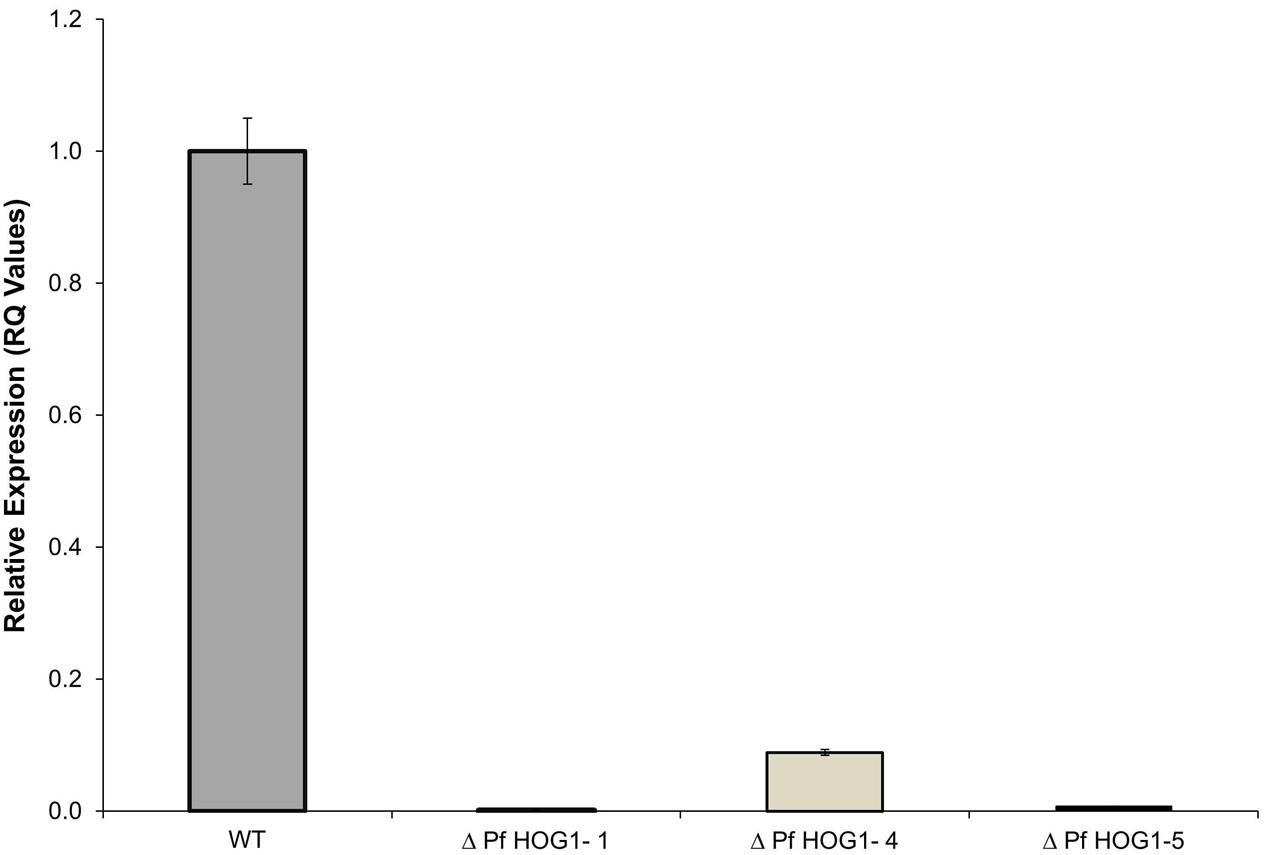
FIGURE 3. Relative expression of PfHog1 gene in the wild type (WT) and silenced mutants P. fijiensis ΔPfHog1 Δ(PfHog1-1, ΔPfHog1-4, and ΔPfHog1-5). Three technical replicates of each mutant and WT control were used in each experiment. The experiment was repeated thrice and data are presented as Mean ± SE.
Response to Osmotic Stress in the PfHog1-Silenced Mutants of P. fijiensis
To study the role of PfHog1 in osmotic stress, assays were performed to investigate tolerance of ΔPfHog1 to high osmotic pressure on PDA medium supplemented with NaCl. Three independent mutants (i.e., ΔPfHog1-1, ΔPfHog1-4, and ΔPfHog1-5) were tested for osmotic stress regulation and similar results were obtained for all mutants tested. There was no difference in mycelial growth between the WT control and ΔPfHog1 on standard PDA medium. However, the mycelial growth of the ΔPfHog1 (i.e., ΔPfHog1-1, ΔPfHog1-4, and ΔPfHog1-5) on PDA medium supplemented with 1 M NaCl was much reduced by 55.46% (p-value = < 0.001, Lsd value = 1.832) at 14 days post-culture on the plates in comparison to WT (Figure 4). This study confirmed that PfHog1 is critical for tolerance to osmotic stress, as 1 M NaCl significantly reduced mycelial growth of the ΔPfHog1 compared to the WT.
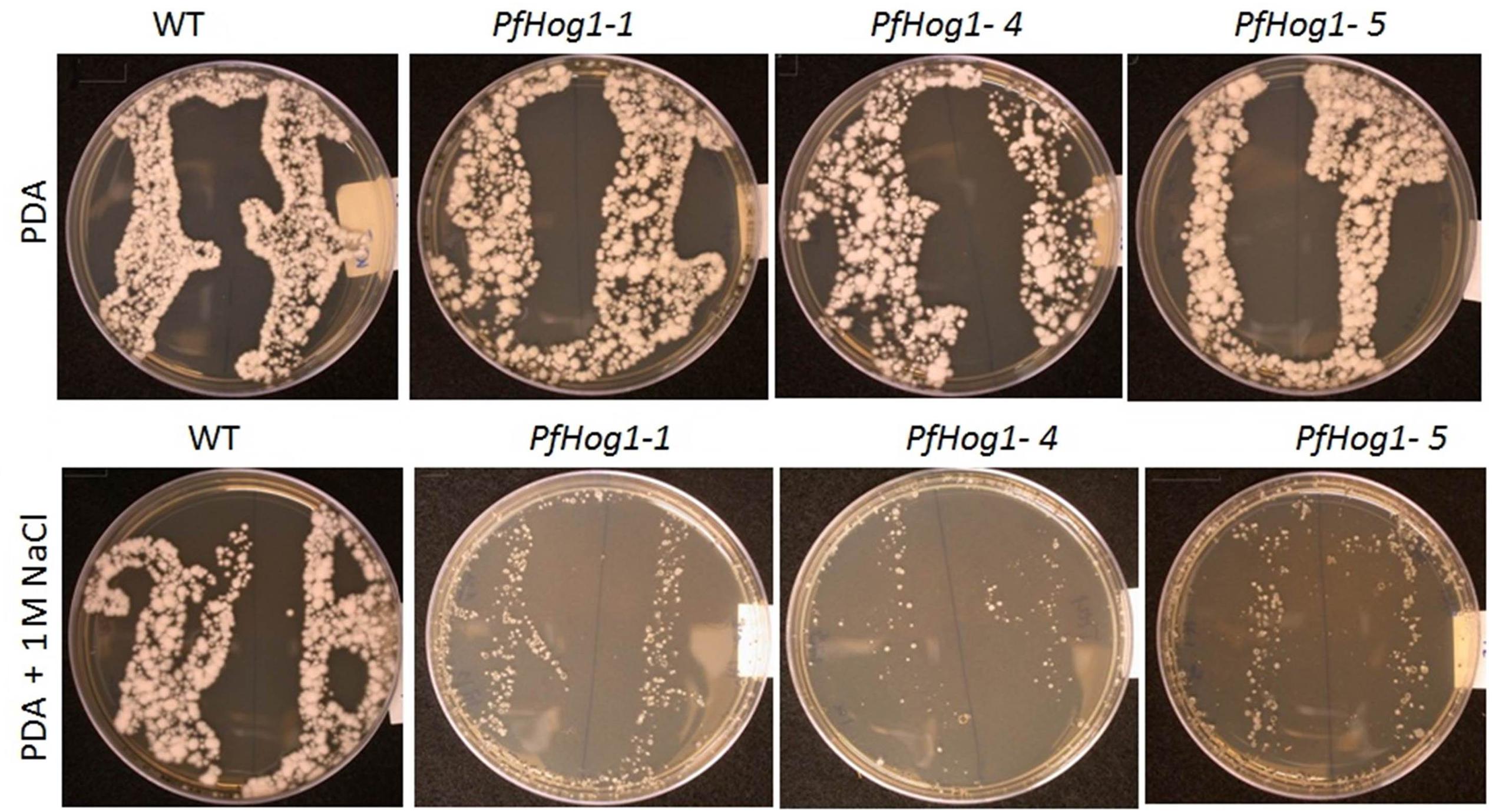
FIGURE 4. Effect of silencing of PfHog1 gene on osmotic stress regulation of P. fijiensis. Growth and development of WT and ΔPf Hog1 mycelium on standard PDA and PDA supplemented with 1 M NaCl.
Virulence of the PfHog1-Silenced Mutant of P. fijiensis on the Banana Host
To determine the role of PfHog1 in virulence of P. fijiensis, banana leaves of cultivar ‘Nakitembe’ were inoculated with mycelia of the ΔPfHog1 mutants (ΔPfHog1-1, ΔPfHog1-4, and ΔPfHog1-5) and WT non-transformed P. fijiensis. The plants inoculated with the WT control strain developed symptoms of black Sigatoka disease in 9 to 10 days post-inoculation (dpi), whereas the development of disease symptoms in plants inoculated with the ΔPfHog1 (ΔPfHog1-1, ΔPfHog1-4, and ΔPfHog1-5) was delayed and started appearing after 15 dpi.
Disease development following the initial appearance of symptoms on the leaves was also much faster in case of the WT control as compared to the ΔPfHog1, as shown by the higher levels of necrosis induced by the former at 45 dpi (Figure 5). Collectively, these results indicate that PfHog1 is an important contributor to virulence of P. fijiensis on its host banana.

FIGURE 5. Effect of silencing of PfHog1 gene on the disease development in banana cultivar ‘Nakitembe’ at 45 days after inoculation with P. fijiensis. Leaves inoculated with 10% rich medium containing 1% tween 20 acted as non-inoculated control. Leaves were inoculated with WT strain of P. fijiensis and three independent mutants ΔPf Hog1 (ΔPfHog1-1, ΔPfHog1-4, and ΔPfHog1-5).
The vegetative growth and development of mycelium of P. fijiensis was monitored in banana leaf tissues. In this experiment, non-infected plant leaf tissues used as negative control was brushed with mixture of 10% rich medium and 1% tween 20 and leaf tissues from plants inoculated with the WT or the ΔPfHog1 were stained with LPCB that binds to fungal chitin (Figure 6). As expected, no fungal growth was detectable in non-infected plant leaves and massive extracellular filamentous fungal growth was detected in leaves from plants inoculated with the WT P. fijiensis. Examination of plant leaves inoculated with the ΔPfHog1 revealed substantially reduced fungal growth as compared to the WT control. No fungal filamentous growth was detected in plant leaves inoculated with the ΔPfHog1, indicating an impaired filamentous growth. ΔPfHog1 remained in cyst or yeast like form within the extracellular space and failed to grow beyond the stomata.
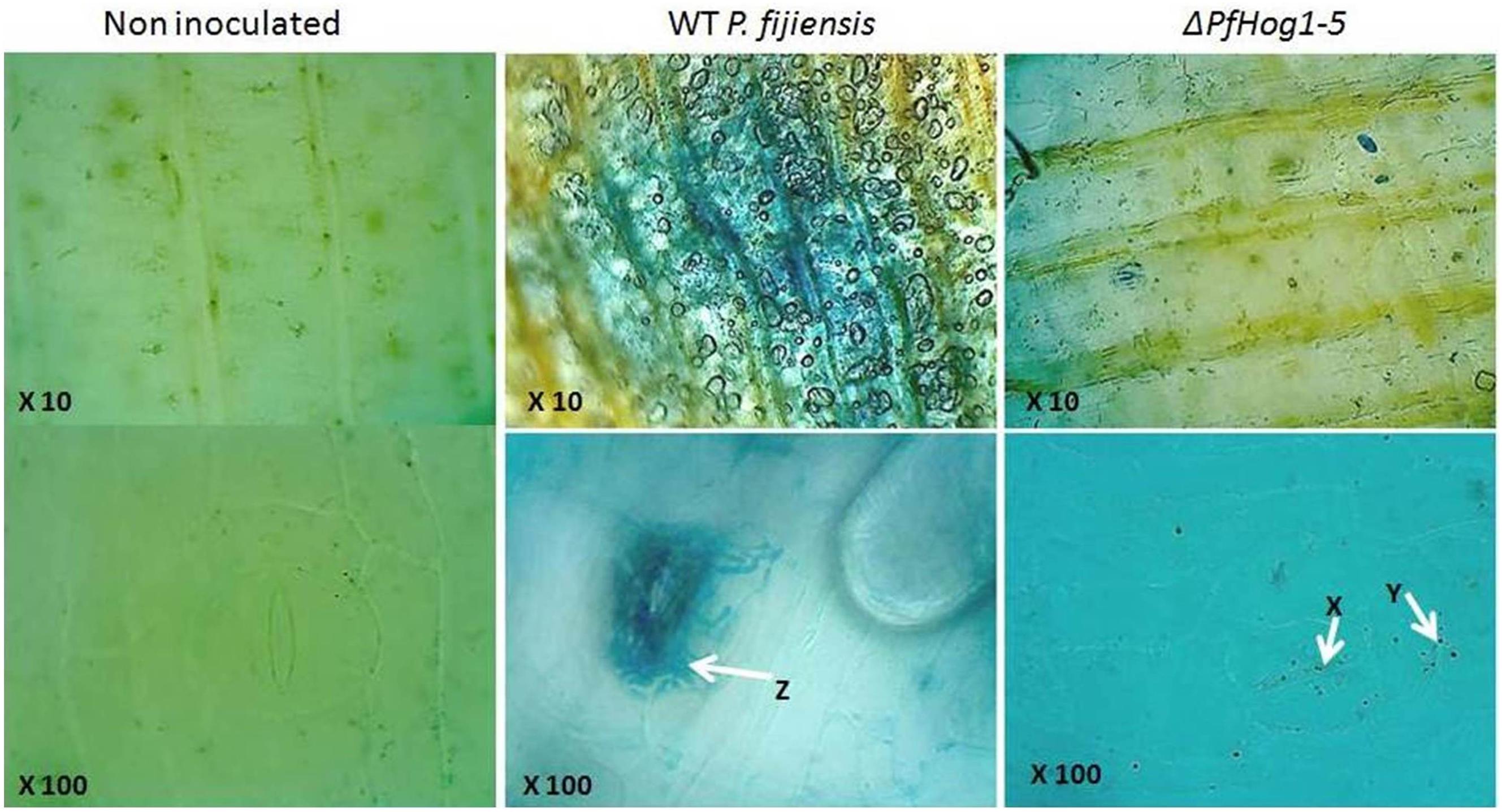
FIGURE 6. Image showing mycelium growth and development in tissue of banana leaf stained with lacto phenol cotton blue. Images were taken at 10X (upper panel) and 100X (lower panel) for non-inoculated banana leaf tissue, leaf inoculated with WT P. fijiensis and leaf inoculated with silenced mutant P. fijiensis (ΔPfHog1-5). Arrow Z is massive mycelium vegetative growth; arrow X, Y non-filamentous or invasive mycelium growth.
Detection and Quantification of P. fijiensis Biomass in Banana Leaves of Infected Plants
The relative growth and biomass of P. fijiensis was quantified in infected leaf tissue by qPCR. Ct values were plotted against the Log10 genomic DNA, to estimate DNA concentration (Y) from samples. A linear regression curve was generated where Y = -0.265X+6.0582. The lowest concentration (0.00062 ng/μl) was detected at Ct value of 33.363, meaning detection above Ct value 33.3 as a negative or noise from amplification. High DNA estimate was obtained in plant leaves inoculated with WT (1.718 ng/g) as compared to leaves inoculated with ΔPfHog1 that had a low DNA estimate (ΔL6 = 0.0517, ΔL8 = 0.0555, ΔL11 = 0.016 corresponding to ΔPfHog1-1, ΔPfHog1-4, and ΔPfHog1-5 mutants, respectively) (Table 1 and Figure 7). This is in agreement with the visual assessment of the virulence assays, where disease severity was lower in plants inoculated with ΔPfHog1 and higher disease severity in plants inoculated with WT.
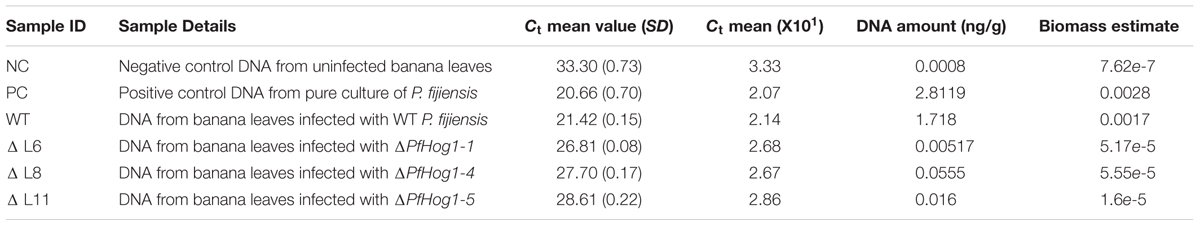
TABLE 1. Estimates of DNA and biomass of P. fijiensis in different banana samples inoculated with wild type (WT) and silenced mutants of P. fijiensis (ΔPfHog1).
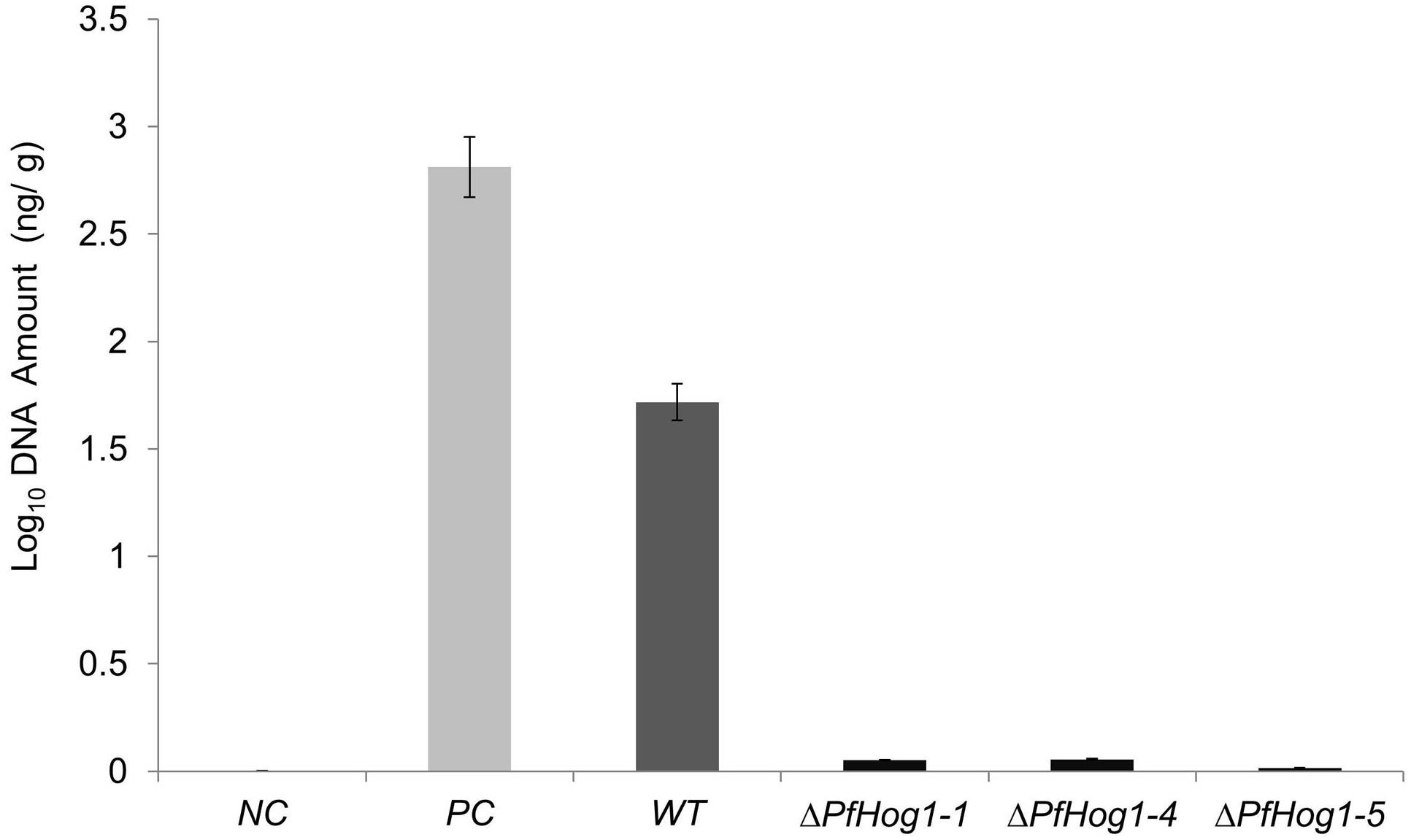
FIGURE 7. The amount of DNA in different samples inoculated with WT and silenced mutant P. fijiensis (ΔPfHog1). NC, Genomic DNA from banana leaf of non-inoculated plant; PC, DNA from pure P. fijiensis culture; WT, DNA from plant inoculated with WT P. fijiensis; ΔPfHog1-1, ΔPfHog1-4, and ΔPfHog1-5, DNA from plants inoculated with mutants. Each experiment has three technical replicates of three mutants and WT. The experiment was replicated twice. All the data are presented as Mean ± SE.
The result confirmed more vegetative growth of P. fijiensis in plants inoculated with the WT non-transformed control in comparison to ΔPfHog1. There was a positive correlation between the amount of DNA in a sample and Ct value. Lower Ct values corresponded to higher amount of DNA/biomass estimate and higher Ct values corresponded to lower amount of DNA/biomass estimate, as seen in Table 1. High relative growth and absolute biomass of P. fijiensis could only be detected in pure culture (positive control) and leaves inoculated with WT P. fijiensis. However, banana leaves inoculated with ΔPfHog1 showed extremely low relative growth and absolute biomass of P. fijiensis. This result confirmed the observation on fungal growth in leaf tissues, the visual virulence assay assessment and leaf staining.
Discussions
We explored the osmotic stress regulation and pathogenicity functions of PfHog1 in P. fijiensis through silencing the targeted gene using RNAi technique. In this study, we used the plasmid vector pKOIISD1, specifically designed for RNAi silencing in fungi. The plasmid pKOIISD1 has nptII as selection marker. The nptII gene or (NeoR/KanR) is the most widely used selection marker. The gene codes for the aminoglycoside 3’-phosphotransferase enzyme, which inactivates by phosphorylation a range of aminoglycoside antibiotics such as kanamycin, neomycin, geneticin (G418), and paromomycin. In this study, transformed P. fijiensis were selected on medium supplemented with geneticin.
The plasmid pKOIISD1 is based on pSilent-Dual1 (pSD1) developed by Nguyen et al. (2008). The pSD1 is an RNA-silencing vector with a convergent dual promoter system that provides a high-throughput platform for functional genomics research in filamentous fungi. Involvement of several calcium signaling related genes in infection related development and pathogenicity of filamentous fungi Magnaporthe oryzae was demonstrated using pSD1 vector system (Nguyen et al., 2008). This provided new evidence that the pSD1 system is more feasible alternative for exploring gene function in fungi. Our study also confirms that pSD1 system is valuable frontline screening tool for gene function before its application in developing transgenic banana with resistance to fungal pathogens. Several post-transcriptional gene silencing studies have been described in protozoa Plasmodium falciparum (Malhotra et al., 2002; McRobert and McConkey, 2002), nematodes Caenorhabditis elegans (Brooks and Isaac, 2002), insects Drosophila melanogaster (Misquitta and Paterson, 1999; Hammond et al., 2001) and filamentous fungi Neurospora crassa (Cogoni et al., 1994). However, little is known about RNAi silencing in P. fijiensis. The ultimate goal of RNAi gene silencing in P. fijiensis is to understand biological function of genes, through phenotyping of silenced P. fijiensis.
Genetic transformation of P. fijiensis has been one of the major challenges in studying pathogenicity genes in this fungus. Several genetic transformation protocols have been developed for P. fijiensis, including using protoplasts (Balint-Kurti et al., 2001) and underwater shock waves with intact conidia (Escobar-Tovar et al., 2015). However, isolating protoplasts from P. fijiensis and stimulation of conidiation in this fungus under artificial conditions is very challenging. P. fijiensis is an economically important pathogen causing black sigatoka disease in banana globally. This pathogen has a number of pathogenicity strategies and the tools that are essential to understand molecular mechanisms are yet to be developed. ATMT have shown to be very efficient in F. oxysporium causing wilt disease in chickpea (Islam et al., 2012) and Colletotrichum acutatum causing anthracnose disease (Talhinhas et al., 2008), however, they used the conidia as compared to mycelium. In this study, a novel RNAi- mediated gene silencing method in P. fijiensis through ATMT of mycelial fragments was developed, which could be used to study function of genes in this fungus. This ATMT protocol helped us to identify mutants for osmolarity and pathogenicity testing, suggesting this technique is applicable for rapid generation of RNAi silenced mutants of P. fijiensis. Further, gene expression analysis for both WT and silenced mutant strains of P. fijiensis, showed that ATMT using mycelia provides the effective method for functional studies of pathogenicity genes.
Our results indicated that PfHog1 is an important regulator of osmotic tolerance in P. fijiensis, similar to previous reports on other filamentous fungi and yeasts (Beever and Laracy, 1986; Schuller et al., 1994; Jin et al., 2012; Babazadeh et al., 2013; Wang et al., 2016). The growth and development of mycelia fragments of ΔPfHog1 mutants were suppressed on PDA medium supplemented with 1 M NaCl; whereas WT P. fijiensis showed normal growth and development of mycelia. Suppression of mycelia growth and development is a result of osmotic stress. Both WT and silenced mutants showed normal mycelia growth and development on PDA medium without 1 M NaCl supplement. This demonstrates that PfHog1 is involved in osmotic stress regulation. Hog1 is well known for its role in osmotic stress regulation and is well studied in Saccharomyces spp. Hog1 mutants of S. cerevisiae showed increased sensitivity to osmotic stress, whereby mutant cells treated with NaCl had reduced colony growth compared to WT (Schuller et al., 1994). Hog1 has also been reported to play an important role in osmotic stress response in Verticillium dahliae (Wang et al., 2016), C. glycerinogenes (Ji et al., 2014) and M. acridum (Jin et al., 2012). NaCl is known to induce osmotic stress. Responses to salt stress are based on osmotic adjustments by osmolyte synthesis and cation transport systems for sodium exclusion (Mager and Siderius, 2002; Yancey, 2005). Therefore, exposing P. fijiensis mycelia in a hyper osmotic environment could lead to a rapid cell dehydration and arrest of cell growth. Under such condition, fungal cells enhance intracellular accumulation of osmolytes, and glycerol cell membranes.
The osmotic stress response seems to be essential during the infection process of P. fijiensis especially during penetration and colonization between plant cells. This is consistent with the observation that Hog1 in Verticillium dahliae (VdHog1) regulates hyperosmotic stress response (Wang et al., 2016). Also the deletion of OSM1 in M. oryzae, a homolog of Hog1, was shown to induce low resistance to osmotic stress, whereby hyperosmotic conditions resulted in morphological defects (Dixon et al., 1999). Similarly, Bipolaris oryzae exhibited reduced growth under hyperosmotic conditions upon deletion of SRM1, which is a homolog of Hog1 (Moriwaki et al., 2007).
We demonstrated that silencing of PfHog1 affects the development of symptoms of black Sigatoka disease. We observed that the banana plants inoculated with the WT P. fijiensis developed symptoms of black Sigatoka disease in 9–10 dpi, however, the plants inoculated with the mutant P. fijiensis showed mild disease symptoms after 15 dpi. Normally, black Sigatoka disease symptoms can appear as early as 10 dpi and could be much severe after 14 dpi under high humidity and high temperatures (Carlier et al., 2000; Marin et al., 2003).
In this study, we demonstrated that silencing of PfHog1 compromised the ability of P. fijiensis to infect banana leaves, due to failing filamentous growth. Indeed ΔPfHog1 failed to switch from yeast like form to filamentous growth in the extracellular space. These results conferred the previous studies in which silencing of Hog1 in Phytophthora sojae impaired zoospore development and growth on both unwounded and wounded soybean (Li et al., 2010). Similarly disruption of MgHog1 led to inability of M. graminicola to infect wheat leaves as a result of failed filamentous growth. In contrast pathogenicity was never compromised in the case of Hog1 mutants of Magnaporthe grisea rice pathogen and C. lagenarium cucumber anthracnose pathogen (Dixon et al., 1999; Kojima et al., 2004). This clearly show that the role of HOG1 pathway in pathogenicity is species specific, meaning only fungal plant pathogens that need to change from yeast- like form to filamentous form are controlled by Hog1 gene. Therefore, here we assigned PfHog1 as a new pathogenicity factor of P. fijiensis since the silenced mutants are impaired in filamentous growth and are unable to penetrate or colonize the host plant tissues as clearly distinguished from the non-mutants which was able to colonize the extracellular space.
Detection and quantification of pathogen biomass in planta is very crucial in studying virulence, growth and development of fungal pathogens. In this study, a novel method of detection and quantification of P. fijiensis in planta was used. The qPCR assay developed here provides the most specific and sensitive technique to quantify P. fijiensis biomass in the banana leaves. β-tubulin was used as reference gene as it is widely and frequently used in qPCR experiments as a representative of constitutively expressed gene in fungi (Faguy and Doolittle, 1998; McKean et al., 2001; Einax and Voigt, 2003). β-tubulin is a good candidate to detection level of DNA and RNA of P. fijiensis. This study is the first report where qPCR was used to detect the relative growth and absolute biomass of P. fijiensis in virulence assay in banana. The amount of DNA detected in nanogram per gram can be converted to microgram per gram to get the biomass of fungal pathogen in the infected leaves. There is no tool at present being used to quantify P. fijiensis biomass in infected banana leaf tissues to determine disease severity. However, disease scores or proportion estimation are commonly being used to determine disease severity for selection of resistance and disease evaluation. This visual evaluation lacks accuracy and precision, where necrosis is low especially amongst the tolerant or resistance cultivars. Lack of well defined estimate compromises effort to understand and define gene regulation pathways and mechanism of resistance in host- pathogen interaction. In this experiment, we could use qPCR assay to quantify the fungal colonization in banana leaf tissue; meaning this study is suitable for quantification or estimation of disease severity of foliar fungal or bacterial pathogens due to its specificity and sensitivity. The results shown here is in line with other studies of relative growth and absolute biomass quantification of fungal pathogens such as F. graminearum in wheat kernel, Botrytis cinerea on grapes, Magnaporthe oryzae in rice and pathogenic fungi in susceptible genotypes of Arabidopsis (Qi and Yang, 2002; Brouwer et al., 2003; Diguta et al., 2010; Horevaj et al., 2011).
In summary, this study concluded that Agrobacterium-mediated transformation of P. fijiensis using mycelium fragments is an easy and efficient way to facilitate functional genomic studies in fungi. RNAi is a potential tool in exploring gene function in Ascomycete and could be an important genetic tool to develop disease resistant crop. This study reports an important effort in developing system for silencing and detecting function of vital fungal genes, which can confer resistance against fungal pathogen in crop plants. Here, we confirm PfHog1 being important in osmotic stress regulation and pathogenicity of P. fijiensis. This study has provided insights for controlling black Sigatoka disease by developing transgenic banana using RNAi technology targeting pathogen gene PfHog1.
Author Contributions
FO conceived and developed the research ideas, performed all experiments, collected data and written manuscript; GT shaped development of research, provided supervision and edited manuscript; L-HC contributed in vector design, gene cloning, and Agrobacterium-mediated transformation of fungus; BF and IS shaped development of research, provided technical guidance and research supervision and edited manuscript; JT supported gene expression studies, microscopy and edited manuscript; WT, JK, and CC provided supervision of research; and LT shaped development of research ideas, provided technical guidance and supervision for conducting research and writing manuscript, critically reviewed and edited manuscript.
Funding
This work was supported in part by research grant from Agricultural Biotechnology Support Project II- USAID feed the Future to National Banana Research Program, NARO Uganda and Norman E. Borlaug Leadership Enhancement in Agriculture Program (LEAP) to University of California Davis, USA.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Thanks to Dr. Andrew Kiggundu for his expert advise, Dr. Richard Omolo for giving access to the quarantine screen house facilities for the virulence assay and Mr. Allan Male for the technical support.
References
Agrawal, N., Dasaradhi, P. V. N., Mohmmed, A., Malhotra, P., Bhatnagar, R. K., and Mukherjee, S. K. (2003). RNA interference: biology, mechanism, and applications. Microbiol. Mol. Biol. Rev. 67, 657–685. doi: 10.1128/MMBR.67.4.657-685.2003
Alonso-Monge, R., Real, E., Wojda, I., Bebelman, J. P., Mager, W. H., and Siderius, M. (2001). Hyperosmotic stress response and regulation of cell wall integrity in Saccharomyces cerevisiae share common functional aspects. Mol. Microbiol. 41, 717–730. doi: 10.1046/j.1365-2958.2001.02549.x
Alvarado-Capo, Y., Leiva-Mora, M., Dita-Rodriguez, M. A., Acosta, M., Cruz, M., Portal, N., et al. (2003). “Early evaluation of black leaf streak resistance by using mycelial suspensions of Mycosphaerella fijiensis,” in Proceedings of the Workshop on Mycosphaerella Leaf Spot Diseases, San José, eds L. Jacome, P. Lepoivre, D. Marin, R. Ortiz, R. Romero, and J. V. Escalant (Montpellier: International Network for the Improvement of Banana and Plantain), 169–175.
Arango Isaza, R. E., Diaz-Trujillo, C., Dhillon, B., Aerts, A., Carlier, J., Crane, C. F., et al. (2016). Combating a global threat to a clonal crop: banana black Sigatoka pathogen Pseudocercospora fijiensis (Synonym Mycosphaerella fijiensis) genomes reveal clues for disease control. PLoS Genet. 12:e1005876. doi: 10.1371/journal.pgen.1005876
Babazadeh, R., Adiels, C. B., Smedh, M., Petelenz-Kurdziel, E., Goksör, M., and Hohmann, S. (2013). Osmostress-induced cell volume loss delays yeast Hog1 signaling by limiting diffusion processes and by Hog1-specific effects. PLoS ONE 8:e80901. doi: 10.1371/journal.pone.0080901
Balint-Kurti, P. J., May, G. D., and Churchill, A. C. L. (2001). Development of a transformation system for Mycosphaerella pathogens of banana: a tool for the study of host/pathogen interactions. FEMS Microbiol. Lett. 195, 9–15. doi: 10.1111/j.1574-6968.2001.tb10490.x
Beever, R. E., and Laracy, E. P. (1986). Osmotic adjustment in the filamentous fungus Aspergillus nidulans. J. Bacteriol. 168, 1358–1365. doi: 10.1128/jb.168.3.1358-1365.1986
Brooks, D. R., and Isaac, R. E. (2002). Functional genomics of parasitic worms: the dawn of a new era. Parasit. Intern. 51, 319–325. doi: 10.1016/S1383-5769(02)00063-6
Brouwer, M., Lievens, B., Hemelrijck, W. V., Ackerveken, G. V. D., Cammue, B. P. A., and Thomma, B. P. H. J. (2003). Quantification of disease progression of several microbial pathogens on Arabidopsis thaliana using real-time fluorescence PCR. FEMS Microbiol. Lett. 228, 241–248. doi: 10.1016/S0378-1097(03)00759-6
Carlier, J., Fouré, E., Gauhl, F., Jones, D. R., Lepoivre, P., Mourichon, X., et al. (2000). Black Leaf Streak. New York, NY: CABI Publishing.
Chen, W., Kastner, C., Nowara, D., Oliveira-Garcia, E., Rutten, T., Zhao, Y., et al. (2016). Host-induced silencing of Fusarium culmorum genes protects wheat from infection. J. Exp. Biol. 67, 4979–4991. doi: 10.1093/jxb/erw263
Cogoni, C., Romano, N., and Macino, G. (1994). Suppression of gene expression by homologous transgenes. Antonie Van Leeuwenhoek 65, 205–209. doi: 10.1007/BF00871948
Diguta, C. F., Rousseaux, S., Weidmann, S., Bretin, N., Vincent, B., Guilloux-Benatier, M., et al. (2010). Development of a qPCR assay for specific quantification of Botrytis cinerea on grapes. FEMS Microbiol. Lett. 313, 81–87. doi: 10.1111/j.1574-6968.2010.02127.x
Dixon, K. P., Xu, J. R., Smirnoff, N., and Talbot, N. J. (1999). Independent signaling pathways regulate cellular turgor during hyperosmotic stress and appressorium-mediated plant infection by Magnaporthe grisea. Plant Cell 11, 2045–2058. doi: 10.1105/tpc.11.10.2045
Einax, E., and Voigt, K. (2003). Oligonucleotide primers for the universal amplification of Beta-tubulin genes facilitate phylogenetic analyses in the regnum Fungi. Organ. Div. Evol. 3, 185–194. doi: 10.1078/1439-6092-00069
Escobar-Tovar, L., Magaña-Ortíz, D., Fernández, F., Guzmán-Quesada, M., Sandoval-Fernández, J. A., Ortíz-Vázquez, E., et al. (2015). Efficient transformation of Mycosphaerella fijiensis by underwater shock waves. J. Microbiol. Methods 119, 98–105. doi: 10.1016/j.funbio.2015.01.002
Faguy, D. M., and Doolittle, W. F. (1998). Cytoskeletal proteins: the evolution of cell division. Curr. Biol. 8, R338–R341. doi: 10.1016/S0960-9822(98)70216-7
FAOSTAT (2013). Agriculture Data. Available at: http://faostat3.fao.org
Gauhl, F., Pasberg-Gauhl, C., and Jones, D. R. (2000). Black Leaf Streak: Disease Cycle and Epidemiology. New York, NY: CABI.
Ghag, S. B., Shekhawat, U. K. S., and Ganapathi, T. R. (2014). Host-induced post-transcriptional hairpin RNA-mediated gene silencing of vital fungal genes confers efficient resistance against Fusarium wilt in banana. Plant Biotechnol. J. 12, 541–553. doi: 10.1111/pbi.12158
Hammond, S. M., Boettcher, S., Caudy, A. A., Kobayashi, R., and Hannon, G. J. (2001). Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293, 1146–1150. doi: 10.1126/science.1064023
Hohmann, S. (2002). Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 66, 300–372. doi: 10.1128/MMBR.66.2.300-372.2002
Horevaj, P., Milus, E. A., and Bluhm, B. H. (2011). A real-time qPCR assay to quantify Fusarium graminearum biomass in wheat kernels. J. Appl. Microbiol. 111, 396–406. doi: 10.1111/j.1365-2672.2011.05049.x
Islam, M. N., Nizam, S., and Verma, P. K. (2012). A highly efficient Agrobacterium mediated transformation system for chickpea wilt pathogen Fusarium oxysporum f. sp. ciceri using DsRed-Express to follow root colonisation. Microbiol. Res. 167, 332–338. doi: 10.1016/j.micres.2012.02.001
Ji, H., Lu, X., Wang, C., Zong, H., Fang, H., Sun, J., et al. (2014). Identification of a novel HOG1 homologue from an industrial glycerol producer Candida glycerinogenes. Curr. Microbiol. 69, 909–914. doi: 10.1007/s00284-014-0670-0
Jin, K., Ming, Y., and Xia, Y. X. (2012). MaHog1, a Hog1-type mitogen-activated protein kinase gene, contributes to stress tolerance and virulence of the entomopathogenic fungus Metarhizium acridum. Microbiology 158, 2987–2996. doi: 10.1099/mic.0.059469-0
Kojima, K., Takano, Y., Yoshimi, A., Tanaka, C., Kikuchi, T., and Okuno, T. (2004). Fungicide activity through activation of a fungal signalling pathway. Mol. Microbiol. 53, 1785–1796. doi: 10.1111/j.1365-2958.2004.04244.x
Li, A., Wang, Y., Tao, K., Dong, S., Huang, Q., DaI, T., et al. (2010). PsSAK1, a stress-activated MAP kinase of Phytophthora sojae, is required for zoospore viability and infection of soybean. Mol. Plant Microbe Interact. 23, 1022–1031. doi: 10.1094/MPMI-23-8-1022i
Mager, H., and Siderius, M. (2002). Novel insights into the osmotic stress response of yeast. FEMS Yeast Res. 2, 251–275. doi: 10.1016/S1567-1356(02)00116-2
Mahuku, G. S. (2004). A simple extraction method suitable for PCR based analysis of plant, fungal and bacterial DNA. Plant Mol. Biol. Rep. 22, 71–81. doi: 10.1007/BF02773351
Malhotra, P., Dasaradhi, P. V. N., Kumar, A., Mohmmed, A., Agrawal, N., Bhatnagar, R. K., et al. (2002). Double-stranded RNA-mediated gene silencing of cysteine proteases (falcipain-1 and -2) of Plasmodium falciparum. Mol. Microb. 45, 1245–1254. doi: 10.1046/j.1365-2958.2002.03105.x
Marin, D. H., Romero, R. A., Guzman, M., and Sutton, T. B. (2003). Black Sigatoka: an increasing threat to banana cultivation. Plant Dis. 87, 208–222. doi: 10.1094/PDIS.2003.87.3.208
McKean, P. G., Vaughan, S., and Gull, K. (2001). The extended tubulin superfamily. J. Cell Sci. 114, 2723–2733.
McRobert, L., and McConkey, G. A. (2002). RNA interference (RNAi) inhibits growth of Plasmodium falciparum. Mol. Biochem. Parasitol. 119, 273–278. doi: 10.1016/S0166-6851(01)00429-7
Mehrabi, R., Zwiers, L.-H., De Waard, M. A., and Kema, G. H. J. (2006). MgHog1 regulates dimorphism and pathogenicity in the fungal wheat pathogen Mycosphaerella graminicola. Mol. Plant Microbe Interact. 19, 1262–1269. doi: 10.1094/MPMI-19-1262
Misquitta, L., and Paterson, B. M. (1999). Targeted disruption of gene function in Drosophila by RNA interference (RNA-i): a role for nautilus in embryonic somatic muscle formation. Proc. Natl. Acad. Sci. U.S.A. 96, 1451–1456. doi: 10.1073/pnas.96.4.1451
Moriwaki, A., Kihara, J., Mori, C., and Arase, S. (2007). A MAP kinase gene, BMK1, is required for conidiation and pathogenicity in the rice leaf spot pathogen Bipolaris oryzae. Microbiol. Res. 162, 108–114. doi: 10.1016/j.micres.2006.01.014
Nguyen, Q. B., Kadotani, N., Kasahara, S., Tosa, Y., Mayama, S., and Nakayashiki, H. (2008). Systematic functional analysis of calcium-signalling proteins in the genome of the rice-blast fungus, Magnaporthe oryzae, using a high-throughput RNA-silencing system. Mol. Microb. 68, 1348–1365. doi: 10.1111/j.1365-2958.2008.06242.x
Nowara, D., Gay, A., Lacomme, C., Shaw, J., Ridout, C., Douchkov, D., et al. (2010). HIGS: Host-Induced Gene Silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell 22, 3130–3141. doi: 10.1105/tpc.110.077040
Ploetz, R. C. (2001). Black sigatoka of banana: The most important disease of a most important fruit. APS Feat. 2, 126. doi: 10.1094/PHI-I-2001-0126-02
Prieto, D., Roman, E., Correia, I., and Pla, J. (2014). The HOG pathway is critical for the colonization of the mouse gastrointestinal tract by Candida albicans. PLoS ONE 9:e87128. doi: 10.1371/journal.pone.0087128
Qi, M., and Yang, Y. (2002). Quantification of Magnaporthe grisea during infection of rice plants using real-time polymerase chain reaction and Northern Blot/Phosphoimaging analyses. Phytopathology 92, 870–876. doi: 10.1094/phyto.2002.92.8.870
Schuller, C., Brewster, J. L., Alexander, M. R., Gustin, M. C., and Ruis, H. (1994). The HOG pathway controls osmotic regulation of transcription via the stress response element (STRE) of the Saccharomyces cerevisiae CTT1 gene. EMBO J. 13, 4382–4389.
Smale, M., Groote, H. D., and Zepeda, F. (2006). “Biosafety and biodiversity risks,” in Genetics Resources Policies Briefs: Promising Crop Biotechnologies for Smallholder Farmers in East Africa: Bananas and Maize. Washington, WA: International Food Policy Research Institute.
Talhinhas, P., Muthumeenakshi, S., Neves-Martins, J. O., Oliveira, H., and Sreenivasaprasad, S. (2008). Agrobacterium-mediated transformation and insertional mutagenesis in Colletotrichum acutatum for investigating varied pathogenicity lifestyles. Mol. Biotechnol. 39, 57–67. doi: 10.1007/s12033-007-9028-1
Thon, M. R., Nuckles, E. M., and Vaillancourt, L. J. (2000). Restriction enzyme-mediated integration used to produce pathogenicity mutants of Colletotrichum graminicola. Mol. Plant Microbe Interact. 13, 1356–1365. doi: 10.1094/MPMI.2000.13.12.1356
Tripathi, L., Tripathi, J. N., Kiggundu, A., Korie, S., Shotkoski, F., and Tushemereirwe, W. K. (2014). Field trial of Xanthomonas wilt disease-resistant bananas in East Africa. Nat. Biotechnol. 32, 868–870. doi: 10.1038/nbt.3007
Wang, Y., Tian, L., Xiong, D., Klosterman, S. J., Xiao, S., and Tian, C. (2016). The mitogen-activated protein kinase gene, VdHog1, regulates osmotic stress response, microsclerotia formation and virulence in Verticillium dahliae. Fungal Genet. Biol. 88, 13–23. doi: 10.1016/j.fgb.2016.01.011
Winkler, A., Arkind, C., Mattison, C. P., Burkholder, A., Knoche, K., and Ota, I. (2002). Heat stress activates the yeast high-osmolarity glycerol mitogen-activated protein kinase pathway, and protein tyrosine phosphatases are essential under heat stress. Eukaryot. Cell 1, 163–173. doi: 10.1128/EC.1.2.163-173.2002
Yancey, H. (2005). Organic osmolytes as compatible, metabolic and counteracting cytoprotectants in high osmolarity and other stresses. J. Exp. Biol. 208, 2819–2830. doi: 10.1242/jeb.01730
Zhang, Y. J., Fan, P. S., Zhang, X., Chen, C. J., and Zhou, M. G. (2009). Quantification of Fusarium graminearum in harvested grain by real-time polymerase chain reaction to assess efficacies of fungicides on Fusarium head blight, deoxynivalenol contamination, and yield of winter wheat. Phytopathology 99, 95–100. doi: 10.1094/Phyto-99-1-0095
Keywords: Agrobacterium tumefaciens, transformation, Pseudocercospora fijiensis, HOG1, osmotic stress, virulence
Citation: Onyilo F, Tusiime G, Chen L-H, Falk B, Stergiopoulos I, Tripathi JN, Tushemereirwe W, Kubiriba J, Changa C and Tripathi L (2017) Agrobacterium tumefaciens-Mediated Transformation of Pseudocercospora fijiensis to Determine the Role of PfHog1 in Osmotic Stress Regulation and Virulence Modulation. Front. Microbiol. 8:830. doi: 10.3389/fmicb.2017.00830
Received: 15 March 2017; Accepted: 24 April 2017;
Published: 16 May 2017.
Edited by:
Hector Mora Montes, Universidad de Guanajuato, MexicoReviewed by:
Luis Castillo, University of La Serena, ChileIrene Castano, Institute for Scientific and Technological Research, Mexico
Copyright © 2017 Onyilo, Tusiime, Chen, Falk, Stergiopoulos, Tripathi, Tushemereirwe, Kubiriba, Changa and Tripathi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Leena Tripathi, l.tripathi@cgiar.org
 Francis Onyilo
Francis Onyilo Geoffrey Tusiime
Geoffrey Tusiime Li-Hung Chen
Li-Hung Chen Bryce Falk
Bryce Falk Ioannis Stergiopoulos
Ioannis Stergiopoulos Jaindra N. Tripathi
Jaindra N. Tripathi Wilberforce Tushemereirwe1
Wilberforce Tushemereirwe1 Leena Tripathi
Leena Tripathi