- 1State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, The First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, China
- 2Key Laboratory of Clinical In Vitro Diagnostic Techniques of Zhejiang Province, Department of Laboratory Medicine, The First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, China
- 3Department of Ultrasound, The First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, China
Analysis of the genotypic characteristics and antimicrobial susceptibility patterns of methicillin-resistant Staphylococcus aureus (MRSA) is essential for the control and treatment of diseases caused by this important pathogen. In this study, MRSA isolates obtained from a tertiary caret hospital in China were subjected to spa typing, SCCmec typing, multiple locus sequence typing (MLST), and PCR targeting of the genes encoding Panton-Valentine leukocidin (PVL). The disk diffusion method was used to test the antimicrobial susceptibility of the isolates to 10 non-beta-lactam antibiotics. Among the 120 MRSA isolates studied, 18 spa types and 15 ST types were identified. The spa t311 type was the most common (a total of 60 isolates; 50%) among the study strains, and nearly all the t311 strains belonged to ST5, which is the most common ST type that was previously reported from China among the t002 isolates. ST5-II/t311 was the major prevalent clone (55, 45.8%), which was followed by ST5-II/t002 (12, 10.0%) and ST59-IV/t437 (11, 9.2%). PVL-encoding genes were found in 6.7% of the isolates. Although the ST5-II/t311 and ST5-II/t002 clones are different spa types, they shared the same resistance profile (clindamycin, erythromycin, and ciprofloxacin). Most isolates of the ST239-III/t037 clone were resistant to clindamycin, erythromycin, ciprofloxacin, gentamicin, tetracycline, and trimethoprim/sulfamethoxazole. By contrast, the MRSA isolates of the ST239-III/t030 clone were more resistant to rifampin, but they were susceptible to trimethoprim/sulfamethoxazole. Our data emphasize the need for ongoing epidemiologic surveillance.
Introduction
Staphylococcus aureus (S. aureus) is a major pathogen that causes diseases ranging from wound infections to life-threatening bacteremia, ventilator-associated pneumonia, and sepsis. Methicillin-resistant S. aureus (MRSA) has emerged as a global pathogen in both hospital and community settings: multiple healthcare-associated MRSA (HA-MRSA) and community-associated MRSA (CA-MRSA) clones have disseminated internationally, which highlights the adaptation of the species to diverse ecological niches (Cuny et al., 2010). The distribution of MRSA clones is dynamic and geographically unique. Information regarding the molecular characteristics and antimicrobial susceptibility patterns of MRSA is essential for controlling and treating diseases caused by this medically important pathogen. As the epidemiology of infections with MRSA has changed, accurate information regarding the scope, and magnitude of MRSA infections is needed. The MRSA problem is not well-recognized in Chinese hospitals and communities. A nationwide, laboratory-based, multicenter surveillance study reported that ST239-III/t030, ST239-III/t037, and ST5-II/t002 were the predominant HA-MRSA clones, but the prevalence rates of these major clones varied markedly in different administrative regions (Xiao et al., 2013). A recent study (Cheng et al., 2013) also showed that these three major clones were associated with two characteristic resistance profiles: gentamicin/ciprofloxacin/rifampicin/levofloxacin for ST239-III/t030 clone and gentamicin/ciprofloxacin/clindamycin/erythromycin/tetracycline/levofloxacin/trimethoprim/sulfamethoxazole for ST239-III/t037 and ST5-II/t002 clones. Considering China's vast territory, the extent to which MRSA clones have spread into Chinese hospitals remains largely unknown. In this study, we determined the genotypes and resistance profiles of MRSA in a tertiary hospital in Eastern China that has 2,600 beds.
Materials and Methods
MRSA Isolates
This study was performed at the First Affiliated Hospital, College of Medicine, Zhejiang University, which is a tertiary hospital in China. A total of 120 non-duplicate clinical MRSA isolates obtained during January of each year from 2010 to 2014 were included in the current study. Identification of S. aureus was confirmed by MALDI-TOF MS (Bruker Daltonics, Bremen, Germany) and phenotypic methicillin resistance was assessed using the cefoxitin disk diffusion method in accordance with the Clinical and Laboratory Standard Institute guidelines (CLSI M100-S26) at our clinical laboratory, which has been accredited by the College of American Pathologists (Clinical Laboratory Standards Institute, 2016). The clinical sources of the isolates included the respiratory tract (n = 51), skin and soft tissue (n = 21), blood culture (n = 30), body fluid (n = 16), and urine (n = 2). A total of 85 (70.8%) isolates were obtained from inpatient cultures obtained >48 h after hospital admission, whereas 35 (29.2%) isolates were obtained from outpatients who had no history of hospitalization or invasive medical procedures in the prior year.
DNA Extraction
All isolates were cultured on blood agar and incubated overnight at 37°C. DNA was isolated using a QIAamp DNA Mini Kit (Qiagen, CA, USA) according to the manufacturer's instructions. The isolated DNA was used as the template for all PCR reactions.
Confirmative Identification of MRSA and Detection of pvl
All S. aureus isolates with phenotypic methicillin resistance were first screened for the presence of the mecA gene, as previously described (Murakami et al., 1991). When mecA-negative isolates were found, further screening for the presence of the mecC gene was performed using a specific PCR method (Cuny et al., 2011). The isolates carrying the mecA or mecC gene were identified as MRSA (Ito et al., 2012). The presence of genes encoding Panton-Valentine leukocidin (PVL) was determined using a previously described PCR strategy (Larsen et al., 2008).
Staphylococcal Protein A (spa) Typing
Spa typing involved PCR amplification and subsequent sequencing of the highly variable X region in the staphylococcal protein A using previously described primers spa-1113f and spa-1514r (Larsen et al., 2008). The spa sequence was analyzed using the spa database website (http://www.ridom.de/spaserver) to assign a unique spa type.
SCCmec Typing
SCCmec typing was determined using a multiplex polymerase chain reaction (PCR) strategy that was previously described as I–V (Boye et al., 2007). The MRSA isolates that had unanticipated fragments or that were lacking fragments by multiplex PCR were defined as non-typeable (NT).
Multiple Locus Sequence Typing (MLST)
MLST was performed on all isolates by sequencing the internal fragments of seven housekeeping genes (arcC, aroE, glpF, gmK, pta, tpi, and yqiL). The sequence profile and sequence type of each allele were determined according to the MLST database (http://saureus.mlst.net). The allelic profiles were assigned by comparing the sequences at each locus with those of the known alleles in the S. aureus MLST database and were accordingly defined as the sequence types (STs). For each locus, distinct allelic variants were assigned an allelic number and each unique combination of seven allele numbers was assigned a novel ST. Bionumerics v.6.06 (http://www.applied-maths.com) was used to generate a minimum spanning tree (MST) from the non-concatenated sequences of seven alleles of the 120 isolates.
Antimicrobial Susceptibility Tests
Antimicrobial susceptibility testing of all the MRSA isolates to 10 non-beta-lactam antibiotics was determined using the disk diffusion method, which was performed according to the CLSI guidelines (M100-S26). The tested antimicrobial agents included erythromycin (ERY), clindamycin (CLI), rifampin (RIF), ciprofloxacin (CIP), gentamicin (GEN), tetracycline (TET), trimethoprim/sulfamethoxazole (SXT), linezolid (LZD), teicoplanin (TEC), and vancomycin (VAN) (Oxoid Ltd., Basingstoke, Hants, Chicago). S. aureus ATCC 25923 was used as a quality control strain.
Definitions
The clone comprising 10% of the isolates was considered the major prevalent clone (Cheng et al., 2013). An isolate was considered multidrug-resistant (MDR) when the isolate was resistant to three or more classes of non-beta-lactam antimicrobial agents (Wang et al., 2008). Antibiotic resistance profiles (ARPs) of MRSA isolates were generated and included antimicrobial agents to which more than 70% of the MRSA isolates were resistant (Cheng et al., 2013).
Ethics Statement
This study was performed in accordance with the recommendations of The First Affiliated Hospital, College of Medicine, Zhejiang University and the Declaration of Helsinki. Written informed consent was obtained from all participating subjects.
Results
Genotyping and pvl Detection
Among the 120 MRSA isolates studied, 15 different STs were identified. The three major STs were ST5 (71, 59.2%), ST239 (18, 15.0%), and ST59 (15, 12.5%). The remaining STs were ST1, ST7, ST22, ST398, ST630, ST764, ST944, ST965, ST1611, ST3355, ST3360, and ST3361, which had frequencies ranging from 0.8 to 1.7% (Figure 1). Among them, ST1611, ST3355, ST3360, and ST3361 were novel and have been deposited in the MLST database. SCCmec typing revealed that SCCmec II (75, 62.5%) was the most common, which was followed by SCCmec III (17, 14.2%), IV (16, 13.3%), and V (5, 4.2%). A total of 7 (5.8%) isolates were defined as NT. Further molecular characterizations showed that most isolates of ST5 carried SCCmec II, whereas ST239 carried SCCmec III and ST59 carried SCCmec IV. In addition, a total of 18 genotypes were identified in the 120 strains by spa typing. The most common spa types were t311 (60, 50.0%), t437 (15, 12.5%), t002 (12, 10.0%), t030 (10, 8.3%), and t037 (7, 5.8%). t311 was a common genotype that had been identified annually in this hospital at a prevalence ranging from 37.0 to 70.0%. The remaining 13 spa types accounted for a total of 13.4% of the isolates, none of which accounted for more than 2%. Five major spa types, including t311 (91.7% belonging to ST5), t437 (73.3%, ST59), t002 (100%, ST5), t030 (100%, ST239), and t037 (85.7%, ST239), prevailed in many departments at this hospital (data not shown). The most common genotype was ST5-II/t311 (55, 45.8%), which was followed by ST5-II/t002 (12, 10.0%), ST59-IV/t437 (11, 9.2%), ST239-III/t030 (10, 8.3%), and ST239-III/t037 (6, 5.0%).
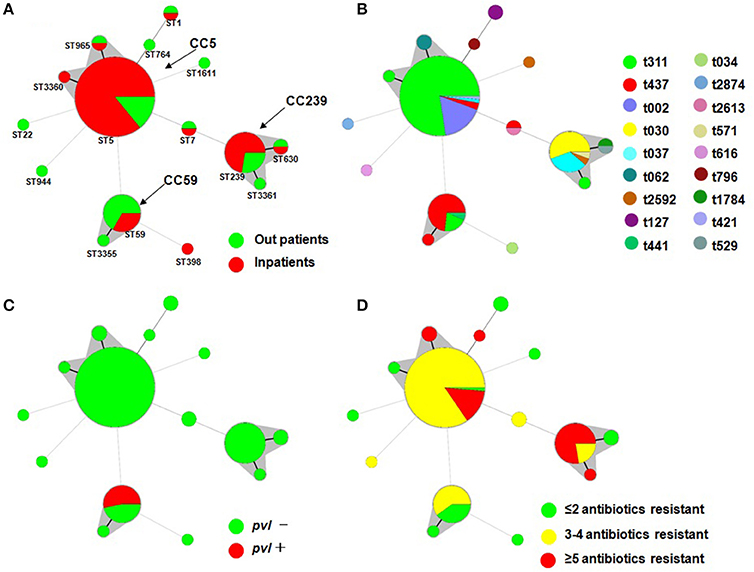
Figure 1. Minimal Spanning Tree (MST) analysis of MRSA strains based on MLST data. Each circle corresponds to an ST. The area of each circle corresponds to the number of isolates. The relationships between the strains are indicated by the connections between the isolates and the lengths of the branches linking them. Black lines connecting pairs of STs indicate that they differ in one allele (thick lines), two alleles (thin lines), or three to seven alleles (dashed lines, refer to ST22-ST5, ST944-ST5, ST59-ST5, ST7-ST5, ST1611-ST5, ST1-ST764, ST398-ST59 and ST7-ST239). Gray zones surround STs belonging to the same clonal complex (the clonal complexes were defined from this collection, and CC5 was predominant). Four MST graphs were separately generated based on the following associations: (A) ST vs. patients; (B) ST vs. spa; (C) ST vs. pvl; and (D) ST vs. multidrug resistance.
Overall, pvl was detected in 8 MRSA isolates (6.7%) that were distributed annually, except for 2014 (Table 1). All pvl-positive isolates belonged to ST59-t437 (Figure 1).
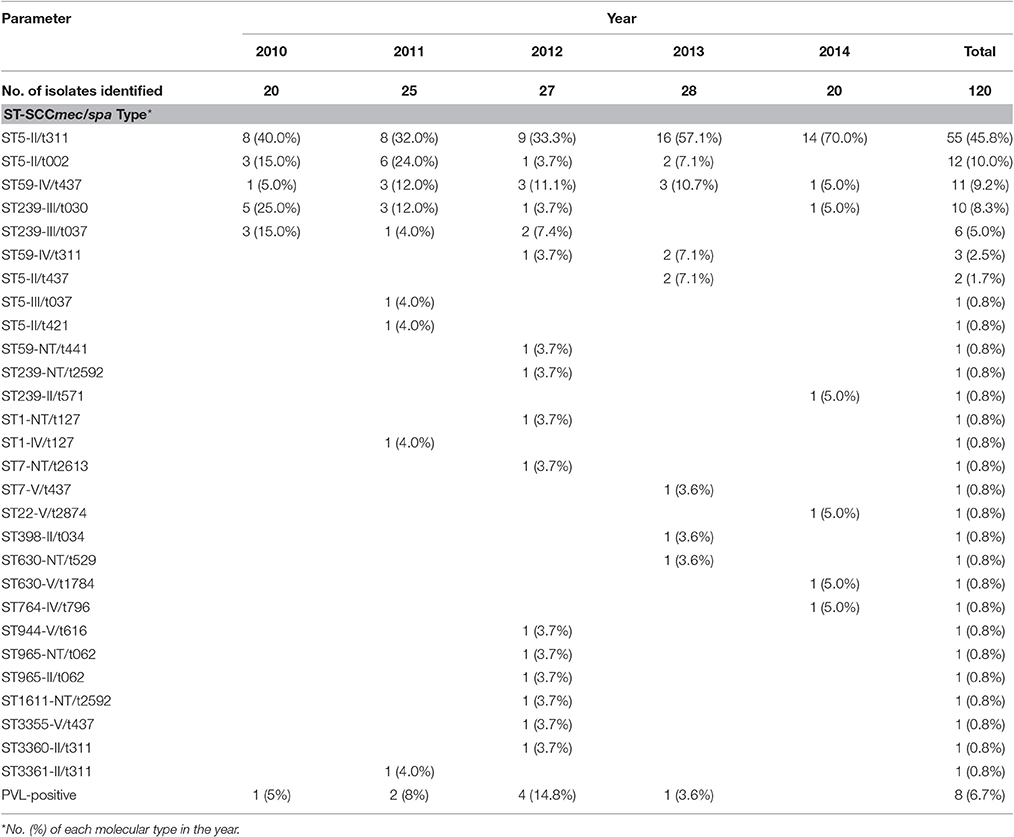
Table 1. Epidemiology of 120 MRSA isolates over 5 years including the distributions of SCCmec, spa, and MLST types.
Antimicrobial Resistance
All the MRSA isolates were susceptible to VAN, LZD, and TEC. The detailed antibiograms of the isolates for the other 7 non-beta-lactam antibiotics stratified by the MLST, SCCmec, and spa types are shown in Table 2. Isolates of the different genetic lineages exhibited distinct patterns of antibiotic susceptibilities. All the ST239 strains were MDR, with resistance to a median of 5 antibiotics for the t030 isolates (range, 4–7) and with a median of 6 antibiotics for the t037 isolates. Interestingly, ST5-II/t311 and ST5-II/t002 isolates shared the same ARPs, and they were frequently resistant to CLI (94.5–100%), ERY (96.7–100%), and CIP (98.2–100%), whereas ST239 isolates were generally resistant to CIP (100%), GEN (90–100%), and TET (100%). The t030 isolates were less likely than the t037 isolates to be resistant to CLI (60 and 100%, respectively) and ERY (60 and 100%, respectively). Notably, ST239-III/t030 isolates showed 100% resistance to RIF and 100% susceptibility to SXT, whereas ST239-III/t037 isolates showed the opposite pattern (Table 2).
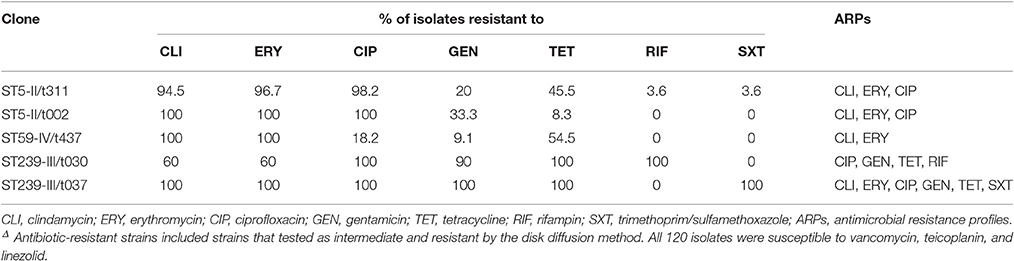
Table 2. Antibiotic resistant rates of 120 methicillin-resistant Staphylococcus aureus isolates by MLST, SCCmec, and spa typesΔ.
MLST Analysis and Phylogenetic Relationship
To determine the clonal relationship between the isolates, we used a Minimal Spanning Tree (MST) algorithm based on the allelic profiles. The 120 sequence-typed isolates were distributed into 9 isolates that did not belong to any clonal complexes (CCs) and 3 groups with a total of 111 isolates corresponding to CCs found in the database (Figure 1). The main detected CC was CC5, which consisted of three STs (ST5, ST965, and ST3360; ST5 was the primary founder). The second most frequently encountered CC, CC239, consisted of three STs (ST239, ST630, and ST3361; ST239 was the primary founder). The third encountered CC, CC59, consisted of two STs (ST59 and ST3355; ST59 was the primary founder). Another group was a doublet in which ST1 and ST764 were linked, and ST7, ST22, ST398, ST944, and ST1611 formed a simple complex (Figure 1).
The MST analysis revealed several interesting relationships with respect to spa type, patients, MDR phenotype and presence of the virulence gene (Figure 1). Three main STs were shared by more than 3 spa types (ST5 by 5, ST239 by 4, and ST59 by 3), and most ST5 and ST239 isolates were MDR and did not carry the pvl gene. It is noteworthy that ST59 consisted of 15 strains, 11 of which were t437, and all 8 pvl-positive strains belonged to t437. A total of 72.2% of ST239 isolates were resistant to more than five antibiotics, and only 14.1% of ST5 isolates were resistant to more than five antibiotics. In addition, 66.7% of the ST59 strains were isolated from outpatients, and 33.3% of ST59, 85.9% of ST5, and 72.2% of ST239 were isolated from inpatients.
Discussion
MRSA is considered the most significant multidrug-resistant organism that causes infections, and the global spread of MRSA has been a major problem worldwide (Song et al., 2011; Klein et al., 2013). Analysis of the genotypic characteristics of MRSA clones is valuable for understanding the evolution and dissemination of MRSA (Stefani et al., 2012). In the present study, we characterized a cluster of MRSA strains obtained from a tertiary hospital in Hangzhou, China. There were limitations to this study. One major bias is the limited number of isolates analyzed. Nevertheless, the high prevalence of ST5-II/t311, a clone that was not previously popular in China or any other Asian country, is significant and striking.
In this study, we found that CC5 was the most common CC, which was followed by CC239 and CC59. This result does not agree with previous reports showing that the predominant CCs were CC239, CC5, and CC59 in China (Cheng et al., 2013; Xiao et al., 2013). Furthermore, ST5 (CC5) was the most predominant ST for SCCmec II isolates, and the most common spa type in the ST5 isolates was t311 (77.5%, 55/71). ST5 is the most common previously reported ST type from China among the t002 isolates (Xiao et al., 2013; Cheng et al., 2013). The repetitive sequence of t311 (26-23-17-34-20-17-12-17-16) is closely related to that of t002 (26-23-17-34-17-20-17-12-17-16), with only one repetition of difference between these two spa types. ST5-II was initially described as the main clone in the USA (Stefani et al., 2012) and Japan (Aires et al., 2000), and it was subsequently detected in several European and Asian countries (Baba et al., 2002; Song et al., 2011). ST5-II was prevalent in 2009 the Chinese cities of Shenyang and Dalian (Liu et al., 2009). ST5-II/t311 has been found in many other countries, such as Brazil, Nigerian, Angola, Argentina, and Oman (Camargo et al., 2013; Kolawole et al., 2013; Conceição et al., 2014; Egea et al., 2014; Udo et al., 2014). From 2010 to 2014, ST5-II/t311 has surpassed ST5-II/t002 as the predominant clone in our hospital; however, its mechanism remains to be further elucidated.
Of note, 15 (12.5%) of the ST59 isolates were identified in our MRSA collection, and more than half of these were PVL-positive. In addition, the ST59 isolates had less antibiotic resistance than ST239 and ST5 isolates. Clones harboring SCCmec IV or V and carrying PVL are usually reported in the context of community-onset infections, and ST59 is known to be the most common CA-MRSA clone among Chinese children (Geng et al., 2010; Wang et al., 2012). In Hong Kong and Taiwan, CA-MRSA clones, including ST59, have spread to hospital settings (Song et al., 2011). Dispersal is also facilitated by the emergence and persistence of MDR clones in hospitals. Our findings further suggest that hospitals in Mainland China are facing the same situation. The findings from this study further support that CA-MRSA strains usually remain susceptible to non-beta-lactam antibiotics and often carry the pvl gene that encodes PVL, a phage-borne leukotoxin (Okuma et al., 2002).
The association of MRSA ARPs with their molecular characteristics can provide useful information for the clinical selection of antibiotics. MRSA isolates also harbor numerous determinants associated with antibiotic resistance (Wendlandt et al., 2013; Yuan et al., 2013). The important finding from this study is that the five major clones were associated with four characteristic ARPs (Table 2). Despite having different spa types, ST5-II/t311 and ST5-II/t002 clones shared the same resistance profile (CLI, ERY, and CIP). Most isolates of ST239-III/t037 clones were resistant to CLI, ERY, CIP, GEN, TET, and SXT. By contrast, the MRSA isolates of the ST239-III/t030 clone were more resistant to RIF, but they were susceptible to SXT, which is concordant with previous reports (Chen et al., 2010; Cheng et al., 2013). This finding suggests that the clinical selection of antibiotics based on typing information is advantageous for treating patients with MRSA infections.
The present study revealed that ST5 was strongly associated with t311 type strains, which has gradually increased in prevalence during the past 5 years. The majority of ST5 strains were resistant to 3 or 4 non-beta-lactam antibiotics and non-ST5 strains harbored the pvl virulence gene. Ultimately, our dataset and analysis demonstrate that ST5-II/t311 is a major prevalent clone that is widespread in the hospital.
Author Contributions
HK and HW designed the experiments, HK and WZ conducted the experiment(s), and XL and FY analyzed the results. All authors reviewed the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by grants from the Department of Science and Technology of Zhejiang Province (2016C33133) and the National Key Basic Research Program (973) of China (2013CB531605).
References
Aires, M., de Lencastre, H., Santos Sanches, S. I., Kikuchi, K., Totsuka, K., and Tomasz, A. (2000). Similarity of antibiotic resistance patterns and molecular typing properties of methicillin-resistant Staphylococcus aureus isolates widely spread in hospitals in New York City and in a hospital in Tokyo, Japan. Microb. Drug Resist. 6, 253–258. doi: 10.1089/mdr.2000.6.253
Baba, T., Takeuchi, F., Kuroda, M., Yuzawa, H., Aoki, K., Oguchi, A., et al. (2002). Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359, 1819–1827. doi: 10.1016/S0140-6736(02)08713-5
Boye, K., Bartels, M. D., Andersen, I. S., Møller, J. A., and Westh, H. (2007). A new multiplex PCR for easy screening of methicillin-resistant Staphylococcus aureus SCCmec types I-V. Clin. Microbiol. Infect. 13, 725–727. doi: 10.1111/j.1469-0691.2007.01720.x
Camargo, C. H., da Cunha Mde, L., Bonesso, M. F., Da Cunha, F., Barbosa, A. N., and Fortaleza, C. M. (2013). Systemic CA-MRSA infection following trauma during soccer match in inner Brazil: clinical and molecular characterization. Diagn. Microbiol. Infect. Dis. 76, 372–374. doi: 10.1016/j.diagmicrobio.2013.03.013
Chen, H., Liu, Y., Jiang, X., Chen, M., and Wang, H. (2010). Rapid change of methicillin-resistant Staphylococcus aureus clones in a Chinese tertiary care hospital over a 15-year period. Antimicrob. Agents Chemother. 54, 1842–1847. doi: 10.1128/AAC.01563-09
Cheng, H., Yuan, W., Zeng, F., Hu, Q., Shang, W., Tang, D., et al. (2013). Molecular and phenotypic evidence for the spread of three major methicillin-resistant Staphylococcus aureus clones associated with two characteristic antimicrobial resistance profiles in China. J. Antimicrob. Chemother. 68, 2453–2457. doi: 10.1093/jac/dkt213
Clinical and Laboratory Standards Institute (2016). Performance Standards for Antimicrobial Susceptibility Testing. Twenty-Sixth Informational Supplement M100-S26. Wayne, PA: Clinical and Laboratory Standards Institute.
Conceição, T., Coelho, C., Santos-Silva, I., de Lencastre, H., and Aires-de-Sousa, M. (2014). Epidemiology of methicillin-resistant and -susceptible Staphylococcus aureus in Luanda, Angola: first description of the spread of the MRSA ST5-IVa clone in the African continent. Microb. Drug Resist. 20, 441–449. doi: 10.1089/mdr.2014.0007
Cuny, C., Friedrich, A., Kozytska, S., Layer, F., Nübel, U., Ohlsen, K., et al. (2010). Emergence of methicillin-resistant Staphylococcus aureus (MRSA) in different animal species. Int. J. Med. Microbiol. 300, 109–117. doi: 10.1016/j.ijmm.2009.11.002
Cuny, C., Layer, F., Strommenger, B., and Witte, W. (2011). Rare occurrence of methicillin-resistant Staphylococcus aureus CC130 with a novel mecA homologue in humans in Germany. PLoS ONE 6:e24360. doi: 10.1371/journal.pone.0024360
Egea, A. L., Gagetti, P., Lamberghini, R., Faccone, D., Lucero, C., Vindel, A., et al. (2014). New patterns of methicillin-resistant Staphylococcus aureus (MRSA) clones, community-associated MRSA genotypes behave like healthcare-associated MRSA genotypes within hospitals, Argentina. Int. J. Med. Microbiol. 304, 1086–1099. doi: 10.1016/j.ijmm.2014.08.002
Geng, W., Yang, Y., Wu, D., Zhang, W., Wang, C., Shang, Y., et al. (2010). Community-acquired, methicillin-resistant Staphylococcus aureus isolated from children with community-onset pneumonia in China. Pediatr. Pulmonol. 45, 387–394. doi: 10.1002/ppul.21202
Ito, T., Hiramatsu, K., Tomasz, A., de Lencastre, H., Perreten, V., et al. (2012). Guidelines for reporting novel mecA gene homologues. Antimicrob. Agents Chemother. 56, 4997–4999. doi: 10.1128/AAC.01199-12
Klein, E. Y., Sun, L., Smith, D. L., and Laxminarayan, R. (2013). The changing epidemiology of methicillin-resistant Staphylococcus aureus in the United States: a national observational study. Am. J. Epidemiol. 177, 666–674. doi: 10.1093/aje/kws273
Kolawole, D. O., Adeyanju, A., Schaumburg, F., Akinyoola, A. L., Lawal, O. O., Amusa, Y. B., et al. (2013). Characterization of colonizing Staphylococcus aureus isolated from surgical wards' patients in a Nigerian university hospital. PLoS ONE 8:e68721. doi: 10.1371/journal.pone.0068721
Larsen, A. R., Stegger, M., and Sørum, M. (2008). spa typing directly from a mecA, spa and pvl multiplex PCR assay-a cost-effective improvement for methicillin-resistant Staphylococcus aureus surveillance. Clin. Microbiol. Infect. 14, 611–614. doi: 10.1111/j.1469-0691.2008.01995.x
Liu, Y., Wang, H., Du, N., Shen, E., Chen, H., Niu, J., et al. (2009). Molecular evidence for spread of two major methicillin-resistant Staphylococcus aureus clones with a unique geographic distribution in Chinese hospitals. Antimicrob. Agents Chemother. 53, 512–518. doi: 10.1128/AAC.00804-08
Murakami, K., Minamide, W., Wada, K., Nakamura, E., Teraoka, H., and Watanabe, S. (1991). Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J. Clin. Microbiol. 29, 2240–2244.
Okuma, K., Iwakawa, K., Turnidge, J. D., Grubb, W. B., Bell, J. M., O'Brien, F. G., et al. (2002). Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J. Clin. Microbiol. 40, 4289–4294. doi: 10.1128/JCM.40.11.4289-4294.2002
Song, J. H., Hsueh, P. R., Chung, D. R., Ko, K. S., Kang, C. I., Peck, K. R., et al. (2011). Spread of methicillin-resistant Staphylococcus aureus between the community and the hospitals in Asian countries: an ANSORP study. J. Antimicrob. Chemother. 66, 1061–1069. doi: 10.1093/jac/dkr024
Stefani, S., Chung, D. R., Lindsay, J. A., Friedrich, A. W., Kearns, A. M., Westh, H., et al. (2012). Meticillin-resistant Staphylococcus aureus (MRSA): global epidemiology and harmonisation of typing methods. Int. J. Antimicrob. Agents 39, 273–282. doi: 10.1016/j.ijantimicag.2011.09.030
Udo, E. E., Al-Lawati, B. A., Al-Muharmi, Z., and Thukral, S. S. (2014). Genotyping of methicillin-resistant Staphylococcus aureus in the Sultan Qaboos University Hospital, Oman reveals the dominance of Panton-Valentine leucocidin-negative ST6-IV/t304 clone. New Microbes New Infect 2, 100–105. doi: 10.1002/nmi2.47
Wang, H., Liu, Y., Sun, H., Xu, Y., Xie, X., and Chen, M. (2008). In vitro activity of ceftobiprole, linezolid, tigecycline, and 23 other antimicrobial agents against Staphylococcus aureus isolates in China. Diagn. Microbiol. Infect. Dis. 62, 226–229. doi: 10.1016/j.diagmicrobio.2008.06.003
Wang, L., Liu, Y., Yang, Y., Huang, G., Wang, C., Deng, L., et al. (2012). Multidrug-resistant clones of community-associated meticillin-resistant Staphylococcus aureus isolated from Chinese children and the resistance genes to clindamycin and mupirocin. J. Med. Microbiol. 61, 1240–1247. doi: 10.1099/jmm.0.042663-0
Wendlandt, S., Lozano, C., Kadlec, K., Gómez-Sanz, E., Zarazaga, M., Torres, C., et al. (2013). The enterococcal ABC transporter gene lsa(E) confers combined resistance to lincosamides, pleuromutilins and streptogramin A antibiotics in methicillin-susceptible and methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 68, 473–475. doi: 10.1093/jac/dks398
Xiao, M., Wang, H., Zhao, Y., Mao, L. L., Brown, M., Yu, Y. S., et al. (2013). National surveillance of methicillin-resistant Staphylococcus aureus in China highlights a still-evolving epidemiology with 15 novel emerging multilocus sequence types. J. Clin. Microbiol. 51, 3638–3644. doi: 10.1128/JCM.01375-13
Keywords: methicillin-resistant Staphylococcus aureus, pvl, genotyping, clonal complexes, multidrug resistance
Citation: Kong H, Yu F, Zhang W, Li X and Wang H (2017) Molecular Epidemiology and Antibiotic Resistance Profiles of Methicillin-Resistant Staphylococcus aureus Strains in a Tertiary Hospital in China. Front. Microbiol. 8:838. doi: 10.3389/fmicb.2017.00838
Received: 25 November 2016; Accepted: 24 April 2017;
Published: 12 May 2017.
Edited by:
Patrick Rik Butaye, Ghent University, BelgiumReviewed by:
Haijian Zhou, National Institute for Communicable Disease Control and Prevention (China CDC), ChinaMaria Angeles Argudín, Université libre de Bruxelles, Belgium
Copyright © 2017 Kong, Yu, Zhang, Li and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongxia Wang, jlzx114@163.com
 Haishen Kong
Haishen Kong Fei Yu
Fei Yu Weili Zhang
Weili Zhang Xuefen Li
Xuefen Li Hongxia Wang
Hongxia Wang