- 1Department of Food Science, Faculty of Agriculture and Food Sciences, Institute of Nutrition and Functional Foods, Laval University, Quebec City, QC, Canada
- 2School of Nutrition Sciences, University of Ottawa, Ottawa, ON, Canada
- 3Department of Microbiology, Faculty of Natural and Life Sciences, Ferhat Abbas University Sétif 1, Sétif, Algeria
Bacterial isolates from raw camel milk were screened for antibacterial activity using the agar diffusion assay. Ten isolates selected for their inhibition of Gram-positive bacteria were identified by 16S sequencing as Enterococcus faecium or durans. An isolate named E. faecium LCW 44 exhibited the broadest antibacterial spectrum with an inhibitory activity against several Gram-positive strains belonging to the genera Clostridium, Listeria, Staphylococcus, and Lactobacillus. E. faecium LCW 44 was shown to produce N-formylated enterocins L50A and L50B, as revealed by mass spectrometry and PCR analyses. This isolate did not harbor any of the virulence factors tested and was shown to be sensitive to all tested antibiotics. It showed high resistance to gastric and intestinal conditions (78 ± 4% survival). Its adhesion index was evaluated at 176 ± 86 and 24 ± 86 on Caco-2 cells and HT-29 cells, respectively, and it significantly reduced adhesion of Listeria monocytogenes by 65 and 49%, respectively. In Macfarlane broth (simulating the nutrient content of the colon), counts of L. monocytogenes were reduced by 2 log10 cycles after 24 h in co-culture with E. faecium LCW 44, compared to the increase of 4 log10 cycles when cultured alone. Comparison with a bacteriocin-non-producing mutant of E. faecium LCW 44 strongly suggests that inhibition of L. monocytogenes was due to bacteriocin production. Altogether, E. faecium LCW 44 thus has potential for use as a probiotic for humans and veterinary medicine.
Introduction
Camel milk is produced mainly in sub-Saharan Africa, where it is a staple food for nomadic peoples (Gerosa and Skoet, 2012). The few published studies on its microbiota have shown that, like other milks, camel milk is a source of lactic acid bacteria, primarily lactobacilli, lactococci, and enterococci (Quigley et al., 2013; Kadri et al., 2015). Nevertheless, differences in the composition of camel milk compared to cow milk (Farah, 1993; Ochirkhuyag et al., 1998; Konuspayeva et al., 2007) and the desert environment away from anthropogenic pressure could underlie significant differences in its microbial ecosystem and the biological characteristics thereof. Camel milk is therefore worth studying as a potential source of yet uncharacterized bacteriocin-producing or otherwise biologically active lactic acid bacteria.
Enterococci, in particular Enterococcus faecalis and Enterococcus faecium, are common in milk (Bhardwaj et al., 2009) and are also ubiquitous in the environment, animal and human gastrointestinal tracts and traditional fermented foods (Moreno et al., 2006). The genus Enterococcus includes a wide range of strains, in some cases suitable as starter cultures or probiotics and in other cases known as spoilage or pathogenic microorganisms (Moreno et al., 2006). Some multidrug-resistant strains, in particular vancomycin-resistant enterococci, are significant causes of nosocomial infections (Arias and Murray, 2012). Their ability to survive adverse conditions, including high-temperature and high-salinity environments, and their adaptability to different growth substrates and conditions, may also allow them to cause spoilage of various foods, especially meat (Björkroth et al., 2005). In contrast, some enterococci play a positive role in various fermented foods, including dairy and vegetable products, apparently through proteolysis, lipolysis, exopolysaccharide production and citrate breakdown (Giraffa, 2003; M’hir et al., 2011). Two strains of Enterococcus are currently recognized as probiotics and are commercially available, namely E. faecium SF68® (NCIMB 10415, Cerbios-Pharma SA, Barbengo, Switzerland) and E. faecalis Symbioflor 1 (SymbioPharm, Herborn, Germany). The efficacy of E. faecium SF68® for the prevention and treatment of diarrhea in humans and other animals has been proven, as well as the use of E. faecalis Symbioflor 1 as an immune regulator in the treatment of recurrent chronic sinusitis or bronchitis (Franz et al., 2011). Therefore, the use of enterococci in foods or as probiotic must be based on case-by-case studies.
Like other lactic acid bacteria, enterococci are known to secrete antimicrobial substances, making them potentially useful for the prevention of bacterial foodborne illness (Franz et al., 2011; Saris, 2014). Many enterococci produce at least one bacteriocin, which is ribosomally synthesized antimicrobial peptide, active against a wide range of foodborne pathogens including Listeria spp. (Nes et al., 2014). According to the classification established by Cotter et al. (2005), most enterocins are class II bacteriocins, defined as non-modified and heat stable. This class includes pediocin-like bacteriocins with a strong anti-Listeria activity (class IIa), two-peptide bacteriocins (class IIb), circular bacteriocins (class IIc), and non-pediocin unmodified peptides (class IId). The reason why enterococci have developed such an ability to produce antimicrobial peptides remains unknown but it is most likely that bacteriocin production could be a beneficial trait in some environments to increase competitiveness of the producing strain and to establish its niche among other microbial groups in a hostile environment (Dobson et al., 2012). It has also been suggested that production of bacteriocin could contribute to probiotic functionality in three ways, as colonizing peptides which facilitate the competition of the producer strain with the resident microbiota, as killing peptides which directly eliminate pathogens, or as signaling peptides toward other bacteria or the immune system (Dobson et al., 2012). For instance, Corr et al. (2007) demonstrated that mice can be protected against Listeria monocytogenes infection by feeding them the bacteriocin-producing strain of Lactobacillus salivarius UCC118, an effect not obtained using a stable non-producing mutant strain. More recently, Kommineni et al. (2015) observed that bacteriocin production by intestinal commensal enterococci in mice reduced vancomycin-resistant enterococci to undetectable levels without significant disruption of the indigenous microbiota. A bacteriocin-encoding plasmid (pPD1) allowed a strain of E. faecalis to colonize the mouse gut, replace indigenous enterococci and outcompete E. faecalis lacking the plasmid (Kommineni et al., 2015). Bacteriocin-producing enterococci thus show promise as probiotics for preventing or treating intestinal infections. The identification of new enterococcal strains with a high probiotic potential must include rigorous evaluation of their behavior, survival and antimicrobial activity under the physiological conditions of the gastrointestinal tract as well as their safety for such use. It should be noted that neither of the commercially sold enterococcal probiotic strains produces any known bacteriocin.
In the present study, we report our characterization of the probiotic properties and safety of a new strain of bacteriocin-producing Enterococcus isolated from camel milk.
Materials and Methods
Bacterial Strains and Culture Conditions
Samples of raw camel milk collected in the month of March in two different regions of Algeria (M’sila and El Oued) were diluted serially (10-fold) in peptone water, plated on MRS agar and incubated for 48 h at 30°C. A total of 59 colonies were selected randomly, purified and stocked at -80°C in MRS broth/40% glycerol solution (1:1). All bacterial strains were cultured for 24 h at 30°C or 37°C in BD Difco media (BD, Sparks, MD, United States) unless otherwise indicated. Listeria, Staphylococcus, and Streptococcus species were grown in tryptic soy broth (TSB) supplemented with 0.6% yeast extract. Escherichia, Salmonella, and Pseudomonas species were grown in LB broth. Clostridium species were grown in RCM medium in an anaerobic chamber (Forma scientific anaerobic system Model 1025). Lactic acid bacteria including Carnobacterium, Enterococcus, Lactobacillus, Lactococcus, and Pediococcus were grown in MRS.
Antibacterial Activity Determination
Agar Well Diffusion Assay
The antibacterial activity in neutralized cell-free supernatants was screened against 23 indicator strains using the agar well diffusion method described by Fernandez et al. (2013). Briefly, optimal medium containing 7.5 g/L of agar was inoculated at approximately 40°C with 150 μL of an overnight culture of the target strain and poured into a sterile Petri dish. Wells 7 mm in diameter were then cut in the solidified agar and filled with 80 μL of the cell-free supernatant previously neutralized at pH 6.5 with 5 M NaOH solution. Inhibition zone diameters were measured after 18 h of incubation under optimal conditions for the strain.
Microtitration Method
Bacteriocin activity was quantified using the microtitration method described by Fernandez et al. (2013). Briefly, twofold serial dilutions (from 125 μL of culture supernatant) in a microtitration plate (96-well microtest, BD Labware, Franklin, NJ, United States) were inoculated with 50 μL of an overnight culture of the target strain diluted to obtain 5 × 104 cfu/well. After 18 h of incubation at the optimal temperature, the OD at 595 nm was recorded using a spectrophotometer (Infinite® F200 PRO, Tecan Inc., Durham, NC, United States). Bacteriocin activity was defined as the highest dilution showing complete inhibition of the strain (OD equal to the uninoculated control) and was calculated as 2n × 1,000/125 where n is the number of inhibited wells and expressed in arbitrary units per milliliter (au/mL).
Molecular Identification of Bacteriocin-Producing Strains
DNA was extracted from an overnight culture of each strain using the wizard genomic DNA purification kit (Promega, Madison, WI, United States) as described by Fernandez et al. (2015). The 16S ribosomal DNA was amplified by PCR (Eppendorf Mastercycler gradient, Hamburg, Germany) with 27f 5′-AGAGTTTGATCMTGGCTCAG-3′ (Gürtler and Stanisich, 1996) and 1390r 5′-GACGGGCGGTGTGTACAA-3′ (Zheng et al., 1996) primers (Invitrogen, Carlsbad, CA, United States). The reaction volume was 50 μL, composed of 1 × Taq buffer (New England Biolabs, Beverly, MA, United States), 0.25 U of Taq DNA polymerase (New England Biolabs), 200 nM of each primer, 200 μM of a dNTP mixture (A, T, C, and G, Invitrogen), and 20 ng of bacterial DNA. The thermal cycle program consisted of an initial cycle of 94°C for 5 min for denaturation and polymerase activation, 35 cycles of 94°C for 45 s, 54°C for 45 s, and 68°C for 60 s, and a final extension step of 5 min at 68°C. PCR products were then subjected to gel electrophoresis (100 V, 1 h) on a 1% agarose gel in Tris-acetate-EDTA 1 × buffer (Ambion, Life Technologies Inc., Burlington, ON, Canada) and visualized by gelRed staining (Biotium, Inc., Hayward, CA, United States). Finally, pure PCR products were sequenced using an ABI 3730XL DNA analyzer (Applied Biosystems, Streetsville, ON, Canada).
Generation of Non-bacteriocin-Producing Mutant E. faecium LCW 44d
Bacteriocin-deficient mutants of Enterococcus faecium LCW 44 were obtained using the method described previously by Criado et al. (2006). MRS broth containing novobiocin (1 μg/mL, Sigma, Oakville, ON, Canada) was inoculated with 105 cfu/mL and incubated for 72 h at 30°C. The culture was then diluted serially (10-fold) and plated on MRS agar. A total of 55 colonies were selected randomly, grown separately in MRS broth for 18 h at 30°C and the activity in the supernatant was assessed using the agar diffusion method against Pediococcus acidilactici UL5 and Listeria ivanovii HPB 28. A non-inhibitory variant thus found was named E. faecium LCW 44d. The presence and the absence of plasmid in E. faecium LCW 44 and LCW 44d, respectively, was confirmed by plasmid profile analysis. Plasmid DNA was isolated using the GeneJET plasmid extraction kit (Thermo Fisher Scientific Inc., Mississauga, ON, Canada) according to the manufacturer’s instructions. Overnight culture (10 mL) was centrifuged at 5,000 × g for 10 min and the cell pellet was lysed for 30 min at 37°C with 40 mg/mL of lysozyme (Sigma) and 200 U/mL of mutanolysin (Sigma). Extraction products and the molecular mass ladder (lambda phage DNA digest by HindIII, Ward’s science, VWR Canada, Mississauga, ON, Canada) were heated for 3 min at 65°C, placed on ice and then resolved immediately by electrophoresis on 0.7% agarose gel in Tris-Acetate-EDTA buffer (1×, Ambion) at 7 V/cm and stained with gelRed (Biotium).
Molecular Identification of Bacteriocins Produced by E. faecium LCW 44
Bacteriocin Purification and Identification
A chemically defined medium (Zhang et al., 2009) inoculated with E. faecium LCW 44 culture at 1% by volume was incubated for 18 h at 30°C. Cell-free supernatant was loaded onto a Sep-Pak C18 cartridge (Waters, Milford, MA, United States) regenerated with 200 mL of methanol and equilibrated with 200 mL of ultrapure water. Adsorbed compounds were eluted with a discontinuous gradient of acetonitrile (0, 25, 50, 75, and 100%) in 5 mM HCl (250 mL per fraction). Acetonitrile was removed from eluted fractions using a rotatory evaporator. The active fraction was loaded onto a preparative C18 column (Luna 10 μm, 250 mm × 21.10 mm, Phenomenex, Torrance, CA, United States) using a Beckman Gold System (Beckman Coulter, Mississauga, ON, Canada). Sample separation was performed at a flow rate of 10 mL/min using a 20–60% linear gradient of acetonitrile in 5 mM HCl and monitoring OD at 214 nm. The most active fraction was concentrated under a continuous flow of nitrogen, injected into a C18 analytical column (AerisTM 3.6 μm, PEPTIDE XB-C18, 250 mm × 4.6 mm, Phenomenex) and eluted with a linear gradient of 40–50% acetonitrile at a flow rate of 1 mL/min and a gradient of 0.5%/min. Peptides were monitored at 214 nm, collected separately and assessed for antibacterial activity. Active peptides were then analyzed by matrix-assisted laser desorption ionization tandem time-of-flight mass spectrometry (MALDI-TOF MS) on an AB SCIEX 4800 Plus MALDI-TOF/TOF instrument as described previously (Hanchi et al., 2016). The raw centroid data were then searched using the Mascot search engine v2.10 (Matrix Science) against a custom database (E. faecium, 267,757 entries). Deamidation of asparagine and glutamine, and oxidation or formylation of methionine were specified in Mascot as variable modifications. Searches were performed with a precursor mass tolerance of 15 ppm and fragment ion tolerances of 0.6 Da.
Genetic Detection of Enterocin Genes
E. faecium LCW 44 and LCW 44d were screened by PCR for enterocin L50A and L50B genes as described by Hanchi et al. (2014) using enterocin-specific primers that amplified both L50A, L50B, 7A, 7B, and MR10 (Liu et al., 2011). The PCR cycle consisted of an initial denaturation at 94°C for 5 min followed by 35 cycles of 94°C for 1 min, annealing at 55°C for 1 min, elongation at 68°C for 1 min and final extension at 68°C for 5 min. PCR products were analyzed by electrophoresis on a 1.5% agarose gel as described above.
Safety Evaluation of E. faecium LCW 44
Antibiotic Susceptibility
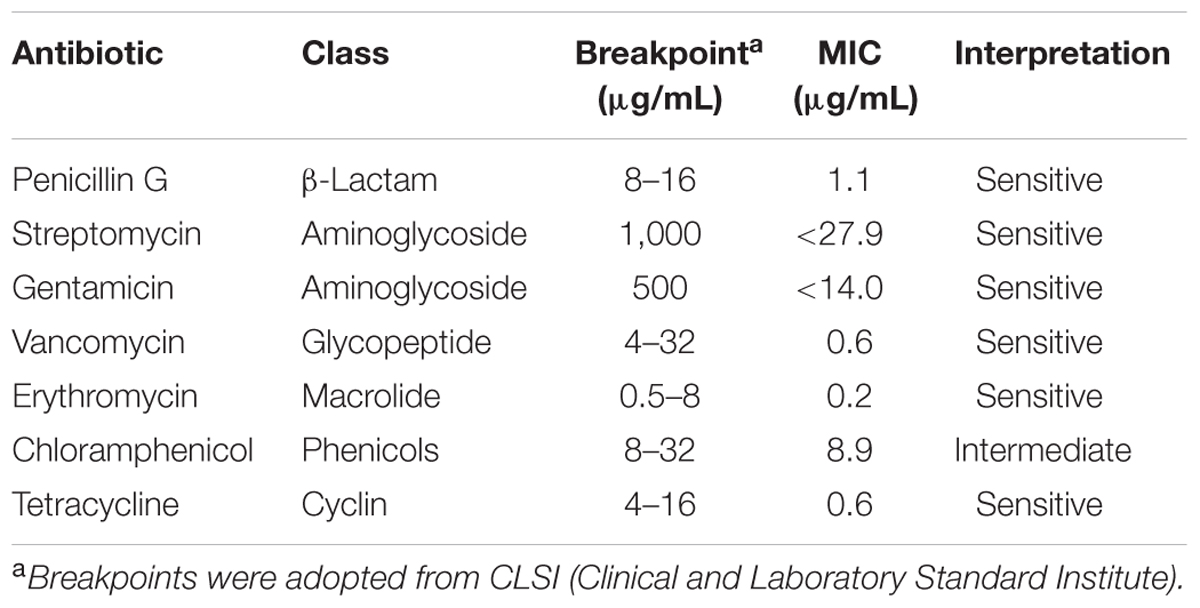
The susceptibility of E. faecium LCW 44 was assessed according to the Clinical and Laboratory Standards Institute guidelines (CLSI), using seven antibiotics from six different classes, including penicillin G, streptomycin, gentamicin, vancomycin, erythromycin, chloramphenicol, and tetracycline, as described by Hanchi et al. (2014).
Presence of Genes Encoding Virulence Factors
E. faecium LCW 44 total DNA was tested by PCR for the presence of any of six genes involved in the pathogenicity of enterococci, namely enterolysin A (EntLysA, Liu et al., 2011), cell wall adhesin (efaAfs, Eaton and Gasson, 2001), enterococcal surface protein involved in immune evasion (esp, Eaton and Gasson, 2001), aggregation substance (asa1, Vankerckhoven et al., 2004), cytolysin (cylA, Vankerckhoven et al., 2004), and gelatinase (gelE, Eaton and Gasson, 2001). DNA from Enterococcus faecalis ATCC 27275 extracted as described above was used as control. All PCR reactions were performed as described by the authors. The presence of the β-hemolysin (cytolysin) and gelatinase were also determined, respectively, by hemolysis test (Semedo et al., 2003) and plating on Todd-Hewitt agar supplemented with 30 g/L of gelatin (Coque et al., 1995).
Assessment of the E. faecium LCW 44 Resistance to Gastrointestinal Stresses
The TIM-1 system (TNO, Nutrition and Food Research Institute, Zeist, Netherlands) was used to assess the behavior of E. faecium LCW 44 under intestinal conditions (pH and enzymes) (Minekus et al., 1995). The model consists of four compartments simulating stomach, duodenum, jejunum, and ileum interconnected in series by computer-controlled peristaltic valve pumps. During digestion, the temperature was kept constantly at 37°C. The pHs in the gastric and small intestine compartments were monitored and controlled, initially at 5.5 in the stomach then gradually decreased to 1⋅8 after 90 min of digestion, at 6.5 in the duodenum, at 6.8 in the jejunum, and at 7.2 in the ileum. Enzymatic secretions consisted of pepsin (Sigma) and lipase in the gastric compartment and trypsin (Sigma), porcine pancreatin (Sigma) and porcine bile extract (Sigma) in the duodenal compartment. Hollow fiber membranes connected to the jejunal and ileal compartments provided dialysis of the contents thereof against small intestinal electrolyte solution. Sample distribution throughout the different compartments of the apparatus was calculated from sensor data and expressed as a function of time by means of computer interface and software. Two independent digestions were performed according the method described previously by Fernandez et al. (2014) with some modifications. Briefly, 300 mL of fermented skim milk was introduced and aliquots of 1 mL were taken in duplicate at 0, 30, 60, 90, and 120 min from the gastric compartment, 30, 60, 90, 120, and 180 min from the duodenal compartment, and 60, 120, 180, 240, and 300 min from the jejunal and ileal compartments for viable count determination. The discharge from the ileum (effluent) and the solution remaining in the duodenal, jejunal, and ileal compartments at the end of the digestion (chyme) were also sampled and taken in account for survival calculation.
Cell Culture
Caco-2 (ATCC HTB-37) and HT-29 (ATCC HTB-38) cells were cultured in MultiCell media (Wisent, Montreal, QC, Canada) at 37°C under a 5% CO2 atmosphere as recommended by DSMZ. Cells were seeded in 24-well plates (Sarstedt, Saint-Léonard, QC, Canada) in medium without antibiotic and used at confluence (>90% estimated by microscopic observation, reached after 4 days) to avoid association to the well.
Adhesion and Competition Assays on Caco-2 and HT-29 Cells
Adhesion and competition assays were adapted from Moroni et al. (2006). After overnight growth, bacterial cultures were washed twice with sterile PBS (5,000 × g for 5 min) and re-suspended at 8 × 108 cfu/mL in cell medium without antibiotic or fetal bovine serum. Confluent cell monolayers were washed twice with sterile PBS and drained prior to contact with bacteria. For adhesion assays, 125 μL of E. faecium LCW 44, LCW 44d, Lactobacillus rhamnosus GG or Listeria monocytogenes LSD 529 suspensions and 125 μL of cell culture medium without antibiotic or fetal bovine serum were added per well to achieve a ratio of bacteria to eukaryotic cells of 100:1. For competition assays, 125 μL of L. monocytogenes LSD 529 suspension and 125 μL of one of the other bacterial suspensions were added per well, reaching a ratio between bacteria of 1:1 while maintaining for each bacterium a ratio bacteria to eukaryotic cells of 100:1. Plates were then incubated for 60 min at 37°C under 5% CO2 atmosphere, drained and rinsed four times with sterile PBS. Cells with adherent bacteria were harvested with 200 μL of trypsin-EDTA for 10 min at 37°C and 300 μL of cell culture medium were added. Adherent lactic acid bacteria were enumerated on MRS agar incubated aerobically at 30°C for 24 h while Listeria was counted on PALCAM agar after 48 h of incubation at 37°C. Results are expressed as the adhesion index, which is the number of bacteria adhering per 100 cells.
Inhibition of L. monocytogenes in Medium Simulating Colonic Nutrients
Macfarlane broth, mimicking the composition of human intestinal contents (Macfarlane et al., 1998), was inoculated at 105 cfu/mL with an overnight culture of L. monocytogenes LSD 530. Overnight cultures of E. faecium LCW 44 or LCW 44d were centrifuged at 5,000 × g for 5 min, re-suspended in 0.1% peptone water to remove residual bacteriocin and added to the Macfarlane broth at 107 cfu/mL. The culture (500 mL) was incubated anaerobically at 37°C for 24 h. Samples were taken every 2 h for the first 12 h and after 24 h. Enterococci and L. monocytogenes were enumerated on MRS and PALCAM agar, respectively. Bacteriocin production was followed using the microtitration method described above.
Statistical Analyses
Adhesion indexes of bacterial strains to intestinal cells were compared among treatments using the one-way analysis of variance (ANOVA) general linear model followed by Tukey’s HSD test. L. monocytogenes counts in Macfarlane broth in the absence or presence of E. faecium were compared using MANOVA for repeated measures (time as repeated measures variable) followed by Tukey’s HSD test. Statistical significance was declared at P < 0.05. All statistical analyses were performed using JMP® software version 11.0 (SAS Institute Inc., Cary, NC, United States).
Results
Screening of Strains for Antibacterial Activity
The 59 bacterial colonies isolated from camel milk were screened against Pediococcus acidilactici UL 5, Listeria ivanovii HPB 28, and Escherichia coli ATCC 25922. The 10 most active isolates were identified as Enterococcus faecium or durans using molecular methods. Table 1 summarizes their inhibition of various Gram-positive and Gram-negative bacteria. Inhibitory activity against Listeria species and/or lactic acid bacteria was variable. E. faecium LCW 44 exhibited the broadest spectrum of activity, inhibiting all tested Listeria species (one L. innocua, one L. ivanovii, and seven L. monocytogenes), most of the lactic acid bacteria, and to a lesser extent Staphylococcus aureus ATCC 6538. This strain identified as E. faecium was chosen for further characterization (GenBank accession number KU299788). The strains LCW 2, 3, 4, 5, 6, 8, and 25 had a spectrum characterized by low activity against 8 of 9 tested Listeria and against lactic acid bacteria. LCW 7 had the narrowest antibacterial activity spectrum, directed against only Lactobacillus leichmannii ATCC 4797 and P. acidilactici UL 5. No activity against tested Gram-negative bacteria was observed.
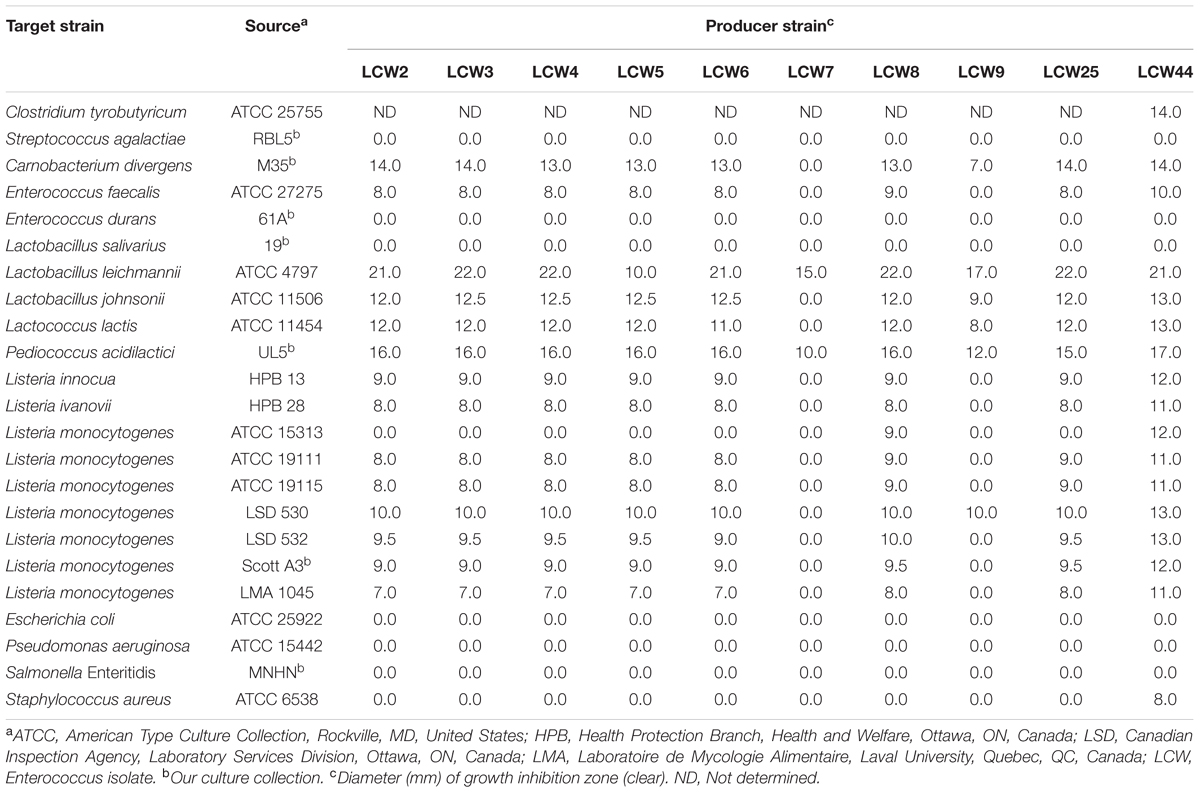
TABLE 1. Antibacterial activity of cell-free culture of enterococci isolated from camel milk, as measured in the standard agar diffusion assay.
E. faecium LCW 44 Produces N-Formylated Enterocins L50A and L50B
Two active peptides, peaks 1 and 2 in Figure 1A, were recovered by analytical HPLC in E. faecium LCW 44 culture supernatant fractionated by reversed-phase chromatography in three consecutive stages. MALDI-TOF spectra revealed single peaks with mono-isotopic masses of 5,203.37 and 5,215.37 Da, respectively (Figures 1B,C). Peptides 1 and 2 were identified by LC-MS/MS, respectively, as formylated enterocin L50B (+28 Da) and formylated enterocin L50A (+28 Da). The experimental b ions closely match those of the theoretically N-formylated forms. Besides, positive amplification with specific primers was observed with E. faecium LCW 44 at the expected length (around 275 bp), but no amplification was recorded with the non-producing mutant of E. faecium LCW 44d (data not shown). In addition, the presence of a single plasmid (a linear fragment about 9,500 bp in length) in E. faecium LCW 44 was confirmed by agarose gel electrophoresis resolution of undigested extracted DNA, and not in E. faecium LCW 44d (data not shown).
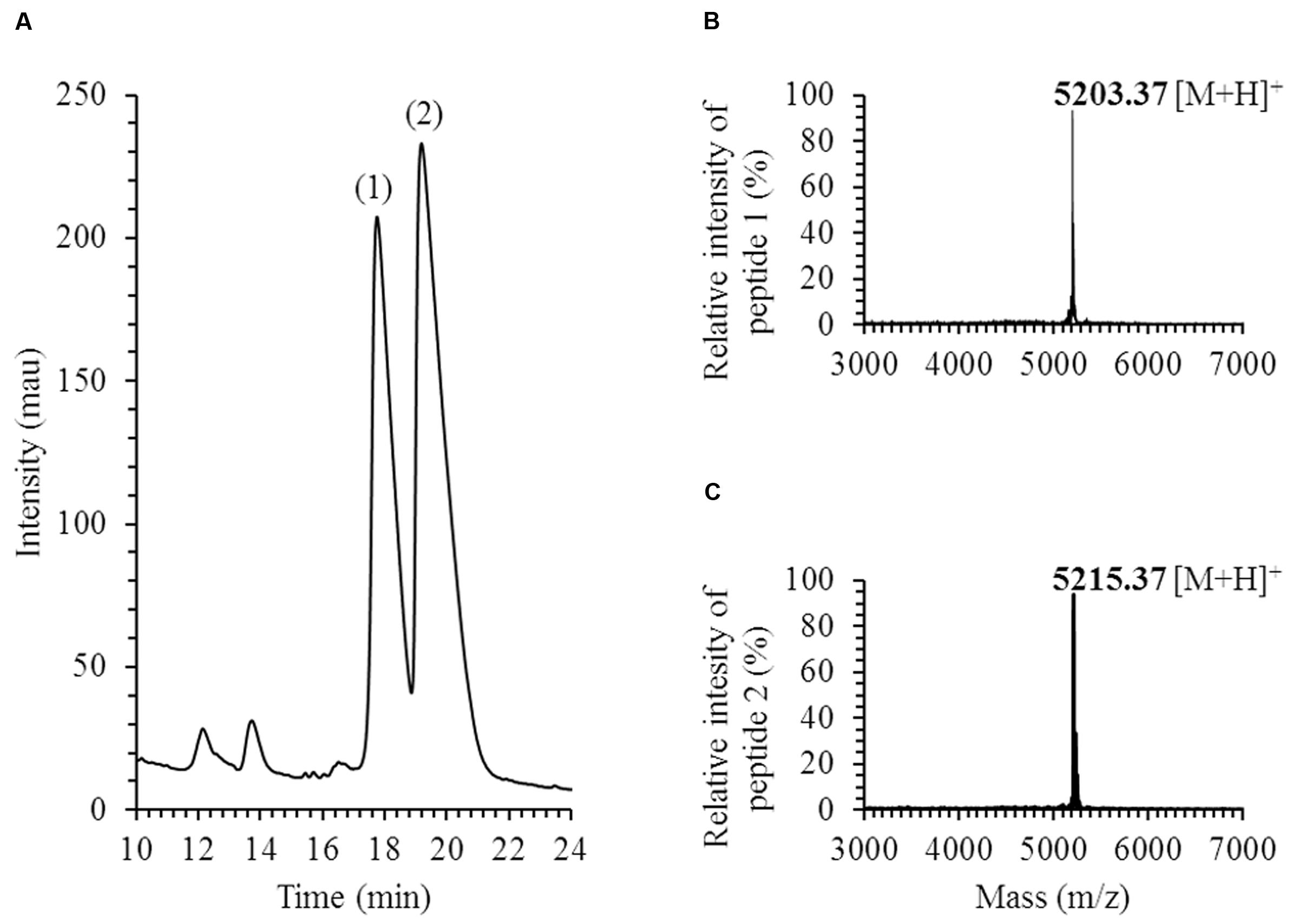
FIGURE 1. Purification and identification of enterocins produced by Enterococcus faecium LCW 44. (A) HPLC elution profile (chromatogram) at 214 nm; (B) MALDI-TOF MS spectrum of enterocin L50B (peptide 1); (C) MALDI-TOF MS spectrum of enterocin L50A (peptide 2); mass is mono-isotopic, in Daltons.
Safety Evaluation of E. faecium LCW 44
E. faecium LCW 44 was sensitive to six antibiotics (Table 2), namely penicillin G, streptomycin, gentamicin, vancomycin, erythromycin, and tetracycline, and showed intermediate sensitivity to chloramphenicol, with a MIC value of 8.9 μg/mL (breakpoint of 8.0 μg/mL). In addition, none of the virulence genes (EntLysA, efaAfs, esp, asa1, cylA, and gelE) was detected in this isolate (Figure 2) and hemolytic and gelatinase activities were absent. In contrast, Enterococcus faecalis ATCC 27275 was positive for enterolysin A (EntLysA), cell wall adhesin (efaAfs), the surface protein involved in immune evasion (esp) and gelatinase (gelE) as described previously for this strain (McBride et al., 2007), and other E. faecalis strains (Hickey et al., 2003; Nueno-Palop and Narbad, 2011). E. faecalis ATCC 27275 was negative for aggregation substance (asa1), cytolysin (cylA), and hemolysis.
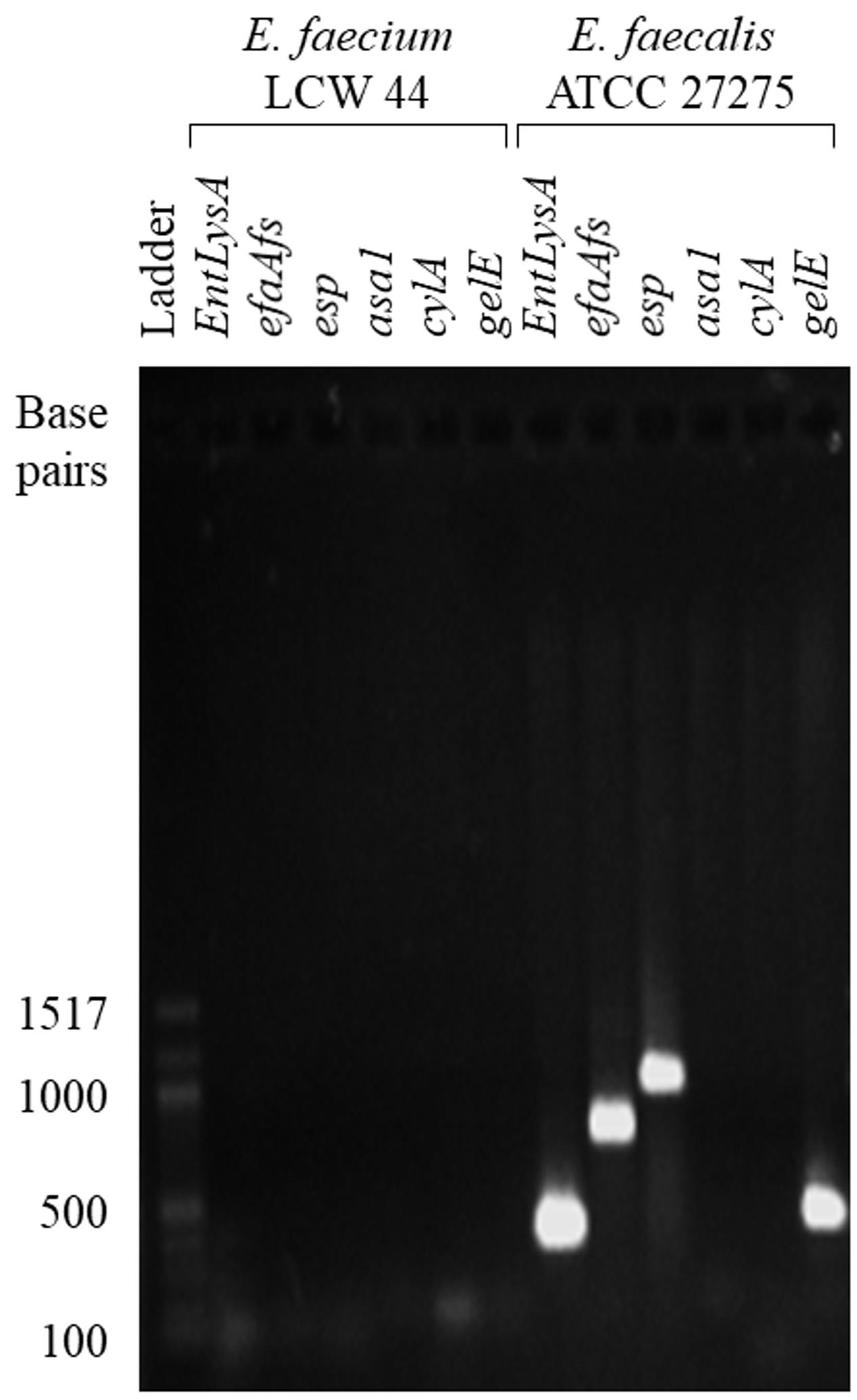
FIGURE 2. PCR fragments generated from total genome of E. faecium LCW 44 and E. faecalis ATCC 27275 using oligonucleotide primers specific for enterolysin A (EntLysA), cell wall adhesin (efaAfs), surface protein involved in immune evasion (esp), aggregation substance (asa1), cytolysin (cylA) and gelatinase (gelE), resolved by electrophoresis on agarose gel.
E. faecium LCW 44 Was Minimally Affected by Digestive Stresses
The ability of E. faecium LCW 44 to survive during passage through the gastric and small intestinal compartments of the TIM-1 model is shown in Figure 3. Sample distribution throughout the different compartments of the apparatus is displayed in Figure 3A. Starting from an initial concentration of 4.6 × 108 cfu/mL, the survival rate was estimated at 78 ± 4 %. Overall, the different stresses had little impact on the viability of E. faecium LCW 44. The most significant decline was observed in the gastric compartment, due most likely to the acidity (pH 1.8).

FIGURE 3. Viability of E. faecium LCW 44 under gastrointestinal conditions (pH and enzymes) in the TIM-1 system. (A) Distribution (%) of the medium (skim milk) in the compartments of the TIM-1 model; (B) Viable count (cfu/mL) determined by plating on agar. Stomach (diamond), duodenum (square), jejunum (circle), ileum (triangle), and efflux (cross). All values are the means of two independent experiments. Error bars indicate the standard error of the mean.
E. faecium LCW 44 Decreased the Adhesion Rate of L. monocytogenes
The adhesion of E. faecium LCW 44 and Lactobacillus rhamnosus GG to undifferentiated Caco-2 and HT-29 cells is shown in Figure 4. No significant difference was observed between E. faecium LCW 44 and its non-bacteriocin-producing mutant E. faecium LCW 44d, which had adhesion indexes of, respectively, 176 ± 86 and 157 ± 86 on Caco-2 cells and 24 ± 86 and 42 ± 86 on HT-29 cells (P > 0.05). These values were significantly lower than those for L. rhamnosus GG, 627 ± 86 and 1020 ± 86, respectively, on Caco-2 and HT-29 cells, which is in agreement with previous reports (Jacobsen et al., 1999). Inhibition of L. monocytogenes LSD 529 adhesion to intestinal cells by E. faecium LCW 44 was evaluated using competition assays (Figure 5). Values of 1204 ± 102 and 655 ± 38, respectively, on Caco-2 and HT-29 cells were measured for L. monocytogenes alone. However, in the presence of E. faecium or L. rhamnosus GG, these numbers significantly dropped by 32–81%, regardless of cell line.
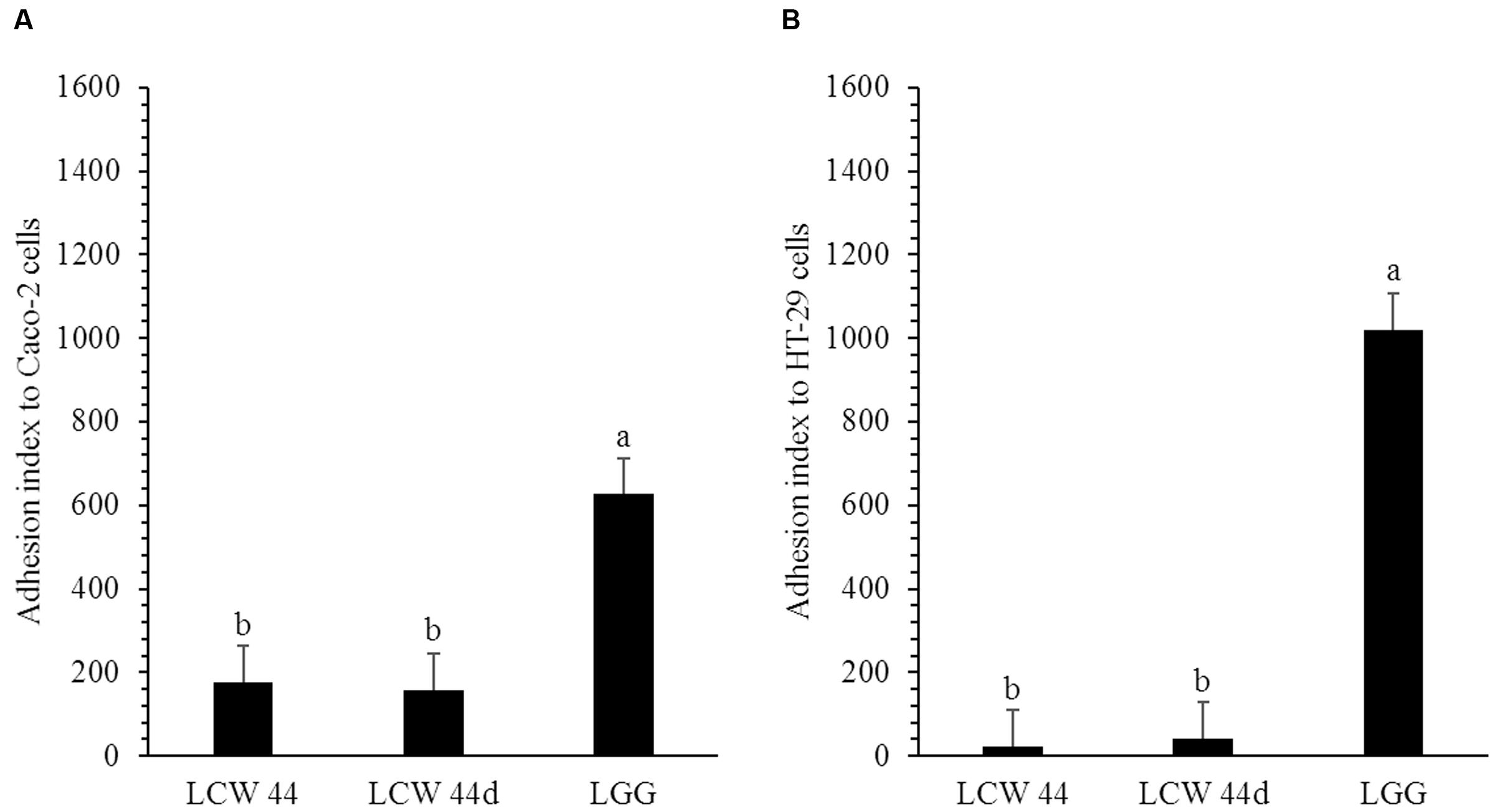
FIGURE 4. Adhesion of E. faecium LCW 44, LCW 44d and L. rhamnosus GG to Caco-2 cells (A) and HT-29 cells (B). Adherent bacteria titer was measured by plating on agar after 1 h of contact. Error bars indicate the standard error of the mean. Different letters indicate significant difference between assays (Tukey’s HSD test, P < 0.05, n = 3).
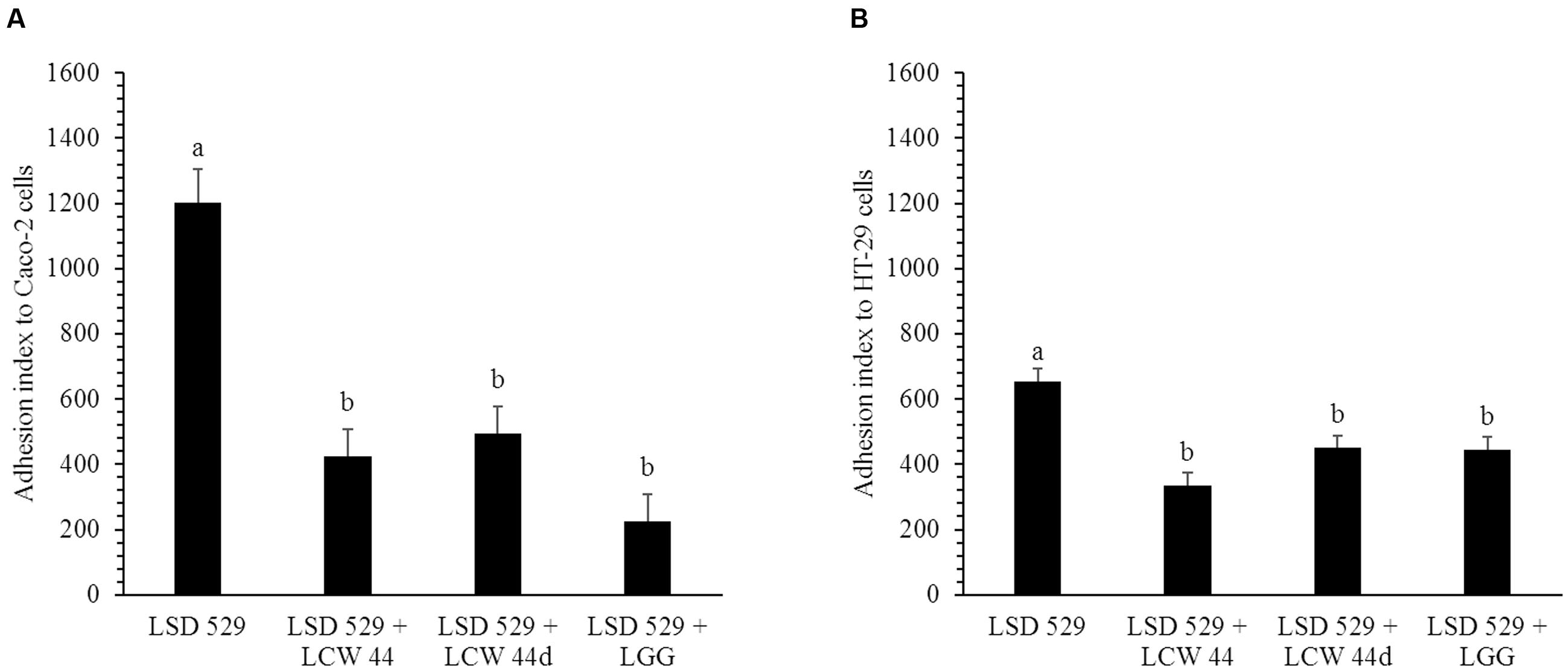
FIGURE 5. Adhesion of L. monocytogenes LSD 529 to Caco-2 cells (A) and HT-29 cells (B) alone and in competition with E. faecium LCW 44, LCW 44d or L. rhamnosus GG. Adherent bacteria titer was measured by plating on agar after 1 h of contact. Error bars indicate the standard error of the mean. Different letters indicate significant difference between assays (Tukey’s HSD test, P < 0.05, n = 3).
Enterocin Production by E. faecium LCW 44 Inhibited L. monocytogenes in Medium Simulating Colonic Nutrients
The inhibition of L. monocytogenes LSD 530 by E. faecium LCW 44 in Macfarlane broth is shown in Figure 6. The variant E. faecium LCW 44d (lacking the plasmid on which enterocins L50A and L50B are encoded) was used as control. Although E. faecium LCW 44 and its variant LCW 44d showed similar growth kinetics, reaching the stationary phase at around 1 × 109 cfu/mL after 4 h of incubation, only E. faecium LCW 44 exhibited significant inhibitory activity against L. monocytogenes, reaching a maximum of 128 au/mL at 4 h and remaining constant for at least 8 h before dropping to 64 au/mL at 24 h (Figure 6A). Growth kinetics of L. monocytogenes cultured alone or in the presence of E. faecium LCW 44 or LCW 44d are shown in Figure 6B. The pure culture reached the stationary phase after 12 h, with 1 × 109 cfu/mL. In presence of E. faecium LCW 44, listerial cell counts were in decline after 4 h and dropped to 2 × 103 cfu/mL after 24 h. In contrast, E. faecium LCW 44d was unable to inhibit L. monocytogenes, but significantly limited its growth to 3 × 107 cfu/mL after 24 h. Inhibition of the growth of L. monocytogenes by E. faecium LCW 44 in Macfarlane broth thus may be due to production of enterocins L50A and L50B.
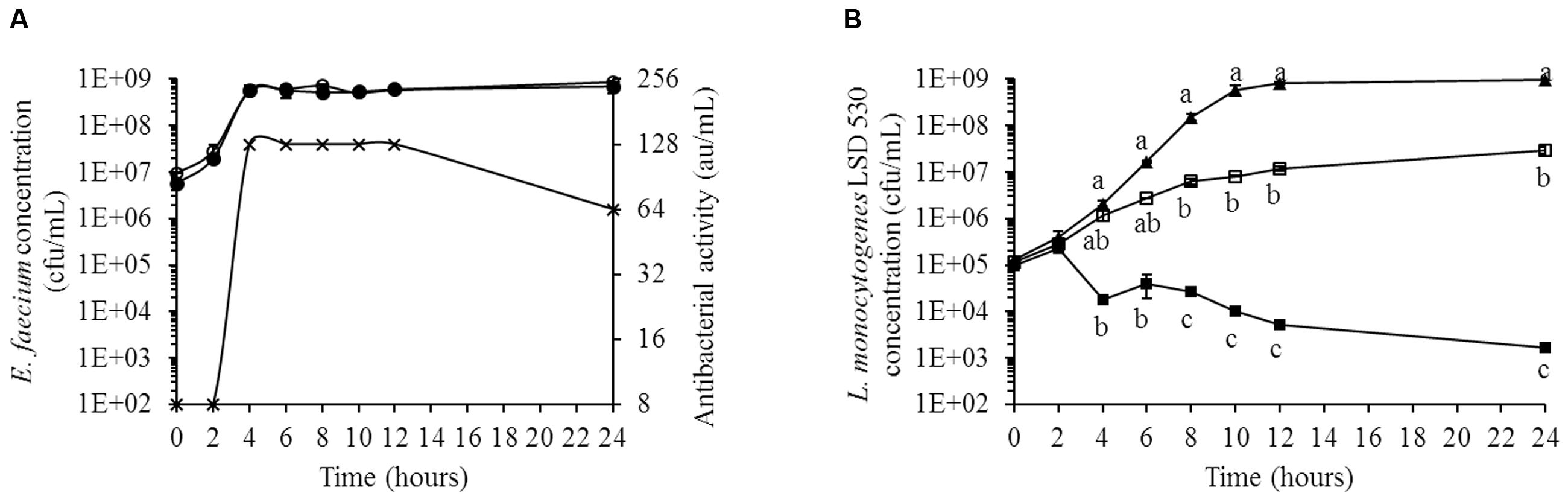
FIGURE 6. Growth kinetics of L. monocytogenes LSD 530 and E. faecium LCW 44 or its non-bacteriocin-producing mutant LCW 44d in Macfarlane medium (simulated intestinal nutrient content). (A) Counts of E. faecium LCW 44 (black circle) and E. faecium LCW 44d (white circle) in co-culture with L. monocytogenes LSD 530, and antibacterial activity (microtitration assay with P. acidilactici UL5) of E. faecium LCW 44 (cross); (B) Growth kinetics of L. monocytogenes LSD 530 alone (triangle, black), in co-culture with E. faecium LCW 44 (black square) or LCW 44d (white square). Error bars indicate the standard error of the mean. Different letters indicate significant difference between assays (Tukey’s HSD test, P < 0.05, n = 3).
Discussion
Enterococci are found in diverse ecological niches including dairy products, where they play an acknowledged role in the organoleptic characteristics (Giraffa, 2003). They are also known to produce one or more bacteriocins that inhibit a wide range of foodborne pathogens including Listeria spp. (Nes et al., 2014). This feature seems to have an impact on niche competition and to contribute to the control of pathogen infections (Corr et al., 2007; Nes et al., 2014).
In this study, a hitherto unexplored niche was searched for new strains of bacteriocin-producing bacteria. Among 59 isolates from raw camel milk, a few were found to have bacteriocin-like activity. One of these in particular, Enterococcus faecium LCW 44, exhibited a broad spectrum of activity against Gram-positive bacteria including Listeria spp. This isolate was shown to produce N-formylated forms of enterocins L50A and L50B, two leaderless class IId bacteriocins of 44 and 43 amino acids, respectively. These enterocins are reportedly encoded in the same non-conjugative plasmid in E. faecium L50 (Nes et al., 2014) and exhibit an antibacterial activity individually or in combination (Iwatani et al., 2011). Such formylated forms have been identified or suggested in culture supernatant of E. faecium isolated from Italian ryegrass (Izquierdo et al., 2008), Spanish dry-fermented sausages (Cintas et al., 1998), and vaginal microbiota in United States (Dezwaan et al., 2007). Enterococcus durans 61A, isolated from artisanal fermented milk in Tunisia, has been shown to produce formylated and non-formylated enterocins L50A and L50B, the formylated peptides being six times more abundant (Hanchi et al., 2016).
Though the genus Enterococcus includes strains with potential as probiotics or food preservatives (Moreno et al., 2006), it also includes some of the most multi-resistant nosocomial pathogens (Arias and Murray, 2012). E. faecium LCW 44 was therefore tested for the factors that contribute most to the emergence of pathogenic strains, namely antibiotic resistance and virulence (Arias and Murray, 2012). It was sensitive to most clinically relevant antibiotics including vancomycin, penicillin G, streptomycin, gentamicin, erythromycin, and tetracycline but showed only moderate susceptibility to chloramphenicol. In comparison, E. faecium L50 has been found resistant to clindamycin, nalidixic acid and streptomycin, and moderately susceptible to erythromycin and kanamycin (Basanta et al., 2008). Secreted virulence factors cytolysin, enterolysin A, and gelatinase were not detected in E. faecium LCW 44, nor were the cell surface determinants including aggregation substance, cell wall adhesion, and enterococcal surface protein involved in immune evasion. In comparison, E. faecium L50 possesses a cell wall adhesin (efaAfm) and a sex pheromone (ccf) (Basanta et al., 2008). The lack of aggregation protein genes in E. faecium LCW 44 corroborates the antibiotic susceptibility since these proteins are encoded in pheromone-responsive plasmids that often also harbor antibiotic resistance genes (Arias and Murray, 2012). The apparent innocuousness of E. faecium LCW 44 might be related to its desert origin, far from well-known antibiotic resistance hotspots, namely medical settings and other environments (e.g., intensive livestock production) that are under anthropogenic pressure (Berendonk et al., 2015). Like the probiotic E. faecium SF68 (Kayser, 2003), E. faecium LCW 44 does not contain the tested virulence genes or the gene for aggregation substance, and thus could be considered as a good candidate for probiotic use in animals or humans. Even though it is expected that conjugation mechanisms would be limited in E. faecium LCW 44 by the absence of aggregation substances (Eaton and Gasson, 2001), further investigation should nevertheless be carried out to establish its ability to acquire virulence determinants.
Enterococci are natural inhabitants of the mammalian gut and especially the terminal part of the small intestine (Walter and Ley, 2011; Lebreton et al., 2014). This intestinal segment is also the entry portal of several pathogenic bacteria such as Salmonella and Listeria (Siebers and Finlay, 1996; Vázquez-Boland et al., 2001). Several groups have examined the possibility of reducing the incidence of these infections using bacteriocin producers such as Enterococcus mundtii CRL 35 (Salvucci et al., 2011), Lactococcus lactis DPC 6520 (Dobson et al., 2011) or Pediococcus acidilactici UL 5 (Dabour et al., 2009). In situ production of bacteriocin does indeed appear to provide protection against pathogenic microorganisms (Corr et al., 2007) and to enhance niche competition in the mammalian gastrointestinal tract (Kommineni et al., 2015). In the present study, we evaluated the capacity of E. faecium LCW 44 to survive during passage through the stomach and small intestine. A reduction of less than 1 log10 cycle was observed, indicating a high capacity for survival under gastrointestinal conditions. Similar resistance has been observed among enterococci including E. faecium SF68 (Lewenstein et al., 1979), Enterococcus faecalis UGRA10 (Cebrián et al., 2012), and E. durans 61A (Hanchi et al., 2014). We evaluated the capacity of E. faecium LCW 44 to adhere to intestinal cells but also its capacity to interfere with pathogen adhesion, in particular Listeria monocytogenes. These adhesion and competition assays were performed with cells in undifferentiated state since they were reported to be more permissive to this pathogen (Gaillard and Finlay, 1996). The adhesion capacity of E. faecium LCW 44 was weak compared to Lactobacillus rhamnosus GG, due likely to the absence of aggregation factors that are involved in adhesion mechanisms (Sartingen et al., 2000; Waar et al., 2002). Comparison with literature is difficult due to the discrepancies regarding the methodology and the expression of results. E. faecium adhesion nevertheless appeared to approach that of other enterococci. Nueno-Palop and Narbad (2011) reported 2.6 × 105 cfu of E. faecalis CP58 adhering to confluent monolayers of Caco-2 cells in 24-well plates, which might correspond to an adhesion index of 26. Sartingen et al. (2000) reported 8.4% of an E. faecalis OG1X inoculum adhering to 1–2 × 106 cells, which might correspond to an adhesion index of 420–840. Despite its weak adhesion, E. faecium LCW 44 decreased significantly the adhesion of L. monocytogenes to both cell lines as much as L. rhamnosus GG did. This probiotic is recognized more for its activity against Salmonella but has been reported to reduce adhesion of L. monocytogenes ATCC 15313 to human intestinal mucus by 53.6 ± 3.6% in a competition assay (Collado et al., 2007). Inhibition of pathogen adhesion by E. faecium LCW 44 was weaker than that measured for E. faecalis UGRA10, which decreased adhesion of L. monocytogenes CECT 4032 to Caco-2 cells by 99.9% (Cebrián et al., 2012). However, it is unclear whether the E. faecalis UGRA10 culture was washed before contact with the cell monolayer, and therefore whether enterocin AS-48 produced during overnight growth contributed to inhibiting Listeria adhesion. The results of pathogen adhesion/inhibition assays are dependent on the lactic acid bacteria and pathogen tested (Gueimonde et al., 2006) and may involve one or several mechanisms including competition for nutrients, immune stimulation, enhancement of barrier function, competitive exclusion and production of antimicrobial substances (O’Toole and Cooney, 2008). The latter two factors are not likely involved in the effect of E. faecium LCW 44, due to its weak adhesion capacity and the fact that the bacteriocin-negative mutant LCW 44d gave similar results.
Finally, E. faecium LCW 44 was able to grow and produce its bacteriocins in Macfarlane medium, meaning that the compounds available in the intestine meet its nutritional requirements for growth and production. Bacteriocin production reached a maximum (128 au/mL) in 4 h, in the early stationary phase. Similar results have been reported previously for L. lactis UL719 and P. acidilactici UL5, which produced their respective bacteriocins in Macfarlane broth after 4 and 6 h, respectively (Fernandez et al., 2013). In the presence of E. faecium LCW 44, L. monocytogenes counts dropped by 5.8 log10 cycles (to 2 × 103 cfu/mL) within 24 h, due likely to production of enterocins L50A and L50B since no such inhibition was not observed in the presence of E. faecium LCW 44d. The bacteriocin-negative mutant did nevertheless limit growth of Listeria by 2 log10 cycles over the same time course. This result is due likely to the greater competitiveness of E. faecium, which reached stationary phase twice as fast as L. monocytogenes. E. faecium LCW 44 might also produce other antimicrobials active against Listeria, since enterococci are known to produce organic acids and multiple bacteriocins (Nes et al., 2007). Inhibition of pathogens by enterococci under intestinal conditions has been reported for E. faecium KH 24, which reduced Salmonella enterica subsp. enterica serovar Enteritidis counts by nearly 1 log cycle in mouse intestines (Bhardwaj et al., 2010). The presence of microbiota induces additional stress that might modulate bacteriocin production. Further studies are needed to confirm that E. faecium LCW 44 inhibits Listeria in the colon in the presence of intestinal microbiota. We have already observed that P. acidilactici UL5 can reduce Listeria counts by 2 log10 cycles after 6 h in Macfarlane broth (Fernandez et al., 2013) but not in a continuous stirred tank reactor containing immobilized human intestinal microbiota (Fernandez et al., 2015). With its short generation time and high adaptability to different growth conditions, E. faecium LCW 44 is expected to compete well with colonic microbiota and to inhibit Listeria under gastrointestinal conditions.
Author Contributions
BF, AV, HD, and IF conceived the research. BF, AV, HD, and AA performed the experiments. BF and AV wrote the manuscript. BF, AV, RH, HD, AA, and IF edited the manuscript. All authors reviewed and accepted the manuscript.
Funding
This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada (NSERC, grant number 203591) and the NSERC-METABIOLAC industrial research chair (grant number IRCPJ 499946-15).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Arias, C. A., and Murray, B. E. (2012). The rise of the Enterococcus: beyond vancomycin resistance. Nat. Rev. Microbiol. 10, 266–278. doi: 10.1038/nrmicro2761
Basanta, A., Sánchez, J., Gómez-Sala, B., Herranz, C., Hernández, P. E., and Cintas, L. M. (2008). Antimicrobial activity of Enterococcus faecium L50, a strain producing enterocins L50 (L50A and L50B), P and Q, against beer-spoilage lactic acid bacteria in broth, wort (hopped and unhopped), and alcoholic and non-alcoholic lager beers. Int. J. Food Microbiol. 125, 293–307. doi: 10.1016/j.ijfoodmicro.2008.04.011
Berendonk, T. U., Manaia, C. M., Merlin, C., Fatta-Kassinos, D., Cytryn, E., Walsh, F., et al. (2015). Tackling antibiotic resistance: the environmental framework. Nat. Rev. Microbiol. 13, 310–317. doi: 10.1038/nrmicro3439
Bhardwaj, A., Gupta, H., Kapila, S., Kaur, G., Vij, S., and Malik, R. K. (2010). Safety assessment and evaluation of probiotic potential of bacteriocinogenic Enterococcus faecium KH 24 strain under in vitro and in vivo conditions. Int. J. Food Microbiol. 141, 156–164. doi: 10.1016/j.ijfoodmicro.2010.05.001
Bhardwaj, A., Kapila, S., Mani, J., and Malik, R. K. (2009). Comparison of susceptibility to opsonic killing by in vitro human immune response of Enterococcus strains isolated from dairy products, clinical samples and probiotic preparation. Int. J. Food Microbiol. 128, 513–515. doi: 10.1016/j.ijfoodmicro.2008.10.010
Björkroth, J., Ristiniemi, M., Vandamme, P., and Korkeala, H. (2005). Enterococcus species dominating in fresh modified-atmosphere-packaged, marinated broiler legs are overgrown by Carnobacterium and Lactobacillus species during storage at 6°C. Int. J. Food Microbiol. 97, 267–276. doi: 10.1016/j.ijfoodmicro.2004.04.011
Cebrián, R., Baños, A., Valdivia, E., Pérez-Pulido, R., Martínez-Bueno, M., and Maqueda, M. (2012). Characterization of functional, safety, and probiotic properties of Enterococcus faecalis UGRA10, a new AS-48-producer strain. Food Microbiol. 30, 59–67. doi: 10.1016/j.fm.2011.12.002
Cintas, L. M., Casaus, P., Holo, H., Hernandez, P. E., Nes, I. F., and Håvarstein, L. S. (1998). Enterocins L50A and L50B, two novel bacteriocins from Enterococcus faecium L50, are related to staphylococcal hemolysins. J. Bacteriol. 180, 1988–1994.
Collado, M. C., Meriluoto, J., and Salminen, S. (2007). In vitro analysis of probiotic strain combinations to inhibit pathogen adhesion to human intestinal mucus. Food Res. Int. 40, 629–636. doi: 10.1016/j.foodres.2006.11.007
Coque, T. M., Patterson, J. E., Steckelberg, J. M., and Murray, B. E. (1995). Incidence of hemolysin, gelatinase, and aggregation substance among enterococci isolated from patients with endocarditis and other infections and from feces of hospitalized and community-based persons. J. Infect. Dis. 171, 1223–1229. doi: 10.1093/infdis/171.5.1223
Corr, S. C., Li, Y., Riedel, C. U., O’Toole, P. W., Hill, C., and Gahan, C. G. M. (2007). Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc. Natl. Acad. Sci. U.S.A. 104, 7617–7621. doi: 10.1073/pnas.0700440104
Cotter, P. D., Hill, C., and Ross, R. P. (2005). Bacteriocins: developing innate immunity for food. Nat. Rev. Microbiol. 3, 777–788. doi: 10.1038/nrmicro1273
Criado, R., Diep, D. B., Aakra,Å., Gutiérrez, J., Nes, I. F., Hernández, P. E., et al. (2006). Complete sequence of the enterocin Q-encoding plasmid pCIZ2 from the multiple bacteriocin producer Enterococcus faecium L50 and genetic characterization of enterocin Q production and immunity. Appl. Environ. Microbiol. 72, 6653–6666. doi: 10.1128/aem.00859-06
Dabour, N., Zihler, A., Kheadr, E., Lacroix, C., and Fliss, I. (2009). In vivo study on the effectiveness of pediocin PA-1 and Pediococcus acidilactici UL5 at inhibiting Listeria monocytogenes. Int. J. Food Microbiol. 133, 225–233. doi: 10.1016/j.ijfoodmicro.2009.05.005
Dezwaan, D. C., Mequio, M. J., Littell, J. S., Allen, J. P., Rossbach, S., and Pybus, V. (2007). Purification and characterization of enterocin 62-6, a two-peptide bacteriocin produced by a vaginal strain of Enterococcus faecium: potential significance in bacterial vaginosis. Microb. Ecol. Health Dis. 19, 241–250. doi: 10.1080/08910600701538240
Dobson, A., Cotter, P. D., Ross, R. P., and Hill, C. (2012). Bacteriocin production: A probiotic trait? Appl. Environ. Microbiol. 78, 1–6. doi: 10.1128/aem.05576-11
Dobson, A., Crispie, F., Rea, M. C., O’Sullivan, O., Casey, P. G., Lawlor, P. G., et al. (2011). Fate and efficacy of lacticin 3147-producing Lactococcus lactis in the mammalian gastrointestinal tract. FEMS Microbiol. Ecol. 76, 602–614. doi: 10.1111/j.1574-6941.2011.01069.x
Eaton, T. J., and Gasson, M. J. (2001). Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl. Environ. Microbiol. 67, 1628–1635. doi: 10.1128/aem.67.4.1628-1635.2001
Farah, Z. (1993). Composition and characteristics of camel milk. J. Dairy Res. 60, 603–626. doi: 10.1017/S0022029900027953
Fernandez, B., Hammami, R., Savard, P., Jean, J., and Fliss, I. (2014). Pediococcus acidilactici UL5 and Lactococcus lactis ATCC 11454 are able to survive and express their bacteriocin genes under simulated gastrointestinal conditions. J. Appl. Microbiol. 116, 677–688. doi: 10.1111/jam.12391
Fernandez, B., Le Lay, C., Jean, J., and Fliss, I. (2013). Growth, acid production and bacteriocin production by probiotic candidates under simulated colonic conditions. J. Appl. Microbiol. 114, 877–885. doi: 10.1111/jam.12081
Fernandez, B., Savard, P., and Fliss, I. (2015). Survival and metabolic activity of pediocin producer Pediococcus acidilactici UL5: its impact on intestinal microbiota and Listeria monocytogenes in a model of the human terminal ileum. Microb. Ecol. 72, 931–994. doi: 10.1007/s00248-015-0645-0
Franz, C. M. A. P., Huch, M., Abriouel, H., Holzapfel, W., and Gálvez, A. (2011). Enterococci as probiotics and their implications in food safety. Int. J. Food Microbiol. 151, 125–140. doi: 10.1016/j.ijfoodmicro.2011.08.014
Gaillard, J.-L., and Finlay, B. B. (1996). Effect of cell polarization and differentiation on entry of Listeria monocytogenes into the enterocyte-like Caco-2 cell line. Infect. Immun. 64, 1299–1308.
Gerosa, S., and Skoet, J. (2012). ESA Working Paper No. 12-01: Milk Availability – Trends in Production and Demand and Medium-Term Outlook. Rome: Food and Agriculture Organization of the United Nations.
Giraffa, G. (2003). Functionality of enterococci in dairy products. Int. J. Food Microbiol. 88, 215–222. doi: 10.1016/S0168-1605(03)00183-1
Gueimonde, M., Jalonen, L., He, F., Hiramatsu, M., and Salminen, S. (2006). Adhesion and competitive inhibition and displacement of human enteropathogens by selected lactobacilli. Food Res. Int. 39, 467–471. doi: 10.1016/j.foodres.2005.10.003
Gürtler, V., and Stanisich, V. A. (1996). New approaches to typing and identification of bacteria using the 16S-23S rDNA spacer region. Microbiology 142, 3–16. doi: 10.1099/13500872-142-1-3
Hanchi, H., Hammami, R., Fernandez, B., Kourda, R., Ben Hamida, J., and Fliss, I. (2016). Simultaneous production of formylated and non-formylated enterocins L50A and L50B, and 61A, a new glycosylated durancin, by Enterococcus durans 61A, a strain isolated from artisanal fermented milk in Tunisia. J. Agric. Food Chem. 64, 3584–3590. doi: 10.1021/acs.jafc.6b00700
Hanchi, H., Hammami, R., Kourda, R., Hamida, J., and Fliss, I. (2014). Bacteriocinogenic properties and in vitro probiotic potential of enterococci from Tunisian dairy products. Arch. Microbiol. 196, 331–344. doi: 10.1007/s00203-014-0978-y
Hickey, R. M., Twomey, D. P., Ross, R. P., and Hill, C. (2003). Production of enterolysin A by a raw milk enterococcal isolate exhibiting multiple virulence factors. Microbiology 149, 655–664. doi: 10.1099/mic.0.25949-0
Iwatani, S., Zendo, T., and Sonomoto, K. (2011). “Class IId or linear and non-pediocin-like bacteriocins,” in Prokaryotic Antimicrobial Peptides, eds D. Drider and S. Rebuffat (New York, NY: Springer), 237–252. doi: 10.1007/978-1-4419-7692-5_13
Izquierdo, E., Bednarczyk, A., Schaeffer, C., Cai, Y., Marchioni, E., Van Dorsselaer, A., et al. (2008). Production of enterocins L50A, L50B, and IT, a new enterocin, by Enterococcus faecium IT62, a strain isolated from Italian ryegrass in Japan. Antimicrob. Agents Chemother. 52, 1917–1923. doi: 10.1128/aac.01409-07
Jacobsen, C. N., Rosenfeldt Nielsen, V., Hayford, A. E., Møller, P. L., Michaelsen, K. F., Pærregaard, A., et al. (1999). Screening of probiotic activities of forty-seven strains of Lactobacillus spp. by in vitro techniques and evaluation of the colonization ability of five selected strains in humans. Appl. Environ. Microbiol. 65, 4949–4956.
Kadri, Z., Spitaels, F., Cnockaert, M., Praet, J., El Farricha, O., Swings, J., et al. (2015). Enterococcus bulliens sp. nov., a novel lactic acid bacterium isolated from camel milk. Antonie van Leeuwenhoek 108, 1257–1265. doi: 10.1007/s10482-015-0579-z
Kayser, F. H. (2003). Safety aspects of enterococci from the medical point of view. Int. J. Food Microbiol. 88, 255–262. doi: 10.1016/S0168-1605(03)00188-0
Kommineni, S., Bretl, D. J., Lam, V., Chakraborty, R., Hayward, M., Simpson, P., et al. (2015). Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature 526, 719–722. doi: 10.1038/nature15524
Konuspayeva, G., Faye, B., Loiseau, G., and Levieux, D. (2007). Lactoferrin and immunoglobulin contents in camel’s milk (Camelus bactrianus, Camelus dromedarius, and hybrids) from Kazakhstan. J. Dairy Sci. 90, 38–46. doi: 10.3168/jds.S0022-0302(07)72606-1
Lebreton, F., Willems, R., and Gilmore, M. (2014). “Enterococcus diversity, origins in nature, and gut colonization,” in Enterococci: From Commensals to Leading Causes of Drug Resistant Infection [Internet], eds M. Gilmore, D. Clewell, Y. Ike, and N. Shankar (Boston, MA: Massachusetts Eye and Ear Infirmary).
Lewenstein, A., Frigerio, G., and Moroni, M. (1979). Biological properties of SF 68, a new approach for the treatment of diarrheal diseases. Curr. Ther. Res. Clin. Exp. 26, 967–981.
Liu, X., Vederas, J. C., Whittal, R. M., Zheng, J., Stiles, M. E., Carlson, D., et al. (2011). Identification of an N-terminal formylated, two-peptide bacteriocin from Enterococcus faecalis 710C. J. Agric. Food Chem. 59, 5602–5608. doi: 10.1021/jf104751v
Macfarlane, G., Macfarlane, S., and Gibson, G. (1998). Validation of a three-stage compound continuous culture system for investigating the effect of retention time on the ecology and metabolism of bacteria in the human colon. Microb. Ecol. 35, 180–187. doi: 10.1007/s002489900072
McBride, S. M., Fischetti, V. A., LeBlanc, D. J., Moellering, R. C. Jr., and Gilmore, M. S. (2007). Genetic diversity among Enterococcus faecalis. PLoS ONE 2:e582. doi: 10.1371/journal.pone.0000582
M’hir, S., Minervini, F., Di Cagno, R., Chammem, N., and Hamdi, M. (2011). Technological, functional and safety aspects of enterococci in fermented vegetable products: a mini-review. Ann. Microbiol. 62, 469–481. doi: 10.1007/s13213-011-0363-x
Minekus, M., Marteau, P., and Havenaar, R. (1995). A multicompartmental dynamic computer-controlled model simulating the stomach and small intestine. Altern. Lab. Anim. 23, 197–209.
Moreno, M. F., Sarantinopoulos, P., Tsakalidou, E., and De Vuyst, L. (2006). The role and application of enterococci in food and health. Int. J. Food Microbiol. 106, 1–24. doi: 10.1016/j.ijfoodmicro.2005.06.026
Moroni, O., Kheadr, E., Boutin, Y., Lacroix, C., and Fliss, I. (2006). Inactivation of adhesion and invasion of food-borne Listeria monocytogenes by bacteriocin-producing Bifidobacterium strains of human origin. Appl. Environ. Microbiol. 72, 6894–6901. doi: 10.1128/aem.00928-06
Nes, I. F., Diep, D. B., and Holo, H. (2007). Bacteriocin diversity in Streptococcus and Enterococcus. J. Bacteriol. 189, 1189–1198. doi: 10.1128/JB.01254-06
Nes, I. F., Diep, D. B., and Ike, Y. (2014). “Enterococcal bacteriocins and antimicrobial proteins that contribute to niche control,” in Enterococci: From Commensals to Leading Causes of Drug Resistant Infection, eds M. Gilmore, D. Clewell, Y. Ike, and N. Shankar (Boston, MA: Massachusetts Eye and Ear Infirmary).
Nueno-Palop, C., and Narbad, A. (2011). Probiotic assessment of Enterococcus faecalis CP58 isolated from human gut. Int. J. Food Microbiol. 145, 390–394. doi: 10.1016/j.ijfoodmicro.2010.12.029
Ochirkhuyag, B., Chobert, J. M., Dalgalarrondo, M., Choiset, Y., and Haertle, T. (1998). Characterization of whey proteins from Mongolian yak, khainak, and bactrian camel. J. Food Biochem. 22, 105–124. doi: 10.1111/j.1745-4514.1998.tb00234.x
O’Toole, P. W., and Cooney, J. C. (2008). Probiotic bacteria influence the composition and function of the intestinal microbiota. Interdiscip. Perspect. Infect. Dis. 2008:175285. doi: 10.1155/2008/175285
Quigley, L., O’Sullivan, O., Stanton, C., Beresford, T. P., Ross, R. P., Fitzgerald, G. F., et al. (2013). The complex microbiota of raw milk. FEMS Microbiol. Rev. 37, 664–698. doi: 10.1111/1574-6976.12030
Salvucci, E., Saavedra, L., Hebert, E. M., Haro, C., and Sesma, F. (2011). Enterocin CRL35 inhibits Listeria monocytogenes in a murine model. Foodborne Pathog. Dis. 9, 68–74. doi: 10.1089/fpd.2011.0972
Saris, P. E. (2014). “Biopreservation by lactic acid bacteria,” in Dairy Microbiology and Biochemistry: Recent Developments, ed. Taylor & Francis Group (Boca Raton, FL: CRC Press), 86.
Sartingen, S., Rozdzinski, E., Muscholl-Silberhorn, A., and Marre, R. (2000). Aggregation substance increases adherence and internalization, but not translocation, of Enterococcus faecalis through different intestinal epithelial cells in vitro. Infect. Immun. 68, 6044–6047. doi: 10.1128/iai.68.10.6044-6047.2000
Semedo, T., Almeida Santos, M., Martins, P., Silva Lopes, M. F., Figueiredo Marques, J. J., Tenreiro, R., et al. (2003). Comparative study using type strains and clinical and food isolates to examine hemolytic activity and occurrence of the cyl operon in enterococci. J. Clin. Microbiol. 41, 2569–2576. doi: 10.1128/jcm.41.6.2569-2576.2003
Siebers, A., and Finlay, B. B. (1996). M cells and the pathogenesis of mucosal and systemic infections. Trends Microbiol. 4, 22–29. doi: 10.1016/0966-842X(96)81501-0
Vankerckhoven, V., Van Autgaerden, T., Vael, C., Lammens, C., Chapelle, S., Rossi, R., et al. (2004). Development of a multiplex PCR for the detection of asa1, gelE, cylA, esp, and hyl genes in enterococci and survey for virulence determinants among european hospital isolates of Enterococcus faecium. J. Clin. Microbiol. 42, 4473–4479. doi: 10.1128/jcm.42.10.4473-4479.2004
Vázquez-Boland, J. A., Kuhn, M., Berche, P., Chakraborty, T., Domínguez-Bernal, G., Goebel, W., et al. (2001). Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14, 584–640. doi: 10.1128/cmr.14.3.584-640.2001
Waar, K., van der Mei, H. C., Harmsen, H. J. M., Degener, J. E., and Busscher, H. J. (2002). Enterococcus faecalis surface proteins determine its adhesion mechanism to bile drain materials. Microbiology 148, 1863–1870. doi: 10.1099/00221287-148-6-1863
Walter, J., and Ley, R. (2011). The human gut microbiome: ecology and recent evolutionary changes. Annu. Rev. Microbiol. 65, 411–429. doi: 10.1146/annurev-micro-090110-102830
Zhang, G., Mills, D. A., and Block, D. E. (2009). Development of chemically defined media supporting high-cell-density growth of lactococci, enterococci, and streptococci. Appl. Environ. Microbiol. 75, 1080–1087. doi: 10.1128/aem.01416-08
Keywords: bacteriocin, enterococci, probiotic, intestinal pathogen, listeriosis, cell adhesion
Citation: Vimont A, Fernandez B, Hammami R, Ababsa A, Daba H and Fliss I (2017) Bacteriocin-Producing Enterococcus faecium LCW 44: A High Potential Probiotic Candidate from Raw Camel Milk. Front. Microbiol. 8:865. doi: 10.3389/fmicb.2017.00865
Received: 28 March 2017; Accepted: 28 April 2017;
Published: 18 May 2017.
Edited by:
Paula Teixeira, Universidade Católica Portuguesa, PortugalReviewed by:
Analia Graciela Abraham, Centro de Investigacion y Desarrollo en Criotecnologia de Alimentos, ArgentinaChiara Montanari, Università di Bologna, Italy
Copyright © 2017 Vimont, Fernandez, Hammami, Ababsa, Daba and Fliss. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ismaïl Fliss, ismail.fliss@fsaa.ulaval.ca Benoît Fernandez, benoitfernandez@outlook.com
†These authors have contributed equally to this work.
 Allison Vimont
Allison Vimont Benoît Fernandez
Benoît Fernandez Riadh Hammami
Riadh Hammami Ahlem Ababsa
Ahlem Ababsa Hocine Daba3
Hocine Daba3 Ismaïl Fliss
Ismaïl Fliss