- 1Department of Environmental Science, Faculty of Agriculture, Bangabandhu Sheikh Mujibur Rahman Agricultural University, Gazipur, Bangladesh
- 2Department of Agricultural Engineering, Faculty of Agriculture, Bangabandhu Sheikh Mujibur Rahman Agricultural University, Gazipur, Bangladesh
- 3Plant Breeding Division, Bangladesh Agricultural Research Institute, Gazipur, Bangladesh
- 4Faculty of Agriculture, Shizuoka University, Shizuoka, Japan
Pectobacterium carotovorum subsp. carotovorum [Pcc (formerly Erwinia carotovora subsp. carotovora)] PC1 causes soft-rot disease in a wide variety of plant species by secreting multiple pathogenicity-related traits. In this study, regulatory mechanism of air-liquid (AL) biofilm formation was studied using a cytR homolog gene deletion mutant (ΔcytR) of Pcc PC1. Compared to the wild type (Pcc PC1), the ΔcytR mutant produced fragile and significantly (P < 0.001) lower amounts of AL biofilm on salt-optimized broth plus 2% glycerol (SOBG), yeast peptone dextrose adenine, and also on King’s B at 27°C after 72 h incubation in static condition. The wild type also produced significantly higher quantities of AL biofilm on SOBGMg– (magnesium deprived) containing Cupper (Cu2+), Zinc (Zn2+), Manganese (Mn2+), Magnesium (Mg2+), and Calcium (Ca2+) compared to the ΔcytR mutant. Moreover, the wild type was produced higher amounts of biofilms compared to the mutant while responding to pH and osmotic stresses. The ΔfliC (encoding flagellin), flhD::Tn5 (encoding a master regulator) and ΔmotA (a membrane protein essential for flagellar rotation) mutants produced a lighter and more fragile AL biofilm on SOBG compared to their wild counterpart. All these mutants resulted in having weak bonds with the cellulose specific dye (Calcofluor) producing lower quantities of cellulose compared to the wild type. Gene expression analysis using mRNA collected from the AL biofilms showed that ΔcytR mutant significantly (P < 0.001) reduced the expressions of multiple genes responsible for cellulose production (bcsA, bcsE, and adrA), motility (flhD, fliA, fliC, and motA) and type III secretion system (hrpX, hrpL, hrpA, and hrpN) compared to the wild type. The CytR homolog was therefore, argued to be able to regulate the AL biofilm formation by controlling cellulose production, motility and T3SS in Pcc PC1. In addition, all the mutants exhibited poorer attachment to radish sprouts and AL biofilm cells of the wild type was resistant than stationary-phase and planktonic cells to acidity and oxidative stress compared to the same cells of the ΔcytR mutant. The results of this study therefore suggest that CytR homolog is a major determinant of Pcc PC1’s virulence, attachment and its survival mechanism.
Introduction
Pectobacterium carotovorum subsp. carotovorum [Pcc (formerly Erwinia carotovora subsp. carotovora)] is a Gram-negative, necrotrophic and opportunistic phytopathogenic enterobacterium. It is responsible for causing soft-rot on a variety of plant species during cultivation and, in many cases, during post-harvest processing and storage of crops, resulting in significant economic loss (Davidsson et al., 2013). Production of plant cell-wall-degrading enzymes, including pectate lyases (Pel), cellulases (Cel), and proteases (Prt) is the most destructive feature of this pathogen (Toth et al., 2003). Its pathogenicity is also known to be influenced by quorum sensing (Charkowski et al., 2012), motility (Matsumoto et al., 2003; Hossain et al., 2005), type III secretion system (T3SS) encoded by the hypersensitive response and pathogenicity (hrp) gene cluster (Rantakari et al., 2007), gluconate metabolism (Mole et al., 2010), the magnesium/nickel/cobalt transport system (Kersey et al., 2012), biosynthesis of pyrimidine, purine, leucine, or serine (Lee et al., 2013) and biofilm formation (Lee et al., 2013). These pathogenicity factors in Pcc have been shown to be controlled by different regulatory proteins, such as KdgR (Liu et al., 1999), PehR-PehS (Flego et al., 2000), ExpI-ExpR (Lee et al., 2013), GacA-GacS (Cui et al., 2001), RsmA-RsmB-RsmC (Chatterjee et al., 2002a), PmrA-PmrB (Hyytiäinen et al., 2003), and CytR (Matsumoto et al., 2003) in a complex manner.
Biofilms are surface-associated microbial communities, and are able to grow in different abiotic and biotic surfaces (Dolan and Costerton, 2002; McDougald et al., 2012). Biofilm development in bacteria can be divided into three distinct stages (Kaplan, 2010) viz., adhesion, multiplication, and dispersion. In the first stage, planktonic bacteria usually adheres to a biotic or abiotic surface either by physical process including van der Waals forces and electrostatic interactions, or by bacterial surface appendages such as flagella and pili (Rosan and Lamont, 2000). In the second stage, bacteria multiply and communicate with each other through cell–cell communication (auto-inducer) signals (McDougald et al., 2012). At this stage, they also secrete a slimy gelatinous material to form a sizable biomass (Kaplan, 2010). This bacterial biomass comprises of exopolysaccharides [EPSs (mainly cellulose)], proteins, and extracellular DNA (Sutherland, 2001; White et al., 2003; Yap et al., 2005; Liang et al., 2010). The contents ultimately determine the architecture and the stability of a biofilm biomass (Koechler et al., 2015) although their quantities vary depending on the bacterial strains and environmental conditions (Zogaj et al., 2001; Solano et al., 2002; Haque et al., 2012). The sessile bacterial biomass are known to have a hetero-dimensional structure (Oliver et al., 2014) with micro-channels and extended limbs to trap nutrients. The whole assembly acts as a protective barrier against toxins, antimicrobial agents, and predators. In the final stage, a mature biofilm often experiences detachment of some of its own biomass. It is signaled either by the bacteria themselves or caused by external forces (Kaplan, 2010). Such dispersal are considered to be a survival mechanism (O’Toole et al., 2000; Solano et al., 2002; Teitzel and Parsek, 2003; Scher et al., 2005; Haque et al., 2012) as the detached biofilms travel to colonize other sites leading to virulence (Elkins et al., 1999; Yildiz and Schoolnik, 1999; van Houdt and Michiels, 2005; Jahn et al., 2008; Yang et al., 2008; Haque et al., 2012).
Cultured bacteria generally form three types of biofilms in the laboratory: solid-air-liquid (SAL) interface biofilm, solid-air (SA) interface biofilm, and air-liquid (AL) interface biofilm popularly known as pellicle (Zogaj et al., 2001; Solano et al., 2002; Yap et al., 2005; Haque et al., 2009). They have separate genetic, chemical and cultural distinctions (Friedman and Kolter, 2004; Yap et al., 2005; Yang et al., 2008; Haque et al., 2012). Formation of AL biofilm is prevalent in numerous Gram-negative aerobic or facultative aerobic bacteria (Armitano et al., 2014). Flagella, different types of pili, curli fimbriae, type I secretion system, T3SS, cellulose, lipopolysaccharide, and quorum sensing are important for AL biofilm formation (Davey and O’Toole, 2000; Yap et al., 2005; Anriany et al., 2006; Jahn et al., 2008; Liang et al., 2010; Haque et al., 2012; Yamamoto et al., 2012). Nutritional (such as media composition, carbon sources and divalent cations particularly, magnesium, calcium, and iron) and environmental conditions (such as temperature, oxygen tension, chemotaxis, pH, and osmolarity) are also important determinants of the development of AL biofilm in bacteria (Yap et al., 2005; Liang et al., 2010; Yamamoto et al., 2011; Haque et al., 2012; Armitano et al., 2013). Furthermore, numerous transcriptional factors such as AgfD in Salmonella typhimurium (Römling et al., 2000), SpoOA in Bacillus subtilis (Branda et al., 2001); SlyA, PhoP-PhoQ two component regulatory system (TCS) in Dickeya dadantii (formerly Erwinia chrysanthemi) 3937 (Haque et al., 2009, 2012), Bcam1359 in Burkholderia cenocepacia H111 (Fazli et al., 2011), QseC in Escherichia coli (Hadjifrangiskou et al., 2012) and RscS in Vibrio fischeri (Yip et al., 2006) were also found to have regulated the AL biofilm formation in response to different environmental and nutritional cues.
CytR (Cytidine Repressor) is known as a transcriptional repressor of nucleoside uptake and catabolism genes in some bacterial species such as E. coli (Valentin-Hansen et al., 1996), Salmonella enterica serovar Typhimurium (Thomsen et al., 1999) and Vibrio cholerae (Haugo and Watnick, 2002). Biofilm formation in V. cholerae is controlled by the CytR through the repression of vps genes, which encode enzymes essential for EPS production (Haugo and Watnick, 2002). Watve et al. (2015), on the other hand, reported that CytR is a global positive regulator of competence, type VI secretion and chitinases in V. cholerae. In case of Pcc PC1 (formerly EC1) however, the CytR homolog has only been partially characterized by Matsumoto et al. (2003). They showed that the ΔcytR mutant is able to reduce the polygalacturonase (Peh) production and increase the production of Pel, Cel, and Prt with respect to its wild counterpart. Swimming motility and the expression of fliA (encoding σ28) and fliC (encoding flagellin) were found to have been dramatically reduced unlike flhD (encoding a master regulator) in the ΔcytR mutant. Consequently, the virulence was radically reduced in the ΔcytR mutant compared to that of the parental strain (Matsumoto et al., 2003). Such discovery was supplemented by Hossain and Tsuyumu (2006) who reported an instance of Pcc PC1 forming SAL biofilm in microtiter plates [made of polyvinyl chloride (PVC)] containing yeast extract peptone broth plus salts of M63 minimal medium at 27°C in static condition. They also showed that SAL biofilm is controlled by motility itself. Despite their efforts, the role of CytR homolog in the formation of AL biofilm in glass test tubes is yet to be quantified under different environmental (i.e., temperature, pH, osmolarity, oxygen tension) and nutritional (i.e., media composition, carbon sources, divalent cations) conditions for Pcc PC1. In addition, the expression of certain genes in this mutant has not been explored with respect to cellulose production.
Cellulose constitutes a gulf of the exopolymeric matrix of AL biofilm in bacteria (Yap et al., 2005; Yang et al., 2008; Haque et al., 2012). It is synthesized by bacterial cellulose synthesis proteins encoded by the bcs operons, such as bcsABCD and bcsEFG (Römling and Galperin, 2015). BcsA is an integral inner membrane protein attached to BcsB, a periplasmic protein. The BcsA contains, among others, a C-terminal fragment that consists of a cyclic-dimeric (3′→5′)-guanosine monophosphate (c-di-GMP) binding PilZ domain (Amikam and Galperin, 2006). The c-di-GMP is known to control numerous cellular functions in bacteria, including biofilm formation, motility and virulence (Yi et al., 2010; Römling et al., 2013). BcsC and BcsD are also required for maximal cellulose production (Saxena et al., 1994). BcsE, BcsF, and BcsG are encoded in the type II bcs operons (Römling and Galperin, 2015) and are essential for optimum cellulose synthesis (Solano et al., 2002). The GIL (GGDEF I-site like) domain of BcsE is able to bind with c-di-GMP result in additional cellulose production (Fang et al., 2014). The Pcc PC1 genome contains all these bcsABCD and bcsEFG operons1. Nonetheless, we are yet to understand if the CytR homolog of Pcc PC1 is also able to regulate the AL biofilm formation by transcriptional control of the bcs genes. The present research aims to explore this area of possibility.
Numerous Gram-negative phytopathogenic bacteria use the T3SS to deliver virulence factors and effectors, such as harpins, avirulence (avr) gene and disease-specific gene products (dsp) from pathogens into host plant cells (Staskawicz et al., 2001). The T3SS is encoded by hrp (hypersensitive response and pathogenicity) and hrc (hypersensitive response conserved) genes. Yap et al. (2005) showed that T3SS regulatory (HrpX, HrpY, HrpS, and HrpL) and effector (HrpA and HrpN) proteins are required for AL biofilm formation in D. dadantii 3937. A more comprehensive study by Yi et al. (2010) showed that T3SS and biofilm formation on plastic are mediated by phosphodiesterases (PDEs) containing GGDEF and EAL-domain proteins that affect c-di-GMP turnover in D. dadantii 3937. The Pcc PC1 genome is known to be containing several GGDEF and EAL-domain proteins1. Therefore, the assumption is that such proteins might regulate the biofilm formation in Pcc PC1. Previous studies in this regard, have shed some light on the regulatory role of these genes in case of SAL biofilm only (Yi et al., 2010). Nonetheless, the scientific communities are yet to find out if these genes are able to regulate the AL biofilm formation, or if the CytR homolog of Pcc PC1 can also be affected. This study will contribute toward understanding the role of CytR homolog in cellulose production, while quantifying the expression of bcs and T3SS genes in Pcc PC1.
Materials and Methods
Bacterial Strains and Growth Media
Pectobacterium carotovorum subsp. carotovorum [formerly E. carotovora subsp. carotovora (Pcc])] PC1 [formerly EC1 (wild type)], its derivative strains, ΔcytR (aflagellated and non-motile mutant), ΔfliC (non-motile and aflagellated mutant), flhD::Tn5 (aflagellated and non-motile mutant) and ΔmotA (flagellated and non-motile mutant) and D. dadantii (formerly Erwinia chrysanthemi) 3937 (wild type) used in this study has been described earlier by Matsumoto et al. (2003), Haque and Tsuyumu (2005), and Hossain et al. (2005). The strains were freshly grown in yeast extract peptone (YP) medium (1% peptone, 0.5% yeast extract, pH 6.8) at 27°C. Nalidixic acid (30 μg/mL) and kanamycin (50 μg/mL) were added to the media when required, and the optical density (OD) of the culture was measured by a spectrophotometer (Intertech, Inc. Tokyo, Japan) at 660 nm.
Media, Carbon Source, and Temperature on AL Biofilm Formation
Yeast extract peptone, Luria-Bertani (LB) medium (1% of tryptone, 0.5% of yeast extract, 0.5% of NaCl, pH 7.0), Salt-optimized broth (SOB) plus 2% of glycerol (SOBG) medium (per liter: 20 g of tryptone, 5 g of yeast extract, 0.5 g of NaCl, 0.186 g of KCl, 2.4 g of MgSO4.7H2O and 2% of glycerol), King’s B (KB) medium (per liter: 10 g of peptone, 1.5 g of K2HPO4, l5 g of MgSO4.7H2O and 15 mL of glycerol), yeast peptone dextrose adenine (YPDA) medium (per liter: 20 g of yeast extract, 40 g of peptone, 40 g of glucose monohydrate, and 80 mg of adenine hemisulfate) and M63 glycerol minimal medium [per liter, 2.5 g of NaCl, 3 g of KH2PO4, 7 g of K2HPO4, 2 g of (NH4)2SO4, 0.5 mg of FeSO4, 2 g of thiamine hydrochloride, and 2 g of glycerol] were used. In order to quantify the impacts of media and temperature on AL biofilm formation, a single colony of the Pcc PC1 and the ΔcytR mutant was grown in shake (180 rpm) culture in YP broth at 27°C until early stationary phase (OD660 at 1.0). Afterward, 50 μl of each culture [ca. 107 colony forming unit (CFU)/mL] were suspended in glass test tubes containing 5 mL of the broth. Two separate suspensions were made and incubated at two different temperatures (27 and 37°C, respectively) for 72 h in static condition. In order to study the role of carbon source on AL biofilm formation, 2% glycerol in SOBG was replaced by 2% of glucose, sucrose, or mannitol. Among the media, the Pcc PC1 produced a thick and robust AL biofilm on SOBG broth at 27°C. The SOBG media and 27°C temperature was therefore taken to study the AL biofilm unless otherwise noted in this manuscript.
Quantification of the Biomass of the AL- and SAL Biofilm
Bacterial strains formed fragile to rigid AL biofilm on SOBG broth. In order to quantify the rigid biomass, samples were prepared as follows: 72 h-old AL biofilms were gently transferred to the fresh test tubes containing 2 mL of distilled water and were vortexed with sterile glass beads. The amount of the biomass in the rigid AL biofilms (OD600) were quantified using an UV spectrophotometer (Ultrospec 3000, Pharmacia Biotech, Cambridge, England). In case of fragile AL biofilm, 1 mL of planktonic culture was carefully collected by the pipette and OD600 was measured. Afterward, each fragile AL biofilm was mixed with 2 mL of planktonic culture and was vortexed. The OD600 of planktonic culture was then subtracted from the OD600 of the planktonic culture plus biomass of the fragile AL biofilm. This would provide the amount of fragile biomass present in the AL biofilm.
The SAL biofilms formed at 37°C were also quantified as described in Haque et al. (2012). In brief, the suspension was carefully removed after 72 h and washed the glass test tubes with distilled water. Afterward, 0.05% (w/v) crystal violet was added and incubated for 30 min followed by rinsing with distilled water. Bound crystal violet was eluted using 95% ethanol and the SAL biofilm was quantified (OD570) using UV spectrophotometer (Ultrospec 3000, Pharmacia Biotech, Cambridge, England).
Congo Red and Calcofluor Binding Assays
Congo red and Calcofluor binding assays were carried out as described in Haque et al. (2009, 2012) with a few modifications. Initially, each bacterial strain (Pcc PC1, ΔcytR, ΔfliC, flhD::Tn5, ΔmotA, and D. dadatii 3937) [used as positive control for the expression of red, dry and rough (rdar) phenotype and expression of cellulose] was grown in YP broth overnight at 27°C under shaking condition (180 rpm). Each bacterial culture was serial diluted 1:100 [ca. 105 CFU/mL] and 2 μL of culture were spotted (four spot in each plate) onto SOBG agar plates containing 40 μg/mL of Congo red (Sigma-Aldrich, St. Louis, MO, United States) or 200 μg/mL of Calcofluor white (Sigma-Aldrich, St. Louis, MO, United States). The plates were then incubated at 27°C in static condition, and photographs were taken after 48 h (for Congo red binding). On the other hand, after 48 h incubation, the plates were placed under UV light (366 nm) and photographs were taken for Calcofluor binding.
Quantification of Cellulose
Amount of cellulose produced in the matrix of the AL biofilms of the bacterial strains (Pcc PC1 ΔcytR, ΔfliC, flhD::Tn5, and ΔmotA) were quantified as described in Anriany et al. (2006) with a few modifications. In brief, strains were grown at 27°C in SOBG broth for 3 days in static condition. Then 3 g (wet weight) of AL biofilm masses were gently collected with sterile spatula and were dried by freeze dryer (Eyela, Freeze dryer, FDU-830, Japan). The dry masses were mixed with 4.5 mL of acetic-nitric reagent (8:2:1 acetic acid: nitric acid: distilled water) and boiled for 20 min. The boiled mixture was then centrifuged and the supernatant was discarded. The pellet was transferred to a Corex centrifuged bottles, washed twice with sterile distilled water and dried in a clean bench. The dried pellet was mixed with 150 μL of concentrated H2SO4 with gentle shaking for 1 h at 27°C. The amount of cellulose was determined by adding 750 μL anthrone (Sigma-Aldrich, St. Louis, MO, United States) reagent (0.2 g in 100 mL H2SO4). The Avicel cellulose (Sigma-Aldrich, St. Louis, MO, United States) was used as standard this case, and the cellulose were quantified at 620 nm.
Isolation of RNA
Wild type (Pcc PC1) and ΔcytR mutant were grown in SOBG broth until AL biofilm was formed. Total RNA was extracted thereafter from the associated bacteria using an RNA isolation kit (Qiagen, Hilden, Germany) and was subjected to DNase I treatment with the TURBO DNase kit (Ambion, Inc., United States) as instructed by the manufacturer. The purity and concentration of RNA was estimated using a Nanodrop ND-100 spectrophotometer (NanoDrop Technologies, Wilmington, DE, United States).
Quantitative Reverse Transcription-PCR
Primers (Table 1) were designed based on Pcc PC1 DNA sequences that can be retrieved from the following address1. cDNA synthesis, verification of the efficiencies of the primers and PCR were carried out as described in Haque et al. (2015). The relative values of transcriptional level were calculated using the ΔΔCT method. The abundance of specific gene was initially normalized using 16S rRNA which was shown to be invariant using best keeper (Matsumoto et al., 2003). The relative expression ratio was calculated as the differences between the cycle threshold (CT) values and was determined using the following equation:
Fold change = 2-ΔΔCT, where ΔΔCT for gene j = (CT,J – CT,16SrRNA)mutant – (CT,J – CT,16SrRNA)wildtype.
Radish Sprouts Attachment Assays
Attachment assays were performed as described in Jahn et al. (2008) with a few modifications. In brief, radish (Raphanus sativus) seed (10 g) was collected from Horticulture Research Centre of Bangladesh Agricultural Research Institute, Gazipur, Bangladesh. Seeds were surface sterilized with 75 mL of 1.5% sodium hypochloride for 30 min on a rotary shaker at 200 rpm and was followed by several washes using sterile distilled water. Single colonies of bacterial strain were inoculated into SOBG broth and were grown overnight at 27°C with agitation. It was then centrifuged before each strain was resuspended in sterile distilled water. Approximately 104 CFU/mL bacteria were used. For attachment assays, seeds were germinated in sterile water for 4 days with regular water changes. The sprouts were transferred in sterile 50 mL conical tubes (10 sprouts/tube) and were incubated in bacterial suspension for 4 h at 27°C with gentle shaking (50 rpm). Sprouts were then rinsed three times with sterile distilled water and homogenized. After serial dilution, the homogenate was plated and incubated at 27°C for 24 h for colonies to be enumerated. The experiment was repeated at least three times with at least seven sprouts analyzed per strain each time.
Stress Tolerance Tests
Stationary (OD660 at 1.5) phase cells (grown in shaking condition) and cells-associated with AL biofilm of the wild type and the ΔcytR mutant were prepared as described in Haque et al. (2012). In order to prepare the planktonic cells (i.e., cells under the AL biofilm), 1 mL of planktonic culture was carefully collected after 72 h incubation followed by centrifugation. The pellet was then re-suspended in sterile distilled water. In order to test the sensitivities, the stationary-phase, the planktonic cells and the AL biofilm cells (ca. 108 CFU/mL) of the wild type and the ΔcytR mutant were separately exposed to acidic pH 4.0 (40 mM citric acid and 20 mM dibasic sodium phosphate, pH 4.0) and oxidative stress (H2O2 at 10 mM) which corresponds to a lethal concentration (Haque et al., 2009). The suspensions were incubated for a further 2 h at 27°C. The number of CFU was determined by serial dilution and plating onto SOBG agar just prior to inoculation and during every 30 min over a 2-h incubation. The survival of treated cells was normalized to the number of CFU at the beginning of the test.
Statistical Analysis
All the experiments were conducted in complete randomized design with three replications and repeated at least three times unless otherwise stated. Data were analyzed using Student’s t-test of the SAS software system 8.02 (SAS Institute, Cary, NC, United States).
Results
Reduced AL Biofilm Formation in the ΔcytR Mutant in Different Growth Media
In order to quantify the impacts of media composition and temperature on AL biofilm formation, the Pcc PC1 and the ΔcytR mutant cells (ca. 107 CFU/mL) were suspended in five different media (SOBG or KB or LB or YP or M63 glycerol minimal media) and was incubated at two different temperatures (27 or 37°C) in static condition. Selection of five different media and two different temperatures was important to generate information that previous researches did not shed light on. A study by Hossain and Tsuyumu (2006) showed that Pcc PC1 produces SAL biofilm on YP broth plus salts of M63 minimal medium in the wells of microtiter plates made of PVC at 27°C in static condition. They also examined two other media such as YP and LB broth but neither of them produced any SAL or AL biofilms in Pcc PC1. The present study particularly deals with AL biofilm formation in glass test tubes under different environmental conditions which were not examined in any other contemporary researchers dealing with Pcc PC1 and its ΔcytR mutant. This study aimed to contribute toward this area of concern, as it was carried out under different media and temperature for a comprehensive result. The results of this study showed that Pcc PC1 formed AL biofilm on SOBG, KB, and YPDA broth after 72 h incubation only at 27°C (Figure 1A). In fact, the AL biofilm in SOBG, KB, and YPDA broth did not continue to grow after 7 days (data not shown). However, the AL biofilm in SOBG broth showed signs of detachment at day 7 (data not shown). The AL biofilms formed by Pcc PC1 on the SOBG broth was quite rigid and was denser compared to those on the KB and YPDA broth. On the other hand, biofilms on YPDA and KB by Pcc PC1 were fragile and hence, were dispersed when the samples were disturbed. AL biofilms also acquired a sizable biomass when the glycerol in SOBG was replaced by glucose, sucrose, and mannitol (data not shown). The ΔcytR mutant, on the other hand, produced thinner, lighter and more fragile AL biofilm on SOBG, YPDA and KB broth (Figure 1A). Quantitative analysis of the biomass at OD600 showed that the wild type produced significantly (P < 0.001) higher biofilm biomass in SOBG (4.18-fold), YPDA (3.18-fold), and KB (3.10-fold) broth compared to the ΔcytR mutant at 27°C (Figure 1B). At 37°C however, the ΔcytR mutant and its wild counterpart formed only SAL biofilms on SOBG, YPDA, and KB broth (Figure 1A). The Pcc PC1 produced a prominent SAL biofilm on SOBG broth compared to those on the YPDA and KB broth (Figure 1A). The other three media (YP, LB, and M63 glycerol minimal) did not produce any SAL or AL biofilm for both of these strains under any of the temperatures (27 or 37°C) (data not shown). Similar to the AL biofilms, the amount of SAL biofilm (OD570) was also significantly (P < 0.001) higher in the wild type on SOBG (2.4-fold) and YPDA (1.9-fold) compared to the ΔcytR mutant (Figure 1B). Initially however, both wild type (Pcc PC1) and ΔcytR mutant cells grew faster in all the broths except in M63 glycerol minimal medium where the growth was slightly delayed both at 27°C (Figures 1C,D) and at 37°C (Figures 1E,F). In general, the cell growth was slightly delayed in ΔcytR mutant compared to that of the wild type in YP broth at both temperatures (Figures 1C–F). These results suggested that biofilm formation controlled by the CytR homolog of Pcc PC1 is regulated by media composition and temperature. Bacterial growth might play an important role in this case, although it is very unlikely to be amongst the major determinants.
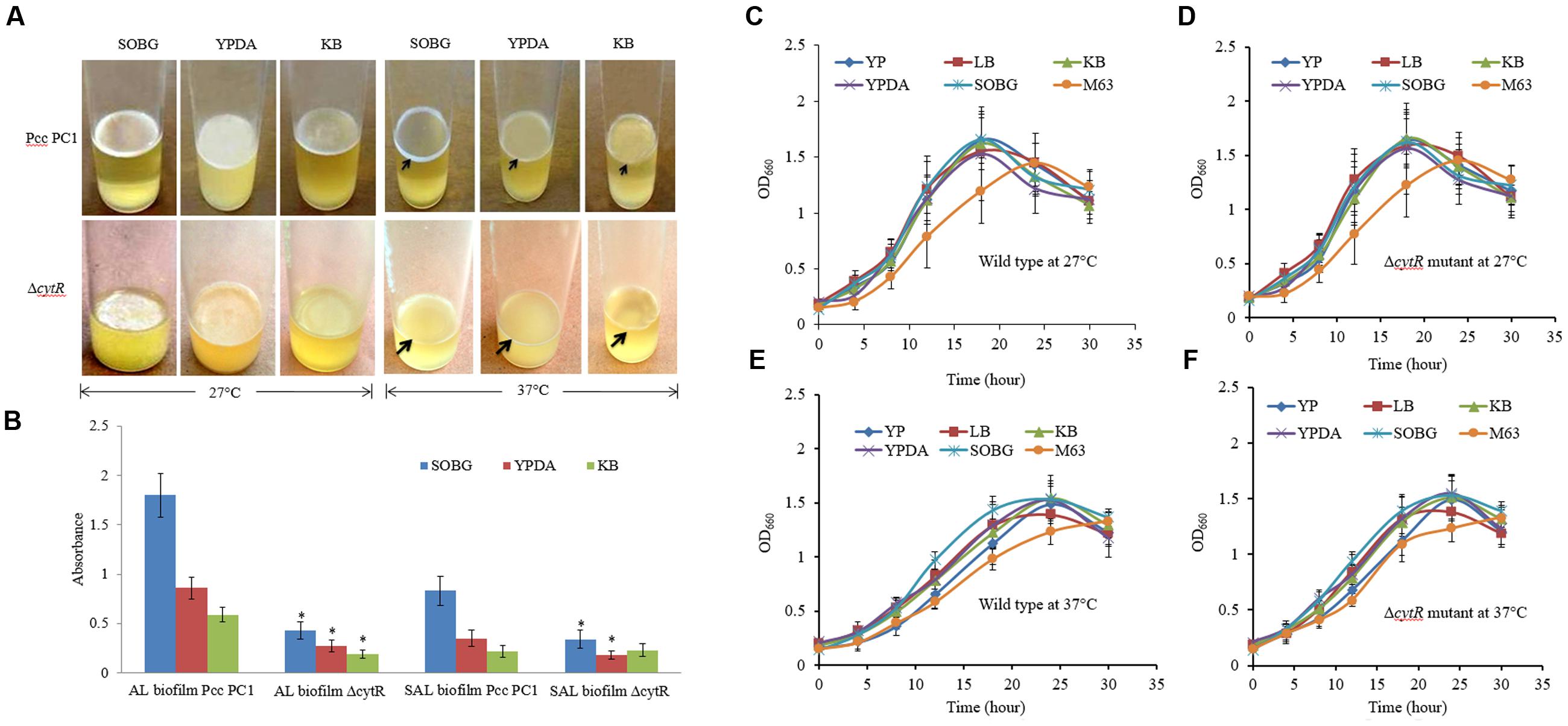
FIGURE 1. Biofilm formation in different growth media. (A) AL- and SAL biofilm formation by the wild type (Pcc PC1) and ΔcytR mutant at 27°C and at 37°C, respectively. (B) Biomass of AL- and SAL biofilms, measured at 600 and 570 nm, respectively. (C–F) Growth rate of the wild type (Pcc PC1) and the ΔcytR mutant in different media in shaking condition at 27 and 37°C, respectively. The values are mean and error bars indicate standard deviations ( ± ) of three independent experiments. Asterisks indicate P < 0.001 (Student’s t-test).
Divalent Cations Induced AL Biofilm Formation Controlled by the CytR Homolog
Divalent cations such as Mg2+, Ca2+, Cu2+, Mn2+, and Zn2+ have been shown to be the inducers and stabilizers of the biofilm formation processes (Song and Leff, 2006; Liang et al., 2010; Haque et al., 2012). However, information regarding the role of such cations in the formation of AL biofilms by Pcc PC1 and its ΔcytR mutant was not available in the literature. In this study, the Pcc PC1 (wild type) and ΔcytR mutant cells were inoculated in SOBGMg– (magnesium deprived) and SOBGMg– with 0.009 M of Mg2+, Ca2+, Mn2+, Cu2+ and Zn2+, were incubated at 27°C under shaking (180 rpm) condition. The growth rate was not significantly different between the wild type (Figure 2A) and the ΔcytR mutant (data not shown) in different incubation period for any divalent cations. Both the wild type and the mutant cells grew quickly and attained its maximum at 18 h of incubation period both in SOBGMg– and SOBGMg– containing 0.009 M Mg2+ and Ca2+ (Figure 2A). In SOBGMg– containing 0.009 M Mn2+, Cu2+, and Zn2+ however, the growth was slow and hence, took 24 h to reach the growth maximum (Figure 2A). Interestingly, it was only after 72 h incubation (in static condition) both the strains developed thinner to denser AL biofilm only on SOBGMg– containing the divalent cations (Figure 2B). Unlike this, the SOBGMg– broth at 27°C did not show any AL biofilms either in the wild type or in the ΔcytR mutant (Figure 2B). Compared to the ΔcytR mutant, wild type built a denser and robust AL biofilm on SOBGMg– containing Mg2+ and Ca2+ and a thinner and more fragile AL biofilm on SOBGMg– containing Cu2+, Mn2+, and Zn2+ (Figure 2B). When quantified, the production of biomass in the ΔcytR mutant was found to be significantly (P < 0.001) lower than that of its wild counterpart (Figure 2C). These results clearly indicate that the CytR homolog may positively regulate AL biofilm formation in Pcc PC1 responding to divalent cations.
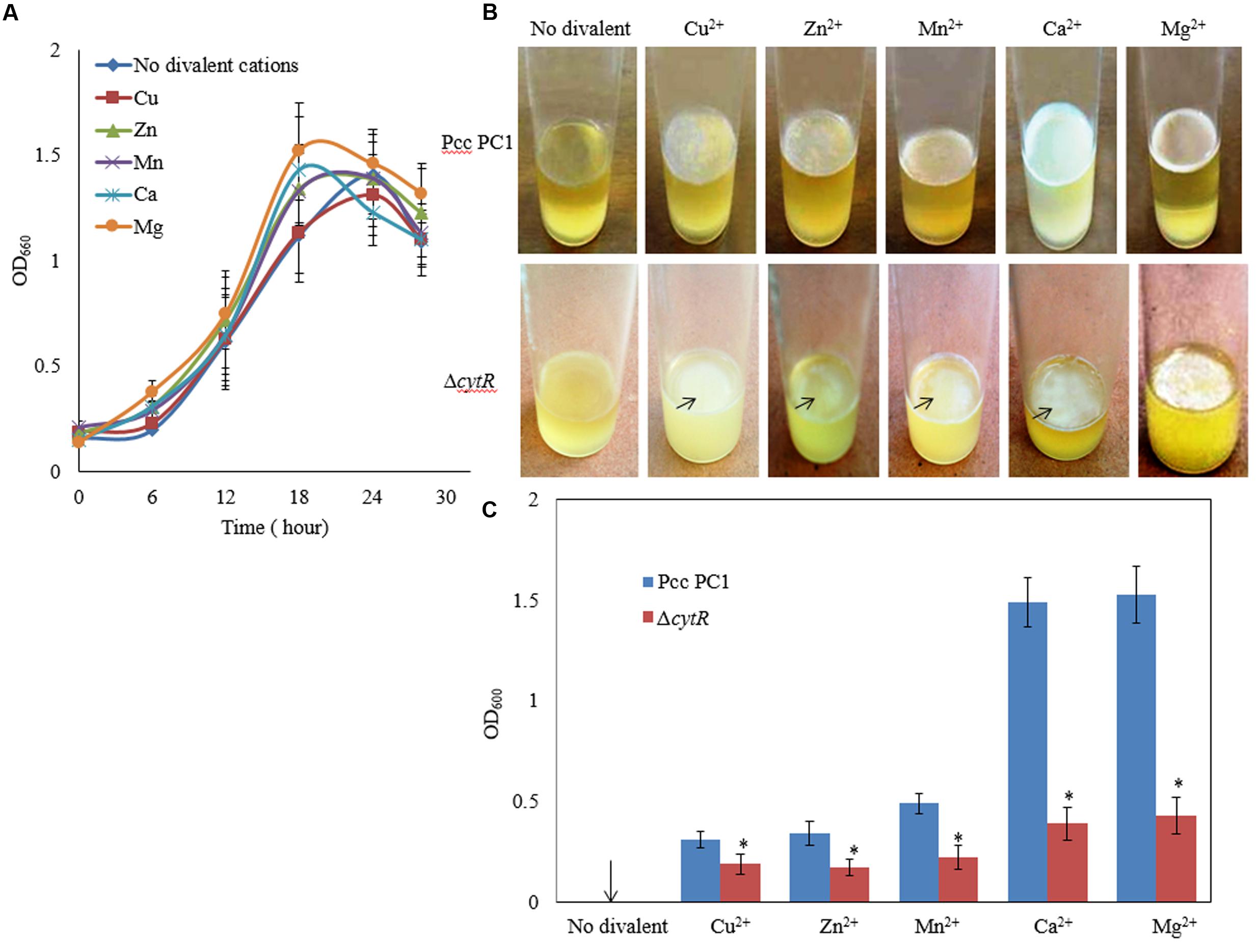
FIGURE 2. Effect of divalent cations on growth and AL biofilm formation at 27°C. (A) Growth of wild type (Pcc PC1) in SOBGMg– (magnesium deprived) and SOBGMg– containing indicated divalent cations. (B) AL biofilm formation by wild type and the ΔcytR mutant. (C) Biomass biofilms. The values are mean and error bars indicate standard deviations ( ± ) of three independent experiments. Asterisks indicate P < 0.001 (Student’s t-test).
Reduced AL Biofilm Formation in the ΔcytR Mutant in Response to pH
During early stages of plant infection, pathogenic bacteria often confronts an acidic pH that ranges from pH 4.5 to 6.5 (Grignon and Sentenac, 1991). In order to assess whether Pcc PC1 and ΔcytR mutant thrives under acidic conditions, these strains were exposed to acidic pH ranges (Figure 3A) using malic acid on SOBG broth at 27°C. The reason behind using malic acid is to stimulate the acidity in plant apoplast that contains malate (Grignon and Sentenac, 1991). The results showed that none of the strains grew at pH 4.5 while showing slower growth at pH 5.0 and pH 5.5 under shaking condition at 27°C. Bacterial cell growth did not differ significantly between the wild type (Figure 3A) and the mutant (data not shown) at different incubation period. The wild type formed a delicate AL biofilm on SOBG broth at pH 5.0 and pH 5.5 after 120 h in static condition (Figure 3B). On the other hand, ΔcytR mutant cells did not grow at the same condition (Figure 3B). After 72 h incubation at 27°C, the wild type formed a thinner AL biofilm on SOBG broth at pH 6.0 compared to pH 7.0 (Figure 3B). In case of ΔcytR mutant, only a few cells aggregated on the top and did not cover the whole surface at pH 6.0 (Figure 3B). Our results indicated that acidic pH delayed the AL biofilm formation. The compounds and/or the genes responsible for AL biofilm formation may be not produced or expressed in acidic conditions leading to AL biofilm formation of Pcc PC1.
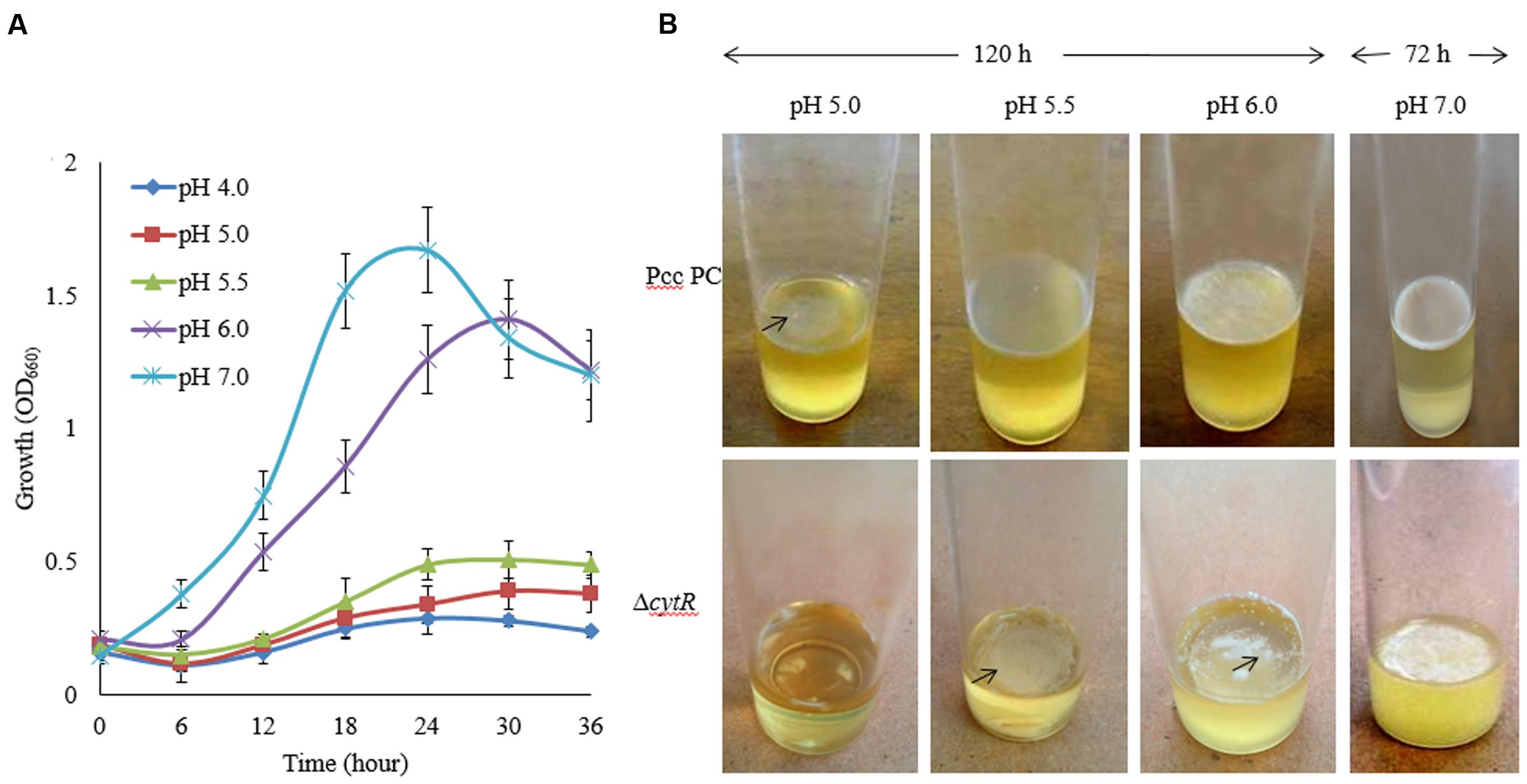
FIGURE 3. Effect of pH on growth and AL biofilm formation in SOBG broth at 27°C. (A) Growth of wild type (Pcc PC1) at indicated pHs. (B) AL biofilm formation by the wild type (Pcc PC1) and the ΔcytR mutant. The values are mean and error bars indicate standard deviations ( ± ) of three independent experiments.
Reduced AL Biofilm Formation in the ΔcytR Mutant in Response to Osmolarity
Osmolarity plays a great role in biofilm formation (Solomon et al., 2005; Rinaudi et al., 2006). In order to assess how osmolarity affects the formation of AL biofilm, NaCl and D-sorbitol were used as osmotic agents in the SOBG broth that contains 0.5 g/L NaCl. We however, tested the growth of bacterial cells under different concentrations (0.05, 0.1, 0.2, 0.3 M) of NaCl in shaking condition at 27°C. The results suggest that the growth (OD660) of both strains was unaffected due to the changes in osmolarity except for 0.3 M NaCl. Compared to the ΔcytR mutant, a firm and intense AL biofilm was developed by the wild type on the SOBG broth containing 0.05 and 0.1 M NaCl after 72 h incubation. Unlike this, 0.3 M NaCl produced small quantities of cells by the wild type and the mutant in shaking condition (Figure 4A) and consequently, a few cells were aggregated on the surface of the standing culture (Figure 4B). Similar results were also observed when 0.3 M NaCl was replaced by 0.3 M D-sorbitol (data not shown). Conversely, when 0.3 M NaCl/D-sorbitol was replaced by 0.3 M sucrose, both wild type and the ΔcytR mutant cells grew rapidly (Figure 4A). At this condition, wild type produced a stronger and thicker AL biofilm compared to the ΔcytR mutant after 72 h incubation in static condition (Figure 4B). These observations suggest that osmotic stress negatively affect AL biofilm formation due to poor growth of the bacterial cells which CytR homolog of Pcc PC1 is able to regulate. This could possibly include the regulation of expression of other components required for AL biofilm formation.
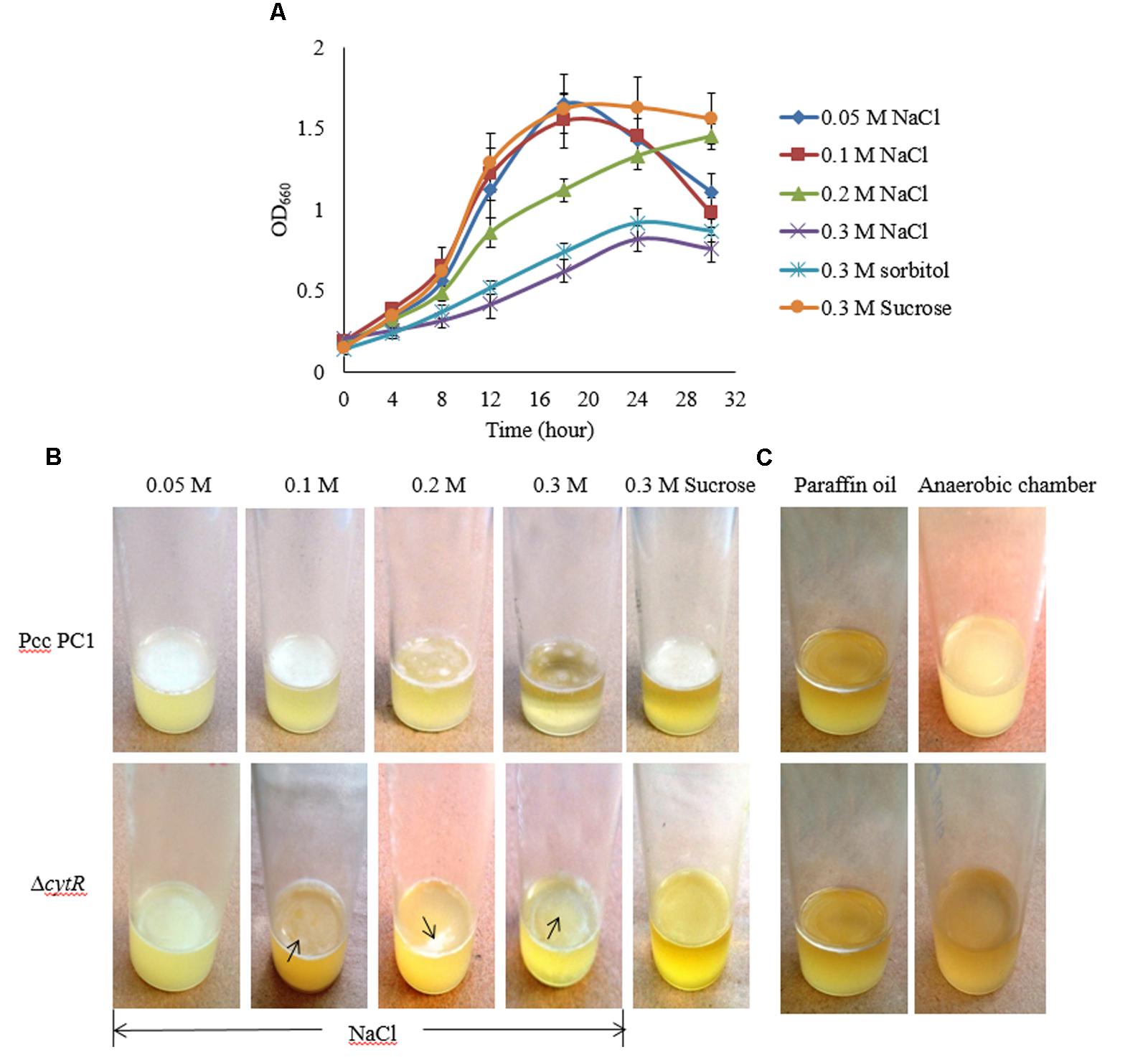
FIGURE 4. Impacts of osmotic stress on growth and AL biofilm formation in SOBG broth at 27°C. (A) Growth of wild type in SOBG containing indicated concentrations of NaCl. (B) AL biofilm formation by the wild type (Pcc PC1) and the ΔcytR mutant in different concentrations of NaCl. (C) No biofilm formed by the wild type and the ΔcytR mutant in anaerobic condition. These experiments were repeated at least three times. The values are mean and error bars indicate standard deviations ( ± ) of three independent experiments.
Oxygen Limits the Formation of AL Biofilm in Pcc PC1 and ΔcytR Mutant
Higher oxygen concentrations play an important role in AL biofilm formation (Gerstel and Römling, 2001; Liang et al., 2010). However, this general statement needs to be tested and quantified for the Pcc PC1 and its ΔcytR mutant. In order to accomplish this, the wild type and the ΔcytR mutant cells (ca. 107 CFU/mL) were suspended in glass test tubes containing 5 mL SOBG broth. It was then sealed with 1.5 mL of sterile liquid paraffin oil and incubated at 27°C for 7 days in static condition. None of the strains formed an AL biofilm although the turbidity of the cells in the broth was increased (Figure 4C). Similar results were also observed when wild type and the ΔcytR mutant cells (ca. 107 CFU/mL) was suspended in glass test tubes containing 5 mL SOBG broth and incubated at 27°C in anaerobic chamber (Thermo, Inc., Portsmouth, NH, United States) for 7 days (Figure 4C). This could be due to the lack of expression of compounds responsible for AL biofilm formation as stated by Gerstel and Römling (2001).
Motility Itself, But Not Presence of Flagella, Is Required for AL Biofilm Formation
Flagella-mediated motility was shown to play an important role in AL biofilm formation (Jahn et al., 2008; Yang et al., 2008; Haque et al., 2012). On the other hand, Hossain and Tsuyumu (2006) showed that motility, but not the presence of flagella, is required for SAL biofilm formation in microtiter plates of Pcc PC1. Because CytR homolog positively regulates flagella-mediated motility (Matsumoto et al., 2003), we examined whether CytR homolog of Pcc PC1 also regulates AL biofilm by controlling the motility itself. In order to confirm whether the motility itself or the presence of flagella is required for AL biofilm formation on SOBG broth at 27°C in stationary condition, we used three aflagellated and non-motile mutants such as ΔcytR, ΔfliC and flhD::Tn5, and one flagellated and non-motile mutant such as ΔmotA (essential for flagellar rotation but not required for flagellar assembly) of Pcc PC1. All the mutants produced a lighter and fragile AL biofilm compared to their wild counterpart after 72 h incubation at 27°C (Figure 5A). The results also showed that AL biofilm was not increased in the mutants even after longer incubation period (data not shown). When the biofilm biomass was quantified, wild type (Pcc PC1) was found to have produced significantly (P < 0.001) more (2.9-, 5.5-, 4.5-, and 9.6-fold) AL biofilms than that of cytR, ΔfliC, flhD::Tn5, and ΔmotA, respectively (Figure 5B). These data, therefore concludes that motility itself, but not the presence of flagella, is required for AL biofilm formation which the CytR homolog of Pcc PC1 is able to control.
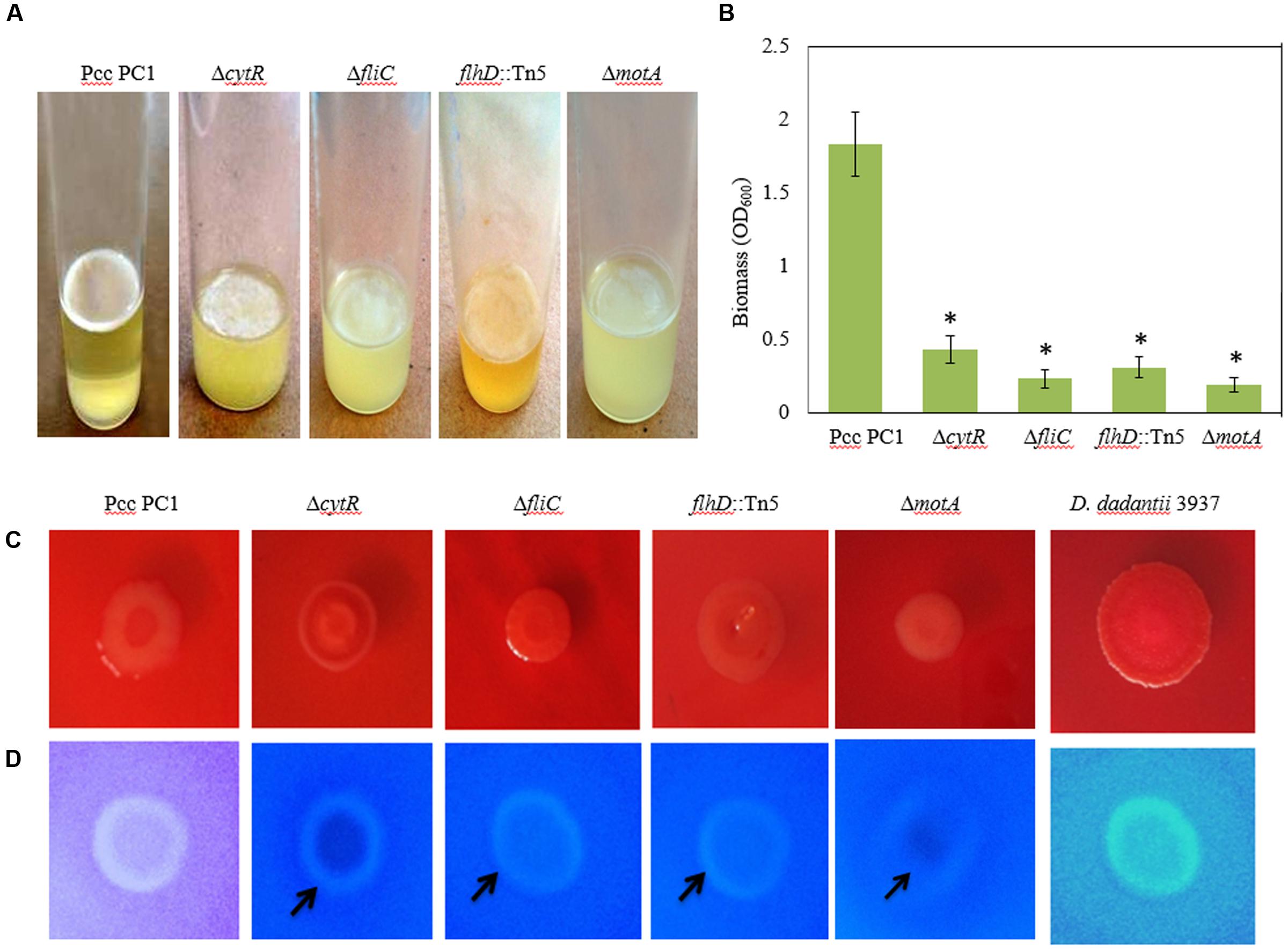
FIGURE 5. AL Biofilm formation by various strains (A). (B) Quantification of the biomass biofilms in the wild type and the mutants. (C) Congo red binding (SOBG agar plates containing 40 μg/mL Congo red) abilities of the wild type and the mutants after 48 h incubation. (D) Calcofluor binding (SOBG agar plates containing 200 μg/mL Calcofluor) capacities of the wild type and the various mutants after 48 h incubation. These experiments were repeated at least three times. The values are mean and error bars indicate standard deviations ( ± ) of three independent experiments. Asterisks indicate P < 0.001 (Student’s t-test).
Reduced Cellulose Production in the ΔcytR, ΔfliC, flhD::Tn5 and ΔmotA Mutant
Biofilm producing bacteria develop red, dry and rough (rdar) phenotype (also known as rugose/wrinkled phenotype) on Congo red agar plates (Römling, 2005; Haque et al., 2012; Milanov et al., 2015). We hypothesized that CytR homolog, FliC, FlhD, and MotA may affect the expression of rdar phenotype. The results of this study show that the expression of rdar phenotype on Congo red agar plates was indistinguishable between the mutants (such as ΔcytR, ΔfliC, flhD::Tn5, and ΔmotA) and the wild type (Figure 5C). Therefore, the rdar phenotype may not be controlled by the CytR homolog, FliC, FlhD, and MotA in Pcc PC1.
It is understood that rdar expressing bacteria usually binds with the cellulose specific dye Calcofluor (Zogaj et al., 2001; Solano et al., 2002; Römling, 2005; Uhlich et al., 2006; Steenackers et al., 2012; Milanov et al., 2015). We therefore evaluated whether wild type Pcc PC1 and the mutants (ΔcytR, ΔfliC, flhD::Tn5, and ΔmotA) also binds to Calcofluor. The results showed that the wild type (Pcc PC1) had induced bright fluorescence similar to D. dadantii 3937, while all the mutants (ΔcytR, ΔfliC, flhD::Tn5, and ΔmotA) weakly induced bright fluorescence (Figure 5D). This result indicated that wild type Pcc PC1 may produce more cellulose-rich EPS compared to the mutants.
Because cellulose production and AL biofilm formation are correlated in bacteria, we quantified cellulose production in the matrix of the biofilms. The wild type was found to have produced more cellulose (49.7 ± 2.3 ng) than the mutants of ΔcytR (21.3 ± 1.7 ng), ΔfliC (13.4 ± 2.3 ng), flhD::Tn5 (24.3 ± 2.7 ng), and ΔmotA (9.8 ± 0.8 ng) mutant. According to these results, the increase in Calcofluor binding seemed to have been reflected in the increase of cellulose production. This indicates that the CytR homolog is capable of regulating the AL biofilm formation by controlling both cellulose production and motility in Pcc PC1.
Reduced Expressions of bcs Genes along with adrA in the ΔcytR Mutant
Bacterial cellulose synthesis (bcs) operons, bcsABZC and bcsEFG, are required for cellulose production leading to AL biofilm formation in E. coli and S. enterica serovar Typhimurium (Römling et al., 2000; Solano et al., 2002; Da Re and Ghigo, 2006). Homology searches revealed that bcsA, bcsB, bcsC, bcsE, bcsF, and bcsG of Pcc PC1 had 67.5, 56.5, 50.5, 41.7, 30, and 62.3 similarity to the translated products of bcsA, bcsB, bcsC, bcsE, bcsF, and bcsG of E. coli K-12, respectively, at the amino acid level. We hypothesized that the CytR homolog may affect expression of bcs genes in Pcc PC1. In order to test this hypothesis, the expression of bcsA (a cellulase synthase) and bcsE (maximal cellulose expresser), was measured by quantitative reverse transcription-PCR using mRNA collected from the AL biofilms formed by the wild type and its ΔcytR mutant. Expressions of bcsA and bcsE were significantly (P < 0.001) reduced (6.3- and 11.1-fold) in the ΔcytR mutant compared to that in the wild type (Figure 6). Therefore, the CytR homolog may positively regulates the transcription of bcs genes in Pcc PC1.
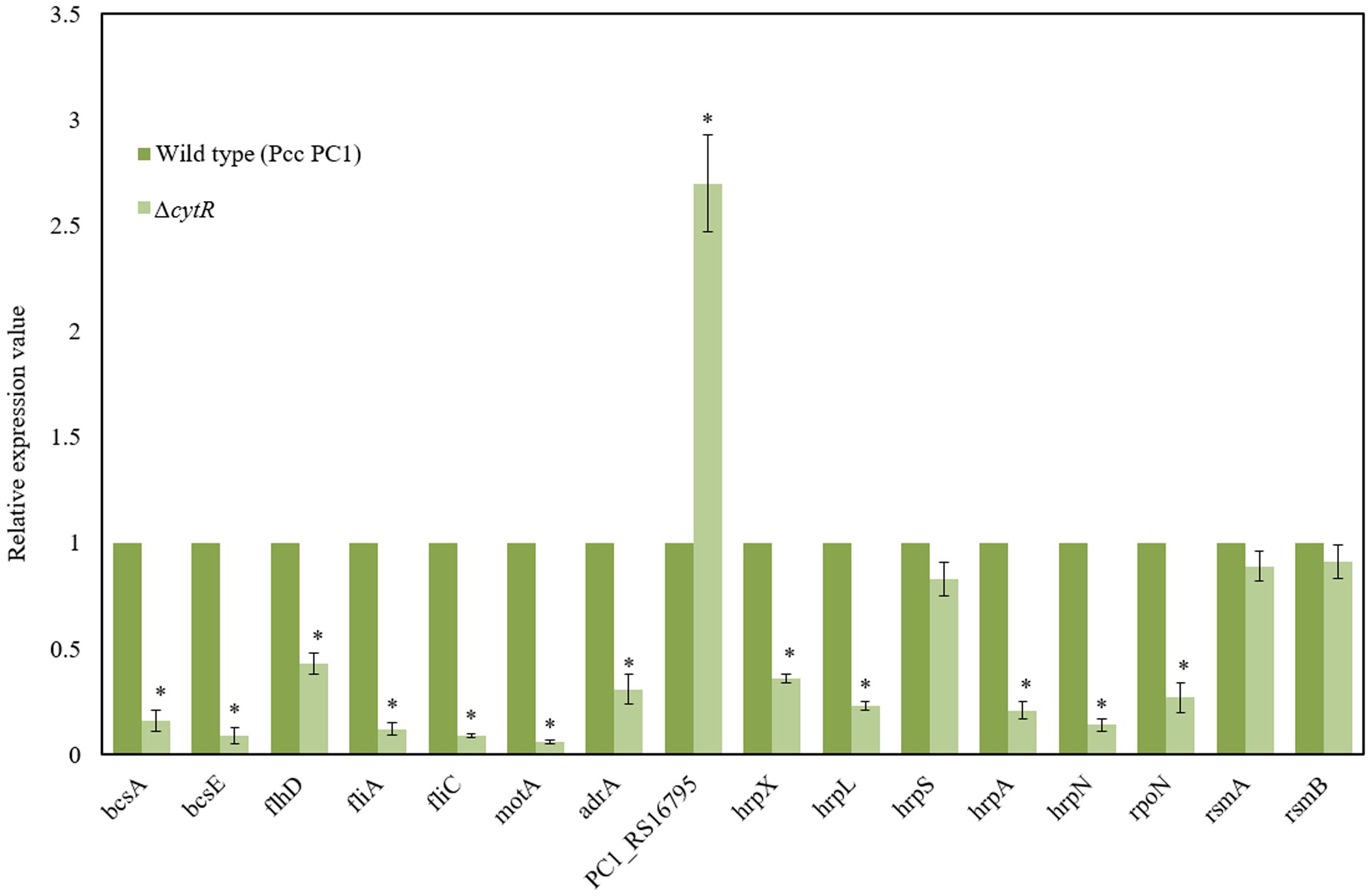
FIGURE 6. Expressions of various genes in the wild type (Pcc PC1) and ΔcytR mutant. Values relative to the mean expression in the wild type were calculated using the ΔΔCT method. Error bars indicate standard deviation ( ± ) for data of three independent experiments. Asterisks indicate P < 0.001 (Student’s t-test).
c-di-GMP is catalyzed by the GGDEF domain proteins present in diguanylate cyclases (DGCs) (Hickman et al., 2005; Ryjenkov et al., 2005) and hydrolyzed by either the EAL or HD-GYP domains present in PDEs (Ryan et al., 2006). The AdrA is a GGDEF domain protein that is able to synthesize the c-di-GMP which binds to the BcsA (Kader et al., 2006; Morgan et al., 2014) and BcsE (Fang et al., 2014). The adrA is also required for cellulose biosynthesis in S. enterica serovar Typhimurium (Römling et al., 2000; Zogaj et al., 2001). In D. dadantii 3937, ecpB (a GGDEF-EAL protein) and ecpC (a EAL protein) were shown to be negatively regulated the biofilm formation (Yi et al., 2010). The Pcc PC1 genome encodes 17 GGDEF domain proteins, six EAL domain proteins and three GGDEF-EAL domain proteins1. In this study, we found that the expression of adrA (a GGDEF domain protein) was considerably lower (3.2-fold) in the ΔcytR mutant (Figure 6) compared to its wild type. However, the expression of PC1_RS16795 (a EAL domain protein) had a dramatic increase (2.7-fold) in the ΔcytR mutant compared to the wild type. Thus, CytR homolog may control the GGDEF (AdrA) and EAL (PC1_RS16795) domain proteins in Pcc PC1.
Reduced Expressions of Motility- and T3SS Genes in the ΔcytR Mutant
Because ΔflhD, ΔfliC, and ΔmotA produced significantly the lower amount of cellulose than their wild counterpart, in this study, expressions of flhD, fliA, fliC, and motA were measured by quantitative reverse transcription-PCR using mRNA collected from the AL biofilms formed by the wild type and its ΔcytR mutant. Expressions of flhD (2.3-fold), fliC (11-fold), fliA (8.3-fold), and motA (16.7-fold) were significantly (P < 0.001) decreased in the ΔcytR mutant than the wild type (Figure 6). These results indicated that CytR homolog positively controls the expressions of flhD, fliA, fliC, and motA in Pcc PC1.
In D. dadantii 3937, T3SS regulatory (hrpX, hrpY, hrpS, and hrpL), structural (hrpA, hrcJ) and effector (hrpN) genes are shown to be required for AL biofilm formation (Yap et al., 2005). GGDEF and EAL domain proteins in D. dadantii 3937 were also shown to be negatively regulated both the T3SS and biofilm formation (Yi et al., 2010). In our study, we found that GGDEF and EAL domain proteins are also controlled by the CytR homolog of Pcc PC1 (Figure 6). Thus, the expression of T3SS genes was examined by quantitative reverse transcription-PCR using mRNA collected from AL biofilms of the wild type and ΔcytR mutant. Compared to the wild type, the expression of hrpX (2.8-fold), hrpL (4.3-fold), hrpA (4.8-fold) and hrpN (7.1-fold) was found to have significantly (P < 0.001) reduced in the ΔcytR mutant (Figure 6). Thus, CytR homolog is positively controlled the hrp genes in Pcc PC1. These results also suggested that CytR homolog is required for hrpL expression, which in turn activates the expression of hrp genes in the HrpL regulon in Pcc PC1 (Chatterjee et al., 2002b; Yap et al., 2005; Yi et al., 2010).
In our experiment however, the expression of hrpS did not differ significantly between the wild type and its ΔcytR mutant (Figure 6). CytR homolog may affect the expression of hrpL through the known hrpL regulators, such as RpoN (σ54), RsmA (a small RNA-binding protein) or RsmB (a regulatory RNA that binds to and sequesters the negative effect of RsmA on hrpL mRNA by forming RsmA–RsmB complex) (Chatterjee et al., 2002b; Yi et al., 2010). The amount of rsmA and rsmB transcripts in ΔcytR mutant was similar to that in the wild type (Figure 6). Compared to the wild type, the expression of rpoN was significantly (P < 0.001) lower (3.7-fold) in the ΔcytR mutant (Figure 6). Therefore, the CytR homolog may be termed as a regulator of hrpL expression which it achieves by altering the expression of rpoN in Pcc PC1.
Reduced Attachments to Radish Sprouts by ΔcytR, ΔfliC, flhD::Tn5, and ΔmotA Mutants
Because CytR homolog can regulate AL biofilm formation, we hypothesized that similar genes were also required for attachment to plant tissues as well. The ΔcytR mutant was significantly reduced (P < 0.001) in attachment to radish sprouts compared with the wild type (Figure 7). We also tested the ΔfliC, flhD::Tn5, and ΔmotA mutants for the attachment. Compared to the wild type, their attachment was dramatically reduced (P < 0.001) in these mutants (Figure 7). Thus, CytR homolog, FliC, FlhD, and MotA may regulate both AL biofilm formation in culture and also during attachment to plants in Pcc PC1.
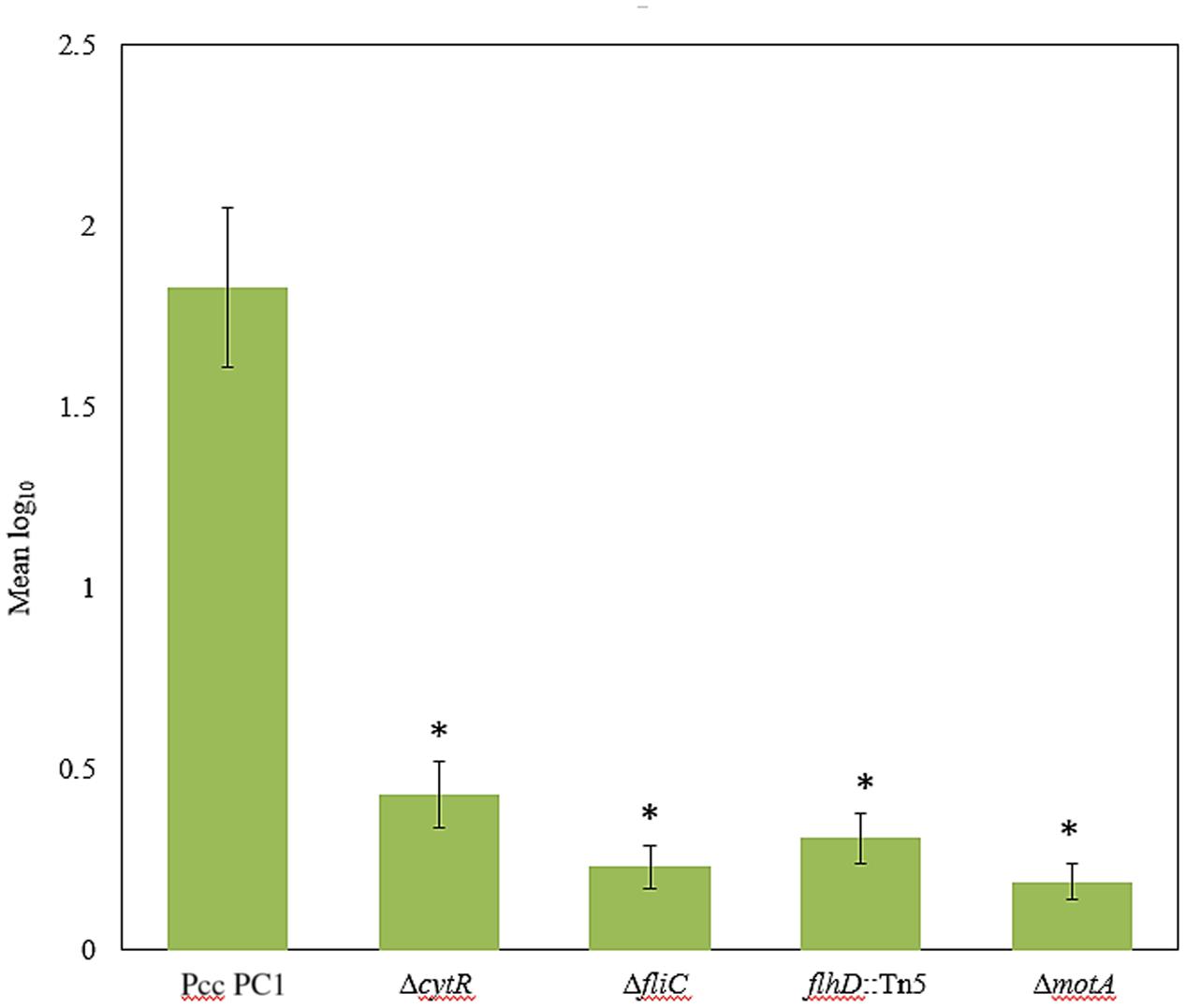
FIGURE 7. Bacterial attachment to radish sprouts. Radish seeds were germinated in sterile water for 4 days then sprouts were transferred in sterile 50 mL conical tubes (10 sprouts/tube) and incubated in bacterial suspension (ca. 104 CFU/mL) for 4 h at 27°C with gentle shaking at 50 rpm. Sprouts were rinsed three times with sterile distilled water then homogenized. After serial dilution, the homogenate was plated and incubated at 27°C for 24 h then the colonies were enumerated. The experiment was repeated at least three times with at least seven sprouts analyzed per strain each time. The values are mean log10 and error bars indicate standard deviation ( ± ) of bacterial cells attached to radish sprouts are presented. Asterisks indicate P < 0.001 (Student’s t-test).
CytR is Required for Survival in Unfavorable Environments
When Pcc PC1 infects a plant, it confronts an unfavorable environment such as acidity and oxidative stresses. These conditions may play a vital role in survival and expression of the virulence factors (Toth et al., 2003) of any organism. Since biofilms play an important role in the survival of bacteria, we compared the sensitivities of AL biofilms and other cells under conditions of acidity (pH 4.0) and oxidative stress generated by 10 mM H2O2. Results suggested that the biofilm cells were more resistant than the planktonic and stationary-phase cells (Figure 8). The wild type (Pcc PC1) was found to have shown more resistance compared to the ΔcytR mutant under acidic pH (Figure 8A). All the planktonic- and stationary-phase cells of the ΔcytR mutant were killed by 10 mM H2O2 within the first 60 min, and only 9% of the bacteria-associated with AL biofilm of the ΔcytR mutant survived within 60-min exposure time (Figure 8B). On the contrary, a 60-min exposure of stationary-phase, planktonic and AL biofilm cells of the wild type (Pcc PC1) against 10 mM H2O2 resulted in 4, 13, and 45% survival, respectively (Figure 8B). Thus, formation of AL biofilm controlled by the CytR homolog may play a great role in the survival of Pcc PC1 under unfavorable environments.
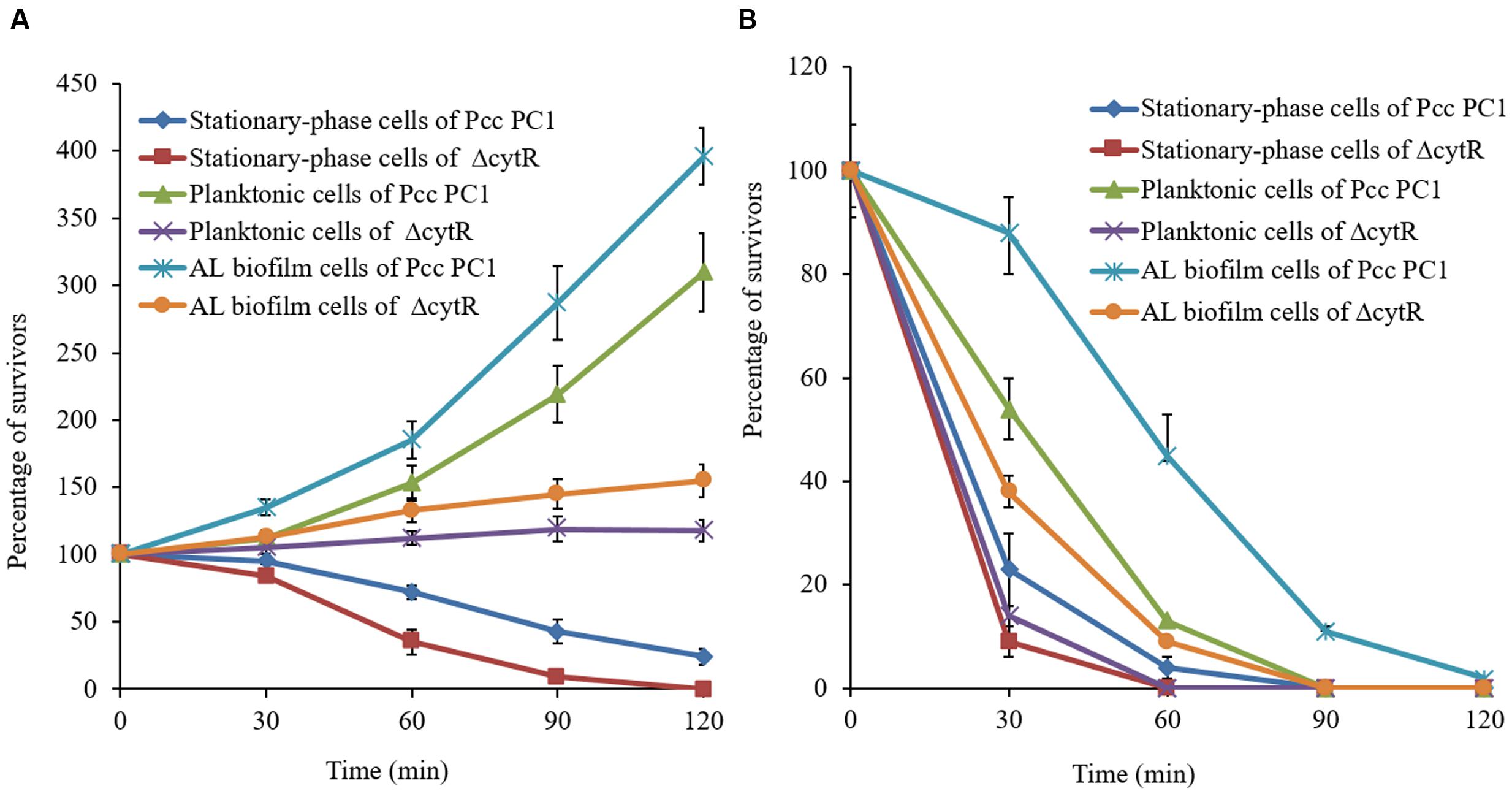
FIGURE 8. Percentage of cells in bacteria-associated with biofilms, planktonic- and stationary-phase cells surviving a 120-min exposure to acidic (4.0) pH (A) and oxidative stress generated by the addition of 10 mM hydrogen peroxide (B). Survival rate of the treated cells was normalized to the number of CFU at the beginning of the challenge. Error bars indicate standard deviation ( ± ) for data of three independent experiments.
Discussion
In this article, we demonstrated that CytR homolog of Pcc PC1 is able to positively regulate the AL biofilm formation while responding to different environmental and nutritional signals (Figures 1–5). The results also showed that the CytR homolog is capable of controlling the expressions of multiple genes required for cellulose biosynthesis, c-di-GMP signaling, motility and T3SS in Pcc PC1 (Figure 6). All the mutants were found to diminish their capacity to attachment to radish sprouts (Figure 7). We also demonstrated that AL biofilm cells of the wild type was resistant than stationary-phase and planktonic cells to acidity and oxidative stress compared to the same cells of the ΔcytR mutant (Figure 8). Therefore it can arguably be said that CytR homolog is able to positively control numerous cellular functions in Pcc PC1.
The ability of a bacteria to form biofilms depend on the nutritional conditions (Yap et al., 2005; Hossain and Tsuyumu, 2006; Rinaudi et al., 2006). Divalent cations, particularly Cu2+, Ca2+, Mn2+ and Zn2+ and Mg2+ play a key role in this process as have been explained for Shewanella oneidensis (Liang et al., 2010), V. cholerae (Kierek and Watnick, 2003), and Pseudomonas fluorescens (Song and Leff, 2006). We also reported that low concentration of Mg2+ significantly increase the AL biofilm formation of D. dadantii 3937 in a PhoP-PhoQ dependent manner (Haque et al., 2012). The present study also confirms that the formation of AL biofilm controlled by the CytR homolog in Pcc PC1 is activated by the different divalent cations, e.g., Mg2+, Ca2+, Cu2+, Zn2+, and Mn2+ (Figure 2B). Biofilm formation also depends on the environmental conditions such as temperature (Yap et al., 2005), osmotic stress and acidity (Rinaudi et al., 2006). In this study, we observed that high temperature, acidic pH, high osmolarity and anaerobic condition negatively affect the AL biofilm formation controlled by the CytR homolog of Pcc PC1 (Figures 1, 3–5). Understandably, lower production of cellulose could lead to such scenario, or the genes required for cellulose synthesis were not expressed under those extreme conditions.
Cellulose and curli are two major components of the AL biofilm in Enterobacteriaece and their traces can be determined by Congo red binding assays (Steenackers et al., 2012; Milanov et al., 2015). For example, bacterial strains expressing both curli and cellulose leads to the rdar phenotype, only cellulose produces the pink, dry and rough (pdar) phenotype, and only curli triggers the brown, dry and rough (bdar) phenotype. A less-prominent phenotype on Congo red agar: ras (red and smooth; curli only), pas (pink and smooth, cellulose only) and bas (brown and smooth; curli only) has also been reported in some E. coli strains (Bokranz et al., 2005). In our study, expression of the rdar phenotype was indistinguishable between the wild type (Pcc PC1) and the mutants (ΔcytR, ΔfliC, flhD::Tn5, and ΔmotA) (Figure 5C). Compared to the wild type, all the mutants (ΔcytR, ΔfliC, flhD::Tn5, and ΔmotA) exhibited weak bond with cellulose specific (Calcofluor) dye (Figure 5D). This study also revealed that cellulose production was significantly reduced in the mutants of ΔcytR, ΔfliC, flhD::Tn5, and ΔmotA compared to their wild counterpart. The present study also showed that expressions of flhD, fliA, fliC, and motA are positively controlled by the CytR homolog of Pcc PC1 (Figure 6). Thus, fragile and lower AL biofilm formation in these mutants may not be due to only the reduction of motility but also the lower production of cellulose.
Bacterial cellulose synthesis is regulated on both transcriptional and post-transcriptional levels. Expressions of bcs genes are controlled by the different regulatory proteins in different bacteria. Transcriptional regulators MlrA and CsgD of S. enterica serovar Typhimurium were shown to modulate cellulose biosynthesis indirectly by regulating expression of c-di-GMP synthases DGCs and phosphodiesterases (PGEs) (Römling, 2005; Römling et al., 2013). In E. coli, AdrA (a GGDEF domain protein) is responsible for cellulose production (Römling et al., 2000; Zogaj et al., 2001). In our present study showed that expressions of bcsA, bcsE, and adrA are positively controlled by the CytR homolog of Pcc PC1 (Figure 6). Thus, CytR homolog may directly regulate cellulose biosynthesis by transcriptional control of bcsA, bcsE, and adrA in Pcc PC1.
AL biofilm formation is regulated by different regulatory proteins in bacteria. Yang et al. (2008) reported a delay in AL biofilm (pellicle) formation in ΔgacA mutant of D. dadantii 3937 due to lower expression of hrpL, dspE (effector protein), hrpA, and hrpN. We, in our previous study, reported that PhoP-PhoQ two component system regulates AL biofilm (pellicle) formation by controlling bcsABCD, adrA and fliC operons but not by controlling the hrp genes in D. dadantii 3937 (Haque et al., 2012). The present study showed that expressions of hrpX, hrpL, hrpA, and hrpN are positively controlled by the CytR homolog of Pcc PC1 (Figure 6). We also observed that the expression of T3SS genes is regulated by HrpL and possibly by RpoN (Figure 6). Based on our results, we therefore, proposed a model in Figure 9. It shows that the expression of motility, T3SS and cellulose producing genes are tightly controlled by the CytR homolog of Pcc PC1 through a sophisticated regulatory cascade (Figure 9). In our study, the expression of adrA (a GGDEF domain protein) is positively controlled by the CytR homolog of Pcc PC1 (Figure 6). It was reported that AdrA directly binds to the cellulose synthase complex to deliver c-di-GMP straight to its BcsA (Kader et al., 2006; Morgan et al., 2014). Recently, it was shown that GIL, a new c-di-GMP binding domain protein binds to BcsE which transfers c-di-GMP to BcsA indirectly (Fang et al., 2014). In line with this idea, our results also suggest a strong possibility that c-di-GMP was synthesized by Pcc PC1 and was controlled by the CytR homolog which lead to the production of cellulose and expression of T3SS (Figure 9). Similar results have been reported regarding D. dadantii 3937 by Yi et al. (2010) who showed that the c-di-GMP turnover was mediated by GGDEF and EAL domain proteins in D. dadantii 3937. However, the exact mechanism of c-di-GMP turnover in Pcc PC1 has yet to be deciphered.
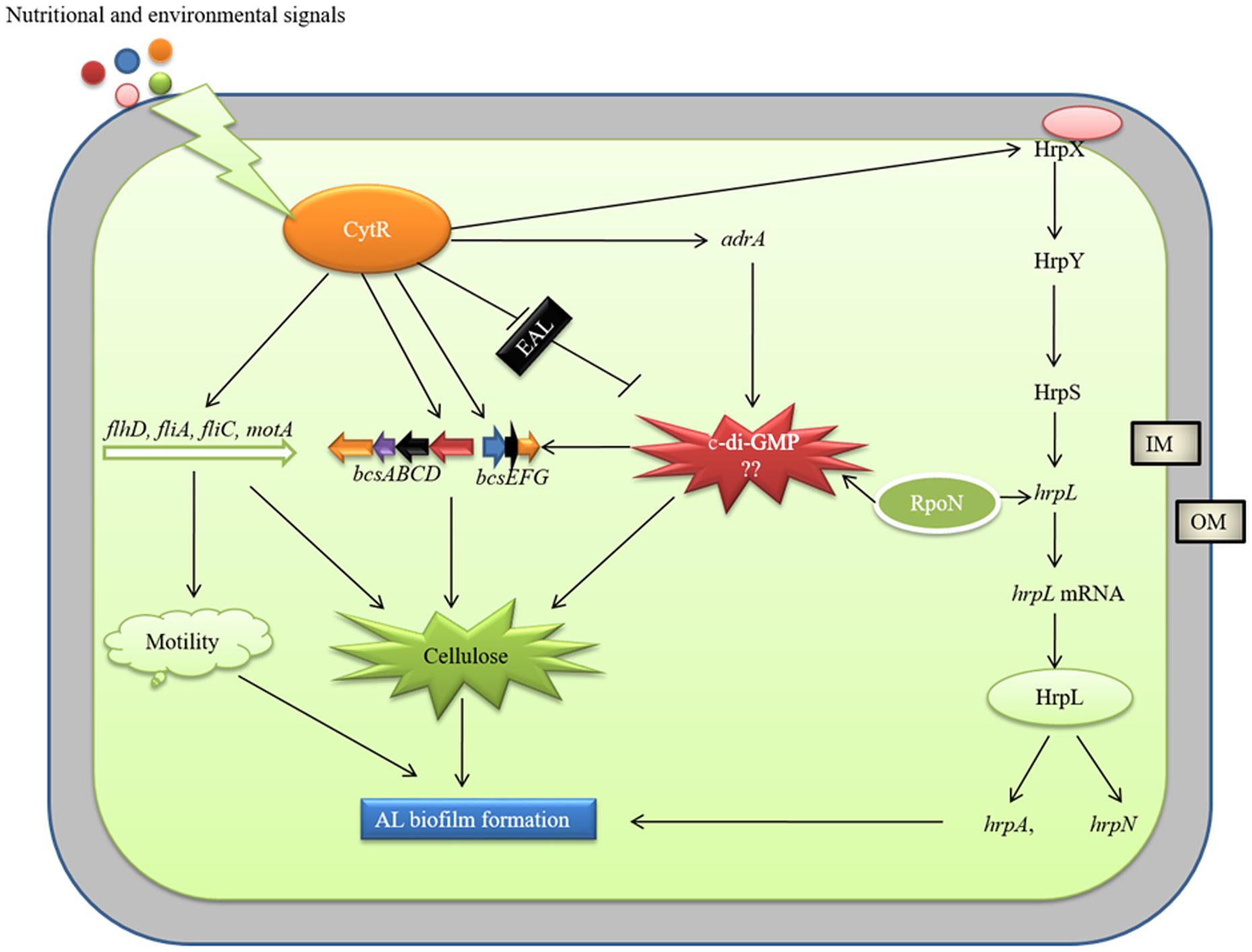
FIGURE 9. Model of CytR homolog regulatory cascade in Pcc PC1. Environmental (temperature, pH, and osmolarity) and nutritional (growth media and divalent cations) signals modulate CytR expression. The CytR homolog directly controls the expressions of bcs operons and adrA (a GGDEF domain protein), responsible for cellulose production. The adrA may produce c-di-GMP that binds to bcsA (Amikam and Galperin, 2006) and bcsE (Fang et al., 2014) leading to increase cellulose production. The CytR homolog also regulates the T3SS, essential for AL biofilm formation. The RpoN and HrpL may play a vital role in c-di-GMP regulation of T3SS. However, molecular mechanism by c-di-GMP regulate cellulose production and T3SS yet to be examined. The CytR homolog positively controls the expressions of flagellar formation and rotation genes. These genes are required for cellulose production and motility.
AL biofilm formation by plant pathogenic bacteria has been shown to be positively related with their virulence in plants (Jahn et al., 2008; Yang et al., 2008; Haque et al., 2009, 2012). Jahn et al. (2011) reported that symptom development and growth in Nicotiana benthamiana are not different between D. dadantii 3937 wild type and the bcsA (a cellulose synthase) mutant. In this experiment, the degree of maceration (in potato tuber) was significantly increased following inoculation of the wild type compared to the ΔcytR mutant (data not shown). However, in planta Pel, Cel, and Prt production were indistinguishable between the wild type and the ΔcytR mutant (data not shown). Thus, CytR homolog may positively regulate the virulence not by controlling the plant cell-wall-degrading enzymes in Pcc PC1.
Several genes have been shown to be important for both AL biofilm formation and bacterial attachment to plants (Barak et al., 2005; Jahn et al., 2008). We used radish sprouts to test bacterial adherence to plants because Pcc PC1 is known to cause sprout rot of radish. The study showed that the ability of mutants, i.e., ΔcytR, ΔfliC, flhD::Tn5, and ΔmotA to adhere themselves to plant tissues was significantly lower than the wild type (Figure 7). It has been reported that cellulose synthesized by pathogens and symbionts is essential for host colonization and survival in stress conditions (Augimeri et al., 2015). We have previously reported that bacteria-associated with pellicle/AL biofilm except for aerobically grown logarithmic- and stationary-phase cells of the D. dadantii 3937 are more resistant to survival in acidic pH (4.0), oxidative stress and high osmolarity (Haque et al., 2012). In this study, we observed that stationary-phase-, planktonic-, and AL biofilm cells of the wild type are more resistant compared to that in the same cells of the ΔcytR mutant under adverse conditions including acidic pH (4.0) and oxidative stress generated by 10 mM H2O2 (Figure 8). These results indicate that cellulose production is linked with the bacterial survival mechanism. Higher cellulose production in the wild type is therefore associated with its increased resistance to acidity and oxidative stress. On the contrary, lower resistance in the ΔcytR mutant is linked with lower production of cellulose. This study also reported that planktonic cells are more resistant than stationary-phase cells (Figure 8). Similar to Matsumoto et al. (2003), we were unable to complement the restoration of the particular phenotype when cytR homolog on pPLAFR3 (tetracycline resistant, low-copy-number plasmid) or on pML122 (gentamicin resistant, low-copy-number plasmid) transformed into ΔcytR mutant of Pcc PC1. Complementation of the phenotypes in this regulatory gene is yet to be confirmed. We do not know why ΔcytR mutant failed to complement the phenotypes. We observed that the CytR homolog of Pcc PC1 regulates numerous cellular functions interacting with different regulatory networks (Figure 9). Therefore, plasmid level of expression might be different at native levels and might cause a no complemented phenomena. Yap et al. (2005) reported that the hrpS mutant of D. dadantii 3937 failed to complement due to a second, unintended spontaneous mutation. It was also reported that subunit interference occurs in homologous proteins leading to failure of complementation (Leonhartsberger et al., 2000).
The AL biofilm formation in Pcc PC1 is an emergent property. We are yet to know what role AL biofilms play in the expression of virulence of Pcc PC1. In this study, we observed that bacteria-associated with AL biofilms are more resistance to acidic pH and oxidative stress. Thus, formation of AL biofilm in Pcc PC1 may protect the cells against unfavorable environment. Moreover, the matrix of AL biofilm of Pcc PC1 is composed of cellulose which is commercially available as a wound dressing material (Abeer et al., 2014). Further research endeavors with Pcc PC1 could look into this area of possibilities.
Conclusion
The present study has demonstrated that CytR homolog of Pcc PC1 positively regulate AL biofilm formation in culture responding to environmental and nutritional signals. This study has clearly demonstrated that CytR homolog of Pcc PC1 positively regulate bacterial attachment to radish sprouts as it arguably controls the expression of numerous genes involved in cellulose production, c-di-GMP signaling, motility and the T3SS. The outcomes of this study are expected to contribute toward understanding the virulence factors, attachment to plant tissues and survival of Pcc PC1 in unfavorable environments.
Author Contributions
All authors listed, have made substantial, direct and intellectual contribution to the work. All authors read and approved the final manuscript.
Funding
This research was partly supported by University Grants Commission of Bangladesh.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Dr. Hiroyuki Matsumoto and Dr. Md. Mijan Hossain, Shizuoka University, Japan for the generation of the mutants used in this study. We also acknowledge the reviewers whose comments and suggestions helped us improve the manuscript.
Footnotes
References
Abeer, M. M., Mohd Amin, M. C., and Martin, C. (2014). A review of bacterial cellulose-based drug delivery system: their biochemistry, current approaches and future prospects. J. Pharm. Pharmacol. 66, 1047–1061. doi: 10.1111/jphp.12234
Amikam, D., and Galperin, M. Y. (2006). PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 22, 3–6. doi: 10.1093/bioinformatics/bti739
Anriany, Y., Sanu, S. N., Wessels, K. R., McCann, L. M., and Joseph, S. W. (2006). Alteration of the roguse phenotype in waaG and ddhC mutants of Salmonella enterica serovar Typhimurium DT104 is associated with inverse production of curli and cellulose. Appl. Environ. Microbiol. 72, 5002–5012. doi: 10.1128/AEM.02868-05
Armitano, J., Méjean, V., and Jourlin-Castelli, C. (2013). Aerotaxis governs floating biofilm formation in Shewanella oneidensis. Environ. Microbiol. 15, 3108–3118. doi: 10.1111/1462-2920.12158
Armitano, J., Méjean, V., and Jourlin-Castelli, C. (2014). Gram-negative bacteria can also form pellicles. Environ. Microbiol. Rep. 6, 534–544. doi: 10.1111/1758-2229.12171
Augimeri, R. V., Varley, A. J., and Strap, J. L. (2015). Establishing a role for bacterial cellulose in environmental interactions: lessons learned from diverse biofilm-producing Proteobacteria. Front. Microbiol. 6:1282. doi: 10.3389/fmicb.2015.01282
Barak, J. D., Gorski, L., Naraghi-Arani, P., and Charkowski, A. O. (2005). Salmonella enterica virulence genes are required for bacterial attachment to plant tissue. Appl. Environ. Microbiol. 71, 5685–5691. doi: 10.1128/AEM.71.10.5685-5691.2005
Bokranz, W., Wang, X., Tschape, H., and Romling, U. (2005). Expression of cellulose and curli fimbriae by Escherichia coli isolated from the gastrointestinal tract. J. Medical Microbiol. 54, 1171–1182. doi: 10.1099/jmm.0.46064-0
Branda, S. S., González-Pastor, J. E., Ben-Yehuda, S., Losick, R., and Kolter, R. (2001). Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. USA 98, 11621–11626. doi: 10.1073/pnas.191384198
Charkowski, A., Blanco, C., Condemine, G., Expert, D., Franza, T., Hayes, C., et al. (2012). The role of secretion systems and small molecules in soft-rot Enterobacteriaceae pathogenicity. Ann. Rev. Phytopathol. 50, 425–450. doi: 10.1146/annurev-phyto-081211-173013
Chatterjee, A., Cui, Y., and Chatterjee, A. K. (2002a). RsmA and the quorum-sensing signal, N-[3-oxohexanoyl]-L-homoserine lactone, control the levels of rsmB RNA in Erwinia carotovora subsp. carotovora by affecting its stability. J. Bacteriol. 184, 4089–4095.
Chatterjee, A., Cui, Y., and Chatterjee, A. K. (2002b). Regulation of Erwinia carotovora hrpLEcc (sigma-LEcc), which encodes an extracytoplasmic function subfamily of sigma factor required for expression of the HRP regulon. Mol. Plant Microbe Interact. 9, 971–980.
Cui, Y., Chatterjee, A., and Chatterjee, A. K. (2001). Effects of the two-component system comprising GacA and GacS of Erwinia carotovora subsp. carotovora on the production of global regulatory rsmB RNA, Extracellular Enzymes, and HarpinEcc. Mol. Plant Microbe Interact. 14, 516–526. doi: 10.1094/MPMI.2001.14.4.516
Da Re, S., and Ghigo, J. M. (2006). A CsgD-independent pathway for cellulose production and biofilm formation in Escherichia coli. J. Bacteriol. 188, 3073–3087. doi: 10.1128/JB.188.8.3073-3087.2006
Davey, M. E., and O’Toole, G. A. (2000). Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64, 847–867. doi: 10.1128/MMBR.64.4.847-867.2000
Davidsson, P. R., Kariola, T., Niemi, O., and Palva, E. T. (2013). Pathogenicity of and plant immunity to soft rot pectobacteria. Front. Plant Sci. 4:191. doi: 10.3389/fpls.2013.00191
Dolan, R., and Costerton, W. (2002). Biofilms: survival mechanisms clinical microorganisms. Clin. Microbiol. Rev. 15, 167–193. doi: 10.1128/CMR.15.2.167-193.2002
Elkins, J. M., Hassett, D. J., Stewart, P. S., Schweizer, H. P., and McDermott, T. R. (1999). Protective role of catalase in Pseudomonas aeruginosa biofilm resistance to hydrogen peroxide. Appl. Environ. Microbiol. 65, 4594–4600.
Fang, X., Ahmad, I., Blanka, A., Schottkowski, M., Cimdins, A., Galperin, M. Y., et al. (2014). GIL, a new c-di-GMP binding protein domain involved in regulation of cellulose synthesis in enterobacteria. Mol. Microbiol. 93, 439–452. doi: 10.1111/mmi.12672
Fazli, M., O’Connell, A., Nilsson, M., Niehaus, K., Dow, J. M., Givskov, M., et al. (2011). The CRP/FNR family protein Bcam1349 is a c-di-GMP effector that regulates biofilm formation in the respiratory pathogen Burkholderia cenocepacia. Mol. Microbiol. 82, 327–341. doi: 10.1111/j.1365-2958.2011.07814.x
Flego, D., Marits, R., Eriksson, A. R., Kõiv, V., Karlsson, M. B., Heikinheimo, R., et al. (2000). A two-component regulatory system, pehR-pehS, controls endopolygalacturonase production and virulence in the plant pathogen Erwinia carotovora subsp. carotovora. Mol. Plant Microbe Interact. 13, 447–455. doi: 10.1094/MPMI.2000.13.4.447
Friedman, L., and Kolter, R. (2004). Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51, 675–690. doi: 10.1046/j.1365-2958.2003.03877.x
Gerstel, U., and Römling, U. (2001). Oxygen tension and nutrient starvation are major signals that regulate agfD promoter activity and expression of the multicellular morphotype in Salmonella typhimurium. Environ. Microbiol. 3, 638–648. doi: 10.1046/j.1462-2920.2001.00235.x
Grignon, C., and Sentenac, H. (1991). pH and ionic conditions in the apoplast. Annu. Rev. Plant Physiol. 42, 103–128. doi: 10.1146/annurev.pp.42.060191.000535
Hadjifrangiskou, M., Gu, A. P., Pinkner, J. S., Kostakioti, M., Zhang, E. W., and Greene, S. E. (2012). Transposon mutagenesis identifies uropathogenic Escherichia coli biofilm factors. J. Bacteriol. 194, 6195–6205. doi: 10.1128/JB.01012-12
Haque, M. M., Hirata, H., and Tsuyumu, S. (2012). Role of PhoP-PhoQ two-component system in pellicle formation, virulence and survival in harsh environments of Dickeya dadantii 3937. J. Gen. Plant Pathol. 78, 176–189. doi: 10.1007/s10327-012-0372-z
Haque, M. M., Hirata, H., and Tsuyumu, S. (2015). SlyA regulates motA and motB, virulence and stress-related genes under conditions induced by the PhoP-PhoQ system in Dickeya dadantii 3937. Res. Microbiol. 166, 467–475. doi: 10.1016/j.resmic.2015.05.004
Haque, M. M., Kabir, M. S., Aini, L. Q., Hirata, H., and Tsuyumu, S. (2009). SlyA, a MarR family transcriptional regulator, is essential for virulence in Dickeya dadantii 3937. J. Bacteriol. 191, 5409–5419. doi: 10.1128/JB.00240-09
Haque, M. M., and Tsuyumu, S. (2005). Virulence, resistance to magainin II and expression of pectate lyase are controlled by the PhoP-PhoQ two-component regulatory system responding to pH and magnesium in Erwinia chrysanthemi 3937. J. Gen. Plant Pathol. 71, 47–53.19. doi: 10.1007/s10327-004-0158-z
Haugo, A. J., and Watnick, P. I. (2002). Vibrio cholerae CytR is a repressor of biofilm development. Mol. Microbiol. 45, 471–483. doi: 10.1046/j.1365-2958.2002.03023.x
Hickman, J. W., Tifrea, D. F., and Harwood, C. S. (2005). A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. U.S.A. 102, 14422–14427. doi: 10.1073/pnas.0507170102
Hossain, M. M., Shibata, S., Aizawa, S.-I., and Tsuyumu, S. (2005). Motility is an important determinant for pathogenesis of Erwinia carotovora subsp. carotovora. Physiol. Mol. Plant Pathol. 66, 134–143. doi: 10.1016/j.pmpp.2005.06.001
Hossain, M. M., and Tsuyumu, S. (2006). Flagella-mediated motility is required for biofilm formation by Erwinia carotovora subsp. carotovora. J. Gen. Plant Pathol. 72, 34–39. doi: 10.1007/s10327-005-0246-8
Hyytiäinen, H., Sjöblom, S., Palomäki, T., Tuikkala, A., and Palva, E. T. (2003). The PmrA-PmrB two-component system responding to acidic pH and iron controls virulence in the plant pathogen Erwinia carotovora ssp. carotovora. Mol. Microbiol. 50, 795–807. doi: 10.1046/j.1365-2958.2003.03729.x
Jahn, C. E., Selimi, D. A., Barak, J. D., and Charkowski, A. O. (2011). The Dickeya dadantii biofilm matrix consists of cellulose nanofibres, and is an emergent property dependent upon the type III secretion system and the cellulose synthesis operon. Microbiology 157, 2733–2744. doi: 10.1099/mic.0.051003-0
Jahn, C. E., Willis, D. K., and Charkowski, A. O. (2008). The flagellar sigma factor FliA is required for Dickeya dadantii virulence. Mol. Plant Microbe Interact. 21, 1431–1442. doi: 10.1094/MPMI-21-11-1431
Kader, A., Simm, R., Gerstel, U., Morr, M., and Römling, U. (2006). Hierarchical involvement of various GGDEF domain proteins in rdar morphotype development of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 60, 602–616. doi: 10.1111/j.1365-2958.2006.05123.x
Kaplan, J. B. (2010). Biofilm dispersal: mechanisms, clinical implications, and potential therapeutic uses. J. Dent. Res. 89, 205–218. doi: 10.1177/0022034509359403
Kersey, C. M., Agyemang, P. A., and Dumenyo, C. K. (2012). CorA, the magnesium/nickel/cobalt transporter, affects virulence and extracellular enzyme production in the soft rot pathogen Pectobacterium carotovorum. Mol. Plant Pathol. 13, 58–71. doi: 10.1111/j.1364-3703.2011.00726.x
Kierek, K., and Watnick, P. I. (2003). The Vibrio cholerae O1390-antigen polysaccharide is essential for Ca2+-depedent biofilm development in sea water. Proc. Natl. Acad. Sci. U.S.A. 100, 14357–14362. doi: 10.1073/pnas.2334614100
Koechler, S., Farasin, J., Cleiss-Arnold, J., and Arséne-Ploetze, F. (2015). Toxic metal resistance in biofilms: diversity of microbial responses and their evolution. Res. Microbiol. 10, 764–773. doi: 10.1016/j.resmic.2015.03.008
Lee, D. H., Lim, J. A., Lee, J., Roh, E., Jung, K., Choi, M., et al. (2013). Characterization of genes required for the pathogenicity of Pectobacterium carotovorum subsp. carotovorum Pcc21 in Chinese cabbage. Microbiology 159, 1487–1496. doi: 10.1099/mic.0.067280-0
Leonhartsberger, S., Ehrenreich, A., and Böck, A. (2000). Analysis of the domain structure and the DNA binding sites of the transcriptional activator FhlA. Eur. J. Biochem. 267, 3672–3684. doi: 10.1046/j.1432-1327.2000.01399.x
Liang, Y., Gao, H., Chen, J., Dong, Y., Wu, L., He, Z., et al. (2010). Pellicle formation in Shewanella oneidensis. BMC Microbiol. 10:291. doi: 10.1186/1471-2180-10-291
Liu, Y., Jiang, G., Cui, Y., Mukherjee, A., Ma, W. L., and Chatterjee, A. (1999). kdgREcc negatively regulates genes for pectinases, cellulase, protease, harpinEcc, and a global RNA regulator in Erwinia carotovora subsp. carotovora. J. Bacteriol. 181, 2411–2422.
Matsumoto, H., Muroi, H., Umehara, M., Yoshitake, Y., and Tsuyumu, S. (2003). Peh production, flagellum synthesis, and virulence reduced in Erwinia carotovora subsp. carotovora by mutation in a homologue of cytR. Mol. Plant Microbe Interact. 16, 389–397. doi: 10.1094/MPMI.2003.16.5.389
McDougald, D., Rice, S. A., Barraud, N., Steinberg, P. D., and Kjelleberg, S. (2012). Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nat. Rev. Microbiol. 10, 39–50. doi: 10.1038/nrmicro2695
Milanov, D. S., Prunic, B. Z., Velhner, M. J., Pajic, M. L., and Cabarkapa, I. S. (2015). Rdar morphotype- a resting stage of some Enterobacteriaceae. Food Feed Res. 42, 43–50. doi: 10.5937/FFR1501043M
Mole, B., Habibi, S., Dangl, J. L., and Grant, S. R. (2010). Gluconate metabolism is required for virulence of the soft-rot pathogen Pectobacterium carotovorum. Mol. Plant Microbe Interact. 23, 1335–1344. doi: 10.1094/MPMI-03-10-0067
Morgan, J. L., McNamara, J. T., and Zimmer, J. (2014). Mechanism of activation of bacterial cellulose synthase by cyclic di-GMP. Nat. Struct. Mol. Biol. 21, 489–496. doi: 10.1038/nsmb.2803
Oliver, M. M. H., Hewa, G. A., and Pezzaniti, D. (2014). Bio-fouling of subsurface type drip emitters applying reclaimed water under medium soil thermal variation. Agril. Water Manage. 133, 12–23. doi: 10.1016/j.agwat.2013.10.014
O’Toole, G., Kaplan, H. B., and Kolter, R. (2000). Biofilm formation as microbial development. Annu. Rev. Microbiol. 54, 49–79. doi: 10.1146/annurev.micro.54.1.49
Rantakari, A., Virtaharju, S., Taira, S., Palva, E. T., Saarilahti, H. T., and Romantschuk, M. (2007). Type III secretion contributes to the pathogenesis of the soft-rot pathogen Erwinia carotovora: partial characterization of the hrp gene cluster. Mol. Plant Microbe Interact. 14, 962–968. doi: 10.1094/MPMI.2001.14.8.962
Rinaudi, L., Fujishige, N. A., Hirsch, A. M., Banchio, E., Zorreguieta, A., and Giordano, W. (2006). Effects of nutritional and environmental conditions on Sinorhizobium meliloti biofilm formation. Res. Microbiol. 15, 867–875. doi: 10.1016/j.resmic.2006.06.002
Römling, U. (2005). Characterization of the rdar morphotype, a multicellular behavior in Enterobacteriaceae. Cell Mol. Life Sci. 62, 1234–1246. doi: 10.1007/s00018-005-4557-x
Römling, U., and Galperin, M. Y. (2015). Bacterial cellulose biosynthesis: diversity of operons, subunits, products and functions. Trends Microbiol. 23, 545–557. doi: 10.1016/j.tim.2015.05.005
Römling, U., Galperin, M. Y., and Gomelsky, M. (2013). Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 77, 1–52. doi: 10.1128/MMBR.00043-12
Römling, U., Rohde, M., Olsen, A., Normark, S., and Reinkoster, J. (2000). agfD, the checkpoint of multicellular and aggregative behaviour in Salmonella typhimurium regulates at least two independent pathways. Mol. Microbiol. 36, 10–23. doi: 10.1046/j.1365-2958.2000.01822.x
Rosan, B., and Lamont, R. J. (2000). Dental plague formation. Microbes Infect. 2, 1599–1607. doi: 10.1016/S1286-4579(00)01316-2
Ryan, R. P., Fouhy, Y., Lucey, J. F., Crossman, L. C., Spiro, S., He, Y. W., et al. (2006). Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc. Natl. Acad. Sci. U.S.A. 103, 6712–6717. doi: 10.1073/pnas.0600345103
Ryjenkov, D. A., Tarutina, M., Moskvin, O. V., and Gomelsky, M. (2005). Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J. Bacteriol. 187, 1792–1798. doi: 10.1128/JB.187.5.1792-1798.2005
Saxena, I. M., Kudlicka, K., Okuda, K., and Brown, R. M. Jr. (1994). Characterization of genes in the cellulose-synthesizing operon (acs operon) of Acetobacter xylinum: implications for cellulose crystallization. J. Bacteriol. 176, 5735–5752. doi: 10.1128/jb.176.18.5735-5752.1994
Scher, K., Römling, U., and Yaron, S. (2005). Effect of heat, acidification, and chlorination on Salmonella enterica serovar Typhimurium cells in a biofilm formed at the air-liquid interface. Appl. Environ. Microbiol. 71, 1163–1168. doi: 10.1128/AEM.71.3.1163-1168.2005
Solano, C., García, B., Valle, J., Berasain, C., Ghigo, J. M., Gamazo, C., et al. (2002). Genetic analysis of Salmonella enteritidis biofilm formation: critical role of cellulose. Mol. Microbiol. 43, 793–808. doi: 10.1046/j.1365-2958.2002.02802.x
Solomon, E. B., Brendan, A. N., Sapers, G. M., and Annous, B. A. (2005). Biofilm formation, cellulose production, and curli biosynthesis by Salmonella originating from produce, animal, and clinical sources. J. Food Protect. 68, 906–912. doi: 10.4315/0362-028X-68.5.906
Song, B., and Leff, L. G. (2006). Influence of magnesium ions on biofilm formation by Pseudomonas fluorescens. Microbiol. Res. 161, 355–361. doi: 10.1016/j.micres.2006.01.004
Staskawicz, B., Mudgett, M. B., Dangl, J. L., and Galan, J. E. (2001). Common and contrasting themes of plant and animal diseases. Science 292, 2285–2289. doi: 10.1126/science.1062013
Steenackers, H., Hermans, K., Vanderleyden, J., and Keersmaecker, D. (2012). An overview on Salmonella biofilms: an overview on occurrence, structure, regulation and eradication. Food Res. Int. 45, 502–531. doi: 10.1016/j.foodres.2011.01.038
Sutherland, I. W. (2001). Biofilm exopolysaccharides: a strong and stick frame-work. Microbiology 147, 3–9. doi: 10.1099/00221287-147-1-3
Teitzel, G. M., and Parsek, M. R. (2003). Heavy metal resistance of biofilm and planktonic Pseudomonas aeruginosa. Appl. Environ. Microbiol. 69, 2313–2320. doi: 10.1128/AEM.69.4.2313-2320.2003
Thomsen, L. E., Pedersen, M., Norregaard-Madsen, M., Valentin-Hansen, P., and Kallipolitis, B. H. (1999). Protein-ligand interaction: grafting of the uridine-specific determinants from the CytR regulator of Salmonella typhimurium to Escherichia coli cytR. J. Mol. Biol. 288, 165–175. doi: 10.1006/jmbi.1999.2668
Toth, I. K., Bell, M. C., Holeva, M. C., and Birch, P. R. J. (2003). Soft rot erwiniae: from genes to genomes. Mol. Plant Pathol. 4, 17–30. doi: 10.1046/j.1364-3703.2003.00149.x
Uhlich, G. A., Cooke, P. H., and Solomon, E. B. (2006). Analyses of the red-dry-rough phenotype of an Escherichia coli O157:H7 strain and its role in biofilm formation and resistance to antimicrobial agents. Appl. Environ. Microbiol. 72, 2564–2572. doi: 10.1128/AEM.72.4.2564-2572.2006
Valentin-Hansen, P., Sogaard-Andersen, L., and Pedersen, H. (1996). A flexible partnership: the CytR anti-activator and the cAMP-CRP activator protein, comrades in transcription control. Mol. Microbiol. 20, 461–466. doi: 10.1046/j.1365-2958.1996.5341056.x
van Houdt, R., and Michiels, C. W. (2005). Role of bacterial cell surface structures in Escherichia coli biofilm formation. Res. Microbiol. 156, 626–633. doi: 10.1016/j.resmic.2005.02.005
Watve, S. S., Thomas, J., and Hammer, B. K. (2015). CytR is a global positive regulator of competence, type VI secretion, and chitinases in Vibrio cholerae. PLoS ONE 10:e0138834. doi: 10.1371/journal.pone.0138834
White, A., Gibson, D. L., Collinson, S. K., Banser, P. A., and Kay, W. W. (2003). Extracellular polysaccharides associated with thin aggregative fimbriae of Salmonella enterica serovar Enteritidis. J. Bacteriol. 185, 5398–5407. doi: 10.1128/JB.185.18.5398-5407.2003
Yamamoto, K., Arai, H., Ishii, M., and Igarashi, Y. (2011). Trade-off between oxygen and iron acquisition in bacterial cells at the air-liquid interface. FEMS Microbiol. Ecol. 77, 83–94. doi: 10.1111/j.1574-6941.2011.01087.x
Yamamoto, K., Arai, H., Ishii, M., and Igarashi, Y. (2012). Involvement of flagella-driven motility and pili in Pseudomonas aeruginosa colonization at the air-liquid interface. Microbes Environ. 27, 320–323. doi: 10.1264/jsme2.ME11322
Yang, S., Peng, Q., Zhang, Q., Yi, X., Choi, C. J., Reedy, R. M., et al. (2008). Dynamic regulation of GacA in type III secretion, pectinase gene expression, pellicle formation, and pathogenicity of Dickeya dadantii (Erwinia chrysanthemi 3937). Mol. Plant Microbe Interact. 21, 133–142. doi: 10.1094/MPMI-21-1-0133
Yap, M.-N., Yang, C.-H., Barak, J. D., Jahn, C. E., and Charkowski, A. O. (2005). The Erwinia chrysanthemi type III secretion system is required for multicellular behavior. Mol. Microbiol. 187, 639–648. doi: 10.1128/jb.187.2.639-648.2005
Yi, X., Yamazaki, A., Biddle, E., Zeng, Q., and Yang, C.-H. (2010). Genetic analysis of two phosphodiesterases reveals cyclic diguanylate regulation of virulence factors in Dickeya dadantii. Mol. Microbiol. 77, 787–800. doi: 10.1111/j.1365-2958.2010.07246.x
Yildiz, F. H., and Schoolnik, G. K. (1999). Vibrio cholerae O1 EI Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc. Natl. Acad. Sci. U.S.A. 96, 4028–4033. doi: 10.1073/pnas.96.7.4028
Yip, E. S., Geszvain, K., DeLoney-Marino, C. R., and Visick, K. L. (2006). The symbiosis regulator rscS controls the syp gene locus, biofilm formation and symbiotic aggregation by Vibrio fischeri. Mol. Microbiol. 62, 1586–1600. doi: 10.1111/j.1365-2958.2006.05475.x
Keywords: CytR, air-liquid biofilm formation, attachment to plants, cellulose production, c-di-GMP signaling, T3SS, motility, Pectobacterium carotovorum subsp. carotovorum PC1
Citation: Haque MM, Oliver MMH, Nahar K, Alam MZ, Hirata H and Tsuyumu S (2017) CytR Homolog of Pectobacterium carotovorum subsp. carotovorum Controls Air-Liquid Biofilm Formation by Regulating Multiple Genes Involved in Cellulose Production, c-di-GMP Signaling, Motility, and Type III Secretion System in Response to Nutritional and Environmental Signals. Front. Microbiol. 8:972. doi: 10.3389/fmicb.2017.00972
Received: 08 March 2017; Accepted: 15 May 2017;
Published: 31 May 2017.
Edited by:
Erh-Min Lai, Academia Sinica, TaiwanReviewed by:
Xiaochen Yuan, University of Wisconsin-Milwaukee, United StatesRonan McCarthy, Animal and Plant Health Agency, United Kingdom
Copyright © 2017 Haque, Oliver, Nahar, Alam, Hirata and Tsuyumu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: M. M. Haque, haque_bes@bsmrau.edu.bd
 M. M. Haque
M. M. Haque M. M. H. Oliver
M. M. H. Oliver Kamrun Nahar3
Kamrun Nahar3