- 1Department of Laboratory Medicine, Civil Aviation General Hospital, Peking University Civil Aviation School of Clinical Medicine, Beijing, China
- 2State Key Laboratory for Infectious Disease Prevention and Control, Chinese Centre for Disease Control and Prevention, National Institute for Communicable Disease Control and Prevention, Beijing, China
- 3Collaborative Innovation Centre for Diagnosis and Treatment of Infectious Diseases, Hangzhou, China
- 4Laboratory of Clinical Microbiology and Infectious Diseases, Department of Pulmonary and Critical Care Medicine, China-Japan Friendship Hospital, Beijing, China
- 5Department of Laboratory Medicine, People's Hospital of Guangxi Zhuang Autonomous Region, Nanning, China
- 6Department of Clinical Laboratory, Affiliated Hospital of Inner Mongolia Medical University, Hohhot, China
- 7Department of Laboratory Medicine, Wuhan Pu Ai Hospital of Huazhong, University of Science and Technology, Wuhan, China
- 8Department of Laboratory Medicine, Henan Provincial People's Hospital, Zhengzhou, China
- 9Department of Laboratory Medicine, Tai'an City Central Hospital (Tai'an), Shandong, China
- 10Department of Laboratory Medicine, First Hospital, Peking University, Beijing, China
- 11Department of Laboratory Medicine, Chengdu First People's Hospital (Chengdu), Sichuan, China
Streptococcus pyogenes, or group A Streptococcus, is a pathogen responsible for a wide range of clinical manifestations, from mild skin and soft tissue infections and pharyngitis to severe diseases. Its epidemiological characteristics should be comprehensively under surveillance for regulating the national prevention and treatment practice. Herein, a total of 140 S. pyogenes, including 38 invasive and 102 noninvasive isolates, were collected from infected patients in 10 tertiary general hospitals from 7 cities/provinces in China during the years 2009–2016. All strains were characterized by classical and molecular techniques for its emm types/subtypes, virulent factors and antibiotic resistance profiling. Of 140 isolates, 15 distinct emm types and 31 subtypes were detected, dominated by emm12 (60 isolates, 42.9%), emm1(43, 30.7%), and emm89 (10, 7.1%), and 8 new emm variant subtypes were identified. All strains, invasive or not, harbored the superantigenic genes, speB and slo. The other virulence genes, smeZ, speF, and speC accounted for 96.4, 91.4, and 87.1% of collected isolates, respectively. Further multilocus sequence typing (MLST) placed all strains into 22 individual sequence types (STs), including 4 newly-identified STs (11, 7.9%). All isolates were phenotypically susceptible to penicillin, ampicillin, cefotaxime, and vancomycin, whereas 131(93.5%), 132(94.2%), and 121(86.4%) were resistant to erythromycin, clindamycin, and tetracycline, respectively. Our study highlights high genotypic diversity and high prevalence of macrolide resistance of S. pyogenes among clinical isolates circulating in China.
Introduction
Streptococcus pyogenes, or Lancefield group A streptococcus, is an essential cause of morbidity and mortality worldwide nowadays, leading to a wide range of human infections, ranging from mild to life-threatening invasive infections (Koutouzi et al., 2015; Plainvert et al., 2016).
A variety of virulence factors contribute to pathogenesis. The M protein, encoded by emm gene, is a surface protein and virulence factor of S. pyogenes, capable of adhering to the epithelium and conferring protection against macrophage killing (Sanderson-Smith et al., 2014). Furthermore, it harbors a highly variable region with the antigen, and the protective role of M-protein type-specific antibody has been confirmed (Baroux et al., 2014; Sanderson-Smith et al., 2014). Moreover, emm gene sequencing is a standard method for typing M protein, and the distribution of emm types demonstrated a high degree of variability in line with geographic location, time, and types of clinical infections (Steer Ac et al., 2009). Therefore, identification of emm type is extremely useful for understanding the local epidemiological characteristics. Moreover, M-type specific antibodies are only responsible for immunity against the homologous M-type, with no effect on infection by heterologous M-types (Sanderson-Smith et al., 2014); therefore, continuous surveillance of emm types is consequently needed to evaluate the possible benefits of an M protein-based S. pyogenes vaccine (Steer Ac et al., 2009). Recently, a new emm-cluster typing system that classifies the many S. pyogenes emm types into 48 discrete emm clusters containing closely related M proteins that share binding and structural properties has been proposed (Sanderson-Smith et al., 2014). This system might predict the M protein vaccine antigen content and serve as a framework to investigate the cross-protection phenomenon between different emm-types (Baroux et al., 2014; Sanderson-Smith et al., 2014).
Besides, streptococcal pyrogenic exotoxins (spe) also play a crucial role in the pathogenesis of S. pyogenes infections by acting as superantigens (speA-C and speF-M). Moreover, streptococcal superantigen A (ssa), streptococcal mitogenic exotoxin Z (smeZ), and other virulence proteins, including C5a peptidase (scpA), streptokinase (ska), streptolysin O (slo), streptolysin S (sagA), and extracellular phospholipase A (sla), also constitute the main epidemiological characteristics of the local S. pyogenes isolates.
Multilocus sequence typing (MLST) has been developed for identifying clones of S. pyogenes isolates as epidemic feature (Plainvert et al., 2016). Finally, previous documents have reported the increased resistance of S. pyogenes strains to erythromycin (Bingen et al., 2004), clindamycin (Bingen et al., 2004), and fluoroquinolone (Lin et al., 2015).
The distribution of the above-mentioned characteristics constitutes the main epidemiology of local S. pyogenes isolates. The changing epidemic characteristics should be under continuous surveillance for effective prevention and treatment (Bingen et al., 2004; Kim et al., 2013; Wajima et al., 2013; Seale et al., 2016). However, scarce data on S. pyogenes infections are available in mainland China, except for a few studies conducted locally with isolates mainly from children with scarlet fever (Liang et al., 2012; Yang et al., 2013). Therefore, the aim of our work is to elucidate the characterization of serotypes, potential virulence factors, evolutionary relationship, and antibiotic resistance of S. pyogenes strains circulating in China. Furthermore, comprehensive genetic relationships deduced from different molecular types will be illustrated.
Materials and Methods
Ethical Statement
This study was exempted from review by the ethics committee of Civil Aviation General Hospital (CAGH), Beijing, because it focused on the epidemical characteristics of S. pyogenes.
Strain Collection and Identification
A total of nonredundant 140 S. pyogenes isolates responsible for human infection were included in current study. The patients enrolled were aged from 2 months to 92 years, with the median age at 19.6 years; 47.1% were female. The isolates were obtained from bloodstream (16 cases), surgical-site, skin/soft tissue infection, and other wound secretions (wound, 18; skin/soft tissue infection, 7), abscess (16), pharyngitis, and/or tonsillitis (pharyngeal swab, 52), respiratory tract (bronchial alveolar lavage fluid, BALF, 3, and phlegm, 10), reproductive tract infection (abscess of vulva, 6), biopsy tissue (1), ear pus (5), pleural effusion (4), urine (1), and pyoderma gangraenosum (1) of patients who visited CAGH (Beijing) during 2009–2016, Peking University First Hospital (Beijing) during 2010–2014, Affiliated hospital of Inner Mongolia medical university (Huhehot, Inner Mongolia Autonomous region) during 2011–2014, Henan Provincial People's Hospital (Zhengzhou, Henan Province) during the period 2014–2015, Wuhan Pu Ai Hospital of Huazhong University of Science and Technology (Wuhan, Hubei Province) during 2013–2015, People's Hospital of Guangxi Zhuang Autonomous Region (Nanning, Guangxi Zhuang Autonomous Region) during 2009–2016, Beijing Tsinghua Chang Gung Hospital, Medical Center of Tsinghua University (Beijing) during 2014–2016, Beijing Tongren Hospital (Beijing) during 2012, Tai'an City Central Hospital (Tai'an, Shandong Province) during the period of 2014–2016, and Mianyan Central Hospital (Mianyang, Sichuan Province) during the period of 2015. The isolates were sent to the Department of Clinical microbiology of CAGH for further confirmation by β-hemolysis on sheep blood agar, grouping of carbohydrate antigen (Streptococcal grouping kit, Oxoid), and 16S rRNA gene sequencing using the primers BAK11w and BAK2, as described previously (Lu et al., 2013).
Case Definition
Definite invasive disease was defined by the isolation of S. pyogenes from a normally-sterile site (e.g., blood, cerebrospinal, pleural, or peritoneal fluid). Noninvasive infection was defined as isolation of the microorganism from a nonsterile site with a clinical syndrome consistent with S. pyogenes infection but that did not meet the probable invasive S. pyogenes disease case definition (Friaes et al., 2013; Baroux et al., 2014).
Emm Typing
All S. pyogenes isolates were subjected to emm typing. The emm gene was amplified by PCR (http://www.cdc.gov/streplab/protocol-emm-type.html), DNA fragments were sequenced and aligned by the comparison of the query sequence with the reference database (ftp://ftp.cdc.gov/pub/infectious_diseases/biotech/tsemm/, Centers for Disease Control and Prevention, Atlanta, GA, USA). The newly-identified sequences defining new emm subtypes were re-sequenced in order to validate the initial findings, then submitted, and assigned to a new subtype. The assignment of emm clusters was based on the CDC database (http://www.cdc.gov/streplab/downloads/distribution-emm-types.pdf) as previously described (Baroux et al., 2014).
MLST
MLST was performed by sequencing seven housekeeping genes (gki, gtr, murI, mutS, recP, xpt, and yqiL; Enright et al., 2001; Wu et al., 2008). After each gene was amplified by PCR, DNA fragments were sequenced. Then, the sequence types (STs) and allelic profiles were performed using MLST database (http://spyogenes.mlst.net/). Each isolate was assigned to an ST. An ST not identified in any cluster was assigned as a singleton. BioNumerics software version 5.1 (Applied Maths, Belgium) was used to create minimum spanning trees to illustrate the relationships between MLST and emm typing. The newly identified alleles and those alleles defining new STs were submitted to the MLST database curator for approval and a number was assigned. The isolates were assigned to one of the clonal complexes (CC) if they shared four or more alleles with the predominant ST.
Virulence Genes
Virulence genes were detected by using conventional PCR amplification method. The target genes included S. pyogenes exotoxin and Streptococcal superantigenic genes (speA–C, speF–M, ssa, and smeZ), and other specific virulence genes (sil, slo, and sla), using primer pairs described previously (Friães et al., 2013; Lu et al., 2016).
Antibiotic Susceptibility Testing and Antimicrobial Resistance Genes
The broth microdilution method was used to determine the susceptibility of all isolates to penicillin G (range of concentration tested: 0.03–4 μg/mL), ampicillin (0.03–4 μg/mL), cefotaxime (0.03–4 μg/mL), erythromycin (0.03–4 μg/mL), clindamycin (0.03–4 μg/mL), levofloxacin (0.25–32 μg/mL), tetracycline (0.255–32 μg/mL), and vancomycin (0.03–4 μg/mL). The cation-adjusted Mueller-Hinton broth with lysed horse blood (2.5–5% v/v) was provided by Tianjing Jinzhang Science and Technology Development, China. The results were interpreted in accordance with the breakpoints set for Streptococcus spp. β-hemolytic group by the Clinical and Laboratory Standards Institute (CLSI, 2016). The macrolide-resistant isolates were further classified as having the cMLSB (constitutive macrolide-lincosamide-streptogramin B resistance), iMLSB (inducible resistance), or M phenotype (macrolide-streptogramin B resistance and lincosamide susceptibility) by the double-disc synergy test.
Genetic determinants of erm (A) (subclass ermTR), erm (B), and mef (A/E) were investigated by PCR in erythromycin-resistant isolates as we described previously (Lu et al., 2016). Furthermore, the resistance genes of tetracycline, including tet (M), tet (K), tet (L), and tet (O), were determined in tetracycline-resistant isolates using PCR as described elsewhere (Lu et al., 2016). PCR products were selected for sequencing and aligned by BLAST software to confirm the correctness. The genes coding DNA gyrase A (gyrA) and topoisomerase IV C (parC) were amplified, sequenced and aligned in all isolates, to determine fluoroquinolone resistance and the sequence substitutions by using the primers previously proposed (Lu et al., 2016). Comparison analyses of sequences were conducted with BioEdit software (Ibis Biosciences, Carlsbad, CA, USA). Clustal-W was used to perform multiple alignments of the nucleotide and predicted amino acid sequences. The reference sequences of gyrA (gyrA-ATCC 700294, AF220945.1) and parC (parC-ATCC 700294, AF220946.1) were accessed from GenBank.
Statistical Method
Chi square (χ2)-test was used to analyze different infection types by year and emm type using SPSS 17.0 software, and significance was defined when the P-value was < 0.05.
Results
Distribution of Sources of S. pyogenes Isolates and emm Distribution
A total of 140 S. pyogenes isolates collected from 10 tertiary hospitals in China were enrolled in the study, including 38(27.1%) of invasive and 102(72.9%) of noninvasive. The isolates were recovered from 66 women and 74 men (ratio 1:1), and 63 (45%) from children (≤14 years). Compared by year, the infection caused by invasive and noninvasive infection was different in different years (χ2 = 19.150, p < 0.008). Details of S. pyogenes isolates recovered are shown in Table 1.
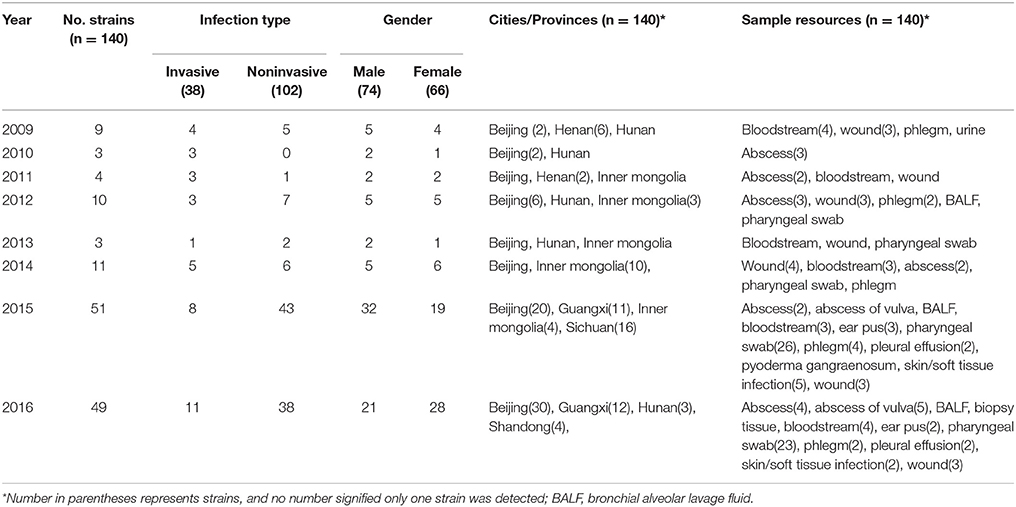
Table 1. Demographic characteristics of patients infected with S. pyogenes during 2009–2016 in China.
The emm type distribution for 140 S. pyogenes strains is demonstrated in Table 2. A total of 15 distinct emm types and 31 subtypes were identified. Overall, the prevalent emm types were emm12 (60, 42.9%), emm1 (43, 30.7%), emm89(10, 7.1%), and emm75 (6, 4.3%), accounting for 85.0% of the total population, as shown in Figure 1, Table 2. However, there was no difference between invasive and noninvasive infections caused by different emm types (χ2 = 23.566, p < 0.073). The emm types in current study belonged to 8 emm clusters. Among them, there were 3 most prevalent clusters, A-C3, A-C4, and E4, accounting for 43 (30.7%), 61 (43.6%), and 15(10.7%), respectively, comprising 85.0% of all isolates. For the 16 isolates causing bloodstream infection, the predominated emm types were emm12 (4 isolates) and 1 (4). In addition, 8 new emm types/subtypes were newly-identified in current study (emm1.85, emm12.40, emm12.93, emm12.94, emm18.43, emm46.3, emm5.141, and emm141.1), those emm sequences were submitted to GenBank under accession number KY697798-KY697805.
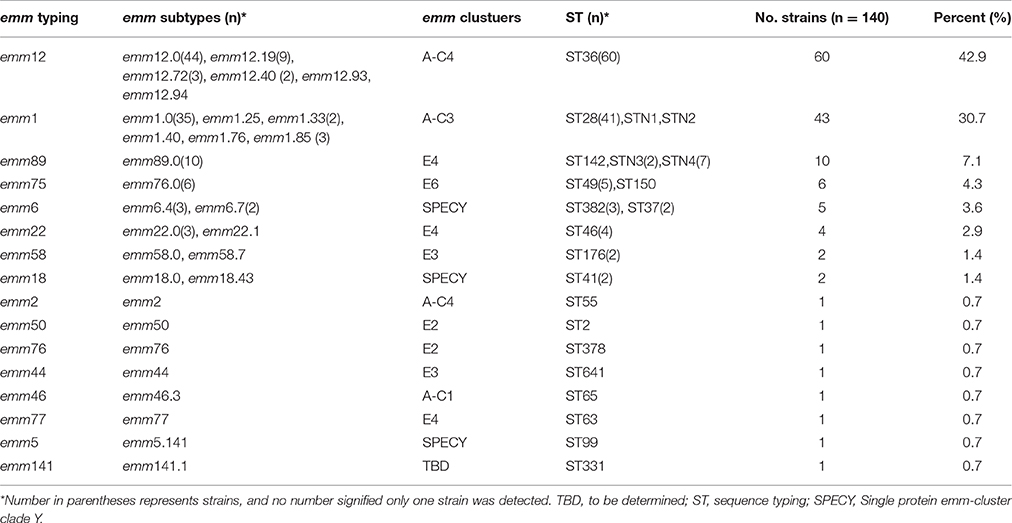
Table 2. Distribution of emm types, emm cluster, and sequence types of S. pyogenes isolated during 2009–2016 in China.
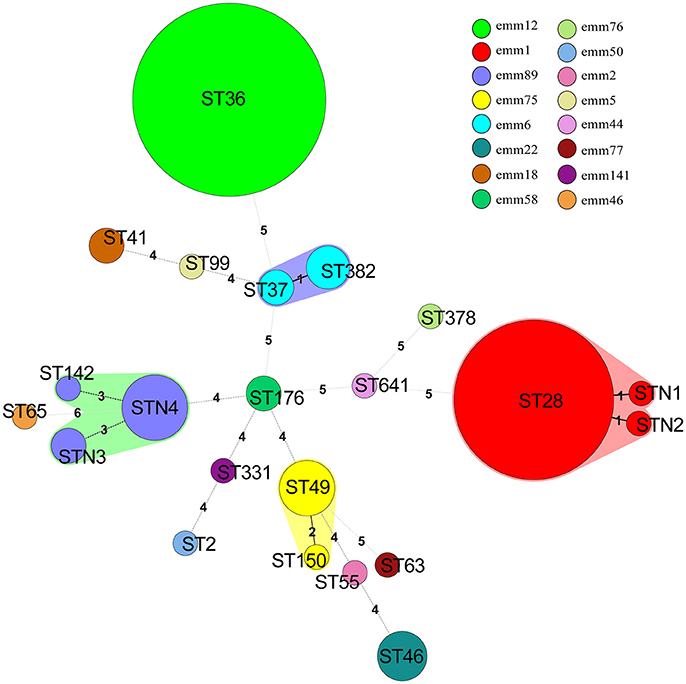
Figure 1. Minimum spanning tree of 140 S. pyogenes isolates in China based on Multilocus sequence typing (MLST). The size of each circle indicates the number of isolates within this particular type. The colors of the halo surrounding the STs denote types that belong to the same clonal complex. Emm types are represented by different colors. The number between different circles represented the number of locus variants.
Distribution of Virulence-Associated Genes
A total of 16 virulence genes were identified by PCR methods. All strains, invasive or not, harbored the 2 superantigenic toxic genes: speB and slo. Of other virulence genes, smeZ, speF and speC were prevalent, accounting for 96.4% (135/140), 91.4% (128/140), and 87.1% (122/140), of collected isolates, respectively. By comparison, the percentage of isolates that harbored sil (1.4%, 2/140), sla (5.7%, 8/140), speL (6.4%, 9/140), and speK (7.9%, 11/140) was extremely low. As demonstrated in Figure 2, the close relationship was notable between emm type and virulence genes, for example, only 2 (1.4%) emm12 S. pyogenes isolates contained speJ, while most emm1(42, 97.7%) harbored it; Only 1 (0.7%) and 5 (3.6%) emm1 S. pyogenes isolates contained speH and speI, but most emm12 harbored them, accounting for 45(75.0%) and 46 (76.7%) isolates, respectively.
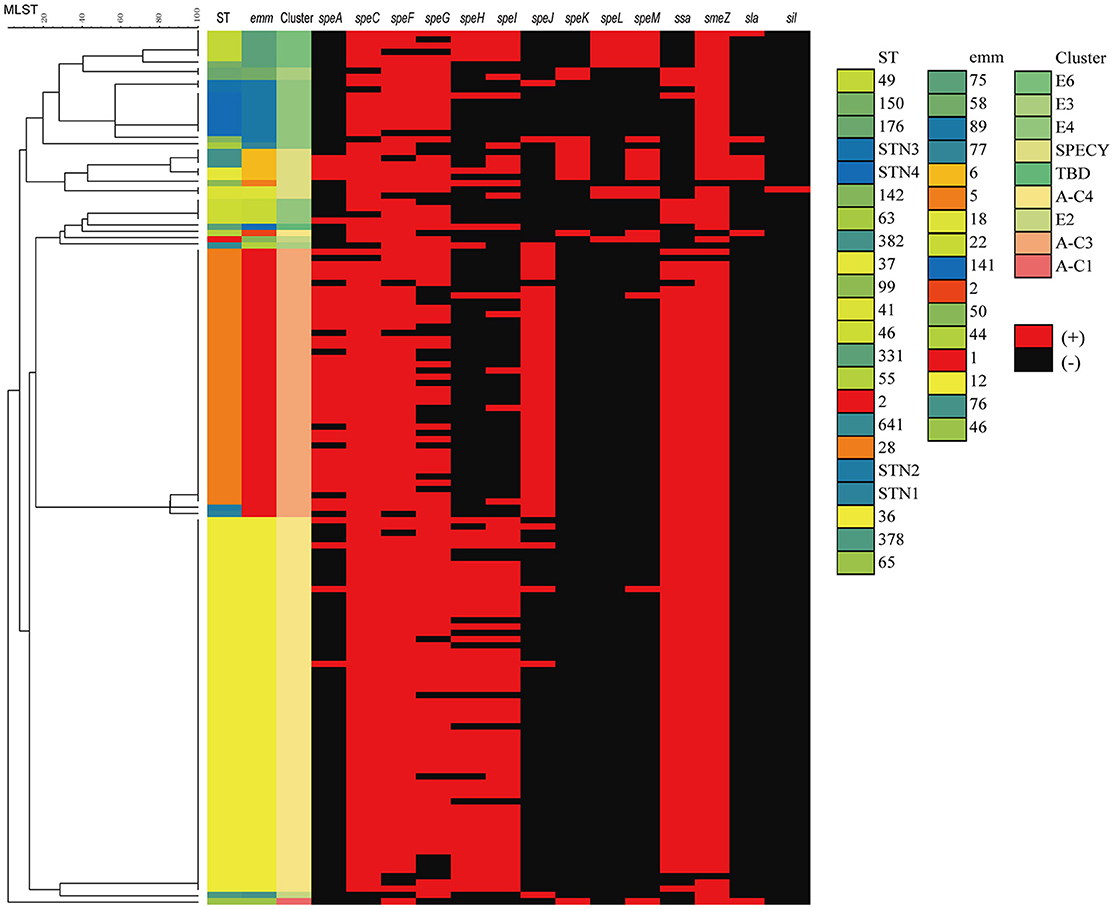
Figure 2. Dendrogram constructed from the multilocus sequence typing (MLST) profiles of seven housekeeping genes of 140 S. pyogenes isolates circulating in 10 hospital settings in China, showing the distributions of virulence genes, emm type and MLST. SPECY, Single protein emm-cluster clade Y. TBD, to be determined.
MLST and Relationship with emm Typing
All 140 isolates were selected for MLST analysis, and a total of 22 individual STs were distinguished (Table 2, Figure 1), including 4 new STs (n = 11, 7.9%) designated as STN1, STN2, STN3, and STN4, respectively. In the alignment of MLST sequence, 3 novel sequences were found, xpt in isolate KT500, recP in KT523, and gtr in both KT205 and KT525, which were designated as xpt1, recP1, and gtr1, respectively. Moreover, 92.1% (129/140) isolates were represented by 10 main STs (having ≧2 isolates), and the two most prevalent STs were ST36 and ST28, accounting for 42.9% (60/140) and 29.3% (41/140), respectively. Besides, MLST analysis indicated that each emm type was almost corresponding to a single ST, or at least, a clonal complex, e.g., emm1 and ST28, emm12, and ST36, as shown in Table 2, Figure 1.
Phenotype and Genetic Profile of Antimicrobial Susceptibility
All isolates collected in current study were phenotypically susceptible to penicillin, ampicillin, cefotaxime, and vancomycin.
Of 140 S. pyogenes isolates, 131 were resistant, 2 intermediate, and 7 susceptible, to macrolide; 132 S. pyogenes isolates were resistant, 1 intermediate, and 7 susceptible, to clindamycin. No M phenotype was detected. Of all erythromycin-resistant cMLSB isolates, cMLSB/erm (B) phenotype/genotype was the most frequently identified (116, 82.9%), while the combination of erm (B)+mef (A/E) and erm (B)+erm (A) were detected only in 6 and 4 isolates. The 5 isolates of iMLSB phenotype harbored exclusively erm (B) gene. Interestingly, all the S. pyogenes isolates in current study, phenotypically susceptible or resistant to macrolide, harbored erm (B) gene.
Of 140 S. pyogenes isolates in current study, 17 were susceptible, 2 intermediate, and 121 resistant to tetracycline. In 121 resistant isolates, no S. pyogenes isolate carried tet (K) gene, only one isolate harbored tet (L) combined with tet (M), and only 9 tetracycline-resistant isolates carried tet (O), singly or combined; comparatively, most isolates (116/121, 95.9%) carried tet (M). Notably, 1 out of the 2 intermediate isolates, and 7 out of 17 susceptible were also positive for tet (M), as shown in Table 3.
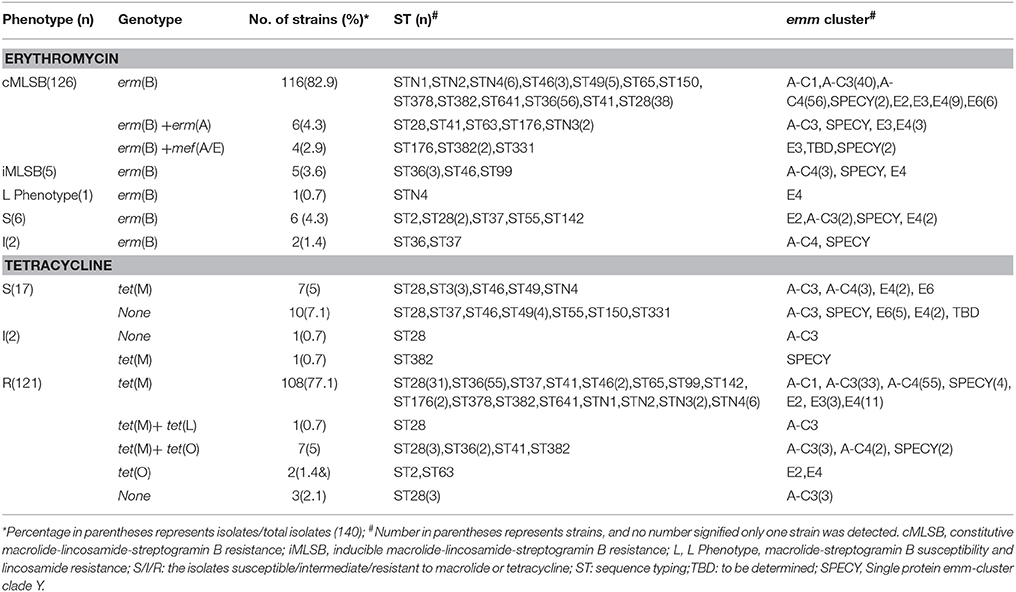
Table 3. Association of erythromycin and tetracycline phenotype and genotype and the distribution of emm and sequence typing; in 140 Streptococcus pyogenes isolated during 2009–2016 in China.
Mutation of gyrA and parC Leading to Resistance to Fluoroquinolone
Fluoroquinolone resistance-related genes, gyrA, and parC, were amplified and sequenced. Regarding to S. pyogenes, the gene gyrA was highly conserved, exhibiting limited variation, and nonsynonymous mutations were only observed in 3 isolates, M99L for isolate KT528, S81F for KT191, and V192I for PCR75 with an MIC at 0.75, >256, and >256 μg/mL, respectively. By comparison, multiple amino acid point mutations were identified in the parC sequences (61.4%, 86/140), both in susceptible and resistant isolates. Among 140 S. pyogenes isolates, 5 were non-susceptible isolates, with details in Table 4. They were obviously sporadically-distributed in various emm-types and infection sites. Moreover, in 135 fluoroquinolone-susceptible S. pyogenes isolates in current study, A121V (59, 43.7%) and D91N (16,11.4%) were the most prevalent mutations. Meanwhile, the close relationship between parC mutation and emm types was notable, for example, all emm12 strains, resistant or not, contained A121V, however, almost no fluoroquinolone-susceptible emm1 strains harbored mutation (except 1 isolate having a mutation of S79-F), emm6 harbored S79A mutation, and emm89 harbored both D91N and S140P mutations, as shown in Table 5.
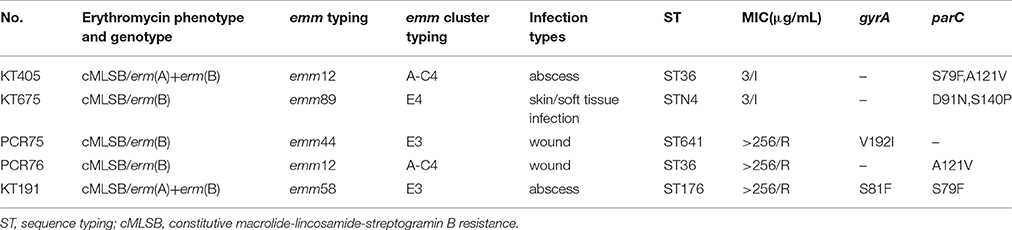
Table 4. Phenotype, genotype, distribution of emm, and other characteristics of 5 fluoroquinolone non-susceptible Streptococcus pyogenes isolates in China.
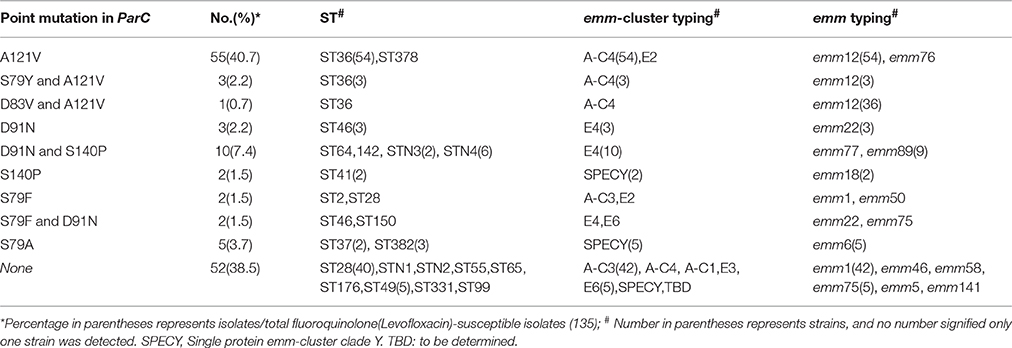
Table 5. Characteristics of amino acid substitutions of ParC in 135 fluoroquinolone(Levofloxacin)-susceptible Streptococcus pyogenes isolates in China.
Discussion
S. pyogenes is an important pathogen involved in a wide variety of human infections and a major cause of morbidity and mortality worldwide, ranging from noninvasive diseases, such as acute pharyngitis, to life-threatening invasive infections, such as sepsis and toxic shock syndrome (Gherardi et al., 2015). Herein, we characterized a collection of 140 S. pyogenes isolates in China during 2009–2016 and compared them with S. pyogenes isolates from around the world.
The distribution of emm genes of S. pyogenes strains should be under continuous surveillance, for its variety in different regions and changing with time make wide implementation of universal M protein vaccine difficult. In present study, 95.7% (134/140) isolates were covered by the 26-valent M-protein-based S. pyogenes vaccine currently demonstrating to be immunogenic and safe in human trials (Steer Ac et al., 2009; Tamayo et al., 2014). Based on above epidemiological features, this vaccine will have extensive coverage of S. pyogenes isolates in China. According to a review regarding to S. pyogenes infection worldwide, the most common emm types were emm1 and emm 12, accounting for 18.3 and 11.1%, followed by emm28 (8.5%), emm3 (6.9%), and emm4 (6.9%) (Steer Ac et al., 2009). Similarly, in current study, the leading emm types were emm12 (42.1%), emm1 (30.7%), emm89 (7.1%), and emm75 (4.3%), accounting for 84.3% of the total population. Our data during a 8-year period is different from that of Greece during 2007–2013 in children [(35 emm types in 1282 strains, with the most prevalent emm types being emm1 (16.7%), emm12 (13.6%), emm77 (10.9%), emm6 (6.8%), and emm89 (6.6%)](Koutouzi et al., 2015), New Caledonia in 2012 [47 emm types among 318 S. pyogenes strains, the 5 most frequent emm types (emm76, emm95, emm25, emm1, and emm93) were responsible for only 51% of the cases] (Baroux et al., 2014), Finland during 2008–2013 [72 emm types in 1122 invasive isolates, of which emm28 (26%), emm89 (12%), and emm1 (12%) were 3 most types)] (Smit et al., 2015), and Portugal during 2006–2009 (the 4 most prevalent emm types, emm1, emm89, emm3, and 6, accounted for 60% of 191 isolates recovered from normally sterile sites; Friães et al., 2013). Furthermore, the distribution of emm types in current study was different from that in the previous study in children with scarlet fever in Beijing, China (647 S. pyogenes isolates, emm12, and 1 accounting for 76.4 and 17.1% respectively; Yang et al., 2013). Changes in the predominant circulating emm types within relatively short time periods in individual regions might be explained by increased populations moving. By comparison, all S. pyogenes isolates in present study were collected from 10 hospitals across China, half patients were adults with clinical confirmed infections, and the majority of them had underlying diseases. Therefore, our results might be more representative of characteristics of S. pyogenes from mainland China. Moreover, the emm types of invasive and noninvasive strains were detected to be insignificantly related, suggesting that no specific types of M proteins were responsible for the invasion of S. pyogenes (Yang et al., 2013).
MLST allowed the comparison of genetic profiles of isolates recovered from various geographic regions. In this study, new gene sequences were identified in xpt, recP, and gtr and 4 novel STs designated. Moreover, isolates within 15 S. pyogenes emm types shared identical or nearly identical STs, demonstrating concordance between the emm type and genetic relatedness. Each emm type was almost exclusively associated with ST, e.g., emm1 with ST28, and emm6 with ST382 (Lin et al., 2015; Plainvert et al., 2016). The horizontal transfer of the emm genes in S. pyogenes seems to be less often.
Virulence factors have a critical role in the pathogencity of S. pyogenes. In current study, all strains, invasive or not, harbored speB and slo. The other virulence genes, smeZ, speC, and speF, were determined in over 90% isolates. By comparison, the percentage of isolates that harbored sil, sla, speL, and speK was <10%, indicating their lose from most S. pyogenes lineages in China. The distribution of superantigens might be varied in accordance with geographic areas. For example, in France, the genes of speA, speC, ssa, and smeZ were detected in 59, 37, 13, and 92% of S. pyogenes in adult with meningitis from 2003 to 2013, respectively (Plainvert et al., 2016). In Australia, speG and smeZ were reported to be present in the majority (90 and 95%), and speC in half, of 107 S. pyogenes isolates, and comparable with our data, the frequency of speL (8%) and speK (9%) were also rare (Commons et al., 2008). However, in Portugal, the speG and smeZ were present in over 90% S. pyogenes isolates, while speJ was found in only 45% of 191 S. pyogenes isolates recovered from normally sterile sites during 2006–2009 (Friães et al., 2013). Furthermore, the distribution of superantigen in current study was different from that found in S. pyogenes isolates from a previous study concerning children with scarlet fever in Beijing, China (Yang et al., 2013), in which almost all emm1 strains had speC, and ssa was the main superantigen of emm1 and emm12 isolates. This might be explained by our isolates having a wide coverage nationwide. Nevertheless, the above study demonstrated an extremely low number of S. pyogenes isolates harbored speK, speL, or speM, consistent to our data (Yang et al., 2013). This might comprise the main characteristics of S. pyogenes isolates in China. In conclusion, the distribution of superantigens varied dramatically, and the isolates with the identical emm type often shared a diverse profile.
Several antimicrobial drugs effectively treat S. pyogenes infections. Herein, all S. pyogenes isolates were susceptible to β-lactams and vancomycin, and the reduced susceptibility to penicillin and cephalosporin was not observed, suggesting that β-lactams were still the first-line antibiotics against S. pyogenes.
Moreover, macrolides have been recommended for patients allergic to β-lactams, and clindamycin is the preferred antibiotic in the treatment of patients with serious soft-tissue infections because of its ability to inhibit the production of several streptococcal virulence factors (Gherardi et al., 2015). Thus, it is critical to conduct continuous surveillance for macrolide- and clindamycin-resistance in S. pyogenes. In present study, approximately 90% S. pyogenes strains were resistant to erythromycin, clindamycin, and tetracycline, and similar resistance rates were also documented in two previous studies conducted in children with scarlet fever in Beijing, China (Liang et al., 2012; Yang et al., 2013). Previously, we have reported a high resistance rate in other two main β-hemolytic Streptococcus, S. agalactiae, and S. dysgalactiae subsp. equisimilis (SDSE), in China (Binghuai Lu et al., 2016; Lu et al., 2016). Put together, erythromycin and clindamycin might be unsuitable in treatment of infections due to β-hemolytic streptococcus in mainland China. However, in contrast, the extent of the resistance of S. pyogenes to the two antibiotics in many other countries has been very low. For example, in Finland, the S. pyogenes isolates collected from blood and skin/soft tissue showed an erythromycin and clindamycin resistance of 1.9 and 0.9% in 2008 and 8.7 and 9.2% in 2013, respectively, though a little increase (Smit et al., 2015). In addition, in current study, both erm and mef (A/E) genes were identified in erythromycin-resistant isolates, with erm (B) dominating. We observed a higher level of discordance between genotype and phenotype in S. pyogenes, and there were erm (B) genes in 9 erythromycin-susceptible isolates, as consistent with previous report in S. agalactiae (Dela Cruz et al., 2007) and SDSE isolates (Lu et al., 2016), hinting an alternate mechanism of resistance or a potential mutation in the 23S Rrna (Dela Cruz et al., 2007). Furthermore, 90.3% (421/466) harbored the erm (B) gene in S. pyogenes strains isolated from Beijing children (Liang et al., 2012). Put together, this might indicate that erm (B) be the predominant resistance genes in β-hemolytic Streptococci, in China (Binghuai Lu et al., 2016; Lu et al., 2016). S. pyogenes clones showing emm types strongly associated with erythromycin resistance may contribute to the overall prevalence of macrolide resistance. In Italy, the most prevalent macrolide resistance mediated by mef (A) was present in 92.2% of emm1 strains (Gherardi et al., 2015). Similarly, in Japan, of 75 S. pyogenes strains typed to emm1, 21.3% contained mef (A) gene (Wajima et al., 2008). However, this is not shown in current research, in which only 6(4.3%) S. pyogenes isolates harboring erm (A), with one strain containing emm1, explained probably by geographic variation. In addition, tet (M) was dominant resistance gene of tetracycline, comparable with our previous report of SDSE and GBS, demonstrating that tet (M) constituted the resistance mechanism of β-hemolytic Streptococci in China (Binghuai Lu et al., 2016; Lu et al., 2016).
The lower levofloxacin-resistant rate was reported in current study, making fluoroquinolone an alternative to β-lactams antibiotics in infections due to S. pyogenes. The development of fluoroquinolone resistance in S. pyogenes is mainly mediated by point mutations in the genes encoding DNA gyrase and topoisomerase IV subunit. High-level fluoroquinolone-resistant S. pyogenes strains are infrequently isolated, and there have been only limited reports from the United States (Biedenbach et al., 2006), Germany (Reinert et al., 2004), Taiwan (Lin et al., 2015), Portugal (Friães et al., 2013), and Japan (Wajima et al., 2013), distributed in emm11, emm12, emm28, emm58, and emm89. Comparatively, the nonsusceptible rate to fluoroquinolones (3.6%) in mainland China was far lower than that in Taiwan (11.1%), where the alterations of S79F and A121V in parC in emm12/ST36 strains were the most frequently mutations (Lin et al., 2015). In current study, there are 3 high-level resistant isolates (MIC > 256 μg/mL) and 2 immediate isolates (MIC = 3 mg/L), comparable to previous studies in China with 3.4 and 0% of them being fluoroquinolone-resistant (Liang et al., 2012; Yang et al., 2013). As previously documented, the mutations usually occur first in parC, conferring low-level fluoroquinolone resistance, then followed by mutations in gyrA that result in high-level fluoroquinolone resistance (Friães et al., 2013; Lin et al., 2015). Nevertheless, in current study, concurrent mutations in parC and gyrA did not exist always in high-level fluoroquinolone-resistant isolates. In 3 high-level resistant isolates distributed in various emm types, the resistance was caused by mutations in parC (A121V), gyrA (V192I), and both gyrA (S81F) and parC (S79F), respectively. Meanwhile, the significant relationship between parC mutation and emm/ST types in China was observed. All emm12 strains, resistant to levofloxacin or not, contained A121V.
Furthermore, our study is limited by a few factors. The sample size is relatively small, recovered only from 10 tertiary general hospitals of 7 cities/provinces in China. Moreover, the temporal and spatial distribution of the S. pyogenes isolates under study was unbalanced. Therefore, continuous studies with larger sample size are still in need to achieve comprehensive epidemic characteristics of S. pyogenes in China.
In summary, the S. pyogenes strains in present study showed some regional characteristics. The high resistance rate to erythromycin and clindamycin, a relatively high frequency of emm 12 and 1, close relationship between the point mutation of A121V in parC and emm12 strains, and high prevalence of erm (B) constitute significant characteristics of our S. pyogenes strains. The current study will contribute to the understanding of molecular characteristics of S. pyogenes circulating in mainland China and the decision making in the context of the M-protein-based vaccine developments.
Author Contributions
YaF, XC, JW, JZ, YL, DL, GZ, LH, HL, FZ, and YC isolated bacteria and performed the laboratory measurements. BL and DW made substantial contributions to conception and design. BL, YuF and DW wrote and revised the manuscript. BL drafted the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was partially supported by Beijing Municipal Science & Technology Commission, PR China (No. Z141107002514036) and Civil Aviation General Hospital Research Funds (Grant no. 2014001).
References
Baroux, N., D'ortenzio, E., Amedeo, N., Baker, C., Ali Alsuwayyid, B., Dupont-Rouzeyrol, M., et al. (2014). The emm-cluster typing system for Group A Streptococcus identifies epidemiologic similarities across the Pacific region. Clin. Infect. Dis. 59, e84–e92. doi: 10.1093/cid/ciu490
Biedenbach, D. J., Toleman, M. A., Walsh, T. R., and Jones, R. N. (2006). Characterization of fluoroquinolone-resistant beta-hemolytic Streptococcus spp. isolated in North America and Europe including the first report of fluoroquinolone-resistant Streptococcus dysgalactiae subspecies equisimilis: report from the SENTRY Antimicrobial Surveillance Program (1997-2004). Diagn. Microbiol. Infect. Dis. 55, 119–127. doi: 10.1016/j.diagmicrobio.2005.12.006
Bingen, E., Bidet, P., Mihaila-Amrouche, L., Doit, C., Forcet, S., Brahimi, N., et al. (2004). Emergence of macrolide-resistant Streptococcus pyogenes strains in French children. Antimicrob. Agents Chemother. 48, 3559–3562. doi: 10.1128/AAC.48.9.3559-3562.2004
Binghuai Lu, X., Junrui, C., Duochunwang, W., Zeng, J., Dong, Y. L., Fengxia, L. I., et al. (2016). Molecular characteristics and antimicrobial resistance in invasive and noninvasive Group B Streptococcus between 2008 and 2015 in China. Diagn. Microbiol. Infect. Dis. 86, 351–357. doi: 10.1016/j.diagmicrobio.2016.08.023
Commons, R., Rogers, S., Gooding, T., Danchin, M., Carapetis, J., Robins-Browne, R., et al. (2008). Superantigen genes in group A streptococcal isolates and their relationship with emm types. J. Med. Microbiol. 57, 1238–1246. doi: 10.1099/jmm.0.2008/001156-0
CLSI (2016). Performance Standards for Antimicrobial Susceptibility Testing, 26th Informational Supplement (M100-S26). Wayne, PA: Clinical and Laboratory Standards Institute.
Dela Cruz, W. P., Richardson, J. Y., Broestler, J. M., Thornton, J. A., and Danaher, P. J. (2007). Rapid determination of macrolide and lincosamide resistance in group b streptococcus isolated from vaginal-rectal swabs. Infect. Dis. Obstet. Gynecol. 2007, 1–6. doi: 10.1155/2007/46581.
Enright, M. C., Spratt, B. G., Kalia, A., Cross, J. H., and Bessen, D. E. (2001). Multilocus sequence typing of Streptococcus pyogenes and the Relationships between emm Type and Clone. Infect. Immun. 69, 2416–2427. doi: 10.1128/iai.69.4.2416-2427.2001
Friaes, A., Lopes, J. P., Melo-Cristino, J., and Ramirez, M. (2013). Changes in Streptococcus pyogenes causing invasive disease in Portugal: evidence for superantigen gene loss and acquisition. Int. J. Med. Microbiol. 303, 505–513. doi: 10.1016/j.ijmm.2013.07.004
Friães, A., Lopes, J. P., Melo-Cristino, J., and Ramirez, M. (2013). Changes in Streptococcus pyogenes causing invasive disease in Portugal: evidence for superantigen gene loss and acquisition. Int. J. Med. Microbiol. 303, 505–513. doi: 10.1016/j.ijmm.2013.07.004
Gherardi, G., Petrelli, D., Di Luca, M. C., Pimentel De Araujo, F., Bernaschi, P., Repetto, A., et al. (2015). Decline in macrolide resistance rates among Streptococcus pyogenes causing pharyngitis in children isolated in Italy. Eur. J. Clin. Microbiol. Infect. Dis. 34, 1797–1802. doi: 10.1007/s10096-015-2414-x
Kim, S. H., Yoon, Y. K., Kim, M. J., and Sohn, J. W. (2013). Clinical impact of time to positivity for Candida species on mortality in patients with candidaemia. J. Antimicrob. Chemother. 68, 2890–2897. doi: 10.1093/jac/dkt256
Koutouzi, F., Tsakris, A., Chatzichristou, P., Koutouzis, E., Daikos, G. L., Kirikou, E., et al. (2015). Streptococcus pyogenes emm types and clusters during a 7-year period (2007 to 2013) in pharyngeal and nonpharyngeal pediatric isolates. J. Clin. Microbiol. 53, 2015–2021. doi: 10.1128/jcm.00301-15
Liang, Y., Liu, X., Chang, H., Ji, L., Huang, G., Fu, Z., et al. (2012). Epidemiological and molecular characteristics of clinical isolates of Streptococcus pyogenes collected between 2005 and 2008 from Chinese children. J. Med. Microbiol. 61, 975–983. doi: 10.1099/jmm.0.042309-0
Lin, J.-N., Chang, L.-L., Lai, C.-H., Huang, Y.-H., Chen, W.-F., Yang, C.-H., et al. (2015). High prevalence of fluoroquinolone-nonsusceptible Streptococcus pyogenes emm12 in Taiwan. Diagn. Microbiol. Infect. Dis. 83, 187–192. doi: 10.1016/j.diagmicrobio.2015.06.018
Lu, B., Fang, Y., Huang, L., Diao, B., Du, X., Kan, B., et al. (2016). Molecular characterization and antibiotic resistance of clinical Streptococcus dysgalactiae subsp. equisimilis in Beijing, China. Infect. Genet. Evol. 40, 119–125. doi: 10.1016/j.meegid.2016.01.030
Lu, B., Shi, Y., Zhu, F., and Xu, X. (2013). Pleuritis due to Brevundimonas diminuta in a previously healthy man. J. Med. Microbiol. 62, 479–482. doi: 10.1099/jmm.0.045013-0
Plainvert, C., Doloy, A., Joubrel, C., Maataoui, N., Dmytruk, N., Touak, G., et al. (2016). Characterization of Streptococcus pyogenes isolates responsible for adult meningitis in France from 2003 to 2013. Diagn. Microbiol. Infect. Dis. 84, 350–352. doi: 10.1016/j.diagmicrobio.2015.12.006
Reinert, R. R., Lütticken, R., and Al-Lahham, A. (2004). High-level fluoroquinolone resistance in a clinical Streptoccoccus pyogenes isolate in Germany. Clin. Microbiol. Infect. 10, 659–662. doi: 10.1111/j.1469-0691.2004.00890.x
Sanderson-Smith, M., De Oliveira, D. M. P., Guglielmini, J., McMillan, D. J., Vu, T., Holien, J. K., et al. (2014). A systematic and functional classification of Streptococcus pyogenes that serves as a new tool for molecular typing and vaccine development. J. Infect. Dis. 210, 1325–1338. doi: 10.1093/infdis/jiu260
Seale, A. C., Davies, M. R., Anampiu, K., Morpeth, S. C., Nyongesa, S., Mwarumba, S., et al. (2016). Invasive group A streptococcus infection among children, rural Kenya. Emerg. Infect. Dis. 22, 224–232. doi: 10.3201/eid2202.151358
Smit, P. W., Lindholm, L., Lyytikainen, O., Jalava, J., Patari-Sampo, A., and Vuopio, J. (2015). Epidemiology and emm types of invasive group A streptococcal infections in Finland, 2008-2013. Eur. J. Clin. Microbiol. Infect. Dis. 34, 2131–2136. doi: 10.1007/s10096-015-2462-2
Steer Ac, L. I., Matatolu, L., Beall, B. W., and Carapetis, J. R. (2009). Global emm type distribution of group A streptococci: systematic review and implications for vaccine development. Lancet Infect. Dis. 9, 611–616. doi: 10.1016/S1473-3099(09)70178-1
Tamayo, E., Montes, M., Garcia-Arenzana, J. M., and Perez-Trallero, E. (2014). Streptococcus pyogenes emm-types in northern Spain; population dynamics over a 7-year period. J. Infect. 68, 50–57. doi: 10.1016/j.jinf.2013.08.013
Wajima, T., Morozumi, M., Chiba, N., Shouji, M., Iwata, S., Sakata, H., et al. (2013). Associations of macrolide and fluoroquinolone resistance with molecular typing in Streptococcus pyogenes from invasive infections, 2010-2012. Int. J. Antimicrob. Agents 42, 447–449. doi: 10.1016/j.ijantimicag.2013.06.022
Wajima, T., Murayama, S. Y., Sunaoshi, K., Nakayama, E., Sunakawa, K., and Ubukata, K. (2008). Distribution of emm type and antibiotic susceptibility of group A streptococci causing invasive and noninvasive disease. J. Med. Microbiol. 57, 1383–1388. doi: 10.1099/jmm.0.2008/002642-0
Wu, H. M., Janapatla, R. P., Ho, Y. R., Hung, K. H., Wu, C. W., Yan, J. J., et al. (2008). Emergence of fluoroquinolone resistance in group B streptococcal isolates in Taiwan. Antimicrob. Agents Chemother. 52, 1888–1890. doi: 10.1128/AAC.00035-08
Keywords: molecular characterization, antibiotic resistance, Streptococcus pyogenes
Citation: Lu B, Fang Y, Fan Y, Chen X, Wang J, Zeng J, Li Y, Zhang Z, Huang L, Li H, Li D, Zhu F, Cui Y and Wang D (2017) High Prevalence of Macrolide-resistance and Molecular Characterization of Streptococcus pyogenes Isolates Circulating in China from 2009 to 2016. Front. Microbiol. 8:1052. doi: 10.3389/fmicb.2017.01052
Received: 11 March 2017; Accepted: 26 May 2017;
Published: 08 June 2017.
Edited by:
Johnan A. R. Kaleeba, Uniformed Services University of the Health Sciences, United StatesReviewed by:
Miklos Fuzi, Semmelweis University, HungaryPeng Yang, Beijing Center for Disease Prevention and Control, China
Konstantin V. Korotkov, University of Kentucky, United States
Copyright © 2017 Lu, Fang, Fan, Chen, Wang, Zeng, Li, Zhang, Huang, Li, Li, Zhu, Cui and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Binghuai Lu, zs25041@126.com
Duochun Wang, wangduochun@icdc.cn
†These authors have contributed equally to this work.
 Binghuai Lu
Binghuai Lu Yujie Fang2,3†
Yujie Fang2,3† Yanyan Fan
Yanyan Fan Ji Zeng
Ji Zeng