
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol. , 26 June 2017
Sec. Virology
Volume 8 - 2017 | https://doi.org/10.3389/fmicb.2017.01169
MicroRNAs (miRNAs) are small non-coding RNAs that have been identified in animals, plants, and viruses. These small RNAs play important roles in post-transcriptional regulation of various cellular processes, including development, differentiation, and all aspects of cancer biology. Rapid-onset T-cell lymphoma of chickens, namely Marek’s disease (MD), induced by Gallid alphaherpesvirus 2 (GaHV2), could provide an ideal natural animal model for herpesvirus-related cancer research. GaHV2 encodes 26 mature miRNAs derived from 14 precursors assembled in three distinct gene clusters in the viral genome. One of the most highly expressed GaHV2 miRNAs, miR-M4-5p, shows high sequence similarity to the cellular miR-155 and the miR-K12-11 encoded by Kaposi’s sarcoma-associated herpesvirus, particularly in the miRNA “seed region.” As with miR-K12-11, miR-M4-5p shares a common set of host and viral target genes with miR-155, suggesting that they may target the same regulatory cellular networks; however, differences in regulatory function between miR-155 and miR-M4-5p may distinguish non-viral and viral mediated tumorigenesis. In this review, we focus on the functions of miR-M4-5p as the viral ortholog of miR-155 to explore how the virus mimics a host pathway to benefit the viral life cycle and trigger virus-induced tumorigenesis.
MicroRNAs (miRNAs) are regulatory non-coding RNAs of approximately 22–24 nt expressed in multicellular organisms and viruses (Kozomara and Griffiths-Jones, 2011). In general, miRNAs regulate post-transcriptional processes through silencing gene expression by binding to the 3′ untranslated region (3′UTR) of mRNA transcripts (Bartel, 2009). miRNAs are involved either in maintaining physiological homeostasis or in facilitating pathogenesis, including virus infection (Bartel, 2009; Skalsky and Cullen, 2010). More than 450 virus-encoded miRNAs have been identified since they were first discovered in 2004 (Pfeffer et al., 2004). The majority of viral miRNAs are encoded by herpesviruses, such as herpes simplex virus (HSV) and human oncogenic Epstein-Barr virus (EBV), Kaposi’s sarcoma-associated herpesvirus (KSHV), and the avian Gallid alphaherpesvirus 2 (GaHV2), highlighting the possible important roles of miRNAs in viral pathogenesis in diverse hosts (Skalsky and Cullen, 2010). Although the detailed mechanisms of EBV- and KSHV-associated disease are unclear, the recently discovered viral miRNAs are potentially powerful determinants of viral pathogenesis (Gottwein and Cullen, 2008; Skalsky and Cullen, 2010); however, a lack of suitable natural animal models has limited the direct functional analyses of viral miRNAs in vivo (Kincaid and Sullivan, 2012).
As an oncogenic Alphaherpesvirus, GaHV2 causes immunosuppression, neurological disease, and rapid-onset T-cell lymphomas in chickens known as Marek’s disease (MD) (Calnek, 2001), which has been proposed as an ideal animal model for herpes virus-related cancer research (Osterrieder et al., 2006). Previous studies have characterized the functions of some GaHV2-specific genes in vitro and/or in vivo (Osterrieder et al., 2006); however, a comprehensive understanding of the molecular mechanisms involved in GaHV2 pathogenesis is lacking. The recent discovery of viral miRNAs encoded in the GaHV2 genome raises the possibility that these tiny non-coding RNAs may be significant for viral pathogenesis, particularly in relation to the virus-induced tumorigenesis (Luo et al., 2010; Yao and Nair, 2014). Interestingly, one GaHV2-encoded miRNA, miR-M4-5p, is a functional miR-155 ortholog (Zhao et al., 2009). Moreover, the “seed region” sequences of miR-M4-5p exhibit high homology to the KSHV-encoded miR-K12-11 (Parnas et al., 2014), suggesting the same, or similar, viral infectious strategies have developed during virus evolution across various viral species. In this review, we focus on relevant research findings that highlight the critical roles of miR-M4-5p in the rapid induction of MD lymphoma.
In miRNA biogenesis (Bartel, 2004; Cullen, 2004; Skalsky and Cullen, 2010), the majority of cellular or viral genomic sequences are first transcribed as primary miRNA (pri-miRNA) in the nucleus by RNA Polymerase II. Subsequently, the RNase III enzyme, Drosha, in concert with the RNA-binding protein, DGCR8, excise pri-miRNA to an approximately 60-nt stem-loop precursor miRNA (pre-miRNA). The pre-miRNA is transferred from the nucleus to the cytoplasm by the Exportin-5 protein directed pathway. In the cytoplasm, the RNase enzyme, Dicer, cleaves the pre-miRNA into a double-stranded of mature miRNA. One of the strands (guide strand), combined with the RNA-induced silencing complex (RISC), orchestrates the regulatory role of targeting and inducing mRNA degradation and/or translational inhibition (Bartel, 2004). The other strand, known as the passenger strand or star miRNA (miRNA∗), is generally thought to be degraded; however, both guide and passenger strands are biofunctional in some cases (Czech et al., 2009; Okamura et al., 2009). Aside from the canonical pathway, there is an alternative, non-canonical generation of pre-miRNA by splicing and disbranching of short hairpin introns, rather than Drosha enzyme processing, known as “mirtrons” (Okamura et al., 2007; Ruby et al., 2007). Most viral miRNAs are processed and matured through the canonical pathway (Cullen, 2011) whereas viral mirtrons have rarely been identified (Rasschaert et al., 2016).
In miRNA-induced gene silencing, the miRNA serves to guide RISC and in binding to target mRNA, whereas the Argonaut (AGO) proteins function as translational inhibition effectors (Flores et al., 2014; Jonas and Izaurralde, 2015). At the 5′-end of the miRNA, 2–7 nt sequences termed as “seed region” are crucial for mRNA target recognition and miRNA-mediated repression function, which usually matches the 3′UTR of the target mRNA. Evolutionary conservation of miRNAs and their 3′UTR binding sites facilitates computational prediction and experimental identification of authentic miRNA targets (Bartel, 2009). Computational analysis suggests that a single miRNA may target hundreds of genes, whereas one gene may be regulated by multiple miRNAs. Over 60% of human protein-coding genes contain at least one conserved miRNA binding site (Friedman et al., 2009), suggesting an important role for miRNA-mediated gene expression regulation.
miR-155 was initially identified as a B-cell integration cluster (bic) gene, which is activated by promoter insertion at a retroviral integration site in avian leukosis virus (ALV)-induced lymphomas (Clurman and Hayward, 1989; Rodriguez et al., 2007). miR-155 is highly conserved in humans, mice, and chickens, particularly the seed region, primarily expressed in lymphocytes of the thymus and spleen, and has regulatory functions in the hematopoietic and immune systems (Rodriguez et al., 2007). Over-expression of miR-155 is related to B cell lymphomas and solid tumors (Tili et al., 2013).
miR-155 is also involved in viral pathogenesis, particularly that of the tumor-related herpesviruses. EBV is a human malignancy related gammaherpesvirus, which induces B-cell lymphomas, including Hodgkin’s and Burkitt’s lymphomas and other types of human cancer (Young and Rickinson, 2004). EBV encodes 25 pre-miRNAs and 44 mature miRNAs (miRBase v21), which play important regulatory roles in virus-induced tumorigenesis (Skalsky and Cullen, 2010; Qiu et al., 2015). Interestingly, EBV-induced cell proliferation, as well as virus latency and reactivation, are associated with elevated miR-155 expression levels (Yin et al., 2008; Linnstaedt et al., 2010; Forte and Luftig, 2011). Importantly, it has been clearly shown that EBV-induced overexpression of cellular miR-155 is essential for transformation by EBV (Linnstaedt et al., 2010).
Kaposi’s sarcoma-associated herpesvirus is another causative agent of human diseases, particularly Kaposi’s sarcoma, primary effusion lymphoma (PEL), and multicentric Castleman’s disease (Ganem, 2006). KSHV encodes 13 pre-miRNAs and 25 mature miRNAs (miRBase v21), which also have important regulatory roles in viral pathogenesis (Gottwein and Cullen, 2008; Kincaid and Sullivan, 2012). It is of great interest that the KSHV-encoded miR-K12-11 functions as a viral ortholog of miR-155 (Gottwein et al., 2007; Skalsky et al., 2007). Both miR-K12-11 and miR-155 are associated with human lymphoma and share a common set of mRNA targets (Gottwein et al., 2007; Skalsky et al., 2007; McClure and Sullivan, 2008). The potentiality of miR-K12-11 compensating for the function of miR-155 has been proved in humanized and transgenic miR-155 knockout mice; however, this is difficult to be confirmed because of the lack of a natural animal model for KSHV infection (Boss et al., 2011; Sin et al., 2013).
The MD associated avian herpesviruses, were previously classified into serotype 1 (MDV-1), serotype 2 (MDV-2) and herpesvirus of Turkeys (HVT), have recently been reclassified as GaHV-2, G. alphaherpesvirus 3 (GaHV-3), and Meleagrid alphaherpesvirus 1 (MeHV1), respectively1. Similar to the other double-stranded DNA herpesviruses (Pfeffer et al., 2004, 2005), all three virus serotypes are confirmed to encode miRNAs (Burnside et al., 2006, 2008; Yao et al., 2007, 2008, 2009; Waidner et al., 2009).
GaHV2-encoded miRNAs were identified in virus-infected chicken embryo fibroblasts (CEFs) and virus-transformed T-lymphoma cells (Burnside et al., 2006, 2008; Yao et al., 2008). Three distinct miRNA clusters in the GaHV2 genome has been confirmed. The first cluster, miR-M9-M4, referred to as the Meq-cluster (Burnside et al., 2006; Yao et al., 2008), is located upstream of the meq oncogene and includes six pre-miRNAs (miR-M9, miR-M5, miR-M12, miR-M3, miR-M2, and miR-M4 in order). Cluster miR-M11-M1, namely the Mid-cluster (Luo et al., 2010), is located downstream of meq and includes three pre-miRNAs (miR-M11, miR-M31, and miR-M1). A third cluster, miR-M8-M10, referred to as the LAT-cluster (Burnside et al., 2006; Yao et al., 2008), is found within the first intron of the GaHV2-encoded 10 kb LAT (latency-associated transcript), and includes five pre-miRNAs (miR-M8, miR-M13, miR-M6, miR-M7, and miR-M10). The sequences of GaHV2 miRNAs from a collection of field and reference strains with various levels of virulence were compared and found to be highly conserved; however, miRNAs expressed from the Meq and Mid- clusters were detected at higher levels in lymphomas caused by a very virulent plus (vv+) strain than in those caused by a less virulent strain. In contrast, expression levels of the miRNAs from the LAT-cluster were equivalent in tumors produced by vv and vv+ strains (Morgan et al., 2008).
Zhao et al. (2011) reported that deletion of the Meq-cluster significantly attenuated the oncogenicity of virulent GaHV2, strongly implicating this cluster as a key regulator of viral pathogenesis. A similar deletion of the Meq-cluster in the very virulent GX0101 strain significantly decreased, rather than abolished, its oncogenicity (Yu et al., 2014). These results suggest that the Meq-cluster is an important, but dispensable, regulator of the development of MD lymphomas. Interestingly, deletion of individual miRNAs of the Meq-cluster produced no effect on virus replication; however, each of the mutants was associated with reduced mortality and gross tumor incidence after infection relative to the parental virus (Zhao et al., 2011; Yu et al., 2014; Teng et al., 2015). The individual miRNA mutants produced variable effects on mortality and gross tumor production in infected chickens, suggesting that they may be involved in different cellular signaling pathways. For example, miR-M3-5p targets and down-regulates the expression of SMAD2, a critical component of the transforming growth factor beta (TGF-β) signaling pathway (Xu et al., 2011). Through suppression of SMAD2, miR-M3-5p significantly promotes cell survival and blocks cisplatin-induced apoptosis, suggesting that viruses may encode miRNAs to directly target cellular factors involved in antiviral processes, including apoptosis, thus proactively creating a cellular environment beneficial to viral latency and oncogenesis (Xu et al., 2011). miR-M4-5p is another member of this cluster which has a specific role in GaHV2 pathogenesis and will be described in more detail below.
The transcription of Mid- and Meq-clusters is driven by a single promoter (prmiRM9M4) during latent phase, while only the Meq-cluster is transcribed during the lytic phase by prmiRM9M4 (Coupeau et al., 2012), implying distinct roles of the Mid-cluster in GaHV2 infection. The Mid-cluster and the associated individual miRNAs are dispensable for GaHV2 replication. However, deletion of miR-M31 decreased the mortality and gross tumor incidence of infected chickens (Teng et al., 2017). miR-221 targets and suppresses the expression of a key cell cycle regulatory protein p27Kip1, which is involved in the induction and progression of T-cell lymphomas (Lambeth et al., 2009). Similar gene regulatory networks may be hijacked by miR-M31-3p, which shares a conserved seed homolog with miR-221 (Morgan et al., 2008). In contrast, deletion of miR-M11 unexpectedly increased viral pathogenicity, and oncogenicity may be partially induced by one member of the mature miRNAs, miR-M11-5p, which targets and suppresses meq expression. Overall, these results suggest that miR-M31-3p may act as an oncogene, while miR-M11-5p may be a potential tumor suppressor during GaHV2 infection (Teng et al., 2017).
The miRNAs in the LAT-cluster are similarly transcribed in various virulent GaHV2-induced tumors (Morgan et al., 2008); however, they exhibit different temporal and spatial transcription profiles during each infectious phase (Luo et al., 2011; Zhao et al., 2015). Interestingly, the GaHV2 immediate-early (IE) genes, ICP4 and ICP27, are putative targets of miR-M7-5p (Strassheim et al., 2012), a member of the LAT-cluster. As a transactivator, ICP4 regulates transcription of early (E) and late (L) genes during virus infection (Pratt et al., 1994; Kato et al., 2002), and it is also necessary for transformation maintenance (Xie et al., 1996), while ICP27 interferes with viral and cellular post-transcriptional splicing (Amor et al., 2011). By targeting these two IE genes, miR-M7-5p may contribute to the establishment and maintenance of latency. These results suggest that, as with other herpesviruses, IE gene-specific miRNAs are common regulatory factors during herpesvirus cytolytic and latent infection (Murphy et al., 2008; Bellare and Ganem, 2009).
miR-M4-5p, which is only encoded by GaHV2, rather than by GaHV3 or MeHV1, has been previously characterized as a viral miR-155 ortholog, sharing an identical seed region with both miR-155 and the KSHV-encoded, miR-K12-11 (Morgan et al., 2008; Zhao et al., 2009; Figure 1A). Using an miRNA target prediction algorithm (Bartel, 2009), candidate cellular mRNA target sites were identified and it was subsequently confirmed that miR-M4-5p and miR-155 down-regulate the expression of a set of shared mRNA targets, including PU.1, CEBPβ, BCL2L13, HIVEP2, PDCD6, Myb, GPM6B, RREB1, and MAP3K7IP2. These targets were confirmed using dual luciferase reporter assays (DLRA) and qRT-PCR analysis, as well as western blotting (Zhao et al., 2009; Muylkens et al., 2010; Figure 1B). In addition, Parnas et al. (2014) performed photoactivatable ribonucleoside-enhanced cross-linking and immunoprecipitation (PAR-CLIP) analysis of RISC binding sites in the GaHV2-transformed MSB-1 cell line to identify targets of miR-M4-5p, and then compared to those of miR-155 and miR-K12-11 similarly identified in EBV-transformed lymphoblastoid cell lines (LCLs) or a KSHV-transformed PEL cell line. The data demonstrated that nine mRNA targets (C1orf103, CSNK1A1, LATS2, MAP3K14, MYB, NR1D2, RORA, RPS6KA3, and WEE1) could be detected in all three cell lines, while four additional mRNA targets (FCHSD2, JARID2, PBEF1, and RAP2A) were only identified in the MSB-1 and LCLs cell lines (Parnas et al., 2014; Figure 1B). These potential mRNA targets were further confirmed by DLRA, the results of which showed that targeting of the 3′UTRs of these genes via the seed regions of miR-155 family members is conserved between humans and chickens, although the data did not demonstrate a statistically significant difference in response to miR-M4-5p and/or miR-155 expression in all cases (Parnas et al., 2014). Among these mRNA targets, WEE1 encodes a kinase that blocks cell-cycle progression and has been associated with inflammation and cancer (Egeland et al., 2016). JARID2 also encodes a cell cycle regulator which is part of a histone methyltransferase complex (Bolisetty et al., 2009). The miR-155/JARID2 axis could regulate cell apoptosis and differentiation in acute myeloid leukemia and abnormal megakaryopoiesis (Norfo et al., 2014; Palma et al., 2014). Three independent research groups identified MYB, a transcription factor involved in the regulation of hematopoiesis and tumorigenesis, as a target for miR-M4-5p, miR-K12-11, and miR-155 (Muylkens et al., 2010; Zhao et al., 2011; Parnas et al., 2014). It is clear that miR-155 could have an important regulatory role in many biological pathways of significance in oncogenesis, hence further investigation is needed to define the in vivo mRNA targets of this miRNA and its viral orthologs.
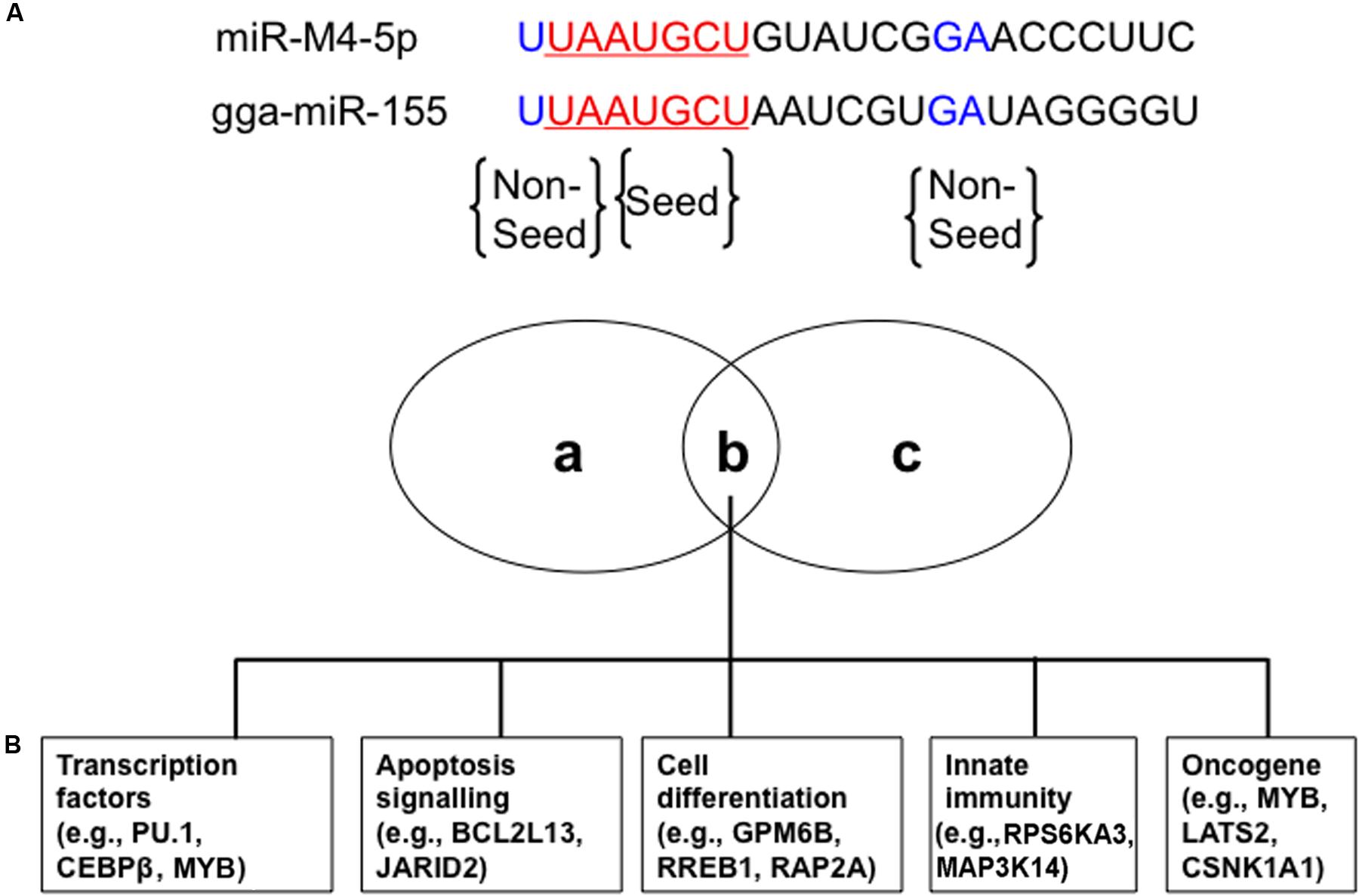
FIGURE 1. Conserved cellular mRNA targets of GaHV2-encoded orthologs of cellular miR-155. (A) miR-M4-5p seed (red) and non-seed (blue) sequences homologous with chicken gga-miR-155. (B) Venn diagram of cellular mRNA targets of both miR-M4-5p and gga-miR-155 involved in different cellular processes with potential relevance to tumorigenesis: (a) and (c) represent miR-M4-5p or gga-miR-155 specific cellular mRNA targets, while (b) represents the shared set of cellular mRNA targets. Adapted from Parnas et al. (2014).
In recent years, bacterial artificial chromosomes (BACs) and mutagenesis techniques have greatly facilitated the introduction of mutations into viral genomes to study GaHV2 gene functions during pathogenesis (Schumacher et al., 2000). Using an infectious BAC clone, mutant viruses with deletions, or two-nucleotide mutations in the miR-M4-5p seed region have been constructed and characterized (Zhao et al., 2011). These mutations had no effect on virus replication; however, compared with the parental oncogenic GaHV2 virus, the mutations abolished its oncogenicity. Interestingly, MD incidence was restored when the viral pre-miRNA, miR-M4, was replaced by gga-miR-155, although tumor induction was slow and detected later than in chickens infected by parental and revertant viruses (Zhao et al., 2011). These observations suggest that miR-M4-5p may be an oncogenic miRNA, the functions of which could be partially restored by miR-155 because the two molecules regulate a common set of mRNA targets through their conserved seed region.
As described above, miR-M4-5p is critical for GaHV2 oncogenicity, and has multiple candidate mRNA targets; however, the details of its precise mechanism of action remain incompletely defined. We have recently identified latent TGF-β binding protein 1 (LTBP1) as a bona fide host mRNA target (Chi et al., 2015). Our data demonstrated that inhibition of LTBP1 expression by miR-M4-5p induced a significant decrease of TGF-β1 secretion and activation, following a significant increase in the expression of c-Myc, a well-known oncogene. It has also been reported that c-Myc regulation in the TGF-β signaling pathway is SMAD-responsive (Yagi et al., 2002). Thus, our data suggest another mechanism by which GaHV2 triggers c-Myc induction in viral oncogenesis. Interestingly, it has also been demonstrated that miR-155 can suppress TGF-β signaling though targeting SMAD2 and SMAD5 in human diseases (Louafi et al., 2010; Rai et al., 2010). As a viral miR-155 ortholog, the KSHV-encoded miR-K12-11 inhibits TGF-β signaling through down-regulation of SMAD5 (Liu et al., 2012). Collectively, these findings indicate that dysregulation of the TGF-β signaling pathway by miR-155 and its viral orthologs may be a common feature shared by oncogenic herpesviruses (Figure 2). Further investigations are required to determine whether there are pathways other than those involving miR-M4-5p that contribute to viral pathogenesis and/or tumorigenesis.
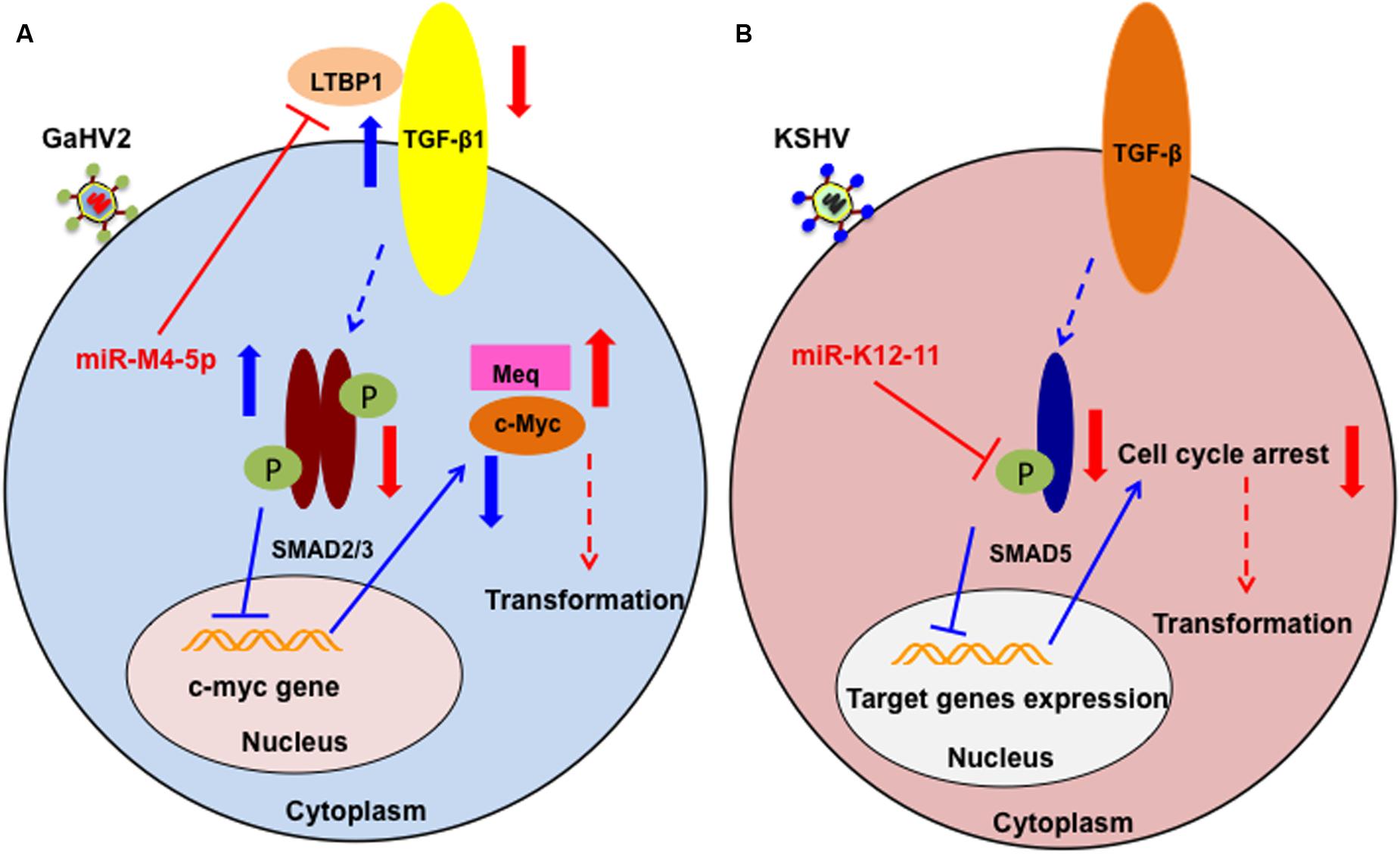
FIGURE 2. Schematic of the potential roles of miR-M4-5p and miR-K12-11 in regulating the TGF-β signaling pathway and promoting tumorigenesis. (A) In normal cell metabolism (blue arrows), c-Myc is down regulated by the phosphorylated SMAD2/3 complex, which is induced by LTBP1 binding to its receptor. During GaHV2 infection (red arrows), miR-M4-5p suppresses the expression levels of LTBP1, leading to reduction of the active SMAD2/3 complex, inducing increased expression of c-Myc. The potential combination of c-Myc and the GaHV2-specific oncoprotein, MEQ, promotes cellular transformation. (B) In normal cell metabolism (blue arrows), activated SMAD5 inhibits target gene expression. During KSHV infection (red arrows), miR-K12-11 inhibits the expression of SMAD5. Through abolishing the TGF-β signaling pathway, miR-K12-11 blocks cell cycle arrest to promote cellular transformation. (A,B) are adapted from Chi et al. (2015) and Liu et al. (2012), respectively.
Herpesvirus-encoded miRNAs play important regulatory roles in both active and latent stages of virus infection, although the molecular mechanisms controlling these activities are largely unexplored. The critical viral miR-155 orthologs, miR-M4-5p and miR-K12-11, share a set of common mRNA targets, suggesting the common viral characteristics of mimicking host miRNAs and aberrant regulation of viral miRNAs may be important in herpesvirus life cycle control, immune evasion, pathogenesis, and even tumorigenesis. The rescue of the regulatory role of miR-M4-5p by miR-155 in vivo by viral genomic locus replacement proves that the two molecules are authentic functional orthologs. Thus, the shared seed region and common set of target mRNAs of miR-M4-5p, miR-K12-11, and miR-155 have facilitated the characterization of miRNA phenotypes in relation to viral tumorigenesis, as in MD. The development of transcriptome-wide identification of miRNA targets has helped to establish the genomic landscape of GaHV2 miRNA functions. However, results generated using the MSB-1 cell line should be treated with caution, as it is a transformed tumor cell line that is simultaneously infected by GaHV2 and GaHV3. Hence, the existence of GaHV3 miRNAs may interfere with the identification of bona fide mRNA targets of GaHV2 miRNAs. Further mRNA target screening and identification should use MD tumor cells, or cell lines only infected by GaHV2. Overall, the next challenge will be to integrate these findings into a better understanding of how the interference mediated by these miRNAs regulates herpesvirus life cycles and their roles in tumorigenesis.
GZ, AS, and MT overview the publications and wrote the manuscript. GZ and JL revised and approved its final version.
This work was supported by the Key Program of NSFC-Henan Joint Fund (no. U1604232); the National Natural Science Foundation of China (no. 31602050 and no. 31372445); and the National Key Research and Development Program of China (no. 2016YFD0500800).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors gratefully acknowledge the critical review by Prof. Norman A. Gregson (ION, UCL, London, United Kingdom).
Amor, S., Strassheim, S., Dambrine, G., Remy, S., Rasschaert, D., and Laurent, S. (2011). ICP27 protein of Marek’s disease virus interacts with SR proteins and inhibits the splicing of cellular telomerase chTERT and viral vIL8 transcripts. J. Gen. Virol. 92(Pt 6), 1273–1278. doi: 10.1099/vir.0.028969-0
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. doi: 10.1016/S0092-8674(04)00045-5
Bartel, D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. doi: 10.1016/j.cell.2009.01.002
Bellare, P., and Ganem, D. (2009). Regulation of KSHV lytic switch protein expression by a virus-encoded microRNA: an evolutionary adaptation that fine-tunes lytic reactivation. Cell Host Microbe 6, 570–575. doi: 10.1016/j.chom.2009.11.008
Bolisetty, M. T., Dy, G., Tam, W., and Beemon, K. L. (2009). Reticuloendotheliosis virus strain T induces miR-155, which targets JARID2 and promotes cell survival. J. Virol. 83, 12009–12017. doi: 10.1128/JVI.01182-09
Boss, I. W., Nadeau, P. E., Abbott, J. R., Yang, Y., Mergia, A., and Renne, R. (2011). A Kaposi’s sarcoma-associated herpesvirus-encoded ortholog of microRNA miR-155 induces human splenic B-cell expansion in NOD/LtSz-scid IL2Rgammanull mice. J. Virol. 85, 9877–9886. doi: 10.1128/JVI.05558-11
Burnside, J., Bernberg, E., Anderson, A., Lu, C., Meyers, B. C., Green, P. J., et al. (2006). Marek’s disease virus encodes microRNAs that map to meq and the latency-associated transcript. J. Virol. 80, 8778–8786. doi: 10.1128/JVI.00831-06
Burnside, J., Ouyang, M., Anderson, A., Bernberg, E., Lu, C., Meyers, B. C., et al. (2008). Deep sequencing of chicken microRNAs. BMC Genomics 9:185. doi: 10.1186/1471-2164-9-185
Calnek, B. W. (2001). Pathogenesis of Marek’s disease virus infection. Curr. Top. Microbiol. Immunol. 255, 25–55. doi: 10.1007/978-3-642-56863-3_2
Chi, J. Q., Teng, M., Yu, Z. H., Xu, H., Su, J. W., Zhao, P., et al. (2015). Marek’s disease virus-encoded analog of microRNA-155 activates the oncogene c-Myc by targeting LTBP1 and suppressing the TGF-beta signaling pathway. Virology 476, 72–84. doi: 10.1016/j.virol.2014.11.027
Clurman, B. E., and Hayward, W. S. (1989). Multiple proto-oncogene activations in avian leukosis virus-induced lymphomas: evidence for stage-specific events. Mol. Cell. Biol. 9, 2657–2664. doi: 10.1128/MCB.9.6.2657
Coupeau, D., Dambrine, G., and Rasschaert, D. (2012). Kinetic expression analysis of the cluster mdv1-mir-M9-M4, genes meq and vIL-8 differs between the lytic and latent phases of Marek’s disease virus infection. J. Gen. Virol. 93(Pt 7), 1519–1529. doi: 10.1099/vir.0.040741-0
Cullen, B. R. (2004). Transcription and processing of human microRNA precursors. Mol. Cell 16, 861–865. doi: 10.1016/j.molcel.2004.12.002
Cullen, B. R. (2011). Viruses and microRNAs: RISCy interactions with serious consequences. Genes Dev. 25, 1881–1894. doi: 10.1101/gad.17352611
Czech, B., Zhou, R., Erlich, Y., Brennecke, J., Binari, R., Villalta, C., et al. (2009). Hierarchical rules for Argonaute loading in Drosophila. Mol. Cell. 36, 445–456. doi: 10.1016/j.molcel.2009.09.028
Egeland, E. V., Flatmark, K., Nesland, J. M., Florenes, V. A., Maelandsmo, G. M., and Boye, K. (2016). Expression and clinical significance of Wee1 in colorectal cancer. Tumour Biol. 37, 12133–12140. doi: 10.1007/s13277-016-5081-3
Flores, O., Kennedy, E. M., Skalsky, R. L., and Cullen, B. R. (2014). Differential RISC association of endogenous human microRNAs predicts their inhibitory potential. Nucleic Acids Res. 42, 4629–4639. doi: 10.1093/nar/gkt1393
Forte, E., and Luftig, M. A. (2011). The role of microRNAs in Epstein-Barr virus latency and lytic reactivation. Microbes Infect. 13, 1156–1167. doi: 10.1016/j.micinf.2011.07.007
Friedman, R. C., Farh, K. K. H., Burge, C. B., and Bartel, D. P. (2009). Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105. doi: 10.1101/gr.082701.108
Ganem, D. (2006). KSHV infection and the pathogenesis of Kaposi’s sarcoma. Annu. Rev. Pathol. 1, 273–296. doi: 10.1146/annurev.pathol.1.110304.100133
Gottwein, E., and Cullen, B. R. (2008). Viral and cellular MicroRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe 3, 375–387. doi: 10.1016/j.chom.2008.05.002
Gottwein, E., Mukherjee, N., Sachse, C., Frenzel, C., Majoros, W. H., Chi, J. T. A., et al. (2007). A viral microRNA functions as an orthologue of cellular miR-155. Nature 450, 1096–1099. doi: 10.1038/nature05992
Jonas, S., and Izaurralde, E. (2015). Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 16, 421–433. doi: 10.1038/nrg3965
Kato, K., Izumiya, Y., Tohya, Y., Takahashi, E., Hirai, K., and Kawaguchi, Y. (2002). Identification and characterization of Marek’s disease virus serotype 1 (MDV1) ICP22 gene product: MDV1 ICP22 transactivates the MDV1 ICP27 promoter synergistically with MDV1 ICP4. Vet. Microbiol. 85, 305–313. doi: 10.1016/S0378-1135(01)00522-3
Kincaid, R. P., and Sullivan, C. S. (2012). Virus-encoded microRNAs: an overview and a look to the future. PLoS Pathog. 8:e1003018. doi: 10.1371/journal.ppat.1003018
Kozomara, A., and Griffiths-Jones, S. (2011). miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 39, D152–D157. doi: 10.1093/nar/gkq1027
Lambeth, L. S., Yao, Y., Smith, L. P., Zhao, Y., and Nair, V. (2009). MicroRNAs 221 and 222 target p27Kip1 in Marek’s disease virus-transformed tumour cell line MSB-1. J. Gen. Virol. 90(Pt 5), 1164–1171. doi: 10.1099/vir.0.007831-0
Linnstaedt, S. D., Gottwein, E., Skalsky, R. L., Luftig, M. A., and Cullen, B. R. (2010). Virally induced cellular microRNA miR-155 plays a key role in B-cell immortalization by Epstein-Barr Virus. J. Virol. 84, 11670–11678. doi: 10.1128/JVI.01248-10
Liu, Y., Sun, R., Lin, X., Liang, D., Deng, Q., and Lan, K. (2012). Kaposi’s sarcoma-associated herpesvirus-encoded microRNA miR-K12-11 attenuates transforming growth factor beta signaling through suppression of SMAD5. J. Virol. 86, 1372–1381. doi: 10.1128/JVI.06245-11
Louafi, F., Martinez-Nunez, R. T., and Sanchez-Elsner, T. (2010). MicroRNA-155 targets SMAD2 and modulates the response of macrophages to transforming growth factor-β. J. Biol. Chem. 285, 41328–41336. doi: 10.1074/jbc.M110.146852
Luo, J., Sun, A. J., Teng, M., Zhou, H., Cui, Z. Z., Qu, L. H., et al. (2011). Expression profiles of microRNAs encoded by the oncogenic Marek’s disease virus reveal two distinct expression patterns in vivo during different phases of disease. J. Gen. Virol. 92(Pt 3), 608–620. doi: 10.1099/vir.0.024158-0
Luo, J., Teng, M., Fan, J., Wang, F., Zhou, L., Deng, R., et al. (2010). Marek’s disease virus-encoded microRNAs: genomics, expression and function. Sci. China Life Sci. 53, 1174–1180. doi: 10.1007/s11427-010-4073-6
McClure, L. V., and Sullivan, C. S. (2008). Kaposi’s sarcoma herpes virus taps into a host microRNA regulatory network. Cell Host Microbe 3, 1–3. doi: 10.1016/j.chom.2007.12.002
Morgan, R., Anderson, A., Bernberg, E., Kamboj, S., Huang, E., Lagasse, G., et al. (2008). Sequence conservation and differential expression of marek’s disease virus microRNAs. J. Virol. 82, 12213–12220. doi: 10.1128/JVI.01722-08
Murphy, E., Vanicek, J., Robins, H., Shenk, T., and Levine, A. J. (2008). Suppression of immediate-early viral gene expression by herpesvirus-coded microRNAs: implications for latency. Proc. Natl. Acad. Sci. U.S.A. 105, 5453–5458. doi: 10.1073/pnas.0711910105
Muylkens, B., Coupeau, D., Dambrine, G., Trapp, S., and Rasschaert, D. (2010). Marek’s disease virus microRNA designated Mdv1-pre-miR-M4 targets both cellular and viral genes. Arch. Virol. 155, 1823–1837. doi: 10.1007/s00705-010-0777-y
Norfo, R., Zini, R., Pennucci, V., Bianchi, E., Salati, S., Guglielmelli, P., et al. (2014). miRNA-mRNA integrative analysis in primary myelofibrosis CD34+ cells: role of miR-155/JARID2 axis in abnormal megakaryopoiesis. Blood 124, E21–E32. doi: 10.1182/blood-2013-12-544197
Okamura, K., Hagen, J. W., Duan, H., Tyler, D. M., and Lai, E. C. (2007). The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 130, 89–100. doi: 10.1016/j.cell.2007.06.028
Okamura, K., Liu, N., and Lai, E. C. (2009). Distinct mechanisms for microRNA strand selection by Drosophila Argonautes. Mol. Cell. 36, 431–444. doi: 10.1016/j.molcel.2009.09.027
Osterrieder, N., Kamil, J. P., Schumacher, D., Tischer, B. K., and Trapp, S. (2006). Marek’s disease virus: from miasma to model. Nat. Rev. Microbiol. 4, 283–294. doi: 10.1038/nrmicro1382
Palma, C. A., Al Sheikha, D., Lim, T. K., Bryant, A., Vu, T. T., Jayaswal, V., et al. (2014). MicroRNA-155 as an inducer of apoptosis and cell differentiation in Acute Myeloid Leukaemia. Mol. Cancer 13:79. doi: 10.1186/1476-4598-13-79
Parnas, O., Corcoran, D. L., and Cullen, B. R. (2014). Analysis of the mRNA targetome of microRNAs expressed by Marek’s disease virus. Mbio 5:e1060-13. doi: 10.1128/mBio.01060-13
Pfeffer, S., Sewer, A., Lagos-Quintana, M., Sheridan, R., Sander, C., Grasser, F. A., et al. (2005). Identification of microRNAs of the herpesvirus family. Nat. Methods 2, 269–276. doi: 10.1038/nmeth746
Pfeffer, S., Zavolan, M., Grasser, F. A., Chien, M., Russo, J. J., Ju, J., et al. (2004). Identification of virus-encoded microRNAs. Science 304, 734–736. doi: 10.1126/science.1096781
Pratt, W. D., Cantello, J., Morgan, R. W., and Schat, K. A. (1994). Enhanced expression of the Marek’s disease virus-specific phosphoproteins after stable transfection of MSB-1 cells with the Marek’s disease virus homologue of ICP4. Virology 201, 132–136. doi: 10.1006/viro.1994.1273
Qiu, J., Smith, P., Leahy, L., and Thorley-Lawson, D. A. (2015). The Epstein-Barr virus encoded BART miRNAs potentiate tumor growth in vivo. PLoS Pathog. 11:e1004561. doi: 10.1371/journal.ppat.1004561
Rai, D., Kim, S. W., McKeller, M. R., Dahia, P. L., and Aguiar, R. C. (2010). Targeting of SMAD5 links microRNA-155 to the TGF-beta pathway and lymphomagenesis. Proc. Natl. Acad. Sci. U.S.A. 107, 3111–3116. doi: 10.1073/pnas.0910667107
Rasschaert, P., Figueroa, T., Dambrine, G., Rasschaert, D., and Laurent, S. (2016). Alternative splicing of a viral mirtron differentially affects the expression of other microRNAs from its cluster and of the host transcript. RNA Biol. 13, 1310–1322. doi: 10.1080/15476286.2016.1244600
Rodriguez, A., Vigorito, E., Clare, S., Warren, M. V., Couttet, P., Soond, D. R., et al. (2007). Requirement of bic/microRNA-155 for normal immune function. Science 316, 608–611. doi: 10.1126/science.1139253
Ruby, J. G., Jan, C. H., and Bartel, D. P. (2007). Intronic microRNA precursors that bypass Drosha processing. Nature 448, 83–86. doi: 10.1038/nature05983
Schumacher, D., Tischer, B. K., Fuchs, W., and Osterrieder, N. (2000). Reconstitution of Marek’s disease virus serotype 1 (MDV-1) from DNA cloned as a bacterial artificial chromosome and characterization of a glycoprotein B-negative MDV-1 mutant. J. Virol. 74, 11088–11098. doi: 10.1128/Jvi.74.23.11088-11098.2000
Sin, S. H., Kim, Y. B., and Dittmer, D. P. (2013). Latency locus complements microRNA 155 deficiency in vivo. J. Virol. 87, 11908–11911. doi: 10.1128/JVI.01620-13
Skalsky, R. L., and Cullen, B. R. (2010). Viruses, microRNAs, and host interactions. Annu. Rev. Microbiol. 64, 123–141. doi: 10.1146/annurev.micro.112408.134243
Skalsky, R. L., Samols, M. A., Plaisance, K. B., Boss, I. W., Riva, A., Lopez, M. C., et al. (2007). Kaposi’s sarcoma-associated herpesvirus encodes an ortholog of miR-155. J. Virol. 81, 12836–12845. doi: 10.1128/JVI.01804-07
Strassheim, S., Stik, G., Rasschaert, D., and Laurent, S. (2012). mdv1-miR-M7-5p, located in the newly identified first intron of the latency-associated transcript of Marek’s disease virus, targets the immediate-early genes ICP4 and ICP27. J. Gen. Virol. 93, 1731–1742. doi: 10.1099/vir.0.043109-0
Teng, M., Yu, Z. H., Sun, A. J., Min, Y. J., Chi, J. Q., Zhao, P., et al. (2015). The significance of the individual Meq-clustered miRNAs of Marek’s disease virus in oncogenesis. J. Gen. Virol. 96, 637–649. doi: 10.1099/jgv.0.000013
Teng, M., Yu, Z. H., Zhao, P., Zhuang, G. Q., Wu, Z. X., Dang, L., et al. (2017). Putative roles as oncogene or tumour suppressor of the Mid-clustered microRNAs in Gallid alphaherpesvirus 2 (GaHV2) induced Marek’s disease lymphomagenesis. J Gen Virol. 98, 1097–1112. doi: 10.1099/jgv.0.000786
Tili, E., Michaille, J. J., and Croce, C. M. (2013). MicroRNAs play a central role in molecular dysfunctions linking inflammation with cancer. Immunol. Rev. 253, 167–184. doi: 10.1111/imr.12050
Waidner, L. A., Morgan, R. W., Anderson, A. S., Bernberg, E. L., Kamboj, S., Garcia, M., et al. (2009). MicroRNAs of Gallid and Meleagrid herpesviruses show generally conserved genomic locations and are virus-specific. Virology 388, 128–136. doi: 10.1016/j.virol.2009.02.043
Xie, Q., Anderson, A. S., and Morgan, R. W. (1996). Marek’s disease virus (MDV) ICP4, pp38, and meq genes are involved in the maintenance of transformation of MDCC-MSB1 MDV-transformed lymphoblastoid cells. J. Virol. 70, 1125–1131.
Xu, S., Xue, C., Li, J., Bi, Y., and Cao, Y. (2011). Marek’s disease virus type 1 microRNA miR-M3 suppresses cisplatin-induced apoptosis by targeting Smad2 of the transforming growth factor beta signal pathway. J. Virol. 85, 276–285. doi: 10.1128/JVI.01392-10
Yagi, K., Furuhashi, M., Aoki, H., Goto, D., Kuwano, H., Sugamura, K., et al. (2002). c-myc is a downstream target of the Smad pathway. J. Biol. Chem. 277, 854–861. doi: 10.1074/jbc.M104170200
Yao, Y., Zhao, Y., Smith, L. P., Lawrie, C. H., Saunders, N. J., Watson, M., et al. (2009). Differential expression of microRNAs in Marek’s disease virus-transformed T-lymphoma cell lines. J. Gen. Virol. 90(Pt 7), 1551–1559. doi: 10.1099/vir.0.009902-0
Yao, Y., Zhao, Y., Xu, H., Smith, L. P., Lawrie, C. H., Watson, M., et al. (2008). MicroRNA profile of Marek’s disease virus-transformed T-cell line MSB-1: predominance of virus-encoded microRNAs. J. Virol. 82, 4007–4015. doi: 10.1128/JVI.02659-07
Yao, Y. X., and Nair, V. (2014). Role of virus-encoded microRNAs in avian viral diseases. Viruses 6, 1379–1394. doi: 10.3390/v6031379
Yao, Y. X., Zhao, Y. G., Xu, H. T., Smith, L. P., Lawrie, C. H., Sewer, A., et al. (2007). Marek’s disease virus type 2 (MDV-2)-encoded microRNAs show no sequence conservation with those encoded by MDV-1. J. Virol. 81, 7164–7170. doi: 10.1128/JVI.00112-07
Yin, Q. Y., McBride, J., Fewell, C., Lacey, M., Wang, X., Lin, Z., et al. (2008). MicroRNA-155 is an Epstein-Barr virus-induced gene that modulates Epstein-Barr virus-regulated gene expression pathways. J. Virol. 82, 5295–5306. doi: 10.1128/JVI.02380-07
Young, L. S., and Rickinson, A. B. (2004). Epstein-Barr virus: 40 years on. Nat. Rev. Cancer 4, 757–768. doi: 10.1038/nrc1452
Yu, Z. H., Teng, M., Sun, A. J., Yu, L. L., Hu, B., Qu, L. H., et al. (2014). Virus-encoded miR-155 ortholog is an important potential regulator but not essential for the development of lymphomas induced by very virulent Marek’s disease virus. Virology 448, 55–64. doi: 10.1016/j.virol.2013.09.017
Zhao, P., Li, X. J., Teng, M., Dang, L., Yu, Z. H., Chi, J. Q., et al. (2015). In vivo expression patterns of microRNAs of Gallid herpesvirus 2 (GaHV-2) during the virus life cycle and development of Marek’s disease lymphomas. Virus Genes 50, 245–252. doi: 10.1007/s11262-015-1167-z
Zhao, Y., Xu, H., Yao, Y., Smith, L. P., Kgosana, L., Green, J., et al. (2011). Critical role of the virus-encoded microRNA-155 ortholog in the induction of Marek’s disease lymphomas. PLoS Pathog. 7:e1001305. doi: 10.1371/journal.ppat.1001305
Keywords: herpesvirus, Marek’s disease virus, GaHV2, miR-155, miR-M4-5p, pathogenesis, tumorigenesis
Citation: Zhuang G, Sun A, Teng M and Luo J (2017) A Tiny RNA that Packs a Big Punch: The Critical Role of a Viral miR-155 Ortholog in Lymphomagenesis in Marek’s Disease. Front. Microbiol. 8:1169. doi: 10.3389/fmicb.2017.01169
Received: 10 April 2017; Accepted: 08 June 2017;
Published: 26 June 2017.
Edited by:
Akio Adachi, Tokushima University, JapanReviewed by:
Mohammad Heidari, Agricultural Research Service (USDA), United StatesCopyright © 2017 Zhuang, Sun, Teng and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Luo, luojun593@aliyun.com
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.