- 1Department of Nursing, Faculty of Human Movement and Quality of Life Sciences, University of Peloponnese, Sparta, Greece
- 2Department of Biopathology and Clinical Microbiology, Athens Medical School, Aeginition Hospital, Athens, Greece
- 3Department of Hygiene, Epidemiology and Medical Statistics, Medical School, University of Athens, Athens, Greece
- 4Department of Clinical Microbiology, “Alexandra” General Hospital, Athens, Greece
Objectives: The symbiosis of Trichomonas vaginalis and Mycoplasma hominis is the first described association between two obligate human parasites. Trichomonas is the niche and the vector for the transmission of M. hominis infection. This clinically significant symbiosis may affect T. vaginalis virulence and susceptibility to treatment. The aims of this study were to investigate the intracellularly present Mycoplasma and Ureaplasma species in T. vaginalis strains isolated from the vaginal discharge of infected women as well as to trace the diversity pattern among the species detected in the isolated strains.
Methods: Hundred pure T. vaginalis cultures were isolated from ~7,500 patient specimens presented with clinical purulent vaginitis. PCR and sequencing for Mycoplasma/Ureaplasma spp. were performed in DNA extracted from the pure cultures. In addition, vaginal discharge samples were cultured for the presence of M. hominis and U. urealyticum. Phylogenetic analysis assisted the identification of interspecies relationships between the Mycoplasma and Ureaplasma isolates.
Results: Fifty four percentage of T. vaginalis isolates were harboring Mycoplasma spp. Phylogenetic analysis revealed three distinct clusters, two with already characterized M. hominis and Ureaplasma spp. (37% of total Mycoplasma spp.), whereas one group formed a distinct cluster matched with the newly identified species Candidatus Mycoplasma girerdii (59.3%) and one or more unknown Mycoplasma spp. (3.7%).
Conclusions: T. vaginalis strains associated with vaginal infection might host intracellular mycoplasmas or ureaplasmas. Intracellular Mollicutes that remain undetected in the extracellular environment when conventional diagnostic methods are implemented may comprise either novel species, such as Candidatus M. giredii, or unknown species with yet unexplored clinical significance.
Introduction
The flagellated protozoan parasite Trichomonas vaginalis is one of the most common causes of non-viral sexually transmitted infections in humans, annually affecting at least 142 million people worldwide (WHO, 2016). Most Trichomonas spp., other than T. vaginalis, are harmless commensals, colonizing the gastrointestinal tract of a broad variety of hosts (Soper, 2004). The majority of infected individuals remains asymptomatic (up to 85%) and the infection might persist for months or years (Sutton et al., 2007; Kissinger, 2015). Clinical manifestations mainly occur in women and vary from mild to severe vaginitis with profuse inflammatory discharge. The course of the infection depends on the individualized host defense mechanisms and the virulence factors of the causative strain (Arroyo et al., 1992; Mendoza-Lopez et al., 2000). Strains of T. vaginalis also present variable susceptibility profiles toward metronidazole and tinidazole, the major antitrichomonal drugs (Sena et al., 2014). Therefore, imponderable etiologic factors along with the reported paucity of therapeutic options impede the treatment of vaginitis.
The clinical importance of the T. vaginalis infections has been reportedly associated with dysbiosis of the urogenital tract microbiome and with the co-existence of the protist with opportunistic microorganisms including the intracellularly hosted Mycoplasma spp. or the T. vaginalis viruses (TVVs; Bar et al., 2015; Margarita et al., 2016). The Trichomonas viruses constitute the first double-stranded RNA (dsRNA) viruses associated with the parasite, and the notion that the trichomonads infected by the TVVs may present prolonged survival within the human host due to immunogenic proteins expression modulation is not new (Goodman et al., 2011).
The partnership between the protozoan and the bacteria or the viruses exerts a modulatory effect on innate immunity and the inflammatory process (Rappelli et al., 2001; Hirt and Sherrard, 2015). T. vaginalis and M. hominis are known to have the same arginine dihydrolase (ADH) pathway, which serves as an alternative source of ATP through arginine scavenging. Subsequent arginine depletion blocks nitric oxide (NO) production by host macrophages, alters host defense mechanism and ameliorates the parasite survival (Morada et al., 2010). Furthermore, the extensively studied symbiotic pattern of M. hominis hosted by T. vaginalis is proved to play a key role in inflammation during trichomoniasis, thus enhancing the severity of the disease (Dessi et al., 2005, 2006; Vancini and Benchimol, 2008; Vancini et al., 2008; Morada et al., 2010; Fiori et al., 2013). T. vaginalis hosting M. hominis have also been found to bear higher phagocytic activity than Mycoplasma–free isolates (Cirillo et al., 1997).
The concept of reductive evolution of the mycoplasmal genome from Gram-positive bacteria, based on comparative genomics, justifies both structural and functional alterations (Razin et al., 1998). Mycoplasma species, belonging in the class of Mollicutes, have developed a small genome which lacks biosynthetic pathways involved in macromolecule building blocks synthesis, therefore explaining the absence of cell wall (Liu et al., 2012). The Mycoplasma genome also carries a minimal set of energy metabolic genes with a restricted adenosine triphosphate (ATP) supply which is essential for their parasitic mode of life (Razin, 1997). Despite the limited genetic redundancy, Mycoplasma spp. present a rather interesting mechanism of self-protection based on their ability to modulate the inflammatory response of the host immune system (Razin et al., 1998).
Among Mycoplasma spp., M. hominis is the most frequently isolated microorganism from the genital tract of both males and females, acting either as part of normal flora or as a putative pathogen responsible for a variety of urogenital infections (Ladefoged, 2000). The Mollicutes represent the most common pathogens associated with T. vaginalis infection with Mycoplasma hominis and Candidatus Mycoplasma girerdii as the predominant strains isolated in trichomoniasis specimens (Koch et al., 1997; Fettweis et al., 2014). The symbiotic phenomenon of M. hominis hosted by the pathogenic protist has received special interest though little is known about the intracellular colonization of T. vaginalis by other Mycoplasma spp. (Martin et al., 2013). The study of the symbiotic relationship between T. vaginalis and M. hominis provides information about (i) the association between two obligate human pathogens and (ii) an additional role of T. vaginalis as a novel niche for M. hominis. Infected T. vaginalis isolates act as M. hominis carriers mediating transmission in other protozoa and human-derived epithelial cells (Rappelli et al., 2001). Mycoplasmas hosted by T. vaginalis have the privilege to evade host immune response, enhance T. vaginalis virulence and affect susceptibility to antibiotic treatment, thus complicating the infection eradication process (Dessi et al., 2005). In particular, a higher rate of phagocytic activity and amoeboid transformation, indicative of increased virulence, is observed in T. vaginalis isolates harboring M. hominis as compared with the mycoplasma-free isolates (Vancini et al., 2008). The endosymbiotic relationship may also alter the host inflammatory respond during trichomoniasis subsequently affecting disease severity (Fiori et al., 2013).
The broad term “mycoplasmas” collectively refers to both the Mycoplasma spp. and the Ureaplasma spp. The latter differentiate to Ureaplasma urealyticum and Ureaplasma parvum which mainly colonize the urogenital tract mucosal surfaces of sexually active women (McIver et al., 2009). U. urealyticum used to be categorized into biovars 1 and 2; however, according to phylogenetic analysis this taxonomic overlap has changed with the identification of the U. parvum formerly known as U. urealyticum biovar 1 (Kong et al., 1999). Both species have been controversial for their participation in human diseases and are generally considered as “low-grade pathogens” (Cox et al., 2016). The arrival of U. parvum at the forefront of research studies has raised questions regarding the virulence and pathogenicity of the microorganism formerly known as a non-pathogenic commensal of the male urethra. In fact, recent studies have proved the participation of the bacterium in pregnancy complications, intrauterine infections, vaginosis, and pelvic disease (McIver et al., 2009; Larsen and Hwang, 2010; Cox et al., 2016). Of great clinical significance is the association of preterm neonates ureaplasmal infection with adverse health outcomes predominantly including bronchopulmonary dysplasia, central nervous system deformities, and necrotizing enterocolitis (Pandelidis et al., 2013; Viscardi, 2014; Glaser and Speer, 2015). To date, Mollicutes including both Mycoplasma and Ureaplasma spp. are known to actively participate in sexually-transmitted infections though the exact role of the latter in trichomonas-associated infections is not widely investigated.
In this study the intracellular presence of Mycoplasma and Ureaplasma species was investigated employing T. vaginalis strains isolated from infected women with vaginitis. The detected bacteria were identified and classified, aiming to disclose whether species other than M. hominis can endosymbiotically colonize T. vaginalis. Ultimately, a major aspect of this study was to surveil both the diversity and the genetic distance among the species of the isolated strains.
Methods
Patients
A total number of 7,500 clinical samples collected from adult women aged 18–64 years was incorporated in this study within a 3-year period (2010–2012). A written informed consent is not required since the clinical isolates do not provide identifying information regarding the participants, as it is in the present study. All patients were presented with purulent vaginitis at the outpatient clinic of the Department of Gynecology of “Alexandra” General Hospital in Athens. From the total samples number, only 100 were confirmed as T. vaginalis positive containing the parasite and were further cultured to investigate the presence of Mycoplasma/Ureaplasma spp.
Sample Collection and Microbiological Processing
Four vaginal swab specimens were collected from each patient for: (i) microscopic examination, (ii) conventional bacteriological culture, (iii) T. vaginalis culture, and (iv) Mycoplasma/Ureaplasma detection. T. vaginalis strains were cultured by daily passages in commercially available Trichosel broth, a modified version of the Simplified Trypticase Serum (STS) Medium (Becton, Dickinson and Company, New Jersey, USA; Kupferberg et al., 1948). Specimens detected to contain T. vaginalis through microscopy, were inoculated into the broth and incubated at 37°C in an aerobic atmosphere. Growth was observed after 2 and 5 days of incubation by wet mount microscopy, and positive samples were subcultured in fresh Trichosel medium in order to enrich the parasite number. The medium contained Chloramphenicol (0.1 g/L) for bacterial growth inhibition including Mycoplasma/Ureaplasma. The initial cultures were followed by four subcultures before harvesting the parasites for further processing. Both primary cultures and subcultures were tested for the presence of extracellular mycoplasmas; an aliquot of each T. vaginalis culture was centrifuged at 500 g for 10 min, and the supernatants were filtered through a 0.45 μm-pore-size filter membrane that only allows small microbial cells such as mycoplasmas to pass through (Dutscher Scientific; Essex, UK). The filtrate was used for DNA extraction and subsequent polymerase chain reaction (PCR) for the detection of Mycoplasma/Ureaplasma spp. Sediments rich in T. vaginalis were stored at −80°C for further handling.
Mycoplasma/Ureaplasma spp. detection in vaginal swabs was performed using conventional bacteriological procedures. Swabs were inoculated in a commercial broth for the detection of M. hominis and U. urealyticum (Mycoplasma U-A broth, CE marked, Bioprepare Microbiology, Attica, Greece). Differential identification was based on a semi-quantitative test fermentation test. More specifically, the kit contains two tubes, one for the M. hominis discrimination based on the arginine fermentation, and one for the U. urealyticum discrimination based on the urea fermentation. The identification of the presence of the Mycoplasma or Ureaplasma spp. is based on the arginine and urea fermentation process leading to an alkaline pH shift and a respective color change from orange to red in the corresponding tube. Broth with a positive reaction was further plated on a commercially available selective medium (Mycotest agar, CE marked, Bioprepare Microbiology, Attica, Greece) that provides all the nutrients needed for the growth of Mycoplasma species along with the appropriate pH value. It also contains antibiotics and antifungal agents that inhibit the growth of most bacteria and fungi. Identification was performed by microscopic characterization of the typical colonies.
DNA Extraction
The DNA extraction from the T. vaginalis rich Trichosel broth culture sediments and the filtered supernatants was performed with the commercially available extraction reagent “Instagene Matrix” (Bio-Rad Laboratories, California, U.S.A.), according to the manufacturer's instructions.
PCR Amplification for T. vaginalis and Mycoplasma/Ureaplasma spp.
The commercial ready mix “Jumpstart Red Taq” (SIGMA Laboratories, St. Louis, U.S.A.) was used for the PCR amplification. The final mix contained 25 μL Jumpstart Red Taq, 1 μM of each primer, 20 μL distilled water and ~300 ng template. Primers for T. vaginalis (TVK-3 and TVK-7) and for Mycoplasma/Ureaplasma (GPO-1 and MGSO) were provided by Invitrogen (Primer design Ltd, Southampton, United Kingdom) according to previously reported design (van Kupperveld et al., 1992; Pillay et al., 2007). The primer final concentration was 10 pmol/μL for each PCR reaction targeting T. vaginalis detection. An aliquot of initial PCR sample was analyzed by conventional 2% agarose gel electrophoresis. DNA was stained with ethidium bromide, visualized and photographed with a computerized transilluminator system (Chemidoc, Bio-Rad Laboratories, California, U.S.A.). Primers and reaction profiles are shown in Tables 1, 2. In positive samples, a distinct 261 bp band was detected for T. vaginalis and a 717 bp band for Mycoplasma/Ureaplasma.
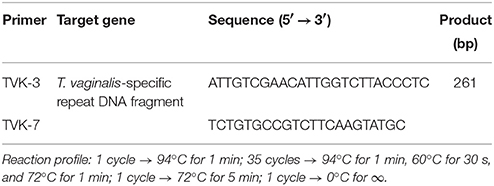
Table 1. Primers and reaction profile for Trichomonas vaginalis (Pillay et al., 2007).

Table 2. Primers and reaction profile for Mycoplasma/Ureaplasma spp. (van Kupperveld et al., 1992).
Quantitative PCR for Mycoplasma/Ureaplasma spp.
For the exclusion of the mycoplasmal DNA presence in the filtered supernatant, a protocol for DNA amplification was performed with a TaqMan probe for M. hominis, M. genitalium, U. urealyticum and U. parvum as described previously (Olsen et al., 2009; Ferandon et al., 2011; Frolund et al., 2014). Primers and reaction profiles are summarized in Table 3. All tests were performed in duplicates with ~300 ng DNA template. Different quantitative real-time PCR standard curves were generated by analyzing 10-fold serial dilutions of purified DNA from M. hominis, M. genitalium, U. urealyticum, and U. parvum as previously described (Campos et al., 2015).
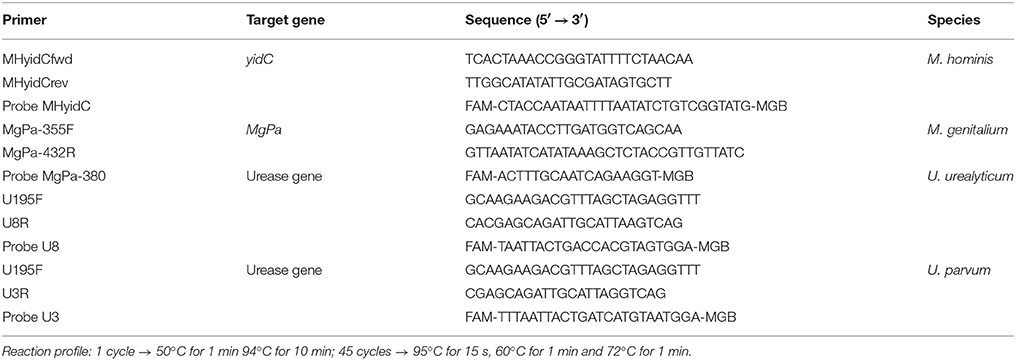
Table 3. Primers and reaction profile for quantitative PCR of M. hominis and M. genitalium, U. urealyticum and U. parvum (Olsen et al., 2009; Ferandon et al., 2011; Frolund et al., 2014).
Sequence Analysis
Capillary Sanger/dideoxy labeling was performed for the sequencing of the PCR products by Macrogen Incorporation (908 World Merdian Center #60–24 Gasan-dong, Geumchum-gu Seoul, Republic of Korea).
Phylogenetic Analysis of Mycoplasma/Ureaplasma spp.
Species definition was accomplished through the phylogenetic analysis of the sequenced samples and the investigation of the phylogenetic relationship between different isolates within the same species. 16S rRNA sequences from each Mycoplasma and Ureaplasma spp. were downloaded (available at GenBank database until March 2015) reaching a total of 302 sequences representing 184 different Mycoplasma species, six different Ureaplasma species and several uncultured and unclassified Mycoplasma and Ureaplasma species. Genetic and phylogenetic analysis was performed with MEGA software (MEGA v6.0), whilst sequences were aligned using the “Muscle” algorithm and were manually edited (Edgar, 2004; Tamura et al., 2013). Genetic distances between the sequences were calculated using the Tamura-Nei model (Tamura and Nei, 1993; Tamura et al., 2011). Phylogenetic trees were constructed using the neighbor-joining method and their reliability was tested by bootstrapping analysis (1,000 replicates) (Saitou and Nei, 1987; Tamura et al., 2011). One cluster was considered significant if it was present in more than 75% of the permuted trees.
Results
Molecular Detection
54% of T. vaginalis isolates were harboring Mycoplasma species. Vaginal discharge cultures revealed one sample infected with M. hominis and one with U. urealyticum (Table 4). All filtered supernatants from T. vaginalis broth cultures, including the two positive vaginal specimens, were negative in quantitative Real-time PCR tests for the presence of M. hominis, M. genitalium, U. urealyticum, and U. parvum DNA.
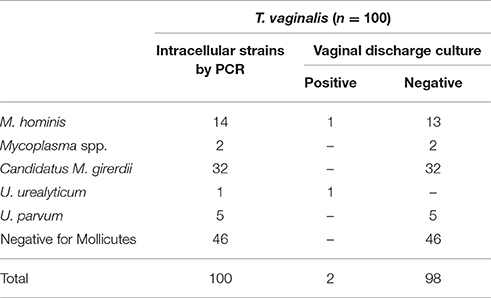
Table 4. Mycoplasma/Ureaplasma spp. intracellularly detected in T. vaginalis isolated from vaginal discharge of infected women by PCR, as well as culture results of the corresponding clinical material.
Isolates Identification
Sequence analysis of T. vaginalis PCR product confirmed the identification of the parasite in all isolates. Sequence analysis of the 54 mycoplasmal PCR products isolated from T. vaginalis revealed 14 M. hominis (25.9%), 1 U. urealyticum (1.9%), 5 U. parvum (9.3%), 32 Candidatus Mycoplasma girerdii (59.3%), and 2 unknown Mycoplasma spp. (3.7%) strains (Table 4).
Species Classification
The phylogenetic analysis classified the various Mycoplasma species, as well as the correlation between multiple isolates of the same species by constructing phylogenetic trees based on the sequence analysis. 302 Mycoplasma and Ureaplasma sequences were deployed to sketch the phylogenetic tree (Figure 1). 14 strains (submitted to the GenBank database with the following accession numbers HS09: KY781839, HS10: KY781841, HS11: KY781842, HS14: KY781845, HS23: KY781852, HS50: KY781873, HS56: KY781876, HS58: KY781878, HS60: KY781880, HS62: KY781882, HS68: KY780615, HS69: KY781885, HS8H: KY781838, HS9H: KY781840) were grouped with two M. hominis reference strains (accession numbers AB680681.1 and AJ002265.1), whereas, within this cluster, two unclassified mycoplasmal strains were also included (EU644475, EU644477; Figure 2).
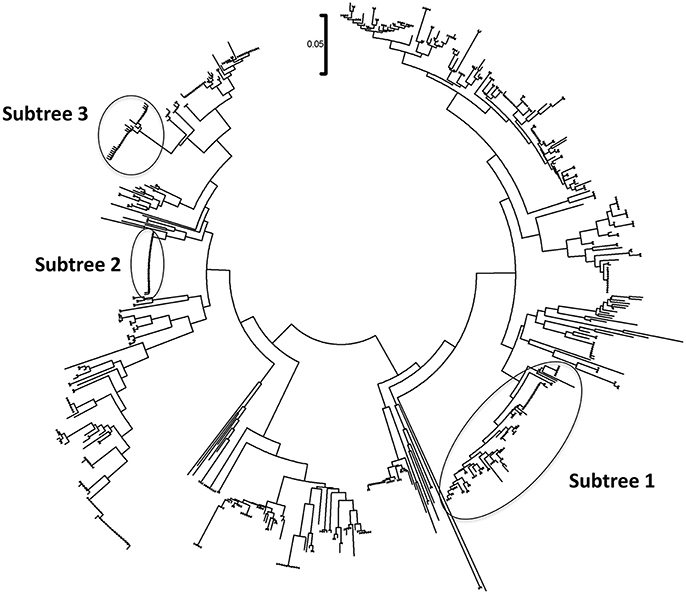
Figure 1. Circular phylogenetic tree including all detected and reference sequences. The evolutionary history was inferred using the Neighbor-Joining method (Saitou and Nei, 1987). The analysis involved 302 nucleotide sequences downloaded from the GenBank database including two representative sequences from each Mycoplasma or Ureaplasma species and all unclassified species. Our sequences clustered into three significant clusters depicting sequences clustering with Mycoplasma hominis strains (Subtree 1), with Candidatus Mycoplasma girerdii strains, and strains of novel Mycoplasma species (Subtree 2) and with Ureaplasma spp. strains (Subtree 3). Accession numbers of the reference sequences used are the following: AB299548.1, AB299549.1, AB558897.1, AB558898.1, AB558899.1, AB576869.1, AB617737.1, AB617738.1, AB680604.1, AB680624.1, AB680625.1, AB680668.1, AB680669.1, AB680678.1, AB680679.1, AB680680.1, AB680681.1, AB680682.1, AB680683.1, AB680684.1, AB680685.1, AB680687.1, AB680688.1, AB680689.1, AB680690.1, AB680691.1, AB680692.1, AB680693.1, AB680694.1, AB725596.1, AB740010.1, AB740012.1, AB758439.1, AB758440.1, AB848713.1, AF001173.1, AF009831.1, AF009832.1, AF042194.1, AF060821.1, AF064062.1, AF125584.1, AF125585.1, AF125587.1, AF125588.1, AF125589.1, AF125592.1, AF125593.1, AF125878.1, AF125879.1, AF132740.1, AF132741.1, AF178676.1, AF212859.1, AF221111.1, AF221112.1, AF221113.1, AF221114.1, AF221115.1, AF221116.1, AF221117.1, AF221118.1, AF221119.1, AF221120.1, AF221121.1, AF261729.1, AF261730.1, AF304323.1, AF304324.1, AF304325.1, AF306346.1, AF538681.1, AF538682.1, AF538683.1, AF538684.1, AF538961.1, AJ002265.1, AJ419900.1, AJ419903.1, AJ419905.1, AM073012.2, AM073013.1, AM073015.1, AM745338.1, AM774638.1, AY050170.1, AY121107.1, AY121108.1, AY150066.1, AY171918.1, AY191226.1, AY366210.1, AY383241.1, AY466443.1, AY529641.1, AY531655.1, AY714305.2, AY756171.1, DQ464424.1, DQ464425.1, DQ641256.1, DQ653410.1, DQ840512.1, DQ840513.1, E02783.1, EF036469.1, EF577506.1, EU646197.1, EU646198.1, EU789558.1, EU789559.1, EU888930.1, FN392885.1, FN392886.1, FN436019.1, FN908083.1, FN908084.1, FN984917.1, GU124613.1, GU124614.1, GU227372.1, GU227388.1, GU227389.1, GU227390.1, GU227391.1, GU227392.1, GU227393.1, GU227394.1, GU227395.1, GU227396.1, GU227397.1, GU227398.1, GU230144.1, GU562823.1, GU569852.1, GU569853.1, GU905011.1, GU905012.1, HM235423.1, HQ634379.1, HQ634380.1, JN214358.1, JN214359.1, JN644767.1, JN644768.1, JN935881.1, JN935892.1, JN935893.1, JQ689949.1, JQ689950.1, JQ897386.1, JQ897387.1, JQ897388.1, KC512403.1, KC512404.1, KF419350.1, L08054.1, L22210.1, L24103.1, L33760.1, L33765.1, M23939.2, M86340.1, NR_024977.1, NR_024978.1, NR_024979.1, NR_024981.1, NR_024982.1, NR_024983.1, NR_024984.1, NR_024985.1, NR_024986.1, NR_024987.1, NR_024988.1, NR_025055.1, NR_025061.1, NR_025062.1, NR_025063.1, NR_025064.1, NR_025065.1, NR_025068.1, NR_025069.1, NR_025070.1, NR_025071.1, NR_025133.1, NR_025134.1, NR_025135.1, NR_025176.1, NR_025177.1, NR_025178.1, NR_025179.1, NR_025180.1, NR_025181.1, NR_025182.1, NR_025183.1, NR_025184.1, NR_025185.1, NR_025186.1, NR_025187.1, NR_025188.1, NR_025896.1, NR_025912.1, NR_025913.1, NR_025914.1, NR_025954.1, NR_025963.1, NR_025964.1, NR_025965.1, NR_025966.1, NR_025968.1, NR_025971.1, NR_025984.1, NR_025985.1, NR_025986.1, NR_025987.1, NR_025988.1, NR_025989.1, NR_026017.1, NR_026034.1, NR_026035.1, NR_026036.1, NR_026037.1, NR_026155.1, NR_029174.1, NR_029175.1, NR_029180.1, NR_029181.1, NR_029183.1, NR_036954.1, NR_037123.1, NR_041844.1, NR_041846.1, NR_042948.1, NR_043138.1, NR_044664.1, NR_044668.1, NR_044673.1, NR_044767.1, NR_044772.1, NR_044811.1, NR_074135.1, NR_074289.1, NR_074301.1, NR_074611.1, NR_074620.1, NR_102477.1, NR_103942.1, NR_104953.1, U02968.1, U04644.1, U04645.1, U04646.1, U04648.1, U04649.1, U04650.1, U04651.1, U04652.1, U04653.1, U04654.1, U04655.1, U04656.1, U09786.1, U09787.1, U09788.1, U15794.1, U15795.1, U16323.1, U16758.1, U16759.1, U16760.1, U19768.1, U22013.1, U22415.1, U26036.1, U26042.1, U26043.1, U26045.1, U26047.1, U26049.1, U26051.1, U26053.1, U26055.1, U29676.1, U44763.1, U44764.1, U44765.1, U44768.1, U44769.1, U44771.1, U56733.1, U58504.1, U58997.1, U67943.1, U67944.1, U67945.1, U67946.1, U83502.1, U83663.1, X00921.1, X62699.1, X76560.1, and Y00149.1.
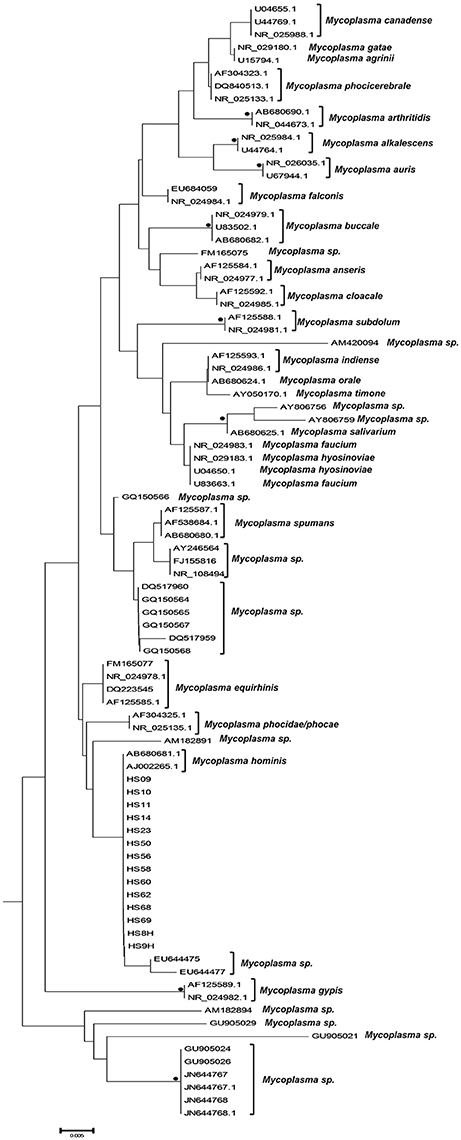
Figure 2. Subtree 1 depicting sequences clustering with Mycoplasma hominis strains. Bullets represent significant clustering as indicated by bootstrapping (>75% of permuted trees). The strains of the present study are designated with “HS” and the respective number.
A bigger group of 34 sequences created a unique cluster with the GenBank accession numbers DS08: KY781830, HS01: KY781831, HS02: KY781832, HS03: KY781833, HS3H: KY781834, HS06: KY781835, HS6H: KY781836, HS08: KY781837, HS12: KY781843, HS13: KY781844, HS16: KY781847, HS18: KY781849, HS19: KY781850, HS21: KY781851, HS25: KY781853, HS27: KY781855, HS30: KY781857, HS31: KY781858, HS32: KY781859, HS33: KY781860, HS35: KY781861, HS37: KY781863, HS39: KY781865, HS40: KY781866, HS41: KY781867, HS42: KY781868, HS43: KY781869, HS44: KY781870, HS46: KY781871, HS47: KY781872, HS52: KY781874, HS57: KY781877, HS59: KY781879, HS67: KY781884 (Figure 3). 32 of them were clustered with six unidentified Mycoplasma strains (HG764212, JX871253, JX508800, HG764211, HG746210, HG746209) and with a new species, Candidatus M. girerdii (CP007711). The high bootstrap support and the alignment between the sequences of these strains compared to the reference sequence, led to the conclusion that they also belong to the same Mycoplasma species. The remaining strains (DS08, HS27) form a distinct cluster that relates distantly with the Candidatus M. girerdii group, suggesting a possible new unidentified Mycoplasma species associated with T. vaginalis infection.
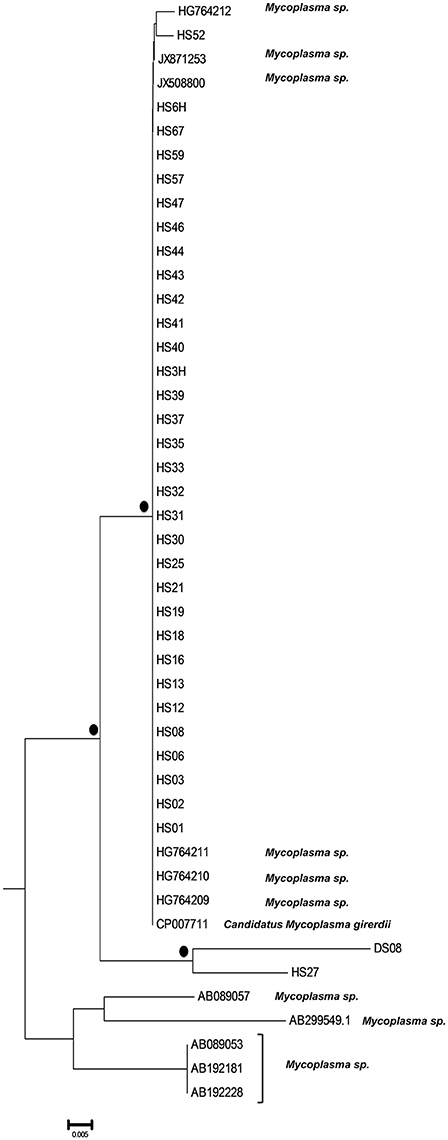
Figure 3. Subtree 2 depicting sequences clustering with Candidatus Mycoplasma girerdii strains, and strains of novel Mycoplasma species. Bullets represent significant clustering as indicated by bootstrapping (>75% of permuted trees). The strains of the present study are designated with “HS” or “DS” and the respective number.
Finally, six strains were clustered with Ureaplasma strains (GenBank accession numbers HS15: KY781846, HS29: KY781856, HS36: KY781862, HS38: KY781864, HS61: KY781881, and HS64: KY798518) of which five were associated with 2 U. parvum serovar three reference strains (NR074176, NR074762) and one with U. urealyticum serovar 10 reference strain (NR102836) (Figure 4).
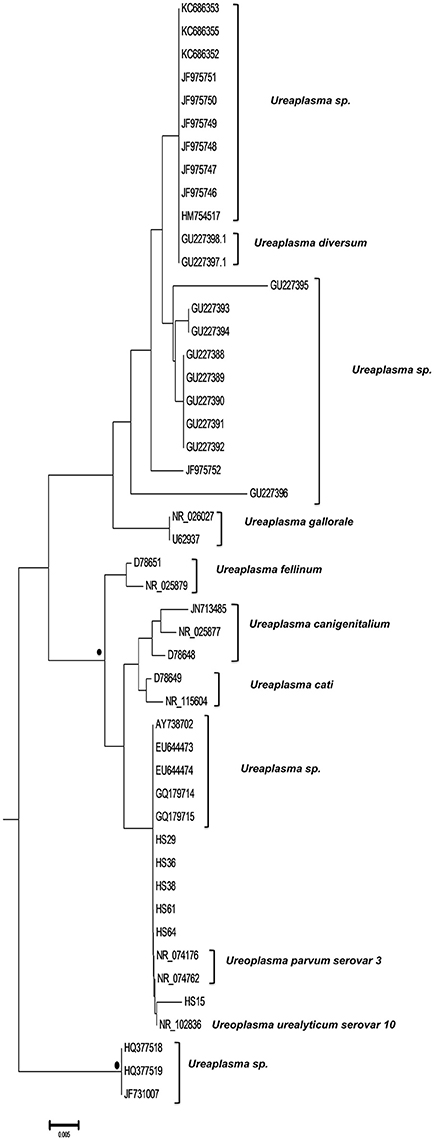
Figure 4. Subtree 3 depicting sequences clustering with Ureaplasma spp. strains. Bullets represent significant clustering as indicated by bootstrapping (>75% of permuted trees). The strains of the present study are designated with “HS” and the respective number.
Discussion
Aspects regarding the pathogenic bacteria symbiosis with protozoan vectors (entry mechanism, virulence alteration, cell cycle) have paved the way toward deciphering symbiotic relationships though the exact principles remain unknown (Catta-Preta et al., 2015). To this end, the presence of Mollicutes within T. vaginalis isolates has been investigated and special interest has been raised regarding the dualism of the beneficial outcome of this co-existence. Are mycoplasmas with the annotated small genome size and the absent cell wall the only ones who benefit from this interplay? To what extent does this symbiosis enhance the existence and favor the virulence of T. vaginalis?
Previous studies have shown that the protist tolerates and even favors the growth of the intracellularly located mycoplasmas, which benefit a safe niche protected from the hostile host environment (Dessi et al., 2005). T. vaginalis is an exclusively human pathogen, primarily transmitted through sexual contact; thus, all the mycoplasmas and the unidentified species of the present study should be of human origin, most probably acquired by the protozoan through consecutive passages in the various human hosts along the transmission chain of the genital tract infection (Johnston and Mabey, 2008; Harp and Chowdhury, 2011).
According to the Trichomonadaceae culture results of our study, only 2% of the T. vaginalis strains detected in the vaginal swabs harbor Mycoplasma/Ureaplasma spp. implying that the extracellular colonization of the vaginal mucosa might be independent from the intracellular. Since the conventional cultural method is solely suitable for the growth and identification of M. hominis and U. urealyticum, the extracellular presence of a Mycoplasma spp. other than M. hominis or U. urealyticum in the vaginal epithelium cannot be excluded. In our case, molecular techniques and strain typing have offered a clearer picture on mycoplasmal biology. However, the presence of abundant genes with high diversity pose technical and methodological limitations regarding the PCR and sequencing analysis leading to non-reliable results when it comes for mixed infections (Kanagawa, 2003). Herein, such a limitation would restrict the identification of more than one Mycoplasma or Ureaplasma species within the same specimen. Next-generation sequencing might contribute in detecting potential T. vaginalis co-infections with multiple bacterial species.
The results from the quantitative PCR of the filtrates indicated the absence of T. vaginalis DNA contamination by extracellularly located mycoplasmas implying that the T. vaginalis rich culture sediments were pure. According to the phylogenetic analysis, the majority of the strains identified belong to species other than M. hominis. Both the unclassified Mycoplasma strains EU644475 and EU644477 detected in our study, have been previously isolated from amniotic fluid and connected with intra-amniotic infection and inflammation leading to preterm birth (PTB; Han et al., 2009).
The novelty of the current study is the confirmation of the ability of Candidatus M. girerdii and other newly reported and previously unknown Mycoplasma spp. to survive and multiply within T. vaginalis. Candidatus M. girerdii has been previously detected in co-infection cases with T. vaginalis, as an emerging Mycoplasma spp. associated with trichomoniasis. However, there is no evidence to confirm the endosymbiotic relationship of the two pathogens (Martin et al., 2013; Fettweis et al., 2014). Candidatus M. girerdii as well as other unknown Mycoplasma spp. have been described in association with T. vaginalis but only as directly detected in vaginal discharge and not in pure T. vaginalis cultures (Martin et al., 2013; Allen-Daniels et al., 2015). Strains HG764209, HG746210, HG746211, HG746212 have been previously isolated as part of the microbiome in low birth-weight infants (Costello et al., 2013), JX871253 has been identified as part of the vaginal microbiome correlating with PTB (Hyman et al., 2014) and JX508800 has been associated with T. vaginalis infection as a member of the Mycoplasma genus (Martin et al., 2013).
With respect to the Ureaplasma spp. most studies have focused on the implication of the bacteria in the antenatal as well as the perinatal period. The absence of reports regarding the symbiosis of T. vaginalis with ureaplasmas sets a serious limitation in understanding the possible relationship between the protozoan and the bacteria (van der Schee et al., 2001; Diaz et al., 2010). In this research, we report for the first time the presence of intracellularly hosted ureaplasmas clustered in both known and unknown species. Interestingly, we identified five unclassified Ureaplasma spp. strains as intracellularly hosted within T. vaginalis isolates of which three have already been reported in the literature and correlated to genital infections as extracellular pathogens. Specifically, AY738702 has been previously isolated from cases of bacterial vaginosis (Oakley et al., 2008), and EU644473 as well as EU644474 have been identified as etiologic agents of intra-amniotic infection and inflammation leading to PTB (Han et al., 2009).
After all, this type of endosymbiosis can trigger challenging-to-handle pathologic phenomena associated with both pathogens involved in the infection. The protozoan-bacterial co-existence with the annotated ability to upregulate the inflammatory process, is reported to be associated with an increased risk for cervical cancer, peripartum complications (chorioamnionitis, PTB, low birth weight), endometritis, pelvic inflammatory disease and complications in women with human immuno-deficiency virus (HIV) infection (Tsai et al., 1995; Huang et al., 2001; Larsen and Hwang, 2010; Djigma et al., 2011; Haggerty and Taylor, 2011; Taylor-Robinson and Lamont, 2011; Thurman and Doncel, 2011; Fraga et al., 2012; Sinkovics, 2012; Fichorova et al., 2013). Another aspect is the clinical importance of the new Mycoplasma spp. and the association with specific pathological entities. As supported by the scientific literature, mycoplasma-infected T. vaginalis strains present inconsistent results regarding the apparition of metronidazole resistance; in the majority of the studies, no statistically significant association links T. vaginalis strains infected by M. hominis with metronidazole resistant phenotypes (Xiao et al., 2006; Butler et al., 2010; da Luz Becker et al., 2015). However, drug resistance acquired when unidentified bacteria are hosted in T. vaginalis sets an interesting yet important burden in the course of the infection which may complicate the currently applied therapeutic practices and needs further exploration. The role of the different T. vaginalis strains in the acquisition or preference of certain bacteria also remains undetermined; however, such a finding would shed light on the clinical impact of this symbiosis within the human host.
As mentioned above, we report for the first time the presence of T. vaginalis endosymbionts other than the widely investigated M. hominis. Other studies investigating the T. vaginalis isolates infection by M. hominis usually perform several methodologies to confirm the symbiotic relationship and clarify the clinical implications of this partnership. The most prevailing techniques applied for this reason include (i) immunofluorescence staining assays which differentiate the intracellularly from the extracellularly located mycoplasmas; (ii) microscopy to identify the cellular composition of the internalized mycoplasmas after being endocytosed by T. vaginalis; (iii) cytochemistry of acid phosphatase to trace the mycoplasmal degradation products, and (iv) PCR analysis for the mycoplasmal DNA detection within the protozoan cells (Dessi et al., 2005; Vancini and Benchimol, 2008). In our case, the intracellular location of the Mollicutes and the absence of cross-contamination or extracellularly-located species were substantiated by (i) daily passages of the T. vaginalis isolates in cultivation media enriched with chloramphenicol, (ii) supernatants filtration and verification of the mycoplasmal DNA absence in the filtered supernatant by quantitative real-time PCR, and (iii) the diversity among the different genera and even within the same species.
One possible limitation of our experimental methodology could be the absence of further evidence supporting the outcomes of this partnership such as the evaluation of the inflammatory process, phagocytosis, and haemolysis following the dual activity of the protozoan and the bacteria (Cirillo et al., 1997; Dessi et al., 2005, 2006; Vancini et al., 2008; Vancini and Benchimol, 2008; Morada et al., 2010; Fiori et al., 2013). Therefore, a future aspect that could follow this study is the confirmation of the endosymbiotic relationship of the protist hosting M. hominis as well as the newly identified Mollicutes with the traditional techniques of staining, microscopy, and immunological profiling of the host-pathogen interaction.
Conclusion
T. vaginalis strains isolated from vaginal infection specimens might host intracellular mycoplasmas, which comes in accordance with our findings presenting an equivocal rate of T. vaginalis strains negative and positive for hosting Mollicutes. The present study aimed to investigate the existence of intracellularly located bacteria as well as the classification of the Mycoplasma and Ureaplasma spp. based upon the phylogenetic analysis. The added value of this work is the confirmation of the relatively new species Candidatus M. girerdii as a T. vaginalis endosymbiont and the first reports on the symbiotic relationship between the protist and the Ureaplasma spp. This work lays the foundation for further investigation of the pathobiology of U. parvum in trichomoniasis and the role that ureaplasmas play in genital infections and not solely in obstetric infections. Finally, a more sophisticated research strategy is needed to verify whether the novel species identified in this study could play a role as endosymbionts in T. vaginalis, act as extracellular commensals or pathogens in human vaginitis. Molecular detection in asymptomatic women and women with non-trichomonas infection could serve as a potent method to answer this question. The symbiotic relationship between T. vaginalis and Mycoplasma/Ureaplasma spp. discloses novel insights in the “pathogen-to-pathogen” interaction providing a paradigm in “microbial socialization” where “unity is strength.”
Author Contributions
AI, MT, and SC contributed to the conception and design of the study, data analysis and interpretation and drafted the manuscript. EM was in charge of the sequence and phylogenetic analyses. KT included patients, and critically reviewed the manuscript. AB drafted and critically reviewed the manuscript. PP, VI, and MM carried out the laboratory work of the study, contributed to the data collection and analysis, and critically reviewed the manuscript. All authors have contributed intellectually during the writing process, have read the final manuscript and approved the contents.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Department of Medical Biopathology and Clinical Microbiology of the Medical School, Aeginition Hospital, National and Kapodistrian University of Athens.
References
Allen-Daniels, M. J., Serrano, M. G., Pflugner, L. P., Fettweis, J. M., Prestosa, M. A., Koparde, V. N., et al. (2015). Identification of a gene in Mycoplasma hominis associated with preterm birth and microbial burden in intraamniotic infection. Am. J. Obstet. Gynecol. 212, 779 e1–79.e13. doi: 10.1016/j.ajog.2015.01.032
Arroyo, R., Engbring, J., and Alderete, J. F. (1992). Molecular basis of host epithelial cell recognition by Trichomonas vaginalis. Mol. Microbiol. 6, 853–862. doi: 10.1111/j.1365-2958.1992.tb01536.x
Bar, A. K., Phukan, N., Pinheiro, J., and Simoes-Barbosa, A. (2015). The interplay of host microbiota and parasitic protozoans at mucosal interfaces: implications for the outcomes of infections and diseases. PLoS Negl. Trop. Dis. 9:e0004176. doi: 10.1371/journal.pntd.0004176
Butler, S. E., Augostini, P., and Secor, W. E. (2010). Mycoplasma hominis infection of Trichomonas vaginalis is not associated with metronidazole-resistant trichomoniasis in clinical isolates from the United States. Parasitol. Res. 107, 1023–1027. doi: 10.1007/s00436-010-1975-y
Campos, G. B., Lobao, T. N., Selis, N. N., Amorim, A. T., Martins, H. B., Barbosa, M. S., et al. (2015). Prevalence of Mycoplasma genitalium and Mycoplasma hominis in urogenital tract of Brazilian women. BMC Infect. Dis. 15:60. doi: 10.1186/s12879-015-0792-4
Catta-Preta, C. M., Brum, F. L., da Silva, C. C., Zuma, A. A., Elias, M. C., de Souza, W., et al. (2015). Endosymbiosis in trypanosomatid protozoa: the bacterium division is controlled during the host cell cycle. Front. Microbiol. 6:520. doi: 10.3389/fmicb.2015.00520
Cirillo, J. D., Falkow, S., Tompkins, L. S., and Bermudez, L. E. (1997). Interaction of Mycobacterium avium with environmental amoebae enhances virulence. Infect. Immun. 65, 3759–3767.
Costello, E. K., Carlisle, E. M., Bik, E. M., Morowitz, M. J., and Relman, D. A. (2013). Microbiome assembly across multiple body sites in low-birthweight infants. MBio 4, e00782–e00713. doi: 10.1128/mbio.00782-13
Cox, C., Saxena, N., Watt, A. P., Gannon, C., McKenna, J. P., Fairley, D. J., et al. (2016). The common vaginal commensal bacterium Ureaplasma parvum is associated with chorioamnionitis in extreme preterm labor. J. Matern. Fetal Neonatal Med. 29, 3646–3651. doi: 10.3109/14767058.2016.1140734
da Luz Becker, D., dos Santos, O., Frasson, A. P., de Vargas Rigo, G., Macedo, A. J., and Tasca, T. (2015). High rates of double-stranded RNA viruses and Mycoplasma hominis in Trichomonas vaginalis clinical isolates in South Brazil. Infect. Genet. Evol. 34, 181–187. doi: 10.1016/j.meegid.2015.07.005
Dessi, D., Delogu, G., Emonte, E., Catania, M. R., Fiori, P. L., and Rappelli, P. (2005). Long-term survival and intracellular replication of Mycoplasma hominis in Trichomonas vaginalis cells: potential role of the protozoon in transmitting bacterial infection. Infect. Immun. 73, 1180–1186. doi: 10.1128/IAI.73.2.1180-1186.2005
Dessi, D., Rappelli, P., Diaz, N., Cappuccinelli, P., and Fiori, P. L. (2006). Mycoplasma hominis and Trichomonas vaginalis: a unique case of symbiotic relationship between two obligate human parasites. Front. Biosci. 11, 2028–2034. doi: 10.2741/1944
Diaz, N., Dessi, D., Dessole, S., Fiori, P. L., and Rappelli, P. (2010). Rapid detection of coinfections by Trichomonas vaginalis, Mycoplasma hominis, and Ureaplasma urealyticum by a new multiplex polymerase chain reaction. Diagn. Microbiol. Infect. Dis. 67, 30–36. doi: 10.1016/j.diagmicrobio.2009.12.022
Djigma, F., Ouedraogo, C., Sagna, T., Ouermi, D., Sanogo, K., Bisseye, C., et al. (2011). HIV-infected women of Burkina Faso: a “reservoir” of mycoplasma infection. J. Infect. Dev. Ctries. 5, 176–181. doi: 10.3855/jidc.950
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Ferandon, C., Peuchant, O., Janis, C., Benard, A., Renaudin, H., Pereyre, S., et al. (2011). Development of a real-time PCR targeting the yidC gene for the detection of Mycoplasma hominis and comparison with quantitative culture. Clin. Microbiol. Infect. 17, 155–159. doi: 10.1111/j.1469-0691.2010.03217.x
Fettweis, J. M., Serrano, M. G., Huang, B., Brooks, J. P., Glascock, A. L., Sheth, N. U., et al. (2014). An emerging mycoplasma associated with trichomoniasis, vaginal infection and disease. PLoS ONE 9:e110943. doi: 10.1371/journal.pone.0110943
Fichorova, R. N., Buck, O. R., Yamamoto, H. S., Fashemi, T., Dawood, H. Y., Fashemi, B., et al. (2013). The villain team-up or how Trichomonas vaginalis and bacterial vaginosis alter innate immunity in concert. Sex. Transm. Infect. 89, 460–466. doi: 10.1136/sextrans-2013-051052
Fiori, P. L., Diaz, N., Cocco, A. R., Rappelli, P., and Dessi, D. (2013). Association of Trichomonas vaginalis with its symbiont Mycoplasma hominis synergistically upregulates the in vitro proinflammatory response of human monocytes. Sex. Transm. Infect. 89, 449–454. doi: 10.1136/sextrans-2012-051006
Fraga, J., Rodriguez, N., Fernandez, C., Mondeja, B., Sariego, I., Fernandez-Calienes, A., et al. (2012). Mycoplasma hominis in Cuban Trichomonas vaginalis isolates: association with parasite genetic polymorphism. Exp. Parasitol. 131, 393–398. doi: 10.1016/j.exppara.2012.05.003
Frolund, M., Bjornelius, E., Lidbrink, P., Ahrens, P., and Jensen, J. S. (2014). Comparison between culture and a multiplex quantitative real-time polymerase chain reaction assay detecting Ureaplasma urealyticum and U. parvum. PLoS ONE 9:e102743. doi: 10.1371/journal.pone.0102743
Glaser, K., and Speer, C. P. (2015). Neonatal CNS infection and inflammation caused by Ureaplasma species: rare or relevant? Expert Rev. Anti Infect. Ther. 13, 233–248. doi: 10.1586/14787210.2015.999670
Goodman, R. P., Ghabrial, S. A., Fichorova, R. N., and Nibert, M. L. (2011). Trichomonasvirus: a new genus of protozoan viruses in the family Totiviridae. Arch. Virol. 156, 171–179. doi: 10.1007/s00705-010-0832-8
Haggerty, C. L., and Taylor, B. D. (2011). Mycoplasma genitalium: an emerging cause of pelvic inflammatory disease. Infect. Dis. Obstet. Gynecol. 2011:959816. doi: 10.1155/2011/959816
Han, Y. W., Shen, T., Chung, P., Buhimschi, I. A., and Buhimschi, C. S. (2009). Uncultivated bacteria as etiologic agents of intra-amniotic inflammation leading to preterm birth. J. Clin. Microbiol. 47, 38–47. doi: 10.1128/JCM.01206-08
Harp, D. F., and Chowdhury, I. (2011). Trichomoniasis: evaluation to execution. Eur. J. Obstet. Gynecol. Reprod. Biol. 157, 3–9. doi: 10.1016/j.ejogrb.2011.02.024
Hirt, R. P., and Sherrard, J. (2015). Trichomonas vaginalis origins, molecular pathobiology and clinical considerations. Curr. Opin. Infect. Dis. 28, 72–79. doi: 10.1097/QCO.0000000000000128
Huang, S., Li, J. Y., Wu, J., Meng, L., and Shou, C. C. (2001). Mycoplasma infections and different human carcinomas. World J. Gastroenterol. 7, 266–269. doi: 10.3748/wjg.v7.i2.266
Hyman, R. W., Fukushima, M., Jiang, H., Fung, E., Rand, L., Johnson, B., et al. (2014). Diversity of the vaginal microbiome correlates with preterm birth. Reprod. Sci. 21, 32–40. doi: 10.1177/1933719113488838
Johnston, V. J., and Mabey, D. C. (2008). Global epidemiology and control of Trichomonas vaginalis. Curr. Opin. Infect. Dis. 21, 56–64. doi: 10.1097/QCO.0b013e3282f3d999
Kanagawa, T. (2003). Bias and artifacts in multitemplate polymerase chain reactions (PCR). J. Biosci. Bioeng. 96, 317–323. doi: 10.1016/S1389-1723(03)90130-7
Kissinger, P. (2015). Epidemiology and treatment of trichomoniasis. Curr. Infect. Dis. Rep. 17:484. doi: 10.1007/s11908-015-0484-7
Koch, A., Bilina, A., Teodorowicz, L., and Stary, A. (1997). Mycoplasma hominis and Ureaplasma urealyticum in patients with sexually transmitted diseases. Wien. Klin. Wochenschr. 109, 584–589.
Kong, F., James, G., Ma, Z., Gordon, S., Bin, W., and Gilbert, G. L. (1999). Phylogenetic analysis of Ureaplasma urealyticum–support for the establishment of a new species, Ureaplasma parvum. Int. J. Syst. Bacteriol. 49(Pt 4), 1879–1889.
Kupferberg, A. B., Johnson, G., and Sprince, H. (1948). Nutritional requirements of Trichomonas vaginalis. Proc. Soc. Exp. Biol. Med. 67, 304–308. doi: 10.3181/00379727-67-16285
Larsen, B., and Hwang, J. (2010). Mycoplasma, Ureaplasma, and adverse pregnancy outcomes: a fresh look. Infect. Dis. Obstet. Gynecol. 2010:521921. doi: 10.1155/2010/521921
Liu, W., Fang, L., Li, M., Li, S., Guo, S., Luo, R., et al. (2012). Comparative genomics of Mycoplasma: analysis of conserved essential genes and diversity of the pan-genome. PLoS ONE 7:e35698. doi: 10.1371/journal.pone.0035698
Margarita, V., Rappelli, P., Dessi, D., Pintus, G., Hirt, R. P., and Fiori, P. L. (2016). Symbiotic association with Mycoplasma hominis can influence growth rate, ATP production, cytolysis and inflammatory response of Trichomonas vaginalis. Front. Microbiol. 7:953. doi: 10.3389/fmicb.2016.00953
Martin, D. H., Zozaya, M., Lillis, R. A., Myers, L., Nsuami, M. J., and Ferris, M. J. (2013). Unique vaginal microbiota that includes an unknown Mycoplasma-like organism is associated with Trichomonas vaginalis infection. J. Infect. Dis. 207, 1922–1931. doi: 10.1093/infdis/jit100
McIver, C. J., Rismanto, N., Smith, C., Naing, Z. W., Rayner, B., Lusk, M. J., et al. (2009). Multiplex PCR testing detection of higher-than-expected rates of cervical mycoplasma, ureaplasma, and trichomonas and viral agent infections in sexually active australian women. J. Clin. Microbiol. 47, 1358–1363. doi: 10.1128/JCM.01873-08
Mendoza-Lopez, M. R., Becerril-Garcia, C., Fattel-Facenda, L. V., Avila-Gonzalez, L., Ruiz-Tachiquin, M. E., Ortega-Lopez, J., et al. (2000). CP30, a cysteine proteinase involved in Trichomonas vaginalis cytoadherence. Infect. Immun. 68, 4907–4912. doi: 10.1128/IAI.68.9.4907-4912.2000
Morada, M., Manzur, M., Lam, B., Tan, C., Tachezy, J., Rappelli, P., et al. (2010). Arginine metabolism in Trichomonas vaginalis infected with Mycoplasma hominis. Microbiol. 156(Pt 12), 3734–3743. doi: 10.1099/mic.0.042192-0
Oakley, B. B., Fiedler, T. L., Marrazzo, J. M., and Fredricks, D. N. (2008). Diversity of human vaginal bacterial communities and associations with clinically defined bacterial vaginosis. Appl. Environ. Microbiol. 74, 4898–4909. doi: 10.1128/AEM.02884-07
Olsen, B., Lan, P. T., Stalsby Lundborg, C., Khang, T. H., and Unemo, M. (2009). Population-based assessment of Mycoplasma genitalium in Vietnam–low prevalence among married women of reproductive age in a rural area. J. Eur. Acad. Dermatol. Venereol. 23, 533–537. doi: 10.1111/j.1468-3083.2009.03117.x
Pandelidis, K., McCarthy, A., Chesko, K. L., and Viscardi, R. M. (2013). Role of biofilm formation in Ureaplasma antibiotic susceptibility and development of bronchopulmonary dysplasia in preterm neonates. Pediatr. Infect. Dis. J. 32, 394–398. doi: 10.1097/INF.0b013e3182791ae0
Pillay, A., Radebe, F., Fehler, G., Htun, Y., and Ballard, R. C. (2007). Comparison of a TaqMan-based real-time polymerase chain reaction with conventional tests for the detection of Trichomonas vaginalis. Sex. Transm. Infect. 83, 126–129. doi: 10.1136/sti.2006.022376
Rappelli, P., Carta, F., Delogu, G., Addis, M. F., Dessi, D., Cappuccinelli, P., et al. (2001). Mycoplasma hominis and Trichomonas vaginalis symbiosis: multiplicity of infection and transmissibility of M. hominis to human cells. Arch. Microbiol. 175, 70–74. doi: 10.1007/s002030000240
Razin, S., Yogev, D., and Naot, Y. (1998). Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 62, 1094–1156.
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Sena, A. C., Bachmann, L. H., and Hobbs, M. M. (2014). Persistent and recurrent Trichomonas vaginalis infections: epidemiology, treatment and management considerations. Expert Rev. Anti Infect. Ther. 12, 673–685. doi: 10.1586/14787210.2014.887440
Sinkovics, J. G. (2012). Molecular biology of oncogenic inflammatory processes. I. Non-oncogenic and oncogenic pathogens, intrinsic inflammatory reactions without pathogens, and microRNA/DNA interactions (Review). Int. J. Oncol. 40, 305–349. doi: 10.3892/ijo.2011.1248
Soper, D. (2004). Trichomoniasis: under control or undercontrolled? Am. J. Obstet. Gynecol. 190, 281–290. doi: 10.1016/j.ajog.2003.08.023
Sutton, M., Sternberg, M., Koumans, E. H., McQuillan, G., Berman, S., and Markowitz, L. (2007). The prevalence of Trichomonas vaginalis infection among reproductive-age women in the United States, 2001–2004. Clin. Infect. Dis. 45, 1319–1326. doi: 10.1086/522532
Tamura, K., and Nei, M. (1993). Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10, 512–526.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Taylor-Robinson, D., and Lamont, R. F. (2011). Mycoplasmas in pregnancy. BJOG 118, 164–174. doi: 10.1111/j.1471-0528.2010.02766.x
Thurman, A. R., and Doncel, G. F. (2011). Innate immunity and inflammatory response to Trichomonas vaginalis and bacterial vaginosis: relationship to HIV acquisition. Am. J. Reprod. Immunol. 65, 89–98. doi: 10.1111/j.1600-0897.2010.00902.x
Tsai, S., Wear, D. J., Shih, J. W., and Lo, S. C. (1995). Mycoplasmas and oncogenesis: persistent infection and multistage malignant transformation. Proc. Natl. Acad. Sci. U.S.A. 92, 10197–10201. doi: 10.1073/pnas.92.22.10197
Vancini, R. G., and Benchimol, M. (2008). Entry and intracellular location of Mycoplasma hominis in Trichomonas vaginalis. Arch. Microbiol. 189, 7–18. doi: 10.1007/s00203-007-0288-8
Vancini, R. G., Pereira-Neves, A., Borojevic, R., and Benchimol, M. (2008). Trichomonas vaginalis harboring Mycoplasma hominis increases cytopathogenicity in vitro. Eur. J. Clin. Microbiol. Infect. Dis. 27, 259–267. doi: 10.1007/s10096-007-0422-1
van der Schee, C., Sluiters, H. J., van der Meijden, W. I., van Beek, P., Peerbooms, P., Verbrugh, H., et al. (2001). Host and pathogen interaction during vaginal infection by Trichomonas vaginalis and Mycoplasma hominis or Ureaplasma urealyticum. J. Microbiol. Methods 45, 61–67. doi: 10.1016/S0167-7012(01)00224-X
van Kupperveld, F. J., van der Logt, J. T., Angulo, A. F., van Zoest, M. J., Quint, W. G., Niesters, H. G., et al. (1992). Genus- and species-specific identification of mycoplasmas by 16S rRNA amplification. Appl. Environ. Microbiol. 207, 2606–2615.
Viscardi, R. M. (2014). Ureaplasma species: role in neonatal morbidities and outcomes. Arch. Dis. Child. Fetal Neonatal Ed. 99, F87–F92. doi: 10.1136/archdischild-2012-303351
WHO (2016). Global Health Sector Strategy on Sexually Transmitted Infections, 2016–2021. Geneva: WHO Press.
Keywords: Trichomonas vaginalis, Mycoplasma, Ureaplasma, Candidatus Mycoplasma girerdii, phylogeny, endosymbiosis, trichomoniasis
Citation: Ioannidis A, Papaioannou P, Magiorkinis E, Magana M, Ioannidou V, Tzanetou K, Burriel AR, Tsironi M and Chatzipanagiotou S (2017) Detecting the Diversity of Mycoplasma and Ureaplasma Endosymbionts Hosted by Trichomonas vaginalis Isolates. Front. Microbiol. 8:1188. doi: 10.3389/fmicb.2017.01188
Received: 17 January 2017; Accepted: 12 June 2017;
Published: 28 June 2017.
Edited by:
Joerg Graf, University of Connecticut, United StatesReviewed by:
Meghan May, University of New England, United StatesSteven S. Witkin, Weill Cornell Medical College, United States
Copyright © 2017 Ioannidis, Papaioannou, Magiorkinis, Magana, Ioannidou, Tzanetou, Burriel, Tsironi and Chatzipanagiotou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stylianos Chatzipanagiotou, schatzipa@gmail.com; schatzi@med.uoa.gr
 Anastasios Ioannidis
Anastasios Ioannidis Panagiota Papaioannou1
Panagiota Papaioannou1 Emmanouil Magiorkinis
Emmanouil Magiorkinis Maria Magana
Maria Magana Angeliki R. Burriel
Angeliki R. Burriel Stylianos Chatzipanagiotou
Stylianos Chatzipanagiotou