- 1Central Veterinary Laboratory of Bingerville, LANADA, Bingerville, Ivory Coast
- 2Department of Bacteriology and Immunology, Veterinary and Agro-chemical Research Centre, Brussels, Belgium
- 3Department of Biomedical Sciences, Institute of Tropical Medicine, Antwerp, Belgium
- 4Research Unit of Epidemiology and Risk Analysis Applied to Veterinary Sciences (UREAR-ULg), Faculty of Veterinary Medicine, Fundamental and Applied Research for Animal and Health Center, University of Liège, Liège, Belgium
Brucellosis is one of the most widespread bacterial zoonotic diseases in the world, affecting both humans and domestic and wild animals. Identification and biotyping of field strains of Brucella are of key importance for a better knowledge of the epidemiology of brucellosis, for identifying appropriate antigens, for managing disease outbreaks and for setting up efficient preventive and control programmes. Such data are required both at national and regional level to assess potential threats for public health. Highly discriminative genotyping methods such as the multiple locus variable number of tandem repeats analysis (MLVA) allow the comparison and assessment of genetic relatedness between field strains of Brucella within the same geographical area. In this study, MLVA biotyping data retrieved from the literature using a systematic review were compared using a clustering analysis and the Hunter-Gaston diversity index (HGDI). Thus, the analysis of the 42 MLVA genotyping results found in the literature on West Africa [i.e., from Ivory Coast (1), Niger (1), Nigeria (34), The Gambia (3), and Togo (3)] did not allow a complete assessment of the actual diversity among field strains of Brucella. However, it provided some preliminary indications on the co-existence of 25 distinct genotypes of Brucella abortus biovar 3 in this region with 19 genotypes from Nigeria, three from Togo and one from Ivory Coast, The Gambia, and Niger. The strong and urgent need for more sustainable molecular data on prevailing strains of Brucella in this sub-region of Africa and also on all susceptible species including humans is therefore highlighted. This remains a necessary stage to allow a comprehensive understanding of the relatedness between field strains of Brucella and the epidemiology of brucellosis within West Africa countries.
Introduction
Brucellosis is one of the most widespread bacterial zoonotic diseases in the world, affecting both humans and domestic and wild animals (Maurin, 2005; Corbel, 2006). The disease is caused by Gram-negative facultative intracellular bacteria of the genus Brucella. According to the World Health Organization (WHO), about 500,000 new cases of human brucellosis are reported annually worldwide (Corbel, 1997; Pappas et al., 2006). In animals, brucellosis is responsible for many economic losses because of abortions, decrease in production (particularly reduced milk production), losses of calves, viable but weak calves, reproductive disorders, and costs of intervention. With its impact on productivity, this disease contributes worsening the deficit of animal protein especially for populations in developing countries, where food needs are continuously increasing (Perry, 2002). In areas where people's livelihood heavily depends on livestock, the impact of brucellosis might therefore exacerbate poverty (Cáceres, 2010).
In spite of its status as a neglected tropical disease, bovine brucellosis remains the most widespread disease in animals and the main concern in Sub-Saharan African countries (Akakpo and Bornarel, 1987; Corbel, 1997; McDermott and Arimi, 2002; Bronsvoort et al., 2009). For a better understanding of the epidemiology of bovine brucellosis, phenotypic, and genotypic knowledge on prevailing Brucella spp. are required in both human and animal hosts. Thus, Brucella causing brucellosis has been investigated throughout the years in different regions of the world including West Africa. In this part of Africa, the presence and the endemicity of brucellosis were confirmed, with Brucella abortus biovar 3 being the most commonly isolated strains in cattle (Sanogo et al., 2013a).
This paper compares and investigates the relatedness between the prevailing field strains of B. abortus biovar 3 from West Africa.
Materials and Methods
Study Area
With a surface area of 5 112 903 km2 representing a fifth of the African continent, West Africa is one of the four major regions of Sub-Saharan Africa. This region includes 14 countries including Benin, Burkina Faso, Cape Verde, The Gambia, Ghana, Guinea, Ivory Coast, Liberia, Mali, Niger, Nigeria, Senegal, Sierra Leone, and Togo (Figure 1). These countries comprise almost 25% of the cattle population of the continent with about 70 million heads of cattle of different types (Bos taurus type, Bos indicus type and crossbreds) (FAO, 2017). These cattle are mostly raised extensively in sedentary herds. This region is also characterized by the existence of frequent livestock movements between countries through transhumance or commercial exchanges.
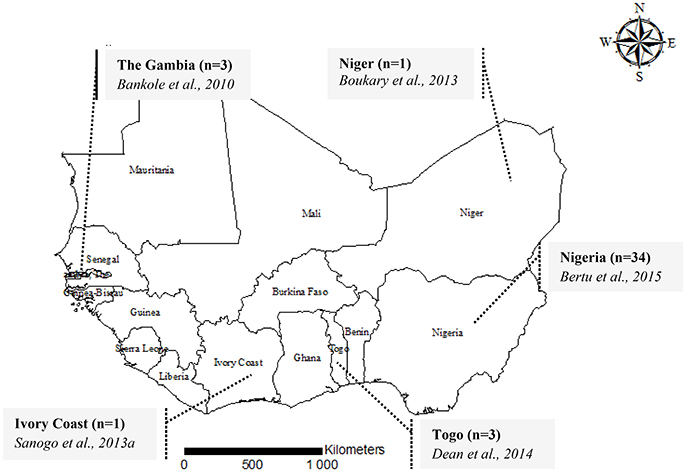
Figure 1. A map showing the geographical origin of MLVA genotyping data of Brucella abortus biovar 3 in West Africa, 2015. For each country, the number of isolates genotyped [i.e., Ivory Coast (n = 1)] is provided followed by the authors and the year of publication (i.e., Sanogo et al., 2013b).
Prevailing Field Strains of Brucella abortus Biovar 3 from West Africa
A Preferred Reporting Items of Systematic reviews and Meta-Analyses (PRISMA) approach (Moher et al., 2009) was used to identify available and accessible information in the literature on typing of prevailing field strains of Brucella in both human and animals through general internet search engines, including Google Scholar and PubMed, with no language and time period restrictions. The search strategy was adapted according to the database. Search terms were composed by combinations of keywords. In Google Scholar, “Brucellosis+Brucella+MLVA+typing+genotyping+Sub-saharan+Africa” was used while in PubMed, the following search algorithm was used: ((((Brucellosis) OR Brucella)) AND (((genotyping) OR typing) OR MLVA)) AND ((Africa) OR sub-Saharan Africa). Firstly, titles and abstracts were screened and available full texts were screened for relevant information. Thus, studies reporting information on genotyping data of field strains of B. abortus biovar 3 from sub-Saharan Africa and especially West Africa were considered and were given a particular focus for final inclusion. When provided, Multiple Locus Variable number of tandem repeats Analysis (MLVA) data [e.g., the number of repeats in a set of variable number of tandem repeats (VNTR) loci] were extracted from the selected paper, summarized, and subjected to further analysis. A flow diagram summarizing the literature search strategy is presented in Figure 2.
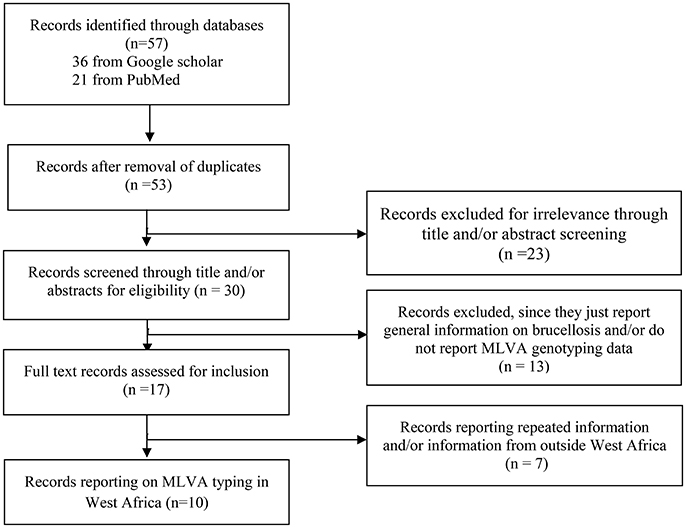
Figure 2. Flow diagram presenting a summary of the literature search on genotyping of field strains of Brucella from West Africa and other parts of Africa.
Multiple Locus Variable Number of Tandem Repeats Analysis
MLVA profiles of field strains of B. abortus biovar 3 isolated from West Africa were used in this study (Figure 1). Briefly, MLVA consists of the assessment of the number of repeats in a set of variable number of tandem repeats (VNTR) loci. In MLVA 16, two sets of VNTRs gathered into 8 microsatellite markers (panel 1: Bruce06, Bruce08, Bruce11, Bruce12, Bruce42, Bruce43, Bruce45, Bruce55) and 8 microsatellite markers (panel 2) comprising two groups (panel 2A: Bruce18, Bruce19, Bruce21; and panel 2B: Bruce04, Bruce07, Bruce09, Bruce16, Bruce30) are examined (Le Flêche et al., 2006; Maquart et al., 2009). The number of repetitions of each locus of each panel, constituting the MLVA profile, is derived from the size of the band of the PCR products (Le Flêche et al., 2006).
Comparison of MLVA Profiles
Diversity and relatedness among field strains of B. abortus biovar 3 from West Africa were assessed by calculating the Hunter-Gaston diversity index (HGDI), a numerical index measuring the probability that two strains consecutively taken from a given population would be placed into different typing groups (Hunter and Gaston, 1988) (http://www.hpa-bioinformatics.org.uk/cgi-bin/DICI/DICI.pl). The relatedness between the distinct MLVA profiles of West African strains and neighbor profiles originating from Africa in the public MLVA Brucella database on MLVAnet (http://mlva.u-psud.fr/brucella/) was also assessed with a Ward hierarchical clustering analysis using the hclust function and the cluster package in R software (http://www.r-project.org). Using results of a Ward linkage clustering analysis of the number of variable tandem repeats, a dendrogram of clustered MLVA profiles of West African strains was also generated. In order to assess potential relatedness with others prevailing strains from sub-Saharan Africa, comparison of the 25 distinct MLVA profiles from West Africa includes three lately published B. abortus biovar 3 MLVA profiles from Tanzania (Mathew et al., 2015) and five other sub-Saharan Africa B. abortus biovar 3 field strains and neighbor profiles from the Brucella MLVA database, namely Kenya (Muendo et al., 2012), Sudan, Uganda, and Chad (Le Flêche et al., 2006) (Table 1).
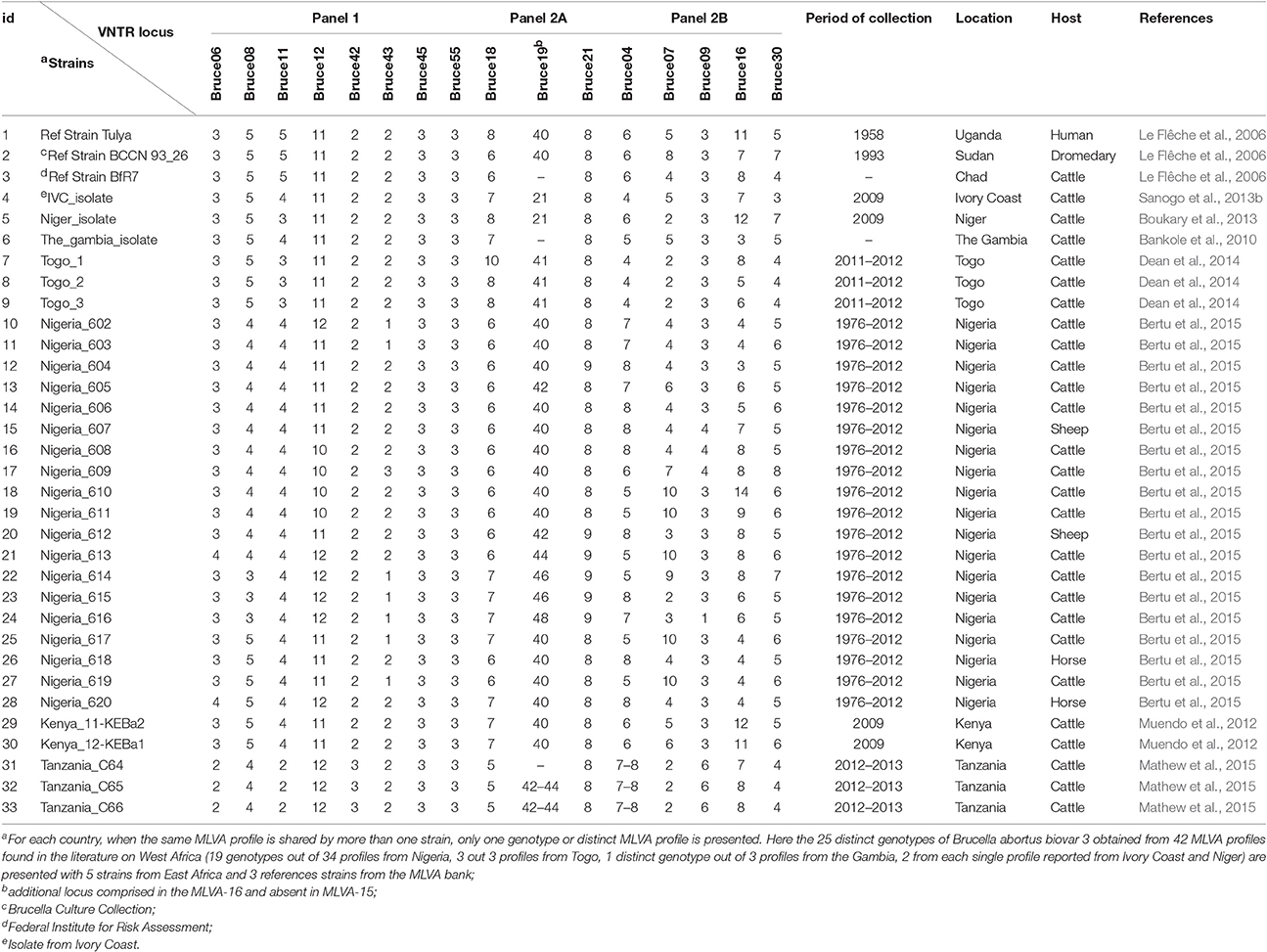
Table 1. Multiple Loci Variable Number Tandem Repeats analysis (MLVA) distinct profiles of West African isolates of B. abortus biovar 3 and some close neighbor profiles from Africa retrieved from literature and from the Brucella MLVA bank.
Results
In order to explore the genetic diversity of field strains of B. abortus biovar 3 from West Africa, available and accessible MLVA genotyping data were retrieved from the literature (Figures 1, 2). Among 57 published papers initially retrieved from the literature search, only 10 papers report MLVA genotyping data of West African B abortus biovar 3 strains. These 10 papers include four review papers reporting already published data covering sub-Saharan Africa in general (Boukary et al., 2013; Ducrotoy et al., 2017) and West Africa in particular (Sanogo et al., 2013a; Dean et al., 2014). Except from Nigeria, where strains came from both imported and autochthonous cattle and from sheep (n = 2) and horse (n = 2), strains originating from other countries were obtained from autochthonous cattle. None of the retrieved MLVA profiles were reported from humans so far. A total of 42 MLVA genotyping results were reported in the literature. Comparison of MLVA profiles of B. abortus biovar 3 field strains reported so far within West Africa revealed the presence of 25 distinct genotypes [e.g., a single genotype from the three strains isolated from The Gambia (Bankole et al., 2010), one from the unique strain from Niger (Boukary et al., 2013), one from the unique strain from Ivory Coast (Sanogo et al., 2013b), three genotypes from the three strains from Togo (Dean et al., 2014), and 19 genotypes from the 34 strains from Nigeria (Bertu et al., 2015)] (Figure 1).
While considering only panel 1 (MLVA 8), which is indicative of the species, diversity indexes of 0.620 (95% CI: 0.532–0.708), 0.580 (95% CI: 0.428–0.732), 0.477 (95% CI: 0.316–0.638), 0.280 (95% CI: 0.085–0.475), and 0.153 (95% CI: 0.000–0.332) were observed, respectively, at locus Bruce08, Bruce12, Bruce43, Bruc11, and Bruc06 with different genotypes. The others loci showed identical number of repeating units among the genotypes observed (e.g., Bruce42: 1; Bruce45: 1; Bruce55: 1) (Table 2). Highest diversity indexes were observed with the set of markers composing panel 2, especially at Bruce16 (HGDI = 0.870, 95% CI: 0.808–0.932), known as one of the most variable locus. Within this panel 2, while considering highly discriminative markers (i.e., Bruce04, Bruce07, Bruce09, Bruce16, and Bruce30), three to nine different alleles were found.
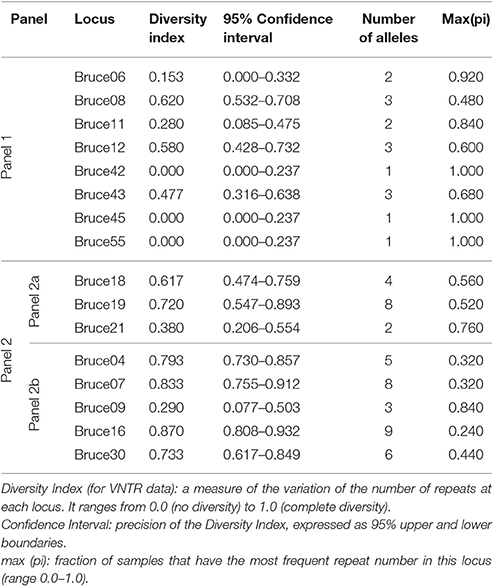
Table 2. The Hunter Gaston Diversity Index for different loci of West African field strains of B. abortus biovar 3 (i.e., from Ivory Coast, Niger, Nigeria, The Gambia, and Togo) based on MLVA 16 data.
Discussion
For many years, Brucella spp. causing bovine brucellosis were characterized using both phenotypic and genotypic methods. While B. abortus biovar 1 have been reported as the most encountered in cattle worldwide (Corbel, 1997), in the USA (Bricker et al., 2003), and in Latin America (Acha and Szyfres, 2003; Lucero et al., 2008; Minharro et al., 2013), B. abortus biovar 3 was predominant in both native cattle and buffalo from eastern Africa and China (Timm, 1982; Domenech et al., 1983). B. abortus biovar 3 was also identified as the most commonly isolated in cattle from West Africa and Sub-Saharan Africa (Sanogo et al., 2013a; Bertu et al., 2015). In West Africa, where only B. abortus was reported so far, field strains of B. abortus biovar 3 were characterized mostly in cattle using a combination of bacteriological phenotypic typing and MLVA genotyping approaches. These West African isolates were mostly characterized from autochthonous cattle and from hygroma fluid samples. Phenotypic methods consisted of bacteriological isolation and identification and relied on a combination of morphological, cultural, serological and biochemical characteristics in order to characterize suspicious colonies (Alton et al., 1988). However, phenotypic typing methods may fail to correctly classify or differentiate some strains as in Nigeria (Bertu et al., 2015). Therefore, conventional bacteriological identification needs to be supplemented by molecular methods such as the VNTR analysis (MLVA). MLVA is a powerful molecular tool for typing and for assessing the potential relationships between Brucella spp. isolates from different sources of infection and from different geographical origins. It is a particularly useful method to study the molecular epidemiology of Brucella where a high discriminatory power is required (Bricker et al., 2003; Cutler et al., 2005; Le Flêche et al., 2006). Wherever possible, more accurate and discriminative typing methods such as the enhanced AMOS-ery PCR and MLVA should be used in complementarity with conventional biotyping methods (Ocampo-Sosa et al., 2005; Bankole et al., 2010; Sanogo et al., 2013b; Dean et al., 2014; Bertu et al., 2015).
Using panel 1 (MLVA8), 10 genotypes were obtained while 18 genotypes were obtained using the combination of panel 1 and 2B (MLVA11). The analysis of the complete MLVA16 (panels 1, 2A and 2B) revealed 25 distinct genotypes. Clustering analysis of the different MLVA profiles suggested the co-existence of distinct clonal complexes (Figure 3). While the three strains isolated from The Gambia shared the same profile, distinct profiles co-existed in Nigeria and Togo. The Togolese strains appeared to be related to many Nigerian strains and isolates from The Gambia. On the other hand, isolates from Niger and Ivory Coast appeared to be genetically related. In Nigeria where distinct profiles also co-exist, some isolates were more related to eastern African isolates originating from Tanzania and Kenya. These observations might suggest a possible relation between African B. abortus biovar 3 strains. Indeed, despite the relative limited number of strains compared, these results provide some preliminary indications on the co-existence of different genetic profiles among the prevailing field strains of B. abortus biovar 3 in this sub-region (Dean et al., 2014; Bertu et al., 2015). This heterogeneity among B. abortus biovar 3 strains originating from Africa was already described, with the North African strains more closely related to European B. abortus biovar 3b strain lineage and the sub-Saharan African strains more related to B. abortus biovar 3a lineage (Ocampo-Sosa et al., 2005; Ica et al., 2008; Bertu et al., 2015; Mathew et al., 2015; Ducrotoy et al., 2017). However, despite the genotypic diversity observed, the closeness of most of sub-Saharan African strains with the human reference Tulya strain from Uganda, put forward the hypothesis of the possible dominance of lineage 3a among West African B. abortus biovar 3 (Bertu et al., 2015; Ducrotoy et al., 2017) and a possible common historical origin of brucellosis in this region. Indeed, this lineage commonly isolated in West Africa is known to be confined in the African continent where B. abortus is believed to originate (Whatmore et al., 2016). Such a hypothesis associated with the observed polymorphisms is in line with unrestricted livestock movements through transhumance and trade among countries composing this sub-region (OECD, 2008), which might favor frequent introduction and reintroduction of the pathogen. So far, data on prevailing strains of Brucella in both animal and human hosts are still scare and irregularly reported (Sanogo et al., 2013a). In order to challenge such hypothesis and allow a better understanding of the epidemiology of brucellosis in West Africa, more molecular typing results are needed. In West Africa, brucellosis (or evidence of its presence) was reported in most of the 14 countries so far (Mangen et al., 2002; Boukary et al., 2013). Adequate and efficient control of brucellosis in this region implies a comprehensive understanding of its epidemiology at West African region scale, in order to include prevailing strains causing the disease and adjust diagnostic tools. Indeed, additional data on prevailing field strain of Brucella are required to identify the sources of infection and to understand the transmission pathways of this infection between animals and from animal to humans (Adone and Pasquali, 2013).
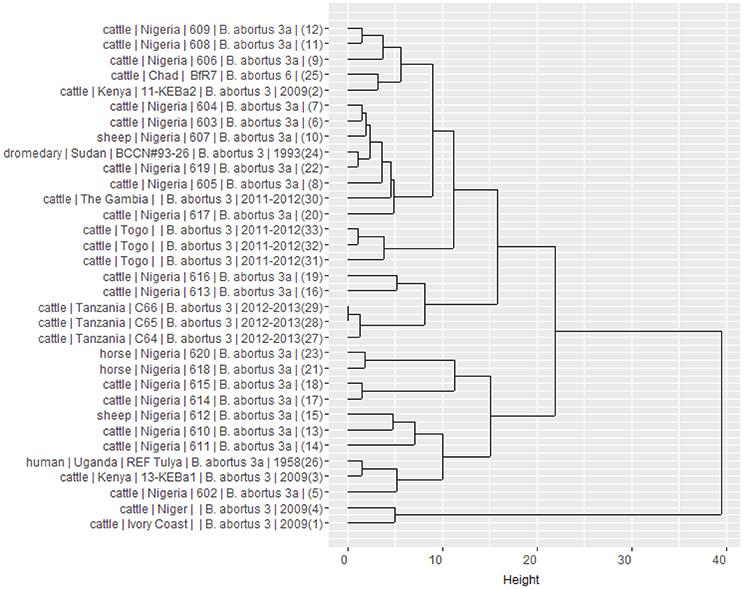
Figure 3. Dendrogram clustered MLVA profiles showing the relation between the 25 West African isolates of B. abortus biovar 3 and eight neighbor profiles from Africa retrieved in the literature and from the Brucella MLVA bank. It is built from results of a Ward linkage cluster analysis of the number of variable tandem repeats (VNTR) of the MLVA 16 loci. For each strain, information on host, country of origin, strain reference, species and biovar, year of isolation (when available) and number of order in the database are provided.
In conclusion, the number of strains analyzed in this study precludes an actual complete and comprehensive assessment of the relatedness of field strains of B. abortus biovar 3 in cattle from West Africa but provides preliminary indications on the co-existence of distinct profiles in this sub-region, in line with other recent findings (Bertu et al., 2015; Ducrotoy et al., 2017). More extended knowledge of prevailing strains in livestock and other hosts remains necessary to actually assess their diversity and to fully understand the molecular epidemiology of Brucella infection, distribution, and transmission patterns within West Africa and across the whole African continent (Godfroid et al., 2013). By allowing comparison among strains, MLVA genotyping methods would also be useful as a surveillance tool of the distribution of brucellosis in West Africa, where frequent movements of livestock between countries are expected to play a role in the spread of Brucellae. So far, MLVA data from West Africa are not available from MLVA public database. It might be therefore suggested that studies publishing MLVA typing results explicitly report, share details profiles and be more informative. This is particularly critical for a sub-region where resources and molecular epidemiological investigations are limited.
Formal collaboration between countries and their respective public health actors is required for sharing available information and for implementing harmonized surveillance and control strategies. Such collaboration coupled with adoption of the concept of “One health approach” would be particularly beneficial in a regional framework, especially in West Africa where national resources and capabilities for prevention, control and surveillance of infectious diseases of public importance such as brucellosis are still scarse (Saegerman et al., 2010, 2012; Marcotty et al., 2013). It is also essential to ensure a sustainable system of data collection on prevailing strains covering the whole West African region with a better coverage of other susceptible domestic and wild animals in order to document sources of human infections and to produce strong molecular evidence informing on the epidemiologic links between strains of Brucella within this region.
Author Contributions
MS performed the literature review and the clustering analyses. MS and CS wrote the manuscript and all authors including DF and ET reviewed, commented and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to the Institute of Tropical Medicine of Antwerp, to the bacteriology and immunology department of the Veterinary Agrochemical Research Centre of Brussels and the University of Liège in Belgium for their respective support. We are also grateful to Marie J. Ducrotoy, Virginie Mick, and Wilson Bertu for providing the MLVA profile from Nigeria.
References
Acha, P. N., and Szyfres, B. (eds.). (2003). “Brucellosis,” in Zoonoses and Communicable Diseases Common to Man and Animals: Vol 1: Bacterioses and Mycoses, 3rd Edn, (Washington, DC: Pan American Health Organization), 40–67.
Adone, R., and Pasquali, P. (2013). Epidemiosurveillance of brucellosis. Rev. Sci. Tech. Off. Int. Epiz. 32, 199–205. doi: 10.20506/rst.32.1.2202
Akakpo, A. J., and Bornarel, P. (1987). Epidémiologie des brucelloses animales en Afrique tropicale: enquêtes clinique, sérologique et bactériologique. Rev. Sci. Tech. Off. Int. Epiz. 6, 981–1027. doi: 10.20506/rst.6.4.313
Alton, G. G., Jones, L. M., Angus, R. D., and Verger, J. M. (1988). Techniques for the Brucellosis Laboratory. Paris: INRA.
Bankole, A. A., Saegerman, C., Berkvens, D., Fretin, D., Geerts, S., Ieven, G., et al. (2010). Phenotypic and genotypic characterisation of Brucella strains isolated from cattle in The Gambia. Vet. Rec. 166, 753–756. doi: 10.1136/vr.b4862
Bertu, W. J., Ducrotoy, M. J., Mu-oz, P. M., Mick, V., Zú-iga-Ripa, A., Bryssinckx, W., et al. (2015). Phenotypic and genotypic characterization of Brucella strains isolated from autochthonous livestock reveals the dominance of B. abortus biovar 3a in Nigeria. Vet. Microbiol. 180, 103–108. doi: 10.1016/j.vetmic.2015.08.014
Boukary, A. R., Saegerman, C., Abatih, E., Fretin, D., Alambédji Bada, R., De Deken, R., et al. (2013). Seroprevalence and potential risk factors for Brucella spp. infection in traditional cattle, sheep and goats reared in urban, periurban and rural areas of Niger. PLoS ONE 8:e83175. doi: 10.1371/journal.pone.0083175
Bricker, B. J., Ewalt, D. R., and Halling, S. M. (2003). Brucella “HOOFPrints”: strain typing by multi-locus analysis of variable number tandem repeats (VNTRs). BMC Microbiol. 3:15. doi: 10.1186/1471-2180-3-15
Bronsvoort, B. M. D., Koterwas, B., Land, F., Handel, I. G., Tucker, J., Morgan, K. L., et al. (2009). Comparison of a flow assay for brucellosis antibodies with the reference cELISA test in West African Bos indicus. PLoS ONE 4:e5221. doi: 10.1371/journal.pone.0005221
Cáceres, S. B. (2010). Comparative Veterinary Capacity in Western Africa: Implications for Livestock Development. Livestock Research for Rural Development. Available online at: http://www.lrrd.org/lrrd22/10/burg22180.htm (Accessed May 26, 2017).
Corbel, M. J. (1997). Brucellosis: an overview (1st International conference on emerging zoonosis). Emerg. Infect. Dis. 3, 213–221. doi: 10.3201/eid0302.970219
Corbel, M. J. (2006). Brucellosis in Humans and Animals. Geneva: World Health Organization (WHO) Press.
Cutler, S. J., Whatmore, A. M., and Commander, N. J. (2005). Brucellosis - new aspects of an old disease. J. Appl. Microbiol. 98, 1270–1281. doi: 10.1111/j.1365-2672.2005.02622.x
Dean, A. S., Schelling, E., Bonfoh, B., Kulo, A. E., Boukaya, G. A., and Pilo, P. (2014). Deletion in the gene BruAb2_0168 of Brucella abortus strains: diagnostic challenges. Clin. Microbiol. Infect. 20, O550–O553. doi: 10.1111/1469-0691.12554
Domenech, J., Corbel, M. J., Thomas, E. L., and Lucet, Ph. (1983). La brucellose bovine en Afrique centrale: VI. Identification et typage des souches isolées au Tchad et au Cameroun. Rev. Elev. Med. Vet. Pays Trop. 36, 19–25.
Ducrotoy, M., Bertu, W. J., Matope, G., Cadmus, S., Conde- Álvarez, R., Gusi, A. M., et al. (2017). Brucellosis in Sub-Saharan Africa: current challenges for management, diagnosis and control. Acta Trop. 165, 179–193. doi: 10.1016/j.actatropica.2015.10.023
FAO (Food And Agricultural Organization) (2017). FAOSTAT. Food and Agricultural Organization Statistic Division Available online at: http://www.fao.org/faostat/fr/#data/QA (Accessed May 26, 2017).
Godfroid, J., Al Dahouk, S., Pappas, G., Roth, F., Matope, G., Muma, J., et al. (2013). A “One Health” surveillance and control of brucellosis in developing countries: moving away from improvisation. Comp. Immunol. Microbiol. Infect. Dis. 36, 241–248. doi: 10.1016/j.cimid.2012.09.001
Hunter, P. R., and Gaston, M. A. (1988). Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26, 2465–2466.
Ica, T., Aydin, F., Erdenlig, S., Guler, L., and Büyükcangaz, E. (2008). Characterization of Brucella abortus biovar 3 isolates from Turkey as biovar 3b. Vet. Rec. 63, 659–661. doi: 10.1136/vr.163.22.659
Le Flêche, P., Jacques, I., Grayon, M., Al Dahouk, S., Bouchon, N. P., Denoeud, F., et al. (2006). Evaluation and selection of tandem repeat loci for a Brucella MLVA typing assay. BMC Microbiol. 6:9. doi: 10.1186/1471-2180-6-9
Lucero, N. E., Ayala, S. M., Escobar, G. I., and Jacob, N. R. (2008). Brucella isolated in humans and animals in Latin America from 1968 to 2006. Epidemiol. Infect. 136, 496–503. doi: 10.1017/S0950268807008795
Mangen, M. J., Otte, J., Pfeiffer, D., and Chilonda, P. (2002). Bovine Brucellosis in sub-Saharan Africa: Estimation of Sero-Prevalence and Impact on Meat and Milk Offtake Potential. Rome: FAO Livestock Information and Policy Branch; AGAL. Livestock Policy Paper n°8.
Maquart, M., Le Flêche, P., Foster, G., Tryland, M., Ramisse, F., Djønne, B., et al. (2009). MLVA-16 typing of 295 marine mammal Brucella isolates from different animal and geographic origins identifies 7 major groups within Brucella ceti and Brucella pinnipedialis. BMC Microbiol. 9:145. doi: 10.1186/1471-2180-9-145
Marcotty, T., Thys, E., Conrad, P., Godfroid, J., Craig, P., Zinsstag, J., et al. (2013). Intersectoral collaboration between the medical and veterinary professions in low-resource societies: the role of research and training institutions. Comp. Immunol. Microbiol. Infect. Dis. 36, 233–239. doi: 10.1016/j.cimid.2012.10.009
Mathew, C., Stokstad, M., Johansen, T. B., Klevar, S., Mdegela, R. H., Mwamengele, G., et al. (2015). First isolation, identification, phenotypic and genotypic characterization of Brucella abortus biovar 3 from dairy cattle in Tanzania. BMC Vet. Res. 11:156. doi: 10.1186/s12917-015-0476-8
Maurin, M. (2005). La brucellose à l'aube du 21ème siècle. Med. Maladies Infect. 35, 6–16. doi: 10.1016/j.medmal.2004.08.003
McDermott, J. J., and Arimi, S. M. (2002). Brucellosis in sub-Saharan Africa: epidemiology, control and impact. Vet. Microbiol. 90, 111–134. doi: 10.1016/S0378-1135(02)00249-3
Minharro, S., Mol, J. P., Dorneles, E. M. S., Barbosa, R. P., Neubauer, H., Melzer, F., et al. (2013). Biotyping and genotyping (MLVA16) of Brucella abortus isolated from cattle in Brazil, 1977 to 2008. PLoS ONE 8:e81152. doi: 10.1371/journal.pone.0081152
Moher, D., Liberati, A., Tetzlaff, J., and Altman, D. G., and The Prisma Group (2009). Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 6:e1000097. doi: 10.1371/journal.pmed.1000097
Muendo, E., Mbatha, P., Macharia, J., Abdoel, T., Janszen, P., Pastoor, R., et al. (2012). Infection of cattle in Kenya with Brucella abortus biovar 3 and Brucella melitensis biovar 1 genotypes. Trop. Anim. Health Prod. 44, 17–20. doi: 10.1007/s11250-011-9899-9
Ocampo-Sosa, A. A., Aguero-Balbin, J., and Garcia-Lobo, J. M. (2005). Development of a new PCR assay to identify Brucella abortus biovars 5, 6 and 9 and the new subgroup 3b of biovar 3. Vet Microbiol. 110, 41–51. doi: 10.1016/j.vetmic.2005.06.007
Organization for Economic Co-operation and Development (OECD) (2008). Livestock and Regional Market in the Sahel and West Africa: Potentials and Challenges. Paris. Available online at: http://www.oecd.org/swac/publications/41848366.pdf
Pappas, G., Papadimitriou, P., Akritidis, N., Christou, L., and Tsianos, E. V. (2006). The new global map of human brucellosis. Lancet Infect. Dis. 6, 91–99. doi: 10.1016/S1473-3099(06)70382-6
Perry, B. (2002). “Chapter 7: animal disease impact on the poor: study results,” in Investing in Animal Research to Alleviate Poverty. Nairobi: International Livestock Research Institute. Available online at: http://ilri.org/infoserv/Webpub/fulldocs/InvestAnim/Book1/media/PDF_chapters/B1_7.pdf
Saegerman, C., Berkvens, D., Godfroid, J., and Walravens, K. (2010). “Bovine brucellosis,” in Infectious and Parasitic Diseases of Livestock, eds P. Lefèvre, J. Blancou, R. Chermette, and G. Uilenberg (Paris: Lavoisier et Commonwealth Agricultural Bureau – International), 971–1001.
Saegerman, C., Dal Pozzo, F., and Humblet, M.-F. (2012). Reducing hazards for humans from animals: emerging and re-emerging zoonoses. Ital. J. Public Health 9, 13–24. doi: 10.2427/6336
Sanogo, M., Abatih, E., Thys, E., Fretin, D., Berkvens, D., and Saegerman, C. (2013a). Importance of identification and typing of Brucellae from West African cattle: a review. Vet. Microbiol. 164, 202–211. doi: 10.1016/j.vetmic.2013.02.009
Sanogo, M., Thys, E., Achi, Y. L., Fretin, D., Michel, P., Abatih, E., et al. (2013b). Bayesian estimation of true prevalence, sensitivity and specificity of Rose Bengal test and indirect ELISA for the diagnosis of bovine brucellosis. Vet. J. 195, 114–120. doi: 10.1016/j.tvjl.2012.06.007
Whatmore, A. M., Koylass, M. S., Muchowski, J., Edwards-Smallbone, J., Gopaul, K. K., and Perrett, L. L. (2016). Extended multilocus sequence analysis to describe the global population structure of the genus brucella: phylogeography and relationship to biovars. Front. Microbiol. 7:2049. doi: 10.3389/fmicb.2016.02049
Keywords: molecular epidemiology, brucellosis, Brucella, Brucella abortus biovar 3, biotyping, MLVA, West Africa
Citation: Sanogo M, Fretin D, Thys E and Saegerman C (2017) Exploring the Diversity of Field Strains of Brucella abortus Biovar 3 Isolated in West Africa. Front. Microbiol. 8:1232. doi: 10.3389/fmicb.2017.01232
Received: 13 December 2016; Accepted: 19 June 2017;
Published: 30 June 2017.
Edited by:
Adrian Whatmore, Animal and Plant Health Agency, United KingdomReviewed by:
Takehiko Kenzaka, Osaka Ohtani University, JapanHeinrich Neubauer, Friedrich Loeffler Institute Greifswald, Germany
Copyright © 2017 Sanogo, Fretin, Thys and Saegerman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Moussa Sanogo, ssanogomoussas@gmail.com
 Moussa Sanogo
Moussa Sanogo David Fretin
David Fretin Eric Thys
Eric Thys Claude Saegerman
Claude Saegerman