- 1Key Laboratory of Dairy Science, Ministry of Education, College of Food Sciences, Northeast Agricultural University, Harbin, China
- 2Food Science and Nutrition Unit, Department of Animal Science, Faculty of Agriculture, University of Benin, Benin City, Nigeria
Lactic acid bacteria play increasingly important roles in the food industry. Streptococcus thermophilus KLDS 3.1003 strain was isolated from traditional yogurt in Inner Mongolia, China. It has shown high antimicrobial activity against selected foodborne and vaginal pathogens. In this study, we investigated and analyzed its complete genome sequence. The S. thermophilus KLDS 3.1003 genome comprise of a 1,899,956 bp chromosome with a G+C content of 38.92%, 1,995 genes, and 6 rRNAs. With the exception of S. thermophilus M17TZA496, S. thermophilus KLDS 3.1003 has more tRNAs (amino acid coding genes) compared to some S. thermophilus strains available on the National Centre for Biotechnology Information database. MG-RAST annotation showed that this strain has 317 subsystems with most genes associated with amino acid and carbohydrate metabolism. This strain also has a unique EPS gene cluster containing 23 genes, and may be a mixed dairy starter culture. This information provides more insight into the molecular basis of its potentials for further applications in the dairy and allied industries.
Introduction
Lactic acid bacteria (LAB) species are recognized globally as an industrially important group of bacteria used for the production of fermented foods such as yogurt, cheese, and butter (Gezginc et al., 2015; Labrie et al., 2015). While some act as commensals and colonizers on the mucosal surface of our gastrointestinal tracts, others are marketed as probiotics used in foods to improve nutrition and health (LAB, 2011; Enujiugha and Badejo, 2015; Canani et al., 2016; Evivie et al., 2017). As the second most important species of LAB, Streptococcus thermophilus has been used for a variety of applications in the dairy and allied industry (Iyer et al., 2010; Kang et al., 2012). While some strains have been shown to produce high amounts of exopolysaccharides (EPS) (Wu et al., 2014; Bai et al., 2016) and bacteriocins (Renye et al., 2016), others have been reported to have a range of probiotic properties which include lowering the effects of diarrhea in young children (Kort et al., 2015), adhesion to intestinal epithelial cells (Kebouchi et al., 2016), anti-inflammatory (Li and Shah, 2016), anti-carcinogenic (Sah et al., 2016), antioxidant (Lee et al., 2015), and bacterial vaginosis-suppressive (Patras et al., 2015) effects.
The need to explore and extensively study microbial strains which have high antimicrobial properties against the spread of notable food pathogens such as bacteria, mold, and yeast can be strategic and novel in the fight to ensure that consumers have safe and nutritious foods. Strains that inhibit the development of vaginal pathogens can also be of high economic value and present new frontiers in the treatment of diverse illnesses (Ankolekar, 2013; Sah et al., 2016). The strain, S. thermophilus KLDS 3.1003 has been shown in recent experiments in our laboratory to possess strong antimicrobial activity (expressed as minimum inhibition zones) against pathogenic Escherichia coli ATCC25922, Staphylococcus aureus ATCC25923 and Gardnerella vaginalis ATCC14018 giving 6.40 ± 0.26, 3.43 ± 2.97, and 5.47 ± 0.04 mm, respectively. The cell-free supernatants (CFS) of this strain were also shown to have antagonistic effects against the above-mentioned pathogens, giving 90.42 ± 0.87, 90.97 ± 0.88, and 90.49 ± 0.62% inhibition, respectively, with catalase treatment (data not shown). Here, the complete genome sequence of S. thermophilus KLDS 3.1003 is reported to give insight on the molecular basis for its various potential industrial applications in the food industry.
Methodology and Bioinformatics of S. thermophilus KLDS 3.1003
Streptococcus thermophilus KLDS 3.1003 was isolated from traditional yogurt culture found in Inner Mongolia, China. The whole genome sequencing of S. thermophilus KLDS 3.1003 was performed using Pacbio RSII (20K library) and Illumina Hiseq 4000 (500 bp PCR-free library) strategies respectively. Then, 402M Hiseq and 556M Pacbio clean data were generated using a refined data filter. PacBio reads were assembled using the protocol in SMRT Analysis v2.3.0 Pipe: RS_HGAP_Assembly3 following the procedure of Chin et al. (2013) and GATK analysis protocol was used to correct single base errors (Li et al., 2009). The genome sequence of this strain was assembled into a contig of 1,899,956 bp and a total of 38,282 polymerase reads were obtained via Pacbio RSII strategy. The assembly of S. thermophilus KLDS 3.1003 was uploaded for annotation using the Metagenomics Rapid Annotation using Subsystem Technology (MG-RAST) (Meyer et al., 2008).
Results
The complete genome sequence of S. thermophulus KLDS 3.1003 was shown to have a G+C content of 38.92%. It has a total of 1,997 genes comprising of 1,731 protein-coding genes, 6 rRNAs, 68 tRNAs, 4 ncRNAs, and 176 pseudo genes (Table 1). These results have also been compared with those of other S. thermophilus strains earlier reported such as ASCC 1275 (Wu et al., 2014), MN-BM-A02 (Shi et al., 2015), ND03 (Sun et al., 2011), CNRZ1066, and LMG18311 (Bolotin et al., 2004) (see S1). Streptococcus thermophilus KLDS 3.1003 has the highest number of tRNA proteins (total of 68) than all the above-mentioned strains. With the exception of S. thermophilus M17TZA496 with a total of 79 tRNA proteins, S. thermophilus KLDS 3.1003 has more amino acid-coding genes than all other sequenced S. thermophilus genome available on the NCBI database till date. The RAST annotation has assigned the genes of this strain into 317 subsystems with most genes associated with amino acids and derivatives metabolism (15.89%), followed by carbohydrates metabolism (12.27%), and then the protein metabolism subsystems (12.21%). No genes were associated with photosynthetic reactions (see Figure 1).
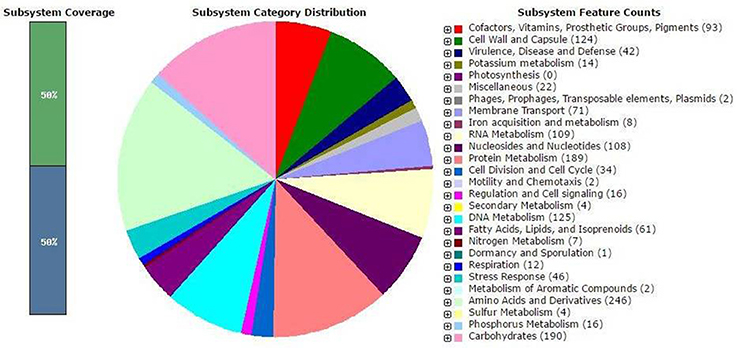
Figure 1. Annotation of Streptococcus thermophilus KLDS 3.1003 as generated by the Metagenome Rapid Annotation using Subsystem Technology (MG-RAST).
The genome of S. thermophilus KLDS 3.1003 has three (3) Comparative Analysis of Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) loci which were classified as types I-A (CRISPR 1), III-A (CRISPR 2), and II-A (CRSIPR 3), respectively. In addition, KLDS 3.1003 has 11, 14, and 1 spacers in CRISPR 1, CRISPR 2, and CRISPR 3 loci, respectively. The HNH-type system, enshrined in the type II-A CRISPR Cas loci, is rich in restriction enzymes as well as being likely responsible for target cleavage (Kleanthous et al., 1999; Garneau et al., 2010). The CRISPR immune system provides an adaptive defense mechanism against phages and other mobile genetic elements (Mojica et al., 2005; Barrangou et al., 2007). In addition, the CRISPR-cas system is important in assessing the ability of bacteria strains to resist food-borne pathogens (Wu et al., 2014; Li P. et al., 2016; Li W. et al., 2016). Given these properties, the host–phage interactions of this strain as well as the mechanism therein may require further studies.
The S. thermophilus KLDS 3.1003 genome has an EPS cluster (from BEN15_RS08500 to BEN15_RS08557 loci) consisting of 23 genes (see S2). In addition to eps1C (BEN15_RS08565), ID (BEN15_RS08560), 2C (BEN15_RS08525), and 2D (BEN15_RS08520), S. thermophilus KLDS3.1003 genome also contains epsA (BEN15_RS08575), epsB (BEN15_RS08570), epsE (BEN15_RS08555), epsF (BEN15_RS08550), epsG (BEN15_RS08540), epsH (BEN15_08530), epsI (BEN15_08530), epsJ (BEN15_RS08530), epsK (BEN15_RS08515), epsL (BEN15_RS08510), epsM (BEN15_RS08505), epsN (BEN15_RS08500), epsO (BEN15_RS0190), epsP (BEN15_RS08475), epsQ (BEN15_RS00260, BEN15_RS00265), and transposase (BEN15_RS08545). Some of these have been earlier reported to be part of the key enzymes necessary for the polymerization and export of EPS (Jolly and Stingele, 2001). Interestingly, the eps1C-ID and eps2C-2D produced by this organism suggests that it is a mixed producer of capsular and ropy EPS, a rare feature similar to high EPS-producing strains like S. thermophilus ASCC 1275 (Wu et al., 2014). This is however subject to further validation as many LABs produce different amounts and types of EPS under different conditions (Caggianiello et al., 2016). Furthermore, eps1C, 1D and eps2C, 2D function in chain length determination and eps A and B facilitate regulation of cell activities (Wu et al., 2014). In comparing the EPS gene cluster of this strain with other prominent EPS-producing strains such as S. thermophilus ASCC 1274 (Wu et al., 2014) via BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) we confirm that the genome of both strains are highly similar, possessing unique properties that confer high potentials for S. thermophilus KLDS 3.1003 in the dairy industry. Furthermore, S. thermophilus KLDS 3.1003 possesses the rare EPS-related enzymes acetyl transferase (BEN15_RS00090) and galactosyl transferase (BEN15_RS08555) which are lacking in all the sequenced S. thermophilus strains earlier compared in this report (see S2). Since EPS is known to play important roles in the improvement of texture and viscosity of yogurt and other fermented milks (Duboc and Mollet, 2001), it would thus be important to determine the amount and composition of EPS produced by this strain. In addition, studies aimed to further elucidate the structure and functionalities of its EPS so as to determine specific areas of application are necessary. Investigating the roles of its EPS in low pH tolerance or storage tolerance using isogenic lines will also provide more information in terms of characterization and possible applications in the food and non-food industries.
In all, information on the whole genome sequence of S. thermophilus KLDS3.1003 presented here provides the research and industrial community with a deeper dataset of the genetic setup for the potential antimicrobial properties and by extension its potential industrial applications.
Nucleotide Sequence Accession Number
The complete genome sequence of S. thermophilus KLDS 3.1003 has been deposited in GenBank under the accession number CP016877. This strain is also available at Dairy Industrial Culture Collection of the Key Laboratory of Dairy Science (KLDS-DICC), Northeast Agricultural University, Harbin, China.
Author Contributions
GH conceived the project idea. SE was assigned the project and worked with all other authors to successfully extract the genome of Streptococcus thermophilus KLDS 3.1003. SE drafted the manuscript. All authors approved the final manuscript draft.
Funding
This study was funded by grants from the National Key Research and Development Program of China (No: 2017YFD0400303) and the National Natural Science Foundation of China (No: 31401512). The Delta State Government of Nigeria is also appreciated for financially supporting SE.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are also grateful to the Beijing Genomic Institute (BGI) Shenzhen, China for sequencing and assembling the genome of S. thermophilus KLDS 3.1003. The Delta State Government of Nigeria is also thanked for financially supporting SE.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01238/full#supplementary-material
References
Ankolekar, C. R. (2013). Lactic Acid Bacteria Based Phenolic Bioactive Modulation from Fruit Systems and Health Benefits. Doctoral thesis, University of Massachusetts, Amberst, MA.
Bai, Y., Sun, E., Shi, Y., Jiang, Y., Chen, Y., Liu, S., et al. (2016). Complete genome sequence of Streptococcus thermophilus MN-BM-A01, a strain with high exopolysaccharides production. J. Biotechnol. 224, 45–46. doi: 10.1016/j.jbiotec.2016.03.003
Barrangou, R., Fremaux, C., Deveau, H., Richards, M., Boyaval, P., Moineau, S., et al. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712. doi: 10.1126/science.1138140
Bolotin, A., Quinquis, B., Renault, P., Sorokin, A., Ehrlich, S. D., Kulakauskas, S., et al. (2004). Complete genome sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus. Nat. Biotechnol. 22, 1554–1558. doi: 10.1038/nbt1034
Caggianiello, G., Kleerebezem, M., and Spano, G. (2016). Current issues and future perspectives of exopolysaccharides produced by lactic acid bacteria. Appl. Microbiol. Biotechnol. 100, 3877–3886. doi: 10.1007/s00253-016-7471-2
Canani, R. B., Sangwan, S., Stefka, A. T., Nocerino, R., Paparo, L., Aitoro, R., et al. (2016). Lactobacillus rhamnosus GG-supplemented formula expands butyrate-producing bacterial strains in food allergic infants. ISME J. 10, 742–750. doi: 10.1038/ismej.2015.151
Chin, C., Alexander, D. H., Marks, P., Klammer, A. A., Drake, J., Heiner, C., et al. (2013). Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569. doi: 10.1038/nmeth.2474
Duboc, P., and Mollet, B. (2001). Applications of exopolysaccharides in the dairy industry. Int. Dairy J. 11, 759–768. doi: 10.1016/S0958-6946(01)00119-4
Enujiugha, V. N., and Badejo, A. A. (2015). Probiotic potentials of cereal-based beverages. Crit. Rev. Food Sci. Nutr. 57, 790–804. doi: 10.1080/10408398.2014.930018
Evivie, S. E., Huo, G. C., Igene, J. O., and Bian, X. (2017). Some current applications, limitations and future perspectives of lactic acid bacteria as probiotics. Food Nutr. Res. 61:1318034. doi: 10.1080/16546628.2017.1318034
Garneau, J. E., Dupuis, M.-È., Villion, M., Romero, D. A., Barrangou, R., Boyaval, P., et al. (2010). The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67–71. doi: 10.1038/nature09523
Gezginc, Y., Topcal, F., Comertpay, S., and Akyol, I. (2015). Quantitative analysis of the lactic acid and acetyldehyde produced by Streptococcus thermophilus and Lactobacillus bulgaricus strains isolated from traditional Turkish yoghurt using HPLC. J. Diary Sci. 98, 1426–1434. doi: 10.3168/jds.2014-8447
Iyer, R., Tomar, S. K., Maheswari, T. U., and Singh, R. (2010). Streptococcus thermophilus strains: multifunctional lactic acid bacteria. Int. Dairy J. 20, 133–141. doi: 10.1016/j.idairyj.2009.10.005
Jolly, L., and Stingele, F. (2001). Molecular organization and functionality of exopolysaccharide gene clusters in lactic acid bacteria. Int. Dairy J. 11, 733–745. doi: 10.1016/S0958-6946(01)00117-0
Kang, X., Ling, N., Sun, G., Zhou, Q., Zhang, L., and Sheng, Q. (2012). Complete genome sequence of Streptococcus thermophilus strain MN-ZLW-002. J. Bacteriol. 194, 4428–4429. doi: 10.1128/JB.00740-12
Kebouchi, M., Galia, W., Genay, M., Soligot, C., Leconte, X., Awussi, A. A., et al. (2016). Implication of sortase-dependent proteins of Streptococcus thermophilus in adhesion to human intestinal epithelial cell lines and bile salt tolerance. Appl. Microbiol. Biotechnol. 100, 3667–3679. doi: 10.1007/s00253-016-7322-1
Kleanthous, C., Kühlmann, U. C., Pommer, A. J., Ferguson, N., Radford, S. E., Moore, G. R., et al. (1999). Structural and mechanistic basis of immunity toward endonuclease colicins. Nat. Struct. Biol. 6, 243–252. doi: 10.1038/6683
Kort, R., Westerik, N., Serrano, M., Douillard, F. P., Gottstein, W., Mukisa, I. M., et al. (2015). A novel consortium of Lactobacillus rhamnosus and Streptococcus thermophilus for increased access to functional fermented foods. Microb. Cell Fact. 14:195. doi: 10.1186/s12934-015-0370-x
LAB (2011). “The 10th LAB Symposium” in Thirty years of Research on Lactic Aci Bacteria, eds. A. Ledeboer, J. Hugenholtz, J. Kok, W. Konings, and J. Wouters (Rotterdam: 24 Media Labs). On behalf of the Stichting Symposium on LAB.
Labrie, S. J., Tremblay, D. M., Plante, P. L., Wasserscheid, J., Dewar, K., Corbeil, J., et al. (2015). Complete genome sequence of Streptococcus thermophilus SMQ-301, a model strain for phage-host interactions. Gen. Announ. 3:e00480-15. doi: 10.1128/genomeA.00480-15
Lee, M., Hong, G. E., Zhang, H., Yang, C. Y., Han, K. H., Mandal, P. K., et al. (2015). Production of the isoflavone aglycone and antioxidant activities of black milk using fermentation with Streptococcus thermophilus S10. Food Sci. Biotechnol. 24, 537–544. doi: 10.1007/s10068-015-0070-7
Li, P., Gu, Q., and Zhou, Q. (2016). Complete genome sequence of Lactobacillus plantarum LZ206, a potential probiotic strain with antimicrobial activity against foodborne pathogens. J. Biotechnol. 238, 52–55. doi: 10.1016/j.jbiotec.2016.09.012
Li, R., Li, Y., Fang, X., Yang, H., Wang, J., Kristiansen, K., et al. (2009). SNP detection for massively parallel whole-genome resequencing. Gen. Res. 19, 1124–1132. doi: 10.1101/gr.088013.108
Li, S., and Shah, N. P. (2016). Characterization, anti-inflammatory and antiproliferative properties of natural and sulfonated exopolysaccharides from Streptococcus thermophilus ASCC 1275. J. Food Sci. 81, 167–176. doi: 10.1111/1750-3841.13276
Li, W., Bian, X., Evivie, S. E., and Huo, G. C. (2016). Comparative Analysis of Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) of Streptococcus thermophilus St-I and its Bacteriophage-Insensitive Mutants (BIM) derivatives. Curr. Microbiol. 73, 393–400. doi: 10.1007/s00284-016-1076-y
Meyer, F., Paarmann, D., D'Souza, M., Olson, R., Glass, E. M., Kubal, M., et al. (2008). The metagenomics RAST server – a public resource for the automatic phylogenetic and functional analysis of metagenomes BMC Bioinfor. 9:386. doi: 10.1186/1471-2105-9-386
Mojica, F. J., Garcı’a-Martı’nez, J., and Soria, E. (2005). Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 60, 174–182. doi: 10.1007/s00239-004-0046-3
Patras, K. A., Wescombe, P. A., Rösler, B., Hale, J. D., Tagg, J. R., and Doran, K. S. (2015). Streptococcus salivarius K12 limits group B Streptococcus vaginal colonization. Infect. Immun. 83, 3438–3444. doi: 10.1128/IAI.00409-15
Renye, J. A. Jr., Somkuti, G. A., Garabal, J. I., and Steinberg, D. H. (2016). Bacteriocin production by Streptococcus thermophilus in complex growth media. Biotech. Lett. 38, 1947–1954. doi: 10.1007/s10529-016-2184-2
Sah, B. N. P., Vasiljevic, T., McKechnie, S., and Donkor, O. N. (2016). Antibacterial and antiproliferative peptides in synbiotic yogurt—release and stability during refrigerated storage. J. Dairy Sci. 99, 4233–4242. doi: 10.3168/jds.2015-10499
Shi, Y., Chen, Y., Li, Z., Yang, L., Chen, W., and Mu, Z. (2015). Complete genome sequence of Streptococcus thermophilus MN-BMA02, a rare strain with a high acid-producing rate and low post acidification ability. Genome Announc. 3, e00979–15. doi: 10.1128/genomeA.00979-15.
Sun, Z., Chen, X., Wang, J., Zhao, W., Shao, Y., Wu, L., et al. (2011). Complete genome sequence of Streptococcus thermophilus strain ND03. J. Bacteriol. 193, 793–794. doi: 10.1128/JB.01374-10
Keywords: Streptococcus thermophilus, genome sequence, EPS, antimicrobial activity, dairy industry
Citation: Evivie SE, Li B, Ding X, Meng Y, Yu S, Du J, Xu M, Li W, Jin D, Huo G and Liu F (2017) Complete Genome Sequence of Streptococcus thermophilus KLDS 3.1003, A Strain with High Antimicrobial Potential against Foodborne and Vaginal Pathogens. Front. Microbiol. 8:1238. doi: 10.3389/fmicb.2017.01238
Received: 31 March 2017; Accepted: 19 June 2017;
Published: 11 July 2017.
Edited by:
Giovanna Suzzi, University of Teramo, ItalyReviewed by:
Giuseppe Spano, University of Foggia, ItalyKonstantinos Papadimitriou, Agricultural University of Athens, Greece
Copyright © 2017 Evivie, Li, Ding, Meng, Yu, Du, Xu, Li, Jin, Huo and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guicheng Huo, gchuo58@126.com
Fei Liu, david.as@163.com
†These authors have contributed equally to this work.
 Smith E. Evivie
Smith E. Evivie Bailiang Li1†
Bailiang Li1† Guicheng Huo
Guicheng Huo