- Department of Microbiology and Molecular Genetics, University of the Punjab, Lahore, Pakistan
Mercury resistant (HgR) Enterobacter sp. AZ-15 was isolated from heavy metal polluted industrial wastewater samples near to districts Kasur and Sheikhupura, Pakistan. 16S rDNA ribotyping and phylogentic analysis showed 98% homology with already reported Enterobacter species. The merF gene encoding transmembrane protein-MerF was amplified from genomic DNA and ligated into pET31b+ vector using restriction endonucleases, SphI and XhoI. The genetic codons of merF gene encoding cysteine residues were mutated into codons, translating into serine residues by site-directed mutagenesis. Ketosteroid isomerase (KSI), a fusion tag which is present in pET31b+ vector, was used in the expression of merFm gene. KSI was used to drive the target peptide (MerFm) into inclusion bodies so that the peptide yield and purity were increased. The stable plasmid pET31b+:merFm was transformed into C43(DE3) E.coli cells. The high expression of uniformly 15N isotopically labeled-MerFm protein was induced with 1 mM IPTG. The purification of 15N-MerFm recombinant protein by Ni-NTA and size exclusion chromatography involved an unfolding/refolding procedure. The two-dimensional HSQC NMR spectra of MerFm protein showed the purity and correct number of resonances for each amide. 1H–15N HSQC NMR experiment also confirmed that no modification of the tryptophan residue occurred during cyanogen bromide cleavage. A small scale reservoir of Luria Bertani (LB) medium supplemented with 20 μg/ml of HgCl2 showed 90% detoxification of Hg by Enterobacter sp. AZ-15. The accumulation of Hg on the cell surface of this strain was visualized by scanning electron microscopy (SEM) which confirmed its potential use in Hg-bioremediation.
Introduction
Mercury toxicity is a worldwide problem to both human and animals. The level of mercury pollution in the environment is being increased day by day due to anthropogenic sources and activities like the discharge of industrial effluent from chlor-alkali industries, mining of metal and incineration of coal (Steenhuisen and Wilson, 2015). It is obvious that both forms of mercury (inorganic and organic) cause cytotoxic and neurotoxic effects to humans and animals (WHO, 2000).
Bacterial detoxification systems have spawned much interest in recent times for their potential usefulness in the bioremediation of environmental contaminants (Silver, 1992). Heavy metals are just one of a variety of contaminants that have appeared in our environment, and quite a few natural resistances to them have already been documented. Plasmid-born resistances to a wide variety of heavy metals have been explored and the genes encoding their resistances have been sequenced (Silver, 1992). The best characterized of these systems is the bacterial mercury detoxification system, the mer operon, and in particular those on transposons Tn21, Tn501, and Tn5053 (Gilbert and Summers, 1988; Dahlberg and Hermansson, 1995).
Bacteria can be used for bioremediation because they take up mercury via membrane potential-dependent sequence-diverged members of the mercuric ion (Mer) superfamily, i.e., a periplasmic Hg-scavenging protein (MerP) and one or more inner membrane-spanning proteins (MerC, MerE, MerF, and MerT), which transport Hg2+ into the cytoplasm (Barkay et al., 2003). All the mer operons have merT and merP, however, some operons, such as transposon Tn21, have merC. The merF is also part of mer operons of Gram negative bacteria and is absent in the mer operons of Gram positive bacteria. The additional merC gene is located between merP and merA. However, it seems not to be essential for Hg2+ resistance since it is absent from Tn501, which confers identical Hg2+ resistance levels (Summers, 1986).
The enzymatic mercury detoxification system is one of the most remarkable because of its level of sophistication. Mercury is toxic at very low levels, as Hg2+ with no known biological benefit. Its toxicity is related to nonspecific reactivity with sulfhydryl groups in proteins and high permeability through biological membranes. Expression of proteins encoding mercury resistance is governed by an operator/promoter region, which interacts with a divergently transcribed MerR biosensor protein. The roles of the gene products designated T (116 amino acids), P (72 amino acids), C (143 amino acids), and A have been worked out using gene deletion studies (Hamlett et al., 1992; Wilson et al., 2000). In detoxification mechanism, the toxic Hg2+ atoms are ‘handed off’ from MerP to MerT, which transports Hg2+ to the reductase, MerA (Ghosh et al., 1999). More recently, a homolog of MerT, designated MerF (81 residues), has been sequenced from Alcaligenes and Pseudomonas genera posited to have the same function (Yeo et al., 1998; Sone et al., 2013).
Integral to this transport system are several pairs of cysteine residues, which are known to bind Hg2+ in a linear bicoordinate manner. In particular, the motifs -CC-, -CXC-, -CXXC-, and -CXXXXXC- are found in MerT/MerF, MerE. MerP, and MerC, respectively (DeSilva et al., 2002). Mutagenesis of the cysteine residues suggests that only one of the cysteine residues in MerP, Cl7, is important, and only the first -CC- pair in MerT and MerF is important (Powlowski and Sahlman, 1999). In MerT mutation of a cysteine residue in the -CXXXXXC- pair is only slightly detrimental to its transport function, while mutation in the vicinal -CC- residues in either MerT or MerF is detrimental in the transportation (Hobman and Brown, 1997).
MerF is predicted to have two membrane-spanning segments. It has been shown definitively to function as a transporter of mercuric ions into the cell by possession of two vicinal pairs of cysteine residues which are involved in the transport of Hg2+ across the membrane and are exposed to the cytoplasm. More importantly, MerF alone is sufficient for the transport of Hg2+ across the cell membrane. NMR studies of integral membrane domain and full length MerF from Escherichia coli (C41 and C43 cells) have been investigated (Das et al., 2012; Lu et al., 2013; Tian et al., 2014).
Inoculation of contaminated sites with selected or engineered bacteria has often not been satisfactory, partly because the introduced metabolic potential was not the limiting factor for pollutant degradation (Cases and de Lorenzo, 2005). This applies also to mercury resistance, which is ubiquitous in soil and water, even in the Arctic (Barkay and Poulain, 2007; Møller et al., 2011). For the treatment of mercury-contaminated groundwater, pilot experiments have been carried out with the aim to establish bio-barriers in the groundwater where sulfate reducing bacteria (SRB) could adsorb mercury and precipitate it as insoluble cinnabar (Wagner-Döbler et al., 2000; Dash and Das, 2012; He et al., 2015).
In the present study, a major transporter protein of bacterial Hg-detoxification system, MerF isolated from Hg-resistant Enterobacter sp. AZ-15 was first time studied and then the potential of selected bacterial strain AZ-15 in the detoxification of Hg (II) was evaluated. These objectives were achieved by (1) the screening of mercury resistant bacteria from polluted natural environment and their 16S rDNA phylogenetic analysis (2) designing the gene construct and expression of mutated merF (merFm) gene (3) purification of MerFm protein by Ni-NTA and size exclusion chromatography (4) NMR studies of 15N isotopically labeled-MerFm protein (5) the analysis of Hg-detoxification potential at lab scale and SEM of selected bacterial cells.
Materials
Enzymes were purchased from New England Biolabs (www.neb.com) unless otherwise noted, and the oligonucleotides were synthesized by Integrated DNA Technologies (www.idtdna.com). GeneJET genomic DNA purification kit and MAX efficiency® DH5α™ competent E. coli cells were purchased from Thermo Fisher© (www.thermofisher.com). Rapid DNA Dephos & ligation kit was obtained from Life Sciences© (www.lifescience.roche.com). Gel extraction kit, miniprep kit for small-scale plasmid preparations and His-tag nickel affinity resin (Ni-NTA) were purchased from Qiagen© (www.qiagen.com). QuikChange lightning site-directed mutagenesis kit was from Agilent Technologies© (www.genomics.agilent.com). Plasmid DNA of pET-31b(+) and bacterial strain OverExpress™ C43(DE3) were purchased from Lucigen© (www.lucigen.com). 15N-ammonium sulfate and d25-sodium dodecyl sulfate were obtained from Cambridge isotope laboratories (www.isotope.com). FPLC Sephacryl S-200 was obtained from Pharmacia LKB (Piscataway, NJ).
Methods—Bacterial Isolates and Growth Conditions
Industrial water samples from different geological areas of districts Kasur and Sheikhupura, Punjab, Pakistan were collected in sterilized polythene bags. All samples were brought to laboratory and different physico-chemical parameters were analyzed within 24–48 h. Bacterial load of the samples was determined by making serial dilutions of 10−1 to 10−4 from 1% initial water sample. In order to isolate the individual colonies, 100 μl of the 10−3 and 100−4 dilutions were spread on LB agar plates containing different concentrations of HgCl2 ranging from 1 to 20 μg/ml. The plates were incubated at 37°C for 24 h. The single isolated and HgCl2 resistant colonies were selected and re-streaked on new LB agar plates without HgCl2 to get purified colonies. All Hg-resistant and purified bacterial cultures were stored as glycerol stocks (30%) at−80°C.
Biochemical and Molecular Characterization
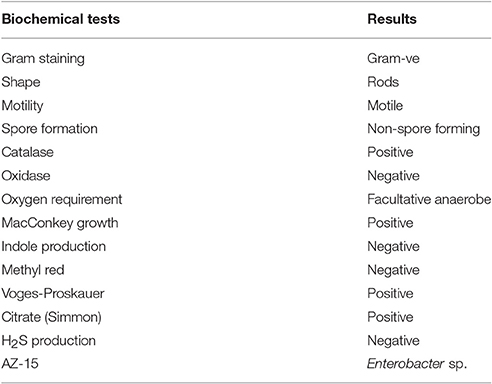
Highly Hg-resistant isolate AZ-15 was biochemically characterized by different tests viz., Gram staining, shape, motility, spore formation, catalase, oxidase, oxygen requirement, MacConkey agar growth, indole production, methyl red, Voges-Proskauer, citrate (Simmon) agar growth, and H2S production by following the protocols of Cappuccino and Sherman (2008). The bacterial genomic DNA of AZ-15 was extracted by genomic DNA purification kit. The universal primers were used for the amplification of 16S rRNA gene using BIOER XP-thermal cycler (Normand, 1995). The amplification conditions for both genes were set as: 5 min at 95°C, 35 cycles of 1 min at 95°C, 1 min at 55°C, and 2 min at 72°C, with a final 5 min chain elongation at 72°C. The sequence results obtained from Macrogen sequencing core facility, Korea were checked through nucleotide blast and submitted to NCBI GenBank.
Cloning of merF Gene into pET31b(+) Vector
The merF gene (wild type) of highly mercury resistant Enterobacter sp. AZ-15 was amplified using the primers given in Table 1. The amplified product of 246 bp was digested with the restriction enzymes Xhol and Sphl, and purified by gel extraction kit. The digested merF gene was ligated with the Xhol-Sphl-cleaved expression vector pET31b(+) (Kuliopulos et al., 1987). The genetic codons encoding cysteine residues in the native sequence of merF gene were mutated into codons encoding serine residues using site directed mutagenesis kit by designing two sets of primers (Table 1). Both sequences of merF gene (merF wild type and mutated merF) along with translated peptide sequences were submitted to the NCBI database (www.ncbi.nlm.nih.gov/) and obtained accession numbers as MF185183 and MF185184, respectively. The designed recombinant plasmid containing mutated merF (merFm) gene was transformed into DH5α competent cells and identified by using same restriction enzymes on agarose gel. The nucleotide sequence of recombinant pET31b(+) containing merFm gene was confirmed by DNA sequencing facility provided marketplace, University of California, San Diego (USA). Then the recombinant plasmid pET31b(+) was transformed into C43(DE3) competent E. coli cells. After transformation, stable C43(DE3): pET31b+merFm clones were screened and stored as glycerol stocks (20% of glycerol) at −80°C.

Table 1. Oligonucleotides for the amplification of 16S rRNA, wild type merF and mutated merF (merFm) genes.
Production of 15N-Labeled Recombinant MerFm
The expression of recombinant merFm gene was optimized in LB medium and unlabeled M9 medium supplemented with different concentrations (0.2, 0.5, and 1 mM) of isopropyl-β-D-thiogalactoside (IPTG) at different time intervals (2, 4, 6, 7 h, and overnight) of incubation. For expression of isotopically labeled MerFm protein, 5 ml of LB medium with final concentration of 50 mg/l of cabenicillin was inoculated with 5 μl of clone C43(DE3): pET31b+merFm glycerol stock. After 5 h incubation at 37°C, 1 ml of starter culture was used to inoculate 50 ml of M9 minimal medium (per liter) containing Na2HPO4 7.0 g, KH2PO43.0 g, NaCl 0.5g, CaCl2 0.1 mM, MgSO4 1 mM, thiamin 50 mg, d-glucose 10 g, and 15N-(NH4)2SO4 1 g containing 50 mg/l of carbenicillin. The culture was kept at 300 rpm at 37°C for overnight and 50 ml culture was poured into 450 ml of same 15N labeled-M9 medium. The cells were allowed to grow at 37°C in shaking incubator until the optical density (O.D600) of 0.6 was achieved. The expression of recombinant plasmid (KSI_MerFm_His-tag) was achieved by adding 1 mM of IPTG as a final concentration and kept on shaking for another 7 h on same conditions. The pellet of C43(DE3) cells containing recombinant protein was obtained by subsequent harvesting at 7,000 rpm for 30 min at 4°C.
Purification of the Recombinant Protein by Ni-NTA Chromatography
The cell pellets were resuspended in 30 ml lysis buffer (50 mM Tris hydrochloride, 15% glycerol (v/v), 1 mM NaN3, pH 8.0). After 10 min of incubation at room temperature, cell lysate was sonicated by a probe sonicator (www.fishersci.com) for 5 min (cycles: 5 s on/10 s off) on ice and then centrifuged at 17,000 rpm for 30 min at 4°C. The pellet at this stage contains purified inclusion bodies containing more than 90% of MerFm fusion protein. The pellet containing inclusion bodies was then resuspended in the nickel column binding buffer (20 mM Tris hydrochloride, 500 mM NaCl, 6 M guanidinium hydrochloride (GndCl), 5 mM imidazole, pH 8.0) by tip sonication, and centrifuged again at 19,000 rpm for 1 h to remove remaining protein, lipids, and lipid-associated debris. The supernatant was loaded onto a Ni-NTA His-Bind Resin column, pre-equilibrated with binding buffer. The column was then washed with washing buffer (20 mM Tris hydrochloride, 500 mM NaCl, 6 M guanidinium hydrochloride (GndCl), 50 mM imidazole, pH 8.0). The fusion protein was eluted with elution buffer (20 mM Tris hydrochloride, 500 mM NaCl, 6 M GndCl, 500 mM imidazole, pH 8.0) and concentrated to 30 ml using an Amicon stirred concentrator cell with YM10 filter membrane and dialyzed against ddH2O in a 10,000 kDa MWCO dialysis membrane with four water changes until the protein precipitates out of solution. The precipitated protein was then lyophilized.
Cyanogen Bromide Cleavage of the Fusion Protein
The lyophilized protein was dissolved in 70% formic acid solution and cleaved by addition of a three-fold excess of cyanogen bromide (CNBr) for 3–5 h in the absence of light. The cleavage reaction was stopped by adding double volume of 1M NaOH. The cleaved polypeptide was dialyzed by using 3,500 kDa MWCO dialysis membrane against water and then lyophilized.
Purification of MerFm by FPLC
Cleaved protein was re-dissolved in 4 ml phosphate SDS buffer (100 mM Na2HPO4, 4 mM SDS, I mM EDTA, lmM NaN3, pH 8.2) and 1 ml 1 N NaOH. Size-Exclusion gel chromatography using a Sephacryl S-200 column equilibrated with phosphate-SDS buffer on a Pharmacia FPLC system used to separate the KSI fusion partner from the pure MerFm protein. Fractions containing MerFm were pooled and concentrated down to 30 ml using an Amicon Stirred cell with a 3,500 MWCO membrane. The pure protein was then dialyzed against ddH2O (with 40 mM β-mercaptoethanol for native MerFm) to remove SDS for six times with time interval of 12 h until the protein precipitates. Each time the protein was centrifuged out of solution, quickly lyophilized, and stored at −20°C for further use, Yields of MerFm were approximately 2–3 mg/l of cell culture. SDS-PAGE was run to monitor the purification, with the final native MerFm protein.
NMR Spectroscopy of MerFm
All NMR samples were prepared by resolubilizing the lyophilized protein in NMR buffer (10 mM Na2HPO4, 500 mM d25-SDS, lmM NaN3, pH 6.0). The protein was then incubated at 40°C for 30 min with intermittent bath sonication and centrifuged at 13,000 rpm for I5 min to remove undissolved protein and other debris. The labeled peptide samples were prepared in different concentrations 10, 40, and 75% of D2O. Two-dimensional heteronuclear single quantum coherence (HSQC) spectra of uniformly 15N-labeled MerFm were recorded on Bruker DMX 600 spectrometer equipped with a triple resonance probe, three-axis pulsed field gradients, a deuterium lock channel and operated at a 1H NMR frequency of 600 MHz with total recording time 7 h. The other parameters for obtaining 15N–1H fast Heteronuclear Single Quantum Cohererance (fHSQC) spectra include pulse sequence with 1,024 points in t2 and 256 points in t1 and 50°C using a 1.5 s recycle delay. The spectra were processed by using Sparky 3 [Goddard and Kneller, University of California San Francisco (UCSF), USA].
Detoxification of Hg2+ by Bacteria and SEM Analysis
An experiment was designed to check the detoxification potential of Enterobacter sp. at a lab scale. Mercury resistant (HgR) bacterial strain AZ-15 and mercury sensitive strain (HgS) Enterobacter cloacae ZA-15 (KJ728671) with equal number of cells (cfu/ml) were inoculated separately in flasks containing 30 mL of LB medium, supplemented each flask with 20 μl/ml of HgCl2 to determine the detoxification efficiency of mercury. The flasks were incubated at 37°C for up to 8 h with 2 h of time intervals at 120 rpm of agitation in triplicates. After incubation, cultures were spun down at 14,000 rpm for 15 min and detoxification of Hg2+ was estimated by dithizone method (Elly, 1973; Khan et al., 2006). For scanning electron microscopy (SEM), one drop of 48 h culture was diluted into 15 ml of ddH2O and dried on carbon surface slide. The topographical, morphological and crystallographic information of bacterial cell membrane was monitored using Scanning Electron Microscope (SEM) (JEOL-JSM-6480).
Statistical Analysis
The phylogenetic analysis of Enterobacter sp. AZ-15 with already reported bacteria on the basis of 16S rRNA gene was done by Mega 6.0 software (Tamura et al., 2013). The results of lab scale mercury detoxification experiment were subjected to mean, standard deviation, analysis of variance (ANOVA) by using SPSS V.20 software (IBM, 2011). The hydropathy plot predicting membrane spanning regions was built by SWISS-MODEL homology (www.expasy.org).
Results
Screening of Hg-Resistant Bacteria
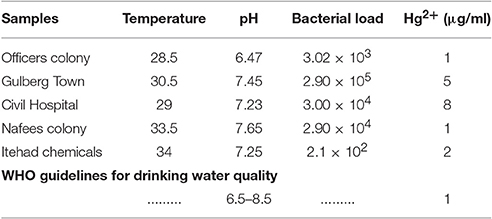
Industrial water samples collected from different geological sites (tanneries) of district Kasur and Itehad chemicals limited, district Sheikhupura, Lahore were checked for physico-chemical properties such as temperature, pH, Hg2+ concentration, bacterial load in the presence of metal stress of HgCl2. Data indicate the level of pH in water sample falls in the range of 6.47–7.65. All samples (100%) were within the pH ranges of WHO drinking water guidelines (WHO, 2004) but the value of HgCl2 which range from 5to 17 μg/ml has crossed limit described by WHO (1 μg/ml) (Javendra, 1995). The bacterial count in these samples is also variable, ranging from 2.1 × 102 to 2.9 × 105 as presented in Table 2. A total 30 bacterial strains including Gram positive and Gram negative were screened on the basis of showing resistance against different concentrations of HgCl2.
Biochemical and 16S rDNA Identification
The most highly Hg-resistant bacterial isolate AZ-15 showing growth at 20 μg/ml of HgCl2, was characterized on the basis of colony morphology and biochemical tests. The results are shown in Table 3. Mercury resistant bacterial isolate was identified by 16S rDNA sequencing (≈1.5 kb) as Enterobacter sp. with accession number KU558920. Other close matches to Enterobacter sp. AZ-15 (KU558920) includes E. cloacae strain E717 (EF059865), Enterobacter sp. BSRA3 (FJ868807), E. cloacae strain NBRC 13535 (NR113615), E. cloacae strain DSM 30054 (NR117679), E. cloacae strain RPR-CCFL3 (KR611993) and Enterobacter sp. Wy2-D9 (JN986806). Enterobacter sp. GJ1-11 (EU139848), Enterobacter sp. BAB-3140 (KF984440), and Enterobacter sp. STJ12 (KC833508) represent as an out group in this phylogenetic analysis (Supplementary Figure 1).
Plasmid Construction
As shown in Figure 1A, the gene construct merFm was designed by ligation of wild type merF gene in pET31b(+) vector. The deduced amino acid sequence of merF gene showed two cysteine residues at the positions 71 and 72. The cysteine residues were mutated into serine residues in merFm gene by site-directed mutagenesis which results no cysteine residue in the final merFm gene construct as shown in Figure 1B. The salient features of construct include T7 promoter, ketosteroid isomerase (KSI) fusion partner, 5′ overhanging sequences compatible with SphI and XhoI restriction enzyme sites, within restriction sites; a transcription initiation codon (ATG) of merFm followed by encoding region of respective protein and a termination codon and a region encoding a hexahistidine amino acid tag and T7 terminator. His-tag sequence is for ease of purification and the KSI peptide sequence is designed to form inclusion bodies. After the transformation of recombinant plasmid into competent DH5α E. coli cells, the plasmid DNA was purified at concentration of 45 mg/ml. The plasmid was then transformed into E. coli C43(DE3) cells for expressing high concentration of MerFm recombinant protein and is thus protected from proteolysis. The fusion protein is non-toxic to the E. coli host cell, and is expressed at levels up to 20% of total cellular protein in E. coli strain C43(DE3). This approach to the production of MerFm in E. coli may be generally applicable to other membrane proteins. We have used it successfully with several other small membrane proteins.
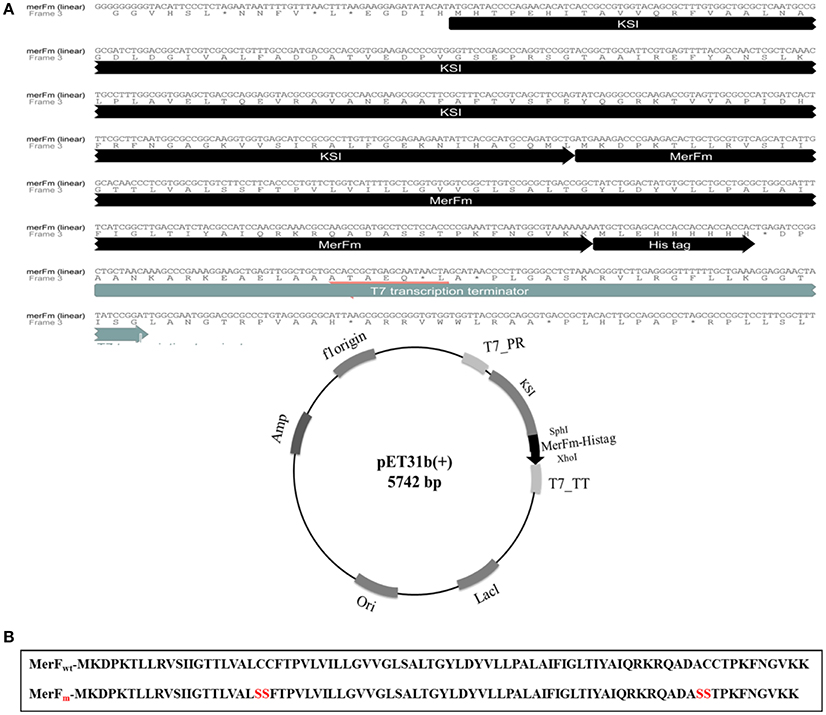
Figure 1. (A) Linear and circular forms of nucleotide and amino acid sequences of pET31b+ plasmid with merFm gene. T7_PR, T7 promoter; T7_TT, T7 terminator; His-tag_KSI, fusion partner; SphI and XhoI, restriction enzyme sites; ori, replication (B) Amide sequences of MerF protein before and after site-directed mutagenesis.
Expression and Purification of 15N-Labeled MerFm Protein
For optimization of recombinant protein MerFm, three different concentrations, 0.2, 0.5, and 1 mM of IPTG were used at different post-induction time (h) viz., 2, 4, 6, 7, and overnight. The high expression of recombinant protein MerFm was achieved with 1 mM of IPTG and 7 h of post-induction time as shown in lane 12 of Supplementary Figure 2. The same conditions were maintained for expression of 15N-labeled MerFm fusion protein. After expression, the separation and purification of 15N-MerFm were performed by following the procedure described in detail in the methodology section. Briefly, the recombinant/fusion protein formed as inclusion bodies was separated from the total cell lysate by centrifugation. The fusion protein was partially purified by nickel affinity chromatography. The data in Supplementary Figure 3, illustrate the expression and isolation of inclusion bodies of full-length MerFm by Ni-NTA column. Lane 1 is showing Mark12™ unstained protein ladder (www.lamdabio.com). After the post-induction period of 7 h, the cells were lysed by sonication and centrifuged. After centrifugation, the supernatant was discarded which contained the total cellular proteins except inclusion bodies is shown in lane 2 of a 12% SDS-PAGE. Lane 3 contains the inclusion bodies solubilized in binding buffer and lanes 4 and 5 show the soluble fraction (flow through) and washing of other proteins with washing buffer, respectively except the targeted fusion protein. The lanes 6–10 show that the insoluble fractions (inclusion bodies) containing primarily the targeted fusion protein. After elution from the nickel column, the KSI fusion partner was cleaved from the MerFm polypeptide in the presence of CNBr at methionine for obtaining purified native MerFm. The lane 2 in Figure 2A shows the cleavage of fusion protein and the lane 3 contains the little bit impurity of KSI fusion partner while lanes 4–8 show the purified fractions 15N-MerFm after size-exclusion chromatography (FPLC). Figure 2B shows the peaks of 15N-MerFm and KSI in FPLC.
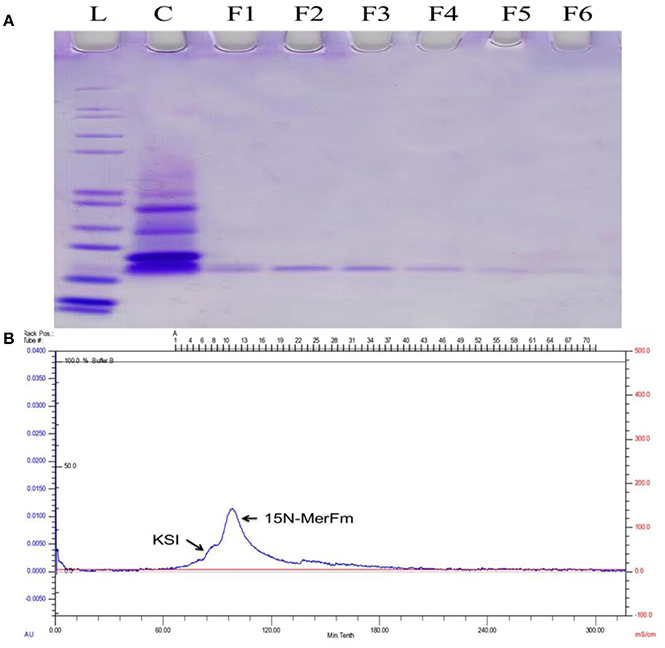
Figure 2. (A) L: Molecular weight (kDa) ladder. C: cleavage of fusion protein. F1–F5: Purified fractions of MerFm from FPLC column. (B) FPLC chromatogram showing peaks of native MerFm and fusion partner KSI.
NMR Spectra of 15N-MerFm
The 1H-15N HSQC spectra of MerFm were obtained with the three concentrations of D2O as shown in Figure 3. The high quality spectrum of 15N-MerfM was obtained with 40% concentration of D20 (Figure 3B). 1H-15N HSQC spectra were recorded under standard solution NMR conditions to confirm the purity and correct folding of the MerFm protein. The two-dimensional HSQC spectrum of 15N enriched MerFm contains the correct number of crosspeaks/resonances for each amide in the protein corresponding to side chain and tryptophan amide groups. This confirms that no modification of the tryptophan residue occurred during CNBr cleavage. Notably, the crosspeaks of six glycines in full-length MerFm are also present in the spectrum (Figure 3B). This is prerequisite for the preparation of well-behaved protein in phospholipid bilayer samples for solid-state NMR studies.
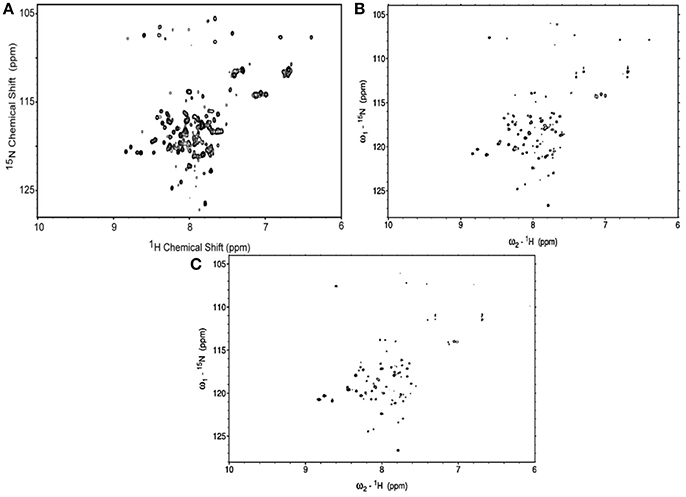
Figure 3. 1H–15N HSQC spectra of uniformly-15N-labeled MerFm in SDS micelles with different concentrations of D2O (A) 10% (B) 40%, and (C) 75%.
Detoxification of Hg2+ and SEM Analysis
In lab scale experiment, the detoxification of Hg2+ by selected bacterial strain indicated 5 (25%), 10 (50%), 18 (90%), and 18 (90%) μg/ml detoxification of Hg2+ out of 20 μg/ml (100%) at 2, 4, 6 and 8 h of incubation, respectively by mercury resistant Enterobacter sp. AZ-15 (p < 0.05). The mercury sensitive strain E. cloacae ZA-15 was used as a negative control (Figure 4A). In SEM analysis, the accumulation of Hg was seen on the cell surface after 8 h of incubation with 20 μg/ml in LB medium. The SEM image showed crystalline structures of Hg deposited on the surface of cell membrane while in negative control, no crystalline structure was observed (Figure 4B). From this experiment, it is evident that Enterobacter sp. AZ-15 can accumulate Hg which confirms its detoxification potential.
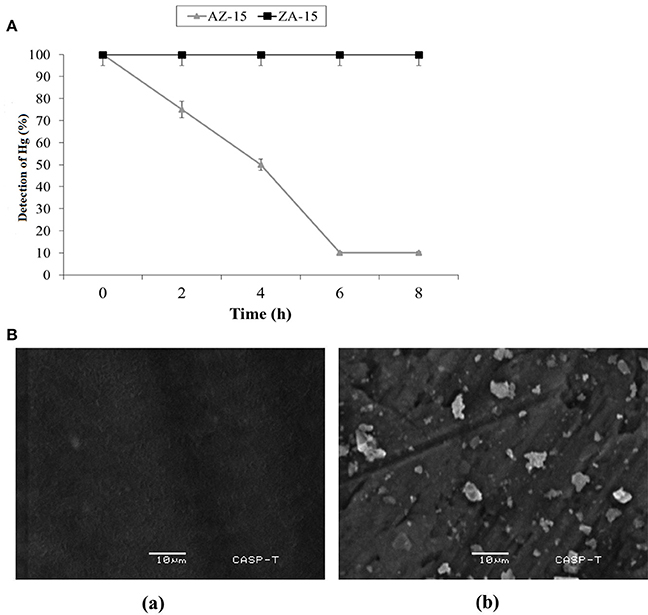
Figure 4. (A) Detoxification of Hg in LB medium. X-axis shows the given incubation time (h) and Y-axis shows the % of Hg detected at different time intervals Ba, SEM analysis of Hg-resistant Enterobacter sp. AZ-15 Bb, Hg-sensitive Enterobacter cloacae ZA-15.
Discussion
Mercury is widespread in our environment. This is not only a result of anthropogenic activities but also due to various geochemical and biological processes. Hence, nature provided a rich toolbox for transformation of mercury. It includes the microbial mercury resistance operon, which carries genes for binding, transport, and detoxification of Hg2+ and organomercurials. Among these tools, the transportation of mercury through microbial membranes plays a crucial role in the bacterial mercury detoxification system (Barkay et al., 2003; Parks et al., 2013).
Four families of integral membrane proteins (IMPs) have been identified in the characterized mer operons, ranging from 2 to 4 predicted transmembrane elements: MerE, MerF, MerT, and MerC. Across the mer operons, MerT, a tri-spanning, 116 residue protein, is considered to be the principal mercury transporter and has been shown to be an essential component in most mer operons capable of bestowing mercuric resistance. The extent to which the other IMPs, MerF and MerE, are capable of functioning independently of MerT has not yet been conclusively determined (Mok et al., 2012).
In previous studies, the full length and truncated structures of MerF have been deeply studied by NMR spectroscopy found in E. coli and in plasmid pMER327/419 of Pseudomonas fluorescens between merP and merA (Veglia and Opella, 2000; Wilson et al., 2000). This is the first study to analyze the gene construction, expression and purification of mutated MerF (MerFm) amplified from Enterobacter sp. AZ-15. The bacterium, Enterobacter sp. AZ-15 isolated from mercury contaminated industrial effluent samples, could resist Hg2+ upto 20 μg/ml. In other reports, the potential of Enterobacter sp. to tolerate Hg2+ varied (Table 4). Phylogenetic analysis of this bacterium AZ-15 showed 98% similarity to already reported Enterobacter species (Supplementary Figure 1). A gene construct of mutated merF was designed and expressed with 1 mM IPTG and 7 h post-induction at 37°C. The cysteine residues in wild type MerF sequence induce aggregation due to disulfide bonds which makes the protein difficult to purify.
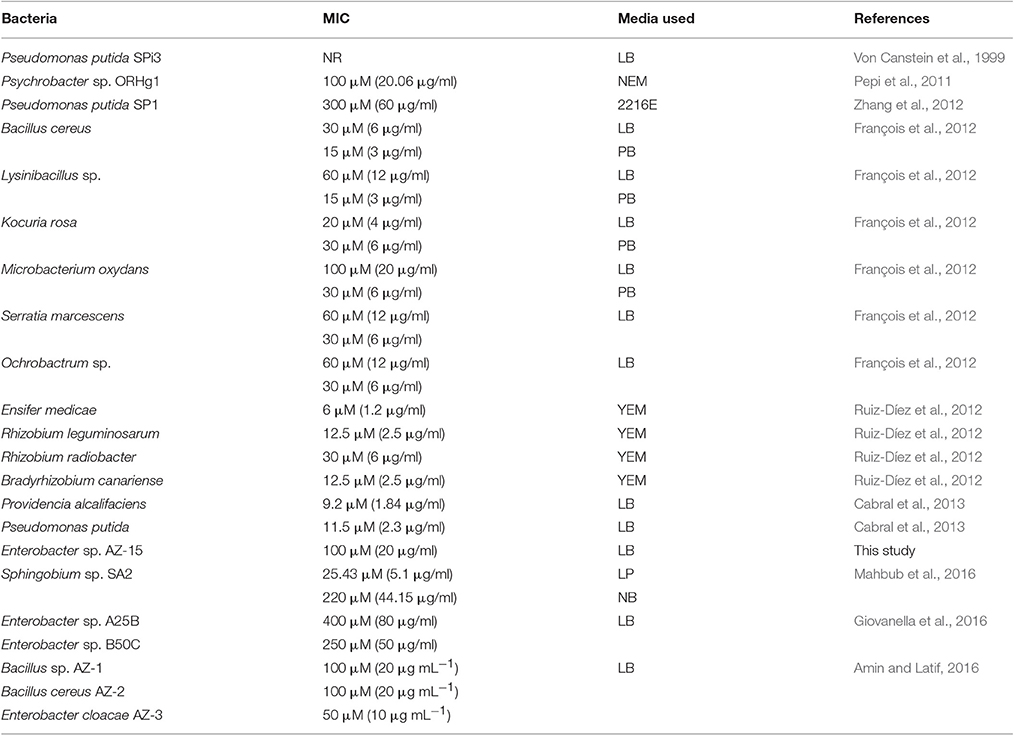
Table 4. Mercury resistant bacteria and their patterns of resistance to different concentrations of Hg2+ in different media.
Moreover, the MerFm was purified by fast protein liquid chromatography (FPLC) but Lu and Opella (2014) used high performance liquid chromatography (HPLC) for purification of wild type and truncated MerF. The high quality spectra of 1H-15N HSQC confirmed full expression of the native sequence of MerFm and each amino acid (with the exception of proline) contains one 1H-15N unit in the backbone. The hydropathy plot (Supplementary Figure 4) suggests that MerF has two hydrophobic trans-membrane helices, although the N-terminal portion of the plot is somewhat atypical. Wild-type MerF has two pairs of vicinal Cys residues (replaced by serines in MerFm), which is unusual, and contrasts with the -CXXC- sequences typically found in proteins that bind mercury and other heavy metals (Opella et al., 2002). The vicinal arrangements of the cysteines have profound consequences for their metal-binding properties and the selectivity for Hg2+ over other metals (DeSilva et al., 2002). With both metal transfer and transport activities, the structural biology of MerF is too elaborate to be explained simply by Hg2+ binding to pairs of cysteines. Therefore, it is essential to determine its three-dimensional structure in phospholipid bilayers, where it executes its functions. Experiments are underway to explore further structural properties of MerFm having natural origin.
The capability of different bacterial species to detoxify Hg2+ varies in different Hg supplemented enriched media as shown in Table 4. The present study also determined that the merF gene in Enterobacter sp. AZ-15 encodes a polypeptide sequence capable of proper folding, NMR studies and also ensures the presence of mer operon. The favorable thermodynamic and spectral characteristics of the recombinant protein will allow the detailed description of this natural protein. During the investigation of Hg-detoxification capability, Enterobacter sp. AZ-15 showed 90% detoxification potential of Hg2+ from HgCl2 supplemented LB medium. The crystalline structures of deposited Hg were visualized by SEM. Sathyavathi et al. (2013) and Mahbub et al. (2016) have previously reported the SEM analysis of same crystalline structures of detoxified Hg.
In conclusion, the expression and purification of 15N-labeled MerFm from mercury resistant Enterobacter sp. AZ-15 (MIC-20 μg/ml), was first time successfully achieved. 1H–15N HSQC NMR spectra confirmed the expression of exact number of all residues of MerFm by producing one cross peak for each 1H–15N pair in a protein. In lab scale experiment, Enterobacter sp. AZ-15 showed 90% bioremediation of Hg from Hg-supplemented LB medium. On the basis of these characteristics, mercury resistant Enterobacter sp. AZ-15 could be used as a potential candidate to deteriorate the toxic effects of Hg2+ from the environment.
Author Contributions
AA conducted the experiments and wrote the paper while ZL proofread and overall supervised the project.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This project was financially supported by Higher Education Commission, Pakistan (Pin No. 112-23128-2BM1-164) and University of the Punjab (Grant No. D/5719/Est.1). The authors are thankful to Prof. Stanley Opella, University of California, San Diego, USA for providing laboratory facilities to conduct research work on MerF protein, Prof. Simon Silver, University of Illinois, USA, and Prof. Ginro Endo, Tohoku Gakuin University, Japan for providing B. megaterium MB1 strain isolated from the sediments of Minamata Bay, as a positive control for Hg-resistance.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01250/full#supplementary-material
References
Amin, A., and Latif, Z. (2016). Screening of mercury-resistant and indole-3-acetic acid producing bacterial-consortium for growth promotion of Cicer arietinum L. J. Basic Microbiol. 57, 204–217. doi: 10.1002/jobm.201600352
Barkay, T., Miller, S. M., and Summers, A. O. (2003). Bacterial mercury resistance from atoms to ecosystems. FEMS Microbiol. Rev. 27, 355–384. doi: 10.1016/S0168-6445(03)00046-9
Barkay, T., and Poulain, A. J. (2007). Mercury (micro) biogeochemistry in polar environments. FEMS Microbiol. Ecol. 59, 232–241. doi: 10.1111/j.1574-6941.2006.00246.x
Cabral, L., Giovanella, P., Gianello, C., Bento, F. M., Andreazza, R., and Camargo, F. A. O. (2013). Isolation and characterization of bacteria from mercury contaminated sites in Rio Grande do Sul, Brazil, and assessment of methylmercury removal capability of a Pseudomonas putida V1 strain. Biodegradation 24, 319–331. doi: 10.1007/s10532-012-9588-z
Cappuccino, J. G., and Sherman, N. (2008). Microbiology: A Laboratory Manual. San Francisco, CA: Pearson/Benjamin Cummings.
Cases, I., and de Lorenzo, V. (2005). Genetically modified organisms for the environment: stories of success and failure and what we have learned from them. Int. Microbiol. 8, 213–222.
Dahlberg, C., and Hermansson, M. (1995). Abundance of Tn3, Tn21, and Tn501 transposase (tnpA) sequences in bacterial community DNA from marine environments. Appl. Environ. Microbiol. 61, 3051–3056.
Das, B. B., Nothnagel, H. J., Lu, G. J., Son, W. S., Tian, Y., Marassi, F. M., et al. (2012). Structure determination of a membrane protein in proteoliposomes. J. Am. Chem. Soc. 134, 2047–2056. doi: 10.1021/ja209464f
Dash, H. R., and Das, S. (2012). Bioremediation of mercury and the importance of bacterial mer genes. Int. Biodeterior. Biodegrad. 75, 207–213. doi: 10.1016/j.ibiod.2012.07.023
DeSilva, T. M., Veglia, G., Porcelli, F., Prantner, A. M., and Opella, S. J. (2002). Selectivity in heavy metal-binding to peptides and proteins. Biopolymers 64, 189–197. doi: 10.1002/bip.10149
Elly, C. T. (1973). Dithizone procedure for mercury analysis. J. Water Pollut. Control Fed. 45, 940–945.
François, F., Lombard, C., Guigner, J.-M., Soreau, P., Brian-Jaisson, F., Martino, G., et al. (2012). Isolation and characterization of environmental bacteria capable of extracellular biosorption of mercury. Appl. Environ. Microbiol. 78, 1097–1106. doi: 10.1128/AEM.06522-11
Ghosh, S., Sadhukhan, P., Chaudhuri, J., Ghosh, D., and Mandal, A. (1999). Purification and properties of mercuric reductase from Azotobacter chroococcum. J. Appl. Microbiol. 86, 7–12. doi: 10.1046/j.1365-2672.1999.00605.x
Gilbert, M. P., and Summers, A. O. (1988). The distribution and divergence of DNA sequences related to the Tn21 and Tn501 mer operons. Plasmid 20, 127–136. doi: 10.1016/0147-619X(88)90015-7
Giovanella, P., Cabral, L., Bento, F. M., Gianello, C., and Camargo, F. A. O. (2016). Mercury (II) removal by resistant bacterial isolates and mercuric (II) reductase activity in a new strain of Pseudomonas sp. B50A. New Biotechnol. 33, 216–223. doi: 10.1016/j.nbt.2015.05.006
Hamlett, N., Landale, E., Davis, B., and Summers, A. (1992). Roles of the Tn21 merT, merP, and merC gene products in mercury resistance and mercury binding. J. Bacteriol. 174, 6377–6385. doi: 10.1128/jb.174.20.6377-6385.1992
He, F., Gao, J., Pierce, E., Strong, P., Wang, H., and Liang, L. (2015). In situ remediation technologies for mercury-contaminated soil. Environ. Sci. Pollut. Res. 22, 8124–8147. doi: 10.1007/s11356-015-4316-y
Hobman, J., and Brown, N. (1997). Bacterial mercury-resistance genes. Met. Ions Biol. Syst. 34, 527.
Khan, H., Ahmed, M. J., and Bhanger, M. I. (2006). A simple spectrophotometric method for the determination of trace level lead in biological samples in the presence of aqueous micellar solutions. J. Spectrosc. 20, 285–297. doi: 10.1155/2006/269568
Kuliopulos, A., Shortle, D., and Talalay, P. (1987). Isolation and sequencing of the gene encoding delta 5-3-ketosteroid isomerase of Pseudomonas testosteroni: overexpression of the protein. Proc. Natl. Acad. Sci. U.S.A. 84, 8893–8897. doi: 10.1073/pnas.84.24.8893
Lu, G. J., and Opella, S. J. (2014). Resonance assignments of a membrane protein in phospholipid bilayers by combining multiple strategies of oriented sample solid-state NMR. J. Biomol. NMR 58, 69–81. doi: 10.1007/s10858-013-9806-y
Lu, G. J., Tian, Y., Vora, N., Marassi, F. M., and Opella, S. J. (2013). The structure of the mercury transporter MerF in phospholipid bilayers: a large conformational rearrangement results from N-terminal truncation. J. Am. Chem. Soc. 135, 9299–9302. doi: 10.1021/ja4042115
Mahbub, K. R., Krishnan, K., Megharaj, M., and Naidu, R. (2016). Bioremediation potential of a highly mercury resistant bacterial strain Sphingobium SA2 isolated from contaminated soil. Chemosphere 144, 330–337. doi: 10.1016/j.chemosphere.2015.08.061
Mok, T., Chen, J. S., Shlykov, M. A., and Saier, M. H. Jr. (2012). Bioinformatic analyses of bacterial mercury ion (Hg2+) transporters. Water Air Soil Pollut. 223, 4443–4457. doi: 10.1007/s11270-012-1208-3
Møller, A. K., Barkay, T., Al-Soud, W. A., Sørensen, S. J., Skov, H., and Kroer, N. (2011). Diversity and characterization of mercury-resistant bacteria in snow, freshwater and sea-ice brine from the High Arctic. FEMS Microbiol. Ecol. 75, 390–401. doi: 10.1111/j.1574-6941.2010.01016.x
Normand, P. (1995). Utilisation des séquences 16S pour le positionnement phylétique d'un organisme inconnu. Oceanis 21, 31–56.
Opella, S. J., DeSilva, T. M., and Veglia, G. (2002). Structural biology of metal-binding sequences. Curr. Opin. Chem. Biol. 6, 217–223. doi: 10.1016/S1367-5931(02)00314-9
Parks, J. M., Johs, A., Podar, M., Bridou, R., Hurt, R. A., Smith, S. D., et al. (2013). The genetic basis for bacterial mercury methylation. Science 339, 1332–1335. doi: 10.1126/science.1230667
Pepi, M., Gaggi, C., Bernardini, E., Focardi, S., Lobianco, A., Ruta, M., et al. (2011). Mercury-resistant bacterial strains Pseudomonas and Psychrobacter spp. isolated from sediments of Orbetello Lagoon (Italy) and their possible use in bioremediation processes. Int. Biodeterior. Biodegrad. 65, 85–91. doi: 10.1016/j.ibiod.2010.09.006
Powlowski, J., and Sahlman, L. (1999). Reactivity of the two essential cysteine residues of the periplasmic mercuric ion-binding protein, MerP. J. Biol. Chem. 274, 33320–33326. doi: 10.1074/jbc.274.47.33320
Ruiz-Díez, B., Qui-ones, M. A., Fajardo, S., López, M. A., Higueras, P., and Fernández-Pascual, M. (2012). Mercury-resistant rhizobial bacteria isolated from nodules of leguminous plants growing in high Hg-contaminated soils. Appl. Microbiol. Biotechnol. 96, 543–554. doi: 10.1007/s00253-011-3832-z
Sathyavathi, S., Manjula, A., Rajendhran, J., and Gunasekaran, P. (2013). Biosynthesis and characterization of mercury sulphide nanoparticles produced by Bacillus cereus MRS-1. Indian J. Exp. Biol. 51, 973–978.
Silver, S. (1992). “Bacterial heavy metal detoxification and resistance systems,” in Biotechnology and Environmental Science: Molecular Approaches, eds. S. Mongkolsuk, P. S. Lovett and J. E. Trempy. (Boston, MA: Springer US), 109–129. doi: 10.1007/978-0-585-32386-2_14
Sone, Y., Nakamura, R., Pan-Hou, H., Itoh, T., and Kiyono, M. (2013). Role of MerC, MerE, MerF, MerT, and/or MerP in resistance to mercurials and the transport of mercurials in Escherichia coli. Biol. Pharm. Bull. 36, 1835–1841. doi: 10.1248/bpb.b13-00554
Steenhuisen, F., and Wilson, S. J. (2015). Identifying and characterizing major emission point sources as a basis for geospatial distribution of mercury emissions inventories. Atmos. Environ. 112, 167–177. doi: 10.1016/j.atmosenv.2015.04.045
Summers, A. O. (1986). Organization, expression, and evolution of genes for mercury resistance. Annu. Rev. Microbiol. 40, 607–634. doi: 10.1146/annurev.mi.40.100186.003135
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tian, Y., Lu, G. J., Marassi, F. M., and Opella, S. J. (2014). Structure of the membrane protein MerF, a bacterial mercury transporter, improved by the inclusion of chemical shift anisotropy constraints. J. Biomol. NMR 60, 67–71. doi: 10.1007/s10858-014-9852-0
Veglia, G., and Opella, S. J. (2000). Lanthanide ion binding to adventitious sites aligns membrane proteins in micelles for solution NMR spectroscopy. J. Am. Chem. Soc. 122, 11733–11734. doi: 10.1021/ja002119d
Von Canstein, H., Li, Y., Timmis, K., Deckwer, W.-D., and Wagner-Döbler, I. (1999). Removal of mercury from chloralkali electrolysis wastewater by a mercury-resistant Pseudomonas putida strain. Appl. Environ. Microbiol. 65, 5279–5284.
Wagner-Döbler, I., Von Canstein, H., Li, Y., Timmis, K. N., and Deckwer, W.-D. (2000). Removal of mercury from chemical wastewater by microoganisms in technical scale. Environ. Sci. Technol. 34, 4628–4634. doi: 10.1021/es0000652
WHO (2000). Air Quality Guidelines for Europe. WHO Regional Publication, European Series Copenhagen: Regional Office for Europe, WHO.
Wilson, J. R., Leang, C., Morby, A. P., Hobman, J. L., and Brown, N. L. (2000). MerF is a mercury transport protein: different structures but a common mechanism for mercuric ion transporters? FEBS Lett. 472, 78–82. doi: 10.1016/s0014-5793(00)01430-7
Yeo, C. C., Tham, J. M., Kwong, S. M., Yiin, S., and Poh, C. L. (1998). Tn5563, a transposon encoding putative mercuric ion transport proteins located on plasmid pRA2 of Pseudomonas alcaligenes. FEMS Microbiol. Lett. 165, 253–260. doi: 10.1111/j.1574-6968.1998.tb13154.x
Keywords: Enterobacter sp. AZ-15, pET31b+, MerF, Size exclusion chromatography, 1H-15N heteronuclear single quantum coherence, Hg-detoxification, scanning electron microscopy (SEM)
Citation: Amin A and Latif Z (2017) Cloning, Expression, Isotope Labeling, and Purification of Transmembrane Protein MerF from Mercury Resistant Enterobacter sp. AZ-15 for NMR Studies. Front. Microbiol. 8:1250. doi: 10.3389/fmicb.2017.01250
Received: 28 April 2017; Accepted: 21 June 2017;
Published: 07 July 2017.
Edited by:
Stéphane Pesce, National Research Institute of Science and Technology for Environment and Agriculture, FranceReviewed by:
Willem J. H. Van Berkel, Wageningen University and Research, NetherlandsWeimin Ma, Shanghai Normal University, China
Copyright © 2017 Amin and Latif. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aatif Amin, aatifamin93@gmail.com
 Aatif Amin
Aatif Amin Zakia Latif
Zakia Latif