- 1Department of Biodiversity and Molecular Ecology, Research and Innovation Centre, Fondazione Edmund Mach, San Michele all'Adige, Italy
- 2Division de Microbiología, Universidad Miguel Hernandez, San Juan de Alicante, Spain
- 3Victorian Bioinformatics Consortium, Monash University, Melbourne, VIC, Australia
- 4Laboratory of Molecular Biology, Department of Plant Science, Wageningen University, Wageningen, Netherlands
- 5Department of Agronomy, Food, Natural Resources, Animals and Environment, University of Padova, Legnaro, Italy
- 6Department of Sustainable Agrobiosystems and Bioresources, Research and Innovation Centre, Fondazione Edmund Mach, San Michele all'Adige, Italy
- 7MOUNTFOR Project Centre, European Forest Institute, San Michele all'Adige, Italy
- 8Institute of Animal Production Systems in Mediterranean Environments-National Research Council, Sassari, Italy
The prominent feature of rhizobia is their molecular dialogue with plant hosts. Such interaction is enabled by the presence of a series of symbiotic genes encoding for the synthesis and export of signals triggering organogenetic and physiological responses in the plant. The genome of the Rhizobium sullae type strain IS123T nodulating the legume Hedysarum coronarium, was sequenced and resulted in 317 scaffolds for a total assembled size of 7,889,576 bp. Its features were compared with those of genomes from rhizobia representing an increasing gradient of taxonomical distance, from a conspecific isolate (Rhizobium sullae WSM1592), to two congeneric cases (Rhizobium leguminosarum bv. viciae and Rhizobium etli) and up to different genera within the legume-nodulating taxa. The host plant is of agricultural importance, but, unlike the majority of other domesticated plant species, it is able to survive quite well in the wild. Data showed that that the type strain of R. sullae, isolated from a wild host specimen, is endowed with a richer array of symbiotic genes in comparison to other strains, species or genera of rhizobia that were rescued from domesticated plant ecotypes. The analysis revealed that the bacterium by itself is incapable of surviving in the extreme conditions that its host plant can tolerate. When exposed to drought or alkaline condition, the bacterium depends on its host to survive. Data are consistent with the view of the plant phenotype as the primary factor enabling symbiotic nitrogen fixing bacteria to survive in otherwise limiting environments.
Introduction
The interaction between rhizobia and legumes in root nodules is an essential element in sustainable agriculture, as this symbiotic association is able to enhance biological fixation of atmospheric nitrogen (N2), and is also a paradigm in plant–microbe signaling (Young et al., 2006; Giraud et al., 2007; Wang et al., 2012). Knowledge of the whole genome would allow the specific features of each rhizobium to be identified. The prominent feature of this group of bacteria is their molecular dialogue with plant hosts, an interaction that is enabled by the presence of a series of symbiotic genes encoding for the synthesis and export of signals triggering organogenetic and physiological responses in the plant (Spaink et al., 1987; Long, 2001). In recent years, significant progress has been made in resolving the complex exchange of signals responsible for nodulation through genome assembly, mutational, and expression analysis, and proteome characterization of legumes (e.g., Sato et al., 2008; Young et al., 2011; Marx et al., 2016) and rhizobia (e.g., Giraud et al., 2007; Tolin et al., 2013; Čuklina et al., 2016; Remigi et al., 2016). In a previous study (Squartini et al., 2002), we described a novel species, R. sullae, that specifically induces symbiotic nodulation in the legume sulla (Hedysarum coronarium L. syn. Sulla coronaria [L.] Medik.; Faboideae; Hedysareae). We had previously provided the first description of the infection process of this legume by its bacterial symbiont and their morphological peculiarities (Squartini et al., 1993). Sulla is found in the Mediterranean basin with a distribution from northern Africa to southern Spain and southern Italy. It is of particular importance in agriculture due to its ability to adapt to drought and coastal conditions (Douglas, 1984), and is therefore an ideal subject for studying salt tolerance (range limit 150–700 mM NaCl), alkaline tolerance (up to pH 9–10.5), and drought stress (ranging from 0.5 to −0.95 MPa for PEG; Fitouri et al., 2012; Issolah et al., 2012). Biochemical and genetic characterization of several bacterial strains nodulating sulla (Struffi et al., 1988; Muresu et al., 2005) allowed us to select R. sullae isolate IS123T (= USDA 4950T = DSM 14623T) as type strain. Phylogenetic analyses (Squartini et al., 2002) suggest it is closely related to the widely-studied congeneric Rhizobium leguminosarum (symbiont of peas) and Rhizobium etli (symbiont of beans).
Focusing, in particular, on the genes ruling the symbiotic association with the host plant, we sequenced the genome of the R. sullae type strain IS123T in order to: (1) compare this genome with other members of the order Rhizobiales, including a conspecific isolate (R. sullae WSM1592, Yates et al., 2015), two congeneric cases (R. leguminosarum bv. viciae and R. etli), and various genera within the legume-nodulating taxa; (2) determine whether or not the type strain of R. sullae, which comes from a plant that still grows in the wild, carries a richer array of genes for the symbiotic interaction with its host; (3) assess whether or not the traits allowing the host plant to endure extreme soil conditions (drought and alkalinity) are also mirrored by appropriate determinants in the bacterial genome.
Materials and Methods
The R. sullae strain IS123T has been previously described as a new species by Squartini et al. (2002). Genomic DNA was isolated from exponentially-growing liquid cultures in Yeast-Mannitol broth. Cultures were lysed and washed, the DNA was extracted using chloroform/phenol, and, after ethanol precipitation, it was purified using a Qiagen DNeasy blood and tissue kit, according to the manufacturer's protocol (Qiagen, Hilden, Germany). Libraries were prepared with the TruSeq DNA Library Preparation Kit (Illumina Inc.), as described by the manufacturer. DNA was sequenced using the Illumina HiSeq platform, and the sequences assembled with an Edena (Exact DE Novo Assembler; Hernandez et al., 2008), which uses an overlap layout consensus algorithm with an overlap cutoff of 47 bases. Assembled contigs were further scaffolded using SSPACE Basic (version 2.0; Boetzer et al., 2011). The scaffolded version of the assembled genome was used for gene prediction and annotation using two independent pipelines: RAST, available at: http://rast.nmpdr.org/ (Aziz et al., 2008), and the Prokka bacterial genome annotation tool (Seemann, 2014), which uses Prodigal (Hyatt et al., 2010) for prokaryotic gene identification. While running the Prokka, the Rhizobiales order was selected to represent the genomes from this class in order to increase the robustness of the annotations. The Prokka and RAST outputs revealed slight differences in the functional annotations, so we manually checked them focusing on the symbiotic nodulation and nitrogen fixation genes. The curated version of Prokka was used as the final annotation of R. sullae. To ascertain the similarity between the assembled genome and previously published genomes of Rhizobium etli and R. sullae WSM1592, we estimated average nucleotide identity (ANI), as previously described by Goris et al. (2007). The ANI analysis was limited to the taxa which were expected to share most identities with the strain under study.
A whole-genome orthology search to identify conserved functions across different organisms was run on six additional rhizobial genomes using the Reciprocal BEST BLAST HIT (RBBH) (Ward and Moreno-Hagelsieb, 2014) with an E-value cutoff of 1E-5. Genes with identity >60% and coverage higher than 60% were considered orthologous.
The MAPLE resource (Metabolic And Physiological potentiaL Evaluator; available at: http://www.genome.jp/tools/maple/help.html) was used to estimate function abundance and evaluate metabolic and physiological potential. The reference database was KEGG (Kyoto Encyclopedia of Genes and Genomes), and R. sullae proteins were mapped and normalized on the ribosomal proteins counts in its pathway database. KAAS (KEGG Automatic Annotation Server) was used for ortholog assignment (KO, Kegg Orthology) and pathway mapping. The PHASTER tool was used to search for sequences of phages and prophages (Arndt et al., 2016). To ascertain whether any genes relevant for rhizobia could be missing, an HMM (Hidden Markow Model) search of the Rhizobiales was carried out to extract the core pan-genome, which was aligned with the genomes of the other rhizobia and scored for conservation percentage. The analysis was carried out using bcgTree, which is a bacterial core gene analyzer (http://www.dna-analytics.biozentrum.uni-wuerzburg.de, Ankenbrand and Keller, 2016).
Results
17,902,513 paired end (PE) reads (2 × 100 bp) were obtained, accounting for a total of 9.1 Gb sequence information.
The draft genome assembly contains 447 contigs with a total assembly size of 7,889,725 bp and an N50 of 72.64 kb, and 317 scaffolds with a total assembly size of 7,889,576 bp and an N50 of 118.24 kb. The longest contig was 296,399 bp, the longest scaffold 488 bp. The whole genome GC content was 59.88% (Table 1). Sequence annotation revealed 7,776 protein coding sequences (CDSs), which is more than those found in the other R. sullae, R. leguminosarum, and R. etli genomes reported so far. A total of 51 tRNA genes and 6 rRNA (rrn) operons were identified.
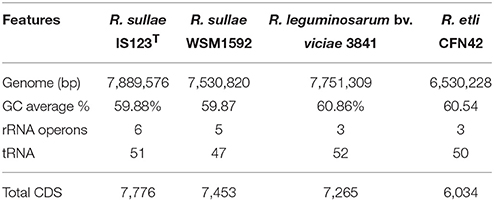
Table 1. Comparative genome features of Rhizobium sullae IS123T, its co-specific R. sullae WSM1592, and its two taxonomically closest relatives R. leguminosarum biovar viciae 3841 and R. etli CFN42.
The raw reads of the R. sullae IS123T genome are available in the European Bioinformatics Institute (EBI) database under project number PRJEB9435. The genome assembly can be accessed under the code ERZ403196 (Sample id: ERS738956, Assembly accession: GCA_900169785, WGS account: FWER01; Scaffold accession range: FWER01000001-FWER01000317).
The gene predictions, CDS FASTA files, and the corresponding proteins are all provided as Supplementary Datasets 2–6.
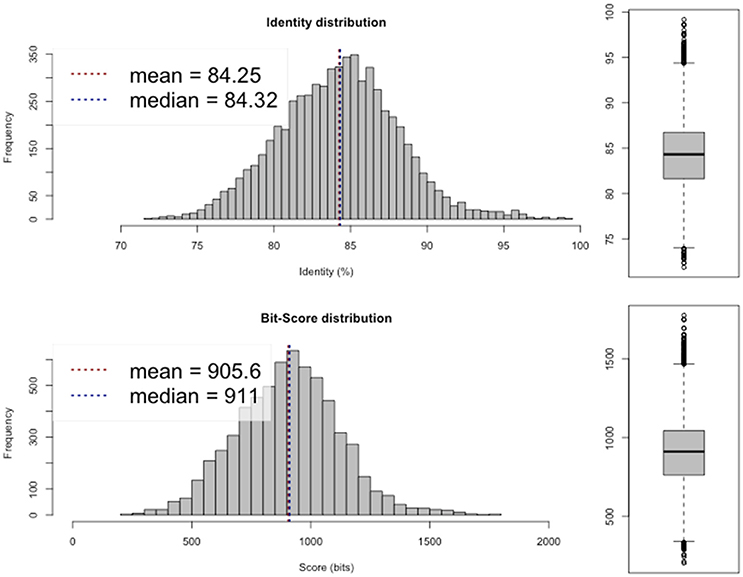
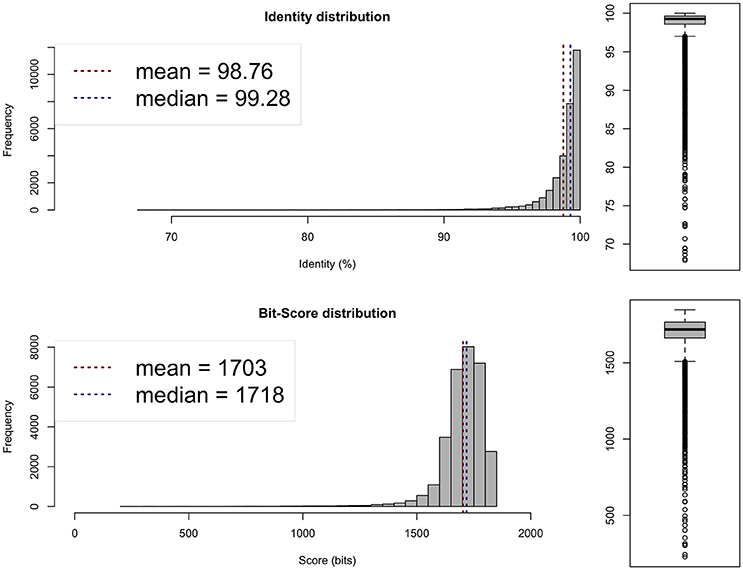
Average nucleotide identity (ANI; Goris et al., 2007; available from http://enve-omics.ce.gatech.edu/ani/) was calculated to assess the similarity between the assembled genome and the genomes of R. sullae WSM1592 and R. etli CFN42. The analysis revealed a high average ANI (97–98%) between the sequenced genome and the genomes of the two afore-mentioned related species (Figures 1, 2), confirming their evolutionary closeness.
In addition, the R. sullae IS123T proteins were mapped against the KEGG pathways database, and the result normalized by the ribosomal proteins. The resulting KEGG orthology assignment of genes and modules, and the corresponding rarefaction curves are shown in Supplementary Material S1, along with an annotated map of a prophage that was found in the R. sullae IS123T genome.
Core pan-genome analysis of the Rhizobiales was carried out to see whether any major genes were missing. The core was found to include 108 genes and the analysis indicated that the R. sullae IS123T genome presented here is more than 99% complete with respect to the core rhizobial pan-genome. This confirms both the trustworthiness of the ortholog analysis and the overall good quality of the genome coverage. The results of the pan-genome analysis are available as Supplementary Dataset 7.
Functional categorization of the genes present in the IS123T strain based on the RAST annotation is shown in Figure 3. While this kind of analysis is often not exhaustive, as the output depends on the accuracy of the gene classification databases, a general picture can be drawn. The majority of its genes are predicted to be involved in carbohydrate metabolism, which is consistent with the rhizobial lifestyle within which exopolysaccharides are important traits involved in host plant recognition, lectin binding, and attachment to root surfaces. In addition, a considerable array of genes relate to membrane transport, and their involvement in the secretion machinery for metabolites and signals is also important in plant–microbe interactions.
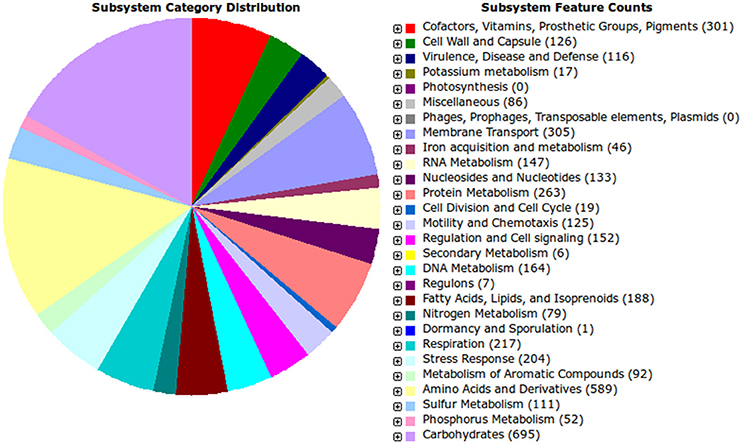
Figure 3. Gene category classification results. The annotation data obtained from the RAST procedure were used to assemble the image.
Sequence annotation confirmed the presence of genes involved in nodulation (nod) and symbiotic N2 fixation (nif and fix). The R. sullae type strain genome displayed all the essential nodulation genes, which are required for early symbiotic nodulation via nod factor production (Baev et al., 1992; Young et al., 2006). Among the nod genes, nodA was present in two copies, the first a short truncated form located in the nodABC operon, the second a supposedly functional full copy upstream of a gene for carbonic anhydrase. Opposite the truncated nodA and its nod box (the NodD regulator-binding cassette for nod genes) in the nodABC operon on the complementary strand, there is a copy of nodD interrupted by the insertion element ISRh1 (Meneghetti et al., 1996). Two other intact copies of nodD are located elsewhere, one of them close to its cognate receptor syrM, while nodB and nodC are present in the afore-mentioned operon, and nodH is immediately downstream of nodC. The nod factor export machinery also appears to be in place, due to the presence of the transport-related nodIJ operon, whose origin has been traced from Beta- to Alpha-Proteobacteria (Aoki et al., 2013). The presence of other defined nod genes means we can predict the following potential characteristics of the corresponding nod factors: regular acylation (presence of nodFE), O-acetylation (nodL and nodX), sulfation (nodH, nodPQ), and a putative O-carbamoylation (nolO). These features, encoding for precise nod factor decorations, may account for the very tight host specificity of this rhizobium for its symbiont plant H. coronarium.
Among the nif genes, which are involved in nitrogen fixation, we identified the structural nitrogenase units nifH, nifD, and nifK, plus regulatory and accessory determinants (nifA, nifB, nifE, nifN, nifS, nifT, nifU, nifW, nifZ, and the 4Fe-4S ferredoxin nitrogenase-associated gene). Among the fix genes, we detected fixL, fixS, and fixC, which potentially encode a symbiotically essential cbb3 high-affinity terminal oxidase, a rate-limiting component for symbiotic N2 fixation (Patschkowski et al., 1996). In fact, ab-initio gene prediction using Prodigal identified three copies of the cbb3 high-affinity terminal oxidase in this R. sullae IS123T genome. In confirmation of the presence of other genes related to symbiosis, we also detected sufBCDE, the gene cluster acting as house-keeping storage genes of FeS clusters (Trotter et al., 2009). Furthermore, we found two copies of gndA gene, which catalyzes 6-phosphogluconate dehydrogenase and is the main source of catabolizing sugars in rhizobial species using the Entner-Doudoroff pathway, as previously reported for R. leguminosarum and R. etli (González et al., 2006; Young et al., 2006).
In order to assess the uniqueness of the genes and the level of conservation of those shared across related taxa, a whole genome orthology search was run on six additional rhizobial genomes. Besides R. sullae IS123T, we used the following species for the analysis (gene counts are indicated in parenthesis): R. sullae WSM1592 (6985), R. leguminosarum bv. viciae 3841 (7281), R. etli CFN42 (5963), Sinorhizobium meliloti 1021 (6217), Mesorhizobium loti MAFF303099 (7283), and Bradyrhizobium japonicum USDA110 (8317).
Occurrences of the corresponding orthologous genes are shown in the “Supplementary Dataset 1, Orthology analysis.” A zero (0) in a cell indicates the absence of an orthologous gene. The second worksheet contains the gene nomenclature and features of all of the genes compared.
On inspecting all the genes known to correspond to symbiotic traits, (nodulation, nitrogen fixation, or their ancillary metabolism), we found that, compared with the type strain R. sullae IS123T, its conspecific R. sullae WSM1592 lacks nifQ (the molybdenum donor for nitrogenase synthesis), the second copy of the structural nifH gene for nitrogenase, a copy of the fixN Cytochrome c oxidase subunit, a copy of the fixL oxygen sensor protein, a copy of the fixK nitrogen fixation regulation protein, and the gene for ferredoxin I, an enzyme linked to the electron flux for N2 reduction to nitrogenase.
The symbiotic genes that are present in R. sullae IS123T but have no orthologs in either of the two species of the same genus and closest to R. sullae according to 16S rRNA taxonomy, namely R. leguminosarum bv. viciae and R. etli, include, in addition to the above: the nodG (fabG) 3-oxoacyl acyl-carrier protein reductase; nodM, encoding functions for efficient Nod signal production; fixJ, the response regulator inducing nif operons in response to microaerobiosis within the nodule tissue; the putative nitrogen fixation protein gene fixT; a copy of the nod factor synthase nodA; a copy of the nodD2 and syrM genes encoding for flavonoid-responsive transcriptional activators; and a copy of the nodU putative carbamoyl transferase.
Interestingly, the ortholog search across the seven taxa, including the three different genera Bradyrhizobium, Mesorhizobium, and Sinorhizobium (Ensifer), revealed the R. sullae IS123T genome to be the one displaying the highest number of genes related to symbiosis, which was 59.
Since the host plant H. coronarium, as mentioned in the introduction, has remarkable properties of tolerance to several environmental stress factors, such as salinity, drought, and alkaline soil pH, and displays regular root nodulation under such conditions, we inspected the genome of its R. sullae microsymbiont to search for genes which could account for corresponding genotypes on the bacterial side.
We identified genes important in maintaining osmoregularity, which included the osmotically inducible protein C. However, in bacteria this gene is reported to be related not to saline conditions, but rather to responses to organic hydroperoxides and reactive oxygen species in general (Lesniak et al., 2002).
An integral membrane protein, YggT, involved in response to extracytoplasmic stress (osmotic shock) is present, but this gene appears to be widespread among bacteria, including Escherichia coli, and is not characteristic of isolates endowed with particular tolerances. The same goes for Aquaporin Z, relevant for osmoregulation but present in E. coli as well (Delamarche et al., 1999). In general, the R. sullae genome does not reveal its distinctiveness in terms of membrane physiology and selectivity, and it contains customary features, such as the osmolarity sensor envZ, the osmosensitive K+ channel histidine kinase KdpD, and the beta-(1–>2) glucan export ATP-binding/permease protein NdvA, which is responsible for the synthesis of osmoregulated periplasmic glucans. These oligosaccharides are common not only to most rhizobia but to the whole Proteobacteria phylum, in which glucan concentration in the periplasm increases in response to a decrease in environmental osmolarity (Bohin, 2000). This finding is again not indicative of any adaptation to salty conditions as the response is instead to the opposite scenario (diluted circulating solution). The presence of this gene is nevertheless worth remarking on, as it is another corollary to the symbiotic proficiency of R. sullae. Indeed, mutations in the same ndvA gene in S. meliloti result in the delayed formation of numerous small white nodules that are not invaded by the mutant bacteria and are consequently unable to fix nitrogen (Ielpi et al., 1990).
Considering that the host plant is able to withstand 700 mM NaCl, and is regularly encountered with well-developed root nodules in the Tuscan pliocenic clays at pH 9.6, the absence of prominent traits of extremophily among the genes of its bacterial symbiont could appear to be in contrast to the habitats where the host plant and bacteria meet. However, it is interesting to recall that in a prior study on the stress tolerance of several strains of R. sullae, including the present type strain IS123T, compared with other rhizobia (Struffi et al., 1988), the R. sullae strains did not perform any better than the other rhizobia. Regarding NaCl, in most strains growth was limited to 0.5%, the same limit in R. leguminosarum bv. trifolii (clover symbiont) and R. leguminosarum bv. viciae (pea symbiont). The limit was 1% in only two strains, but this was still lower than in S. meliloti (symbiont of alfalfa), which tolerated values up to 3%. As for alkali tolerance, R. sullae did not perform very differently from the others, and R. sullae IS123T, in particular, was able resist up to pH 8.5, the same level attained by the pea and clover symbionts, and lower than that recorded for the alfalfa symbiont (Struffi et al., 1988). These data are consistent with the lack of obvious stress tolerance traits in R. sullae emerging from the present genome analysis, and allow for an interpretation focused on the ecological coexistence of tolerant hosts and non-tolerant microorganisms. In our earlier experiment with R. leguminosarum bv. trifolii (Casella et al., 1988), we demonstrated that the ability to survive in the presence of increasing doses of different heavy metals was dependent on the plant's phenotype, irrespective of the bacterial phenotype. For example, at chromium levels which were still tolerated by clover but high above the minimum inhibitory concentration for the rhizobium, the latter was nevertheless able to effectively nodulate the plant. Interestingly, we also noticed that on curing those rhizobia of their large symbiotic plasmids, they lost the ability to induce and invade root nodules, but at the same time their intrinsic level of chromium tolerance increased. These data, along with the afore-mentioned absence of either genomic or phenotypic evidence for salt- or alkali-tolerance traits in R. sullae, lead us to the view that in these endophytic plant–microbe interactions, of the two partners the plant host is the one that is critically endowed with the ability to occupy challenging environments. In this sense, the symbiosis assumes the role of a shelter for the hosted microorganisms, which could otherwise not endure the same stress factors when facing them as free living cells. In the case of heavy metals, as the study cited showed, it was also rather revealing to see that rhizobia could “opt” between entering the plant, which offers a shielded niche, or renounce the whole symbiotic relationship by dropping an extrachromosomal replicon. The consequent loss of interactivity, due to the absence of the plasmid-borne genes, accounts for several membrane permeability changes, which are related to overall increased resistance to extracytoplasmic stress factors.
In the particular case of R. sullae, it should be added that its host, H. coronarium, has unique features of environmental adaptation, which, as far as is known, have not been encountered in any other plant. As we described in an earlier report (Tola et al., 2009), this genus is capable of forming modified lateral roots (called shovel roots) that accumulate calcium crystals, a mechanism that explains the plant's ability to grow, often as a unique vegetation form, in calcareous soils of extreme alkalinity. The scavenging of Ca2+ ions locally affects the chemical equilibrium of the soil carbonate buffer and allows efficient acidification of the rhizosphere, even in limestone-rich, highly basic soils. In the same report, we showed that H. coronarium is able to change the pH of its surrounding solution, even from alkalinity to acidity, while other legumes are only able to exert the reverse effect. These findings further support the evidence that it is the plant's task to withstand the harsh conditions characterizing its habitat. At the same time, the plant is able to improve the conditions in the root microenvironment, allowing its microbial partner to multiply in a situation it would otherwise not be able to withstand, and eventually to be fully rescued within the endophytic and symbiotic domains. The picture of this interaction is thus reconciled with the genome annotations of R. sullae, which, as mentioned above, featured genes that were not very different from those of most average bacteria in terms of coping with environmental stress, yet also featured a plethora of genes for symbiotic interactions.
The genome of R. sullae IS123T presented here draws attention to the way it differs from the available genome of the cognate strain R. sullae WSM1592, and has shown how the type strain appears to be endowed with a richer array of genes pertaining to the symbiotic phenotypes of nodulation and nitrogen fixation. Moreover, there are certain ecological aspects that allow us to make further inferences. These concern the migratory path of the host plant on its colonization route into Europe from the North African plains, which is presumed to coincide with its domestication and subsequent cropping. The type strain sequenced here was, in fact, isolated in the early seventies from a wild stand of its host in Cadiz province, the southernmost part of Spain, facing the Straight of Gibraltar and Africa. Strain WSM1592, instead, was recovered in 1995 from cultivated Sulla plants grown at the Ottava experimental station on the Italian island of Sardinia. The sites where the two strains were isolated within their host home range are shown on the map of the western Mediterranean area in Figure 4. Some geographical as well as ecological variables may account for the observed inter-strain variations between IS123T and WSM1592. In this regard, it is worth remarking that H. coronarium is one of the few plant species which still exist in both wild and cultivated conditions. This unique feature makes it possible to sample and compare the genomes of two strains, one from the wild (IS123T), the other from an agricultural context (WSM1592), and to investigate the subtle ways in which they differ in spite of an overall conserved genome. The host plant is considered to be native to Algeria, Tunisia and Morocco, as well as Spain (http://www.ildis.org/), where it is found essentially in the southernmost region. The presumed origin of the plant species in the north-western African belt above the Saharan Atlas range is supported by the high frequency of sites in which H. coronarium is encountered in natural populations in that area compared with European countries, and by the fact that the related species Hedysarum flexuosum is the only legume nodulated by rhizobia, whose 16S rRNA sequence displays a >99% similarity with the R. sullae type strain IS123T (Aliliche et al., 2016). These nodules are ineffective, as R. sullae is, as far as is currently known, fully symbiotic only with H. coronarium.
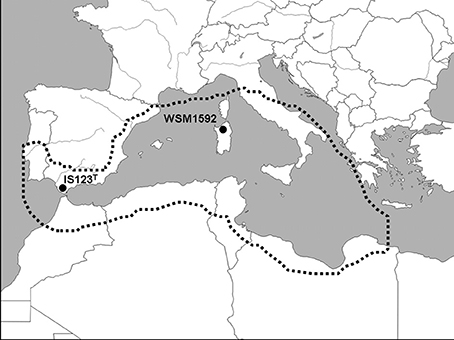
Figure 4. Map of the Western Mediterranean region showing the isolation sites of Rhizobium sullae IS123T and WSM1592. The dotted line encompasses sites where the host plant Hedysarum coronarium L. (Sulla coronaria Medik) is currently distributed, and includes areas where the plant has been attributed the status “native” (Algeria, Morocco, Tunisia, Spain), or “uncertain origin,” i.e., possibly introduced (Corsica, Sardinia, Continental Italy, Sicily, Lybia, Egypt). The plant is also naturalized or cropped in other areas, where it has been intentionally introduced, including the Balearic Islands, Portugal, former Yugoslavia, Malta, Lebanon, and Syria. Source of the distribution data: International Legume Database and Information Service (http://www.legumes-online.net/ildis/aweb/database.htm).
In conclusion, the present study enabled us to verify ecological correspondences between host–plant lifestyles and bacterial symbiont genotypes. In addition to the interest in comparing genomic features of isolates from cultivated vs. wild Sulla, the present genome, a strain collected from a wild specimen thriving in an arid region of the Mediterranean, provided us with the possibility to investigate, and to rule out, the presence of genes related to drought and salt tolerance, which are two major characteristics of the naturally-occurring ecotypes of its plant host. Its range extends throughout the near-desert belt in Northern Africa, and its invasion of Mediterranean Europe has apparently followed a route via Gibraltar, the site where R. sullae IS123T was isolated and the transcontinental crossing point.
Author Contributions
AS and RM: conceived the project. Rv, RG, AG, and EP: performed the sequencing GS, TS, NL, and RR: were responsible of bioinformatics analysis. AS: wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
GS thanks the Department of Biodiversity and Molecular Ecology, Research and Innovation Centre, Fondazione Edmund Mach, San Michele all'Adige, Italy, for computational support. Tessa Say is gratefully acknowledged for revising the English language style throughout the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01348/full#supplementary-material
References
Aliliche, K., Beghalem, H., Landoulsi, A., and Chriki, A. (2016). Molecular phylogenetic analysis of Rhizobium sullae isolated from Algerian Hedysarum flexuosum. Antonie Van Leeuwenhoek 109, 897–906. doi: 10.1007/s10482-016-0688-3
Ankenbrand, M. J., and Keller, A. (2016). bcgTree: automatized phylogenetic tree building from bacterial core genomes. Genome 59, 783–791. doi: 10.1139/gen-2015-0175
Aoki, S., Ito, M., and Iwasaki, W. (2013). From β-to α-proteobacteria: the origin and evolution of rhizobial nodulation genes. nodIJ. Mol. Biol. Evol. 30, 2494–2508. doi: 10.1093/molbev/mst153
Arndt, D., Grant, J., Marcu, A., Sajed, T., Pon, A., Liang, Y., et al. (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21. doi: 10.1093/nar/gkw387
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Baev, N., Schultze, M., Barlier, I., Ha, D. C., Virelizier, H., Kondorosi, E., et al. (1992). Rhizobium nodM and nodN genes are common nod genes: nodM encodes functions for efficiency of nod signal production and bacteroid maturation. J. Bacteriol. 174, 7555–7565. doi: 10.1128/jb.174.23.7555-7565.1992
Boetzer, M., Henkel, C. V., Jansen, H. J., Butler, D., and Pirovano, W. (2011). Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27, 578–579. doi: 10.1093/bioinformatics/btq683
Bohin, J.-P. (2000). Osmoregulated periplasmic glucans in Proteobacteria. FEMS Microbiol. Lett. 186, 11–19. doi: 10.1111/j.1574-6968.2000.tb09075.x
Casella, S., Frassinetti, S., Lupi, F., and Squartini, A. (1988). Effect of cadmium, chromium and copper on symbiotic and free-living Rhizobium leguminosarum biovar trifolii. FEMS Microbiol. Lett. 49, 343–347. doi: 10.1111/j.1574-6968.1988.tb02754.x
Čuklina, J., Hahn, J., Imakaev, M., Omasits, U., Förstner, K. U., Ljubimov, N., et al. (2016). Genome-wide transcription start site mapping of Bradyrhizobium japonicum grown free-living or in symbiosis - a rich resource to identify new transcripts, proteins and to study gene regulation. BMC Genomics 17:302. doi: 10.1186/s12864-016-2602-9
Delamarche, C., Thomas, D., Rolland, J.-P., Froger, A., Gouranton, J., Svelto, M., et al. (1999). Visualization of AqpZ-mediated water permeability in Escherichia coli by cryoelectron microscopy. J. Bacteriol. 181, 4193–4197.
Douglas, G. B. (1984). Seed production of sulla: a plant for soil conservation. Proc. N.Z. Grassland Assoc. 45, 239–242.
Fitouri, S. D., Trabelsi, D., Saïdi, S., Zribi, K., Jeddi, F. B., and Mhamdi, R. (2012). Diversity of rhizobia nodulating sulla (Hedysarum coronarium L.) and selection of inoculant strains for semi-arid Tunisia. Ann. Microbiol. 62, 77–84. doi: 10.1007/s13213-011-0229-2
Giraud, E., Moulin, L., Vallenet, D., Barbe, V., Cytryn, E., Avarre, J. C., et al. (2007). Legumes symbioses: absence of Nod genes in photosynthetic bradyrhizobia. Science 316, 1307–1312. doi: 10.1126/science.1139548
González, V., Santamaría, R. I., Bustos, P., Hernández-González, I., Medrano-Soto, A., Moreno-Hagelsieb, G., et al. (2006). The partitioned Rhizobium etli genome: genetic and metabolic redundancy in seven interacting replicons. Proc. Natl. Acad. Sci. U.S.A. 103, 3834–3839. doi: 10.1073/pnas.0508502103
Goris, J., Konstantinidis, K. T., Klappenbach, J. A., Coenye, T., Vandamme, P., and Tiedje, J. M. (2007). DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91. doi: 10.1099/ijs.0.64483-0
Hernandez, D., François, P., Farinelli, L., Osterâs, M., and Schrenzel, J. (2008). De novo bacterial genome sequencing: millions of very short reads assembled on a desktop computer. Genome Res. 18, 802–809. doi: 10.1101/gr.072033.107
Hyatt, D., Chen, G. L., Locascio, P. F., Land, M. L., Larimer, F. W., and Hauser, L. J. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119
Ielpi, L., Dylan, T., Ditta, G. S., Helinski, D. R., and Stanfield, S. W. (1990). The ndvB locus of Rhizobium meliloti encodes a 319-kda protein involved in the production of beta-(1-2)-glucan. J. Biol. Chem. 15, 2843–2851.
Issolah, R., Tahar, A., Derbal, N., Zidoun, F., Ait Meziane, M. Z., Oussadi, A., et al. (2012). Ecological characterization of the natural habitat of Sulla (Fabaceae) in Northeastern Algeria. Rev. D' Ecol. 67, 295–304. Available online at: http://hdl.handle.net/2042/55922
Lesniak, J., Barton, W. A., and Nikolov, D. B. (2002). Structural and functional characterization of the Pseudomonas hydroperoxide resistance protein Ohr. EMBO J. 21, 6649–6665. doi: 10.1093/emboj/cdf670
Long, S. R. (2001). Genes and signals in the Rhizobium-legume symbiosis. Plant Physiol. 125, 69–72. doi: 10.1104/pp.125.1.69
Marx, H., Minogue, C. E., Jayaraman, D., Richards, A. L., Kwiecien, N. W., Sihapirani, A. F., et al. (2016). A proteomic atlas of the legume Medicago truncatula and its nitrogen-fixing endosymbiont Sinorhizobium meliloti. A proteomic atlas of the legume Medicago truncatula and its nitrogen-fixing endosymbiont Sinorhizobium meliloti. Nat. Biotechnol. 34, 1198–1205 doi: 10.1038/nbt.3681
Meneghetti, F., Alberghini, S., Tola, E., Giacomini, A., Ollero, F. J., Squartini, A., et al. (1996). Presence of unique repeated insertion sequences in nodulation genes of Rhizobium ‘hedysari.’ Plant Soil 186, 113–120. doi: 10.1007/BF00035064
Muresu, R., Sulas, L., Polone, E., and Squartini, A. (2005). PCR primers based on different portions of insertion elements can assist genetic relatedness studies, strain fingerprinting and species identification in rhizobia. FEMS Microbiol. Ecol. 54, 445–453. doi: 10.1016/j.femsec.2005.05.008
Patschkowski, T., Schluter, A., and Priefer, U. B. (1996). Rhizobium leguminosarum bv viciae contains a second fnr/fixK-like gene and an unusual fixL homologue. Mol. Microbiol. 21, 267–280. doi: 10.1046/j.1365-2958.1996.6321348.x
Remigi, P., Zhu, J., Young, J. P. W., and Masson-Boivin, C. (2016). Symbiosis within symbiosis: evolving nitrogen-fixing legume symbionts. Trends Microbiol. 24, 63–75. doi: 10.1016/j.tim.2015.10.007
Sato, S., Nakamura, Y., Kaneko, T., Asamizu, E., Kato, T., Nakao, M., et al. (2008). Genome structure of the legume, Lotus japonicus. DNA Res. 15, 227–239. doi: 10.1093/dnares/dsn008
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Spaink, H. P., Wijffelman, C. A., Pees, E., Okker, R. J. H., and Lugtenberg, B. J. J. (1987). Rhizobium nodulation gene nodD as a determinant of host specificity. Nature 328, 337–340. doi: 10.1038/328337a0
Squartini, A., Dazzo, F. B., Casella, S., and Nuti, M. P. (1993). The root nodule symbiosis between Rhizobium ‘hedysari’ and its drought-tolerant host Hedysarum coronarium. Symbiosis 15, 227–238.
Squartini, A., Struffi, P., Döring, H., Selenska-Pobell, S., Tola, E., Giacomini, A., et al. (2002). Rhizobium sullae sp. nov., (formerly 'Rhizobium hedysari'), the root-nodule microsymbiont of Hedysarum coronarium L. Int. J. Syst. Evol. Microbiol. 52, 1267–1276. doi: 10.1099/00207713-52-4-1267
Struffi, P., Corich, V., Giacomini, A., Benguedouar, A., Squartini, A., Casella, S., et al. (1988). Metabolic properties, stress tolerance and macromolecular profiles of rhizobia nodulating Hedysarum coronarium. J. Appl. Microbiol. 84, 81–89. doi: 10.1046/j.1365-2672.1997.00318.x
Tola, E., Henriquez-Sabà, J. L., Polone, E., Dazzo, F. B., Concheri, G., Casella, S., et al. (2009). Shovel roots: a unique stress-avoiding developmental strategy of the legume plant Hedysarum coronarium L. Plant Soil 322, 25–37. doi: 10.1007/s11104-008-9861-4
Tolin, S., Arrigoni, G., Moscatiello, R., Masi, A., Navazio, L., Sablok, G., et al. (2013). Quantitative analysis of the naringenin-inducible proteome in Rhizobium leguminosarum by isobaric tagging and mass spectrometry. Proteomics 13, 1961–1972. doi: 10.1002/pmic.201200472
Trotter, V., Vinella, D., Loiseau, L., De Choudens, S. O., Fontecave, M., and Barras, F. (2009). The CsdA cysteine desulphurase promotes Fe/S biogenesis by recruiting Suf components and participates to a new sulphur transfer pathway by recruiting CsdL (ex-YgdL), a ubiquitin-modifying-like protein. Mol. Microbiol. 74, 1527–1542. doi: 10.1111/j.1365-2958.2009.06954.x
Wang, D., Yang, S. M., Tang, F., and Zhu, H. Y. (2012). Symbiosis specificity in the legume - rhizobial mutualism. Cell. Microbiol. 14, 334–342. doi: 10.1111/j.1462-5822.2011.01736.x
Ward, N., and Moreno-Hagelsieb, G. (2014). Quickly finding orthologs as reciprocal best hits with BLAT, LAST, and UBLAST: How much do we miss? PLoS ONE 9:e101850. doi: 10.1371/journal.pone.0101850
Yates, R., Howieson, J., De Meyer, S. E., Tian, R., Seshadri, R., Pati, A., et al. (2015). High-quality permanent draft genome sequence of Rhizobium sullae strain WSM1592; a Hedysarum coronarium microsymbiont from Sassari, Italy. Stand. Genomic Sci. 10, 1–6. doi: 10.1186/s40793-015-0020-2
Young, J. P. W., Crossman, L., Johnston, A., Thomson, N., Ghazoui, Z., Hull, K., et al. (2006). The genome of Rhizobium leguminosarum has recognizable core and accessory components. Genome Biol. 7:R34. doi: 10.1186/gb-2006-7-4-r34
Keywords: Rhizobium sullae, type strain, Hedysarum coronarium, Sulla coronaria, nod genes, host-specific symbiotic adaptation, root-nodule bacteria, nitrogen fixation
Citation: Sablok G, Rosselli R, Seeman T, van Velzen R, Polone E, Giacomini A, La Porta N, Geurts R, Muresu R and Squartini A (2017) Draft Genome Sequence of the Nitrogen-Fixing Rhizobium sullae Type Strain IS123T Focusing on the Key Genes for Symbiosis with its Host Hedysarum coronarium L. Front. Microbiol. 8:1348. doi: 10.3389/fmicb.2017.01348
Received: 17 March 2017; Accepted: 03 July 2017;
Published: 26 July 2017.
Edited by:
Vasco Ariston De Carvalho Azevedo, Universidade Federal de Minas Gerais, BrazilReviewed by:
Luis Carlos Guimarães, Federal University of Pará, BrazilAlice Rebecca Wattam, Virginia Tech, United States
Siomar De Castro Soares, Universidade Federal do Triângulo Mineiro, Brazil
Copyright © 2017 Sablok, Rosselli, Seeman, van Velzen, Polone, Giacomini, La Porta, Geurts, Muresu and Squartini. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gaurav Sablok, sablokg@gmail.com
Andrea Squartini, squart@unipd.it
 Gaurav Sablok
Gaurav Sablok Riccardo Rosselli
Riccardo Rosselli Torsten Seeman3
Torsten Seeman3 Robin van Velzen
Robin van Velzen Alessio Giacomini
Alessio Giacomini Nicola La Porta
Nicola La Porta Rene Geurts
Rene Geurts Rosella Muresu
Rosella Muresu Andrea Squartini
Andrea Squartini