- Department of Botany, Institute of Science, Banaras Hindu University, Varanasi, India
Alternaria is an important fungus to study due to their different life style from saprophytes to endophytes and a very successful fungal pathogen that causes diseases to a number of economically important crops. Alternaria species have been well-characterized for the production of different host-specific toxins (HSTs) and non-host specific toxins (nHSTs) which depend upon their physiological and morphological stages. The pathogenicity of Alternaria species depends on host susceptibility or resistance as well as quantitative production of HSTs and nHSTs. These toxins are chemically low molecular weight secondary metabolites (SMs). The effects of toxins are mainly on different parts of cells like mitochondria, chloroplast, plasma membrane, Golgi complex, nucleus, etc. Alternaria species produce several nHSTs such as brefeldin A, tenuazonic acid, tentoxin, and zinniol. HSTs that act in very low concentrations affect only certain plant varieties or genotype and play a role in determining the host range of specificity of plant pathogens. The commonly known HSTs are AAL-, AK-, AM-, AF-, ACR-, and ACT-toxins which are named by their host specificity and these toxins are classified into different family groups. The HSTs are differentiated on the basis of bio-statistical and other molecular analyses. All these toxins have different mode of action, biochemical reactions and signaling mechanisms to cause diseases. Different species of Alternaria produced toxins which reveal its biochemical and genetic effects on itself as well as on its host cells tissues. The genes responsible for the production of HSTs are found on the conditionally dispensable chromosomes (CDCs) which have been well characterized. Different bio-statistical methods like basic local alignment search tool (BLAST) data analysis used for the annotation of gene prediction, pathogenicity-related genes may provide surprising knowledge in present and future.
Introduction
Fungal kingdom is very interesting in both useful and harmful point of view, which includes more than 1.5 million species, but only 100,000 species have been described, out of them 15,000 species cause disease in plants (Maharshi and Thaker, 2012). Due to increasing plants and fungal diversity, the complexity of pathogenic mechanism also increases between them on the morphologically level by forming a highly specialized structure of infections (Hawkswort, 1991; Horbach et al., 2011). Fungi produce various secondary metabolites (SMs) which affect their host plants at different stages of pathogenesis (Berestetskiy, 2008; Friesen et al., 2008a,b; Meena et al., 2015). The fungal pathogenic SMs are regarded as not essential for life, but their roles are quite versatile (Stergiopoulos et al., 2013; Pusztahelyi et al., 2015; Meena et al., 2016a). The genetically coded possibilities for the production of secondary metabolites, stimuli and the various phytotoxins generally predict the fungal-host plant interactions and pathogenic behavior of fungi.
The plant pathogenic fungi are divided into biotrophic, hemibiotrophic, and necrotrophic pathogens. These different pathogenic life styles require different molecular weaponry. Necrotrophic fungi infect and kill host tissue and extract nutrients from dead host cells. Biotrophic fungi colonize living host tissue and obtain nutrients from living tissue; whereas hemibiotrophic fungi display two phases during the infection process; first is an initial biotrophic phase followed by a necrotrophic stage (Lo Presti et al., 2015). Necrotrophic and hemibiotrophic fungal species basically show the contrasting mechanistic process of promoting disease, and many HSTs and proteins are the examples of effectors which fundamentally overlap (Condon et al., 2013). These life styles of plant pathogenic fungi provide general information about their interaction with the host, although the distinction between biotrophic and hemibiotrophic mode of action is still not so clear.
Alternaria species have shown different life styles i.e., from saprophytes to endophytes to pathogen (Thomma, 2003; Dang et al., 2015). They are very successful pathogenic genus that causes disease in large number of economically important plants, including apple, broccoli, cauliflower, potato, tomato, citrus, pear, strawberry, tobacco, etc. (Meena et al., 2016a). Alternaria creates large economic losses due to their host range and their worldwide distribution. Approximately, 300 species of genus Alternaria have been identified worldwide which includes Alternaria alternata, Alternaria tenuissima, Alternaria arborescense, Alternaria brassicicola, Alternaria infectoria, and Alternaria solani (Lee et al., 2015). These Alternaria species have been reported to cause diseases in nearly 400 plant species, in which A. alternata infects almost 100 plant species. It is also responsible for post-harvested diseases in various crops (Coates and Johnson, 1997; Woudenberg et al., 2015; Meena et al., 2017c; Sajad et al., 2017) causing asthma and infection of upper respiratory tract in humans (Kurup et al., 2000). The reasons behind pathogenicity are the production of diverse phytotoxins.
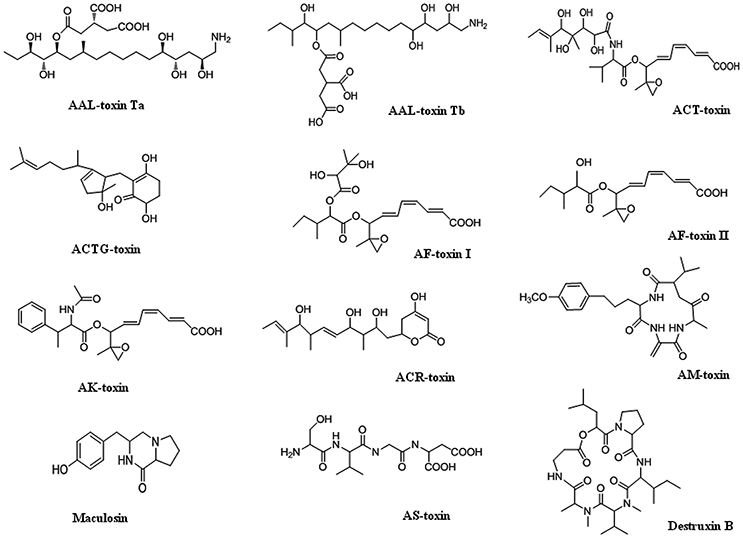
Alternaria mycotoxins have been frequently isolated and reported in fruits and vegetables, such as tomatoes, citrus fruits, Japanese pears, prune nectar, red currant, carrots, barley, oats, olives, mandarins, melons, peppers, apples, raspberries, cranberries, grape, sunflower seeds, oilseed rape meal, flax seed, linseed, pecans, melon, lentils, wheat, and other grains (Patriarca et al., 2007; Ostry, 2008; Logrieco et al., 2009; Andersen et al., 2015; Woudenberg et al., 2015; Meena et al., 2016a,b). More than 70 phytotoxins produced by species of Alternaria have been characterized, and include virulence factors that have both non-specific and specific host interactions. Several Alternaria SMs have been evaluated by the European Food Safety Authority (EFSA) as potentially causing risks to human health, including alternariol (AOH), alternariol monomethyl ether (AME), tenuazonic acid (TeA), altenuene (ALT), and tentoxin (TEN) [(EFSA Panel on Contaminants in the Food Chain (CONTAM), 2011; Rychlik, 2013)]. Alternaria produces host-specific toxins as well as non-host specific toxins (nHSTs). Generally, non-host-specific toxins have relatively mild phytotoxic effects, affect a broad spectrum of plant species and are thought to be an additional factor of disease alongside, for instance, penetration mechanisms and enzymatic processes. Although, they generally act as virulence factors and intensify disease symptom severity, they are not absolutely required for establishing disease since they are also toxic to plant species outside the host range of the pathogen. In Alternaria, many host-specific toxins have been identified, although the precise action of only a few has been studied in detail. Structures of different toxins are given in Figure 1. Brefeldin A (dehydro-), curvularin, tenuazonic acid, tentoxin, and zinniol are examples of non-host specific toxins that are produced by several Alternaria species (Thomma, 2003; Meena et al., 2017a,b).
Pathogenicity of Alternaria Species
The direct and indirect relationship hypothesis of pathogenicity proposed by Andrew et al. (2009), in which he expected that the genes of fungal genome database are responsible for pathogenicity and geographical distribution. The intensive study of disease pathogenesis via rDNA analysis has enabled phylogenetic classifications and investigations into the extent of DNA mutation (Lv et al., 2012). Further, there is no direct correlation observed between rDNA variant distribution and host-specific pathogenicity in toxins producing fungi (Koch et al., 1991). However, the pathogenic specialization of A. alternata might be controlled by some small number of genes which is helpful in HSTs toxin biosynthesis. Repeated DNA sequencing patterns of fungal supernumerary chromosomes suggest their different evolutionary history from essential chromosomes in the same genome and they may have been introduced into the fungal genome through horizontal gene transfer (HGT) from another species (Rosewich and Kistler, 2000). DNA-DNA reassociation and 16S rRNA sequence analysis are successful in bacterial taxonomy; therefore, these technologies are also applying in fungal taxonomy (Bruns et al., 2003). DNA-DNA analysis has suggested a close relationship between HSTs producing Alternaria fungal species and non-pathogenic A. alternata (Kuninaga and Yokozawa, 1987).
The question arises that how Alternaria does actually causes disease, and which symptoms result? The fungus on the attack of host plants secretes some hydrolytic enzyme during penetration process to gain entry into plant tissue and at that time plants also secrete some chemical compounds. In this way, there is a generation of cell fragmentation of plants and fungi. These oligosaccharides compounds can elicit broad host range defense responses that slow pathogen ingress. Thus, the rapid elicitation of plant's defense responses mandates the successful pathogenic fungi must have developed strategies to suppress the response or avoid the host's potential responses (Jackson and Taylor, 1996; Knogge, 1996). The effect of these fungi is mainly on genetics, mode of action, and biosynthesis. The low weight SMs have their target on the biochemical pathway, and their action that can have pleiotropic effects on host plant metabolism.
Toxin Production by Alternaria Species
Non-host Specific Toxins (nHSTs) by Different Alternaria Species
Approximately, 30 nHSTs secondary metabolites are known and characterized in which alternariol (AOH), alternuene (ALT), zinniol, tenuazonic acid (TeA), alteriol monomethyl ether (AME), brefeldin A (dehydro-), tentoxin (TEN), curvularin, and alterotoxin (ATX) I, II, III, are some of the known toxins produced by different Alternaria species (Andersen et al., 2015; Lee et al., 2015). Fujiwara et al. (1988) suggested that brefeldin A actions on Golgi-complex cause disassembly and act as an inhibitor of its secretion, while curvularin inhibits cell division by microtubule assembly disturbance (Robeson and Strobel, 1981) and tenuazonic acid inhibits process of protein synthesis (Meronuck et al., 1972). Zinniol disturbed membrane permeabilization (Thuleau et al., 1988), and tentoxin inhibits chloroplast photo-phosphorylation through binding to ATP synthase resulting in the ATP hydrolysis and ATP synthesis inhibition (Steele et al., 1978). Since, these toxins often target basic cellular processes, so these are regarded as potent mycotoxins. By UV/Vis and MS/MS spectra comparison, alternarionic acid, dehydrocurvularin, pyrenochaetic acid, and altechromone A were identified from species of Alternaria and other fungal SMs. Most of these are derivatives of AOH and TEN (Andersen et al., 2015).
Host-Specific Toxins by Different Alternaria Species
On the basis of scientific observation, there are seven HSTs have been identified from A. alternata. These HSTs are families of closely related diverse group low molecular weight natural compounds (Tsuge et al., 2013). These families are: (1) Epoxy-decatrienoic acid (EDA)—AK-, AF-, and ACT- are the known toxic compound in this family, (2) Cyclic depsipeptide (cyclic tetrapeptide)—AM toxin also known as alternariolide, (3) Amino pento/polyketide (these are sphinganine analog compound)—AAL-toxin, and (4) Polyketide—ACR-toxin (syn. ACRL-toxin).
Interestingly, another HSTs is HC-toxin (a cyclic tetrapeptide), a well-known produced toxin by the plant pathogenic fungus Cochliobolus carbonum, has also been discovered in the species of Alternaria jesenskae, pathotype of Fumana procumbens, and the gene responsible for this toxin is AjTox2 identified by genomic sequencing. Culture filtrate confirmed HC-toxin production through reverse phase HPLC and TLC assay (Wight et al., 2013), while its taxonomic identity confirmed by sequencing of ITS regions (Labuda et al., 2008).
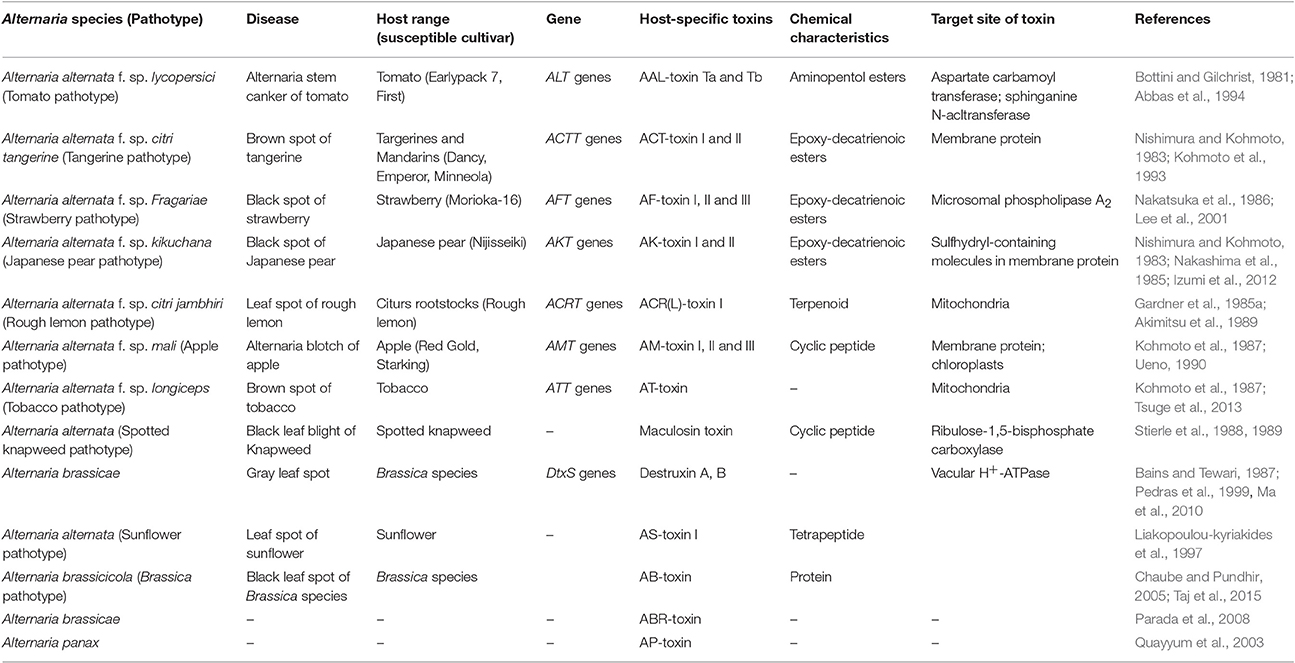
These toxins are involved in the development of few destructive diseases. Alternaria pathotypes produce HSTs which are diverse ranged chemical compounds ranging from low molecular weighted peptides to cyclic peptides. Wolpert et al. (2002) reported that HSTs were biologically produced on their diverse specific plants species, so that it determines the host range of toxin-producing pathogens. Often, not all genotypes of a host plant species are sensitive to the toxin, and similarly, not all isolates of a pathogen species produce the toxin. Table 1 showing host-specific toxins (HSTs) produced by different species of Alternaria.
These HSTs produced several effects on a narrow species that serve as host to the fungi and are necessary for disease. In contrast to the HSTs, the dominant properties of less virulent small-spored species of Alternaria producing host-specific toxins are mainly due to strong selection pressure corresponding from modern monocrop agriculture and newly developed susceptible genotypes (Chou and Wu, 2002). Till now, 13 HSTs are identified by the different variants of Alternaria species and most of these variants are considered as A. alternata pathotypes (Table 1). In HSTs producing and nHSTs producing Alternaria species, all pathogenic species have some extra chromosomes, while these extra chromosomes are not carried by non-pathogenic species (Akamatsu et al., 1999; Akamatsu, 2004). The extra chromosomes are not required for normal growth but have an adaptive advantage in some habitats to the individual species. These are referred as conditionally dispensable chromosomes (CDCs).
Mode of Action and Role of Host-Specific Toxin
The plant pathogen fungi undergo several complex and crucial processing steps of pathogenesis viz. attachment to the plant surface, germination and formation of infectious structures, penetration, and colonization into host cell, which are crucially important steps to cause disease. Generally, plasma membrane, chloroplast, mitochondria, and some important enzymes are the inhibitory sites for the action of HSTs of A. alternata but the other target sites are ER, nucleus, vacuole, and Golgi bodies (Tsuge et al., 2013). All target sites are identified on the basis of different scientific studies (Tsuge et al., 2013; Meena et al., 2016a; Zhang et al., 2016). HSTs produce their effects on different targeted cell organelles and induce cell death in hosts (Figure 2). HSTs generally cause suppression of defense responses of genotype and also disturb other metabolic signaling pathway. HSTs have both properties such as necrosis and suppression of defense process of susceptible hosts (Wolpert et al., 2002). Kohmoto et al. (1989) observed the differences between pathogenicity and host cell death when interactions occur between AM-toxin producing isolates with apple and AK-toxin producing isolates with Japanese pear.
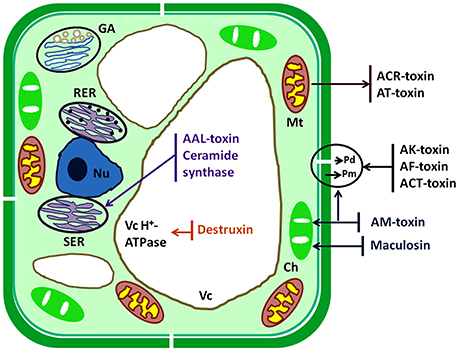
Figure 2. Schematic diagram of target sites of HSTs produced by Alternaria species. Ch, chloroplast; ER, endoplasmic reticulum; GA, Golgi apparatus; Mt, mitochondrion; Nu, nucleus; Pd, plasmodesma; Pm, plasma membrane; Vc, vacuole.
Alternaria HSTs play a vital role as an effector which determines the pathogenicity. Many scientists have reported that simultaneous treatment with HSTs and conidia of non-pathogenic A. alternata strains lead to the initiation of infection (Akimitsu et al., 1989; Yamagishi et al., 2006). The controversial finding of many scientists states that the gene silencing causing loss of HSTs production which leads to pathogenicity disappearance (Harimoto et al., 2008; Miyamoto et al., 2008). A hypothetical idea based on HST-receptor mediated model of pathogenesis was given by Pringle and Scheffer (1964). Before penetration fungal spp. produces a signal molecule to recognize host, and after that release fungal HSTs which specifically binds to host receptor site, and finally, these suppress the host defense against fungus and leads to cell death (Scheffer and Livingston, 1984; Kohmoto and Otani, 1991).
Relation of HSTs, Conditionally Dispensable Chromosomes (CDCs), and Pathogenicity
The CDCs have some characteristic genes responsible for the production of specific HSTs. Several small chromosomes having HSTs genes and transposons like sequences in fungi which have been identified as CDCs, and are present in several small-spored Alternaria species (Hatta et al., 2002). Molecular studies suggested that these HSTs biosynthetic gene clusters reside on a single small chromosome which is <2 Mb in size (Akamatsu et al., 1999; Tsuge et al., 2013). These HSTs genes are located on CDCs as gene clusters and control the production of the toxins viz. AM-toxin from apple, AF- toxin from strawberry, AK-toxin from Japanese pear, ACT-toxin from tangerine, and AAL-toxin from tomato pathotypes and so on (Hu et al., 2012).
Predictions of CDCs Genes
Two methods are commonly used to predict the residential CDCs genes function, (I) use of BLAST against national center for biotechnology information (NCBI) non-redundant database, Pfam (Bateman et al., 2004), and NCBI for functional domains search (Marchler-Bauer et al., 2005), and (II) scanning to identify transcription factors, pathogenicity related genes, PKS genes, NRPS genes, and P450 transporters (Hu et al., 2012). Hu et al. (2012) used marker assisted contigs identification in Alternaria to identify CDCs genes which carrying two Alt1 and AaMSAS cluster of toxin biosynthetic genes (Yamagishi et al., 2006). Among 209 predicted proteins sequencing data, some CDCs genes were identified and characterized, in which 31 having PKS domains and 2 proteins have high modulator protein domains: KS-AT-KR-ACP on CDC_141 and KS-AT-DH-ER-KR-ACP on CDC_165 and remaining 29 PKS protein carrying over two ACPs (Acyl carrier proteins) domains, 7 NRPS domains proteins were predicted having 3 Enterobactin domains, 2 Bacitracin domain, 1 Pyochelin domain and 1 CDA1 domain, 7 protein for P450 monooxygenase, 24 for transcription factor and 37 were identified for pathogenicity during plant-pathogen interactions (Baldwin et al., 2006; Proctor et al., 2009; Hu et al., 2012).
Most recently, another method was applied to genome annotation of Alternaria and comparison data has facilitated functional genomics studies of the fungus in the context of plant and human pathogenicity (Dang et al., 2015). Dang et al. (2015) also provided sequential genome's annotation and comparisonal data of 25 Alternaria species which were analyzed by using multiple computational and comparisonal genomes incorporated tools. Researchers analyzed the sequences through multiple annotation modules including repetitive sequence annotation, gene prediction, protein function and domain structure, while whole genome alignment and homology analysis methods were performed for comparative genomics (Koonin and Galperin, 2003; Bihon et al., 2016). They used InterPro database (Hunter et al., 2012) and Pfam (Finn et al., 2013) for protein domain and family annotation, Blast2GO (Gotz et al., 2008) and InterPro for gene annotation, signal P (Bendtsen et al., 2004), WOLF-Psort (Horton et al., 2007) and Phobius (Kall et al., 2007) for signal peptides, TMHMM (Krogh et al., 2001) for trans-membrane proteins, BLAST search against PHI-base (Winnenburg et al., 2008) for pathogenicity-related candidate genes, CAZY database (Cantarel et al., 2009) and dbCAN (Yin et al., 2012) for active carbohydrates enzymes, BLAST based homology searches and Allerdictor for the identity of allergens (Dang and Lawrence, 2014), batched BLAST search tools from MEROPS database (Rawlings et al., 2012) for protease annotation and SMURF were used for SMs (Khaldi et al., 2010). They reported the Alternaria annotation and comparisonal genomes data sequences to Ensembl database schema using a self-developed tool (EnsImport). EnsImport supports multiple standard file formats such as FASTA, AGP, GFF3, and XMFA, and outputs from widely-used tools such as BLAST, InterPro, RepeatMasker, OrthoMCL, and Blast2GO.
Genetics of Alternaria alternata Produced HSTs
HSTs and nHSTs produced by Alternaria species play an important role as virulence factors during plant pathogenesis. Many known and unknown genes are responsible for the production of these SMs (Dang et al., 2015). The gene sequences identified as CDCs are known as extended families of transposon like sequences (Hatta et al., 2006). The term commonly used is HGT, which is the movement of genetic material without any recombination (Syvanen, 1985). The fungal HGT, the movement of plasmids, mycoviruses, gene clusters, transposable elements and sometime whole chromosomes have expressed between the individual species (Rosewich and Kistler, 2000). HGT in Alternaria occurs frequently because of its wide host range transportable pathogenicity of chromosome may increase pathogens adaptation to the environment, asexual reproduction, and loss of CDC in the absence of a host and cause reduction of carrying extra genome content (Hu et al., 2012). The genetic analysis observation indicated that the fungal toxin protein virulence patterns expected to match with the host sensitive proteins in some manner which is identified in some fungal races (Friesen et al., 2007, 2008b). So, directly or indirectly both the pathogen HST gene and a host susceptible gene interaction between their genes products are required for disease, which inverse situation of gene-for-gene interaction (Wolpert et al., 2002). HSTs genes functions as virulence factors with largely additive effects on disease development and used to influence host plant physiology (Horbach et al., 2011).
HSTs producing A. alternata fungus becomes a good model organism for fungal developmental studies due to its characterized effects on the host plant. Each pathotype toxin and disease on a particular host can be identified by its specificity and can be distinguished by their necrotic symptoms that developed after inoculation with the disease causing pathotype or after HSTs treatment by pathotype (Izumi et al., 2012). Different studies have suggested that RNA silencing and homologous recombination-mediated gene destruction are essential for particular HSTs productions and pathogenicity (Tanaka and Tsuge, 2000; Izumi et al., 2012). Molecular assessment basis of HSTs have identified the gene clusters for pathogenic specialization (Tsuge et al., 2013).
Important HSTs Produced by Alternaria Species
AAL-Toxin
AAL-toxins are chemically propane 1,2,3-tricarboxylic acid (PTCA) which is esterified to 1-amino-11,15-dimethylheptadeca-2,4,5,13,14-pentol. These are structurally sphingosine and sphinganine analogs (Bottini and Gilchrist, 1981; Brandwagt et al., 2001). The scientist identified five types of AAL-toxin related molecules viz. sphingosine (TA), phytosphingosine (TB), sphinganine (TC), tetra-acetyl-phytosphingosine N- Lignoceroyl-d (TD), and L- sphinganine, each consisting with two isomers (Caldas et al., 1994). TA and TB analogs of AAL-toxins showing related specific action and showing 30–400 times higher activity than the other form analogs TC, TD, and TE (Caldas et al., 1994). So, these two toxins are taken into consideration and referred as AAL-toxins. These toxins are produced by Alternaria alternata f. sp. lycopersici, a pathogen causing stem canker in tomato (Lycopersicon esculentum) which exhibits high degree of host specificity and plays a major role in pathogenesis by causing leaf necrosis (Prasad and Upadhyay, 2010). TA type of toxin is most active and highly produced which has molecular mass of 522 kb. AAL-toxin sensitivity and insensitivity to plant tissue have been revealed (Spassieva et al., 2002). The necrotic symptoms of AAL-toxin can be detected from a susceptible line of detached leaflets (Brandwagt et al., 2001). Gilchrist (1997) assumed that AAL-toxin inhibits the activity of aspartate carbamoyl transferase (ACTase) on disruption of pyrimidine metabolism but it could not be confirmed experimentally. AAL-toxins isolated from Alternaria alternata f. sp. lycopersici inhibited ceramide synthase and induced programmed cell death (PCD; Michaelson et al., 2016).
Kawaguchi et al. (1991) observed that ethanolamine (EA), phosphoethanolamine (PEA) and five other related chemicals accumulation occur in AAL-toxin treated leaves. It was also found that cell free ACTase preparation of homozygous susceptible and homozygous resistant genotypes of host and non-host specific sources express differential AAL-toxin sensitivity. Generally, AAL-toxins have its effect on mitochondria but their exact target site is still uncertain. The possible AAL-toxin biosynthetic pathway was in under investigation because EA and PEA are primary and secondary intermediate metabolites of biosynthetic pathways.
Orolaza et al. (1992) suggested that 14C labeling of ethanolamine to susceptible AAL-toxin treated leaf discs have shown strong inhibition of EA incorporated into phosphatidyl ethanolamine (PtdEA). Therefore, phospholipid pathway involved enzymes were suggested as potential biochemical targets for AAL-toxins. AAL-toxins have structural similarities to fumonisins because AAL-toxins have one PTCA and fumonisins with two PTCA side chains esterified to aminopentol back bones (Brandwagt et al., 2000). Gilchrist (1998) referred both AAL-toxin and fumonisin collectively as sphinganine analog mycotoxins (SAMS) due to their structural and functional similarities, and toxicity to plant and mammalian cells. They also show inhibitory action to sphingolipid biosynthesis and induced PCD in both plant and mammalian cells (Abbas et al., 1994; Spassieva et al., 2002, 2006; Tsuge et al., 2013). The sensitivity and disease resistance in tomato to AAL-toxin are regulated by Asc1 (Alternaria stem canker resistance gene 1) gene locus which encodes a host recognition factor (Egusa et al., 2009). These plant-pathogen interactions come under direct or indirect interaction between AAL-toxin and Asc locus products in which Asc action is linked to sucrose transport, ethylene biosynthesis, pyrimidine metabolism, and cell death (Moussatos et al., 1994).
According to Zhang et al. (2011), AAL-toxin induced PCD in tomato leaves is promoted by both jasmonic acid and ethylene by disrupting sphingolipid metabolism. While, the experimental finding of Akamatsu et al. (1997) states that AAL-toxin deficient REMI mutants are non-pathogenic to sensitive tomato plants. AAL-toxins also induce apoptotic-like responses. The induced PCD was taken place by DNA laddering, TUNEL-positive cells and the formation of apoptotic like bodies (Tsuge et al., 2013). AAL-toxin induced PCD involves ceramide signaling and cell cycle disruption (Wolpert et al., 2002). The physiological effects of AAL-toxins represent development of necrotic lesions on fruits and leaves, inhibition of in-vitro development of calli, pollen, roots and shoots, and also reduce the viability of protoplasts and suspension cells (Ismaiel and Papenbrock, 2015).
AM-Toxin
AM-toxin is other type of HSTs, which is responsible for causing Alternaria blotch on apple, a worldwide distributed disease. It is cyclic depsipeptide of alternariolide (Ueno et al., 1975, 1977). This type of cyclic depsipeptide chemical structural compound is also found in other toxins of plant pathogens i.e., HC-toxin from Cochliobolus carbonum race1 (Gross et al., 1982) and tentoxin from Alternaria tenuis (Mayer et al., 2001). Cyclic peptides are synthesized by non-ribosomal pathways by large multifunctional enzymes called non-ribosomal peptide synthetase (NRPS), and also by polymerase chain reaction (PCR) based cloning with primers having highly conserved domains of fungal NRPS genes (Keller et al., 2005; Tsuge et al., 2013). AMT1 and AMT2 are two biosynthetic genes for AM-toxin (Johnson et al., 2000; Harimoto et al., 2007). AMT1 encodes non-ribosomal peptide synthetase attaching with four catalytic domains, which is responsible for activation of each residue in AM-toxin whereas AMT2 encodes an aldo-keto reductase enzyme required for biosynthesis of 2-hydroxy-isovaleric acid i.e., one of the AM-toxin residues (Harimoto et al., 2007, 2008). Chloroplasts are the important cellular organelles which serve as the primary site for AM-toxins. On the basis of AM-toxin acting site and known pathogenesis, it is identified that Alternaria alternata f. sp. mali pathogen may secrete AM-toxins that act on the susceptible leaf cells causing tissues damage (Zhang et al., 2015).
Alternaria alternata f. sp. mali (apple pathotype) has small chromosomes having <1.9 Mb size which is not found in non-pathogenic strains of A. alternata. It suggests strongly that AMT1 resides on small chromosome of 1.1–1.8 Mb of apple pathotype strains (Johnson et al., 2001). The apple pathotype mutant strains have 1.1 Mb CDCs encoding AMT genes which are responsible for AM-toxin biosynthesis and its pathogenicity to susceptible apple strains (Johnson et al., 2001). According to Covert (1998), fungal supernumerary chromosomes are not required for growth but it has advantages for colonizing certain ecological niches (Harimoto et al., 2007).
EST (expressed sequence tag) analysis of AMT genes encoded on 1.4 Mb chromosome in the apple pathotype strain IFO 8984 was performed to identify the structure, function, and origin of Alternaria alternata, in which CDCs responsible for HSTs biosynthesis (Harimoto et al., 2007). Harimoto et al. (2007) generated a cDNA library from AM-toxin producing culture and identified 80 unigenes from 40,980 clones with 1.4 Mb chromosome probe from 196 ESTs. The sequence analysis of these genes showed that most of the small chromosomes encoding these genes with unknown function and also most of the genes expressed at remarkably low level under testing condition. Comparison of the transcription levels of the genes in toxin-producing and non-producing cultures identified 21 genes, including AMT1 and AMT2, that were up-regulated (>10 fold) in toxin producing cultures. Sequence analysis suggested that the up-regulated genes include candidates for novel AM-toxin biosynthetic genes. Disruption of three genes, AMT2, AMT3, and AMT4 also upregulated in toxin-producing cultures IFO8984 having multiple copies of the genes in the genome, and all showed similarities in structures (Tsuge et al., 2013).
AF-Toxin
AF-toxins are other types of A. alternata produced HSTs toxins having <2 Mb sized CDCs encoding AFT genes (Hatta et al., 2002). AF-toxin produced by A. alternata has three related molecular species types viz. AF-toxin I, II, and III. AF-toxin I is highly toxic to both strawberry and pear (Nishimura and Nakatsuka, 1989; Tsuge et al., 2013). Another, AF-toxin II is toxic to only pear while toxin III is highly toxic to strawberry and slightly to pear (Maekawa et al., 1984). AF-toxin I and III are valine derivatives of 2,3-dyhydroxy-isovaleric acid and 2-hydroxy isovaleric acid respectively, while AF II is an isoleucine derivative of 2-hydroxy valeric acid (Tsuge et al., 2013).
Hatta et al. (2006) concluded structure of AF-toxin, on the basis of 1.0 Mb chromosomal strain of NAF8 and found 2–7 copies of 20 AFT regions. They also found many transposon-like sequences and most of which were inactive transposon fossils. On cellular level, AF-toxin affects plasma membrane of susceptible cells and causes a sudden increase in loss of K+ after a few minutes of toxin treatment (Park and Ikeda, 2008). These toxin-induced dysfunctions of plasma membrane which were confirmed by microscopic study with no other cell organelles (Tsuge et al., 2013). Electrophysiological studies of AF-toxin showed that plasma membrane becomes irreversibly depolarised (Namiki et al., 1986; Otani et al., 1989). A polarization occurs mostly in the respiration-dependent component of membrane potential which is sustained by H+ pump. AF-toxin possibly affects the plasma membrane H+ATPase (Tsuge et al., 2013). However, no direct effect of AF-toxin was observed on isolated susceptible host cell plasma membrane ATPase activity (Akimitsu et al., 2013).
AK-Toxin
AK-toxins are the esters of 9,10-epoxy 8-hydroxy 9-methyldecatrienoic acid (EDA) produced by Japanese pear pathotype of A. alternata and was first reported in the Japanese pear black spot disease (Nakashima et al., 1985; Nakatsuka et al., 1986; Tsuge et al., 2013). EDA is an intermediate for toxin biosynthetic pathways. H3-labeled EDA when added in a growing liquid culture of the Japanese pear pathotype strain, converted to AK-toxin. Tanaka et al. (1999) used restriction mediated-integration transformation (REMI) to isolate AK-toxin-minus (or AK-toxin lack) mutant (Akimitsu et al., 2013). They observed that mutants affected essential AK-toxin biosynthetic genes and structural and functional analysis of clones containing tagged site represented six AK-toxin biosynthetic genes i.e., AKT1, AKT2, AKT3, AKT4, AKTR, and AKTS1 (Tanaka and Tsuge, 2000). AKTR encodes transcription regulator having a zinc binuclear cluster DNA binding domain, a protein type of fungal Zn(II)2 Cys6 family (Tanaka et al., 1999; Tanaka and Tsuge, 2000) and other fungal genes encoding proteins showed similarity with many enzymes (Tsuge et al., 2013). So, it acts as regulatory genes for AK-toxin biosynthetic enzymes.
Japanese pear pathotype strains also have multiple copies of functional or non-functional homologous of the AKT genes (Tanaka et al., 1999; Tanaka and Tsuge, 2000). AKTS1 also found unique in this pathotype in DNA blot analysis when these techniques used to assess AKT homologous distribution in A. alternata pathogen (Tanaka et al., 1999). These AKTS1 genes were also present in the tangerine and strawberry pathotype and all three pathotype share common genes required for EDA biosynthesis.
REMI techniques used for AKT protein and AKT1 gene tagging to mutagenize which is required for biosynthesis of AK-toxin and pathogenicity of the Japanese pear pathotype (Tanaka et al., 1999; Wolpert et al., 2002). Another experimental observation of Masunaka et al. (2000) stated that AKT1 and AKT2 were also present in tangerine and strawberry pathotype. The two other AKT3 and AKTR genes are also required for AK-toxin biosynthesis. AKT1, AKT2, AKT3, and AKTR and all their homologous are present on a single chromosome (Tanaka and Tsuge, 2000). Imazaki et al. (2010) reported that enzymes of AK-toxin biosynthesis are localized in peroxisomes and peroxisomal targeting signal type1 (PS1)- like tripeptides at their C-terminal ends. Mutation in AaPEX6, which encodes a peroxin protein required for peroxisome biogenesis from the Japanese pear pathotype. Lack of this peroxisome function, AK-toxin production and pathogenicity become loss completely.
ACR-Toxin (Syn. ACRL-Toxin)
ACR-toxin, a polyketide of long fatty acid (Gardner et al., 1985a,b) causes rough lemon leaf spot disease. The target site of this toxin is mitochondria which makes it dysfunctional (Kohmoto et al., 1984; Akimitsu et al., 1989). ACR-toxin causes uncoupling of oxidative phosphorylation and causes leakage of NAD+ cofactor from tricarboxylic acid cycle (TCA) and loss of membrane potential (Akimitsu et al., 1989).
The clustered biosynthetic genes of ACR-toxin have generally occurred on <2 Mb small sized chromosomes (Ito et al., 2004; Miyamoto et al., 2008, 2009, 2010; Ajiro et al., 2010; Izumi et al., 2012). Masunaka et al. (2005) observed that lemon pathotype strains also carried a small chromosome of ~1.5 Mb which is correlated with the toxin production and pathogenicity to rough lemon.
To investigate the relationship of ACRS to sensitivity to ACR-toxin and hence susceptibility to A. alternata rough lemon pathotype, Ohtani et al. (2002) sequenced this region of the mitochondrial genome from resistant cultivars and 13 species of citrus and found that the regions in the resistant citrus are identical to that of rough lemon (Ohtani et al., 2002). However, examination of ACRS transcripts demonstrated that sensitivity to the toxin is not controlled by the presence or absence of ACRS but rather by post-transcriptional modification of the ACRS transcripts (Figure 3). The peptide encoded by ACRS was detected by immunoblotting only in rough lemon mitochondria, but not in toxin-insensitive citrus mitochondria, and the peptide appeared to consist of sodium dodecyl sulfate (SDS)-resistant oligomers that have been reported for many pore-forming transmembrane proteins (Figure 3; Ohtani et al., 2002).
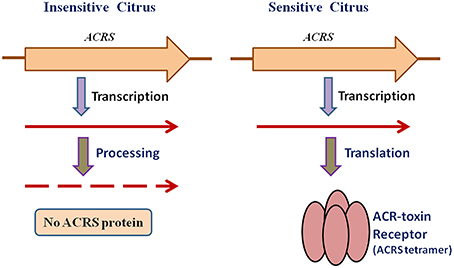
Figure 3. Mechanism of ACR-toxin sensitivity controlled by receptor transcript processing in mitochondria.
ACRTS1 and ACRTS2 are the two genes required for ACR-toxin production. Hutchinson and Fujii (1995) classified ACRTS2 as types I enzyme, is a single large and multifunctional polypeptide having all necessary enzymatic domains for multiple cycles of condensation and β-keto processing. ACRTS1 has more than three copies in the rough lemon pathotype genome. However, combination of both homologous recombination-mediated gene disruption and RNA silencing functionally suppressed all paralogs. Therefore, this gene is required for ACR-toxin biosynthesis and pathogenicity (Izumi et al., 2012), but there are some limitations of homologous recombination-mediated gene disruption. It is difficult to disrupt entire duplicated and multiple paralog functional copies of genes. RNA silencing solves this problems by knockdown all transcripts from all functional copies in HST- producing A. alternata (Miyamoto et al., 2008).
RNA silencing method was first applied to tangerine pathotype HST biosynthesis gene ACTT2, a polyketide synthase (PKS) which is similar to rough lemon pathotypes. This is silenced by transforming the pathotype strain with a plasmid construct expressing hairpin ACRT2 RNA. The ACRT2-silenced transformants in which ACRT2 transcripts were not detectable, ACR-toxins were lost the production and pathogenicity, indicating that this gene encodes a PKS essential for ACR-toxin biosynthesis and hence pathogenicity (Izumi et al., 2012).
ACT-Toxin
Polyketides are the largest families of SMs synthesized by fungi, microbes and plants (Anand et al., 2010). ACT-toxin is one of the types of polyketides HSTs of tangerine pathotype. A. alternata causes brown spot disease (Timmer et al., 2000; Miyamoto et al., 2009). ACT-toxin action appears to be complex in the cell and the primary site of action for this toxin is plasma membrane (Kohmoto et al., 1993; Miyamoto et al., 2008). Kohmoto et al. (1993) detected ACT-toxins ACTT1 and ACTT2 from purified germinating conidia fluids of tangerine pathotype and expected that it participates in the biosynthesis of ACT-, AK- and AF-toxins due to their related common chemical structure (Tanaka et al., 1999; Masunaka et al., 2000, 2005).
Molecular analysis by Masunaka et al. (2000, 2005) suggests that both ACT-toxins are present on a single chromosome with a size of 1.1–1.9 and ACTT2 toxins exists in the genome as multiple copies form. Harimoto et al. (2008) used repeated round of homologous recombination-mediated gene disruption which is essential to knockout all copies of a particular target gene but it is difficult to disrupt entire copies of ACTT1 genes. Homologous gene disruption has some limitations like lack of selectable marker for multiple transformants, low efficiency etc. Harimoto et al. (2008) also overcome these limitations by RNA silencing and beneficial to knock down all functional ACTT1 genes. Since, RNA silencing induced double stranded RNA (dsRNA) post-transcriptional gene silencing phenomenon. It is cleaved by Dicer (a nuclease of the RNA III family) into small interfering RNAs (siRNA) to a ribonucleo-protein complex [RNA-induced-silencing-complex (RISC)] (Bernstein et al., 2001; Hammond et al., 2001). RISC recognizes and degrades homologous mRNAs by complementary base pairing (Elbashir et al., 2001). The RNA silencing used vectors generate hairpin structure of RNA. This structure have sense and antisense sequenced target gene with an intron sequence-based hairpin head spacer which are effective and relatively stable for sequencing, which will be useful for HSTs genes function in the other pathotype of A. alternata as well as other filamentous fungi producing HSTs (Liu et al., 2002; Kadotani et al., 2003; Miyamoto et al., 2008).
RNA silencing has never been demonstrated in Alternaria spp., so Isshiki et al. (2003) had constructed a vector for generating hairpin RNA of the green fluorescent protein (GFP) gene in A. alternata. The silencing genes were over-expressed with green fluorescence protein for determining the gene function. In addition, they analyzed the function of ACTT2 gene in ACT biosynthesis. The ACTT2 gene having multiple copies and high sequence identity were silenced by introducing a vector generated ACTT2 hairpin RNA. The sequence identity of ACTT2 and ACTT1 is 98.8% while ACTT2 homologs identity from dual toxin producing A. alternata strain also expressed similarity of 99% for ACTT2 and 98.9% ACTT3 (Miyamoto et al., 2008).
Conclusions
Genomic and transcriptomic comparisons are now taken in use to obtain genetic features of fungal pathogen to survive successfully in various stressful ecological habitats and to survive itself in different pathogenic life styles. The deep knowledge of the plant pathogen may be helpful for the production of new resistant variety of plant cultivars against different biotic stresses. Environmental factors such as heat or drought also affect the plant-fungal interactions as well as SMs production.
Toxins are recognized as important determinant of pathogenicity in different species of Alternaria. Host-specific toxins of Alternaria spp. plant pathogens play an important role in pathogenesis and could be applied as selective agents in in-vitro selection at the cellular level for disease resistance. The role of a toxin as a disease determinant is proved by the occurrence of the toxin in infected plants and the ability of the toxin alone to elicit at least part of the symptoms of the disease.
Genome sequencing and genome analysis comparison of pathogenic variation in the different species of Alternaria have fascinated to understanding of its evolutionary relationship with other fungi and identification of pathogenicity associated candidate genes, and provide important information for understanding its virulence variation and mechanism under its interaction with the host.
During last decades, the molecular events at an ever-increasing rate applying to know the gene data history and cellular processes responsible for the specific role of toxins. In future, these combined analytical approaches with new insight and revised concepts of new research in fungal genetics and biochemistry will provide a surprising knowledge of SMs corresponding biosynthetic genes and its effect on cellular processes.
Author Contributions
MM provided the general concept and drafted part of the manuscript. MM, SG, PS, and AZ wrote the manuscript. MD and RU also helped in preparation of the manuscript as required by the journal guideline. All authors revised and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer AMR and handling Editor declared their shared affiliation, and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
Acknowledgments
The work was supported in part by Rajiv Gandhi National Fellowship granted by University grants commission (UGC), New Delhi, India to MM. The author is also thankful to Dr. Anand Vikram Singh, Department of Botany, Banaras Hindu University, Varanasi for helping in draw chemical structure of toxins by ChemBioDraw Ultra 12.0 software.
Abbreviations
SMs, Secondary metabolites; HSTs, Host-specific toxins; nHSTs, Non-host specific toxins; CDCs, Conditionally dispensable chromosomes; HGT, Horizontal gene transfer; PCD, Programmed cell death; PCR, Polymerase chain reaction; TCA, Tricarboxylic acid cycle; BLAST, Basic local alignment search tool; NCBI, National center for biotechnology information.
References
Abbas, H. K., Tanaka, T., Duke, S. O., Porter, J. K., Wray, E. M., Hodges, L., et al. (1994). Fumonisin- and AAL-toxin induced disruption of sphingolipid metabolism with accumulation of free sphingoid bases. Plant Physiol. 106, 1085–1093. doi: 10.1104/pp.106.3.1085
Ajiro, N., Miyamoto, Y., Masunaka, A., Tsuge, T., Yamamoto, M., Ohtani, K., et al. (2010). Role of the host-specific ACT-toxin synthesis gene ACTTS2 encoding an enoyl-reductase in pathogenicity of the tangerine pathotype of Alternaria alternata. Phytopathology 100, 120–126. doi: 10.1094/PHYTO-100-2-0120
Akamatsu, H. (2004). Molecular biological studies on the pathogenicity of Alternaria alternata tomato pathotype. Jpn. J. Phytopathol. 70:160. doi: 10.3186/jjphytopath.70.160
Akamatsu, H., Itoh, Y., Kodama, M., Otani, H., and Kohmoto, K. (1997). AAL-toxin deficient mutants of Alternaria alternata tomato pathotype by restriction enzyme-mediated integration. Phytopathology 87, 967–972. doi: 10.1094/PHYTO.1997.87.9.967
Akamatsu, H., Taga, M., Kodama, M., Johnson, R., Otani, H., and Kohmoto, K. (1999). Molecular karyotypes for Alternaria plant pathogens known to produce host-specific toxins. Curr. Genet. 35, 647–656. doi: 10.1007/s002940050464
Akimitsu, K., Kohmoto, K., Otani, H., and Nishimura, S. (1989). Host-specific effect of toxin from the rough lemon pathotype of Alternaria alternata on mitochondria. Plant Physiol. 89, 952–931. doi: 10.1104/pp.89.3.925
Akimitsu, K., Tsuge, T., Kodama, K., Yamamoto, M., and Otani, H. (2013). Alternaria host-selective toxins: determinant factors of plant disease. J. Gen. Plant Pathol. 80, 109–122. doi: 10.1007/s10327-013-0498-7
Anand, S., Prasad, M. V. R., Yadav, G., Kumar, N., Shehara, J., Ansari, M. Z., et al. (2010). SBSPKS: structure based sequence analysis of polyketide synthases. Nucleic Acids Res. 38, 487–496. doi: 10.1093/nar/gkq340
Andersen, B., Nielson, K. F., Fernandez, P. V., and Patriarca, A. (2015). Characterization of Alternaria strains from Argentinean blackberry, tomato, walnut and wheat. Int. J. Food Microbiol. 196, 1–10. doi: 10.1016/j.ijfoodmicro.2014.11.029
Andrew, M., Peever, T. L., and Pryor, B. M. (2009). An expanded multilocus phylogeny does not resolve morphological species within the small-spored Alternaria species complex. Mycologia 101, 95–109. doi: 10.3852/08-135
Bains, P. S., and Tewari, J. P. (1987). Purification, chemical charecterization and host-specficty of the toxin produced by Alternaria brassicae. Physiol. Mol. Plant Pathol. 30, 259–371. doi: 10.1016/0885-5765(87)90039-7
Baldwin, T. K., Wimenburg, R., Urban, M., Rawlings, C., Koehler, J., and Hammond-Kosack, K. S. (2006). The pathogen host interaction database (PHI base) provides insights into genetic and novel themes of pathogenicity. Mol. Plant Microbe Intract. 19, 1451–1462. doi: 10.1094/MPMI-19-1451
Bateman, A., Coin, L., Durbin, R., Finn, R. D., Hollich, V., Griffiths-Jones, S., et al. (2004). The Pfam protein families database. Nucleic Acids Res. 32, D138–D141. doi: 10.1093/nar/gkh121
Bendtsen, J. D., Nielsen, H., Von, H. G., and Brunak, S. (2004). Improved prediction of signal peptides: signal 3.0. J. Mol. Biol. 340, 783–795. doi: 10.1016/j.jmb.2004.05.028
Berestetskiy, A. O. (2008). A review of fungal phytotoxins: from basic studies to practical use. Appl. Biochem. Microbiol. 44, 453–465. doi: 10.1134/S0003683808050013
Bernstein, E., Caudy, A. A., Hammond, S. M., and Hannon, G. J. (2001). Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409, 363–366. doi: 10.1038/35053110
Bihon, W., Cloete, M., Gerrano, A. S., Oelofse, D., and Adebola, P. (2016). Draft genome sequence of Alternaria alternata isolated from onion leaves in South Africa. Genome Announc. 4, e01022–e01016. doi: 10.1128/genomeA.01022-16
Bottini, A. T., and Gilchrist, D. G. (1981). Phytotoxins I. A 1-aminodimethyl-heptadecapentol from Alternaria alternata f. sp. lycopersici. Tetrahedron Lett. 22, 2719–2722. doi: 10.1016/S0040-4039(01)90534-9
Brandwagt, B. F., Kneppers, T. J. A., Van der Weerden, G. M., Nijkamp, H. J. J., and Hille, J. (2001). Most AAL-toxin-sensitive Nicotiana species are resistant to the tomato fungal pathogen Alternaria alternata f. sp. lycopersici. Mol. Plant Microbe Interact. 4, 460–470. doi: 10.1094/MPMI.2001.14.4.460
Brandwagt, B. F., Mesbah, L. A., Takken, F. L. W., Laurent, P. L., Kneppers, T. J. A., Hille, J., et al. (2000). A longevity assurance gene homolog of tomato mediates resistance to Alternaria alternata f. sp. lycopersici toxins and fumonisin B-1. Proc. Natl. Acad. Sci. U.S.A. 97, 4961–4966. doi: 10.1073/pnas.97.9.4961
Bruns, A., Philipp, H., Cypionka, H., and Brinkhoff, T. (2003). Aeromicrobium marinum sp. nov., an abundant pelagic bacterium isolated from the German Wadden Sea. Int. J. Syst. Evol. Microbiol. 53, 1917–1923. doi: 10.1099/ijs.0.02735-0
Caldas, E. D., Jones, A. D., Ward, B., Winter, C. K., and Gilchrist, D. G. (1994). Structural characterization of three new AAL-toxins produced by Alternaria alternata f. sp. lycopersici. J. Agric. Food Chem. 42, 327–333. doi: 10.1021/jf00038a018
Cantarel, B. L., Coutinho, P. M., Rancurel, C., Bernand, T., Lombard, V., and Henrissat, B. (2009). The carbohydrates active enzyme database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37, D233–D238. doi: 10.1093/nar/gkn663
Chaube, H. S., and Pundhir, V. S. (2005). Crop Diseases and Their Management. Prentice-Hall of India Private Limited.
Chou, H.-H., and Wu, W.-S. (2002). Phylogenetic analysis of internal transcribed spacer regions of the genus Alternaria, and the significance of filament-beaked conidia. Mycol. Res. 106, 164–169. doi: 10.1017/S0953756201005317
Coates, L., and Johnson, G. (1997). “Postharvest diseases of fruit and vegetables,” in Plant Pathogens and Plant Diseases, eds J. F. Brown and H. J. Ogle (Armidale, NSW: Rockvale Publications), 533–548.
Condon, B. J., Leng, Y., Wu, D., Bushley, K. E., Ohm, R. A., Otillar, R., et al. (2013). Comparative genome structure, secondary metabolites and effector coding capacity across Cochliobolus pathogens. PLoS Gent. 9:e1003233. doi: 10.1371/journal.pgen.1003233
Covert, S. F. (1998). Supernumerary chromosomes in filamentous fungi. Curr. Genet. 33, 311–319. doi: 10.1007/s002940050342
Dang, H. X., and Lawrence, C. B. (2014). Allerdictor: fast allergen prediction using text classification techniques. Bioinformatics 30, 1120–1128. doi: 10.1093/bioinformatics/btu004
Dang, H. X., Pryor, B., Peever, T., and Lawerence, C. B. (2015). The Alternaria genomes database: a comprehensive resource for a fungal genus comparised of saprophytes, plant pathogens, and allergic species. BMC Genomics 16:239. doi: 10.1186/s12864-015-1430-7
EFSA Panel on Contaminants in the Food Chain (CONTAM) (2011). Scientific opinion on the risks for animal and public health related to the presence of Alternaria toxins in feed and food. EFSA J. 9:2407. doi: 10.2903/j.efsa.2011.2407
Egusa, M., Akamatsu, H., Tsuge, T., Otani, H., and Kodama, M. (2009). Induced resistance in tomato plants to the toxin-dependent necrotrophic pathogen Alternaria alternata. Physiol. Mol. Plant Pathol. 73, 67–77. doi: 10.1016/j.pmpp.2009.02.001
Elbashir, S. M., Lendeckel, W., and Tuschl, T. (2001). RNA interference is mediated by 21- and 22- nucleotide RNAs. Genes Dev. 15, 188–200. doi: 10.1101/gad.862301
Finn, R. D., Bateman, A., Clements, J., Coggill, P., Ebenhardt, R. Y., Eddy, S. R., et al. (2013). Pfam: the protein families database. Nucleic Acids Res. 42, D202–D230. doi: 10.1093/nar/gkt1223
Friesen, T. L., Faris, J. D., Solomon, P. S., and Oliver, R. P. (2008a). Host-specific toxins: effectors of necrotrophic pathogenicity. Cell Microbiol. 10, 1421–1428. doi: 10.1111/j.1462-5822.2008.01153.x
Friesen, T. L., Meinhardt, S. W., and Faris, J. D. (2007). The Stagonospora nodorum-wheat pathosystem involves multiple proteineous host-selective toxins and corresponding host-selective genes that interact in an inverse gene for gene manner. Plant J. 51, 681–692. doi: 10.1111/j.1365-313X.2007.03166.x
Friesen, T. L., Zhang, Z. C., Solomon, P. S., Oliver, R. P., and Faris, J. D. (2008b). Characterization of the interaction of a novel Stagonospora nodorum host-selective toxin with a wheat susceptibility gene. Plant Physiol. 146, 682–693. doi: 10.1104/pp.107.108761
Fujiwara, T., Oda, K., Yokota, S., Takatsuki, A., and Ikehara, Y. (1988). Brefeldin A causes disassembly of the Golgi complex and accumulation of secretory proteins in the endoplasmic reticulum. J. Biol. Chem. 263, 18545–18552.
Gardner, J. M., Kono, Y., Tatum, J. H., Suzuki, Y., and Takeuchi, S. (1985a). Structure of the major component of ACRL-toxins, host-specific pathotoxic compound produced by Alternaria citri. Agric. Biol. Chem. 49, 1235–1238. doi: 10.1271/bbb1961.49.1235
Gardner, J. M., Kono, Y., Tatum, J. H., Suzuki, Y., and Takeuchi, S. (1985b). Plant pathotoxins from Alternaria citri: the major toxin specific for rough lemon plants. Phytochemistry 24, 2861–2867. doi: 10.1016/0031-9422(85)80015-7
Gilchrist, D. G. (1997). Mycotoxins reveal connections between plants and animals in apoptosis and ceramide signaling. Cell Death Differ. 4, 1312–1317. doi: 10.1038/sj.cdd.4400312
Gilchrist, D. G. (1998). Programmed cell death in plant disease: the purpose and promise of cellular suicide. Annu. Rev. Phytopathol. 36, 393–414. doi: 10.1146/annurev.phyto.36.1.393
Gotz, S., Garcia-Gomez, J. M., Terol, J., Williams, T. D., Nagaraj, S. H., Nueda, M. J., et al. (2008). High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36, 3420–3435. doi: 10.1093/nar/gkn176
Gross, M. L., McCrery, D., Crow, F., Tomer, K. B., Pope, M. R., Ciuffetti, L. M., et al. (1982). The structure of the toxin from Helminthosporium carbonum. Tetrahedron Lett. 23, 5381–5384. doi: 10.1016/0040-4039(82)80135-4
Hammond, S. M., Caudy, A. A., and Hannon, G. J. (2001). Post-transcriptional gene silencing by double stranded RNA. Nat. Rev. 2, 100–119. doi: 10.1038/35052556
Harimoto, T., Hatta, R., Kodama, M., Yamamoto, M., Otani, H., and Tsuge, T. (2007). Expression profiles of genes encoded by the supernumerary chromosome controlling AM-toxin biosynthesis and pathogenicity in the apple pathotype of Alternaria alternata. Mol. Plant Microbe Interact. 12, 1463–1476. doi: 10.1094/MPMI-20-12-1463
Harimoto, T., Tanaka, T., Kodama, M., Yamamoto, M., Otani, H., and Tsuge, T. (2008). Multiple copies of AMT2 are prerequisite for the apple pathotype of Alternaria alternata to produce enough AM-toxin for expressing pathogenicity. J. Gen. Plant Pathol. 74, 222–229. doi: 10.1007/s10327-008-0089-1
Hatta, R., Ito, K., Hosaki, Y., Tanaka, T., Tanaka, A., Yamamoto, M., et al. (2002). A conditionally dispensable chromosome controls host-specific pathogenicity in the fungal plant pathogen Alternaria alternata. Genetics 161, 59–70. Available online at: http://www.genetics.org/content/161/1/59.long
Hatta, R., Shinjo, A., Ruswandi, S., Kitani, K., Yamamoto, M., Akimitsu, K., et al. (2006). DNA transposon fossils present on the conditionally dispensable chromosome controlling AF-toxin biosynthesis and pathogenicity of Alternaria alternata. J. Gen. Plant Pathol. 72, 210–219. doi: 10.1007/s10327-006-0277-9
Hawkswort, D. L. (1991). The fungal dimension of biodiversity: magnitude, significance, and conservation. Mycol. Res. 95, 641–655. doi: 10.1016/S0953-7562(09)80810-1
Horbach, R., Navarro-Quesada, A. R., Knogge, W., and Deising, H. B. (2011). When and how to kill a plant cell: infection strategies of plant pathogenic fungi. J. Plant Physiol. 168, 51–62. doi: 10.1016/j.jplph.2010.06.014
Horton, P., Park, K. J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J., et al. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35, W585–W587. doi: 10.1093/nar/gkm259
Hu, J., Chen, C., Peever, T., Dang, H., Lawerence, C., and Mitchell, T. (2012). Genomic characterizations of conditionally dispensable chromosome in Alternaria arborescence provide evidence for horizontal gene transfer. BMC Genomics 13:171. doi: 10.1186/1471-2164-13-171
Hunter, S., Jones, P., Mitchell, A., Apweiler, R., Attwood, T. K., Bateman, A., et al. (2012). InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 40, D306–D312. doi: 10.1093/nar/gks456
Hutchinson, C. R., and Fujii, I. (1995). Polyketide synthase gene manipulation: a structure function approach in engineering novel antibiotics. Ann. Rev. Microbial. 49, 201–238. doi: 10.1146/annurev.mi.49.100195.001221
Imazaki, A., Tanaka, A., Harimoto, Y., Yamamoto, M., Akimitsu, K., Park, P., et al. (2010). Contribution of peroxisomes to secondary metabolism and pathogenicity in the fungal plant pathogen Alternaria alternata. Eukaryot. Cell 9, 682–694. doi: 10.1128/EC.00369-09
Ismaiel, A. A., and Papenbrock, J. (2015). Mycotoxins: producing fungi and mechanisms of phytotoxicity. Agriculture 5, 492–537. doi: 10.3390/agriculture5030492
Isshiki, A., Ohtani, K., Kyo, M., Yamamoto, H., and Akimitsu, K. (2003). Green fluorescent detection of fungal colonization and endopolygalacturonase gene expression in the interaction of Alternaria citri with citrus. Phytopathology 93, 768–773. doi: 10.1094/PHYTO.2003.93.7.768
Ito, K., Tanaka, A., Hatta, R., Yamamoto, M., Akimitsu, K., and Tsuge, T. (2004). Dissection of the host range of the fungal plant pathogen Alternaria alternata by modification of secondary metabolism. Mol. Microbiol. 52, 399–411. doi: 10.1111/j.1365-2958.2004.04004.x
Izumi, Y., Ohtani, K., Miyamoto, Y., Masunaka, A., Fukumoto, T., Gomi, K., et al. (2012). A polyketide synthase gene, ACRTS2, is responsible for biosynthesis of host-selective ACR-toxin in the rough lemon pathotype of Alternaria alternata. Mol. Plant Microbe Interact. 25, 1419–1429. doi: 10.1094/MPMI-06-12-0155-R
Jackson, A. O., and Taylor, C. B. (1996). Plant-microbe interactions: life and death at the interface. Plant Cell 8, 1651–1668. doi: 10.1105/tpc.8.10.1651
Johnson, L., Johnson, R. D., Akamatsu, H., Salamiah, A., Otani, H., Kohmoto, K., et al. (2001). Spontaneous loss of a conditionally dispensable chromosome from Alternaria alternata apple pathotype leads to loss of toxin production and pathogenicity. Curr. Genet. 40, 65–72. doi: 10.1007/s002940100233
Johnson, R. D., Johnson, L., Itoh, Y., Kodama, M., Otani, H., and Kohmoto, K. (2000). Cloning and characterization of a cyclic peptide synthetase gene from Alternaria alternata apple pathotype whose product is involved in AM-toxin synthesis and pathogenicity. Mol. Plant Microbe Interact. 13, 742–753. doi: 10.1094/MPMI.2000.13.7.742
Kadotani, N., Nakayashiki, H., Tosa, Y., and Mayama, S. (2003). RNA silencing in the phytopathogenic fungus Magnaporthe oryzae. Mol. Plant Microbe Interact. 16, 769–776. doi: 10.1094/MPMI.2003.16.9.769
Kall, L., Krogh, A., and Sonnhammer, E. L. L. (2007). Advantages of combined trans-membrane topology and signal peptide prediction-the Phobius web server. Nucleic Acid Res. 35, W429–W432. doi: 10.1093/nar/gkm256
Kawaguchi, I., Orozala, N. P., Tsuge, T., Nishimura, S., and Doke, N. (1991). Changes in amino contents of susceptible tomato cultivars infected with Alternaria alternata tomato. Ann. Phytopath. Soc. Jpn. 57, 526–533. Available online at: https://www.jstage.jst.go.jp/article/jjphytopath1918/57/4/57_4_526/_pdf
Keller, N. P., Turner, G., and Bennett, J. W. (2005). Fungal secondary metabolism—biochemistry to genomics. Nat. Rev. Microbiol. 3, 937–947. doi: 10.1038/nrmicro1286
Khaldi, N., Seifuddin, F. T., Turnner, G., Haft, D., Nierman, W. C., Wolfe, K. H., et al. (2010). SMURF: genomic mapping of fungal secondary metabolites clusters. Fungal Genet. Biol. 47, 736–741. doi: 10.1016/j.fgb.2010.06.003
Koch, R., Song, K., Osborn, T. C., and Williams, P. H. (1991). Relationship between pathogenicity and phylogeny based on restriction fragment length polymorphism in Leptosphaeria maculans. Mol. Plant Microbe Interact. 4, 341–349. doi: 10.1094/MPMI-4-341
Kohmoto, K., Itoh, Y., Shimomura, N., Kondoh, Y., Otani, H., Kodama, M., et al. (1993). Isolation and biological activities of two host-specific toxins from the tangerine pathotype of Alternaria alternata. Phytopathology 83, 495–502. doi: 10.1094/Phyto-83-495
Kohmoto, K., Kondoh, Y., Kohguchi, T., Otani, H., Nishimura, S., and Scheffer, R. P. (1984). Ultrastructural changes in host leaf cells caused by host-selective toxin of Alternaria alternata from rough lemon. Can. J. Bot. 62, 2485–2492. doi: 10.1139/b84-339
Kohmoto, K., and Otani, H. (1991). Host recognition by toxigenic plant pathogens. Experientia 47, 755–764. doi: 10.1007/BF01922454
Kohmoto, K., Otani, H., Kodama, M., and Nishimura, S. (1989). “Host-recognition: can accessibility to fungal invasion be induced by host-specific toxins without necessitating necrotic cell death?” in Phytotoxins and Plant Pathogensis, eds A. Graniti, R. D. Durbin, and A. Ballio (Berlin; Heidelberg; New York, NY: Springer), 249–265.
Kohmoto, K., Otani, H., and Nishimura, S. (1987). “Primary action sites for host-specific toxins produced by Alternaria species,” in Molecular Determinants of Plant Diseases, ed S. Nishimura (Tokyo; Berlin: Japan Scientific Societies Press; Springer-Verlag), 127–143.
Koonin, E. V., and Galperin, M. Y. (2003). Sequence-Evolution-Function: Computational Approaches in Comparative Genomics. New York, NY: Springer Science and Business Media, Kluwer Academic Publishers.
Krogh, A., Larsson, B., Von Heijne, G., and Sonnhammer, E. L. L. (2001). Predicting transmembrane protein topology with hidden morkov model: application to complete genomes. J. Mol. Biol. 305, 567–580. doi: 10.1006/jmbi.2000.4315
Kuninaga, S., and Yokozawa, R. (1987). Taxonomic study of plant pathogens on the basis of DNA sequence homology. I. Genetic relatedness among species of the genus Alternaria. Ann. Phytopathol. Soc. Jpn. 53, 368–369.
Kurup, V. P., Shen, H. D., and Banergee, B. (2000). Respiratory fungal allergy. Microb. Infect. 2, 1101–1110. doi: 10.1016/S1286-4579(00)01264-8
Labuda, R., Elias, P. Jr., and Sterflinger, K. (2008). Alernaria jesensekae sp. nov., a new species from Slovakia on Fumana procumbens (Cistaceae). Microbiol. Res. 163, 208–214. doi: 10.1016/j.micres.2006.05.004
Lee, H. B., Kim, C. J., and Yu, S. H. (2001). First report of strawberry fruit rot caused by Alternaria tenuissima in Korea. Plant Dis. 85:563. doi: 10.1094/PDIS.2001.85.5.563B
Lee, H. B., Patriarca, A., and Magan, N. (2015). Alternaria in food: ecophysiology, mycotoxin production and toxicology. Mycobiology 43, 93–106. doi: 10.5941/MYCO.2015.43.2.93
Liakopoulou-kyriakides, M., Lagopodi, A. L., Thanassoulopoulos, C. C., Stavropoulos, G. S., and Magafa, V. (1997). Isolation and synthesis of a host-selective toxin produced by Alternaria alternata. Phytochemistry 45, 37–40. doi: 10.1016/S0031-9422(96)00789-3
Liu, H., Cottrell, T. R., Pierini, L. M., Goldman, W. E., and Doering, T. L. (2002). RNA interference in the pathogenic fungus Cryptococcus neoformans. Genetics 160, 463–470. Available online at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1461992/
Logrieco, A., Moretti, A., and Solfrizzo, M. (2009). Alternaria toxins and plant diseases: an overview of origin and occurrence. World Mycotoxin J. 2, 129–140. doi: 10.3920/WMJ2009.1145
Lo Presti, L., Lanver, D., Schweizer, G., Tanaka, S., Liang, L., Tollot, M., et al. (2015). Fungal effectors and plant susceptibility. Annu. Rev. Plant Biol. 66, 513–545. doi: 10.1146/annurev-arplant-043014-114623
Lv, D., Zhang, J. Y., Zhang, Z., Zhou, Z. Q., Chen, X. K., Du, X. L., et al. (2012). The relationship between rDNA-ITS sequences and biological characteristics of the apple ring rot pathogen Botryosphaeria berengeriana de Not f. sp. piricola (Nose). Fungal Genomics Biol. 2:104. doi: 10.4172/2165-8056.1000104
Ma, Y.-T., Qiao, L.-R., Shi, W.-Q., Zhang, A.-L., and Gao, J.-M. (2010). Metabolites produced by an endophyte Alternaria alternata isolated from Maytenus hookeri. Chem. Nat. Compd. 46, 504–506. doi: 10.1007/s10600-010-9662-x
Maekawa, N., Yamamoto, M., Nishimura, S., Kohmoto, K., Kuwada, K., and Watanabe, Y. (1984). Studies on host-specific AF-toxins produced by Alternaria alternata strawberry pathotype causing Alternaria black spot of strawberry. (1) Production of host-specific toxins and their biological activities. Ann. Phytopathol. Soc. Jpn. 50, 600–609. doi: 10.3186/jjphytopath.50.600
Maharshi, A. R., and Thaker, V. S. (2012). Growth and development of plant pathogenic fungi in define media. Eur. J. Exp. Biol. 2, 44–54. Available online at: http://www.imedpub.com/articles/growth-and-development-of-plant-pathogenic-fungi-in-define-media.pdf
Marchler-Bauer, A., Anderson, J. B., Cherukuri, P. F., Deweese-Scotte Geer, L. Y., Gwadz, M., He, S., et al. (2005). CDD: a consumed domain database for protein classification. Nucleic Acid Res. 33, D192–D196. doi: 10.1093/nar/gki069
Masunaka, A., Ohtani, K., Peever, T. L., Timmer, L. W., Tsuge, T., Yamamoto, M., et al. (2005). An isolate that is pathogenic to both tangerines and rough lemon and produces two host-selective toxins, ACT- and ACR-toxins. Phytopathology 95, 241–247. doi: 10.1094/PHYTO-95-0241
Masunaka, A., Tanaka, A., Tsuge, T., Peever, T. L., Timmer, L. W., Yamamoto, M., et al. (2000). Distribution and characterization of AKT homologs in the tangerine pathotype of Alternaria alternata. Phytopathology 90, 762–768. doi: 10.1094/PHYTO.2000.90.7.762
Mayer, A. M., Staples, R. C., and Gilad, N. L. (2001). Mechanisms of survival of necrotrophic fungal plant pathogens in hosts expressing the hyper-sensitive response. Phytochemistry 58, 33–41. doi: 10.1016/S0031-9422(01)00187-X
Meena, M., Prasad, V., and Upadhyay, R. S. (2017a). Evaluation of biochemical changes in leaves of tomato infected with Alternaria alternata and its metabolites. Vegetos 30:2. doi: 10.4172/2229-4473.2017.00020.9
Meena, M., Prasad, V., and Upadhyay, R. S. (2017b). Evaluation of Alternaria alternata isolates for metabolite production isolated from different sites of Varanasi, India. J. Agric. Res. 2:000124.
Meena, M., Prasad, V., Zehra, A., Gupta, V. K., and Upadhyay, R. S. (2015). Mannitol metabolism during pathogenic fungal–host interactions under stressed conditions. Front. Microbiol. 6, 1019–1026. doi: 10.3389/fmicb.2015.01019
Meena, M., Zehra, A., Dubey, M. K., Aamir, M., Gupta, V. K., and Upadhyay, R. S. (2016a). Comparative evaluation of biochemical changes in tomato (Lycopersicon esculentum Mill.) infected by Alternaria alternata and its toxic metabolites (TeA, AOH, and AME). Front. Plant Sci. 7:1408. doi: 10.3389/fpls.2016.01408
Meena, M., Zehra, A., Dubey, M. K., and Upadhyay, R. S. (2016b). Mannitol and proline accumulation in Lycopersicum esculentum during infection of Alternaria alternata and its toxins. Int. J. Biomed. Sci. Bioinform. 3, 64–68.
Meena, M., Zehra, A., Swapnil, P., Dubey, M. K., Patel, C. B., and Upadhyay, R. S. (2017c). Effect on lycopene, β-carotene, ascorbic acid and phenolic content in tomato fruits infected by Alternaria alternata and its toxins (TeA, AOH and AME). Arch. Phytopathol. Plant Protect. 50, 317–329. doi: 10.1080/03235408.2017.1312769
Meronuck, R. A., Steele, J. A., Mirocha, C. J., and Christensen, C. M. (1972). Tenuazonic acid, a toxin produced by Alternaria alternata. Appl. Microbiol. 23, 613–617.
Michaelson, L. V., Napier, J. A., Molina, D., and Faure, J. D. (2016). Plant sphingolipids: their importance in cellular organization and adaption. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1861, 1329–1335. doi: 10.1016/j.bbalip.2016.04.003
Miyamoto, Y., Ishii, Y., Honda, A., Masunaka, A., Tsuge, T., Yamamoto, M., et al. (2009). Function of genes encoding acyl-CoA synthetase and enoyl-CoA hydratase for host-selective ACT-toxin biosynthesis in the tangerine pathotype of Alternaria alternata. Phytopathology 99, 369–377. doi: 10.1094/PHYTO-99-4-0369
Miyamoto, Y., Masunaka, A., Tsuge, T., Yamamoto, M., Ohtani, K., Fukumoto, T., et al. (2008). Functional analysis of a multicopy host-selective ACT-toxin biosynthesis gene in the tangerine pathotype of Alternaria alternata using RNA silencing. Mol. Plant Microbe Interct. 2, 1591–1599. doi: 10.1094/MPMI-21-12-1591
Miyamoto, Y., Masunaka, A., Tsuge, T., Yamamoto, M., Ohtani, K., Fukumoto, T., et al. (2010). ACTTS3 encoding a polyketide synthase is essential for the biosynthesis of ACT-toxin and pathogenicity in the tangerine pathotype of Alternaria alternata. Mol. Plant Microbe Interact. 23, 406–414. doi: 10.1094/MPMI-23-4-0406
Moussatos, V. V., Yang, S. F., War, B., and Gilchrist, D. G. (1994). AAL-toxin- induced physiological changes in Lycopersicon esculentum Mill: roles of ethylene and pyrimidine biosynthesis intermediates in the necrosis response. Physiol. Mol. Plant Pathol. 35, 1223–12276. doi: 10.1016/S0885-5765(05)80101-8
Nakatsuka, S., Ueda, K., Goto, T., Yamamoto, M., Nishimura, S., and Kohmoto, K. (1986). Structure of AF-toxin II, one of the host-specific toxins produced by Alternaria alternata strawberry pathotype. Tetrahedron Lett. 27, 2753–2756. doi: 10.1016/S0040-4039(00)84635-3
Nakashima, T., Ueno, T., Fukami, H., Taga, T., Masuda, H., Osaki, K., et al. (1985). Isolation and structureof AK-toxin I and II, host specific phytotoxic metabolites produced by Alternaria alternata Japanese pear pathotype. Agric. Biol. Chem. 49, 807–815.
Namiki, F., Okamoto, H., Katou, K., Yamamoto, M., Nishimura, S., Nakatsuka, S., et al. (1986). Studies on host-specific toxins produced by Alternaria alternata strawberry pathotype causing Alternaria black spot of strawberry (5) Effect of toxins on membrane potential of susceptible plants by means of electrophysiological analysis. Ann. Phytopathol. Soc. Jpn. 52, 610–619. doi: 10.3186/jjphytopath.52.610
Nishimura, S., and Kohmoto, K. (1983). Host-specific toxins and chemical structures from Alternaria species. Ann. Rev. Phytopathol. 21, 87–116. doi: 10.1146/annurev.py.21.090183.000511
Nishimura, S., and Nakatsuka, S. (1989). “Trends in host-selective toxin research in Japan,” in Host-Specific Toxins: Recognition and Specificity Factors in Plant Disease, eds K. Kohmoto and R. D. Durbin (Tottori: Tottori University Press), 19–31.
Ohtani, K., Yamamoto, H., and Akimitsu, K. (2002). Sensitivity to Alternaria alternata toxin in citrus because of altered mitochondrial RNA processing. Proc. Natl. Acad. Sci. U.S.A. 99, 2439–2444. doi: 10.1073/pnas.042448499
Orolaza, N. P., Kawakita, K., and Doke, N. (1992). Inhibitory effect of AL-toxin produced by Alternaria alternata tomato pathotype on the biosynthesis of phosphatidylethanolamine in tomato leaves susceptible to the fungus. Ann. Phytopath. Soc. Jpn. 58, 719–725. doi: 10.3186/jjphytopath.58.719
Ostry, V. (2008). Alternaria mycotoxins: an overview of chemical characterization, producers, toxicity, analysis and occurrence in foodstuffs. World Mycotoxin J. 1, 175–188. doi: 10.3920/WMJ2008.x013
Otani, H., Tomiyama, K., Okamoto, H., Nishimura, S., and Kohmoto, K. (1989). Effect of AK-toxin produced by Alternaria alternata Japanese pear pathotype on membrane potential of pear cells. Ann. Phytopathol. Soc. Jpn. 55, 466–468. doi: 10.3186/jjphytopath.55.466
Parada, R. Y., Sakuna, E., Mori, N., Oka, K., Egusa, M., Kodoma, M., et al. (2008). Alternaria brassicae produces a host-specific protein toxin from germinating spores on host leaves. Phytopathology 98, 458–463. doi: 10.1094/PHYTO-98-4-0458
Park, P., and Ikeda, K. (2008). Ultrastructural analysis of responses of host and fungal cells during plant infection. J. Gen. Plant Pathol. 74, 2–14. doi: 10.1007/s10327-007-0042-8
Patriarca, A., Azcarate, M. P., Terminiello, L., and Pinto, F. V. (2007). Mycotoxin production by Alternaria strains isolated from Argentinean wheat. Int. J. Food Microbiol. 119, 219–222. doi: 10.1016/j.ijfoodmicro.2007.07.055
Pedras, M. S. C., Zaharia, I. L., Gai, Y., Smith, K. C., and Ward, D. E. (1999). Metabolism of the host-selective toxins destruxin B and homodestruxin B: probing a plant disease resistance trait. Organ. Lett. 1, 1655–1658. doi: 10.1021/ol991042z
Prasad, V., and Upadhyay, R. S. (2010). Alternaria alternata f. sp. lycopersici and its toxin trigger production of H2O2 and ethylene in tomato. J. Plant Pathol. 92, 103–108. doi: 10.4454/jpp.v92i1.19
Pringle, R. B., and Scheffer, R. P. (1964). Host-specific plant toxins. Annu. Rev. Phytopathol. 2, 133–156. doi: 10.1146/annurev.py.02.090164.001025
Proctor, R. H., McCormick, S. P., Alexander, N. J., and Desjardins, A. E. (2009). Evidence that a secondary metabolic biosynthetic gene cluster has grown by gene relocation during evolution of the filamentous fungus Fusarium. Mol. Microbiol. 74, 1128–1142. doi: 10.1111/j.1365-2958.2009.06927.x
Pusztahelyi, T., Holb, I. J., and Pocsi, I. (2015). Secondary metabolites in fungus-plant interactions. Front. Plant Sci. 6:573. doi: 10.3389/fpls.2015.00573
Quayyum, H. A., Gijzen, M., and Tranquair, J. A. (2003). Purification of a necrosis-inducing, host specific protein toxin from spore germination fluid of Alternaria panax. Phytopathology 93, 323–328. doi: 10.1094/PHYTO.2003.93.3.323
Rawlings, N. D., Barrett, A. J., and Bateman, A. (2012). MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 12, D343–D350. doi: 10.1093/nar/gkr987
Robeson, D. J., and Strobel, G. A. (1981). Alpha-beta-dehydrocurvularinm and curvularin from Alternaria cinerariae. Z. Naturforsch. 36, 1081–1083.
Rosewich, U. L., and Kistler, H. C. (2000). Role of horizontal gene transfer in the evolution of fungi. Annu. Rev. Phytopathol. 38, 325–363. doi: 10.1146/annurev.phyto.38.1.325
Rychlik, A. M. (2013). Potential health hazards due to the occurrence of the mycotoxin tenuazonic acid in infant food. Eur. Food. Res. Technol. 236, 491–497. doi: 10.1007/s00217-012-1901-x
Sajad, A. M., and Jamaluddin Abid, H. Q. (2017). Fungi associated with the spoilage of post harvest tomato fruits and their frequency of occurrences in different markets of Jabalpur, Madhya-Pradesh, India. Int. J. Cur. Res. Rev. 9, 12–16. Available online at: ijcrr.com/uploads/118_pdf.pdf
Scheffer, R. P., and Livingston, R. S. (1984). Host-selective toxins and their role in plant diseases. Science 223, 17–21. doi: 10.1126/science.223.4631.17
Spassieva, S. D., Markham, J. E., and Hille, J. (2002). The plant disease resistance gene Asc-1 prevents disruption of sphingolipid metabolism during AAL-toxin-induced programmed cell death. Plant J. 32, 561–572. doi: 10.1046/j.1365-313X.2002.01444.x
Spassieva, S. D., Seo, J. G., Jiang, J. C., Bielawski, J., Alvarez-Vasquez, F., Jazwinski, S. M., et al. (2006). Necessary role for the Lag1p motif in (dihydro)ceramide synthase activity. J. Biol. Chem. 281, 33931–33938. doi: 10.1074/jbc.M608092200
Steele, J. A., Durbin, R. D., Uchytil, T. F., and Rich, D. H. (1978). Tentoxin–uncompetitive inhibitor of lettuce chloroplast coupling factor-1. Biochim.Biophys. Act. 501, 72–82. doi: 10.1016/0005-2728(78)90096-8
Stergiopoulos, I., Collemore, J., Mehrabi, R., and De Wit, P. J. G. M. (2013). Phytotoxic secondary metabolites and peptides produced plant pathogenic Dothideomycete fungi. FEMS Microbiol Rev. 37, 67–93. doi: 10.1111/j.1574-6976.2012.00349.x
Stierle, A. C., Cardellina, I. I., and Strobel, G. A. (1989). Phytotoxins from Alternaria alternata, a pathogen of spotted knapweed. J. Nat. Prod. 52, 42–47. doi: 10.1021/np50061a003
Stierle, A., Cardellina, J. H., and Strobel, G. A. (1988). Maculosin, a host-specific phytotoxin for spotted knapweed from Alternaria alternata. Proc. Natl. Acad. Sci. U.S.A. 85, 8008–8013. doi: 10.1073/pnas.85.21.8008
Syvanen, M. (1985). Cross gene transfer; implications for a new theory of evolution. J. Theor. Biol. 112, 333–343. doi: 10.1016/S0022-5193(85)80291-5
Taj, G., Meena, P. D., Giri, P., Pandey, D., Kumar, A., and Kumar, A. (2015). Pathogenesis mechanisms employed by Alternaria species. J. Oilseed Brassica 6, 213–240. Available online at: http://www.srmr.org.in/ojs/index.php/job/article/view/11/10
Tanaka, A., Shiotani, H., Yamamoto, M., and Tsuge, T. (1999). Insertional mutagenesis and cloning of the genes required for biosynthesis of the host-specific AK-toxin in the Japanese pear pathotype of Alternaria alternata. Mol. Plant Microbe Interact. 12, 691–702. doi: 10.1094/MPMI.1999.12.8.691
Tanaka, A., and Tsuge, T. (2000). Structural and functional complexity of the genomic region controlling AK-toxin biosynthesis and pathogenicity in the Japanese pear pathotype of Alternaria alternata. Mol. Plant Microbe Interact. 13, 975–986. doi: 10.1094/MPMI.2000.13.9.975
Thomma, B. P. (2003). Alternaria spp.: from general saprophyte to specific parasite. Mol. Plant Pathol. 4, 225–236. doi: 10.1046/j.1364-3703.2003.00173.x
Thuleau, P., Graziana, A., Rossignol, M., Kauss, H., Auriol, P., and Ranjeva, R. (1988). Binding of the phytotoxin zinniol stimulates the entry of calcium into plant-protoplasts. Proc. Natl. Acad. Sci. U.S.A. 85, 5932–5935. doi: 10.1073/pnas.85.16.5932
Timmer, L. W., Solel, Z., and Orozco-Santos, M. (2000). “Alternaria brown spot of mandarins,” in Compendium of Citrus Diseases, eds L. W. Timmer, S. M. Garnsey, and J. H. Graham (St. Paul, MN: The American Phytopathological Society Press), 19–21.
Tsuge, T., Harimotao, Y., Akamatsu, K., Ohtani, K., Kodama, M., Akagi, Y., et al. (2013). Host–selective toxins produced by the plant pathogenic fungus Alternaria alternata. FEMS Microbiol. Rev. 37, 44–66. doi: 10.1111/j.1574-6976.2012.00350.x
Ueno, T. (1990). Secondary metabolites related to host selection by plant pathogenic fungi. Pure Appl. Chem. 62, 1347–1352. doi: 10.1351/pac199062071347
Ueno, T., Nakashima, T., Hayashi, Y., and Fukami, H. (1975). Structures of AM-toxin I and II host specific phytotoxic metabolites produced by Alternaria mali. Agric. Biol. Chem. 39, 1115–1122. doi: 10.1271/bbb1961.39.1115
Ueno, T., Nakashima, T., Uemoto, M., Fukami, H., Lee, S. N., and Izumiya, N. (1977). Mass spectrometry of Alternaria mali toxins and related cyclodepsipeptides. Biomed. Mass Spectrom. 4, 134–142. doi: 10.1002/bms.1200040303
Wight, W. D., Labuda, R., and Walton, J. D. (2013). Conservation of the genes for HC-toxin biosynthesis in Alternaria jesenskae. BMC Microbiol. 13:165. doi: 10.1186/1471-2180-13-165
Winnenburg, R., Urban, M., Beacham, A., Baldwin, T. K., Holland, S., Lindeberg, M., et al. (2008). PHI-base updates: addition to the pathogen host interaction database. Nucleic Acids Res. 36, D572–D576. doi: 10.1093/nar/gkm858
Wolpert, T. J., Dunkle, L. D., and Ciuffetti, L. M. (2002). Host-selective toxins and avirulence determinants: what's in a name? Annu. Rev. Phytopathol. 40, 251–285. doi: 10.1146/annurev.phyto.40.011402.114210
Woudenberg, J. H. C., Seidl, M. F., Groenewald, J. Z., de Vries, M., Stielow, J. B., Thomma, B. P., et al. (2015). Alternaria section Alternaria: species, formae specials or pathotypes? Stud. Mycol. 82, 1–21. doi: 10.1016/j.simyco.2015.07.001
Yamagishi, D., Akamatsu, H., Otani, H., and Kodama, M. (2006). Pathological evaluation of host-specific AAL-toxins and fumonisin mycotoxins produced by Alternaria and Fusarium species. J. Gen. Plant Pathol. 72, 323–326. doi: 10.1007/s10327-006-0291-y
Yin, Y., Mao, X., Yang, J., Chen, X., Mao, F., and Xu, Y. (2012). dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 40, W445–W451. doi: 10.1093/nar/gks479
Zhang, C. X., Tian, Y., and Cong, P. H. (2015). Proteome analysis of pathogen-responsive proteins from apple leaves induced by the Alternaria blotch Alternaria alternata. PLoS ONE 10:e0122233. doi: 10.1371/journal.pone.0122233
Zhang, L., Jia, C., Liu, L., Li, C., and Wang, Q. (2011). Involvement of jasmonates and ethylene in Alternaria alternata f. sp. lycopersici toxin-induced tomato cell death. J. Exp. Bot. 15, 5405–5418. doi: 10.1093/jxb/err217
Keywords: Alternaria species, secondary metabolites, host-specific toxins, non-host specific toxins, conditionally dispensable chromosomes, pathogenicity
Citation: Meena M, Gupta SK, Swapnil P, Zehra A, Dubey MK and Upadhyay RS (2017) Alternaria Toxins: Potential Virulence Factors and Genes Related to Pathogenesis. Front. Microbiol. 8:1451. doi: 10.3389/fmicb.2017.01451
Received: 19 May 2017; Accepted: 18 July 2017;
Published: 08 August 2017.
Edited by:
Helio K. Takahashi, Federal University of São Paulo, BrazilReviewed by:
Luciana Lopes Guimaraes, Universidade Santa Cecilia, BrazilAnderson Messias Rodrigues, Federal University of São Paulo, Brazil
Copyright © 2017 Meena, Gupta, Swapnil, Zehra, Dubey and Upadhyay. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mukesh Meena, mukeshmeenabhu@gmail.com
 Mukesh Meena
Mukesh Meena Sanjay K. Gupta
Sanjay K. Gupta Prashant Swapnil
Prashant Swapnil Andleeb Zehra
Andleeb Zehra Manish K. Dubey
Manish K. Dubey Ram S. Upadhyay
Ram S. Upadhyay