- 1Key Laboratory of Marine Environmental Corrosion and Biofouling, Institute of Oceanology, Chinese Academy of Sciences, Qingdao, China
- 2College of Marine Life Sciences, Ocean University of China, Qingdao, China
Metal corrosion is of worldwide concern because it is the cause of major economic losses, and because it creates significant safety issues. The mechanism of the corrosion process, as influenced by bacteria, has been studied extensively. However, the bacterial communities that create the biofilms that form on metals are complicated, and have not been well studied. This is why we sought to analyze the composition of bacterial communities living on steel structures, together with the influence of ecological factors on these communities. The corrosion samples were collected from rust layers on steel plates that were immersed in seawater for two different periods at Sanya and Xiamen, China. We analyzed the bacterial communities on the samples by targeted 16S rRNA gene (V3–V4 region) sequencing using the Illumina MiSeq. Phylogenetic analysis revealed that the bacteria fell into 13 phylotypes (similarity level = 97%). Proteobacteria, Firmicutes and Bacteroidetes were the dominant phyla, accounting for 88.84% of the total. Deltaproteobacteria, Clostridia and Gammaproteobacteria were the dominant classes, and accounted for 70.90% of the total. Desulfovibrio spp., Desulfobacter spp. and Desulfotomaculum spp. were the dominant genera and accounted for 45.87% of the total. These genera are sulfate-reducing bacteria that are known to corrode steel. Bacterial diversity on the 6 months immersion samples was much higher than that of the samples that had been immersed for 8 years (P < 0.001, Student’s t-test). The average complexity of the biofilms from the 8-years immersion samples from Sanya was greater than those from Xiamen, but not significantly so (P > 0.05, Student’s t-test). Overall, the data showed that the rust layers on the steel plates carried many bacterial species. The bacterial community composition was influenced by the immersion time. The results of our study will be of benefit to the further studies of bacterial corrosion mechanisms and corrosion resistance.
Introduction
Structural steel is widely used in marine environments because it is strong, readily available, easy to fabricate, and cost-effective, overall. However, steel is subject to corrosion. This is a serious worldwide problem and has a great social and economic impact (Hou et al., 2017). Corrosion is caused by complex chemical, physical and biological processes (Kip and Veen, 2015). Biological (in fact, microbiological influenced corrosion MIC) plays a critical role (Baboian, 2005). MIC is caused by electrochemical reactions created by those microorganisms that form ‘biofilms’ on immersed steel structures (Hamilton, 1991). Fungi are closely associated to this process (e.g., Arthrinium phaeospermum, Aspergillus niger, Chrysosporium merdarium and acidotolerant black yeast) (Lugauskas et al., 2009; Leo et al., 2013). Lugauskas et al. (2009) found that various strains of the same fungal species have different influences on submerged metal surfaces. However, bacteria are the main component of the biofilms, and contribute most to MIC (Bermont-Bouis et al., 2007) and the formation and transformation of corrosion products (Sun H. et al., 2014). The metabolic activities of bacterial communities within the biofilms interact with environmental factors, such as dissolved oxygen, pH, organic, and inorganic compounds, etc., to influence the electrochemical state of the metal and influence the rate of corrosion (Beech, 2004; Beech and Sunner, 2004; Coetser and Cloete, 2005; Videla and Herrera, 2005). It is also known that the bacterial surface associations within biofilms influence the electrochemical reaction rate (Dang and Lovell, 2016). Diverse bacterial populations can coexist in biofilms and often form synergistic communities (consortia) which contribute to the electrochemical processes via cooperative metabolic processes (Gonzalez-Rodrıguez et al., 2008; Korenblum et al., 2008).
Some of the bacteria species that are associated with steel corrosion have been identified. They includes sulphate-reducing bacteria (SRB), sulphur-oxidizing bacteria (SOB), iron-reducing bacteria (IRB), and iron-oxidizing bacteria (IOB) (Sun J. et al., 2014), etc. SRB are regarded as the most influential (Duan et al., 2008), and are regarded as the main corrosion-accelerating factor in the context of the MIC of metals in marine environments (Angell and Urbanic, 2000). Other types of bacteria may also play an important role, e.g., methanogens and metal reducing-bacteria (Zhu et al., 2003; Gonzalez-Rodrıguez et al., 2008). Moreover, what is interesting is that bacteria not only cause corrosion but can also inhibit or protect against corrosion, which is termed as MIC inhibition (MICI) (Zuo, 2007). There is currently a focus on exploiting bacteria and their metabolic by-products, including biofilm and extracellular polymeric substances (EPSs), to reduce MIC. The aim is to replace the biocides and toxic evaporative, organic compounds that are currently employed as rust retardants (Grooters et al., 2007). The mechanisms of MIC and MICI are not completely understood. They cannot be connected with a single biochemical reaction or a single bacterial species or cluster (Kip and Veen, 2015). It is therefore necessary, in this context, to learn more about the nature of the species complexes that form on corroding steel and rust that is immersed in seawater, so as to learn how to protect steel structures in marine environments.
Analyses of the bacterial communities of early developing biofilms in the rust layers of steel originally relied upon plate culturing techniques (Bermont-Bouis et al., 2007), which is laborious, imprecise, and time-consuming. Significantly, nearly all of the bacterial species from this environment do not reproduce on culture plates (Dunbar et al., 1999). Advances in molecular biology now permit us to analyze bacterial communities with considerable more precision. The techniques we adopted to investigate the composition of the bacterial communities were terminal restriction fragment length polymorphism (T-RFLP), denaturant gradient gel electrophoresis (DGGE), fluorescence in situ hybridization (FISH), and 16S rRNA gene libraries. Proteobacteria was recognized as the dominant bacterial group during the first 36 h of biofilm formation by using 16S rRNA gene libraries and T-RFLP (Lee et al., 2008). Citrobacter spp., Enterobacter spp. and Halanaerobium spp. were identified as the dominant bacteria of biofilms after 40-days immersion by ribosomal library and DGGE. FISH analysis was also used in the study of bacterial community composition, and the results showed that Alphaproteobacteria was the dominant community during the first few weeks of biofilm growth. In addition, it became apparent that the combination of FISH and confocal microscopy was of critical importance. It allowed us to define the relative importance of different bacteria in causing corrosion, and provided information both about the spatial structure of the corrosion biofilms, and quantitative information about the bacteria (Dang and Lovell, 2002a,b). Recently, high-throughput Illumina sequencing has been frequently used to investigate the bacterial community composition of various environments (Moreau et al., 2014; Sun J. et al., 2014; Chao et al., 2015). and allowed us to gain deeper insight into the bacterial community composition of the samples (Bokulich and Mills, 2012; Mayo et al., 2014). Our research was greatly enhanced by access to MiSeq sequencing which allowed us to obtain comprehensive information covering the composition of the bacterial communities we targeted. This follows Vigneron et al. (2016) who adopted this technique to reveal that Desulfovibrio species was the dominant bacteria on an offshore oil production facility. We consider that the application of this technology in the current area of research is in its infancy.
In this study we characterized the composition of the bacterial communities in corrosion samples that had been collected from rust layers on steel plates that had been immersed in seawater, by means of high-throughput Illumina MiSeq sequencing. In addition, we analyzed the influence of ecological factors on the bacterial communities. The results of our study have important implications for further study of bacterial corrosion mechanisms and anti-corrosion.
Materials and Methods
Sample Sites and Collection
The plates of steel had the following composition (wt. %): C 0.16, Si 0.12, Mn 0.45, S 0.029, and P 0.019. Nine samples were collected in December 2014 for this study. Among them, six samples (SE1, SE2, SE3, SE4, SE5, and SOH) were collected from the coastal zone of the Hongtang Bay which is located in Sanya City, Hainan Province. The sample identified as SOH provided us with rust layers from steel plates that had been immersed in seawater for 6 months. Samples identified as XE4, XE5 and XE6 were collected from the rust layers of steel plates that had been immersed in seawater for 8 years in a coastal zone of the island of Gulang, which is situated in Xiamen City, Fujian Province.
Large fouling organisms were removed with sterile forceps in sterile conditions from the steel plates as soon as they were removed from the sea. The surface of the test material was gently rinsed in sterilized seawater to remove unattached bacteria. The deposits were sampled with metallic spatulas, taking care not to crush the samples or expose them to air for too long. They were immediately placed in 10 ml sterile plastic centrifuge tubes, transported to the laboratory on dry ice, and were stored at -80°C pending analysis (Païssé et al., 2013). Meanwhile, the salinity, temperature and pH of the seawater were measured by multiparameter water quality detector (CTD90M, Germany).
DNA Extraction
The total community genomic DNA of each sample was extracted according to the method of Zhou et al. (1996). Five microliter of each genomic DNA were subjected to 1% agarose gel electrophoresis to examine its integrity. The concentration of the DNA was measured with a UV-vis spectrophotometer (NanoDrop 2000c, United States) to identify that adequate amounts of high-quality total genomic DNA were extracted.
16S rRNA Gene Amplification by PCR
V3–V4 region of the bacterial 16S rRNA gene was amplified by PCR (95°C for 3 min followed by 27 cycles of 95°C for 30 s, 55°C for 30 s and 72°C for 45 s and a final extension at 72°C for 10 min using the primers 338F 5′-barcode-ACTCCTACGGGAGGCAGCAG-3′ and 806R 5′-GGACTACHVGGGTWTCTAAT-3′ (Dennis et al., 2013), where the barcode was an eight-base sequence that was unique to each sample. The PCR reactions were performed in triplicate in 20 μl reactions, containing 2 μl of 10× Ex Taq buffer, 2 μl of 2.5 mM dNTPs, 0.8 μl of each primer (5 μM), 0.2 μl Ex Taq polymerase, 0.2 μl of BSA, 14 μl of ddH2O and 10 ng of template DNA.
Illumina MiSeq Sequencing
The amplicons were extracted from 2% agarose gels and purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, United States) according to the manufacturer’s instructions. The purified amplicons were quantified using QuantiFluorTM -ST (Promega, United States), pooled in equimolar ratios and subjected to paired-end sequencing (2 × 250) on an Illumina Miseq platform according to standard protocols. The raw reads were deposited into the NCBI Sequence Read Archive (SRA) database.
Processing Sequencing Data
The raw fastq files were demultiplexed and quality-filtered using QIIME (version 1.9.1) (Caporaso et al., 2010) with the following criteria: (i) The output data (reads) were truncated at any site receiving an average quality score < 20 over a 50 base pair (bp) sliding window. (ii) Primers were matched exactly allowing a two nucleotide mismatching, and reads containing ambiguous bases were removed. (iii) Sequences whose overlap was longer than 10 bp were merged according to their overlap sequence. Operational taxonomic units (OTUs) were clustered with a 97% similarity cut-off using UPARSE version 7.11 (Edgar, 2013). The normalization process followed OTU clustering. Chimeric sequences were identified and removed using UCHIME (Edgar, 2010; Edgar et al., 2011). The taxonomy of each 16S rRNA gene sequence was analyzed with RDP Classifier2 (Wang et al., 2007) against the Silva (SSU128) 16S rRNA database using a confidence threshold of 70% (Quast et al., 2013).
The relative abundances of the phylum, class and genus levels were plotted as a bar graph. Heatmaps based on the relative abundance of OTUs at the phylum and genus levels were also generated with R program (R Development Core Team, 2013). A venn diagram was created using Mothur v.1.30.1 (Schloss et al., 2009) to identify the similarities and differences of the communities in the three kinds of samples (sample SOH, samples from Sanya, and samples from Xiamen). In alpha diversity analysis, alpha diversity parameters such as Chao, Ace, Simpson, and Shannon were estimated using mothur (version v.1.30.13) with a 97% similarity cut-off (Schloss et al., 2009). They provided a means of evaluating the potential total number of OTU and an estimate of the level of diversity in each sample. Rarefaction curves based on these metrics were generated. In beta diversity analysis, differences in the bacterial communities among the nine samples were preformed by a hierarchical cluster tree created using the unweighted pair-group method with arithmetic mean (UPGMA). A principal co-ordinates analysis (PCoA) plot was also obtained using Mothur with the calculation of Bray–Curtis (Schloss et al., 2009).
Data Accession Number
The obtained raw sequences were deposited in the NCBI database (Accession Number: PRJNA396473).
Results
Diversity Analysis and Richness of OTUs
A total 558,632 high-quality bacterial V3–V4 Illumina sequences, ranging from 47,920 to 77,230, were obtained for further analysis (Table 1). Data were normalized by subsampling the 16S rDNA data at 45,530 reads per sample to correct for unequal sequencing depth. The average length of the high-quality sequences from the nine corrosion samples was 441 bp. After random re-sampling at the 0.03 distance level, the average number of OTUs in the 8 years samples was 1,695. However, there were 6,020 OTUs in the sample immersed for 6 months. For the 8 years samples the average numbers were: OTUs 1,695, ACE 1,955 Chao1 index 1826, Shannon and Simpson index 4.70 and 0.0378. For the 6 months sample the numbers were of OTUs 6,020, ACE 7,026 and Chao1 index 6,422, Shannon and Simpson index 7.11 and 0.0050 (Table 1). The rarified Chao1 and Shannon diversity indexes showed remarkable differences across the 8 years samples and the 6 months sample (P < 0.001, Student’s t-test). Bacterial diversity and richness were higher in the 6 months immersion sample compared to the 8 years immersion samples, as described by the Shannon and Simpson diversity indices, ACE and Chao1 index. This was also confirmed by rarefaction curve analyses of the OTUs (Figure 1). Meanwhile, the average richness of 8 years immersion samples from Sanya (ACE index 2,249 and Chao1 index 2,092) was higher than that from Xiamen (ACE index 1,463 and Chao1 index 1,383). However, both of the rarified Chao1 and Shannon diversity indexes were not significantly different (P > 0.05, Student’s t-test) across the 8 years samples from Sanya and Xiamen.
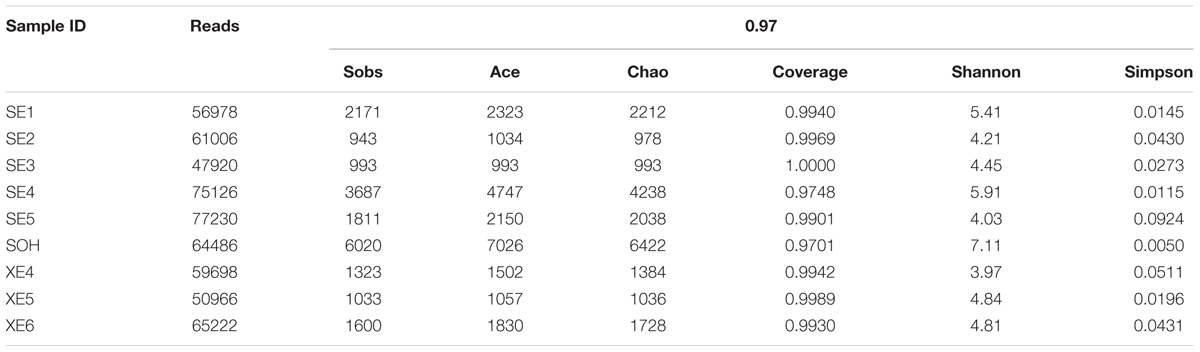
TABLE 1. Number of sequences analyzed, OTUs, estimated community richness indices (Chao and Ace), coverage, and community diversity indices (Shannon and Simpson) of the 16S rRNA libraries of the corrosion samples.
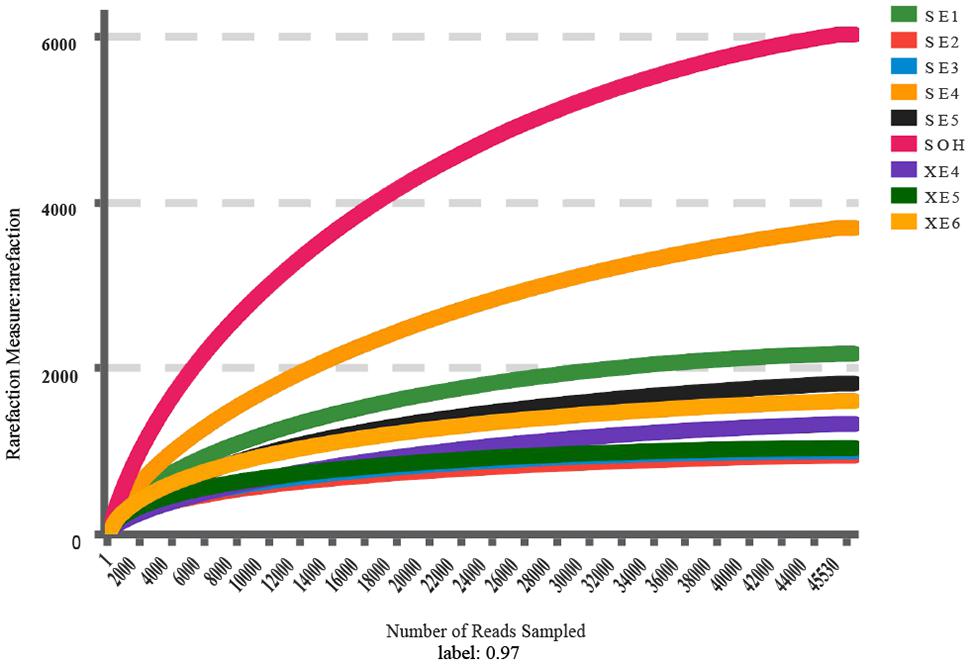
FIGURE 1. Rarefaction analysis of the V3/V4 MiSeq sequencing reads of the 16S rRNA gene from the nine corrosion samples at a 97% sequence similarity cutoff value.
The Venn diagram showed that SE (8 years immersion samples from Sanya) and XE (8 years immersion samples from Xiamen) shared 1,180 OTUs, SE and SOH shared 1,887 OTUs, XE and SOH shared 622 OTUs and 480 OTUs were shared by all nine samples (Figure 2). The average number of OTUs in the 8 years samples from Sanya and Xiamen were 1,387 and 1,013, respectively. In addition, the Good’s coverage values (Table 1) and the rarefaction curves of all corrosion samples (Figure 1) indicated that the 16S rRNA gene sequences derived from these corrosion samples could represent the total bacterial community in this study.
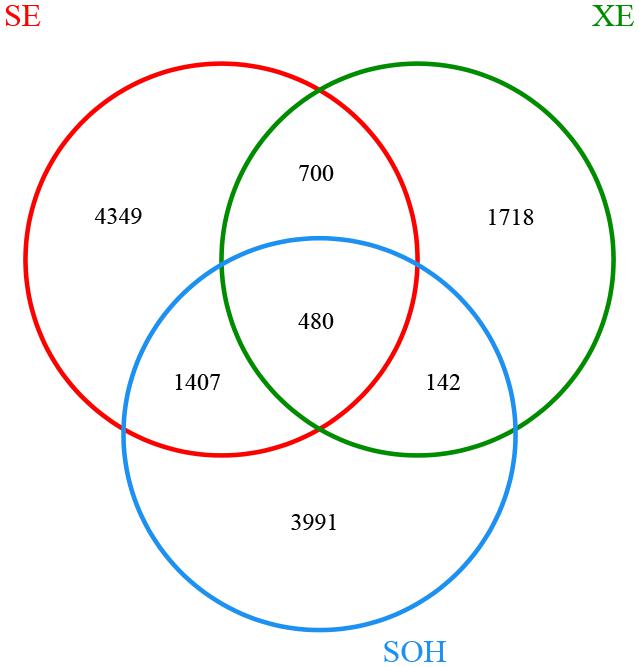
FIGURE 2. Venn diagram was at a distance of 0.03. There were 6,936 OTUs in sample SE (five corrosion samples that were collected from Sanya and had been immersed in seawater for 8 years). There were 3,040 OTUs in sample XE (three corrosion samples that were collected from Xiamen and had been immersed in seawater for 8 years). There were 6,020 OTUs in sample SOH.
Analysis of Bacterial Communities
At the phylum level, more than 13 prokaryotic phyla were found in the nine samples accounting for 95.35% of the total community, namely Proteobacteria (63.44%), Firmicutes (19.12%), Bacteroidetes (6.28%), Tenericutes (1.57%), Actinobacteria (0.99%), Chloroflexi (0.86%), Thermotogae (0.81%), Cyanobacteria (0.54%), Acidobacteria (0.49%), Planctomycetes (0.36%), Spirochaetae (0.32%), Nitrospirae (0.34%) and lgnavibacteriae (0.23%) (Figure 3 and Table 2). Proteobacteria, Firmicutes and Bacteroidetes were the core phyla, accounting for nearly 88.84% of the total. For the majority of corrosion samples, Proteobacteria was the dominant phylum, ranging from 26.82 to 82.93% of the total number of phyla. Firmicutes was the second most represented phylum, ranging from 0 to 62.14% of the total number of phyla. Bacteroidetes was the third most dominant phylum, ranging from 2.39 to 12.00% of the total number of phyla. However, in SE5, Firmicutes (62.14%) and Proteobacteria (26.82%) were the first and second most abundant phyla, which was markedly different to the distribution in the other samples. The remaining 10 phyla were represented at a low level on individual samples. Furthermore, the hierarchical clustering heat map of the in-depth taxonomic analysis was plotted to compare the membership and structure of each sample at the phylum level. It also indicated that Proteobacteria, Bacteroidetes and Firmicutes were the three dominant bacterial communities (Figure 4).
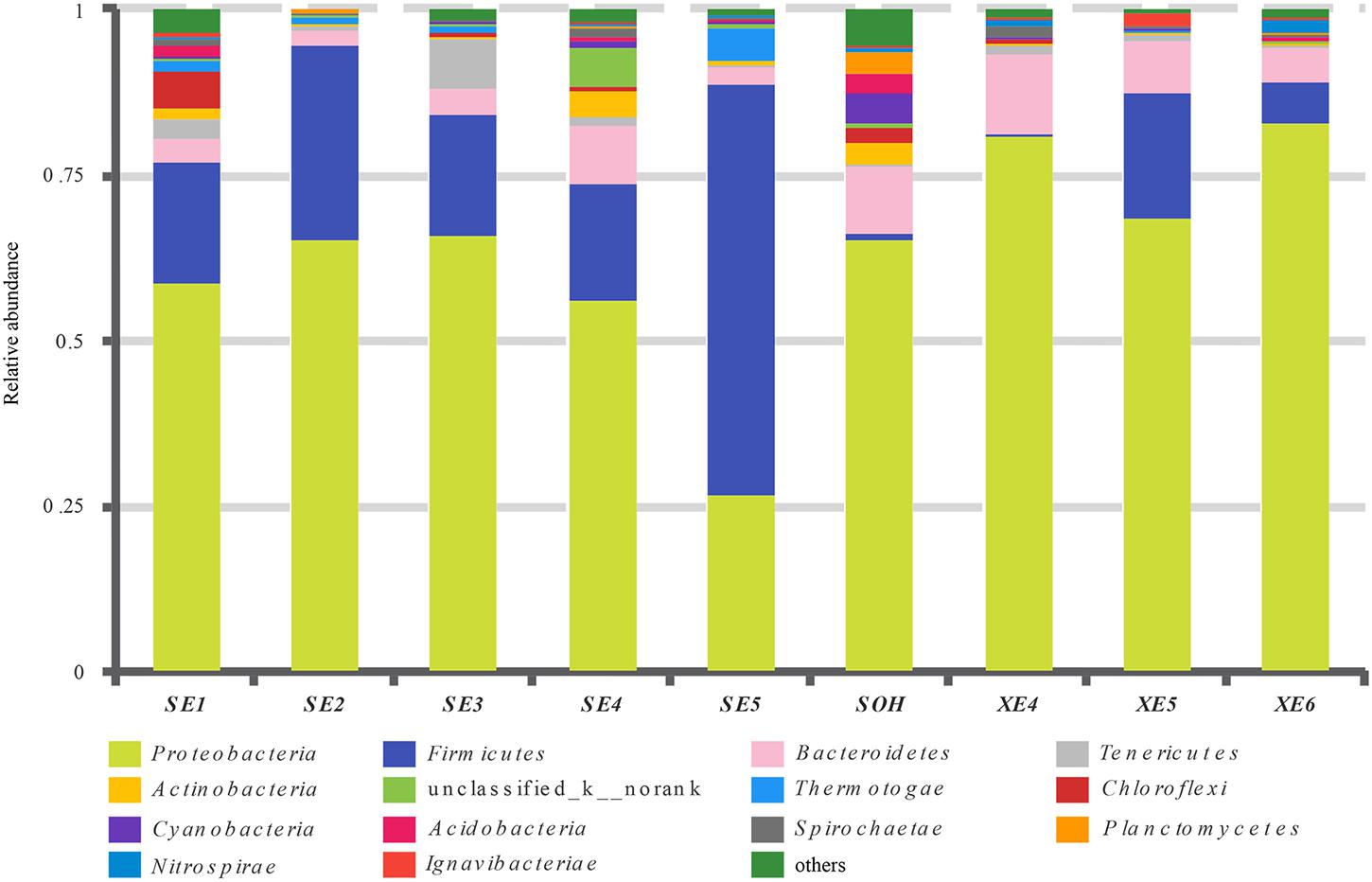
FIGURE 3. Relative abundances of bacterial 16S rRNA gene sequences from the corrosion samples presented at the phylum level.
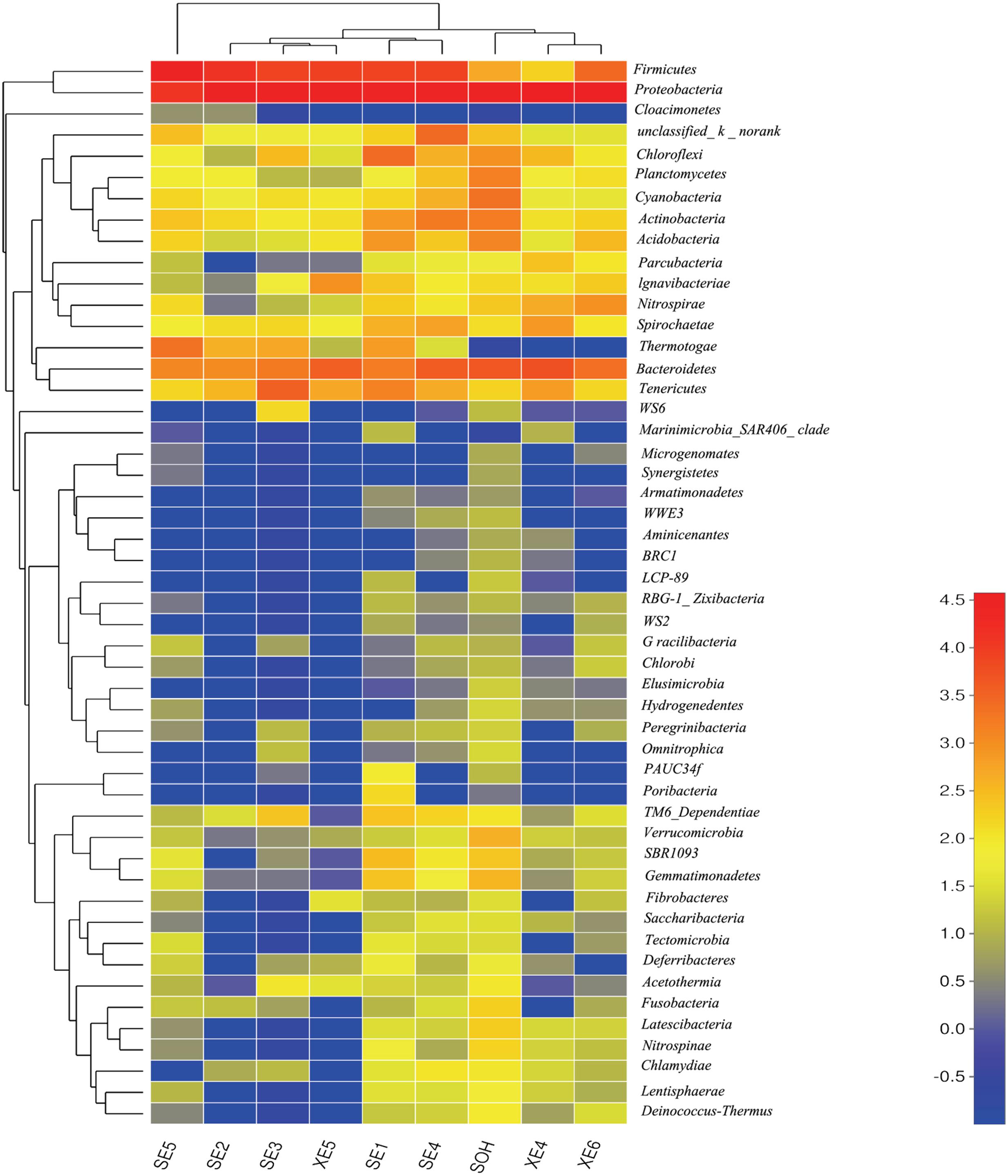
FIGURE 4. The bacterial community distributions among the nine corrosion samples at the phylum level.
At the class level, more than 21 classes of prokaryote were found overall and accounted for 86.80% of the total community (Figure 5 and Table 3). For the majority of corrosion samples, Deltaproteobacteria was the most abundant class, ranging from 16.50 to 71.56% according to the samples. Clostridia came second and ranged from 0 to 61.85% of the whole community. Gammaproteobacteria was the third most dominant class, ranging from 1.78 to 22.0% of the whole bacterial community. Some other classes (e.g., Alphaproteobacteria and Bacteroidia) also occupied a relatively large proportion of the bacterial community composition, based on the average abundance analysis (Figure 5 and Table 3). In addition, some classes, which occupied a relatively small proportion of the community composition, but which have been associated with corrosion (such as Zetaproteobacteria), were also found in this study (Figure 5). However, the community composition of some samples was unique at the class level. For example, Clostridia (61.85%) and Deltaproteobacteria (16.50%) were the first and second dominant classes in SE5. Bacteroidia (9.06%) and Gammaproteobacteria (12.59%) were the second dominant bacterial class in XE4 and XE6, respectively. Alphaproteobacteria was the third dominant bacterial classes in SE4 (15.48%) and SOH (19.91%). Furthermore, the hierarchical clustering heat map was also plotted to compare the membership and structure of each sample at the class level. It also indicated that Deltaproteobacteria, Clostridia and Gammaproteobacteria were the dominant three bacterial communities among the top 50 classes across all the samples (Figure 6).
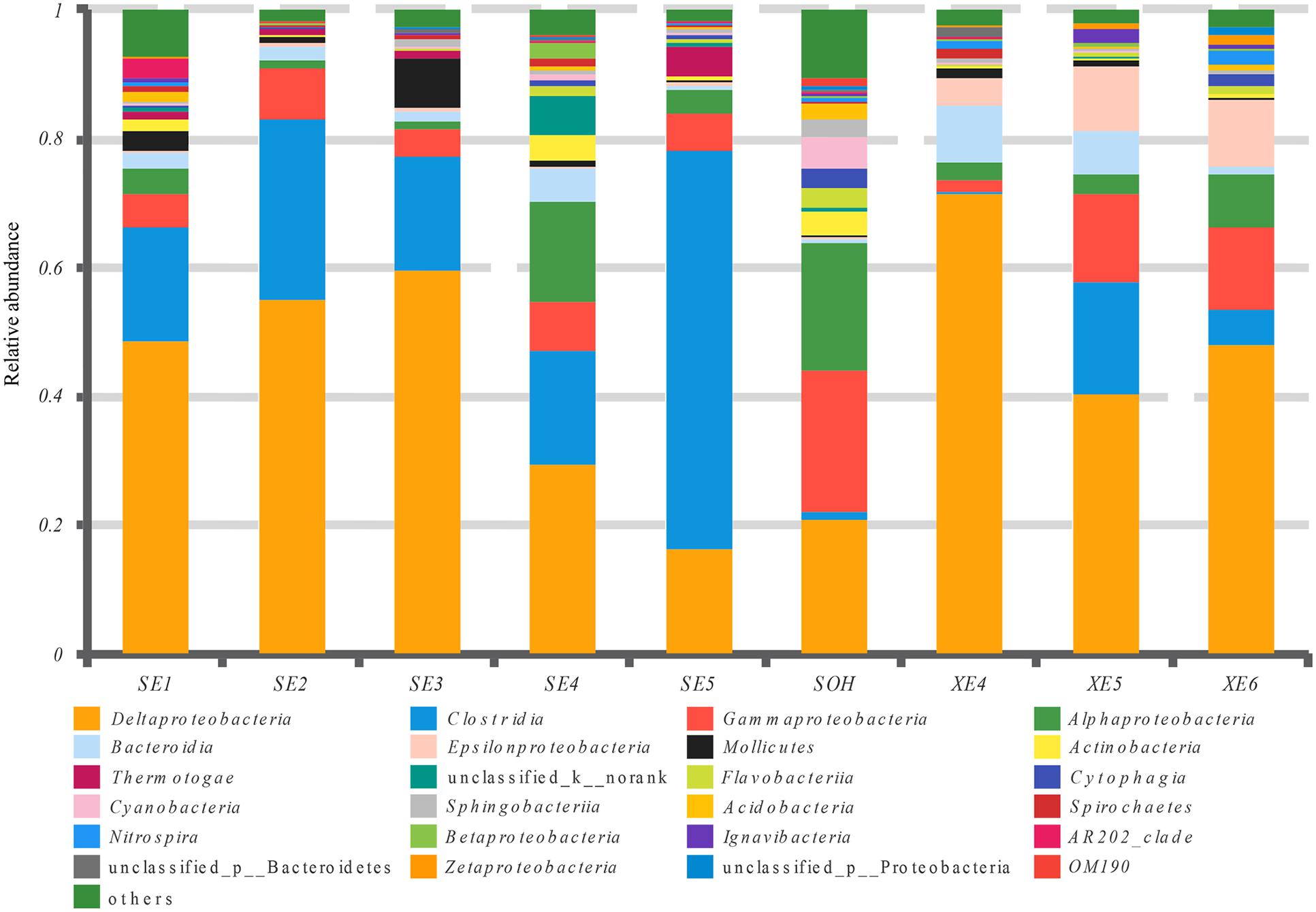
FIGURE 5. Relative abundances of bacterial 16S rRNA gene sequences from the corrosion samples presented at the class level.
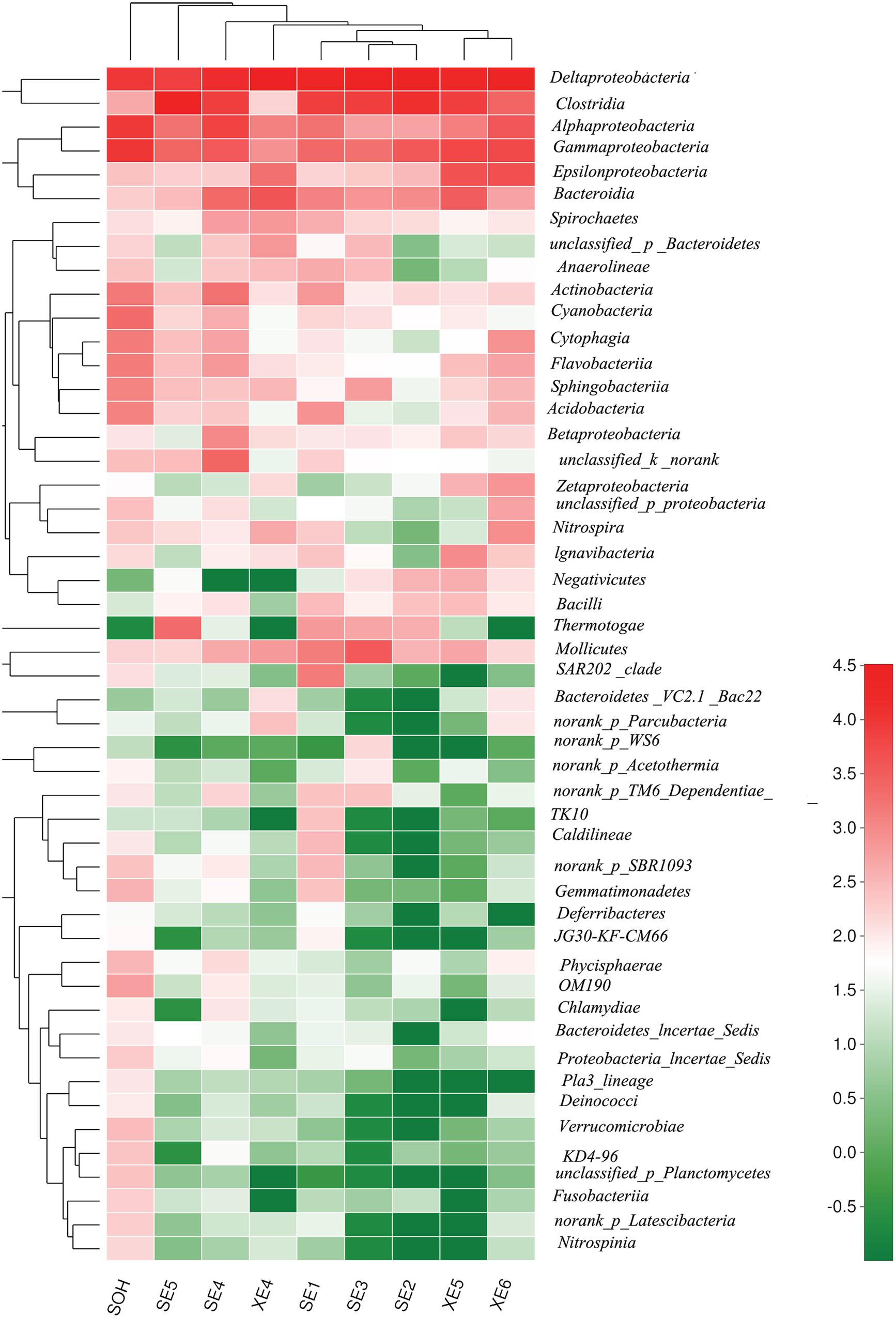
FIGURE 6. The bacterial community distributions among the nine corrosion samples at the class level.
More than 56 genera were identified (Figure 7 and Table 4). For the majority of corrosion samples, Desulfovibrio was the most abundant, ranging from 3.59 to 42.04% of the total number of genera. Desulfobacter came second (2.70–18.75%), and Desulfotomaculum was the third (0–56.04%). Other genera were well represented, based on the average abundance analysis (e.g., Sulfurimonas and Desulfonatronum) (Figure 7 and Table 4). The generic profile of some samples was unique. For example, Desulfotomaculum (56.04%) and Desulfobacter (18.01%) were the dominant genera in SE5 and XE5, respectively. Sulfurimonas (10.31%) was the second most dominant genus in samples XE6. The hierarchical clustering heat map indicated that Desulfovibrio, Desulfobacter and Firmicutes were the dominant three bacterial genera among the top 100 genera (Figure 8).
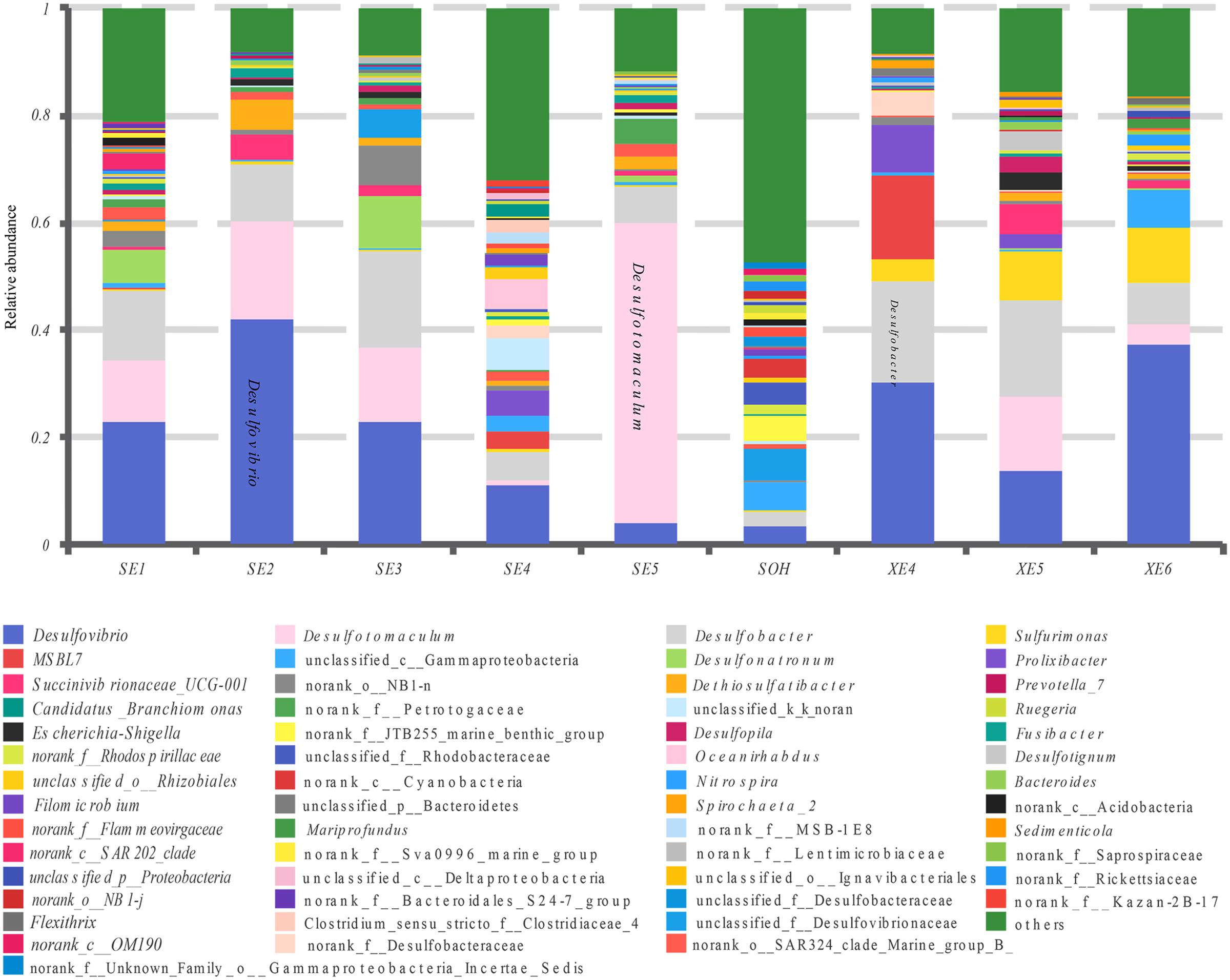
FIGURE 7. Relative abundances of bacterial 16S rRNA gene sequences from the corrosion samples presented at the genus level.
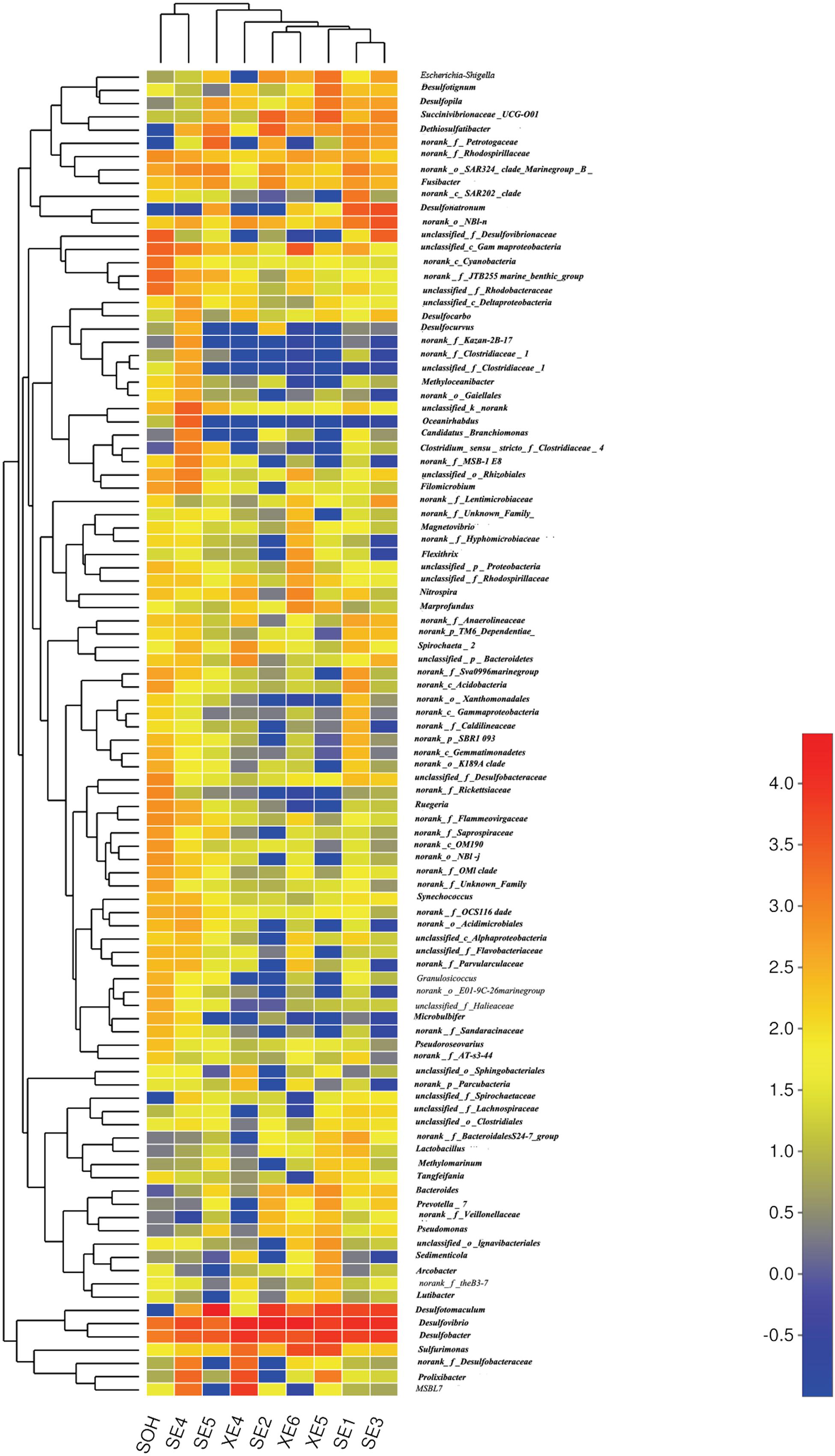
FIGURE 8. The bacterial community distributions among the nine corrosion samples at the genus level.
Beta Diversity Analysis of the Nine Corrosion Samples
Two methods were adopted to analyze the beta diversity of the nine samples (Figures 9, 10). Firstly, a hierarchical cluster tree of the bacterial communities was constructed by means of the UPGMA at a 97%-similarity OTU level. This showed that the data were clustered in two distinct groups (Figure 9). Group 1 contained the 6 months immersion sample (SOH) and one 8 years immersion sample (SE4). Group 2 included the other 8 years immersion samples. Afterwards, a principal coordinates analysis (PCoA) then targeted major bacterial clades, and confirmed the output of the first method, and explained 51.06% of the observed variation (Figure 10). Eight years immersion samples (except SE4) were grouped to the right of the graph along PC1. SOH was separated from the 8-years immersion samples and grouped to the left of the graph along PC1. Whereas SE4 was grouped in the middle of the graph between SOH and the other 8-year immersion samples. There was a clear distinction between SOH and the other corrosion samples along the first axis. Furthermore, bacterial communities were separated by the second axis. The results of the two methods indicated that the bacterial diversity (bacterial community composition) was clearly correlated to the immersion period. The sea area had no influence on the composition of the bacterial community.
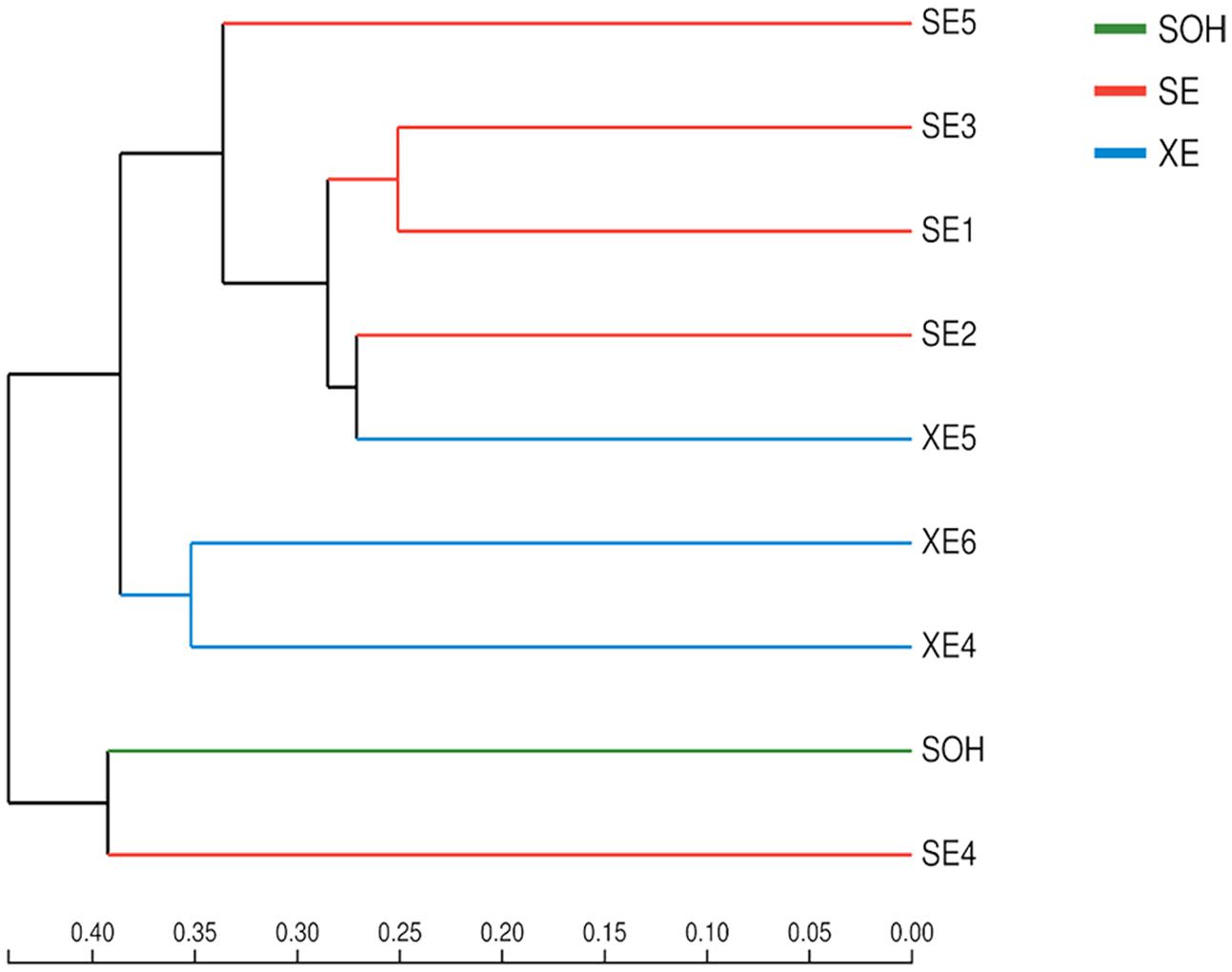
FIGURE 9. A hierarchical cluster tree created using UPGMA with Bray–Curtis at the level of OTU. Microbial community distribution patterns at a 97%-similarity OTU level.
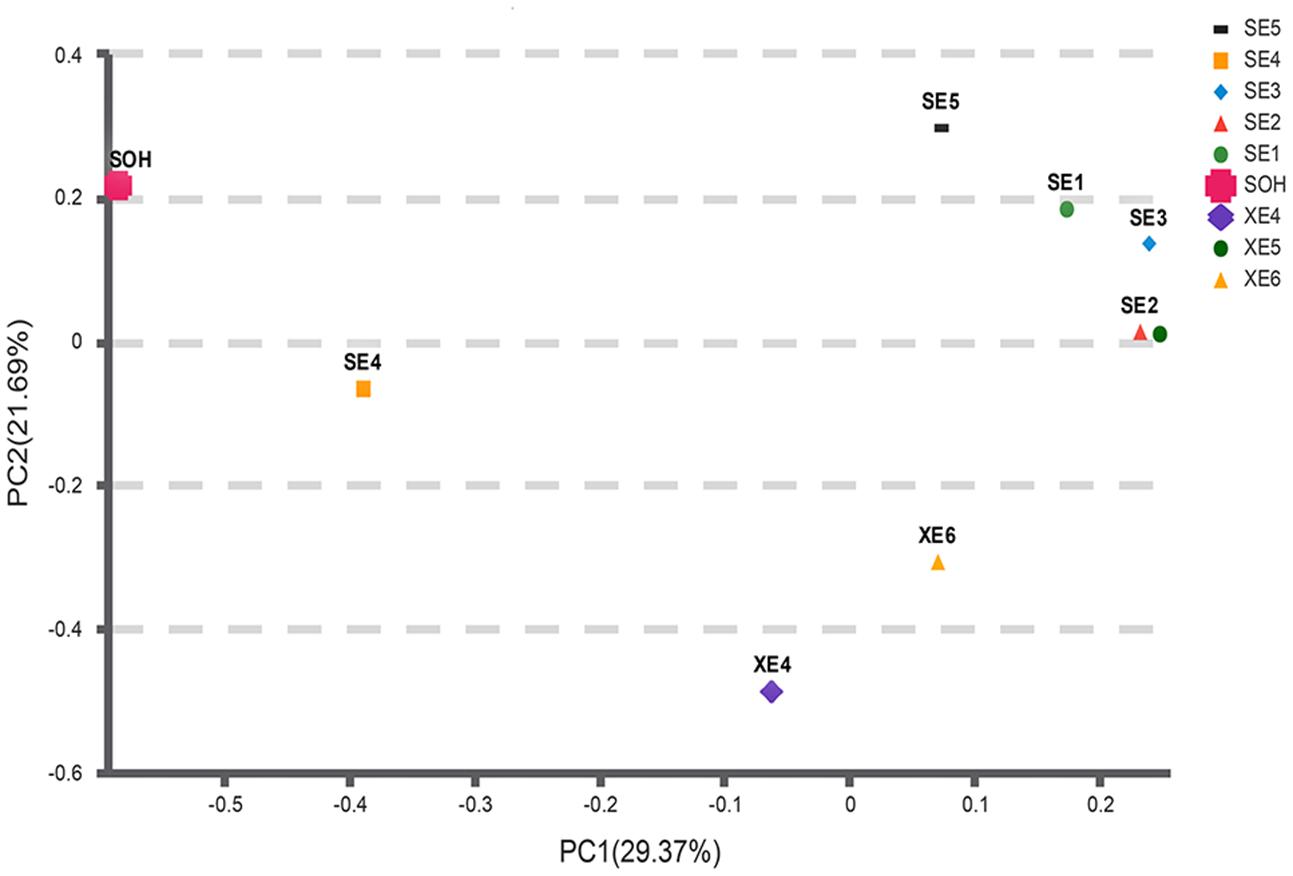
FIGURE 10. A PCoA scatter plot was calculated with Bray-Curtis. The first two components PC1 and PC2 explain 29.37 and 21.69% of the variations, respectively. Microbial community distribution patterns at a 97%-similarity OTU level.
Discussion
Analysis of Composition of the Bacterial Community of the Corrosion Samples
Compared with samples from other environments, such as marine sediments (Liu et al., 2015), and seawater samples (Suh et al., 2015; Yang et al., 2015), the composition of the bacterial communities on the corrosion samples was similar at the phylum level, but significantly different at the genus level. In this study, Proteobacteria, Bacteroidetes and Firmicutes were the three core phyla in all samples. Proteobacteria was dominant in the majority of samples. This was also observed by Vigneron et al. (2016). They analyzed the bacterial community composition of corrosion samples taken from an offshore oil production facility. Proteobacteria also emerged as the dominant bacterial phylum in the initial stage of biofilm formation on carbon steel (Bermont-Bouis et al., 2007; Jones et al., 2007; Lee et al., 2008; Dang et al., 2011; McBeth and Emerson, 2016). We can point to a number of reasons for the dominance of Proteobacteria in rust samples. Members of this phylum are pioneer surface colonizers and important biofilm ‘builders.’ The ‘facilitation’ of biofilm formation is an important step in the further development of diverse populations, and their on-going stability (Slightom and Buchan, 2009; Dang et al., 2008, 2011). It is noteworthy that Proteobacteria is also the largest bacterial phylum and the most abundant across a range of environmental conditions (Liu et al., 2015; Qi et al., 2016) and in seawater (Suh et al., 2015; Yang et al., 2015; Mancuso et al., 2016).
Bacteroidetes was the second most abundant phylum. It was found to be dominant in biofilms formed on steel plates immersed in the sea for 40 days (Dang et al., 2011; McBeth and Emerson, 2016). Bacteroidetes is a dominant phylum in marine environments (Kirchman, 2002), and has also been linked to biological corrosion. Bacteroidetes can also contribute to the survival of other of surface colonizers and the formation and development of biofilms (Dang et al., 2011). The composition and abundance of surface-associated bacterial colonies may be influenced by ‘predatory’ members of the phylum Bacteroidetes (Dang and Lovell, 2016). Bacteroidetes members are known to degrade complex biopolymers (Kirchman, 2002), which may assist in the creation of an aerobic environment with a biofilm, that is conducive to the growth of colonizing bacteria. Firmicutes was the third most dominant phylum in the majority of corrosion samples. It was found to be abundant in biofilms in the rust layer, based on 16S rRNA gene (Zhang and Fang, 2001; Luan et al., 2012). The presence of a Firmicutes member, Tindallia texcoconensis, isolated from lake Texcoco, Mexico by Alazard et al. (2007), was associated with hydrogen production, that provided for SRB. Some members of this phylum generate H2S and organic acids that can cause corrosion. For example, Acetobacterium carbinolicum produces acetic acid which can corrode steel (Paarup et al., 2006).
At the class level, Deltaproteobacteria was the dominant class in the majority of corrosion samples. This observation parallels information from studies of the bacterial communities of samples collected from water-flooded petroleum reservoirs, water injection systems of Brazilian offshore oil platforms, and corrosive petroleum reservoirs in Yangzhou (Korenblum et al., 2010; Li et al., 2016; Tian et al., 2017). There are many important SRB groups belonging to this class, e.g., Desulfovibrio, Desulfobacter and Desulfonatronum. Some species of SRB in the Deltaproteobacteria can promote the production of corrosive hydrogen sulfide from metallic sulfates (Kan et al., 2011). Clostridia was the second most abundant class in the majority of corrosion samples. This was similar observation to the results of a study of the composition of the bacterial community composition of biofilms from metal surfaces of an alkaline district heating system, and samples collected from water-flooded petroleum reservoirs (Kjeldsen et al., 2007; Tian et al., 2017). Some important SRB groups also belong to this class, for instance Desulfotomaculum. Some Clostridia produce acetic, butyric, or formic acids so that their presence may also lead to corrosion (Broda et al., 2000). Some are homoacetogens meaning that they convert carbon dioxide and hydrogen into acetate and propionate (Boga and Brune, 2003). The third most abundant class was the Gammaproteobacteria. Dang et al. (2011) reported that Gammaproteobacteria (mainly Alteromonadales and Oceanospirillales) are pioneer and long term surface colonizers, and can also contribute to the initiation and on-going development of biofilms. There are some other classes that were identified by this study that are known to contribute to steel corrosion, for instance Epsilonproteobacteria and Zetaproteobacteria (Dang et al., 2011; McBeth et al., 2011). Related research showed that Epsilonproteobacteria were the possible cause of microbial corrosion in pipelines injected with bisulfite (An et al., 2015).
At the genus level, three SRBs genera, Desulfovibrio spp., Desulfotomaculum spp. and Desulfobacter spp. formed a large proportion of the bacterial communities that were analyzed in this study. Desulfovibrio spp. were the most abundant. This complies with the data from a study of the corrosive marine biofilms of carbon steels immersed in seawater for 8 months (Bermont-Bouis et al., 2007). Desulfovibrio was also the most abundant bacterial genus in corrosion samples from oil pipelines in the Southeast of Mexico (Zhang and Fang, 2001; Neria-González et al., 2006; Vigneron et al., 2016). This agrees with previous studies that show that this genus is often the main cause of bacteria related corrosion (Miranda et al., 2006; Ilhan-Sungur et al., 2007). Members of genus Desulfovibrio are metabolically diverse and can reduce iron sulfate and, with hydrogen, produce H2S (Dinh et al., 2004). Significantly, the pH of an aquatic environment is modified by the presence of H2S, leading to the generation of a corrosion product, FeS in the presence of iron. The steel corrosion capacity of Desulfovibrio spp., such as D. vulgaris (Zhang et al., 2015, 2016), D. alaskensis (Wikieł et al., 2014) and D. desulfuricans (Lopes et al., 2006) has been extensively studied in laboratory experiments. Different mechanisms of corrosion development caused by Desulfovibrio spp. have been described and show that members of this genus have a interact with Clostridium species (Zhang and Fang, 2001), which was the second most abundant bacterial genus in sample SE4.
Desulfotomaculum spp. (the second most abundant genus) is a gram-positive SRB and is thermophilic. It plays an important role in MIC (Cetin et al., 2007) by accelerating cathodic depolarization and decelerating anodic depolarization (Cetin and Aksu, 2009). The ability of members of this genus to corrode steel has been studied extensively in laboratory experiments: D. nigrificans (Mystkowska et al., 2015), D. orientis (Ren and Wood, 2004), and D. kuznetsovii (Anandkumar et al., 2015). Members of this genus are usually associated with oil, and have been isolated from the crude oil field, oil production wells, or even the cooling towers of a petroleum refinery (Cetin et al., 2007; Cetin and Aksu, 2009; Anandkumar et al., 2015). Desulfobacter spp., is a mesophilic, gram-negative genus with an oval morphology in the marine environment. Its ability to oxidize acetic acid is a characteristic (Widdel, 1988). It can also reduce organic substrates to CO2 in a strictly anaerobic environment. However, the roles that Desulfobacter spp. play in steel corrosion are still unknown. Further study of its corrosive properties are needed. Some bacteria were found to inhibit MIC by the formation of a biofilm on the surface of steel. They included gramicidin-producing Bacillus brevis (Nikolaev and Plakunov, 2007), although Vibrio neocaledonicus may have the highest known level of corrosion inhibition (Moradi et al., 2015a,b). They did not appear in our results, but this might mean that they were present but at levels that were too low for detection, or that they were present at higher levels but were not detectable by techniques we adopted.
Comparative Analysis of Bacterial Community Composition
It is well-known that biofilm maturity significantly affects the bacterial communities of biofilms (Neria-González et al., 2006). The succession pattern of these bacterial communities is tied to the immersion time of the steel. The steel could be exposed to local acidification with a decrease in the redox potential over time. This might stabilize the conditions so that the anaerobes are better accommodated. Also, the increase in the local concentration of dissolved iron salts may affect the biofilm community. Although SRB were dominant in all of our samples, the bacterial community composition of samples immersed for 8 years was significantly different to that of the sample immersed for 6 months. The bacterial diversity of the 6-months sample was higher than that the others. This result was consistent with previous studies. Bermont-Bouis et al. (2007) reported that there was a big difference between the bacterial community composition of 8 months immersion samples and 1 month immersion samples. They found that SRB were also the dominant population in mature biofilms after an 8 months immersion, but Vibrio spp. (Gammaproteobacteria) was the main component in samples that had been immersed for 1 month (Bermont-Bouis et al., 2007). This may be because biofilms form in a consistent series of discrete steps, or as a time series, each being associated with a different bacterial community (Stoodley et al., 2002).
Oxygen is consumed throughout the formation of biofilms. For instance, members of the Bacteroidetes may contribute to a decrease the quantity of oxygen emitted: these bacteria degrade high-molecular weight organic matter (Kirchman, 2002). The reduction of oxygen in the biofilms generates the anaerobic environment, which is needed to induce SRB growth. Over time, an increasingly acidic and anaerobic environment develops, and this is believed to result in the succession of membership of the biofilm community. In this study, the bacterial diversity of the samples immersed for 6 months was higher than that of the other samples. We believe that the anaerobic environment formed after 8 years was more suitable for the growth of SRB than that of the samples that had been immersed for 6 months. This implies that the anaerobic environment formed after 8 years was clearly unsuitable for the aerobic bacteria (the early colonizers), which is the reason for there being a reduction in bacterial diversity over time. That SRB may be only a minor component at the initial stage of biofilms is supported by Dang et al. (2011). Earlier studies have shown that the anaerobic zone will form underneath the upper aerobic layer when it is 10–25 mm thick (Coulter and Russell, 1976). At that point, the biofilm is clearly composed of a complex consortium of aerobic and anaerobic bacteria (Baker et al., 2003; Zhang et al., 2003). In addition, the bacterial community composition of biofilms is also changed most at the beginning of immersion (Dang and Lovell, 2000; Lee et al., 2008).
The methods of analysis can also have an impact on our understanding of the structure of bacterial communities. Proteobacteria was the dominant group for all corrosion samples no matter what methods were used. But the numbers of phyla and genera obtained from the corrosion samples were influenced by the methodology. We obtained more than 50 phyla and 100 genera by high-throughput Illumina sequencing (Figures 4, 8): that is many more than have previously been detected (Luan et al., 2012; Chen et al., 2014). Equally important, the number of OTUs obtained by high-throughput sequencing was greater than had been revealed by the traditional methods (plating) and T-RFLP technique. 19,581 OTUs were found in the nine corrosion samples by high-throughput Illumina sequencing in this study, whereas only 64 OTUs and 24 OTUs were previously identified in corrosion samples by PCR-RFLP (Luan et al., 2012; Chen et al., 2014). The high-throughput Illumina sequencing method is clearly ideal because we were able to achieve in depth quantitative analyses of microbial communities (Bokulich and Mills, 2012; Mayo et al., 2014).
Many environmental factors can affect the composition of bacterial communities. In this study, the richness of immersed steel was related to the sea location. The average richness of 8-years immersion samples from Xiamen was numerically lower than that of Sanya, although the difference was not significant. Among the environmental factors, salinity, pH and temperature generally have a significant effect on the bacterial community composition. Parallel research has shown that saline water irrigation can change bacterial metabolic activities and community structures (Chen et al., 2017). Cell growth rate was inhibited by high salinity, but the viability and integrity of the bacterial membrane were increased (Kim and Chong, 2017). The functional structure of a bacterial community was significantly affected by pH (Joshi et al., 2017). However, in our study, there was little difference in salinity at the two locations (Sanya 33.97aaa and Xiamen 31.96aaa) and pH (Sanya 8.48 and 8.56). As the seawater temperature at Sanya (25.14°C) was higher than that of Xiamen (19.27°C), we speculate that temperature caused the small difference. Many studies have shown that temperature is a major influence on the composition of bacterial communities in the marine environment. The composition of the bacterial community of crude oil-contaminated marine sediments or seawaters were shown by Bargiela et al. (2015) and Meng et al. (2016) to be strongly linked to temperature. Even more important, studies have shown that temperature is also related to corrosion levels. In the aquatic system, temperature plays an important role in the changes of most biofilm parameters, and in their propagation and metabolism (Bott, 1996; Rao, 2010). In addition, the amount of bacteria (whether aerobic or anaerobic bacteria) in biofilms was also temperature dependant (Bott, 1996; Guo et al., 2006). It has been reported that the amount of bacteria in the rust layer of immersed carbon steel in Yulin station was more abundant than that of Qingdao station because of the different temperature (Guo et al., 2006). In this study, the seawater temperature at Sanya was nearly 5°C higher than at Xiamen. The temperature of the Sanya coast was much more appropriate for the growth of bacteria. The degree of corrosion was enhanced by the presence of many more large fouling organisms in the warmer water. This resulted in the provision of higher levels of soluble nutrients provided by the decomposition of the other organisms. However, except for them, many other factors (like nutrients, dissolved oxygen and so on) could also influence the bacterial communities. The difference of the diversity was probably the result of integrated effects of the multiple factors. So far, it is hard to explain how the 6 months immersion sample (SOH) and 8 years immersion sample (SE4) were clustered in the same group in the multi-sample dendrogram. Our next step is to study the bacterial communities from samples from different substrates with a wider range of immersion times in a wider range of seawaters. This will greatly improve our knowledge of the relationships between environmental factors and bacterial community structure.
Conclusion
The bacterial community composition of corrosion samples collected from rust layers of steel plates immersed in seawater for 6 months and 8 years at Sanya and Xiamen was revealed by means of Illumina MiSeq sequencing. We identified members of 13 phyla. Proteobacteria, Firmicutes and Bacteroidetes three dominated and accounted for nearly 89.03% of the total. Desulfovibrio spp., Desulfotomaculum spp. and Desulfobacter spp. were the core genera. The bacterial diversity from steel plate that has been immersed for 6 months was significantly higher than that taken from plates that had been immersed for 8 years. The average richness of biofilms removed from steel plates immersed for 8 years from Sanya was numerically but not significantly higher in similar samples taken from Xiamen at the same time. We identified bacteria that had not previously been found in this niche, although we do not know if they are involved in the corrosion of steel.
Author Contributions
The article and experiment done by XL, YL took part in the experiment, experimental design done by JD and HX. The other authors took part in the sample collection.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Basic Research Program (No. 2014CB643304) and national natural science foundation of China (No. 41576080). We thank the Qingdao Research Institute for Marine Corrosion for providing 8-year immersion samples.
Footnotes
- ^ http://drive5.com/uparse/
- ^ http://rdp.cme.msu.edu/
- ^ http://www.mothur.org/wiki/Schloss_SOP\#Alpha_diversity
References
Alazard, D., Badillo, C., Fardeau, M. L., Cayol, J. L., Thomas, P., Roldan, T., et al. (2007). Tindallia texcoconensis, sp. nov. a new haloalkaliphilic bacterium isolated from lake texcoco, mexico. Extremophiles 11, 33–39. doi: 10.1007/s00792-006-0006-5
An, D., Dong, X., An, A., Park, H. S., Strous, M., and Voordouw, G. (2015). Metagenomic analysis indicates epsilonproteobacteria as a potential cause of microbial corrosion in pipelines injected with bisulfite. Front. Microbiol. 7:28. doi: 10.3389/fmicb.2016.00028
Anandkumar, B., Choi, J., Venkatachari, G., and Maruthamuthu, S. (2015). Molecular characterization and corrosion behavior of thermophilic (55°c) srb desulfotomaculum kuznetsovii isolated from cooling tower in petroleum refinery. Mater. Corros. 60, 730–737. doi: 10.1002/maco.200805177
Angell, P., and Urbanic, K. (2000). Sulphate-reducing bacterial activity as a parameter to predict localized corrosion of stainless alloys. Corros. Sci. 42, 897–912. doi: 10.1016/S0010-938X(99)00116-X
Baboian, R. (ed.) (2005). Corrosion Tests and Standards: Application and Interpretation. West Conshohocken PA: ASTM International, 367.
Baker, P. W., Ito, K., and Watanabe, K. (2003). Marine prosthecate bacteria involved in the ennoblement of stainless steel. Environ. Microbiol. 5, 925–932. doi: 10.1046/j.1462-2920.2003.00489.x
Bargiela, R., Mapelli, F., Rojo, D., Chouaia, B., Tornés, J., Borin, S., et al. (2015). Bacterial population and biodegradation potential in chronically crude oil-contaminated marine sediments are strongly linked to temperature. Sci. Rep. 5:11651. doi: 10.1038/srep11651
Beech, I. B. (2004). Corrosion of technical materials in the presence of biofilms - current understanding and state-of-the art methods of study. Int. Biodeterior. Biodegradation 53, 177–183. doi: 10.1016/S0964-8305(03)00092-1
Beech, I. B., and Sunner, J. (2004). Biocorrosion: towards understanding interactions between biofilms and metals. Curr. Opin. Biotechnol. 15, 181–186. doi: 10.1016/j.copbio.2004.05.001
Bermont-Bouis, D., Janvier, M., Grimont, P., Dupont, I., and Vallaeys, T. (2007). Both sulfate-reducing bacteria and enterobacteriaceae take part in marine biocorrosion of carbon steel. J. Appl. Microbiol. 102, 161–168. doi: 10.1111/j.1365-2672.2006.03053.x
Boga, H. I., and Brune, A. (2003). Hydrogen-dependent oxygen reduction by homoacetogenic bacteria isolated from termite guts. Appl. Environ. Microbiol. 69, 779–786. doi: 10.1128/AEM.69.2.779-786.2003
Bokulich, N. A., and Mills, D. A. (2012). Next-generation approaches to the microbial ecology of food fermentations. BMB Rep. 45, 377–389. doi: 10.5483/BMBRep.2012.45.7.148
Bott, T. R. (1996). 96/03046 - fouling of heat exchangers. Fuel. Energ. Abstr. 37, 211. doi: 10.1016/0140-6701(96)88959-4
Broda, D. M., Saul, D. J., Bell, R. G., and Musgrave, D. R. (2000). Clostridium algidixylanolyticum sp. nov., a psychrotolerant, xylan-degrading, spore-forming bacterium. Int. J. Syst. Evol. Microbiol. 50, 623–631. doi: 10.1099/00207713-50-2-623
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). Qiime allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Cetin, D., and Aksu, M. L. (2009). Corrosion behavior of low-alloy steel in the presence of desulfotomaculum, sp. Corros. Sci. 51, 1584–1588. doi: 10.1016/j.corsci.2009.04.001
Cetin, D., Bilgic, S., and Donmez, G. (2007). Biocorrosion of low alloy steel by desulfotomaculum sp. and effect of biocides on corrosion control. ISIJ Int. 47, 1023–1028. doi: 10.2355/isijinternational.47.1023
Chao, Y., Mao, Y., Wang, Z., and Zhang, T. (2015). Diversity and functions of bacterial community in drinking water biofilms revealed by high-throughput sequencing. Sci. Rep. 5:10044. doi: 10.1038/srep10044
Chen, L., Li, C., Feng, Q., Wei, Y., Zheng, H., Zhao, Y., et al. (2017). Shifts in soil microbial metabolic activities and community structures along a salinity gradient of irrigation water in a typical arid region of china. Sci. Total Environ. 598, 64–70. doi: 10.1016/j.scitotenv.2017.04.105
Chen, Y. W., Luan, X., and Duan, J. Z. (2014). Diversity of bacterial community on stainless steel surface immersed in seawater. Oceanol. Limnol. Sin. 45, 1064–1070. doi: 10.1007/BF00118994
Coetser, S. E., and Cloete, T. E. (2005). Biofouling and biocorrosion in industrial water systems. Crit. Rev. Microbiol. 31, 213–232. doi: 10.1080/10408410500304074
Coulter, W. A., and Russell, C. (1976). Ph and eh in single and mixed culture bacterial plaque in an artificial mouth. J. Appl. Microbiol. 40, 73–87. doi: 10.1111/j.1365-2672.1976.tb00593.x
Dang, H., Chen, R., Wang, L., Shao, S., Dai, L., Ye, Y., et al. (2011). Molecular characterization of putative biocorroding microbiota with a novel niche detection of Epsilon- and Zetaproteobacteria in Pacific Ocean coastal seawaters. Environ. Microbiol. 13, 3059–3074. doi: 10.1111/j.1462-2920.2011.02583.x
Dang, H., Li, T., Chen, M., and Huang, G. (2008). Cross-ocean distribution of rhodobacterales bacteria as primary surface colonizers in temperate coastal marine waters. Appl. Environ. Microbiol. 74, 52–60. doi: 10.1128/AEM.01400-07
Dang, H., and Lovell, C. R. (2000). Bacterial primary colonization and early succession on surfaces in marine waters as determined by amplified rrna gene restriction analysis and sequence analysis of 16s rrna genes. Appl. Environ. Microbiol. 66, 467–475. doi: 10.1128/AEM.66.2.467-475.2000
Dang, H. Y., and Lovell, C. R. (2002a). Numerical dominance and phylotype diversity of marine Rhodobacter species during early colonization of submerged surfaces in coastal marine waters as determined by 16S ribosomal DNA sequence analysis and fluorescence in situ hybridization. Appl. Environ. Microbiol. 68, 496–504. doi: 10.1128/AEM.68.2.496-504.2002
Dang, H., and Lovell, C. R. (2002b). Seasonal dynamics of particle-associated and free-living marine Proteobacteria in a salt marsh tidal creek as determined using fluorescence in situ hybridization. Environ. Microbiol. 4, 287–295. doi: 10.1046/j.1462-2920.2002.00295.x
Dang, H., and Lovell, C. R. (2016). Microbial surface colonization and biofilm development in marine environments. Microbiol. Mol. Biol. Rev. 80, 91–138. doi: 10.1128/MMBR.00037-15
Dennis, K. L., Wang, Y., Blatner, N. R., Wang, S., Saadalla, A., Trudeau, E., et al. (2013). Adenomatous polyps are driven by microbe-instigated focal inflammation and are controlled by 1L-10-producing T cells. Cancer Res. 73, 5905–5913. doi: 10.1158/0008-5472.CAN-13-1511
Dinh, H. T., Kuever, J., Muszmann, M., Hassel, A. W., Stratmann, M., and Widdel, F. (2004). Iron corrosion by novel anaerobic microorganisms. Nature 427, 829–832. doi: 10.1038/nature02321
Duan, J., Wu, S., Zhang, X., Huang, G., Du, M., and Hou, B. (2008). Corrosion of carbon steel influenced by anaerobic biofilm in natural seawater. Electrochim. Acta 54, 22–28. doi: 10.1016/j.electacta.2008.04.085
Dunbar, J., Takala, S., Barns, S. M., Davis, J. A., and Kuske, C. R. (1999). Levels of bacterial community diversity in four arid soils compared by cultivation and 16s rrna gene cloning. Appl. Environ. Microbiol. 65, 1662–1669.
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Edgar, R. C. (2013). Uparse: highly accurate otu sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). Uchime improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Gonzalez-Rodrıguez, C. A., Rodríguez-Gómez, F. J., and Genescá-Llongueras, J. (2008). The influence of desulfovibrio vulgaris, on the efficiency of imidazoline as a corrosion inhibitor on low-carbon steel in seawater. Electrochim. Acta 54, 86–90. doi: 10.1016/j.electacta.2008.02.119
Grooters, M., Harneit, K., Wöllbrink, M., Sand, W., Stadler, R., and Fürbeth, W. (2007). Novel steel corrosion protection by microbial extracellular polymeric substances (eps) – biofilm-induced corrosion inhibition. Adv. Mater. Res. 20-21, 375–378. doi: 10.4028/www.scientific.net/AMR.20-21.375
Guo, P., Yan, M., Huang, G., Du, M., and Peng, Y. (2006). A study on microbiologically influenced corrosion of a carbon steel in seawater. Corros. Sci. Prot. Technol. 18, 410–413.
Hamilton, W. A. (1991). “Sulphate-reducing bacteria and their role in biocorrosion,” in Biofouling and Biocorrosion in Industrial Water Systems, eds H. C. Flemming and G. G. Geesey (Berlin: Springer), 187–193.
Hou, B. R., Li, X. G., Ma, X. M., Du, C. W., Zhang, D. W., Zheng, M., et al. (2017). The cost of corrosion in China. npj Mater. Degrad. 1, 4. doi: 10.1038/s41529-017-0005-2
Ilhan-Sungur, E., Cansever, N., and Cotuk, A. (2007). Microbial corrosion of galvanized steel by a freshwater strain of sulphate reducing bacteria (Desulfovibrio sp.). Corros. Sci. 49, 1097–1109. doi: 10.1016/j.corsci.2006.05.050
Jones, P. R., Cottrell, M. T., Kirchman, D. L., and Dexter, S. C. (2007). Bacterial community structure of biofilms on artificial surfaces in an estuary. Microb. Ecol. 53, 153–162. doi: 10.1007/s00248-006-9154-5
Joshi, D. R., Zhang, Y., Gao, Y., Liu, Y., and Yang, M. (2017). Biotransformation of nitrogen- and sulfur-containing pollutants during coking wastewater treatment: correspondence of performance to microbial community functional structure. Water Res. 121, 338–348. doi: 10.1016/j.watres.2017.05.045
Kan, J., Chellamuthu, P., Obraztsova, A., Moore, J. E., and Nealson, K. H. (2011). Diverse bacterial groups are associated with corrosive lesions at a granite mountain record vault (gmrv). J. Appl. Microbiol. 111, 329–337. doi: 10.1111/j.1365-2672.2011.05055.x
Kim, L. H., and Chong, T. H. (2017). Physiological responses of salinity-stressed vibrio sp. and the effect on biofilm formation on nanofiltration membrane. Environ. Sci. Technol. 51, 1249–1258. doi: 10.1021/acs.est.6b02904
Kip, N., and Veen, J. A. V. (2015). The dual role of microbes in corrosion. ISME. J. 9, 542–551. doi: 10.1038/ismej.2014.169
Kirchman, D. L. (2002). The ecology of cytophaga-flavobacteria in aquatic environments. FEMS Microbiol. Ecol. 39, 91–100. doi: 10.1111/j.1574-6941.2002.tb00910.x
Kjeldsen, K. U., Kjellerup, B. V., Egli, K., Frølund, B., Nielsen, P. H., and Ingvorsen, K. (2007). Phylogenetic and functional diversity of bacteria in biofilms from metal surfaces of an alkaline district heating system. FEMS Microbiol. Ecol. 61, 384–397. doi: 10.1111/j.1574-6941.2006.00255.x
Korenblum, E., Sebastián, G. V., Paiva, M. M., Coutinho, C. M. L. M., Magalhães, F. C. M., Peyton, B. M., et al. (2008). Action of antimicrobial substances produced by different oil reservoir bacillus, strains against biofilm formation. Appl. Microbiol. Biotechnol. 79, 97–103. doi: 10.1007/s00253-008-1401-x
Korenblum, E., Valoni,É, Penna, M., and Seldin, L. (2010). Bacterial diversity in water injection systems of brazilian offshore oil platforms. Appl. Microbiol. Biotechnol. 85, 791–800. doi: 10.1007/s00253-009-2281-4
Lee, J. W., Nam, J. H., Kim, Y. H., Lee, K. H., and Lee, D. H. (2008). Bacterial communities in the initial stage of marine biofilm formation on artificial surfaces. J. Microbiol. 46, 174–182. doi: 10.1007/s12275-008-0032-3
Leo, F. D., Campanella, G., Proverbio, E., and Urzì, C. (2013). Laboratory tests of fungal biocorrosion of unbonded lubricated post-tensioned tendons. Constr. Build. Mater. 49, 821–827. doi: 10.1016/j.conbuildmat.2013.08.071
Li, X. X., Liu, J. F., Yao, F., Wu, W. L., Yang, S. Z., Mbadinga, S. M., et al. (2016). Dominance of desulfotignum, in sulfate-reducing community in high sulfate production-water of high temperature and corrosive petroleum reservoirs. Int. Biodeterior. Biodegradation 114, 45–56. doi: 10.1016/j.ibiod.2016.05.018
Liu, J., Liu, X., Wang, M., Qiao, Y., Zheng, Y., and Zhang, X. H. (2015). Bacterial and archaeal communities in sediments of the north chinese marginal seas. Microb. Ecol. 70, 1–13. doi: 10.1007/s00248-014-0553-8
Lopes, F. A., Morin, P., Oliveira, R., and Melo, L. F. (2006). Interaction of desulfovibrio desulfuricans biofilms with stainless steel surface and its impact on bacterial metabolism. J. Appl. Microbiol. 101, 1087–1095. doi: 10.1111/j.1365-2672.2006.03001.x
Luan, X., Duan, J. Z., and Chen, Z. M. (2012). Diversity of bacterial community on the surface of Steel in temperate coastal marine waters. Period. Ocean Univ. China 42, 107–115.
Lugauskas, A., Prosyčevas, I., Ramanauskas, R., Grigucevičienė, A., Selskienė, A., and Pakštas, V. (2009). The influence of micromycetes on the corrosion behaviour of metals(steel, al) under conditions of the environment polluted with organic substances. Mat. Sci. 15, 224–235.
Mancuso, F. P., D’Hondt, S., Willems, A., Airoldi, L., and Clerck, O. D. (2016). Diversity and temporal dynamics of the epiphytic bacterial communities associated with the canopy-forming seaweedcystoseira compressa(esper) gerloff and nizamuddin. Front. Microbiol. 7:230. doi: 10.3389/fmicb.2016.00476
Mayo, B., Rachid, C. T., Alegría,Á, Leite, A. M., Peixoto, R. S., and Delgado, S. (2014). Impact of next generation sequencing techniques in food microbiology. Curr. Genomics 15, 293–309. doi: 10.2174/1389202915666140616233211
McBeth, J. M., and Emerson, D. (2016). In situmicrobial community succession on mild steel in estuarine and marine environments: exploring the role of iron-oxidizing bacteria. Front. Microbiol. 7:231. doi: 10.3389/fmicb.2016.00767
McBeth, J. M., Little, B. J., Ray, R. I., Farrar, K. M., and Emerson, D. (2011). Neutrophilic iron-oxidizing “zetaproteobacteria” and mild steel corrosion in nearshore marine environments. Appl. Environ. Microbiol. 77, 1405–1412. doi: 10.1128/AEM.02095-10
Meng, L., Liu, H., Bao, M., and Sun, P. (2016). Microbial community structure shifts are associated with temperature, dispersants and nutrients in crude oil-contaminated seawaters. Mar. Pollut. Bull. 111, 203–212. doi: 10.1016/j.marpolbul.2016.07.010
Miranda, E., Bethencourt, M., Botana, F. J., Cano, M. J., Sánchez-Amaya, J. M., Corzo, A., et al. (2006). Biocorrosion of carbon steel alloys by an hydrogenotrophic sulfate-reducing bacterium desulfovibrio capillatus, isolated from a mexican oil field separator. Corros. Sci. 48, 2417–2431. doi: 10.1016/j.corsci.2005.09.005
Moradi, M., Song, Z., and Tao, X. (2015a). Introducing a novel bacterium, vibrio neocaledonicus, sp. with the highest corrosion inhibition efficiency. Electrochem. Commun. 51, 64–68. doi: 10.1016/j.elecom.2014.12.007
Moradi, M., Xiao, T., and Song, Z. (2015b). Investigation of corrosion inhibitory process of marine vibrio neocaledonicus, sp. bacterium for carbon steel. Corros. Sci. 100, 186–193. doi: 10.1016/j.corsci.2015.07.030
Moreau, M. M., Eades, S. C., Reinemeyer, C. R., Fugaro, M. N., and Onishi, J. C. (2014). Illumina sequencing of the v4 hypervariable region 16s rrna gene reveals extensive changes in bacterial communities in the cecum following carbohydrate oral infusion and development of early-stage acute laminitis in the horse. Vet. Microbiol. 168, 436–441. doi: 10.1016/j.vetmic.2013.11.017
Mystkowska, J., Ferreira, J. A., Leszczyńska, K., Chmielewska, S., Dbbbabrowski, J. R., Wieciński, P., et al. (2015). Biocorrosion of 316lv steel used in oral cavity due to desulfotomaculum nigrificans bacteria. J. Biomed. Mater. Res. B 105, 222–229. doi: 10.1002/jbm.b.33518
Neria-González, I., Wang, E. T., Ramírez, F., Romero, J. M., and Hernández-Rodríguez, C. (2006). Characterization of bacterial community associated to biofilms of corroded oil pipelines from the southeast of mexico. Anaerobe 12, 122–133. doi: 10.1016/j.anaerobe.2006.02.001
Nikolaev, Y. A., and Plakunov, V. K. (2007). [biofilm–”city of microbes” or an analogue of multicellular organisms?]. Microbiology 76, 149–163. doi: 10.1134/S0026261707020014
Paarup, M., Friedrich, M. W., Tindall, B. J., and Finster, K. (2006). Characterization of the psychrotolerant acetogen strain syra5 and the emended description of the species acetobacterium carbinolicum. Antonie Van Leeuwenhoek 89, 55–69. doi: 10.1007/s10482-005-9009-y
Païssé, S., Ghiglione, J. F., Marty, F., Abbas, B., Gueuné, H., Amaya, J. M. S., et al. (2013). Sulfate-reducing bacteria inhabiting natural corrosion deposits from marine steel structures. Appl. Microbiol. Biotechnol. 97, 7493–7504. doi: 10.1007/s00253-012-4464-7
Qi, Y., Ying, W., Zhu, Z., Wang, X., Li, Z., and Jing, Z. (2016). Bacterial diversity in the surface sediments of the hypoxic zone near the changjiang estuary and in the east china sea. Microbiologyopen 5, 323–339. doi: 10.1002/mbo3.330
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The silva ribosomal rna gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
R Development Core Team (2013). R: A Language and Environment for Statistical Computing. Vienna: The R Foundation for Statistical Computing.
Rao, T. S. (2010). Comparative effect of temperature on biofilm formation in natural and modified marine environment. Aquat. Ecol. 44, 463–478. doi: 10.1007/s10452-009-9304-1
Ren, D., and Wood, T. K. (2004). (5z)-4-bromo-5-(bromomethylene)-3-butyl-2(5h)-furanone reduces corrosion from desulfotomaculum orientis. Environ. Microbiol. 6, 535–540. doi: 10.1111/j.1462-2920.2004.00587.x
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Slightom, R. N., and Buchan, A. (2009). Surface colonization by marine roseobacters: integrating genotype and phenotype. Appl. Environ. Microbiol. 75, 6027–6037. doi: 10.1128/AEM.01508-09
Stoodley, P., Sauer, K., Davies, D. G., and Costerton, J. W. (2002). Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56, 187–209. doi: 10.1146/annurev.micro.56.012302.160705
Suh, S. S., Park, M., Hwang, J., Kil, E. J., Jung, S. W., Lee, S., et al. (2015). Seasonal dynamics of marine microbial community in the south sea of korea. PLOS ONE 10:e0131633. doi: 10.1371/journal.pone.0131633
Sun, H., Shi, B., Lytle, D. A., Bai, Y., and Wang, D. (2014). Formation and release behavior of iron corrosion products under the influence of bacterial communities in a simulated water distribution system. Environ. Sci. Proces. Impacts 16, 576–585. doi: 10.1039/c3em00544e
Sun, J., Zhang, Q., Zhou, J., and Wei, Q. (2014). Illumina amplicon sequencing of 16s rrna tag reveals bacterial community development in the rhizosphere of apple nurseries at a replant disease site and a new planting site. PLOS ONE 9:e111744. doi: 10.1371/journal.pone.0111744
Tian, H., Gao, P., Chen, Z., Li, Y., Li, Y., Wang, Y., et al. (2017). Compositions and abundances of sulfate-reducing and sulfur-oxidizing microorganisms in water-flooded petroleum reservoirs with different temperatures in china. Front. Microbiol. 8:143. doi: 10.3389/fmicb.2017.00143
Videla, H. A., and Herrera, L. K. (2005). Microbiologically influenced corrosion: looking to the future. Int. Microbiol. 8, 169–180.
Vigneron, A., Alsop, E. B., Chambers, B., Lomans, B. P., Head, I. M., and Tsesmetzis, N. (2016). Complementary microorganisms in highly corrosive biofilms from an offshore oil production facility. Appl. Environ. Microbiol. 82, 2545–2554. doi: 10.1128/AEM.03842-15
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naïve bayesian classifier for rapid assignment of rrna sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Widdel, F. (1988). “Microbiology and ecology of sulfate- and sulfur-reducing bacteria,” in Biology of Anaerobic Microorganisms, ed. A. J. B. Zehnder (New York, NY: John Wiley & Sons), 469–585.
Wikieł, A. J., Datsenko, I., Vera, M., and Sand, W. (2014). Impact of desulfovibrio alaskensis biofilms on corrosion behaviour of carbon steel in marine environment. Bioelectrochemistry 97, 52–60. doi: 10.1016/j.bioelechem.2013.09.008
Yang, C., Yi, L., Zhou, B., Zhou, Y., Wei, Z., Yun, T., et al. (2015). Illumina sequencing-based analysis of free-living bacterial community dynamics during an akashiwo sanguine bloom in xiamen sea, china. Sci. Rep 5:8476. doi: 10.1038/srep08476
Zhang, P., Xu, D., Li, Y., Yang, K., and Gu, T. (2015). Electron mediators accelerate the microbiologically influenced corrosion of 304 stainless steel by the desulfovibrio vulgaris biofilm. Bioelectrochemistry 101, 14–21. doi: 10.1016/j.bioelechem.2014.06.010
Zhang, T., and Fang, H. H. (2001). Phylogenetic diversity of a srb-rich marine biofilm. Appl. Microbiol. Biotechnol. 57, 437–440. doi: 10.1007/s002530100770
Zhang, T., Fang, H. H., and Ko, B. C. (2003). Methanogen population in a marine biofilm corrosive to mild steel. Appl. Microbiol. Biotechnol. 63, 101–106. doi: 10.1007/s00253-003-1396-2
Zhang, Y., Pei, G., Chen, L., and Zhang, W. (2016). Metabolic dynamics of desulfovibrio vulgaris biofilm grown on a steel surface. Biofouling 32, 725–736. doi: 10.1080/08927014.2016.1193166
Zhou, J., Bruns, M. A., and Tiedje, J. M. (1996). Dna recovery from soils of diverse composition. Appl. Environ. Microbiol. 62, 316–322.
Zhu, X. Y., Lubeck, J., and Nd, K. J. (2003). Characterization of microbial communities in gas industry pipelines. Appl. Environ. Microbiol. 69, 5354–5363. doi: 10.1128/AEM.69.9.5354-5363.2003
Keywords: bacterial community, MIC, carbon steel, Illumina MiSeq sequencing, 16S rRNA gene
Citation: Li X, Duan J, Xiao H, Li Y, Liu H, Guan F and Zhai X (2017) Analysis of Bacterial Community Composition of Corroded Steel Immersed in Sanya and Xiamen Seawaters in China via Method of Illumina MiSeq Sequencing. Front. Microbiol. 8:1737. doi: 10.3389/fmicb.2017.01737
Received: 22 May 2017; Accepted: 25 August 2017;
Published: 12 September 2017.
Edited by:
Hongyue Dang, Xiamen University, ChinaReviewed by:
Filomena De Leo, Università degli Studi di Messina, ItalyMalin Bomberg, VTT Technical Research Centre of Finland, Finland
Copyright © 2017 Li, Duan, Xiao, Li, Liu, Guan and Zhai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jizhou Duan, jizhouduan@163.com Hui Xiao, xiaoh28@163.com
 Xiaohong Li
Xiaohong Li Jizhou Duan1*
Jizhou Duan1* Fang Guan
Fang Guan