- 1College of Life Sciences, Fujian Agriculture and Forestry University, Fuzhou, China
- 2Key Laboratory of Crop Ecology and Molecular Physiology, Fujian Agriculture and Forestry University, Fuzhou, China
- 3College of Crop Science, Fujian Agriculture and Forestry University, Fuzhou, China
- 4Fujian Provincial Key Laboratory of Agroecological Processing and Safety Monitoring, Fujian Agriculture and Forestry University, Fuzhou, China
Radix pseudostellariae is a perennial tonic medicinal plant, with high medicinal value. However, consecutive monoculture of this plant in the same field results in serious decrease in both yield and quality. In this study, a 3-year field experiment was performed to identify the inhibitory effect of growth caused by prolonged monoculture of R. pseudostellariae. DGGE analysis was used to explore the shifts in the structure and diversity of soil Fusarium and Pseudomonas communities along a 3-year gradient of monoculture. The results demonstrated that extended monoculture significantly boosted the diversity of Fusarium spp., but declined Pseudomonas spp. diversity. Quantitative PCR analysis showed a significant increase in Fusarium oxysporum, but a decline in Pseudomonas spp. Furthermore, abundance of antagonistic Pseudomonas spp. possessing antagonistic ability toward F. oxysporum significantly decreased in consecutively monocultured soils. Phenolic acid mixture at the same ratio as detected in soil could boost mycelial and sporular growth of pathogenic F. oxysporum while inhibit the growth of antagonistic Pseudomonas sp. CJ313. Moreover, plant bioassays showed that Pseudomonas sp. CJ313 had a good performance that protected R. pseudostellariae from infection by F. oxysporum. In conclusion, this study demonstrated that extended monoculture of R. pseudostellariae could alter the Fusarium and Pseudomonas communities in the plant rhizosphere, leading to relatively low level of antagonistic microorganisms, but with relatively high level of pathogenic microorganisms.
Introduction
As much as 70% of medicinal plants suffer from consecutive monoculture problem, also known as replant disease or soil sickness. These problems are commonly observed in the production of many Chinese medicinal herbs, including Radix pseudostellariae, Rehmannia glutinosa, Panax notoginseng, etc (Zhang and Lin, 2009). R. pseudostellariae, a perennial tonic medicinal plant, belongs to the family Caryophyllaceae with extremely high medicinal value (Zhao W.O. et al., 2015). Consecutive monoculture of this plant in the same field leads to a serious decrease in both quality and yield of roots along with poor plant performance, which severely limited production and utilization of its medicinal plant virtues (Lin et al., 2015). Therefore, it is necessary to explore the mechanism of consecutive monoculture problems affecting the plant and develop effective control strategies for R. pseudostellariae.
Fusarium species is one of the most abundant, prevalent, and important soil fungi (Damicone and Manning, 1985). It is notorious due to the ability of attacking diversity of host plants and bring upon them diseases like vascular wilts, seedling damping off and rots of stem (Pietro et al., 2003; Punja and Parker, 2009; Chakravarty and Hwang, 2010). Similarly, the soil-borne disease caused by F. oxysporum in R. pseudostellariae fields were reported (Zhao Y.P. et al., 2015), however, the other Fusarium species are often overlooked. Therefore, in order to develop the full potential of the disease-suppressive microbial community in the biological control, we need more information to unravel the different roles of this potentially important species.
In recent years, more attentions were paid to develop the environment friendly and good agriculture practices for disease control. It has become important to explore the nature of microbial diversities in the soil, particularly Pseudomonas in different cropping periods or regime (Mendes et al., 2011). Pseudomonas species were reported to have a wide range of functional groups, such as plant pathogens (Samson et al., 1998), xenobiotic degraders (Clausen et al., 2002) and plant growth promoters (Patten and Glick, 2002). In addition, Pseudomonas species can be used as biological control agents for soil-borne pathogens, including black rot of tobacco, disease of wheat and Fusarium wilt (Raaijmakers and Weller, 1998; Patten and Glick, 2002; Mendes et al., 2011).
Recently, the increasing evidences suggest that plant–microbial interactions play many pivotal roles in soil quality and plant health (Lakshmanan et al., 2014; Macdonald and Singh, 2014). Li et al. (2014) reported that the peanut root exudates can selectively inhibit certain communal bacteria, such as Gelria glutamica, Mitsuaria chitosanitabida, and Burkholderia, but stimulate the bacterial taxon of Desulfotomaculum ruminis and the fungal taxa F. oxysporum in soil. Wu et al. (2016c) found that the amount of two pathogenic fungi (F. oxysporum and Aspergillus flavus) in the rhizosphere significantly increased after Rehmannia glutinosa monoculture. Wu et al. (2015) indicated that long-term continuous cropping of black pepper (Piper nigrum L.) could lead to a significant decrease in soil bacterial content, especially the Pseudomonas spp., suggesting that the soil microbes might be responsible for soil health.
Denaturing gradient gel electrophoresis (DGGE) is considered as an effective technique to directly analyze the structural and diversity of microbial communities (Kozdrój and van Elsas, 2001). The traditional method of assessing the diversity of Fusarium is based on enumeration and isolation of strains which were grown on selective media (Vujanovic et al., 2002). However, morphological identification of Fusarium species is a time-consuming and formidable task. Yergeau et al. (2005) described a PCR-DGGE method to detect the presence of multiple Fusarium spp. from environmental samples. The method is based on the specific amplification and separation of the transcription elongation factor-1α (Ef1α) gene. Similarly, Widmer et al. (1998) designed a primer set (PsR and PsF) which was based on the 16S rDNA gene of Pseudomonas spp. in 1998. When combining the PsR and PsF primers, Evans et al. (2004) developed a semi-nested PCR and DGGE to rapidly study the diversity within the genus Pseudomonas. Therefore, the role of soil microbial ecology in the prevention and control of plant diseases has been given more attention (Philippot et al., 2013; Cha et al., 2015). However, few studies have been carried out to understand the relationship between Pseudomonas and Fusarium of R. pseudostellariae, and the approaches to overcome diseases associated with this plant.
In this study, DGGE combined with qPCR technique was used to analyze the shifts of Pseudomonas and Fusarium communities in rhizosphere soil under R. pseudostellariae monoculture. Several microorganisms closely related to the problem of prolonged monoculture were isolated and performed for plant-microbe interactions study. Our study can help to illustrate the effects of ecological environment and root exudates on the selection of soil microbes in rhizosphere soil, and provide useful information on potential indigenous microflora for soil remediation and improvement.
Materials and Methods
Field Experiment
In this study, the R. pseudostellariae cultivar ‘Zheshen 2’ was used as the test material. The experiment was carried out at the experimental station of Fuding City, Fujian Province (27°26′ N, 120°04′ E). The experimental field which previously planted Oryza sativa was performed for this study with four treatments: (1) control with no R. pseudostellariae cultivation (CK), (2) the newly planted R. pseudostellariae cultivation (FP), (3) 2-year consecutive monoculture (SP), (4) 3-year consecutive monoculture (TP). The physical and chemical properties of the soil were detected before the experiment was initiated: total nitrogen of 1.83g kg-1, available nitrogen of 26.23 mg kg-1, total phosphorus of 0.47 g kg-1, and available phosphorus of 96.34 mg kg-1, total K of 8.46 g kg-1, and available K of 365.21 mg kg-1. The station has a subtropical oceanic monsoon climate, annual mean temperature at 18.4°C. All treatments were treated with the same fertilization and field management during the experiment.
Soil Sampling and DNA Extraction
The above ground or below ground biomass of R. pseudostellariae become significantly different after 5 months of planting (Figure 1A), according to our previously study (Wu et al., 2016b). Therefore, soil samples were randomly collected from five different points at each field on April 22nd, 2015. Additionally, we harvested the plants for yield determination on July 2nd, 2015 (Figure 1B).
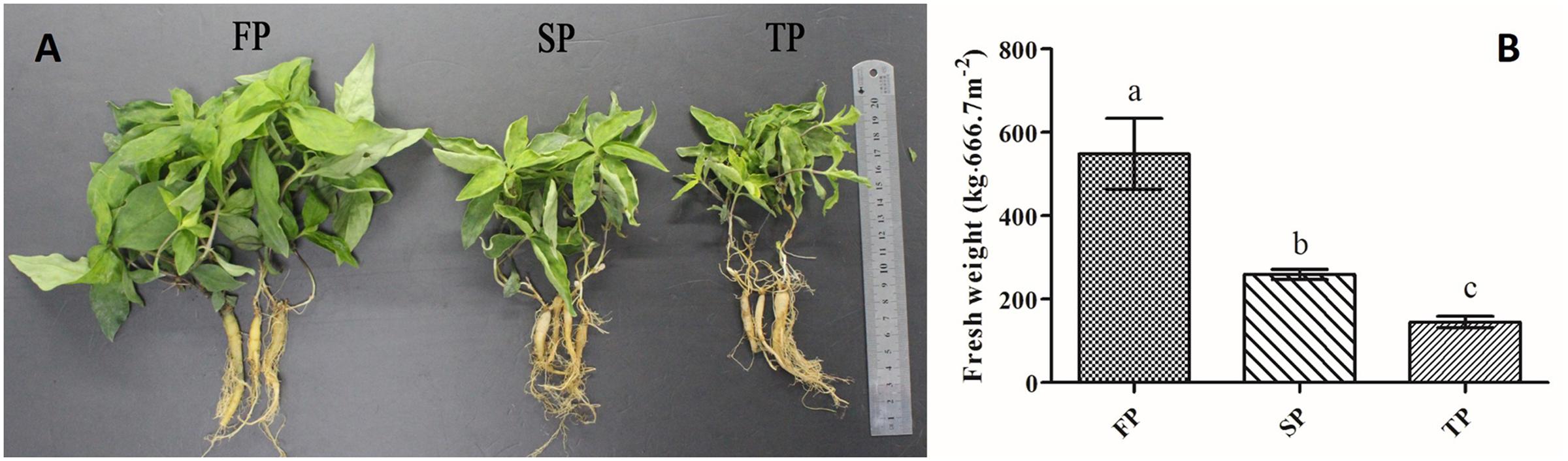
FIGURE 1. (A) Photographs of above and below ground components of R. pseudostellariae under 1-year, 2-year, and 3-year consecutive monoculture. (B) Yield of R. pseudostellariae under 1-year (FP), 2-year (SP) and 3-year consecutive monoculture (TP).
Soil samples were collected after digging the plant samples. Firstly, the loosely adhering soil was shaken off, then scraping the soil that was still attached to the root as rhizosphere soil. DNA was immediately extracted from 0.5 g soil sample per treatment using Biofast Soil Genomic DNA Extraction Kit (BioFlux, Hangzhou, China) according to the manufacturer’s protocols. We further determined the DNA concentration using Nanodrop 2000C Spectrophotometer (Thermo Scientific, United States) and then diluted it to 20 ng μL-1.
PCR-DGGE and Analysis
Fusarium-specific PCR was performed according to the nested amplification of the Ef1α gene. The first round of PCR reactions was performed by the Ef-1 and Ef-2 primers (O’Donnell et al., 1998). It was carried out in 50 μl volumes containing 25 μl of 2 × EasyTaq PCR SuperMix (Transgen Biotech, Beijing, China), 1 μl of each primer and 40 ng template soil DNA. The program of PCR was performed by the following protocol: 95°C for 5 min, 30 cycles of denaturation (95°C for 1 min), annealing (55°C for 1 min), extension (72°C for 1 min), and 1 cycle of final extension (72°C for 10 min). The amplicons were subsequently diluted (1:20) and used for the second PCR reaction via Alfie1-GC and Alfie2 (Yergeau et al., 2005) primers. Second round PCR protocol was similar to the method of the first reaction, except for the annealing (57°C for 50 s) and extension (72°C for 50 s).
Pseudomonas-specific PCR was based on the nested amplification of the V6/V7 region of Pseudomonas spp. The first round of PCR reactions was used the PsF and PsR primers (Tan and Ji, 2010). PCR reaction was carried out in 50 μl volumes containing 25 μl of 2× EasyTaq PCR SuperMix (Transgen Biotech, Beijing, China), 1 μl of each primer and 20 ng template soil DNA. The program of PCR was performed by the following protocol: 95°C for 5 min, 30 cycles of denaturation (95°C for 1 min), annealing (64°C for 1 min), extension (72°C for 1 min), and 1 cycle of final extension (72°C for 10 min). The PCR products of first round was used to perform the second PCR reaction and the primers F968-GC1 and PsR were used (Garbeva et al., 2004). The following cycling protocol was performed for the second PCR: 1 cycle of initial denaturation at 94°C for 5 min, 10 cycle of denaturation (94°C for1 min), 1 min at 60°C (every subsequent one using a 0.5°C lower annealing temperature), and 2 min at 72°C, 1 cycle of 95°C for 5 min, 30 cycles of denaturation (95°C for 1 min), annealing (55°C for 1 min), and 1 cycle of final extension (72°C for 10 min). All PCR products were detected using 1.2% agarose gel and purified using a Gel Extraction Kit (OMEGA Bio-Tek, United States) according to the manufacturer’s instructions. The purified PCR products were used to perform DGGE experiments.
DGGE Analysis
We performed DGGE by using an 8% (w/v) polyacrylamide gel with 35–55% and 45–60% denaturant gradients for Fusarium-specific and Pseudomonas-specific communities, respectively, using the Junyi JY-TD331A system (JUNYI, Beijing, China). DGGE was carried out at 80 V and 60°C for 12 h and 15 h in 1x TAE buffer. After electrophoresis, gels were stained with silver stain. For analysis of the molecular community profiles, gels were digitized by using the Quantity One 4.0 software (BioRad). When bands were identified, they were excised from the DGGE gel by using a sterile scalpel. After incubation overnight at 4°C, DNA was eluted from the gel. The amplicons were amplified by using the Alfie1-GC/Alfie2 and F968-GC/Psr primer sets (as mentioned before). PCR amplicons were cloned into the pEASY-T1 Cloning vector (Transgen Biotech, Beijing, China) by using manufacturer’s instructions. Sequences were compared to the sequences on GenBank of NCBI using the BlastN search method.
Quantitative PCR for Fusarium oxysporum and Pseudomonas spp.
The fragments of F. oxysporum and Pseudomonas were cloned into the pEASY-T1 Cloning vector (TransGen Biotech Co., Beijing, China). Two plasmids were purified as described above. After determining DNA concentration, it was immediately diluted into 2, 1, 0.5, 0.1, 0.05, 0.01, 0.005, and 0.001 ng ml-1. The reaction of standard curve was performed following the qPCR amplification protocol as described in Supplementary Table S1. In addition, the standard curve was generated by log10 value against the threshold cycle (Ct) value.
We further performed real-time PCR quantifications of F. oxysporum (primer sets ITS1F and AFP308R) and Pseudomonas (primer sets PsF and PsR) in four soil samples, and amplification protocol as described in Supplementary Table S1. Reaction of qPCR was performed in 15 μl mixture, containing 7.5 μl TransStart Green qPCR SuperMix (Transgen Biotech, Beijing, China), 0.6 μl of each primer (10 μ M) and 20 ng DNA.
Isolation of Fusarium spp.
For isolation of Fusarium spp., potato dextrose agar (PDA) was used to isolate and subculture the fungus. Soil suspensions were prepared by adding 10 g of fresh soil in a flask containing 90 ml of sterile water (10-1 g l-1), 100 μl soil suspensions were plated onto PDA. Plates were incubated at 30°C for 18 h, and then each single colony was isolated and purified. Fusarium genomic DNA extraction was done by using CTAB-based method as described by Rogers and Bendich (1985). The primer sets ITS1F and ITS4 (Supplementary Table S2) were used for ITS amplification. PCR amplicons were sent to Shanghai BoShang for sequencing. We further used BlastN search method to compare sequences to the GenBank database.
Isolation and Counting of Pseudomonas spp. with Antagonistic Activity toward Fusarium oxysporum
For isolation of Pseudomonas spp., Pseudomonas selective isolation agar (PSIA) (Krueger and Sheikh, 1987) was used. As described above, each soil suspensions was prepared (10-1 g l-1), after serial dilution, 60 μl soil suspensions (10-3 g l-1) were plated onto PSIA, incubated at 30°C for 30 h, and then each single colony was purified. Results were descripted as the numbers of CFU per g-1 (dry weight) soil.
For in vitro antagonism assays, we inoculated F. oxysporum to the center of the PDA plates and Pseudomonas isolates to the side of the plates at the same time. The results of antagonistic activity against F. oxysporum were recorded after 5 days of incubation at 30°C.
After incubation, we selected Pseudomonas isolates that had antagonistic activity against F. oxysporum for DNA extraction. Pseudomonas genomic DNA was extracted using the Bacteria Genomic DNA kit (CWbiotech, Beijing, China). The primer sets 27F and 1522R were used for 16S rRNA amplification. The thermal conditions are listed in the Supplementary Table S2. PCR amplicons were sent to Shanghai BoShang for sequencing. Finally, we used BlastN search method to compare sequences to the GenBank database for the identification purpose. Sequences were used Clustal X to align, and then phylogenetic trees were constructed with MEGA6.06 using a neighbor joining approach.
Evaluation of the Pathogenicity of Fusarium oxysporum and Biocontrol Effects of Pseudomonas sp. CJ313
Radix pseudostellariae were planted in plastic pots and placed in a green house on December 15, 2015. The spore suspension of isolated F. oxysporum was added to the soil through pipette for observing the effects of Fusarium wilt in R. pseudostellariae after 5 months of planting. In order to assess biocontrol potential of Pseudomonas spp., the effect of isolated strain CJ313 was examined after 15 days of its exogenous addition. We added equal amount of LB as a control (CK) at the same time. Each treatment has three replicates. After 16 days, we collected rhizospheric soil from two treatments, then soil samples were immediately used to extract DNA and qPCR of F. oxysporum and Pseudomonas spp. as described above.
The Effect of Phenolic Acids on the Growth of Isolated Fusarium oxysporum, Pseudomonas sp. CJ313 and Pseudomonas sp. CJ361
Based on our previous HPLC results of phenolic acids in the R. pseudostellariae rhizosphere (Wu et al., 2016a), we prepared the solutions of eight phenolic acids (p-hydroxybenzoic acid, gallic acid, coumaric acid, syringic acid, vanillic acid, ferulic acid, vanillin and benzoic acid) and their mixture to assess its effect on the growth of isolated F. oxysporum. The ratio of their mixtures was the same as detected in the soil. We prepared the 10-fold dilution of soil extract agar medium (SEM), and added the phenolic acids into the SEM to reach final concentrations 30, 60, 120, 240, 480, 960 μ mol L-1. We inoculated isolated F. oxysporum onto the SEM plates to assess the mycelium growth mediated by phenolic acids. There were three replicates for each treatment. After incubation at 28°C for 8 days, we recorded the mycelium diameter. Likewise, isolated F. oxysporum was inoculated into 10-fold dilution of SEM by adding the phenolic acid mixtures, and solution was incubated at 200 rpm and 30°C for 7 days. F. oxysporum spores were counted by a hemocytometer.
We also detected the effects of eight phenolic acids and their mixtures on the growth of isolated Pseudomonas sp. CJ313 and CJ361. Specifically, the isolated Pseudomonas sp. CJ313 and CJ361 were determined by adding the phenolic acids to a LB medium with 8-fold dilution. After 8–10 h incubation at 200 rpm and 30°C, we determined the bacterial density at 600 nm using a microplate reader (Thermo Scientific Multiskan MK3, Shanghai, China).
Statistical Analyses
For all parameters, multiple comparison was carried out by one-way analysis of variance (ANOVA) followed by LSD’s test (P ≤ 0.05) using DPS 7.05 software. PCA analysis was performed by SPSS 20.0 software. DGGE for detecting the band was performed with the Quantity one v4.6.2 software.
Results
The Morphology and Yield of R. pseudostellariae under Consecutive Monoculture
We observed that plants of FP displayed more aboveground biomass and less adventitious roots relative to continuously monocultured plants of SP and TP (Figure 1A). Moreover, our results revealed that the yield of newly planted R. pseudostellariae roots (FP) was significantly (P ≤ 0.05) higher that of 2-year consecutive monoculture (SP) and 3-year consecutive monoculture (TP) (Figure 1B).
Fusarium-Specific DGGE
Fusarium-specific PCR-DGGE analysis showed that the rhizosphere Fusarium community structures changed with the increasing years of monoculture (Figure 2). The principal component analysis (PCA) of the DGGE profile was performed to demonstrate the relative position of four soil samples. In PCA, first principal component explained 57.70% of variance and second principal component 28.44% of total variance (Figure 3A). Furthermore, PCA showed that the Fusarium community in CK, FP and SP was separated from TP by the first principal component, and FP was separated from SP by the second principal component (Figure 3A).
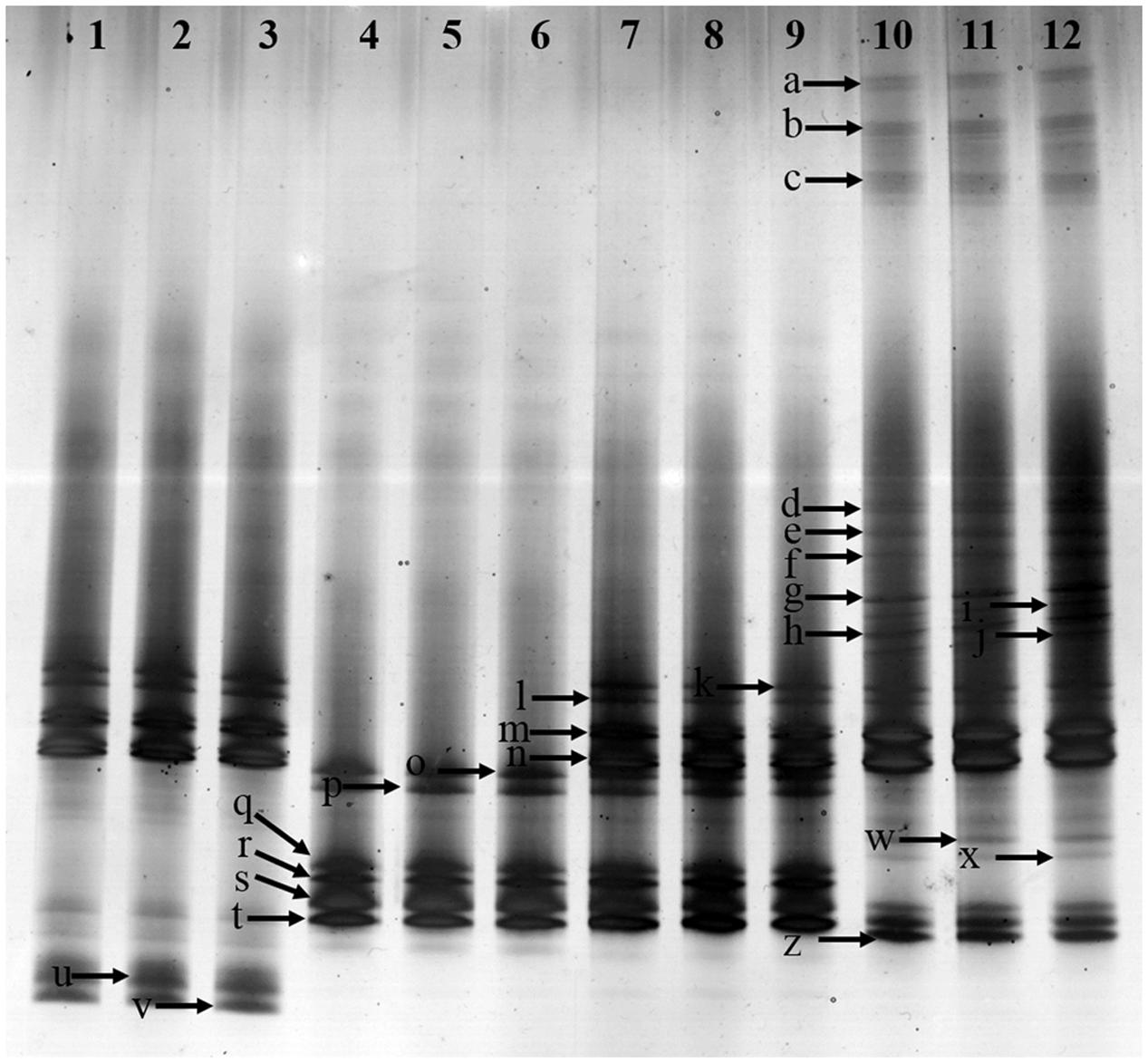
FIGURE 2. Denaturing gradient gel electrophoresis (DGGE) gel showing community structures of Fusarium elongation factor genotypes (EF1α) amplified from soil extracted DNA. Lanes 1, 2 and 3-control with no Radix pseudostellariae cultivation (CK), lanes 4, 5 and 6- the newly planted R. pseudostellariae cultivation(FP), lanes 7, 8, and 9- 2-year consecutive monoculture(SP), lanes 10, 11, and 12- 3-year consecutive monoculture (TP). Band positions are marked with an arrow and correspond to data in Table 2.
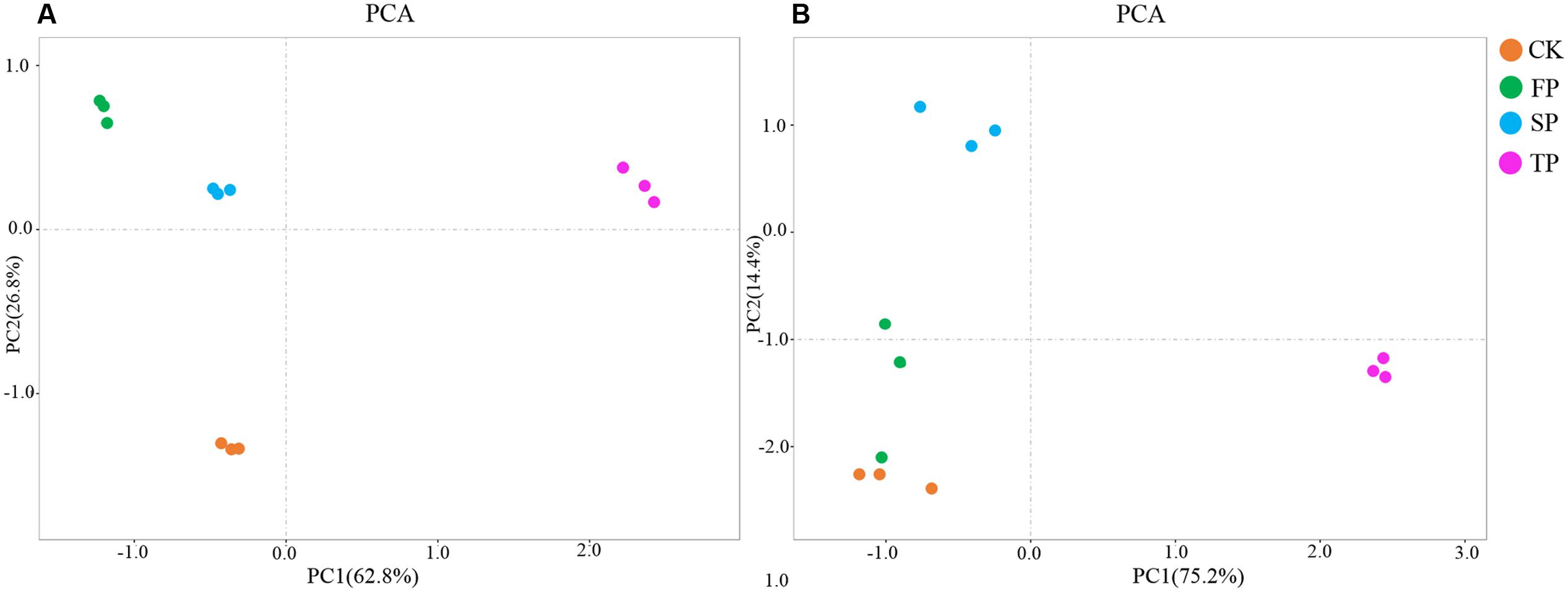
FIGURE 3. Principal component analysis of Fusarium DGGE results (A) Pseudomonas DGGE results (B). CK, control with no R. pseudostellariae cultivation. FP, the newly planted Radix. pseudostellariae cultivation. SP, 2-year consecutive monoculture. TP, 3-year consecutive monoculture.
The diversity of Fusarium-specific DGGE was also determined. The study revealed that Simpson, Shannon, evenness and Brillouin’s index of Fusarium communities significantly increased with prolonged or increasing years of monoculture (P ≤ 0.05) (Table 1).
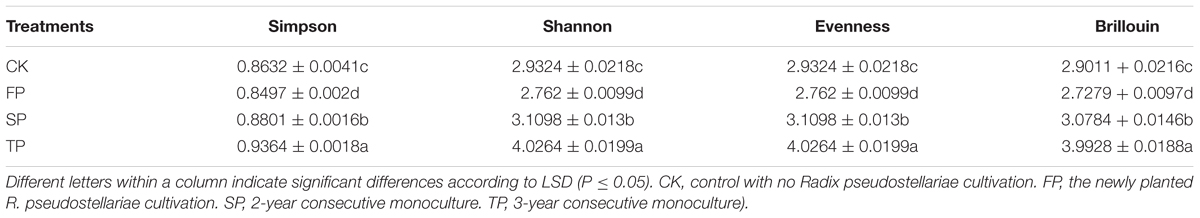
TABLE 1. Estimated Simpson, Shannon, Evenness and Brillouin’s indices for all the samples using Fusarium-specific DGGE.
Analysis of the DGGE Bands of Fusarium spp.
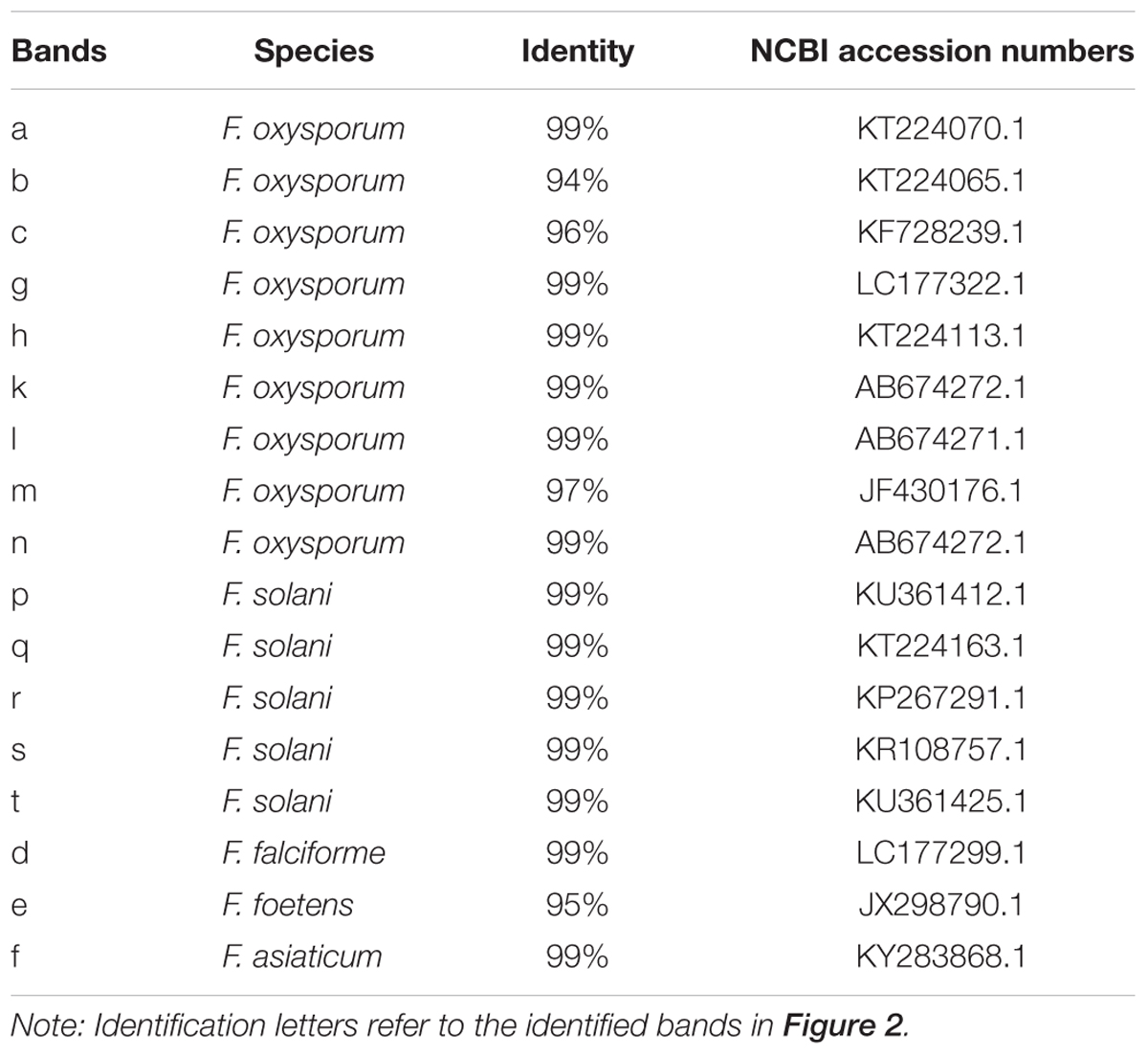
In order to further extract more detailed information from the DGGE bands in this study, excised bands from DGGE were sequenced. A total of 17 bands were identified in rhizospheric soil (Table 2). The Fusarium spp. belonged to 5 species, e.g., F. oxysproum (band a, b, c, g, h, k, l, m and n), F. solani (band p, q, r, s and t), F. asiaticum (band d), F. falciforme (band e), F. foetens (band f). Specifically, the bands of F. oxysporum significantly increased along with years of continuous cropping.
Pseudomonas-Specific DGGE
Pseudomonas-specific PCR-DGGE analyses showed significantly changed Pseudomonas community structures in the rhizosphere with increasing years of monoculture (Figure 4). Likewise, we performed PCA to demonstrate the relative position of four soil samples. In PCA, the first principal component explained 83.30% of variance and second principal component 9.0% of total variance (Figure 3B). Furthermore, PCA showed the Pseudomonas community in CK, FP and SP were separated from the microbial community in TP by principal component 1, and the community in FP and CK was separated from the microbial communities in SP and TP by principle component 2 (Figure 3B).
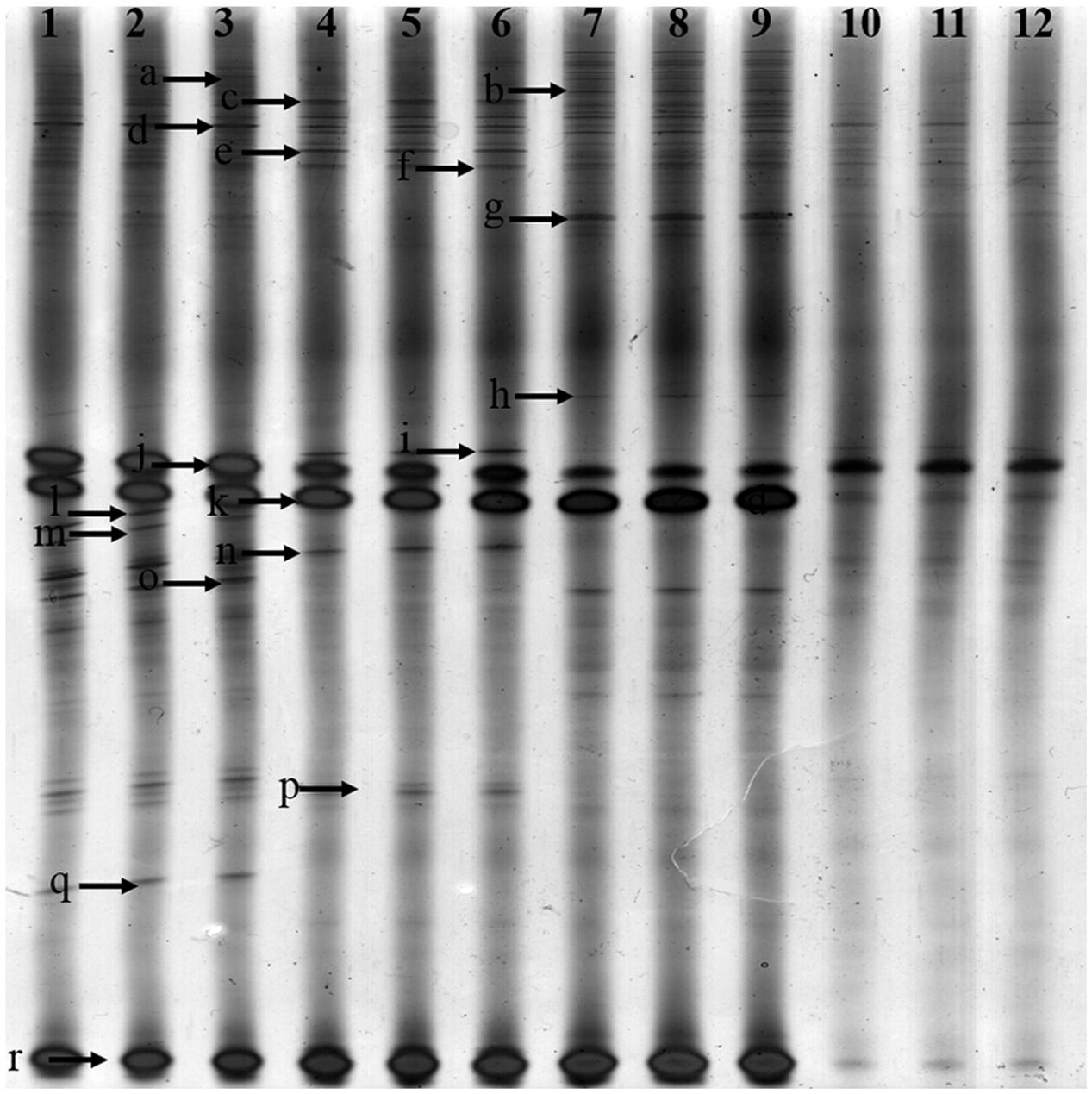
FIGURE 4. Denaturing gradient gel electrophoresis gel showing community structures of Pseudomonas amplified from soil extracted DNA. Lanes 1, 2, and 3-control with no Radix pseudostellariae cultivation (CK), lanes 4, 5, and 6- the newly planted R. pseudostellariae cultivation(FP), lanes 7, 8, and 9- 2-year consecutive monoculture(SP), lanes 10, 11, and 12- 3-year consecutive monoculture(TP). Band positions are marked with an arrow and correspond to data in Table 4.
The diversity of visible bands, Shannon and Brillouin’ index of Pseudomonas community significantly decreased with increasing years of monoculture (P ≤ 0.05). However, the opposite was true for the Simpson’ index of the Pseudomonas community. There was no significant difference in evenness index among the four samples (Table 3).
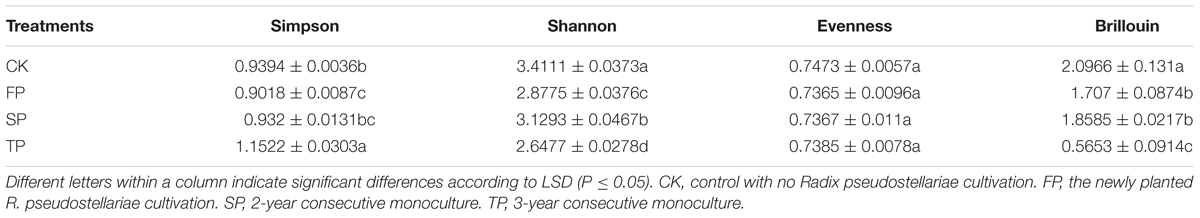
TABLE 3. Estimated Simpson, Shannon, Evenness and Brillouin’s indices for all the samples using Pseudomonas-specific DGGE.
Analysis of the DGGE Bands of Pseudomonas spp.
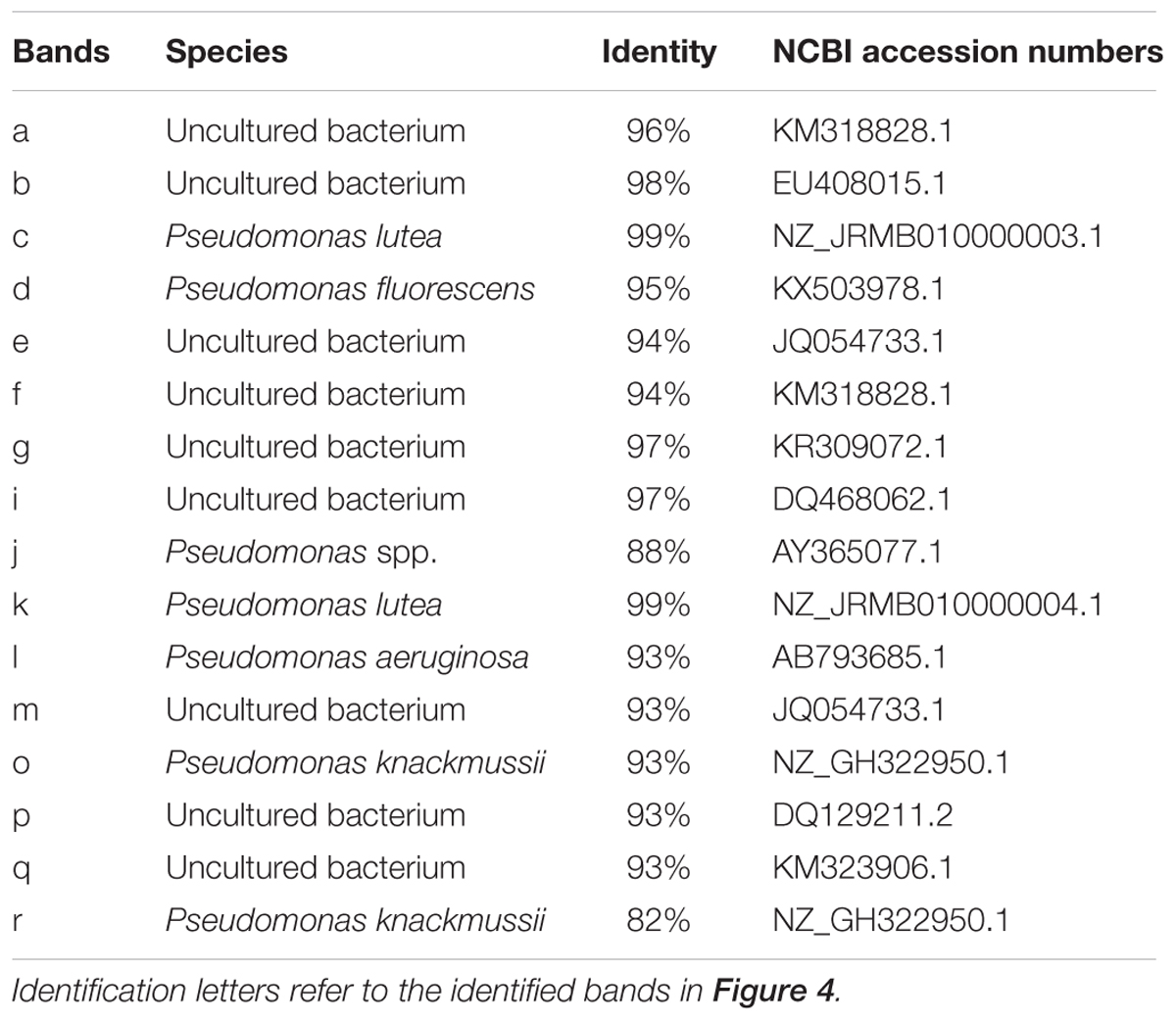
To further extract more detailed information from the DGGE bands, we excised and sequenced bands from DGGE. A total of 15 bands were identified in rhizospheric soil (Table 4). Pseudomonas spp. could be further divided into five species, e.g., Pseudomonas lutea (band c and k), Pseudomonas fluorescens (band d), Pseudomonas aeruginosa (band l), Pseudomonas knackmussii (band o and r), Pseudomonas sp. (band j) and uncultured bacterium (a, b, f, g, i, m, p and q).
Abundance of Pseudomonas and Fusarium oxysporum by Quantitative PCR
First, standard curves of y = -0.2487x + 9.898 (R2 = 0.997) and y = -0.271x + 9.8309 (R2 = 0.990) were developed for Pseudomonas and F. oxysporum qPCR analyses respectively. The amount of F. oxysporum was significantly (P ≤ 0.05) higher in continuous monoculture soils (SP and TP) than in control (CK) and the newly planted soils (FP) (Figure 5A). The result of qPCR was consistent with the Fusarium-specific DGGE results (Table 1). However, the opposite was true for the qPCR result of Pseudomonas (Figure 5B).
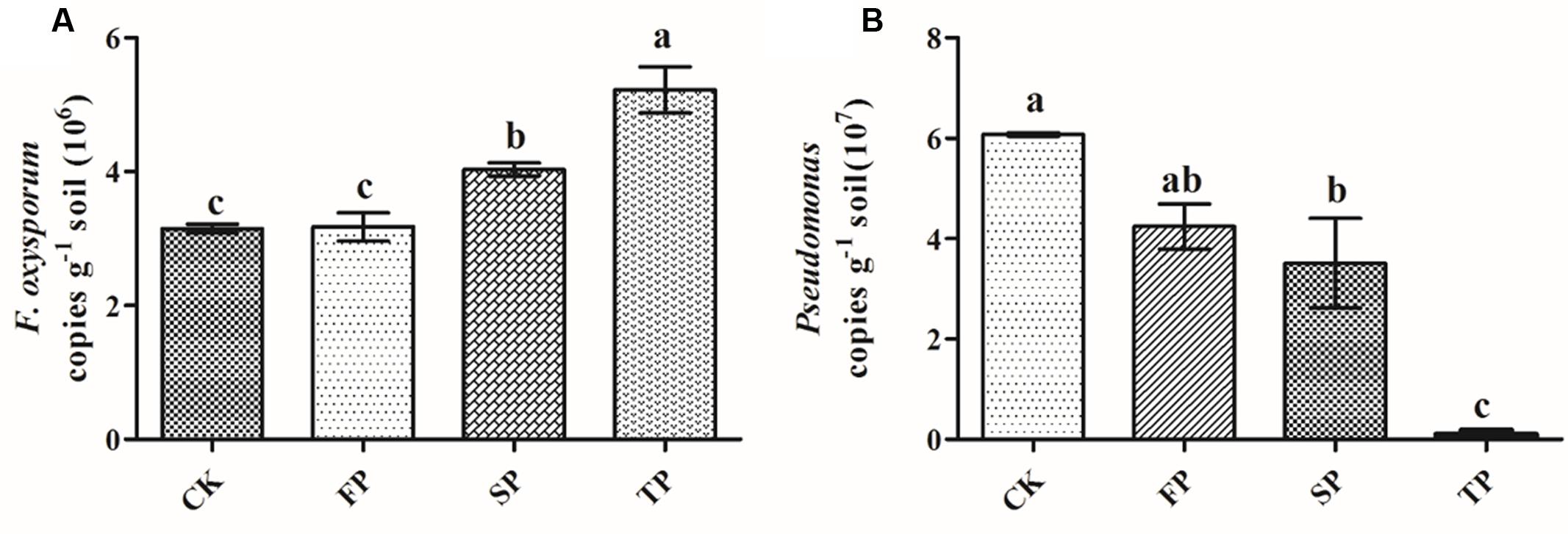
FIGURE 5. Quantification of Fusarium oxysporum (A) and Pseudomonas (B) in control (CK), the newly planted Radix pseudostellariae cultivation (FP), 2-year consecutive monoculture (SP), and 3-year consecutive monoculture (TP) by quantitative PCR. Data are means ± standard errors (one-way analysis of variance, n = 4).
Isolation and Screening for F. oxysporum with High Pathogenicity
In our study, we separated and sequenced one strain of F. oxysporum. We found that the isolated F. oxysporum quickly led to wilt disease on the tissue culture of R. pseudostellariae (Figure 7A), and it also occurred in pots with F. oxysporum (Figure 7B). These results demonstrated that isolated F. oxysporum had the high pathogenicity on R. pseudostellariae.
Screening for Pseudomonas Isolates with Antagonistic Activity toward F. oxysporum
For in vitro antagonism assays, we screened a total of 317 Pseudomonas isolates from four different soils. The results showed that the isolation frequencies of Pseudomonas were significantly higher in FP than SP and TP. The highest isolation frequencies were found in the newly planted (FP) soil (Figure 6A). In vitro antagonism assays, the number of Pseudomonas spp. with antagonistic activity toward F. oxysporum significantly declined with prolonged monoculture (Figure 6B). These isolation frequencies were similar to results of Pseudomonas obtained by qPCR. Approximately 17.4% (87 of 317) of all isolates showed the antagonistic activity. Strain 313 and 361 but not 117 had antagonistic activity against F. oxysporum (Figure 6C). The sequences of Pseudomonas sp. CJ313 and Pseudomonas sp. CJ361 isolates were obtained to perform phylogenetic tree analysis. The neighbor-joining method generated a dendrogram with two main branches, where the first branch included Pseudomonas sp. CJ361 and the second branch comprised Pseudomonas sp. CJ313 (Supplementary Figure S1).
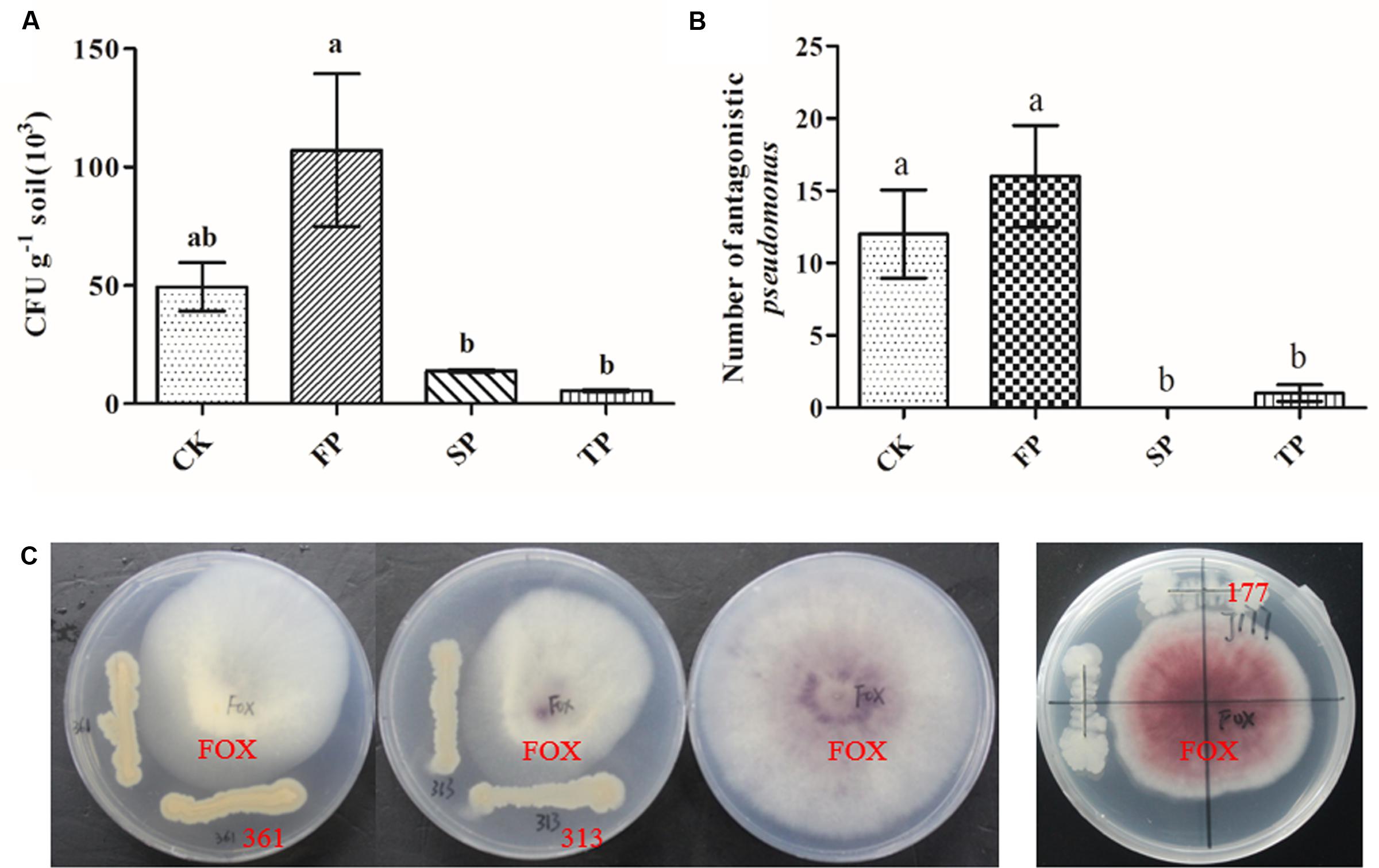
FIGURE 6. Pseudomonas populations (CFU g-1 of dry soil) in rhizosphere soils under four different samples (A), Number of Pseudomonas spp. with antagonistic activity against F. oxysporum in four different samples (B). Petri plates used for evaluation of antagonistic activity of Pseudomonas strains 313, 361 and 117 against F. oxysporum (C). CK, FP, SP and TP represent the control, newly planted, 2-year, and 3-year consecutively monoculture soils, respectively. Data are means ± standard errors (one-way analysis of variance, n = 3).
Biocontrol Effects of Pseudomonas sp. CJ313
We further evaluated the antagonism of Pseudomonas CJ313 to F. oxysporum. In the pot experiment, we found that the isolated Pseudomonas CJ313 significantly inhibited the growth of F. oxysporum, and the R. pseudostellariae grew well without disease symptoms during the period of experiment (Figure 7B). Moreover, qPCR indicated that the abundance of Pseudomonas was significantly higher in Pseudomonas. CJ313 treatment than in control (CK), whereas F. oxysporum showed the opposite trend (Figure 7C). The results clearly showed that strain Pseudomonas. CJ313 has the potential of biological control. The results further suggested that exogenous antagonism of Pseudomonas could be effective against F. oxysporum infection. In addition, the results also demonstrated that the imbalance of these two strains (Pseudomonas sp. CJ313 and F. oxysporum) could be an important cause of the continuous cropping related diseases.
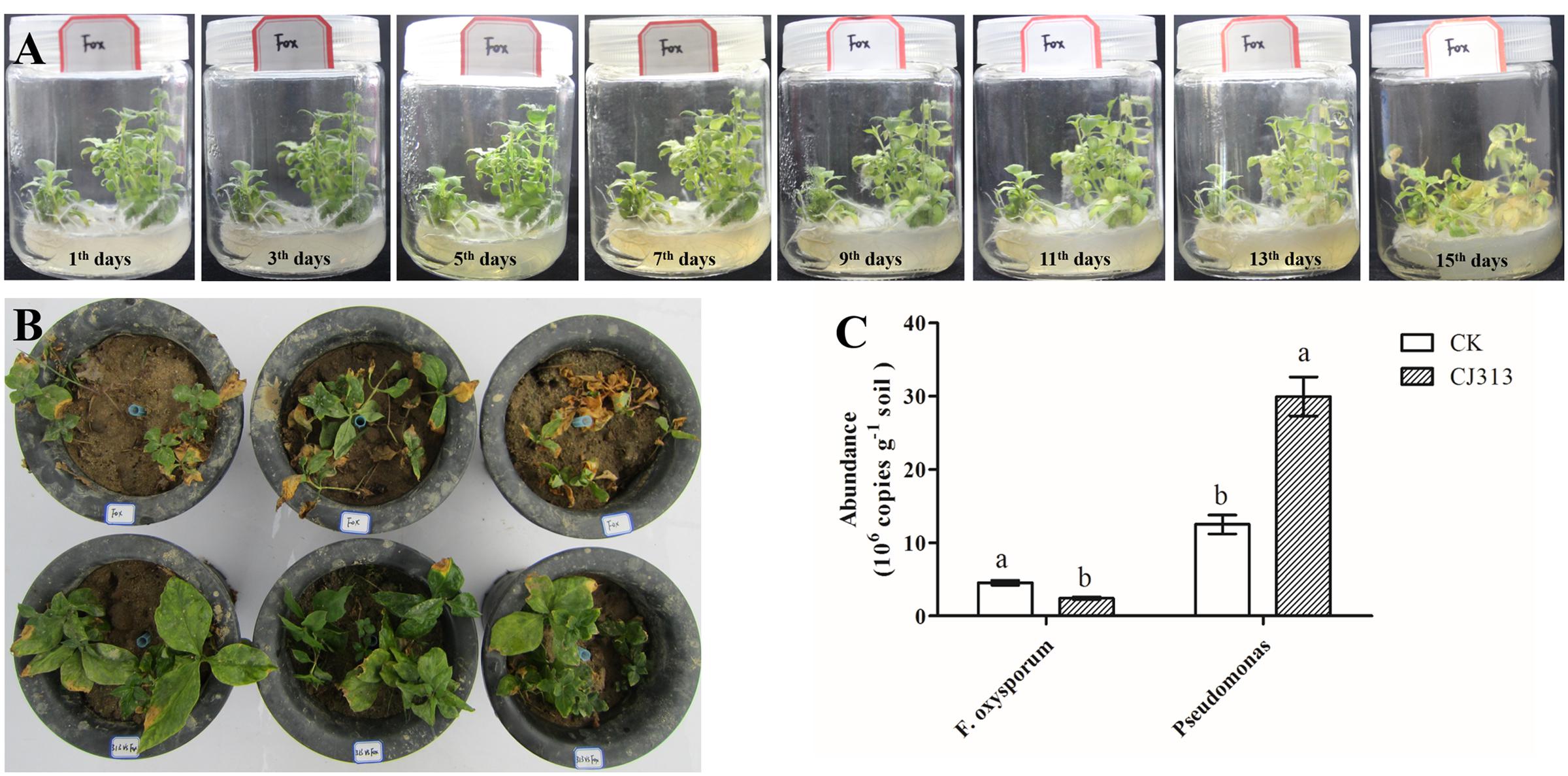
FIGURE 7. Assessment of the pathogenic potential of isolated F. oxysporum (A), the biocontrol potential of Pseudomonas sp. CJ313 against F. oxysporum (FOX) (B), Quantification of F. oxysporum and Pseudomonas spp. from two samples. Data are means ± standard errors (one-way analysis of variance, n = 4).
The Effect of Phenolic Acids on the Growth of Isolated Fusarium oxysporum, Pseudomonas spp.
The results showed that mycelial and sporular growth of F. oxysporum was significantly promoted by phenolic acid mixture (Figures 8A,B). Further analysis showed that p-hydroxybenzoic acid, vanillin, coumaric acid and ferulic acid could significantly promoted mycelial growth of F. oxysporum among the eight phenolic compounds (Supplementary Figure S2). The results also indicated that the growth promotion by mixture was more than that of single phenolic acid on F. oxysporum (Figure 8A). However, the mixture significantly inhibited Pseudomonas sp. CJ313 (Figure 8C) and Pseudomonas sp. CJ361 (Figure 8D) growth. Among them, vanillic acid and syringic acid has the more inhibitory effects on Pseudomonas sp. CJ313 than others (Supplementary Figure S3). Likewise, coumaric acid, ferulic acid syringic acid had the greatest inhibitory effect on Pseudomonas sp. CJ361 (Supplementary Figure S4). The results indicated that certain allelochemicals of R. pseudostellariae root exudates possessed the selective effects on rhizosphere microbes.
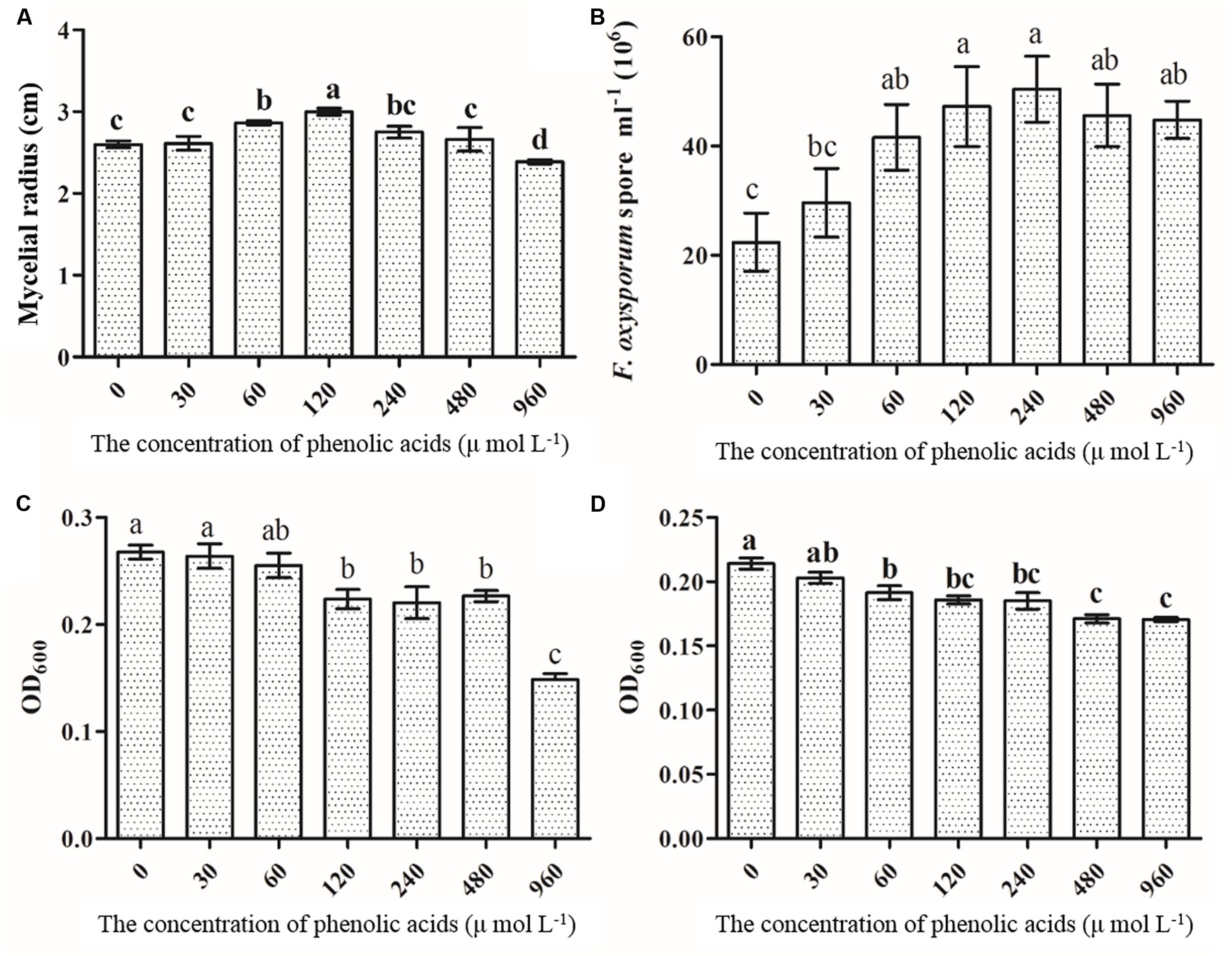
FIGURE 8. The effects of phenolic acid mixture on the growth of F. oxysporum (A), sporulation of F. oxysporum (B), Pseudomonas sp. CJ313 (C), Pseudomonas sp. CJ361 (D). The proportion of phenolic acids was the same as the ratio detected in the rhizosphere soil of Radix pseudostellariae. CK, FP, SP and TP represent the control, newly planted, 2-year, and 3-year consecutively monoculture soils, respectively. Data are means ± standard errors (one-way analysis of variance, n = 4).
Discussion
Our studies presented a significant decline in the yield of R. pseudostellariae along with less aboveground biomass in consecutive monoculture field (Figure 1). Recently, researchers have focused on the biological relationships between plants and rhizosphere microorganisms, which are essential for plant growth and health (Haney and Ausubel, 2015; Lebeis et al., 2015). The study of F. oxysporum has become common due to its ability to cause diseases of important economic crops (Gordon et al., 1989; Gordon and Martyn, 1997). DGGE results revealed significant changes in Pseudomonas and Fusarium communities in the rhizosphere of R. pseudostellariae with prolonged monoculture (Figures 2, 4). Based on the DGGE analysis of Fusarium, we indicated that prolonged monoculture of R. pseudostellariae led to a significant increase in Fusarium species, especially F. oxysporum (Table 1). Quantitative PCR assay confirmed the increase in F. oxysporum with the increasing years of monoculture (Figure 3). These results are supported by the work of different researchers as stated that F. oxysporum is one of main pathogenic species to plants under monoculture regime (Wu et al., 2016b,c).
Due to an extensive distribution of Pseudomonas species in the environment, several studies reported an abundance of antagonistic Pseudomonas species, which controls specialized pathogens that are responsible for disease suppression in soils (Gorlach-Lira and Stefaniak, 2009; Mendes et al., 2011). Our study of Pseudomonas-DGGE revealed that the diversity of Pseudomonas spp. significantly declined with the prolonged monoculture. More importantly, it was found that the relative abundances of antagonistic Pseudomonas spp. declined in soils under consecutive monoculture, and a similar tendency was recorded for other Pseudomonas species studied in the selective medium assay. Similar effects of plants on the abundance of antagonistic Pseudomonas spp. under monoculture were found by Gorlach-Lira and Stefaniak (2009). Hence, the abundance of the Pseudomonas populations in soil of R. pseudostellariae were seriously affected by monoculture. In addition, it was also found that the abundance of Pseudomonas spp. having antagonistic activities against F. oxysporum significantly decreased with the increasing years of monoculture, and this was confirmed by the in vitro antagonism assays. This important antagonistic interaction effects between Pseudomonas and F. oxysporum need particular attention in disease management under a clear cropping system of R. pseudostellariae. Therefore, it is necessary to make robust inferences about balance between Pseudomonas communities and Fusarium of R. pseudostellariae.
Our previous study revealed that most phenolic acids of R. pseudostellariae from rhizosphere soil indicated no direct autotoxicity toward tissue culture seedlings of R. pseudostellariae (Wu et al., 2016a). Besides, many researchers did not support the assumption that the concentrations of allelochemicals in the soil were sufficient to directly influence the development of host plants or neighboring plants (Ehlers, 2011; Weidenhamer et al., 2013). A growing number of researchers reported that the microflora disorder mediated by plant root exudates was the crucial factor leading to plant consecutive monoculture problems (Wu et al., 2014). Root exudates have selective effects on certain microorganisms in the soil and can promote or inhibit the growth of a certain population (Haichar et al., 2008; Hartmann et al., 2009). In this study, the results indicated that the phenolic acid mixture had a significant improvement on the growth of mycelial and spore of pathogenic F. oxysporum (Figures 8A,B). However, phenolic acid mixture could greatly inhibit the growth of antagonistic Pseudomonas sp. CJ313 and CJ361 (Figures 8C,D). Zhou and Wu (2012) observed that the abundance of F. oxysporum in soil was significantly increased by p-coumaric acid, which led to the severity of Fusarium wilt in field conditions. Wu et al. (2016a) reported that phenolic acid, such as syringic acid, significantly promoted the growth of Talaromyces helicus and Kosakonia sacchari, and inhibited growth of Bacillus pumilus. Bais et al. (2002) found that rosmarinic acid had a significant and deleterious effect on Pseudomonas aeruginosa. Furthermore, plant bioassays with representative isolates of Pseudomonas showed that Pseudomonas CJ313 had a good performance that protected R. pseudostellariae from infection by F. oxysporum (Figure 7B). Combined with above-mentioned results, we can draw robust inferences that the imbalance of belowground microbial community resulted in the poor growth of monocultured R. pseudostellariae by root exudates.
Conclusion
Based on multifaceted approaches, such as cultural-independent and culture-dependent analyses, this study indicated that R. pseudostellariae biomass decreased under 3-year extended monoculture resulted from two important factors: (i) the decrease of antagonistic microorganisms (Pseudomonas sp. CJ313 and CJ361) against pathogens (F. oxysporum) might be due to selective inhibitory effect of root exudates, especially phenolic compounds and (ii) an increase of F. oxysporum which significantly induced the poor growth of R. pseudostellariae at a time when pathogenic microbes (F. oxysporum) have become dominant. (iii) Isolated Pseudomonas CJ313 of its exogenous addition could protected R. pseudostellariae from infection by F. oxysporum. These results are very important in the development of potential management approaches to solve the R. pseudostellariae problems under consecutive monoculture. However, additional works are still needed to explore the relationship between aboveground plant performance and belowground microbial diversity.
Author Contributions
WL and JC conceived the study; JC wrote the paper; JC, and LW performed experiments; JC, SL, and ZX performed the statistical analyses; HW, XQ, YW, XW, and JW were involved in field management. MK assisted in English correction. All authors discussed the results and commented on the manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (No. 81573530, 31271670, 31401306), Natural Science Foundation of Fujian Province (2017J01803).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are thankful to Major agricultural extension services (Major agricultural extension services, KNJ-153015, Fujian Province, China) for providing the funds in this work. We are also thankful to Dr. Komivi Senyo Akutse and Professor Christopher Rensing for assistance in English correction.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01748/full#supplementary-material
References
Bais, H. P., Walker, T. S., Schweizer, H. P., and Vivanco, J. M. (2002). Root specific elicitation and antimicrobial activity of rosmarinic acid in hairy root cultures of Ocimum basilicum. Plant Physiol. Biochem. 40, 983–995. doi: 10.1016/s0981-9428(02)01460-2
Cha, J. Y., Han, S., Hong, H. J., Cho, H., Kim, D., Kwon, Y., et al. (2015). Microbial and biochemical basis of a Fusarium wilt-suppressive soil. ISME J. 10, 119–129. doi: 10.1038/ismej.2015.95
Chakravarty, P., and Hwang, S. F. (2010). Effect of an ectomycorrhizal fungus, Laccaria laccata, on Fusarium damping-off in Pinus banksiana seedlings. For. Pathol. 21, 97–106. doi: 10.1111/j.1439-0329.1991.tb00949.x
Clausen, G. B., Larsen, L., Johnsen, K., Julia, R. D. L., and Aamand, J. (2002). Quantification of the atrazine-degrading Pseudomonas sp. strain ADP in aquifer sediment by quantitative competitive polymerase chain reaction. FEMS Microbiol. Ecol. 41, 221–229. doi: 10.1111/j.1574-6941.2002.tb00983.x
Damicone, J. P., and Manning, W. J. (1985). Frequency and pathogenicity of Fusarium spp. isolated from first-year asparagus grown from transplants. Plant Dis. 69, 413–416.
Ehlers, B. K. (2011). Soil microorganisms alleviate the allelochemical effects of a thyme monoterpene on the performance of an associated grass species. PLOS ONE 6:e26321. doi: 10.1371/journal.pone.0026321
Evans, F. F., Seldin, L., Sebastian, G. V., Kjelleberg, S., Holmström, C., and Rosado, A. S. (2004). Influence of petroleum contamination and biostimulation treatment on the diversity of Pseudomonas spp. in soil microcosms as evaluated by 16S rRNA based-PCR and DGGE. Lett. Appl. Microbiol. 38, 93–98. doi: 10.1111/j.1472-765x.2003.01455.x
Garbeva, P., Veen, J. A., and Elsas, J. D. (2004). Assessment of the diversity, and antagonism towards Rhizoctonia solani AG3, of Pseudomonas species in soil from different agricultural regimes. FEMS Microbiol. Ecol. 47, 51–64. doi: 10.1016/s0168-6496(03)00234-4
Gordon, T. R., and Martyn, R. D. (1997). The evolutionary biology of Fusarium oxysporum. Annu. Rev. Phytopathol. 35, 111–128. doi: 10.1146/annurev.phyto.35.1.111
Gordon, T. R., Okamoto, D., and Jacobson, D. J. (1989). Colonization of muskmelon and nonsusceptible crops by Fusarium oxysporum f. sp. melonis and other species of Fusarium. Phytopathology 79, 1095–1100. doi: 10.1094/phyto-79-1095
Gorlach-Lira, K., and Stefaniak, O. (2009). Antagonistic activity of bacteria isolated from crops cultivated in a rotation system and a monoculture against Pythium debaryanum and Fusarium oxysporum. Folia Microbiol. 54, 447–450. doi: 10.1007/s12223-009-0062-1
Haichar, F. Z., Marol, C., Berge, O., Rangel-Castro, J. I., Prosser, J. I., Balesdent, J., et al. (2008). Plant host habitat and root exudates shape soil bacterial community structure. ISME J. 2, 1221–1230. doi: 10.1038/ismej.2008.80
Haney, C. H., and Ausubel, F. M. (2015). Plant microbiome blueprints. Science 349, 788–789. doi: 10.1126/science.aad0092
Hartmann, A., Schmid, M., Tuinen, D. V., and Berg, G. (2009). Plant-driven selection of microbes. Plant Soil 321, 235–257. doi: 10.1007/s11104-008-9814-y
Kozdrój, J., and van Elsas, J. D. (2001). Structural diversity of microorganisms in chemically perturbed soil assessed by molecular and cytochemical approaches. J. Microbiol. Methods 43, 197–212. doi: 10.1016/s0167-7012(00)00197-4
Krueger, C. L., and Sheikh, W. (1987). A new selective medium for isolating Pseudomonas spp. from water. Appl. Environ. Microbiol. 53, 895–897.
Lakshmanan, V., Selvaraj, G., and Bais, H. P. (2014). Functional soil microbiome: belowground solutions to an aboveground problem. Plant Physiol. 166, 689–700. doi: 10.1104/pp.114.245811
Lebeis, S. L., Paredes, S. H., Lundberg, D. S., Breakfield, N., Gehring, J., Mcdonald, M., et al. (2015). Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 349, 860–864. doi: 10.1126/science.aaa8764
Li, X. G., Ding, C. F., Hua, K., Zhang, T. L., Zhang, Y. N., Zhao, L., et al. (2014). Soil sickness of peanuts is attributable to modifications in soil microbes induced by peanut root exudates rather than to direct allelopathy. Soil Biol. Biochem. 78, 149–159. doi: 10.1016/j.soilbio.2014.07.019
Lin, S., Huangpu, J. J., Chen, T., Wu, L. K., Zhang, Z. Y., and Lin, W. X. (2015). Analysis of soil microbial community structure and enzyme activities associated with negative effects of Pseudostellaria heterophylla consecutive monoculture on yield. Pak. J. Bot. 47, 761–769.
Macdonald, C., and Singh, B. (2014). Harnessing plant-microbe interactions for enhancing farm productivity. Bioengineered 5, 5–9. doi: 10.4161/bioe.25320
Mendes, R., Kruijt, M., de Bruijn, I., Dekkers, E., van der Voort, M., Schneider, J. H., et al. (2011). Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332, 1097–1100. doi: 10.1126/science.1203980
O’Donnell, K., Kistler, H. C., Cigelnik, E., and Ploetz, R. C. (1998). Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. U.S.A. 95, 2044–2049. doi: 10.1073/pnas.95.5.2044
Patten, C. L., and Glick, B. R. (2002). Role of Pseudomonas putida indoleacetic acid in development of the host plant root system. Appl. Environ. Microbiol. 68, 3795–3801. doi: 10.1128/aem.68.8.3795-3801.2002
Philippot, L., Raaijmakers, J. M., Lemanceau, P., and van der Putten, W. H. (2013). Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 11, 789–799. doi: 10.1038/nrmicro3109
Pietro, A. D., Madrid, M. P., Caracuel, Z., Delgado-Jarana, J., and Roncero, M. I. (2003). Fusarium oxysporum: exploring the molecular arsenal of a vascular wilt fungus. Mol. Plant Pathol. 4, 315–325. doi: 10.1046/j.1364-3703.2003.00180.x
Punja, Z. K., and Parker, M. (2009). Development of fusarium root and stem rot, a new disease on greenhouse cucumber in British Columbia, caused by Fusarium oxysporum f. sp. radicis-cucumerinum. Can J. Plant Pathol. 22, 349–363. doi: 10.1080/07060660009500453
Raaijmakers, J. M., and Weller, D. M. (1998). Natural plant protection by 2,4-Diacetylphloroglucinol-Producing Pseudomonas spp. in take-all decline soils. Mol. Plant Microbe Interact. 11, 144–152. doi: 10.1094/mpmi.1998.11.2.144
Rogers, S. O., and Bendich, A. J. (1985). Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Mol. Biol. 2, 69–76. doi: 10.1007/BF00020088
Samson, R., Shafik, H., Benjama, A., and Gardan, L. (1998). Description of the bacterium causing blight of leek as Pseudomonas syringae pv. porri (pv. nov.). Phytopathology 88, 844–850. doi: 10.1094/phyto.1998.88.8.844
Tan, Y., and Ji, G. (2010). Bacterial community structure and dominant bacteria in activated sludge from a 70 degrees C ultrasound-enhanced anaerobic reactor for treating carbazole-containing wastewater. Bioresour. Technol. 101, 174–180. doi: 10.1016/j.biortech.2009.08.044
Vujanovic, V., Hamel, C. H. S., and St, A. M. (2002). Development of a selective myclobutanil agar (MBA) medium for the isolation of Fusarium species from asparagus fields. Can. J. Microbiol. 48, 841–847. doi: 10.1139/w02-082
Weidenhamer, J. D., Li, M., Allman, J., Bergosh, R. G., and Posner, M. (2013). Evidence does not support a role for gallic acid in Phragmites australis invasion success. J. Chem. Ecol. 39, 323–332. doi: 10.1007/s10886-013-0242-y
Widmer, F., Seidler, R. J., Gillevet, P. M., Watrud, L. S., and Giovanni, G. D. D. (1998). A highly selective pcr protocol for detecting 16S rRNA genes of the genus Pseudomonas (Sensu Stricto) in environmental samples. Appl. Environ. Microbiol. 64, 2545–2553.
Wu, H., Wu, L., Wang, J., Zhu, Q., Lin, S., Xu, J., et al. (2016a). Mixed phenolic acids mediated proliferation of pathogens Talaromyces helicus and Kosakonia sacchari in continuously monocultured Radix Pseudostellariae rhizosphere soil. Front. Microbiol. 7:335. doi: 10.3389/fmicb.2016.00335
Wu, L. K., Chen, J., Wu, H. M., Wang, J. Y., Wu, Y. H., Lin, S., et al. (2016b). Effects of consecutive monoculture of Pseudostellaria heterophylla on soil fungal community as determined by pyrosequencing. Sci. Rep. 6:26601. doi: 10.1038/srep26601
Wu, L. K., Wu, H. M., Chen, J., Wang, J. Y., and Lin, W. X. (2016c). Microbial community structure and its temporal changes in Rehmannia glutinosa rhizospheric soils monocultured for different years. Eur. J. Soil Biol. 72, 1–5. doi: 10.1016/j.ejsobi.2015.12.002
Wu, L. K., Lin, X. M., and Lin, W. X. (2014). Advances and perspective in research on plant-soil-microbe interactions mediated by root exudates. Chin. J. Plant Ecol. 38, 298–310. doi: 10.3724/sp.j.1258.2014.00027
Wu, X., Li, Z. G., Liu, H. J., Chao, X., Zhang, R. F., Wu, H. S., et al. (2015). The effect of long-term continuous cropping of Black Pepper on soil bacterial communities as determined by 454 pyrosequencing. PLOS ONE 10:e0136946. doi: 10.1371/journal.pone.0136946
Yergeau, E., Filion, M. V., and St-Arnaud, M. (2005). A PCR-denaturing gradient gel electrophoresis approach to assess Fusarium diversity in asparagus. J. Microbiol. Methods 60, 143–154. doi: 10.1016/j.mimet.2004.09.006
Zhang, Z. Y., and Lin, W. X. (2009). Continuous cropping obstacle and allelopathic autotoxicity of medicinal plants. Chin. J. EcoAgric. 17, 189–196. doi: 10.3724/sp.j.1011.2009.00189
Zhao, W. O., Pang, L., Dong, N., and Yang, S. (2015). LC-ESI-MS/MS analysis and pharmacokinetics of heterophyllin B, a cyclic octapeptide from Pseudostellaria heterophylla in rat plasma. Biomed. Chromatogr. 29, 1693–1699. doi: 10.1002/bmc.3481
Zhao, Y. P., Wu, L. K., Chu, L. X., Yang, Y. Q., Li, Z. F., Azeem, S., et al. (2015). Interaction of Pseudostellaria heterophylla with Fusarium oxysporum f. sp. heterophylla mediated by its root exudates in a consecutive monoculture system. Sci. Rep. 5:8197. doi: 10.1038/srep08197
Keywords: Radix pseudostellariae, DGGE, Fusarium, Pseudomonas, quantitative PCR
Citation: Chen J, Wu L, Xiao Z, Wu Y, Wu H, Qin X, Wang J, Wei X, Khan MU, Lin S and Lin W (2017) Assessment of the Diversity of Pseudomonas spp. and Fusarium spp. in Radix pseudostellariae Rhizosphere under Monoculture by Combining DGGE and Quantitative PCR. Front. Microbiol. 8:1748. doi: 10.3389/fmicb.2017.01748
Received: 17 April 2017; Accepted: 28 August 2017;
Published: 15 September 2017.
Edited by:
Aurelio Ciancio, Consiglio Nazionale Delle Ricerche (CNR), ItalyReviewed by:
Munusamy Madhaiyan, Temasek Life Sciences Laboratory, SingaporeMaria Ludovica Saccà, Consiglio per la Ricerca in Agricoltura e l’Analisi dell’Economia Agraria (CREA), Italy
Copyright © 2017 Chen, Wu, Xiao, Wu, Wu, Qin, Wang, Wei, Khan, Lin and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenxiong Lin, lwx@fafu.edu.cn
 Jun Chen
Jun Chen Linkun Wu
Linkun Wu Zhigang Xiao
Zhigang Xiao Yanhong Wu
Yanhong Wu Hongmiao Wu
Hongmiao Wu Xianjin Qin
Xianjin Qin Juanying Wang
Juanying Wang Xiaoya Wei
Xiaoya Wei Muhammad U. Khan
Muhammad U. Khan Sheng Lin
Sheng Lin Wenxiong Lin
Wenxiong Lin