- Key Laboratory of Dairy Biotechnology and Engineering, Ministry of Education, Key Laboratory of Dairy Products Processing, Ministry of Agriculture, Inner Mongolia Agricultural University, Hohhot, China
Teat disinfection pre- and post-milking is important for the overall health and hygiene of dairy cows. The objective of this study was to evaluate the efficacy of a novel probiotic lactobacilli-based teat disinfectant based on changes in somatic cell count (SCC) and profiling of the bacterial community. A total of 69 raw milk samples were obtained from eleven Holstein-Friesian dairy cows over 12 days of teat dipping in China. Single molecule, real-time sequencing technology (SMRT) was employed to profile changes in the bacterial community during the cleaning protocol and to compare the efficacy of probiotic lactic acid bacteria (LAB) and commercial teat disinfectants. The SCC gradually decreased following the cleaning protocol and the SCC of the LAB group was slightly lower than that of the commercial disinfectant (CD) group. Our SMRT sequencing results indicate that raw milk from both the LAB and CD groups contained diverse microbial populations that changed over the course of the cleaning protocol. The relative abundances of some species were significantly changed during the cleaning process, which may explain the observed bacterial community differences. Collectively, these results suggest that the LAB disinfectant could reduce mastitis-associated bacteria and improve the microbial environment of the cow teat. It could be used as an alternative to chemical pre- and post-milking teat disinfectants to maintain healthy teats and udders. In addition, the Pacific Biosciences SMRT sequencing with the full-length 16S ribosomal RNA gene was shown to be a powerful tool for monitoring changes in the bacterial population during the cleaning protocol.
Introduction
Bovine mastitis, an inflammation of the mammary gland, causes physical, chemical, and usually, bacteriological changes in milk and pathological changes in the glandular tissues of the udder that affect the quality and quantity of milk (Sharma and Jeong, 2013). It has a high incidence and prevalence in dairy cows, affecting the net earnings of milk producers worldwide (Frola et al., 2011). Generally, clinical mastitis is easily diagnosed by visible clinical manifestations, such as red, hot, and swollen mammary glands (Sharma et al., 2007). Subclinical mastitis has no visible clinical symptoms in mammary glands and in milk, but milk production decreases, somatic cell count (SCC) increases, pathogens are present in the secretion, and the milk composition is altered (Lafarge et al., 2004). Therefore, development of effective, reliable, and safe mastitis prevention and treatment strategies has become a focus of research.
In China, a teat disinfectant is usually applied to prevent mastitis in pasture. Teat disinfection pre- and post-milking is important not only to reduce the possibility of mastitis, but also to reduce the risk of bacterial contamination of milk (Suriyasathaporn and Chupia, 2011; Zucali et al., 2011). Currently, various teat cleaning disinfectants, including iodophor solution, iodine based gel, sodium hypochlorite, dodecyl benzene sulfonic acid, chlorine, chlorhexidine, phenolic compounds, alcohol, and guava leaf extract, have been used for pre-milking teat dipping (Ingawa et al., 1992; Foret et al., 2005; Gibson et al., 2008). These chemical disinfectants can reduce major pathogen infections; however, the high concentration of chemical substances has raised the concern of potential residues in milk (Galton et al., 1986). As a result, a natural substance that exhibits inhibitory activity against bacteria was developed as a teat disinfectant to minimize bacterial contamination in raw milk. A previous study showed that methanol extracts from guava leaves used as a pre-milking teat disinfectant could significantly reduce teat-end bacterial loads compared with routine udder sanitization without teat dipping (Kummee et al., 2015).
The application of probiotic lactic acid bacteria (LAB) is now considered the best choice for the treatment of many infectious human diseases and for the control of bovine mastitis (Tagg and Dierksen, 2003; Soleimani et al., 2010). It is well known that these bacteria are safe, non-pathogenic, and exhibit many properties that prevent spoilage and the growth of pathogenic bacteria (Mojgani and Ashtiani, 2006; Yezli et al., 2015). The Lactobacillus (L.) plantarum strains IMAU 80065 and IMAU 10155 were isolated from pickles and fermented camel milk from Sichuan and Inner Mongolia, China, respectively (Yu et al., 2012). They were screened from 347 isolates and produce bacteriocins that significantly inhibit the growth of Escherichia (E.) coli, Staphylococcus (S.) aureus, Salmonella, Shigella, and Listeria (Yu et al., 2015). The objective of this study was to compare the effectiveness of a novel teat disinfectant containing two probiotic L. plantarum strains with a commercial disinfectant (CD). We analyzed the SCC, the number of pathogenic bacteria, and the bacterial community of the raw milk. Furthermore, we used Pacific Biosciences (PacBio) single molecule, real-time sequencing technology (SMRT) based on the full-length 16S ribosomal RNA (rRNA) gene to accurately and systematically depict the bacterial profiles of cow milk.
Materials and Methods
Ethics Statement
The study protocol was approved by the Ethical Committee of Inner Mongolia Agricultural University (Hohhot, China) and was permitted by the owners of the sampled dairy farm. Every effort was made to minimize animal suffering.
Treatment and Sample Collection
This study was conducted with 25 lactating Holstein-Friesian dairy cows (2- to 4-years-old) on the Mengele dairy farms of Inner Mongolia, China in May, 2016 when the cows were kept indoors at all times. All cows were fed a total mixed ration (hay 5 kg/d, corn silage 25 kg/d, mixed concentrate 10 kg/d, and sodium bicarbonate 0.15 kg/d) according to standard practice and were kept in a tied housing system. The average milk yield of each dairy cow was 30 kg. Milk samples were collected during the morning milking by members of the research team. At the beginning of the experiment, primary udder hygiene was performed by washing with sterile water and wiping with disposable cloths. Following surface cleaning, several streams of foremilk were then removed prior to sample collection. An approximately 100-mL milk sample was collected in a sterile bottle for determination of SCC. The SCC of raw milk was measured using a laser beam Bentley FTS/FCM400 Combi Instrument (Chaska, MN, United States). A threshold of 200,000 cells/mL is indicative of subclinical mastitis (Dervishi et al., 2016). Eleven cows used in this study were diagnosed with subclinical mastitis because the SCC in a majority of milk samples was ≥200,000 cells/mL. Sterile gloves were used throughout the sampling procedures.
The left and right two teats were treated separately as individual groups. Two teat treatments were applied to the two groups within each sampling phase. After the continued application of the primary hygiene protocol, the left two teats were immersed (dipping treatment) before and after milking with LAB disinfectant containing 5 × 1010 colony-forming units/mL LAB (LAB group). The control (CD) group (right) was treated with the CD (Dipal Concentrate 1+4, Delaval, Tianjin). A total of 50 mL of foremilk and milk was aseptically collected from a quarter and then the samples from the two quarters in the same group were mixed for further study. Cows were individually sampled before treatment (day 0), and after 1, 6, and 10 days of continued cleaning. After 10 days, five cows from each group were randomly selected to continue washing with sterile water, instead of the LAB and CD, and milk was collected 2 days later. A total of 76 raw milk samples were obtained from 11 cows. These samples were kept on ice and transferred to our laboratory. Seven samples may have been contaminated with fecal matter and were removed from our analysis. Detailed information about the samples and the corresponding cows is provided in Table 1. All analyses were performed in duplicate.
DNA Extraction and Quantitative Polymerase Chain Reaction (qPCR)
A 3-mL milk sample was centrifuged at 7000 × g for 20 min in an Eppendorf 5810R centrifuge and the supernatant was removed. The remaining pellet was resuspended in 500 mL of sterile water and 500 mg of lysozyme were added prior to incubation for 12 h at 37°C to maximize bacterial DNA extraction. Genomic DNA was extracted using the DNeasy mericon Food Kit (69514, Qiagen, Germany) according to the manufacturer’s instructions. Finally, 100 μL of elution buffer were added, and elution was performed following a 20-min incubation at room temperature. DNA concentration and purity were evaluated using 1.0% agarose gel electrophoresis and optical density using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, United States). All extracted DNA samples were stored at -20°C until further use.
To accurately quantify pathogenic bacteria and lactobacilli in raw milk, E. coli, S. aureus, Streptococcus (S.) agalactiae, and Lactobacillus were chosen as targets for qPCR analysis with primers designed with the Primer Premier 5.0 program (Premier Biosoft International, Palo Alto, CA, United States) (Supplementary Table S1). qPCR was performed using a one-step real-time PCR System (Applied Biosystems, Foster City, CA, United States) and a SYBR Premix Ex Taq II kit (Takara Bio Inc., Japan). The amplification reaction mixture and program were prepared as previously described (Ma et al., 2015). Corresponding strains were used for standard curve construction, the coefficient of determination (R2 value) of the q-PCR was greater than 0.990, and the efficiency value ranged from 96.3 to 101.8%. PAST software was used to perform statistical analyses (Hammer and Harper, 2009). Data generated by qPCR were expressed as log gene copy number per mL of sample. Bacterial amounts were expressed as means ± standard error and presented as box-plots. The Mann–Whitney test with Bonferroni correction for multiple testing was used to evaluate the difference between samples in a pairwise manner. Sample groups with corrected p-values < 0.05 were considered significantly different.
PCR Amplification and SMRT Sequencing
Genomic DNA was used as a template for PCR amplification of the 16S rRNA gene with primers 27F (5′-GAGAGTTTGATCCTGGCTCAG-3′) and 1541R (5′-AAGGAGGTGATCCAGCCGCA-3′), which contained a set of 16-nucleotide barcodes for barcoded SMRT sequencing of the full-length 16S rRNA gene. The PCR conditions were as follows: 95°C for 4 min and 30 cycles at 95°C for 1 min, 60°C for 45 s, and 72°C for 1 min, followed by a final cycle at 72°C for 7 min (2720 Thermal Cycler, Applied Biosystems) (Liu et al., 2015). Amplicons were sequenced using P6-C4 chemistry on a PacBio RS II instrument (Pacific Biosciences, Menlo Park, CA, United States). Verification of the amplicons and sequence preprocessing were performed as previously described (Mosher et al., 2013).
Data Analysis
Raw sequence data were processed using the RS_ReadsOfinsert.1 protocol available in the SMRT Portal (version 2.7) (Hou et al., 2015). The extracted high-quality sequences were analyzed using the Quantitative Insights into Microbial Ecology (QIIME) package (version 1.7) and alignment of high-quality sequences under 100% clustering of sequence identity was performed using PyNAST and UCLUST softwares (Caporaso et al., 2009; Edgar, 2010). The unique sequence set was classified into operational taxonomic units (OTUs) under a threshold of 98.6% identity using UCLUST after selection of the representative sequences (Lozupone et al., 2006). Each OTU representative sequence was identified using the ribosomal database project (RDP) II database with a minimum bootstrap threshold of 80% (Cole et al., 2007). The de novo taxonomic tree was constructed based on the representative OTU set in FastTree software (Price et al., 2009) for downstream analysis, including the beta diversity calculation. The Shannon–Wiener, Simpson’s diversity, Chao1, and rarefaction estimators were calculated to evaluate alpha diversity. UniFrac distance was calculated based on the phylogenetic tree (Lozupone and Knight, 2005). Both weighted and unweighted calculations were performed for principal coordinate analysis (PCoA). Graphs were generated with the R package (version 3.1.2) and Origin software (version 8.5).
Results
SCC Determination
The SCC of 69 milk samples ranged from 6.7 to 102 × 104 cells/mL during the cleaning protocol (Figure 1). The SCC gradually decreased over the course of the experiment and the SCC of the LAB group was slightly lower than that of the CD group. However, the SCC increased significantly after dipping with sterile water suggesting that LAB and CDs may help prevent mastitis.
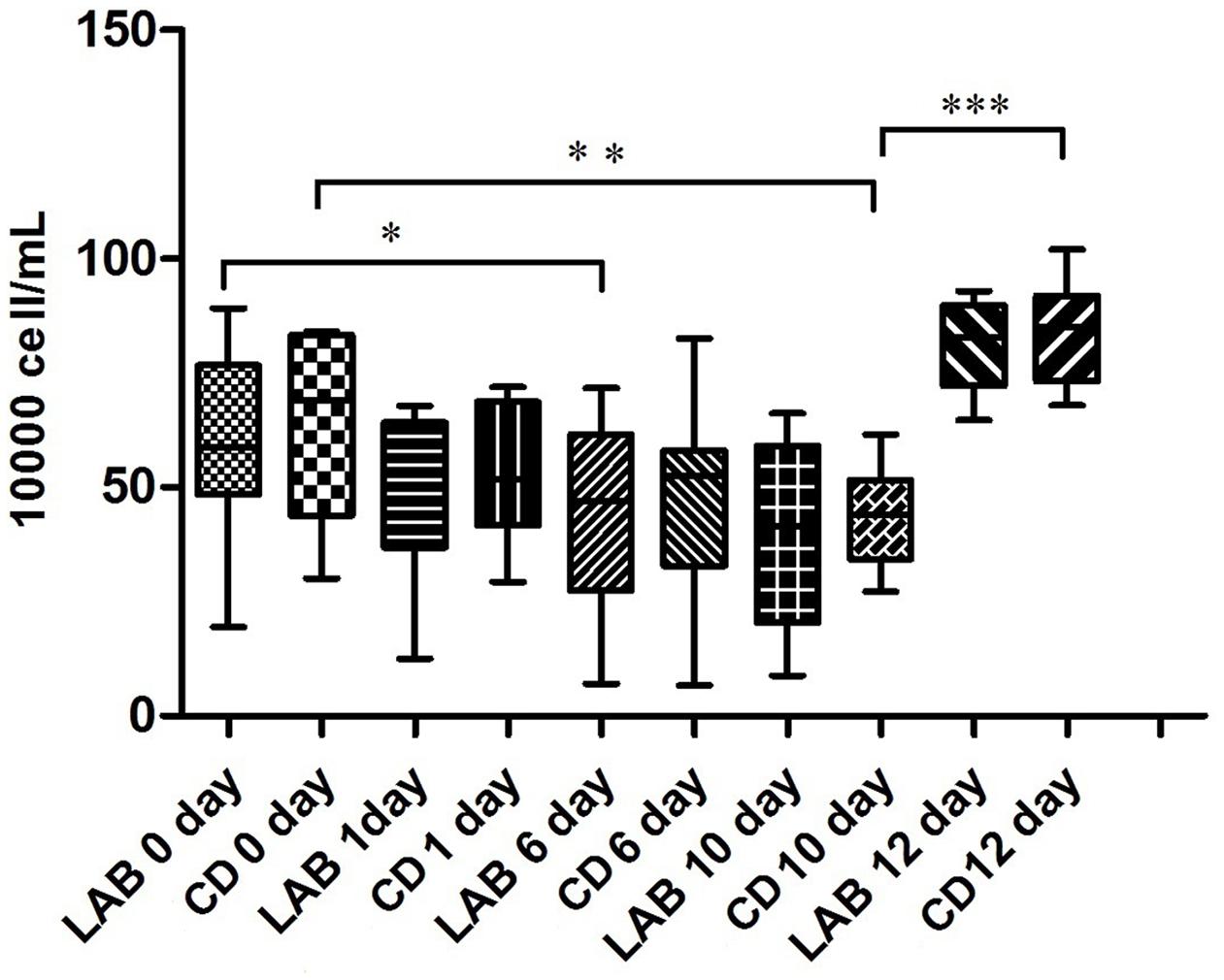
FIGURE 1. Changes in SCC in cow milk during the cleaning process with LAB or chemical disinfectant (CD). Significant differences between sample pairs were evaluated by pairwise Mann–Whitney test with Bonferroni correction; ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Quantitative PCR of the Lactobacillus Genus and Pathogenic Bacteria
The Lactobacillus genus and common pathogenic bacteria in raw milk, including E. coli, S. aureus, and S. agalactiae, were quantified by qPCR, and the bacterial composition over time is presented in Figure 2 and Supplementary Table S2. The mean amounts of E. coli, S. aureus, and S. agalactiae in the LAB group after 10 days of the cleaning protocol were significantly lower than the other groups, while the Lactobacillus genus in the LAB group after 10 days was significantly higher than in the other groups (p < 0.05). In particular, after dipping with sterile water, the amount of S. agalactiae in the CD group was significantly higher than in the LAB group (p < 0.001). Our results show that both LAB and CDs can inhibit these pathogenic bacteria and the LAB disinfectant exhibits long-term effectiveness against S. agalactiae.
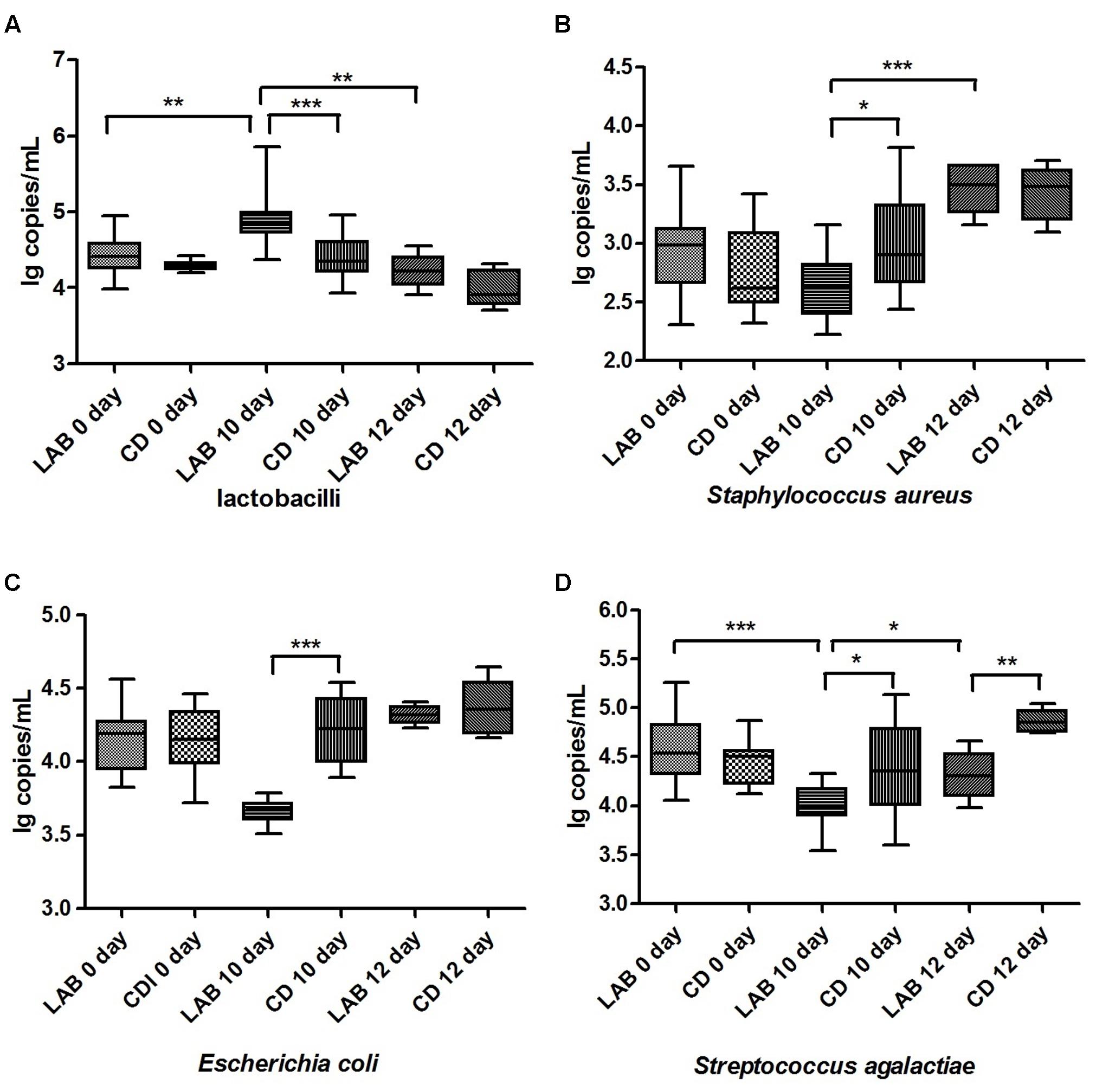
FIGURE 2. Quantification of Lactobacillus (A), Staphylococcus aureus (B), Escherichia coli (C) and Streptococcus agalactiae (D) in cow milk. Bacterial amounts are expressed in log copy number per milliliter of milk sample. Boxes show the median and 25th and 75th percentiles; the lower and upper adjacent hinges show the minimum and maximum values. Significant differences between sample pairs were evaluated by pairwise Mann–Whitney test with Bonferroni correction; ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Sequence Abundance and Diversity
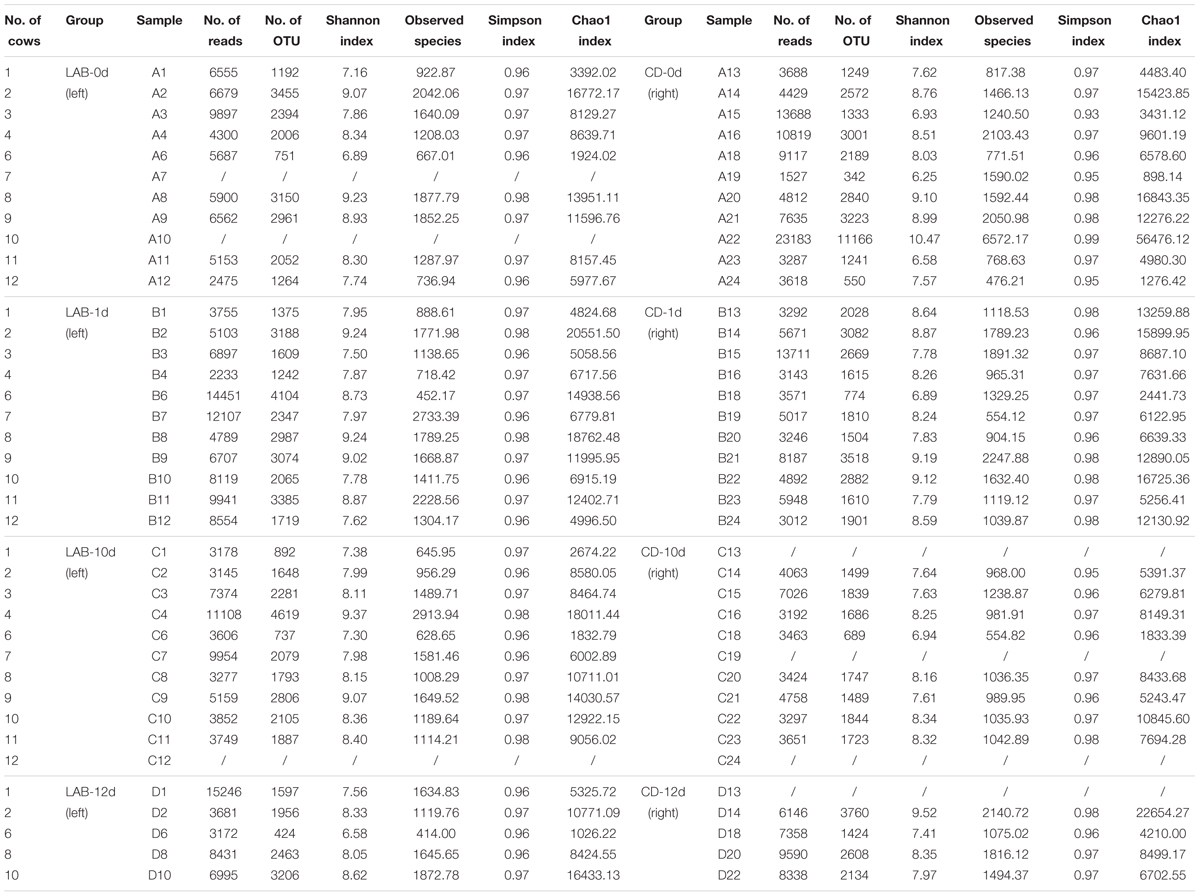
We performed SMRT sequencing of the full-length 16S rRNA gene to obtain accurate bacterial profiles of raw milk at the species level. A total of 503,162 raw reads were generated from 69 milk samples, with an average of 6,341 reads per sample. The total number of unique and classifiable representative bacterial OTU sequences was 15,274 (average = 2213.25 OTUs per sample, range = 342–11166, standard deviation = 913.65). The Shannon index, Simpson diversity index, Chao1, and observed species of each sample were used to evaluate species richness and diversity (Table 1). These values indicated that the majority of samples exhibited a high level of bacterial biodiversity. The Shannon diversity curves indicated that the sequence depth obtained was adequate for all samples (Figure 3).
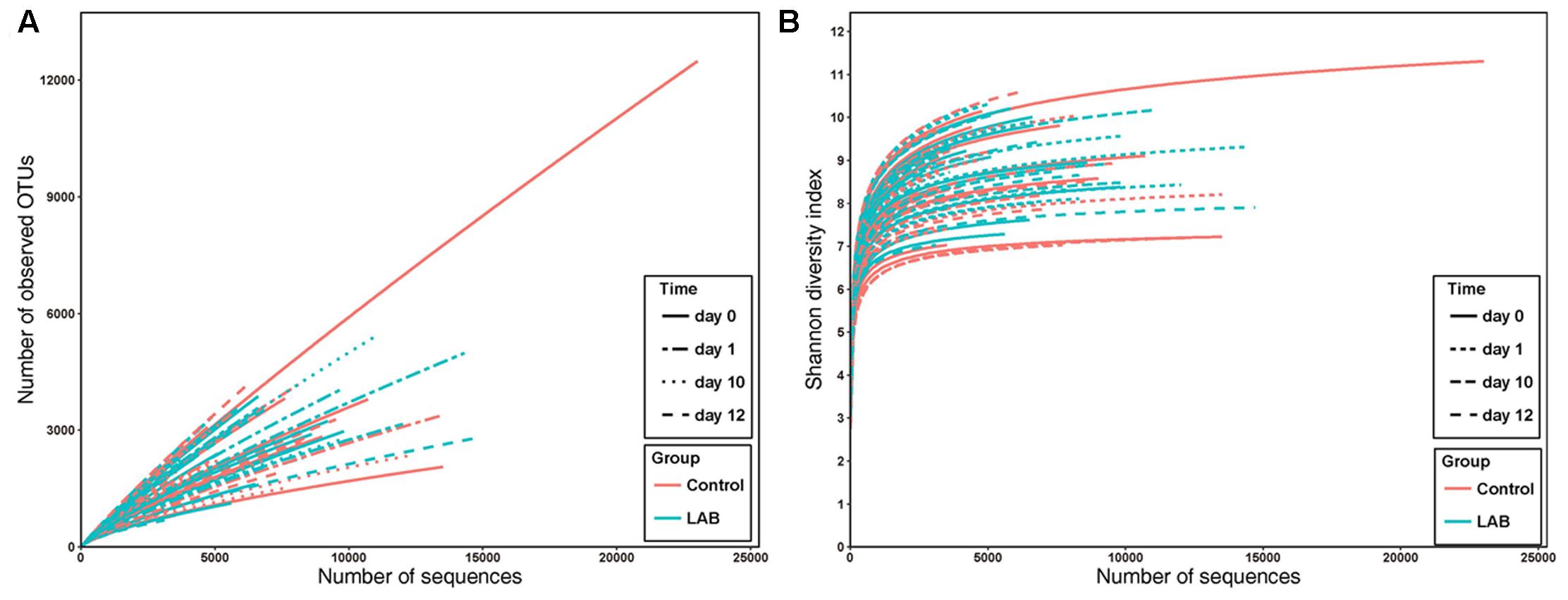
FIGURE 3. Rarefaction analysis (A) and Shannon diversity (B) estimates of the SMRT sequencing reads of bacteria in cow milk. Lines with colors represent samples from different groups. Lines with different textures represent samples from different times.
Change in Bacterial Composition Following Cleaning
Based on the homologous sequence alignment and clustering with the information extracted from the RDP and Greengenes (version 13_8) databases, we determined the lowest level of taxonomy of the identified OTUs. A total of 2.55% of the bacterial sequences could not be identified at the genus level (Supplementary Table S3). From the 69 samples, 25 phyla, 373 genera, and 796 species were identified. Firmicutes (91.14%) and Proteobacteria (7.96%) were the most prevalent phyla, which exhibited no significant differences in the LAB and CD groups during the cleaning process.
At the genus level, the major bacterial genera with relative abundances >1% belonged to Bacillus sp. (average 76.17%), Pseudomonas sp. (average 4.05%), Lactococcus sp. (average 3.55%), Oceanobacillus sp. (average 2.80%), and Lactobacillus sp. (average 1.49%) (Supplementary Table S3). Over the course of the experiment, the relative abundance of Bacillus sp. increased, although its relative abundance gradually decreased after dipping with sterile water. In the LAB and CD groups, Bacillus sp. showed no significant differences during the cleaning protocol. The relative abundance of Pseudomonas sp. was sharply decreased after cleaning with LAB or chemical disinfectant, but was significantly increased in the CD group after dipping with sterile water. The relative abundance of Lactococcus sp. was increased after cleaning with LAB, but decreased after cleaning with chemical disinfectant. This difference was significant in the two groups. After cleaning with sterile water, the amount of Lactococcus sp. in the LAB group decreased and was equal to that of the CD group. Lactobacillus sp. in the LAB group rapidly increased from 0 to 12 days, while there was almost no change in Lactobacillus sp. in the CD group.
In both the LAB and CD groups, the bacterial species primarily included Bacillus (B.) cereus (average 28.43%), B. flexus (average 27.94%), Oceanobacillus (O.) profundus (average 2.74%), the B. pumilus group (average 5.28%), Lactococcus piscium (average 2.74%), Pseudomonas fragi (average 1.67%), Lactococcus lactis (average 0.81%), and the L. plantarum group (average 0.96%) (Supplementary Table S3). As shown in Figure 4, the relative abundance of B. flexus increased during the cleaning protocol. Bacillus cereus increased slightly and then decreased after dipping with sterile water. The relative abundances of the B. pumilus group and Pseudomonas stutzeri were stable over the 12 days. The three species in the Bacillus genus exhibited no significant differences between the LAB and CD groups. The L. plantarum group increased sharply during the cleaning protocol with LAB, while there were no significant changes in the CD group. Oceanobacillus profundus increased and was significantly higher in the LAB group than in the CD group.
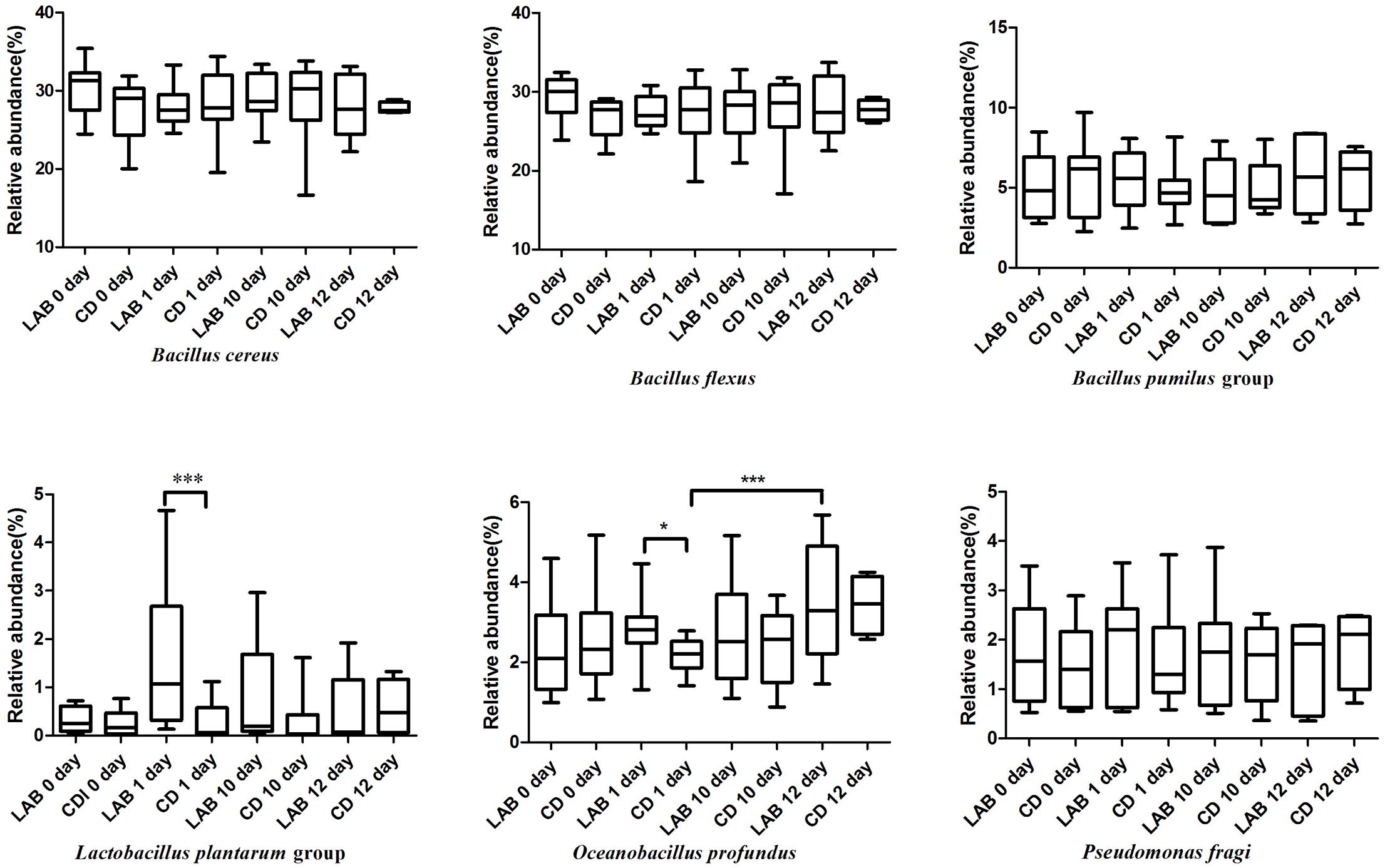
FIGURE 4. Boxplots of the relative abundance of six high abundance (>1%) bacteria detected in cow milk samples at the species level with significant differences.
The relative abundance of some low abundance (<1%) species changed significantly during the cleaning protocol (Figure 5). These species included Acinetobacter schindleri, Acidovorax radicis, Psychrobacter faecalis, Limnobacter thiooxidans, Massilia (M.) timonae, Naxibacter (N.) varians, Paenibacillus validus, Deinococcus (D.) grandis, and Pseudomonas alcaligenes. Acinetobacter schindleri, Acidovorax radicis, Limnobacter thiooxidans, N. varians, and Pseudomonas alcaligenes were detected at 0 day and their relative abundances decreased to almost zero following cleaning. Deinococcus grandis increased during the cleaning protocol, while M. timonae decreased sharply at 6 days, but its relative abundance increased gradually after dipping with sterile water. The abundance of Psychrobacter faecalis initially increased then subsequently decreased.
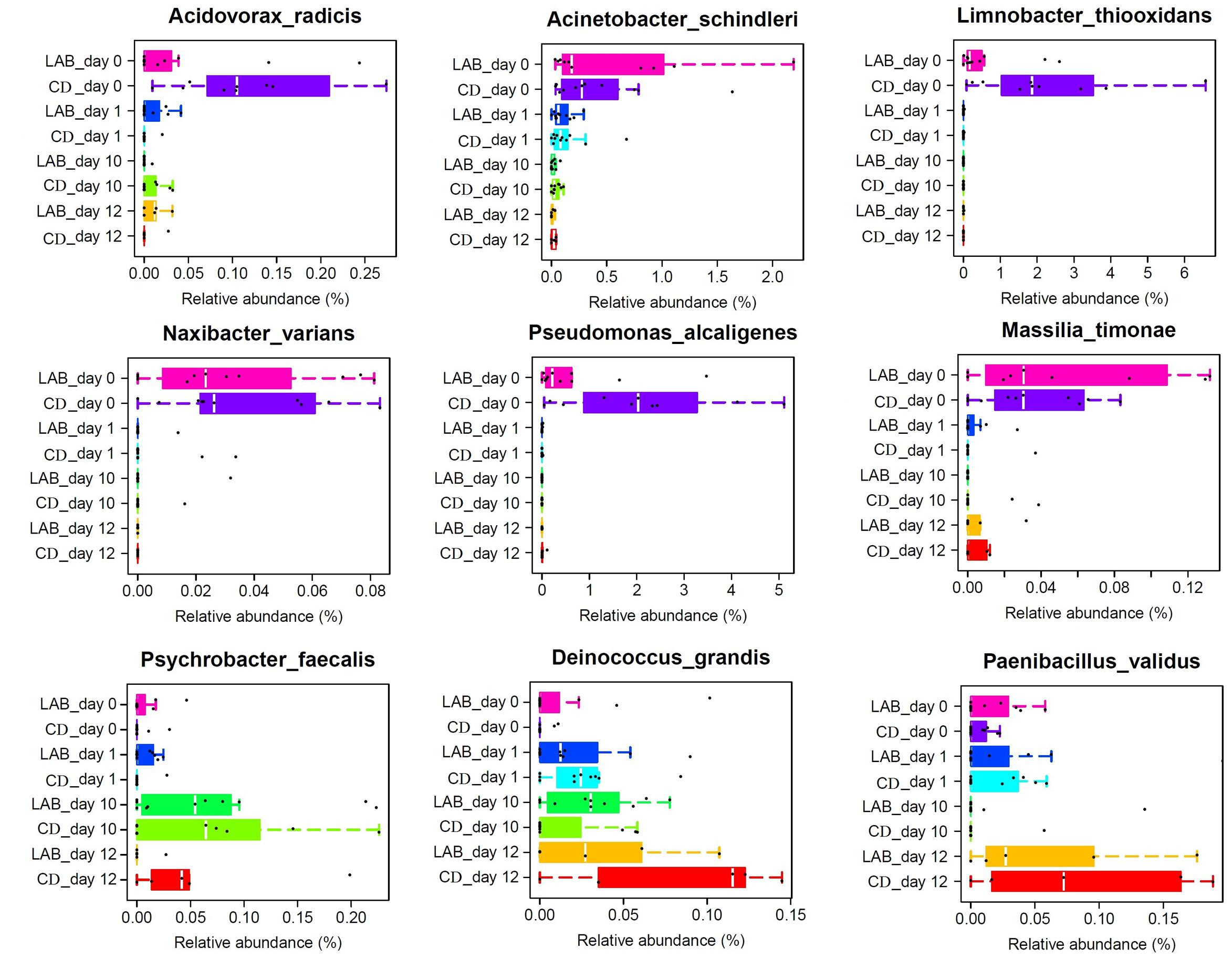
FIGURE 5. Boxplots of the relative abundance of nine low abundance (<1%) bacteria detected in cow milk samples at the species level with significant differences.
Comparison of the Bacterial Community Structure
To compare the structure of the bacterial community in all groups, we performed weighted UniFrac PCoA based on the OTU abundance table. Principal coordinates based on the weighted (PC1 and PC3 accounted for 35.05 and 9.59% of the total variance, respectively) (Figure 6A) and (PC2 and PC3 accounted for 23.87 and 9.59% of the total variance, respectively) (Figure 6B) UniFrac distances revealed apparent bacterial structural differences, as the symbols representing samples of the CD groups at 0 and 10 days were separated on both PCoA score plots with only minor overlap. However, the symbols representing samples of the other five groups overlapped. Results from MANOVA based on unweighted (p < 0.05) UniFrac distances further confirmed the structural differences in bacterial composition between the two sample groups at 10 days.
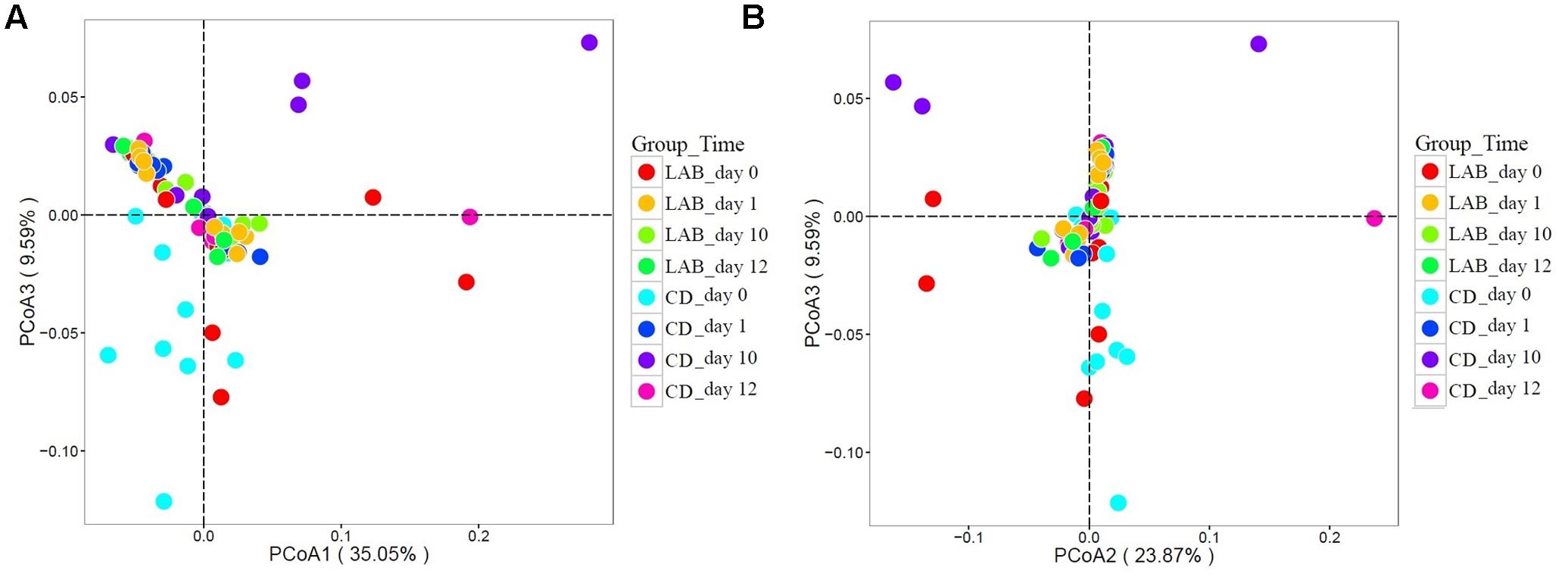
FIGURE 6. Weighted UniFrac principal coordinate analysis (PCoA) of the bacterial communities in cow milk samples. (A) PCoA scores plot based on weighted UniFrac principal components 1 and 3. (B) PCoA scores plot based on weighted UniFrac principal components 2 and 3. Each symbol represents the cow milk microbiota of one sample; sample group is represented by the respective color.
Discussion
Microorganisms in raw milk directly impact the flavor, quality, and shelf life of milk products. Moreover, the presence of pathogens in raw milk can lead to severe illness (Oliver et al., 2009; Quigley et al., 2013). The application of teat disinfection pre- and post-milking is recommended to reduce the number of bacteria on the teat skin and in milk (Oliver et al., 1993). The aims of this study were to harness the PacBio SMRT sequencing technology to investigate the influence that LAB teat disinfectant and CD have on teat milk microbiota from individual cows. Our findings confirm the efficacy of LAB disinfectant in inhibiting pathogenic bacterial growth and improving the bacterial populations of raw milk during the cleaning process.
Currently, the SCC and the California mastitis test (CMT) are used to diagnose mastitis. Somatic cells are naturally present in milk, which is widely used to distinguish healthy quarters from quarters with an inflammatory response most likely due to an intramammary infection (Schukken et al., 2003; Oikonomou et al., 2014). Usually, an SCC of <100,000 cells/mL is considered healthy, and a threshold of 200,000 cells/mL was shown to have high sensitivity and specificity as an identifier of subclinical mastitis (Hillerton, 1999; Diepers et al., 2016). In this study, cows were selected based on this criterion. During the course of teat disinfectant treatment, the SCC gradually decreased and the SCC of the LAB group was slightly lower than that of the CD group. Although the SCC did not decrease to a healthy level, changes were seen in the microfloral structure of teat milk. Previous research also showed that the microfloral structure of bulk tank milk is associated with the SCC (Rodrigues et al., 2017).
Generally, S. aureus, S. uberis, and S. agalactiae are recognized as major virulent mastitis-causing bacteria (Kuehn et al., 2013). Therefore, researchers have focused on developing a safe and effective alternative to the use of chemicals to inhibit these pathogenic bacteria. Previous studies demonstrated the inhibitory effects of secreted bacteriocins, like nisin and lacticin 3147 (Cao et al., 2007; Klostermann et al., 2010), and live probiotic microorganisms, in in vitro and in vivo approaches (Espeche et al., 2009; Diepers et al., 2016). Interestingly, Oikonomou et al. (2014) reported that these bacterial species were present in small quantities in healthy cow milk and hypothesized that they are part of the normal bacterial flora of the mammary gland. In our study, these bacterial species were also detected in lower quantities in the majority of milk samples by SMRT sequencing and qPCR. However, the mean amounts of S. aureus and S. agalactiae gradually decreased during the cleaning protocol, particularly in the LAB group, after 10 days (Figure 2). This result confirms that our LAB disinfectant possesses antibacterial activity primarily against mastitis-causing bacteria in vivo. Moreover, the antibacterial effect of LAB was marginally better than that of the chemical disinfectant.
Probiotic products against udder diseases have been developed over the past few decades and comprise bacteriocins for external application, intramammarily applied bacteriocins, and live probiotic microorganisms. Encouraging results were obtained with the injection of live cultures of selected LAB strains into the bovine udder, which were able to inhibit several mastitis pathogens (Crispie et al., 2008; Frola et al., 2011). Results have shown that preparations of the live culture were as effective as commonly used antibiotics for the treatment of intramammary infections and did not show adverse effects on mammary tissue. Moreover, the bovine teat surface can contain a high diversity of bacteria (Verdiermetz et al., 2012). Monsallier et al. (2012) proposed that the teat skin was a source of microbial populations in raw milk and that farm management and animal grazing practices influenced the diversity and microbiota of raw milk. Another study also highlights that the teat surface and herd habitats are significant drivers of milk microbiota composition (Doyle et al., 2017). Hence, it is beneficial to use a probiotic LAB teat disinfectant as a protective barrier to inhibit pathogens and improve the microbial balance of the teat. Our data provide new insights into probiotic LAB regulation of the bacterial composition of the teat and suggest a role for LAB in the prevention of mastitis. The results revealed differences in the bacterial composition of milk during the cleaning process with two disinfectants. These differences were reflected in the beta-diversity measurements, as illustrated in the PCoA analysis of the UniFrac distance (Figure 6). The UniFrac PCoA showed that the non-cleaned (0 day) and cleaned, LAB cleaned, and chemical cleaned samples mostly fell within separate clusters. That means the LAB teat disinfectant can alter the microbial structure of teat milk.
Analysis of the bacterial composition of the milk samples after 1, 6, 10, and 12 days of teat dipping in the same individuals revealed bacterial species diversity (Figure 4 and Supplementary Figure S1). The relative abundance of the L. plantarum group in the LAB group following cleaning was slightly higher than in the CD group. This demonstrates that the LAB disinfectant could be transferred through the milk ducts into the teat and mammary glands to further regulate the bacterial community of the teat. In addition to its presence in raw milk, O. profundus has been found in deep-sea sediment core samples and wheat rhizospheric soil (Kim et al., 2007; Aislabie et al., 2008; Ranieri et al., 2012). The relatively high abundance of O. profundus in our samples may have been caused by potential contamination from the feeding environment and forage grass. It is worth noting that the relative abundance of O. profundus in the LAB group was significantly higher than in the CD group, suggesting that further research is needed to determine the role O. profundus may play in the microbiota of cow teats.
The relative abundances of Acinetobacter schindleri, Acidovorax radicis, Limnobacter thiooxidans, N. varians, M. timonae, Paenibacillus validus, and Pseudomonas alcaligenes gradually decreased following cleaning (Figure 5). With the exception of Pseudomonas alcaligenes, these species are not commonly found in milk. Acinetobacter schindleri was first found in the urine of a male outpatient with cystitis (Nemec et al., 2001) and the Acinetobacter genus is widely distributed in nature and frequently found in milk, soil, and water (Dortet et al., 2006; Li et al., 2016). Acidovorax radicis is a wheat root-colonizing bacterium that was previously identified from surface-sterilized wheat roots (Li et al., 2011). Limnobacter thiooxidans is a thiosulfate-oxidizing bacterium that was isolated from freshwater lake sediment (Spring et al., 2001). Massilia timonae was isolated from patient blood and soil (Lindquist et al., 2003) and N. varians was identified in clinical specimens and water (Kämpfer et al., 2008). Pseudomonas alcaligenes is a human pathogen, but occurrences are rare. Previous studies have shown that Pseudomonas spp. are responsible for the spoilage of milk and dairy products because of their ability to produce heat-resistant proteolytic and lipolytic enzymes at low temperatures (Ercolini et al., 2009; Zhang et al., 2015). The presence of these species may originate from components of the environment in contact with the teat skin. Our results suggest that teat dipping with LAB and CDs have identical efficacy in the reduction of these contaminating species. Paenibacillus validus was decreased at 10 days with both disinfectants, but increased sharply after cleaning with water for 2 days. This species is abundant in soil but rarely found in milk (Hildebrandt et al., 2006), while other Paenibacillus species have been detected in cow’s milk and various milk products. Paenibacillus spp. are of interest as they have been associated with the biocontrol of pathogenic bacteria and a decreased shelf life of milk products (Ziarno and Zareba, 2010; Debois et al., 2013). In particular, Ranieri et al. (2012) suggested that Paenibacillus spp. could represent an important tool for determining the quality of raw milk, as well as for identifying potential contamination sites at the farm and processing facility. Obviously, both LAB and CDs have the ability to decrease the relative abundance of Paenibacillus validus, although LAB disinfectants are more durable.
Deinococcus grandis and Psychrobacter faecalis increased during the cleaning protocol. Deinococcus grandis is a radioresistant bacterium that was initially isolated from freshwater fish and animal feces (Oyaizu et al., 1987). Some species in the Deinococcus genus are found in milk products. Members of the Deinococcus genus are also used as ingredients in food to ferment soymilk and as a feed additive for hens to enhance yolk coloration to meet customer demand (Wu et al., 2016). Psychrobacter faecalis was first isolated from pigeon feces and has not been detected in milk. While Psychrobacter spp. have been found at a relatively high level in the raw milk of healthy cows compared with cows with mastitis (Kuehn et al., 2013), these bacteria were not previously associated with mastitis (Vacheyrou et al., 2011). Given the change in the amounts of these species during the cleaning protocol, we favor the explanation that in small amounts they are part of the normal bacterial flora of the teat and may encourage teat microflora balance. Future research should aim to investigate the origin of these species and their role in teat microflora.
Three species of Bacillus were dominant in our samples. Previous studies have reported the presence of Bacillus spp. in raw milk (Hou et al., 2015) because of their heat resistance properties and intrinsic antagonism toward other microbes (Griffiths and Phillips, 1990). The season (summer, approximately 30°C) and sanitary conditions of the location of the sample collection may have resulted in the higher relative abundance of Bacillus spp. in the teat, which did not exhibit significant changes during the cleaning process. This suggests that these two different cleaning disinfectants were not able to inhibit Bacillus spp.; hence, it will be important to screen additional LAB strains and develop novel disinfectants aimed specifically at Bacillus spp.
In this study, we used the latest PacBio SMRT sequencing technology to describe the changes in the bacterial community during the cleaning protocol based on the full-length 16S rRNA gene. Compared with previous sequencing approaches, the PacBio SMRT technology has a high capacity for the production of long reads and has been shown to offer higher taxonomic resolution in the profiling of bacterial communities in dairy product samples to the species level (Hou et al., 2015; Li et al., 2016; Zheng et al., 2016). It was therefore not surprising to find more uncommon species and numerous unidentified sequences or sequences identified as uncultured bacteria in our study. Previous data on the bacterial profile of dairy products were generated largely by second-generation sequencing technology, which generates high quality, though short, sequence reads that preclude the accurate assignment of DNA sequences at the species level and even limits classification at the genus level (Amir et al., 2013). Furthermore, PacBio SMRT technology can greatly shorten the time required to detect and analyze the overall bacterial profile of a sample to 24 h (Zheng et al., 2016). By employing SMRT sequencing technology coupled with full-length 16S rRNA sequence determination in this study, clear bacterial population structural differences in the teat were revealed in 69 raw milk samples during the cleaning protocol using LAB or chemical disinfectant.
Rainard (2017) was skeptical about the potential of probiotics as a potential preventive solution for mastitis control. However, the present study demonstrates that teat dips containing LAB possess the ability to reduce both the SSC and pathogenic bacteria, thus regulating the bacterial composition of the teat. Moreover, the ability of LAB to increase beneficial microbes was relatively higher than chemical disinfectants. Overall, our findings indicate that LAB as a teat disinfectant is antimicrobial, non-irritating, and highly efficient and can be used for teat dipping following a routine udder sanitization procedure. Previous studies have been devoted to the development of novel disinfectants, such as plant extracts, to replace chemical disinfectants (Zeedan et al., 2014; Kummee et al., 2015).
This is the first report on the use of PacBio SMRT sequencing technology to evaluate the impacts of LAB disinfectant on the bacterial composition of the teat and comparison with a chemical disinfectant. Our results indicate that the teat bacterial community is highly diverse and complex. Both LAB and CD altered the bacterial composition of the teat and, in particular, reduced the number of mastitis-causing bacteria. Teat dipping with LAB has advantages over chemical disinfectant in that it is harmless to the milker’s health and non-toxic to milk consumers. Therefore, LAB disinfectant could be used as an alternative to chemical pre- and post-milking teat disinfectants to maintain udder health. Some species showed significant differences between cleaning and non-cleaning, suggesting these species could also be used as markers of udder health. In addition, the PacBio SMRT sequencing technology, a powerful and valuable tool, provided accurate microbiota profiling data in our study.
Accession Number
Sequencing data for the 16S rRNA sequences have been deposited in the Metagenomics Rapid Annotation Server with the accession number MGP80281.
Author Contributions
JY and HZ designed the experiments. JY, YR, XX, and WH performed the experiments. JY, YR, and HZ drafted the manuscript. All authors read and approved the final manuscript.
Funding
This research was supported by the China Agriculture Research System (Grant CARS-36 to HZ). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are indebted to the members of the Key Laboratory of Dairy Biotechnology and Engineering, Inner Mongolia Agricultural University, Hohhot, for their technical supports.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01782/full#supplementary-material
References
Aislabie, J. M., Jordan, S., and Barker, G. M. (2008). Relation between soil classification and bacterial diversity in soils of the Ross Sea region, Antarctica. Geoderma 144, 9–20. doi: 10.1016/j.geoderma.2007.10.006
Amir, A., Zeisel, A., Zuk, O., Elgart, M., Stern, S., Shamir, O., et al. (2013). High-resolution microbial community reconstruction by integrating short reads from multiple 16S rRNA regions. Nucleic Acids Res. 41:e205. doi: 10.1093/nar/gkt1070
Cao, L. T., Wu, J. Q., Xie, F., Hu, S. H., and Mo, Y. (2007). Efficacy of nisin in treatment of clinical mastitis in lactating dairy cows. J. Dairy Sci. 90, 3980–3985. doi: 10.3168/jds.2007-0153
Caporaso, J. G., Bittinger, K., Bushman, F. D., Desantis, T. Z., Andersen, G. L., and Knight, R. (2009). PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26, 266–267. doi: 10.1093/bioinformatics/btp636
Cole, J. R., Chai, B., Farris, R. J., Wang, Q., Kulamsyedmohideen, A. S., Mcgarrell, D. M., et al. (2007). The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 35(Suppl. 1), 169–172. doi: 10.1093/nar/gkl889
Crispie, F., Alonsogómez, M., O’Loughlin, C., Klostermann, K., Flynn, J., Arkins, S., et al. (2008). Intramammary infusion of a live culture for treatment of bovine mastitis: effect of live lactococci on the mammary immune response. J. Dairy Res. 75, 374–384. doi: 10.1017/S0022029908003385
Debois, D., Ongena, M., Cawoy, H., and De, P. E. (2013). MALDI-FTICR MS imaging as a powerful tool to identify Paenibacillus antibiotics involved in the inhibition of plant pathogens. J. Am. Soc. Mass Spectrom. 24, 1202–1213. doi: 10.1007/s13361-013-0620-2
Dervishi, E., Zhang, G., Dunn, S. M., Mandal, R., Wishart, D. S., and Ametaj, B. N. (2016). GC-MS metabolomics identifies metabolite alterations that precede subclinical mastitis in the blood of transition dairy cows. J. Proteome Res. 16, 433–446. doi: 10.1021/acs.jproteome.6b00538
Diepers, A. C., Krömker, V., Zinke, C., Wente, N., Pan, L., Paulsen, K., et al. (2016). In vitro ability of lactic acid bacteria to inhibit mastitis-causing pathogens. Sustain. Chem. Pharm. 5, 84–92. doi: 10.1016/j.scp.2016.06.002
Dortet, L., Legrand, P., Soussy, C.-J., and Cattoir, V. (2006). Bacterial identification, clinical significance, and antimicrobial susceptibilities of Acinetobacter ursingii and Acinetobacter schindleri, two frequently misidentified opportunistic pathogens. J. Clin. Microbiol. 44, 4471–4478. doi: 10.1128/JCM.01535-06
Doyle, C. J., Gleeson, D., O’Toole, P. W., and Cotter, P. D. (2017). Impacts of seasonal housing and teat preparation on raw milk microbiota: a high-throughput sequencing study. Appl. Environ. Microbiol. 83, e02694–16. doi: 10.1128/AEM.02694-16
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Ercolini, D., Russo, F., Ferrocino, I., and Villani, F. (2009). Molecular identification of mesophilic and psychrotrophic bacteria from raw cow’s milk. Food Microbiol. 26, 228–231. doi: 10.1016/j.fm.2008.09.005
Espeche, M. C., Otero, M. C., Sesma, F., and Nader-Macias, M. E. (2009). Screening of surface properties and antagonistic substances production by lactic acid bacteria isolated from the mammary gland of healthy and mastitic cows. Vet. Microbiol. 135, 346–357. doi: 10.1016/j.vetmic.2008.09.078
Foret, C. J., Corbellini, C., Young, S., and Janowicz, P. (2005). Efficacy of two iodine teat dips based on reduction of naturally occurring new intramammary infections. J. Dairy Sci. 88, 426–432. doi: 10.3168/jds.S0022-0302(05)72704-1
Frola, I. D., Pellegrino, M. S., Espeche, M. C., Giraudo, J. A., Nadermacias, M. E., and Bogni, C. I. (2011). Effects of intramammary inoculation of Lactobacillus perolens CRL1724 in lactating cows’ udders. J. Dairy Res. 79, 84–92. doi: 10.1017/S0022029911000835
Galton, D. M., Petersson, L. G., and Erb, H. N. (1986). Milk iodine residues in herds practicing iodophor premilking teat disinfection. J. Dairy Sci. 69, 267–271. doi: 10.3168/jds.S0022-0302(86)80397-6
Gibson, H., Sinclair, L. A., Brizuela, C. M., Worton, H. L., and Protheroe, R. G. (2008). Effectiveness of selected premilking teat-cleaning regimes in reducing teat microbial load on commercial dairy farms. Lett. Appl. Microbiol. 46, 295–300. doi: 10.1111/j.1472-765X.2007.02308.x
Griffiths, M. W., and Phillips, J. D. (1990). Incidence, source and some properties of psychrotrophic Bacillus spp found in raw and pasteurized milk. Int. J. Dairy Technol. 43, 62–66. doi: 10.1111/j.1471-0307.1990.tb02446.x
Hammer,Ø, and Harper, D. A. T. (2009). Past: paleontological statistics software package for educaton and data anlysis. Palaeontol. Electron 4, 1–9.
Hildebrandt, U., Ouziad, F., Marner, F. J., and Bothe, H. (2006). The bacterium Paenibacillus validus stimulates growth of the arbuscular mycorrhizal fungus Glomus intraradices up to the formation of fertile spores. FEMS Microbiol. Lett. 254, 258–267. doi: 10.1111/j.1574-6968.2005.00027.x
Hillerton, J. E. (1999). Redefining Mastitis Based on Somatic Cell Count. Brussels: International Dairy Federation, 4–6.
Hou, Q., Xu, H., Zheng, Y., Xi, X., Kwok, L. Y., Sun, Z., et al. (2015). Evaluation of bacterial contamination in raw milk, ultra-high temperature milk and infant formula using single molecule, real-time sequencing technology. J. Dairy Sci. 98, 8464–8472. doi: 10.3168/jds.2015-9886
Ingawa, K. H., Adkinson, R. W., and Gough, R. H. (1992). Evaluation of a gel teat cleaning and sanitizing compound for premilking hygiene. J. Dairy Sci. 75, 1224–1232. doi: 10.3168/jds.S0022-0302(92)77871-0
Kämpfer, P., Falsen, E., and Busse, H. J. (2008). Naxibacter varians sp. nov. and Naxibacter haematophilus sp. nov., and emended description of the genus Naxibacter. Int. J. Syst. Evol. Microbiol. 58, 1680–1684. doi: 10.1099/ijs.0.65516-0
Kim, Y. G., Choi, D. H., Hyun, S., and Cho, B. C. (2007). Oceanobacillus profundus sp. nov., isolated from a deep-sea sediment core. Int. J. Syst. Evol. Microbiol. 57, 409–413. doi: 10.1099/ijs.0.64375-0
Klostermann, K., Crispie, F., Flynn, J., Meaney, W. J., Paul, R. R., and Hill, C. (2010). Efficacy of a teat dip containing the bacteriocin lacticin 3147 to eliminate Gram-positive pathogens associated with bovine mastitis. J. Dairy Res. 77, 231–238. doi: 10.1017/S0022029909990239
Kuehn, J. S., Gorden, P. J., Munro, D., Rong, R., Dong, Q., Plummer, P. J., et al. (2013). Bacterial community profiling of milk samples as a means to understand culture-negative bovine clinical mastitis. PLOS ONE 8:e61959. doi: 10.1371/journal.pone.0061959
Kummee, P., Borisutpeth, M., Chanlun, S., Kanbutra, P., and Chanlun, A. (2015). Efficacy of guava leaf extract as alternative pre-milking teat dipping in reducing teat–end bacterial load of milking dairy cows. Int. J. Pharm. Pharm. Sci. 7, 434–438.
Lafarge, V., Ogier, J. C., Girard, V., Maladen, V., Leveau, J. Y., Gruss, A., et al. (2004). Raw cow milk bacterial population shifts attributable to refrigeration. Appl. Environ. Microbiol. 70, 5644–5650. doi: 10.1128/AEM.70.9.5644-5650.2004
Li, D., Rothballer, M., Schmid, M., Esperschütz, J., and Hartmann, A. (2011). Acidovorax radicis sp. nov., a wheat-root-colonizing bacterium. Int. J. Syst. Evol. Microbiol. 61, 2589–2594. doi: 10.1099/ijs.0.025296-0
Li, L., Renye, J. A. Jr., Feng, L., Zeng, Q., Tang, Y., Huang, L., et al. (2016). Characterization of the indigenous microflora in raw and pasteurized buffalo milk during storage at refrigeration temperature by high-throughput sequencing. J. Dairy Sci. 99, 7016–7024. doi: 10.3168/jds.2016-11041
Lindquist, D., Murrill, D., Burran, W. P., Winans, G., Janda, J. M., and Probert, W. (2003). Characteristics of Massilia timonae and Massilia timonae-like isolates from human patients, with an emended description of the species. J. Clin. Microbiol. 41, 192–196. doi: 10.1128/JCM.41.1.192-196.2003
Liu, W., Zheng, Y., Kwok, L. Y., Sun, Z., Zhang, J., Guo, Z., et al. (2015). High-throughput sequencing for the detection of the bacterial and fungal diversity in Mongolian naturally fermented cow’s milk in Russia. BMC Microbiol. 15:45. doi: 10.1186/s12866-015-0385-9
Lozupone, C., Hamady, M., and Knight, R. (2006). UniFrac – An online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7:371. doi: 10.1186/1471-2105-7-371
Lozupone, C., and Knight, R. (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71, 8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005
Ma, C., Zhao, J., Xi, X., Ding, J., Wang, H., Zhang, H., et al. (2015). Bovine mastitis may be associated with the deprivation of gut Lactobacillus. Benef. Microbes 7, 95–102. doi: 10.3920/BM2015.0048
Mojgani, N., and Ashtiani, M. P. (2006). In vitro inhibition of mastitis pathogens by bacteriocin RN 86 produced by an indigenous Lactobacillus casei isolate. J. Appl. Sci. 6, 2629–2634. doi: 10.3923/jas.2006.2629.2634
Monsallier, F., Verdier-Metz, I., Agabriel, C., Martin, B., and Montel, M. C. (2012). Variability of microbial teat skin flora in relation to farming practices and individual dairy cow characteristics. Dairy Sci. Technol. 92, 265–278. doi: 10.1007/s13594-012-0064-7
Mosher, J. J., Bernberg, E. L., Shevchenko, O., Kan, J., and Kaplan, L. A. (2013). Efficacy of a 3rd generation high-throughput sequencing platform for analyses of 16S rRNA genes from environmental samples. J. Mirobiol. Methods 95, 175–181. doi: 10.1016/j.mimet.2013.08.009
Nemec, A., De Baere, T., Tjernberg, I., Vaneechoutte, M., van der Reijden, T. J., and Dijkshoorn, L. (2001). Acinetobacter ursingii sp. nov. and Acinetobacter schindleri sp. nov., isolated from human clinical specimens. Int. J. Syst. Evol. Microbiol. 51, 1891–1899. doi: 10.1099/00207713-51-5-1891
Oikonomou, G., Bicalho, M. L., Meira, E., Rossi, R. E., Foditsch, C., Machado, V. S., et al. (2014). Microbiota of cow’s milk; distinguishing healthy, sub-clinically and clinically diseased quarters. PLOS ONE 9:e85904. doi: 10.1371/journal.pone.0085904
Oliver, S. P., Boor, K. J., Murphy, S. C., Murinda, S. E., and Callaway, T. R. (2009). Food safety hazards associated with consumption of raw milk. Foodborne Pathog. Dis. 6, 793–806. doi: 10.1089/fpd.2009.0302
Oliver, S. P., Lewis, M. J., Ingle, T. L., Gillespie, B. E., Matthews, K. R., and Dowlen, H. H. (1993). Premilking teat disinfection for the prevention of environmental pathogen intramammary infections. J. Food Protect. 56, 852–855. doi: 10.4315/0362-028X-56.10.852
Oyaizu, H., Stackebrandt, E., Schleifer, K. H., Ludwig, W., Pohla, H., Hirata, A., et al. (1987). A radiation-resistant rod-shaped bacterium, Deinobacter grandis gen. nov., sp. nov., with peptidoglycan containing ornithine. Int. J. Syst. Bacteriol. 37, 62–67. doi: 10.1099/00207713-37-1-62
Price, M. N., Dehal, P. S., and Arkin, A. P. (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650. doi: 10.1093/molbev/msp077
Quigley, L., O’Sullivan, O., Stanton, C., Beresford, T. P., Ross, R. P., Fitzgerald, G. F., et al. (2013). The complex microbiota of raw milk. FEMS Microbiol. Rev. 37, 664–698. doi: 10.1111/1574-6976.12030
Rainard, P. (2017). Mammary microbiota of dairy ruminants: fact or fiction? Vet. Res. 48, 25. doi: 10.1186/s13567-017-0429-2
Ranieri, M. L., Ivy, R. A., Mitchell, W. R., Call, E., Masiello, S. N., Wiedmann, M., et al. (2012). Real-Time PCR detection of Paenibacillus spp. in raw milk to predict shelf life performance of pasteurized fluid milk products. Appl. Environ. Microbiol. 78, 5855–5863. doi: 10.1128/AEM.01361-12
Rodrigues, M. X., Lima, S. F., Canniatti-Brazaca, S. G., and Bicalho, R. C. (2017). The microbiome of bulk tank milk: characterization and associations with somatic cell count and bacterial count. J. Dairy Sci. 100, 2536–2552. doi: 10.3168/jds.2016-11540
Schukken, Y. H., Wilson, D. J., Welcome, F., Garrison-Tikofsky, L., and Gonzalez, R. N. (2003). Monitoring udder health and milk quality using somatic cell counts. Vet. Res. 34, 579–596. doi: 10.1051/vetres:2003028
Sharma, N., Gupta, S. K., Sharma, U., and Hussain, K. (2007). Treatment of clinical mastitis in buffalo-A case report. Buffalo Bull. 26, 56–58.
Sharma, N., and Jeong, D. K. (2013). Stem cell research: a novel boulevard towards improved bovine mastitis management. Int. J. Biol. Sci. 9, 819–829. doi: 10.7150/ijbs.6901
Soleimani, N. A., Kermanshahi, R. K., Yakhchali, B., and Sattari, T. N. (2010). Antagonistic activity of probiotic lactobacilli against Staphylococcus aureus isolated from bovine mastitis. Afr. J. Microbiol. Res. 420, 2169–2173.
Spring, S., Kämpfer, P., and Schleifer, K. H. (2001). Limnobacter thiooxidans gen. nov., sp. nov., a novel thiosulfate-oxidizing bacterium isolated from freshwater lake sediment. Int. J. Syst. Evol. Microbiol. 51, 1463–1470. doi: 10.1099/00207713-51-4-1463
Suriyasathaporn, W., and Chupia, V. (2011). Reduction in numbers of bacteria after pre-milking teat dipping in milking dairy cows. CMU. J. Nat. Sci. 10, 301–306.
Tagg, J. R., and Dierksen, K. P. (2003). Bacterial replacement therapy: adapting ‘germ warfare’ to infection prevention. Trends Biotechnol. 21, 217–223. doi: 10.1016/S0167-7799(03)00085-4
Vacheyrou, M., Normand, A. C., Guyot, P., Cassagne, C., Piarroux, R., and Bouton, Y. (2011). Cultivable microbial communities in raw cow milk and potential transfers from stables of sixteen French farms. Int. J. Food Microbiol. 146, 253–262. doi: 10.1016/j.ijfoodmicro.2011.02.033
Verdiermetz, I., Gagne, G., Bornes, S., Monsallier, F., Veisseire, P., Delbèspaus, C., et al. (2012). Cow teat skin, a potential source of diverse microbial populations for cheese production. Appl. Environ. Microbiol. 78, 326–333. doi: 10.1128/AEM.06229-11
Wu, S. Y., Li, I. C., Lin, Y. C., and Chen, C. C. (2016). Characterization and safety evaluation of Deinococcus grandis as feed additive for hens. Regul. Toxicol. Pharm. 76, 121–127. doi: 10.1016/j.yrtph.2016.01.019
Yezli, W., Zebboudj-Yezli, N., Hamini-Kadar, N., Kihal, M., and Henni, J. E. (2015). An in vitro antagonistic activity evaluation of rhizobacteria against Fusarium oxysporum f. sp. radicis-lycopersici (Forl) isolated from the Algerian west. Int. J. Biosci. 7, 95–103. doi: 10.12692/ijb/7.1.95-103
Yu, H. J., Chen, Y. F., Yang, H. J., Yang, J., Xue, J. G., Li, C. K., et al. (2015). Screening for Lactobacillus plantarum with potential inhibitory activity against enteric pathogens. Ann. Microbiol. 65, 1–9. doi: 10.1007/s13213-014-0963-3
Yu, J., Gao, W., Qing, M., Sun, Z., Wang, W., Liu, W., et al. (2012). Identification and characterization of lactic acid bacteria isolated from traditional pickles in Sichuan, China. J. Gen. Appl. Microbiol. 58, 163–172. doi: 10.2323/jgam.58.163
Zeedan, G. S. G., Abdalhamed, A. M., Abdeen, E., Ottai, M. E., and Abdelshafy, S. (2014). Evaluation of antibacterial effect of some Sinai medicinal plant extracts on bacteria isolated from bovine mastitis. Vet. World 7, 991–998. doi: 10.14202/vetworld.2014.991-998
Zhang, R., Huo, W., Zhu, W., and Mao, S. (2015). Characterization of bacterial community of raw milk from dairy cows during subacute ruminal acidosis challenge by high-throughput sequencing. J. Sci. Food Agric. 95, 1072–1079. doi: 10.1002/jsfa.6800
Zheng, Y., Xi, X., Xu, H., Hou, Q., Bian, Y., Yu, Z., et al. (2016). Using PacBio long-read high-throughput microbial gene amplicon sequencing to evaluate infant formula safety. J. Agric. Food Chem. 64, 6993–7001. doi: 10.1021/acs.jafc.6b01817
Ziarno, M., and Zareba, D. (2010). Characteristics and occurrence of Paenibacillus bacteria in milk and milk products. Med. Weter 66, 600–603.
Keywords: subclinical mastitis, lactic acid bacteria, teat disinfectant, SMRT sequencing, bacterial community
Citation: Yu J, Ren Y, Xi X, Huang W and Zhang H (2017) A Novel Lactobacilli-Based Teat Disinfectant for Improving Bacterial Communities in the Milks of Cow Teats with Subclinical Mastitis. Front. Microbiol. 8:1782. doi: 10.3389/fmicb.2017.01782
Received: 13 June 2017; Accepted: 04 September 2017;
Published: 26 September 2017.
Edited by:
Giovanna Suzzi, Università di Teramo, ItalyReviewed by:
Jorge Reinheimer, National University of the Littoral, ArgentinaBeatrix Stessl, Veterinärmedizinische Universität Wien, Austria
Copyright © 2017 Yu, Ren, Xi, Huang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Heping Zhang, hepingdd@vip.sina.com
†These authors have contributed equally to this work.
 Jie Yu
Jie Yu Yan Ren
Yan Ren XiaoXia Xi
XiaoXia Xi