- 1Key Lab of Molecular Virology, Institute of Medical Microbiology, Department of Infectious Diseases, Huashan Hospital, Fudan University, Shanghai, China
- 2Department of Molecular Microbiology and Immunology, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, MD, United States
Persisters comprise a group of phenotypically heterogeneous metabolically quiescent bacteria with multidrug tolerance and contribute to the recalcitrance of chronic infections. Although recent work has shown that toxin-antitoxin (TA) system HipAB depends on stringent response effector (p)ppGppin persister formation, whether other persister pathways are also dependent on stringent response has not been explored. Here we examined the relationship of (p)ppGpp with 15 common persister genes (dnaK, clpB, rpoS, pspF, tnaA, sucB, ssrA, smpB, recA, umuD, uvrA, hipA, mqsR, relE, dinJ) using Escherichia coli as a model. By comparing the persister levels of wild type with their single gene knockout and double knockout mutants with relA, we divided their interactions into five types, namely A “dependent” (dnaK, recA), B “positive reinforcement” (rpoS, pspF, ssrA, recA), C “antagonistic” (clpB, sucB, umuD, uvrA, hipA, mqsR, relE, dinJ), D “epistasis” (clpB, rpoS, tnaA, ssrA, smpB, hipA), and E “irrelevant” (dnaK, clpB, rpoS, tnaA, sucB, smpB, umuD, uvrA, hipA, mqsR, relE, dinJ). We found that the persister gene interactions are intimately dependent on bacterial culture age, cell concentrations (diluted versus undiluted culture), and drug classifications, where the same gene may belong to different groups with varying antibiotics, culture age or cell concentrations. Together, this study represents the first attempt to systematically characterize the intricate relationships among the different mechanisms of persistence and as such provide new insights into the complexity of the persistence phenomenon at the level of persister gene network interactions.
Introduction
Chronic and recalcitrant biofilm infections tolerant to antibacterial treatment pose a major medical problem, since they can cause considerable morbidity and frequently require multiple courses of antibiotic treatment, which in turn may contribute to the emergence of stable antibiotic resistance. Persisters are considered to play a predominant role in the recalcitrance of chronic bacterial infections by Mycobacterium tuberculosis and Escherichia coli (Zhang et al., 2012; Cui et al., 2016). They are well known for their survival in supra-lethal dose of multiple antibiotic and other environmental stresses. This phenomenon is widely present in virtually all microbes (Allison et al., 2011; Zhang et al., 2012; Feng et al., 2015; Xu et al., 2016). In contrast to antibiotic resistance, bacterial persister cells constitute a subpopulation of metabolically quiescent slow-growing or growth arrested cells with no heritable resistance mutations or increased minimal inhibitory concentration (MIC) compared to wild–type cells.
Although the exact mechanisms underlying persistence have yet to be uncovered, several pathways have been identified to be implicated in the formation of bacterial persisters (Zhang, 2014; Harms et al., 2016; Kaldalu et al., 2016; Van den Bergh et al., 2017). Since the discovery of hipA7 strain having a 100∼1000-fold increase in persister level (Moyed and Bertrand, 1983), at least 10 type II TA models have been reported to have an intimate association with persistence in E. coli (Lewis, 2010; Wang and Wood, 2011; Maisonneuve et al., 2013). However, the role of TA models in persisters is being challenged by some recent papers (Ramisetty et al., 2016; Van Melderen and Wood, 2017). In addition to TA genes, SOS response is required in persister formation under DNA damaging conditions, such as fluoroquinolones (Dorr et al., 2009). And indole also mediates persistence under nutrient-limiting conditions (Vega et al., 2012). Our previous studies have discovered energy production genes sucB/ubiF, trans-translation genes ssrA/smpB, and phosphate metabolism regulating gene phoU, being important for pesister formation (Li and Zhang, 2007; Ma et al., 2010; Li et al., 2013). (p)ppGpp, which is synthesized by RelA/SpoT from GDP or GTP (Hauryliuk et al., 2015) and rapidly accumulates during the stringent response (SR) under amino acid starvation, has been shown to be a critical metabolic mediator of persisters (Korch et al., 2003; Fung et al., 2010; Maisonneuve et al., 2013; Amato and Brynildsen, 2015; Germain et al., 2015). The SR orchestrates accommodation to various conditions (Steinchen and Bange, 2016) and its second messenger (p)ppGpp can lead to alteration of many cellular activities by downregulation of genes or enzymes for rapid growth and upregulation of genes for stress survival. Korch et al. (2003) first observed that TA module toxin hipA7 produced a high level of persistence which is dependent on (p)ppGpp synthesized by relA when exposed to antibiotics. Recently, it has been shown that not all type II TA modules involved in persistence require the activation of stringent response (Germain et al., 2015). However, it remains unclear whether other persister genes require (p)ppGpp to exhibit their persistence phenotype.
Although many persister genes have been identified so far, their impacts on persister levels differ considerably depending on different times and antibiotics being used (Wu et al., 2015). We have shown that different genes have different roles under the same antibiotic exposure (Wu et al., 2015). In addition, the same gene could exhibit varying importance to different antibiotics. The purpose of the present work is to investigate the interactions of the known persister genes with the (p)ppGpp pathway. By studying the differential phenotype of single gene knockout strains and double knockout strains with relA, we unraveled that different persister genes have distinct relationships with (p)ppGpp, which led to changing or even reversal in persister levels. Of the 15 persister genes (dnaK, clpB, rpoS, pspF, tnaA, sucB, ssrA, smpB, recA, umuD, uvrA, hipA, mqsR, relE, dinJ) we evaluated, only ΔdnaK and ΔrecA (gentamicin and ampicillin) mutant strains were dependent on (p)ppGpp in terms of inhibiting persister formation while the other 13 persister genes fell in either synergistic, antagonistic, overtaking or irrelevant categories (see Figures 6–8 and Supplementary Table S1). Our findings shed new light on the complex interactions of persister genes with the (p)ppGpp pathway.
Materials and Methods
Bacterial Strains and Growth Conditions
The strains used in this study were derived from wild type E. coli K12 strain W3110. Cells were routinely cultured in Luria-Bertani (LB) broth (10 g Bacto-tryptone, 5 g yeastextract, and 10 g NaCl/liter). Cells from -80°C stock were grown overnight to 109 CFU/ml in LB in 17- by 100-mm polypropylene tubes, diluted 1:1000 into 4 mL LB broth, and grown to stationary phase (109 CFU/ml) or log phase (108 CFU/ml) at 37°C with shaking (200 rpm) unless otherwise stated.
Knockout Mutant Construction
Deletion of persister genes made in the E. coli W3110 background was achieved by using the λ Red recombination system, as described by Datsenko and Wanner (2000). The deleted genes were stably replaced with a chloramphenicol resistance gene and this selectable marker was removed using pCP20 when needed. All mutants and plasmid insertions were confirmed by PCR and sequencing (Biosune). Further details of primers designed for this purpose and additional external primers used to verify the correct integration of the PCR fragments by homologous recombination are described in our previous work(Wu et al., 2015).
Persister Assay
Persistence was measured by determining the bacterial survival as colony-forming units (CFUs) per 1mL after exposure to 200 μg/ml ampicillin, or 8 μg/ml norfloxacin, or 40 μg/ml gentamicin for stationary phase cultures undiluted as well as diluted (1:100) in some cases. Following overnight growth, 1 ml undiluted cultures or 1 ml diluted cultures (10 μl cultures and 990 μl LB) were transferred to a 1.5 ml Eppendorf tube and immediately treated with the above antibiotics and incubated at 37°C without shaking for different times. Stationary or log phase cultures (1 ml) without antibiotic exposure were included as controls in the persister assay. The initial cell number was checked by sampling 10 μl and serially diluting and plating on LB agar. The cell viability was measured by samples withdrawn at the desired time points, washed and serially diluted in PBS, followed by inoculation onto LB agar without antibiotics. The CFU counts were measured after overnight incubation at 37°C.
Results
Since the well-known persister gene hipA was shown to depend on (p)ppGpp to mediate persistence, a growing number of persister studies have shifted their emphasis toward the role of (p)ppGpp and its correlation with other persister genes. However, it is still unclear to what extent other persister genes are dependent on (p)ppGpp. To address this question, we constructed double knockout mutants of 15 known persister genes (dnaK, clpB, rpoS, pspF, tnaA, sucB, ssrA, smpB, recA, umuD, uvrA, hipA, mqsR, relE, dinJ) with relA which encodes (p)ppGpp synthetase. Theoretically, if a single-gene knockout mutant that affects persister level is dependent on (p)ppGpp, then its double knockout mutant with relA will display a similar persister phenotype as the wild type will do. With this assumption, we determined the persister levels of the single and double knockout mutants as well as ΔrelA mutant and the parent strain W3110. Early stationary phase cultures of the wild type and mutants were treated with ampicillin (200 μg/ml), norfloxacin (8 μg/ml) or gentamicin (40 μg/ml), and the persister levels were measured at different time points for each antibiotic. We found that among the 15 persister genes analyzed, only ΔdnaK and ΔrecA were dependent on (p)ppGpp in terms of their effect on persister levels, while the other 13 persister genes (clpB, rpoS, pspF, tnaA, sucB, ssrA, smpB, recA, umuD, uvrA, mqsR, relE, dinJ) did not seem to be dependent on (p)ppGpp because their double knockout mutants exhibited different persister numbers compared with the parent strain W3110. However, in the case of hipA as a well-known example of ppGpp-dependent gene (Korch et al., 2003), the hipA7 allele confers persistence in a manner that is dependent on ppGpp since lack of ppGpp due to relA mutation diminished the high persistence phenotype in the hipA7strain. However, in the case of dnaK and recA mutant cells, we found ppGpp is required for the opposite effect (i.e., lower persistence).
DnaK and RecA Are Implicated in Persistence to Gentamicin and Ampicillin and Their Persistence Levels Are Dependent on (p)ppGpp
We found that addition of three different antibiotics to early stationary phase culture of the ΔdnaK mutant indeed produced dramatically lower persister numbers compared with its parent strain W3110. The ratio was < 10-6 for gentamicin and ampicillin (see Figures 1A,B) and about 1:10 for norfloxacin, respectively. The ΔrelA mutant also had a 103∼104 -fold lower (gentamicin and ampicillin) (see Figures 1A,B) and 10∼100-fold lower (norfloxacin) persister numbers than W3110. Interestingly, the ΔrelAΔdnaK mutant demonstrated a 103∼106 -fold higher persistence phenotype compared with either ΔdnaK or ΔrelA and was similar to the level of the parent strain with gentamicin and ampicillin exposure when the cultures were taken from early stationary phase (see Figures 1A,B). However, with norfloxacin exposure, the persister numbers produced by ΔrelAΔdnaK remained the same as ΔdnaK when the cultures were taken from early stationary phase (5 h) (∼10 –fold decrease of either ΔrelAΔdnaK or ΔdnaK compared with W3110) (see Figure 8). At late stationary phase (18 h), the persister numbers of ΔdnaK kept at significant low levels for all the three antibiotics (106∼108 –fold lower for gentamicin and norfloxacin, ∼103 –fold lower for ampicillin compared with W3110), but ΔrelA showed a similar persister number as W3110 for all the antibiotics we tested. Although the persister levels of the ΔrelAΔdnaK mutant were the same as those of W3110 or ΔrelA in the presence of gentamicin and ampicillin, and only < 10 –fold decrease compared with W3110 or ΔrelA for norfloxacin, it was difficult to discriminate whether ΔrelAΔdnaK reverted back to W3110 or was the same as ΔrelA. To address this, we chose the early stationary phase inocula for our subsequent test for all the 15 persister genes we tested.
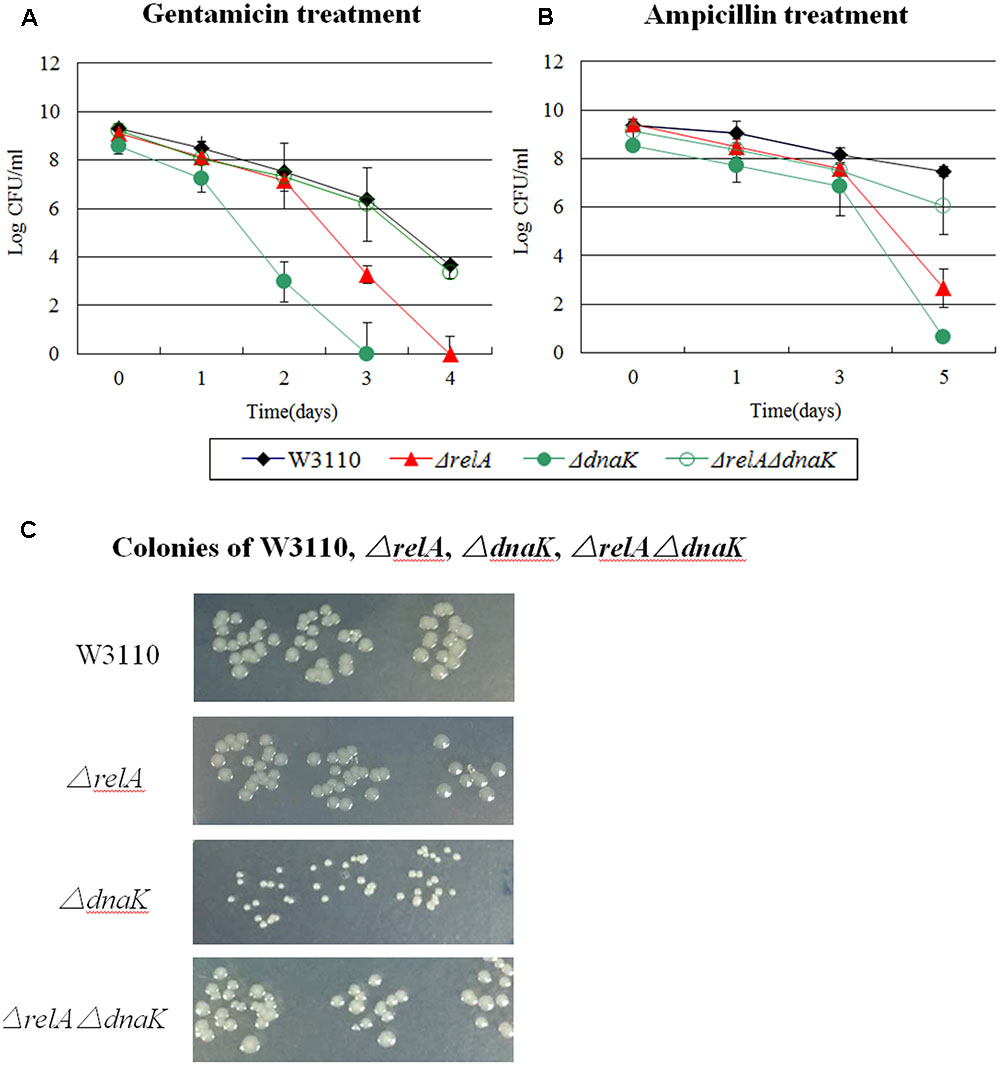
FIGURE 1. Effect of dnaK single and double-gene knockout mutations on E. coli persister formation. Cultures of W3110 and persister gene deletion mutants grown to stationary phase and immediately treated with (A) gentamicin (40 μg/ml) and (B) ampicillin (200 μg/ml). (C) Colonies of W3110, ΔrelA, ΔdnaK and ΔrelAΔdnaK after 16 h incubation at 37°C on LB plates without antibiotics. Data shown are from three independent experiments with 3 replicates per experiment and error bars indicate the standard deviations.
It is important to note that the ΔdnaK mutant had a growth defect with characteristic small colonies on agar plates. However, this characteristic was diminished and reverted back to that of wild type strain W3110 in the case of ΔrelAΔdnaK (see Figure 1C). Together, the above results indicated the DnaK is an important factor for all the three antibiotic related persister formation pathways. The decreased persistence phenotype as well as its growth defect of ΔdnaK seemed to depend on functional (p)ppGpp. And this dependence had intimate relationship with the age of inocula and antibiotic classification. At early stationary phase, the ΔrelAΔdnaK mutant did not revert back to a phenotype similar to W3110 with norfloxacin exposure as it did with gentamicin and ampicillin. As for the age of inocula, we observed the reverted phenotype of ΔrelAΔdnaK only at early stationary phase, but with late stationary phase inocula, ΔrelA and W3110 exhibited the same persister level. However, the assumption conflicted with the fact (p)ppGpp can trigger cells to enter persistent state. Because low (p)ppGpp concentration in ΔrelA is supposed to accelerate the decrease of persisters in ΔdnaK, but instead ΔrelA had the opposite effect in the ΔdnaK background.
The mutant strain of ΔrecA which is involved in SOS response also exhibited the same phenomenon as ΔdnaK with gentamicin and ampicillin exposure in the early stationary phase. We observed that after 4 days of gentamicin treatment, the persister levels of ΔrelA and ΔrecA decreased below the limit of detection, but ΔrelAΔrecA still had ∼102 CFU/ml viable cells left although this number was lower (∼104-fold decrease) than that of W3110 (∼106) (see Figure 6). On the 5th day of ampicillin treatment, ΔrelAΔrecA had almost 105 CFU number while ΔrelA and ΔrecA had only 102∼103 CFU (see Figure 7).
The dnaK mutant was temperature sensitive and had a deficient growth even at 30°C. Therefore, to exclude the impact of impaired growth inherent in ΔdnaK, we analyzed the persister levels of W3110, ΔrelA, ΔdnaK and ΔrelAΔdnaK at 25°C. As a result, the same phenomenon was observed for all the three antibiotics. That is, the persister level of ΔrelAΔdnaK was similar to that of the parent strain W3110 for gentamicin and ampicillin and was the same as ΔdnaK for norfloxacin (data not shown). Another evidence which supported that ΔdnaK did not obtain compensatory mutations was the small colonies during the period of persister analysis.
(p)ppGpp or Other Persister Pathways Are Insufficient Alone and a Positive Reinforcement Is Necessary to Eliminate Persister Formation
Previously we have shown the hierarchy in importance of various persister genes (Wu et al., 2015). PspF is an enhancer-binding protein and a transcriptional activator of phage shock (Psp) system (Osadnik et al., 2015). While Psp pathway is involved in indole-induced persister formation (Vega et al., 2012), pspF was demonstrated to be less important than other persister genes (Wu et al., 2015). Here, we confirmed the results of our previous study, and found that the ΔpspF had a negligible impact on persister levels of the mutant for gentamicin and ampicillin exposure, and showed only a 10 fold defect in norfloxacin exposure. Unexpectedly, the double knockout mutant of relA and pspF significantly decreased the persister numbers (105∼107 -fold) compared with ΔpspF alone and was also more prominent than ΔrelA (102∼103-fold decrease for gentamicin and ampicillin and >105-fold reduction for norfloxacin) (see Figures 2A–C). In addition, we found that the colonies formed by ΔrelAΔpspF were smaller in size and grew more slowly than W3110 (the maximum CFU number of ΔrelAcccpspF was only 108). This is in contrast to dnaK, as the ΔrelAΔpspF double mutant caused considerable persister defect, and ΔrelAΔpspF mutant was rapidly killed by all the three antibiotics tested regardless of the stage of stationary phase.
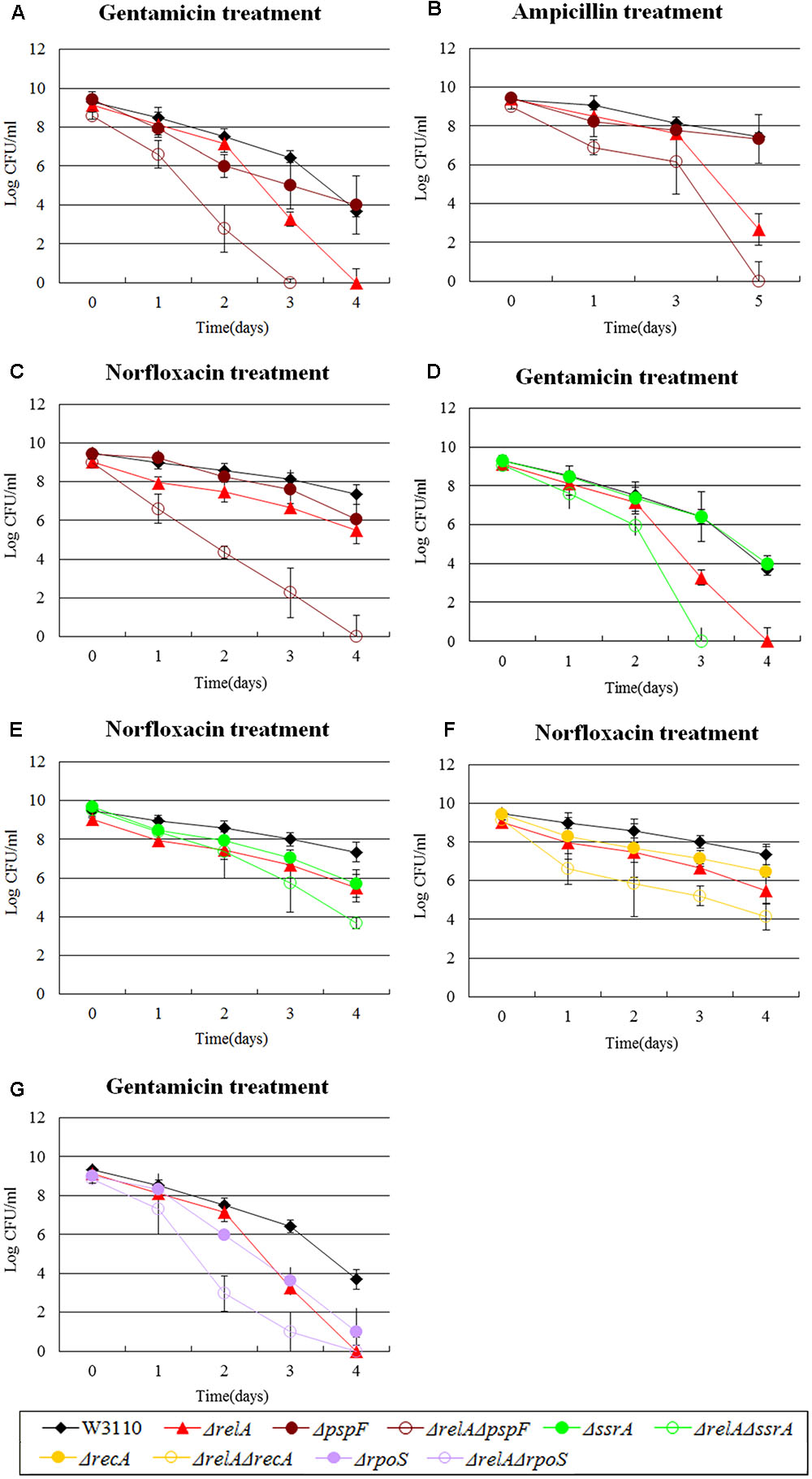
FIGURE 2. Effect of single-gene knockout mutations of pspF, ssrA, recA and rpoS and their double-gene knockout mutations with relA on E. coli persister formation. Early stationary phase cultures of W3110, ΔrelA and (A–C) pspF, (D,E) ssrA knockout mutations, (F) recA and (G) rpoS knockout mutations were exposed to gentamicin (40 μg/ml), norfloxacin (8 μg/ml) and ampicillin (200 μg/ml) for 4–5 days at 37°C without shaking. Cells (10 or 200 μl) were removed, washed and plated to determine the persister numbers at the indicated time points. Data shown are from three independent experiments and error bars indicated the standard deviations.
Among the genes we tested, there were other three genes analogous to pspF. For example, ssrA, a trans-translation gene, mediates tolerance to multiple antibiotics (Li et al., 2013). Interestingly, ΔrelAΔssrA exhibited a more significant deficiency in persister numbers when exposed to gentamicin and norfloxacin compared with W3110, and >106-fold and >100-fold decrease were observed for ΔrelA or ΔssrA in gentamicin and norfloxacin treatment, respectively (see Figures 2D,E). Similar behavior was observed for recA (SOS response) and rpoS (global regulator). Their double knockout mutants with relA were more susceptible to just one antibiotic (norfloxacin or gentamicin), with 10∼100-fold decrease for ΔrelAΔrecA and >1000-fold decrease for ΔrelAΔrpoS (at day 2∼3) compared to W3110 and their respective single gene deletion mutants (see Figures 2F,G). These findings demonstrated that a synergistic effect existing between these genes (pspF, ssrA, recA, and rpoS) and (p)ppGpp in undermining the formation of persisters, because neither ΔrelA nor the single gene knockout mutants of the four genes could produce such significant persister deficiency. Although our data shown above indicated the persister level of ΔrecA might be dependent on (p)ppGpp when challenged with gentamicin and ampicillin, it is not the case in the presence of norfloxacin. The phenomenon once again confirmed our previous study that the persistence phenomenon is not fixed but is in a dynamic state (Zhang, 2014; Wu et al., 2015).
An Antagonistic Effect between (p)ppGpp and Some Persister Genes
In our study, the persister levels of 8 double knockout mutants (ΔrelAΔclpB, ΔrelAΔhipA, ΔrelAΔmqsR, ΔrelAΔrelE, ΔrelAΔdinJ, ΔrelAΔumuD, ΔrelAΔuvrA, ΔrelAΔsucB) fell between those of ΔrelA and their own single gene deletion mutants. Based on the previous research, the high persistence of hipA7 mutant was eliminated when relA was deleted during treatment of penicillin. Therefore, we explored whether ΔhipA had the same connection with ΔrelA as hipA7 did. Data shown here indeed supported our assumption in the treatment of ampicillin, where ΔrelAΔhipA decreased the persister number (∼10 -fold change) compared with W3110 and ΔhipA but still higher than that of ΔrelA (∼104-fold higher) (see Figure 3A). However, ΔhipA did not exibit an obvious change in persister numbers compared to W3110 when challenged to ampicillin. The results were not unexpected, because previous work in our lab have found the impact of ΔhipA on persisters was not obvious (Wu et al., 2015). We attributed this distinction to different assay methods and conditions. Another heat shock protein clpB, also exhibited the similar phenomenon where the persister level of ΔrelAΔclpB was between those of ΔrelA and ΔclpB in the presence of ampicillin (see Figure 3B).
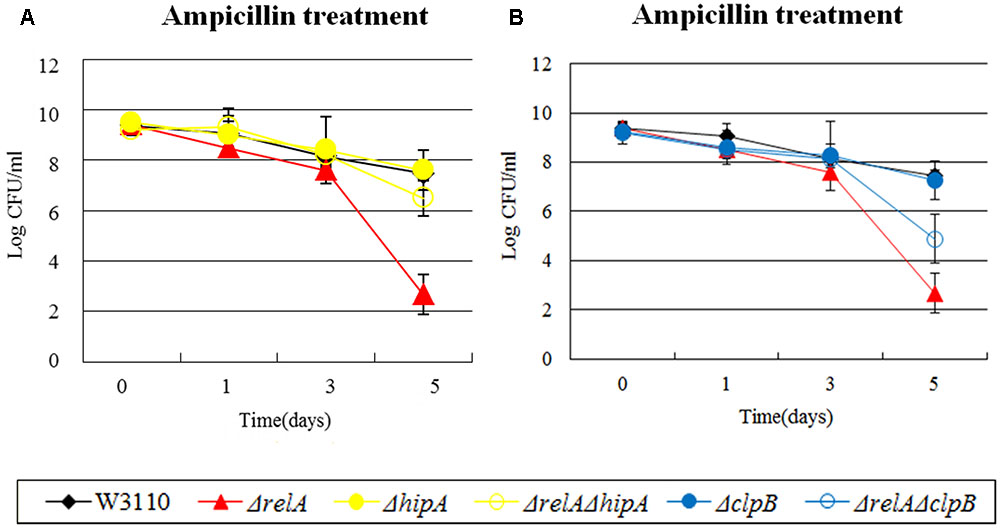
FIGURE 3. Effect of single-gene knockout mutations of hipA and clpB and their double-gene knockout mutations with relA on E. coli persister formation under ampicillin treatment. Early stationary phase cultures of W3110, ΔrelA and (A) hipA or (B) clpB knockout mutations were exposed to ampicillin (200 μg/ml) for 4–5 days at 37°C without shaking. Cells (10 or 200 μl) were removed, washed and plated to determine the persister number at the indicated time points. Data shown are from three independent experiments and error bars indicate the standard deviations.
The Impact of (p)ppGpp on Persistence Far Outweighs That of Some Persister Genes
For some genes, we discovered that the impact of their double knockout mutants on persistence was all but similar to ΔrelA when exposed to certain antibiotics. For example, in the presence of gentamicin and ampicillin, a ∼102-fold lower persister number was observed for ΔtnaA than W3110 while ΔrelAΔtnaA had a defect which was the same as ΔrelA (104∼105-fold decrease compared with W3110) (see Figures 4A,B). Likewise, the extent of decrease in persister levels of ΔrelAΔssrA, ΔrelAΔclpB and ΔrelAΔhipA was almost identical to ΔrelA on exposure to ampicillin and (or) gentamicin (see Figures 4C–E). rpoS and smpB were also in this group when challenged with norfloxacin or ampicillin.
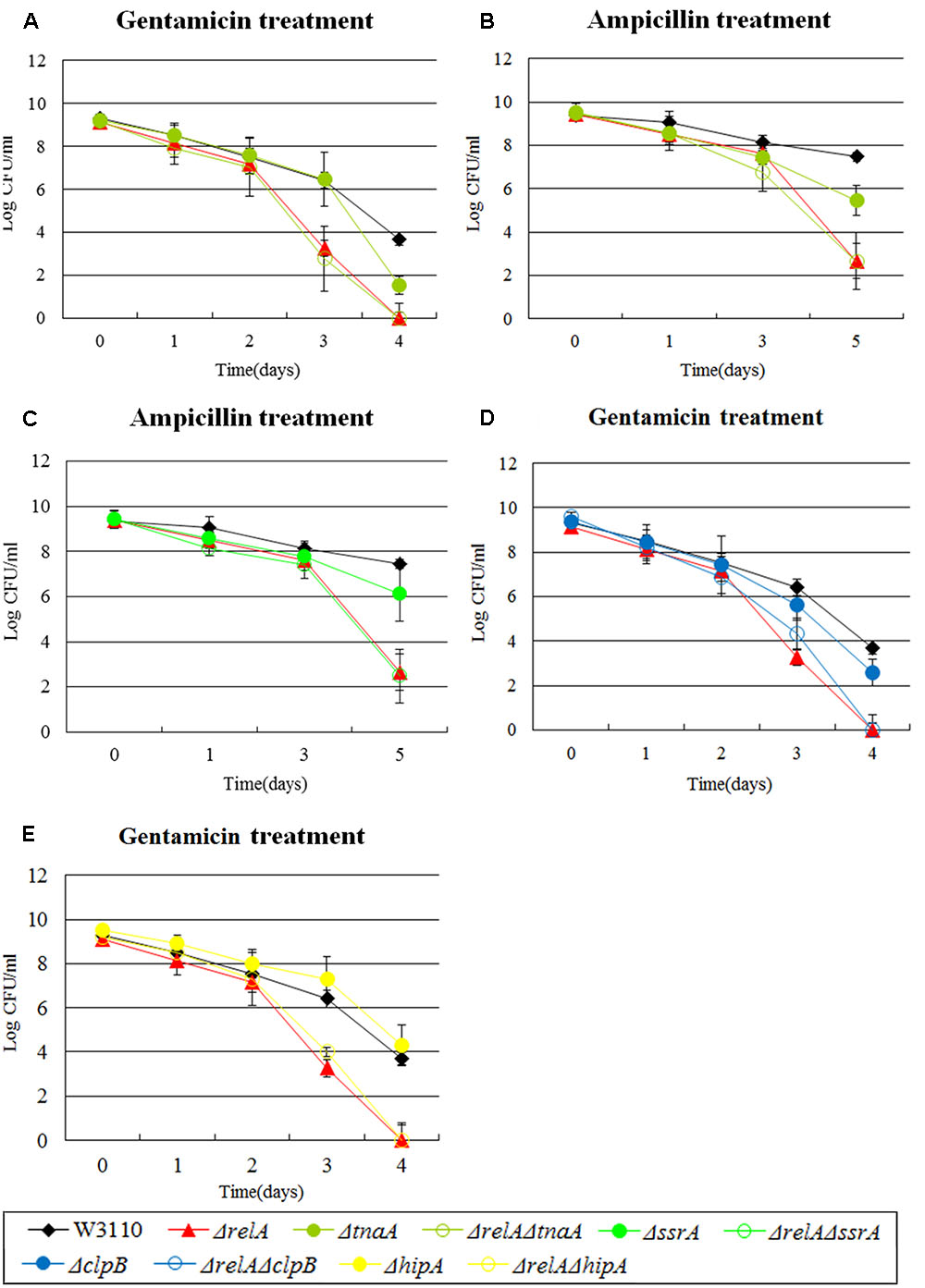
FIGURE 4. Effect of single-gene knockout mutations of tnaA, ssrA, clpB and hipA and their double-gene knockout mutations with relA on E. coli persister formation under gentamicin or ampicillin treatment. Early stationary phase cultures of W3110, ΔrelA and (A,B) tnaA, (C) ssrA, (D) clpB and (E) hipA knockout mutations were exposed to gentamicin (40 μg/ml) or ampicillin (200 μg/ml) for 4–5 days at 37°C without shaking. Cells (10 or 200 μl) were removed, washed and plated to determine the persister number at the indicated time points. Data shown are from three independent experiments and error bars indicate the standard deviations.
The Persister Levels of Some Persister Gene Mutants Are Independent of (p)ppGpp
In our study, we found persister levels of 12 persister genes (dnaK, clpB, rpoS, tnaA, sucB, smpB, umuD, uvrA, hipA, mqsR, relE, dinJ) were not affected by (p)ppGpp in the presence of at least one antibiotic. That is, the double knockout mutants showed a similar persister level to their single-gene knockout mutants (see Supplementary Table S1 and Figures 6–8). Although we considered that the low persister levels of ΔdnaK in gentamicin and ampicillin was dependent on ppGpp as ΔrelAΔdnaK had a similar persister phenotype to W3110, it seemed dnaK was independent of (p)ppGpp in the treatment of norfloxacin for the similar persister numbers of ΔdnaK and ΔrelAΔdnaK. For clpB, rpoS, tnaA, sucB, smpB, umuD, uvrA, hipA, mqsR, relE and dinJ, they also occupied at least two categories (A, B, C, D, or E). This indicated that they could not only be “positive reinforcement”/“antagonistic”/“epistasis” with (p)ppGpp but also can be independent of (p)ppGpp in the presence of some specific antibiotics.
Stationary Phase Culture Are More Suitable for Analyzing the Interaction of Persister Genes than Log Phase Cultures
Diverse methods have been applied for persister measurement by many research groups (e.g., diluted or undiluted cultures, log phase or stationary phase cultures, shaking or without shaking and CFU or OD adjusted) (Orman and Brynildsen, 2015; Volzing and Brynildsen, 2015; Wu et al., 2015; Chowdhury et al., 2016; Shan et al., 2017). However, each method had its limitations/shortcomings. For example, in the case of diluted cultures, bacteria would become so sensitive to antibiotics that the persister levels often failed to be detected. The problem with log phase culture is the discordant growth rates of different mutants which could result in different initial CFU for different mutant strains. Here, to avoid the above potential issues, we chose stationary phase cultures without dilution to enrich persisters to determine the persister levels in our mutant strains. Although biphasic killing curves are often used to demonstrate the persister phenomenon, they were not seen for most strains (see Figures 1–4). To ensure the appropriateness of our methods utilizing stationary phase cultures, we randomly selected 5 pairs of single and double gene knockout mutants and subjected them to killing curve analysis along with W3110 and ΔrelA using exponential phase cultures typically used in biphasic killing curve as demonstration of persister phenomenon. To exclude the changes in bacterial density, the survival curves of these strains were performed in the absence of antibiotics simultaneously. As expected, no significant decrease in cell viability were observed for the 10 mutant strains (data not shown). From Figures 5A–F, we observed in the treatment of gentamicin or norfloxacin, only few displayed atypical biphasic hallmark (ΔrelAΔsmpB, ΔrelAΔpspF, ΔrelAΔssrA in norfloxacin treatment and ΔdnaK in gentamicin treatment), while the majority of strains showed typical biphasic killing curves. However, in the presence of ampicillin treatment, the killing curves of six of the mutant strains did not show the biphasic characteristic (ΔrelA, ΔrelAΔdnaK, ΔrelAΔsmpB, ΔrelAΔpspF, ΔrelAΔssrA, ΔrelAΔtnaA) (see Figures 5G–I). In addition, the interaction categories in log phase were also different from those in stationary phase (see Table 1). For example, when withdrawn from log phase, ssrA and tnaA fell in type E while they were categorized as type B or D in stationary phase in gentamicin treatment. In the presence of ampicillin, instead of depending on (p)ppGpp, dnaK was classified as “epistasis” group when cultures were withdrawn from log phase. Categories of tnaA and smpB also underwent changes from type D in stationary phase to type C in log phase. When exposed to norfloxacin, category changes were only seen in tnaA (type C in log phase and type E in stationary phase).
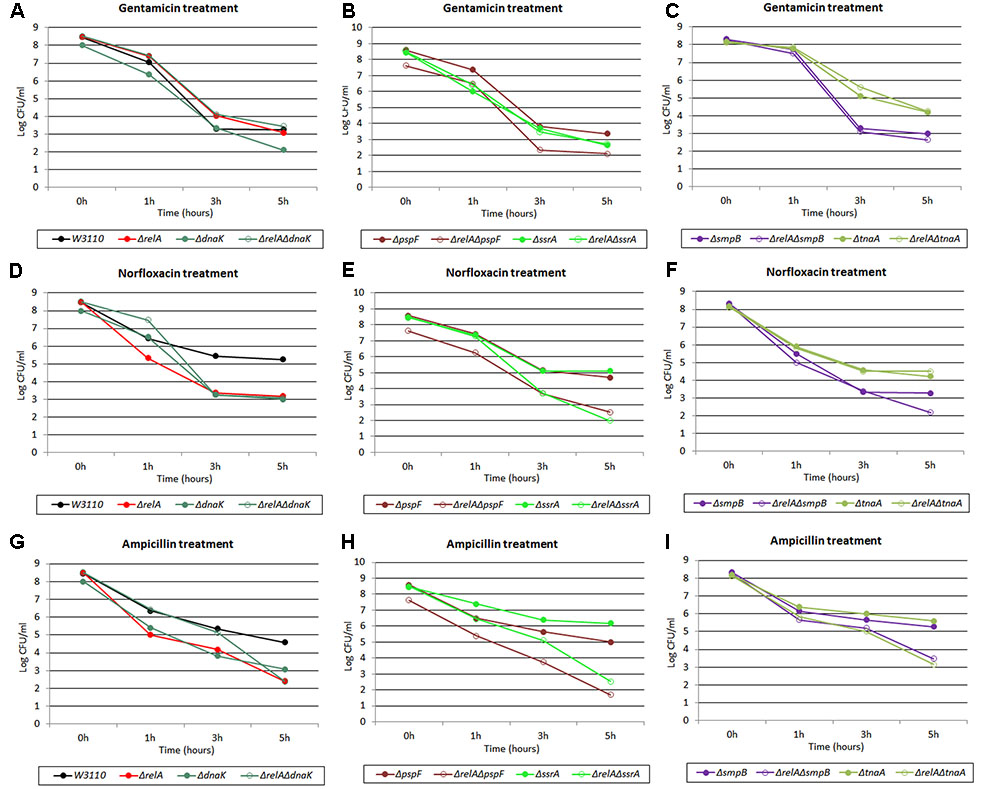
FIGURE 5. Killing curves of W3110, ΔrelA, ΔdnaK, ΔrelAΔdnaK, ΔpspF, ΔrelAΔpspF, ΔssrA, ΔrelAΔssrA, ΔtnaA, ΔrelAΔtnaA, ΔsmpB and ΔrelAΔsmpB under three antibiotics treatment in log phase stage. Log phase cultures of the 10 strains were exposed to (A–C) gentamicin (30 μg/ml), (D–F) norfloxacin (8 μg/ml) and (G–I) ampicillin (150 μg/ml) for 5 h at 37°C without shaking. Cells (10 or 200 μl) were removed, washed and plated to determine the persister number at the indicated time points.
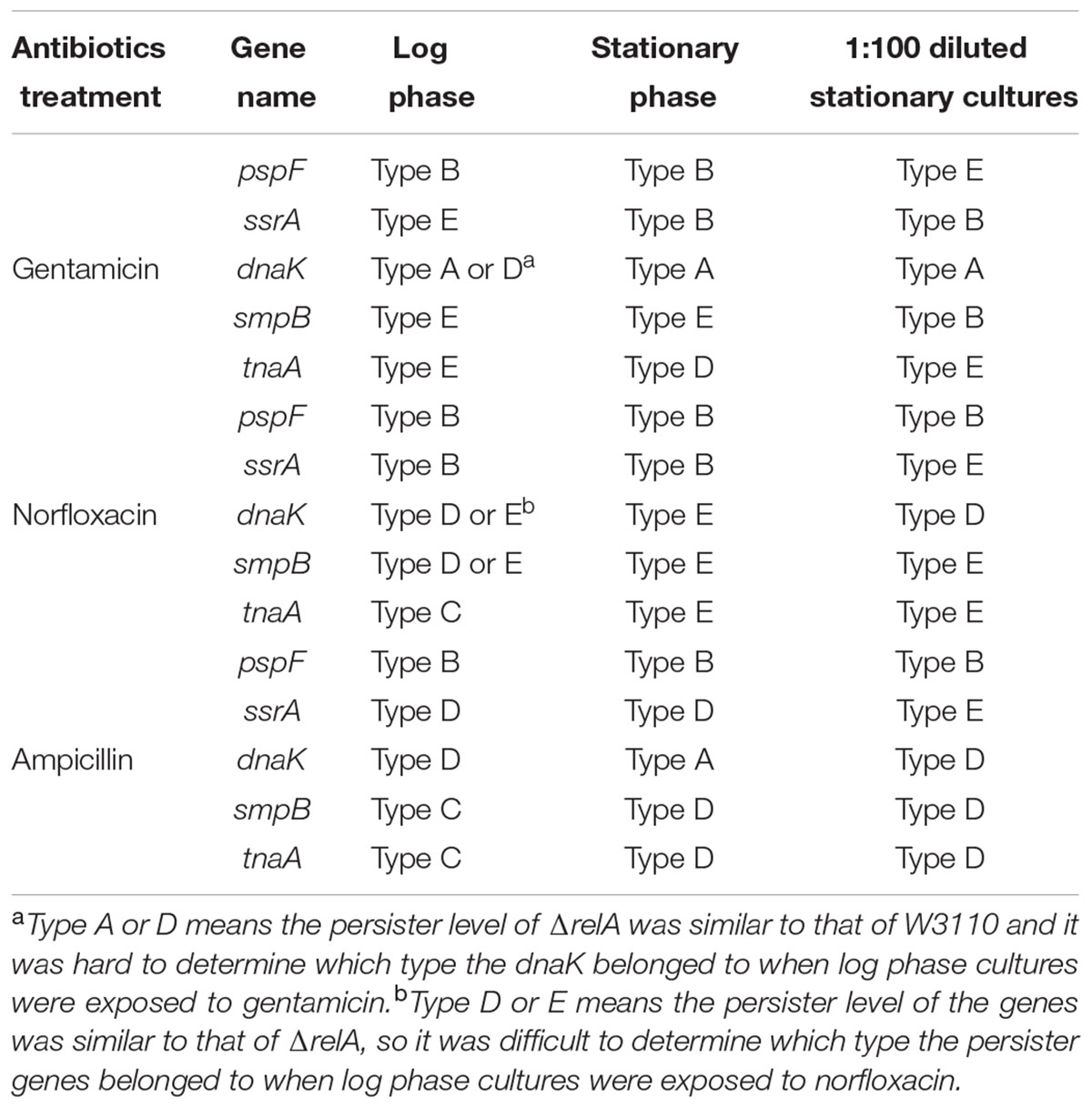
TABLE 1. The effect of the interaction of persister genes pspF, ssrA, dnaK, smpB, and tnaA with (p)ppGpp varies in different cultures in the presence of three cidal antibiotics.
Using Undiluted Cultures for Persister Assay Is More Beneficial for Identifying Genes that Interact with (p)ppGpp
Persister levels of E. coli parent strain W3110 and 11 randomly selected mutant strains (dnaK, clpB, pspF, tnaA, sucB, ssrA, smpB, recA, hipA, mqsR, relE) were also measured using (1:100) diluted cultures as described in “Materials and Methods.” A comparison of the results obtained from the two assay methods (undiluted and diluted) revealed that the interaction categories were also intimately related to experimental methods (see Table 1 and Supplementary Tables S1, S2). For example, instead of being dependent on (p)ppGpp under gentamicin and ampicillin treatment, recA was independent of (p)ppGpp for ampicillin (type E) and had a reinforcement effect with (p)ppGpp for gentamicin (type B). pspF, which belongs to type B (positive reinforcement) under all the three antibiotics when tested using undiluted cultures, was independent of (p)ppGpp for gentamicin when diluted cultures were used. Other genes, such as tnaA and sucB, also turned out to be independent of (p)ppGpp under gentamicin treatment. Taken together, our results indicated when cultures were diluted, the relationships (type A/B/C/D) of some genes with (p)ppGpp tended to become type E (irrelevant). This suggests that more genes dependent on (p)ppGpp would be revealed when undiluted cultures are used.
Discussion
Bacteria often reside in environments with myriad stresses that require the immediate switching to proper physiological state in response to new conditions. Previous studies have revealed multiple genes belonging to different pathways being involved in the formation of persisters (Zhang, 2014), however, they mainly focus on one single gene or one pathway at a time and their epistatic interactions and relationships in the context of persister gene/pathway network are mostly unknown. Our findings presented here provide new insights about the mechanisms of persisters. Although previous studies have carried out research on the role of (p)ppGpp in persistence, the majority of them involved starvation stress or only one antibiotic. For example, Korch and colleagues showed hipA7 mutant had increased persister cells dependent on (p)ppGpp synthesis using one antibiotic (penicillin) (Korch et al., 2003). It has also been shown that loss of trans-translation genes (ssrA/smpB) and clp system decreased persister formation through (p)ppGpp in ampicillin treatment and diauxie (Amato and Brynildsen, 2015). Here we systematically addressed the interactions of different persister genes with the (p)ppGpp pathway using three classical bactericidal antibiotics (gentamicin, ampicillin and norfloxacin) in persister assays using stationary phase cultures. Unexpectedly, most of the 15 common persister genes we tested had relationships with (p)ppGpp to at least one of the antibiotics for early stationary phase (5 h) cultures and their connections varied according to the antibiotics. Furthermore, we found the connections of (p)ppGpp with some genes in certain antibiotic exposures completely disappeared when the inocula were taken from stationary phase (18 h). Our findings suggest that the interactions between the common persister genes and (p)ppGpp are drug-specific and culture age-dependent.
Based on our results, the persister genes we tested have complex interactions with ppGpp and could be divided into five categories for all the three antibiotics according to their relationships with (p)ppGpp (see Figures 6–8). The first group includes genes that affect persister level in a (p)ppGpp dependent manner. Among the genes (dnaK, clpB, rpoS, pspF, tnaA, sucB, ssrA, smpB, recA, umuD, uvrA, hipA, mqsR, relE, dinJ) we tested, only dnaK (global regulator) and recA (SOS response) belong to this group. Although the dnaK mutant was shown to have (p)ppGpp accumulation at high temperatures (Brown et al., 2002), it was not tested under antibiotic conditions for persistence phenotype. Therefore, we hypothesized there might be also an increasing concentration of (p)ppGpp in ΔdnaK under antibiotics treatment. If it is true, the double knockout mutant with relA will exhibit a lower pesister level than ΔdnaK did. However, contrary to our assumption, instead of reducing persister level of ΔdnaK further, ΔrelAΔdnaK exhibited a higher persister level than both ΔdnaK and ΔrelA as it even reverted to the level of wild type strain W3110. It seems that the persister defect of ΔdnaK depended on a high concentration of (p)ppGpp. But this assumption contradicts with the well-known fact that high (p)ppGpp levels induce persister formation and drug tolerance. Another possibility is that there may be other pathways to mediate the regulation of ΔrelAΔdnaK. These pathways may be activated only when dnaK expression and (p)ppGpp are both at low levels, and their role is to maintain survival of bacteria. Therefore, further investigation of the mechanism is needed in future studies.
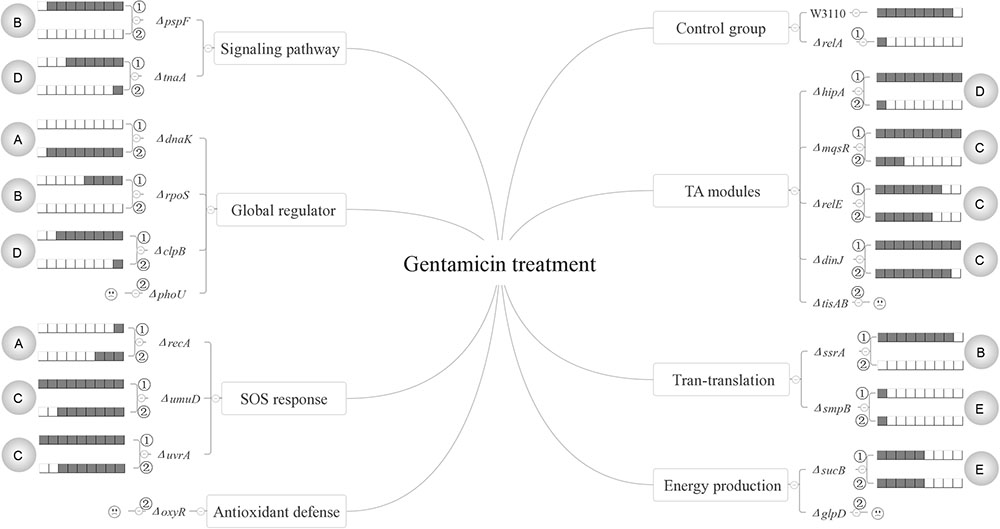
FIGURE 6. Models of the five relationships of (p)ppGpp and 15 persister genes in the presence of gentamicin. The gray circles on both flanks of the figures represent the 5 classifications of the 15 persister genes, A “dependent,” B “positive reinforcement,” C “antagonistic,” D “epistasis” and E “irrelevant”; The relative persister levels of W3110, ΔrelA and single or double-gene knockout mutant strains of the 15 persister genes are presented as the gray part of the columns;  and
and  refer to single and double-gene knockout mutant strains, respectively. The “
refer to single and double-gene knockout mutant strains, respectively. The “ ” symbolized the double-gene knockout mutant strains of four persister genes (phoU, oxyR, tisAB, glpD) failed to be constructed.
” symbolized the double-gene knockout mutant strains of four persister genes (phoU, oxyR, tisAB, glpD) failed to be constructed.
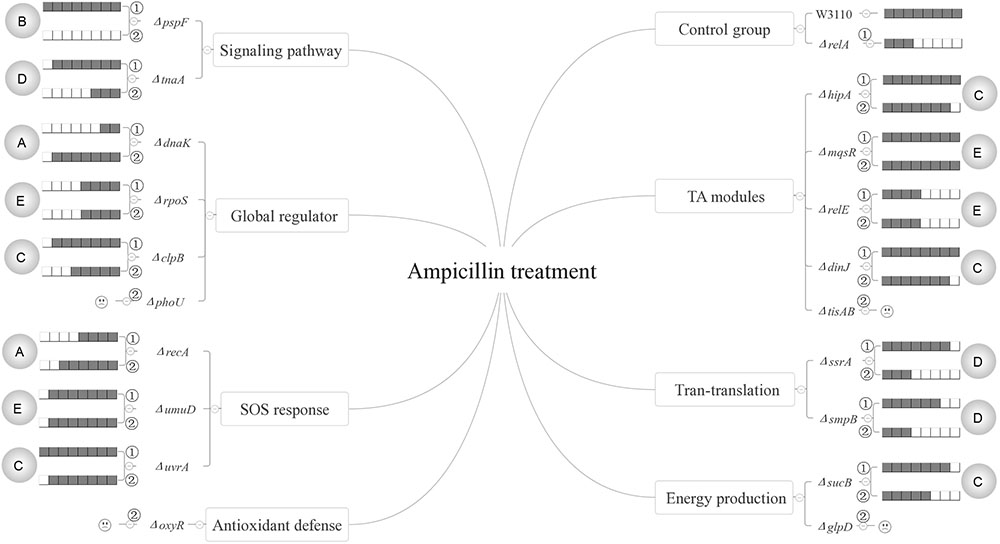
FIGURE 7. Models of the five relationships of (p)ppGpp and 15 persister genes in the presence of ampicillin. The gray circles on both flanks of the figures represent the five classifications of the 15 persister genes, A “dependent,” B “positive reinforcement,” C “antagonistic,” D “epistasis” and E “irrelevant”; The relative persister levels of W3110, ΔrelA and single or double-gene knockout mutant strains of the 15 persister genes are presented as the gray part of the columns;  and
and  refer to single and double-gene knockout mutant strains, respectively. The “
refer to single and double-gene knockout mutant strains, respectively. The “ ” symbolized the double-gene knockout mutant strains of four persister genes (phoU, oxyR, tisAB, glpD) failed to be constructed.
” symbolized the double-gene knockout mutant strains of four persister genes (phoU, oxyR, tisAB, glpD) failed to be constructed.
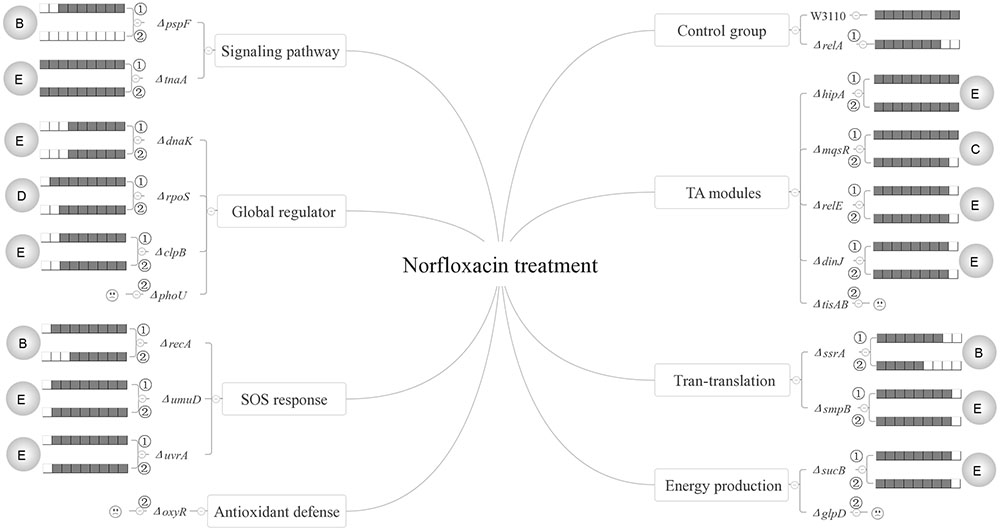
FIGURE 8. Models of the five relationships of ppGpp and 15 persister genes in the presence of norfloxacin. The gray circles on both flanks of the figures represent the five classifications of the 15 persister genes, A “dependent,” B “positive reinforcement,” C “antagonistic,” D “epistasis” and E “irrelevant”; The relative persister levels of W3110, ΔrelA and single or double-gene knockout mutant strains of the 15 persister genes are presented as the gray part of the columns;  and
and  refer to single and double-gene knockout mutant strains, respectively. The “
refer to single and double-gene knockout mutant strains, respectively. The “ ” symbolized the double-gene knockout mutant strains of four persister genes (phoU, oxyR, tisAB, glpD) failed to be constructed.
” symbolized the double-gene knockout mutant strains of four persister genes (phoU, oxyR, tisAB, glpD) failed to be constructed.
The second category is that persister genes have positive reinforcement effect with (p)ppGpp in persister formation. We defined it as “positive reinforcement effect” because their double knockout mutants had lower persister levels than either their own single gene mutants or ΔrelA. Genes in this group are involved in pathways of trans-translation (ssrA), SOS response (recA), a global regulator (rpoS), and indole signaling pathways (pspF). Of these genes, pspF showed such a relationship with (p)ppGpp for all the three antibiotics while ssrA did so for gentamicin and norfloxacin. For recA and rpoS, we observed this effect only in the presence of norfloxacin or gentamicin. The results presented here may have two explanations. The first is that the two pathways affect persister formation independently and consequently lead to the lower persister levels. But this explanation does not seem to apply for pspF in ampicillin treatment and ssrA in gentamicin treatment. Because ΔpspF and ΔssrA produced similar persister levels as W3110, the persister levels of their double-gene knockout mutants with relA were expected to be the same as ΔrelA accordingly, but surprisingly the two double knockout mutants exhibited much lower persister levels than ΔrelA did. Because Psp pathway is involved in indole-induced persister formation (Vega et al., 2012), and this suggests that there may be an interaction between stringent response and indole signaling pathways. We propose that there may be an induction of (p)ppGpp in ΔpspF which maintains its persister level. When we delete relA in ΔpspF, the (p)ppGpp induction is impaired which results in the significantly low persister level. Or another explanation is that the higher persister levels in single-gene knockout mutants (ΔpspF, ΔssrA, ΔrecA and ΔrpoS) compared with their double-gene knockout strains required the production of (p)ppGpp, so when relA was deleted, (p)ppGpp production sharply decreased and resulted in the low persister levels of the double knockout mutants. And it is readily adaptable for rpoS. Previous work showed the induction of (p)ppGpp by IPTG may positively regulate RpoS in MG1655 via dksA (Brown et al., 2002). Besides, (p)ppGpp is a plausible participant in stresses associated with RpoS regulation (Lange and Hengge-Aronis, 1994; Cassels et al., 1995; Hengge-Aronis, 1996; Loewen et al., 1998). Kayama and colleagues also revealed that in Pseudomonas aeruginosa, rpoS implemented its role in ofloxacin tolerance through (p)ppGpp although the exact mechanism involved in ofloxacin tolerance was not elucidated (Kayama et al., 2009). Based on the above reasons, we proposed that in terms of persister formation of E. coli, global regulators RpoS and (p)ppGpp are also interrelated and interact with each other, they may be both induced to maintain bacterial survival when exposed to antibiotic (gentamicin). When either of the two genes is deleted, the surviving cells will decrease. However, the extent of decrease is less than that when they are both deleted. As for single-gene deletion mutant, another gene can still function in other pathways involved in persistence. It is also possible that the four genes (pspF, ssrA, recA and rpoS) may play a key role in maintaining the low concentration of (p)ppGpp in ΔrelA via SpoT, another (p)ppGpp synthetase with a weak synthetic activity. Deletion of them can severely affect the production of ppGpp, and consequently lead to the lower persister levels. This will be tested in future studies.
Genes in the third category have a common feature where their persister levels of double knockout mutants lie between the single gene deletion mutants and ΔrelA. So genes in this group are termed “antagonistic” with (p)ppGpp. Pathways involved in this category are SOS response (umuD/uvrA), energy production (sucB), TA model (hipA/mqsR/relE/dinJ) and heat shock protein (clpB). Of the 8 double knockout mutants (ΔrelAΔumuD, ΔrelAΔuvrA, ΔrelAΔsucB, ΔrelAΔhipA, ΔrelAΔmqsR, ΔrelAΔrelE, ΔrelAΔdinJ, ΔrelAΔclpB), they all exhibited lower persister levels than their own single gene knockout strains. It is worthwhile to note that although previous studies have reported ectopic expression of hipA can lead to increased persister numbers by inducing (p)ppGpp production via RelA in the treatment of β-lactam or quinolone antibiotic(Korch et al., 2003; Bokinsky et al., 2013; Germain et al., 2013). However, all the research focused on the induction of (p)ppGpp by HipA overexpression. Here we attempted to explore if (p)ppGpp is related with hipA of W3110 by observing the persister levels in ΔhipA and ΔrelAΔhipA. The fact that ΔhipA did not exhibit obvious low persister levels relative to W3110 in the treatment of ampicillin is consistent with our previous work. However, there was discrepancy between our present and previous work (Wu et al., 2015) in the presense of gentamicin (ΔhipA exhibited a little higher persister level than W3110 in this study). The assay methods and experimental conditions may be the major contributing factors because cultures we used in this study were not diluted as our previous research did. Besides, additional deletion of relA in ΔhipA had a lower persister level compared with ΔhipA and W3110. This phenomenon indicates that HipA may have an antagonistic effect with (p)ppGpp in persister formation under ampicillin treatment. Further exploration is required to elucidate whether other genes in this group would impact the concentration of (p)ppGpp.
The fourth group of rpoS (norfloxacin), clpB (gentamicin), ssrA/smpB (ampicillin), tnaA (gentamicin and ampicillin) and hipA (gentamicin) are supposed to be controlled by or dependent on (p)ppGpp or (p)ppGpp regulated genes. The last group contains most of the genes (dnaK, clpB, rpoS, tnaA, sucB, smpB, mqsR, umuD, uvrA, hipA, relE, dinJ). Genes in this category may be irrelevant to (p)ppGpp because additional deletion of relA did not bring about different persister levels compared with their single-gene mutants.
Notably, based on our data, only two genes displayed (p)ppGpp-dependent behavior while most persister genes we tested did not depend on (p)ppGpp in our persister assay. These results indicate that (p)ppGpp might not be so important, which challenges the current thinking (Amato et al., 2013; Amato and Brynildsen, 2015). Future studies with more persister genes and stresses should be performed to validate this finding.
Furthermore, by analyzing the killing curves of 10 selected mutants as well as W3110 and ΔrelA in both log phase and stationary phase cultures, we found that hallmark of persistence as commonly shown with biphasic killing curve may not be applicable to cultures from stationary phase when challenged with the three antibiotics without culture dilution. Besides, while the majority of log phase cultures of the mutants displayed the characteristic biphasic killing in the presence of gentamicin and norfloxacin, 50% of the 12 persister gene mutant strains we tested did not show typical biphasic killing kinetics when challenged with ampicillin. Thus, it would appear that the biphasic killing hallmark of persistence is related to the bacterial growth stage and antibiotics, which means cultures withdrawn from stationary phase or cultures treated with ampicillin would not have biphasic killing curves. We therefore emphasize that there is limitation of solely relying on biphasic killing as the standard for persister demonstration, as is commonly believed or practiced in the field. This non-biphasic killing curves in persister assay are also commonly used by other research groups. For example, when Iris Keren analyzed the persister levels of M. tuberculosis under different concentrations of streptomycin, ciprofloxacin or rifampin, most killing curves did not display biphasic characteristic (Keren et al., 2011). Had we relied on the biphasic killing as the sole criterion for persister demonstration using log phase, we would have missed detection of significant relationships of persister genes with (p)ppGpp that are only observed in stationary phase instead of log phase used for biphasic killing demonstration. Besides, a comparison of two different persister assay methods (undiluted or diluted) in our study revealed that in order to exploit more (p)ppGpp-dependent genes, undiluted cultures may be a better choice. In addition, while previous studies indicate that culture age, inoculum size, type of antibiotics all affect persister levels for single persister genes (Li and Zhang, 2007; Luidalepp et al., 2011), an important observation of the current study is that these factors also affect the persister gene interactions and their types, suggesting a variable and plastic or adaptive persister gene interaction network in response to these changes. Future studies on more genes in different persister pathways are needed to gain a more comprehensive understanding of the persister gene interaction network. Such improved understanding will be important for developing more effective drugs killing persisters for improved treatment of persistent bacterial infections.
Author Contributions
YZ, WZ, and SL conceived and designed the experiments. SL, NW, SZ, and YY performed the gene knockout experiments. SL performed the data analysis. SL wrote the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (81572046).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Peng Cui, Tao Xu, and Jing Wu, the members of the Infectious Disease Department at Huashan Hospital for their helpful advice on the experiments.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01795/full#supplementary-material
References
Allison, K. R., Brynildsen, M. P., and Collins, J. J. (2011). Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature 473, 216–220. doi: 10.1038/nature10069
Amato, S. M., and Brynildsen, M. P. (2015). Persister heterogeneity arising from a single metabolic stress. Curr. Biol. 25, 2090–2098. doi: 10.1016/j.cub.2015.06.034
Amato, S. M., Orman, M. A., and Brynildsen, M. P. (2013). Metabolic control of persister formation in Escherichia coli. Mol. Cell 50, 475–487. doi: 10.1016/j.molcel.2013.04.002
Bokinsky, G., Baidoo, E. E., Akella, S., Burd, H., Weaver, D., Alonso-Gutierrez, J., et al. (2013). HipA-triggered growth arrest and beta-lactam tolerance in Escherichia coli are mediated by RelA-dependent ppGpp synthesis. J. Bacteriol. 195, 3173–3182. doi: 10.1128/jb.02210-12
Brown, L., Gentry, D., Elliott, T., and Cashel, M. (2002). DksA affects ppGpp induction of RpoS at a translational level. J. Bacteriol. 184, 4455–4465. doi: 10.1128/jb.184.16.4455-4465.2002
Cassels, R., Oliva, B., and Knowles, D. (1995). Occurrence of the regulatory nucleotides ppGpp and pppGpp following induction of the stringent response in staphylococci. J. Bacteriol. 177, 5161–5165.
Chowdhury, N., Kwan, B. W., and Wood, T. K. (2016). Persistence increases in the absence of the alarmone guanosine tetraphosphate by reducing cell growth. Sci. Rep. 6:20519. doi: 10.1038/srep20519
Cui, P., Niu, H., Shi, W., Zhang, S., Zhang, H., Margolick, J., et al. (2016). Disruption of membrane by colistin kills uropathogenic Escherichia coli persisters and enhances killing of other antibiotics. Antimicrob. Agents Chemother. 60, 6867–6871. doi: 10.1128/aac.01481-16
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645. doi: 10.1073/pnas.120163297
Dorr, T., Lewis, K., and Vulic, M. (2009). SOS response induces persistence to fluoroquinolones in Escherichia coli. PLOS Genet. 5:e1000760. doi: 10.1371/journal.pgen.1000760
Feng, J., Shi, W., Zhang, S., and Zhang, Y. (2015). Persister mechanisms in Borrelia burgdorferi: implications for improved intervention. Emerg. Microbes Infect. 4:e56. doi: 10.1038/emi.2015.56
Fung, D. K., Chan, E. W., Chin, M. L., and Chan, R. C. (2010). Delineation of a bacterial starvation stress response network which can mediate antibiotic tolerance development. Antimicrob. Agents Chemother. 54, 1082–1093. doi: 10.1128/aac.01218-09
Germain, E., Castro-Roa, D., Zenkin, N., and Gerdes, K. (2013). Molecular mechanism of bacterial persistence by HipA. Mol. Cell 52, 248–254. doi: 10.1016/j.molcel.2013.08.045
Germain, E., Roghanian, M., Gerdes, K., and Maisonneuve, E. (2015). Stochastic induction of persister cells by HipA through (p)ppGpp-mediated activation of mRNA endonucleases. Proc. Natl. Acad. Sci. U.S.A. 112, 5171–5176. doi: 10.1073/pnas.1423536112
Harms, A., Maisonneuve, E., and Gerdes, K. (2016). Mechanisms of bacterial persistence during stress and antibiotic exposure. Science 354:aaf4268. doi: 10.1126/science.aaf4268
Hauryliuk, V., Atkinson, G. C., Murakami, K. S., Tenson, T., and Gerdes, K. (2015). Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat. Rev. Microbiol. 13, 298–309. doi: 10.1038/nrmicro3448
Hengge-Aronis, R. (1996). Back to log phase: sigma S as a global regulator in the osmotic control of gene expression in Escherichia coli. Mol. Microbiol. 21, 887–893.
Kaldalu, N., Hauryliuk, V., and Tenson, T. (2016). Persisters-as elusive as ever. Appl. Microbiol. Biotechnol. 100, 6545–6553. doi: 10.1007/s00253-016-7648-8
Kayama, S., Murakami, K., Ono, T., Ushimaru, M., Yamamoto, A., Hirota, K., et al. (2009). The role of rpoS gene and quorum-sensing system in ofloxacin tolerance in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 298, 184–192. doi: 10.1111/j.1574-6968.2009.01717.x
Keren, I., Minami, S., Rubin, E., and Lewis, K. (2011). Characterization and transcriptome analysis of Mycobacterium tuberculosis persisters. mBio 2:e00100-11. doi: 10.1128/mBio.00100-11
Korch, S. B., Henderson, T. A., and Hill, T. M. (2003). Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Mol. Microbiol. 50, 1199–1213.
Lange, R., and Hengge-Aronis, R. (1994). The cellular concentration of the sigma S subunit of RNA polymerase in Escherichia coli is controlled at the levels of transcription, translation, and protein stability. Genes Dev. 8, 1600–1612.
Lewis, K. (2010). Persister cells. Annu. Rev. Microbiol. 64, 357–372. doi: 10.1146/annurev.micro.112408.134306
Li, J., Ji, L., Shi, W., Xie, J., and Zhang, Y. (2013). Trans-translation mediates tolerance to multiple antibiotics and stresses in Escherichia coli. J. Antimicrob. Chemother. 68, 2477–2481. doi: 10.1093/jac/dkt231
Li, Y., and Zhang, Y. (2007). PhoU is a persistence switch involved in persister formation and tolerance to multiple antibiotics and stresses in Escherichia coli. Antimicrob. Agents Chemother. 51, 2092–2099. doi: 10.1128/aac.00052-07
Loewen, P. C., Hu, B., Strutinsky, J., and Sparling, R. (1998). Regulation in the rpoS regulon of Escherichia coli. Can. J. Microbiol. 44, 707–717.
Luidalepp, H., Joers, A., Kaldalu, N., and Tenson, T. (2011). Age of inoculum strongly influences persister frequency and can mask effects of mutations implicated in altered persistence. J. Bacteriol. 193, 3598–3605. doi: 10.1128/jb.00085-11
Ma, C., Sim, S., Shi, W., Du, L., Xing, D., and Zhang, Y. (2010). Energy production genes sucB and ubiF are involved in persister survival and tolerance to multiple antibiotics and stresses in Escherichia coli. FEMS Microbiol. Lett. 303, 33–40. doi: 10.1111/j.1574-6968.2009.01857.x
Maisonneuve, E., Castro-Camargo, M., and Gerdes, K. (2013). (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell 154, 1140–1150. doi: 10.1016/j.cell.2013.07.048
Moyed, H. S., and Bertrand, K. P. (1983). hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 155, 768–775.
Orman, M. A., and Brynildsen, M. P. (2015). Inhibition of stationary phase respiration impairs persister formation in E. coli. Nat. Commun. 6:7983. doi: 10.1038/ncomms8983
Osadnik, H., Schöpfel, M., Heidrich, E., Mehner, D., Lilie, H., Parthier, C., et al. (2015). PspF-binding domain PspA1-144 and the PspA⋅F complex: new insights into the coiled-coil-dependent regulation of AAA+ proteins. Mol. Microbiol. 98, 743–759. doi: 10.1111/mmi.13154
Ramisetty, B. C., Ghosh, D., Roy Chowdhury, M., and Santhosh, R. S. (2016). What is the link between stringent response, endoribonuclease encoding Type II toxin-antitoxin systems and persistence? Front. Microbiol. 7:1882. doi: 10.3389/fmicb.2016.01882
Shan, Y., Brown Gandt, A., Rowe, S. E., Deisinger, J. P., Conlon, B. P., and Lewis, K. (2017). ATP-dependent persister formation in Escherichia coli. mBio 8:e02267-16. doi: 10.1128/mBio.02267-16
Steinchen, W., and Bange, G. (2016). The magic dance of the alarmones (p)ppGpp. Mol. Microbiol. 101, 531–544. doi: 10.1111/mmi.13412
Van den Bergh, B., Fauvart, M., and Michiels, J. (2017). Formation, physiology, ecology, evolution and clinical importance of bacterial persisters. FEMS Microbiol. Rev. 41, 219–251. doi: 10.1093/femsre/fux001
Van Melderen, L., and Wood, T. K. (2017). Commentary: what is the link between stringent response, endoribonuclease encoding Type II toxin-antitoxin systems and persistence? Front. Microbiol. 8:191. doi: 10.3389/fmicb.2017.00191
Vega, N. M., Allison, K. R., Khalil, A. S., and Collins, J. J. (2012). Signaling-mediated bacterial persister formation. Nat. Chem. Biol. 8, 431–433. doi: 10.1038/nchembio.915
Volzing, K. G., and Brynildsen, M. P. (2015). Stationary-phase persisters to ofloxacin sustain DNA damage and require repair systems only during recovery. mBio 6:e00731-15. doi: 10.1128/mBio.00731-15
Wang, X., and Wood, T. K. (2011). Toxin-antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl. Environ. Microbiol. 77, 5577–5583. doi: 10.1128/aem.05068-11
Wu, N., He, L., Cui, P., Wang, W., Yuan, Y., Liu, S., et al. (2015). Ranking of persister genes in the same Escherichia coli genetic background demonstrates varying importance of individual persister genes in tolerance to different antibiotics. Front. Microbiol. 6:1003. doi: 10.3389/fmicb.2015.01003
Xu, T., Han, J., Zhang, J., Chen, J., Wu, N., Zhang, W., et al. (2016). Absence of protoheme IX farnesyltransferase CtaB causes virulence attenuation but enhances pigment production and persister survival in MRSA. Front. Microbiol. 7:1625. doi: 10.3389/fmicb.2016.01625
Zhang, Y. (2014). Persisters, persistent infections and the Yin-Yang model. Emerg. Microbes Infect. 3:e3. doi: 10.1038/emi.2014.3
Keywords: persistence, persister gene, ppGpp, knockout mutant, interactions
Citation: Liu S, Wu N, Zhang S, Yuan Y, Zhang W and Zhang Y (2017) Variable Persister Gene Interactions with (p)ppGpp for Persister Formation in Escherichia coli. Front. Microbiol. 8:1795. doi: 10.3389/fmicb.2017.01795
Received: 02 June 2017; Accepted: 05 September 2017;
Published: 20 September 2017.
Edited by:
Hao Shen, Perelman School of Medicine, United StatesReviewed by:
Ewa Laskowska, University of Gdansk, PolandMario M. D’Elios, University of Florence, Italy
Copyright © 2017 Liu, Wu, Zhang, Yuan, Zhang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying Zhang, yzhang@jhsph.edu Wenhong Zhang, zhangwenhong@fudan.edu.cn
 Shuang Liu
Shuang Liu Nan Wu1
Nan Wu1 Wenhong Zhang
Wenhong Zhang Ying Zhang
Ying Zhang