- Department of Food Science, Cornell University, Ithaca, NY, United States
Among Listeria monocytogenes' four alternative σ factors, σB controls the largest regulon. As σB-dependent transcription of some genes may be masked by overlaps among regulons, and as some σB-dependent genes are expressed only under very specific conditions, we hypothesized that the σB regulon is not yet fully defined. To further extend our understanding of the σB regulon, we used RNA-seq to identify σB-dependent genes in an L. monocytogenes strain that expresses σB following rhamnose induction, and in which genes encoding the other alternative sigma factors have been deleted. Analysis of RNA-seq data with multiple bioinformatics approaches, including a sliding window method that detects differentially transcribed 5′ untranslated regions (UTRs), identified 105 σB-dependent transcription units (TUs) comprising 201 genes preceded by σB-dependent promoters. Of these 105 TUs, 7 TUs comprising 15 genes had not been identified previously as σB-dependent. An additional 23 genes not reported previously as σB-dependent were identified in 9 previously recognized σB-dependent TUs. Overall, 38 of these 201 genes had not been identified previously as members of the L. monocytogenes σB regulon. These newly identified σB-dependent genes encode proteins annotated as being involved in transcriptional regulation, oxidative and osmotic stress response, and in metabolism of energy, carbon and nucleotides. In total, 18 putative σB-dependent promoters were newly identified. Interestingly, a number of genes previously identified as σB-dependent did not show significant evidence for σB-dependent transcription in our experiments. Based on promoter analyses, a number of these genes showed evidence for co-regulation by σB and other transcriptional factors, suggesting that some σB-dependent genes require additional transcriptional regulators along with σB for transcription. Over-expression of a single alternative sigma factor in the absence of all other alternative sigma factors allowed us to: (i) identify new σB-dependent functions in L. monocytogenes, such as regulation of genes involved in 1,2-propanediol utilization (LMRG_00594-LMRG_00611) and biosynthesis of pyrimidine nucleotides (LMRG_00978-LMRG_00985); and (ii) identify new σB-dependent genes involved in stress response and pathogenesis functions. These data further support that σB not only regulates stress response functions, but also plays a broad role in L. monocytogenes homeostasis and resilience.
Introduction
Listeria monocytogenes is a Gram-positive foodborne pathogen that causes the serious invasive disease listeriosis, predominantly in susceptible populations such as the immunocompromised, pregnant women, and adults over 65 years old (Goulet et al., 2012). In the US, L. monocytogenes causes around 1,600 human listeriosis cases resulting in ~260 deaths annually (Scallan et al., 2011). L. monocytogenes' ability to rapidly respond to changing environmental conditions enables it to survive under a wide range of circumstances such as those that may be encountered during food processing as well as in animal or human hosts (Gray et al., 2006). Alternative sigma factors represent one key regulatory mechanism that allows bacteria to adjust rapidly to different environments. Differential association between alternative sigma factors and core RNA polymerase allows the RNA polymerase to recognize specific promoter sequences and initiate transcription of targeted genes under specific conditions. L. monocytogenes has up to four alternative sigma factors (σB, σC, σH, and σL) in addition to the housekeeping sigma factor σA. The four alternative sigma factors regulate transcription of genes important for virulence and for response to various stress and growth conditions (Chaturongakul et al., 2011). To date, the general stress response regulator σB is the most extensively studied alternative sigma factor in L. monocytogenes; σB has been shown to control a regulon of more than 180 genes (Raengpradub et al., 2008). Specifically, σB plays important roles in virulence and stress response, including transition to stationary phase and resistance to acid, osmotic, arsenate, oxidative, and cold stresses (O'byrne and Karatzas, 2008; Mujahid et al., 2013b).
Regulons controlled by other L. monocytogenes alternative sigma factors are less well defined. σC, which is present only in lineage II strains of L. monocytogenes, appears to have a small regulon (<10 genes) that contributes to thermal resistance (Zhang et al., 2005). σL, which is a member of the σ54 family, has a regulon of >70 genes involved in carbon and amino acid metabolism and cold stress (Raimann et al., 2009). σH appears to have the second largest regulon among the alternative sigma factors (>150 genes) (Chaturongakul et al., 2011) and has been reported to contribute to growth in minimal media, alkaline stress response, virulence (Rea et al., 2004), and competence (Liu et al., 2016; Medrano Romero and Morikawa, 2016). Previous studies in L. monocytogenes have identified considerable overlaps among the regulons controlled by these four alternative sigma factors (Chaturongakul et al., 2011; Mujahid et al., 2013a). Redundant regulation of a gene by multiple sigma factors may make it difficult to classify a gene as a member of a specific regulon following deletion of a single alternative sigma factor. Hence the commonly used approach of comparing transcript levels between a wild type (WT) strain and a corresponding isogenic mutant bearing a deletion of a targeted alternative sigma factor gene may not provide a complete picture of the regulon for a given alternative sigma factor as the approach may fail to identify all genes that are co-regulated by multiple transcriptional regulators. Furthermore, many of the previous transcriptional studies of the σB regulon (Chatterjee et al., 2006; Ollinger et al., 2008, 2009; Raengpradub et al., 2008; Toledo-Arana et al., 2009; Oliver et al., 2010; Chaturongakul et al., 2011; Ribeiro et al., 2014) have used microarray technology, which is heavily affected by extrinsic noise and is limited to annotated open reading frames (ORFs). In order to provide a more complete definition of the L. monocytogenes σB regulon, we performed RNA sequencing (RNA-seq) using an L. monocytogenes strain that expresses σB under an inducible promoter as the only active alternative sigma factor, thereby removing potential redundancies in regulation by σC, σH, and σL.
Materials and Methods
Bacterial Strains, Mutant Construction
The quadruple alternative σ factor mutant (ΔBCHL; FSL C3-0135) of L. monocytogenes strain 10403S (Mujahid et al., 2013a), which was used here as the background strain, was modified further to overexpress sigB from a rhamnose inducible promoter. The strain was constructed using methods analogous to those previously reported for construction of a strain overexpressing σH (Liu et al., 2016). Briefly, the sigB ORF was amplified from L. monocytogenes 10403S by PCR and cloned downstream of the rhamnose inducible promoter Prha in plasmid pLF1 (Fieseler et al., 2012); this construct was transformed into E. coli strain SM10 λpir to allow for subsequent conjugation of the plasmid into L. monocytogenes 10403S ΔBCHL, followed by chromosomal integration of the Prha-sigB construct in an arginine tRNA gene locus (yielding strain 10403S::ΔBCHL Prha-sigB; FSL B2-0425) (Lauer et al., 2002). A control strain (ΔBCHL-Prha; FSL B2-0429) was constructed by introducing plasmid pLF1 into the chromosome of L. monocytogenes 10403S ΔBCHL through conjugation and chromosomal integration (see Supplementary Table 1 for strains, plasmids and primers used).
Growth Conditions and Rhamnose Induction
Strains were streaked from frozen Brain Heart Infusion (BHI) glycerol stocks onto a BHI agar plate, followed by incubation at 37°C for 24 h. A single colony was subsequently inoculated into 5 ml of BHI broth in a 10 ml tube and incubated at 37°C for 18 h with shaking (220 rpm). After incubation, 50 μl BHI culture was inoculated into fresh 5 ml BHI broth in a 10 ml tube and grown to OD600 0.4–0.5 at 37°C with shaking. Induction of sigB transcription was performed by adding 250 μl of 1 M rhamnose stock solution to 5 ml OD600 0.4–0.5 bacterial cultures (for a final concentration of 50 mM rhamnose), followed by incubation at 37°C for an additional 30 min. Induction with rhamnose was performed for both 10403S::ΔBCHL Prha-sigB and ΔBCHL-Prha. qRT-PCR using the SYBR Green Master Mix Reagent (Life Technologies) and the ABI Prism 7000 Sequence Detection System (Applied Biosystems, Foster City, CA) was used to determine 50 mM rhamnose as the optimal rhamnose concentration for sigB induction (Supplementary Figure 1). For these experiments, transcript levels were determined for sigB and the housekeeping gene rpoB in strain 10403S::ΔBCHL Prha-sigB. Expression level differences were determined by the ΔΔCt method using the housekeeping gene rpoB as reference gene (Livak and Schmittgen, 2001).
RNA Isolation and Sequencing
To define the σB regulon, strains 10403S::ΔBCHL Prha-sigB and ΔBCHL-Prha were grown to log phase, followed by induction with 50 mM rhamnose for 30 min, as previously described (Liu et al., 2016). RNA isolation and cDNA library construction were also performed as previously described (Liu et al., 2016). All experiments were performed in three biological replicates. Indexed RNA-seq libraries were quantified by digital PCR and sequencing was carried out on a Hiseq 2500 (single-end, 150-bp per read) at the Cornell Core Facility for RNA-sequencing. RNA-seq data are accessible through GEO Series accession number GSE94284.
RNA-Seq Alignment, Coverage and Differential Expression Analysis
RNA-seq data analysis was performed as previously described (Liu et al., 2016). Briefly, sequence reads were aligned to a 10403S genome using the BWA-mem algorithm (Li and Durbin, 2010) and the data for coverage per base on sense and antisense strands were obtained separately using samtools (Li et al., 2009). Differential expression of genes between the two strains (ΔBCHL::Prha and ΔBCHL::Prha-sigB) was initially assessed using the Bayseq package for R version 2.2.0 (Hardcastle and Kelly, 2010). Genes were considered differentially expressed (DE) if the FDR (False Discovery Rate) was <0.05 and the Fold Change (FC) was more than 2.0 or <0.5. Genes with significantly higher transcript levels in the ΔBCHL-Prha-sigB strain were identified as upregulated by σB (FC > 2.0) while genes with significantly higher transcript levels in ΔBCHL-Prha were identified as downregulated (FC < 0.5). A sliding window method described previously (Liu et al., 2016) was also applied to identify significant differential expression of fragments along the chromosome; this method provides more sensitive identification of σB-dependent genes, particularly if genes are preceded by multiple promoters (as described in Raimann et al., 2009 and Liu et al., 2016). Briefly, the 10403S genome was divided into windows of 51 nt (window size) with 25 nt overlap (sliding window) and the RNA-Seq coverage was obtained for each window. Bayseq was then used, as described above, to identify windows with significant differential expression (FDR < 0.05 and FC > 2.0) between the ΔBCHL::Prha-sigB and the ΔBCHL::Prha strains. Overlapping windows were considered as one fragment, and fragments mapped to the same transcription unit (TU) were considered to comprise one TU. TUs are defined as all of the genes and/or noncoding RNAs (ncRNAs) that are included in a transcript initiated from a specific promoter. These analyses used an L. monocytogenes 10430S genome annotation where TUs were designated based on an RNA-seq analysis of the 10403S WT strain (Oliver et al., 2009); a file with this genome annotation in Genbank format is available from the authors upon request.
Comparison between Previously Identified σB-Dependent Genes and Genes Identified in this Study
To allow us to define newly identified σB-dependent genes, we compiled data on the L. monocytogenes σB regulon from 12 independent studies that used genome-wide transcriptomics and proteomics approaches to compare different L. monocytogenes WT strains and corresponding isogenic sigB deletion mutants (Wemekamp-Kamphuis et al., 2004; Chatterjee et al., 2006; Abram et al., 2008a,b; Ollinger et al., 2008, 2009; Orsi et al., 2008; Raengpradub et al., 2008; Oliver et al., 2009, 2010; Toledo-Arana et al., 2009; Chaturongakul et al., 2011; Ribeiro et al., 2014). Combined, these studies identified a total of 902 different genes as σB-dependent; only genes that were identified here, but not among these previously reported 902 genes were classified as “newly identified σB-dependent genes”. We also defined a “high confidence” core σB regulon for L. monocytogenes lineage II (which includes strain 10403S, used here), which contained 184 genes that have been reported as σB-dependent in at least three independent experiments among the 12 studies detailed above. This “high confidence” core σB regulon was used to define the previously identified σB-dependent genes that were not found to be differentially expressed (NDE) in our study reported here. The datasets supporting the conclusions of this article are available in the GEO repository (Private link for reviewers: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=gbktssowflaften&acc=GSE94284).
Promoter Search and Consensus
The transcription start sites (TSS) and 5′ untranslated regions (UTRs) of the differentially expressed TUs were manually annotated based on visual inspection of the locations where RNA-seq coverage data abruptly increased. Subsequently, the 5-50 nt regions upstream of the TSSs were manually scanned for σB-dependent promoter consensus sequences (GTTT-N12−16-GGGTAT). A consensus sequence logo of the newly identified σB-dependent promoters was generated using the WebLogo generator (Crooks et al., 2004).
5′ Rapid Amplification of cDNA Ends (5′ RACE) Analysis
Six newly identified σB-dependent promoters were selected to map promoter regions with the 5′ RACE system (ThermoFisher); these experiments were performed in three biological replicates. Briefly, isolated RNA was reverse transcribed into cDNA with gene-specific primers and cDNA was tailed with dCTP by terminal transferase. The products were then amplified with a nested gene-specific primer and an abridged anchored primer in a touchdown PCR with DreamTaq PCR master mix (ThermoFisher). PCR products were visually analyzed with agarose gel electrophoresis.
Go Term Enrichment Analysis
Gene Ontology (GO) enrichment analysis to identify GO terms enriched among all 201 genes identified in the 105 σB-dependent TUs was performed using the GOseq 1.24.0 package for R (Young et al., 2010) as previously described (Tang et al., 2015). All GO terms containing more than five genes were analyzed for enrichment. FDR correction for multiple testing was applied and only GO terms with FDR < 0.05 were considered significant.
Comparative Genomics of Newly Identified σB-Dependent Promoters
Newly identified σB-dependent promoters were searched against a database containing 27 finished genomes representing L. monocytogenes lineage I (n = 10), L. monocytogenes lineage II (n = 9), L. monocytogenes lineage III (n = 4), Listeria innocua (n = 1), Listeria seeligeri (n = 1), Listeria ivanovii (n = 1), and Listeria welshimeri (n = 1) using BLAST [32]. Matches with coverage >70% and identity >60% were considered significant and the promoters were, therefore, considered to be present in the respective genome.
Results
Combined Bioinformatics Analyses Identify a Number of New σB-Dependent Genes That Showed Significant Differential Transcript Levels in the Respective ORF
Using RNA-seq data from an L. monocytogenes strain with deletions in genes encoding all four alternative sigma factors (ΔBCHL) as well as from a corresponding isogenic strain where sigB was re-introduced under rhamnose regulation (ΔBCHL-Prha-sigB), we employed two separate bioinformatics approaches to identify σB-dependent genes. These bioinformatics approaches included (i) Bayseq analysis that assessed differential expression using RNA-seq coverage for the full length of all annotated genes (“the traditional Bayseq approach”) and (ii) Bayseq analysis that assessed differential expression using RNA-seq coverage for sliding windows that covered the complete L. monocytogenes chromosome (“the sliding window approach”).
With the traditional Bayseq approach, we initially identified 141 genes, including 7 ncRNAs, that showed significantly higher transcripts levels (FDR < 0.05; FC > 2.0) in the L. monocytogenes strain overexpressing σB (ΔBCHL-Prha-sigB), as compared to the ΔBCHL strain that does not express any alternative sigma factors (Table 1). Among the 141 genes with higher transcript levels, 117 were preceded by upstream σB-dependent promoters. When the sliding window approach was applied, we identified 299 fragments that showed significantly higher transcripts levels (FDR < 0.05; FC > 2.0) in the L. monocytogenes strain overexpressing σB. These fragments represented 177 genes, including 7 ncRNAs; 124 of these genes (including all 7 ncRNAs) were preceded by σB-dependent promoters (Supplementary Table 2). Together, these two analyses identified 193 genes as upregulated by σB; 133 of these genes (including 7 noncoding RNAs) were preceded by σB-dependent promoters (Table 1). These 133 genes are located in 93 TUs that each includes a σB-dependent promoter. Overall, 141 of the 193 upregulated genes identified here had been reported as σB-dependent in previous microarray, RNA-seq, or proteomics studies in L. monocytogenes (Wemekamp-Kamphuis et al., 2004; Chatterjee et al., 2006; Abram et al., 2008a,b; Ollinger et al., 2008, 2009; Raengpradub et al., 2008; Oliver et al., 2009, 2010; Toledo-Arana et al., 2009; Chaturongakul et al., 2011; Ribeiro et al., 2014). Among the 52 genes newly identified, with these approaches, as upregulated by σB, 16 genes (representing 10 TUs) were preceded by a putative σB-dependent promoter, while 36 did not include an upstream σB-dependent promoter. In the subsequent sections, we will only focus on those genes that showed both evidence for higher transcript levels in the presence of σB and have an upstream σB-dependent promoter.
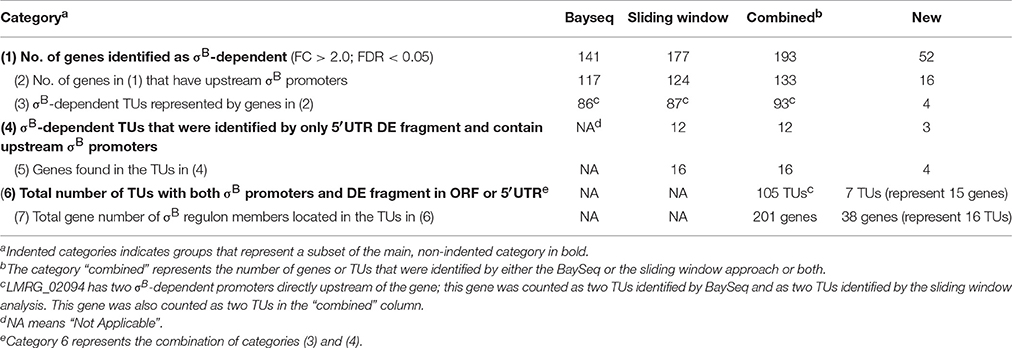
Table 1. Summary of traditional Bayseq and sliding window analyses results for upregulated genes and TUs.
With the traditional Bayseq approach, we also identified 18 genes that showed significantly lower transcripts levels (FDR < 0.05; FC < 0.5; see Supplementary Table 3) in the L. monocytogenes strain overexpressing σB (ΔBCHL-Prha-sigB), as compared to the ΔBCHL strain. Among these 18 downregulated genes, 9 had been reported as downregulated in previous studies (Wemekamp-Kamphuis et al., 2004; Chatterjee et al., 2006; Abram et al., 2008a,b; Ollinger et al., 2008, 2009; Raengpradub et al., 2008; Oliver et al., 2009, 2010; Toledo-Arana et al., 2009; Chaturongakul et al., 2011; Ribeiro et al., 2014). Most of the nine genes newly identified as downregulated by σB encode transport proteins, including (i) LMRG_01249 and LMRG_01248, which encode subunits of a PTS galactitol transporter; (ii) LMRG_01581, which encodes ArpJ, an amino acid ABC transporter permease; and (iii) LMRG_01595, which encodes an MFS transporter permease. Other downregulated genes include LMRG_01332, which encodes Listeria adhesion protein (Lap), an alcohol acetaldehyde dehydrogenase involved in pathogenesis (Jagadeesan et al., 2010); LMRG_00198, encoding a phosphoglycerate mutase; LMRG_01596, encoding a shikimate 5-dehydrogenase, and LMRG_01597 encoding an NADH oxidase. Interestingly, one of the nine newly identified σB-downregulated genes was LMRG_01250, which encodes a putative transcriptional regulator.
Sliding Window Analyses of 5′UTR Regions Identified 12 Additional TUs with σB-Dependent Transcription and Upstream σB-Dependent Promoters in the Absence of Significant Differential Transcript Levels in the Respective ORF
The sliding window approach used here not only identified differentially regulated genes via identification of differentially transcribed fragments that mapped within an ORF (described above), but also identified differentially transcribed fragments that mapped into different 5′UTR, even if the associated downstream genes did not show evidence for differential transcription. This approach thus identified additional TUs (which may include one or multiple genes) as σB-dependent. For example, in the intergenic region upstream of srtA we identified a fragment with an FC of 3.77 located upstream of the srtA σA-dependent promoter (Figure 1). Visual inspection of the region upstream of this fragment identified a σB-dependent promoter. Hence, using the sliding window approach, we identified srtA as a new σB-dependent TU. Overall, this approach identified 12 additional TUs (Supplementary Table 4) with σB-dependent transcription and upstream σB-dependent promoters. For each of these TUs, the associated genes located in the TU did not show significant evidence for higher transcript levels in the strain overexpressing σB.
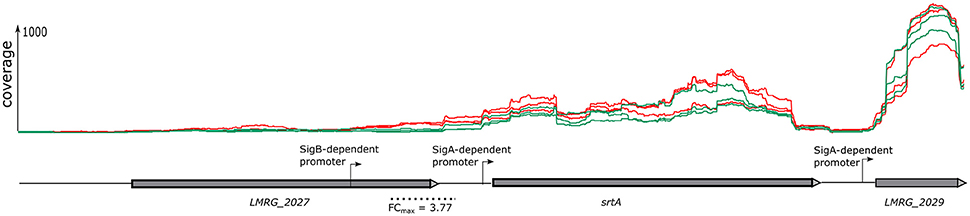
Figure 1. Gene co-transcribed from both σB-dependent and a σA-dependent promoters. RNA-seq coverage is shown for the ΔBCHL Prha-sigB strain (red) and ΔBCHL-Prha strain (green). Transcription start sites (TSS) are indicated by arrows labeled with the regulating sigma factor. The dotted line below the schematic represents the region found to be differentially expressed (FDR < 0.05) with the sliding window approach. The highest window fold change (FC) is shown under the dotted line.
Overall Analyses Reveal a Total of 105 σB-dependent TUs That Cover 201 Genes, Including a Number of Genes Not Previously Identified as Part of the σB Regulon
Overall, our analyses identified 105 σB-dependent TUs (i.e., TUs upregulated in the presence of σB) that are preceded by σB-dependent promoters; this includes (i) 93 TUs that showed significant differential transcript levels in one or multiple ORFs that are part of a TU and (ii) 12 TUs that showed significant differential transcript levels in a fragment located in the 5′UTR. Seven of these 105 TUs had not been previously identified as σB-dependent (meaning none of the 15 genes in these seven TUs had been identified as σB-dependent in previous studies (Wemekamp-Kamphuis et al., 2004; Chatterjee et al., 2006; Abram et al., 2008a,b; Ollinger et al., 2008, 2009; Raengpradub et al., 2008; Oliver et al., 2009, 2010; Toledo-Arana et al., 2009; Chaturongakul et al., 2011; Ribeiro et al., 2014). Overall, the 105 σB-dependent TUs include 201 different genes; 38 of these 201 genes had not been previously identified as members of the L. monocytogenes σB regulon (Tables 1, 2). While additional genes were newly identified as σB-dependent through our bioinformatics approaches (see Table 1), not all of these genes met the dual criteria of having both an upstream σB-dependent promoter and differential upregulation in the ΔBCHL-Prha-sigB strain.
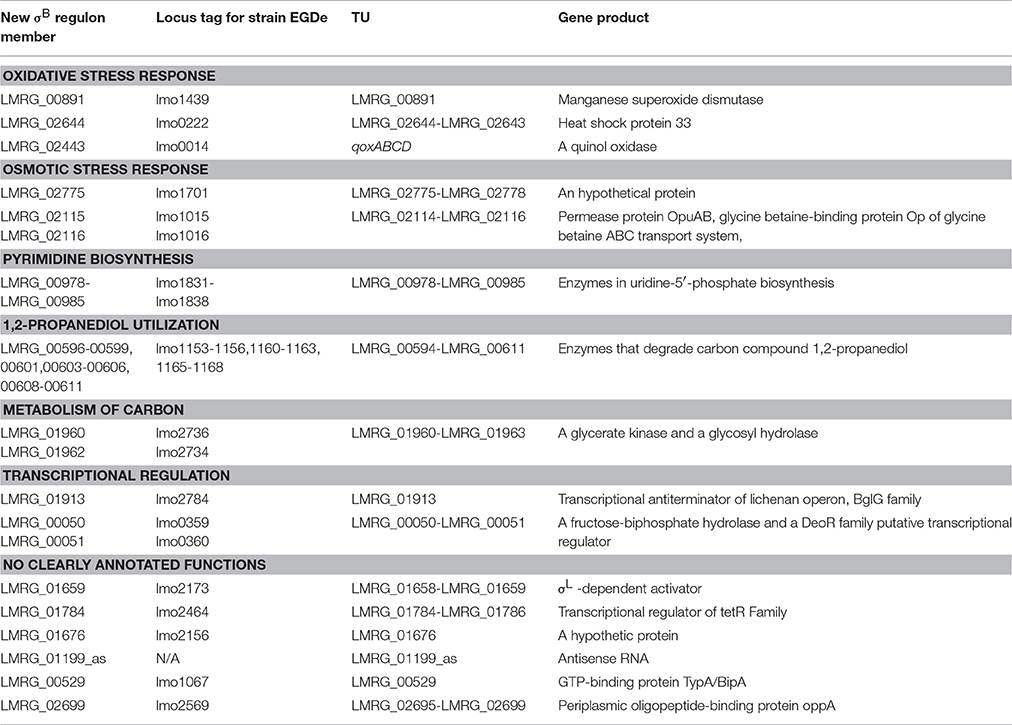
Newly Identified σB Regulon Members Include Genes Involved in Stress Response, Metabolism of Carbon and Nucleotides, and Virulence
The 38 genes newly identified as members of the σB regulon (Table 2) encode proteins involved in oxidative and osmotic stress response, metabolism of energy, carbon and nucleotides, transcriptional regulation, and other functions of L. monocytogenes. Newly identified σB-dependent genes involved in oxidative stress response include LMRG_00891, LMRG_02644, and LMRG_02443, which is the second gene in the qoxABCD operon. Our data newly identified σB-dependent promoters upstream of all of these operons. We also newly identified LMRG_02775 as a member of the σB regulon. While the other members of the LMRG_02775-LMRG_02778 operon had previously been identified as σB-dependent (Oliver et al., 2010; Chaturongakul et al., 2011), we newly identified a σB-dependent promoter upstream of this operon, suggesting direct σB-dependent transcription of this 4-gene operon, which encodes four proteins, including one OsmC/Ohr family protein. While the functions of the genes in this operon have not yet been elucidated, OsmC proteins are induced by ethanol and osmotic stresses and Ohr proteins are induced by organic peroxide and are involved in organic hydroperoxide detoxification in Gram-negative bacteria (Atichartpongkul et al., 2001). In other intracellular pathogens such as Mycobacterium tuberculosis, OsmC proteins have been reported to protect against organic hydroperoxide stress (Saikolappan et al., 2011), suggesting this operon may contribute to oxidative or osmotic stress response in L. monocytogenes. We also newly identified two members of the LMRG_02114-LMRG_02116 operon as part of the σB regulon and newly identified a σB-dependent promoter upstream of this operon. This operon encodes the OpuAA, OpuAB, and OpuAC subunits of the glycine betaine ABC transport system. Expression of the opuA operon is under osmotic control in B. subtilis, and a B. subtilis mutant strain lacking the OpuA transport system showed a considerably decreased ability to uptake the osmoprotectant glycine betaine (Kempf and Bremer, 1995).
Our study also identified a number of new members of the σB regulon involved in energy, carbon and nucleotide metabolism. Specifically, we newly identified LMRG_00978-LMRG_00985 as a σB-dependent TU; genes in this TU encode a set of enzymes involved in uridine-5′-phosphate biosynthesis, a pyrimidine ribonucleotides de novo biosynthesis pathway (Figure 2). Importantly, our data also indicate that all genes in the LMRG_00594-LMRG_00611 operon are part of the σB regulon; this operon encodes all enzymes involved in utilization of carbon compound 1,2-propanediol, which is an important carbon source during infection for bacterial pathogens such as S. enterica (Bobik et al., 1999) and was reported to be degraded by L. innocua (Xue et al., 2008). In addition to providing direct RNA-seq evidence for significant differential transcript levels for 6 genes in this operon, which had not been previously identified as σB-dependent, we also identified a σB-dependent promoter upstream of this operon; in previous studies, only 5 genes in this 18-gene operon had been identified as σB-dependent (Wemekamp-Kamphuis et al., 2004; Chatterjee et al., 2006; Abram et al., 2008a,b; Ollinger et al., 2008, 2009; Raengpradub et al., 2008; Oliver et al., 2009, 2010; Toledo-Arana et al., 2009; Chaturongakul et al., 2011; Ribeiro et al., 2014). Our data also newly identified a σB-dependent promoter upstream of the three-gene operon LMRG_01960-LMRG_01962 with LMRG_01960 and LMRG_01962 newly identified as σB-dependent. We also newly identified a σB-dependent promoter upstream of LMRG_01432-LMRG_01431, suggesting direct σB regulation of this operon.
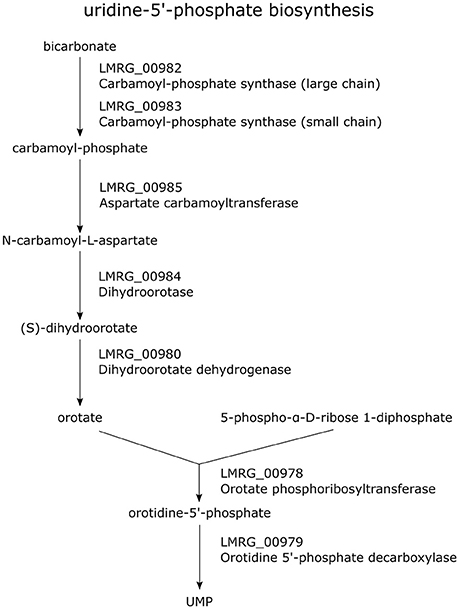
Figure 2. Pathway of uridine-5′-phosphate biosynthesis. Genes encoding the enzymes involved in this pathway are labeled in purple. All genes in the TU LMRG_00985-LMRG_00978 except LMRG_00981 are shown in this pathway.
We also identified new members of the σB regulon that encode transcriptional regulators, further supporting that σB is part of a complex regulatory network in L. monocytogenes. Specifically, we newly identified LMRG_01913, which encodes a lichenan operon transcriptional antiterminator as a σB-dependent TU. In addition, we newly identified LMRG_00050-LMRG_00051 as a σB-dependent TU, in which LMRG_00051 encodes a putative transcriptional regulator. LMRG_01659, encoding a σ54-dependent activator (Arous et al., 2004) and LMRG_01784, encoding a TetR family transcriptional regulator, were also newly identified as members of the σB regulon. While the other genes in TUs LMRG_01658-LMRG_01659 and LMRG_01784-LMRG_01786 had previously been reported as σB-dependent (Raengpradub et al., 2008; Oliver et al., 2009, 2010; Ollinger et al., 2009; Toledo-Arana et al., 2009; Chaturongakul et al., 2011; Ribeiro et al., 2014), we newly identified σB-dependent promoters upstream of both operons.
Our data also identified a σB-dependent promoter upstream of a TU including a single gene encoding sortase A, srtA. SrtA is involved in proteolysis and virulence in L. monocytogenes (Garandeau et al., 2002). Additional newly identified σB-dependent TUs (where none of the genes in a TU had previously been identified as σB-dependent) included LMRG_00529 and two TUs (LMRG_01676 and LMRG_01199_as) with no clearly annotated functions.
Gene Ontology Analyses Identify a Role for σB in Regulating Genes Encoding Functions Involved in Transport, Homeostasis, Pathogenesis, and Nucleotide Metabolism
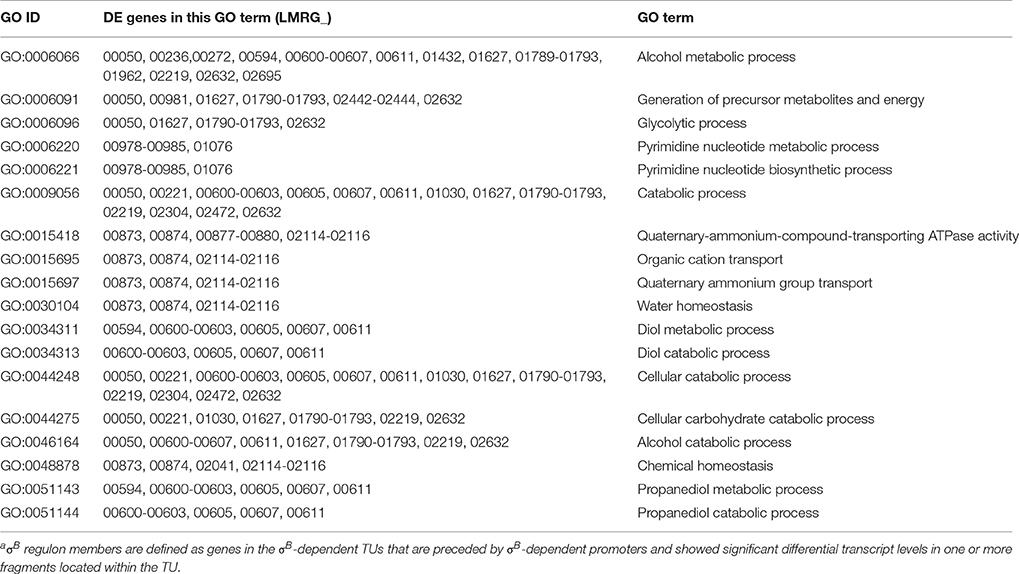
Gene ontology (GO) enrichment analysis performed for the σB regulon (i.e., all 201 genes that were part of the 105 σB-dependent TUs identified here) found 18 GO terms to be overrepresented (see Table 3), including GO terms related to transport, homeostasis, metabolic and catabolic processes. In addition, GO terms related to “pyrimidine nucleotide biosynthetic and metabolic processes”; “glycolysis, metabolic and catabolic processes of propanediol, diol and alcohol,” were also over-represented, suggesting a role for σB in regulating general metabolic functions.
Conservation of Newly Identified σB-Dependent Promoters in L. monocytogenes and Listeria spp.
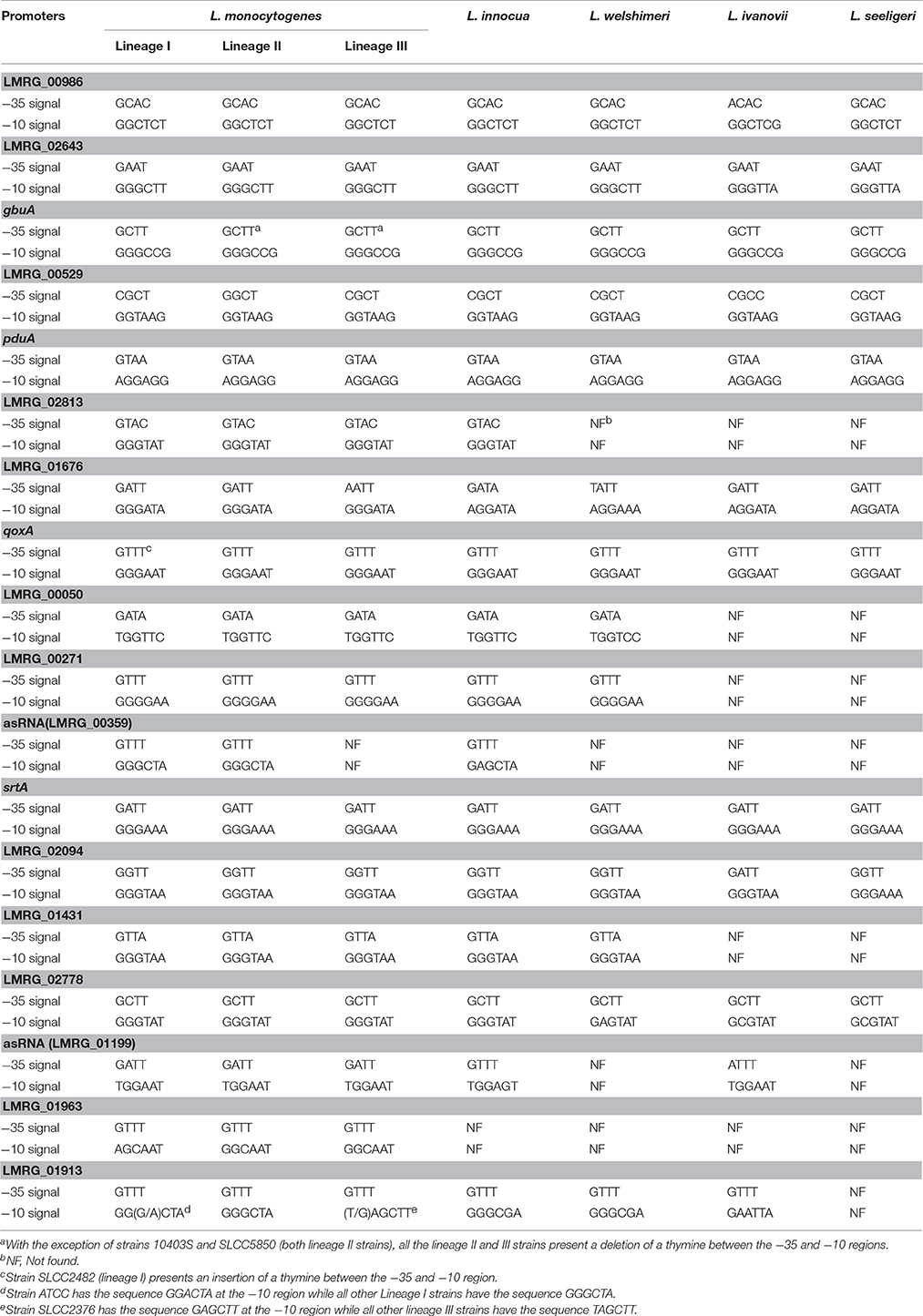
Overall, we newly identified 18 putative σB-dependent promoters here (see Figure 3 for a consensus sequence). 5′ RACE performed on six of these promoters confirmed five of these promoters as σB-dependent (Figure 4). For pduA, where 5′RACE did not detect a clear σB-dependent transcript, a putative σA-dependent promoter was found 14 nt upstream of the putative σB-dependent promoter; transcripts from this σA-dependent promoter may have masked the σB-dependent transcript. Across all 18 newly identified putative σB-dependent promoters, the first three nucleotides of the −10 signal sequence (GGG) are generally conserved among both these newly identified putative σB-dependent promoters and the previously published σB-dependent promoter sequences (Oliver et al., 2009), the last three nucleotides appear to be less conserved among the new promoters identified here, as compared to the previously reported σB-dependent promoter sequences (Figure 3). Through comparative genomics, we assessed that all but one of the 18 σB-dependent promoters newly identified in L. monocytogenes 10403S were present in the 23 additional L. monocytogenes genomes analyzed here; the σB-dependent promoter upstream of the anti-sense RNA (asRNA), which overlaps LMRG_00359 in the opposite strand, was not found in the four lineage III genomes analyzed (Table 4). Overall, −35 and −10 promoter regions were highly conserved among different strains and lineages, even though some promoters show lineage- or strain-specific sequence features (Table 4). Further analysis showed that only 10 of the 18 σB-dependent promoters newly identified in L. monocytogenes were found in all 5 Listeria species analyzed (i.e., L. monocytogenes, L. innocua, L. ivanovii, L. welshimeri, and L. seeligeri) (Table 4). While these comparisons suggest that we identified new σB-dependent functions that are largely conserved in L. monocytogenes and to a lesser extent in other Listeria sensu strictu species, future comparative genomics analyses that utilize additional closed genomes as they become available will help to further define lineage-, strain-, and species-specific members of the SigB regulon.
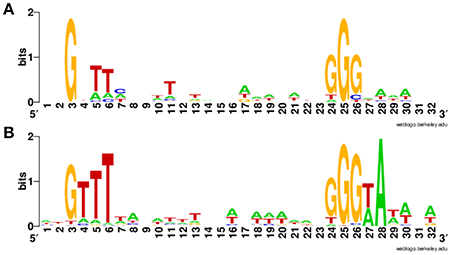
Figure 3. Sequence logo for newly identified putative σB-dependent promoters. (A) Sequence logo for the 18 σB-dependent promoters associated with newly identified σB-dependent genes. (B) Sequence logo for σB-dependent promoters published in previous study (Oliver et al., 2009).
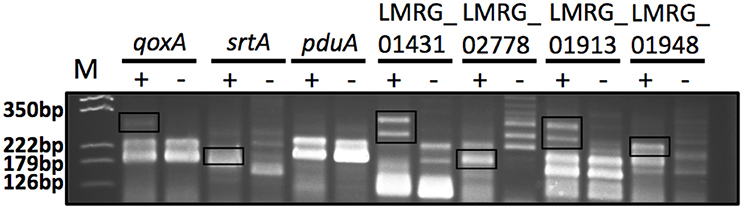
Figure 4. 5′RACE PCR confirmation of six putative σB-dependent promoters newly identified here by RNA-seq. The six putative σB-dependent promoters selected for confirmation are located upstream of qoxA, srtA, pduA, LMRG_01431, LMRG_02778, and LMRG_01913. LMRG_01948 was used as a positive control, as it was reported to have a strong σB-dependent promoter (Abram et al., 2008b). The image shows the 5′RACE PCR products in a 3% agarose gel; the two lanes for each target represent reaction performed with RNA isolated from the ΔBCHL Prha-sigB strain, which expresses σB (+) and the ΔBCHL-Prha strain, which does not express σB (−). The σB-dependent transcript bands are shown in rectangles. Multiple 5′RACE bands were expected for these genes as they were all initially characterized by having a putative σB-dependent promoter as well as an additional σA-dependent promoter. Five of six promoters selected for confirmation displayed clear σB-dependent transcript bands, supporting the existence of σB-dependent promoters. 5′RACE PCR for pduA did not yield a band that confirmed the putative σB-dependent promoter that was identified by RNA-seq; a putative σA-dependent promoter was found 14 nt upstream of the putative σB-dependent promoter; transcripts from this σA-dependent promoter may have masked the σB-dependent transcript. The expected sizes of the σB-dependent transcript bands are around 250 bp for qoxA, 170 bp for srtA, 165 bp for pduA, 275 bp for LMRG_01431, 185 bp for LMRG_02778, 275 bp for LMRG_01913, and 210 bp for LMRG_01948. The image was adjusted for contrast, intensity levels and saturation using Photoshop; manipulations were performed on the whole picture and no specific bands were enhanced or modified. The results shown here are representative of three biological replicates.
Comparison with Previously Reported σB-Dependent Regulon Members Identifies a Number of σB-Dependent Genes That Are Also Regulated by σA
In order to compare the σB-dependent genes identified here to previously reported σB-dependent genes, we used data from previous microarray, RNA-seq and proteomic studies in different strains and under different stress conditions (Wemekamp-Kamphuis et al., 2004; Chatterjee et al., 2006; Abram et al., 2008a,b; Ollinger et al., 2008, 2009; Raengpradub et al., 2008; Oliver et al., 2009, 2010; Toledo-Arana et al., 2009; Chaturongakul et al., 2011; Ribeiro et al., 2014), to define a core σB regulon for L. monocytogenes lineage II, which includes the strain we used here (10403S). Overall, 184 genes have been reported as σB-dependent at least three times in prior publications (representing high confidence for each as a member of the σB regulon). Our data showed that 110 of the 193 genes that were identified as significantly upregulated by σB in our study also were included in this “high confidence regulon” of 184 genes. Therefore, overexpression of σB in ΔBCHL failed to identify 74 genes previously reported as upregulated by σB (Supplementary Table 5).
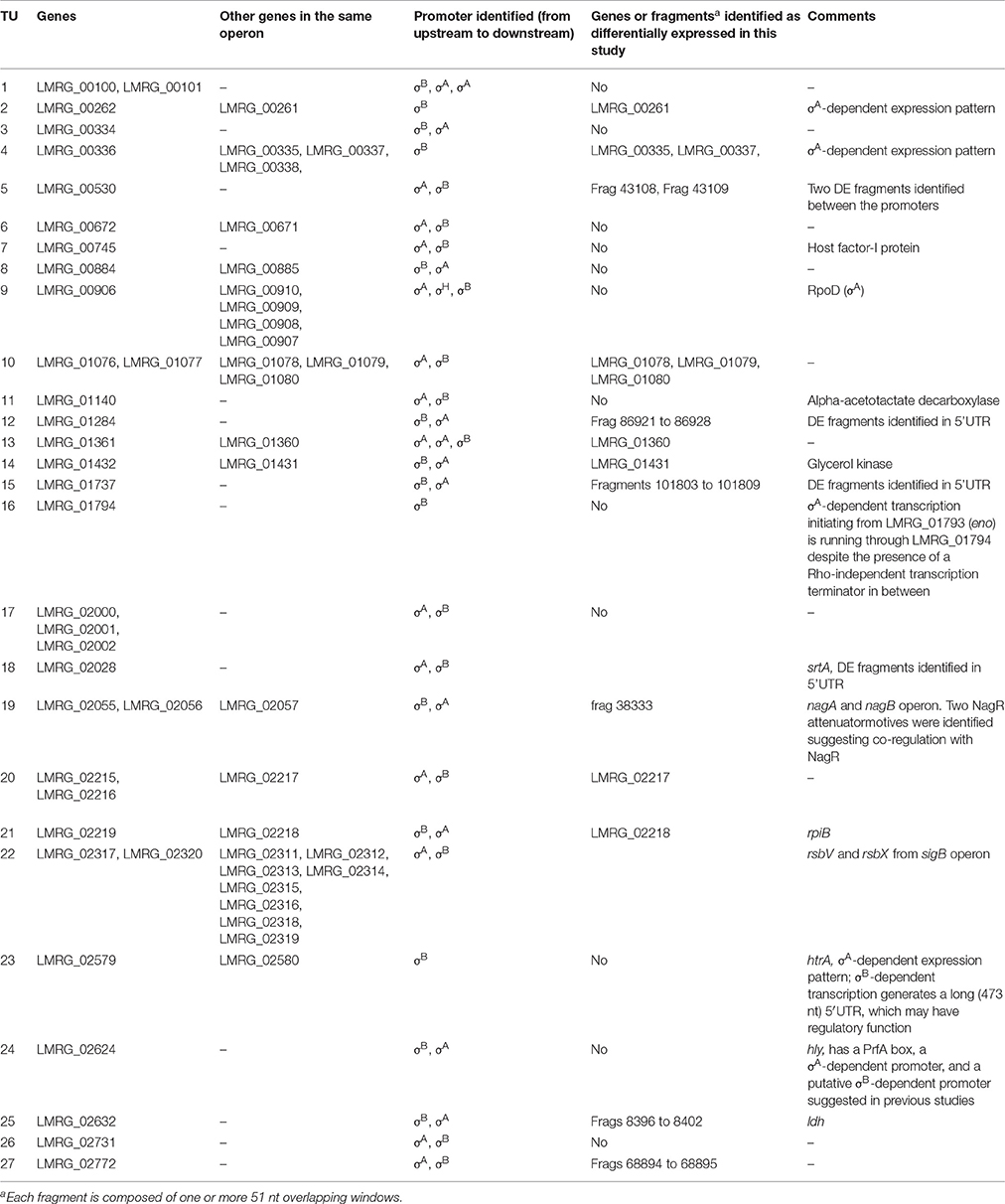
Promoter searches upstream of the 5′ UTR of these 74 genes identified upstream σB-dependent promoters for 35 of these genes; these 35 genes represent 28 TUs with corresponding σB-dependent promoters. Twenty-three of these 28 TUs had both σA- and σB-dependent promoters (Table 5). Therefore, it is possible that expression from the σA-dependent promoter masked expression from the σB-dependent promoter in these genes, not allowing their identification as σB-dependent in our study. While no clear σA-dependent promoters were identified for the other five TUs, RNA-seq coverage patterns in the quadruple mutant, which only expresses σA, suggested that four of these TUs may be co-regulated by σA. These four TUs were LMRG_00261-LMRG_00262, LMRG_00335-LMRG_00338, LMRG_01794, and htrA (LMRG_02579) whose σB-dependent transcription generates a long (473 nt) 5'UTR, which may have a regulatory function.
Some σB-Dependent TUs Are Also Regulated by Other Transcriptional Regulators
Three TUs previously reported as σB-dependent, but not identified as such in our study reported here, were found to be co-regulated by σB-dependent promoters and other alternative sigma factors and transcriptional factors. LMRG_02294, encoding a hypothetical protein, is co-transcribed with LMRG_02293, a gene encoding a putative amidohydrolase. A σB-dependent−10 region and a σL-dependent promoter in close proximity have been identified for this TU, suggesting co-regulation of transcription (Supplementary Table 6). As the strains used in this study lack an active σL, overexpression of σB itself might not be sufficient to significantly upregulate the TU. LMRG_02294 only showed a non-significant upregulation in the strain expressing σB.
LMRG_00906 (rpoD, encoding σA), which is part of the LMRG_00911-LMRG_00906 TU, is co-transcribed from an upstream σB-dependent promoter, a σH-dependent promoter upstream of LMRG_00908 and a σA-dependent promoter upstream of LMRG_00911. Strong expression from the σA-dependent promoter as well as the absence of σH may explain why no significant differential expression of rpoD was observed in our study. LMRG_02624 (hly) has a PrfA box, a σA-dependent promoter, and a putative σB-dependent promoter suggested in previous studies (Orsi et al., 2008; Tsai et al., 2011). hly was only reported as σB-dependent with active expression of PrfA (Ollinger et al., 2009; Toledo-Arana et al., 2009; Ribeiro et al., 2014), which was not shown to be significantly differentially expressed in our study.
Discussion
A number of studies have analyzed L. monocytogenes alternative-sigma-factor-dependent expression profiles at the transcriptional and protein levels based on comparisons between WT and single deletion mutant strains. However, regulon redundancy and overlaps in regulation by multiple sigma factors is common, as has been shown in Listeria (Chaturongakul et al., 2011), B. subtilis (Mascher et al., 2007), and in other bacteria (Nuss et al., 2010). We thus used an experimental design that enabled us to study the functions and genes regulated by σB in the absence of all other alternative sigma factors in order to further explore the L. monocytogenes σB regulon and σB-dependent gene regulation. Over-expression of a single alternative sigma factor in the absence of all other alternative sigma factors, the specific strategy used here, has previously been shown to eliminate redundant regulation by multiple alternative sigma factors, thus allowing improved insights into both the function of a single sigma factor and coregulation by different sigma factors (Mujahid et al., 2013a; Wang et al., 2014; Liu et al., 2016). As detailed above, this approach allowed us to (i) identify new σB-dependent functions in L. monocytogenes, such as regulation of genes involved in 1,2-propanediol utilization and biosynthesis of pyrimidine nucleotides; (ii) identify new σB-dependent genes involved in stress response and virulence functions; and (iii) further define a role for σB in L. monocytogenes homeostasis.
L. monocytogenes σB Directly Upregulates >100 TUs and >200 Genes
Our approach shows that the σB regulon includes >100 TUs and >200 genes that are directly up-regulated by σB. As RNA-seq has been well documented, in both L. monocytogenes (Oliver et al., 2009) and other organisms (Vivancos et al., 2010), to provide quantitative data that are well correlated with qPCR data, qPCR confirmation of genes newly identified as σB-dependent was not deemed necessary; rather σB-dependent transcriptional start sites were confirmed for selected genes via 5′ RACE. By comparison, previous studies have reported the σB regulon in 10403S as including >110 genes and >80 promoters (Raengpradub et al., 2008; Abram et al., 2008a,b; Oliver et al., 2009; Ollinger et al., 2009; Chaturongakul et al., 2011). Combined results from previous microarray, RNA-seq and proteomic studies in different L. monocytogenes strains under different stress conditions show a large σB pan-regulon of up to 902 genes (Wemekamp-Kamphuis et al., 2004; Chatterjee et al., 2006; Abram et al., 2008a,b; Ollinger et al., 2008, 2009; Raengpradub et al., 2008; Oliver et al., 2009, 2010; Toledo-Arana et al., 2009; Chaturongakul et al., 2011; Ribeiro et al., 2014) and a core regulon in lineage II strains of 184 genes, as supported by three or more previous experiments. Overall, our data further confirm that σB is the alternative sigma factor that regulates the largest regulon; other regulators that directly regulate large gene sets in L. monocytogenes include σH, which directly regulates more than 60 genes (Chaturongakul et al., 2011; Liu et al., 2016).
Newly Identified σB-Dependent Promoters Reveal a Novel Role for σB in Oxidative Stress, 1,2-Propanediol Utilization and Biosynthesis of Pyrimidine Nucleotides
While L. monocytogenes σB clearly has been shown to be important for survival under many stress conditions, such as exposure to heat, bile, osmotic and acid stress (Ferreira et al., 2001, 2003; Sue et al., 2004; Seifart Gomes et al., 2011), the role of σB in oxidative stress response is less clear. Previous studies have reported results ranging from hypersensitivity (Ferreira et al., 2001; Oliver et al., 2010) to hyperresistance (Moorhead and Dykes, 2003; Boura et al., 2016) to oxidative stress for σB null mutants. For example, a previous study reported that viability of a 10403S ΔsigB mutant was 100-fold lower than the WT under oxidative stress induced by cumene hydroperoxide in stationary phase (Ferreira et al., 2001). However, a recent study using L. monocytogenes 10403S and EGD-e showed that deletion of sigB led to hyperresistance to H2O2 in stationary phase cells grown under aerobic conditions (Boura et al., 2016). Further, while studies in B. cereus showed that loss of σB or σB regulon members showed impaired glucose-starvation-induced resistance to H2O2 (Engelmann and Hecker, 1996; Antelmann et al., 1997; Zuber, 2009), the ΔsigB mutant showed either a similar degree of resistance (Engelmann and Hecker, 1996) or hyperresistance to H2O2 induced oxidative stress (Van Schaik et al., 2005). Interestingly, we found both hslO and the qoxABCD operon to be transcribed from a σB-dependent promoter, suggesting a direct role of σB in resistance to oxidative stress. Among heat shock proteins, HslO has a unique chaperone activity because it is redox regulated and protects both thermally and oxidatively damaged proteins from irreversible aggregation (Kim et al., 2001). The importance of qoxABCD in oxidative stress response is also supported by observations in B. subtilis where a qoxABCD mutant shows reduced growth under aerobic conditions (Winstedt and Von Wachenfeldt, 2000). Upregulation of qox genes in L. monocytogenes in the presence of glycerol further indicates their involvement in adaption to an aerobic environment (Joseph et al., 2008). Our results suggest that the operon qoxABCD should be a focus for future studies on the molecular mechanisms behind oxidative stress resistance. As the exact role of σB in oxidative stress response in L. monocytogenes has not been elucidated to date, identification of these genes as regulated by σB-dependent promoters may provide fresh insight into the role of σB in oxidative stress response.
Interestingly, our data also found the propanediol utilization (pdu) operon to be transcribed from an upstream σB-dependent promoter, revealing a new role for σB in the utilization of 1,2-propanediol. 1,2-propanediol is produced during the catabolism of rhamnose and fucose, two sugars that are abundant in mammalian glycoconjugates (Staib and Fuchs, 2014). In addition, 1,2-propanediol is also used in various food products as a direct food additive (Cameron et al., 1998). The metabolism of 1,2-propanediol has been well studied in Salmonella; this organism is not only able to use 1,2-propanediol as a carbon source, but the pdu operon has also been shown to be important for virulence in Salmonella (Bobik et al., 1999). So far, studies in L. monocytogenes showed that transcription of some of the pdu genes increases under certain conditions, including lack of glucose (Nilsson et al., 2005), during intracellular growth (Joseph et al., 2006), and during growth on the surface of cold smoked salmon slices (Tang et al., 2015). In L. innocua, the pduD gene is required for 1,2-propanediol metabolism and 17 genes within the pduA-to-pduF gene cluster are induced when cells are grown in a medium containing 1,2-propanediol (Xue et al., 2008). The studies detailed above suggest the importance of 1,2-propanediol utilization in Listeria during growth in foods and in the host environment; our study reveals evidence for a regulatory role for σB in 1,2-propanediol utilization. This finding further illustrates important regulatory roles for σB in adaptation and transition to food and host environments.
Another σB-dependent promoter newly identified here is upstream of the LMRG_00978 to LMRG_00986 operon, which encodes proteins involved in pyrimidine ribonucleotide biosynthesis. Previous studies showed that genes involved in the de novo synthesis of purines (purA, purQ, lmo1771) and pyrimidines (pyrE) are required for intracellular proliferation of L. monocytogenes, suggesting that these bases and nucleotides are not provided by the host cell, but must be synthesized by the bacterium (Schauer et al., 2010). Our study supports σB as one of the key regulators for nucleotide metabolism, which is essential for L. monocytogenes to succeed in the intracellular environment.
Newly Identified σB-Dependent Promoters Reveal Additional Mechanisms for σB-Dependent Roles in Osmotic Stress, Glycerol Utilization, and Virulence
In addition to identification of new σB-dependent functions, as detailed above, our data also newly identified σB-dependent genes involved in functions previously linked to σB regulation, including oxidative stress, osmotic stress, glycerol utilization, and virulence. The response to osmotic stress is one of the first reported specific functions for σB in L. monocytogenes (Becker et al., 1998). Specific genes involved in osmotic stress response, such as opuCA, have a confirmed σB-dependent promoter (Kazmierczak et al., 2003) and are activated by osmotic stress in a σB-dependent manner (Sue et al., 2004). The osmotic activation of σB in L. monocytogenes is rapid, transient, and proportional to the magnitude of the osmotic stress applied (Utratna et al., 2011). Here, two operons with a role in osmotic stress response were newly identified as being directly regulated by σB, including (i) LMRG_02114-LMRG_02116 and (ii) LMRG_02775-LMRG_02778, which includes one gene encoding an OsmC/Ohr family protein. Previous studies have found that one of two genes encoding OsmC/Ohr family proteins is a member of the σB regulon in B. subtilis (Kempf and Bremer, 1995; Volker et al., 1998), suggesting a general role for σB-dependent regulation of OsmC/Ohr family proteins. These osmotic stress response proteins, newly identified to be σB-dependent, broaden our knowledge of how σB fine-tunes gene regulation to support chemical and water homeostasis in L. monocytogenes. We hypothesize that these σB-dependent genes could play an important role under high osmotic stress conditions such as those encountered by L. monocytogenes during gastrointestinal passage (Sue et al., 2004).
Glycerol is used as an alternative carbon source by intracellular L. monocytogenes (Eylert et al., 2008). Previous publications also suggested the importance of the ability to metabolize a series of carbon sources for successful infection by intracellular human pathogens (Olive and Sassetti, 2016) such as M. tuberculosis (de Carvalho et al., 2010). In L. monocytogenes, the mutant defective in the uptake and metabolism of glycerol showed impaired intracellular growth (Eylert et al., 2008). In a previous proteomics study, σB was found to regulate the LMRG_02000-LMRG_02002 operon, which encodes subunits of dihydroxyacetone kinase, an enzyme involved in glycerol metabolism in L. monocytogenes (Abram et al., 2008b). In addition, a sigB mutant had a diminished ability to use glycerol as a sole carbon source (Abram et al., 2008b). In our study, the LMRG_01432-LMRG_01431 operon, which encodes a glycerol kinase and a glycerol uptake facilitator protein, was found to have a σB-dependent promoter, further supporting an important role for σB in regulation of glycerol metabolism. As growth in glycerol also upregulates PrfA activity (Joseph et al., 2008; Stoll et al., 2008), this newly revealed role for σB in glycerol metabolism further supports a link between carbohydrate metabolism and pathogenesis in L. monocytogenes.
While σB previously has been recognized as a transcriptional regulator of a number of virulence genes, including prfA, inlA, inlC2, and inlD (Mcgann et al., 2007; Guldimann et al., 2016), our study revealed that another virulence gene, srtA, which is involved in proteolysis and processing of internalin proteins (Garandeau et al., 2002), is also directly regulated by σB. SrtA is required for the cell wall anchoring of InlA and, presumably, for the anchoring of other LPXTG-containing proteins that are involved in listerial infections (Bierne et al., 2002). A recent study also reported that inhibition of sortase A by chalcone could prevent L. monocytogenes infection (Li et al., 2016). In S. aureus, a number of sortase substrate proteins were previously observed to have higher or lower expression in a the ΔsigB mutant (Hempel et al., 2010). Our finding confirms and extends the role σB plays in pathogenesis, especially in the interaction between pathogen and host surface molecules during host cell invasion.
Co-regulation of Genes by σB and σA as Well as Other Transcriptional Regulators Supports Importance of σB in Resilience and Homeostasis
While L. monocytogenes σB has previously been well defined as a regulator of stress response and virulence functions, our data reported here further support a broader role for σB in resilience and homeostasis. The enriched GO terms related to general transport and homeostasis confirmed contributions of σB in coordinating complex networks responsive to changing environmental conditions.
Importantly, we also found a number of σB-dependent TUs and genes preceded by both σB and σA-dependent promoters (Supplementary Table 7). Key genes in this category include prfA, rsbV, qoxABCD (aerobic respiration), mogR (repressor of genes involved in flagella), phoU (phosphate transport regulation), cggR (central glycolytic genes regulator), ltrC (response to cold) and others. Genes preceded by both a promoter regulated by a housekeeping sigma factor and a promoter regulated by an alternative sigma factor have been identified in other bacteria. For example, in B. subtilis, clpC is preceded by two promoters (σA and σB) (Kruger et al., 1996) and expression of the ureABC operon, which encodes urease, is dependent on σA and σH (Wray et al., 1997). Additionally, the Extracytoplasmic Function (ECF) sigma factors of B. subtilis are known to co-regulate promoter regions with an existing σA promoter (Eiamphungporn and Helmann, 2008; Kingston et al., 2011). In E. coli, extensive overlaps between promoters of the primary sigma factor σ70 and alternative sigma factors such as σ32 and others also suggest coregulation of gene expression by multiple sigma factors under various growth conditions (Wade et al., 2006).
Seventy-four genes identified in previous studies as σB-dependent were not differentially expressed as σB-dependent in this study. A considerable number of these genes were co-regulated by more than one sigma factor, predominantly by σB and σA. A likely explanation regarding why some previously identified σB-dependent genes showed no differential expression in this study may reflect the growth conditions under which the experiments were conducted: the cells were in logarithmic growth with no additional imposed stress conditions. Previous studies used stress conditions such as stationary phase and salt stress to induce σB expression. In contrast, our experiment used artificial induction of σB expression by addition of rhamnose to log-phase cells. The transcriptional activity of the σA housekeeping sigma factor has been found to be diminished under stress conditions (Sharma and Chatterji, 2010; Delumeau et al., 2011). Fast-growing cells depend on σA to regulate expression of housekeeping genes such as those involved in protein biosynthesis, DNA replication and structural proteins. Therefore, under log-phase, σA is usually highly active to support the reproductive needs of the cells (Delumeau et al., 2011). Under the conditions used in this study, genes co-transcribed by both σA and σB under log-phase might have very little expression originating from the sometimes weaker σB promoter if the stronger σA promoter is located upstream or very close to the σB promoter.
Conclusions
In this study, we used RNA-seq to explore the role of σB in L. monocytogenes by overexpressing σB in a strain where genes encoding all other alternative sigma factors had been deleted. Combined with prior data revealing important roles for σB in pathogenesis and stress response, identification of new putative σB-dependent promoters upstream of a number of genes indicates a broader regulon for this alternative sigma factor, which also appears to contribute to cellular homeostasis.
Transcriptomic approaches such as RNA-seq and microarrays as well as proteomic approaches are powerful tools to explore the regulation of sigma factors, however, they have limited abilities to distinguish indirectly regulated genes from directly regulated genes. Further experiments with approaches such as ChIP-seq may allow a better definition of the direct regulons of sigma factors.
Author Contributions
YL performed the RNA-seq experiments and initial data analyses and was a major contributor in writing the manuscript; RO performed sliding window analysis of RNA-Seq data and promoter identification; KB, MW, and VG co-wrote the manuscript and conceived the study. All authors read and approved the final manuscript.
Funding
This project was supported by a grant from the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health (NIH) (2 RO1 AI052151-05A1).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Martin J. Loessner at ETH Zurich for the kind gift of plasmid pLF1.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.01910/full#supplementary-material
Supplementary Figure 1. qRT-PCR results of rhamnose induction of sigB and the σB-dependent gene LMRG_01602. Expression of σB under rhamnose induction was tested by qRT-PCR. LMRG_01602 was used as the targeted σB-dependent gene and rpoB as the reference gene; error bars show the standard deviation for the 2 biological replicates; Y axis represents the fold change of the gene expression level. Fold change is calculated as −ΔCt = CtrpoB − Cttarget gene. The σB-dependent gene referred in second graph is LMRG_01602.
Supplementary Table 1. Plasmid, primers and strains used in this study.
Supplementary Table 2. Genes and TUs upregulated by σB identified by traditional BaySeq and sliding window analyses.
Supplementary Table 3. Genes downregulated by σB identified by traditional BaySeq.
Supplementary Table 4. σB-dependent TUs identified by sliding window analyses, but not by the traditional BaySeq approach.
Supplementary Table 5. List of NDE σB-dependent genes.
Supplementary Table 6. 5′UTRs of the NDE σB-dependent genes.
Supplementary Table 7. TUs with σB-dependent promoters.
Abbreviations
asRNA, anti-sense RNA; BHI, Brain Heart Infusion; BWA, Burrows-Wheeler Aligner; DE, differentially expressed; ECF, Extracytoplasmic Function; FC, fold change; FDR, false discovery rate; GEO, Gene Expression Omnibus; GO, Gene Ontology; ncRNA, noncoding RNA; NDE, not differentially expressed; NEB, New England Biolabs; NIAID, National Institute of Allergy and Infectious Diseases; NIH, National Institutes of Health; NRC, normalized RNA-seq coverage; ORF, open reading frame; 5′ RACE, 5′ Rapid amplification of cDNA ends; RNA-seq, RNA sequencing; TU, transcription unit; UTR, untranslated region; WT, wild type; TSS, transcription start sites.
References
Abram, F., Starr, E., Karatzas, K. A., Matlawska-Wasowska, K., Boyd, A., Wiedmann, M., et al. (2008a). Identification of components of the Sigma B regulon in Listeria monocytogenes that contribute to acid and salt tolerance. Appl. Environ. Microbiol. 74, 6848–6858. doi: 10.1128/AEM.00442-08
Abram, F., Wan-Lin, S., Wiedmann, M., Boor, K. J., Coote, P., Botting, C., et al. (2008b). Proteomic analyses of a Listeria monocytogenes mutant lacking σB identify new components of the σB regulon and highlight a role for σB in the utilization of glycerol. Appl. Environ. Microbiol. 74, 594–604. doi: 10.1128/AEM.01921-07
Antelmann, H., Engelmann, S., Schmid, R., Sorokin, A., Lapidus, A., and Hecker, M. (1997). Expression of a stress- and starvation-induced dps/pexB-homologous gene is controlled by the alternative sigma factor SigmaB in Bacillus subtilis. J. Bacteriol. 179, 7251–7256. doi: 10.1128/jb.179.23.7251-7256.1997
Arous, S., Buchrieser, C., Folio, P., Glaser, P., Namane, A., Hebraud, M., et al. (2004). Global analysis of gene expression in an rpoN mutant of Listeria monocytogenes. Microbiology 150, 1581–1590. doi: 10.1099/mic.0.26860-0
Atichartpongkul, S., Loprasert, S., Vattanaviboon, P., Whangsuk, W., Helmann, J. D., and Mongkolsuk, S. (2001). Bacterial Ohr and OsmC paralogues define two protein families with distinct functions and patterns of expression. Microbiology 147, 1775–1782. doi: 10.1099/00221287-147-7-1775
Becker, L. A., Cetin, M. S., Hutkins, R. W., and Benson, A. K. (1998). Identification of the gene encoding the alternative sigma factor SigmaB from Listeria monocytogenes and its role in osmotolerance. J. Bacteriol. 180, 4547–4554.
Bierne, H., Mazmanian, S. K., Trost, M., Pucciarelli, M. G., Liu, G., Dehoux, P., et al. (2002). Inactivation of the srtA gene in Listeria monocytogenes inhibits anchoring of surface proteins and affects virulence. Mol. Microbiol. 43, 869–881. doi: 10.1046/j.1365-2958.2002.02798.x
Bobik, T. A., Havemann, G. D., Busch, R. J., Williams, D. S., and Aldrich, H. C. (1999). The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B12-dependent 1, 2-propanediol degradation. J. Bacteriol. 181, 5967–5975.
Boura, M., Keating, C., Royet, K., Paudyal, R., O'donoghue, B., O'byrne, C. P., et al. (2016). The presence of SigB in Listeria monocytogenes strains EGD-e and 10403S leads to hypersensitivity to hydrogen peroxide in stationary phase under aerobic conditions. Appl. Environ. Microbiol. 82, 4584–4591. doi: 10.1128/AEM.00709-16
Cameron, D. C., Altaras, N. E., Hoffman, M. L., and Shaw, A. J. (1998). Metabolic engineering of propanediol pathways. Biotechnol. Prog. 14, 116–125. doi: 10.1021/bp9701325
Chatterjee, S. S., Hossain, H., Otten, S., Kuenne, C., Kuchmina, K., Machata, S., et al. (2006). Intracellular gene expression profile of Listeria monocytogenes. Infect. Immun. 74, 1323–1338. doi: 10.1128/IAI.74.2.1323-1338.2006
Chaturongakul, S., Raengpradub, S., Palmer, M. E., Bergholz, T. M., Orsi, R. H., Hu, Y., et al. (2011). Transcriptomic and phenotypic analyses identify coregulated, overlapping regulons among PrfA, CtsR, HrcA, and the alternative sigma factors σB, σC, σH, and σL in Listeria monocytogenes. Appl. Environ. Microbiol. 77, 187–200. doi: 10.1128/AEM.00952-10
Crooks, G. E., Hon, G., Chandonia, J. M., and Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
de Carvalho, L. P., Fischer, S. M., Marrero, J., Nathan, C., Ehrt, S., and Rhee, K. Y. (2010). Metabolomics of Mycobacterium tuberculosis reveals compartmentalized co-catabolism of carbon substrates. Chem. Biol. 17, 1122–1131. doi: 10.1016/j.chembiol.2010.08.009
Delumeau, O., Lecointe, F., Muntel, J., Guillot, A., Guedon, E., Monnet, V., et al. (2011). The dynamic protein partnership of RNA polymerase in Bacillus subtilis. Proteomics 11, 2992–3001. doi: 10.1002/pmic.201000790
Eiamphungporn, W., and Helmann, J. D. (2008). The Bacillus subtilis σM regulon and its contribution to cell envelope stress responses. Mol. Microbiol. 67, 830–848. doi: 10.1111/j.1365-2958.2007.06090.x
Engelmann, S., and Hecker, M. (1996). Impaired oxidative stress resistance of Bacillus subtilis sigB mutants and the role of katA and katE. FEMS Microbiol. Lett. 145, 63–69. doi: 10.1111/j.1574-6968.1996.tb08557.x
Eylert, E., Schar, J., Mertins, S., Stoll, R., Bacher, A., Goebel, W., et al. (2008). Carbon metabolism of Listeria monocytogenes growing inside macrophages. Mol. Microbiol. 69, 1008–1017. doi: 10.1111/j.1365-2958.2008.06337.x
Ferreira, A., O'byrne, C. P., and Boor, K. J. (2001). Role of σB in heat, ethanol, acid, and oxidative stress resistance and during carbon starvation in Listeria monocytogenes. Appl. Environ. Microbiol. 67, 4454–4457. doi: 10.1128/AEM.67.10.4454-4457.2001
Ferreira, A., Sue, D., O'byrne, C. P., and Boor, K. J. (2003). Role of Listeria monocytogenes σB in survival of lethal acidic conditions and in the acquired acid tolerance response. Appl. Environ. Microbiol. 69, 2692–2698. doi: 10.1128/AEM.69.5.2692-2698.2003
Fieseler, L., Schmitter, S., Teiserskas, J., and Loessner, M. J. (2012). Rhamnose-inducible gene expression in Listeria monocytogenes. PLoS ONE 7:e43444. doi: 10.1371/journal.pone.0043444
Garandeau, C., Reglier-Poupet, H., Dubail, I., Beretti, J. L., Berche, P., and Charbit, A. (2002). The sortase SrtA of Listeria monocytogenes is involved in processing of internalin and in virulence. Infect. Immun. 70, 1382–1390. doi: 10.1128/IAI.70.3.1382-1390.2002
Goulet, V., Hebert, M., Hedberg, C., Laurent, E., Vaillant, V., De Valk, H., et al. (2012). Incidence of listeriosis and related mortality among groups at risk of acquiring listeriosis. Clin. Infect. Dis. 54, 652–660. doi: 10.1093/cid/cir902
Gray, M. J., Freitag, N. E., and Boor, K. J. (2006). How the bacterial pathogen Listeria monocytogenes mediates the switch from environmental Dr. Jekyll to pathogenic Mr. Hyde. Infect. Immun. 74, 2505–2512. doi: 10.1128/IAI.74.5.2505-2512.2006
Guldimann, C., Boor, K. J., Wiedmann, M., and Guariglia-Oropeza, V. (2016). Resilience in the face of uncertainty: sigma B fine-tunes gene expression to support homeostasis in Gram-positive bacteria. Appl. Environ. Microbiol. 82, 4456–4469. doi: 10.1128/AEM.00714-16
Hardcastle, T. J., and Kelly, K. A. (2010). BaySeq: empirical Bayesian methods for identifying differential expression in sequence count data. BMC Bioinformatics 11:422. doi: 10.1186/1471-2105-11-422
Hempel, K., Pane-Farre, J., Otto, A., Sievers, S., Hecker, M., and Becher, D. (2010). Quantitative cell surface proteome profiling for SigB-dependent protein expression in the human pathogen Staphylococcus aureus via biotinylation approach. J. Proteome Res. 9, 1579–1590. doi: 10.1021/pr901143a
Jagadeesan, B., Koo, O. K., Kim, K. P., Burkholder, K. M., Mishra, K. K., Aroonnual, A., et al. (2010). LAP, an alcohol acetaldehyde dehydrogenase enzyme in Listeria, promotes bacterial adhesion to enterocyte-like Caco-2 cells only in pathogenic species. Microbiology 156, 2782–2795. doi: 10.1099/mic.0.036509-0
Joseph, B., Mertins, S., Stoll, R., Schar, J., Umesha, K. R., Luo, Q., et al. (2008). Glycerol metabolism and PrfA activity in Listeria monocytogenes. J. Bacteriol. 190, 5412–5430. doi: 10.1128/JB.00259-08
Joseph, B., Przybilla, K., Stuhler, C., Schauer, K., Slaghuis, J., Fuchs, T. M., et al. (2006). Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J. Bacteriol. 188, 556–568. doi: 10.1128/JB.188.2.556-568.2006
Kazmierczak, M. J., Mithoe, S. C., Boor, K. J., and Wiedmann, M. (2003). Listeria monocytogenes σB regulates stress response and virulence functions. J. Bacteriol. 185, 5722–5734. doi: 10.1128/JB.185.19.5722-5734.2003
Kempf, B., and Bremer, E. (1995). OpuA, an osmotically regulated binding protein-dependent transport system for the osmoprotectant glycine betaine in Bacillus subtilis. J. Biol. Chem. 270, 16701–16713. doi: 10.1074/jbc.270.28.16701
Kim, S. J., Jeong, D. G., Chi, S. W., Lee, J. S., and Ryu, S. E. (2001). Crystal structure of proteolytic fragments of the redox-sensitive Hsp33 with constitutive chaperone activity. Nat. Struct. Biol. 8, 459–466. doi: 10.1038/87639
Kingston, A. W., Subramanian, C., Rock, C. O., and Helmann, J. D. (2011). A σW-dependent stress response in Bacillus subtilis that reduces membrane fluidity. Mol. Microbiol. 81, 69–79. doi: 10.1111/j.1365-2958.2011.07679.x
Kruger, E., Msadek, T., and Hecker, M. (1996). Alternate promoters direct stress-induced transcription of the Bacillus subtilis clpC operon. Mol. Microbiol. 20, 713–723. doi: 10.1111/j.1365-2958.1996.tb02511.x
Lauer, P., Chow, M. Y. N., Loessner, M. J., Portnoy, D. A., and Calendar, R. (2002). Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J. Bacteriol. 184, 4177–4186. doi: 10.1128/JB.184.15.4177-4186.2002
Li, H., Chen, Y., Zhang, B., Niu, X., Song, M., Luo, Z., et al. (2016). Inhibition of sortase A by chalcone prevents Listeria monocytogenes infection. Biochem. Pharmacol. 106, 19–29. doi: 10.1016/j.bcp.2016.01.018
Li, H., and Durbin, R. (2010). Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595. doi: 10.1093/bioinformatics/btp698
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Liu, Y., Orsi, R. H., Boor, K. J., Wiedmann, M., and Guariglia-Oropeza, V. (2016). An advanced bioinformatics approach for analyzing RNA-seq data reveals sigma H-dependent regulation of competence genes in Listeria monocytogenes. BMC Genomics 17:115. doi: 10.1186/s12864-016-2432-9
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Mascher, T., Hachmann, A. B., and Helmann, J. D. (2007). Regulatory overlap and functional redundancy among Bacillus subtilis extracytoplasmic function sigma factors. J. Bacteriol. 189, 6919–6927. doi: 10.1128/JB.00904-07
Mcgann, P., Wiedmann, M., and Boor, K. J. (2007). The alternative sigma factor σB and the virulence gene regulator PrfA both regulate transcription of Listeria monocytogenes internalins. Appl. Environ. Microbiol. 73, 2919–2930. doi: 10.1128/AEM.02664-06
Medrano Romero, V., and Morikawa, K. (2016). Listeria monocytogenes SigmaH contributes to expression of competence genes and intracellular growth. J. Bacteriol. 198, 1207–1217. doi: 10.1128/JB.00718-15
Moorhead, S. M., and Dykes, G. A. (2003). The role of the sigB gene in the general stress response of Listeria monocytogenes varies between a strain of serotype 1/2a and a strain of serotype 4c. Curr. Microbiol. 46, 461–466. doi: 10.1007/s00284-002-3867-6
Mujahid, S., Orsi, R. H., Boor, K. J., and Wiedmann, M. (2013a). Protein level identification of the Listeria monocytogenes Sigma H, Sigma L, and Sigma C regulons. BMC Microbiol. 13:156. doi: 10.1186/1471-2180-13-156
Mujahid, S., Orsi, R. H., Vangay, P., Boor, K. J., and Wiedmann, M. (2013b). Refinement of the Listeria monocytogenes σB regulon through quantitative proteomic analysis. Microbiology 159, 1109–1119. doi: 10.1099/mic.0.066001-0
Nilsson, L., Hansen, T. B., Garrido, P., Buchrieser, C., Glaser, P., Knochel, S., et al. (2005). Growth inhibition of Listeria monocytogenes by a nonbacteriocinogenic Carnobacterium piscicola. J. Appl. Microbiol. 98, 172–183. doi: 10.1111/j.1365-2672.2004.02438.x
Nuss, A. M., Glaeser, J., Berghoff, B. A., and Klug, G. (2010). Overlapping alternative sigma factor regulons in the response to singlet oxygen in Rhodobacter sphaeroides. J. Bacteriol. 192, 2613–2623. doi: 10.1128/JB.01605-09
O'byrne, C. P., and Karatzas, K. A. (2008). The role of Sigma B (σB) in the stress adaptations of Listeria monocytogenes: overlaps between stress adaptation and virulence. Adv. Appl. Microbiol. 65, 115–140. doi: 10.1016/S0065-2164(08)00605-9
Olive, A. J., and Sassetti, C. M. (2016). Metabolic crosstalk between host and pathogen: sensing, adapting and competing. Nat. Rev. Microbiol. 14, 221–234. doi: 10.1038/nrmicro.2016.12
Oliver, H. F., Orsi, R. H., Ponnala, L., Keich, U., Wang, W., Sun, Q., et al. (2009). Deep RNA sequencing of L. monocytogenes reveals overlapping and extensive stationary phase and Sigma B-dependent transcriptomes, including multiple highly transcribed noncoding RNAs. BMC Genomics 10:641. doi: 10.1186/1471-2164-10-641
Oliver, H. F., Orsi, R. H., Wiedmann, M., and Boor, K. J. (2010). Listeria monocytogenes σB has a small core regulon and a conserved role in virulence but makes differential contributions to stress tolerance across a diverse collection of strains. Appl. Environ. Microbiol. 76, 4216–4232. doi: 10.1128/AEM.00031-10
Ollinger, J., Bowen, B., Wiedmann, M., Boor, K. J., and Bergholz, T. M. (2009). Listeria monocytogenes σB modulates PrfA-mediated virulence factor expression. Infect. Immun. 77, 2113–2124. doi: 10.1128/IAI.01205-08
Ollinger, J., Wiedmann, M., and Boor, K. J. (2008). Sigma(B)- and PrfA-dependent transcription of genes previously classified as putative constituents of the Listeria monocytogenes PrfA regulon. Foodborne Pathog. Dis. 5, 281–293. doi: 10.1089/fpd.2008.0079
Orsi, R. H., Maron, S. B., Nightingale, K. K., Jerome, M., Tabor, H., and Wiedmann, M. (2008). Lineage specific recombination and positive selection in coding and intragenic regions contributed to evolution of the main Listeria monocytogenes virulence gene cluster. Infection Genetics and Evolution 8, 566–576. doi: 10.1016/j.meegid.2008.04.006
Raengpradub, S., Wiedmann, M., and Boor, K. J. (2008). Comparative analysis of the σB-dependent stress responses in Listeria monocytogenes and Listeria innocua strains exposed to selected stress conditions. Appl. Environ. Microbiol. 74, 158–171. doi: 10.1128/AEM.00951-07
Raimann, E., Schmid, B., Stephan, R., and Tasara, T. (2009). The alternative sigma factor σL of L. monocytogenes promotes growth under diverse environmental stresses. Foodborne Pathog. Dis. 6, 583–591. doi: 10.1089/fpd.2008.0248
Rea, R. B., Gahan, C. G., and Hill, C. (2004). Disruption of putative regulatory loci in Listeria monocytogenes demonstrates a significant role for Fur and PerR in virulence. Infect. Immun. 72, 717–727. doi: 10.1128/IAI.72.2.717-727.2004
Ribeiro, V. B., Mujahid, S., Orsi, R. H., Bergholz, T. M., Wiedmann, M., Boor, K. J., et al. (2014). Contributions of σB and PrfA to Listeria monocytogenes salt stress under food relevant conditions. Int. J. Food Microbiol. 177, 98–108. doi: 10.1016/j.ijfoodmicro.2014.02.018
Saikolappan, S., Das, K., Sasindran, S. J., Jagannath, C., and Dhandayuthapani, S. (2011). OsmC proteins of Mycobacterium tuberculosis and Mycobacterium smegmatis protect against organic hydroperoxide stress. Tuberculosis (Edinb) 91(Suppl. 1), S119–S127. doi: 10.1016/j.tube.2011.10.021
Scallan, E., Hoekstra, R. M., Angulo, F. J., Tauxe, R. V., Widdowson, M.-A., Roy, S. L., et al. (2011). Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 17, 7–15. doi: 10.3201/eid1701.P11101
Schauer, K., Geginat, G., Liang, C., Goebel, W., Dandekar, T., and Fuchs, T. M. (2010). Deciphering the intracellular metabolism of Listeria monocytogenes by mutant screening and modelling. BMC Genomics 11:573. doi: 10.1186/1471-2164-11-573
Seifart Gomes, C., Izar, B., Pazan, F., Mohamed, W., Mraheil, M. A., Mukherjee, K., et al. (2011). Universal stress proteins are important for oxidative and acid stress resistance and growth of Listeria monocytogenes EGD-e in vitro and in vivo. PLoS ONE 6:e24965. doi: 10.1371/journal.pone.0024965
Sharma, U. K., and Chatterji, D. (2010). Transcriptional switching in Escherichia coli during stress and starvation by modulation of sigma activity. FEMS Microbiol. Rev. 34, 646–657. doi: 10.1111/j.1574-6976.2010.00223.x
Staib, L., and Fuchs, T. M. (2014). From food to cell: nutrient exploitation strategies of enteropathogens. Microbiology 160, 1020–1039. doi: 10.1099/mic.0.078105-0
Stoll, R., Mertins, S., Joseph, B., Müller-Altrock, S., and Goebel, W. (2008). Modulation of PrfA activity in Listeria monocytogenes upon growth in different culture media. Microbiology 154, 3856–3876. doi: 10.1099/mic.0.2008/018283-0
Sue, D., Fink, D., Wiedmann, M., and Boor, K. J. (2004). σB-dependent gene induction and expression in Listeria monocytogenes during osmotic and acid stress conditions simulating the intestinal environment. Microbiology 150, 3843–3855. doi: 10.1099/mic.0.27257-0
Tang, S., Orsi, R. H., Den Bakker, H. C., Wiedmann, M., Boor, K. J., and Bergholz, T. M. (2015). Transcriptomic analysis of Listeria monocytogenes adaptation to growth on vacuum-packed cold smoked salmon. Appl. Environ. Microbiol. 81, 6812–6824. doi: 10.1128/AEM.01752-15
Toledo-Arana, A., Dussurget, O., Nikitas, G., Sesto, N., Guet-Revillet, H., Balestrino, D., et al. (2009). The Listeria transcriptional landscape from saprophytism to virulence. Nature 459, 950–956. doi: 10.1038/nature08080
Tsai, Y. H. L., Maron, S. B., Mcgann, P., Nightingale, K. K., Wiedmann, M., and Orsi, R. H. (2011). Recombination and positive selection contributed to the evolution of Listeria monocytogenes lineages III and IV, two distinct and well supported uncommon L. monocytogenes lineages. Infect. Genet. Evol. 11, 1881–1890. doi: 10.1016/j.meegid.2011.08.001
Utratna, M., Shaw, I., Starr, E., and O'byrne, C. P. (2011). Rapid, transient, and proportional activation of σB in response to osmotic stress in Listeria monocytogenes. Appl. Environ. Microbiol. 77, 7841–7845. doi: 10.1128/AEM.05732-11
Van Schaik, W., Zwietering, M. H., De Vos, W. M., and Abee, T. (2005). Deletion of the sigB Gene in Bacillus cereus ATCC 14579 leads to hydrogen peroxide hyperresistance. Appl. Environ. Microbiol. 71, 6427–6430. doi: 10.1128/AEM.71.10.6427-6430.2005
Vivancos, A. P., Guell, M., Dohm, J. C., Serrano, L., and Himmelbauer, H. (2010). Strand-specific deep sequencing of the transcriptome. Genome Res. 20, 989–999. doi: 10.1101/gr.094318.109
Volker, U., Andersen, K. K., Antelmann, H., Devine, K. M., and Hecker, M. (1998). One of two osmC homologs in Bacillus subtilis is part of the SigmaB-dependent general stress regulon. J. Bacteriol. 180, 4212–4218.
Wade, J. T., Castro Roa, D., Grainger, D. C., Hurd, D., Busby, S. J., Struhl, K., et al. (2006). Extensive functional overlap between sigma factors in Escherichia coli. Nat. Struct. Mol. Biol. 13, 806–814. doi: 10.1038/nsmb1130
Wang, S. Y., Orsi, R. H., Tang, S. L., Zhang, W., Wiedmann, M., and Boor, K. J. (2014). Phosphotransferase system-dependent extracellular growth of Listeria monocytogenes is regulated by alternative sigma factors σL and σH. Appl. Environ. Microbiol. 80, 7673–7682. doi: 10.1128/AEM.02530-14
Wemekamp-Kamphuis, H. H., Wouters, J. A., De Leeuw, P. P., Hain, T., Chakraborty, T., and Abee, T. (2004). Identification of sigma factor Sigma B-controlled genes and their impact on acid stress, high hydrostatic pressure, and freeze survival in Listeria monocytogenes EGD-e. Appl. Environ. Microbiol. 70, 3457–3466. doi: 10.1128/AEM.70.6.3457-3466.2004
Winstedt, L., and Von Wachenfeldt, C. (2000). Terminal oxidases of Bacillus subtilis strain 168: one quinol oxidase, cytochrome aa3 or cytochrome bd, is required for aerobic growth. J. Bacteriol. 182, 6557–6564. doi: 10.1128/JB.182.23.6557-6564.2000
Wray, L. V., Ferson, A. E., and Fisher, S. H. (1997). Expression of the Bacillus subtilis ureABC operon is controlled by multiple regulatory factors including CodY, GlnR, TnrA, and Spo0H. J. Bacteriol. 179, 5494–5501. doi: 10.1128/jb.179.17.5494-5501.1997
Xue, J., Murrieta, C. M., Rule, D. C., and Miller, K. W. (2008). Exogenous or L-rhamnose-derived 1,2-propanediol is metabolized via a PduD-dependent pathway in Listeria innocua. Appl. Environ. Microbiol. 74, 7073–7079. doi: 10.1128/AEM.01074-08
Young, M. D., Wakefield, M. J., Smyth, G. K., and Oshlack, A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11:R14. doi: 10.1186/gb-2010-11-2-r14
Zhang, C., Nietfeldt, J., Zhang, M., and Benson, A. K. (2005). Functional consequences of genome evolution in Listeria monocytogenes: the lmo0423 and lmo0422 genes encode sigmaC and LstR, a lineage II-specific heat shock system. J. Bacteriol. 187, 7243–7253. doi: 10.1128/JB.187.21.7243-7253.2005
Keywords: RNA-seq, Listeria monocytogenes, sigma B, overlapping regulons, promoter
Citation: Liu Y, Orsi RH, Boor KJ, Wiedmann M and Guariglia-Oropeza V (2017) Home Alone: Elimination of All but One Alternative Sigma Factor in Listeria monocytogenes Allows Prediction of New Roles for σB. Front. Microbiol. 8:1910. doi: 10.3389/fmicb.2017.01910
Received: 24 June 2017; Accepted: 19 September 2017;
Published: 11 October 2017.
Edited by:
Avelino Alvarez-Ordóñez, Universidad de León, SpainReviewed by:
Weili Liang, Chinese Center for Disease Control and Prevention, ChinaKathrin Rychli, Veterinärmedizinische Universität Wien, Austria
Copyright © 2017 Liu, Orsi, Boor, Wiedmann and Guariglia-Oropeza. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Veronica Guariglia-Oropeza, vg93@cornell.edu
 Yichang Liu
Yichang Liu Renato H. Orsi
Renato H. Orsi Kathryn J. Boor
Kathryn J. Boor Martin Wiedmann
Martin Wiedmann Veronica Guariglia-Oropeza
Veronica Guariglia-Oropeza